 Open Access
Open Access
ARTICLE
Graph Ranked Clustering Based Biomedical Text Summarization Using Top k Similarity
Department of Computer Science and Engineering, National Institute of Technology, Raipur, 492001, Chhattisgarh, India
* Corresponding Author: Supriya Gupta. Email:
Computer Systems Science and Engineering 2023, 45(3), 2333-2349. https://doi.org/10.32604/csse.2023.030385
Received 25 March 2022; Accepted 01 July 2022; Issue published 21 December 2022
Abstract
Text Summarization models facilitate biomedical clinicians and researchers in acquiring informative data from enormous domain-specific literature within less time and effort. Evaluating and selecting the most informative sentences from biomedical articles is always challenging. This study aims to develop a dual-mode biomedical text summarization model to achieve enhanced coverage and information. The research also includes checking the fitment of appropriate graph ranking techniques for improved performance of the summarization model. The input biomedical text is mapped as a graph where meaningful sentences are evaluated as the central node and the critical associations between them. The proposed framework utilizes the top k similarity technique in a combination of UMLS and a sampled probability-based clustering method which aids in unearthing relevant meanings of the biomedical domain-specific word vectors and finding the best possible associations between crucial sentences. The quality of the framework is assessed via different parameters like information retention, coverage, readability, cohesion, and ROUGE scores in clustering and non-clustering modes. The significant benefits of the suggested technique are capturing crucial biomedical information with increased coverage and reasonable memory consumption. The configurable settings of combined parameters reduce execution time, enhance memory utilization, and extract relevant information outperforming other biomedical baseline models. An improvement of 17% is achieved when the proposed model is checked against similar biomedical text summarizers.Keywords
Enormous medical records, web articles, electronic health records (EHR), and clinical reports are available online, containing important information related to the biomedical field. The trending demand for information retrieval and hypothesis interpretation from the vast collection of biomedical documents inspires the usage of automated text summarization [1]. The modern automatic summarizers can ease the retrieval of informative content from biomedical literature to form a frame for readers, researchers, and medical practitioners [2]. Original text documents can be represented by summaries which are a brief and consolidated snapshot of informative data [3]. Biomedical text summarization frameworks play a critical role in analyzing biomedical text literature and assist domain specialists in data visualization [4]. Until this point, the summarization is done using various probabilistic, linguistic; graph and ML based techniques [3,5]. The past research demonstrates that the graph-dependent methodology can be fruitful for the evaluation of generic as well as domain-specific summarization [6–9]. There are a couple of challenges in graph-based implementation. First, the relationships and connections among individual sentences must be precisely evaluated after identifying root nodes within the graph via ranking with an effective strategy. The other challenge is to make a summary with subthemes with non-redundant data. In our work, the above fundamental difficulties of graph ranking strategies are addressed and clubbed probabilistic clustering to the proposed approach regarding biomedical text summarization.
The suggested framework utilizes specific text embeddings dependent on the domain so that the connectivity among inter-sentences can be precisely evaluated. Additionally, context-oriented words are used for pre-training the model empowered by BERT [10]. Bio-BERT pre-trained model assigns tokens and forms word vectors dynamically. Then, the proposed method applies the probabilistic clustering, the Sampled-Dirichlet Process Mixture model (S-DPMM), for clustering the sentences derived from Gibb’s Dirichlet process mixture model. The input biomedical text clusters are demonstrated in the form of a graph consisting of nodes and edges. The sentences are converted into vectors to estimate the association power between them. The proposed model performance is analyzed with three graph-based ranking strategies, PageRank (PR), Hyperlink Induced Topic Search (HITS), and Positional Power Function (PPF) in this study [11–13]. HITS algorithm utilizes hyper-linking of nodes and is characterized by authority and hub parameters. Whereas PPF implies power evaluation of high valued vertex in the graph by validating local and global information. Sentences ranked higher hold vital information related to the biomedical domain and the proposed summarizer collates them in the form of a summary. The generated output is then checked with the help of ROUGE and objective function measurements [14]. The introduced summarization framework outcome depicts improved performance when it uses an appropriate blend of Bio-BERT word embeddings, S-DPMM clustering, and different ranking algorithms.
The remainder of this work is outlined in various sections listed below. Section 2 highlights the background work done in the past. The proposed method of summarization process with different flavors is explained in Section 3. The experimental analysis and quantitative evaluations are represented by Section 4. At last, Section 5 gives attention to concluding the theme and defines the future boundaries of the proposed work.
Text summarizers can be classified into single or multi-document, extractive or abstractive, and generic or query oriented per different strategies. The summarization strategy introduced in this paper is generic single biomedical document extractive in nature. Until this point, numerous text summarization frameworks have been proposed, like item-set-based, graph-based, machine learning-based and deep learning based and other hybrid approaches [3,5,15]. The statistical summarization process uses term recurrence, or word frequency occasionally blended with a theme-based, to find ideal subsets of useful sentences [3,4,15]. Probabilistic strategies influence the probability dissemination of words, concepts, and topics inside the text to estimate new possibility dispersions for the expected summary. The possibility of the final summary dispersion is changed per the input text or follows the appropriation of primary concepts and themes [4,16]. Semantic jobs, archive structure, topic modeling, and thematic presentation design are among the fundamental strategies that address the summary issue by linguistic properties of the text [3,17]. Machine learning AI strategies have been broadly examined with regard to context based summarization. Various difficulties of summarization have been tended to by a wide scope of AI and ML approaches, including clustering [2,18], classification [4], itemset and ruleset mining [12,19], optimization [20,21], and neural organizations [22,23]. The hybrid methods with the collaboration of two or three different approaches are also studied, and it is observed that more efforts are required to analyze the biomedical text [1,24,25]. Past research tended to the summarization of various sorts of biomedical archives, for example, preliminary clinical reports [26], medical records [27], clinical notes [28], patient-explicit proposals [29], and biomedical articles [18]. A few techniques influence archives present for the biomedical theme and portray it semantically [4,11,18]. Unlike standard biomedical summarization tools, the presented strategy uses unique area straight keyword collection sets. This permits the evaluation of content-relatedness dependent on contextual regularities, and linguistics.
Ongoing NLP exploration specific to the biomedical area is beneficial through continuous word presentation by neural network models. In a recent study, the Bi-LSTM and BILU-NEMH neural network-based NLP models are used to generate hyper-graphs and classify nested mention entities in the present text. Multiple tasks for sequence labeling are analyzed in the attention-specific and self-attention neural network models applied at a term and individual letter levels. Similarly, the authors describe latent variable conditional random field models LVCRF-I and LVCRF-II for improved sequence labeling [30–34]. The drug discovery research word2vec embedding used by Nelson et al. [35] finds the gene associated with disease from the PubMed articles. Erkan et al. [36] fostered a unique big data design framework that utilizes the Word2vec tool o facilitate semantic resemblance assessment. Saggion [37] explored the handiness of the Word2vec technique in estimating the relatedness of key biomedical concepts. Blagec et al. [38] checked the capacity of various techniques involved for neural-specific text component sets relatedness assessment. The proposed work uses the biomedical dataset that is contextual data and tough to summarize rather than context-free data. The analysis is done on the different contextual, and context-free data for the summarization [6–12] where GraphSum [12] makes the extractive summary by creating nodes and similarity edges to find the correlations of the nodes, and the highest score sentences make final summary. Similarly, Brin et al. [11] provided the semantic graph on the biomedical data using biomedical concepts in the form of nodes and their correlations with the sub-themes using the edges similarity. The other pre-trained SOTA NLP models released in recent years are GPT-2, which has only decoder blocks, and T5, which has both encoder and decoder blocks. Although GPT-2 is very smart in generating coherent language terms but is very heavyweight and is suitable for providing context-specific reviews. T5 is the most advanced model and is created by Google on top of the transformers. It is pre-trained on 7 TB of dataset and is very powerful for summarization and reading comprehension. But T5 is a costly model and can cause memory and a space issue since it is pre-trained on a large dataset. Compared to GPT and T5 models, BERT consumes less space [39,40].
Presently, the itemset mining approach with frequent pattern text analysis focuses on the graph-based summarization [10] where the medical sentences are represented as nodes and edges denote the similarity measure [41]. Earlier DUC 2004 is mostly used for evaluating the extractive text summarization which contains the news outline with synopsis. The existing techniques perform R1 score .33, R2 score of .12 in DUC 2004. The benchmark dataset is CNN/Daily Mail dataset to evaluate extractive summaries with the R-1 along with R-2 values of .44 and .2, respectively [27]. The Gigaword [36] and X-Sum [37] are alternative options for assessing extractive summary with R-1 values somewhere in the range of 0.4 and 0.48, and R-2 scores somewhere in the range of 0.2 and 0.25 were accounted for by best in class techniques on these two datasets [37,38]. The score depends on the input dataset type (dependent and independent) and the summary size.
The previous studies lacked to provide the contextual meaning of biomedical keywords, which we have covered by using UMLS and Bio-BERT in this work. Also, coverage, non-redundancy, and information retention were on the lower side, improving clustering and top-k similarity-based graph preparation. This paper mainly focuses on word embeddings to evaluate the semantic theme analysis in the graph-based biomedical text summarizer. The proposed method combines word embedding with the top k similarity edges and the clustering of the document. We assess our biomedical domain explicit summarization framework over a collection of PubMed archives. The suggested apparatus is tried and analyzed in parallel with benchmark biomedical summarizers.
This proposed method contains article preprocessing, BioBERT word embeddings, and probabilistic clustering with a graph ranking approach for making the biomedical summary. The major sub-modules of suggested framework are depicted in Fig. 1. The fundamental building blocks include data preparation and cleansing via NLP, identifying domain-centric vital keywords through UMLS, forming peculiar word embeddings by BERT, dual-mode execution support for clustering, and graph-modeling with multiple ranking approaches. The execution flow from initiation to summary finalization for the bio-medical article is clearly evident from the framework model.
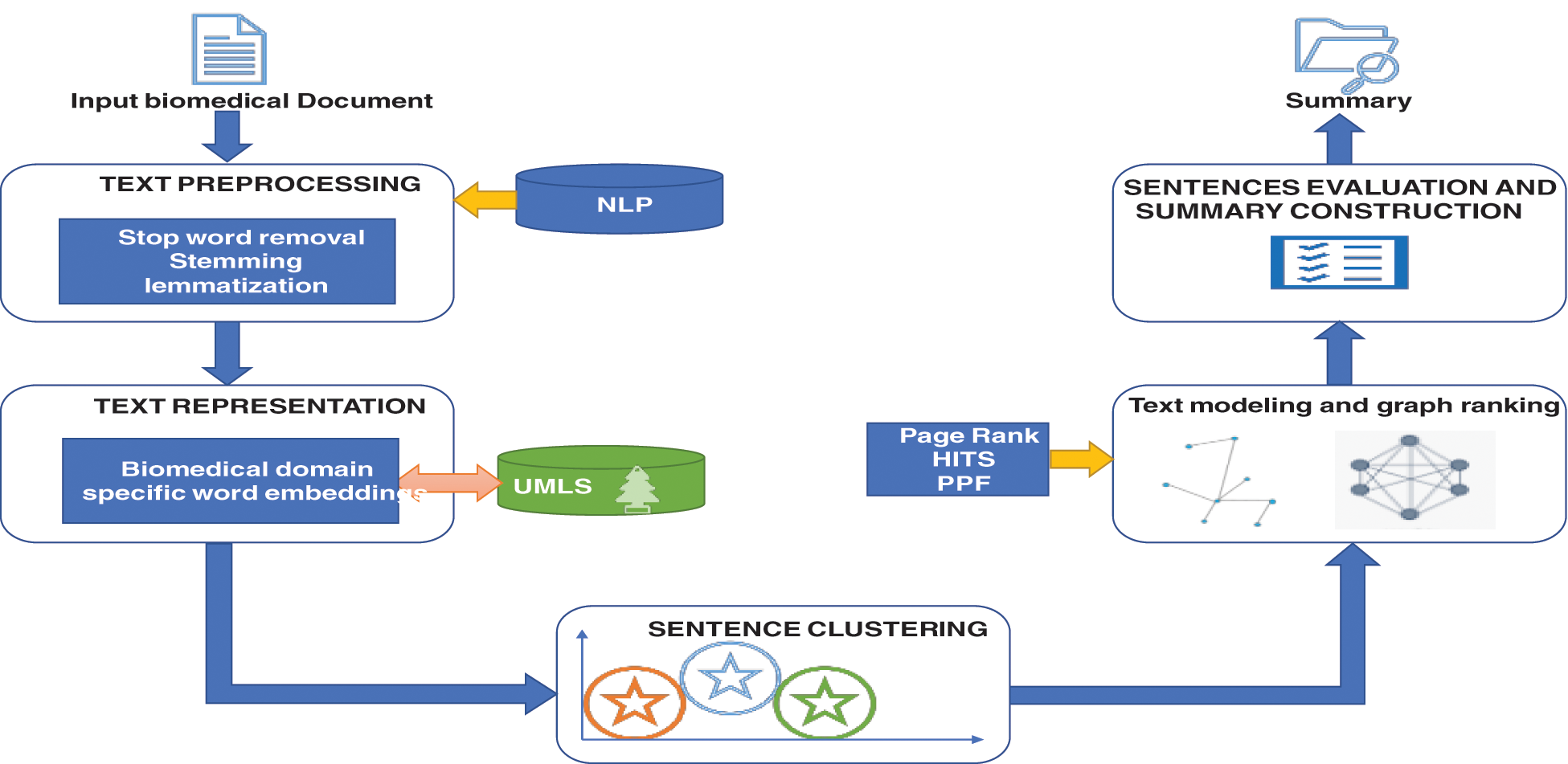
Figure 1: The structure of biomedical text summarization using clustering and graph-based ranking
For the most part, an introductory biomedical document in its raw form is inadmissible to process. Consequently, this requires fundamental modification of inbound documents through the new framework. The preliminary thing is isolating individual text documents from the substantial biomedical archive via an appropriate parser. Significant crude information is skimmed from the rest of author’s reference, pictorial, and numeric details producing unadulterated text data. The processed file is then divided into separate lines and terms, which are handled and denoted via:
where each sentence or line
The non-useful substance and unimportant data are observed as wastage; for example punctuations, unnecessary articles and stop words are taken out atomically from the assortment as displayed in Eqs. (1) and (2). Terms with the normal forms are converted into simple structures through stemming. These text records are further available for imminent action.
Biomedical ontologies and representative vocabularies like UMLS, SNOMED-CT, and MeSH in the medical field enable remarkable collection of tools and files to increase interoperability among computing devices. The objective of using these ontologies is to extract domain explicit features or concept identification through well-defined medical dictionaries. UMLS vocab assists in scrutinizing crucial semantics, whereas BERT helps in relevant plotting. The indexing information of various domain elements with their equivalent words is listed in UMLS through more than 200 dictionaries [36]. A vital part assumes building connections among shortlisted elements to form a semantic grid [42]. Notwithstanding the above listed embeddings play a crucial role in forming associations among recognized vocabularies. The immaterial ideas get sifted, and domain-specific significant highlights having actual weights are saved for additional treatment.
3.3 Biomedical Word Embeddings
Word embeddings can be represented as the key terms and extracted concepts which are arranged as real value word vectors of precise length carrying specific context. The different word embeddings formation techniques are layers in the conjugation of neural networks, Word2Vec and GloVe. The context-free embeddings are deficient in linguistic context modeling and unable to catch the semantic and syntactic regularities. These embeddings provide singleton interpretation of every term despite the order and environment of appearance. Rather, context-sensitive frameworks change dynamically according to the word appearance with the target words to give contextual representation. One key linguistic sub-framework used to illustrate semantics in dual directions is BERT [9]. The count of different components is variable in various types of BERT versions. The Bio-BERT variation is created via pre-trained basic BERT over a huge biomedical repository of documents [10]. The proposed model uses Bio-BERT to establish plotting among inbound data with semantic embeddings. Proposed model uses the pre-trained Bio-BERT on PubMed archives [43,44]. The features are extracted and linked to the generated embeddings. Specific sentences are characterized using vector notations obtained from averaging each word concerning the corresponding sentences.
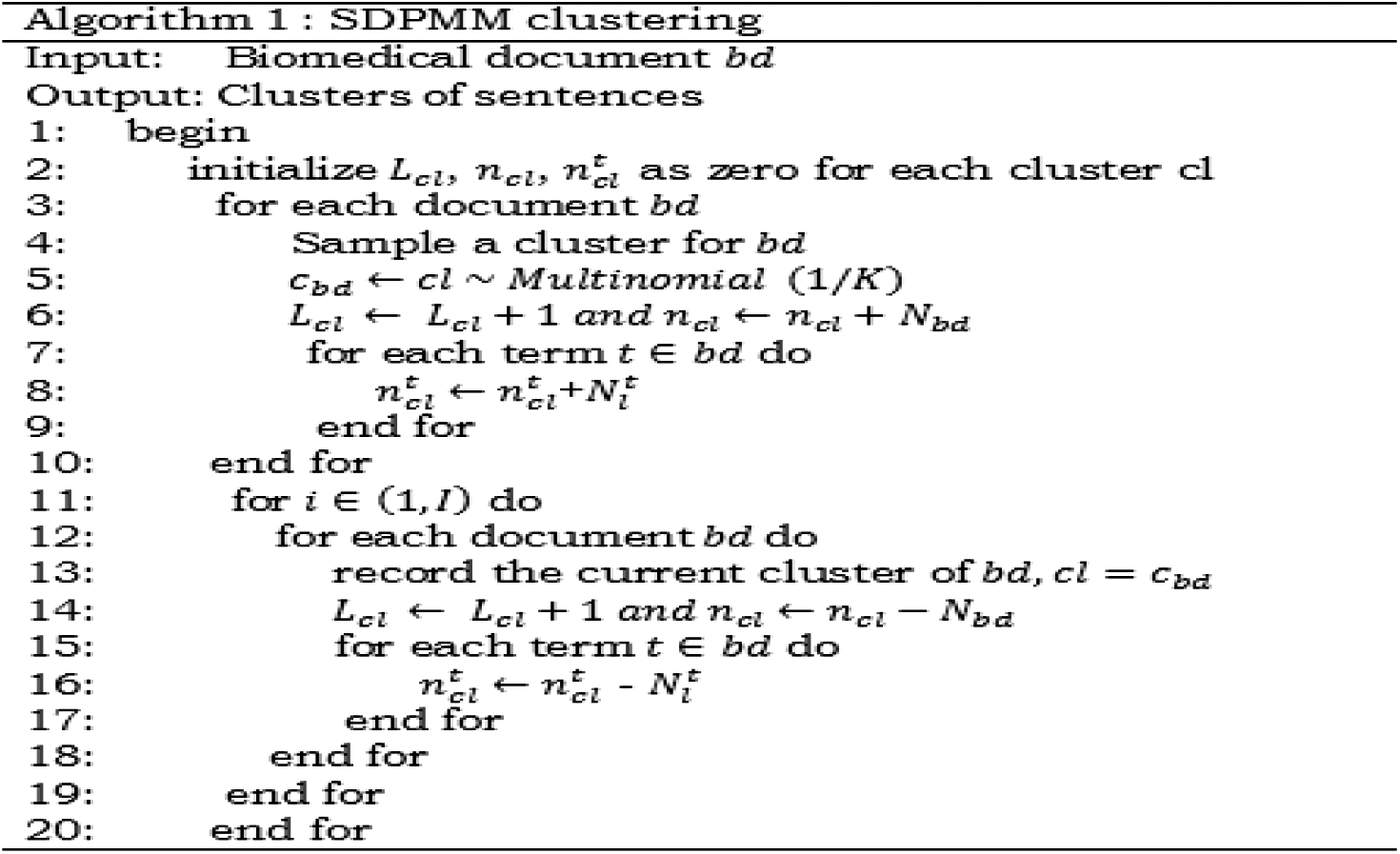
This proposed method uses the Sampled-Dirichlet Process Mixture model (S-DPMM) to cluster the subthemes from the biomedical records. The proposed model generates the clusters using a multinomial probability of the terms.
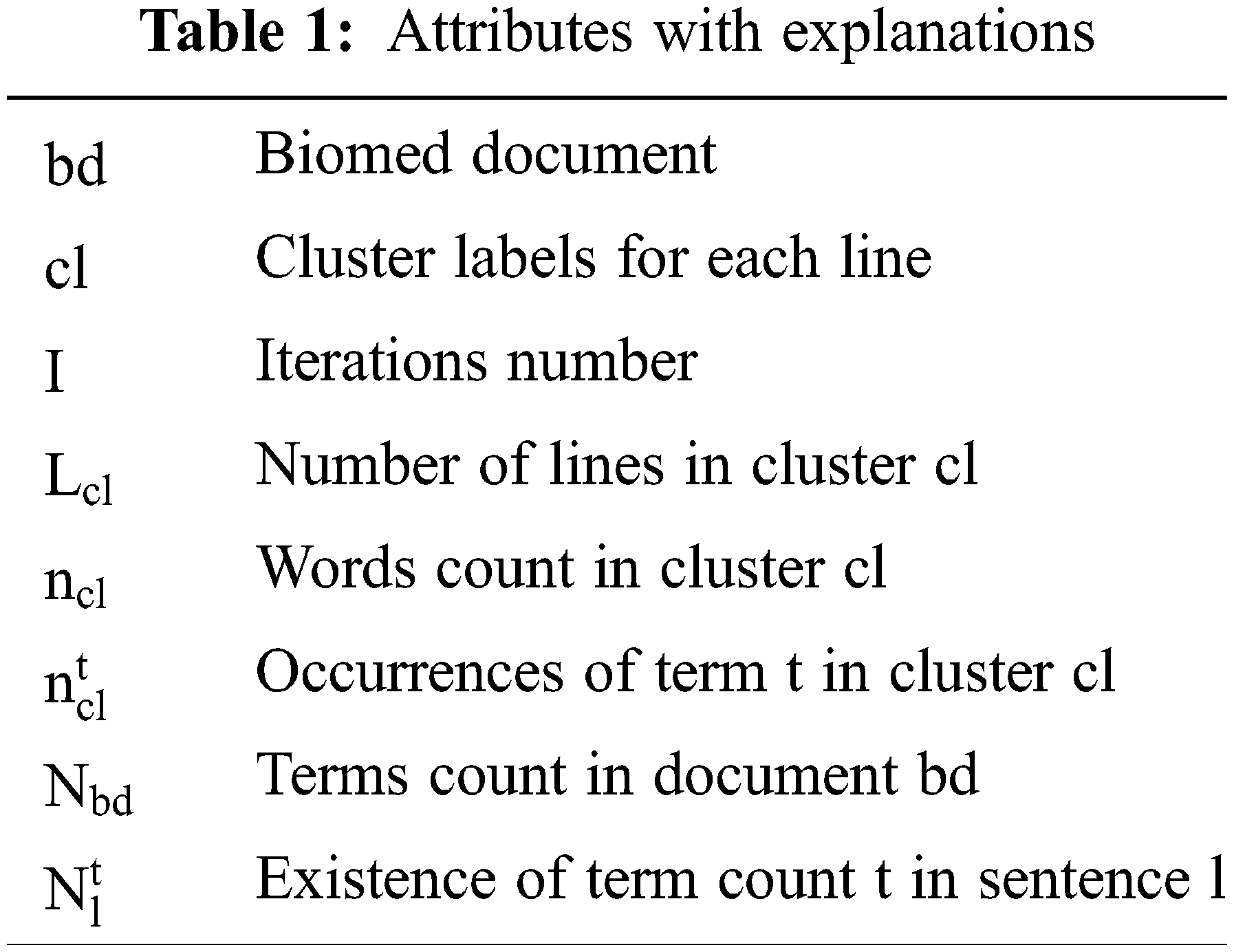
The vital parameters which are used for clustering are mentioned in Tab. 1. There are composite parts that ought to be seen like individual clusters. Under the DPMM strategy, the frequent probability of term ‘t’ concerning line ‘l’ residing in the document is denoted as p(t|l) along with p(t|cl), and the likelihood of terms in the cl cluster is checked [45]. Pick k, count of the cluster according to loads signified via p(cl = k). Selective items having common characteristics are grouped and demonstrated via p(l│cl = k). Hence, the likelihood of any line in any specific cluster is characterized via Eq. (3).
Also, the probability of line l to be included in cluster k can be presented in Eq. (4).
Nigam et al. portray clusters as a polynomial appropriation of terms,
DPMM model presents Gibb’s sample approach where the method allots the cluster for each sentence or line l as indicated by
3.5 Graph-Based Sentence Ranking
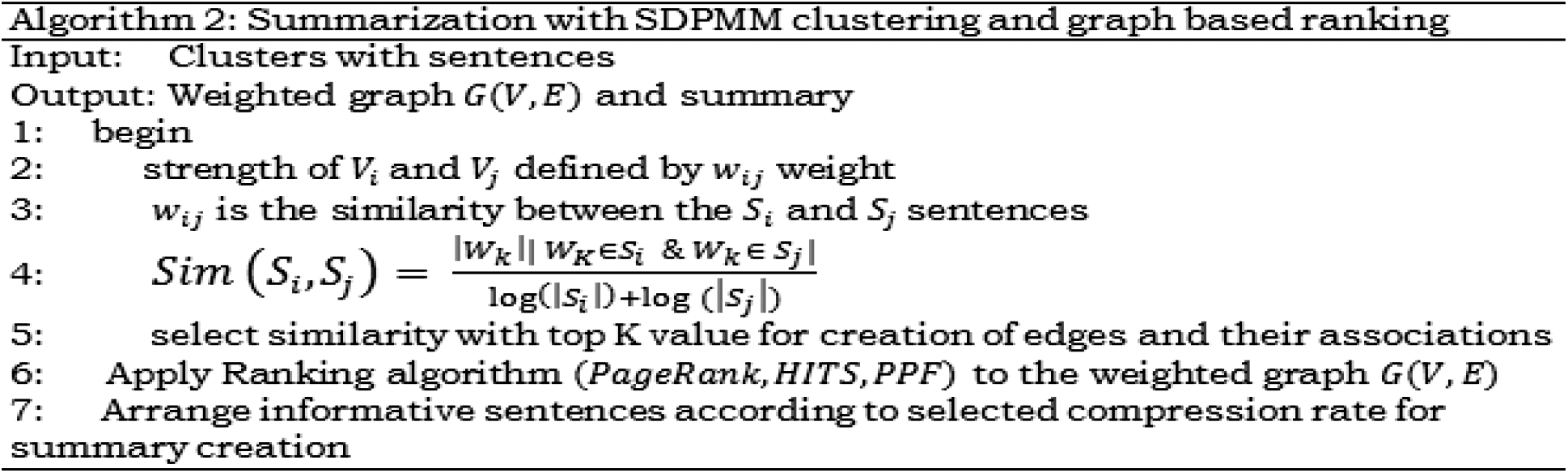
The proposed assembly transforms the original text article into a vectorized graph with weightage to capture connections among the lines. The lines and their associations are portrayed as a vectorized graph with subjective edges. The widely used measure is Cosine similarity for evaluating vector space between two objects which estimates similitude with the angle measure between the vectors.
Let
As explained in Algorithm 2, the summarizer processes vector sets to calculate cosine measures and arranges them from higher to lower values. The
The proposed methodology separately evaluates these three graph based ranking algorithms for the biomedical article summarization model. PageRank (PR) algorithm is a famous method to evaluate the significance of multiple graph vertex and their connections. The highly important apexes have the highest score rank, which shows strong connection quality between the vertices. The evaluation of the PR value for all vertices;
To promote the centrality of nodes and prevent clique attack, the damping factor
HITS algorithm represents a link-oriented method that measures the node importance with the authority and hub property. The node contented with the incoming links shows the authority score, whereas hub values present the associations between nodes corresponding to outgoing-links. The proposed model’s undirected structure obtained from biomedical articles and vertex sets of incoming, outgoing links are indistinguishable. A single value evaluated by combining authority as well as hub property is provided in Eq. (7):
PPF algorithm is the ranking-based process that evaluates the command of a node with axiomatic and iterative property combinations to gather the local and global data in the structured graph. The node weightage and count of links can be iterated and mixed with calculating the power score to each vertex
The score of each vertex is arranged from high to low order for the construction of a graph. The summarizer utilizes compression rate to form the desired output summary with size
An assortment of 100 biomedical writing archives from PubMed acts as inbound data for the trial examination. The records belonging to the biomedical domain are sanitized and transformed before processing, as explained earlier in Section 3.1. The organization of data inside the corpus under test is given in Tab. 2. Information categories are presented in Tab. 3 separately. The insignificant component-plain subtleties, references, graphical representation, and catalog is physically isolated from unique text records. There is always a challenge to procuring standard datasets related to the biomedical area; thus abstracts are considered model rundowns for the inbound articles.

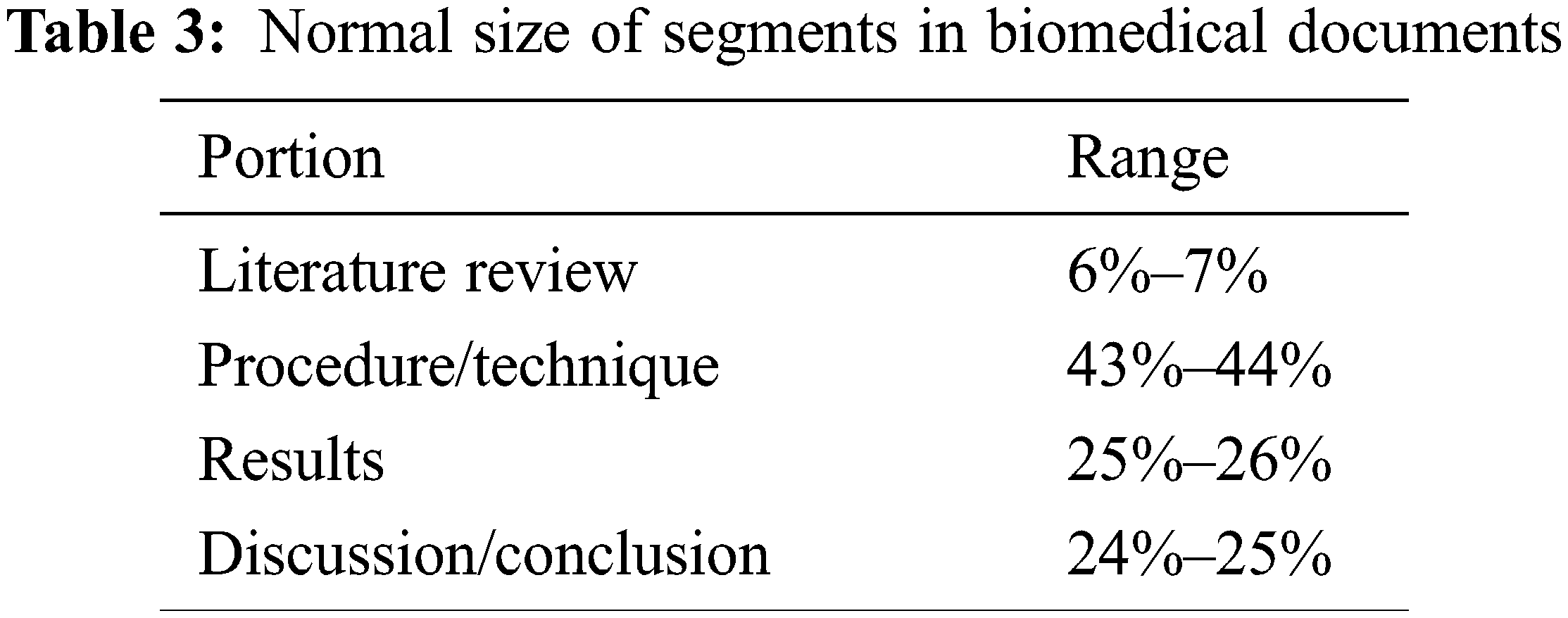
The outbound outline article can be checked via the ROUGE apparatus [14] as it gives a significant connection with human calculation. This assesses and delivers the model rundowns and evaluates the common substance by giving various scores. Higher substance cross-over between produced rundown and model synopses gives higher scores. In this paper, ROUGE-1/2 (R-1/2), Skip-gram (R-SU4), and longest common subsequence (R-L) are utilized to score for showing high levels of connection with human evaluation [14]. R-1 and bigrams gauge the unigrams substance cross-over are taken for R-2 evaluation.
In this stage, the proposed summarizer has different subject groups with their most normal successive ideas in bunches. In this way, the reasonable sentence determination approach should cover every one of the critical biomedical sub-subjects from the report and remove any dull concentrates coming from solitary topics. For accomplishing this evenhanded, the sentence choice consolidates data from all groups according to the given size and compression ratio. According to the information present in document
The
For comparison eight graph-based methods and two baseline summarizers have been taken. The technique covers probabilistic, graph/feature-based methodologies, and machine learning. The evaluation is done with the domain explicit and domain-independent methods. A short depiction of the comparison process is specified below.
Biomedical Graph-based Summarizer (BGSumm) [46] uses frequent itemset mining with the combination of UMLS concepts to make the final summary with meaningful topics and essential themes. The node ranking approach provides the final summary by choosing the high score sentences.
Conceptual graph-based summarizer [47] is biomedical article summarizer which utilizes the hyponymy relation with the node and edge weight. The clustering finds the related topic within the article and UMLS maps each concept of the sentences and finally constructs a graph with a voting mechanism and makes the final summary with extracted topics.
Clustering and Itemset-based Biomedical Summarizer (CIBS) [15] utilizes a mining strategy on the extricated biomedical item sets to find the fundamental thoughts and employs clustering to select peculiar, related sentences from each cluster to create the output summary.
Bayesian biomedical summarizer [4] gives the probability distribution of concepts with the input document and important concepts extracted by the data mining and statistical methods. For final summary construction, the model utilized the odd posterior ratio to extract sentences. TextRank [27] uses similarity measure to find the connection weights between the nodes (sentences). It makes the summary with the influential nodes.
LexRank [36] is the primary graph-based text summarizer method where the graph is a combination of nodes and the edges with weights that uses TF-IDF and cosine similarity for measurement. It considers eigenvector and connection matrices for picking the highest centrality sentences for generating the final summary.
Enhanced SUMMA (E-SUMMA) is built on the SUMMA summarizer [37] that process POS and TF-IDF for sentence extraction and uses frequency and similarity as evaluation features.
TexLexAn summarizer uses common features like keywords, cue-phrases, and frequency to find the score of sentences and the highest score sentences to make the final outline [46].
Lead baseline shortlists initial N sentences, whereas Random baseline considers the arbitrary scrutinized sentences.The trial and hit method garner the optimum values for different parameters. Usually, the compression ratio benchmark ranges from 0.15–0.35 [27]. A compression value of 0.3 is considered throughout the trials.
The analysis and outcome of conducted experiments are examined and highlighted in this part of the paper. Tabular and graphical depiction of parameter configuration is briefed along with a quality review of the outcome. The outbound system values are paralleled between proposed and baseline summarizers.
Experimental analysis is a critical technique that helps analysts evaluates different hypotheses identified during research. Multiple experiments revolving around several of clusters, compression rates and for different k values have been performed. The primary objective is to check our model’s fitment of the appropriate graph ranking algorithm. The impact of varying parameters on the model performance can also reveal some exciting points through this analysis.
The initial setup is employed to evaluate ROUGE parameters when the cluster number is set to 5; compression rate defaulted to 30%, and variation of the similarity value k. The experiments are one by one performed using individual graph ranking algorithms i.e., page rank, HITS, and PPF. The evaluated Rouge values are found to be slightly better in case of the PR and PPF algorithms as compared to HITS, as shown in Tab. 4.
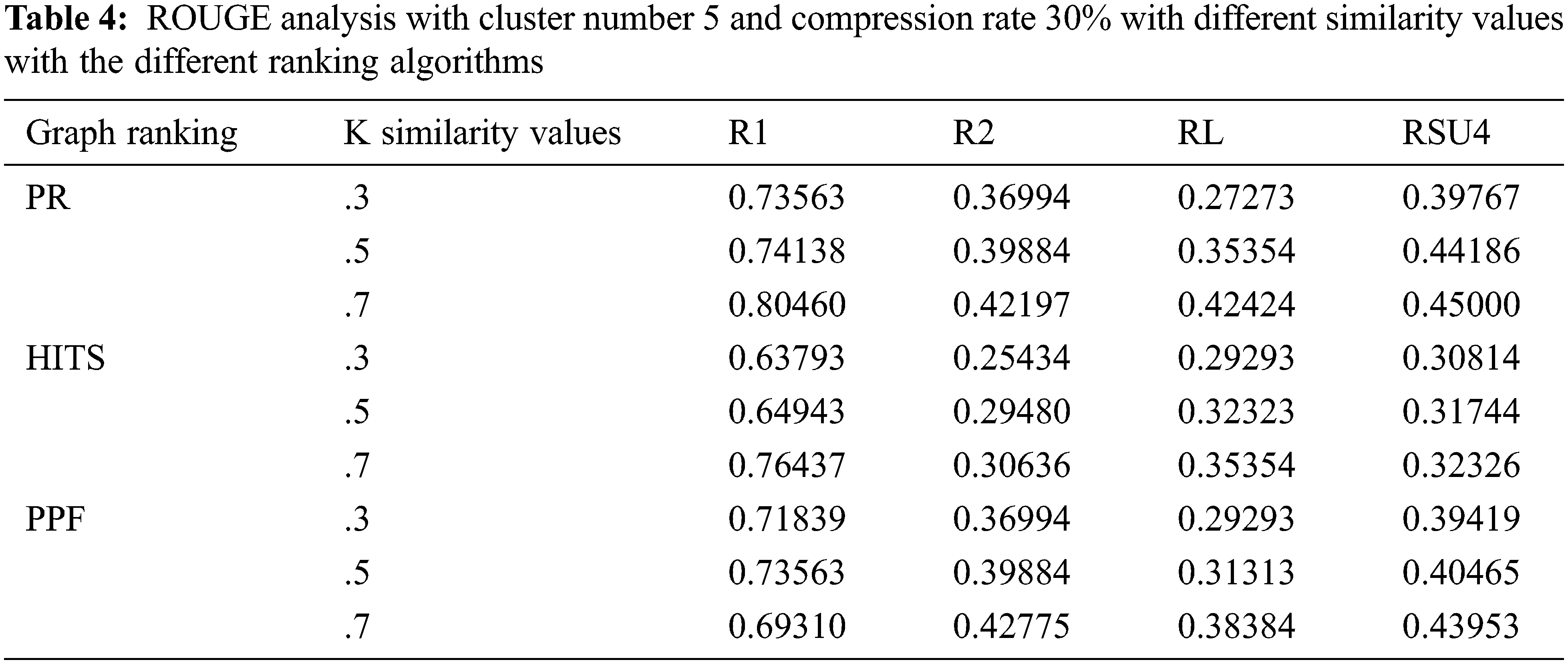
As depicted in Tab. 5, the subsequent setup analysis indicates the ROUGE performance when the cluster number is adjusted to 10 on the specific compression rate of 30%. The similarity k values are varied, and through the experiments, it can be implied that optimum ROUGE scores are observed when k value is kept in the range of 0.5 to 0.7.
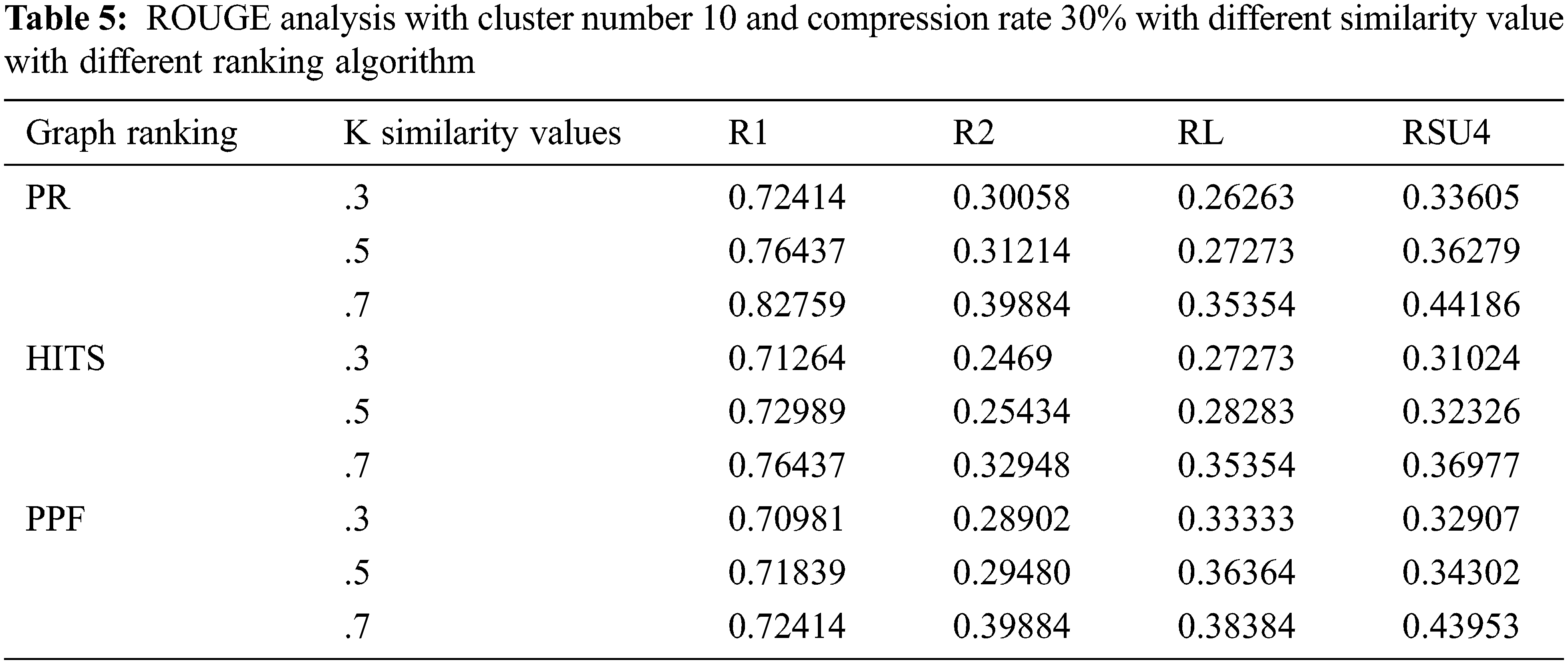
The other setup included the study of varying compression factors when the number of clusters is set to 5 and the k similarity value is held at 0.7. The compression factors of 20%, 30%, and 40% are used for individual graph ranking PR, HITS, and PPF algorithms, as shown in Tab. 6. The model performance improved when the compression rate is increased, so we have fixed it at 30% CR, which is good enough to make an informative summary. The subsequent experiment reflects how the model performed when executed with and without clustering modes with a .7 top k similarity value and cluster numbers set to 5 when the other parameters are kept according to the outcome of earlier experiments. The C+ row depicts with clustering and C− row indicates without clustering, as illustrated in Fig. 2. The results show that the model performed better in the non-clustering mode with improved ROUGE scores.
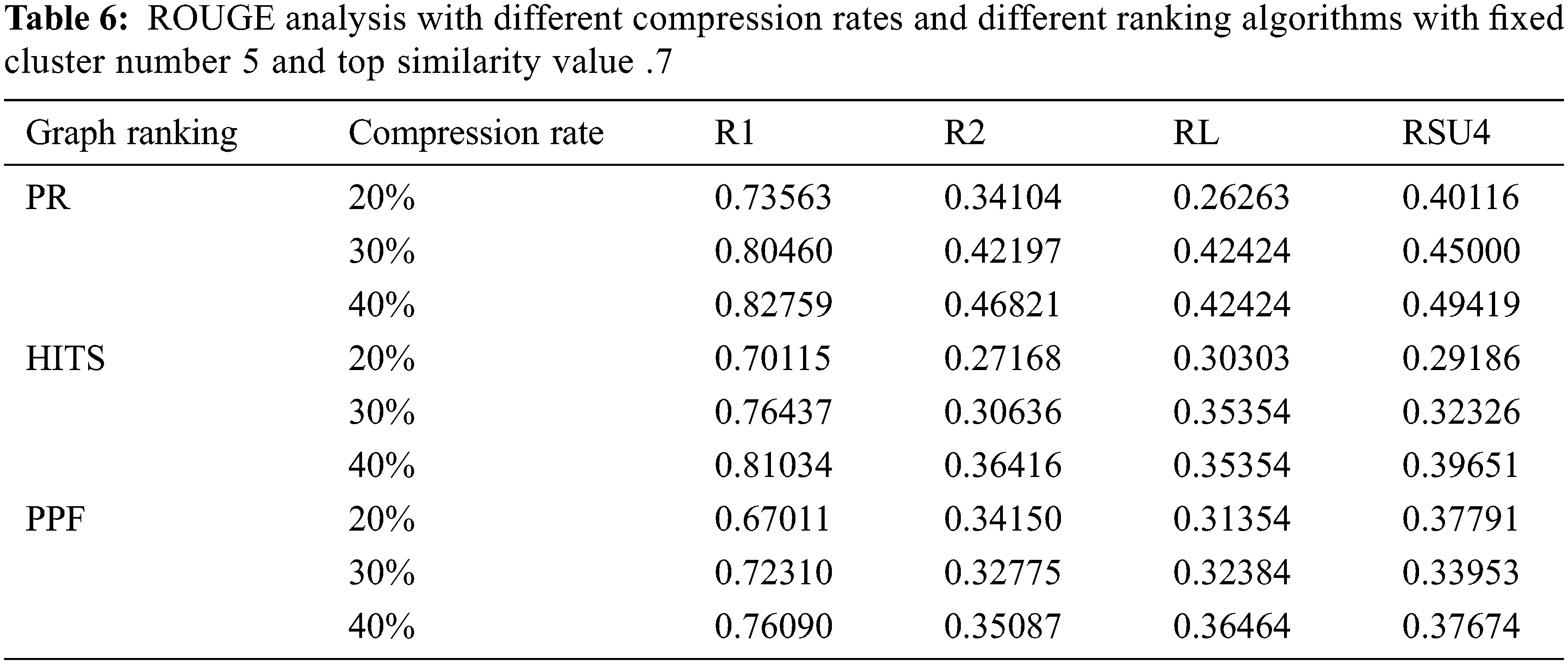
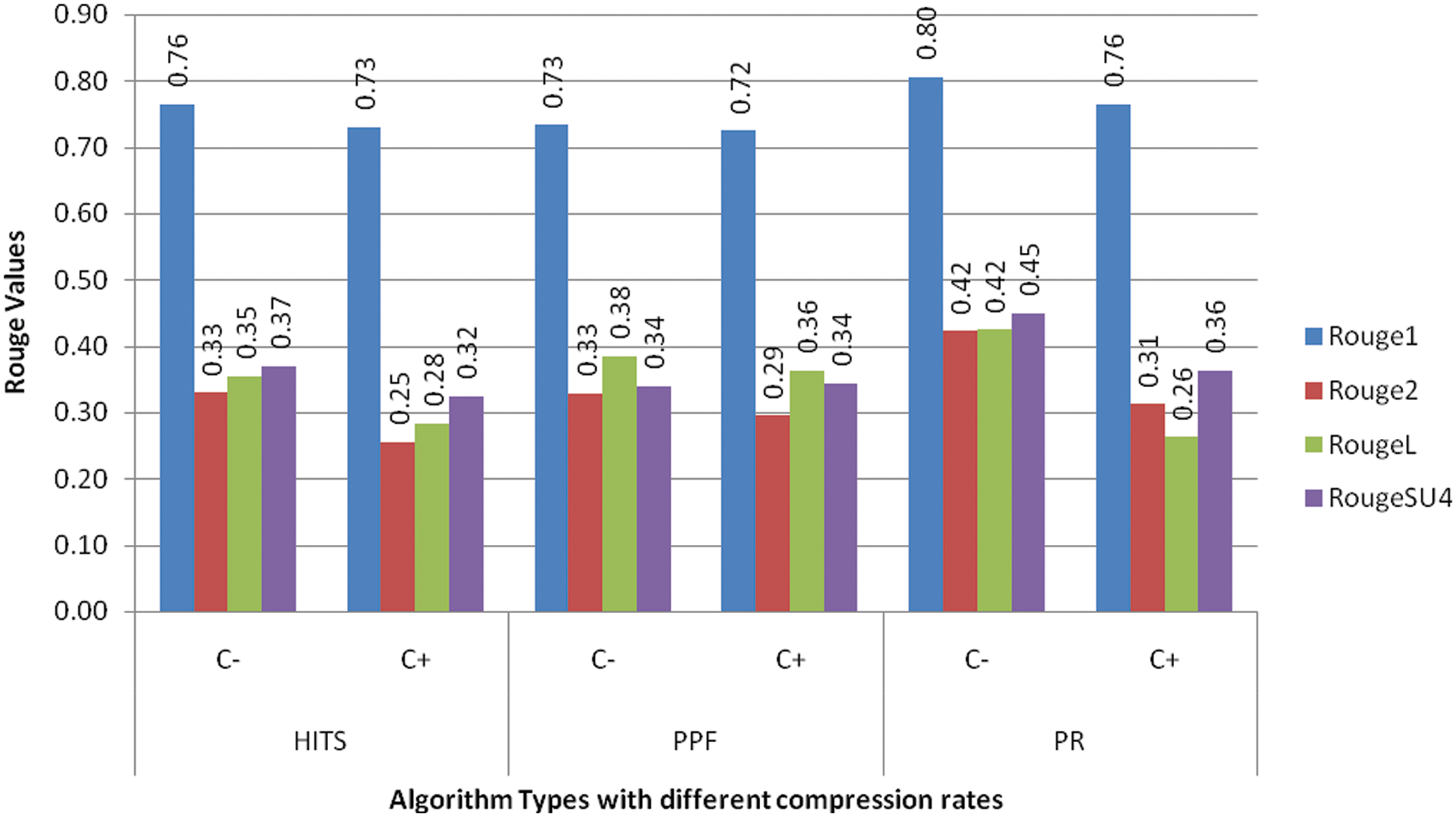
Figure 2: ROUGE analysis of biomedical summaries with dual-mode execution with clustering (+C) and without clustering (−C) with fixed cluster number 5 top K similarity value .7 and compression rate 30%
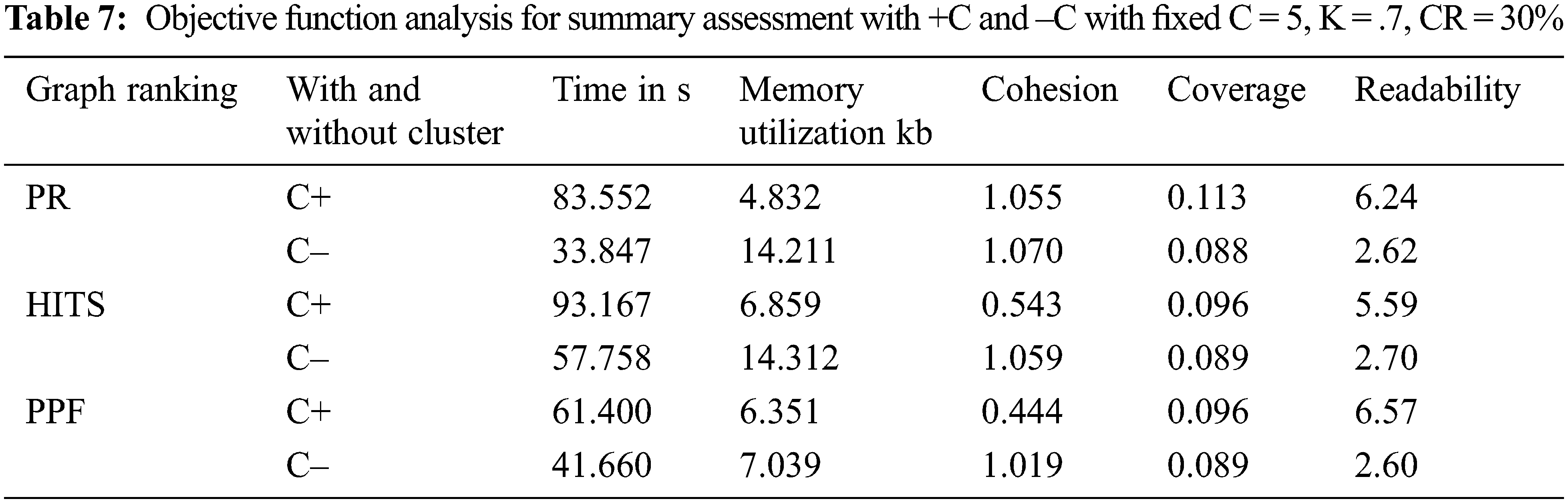
The findings of the previous experiment forced us to check whether clustering should be used over non-clustering mode or not. Therefore, we have reviewed the output of multiple objective functions, which can help in making an informed decision. The values obtained with clustering mode, as shown in Tab. 7, reflect improved coverage and readability of the system evaluated summaries. Although, the timeframe for summary evaluation is a bit longer than the non-clustering mode, the savings in memory consumption balance it out when the output summary is more readable and covers essential information through clustering mode. The model can be used in dual-mode per the specific requirement.
The different experiments are performed in the proposed method utilizing different compression rates, K similarity values and the cluster numbers with the different graph-based ranking algorithms Page Rank, HITS, and PPF. Edges relate to nodes in structured representation through the weight scores derived from explicit similarity values. It uses the Bio-BERT approach for semantic embeddings to find the semantics word mapping with the UMLS base. The proposed system uses clustering and Bio-BERT biomedical summarization to acquire the ROUGE scores. For making a precise and informative summary, selecting the best K similarity value with the combination of word embedding and ranking techniques is essential. In the proposed model, the experiment shows better results when top K similarity values are set in a range of .5 to .7. The increased count of K can produce a large number of edges, and their weights could impact the graph node rank after evaluating scores. These additional edges can deceive the ranking by avoiding the inclusion of significant sentences, which is also apparent via ROUGE scores. This causes reduction in information retention. On contrary, low K values of 0.3 or 0.4 results in fewer edges leading to insufficient information on the linkage of nodes. This affects the summarizer performance with low ROUGE values due to inappropriate short listing of correlated sentences.
The contextualized BioBERT is implemented with dual modes of with and without clustering. In this model, the graph for related sentences is constructed using PageRank, PPF, and HITS algorithms. The generated summary outcome via PageRank provides improved results including retention rate as shown in Fig. 3 compared to PPF and HITS. PageRank algorithm output represents both the relatedness and importance of the evaluated sentences, which is missing in PPF and HITS. PPF employs a much simpler rank technique than PageRank. HITS is more suitable when the inwards/outwards nodes are well-known, which is not the case in the model’s undirected graph representation.
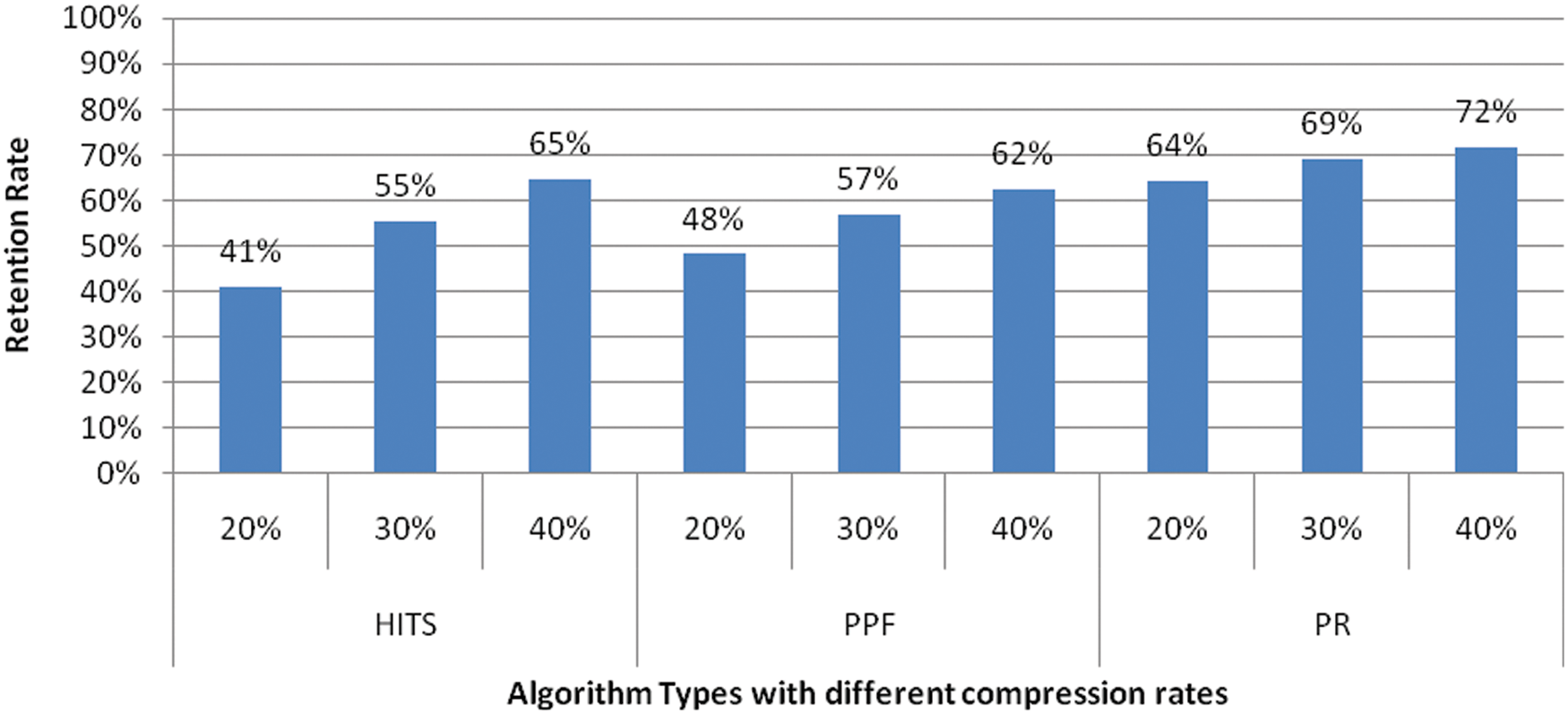
Figure 3: Retention rate for different graph ranking approach with clustering at different compression rate
5.3 Comparison with Baseline and Benchmarks
There are many baselines for text summarization for domain-specific and domain-independent. The proposed method includes the word conceptual embeddings BioBERT for semantic analysis of biomedical documents; then, the SDPMM clustering makes the clusters of sentences with the different themes of biomedical literature. Each cluster makes the graph with the crucial sentences as nodes and connections of sentences as edges with the different graph based ranking algorithms (PR, HITS, and PPF). The given comparison is between the proposed method and baselines.
As shown in Tab. 8, the proposed method with BioBERT and SDPMM-based graph ranking summarization perform better than the baselines on biomedical documents. The proposed method shows 17% improvement for summarization, and the page rank shows a good improvement with clustering and contextual embeddings for the summary. The generated summary has many enhancements in ROUGE, memory utilization, coverage, and cohesion with a fixed compression rate. Non-redundant sentence makes the summary quality good and shows a good enough retention rate for the generated summary.
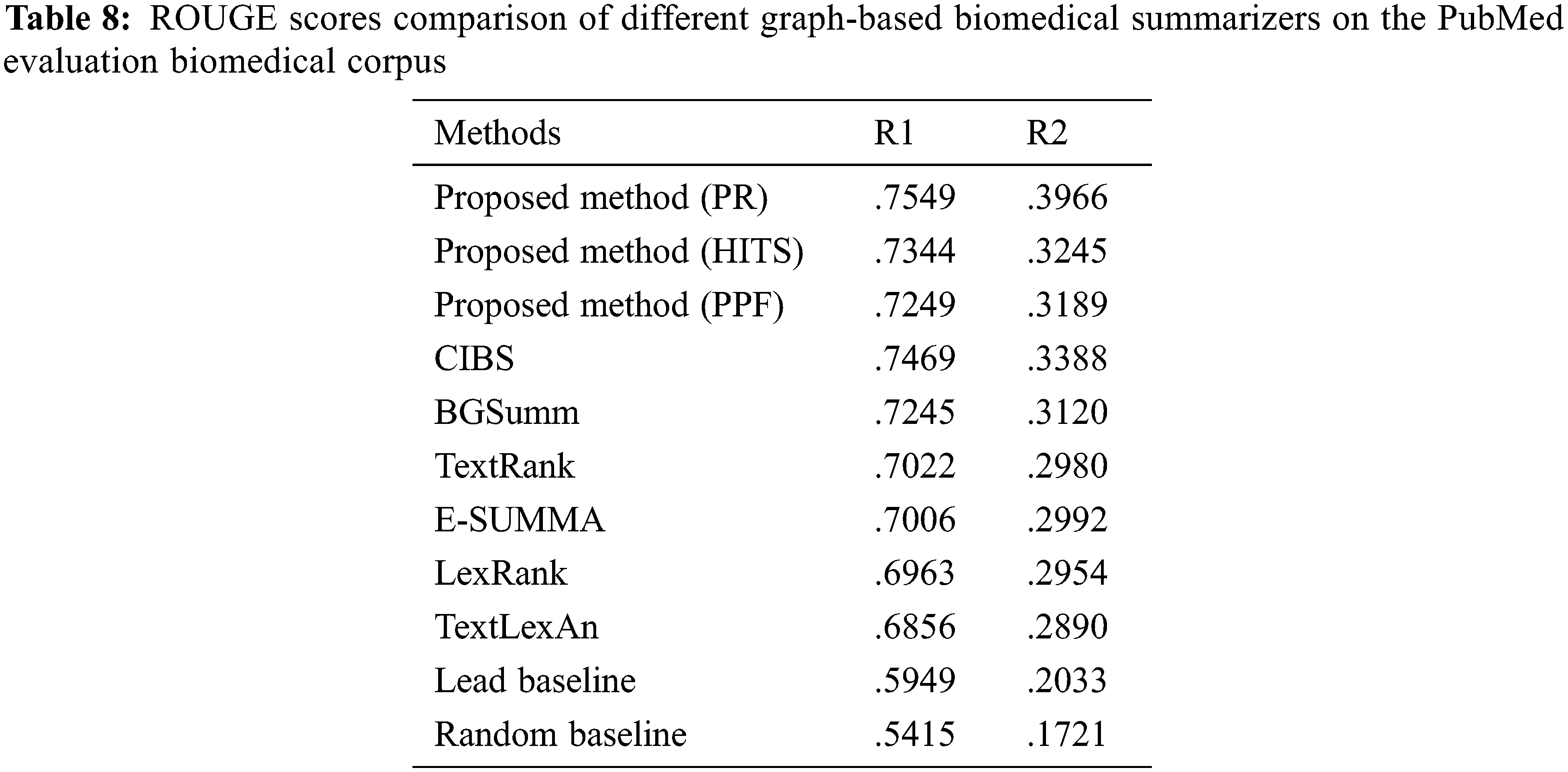
The presented method is a summarization strategy specifically designed for bio-med archives via semantic embeddings. The method covers the demonstration of inbound biomedical text as a graph along with different weights. The vector transformation aids in the evaluation of sentence closeness with each other. The objective considered selection of applicable sentences and to achieve this, a variety of mostly known graph-based ranking approaches are embraced. The outcome exhibited an improved performance of 17% when the summarizer utilized a legitimate blend of probabilistic clustering (SDPMM) and contextualized embeddings (BioBERT) alongside a proficient algorithm for rank calculation. The selective scrutiny of impactful sentences and effective portrayal of bio-med text is important for assessing of summary quality. The inter-dependencies and closeness of sentences are found via semantics, linguistics, and spot-on information given through the model. The improved sentence representation expands the quality of the summary by including important unique information as displayed by the ROUGE scores. The model is tested with multiple compression rates and information retention, along many more objective function values like cohesion, coverage, and readability are assessed. Proposed method also analyzed the dual-mode operation with and without clustering (C+, C−) and showcased improved results. The memory utilization with clustering mode is more efficient for the generated summary than without clustering. It is also observed that optimum results are obtained when top k similarity and cluster number are set at 0.7 and 5, respectively. The most suitable memory utilization is 4.832 Kb when the top-k summarizer is executed with the PageRank method in clustering mode. Similarly, the best information retention of 72% is found with the PageRank technique using the proposed model. The suggested word embedding and clustering framework combined with page rank graph-based ranking algorithm give good memory savings, effective summarization, and best summary scores. The presented work is beneficial for medical scholars and practitioners in the analysis of precision medicine, drug discovery, and establishing biomedical relationships like protein gene mapping and other biomedical research areas.
Acknowledgement: I acknowledge my guide and co-guide and my organization National Institute of Technology Raipur, for helping me carry out this research work.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. R. Mishra, J. Bian, M. Fiszman, C. R. Weir, S. Jonnalagadda et al., “Text summarization in the biomedical domain: A systematic review of recent research,” Journal of Biomedical Informatics, vol. 52, pp. 457–467, 2014. [Google Scholar]
2. O. Rouane, H. Belhadef and M. Bouakkaz, “Combine clustering and frequent itemsets mining to enhance biomedical text summarization,” Expert Systems with Applications, vol. 135, pp. 362–373, 2019. [Google Scholar]
3. M. Gambhir and V. Gupta, “Recent automatic text summarization techniques: A survey,” Artificial Intelligence, vol. 47, pp. 1–66, 2016. [Google Scholar]
4. M. Moradi and N. Ghadiri, “Different approaches for identifying important concepts in probabilistic biomedical text summarization,” Artificial Intelligence in Medicine, vol. 84, pp. 101–116, 2018. [Google Scholar]
5. J. -G. Yao, X. Wan and J. Xiao, “Recent advances in document summarization,” in Knowledge and Information Systems, Springer London, UK, pp. 1–40, 2017. [Google Scholar]
6. H. Van Lierde and T. W. S. Chow, “Learning with fuzzy hypergraphs: A topical approach to query-oriented text summarization,” Information Science, vol. 496, pp. 212–224, 2019. [Google Scholar]
7. M. A. Mosa, A. Hamouda and M. Marei, “Graph coloring and ACO based summarization for social networks,” Expert Systems with Applications, vol. 74, pp. 115–126, 2017. [Google Scholar]
8. G. Glavaš and J. Šnajder, “Event graphs for information retrieval and multi-document summarization,” Expert Systems with Applications, vol. 41, pp. 6904–6916, 2014. [Google Scholar]
9. H. Van Lierde and T. W. S. Chow, “Query-oriented text summarization based on hypergraph transversals,” Information Processing and Management, vol. 56, pp. 1317–1338, 2019. [Google Scholar]
10. J. Lee, W. Yoon, S. Kim, D. Kim, S. Kim et al., “BioBERT: Pre-trained biomedical language representation model for biomedical text mining,” arXiv preprint arXiv: 1901.08746, 2019. [Google Scholar]
11. S. Brin and L. Page, “The anatomy of a large-scale hypertextual web search engine,” Computer Networks and ISDN Systems, vol. 30, pp. 107–117, 1998. [Google Scholar]
12. J. M. Kleinberg, “Authoritative sources in a hyperlinked environment,” Journal of the ACM (JACM), vol. 46, pp. 604–632, 1999. [Google Scholar]
13. P. J. -J. Herings, G. V. D. Laan and D. Talman, “The positional power of nodes in digraphs,” Social Choice and Welfare, vol. 24, pp. 439–454, 2005. [Google Scholar]
14. C. -Y. Lin, “Rouge: A package for automatic evaluation of summaries in text summarization branches out,” in Proc. of the ACL-04 Workshop, Barcelona, Spain, 2004. [Google Scholar]
15. M. Moradi, “CIBS: A biomedical text summarizer using topic-based sentence clustering,” Journal of Biomedical Informatics, vol. 88, pp. 53–61, 2018. [Google Scholar]
16. M. Moradi and N. Ghadiri, “Quantifying the informativeness for biomedical literature summarization: An itemset mining method,” Compute Methods Programs Biomed, vol. 146, pp. 77–89, 2017. [Google Scholar]
17. M. Yousefi-Azar and L. Hamey, “Text summarization using unsupervised deep learning,” Expert Systems with Applications, vol. 68, pp. 93–105, 2017. [Google Scholar]
18. A. Joshi, E. Fidalgo, E. Alegre and L. Fernández-Robles, “SummCoder: An unsupervised framework for extractive text summarization based on deep auto-encoders,” Expert Systems with Applications, vol. 129, pp. 200–215, 2019. [Google Scholar]
19. J. M. Sanchez-Gomez, M. A. Vega-Rodríguez and C. J. Pérez, “Extractive multi-document text summarization using a multi-objective artificial bee colony optimization approach,” Knowledge-Based Systems, vol. 159, pp. 1–8, 2018. [Google Scholar]
20. S. Afantenos, V. Karkaletsis and P. Stamatopoulos, “Summarization from medicaldocuments: A survey,” Artificial Intelligence in Medicine, vol. 33, pp. 157–177, 2005. [Google Scholar]
21. L. H. Reeve, H. Han and A. D. Brooks, “The use of domain-specific concepts in biomedical text summarization,” Information Processing and Management, vol. 43, pp. 1765–1776, 2007. [Google Scholar]
22. M. A. Mosa, A. S. Anwar and A. Hamouda, “A survey of multiple types of text summarization with their satellite contents based on swarm intelligence optimization algorithms,” Knowledge-Based Systems, vol. 163, pp. 518–532, 2019. [Google Scholar]
23. P. Mehta and P. Majumder, “Effective aggregation of various summarization techniques,” Information Processing and Management, vol. 54, pp. 145–158, 2018. [Google Scholar]
24. H. Moen, L. -M. Peltonen, J. Heimonen, A. Airola, T. Pahikkala et al., “Comparison of automatic summarisation methods for clinical free text notes,” Artificial Intelligence in Medicine, vol. 67, pp. 25–37, 2016. [Google Scholar]
25. G. Del Fiol, J. Mostafa, D. Pu, R. Medlin, S. Slager et al., “Formative evaluation of a patient-specific clinical knowledge summarization tool,” International Journal of Medical Informatics, vol. 86, pp. 126–134, 2016. [Google Scholar]
26. R. Pivovarov and N. Elhadad, “Automated methods for the summarization of electronic health records,” Journal of American Medical Informatics Association, vol. 22, pp. 938–947, 2015. [Google Scholar]
27. R. Mihalcea and P. Tarau, “TextRank: Bringing order into texts,” Proceedings of the 2004 conference on empirical methods in natural language processing, pp. 404–411, 2004. [Google Scholar]
28. Y. Zhu, E. Yan and F. Wang, “Semantic relatedness and similarity of biomedical terms: Examining the effects of recency, size, and section of biomedical publications on the performance of word2vec,” BMC Medical Informatics and Decision Making, vol. 17, pp. 95, 2017. [Google Scholar]
29. K. Blagec, H. Xu, A. Agibetov and M. Samwald, “Neural sentence embedding models for semantic similarity estimation in the biomedical domain,” BMC Bioinformatics, vol. 20, pp. 178, 2019. [Google Scholar]
30. J. C. W. Lin, Y. Shao, Y. Zhou, M. Pirouz and H. -C. Chene, “A bi-LSTM mention hypergraph model with encoding schema for mention extraction,” Engineering Applications of Artificial Intelligence, vol. 85, no. 1, pp. 175–181, 2019. [Google Scholar]
31. J. C. W. Lin, Y. Shao, J. Zhang and U. Yun, “Enhanced sequence labeling based on latent variable conditional random fields,” Neurocomputing, vol. 403, no. 1, pp. 431–440, 2020. [Google Scholar]
32. J. C. W. Lin, Y. Shao, Y. Djenouri and U. Yun, “ASRNN: A recurrent neural network with an attention model for sequence labeling,” Knowledge-Based Systems, vol. 212, no. 1, pp. 106548, 2021. [Google Scholar]
33. Y. Shao, J. C. W. Lin, G. Srivastava, A. Jolfaei, D. Guo et al., “Self-attention-based conditional random fields latent variables model for sequence labeling,” Pattern Recognition Letters, vol. 145, no. 1, pp. 157–164, 2021. [Google Scholar]
34. J. C. W. Lin, Y. Shao, P. F. Viger and F. Hamido, “BILU-NEMH: A BILU neural-encoded mention hypergraph for mention extraction,” Information Sciences, vol. 496, no. 1, pp. 53–64, 2019. [Google Scholar]
35. S. J. Nelson, T. Powell and B. Humphreys, “The unified medical language system (UMLS) project,” Encyclopedia of Library and Information Science, New York: Marcel Dekker, Inc. pp. 369–378, 2002. [Google Scholar]
36. G. Erkan and D. R. Radev, “LexRank: Graph-based lexical centrality as salience in text summarization,” Journal of Artificial Intelligence Research, vol. 22, pp. 457–479, 2004. [Google Scholar]
37. H. Saggion, “SUMMA: A robust and adaptable summarization tool,” Traitement Automatique Design Langues, vol. 49, pp. 103–125, 2008. [Google Scholar]
38. R. Kiros, Y. Zhu, R. R. Salakhutdinov, R. Zemel, R. Urtasun et al., “Skipthought vectors,” in Advance in Neural Information Processing System, pp. 3294–3302, 2015. [Google Scholar]
39. D. B. Abeywickrama, N. Bicocchi, M. Mamei and F. Zambonelli, “The SOTA approach to engineering collective adaptive systems,” International Journal on Software Tools for Technology Transfer, vol. 22, no. 1, pp. 399–415, 2020. [Google Scholar]
40. S. Babichev, V. Lytvynenko, J. Skvor and J. Fiser, “Model of the objective clustering inductive technology of gene expression profiles based on SOTA and DBSCAN clustering algorithms,” Advances in Intelligent Systems and Computing, vol. 689, no. 1, pp. 21–39, 2017. [Google Scholar]
41. L. H. Reeve, H. Han, S. V. Nagori, J. C. Yang, T. A. Schwimmer et al., “Concept frequency distribution in biomedical text summarization,” in Proc. of the 15th ACM Int. Conf. on Information and Knowledge Management, Arlington, VA, USA, pp. 604–611, 2006. [Google Scholar]
42. L. Plaza, A. Díaz and P. Gervás, “A semantic graph-based approach to biomedical summarization,” Artificial Intelligence in Medicine, vol. 53, pp. 1–14, 2011. [Google Scholar]
43. J. Devlin, M. W. Chang, K. Lee and K. Toutanova, “Bert: Pre-training of deep bidirectional transformers for language understanding,” arXiv preprint arXiv: 1810.04805, 2018. [Google Scholar]
44. V. Gupta and G. S. Lehal, “A survey of text summarization extractive techniques,” Journal of Emerging Technologies in Web Intelligence, vol. 2, pp. 258–268, 2010. [Google Scholar]
45. D. Ding and G. Karabatsos, “Dirichlet process mixture models with shrinkage prior,” Wiley, pp. 71, 2021. https://doi.org/10.1002/sta4.3. [Google Scholar]
46. M. Mohamed and M. Oussalah, “SRL-ESA-TextSum: A text summarization approach based on semantic role labeling and explicit semantic analysis,” Information Processing and Management, vol. 56, pp. 1356–1372, 2019. [Google Scholar]
47. E. Baralis, L. Cagliero, N. Mahoto and A. Fiori, “GRAPHSUM: Discovering correlations among multiple terms for graph-based summarization,” Information Science, vol. 249, pp. 96–109, 2013. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools