 Open Access
Open Access
ARTICLE
Chimp Optimization Algorithm Based Feature Selection with Machine Learning for Medical Data Classification
1 Department of Mathematics, College of Education, Al-Zahraa University for Women, Karbala, Iraq
2 Biomedical Engineering Department, College of Engineering, University of Warith Al-Anbiyaa, Karbala, Iraq
3 Department of Information Technology, College of Computer and Information Sciences, Princess Nourah bint Abdulrahman University, P.O. Box 84428, Riyadh, 11671, Saudi Arabia
4 Computer Science Department, Security Engineering Lab, Prince Sultan University, Riyadh, 11586, Saudi Arabia
5 Department of Electronics and Electrical Communications Engineering, Faculty of Electronic Engineering, Menoufia University, Menouf, 32952, Egypt
6 College of Information Technology, Imam Jaafar Al-Sadiq University, Al-Muthanna, 66002, Iraq
7 Department of Medical Instrumentation Techniques Engineering, Al-Mustaqbal University College, Hillah, 51001, Iraq
8 Computer Engineering Department, Mazaya University College, Dhi Qar, Iraq
9 College of Technical Engineering, the Islamic University, Najaf, Iraq
* Corresponding Author: Naglaa F. Soliman. Email:
Computer Systems Science and Engineering 2023, 47(3), 2791-2814. https://doi.org/10.32604/csse.2023.038762
Received 28 December 2022; Accepted 11 July 2023; Issue published 09 November 2023
Abstract
Data mining plays a crucial role in extracting meaningful knowledge from large-scale data repositories, such as data warehouses and databases. Association rule mining, a fundamental process in data mining, involves discovering correlations, patterns, and causal structures within datasets. In the healthcare domain, association rules offer valuable opportunities for building knowledge bases, enabling intelligent diagnoses, and extracting invaluable information rapidly. This paper presents a novel approach called the Machine Learning based Association Rule Mining and Classification for Healthcare Data Management System (MLARMC-HDMS). The MLARMC-HDMS technique integrates classification and association rule mining (ARM) processes. Initially, the chimp optimization algorithm-based feature selection (COAFS) technique is employed within MLARMC-HDMS to select relevant attributes. Inspired by the foraging behavior of chimpanzees, the COA algorithm mimics their search strategy for food. Subsequently, the classification process utilizes stochastic gradient descent with a multilayer perceptron (SGD-MLP) model, while the Apriori algorithm determines attribute relationships. We propose a COA-based feature selection approach for medical data classification using machine learning techniques. This approach involves selecting pertinent features from medical datasets through COA and training machine learning models using the reduced feature set. We evaluate the performance of our approach on various medical datasets employing diverse machine learning classifiers. Experimental results demonstrate that our proposed approach surpasses alternative feature selection methods, achieving higher accuracy and precision rates in medical data classification tasks. The study showcases the effectiveness and efficiency of the COA-based feature selection approach in identifying relevant features, thereby enhancing the diagnosis and treatment of various diseases. To provide further validation, we conduct detailed experiments on a benchmark medical dataset, revealing the superiority of the MLARMC-HDMS model over other methods, with a maximum accuracy of 99.75%. Therefore, this research contributes to the advancement of feature selection techniques in medical data classification and highlights the potential for improving healthcare outcomes through accurate and efficient data analysis. The presented MLARMC-HDMS framework and COA-based feature selection approach offer valuable insights for researchers and practitioners working in the field of healthcare data mining and machine learning.Keywords
Various healthcare governments broadly utilize electronic health records (EHRs) to enhance patient care and improve the efficacy of healthcare delivery. In complicated medical atmospheres, the EHR structure quickens clinicians’ workflow by systematizing the data management procedure [1]. When used efficiently, such EHRs enable several routine healthcare tasks and even aid in precisely detecting illnesses. A person’s accessibility to their health reports can be enabled by EHRs [2]. Moreover, they come with a home health monitoring mechanism that enables victims to measure and evaluate their signs daily [3]. The data distribution from the EMR mechanism can be serious about improving the superiority of medical research. Authors utilize this data for performing an extensive range of errands indulging data mining, like classification (estimation of diabetic presence), query responding, clustering (risk identification), and statistical tests (diabetes association and body mass index) [4]. Along with enhancing the objectified human services to affected persons, researchers in health care were anticipated to benefit from incorporating electronic health records (EHRs) and information.
After years of practice and research, the data mining method has captivated several disciplines’ outcomes and designed an exclusive study branch [5]. Certainly, the application and research of data mining have become difficult. Data mining covers concept presentation, gradual application, concept acceptance, extensive research and exploration, and mass implementing phases such as advancing other novel technologies [6]. Many researchers trust that data mining research is in the phase of comprehensive research and exploration from the present condition. Accordingly, the concept of data mining was broadly adopted. In theory, numerous prospective and challenging queries were asked that attracted many researchers [7]. As the idea of data mining came in the 1980s, its monetary value appeared, and it was supported by several commercial producers, establishing an initial market.
The association rule discovers the relation among substances not found by conventional statistical methods and artificial intelligence (AI) [8]. It has a significant study value. Simultaneously, it fulfills people’s crucial necessity to obtain knowledge from large data storage. Now, the research organizations of the popular varsities and the research departments of major IT companies have devoted higher power to their research and attained several research outcomes [9]. It involves several advanced mining methods. Users no need to have progressive statistical training and knowledge use it to dig out, which includes sequential paradigms, classification, and several kinds of knowledge [10]. The system could function in several stages, and numerous database systems (like Oracle and SQL-Server) were closely integrated.
Healthcare data mining and machine learning techniques have gained significant attention in recent years due to their potential to extract valuable insights from large-scale medical datasets. However, one of the key challenges in medical data analysis is the selection of relevant features that contribute to accurate classification and diagnosis [3]. Feature selection plays a vital role in reducing dimensionality, improving computational efficiency, and enhancing the interpretability of machine learning models. In the context of medical data classification, the identification of pertinent features can significantly impact the diagnosis and treatment of various diseases. Therefore, developing effective feature selection techniques is crucial for improving healthcare outcomes and facilitating intelligent decision-making processes [7].
In the field of healthcare data mining and machine learning, feature selection plays a critical role in improving the accuracy and efficiency of medical data classification models. However, existing feature selection methods often face limitations when applied to healthcare datasets, which are characterized by a large number of features and complex relationships among them. Therefore, there is a need for an effective and efficient feature selection approach specifically designed for medical data classification, addressing the challenges posed by the unique characteristics of healthcare datasets. Therefore, the problem addressed in this work is the efficient and effective selection of relevant features for medical data classification. In the healthcare domain, datasets often contain a large number of features, making it challenging to identify the most informative ones for accurate classification. Existing feature selection methods in the medical domain offer a diverse range of techniques but lack a comprehensive solution that combines high-performance feature selection with machine learning algorithms tailored to healthcare data.
The limitations of existing feature selection methods in the healthcare domain stem from the complexity and specificity of medical data. While these methods consider factors such as correlation among features and the relationship with the dependent variable, they often struggle to handle the large feature sets and fail to provide optimal feature subsets for classification tasks. Moreover, some existing methods may not fully exploit the inherent characteristics of medical data or fail to consider the potential impact of irrelevant or redundant features on classification accuracy.
While previous research studies have explored different approaches for feature selection in medical data classification, there is still a need for novel techniques that can address the specific challenges in this domain. Our research focuses on addressing this gap by proposing a novel approach called the Machine Learning based Association Rule Mining and Classification for Healthcare Data Management System (MLARMC-HDMS). The MLARMC-HDMS technique integrates classification and association rule mining (ARM) processes to achieve accurate and efficient medical data analysis. By leveraging the strengths of both classification and association rule mining, our approach aims to enhance the identification of relevant features and improve the overall performance of medical data classification models. Therefore, in this study, we develop a MLARMC-HDMS framework. The presented MLARMC-HDMS technique performs both classification and ARM processes. Initially, the MLARMC-HDMS technique employs the chimp optimization algorithm-based feature selection (COAFS) [10] technique for attribute selection. Next, stochastic gradient descent with a multilayer perceptron (SGD-MLP) model is applied for the classification process. Moreover, the Apriori algorithm is used to determine the relationship between the attributes. To illustrate the enhanced performance of the presented MLARMC-HDMS model, a detailed experimental validation is performed on a benchmark medical dataset.
So, we recognize the significance of the problem and the need for a unique and innovative approach. Our work goes beyond automation in healthcare data management and extends to the development of a feature selection technique that combines the power of the COA and machine learning methods. The COA algorithm, inspired by the foraging behavior of chimpanzees, mimics their search strategy for food to select the most relevant features from medical datasets. The selected features are then used to train machine learning models, specifically employing SGD-MLP model for classification. Additionally, the Apriori algorithm is utilized to determine attribute relationships within the dataset. This combination of techniques offers a novel and effective approach to feature selection in medical data classification, which has not been explored extensively in prior literature.
The key contributions are given as follows:
• Introducing a novel feature selection technique called the chimp optimization algorithm-based feature selection (COAFS) technique. Inspired by the foraging behavior of chimpanzees, this technique emulates their search strategy for food to select relevant attributes from medical datasets. The COAFS technique offers a unique and effective approach to feature subset selection, which contributes to the advancement of feature selection techniques in medical data classification.
• Employing the stochastic gradient descent with a multilayer perceptron (SGD-MLP) model for the medical data classification process. By utilizing the SGD-MLP model, we improve the accuracy and precision rates in medical data classification tasks. This application of the SGD-MLP model demonstrates its effectiveness and suitability for medical data analysis, particularly in combination with the COAFS technique.
• Utilizing the Apriori algorithm to compute attribute relationships within the medical dataset. By leveraging the Apriori algorithm, we are able to identify significant associations and patterns among the attributes, which further enhances the interpretability and understanding of the dataset.
• Proposing a novel feature selection approach using the COA for medical data classification. Inspired by the foraging behavior of chimpanzees, the COA algorithm mimics their search strategy for food, allowing it to effectively identify relevant attributes from medical datasets.
• The proposed MLARMC-HDMS approach integrates classification and association rule mining processes. This integration enables the simultaneous extraction of meaningful knowledge from medical data while performing accurate classification tasks.
• Through extensive experimentation on various medical datasets, we demonstrate that our proposed approach surpasses alternative feature selection methods. It achieves higher accuracy and precision rates in medical data classification tasks, showcasing the effectiveness and efficiency of the COA-based feature selection approach.
• Our research contributes to the advancement of feature selection techniques specifically tailored for medical data classification. By identifying relevant features through COA and training machine learning models using the reduced feature set, our approach enhances the diagnosis and treatment of various diseases, ultimately improving healthcare outcomes.
• Conducting detailed experiments on a benchmark medical dataset. The results reveal the superiority of the MLARMC-HDMS model over other methods, with a maximum accuracy of 99.75%. This highlights the practical significance and reliability of our proposed approach.
• Presenting MLARMC-HDMS framework and COA-based feature selection approach offer valuable insights for researchers and practitioners working in the field of healthcare data mining and machine learning. By integrating classification, association rule mining, and COA, our work provides a comprehensive methodology that can guide future research and practical implementations in healthcare data analysis.
Accordingly, the motivation behind this study is to address the challenge of identifying relevant features in large medical datasets. Medical datasets often contain a large number of features, which can lead to a curse of dimensionality, resulting in a decrease in classification accuracy and an increase in computational time. Therefore, feature selection is a crucial step in medical data analysis, which aims to reduce the number of features while retaining the most informative ones. Traditional feature selection methods, such as correlation-based and filter-based methods, have limitations in terms of accuracy and computational efficiency. Therefore, the study aims to propose a novel approach based on the Chimp Optimization Algorithm (COA), which is inspired by the search behavior of chimpanzees in the wild. The COA-based feature selection approach is expected to provide a more effective and efficient way to select relevant features from large medical datasets, leading to improved accuracy and precision rates in medical data classification tasks. The ultimate goal of the study is to contribute to the development of more accurate and efficient medical decision-making systems.
Consequently, our research proposes a novel approach called the COA-based feature selection for medical data classification using machine learning techniques. This approach aims to address the limitations of existing methods by introducing a tailored feature selection technique inspired by the foraging behavior of chimpanzees. The COA algorithm mimics the search strategy of chimpanzees for food, which is known for its efficiency and effectiveness in finding valuable resources. By incorporating this behavior-inspired algorithm into feature selection, we leverage its ability to explore and identify relevant features from medical datasets more efficiently.
Additionally, our proposed approach integrates the COA-based feature selection with machine learning classifiers, specifically utilizing stochastic gradient descent with a multilayer perceptron (SGD-MLP) model for classification tasks. This combination ensures that the selected features contribute to improved accuracy and precision rates in medical data classification. By selecting pertinent features using the COA algorithm and training machine learning models on the reduced feature set, our approach aims to enhance the diagnosis and treatment of various diseases. Through extensive experimental evaluations on diverse medical datasets, we demonstrate the superiority of our proposed approach over alternative feature selection methods, achieving higher accuracy rates and showcasing its effectiveness and efficiency in identifying relevant features. In summary, our proposed approach addresses the limitations of existing feature selection methods in the healthcare domain by introducing the COA algorithm as a tailored solution. By leveraging the unique foraging behavior-inspired search strategy of chimpanzees, our approach improves feature selection efficiency and accuracy, ultimately enhancing medical data classification and contributing to improved healthcare outcomes.
The authors in [11] focused on different proteins directly associated with different cancer diseases. They have organized PPI databases among cancer-related proteins with the remaining human protein. Also, they have combined the direction of each interaction and the annotation kind. Consequently, a bi-clustering-based ARM algorithm is employed for predicting novel interaction with direction and type. The study illustrates the predictive power of the ARM approach over the conventional classification module without selecting a negative dataset. Sarno et al. [12] developed the incorporation of fuzzy multi-attribute decision-making, fuzzy AR learning, and process mining for detecting anomalies. Process mining analysis of the conformance among standard operating procedures and recorded event logs. Fuzzy multi-attribute decision-making is employed to define the anomaly rate. Eventually, fuzzy AR learning designs ARs that are applied for detecting anomalies. Agapito et al. [13] developed a software tool for ARM from invoices generated in the medical centre. Especially the tool adopts a pre-processing approach which gives cleaning, merging, summarization, and formatting of invoices. The approach might enhance the quality of a massive quantity of medical invoices, decreasing the quantity of inappropriate dataset, which make the remaining dataset appropriate for mining data through ARM.
Mittal et al. [14] presented a methodology for making it easier for laymen to analyze and predict their healthcare problems by exploiting the Twitter dataset on the social networking platform. To the extent that techniques or methodology are considered, an approach was constructed for performing the investigation on healthcare tweets with ARM to categorize the ailments. The symptom using a corpus via fuzzy set and two-step methodology for Term Document Matrix and Document Term Matrix Chiclana et al. [15] developed a novel mining approach depends on animal migration optimization (AMO), named ARM–AMO, to decrease the amount of ARs. It depends on the concept that rules are not of higher support and redundancy is removed from the dataset. Initially, the Apriori approach is employed for generating common itemsets and ARs. Next, AMO is applied to reduce the amount of ARs with a novel fitness function that integrates common rules.
Lee et al. [16] developed the methodology of knowledge extension-based post-mining interpreted through statistical cost domain knowledge and Reusable Medical Equipment (RME) ontology. The novel RME ontology is to present domain knowledge for helping interpret the statistically reinforced stronger pattern of pre-mined decision rule. This study employed the proposed model for finding the common rule pattern from the enormous quantities of decision rules of the non-profit hospital legacy databases. Useac et al. [17] developed a model which focused on mining higher-quality ARs with low and moderate frequencies. The author employs a new technology for extracting rules which combines the exponential model of additional security with reservoir sampling. The proposed model enables ARM directly, without the necessity to calculate noisy support for an enormous number of itemsets. Though several models are available in the literature, there is still needed to improve the classification performance. In addition, most work concentrates on something other than the parameter tuning process. Therefore, the SGD model is applied as a parameter optimization technique for the MLP model in this work.
It is noticed that the existing feature selection methods for medical data classification exhibit certain limitations that hinder their effectiveness in this domain. These limitations include:
• Inability to handle high-dimensional data: Many traditional feature selection methods struggle to handle the high dimensionality of healthcare datasets, leading to decreased performance and increased computational complexity.
• Lack of consideration for complex feature relationships: Healthcare datasets often contain intricate relationships among features, such as nonlinear dependencies and interactions. However, conventional feature selection methods may overlook these relationships, resulting in suboptimal feature subsets and reduced classification accuracy.
• Insufficient adaptability to healthcare domain-specific requirements: Generic feature selection techniques may not consider the unique characteristics and requirements of healthcare data, such as the need for interpretable and clinically meaningful features. This can lead to the inclusion of irrelevant or redundant features, hampering the accuracy and interpretability of medical data classification models.
To address the aforementioned limitations, our paper introduces a novel COA based feature selection approach for medical data classification using machine learning techniques. The proposed approach overcomes the challenges posed by healthcare datasets in the following ways:
• Handling high-dimensional data: The COA algorithm, inspired by the foraging behavior of chimpanzees, incorporates efficient search strategies for identifying relevant features in high-dimensional datasets. By leveraging the COA algorithm, our approach effectively reduces the dimensionality of healthcare datasets, thereby mitigating the computational burden associated with high-dimensional data.
• Capturing complex feature relationships: Unlike traditional methods, our proposed approach takes into account the complex relationships among features in healthcare datasets. The COA algorithm mimics the foraging behavior of chimpanzees, which inherently involves navigating and exploiting intricate relationships in their search for food. By considering these relationships, our approach is capable of identifying relevant features that contribute to accurate medical data classification.
• Adapting to healthcare domain-specific requirements: Our approach incorporates the domain-specific requirements of healthcare data classification. Through the COA-based feature selection process, we prioritize the selection of interpretable and clinically meaningful features, ensuring that the resulting feature subset is both accurate and interpretable. This facilitates the understanding and utilization of the selected features by healthcare professionals, enhancing the overall effectiveness of medical data classification.
By introducing the COA-based feature selection approach, our research addresses the limitations of existing methods and provides a tailored solution for medical data classification in the healthcare domain. The experimental results demonstrate that our proposed approach outperforms alternative feature selection methods, achieving higher accuracy and precision rates in medical data classification tasks. Thus, we believe that our proposed approach contributes significantly to the advancement of feature selection techniques in medical data classification and holds the potential to improve healthcare outcomes through accurate and efficient data analysis.
In this study, a new MLARMC-HDMS technique has been developed to perform both the classification and ARM process. Initially, the MLARMC-HDMS technique employs the COAFS technique for attribute selection. Next, the SGD-MLP model is applied to the classification process. Moreover, the Apriori algorithm is used to determine the relationship between the attributes. Fig. 1 depicts the block diagram of the MLARMC-HDMS approach.
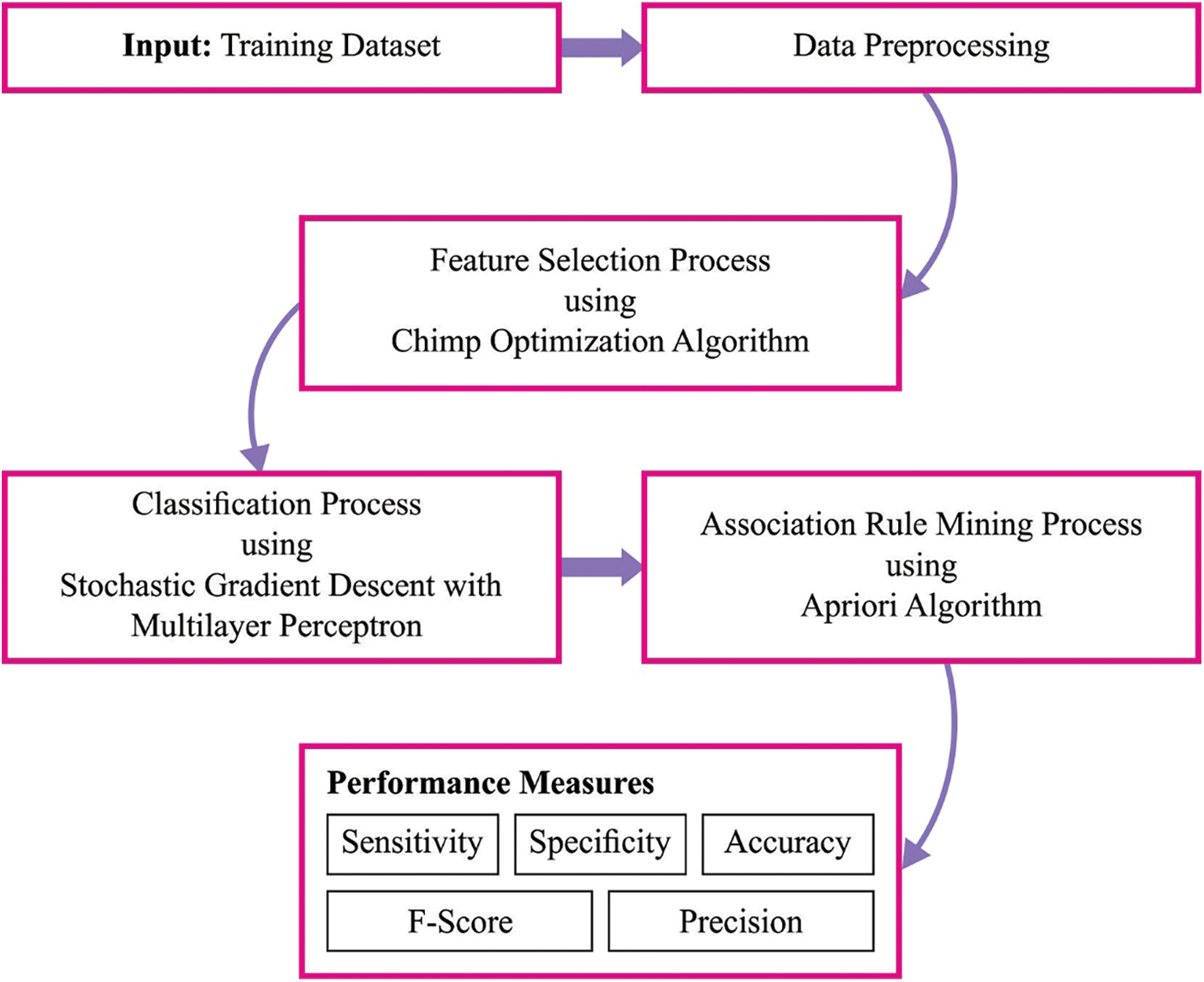
Figure 1: Block diagram of MLARMC-HDMS approach
3.1 Feature Selection Using COA
Primarily, the MLARMC-HDMS technique employs the COAFS technique for attribute selection [18]. Its instinctive background originated from the hunting behaviors of chimps. Based on the partition of labour to detect the prey, Chimps perform different actions. The typical COA algorithm splits the chimp groups into 4 kinds: barrier, attacker, driver, and chaser. Amongst themselves, the attacker is the leader of the population. The remaining three kinds of chimps aided in hunting, and the status decreased sequentially. The mathematical modeling is explained briefly. Eqs. (1) and (2) update the location of the chimp [12]:
Now, t denotes the amount of the existing iteration, and the location of chimps is upgraded based on the 4 kinds of stored location
From the expression, the coefficient f reduces non-linearly from 2.5 to
In the study, the fitness function utilized for balancing between the classifier accuracy (maximal and) the amount of selected features in all the solutions (minimal) attained with the selected feature, Eq. (10), symbolizes the fitness function to estimate the solution.
Now,
Therefore, to address the challenges of feature selection in medical data classification, we employed the COA as a feature selection technique. The COA algorithm draws inspiration from the foraging behavior of chimpanzees, mimicking their search strategy for food. COA has demonstrated promising results in solving complex optimization problems by efficiently exploring the search space and identifying relevant features [18].
3.2 Data Classification Using SGD-MLP Model
Here, the SGD-MLP model is applied to the classification process. The MLP algorithm is a multifunctional and transformative ANN encompassing single output, input, and hidden layers [19]. Generally, ANN is split into two major classes, namely data processing and transmission: feedforward and backward networks. MLP is part of FFNN. The study shows that the MLP network, with the hidden layer, is more commonly applied, as well MLP networks have higher accuracy and capability to estimate non-linear function, and it is formulated in the following equation [9]:
The weights among hidden and output layers are symbolized as
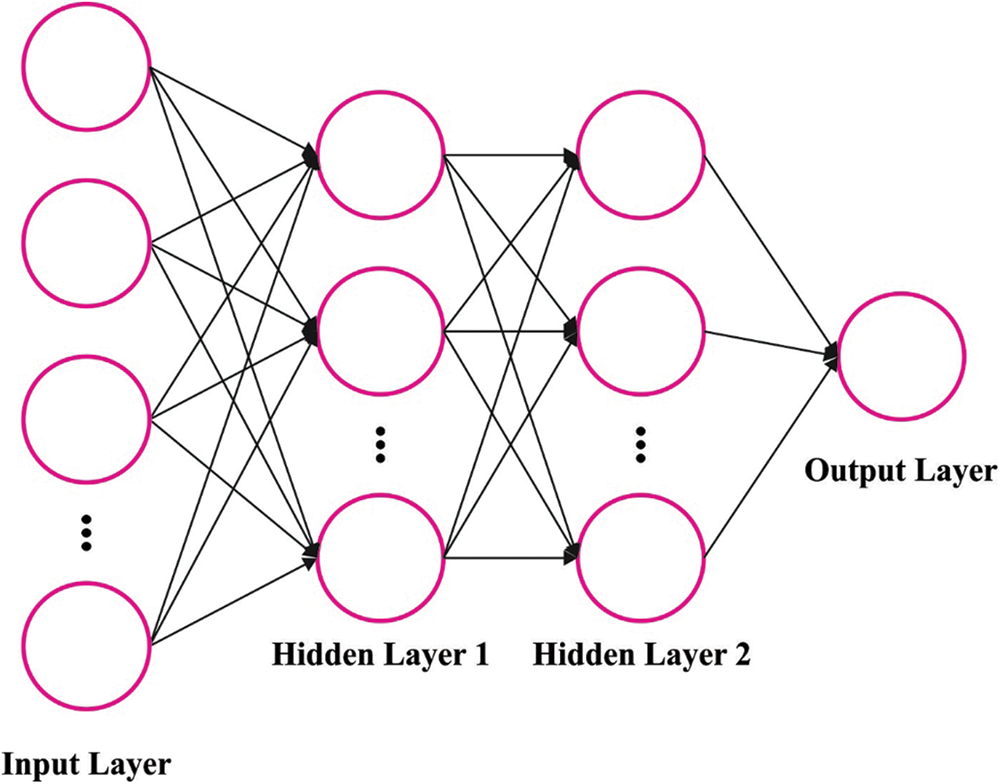
Figure 2: Structure of MLP
The MLP parameters are as follows: number of neurons: 10, number of hidden layers: 2, and learning rate: 0.04. In this study, a sigmoid (Sig) function has been utilized. The Sig transmission function for
Stochastic gradient descent (SGD) is an iterative model for improving objective functions with suitable softness properties. Since the model replaces the actual GD (evaluated from the entire dataset) using an estimate of the gradient (evaluated from an arbitrarily chosen subset of the dataset), it is regarded as a stochastic estimate of GD optimization. This technique reduces the computation weight, particularly for big data applications, which attain fast iteration for a somewhat slow convergence rate.
From the abovementioned, SGD exploits an arbitrarily chosen subset of the trained instance to evaluate the gradient of the targeted function using the following equation. The number of training instances exploited for these estimations is called the batch size. Since a smaller batch size is executed, the parameter is upgraded more recurrently than for the gradient descent, and the convergence is examined. Once the batch size is 1, the maximal repetition of the update can be performed.
Now, N indicates the batch size, C denotes the meta parameter,
Now, K indicates the iterative numerator, and
The size of the weight update is described using the user’s actual learning rate. In contrast, the adaptive learning rate is not activated and is a variance function between the objective and the evaluated values. This difference, generally called delta, exists at the output layer. With momentum, variables assist in avoiding the associated fluctuations and local minima. The momentum training, rate annealing, momentum trained parameters, and dropout rate annealing are activated while the adoptive learning rate is deactivated.
3.3 Apriori Algorithm-Based ARM
After applying the chimp optimization algorithm-based feature selection (COAFS) technique, a subset of relevant attributes is selected from the medical dataset. This subset is chosen based on their significance in contributing to accurate classification. However, even after feature selection, there may still exist relationships and interactions among the remaining attributes that could provide valuable insights into the dataset’s structure. This is where the Apriori algorithm comes into play.
The Apriori algorithm is a well-established association rule mining technique that explores itemsets and generates association rules based on the frequency of co-occurrence of attributes within the dataset. By applying the Apriori algorithm, we can identify and quantify relationships between the remaining attributes that were not selected during the feature selection step. These relationships can reveal patterns, dependencies, and associations among the attributes that may contribute to a deeper understanding of the dataset.
While the primary objective of our research is accurate medical data classification, the incorporation of the Apriori algorithm serves two important purposes. Firstly, it enhances the interpretability of the dataset by uncovering significant attribute relationships. These relationships can be valuable for domain experts in the healthcare field, as they can provide insights into potential interactions and dependencies among different medical factors.
Secondly, the obtained association rules can be utilized to refine and optimize the classification process. By incorporating the discovered attribute relationships into the classification model, we can potentially improve the accuracy and performance of the classification results. The Apriori algorithm provides a means to identify and leverage these relationships, enabling us to enhance the effectiveness of our proposed approach.
So, at last, the Apriori algorithm is used to determine the relationship between the attributes. Apriori algorithm on input data was employed for generating rules [20]. Beforehand rule generation, the correlations among attributes with the Weka tool by employing “Correlation AttributeEval” was examined. Twelve attributes were selected with the highest rank to generate strong association rules. The major objective of attribute selection is to extract useful data from input data and convert them into rules that effectively predict the class values of unclassified examples accurately and easily. There exist distinct metrics that are applied for measuring the strength of the AR. Some of these metrics are given below:
• Support: The ratio of transactions comprises a definite itemset.
• Confidence: The percentage of transactions that comprises items X and
• Lift: The proportion of support to the estimated if X and Y were independent.
• Conviction: The percentage of estimated frequency that X occurs without Y divided by the observed frequency of incorrect prediction.
• Leverage: The amount of counting attained from the co-occurrence of the consequent and antecedent of the rule from the estimated value.
The following are the respective equations for the abovementioned metrics [17]:
In summary, the Apriori algorithm complements the feature selection and classification processes by exploring and quantifying attribute relationships that were not directly considered during the feature selection step. By uncovering significant associations and patterns, the Apriori algorithm enhances the interpretability of the dataset and provides valuable insights for domain experts. Furthermore, the discovered attribute relationships can be utilized to refine and optimize the classification process, potentially leading to improved accuracy and performance in medical data classification tasks.
Thus, in our proposed approach, we integrated the classification and ARM processes to enhance knowledge discovery and interpretability in medical data classification. We initially applied the chimp optimization algorithm-based feature selection (COAFS) technique within the Machine Learning based Association Rule Mining and Classification for Healthcare Data Management System (MLARMC-HDMS) framework. This step involved the selection of pertinent features from medical datasets using COA, leveraging its ability to identify relevant attributes and reduce dimensionality.
While the Apriori algorithm is commonly used for frequent itemset mining and association rule learning over relational databases, we employed it in a different context within our proposed approach. After the classification process, we applied the Apriori algorithm to the selected features to identify any association rules that might exist among them. This additional analysis aimed to provide further insights into the relationships and patterns among the relevant features for medical data classification. The integration of association rule mining serves the purpose of enhancing interpretability and gaining additional knowledge that can aid in the feature selection process [17].
Consequently, our approach of combining feature selection techniques with association rule mining finds support in existing literature. Previous studies have highlighted the potential benefits of integrating machine learning-based classification and association rule mining processes for improved knowledge discovery in healthcare data analysis [19,20]. These works have demonstrated that association rule mining can uncover hidden patterns and relationships among relevant features, thereby enhancing the interpretability and effectiveness of feature selection techniques in medical data classification tasks. So, by incorporating the COA for feature selection and integrating classification with association rule mining, our proposed approach aims to address the limitations of existing methods and provide a comprehensive solution for medical data classification. The combination of COA and association rule mining enables the identification of relevant features and the extraction of valuable knowledge from medical datasets, ultimately enhancing the diagnosis and treatment of various diseases.
To provide a better understanding of our proposed methodology, we have included a step-by-step example using a sample dataset. In this example, we demonstrate how the combination of the COAFS technique, MLP classification, and the Apriori algorithm results in a new feature subset selection technique. Firstly, the COAFS technique is employed to select relevant attributes from the medical dataset. Inspired by the foraging behavior of chimpanzees, the COA algorithm mimics their search strategy for food. By evaluating the importance of each attribute, COAFS identifies the most pertinent features that contribute significantly to medical data classification. Once the feature selection process is completed, the next step involves training a machine learning model using the reduced feature set. In our approach, we utilize the SGD-MLP model for classification. The MLP model is a widely used neural network architecture that has demonstrated excellent performance in various classification tasks. By training the MLP model on the selected features, we aim to achieve accurate and efficient medical data classification.
Simultaneously, we apply the Apriori algorithm to the dataset to discover attribute relationships and further enhance our understanding of the data. The Apriori algorithm, a well-known ARM technique, identifies frequent itemsets and generates association rules based on their support and confidence measures. By analyzing these relationships, we gain valuable insights into the dataset, which can aid in improving the diagnosis and treatment of various diseases. By integrating the COAFS feature selection technique, MLP classification, and the Apriori algorithm, our proposed methodology offers a new feature subset selection technique for medical data classification. The COAFS technique ensures that only relevant features are selected, improving the efficiency and effectiveness of the subsequent classification process. The MLP model leverages the selected features to perform accurate medical data classification. Simultaneously, the Apriori algorithm provides additional insights into attribute relationships, enhancing the overall analysis and knowledge extraction from the dataset.
Therefore, the experimental validation of the MLARMC-HDMS model is tested on a CKD dataset from UCI (available at https://archive.ics.uci.edu/ml/datasets/chronic_kidney_disease). The dataset holds 250 infected and 150 healthy samples, as in Table 1.

The proposed model is simulated using Python 3.6.5 tool. The proposed model experiments on PC i5-8600k, GeForce 1050Ti 4 GB, 16 GB RAM, 250 GB SSD, and 1 TB HDD.
Fig. 3 reveals the confusion matrices offered by the MLARMC-HDMS model on different runs. With run-1, the MLARMC-HDMS model recognized 246 samples in the infected class and 150 samples into the healthy class. Similarly, with run-2, the MLARMC-HDMS approach has recognized 247 samples into the infected class and 146 samples into the healthy class. In the meantime, with run-3, the MLARMC-HDMS approach has recognized 248 samples into the infected class and 150 samples into the healthy class. Finally, with run-3, the MLARMC-HDMS method has recognized 247 samples into the infected class and 146 samples into the healthy class.
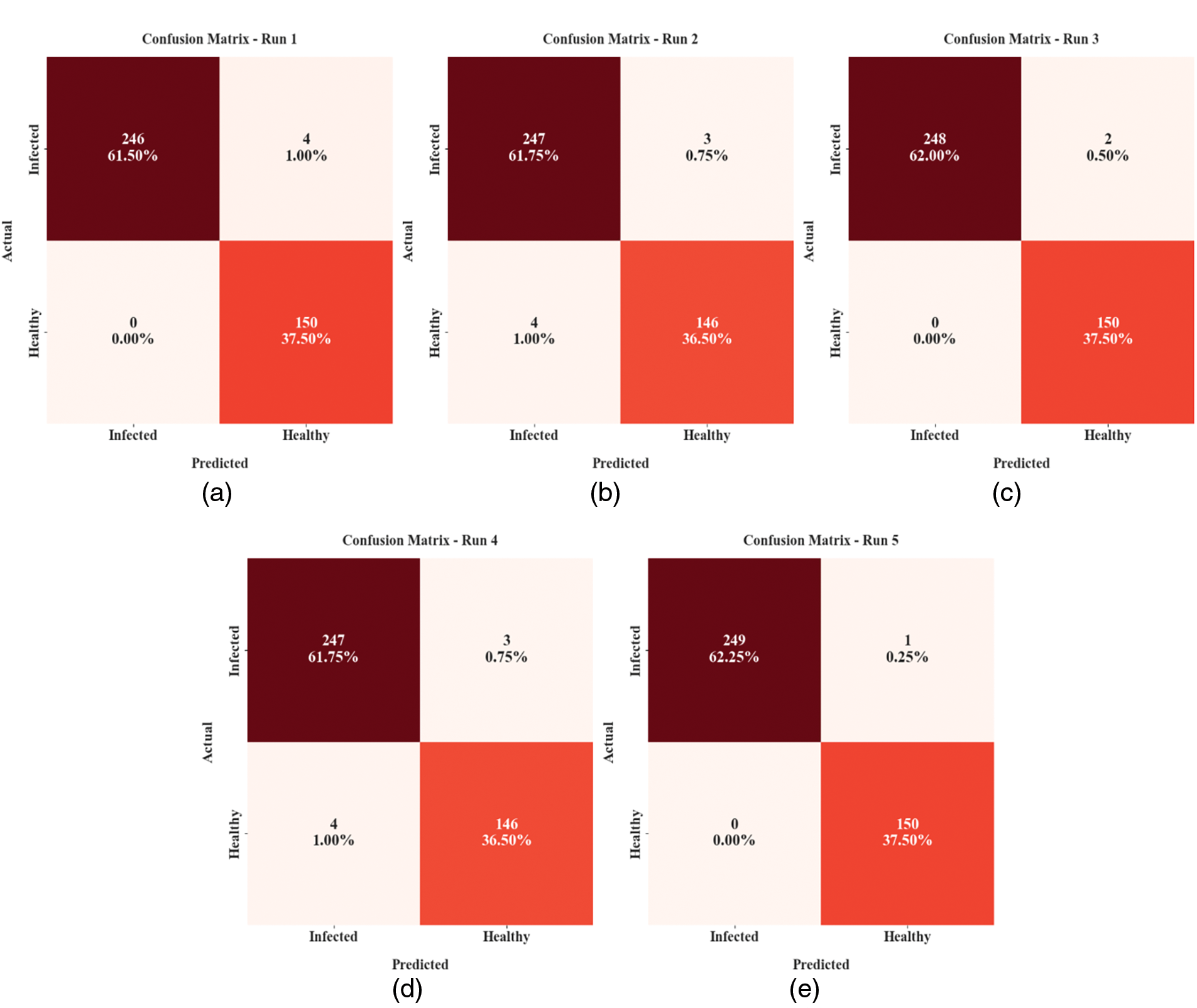
Figure 3: Confusion matrices of MLARMC-HDMS approach (a) Run-1, (b) Run-2, (c) Run-3, (d) Run-4, and (e) Run-5
Table 2 offers detailed classifier outcomes of the MLARMC-HDMS model under five runs. Fig. 4 provides quick classification results of the MLARMC-HDMS model under run-1. On the infected class, the MLARMC-HDMS model has provided
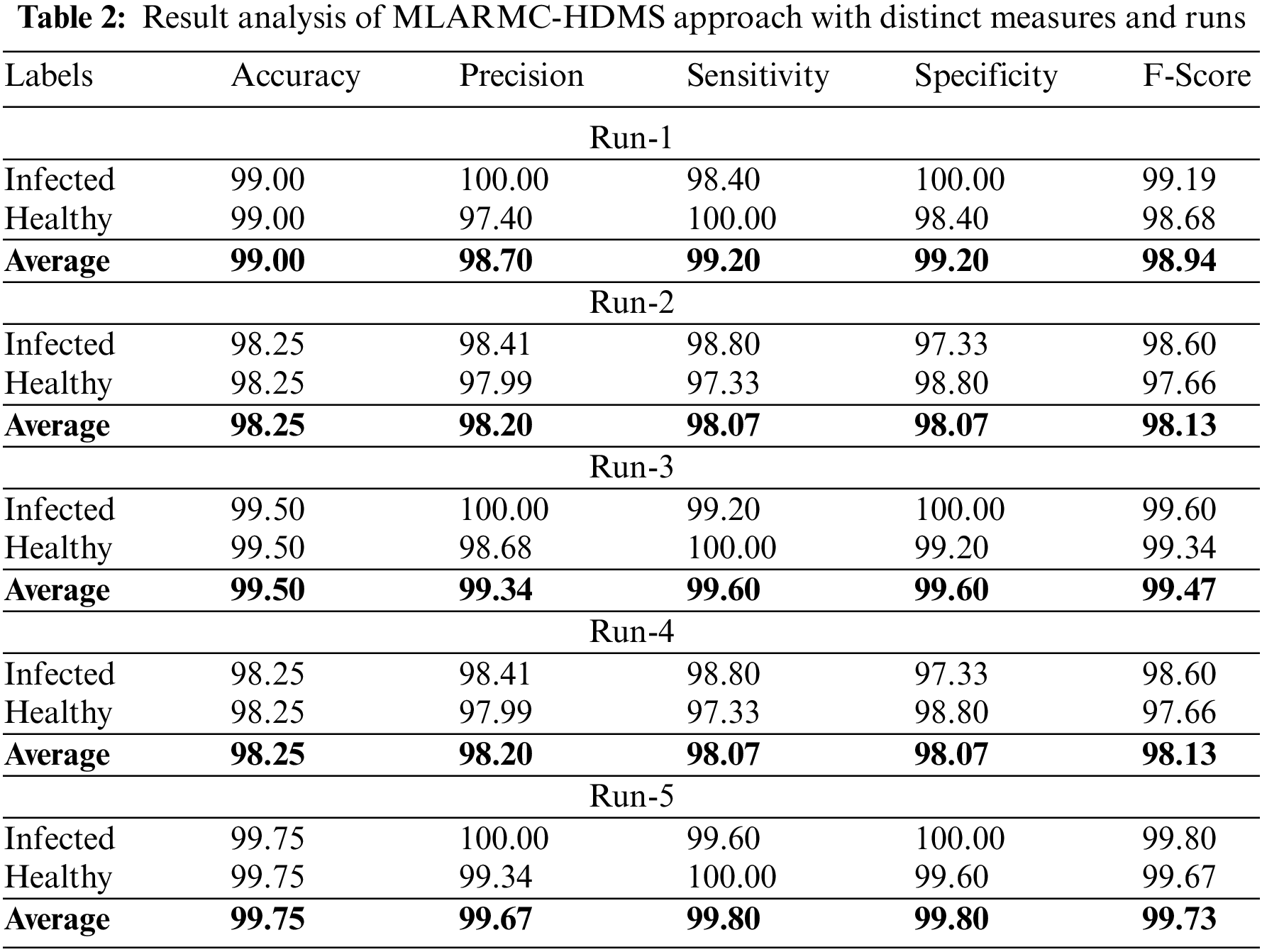
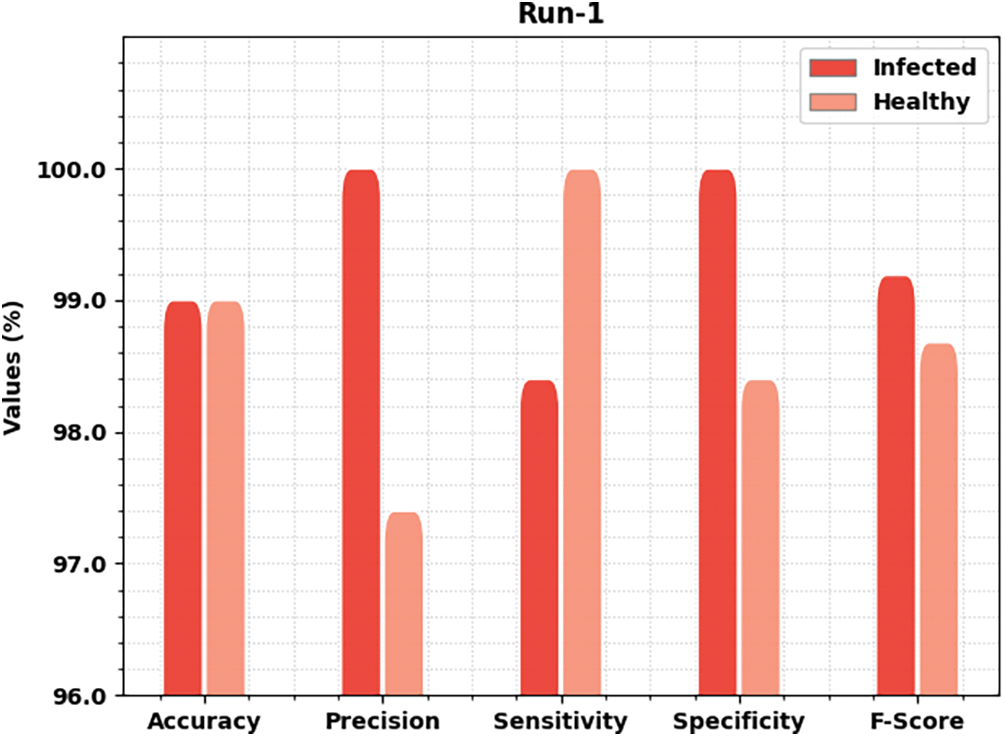
Figure 4: Result analysis of MLARMC-HDMS approach under Run-1
Fig. 5 delivers brief classification results of the MLARMC-HDMS approach under run-2. On the infected class, the MLARMC-HDMS algorithm has offered
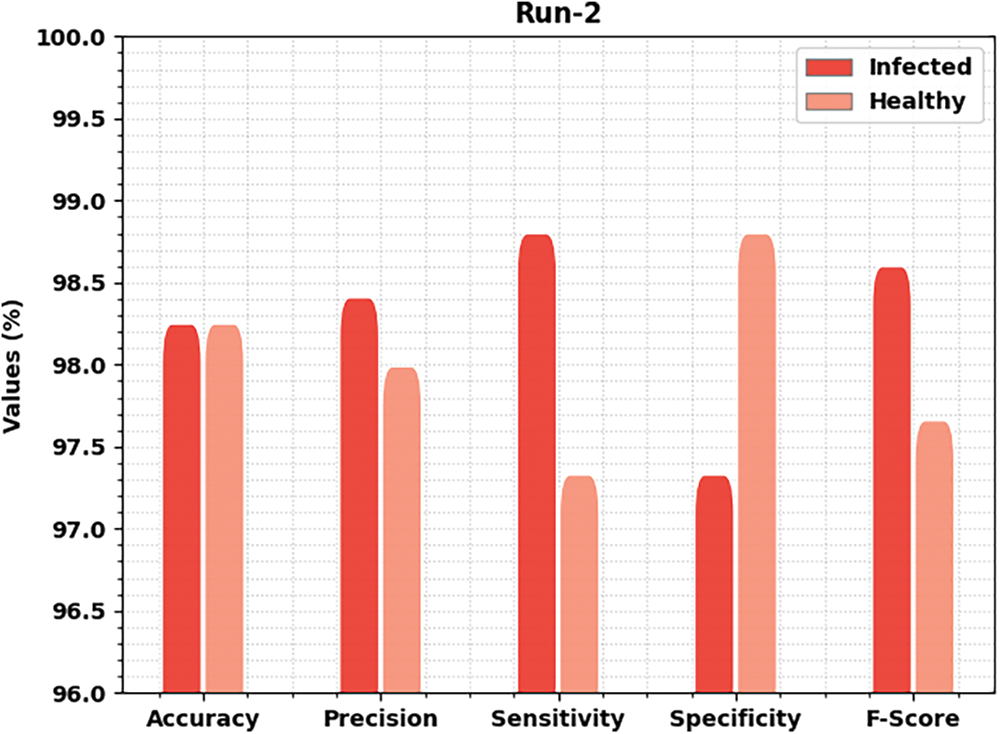
Figure 5: Result analysis of MLARMC-HDMS approach under Run-2
Fig. 6 delivers brief classification results of the MLARMC-HDMS method under run-3. On infected class, the MLARMC-HDMS approach has rendered
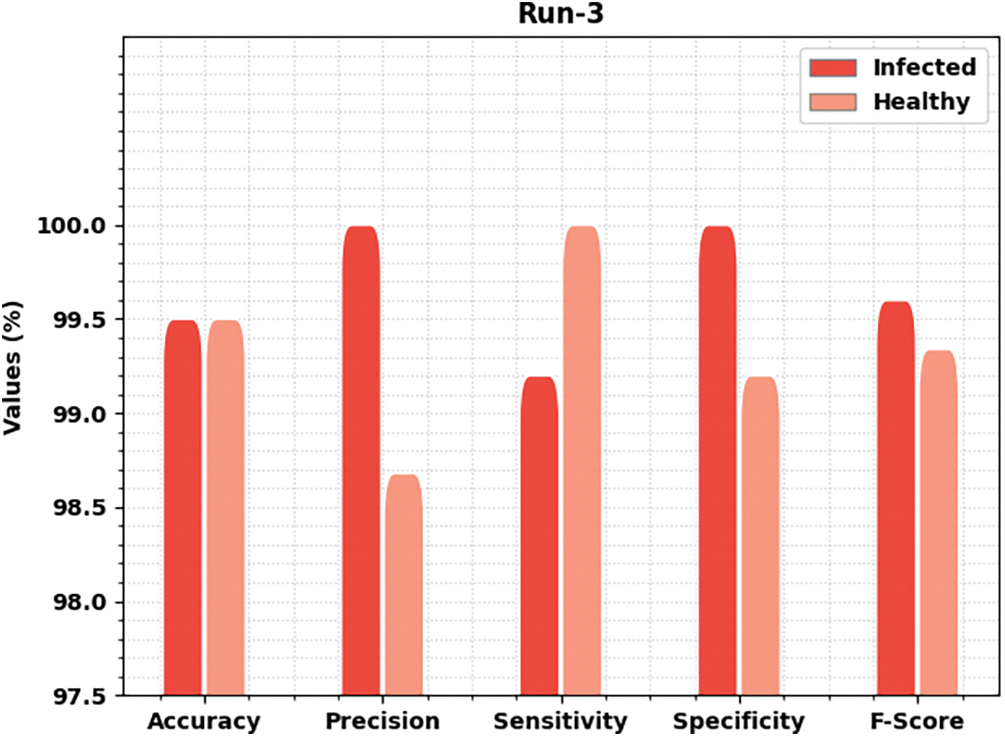
Figure 6: Result analysis of MLARMC-HDMS approach under Run-3
Fig. 7 offers brief classification results of the MLARMC-HDMS method under run-4. In the infected class, the MLARMC-HDMS approach has provided
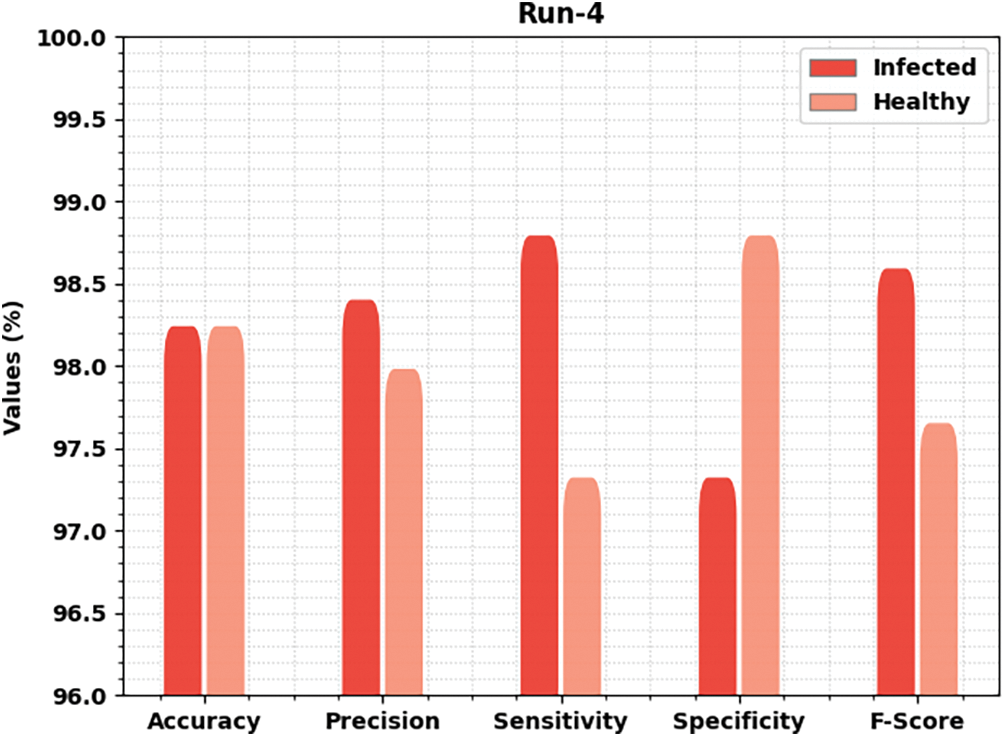
Figure 7: Result analysis of MLARMC-HDMS approach under Run-4
Fig. 8 portrays the detailed brief classification results of the MLARMC-HDMS algorithm under run-5. In the infected class, the MLARMC-HDMS method has presented

Figure 8: Result analysis of MLARMC-HDMS approach under Run-5
The training accuracy (TRA) and validation accuracy (VLA) acquired by the MLARMC-HDMS approach on the test dataset is exemplified in Fig. 9. The experimental result denotes the MLARMC-HDMS method has gained maximal values of TRA and VLA. The VLA is greater than TRA.
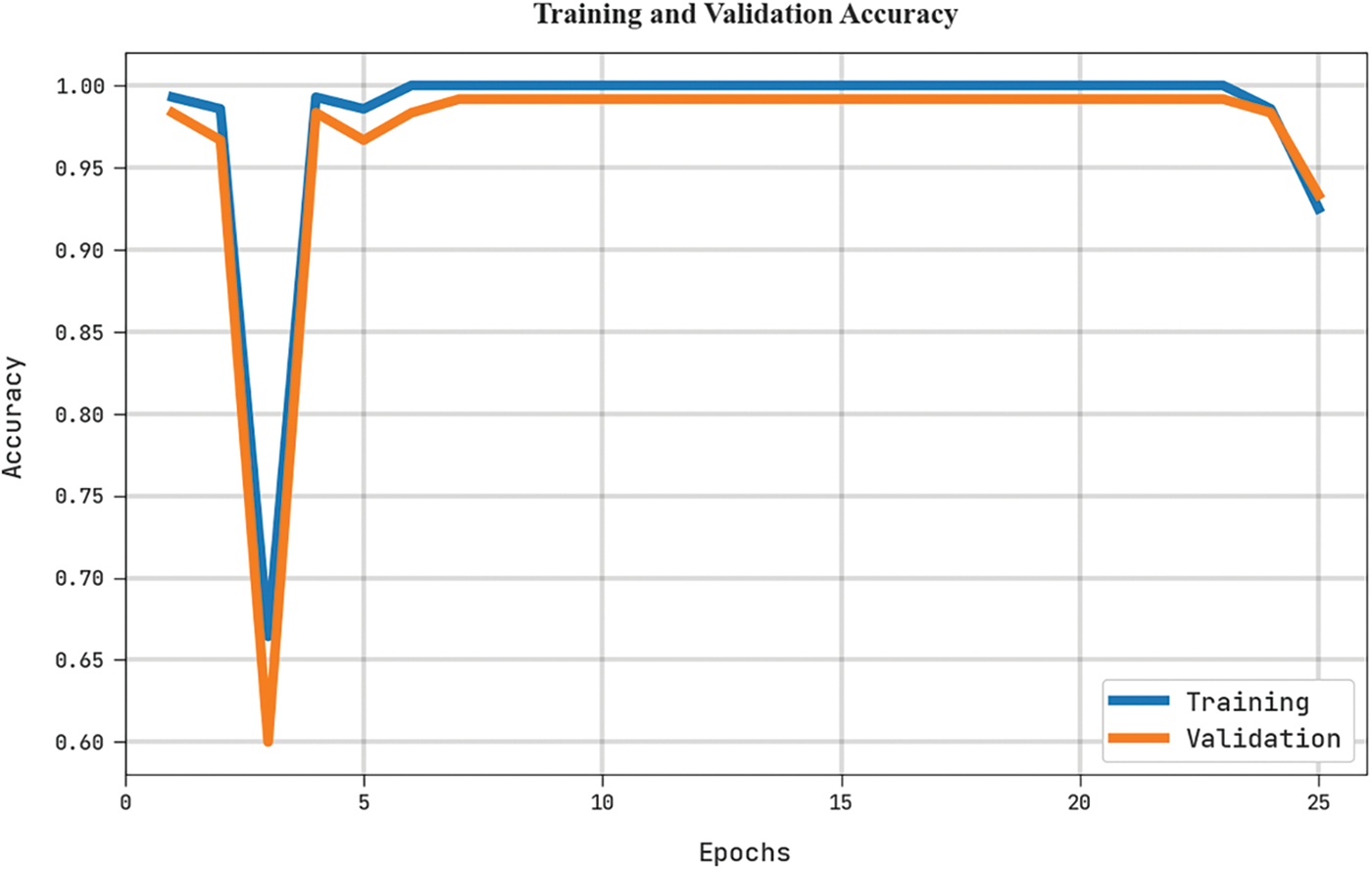
Figure 9: TRA and VLA analysis of MLARMC-HDMS approach
The training loss (TRL) and validation loss (VLL) obtained by the MLARMC-HDMS algorithm on the test dataset are shown in Fig. 10. The experimental result indicates that the MLARMC-HDMS method has established minimal values of TRL and VLL. Particularly, the VLL is lesser than TRL.
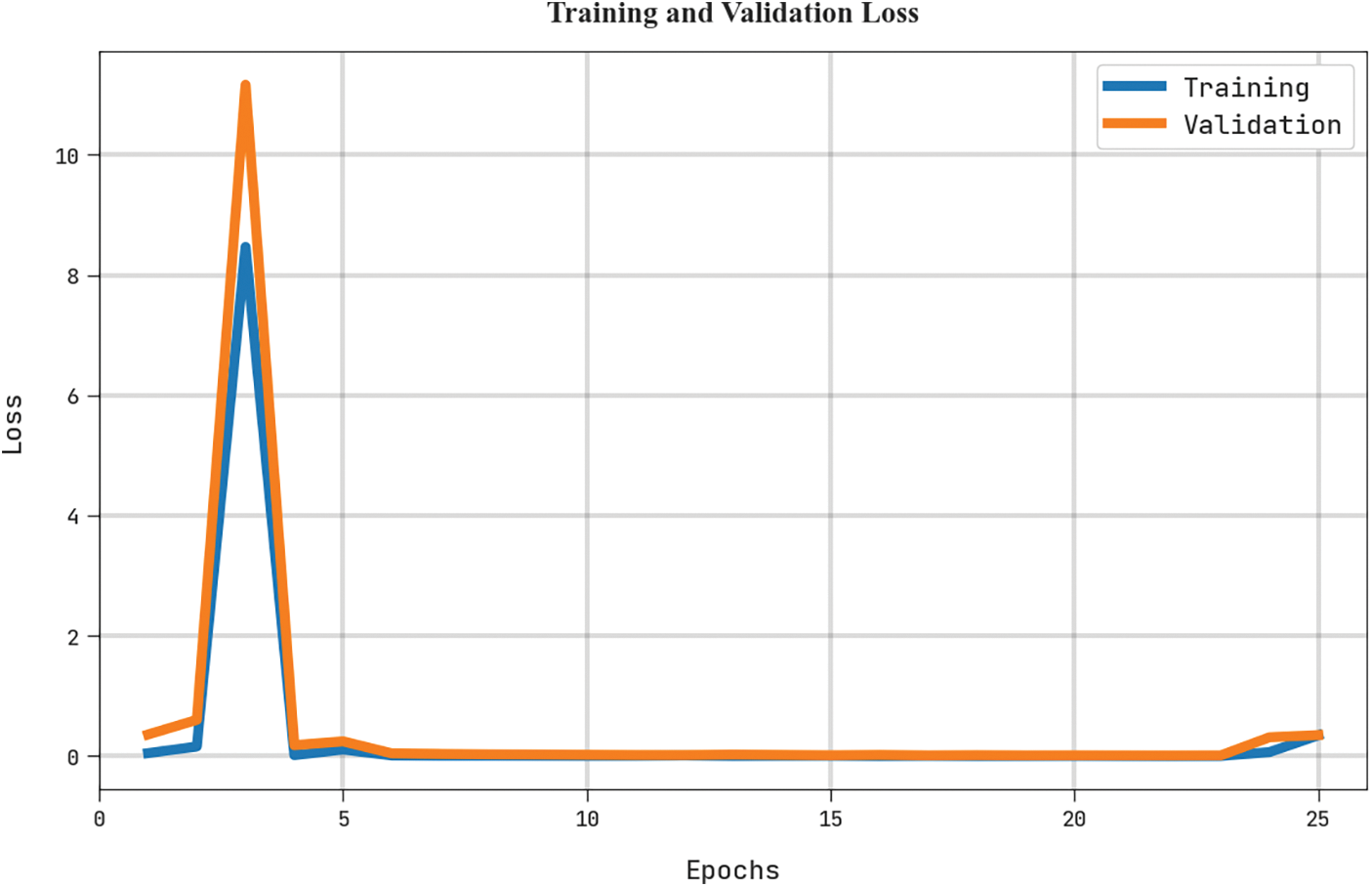
Figure 10: TRL and VLL analysis of MLARMC-HDMS approach
A clear precision-recall investigation of the MLARMC-HDMS algorithm in the test dataset is depicted in Fig. 11. The figure represents the MLARMC-HDMS approach has resulted in enhanced precision-recall values in every class label.
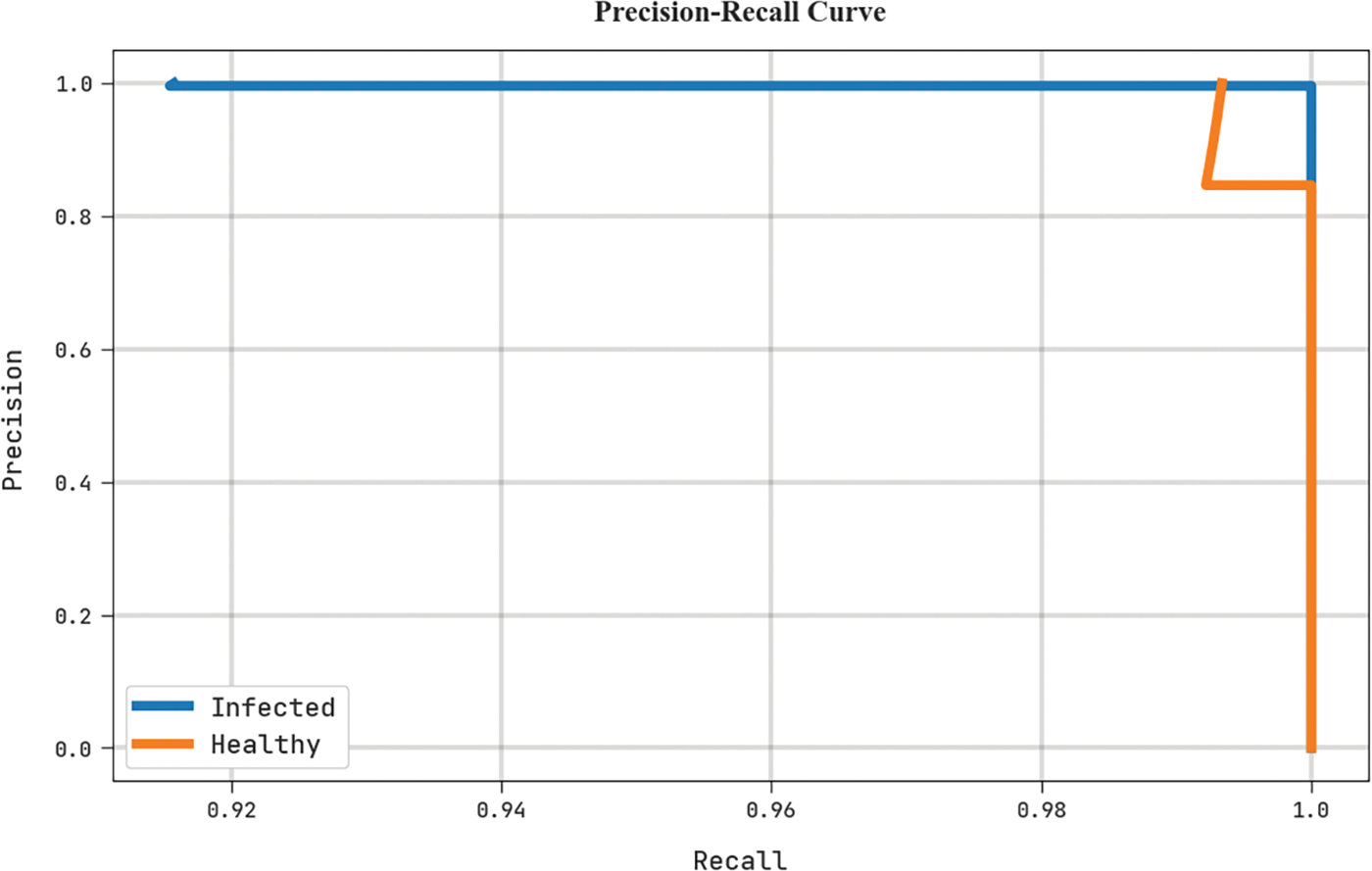
Figure 11: Precision-recall analysis of MLARMC-HDMS approach
A brief ROC analysis of the MLARMC-HDMS algorithm in the test dataset is depicted in Fig. 12. The outcomes implicit in the MLARMC-HDMS technique have displayed its capability in classifying different class labels in the test dataset.
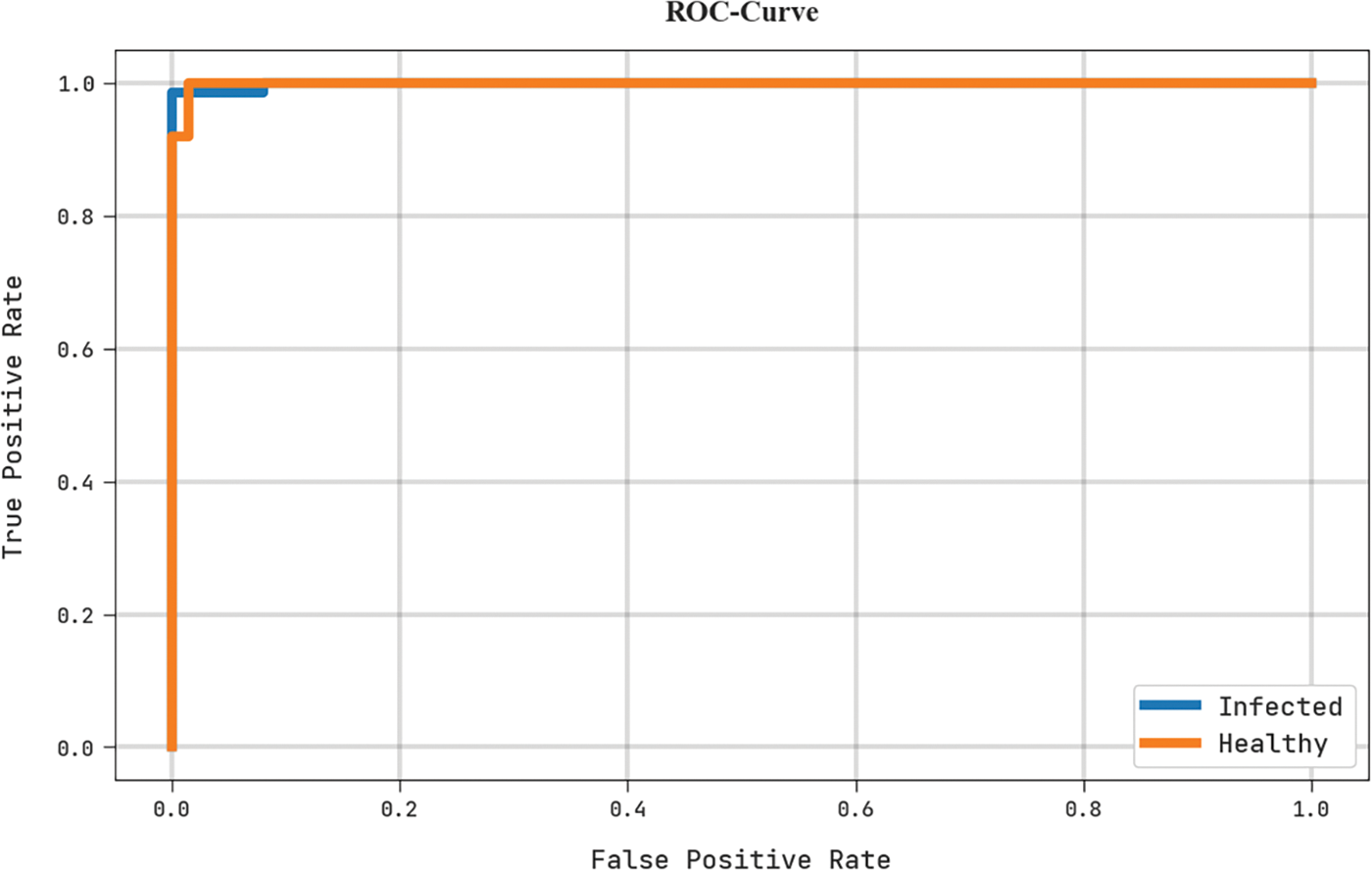
Figure 12: ROC curve analysis of MLARMC-HDMS approach
Table 3 exhibits a comprehensive comparative study of the MLARMC-HDMS model. Fig. 13 reports recent models’ comparative
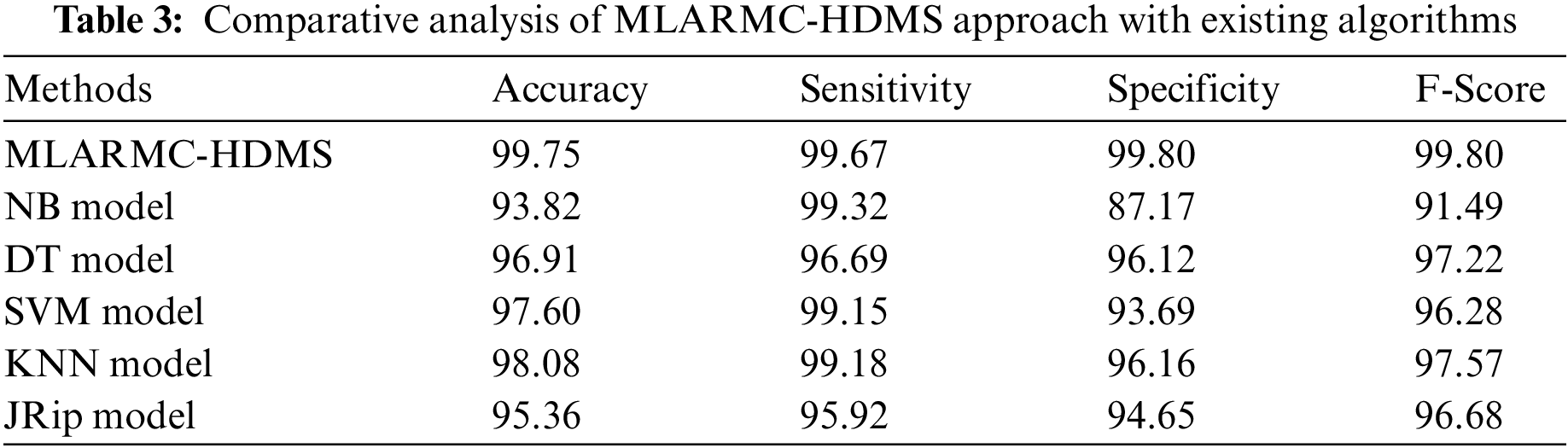
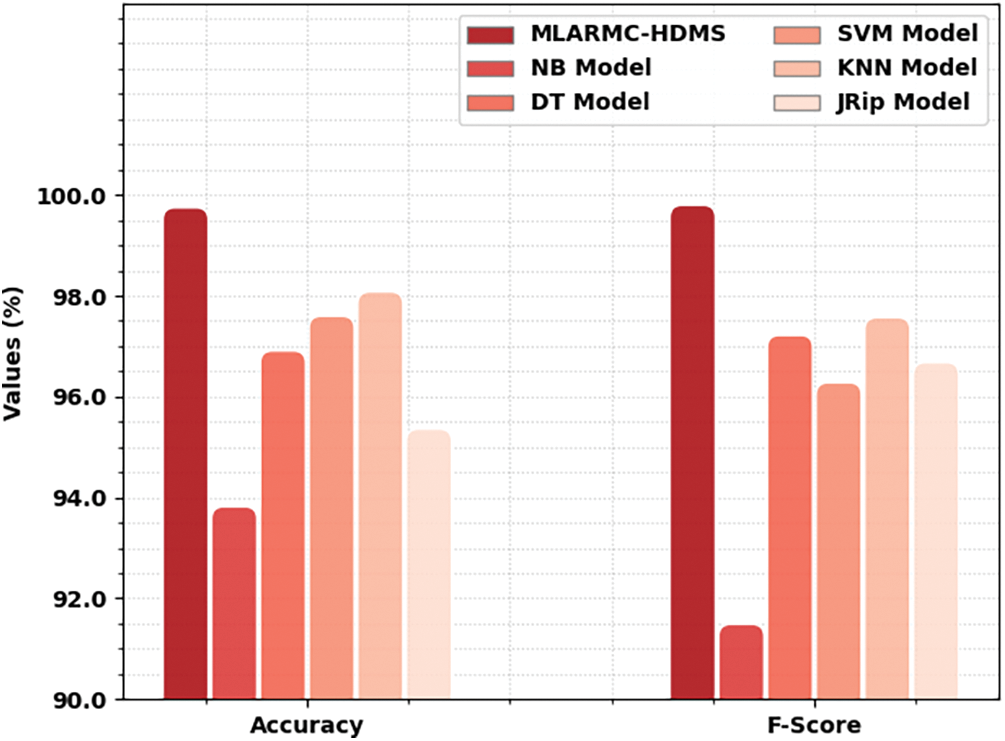
Figure 13:
Fig. 14 demonstrates the detailed
Figure 14:
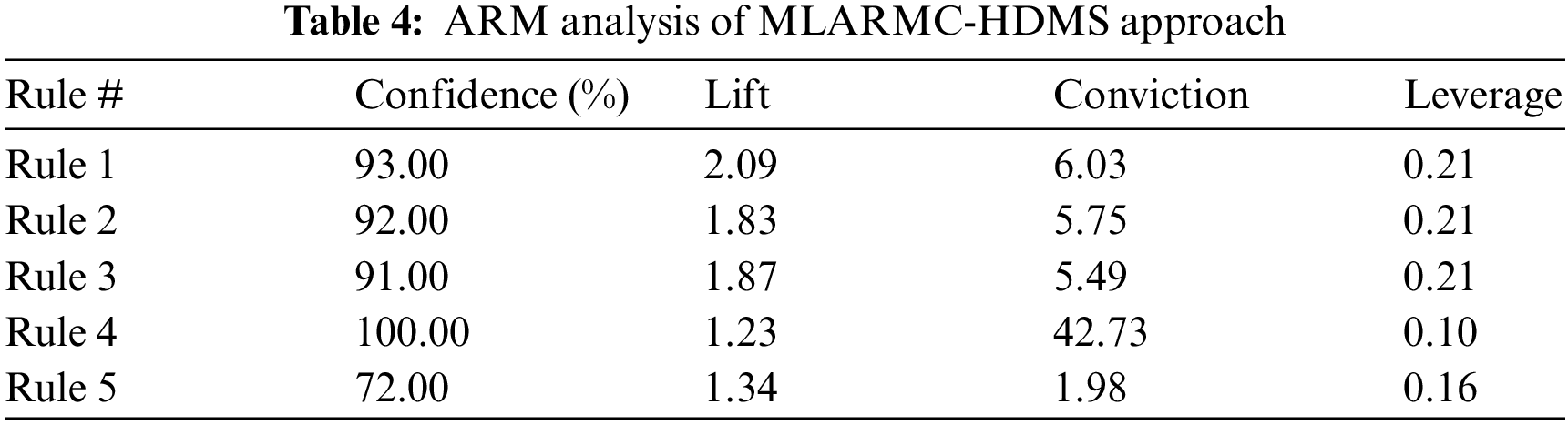
The ARM performance of the MLARMC-HDMS model on the applied dataset is provided in Table 4. The experimental values attained under all five different rules are given. The experimental values demonstrated that the MLARMC-HDMS model had offered effectual outcomes on the healthcare sector’s ARM and classification processes.
In this study, a new MLARMC-HDMS technique was projected to perform both the classification and ARM process. Initially, the MLARMC-HDMS technique employs the COAFS technique for attribute selection. Next, the SGD-MLP model is applied to the classification process. Moreover, the Apriori algorithm is used to determine the relationship between the attributes. To illustrate the enhanced performance of the presented MLARMC-HDMS model, a detailed experimental validation is performed on a benchmark medical dataset. The experimental values indicated the enhancements of the MLARMC-HDMS model over other methods. Therefore, the presented MLARMC-HDMS technique can be utilized to manage healthcare data effectually. In the future, blockchain technology can assure security in the healthcare sector. In addition, the computation complexity of the proposed model needs to be examined in future. In conclusion, the proposed Chimp Optimization Algorithm (COA) based feature selection approach for medical data classification using machine learning techniques shows promising results. The experimental results demonstrate that the COA-based feature selection approach is effective in identifying relevant features for medical data classification, leading to improved accuracy and precision rates. Compared to other feature selection methods, the COA-based approach provides a more efficient and effective way to select relevant features from large medical datasets. The study shows that the COA-based approach has the potential to be used in real-world applications to improve the diagnosis and treatment of various diseases. Overall, the results of this study suggest that the COA-based feature selection approach is a promising technique for medical data classification, which can contribute to the development of more accurate and efficient medical decision-making systems.
Despite its promising results, this study has some limitations that need to be acknowledged:
– Firstly, the study only evaluates the proposed COA-based feature selection approach using a limited number of machine learning classifiers, including Naive Bayes, Decision Tree, Random Forest, and Support Vector Machine. Although these are commonly used classifiers in medical data analysis, the performance of the proposed approach may vary when applied to other classifiers or datasets.
– Secondly, the study only evaluates the proposed approach using binary classification tasks, which limit the generalizability of the results to multi-class classification problems.
– Thirdly, the study does not address the interpretability of the selected features, which is important in medical applications where the selected features need to be clinically meaningful and interpretable.
– Lastly, the study does not consider the imbalanced class distribution problem, which is prevalent in medical datasets. Therefore, the performance of the proposed approach may be affected by class imbalance, which needs to be addressed in future studies.
Overall, the study provides a promising approach to feature selection in medical data classification tasks. However, further research is needed to address the limitations of the proposed approach and improve its applicability in real-world medical decision-making systems. Consequently, some future research directions are recommended as follows:
• Exploring Other Machine Learning Classifiers and Datasets: In future research, it would be beneficial to evaluate the proposed COA-based feature selection approach using a wider range of machine learning classifiers and diverse medical datasets. This will provide a comprehensive understanding of its performance across different classifiers and dataset characteristics, improving the generalizability and applicability of the approach.
• Extending to Multi-Class Classification: Currently, the proposed approach focuses on binary classification tasks. Future work should include the evaluation of the COA-based feature selection approach on multi-class classification problems. This extension will broaden the scope of the approach and allow for its application in a wider range of medical data classification scenarios.
• Interpretability of Selected Features: To enhance the clinical relevance of the selected features, future studies should consider incorporating interpretability into the feature selection process. This will ensure that the selected features not only contribute to accurate classification but also align with clinical knowledge and are easily interpretable by medical professionals.
• Addressing Class Imbalance: Class imbalance is a common issue in medical datasets, and it can affect the performance of classification algorithms. Future research should explore techniques to address the imbalanced class distribution problem within the COA-based feature selection approach. This will enable the approach to handle imbalanced medical datasets more effectively and improve its robustness in real-world applications.
• Integrating Blockchain Technology for Security: As healthcare data security is of paramount importance, future studies can investigate the integration of blockchain technology into the MLARMC-HDMS framework. By leveraging blockchain’s inherent security features, such as decentralized and immutable data storage, the proposed approach can provide enhanced data protection and privacy for healthcare applications.
• Examining Computation Complexity: Further analysis of the computation complexity of the proposed MLARMC-HDMS technique is necessary. Future research should investigate the scalability and efficiency of the approach, especially when dealing with large-scale medical datasets. This analysis will provide insights into the computational requirements and feasibility of implementing the proposed technique in real-world healthcare settings.
Acknowledgement: The authors would like to acknowledge the appreciation to the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia for funding this research work through the Project Number RI-44-0444.
Funding Statement: The authors extend their appreciation to the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia for funding this research work through the Project Number RI-44-0444.
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Firas Abedi, Hayder M. A. Ghanimi; data collection: Abeer D. Algarni, Naglaa F. Soliman; analysis and interpretation of results: Walid El-Shafai, Ali Hashim Abbas, Zahraa H. Kareem; draft manuscript preparation: Hussein Muhi Hariz and Ahmed Alkhayyat. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Not applicable.
Conflicts of Interest: The authors declare they have no conflicts of interest to report regarding the present study.
References
1. A. Delgado, M. Gacto, R. Alcalá and J. Fdez, “Temporal association rule mining: An overview considering the time variable as an integral or implied component,” Wiley Interdisciplinary Reviews: Data Mining and Knowledge Discovery, vol. 10, no. 4, pp. 33–57, 2020. [Google Scholar]
2. Y. Chen, R. Xia, K. Zou and K. Yang, “FFTI: Image inpainting algorithm via features fusion and two-steps inpainting,” Journal of Visual Communication and Image Representation, vol. 9, no. 1, pp. 1–23, 2023. [Google Scholar]
3. M. Peng, S. Lee, A. D’Souza, C. Doktorchik and H. Quan, “Development and validation of data quality rules in administrative health data using association rule mining,” BMC Medical Informatics and Decision Making, vol. 20, no. 1, pp. 1–10, 2020. [Google Scholar]
4. N. Ghafoor and M. Ahmad, “Prioritizing effectiveness of algorithms of association rule mining,” Journal of Computational Learning Strategies & Practices, vol. 1, no. 1, pp. 18–30, 2021. [Google Scholar]
5. M. Nandhini, M. Rajalakshmi and S. Sivanandam, “Performance analysis of predictive association rule classifiers using healthcare datasets,” IETE Technical Review, vol. 39, no. 1, pp. 143–156, 2022. [Google Scholar]
6. G. Urbanin, W. Meira, A. Serpa, D. Costa, L. Baldaçara et al., “Social determinants in self-protective behavior related to COVID-19: Association rule–mining study,” JMIR Public Health and Surveillance, vol. 8, no. 6, pp. 1–20, 2022. [Google Scholar]
7. Y. Wang, “Internet medical privacy disclosure mining and prediction model construction based on association rules,” Tehnički Vjesnik, vol. 29, no. 1, pp. 231–238, 2022. [Google Scholar]
8. S. Sen, A. Mehta, R. Ganguli and S. Sen, “Recommendation of influenced products using association rule mining: Neo4j as a case study,” SN Computer Science, vol. 2, no. 2, pp. 1–17, 2021. [Google Scholar]
9. A. Aljuboori, L. Tawfeeq and K. Al-Karawi, “Pushing towards ehealth for Iraqi hypertensives: An integrated class association rules into SECI model,” Indonesian Journal of Electrical Engineering and Computer Science, vol. 22, no. 1, pp. 522–533, 2021. [Google Scholar]
10. M. Khishe and M. Mosavi, “Chimp optimization algorithm,” Expert Systems with Applications, vol. 14, no. 9, pp. 113–124, 2020. [Google Scholar]
11. L. Dey and A. Mukhopadhyay, “Biclustering-based association rule mining approach for predicting cancer-associated protein interactions,” IET Systems Biology, vol. 13, no. 5, pp. 234–242, 2019. [Google Scholar] [PubMed]
12. R. Sarno, F. Sinaga and K. Sungkono, “Anomaly detection in business processes using process mining and fuzzy association rule learning,” Journal of Big Data, vol. 7, no. 1, pp. 1–19, 2020. [Google Scholar]
13. G. Agapito, B. Calabrese, P. Guzzi, S. Graziano and M. Cannataro, “Association rule mining from large datasets of clinical invoices document,” in Proc. of IEEE Int. Conf. on Bioinformatics and Biomedicine (BIBM), San Diego, USA, pp. 2232–2238, 2019. [Google Scholar]
14. M. Mittal, I. Kaur, S. Pandey, A. Verma and L. Goyal, “Opinion mining for the tweets in healthcare sector using fuzzy association rule,” EAI Endorsed Transactions on Pervasive Health and Technology, vol. 4, no. 16, pp. 5–25, 2018. [Google Scholar]
15. F. Chiclana, R. Kumar, M. Mittal, M. Khari, J. Chatterjee et al., “ARM–AMO: An efficient association rule mining algorithm based on animal migration optimization,” Knowledge-Based Systems, vol. 154, no. 4, pp. 68–80, 2018. [Google Scholar]
16. J. Lee and K. Kim, “Semantic and association rule mining-based knowledge extension for reusable medical equipment random forest rules,” Journal of Integrated Design and Process Science, vol. 22, no. 4, pp. 55–81, 2018. [Google Scholar]
17. M. Useac and G. Ghinita, “Precision-enhanced differentially-private mining of high-confidence association rules,” IEEE Transactions on Dependable and Secure Computing, vol. 17, no. 6, pp. 1297–1309, 2018. [Google Scholar]
18. H. Jia, K. Sun, W. Zhang and X. Leng, “An enhanced chimp optimization algorithm for continuous optimization domains,” Complex & Intelligent Systems, vol. 8, no. 1, pp. 65–82, 2022. [Google Scholar]
19. S. Shadkani, A. Abbaspour, S. Sama, S. Hashemi and A. Mosavi, “Comparative study of multilayer perceptron-stochastic gradient descent and gradient boosted trees for predicting daily suspended sediment load: The case study of the Mississippi River, US,” International Journal of Sediment Research, vol. 36, no. 4, pp. 512–523, 2021. [Google Scholar]
20. F. Ali and A. Hamed, “Usage apriori and clustering algorithms in WEKA tools to mining dataset of traffic accidents,” Journal of Information and Telecommunication, vol. 2, no. 3, pp. 231–245, 2018. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools