 Open Access
Open Access
ARTICLE
Covid-19 Diagnosis Using a Deep Learning Ensemble Model with Chest X-Ray Images
Computer Engineering, Cankiri Karatekin University, Cankiri, 18100, Turkey
* Corresponding Author: Fuat Türk. Email:
Computer Systems Science and Engineering 2023, 45(2), 1357-1373. https://doi.org/10.32604/csse.2023.030772
Received 01 April 2022; Accepted 16 May 2022; Issue published 03 November 2022
Abstract
Covid-19 is a deadly virus that is rapidly spread around the world towards the end of the 2020. The consequences of this virus are quite frightening, especially when accompanied by an underlying disease. The novelty of the virus, the constant emergence of different variants and its rapid spread have a negative impact on the control and treatment process. Although the new test kits provide almost certain results, chest X-rays are extremely important to detect the progression and degree of the disease. In addition to the Covid-19 virus, pneumonia and harmless opacity of the lungs also complicate the diagnosis. Considering the negative results caused by the virus and the treatment costs, the importance of fast and accurate diagnosis is clearly seen. In this context, deep learning methods appear as an extremely popular approach. In this study, a hybrid model design with superior properties of convolutional neural networks is presented to correctly classify the Covid-19 disease. In addition, in order to contribute to the literature, a suitable dataset with balanced case numbers that can be used in all artificial intelligence classification studies is presented. With this ensemble model design, quite remarkable results are obtained for the diagnosis of three and four-class Covid-19. The proposed model can classify normal, pneumonia, and Covid-19 with 92.6% accuracy and 82.6% for normal, pneumonia, Covid-19, and lung opacity.Keywords
Towards the end of 2019, a major pandemic occurred in China due to a new virus spreading there [1,2]. This virus, named Covid19, can show various symptoms that can lead to chronic respiratory disease and fatal acute respiratory distress syndrome [3,4]. Covid-19 virus, which is rapidly transmitted from person to person with its changing variants, has been detected in more than 350 million people as of January 2022. It has also directly caused the death of more than 5 million people [5]. Given the possibility of rapid transmission of Covid-19, a state of emergency is declared by the World Health Organization (WHO) in 2020. Polymerase chain reaction (PCR) testing can be considered the standard diagnostic test for Covid-19 [6]. However, limited supplies and requirements for a fully equipped laboratory hinder accurate diagnosis of Covid-19 patients and delay cessation of pandemic infection. Clinically, the best method is computed tomography (CT) of the chest, which allows rapid diagnosis. In addition, the clinical diagnosis of Covid-19 patients can be made more confidently based on the person’s general health, symptoms, travel history, and laboratory tests. In other words, the diagnosis of Covid-19 based on chest radiographs is very important factor and is used as a reference in many studies [7,8]. With evolving technological capabilities, artificial intelligence methods that help physicians diagnose many diseases are a topic that all researchers are eagerly interested in Covid-19. However, the lack of sufficient and easily accessible datasets makes it difficult to make diagnoses using artificial intelligence methods. For this reason, it is significant to build a worldwide network of public datasets. In addition, the fact that artificial intelligence, especially deep learning methods, produces more successful results with large data sets should not be ignored.
In recent years, Deep Learning methods have become popular in medical image processing and diagnostic problems [9]. In the first study reviewed in this context, Gour et al. [10] performed image classification using an uncertainty-sensitive convolutional neural network with X-ray images for the diagnosis of Covid-19. In the study, an uncertainty-sensitive convolutional neural network model called UA-ConvNet was proposed for automatic detection of Covid-19 disease from CXR images in the predictions of the model. This approach uses the EfficientNet-B3 model and the Monte Carlo (MC) dropout method. The proposed ConvNet model achieves a G-mean of 98.02% and a sensitivity of 98.15% for the multi-class classification task on the Covid-19 Chest X-ray (CXR) dataset.
Malhotraa et al. [11] proposed a multitasking method for the diagnosis of Covid-19 on radiographs. This study has two main contributions. First, the Covid-19 multitasking model (COMiT-Net) was presented. The proposed model predicts whether the CXR has Covid-19 features, as well as performs segmentation of the relevant regions to make the model explainable. Second, they manually added lung regions and semantic segmentation of Covid-19 symptoms in CXRs from the Covid-19 dataset with the help of medical experts. Experiments with more than 2500 preliminary images show that the proposed model provides a sensitivity of 96.80% with a specificity of 90%.
Liua et al. [12] emphasized that segmentation from CT scans is important for accurate diagnosis Covid-19. In their study, they stated that although the Convolutional Neural Network (CNN) has great capability to perform the segmentation task, the existing Deep Learning segmentation methods were time consuming to train and require additional explanations. In this paper, a new low-supervision segmentation method for Covid-19 infections in CT slices was proposed using uncertainty-sensitive self-assembly and transformation-consistent techniques. The proposed method was evaluated on a total of three data sets. The results indicate that this method was more effective than other supervised methods and performs close to fully supervised methods.
Deb et al. [13] proposed a Deep Learning-based ensemble model for the diagnosis of Covid-19. The proposed model was applied to two and three separate classes using the Covid-19 dataset. The community model was related to four convolutional networks: VGGNet, GoogleNet, DenseNet and NASNet. They achieved 88.98% accuracy for three-class classification and 98.58% for two-class classification in the publicly available dataset.
Ghosh et al. [14] developed a deep learning model called ENResNet and this model was used to diagnose Covid-19. In this study, they emphasized that it is difficult to distinguish normal pneumonia from Covid-19 because it has similar symptoms. They succeeded in testing the model they developed to overcome this difficulty on three classes: covid-normal and covid-normal-pneumonia. Compared to other methods, the proposed ENResNet achieved a classification accuracy of 99.7% and 98.4% for binary classification and multi-class detection, respectively.
Ayan et al. [15] proposed a Deep Learning-based ensemble model for the diagnosis of pediatric pneumonia. In this study, they stated that pneumonia was a fatal disease that occurs in the lungs and is caused by a viral or bacterial infection. They also emphasized that diagnosis from Chest X-rays could be difficult and prone to error because it resembles other infections in the lungs. For this reason, they were able to test their developed ensemble model on a data set with two and three separate class labels. Their proposed ensemble method achieved successful results with 95.21% accuracy and 97.76% sensitivity in the test data. In addition, the proposed ensemble method was able to achieve a classification accuracy of 90.71% for normal, viral, and bacterial pneumonia in chest radiographs.
Lafraxo et al. [16] developed a Deep Learning-based architecture they call CoviNet. In their studies, they talked about the dangers of Covid-19 disease and the difficulty of diagnosing it. They explained that the architecture they developed would help overcome these difficulties. The study, which they tested on a public dataset, achieved 98.62% accuracy for binary classification and 95.77% for multi-class classification.
Linda et al. [17] recommended COVID-Net for chest classification. Radiological images were divided into three and four classes. They proposed a proprietary model and created one of the first designs for Covid-19 CXR recognition. As a result of their studies, they achieved for three class 92.4 and four class 83.5% accuracy.
Shastri et al. [18] developed a deep learning model called CheXImageNet. In this study, they mentioned that although there were some methods made on respiratory samples for the detection of Covid-19 patients, radiological imaging or X-ray images are needed for treatment. They stated that the use of Deep Neural Networks-based methodologies was beneficial for the accurate identification of this disease and can also help to overcome the shortage of competent physicians in a wide range of fields. To compare the effectiveness of the proposed study, experiments were conducted for both dual-class and multi-class. 100% accuracy was reported for both dual classification (Covid-19 and Normal X-Ray cases) and classification for three classes (Covid-19, Normal X-Ray and Pneumonia cases), respectively.
Lascu [19] proposed a Deep Learning-based architecture to classify Covid-19 disease and differentiate it from other lung diseases. In addition, in this study, he tried to show the difference in the use of transfer learning. It was emphasized that there is a need for segmentation of diseased parts of the lung for the classification study. As a result of the study, it was able to make a successful classification with an accuracy value of 94.9%.
Jamil et al. [20] combined a structure they call Bag-of-Features (BoF) with a deep learning model for Covid-19 screening. It has been emphasized that the newly developed model is more successful than other models in detecting the Covid-19 disease. As a result of the study, the accuracy rate was obtained as 99.5%.
Wang et al. stated that the Covid-19 classification task is difficult. In order to overcome this difficulty, they developed a model they call MAI-NeT. This model has a smaller number of layers and parameters than other models. For this reason, they have said that covid-19 disease can be detected more efficiently. According to the experimental results, they were able to detect covid-19 disease with an accuracy of 96.42% [21]. Wang et al., with another model called CFW-Net, were able to detect covid-19 with an accuracy value of 94.35% [22].
As can be seen from the literature review, most of the studies on the diagnosis and classification of Covid-19 are based on a single data set. Moreover, ensemble models often contain the same architectural structures. This paper presents a new option for the dataset and ensemble model outside the known patterns. The innovations that this study contributed to science can be summarized as follows.
• In order to contribute to the literature, a new dataset with balanced number of cases with three and four classes is proposed. For this dataset, Deb et al., Cohen et al. [23] and datasets used in the Covid-19 grand challenge competition accepted in previous scientific studies and competitions are examined. From these datasets, a large dataset is created, containing four separate class labels: Normal, Covid-19, Pneumonia, and Lung Opacity. Meanwhile, pneumonia cases (including viral pneumonia, bacterial and viral strains) are selected in proportion to their numbers. The data that may be equal or wrong are examined and removed one by one.
• In the datasets used in previous Covid-19 deep learning studies, the results could not be evaluated decisively enough, as the amount of data between classes was inconsistent. With this data set, it can be possible to make stable inferences for multiple classification.
• Four of the most appropriate deep learning methods for the dataset are selected for the ensemble model and supported by the architecture of ResNet + + (Türk et al. [24]), which has never been done before in these studies.
• With the proposed ensemble model algorithm, a successful classification process has been performed in distinguishing lung opacity (benign cystic structures) data, which was often ignored in previous studies.
The remainder of this paper is organized as follows: Section 2 describes the data set and the structure of the material and methods. The experiment results are described in Section 3. The paper is concluded in Section 4.
2.1 Description of the Data Sets
In the study, three different data sets are examined in detail. The data set designations are implemented as follows.
• Normal: individuals without signs of disease on chest images.
• Covid-19: individuals who showed the spread of Covid-19 virus on chest images and were diagnosed by physicians
• Pneumonia: individuals who have shown pneumonia (with its subtypes) on chest images and have been diagnosed by physicians
• Lung opacity: individuals with types of cysts that become lodged in the lungs after finding an environment in which to become lodged and cannot be detected by specialists. Fig. 1 images with examples of four different classes are also included.
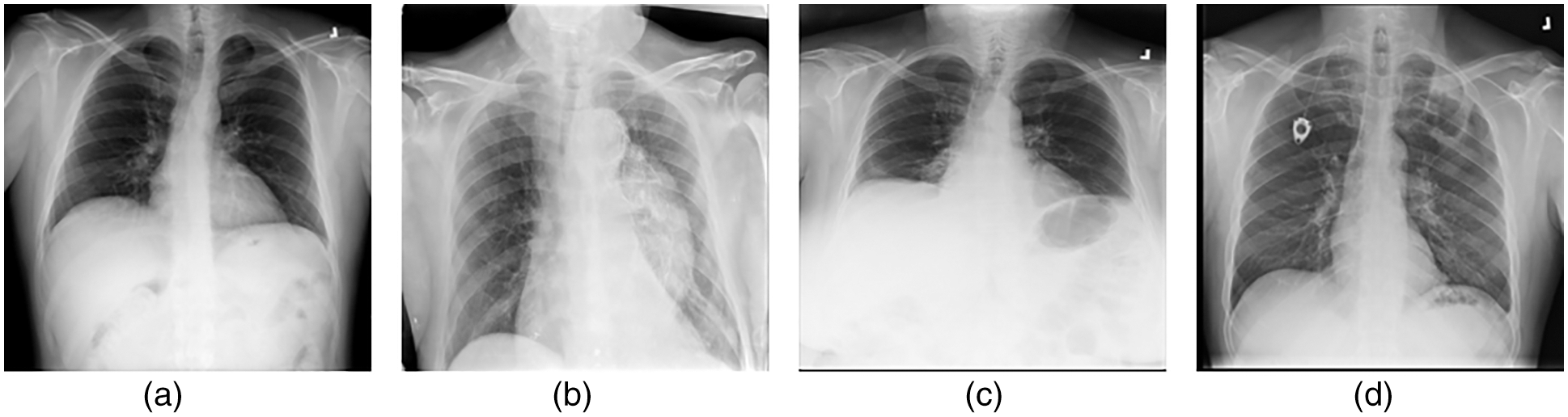
Figure 1: (a) Normal (b) Covid-19 (c) Pneumonia (d) Lung opacity
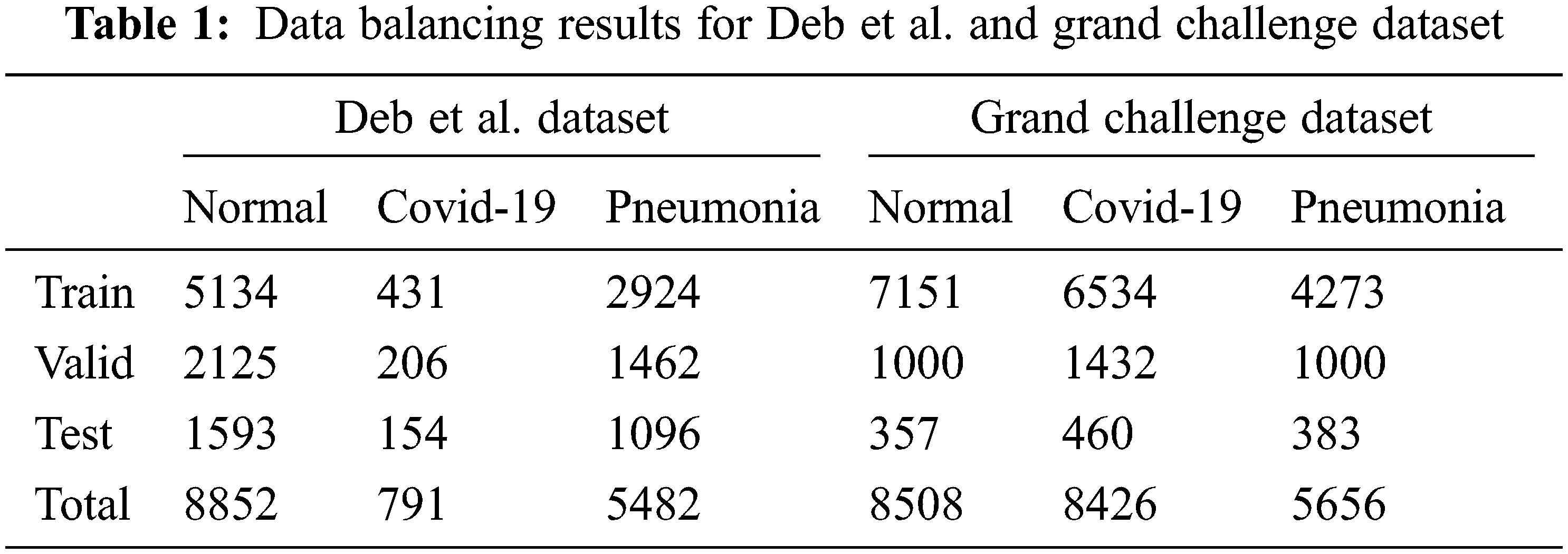
Existing class data are divided into training, testing, and validation sets to develop and tune detection/classification algorithms. First, Deb et al. data set contains three separate classes. Second, the data set used in the Covid-19 Grand Challenge battle is examined, some of which are publicly available. This data set includes 3 different classes The final version resulting from the updates is shown in Tab. 1.
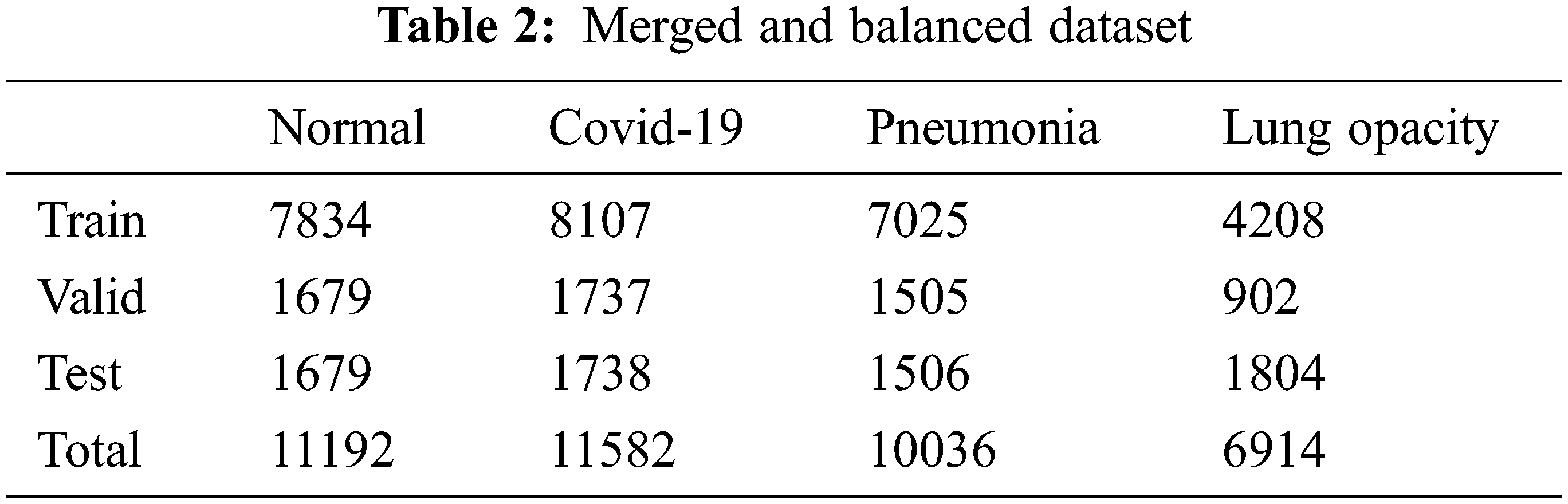
Finally, in addition to the two datasets, a new dataset is created with samples of lung opacity from the Cohen et al. dataset. There are four different classes in this dataset. Addressing imbalances in sub selection between training, test, and validation samples for normal and Covid-19 data. For the pneumonia data class, viral pneumonia, bacterial and viral subtypes are equally distributed. This is for avoid inconsistencies in classification. Images belonging to the pulmonary opacity class are evenly distributed. The final state of the dataset as a result of the updates is shown in Tab. 2.
In the training phase, image pre-processing is applied in order to better examine the existing images and interpret them faster. So, the OpenCV library is used in Python programming language. First, the value is set to 255 if the pixel density is greater than the specified threshold and 0 (black), otherwise. Then, a noise removal technique is used to keep the edges sharp. Finally, to extend the intensity range, contrast enhancement is performed on the image. Fig. 2 shows the images before and after image preprocessing.
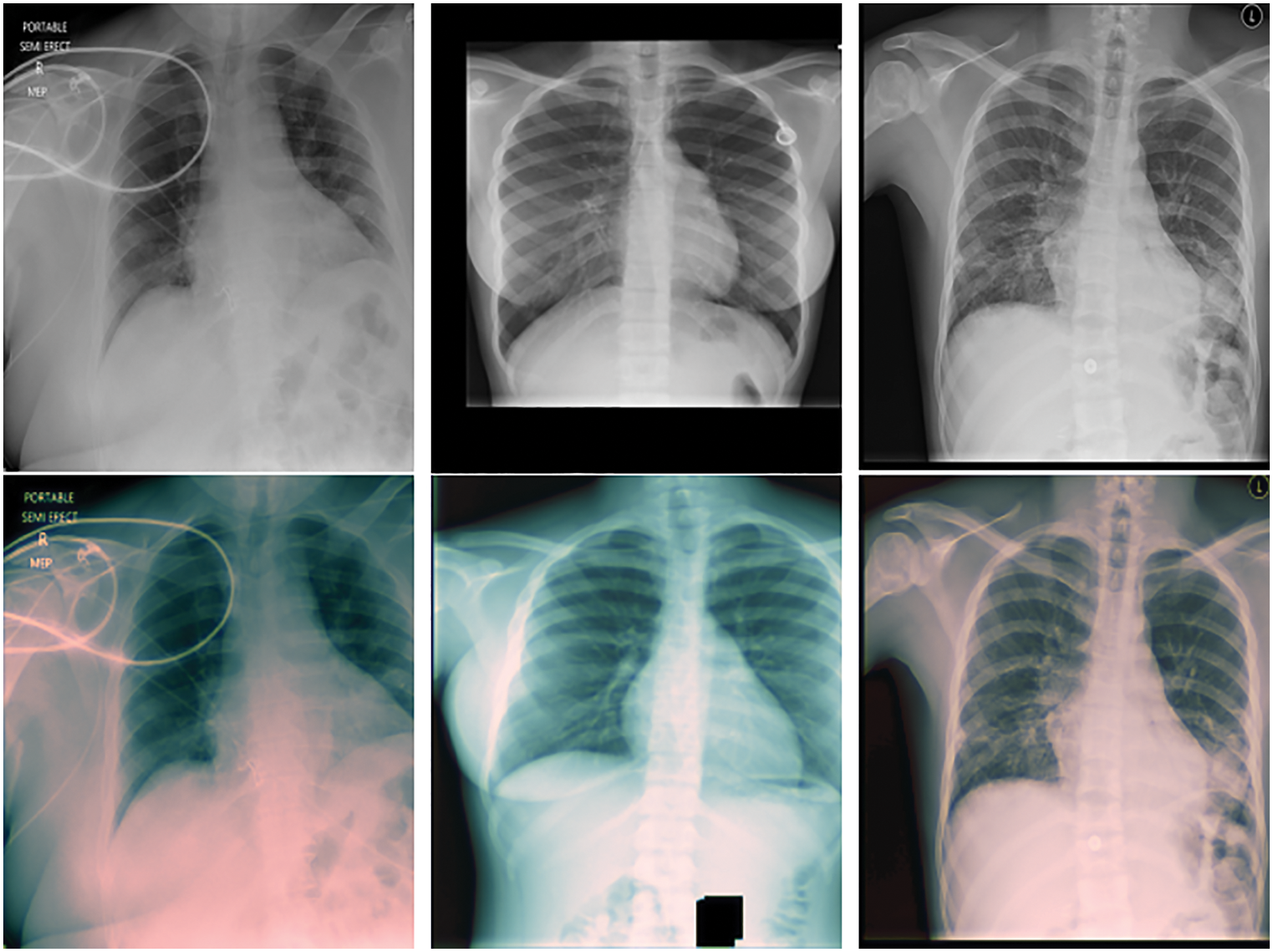
Figure 2: Input image and preprocessing phase
While selecting the appropriate architectures for the model, the ensemble model is created by working together four of them that gave the best results in the TensorFlow library. At this stage, the newly created dataset is taken as a reference.
NasNet-Mobile: NasNet is a scalable CNN architecture (built by neural architecture research) consisting of basic building blocks (cells) optimized using reinforcement learning. A cell consists of only a few operations (several separable convolutions and pooling) and is repeated many times according to the required capacity of the network. The mobile version (Nasnet-Mobile) consists of 12 cells with 5.3 million parameters and 564 million multi-accumulations (MACs) [25].
DenseNet: The densely connected Convolutional Network, developed by Huang et al. [26] in 2018, is formed by adding each layer of a network to every other layer in a feed-forward manner. This important work has allowed the development of a deeper and more accurate CNN.
Inception V3: CNN [27–30] is a neural network designed specifically for image recognition problems. It mimics the multi-layered process of human image recognition: pupils swallowing pixels; initial processing of specific cells in the cerebral cortex finding shape edges and directions; abstract identification of shapes (such as circles and squares); and more abstract decisions. CNNs typically contain five layers. These are referred to as Input, Convolutional, Pooled, Fully Connected, and Output.
VGG19: Proposed by Oxford College in 2014. The simplicity and applicability advantages of this model, which is a convolutional neural network, are terrific. It can also show high performance in image classification and target recognition [31]. As its name suggests, VGG19 includes 19 layers. Instead of using large filters, the VGG19 model uses multiple 3 × 3 filters per layer, with a size of 11 × 11 like Alexnet [32,33].
ResNet + +: This architecture can be thought of as two nested ResNet blocks. Unlike a regular ResNet model, this structure combines the output layer with the two previous layers. This allows small residual blocks of the first output to be captured. Adding this layer to all blocks slows down the network significantly, so adding it to the correct layer is extremely important. Usually, better results are obtained in the last layer [24]. Eqs. (1)–(3) express the application of the architectural structure.
Transfer Learning: Training a network from scratch using the entire dataset for training causes labour and time loss. As is known, with transfer learning, models that show higher success and learn faster are obtained with less training data by using previous knowledge. Based on this inference, this method is used in order to increase the learning success and speed up the education process. Therefore, a method of re-using a previously created network with transfer learning was preferred in training.
First, separate CNN constructs are used for feature extraction. The extracted features are transferred to a fully linked layer for final classification. Next, a combination of the four best performing CNN structures (InceptionV3, DenseNet, VGG19, NasNet Mobile) is used for feature extraction. Finally, the ResNet + + architectural structure that used in the U-Net and V-Net models was added to the output block.
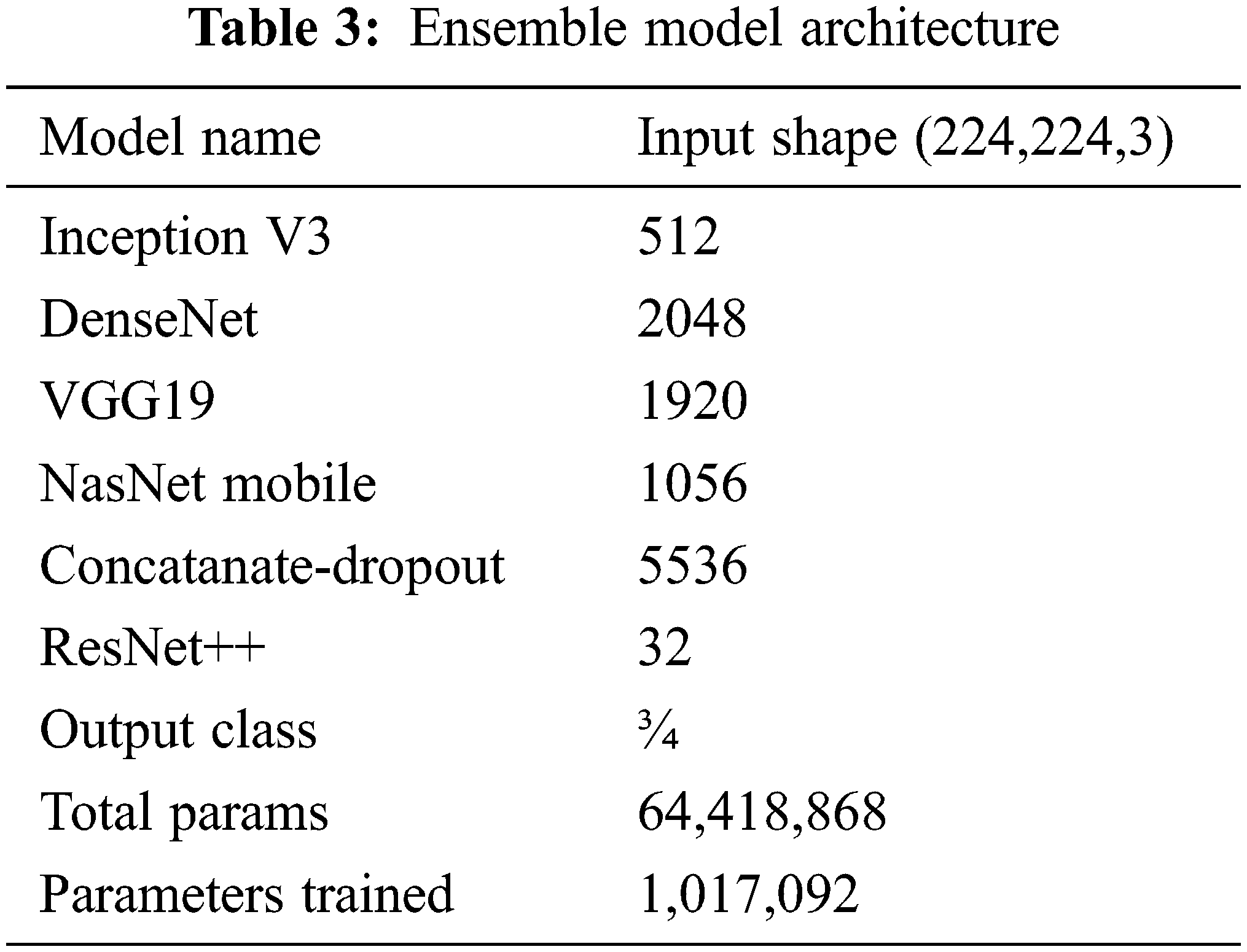
It was clearly found that using the designed ensemble model improves the classification performance. Lung opacity requires 1 to 3 years of follow-up. In addition, other classes require a certain period of follow-up under the control of a physician, depending on the degree of disease [34]. At the end of these periods, new CT images can be taken and tested on the same model. In this way, the long-term reliability of the developed model will be checked. The ensemble model developed along with the above models is shown in Fig. 3. The details of the parameters and architecture are given in Tab. 3 so that the model can be fitted.
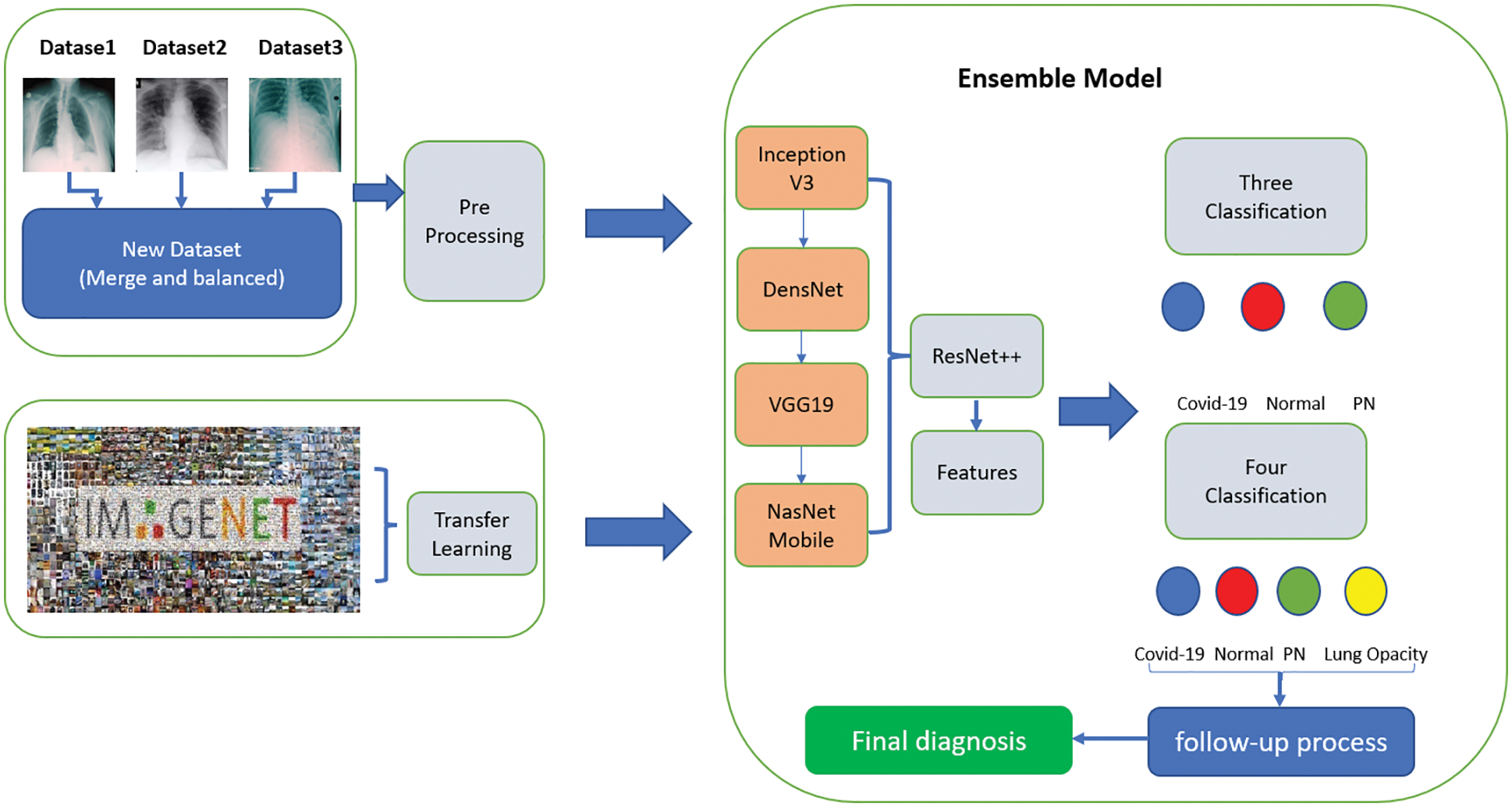
Figure 3: Proposed ensemble model
All resources related to training and dataset are available in the account on TUBITAK-TRUBA servers. The results related to the process are given below as stages of three different classes. Also, the dataset and the source codes are available on the account of GitHub (for codes: https://github.com/turkfuat/covid19-multiclass). The training phase is conducted in three different stages. These are carried out in three classes, four classes, and suitable for literature comparison, respectively.
3.1 Training Stage Merged and Balanced Data Sets for Three/Four Classes
In the first stage, the ensemble model is trained and validated for three classes using our self-created merged and balanced dataset. This phase is performed for pneumonia, normal and Covid-19. In the second stage, the ensemble model is trained and validated for four classes, using datasets created by ourselves. This phase is performed for pneumonia, normal, lung opacity and Covid-19. The table shows the accuracy values of the four models and the ensemble model in the training result. The Tab. 4 shows the accuracy values of the three/four models and the ensemble model in the training result. In Fig. 4 and in Fig. 5, accuracy and loss graphs are shown as a result of the training.
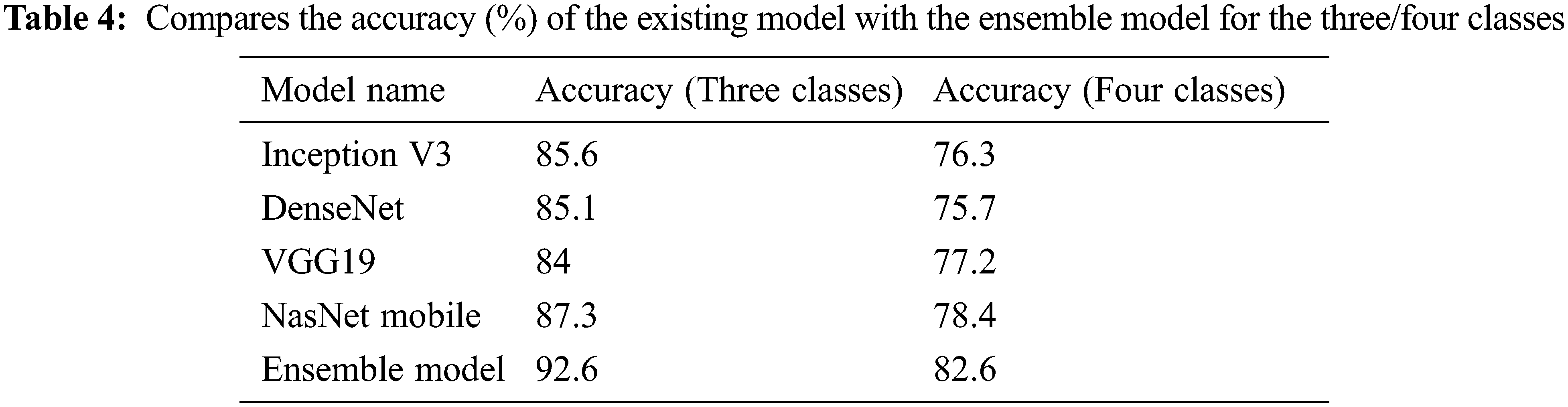
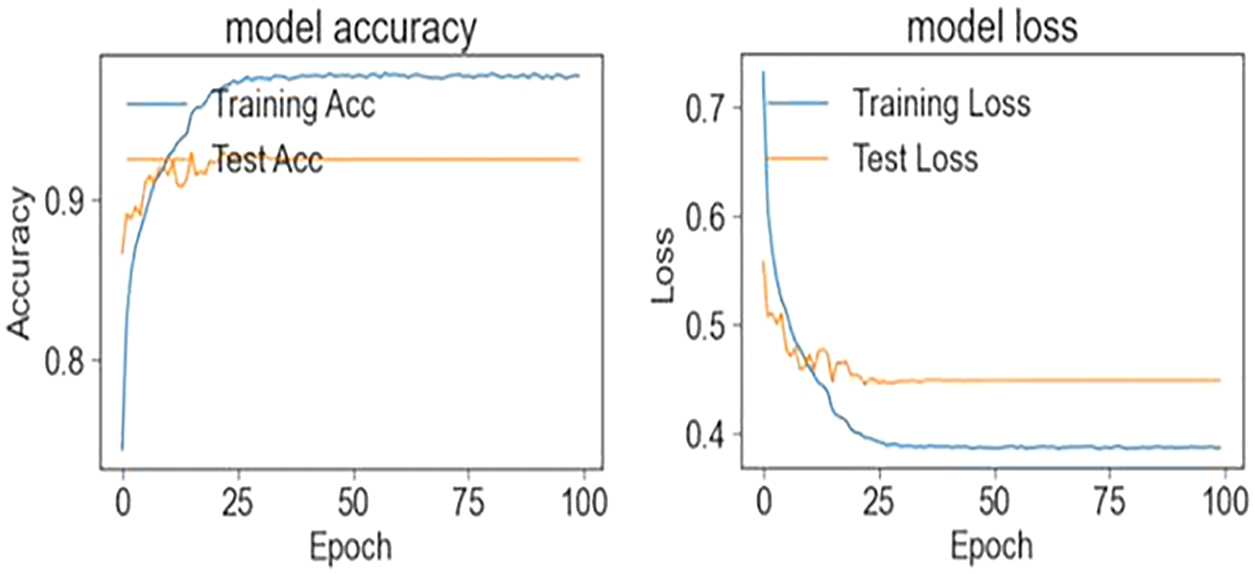
Figure 4: Train and test accuracy/loss graph (three classes)
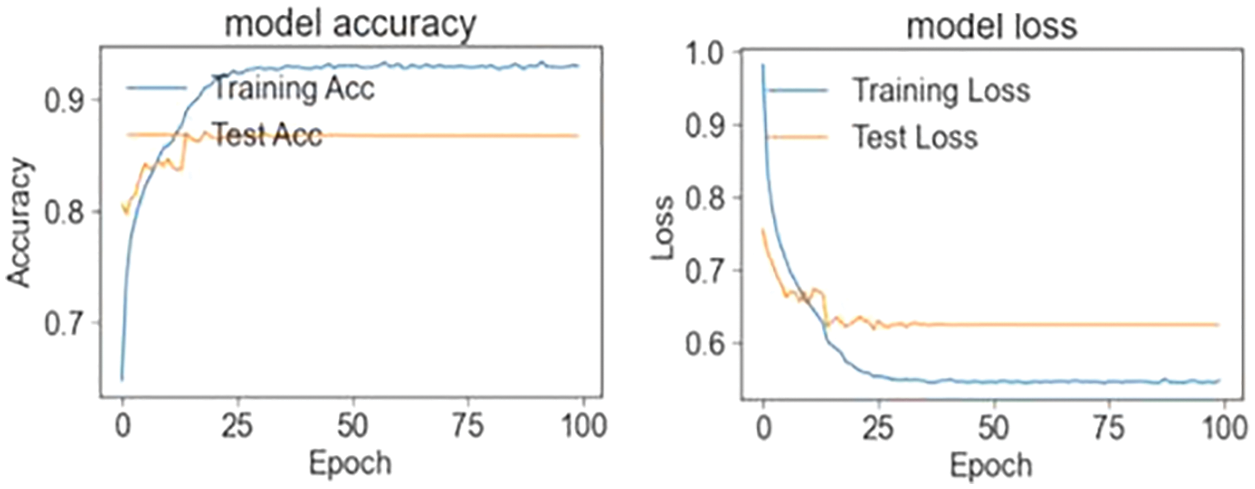
Figure 5: Train and test accuracy/loss graph (four classes)
3.2 Training Stage Cohen et al. Data Sets for Three/Four Classes
In the third stage, the data set of Cohen et al. is studied to compare the model with the studies in the literature. This level is for pneumonia, normal, lung opacity and Covid-19. The Tab. 5 shows the accuracy values of the four models and the ensemble model in the training result. Figs. 6 and 7 shows the accuracy and loss plots as a result of the training.
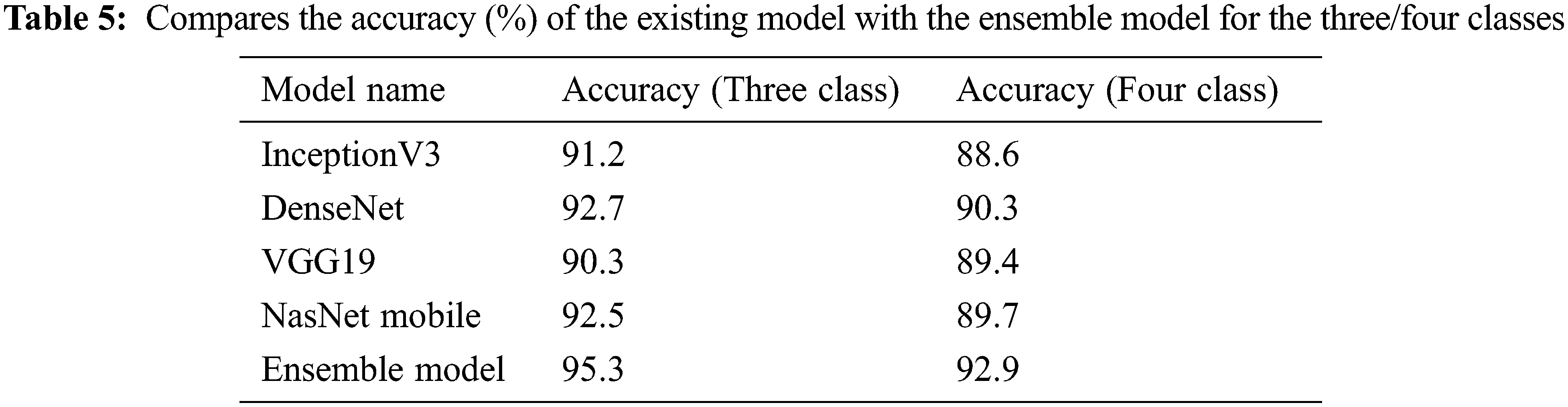
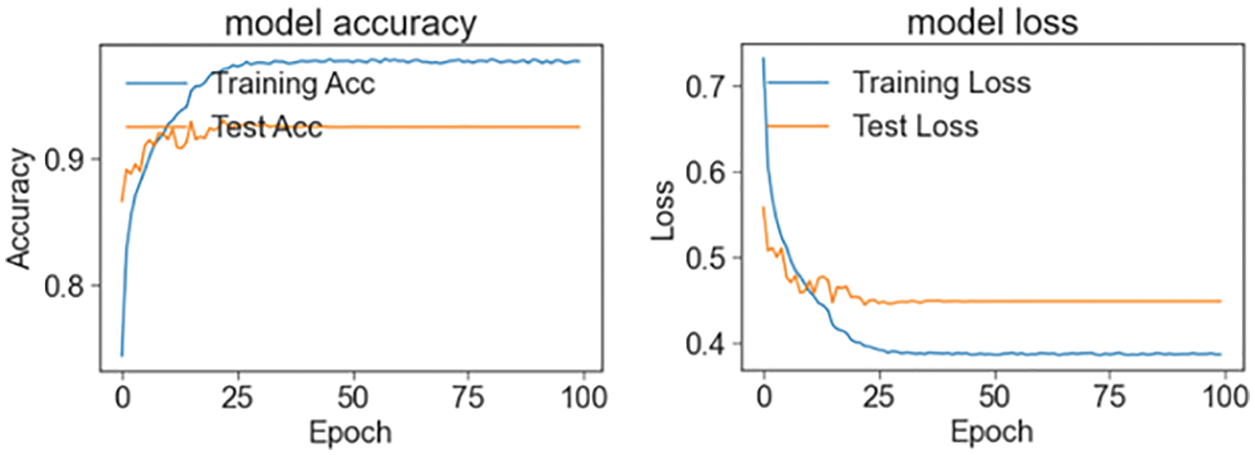
Figure 6: Train and test accuracy/loss graph (Cohen et al. three classes)
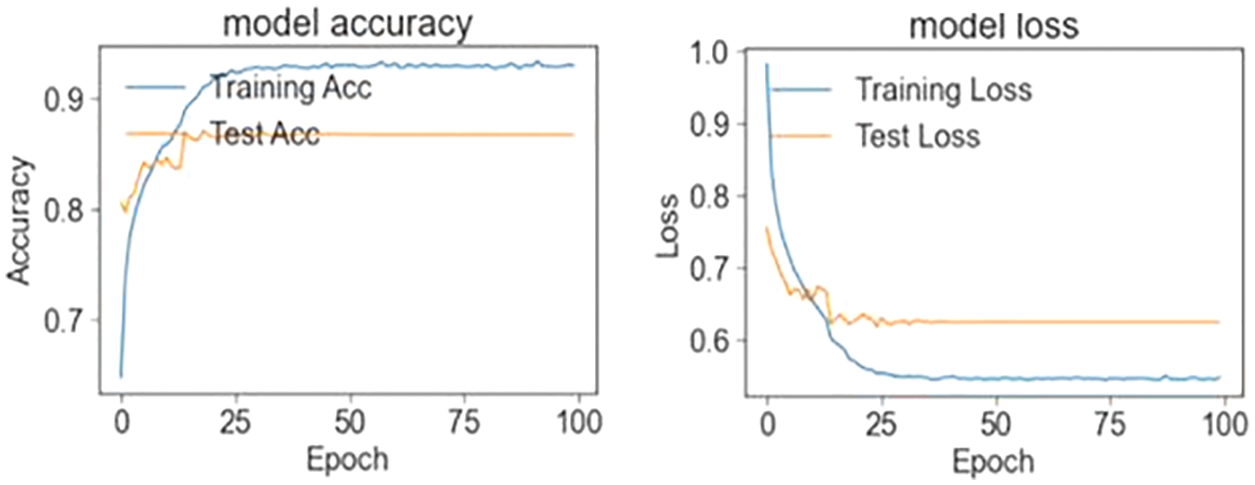
Figure 7: Train and test accuracy/loss graph (Cohen et al. four classes)
3.3 Evaluation Merged and Balanced Data Sets for Three/Four Classes
Precision, recall and F-measure are the main metrics to measure the performance of the classification algorithms. Precision shows how many of the values predicted as Positive are actually Positive. Recall is a metric that shows how many of the transactions that should have been predicted as Positive were predicted as Positive. F1 Score value shows the harmonic mean of the Precision and Recall values. In addition, the precision, recall and F1-score values are shown in Tab. 6. Since lung opacity is the hardest class to prediction, precision value was found to be lower than other data. Similarly, the F1 score is related to the precision value, so it is calculated lower than the other classes. Subsequently, the values of the confusion matrix are shown in Fig. 8 and the samples misdiagnosed by the ensemble model are shown in Fig. 9. According to the results, it can be said that the ensemble model is successful for three different classifications.
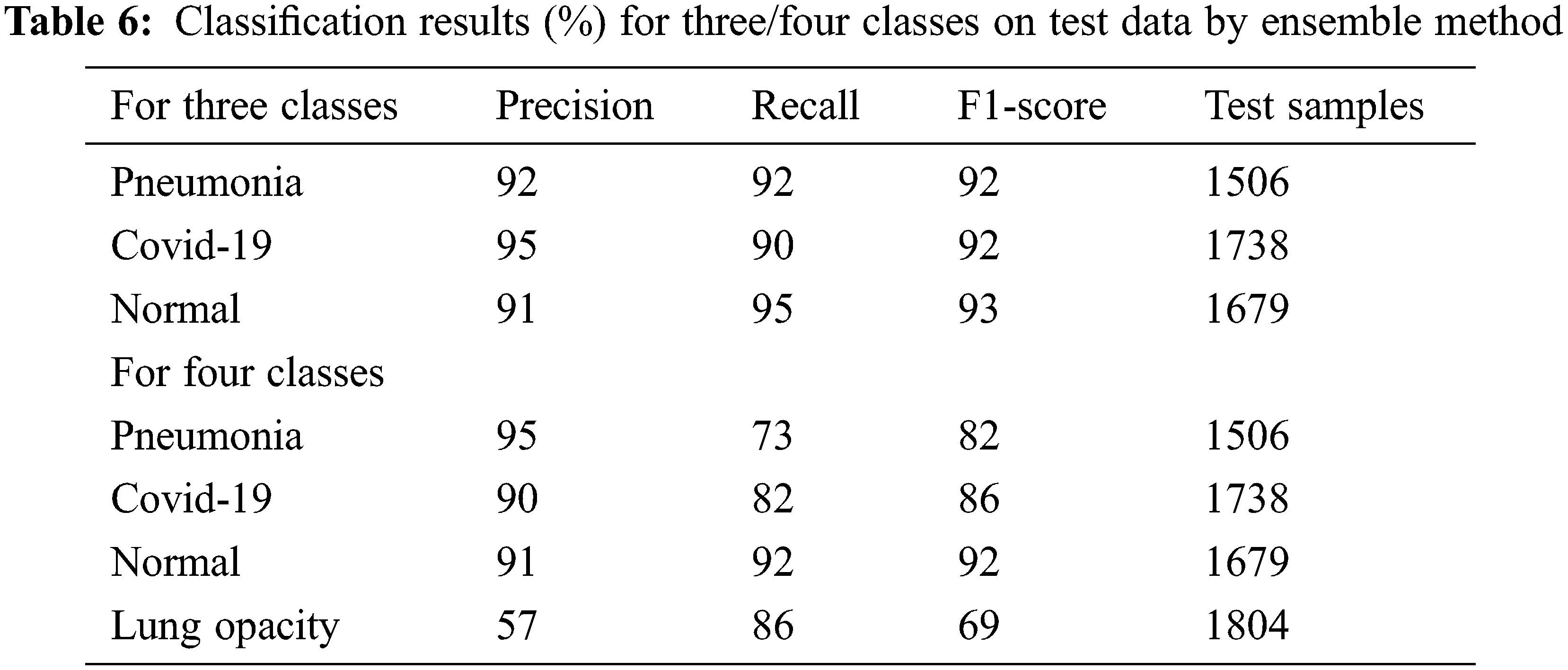
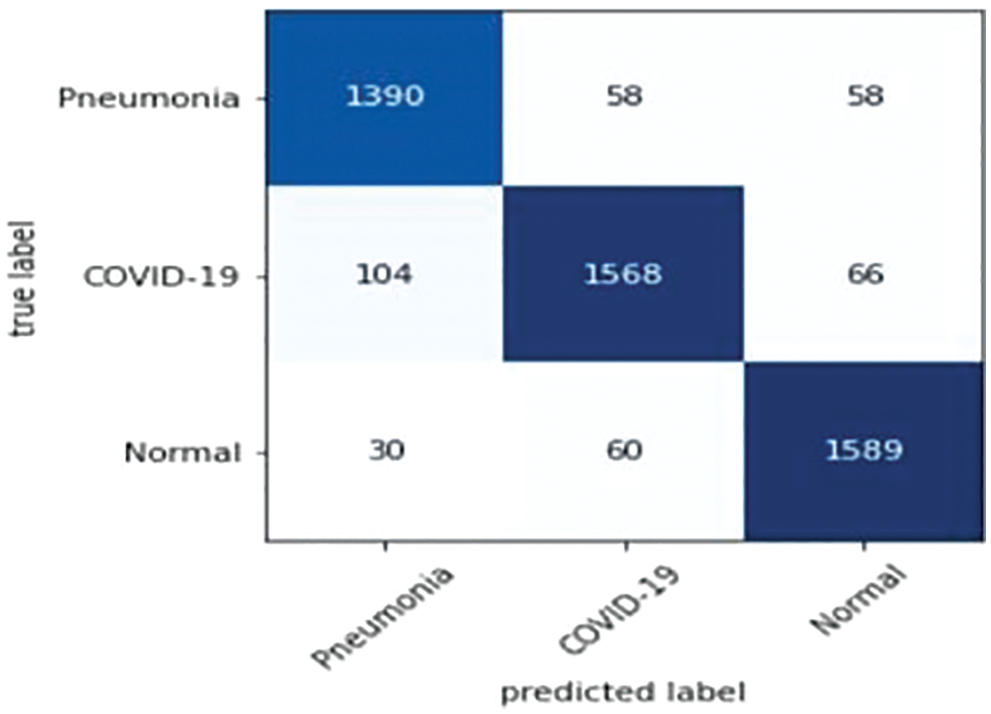
Figure 8: Confusion matrix: Ensemble model trained by three classes data
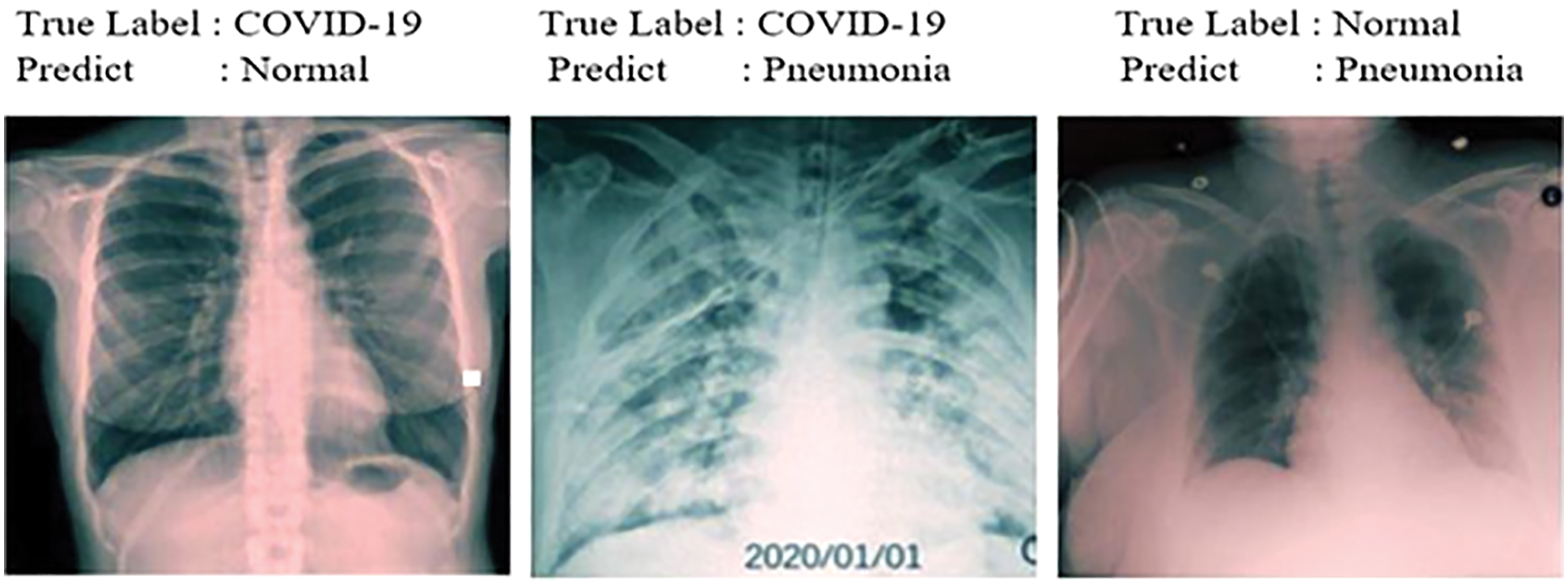
Figure 9: Samples misdiagnosed by the ensemble model for three classes
Finally, the values of the confusion matrix are shown in Fig. 10 and the samples misdiagnosed by the ensemble model are shown in Fig. 11. According to the results, it can be said that the ensemble model is successful for four different classifications. Diagnosis of the lung opacity class reduces the accuracy values to a bit at this stage, especially since it has similarities with Covid-19 and pneumonia types.
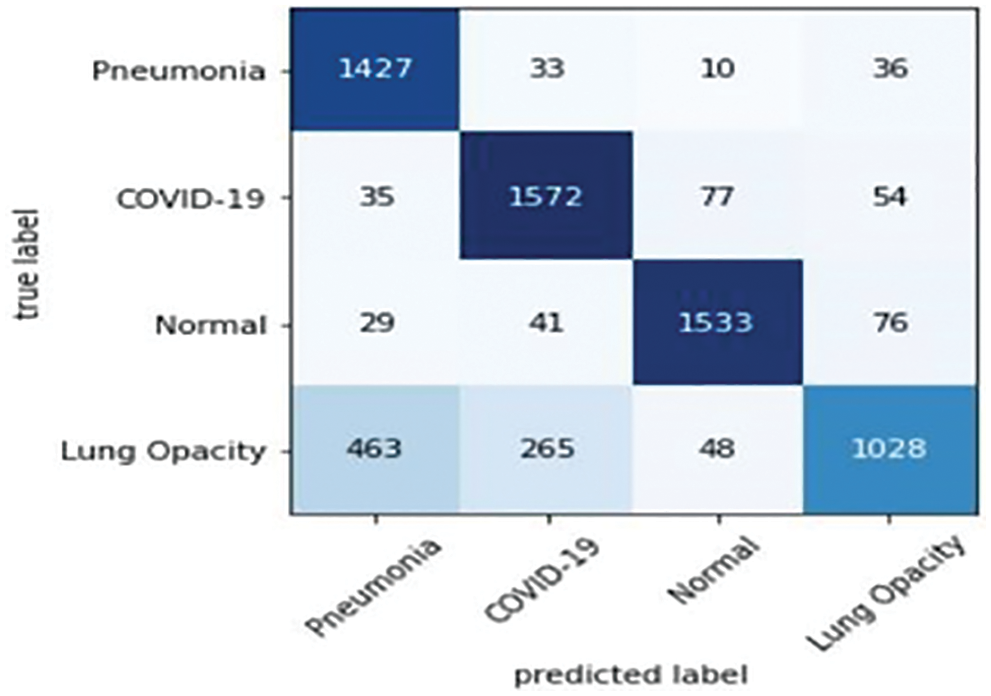
Figure 10: Confusion matrix: Ensemble models trained by four classes data
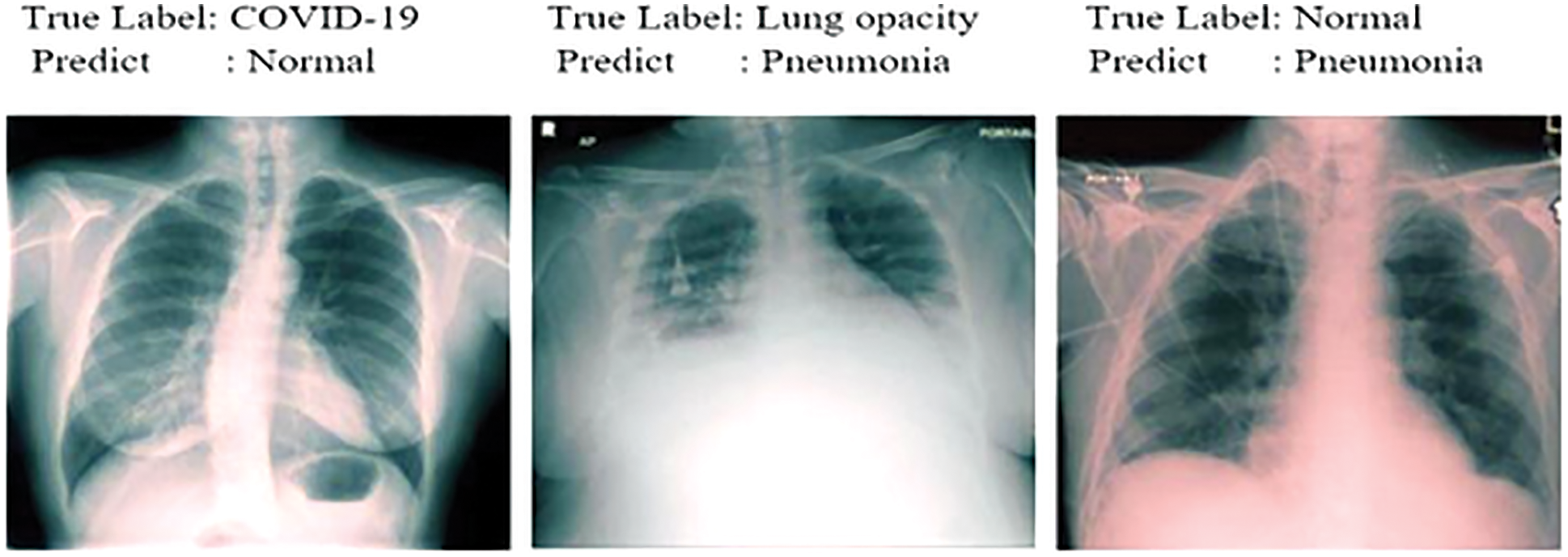
Figure 11: Samples misdiagnosed by the ensemble model for four classes
3.4 Evaluation Cohen et al. Data Sets for Three/Four Classes
The precision, recall, and F1 score values are shown in Tab. 7. Finally, the values of the confusion matrix are shown in Fig. 12.
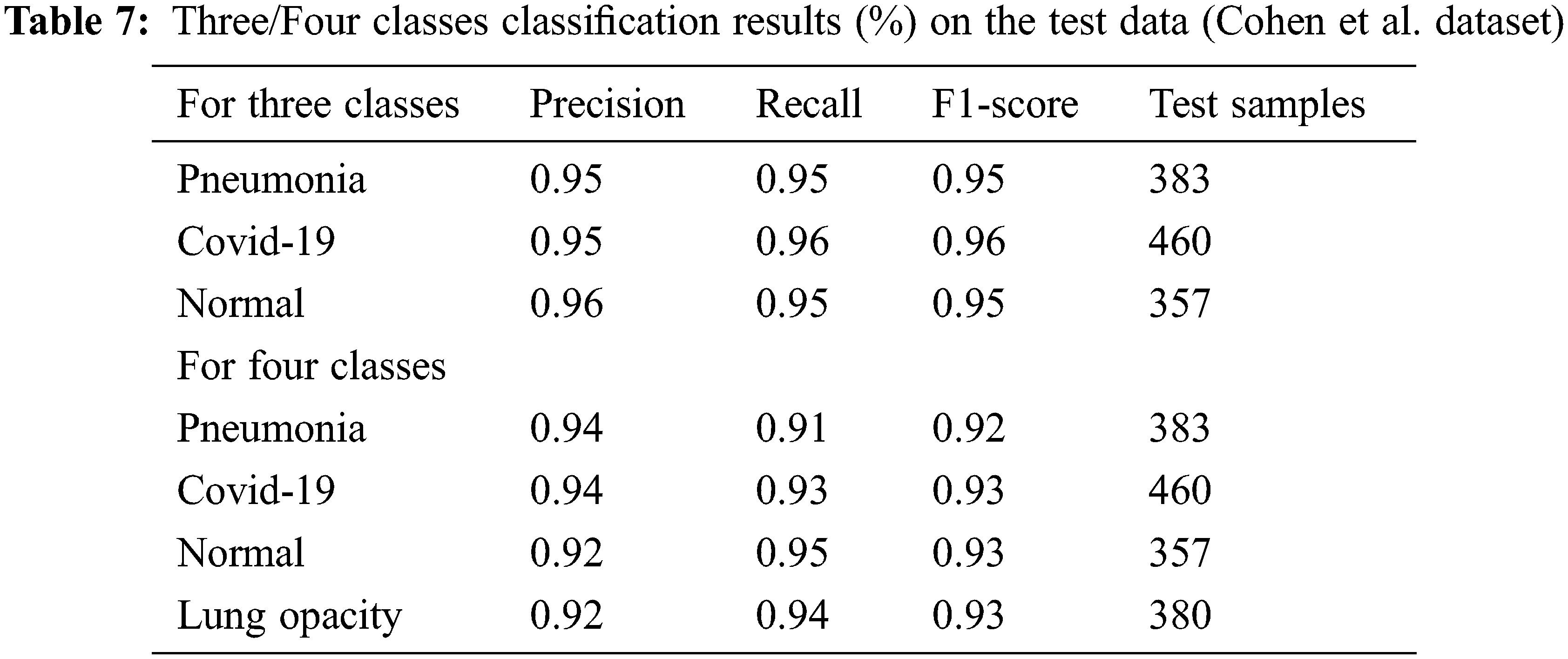
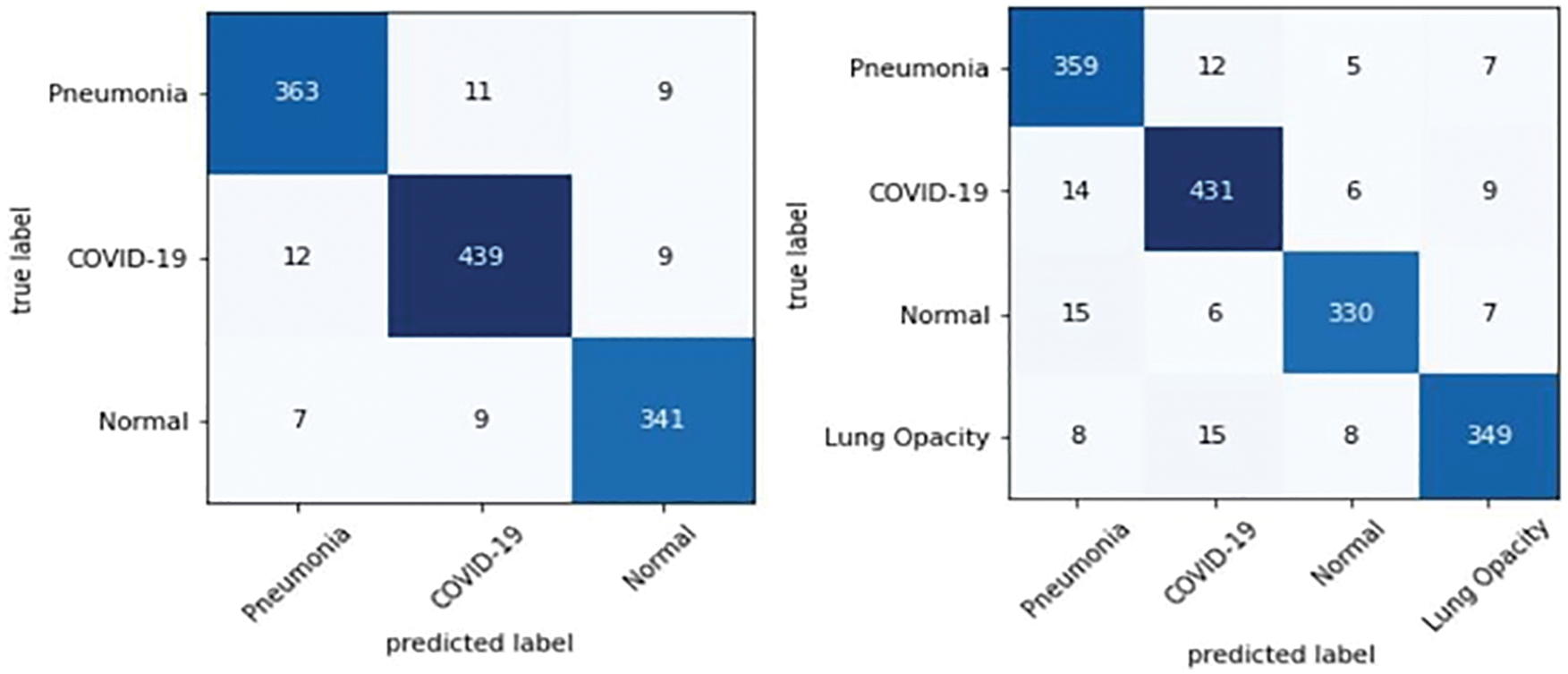
Figure 12: Confusion matrix: Ensemble model trained by three/four class data (Cohen et al. dataset)
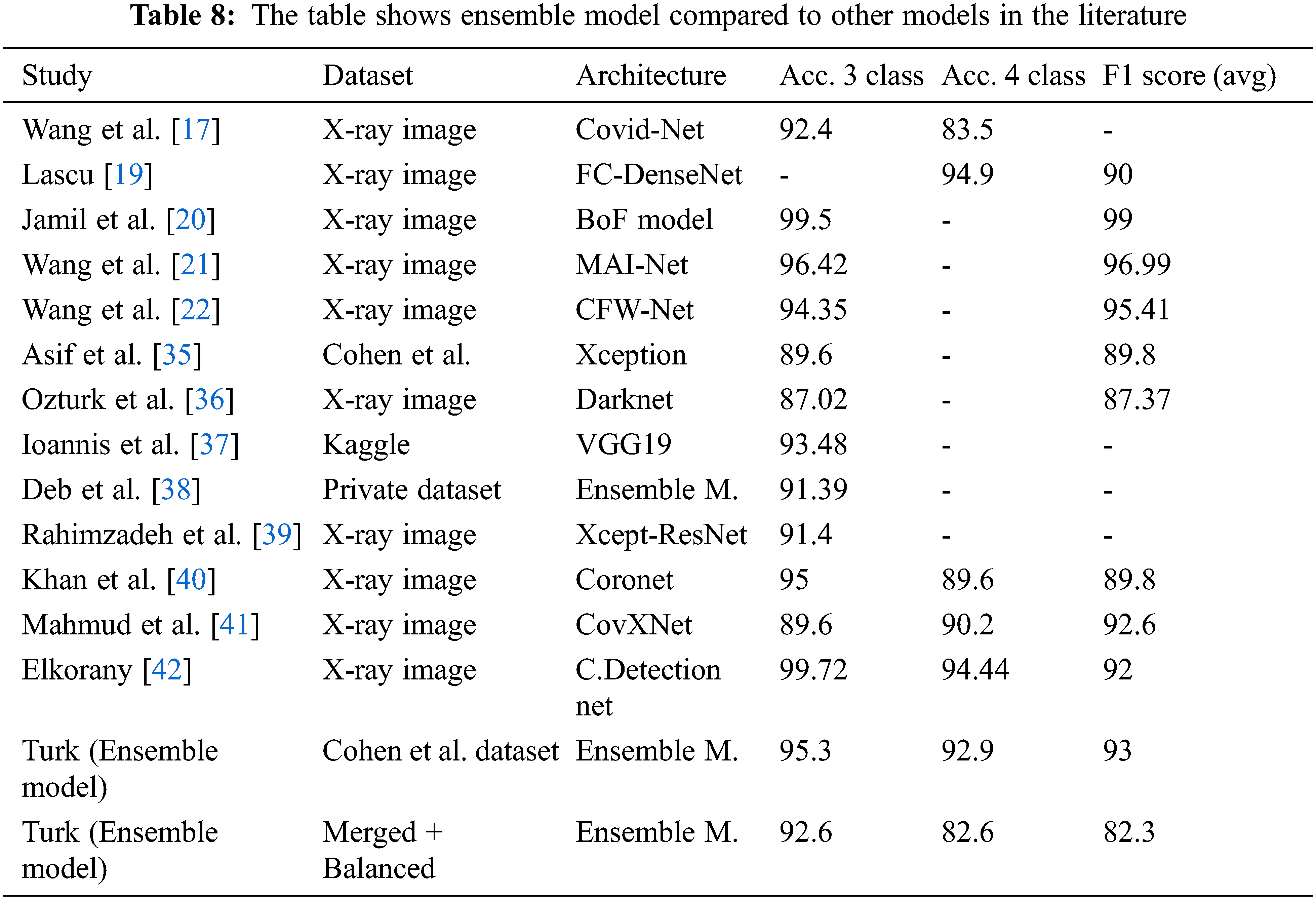
Tab. 8 shows an overall comparison of the results of the other Covid-19 studies in the literature with the results of this study. It can be said that the ensemble model developed at this stage provides successful results when compared with the studies conducted on the same dataset as well as on the other datasets. Moreover, it is reasonable to say that sub-class types are also important in multiclass studies. This is because comparing pneumonia and its sub-types with images of pulmonary opacities is not the same. Even small differences in lung images can lead to erroneous results in classification. Considering these details, it can be said that the ensemble model has a remarkable success in classification problems. Also, it shows the comparison of the classification accuracy (%) of the proposed ensemble model with the other existing approaches.
Linda et al. [17] achieved 92.4% accuracy for a three-class and 83.5% for a four-class study using the Covid-Net model and the X-ray images dataset.
Lascu [19] achieved 94.9% accuracy and of 90% F1 score for three-class a study with a model based on FC-DenseNet.
Jamil et al. [20] achieved 99.5% accuracy and 99% F1 score for a three-class a study with the model they called BoF Framework.
Wang et al. [21] achieved 96.42% accuracy and 96.99% F1 score for a three-class a study with the model they called MAI-Net.
Wang et al. [22] achieved 94.35% accuracy and 95.41% F1 score for a three-class a study with the model they called CFW-Net.
Khan et al. [35] used the Xception-based model they call CoroNet, achieved 89.6% accuracy and 89.8% F1 score for a three-class study with the Cohen et al. dataset.
Ozturk et al. [36] used the model they call Darknet, achieved an accuracy value of 87.02% and 87.37% F1 score for a three-class study.
Apostolopoulos et al. [37] achieved 93.48% accuracy in a three-class study with a Kaggle-based dataset using the VGG19 model.
Deb et al. [38] achieved 91.39% accuracy for a three-class study with the dataset they created using the Ensemble model (based on four pre-trained DCNN).
Rahimzadeh et al. [39] achieved 91.4% accuracy for a three-class study using the Concatenation model (Xception + ResNet50V2).
Khan et al. [40] achieved 95% accuracy in a three-class study, 89.6% accuracy and 89.8% F1 score for four-class study with the (Xpection) based model they called Coronet.
Mahmud et al. [41] achieved 89.6% accuracy for a three-class study, 90.2% accuracy and 92.6% F1 score for four-class study with the model they called CovXNet.
Elkoranyand et al. [42] achieved 99.72% accuracy for a three-class study, 94.44% accuracy and 92% F1 score for a four-class study using a set of 1200 lung images. It is useful to remember that the study characterized pneumonia (viral and bacterial) subtypes for four classes.
With the ensemble model used in this study with Cohen et al. data set, it was able to achieved an accuracy of 95.3% for a three-class study and 92.9% for a four classes study. In addition, the F1 score value was calculated as 93% on average.
With the ensemble model used in this study, it was able to achieved an accuracy of 92.6% for a three classes study and 82.6% for a four-classes study with the merged and balanced dataset. In addition, the F1 score value was calculated as 82.3% on average. It is useful to remind the difficulty of classifying the Lung opacity data (A class that can generally be confused with pneumonia and Covid-19). Therefore, it is acceptable that the accuracy and F1 score values are lower than some studies.
This research has provided a framework for diagnosing Covid-19 diseases with lung images. Furthermore, an appropriate system has been developed not to confuse pneumonia with lung opacity. Besides pre-trained CNN models, the ensemble method supported by the ResNet++ architecture can help radiologists interpret critical situations related to Covid-19 more deeply. Moreover, the dataset created can be a guide for new Covid-19 classification studies. Through to this dataset, it is thought that multi-class studies on Covid-19 can be apply. Also, in the future, current study can be advanced by designing automated systems based on a deep learning model that covers any disease image using CT scan and X-ray imaging evaluated by radiologists. More features need to be learned to properly classify neural networks for big data. This is only possible with datasets that contain balances and large numbers of samples. The model has shown promising results in both the previously used dataset and the dataset which have created. For the future studies, it can be said that the model will work with the same stability and give good results.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. C. Huang, Y. Wang, X. Li, L. Ren, J. Zhao et al., “Clinical features of patients infected with 2019 novel coronavirus in wuhan, China,” The Lancet, vol. 395, pp. 497–506, 2020. [Google Scholar]
2. H. Lu, C. W. Stratton and Y. W. Tang, “Outbreak of pneumonia of unknown etiology in Wuhan China: The mystery and the miracle,” Journal of Medical Virology, vol. 92, no. 4, pp. 401–402, 2020. [Google Scholar]
3. N. Chen, M. Zhou, X. Dong, J. Qu, F. Gong et al., “Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study,” The Lancet, vol. 395, pp. 507–513, 2020. [Google Scholar]
4. E. Lefkowitz, D. Dempsey, R. Hendrickson, R. Orton, S. Siddell et al., “Virus taxonomy: The database of the international committee on taxonomy of viruses (ICTV),” Nucleic Acids Research, vol. 46, pp. 708–717, 2020. [Google Scholar]
5. Covid-19 data portal Turkey. 2022. Available: https://covid19.tubitak.gov.tr. [Google Scholar]
6. F. Albogamy, M. Faisal, M. Arafah and H. ElGibreen, “Covid-19 symptoms periods detection using transfer-learning techniques,” Intelligent Automation Soft Computing, vol. 32, no. 3, pp. 1921–1937, 2022. [Google Scholar]
7. W. Guan, Z. Ni, Y. Hu, W. Liang, C. Ou et al., “Clinical characteristics of 2019 novel coronavirus infection in China,” MedRxiv, vol. 28, pp. 394, 2020. [Google Scholar]
8. J. Lei, J. Li and X. Qi, “CT imaging of the 2019 novel coronavirus (2019-nCoV) pneumonia,” Radiology, vol. 295, pp. 18, 2020. [Google Scholar]
9. T. Babu, T. Singh, D. Gupta and S. Hameed, “Optimized cancer detection on various magnified histopathological colon images based on DWT features and FCM clustering,” Turk J. Elec. Eng. Comp. Sci., vol. 30, pp. 1–17, 2020. https://doi.org/10.3906/elk-2108-23. [Google Scholar]
10. M. Gour and S. Jain, “Uncertainty-aware convolutional neural network for covid-19 X-ray images classification,” Computers in Biology and Medicine, vol. 140, pp. 105047, 2022. https://doi.org/10.1016/j.compbiomed.2021.105047. [Google Scholar]
11. A. Malhotraa, S. Mittal, P. Majumdar, S. Chhabraa, K. Thakral et al., “A. multi-task driven explainable diagnosis of covid-19 using chest X-ray images,” Pattern Recognition, vol. 122, pp. 108243, 2022. https://doi.org/10.1016/j.patcog.2021. [Google Scholar]
12. X. Liua, Q. Yuana, Y. Gaoc, K. He, S. Wanga et al., “Weakly supervised segmentationof COVID19 infection with scribble annotation on CT images,” Pattern Recognition, vol. 122, pp. 108341, 2022. https://doi.org/10.1016/j.patcog.2021. [Google Scholar]
13. S. D. Deb, R. Kumar, K. Jha and P. A. Tripathi, “Multi model ensemble based deep convolution neural net-work structure for detection of COVID19,” Biomedical Signal Processing and Control, vol. 71, pp. 103126, 2022. https://doi.org/10.1016/j.bspc.2021.103126. [Google Scholar]
14. S. Ghosh and A. Ghosh, “ENResNet: A novel residual neural network for chest X-ray enhancement based covid-19 detection,” Biomedical Signal Processing and Control, vol. 72, pp. 103286, 2022. https://doi.org/10.1016/j.bspc.2021.103286. [Google Scholar]
15. E. Ayan, B. Karabulut and H. M. Unver, “Diagnosis of pediatric pneumonia with ensemble of deep convolutional neural networks in chest X ray images,” Arabian Journal for Science and Engineering, vol. 47, pp. 2123–2139, 2021. [Google Scholar]
16. S. Lafraxo and M. Ansari, “CoviNet: Automated covid-19 detection from X-rays using deep learning techniques,” 6th IEEE Congress on Information Science and Technology (CiSt), vol. 978, pp. 6646, 2021. https://doi.org/10.1109/CIST49399. [Google Scholar]
17. W. Linda, Z. Quiu and A. Wong, “Tailored deep convolutional neural network design for detection of covid-19 cases from chest radiography images,” Journal of Network and Computer Applications, vol. 10, pp. 19549, 2020. [Google Scholar]
18. S. Shastri, I. Kansal, S. Kumar, S. Kuljeet, P. Renu et al., “CheXImageNet: A novel architecture for accurate classification of covid-19 with chest x-ray digital images using deep convolutional neural networks,” Health and Technology, vol. 12, pp. 193–204, 2020. https://doi.org/10.1007/s12553-021-00630-x. [Google Scholar]
19. M. R. Lascu, “Deep learning in classification of covid-19 coronavirus, pneumonia and healthy lungs on CXR and CT images,” Journal of Medical and Biological Engineering, vol. 41, pp. 514–522, 2021. [Google Scholar]
20. S. Jamil, M. S. Abbas, M. Ahsan, M. T. Ejaz, “A bag-of-features (BoF) based novel framework for the detection of COVID-1,” in 15th. Int. Conf. on Open-Source Systems and Technologies (ICOSST), Lahore, Pakistan, 2021. [Google Scholar]
21. W. Wang, X. Huang, J. Li, P. Zhang and X. Wang, “Detecting COVID-19 patients in X-ray images based on MAI-nets,” International Journal of Computational Intelligence Systems, vol. 14, no. 1, pp. 1607–1616, 2021. [Google Scholar]
22. W. Wang, H. Liu, J. Li, H. Nie and X. Wang, “Using CFW-net deep learning models for X-ray images to detect COVID-19 patients,” International Journal of Computational Intelligence Systems, vol. 14, no. 1, pp. 199–207, 2021. [Google Scholar]
23. J. P. Cohen, P. Morrison, L. Dao, K. Roth, T. Q. Duong et al., “Covid-19 image data collection: Prospective predictions are the future,” arXiv preprint arXiv: 2006.11988, 2020. [Google Scholar]
24. F. Türk, M. Lüy and N. Barıscı, “Kidney and renal tumor segmentation using a hybrid V-net-based model,” MDPI Mathematics, vol. 8, no. 10, pp. 1772, 2020. https://doi.org/10.3390/math8101772. [Google Scholar]
25. B. Zoph, G. Brain, V. Vasudevan, J. Shlens and V. L. Quoc, “Learning transferable architectures for scalable,” Image Recognition, pp. 8697–8710, 2018. [Google Scholar]
26. G. Huang, Z. Liu, L. Maaten and K. Q. Weinberger, “Densely connected convolutional networks,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, Honolulu, Hawaii, pp. 4700–4708, 2017. [Google Scholar]
27. S. Matiz and K. E. Barner, “Inductive conformal predictor for convolutional neural networks: Applications to active learning for image classification,” Pattern Recognition, vol. 90, pp. 172–182, 2018. [Google Scholar]
28. Y. Wang, Y. T. Chen, N. N. Yang, L. F. Zheng, N. Dey et al., “Classification of mice hepatic granuloma microscopic images based on a deep convolutional neural network,” Appied Soft Computing, vol. 74, pp. 40–50, 2019. [Google Scholar]
29. N. Meng, E. Y. Lam, K. K. Tsia and H. K. So, “Large-scale multi-class image-based cell classification with deep learning,” IEEE Journal of Biomedical and Health Informatics, vol. 23, no. 5, pp. 2091–2098, 2019. [Google Scholar]
30. N. Dong, L. Zhao, C. H., Wu and J. F. Chang, “Inception v3 based cervical cell classification combined with artificially extracted features,” Applied Soft Computing, vol. 93, pp. 106311, 2020. https://doi.org/10.1016/j.asoc.2020.106311. [Google Scholar]
31. K. Simonyan and A. Zisserman, “Very deep convolutional networks for large-scale image recognition,” arXiv:1409.1556, 2014. [Google Scholar]
32. S. Kavitha and B. Dhanapriya, “Neural style transfer using VGG19 and alexnet,” in Int. Conf. on Advancements in Electrical, Electronics, Communication, Computing and Automation (ICAECA), Tamilnadu, India, 2021. https://doi.org/10.1109/ICAECA52838. [Google Scholar]
33. Y. Xia, M. Cai, N. Chuankun, C. Wang, E. Shiping et al., “Switch state recognition method based on improved VGG19 network IEEE 4th advanced information technology,” in Electronic and Automation Control Conf., Beijing Shi, China, 2019. https://doi.org/10.1109/IAEAC47372.2019.8998029. [Google Scholar]
34. C. T. Lee, “What do we know about ground-glass opacity nodules in the lung?” National Library of Medicine, vol. 4, no. 5, pp. 656–659, 2015. [Google Scholar]
35. A. L. Khan, J. L. Shah and M. M. Bhat, “Coronet: A deep neural network for detection and diagnosis of covid-19 from chest x-ray images,” Computer Methods and Programs in Biomedicine, vol. 196, pp. 105581, 2020. https://doi.org/10.1016/j.cmpb. [Google Scholar]
36. T. Ozturk, M. Talo, E. A. Yildirim, U. B. Baloglu, O. Yildirim et al., “Automated detection of covid-19 cases using deep neural networks with x-ray images,” Computers in Biology and Medicine, vol. 121, pp. 103792, 2020. https://doi.org/10.1016/j.compbiomed.2020.10379. [Google Scholar]
37. I. D. Apostolopoulos and T. A. Mpesiana, “Covid-19: Automatic detection from x-ray images utilizing transfer learning with convolutional neural networks,” Physical and Engineering Sciences in Medicine, vol. 43, pp. 635–640, 2020. [Google Scholar]
38. S. D. Deb and R. K. Jha, “Covid-19 detection from chest x-ray images using ensemble of cnn models,” in Int. Conf. on Power, Instrumentation, Control and Computing (PICC), pp. 1–5, 2020. https://doi.org/10.1109/PICC51425.2020.9362499. [Google Scholar]
39. M. Rahimzadeh and A. Attar, “Modified deep convolutional neural network for detecting covid-19 and pneumonia from chest X-ray images based on the concatenation of xception and resnet50v2,” Informatics in Medicine Unlocked, vol. 19, pp. 100360–100360, 2020. https://doi.org/10.1016/j.imu.2020.100360. [Google Scholar]
40. A. I. Khan, J. L. Shah and M. M. Bhat, “CoroNet: A deep neural network for detection and diagnosis of covid-19 from chest x-ray images,” Comput. Methods Programs Biomed., vol. 196, pp. 105581, 2020. https://doi.org/10.1016/j.cmpb.2020.105581. [Google Scholar]
41. T. Mahmud, M. A. Rahman and S. A. Fattah, “CovXNet: A multi-dilation convolutional neural network for automatic covid-19 and other pneumonia detection from chest X-ray images with transferable multi-receptive feature optimization,” Computers in Biology and Medicine, vol. 122, pp. 103869–103869, 2020. [Google Scholar]
42. A. Elkoranyand and Z. F. Elsharkawy, “COVIDetection-net: A tailored covid-19 detection from chest radiography images using deep learning,” Optik, vol. 231, pp. 166405, 2020. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools