 Open Access
Open Access
ARTICLE
Ensemble Learning for Fetal Health Classification
1 Department of Computer Science, College of Computer Engineering and Sciences, Prince Sattam bin Abdulaziz University Al-Kharj, 16273, Saudi Arabia
2 Department of Computer Science, COMSATS University, Islamabad, 53000, Pakistan
3 Faculty of Computers & Information Technology, Computer Science Department, University of Tabuk, Tabuk, 71491, Saudi Arabia
4 Department of Computer Engineering and Networks, College of Computer and Information Sciences, Jouf University, Sakaka, 72388, Saudi Arabia
5 Information Systems Department, Faculty of Management Comenius University in Bratislava, Odbojárov 10, 82005 Bratislava 25, Slovakia
6 Department of Computer Science, Kinnaird College for Women, Lahore, 54000, Pakistan
* Corresponding Authors: Mesfer Al Duhayyim. Email: ; Natalia Kryvinska. Email:
Computer Systems Science and Engineering 2023, 47(1), 823-842. https://doi.org/10.32604/csse.2023.037488
Received 05 November 2022; Accepted 07 April 2023; Issue published 26 May 2023
Abstract
: Cardiotocography (CTG) represents the fetus’s health inside the womb during labor. However, assessment of its readings can be a highly subjective process depending on the expertise of the obstetrician. Digital signals from fetal monitors acquire parameters (i.e., fetal heart rate, contractions, acceleration). Objective:: This paper aims to classify the CTG readings containing imbalanced healthy, suspected, and pathological fetus readings. Method:: We perform two sets of experiments. Firstly, we employ five classifiers: Random Forest (RF), Adaptive Boosting (AdaBoost), Categorical Boosting (CatBoost), Extreme Gradient Boosting (XGBoost), and Light Gradient Boosting Machine (LGBM) without over-sampling to classify CTG readings into three categories: healthy, suspected, and pathological. Secondly, we employ an ensemble of the above-described classifiers with the over-sampling method. We use a random over-sampling technique to balance CTG records to train the ensemble models. We use 3602 CTG readings to train the ensemble classifiers and 1201 records to evaluate them. The outcomes of these classifiers are then fed into the soft voting classifier to obtain the most accurate results. Results:: Each classifier evaluates accuracy, Precision, Recall, F1-scores, and Area Under the Receiver Operating Curve (AUROC) values. Results reveal that the XGBoost, LGBM, and CatBoost classifiers yielded 99% accuracy. Conclusion:: Using ensemble classifiers over a balanced CTG dataset improves the detection accuracy compared to the previous studies and our first experiment. A soft voting classifier then eliminates the weakness of one individual classifier to yield superior performance of the overall model.Keywords
A fetus is susceptible to several complications during the gestation period. Long-term neonatal neurological disorders, stillbirths, and intrapartum fetal death are just a few of the disastrous outcomes of obstetric negligence [1]. Annually over 1.3 million fetal deaths occur during labor [2]. One of the primary causes of fetal mortality is birth asphyxia. Birth asphyxia occurs when there is an interruption in the blood flow towards the placenta, causing low oxygen supply to the fetus’s brain, i.e., hypoxia [3]. Lifelong physical and mental abnormalities like speech disorders, hearing and vision impairment, epilepsy, and cerebral palsy are direct outcomes of birth asphyxia resulting from hypoxia [4]. Conventional childbirth techniques, non-computerized CTG, and the dependency on the expertise of the medical practitioner have been a few of the primary contributors to fetal hypoxia that can result in such permanent brain damage and fetal death before or during labor [5].
Computerized CTG has been an effective tool to help monitor a mother and fetus’s well-being. It helps monitor fetal heart rate, uterine contractions, acceleration, decelerations, variability, and fetal movement and clearly represents the mother and child’s health. Unfortunately, the subjectivity of the clinical assessment of CTG outcomes has been unable to significantly reduce fetal complications [6]. The assessment of physicians is based on the degree of hypoxia of the fetus to classify the fetal heart rate into normal, suspicious, or pathological [5]. This subjective assessment can not only be erroneous but can be highly dependent on the expertise and experience of the physician, thus increasing the associated economic burden. For decades, new and improved techniques have been researched in medical diagnostics to automate the disease/symptom prediction process. A few state-of-the-art AI-supervised and unsupervised approaches that have been employed in the field of medicine include ML classification techniques [7], evolutionary and genetic algorithms [8], and CNN and DNN-based classification approaches [9]. Furthermore, researchers have employed ensemble classification techniques to optimize the outcome of the ML model to yield a higher prediction accuracy.
Artificial intelligence to automate the CTG evaluation process is being widely researched and integrated with promising machine learning tools. An automated CTG assessment has proven to be much more accurate and can allow for timely intervention. Researchers have employed several ML and DL [10] classifiers for classification tasks. Classifiers like Logistic Regression, Support Vector Machines (SVM) [11] and [12], Gaussian Naive Bayes [12], and Neural Networks [13], etc. have been used to classify CTG features and predict the fetal health status. The proposed work uses minority resampling techniques and ensemble learning to classify fetal health. A single classifier is limited by the algorithm approach and can yield a biased output. Ensemble learning combines various decision-making algorithms and does not rely on the outcome of one classifier, hence improving prediction accuracy. We have employed the AdaBoost, CatBoost, Random Forest Classifier, XGBoost, and LGBM to evaluate the CTG data obtained from the publicly available Fetal Health dataset. A soft voting classifier then summarizes the ensemble learners’ outcomes. A 99% accuracy is achieved in predicting the three classes normal fetus, suspected abnormal, and pathological (requiring immediate attention). The accuracies and outcomes have been compared to assess the performance of each ensemble classifier.
The motivation to conduct this research is to automate the fetal health classification process to obtain a timely and accurate fetal and maternal health diagnosis. Since CTG abnormalities are life-threatening medical emergencies, diagnosis and interpretation require absolute precision. While automating the fetal health classification process, a high level of accuracy can only be achieved when the dataset is balanced and the prediction models are strong. The proposed approach will remove the dataset imbalance and use ensemble classifiers to classify the CTG records into healthy, suspected, and pathological. The paper aims to highlight the superior performance of the ensemble classifiers and the resulting superior accuracy achieved by applying the soft voting classifier that gives the average outcome of all ensemble classifiers. The proposed model will not only allow for a timely diagnosis and reduce fetal mortality and birth complications but will enable medical practitioners to integrate the CTG automation process into the physical exam resulting in a much more accurate and faster diagnosis procedure.
This paper aims to contribute to research in the following aspects:
• Proposes an approach to classify fetal health by evaluating the CTG features like heart rate, fetal movements, acceleration, deceleration, and uterine contractions into three categories: healthy fetus, suspected abnormal fetus, and pathological state that requires immediate attention.
• Present a comparison between different individual classifiers and state-of-the-art to evaluate the best model and provide a basis for future research.
• Employ a soft voting classifier to summarize the ensemble models’ outcome and classify CTG data into three categories.
• Predict fetal health accurately and timely so that fetal mortality and life-long neurological disorders can be minimized.
The rest of the paper is structured as follows. Section 2 is a literature review of the work of researchers to automate the classification of CTG features. Section 3 elaborates on the methodology employed by the proposed research, including a discussion of the dataset and the algorithms and evaluators employed. The results and findings of the proposed work are presented in Section 4. Section 5 presents a discussion of our findings and provides a comparative analysis of the ensemble classifiers’ results. Finally, Section 5 summarizes and concludes the work by highlighting the main findings.
CTG data has been the focus of attention of many researchers to assess fetal health using different AI techniques. A few researchers have used Neural Networks to predict fetal health using fetal heart rate data. The authors developed a CNN-based model in [14] to automatically detect Fetal Acidosis using fetal heart rate. The model was able to achieve a 99.09% accuracy. Another research [15] used CNN to classify fetal heart rates obtained from electronic fetal monitors. The proposed model was evaluated on three models for comparison. The resulting classification accuracies of the SVM, Multilayer Perceptron, and CNN were 79.66%, 85.98%, and 93.24%, respectively. The authors in [16] have proposed an 8-layer Deep-CNN to automate the prediction of fetal acidosis using fetal heart rate signals by applying the wavelet transform. The proposed model achieved a 10-fold cross-validation accuracy of 98.34%.
A novel approach was proposed by the authors in [17] whereby a CTG image-based time-frequency (IBTF) analysis was conducted that included Short-Time Fourier transform (STFT) and gray level co-occurrence matrix (GLCM). Feature subsets were extracted and given as input to the least square SVM (LS-SVM) classifier. Genetic algorithms were then used to obtain sensitivity and specificity of 63.45% and 65.88%, respectively. Many researchers have employed ML models. The authors in [18] used twelve different ML classifiers to classify CTG data and then used a soft voting classifier to obtain a 95.9% model accuracy. Another research [19] has used SVM, random forest, multilayer perceptron, and K-Nearest Neighbor (KNN) to evaluate CTG data. Many researchers have employed a combination of approaches to create the most accurate model for fetal health classification.
The author in [20] has employed a Decision Tree, SVM, stochastic gradient descent, Naive Bayes, and Deep learning models like CNN and DenseNet to evaluate the CTG data to predict fetal hypoxia. The authors have also used Bagging Tree, AdaBoost, and Voting Classifier to predict fetal hypoxia status accurately and have obtained the best results from the Bagging Tree method using the Naive Bayes classifier. A bagging ensemble machine learning algorithm was used by [21] to evaluate fetal heart rates and predict whether they are normal or abnormal. The authors have evaluated the performance of SVM, Artificial Neural Networks, KNN, Random Forest, and Decision trees using the bagging approach and concluded that the bagging approach had yielded promising results, especially the bagging approach along with random forest with accuracy up to 99.02%.
In another work [22], the authors have proposed an intelligent classification model using the Deep Forest algorithm to detect fetal abnormality accurately. Random Forest, Weighted Random Forest, Completely Random Forest, and Gradient Boosting Decision Tree were integrated to yield the best detection/prediction accuracy. In [12], the authors have explored resampling techniques to classify imbalanced CTG data and employed several ML and ensemble models to evaluate fetal health. Seven ML and ensemble models have been used to classify fetal health conditions into one of three categories, i.e., normal, suspected, or pathological, by the authors in [23]. Models include SVM, KNN, Neural Networks, Long Short-Term Memory (LSTM), XGBoost, LGBM, and Random Forest. Almost all the models yielded accuracies between 89–99% in classifying fetal health conditions into one of the three categories.
The proposed model employed five ensemble learners, Random Forest, AdaBoost, XGBoost, CatBoost, and LGBM, on the Fetal Health Classification dataset comprising CTG records. Their accuracy, recall, F1-score, and AUCROC are recorded and compared. A soft voting classifier is then used, the input to which are the outcomes of the five ensemble learners, illustrated in Fig. 1. The average voting from all classifiers are used to classify the records into healthy, suspected, and pathological. Finally, the outcome of our soft voting classifier is shared.
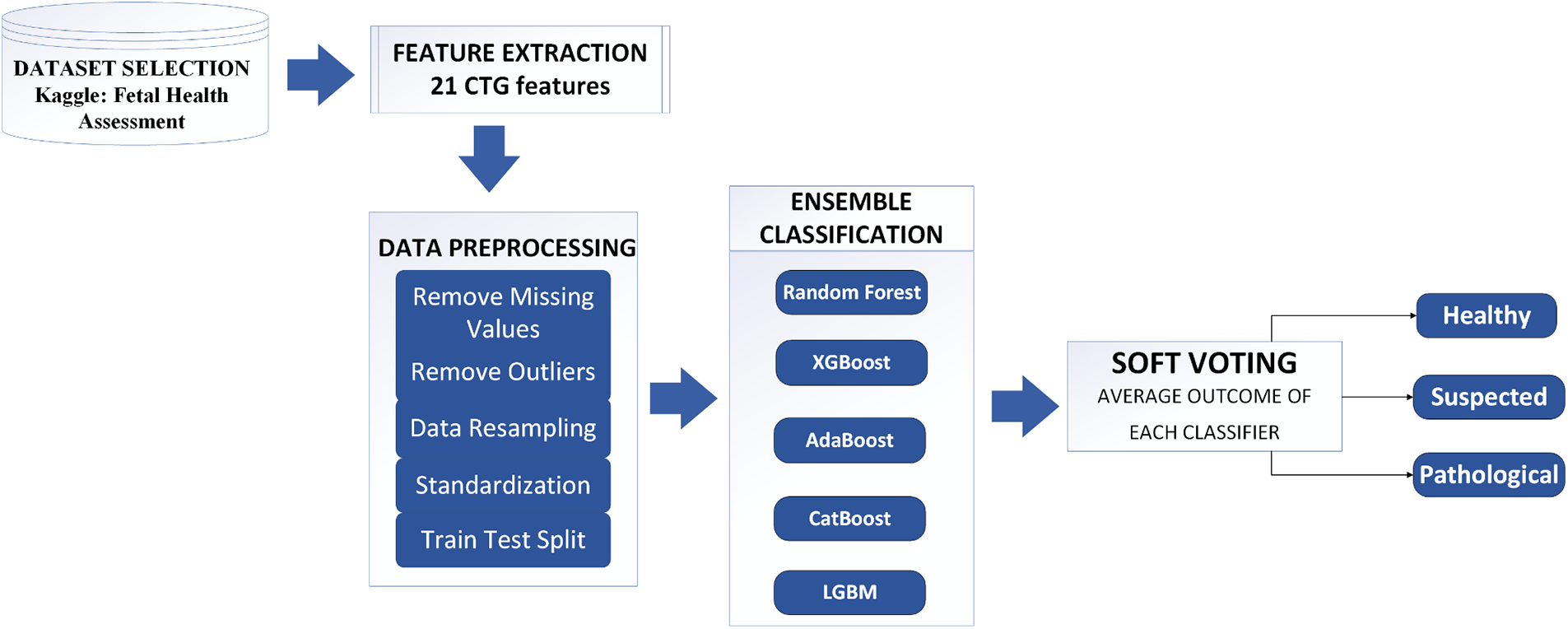
Figure 1: The proposed methodology of fetal health assessment
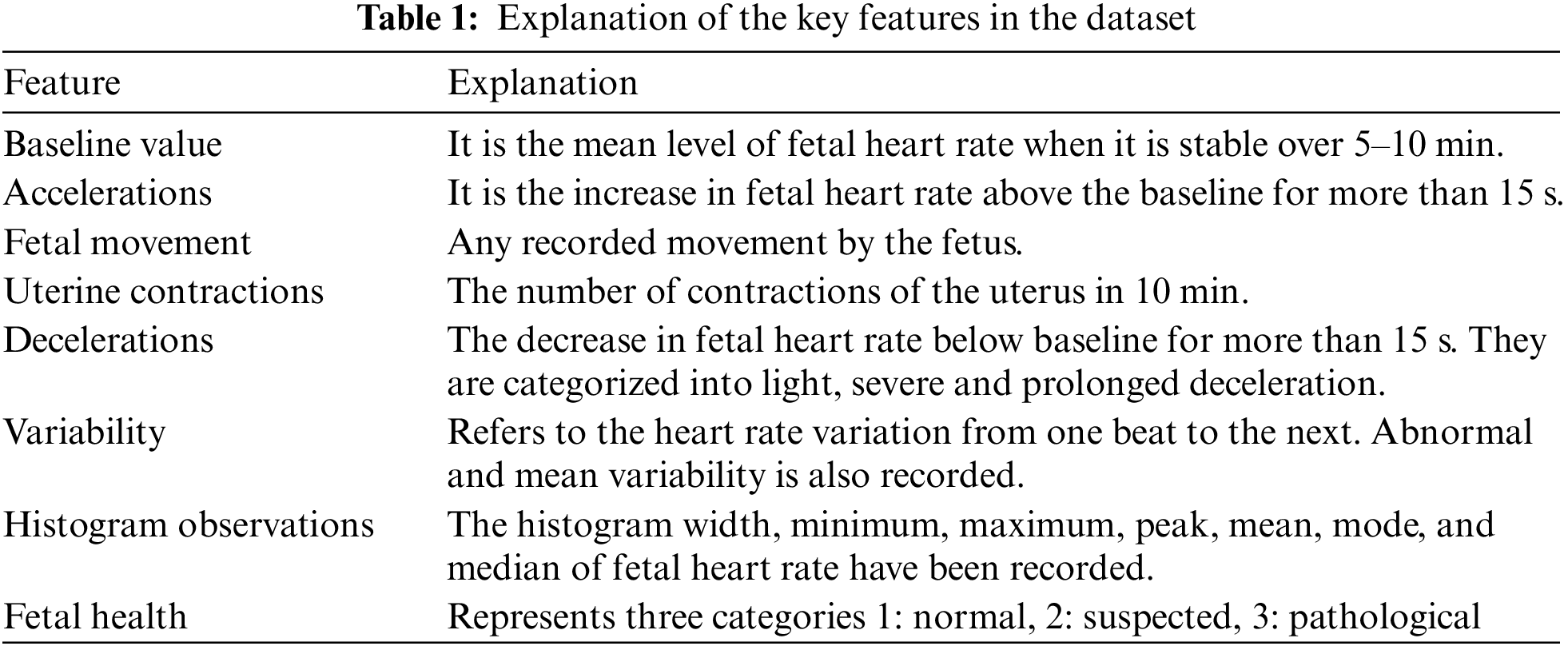
The dataset is obtained from Kaggle Fetal Health Assessment [24] and [25]. It comprises 2126 records obtained from CTG readings from pregnant ladies at 29–42 gestational weeks. The author in [25] developed a program for automated analysis of CTG readings. The SisPorto 2.0 system gathered signals from Sonicaid and Hewlett-Packard fetal monitors tested on over 6000 pregnancies. The three experts’ monitor readings and input established baseline fetal heart, acceleration, deceleration, contractions, and variability readings. Hence the acquired dataset is part of a study conducted to establish the validity of the SisPorto 2.0. The dataset consists of 21 decimal value features and a label for each record, categorizing the record according to the Fetal health status into ‘Healthy,’ ‘Suspected,’ and ‘Pathological.’ Table 1 discusses some of the significant features.
The CTG data is preprocessed to check for missing values and outliers.
Missing values: There are no missing values in the dataset. The minimum value observed against some features is 0. 0 and is not considered null.
Outliers: A box plot analysis of each feature is observed to find the outliers in our dataset. The values marked by the upper and lower bar outside the box are categorized as outliers. Fig. 2 is an illustration of our findings.
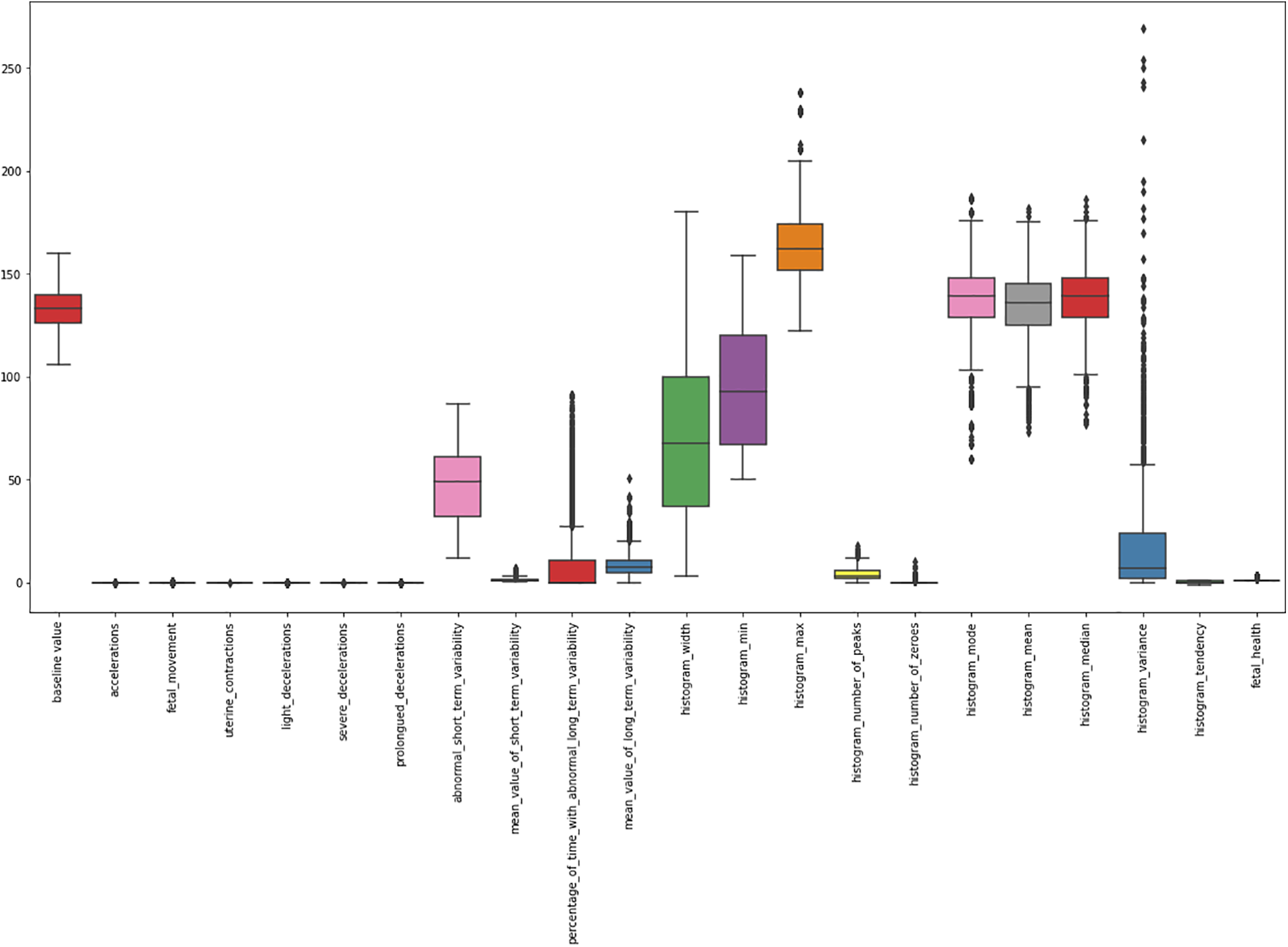
Figure 2: A box plot of CTG data
Outliers are observed in the following features: Histogram Variance, Histogram Median, Histogram Mean, Histogram Mode, Percentage of time with abnormal long-term variability, and Mean value of short-term variability. An outlier cannot be ignored and can significantly alter the results of an AI model. The outliers are therefore removed by setting an upper and lower threshold. All values beyond the threshold are removed from the dataset. To compute the upper boundary and lower boundary of each attribute, the following Eqs. (1) and (2) are used:
The attribute mean signifies the average value obtained from each feature. Standard deviation is the variation amongst the values for each feature. All values more than or less than 3 times the standard deviations from the mean are excluded. These values indicated they were outliers and varied from the average values obtained during CTG observations. A total of 182 outliers are discovered, and hence they are removed from the dataset. The resulting dataset contained 1944 instances.
CTG data contains readings of random gestation and labor cases. Although the chance of abnormality in CTG readings is significantly low, they are the most significant findings that cannot be ignored. Our Fetal Health Assessment dataset contains imbalanced data with a relatively higher ratio of the healthy fetus (1601 instances) as compared to the suspected (283 instances) and pathological (60 instances). Training a model on imbalanced data can generate misleading results since the model will be trained with the maximum number of healthy instances instead of suspected and pathological ones. To obtain an accurate prediction model, it is essential to balance the dataset first. Several researchers have proposed different techniques to balance an unbalanced dataset, the most commonly used of which are random over-sampling, random under-sampling, imbalanced resampling, and SMOTE (Synthetic Minority Over-sampling Technique), etc. We used the “Random over-sampling with replacement” technique to balance our dataset. The principle behind this technique is to increase the minority class by duplicating randomly selected records from the minority class. When using it with replacement, the probability of the record being reselected for duplication remains the same every time. Replacing each entry after it is selected for duplication also allowed for a bigger pool for selection. Over-sampling with replacement is not only a straightforward technique to balance the dataset but also a good choice for our dataset because the CTG records don’t have a very high degree of variation within a class. Secondly, over-sampling, as opposed to under-sampling, preserves the data integrity and allows for a larger dataset that can improve the generalization of our model. To balance our dataset, the minority classes, i.e., the suspected and the pathological class instances, are oversampled. 1601 instances of both minority classes are generated to match the majority class. All three classes, i.e., healthy, suspected, and pathological, had 1601 instances each. A total of 4803 CTG records are used to train and test the models.
The CTG data is normalized using the Standard Scaler python library. The Standard scaler normalizes the data by computing the z-score and bringing it within the same range, making it easy to compare and compute.
Ensemble learning is an optimized approach that combines multiple ML learners to reduce the error of a single learner to achieve superior prediction outcomes [26]. Ensemble-based classification can prove especially useful to avoid overfitting since it averages the outcome of single ML learners and provides an optimal search space, an extension of the individual ML classifier’s search space resulting in a better representation of the data [27] and [28]. We chose 5 of the most used dependent ensemble models where the outcome of one model is used to improve the further models, i.e., 1) Random Forest, 2) XGBoost, 3) AdaBoost, 4) CatBoost, and 5) LGBM.
4 Experimental Analysis and Results
The proposed work aims to classify the fetal health dataset into 3 classes: Healthy, Suspected and Pathological so that a healthy and distressed fetus can be distinguished. The experimental evaluation of different models uses Python 3.6 on the online Jupyter notebook, i.e., ‘Google Colab’. Five ensemble classifiers are trained on the given dataset and then tested on the test dataset. The data is split in the ratio 75:25 for training and testing, respectively. We ran two experiments: classification with imbalanced data and balanced data. This paper considers several evaluation metrics commonly used to assess performance, including Accuracy, Precision, Recall, the F-measure, and the Confusion Matrix (Eqs. (3)−(6)). It is possible to calculate True Positives (TP), True Negatives (TN), False Positives (FP), and False Negatives (FN) by analyzing the ensuing confusion matrices. AUROC represents the degree or measure of separability. Further, it describes how much the AI-based model can distinguish between class labels. The following equations can be used to determine values for Accuracy, Precision, Recall, and F1-score:
4.1 Classification with Imbalanced Data
We first run multiple experiments with imbalanced data to understand the rationale behind balancing the data. This experiment’s data contains 1601 healthy, 283 suspected, and 60 pathological instances. We utilize the same algorithms for all experiments throughout the research.
4.1.1 Random Forest (RF) Classifier
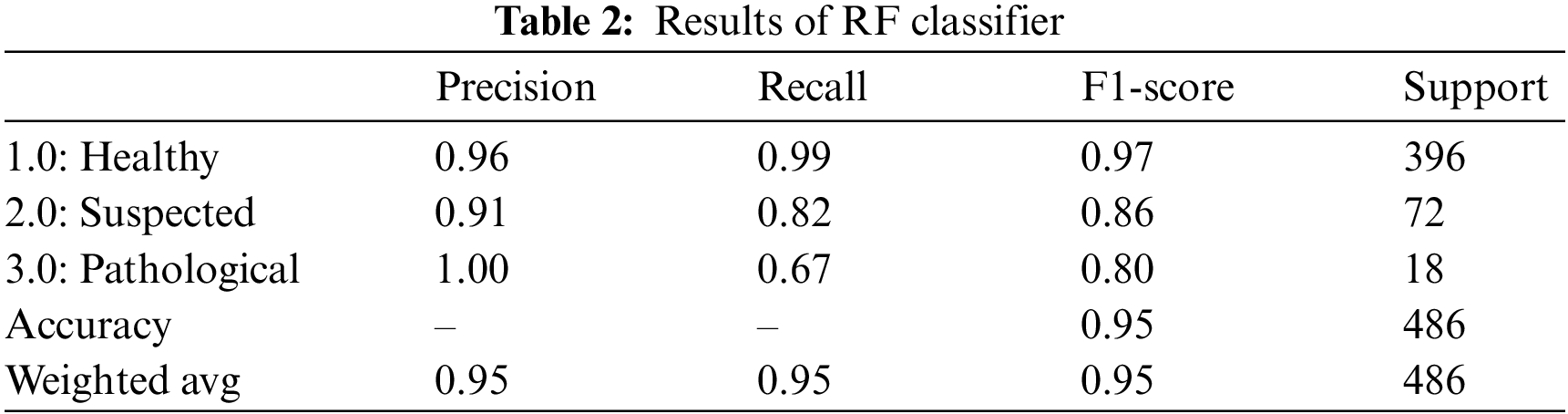
Random Forest classifier is a supervised learning technique using a hierarchy of decision trees to present an aggregated result using the bagging approach [29]. RF classifier yielded an overall accuracy of 95.0%. Table 2 summarizes the outcomes of the RF classifier for fetus health classification. The precision, recall and F1-score outperform all three classes for the CTG data. The precision for predicting healthy cases is 96%, while for the suspected cases is the lowest, 91.0%, and for the pathological fetus is 100%. By observing the recall value, 99.0% of Healthy cases are predicted correctly; Suspected cases are 82% successfully predicted, 67.0% of Pathological records are predicted correctly, and the rest are falsely predicted cases. Similarly, for the F1-score, 97.0% of Healthy fetus cases, 86.0% of Suspected cases and 80.0% of pathological fetus cases are predicted successfully.
4.1.2 Extreme Gradient Boosting (XGBoost) Classifier
XGBoost algorithm, also known as the gradient-boosted decision tree classifier, is an extension of the random forest classifier built upon multiple decision trees. The outcome is improved with each iteration by building upon the residual error from the previous model [30]. The outcomes of the XGBoost classifier for fetus health classification are summarized in Table 3. XGBoost classifier gave an overall accuracy of 95.0%. For all three classes, precision, recall and F1-score performed outclassed. The precision of the health fetus cases prediction is 96.0%, the Suspected fetus cases are predicted at 89.0%, and the Pathological fetus cases are at 81.0% predicted accurately. Similarly, for recall, 98.0% of Healthy fetus cases, 78.0% of Suspected cases and 94.0% of Pathological fetus cases are predicted successfully. By observing the F1-score, we see that 97.0% of Healthy fetus cases out of 396 are predicted correctly; 83.0% of Suspected cases and 87.0% of Pathological fetus cases are successfully predicted, while the rest of the cases are misdiagnosed.
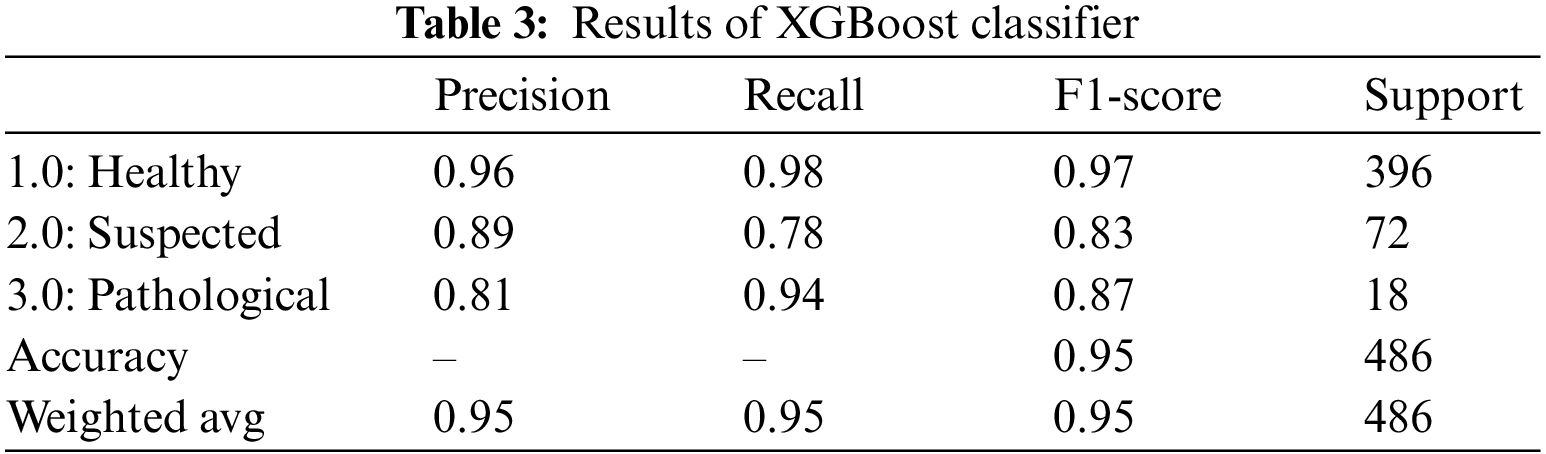
4.1.3 Adaptive Boosting (AdaBoost) Classifier
Another boosting classifier that tweaks the outcomes from the previous weak learners and adapts the new learner to yield better and more robust results is the AdaBoost classifier [31]. Table 4 summarizes the AdaBoost classifier outcomes for fetus health classification. For the CTG dataset, the AdaBoost classifier provided an overall accuracy of 90.0%. The precision for the Healthy fetus cases is 94.0%, and 74.0% for Suspected cases is predicted correctly, while the rest cases are predicted negatively. Similarly, 95.0% of Healthy cases records and 71.0% of Suspected cases are diagnosed accurately for the recall. In contrast, the F1-score of the Healthy cases is 95.0%, Suspected cases are 72.0%, and overall, 67.0% of the Pathological cases are predicted successfully.
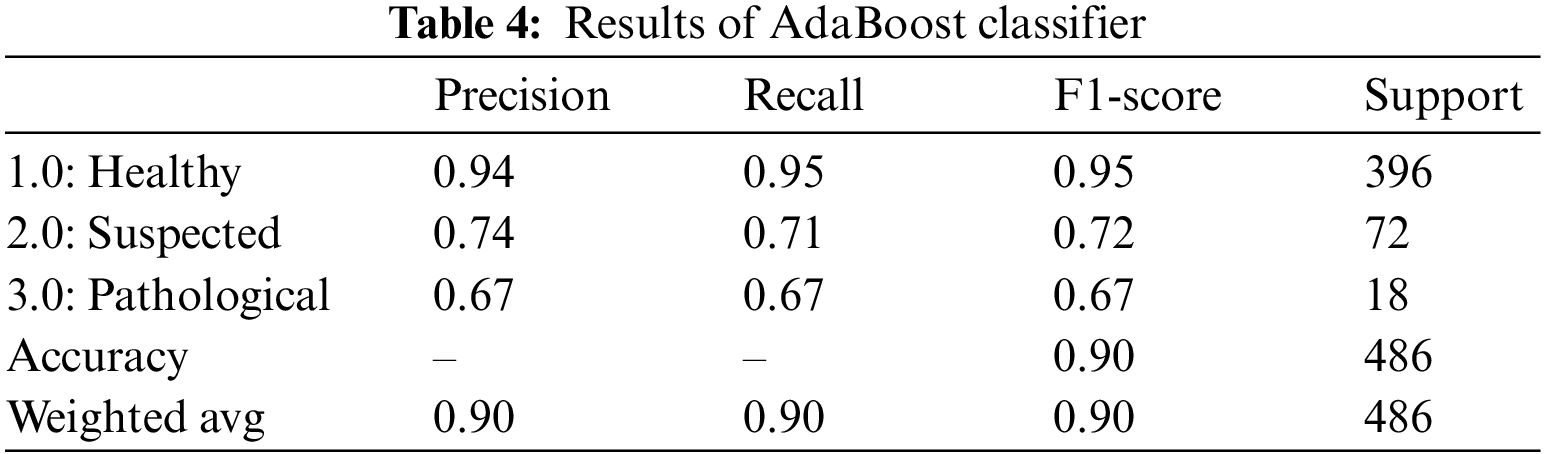
4.1.4 Categorical Boosting (CatBoost) Classifier
CatBoost classifier is another gradient boosting algorithm that, although very similar to the CatBoost algorithm, directly uses categorical features rather than encoding them first [32]. The CatBoost classifier yielded an overall accuracy of 96.0%. Table 5 summarizes the outcomes of the CatBoost classifier for fetus health classification. The precision, recall and F1-score outclass all three classes. For the CTG data, the precision of the Healthy fetus cases is 97.0%; Suspected cases are 88.0%, and Pathological fetus cases are 94.0% predicted correctly. By observing the recall value, 98.0% were Healthy fetus cases, 83.0% were Suspected cases, and 89.0% were Pathological fetus cases successfully diagnosed. Similarly, the Healthy fetus cases 98.0%, Suspected cases 86.0%, and Pathological fetus cases 91.0% F1-score were predicted positively, and the rest were falsely negative.
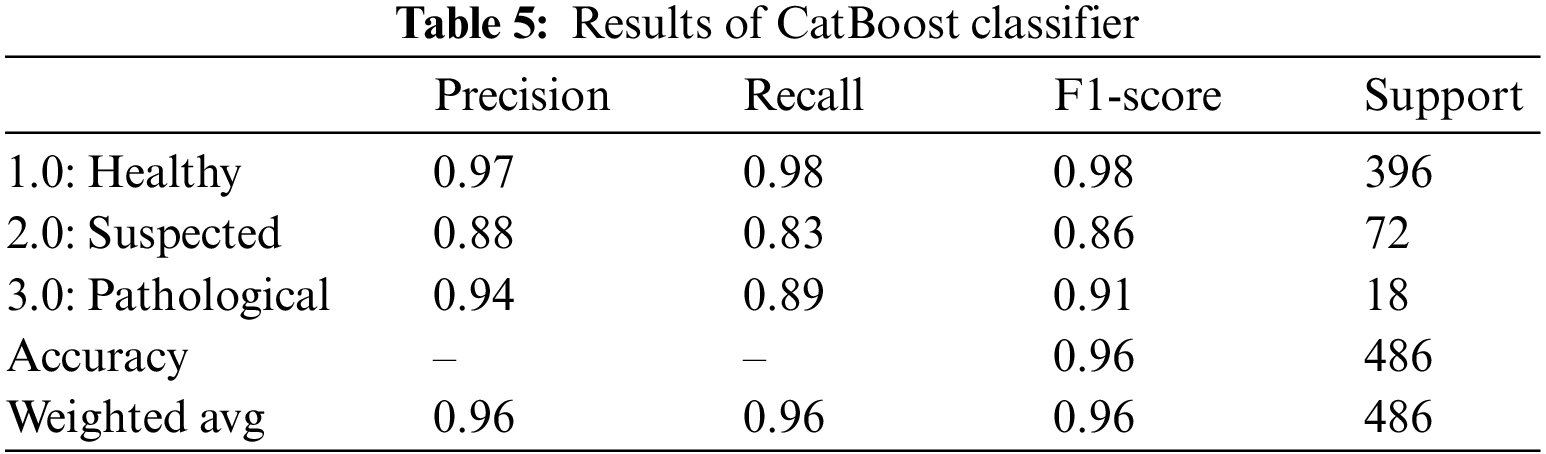
4.1.5 Light Gradient Boosting Machine (LGBM)
The LGBM is by far the fastest and one of the most effective gradient-boosting algorithms. Rather than a depth-wise data split, the LGBM classifier performs a leaf-wise split [33]. The outcomes of the LGBM classifier for fetus health classification are presented in Table 6. LGBM classifier provided an overall accuracy of 96.0%. For all three classes, precision, recall and F1-score outperformed. The precision of the health fetus cases prediction is 97.0%, the Suspected fetus cases are predicted at 89.0%, and the Pathological fetus cases are at 88.0% predicted accurately. Similarly, 99.0% of Healthy fetus cases, 82.0% of Suspected cases and 83.0% of Pathological fetus cases are predicted successfully for recall. By observing the F1-score, we see that 98.0% of Healthy fetus cases out of 396 are predicted correctly; 86.0% of Suspected and Pathological fetus cases are successfully predicted, while the rest are predicted negatively.
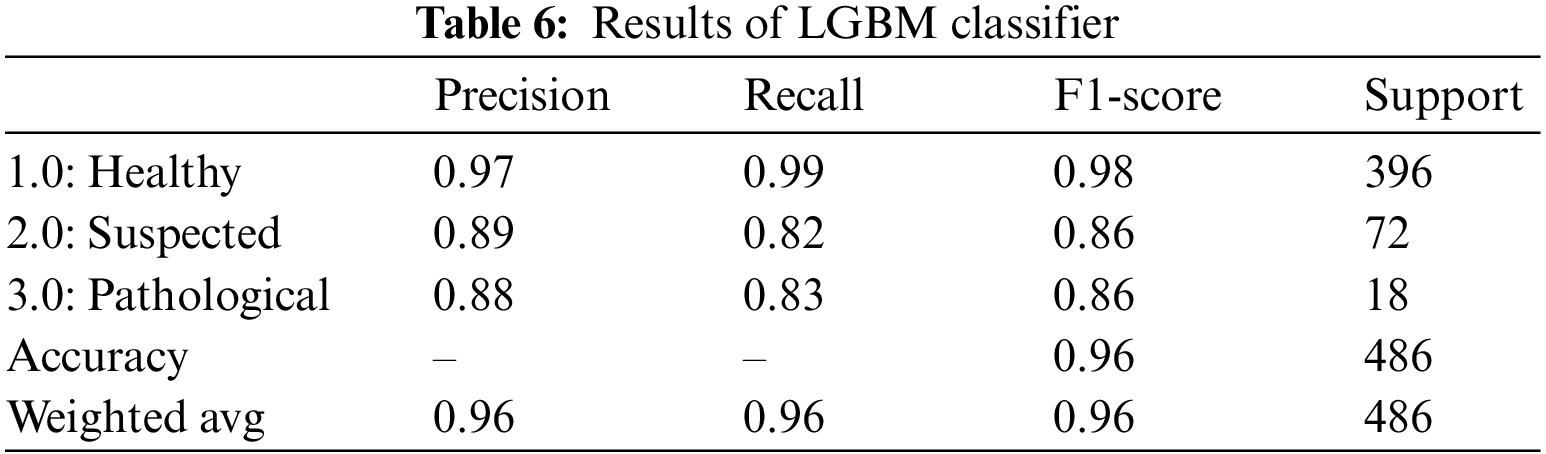
The voting classifier, also known as a meta classifier, uses the outcome from the RF, XGBoost, AdaBoost, CatBoost, and LGBM to classify the CTG data [34] and [35]. Machine learning and other approaches have proven useful for fetal health problems [36–39]. VC classifier gave an overall accuracy of 96.0%. Table 7 summarizes the outcomes of the VC classifier for fetus health classification. For the CTG data, the precision, recall and F1-score outclassed all three classes. The precision for predicting healthy cases is 97%, while for the suspected cases is the lowest, 88.0%, and for the pathological fetus is 94.0%. By observing the recall value, 98.0% of Healthy cases are predicted correctly; Suspected cases and Pathological fetus cases are 83% successfully predicted, and the rest are falsely predicted cases. Similarly, for the F1-score, 98.0% of Healthy fetus cases, 86.0% of Suspected cases and 88.0% of pathological fetus cases are accurately predicted.
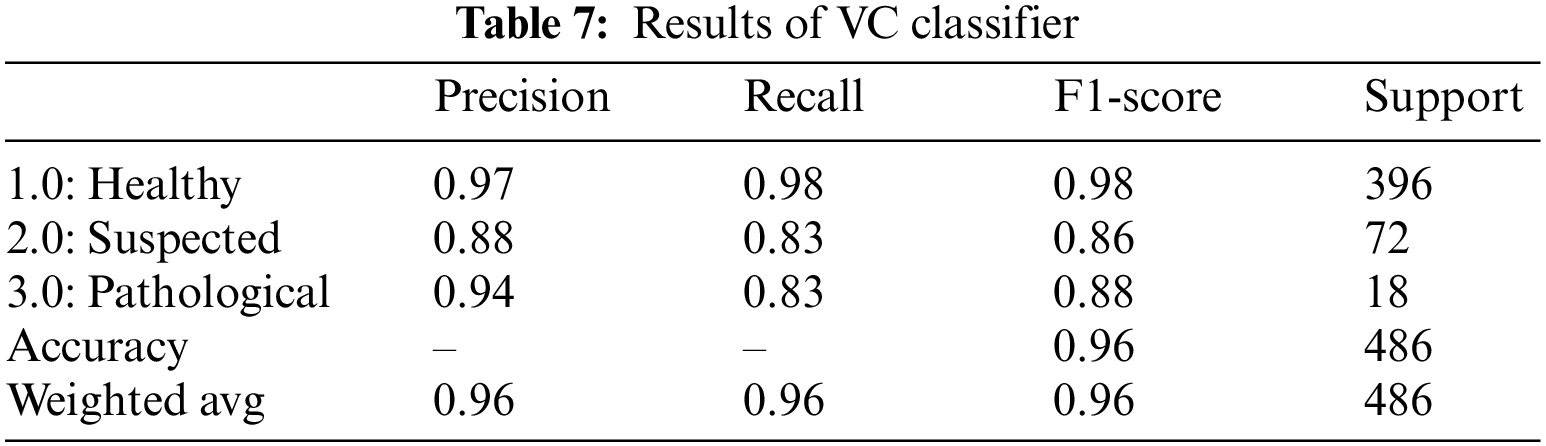
The confusion matrix in Fig. 3a shows that out of a total of 396 healthy fetus instances, the RF classifier could predict 392 instances successfully. Similarly, out of 72 suspected fetus cases, 59 are predicted accurately, and out of 18 pathological instances, 12 are predicted successfully. From a total of 486 CTG readings, 23 are misdiagnosed. The confusion matrix of the XGBoost classifier displayed in Fig. 3b indicates that 387 instances are predicted successfully, whereas 9 instances are misdiagnosed out of 396 total instances. Similarly, out of 72 suspected fetus cases, 56 are predicted accurately, 16 are misdiagnosed, and out of 18 pathological instances, 17 are predicted successfully. From a total of 486 CTG readings, only 26 are misdiagnosed. The AdaBoost confusion matrix shown in Fig. 3c indicates that out of 396 healthy fetus instances, 376 instances are predicted successfully, whereas 20 instances are misdiagnosed. Similarly, out of 72 suspected fetus cases, 51 are predicted accurately, and 21 are misdiagnosed. The 12 pathological instances are predicted successfully out of 18 total instances. From a total of 486 CTG readings, 47 are misdiagnosed. The confusion matrix of the CatBoost classifier is provided in Fig. 3d, which displays the healthy fetus instances. Of 396 total samples, 389 are predicted accurately, whereas 7 are misdiagnosed. Similarly, 60 suspected fetus cases are predicted successfully out of 72 cases, and 12 cases are misdiagnosed. There are 18 pathological instances where 16 are successfully predicted, and only 2 are misdiagnosed. From a total of 486 CTG readings, only 21 are misdiagnosed. The confusion matrix in Fig. 3e shows that out of 396 healthy fetus instances, the LGBM classifier could predict 391 samples successfully, while 5 were misdiagnosed. Similarly, out of 72 suspected fetus cases, 59 are predicted accurately, whereas 13 cases are misdiagnosed, and out of 18 pathological instances, 15 are predicted successfully, and 3 are misdiagnosed. From a total of 486 CTG readings, only 21 are misdiagnosed. The confusing matrix for the voting classifier of CTG records displayed in Fig. 3f indicates 396 healthy fetus instances; the VC classifier could predict 390 instances successfully, whereas 6 instances are misdiagnosed. Similarly, out of 72 suspected fetus cases, 60 are predicted accurately, and 12 are misdiagnosed. The 15 pathological instances out of 18 are predicted successfully. From a total of 486 CTG readings, only 21 are misdiagnosed.
Figure 3: Confusion matrix of selected classifier on imbalanced data
4.2 Classification with Balanced Data
Regarding balancing data, 2126 records are increased to 4803 CTG records via oversampling to balance the dataset. 3602 records are used for training, and 1201 for testing the models.
4.2.1 Random Forest (RF) Classifier
Table 8 summarizes the outcomes of the RF classifier for fetus health classification. When applied to the CTG data, the RF classifier yielded an overall accuracy of 98.0% and an AUROC equal to 98.6%, shown in Table 14. The precision and recall scores varied between 95% and 100% for all three classes. The precision and recall values clearly show that out of the 400 suspected fetuses, 95% are correctly identified as positive, whereas 99% and 100% are correctly identified as positive for the healthy and pathological classes, respectively.
4.2.2 Extreme Gradient Boosting (XGBoost) Classifier
XGBoost classifier applied to the CTG data yielded promising results with an overall accuracy of 0.99. Precision, F1-score, and Recall all showed promising outcomes for all three classes; as seen from Table 9.100% of the classified healthy and pathological instances are actually positive too. This indicates that XGBoost did not have any false positive classification for healthy and pathological class, i.e., did not falsely classify any instance as healthy or pathological. But this is different for the suspected class since 96% of those classified as suspected fetuses are correct. The rest belonged to another class. The recall also indicates that out of 400 suspected and 387 pathological fetuses, the XGBoost classifier can classify all positive instances correctly and did not misclassify these two classes as negative for any instance. Still, for the healthy class, only 96% of those that are actually healthy are classified as healthy; the remaining are misclassified. The AUROC indicated a 99.0% value in Table 14 which also shows an excellent categorical classification.
4.2.3 Adaptive Boosting (AdaBoost) Classifier
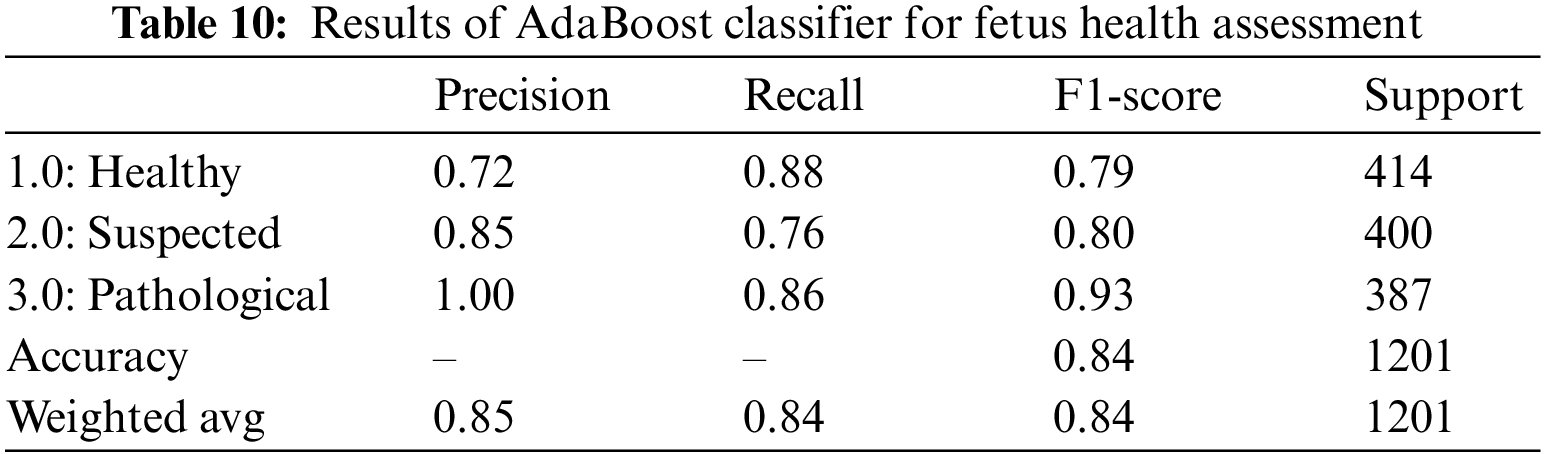
When applied to CTG data, the AdaBoost classifier yielded an overall accuracy of 84%. It is observed that the precision for healthy sample prediction is the lowest, i.e., 72%, while the precision of the prediction of the pathological fetus is 100%. The precision, recall, and F1-score for AdaBoost over CTG data are lower than other classifiers, and the AUROC value of 88.0% indicates that categorical classification is 88% effective. Hence, we observe that only 72% of those instances are classified as healthy, 85% of those classified as suspected are actually true for the class, and the remaining are wrongly classified. Similarly, by observing the recall value, we can see that 88% of 414 healthy records, 76% of the 40 suspected records, and 86% of the 387 pathological records are correctly classified; the rest are all falsely classified as negative. Table 10 is the classification report for the AdaBoost classifier for CTG data.
4.2.4 Categorical Boosting (CatBoost) Classifier
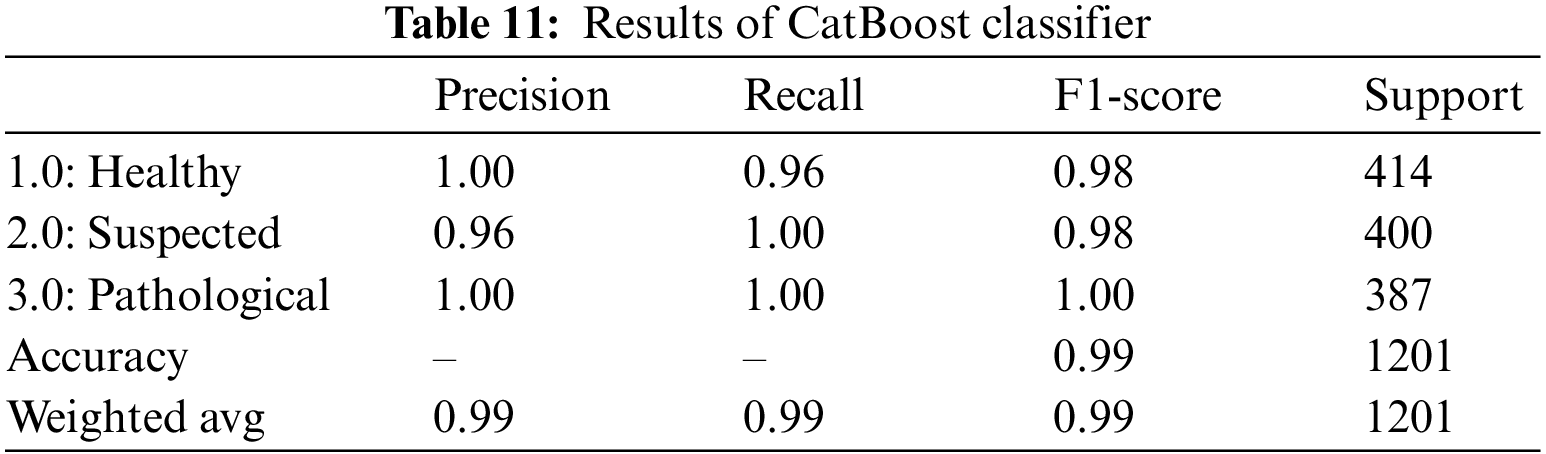
For CTG data CatBoost has shown quite promising results by yielding an overall accuracy of 99%. The classifier achieved a precision of 100% for healthy and pathological cases and a 100% recall rate for suspected and pathological cases, as shown in Table 11. These values represent the model’s accuracy in classifying all the actual positive instances of healthy and pathological. 96% of those instances classified as suspected fetuses belonged to the class, and the rest were falsely classified as “Suspected”. By observing the recall, we see that 100% of the actual suspected and pathological instances are correctly classified, but only 96% of the 414 healthy instances are correctly classified.
4.2.5 Light Gradient Boosting Machine (LGBM)
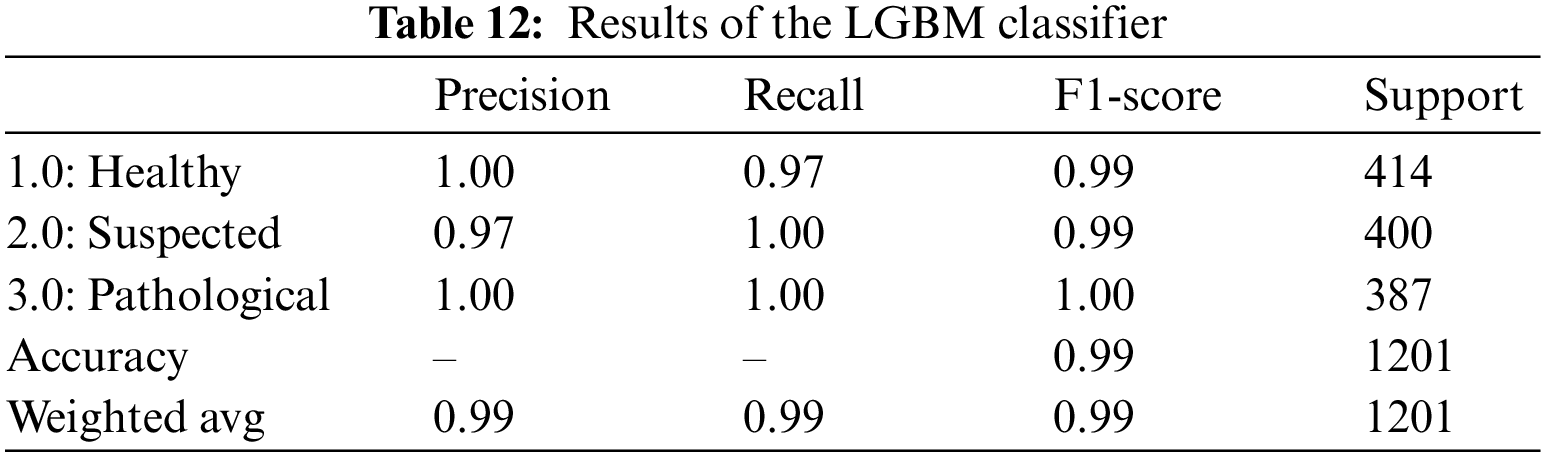
For CTG data, the LGBM classifier gave an overall 99% accuracy and AUROC value in Table 14. Precision, Recall, and F1-Score for all three classes are reasonably high, ranging from 97% to 100%. The precision value shows that all the instances marked as healthy and pathological are true for that class. In contrast, 97% of the instances predicted as suspected fetuses belonged to the suspected class; the rest are misclassified. Recall also shows that 97% of the actual healthy instances are classified correctly, and the rest are misclassified. Table 12 is the classification report for the model applied to CTG data.
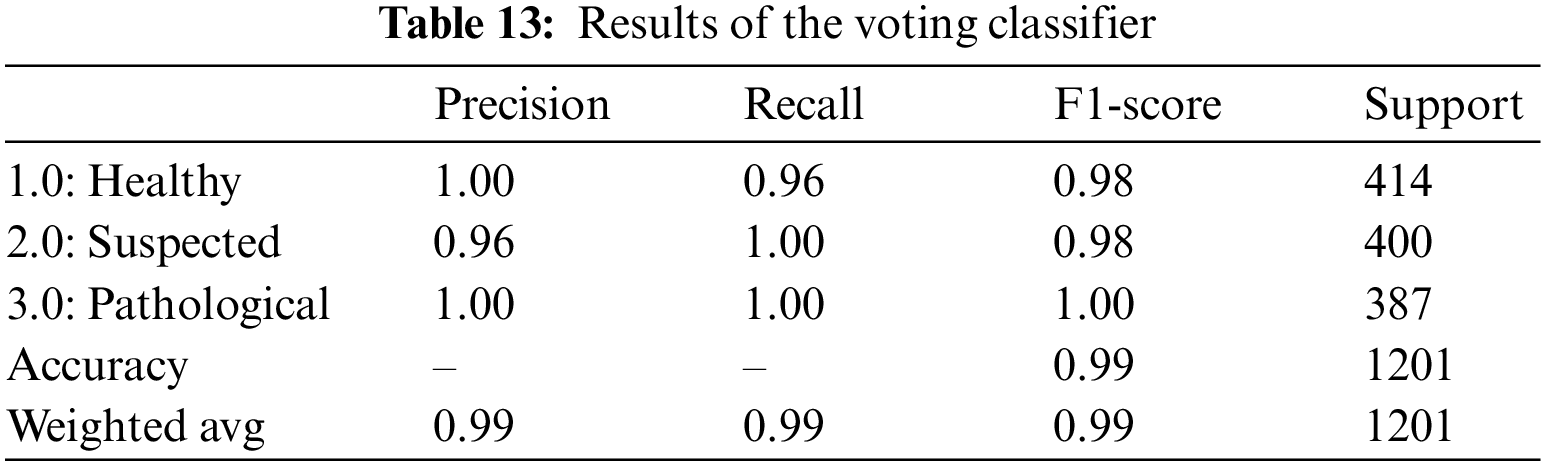
A soft voting mechanism is employed where the average outcome of all ensemble classifiers is used to classify CTG data. A soft voting classifier can be handy where different ensemble learners perform equally well. Averaging their performance can ensure that individual ensemble learners’ weaknesses can be balanced. As opposed to the hard-voting mechanism, which selects the prediction based on the majority votes, a soft-voting classifier incorporates further information, which includes the ability and inabilities of each learner’s classification techniques. The soft-voting classifier yielded an overall accuracy of 99%, shown in Table 13. It uses the outcome from all five ensemble learners to classify a record and yields the average of all 5 learners to predict the final classification. The precision of healthy and pathological and the recall for suspected and pathological CTG records is 100%. These values indicate that all those classified as healthy and pathological are true, but 96% of those classified as suspected are true for the class. Similarly, from the recall, we observe that all classified as suspected and pathological fetuses are correctly classified. Still, for the healthy class, 96% of those classified are only true, and the rest are wrongly classified. A 99.0% AUROC, shown in Table 14, is achieved via the voting classifier, establishing that the soft voting classifier gives an excellent categorical classification.
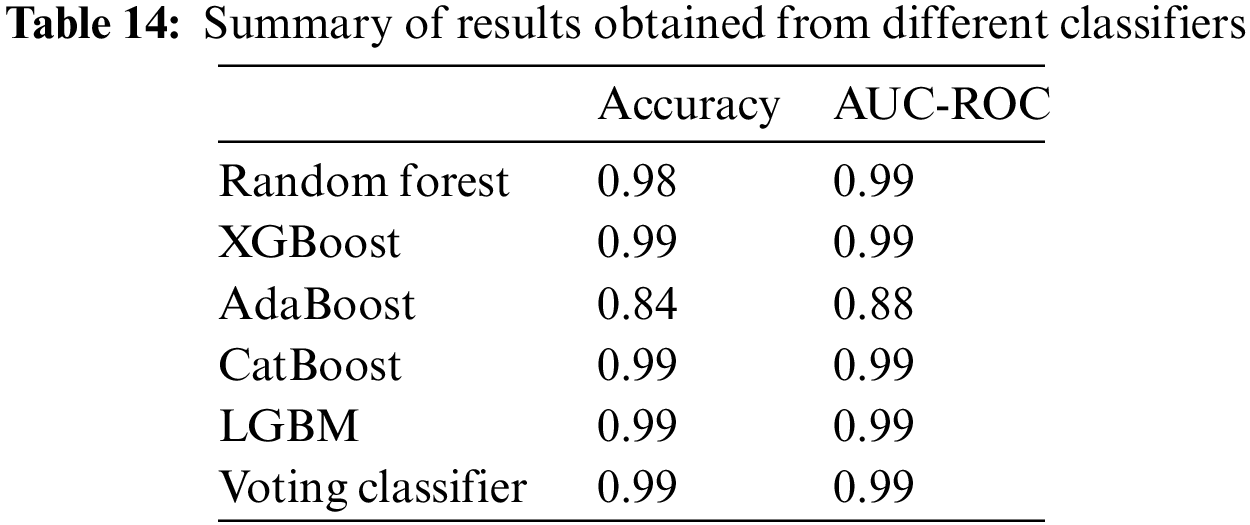
The confusion matrix in Fig. 4a shows that out of a total of 414 healthy fetus instances, the RF classifier could predict 393 instances successfully. Similarly, out of 400 suspected fetus cases, 398 are predicted accurately, and all pathological instances are predicted successfully. From a total of 1201 CTG readings, only 23 are misdiagnosed. The Confusion Matrix for XGBoost-based classification of CTG data displayed in Fig. 4b indicates that for the healthy class, the classifier predicted 96% of the instances correctly and misclassified only 16 out of a total of 414 healthy samples. It achieved 100% accuracy for the pathological and suspected cases and did not misclassify any instance. The AdaBoost classifier couldn’t classify any class values with 100% accuracy, as shown in the confusion matrix in Fig. 4c. 365 out of 414 healthy fetus instances are classified accurately, whereas 49 are misclassified. 24% of the suspected cases are misclassified, and only 304 out of 400 cases are correctly classified. 334 out of the 387 pathological instances are predicted accurately. The CatBoost classifier has misclassified only 18 out of 1201 CTG records, as seen in Fig. 4d. All suspected and pathological cases are correctly classified. The confusion matrix shows the almost 100% accurate prediction of all classes in Fig. 4e. The classifier misclassified 11 instances from a total of 1201 CTG records. All the cases for suspected CTG and pathological records are accurately classified into their respective classes. The confusion matrix for the voting classifier in Fig. 4f shows that it could accurately classify all the instances of suspected and pathological CTG records. The model can classify 96% of the CTG records as healthy, with only 17 out of 1201 instances that are misclassified.
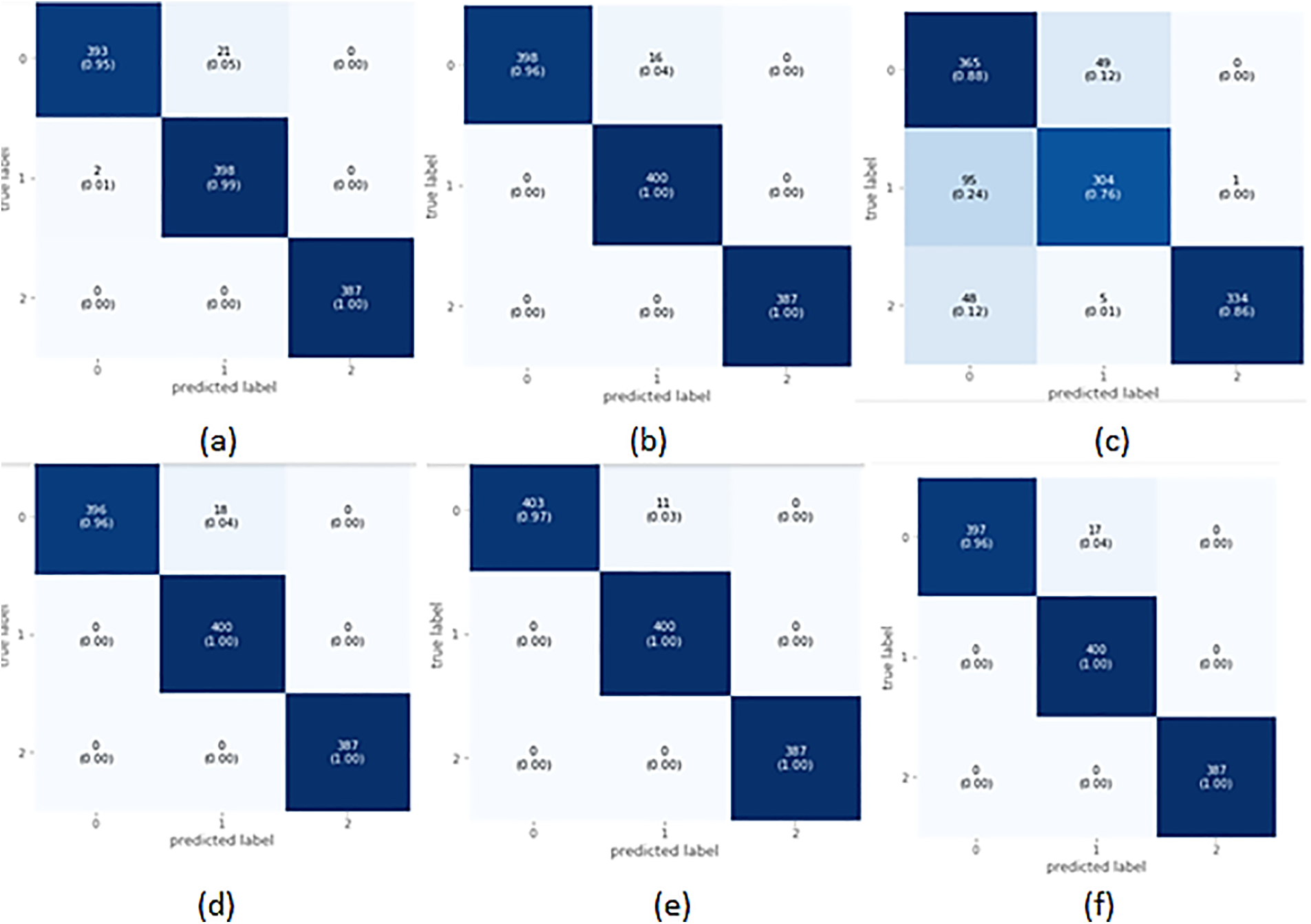
Figure 4: Confusion matrix of selected classifiers on balanced data
Fig. 5 illustrates the learning curve of the voting classifier. It demonstrates the effect of increasing training examples on the training and validation scores. There is no significant impact on the training and validation scores when the size of the dataset is increased beyond 3000. The voting classifier perfectly fit the training data and showed reasonably good accuracy for the validation data. The AUROC curve for the voting classifier, as seen in Fig. 6, shows an AUROC of 99.0%, indicating that our voting classifier has been very effective in classifying the data into the 3 categories of healthy, suspected abnormal, and pathological CTG readings. Since there is no significant difference in the outcomes of all the ensemble classifiers, our soft voting classifier, which yields the average of the results from the ensemble learners, also achieved a 99% accuracy and a 99.0% AUROC. It is also observed that most misclassifications occur when a healthy CTG record is labeled suspected. Due to the nominal difference between the two classes, a healthy record can easily be misclassified. The preprocessed and balanced dataset yielded excellent outcomes when passed to different ensemble learners. Almost all classifiers, except for the AdaBoost classifier, achieved a 99% accuracy and an AUROC of 99.0%, which can be observed in Table 14.
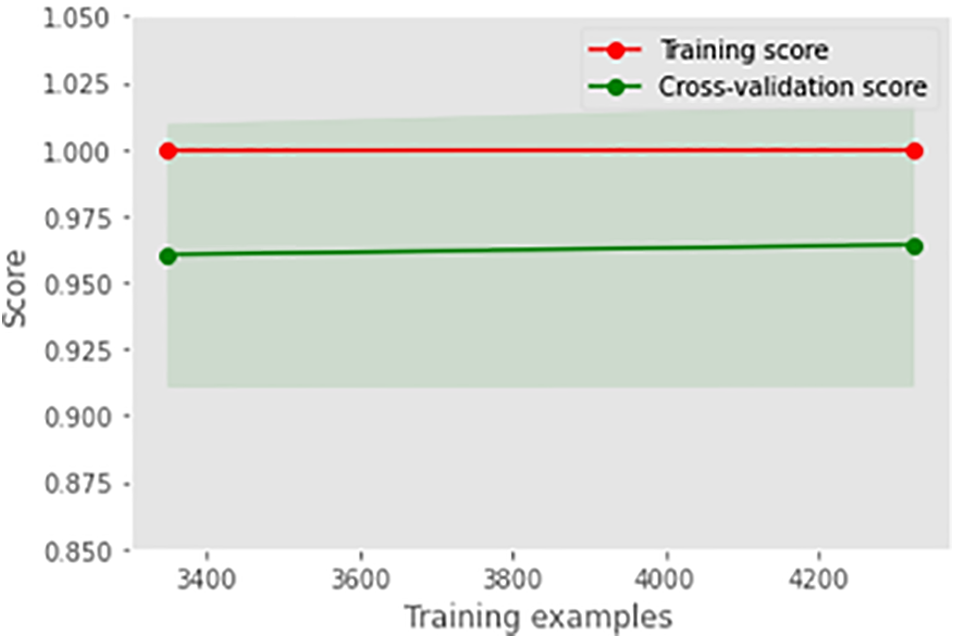
Figure 5: Learning curve of voting classifier
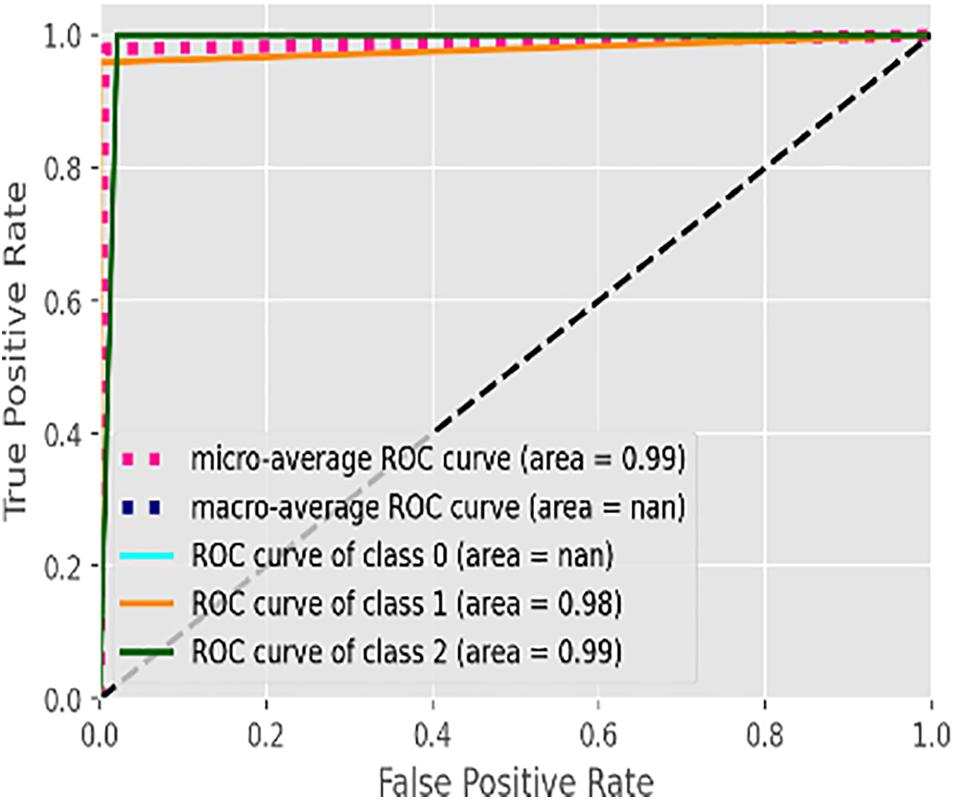
Figure 6: AUROC of voting classifier
Table 15 compares our results with state-of-the-art [40,18]. Authors in [40] achieved the best results of 94.5% using the SVM classifier. Similarly, authors in [18] achieved the best results of 95.9% using the blender model. It is noticed that the proposed approach works better in comparison with state-of-the-art studies.

We classified CTG records of pregnant women to monitor the fetus’s health and accurately and timely diagnose fetal stress to reduce lifelong fetal abnormalities and mortality. We proposed ensemble-based classification along with soft-voting classification. We employed the Random Forest, AdaBoost classifier, XGBoost classifier, CatBoost classifier, and LGBM classifier and compared their accuracies, AUC-ROC, and classification reports. It is observed that ensemble classifiers could yield promising results provided with accurate data. Results revealed that, except for the AdaBoost classifier achieved a 99% accuracy and an AUROC of 99.0%. Since there is no significant difference in the outcomes of all the ensemble classifiers, our soft voting classifier, which yields the average of the results from the ensemble learners, also achieved a 99% accuracy and a 99.0% AUC-ROC. It is also observed that most misclassifications occur where a healthy CTG record is labeled suspected. Due to the nominal difference between the two classes, a healthy record can easily be misclassified. We conclude that over-sampling to balance the CTG data provided the necessary accuracy to our dataset, making it easy to train and classify. Our ensemble models are quite precise in classifying the dataset, predicting whether a CTG record corresponds to healthy reading, suspected abnormal, or pathological. This work will help future researchers develop better models to differentiate between healthy and suspected CTG readings. A larger dataset will help train the models further and improve accuracy.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. K. Yammine, E. Mirel and E. Chahin, “Variations and morphometrics of palmaris longus in fetuses: A meta-analysis of cadaveric studies,” Surgical and Radiologic Anatomy, vol. 42, no. 3, pp. 281–287, 2020. [Google Scholar] [PubMed]
2. K. McNamara, S. Meaney and K. O’Donoghue, “Intrapartum fetal death and doctors: A qualitative exploration,” Acta Obstetricia et Gynecologica Scandinavica, vol. 97, no. 7, pp. 890–898, 2018. [Google Scholar] [PubMed]
3. S. Zhang, B. Li, X. Zhang, C. Zhu and X. Wang, “Birth asphyxia is associated with increased risk of cerebral palsy: A meta-analysis,” Frontiers in Neurology, vol. 11, pp. 704, 2020. [Google Scholar] [PubMed]
4. O. E. Shaw and J. Y. Yager, “Preventing childhood and lifelong disability: Maternal dietary supplementation for perinatal brain injury,” Pharmacological Research, vol. 139, pp. 228–242, 2019. [Google Scholar] [PubMed]
5. S. Das, H. Mukherjee, K. Santosh, C. K. Saha and K. Roy, “Periodic change detection in fetal heart rate using cardiotocograph,” in 2020 IEEE 33rd Int. Symp. on Computer-Based Medical Systems (CBMS), Rochester, MN, USA: IEEE, pp. 104–109, 2020. [Google Scholar]
6. A. A. Lovers, A. Ugwumadu and A. Georgieva, “Cardiotocography and clinical risk factors in early term labor: A retrospective cohort study using computerized analysis with oxford system,” Frontiers in Pediatrics, pp. 64, 2022. [Google Scholar]
7. P. Guleria, S. Ahmed, A. Alhumam and P. N. Srinivasu, “Empirical study on classifiers for earlier prediction of covid-19 infection cure and death rate in the Indian states,” Healthcare, vol. 10, no. 1, pp. 85, 2022. [Google Scholar] [PubMed]
8. M. Behera, A. Sarangi, D. Mishra, P. K. Mallick, J. Shafi et al., “Automatic data clustering by hybrid enhanced firefly and particle swarm optimization algorithms,” Mathematics, vol. 10, no. 19, pp. 3532, 2022. [Google Scholar]
9. S. Ahmed, P. Naga Srinivasu, A. Alhumam and M. Alarfaj, “Aal and internet of medical things for monitoring type-2 diabetic patients,” Diagnostics, vol. 12, no. 11, pp. 2739, 2022. [Google Scholar] [PubMed]
10. M. Asam, S. H. Khan, A. Akbar, S. Bibi, T. Jamal et al., “Iot malware detection architecture using a novel channel boosted and squeezed cnn,” Scientific Reports, vol. 12, no. 1, pp. 1–12, 2022. [Google Scholar]
11. A. K. Pradhan, J. K. Rout, A. B. Maharana, B. K. Balabantaray and N. K. Ray, “A machine learning approach for the prediction of fetal health using ctg,” in 2021 19th OITS Int. Conf. on Information Technology (OCIT), Bhubaneswar, India: IEEE, pp. 239–244, 2021. [Google Scholar]
12. J. Piri and P. Mohapatra, “Imbalanced cardiotocography data classification using resampling techniques,” in Proc. of Int. Conf. on Machine Intelligence and Data Science Applications, Springer, pp. 681–692, 2021. [Google Scholar]
13. J. Ogasawara, S. Ikenoue, H. Yamamoto, M. Sato, Y. Kasuga et al., “Deep neural network-based classification of cardiotocograms outperformed conventional algorithms,” Scientific Reports, vol. 11, no. 1, pp. 1–9, 2021. [Google Scholar]
14. N. Baghel, R. Burget and M. K. Dutta, “1D-fhrnet: Automatic diagnosis of fetal acidosis from fetal heart rate signals,” Biomedical Signal Processing and Control, vol. 71, no. 1, pp. 102794, 2022. [Google Scholar]
15. J. Li, Z. -Z. Chen, L. Huang, M. Fang, B. Li et al., “Automatic classification of fetal heart rate based on convolutional neural network,” IEEE Internet of Things Journal, vol. 6, no. 2, pp. 1394–1401, 2018. [Google Scholar]
16. Z. Zhao, Y. Deng, Y. Zhang, Y. Zhang, X. Zhang et al., “Deepfhr: Intelligent prediction of fetal acidemia using fetal heart rate signals based on convolutional neural network,” BMC Medical Informatics and Decision Making, vol. 19, no. 1, pp. 1–15, 2019. [Google Scholar]
17. Z. Cömert, A. F. Kocamaz and V. Subha, “Prognostic model based on image-based time-frequency features and genetic algorithm for fetal hypoxia assessment,” Computers in Biology and Medicine, vol. 99, no. 1, pp. 85–97, 2018. [Google Scholar]
18. J. Li and X. Liu, “Fetal health classification based on machine learning,” in 2021 IEEE 2nd Int. Conf. on Big Data, Artificial Intelligence and Internet of Things Engineering (ICBAIE), Nanchang, China: IEEE, pp. 899–902, 2021. [Google Scholar]
19. A. Mehbodniya, A. J. P. Lazar, J. Webber, D. K. Sharma, S. Jayagopalan et al., “Fetal health classification from cardiotocographic data using machine learning,” Expert Systems, vol. 39, no. 6, pp. e12899, 2022. [Google Scholar]
20. P. R. D. Intan, M. A. Maâsum, N. Alfiany, W. Jatmiko, A. Kekalih et al., “Ensemble learning versus deep learning for hypoxia detection in ctg signal,” in 2019 Int. Workshop on Big Data and Information Security (IWBIS). Bali, Indonesia: IEEE, pp. 57–62, 2019. [Google Scholar]
21. A. Subasi, B. Kadasa and E. Kremic, “Classification of the cardiotocogram data for anticipation of fetal risks using bagging ensemble classifier,” Procedia Computer Science, vol. 168, pp. 34–39, 2020. [Google Scholar]
22. Y. Chen, A. Guo, Q. Chen, B. Quan, G. Liu et al., “Intelligent classification of antepartum cardiotocography model based on deep forest,” Biomedical Signal Processing and Control, vol. 67, no. 2, pp. 102555, 2021. [Google Scholar]
23. N. Rahmayanti, H. Pradani, M. Pahlawan and R. Vinarti, “Comparison of machine learning algorithms to classify fetal health using cardiotocogram data,” Procedia Computer Science, vol. 197, pp. 162–171, 2022. [Google Scholar]
24. Kaggle, “Fetal health classification,” 2000. [Online]. Available: https://www.kaggle.com/datasets/andrewmvd/ [Google Scholar]
25. D. Ayres-de Campos, J. Bernardes, A. Garrido, J. Marques-de Sa and L. Pereira-Leite, “Sisporto 2.0: A program for automated analysis of cardiotocograms,” Journal of Maternal-Fetal Medicine, vol. 9, no. 5, pp. 311–318, 2000. [Google Scholar] [PubMed]
26. X. Dong, Z. Yu, W. Cao, Y. Shi and Q. Ma, “A survey on ensemble learning,” Frontiers of Computer Science, vol. 14, no. 2, pp. 241–258, 2020. [Google Scholar]
27. O. Sagi and L. Rokach, “Ensemble learning: A survey,” Wiley Interdisciplinary Reviews: Data Mining and Knowledge Discovery, vol. 8, no. 4, pp. e1249, 2018. [Google Scholar]
28. A. Campagner, D. Ciucci and F. Cabitza, “Aggregation models in ensemble learning: A large-scale comparison,” Information Fusion, vol. 90, pp. 241–252, 2023. [Google Scholar]
29. V. Jackins, S. Vimal, M. Kaliappan and M. Y. Lee, “Ai-based smart prediction of clinical disease using random forest classifier and naive bayes,” The Journal of Supercomputing, vol. 77, no. 5, pp. 5198–5219, 2021. [Google Scholar]
30. R. -C. Chen, R. E. Caraka, N. E. G. Arnita, S. Pomalingo, A. Rachman et al., “An end to end of scalable tree boosting system,” Sylwan, vol. 165, no. 1, pp. 1–11, 2020. [Google Scholar]
31. F. Wang, Z. Li, F. He, R. Wang, W. Yu et al., “Feature learning viewpoint of adaboost and a new algorithm,” IEEE Access, vol. 7, pp. 149 890–149 899, 2019. [Google Scholar]
32. G. Huang, L. Wu, X. Ma, W. Zhang, J. Fan et al., “Evaluation of catboost method for prediction of reference evapotranspiration in humid regions,” Journal of Hydrology, vol. 574, pp. 1029–1041, 2019. [Google Scholar]
33. B. S. Ahamed and M. S. Arya Dr., “Prediction of type-2 diabetes using the lgbm classifier methods and techniques,” Turkish Journal of Computer and Mathematics Education (TURCOMAT), vol. 12, no. 12, pp. 223–231, 2021. [Google Scholar]
34. S. Kumari, D. Kumar and M. Mittal, “An ensemble approach for classification and prediction of diabetes mellitus using soft voting classifier,” International Journal of Cognitive Computing in Engineering, vol. 2, pp. 40–46, 2021. [Google Scholar]
35. D. Agnihotri, K. Verma, P. Tripathi and B. K. Singh, “Soft voting technique to improve the performance of global filter based feature selection in text corpus,” Applied Intelligence, vol. 49, no. 4, pp. 1597–1619, 2019. [Google Scholar]
36. F. Cabitza and A. Campagner, “The need to separate the wheat from the chaff in medical informatics: Introducing a comprehensive checklist for the (self)-assessment of medical ai studies,” International Journal of Medical Informatics, vol. 153, pp. 104510, 2021. [Google Scholar] [PubMed]
37. K. Yammine, E. Mirela and C. Assi, “Variations and morphometrics of palmaris longus in fetuses: A meta-analysis of cadaveric studies,” Surgical and Radiologic Anatomy, vol. 42, no. 3, pp. 281–287, 2020. [Google Scholar] [PubMed]
38. F. Srour and S. Karkoulian, “Exploring diversity through machine learning: A case for the use of decision trees in social science research,” International Journal of Social Research Methodology, vol. 25, no. 6, pp. 725–740, 2022. [Google Scholar]
39. A. Khazzaka, R. Elie, Z. Sleiman, S. Boussios and N. Pavlidis, “Systematic review of fetal and placental metastases among pregnant patients with cancer,” Cancer Treatment Reviews, pp. 102356, 2022. [Google Scholar]
40. A. Mehbodniya, A. J. P. Lazar, J. Webber, D. K. Sharma, S. Jayagopalan et al., “Fetal health classification from cardiotocographic data using machine learning,” Expert Systems, vol. 39, no. 6, pp. e12899, 2022. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools