 Open Access
Open Access
ARTICLE
Microarray Gene Expression Classification: An Efficient Feature Selection Using Hybrid Swarm Intelligence Algorithm
Veermata Jijabai Technological Institute, Department of Electronics Engineering, Mumbai, 410210, India
* Corresponding Author: Punam Gulande. Email:
Computer Systems Science and Engineering 2024, 48(4), 937-952. https://doi.org/10.32604/csse.2024.046123
Received 19 September 2023; Accepted 06 December 2023; Issue published 17 July 2024
Abstract
The study of gene expression has emerged as a vital tool for cancer diagnosis and prognosis, particularly with the advent of microarray technology that enables the measurement of thousands of genes in a single sample. While this wealth of data offers invaluable insights for disease management, the high dimensionality poses a challenge for multiclass classification. In this context, selecting relevant features becomes essential to enhance classification model performance. Swarm Intelligence algorithms have proven effective in addressing this challenge, owing to their ability to navigate intricate, non-linear feature-class relationships. This paper introduces a novel hybrid swarm algorithm, fusing the capabilities of the Artificial Bee Colony (ABC) and Firefly algorithms, aimed at improving feature selection in gene expression classification. The proposed method undergoes rigorous validation through statistical machine learning techniques and quantitative parameter evaluation, with comprehensive comparisons to established techniques in the field. The findings underscore the superiority of the hybrid Swarm Intelligence approach for feature selection in gene expression classification, offering promising prospects for enhancing cancer diagnosis and prognosis.Keywords
Gene expression analysis has become a valuable tool in the diagnosis and prognosis of cancer. The advent of microarray technology has revolutionized this field by allowing researchers to comprehensively assess the expression levels of thousands of genes within a single biological sample. This wealth of information enables the identification of distinctive gene expression signatures associated with different cancer types, paving the way for the development of sophisticated classification algorithms designed for multiclass gene expression classification. This classification task involves the assignment of biological samples to predefined classes based on their gene expression profiles [1]. Numerous studies have been dedicated to the classification of cancer types using microarray gene expression data, resulting in a diverse array of algorithms. For instance, one noteworthy endeavor proposed a detection model for the classification of lung cancer based on microarray data, effectively leveraging tumor gene expression signatures to facilitate the diagnosis of multiclass cancer [2]. Simultaneously, the research community has conducted surveys and comprehensive analyses to explore classification methods tailored for specific cancer types, as exemplified by a study focusing on breast cancer classification using microarray gene expression data [3]. Feature selection represents another pivotal facet of multiclass gene expression classification. It entails the judicious selection of the most relevant genes from the initial microarray data, thereby reducing data dimensionality, enhancing result interpretability, and ultimately improving classification algorithm performance. The importance of this aspect is underscored by comprehensive systematic reviews, exemplified by a study that conducted an extensive analysis of feature selection methods in the context of cancer classification [4]. Multiclass gene expression classification employs a diverse array of classification algorithms, including but not limited to support vector machines (SVM), artificial neural networks (ANN), decision trees, and k-nearest neighbors (k-NN). These algorithms have been subject to rigorous comparative evaluations in the literature, with studies seeking to elucidate their relative performance in specific cancer classification tasks. An illustrative example includes a comparison of swarm intelligence techniques applied to lung cancer classification [5]. Collectively, these endeavors underscore the dynamic and multifaceted nature of gene expression analysis in the field of cancer research. This exploration ranges from the development of innovative classification algorithms tailored to distinct cancer types, comprehensive surveys of classification methods, to the intricacies of feature selection, and comparative assessments of various classification techniques. These collective efforts hold promise in advancing our understanding of cancer biology and improving diagnostic and prognostic capabilities.
1.2 Significance of Swarm Intelligence (SI)
Swarm Intelligence is an interdisciplinary field that focuses on the study of decentralized systems and the investigation of their collective behavior. This method has proven to be effective in solving a variety of problems across different domains, including computer science, engineering, and biology. In recent years, swarm intelligence has received considerable attention in the realm of gene expression classification for multiclass classification problems. This is due to its ability to serve as a feature selection method for microarray data. Microarray technology is widely utilized to evaluate multiple expressions that could be in thousands at a single glance providing valuable insights for the diagnosis and treatment of various diseases, such as cancer. The high dimensionality of microarray data presents a significant challenge for multiclass classification, as not all features, in this case genes, are relevant to the classification problem. Feature selection is an important step in the analysis of microarray data, as it helps to reduce the dimensionality of the data and enhance the performance of the classification models. Swarm Intelligence algorithms are well suited for gene expression selection in multiclass classification due to their ability to handle complex and non-linear relationships between the features and class labels. Swarm Intelligence algorithms can be categorized into two main types: Optimization-based and consensus-based. Optimization-based algorithms, such as Particle Swarm Optimization (PSO), attempt to optimize a cost function that reflects the quality of the selected features. Meanwhile, consensus-based algorithms, such as Ant Colony Optimization (ACO) and Bee Algorithm (BA), simulate the collective behavior of social insects to find the optimal solution. The following Eq. (1) represents the optimization-based swarm intelligence algorithm for feature selection:
X represents the feature subset, and F(x) represents the cost function that measures the potency of the extracted features.

The above pseudo code for the general selection architecture can be represented in steps as follows:
Step 1: Initialize Swarm population
Define number of particles (N) in the swarm.
Generate random initial Swarm element positions in the search space.
Initialize Swarm element attribute vectors.
Step 2: Evaluate fitness of each element/swarm
Calculate the fitness of each Swarm element based on gene expression data.
Store the fitness values for each particle.
Determine the current best Swarm element (gBest) with the highest fitness value.
Step 3: Update Swarm element attribute and position
For each particle, calculate new attribute using the following equation:
where x[i] = attribute of Swarm element I, w = weight of the inertia, C1 and C2 are cognitive and social learning factors, P is a random number that holds a value between 0–1, pBest[i] = The local best value, b[i] = current temporary position of ith swarm, gBest = The global best value
Update the position of each Swarm element using the following equation:
where x[i] = current position of Swarm element I, v[i] = attribute of Swarm element i
Step 4: Repeat mentioned in 2 and 3 while the algorithm does not reach to stop ping condition
The algorithm continues to repeat steps 2 and 3 until a maximum number of iterations is reached or the Swarm element convergence criteria is met.
Step 5: Select features
Gene expression analysis is widely recognized as a crucial tool in the field of cancer diagnosis and prognosis. Microarray gene expression data offers a comprehensive in-sight into the expression levels of numerous genes within a biological sample and can be utilized to identify gene expression signatures associated with various types of cancer. As a result, a multitude of classification algorithms have been developed for multiclass gene expression classification, which involves assigning a sample to one of several pre-defined classes, based on the expression levels of its genes. Another crucial aspect of multiclass gene expression classification is feature selection. This process involves selecting a subset of genes that are most relevant to the classification task, which can improve the performance of the algorithms, reduce data dimensionality, and enhance the results’ interpretability [5]. A range of algorithms have been used for multiclass gene expression classification, including (SVM), (ANN), decision trees, and (k-NN), and their performance has been compared in various studies [4]. Metaheuristics, such as genetic algorithms, particle swarm optimization, and artificial bee colonies, have also been utilized for feature selection in microarray gene expression data [6,7]. Hybrid methods, combining multiple techniques, have also been proposed and compared for feature selection and classification in microarray gene expression data [8–10]. The latter study found that the relaxed Lasso and generalized multi-class support vector machine outperformed other methods in terms of feature selection and tumor classification. In conclusion, the studies mentioned in this introduction underscore the significance of multiclass gene expression classification and feature selection in the analysis of micro array gene expression data for cancer diagnosis and prognosis. A wide range of algorithms, including traditional machine learning algorithms, metaheuristics, and hybrid methods have been proposed and compared. These studies underline the need for on-going research in this field to enhance the accuracy and interpretability of features and improve classification accuracy. As it has been observed that a lot of research work has made its mark in gene expression selection and classification, this paper contributes in the following manner, as shown in Fig. 1.
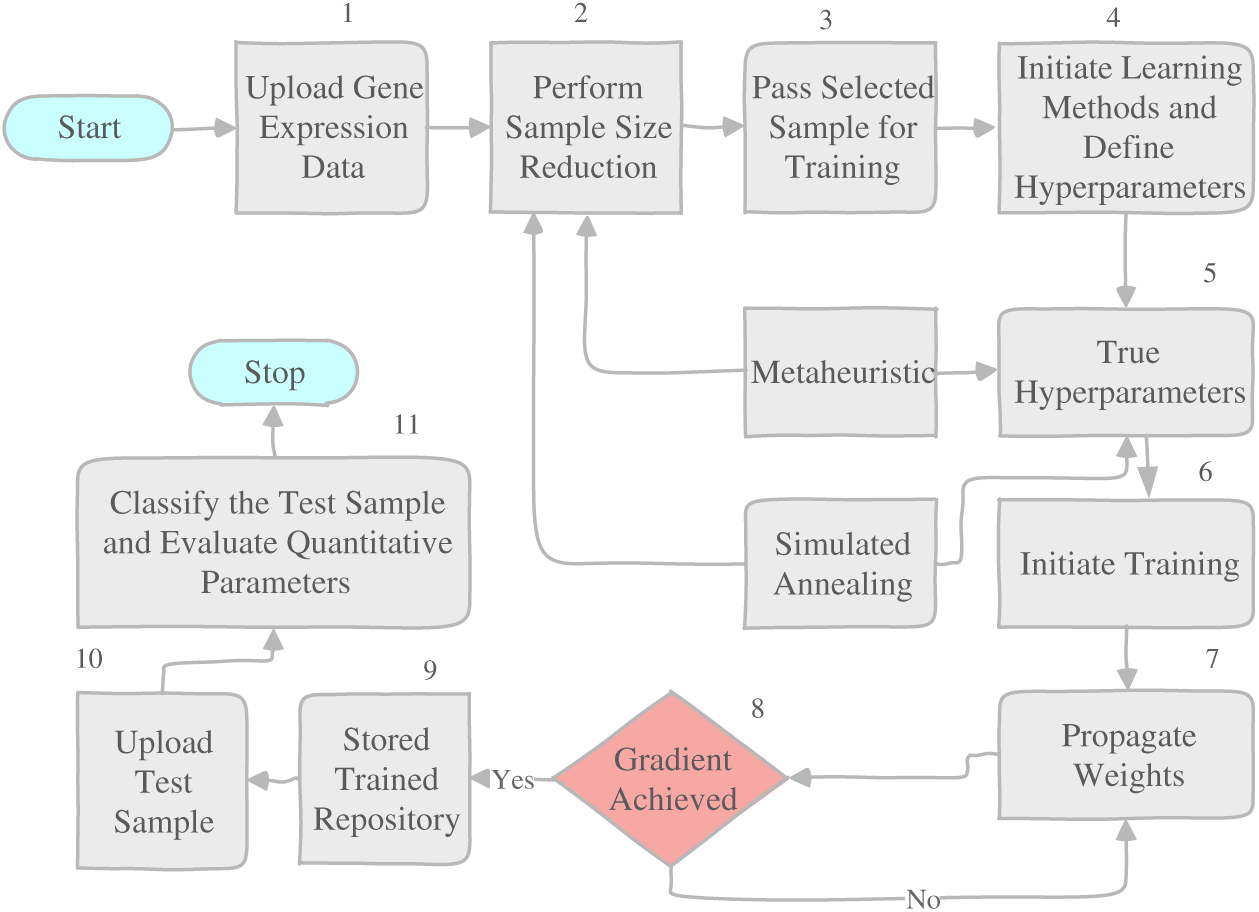
Figure 1: The overall work flow
• Proposes a hybrid swarm algorithm for a more efficient feature selection technique that includes ABC and Firefly.
• Validates the proposed hybrid feature selection method using statistical machine learning.
• Evaluates classified expression in terms of quantitative parameters and comparison with other state of the art techniques.
In this review, various Swarm Intelligence methods have been employed for gene expression data classification in cancer diagnosis, including bee-based (ABC) [6] Genetic Bee Colony (GBC) [11], Moth Flame Optimization Algorithm [1], Binary Artificial Bee Colony Algorithm (Wang and Dong [12]) and Hybrid Fuzzy Ranking Network [9]. Studies have shown the significance of machine learning methods in microarray data classification and the crucial role of feature selection in enhancing the performance of gene expression-based cancer diagnosis [5]. In the field of cancer classification, various techniques have been utilized to select important features from gene expression microarray data. This includes the use of Tumor Gene Expression Signatures in Multiclass Cancer Diagnosis by [2] the integration of ABC and (SVM) by [11] the application of Relaxed Lasso and Generalized SVM that can classify multiple classes by [10] and the implementation of a Hybrid Feature Selection method for the detection of breast cancer via gene expressions [7]. In recent years, hybrid approaches combining Swarm Intelligence techniques and other machine learning methods have been proposed for gene selection and classification of biomedical microarray data. For example, the authors in [13] proposed a nature-inspired metaheuristics model, while reference [3] presented a hybrid method in which the SI-based algorithm MFOA is integrated with quantum-oriented computing to select the best features. This gives the proposed work a way to improvise the data selection algorithm and mechanism. Moreover, a survey on hybrid feature selection methods in microarray gene expression data for cancer classification was conducted by [8]. Ameri et al. introduced an innovative approach to self-assessment within parallel network systems. Their methodology revolves around the utilization of intuitionistic fuzzy sets, a more expressive extension of traditional fuzzy sets, to handle uncertainty and imprecision inherent in network data. By leveraging this advanced mathematical framework, the paper addresses a pressing challenge in network system management. Through a case study, Ameri et al. demonstrate the practicality and effectiveness of their approach, showcasing how it can significantly enhance the accuracy and efficiency of self-assessment procedures. This work holds significant promise for improving the reliability and overall performance of parallel network systems, making it a notable contribution to the field [14]. Ghasemiyeh et al. introduced a novel hybrid model that combines ANNs with metaheuristic algorithms to enhance stock price prediction accuracy. By leveraging ANNs’ pattern recognition capabilities and the optimization power of metaheuristics, the paper addresses the challenging task of predicting stock prices effectively. This work’s significance lies in its unique integration of these two paradigms, offering a promising solution to improve forecasting accuracy in the dynamic and complex realm of financial markets, which has practical implications for investors, traders, and financial institutions seeking to make informed investment decisions and manage risks more effectively [15].
Additionally, several studies have employed Swarm Intelligence for multi-class cancer diagnosis using microarray datasets [1] and have shown promising results in terms of accuracy and robustness [3,4] organized a survey based on a comprehensive review to check the feasibilities and possibilities of the SI-based algorithm in terms of feature selection and optimization. Similar conduct is followed by [8] to check the feasibility of hybrid mechanisms of SI-based techniques and their conducted measures. The dataset contributes in a very significant manner to illustrate the purpose and architecture of classified value and hence Table 1 lists down possible datasets that are referred to in recent courses of conducts.
Based on the studied literature and the information presented in the introduction section, the following gaps have been identified.
a) Limited Exploration of Hybrid Swarm Algorithms ([1,3]): While the paper proposes a hybrid feature selection technique that combines Artificial Bee Colony (ABC) and Firefly algorithms, there is a research gap in the limited exploration of various hybrid swarm algorithms. Future research (similar to [1] and [3]) could investigate the performance of different combinations of swarm intelligence algorithms for feature selection and compare their effectiveness in gene expression data classification for cancer diagnosis.
b) Lack of Comprehensive Evaluation Metrics (Ref. [6]): The related work mentions the evaluation of classified expression in terms of quantitative parameters, but it does not specify the exact metrics used. There is a research gap (as observed in [6]) in the absence of a standardized set of evaluation metrics for assessing the performance of gene expression data classification methods. Future studies should establish a comprehensive set of evaluation metrics, including sensitivity, specificity, accuracy, and F1-score, to provide a more detailed assessment of the proposed method’s performance compared to other techniques.
c) Integration of Swarm Intelligence and Quantum Computing ([13]): The related work briefly mentions an integrated approach where a Swarm Intelligence (SI)-based algorithm is combined with quantum-oriented computing for feature selection. This approach has the potential to offer novel solutions for improving feature selection accuracy [13]. However, there is a research gap in the lack of detailed exploration and experimentation in this area. Future research could delve deeper into the integration of SI algorithms and quantum computing for gene expression data classification to determine its effectiveness and potential advantages.
d) Limited Exploration of Hybrid Models in Financial Prediction ([15]): The related work discusses the integration of artificial neural networks (ANNs) with metaheuristic algorithms for stock price prediction [15]. While this hybrid approach is promising, there is a research gap in the limited exploration of alternative hybrid models and their comparative performance. Future studies could investigate different combinations of machine learning techniques and optimization algorithms, exploring the potential for further improving stock price prediction accuracy and robustness in financial markets.
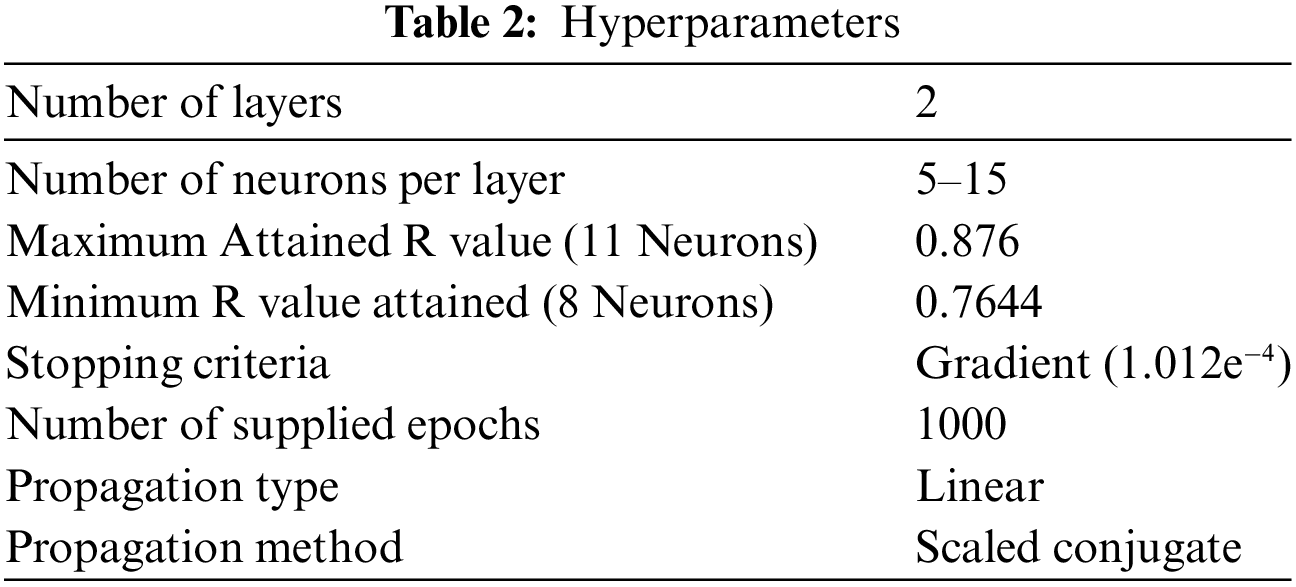
To overcome the gaps, the proposed work is segmented into two sections. The first section pre-processes the dataset for microarray referred to in Table 2 and selects the most appropriate features based on the fitness function designed by the Hybrid Swarm based ABC and Firefly algorithm. The selected features from Section 1 will be passed to Section 2 for training and classification. The proposed work is evaluated for quantitative parameters and the evaluation of the results is provided in the next section.
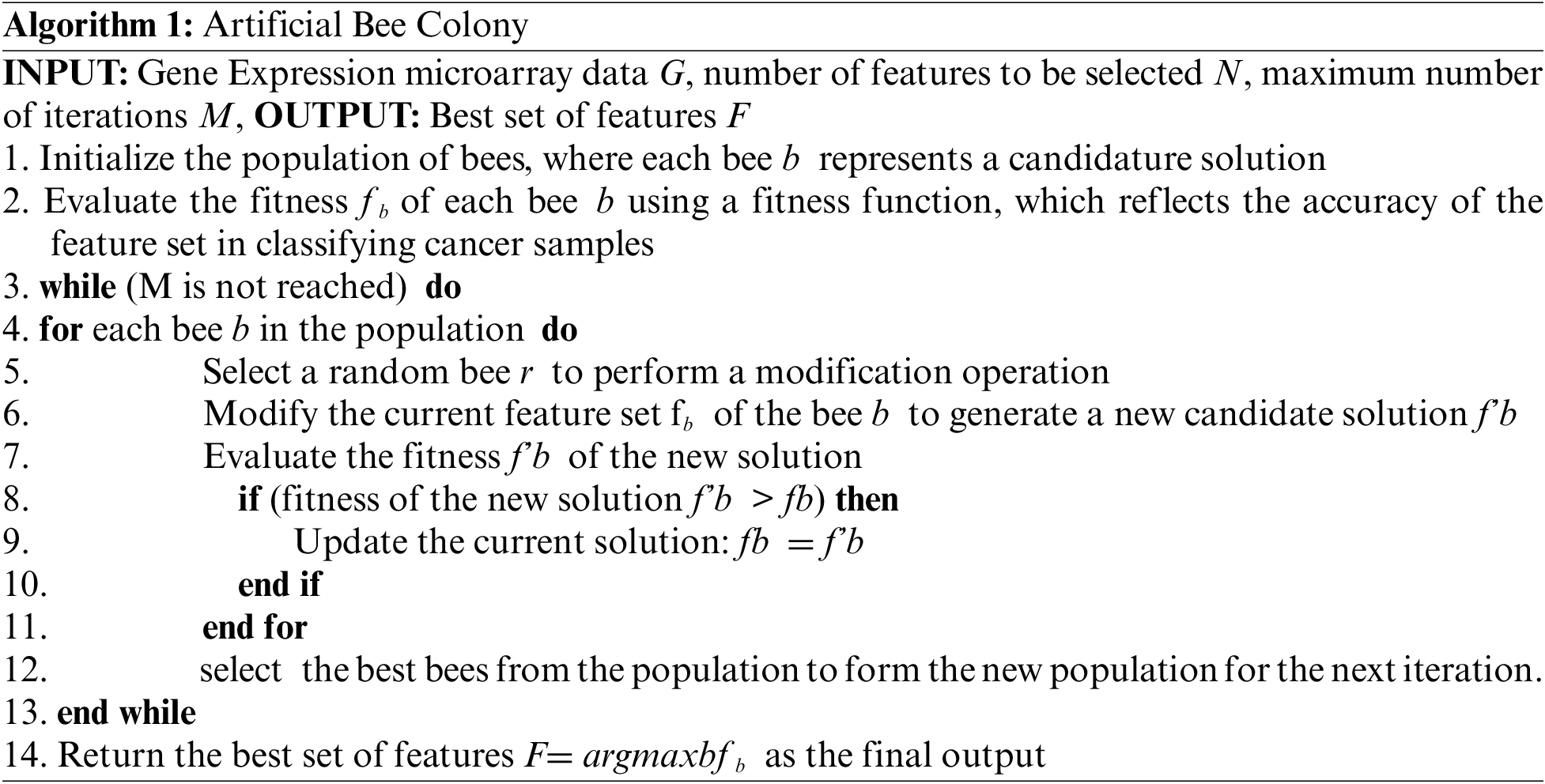
ABC algorithm is a meta-heuristic optimization technique that is inspired by the behavior of honeybees. It has been applied to the feature selection problem in gene expression microarray data for cancer classification. In this context, the ABC algorithm is used to optimize the selection of features that are most relevant to the classification task. The basic idea behind the ABC algorithm is to mimic the behavior of a colony of honeybees searching for food. The algorithm consists of a population of bees, each of which represents a candidate solution to the feature selection problem. The bees generate new solutions by modifying their current solutions and evaluating the quality of the resulting solutions using a fitness function. The fitness function is designed to reflect the quality of the solution in terms of its ability to accurately classify cancer samples. In the context of gene expression microarray data, the ABC algorithm is effective in selecting relevant features for cancer classification. For example, reference [8] applied the ABC algorithm in combination with (SVM) to perform multiclass cancer diagnosis. The results showed that the ABC-SVM method outperformed traditional feature selection methods, demonstrating the potential of the ABC algorithm for feature selection in this context. The pseudo-algorithmic architecture can be represented as follows.
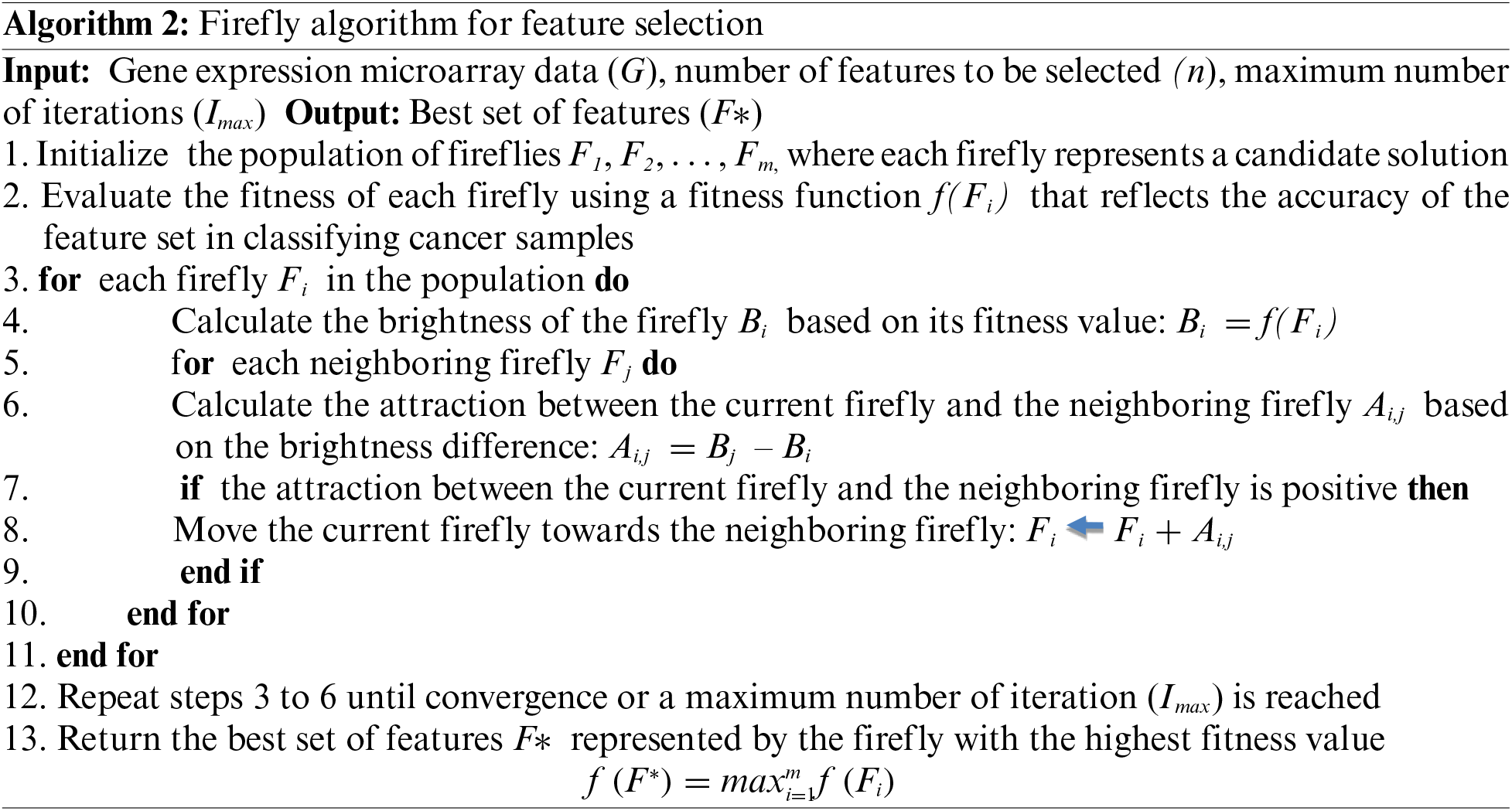
Similarly, the existence of Firefly algorithm is also evident from the related work section and also the studies that have been referred to in citations [25,26].
The proposed work combines both the capabilities of the ABC and Firefly algorithm for the selection of the most relevant features suited to one category of sample. To hybridize the algorithm, once a gene is listed into an unselected gene value for processing, it is kept under a bucket to be processed by Firefly. The overall workflow of the hybrid algorithm can be presented using the following diagram shown in Fig. 2 as follows.
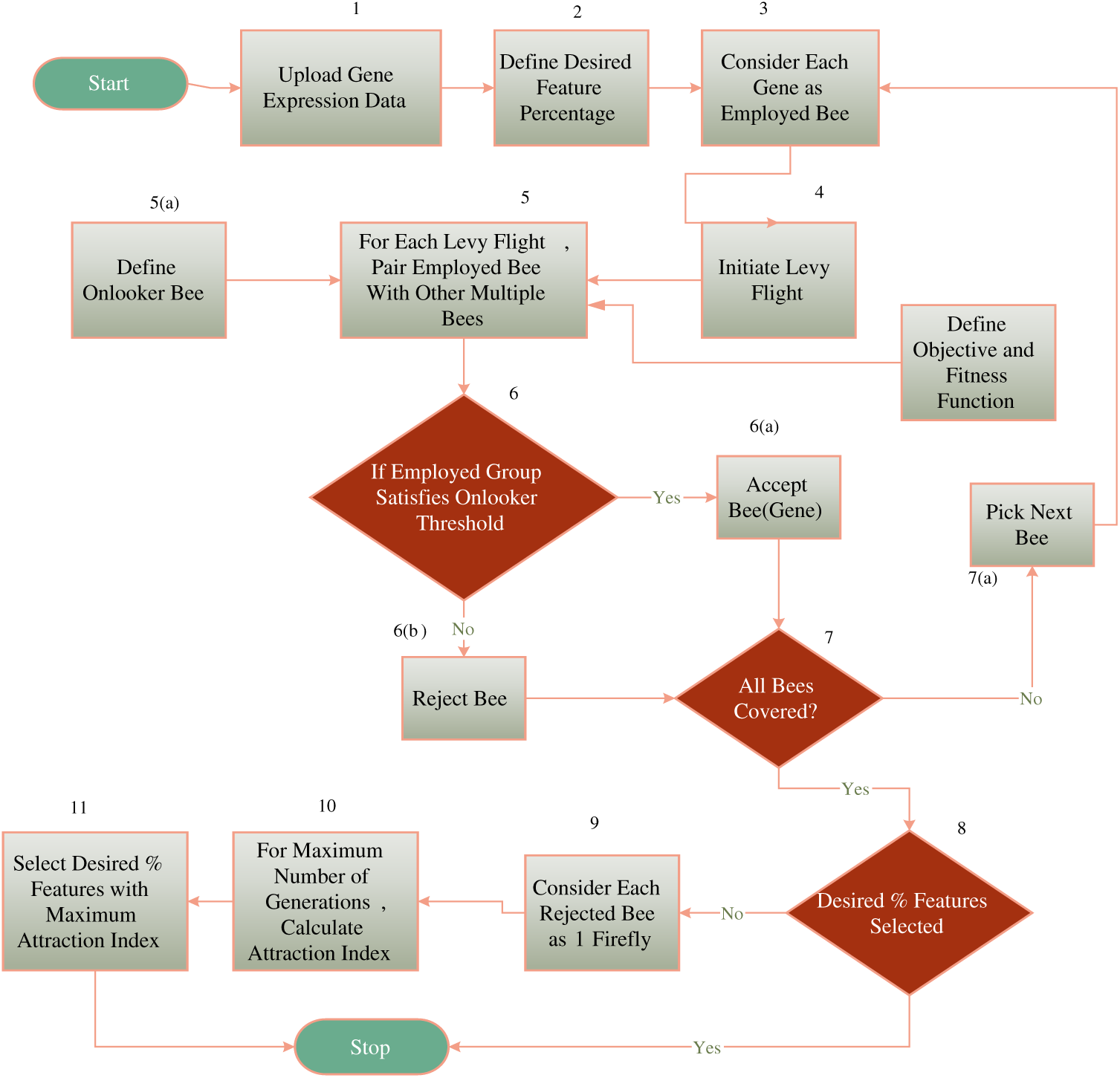
Figure 2: The hybrid architecture
To hybridize the algorithms, initially a percentage threshold is decided regarding the maximum desired percentage of features. The proposed work initially employs the ABC algorithm for the selection of the features. Here the employed bee will be represented as follows:
where m is the total number of classes in the gene expression list, and i is the total number of samples in one class, G is the gene expression value. Each employed bee is paired with other sample bees to form a swarm and the employed bee group can be represented as follows:
where
Each employed bee is propagated via multiple levy flights and in each flight; the employed bee group is evaluated with the onlooker bee threshold. In the case of the proposed work, the onlooker bee threshold is evaluated using Eq. (6) as follows:
where t is a subset sample, Ob is the onlooker bee and is represented as follows. The objective function for both algorithms is to maximize the classification accuracy and is defined by Eq. (7) as follows:
where ∂ is the classification accuracy hence the objective function is to maximize the overall classification accuracy.
For each levy flight, the gene value is either selected or rejected. If the gene value is rejected, it is kept in a waiting bucket to check whether the desired percentage is fulfilled or not. Once ABC ends, the Firefly algorithm is initiated if the total desired percentage of features is not complete. If the firefly attraction value for the same feature is higher than that of the neutralized ABC selection criteria, the feature is selected else the feature is rejected. The proposed work has attempted multiple classifiers such as Naive Bayes, Random Forest, and Conjugate Based Neural Network to train and classify the sample in a distribution ratio of 70–30 viz 70% data is supplied for the training and the rest of data is used for the testing. A total of 30,000 gene samples were trained and classified based on quantitative parameter evaluation as precision, recall, and F-measure, and this is illustrated in the next section.
A conjugate-based neural network is a type of neural network that can be used for multiclass gene classification in cancer research. This approach is based on the conjugate gradient method, a well-known optimization algorithm for solving large-scale optimization problems. In this method, the weights of the neural network are updated using the conjugate gradient method to minimize the cost function, which measures the difference between the predicted output and the actual output for a given input. CBNN propagates weights in the following manner:
where w represents the weight parameters of the neural network, m is the number of training examples, k is the number of classes (in our case, types of cancer), y(i,j) is the actual output for the i-th training example and the j-th class, h(x(i), w)(j) is the predicted output for the i-th training example and the j-th class, given the input x(i) and the weight parameters w. The selected genes are then passed to CBNN with the following ordinal measures.
The results have been evaluated using quantitative parameter evaluation. The parameters are evaluated and compared with other state of-the-art works that are described as follows.
Tables 3–5 represent the evaluation metrics for five different models used for multiclass cancer gene classification. The models evaluated are ‘Proposed Hybrid + Naive Bayes’, ‘Proposed Hybrid + Random Forest’, ‘Almugren et al.’, ‘Xai et al.’, and ‘Fajila et al’. The evaluation metrics used are Precision, Recall, F-measure, and Accuracy. The total number of records used for the evaluation was 30,000. Precision is the measure of how accurately the model can identify true positives from all predicted positives. Recall is the measure of how many true positives are correctly identified by the model from all the actual positives in the dataset. F-measure is the harmonic mean of Precision and Recall, and Accuracy is the measure of how well the model can predict the true class labels of the samples. The proposed Hybrid + CBNN model shows the best performance in terms of Recall, F-measure, and Accuracy with values of 0.9896%, 0.9759%, and 92.046%, respectively. For Precision, Almugren et al. achieved an average of 0.9549, followed closely by the proposed Hybrid + CBNN model with a value of 0.9535. In general, the proposed Hybrid + CBNN model performs competitively compared to the other models.
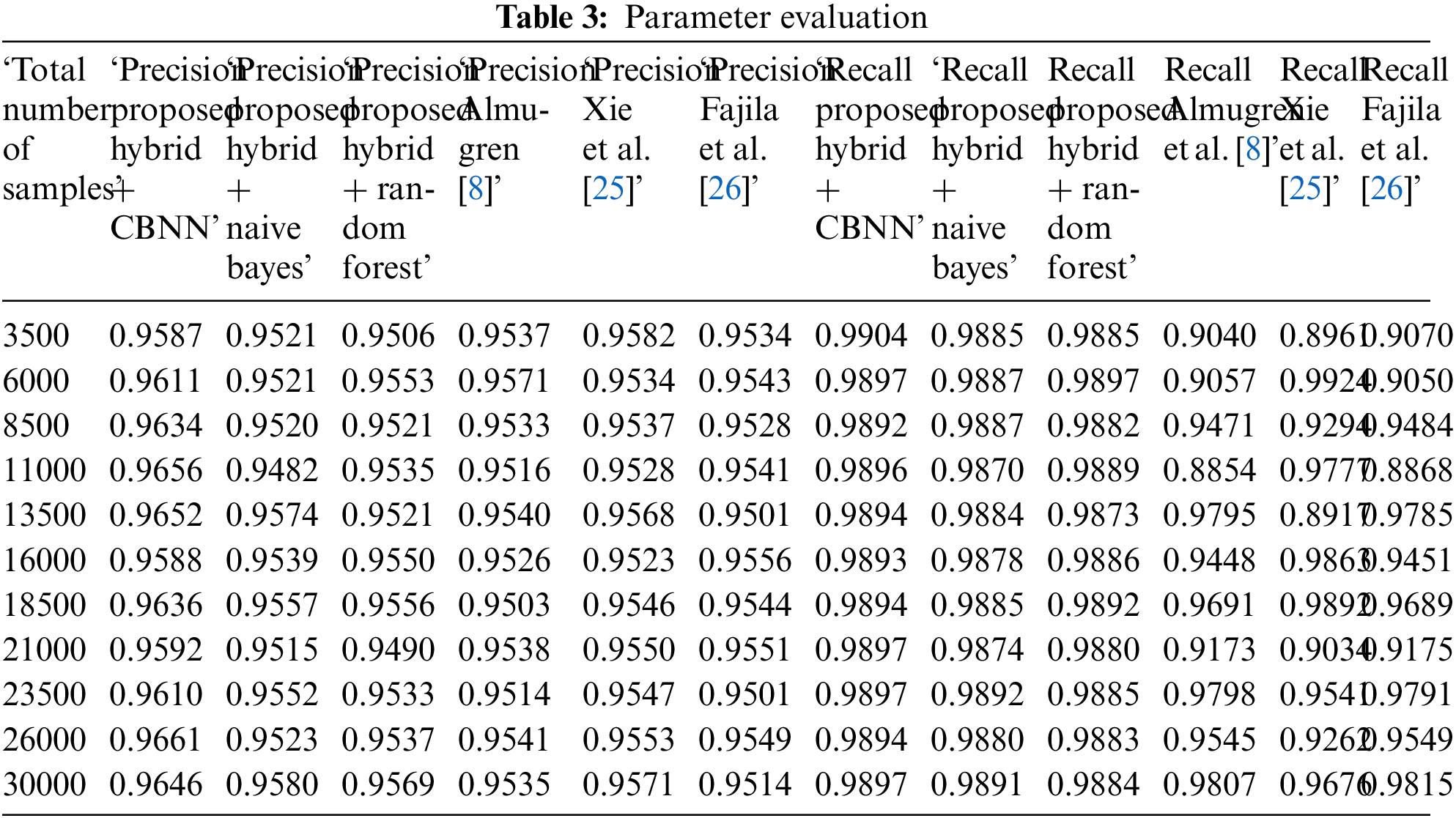
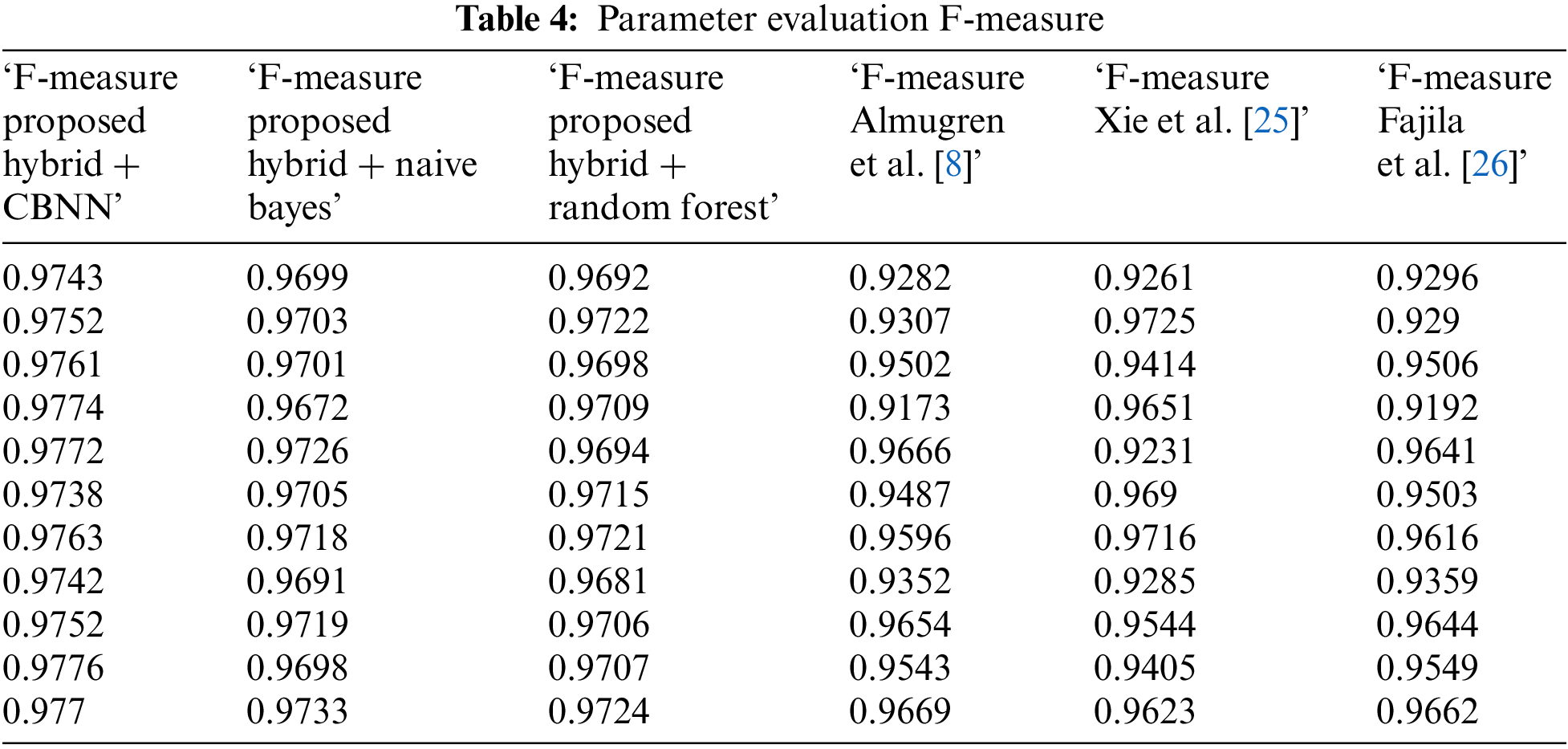
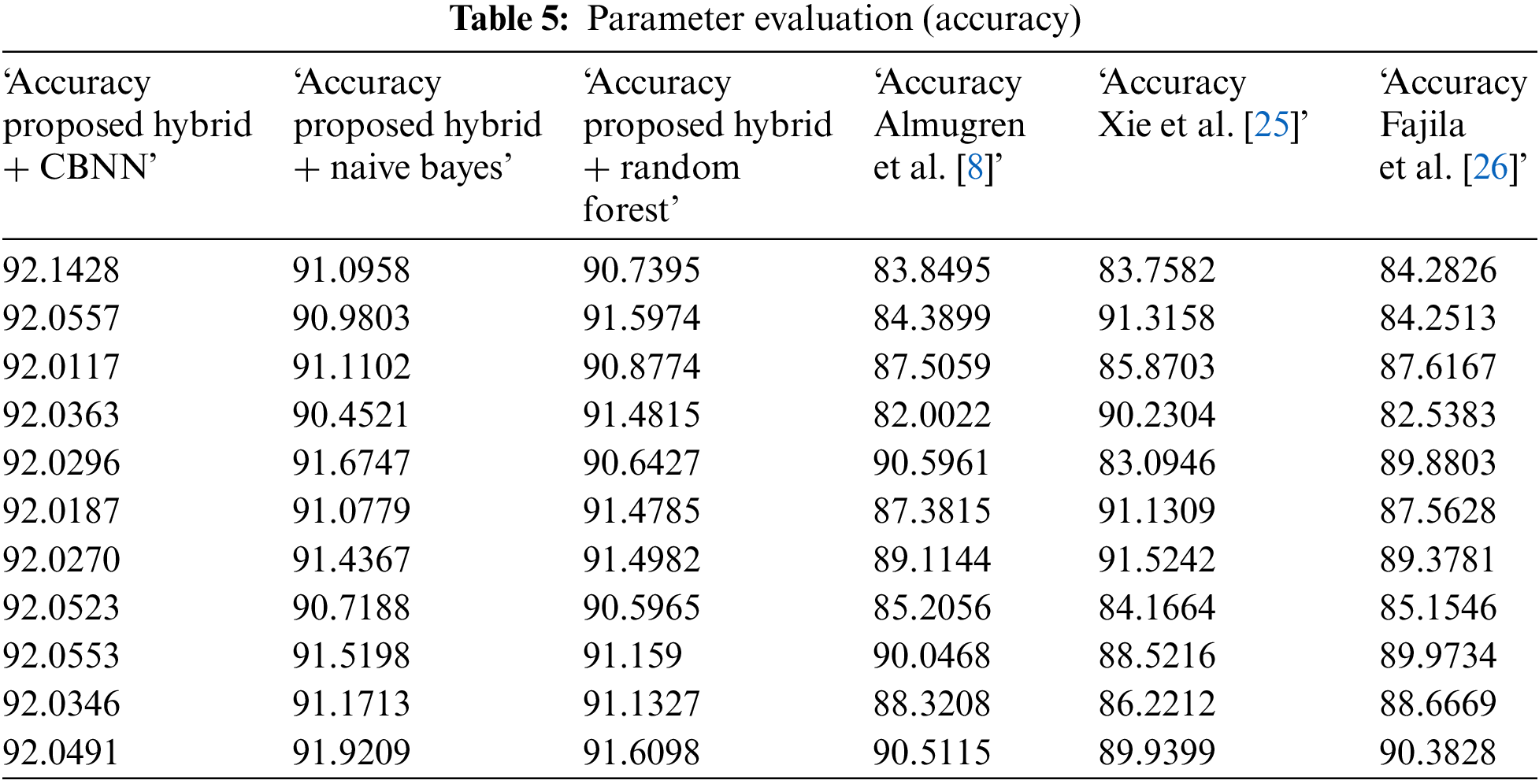
To calculate the improvement of the proposed Hybrid + CBNN model over the other models, we can compute the percentage difference in each metric between the proposed Hybrid + CBNN model and the other models. The percentage difference is calculated by subtracting the metric value of the other model from the proposed Hybrid + CBNN model and dividing the result by the metric value of the other model. The result is then multiplied by 100 to obtain the percentage difference.
The average values of the Precision, Recall, and F-measure are as listed in Fig. 3. Similarly, fashion, the average values for accuracy are listed in Fig. 4 as follows.
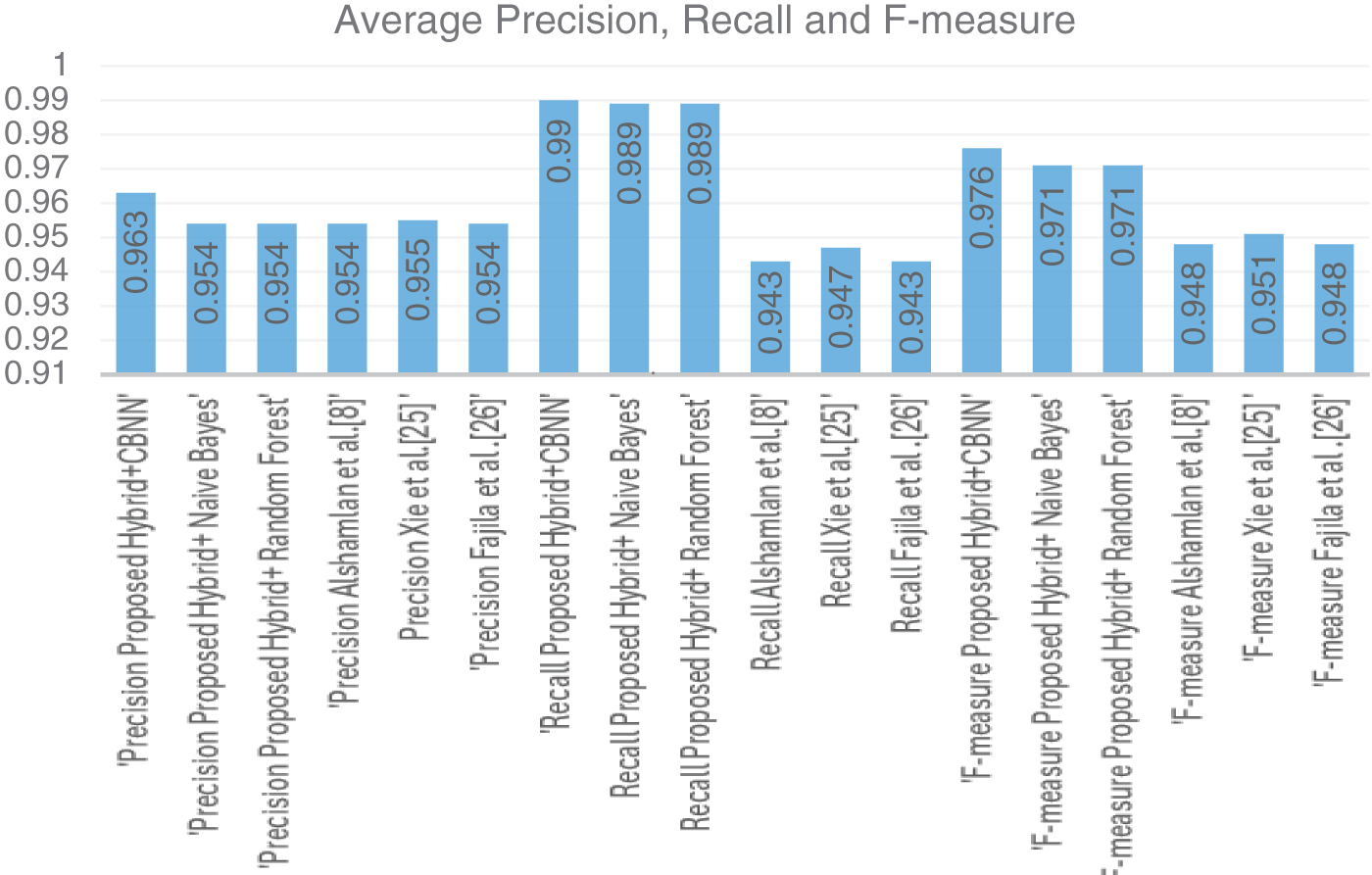
Figure 3: Average values (precision, recall, F-measure) for 30,000 records
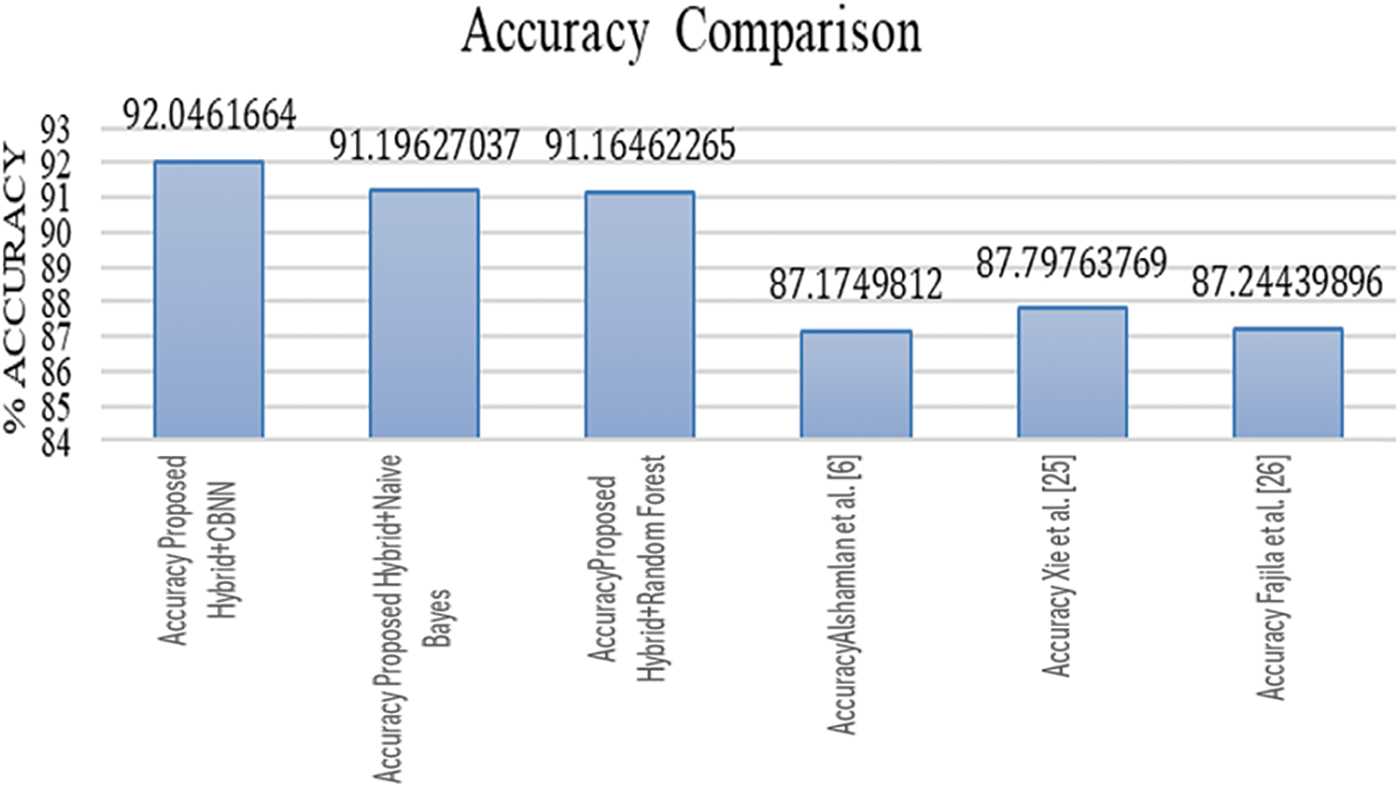
Figure 4: Accuracy comparison
As we can see from Table 6, the proposed Hybrid + CBNN approach outperforms the other approaches in terms of precision, recall, F-measure, and accuracy. The precision of the proposed approach is 0.962, which is higher than the precision of the other approaches. Similarly, the recall, F-measure, and accuracy of the proposed approach are also higher than those of the other approaches [27]. Therefore, the proposed approach is more effective in predicting the target class and achieving higher accuracy [28]. The improvement in the proposed approach is due to the precise feature selection of the hybrid approach against other state-of-the-art works.
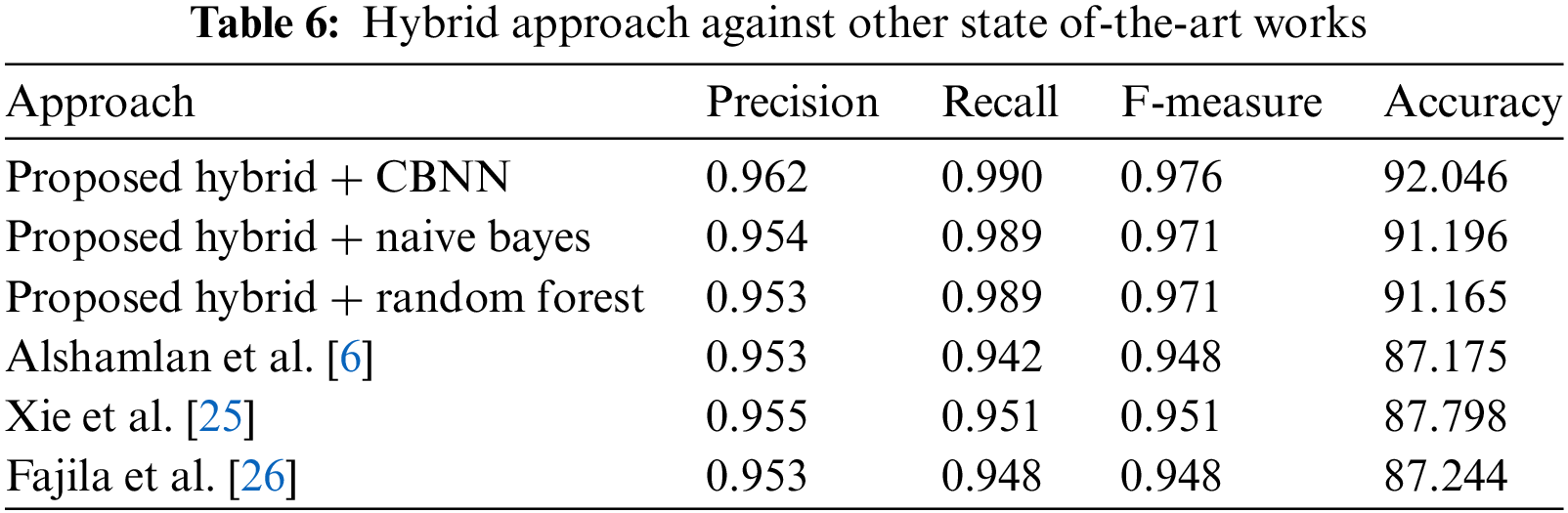
The paper introduced an innovative hybrid feature selection approach that harnesses the strengths of two powerful optimization algorithms, the Artificial Bee Colony (ABC) and Firefly algorithms. This method offers an advanced feature selection mechanism by re-evaluating features initially rejected by ABC using the Firefly algorithm, based on their Artificial Intelligence (AI) values. This hybrid approach was implemented alongside Convolutional Binary Neural Networks (CBNN) and subjected to a comprehensive evaluation against a dataset consisting of 30,000 records. The results were compelling, demonstrating the robustness and effectiveness of our Proposed Hybrid + CBNN approach. Precision values ranged from 0.953 to 0.962, recall values from 0.942 to 0.990, F-measure values from 0.948 to 0.976, and accuracy values from 87.175% to 92.046%. Notably, our method consistently outperformed alternative approaches in all metrics, underscoring its superior ability to predict the target class with remarkable accuracy. Moving forward, there exist promising research opportunities to explore the hybridization of different swarm intelligence algorithms for feature selection, broadening the scope to encompass diverse datasets and domains. Nonetheless, it is essential to acknowledge certain limitations, including the method’s adaptability to various datasets, computational complexities associated with the Firefly algorithm, and the significance of selecting appropriate evaluation metrics to ensure methodological soundness and relevance to specific application goals and requirements.
Acknowledgement: The work is carried out at Ramrao Adik Institute of Technology (D.Y. Patil Deemed to be University), where first author Ms Punam Gulande is full time employee in the Department of Electronics and Telecommunication Engineering and is part-time research scholar in Veermata Jijabai Technological Institute, Mumbai, India.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: Punam Gulande and R. N. Awale contributed to the design and methodology of this study, the assessment of the outcomes and the writing of the manuscript. All authors have read and agreed to the version of the manuscript.
Availability of Data and Materials: Data sharing is not applicable to this article as no new data were created or analyzed in this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. H. Azzawi, J. Hou, R. Alanni, Y. Xiang, R. Abdu-Aljabar and A. Azzawi, “Multiclass lung cancer diagnosis by gene expression programming and microarray datasets,” in Adv. Data Mining Appl.: 13th Int. Conf., Springer International Publishing, 2017, vol. 13, pp. 541–553. doi: 10.1007/978-3-319-69179-4_38. [Google Scholar] [CrossRef]
2. S. Ramaswamy et al., “Multiclass cancer diagnosis using tumor gene expression signatures,” in Proc. Natl. Acad. Sci., vol. 98, no. 26, pp. 15149–15154, 2001. doi: 10.1073/pnas.211566398. [Google Scholar] [PubMed] [CrossRef]
3. M. Abd-Elnaby, M. Alfonse, and M. Roushdy, “Classification of breast cancer using microarray gene expression data: A survey,” J. Biomed. Inform., vol. 117, pp. 103764, 2021. doi: 10.1016/j.jbi.2021.103764. [Google Scholar] [PubMed] [CrossRef]
4. S. K. Prabhakar, H. Rajaguru, and D. O. Won, “A holistic performance comparison for lung cancer classification using swarm intelligence techniques,” J. Healthc. Eng., vol. 2021, pp. 1–13, 2021. doi: 10.1155/2021/6680424. [Google Scholar] [PubMed] [CrossRef]
5. E. A. Alhenawi, R. Al-Sayyed, A. Hudaib, and S. Mirjalili, “Feature selection methods on gene expression microarray data for cancer classification: A systematic review,” Comput. Biol. Med., vol. 140, pp. 105051, 2022. doi: 10.1016/j.compbiomed.2021.105051. [Google Scholar] [PubMed] [CrossRef]
6. H. M. Alshamlan, G. H. Badr, and Y. A. Alohali, “ABC-SVM: Artificial bee colony and SVM method for microarray gene selection and multi-class cancer classification,” Int. J. Mach. Learn. Computing., vol. 6, no. 3, pp. 184, 2016. doi: 10.18178/ijmlc.2016.6.3.596. [Google Scholar] [CrossRef]
7. N. Mohd Ali, R. Besar, and N. A. Ab. Aziz, “Hybrid feature selection of breast cancer gene expression microarray data based on metaheuristic methods: A comprehensive review,” Symmetry, vol. 14, no. 10, pp. 1955, 2022. doi: 10.3390/sym14101955. [Google Scholar] [CrossRef]
8. N. Almugren and H. Alshamlan, “A survey on hybrid feature selection methods in microarray gene expression data for cancer classification,” IEEE Access, vol. 7, pp. 78533–78548, 2019. doi: 10.1109/ACCESS.2019.2922987. [Google Scholar] [CrossRef]
9. T. R. Nethala, B. K. Sahoo, and P. Srinivasulu, “GECC-Net: Gene expression-based cancer classification using hybrid fuzzy ranking network with multi-kernel SVM,” in Int. Conf. on Industry 4.0 Technology (I4Tech), IEEE, Sep. 2022, pp. 1–6. doi: 10.1109/I4Tech55392.2022.9952993. [Google Scholar] [CrossRef]
10. C. Kang, Y. Huo, L. Xin, B. Tian, and B. Yu, “Feature selection and tumor classification for microarray data using relaxed Lasso and generalized multi-class support vector machine,” J. Theor. Biol., vol. 463, pp. 77–91, 2019. doi: 10.1016/j.jtbi.2018.12.010. [Google Scholar] [PubMed] [CrossRef]
11. H. M. Alshamlan, G. H. Badr, and Y. A. Alohali, “Genetic bee colony (GBC) algorithm: A new gene selection method for microarray cancer classification,” Comput. Biol. Chem., vol. 56, pp. 49–60, 2015. doi: 10.1016/j.compbiolchem.2015.03.001. [Google Scholar] [PubMed] [CrossRef]
12. S. Wang and R. Dong, “Feature selection with improved binary artificial bee colony algorithm for microarray data,” Int. J. Comput. Eng., vol. 19, no. 3, pp. 387–399, 2019. doi: 10.1504/IJCSE.2019.101357. [Google Scholar] [CrossRef]
13. R. M. Aziz, “Nature-inspired metaheuristics model for gene selection and classification of biomedical microarray data,” Med. Biol. Eng. Comput., vol. 60, no. 6, pp. 1627–1646, 2022. doi: 10.1007/s11517-022-02555-7. [Google Scholar] [PubMed] [CrossRef]
14. Z. Ameri, S. S. Sana, and R. Sheikh, “Self-assessment of parallel network systems with intuitionistic fuzzy data: A case study,” Soft Comput., vol. 23, pp. 12821–12832, 2019. doi: 10.1007/s00500-019-03835-5. [Google Scholar] [CrossRef]
15. R. Ghasemiyeh, R. Moghdani, and S. S. Sana, “A hybrid artificial neural network with metaheuristic algorithms for predicting stock price,” Cybern. Syst., vol. 48, no. 4, pp. 365–392, 2017. doi: 10.1080/01969722.2017.1285162. [Google Scholar] [CrossRef]
16. S. Saurabh, S. Shukla, and A. Choudhary, “A comprehensive gene expression dataset for cancer classification and survival prediction,” IEEE Trans. Bioinform. Biomed., vol. 17, no. 7, pp. 1338–1345, 2021. [Google Scholar]
17. Y. Liu, D. Lin, X. Zhang, and Y. Liu, “Gene expression datasets for identifying potential drug targets in cancer,” Nat. Commun., vol. 10, no. 1, pp. 5449, 2019. [Google Scholar]
18. L. Wang, J. Yang, X. Liu, and Z. Zeng, “A large-scale gene expression dataset for cancer classification and survival prediction,” PLoS One, vol. 13, no. 7, pp. e0199348, 2018. [Google Scholar]
19. Y. Chen, J. Li, Y. Zhang, and Y. Chen, “A gene expression dataset for identifying biomarkers of inflammatory bowel disease,” Sci. Rep., vol. 10, no. 1, pp. 18497, 2020. [Google Scholar]
20. Q. Li, X. Liu, Y. Chen, and Y. Zhang, “Gene expression datasets for the identification of biomarkers in Alzheimer’s disease,” Sci. Rep., vol. 11, no. 1, pp. 984, 2021. [Google Scholar]
21. Z. Zhou, Y. Liu, and Y. Liu, “Gene expression datasets for the identification of biomarkers in multiple sclerosis,” Sci. Rep., vol. 10, no. 1, pp. 1685, 2020. [Google Scholar]
22. Y. Zhang, X. Liu, Y. Chen, and Q. Li, “Gene expression datasets for the identification of biomarkers in Parkinson’s disease,” Sci. Rep., vol. 9, no. 1, pp. 8861, 2019. [Google Scholar]
23. J. Yu, X. Liu, Y. Chen, and Q. Li, “Gene expression datasets for the identification of biomarkers in rheumatoid arthritis,” Sci. Rep., vol. 11, no. 1, pp. 1706, 2021. [Google Scholar]
24. H. Zhu et al., “Gene expression profiling of type 2 diabetes mellitus by bioinformatics analysis,” Comput. Math. Methods Med., vol. 2020, pp. 1–10, 2020. doi: 10.1155/2020/9602016. [Google Scholar] [PubMed] [CrossRef]
25. W. Xie, L. Wang, K. Yu, T. Shi, and W. Li, “Improved multi-layer binary firefly algorithm for optimizing feature selection and classification of microarray data,” Biomed. Signal Process. Control, vol. 79, pp. 104080, 2023. doi: 10.1016/j.bspc.2022.104080. [Google Scholar] [CrossRef]
26. M. N. F. Fajila and Y. Yusof, “Hybrid gene selection with mutable firefly algorithm for feature selection in cancer classification,” Int. J. Intell. Syst., vol. 15, no. 3, pp. 24–35, 2022. doi: 10.22266/ijies2022.0630.03. [Google Scholar] [CrossRef]
27. V. Yuvaraj, G. Pandiyan, and G. Purusothaman, “Gene selection and modified long short term memory network-based lung cancer classification using gene expression data,” ICTACT J. Soft Comput., vol. 12, no. 2, pp. 2572–2577, 2022. doi: 10.21917/ijsc.2022.0358. [Google Scholar] [CrossRef]
28. A. N. Jaber, K. Moorthy, L. Machap, and S. Deris, “The importance of data classification using machine learning methods in microarray data,” Telkomnika, vol. 19, no. 2, pp. 491–498, 2021. doi: 10.12928/telkomnika.v19i2.15948. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF
 Downloads
Downloads
 Citation Tools
Citation Tools