 Open Access
Open Access
ARTICLE
Modelling a Fused Deep Network Model for Pneumonia Prediction
Department of Computer Science and Engineering, Karpagam Academy of Higher Education, Coimbatore, India
* Corresponding Author: M. A. Ramitha. Email:
Computer Systems Science and Engineering 2023, 46(3), 2725-2739. https://doi.org/10.32604/csse.2023.030504
Received 28 March 2022; Accepted 24 June 2022; Issue published 03 April 2023
Abstract
Deep Learning (DL) is known for its golden standard computing paradigm in the learning community. However, it turns out to be an extensively utilized computing approach in the ML field. Therefore, attaining superior outcomes over cognitive tasks based on human performance. The primary benefit of DL is its competency in learning massive data. The DL-based technologies have grown faster and are widely adopted to handle the conventional approaches resourcefully. Specifically, various DL approaches outperform the conventional ML approaches in real-time applications. Indeed, various research works are reviewed to understand the significance of the individual DL models and some computational complexity is observed. This may be due to the broader expertise and knowledge required for handling these models during the prediction process. This research proposes a holistic approach for pneumonia prediction and offers a more appropriate DL model for classification purposes. This work incorporates a novel fused Squeeze and Excitation (SE) block with the ResNet model for pneumonia prediction and better accuracy. The expected model reduces the human effort during the prediction process and makes it easier to diagnose it intelligently as the feature learning is adaptive. The experimentation is carried out in Keras, and the model’s superiority is compared with various advanced approaches. The proposed model gives 90% prediction accuracy, 93% precision, 90% recall and 89% F1-measure. The proposed model shows a better trade-off compared to other approaches. The evaluation is done with the existing standard ResNet model, GoogleNet+ResNet+DenseNet, and different variants of ResNet models.Keywords
With the emergence of society, environmental conditions, living habits, and peoples’ lifestyles gradually increase the severity of various issues indirectly over other connected diseases [1]. Major diseases like cardiovascular disease, brain damage, diabetes, cancer and vision impairments, etc., have a crucial impact worldwide [2]. While considering diabetes alone, roughly 422 million people are affected all over the world, and of which about 90% have Type II diabetes. With age, the loss of function and heart senescence leads to increased heart disease risk. Nearly 90% of global death is due to heart disease [3]. These sorts of global diseases seriously influence the human health condition and diminish personal productivity. Similarly, it worsens the social pressure and triggers the medical expenditure on health conditions [4]. The ultimate objective is to identify the risk probability of any individual suffering from a particular disease [5]. Many factors influence various conditions among the vast populations. By considering the different feature sets with extensive and more comprehensive dimensions, the individual, complexity, and disease variations must be identified [6]. Simply handling all these essential tasks manually is not only complex, but it also consumes vast financial and human resources. The enormous development of society has endorsed technological progression. It leads to more rapid medical data acquisition. The medical institutions worldwide are acquiring diverse health-related data and continuously expanding the scaling process [7]. These sorts of medical data include basic patient information, i.e., electronic medical records (EMR) and electronic health records (HER), instrument (medical) data, image data and so on [8]. This data is highly vulnerable to various factors. For instance, redundant or incomplete data is identified due to timeliness or personal security issues that make the data fallacious and privacy-based issues. Additionally, medical data is divergent from ordinary data due to its high dimensionality, complexity, irregularity, and heterogeneity and includes more unstructured data [9]. It is highly complex to attain or handle these data manually. However, in reality, medical diagnosis is constrained by medical conditions, diagnostician level, and patient differences. Based on these issues, some people urgently require various auxiliary techniques to assist in disease identification and prediction [10].
With advanced technologies in medical image processing [11] and other applications [12], a lot of work has been put into creating automated algorithms modelled over the past few years. These approaches often use images to forecast disease occurrence based on intensity, such as existing prediction, simply a reference process, or a referable perception process, among other things. In contrast, Chen et al. [13] used a transfer learning method to identify disease in the earlier stage. Azarang et al. [14] suggested utilizing sparse representation and unbalanced data to categorize the image as usual or malicious using a multi-kernel cross-learning approach. To detect referable images, Wei et al. [15] used an assembly of nine GoogleNets. For identifying imagination as simply a reference image, Zhou et al. [16] created a convolutional neural network. These approaches often provide excellent clinical effectiveness before determining the seriousness by creating optimum models utilizing many datasets. They are, however, frequently criticized for their lack of readability because they merely offer a unique number as a medical result for an input image, leaving no intelligible evidence (such as the appearance of particular disease-related characteristics) to back up the diagnosis. The medical data offered by these technologies are complicated for doctors to comprehend and adopt, completely restricting their use in practice.
The severity levels of the disease are determined using several severity classification methods [17–19] based on the presence of distinct indicators (i.e., disease-associated characteristics) in the provided input image. The disease-related characteristics are present, and it will considerably improve the performance of the diagnostic conclusion by giving a complete identified section and corroboration for the complex audits and inspection results. The author studied the potential of establishing autonomous computerized programs for diagnosing risk levels and the existence of disease-related characteristics in endoscopic images [20]. The existence of disease-related characteristics in endoscopic images is first discovered, and afterwards, the extent of disease follows options on this. The findings show that utilizing the presence of disease-related traits as a predictor makes it possible to attain excellent diagnostic ability for severity [21]. A hierarchical architecture is intended to combine the attributes of the disease critical aspects taken from the image for severity detection due to the causal link between related functions and harshness. Finally, the suggested computational intelligence project has three distinct benefits. First, introducing extra information regarding related aspects via a hierarchy organization makes it feasible to enhance the productivity of severity evaluation [22]. Second, the production of associated characteristics might make severity diagnostics results more interpretable. Using two autonomous sample sets obtained in disease diagnostics, we examine the suggested technique for categorizing various severity and likelihood and identifying simply a reference, which is critical for diagnosis and screening [23]. The author has quantitatively measured the suggested approach’s efficacy in several expert systems.
Deep Learning works effectually using Graphical Processing Unit (GPU) systems. GPUs are effective for various basic DL approaches with parallel-computing operations like matrix multiplication, activation functions, and convolutions. Stacked memory and updated GPU models improve the bandwidth significantly [24]. The improvements facilitate various primitives to use every computational resource effectually over the available GPUs. The GPU performs more efficiently than CPU, i.e., 10–20:1 connected to dense linear operations. The maximal parallel processing is considered the base programming. For instance, it includes 64 computational units. It possesses four engines per computational layer and every engine has 16 floating-point computational lanes. GPU performance is attained only with the addition and multiplication functions to merge the instructions for primitive matching associated with the matrix function [25]. Thus, it is known that deep learning execution over GPU gives expected results and reduces computational complexity. The proposed model is effectually used in medical applications.
This research concentrates on modelling an efficient approach for pneumonia prediction. Thus, faster and more accurate pneumonia prediction means a lot. There is a need to provide timely access to the treatment process, and saving money is substantial. Various existing research works are carried out; however, they fail to give a better prediction. This has laid the path to perform this research, and the following are the significant research contributions: (1) To acquire datasets from the publicly available online resources for pneumonia prediction; (2) To fuse the well-known DL approaches, i.e., Squeeze and Excitation network (SE) and ResNet model for disease prediction; (3) To perform training, testing and validation using Keras; (4) To compare the anticipated fused model with the existing approach to project the significance of the anticipated model. The work is structured as follows: In Section 2, various DL approaches are analyzed for their pros and cons with the disease prediction; Section 3 portrays the detailed analysis of the fused model. The numerical results and discussions related to the fused model are demonstrated in Section 4. A summary of the work and future research ideas are provided in Section 5.
Deep Learning (DL) is a category of Artificial Intelligence (AI) approaches based on neural networks and motivated by the brain’s structures. DL is a term that refers to approaches for automatically recognizing the graphical model of data’s underlying and fundamental connections [26]. Unlike standard machine learning approaches, deep learning approaches demand far less operator direction, so they do not rely on the development of finger features, which may be moment and challenging, but instead learn relevant feature representations [27]. Furthermore, when the number of data grows, DL approaches scale markedly improved to classical Machine Learning. This segment provides a short overview of some key DL concepts [28]. A Deep (Feed-Forward) Naive Bayes (DNN), on the other hand, has far more than one hidden layer [29]. Every input network and two hidden clusters are joined to each hidden neuron by networks in a city, so each is concealed and outputting models of many biological neurons. Furthermore, because these circuits only accept one matrix as input, they cannot be employed directly with neuroimaging studies. Convolutional Neural Networks (CNNs), which take 2D arrays as input rather than external neural networks, are influenced by biological perception and are based on an essential mathematical process called “transform”. The fundamental distinction between a CNN and a DNN is that, all synapses in one layer input to calculate the outcome of every hidden neuron in a second. In contrast, in the former, this is not the situation [30]. Instead, a CNN computes convolution layers by moving over a portion of the top image to build a depth map using filtration or loudspeakers. The former learns photographic characteristics, converted from one array sent into the later, effectively a Deep Neural System, to categorize the input picture using the created attributes [31]. Because of their capacity to retain the structural stability of the image, UNet [32] designs are more suited for text categorization than regular CNNs. They comprise process parameters that capture the essential framework and asymmetrically expansion path. It allows accurate and consistent categorization. Furthermore, because it analyses the picture in one pass instead of several areas in a template matching technique, a UNet design has fewer parameters. It is quicker than typical CNNs, so such structures are termed “Fully Convolutional Networks” (FCN). At last, it requires far less information to accomplish a feature extraction than classical CNNs. It is critical for computer-aided diagnosis, where the quantity of information is much smaller than other applications in image processing [32]. Implementations are better suited for text categorization than classical CNNs, due to their capability to maintain the image’s structural strength. They have a shrinking path that captures the practical detail and an asymmetric growing way that allows for precise categorization. Furthermore, because it analyses the picture in one shot or several areas in a template matching technique, a UNet design has lower complexity. It is quicker than typical CNNs, so these structures are termed “Fully Convolutional Networks” (FCN). Secondly, it uses far less input to do a feature extraction than typical CNNs, which is critical for medical image processing since data availability is significantly lower than in other disciplines of machine vision.
Generative Adversarial Network (GAN) [33] is an essential class of convolutional layers. The generating networks, which create candidates sampling based on the old given data, and the classification algorithm, which tries to discriminate the produced candidates sampling from the actual dataset, make up a typical GAN. The manufacturer can provide candidate sampling closely matched to the genuine distribution of the data using such a training method. Picture amazingly (i.e., generating high-resolution copies of the input image), art creation and picture translating (e.g., converting a day image to its night equivalent) are all applications of GANs [34]. Retraining a deep convolutional neural network is highly time-consuming in computer systems and data. ImageNET [35], the world’s most extensive item identification collection, contains approximately 14 million real-life photos of animals, equipment, food, people, and automobiles. Supervised Methods, which denotes mining several models (i.e., base models) to provide more remarkable prediction outcomes than the existing models, is another critical area of AI study [36]. A diversified group of base models is used in the community knowledge modelling framework to provide single estimates from a composite of their unique outcomes on the same information or a portion of the offered accessible data. It is a learning method that reduces the model’s classification error and efficiently combines input from different modalities. Given that each model that has been learned is the same across models, numerous group approaches are applied to all potential models [37]. However, some fundamental obstacles must be overcome to promote AI’s use in medical settings [38,39]. Kuenzi et al. [40] anticipated a novel weighted classifier-based model that merges the prediction weight attained from DenseNet, MobileNetV3, Xception, Inception V3, and ResNet 18 models. The author shows that the final classifier model achieves 90% over the testing set [41–48]. However, these models lead to high processing time. Thus, there is no suitable method for the proposed model. Moreover, the anticipated lightweight model is adequate for pneumonia prediction. But there are some research constraints and challenges that need to be addressed. The present DL approaches for pneumonia prediction show some limitations in selecting the hyper-parameters to construct an accurate and lightweight model. These sorts of constraints are addressed in the proposed research work. Table 1 depicts the comparison of various deep learning approaches.

This research includes two major phases: (1) dataset acquisition for predicting chest X-rays and (2) designing a fused SE and ResNet for disease prediction. The performance of the anticipated model is estimated with metrics such as prediction accuracy, precision, recall and F1-score.
This research considers the Pneumonia (chest X-ray) dataset, a publicly available online dataset. It includes 540 normal samples for training, 3882 abnormal samples for training, 8 normal and abnormal samples for validation, 234 normal samples for testing and 390 abnormal samples for training.
This section discusses the design flow of the anticipated model. The flow of the anticipated model is given in Fig. 1. It comprises inner blocks like convolutional layers, fully connected (FC) layers and global average pooling (GAP) layers. The figure represents the convolutional blocks with several SE and ResNet blocks.

Figure 1: Flow of ensemble classifier model
3.2.1 Squeeze and Xcitation Blocks (SE)
SE block is a computational unit modelled for any transformation
Here,

Figure 2: SE-ResNet model

Figure 3: Proposed SE-ResNet architecture
3.2.2 Squeezing with Global Information
The filter works with the local receptive field, and the converted output is not competent to share the essential data with successive channels. The channel dependency is measured as an issue that turns out to be severe in the deep learning network model. It turns out to be a crucial issue in smaller receptive field sizes. A global summary for the channel is noted as channel statistics attained with squeeze global spatial information to solve this issue better. The adoption of GPA pools is probably the essential feature with channel descriptors. The spatial dimensions
Here,
3.2.3 Adaptive Recalibration With Excitation
Here, the simple gated function is performed with sigmoid activation to attain a suitable objective that fulfils two criteria: (1) the model captures channel non-linearity, and (2) it needs to predict the non-mutual relationship among the multiple channels and facilitate to emphasize the activation. The channel-wise values are attained from the prior squeeze operation, which completely captures the feature dependencies during excitation operation. It is mathematically expressed as in Eq. (3):
Here,
Here,
During the neural network training phase, the model must learn features and random noise from the provided training data and is over-fitted. The model performs poorly during the testing phase with some unknown datasets. Specifically, poor performance is observed when the network possesses enormous neurons with independent parameters. It is a technique where some neuron percentages are deleted during the network training phase. The neuron that intends to be removed has to be chosen randomly to assist the model with more generalization. It predicts the unseen dataset well. The dropout approaches need to address the over-fitting issues in the anticipated model.
The model is constructed with SE and ResNet blocks (See Fig. 3a), where the original ResNet blocks are fused entirely with special computational SE. The block is constructed with some GAP layer followed by an FC non-linearity model. The proposed system consists of six ResNet blocks and one SENet. The six ResNet blocks are connected back to back as shown in Fig. 3. The side outputs are taken from the last four ResNet blocks because the feature map size attained from the first two blocks is larger. The first two blocks learn lower-level visual features that are not so appropriate for classification purposes. The inner model specifies the total SE and ResNet block splits where the output blocks are fed into the concatenated model. The outer model specifies the number of SE and ResNet blocks that forms the fused network model. The input chest X-ray image is fed into the Conv1 layer, followed by the six ResNet layers, while the output from the third ResNet layer is fed to the ResNet layer and extracted as a side output. This process is carried out until the last (sixth ResNet) layer. The fused results of all these sides are provided for scaling output and global pooling. The layer functionality is described above. However, the output from the sigmoid layer is also given to the scaling layer. Based on this functionality, the images are tested and trained. However, the complex nature of the anticipated model leads to computational complexity. When the learning model gets deeper, it becomes extremely difficult for the layers to propagate the information from shallow layers, and the information is lost. It is known as the degradation problem.
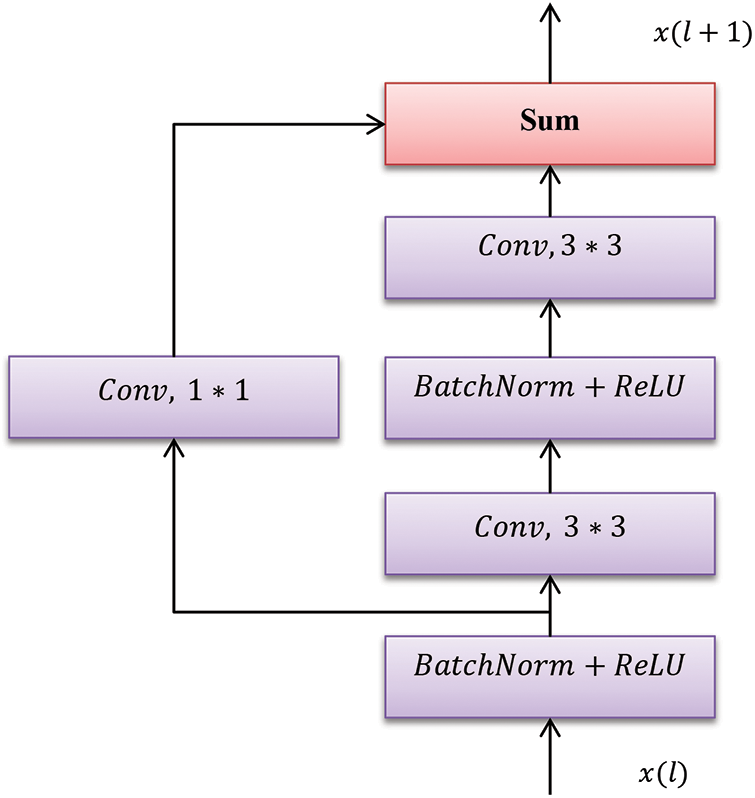
Figure 3(a): One ResNet Unit
4 Numerical Results and Discussion
The anticipated deep network model is executed in the Keras simulation environment. The outcomes are validated with accuracy, recall, precision, and F-measure. The deep network model is known as an ‘inducer’. For deep classification, inducers are used for provisioning the system performance. It provides superior outcomes compared to other techniques. The initial step is to construct multiple subsets from the available training set. Next, the classifiers are merged with SE blocks and ResNet, where outcomes are attained with superior predictive function. Here, the available online dataset is considered, and evaluation is done with various metrics and expressed in Eqs. (5)–(8):
True Positive (TP)–the model intends to identify the right features as appropriate ones;
True Negative (TN)–the model intends to identify the incorrect features as wrong ones;
False Positive (FP)–the model intends to predict the incorrect features as the right one;
False Negative (FN)–the model intends to predict the right features as the wrong ones;
The provided images are resized and transformed to grey-scale to feed the anticipated SE and ResNet model input. The batch size must be chosen during the training process, and the total iterations are set to 100 epochs. Some ten iterations are chosen to get an average result during the testing process. Here, batch normalization accelerates the quick convergence and training process before provisioning the input to the successive layers. Batch normalization is performed to attain superior enhancement during the training process. The experimental result demonstrates that the model works superiorly during the testing and training phases by handling the over-fitting problem. Some dropout techniques are provided to establish the generalized model to handle this issue. The loss function is reduced gradually during the testing and training process. The training loss is zero during a stable state, and the testing loss is nearer to zero, where the difference between the training and testing loss is nearer to zero. It shows that the anticipated model shows better generalization and no over-fitting issues, i.e.,
Here, metrics like precision, recall, F1-measure, and accuracy are evaluated and compared. Table 2 compares the anticipated deep SE-ResNet model with the existing standard ResNet model. The anticipated model shows 90% prediction accuracy, which is 25% higher than the standard model. The precision is 93%, which is 23% higher than the ResNet model, and recall is 90%, 35% higher than the standard ResNet model. F measure is 89% which is 29% higher than the traditional approach (see Fig. 4). Similarly, the outcomes of the individual ResNet-50 and DenseNet are lesser than the proposed model. Figs. 5 and 6 depict the training accuracy, validation accuracy and training loss and validation loss.


Figure 4: Comparison of various performance metrics

Figure 5: Training and validation accuracy

Figure 6: Training and validation loss
It is recorded that the class accuracy is higher, i.e., 100% which is complex comparatively and like other classes. The execution speed is roughly 12 ms for predicting the incoming image features and depicts the comparative performance of the anticipated model with the general approaches. From the above figures, the model shows clear dominance over the prevailing methods. The standard ResNet predicts the complexity of the correlation when the input image is enormous (data volume) and complex. However, the model does not use the higher spatial information without entirely relying on the spectral signature. Some existing approaches need many parameters as the layers are FC. Therefore, connection weights proliferate and turn uncontrollably, leading to over-fitting, inefficiency, redundancy, and lack of generalization ability. Moreover, some complexity are caused by differentiating the features, which is noise sensitive, tedious, and time-consuming. Some deep CNN models show superior performance when compared to conventional learning approaches but lack the fine-tuning of parameters. Generally, ResNet possesses huge parameters and complex network architecture and is computationally expensive. Stacking convolutional layers provide a more profound architecture that does not increment generalization and validation accuracy. However, it ends with the adoption of more parameters with increased training time. While comparing the ResNet-18 and ResNet-50, it is observed that the accuracy of the proposed SE-ResNet is promising compared to the existing approaches. In some cases, a dense network model performs more efficiently than the available ResNet model with respect to accuracy. However, this dense network model consumes more memory than another ResNet model from various concatenated layers.
Here, the deep SE and ReLU are skipped connections to handle the vanishing gradient efficiently and facilitate smooth gradient flow. Finally, global pooling is adopted before the final fully-connected layers, inherent in the prevailing CNN model. The similarities and relations are easily predicted among the feature maps and output. The proposed deep SE-ResNet model shows two potential benefits with the deep network model. Specifically, the process is performed concurrently with going deep. It is also known as the split and merges technique and needs some hyper-parameters known as cardinality to determine how the parallel path and division are necessary for the model. So, the anticipated model requires only one hyper-parameter known as cardinality indeed of huge hyper-parameters needed by the conventional deep CNN model. The expected SE block is authenticated to learn the complex features using feature independencies. Similarly, the model comprises skip connections over the ResNet block, eliminating vanishing gradients. Therefore, the method provides superior outcomes compared to other approaches. However, there are some misclassifications during the testing process. These misclassifications are due to a lack of variations, insufficient datasets, and similarity patterns. Moreover, the enhancements in the model architecture are due to the fused nature of the model. The existing approaches show some misclassification rates compared to others.
This work calibrates the SE and ResNet-based deep network model, composed of the SE computational layer over the ResNet model, to recognize the disease. With the initiation of the proposed model, the work provides conclusive outcomes drawn with the reduction of complexity and hyper-parameter and an increase in the ability to learn highly complex features. It is known that the strategy is essential for the extraction of more robust discriminative features. Moreover, adopting batch normalization provides the model with a faster training capacity, even in the case of a large dataset. The anticipated model is scalable owing to the faster convergence rate and lower complexity. It attains an average prediction accuracy of 90%, the precision of 93%, recall of 90%, and F-measure of 89%, respectively, with the chest X-ray dataset. The knowledge extracted from this work shows better prediction accuracy with the provided dataset. The outcomes demonstrate that the anticipated model offers a superior breakthrough in modelling a deep network model. The simulation is done in Keras, and the anticipated model shows a better trade-off than other approaches. The primary research constraint is the acquisition of the dataset and the training process for 100 epochs. This work will be extended with the adoption of the complex dataset, and this model can be tested over the other learning approaches to evaluate the results with further enhancement. Also, pre-trained network models are adopted for further analysis.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. B. Shickel, P. J. Tighe, A. Bihorac and P. Rashidi, “Deep EHR: A survey of recent advances in deep learning techniques for electronic health record (EHR) analysis,” IEEE Journal of Biomedical and Health Informatics, vol. 22, no. 5, pp. 1589–1604, 2017. [Google Scholar] [PubMed]
2. C. Cao, F. Liu, H. Tan, D. Song, W. Shu et al., “Deep learning and its applications in biomedicine,” Genomics Proteomics & Bioinformatics, vol. 16, no. 1, pp. 17–32, 2018. [Google Scholar]
3. T. Kaur and T. K. Gandhi, “Automated brain image classification based on VGG-16 and transfer learning,” in Proc. of IEEE Int. Conf. on Information Technology, Bhubaneswar, India, pp. 94–98, 2019. [Google Scholar]
4. S. Tang, S. Yuan and Y. Zhu, “Convolutional neural network in intelligent fault diagnosis toward rotatory machinery,” IEEE Access, vol. 8, no. 1, pp. 86510–86519, 2020. [Google Scholar]
5. R. Siddiqi, “Automated pneumonia diagnosis using a customized sequential convolutional neural network,” in Proc. of 3rd Int. Conf. on Deep Learning Technologies (ICDLT), New York, USA, pp. 64–70, 2019. [Google Scholar]
6. M. S. Hossain, S. U. Amin, M. Alsulaiman and G. Muhammad, “Applying deep learning for epilepsy seizure detection and brain mapping visualization,” ACM Transactions on Multimedia Computing, Communications, and Applications (TOMM), vol. 15, no. 1s, pp. 1–17, 2019. [Google Scholar]
7. A. Sherstinsky, “Fundamentals of recurrent neural network (RNN) and long short-term memory (LSTM) network,” Physica D: Nonlinear Phenomena, vol. 404, no. 2, pp. 132306, 2020. [Google Scholar]
8. H. M. Lynn, S. B. Pan and P. Kim, “A deep bidirectional GRU network model for biometric electrocardiogram classification based on recurrent neural networks,” IEEE Access, vol. 7, no. 1, pp. 145395–145405, 2019. [Google Scholar]
9. Y. Cui, S. Zhao, H. Wang, L. Xie, Y. Chen et al., “Identifying brain networks at multiple time scales via deep recurrent neural network,” IEEE Journal of Biomedical and Health Informatics, vol. 23, no. 6, pp. 2515–2525, 2018. [Google Scholar] [PubMed]
10. I. Banerjee, Y. Ling, M. C. Chen, S. A. Hasan, C. P. Langlotz et al., “Comparative effectiveness of convolutional neural network (CNN) and recurrent neural network (RNN) architectures for radiology text report classification,” Artificial Intelligence in Medicine, vol. 97, no. 2, pp. 79–88, 2019. [Google Scholar] [PubMed]
11. G. Liang, H. Hong, W. Xie and L. Zheng, “Combining convolutional neural network with recursive neural network for blood cell image classification,” IEEE Access, vol. 6, no. 4, pp. 36188–36197, 2018. [Google Scholar]
12. J. C. Kim and K. Chung, “Multi-modal stacked denoising autoencoder for handling missing data in big healthcare data,” IEEE Access, vol. 8, no. 1, pp. 104933–104943, 2020. [Google Scholar]
13. M. Chen, X. Shi, Y. Zhang, D. Wu and M. Guizani, “In-depth features are learned for medical image analysis with a convolutional autoencoder neural network,” IEEE Transactions on Big Data, vol. 1, no. 2, pp. 1, 2017. [Google Scholar]
14. A. Azarang, H. E. Manoochehri and N. Kehtarnavaz, “Convolutional autoencoder-based multispectral image fusion,” IEEE Access, vol. 7, no. 3, pp. 35673–35683, 2019. [Google Scholar]
15. R. Wei and A. Mahmood, “Recent advances in variational autoencoders with representation learning for biomedical informatics: A survey,” IEEE Access, vol. 9, no. 4, pp. 4939–4956, 2021. [Google Scholar]
16. Z. Zhou, Y. Wang, Y. Guo, Y. Qi and J. Yu, “Image quality improvement of hand-held ultrasound devices with a two-stage generative adversarial network,” IEEE Transactions on Biomedical Engineering, vol. 67, no. 1, pp. 298–311, 2019. [Google Scholar] [PubMed]
17. X. Bing, W. Zhang, L. Zheng and Y. Zhang, “Medical image super-resolution using improved generative adversarial networks,” IEEE Access, vol. 7, no. 1, pp. 145030–145038, 2019. [Google Scholar]
18. Y. Li, J. Zhao, Z. Lv and J. Li, “Medical image fusion method by deep learning,” International Journal of Cognitive Computing in Engineering, vol. 2, no. 2, pp. 21–29, 2021. [Google Scholar]
19. Z. Yang, Y. Chen, Z. Le, F. Fan and E. Pan, “Multi-source medical image fusion based on wasserstein generative adversarial networks,” IEEE Access, vol. 7, no. 4, pp. 175947–175958, 2019. [Google Scholar]
20. C. Wang, C. Xu, X. Yao and D. Tao, “Evolutionary generative adversarial networks,” IEEE Transactions on Evolutionary Computation, vol. 23, no. 6, pp. 921–934, 2019. [Google Scholar]
21. A. H. Al-Fatlawi, M. H. Jabardi and S. H. Ling, “Efficient diagnosis system for Parkinson’s disease using deep belief network,” in Proc. of IEEE Congress on Evolutionary Computation, Vancouver, Canada, pp. 1324–1330, 2016. [Google Scholar]
22. S. Khan, N. Islam, Z. Jan, I. U. Din and J. J. C. Rodrigues, “A novel deep learning-based framework for the detection and classification of breast cancer using transfer learning,” Pattern Recognition Letters, vol. 125, no. 1, pp. 1–6, 2019. [Google Scholar]
23. Z. Hu, J. Tang, Z. Wang, K. Zhang, L. Zhang et al., “Deep learning for image-based cancer detection and diagnosis−A survey,” Pattern Recognition, vol. 83, no. 2, pp. 134–149, 2018. [Google Scholar]
24. M. Nishiga, D. W. Wang, Y. Han, D. B. Lewis and J. C. Wu, “COVID-19 and cardiovascular disease: From basic mechanisms to clinical perspectives,” Nature Review Cardiology, vol. 17, no. 9, pp. 543–558, 2020. [Google Scholar]
25. D. Fan, J. Yang, J. Zhang, Z. Lv, H. Huang et al., “Effectively measuring respiratory flow with portable pressure data using a backpropagation neural network,” IEEE Journal of Translational Engineering in Health and Medicine, vol. 6, pp. 1–12, 2018. [Google Scholar]
26. P. Rajpurkar, J. Irvin, K. Zhu, B. Yang, H. Mehta et al., “Radiologist-level pneumonia detection on chest X-rays with deep learning,” arxiv preprint, arXiv: 1711.05225, 2017. [Google Scholar]
27. M. Raghu, C. Zhang, J. Kleinberg and S. Bengio, “Transfusion: Understanding transfer learning for medical imaging,” arXiv preprint, arXiv: 1902.07208, 2019. [Google Scholar]
28. C. J. Saul, D. Y. Urey and D. C. Taktakoglu, “Early diagnosis of pneumonia with deep learning,” arXiv preprint, arXiv: 1904.00937, 2019. [Google Scholar]
29. A. Timmis, N. Townsend, C. P. Gale, A. Torbica, M. Lettino et al., “European society of cardiology: Cardiovascular disease statistics 2019,” European Heart Journal, vol. 41, no. 1, pp. 12–85, 2020. [Google Scholar] [PubMed]
30. H. Shi, H. Wang, C. Qin, L. Zhao and C. Liu, “An incremental learning system for atrial fibrillation detection based on transfer learning and active learning,” Computer Methods and Programs in Biomedicine, vol. 187, no. 5, pp. 105219, 2020. [Google Scholar] [PubMed]
31. H. Shi, H. Wang, Y. Huang, L. Zhao, C. Qin et al., “A hierarchical method based on extreme weighted gradient boosting in ECG heartbeat classification,” Computer Methods and Programs in Biomedicine, vol. 171, no. 2, pp. 1–10, 2019. [Google Scholar] [PubMed]
32. Y. Jin, C. Qin, J. Liu, K. Lin, H. Shi et al., “A novel domain adaptive residual network for automatic atrial fibrillation detection,” Knowledge-Based Systems, vol. 203, no. 1, pp. 106122, 2020. [Google Scholar]
33. J. W. Kuang, H. Z. Yang, J. J. Liu and Z. J. Yan, “Dynamic prediction of cardiovascular disease using improved LSTM,” International Journal of Crowd Science, vol. 3, no. 1, pp. 14–25, 2019. [Google Scholar]
34. H. Shi, C. Qin, D. Xiao, L. Zhao and C. Liu, “Automated heartbeat classification based on a deep neural network with multiple input layers,” Knowledge-Based Systems, vol. 188, no. 2, pp. 105036, 2020. [Google Scholar]
35. P. Lu, S. Guo, H. Zhang, Q. Li, Y. Wang et al., “Research on improved depth belief network-based prediction of cardiovascular diseases,” Journal of Healthcare Engineering, vol. 2018, no. 3, pp. 1–9, 2018. [Google Scholar]
36. Y. Ding, J. H. Sohn, M. G. Kawczynski, H. Trivedi, R. Harnish et al., “A deep learning model to predict a diagnosis of alzheimer disease by using 18F-for PET of the brain,” Radiology, vol. 290, no. 2, pp. 456–464, 2019. [Google Scholar] [PubMed]
37. S. Ji, W. Xu, M. Yang and K. Yu, “3D convolutional neural networks for human action recognition,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 35, no. 1, pp. 221–231, 2012. [Google Scholar]
38. A. Tiwari, S. Srivastava and M. Pant, “Brain tumour segmentation and classification from magnetic resonance images: Review of selected methods from 2014 to 2019,” Pattern Recognition Letters, vol. 131, no. 1, pp. 244–260, 2020. [Google Scholar]
39. L. Tong, J. Mitchel, K. Chatlin and M. D. Wang, “Deep learning-based feature-level integration of multi-omics data for breast cancer patients survival analysis,” BMC Medical Informatics and Decision Making, vol. 20, no. 1, pp. 1–12, 2020. [Google Scholar]
40. B. M. Kuenzi, J. Park, S. H. Fong, K. S. Sanchez, J. Lee et al., “Predicting drug response and synergy using a deep learning model of human cancer cells,” Cancer Cell, vol. 38, no. 5, pp. 672–684, 2020. [Google Scholar] [PubMed]
41. Rohit, “Pneumonia detection in chest X-ray images using an ensemble of deep learning models,” PLOS ONE, vol. 16, no. 9, pp. e0256630, 2021. [Google Scholar]
42. M. B. Sudhan, M. Sinthuja, S. P. Raja, J. Amutharaj, G. C. P. Latha et al., “Segmentation and classification of glaucoma using U-Net with deep learning model,” Journal of Healthcare Engineering, vol. 2022, no. 1601354, pp. 1–10, 2022. [Google Scholar]
43. S. Sridhar, J. Amutharaj, V. Prajoona, B. Arthi, S. Ramkumar et al., “A torn ACL mapping in knee MRI images using deep convolution neural network with inception-v3,” Journal of Healthcare Engineering, vol. 2022, no. 7872500, pp. 1–9, 2022. [Google Scholar]
44. S. Rinesh K. Maheswari, B. Arthi, P. Sherubha, A. Vijay et al., “Investigations on brain tumor classification using hybrid machine learning algorithms,” Journal of Healthcare Engineering, vol. 2022, no. 2761847, pp. 1–9, 2022. [Google Scholar]
45. P. Manimegalai and P. K. Jayalakshmi,“A study on diabetic retinopathy detection using image processing,” Journal of Computational Science and Intelligent Technologies, vol. 2, no. 1, pp. 21–26, 2021. [Google Scholar]
46. N. N. Anjum, “A study on segmenting brain tumor MRI images,” Journal of Computational Science and Intelligent Technologies, vol. 2, no. 1, pp. 1–6, 2021. [Google Scholar]
47. T. Rajendran, K. P. Sridhar and S. Manimurugan,“Recent innovations in soft computing applications,” Current Signal Transduction Therapy, vol. 14, no. 2, pp. 129–130, 2019. [Google Scholar]
48. T. Rajendran, K. P. Sridhar, S. Manimurugan and S. Deepa,“Advanced algorithms for medical image processing,” The Open Biomedical Engineering Journal, vol. 13, no. 1, pp. 102, 2019. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools