 Open Access
Open Access
REVIEW
A Survey of Lung Nodules Detection and Classification from CT Scan Images
1 Faculty of Computing and Technology, IQRA University, Islamabad, 44000, Pakistan
2 Faculty of Engineering and Computer Science, National University of Modern Languages, Islamabad, 44000, Pakistan
3 Faculty of Computing and Informatics, Multimedia University, Cyberjaya, 63100, Malaysia
4 Faculty of Computing, Riphah International University, Islamabad, 44000, Pakistan
5 Department of Information Sciences, Division of Sciences and Technology, University of Education, Lahore, 54770, Pakistan
* Corresponding Author: Mazliham Mohd Su’ud. Email:
Computer Systems Science and Engineering 2024, 48(6), 1483-1511. https://doi.org/10.32604/csse.2024.053997
Received 15 May 2024; Accepted 13 August 2024; Issue published 22 November 2024
Abstract
In the contemporary era, the death rate is increasing due to lung cancer. However, technology is continuously enhancing the quality of well-being. To improve the survival rate, radiologists rely on Computed Tomography (CT) scans for early detection and diagnosis of lung nodules. This paper presented a detailed, systematic review of several identification and categorization techniques for lung nodules. The analysis of the report explored the challenges, advancements, and future opinions in computer-aided diagnosis CAD systems for detecting and classifying lung nodules employing the deep learning (DL) algorithm. The findings also highlighted the usefulness of DL networks, especially convolutional neural networks (CNNs) in elevating sensitivity, accuracy, and specificity as well as overcoming false positives in the initial stages of lung cancer detection. This paper further presented the integral nodule classification stage, which stressed the importance of differentiating between benign and malignant nodules for initial cancer diagnosis. Moreover, the findings presented a comprehensive analysis of multiple techniques and studies for nodule classification, highlighting the evolution of methodologies from conventional machine learning (ML) classifiers to transfer learning and integrated CNNs. Interestingly, while accepting the strides formed by CAD systems, the review addressed persistent challenges.Keywords
Cancer can be defined as an illness that occurs in different forms. Usually, it involves clusters of abnormal cells. These cells lead to ungoverned multiplication and division, leading to the formation of tumors [1]. In 2020, the World Health Organization (WHO) released a report that cancer was one of the major sources of mortality, amounting to 10 million cases, which equals one out of every six deaths [2]. According to the American Cancer Society (ACS), in 2023, 238,340 new cases of lung cancer and 127,070 fatalities were estimated in the United States (US). Fig. 1 below shows the estimated lung cancer data in the US for the last six years (2018–2023) [3]. Most patients in developing nations are diagnosed with lung cancer when it has already progressed to a later stage, without effective or proper cures. Research indicates that among Chinese patients with advanced lung cancer, the 5 years survival rate is only 16%. In contrast, the survival rate could increase to 70% if the cancer is identified in its early stages [4].
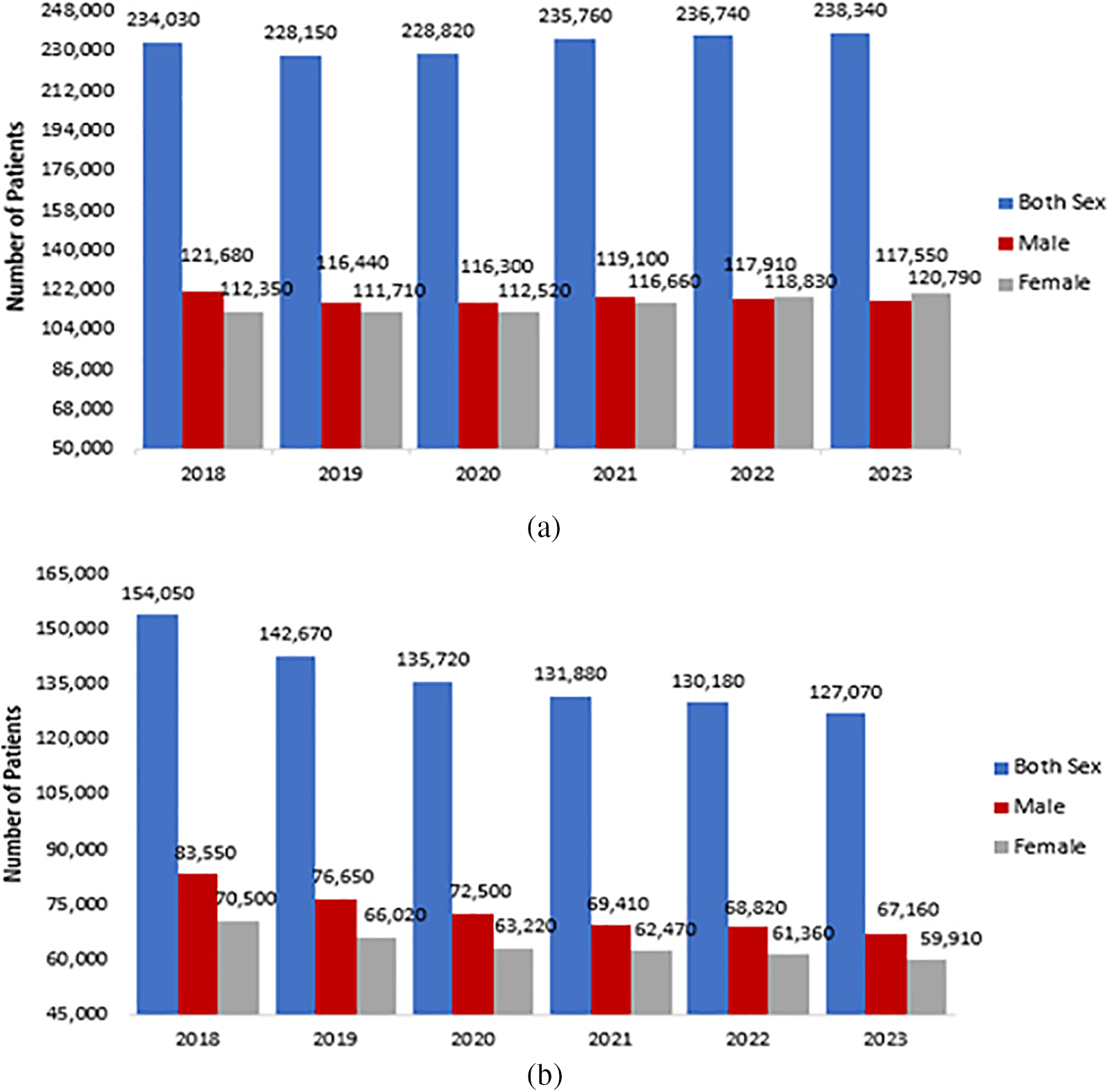
Figure 1: ACS last six years lung cancer data in the US: (a) Estimated numbers of new lung cases; (b) Estimated numbers of expired patients US [3]
Some of the leading causes of pulmonary cancer are consuming arsenic-containing drinking water, smoking harmful particles, drinking alcohol, outdoor air pollution, eating red meat, etc., all contributing to pulmonary cancer fatalities and illnesses. On the other hand, lung cancer treatment is too expensive. The health system is searching for new cancer cures or treatments for lung patients. To remove financial and psychological hurdles, lung cancer interventions such as initial-stage prevention, screening tests, monitoring, and early-stage nodule detection remain a high focus [5]. Lung nodule intelligence examination is an excellent cancer avoidance strategy that includes both a detection and a classification stage. Lung cancer is categorized into two primary types: non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC). NSCLC accounts for around 85% of all lung cancer occurrences, while SCLC is responsible for the remaining 15% and is known for its high malignancy [6]. Recent statistics show that in 2022, there were 44,213 new pulmonary cancer cases, and 38,292 deaths were reported in Brazil [7]. Lung diseases are also categorized and ranked with respect to genders, such as the 5th most frequently found type of lung cancer in females and the 3rd in number for males, according to the statistics provided by the Brazilian National Cancer Institute.
Generally, pulmonary nodules are small, rounded, or geometric shapes with approximately 3–30 mm diameters. These nodules exist in various sizes, densities, locations, and surroundings. Micro-nodules are nodules with diameters <3 mm. Non-cancerous nodules, such as the bronchi barrier and blood vessels, generate false-positive results in the detection process due to their similar appearance to cancerous nodules. Pulmonary nodules are classified into four types based on their location. The first is a well-circumscribed nodule in the middle of the lung with no connections to the vasculature. The second type is a juxta-vascular nodule, which connects to the vessels and is even in the middle of the lung. The third type is the pleural tail, located on the pleural surface and connected by a thin structure. The fourth lesion is a juxta-pleural nodule firmly attached to the pleural surface [8]. It is a simple classification technique for labeling a nodule to analyze its different parameters, such as position and connections, which can help identify lung cancer. Fig. 2 shows an example image of different nodules. It is more challenging to diagnose pulmonary nodule lesions since they usually have no symptoms. However, studies show that nodules with a diameter greater than 8 mm, which are sub-solid, speculated, and lobulated, are more likely to be malignant [9,10].
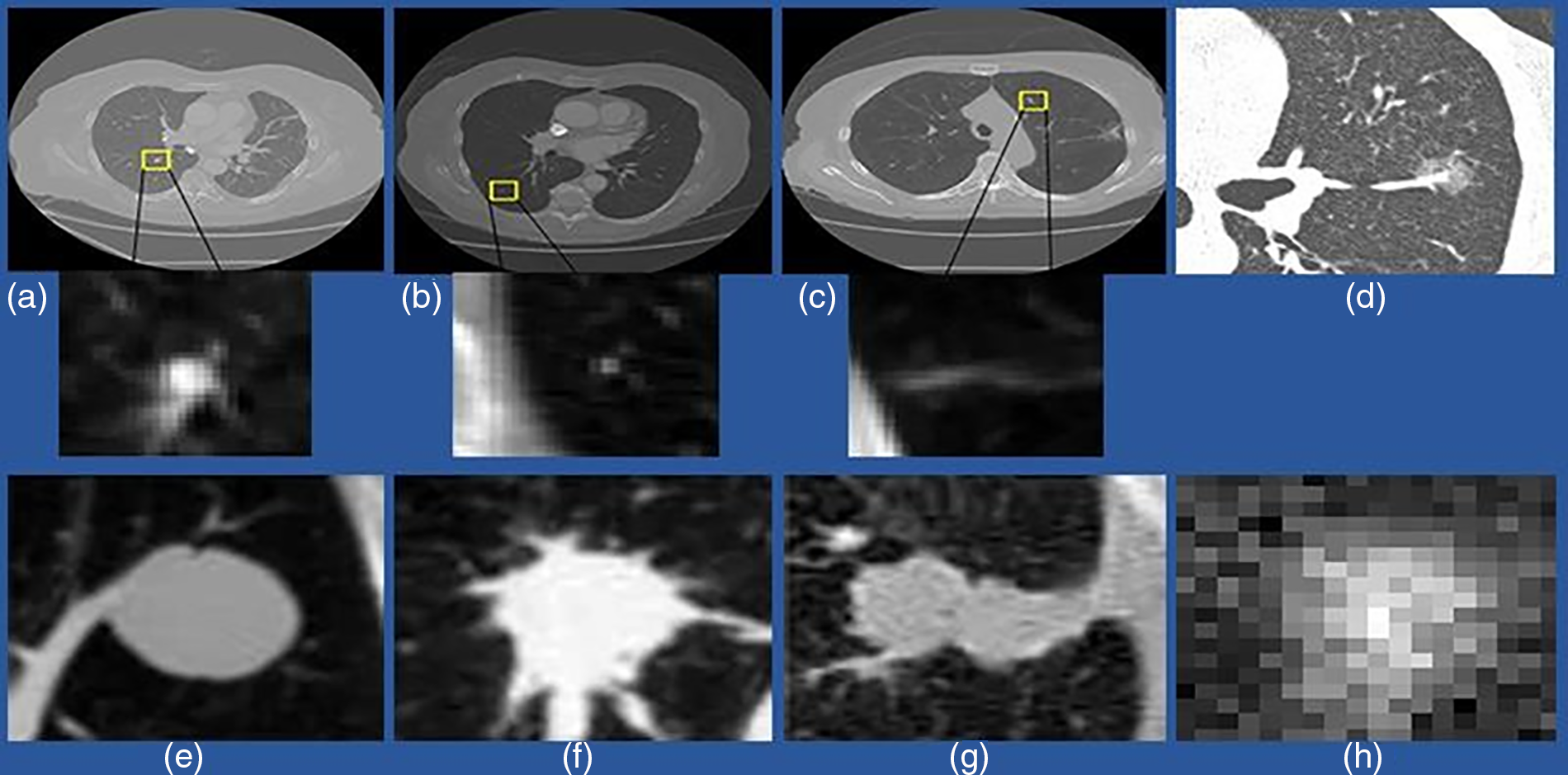
Figure 2: An example of a different variety of lung nodule candidates using a CT scan. (a) Isolated nodules with a diameter >3 mm; (b) Micro-nodules having a diameter <3 mm; (c) Non-nodule with a diameter >3 mm; (d) indicates ground-glass nodules (GGN); (e) Isolated solid nodules; (f) Solid irregular and Juxtavascular nodules; (g) Juxta-pleural nodule (h) Sub-solid and isolated nodules
CT is considered the primary imaging modality for detecting and diagnosing lung nodules (LNs), providing superior resolution and cost-efficiency. Such imaging techniques recognize nodules and become helpful in their characterization and monitoring development. Besides advantages, studies [11–13] have also presented some limitations of CT. On the other hand, such deficiencies could be efficiently mitigated by executing a computer-aided detection (CADe) system, which effectively complements conventional CT scans by retrieving pertinent information about lung nodules. It incorporates extensive growth patterns, features and characteristics, and other data, which probably remained invisible instantly with the help of traditional imaging alone. The integration of CADe into the diagnostic phenomenon is especially beneficial because it helps radiologists establish more diagnoses in a timely and accurate manner. By employing modern ML algorithms, CADe elevates the diagnostic workflow by offering radiologists extra information and pointing out areas of concern. The collaborative approach enhances nodule efficiency detection and categorization, as well as plays a role in more informed decision-making on account of patient care. Moreover, the CADe incorporation into lung nodule diagnosis mirrors the persistent synergy between medical imaging and technological advancements. As clinical professionals persistently struggle to improve accurate diagnosis and expedite the diagnosis process, CADe is a significant tool in the pulmonary imaging realm; thus, it is an effective avenue for elevated patient consequences and more efficient disease management.
