 Open Access
Open Access
ARTICLE
Performance of Deep Learning Techniques in Leaf Disease Detection
1 Centre of Real Time Computer Systems, Kaunas University of Technology, Kaunas, 51386, Lithuania
2 Department of Computer Science, Air University, Islamabad, 44000, Pakistan
3 Department of Computer Science, City University, Peshawar, 25000, Pakistan
* Corresponding Author: Robertas Damasevicius. Email:
Computer Systems Science and Engineering 2024, 48(5), 1349-1366. https://doi.org/10.32604/csse.2024.050359
Received 04 February 2024; Accepted 24 July 2024; Issue published 13 September 2024
Abstract
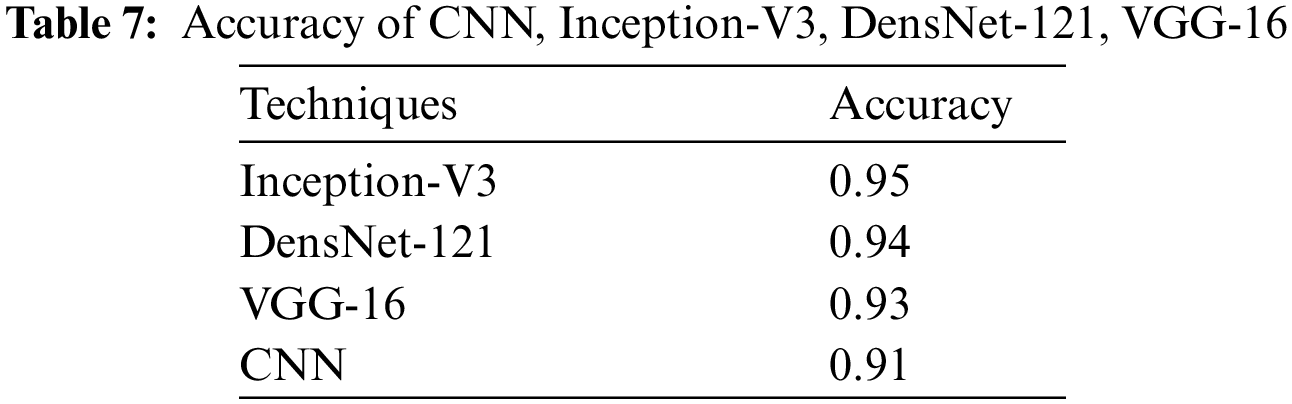
Plant diseases must be identified as soon as possible since they have an impact on the growth of the corresponding species. Consequently, the identification of leaf diseases is essential in this field of agriculture. Diseases brought on by bacteria, viruses, and fungi are a significant factor in reduced crop yields. Numerous machine learning models have been applied in the identification of plant diseases, however, with the recent developments in deep learning, this field of study seems to hold huge potential for improved accuracy. This study presents an effective method that uses image processing and deep learning approaches to distinguish between healthy and infected leaves. To effectively identify leaf diseases, we employed pre-trained models based on Convolutional Neural Networks (CNNs). There are four deep neural networks approaches used in this study: Convolutional Neural Network (CNN), Inception-V3, DenseNet-121, and VGG-16. Our focus was on optimizing the hyper-parameters of these deep learning models with prior training. For the evaluation of these deep neural networks, standard evaluation measures are used, such as F1-score, recall, precision, accuracy, and Area Under Curve (AUC). The overall outcomes show the better performance of Inception-V3 with an achieved accuracy of 95.5%, as well as the performance of DenseNet-121 with an accuracy of 94.4%. VGG-16 performed well as well, with an accuracy of 93.3%, and CNN achieved an accuracy of 91.9%.Keywords
The agriculture experiences a global decline in profitability as production declines. By 2050, human civilization will need to raise food production by an estimated 70% to feed the world’s anticipated 9 billion inhabitants. The quantity and quality of crops can be negatively impacted by infections and diseases that affect plants and crops [1]. Currently, infectious illnesses limit potential output by 40% on average, with yield losses reaching 100% for many farmers in underdeveloped nations. One of the most important aspects of a good farming system is the identification of diseases. In an effort to boost food production, the agriculture sector is looking for effective ways to prevent crop damage. Precision agriculture research has focused a lot of attention on the use of digital leaf images to diagnosis plant diseases. The use of precise plant protection and growth can be made possible by recent developments in artificial intelligence (AI), image processing, and graphical processing units (GPUs), which build upon and enhance current technology [2].
Plant diseases significantly impact global agricultural productivity, stemming from a complex interplay of pathogens, environmental factors, and host susceptibility. These diseases are primarily caused by fungi, bacteria, and viruses. For instance, fungal diseases like powdery mildew and rust typically manifest as spore-producing lesions on plant surfaces, leading to defoliation, stunted growth, and reduced photosynthetic capacity [3]. Bacterial infections, exemplified by blight and wilt, can cause rapid cell death and systemic infections that result in damp, rotted tissue [4]. Viral diseases, like tobacco mosaic virus, lead to mosaic patterns on leaves, stunted growth, and yield reduction [5]. Each type of disease presents unique diagnostic and management challenges, often requiring targeted strategies that may include chemical treatments, genetic resistance, and cultural practices to prevent and mitigate their spread. Understanding the specific visible characteristics of these pathogens and their early detection using AI models is crucial for developing effective control measures and ensuring plant health and productivity [6]. AI can enhance the detection and management of plant diseases by automating the recognition of symptoms and suggesting timely interventions.
Traditionally, the process of checking the health condition of plant leaves involves manual observation using the unaided eye. Manual plant disease detection is a tedious process that necessitates ongoing monitoring and plant pathology expertise. Due to its time-consuming nature, this visual assessment is not practical for covering huge areas [7]. The AI techniques have significantly trans-formed plant disease detection and identification tasks from conventional methods, and they have also significantly increased in accuracy. Deep learning is currently the most effective method for using computers to identify plant diseases. Fulari et al. [8] presented an effective method that uses image processing and machine learning approaches to distinguish between healthy and damaged or infected leaves. Diseases of the leaves, such as fungus, bacteria, and viruses, can cause sickness and form patches, making it difficult to determine if the source is fungal, bacterial, or virus.
Many diseases can result in various losses or fruit decay [9]. Many agriculture professionals devote a significant amount of work to determining the diseases of a class (bacteria, fungus, viruses). Predicting leaf disease is critical for agricultural organizations to make the best possible leaf care decisions [10]. Incorrect judgments are likely to result in plant loss or delays in plant treatment. Furthermore, predicting the correct plant disease has long been considered a vital problem. To handle this sort of agricultural care challenge, machine learning (ML) approaches have previously been created. Neural networks have recently proved effective in a range of applications, including aiding in the treatment of leaves [11]. Through the training of a finite number of neural networks and then integrating their outputs, neural network ensembles can dramatically increase the generalization capacity of learning systems. Some agricultural professionals believe it is difficult to make disease-related judgments because they lack expertise in all agricultural areas.
Famine and starvation can result from plant disease losses, particularly in less developed countries where disease-control tools are limited and annual losses of 30% to 50% of essential crops are not uncommon. Losses can be much larger in some years, which has severe effects on individuals who depend on the crop for their livelihood [12]. Large-scale disease outbreaks among food crops have historically caused famines and vast migrations. In 1845, a severe outbreak of potato bacteria erupted in Europe. Due to its capacity to extract features, deep learning models play a vital role in detecting a variety of plant diseases [13]. Multimodal analyses are automatically analyzed using DL models. Computer vision demonstrates how a computer obtains information from image and video processing using deep learning algorithms. CNNs, VGG-16, Inception-V3, and DenseNet-121 are some of the most advanced approaches currently available.
To address this problem, it is critical to establish a plant disease detection system that integrates agricultural knowledge with integrated system that deliver better and more accurate findings which benefits the society [14,15]. This study aims to demonstrate the effectiveness of several widely used Deep learning techniques by showing how they may be used for a variety of diverse but related tasks. The suggested models was compared in this study to some of the most widely used Deep learning techniques currently on the market, including as Inception-v3, DenseNet-121, VGG-16. The goal is to identify a model that can identify whether a leaf is healthy or suffering from a disease. This study employs accuracy, precision, recall, F1-score, and AUC as assessment measures to evaluate existing approaches. The Inception-V3 model performed better than the DenseNet-121, VGG-16 according to the results.
This study introduces a robust approach to the identification of leaf diseases using deep learning given the pressing need to enhance agricultural yield efficiencies in response to global food demand. By evaluating and comparing four advanced deep learning architectures—CNN, Inception-V3, DenseNet-121, and VGG-16—across various performance metrics, this study identifies the superior performance of the Inception-V3 model. This contribution provides both theoretical insights and practical applications that can be integrated into precision agriculture practices, thus supporting the optimization of plant health management and contributing to sustainable agricultural outputs.
Machine learning and deep learning can assist in health status identification of plants [1], leaves [16] or seeds [17]. Plant diseases result from bacterial or viral infections that interfere with a plant’s ability to grow normally. Effects on plant leaves might range from dying to discolored. Numerous bacteria are also harmful and have the ability to infect humans, plants, and other living things [2]. Plant pathology knowledge and continuous monitoring are required for the laborious manual process of plant disease detection [7]. Identification of diseases is one of the most important components of a productive farming system. Farmers typically use visual observations to spot disease symptoms in plants that require ongoing care. They demonstrated how a convolutional neural network (CNN) can accurately identify a wide range of disorders [8].
Arsenovic et al. [10] suggested a method to detect disease in plants by training a Modified LeNet VGG, VGG-16, Inception-V3, and Modified AlexNet. The proposed CNN model identifies healthy and affected plants of 42 diseases. These models achieved an accuracy Modified ALexNet 92.88% VGG 99.53%, VGG-16 90.40%, Inception-V3 99.76%, Modified AlexNet 97.62%. Li et al. [11] presented a technique for identifying plant diseases by using a photo of the affected leaf that was taken. Resnet, Inception-V3, VGG-16, and VGG-19 are trained to discriminate between healthy and pathological samples by properly selecting feature values. A cultivating farm’s output can be increased and the quality of the plants can be guaranteed by using this image analysis technology to remove good, healthy pepper plants. By examining the visual symptoms present on the plant’s leaves, this algorithm aids in detecting the presence of diseases [15].
Marzougui et al. [12] proposed a method that works based on “resnet” criteria. The accuracy of 94.80% after training for 10 epochs. After performing implementation with proposed “ResNet” architecture, the fully detection performance reaches 98.96%. In Jiang et al.'s research [13] incorporating the GoogLeNet Inception structure, an innovative apple leaf disease recognition model based on deep-CNNs is suggested. Lastly, the hold-out testing dataset, which comprises 26, 377 photos of diseased apple leaves, is used to train the suggested INAR-SSD model to recognise these five common apple leaf illnesses.
Francis et al. [18] developed a method to detect diseases in the plant using image processing of the captured image of the diseased leaf. To identify leaf diseases, the leaves of the pepper plant are used as a set of leaves. Healthy and diseased plants can be distinguished using this method, which also yields better results. Manual observation with the unaided eye has been used to assess the health of leaves or seeds [17]. Chugh et al. [19] proposed a work in which the author used a plant village dataset which has about 1000 leaf images of early blight and 152 images of healthy plants. For this model, the dataset has been divided into 2 parts, which are the training set and the test set. The dataset contains 0.2 split ratio. The pre-trained model used on this dataset for feature extraction is Inception-V3. For classification, CNN model provides an accuracy of about 90% based on the training and testing done on it.
Ouhami et al. [20] introduced a method which aims to identify the best machine-learning model for detecting tomato crop disease in RGB photos. The various deep learning model used to detect tomato crop disease. DenseNet-161 and DenseNet-121 and VGG-16 with transfer learning to solve this problem. The infected images of plants were separated into six categories: infections, insect assaults, and plant illnesses. The findings show an accuracy up to 95.65% for the DenseNet-161 which is higher than other models used in this study, 94.93% for DenseNet-121, and 90.58% for the VGG-16 model which shows the lower accuracy to detect the plant disease.
Deep Learning models AlexNet and SqueezeNet v1.1 were used to classify tomato plant diseases, with AlexNet proving to be the most accurate deep learning model [21]. According to Sakshi Raina [22], various scholars proposed the fundamentals of plant disease detection strategies. Based on diverse datasets and their preferences, tabular analysis provides identification, segmentation, and classification techniques. The recognition and learning rates of GPDCNN are greater. GANs have stronger evidence of information appropriation (more refined and clear images). A Multilayered Convolutional Neural Network that has deviated from its paradigms has the benefit of being able to recognize essential aspects without the need for human intervention. To put it another way, we gave a summary of the possible methodologies for solving the problem as well as the datasets.
Ajra et al. [23] presented a system obtains a total accuracy of 96.5% with AlexNet for the categorization of healthy and infected leaves, whereas ResNet-50 achieves an overall accuracy of 97%. On the other side, for leaf disease identification, ResNet-50 has an overall accuracy of 96.1%, whereas AlexNet has an overall accuracy of 95.3%. From all of the comparisons, it has been determined that ResNet-50 outperforms AlexNet.
Patil et al. [24] described how transfer learning is used to identify diseases in mango leaf image datasets. Deep convolutional neural network training was carried out using a deep residual architecture called residual neural network (ResNET). ResNETs can attain higher accuracy and are simple to tune. The accuracies range from 94% to 98% according to the experimental results from ResNET architectures, such as ResNet-18, ResNet-34, ResNet-50, and ResNet-101.
Rahman et al. [25] suggested an automatic image processing method that involves the farmer taking a image of a tomato leaf using the Leaf Disease Identification system app for Android. The suggested method uses the Gray Level Co-Occurrence Matrix (GLCM) algorithm to calculate 13 distinct statistical variables from tomato leaves. Support Vector Machine (SVM) is used to classify the acquired features into various diseases. The suggested approach is put into practice as a mobile application.
Atila et al. [26] used EfficientNet deep learning architecture for classifying plant leaf diseases. Using the PlantVillage dataset, models were trained on both original and augmented datasets, containing 55,448 and 61,486 images, respectively. Experiments demonstrated that the B5 and B4 models of the EfficientNet architecture outperformed other models, achieving the highest accuracy rates of 99.91% and 99.97% on the original and augmented datasets, respectively.
CNNs are a fairly common type of neural network design that find extensive application in a wide range of computer vision applications. The CNN architecture and its variants have been employed by researchers to categorise and identify plant diseases [27]. The Deep CNN model carried out an activity to identify the category of symptoms for four different cucumber leaf illnesses. The architecture used is a simple and quick method for identifying photos that are modest in size [28]. Agarwal et al. [29] suggested the Tomato Leaf Disease Detection (ToLeD) model, a model which is based on the architecture of CNN which achieves 91.2% accuracy rate for classifying target 10 leaf diseases using tomato leaves dataset.
The literature on plant disease detection using deep learning methods reveals several notable gaps. Numerous studies have applied CNNs to classify and detect a variety of plant diseases. There is an apparent emphasis on achieving high accuracy, with less consideration given to understanding of why certain predictions are made by the models remains under-discussed, which is crucial for trust and practical application in agricultural settings. Studies that used sophisticated architectures like Inception-V3 and EfficientNet primarily utilize supervised learning techniques with pre-labeled datasets. There is a knowledge gap in unsupervised or semi-supervised learning approaches, which could potentially reduce the dependency on extensively annotated datasets that are ex-pensive and time-consuming to create. While current studies showcase the use of transfer learning to adapt pre-trained models to plant disease detection, the adaptation to cross-species or cross-pathogen applications without substantial retraining is not well-covered. This indicates a need for more robust models that can generalize across different types of plants and diseases more effectively.
This study examines methods for recognising viral, fungus, and bacterial plant leaf diseases. The procedure for detecting leaf disease using deep learning approaches is shown in Fig. 1.
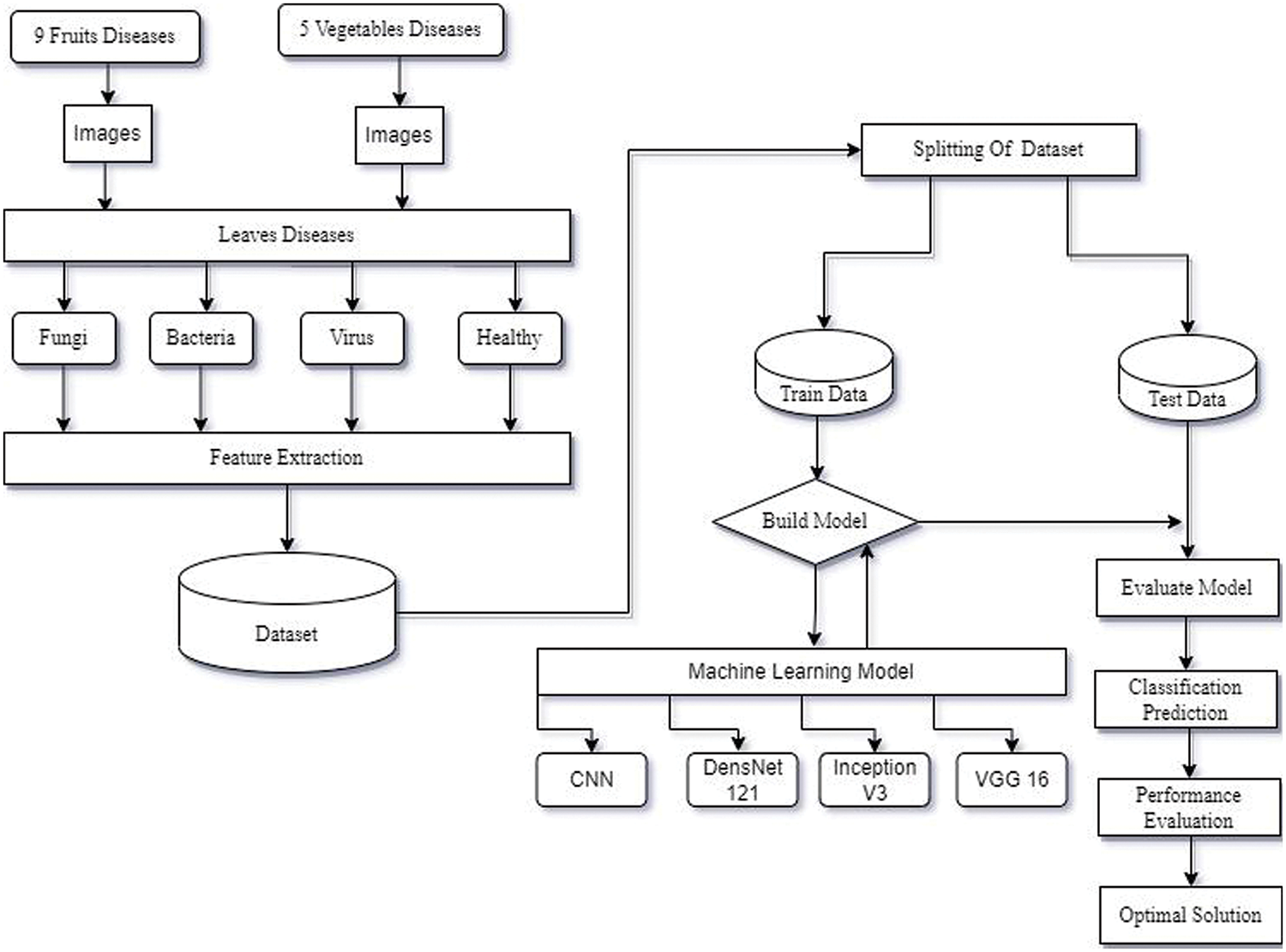
Figure 1: The methods used to detect leaf disease
Images of both healthy and diseased plant tissue were taken from a plant community dataset. A dataset contains 50,000 selected photos of both healthy and diseased leaves of agricultural plants [30]. Bacterial, fun-gal, and viral diseases affecting plant leaves include scab, blight, rust, scorch, mites, mosaic, leaf curl, albication, bacterial spot and wilting. The classification of bacterial, fungal, and viral illnesses is shown in Fig. 2. The main goal of this study is to correctly classify leaves into the following four categories (healthy, fungus, virus and bacteria).
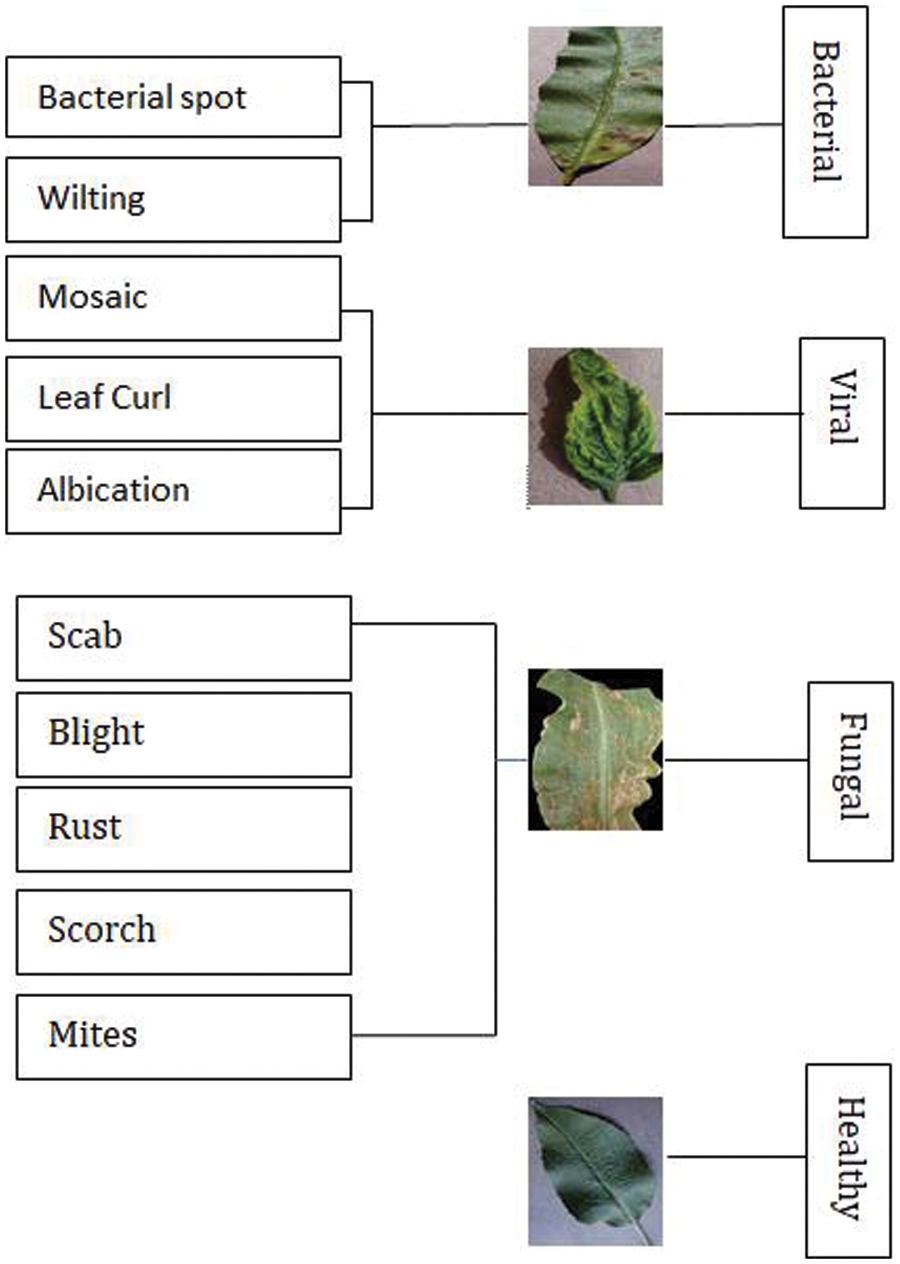
Figure 2: Disease in leaf with their categories in various crops
The dataset used in this study is publicly accessible and taken from PlantVillage dataset, hosted on Kaggle1. The PlantVillage dataset is a comprehensive collection designed to support the development and evaluation of machine learning models in the identification and diagnosis of plant diseases. It encompasses a wide array of healthy and diseased plant leaves images, classified across numerous species and disease conditions. This dataset serves as a pivotal resource for researchers and practitioners aiming to advance agricultural practices through the application of artificial intelligence, enhancing the ability to detect, categorize, and address plant health issues efficiently.
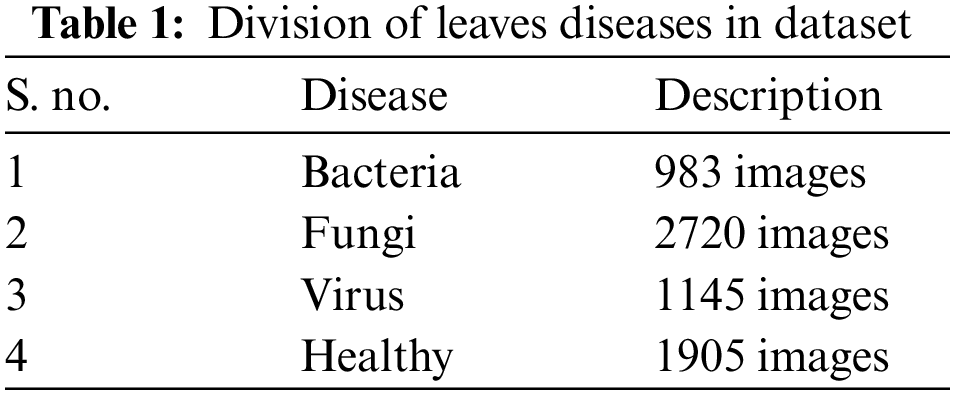
In this work, we use the 6753 leaf disease images which contain the healthy, virus, bacteria, and fungi. The database comprises 3 categories in total. Each one stands for either a plant that is disease-free or one that is not. We have different leaf diseases in the agricultural area. But we mainly focused on leaves with major classes of bacteria, fungi, and viruses, in which we have 9 affected fruit leaf images caused by bacteria, fungi, or viruses and 5 affected vegetable leaf images The detailed division of Diseases is presented in Table 1.
In this study, a 5-folds cross-validation approach was employed to ensure the robustness and unbiased evaluation of the deep learning models developed for leaf disease classification. This technique involves partitioning the data into 5 equal subsets, where each subset serves once as a test set while the others form the training set (80% was used for training and 20% was used for testing and validation). This process is repeated 5 times, with each of the 5 subsets used exactly once as the test data. The key advantage of using k-folds cross-validation is that it mitigates the risk of model overfitting and provides a comprehensive insight into how the model is expected to perform in general, rather than on a specific set of data. This methodology is especially crucial in fields like plant pathology, where ensuring the model’s ability to generalize across different types of plant diseases is fundamental for deploying such models in real-world agricultural settings. The results from each fold are then averaged to produce a single estimation, providing a more reliable assessment of model performance than a simple train-test split. This validation approach enhances the credibility of the model’s predictive power across diverse conditions and plant species.
Four classification systems, including CNN, VGG-16, DenseNet-121, and Inception-V3, have been utilized to identify methods with higher accuracy and reduced error rates. Each strategy is briefly described in the subsection.
4.1 Convolutional Neural Networks
Several agricultural image categorization projects use deep CNN, a feed-forward artificial neural network (ANN) [30]. When it comes to image categorization, using Deep CNN can significantly cut down on the amount of time and effort required for feature engineering. Convolutional neural networks are composed of several artificial neuronal layers. Artificial neurons are mathematical operations that determine an activation value by computing the weighted sum of many inputs.
Its multi-layered structure makes it effective in analyzing images and extracting the key elements. Multiple layers which is hidden in a convolution neural network is used to extract information extraction from images. The input image, convolutional layer and pooling layer, fully connected layers, and the final output consists of four layers that make up a CNN, as seen in Fig. 3 [31].
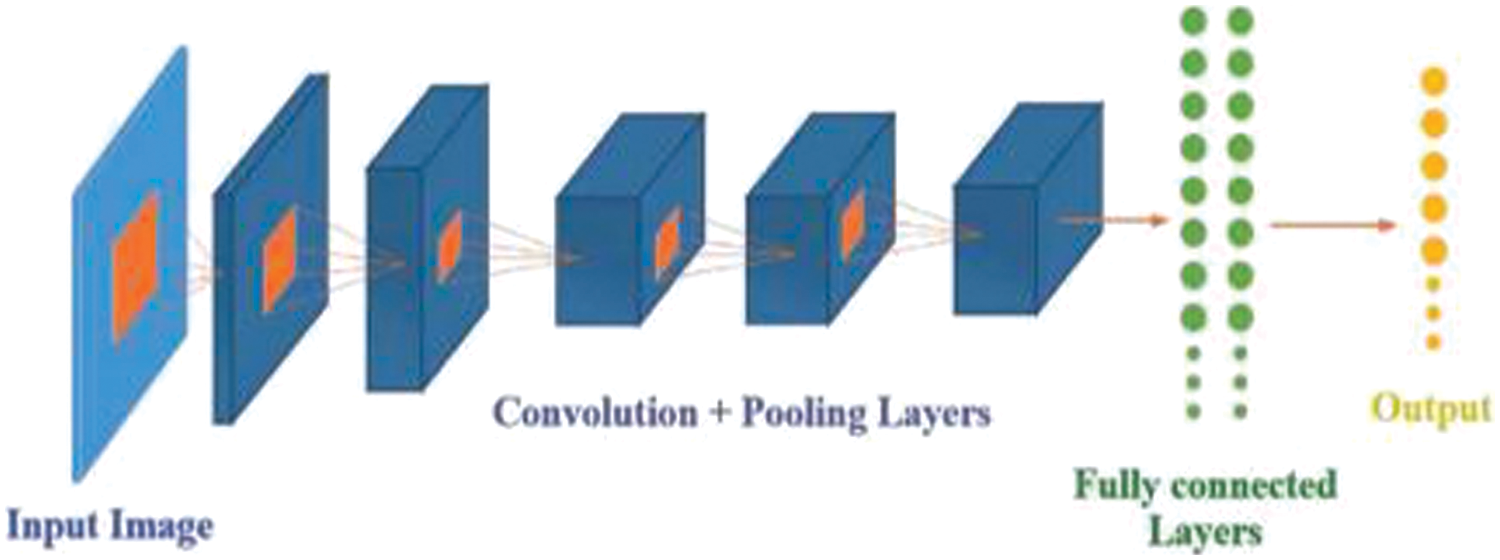
Figure 3: Architecture of convolutional neural network (Reprinted with permission from reference [31]. 2019, IEEE)
A convolutional layer is always the initial layer of a convolutional neural network. Convolutional layers send the result to the following layer after applying a convolution operation on the input. Every pixel in a convolution’s receptive area is combined into a single value. In a convolution layer, a number of filters combine to carry out the convolution operation [32]. Pixel values are seen as a matrix in every image. Examine the subsequent 5 × 5 image, where each pixel has a value of either 0 or 1. Additionally, a 3 × 3 filter matrix is present. To obtain the convolved feature matrix, move the filter matrix across the image and calculate the dot product as shown in Fig. 4.
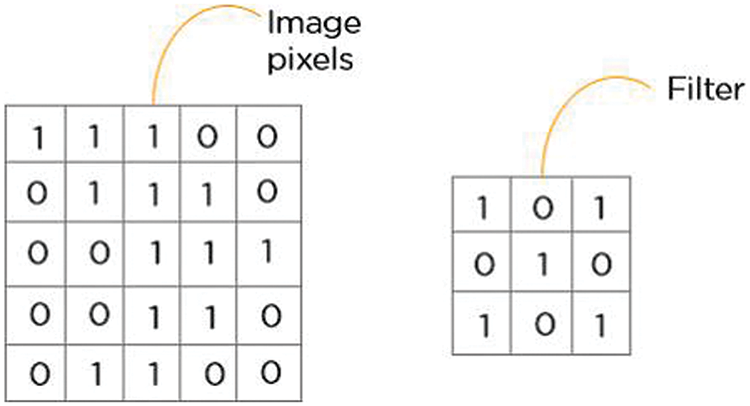
Figure 4: 5 × 5 image pixel value into 3 × 3 filter matrix
This layer decreases the size of the neurons in the downsampling layer and lessens overfitting. This layer minimises the size of the feature map, lowers the number of parameters, speeds up computation, shortens training times, and manages overfitting [33]. A model that achieves 100% on the training dataset and 50% on the test data is said to be overfitting. An illustration of the pooling procedure is shown in Fig. 5. The feature map dimensions were reduced by using ReLU and max pooling.
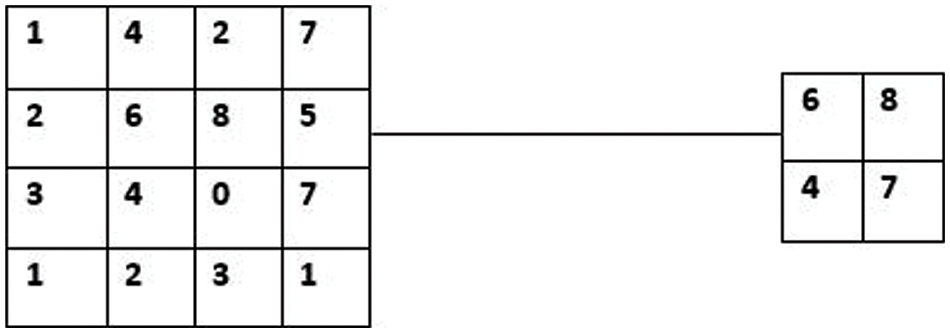
Figure 5: Max pooling with 2 × 2 filters and stride 2
Each convolution layer makes use of a non-linear ReLU (Rectified Linear Unit) activation layer. This layer also applies the dropout layers that are used to avoid overfitting.
This layer is used to analyse the class probabilities and the output is the input of the classifier. In this layer, leaf disease are recognized and classified using the well-known Softmax classifier as the input classifier. The convolutional layer, which carries out the convolution operation, receives the image’s pixels. The outcome is a fragmented map. The rectified feature map is produced by applying the convolved map to a ReLU function. Multiple convolutions and ReLU layers are applied to the image in order to locate the features. Various pooling layers employing different filters are employed to recognise distinct areas within the image. Flattening the pooled feature map and given to a layer which is fully connected in order to achieve the final result. The CNN recognizes a leaf disease shown in Fig. 6.
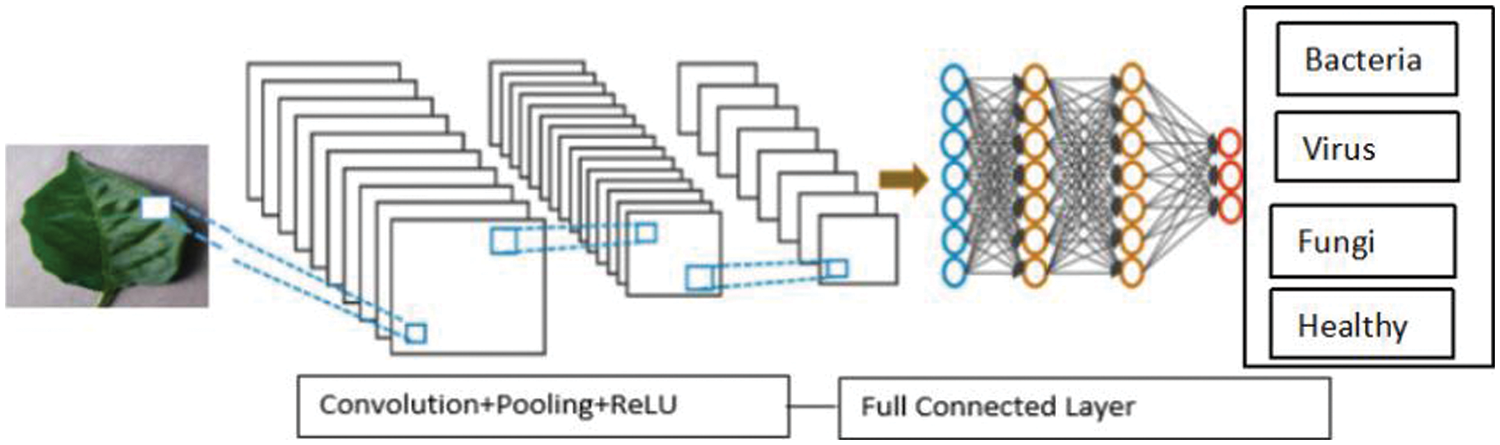
Figure 6: Leaf disease detection using CNN
One of the most often used CNN architectures is VGG-16, mostly because it integrates nicely with ImageNet, a sizable project used for visual object detection [34]. There are 16 layers. An image with dimensions is used as the network’s input (224, 224, 3). Following a layer with 2 convolution layers that have 256 filter size and 3, 3 paddings, there are two layers with 64 channels of 3 × 3 filter size and the same padding, and then the (2, 2) stride. The image is then split into two sets of layers: a max pool layer and a set of three convolution layers. The resulting image is transmitted to the other layers after each of these has applied 512 filters of the same size. The size of the filters in these two layers differs in that AlexNet uses filters that is 3 by 3 while ZF-Net uses filters that are 7 by 7 in size. The network manipulates the number of inputs in some of the layers using a 1 × 1 pixel. To stop the spatial feature from impacting the image, padding is introduced after each convolution layer. Furthermore, it doesn’t increase accuracy overall. The VGG-16 recognizes a leaf disease shown in Fig. 7.
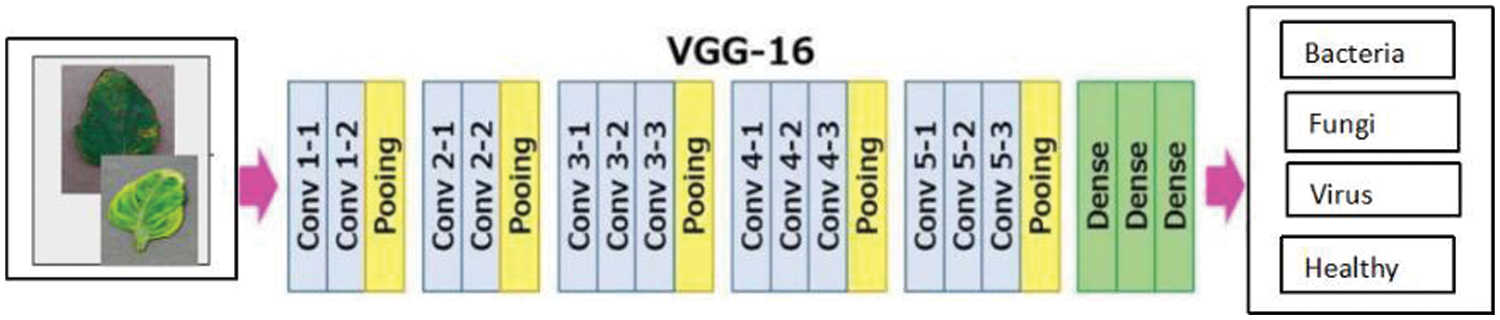
Figure 7: Leaf disease detection using VGG-16
The convolution and max-pooling layers were stacked to produce a (7, 7, feature map. This result is flattened to produce a (1, 25088) feature vector. All three of the ensuing layers are completely connected. Taking the final feature vector as input, the first creates a (1, 4096) vector; the second does the same; and the third creates a vector of 1000 channels for 1000 ILSVRC classes. The soft-max layer receives the output of the third fully connected layer after which the classification vector is normalized. Once the classification vector has been constructed, the top five categories require to be evaluated. All layers which is hidden have ReLU as their activation function. Generally, VGG does not use Local Response Normalization (LRN) because it increases training times and memory requirements [35]. There are two versions of VGG-16 (C and D). VGG utilizes (1, 1) filter size convolution rather than (3, 3) filter size convolution, which is the only significant difference between them. These two have parameters of 134 million and 138 million, respectively.
It is one of the most recent advancements in visual object recognition using neu-ral networks. Swaminathan et al. [36]. debuted DenseNet-121, a well-liked CNN architecture, as a member of the DenseNet family in 2017. Dense connections between layers, which enable information to move through the network more quickly and accurately, are the foundation of the network. Each of the several dense blocks that make up the DenseNet-121 design has a set of convolutional layers that are densely coupled to every other layer. Although there are some significant differences, DenseNet and ResNet are relatively comparable to one another. ResNet utilizes an approach (+) to add the previous layer (identity) with the upcoming layer, whereas DenseNet uses an approach (.) to concatenate the output of the previous layer with the output of the subsequent layer [37]. The DenseNet-121 recognizes a leaf disease shown in Fig. 8.
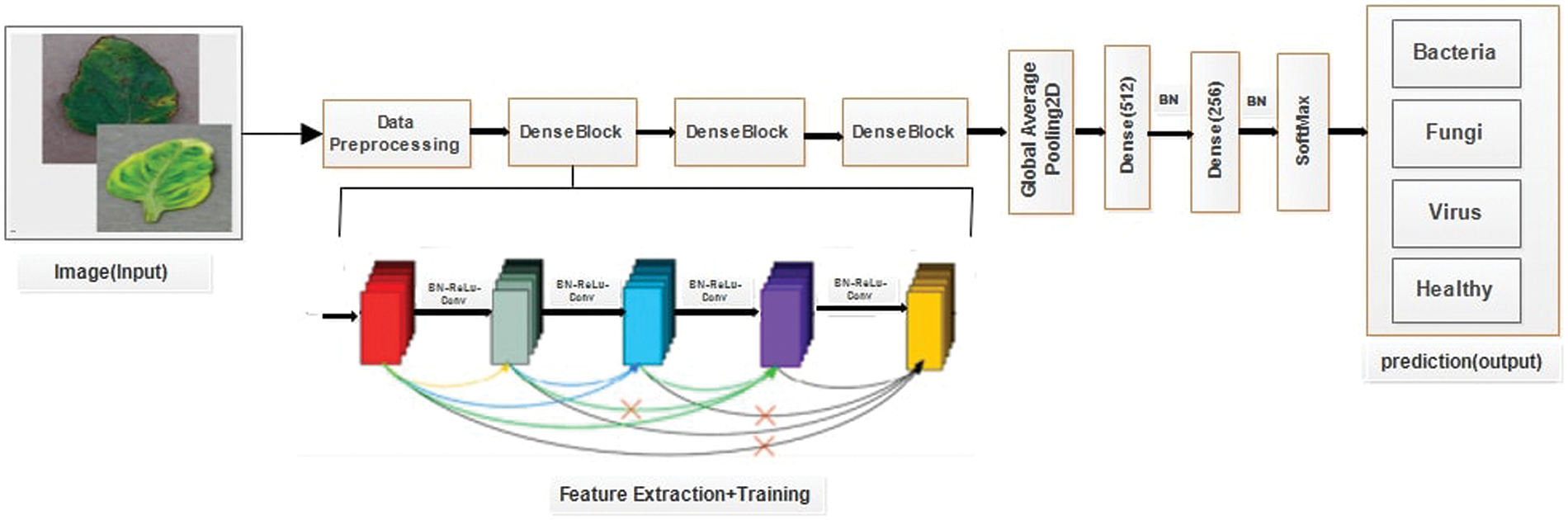
Figure 8: Leaf disease detection using DenseNet-121
In DenseNet, an output from the first layer becomes an input for the second layer when the composite function operation is used. The convolution layer, batch normalization layer, pooling layer, and non-linear activation layer make up this composite process. As a result of these links, the network contains L(L + 1)/2 direct connections. The letter L stands for the number of levels in the architecture. DenseNet is available in many different configurations, including DenseNet-121, DenseNet-160, DenseNet-201, and others.
Inception-V3 is a more advanced version of the core model Inception-V1, which was initially released in 2015. Additionally, batchnorm is applied to activation inputs and used throughout the model. Feature extraction facilitates the model’s ability to discern between each of the image’s attributes and com-prehend them for further interpretation [38]. The “Factorization into smaller convolutions” technique, which divides the convolution process into multiple smaller convolution operations, is another new feature of Inception-V3. This facilitates the reduction of the number of parameters that must be computed and expedites the process of inference and training. The Inception-V3 recognizes a leaf disease shown in Fig. 9.
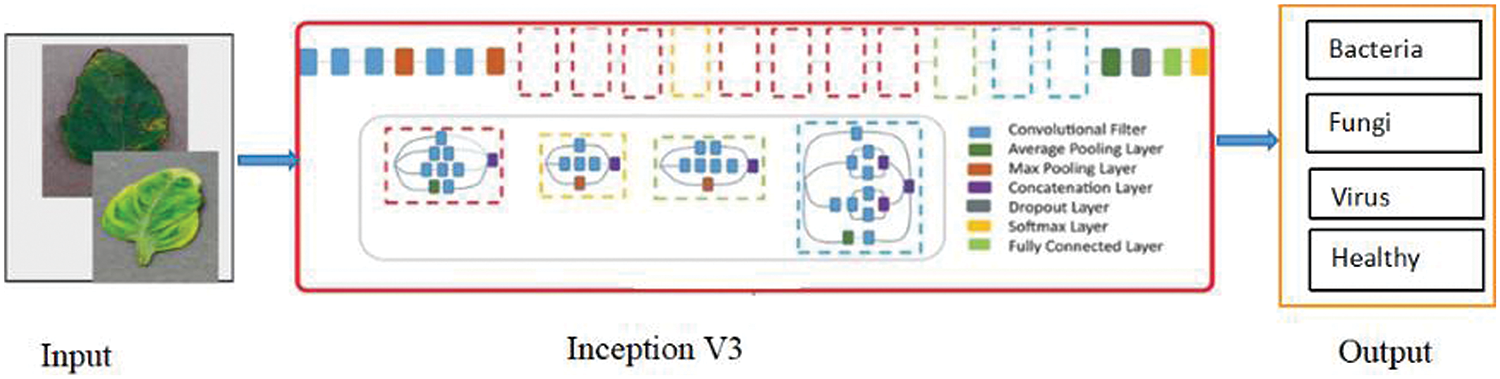
Figure 9: Leaf disease detection using Inception-V3
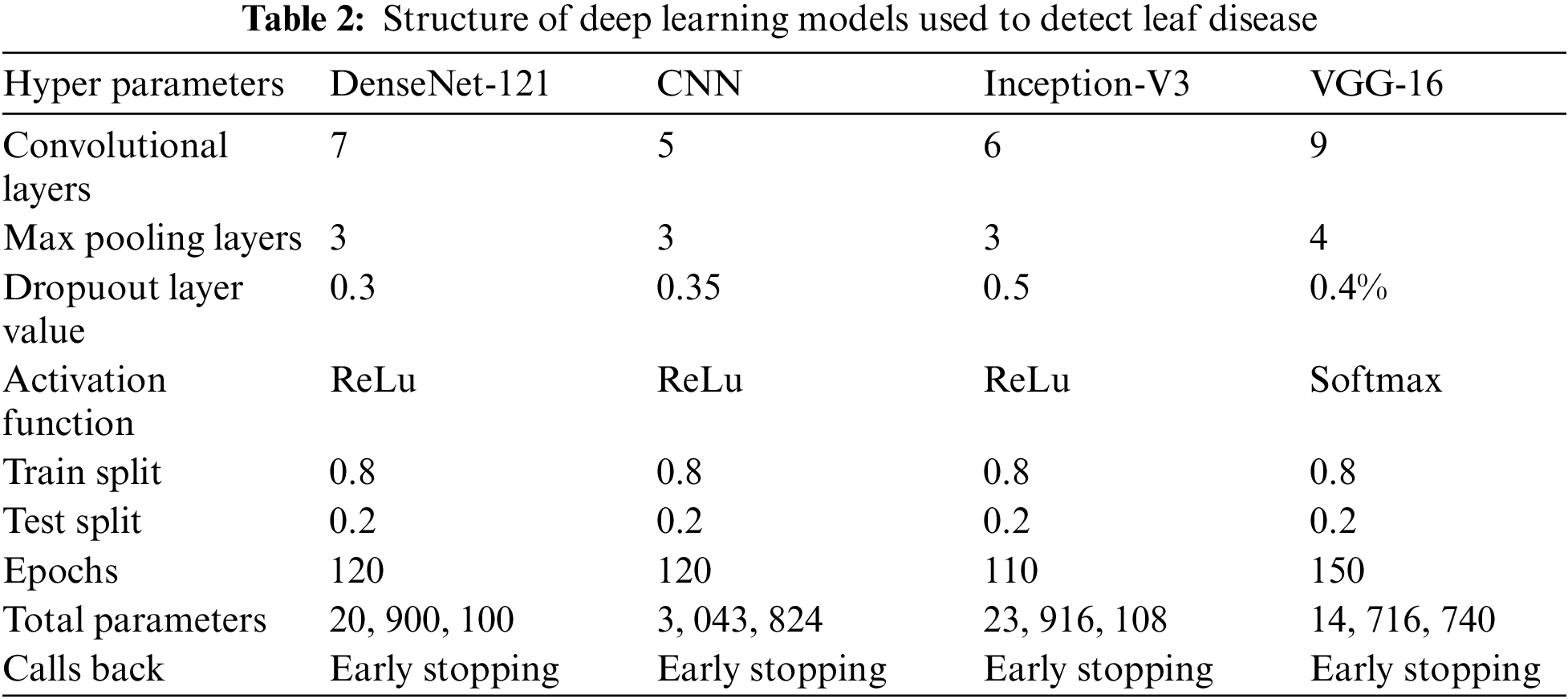
First, the performance of deep learning techniques is examined in terms of precision, recall, F1-score, and accuracy. The results are analyzed to determine which strategy performed better and why. The hyperparametrs of the deep learning models are shown in Table 2.
We used Python3 to create and train our model. For our purpose, we leveraged the capabilities of multiple frameworks and libraries, including Numpy, Pandas, Tensorflow, and Keras, in addition to Python. The results from the examination of classification algorithms are presented in this section. The following metrics were used to rate these classifiers: precision, recall, F1-score,
AUC, and accuracy. CNN, DenseNet-121, VGG-16, and Inception-V3 were four classifiers evaluated on bacteria, fungi, viruses diseases, and healthy plant leaves.
An important factor in assessing the classifier is the performance metric. Furthermore, it serves as a guide for classifier modelling. Four metrics are used to evaluate the performance of our proposed model and each pre-trained model separately: Validation Accuracy, Positive Predictive Value (PPV), some-times called Precision, Recall, and F1-score. Accuracy is the proportion of correct forecasts to all of the model’s predictions. The precision of a classifier can be calculated by dividing its anticipated number of positive results by the number of correct positive outcomes. The ratio of accurate positive results to the total number of relevant samples—that is, all samples that ought to have been classified as positive is known as recall. Precision and Recall are harmonic means that make up the F1-score.
We present the outcomes of the models used in this study. The accuracy of models at target leaf diseases are shown in Table 3. Observations show that Inception-V3 classifies most accurately on bacteria and virus disease. DenseNet-121 most accurately on fungi leaf disease. The accuracy for identifying healthy leaves is exceptionally high across all models. This could imply that healthy leaves have distinct characteristics that are easily distinguishable by these models. Tables 4–6 show the precision, recall and F1-score of the models used to detect leaf disease. It is observed that Inception-V3 precision, recall and F1-score value is higher than CNN, DenseNet-121, and VGG-16. The confusion matrix in Figs. 10, 11 shows how well the models employed in this study performed on the plant dataset. It has been noted that the Inception-V3 performs well at early leaf disease detection. We use the area under score to tabulate the following numerical data in Figs. 12 and 13 of the employed techniques CNN, VGG-16, Inception-V3, and DenseNet-121 respectively show the area under curve score. To determine the accuracy of each classifier, experiments were conducted on a dataset of images of bacteria, fungi, viruses, and healthy leaves. Table 7 shows the accuracy of CNN, Inception-V3, DenseNet-121, and VGG-16 calculated throughout the experiments.
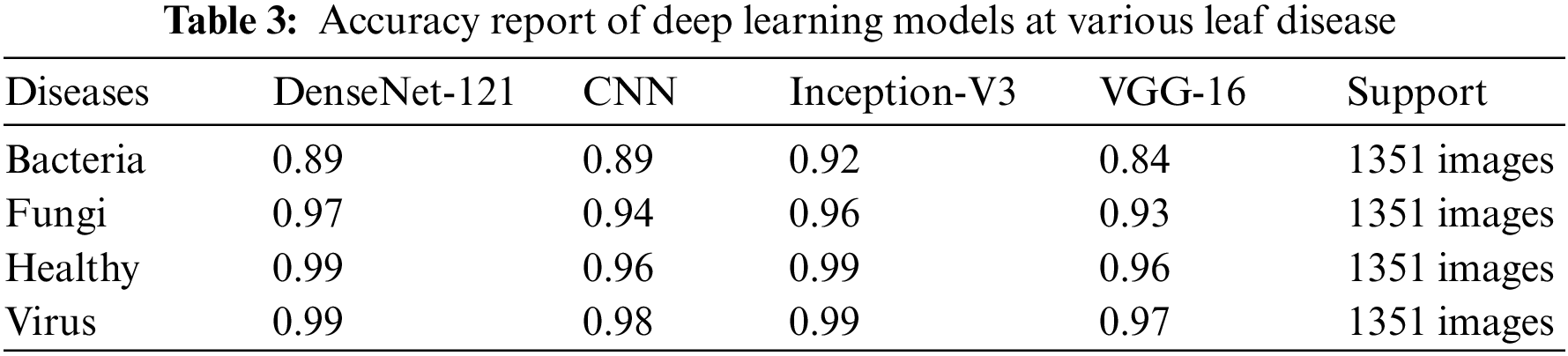
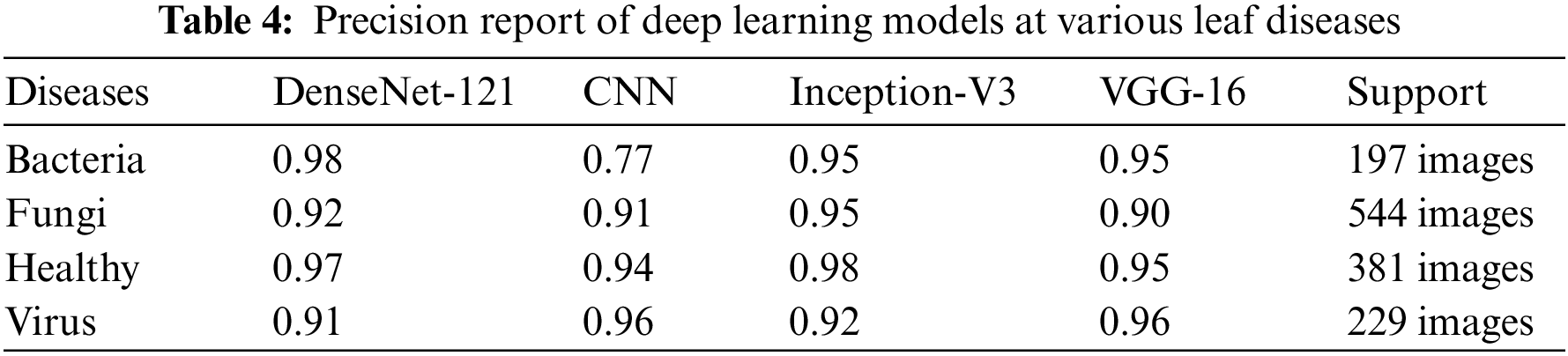
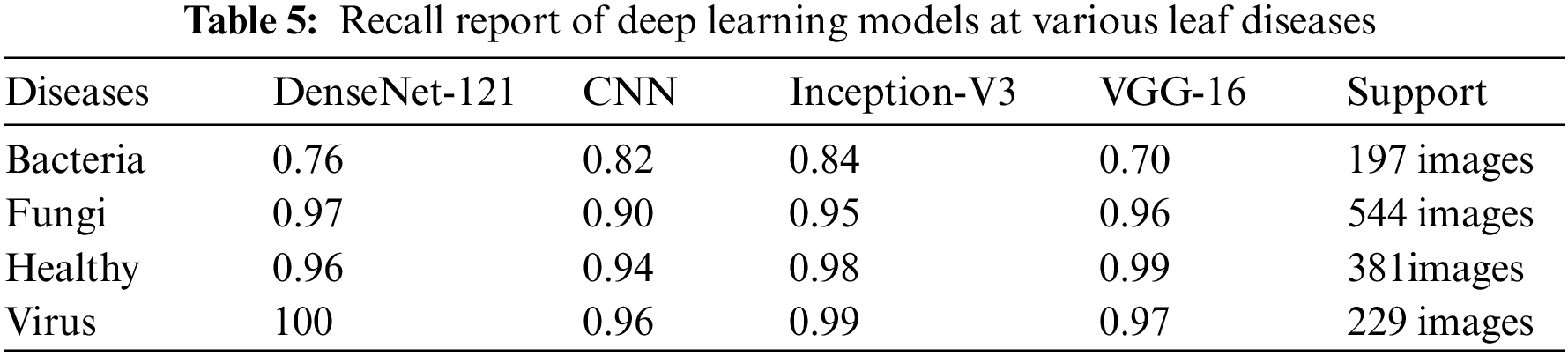
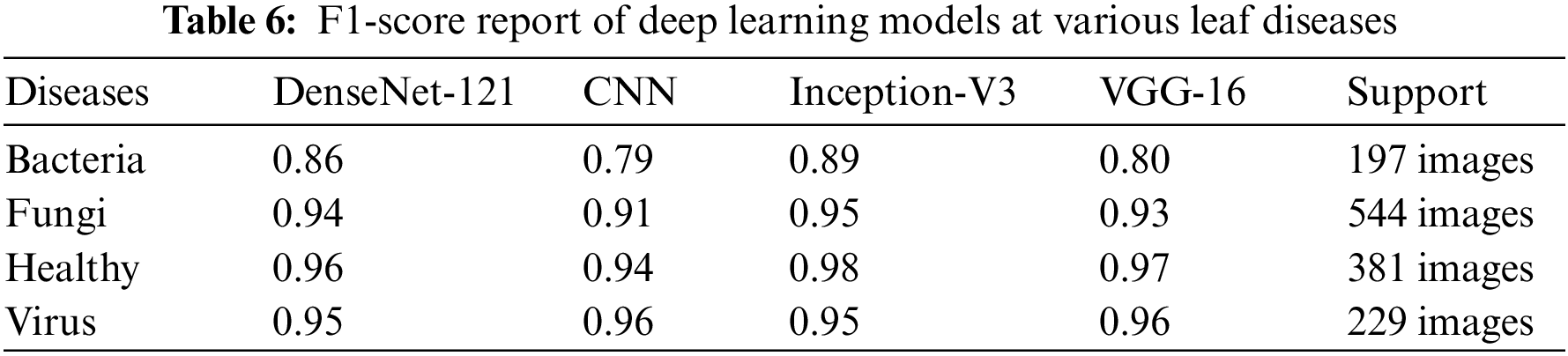
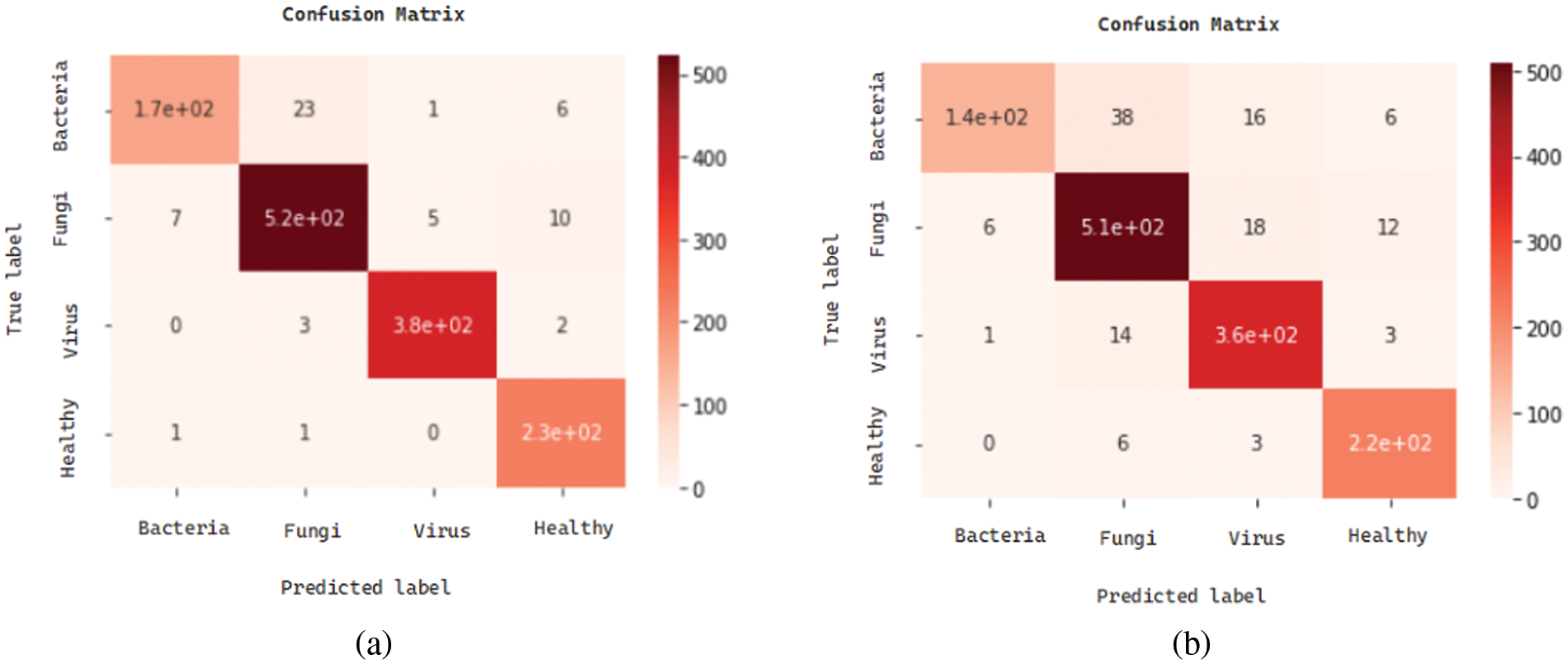
Figure 10: Confusion matrices: (a) confusion matrix of Inception-V3, (b) confusion matrix of VGG-16
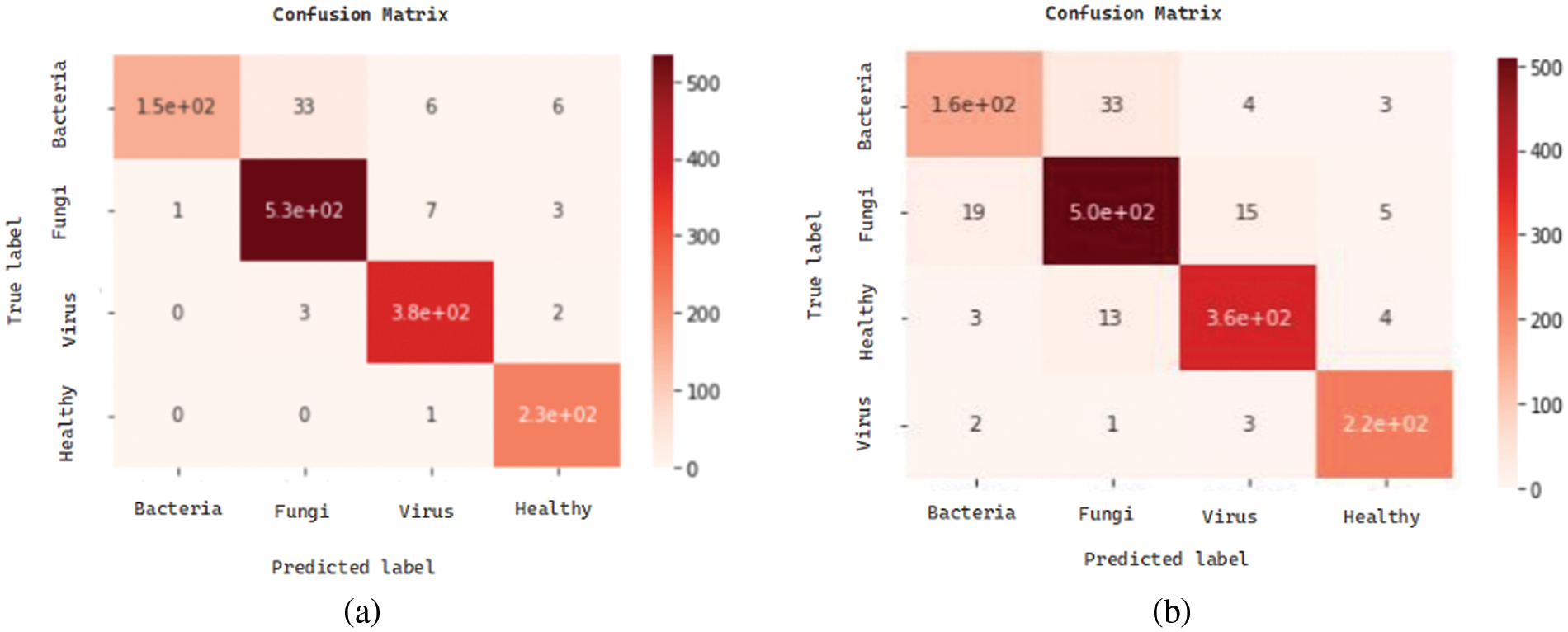
Figure 11: Confusion matrices: (a) confusion matrix of DenseNet-121, (b) confusion matrix of CNN
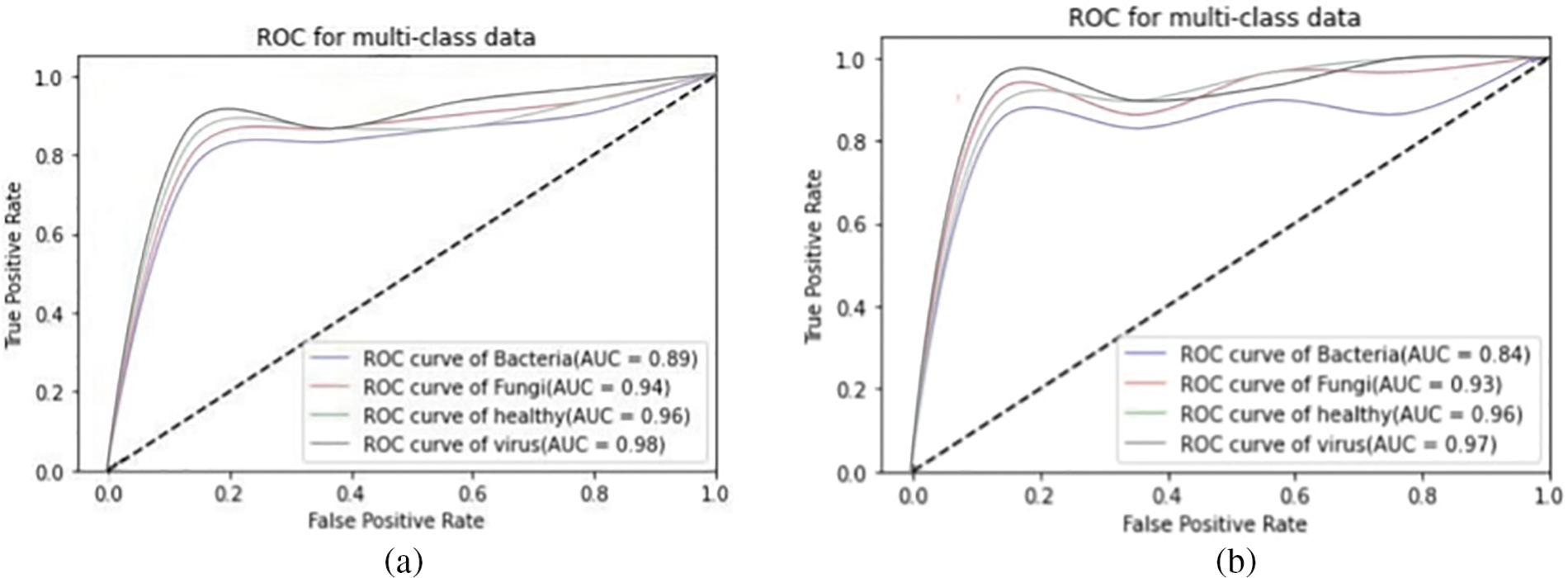
Figure 12: AUC-ROC of (a) CNN, and (b) VGG-16
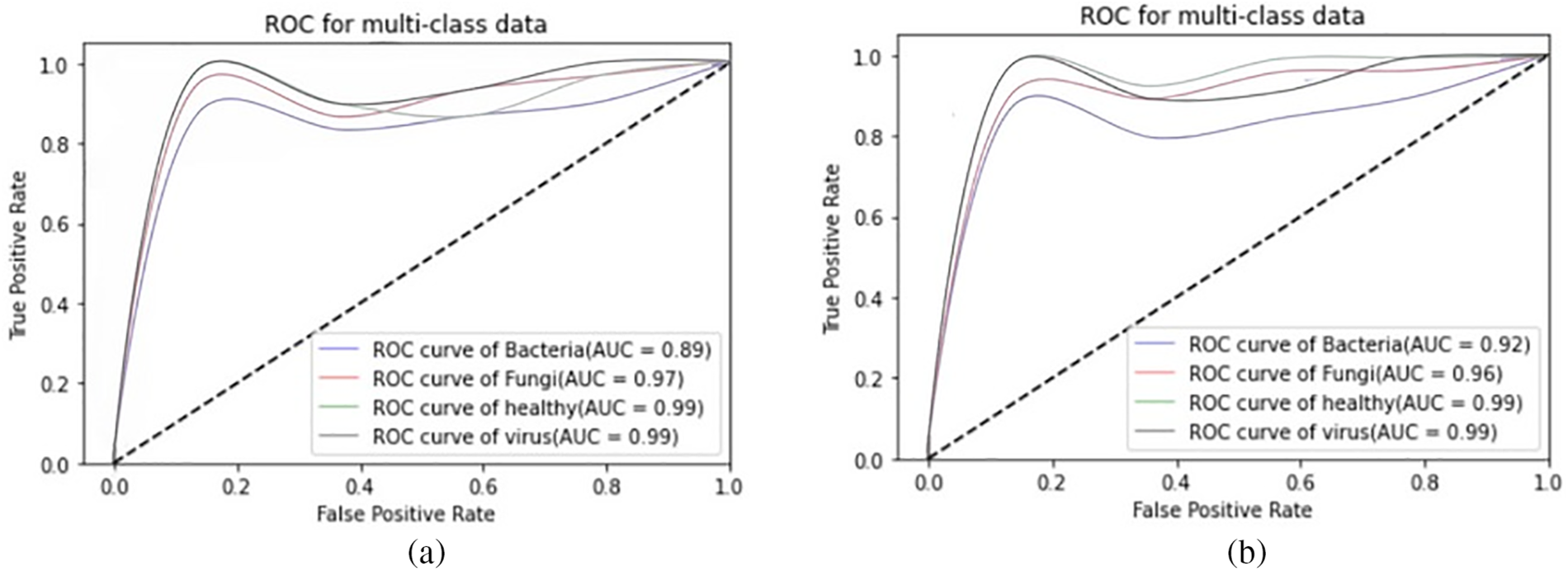
Figure 13: AUC-ROC plots: (a) AUC-ROC of DenseNet-121, (b) AUC-ROC of Inception-V3
The study shows that Inception-V3 outperformed other approaches in each of the aforementioned diseases.
DenseNet, and VGG-16 all produced results that were slightly better. In addition to achieving higher results in terms of accuracy, Inception-V3 also did so in terms of precision, recall, F1-score, and AUC. Inception-V3 consistently shows high accuracy across all categories of leaf diseases, particularly excelling in identifying bacteria, virus, fungi and healthy leaves. This suggests that Inception-V3 is quite effective in distinguishing between these specific conditions, possibly due to its more sophisticated architecture. DenseNet-121’s highest accuracy is noted in detecting virus in leaf disease. This might indicate that the features of virus diseases are more effectively captured by the DenseNet-121 architecture. Inception-V3 not only leads in accuracy but also in precision, recall, and F1-scores. The ability to persist to detect leaf disease is highlighted by its consistency across several parameters. It’s essential to maintain a balance between recall and precision. For example, DenseNet-121 exhibits a high precision of 98% for detecting bacteria, but its recall is much lower, at 76%. Although it misses a large proportion of real cases, it is quite accurate at identifying bacteria. However, its precision is higher compared to Inception-V3 and VGG-16. The lower recall value can be attributed to the model’s tendency to miss certain true positives, effectively failing to identify all instances of disease present in the dataset. Such a phenomenon may arise due to the inherent complexity and variability of disease symptoms across different plant species, which can be challenging for the model to uniformly recognize, especially under limited or imbalanced training conditions. Every model shows improvements in many domains. For instance, DenseNet-121 is excellent at detecting, whereas VGG-16 has great recall and precision in specific categories, but Inception-V3 performs well overall. All things considered, the Inception-V3 model is remarkable for its outstanding outcomes in a wide range of evaluations and diseases. However, the specific choice of model might depend on the leaf disease being targeted and the balance between precision and recall that is desired. The results also emphasize the importance of using a diverse range of models for comprehensive disease detection in leaves.
We presented an ensemble of pre-trained deep learning models (DenseNet-121, CNN, VGG-16 and Inception-V3) and we examined how well they performed using a plant dataset. The main drawback of our work is that we only examined four classes (bacteria, fungi, viruses, and healthy). The performance was examined with some widely used assessment metrics, and the four models’ scores individually for a variety of criteria, including F1, accuracy, precision, and recall. Outcomes show the better performance of Inception-V3 in achieving high accuracy. As well as that, CNN, VGG-16, and DenseNet-121 also have better performance, slightly less than Inception-V3. The accuracy of Inception-V3 is 95%. With this level of image precision, growers may automate the disease classification process using this method. This research was to cover a comparative analysis of deep learning models used for detecting leaf disease. The literature shows that a variety of study fields have produced prediction systems, although improving accuracy remains a challenging task. Despite the wide research done in this field, there is still room for improvement. This study focuses on improving the accuracy rate of prediction. The automatic identification of plant leaf disease will benefit from this effort, and early disease detection will boost agricultural productivity. We hope to add more kinds of diseases to our model’s target classes in subsequent research and higher performance levels are anticipated from more deep learning techniques in the future.
Acknowledgement: None.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Robertas Damasevicius, Faheem Mahmood, Yaseen Zaman, Sobia Dastgeer, Sajid Khan; data collection: Robertas Damasevicius, Faheem Mahmood, Yaseen Zaman, Sobia Dastgeer, Sajid Khan; analysis and interpretation of results: Robertas Damasevicius, Faheem Mahmood, Yaseen Zaman, Sobia Dastgeer, Sajid Khan; draft manuscript preparation: Robertas Damasevicius, Faheem Mahmood, Yaseen Zaman, Sobia Dastgeer, Sajid Khan. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The PlantVillage dataset used in this study is available at https://www.kaggle.com/datasets/abdallahalidev/plantvillage-dataset (accessed on 25 January 2024).
Ethics Approval:: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1https://www.kaggle.com/datasets/abdallahalidev/plantvillage-dataset (accessed on 25 January 2024).
References
1. J. A. Wani, S. Sharma, M. Muzamil, S. Ahmed, S. Sharma and S. Singh, “Machine learning and deep learning based computational techniques in automatic agricultural diseases detection: Methodologies, applications, and challenges,” Arch. Comput. Methods Eng., vol. 29, no. 1, pp. 641–677, 2022. doi: 10.1007/s11831-021-09588-5. [Google Scholar] [CrossRef]
2. P. Rani, S. Kotwal, J. Manhas, V. Sharma, and S. Sharma, “Machine learning and deep learning based computational approaches in automatic microorganisms image recognition: Methodologies, challenges, and developments,” Arch. Comput. Methods Eng., vol. 29, no. 3, pp. 1801–1837, 2022. doi: 10.1007/s11831-021-09639-x. [Google Scholar] [PubMed] [CrossRef]
3. G. C. Percival and G. A. Fraser, “The influence of powdery mildew infection on photosynthesis, chlorophyll fluorescence, leaf chlorophyll and carotenoid content of three woody plant species,” Arboric. J., vol. 26, no. 4, pp. 333–346, 2002. doi: 10.1080/03071375.2002.9747348. [Google Scholar] [CrossRef]
4. M. Khater, A. de la Escosura-Muñiz, and A. Merkoçi, “Biosensors for plant pathogen detection,” Biosens. Bioelectron., vol. 93, pp. 72–86, 2017. doi: 10.1016/j.bios.2016.09.091. [Google Scholar] [PubMed] [CrossRef]
5. R. T. Plumb, “Viruses and plant disease,” Nature, vol. 158, pp. 885–886, 2001. doi: 10.1038/npg.els.0000770. [Google Scholar] [CrossRef]
6. M. Kumar, A. Kumar, and V. S. Palaparthy, “Soil sensors-based prediction system for plant diseases using exploratory data analysis and machine learning,” IEEE Sens. J., vol. 21, no. 17, pp. 17455–17468, 2021. doi: 10.1109/JSEN.2020.3046295. [Google Scholar] [CrossRef]
7. H. Alaa, K. Waleed, M. Samir, T. Mohamed, H. Sobeah and M. A. Salam, “An intelligent approach for detecting palm trees diseases using image processing and machine learning,” Int. J. Adv. Comput. Sci. Appl., vol. 11, no. 7, 2020. doi: 10.14569/issn.2156-5570. [Google Scholar] [CrossRef]
8. U. N. Fulari, R. K. Shastri, and A. N. Fulari, “Leaf disease detection using machine learning,” J. Seybold Rep., vol. 1533, pp. 9211, 2020. [Google Scholar]
9. O. O. Abayomi-Alli, R. Damasevicius, S. Misra, and A. Abayomi-Alli, “FruitQ: A new dataset of multiple fruit images for freshness evaluation,” Multimed. Tools Appl., vol. 83, no. 4, pp. 11433–11460, 2024. doi: 10.1007/s11042-023-16058-6. [Google Scholar] [CrossRef]
10. M. Arsenovic, M. Karanovic, S. Sladojevic, A. Anderla, and D. Stefanovic, “Solving current limitations of deep learning based approaches for plant disease detection,” Symmetry, vol. 11, no. 7, pp. 939, 2019. doi: 10.3390/sym11070939. [Google Scholar] [CrossRef]
11. L. Li, S. Zhang, and B. Wang, “Plant disease detection and classification by deep learning—A review,” IEEE Access, vol. 9, pp. 56683–56698, 2021. [Google Scholar]
12. F. Marzougui, M. Elleuch, and M. Kherallah, “A deep CNN approach for plant disease detection,” in 2020 21st Int. Arab Conf. Inform. Technol. (ACIT), Giza, Egypt, 2020, pp. 1–6. [Google Scholar]
13. P. Jiang, Y. Chen, B. Liu, D. He, and C. Liang, “Real-time detection of apple leaf diseases using deep learning approach based on improved convolutional neural networks,” IEEE Access, vol. 7, pp. 59069–59080, 2019. doi: 10.1109/ACCESS.2019.2914929. [Google Scholar] [CrossRef]
14. F. Adenugba, S. Misra, R. Maskeliunas, R. Damasevicius, and E. Kazanavicius, “Smart irrigation system for environmental sustainability in Africa: An internet of everything (IoE) approach,” Math. Biosci. Eng., vol. 16, no. 5, pp. 5490–5503, 2019. doi: 10.3934/mbe.2019273. [Google Scholar] [PubMed] [CrossRef]
15. Z. U. Rehman et al., “Recognizing apple leaf diseases using a novel parallel real-time processing framework based on mask RCNN and transfer learning: An application for smart agriculture,” Inst. Eng. Technol. (IET) Image Process., vol. 15, no. 10, pp. 2157–2168, 2021. doi: 10.1049/ipr2.12183. [Google Scholar] [CrossRef]
16. K. Kayaalp, “Classification of medicinal plant leaves for types and diseases with hybrid deep learning methods,” Inf. Technol. Control., vol. 53, no. 1, pp. 19–36, 2024. doi: 10.5755/j01.itc.53.1.34345. [Google Scholar] [CrossRef]
17. C. Qiu, F. Ding, X. He, and M. Wang, “Apply physical system model and computer algorithm to identify osmanthus fragrans seed vigor based on hyper-spectral imaging and convolutional neural network,” Inf. Technol. Control., vol. 52, no. 4, pp. 887–897, 2023. doi: 10.5755/j01.itc.52.4.34476. [Google Scholar] [CrossRef]
18. J. Francis, A. S. Dhas, and B. Anoop, “Identification of leaf diseases in pepper plants using soft computing techniques,” Int. J. Comput. Bus. Res. (IJCBR), vol. 6, no. 2, pp. 1–12, 2016. doi: 10.1109/ICEDSS.2016.7587787. [Google Scholar] [CrossRef]
19. G. Chugh, A. Sharma, P. Choudhary, and R. Khanna, “Potato leaf disease detection using inception v3,” Int. Res. J. Eng. Technol. (IRJET), vol. 7, no. 2, pp. 1–12, 2020. [Google Scholar]
20. M. Ouhami, Y. Es-Saady, M. E. Hajji, A. Hafiane, R. Canals and M. E. Yassa, “Deep transfer learning models for tomato disease detection,” in Image and Signal Process., 2019, vol. 12119, pp. 65–73. doi: 10.1007/978-3-030-51935-3_7. [Google Scholar] [CrossRef]
21. M. H. Saleem, J. Potgieter, and K. M. Arif, “Plant disease detection and classification by deep learning,” Plants, vol. 8, no. 11, pp. 468, 2019. doi: 10.3390/plants8110468. [Google Scholar] [PubMed] [CrossRef]
22. D. A. G. Sakshi Raina, “A study on various techniques for plantleaf disease detection using leaf image,” IEEE Xplore, vol. 7, no. 3, pp. 20, 2021. [Google Scholar]
23. H. Ajra, M. K. Nahar, L. Sarkar, and M. S. Islam, “Disease detection of plant leaf using image processing and CNN with preventive measures,” in 2020 Emerg. Technol. Comput., Commun. Electron. (ETCCE), 2020, pp. 1–6. doi: 10.1109/ETCCE51779.2020.9350890. [Google Scholar] [CrossRef]
24. R. Y. Patil, S. Gulvani, V. B. Waghmare, and I. K. Mujawar, “Image based anthracnose and redrust leaf disease detection using deep learning,” TELKOMNIKA (Telecommun. Comput. Electron. Control), vol. 20, no. 6, pp. 1256–1263, 2022. [Google Scholar]
25. S. U. Rahman, F. Alam, N. Ahmad, and S. Arshad, “Image processing based system for the detection, identification and treatment of tomato leaf diseases,” Multimed. Tools Appl., vol. 82, no. 6, pp. 9431–9445, 2023. doi: 10.1007/s11042-022-13715-0. [Google Scholar] [CrossRef]
26. U. Atila, M. Uçar, K. Akyol, and E. Uçar, “Plant leaf disease classification using efficientnet deep learning model,” Ecol. Inform., vol. 61, pp. 101182, 2021. doi: 10.1016/j.ecoinf.2020.101182. [Google Scholar] [CrossRef]
27. M. E. Chowdhury et al., “Automatic and reliable leaf disease detection using deep learning techniques,” AgriEngineering, vol. 3, no. 2, pp. 294–312, 2021. doi: 10.3390/agriengineering3020020. [Google Scholar] [CrossRef]
28. M. Nagaraju and P. Chawla, “Systematic review of deep learning techniques in plant disease detection,” Int. J. Syst. Assur. Eng. Manage., vol. 11, pp. 547–560, 2020. doi: 10.1007/s13198-020-00972-1. [Google Scholar] [CrossRef]
29. M. Agarwal, A. Singh, S. Arjaria, A. Sinha, and S. Gupta, “ToLeD: Tomato leaf disease detection using convolution neural network,” Procedia Comput. Sci., vol. 167, pp. 293–301, 2020. doi: 10.1016/j.procs.2020.03.225. [Google Scholar] [CrossRef]
30. Kaggle, PlantVillage Dataset. 2020. Accessed: May 8, 2022. [Online]. Available: https://www.kaggle.com/datasets/abdallahalidev/plantvillage-dataset [Google Scholar]
31. S. V. Militante, B. D. Gerardo, and N. V. Dionisio, “Plant leaf detection and disease recognition using deep learning,” in 2019 IEEE Eurasia Conf. IOT, Commun. Eng. (ECICE), Yunlin, Taiwan, 2019, pp. 579–582. doi: 10.1109/ECICE47484.2019.8942686. [Google Scholar] [CrossRef]
32. S. Albawi, T. A. Mohammed, and S. Al-Zawi, “Understanding of a convolutional neural network,” in 2017 Int. Conf. Eng. Technol. (ICET), Antalya, Turkey, 2017, pp. 1–6. doi: 10.1109/ICEngTechnol.2017.8308186. [Google Scholar] [CrossRef]
33. A. Krizhevsky, I. Sutskever, and G. E. Hinton, “ImageNet classification with deep convolutional neural networks,” Commun. ACM, vol. 60, no. 6, pp. 84–90, 2017. doi: 10.1145/3065386. [Google Scholar] [CrossRef]
34. A. A. Alatawi, S. M. Alomani, N. I. Alhawiti, and M. Ayaz, “Plant disease detection using ai based VGG-16 model,” Int. J. Adv. Comput. Sci. Appl., vol. 13, no. 4, 2022. doi: 10.14569/issn.2156-5570. [Google Scholar] [CrossRef]
35. M. S. Alzahrani and F. W. Alsaade, “Transform and deep learning algorithms for the early detection and recognition of tomato leaf disease,” Agronomy, vol. 13, no. 5, pp. 1184, 2023. doi: 10.3390/agronomy13051184. [Google Scholar] [CrossRef]
36. A. Swaminathan et al., “Multiple plant leaf disease classification using DenseNet-121 architecture,” Int. J. Electr. Eng. Technol., vol. 12, pp. 38–57, 2021. [Google Scholar]
37. W. Liu and K. Zeng, “SparseNet: A sparse DenseNet for image classification,” arXiv preprint arXiv:1804.05340, 2018. [Google Scholar]
38. A. Kumar, R. Razi, A. Singh, and H. Das, “Res-VGG: A novel model for plant disease detection by fusing VGG16 and ResNet models,” in Int. Conf. Mach. Learn., Image Process., Netw. Secur. Data Sci., Singapore, Springer, 2020, pp. 383–400. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools