 Open Access
Open Access
ARTICLE
Horizontal Voting Ensemble Based Predictive Modeling System for Colon Cancer
1 Department of Electronics and Communication Engineering, Infant Jesus College of Engineering, Tuticorin, Tamil Nadu, India
2 Department of Computer Science and Engineering, Infant Jesus College of Engineering, Tuticorin, Tamil Nadu, India
* Corresponding Author: Ushaa Eswaran. Email:
Computer Systems Science and Engineering 2023, 46(2), 1917-1928. https://doi.org/10.32604/csse.2023.032523
Received 20 May 2022; Accepted 28 October 2022; Issue published 09 February 2023
Abstract
Colon cancer is the third most commonly diagnosed cancer in the world. Most colon AdenoCArcinoma (ACA) arises from pre-existing benign polyps in the mucosa of the bowel. Thus, detecting benign at the earliest helps reduce the mortality rate. In this work, a Predictive Modeling System (PMS) is developed for the classification of colon cancer using the Horizontal Voting Ensemble (HVE) method. Identifying different patterns in microscopic images is essential to an effective classification system. A twelve-layer deep learning architecture has been developed to extract these patterns. The developed HVE algorithm can increase the system’s performance according to the combined models from the last epochs of the proposed architecture. Ten thousand (10000) microscopic images are taken to test the classification performance of the proposed PMS with the HVE method. The microscopic images obtained from the colon tissues are classified into ACA or benign by the proposed PMS. Results prove that the proposed PMS has ~8% performance improvement over the architecture without using the HVE method. The proposed PMS for colon cancer reduces the misclassification rate and attains 99.2% of sensitivity and 99.4% of specificity. The overall accuracy of the proposed PMS is 99.3%, and without using the HVE method, it is only 91.3%.Keywords
The sequential mutations in the colonic epithelial cells are the primary reason for colon cancer. These mutations transform the normal epithelial cells into AdenoCAarcinomas (ACA) with increasing dysplasia. Fig. 1 shows the cancer statistics in 2020 by the International agency for research on cancer. It indicates that 10% of cancers worldwide are colon cancers, the third amongst others, next to breast and lung cancers. To ensure good classification results for colon cancer using microscopic images, many Computer Aided Diagnosis (CAD) systems are designed.
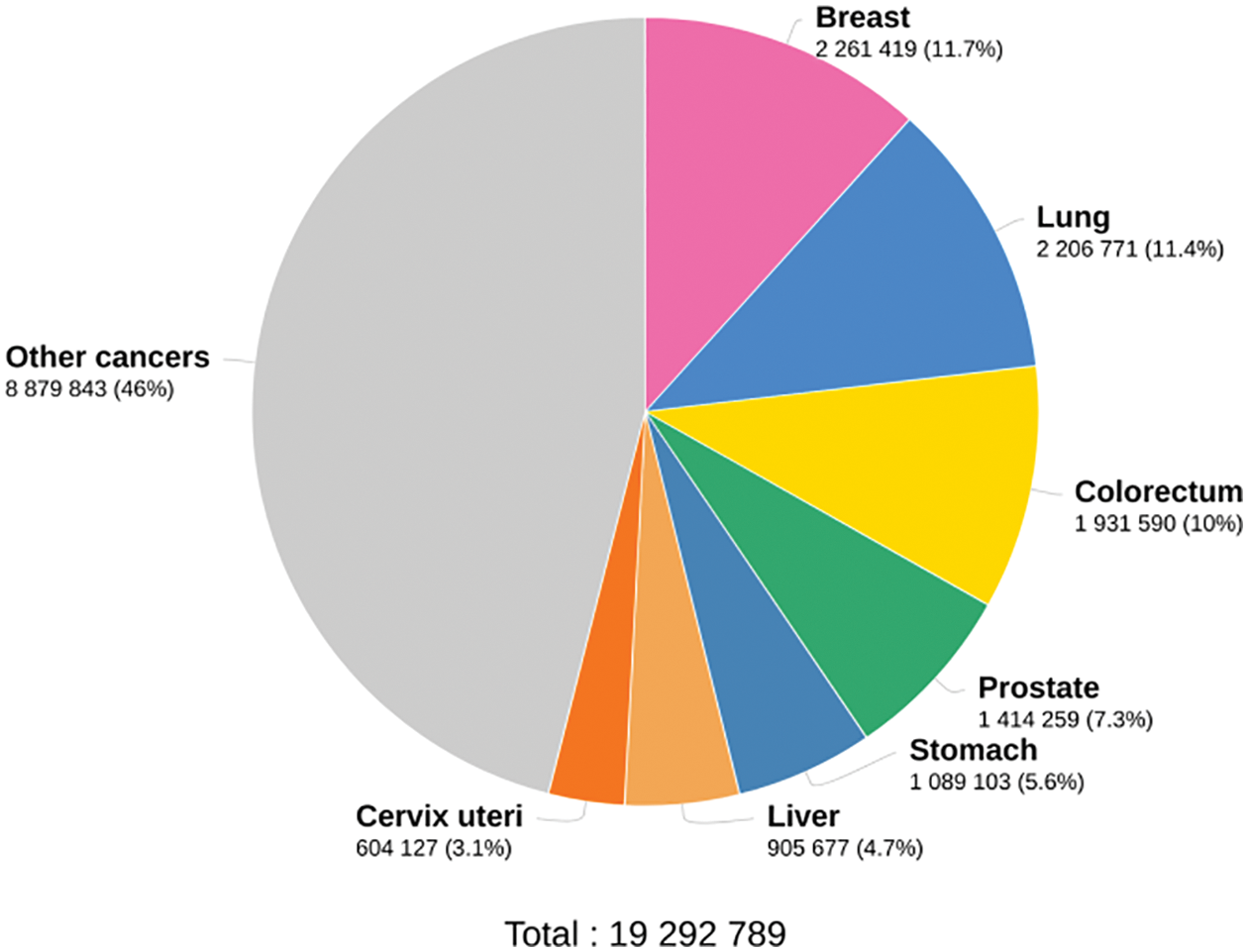
Figure 1: Worldwide cancer statistics in 2020
Numerous deep learning architectures have been developed for image based classification in the medical domain for disease classification. Their performances depend on the arrangement of convolution filters, pooling layers, and fully connected layers with fine-tuned parameters. With the varying benefits of the numerous analyses, it’s hard to choose the best one. Recently, researchers have developed hybrid and ensemble supervised CAD approaches. By combining strategies to handle distinct CAD components, they expect to overcome some of a technique's shortcomings while keeping any benefits. To achieve this, Horizontal Voting Ensemble (HVE) uses various models from the twelve-layer deep learning architecture model’s most recent epochs.
The objective is to develop and test a CAD system for microscopic images with high sensitivity and specificity for colon cancer. The salient feature of the Predictive Modeling System (PMS) is the development of a HVE system for effective classification. The rest of this paper is as follows: Section 2 discusses the related literature for colon cancer classification. Section 3 discusses the design of the proposed PMS with the HVE method for colon cancer classification. Section 4 contains the results obtained in this work briefly, and the significance of the results is also discussed. Section 5 gives the overall conclusion of the proposed system.
A pre-trained AlexNet is employed in [1] for colon and lung cancer classification from microscopic images. The transfer learning technique is employed by modifying the last four layers of AlexNet. The images are enhanced using class-selective image processing before training. A Weakly-Supervised Learning (WSL) based system is described in [2] for colon cancer classification. Instead of using common classification losses, Kullback-Leibler (KL) divergence losses are employed for better accuracy.
A Shearlet transform based colon cancer classification system is discussed in [3]. It uses multi-scale features extracted from different shearlet decomposition levels, directions and Convolution Neural Network (CNN). The obtained multi-scale features and the original image are fed to CNN for classification. An efficient mask region CNN system for colon cancer classification by detecting benign polyps is described in [4]. Initially, an image filter is employed to filter the low-quality image data, and then the filtered images are grouped. This system generates an individual model for each patient for better analysis.
An unsupervised feature extraction approach is described in [5] for colon cancer classification. It has three parts; salient sub-region identification, quantization of identified sub-regions by a set of features, and a deep belief network for classification using restricted Boltzmann machines. A detection-identification combined system is discussed in [6] for colon cancer classification using microscopic images. At first, the nuclei positions are detected and enhanced by a cascade residual fusion block. Then, the identification of cell nuclei is made by a multi-cropping network around the center of the nucleus.
A locality-sensitive deep learning architecture is discussed in [7] for colon cancer classification. This system tries to identify the center of the nucleus using a semantic approach. Then, the nuclei are classified using a CNN combined with a neighbouring ensemble predictor. A Markovian model is developed in [8] for diagnosing colon cancer. It uses a re-sampling framework to generate more microscopic images from an image and then models them by a Markov process. Different techniques for colon cancer classification are reviewed in [9]. Many texture-based systems, object-oriented texture analysis systems, and hyperspectral-based systems are discussed.
Yolov5 architecture is used in [10] to classify cancer tissue images. Yolov5 architecture is initially designed for object detection. It uses a cross stage partial network as a backbone with spatial pyramid pooling. Data augmentation with four CNNs is discussed in [11] for colon cancer classification. It uses baseline CNN and CNN with a different number of blocks (two, three, and three with augmented images) for the classification. The conventional handcrafted features are utilized in [12] with the help of CNN for colon cancer classification. Features based on structure, colour, shape, and texture are extracted and conventional classifiers such as support vector machines and random forest classifiers are used. Features from the DenseNet architecture are also employed with the traditional classifiers to improve performance.
A deep learning approach is discussed in [13], using whole slide images for colon cancer classification. It uses pre-trained architectures such as AlexNet, DenseNet, VGG, and ResNet with transfer learning. Pixel-wise segmentation is also employed after classification using the UNet and SegNet architectures. Multiple weighted semi-supervised system is described in [14] for colon cancer classification using whole slide images. A transferred pre-trained network is utilized for extracting high-level features at first from the sampled patches only. These features are combined with the label-rich features, and multiple weighted loss functions are employed for the classification.
The design of pattern recognition systems is challenging to deal with difficulties in defining significant features that distinguish a given pattern from another and variability among patterns belonging to the same class or category. The different classes may overlap or have large variances. Fig. 2 shows a typical pattern recognition system.

Figure 2: Typical pattern recognition system
In a typical pattern recognition system, the image/signal acquisition system is used to make a representation of objects. For example, in the proposed system, microscopy takes histopathological images of the affected tissues in the large intestine. Each image constitutes a pattern that the pattern recognition systems can classify. The images obtained by this step form the pattern space. Each image contains a considerable amount of redundant data, and thus the feature extraction process is introduced to extract the relevant information (feature space) that is necessary for classification. Finally, the decision algorithm transforms the feature space into the classification space. In this work, the classification space has two dimensions (binary classification), which represent the result of the decision algorithm. There are two different approaches; supervised and unsupervised can be used for designing a classifier. Supervised classification is adopted in this work. The training samples are labeled by their actual class (ACA or Benign) in a supervised learning approach. The labels are then used to guide or supervise the classifier during the learning process.
Deep learning architecture is employed in many medical image analysis systems such as pneumonia classification [15], mammogram classification [16], skin cancer [17,18], Covid-19 diagnosis [19], and vascular tissue simulation model [20]. They use a neural network for the classification and convolution and max pooling layer to extract deep features. Neural network models are inspired by the activities of the human brain and attempt to reproduce the same by using simple processing units called “neurons” with high processing power and speed. The neurons in the neural networks are connected by weights. These weights are updated in order to achieve a good classification result. An neural network learns from experience (supervised), and the final prediction is performed from the learned representations or trained network.
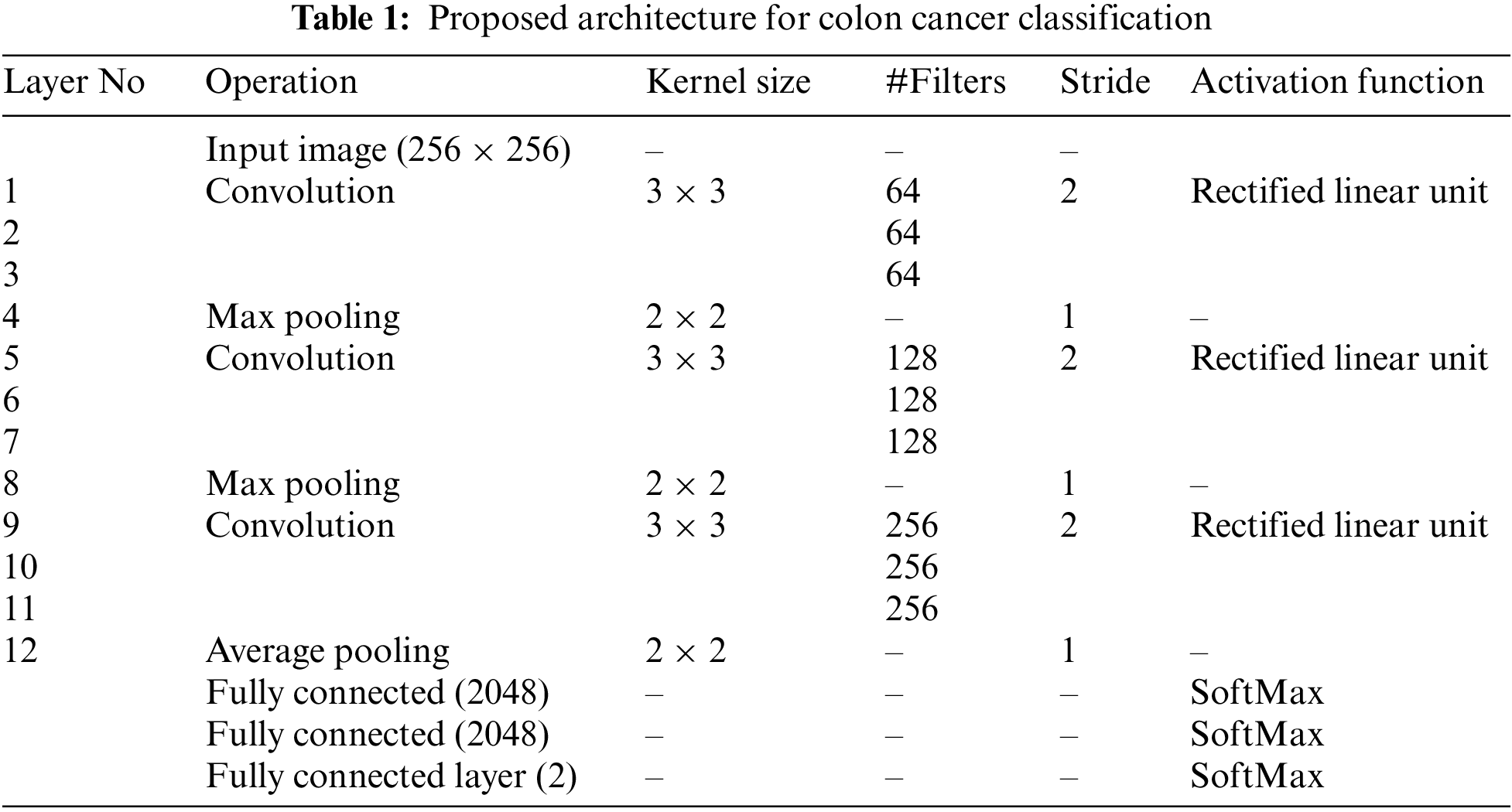
Table 1 shows the proposed deep learning architecture for classifying microscopic images for colon cancer classification. The number of pixels shifted across the input matrix is called the stride. When the stride equals one, the filters forward one pixel at a time. If the stride is 2, we simultaneously move filters by 2 pixels. The rectified linear unit is the most popular activation function, and it is generally utilized in many deep learning and convolutional neural networks. It is half rectified (from bottom) and f(z) equals z when z is above or equal to zero. It is defined by
The proposed architecture is exclusively concerns the decision-theoretic approach applied to recognizing and classifying microscopic images of colon tissues. The neural network used in this work is known as feed-forward with two hidden layers and back-propagation as the learning algorithm. Fig. 3 shows a simple feed-forward network.
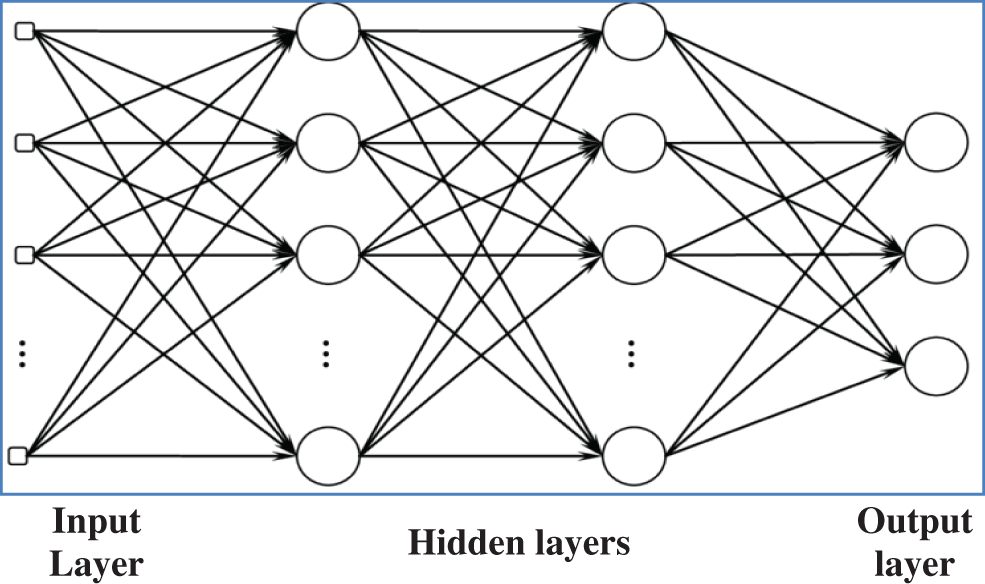
Figure 3: Simple feed forward network
After extracting deep features using convolution and max pooling layer, the fully connected layer uses them for classification using a neural network classifier. Before applying HVE methods, a single model is developed. The model uses the Adam optimizer with a learning rate of 0.01, and the cross entropy loss is employed to update the model’s weights. In machine learning and statistics, the learning rate controls the step size at each iteration while minimizing the loss function. The backpropagation algorithm requires that the proposed network model be trained for a predefined number of epochs (200). The input (microscopic images) and output patterns (ACA or benign) pairs are called batches. Using batch size (number of batches = 10), the efficiency of the network is also optimized, and it avoids having many input patterns loaded at a time. No zero padding is employed in this work. Once the network is well trained, it can be used to test the unknown samples. In order to employ the proposed HVE method, the trained model is stored for the last 15 epochs. The cross-entropy loss for n-class classification system is defined by
where
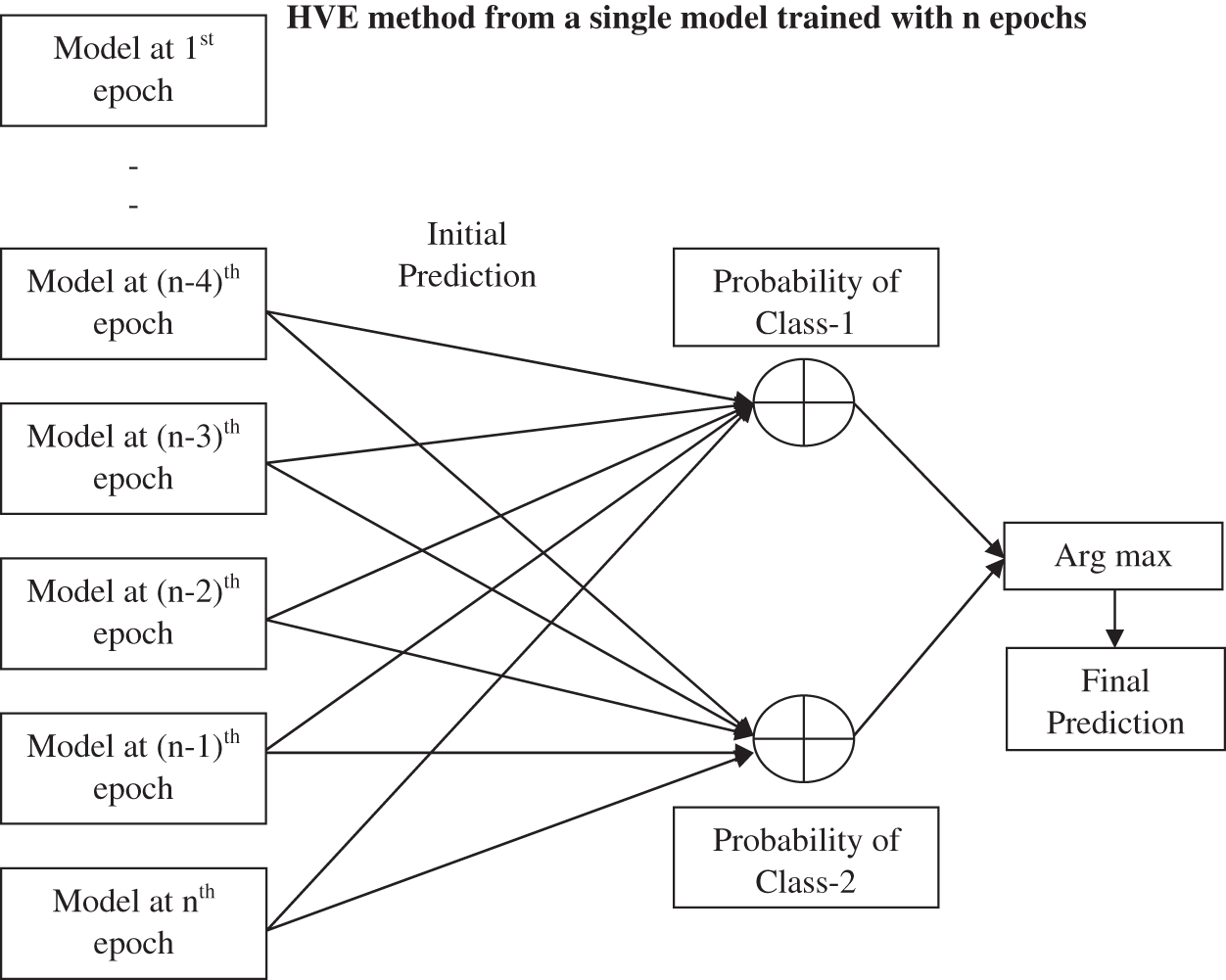
Figure 4: Proposed HVE method
Before the end of the training, the proposed HVE method generates multiple models from the predefined number of epochs. In this work, multiple models are developed from the last 5, 10, and 15 epochs, and their performances are evaluated by an ensemble approach using the argmax function. It is commonly used in many pattern recognition problems to find the class of the testing sample with maximum probabilities. It returns the argument(x) to the function f(.) that results in the maximum value. It is defined as
From the result of argmax function in Eq. (3), the class of the given test sample is assigned. Ten thousand (10000) microscopic images are used for the performance analysis. These images are taken from [21,22]. This database has microscopic images obtained from colon tissues with two classes; ACA and benign. Each class has 5000 images with 768 × 768 pixels. Fig. 5 shows samples from each class.
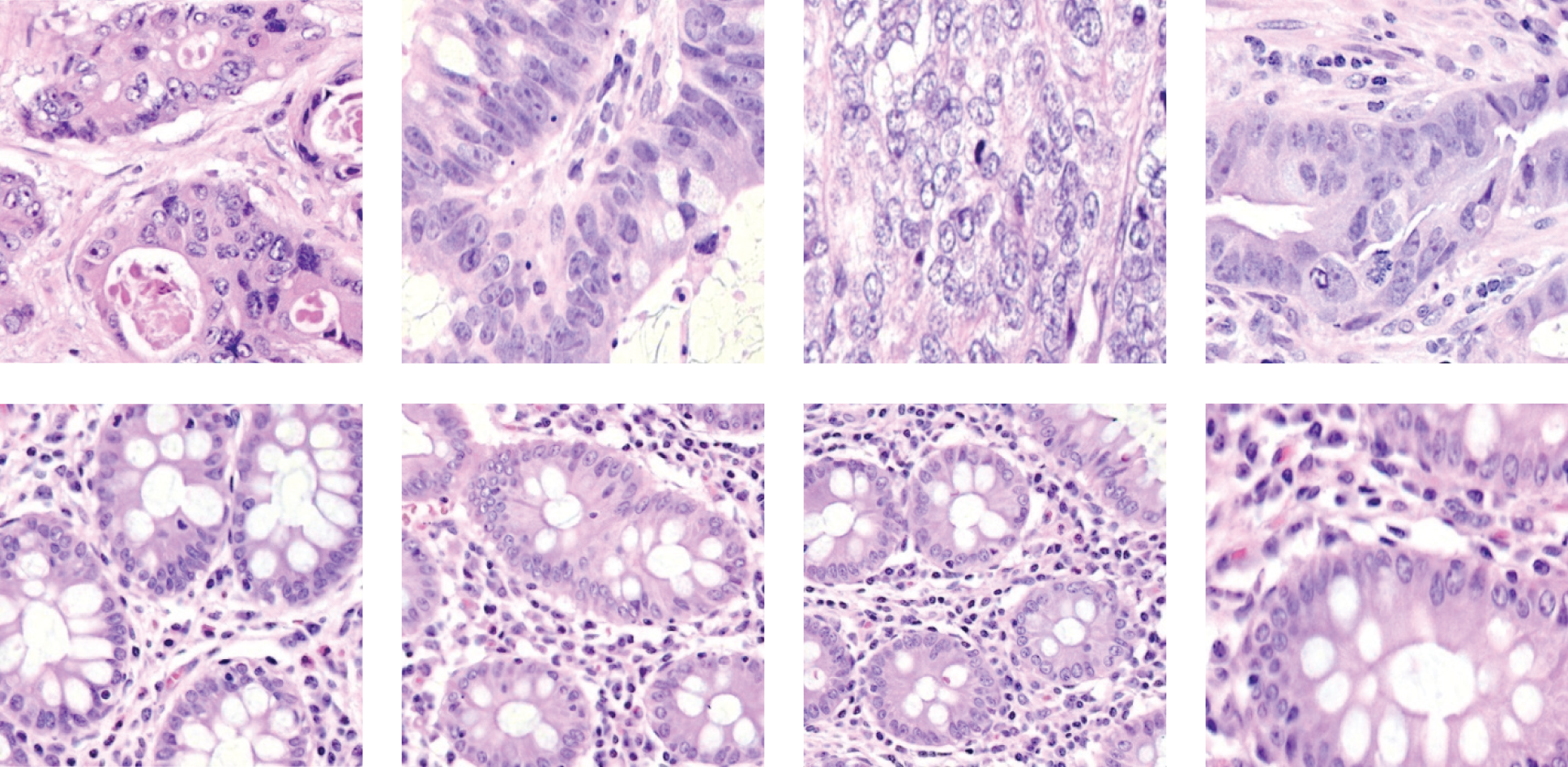
Figure 5: Colon cancer microscopic images; colon ACA (top row) colon benign tissue (bottom row)
The proposed PMS system is evaluated in terms of five performance metrics; Sensitivity, Positive Predictive Value (PPV), Specificity, Negative Predictive Value (NPV), and Accuracy. To find these performance metrics, a matrix with two rows and two columns is formed from the outputs of the proposed system. This matrix is called the confusion matrix, and is shown in Table 2.
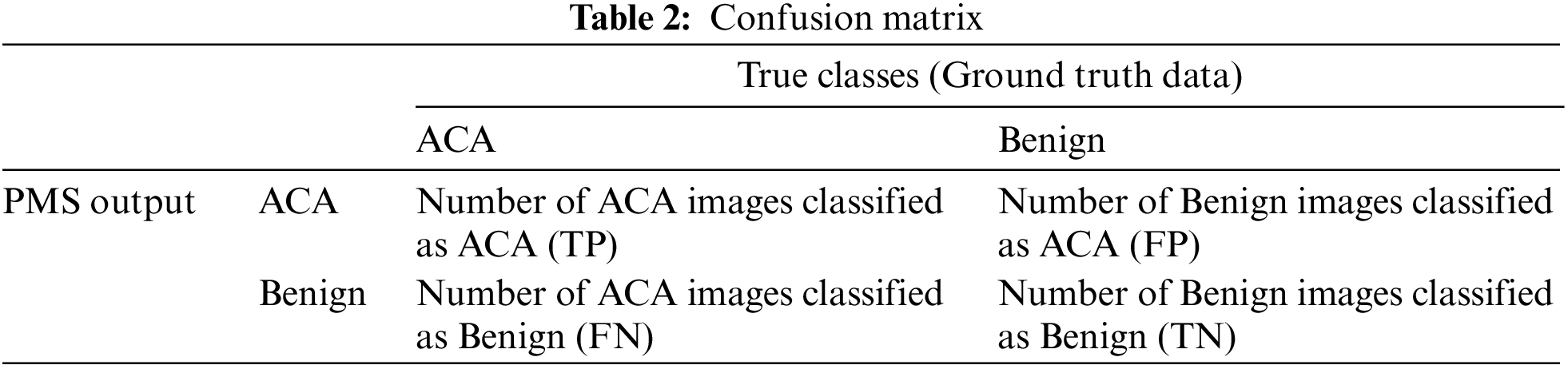
The definitions for the performance metrics used in this work are as follows:
1. Sensitivity =
2. PPV =
3. Specificity =
4. NPV =
5. Accuracy =
A random split approach (60:40) is used to provide training images to train the proposed deep learning architecture. Thus the system is trained with 6000 images (3000 ACA and 3000 Benign) and tested with 4000 images (2000 ACA and 2000 Benign).
This section discusses the performance of the proposed PMS for diagnosing colon cancer. Fig. 6 shows the obtained outputs in a confusion matrix form for the proposed architecture with the HVE system for the last 5, 10, and 15 epochs, and without introducing the HVE system.
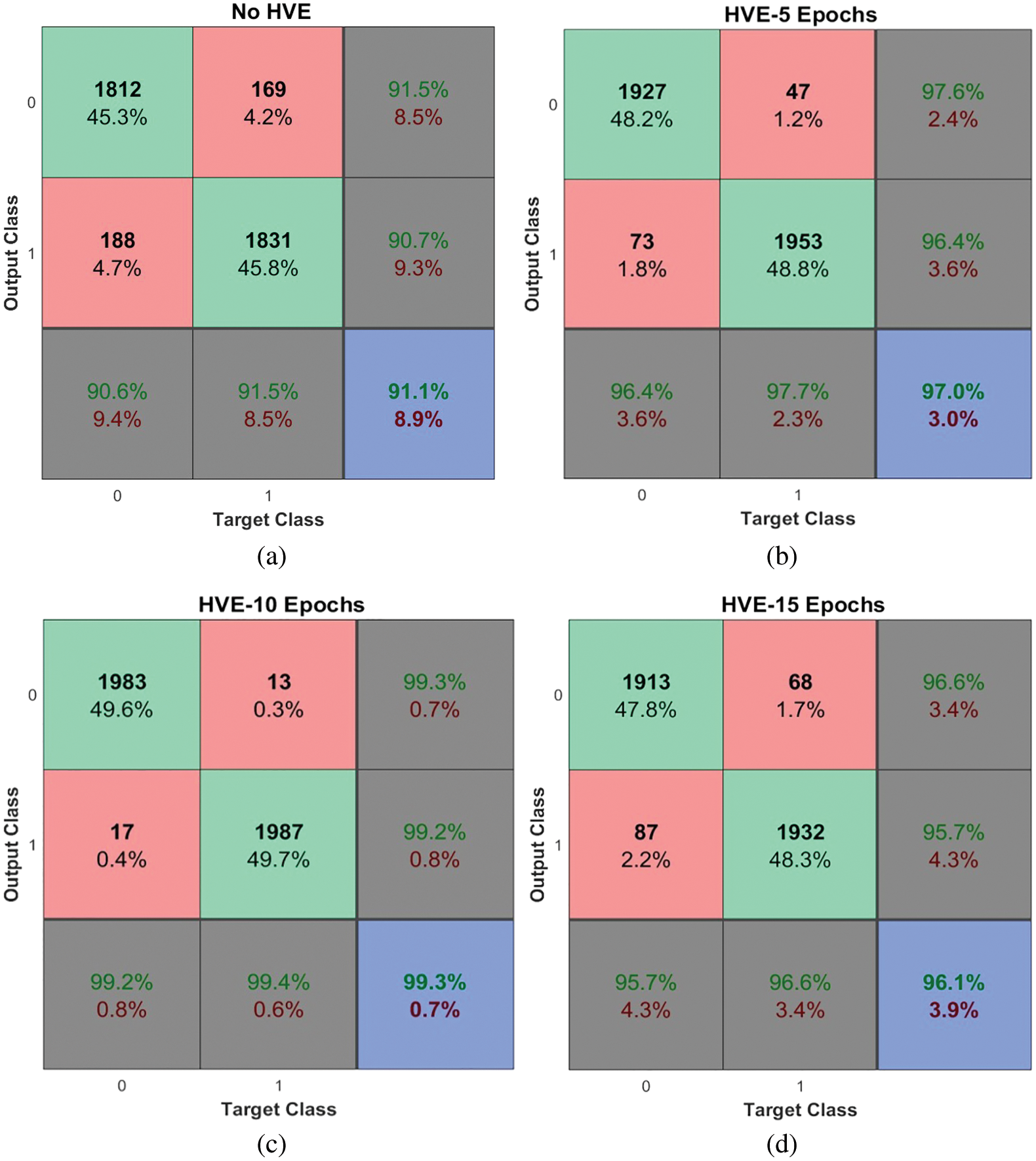
Figure 6: Performances of the proposed PMS for colon cancer classification system using HVE method
It is observed from Fig. 6 that the PMS achieves 99.3% accuracy when using HVE methods with the last ten epochs of the deep learning model. The PMC achieves approximately 8% performance improvement compared to the proposed deep learning architecture without the HVE method (91.1%). It is noted that only 17 ACA images and 13 benign images are misclassified and achieve 99.2% and 99.4% sensitivity and specificity respectively. The performance metrics are summarized below in Table 3.
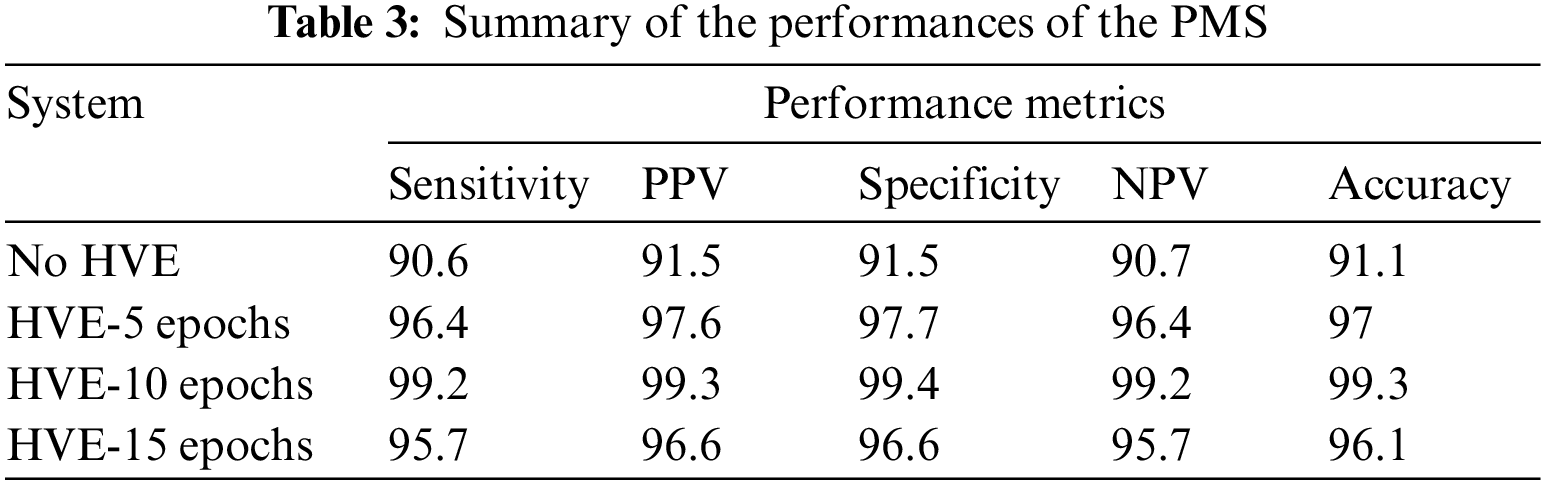
Fig. 7 shows the Receiver Operating Characteristic (ROC) of the proposed PMS for colon cancer classification. The ROC is drawn between the two important measures, such as sensitivity (y-axis) and 1-specificity (x-axis). The main advantage of the ROC curve is that the visual representation (Fig. 7) clearly shows the performance of the classification system.
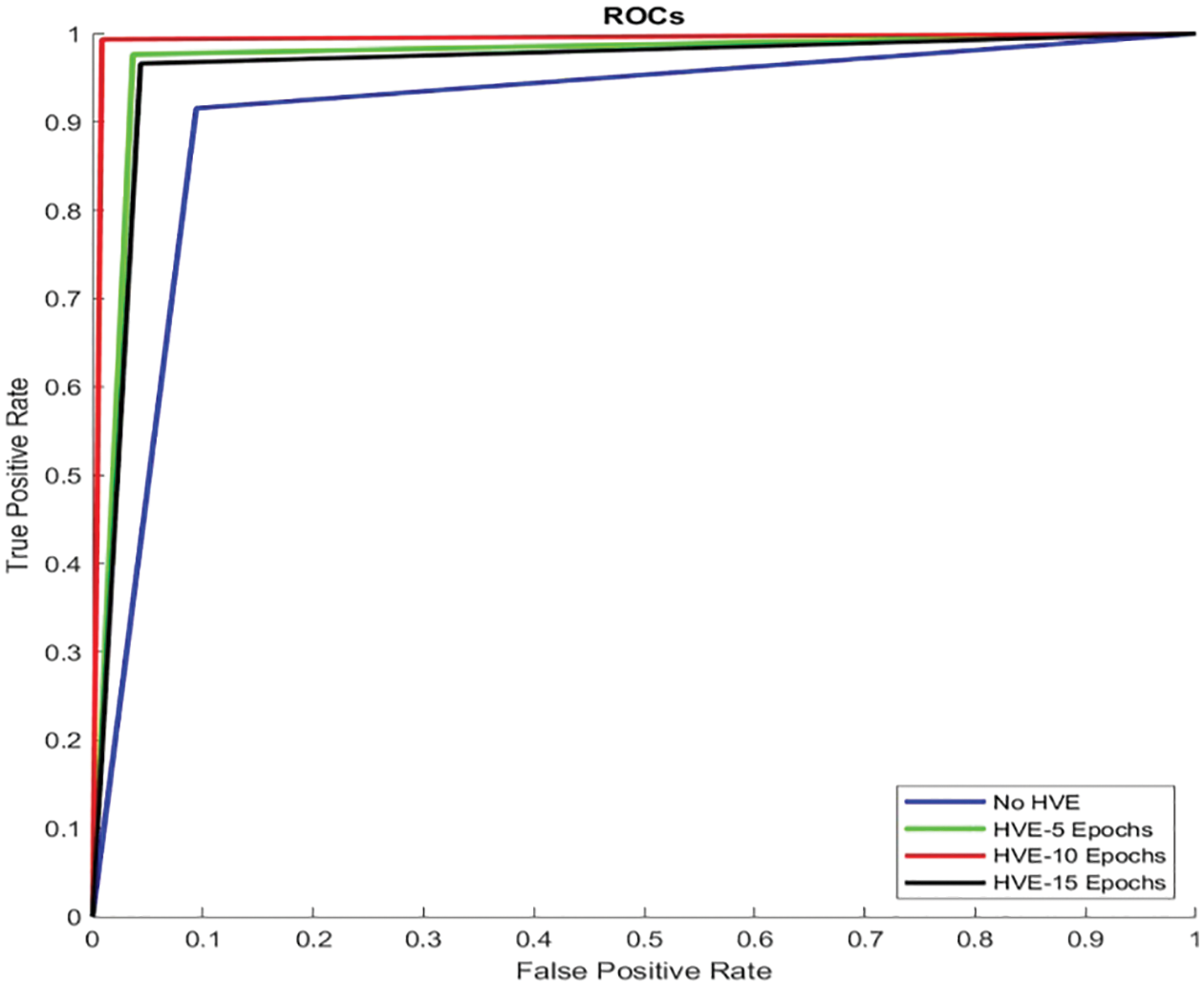
Figure 7: ROCs of the proposed PMS for colon cancer classification
It can be seen from Fig. 7 that the ROC of HVE-10 Epochs is very close to the y-axis and touches the top border of the plot. It means that this system has a low false positive rate (0.6%) and a high true positive rate (99.2%) than other systems’ performances, such as HVE-5 Epochs, HVE-15 Epochs, and No HVE. The area occupied by this curve (HVE-10 Epochs) is 0.993. The area occupied by other curves is 0.911 (No HVE), 0.97 (HVE-5 Epochs), and 0.961 (HVE-15 Epochs). Fig. 8 shows the graphical representation of performances attained by the proposed PMS for colon cancer classification.
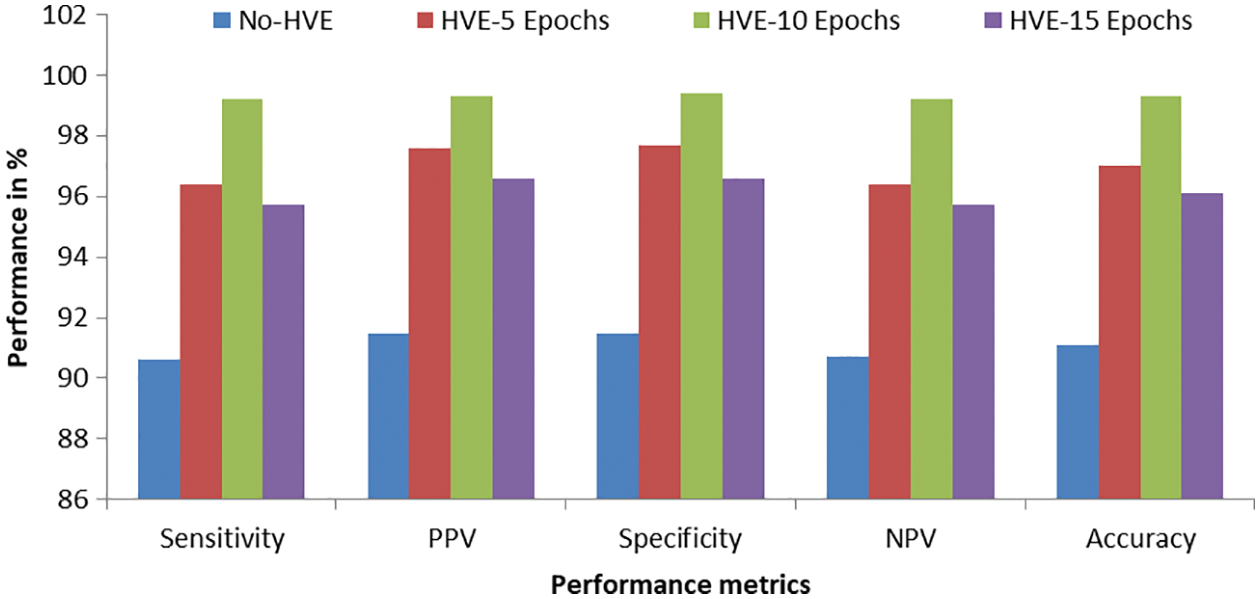
Figure 8: Graphical representation of performances attained by the proposed PMS for colon cancer classification
It can be seen from Fig. 8 that the proposed PMS with the HVE-10 epochs outperforms other combinations such as HVE-5 and HVE-15 epochs. Also, the PMS’s performances are better for colon cancer classification compared to those without HVE in the deep learning architecture. It is well known that the HVE approach combines the qualities of a pre-defined number of models and thus provides better performance than a single model. The significant improvements of the HVE approach in terms of accuracy, sensitivity, and specificity can be observed in Figs. 6 to 8.
In this work, the concept of the HVE method is developed for the effectively classifying colon cancer using microscopic images. In the HVE method, multiple models are selected from the pre-defined number of last epochs. Then, these models are combined to achieve better performances for colon cancer classification using an ensemble approach. The proposed system will be equivalent to supervised learning, where the data within the meta-database defines the predefined classes. It can also be considered as a decision-theoretic pattern recognition system with the help of a twelve-layer deep learning architecture for colon cancer classification. The proposed deep learning architecture enables classification and extracts optimal texture features simultaneously. The obtained results provide very effective classification when tested with 10000 microscopic images. The use of the HVE-10 Epochs in deep learning gives an overall accuracy of 99.3% for colon cancer classification.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. S. Mehmood, T. M. Ghazal, M. A. Khan, M. Zubair, M. T. Naseem et al., “Malignancy detection in lung and colon histopathology images using transfer learning with class selective image processing,” IEEE Access, vol. 10, pp. 25657–25668, 2022. [Google Scholar]
2. S. Belharbi, J. Rony, J. Dolz, I. B. Ayed, L. Mccaffrey et al., “Deep interpretable classification and weakly-supervised segmentation of histology images via max-min uncertainty,” IEEE Transactions on Medical Imaging, vol. 41, no. 3, pp. 702–714, 2022. [Google Scholar]
3. M. Liang, Z. Ren, J. Yang, W. Feng and B. Li, “Identification of colon cancer using multi-scale feature fusion convolutional neural network based on shearlet transform,” IEEE Access, vol. 8, pp. 208969–208977, 2020. [Google Scholar]
4. X. Yang, Q. Wei, C. Zhang, K. Zhou, L. Kong et al., “Colon polyp detection and segmentation based on improved MRCNN,” IEEE Transactions on Instrumentation and Measurement, vol. 70, pp. 1–10, 2021. [Google Scholar]
5. C. T. Sari and C. Gunduz-Demir, “Unsupervised feature extraction via deep learning for histopathological classification of colon tissue images,” IEEE Transactions on Medical Imaging, vol. 38, no. 5, pp. 1139–1149, 2019. [Google Scholar]
6. X. Li, W. Li and R. Tao, “Staged detection-identification framework for cell nuclei in histopathology images,” IEEE Transactions on Instrumentation and Measurement, vol. 69, no. 1, pp. 183–193, 2020. [Google Scholar]
7. K. Sirinukunwattana, S. E. A. Raza, Y. W. Tsang, D. R. J. Snead, I. A. Cree et al., “Locality sensitive deep learning for detection and classification of nuclei in routine colon cancer histology images,” IEEE Transactions on Medical Imaging, vol. 35, no. 5, pp. 1196–1206, 2016. [Google Scholar]
8. E. Ozdemir, C. Sokmensuer and C. Gunduz-Demir, “A resampling-based markovian model for automated colon cancer diagnosis,” IEEE Transactions on Biomedical Engineering, vol. 59, no. 1, pp. 281–289, 2012. [Google Scholar]
9. S. Rathore, M. Hussain, A. Ali and A. Khan, “A recent survey on colon cancer detection techniques,” IEEE/ACM Transactions on Computational Biology and Bioinformatics, vol. 10, no. 3, pp. 545–563, 2013. [Google Scholar]
10. R. K. Ramesh and C. N. Savithri, “Colon cancer detection using YOLOv5 architecture,” in Int. Conf. on Communication, Computing and Internet of Things, Chennai, India, pp. 1–5, 2022. [Google Scholar]
11. A. Kumar, A. Vishwakarma, V. Bajaj, A. Sharma and C. Thakur, “Colon cancer classification of histopathological images using data augmentation,” in Int. Conf. on Control, Automation, Power and Signal Processing, Jabalpur, India, pp. 1–5, 2021. [Google Scholar]
12. N. Kumar, M. Sharma, V. P. Singh, C. Madan and S. Mehandia, “An empirical study of handcrafted and dense feature extraction techniques for lung and colon cancer classification from histopathological images,” Biomedical Signal Processing and Control, vol. 75, no. 1, pp. 1–18, 2022. [Google Scholar]
13. A. B. Hamida, M. Devanne, J. Weber, C. Truntzer, V. Derangère et al., “Deep learning for colon cancer histopathological images analysis,” Computers in Biology and Medicine, vol. 136, no. 7, pp. 1–18, 2021. [Google Scholar]
14. P. Wang, P. Li, Y. Li, J. Xu and M. Jiang, “Classification of histopathological whole slide images based on multiple weighted semi-supervised domain adaptation,” Biomedical Signal Processing and Control, vol. 73, no. 2016, pp. 1–16, 2022. [Google Scholar]
15. M. A. Ramitha and N. Mohanasundaram, “Classification of pneumonia by modified deeply supervised ResNet and SENet using chest x-ray images,” International Journal of Advances in Signal and Image Sciences, vol. 7, no. 1, pp. 30–37, 2021. [Google Scholar]
16. G. Jayandhi, J. Leena Jasmine and S. Mary Joans, “Mammogram learning system for breast cancer diagnosis using deep learning SVM,” Computer Systems Science and Engineering, vol. 40, no. 2, pp. 491–503, 2022. [Google Scholar]
17. R. Thamizhamuthu and D. Manjula, “Skin melanoma classification system using deep learning,” Computers, Materials & Continua, vol. 68, no. 1, pp. 1147–1160, 2021. [Google Scholar]
18. S. Justin and M. Pattnaik, “Skin lesion segmentation by pixel by pixel approach using deep learning,” International Journal of Advances in Signal and Image Sciences, vol. 6, no. 1, pp. 12–20, 2020. [Google Scholar]
19. X. Zhang, J. Zhou, W. Sun and S. Kumar Jha, “A lightweight cnn based on transfer learning for covid-19 diagnosis,” Computers, Materials & Continua, vol. 72, no. 1, pp. 1123–1137, 2022. [Google Scholar]
20. X. Zhang, H. Wu, W. Sun, A. Song and S. Kumar Jha, “A fast and accurate vascular tissue simulation model based on point primitive method,” Intelligent Automation & Soft Computing, vol. 27, no. 3, pp. 873–889, 2021. [Google Scholar]
21. A. A. Borkowski, M. M. Bui, L. B. Thomas, C. P. Wilson, L. A. DeLand et al., “Lung and colon cancer histopathological image dataset (LC25000),” in Proc. in Computer Vision and Pattern Recognition, Cornell University, NY, USA, pp. 1–2, 2019. [Google Scholar]
22. LC25000 Lung and Colon Cancer Histopathological Image Dataset: 2019. [Online]. Available: https://github.com/tampapath/lung_colon_image_set/blob/master/README.md. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools