 Open Access
Open Access
ARTICLE
A Framework of Deep Learning and Selection-Based Breast Cancer Detection from Histopathology Images
1 Department of Computer Science, COMSATS University Islamabad, Wah Campus, Wah Cantt, Pakistan
2 Computer Sciences Department, College of Computer and Information Sciences, Princess Nourah Bint Abdulrahman University (PNU), P.O. BOX 84428, Riyadh, 11671, Saudi Arabia
3 College of Computer Engineering and Science, Prince Sattam Bin Abdulaziz University, Al-Kharaj, 11942, Saudi Arabia
4 Department of Computer Science, Hanyang University, Seoul, 04763, Korea
5 Center for Computational Social Science, Hanyang University, Seoul, 04763, Korea
* Corresponding Author: Byoungchol Chang. Email:
Computer Systems Science and Engineering 2023, 45(2), 1001-1016. https://doi.org/10.32604/csse.2023.030463
Received 26 March 2022; Accepted 17 May 2022; Issue published 03 November 2022
Abstract
Breast cancer (BC) is a most spreading and deadly cancerous malady which is mostly diagnosed in middle-aged women worldwide and effecting beyond a half-million people every year. The BC positive newly diagnosed cases in 2018 reached 2.1 million around the world with a death rate of 11.6% of total cases. Early diagnosis and detection of breast cancer disease with proper treatment may reduce the number of deaths. The gold standard for BC detection is biopsy analysis which needs an expert for correct diagnosis. Manual diagnosis of BC is a complex and challenging task. This work proposed a deep learning-based (DL) solution for the early detection of this deadly disease from histopathology images. To evaluate the robustness of the proposed method a large publically available breast histopathology image database containing a total of 277524 histopathology images is utilized. The proposed automatic diagnosis of BC detection and classification mainly involves three steps. Initially, a DL model is proposed for feature extraction. Secondly, the extracted feature vector (FV) is passed to the proposed novel feature selection (FS) framework for the best FS. Finally, for the classification of BC into invasive ductal carcinoma (IDC) and normal class different machine learning (ML) algorithms are used. Experimental outcomes of the proposed methodology achieved the highest accuracy of 92.7% which shows that the proposed technique can successfully be implemented for BC detection to aid the pathologists in the early and accurate diagnosis of BC.Keywords
BC is a most dangerous sort of cancerous malady that may lead to death if not detected and handles properly with medical treatment at its initial stage [1]. IDC is the most ordinary kind of cancer disease mostly present in middle-aged women and is being increased in the past two decades [2]. The BC positive newly diagnosed cases in 2018 reached 2.1 million around the world with a death rate of 11.6% of total cases [3]. In the IDC type of breast tumor, the cancerous cells originated from the skin spread to their neighbor cells that may spread to the other body organs over time. The expected growth of breast cancer in the United States with IDC is presented in [4]. Early detection of this cancer with proper treatment may increase the survival rate by 80% [5]. The current gold standard for pathology analysis is still tissue biopsy despite the rapid development of imaging technology such that x-ray and magnetic resonance imaging (MRI) [6,7]. The microscopic structure of the breast cancerous tumor is analyzed with the help of a biopsy that needs expert pathologists. Histology images are being utilized to differentiate the different types of breast cancer for the early detection of cancer [8]. Manual analysis of BC detection is a very challenging and laborious process that requires the knowledge of professionals to study large scanning images for diagnosis. To overcome these issues computer-aided diagnosis is gaining increased intention for early detection of BC [9].
ML is a well-known subtype of artificial intelligence (AI) [10] that is being utilized to automate the physical diagnosis and detection of different maladies [11–13]. In ML the feature extraction process is carried out with manual hand engineering of different features. After successful manual extraction of features in the next step the classification task is carried out by using different ML algorithms [14]. The handcrafted feature such that shape and texture feature is implemented for BC detection at a large scale. Wang et al. presented an automatic ML-based solution for BC diagnosis with the help of cell pathological images by using shape and color texture features. In their study for the classification task, an support vector machine (SVM) classifier is utilized [15]. Jin et al. presented a study to automatically diagnose breast tumors as benign and malignant from breast cytology images by using a Naïve Bayes classifier [16]. Another ML-based solution for BC detection is presented by Filipczuk et al. In their study different ML algorithms such that k-nearest neighbor (KNN), SVM, decision tree, and Naïve Bayes are utilized for automatic detection of breast tumors with the assistance of microscopy images [17]. Deep convolutional neural network (CNN) models are being applied at a large scale for different disease detection because of automatic feature learning. DL achieved promising results in medical image analysis, particularly in BC diagnosis. Cruz-Rao proposed a deep CNN model for IDC disease detection from whole slide images and reported a balanced average accuracy of 84.23% [18]. A transfer learning-based automatic diagnosis system for BC detection using the BreaKHis dataset is presented by Bhuiyan et al. [19] After successful deep feature extraction using transfer learning for classifying the BC positive and negative malady different ML classifiers are used including SVM and KNN.
This study presented a thirty-layer deep CNN model and novel FS framework for automatic diagnosis of BC by using histopathology images. For training and testing of the proposed method, a publically available large dataset of histopathology images is utilized. The proposed methodology achieved the highest accuracy results. The major contributions of the proposed deep learning-based BC classification methodology are presented below in bullets.
• A deep CNN network consisting of a total of 30 layers is proposed for BC classification into IDC and no-IDC tumors.
• The deep learning network is trained from scratch at the CIFAR-100 dataset for feature learning.
• The proposed CNN model is utilized as a feature extractor by using transfer learning, for this purpose a large public dataset of BC is utilized.
• A novel feature-level fusion-based FS framework is proposed for the best FS.
• Validation of selected FV is performed with different ML algorithms and the best results are also compared with existing methods in the literature for BC classification.
In the last decade, different ML and DL-based techniques for BC detection and classification from histopathology images have been proposed. Many innovative computers aided diagnostic systems (CADs) for breast cancer detection with handcrafted feature extraction techniques are proposed by the researchers. Dundar et al. present an approach for BC classification by using the statistical and shape features [20]. A local binary pattern-based solution for BC diagnosis using the MIAS dataset is presented in [21]. Another handcrafted feature extraction-based automated solution for BC detection is presented by Yasiran et al. [22]. In their study, hydraulic texture features and SVM are utilized for the classification task. Niwas et al. presented a Log-Gabor wavelet feature extraction mechanism from needle biopsy samples for the classification of breast cancer with the help least square SVM classifier [23]. A web-based service for BC diagnosis using cytological images is proposed by George et al. [24]. Shukla et al. [25] presented a study for the categorization of healthy and non-healthy cells by extracting the shape and morphological features and reported an accuracy of 85.5%.
Recently, numerous CNN-based methods have been presented for the early detection of BC from image datasets. Narayanan et al. [26] presented a DL solution for the classification of IDC positive and IDC negative breast tumors from a publically available large dataset of breast cancer detection containing BC histopathology images. Their model is comprised of a total of five convolution layers and a fully connected (FC) layer. A softmax layer is utilized for the classification task. Initially, the original images were down-sampled for better performance and processed through a histogram equalization method and color consistency technique. After preprocessing processed images were then supplied as input to the deep CNN model for the classification task. Results concluded that preprocessing with the color consistency technique yielded higher performance than the histogram equalization method. BC detection using mammography images is presented by Debelee et al. [27]. In their work, a deep CNN network is proposed for deep feature extraction and principal component analysis (PCA) is applied for FS. After that, the selected FV is further used as input to the different ML models for normal and abnormal mammography image classification. In another work, the same task is carried out by using transfer learning with Inception-v3 by Debelle et al. [28] for performance evaluation local and public datasets were utilized. A feature fusion was also carried out and compared in their work. A DL-based study for IDC classification is presented by Romano and Hernandez [29]. They proposed an improved CNN model containing two convolution layers, and accept reject the new pool layer. After preprocessing phase, the input images were supplied to the CNN model for classification. Their model yielded an accuracy of 85.41%.
In another work, a DL-based CAD system for BC detection using a large dataset was presented by Rahman et al. [30]. For solving the problem of class imbalance in the input dataset the images were selected randomly with an equal number for each class. The data augmentation method was utilized to overcome the problem of model overfitting. The average accuracy of 89% was yielded by their model. Wang et al. [31] presented four deep CNN architectures for BC classification by using a large publically available dataset. For the classification of IDC tumors, their study reported an accuracy of 89% without performing data augmentation. The attention-based deep CNN model for BC detection is proposed by Sanyal et al. [32]. In their study different image patches that contained higher information gained higher attention for further processing of detecting cancer for this purpose histopathology images with high resolution was utilized. In another study, Sanyal et al. perform the same task by using different CNN models by the hybrid ensemble of deep models [33]. Recently, Chapala and Sujatha proposed a transfer learning-based method for BC detection. Two pre-trained models resnet34 and resnet50 were utilized to transfer the learned feature for classification tasks with different training testing ratios of the dataset [34]. For more understanding of BC classification and segmentation interested readers are referred to the latest studies found in [35–38].
This section discussed the proposed technique for BC classification from histology images in detail. The general workflow of the proposed methodology for BC detection from the histopathology dataset is presented in Fig. 1. The proposed methodology for BC detection is comprised of three major steps. In the initial step, a convolution neural network is proposed for automatic deep feature extraction. For this purpose, the proposed deep CNN model is pre-trained at a publically available CIFAR-100 [39] dataset that contained 60 thousand images of hundred different classes. The deep CNN model is trained from the scratch at 60 e pouches with a mini-batch of 128, an initial learner rate of 0.01, and an ‘sgdm’ optimizer. In the second step, the proposed pre-trained network is further used for deep feature extraction. In this step breast cancer IDC positive and negative images from the database are fed to the proposed pre-trained deep learning model for deep features extraction by using the simple transfer learning concept. After extraction of deep features (DF), in the third step, the extracted FVs are passed to a novel FS framework for the best FS. The FS framework utilized the serial feature fusion strategy for selection. The input FV is fed to two different filter-based selection algorithms including R-Relief and PCC (Pearson correlation coefficient) in parallel. The output selected FV of both algorithms is fed as input to PCA to get the one final selected FV for further processing. Finally, for the classification task different ML algorithms are trained to automatically detect breast cancer. Complete detail of each section of the proposed architecture and data source are presented below.
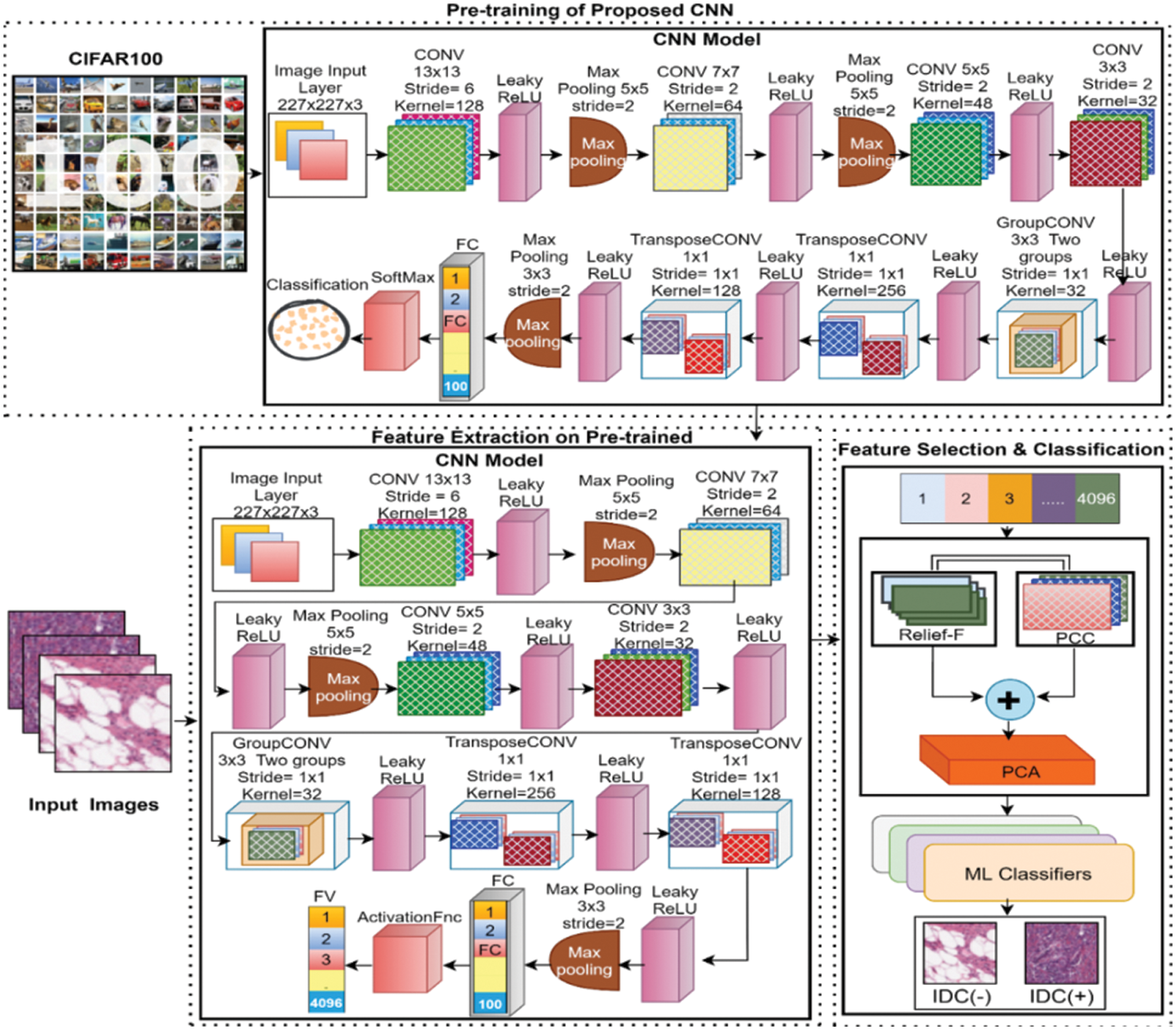
Figure 1: General workflow of the proposed methodology for BC detection using histopathology images
For high-level image feature extraction, CNNs are being used at a large scale in different fields such as computer vision and medical. CNN belongs to a class of DL methods that are commonly utilized for analyzing visual imagery. To utilize the power of DL, deep feature extraction is the fast and easiest way because features can be extracted without consuming much time in training a full CNN model. Transfer learning is a term that is used to take out learned image features from pre-trained networks. In CNN models a single pass through a network is required for feature extraction. In this study, a thirty-layer proposed deep learning algorithm is used for feature extraction purposes. The detail of the network is presented below.
3.2 Convolutional Neural Networks (CNN)
In this study, a thirty-layered CNN model for deep feature extraction is proposed. The complete architecture of the layers in the proposed deep CNN is presented in Fig. 2. The input size of the proposed model is 227 × 227 × 3 with zero center normalization. The next second layer is a two-dimensional (2D) convolution layer (CL) that has 128 kernels with the size of 13 × 13 × 3 and a stride size of 6 with the same padding strategy. A leaky rectified linear unit (ReLU) with a scale size of 0.01 is utilized as an activation function (AF) after CL. After that, a batch normalization layer is applied with 128 channels for normalization purposes. A max-pooling layer of size 5 × 5 and stride 2 with the same padding is utilized after normalization. The next layer is a second CL-that contained 64 kernels of size 7 × 7 × 128 with the same padding and stride size of 2. This layer is succeeded by batch normalization, leaky ReLU, and max pooling. The next third CL is composed of 48 kernels of size 5 × 5 × 64 with the same padding and stride of 2. After the third convolution layer fourth leaky ReLU layer is implemented. The fourth CL has 32 kernels of size 3 × 3 × 48 with the same padding and stride of 2 succeeded by a leaky ReLU function. The fifth CL is a grouped convolution layer that has two groups of 32 kernels of size 3 × 3 × 16 with the same padding and stride size of 1 followed by a leaky ReLU layer of scale 0.01. The 6th convolution layer is transposed CL with a stride size of one and a cropping size of zero. The number of kernels in transposed convolution layer is 256 with a size of 3 x 3 x 64 and after this layer, a leaky ReLU layer is applied as an AF. The 7th convolution layer is also a transposed convolution layer of size 3 × 3 × 256 with 128 kernels and a stride size of 1. The proposed CNN is comprised of a total of seven convolution layers four of which are 2D convolution layers, one grouped convolution layer, and two transposed CLs. The 7th CL is succeeded by the leaky ReLU layer, a batch normalization of 128 channels, and pooling of size 3 x 3 with a stride of size 2. After the last pooling layer, an FC layer of size 4096 followed by an activation layer is utilized. A dropout layer is utilized prior to the second FC layer with a 50% drop for performance enhancement. The second fully connected layer also has the 4096 number of connections and is followed by the leaky ReLU and 50% dropout layer respectively. The third FC layer is designed according to the training dataset that has a hundred different classes so, the total number of connections in the last FC layer is 100. After this fully connected layer, a flattened layer of softmax is utilized. Finally, for the classification task, a classification layer is added as the last layer.
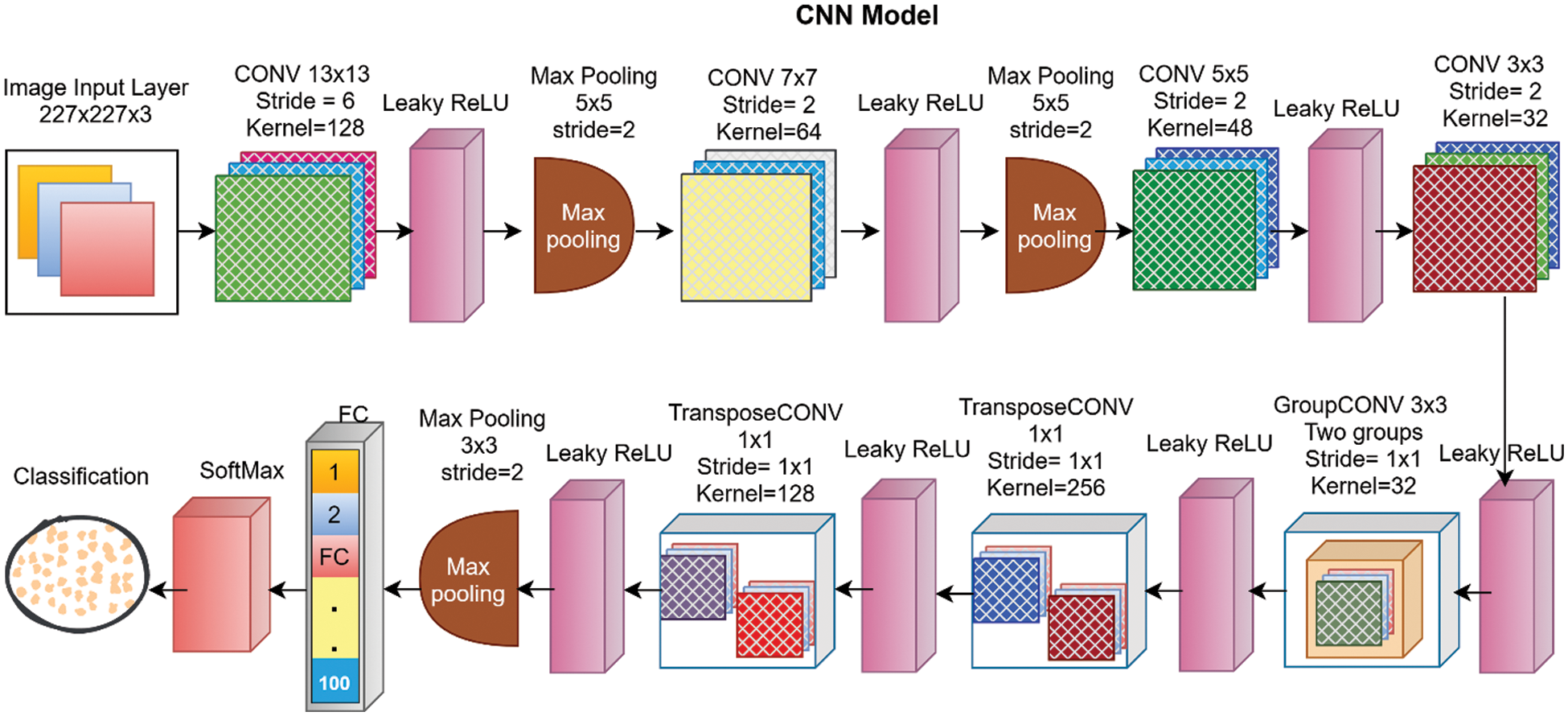
Figure 2: Proposed DL network with detail of each layer
In this work, the proposed network is trained at the CIFAR-100 dataset for feature learning. The CIFAR-100 dataset is a publically available dataset that contains 60 thousand images of hundred different classes. After successful training of the proposed CNN model the trained network is further utilized for the feature extraction. The feature extraction task is accomplished by using a simple transfer learning methodology. For this purpose, the breast cancer IDC dataset is passed to the trained model for feature learning. The FV is extracted from the second FC layer namely FC2, as a result, an FV of size N × 4096 is collected for the further classification task. After successful extraction of the FV, the extracted FV is passed to a proposed hybrid FS framework. Finally, the selected FV is inputted into different ML algorithms for classification. The proposed FS framework is presented in detail in the next section of this article.
3.3 Features Selection Network
FS is a key task in ML and computer vision that is being used for the higher performance of classifiers. The redundant and irrelevant information in the feature set is a burden for classification tasks that may lead to misclassification and can lower the classification accuracy with a higher error rate. To overcome these issues different FS methods have been proposed for optimal FS. This research work presented a feature-level fusion-based FS methodology for top FS. The proposed hybrid selection algorithm is presented in Algorithm 1. The proposed FS framework is mainly comprised of 3 steps. Initially, the extracted vector is pipelined as input to the two filter-based FS methods including PCC [40], and Relief-F [41]. In the second step, both selected FVs are fused horizontally for further selection. In the last step, the fused selected FV is passed to the PCA for the best top FS. The proposed FS methodology is presented in Fig. 3. Both the PCC FS algorithm and Relief-F FS algorithm are filter-based selection algorithms that are much faster and functionally independent. In this study for PCC FS, the input FV is passed to the PCC algorithm that gives an output FV of size N × 1000 as the number of selected feature parameters was set to N × 1000. The mathematical formulation of the PCC algorithm is presented in Eq. (1).
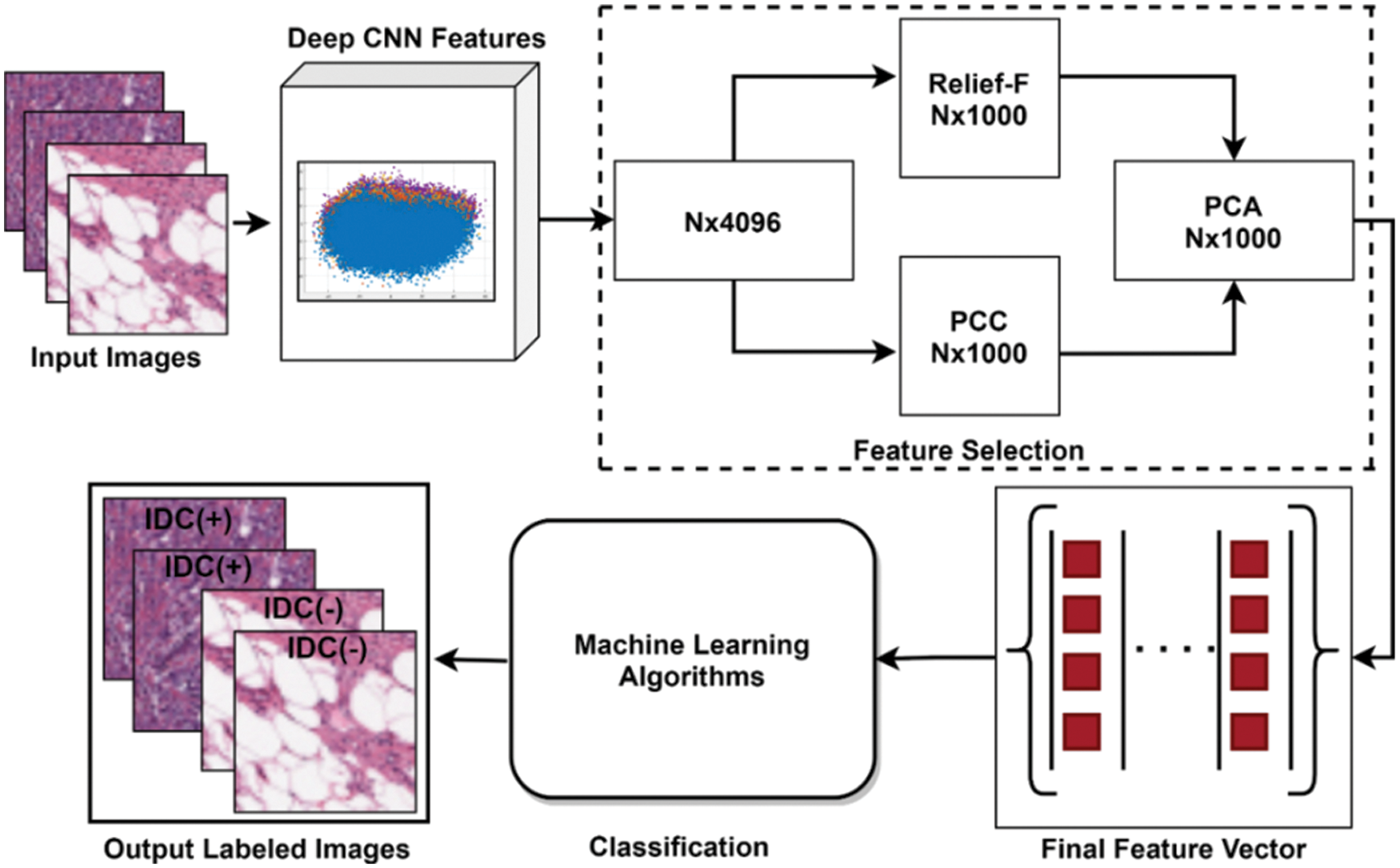
Figure 3: Proposed FS framework
In the second FS algorithm Relief-F, the same FV is passed as an input for FS and as an output, a selected FV of size N x 1000 is acquired. Relief-F is suitable for multiclass selection problems in which one near miss for each of the different classes is calculated and averaged for updating estimates weight [M] instead of calculating it for all different classes. The mathematics behind the selection of features using Relief-F is presented in Eq. (2).
After successful implementation of filter-based FS methods, the two selected FVs are fused horizontally and passed to the PCA for further selection. PCA is a linear features selection procedure that is widely used in the field of ML for FS [42].

4 Experimental Results and Discussion
This study utilized a very large publically available dataset containing a total of 277524 histopathology images 78687 of which are IDC positive images and the remaining are non-IDC images. Dataset can be accessed at the Kaggle site for research purposes only [43]. All the images were collected in PNG format from a total of 162 whole slide images. During the collection process, all the BC specimens were scanned at 40×. The examples of dataset images from the large publically available dataset are depicted in Fig. 4.
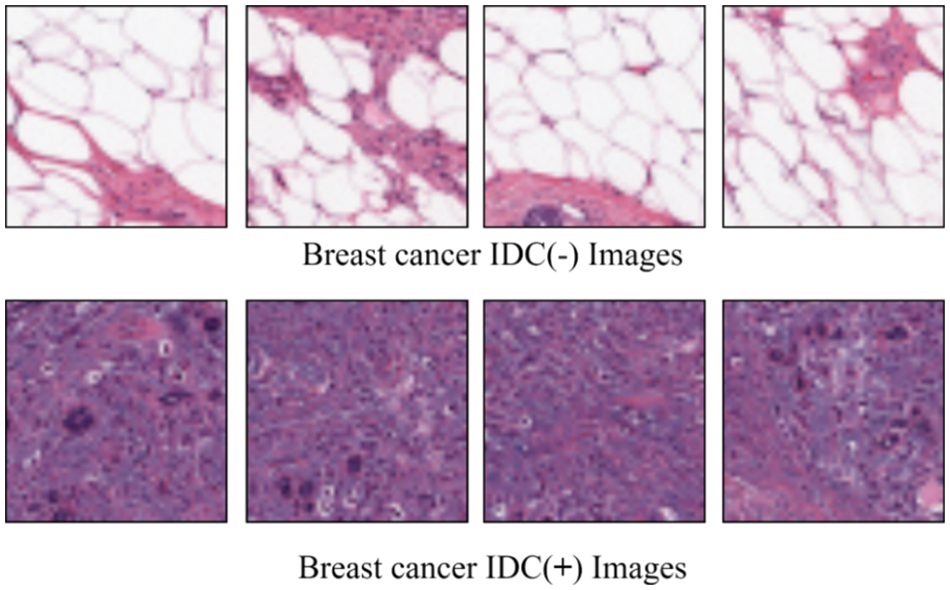
Figure 4: The dataset images are taken from a large publically available IDC dataset [43]
Experiment 1: In this experiment, the results are computed before applying the proposed FS framework. The proposed deep CNN model is utilized for the feature extraction process with the help of the activation function. After the successful extraction of deep features, the extracted FV is inputted to diverse ML classifiers including Linear (LSVM), Cubic (CSVM), Fine Tree (FT), Ensemble Boosted Tree (EBT), Ensemble Subspace Discriminant (ESD), Medium Gaussian (MGSVM), Coarse Gaussian (CGSVM), and Quadratic (QSVM). The results were computed by using 5-fold and 10-fold validation techniques. The experimental outcomes of the proposed method with 10-fold are tabulated in Tab. 1.
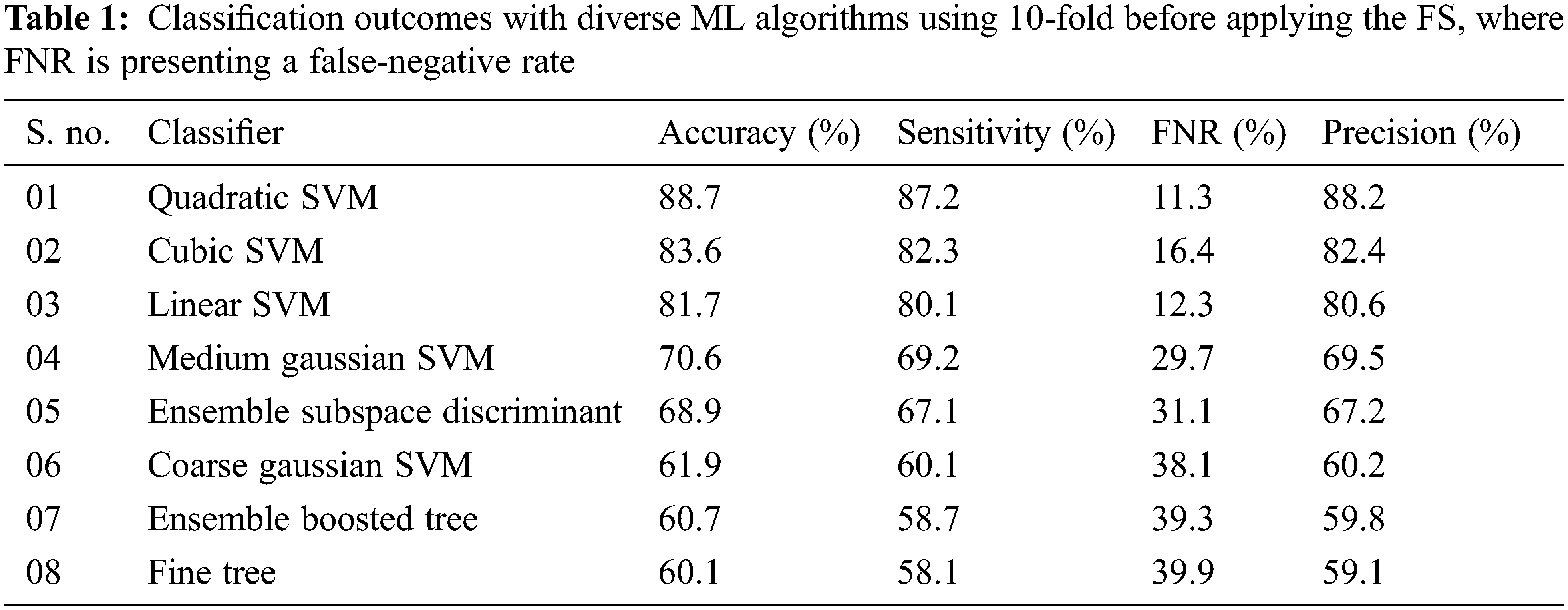
In the second scenario, the results are generated by using a 5-fold validation strategy where data is divided into five different parts. The results of using the 5-fold method are presented in Tab. 2. The area plot comparison of both folds is presented in Fig. 5. The comparison results show that the 10-fold validation method achieved better performance as compared to the 5-fold validation.
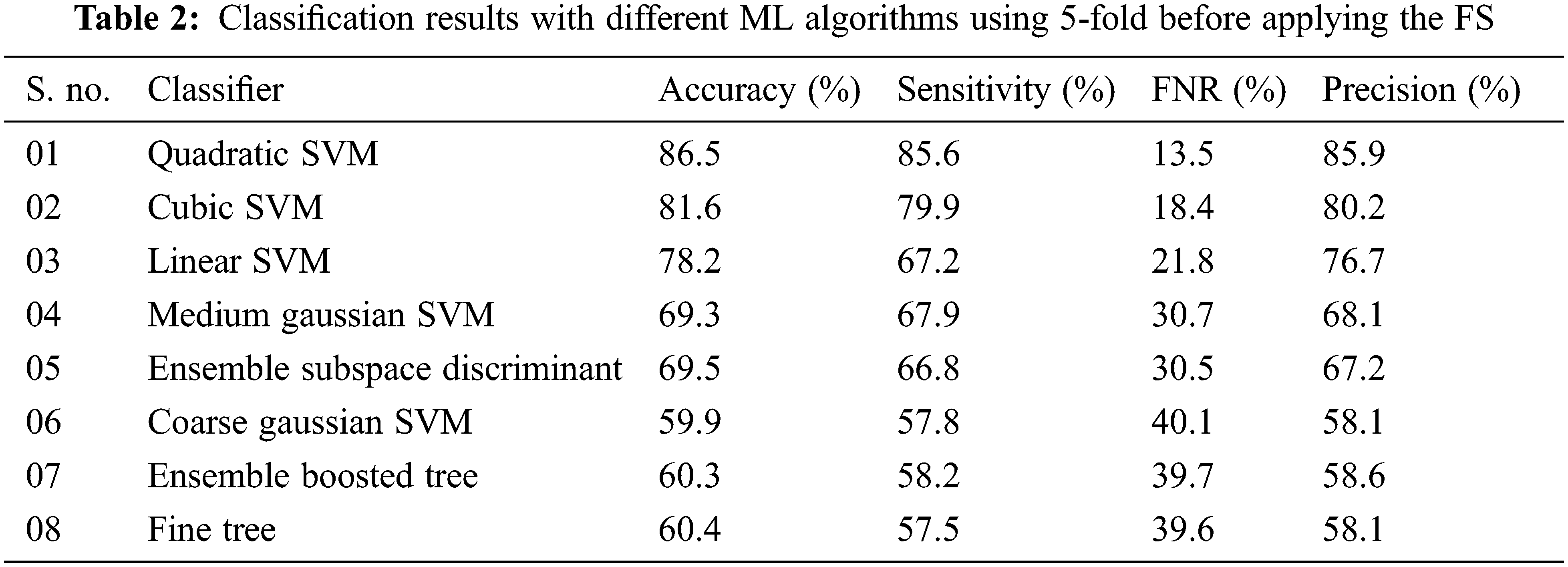
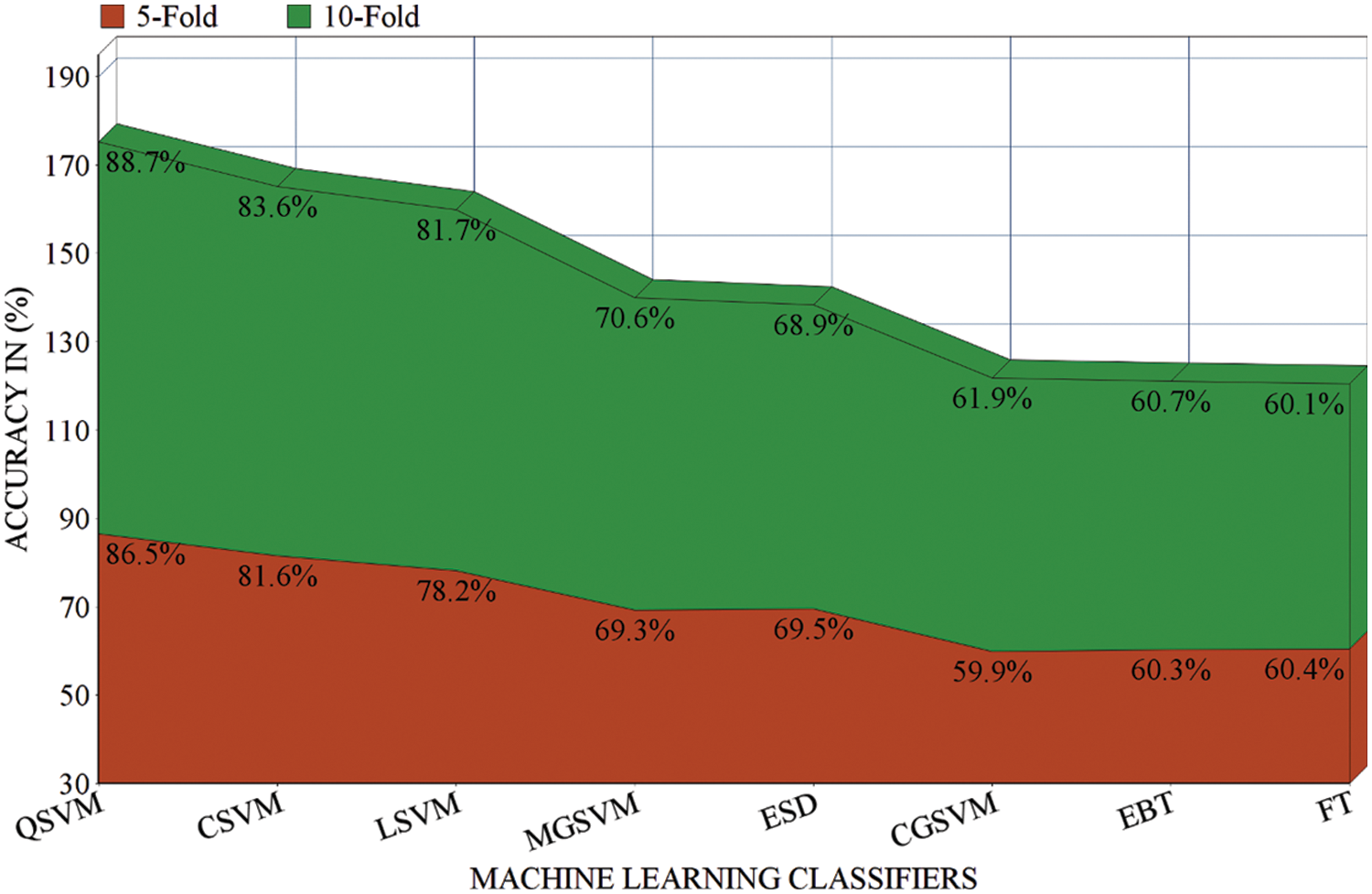
Figure 5: Results comparison of different ML classifiers before applying the FS in terms of 5-fold and 10-fold
Experiment 2: In this experiment, the proposed DL network is utilized for feature extraction purposes by using a simple transfer learning concept. After successful extraction of the deep features the extracted FV is passed through the proposed FS framework as a result, the top N × 1000 features are selected for further processing. This labeled FV is inputted into the ML algorithms for the IDC classification task. Results by using a 10-fold methodology are given in Tab. 3.
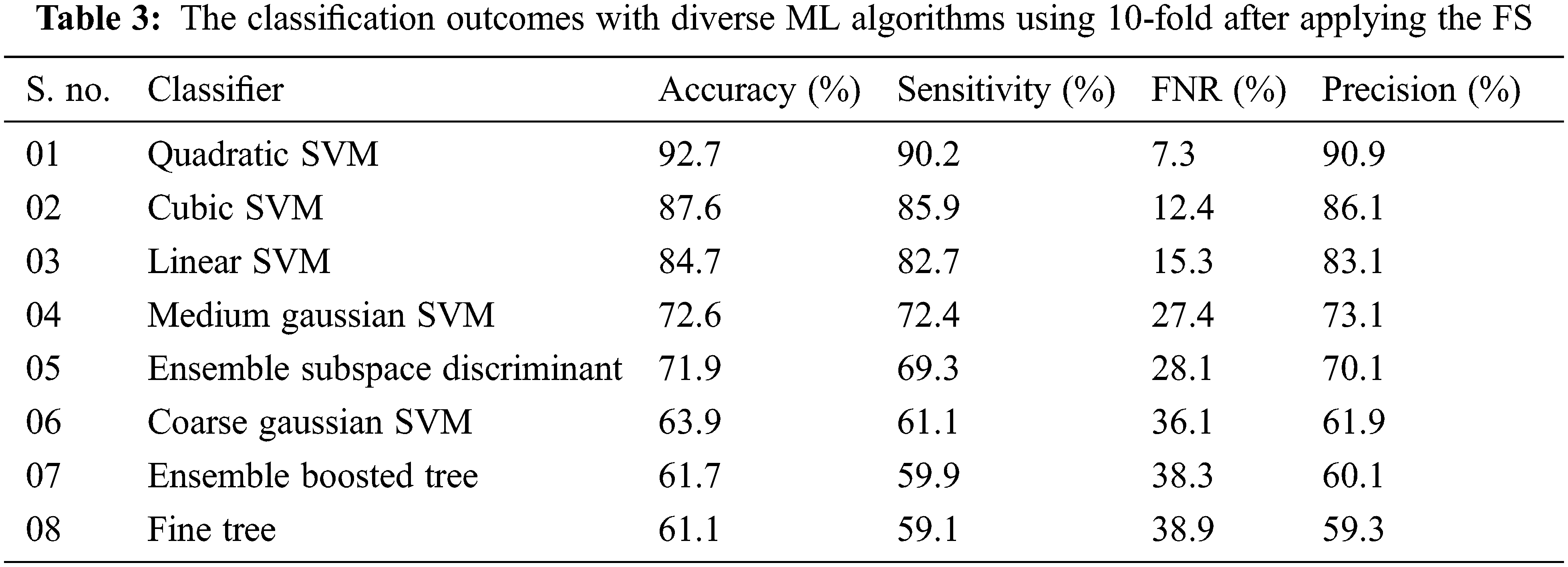
For performance evaluation average accuracy, FNR, sensitivity, and average precision are calculated. With 10-Fold cross-validation (CV) the highest average accuracy of 92.7% is attained on the quadratic SVM classifier and the worst average accuracy classification is achieved on the fine tree with an accuracy of 61.1% while the cubic SVM gives the average validation accuracy of 87.6%. The accuracy of the ML classifiers including LSVM, MGSVM, CGSVM, EBT, and ESD is 84.7%, 72.6%, 63.9%, 61.7%, and 71.9% respectively. The FNR of each classifier such that LSVM, CSVM, QSVM, MGSVM, CGSVM, FT, EBT, and ESD is calculated as 15.3%, 12.4%, 7.3%, 27.7%, 36.1%, 38.9%, 38.3%, and 28.1% respectively. Precision and training time in second for each classifier is also calculated the model training time in second of each classier with a 10-fold cross-validation method in 8285, 31687, 3160, 3197, 26.77, 203, 4314, and 642 s respectively. In the 10-CV method highest accuracy is achieved by the quadratic SVM classifier and the lowest training time is observed on the FT classifier. In the second scenario testing of the proposed methodology is performed by using a 5-fold procedure where data is randomly split into five chunks first four chunks are used for training purposes and the last one is used for testing in the first fold. Similarly, at the end of the last fold, whole data was utilized for training as well as testing purposes and average accuracy is calculated. The results by using 5-fold-CV for IDC classification are tabulated in Tab. 4.
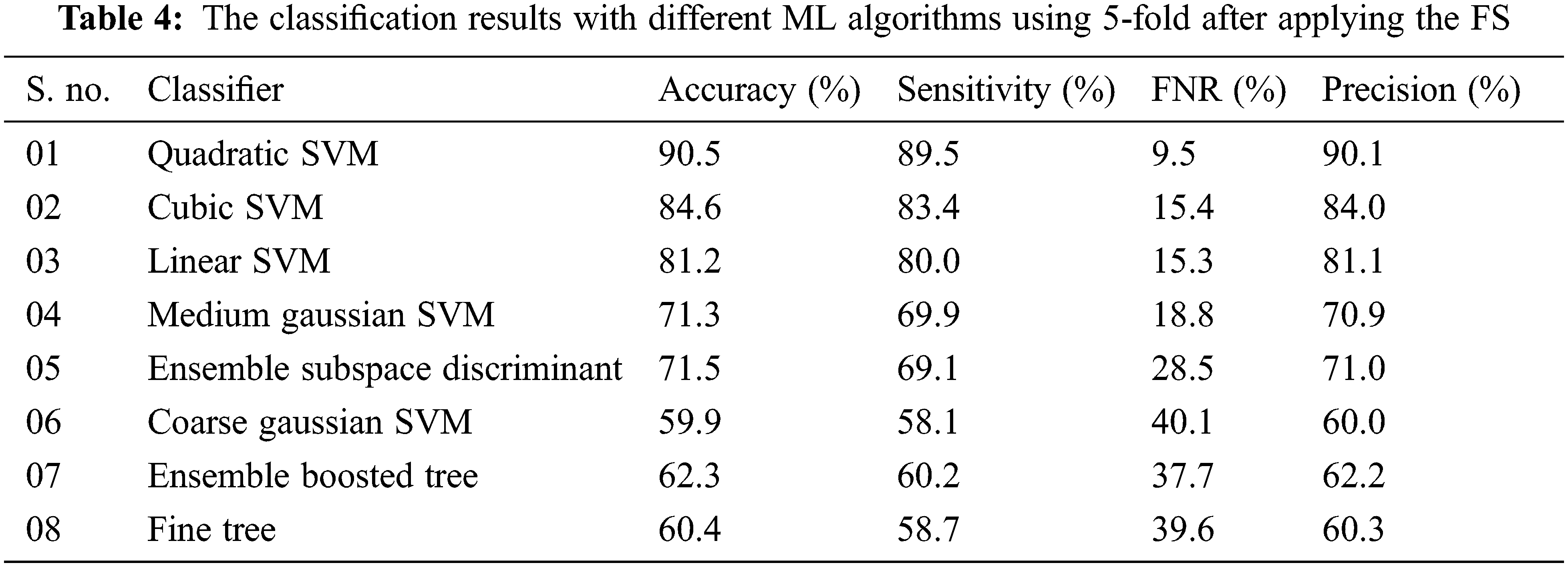
The highest accuracy of 90.5% was achieved in a 5-fold validation procedure with the QSVM algorithm which takes 1065 s in the training process while FNR and precision are reported as 9.5% and 90.1% respectively. The CSVM classifier gives the second-highest accuracy that is 84.6% and the worst accuracy of IDC classification was achieved by the CGSVM classifier which gives an accuracy of 59.9%. The precision score for each classifier including LSVM, CSVM, QSVM, MGSVM, CGSVM, FT, EBT, and ESD are also computed that are 81.1%, 84.0%, 90.1%, 70.9%, 60.0%, 60.3%, 62.2%, and 70.1% respectively. The contribution of the proposed work in the form of results compared with existing approaches is also carried out. Wold et al. [44] proposed a CNN for IDC classification and reported an accuracy of 89% while Yadavendra and Chand [45] proposed different methods for BC classification and reported an accuracy of 90% with 88% precision. Recently Alanazi et al. [46], Zhang et al. [47], Roy et al. [48], and Yang et al. [42] proposed different studies for IDC classification and reported 87%, 87%, and 92% accuracies respectively. In the proposed methodology for IDC breast cancer classification, the highest accuracy index of 92.7% is achieved, a detailed comparison of our architecture with different existing methodologies in literature is presented in Tab. 5. A comparison of both folds is also performed with the observation that in the case of the 10-fold procedure the accuracy index is higher than the other but the classifiers training time is also higher as compared to the 5-fold cross-validation method, bar chart comparison of accuracies of both folded cross-validation is presented in Fig. 6. A line graph comparison of all the classifiers before and after implementation of selection with both folds is presented in Fig. 7.
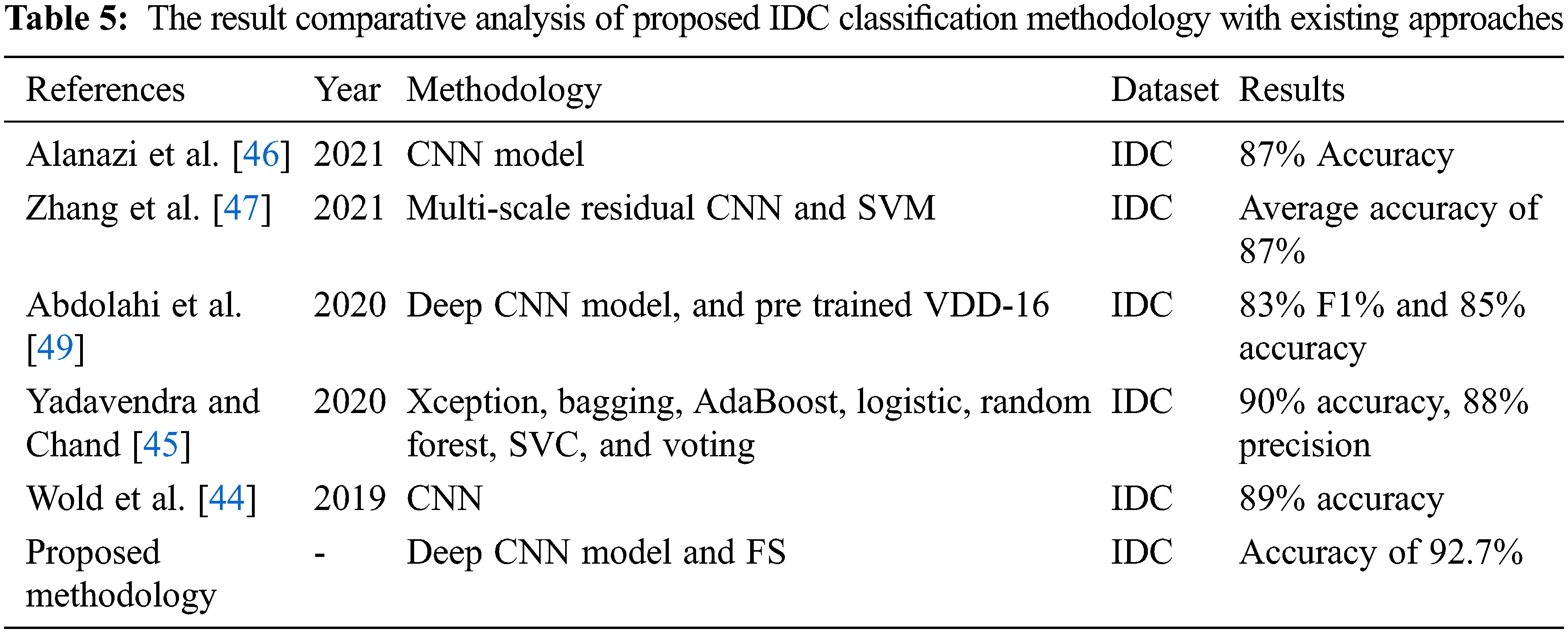
Figure 6: Results comparison of different ML classifiers after applying FS in terms of 5-fold and 10-fold
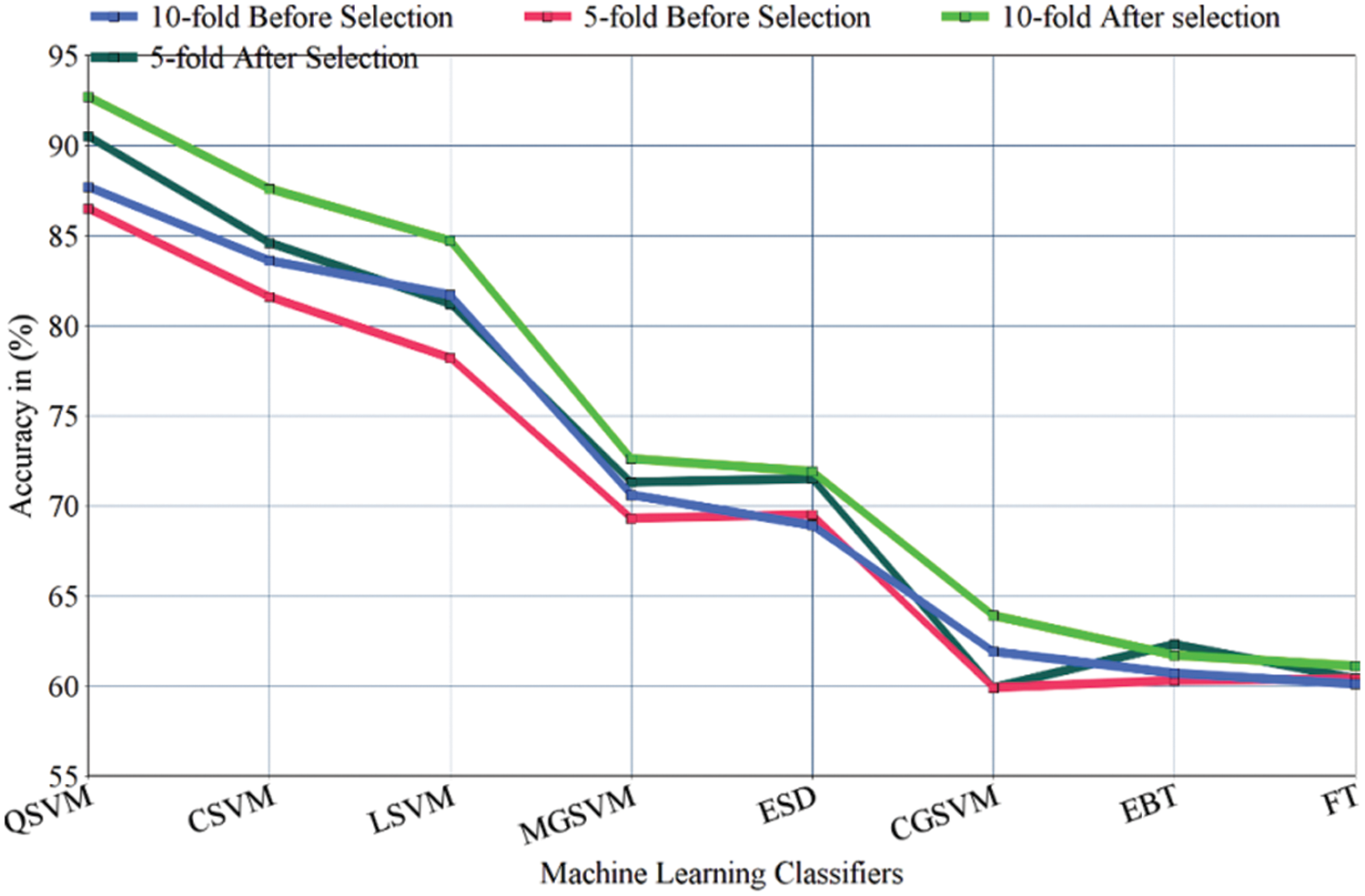
Figure 7: A line graph comparison of ML classifiers before and after implementation of FS with 5&10-folds
The detailed analysis and discussion of the proposed methodology are presented in this section. Breast cancer detection in IDC form is a promising task many techniques have been proposed for IDC classification as was described above in the introduction and literature review sections of this paper. This study proposed a novel hybrid features level fusion-based technique to resolve the problem of IDC classification. The features selection framework for best FS is also applied for better performance of the classifiers. In this work, two different experiments were performed by using eight ML classifiers including LSVM, CSVM, QSVM, MGSVM, CGSVM, FT, EBT, and ESD classifiers for IDC classification with 5-fold and 10-fold cross-validation methods. IDC histopathology images are classified into IDC positive and IDC negative classes by using the proposed classification methodology. In the first experiment, the results were calculated before applying the FS method in this setup proposed method achieved 88.7% accuracy with 10-fold implementation. In the second evaluation test of the proposed method, the results were computed after applying the proposed FS framework and attained 92.7% accuracy by using 10-fold validation and 90.5% with a 5-fold validation procedure, in both folds, the highest accuracy is achieved with QSVM classifiers. The second highest accuracy was achieved on the CSVM classifier which is 87.6% and 84.6% by 10-fold and 5-fold cross-validation methods respectively. Experimental outcomes infer that the highest average accuracy is produced by using the proposed FS framework. A comparison of feature level fusion-based proposed methodology with existing methodologies is also carried out and reported in Tab. 5. Experimental outcomes concluded that this work performed better and achieved 92.7% accuracy hence can successfully be utilized for the early and accurate diagnosis of BC. This study proposed BC detection from histopathology images. Some limitations of this work are presented in this section. This work only utilized the deep features for the classification task, classical feature fusion with deep features can also be applied for performance enhancement. This study did not introduce any data augmentation method or pre-processing step. In the future, this work may be extended to utilize the different pre-processing steps with a fusion of features from different domains to get higher accuracy.
This study presented a deep CNN model and FS framework for BC detection and classification. After pre-training of the proposed network, the trained CNN network was further employed as the feature pullout process of the IDC classification problem. The deep features collection process is carried out by simple transfer learning. For FS, a novel hybrid features level fusion-based selection framework for BC detection from histopathology images is proposed. In the proposed methodology after successful deep feature extraction. The extracted FV is then supplied to the proposed FS framework for the best FS. Finally, the labeled selected FV is passed to the different ML classifiers for the classification of IDC tumors. The validation results presented that our methodology achieved the highest accuracy of 92.7%. From the discussion of all the above outcomes, it can be summed up that the proposed hybrid feature level fusion selection method selected the robust features for BC classification. In the future, a new method will be adopted to develop an intelligent CAD system for IDC classification by implementing the latest CNN models like Dense Net and Capsule Net with different features fusion and selection methods.
Funding Statement: This work was supported by the “Human Resources Program in Energy Technology” of the Korea Institute of Energy Technology Evaluation and Planning (KETEP), granted financial resources from the Ministry of Trade, Industry & Energy, Republic of Korea. (No. 20204010600090).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. K. Jabeen, M. Alhaisoni, U. Tariq, Y. D. Zhang and A. Hamza, “Breast cancer classification from ultrasound images using probability-based optimal deep learning feature fusion,” Sensors, vol. 22, no. 4, pp. 807, 2022. [Google Scholar]
2. S. Zahoor, I. U. Lali, K. Javed and W. Mehmood, “Breast cancer detection and classification using traditional computer vision techniques: A comprehensive review,” Current Medical Imaging, vol. 16, no. 2, pp. 1187–1200, 2020. [Google Scholar]
3. R. L. Siegel, K. D. Miller and A. Jemal, “Cancer statistics, 2017,” CA: A Cancer Journal for Clinicians, vol. 67, no. 4, pp. 7–30, 2017. [Google Scholar]
4. A. J. White, “Invited perspective: Air pollution and breast cancer risk: Current state of the evidence and next steps,” Environmental Health Perspectives, vol. 129, no. 6, pp. 051302, 2021. [Google Scholar]
5. K. Muhammad, M. Sharif, T. Akram and S. Kadry, “Intelligent fusion-assisted skin lesion localization and classification for smart healthcare,” Neural Computing and Applications, vol. 11, no. 2, pp. 1–16, 2021. [Google Scholar]
6. M. Alhaisoni, U. Tariq, N. Hussain, A. Majid and R. Damaševičius, “COVID-19 case recognition from chest CT images by deep learning, entropy-controlled firefly optimization, and parallel feature fusion,” Sensors, vol. 21, no. 3, pp. 7286, 2021. [Google Scholar]
7. K. S. Manic, R. Biju, W. Patel and S. Uma, “Extraction and evaluation of corpus callosum from 2D brain MRI slice: A study with cuckoo search algorithm,” Computational and Mathematical Methods in Medicine, vol. 2021, no. 3, pp. 1–18, 2021. [Google Scholar]
8. L. He, L. R. Long, S. Antani and G. R. Thoma, “Histology image analysis for carcinoma detection and grading,” Computer Methods and Programs in Biomedicine, vol. 107, no. 4, pp. 538–556, 2012. [Google Scholar]
9. J. Bai, R. Posner, T. Wang, C. Yang and S. Nabavi, “Applying deep learning in digital breast tomosynthesis for automatic breast cancer detection: A review,” Medical Image Analysis, vol. 71, no. 4, pp. 102049, 2021. [Google Scholar]
10. I. Ashraf, M. Alhaisoni, R. Damaševičius, R. Scherer and A. Rehman, “Multimodal brain tumor classification using deep learning and robust feature selection: A machine learning application for radiologists,” Diagnostics, vol. 10, no. 2, pp. 565, 2020. [Google Scholar]
11. M. J. Borst and J. A. Ingold, “Metastatic patterns of invasive lobular versus invasive ductal carcinoma of the breast,” Surgery, vol. 114, no. 21, pp. 637–642, 1993. [Google Scholar]
12. X. Zhang, X. Sun, W. Sun, T. Xu and P. Wang, “Deformation expression of soft tissue based on BP neural network,” Intelligent Automation, vol. 32, no. 3, pp. 1041–1053, 2022. [Google Scholar]
13. M. J. Umer, J. Amin, M. Sharif, M. A. Anjum and F. Azam, “An integrated framework for COVID-19 classification based on classical and quantum transfer learning from a chest radiograph,” Concurrency Computation: Practice Experience, vol. 4, no. 1, pp. e6434, 2021. [Google Scholar]
14. M. Nawaz, T. Nazir, A. Javed, U. Tariq and H. S. Yong, “An efficient deep learning approach to automatic glaucoma detection using optic disc and optic cup localization,” Sensors, vol. 22, no. 1, pp. 434, 2022. [Google Scholar]
15. P. Wang, X. Hu, Y. Li, Q. Liu and X. Zhu, “Automatic cell nuclei segmentation and classification of breast cancer histopathology images,” Signal Processing, vol. 122, no. 26, pp. 1–13, 2016. [Google Scholar]
16. Z. Jan, S. U. Khan, N. Islam, M. A. Ansari and B. Baloch, “Automated detection of malignant cells based on structural analysis and naive Bayes classifier,” Sindh University Research Journal-SURJ (Science Series), vol. 48, pp. 1–13, 2016. [Google Scholar]
17. P. Filipczuk, T. Fevens, A. Krzyzak and R. Monczak, “Computer-aided breast cancer diagnosis based on the analysis of cytological images of fine needle biopsies,” IEEE Transactions on Medical Imaging, vol. 32, no. 21, pp. 2169–2178, 2013. [Google Scholar]
18. A. Cruz-Roa, A. Basavanhally, F. González, H. Gilmore and S. Ganesan, “Automatic detection of invasive ductal carcinoma in whole slide images with convolutional neural networks,” Methods, vol. 14, no. 2, pp. 1–9, 2014. [Google Scholar]
19. M. N. Q. Bhuiyan, M. Shamsujjoha, S. H. Ripon, F. H. Proma and F. Khan, “Transfer learning and supervised classifier based prediction model for breast cancer,” in Big Data Analytics for Intelligent Healthcare Management, NY, USA: Elsevier, 2019, pp. 59–86. [Google Scholar]
20. M. M. Dundar, S. Badve, G. Bilgin, V. Raykar and R. Jain, “Computerized classification of intraductal breast lesions using histopathological images,” IEEE Transactions on Biomedical Engineering, vol. 58, no. 13, pp. 1977–1984, 2011. [Google Scholar]
21. P. Král and L. Lenc, “LBP features for breast cancer detection,” in 2016 IEEE Int. Conf. on Image Processing (ICIP), New Delhi, India, 2016, pp. 2643–2647. [Google Scholar]
22. S. S. Yasiran, S. Salleh and R. Mahmud, “Haralick texture and invariant moments features for breast cancer classification,” Methods, vol. 3, no. 5, pp. 020022, 2016. [Google Scholar]
23. S. I. Niwas, P. Palanisamy, W. J. Zhang, N. A. M. Isa and R. Chibbar, “Log-gabor wavelets based breast carcinoma classification using least square support vector machine,” Methods, vol. 3, no. 5, pp. 219–223, 2011. [Google Scholar]
24. Y. M. George, H. H. Zayed, M. I. Roushdy and B. M. Elbagoury, “Remote computer-aided breast cancer detection and diagnosis system based on cytological images,” IEEE Systems Journal, vol. 8, no. 3, pp. 949–964, 2014. [Google Scholar]
25. K. K. Shukla, A. Tiwari and S. Sharma, “Classification of histopathological images of breast cancerous and non cancerous cells based on morphological features,” Biomedical and Pharmacology Journal, vol. 10, no. 2, pp. 353–366, 2017. [Google Scholar]
26. B. N. Narayanan, V. Krishnaraja and R. Ali, “Convolutional neural network for classification of histopathology images for breast cancer detection,” Sensors, vol. 19, no. 2, pp. 291–295, 2019. [Google Scholar]
27. T. G. Debelee, M. Amirian, A. Ibenthal, G. Palm and F. Schwenker, “Classification of mammograms using convolutional neural network based feature extraction,” Sensors, vol. 19, no. 2, pp. 89–98, 2017. [Google Scholar]
28. T. G. Debelee, A. Gebreselasie, F. Schwenker, M. Amirian and D. Yohannes, “Classification of mammograms using texture and cnn based extracted features,” Journal of Biomimetics, Biomaterials and Biomedical Engineering, vol. 42, pp. 79–97, 2019. [Google Scholar]
29. A. M. Romano and A. A. Hernandez, “Enhanced deep learning approach for predicting invasive ductal carcinoma from histopathology images,” Sensors, vol. 19, no. 2, pp. 142–148, 2019. [Google Scholar]
30. M. J. U. Rahman, R. I. Sultan, F. Mahmud, S. Al Ahsan and A. Matin, “Automatic system for detecting invasive ductal carcinoma using convolutional neural networks,” Applied Sciences, vol. 11, no. 4, pp. 0673–0678, 2018. [Google Scholar]
31. J. L. Wang, A. K. Ibrahim, H. Zhuang, A. M. Ali and A. Y. Li, “A study on automatic detection of IDC breast cancer with convolutional neural networks,” Sensors, vol. 12, no. 5, pp. 703–708, 2018. [Google Scholar]
32. R. Sanyal, M. Jethanandani and R. Sarkar, “DAN: Breast cancer classification from high-resolution histology images using deep attention network,” in Innovations in Computational Intelligence and Computer Vision, Toranto, Canada: Springer, 2021, pp. 319–326. [Google Scholar]
33. R. Sanyal, D. Kar and R. Sarkar, “Carcinoma type classification from high-resolution breast microscopy images using a hybrid ensemble of deep convolutional features and gradient boosting trees classifiers,” IEEE/ACM Transactions on Computational Biology and Bioinformatics, vol. 21, no. 2, 2021. [Google Scholar]
34. H. Chapala and B. Sujatha, “ResNet: Detection of invasive ductal carcinoma in breast histopathology images using deep learning,” Journal of Biomimetics, Biomaterials and Biomedical Engineering, vol. 12, no. 4, pp. 60–67, 2020. [Google Scholar]
35. B. Mughal, M. Sharif and N. Muhammad, “A novel classification scheme to decline the mortality rate among women due to breast tumor,” Microscopy Research Technique, vol. 81, no. 6, pp. 171–180, 2018. [Google Scholar]
36. B. Mughal and M. Sharif, “Bi-model processing for early detection of breast tumor in CAD system,” The European Physical Journal Plus, vol. 132, no. 7, pp. 1–14, 2017. [Google Scholar]
37. M. J. Umer, M. Sharif and S. Kadry, “Multi-class classification of breast cancer using 6B-net with deep feature fusion and selection method,” Journal of Personalized Medicine, vol. 12, no. 6, pp. 683, 2022. [Google Scholar]
38. M. Yasmin and M. Sharif, “Survey paper on diagnosis of breast cancer using image processing techniques,” Science, vol. 2277, no. 3, pp. 2502, 2013. [Google Scholar]
39. M. A. Khan, Y. D. Zhang, S. A. Khan, M. Attique and S. Seo, “A resource conscious human action recognition framework using 26-layered deep convolutional neural network,” Multimedia Tools and Applications, vol. 21, no. 5, pp. 1–23, 2020. [Google Scholar]
40. P. Chen, F. Li and C. Wu, “Research on intrusion detection method based on Pearson correlation coefficient feature selection algorithm,” Journal of Physics: Conference Series, vol. 1757, no. 9, pp. 012054, 2021. [Google Scholar]
41. R. J. Urbanowicz, M. Meeker, W. La Cava and R. S. Olson, “Relief-based feature selection: Introduction and review,” Journal of Biomedical Informatics, vol. 85, no. 4, pp. 189–203, 2018. [Google Scholar]
42. J. Yang, D. Zhang and A. F. Frangi, “Two-dimensional PCA: A new approach to appearance-based face representation and recognition,” IEEE Transactions on Pattern Analysis Machine Intelligence, vol. 26, no. 5, pp. 131–137, 2004. [Google Scholar]
43. A. Aby, “Breast histopathology images,” Open Source, vol. 1, no. 1, pp. 1–1, 2022. [Google Scholar]
44. S. Wold, P. Geladi, K. Esbensen and J. Öhman, “Multi-way principal components-and PLS-analysis,” Journal of Chemometrics, vol. 1, no. 2, pp. 41–56, 1987. [Google Scholar]
45. Yadavendra and S. Chand, “A comparative study of breast cancer tumor classification by classical machine learning methods and deep learning method,” Machine Vision and Applications, vol. 31, no. 11, pp. 46, 2020. [Google Scholar]
46. S. A. Alanazi, M. M. Kamruzzaman, M. N. Islam Sarker, M. Alruwaili and N. Alshammari, “Boosting breast cancer detection using convolutional neural network,” Journal of Healthcare Engineering, vol. 2021, no. 3, pp. 1–11, 2021. [Google Scholar]
47. J. Zhang, X. Guo, B. Wang and W. Cui, “Automatic detection of invasive ductal carcinoma based on the fusion of multi-scale residual convolutional neural network and SVM,” IEEE Access, vol. 9, no. 5, pp. 40308–40317, 2021. [Google Scholar]
48. S. D. Roy, S. Das, D. Kar, F. Schwenker and R. Sarkar, “Computer aided breast cancer detection using ensembling of texture and statistical image features,” Sensors, vol. 21, no. 13, pp. 3628, 2021. [Google Scholar]
49. M. Abdolahi, M. Salehi, I. Shokatian and R. Reiazi, “Artificial intelligence in automatic classification of invasive ductal carcinoma breast cancer in digital pathology images,” Medical Journal of the Islamic Republic of Iran, vol. 56, no. 2, 2020. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools