DOI:10.32604/csse.2022.021812

| Computer Systems Science & Engineering DOI:10.32604/csse.2022.021812 |  |
| Article |
X-ray Image-Based COVID-19 Patient Detection Using Machine Learning-Based Techniques
1Department of Information Technology, College of Computer, Qassim University, Buraidah, 52571, Saudi Arabia
2Department of Electrical Engineering, College of Engineering & Information Technology, Onaizah Colleges, Onaizah, 51911, Saudi Arabia
3Digital Image Processing Lab, Islamia College, Peshawar, 25120, KPK, Pakistan
4Department of Computer Science, College of Computer and Information Sciences, Prince Sultan University, Riyadh, 11586, Saudi Arabia
5Department of Electrical Engineering, National University of Computer and Emerging Sciences, Peshawar, Pakistan
*Corresponding Author: Muhammad Islam. Email: m.islam@oc.edu.sa
Received: 15 July 2021; Accepted: 26 September 2021
Abstract: In early December 2019, the city of Wuhan, China, reported an outbreak of coronavirus disease (COVID-19), caused by a novel severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). On January 30, 2020, the World Health Organization (WHO) declared the outbreak a global pandemic crisis. In the face of the COVID-19 pandemic, the most important step has been the effective diagnosis and monitoring of infected patients. Identifying COVID-19 using Machine Learning (ML) technologies can help the health care unit through assistive diagnostic suggestions, which can reduce the health unit's burden to a certain extent. This paper investigates the possibilities of ML techniques in identifying/detecting COVID-19 patients including both conventional and exploring from chest X-ray images the effect of viral infection. This approach includes pre-processing, feature extraction, and classification. However, the features are extracted using the Histogram of Oriented (HOG) and Local Binary Pattern (LBP) feature descriptors. Furthermore, for the extracted features classification, six ML models of Support Vector Machine (SVM) and K-Nearest Neighbor (KNN) is used. Experimental results show that the diagnostic accuracy of random forest classifier (RFC) on extracted HOG plusLBP features is as high as 94% followed by SVM at 93%. The sensitivity of the K-nearest neighbour model has reached an accuracy of 88%. Overall, the predicted approach has shown higher classification accuracy and effective diagnostic performance. It is a highly useful tool for clinical practitioners and radiologists to help them in diagnosing and tracking the cases of COVID-19.
Keywords: Image pre-processing; detection; classification; x-ray images; filter
Since the beginning of the COVID-19 pandemic and its spread around the world, several researchers from different research fields have participated in order to accelerate the recovery of the infected and the safety of the uninfected humans from the Coronavirus. Yadav et al. [1] have proposed a novel framework for classifying different coronaviruses using Support Vector Regression (SVR) to perform different tasks. These tasks include detecting transmission, the growth rate of COVID-19, advising the end of its spread, the impact of weather on the coronavirus, analysing the transmission rate of the virus, and comparing the performance of SVR with others such as Linear regression and polynomial regression models. On the basis of model accuracy comparison, they have found SVR to be the best model.
Though related to SARS and Middle East respiratory syndrome (MERS), COVID-19 shows some peculiar features which, to date, are not completely understood. However, the fatality rate of COVID-19 of 2.12% is lower than that of SARS (9.5%) and much lower than that of MERS (34.4%) [2]. To detect from chest X-ray images using a machine learning model, Mahdy et al. [3] have proposed a machine learning-based framework to classify patients infected with SARS and MERS. They have used a machine learning model referred to as Support Vector Machine (SVM) for classification. The input image size of their model is 512 × 512 in JPEG format. They have achieved 97.48% overall accuracy using their trained model for SARS. The contagious coronavirus due to COVID-19 is spreading rapidly among humans and causing severe acute respiratory illness in humans. However, to identify an infected patient with symptoms, Wu et al. [4] have proposed a method to accurately identify symptoms from Computerised Tomography (CT) X-ray images of coronavirus. They collected 253 samples from different sources of COVID-19 infected patients. Their proposed work achieved 95% accuracy in identifying infected patients. There are several different symptoms of COVID-19 but the most common include fever, shortness of breath, and cough.
Statistically, according to the World Health Organization (WHO), there have been 200,840,180 confirmed cases globally of COVID-19, including 4,265,903 deaths reported with 3,984,596,440 vaccine doses administered as of 8th August 2021. Furthermore, due to the massive spreading of the pandemic in many developed and developing countries, the health systems have since registered an increase in demand for special care. Cases of the COVID-19 pandemic cases are constantly reported across the globe [5]. The total statistics regarding confirmed COVID-19 cases, including deaths by regions, are as shown in Fig. 1.
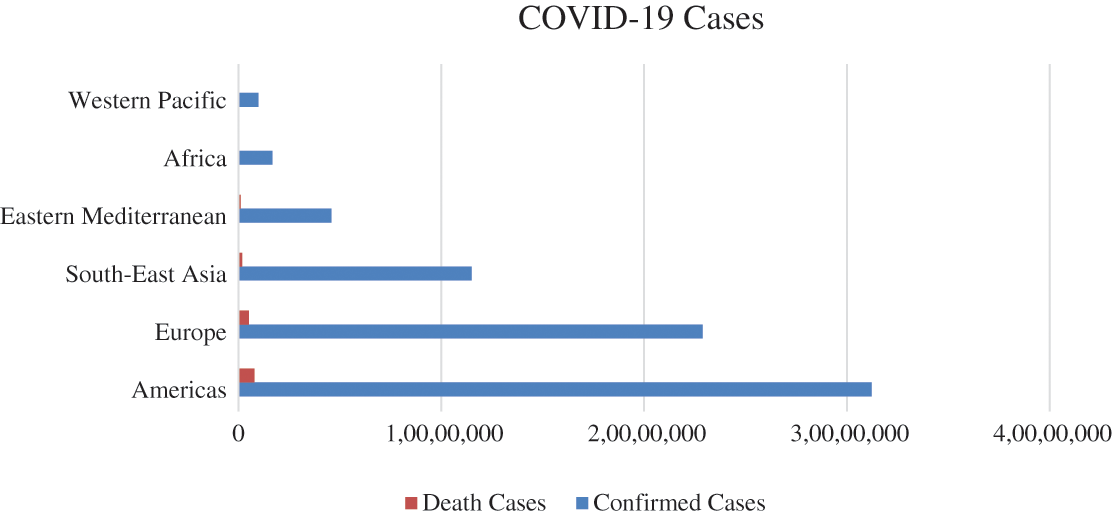
Figure 1: COVID-19 pandemic region-wise confirmed and death cases
The spread of coronavirus (COVID-19) increased exponentially from human to human outside China. The highest number of cases are reported from the United States of America (USA), India, Brazil, Russia, France, United Kingdom (UK), Italy, Spain, Argentina, Colombia, Germany, and Mexico. According to the latest statistics on COVID-19, the USA currently has the largest number of confirmed cases; that is, 16,446,844 and this number is continuing to rise. On the other hand, WHO has confirmed that the increase in death ratio of 301,536 cases up to date (17th December 2020). All the reported are confirmed, and the death cases of different countries are shown in Fig. 2.
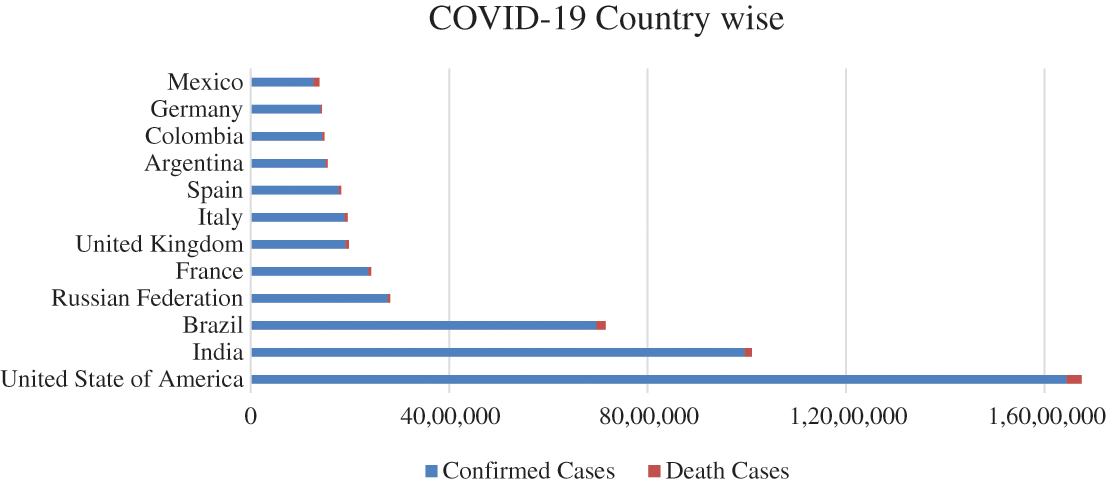
Figure 2: COVID-19 pandemic country-wise conformed and death cases
Most of the symptoms of COVID-19 include fever, sore throat, dry cough, shortness of breath, muscle aches and pains, tiredness, diarrhoea, and nausea as per the WHO information report [5]. It appears to be the careless nature of humans that is causing the spread of the current pandemic all over the world. The WHO has recommended that all people protect/secure themselves from the COVID-19 pandemic by using sanitiser and washing their hands every 20 min, wearing a face mask, maintaining a social distance of at least two metres, especially from others in the vicinity irrespective of the sneezing and coughing symptoms [5].
Research studies have shown that the use of imaging technology (X-ray or CT) to diagnose and screen COVID-19 has higher sensitivity and it may be used as a more accurate alternative to RT-PCR [6]. Usually, CT imaging takes longer than X-ray imaging, and a period of X-ray imaging can considerably accelerate the speed of malady screening.
However, some developing countries may not have the capacity or ability to secure adequate high-quality CT imaging scanners. Therefore, X-ray instrumentality, with its low effective cost and easy operability, is an ideal option. Moreover, most clinics and medical establishments have deployed X-ray equipment as necessary imaging equipment. Compared to CT imaging, X-ray imaging is the most common and the most widely used diagnostic imaging technology playing an important role in clinical nursing and epidemiologic analysis [7]. Thus, this study has designated the chest X-ray pictures for analysis in this paper. However, radiologists and consultants chiefly interpret images supported by personal clinical expertise once analysing X-ray images [8]. Some images are hard to explore for identifying features due to image quality, scanner quality or environmental conditions. Such images aren't entirely consistent, because of their inconclusive clear conclusions. Additionally, the work of the interpretation of images is vast, and doctors are susceptible to misdiagnosis. Therefore, there is the need for an imperative Artificial Neural Network (ANN) approach within a computer-aided designation system to aid radiologists in interpreting the images more rapidly and accurately.
To identify the infected human from the symptoms Özkaya et al. [9] have proposed a framework where they have extracted features from the subset-1 and subset-2 datasets using a deep learning technique called Convolution Neural Network (CNN), and for the classifications of these features, machine learning model SVM is used. The proposed model has achieved almost 95.60% accuracy on subset-1% and 98.27% accuracy on the subset-2 dataset.
To prevent the COVID-19 from spreading around the globe requires its early-stage detection and diagnosis of infected patients. Abbas et al. [10] have proposed the technique called Transfer and Compose (TraC) instead of using the CNN model. The authors have classified normal and infected persons by using X-ray chest images achieving around 95.12% accuracy in consequence.
Up to date (17th December 2020), there has been no drug or vaccine fully available to the public for the COVID-19 pandemic. However, to identify an infected coronavirus patient in real-time Al-Karawi et al. [11] have proposed a machine learning-based model for detecting an infected person from CT scans. They have used an adaptive winner filter as a pre-processing step for noise reduction, Fast Fourier Transform (FFT) spectrum is used to extract identification features, and SVM is used to classify features extracted from COVID-19 CT scan images. To provide the result in real-time, an FFT-Gabor scheme is used. The proposed model has achieved almost 95.37% accuracy, 95.99% sensitivity, and 94.76% specificity.
According to WHO, X-ray and CT images are two different imaging modalities that can be considered to detect and identify COVID-19. To classify and segment the infected areas, Wan et al. [12] have introduced the Inception Recurrent Residual Convolutional Neural Network (IRRCNN) and NABLA-3 network models for segmentation and classification of these images. The proposed algorithms on X-ray and CT images have achieved 84.6% and 98.78% accuracy, respectively. According to WHO, quarantine and appropriate treatment are the most suitable ways to contain the virus from spreading along with treating the affected patients. To detect COVID-19 using chest X-ray images, Alqudah et al. [13] have proposed an Artificial intelligence (AI) based method in which the authors have used an SVM-based RFC, ML-model of using CNN to classify images. They have used CNN to extract graphical features and categorised these features using the SoftMax tool. The proposed method has achieved a higher accuracy of 95.2% than its counterpart.
