 Open Access
Open Access
ARTICLE
Brain Tumor Identification Using Data Augmentation and Transfer Learning Approach
1 Department of Electronics and Communication Engineering, Kongu Engineering College, 638060, Tamil Nadu, India
2 Department of Electronics and Communication Engineering, Sona College of Technology, 636005, Tamil Nadu, India
3 Department of Computer Science and Engineering, Sri Sairam Institute of Technology, Chennai, 600044, Tamil Nadu, India
4 Department of Electrical and Electronics Engineering, Sri Sairam Institute of Technology, Chennai, 600044, Tamil Nadu, India
5 Department of Computer Science and Engineering, Sri Venkateswara College of Engineering, Sriperumbudur, 602117, Tamil Nadu, India
6 Department of Electrical and Electronics Engineering, Sri Venkateswara College of Engineering, Sriperumbudur, 602117, Tamil Nadu, India
7 Department of Computer Science, College of Computing, Khon Kaen University, KhonKaen, 40002, Thailand
8 School of Telecommunication Engineering, Suranaree University of Technology, Nakhon Ratchasima, 30000, Thailand
9 Department of Computer Science, College of Computer and Information Sciences, Majmaah University, Al-Majmaah, 11952, Saudi Arabia
10 Department of Computer Engineering, College of Computer and Information Sciences, Majmaah University, Al-Majmaah, 11952, Saudi Arabia
* Corresponding Author: Chitapong Wechtaisong. Email:
Computer Systems Science and Engineering 2023, 46(2), 1845-1861. https://doi.org/10.32604/csse.2023.033927
Received 01 July 2022; Accepted 04 November 2022; Issue published 09 February 2023
Abstract
A brain tumor is a lethal neurological disease that affects the average performance of the brain and can be fatal. In India, around 15 million cases are diagnosed yearly. To mitigate the seriousness of the tumor it is essential to diagnose at the beginning. Notwithstanding, the manual evaluation process utilizing Magnetic Resonance Imaging (MRI) causes a few worries, remarkably inefficient and inaccurate brain tumor diagnoses. Similarly, the examination process of brain tumors is intricate as they display high unbalance in nature like shape, size, appearance, and location. Therefore, a precise and expeditious prognosis of brain tumors is essential for implementing the of an implicit treatment. Several computer models adapted to diagnose the tumor, but the accuracy of the model needs to be tested. Considering all the above mentioned things, this work aims to identify the best classification system by considering the prediction accuracy out of AlexNet, ResNet 50, and Inception V3. Data augmentation is performed on the database and fed into the three convolutions neural network (CNN) models. A comparison line is drawn between the three models based on accuracy and performance. An accuracy of 96.2% is obtained for AlexNet with augmentation and performed better than ResNet 50 and Inception V3 for the 120th epoch. With the suggested model with higher accuracy, it is highly reliable if brain tumors are diagnosed with available datasets.Keywords
A brain tumor is an abnormal increase of cells in the brain or skull. Tumors can develop from the brain tissue (primary), or most cancers from elsewhere within the body can spread to the brain (secondary). MRI is a medical imaging technique that produces three-dimensional clear physical images [1]. Neurologists manually categorize the brain MR images using the World Health Organization (WHO) classification [2]. Doctors and researchers proceed to refine MRI methods to help in scientific methodology and research. The improvement of MRI revolutionized medicine. It is mainly used in disease detection, analysis, and remedy monitoring. It is based on sophisticated innovation that energizes and detects the amendment inside the course of the rotational axis of protons in the water that produces dwelling tissues. The MRI suits well for the non-bony portion of the body’s soft tissue. MRI can separate white matter and grey matter inside the brain and can be used to analyze aneurysms and tumors. It is used to improve the grasp of the brain organization and gives a probably new widespread for measuring neurological status and neurosurgical likelihood.
Meninges is a three-layered membrane with the outer layer being the dura mater, the middle layer being arachnoid and the inner layer being pia matter. It is found just below the skull covering the brain and spinal cord. Meningioma tumors grow on the surface of the arachnoid cell in the meninges. This tumor grows inward or outward, causing pressure on the brain or thickening of the skull. These tumors grow slowly and are benign, and less than 10% may be malignant. It is found among people between the age of 40 and 60 years. The white matter in the brain cell is called glial cells. The tumor that originates in this cell is called a glioma tumor. Astrocytoma, ependymomas, and oligodendrogliomas are the three types of glial cells present in the brain. Glioma tumor is classified based on the type of cell affected. Since this tumor originates in the brain, it is called a primary brain tumor. It accounts for 30% of brain tumors and 80% of malignant brain tumors. Pituitary glands are located at the back of the nose, just below the brain. Abnormal growth in the pituitary gland produces pituitary tumors. This gland controls growth, heart rate, and ability of a person. It is also responsible for controlling the number of hormones produced. It accounts for 12% to 19% of primary brain tumors, but most of them are benign.
MRI is usually influenced by numerous artifacts and noise sources [3]. In MRI, the complicated value of raw data are obtained in spatial frequency space and are corrupted by using Gaussian distribution noise. To calculate signal magnitude, one can first carry out a magnitude operation on the dataset and calculate the signal amplitude from the image of the received magnitude. Hence, noise restoration in MRI images is complicated due to its dispersions variants [4]. The Rician distribution tends to Rayleigh distribution in low-intensity areas, whereas it approaches a Gaussian distribution in immoderate-intensity areas.
The benefits of MRI are that the images are received invasive except inflicting any radiation on the patients, by way of growing actual 3D images with an excessive solution, and high Signal to noise ratio, and additionally refers to distinction mechanism for imaging several kinds of tissue and tissue residence which are in particular beneficial for soft tissues. Also, MRI is a better option for soft tissues, particularly for the research of brain tumors and the most notable difference is that MRIs do not use X-rays, while CT scans use X-rays which produce radiation and are harmful. However, some of the disadvantages of CNN are listed below:
• A Convolutional neural network is significantly slower due to an operation such as max pool.
• If the CNN has several layers, then the training process takes a lot of time if the computer lacks a good Graphics processing unit (GPU).
• A ConvNet requires a large Dataset to process and train the neural network.
There is abundant evidence that researchers have focused on brain tumor diagnosis by classification schemes using MRI images for the last few decades. The researchers demonstrated several techniques to increase the computation accuracy of brain tumors and their classifications. Exceptionally, several research works are related to Machine learning (ML) algorithms is adapted for the segmentation of medical images. A few of the recent works are discussed below:
Kumar et al. [5] proposed a hybrid approach to classify brain MRI tumor images. The term hybrid approach represents the adaptation of discrete wavelet transformation for feature extraction and genetic algorithm to reduce the number of features and support vector machine (SVM) for brain tumor classification. Further, Gu et al. [6] suggested an approach based on Glioma grading conventional MR images using deep learning study with transfer learning. In this work, photographs of the tumor represent about 80% of the tumor and are segmented with a rectangular area of interest (ROI). Then, 20% of the data was randomly chosen as a test dataset. A broad natural image database, AlexNet, and GoogLeNet, were learned and fine-tuned from pre-trained models on ImageNet and MRIs. Also, Deniz et al. [7] recommended a transfer learning-based histopathologic image classification for breast cancer detection classifier on histopathologic images for breast cancer. For extraction functionality, the authors used pre-trained Visual Geometry Group (VGG)-16 models and a fine-tuned AlexNet, which were then categorized using SVM. The assessment revealed that transfer learning provided better results than deep extraction of features and SVM classification.
Moreover, Hussein et al. [8] proposed lung and pancreatic tumor characterization in the deep learning era using novel supervised and unsupervised learning approaches. 3D convolutionary neural network and transfer learning is considered, and this is based on the supervised learning for which significant gains have been demonstrated with deep learning algorithms. An unmonitored learning algorithm is explored to overcome the restricted supply of labeled training knowledge and typical problems in medical imaging applications. To classify tumors, they used proportion-support vector machines. They tested the suggested supervised and unsupervised learning algorithms on two tumor detection challenges, lung with 1018 Computed Tomography (CT) and pancreas with 171 MRI scans. They obtained the findings of sensitivity and specificity in both problems.
Deep transfer learning on brain abnormality classification achieved significant classification efficiency. Additionally, Talo et al. [9] suggested the application of deep transfer learning for automated brain abnormality classification using MR images. With data augmentation and fine-tuned transfer learned model, they used ResNet-34 & also trained modified dense layers. The experimental findings concluded that a deep transfer-learned paradigm could be best applied to medical image classification with minimal pre-processing. Transfer learning is the technique by which the knowledge gained by an already trained model is used to learn another data set. Transfer learning (TL) allows using a pre-trained convolution neural network (CNN) model, which was developed for another related application. Transfer learning has shown its potential in the computer-aided diagram (CAD) of medical problems. The main objective of the TL is to enhance the learning capacity in the target domain using the knowledge of the source domain, the source learning task in the same domain, and the associated learning task.
Furthermore, Jain et al. [10] recommended convolutional neural network-based Alzheimer’s disease classification from magnetic resonance brain images. To detect Alzheimer’s disease through MRI, a pre-trained VGG-16 network is used. Transfer learning is applied to brain tumors in content-based image retrieval (CBIR). The assessment is carried out on a dataset that was freely accessible, and positive findings were obtained. For the validation range, the accuracy of the 3-way classification using the approach mentioned is 95.73 percent. Eventually, Swati et al. [11] proposed content-based brain tumor retrieval from MR images using transfer learning. To quantify the comparison between the query image and base image, it uses a deep convolutional neural network VGG19-based novel feature extraction method and implements closed-form metric learning. They embraced transfer learning and suggested a block-wise fine-tuning approach to increase retrieval efficiency. Extensive studies were conducted on a publicly accessible contrast-enhanced (CE)-MRI dataset comprising three types of brain tumors (i.e., glioma, meningioma, and pituitary tumor) obtained from 233 patients in axial and coronal and sagittal views with a total of 3064 images. The CE-MRI dataset attained a fivefold mean accuracy of 96.13 percent, which outperforms the state-of-the-art programs. Also, it is reported that an adaptive independent subspace analysis of Brain Magnetic Resonance imaging data, and is used to recognize the noticeable movement in the MRI scan data, and the findings are evaluated using the grey-level co-occurrence matrix, grey-level size-zone matrix, grey-level run-length matrix, and adjacent grey-tone discrepancy matrix and texture characteristics. Eventually, Saman et al. [12] suggested a survey on brain tumor segmentation and feature extraction of Magnetic Resonance (MR) images. In this work, various MRI segmentation and feature extraction methods are overviewed, and their features, merits, and demerits of the significant number of procedures are condensed. It offers a different segmentation algorithm from the basic threshold approach to the high-level segmentation method, enhancing the precision of the classification by selecting the most efficient characteristics and increasing the collection of training results. The extraction of the function gives the tumor characteristics, and it is advantageous to use it as an input to the classifier. In this, texture-based features, intensity-based features, shape-based features, and procedures for extracting deep learning-based features were investigated. Some of the notable literature reports are illustrated in Table 1.
Still, the available automated tumor detection systems are not providing satisfactory output, and there is a significant demand for a robust automated computer-aided diagnosis system for brain tumor detection. Conventional machine learning-based algorithms and models require domain-specific expertise and experience. These methods need efforts for segmentation and manual extraction of structural or statistical features, which may result in degradation of accuracy and efficiency of the system performance.
Based on the literature report, the objectives of this work are proposed as follows:
– To perform the comparative evaluation of CNN approaches to attain the best brain tumor classification accuracy rate.
– By implementing AlexNet, ResNet 50, and Inception V3 approaches, their performances are evaluated.
– To integrate the concept of augmentation to build the MR images in higher numbers considering the limited source images.
– To design the transfer learning approach for best accuracy prediction from the classifications.
– To obtain a higher accuracy rate of brain tumor classification using augmented MR images.
The rest of the article is illustrated as follows: Section 3 describes the various methodologies adopted for this study to classify the brain tumors, Section 4 demonstrates the results and discussions of the proposed models with a comparative study, and Section 5 concludes the work based on the attained outcomes.
A convolution neural network can robustly classify objects as invariance properties if put in different orientations. More precisely, translation, perspective, size, or illumination can be invariant to a CNN. This is essentially the premise of the augmentation of results. A dataset of images are taken under a small set of conditions in a real-world scenario. But, our target application can exist in several circumstances, such as various orientations, brightness, scale, location, etc. By training the network with more synthetically made data, it is accounted for these situations:
3.1 Data Collection and Sorting
CNN is a multi-neural layers network that perceives the pixel values with minimal processing. The CNN models used in this work are AlexNet, ResNet 50, and Inception V3. Analaysing these models, a database of 8487 images was used for training, validating, and testing. The most perplexing concern in medical imaging investigation is segmenting neurological syndromes from wide-ranging imaging datasets because a clot varies in size and type. MRI scan is employed to accumulate proof, and the images in the dataset have been labeled by skilled neurologists manually.
Moreover, the accuracy of the models differs from the existing models as data augmentation has been done. Machine learning becomes more effective when it is trained on a large dataset. In reality, it is challenging to acquire a large dataset. Hence, Data augmentation is performed. It uses various features like rotation range, sheer transform, zoom, fill up, flips, crop, and translation. Shifting the images left, right, up, or down to remove positional bias has proved to be quite helpful.
The overall framework of the suggested classification system is shown in Fig. 1. The input datasets are considered training, test, and class labels of the training datasets and act as input for the classifier model. The suggested model predicts the class labels and forms an output for the considered test datasets.
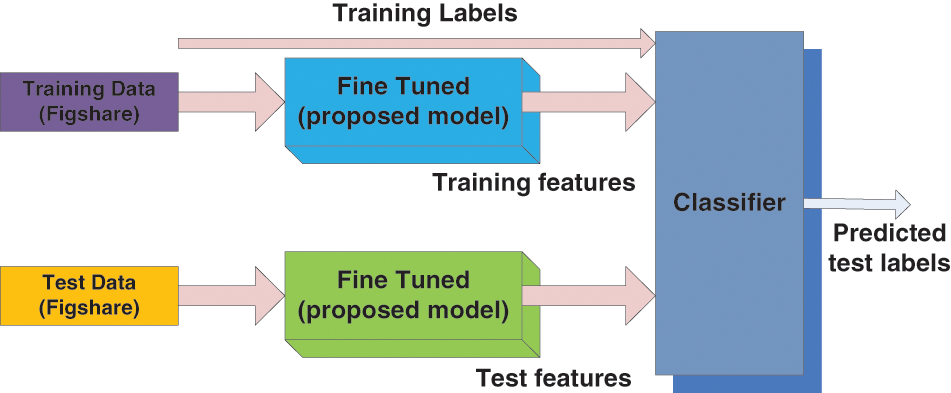
Figure 1: Overall framework for the proposed classification systems
AlexNet has a greater capacity to attain a higher accuracy rate even during challenging datasets. The overall performance of the AlexNet can be intensively harmed when any of the convolutional layers is removed [26,27]. Also, it is a leading framework for any element recognition. It has an assortment of features in the ML domain of computer vision. AlexNet would be increased for image processing tasks than any CNN models. It encompasses five wholly linked layers and three convolutional layers. The term ‘convolutional Kernels’ is called filters that sample significant features from the available image dataset.
Further, the consequence of the fifth convolutional level is nursed into an arrangement of two wholly linked layers via the Overlapping max-pooling layer. This function permits a bigger model for the training; nonetheless, it also decreases the training period. It enables to development of multi-GPU using half brains models on solitary GPUs and the lasting partition on alternative GPUs. The full exertion flow of the AlexNet framework is shown in Fig. 2.
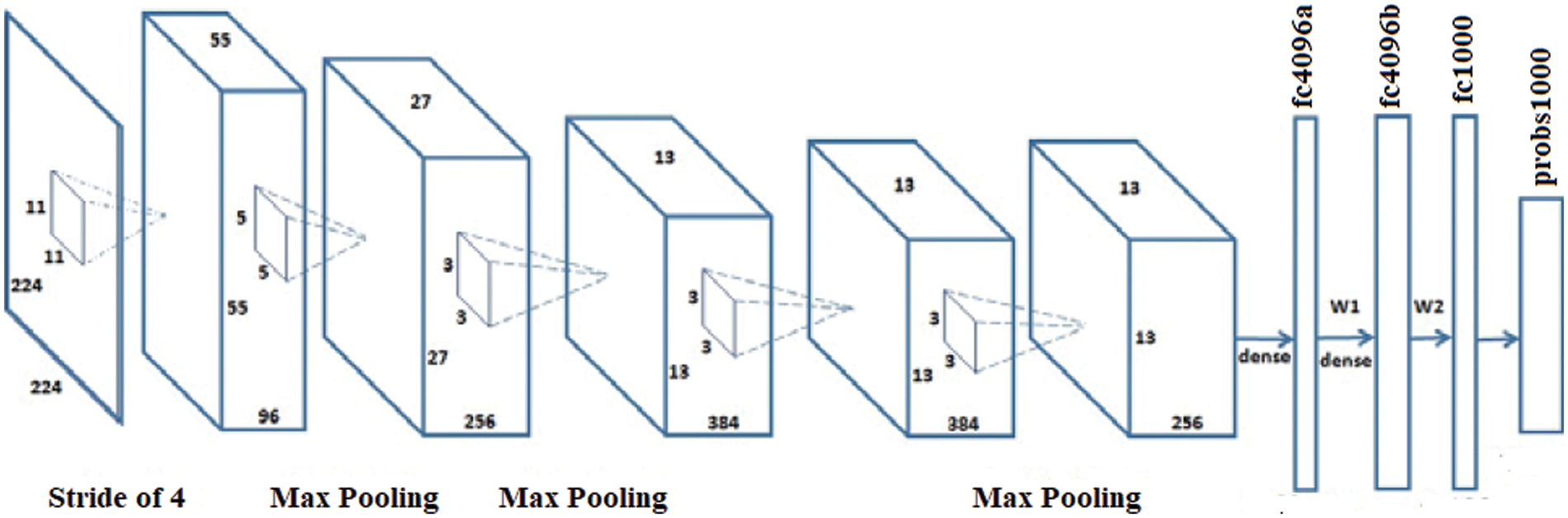
Figure 2: Architecture of the AlexNet model
This framework comprises four stages of operation, which have input image-sized multiples of 32 and color. It implements the preliminary convolution adapting 7 × 7 and Message passing library (MPL) considering 3 × 3 kernel sizes [28]. The first convolutional layer applies 96 filters of 7 × 7 on the input images at stride4 in Conv1, whereas 3 × 3 filters are applied at stride 2 in pool1 to increase the efficiency of the network. Subsequently, it switches the initial stage of the grid using three residual blocks (each encompassing three layers). Different adapted kernel sizes, likely 64, 64, and 128, implement the convolution in 3 layers (stage 1 block). The stability of ResNet 50 is decreased with increasing epochs because of over fitting issues. Next, it is essential to proceed from the first stage to the second, therefore, the input sizes are bridged into halves, and the width gets doubled. For every residual task (F), three layers (1 × 1, 3 × 3, 1 × 1) are arranged in sandwich mode. This network comprises a mean pooling level in the final stage straggled by 1000 neurons. The layered framework on the ResNet50 system is presented in Fig. 3 [28]. The first convolutional layer applies 96 filters of 11 × 11 on the input images at stride4 in Conv1, whereas 3 × 3 filters are applied at stride 2 in pool1. Likewise, the second convolutional layer applies 256 filters of 5 × 5. Similarly, 3 × 3 filter size is used in the third, fourth, and fifth layers with the filters 384, 384, and 256. The ReLU activation function is applied in each convolution layer.
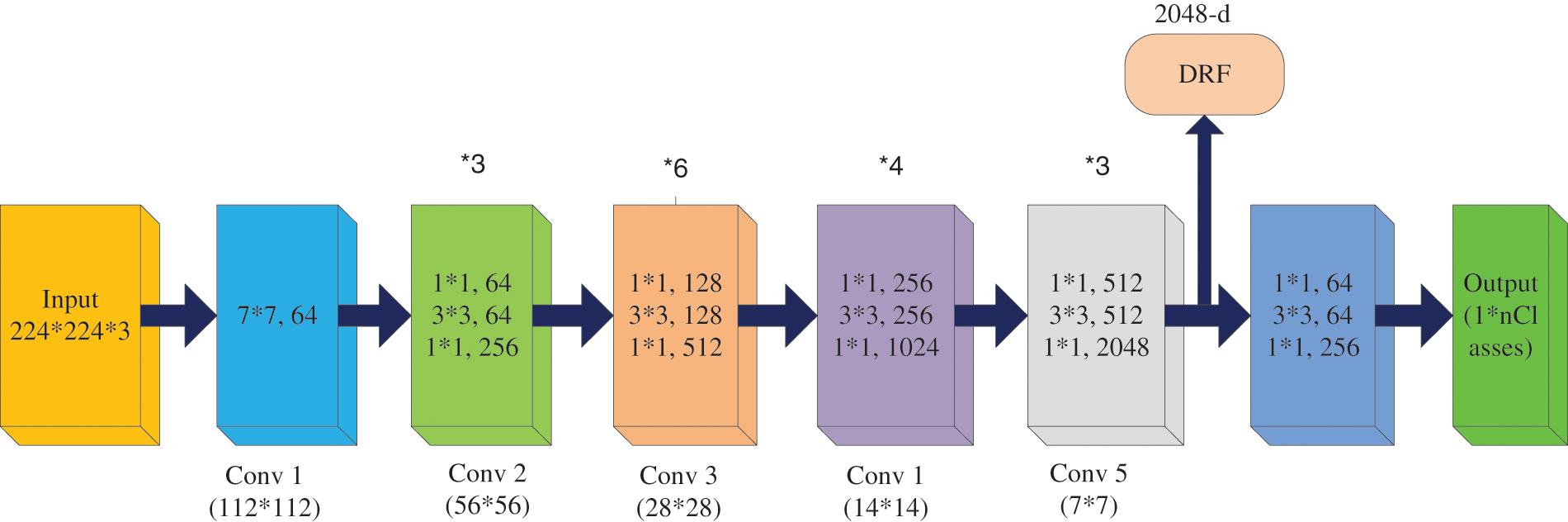
Figure 3: Architecture of ResNet 50
Inception v3 is a CNN model that achieves higher accuracy on the ImageNet dataset. It establishes one of the significant stages in the expansion of CNN framework and comprises four different versions with variances in enactment and accuracy [29]. It contains 42 layers along with earlier versions, and some of them are normalization for auxiliary classifiers, 7 × 7 convolution, label smoothing, and RMS Prop optimizer. This model is the pinnacle of different ideas developed by many researchers over the decades. The original work was based on the Rethinking the Inception Architecture for Computer Vision suggested by Szegedy et al. This model is made of symmetric and asymmetric building blocks, including average pooling, convolutions, and dropouts max pooling, concerts, and fully connected layers. The batch norm is used widely throughout the model and is applied to the activation inputs. Fig. 4 depicts the architecture of Inception V3, and the Softmax facilitates the loss computation.
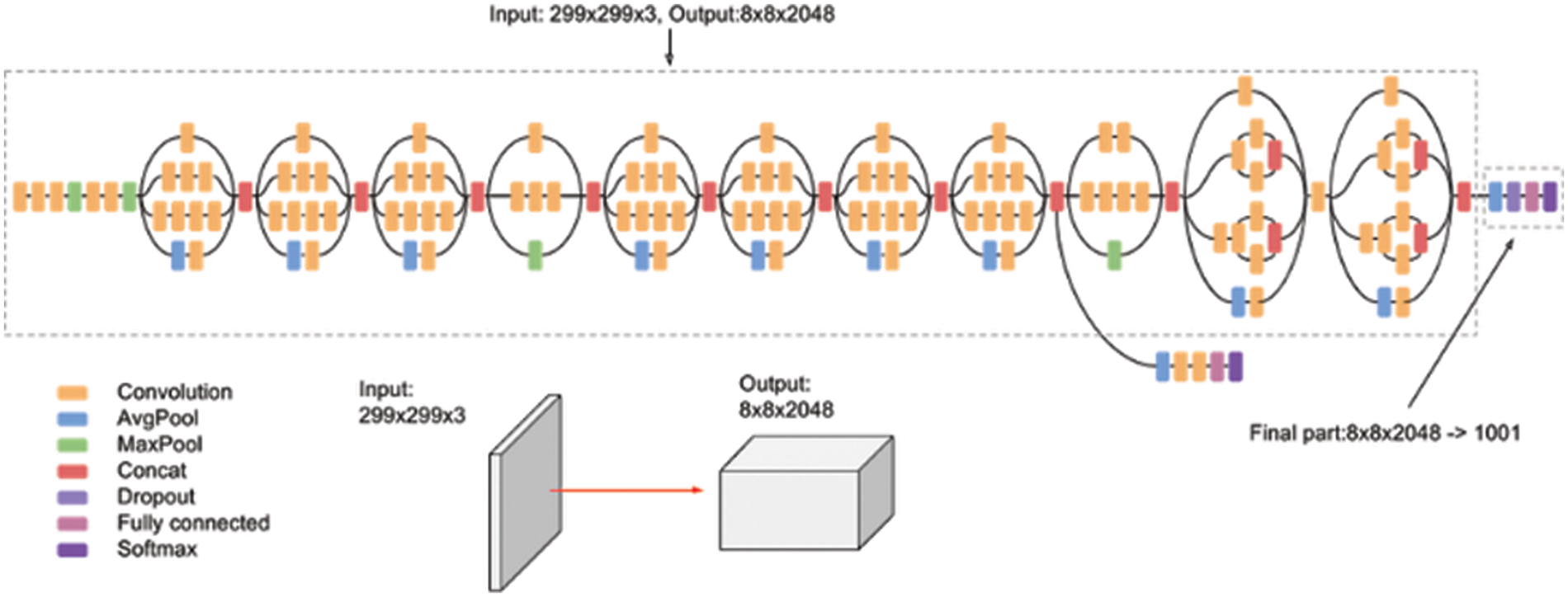
Figure 4: Inception V3 architecture
3.3 Transfer Learning (TL) in Inductive Setting
TL is a form of machine learning where a model built for a function is reused on a second task as the beginning of a model. It is a standard method where pre-trained models are used as the beginning for the computer vision and natural language processing tasks, for the ample computational and time resources needed to be constructed for neural network models on these issues. TL is the knowledge attained using a previously trained model that can be adapted to study other datasets [26]. The main objective of the TL is to enhance the learning capacity in the target domain (Dt) using the knowledge of the source domain (Ds), source learning task (Ts) in the same domain, and the associated learning task (Tt), the various settings are described for TL, and it is based on task type and the nature of the dataset presented at the Dt and Ds [30]. Generally, the TL method is called inductive TL because characterized data is accessible in both domains, i.e., source and target, for a classification assignment [31]. Considering this statement, now the domain is signified as D = (Ai Bi) ∀i (Where ‘Ai’ denotes the feature vector and ‘Bi’ means the class label, and ‘i’ represents the training sample iteration. Fig. 5 illustrates the framework of the TL setting for the proposed model adapted for this work.
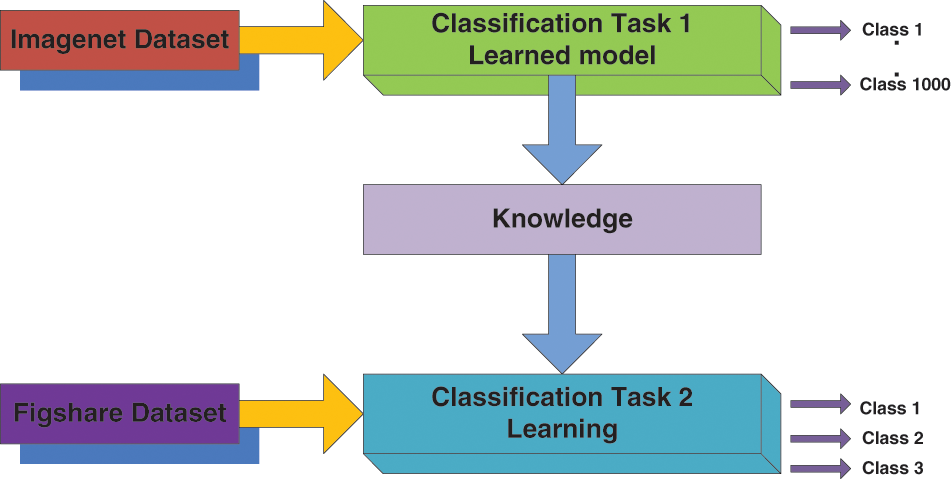
Figure 5: Transfer learning model setting
Transfer learning is the technique by which the knowledge gained by an already trained model is used to learn another data set. Transfer learning allows using a pre-trained CNN model, which was developed for another related application. Transfer learning has shown its potential in CAD of medical problems. The main objective of the TL is to enhance the learning capacity in the target domain using the knowledge of the source domain, the source learning task in the same domain, and the associated learning task.
The suggested classification models are demonstrated in the MATLAB virtual platform, and the observed outcomes are illustrated in the following sections.
DA is the best way to make an efficient classification model that can be used to train the available data into more data. The number of images for training and testing the datasets is limited. One way to solve this issue is to build duplicate data and add it to the training set. It is reasonable to build duplicate data for some classification tasks, which primarily postulates data augmentation. In practice, the dataset is limited, but the target application may have asymmetric conditions like, as different orientations, brightness, scale, location, etc. These situations are accounted for by training the CNN with supplementary modified data. There are several features adapted for augmentation, as follows:
– Rotation_range is in degrees (0–180), within which the images randomly rotate to pictures.
– Height_shift and Width_shift are ranges within which a part or entire image randomly translates horizontally or vertically.
– Shear_range is for the random application of shear transforms.
– Zoom_range is for the random application of zoom-in pictures.
– Horizontal_flip is for the random flipping of half image horizontally (with no horizontal symmetry).
– Fill_mode is to fill in the newly created pixels after the rotation or shift.
Further, the accuracy of the network can be enhanced through augmentation as the class meningioma have the smallest number of samples in the training set, and the most affected class in terms of misclassifications and Overfitting could be avoided by data augmentation. The following figure shows the original image (Fig. 6a) and the augmented images (Fig. 6b). The original image is increased in the ratio of 1:9 using features like rotation range (15 degrees), sheer transform, zoom in/out, flip, width shift, and height shift. Then the final augmented images are derived, as shown in the figure. The original database comprises 843 images which were then augmented to 8487 images, and these augmented images are used for training, validation, and testing for other processes.
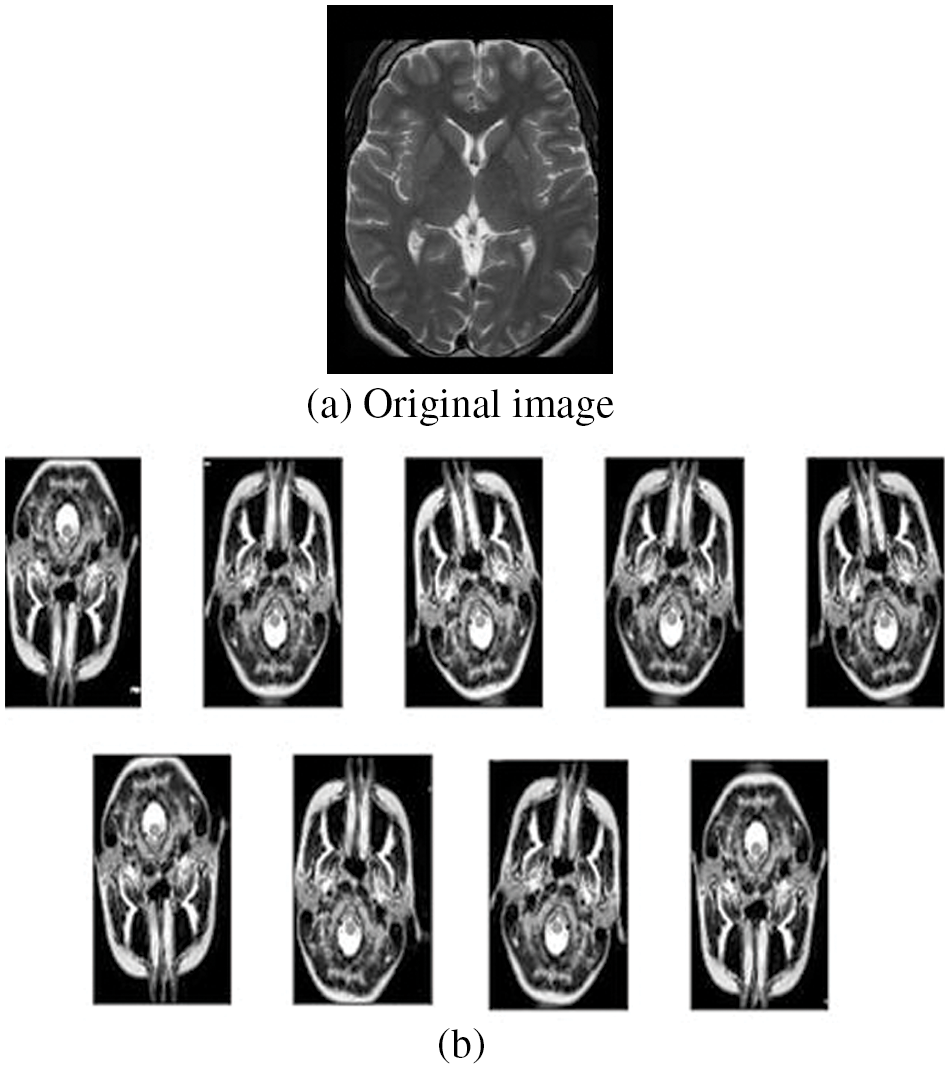
Figure 6: Original and augmented images
4.2 Pre-processing and Datasets
The datasets are taken from the figshare platform uploaded by Cheng [32], which is flexibly accessible and is generally adapted for assessing the classification and retrieval systems. From the dataset, brain tumors containing 843 T1-weighted contrast-enhanced images (taken from Philips medical systems: GyroscanIntera 1.5 T) from 233 patients with three kinds of brain tumor: meningioma (300 slices), glioma (343 slices), and pituitary tumor (200 slices) and then augmented into 8487 brain MRI images. The T1-weighted pictures are produced using short TE and TR times. The contrast and brightness of the image are significantly assessed by the T1 properties of tissue. The T1 parameters are defined as: repetition time = 9.813, echo time = 4.603, number of phase encoding steps = 192, echo train length = 0, reconstruction diameter = 240, and flip angle = 8. The dataset comprises images of the MRI modality that encompasses three views: coronal, sagittal, and axial. The executed images are transferred into MATLAB files with 512 × 512 sizes. The MRI pictures from the data sets are pre-processed for evaluation, as described in Fig. 7, and the images are standardized in intensity ranges.

Figure 7: Dataset pre-processing method
The test images were acquired using a 3 Tesla Siemens Magnetom Spectra MR machine. The total numbers of slices for all channels were 15, which leads to a total of 135 images at nine slices or images per patient with a field of view of 200 mm, an interslice gap of 1 mm, and a voxel of size 0.78 mm × 0.78 mm × 0.5 mm. The proposed methodology is applied to a real dataset, including brain MR images of 512 × 512 pixel size, and was converted into grayscale before further processing. The images are re-sized using min-max normalization practice considering the intensity scales between 0 and 1. Whole datasets are divided into several subsets. Some subsets can be adapted for testing, and others are considered for the training set. Further, this can be recurrent to warrant that all subsets were tested at least once. The possible methods to reduce the systematic are bootstrap aggregation, boosting ensemble, and stacking ensemble approaches. The training set is split into five smaller sets. A model is trained using 4 of the folds as training data, and a fold of data is used for testing. The same procedure is repeated until all folds are tested.
The accuracy curves for all three models were obtained, and the results were illustrated. Fig. 8 shows the accuracy of the AlexNet model for 20 epochs. As the epochs increase, the accuracy is seen to be increased in both training and validation. The accuracy of the model is evaluated to be 95.2% which is comparatively more significant and less stable.
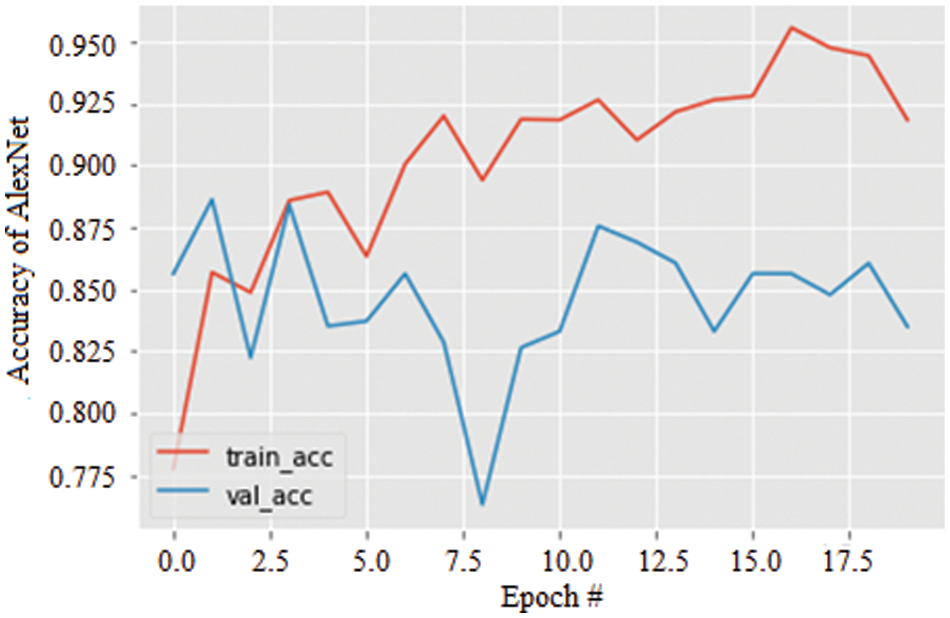
Figure 8: Accuracy graph of AlexNet
Specifically, the stability of the accuracy can be validated by the variation in the data augmentation and found to be excellent compared with the AlexNet model. Further, Fig. 9 shows the accuracy of the ResNet 50 model for 20 epochs. As the epochs increase, the accuracy is seen to be increased in both training and validation by about 91.27%.
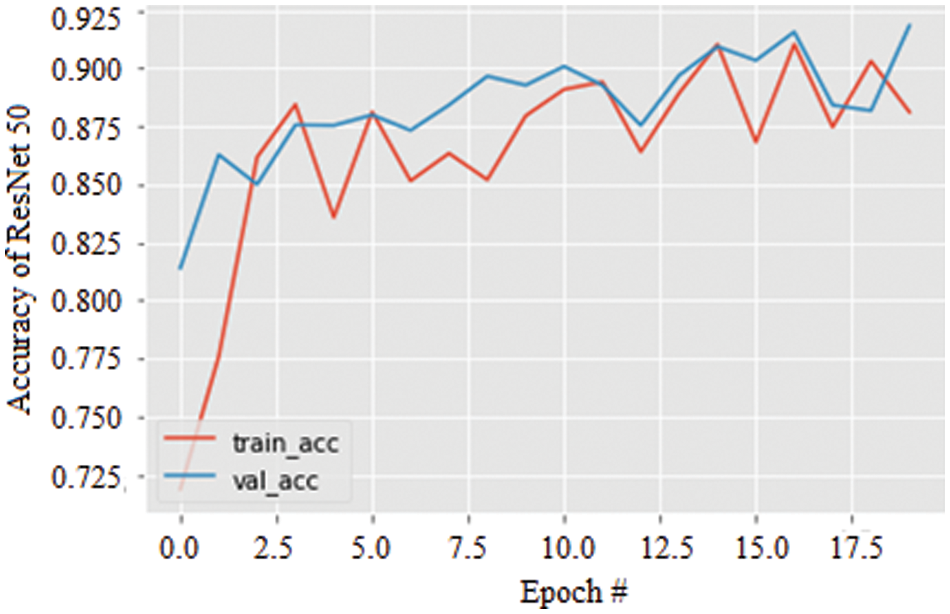
Figure 9: Accuracy graph of ResNet 50
Lastly, the accuracy of the Inception V3 model for 20 epochs is simulated and illustrated in Fig. 10. Again, increasing epochs augment the model’s accuracy in both training and validation and are observed to be 89.52%. However, similar to AlexNet, the stability of the model is not significant compared with the ResNet 50 model.
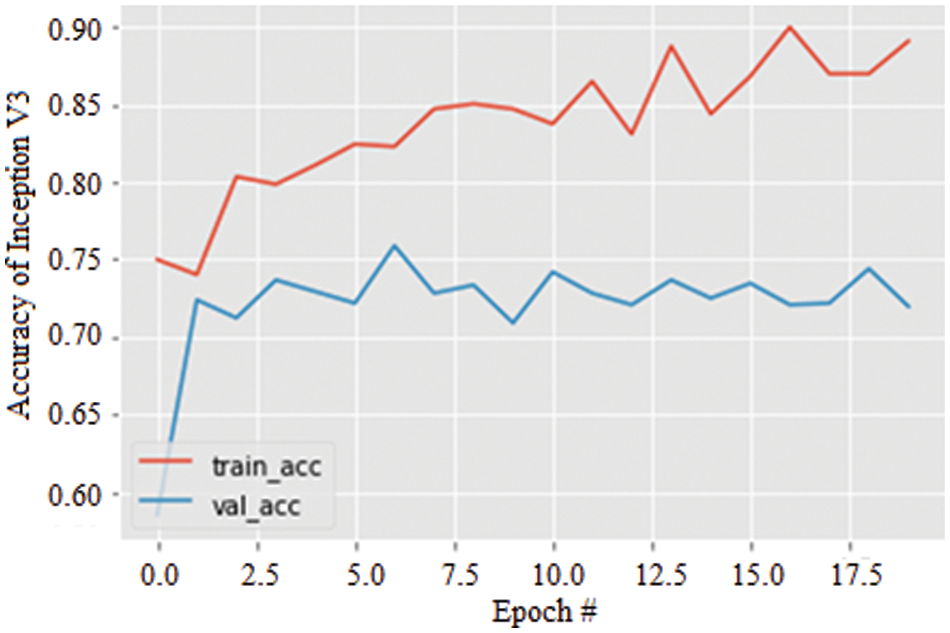
Figure 10: Accuracy graph of Inception V3
With further increase of epoch, the accuracy of all three models is somewhat enhanced. At the 50th epoch, the accuracy is improved significantly and found to be great for all three models, i.e., AlexNet, ResNet 50, and Inception V3. Finally, the maximum accuracy is found at the 120th epoch, and the complete behavior is illustrated in Fig. 11. Further increase of epoch, i.e., beyond 120, the accuracy tends to take a declining trend. Henceforth, the 120th epoch is the peak accuracy when the data augmentation is performed on the data set.
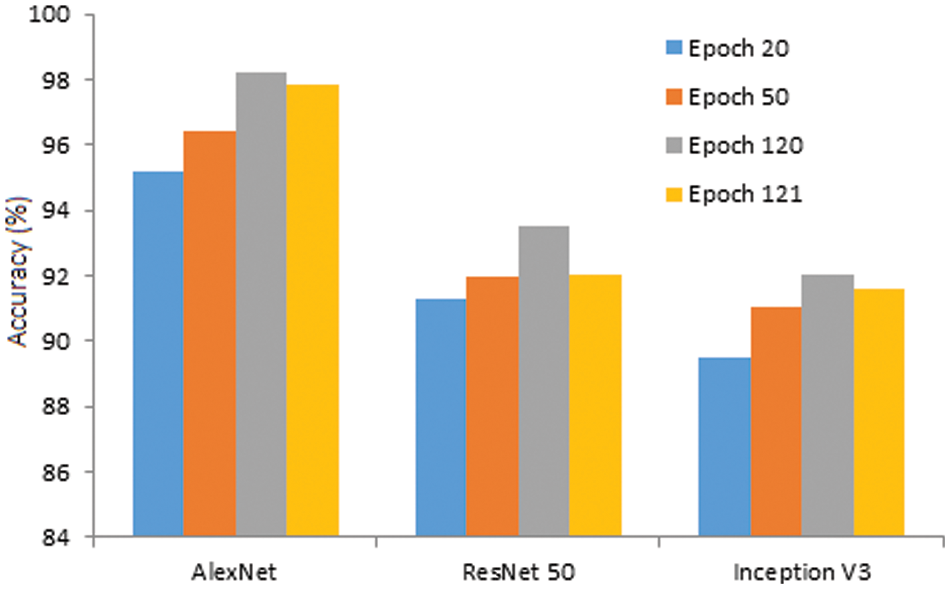
Figure 11: Accuracy of the model against epoch
Further, the same models are simulated using original image datasets, and the observed results are compared with augmented results for 120 epochs in Fig. 12. The accuracy of AlexNet, ResNet 50, and Inception V3 is reduced from 98.24% to 89.56%, 93.51% to 85.34%, and 92.07% to 83.21%, respectively. The results ensure that the augmented datasets effectively identify brain tumors with a higher accuracy rate.
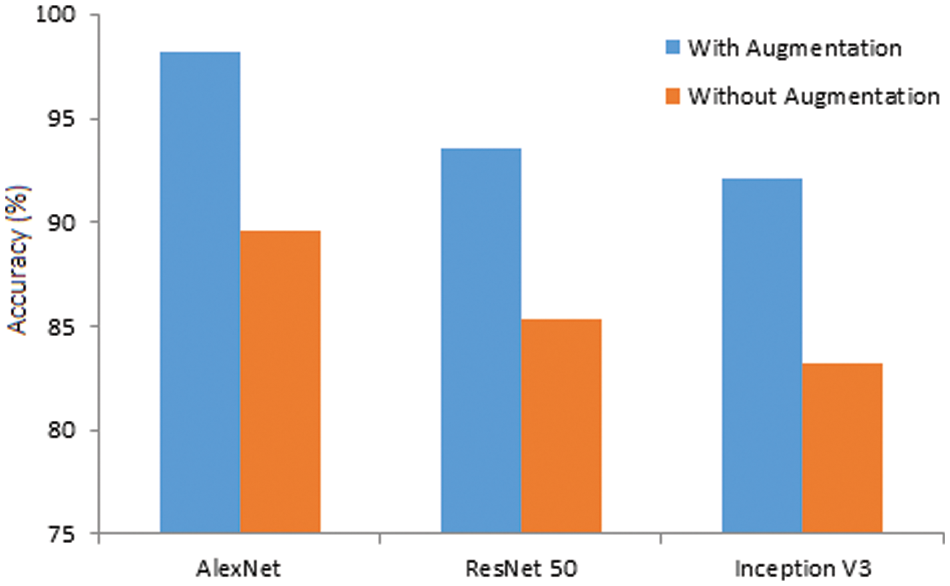
Figure 12: Accuracy of the models (with and without augmentation)
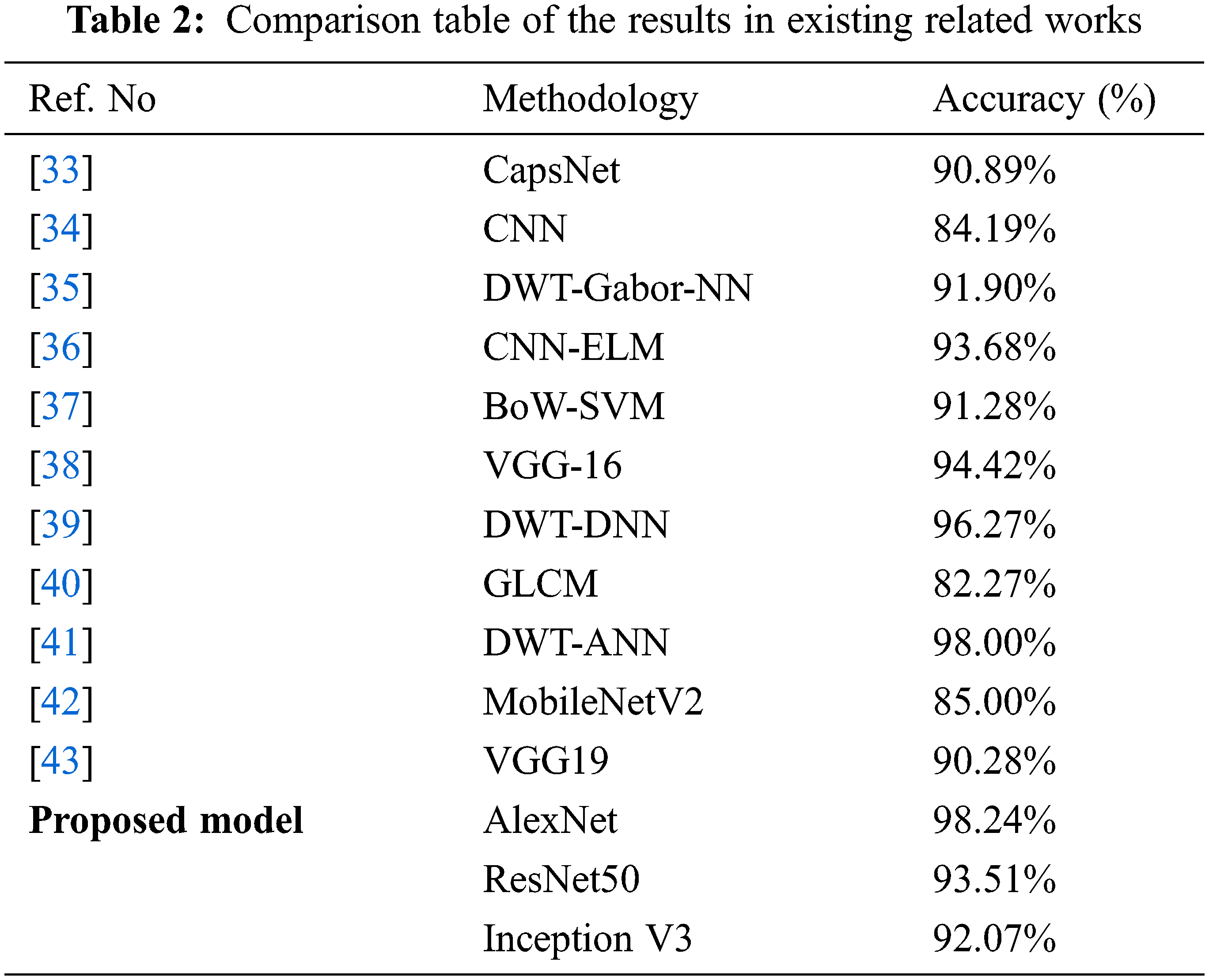
Furthermore, the outcomes of the proposed models are compared with existing reports, which is illustrated in Table 2.
The above table proved that the proposed models with augmentation offered the best brain tumor prediction accuracy compared with other related works. AlexNet outperforms other models among the proposed models due to its significant and relative accuracy scale. This method issued for brain tumor identification using MR images effectively.
This work demonstrates the effectiveness of the CNN architecture in recognizing the brain tumor using augmented MRI images by adapting various approaches such as Alexnet, ResNet50, and Inception V3. Based on the observed results, the following conclusions are made:
• AlexNet performs better than Resnet 50 & Inception V3 for the particular data set given based on better symmetric scale. However, the accuracy is slightly more stable in ResNet 50.
• As the number of epochs increased, the accuracy improved for all three models. The obtained accuracy at the 120th epoch for AlexNet is 98.24% with data augmentation. Relatively, ResNet 50 & Inception V3 are 93.51% and 92.07%, respectively.
• With data augmentation, AlexNet outperforms both ResNet 50 and Inception V3. Hence, it is concluded that AlexNet is the most preferred out of all the three CNN models used in this work when data augmentation is applied.
• Without augmentation, prediction accuracy obtained by the three models diminished significantly. Therefore, the AlexNet model can be effectively used for brain tumor classification with data augmentation.
• Though the accuracy of the AlexNet shows best scale comaratively, it has the limitation of particularly highly unstable characteristics.
To sum up, the AlexNet model offers the best accuracy scale compared with the other two considered models, still the stability of the accuracy needs to be maintained for the increasing epoch. Also, to ensure higher reliability of the AlexNet, it is essential to demonstrate the sensitivity and specificity of the proposed model. Therefore, the extension of this work will focus on these gaps, i.e., instability, and sensitivity and specificity assessment of the suggested model [44–52].
Acknowledgement: Ahmed Alhussen would like to thank the Deanship of Scientific Research at Majmaah University for supporting this work under Project No. R-2022-####.
Funding Statement: Ahmed Alhussen would like to thank the Deanship of Scientific Research at Majmaah University for supporting this work under Project No. R-2022-####.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. N. Mushtaq, A. A. Khan, F. A. Khan, M. J. Ali, M. M. Ali Shahid et al., “Brain tumor segmentation using multi-view attention based ensemble network,” Computers, Materials & Continua, vol. 72, no. 3, pp. 5793–5806, 2022. [Google Scholar]
2. B. Subhashis, “Brain tumor detection and classification from multi-channel MRIs using deep learning and transfer learning,” Computer Science, vol. 1, no. 1, pp. 1898–5338, 2017. [Google Scholar]
3. R. Hao, K. Namdar, L. Liu and F. Khalvati, “A transfer learning–based active learning framework for brain tumor classification,” Frontiers in Artificial Intelligence, vol. 4, pp. 635766, 2021. [Google Scholar]
4. G. Allah, A. M. Sarhan and A. M. Elshennawy, “Classification of brain MRI tumor images based on deep learning PGGAN augmentation,” Diagnostics, vol. 11, no. 12, pp. 2343, 2021. [Google Scholar]
5. S. Kumar, C. Dabasand and S. Godara, “Classification of brain MRI tumor images: A hybrid approach,” Procedia Computer Science, vol. 122, pp. 510–517, 2017. [Google Scholar]
6. Y. Gu, X. Lu, L. Yang, B. Zhang, D. Yu et al., “Automatic lung nodule detection using a 3D deep convolutional neural network combined with a multiscale prediction strategy in chest CTs,” Computer Biology Medicine, vol. 103, pp. 220–231, 2018. [Google Scholar]
7. E. Deniz, A. Şengür, Z. Kadiroğlu, Y. Guo, V. Bajaj et al., “Transfer learning based histopathologic image classification for breast cancer detection,” Health Information Science System, vol. 6, no. 1, pp. 96–104, 2018. [Google Scholar]
8. S. Hussein, P. Kandel, C. W. Bolan, M. B. Wallace and U. Bagci, “Lung and pancreatic tumor characterization in the deep learning era: Novel supervised and unsupervised learning approaches,” IEEE Transactions on Medical Imaging, vol. 6, pp. 209–222, 2019. [Google Scholar]
9. M. Talo, U. B. Baloglu and U. R. Acharya, “Application of deep transfer learning for automated brain abnormality classification using MR images,” Cognitive System Resources, vol. 54, pp. 176–188, 2019. [Google Scholar]
10. R. Jain, N. Jain, A. Aggarwal and D. J. Hemanth, “Convolutional neural network based Alzheimer’s disease classification from magnetic resonance brain images,” Cognitive System Research, vol. 52, no. 2, pp. 45–57, 2019. [Google Scholar]
11. Z. N. K. Swati, Q. Zhao, M. Kabir, F. Ali, A. Zakir et al., “Content-based brain tumor retrieval for MR images using transfer learning,” IEEE Access, vol. 7, pp. 17809–17822, 2019. [Google Scholar]
12. S. Saman and J. Narayanan, “Survey on brain tumor segmentation and feature extraction of MR images,” Springer Journal of Multimedia Information Retrieval, vol. 8, no. 2, pp. 19–99, 2012. [Google Scholar]
13. M. Shahajad, D. Gambhir and R. Gandhi, “Features extraction for classification of brain tumor MRI images using support vector machine,” in 11th Int. Conf. on Cloud Computing, Data Science Engineering (Confluence), Noida, India, pp. 767–772, 2021. [Google Scholar]
14. M. Bhuvaneswari, “Automatic segmenting technique of brain tumors with convolutional neural networks in MRI images,” in 6th Int. Conf. on Inventive Computation Technologies (ICICT), Tamil Nadu, India, pp. 759–764, 2021. [Google Scholar]
15. N. Noreen, S. Palaniappan, A. Qayyum, I. Ahmad, M. Imran et al., “A deep learning model based on concatenation approach for the diagnosis of brain tumor,” IEEE Access, vol. 8, pp. 55135–55144, 2020. [Google Scholar]
16. Y. Bhanothu, A. Kamalakannan and G. Rajamanickam, “Detection and classification of brain tumor in MRI images using deep convolutional network,” in 6th Int. Conf. on Advanced Computing and Communication Systems (IC-ACCS), Bombai, India, vol. 1, pp. 248–252, 2020. [Google Scholar]
17. M. R. Ismael and I. Abdel-Qader, “Brain tumor classification via statistical features and back-propagation neural network,” in IEEE Int. Conf. on Electro/Information Technology (EIT), New Delhi, India, vol. 1, pp. 0252–0257, 2018. [Google Scholar]
18. S. Deepak and P. Ameer, “Brain tumor classification using deep CNN features via transfer learning,” Computers in Biology and Medicine, vol. 111, pp. 103345, 2019. [Google Scholar]
19. H. T. Zaw, N. Maneeratand and K. Y. Win, “Brain tumor detection based on naive Bayes classification,” in 5th Int. Conf. on Engineering, Applied Sciences and Technology (ICEAST), Florida, USA, pp. 1–4, 2019. [Google Scholar]
20. S. Irsheidat and R. Duwairi, “Brain tumor detection using artificial convolutional neural networks,” in 11th Int. Conf. on Information and Communication Systems (ICICS), Irbid, Jordan, pp. 197–203, 2020. [Google Scholar]
21. C. L. Choudhury, C. Mahanty, R. Kumar and B. K. Mishra, “Brain tumor detection and classification using convolutional neural network and deep neural network,” in Int. Conf. on Computer Science, Engineering and Applications (ICCSEA), Ganupur, India, vol. 1, pp. 1–4, 2020. [Google Scholar]
22. V. Singh, M. K. Gourisaria, S. S. Rautaray, M. Pandey, M. Sahni et al., “Diagnosis of intracranial tumors via the selective CNN data modeling technique,” Applied Science, vol. 12, no. 2, pp. 2900, 2020. [Google Scholar]
23. G. Allah, A. M. Sarhan and N. Melshennawy, “Classification of brain MRI tumor images based on deep learning PGGAN augmentation,” Diagnostics, vol. 11, no. 12, pp. 2343, 2021. [Google Scholar]
24. C. Srinivas, N. Prasad, M. Zakariah, Y. Ajmialothaibi, K. Shaukat et al., “Deep transfer learning approaches in performance analysis of brain tumor classification using MRI images,” Journal of Healthcare Engineering, vol. 2022, pp. 3264367, 2022. [Google Scholar]
25. A. Naseer, T. Yasir, A. Azhar, T. Shakeel and K. Zafar, “Computer-aided brain tumor diagnosis: Performance evaluation of deep learner CNN using augmented brain MRI,” International Journal of Biomedical Imaging, vol. 2021, pp. 5513500, 2021. [Google Scholar]
26. S. Deepak and P. M. Ameer, “Brain tumor classification using deep CNN features via transfer learning,” Computers in Biology and Medicine, vol. 111, pp. 103345, 2019. [Google Scholar]
27. M. Sunita, S. Kulkarni and G. Sundari, “Transfer learning using convolutional neural network architectures for glioma classification from MRI images,” International Journal of Computer Science and Network Security, vol. 21, no. 2, pp. 198–204, 2021. [Google Scholar]
28. M. Sunita,S. Kulkarni and G. Sundari, “Comparative analysis of performance of deep CNN based framework for brain MRI classification using transfer learning,” Journal of Engineering Science and Technology, vol. 16, no. 4, pp. 2901–2917, 2021. [Google Scholar]
29. B. Premamayudu, A. Lakshmipathi and S. Peram, “Deep neural networks with transfer learning model for brain tumors classification,” Traitement du Signal, vol. 37, no. 4, pp. 593–601, 2020. [Google Scholar]
30. L. Shao, F. Zhu and X. Li, “Transfer learning for visual categorization: A survey,” IEEE Transactions on Neural Network Learning System, vol. 26, no. 5, pp. 1019–1034, 2015. [Google Scholar]
31. S. J. Pan and Q. Yang, “A survey on transfer learning,” IEEE Transactions on Knowledge Data Engineering, vol. 22, no. 10, pp. 1345–1359, 2010. [Google Scholar]
32. J. Cheng, “Brain tumor dataset,” Figshare Dataset, vol. 12, pp. 101–111, 2017. [Google Scholar]
33. P. Afshar, K. N. Plataniotis and A. Mohammadi, “Capsule networks for brain tumor classification based on MRI images and course tumor boundaries,” in IEEE Int. Conf. on Acoustics, Speech and Signal Processing, Brighton, UK, pp. 1368–1372, 2019. [Google Scholar]
34. N. Abiwinanda, M. Hanif, S. T. Hesaputra, A. Handayani and T. R. Mengko, “Brain tumor classification using convolutional neural network,” Springer World Congress on Medical Physics and Biomedical Engineering, vol. 12, no. 3, pp. 183–189, 2018. [Google Scholar]
35. M. R. Ismael and I. Abdel-Qader, “Brain tumor classification via statistical features and back-propagation neural network,” in IEEE Int. Conf. on Electro/Information Technology, Oakland, USA, pp. 0252–0257, 2018. [Google Scholar]
36. A. Pashaei, H. Sajedi and N. Jazayeri, “Brain tumor classification via convolutional neural network and extreme learning machines,” in IEEE 8th Int. Conf. on Computer and Knowledge Engineering, Mashhad, Iran, pp. 314–319, 2018. [Google Scholar]
37. J. Cheng, W. Huang, S. Cao, R. Yang, W. Yang et al., “Enhanced performance of brain tumor classification via tumor region augmentation and partition,” PLoS One, vol. 10, no. 10, pp. 14–38, 2015. [Google Scholar]
38. M. R. Beyin, “Performance comparison of different pre-trained deep learning models in classifying brain MRI images,” Acta Infologica, vol. 5, no. 1, pp. 141–154, 2021. [Google Scholar]
39. H. Mohsen, “Classification using deep learning neural networks for brain tumors,” Future Computing and Informatics Journal, vol. 3, pp. 68–71, 2018. [Google Scholar]
40. W. Widhiarso, Y. Yohannes and C. Prakarsah, “Brain tumor classification using gray level co-occurrence matrix and convolutional neural network,” Indonesian Journal of Electronics and Instrumentation Systems, vol. 8, no. 2, pp. 179–190, 2018. [Google Scholar]
41. M. Sasikala and N. Kumaravel, “A wavelet-based optimal texture feature set for classification of brain tumours,” Journal of Medical Engineering & Technology, vol. 32, no. 3, pp. 198–205, 2008. [Google Scholar]
42. E. I. Zacharaki, S. Wang, S. Chawla, D. Soo Yoo and E. R. Davatzikos, “Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme,” Magnetic Resonance in Medicine, vol. 62, no. 6, pp. 1609–1618, 2009. [Google Scholar]
43. J. Cheng, W. Huang and S. Cao, “Enhanced performance of brain tumor classification via tumor region augmentation and partition,” PLoS One, vol. 10, no. 10, pp. 140–154, 2015. [Google Scholar]
44. A. Jawaharlalnehru, S. Thalapathiraj, R. Dhanasekar, L. Vijayaraja, R. Kannadasan et al., “Machine vision-based human action recognition using spatio-temporal motion features (STMF) with difference intensity distance group pattern (DIDGP),” Electronics, vol. 11, no. 15, pp. 2363, 2022. [Google Scholar]
45. Y. C. Shekhar, J. Singh, A. Yadav, H. S. Pattanayak, R. Kumar et al., “Malware analysis in IoT & android systems with defensive mechanism,” Electronics, vol. 11, no. 15, pp. 2354, 2022. [Google Scholar]
46. J. Arunnehru, T. Sambandham, V. Sekar, D. Ravikumar, V. Loganathan et al., “Target object detection from unmanned aerial vehicle (UAV) images based on improved YOLO algorithm,” Electronics, vol. 11, no. 15, pp. 2343, 2022. [Google Scholar]
47. L. Kuruva, R. Kaluri, N. Gundluru, Z. S. Alzamil, D. S. Rajput et al., “A review on deep learning techniques for IoT data,” Electronics, vol. 11, no. 10, pp. 1604, 2022. [Google Scholar]
48. K. M. AnulHaq, U. Garg, M. Abdul Rahim Khan and V. Rajinikanth, “Fusion-based deep learning model for hyperspectral images classification,” Computers, Materials & Continua, vol. 72, no. 1, pp. 939–957, 2022. [Google Scholar]
49. M. AnulHaq, “Cdlstm: A novel model for climate change forecasting,” Computers, Materials & Continua, vol. 71, no. 2, pp. 2363–2381, 2022. [Google Scholar]
50. M. AnulHaq, “Smotednn: A novel model for air pollution forecasting and aqi classification,” Computers, Materials & Continua, vol. 71, no. 1, pp. 1403–1425, 2022. [Google Scholar]
51. M. AnulHaq and M. Abdul Rahim Khan, “Dnnbot: Deep neural network-based botnet detection and classification,” Computers, Materials & Continua, vol. 71, no. 1, pp. 1729–1750, 2022. [Google Scholar]
52. M. AnulHaq, M. Abdul Rahim Khan and T. Al-Harbi, “Development of pccnn-based network intrusion detection system for edge computing,” Computers, Materials & Continua, vol. 71, no. 1, pp. 1769–1788, 2022. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF
 Downloads
Downloads
 Citation Tools
Citation Tools