Nowadays, technologies are updated regularly, and CAD systems significantly facilitate medical doctors in the detection of lung cancer tumors. A CAD system has two main types: a computer-aided detection (CADe) system and a computer-aided diagnosis (CADx) system. By specifically stressing the adoption of deep learning algorithms, CAD systems have evolved as an effective tool for automated detection and lung nodule classification in CT scan images [14]. The purpose of the CADe system is to identify regions of interest in pulmonary CT scans in order to find abnormal lesions. Some studies have highlighted the tendency of CADe systems to retrieve usefulness and pertinent data on lung nodules–this plays potential roles in the diagnosis process. As Khan et al. [13], CADe assists in identifying key lesions along with demonstrating proficiency in the discrimination between non-nodules and nodules, which incorporates consolidations and blood vessels. Such discrimination is essential to overcome false positives and strengthen the nodule detection specificity [15,16]. Besides, the CADx systems integrated into the diagnostic setting add an additional layer of sophistication [17]. As Han et al. [18], CADx systems contribute integrally to the characterization of lesions by facilitating insights into their forms, phases, and progression. Such systems go beyond mere detection, intending to classify lesions as either benign or malignant nodules, thus helping radiologists form a more accurate and extensive diagnosis.
The primary purpose of CAD systems is to improve the radiologists’ ability to diagnose cancer, as well as their accuracy while shortening the time it takes to interpret CT scans [14]. In this way, the CAD system plays a vital role in assisting medical doctors in their decision-making; as the technology integration in health and clinical care emerges consistently, it becomes more integral for assessing the contemporary setting of lung nodule detection methodologies and gaining insights into the challenges and advancement in the field [17,19]. In the wake of the effectiveness and challenges, there is a need to develop a more synthesized view of the information, particularly in the false positives (FPs) setting as well as the need for persistent algorithm refinements for accommodating different patient populations. Research by [20,21] demonstrated the persistent endeavor to enhance the performance of the CADe system; thus, the usefulness of the iterative development and processes of validations is emphasized. The increasing prominence of CAD systems in assessing and diagnosing lung nodules initially is not deniably transformative for the radiology field, which provides efficiencies, overcoming interpretation time potentially for radiologists probing CT images. The paradigm shift has significantly streamlined the processes of lung cancer screening, causing an estimated compound growth rate of 7.7% yearly across the world for the CAD market by 2031, with 23% of such systems known for detecting lung cancer [22].
On the other hand, the CAD system’s emergence is noted by the shift from conventional and handcrafted characteristics retrieval approaches to additional current deep learning approaches, mainly CNNs [14]. While traditional CAD systems depend on engineered features (including shape and texture analysis) manually, their effectiveness is hurdled by restrictions. Such restrictions involve medically accelerated abstraction from suboptimal image samples, insufficient normalization, universality, uniformity, and the substantial time investment needed. Considering the loopholes, a prominent transition has occurred toward deep learning methodologies, particularly CNNs. Studies by Zhu et al. [23] and Halder et al. [24] pointed out CNN’s superiority regarding time efficiency and accuracy compared to traditional CAD systems. In these studies, the CNN approaches mitigate multiple challenges, such as extracting related characteristics and features as well as concise, objective identification. The success increases beyond the detection of lung nodules–this exhibits effective and outstanding performance in domains like image analysis medically. Along with adopting CNNs in CAD systems, it is considered highly effective and adequate for approaching the shift critically. The research performed by Mehmood et al. [25] stated the need for considerable validation and benchmarking processes to ensure the CNN-based CAD system’s reliability and generalisability throughout different groups of patients and imaging situations. In addition, persistent endeavors are needed to mitigate fundamental biases in data training and optimize the algorithm’s performance.
However, several reviews have been available previously that provide information about the detection, diagnosis, and analysis of pulmonary nodules with CT scans. The aim of those studies is to provide the best technique that has been developed at that time and compare the experimental results. However, with the developments and enhancement of new methods, techniques, and medical equipment, a DL-based solution for nodule diagnosis has just recently been introduced. Therefore, a comprehensive, up-to-date review of the time and research is needed as well, and this is the primary outcome and impact of this exploratory work.
This research paper is divided into different sections. Each section contains an explanation of the related topic. Section 2 covers the experimental benchmark, including publicly available datasets of CT scans and related competitions. Section 3 explains the complete workflow of CAD systems along with effective techniques related to every four main components. In Section 4, discussion is presented. Section 5 outlines the current challenges as well as the future prospective directions. The conclusion and results of the research are mentioned in the final Section 6.
To build an effective CAD method to diagnose and categorize lung nodules, the researchers should focus on these experimental benchmarks, including databases and large-scale competition. CAD systems require huge, CT images to train detection models. Hence, gathering publicly available datasets is essential. Appropriate evaluation measures are needed to validate the performance of different techniques accurately. Furthermore, large-scale diagnosis competitions continuously have updated techniques and methods trained on a uniform database and performance standard criteria.
2.1 Open Datasets of Pulmonary Nodule
CT images are acquired for CAD system evaluation. Most images inside nodule detection and classification are 2-D and 3-D slices with 512 × 512 pixels. All images are kept in the DICOM (Digital Imaging and Communications in Medicine) file type. Higher resolution CT copies are available from a limited number of openly available databases, such as the Lung Image Database Consortium (LIDC) and Automatic Nodule Detection (ANODE), and from other hospitals (private databases). The following list includes a few well-known publicly accessible datasets.
The National Lung Screening Trial (NLST) was one of the clinical experiments carried out through randomization for screening of lung swelling cancer. In this study, approximately 53,454 people have enrolled whose ages ranged from 33–74 years. The duration was August 2002 and April 2004. Information was gathered on cancer detection and mortality, and it took place until 31 December, 2009. The dataset contains 26,254 low-dose CT images, 26,254 chest X-rays, and digitized histopathology images from 451 participants. More than 75,000 CT screening examinations are available from the 2 lac imaging series, which include information about participant attributes, diagnostic procedures, screening examination results, lung cancer, and death rates [26].
A dataset was created by the research teams including Vision and Image Analysis (via) and International Early Lung Cancer Action Program (I-ELCAP) to analyze the functioning of various CAD methods. The developed database contains an image set of 50 dose computed tomography whose thickness will be 1.25 mm, types, locations, and sizes of nodules are also given. The sizes of pulmonary nodules are small and accessible to the general public [27].
The LIDC-IDRI database is the most extensive, freely accessible reference resource for the early detection of lung cancer [28]. The LIDC dataset has 1018 cases of CT scan images, along with corresponding XML files that contain two-stage image annotations from 4 qualified medical radiologists. Even annotation also includes information related to types, positions, and characteristics of nodules. Lung Nodules are categorized into three parts-nodule >=3 mm, nodule <3 mm, and non-nodule >=3 mm. All images were stored in the DICOM format.
ANODE09 dataset originates from 118 NELSON. It is a freely accessible dataset consisting of 55 thoracic CT images interpreted by two radiologists. Five of these images are being used as part of the training process, while the other 50 are for testing. Some more details related to this database are mentioned in Van Ginneken et al. [29] research study. For research on lung nodules, additional databases such as Lung CT-Diagnosis, NELSON, QIN LUNG CT, Reference Image Database to Evaluate Therapy Response (RIDER) lung CT, ACRIN-NSCLC-FDG-PET, and RIDER lung PET-CT are also freely accessible in addition to NLST, via/I-ELCAP, NELSON, LIDC-IDRI, and ANODE09. All these datasets are smaller in size than LIDC-IDRI. Table 1 represents summarized information of all available data sets. Table 2 represents multiple grand competitions and challenges of the CAD System.
2.2 Articles Search Strategies
This research collected information from different relevant secondary sources, which became possible through employing different search strategies. For exploring lung nodule detection and categorization from images of CT scans, it is useful to initiate by recognizing key concepts crucial to the research. Such concepts seldom involve “CAD systems”, “CT scan images”, “Lung Nodule Detection”, and “CT Scan images”. Once such concepts are recognized, the next step includes related key terms and phrases such as “Computed tomography for lung lesions”, “Pulmonary nodule identification”, and “Automated lung cancer diagnosis”. To guarantee the search was utmost, it is useful to use synonyms and relevant terms for every key concept. Such expanded vocabulary tends to enhance search inclusivity. For example, “CT scan*AND CAD system*” widens the automated diagnosis scope, and “Lung nodule*” captures differences in nodule terminology. Logical key terms combination is integral while adopting Boolean operators, including OR, AND, and NOT, aids in structuring the search efficiently. Integrating “The CAD system” and “Lung nodule detection” guarantees an emphasis on studies at the two critical elements’ intersection.
Relevant and specialized databases such as IEEE Xplore, PubMed, and Google Scholar are employed for targeted searches in the computer science and medical imaging domains. Moreover, the examination of the citation lists of selected papers also presented seminal works and instructed researchers in basic studies in the field. In consideration of the dynamic nature of the study, filters for publication date and language are set to guarantee the inclusion of recent and related studies. Collectively, these search strategies allow researchers to cast a wide net so that both relevance and specificity in the research retrieval ensure that it pertains to lung nodule detection and categorization from the CT scan images.
Numerous research has been conducted throughout the years to increase CAD system efficiency. A CAD system is based on four basic elements: (1) preprocessing, (2) lung segmentation, (3) nodule detection, and (4) classification. All these basic elements have been accurately summarized by considering the literature study that has been done so far. A workflow diagram of a nodule CAD system is presented in Fig. 3:
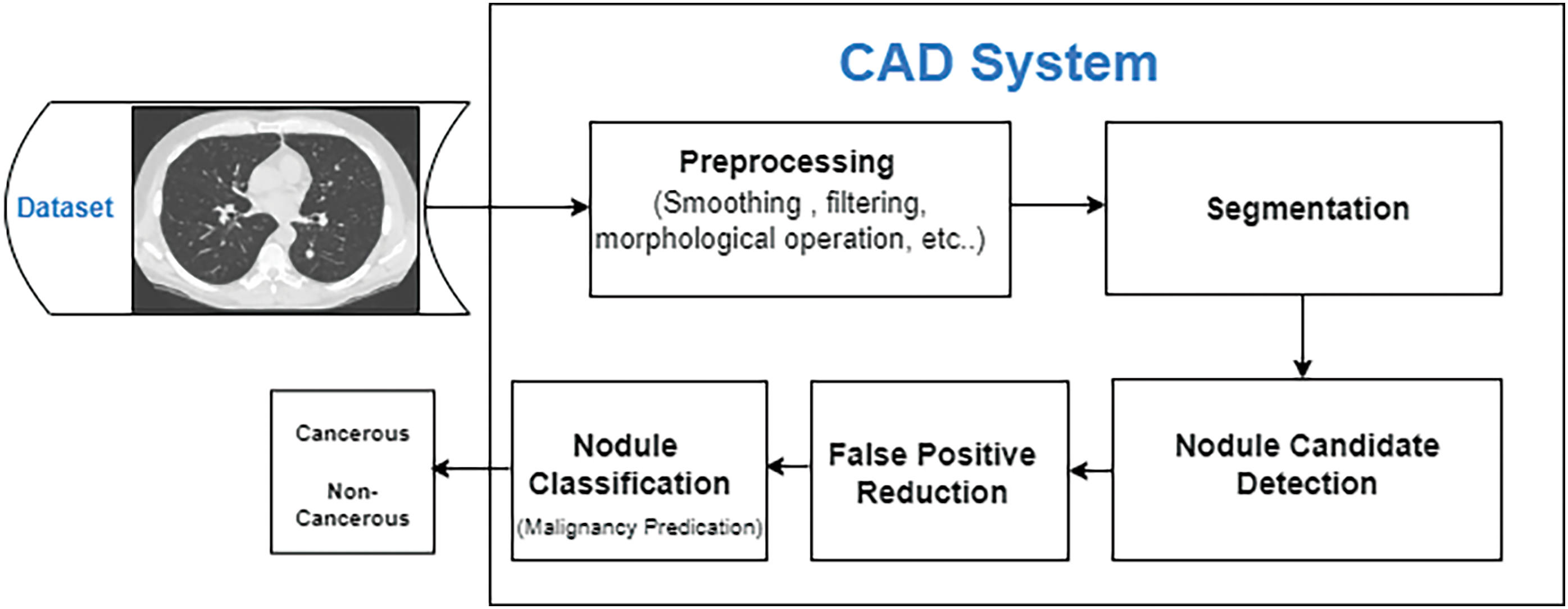
Figure 3: CAD system complete process flow diagram
The preprocessing step in the investigation of lung CT scans is highly critical. Its purpose is to refine the resolution and quality of the raw images and remove any unwanted information (noise, artifacts, etc.) from the images to achieve better results. Additionally, preprocessing increases the effectiveness of nodule detection and simplifies valuable information. Different methods and techniques are in practice for medical imaging preprocessing, such as thresholding, contrast enhancement, component analysis, morphological process, and image filtering [42–45]. Besides these approaches, thresholding and component analysis appear proficient ways to isolate lung dimensions from disrupting elements. The region expanding can be used to determine lung volume from the bronchi. After calculating the lung volume, erosion and enlargement techniques can be used to obtain a well-defined lung. To eliminate noise and improve image quality, a large number of different filters are used, including the Gaussian smooth filter, enhancement filter, smooth filtering, and the arithmetic mean filter. All these approaches can be applied and joined in different ways to gain several segmentation effects and outcomes. A research study [42] evaluated lung regions using various preprocessing techniques, including connected component labeling, hole filling, and the best gray-level thresholding. To reduce delay and eliminate noise in the processing of images, the authors applied a Gaussian filter [43]. Another paper [44] discussed three preprocessing methods for medical images: top-hat transformation, median, and adaptive bilateral filter. Among these, the adaptive bilateral filter yielded the most impressive results. A thorough description of the preprocessing is illustrated in Fig. 4.
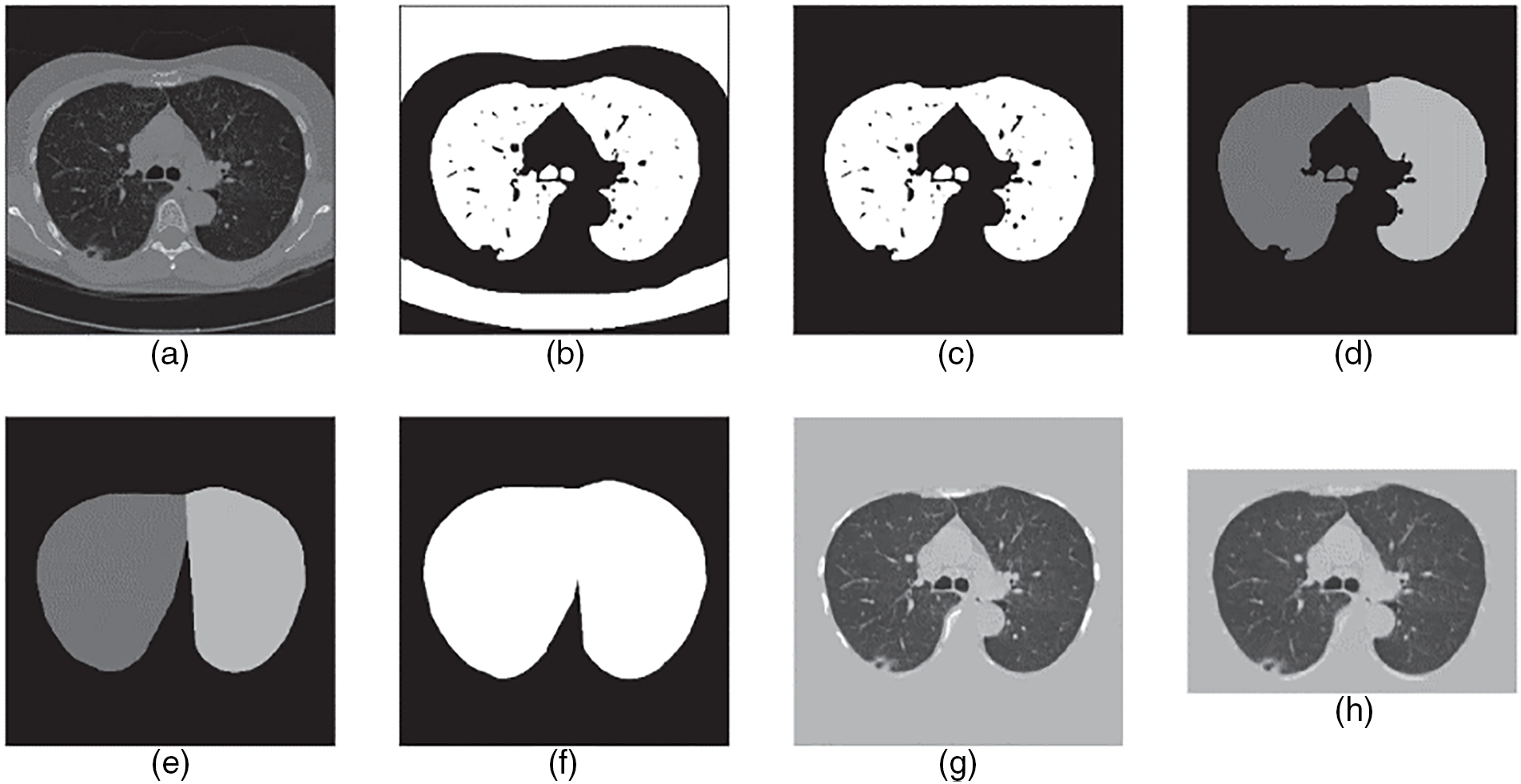
Figure 4: The step-by-step preprocessing explanation. (a) Convert the computed tomography scan into HU form. (b) Applied thresholding method to binarize the image. (c) Select the relevant lung volume region. (d) Split lungs into two groups: left lung and right lung. (e) Compute each lung’s convex hull. (f) Combine and expand 2 masks. (g) Multiply the image by the mask, then add tissue luminance to the masked area before converting the image to UINT8. (h) Resize the image into a particular size by cropping it [45]
The second stage in detecting lung nodules from CT scans is segmentation to isolate lung parenchyma from other organs. Lung nodules (LNs) are located in the lung parenchyma, so their detection allows for decreasing the number of false-positive nodules outside the lung. However, the complex lung structure makes this step extremely difficult. Soliman et al. [46] used region growth and component analysis to determine the context of all 3D pulmonary scans. The researchers then adopted a hybrid 3D Markov-Gibbs Random Field (MGRF) framework to identify the suspicious parts of the lung’s volumes and segment them as benign or cancerous nodules.
Extending above, Aresta et al. [47] presented iW-Net, a DL model with nodule dissection and user intrusion features. However, when nodules of large size used here, the proposed system worked efficiently, and the segmentation quality increased even more when user input included. Supporting this, Nemoto et al. [48] used 2D and 3D U-Net frameworks for semantic dissection of lung CT scans and compared the efficiency result with smart segmentation tools. Different algorithms for medical image segmentation based on DL are suggested. Another study [49] discussed a well-functioning CNN to segment different lung diseases in CT images. Additionally, a multi-instance loss and adversarial loss technique incorporated with the neural network to tackle the problem of segmentation in other medical cases. The approach tested on LIDC, CLEF, and HUG datasets. In this paper [50], the authors presented a recurrent U-Net (RU-NET) model and a recurrent residual U-Net (R2U-Net) model for medical image segmentation. This framework outperforms other network parameters in terms of performance. In this work [51], the authors presented a lung nodule segmentation network that incorporated multiple CNN blocks to adjust the U-Net algorithm. The study found that the effectiveness of a nodule segmentation approach can be measured based on its accuracy, efficiency, dependability, constancy, and level of automation.
It aims to locate and detect all LNs correctly in the CT scan and reduce the FP result to achieve maximum sensitivity. It’s usually divided into candidate nodule detection (CNDD) and false-positive reduction (FPRD). In the last three decades, numerous methodologies have been presented for detecting multiple LNs due to the complexities of nodule texture, size, shape, location, etc. These methods can be divided into conventional and deep neural network (DNN) approaches. The traditional methods include image processing and ML classifier techniques to recognize nodules and improve the false-positive ratio by increasing the correlation between the feature profiles and suspicious regions. Alternatively, DNN-based approaches include CNN-based methods that extract implicit features from medical images and automatically tune system performance. In the next sub-section, some effective CNDD and FPRD algorithms are presented. In Tables 3 and 4, all the useful algorithms proposed from 2018 to 2024 are listed and summarized below.
3.3.1 Candidate Nodule Detection
It is a technique to detect suspicious areas of abnormal tissues and predict their position and probability. In this step, CAD identifies as many candidate nodules (CNs) as possible to enhance the chance of a patient’s long life. For decades, various traditional methods based on hand-crafted features, i.e., thresholding, region growing, morphological operation, template matching, and shaped-based, have been commonly deployed for CNDD. Narayanan et al. [52] have detected and segmented CNs concurrently using intensity thresholding in conjunction with morphological processing. In [53], Otsu’s threshold method for nodule detection opted. Solid nodules detected using Otsu’s thresholding, included in a fixed-width sliding window. Finally, the logical OR operation used to merge all windows. Multi-layered Laplacian-of Gaussian (LoG) filters and Otsu’s used to detect sub-solid and non-solid nodules. A 3-D region-growing algorithm suggested in [54] to segment nodules accurately. The candidate nodule initially detected by using a selective enhancement filter and thresholding method. The region-based method gives a degraded performance as compared with a threshold-based CNDD.
In [11], a 3-D mass-spring model for nodule detection suggested. They applied both region-growing and morphological techniques for precise segmentation of lung volume. Same as a CAD system for nodule detection and classification is developed by Namin et al. [55]. Initially, the segmentation of LNs done using a thresholding approach—the next step involved enhancing the nodules and removing noise with a Gaussian filter. Then, the intensity and volumetric shape index (SI) utilized to find potential nodules. Finally, a Fuzzy K-Nearest Neighbor (KNN) approach employed to categorize the nodules into benign and malignant types.
Deep learning is gaining popularity due to its high success rate, and more and more detection techniques based on DNNs are being presented. Many DDN-based algorithms, especially convolutional neural networks (CNNs), are utilized to extract both low-and high-level internal features from nodules and significantly increase sensitivity and accuracy detection rates. As a result, candidate nodules can be identified with ease. The models most commonly used for nodule detection include CNN, U-Net, feature pyramid network (FPN), region proposal network (RPN), residual network (ResNet), visual geometry group (VGG-net), densely connected convolutional network (Dense-net), Inception Res-net, Alexa-net, Google-net, and Retina-Net. Essential components such as convolutions and pooling are almost identical in all these network architecture.
Nguyen et al. [56] designed a CAD system with adaptive anchor boxes based on the Faster R-CNN framework for CNDD. A mean-shift clustering approach used to automatically learn anchor boxes using the ground truth nodule size of the training data. Learning anchors improve the performance of fast R-CNNs in node detection. Another study [57] presented a new method for speedy nodule candidate detection. We applied a nodule size fitting model that complies with a Faster R-CNN algorithm to detect various nodule types, locations, and sizes. After detection, candidates can be located based on their bounding box. In addition, several convergent networks have been developed that combine the performance of multiple networks using multi-stream architectures [58–60].
Zheng et al. [58] developed a computerized system using CNN-based maximum intensity projection (MIP). The proposed architecture included four merged 2DCNNs based on U-Net and one 3DCNNs based on VGG-NET. Component analysis and binary morphology used to segment lung volume thresholds. The authors then used 2D-CNN to detect potential nodules in four streams using MIP images with various plate thicknesses as input. The proposed system tested on LUNA16 and LIDC-IDRI datasets and achieved high sensitivity. A DL base approach developed in [60], to enhance the performance of pulmonary nodule detection. The authors applied a CNN of five convolutional layers with an 11-layer CNN (5 convolutional layers) to extract nodule features. The system tested and trained on LIDC/IDRI datasets.
3.3.2 False Positive Reduction
Even after the CNDD stage, a high number of FPs per scan reduces the accuracy of node detection. Too many FPs lead to overdiagnosis and treatment, as well as a waste of medical resources and time. Thus, reduce in the number of FPs is required to enhance the accuracy of nodule detection. FPRD primarily focuses on identifying true nodules from the extracted candidates and removing false nodules during the detection phase corresponding to the binary classification task. Many studies have focused on FPRD. Numerous studies have devoted their attention to FPRD [24,61,62].
In the FPRD stage, a number of parameters based on intensity, texture, and shape must be extracted from potential nodule CT images and used as input for classifiers to distinguish between nodules and non-nodules. Several ML classifiers are frequently employed to recognize true and false nodules, such as support vector machine (SVM), KNN classifiers, linear discriminant classifiers, and various boosting classifiers. One of the most difficult aspects of this phase is to find the proper balance between sample size and nodule type. Due to the complexity of the location, different sizes and types can confuse the classifier. For instance, a study [62] introduced a type-II fuzzy approach to enhance the quality of raw CT scan images. The researcher next applied a revised spatial kernelized fuzzy c-means clustering algorithm for inner structure segmentation. Finally, morphological operations such as opening, closing, and filling utilized for nodule detection. In order to determine if the nodule candidate is malignant or benign and has lower FPs, three classifiers—MLP, KNN, and SVM are used. In [24], the authors presented an automatic CAD system for finding lung nodules. First, they implemented the gaussian mixture model (GMM) for nodule segmentation and then worked on a morphology filter to remove the vessels. For CNDD, they used various intensity and shape-based traits. To cut down on false positives, they employed an SVM classifier with 10-fold cross-validations and obtained a sensitivity rate of 89.77 with the LIDC/IDRC dataset. Table 3 shows the summary of candidate nodule detection, FP reduction, and information.
In the past, DL networks found to be more effective than traditional methods for classifying and identifying images. Currently, it is the preferred technology due to its fast training, adaptable structures, diverse usage, and accuracy. As a result, researchers have put forth a range of DL algorithms for improved detection and classification performance [71–73,80]. Elaborating thus, Ozdemir et al. [71] presented a novel diagnostic system that accurately assesses probabilities on low-dose computed tomography images of lung cancer. This system utilizes a 3D-CNN to identify and classify malignant LNs. The system tested on two available datasets, the Kaggle data science bowl challenge and Luna16. To reduce the FP rate and improve accuracy, the authors highlighted the importance of the coupling between the diagnosis and detection components. For the first time, the CT images analyzed using DL systems, and model uncertainty used to determine calibrated probabilities for the classification process. This combination of calibrated probabilities and model uncertainty leads to risk-based decision-making for the diagnosis phase and helps to increase accuracy.
Masood et al. [73] proposed a cloud-based automated CADe system based on a 3D-CNN model in this research. Median Intensity Projection (MeIP) generates MeIP images, after which traditional procedures utilized to extract multi-scale, multi-angle, and multi-view patches from the MeIP images. In the nodule detection part, a multi-region proposal network (mRPN) model designed with a modified VGG-16 backbone to identify potential nodules from the extracted patches. Finally, a 3D-CNN with a modified ResNet-10 used to decrease FPs. The proposed system underwent a thorough evaluation using LUNA16, ANODE09, and LIDC-IDRI data, achieving sensitivities of 0.98, 0.976, and 0.988 respectively with 1.97, 2.3, and 1.97 FPs/scan.
Advanced image processing algorithms for CAD systems have mainly developed the innovative use of deep learning in detecting canal nodules, improving lung cancer diagnosis confidently and accurately. One of the modern developments is you only look once (YOLO) [80], which has recently been attributed to being revolutionary, as it allows for the real-time analysis of lung CT scans by predicting the most appropriate boxes for localization while offering class probabilities. As for the presented research, although YOLO increases the speed and reliability of detection, the ability of the method to detect small and faint nodules, which are essential when diagnosing cancer at an early stage, still necessitates independent clinical testing.
Cao et al. [72] addressed the problem of detecting LNs due to its complexity and environmental heterogeneity by proposing a two-level convolutional neural network (TSCNN). A modified/extended U-Net segmentation network has been used to emphasize the initial presence of nodules at the first stage. They proposed an advanced training sampling technique to achieve higher recall and minimize the introduction of excessive FP nodes. This phase also provides a two-phase forecasting approach, which is a method of predicting future instances based on past and present data. In the final stage, we developed three 3D CNN-based algorithms to eliminate false-positive LNs based on SeResNet, DenseNet, and InceptionNet classification networks. According to [78], detecting small nodules from lung medical images is very challenging. The suggested work develops a unique CASP YOLOv5 model that expands the experimental field of the ASPP Module and combines and improves CBAM YOLOv5s and the detection head process for lung nodule prediction. Here, experiments performed on two datasets named LUNA16 and X-Nodule. The results of the experiment indicate that for LUNA16, the F1 score is 0.720, the mean average precision is 0.740, and for X-Nodule, it is 0.794 and 0.766, respectively. In [79], an analysis of lung cancer done. Here, DL-based methods have been applied to classify and detect lung nodules. The proposed system provides a comprehensive analysis of various fundamental architectures related to the classification of pulmonary nodules (Mobillenetv3, Resnet101, VGG-16) and for lung nodule position detection, including Faster-RCNN, YOLOv3, and SSD. For detecting the position of nodules, the results show that a good average accuracy rate lies between 0.92 and 0.95, with an average detection accuracy range of 0.93 to 0.94 for detecting the position of nodules.
The final CAD step system is nodule classification, involving FP reduction, which can increase classification efficiency. The majority of LNs are benign; however, it could be a sign of lung cancer in its early stages. Early diagnosis of a cancerous lesion can increase the patient’s survival rate. Many CAD systems have been developed to assist doctors in differentiating malignant lesions from benign ones, but few of them have been designed for nodule-type classification [21]. For instance, the patient’s age, sex, smoke, toxic particles, and alcohol are leading causes of nodule lesions. Although malignant nodules are large in size having diameters greater than 8 mm with complex surface structures and characteristics. Estimating the malignancy probability is mainly dependent on analyzing the measurements of the nodule’s size and the way it looks. Table 4 represents nodule classification techniques.
Different classification methods are used: (1) ML classifiers, such as SVM, Decision Trees, Principal Component Analysis (PCA), KNN classifiers, Bayesian classifiers, and optimal linear classifiers [69,89]; (2) Deep Neural Network include CNNs [72,82,84]; (3) CNN integrated with ML classifiers [23]; (4) CNNs trained with transfer learning algorithms [70,96].
In this study [69], various ML algorithms, including PCA, KNN, SVM, Naïve Bayes, Decision Trees, and Artificial Neural Networks (ANN), are opted to identify lung cancer nodules. All the techniques are compared, and ANN and Decision Tree show the best results, with 82, 93% accuracy with preprocessing and 43, 24% without preprocessing. A study [84] presented a lightweight multi-section CNN method to analyze cross-sections of nodules and classify them into either cancerous/non-cancerous. The validity of this approach tested on the LIDC data and obtained an average accuracy of 93.18%. In [82], authors presented a new approach based on the CNN algorithm called Gated-Dilated (GD) networks. The GD network used two dilated convolutions as an alternative to max-polling to accurately identify nodules of varying sizes as either benign or malignant. Moreover, a Context-Aware sub-network used to direct the features among the two dilated convolutions. The technique tested on the LIDC-IDRI data and achieved remarkable results of 92.57% accuracy with a sensitivity of 92.67%. In [95], the authors developed a novel DL-based model that automatically detected and analyzed CT images of lung nodules. In this work, the main focus is on nodule classification and segmentation. MRKM clustering applied to perform segmentation, and an enhanced CNN model utilized for classification. For feature accuracy enhancement, the ATSO algorithm has been used to select CNN classifier parameters in an optimized way and achieved a maximum accuracy of 96.5% with a dice similarity coefficient of 94.5%.
A recurrent residual CNN that based on the U-Net approach presented by Xia et al. [12]. Then, to categorize nodules as either IA or non-IA, we integrated DL with radiomics characteristics, and as a result, we achieved an AUC value of 0.90 plus or minus 0.03. Authors [23] proposed an automated diagnosis system called DeepLung. DeepLung is separated into nodule detection and classification stages. Initially, preprocessing methods applied to segment lung volume. Then, to identify potential nodules, a 3D Faster R-CNN with a 3D dual-path block and an encoder-decoder structure built, similar to a U-Net. Afterward, 3D-DPN utilized to extract suspicious characteristics of the candidate nodule. Finally, a gradient boosting machine (GBM) in conjunction with DPN applied to decide if the LNs are cancerous. Testing the LIDC datasets with this method led to an accuracy of 87.5% and an error rate of 12.5%.
Based on CT scans, the authors [91] presented a novel technique for diagnosing LNs that combines deep transfer CNN with an extreme learning machine. They used an extreme learning machine classifier after using a DL transfer CNN to extract high-level characteristics of lung nodules. It shown to generate results that superior to those obtained using other approaches considered state-of-the-art approaches. In their most recent article [70], a DL-based method to lower the false positive rate of nodule recognition suggested to increase networks’ effectiveness for nodule detection. They changed parameters in fully linked layers and fine-tuned VGG-16 using transfer learning. Then, they used these finely tuned networks as feature extractors to pull out characteristics of lung nodules, after which they trained SVM to classify the nodules.
A comprehensive examination of CADx systems has reflected a foremost advancement in automated lung nodule analysis. Neural Network-based algorithms such as CNNs are utilized to enhance the measures of accuracy, sensitivity, specificity, and FP rate per scan in nodule detection and classification processes, making CAD systems more efficient at identification and analyzing lung cancer in its initial stages. Although CT scanning methods and DL techniques are growing in popularity, more intelligent CAD systems are being developed, yet there are still issues and difficulties that need to be tackled. The research findings demonstrated that optimizing DL algorithms for CAD of LNs includes mitigating challenges for elevating accuracy, interpretability, and initial-phase detection while enhancing the classification of lung cancer. An integral component is the refinement of the DL model architecture to enhance interpretability. CNNs have exhibited success; however, attention must be offered to architecture such as XAI or attention mechanisms–it allows the model to point out regions impacting its decisions. The attention of attention maps and salience approaches offer support in gaining better insights into the features that are useful to nodule detection. Additionally, mitigating data challenges is vital. Annotating immense datasets with different nodule features guarantees the model witnesses an extensive scenarios range so that generalization is boosted. In link with this, transfer learning from pre-trained frameworks on comprehensive datasets tends to elevate performance, particularly in scenarios with restricted labeled information. The stress on data augmentation methods guarantees effectiveness by presenting the model to differences in nodule appearances. In accuracy terms, fine-tuning hyperparameters remains useful. For instance, as per this research findings, holistic experimentations with learning rates, optimizer choices, and batch sizes allow resulting configurations, heightening overall accuracy and model convergence. Additionally, incorporating ensemble approaches that combine predictions from various models elevates generalization. The CNN integration has advanced CAD systems for detecting lung nodules in chest CT scans. The hybrid technique integrates CNN’s strengths in learning considerable patterns with conventional radiomic features; thus, elevating interpretability. Mitigating false positives, the emphasis on the refinement of FPRD employing modern techniques, including deep reinforcement learning and 3D CNNs, elevates accuracy. In the classification of lung cancer, hybrid models integrating DL with conventional ML classifiers, augmented by SHAP values, are explainability techniques that synergize strengths for efficient decision-making in medical aspects.
The above research has been supported by several past studies that considered CT scans as an adequate standard for exploring lung parenchyma [14,86,96]. As Huang et al. [91], DL has become prominent and substantial attention, and numerous CNN architectures have been executed progressively in CAD schemes for diagnosing lungs. The CNN procedure is integrally classified by its tendency to acquire multiple ascertained data. Moreover, it employs restricted direct supervision to maximize categorization. CNN is also capable of auto-defining unfamiliar features. Some techniques exhibit compelling outcomes with either high rates of sensitivity or low rates of FP. Maintaining high sensitivity rate linked with a low level of FP is essential for building robust CADe systems and proving their usefulness in discriminatory pulmonary nodules. Khan et al. [13] and Nasrullah et al. [86] executed a DL architecture, regarded as Multi-Crop CNN, enabling the cropping of nodule regions with salient data from convolutional feature maps and implementing numerous max pooling.
Past studies [14,17,56] illustrated the substantial progress made in CAD systems for detecting and classifying lung nodules in CT scans. Primarily, the emphasis lies in applying DL, especially CNNs because of their tendency to retrieve automatically features from input information–this eliminates the requirement for feature engineering manually [67]. The transition towards deep learning has been accelerated by its tendency to learn patterns and spatial data–it is imperative for the CT scans’ 3D nature. Comparing MTANNs (massive-training artificial neural networks) and CNN models presented the MTANN’s efficiency in the detection and classification of nodules–this stresses their adeptness at controlling semantic features of the lower level. Studies seldom integrated numerous CNN architectures or employed ensembled approaches for elevating system performance [55,56]. Approaches ranged from employing different CNNs with numerous patch sizes for nodule detection of the candidates to embedding deep features retrieved by networks such as AlexNet into cascade classifiers [17,44].
Notably, the integration of handcrafted characteristics with deep features in some cases outperformed employing either CNN or SVM alone for the classification of lung nodules. The hybrid strategy intended to use paradigms strengths [4,22]. The 3D CNNs choice over 2D CNNs pointed out as more suited for the CT scans 3D nature–this offers effective spatial data modeling [97]. Nevertheless, despite the benefits of 3D CNNs, most of the chosen works (approximately 91%) employed 2D CNN architectures, illustrating a prevailing preference. Zhao et al. [30] and Wang et al. [75] presented the integral contribution to deep learning, particularly CNNs, in surpassing the constraints of conventional CAD approaches and other ML approaches. Computer-aided detection (CAD) exhibited a substantial advancement by producing related features automatically with the help of pooling and convolution layers, overcoming dependency on feature extraction manually [96,98]. CNNs’ iterative self-learning approach played a role in the persistent improvement in detecting lung nodule rates. Comparing traditional CAD and deep learning methods pointed out the benefits of CNNs such as their tendency to learn directly from images without the demand for segmentation of nodules [82,85]. It does not merely preserve more data but also eradicates the losing imperative details risks at the time of segmentation. Obtained deep features from massive training information explored for having a superior representational capability in comparison to handcrafted characteristics, which are seldom dependent on expert analysis subjectively [20].
5 Challenges and Future Perspectives
There is a need for a sizable amount of precise and well-categorized datasets for effective deep-learning models to research and analyze medical images. Unfortunately, the existing publicly available database of lung CT scan images is not labeled correctly, leading to mixed-up annotations for the different datasets. Consequently, gathering thoracic CT images with precise labels is a challenging task. Conversely, confidentiality can be the most difficult complication in collecting individual lung CT scans. There are also various hospital protocols and national strategies to protect personal data.
To tackle the data shortage issue, data augmentation techniques are used, which include scaling, cropping, flipping, and rotation of CT scan images. Additionally, Generative Adversarial Networks (GAN) are implemented to generate additional artificial samples [18]. On the other hand, CAD systems mainly rely on supervised learning (SL) for nodule detection and classification. However, SL is expensive and time-consuming, requiring huge, labeled data. Semi/Unsupervised learning methods, like transfer learning or fine-tuning, may be more appropriate in such situations to reduce FPs.
In the case of SL, for example, the fact that there is usually very little labeled data is often accompanied by overfitting, where the model learns noise rather than the phenomena that are looked at in pattern recognition [12]. This is further magnified when the complexity of the model is higher than the amount of information incorporated in the development of the model by performing poorly in unseen examples. Also, convergence issues arise when the models fail to learn optimal parameters for their corresponding models, giving rise to unstable training dynamics or sub-optimality. Other related approaches, such as cross-validation, early stopping, and learning rate scheduling, can go a long way in reducing overfitting but cannot fully overcome the problems from limited data. This requires moving toward approaches for learning from the data that incorporate the natural structure of the data without necessarily having labeled instances.
There are better ways of improving the model component that can reduce sensitivity to noise when trained with limited data, including unsupervised learning. Methods such as GANs and autoencoders involve creating new training examples or embedding vectors in order to represent essential elements of the original data. These methods help to reduce overfitting associated with stout lar possibility due to an insufficient number of labeled samples in a training dataset by increasing the size of the training data set or by selecting meaningful features. Moreover, self-supervised learning and the transfer learning model apply to readymade data association or ready models to amend the task-oriented models, and this eliminates the possibility of getting trapped in local optima as it starts at a more stable point of convergence. Although, indeed, these approaches are not without their issues–for one, synthetic data may deviate from reality to avoid compromising their sources, or, when employing transfer learning, starting with a designated domain might prove difficult–these are novel concepts for overcoming the limitations of scarcity in data for the improvement of the supervisory training of learning models.
A CNN-based neural network model trained using a black box manner automatically recognizes and categorizes LNs but does not explain how they develop. For radiologists to identify the primary root of the disease, the interpretability of a DL model is crucial. For this more study of neural networks is needed to extend the interpretability of the neural network models, because if there is a difference between doctor and CAD system assessment, it will help the radiologists to find the main reason and to overcome the doubts and concerns.
5.4 Lack of Continual Learning Ability
Radiologists require a proficient CAD system in order to deliver the most suitable clinical decisions when faced with unexpected samples. Consequently, the CAD system must possess the capability to learn and be trained to identify new medical images. However, the current CAD systems are typically developed and executed using trained models, indicating that they work correctly in static conditions rather than varying conditions. As a result, they are unable to detect a few untrained samples resulting in inaccurate diagnosis [99]. Thus, it is essential to create and construct a CAD system that has the capacity to continually learn and adapt to the varying conditions that happen in real-time contexts.
Implementing a novel CNN architecture employing cloud computing techniques is an effective way to construct systems that can learn continually. Using this approach, the nodule recognition archives can be stored on the cloud to upgrade the datasets, thus enabling the presented deep learning network to be trained on a cloud for adjusting real-time environment variations without interruption [73].
5.5 Uncertain Quantification (UQ)
The practice of UQ in DNN can enhance the validity of the semi-supervised medical image segmentation process [20]. This could be a useful area of research study for pulmonary nodule CAD systems in the future. A significant number of resources and stringent regulations will be mandatory for lung nodule screening to guarantee substantial gains and lessen FPs. The design and testing of more straightforward nodule evaluation algorithms using new diagnostic modalities like molecular signatures, biomarkers, and liquid biopsies should also be the focus of future research [97]. Machine learning and DL algorithms are the best options for this endeavor and could lead to remarkable advancements in the field of radiology due to their ability to automatically detect and classify nodules with greater accuracy.
The holistic review of CADx systems for lung nodule detection and categorization pointed out multiple implications of the research. Most importantly, the dataset scarcity challenge makes it necessary to emphasize the creation of a standardized and well-annotated dataset for CT scans of the lung that mitigates challenges of data security and confidentiality with the help of collaboration between medical institutions. In addition, the challenge of overfitting in SL becomes appropriate for researchers investigating unsupervised learning approaches, such as transfer learning, to enhance the CAD system generalization capability with restricted labeled data. For future research, the lack of interpretability urges the enhancement of interpretability that facilitates CAD systems and radiologists’ collaboration by probing how models make decisions and how methods are developed to interpret and explain their outputs.
With respect to research findings’ implications, the insufficiency of continual learning capabilities in contemporary CAD systems implies that the study efforts must be directed towards developing systems with the tendency for continual learning, significantly through innovative CNN designs and cloud-based approaches. Besides, the integration of uncertain quantification techniques, particularly in semi-supervised segmentation of medical images, hints at an effective avenue for more clinically applicable and reliable systems of CAD. In the future, researchers must investigate the design and testing of CAD algorithms, which includes information from new diagnostic modalities like liquid biopsies and molecular signatures, for more accurate and extensive assessments of lung nodules. Mitigating regulatory and ethical considerations is imperative, so researchers in the future must consider the development of standards and guidelines for regulatory and ethical considerations while using CAD systems in lung nodule screening. Finally, investigating hybrid approaches (which integrate conventional ML methods with DL algorithms) could elevate the CAD system’s reliability and performance for lung nodule analysis. Thus, the implication of this clinical research can play a role in the CAD systems advancement so that more interpretable, adaptable, and effective for emerging medical practices.
Technology is being used extensively in the healthcare sector to diagnose chronic diseases. As early detection can surge the endurance rate of patients, therefore, continuous advancement in CAD system is the need of an hour. Researchers are constantly trying to devise new methods and, at the same time, making efforts to address the shortfalls in existing detection and diagnostic procedures of lung nodules. In this paper, we have discussed the major causes and types of lung nodules. Further, we have compared the reported detection and classification nodule algorithms based on the different datasets and various performance metrics.
Based on the research findings, the CAD field for lung nodule analysis has seen prominent advancements, especially with the integration of neural network algorithms such as CNNs. Such algorithms have enhanced the sensitivity, accuracy, and false-positive rates, as well as detected and categorized nodules’ specificity. The ability of CAD systems to recognise and analyse lung cancers in their initial phases has been enhanced. Despite the advancements, challenges and opportunities for improvement remain. DL optimization is integral to dealing with issues regarding early detection, interpretability, and accuracy. DL model architecture refinement including employing Explainable AI (XAI) or attention mechanisms that can elevate interpretability by pointing out areas influencing model decisions. The integration of attention maps and saliency approaches can form a better understanding of the attributes impacting nodule detection.
Dealing with data challenges is integral. Annotating an extensive dataset with numerous nodule attributes ensures the model deals with a wide number of scenarios, elevating generalization. Fine-tuning hyperparameters is vital to acquiring accuracy. Practical experimentation with learning rates (LR), optimizer selection, and batch sizes assist in demonstrating configurations, which elevate model convergence and accuracy. Ensemble methods can be used to enhance generalization when combining predictions from diverse models. Clinical acceptance has made interpretable features considerable. Integrating radionics features, as well as the DL model, allows emphasizing shape, intensity, and texture that helps in comprehending decision-making approaches. The hybrid technique capitalizes on the DL strengths in learning substantial patterns while retaining the conventional radionics features’ interpretability.
For further mitigating FPs and enhancing the initial detection phase, a concerted effort must be given toward the refinement of the FPRD phase. Using modern techniques, including 3D CNNs, while deep reinforcement learning for FPRD enhances the ability of the model to differentiate true nodules from false positives so that the nodule detection accuracy is elevated. In the lung cancer categorization setting, the emphasis must divert towards developing hybrid models, which combine DL algorithms with other categorization techniques like ML classifiers. The approach causes accurate classifications of lung cancer. Thus, advancements in CADx are effective, and continued research and development (R&D) endeavors are required to minimize current challenges and make such systems even more proficient tools for initial detecting and diagnosing lung cancer.
Acknowledgement: The authors are grateful to all the editors and anonymous reviewers for their comments and suggestions and thank all the members who have contributed to this work with us.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: Study conception and design: Salman Ahmed; Data collection and analysis: Salman Ahmed; Enhance the scientific rigor and readability of the review: Mazliham Mohd Su’ud, Muhammad Mansoor Alam; Draft manuscript preparation: Salman Ahmed; Review and editing: Fazli Subhan, Adil Waheed. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Not applicable.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. “What is cancer? Differences between cancer cells and normal cells,” National Cancer Institute, Oct. 2021. Accessed: Apr. 15, 2024. [Online]. Available: https://www.cancer.gov/about-cancer/understanding/what-is-cancer [Google Scholar]
2. “Cancer,” Feb. 2022. Accessed: Apr. 15, 2024. [Online]. Available: https://www.who.int/news-room/fact-sheets/detail/cancer [Google Scholar]
3. “Key statistics for lung cancer,” The American Cancer Society, Jan. 2024. vol. 5, Accessed: Apr. 15, 2024. [Online]. Available: https://www.cancer.org/cancer/lung-cancer/about/key-statistics.html [Google Scholar]
4. N. Camarlinghi, “Automatic detection of lung nodules in computed tomography images: Training and validation of algorithms using public research databases,” Eur. Phys. J. Plus., vol. 128, no. 9, Sep. 2013, Art. no. 110. doi: 10.1140/epjp/i2013-13110-5. [Google Scholar] [CrossRef]
5. S. Cressman et al., “The cost-effectiveness of high-risk lung cancer screening and drivers of program efficiency,” J. Thorac. Oncol., vol. 12, no. 8, pp. 1210–1222, 2017. doi: 10.1016/j.jtho.2017.04.021. [Google Scholar] [PubMed] [CrossRef]
6. C. S. Dela Cruz, L. T. Tanoue, and R. A. Matthay, “Lung cancer: Epidemiology, etiology, and prevention,” Clin. Chest. Med., vol. 32, no. 4, pp. 605–644, 2011. doi: 10.1016/j.ccm.2011.09.001. [Google Scholar] [PubMed] [CrossRef]
7. “Statistics at a glance, 2022 Top 5 most frequent cancers,” Global Cancer Observatory. 2022. Accessed: Apr. 15, 2024. [Online]. Available: https://gco.iarc.who.int/media/globocan/factsheets/populations/76-brazil-fact-sheet.pdf [Google Scholar]
8. W. J. Kostis, A. P. Reeves, D. F. Yankelevitz, and C. I. Henschke, “Three-dimensional segmentation and growth-rate estimation of small pulmonary nodules in helical CT images,” IEEE Trans. Med. Imaging, vol. 22, no. 10, pp. 1259–1274, 2003. doi: 10.1109/TMI.2003.817785. [Google Scholar] [PubMed] [CrossRef]
9. H. Macmahon et al., “Guidelines for management of incidental pulmonary nodules detected on CT images: From the Fleischner Society,” Radiology, vol. 284, no. 1, pp. 228–243, 2017. doi: 10.1148/radiol.2017161659. [Google Scholar] [PubMed] [CrossRef]
10. P. Monkam, S. Qi, H. Ma, W. Gao, Y. Yao and W. Qian, “Detection and classification of pulmonary nodules using convolutional neural networks: A survey,” IEEE Access, vol. 7, pp. 78075–78091, 2019. doi: 10.1109/ACCESS.2019.2920980. [Google Scholar] [CrossRef]
11. D. Cascio, R. Magro, F. Fauci, M. Iacomi, and G. Raso, “Automatic detection of lung nodules in CT datasets based on stable 3D mass-spring models,” Comput. Biol. Med., vol. 42, no. 11, pp. 1098–1109, 2012. doi: 10.1016/j.compbiomed.2012.09.002. [Google Scholar] [PubMed] [CrossRef]
12. X. Xia et al., “Comparison and fusion of deep learning and radiomics features of ground-glass nodules to predict the invasiveness risk of stage-I lung adenocarcinomas in CT scan,” Front. Oncol., vol. 10, Mar. 2020, Art. no. 43. doi: 10.3389/fonc.2020.00418. [Google Scholar] [PubMed] [CrossRef]
13. S. A. Khan, S. Hussain, S. Yang, and K. Iqbal, “Effective and reliable framework for lung nodules detection from CT scan images,” Sci. Rep., vol. 9, no. 1, Dec. 2019, Art. no. 248. doi: 10.1038/s41598-019-41510-9. [Google Scholar] [PubMed] [CrossRef]
14. J. Zhang, Y. Xia, H. Cui, and Y. Zhang, “Pulmonary nodule detection in medical images: A survey,” in Biomedical Signal Processing and Control. Elsevier Ltd., May 1, 2018, vol. 43, pp. 138–147. doi: 10.1016/j.bspc.2018.01.011. [Google Scholar] [CrossRef]
15. I. W. Harsono, S. Liawatimena, and T. W. Cenggoro, “Lung nodule detection and classification from Thorax CT-scan using RetinaNet with transfer learning,” J. King Saud Univ.–Comput. Inf. Sci., vol. 34, no. 3, pp. 567–577, 2022. doi: 10.1016/j.jksuci.2020.03.013. [Google Scholar] [CrossRef]
16. B. K. J. Veronica, “An effective neural network model for lung nodule detection in CT images with optimal fuzzy model,” Multimed. Tools Appl., vol. 79, no. 19–20, pp. 14291–14311, 2020. doi: 10.1007/s11042-020-08618-x. [Google Scholar] [CrossRef]
17. I. R. S. Valente, P. C. Cortez, E. C. Neto, J. M. Soares, V. H. C. de Albuquerque and J. M. R. S. Tavares, “Automatic 3D pulmonary nodule detection in CT images: A survey,” Comput. Methods Programs Biomed., vol. 124, pp. 91–107, 2016. doi: 10.1016/j.cmpb.2015.10.006. [Google Scholar] [PubMed] [CrossRef]
18. C. Han et al., “Synthesizing diverse lung nodules wherever massively: 3D multi-conditional GAN-based CT image augmentation for object detection,” presented at the 2019 Proc. Int. Conf. 3D Vis., 3DV, Quebec City, QC, Canada, Sep. 9–12, 2019, pp. 729–737. [Google Scholar]
19. R. Mastouri, N. Khlifa, H. Neji, and S. Hantous-Zannad, “Deep learning-based CAD schemes for the detection and classification of lung nodules from CT images: A survey,” J. Xray Sci. Technol., vol. 28, no. 4, pp. 591–617, 2020. doi: 10.3233/XST-200660. [Google Scholar] [PubMed] [CrossRef]
20. M. Abdar et al., “A review of uncertainty quantification in deep learning: Techniques, applications and challenges,” Inf. Fusion, vol. 76, pp. 243–297, 2021. doi: 10.1016/j.inffus.2021.05.008. [Google Scholar] [CrossRef]
21. F. Ciompi et al., “Towards automatic pulmonary nodule management in lung cancer screening with deep learning,” Sci. Rep., vol. 7, pp. 1–11, 2017. doi: 10.1038/srep46479. [Google Scholar] [PubMed] [CrossRef]
22. S. Singh and R. Deshmukh. “Lung cancer screening market size, share, competitive landscape and trend analysis report, by type, by age group, by end user: Global opportunity analysis and industry forecast, 2021–2031,” Oct. 1, 2022. Accessed: Apr. 15, 2024. [Online]. Available: https://www.alliedmarketresearch.com/lung-cancer-screening-marketA31460 [Google Scholar]
23. W. Zhu, C. Liu, W. Fan, and X. Xie, “DeepLung: Deep 3D dual path nets for automated pulmonary nodule detection and classification,” presented at the IEEE Winter Conf. Appl. Comput. Vis., WACV 2018, Lake Tahoe, NV, USA, Mar. 12–15, 2018, pp. 673–681. [Google Scholar]
24. A. Halder, S. Chatterjee, and D. Dey, “Morphological filter aided GMM technique for lung nodule detection,” presented at the IEEE Appl. Signal Process. Conf., ASPCON 2020, 2020, pp. 198–202. doi: 10.1109/ASPCON49795.2020.9276715. [Google Scholar] [CrossRef]
25. S. Mehmood et al., “Malignancy detection in lung and colon histopathology images using transfer learning with class selective image processing,” IEEE Access, vol. 10, pp. 25657–25668, 2022. doi: 10.1109/ACCESS.2022.3150924. [Google Scholar] [CrossRef]
26. “The National Lung Screening Trial (NLST) datasets and data dictionaries,” Dec. 31, 2009, pp. 29–30. Accessed: Apr. 15, 2024. [Online]. Available: https://cdas.cancer.gov/datasets/nlst/ [Google Scholar]
27. “ELCAP Public Lung Image Database Welcome to the VIA/I-ELCAP Public Access Research Database,” Dec. 20, 2003. Accessed: Apr. 15, 2024. [Online]. Available: http://www.via.cornell.edu/databases/lungdb.html [Google Scholar]
28. S. G. Armato et al., “The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRIA completed reference database of lung nodules on CT scans,” Med. Phys., vol. 38, no. 2, pp. 915–931, 2011. doi: 10.1118/1.3528204. [Google Scholar] [PubMed] [CrossRef]
29. B. Van Ginneken et al., “Comparing and combining algorithms for computer-aided detection of pulmonary nodules in computed tomography scans: The ANODE09 study,” Med. Image Anal., vol. 14, no. 6, pp. 707–722, 2010. doi: 10.1016/j.media.2010.05.005. [Google Scholar] [PubMed] [CrossRef]
30. Y. R. Zhao, X. Xie, H. J. De Koning, W. P. Mali, R. Vliegenthart and M. Oudkerk, “NELSON lung cancer screening study,” Cancer Imaging, vol. 11, pp. 79–84, 2011. doi: 10.1102/1470-7330.2011.9020. [Google Scholar] [PubMed] [CrossRef]
31. S. G. Armato et al., “LIDC-IDRI | Data from the Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRIA completed reference database of lung nodules on CT scans,” Cancer Imaging Arch., pp. 4–8, 2012. doi: 10.7937/K9/TCIA.2015.LO9QL9SX. [Google Scholar] [CrossRef]
32. O. Grove et al., “Quantitative computed tomographic descriptors associate tumor shape complexity and intratumor heterogeneity with prognosis in lung adenocarcinoma,” PLoS One, vol. 10, no. 3, pp. 1–14, 2015. doi: 10.1371/journal.pone.0118261. [Google Scholar] [PubMed] [CrossRef]
33. R. Goldgof et al., “Data from QIN_LUNG_CT,” Cancer Imaging Arch., no. 2015, pp. 28–29, 2021. doi: 10.7937/K9/TCIA.2015.NPGZYZBZ. [Google Scholar] [CrossRef]
34. B. Zhao, L. H. Schwartz, M. G. Kris, and G. J. Riely, “Coffee-break lung CT collection with scan imagesreconstructed at multiple imaging parameters (Version 3),” Cancer Imaging Arch., 2015. doi: 10.7937/k9/tcia.2015.u1x8a5nr. [Google Scholar] [CrossRef]
35. P. Muzi, M. Wanner, and P. Kinahan, “Data from RIDER Lung PET-CT,” Cancer Imaging Arch., pp. 5–6, 2015. doi: 10.7937/k9/tcia.2015.ofip7tvm. [Google Scholar] [CrossRef]
36. M. Machtay et al., “Prediction of survival by [18F] fluorodeoxyglucose positron emission tomography in patients with locally advanced non-small-cell lung cancer undergoing definitive chemoradiation therapy: Results of the ACRIN 6668/RTOG, 0235 trial,” J. Clin. Oncol., vol. 31, no. 30, pp. 3823–3830, 2013. doi: 10.1200/JCO.2012.47.5947. [Google Scholar] [PubMed] [CrossRef]
37. “ANODE09—Grand Challenge,” 2009. Accessed: Apr. 15, 2024. [Online]. Available: https://anode09.grand-challenge.org/Results/ [Google Scholar]
38. “LUNA16—Grand Challenge,” 2016. Accessed: Apr. 15, 2024. [Online]. Available: https://luna16.grand-challenge.org/Results/ [Google Scholar]
39. “Data Science Bowl,” 2017. Accessed: Apr. 15, 2024. [Online]. Available: https://www.kaggle.com/c/data-science-bowl-2017 [Google Scholar]
40. “CodaLab—Competition,” 2018. Accessed: Apr. 15, 2024. [Online]. Available: http://isbichallenges.cloudapp.net/competitions/15#learn_the_details-overview [Google Scholar]
41. “LNDb challenge—Grand challenge,” 2019. Accessed: Apr. 15, 2024. [Online]. Available: https://lndb.grand-challenge.org/TestRanking/ [Google Scholar]
42. S. M. Naqi, M. Sharif, and I. U. Lali, “A 3D nodule candidate detection method supported by hybrid features to reduce false positives in lung nodule detection,” Multimed. Tools Appl., vol. 78, no. 18, pp. 26287–26311, 2019. doi: 10.1007/s11042-019-07819-3. [Google Scholar] [CrossRef]
43. R. Manickavasagam and S. Selvan, “Automatic detection and classification of lung nodules in CT image using optimized neuro fuzzy classifier with cuckoo search algorithm,” J. Med. Syst., vol. 43, no. 3, p. 727, 2019. doi: 10.1007/s10916-019-1177-9. [Google Scholar] [PubMed] [CrossRef]
44. P. Kavitha and S. Prabakaran, “A novel hybrid segmentation method with particle swarm optimization and fuzzy c-mean based on partitioning the image for detecting lung cancer,” Int. J. Eng Adv Technol., vol. 8, no. 5, pp. 1223–1227, 2019. doi: 10.20944/preprints201906.0195.v1. [Google Scholar] [CrossRef]
45. F. Liao, M. Liang, Z. Li, X. Hu, and S. Song, “Evaluate the malignancy of pulmonary nodules using the 3-D deep leaky noisy-OR network,” IEEE Trans. Neural Netw. Learn Syst., vol. 30, no. 11, pp. 3484–3495, 2019. doi: 10.1109/TNNLS.2019.2892409. [Google Scholar] [PubMed] [CrossRef]
46. A. Soliman et al., “Accurate lungs segmentation on CT chest images by adaptive appearance-guided shape modeling,” IEEE Trans. Med. Imaging, vol. 36, no. 1, pp. 263–276, 2017. doi: 10.1109/TMI.2016.2606370. [Google Scholar] [PubMed] [CrossRef]
47. G. Aresta et al., “iW-Net: An automatic and minimalistic interactive lung nodule segmentation deep network,” Sci. Rep., vol. 9, no. 1, pp. 1–9, 2019. doi: 10.1038/s41598-019-48004-8. [Google Scholar] [PubMed] [CrossRef]
48. T. Nemoto et al., “Efficacy evaluation of 2D, 3D U-Net semantic segmentation and atlas-based segmentation of normal lungs excluding the trachea and main bronchi,” J. Radiat. Res., vol. 61, no. 2, pp. 257–264, 2020. doi: 10.1093/jrr/rrz086. [Google Scholar] [PubMed] [CrossRef]
49. T. Zhao, D. Gao, J. Wang, and Z. Tin, “Lung segmentation in CT images using a fully convolutional neural network with multi-instance and conditional adversary loss,” in Int. Symp. Biomed. Imaging, Washington, DC, USA, IEEE. 2018. doi: 10.1109/ISBI.2018.8363626. [Google Scholar] [CrossRef]
50. M. Z. Alom, C. Yakopcic, M. Hasan, T. M. Taha, and V. K. Asari, “Recurrent residual U-Net for medical image segmentation,” J. Med. Imaging, vol. 6, no. 1, 2019. doi: 10.1117/1.JMI.6.1.014006. [Google Scholar] [PubMed] [CrossRef]
51. G. Pezzano, V. R. Ripoll, and P. Radeva, “CoLe-CNN: Context-learning convolutional neural network with adaptive loss function for lung nodule segmentation,” Comput. Methods Programs Biomed., vol. 198, pp. 4–7, 2021. doi: 10.1016/j.cmpb.2020.105792. [Google Scholar] [PubMed] [CrossRef]
52. B. N. Narayanan, R. C. Hardie, T. M. Kebede, and M. J. Sprague, “Optimized feature selection-based clustering approach for computer-aided detection of lung nodules in different modalities,” Pattern Anal. Appl., vol. 22, no. 2, pp. 559–571, 2019. doi: 10.1007/s10044-017-0653-4. [Google Scholar] [CrossRef]
53. G. Aresta, A. Cunha, and A. Campilho, “Detection of juxta-pleural lung nodules in computed tomography images,” Med. Comput. Sci., 2017. doi: 10.1117/12.2252022. [Google Scholar] [CrossRef]
54. J. J. Suárez-Cuenca, W. Guo, and Q. Li, “Automated detection of pulmonary nodules in CT: False positive reduction by combining multiple classifiers,” Med. Comput. Sci., vol. 7963, no. March 2011, p. 796338, 2011. doi: 10.1117/12.878793. [Google Scholar] [CrossRef]
55. S. T. Namin, H. A. Moghaddam, R. Jafari, M. Esmaeil-Zadeh, and M. Gity, “Automated detection and classification of pulmonary nodules in 3D thoracic CT images,” presented at the Proc. IEEE Int. Conf. Syst. Man Cybern., Istanbul, Turkey, Oct. 10–13, 2010, pp. 3774–3779. doi: 10.1109/ICSMC.2010.5641820. [Google Scholar] [CrossRef]
56. C. C. Nguyen, G. S. Tran, V. T. Nguyen, J. C. Burie, and T. P. Nghiem, “Pulmonary nodule detection based on faster R-CNN with adaptive anchor box,” IEEE Access, vol. 9, pp. 154740–154751, 2021. doi: 10.1109/ACCESS.2021.3128942. [Google Scholar] [CrossRef]
57. J. Wang et al., “Pulmonary nodule detection in volumetric chest CT scans using CNNs-based nodule-size-adaptive detection and classification,” IEEE Access, vol. 7, pp. 46033–46044, 2019. doi: 10.1109/ACCESS.2019.2908195. [Google Scholar] [CrossRef]
58. S. Zheng, J. Guo, X. Cui, R. N. J. Veldhuis, M. Oudkerk and P. M. A. Van Ooijen, “Automatic pulmonary nodule detection in CT scans using convolutional neural networks based on maximum intensity projection,” IEEE Trans. Med. Imag., vol. 39, no. 3, pp. 797–805, 2020. doi: 10.1109/TMI.2019.2935553. [Google Scholar] [PubMed] [CrossRef]
59. S. Liu et al., “No surprises: Training robust lung nodule detection for low-dose CT scans by augmenting with adversarial attacks,” IEEE Trans. Med. Imaging, vol. 40, no. 1, pp. 335–345, 2021. doi: 10.1109/TMI.2020.3026261. [Google Scholar] [PubMed] [CrossRef]
60. R. Manickavasagam, S. Selvan, and M. Selvan, “CAD system for lung nodule detection using deep learning with CNN,” Med. Biol. Eng. Comput., vol. 60, no. 1, pp. 221–228, 2022. doi: 10.1007/s11517-021-02462-3. [Google Scholar] [PubMed] [CrossRef]
61. S. M. Naqi, M. Sharif, and A. Jaffar, “Lung nodule detection and classification based on geometric fit in parametric form and deep learning,” Neural Comput. Appl., vol. 32, no. 9, pp. 4629–4647, 2020. doi: 10.1007/s00521-018-3773-x. [Google Scholar] [CrossRef]
62. F. V. Farahani, A. Ahmadi, and M. H. F. Zarandi, “Hybrid intelligent approach for diagnosis of the lung nodule from CT images using spatial kernelized fuzzy c-means and ensemble learning,” Math. Comput. Simul., vol. 149, pp. 48–68, 2018. doi: 10.1016/j.matcom.2018.02.001. [Google Scholar] [CrossRef]
63. W. Zhang, X. Wang, X. Li, and J. Chen, “3D skeletonization feature based computer-aided detection system for pulmonary nodules in CT datasets,” Comput. Biol. Med., vol. 92, pp. 64–72, 2018. doi: 10.1016/j.compbiomed.2017.11.008. [Google Scholar] [PubMed] [CrossRef]
64. H. Tang, D. R. Kim, and X. Xie, “Automated pulmonary nodule detection using 3D deep convolutional neural networks,” in Int. Symp. Biomed. Imaging, Washington, DC, USA, pp. 523–526, 2018. doi: 10.1109/ISBI.2018.8363630. [Google Scholar] [CrossRef]
65. H. Tang, X. Liu, and X. Xie, “An end-to-end framework for integrated pulmonary nodule detection and false positive reduction,” in Int. Symp. Biomed. Imaging, Venice, Italy, pp. 859–862, 2019. doi: 10.1109/ISBI.2019.8759244. [Google Scholar] [CrossRef]
66. S. A. El-Regaily, M. A. M. Salem, M. H. Abdel Aziz, and M. I. Roushdy, “Multi-view convolutional neural network for lung nodule false positive reduction,” Expert. Syst. Appl., vol. 162, 2020, Art. no. 113017. doi: 10.1016/j.eswa.2019.113017. [Google Scholar] [CrossRef]
67. J. Liu, L. Cao, O. Akin, and Y. Tian, “Accurate and robust pulmonary nodule detection by 3D feature pyramid network with self-supervised feature learning,” 2019, arXiv:1907.11704. [Google Scholar]
68. H. Cheng, Y. Zhu, and H. Pan, “Modified U-Net block network for lung nodule detection,” presented at the 2019 in Proc. IEEE 8th Joint Int. Inform. Technol. Artif. Intell. Conf., ITAIC, Chongqing, China, May 24–26, 2019, pp. 599–605. doi: 10.1109/ITAIC.2019.8785445. [Google Scholar] [CrossRef]
69. Ö. Günaydin, M. Günay, and Ö. Şengel, “Comparison of lung cancer detection algorithms,” in 2019 Sci. Meet. Elect.-Electron. Biomed. Eng. Comput. Sci. (EBBT), Istanbul, Turkey, 2019, pp. 1–4. doi: 10.1109/EBBT.2019.8741826. [Google Scholar] [CrossRef]
70. Z. Shi et al., “A deep CNN based transfer learning method for false positive reduction,” Multimed. Tools Appl., vol. 78, no. 1, pp. 1017–1033, 2019. doi: 10.1007/s11042-018-6082-6. [Google Scholar] [CrossRef]
71. O. Ozdemir, R. L. Russell, and A. A. Berlin, “A 3D probabilistic deep learning system for detection and diagnosis of lung cancer using low-dose CT scans,” IEEE Trans. Med. Imag., vol. 39, no. 5, pp. 1419–1429, 2020. doi: 10.1109/TMI.2019.2947595. [Google Scholar] [PubMed] [CrossRef]
72. H. Cao et al., “A two-stage convolutional neural networks for lung nodule detection,” IEEE J. Biomed. Health Inform., vol. 24, no. 7, pp. 2006–2015, 2020. doi: 10.1109/JBHI.2019.2963720. [Google Scholar] [PubMed] [CrossRef]
73. A. Masood et al., “Cloud-based automated clinical decision support system for detection and diagnosis of lung cancer in chest CT,” IEEE J. Transl. Eng. Health Med., vol. 8, pp. 1–13, 2020. doi: 10.1109/JTEHM.2019.2955458. [Google Scholar] [PubMed] [CrossRef]
74. S. Zheng et al., “Deep convolutional neural networks for multiplanar lung nodule detection: Improvement in small nodule identification,” Med. Phys., vol. 48, no. 2, pp. 733–744, 2021. doi: 10.1002/mp.14648. [Google Scholar] [PubMed] [CrossRef]
75. D. Wang, Y. Zhang, K. Zhang, and L. Wang, “FocalMix: Semi-supervised learning for 3D medical image detection,” presented at the IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit., Seattle, WA, USA, Jun. 13–19, 2020, pp. 3950–3959. doi: 10.1109/CVPR42600.2020.00401. [Google Scholar] [CrossRef]
76. Y. Su, D. Li, and X. Chen, “Lung nodule detection based on faster R-CNN framework,” Comput. Methods Programs. Biomed., vol. 200, 2021, Art. no. 105866. doi: 10.1016/j.cmpb.2020.105866. [Google Scholar] [PubMed] [CrossRef]
77. D. Zhao, Y. Liu, H. Yin, and Z. Wang, “A novel multi-scale CNNs for false positive reduction in pulmonary nodule detection,” Expert. Syst. Appl., vol. 207, pp. 21–23, 2022. doi: 10.1016/j.eswa.2022.117652. [Google Scholar] [CrossRef]
78. Z. Ji et al., “Lung nodule detection in medical images based on improved YOLOv5s,” IEEE Access, vol. 11, pp. 76371–76387, 2023. doi: 10.1109/ACCESS.2023.3296530. [Google Scholar] [CrossRef]
79. I. Ahmed, A. Chehri, G. Jeon, and F. Piccialli, “Automated pulmonary nodule classification and detection using deep learning architectures,” IEEE/ACM Trans. Comput. Biol. Bioinform., vol. 20, no. 4, pp. 2445–2456, Jul. 2023. doi: 10.1109/TCBB.2022.3192139. [Google Scholar] [PubMed] [CrossRef]
80. L. Goel and P. Patel, “Improving YOLOv6 using advanced PSO optimizer for weight selection in lung cancer detection and classification,” Multimed. Tools Appl., vol. 83, pp. 78059–78092, 2024. doi: 10.1007/s11042-024-18441-3. [Google Scholar] [CrossRef]
81. J. Uthoff et al., “Machine learning approach for distinguishing malignant and benign lung nodules utilizing standardized perinodular parenchymal features from CT,” Med. Phys., vol. 46, no. 7, pp. 3207–3216, 2019. doi: 10.1002/mp.13592. [Google Scholar] [PubMed] [CrossRef]
82. M. Al-Shabi, H. K. Lee, and M. Tan, “Gated-dilated networks for lung nodule classification in CT scans,” IEEE Access, vol. 7, pp. 178827–178838, 2019. doi: 10.1109/ACCESS.2019.2958663. [Google Scholar] [CrossRef]
83. M. Al-Shabi, B. L. Lan, W. Y. Chan, K. H. Ng, and M. Tan, “Lung nodule classification using deep local-global networks,” Int. J. Comput. Assist. Radiol. Surg., vol. 14, no. 10, pp. 1815–1819, 2019. doi: 10.1007/s11548-019-01981-7. [Google Scholar] [PubMed] [CrossRef]
84. P. Sahu, D. Yu, M. Dasari, F. Hou, and H. Qin, “A lightweight multi-section CNN for lung nodule classification and malignancy estimation,” IEEE J. Biomed. Health Inform., vol. 23, no. 3, pp. 960–968, 2019. doi: 10.1109/JBHI.2018.2879834. [Google Scholar] [PubMed] [CrossRef]
85. Y. Ren et al., “A manifold learning regularization approach to enhance 3D CT image-based lung nodule classification,” Int. J. Comput. Assist. Radiol. Surg., vol. 15, no. 2, pp. 287–295, 2020. doi: 10.1007/s11548-019-02097-8. [Google Scholar] [PubMed] [CrossRef]
86. N. Nasrullah, J. Sang, M. S. Alam, M. Mateen, B. Cai and H. Hu, “Automated lung nodule detection and classification using deep learning combined with multiple strategies,” Sensors, vol. 19, 2019, Art. no. 3722. doi: 10.3390/s19173722. [Google Scholar] [PubMed] [CrossRef]
87. S. Chen, K. Ma, and Y. Zheng, “Med3D: Transfer learning for 3D medical image analysis,” 2019. doi: 10.48550/arXiv.1904.00625. [Google Scholar] [CrossRef]
88. S. Hussein, P. Kandel, C. W. Bolan, M. B. Wallace, and U. Bagci, “Lung and pancreatic tumor characterization in the deep learning era: Novel supervised and unsupervised learning approaches,” IEEE Trans. Med. Imag., vol. 38, no. 8, pp. 1777–1787, 2019. doi: 10.1109/TMI.2019.2894349. [Google Scholar] [PubMed] [CrossRef]
89. H. Shakir, H. Rasheed, and T. M. Rasool Khan, “Radiomic feature selection for lung cancer classifiers,” J. Intell. Fuzzy Syst., vol. 38, no. 5, pp. 5847–5855, 2020. doi: 10.3233/JIFS-179672. [Google Scholar] [CrossRef]
90. D. R. Baldwin et al., “External validation of a convolutional neural network artificial intelligence tool to predict malignancy in pulmonary nodules,” Thorax, vol. 75, no. 4, pp. 306–312, 2020. doi: 10.1136/thoraxjnl-2019-214104. [Google Scholar] [PubMed] [CrossRef]
91. X. Huang, Q. Lei, T. Xie, Y. Zhang, Z. Hu and Q. Zhou, “Deep transfer convolutional neural network and extreme learning machine for lung nodule diagnosis on CT images,” Knowl. Based Syst., vol. 204, 2020, Art. no. 106230. doi: 10.1016/j.knosys.2020.106230. [Google Scholar] [CrossRef]
92. P. Ardimento, L. Aversano, M. L. Bernardi, and M. Cimitile, “Deep neural networks ensemble for lung nodule detection on chest CT scans,” presented at the Int. Joint Conf. Neural Netw., Shenzhen, China, Jul. 18–22, 2021. doi: 10.1109/IJCNN52387.2021.9534176. [Google Scholar] [CrossRef]
93. G. Zhang, L. Lin, and J. Wang, “Lung nodule classification in CT images using 3D DenseNet,” J. Phys. Conf. Ser., vol. 1827, no. 1, 2021, Art. no. 012155. doi: 10.1088/1742-6596/1827/1/012155. [Google Scholar] [CrossRef]
94. F. Shi et al., “Semi-supervised deep transfer learning for benign-malignant diagnosis of pulmonary nodules in chest CT images,” IEEE Trans. Med. Imaging, vol. 41, no. 4, pp. 771–781, 2022. doi: 10.1109/TMI.2021.3123572. [Google Scholar] [PubMed] [CrossRef]
95. M. Dhasny Lydia and Dr. M. Prakash, “An improved convolution neural network and modified regularized K-means-based automatic lung nodule detection and classification,” J. Digit. Imaging, vol. 36, no. 4, pp. 1431–1446, Aug. 2023. doi: 10.1007/s10278-023-00809-w. [Google Scholar] [PubMed] [CrossRef]
96. S. Tang, M. Yang, and J. Bai, “Detection of pulmonary nodules based on a multiscale feature 3D U-Net convolutional neural network of transfer learning,” PLoS One, vol. 15, pp. 1–27, 2020. doi: 10.1371/journal.pone.0235672. [Google Scholar] [PubMed] [CrossRef]
97. K. Loverdos, A. Fotiadis, C. Kontogianni, M. Iliopoulou, and M. Gaga, “Lung nodules: A comprehensive review on current approach and management,” Ann. Thorac. Meed., vol. 14, no. 4, pp. 226–238, 2019. doi: 10.4103/atm.ATM_110_19. [Google Scholar] [PubMed] [CrossRef]
98. A. Masood et al., “Computer-assisted decision support system in pulmonary cancer detection and stage classification on CT images,” J. Biomed. Inform., vol. 79, pp. 117–128, 2018. doi: 10.1016/j.jbi.2018.01.005. [Google Scholar] [PubMed] [CrossRef]
99. B. Liu et al., Evolving the Pulmonary Nodules Diagnosis from Classical Approaches to Deep Learning-Aided Decision Support: Three Decades’ Development Course and Future Prospect, Berlin Heidelberg: Springer, 2020, vol. 146, no. 1. doi: 10.1007/s00432-019-03098-5. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF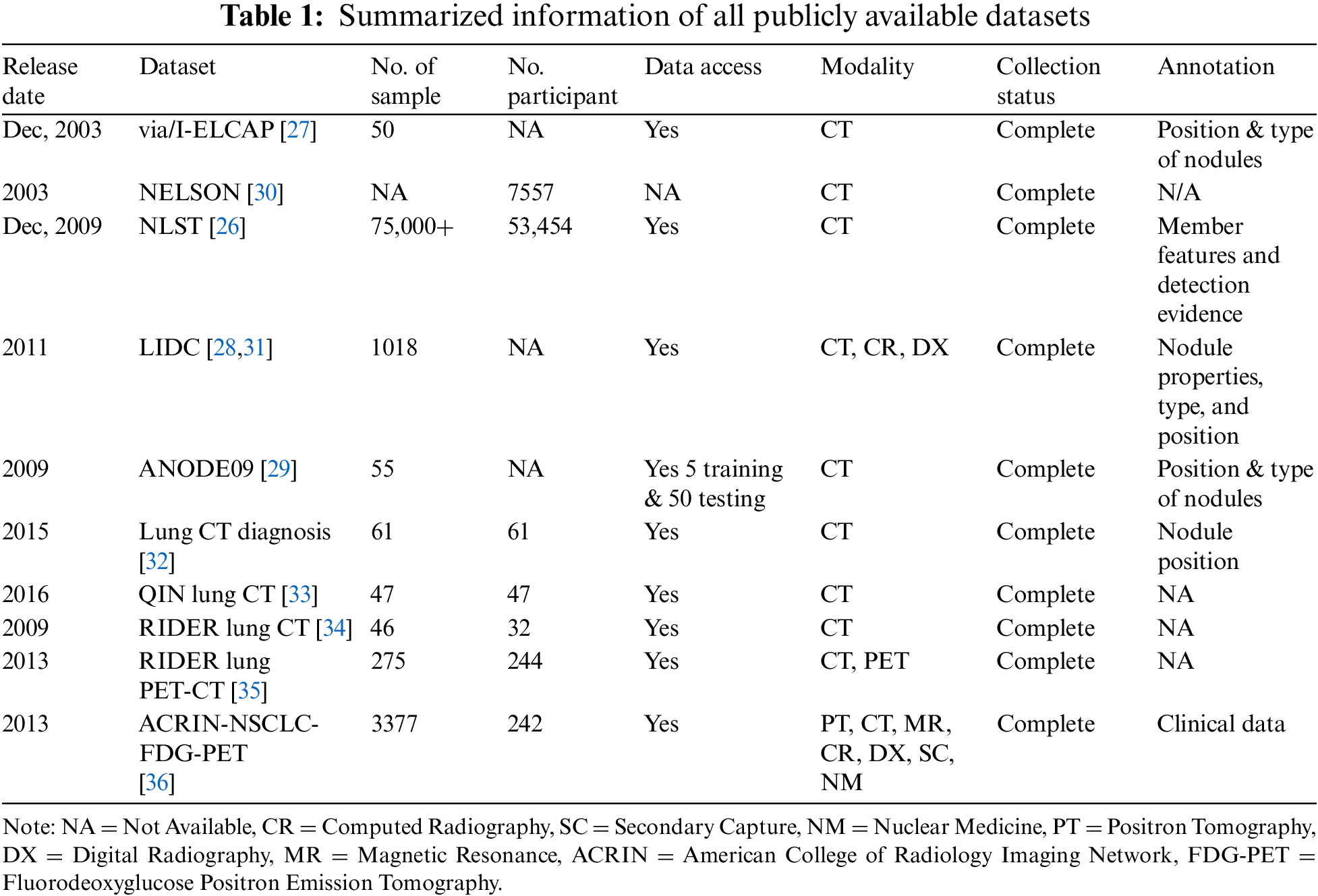
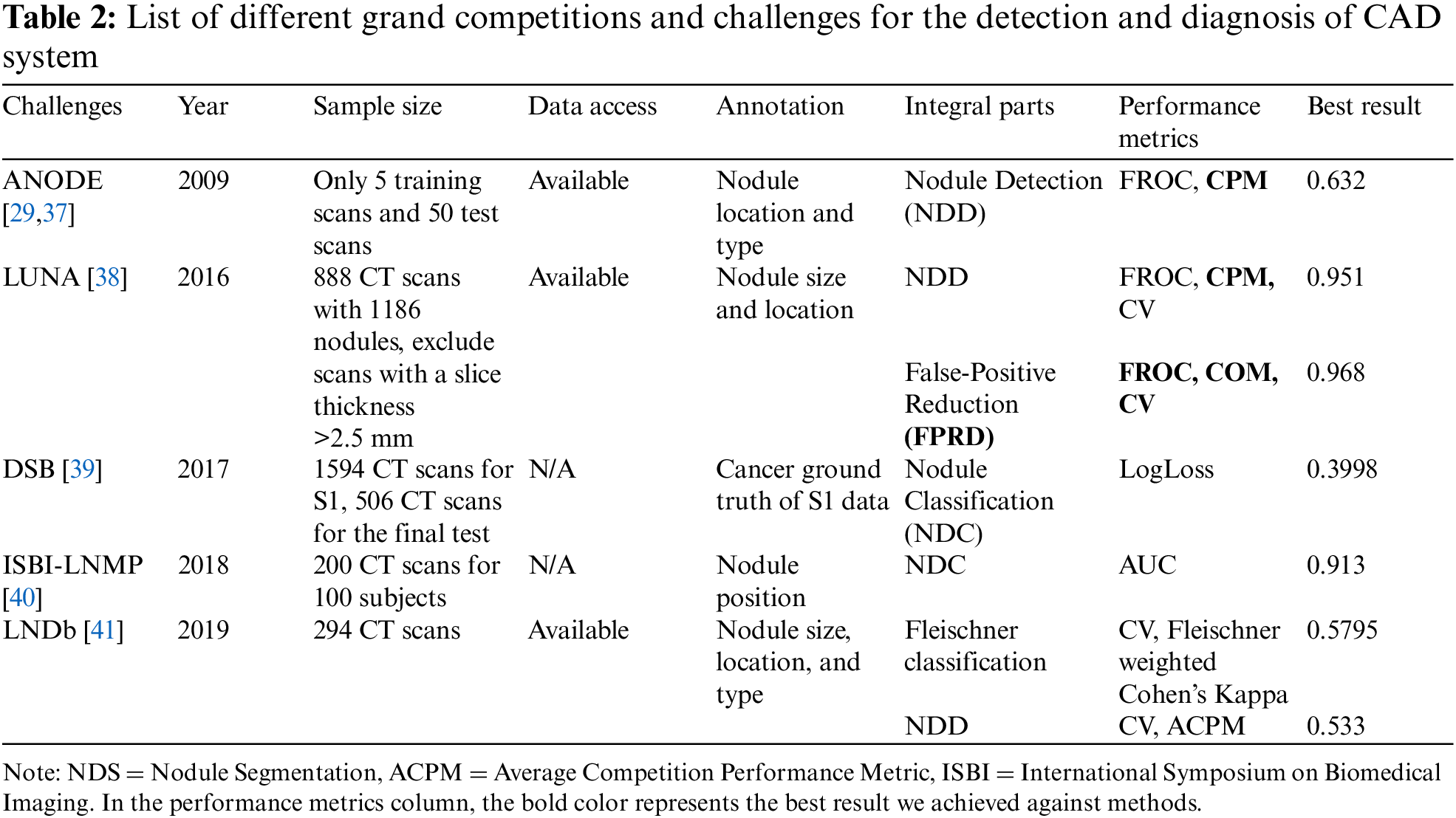
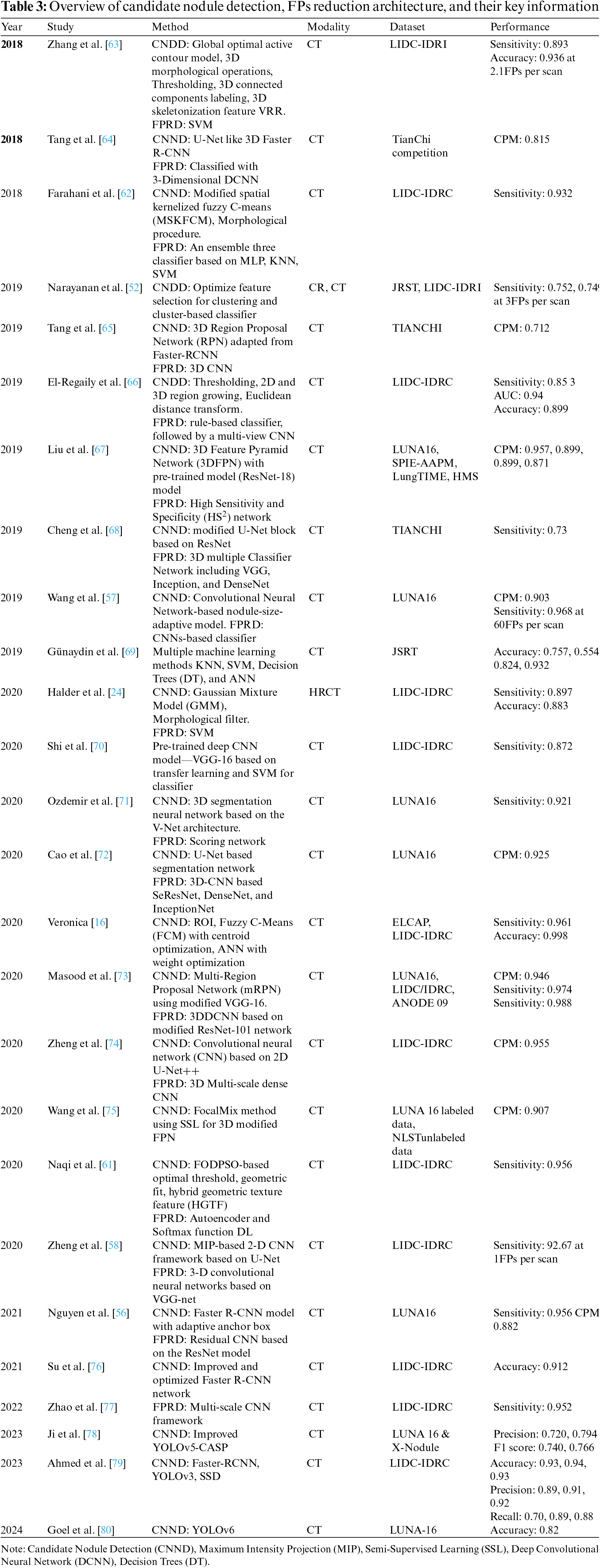
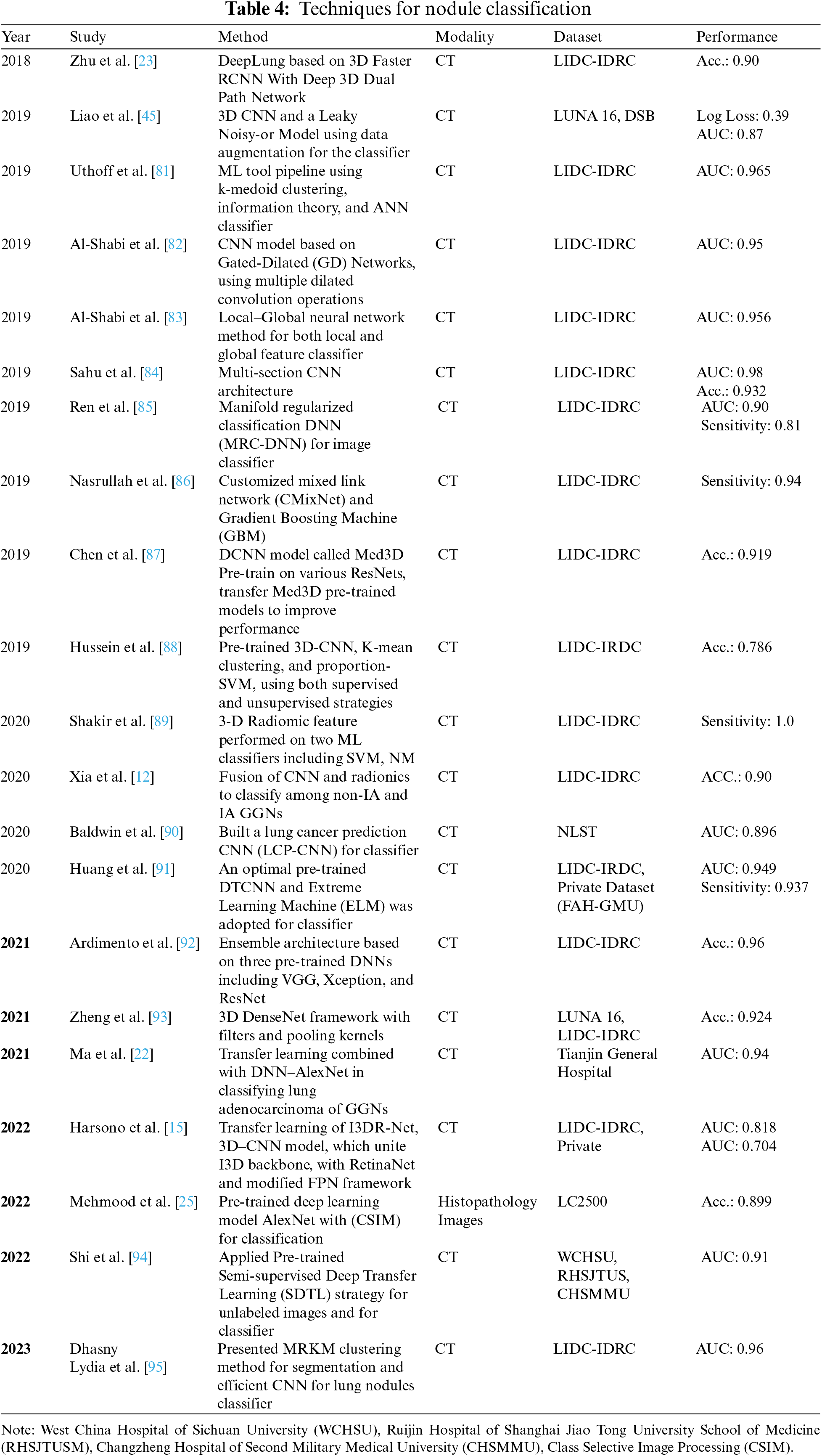
 Downloads
Downloads
 Citation Tools
Citation Tools