According to WHO, the human fatality rate from the COVID-19 pandemic is almost 2.12% with the US leading the entire world, while Africa with the countries of the Western Pacific is still at a minimum. However, it has required extensive health care and quick detection based on symptoms to protect people. For the classification of patients based on symptoms, Sun et al. [14] have proposed a machine-learning-based method in which they collected 220 clinical symptoms records and classified them into age, GSH, CD3, and protein using SVM. The overall accuracy of their proposed model is 77.5% and the specificity is 78.4%. Further, the area under the receiver operating characteristic (AUROC) has an accuracy of 99.96% on the training and 93.33% on the test dataset. On the other hand, to identify COVID-19 using X-ray images, the authors in [15] have proposed a framework called deep learning in which they have used restnet50 and SVM to classify the images. They have also used the Kaggle chest X-ray images dataset that achieved 95% accuracy. Furthermore, to detect the pandemic virus through radiography and radiology images. Minaee et al. [16] have proposed deep learning techniques to detect the pandemic virus using Chest X-ray images. The authors have used four pre-trained deep learning models of ResNet-18. ResNet-50, Squeeze Net, and DencNet-121. They have combined two datasets of Chest X-ray images in which to include nearly 5000 chest X-ray images. The highest accuracy achieved is 97% sensitivity and 90% specificity with all the pre-trained models. Furthermore, COVID-19 can be quickly detected by performing a rapid COVID-19 polymerase chain reaction (PCR) test. Rapid virus detection using the chest X-ray images, Tartaglione et al. [17] have proposed a deep learning framework for virus detection using image samples during testing. They have collected the CORDA, RSNA, COVID-Chest X-ray, Chest X-ray datasets from the Northern Italy medical centre for model training. However, to classify the images of the collected dataset, they have used Resnet-50 and ResNet-18 to achieve 85% and 100% accuracy respectively in the X-ray chest images dataset. To distinguish between pneumonia and coronavirus, Wenqi et al. [18] have proposed a deep learning model to identify normal, COVID-19-infected and pneumonia-affected individuals from chest X-ray images. Layer-wise Relevance Propagation (LRP) and Gradient-Guided Class Activation Maps (Grad-CAM++) are used as pre-processing techniques to highlight the affected regions in the X-ray images. They have trained a DNN on the pre-processed data with an achievement of 89.1% as positive expectancy and 83% accuracy for recall.
For the early diagnosis of COVID-19 patients, Farid et al. [19] have proposed a method that uses a four-image filtering technique with hybrid feature selection. They have extracted features from CT images via statistical and ML-based techniques.
The key contributions of this paper include augmentation on the dataset to make variations within the category of the COVID-19 dataset to achieve high accuracy, rather than exploiting just as one attribute descriptor. However, two feature descriptors will be used to extract features and mix the features of these into a single list to train the ML model with more significant features. Furthermore, the authors have categorised three classes of high-resolution X-ray images compared to the current state of the art technology using six classifiers that appear in the first classifier due to high performance during testing. The paper is structured as follows: the proposed work is mentioned in Second 2; Section 3 introduces the methodology employed; Section 4 provides the results and discussion, and Section 5 ends the paper with a conclusion.
Fig. 3 illustrates the methodology of the proposed framework which is divided into five subsections including i) dataset, as discussed here in this proposed framework, ii) pre-processing, which includes, the details of the data cleaning and data preparation techniques, iii) feature extraction, which contains the description of feature extraction techniques, iv) training, which contains details of machine learning models are trained on the features extracted, and v) evaluation, that contains testing of the trained model on the testing data. The pre-processing steps contain noise removal, augmentation and resizing. Furthermore, the training steps extract features that involve training six ML models. Finally, testing on the new data step includes data equation, pre-processing, feature extraction and prediction of the trained model.
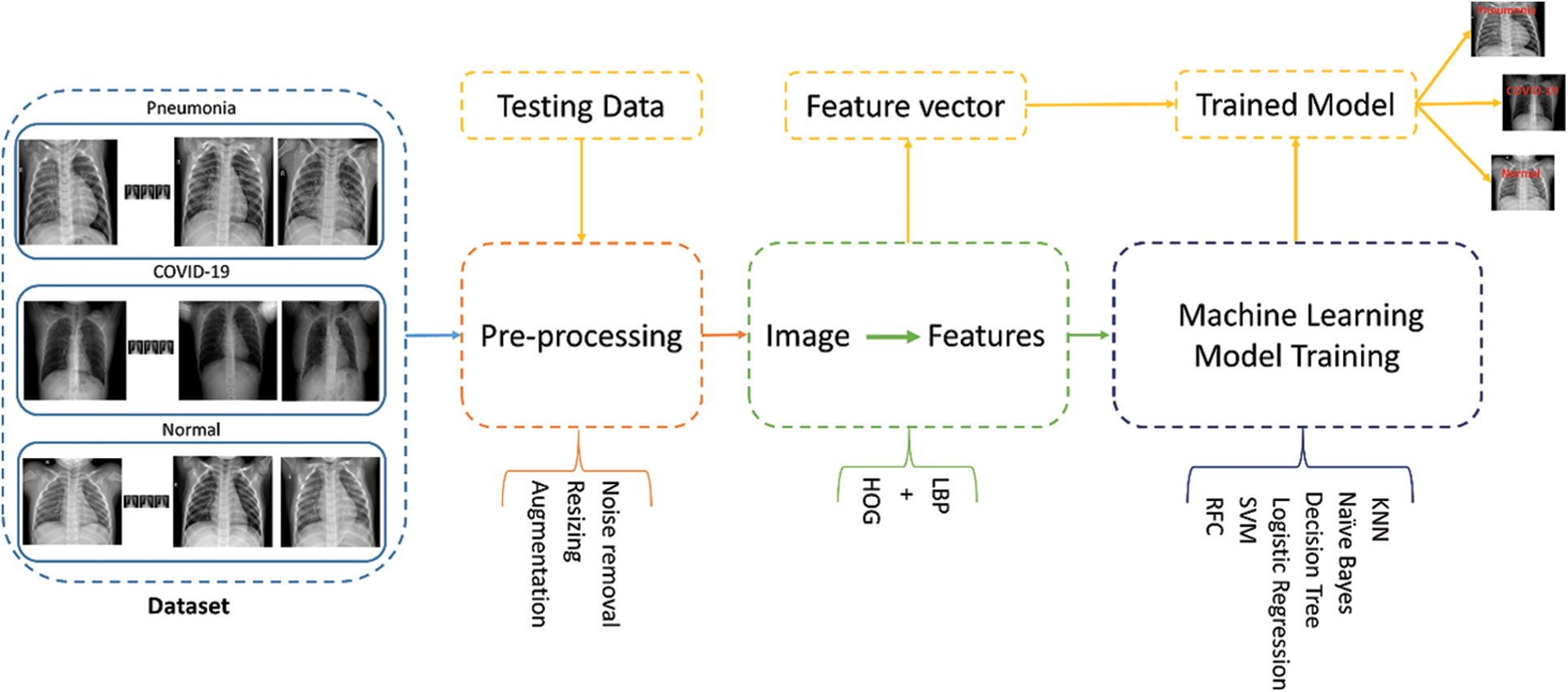
Figure 3: Proposed framework: Dataset consist of three classes which include, COVID-19, normal and pneumonia classes of X-ray chest images
A dataset is a necessary and initial step in training machine learning models. The authors have chosen the COVID-19 dataset using the open-source Kaggle repository because it is a dataset that has been verified by different experts. Samples of chest X-ray images based on the specified dataset are as shown in Fig. 4. The original dataset consists of 2800 images. The first row contains the images of human chests infected by COVID-19, the second row contains Normal, and the third one contains X-ray images of patients with Pneumonia. However, augmentation is performed on the COVID-19 category by the authors to equalise the images and avoid the data difference between categories and the details of the augmentation. However, as shown in Fig. 4, the dataset contains a total of three categories of Normal, COVID-19, and Pneumonia with 1000 chest X-ray images of each category. Furthermore, the authors have divided the whole dataset into evaluation sets (80%) and training dataset (20%) as shown in Fig. 5.
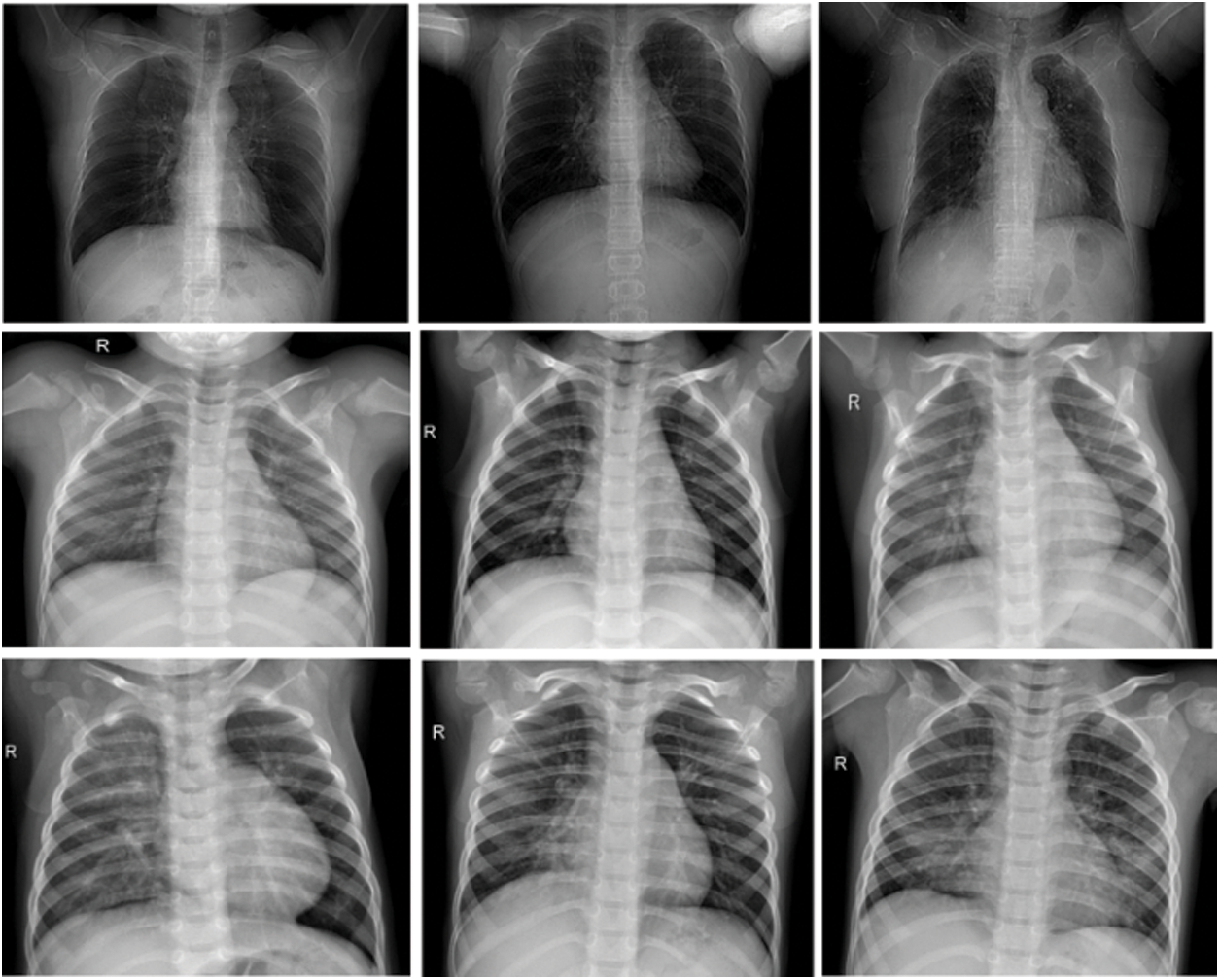
Figure 4: Samples images from used COVID-19 dataset
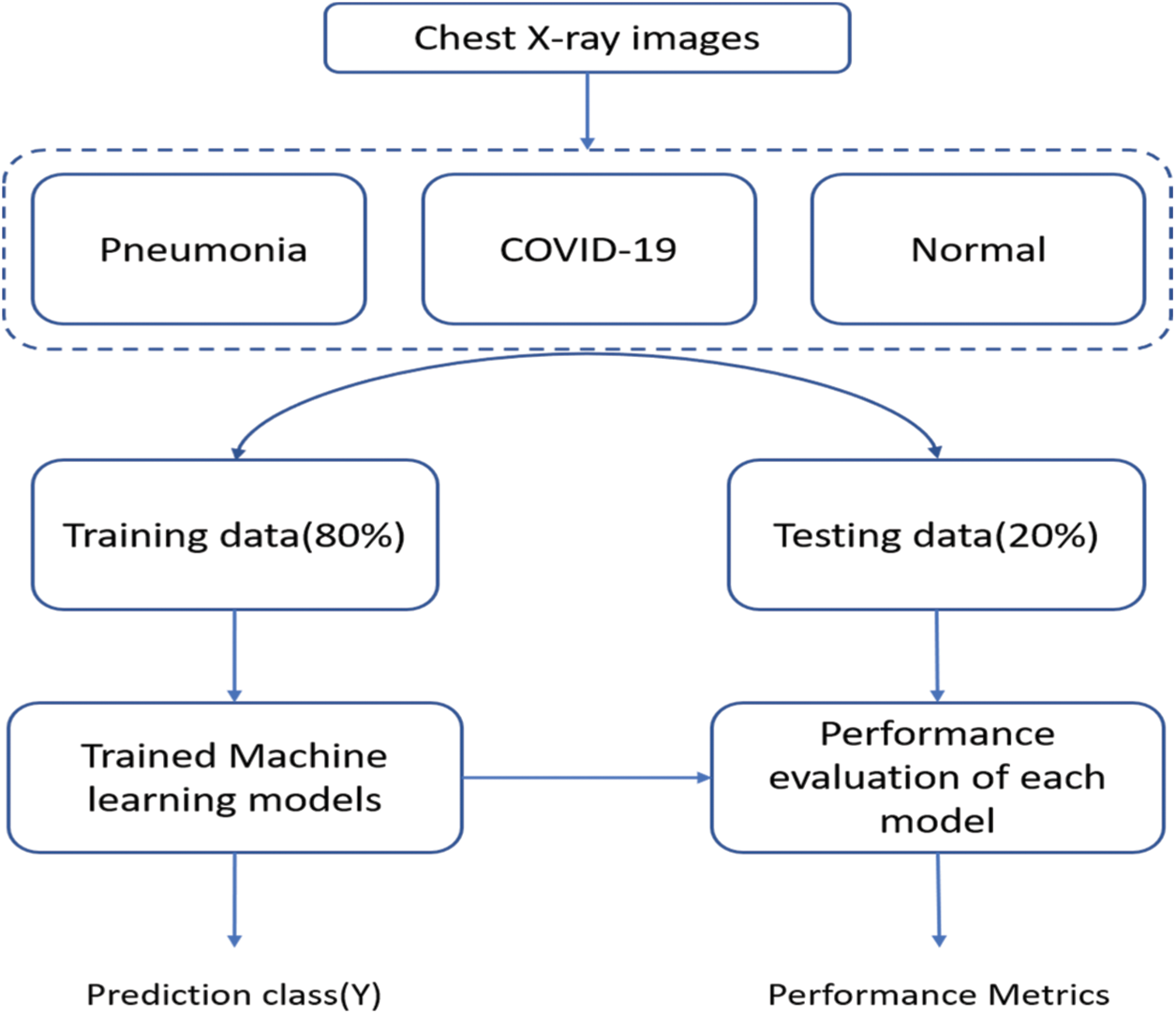
Figure 5: Flowchart of the proposed model
The flowchart of the proposed model is as shown in Fig. 5. The X-ray images are taken from the chest to complete the dataset. The dataset has three categories which include pneumonia, COVID-19 and normal. Further, the data has been divided into training (80%) and testing (20%). Finally, when training ML models on the training data the performance of the individual model on testing data is evaluated to obtain the performance metrics.
The pre-processing step also called the data preparation and cleaning step, is a crucial step that follows data acquisition. The data is used in an augmentation as the first step in creating possible variations in the dataset. Then, the data is augmented by creating different brightness variants of the existing COVID-19 class. However, to reduce the computational time, the images are resized up to 200 × 200. Additionally, to clean the data from possible piper and salt noise, the median and mean filter on the howl dataset of X-ray images is applied.
2.3 Feature Extraction Techniques
Feature extraction is the process of extracting meaningful information from the input data. As shown in Fig. 3, in the proposed framework, features are extracted from input data using Histogram of Oriented Gradient HOG directed gradient graph proposed by Dalal et al. [20] and Local Binary Pattern (LBP) in [21]. The detail of these extraction models are as follows:
A Decision Tree (DT) is a supervised ML algorithm presented in [22] which is being used for continuous and categorical problem-solving. The Decision Tree uses a two-step method to learn the extracted features from the data in minimising the error while handling the classification problem. In the first step, the DT learns from data and in the second step the model provides a prediction on the testing sample of data. We have used the DT for the classification of three classes of X-ray chest images while performance-rated discussion of the DT in the results section.
Naive Bayes is an ML model that was introduced in the 1960s under the method of text retrieval problem. The Naïve Bayes algorithm works on the basis of Bayes theorem which is mostly used in the solution of the classification problem. Bayes theorem provides to calculate the posterior probability denoted by
2.3.3 Random Forest Technique (RFC)
RFC is an ML algorithm proposed by Ho [23] using the regression and classification of problems in supervised ML. RFC contains decision trees which are taking a random sample from the dataset, each decision tree makes a prediction for the sample of the class. If for data the class has more votes, it becomes a prediction of the ML models.
2.3.4 Support Vector Machine Technique (SVM)
Support Vector Machine (SVM) is a supervised-based machine learning model proposed in [24]. The SVM model uses a hyper plane or hyper line to classify binary or multiclass data into different classes. The SVM selects the hyper plane having less classification error. In our proposed framework, the SVM is used after feature extraction from the X-ray chest images dataset to classify the data into three classes of normal, pneumonia, and COVID-19. The performance of the models is as shown in Fig. 7.
Logistic Regression (LR) is an ML model which is used mostly for binary classification of the data [24], which enables the LR to handle the multiclass classification. The LR makes the prediction on the input data and after calculating the cost function adjusting its weight accordingly using the formula of gradient descent as shown in Eq. (2). So, this process is continuously going on until to get a local or global minimum.
2.3.6 K-Nearest Neighbor (KNN)
KNN is an ML algorithm [25] that is used for regression as well as for the classification of dataset purposes. KNN works on the basis of finding similarities between the input data and training instances to predict labels. In the KNN, each object votes for a class, and the class with the most votes becomes a prediction by the model.
After the pre-processing and feature extraction steps, as shown in the proposed framework, the authors have used various ML models such as decision tree, Naïve Bayes, Random Forest Classification (RFC), SVM, Logistic regression, and KNN to perform the training of the classification of features extracted from X-ray chest images dataset.
The model has been evaluated on the unseen data after the training of the model as shown in Fig. 3 in what is called the testing phase. For the evaluation of the machine learning models research community is using different techniques of ROC and confusion matrix. The confusion matrix contains True Positive (TP), True Negative (TN), False Positive (FP) and False Negative values. The TP shows how often the model correctly has predicted the positive class. The TN, on the other hand, shows the rate of incorrect classification of the positive class. Further, FP shows the rate of correct classification of negative class however FN refers to the value of incorrect classification on negative class. For better evaluation, we have used the following equations which using the upper discussed four important values to calculate accuracy, precision, recall and F1 score.
3 Experimental Results and Discussion
In this section, different aspects were evaluated including the augmentation results and the performance of the classification trained models of the extracted features from the chest X-ray images on the use of raw data. In the original dataset, 1300 images have been shown as normal, and 200 images have been shown as infected of pneumonia, and as such these provide an inaccurate prediction of the COVID-19 sample while testing due to data differences. Fig. 6 shows a screenshot of multiple chest X-ray images based on patients. The pre-processing through deep learning API Keras technique, however, has been used for equalisation and creating variations in brightness in order to make the COVID-19 affected images distinctively clear for understanding. The ultimate results of pre-processing have been to make the conclusive understanding clearer to none experts like clinicians or technicians.
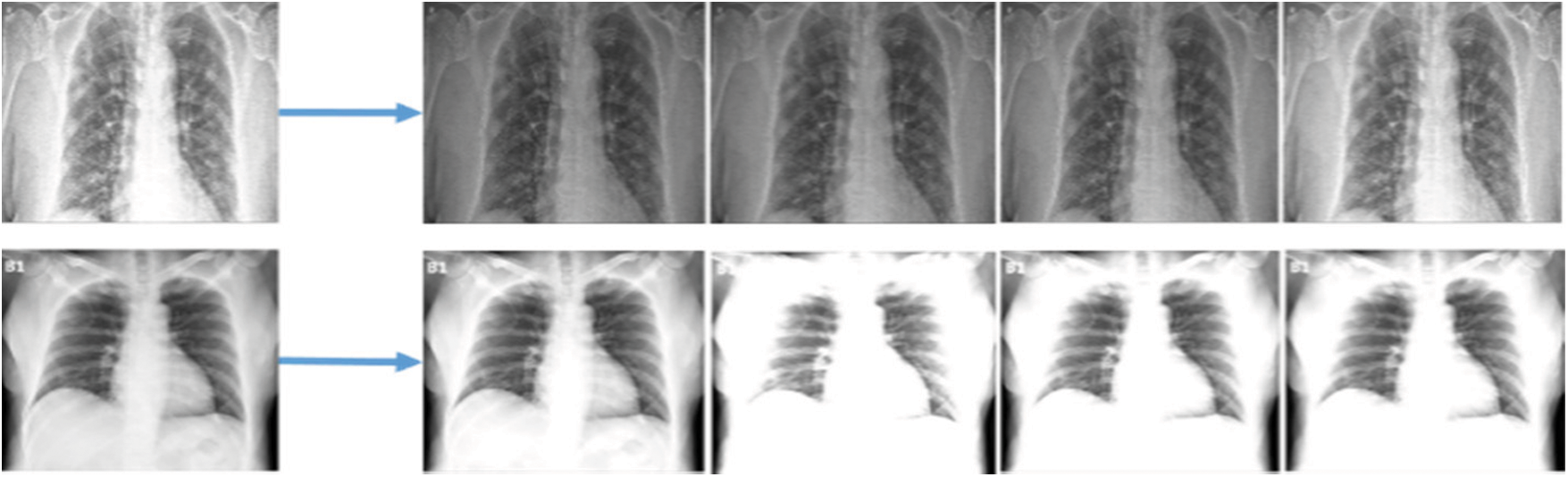
Figure 6: Screenshots of data augmentation of different types of images
In this research paper, the authors have trained different state-of-the-art ML models on the extracted feature which include, Decision tree, Random Forest Classifier, Naïve Bayes, SVM, and Logistic regression and KNN. However, to evaluate the performance of the trained model, the authors have additionally and more precisely evaluated the model by calculating the accuracy of each model which shows the accurate prediction of the model on testing samples. The evaluation performance of the proposed model is as shown in Fig. 7, which shows RFC is with accuracy and precision, Recall and F1-Score values each of 94%. However, SVM has 93% accuracy and precision. Recall and F1-score are 94% and 93%, respectively, indicating the performances of the model on the testing set of X-ray chest images.
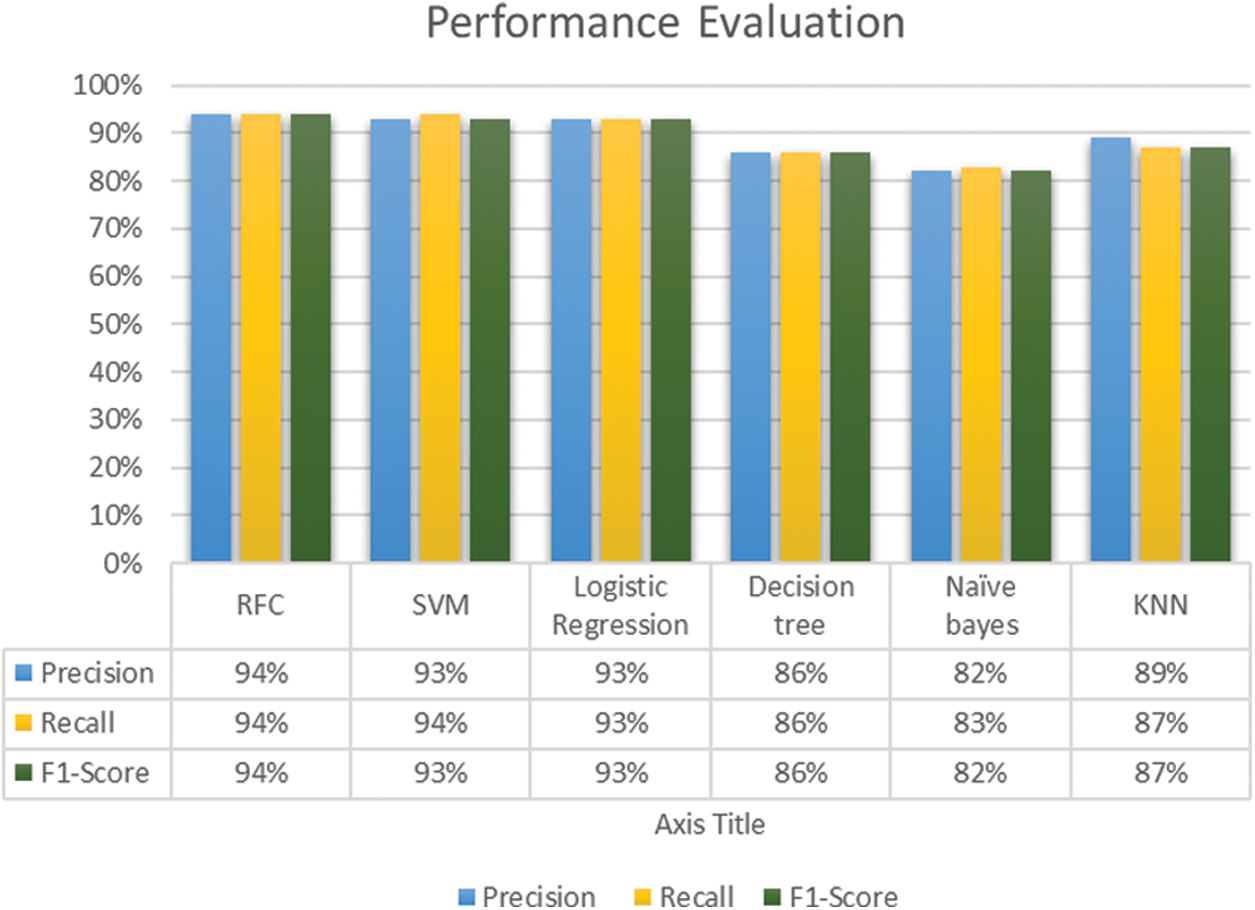
Figure 7: Performance evaluation graph of models
The performance of the models has been evaluated purely on the basis of accuracy as shown in Fig. 8. RFC has the highest accuracy of 94% more than other trained models followed by SVM with 93% accuracy. However, the lowest accuracy of 82% in these models has been achieved by the Naïve Bayes.
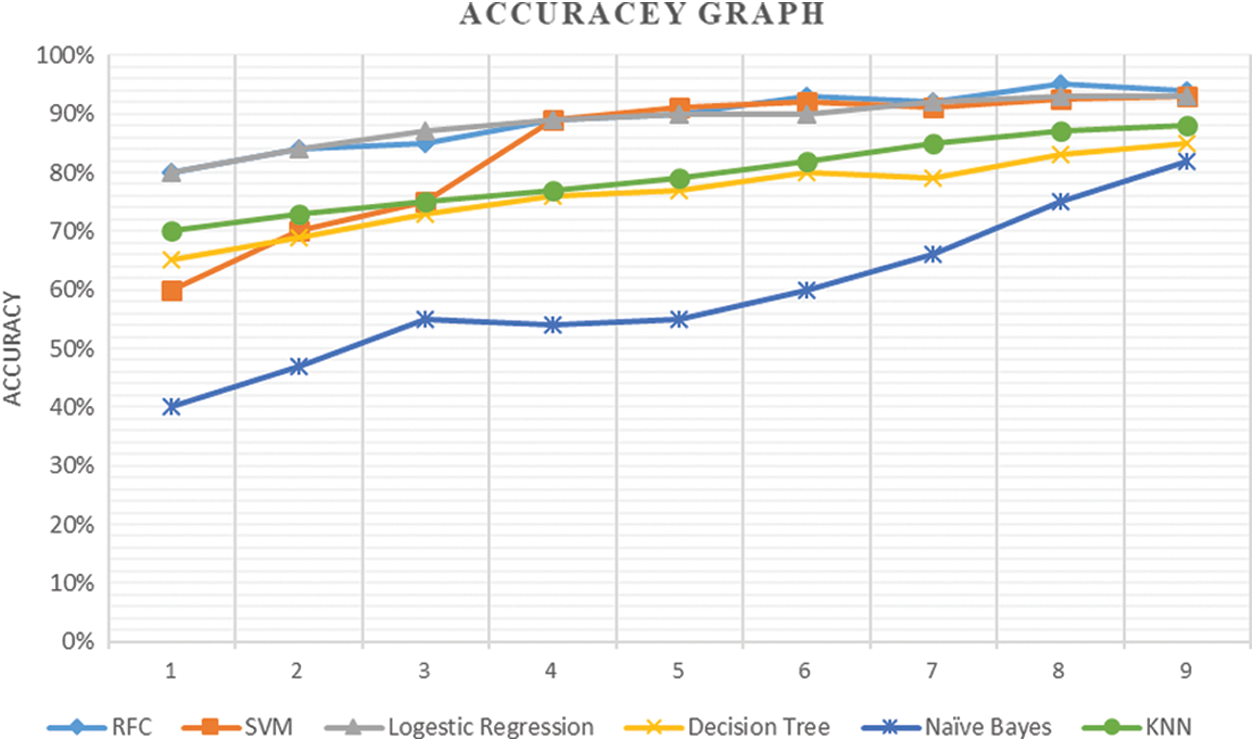
Figure 8: Accuracy of each model start from the initial point on the first iteration with stable condition
The COVID-19 pandemic is spreading across various regions, causing serious cardiovascular disease in humans. Early disease diagnosis and care can help to eliminate and contain the pandemic from further spreading. This proposed research has investigated the potential of ML to differentiate between the COVID-19, viral pneumonia infected, and normal by exploring chest X-ray images for the effect of viral infection. The experimental results have shown that the performance of the ML models on HOG and LBP features. Furthermore, in the following techniques, the RFC technique has obtained the highest accuracy of 94% followed by the SVM with 93% accuracy on the HOG features, respectively. Moreover, it is highly recommended for future work to seek and use more datasets from medical centres to classify the resulting data by using deep learning techniques in order to improve the accuracy of the model and helps the doctors to detect the COVID-19 viruses for a better world.
Funding Statement: This work is supported by the Information Technology Department, College of Computer, Qassim University, 6633, Buraidah 51452, Saudi Arabia.
Conflicts of Interest: The author declares to have no conflicts of interest in reporting this study.
1. M. Yadav, M. Perumal and M. Srinivas, “Analysis on novel coronavirus (COVID-19) using machine learning methods,” Chaos, Solitons & Fractals, vol. 139, 110050, 2020. [Google Scholar]
2. N. Petrosillo, G. Viceconte, O. Ergonul, G. Ippolito and E. Petersen, “COVID-19, SARS and MERS: Are they closely related,” Clinical Microbiology and Infection, vol. 26, no. 6, pp. 729–734, 2020. [Google Scholar]
3. L. N. Mahdy, K. A. Ezzat, H. H. Elmousalami, H. A. Ella and A. E. Hassanien, “Automatic x-ray covid-19 lung image classification system based on multi-level thresholding and support vector machine,” MedRxiv, 2020. https://doi.org/10.1101/2020.03.30.20047787. [Google Scholar]
4. J. Wu, P. Zhang, L. Zhang, W. Meng, J. Li et al., “Rapid and accurate identification of COVID-19 infection through machine learning based on clinical available blood test results,” MedRxiv, 2020. https://doi.org/10.1101/2020.04.02.20051136. [Google Scholar]
5. W. H. WHO. “Coronavirus disease (COVID-19) situation report,” Haziran, vol. 25, 141, 2020. [Google Scholar]
6. M. L. Giger, “Machine learning in medical imaging,” Journal of the American College of Radiology, vol. 15, no. 3, pp. 512–520, 2018. [Google Scholar]
7. N. Khan, F. Ullah, M. A. Hassan and A. Hussain, “COVID-19 classification based on chest X-ray images using machine learning techniques,” Journal of Computer Science and Technology Studies, vol. 2, no. 2, pp. 1–11, 2020. [Google Scholar]
8. S. Öztürk, U. Özkaya and M. Barstuğan. “Classification of coronavirus (COVID-19) from x-ray and CT images using shrunken features,” International Journal of Imaging Systems and Technology, vol. 31, no. 1, pp. 5–15, 2021. [Google Scholar]
9. U. Özkaya, Ş Öztürk and M. Barstugan, “Coronavirus (COVID-19) classification using deep features fusion and ranking technique,” in Big Data Analytics and Artificial Intelligence Against COVID-19: Innovation Vision and Approach, Cham: Springer, pp. 281–295, 2020. [Google Scholar]
10. A. Abbas, M. M. Abdelsamea and M. M. Gaber, “Classification of COVID-19 in chest x-ray images using DeTraC deep convolutional neural network,” Applied Intelligence, vol. 51, no. 2, pp. 854–864, 2021. [Google Scholar]
11. D. Al-Karawi, S. Al-Zaidi, N. Polus and S. Jassim, “Machine learning analysis of chest CT scan images as a complementary digital test of coronavirus (COVID-19) patients,” MedRxiv, 2020. https://doi.org/10.1101/2020.04.13.20063479. [Google Scholar]
12. Y. Wan, H. Zhou and X. Zhang, “An interpretation architecture for deep learning models with the application of COVID-19 diagnosis,” Entropy, vol. 23, no. 2, pp. 204, 2021. [Google Scholar]
13. M. A. Alqudah, S. Qazan, H. Alquran, I. A. Qasmieh and A. Alqudah, “Covid-2019 detection using x-ray images and artificial intelligence hybrid systems,” 2020. https://doi.org/10.5455/jjee.204-158531224. [Google Scholar]
14. L. Sun, F. Song, N. Shi, F. Liu, S. Li et al., “Combination of four clinical indicators predicts the severe/critical symptom of patients infected COVID-19,” Journal of Clinical Virology, vol. 128, no. 104431, 2020. https://doi.org/10.1016/j.jcv.2020.104431 [Google Scholar]
15. A. Narin, C. Kaya and Z. Pamuk, “Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks,” Pattern Analysis and Applications, pp. 1–14, 2021. https://10.1007/s10044-021-00984-y. [Google Scholar]
16. S. Minaee, R. Kafieh, M. Sonka, S. Yazdani and G. J. Soufi, “Deep-covid: Predicting covid-19 from chest x-ray images using deep transfer learning,” Medical Image Analysis, vol. 65, no. 101794, 2020. https://doi.org/10.1016/j.media.2020.101794. [Google Scholar]
17. E. Tartaglione, C. A. Barbano, C. Berzovini, M. Calandri and M. Grangetto, “Unveiling covid-19 from chest x-ray with deep learning: A hurdles race with small data,” International Journal of Environmental Research and Public Health, vol. 17, no. 18, 6933. 2020. https://doi.org/10.3390/ijerph17186933. [Google Scholar]
18. S. Wenqi, L. Tong, Y. Zhu and M. D. Wang, “COVID-19 automatic diagnosis with radiographic imaging: explainable AttentionTransfer deep neural networks,” IEEE Journal of Biomedical and Health Informatics, vol. 25, no. 7, pp. 2376–2387, 2021. [Google Scholar]
19. A. A. Farid, G. I. Selim, H. Awad and A. Khater, “A novel approach of CT images feature analysis and prediction to screen for corona virus disease (COVID-19),” International Journal of Scientific & Engineering Research, vol. 11, no. 3, 2020. https://doi.org/10.20944/preprints202003.0284.v1. [Google Scholar]
20. N. Dalal and B. Triggs, “Histograms of oriented gradients for human detection,” in IEEE Computer Society Conf. on Computer Vision and Pattern Recognition (CVPR'05), San Diego, CA, USA, vol. 1, pp. 886–893, 2005. [Google Scholar]
21. M. Heikkilä, M. Pietikäinen and C. Schmid, “Description of interest regions with local binary patterns,” Pattern Recognition, vol. 42, no. 3, pp. 425–436, 2009. [Google Scholar]
22. S. R. Safavian and D. Landgrebe, “A survey of decision tree classifier methodology,” IEEE Transactions on Systems, man, and Cybernetics, vol. 21, no. 3, pp. 660–674, 1991. [Google Scholar]
23. T. K. Ho, “The random subspace method for constructing decision forests,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 20, no. 8, pp. 832–844, 1998. [Google Scholar]
24. S. W. Noble, “What is a support vector machine,” Nature Biotechnology, vol. 24, no. 12, pp. 1565–1567, 2006. [Google Scholar]
25. A. Y. Ng and M. I. Jordan, “On discriminative vs. generative classifiers: A comparison of logistic regression and naive Bayes.” in 14th Int. Conf. on Neural Information Processing Systems: Natural and Synthetic, ACM Digital Library, pp. 841–848, 2001. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |