 Open Access
Open Access
ARTICLE
MDEV Model: A Novel Ensemble-Based Transfer Learning Approach for Pneumonia Classification Using CXR Images
1 Department of Software Engineering, Mehran University of Engineering and Technology, Jamshoro, Pakistan
2 College of Software, Sungkyunkwan University, Suwon, Korea
3 Department of Computer Systems, Mehran University of Engineering and Technology, Jamshoro, Pakistan
4 Department of Computer Education, Sungkyunkwan University, Seoul, Korea
* Corresponding Authors: Jahwan Koo. Email: ; Nawab Muhammad Faseeh Qureshi. Email:
Computer Systems Science and Engineering 2023, 46(1), 287-302. https://doi.org/10.32604/csse.2023.035311
Received 16 August 2022; Accepted 10 October 2022; Issue published 20 January 2023
Abstract
Pneumonia is a dangerous respiratory disease due to which breathing becomes incredibly difficult and painful; thus, catching it early is crucial. Medical physicians’ time is limited in outdoor situations due to many patients; therefore, automated systems can be a rescue. The input images from the X-ray equipment are also highly unpredictable due to variances in radiologists’ experience. Therefore, radiologists require an automated system that can swiftly and accurately detect pneumonic lungs from chest x-rays. In medical classifications, deep convolution neural networks are commonly used. This research aims to use deep pre-trained transfer learning models to accurately categorize CXR images into binary classes, i.e., Normal and Pneumonia. The MDEV is a proposed novel ensemble approach that concatenates four heterogeneous transfer learning models: MobileNet, DenseNet-201, EfficientNet-B0, and VGG-16, which have been finetuned and trained on 5,856 CXR images. The evaluation matrices used in this research to contrast different deep transfer learning architectures include precision, accuracy, recall, AUC-roc, and f1-score. The model effectively decreases training loss while increasing accuracy. The findings conclude that the proposed MDEV model outperformed cutting-edge deep transfer learning models and obtains an overall precision of 92.26%, an accuracy of 92.15%, a recall of 90.90%, an auc-roc score of 90.9%, and f-score of 91.49% with minimal data pre-processing, data augmentation, finetuning and hyperparameter adjustment in classifying Normal and Pneumonia chests.Keywords
In the area of artificial intelligence, known as computer vision, objects are reliably identified and categorized by computers using deep learning models. Many AI-based technologies are currently employed to deal with healthcare domains such as biological issues. Pneumonia is one of the fatal diseases that cause morbidity and mortality. It is a primary cause of hospitalization in the world, said by the Centers for Disease Control and Prevention [1]. In the prediction of disease, AI models provide accuracy comparable to that of a single radiologist [2]. A technique with early detection and high accuracy outcomes is required to help radiologists identify Pneumonia from CXR images. CT scans of the lungs, Chest X-rays, chest MRIs, chest radiographs, chest ultrasounds, and lung biopsies can all be used to detect Pneumonia. Chest x-rays are frequently utilized by medical professionals due to their accessibility and affordability [3]. Therefore, this study primarily focuses on CXR images. Deep convolutional neural networks (CNN) perform remarkably well when working with images [4]. ImageNet dataset models that are trained on 14 million images for 20,000 classes are extremely successful transfer learning models because of their quick, fast and accurate training processes. Rather than training a CNN from scratch, which requires a long time, high computational cost, and expensive hardware, a more practical technique is to utilize transfer learning models that are already trained on a huge dataset [5]. Saul et al. [6] used convolutional with residual networks trained with data augmentation and pre-processing techniques to classify pneumonia images from check x-ray images and yield 78.73% accuracy, whereas Luján-García et al. [7] implement an XceptionNet transfer learning model to categorize check x-ray images of patients without pulmonary disease (named Normal) and people with pulmonary disease (specified Pneumonia). The findings are precision 84%, F-measure 91%, recall 99%, and AUC-ROC curve 97%. Comparing these two publications revealed that deep transfer learning models produced superior outcomes to the state-of-art methods.
CNN-based deep learning models have recently become the standard choice for medical image classifications, according to recent studies [8]. Researchers used several deep learning techniques to detect pneumonia disease using CXR images. However, they have not worked on the models selected for this study along with the finetuning in combination with the concatenation of different models, as concatenating multiple different models results in a combinational effect with good accuracy results. The purpose of this research is to use transfer learning models in combination with data pre-processing and data augmentation techniques to precisely classify CXR images into the Normal and Pneumonia classes. This paper introduced a novel concept of the MDEV model to classify pneumonia patients from CXR images using deep ensemble-based transfer learning models that concatenate four heterogeneous models namely, MobileNet, DenseNet-201, EfficientNet-B0, and VGG-16. A different combination of four of other many models were assumed, and a set of selected models results better in the available dataset and resources. Moreover, this research is limited to four base models due to fewer available resources.
All these four pre-trained models are finetuned and concatenated following the baseline of the stacking ensemble that trains a meta-learner to predict the output. The MDEV model could be useful to the medical field that aid radiologists to quickly diagnosing and confirming pneumonia cases with the potential to contribute to early detection with high accuracy significantly.
This research is divided into five sections. The related work is covered in section two, which contains the contributions of state-of-the-art studies and emphasis on the proposed key work. Section three covers the material and methods of this study, describing the dataset used and the background of transfer learning architectures based on the convolutional neural networks and the proposed MDEV model. Comparative analysis in section four. Finally, a conclusion and suggestions for additional research are included in section five.
In the medical world, detecting Pneumonia with CXR images is a challenging process that requires much expertise. Researchers are working on this subject to find the best methods and results for detecting Pneumonia. Today, CNN-based approaches produce great results and are commonly used [9,10]. Working with a huge dataset can take a long time for a model to learn. Here comes the concept of transfer learning: the learnings are transferred from one model trained on millions of images to another model effectively [11]. A quick and cost-effective approach to identifying patients with lung diseases is to use CXR images. Fan et al. [12] employ two deep pre-trained models, DenseNet121 and ResNet50, to identify CXR images and classify them into fourteen clinical classes. The two pretrained networks were compared, and it was determined that adopting DenseNet121 as the foundational model with data augmentation approaches resulted in superior generalization capacity on the test datasets. To increase the dataset for the analysis, a deep convolutional Generative Adversarial Network is applied [13], after which a pre-trained VGG19 model is applied to the larger dataset, and the accuracy of the original and augmented datasets are compared for the identical task of pneumonia classification. To automatically classify pneumonia images using binary classification, In the study [14], retraining a baseline CNN is contrasted with improved versions of (DenseNet201, VGG16, VGG19, Inception, InceptionV3, Xception, Resnet50, ResNetV2, and MobileNetV2). The modified version of Resnet50 is said to perform brilliantly, with an accuracy of above 96%. Three CNN models are ensembled in the paper [15] namely DenseNet-121, ResNet-18, and GoogLeNet. Predictions of the ensemble learning model perform better than those of any of its base learners because it combines the discriminative information of all of its constituent models. The outputs of the three models are aggregated to get an overall average weight. Two open-source datasets are used to compare the accuracy of the ensembled result. Reference [16] uses four transfer learning techniques (DenseNet121, VGG19, ResNet50, and Xception) to diagnose pneumonia disease in chest X-ray images of 1–5 years children. The performance evaluation of these models revealed that DenseNet121 and Xception have the best automated pneumonia classification ability. The three classification systems of normal, bacterial Pneumonia and viral Pneumonia are employed by Rahman et al. [17]. AlexNet, ResNet18, DenseNet201, and SqueezeNet, these four pre-trained models, are evaluated, and their accuracy is compared in these categories. Chouhan et al. perform an ensemble on the prediction of five transfer learning architectures, namely AlexNet, DenseNet121, Inception V3, GoogLeNet, and ResNet18, against the normal dataset bacterial and viral Pneumonia. The findings show that the ensemble model outperformed all other models [18].
The related work compares cutting-edge transfer learning techniques for pneumonia disease classification from CXR images. They perform different transfer learning patterns with different approaches and datasets and obtain variations in results. It is observed that there is no combined research done on fine-tuned models and ensemble approaches. As ensemble learning results in a combinational effect, and finetuning results in performance by modifying layers.
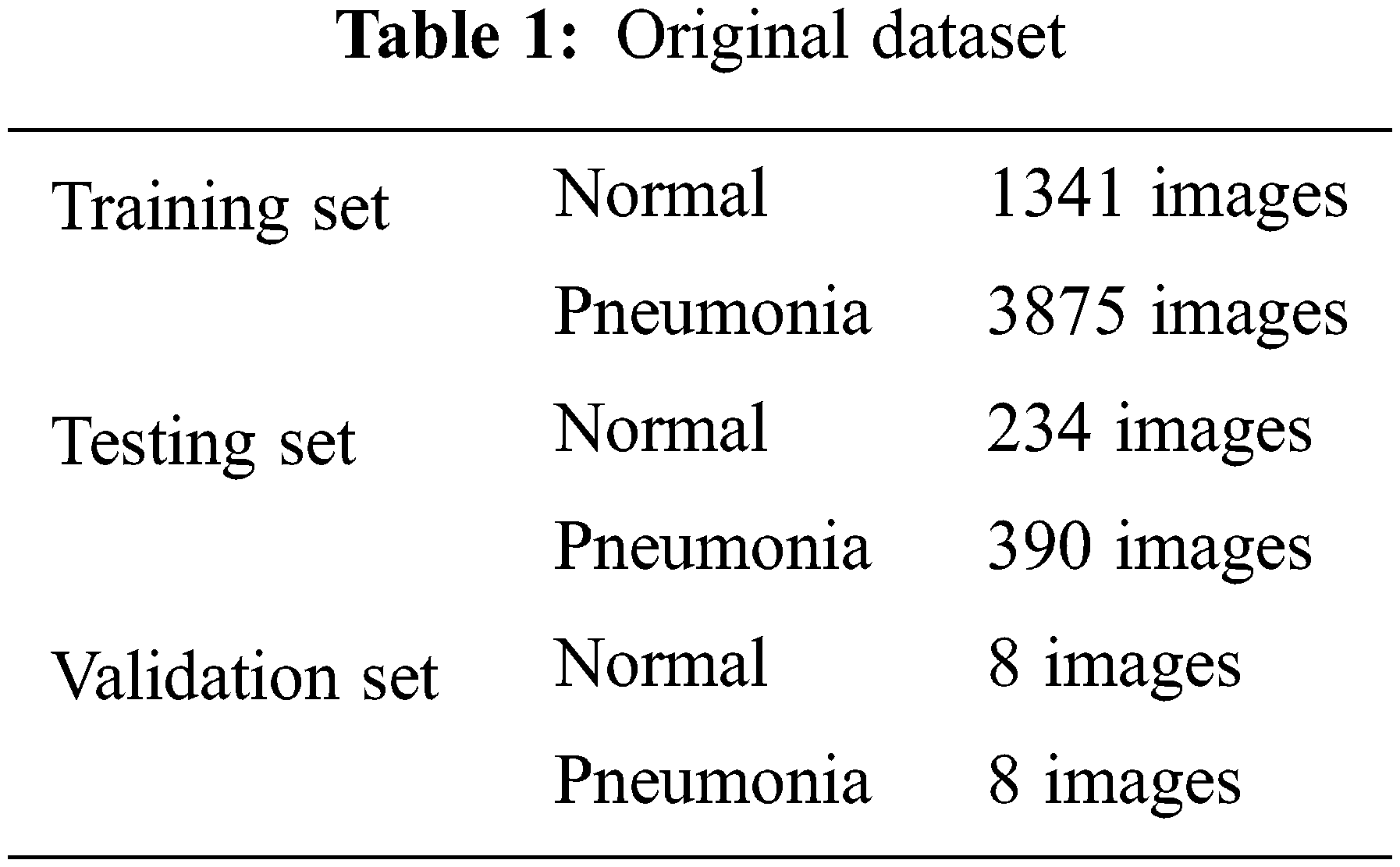
The source of data is Kaggle.com. Paul M-Kaggle dataset of chest X-ray images (Pneumonia) [19]. The network is fed with images of chest X-rays, as shown in Fig. 1. There are 5,856 images divided into two categories: Normal and Pneumonia, as shown in Table 1. Every chest radiograph was checked for quality control, and any scans that were subpar were discarded or indecipherable being discarded for further examination of CXR images and then graded by a physician before being fed into a deep learning network.
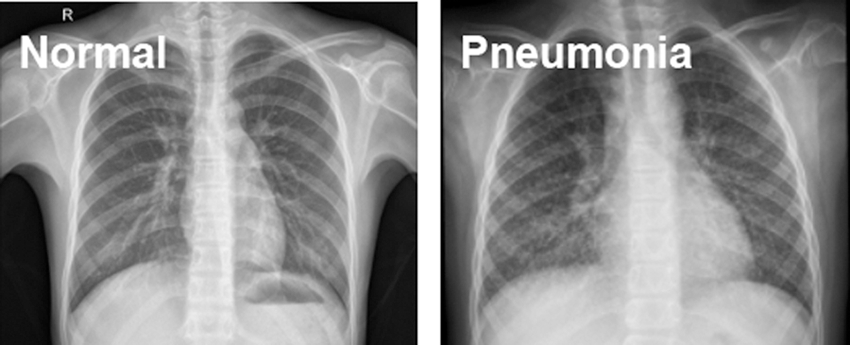
Figure 1: Normal and Pneumonia CXR images
The dataset’s image resolution ranges from 400 p to 2000 p; thereby, all images are scaled to a set size of 224 × 224 (standard choice) before feeding to the CNN because neural networks only take inputs of the same size and the smaller the image size, the better the model training.
3.2.2 Dataset Append and Segment
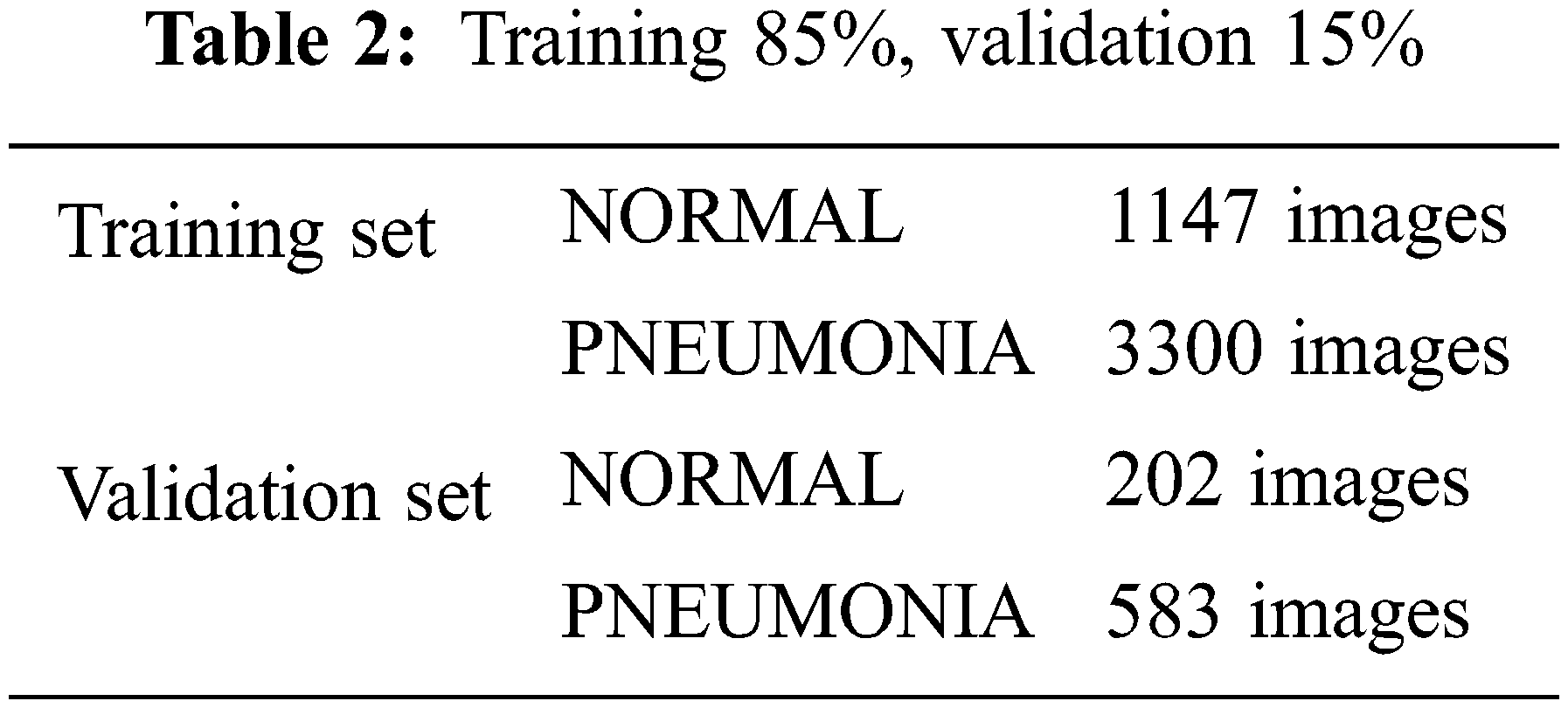
In the opensource dataset, validation set has a smaller number of images which is not suitable to validate the model. Training and validation data sets are appended and segmented into 85% training and 15% validation. Combining training and validation sets results in 3883 Pneumonia and 1349 Normal images. 85% training set results in 3300 Pneumonia and 1147 Normal images. 15% validation set results in 583 Pneumonia and 202 Normal images, as shown in Table 2.
The performance of classifiers suffers greatly when they are presented with datasets that are imbalanced, meaning that there are disproportionately more negative examples than positive ones. Because the Pneumonia images in the training set are higher than the Normal images, weights are balanced with the help of Eqs. (1) and (2).
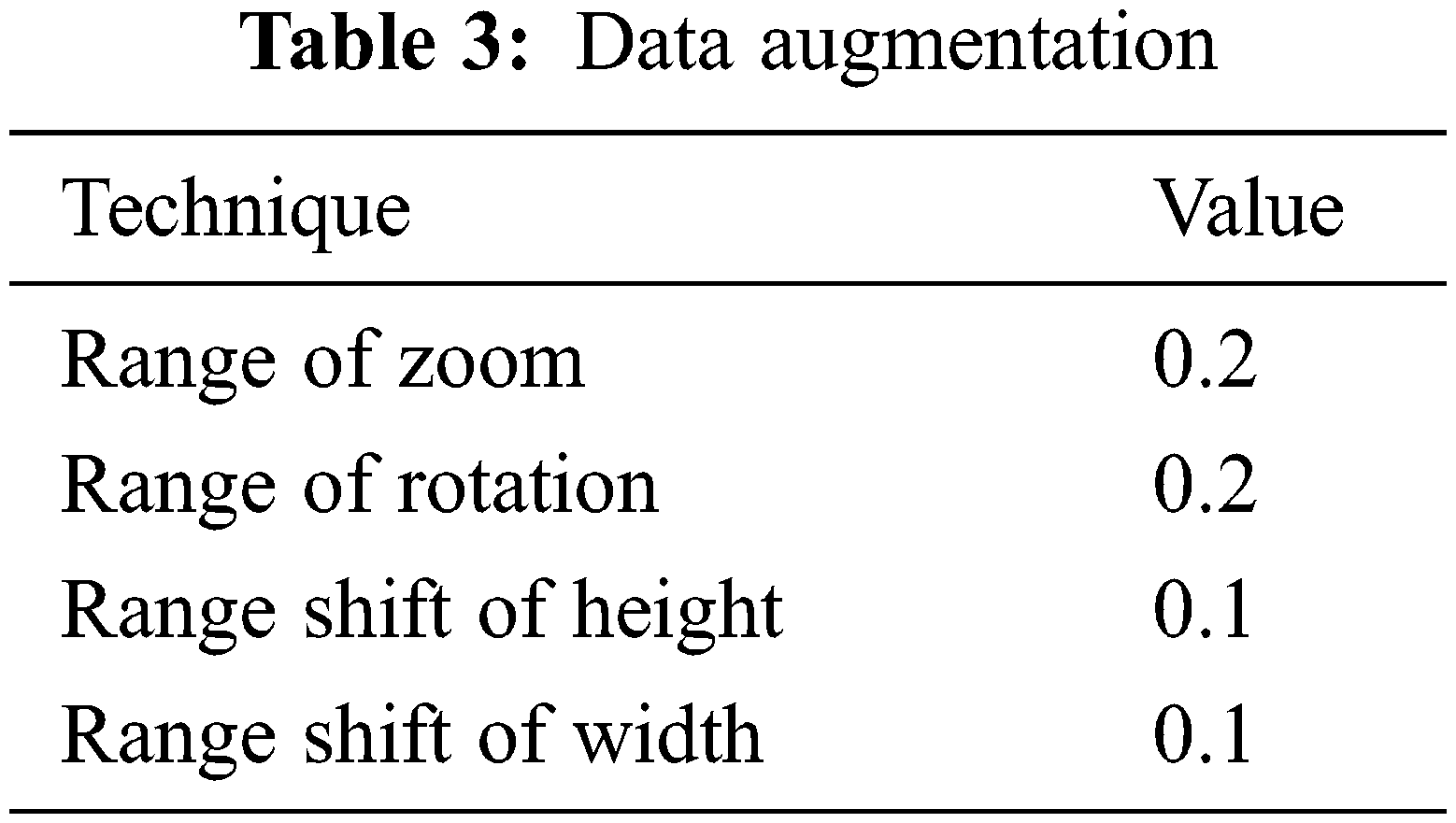
Data augmentation increases CNN performance, avoids overfitting, and is simple to use. The data augmentation techniques utilized in this research are zoom, rotate, height and width shift as shown in Table 3. Different values were examined with different data augmentation techniques, such as cropping, and brightness were also examined but the outcomes were not any better, this may vary from dataset to dataset due to original resolutions.
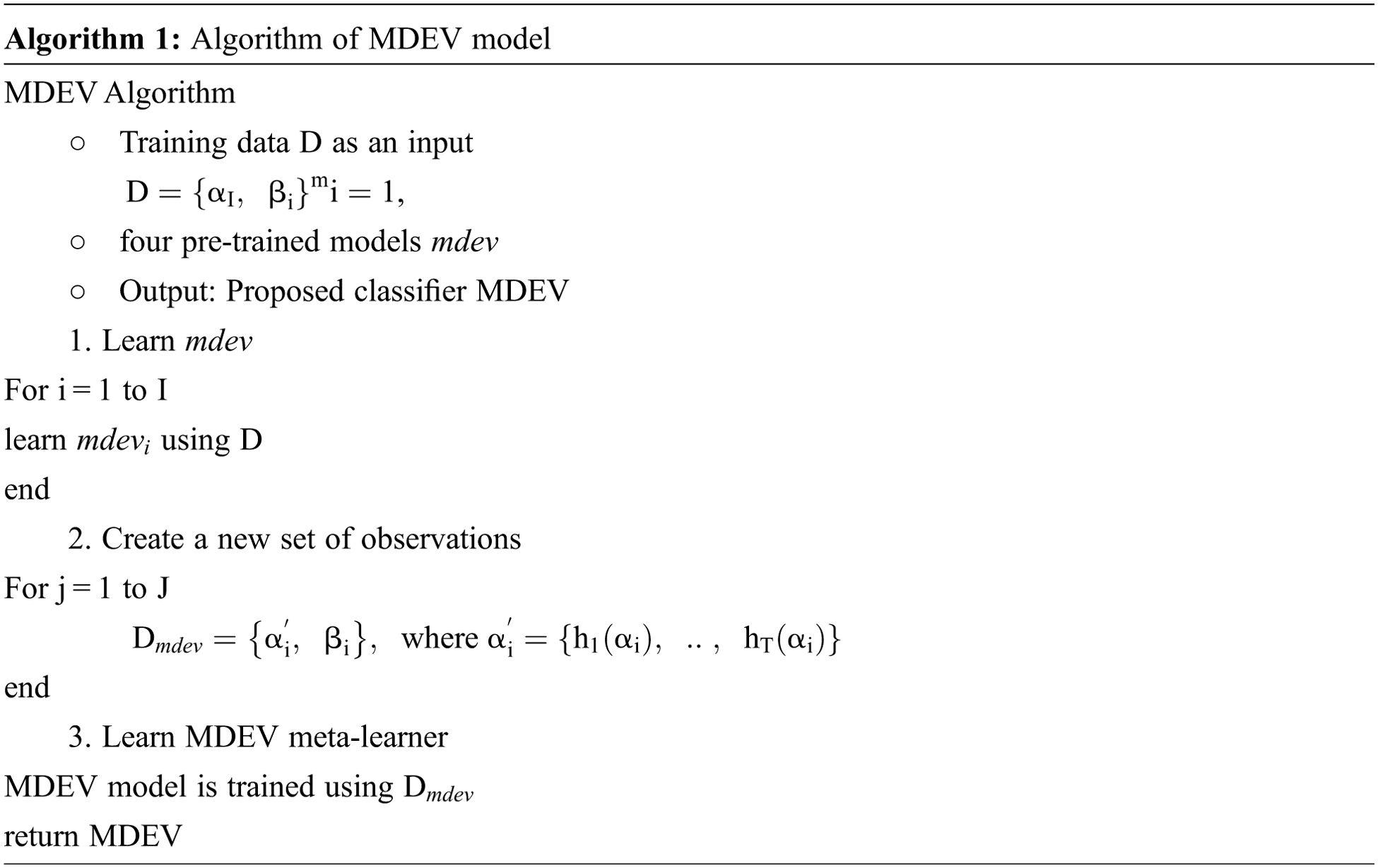
In this paper, the novel MDEV model is proposed that uses ensemble learning of pre-trained models namely, MobileNet, DenseNet-102, EfficientNet-B0, VGG-16. All these models are finetuned and coupled with image pre-processing and augmentation techniques to divide chest x-ray images in binary classes i.e., Normal or Pneumonia as shown in Fig. 2. Initially, samples were pre-processed to the 224 × 224 dimension specified in CXR dataset. Since noise in the dataset is the main cause of prediction issues in learning models, data pre-processing and augmentation are crucial tasks. Input the pre-processed images into four deep pre-trained neural networks that use distinct paradigms to extract information from latent variables. After evaluating the retrieved feature vectors, the outputs of four transfer learning models are concatenated to build a stacked ensemble in which a new model learns to combine the predictions of several different individual base models to classify the feedback into a binary class in the most efficient way. Individual models are referred to as first-level learners, whereas the combiner is referred to as a second-level learner, or meta-learner, as shown in Fig. 3 and the algorithm in Algorithm 1. The weights of each class must be balanced when combining predictions; otherwise, the meta-learner will learn more from the class with the higher weight.
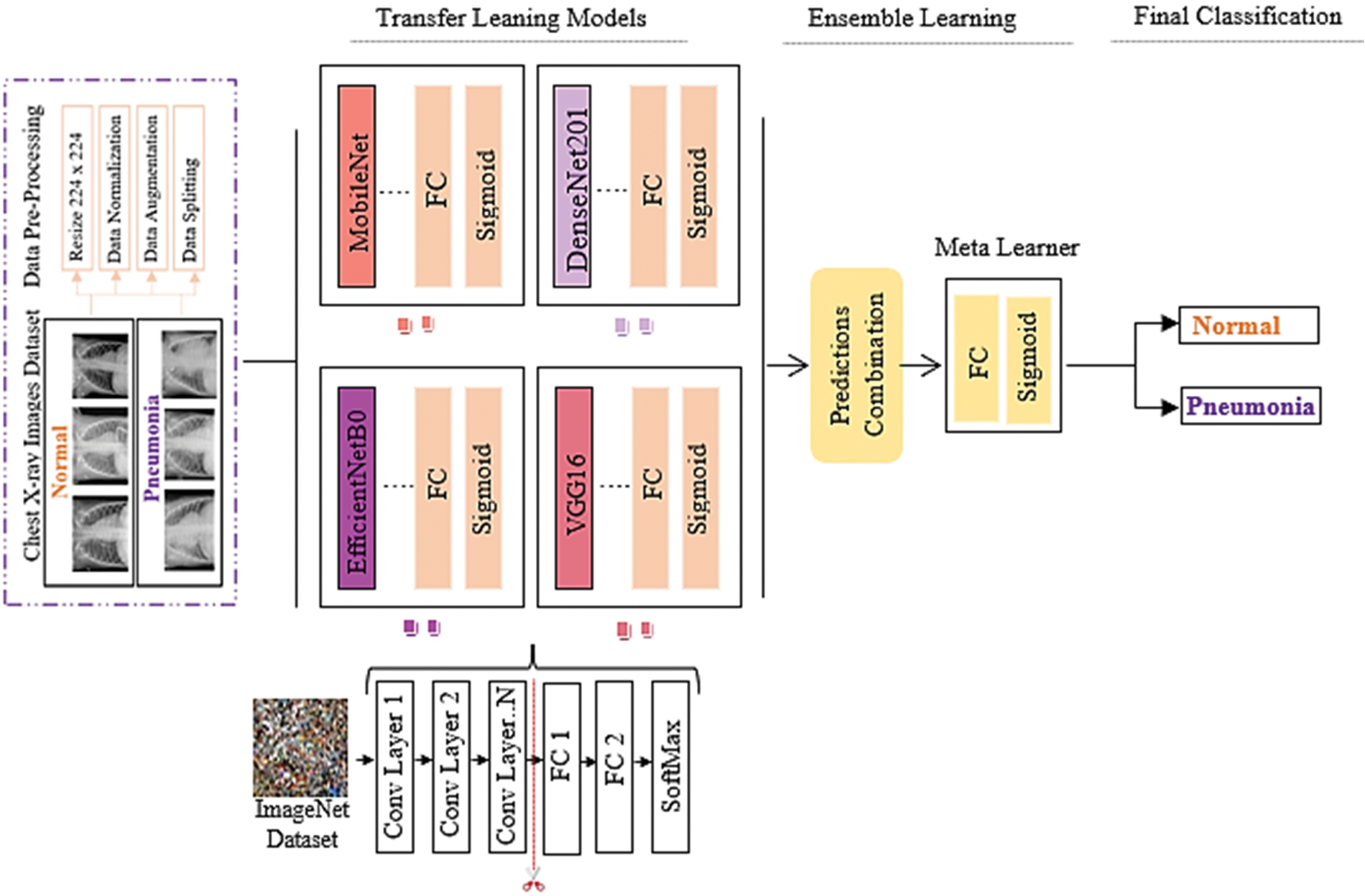
Figure 2: MDEV model

Figure 3: Flow of stacking ensemble learning
Sequential CNN is a generic model in which a convolutional layer extracts salient features from input images, a pooling layer reduces the parameters while constraining spatial dependencies and improving performance, and a fully connected layer acts as a classifier based on the feature maps extracted from the pooling layers [20], as shown in Fig. 4.
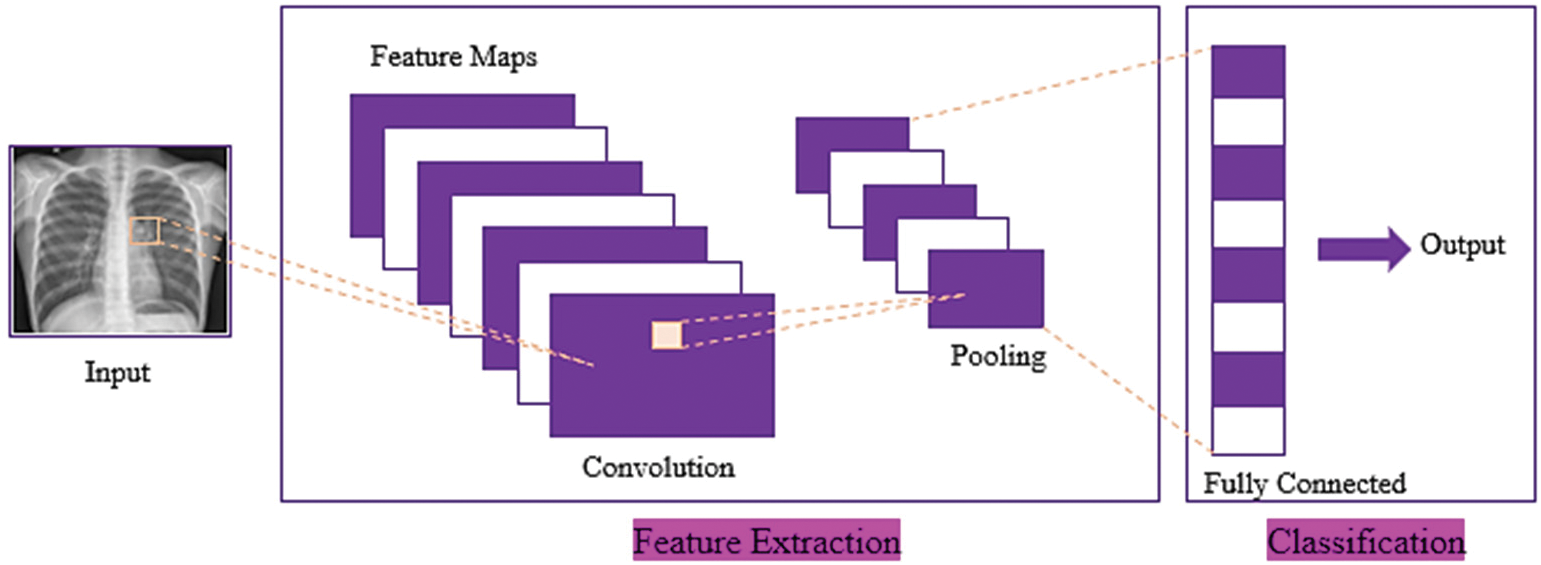
Figure 4: Basic architecture of convolution neural network
The input images dimension (I) convolutional function using the input provided and filter Conv (I, K) are represented using the following equations:
where nH is the Height, nW is the Width, nC is the Channels, the input image is I, and the kernel filter is K.
The training and validation loss is 0.3939 and the overall accuracy is 85.90%, precision 88.28%, recall 82.14%, and F-score 83.85%, as shown in Fig. 5. From 234 test images, 157 were correctly classified as Normal, and 77 were found to be wrongly classified. From 390 images, 379 were correctly categorized as Pneumonia, whereas 11 were wrongly classified. It gives 88 wrong predictions.
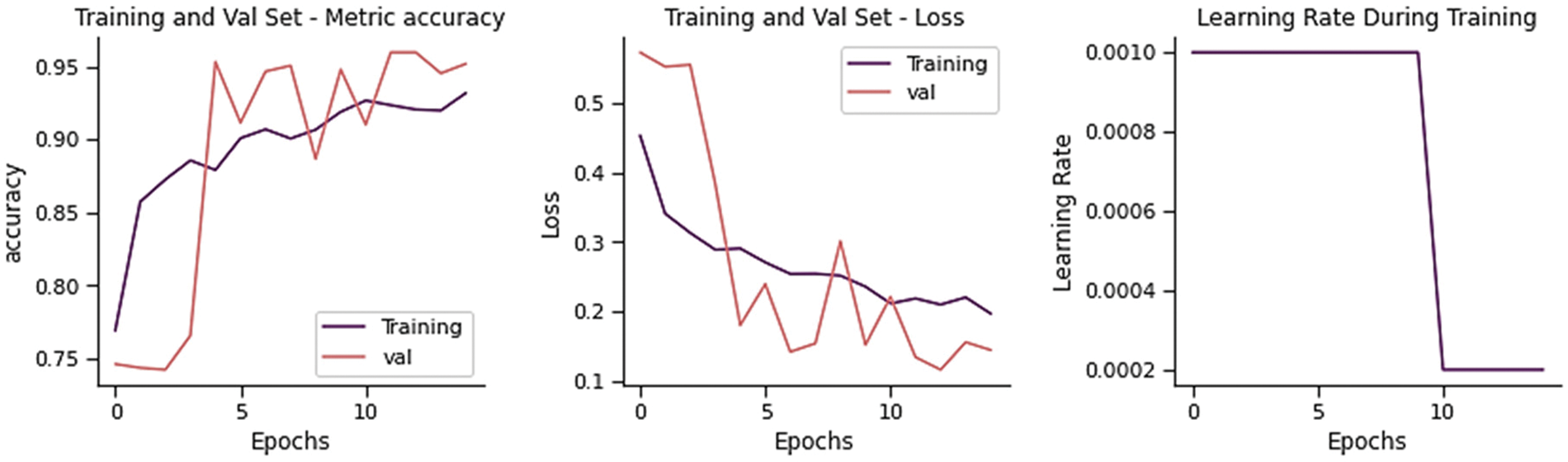
Figure 5: Training performance of sequential CNN on chest x-ray dataset
Applying information from one task to a different but related task is “transfer learning.” ImageNet, a big dataset with roughly 14 million images trained across 21000 classes, is used to train TL models. This method reduces both the training time and the generalization error. It extracts layers from a trained model, freezes them to prevent data loss during later training rounds, place some new trainable layers on top of frozen layers to make predictions from previously mentioned features to a new dataset, and trains the new layers on the dataset [21].
The description of transfer learning models is provided by the Keras Applications [22], as shown in Table 4. Using the ImageNet validation dataset, the model’s performance is correlated with the up-1 and up-5 accuracy. The network’s topological size is referred to as deep. It contains layers such as activation, batch normalization, and so on. The average time for each inference step (CPU or GPU) is 30 batches and ten iterations.
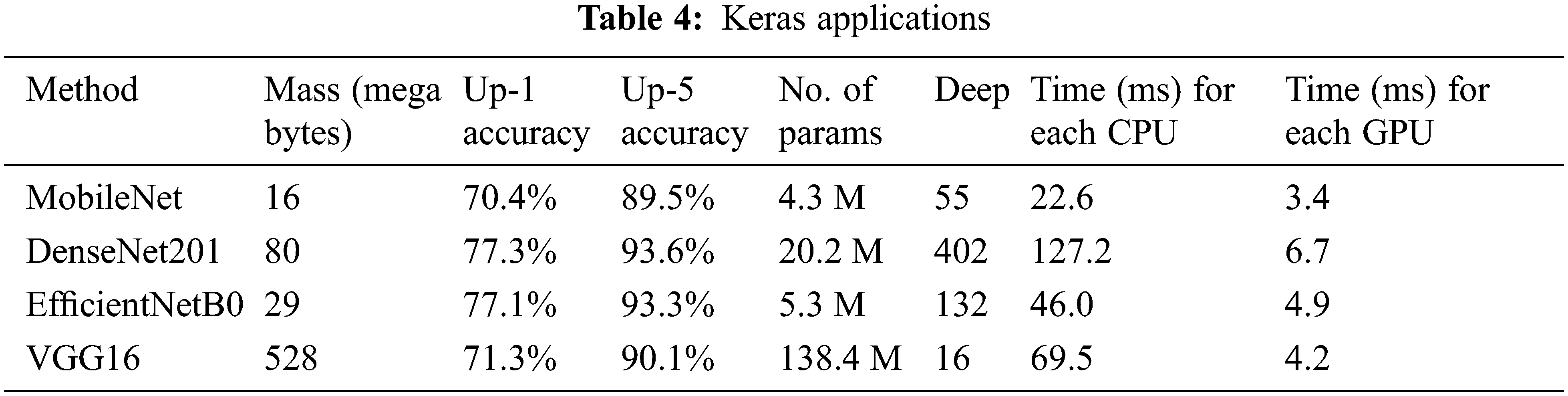
Mobile Nets were created to support mobile applications in a limited phenomenon. The network’s main benefit is the collection of linear bottlenecks and inverted residual layers. A low-dimensional input accumulates in the deep network, which increases by elevating dimensional space. The elevated properties are filtered using depth-wise separable CNNs before being projected into a low-dimensional environment using linear CNNs [23]. This improvement lowers the requirement to access the main memory of the mobile application, resulting in faster computations owing to the usage of cache memory. MobileNet’s architecture is depicted in Fig. 6.
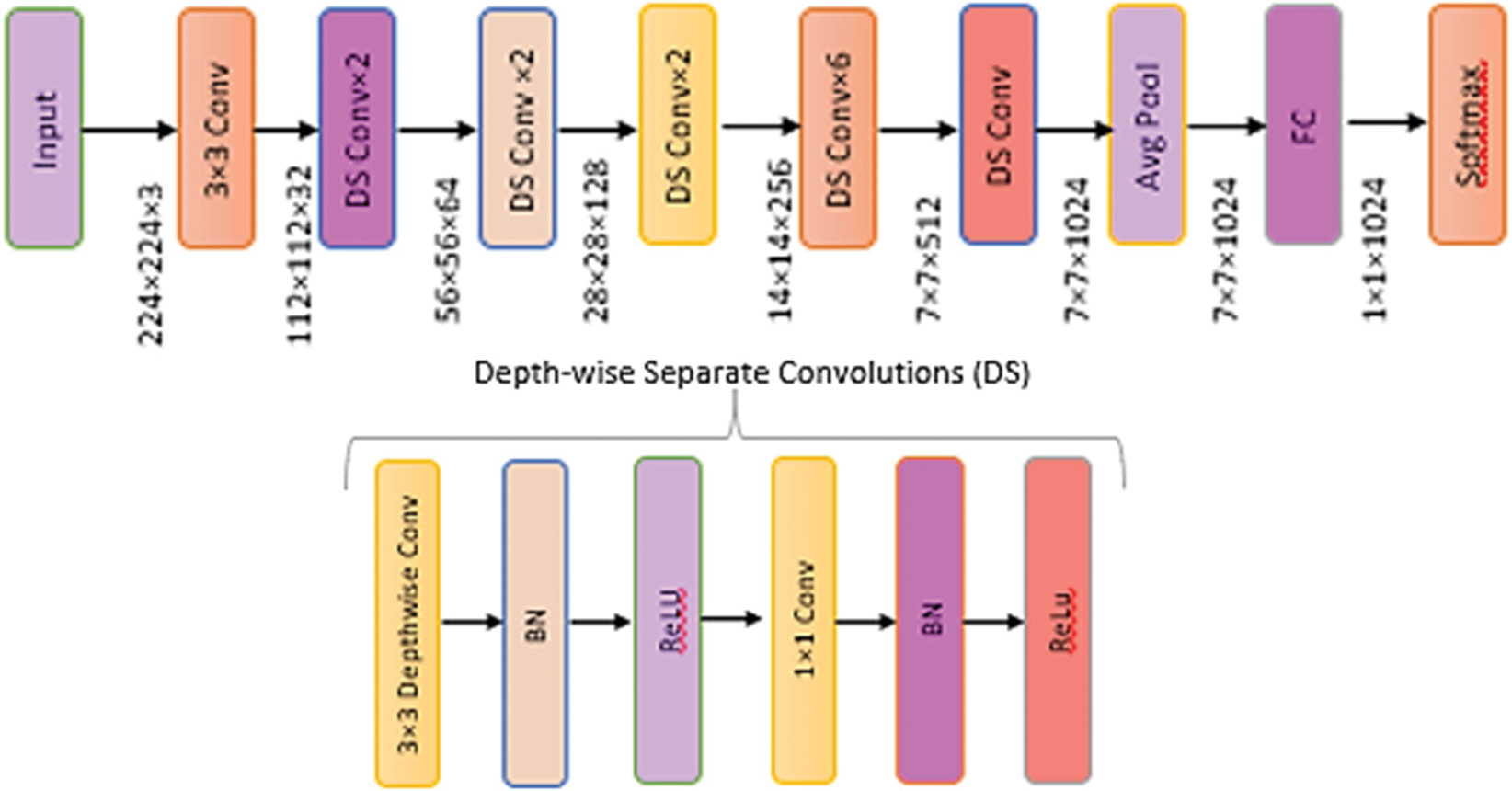
Figure 6: Architecture of MobileNet model
The training and validation loss is 0.4026, and the overall accuracy is 78.85%, precision 79.47%, recall 81.37, and F-score 78.62%, as shown in Fig. 7. From 234 test images, 214 were correctly classified as Normal, and 20 were found to be wrongly classified. From 390 images, 278 were correctly categorized as Pneumonia, whereas 112 was wrongly classified. It gives 132 wrong predictions.
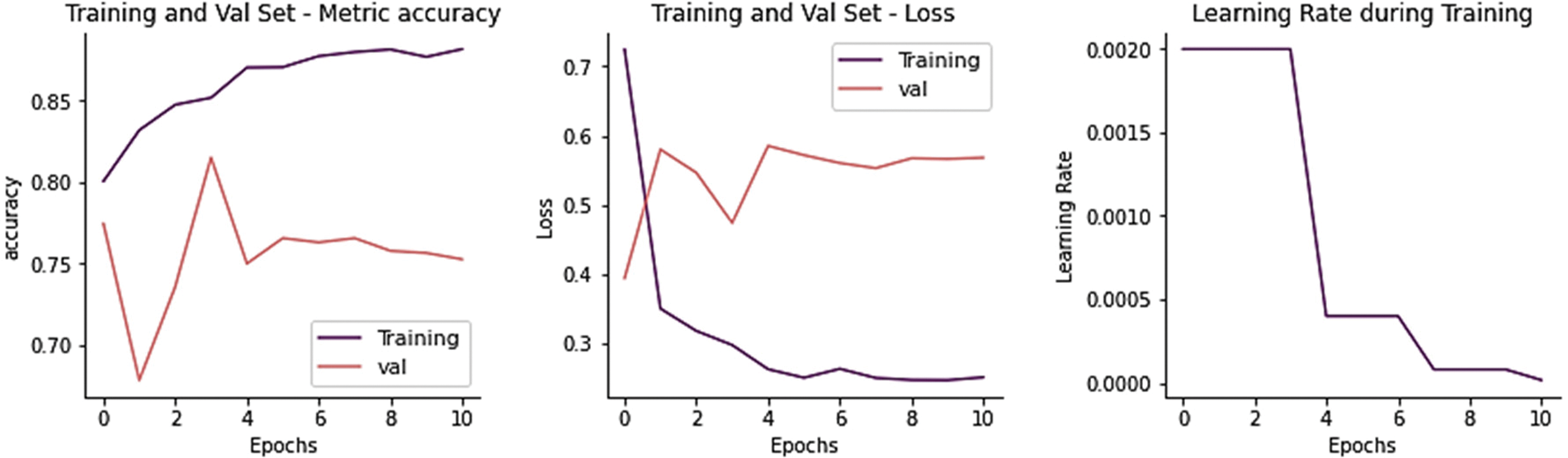
Figure 7: Training performance of MobileNet model on chest x-ray dataset
The densely linked CNNs were motivated by Res-Nets’ residual connections, which forced thick blocks to have long-chained residual connections. For N layers, Dense-Nets have N(N + 1)/2 connections, regarding the residual connections, which improve the network’s capacity to extract detailed attributes while degrading image quality. The dense and transition blocks are arranged sequentially to offer a knowledge collection and a 3 × 3 bottleneck receptive field that improves processing efficiency [24]. Finetuning greater weights increases generalization in deeper networks with 121 to 201 layers. DenseNet’s architecture is seen in Fig. 8.
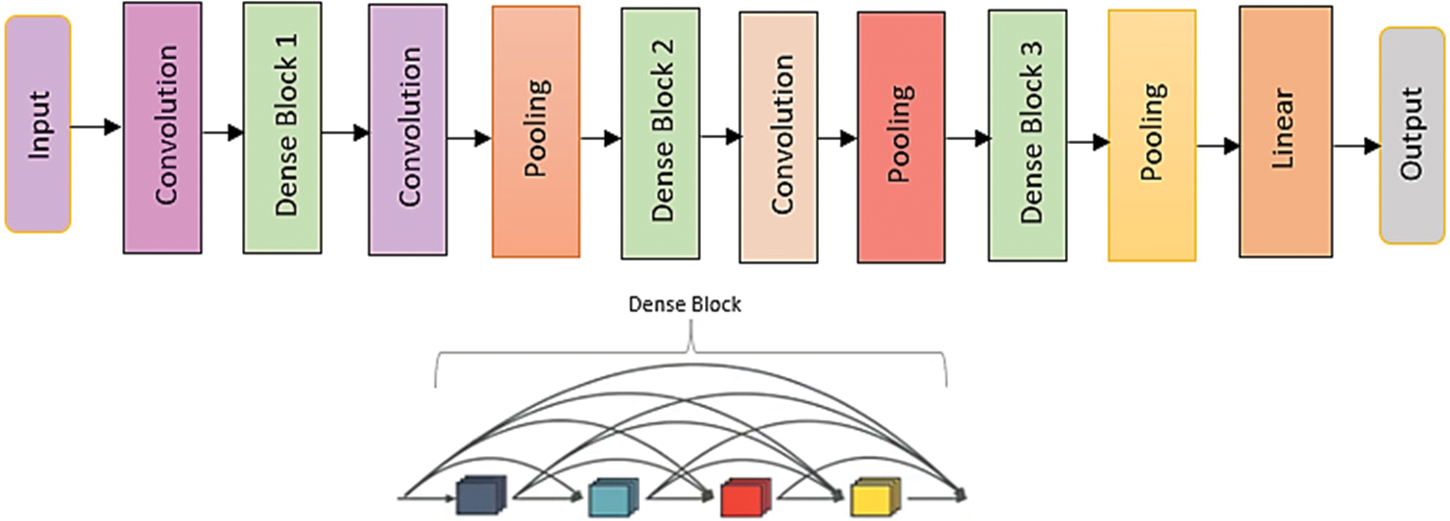
Figure 8: Architecture of DenseNet model
The training and validation loss is 0.4763 and the overall accuracy is 83.65%, precision 83.40%, recall 81.20, and F-score 82.02% as shown in Fig. 9. From 234 test images, 167 were correctly classified as Normal, and 67 were found to be wrongly classified. From 390 images, 355 were correctly categorized as Pneumonia, whereas 35 was wrongly classified. It gives 102 wrong predictions.
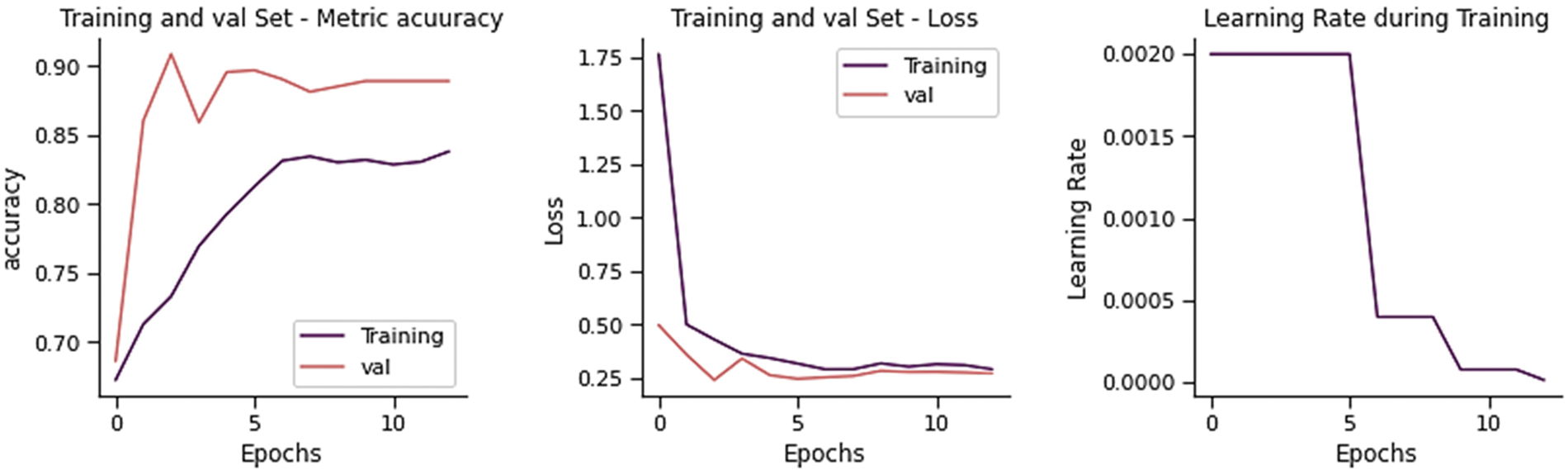
Figure 9: Training performance of DenseNet-201 model on chest x-ray dataset
The collection of EfficientNets models (EfficientNet-B0 to EfficientNet-B7) that scale up from the whole network (commonly referred to as EfficientNet-B0). By reducing parameters and Floating-Point Operations Per Second (FLOPS), EfficientNets improves the model’s accuracy and efficacy [25]. The EfficientNet-B0 baseline architecture’s feature extraction comprises a collection of MBConv blocks for mobile inverted bottleneck convolution with squeeze-and-excitation, batch normalization, and swish activation, as shown in Fig. 10.
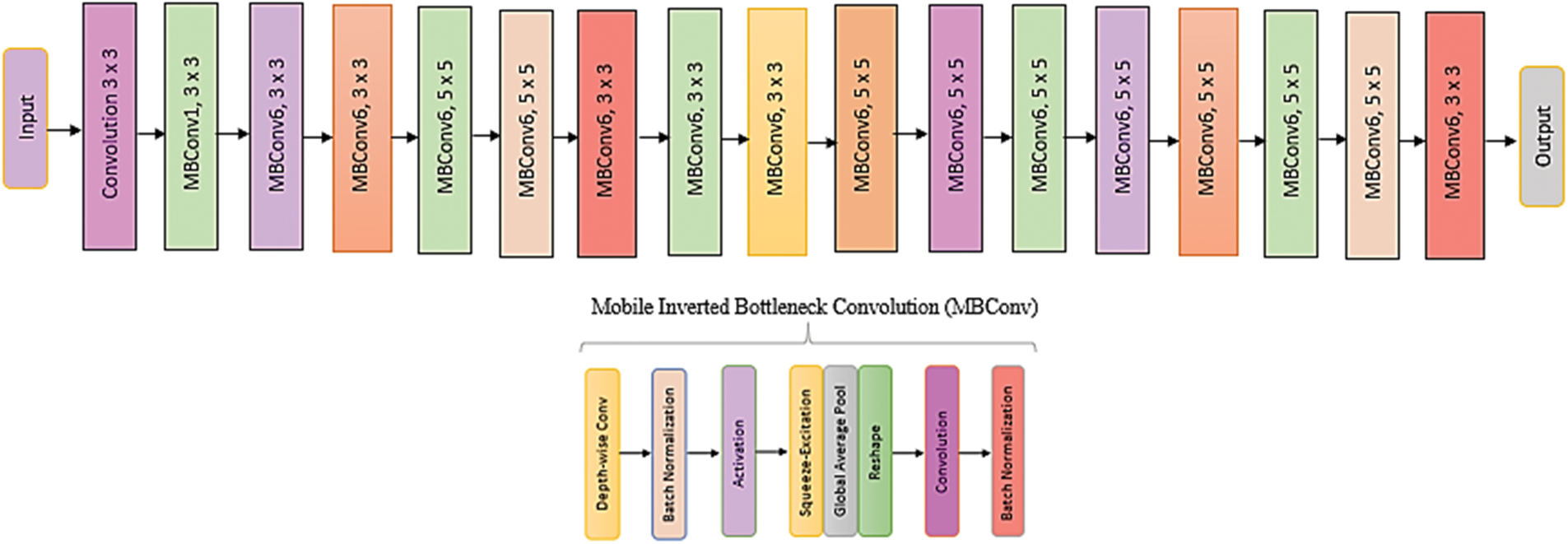
Figure 10: Architecture of EfficientNet model
The training and validation loss is 0.2110, and the overall accuracy is 90.38%, precision 90.64%, recall 88.72, and F-score 89.51%, as shown in Fig. 11. From 234 test images, 192 were correctly classified as Normal, and 42 were found to be wrongly classified. From 390 images, 372 were correctly categorized as Pneumonia, whereas 18 were wrongly classified. It gives 60 wrong predictions.
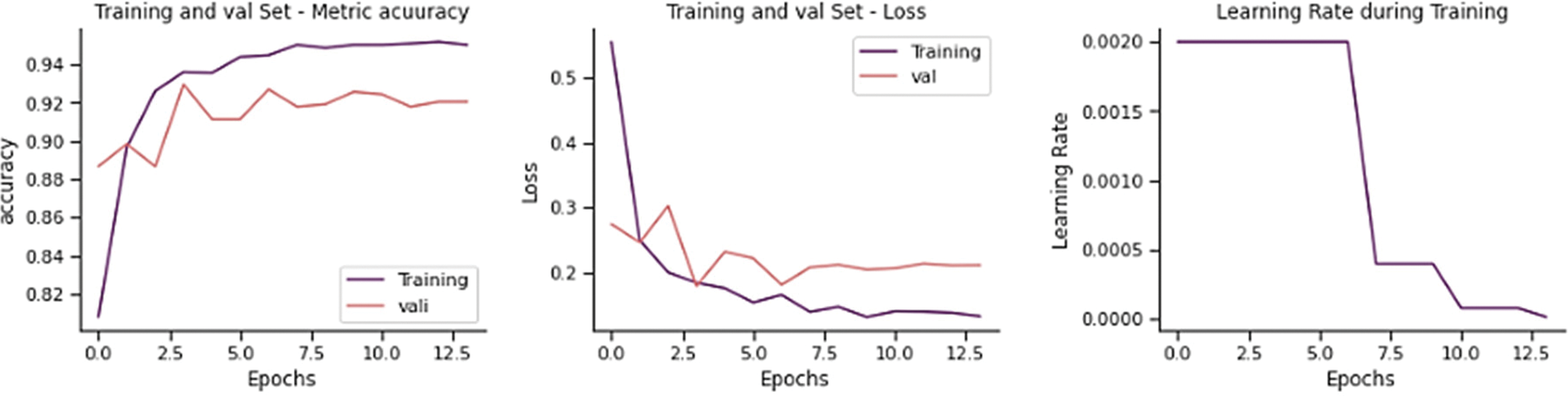
Figure 11: Training performance of EfficientNet-B0 model on chest x-ray dataset
VGG model is a type of CNN. This network is simple as it only contains 3 × 3 convolutional layers on top of each layer, as shown in Fig. 12. The size can be lowered using max pooling. After that, two layers are completely connected with 4,094 nodes each, preceded by a SoftMax [26,27]. VGG-16 and VGG-19 are ConvNets with deep 16 and 19 layers, respectively.
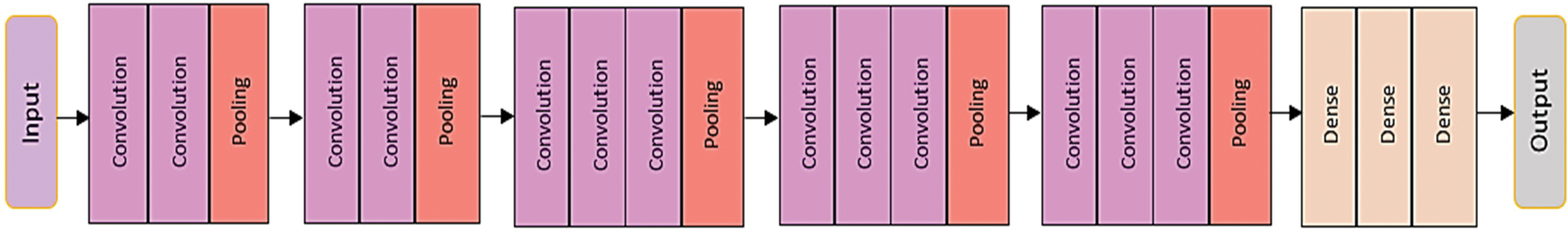
Figure 12: Architecture of VGG model
The training and validation loss is 0.2871, and the overall accuracy is 87.82%, precision 86.76%, recall 88.21, and F-score 87.29%, as shown in Fig. 13. From 234 test images, 210 were correctly classified as Normal, and 24 were found to be wrongly classified. From 390 images, 338 were correctly categorized as Pneumonia, whereas 52 were wrongly classified. It gives 76 wrong predictions.
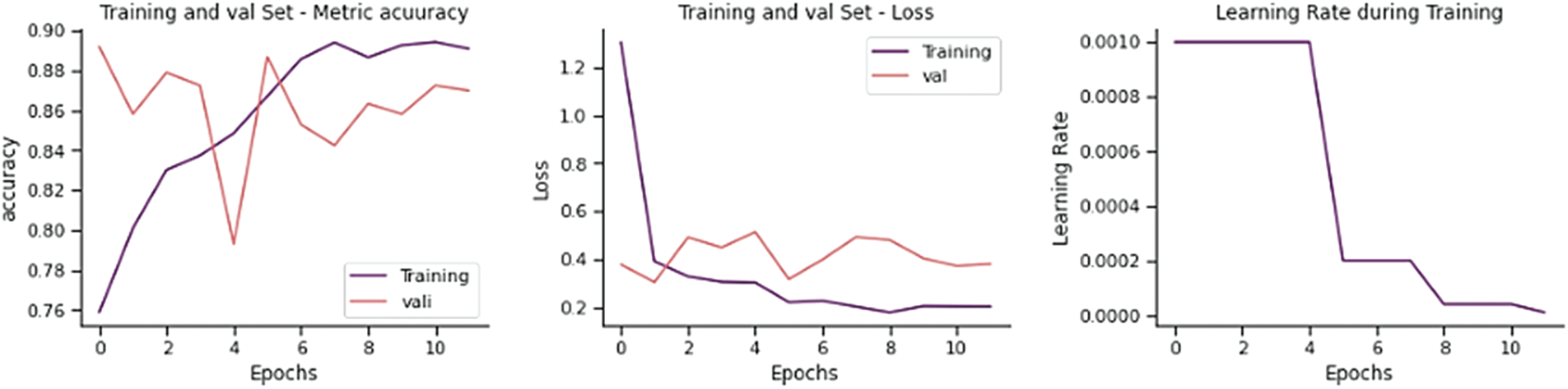
Figure 13: Training performance of VGG-16 model on chest x-ray dataset
The MDEV model is diagrammatically represented in Fig. 14.
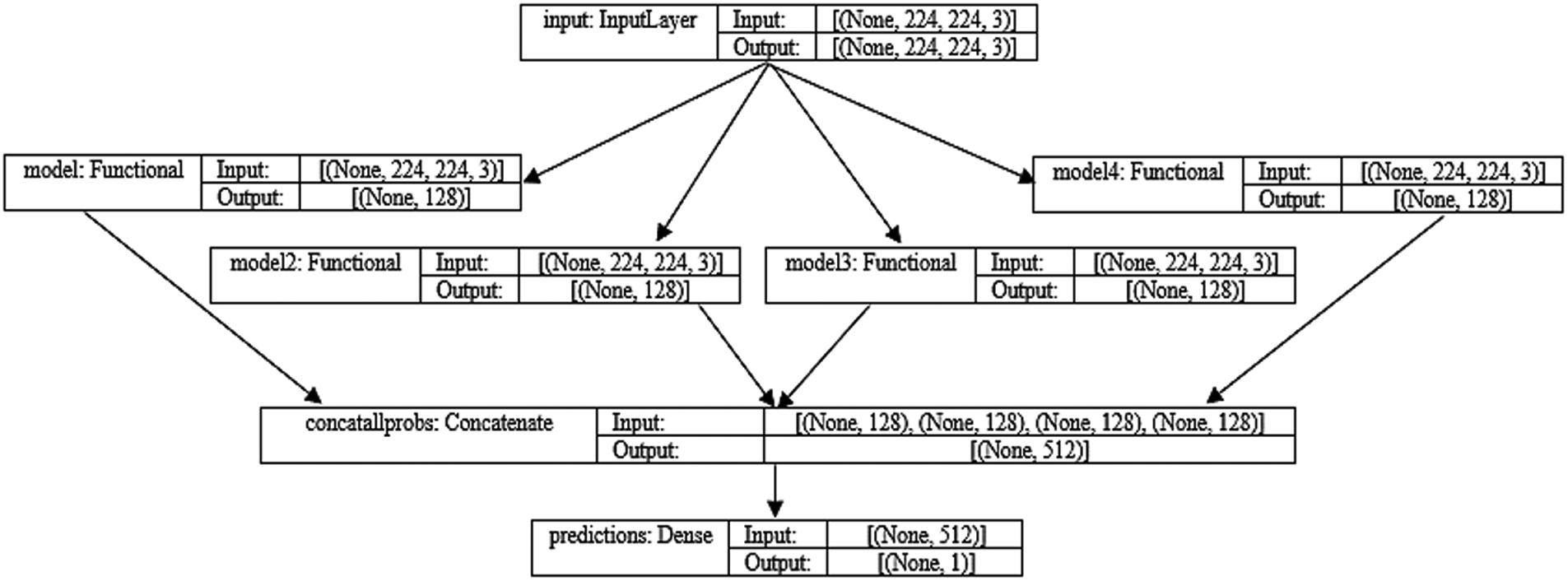
Figure 14: MDEV model architecture and four flows of input
The training and validation loss is 0.2041, and the overall accuracy is 92.15%, precision 92.26%, recall 90.90, and F-score 91.49%, as shown in Fig. 15. From 234 test images, 201 were correctly classified as Normal, and 33 were found to be wrongly classified. From 390 images, 374 were correctly categorized as Pneumonia whereas 16 were wrongly classified. It gives 49 wrong predictions.
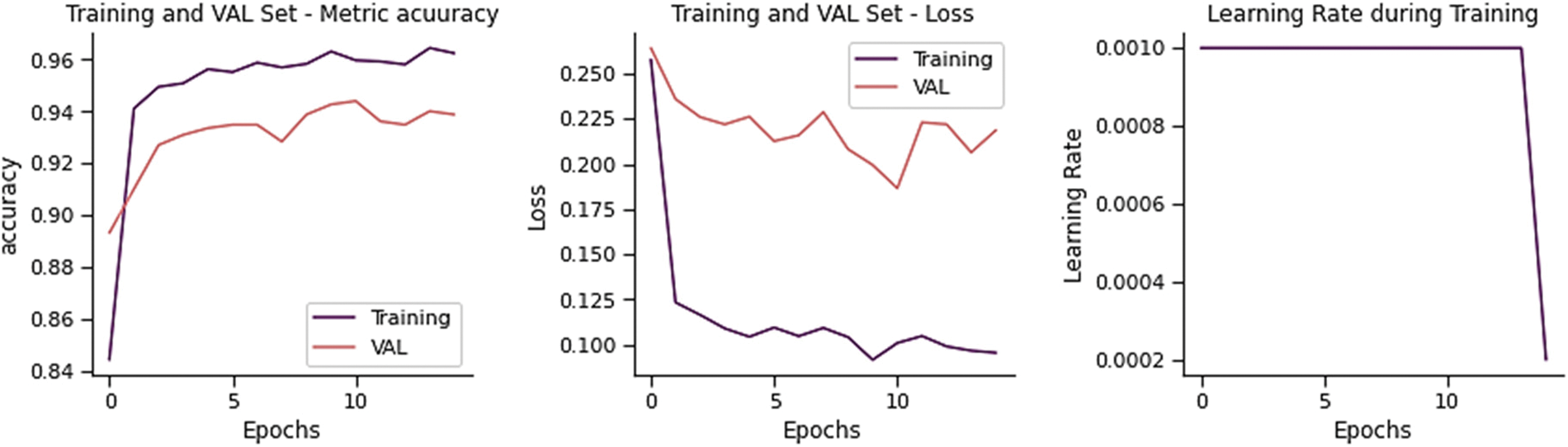
Figure 15: Training performance of MDEV model on chest x-ray dataset
Four transfer learning models are used in this research to classify Pneumonia and Normal images on input CXR images. Stacking the ensemble classification technique is employed, which combines predictions from many models to create a new model which employs a meta-learning method to figure out how to combine predictions from two or more base models to produce test image prediction results. The confusion matrix of all models is shown in Fig. 16.
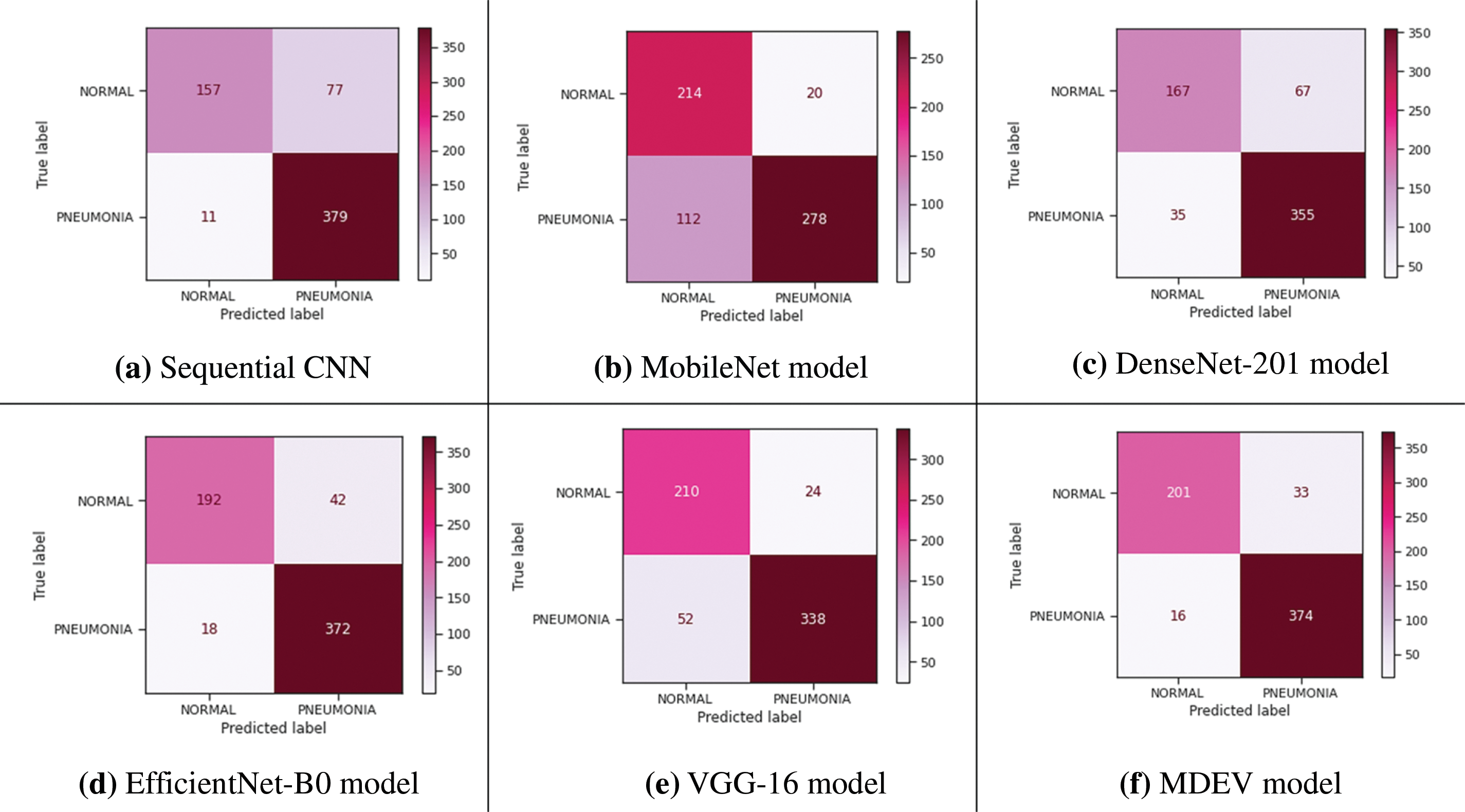
Figure 16: Confusion matrix of all models
The transfer learning models against generic convolution neural network (CNN) are compared, and the results are that CNN gives good accuracy results, but it has some limitations of speed, size, etc., for which transfer learning models outperform. MobileNet, DenseNet, EfficientNet, and VGG are four transfer learning models that are compared in terms of precision, accuracy, recall, AUC-roc, and f1-score, and the result is that EfficientNet gives high accuracy among all others. Finally, if transfer learning models are compared with the proposed MDEV model, it results that MDEV outperforms that of individual base models as it gives the combinational effect as shown in Table 5, and AUC-ROC curve is shown in Fig. 17 depicts how well models can differentiate between Normal and Pneumonia classifications.
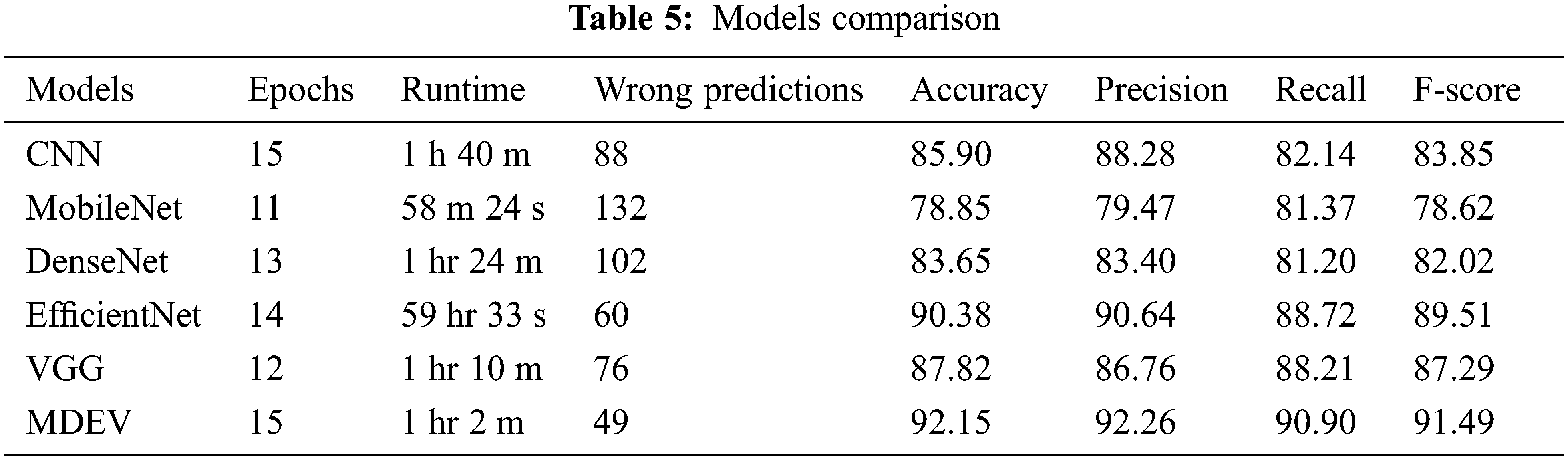
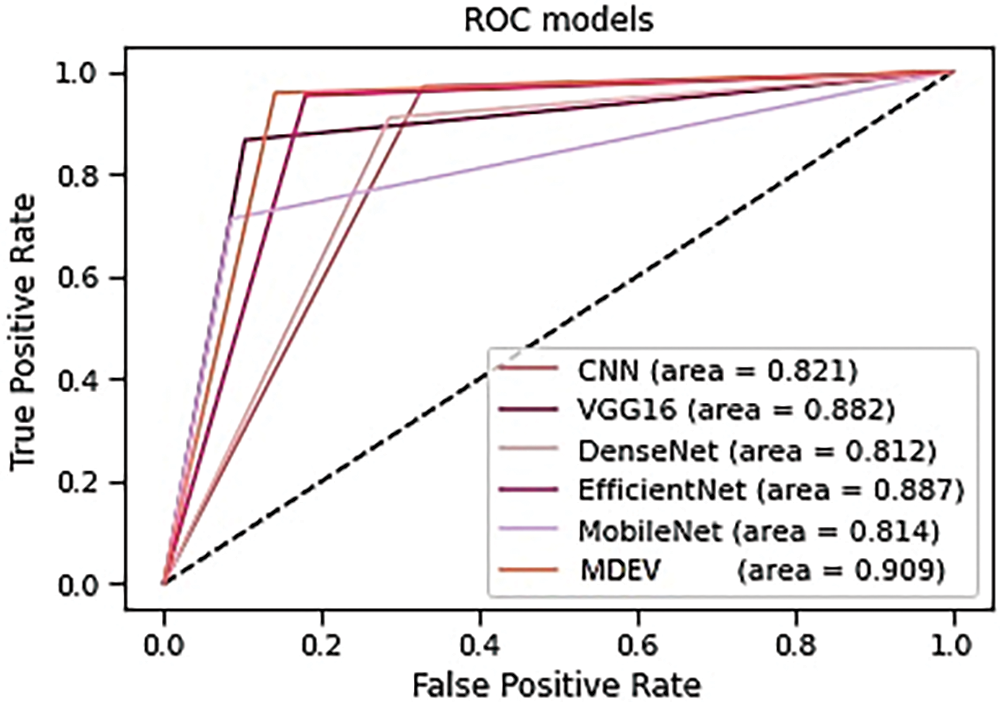
Figure 17: AUC-ROC curve of models
The findings of the suggested MDEV model for pneumonia disease classification are discussed in this section. Python is the chosen programming language for data analysis that could be used to solve deep-learning-based difficulties. Anaconda Navigator, Jupyter Notebook, and other tools using Python 3.9.7 are implemented and evaluated in the proposed model. The models are trained and validated on an Intel Core i5 7th generation x64-based processor with 8 GB RAM, 256 SSD, and HD Graphics 620.
The publicly available dataset from Kaggle is used to apply data pre-processing and data augmentation techniques. The data input size of images was fixed to 224 × 224 × 3 to make training easier, and data augmentation techniques such as zoom range 0.2, rotation range 0.2, height shift range 0.1, and width shift range 0.1 were used. The data was split into 85% training and 15% testing. AveragePooling2D, flattened, dense, and dropout layers were used to alter the underlying models. Following that, all models were compiled using Adam optimizer, Sigmoid and ReLU activation functions, 15 epochs, batch size 32, dropout 0.5, learning rate 0.002, GPU computations, and binary log loss function was trained to lower the dimension of the extracted feature. To regulate the number of iterations/epochs, the LearningRateScheduler call-back function is used. The new learning rate is returned by a function that accepts an epoch index as input and returns an epoch index as output. Based on the epoch, the learning rate is adjusted. The ModelCheckpoint, ReduceLRonPlateau, and EarlyStopping call-back functions are also employed where patience is set to 10 and validation loss is noticed. After the training phase, the test dataset uses certain evaluation metrics to see how good the models are.
The proposed ensemble model concatenates the four deep base models used to categorize Normal and Pneumonic chest x-ray images. The MobileNet gives overall accuracy of 78.85%, precision 79.47%, recall 81.37, F-score 78.62% and auc-roc 81.4%. DenseNet-201 gives accuracy 83.65%, precision 83.40%, recall 81.20, F-score 82.02% and auc-roc 81.2%. EfficientNet-B0 gives accuracy of 90.38%, precision 90.64%, recall 88.72, F-score 89.51% and auc-roc 88.7%. Vgg-16 gives accuracy 87.82%, precision 86.76%, recall 88.21, F-score 87.29% and auc-roc 88.2%. The four transfer learning models have also been compared against generic sequential convolution neural networks, and results show that transfer learning models perform far beyond state-of-the-art CNNs. Comparing the ensemble MDEV model with base models resulted in the ensemble performing excellently.
This study proposes a novel MDEV model for pneumonia disease classification using CXR images. Each transfer learning model takes around an hour to train and validate. All models are trained using the Adam optimizer, ReLU, and Sigmoid activation, early stopping, and binary log loss function. 0.002 is the learning rate, while 0.5 is the dropout rate. All models undergo 15 iterations/epochs of training. On the test set, our best MDEV model has achieved 92.15% accuracy, a precision of 92.26%, a recall of 90.90%, an f-score of 91.49%, and an AUROC score of 90.9% with minimal pre-processing and hyperparameter adjustment. Furthermore, a comparative analysis of four transfer learning models and sequential convolution neural network with the proposed model is compared for pneumonia classification on 5,856 CXR datasets. The proposed method can be used to detect pneumonia cases as an alternative to traditional diagnostic methods and will greatly assist the radiologist in capturing more clinically valuable images and swiftly determining the type of Pneumonia. The MDEV model can be translated into software, deployed on lab computers, or used in remote locations where scarce radiologists are. Asthma, bronchiectasis, emphysema, coronavirus, viral and bacterial Pneumonia, and tuberculosis can all be detected using the MDEV model; just dataset needs to be changed. The model’s accuracy will improve as more chest images are fed into it. This quick classification will allow computer-aided technology to be used in novel ways, such as screening pneumonia patients at airports. The weakness of the MDEV model is that it is more complex, harder to interpret, high computational power than that of light-weighted base models.
Future studies can increase the number of datasets, apart from the Kaggle dataset; real-time inputs could also be considered and improve the transfer learning architecture by altering hyperparameters and transfer learning combinations for a better complex network structure.
Acknowledgement: This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2021R1I1A1A01052299).
Funding Statement: This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2021R1I1A1A01052299).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. P. Schulz, S. Moore, D. Smith, J. Javed and A. Wilde, “Missed pneumococcal vaccination opportunities in adults with invasive pneumococcal disease in a community health system,” Open Forum Infectious Diseases, vol. 9, no. 4, pp. 1–3, 2022. [Google Scholar]
2. N. Hamza Shah, M. Junaid, N. Muhammad Faseeh and D. Shin, “A multi-blocked image classifier for deep learning,” Mehran University Research Journal of Engineering and Technology, vol. 39, no. 3, pp. 583–594, 2020. [Google Scholar]
3. A. Alimadadi, S. Aryal, I. Manandhar, P. Munroe, B. Joe et al., “Artificial intelligence and machine learning to fight COVID-19,” Physiological Genomics, vol. 52, no. 4, pp. 200–202, 2020. [Google Scholar]
4. B. Sheri, P. Kumari, I. F. Siddiqui and H. Norman, “Artificial intelligence-based memory stash Alzheimer’s aid,” in Int. Conf. on Information Science and Communication Technology (ICISCT), Karachi, Pakistan, pp. 1–5, 2020. [Google Scholar]
5. M. Hashmi, S. Katiyar, A. Keskar, N. Bokde and Z. Geem, “Efficient pneumonia detection in chest x-ray images using deep transfer learning,” Diagnostics, vol. 10, no. 6, pp. 417, 2020. [Google Scholar]
6. C. J. Saul, D. Urey and C. Taktakoglu, “Early diagnosis of pneumonia with deep learning,” arXiv.org, 2022. [Online]. Available: https://doi.org/10.48550/arXiv.1904.00937. [Google Scholar]
7. J. Luján-García, C. Yáñez-Márquez, Y. Villuendas-Rey and O. Camacho-Nieto, “A transfer learning method for pneumonia classification and visualization,” Applied Sciences, vol. 10, no. 8, pp. 8–14, 2020. [Google Scholar]
8. Z. Baloch, F. Shaikh and M. Ulnar, “A Context-aware data fusion approach for health-IoT,” International Journal of Information Technology, vol. 10, no. 3, pp. 241–245, 2018. [Google Scholar]
9. D. Han, Q. Liu and W. Fan, “A new image classification method using CNN transfer learning and web data augmentation,” Expert Systems with Applications, vol. 95, pp. 43–56, 2018. [Google Scholar]
10. U. Khuhawar, I. Siddiqui, Q. Arain, M. Siddiqui and N. Qureshi, “On-ground distributed COVID-19 variant intelligent data analytics for a regional territory,” Wireless Communications and Mobile Computing, vol. 2021, pp. 1–19, 2021. [Google Scholar]
11. Z. Khademi, F. Ebrahimi and H. Kordy, “A transfer learning-based CNN and LSTM hybrid deep learning model to classify motor imagery EEG signals,” Computers in Biology and Medicine, vol. 143, pp. 105288, 2022. [Google Scholar]
12. R. Fan and S. Bu, “Transfer-learning-based approach for the diagnosis of lung diseases from chest x-ray images,” Entropy, vol. 24, no. 3, pp. 1–13, 2022. [Google Scholar]
13. T. Rajasenbagam, S. Jeyanthi and J. Pandian, “Detection of pneumonia infection in lungs from chest x-ray images using deep convolutional neural network and content-based image retrieval techniques,” Journal of Ambient Intelligence and Humanized Computing, 2021. Available: https://link.springer.com/article/10.1007/s12652-021-03075-2. [Google Scholar]
14. K. El Asnaoui, Y. Chawki and A. Idri, “Automated methods for detection and classification pneumonia based on x-ray images using deep learning,” Studies in Big Data, vol. 90, pp. 257–284, 2021. Available: https://doi.org/10.1007/978-3-030-74575-2_14. [Google Scholar]
15. R. Kundu, R. Das, Z. Geem, G. Han and R. Sarkar, “Pneumonia detection in chest X-ray images using an ensemble of deep learning models,” PloS One, vol. 16, no. 9, pp. 1–29, 2021. [Google Scholar]
16. M. Salehi, R. Mohammadi, H. Ghaffari, N. Sadighi and R. Reiazi, “Automated detection of pneumonia cases using deep transfer learning with pediatric chest x-ray images,” The British Journal of Radiology, vol. 94, no. 1121, pp. 1–8, 2021. [Google Scholar]
17. T. Rahman, M. E. H. Chowdhury, A. Khandakar, K. R. Islam, K. F. Islam et al., “Transfer learning with deep convolutional neural network (CNN) for pneumonia detection using chest x-ray,” Applied Sciences, vol. 10, no. 9, pp. 10–14, 2020. [Google Scholar]
18. V. Chouhan, S. K. Singh, A. Khamparia, D. Gupta, P. Tiwari et al., “A novel transfer learning based approach for pneumonia detection in chest x-ray images,” Applied Sciences, vol. 10, no. 2, pp. 5–14, 2020. [Google Scholar]
19. “Chest X-ray images (Pneumonia),” Kaggle.com, 2022. [Online]. Available: https://www.kaggle.com/datasets/paultimothymooney/chest-xray-pneumonia. [Google Scholar]
20. M. Monti, J. Poon, V. Chung and F. Monshi, “Covidxraynet: Optimizing data augmentation and CNN hyperparameters for improved COVID-19 detection from CXR,” Computers in Biology and Medicine, vol. 133, pp. 2–13, 2021. [Google Scholar]
21. H. Kim, A. Cosa-Linan, N. Santhanam, M. Jannesari, M. Maros et al., “Transfer learning for medical image classification: A literature review,” BMC Medical Imaging, vol. 22, no. 1, pp. 1–13, 2022. [Google Scholar]
22. K. Team, “Keras documentation: Keras applications,” Keras.io, 2022. [Online]. Available: https://keras.io/api/applications/. [Google Scholar]
23. Z. Yue, L. Ma and R. Zhang, “Comparison and validation of deep learning models for the diagnosis of pneumonia,” Computational Intelligence and Neuroscience, vol. 2020, pp. 1–8, 2020. [Google Scholar]
24. A. Alhudhaif, K. Polat and O. Karaman, “Determination of COVID-19 pneumonia based on generalized convolutional neural network model from chest X-ray images,” Expert Systems with Applications, vol. 180, pp. 2–9, 2021. [Google Scholar]
25. E. Ayan and H. M. Ünver, “Diagnosis of pneumonia from chest x-ray images using deep learning,” Scientific Meeting on Electrical-Electronics & Biomedical Engineering and Computer Science (EBBT), pp. 1–5, 2019. [Google Scholar]
26. Y. Khurana and U. Soni, “Leveraging deep learning for COVID-19 diagnosis through chest imaging,” Neural Computing and Applications, vol. 34, no. 16, pp. 14003–14012, 2022. [Google Scholar]
27. S. A. Wagan, J. Koo, I. F. Siddiqui, M. Attique, D. R. Shin et al., “Internet of medical things and trending converged technologies: A comprehensive review on real-time applications,” Journal of King Saud University-Computer and Information Sciences, 2022. Available: https://www.sciencedirect.com/science/article/pii/S1319157822003263. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools