 Open Access
Open Access
ARTICLE
Real-Time Multi-Feature Approximation Model-Based Efficient Brain Tumor Classification Using Deep Learning Convolution Neural Network Model
1 Department of Computer Science and Engineering, School of Computing, SRM Institute of Science and Technology, Kattankulathur, Chengalpattu, Chennai, 603203, Tamilnadu, India
2 Department of Information Technology, MLR Institute of Technology, Hyderabad, 500043, Telangana, India
3 Department of Computing Technologies, School of Computing, SRM Institute of Science and Technology, Kattankulathur, Chengalpattu, Chennai, 603203, Tamilnadu, India
* Corresponding Author: M. Baskar. Email:
Computer Systems Science and Engineering 2023, 46(3), 3883-3899. https://doi.org/10.32604/csse.2023.037050
Received 21 October 2022; Accepted 24 February 2023; Issue published 03 April 2023
Abstract
The deep learning models are identified as having a significant impact on various problems. The same can be adapted to the problem of brain tumor classification. However, several deep learning models are presented earlier, but they need better classification accuracy. An efficient Multi-Feature Approximation Based Convolution Neural Network (CNN) model (MFA-CNN) is proposed to handle this issue. The method reads the input 3D Magnetic Resonance Imaging (MRI) images and applies Gabor filters at multiple levels. The noise-removed image has been equalized for its quality by using histogram equalization. Further, the features like white mass, grey mass, texture, and shape are extracted from the images. Extracted features are trained with deep learning Convolution Neural Network (CNN). The network has been designed with a single convolution layer towards dimensionality reduction. The texture features obtained from the brain image have been transformed into a multi-dimensional feature matrix, which has been transformed into a single-dimensional feature vector at the convolution layer. The neurons of the intermediate layer are designed to measure White Mass Texture Support (WMTS), Gray Mass Texture Support (GMTS), White Mass Covariance Support (WMCS), Gray Mass Covariance Support (GMCS), and Class Texture Adhesive Support (CTAS). In the test phase, the neurons at the intermediate layer compute the support as mentioned above values towards various classes of images. Based on that, the method adds a Multi-Variate Feature Similarity Measure (MVFSM). Based on the importance of MVFSM, the process finds the class of brain image given and produces an efficient result.Keywords
Human society faces different health challenges. Several diseases are identified every year. Among them, most of the diseases are seasonal, but some of them are not. Also, most infections are curable by providing a set of treatments. However, a few diseases are identified as dangerous and harmful to health. Brain tumors are the most challenging diseases that have been identified and affect the health of humans. It has been recognized by monitoring and analyzing the brain images captured by several devices like Cathode Tomography (CT) and MRI.
The brain is the master component of the human body that controls the activity of the entire human body. It is identified as a collection of cells or neurons that have specific operations. It has been occupied by limited space in the skull. The presence of tumors in the brain is identified by identifying the presence of foreign bodies in and around the brain cells. The tumor is generally classified as malignant or benign, where malignant shows cancer cells, but harmless shows just damaged cells.
The tumor starts from specific cells in the brain and migrates to other cells as metastatic. The tumor will spread to the spinal cord in a few cases. A brain tumor diagnosis is performed by capturing CT and MRI images of the head, spine, and brain. In some patients, the cancer tumor will also spread to the spinal cord. Brain cancer treatment will depend on the size or type of tumor and its growth. Treatment includes surgery, radiation therapy, chemotherapy, targeted biological therapy, or a combination there of.
The presence of a brain tumor is identified by using several techniques. Still, the applications of image processing techniques are enormous, and they have been adapted for several problems. Several approaches are available and adapted to brain tumor image classification problems. Clustering and segmentation techniques, for example, are widely applied to the problem of brain image classification. Support Vector Machines (SVM), Neural Networks (NN), Convolution Networks, and Decision Trees (DT) are used on several occasions for image classification. Still, the classification performance is greatly based on the method of similarity measurement and the features used.
Deep learning models have recently been identified as solutions to classification problems. It supports high dimensionality, big data, and dimension reduction. The robust support system would use images from various regions and patients for brain image classification. This increases the volume and size of images, which challenges the classification process. The deep learning models are better at handling such big data with higher dimensionality.
The brain tumor is named an anomaly, representing a tumor’s presence in the brain cells. This research focuses on anomaly detection, which may lead to cancer or any death-causing disease. The detection of such an anomaly has the potential to reduce the radiologist’s workload. Attention deficit and learning disorders, autism, brain tumors, cerebral palsy and movement disorders, epilepsy, and sleep disorders can all cause such anomalies. These anomalies can be found through many imaging techniques like CT or MRI images.
This article discusses a novel multi-feature approximation-based brain tumor classification approach with CNN. The problem of brain image classification can be handled by approximating feature values of various features towards different classes of the brain image. The method is focused on considering various features like grey mass, white mass, texture, and shape features. By approximating the values of such features, the process would compute the value of the Multi Variant Feature similarity Measure (MVFSM), which has been measured by computing White Mass Texture Support (WMTS), Gray Mass Texture Support (GMTS), White Mass Covariance Support (WMCS), Gray Mass Covariance Support (GMCS), and Class Texture Adhesive Support (CTAS). The value of MVFSM has been used in identifying the class of brain images. The detailed approach is discussed in detail in this section.
Researchers recommend several approaches for brain tumor classification and anomaly detection. Such methods for brain image classification are detailed in this section.
The tumors in T1-weighted contrast-enhanced MRI images are approached with a CNN-based model, which preprocesses the images using CNN and extracts features to perform classification [1]. To improve the classification accuracy, a Deep CNN-based model is presented, which finds and predicts the status of gliomas at different mutations [2]. In [3], an IoT-based deep learning model is adapted for detecting brain tumors. It combines CNN and Short Term Memory (CNN-LSTM) in extracting the features and classifying them into various classes. In [4], the author presents a metastasis segmentation scheme with a CNN model classification, which segments the image for metastases, and CNN is used towards classification. A multi-scale CNN model is presented for Gilomas brain tumor classification, which preprocesses the image by applying the intensity normalization technique and enhances the image contrast. Further, the image features are trained with the 3d network, and data augmentation schemes are used for brain image classification [5].
A novel semantic segmentation-based classification model with CNN is presented to support the classification of brain cells and images in [6], which segments the image according to the semantic segmentation network and classifies the image using Googles-Net. Similarly, a DCNN-based tumor segmentation and classification scheme with SVM is named (DCNN-F-SVM). The network is trained with the features initially, and the predicted values of samples are used for classification with the SVM [7]. In [8], a deep auto-encoder-oriented classification model is presented, which uses spectral data augmentation. The method applies Discrete Wavelet Transform (DWT) in data reduction and extracts the features for classification.
The classification problem is approached with CNN in [9], which enhances the image by equalizing the histogram and segments the image using U-Net. Furthermore, the classification is performed using the 3D-CNN model. In [10], the author presents a detailed review and identifies the several challenges in classifying brain images into various classes. In [11], the author introduced a minutiae propagation-based forgery detection scheme which classifies the fingerprint images according to the minutiae features like joints, bifurcation, etc. Forgery detection has been performed according to the features extracted [12] presents an efficient approach for classifying brain images that apply DWT in normalizing the image. Further, principle component analysis is applied towards feature selection where classification is performed with Feed Forward Neural Networks (FF-ANN) and Back-Propagation Neural Networks (BPNN). The classifiers have been used to classify subjects as normal or abnormal MRI brain images. In [13], the author presented a human face detection and recognition scheme which uses contour features and performs classification according to the similarity of the contour features.
In [14], an artificial intelligence-based tumor classification approach is discussed, which normalizes the brain image and applies an (Advanced Driver Assistance Systems) ADAS optimization function with Deep CNN towards classification. In [15], a multi-source correlation network is presented to learn the multi-source correlation towards handling missing data in brain image classification. The model uses a multi-source correlation network that understands the correlated features to improve the segmentation, which improves the classification performance. In [16], a Generative Adversarial Network (GAN)-based augmentation technique called BrainGAN is discussed, automatically checking the augmented images for the presence of brain tumors.
In [17], a deep learning model called DenseNet 201 is presented, which has been combined with Support Vector Machines (SVM). In [18], they focused on classifying images against three classes of tumors, which adopt the concept of deep transfer learning and use a pre-trained GoogLeNet to extract features from brain MRI images. According to the features extracted, classification is performed with traditional classifiers. The problem of brain image classification is approached with a multi-view dynamic fusion framework in [19], which constructs a deep neural network with the outcome of segmentation results.
In [20], they use a pre-trained deep CNN model and propose a block-wise fine-tuning strategy based on transfer learning. In [21], an efficient automated segmentation mathematical model is presented over a Deep Neural Network, which uses U-Net in segmenting the images to support classification. In [22], they apply transfer learning techniques towards brain tumor classification, which trains the features with CNN, and classification is performed with SVM. In [23], an efficient Optimized Threshold Difference (OTD) and Rough Set Theory (RST) based automatic segmentation model is presented. The ROI is detected with a two-level segmentation algorithm. Features are extracted from the segmented images using the Gray-Level Co-occurrence Matrix (GLCM). RST is employed with the extracted features towards classification. A Bat with Fuzzy C-ordered Means (BAFCOM) based approach is discussed in [24] towards classifying the brain images. A Mutual Information Accelerated Singular Value Decomposition (MI-ASVD) model is presented in [25], which produces Grey-Level Run-Length Matrix (GLRLM), texture, and color intensity features towards classification with a neural network.
All the above-discussed approaches fail to achieve the expected accuracy in classifying the brain images against the different classes considered.
3 Multi-Feature Approximation with CNN-Based Brain Tumor Classification Model (MFA-CNN)
The proposed multi-feature white mass similarity-based brain tumor classification model starts with the input 3D MRI images and applies Gabor filters at multiple levels. The application of the Gabor filter eliminates the noise introduced by the capturing device. Further, the quality of the image is improved by equalizing the histogram. The grey and white mass features, the tumor’s texture, and the tumor’s shape features are extracted from the improved image. Such features that are removed are trained with the convolution neural network. The neurons estimate White Mass Texture Support (WMTS), Gray Mass Texture Support (GMTS), White Mass Covariance Support (WMCS), Gray Mass Covariance Support (GMCS), and Class Texture Adhesive Support (CTAS). Using these values, the method computes Multi Variant Feature Support Measure (MVFSM) to identify the class of brain image given. The detailed approach is discussed in this section.
The MFA-CNN model architecture is presented in Fig. 1, and the functional modules are described in detail in this section.
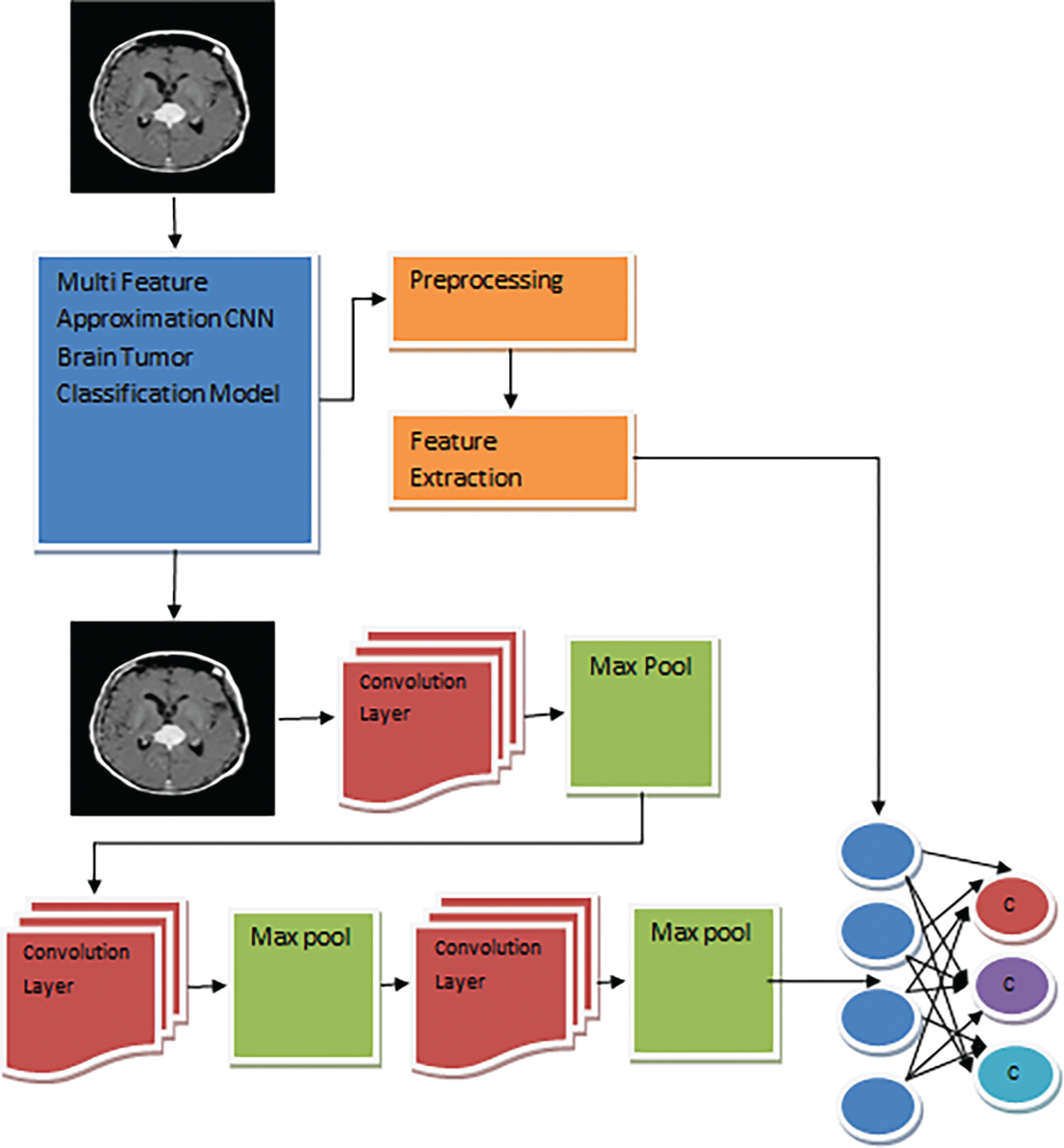
Figure 1: MFA-CNN model architecture
The brain image data set has been used for training and testing. At this stage, the method reads the brain image and applies the Gabor filter at multiple levels. Applying the Gabor filter eliminates the noise from the image and applies histogram equalization. The histogram equalization process improves the quality of the image, which supports classification accuracy.
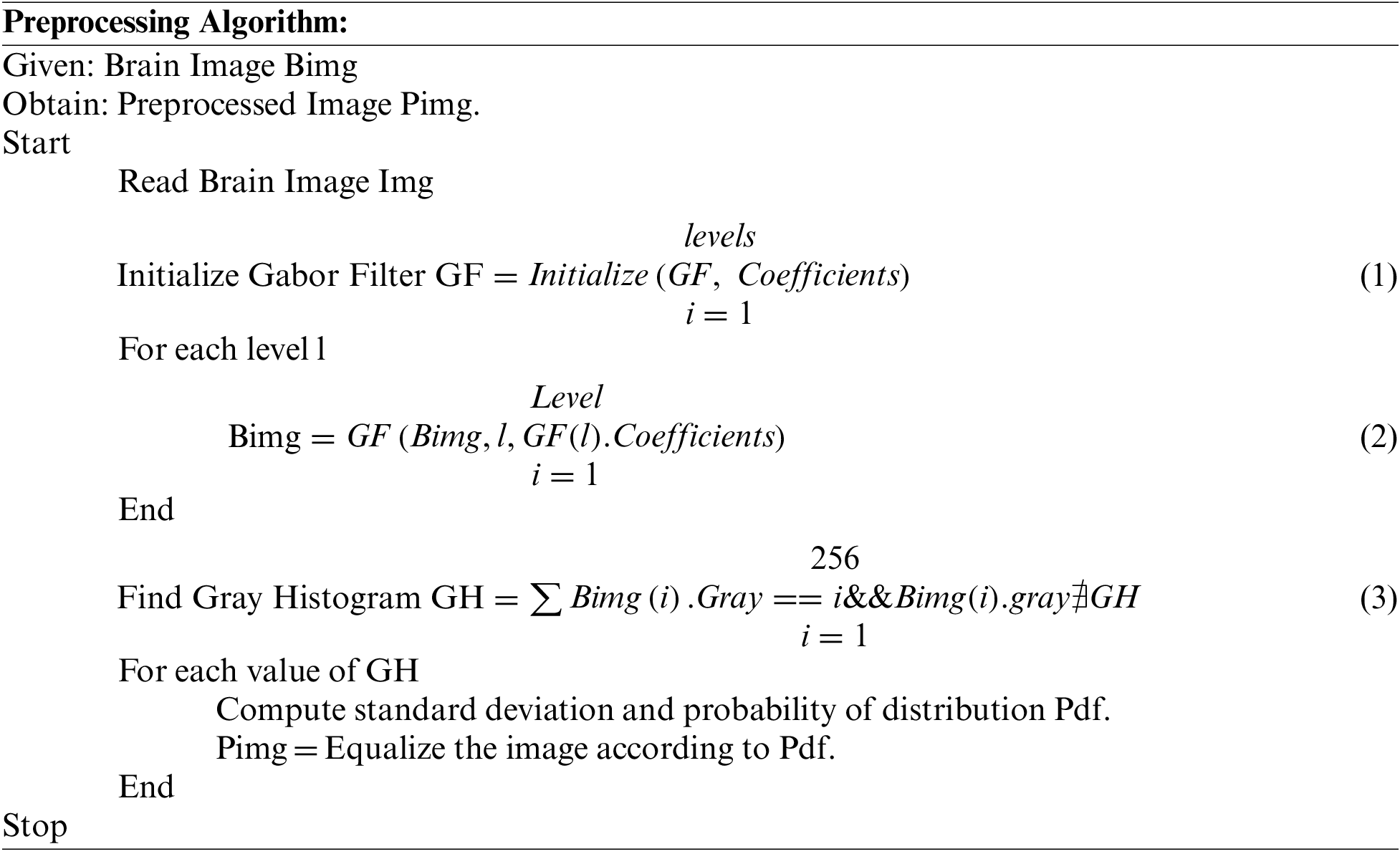
The preprocessing scheme reads the brain image and applies the Gabor filter at multiple levels of the picture. Further, to improve the quality of the image, the histogram of the image is equalized.
The features of the brain image are extracted in multiple forms. First, the method applies the Gray Threshold Segmentation technique to perform this. The Gray threshold segmentation approach first identifies the maximum white pixel value according to the histogram generated. From the histogram values, the top 3 levels are selected from the histograms. Now, according to the importance of the histogram and the greyscale, the method groups the pixels to perform segmentation. The method extracts the shape, texture, white mass, and grey mass values from the segmented region. Extracted features are used to train the network and to perform brain image classification.

The feature extraction scheme discussed above explains the ways of feature extraction applied to the brain image. The method uses the grey threshold segmentation technique, which groups the pixels according to the grey scale values. The process extracts the shape and texture features to generate the feature vector according to the segmentation result. A generated feature vector has been used to perform brain image classification.
The problem of brain image classification is approached according to the deep learning model. The proposed model with CCN has several convolution layers and pooling layers. It has three layers for convolving the features and three for pooling them. First, the method generates the neural network with a specified number of layers. First, the features of the image are extracted according to the feature extraction model given.
Further, the shape and texture features are fed through the model design, which applies convolve and max-pooling operations to reduce shape and texture features. Both texture and shape features are reduced in their dimension and converted into one-dimensional arrays with a size of 11000. Such reduced features are given to the neural network layers of neurons. Each brain image of various classes is convolved and given to the neurons at the training phase, which is also combined with the grey and white mass values. Such a trained network has been used to perform testing in the classification phase.

The above discussed represents how the CNN training has been performed. The model extracts the features and initializes the network. According to the network, the method applies convolution and pooling to reduce the size of the feature vector to support the classification problem. The process involves the filters in varying numbers from 32 to 128. This reduces the size of features to a great extent, whereas the size of the filter is the same at each level.
The testing phase covers most of the procedure, which reads the input test image and preprocesses the image to eliminate the noise and improve the image quality. As the MRI image is three-layered, the method extracts the features of the image as shape, texture, and mass values. Further, the process applies convolution and max pooling by using different filters, which reduce the size of features and convert them into a single array of a thousand indexes. The neurons at the intermediate layer compute the values of White Mass Texture Support (WMTS), Gray Mass Texture Support (GMTS), White Mass Covariance Support (WMCS), Gray Mass Covariance Support (GMCS), and Class Texture Adhesive Support (CTAS). Using these values, the method estimates the Multi-Feature Similarity Measure (MFSM). Based on the importance of MFSM, a single class is identified and labeled as a result. According to the value of MFSM, the method classifies the input test image.
Consider the test image given as T. Then the features extracted are Tf, Sf, Gm, and Wm, where Tf is the texture feature, sf is the shape feature, Gm is the grey mass, and Wm is the white mass value. Also, the testing scheme trains the network with convolution and produces a single feature vector Fv of size 11000 by applying different convolution and pooling operations. The method estimates the weight of the given test image in the testing phase. With the weight measures, the process uses the feature vector generated. Now, the method computes the value of similarity in various features for different classes of brain tumors.
First, the method computes the value of White Mass Texture Support (WMTS) as follows:
To perform this, first, the method computes White Mass Similarity (WMS) as follows:
Now the value of WMTS is measured as follows:
Second, the method computes the value of Gray Mass Texture Support (GMTS) as follows:
To measure the value of GMTS, the method first computes the value of grey mass similarity (GMS) as follows:
Now the value of GMTS is measured as follows:
Third, the method computes the value of Gray Mass Covariance Support (GMCS) as follows:
The value of gray mass covariance support is measured by computing the variance of pixels with the required value. It has been measured as follows:
Fourth, the method computes the value of White Mass Covariance Support (WMCS).
First, the value of Class Texture Adhesive Support (CTAS) is measured as follows:
To measure the value of CTAS, the method first computes the TFS (Texture Feature Similarity) value as follows:
The value of Texture feature similarity (TFS) is measured as follows:
Similarly, the value of Shape feature similarity (SFS) is measured as follows:
Using all these measures, the method computes the value of MFSM as follows:
According to the value of MFSM, the method classifies the brain tumor image into various classes.
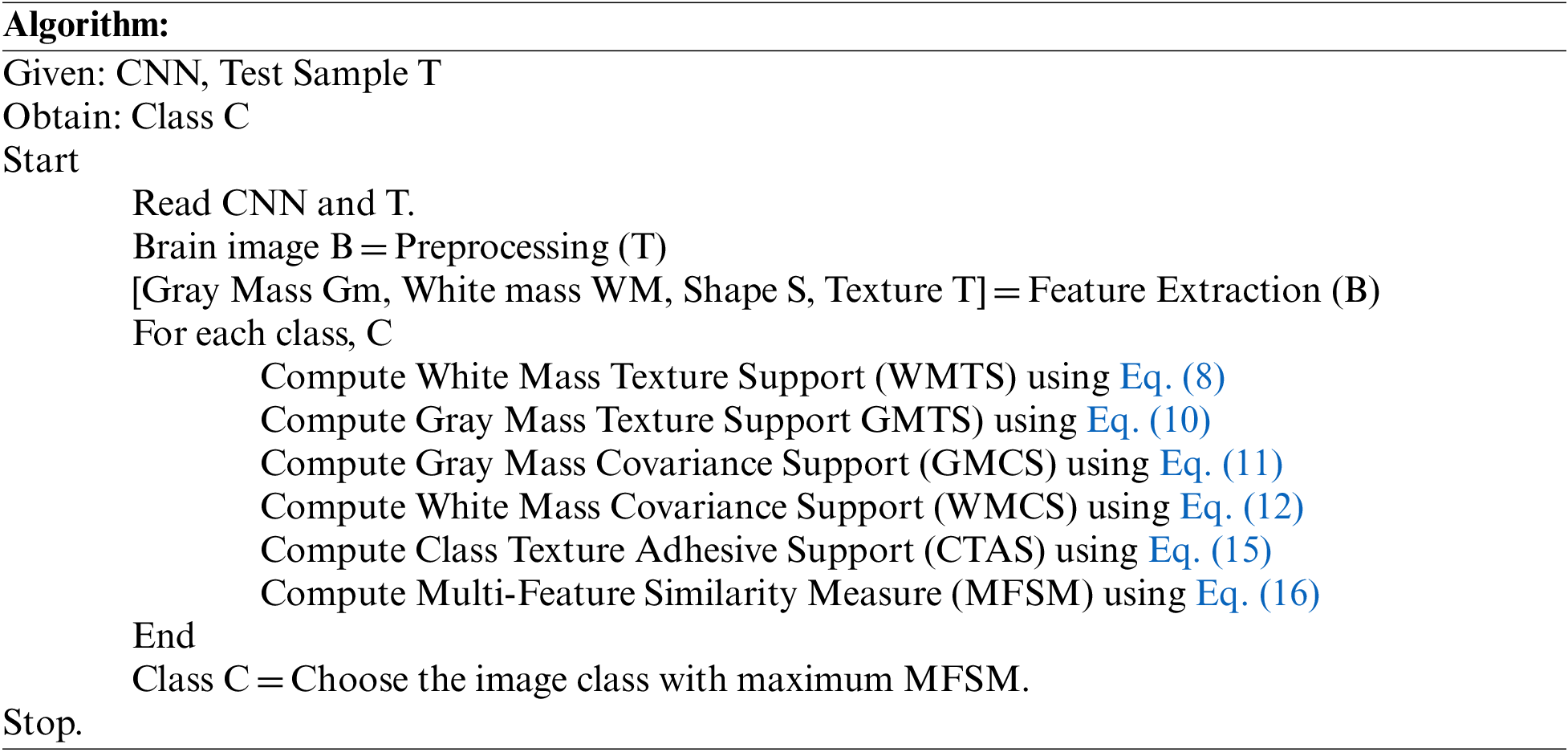
The above-discussed algorithm estimates various measures on different features to compute an MFSM value, based on which a single class with maximum MFSM is selected.
4 Experimental Results and Analysis
The proposed MFA-CNN model for brain image classification has been implemented in Python. The method has been evaluated for its performance in various parameters. The process has been measured for efficiency under multiple parameters using a brain tumor data set. This section presents the evaluation results in detail.
The details of the data set and tools used to evaluate the performance of the brain image classification algorithm are presented in Table 1. The Figshare data set has a total of 3064 images of T1-weighted contrast-enhanced photos belonging to three classes of brain tumors: meningioma, glioma, and pituitary tumor. The performance is measured on different parameters and presented in this section.
The result of segmentation and classification produced by the proposed approach is presented in Table 2, which exactly classifies the brain images with higher accuracy.
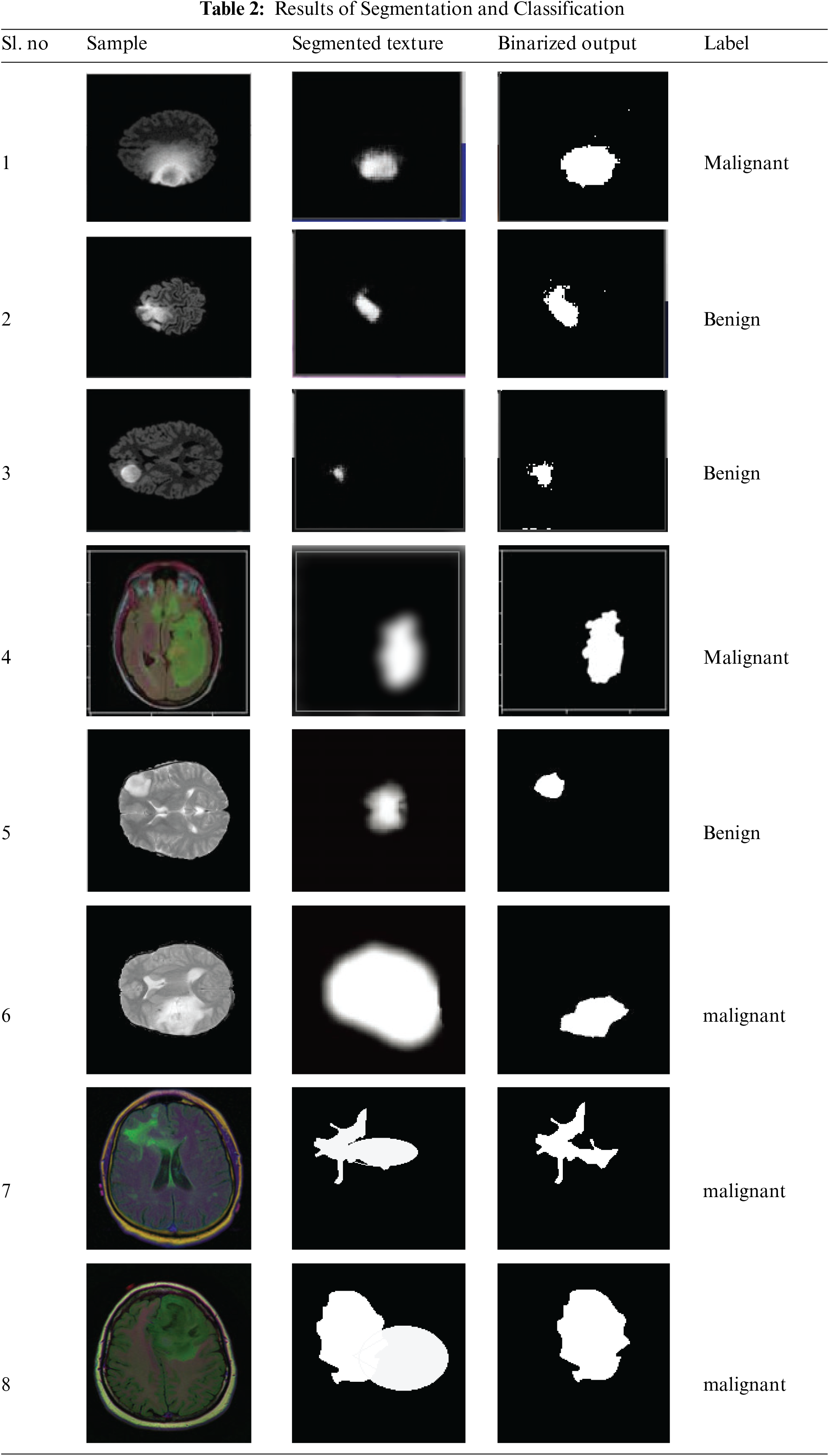
The performance of methods detecting the anomaly from the brain images of different classes is measured and presented in Table 3. The proposed MFA-CNN algorithm has produced higher accuracy in anomaly detection in each brain tumor class.
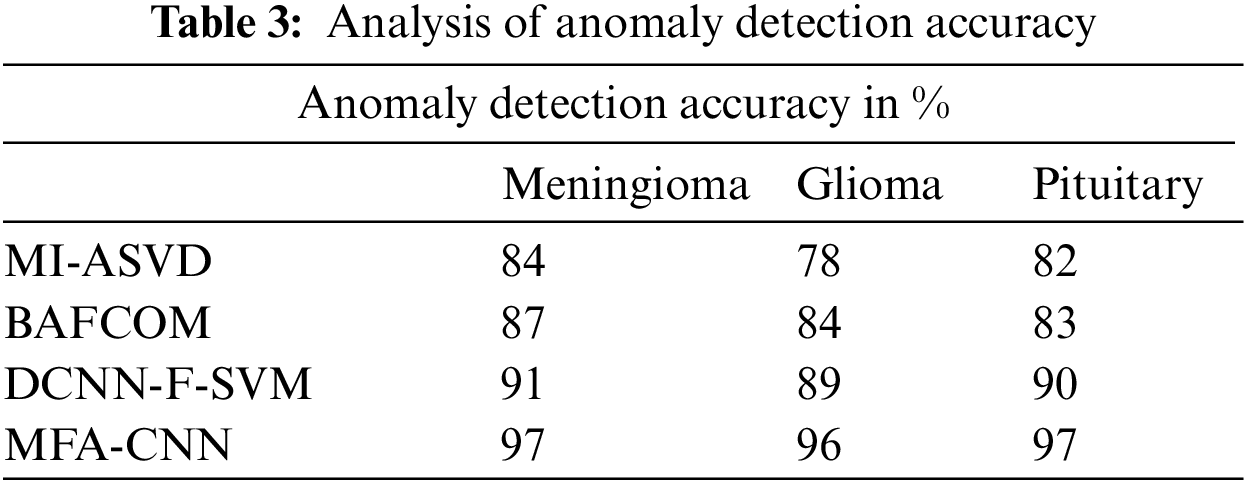
The accuracy in detecting the anomaly from the 3D MRI image has been presented in Fig. 2, which measures the performance of various methods and compares them in the above chart. From the analysis, it is clear that the proposed MFA-CNN model has achieved greater effectiveness than the rest of the models in detecting the anomaly from the brain tumor image.
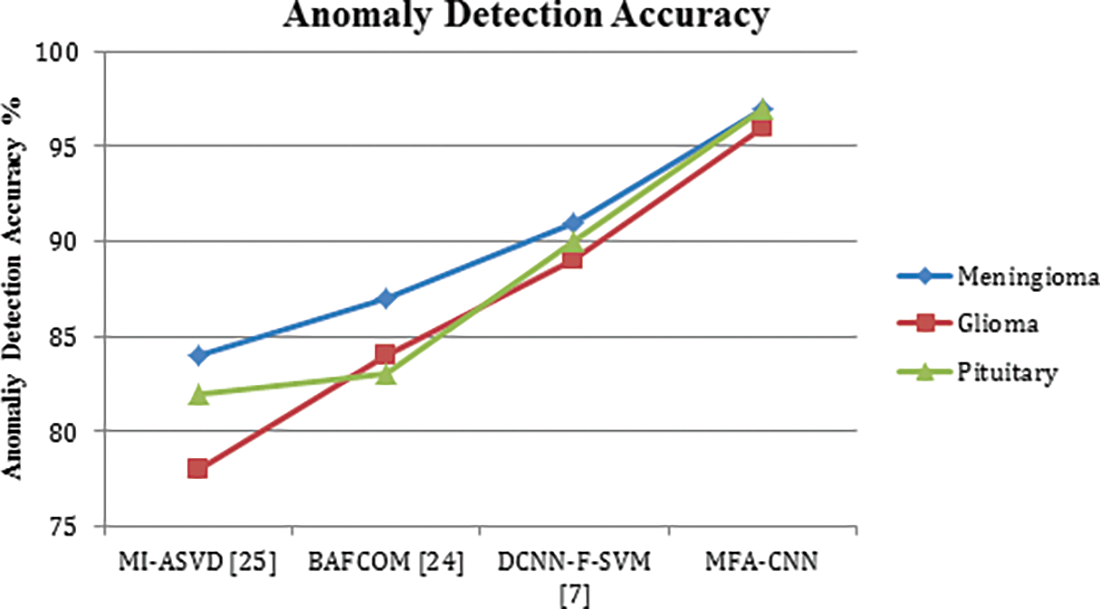
Figure 2: Analysis of anomaly detection accuracy
The ratio of false classification in anomaly detection has been measured for various methods towards three different classes of tumors and compared in Table 4. In each case, the proposed MFA-CNN has produced the lowest false ratio compared to other approaches.
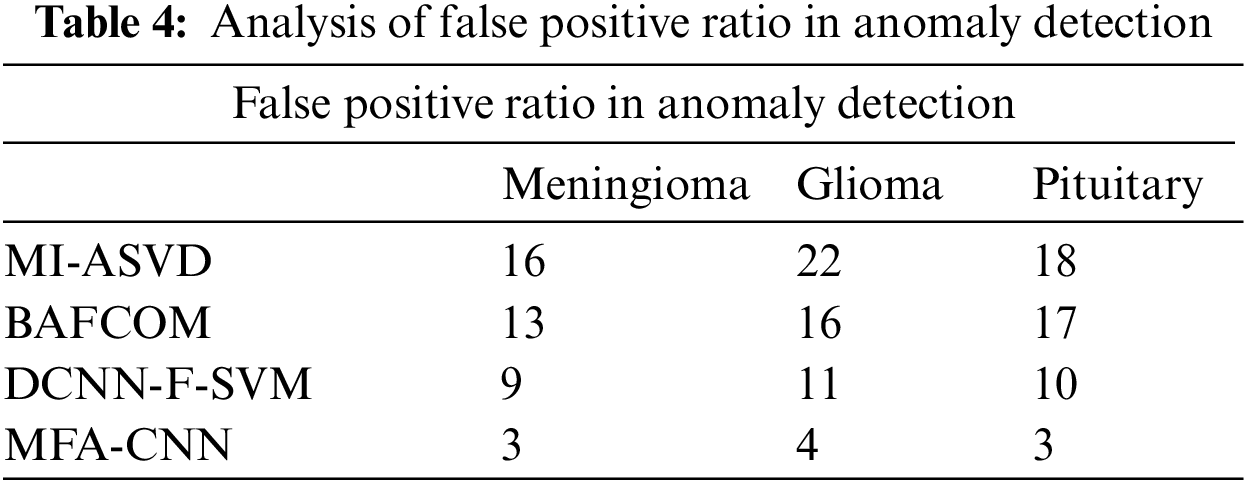
The false positive ratio introduced by different methods in detecting the anomaly has been presented in Fig. 3, where the proposed MFA-CNN model has produced a lower false positive ratio compared to all the other approaches.
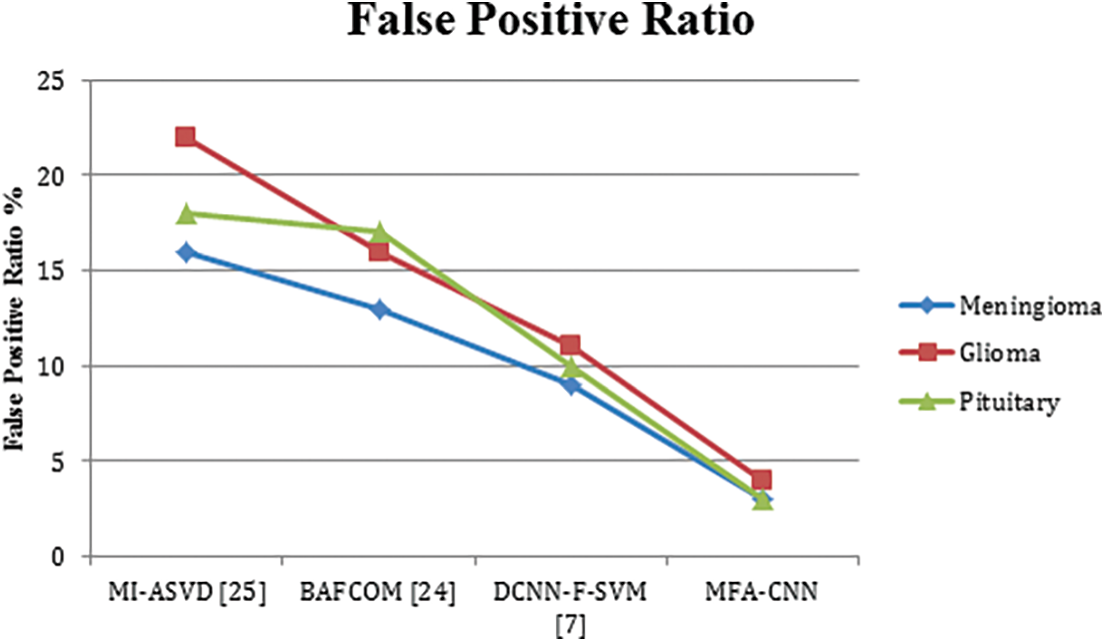
Figure 3: Analysis of false positive ratio
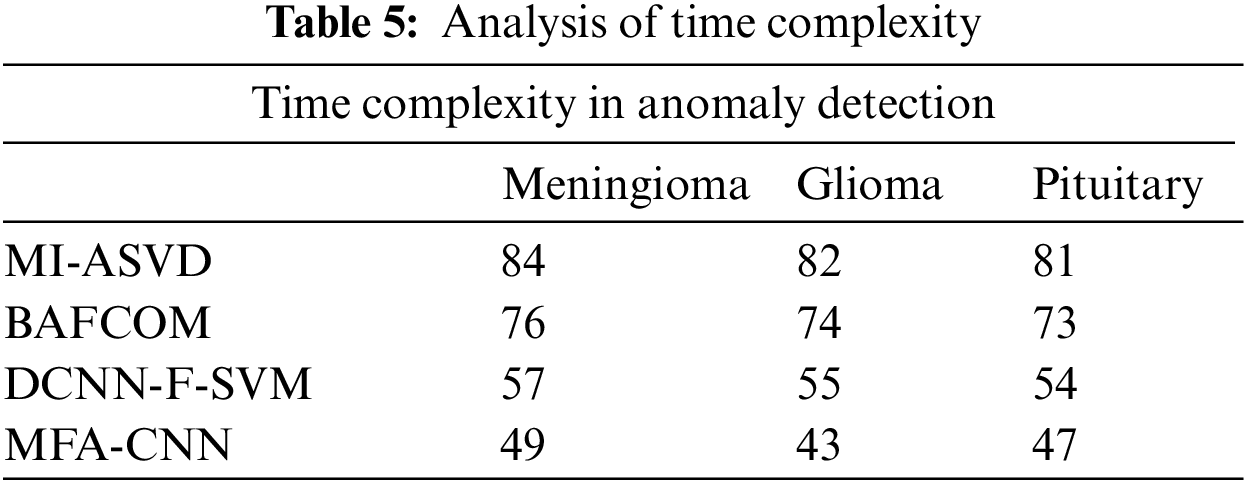
The performance of the methods in terms of time complexity has been measured and presented in Table 5, where the proposed MFA-CNN has produced less time complexity in all the classes than other approaches.
The value of time complexity produced by different methods in detecting the anomaly from the input image has been presented in Fig. 4, where the proposed MFA-CNN classifier has achieved less time complexity compared to other methods.
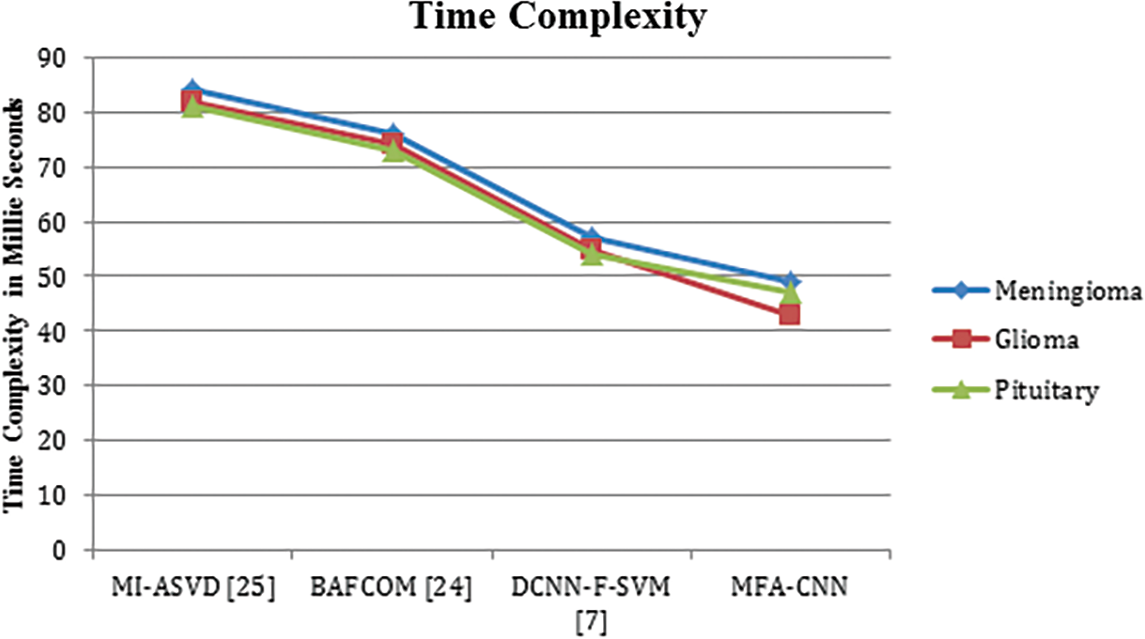
Figure 4: Analysis of time complexity in millie seconds
This paper presented a multi-feature approximation with the CNN model (MFA-CNN) toward brain image classification. The method preprocesses the input brain image and extracts various features from the image. Also, the image has been convolved to reduce the features. The process trains the network by using the reduced features and features like texture, shape, and mass values of white and gray. In the testing phase, the same operations are carried out to measure different mass similarity values on texture, shape, and white mass features. The neurons estimate White Mass Texture Support (WMTS), Gray Mass Texture Support (GMTS), White Mass Covariance Support (WMCS), Gray Mass Covariance Support (GMCS), and Class Texture Adhesive Support (CTAS). Using these values, the method estimates the Multi-Feature Similarity Measure (MFSM). Based on that, the technique performs brain image classification. The proposed approach improves the performance of brain image classification and reduces the false ratio with the least time complexity.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. J. Seetha and R. Selvakumar, “Brain tumor classification using convolutional neural network,” Biomedical and Pharmacology Journal, vol. 11, no. 3, 2018. https://doi.org/10.13005/bpj/1511 [Google Scholar] [CrossRef]
2. P. Chang, B. W. J. Grinband, M. K. M. Bardis, G. Cadena, M. Y. Su et al., “Deep-learning convolutional neural networks accurately classify genetic mutations in gliomas,” American Journal of Neuro Radiology, vol. 39, no. 7, pp. 1201–1207, 2018. [Google Scholar] [PubMed]
3. R. Vankdothu, M. AbdulHameed and H. Fatima,“A brain tumor identification and classification using deep learning based on CNN-LSTM method,” Computers and Electrical Engineering, vol. 101, no. 107960, 2022. https://doi.org/10.1016/j.compeleceng.2022.107960 [Google Scholar] [CrossRef]
4. Y. Liu, S. Stojadinovic, B. Hrycushko, Z. Wardak, L. Steven et al., “A deep convolutional neural network-based automatic delineation strategy for multiple brain metastases stereotactic radiosurgery,” PLoS One, vol. 12, no. 10, pp. 1–17, 2017. [Google Scholar]
5. H. Mzoughi, I. Njeh, A. Wali, M. B. Slimaand, A. B. Hamida et al., “Deep multi-scale 3D convolutional neural network (CNN) for MRI gliomas brain tumor classification,” Journal of Digital Imaging, vol. 33, no. 4, pp. 903–915, 2020. [Google Scholar] [PubMed]
6. T. Ruba, R. Tamilselvi, M. P. Beham and N. Aparna, “Accurate classification and detection of brain cancer cells in MRI and CT images using nano contrast agents,” Journal of Biomedical Pharmacology, vol. 13, no. 3, 2020. https://bit.ly/38S9IO4 [Google Scholar]
7. W. Wentao, D. Jiaoyang, G. Xiangyu, W. Gu, F. Zhao et al., “An intelligent diagnosis method of brain MRItumor segmentation using deep convolutional neural network and SVM algorithm,” Computational and Mathematical Methods in Medicine, 2020. https://doi.org/10.1155/2020/6789306 [Google Scholar] [PubMed] [CrossRef]
8. D. R. Nayak, N. Padhy, P. Mallick and K. Ashish Singh, “A deep auto encoder approach for detection of brain tumor images,” Computers and Electrical Engineering, vol. 102, no. 108238, 2022. https://doi.org/10.1016/j.compeleceng.2022.108238 [Google Scholar] [CrossRef]
9. M. A. Sameer, O. Bayat and H. J. Mohammed, “Brain tumor segmentation and classification approach for MR images based on convolutional neural networks,” in IEEE Conf. on Information Technology to Enhance E-Learning and Other Application (IT-ELA), pp. 138–143, Baghdad, Iraq, 2020. https://doi.org/10.1109/IT-ELA50150.2020.9253111 [Google Scholar] [CrossRef]
10. M. Waqasnadeem, A. Mohammed, A. Ghamdi, M. Hussain, A. Khan et al., “Brain tumor analysis empowered with deep learning: A review, taxonomy, and future challenges,” MDPI, Brain Sciences, vol. 10, no. 2, pp. 118, 2020. https://doi.org/10.3390/brainsci10020118 [Google Scholar] [PubMed] [CrossRef]
11. M. Baskar, R. Renukadevi and J. Ramkumar, “Region centric minutiae propagation measure orient forgery detection with finger print analysis in health care systems,” Neural Process Letter, 2021. https://doi.org/10.1007/s11063-020-10407-4 [Google Scholar] [CrossRef]
12. K. D. Kharat, P. Kulkarni and M. B. Nagori, “Brain tumor classification using neural network based methods,” International Journal of Computer Science and Informatic, vol. 2, no. 9, 2012. https://doi.org/10.47893/IJCSI.2012.1075 [Google Scholar] [CrossRef]
13. T. S. Arulananth, M. Baskar and R. Sateesh, “Human face detection and recognition using contour generation and matching algorithm,” Indonesian Journal of Electrical Engineering and Computer Science, vol. 16, no. 2, pp. 709–714, 2019. [Google Scholar]
14. A. Bardhan, T. Han-Trong, H. Nguyen Van, H. Nguyen ThiThanh, V. TranAnh et al., “An efficient method for diagnosing brain tumors based on MRI images using deep convolutional neural networks,” Applied Computational Intelligence and Soft Computing, vol. 2022, 2022. https://doi.org/10.1155/2022/2092985 [Google Scholar] [CrossRef]
15. T. Zhou, P. Vera, S. Canu and S. Ruan, “Missing data imputation via conditional generator and correlation learning for multimodal brain tumor segmentation,” Pattern Recognition Letters, vol. 158, no. 2022, pp. 125–132. https://doi.org/10.1016/j.patrec.2022.04.019 [Google Scholar] [CrossRef]
16. H. H. N. Alrashedy, A. FahadAlmansour, D. M. Ibrahim and M. A. Hammoudeh, “Brain MRI image generation and classification framework using GAN architectures and CNN models,” Sensors, vol. 22, no. 11, pp. 4297, 2022. https://doi.org/10.3390/s22114297 [Google Scholar] [PubMed] [CrossRef]
17. A. E. Minarno, I. S. Kantomo, F. D. SetiawanSumadi, H. A. Nugroho and Z. Ibrahim, “Classification of brain tumors on MRI images using densenet and support vector machine,” International Journal on Informatics Visualization, vol. 6, no. 2, 2022. https://doi.org/10.30630/joiv.6.2.991 [Google Scholar] [CrossRef]
18. S. Deepak and P. M. Ameer, “Brain tumor classification using deep CNN features via transfer learning,” Computers in Biology and Medicine, vol. 111, pp. 1033–1045, 2019. https://doi.org/10.1016/J.COMPBIOMED.2019.103345 [Google Scholar] [PubMed] [CrossRef]
19. Y. Ding, W. Zheng, J. Geng, Z. Qin, K. K. R. Choo et al., “A Multi-view dynamic fusion framework for multimodal brain tumor segmentation,” IEEE Journal of Biomedical and Health Informatics, vol. 26, no. 4, pp. 1570–1581, 2022. https://doi.org/10.1109/JBHI.2021.3122328 [Google Scholar] [PubMed] [CrossRef]
20. Z. N. K. Swati, Q. Zhao, M. Kabir, F. Ali, Z. Ali et al., “Brain tumor classification for MR images using transfer learning and fine-tuning,” Computerized Medical Imaging Graphics, vol. 75, pp. 34–46, 2019. https://doi.org/10.1016/J.COMPMEDIMAG.2019.05.001 [Google Scholar] [PubMed] [CrossRef]
21. A. S. Ladkat, S. L. Bangare, V. Jagota, S. Sanober, S. M. Beram et al., “Deep neural network-based novel mathematical model for 3D brain tumor segmentation,” Computational Intelligence and Neuroscience, vol. 2022, 2022. https://doi.org/10.1155/2022/4271711 [Google Scholar] [PubMed] [CrossRef]
22. N. Ullah, J. A. Khan, M. S. Khan, W. Khan, I. Hassan et al., “An effective approach to detect and identify brain tumors using transfer learning,” MDPI, Applied Sciences, vol. 12, no. 5645, 2022. https://doi.org/10.3390/app12115645 [Google Scholar] [CrossRef]
23. D. M. Toufiq, A. Makki Sagheer and H. Veisi, “Brain tumor segmentation from magnetic resonance image using optimized thresholded difference algorithm and rough set,” TEM Journal, vol. 11, no. 2, pp. 631–638, 2022. https://doi.org/10.18421/TEM112 [Google Scholar] [CrossRef]
24. A. M. Alhassan and W. M. N. W. Zainon, “BAT algorithm with fuzzy c-ordered means (BAFCOM) clustering segmentation and enhanced capsule networks (ECN) for brain cancer MRI images classification,” IEEE Access, vol. 8, pp. 201741–201751, 2020. https://doi.org/10.1109/ACCESS.2020.3035803 [Google Scholar] [CrossRef]
25. Z. A. A. Saffar and T. Yildirim, “A novel approach to improving brain image classification using mutual information-accelerated singular value decomposition,” IEEE Access, vol. 8, pp. 52575–52587, 2020. https://doi.org/10.1109/ACCESS.2020.2980728 [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools