DOI:10.32604/csse.2022.022017

| Computer Systems Science & Engineering DOI:10.32604/csse.2022.022017 |  |
| Article |
Rice Leaves Disease Diagnose Empowered with Transfer Learning
1College of Computer and Information Sciences, Jouf University, Sakaka, 72341, Saudi Arabia
2Department of Computer Science, Faculty of Computers and Artificial Intelligence, Cairo University, 12613, Egypt
3School of Computer Science, National College of Business Administration and Economics, Lahore, 54000, Pakistan
4Department of Computer Science, Lahore Garrison University, Lahore, 54000, Pakistan
5Pattern Recognition and Machine Learning Lab, Department of Software, Gachon University, Seongnam, 13557, Korea
6Department of Information System, Faculty of Computing and Information Technology, King Abdulaziz University, Jeddah, 21589, Saudi Arabia
*Corresponding Author: Muhammad Adnan Khan. Email: adnan@gachon.ac.kr
Received: 23 July 2021; Accepted: 24 August 2021
Abstract: In the agricultural industry, rice infections have resulted in significant productivity and economic losses. The infections must be recognized early on to regulate and mitigate the effects of the attacks. Early diagnosis of disease severity effects or incidence can preserve production from quantitative and qualitative losses, reduce pesticide use, and boost ta country’s economy. Assessing the health of a rice plant through its leaves is usually done as a manual ocular exercise. In this manuscript, three rice plant diseases: Bacterial leaf blight, Brown spot, and Leaf smut, were identified using the Alexnet Model. Our research shows that any reduction in rice plants will have a significant beneficial impact on alleviating global food hunger by increasing supply, lowering prices, and reducing production's environmental impact that affects the economy of any country. Farmers would be able to get more exact and faster results with this technology, allowing them to administer the most acceptable treatment available. By Using Alex Net, the proposed approach achieved a 99.0% accuracy rate for diagnosing rice leaves disease.
Keywords: Rice; bacterial leaf blight; brown spot; leaf smut; machine learning; alexnet
Rice plants are susceptible to stress due to both abiotic and biotic causes. Non-living forces such as wind and water are classified as abiotic, while biotic species include plants, animals, bacteria, and fungi. Stresses in rice plants manifest themselves in a variety of ways. Plant water stress, for example, can impede photosynthesis, decrease evapotranspiration, and elevate leaf surface temperature. Other morphological abnormalities, such as discoloration of leaves, wilting, and plants that do not produce panicles or produce empty panicles, can be caused by bacteria, viruses, or fungus [1].
Rice is one of the world’s most significant food crops. Almost half of the world's population eats it as a staple diet. As the world’s population and food supply growth so does the need for rice. Reduced yield due to pests, poor input management, uneven nutrient use, inadequate irrigation water utilization, and environmental degradation are the most common agronomic issues in rice agriculture. Furthermore, several of these issues may exacerbate existing problems. For example, plant health suffers due to unequal nutrient utilization caused by inappropriate fertilizer application. As a result, the sensitivity of plants to pests and diseases is increased. As a result of these issues, rice yield and quality have decreased significantly [2].
Plant diseases are one of the factors that contribute to the loss of agricultural product quality and quantity. Both of these factors can directly impact a country’s overall crop yield [3]. The main issue is that the plants aren’t being continuously monitored. New farmers aren’t always aware of diseases or when they occur. Diseases can affect any plant at any time, according to the general rule. Continuous monitoring, on the other hand, might be able to keep disease infection at bay. Therefore, one of the important research topics in the agricultural domain is detecting plant diseases [4].
Rice is an essential food source for rural people, and it is also the world's second most-produced cereal crop. Rice belongs to the Poaceae family and is divided into two subspecies: Japonica and Indica. Rice is Asia’s most well-known low-cost and nutrient-dense cuisine. Rice is grown in five different parts of the world: Asia, Africa, America, Europe, and Oceania. According to the Food and Agriculture Organization of the United Nations (FAOSTAT) report, Asian countries deliver and consume 91.05% of the world’s rice. The remainder of the rice harvest is divided among other world regions, with Africa receiving 2.95%, America 5.19%, Europe 0.67%, and Oceania 0.15% [5]. According to the World Bank, rice consumption demand is expected to rise by 51% by 2025, far outpacing the population growth shown in Fig. 1.
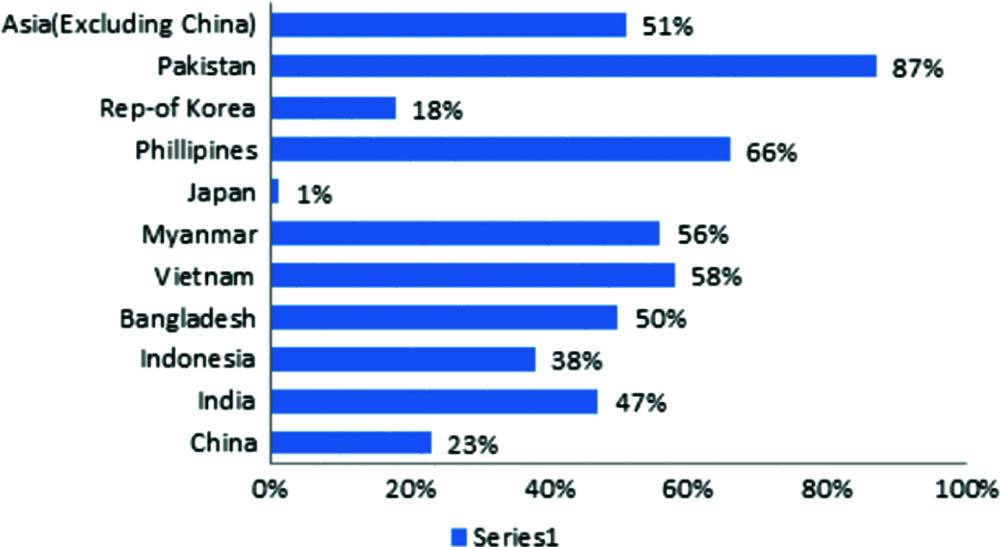
Figure 1: Depicts rice consumption projections for major asian countries from 1995 to 2015
This study uses machine learning and image processing techniques to solve autonomous disease detection in rice plants, one of the essential foods in any country. Diseases are caused by bacteria, fungi, and viruses on any plant. Bacterial leaf blight, Brown spot, and Leaf smut are the most frequent diseases that affect rice plants. The Characteristics of different rice leaves diseases shown in Tab. 1.

The External appearances of diseased plants can be subjected to image processing processes. A disease sign, on the other hand, varies depending on the plant. Some diseases are brown in hue, while others are yellow. Each disease has its distinct features. The form, size, and colour of disease symptoms vary by condition. Some diseases are the same colour but have different shapes, while others are the same colour but have distinct forms. As a result, farmers are occasionally perplexed and unable to make informed pesticide decisions [6].
The traditional method of identifying rice infections is frequently done manually, and it has shown to be unreliable, expensive, and time-consuming. In addition, although the machine learning methodology for detecting rice diseases is relatively straightforward, it is easy to mistake some similar diseases that have detrimental effects on rice growth [7].
Agriculture research aims to increase productivity and food quality while lowering costs and increasing profits, and it has recently gained traction. Accurate diagnosis and quick resolution of the field problem are essential components of crop management.
Prajapati et al. used 40 images of “Brown spot, bacterial leaf blight, and Leaf smut” in their study. They used K-means and extracted 88 features from three categories for disease segmentation, including colour, texture, and shape. Rice leaf diseases are classified using SVM. Another recent study by Prajapati et al. included a detailed analysis and survey of cotton leaf diseases. Cotton leaf diseases include“Alternaria leaf spot,” “red spot,” and “bacterial blight Cercospora leaf spot.” The image’s background is removed during the pre-processing stage. After that, the Otsu thresholding method is used to segment the data. Finally, colour, shape, and texture were extracted from the segmented images, and the extracted features were used as input to the classifiers [6].
Phadikar & Sil proposed a rice disease identification approach in which diseased rice images were classified using Self organizing map (SOM) (via neural network). The train images were obtained by extracting the features of the infected parts of the leave. In contrast, four different types of images were used for testing purposes in an earlier study [7].
Back propagation neural networks were used by Kahar et al. to propose a work. Three output nodes were used to detect three illnesses (Leaf blight, Leaf blast, and Sheath blight). In addition, rice leaf disease samples were gathered at three different stages (early, middle, and late). In a neural network, the grouping process employs the sigmoid activation function [8].
Shah et al. provide an overview of numerous ways to detect and classify rice plant illness and address several machine learning and image processing techniques. Prajapati, Shah, and Dabhi used the Support vector machine (SVM) methodology for multi-class classification to detect three types of rice illnesses (bacterial leaf blight, brown spot, and leaf smut). Images of diseased rice plants were collected using a digital camera in a rice field, with a training dataset accuracy of 93.33% and a test dataset accuracy of 73.33% [9].
The SVM classifier was used by Singh et al. to suggest two classification methods for ordinary and sick rice leaves. First, the partition of images is done using K-means segmentation, which has a classification accuracy of 82% [10].
Colourtrans formation structure was proposed by Khirade et al., and then device-independent colour space transformation was used. Finally, methods such as clipping, histogram equalization, image smoothing, and image enhancement were employed [11].
Zhou et al. employed a fuzzy C-means algorithm to categorise locations into one of four classes: no infestation, mild infestation, considerable infestation, and severe infestation. According to their findings, the accuracy in distinguishing cases where rice plant-hopper infestation had occurred or not reached 87%, while the accuracy in distinguishing four groups was 63.5% [12].
Sanyal et al. suggested a method for detecting and classifying six types of mineral deficiencies in rice crops using a Multi-layer perceptron (MLP) based neural network for each type of feature (texture and colour). Both networks have a hidden layer with a variable number of neurons in the hidden layer (40 for texture and 70 for colour), where 88.56% of the pixels were correctly categorized. Similarly, the same authors presented another study in which two types of rice-related disorders (blast and brown spots) were effectively detected [13].
Using fractal Fourier analysis, Asfarian et al. developed a new texture analysis method to identify four rice illnesses (bacterial leaf blight, blast, brown spot, and tungro virus). The image of the rice leaf was transformed to CIELab colour space in their proposed study, and the system achieved an accuracy of 92.5% [14].
Rahman et al. established a CNN methodology for detecting illnesses and pests from rice plant photos (five classes of diseases, three classes of pests, and one healthy plant and others). A total of 1,426 images were acquired, with the system achieving a mean validation accuracy of 94.33% utilizing four distinct types of cameras [15].
Deep & Machine learning arose over the last two decades from the increasing capacity of computers to process large amounts of data. Computational Intelligence approaches like Swarm Intelligence [16], Evolutionary Computing [17] like Genetic Algorithm [18], Neural Network [19], Deep Extreme Machine learning [20] and Fuzzy system [21–27] are strong candidate solution in the field of smart city [28–30], smart health [31–33], and wireless communication [34,35], etc.
These are the flaws in previous techniques is as follows:
• Performance affected when the data set contains additional noise, such as when target classes overlap [6].
• Because classification works by placing data points above and below the classifying hyperplane, there is no probabilistic explanation [7].
• A combination of statistical and structural characteristics should be used [14].
The proposed strategy makes the model more dependable by using a diversified and considerable number of classes and images. The following main contributions are presented in this paper:
• In our recommended approach, we acquire the most significant outcomes without augmentation.
• The Alex net layers were tweaked in this method to achieve better results.
In this study, the AlexNet model is utilized to detect disease in rice leaves. We use AlexNet rather than any other pre-trained model since we want to work with a simple model and assess performance without sacrificing memory or time. Because AlexNet was taught faster by efficiently implementing the GPU of the convolution and all other processing in the CNN training, the layers (Conv., max-out, BN, MXP), (Conv. and max-out), and (FC, max-out, Dropout) were repeated twice in the AlexNet model [36].
Fig. 2 shows the application of our proposed approach, in which affected rice leaves data, is collected from fields through handy camera. Collected data is send to the cloud where database store the data and by using Alex net to check the result, which disease they have. Then store this in to the database. Then disease application is used to see the results.
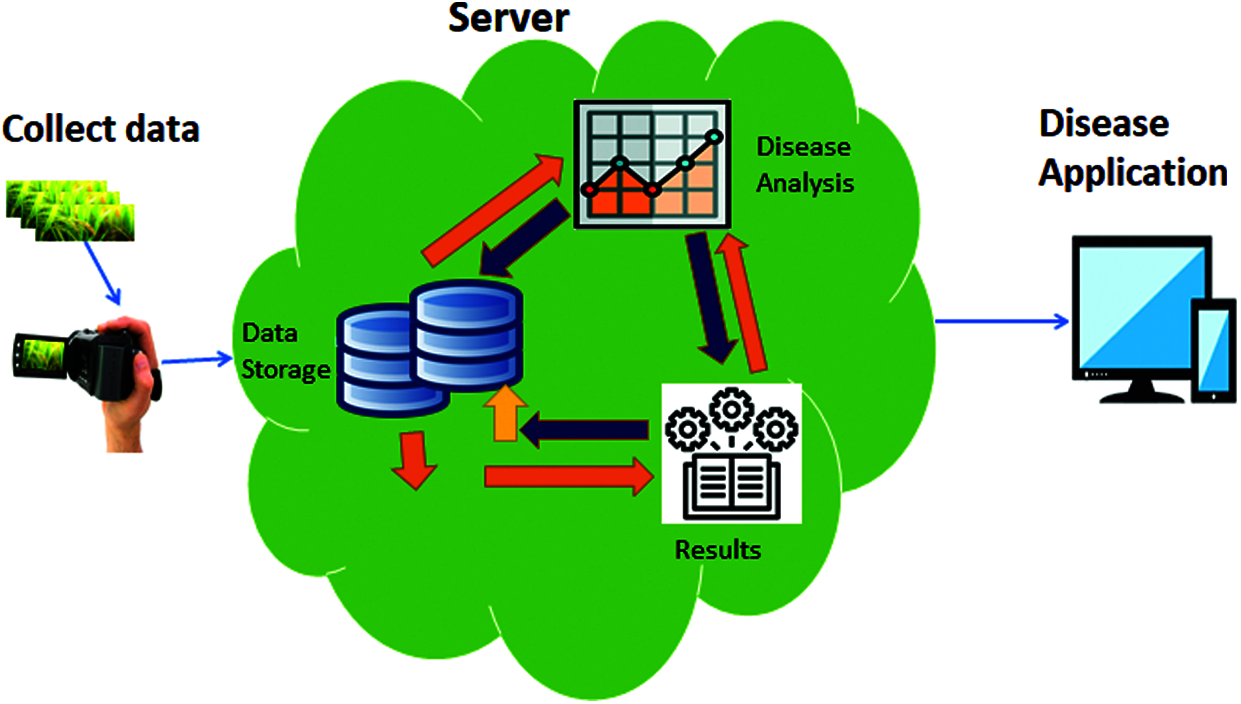
Figure 2: Application of automatic diagnose disease in rice leaves using machine learning
On many datasets, the AlexNet model may produce high-performance accuracy measurements. Detaching any of the convolutional layers, on the other hand, must dramatically reduce AlexNet’s. An RBG image of size 227 × 227 must be used as input. AlexNet would have been forced to employ significantly lower network layers if it didn’t have this image size. The input image is converted to RGB if it isn’t already. If the size of the input image is not 227 × 227, it will be transformed. Convolution and max-pooling with BN are performed by the first convolution layer, which has 96 different 11 × 11 size filters. Consider a 227 × 227 × 3 input image applied to a convolution layer 1 with a square filter size of 11 × 11 and 96 output maps (channels). Then there’s layer 1:
• Which has (227 × 227 × 96) output neurons, one for each of the 227 × 227 “pixels” in the input and across the 96 output maps.
• There are (11 × 11) (3 × 96) weights per filter (the input size ran through the kernel), for a total of 96 kernels (one for each output channel).
• There are connections for (227 × 227 × 3 × 11 × 11 × 96). Each of the (227 × 227 × 96) output units has a single filter that processes (11 × 11 × 3) values in the input.
The next four convolution layers follow the same pattern. Each layer has its own input, filter, and output map sizes, as well as the matching number of filters. On the other side, there are two completely connected layers that were exercised with dropout and Softmax at the conclusion of the model to act as the discriminant.
Fig. 3 shows the functioning procedures. It describes the data for this challenge came from a kaggle competition called rice leaf disease detection. Three picture files of rice leaf diseases were included in this dataset: bacterial leaf blight, brown spot, and leaf smut. There were 40 photos in each file. We obtained 120 photos in total from this dataset, which corresponded to the three rice leaf diseases. Our dataset was divided into two parts: training data and test data. Data from our dataset was used as training and test data in 80 percent and 20 percent of cases, respectively. As a result, we had 96 training photos and 24 test images in our dataset. Finally, the AlexNet neural network was used to predict rice leaf disease classification.
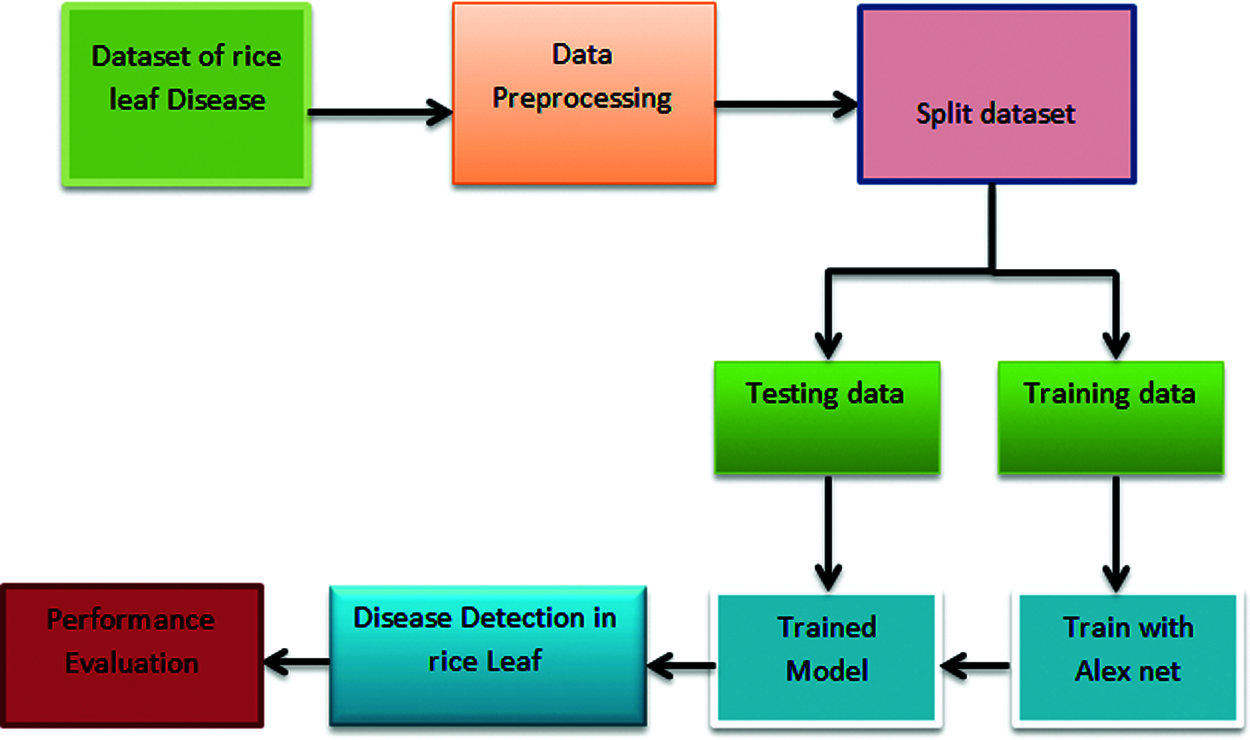
Figure 3: Framework of automatic diagnose disease in rice leaves using transfer learning
Fig. 4 explains that the information for this challenge came from a kaggle competition named rice leaf disease detection. The bacterial leaf blight, brown spot, and leaf smut photo files were included in this collection. Each file contained forty images. This dataset yielded a total of 120 pictures, which related to the three rice leaf diseases. There were two portions to our dataset: training data and test data. In 80% and 20% of cases, data from our dataset was used as training and test data, respectively. As a result, our dataset had 96 training images and 24 test images. Finally, to predict rice leaf disease, the AlexNet neural network is deployed.
There are five convolutional layers and three fully linked layers in Alex net network. If we delete any of our network’s convolutional layers, its performance will suffer significantly. The picture input layer was the initial layer, and it utilized input color images with dimensions of 22p by 227 by 3.
Alexnet’s final three levels are made up of three classes. All layers except the last three were pulled from our AlexNet neural network and tweaked for rice leaf disease detection. In our research, we replaced the last three layers with a fully connected layer, a softmax layer, and a classification output layer to transfer classification and detection duties. Along with the additional data, the possibilities for the new fully connected layer were listed. Finally, our model correctly identified the three most common rice leaf diseases and predicted the accuracy of detection. The Proposed Model of Automatic Disease Diagnosis in Rice Leaves Using Alex Net is shown in Fig. 4.
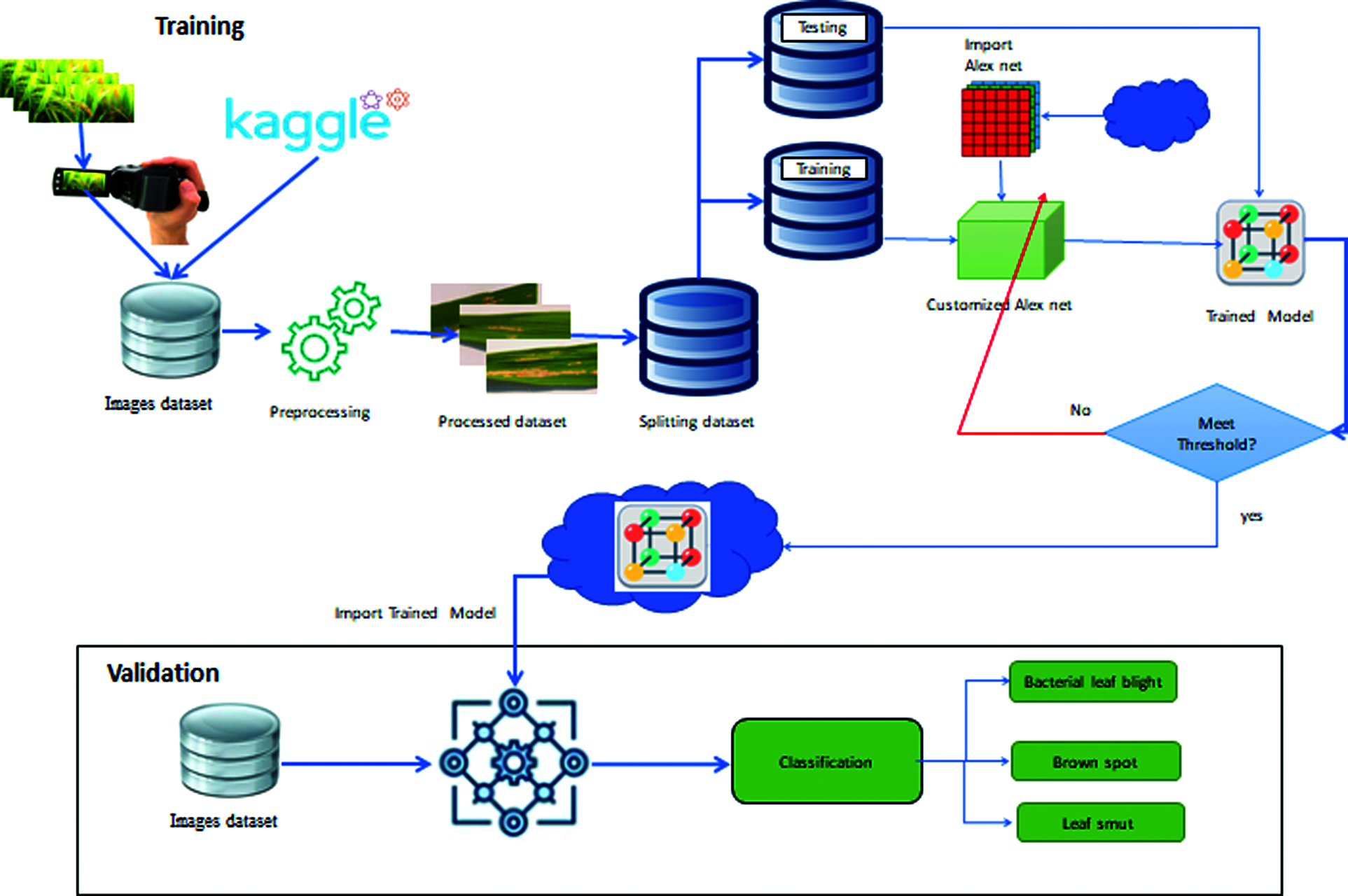
Figure 4: Proposed model of automatic diagnose disease in rice leaves using transfer learning
The data for this challenge came from a kaggle competition called rice leaf disease detection. Three picture files of rice leaf diseases were included in this dataset: bacterial leaf blight, brown spot, and leaf smut. There were 40 photos in each file. We obtained 120 photos in total from this dataset, which corresponded to the three rice leaf diseases. Our dataset was divided into two parts: training data and test data. Data from our dataset was used as training and test data in 80 percent and 20 percent of cases, respectively. As a result, we had 96 training photos and 24 test images in our dataset. Finally, the AlexNet neural network was used to predict rice leaf disease classification.
Figs. 5 and 6 shown the accuracy and loss rate of our proposed approach. For constant learning in the transferred layers, use a learning rate of 0.0001. On our complete training dataset, we defined an epoch of size 50 as a full training cycle, and the validation frequency was every three iterations during training. Due to a hardware limitation, the elapsed time in our work was 8 min and 9 s. Because our dataset contains colored images with high resolution, our model is forced to use a GPU (Graphics Processing Unit) by default. However, our device lacked a GPU. Due to the lack of a GPU, our model relied on the CPU (Central Processing Unit), which required a long time to complete our assignment. This elapsed time may differ between devices. Following formulas used for calculate precision, recall.
In above Eqs. (1) and (2) Ψi, ξi & ξj represents the true positive, the false positive and the true Negative respectively.
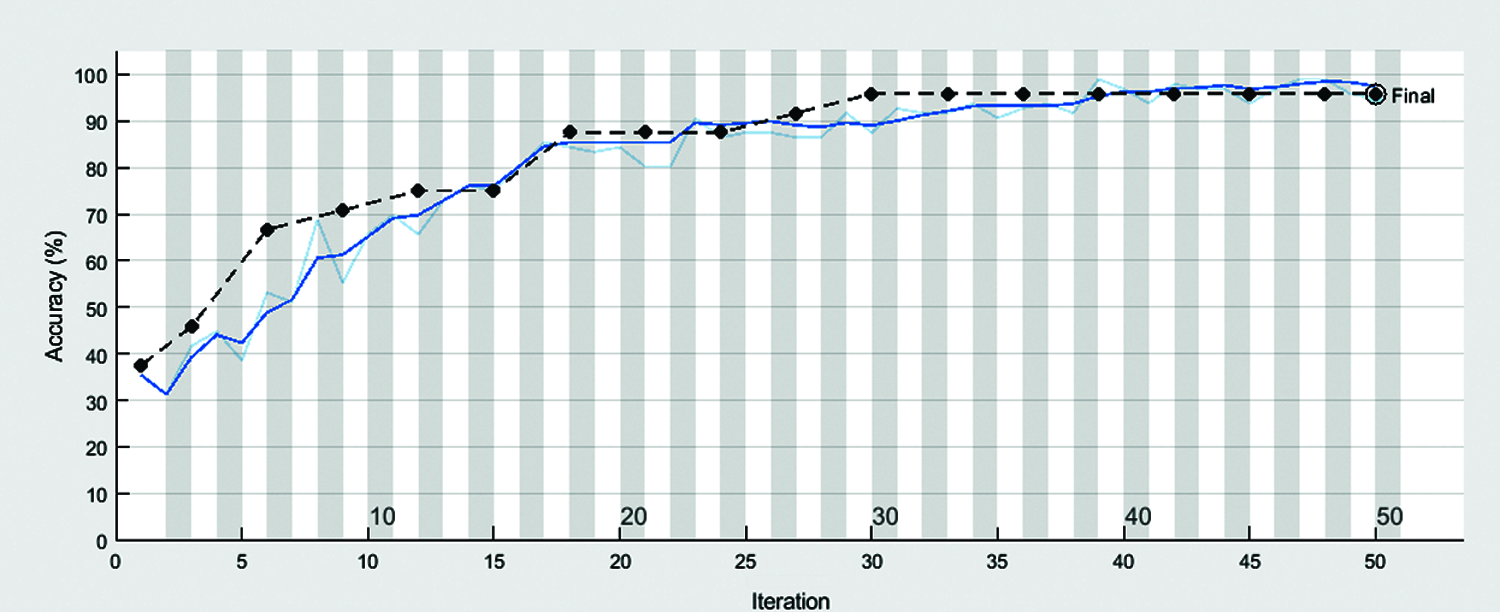
Figure 5: Accuracy of automatic diagnose disease in rice leaves using transfer learning
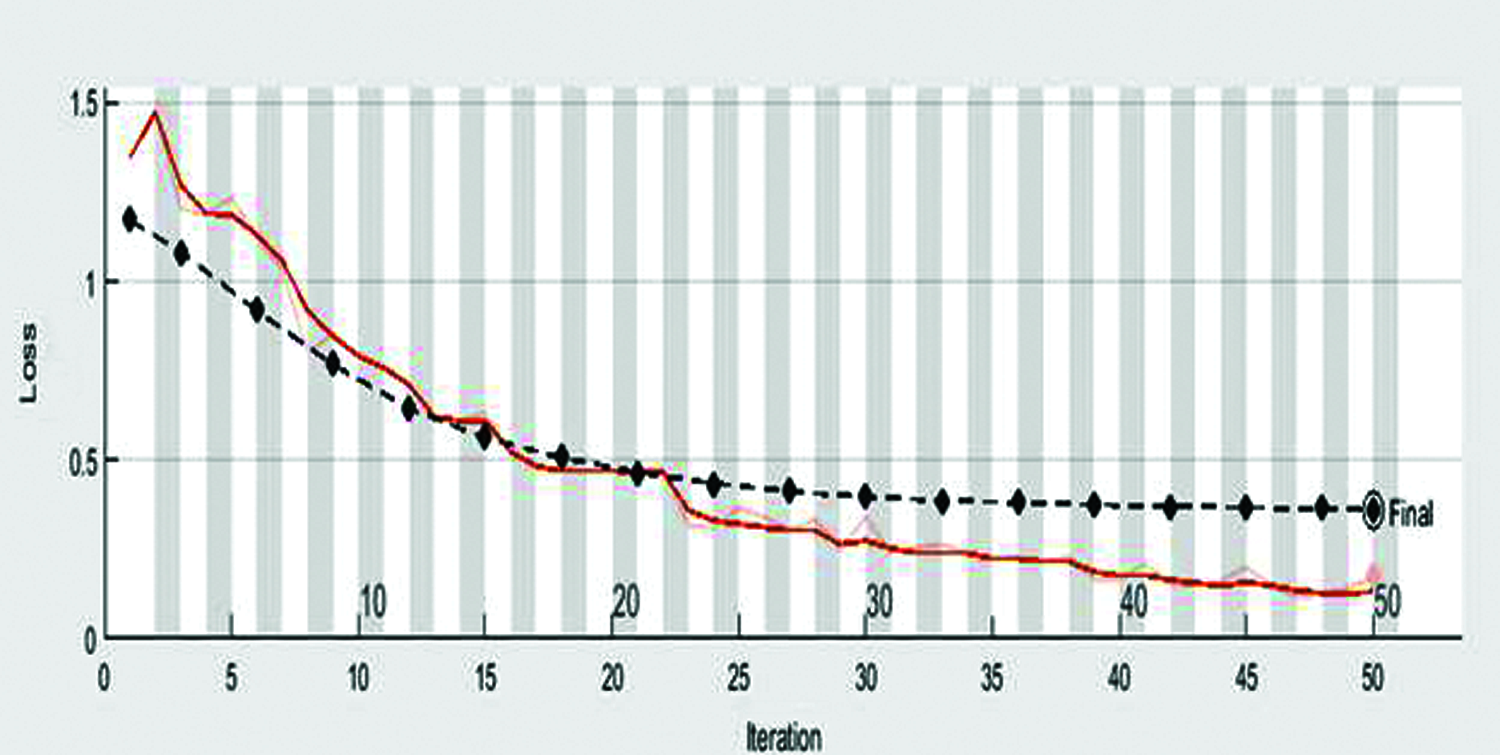
Figure 6: Loss rate of automatic diagnose disease in rice leaves using transfer learning
Tab. 2 describes the training results of confusion matrix by using 96 samples of data, included the following parameters:
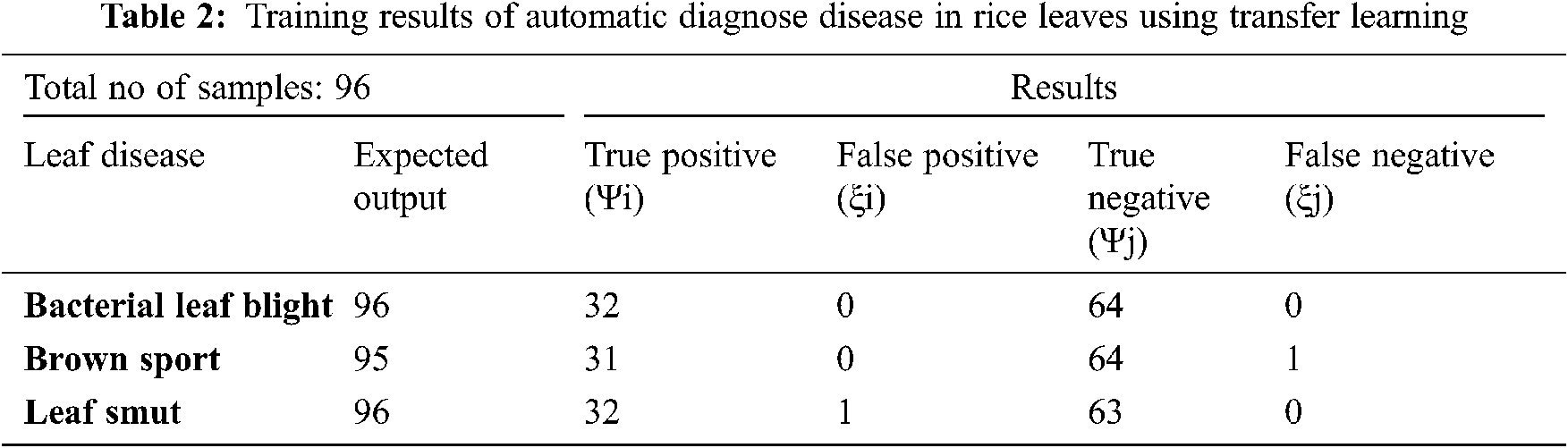
True Positive (Ψi): The model correctly classified 32 bacterial leaf blight, 31 brown sport and 32 leaf smut data points in the positive class, meaning that the model correctly identified data points in the positive class.
False Positive (ξi): As a result, the model misidentified 0 bacterial leaf blight, 0 brown sport and 1 leaf smut pieces of negative class data as positive class data.
True Negative (Ψj): The model correctly classified 64 bacterial leaf blight, 64 brown sport and 63 leaf smut data items from the negative class.
False Negative (ξj): As a result, the model misidentified 0 bacterial leaf blight, 1 brown sport and 0 leaf smut pieces of positive class data as negative class data.
Tab. 3 describes the validation results of confusion matrix by using 24 samples of data, included the following parameters:
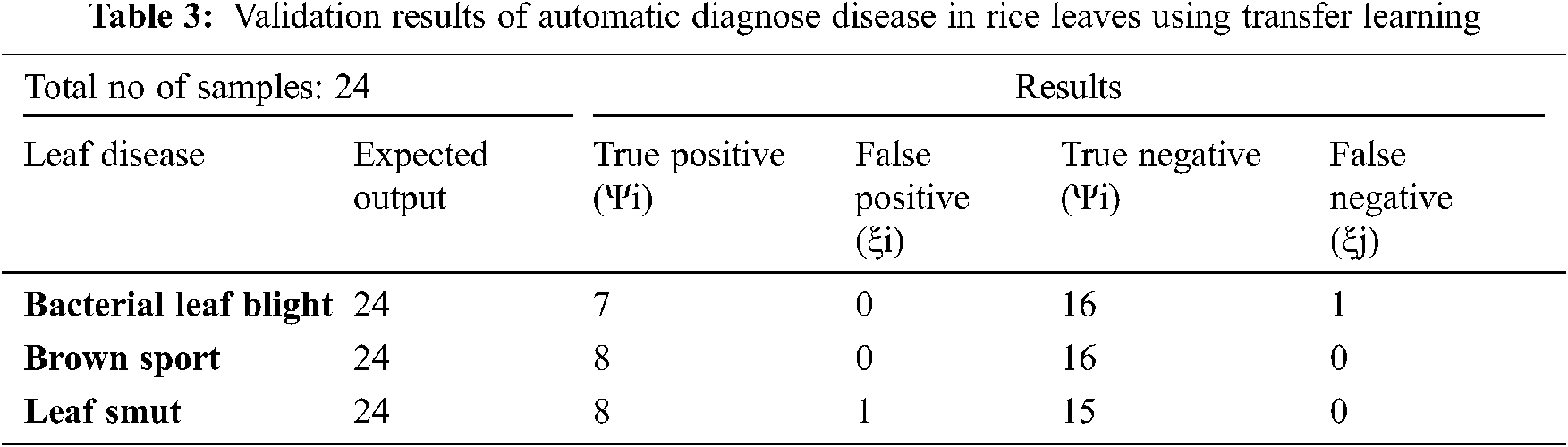
True Positive (Ψi): The model correctly classified 7 bacterial leaf blight, 8 brown sport and 8 leaf smut data points in the positive class, meaning that the model correctly identified data points in the positive class.
False Positive (ξi): As a result, the model misidentified 0 bacterial leaf blight, 0 brown sport and 1 leaf smut pieces of negative class data as positive class data.
True Negative (Ψj): The model correctly classified 16 bacterial leaf blight, 16 brown sport and 15 leaf smut data items from the negative class.
False Negative (ξj): As a result, the model misidentified 1 bacterial leaf blight, 0 brown sport and 0 leaf smut pieces of positive class data as negative class data.
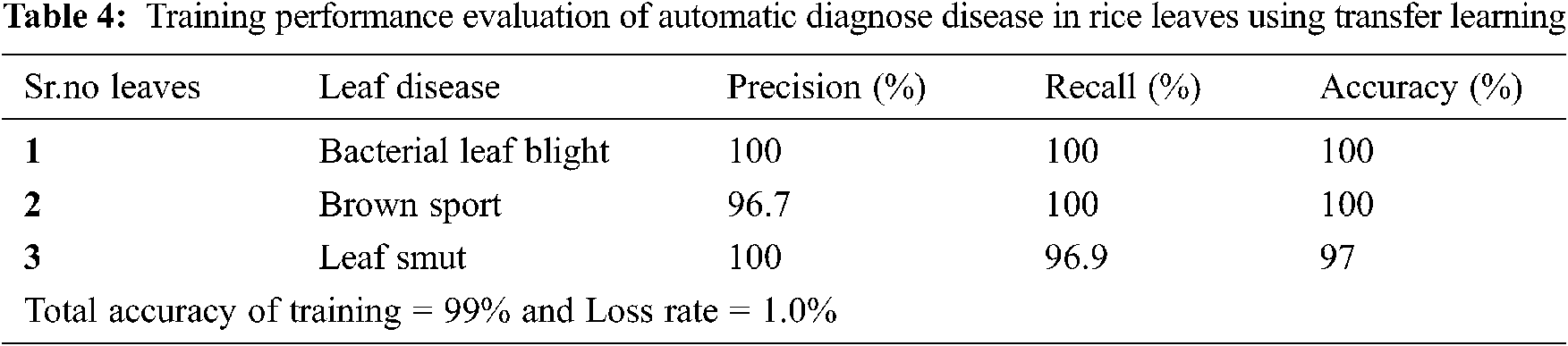
Tab. 4 describes the training performance evaluation of our proposed system in which precision is 100% in bacterial leaf blight, 96.7% in brown sport and 100% in leaf smut. Recall is 100% in bacterial leaf blight, 100% in brown sport and 96.9% in leaf smut. Accuracy is 100% in bacterial leaf blight, 100% in brown sport and 97% in leaf smut.
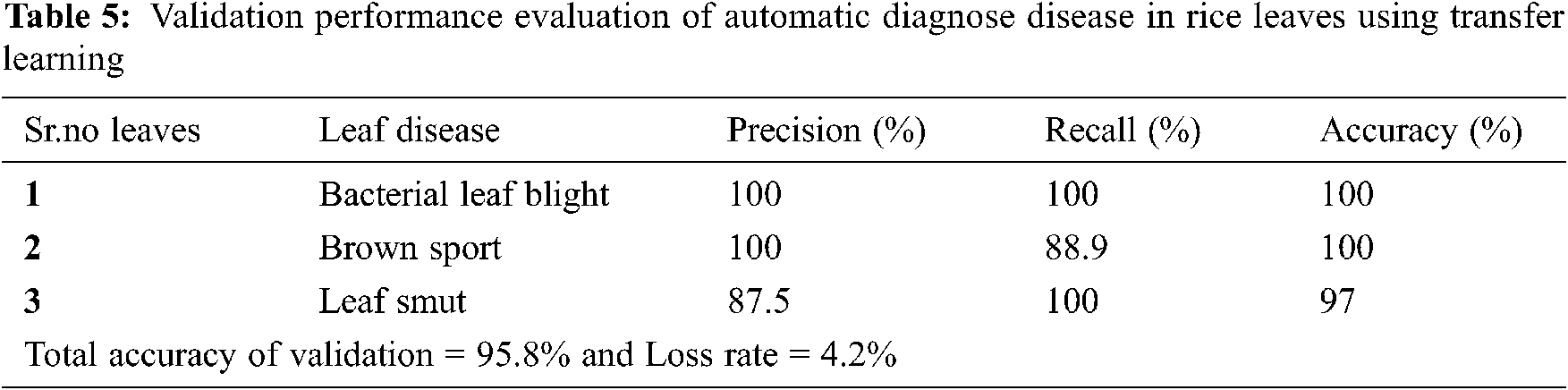
Tab. 5 describes the validation performance evaluation of our proposed system in which precision is 100% in bacterial leaf blight, 100% in brown sport and 87.5% in leaf smut. Recall is 100% in bacterial leaf blight, 88.9% in brown sport and 100% in leaf smut. Accuracy is 100% in bacterial leaf blight, 100% in brown sport and 97% in leaf smut.
We categorized the validation images using the adjusted AlexNet approach, as seen by this training progress. Our model was trained with a small number of data in the first epoch, which explains why the accuracy based on test data was poor and there was a lot of loss. However, as the number of epochs was steadily increased, the accuracy improved as the trained data was expanded, and the validation loss decreased. Below figure shows the accuracy of all related work, then we analysis that our accuracy is greater from all others.
Fig. 7 shown the comparison of proposed system accuracy with related work. We have attempted to work with the same issue area using a different methodology called AlexNet, with high expectations. Finally, we achieved 99% accuracy in detecting rice leaf disease, as predicted by our network.
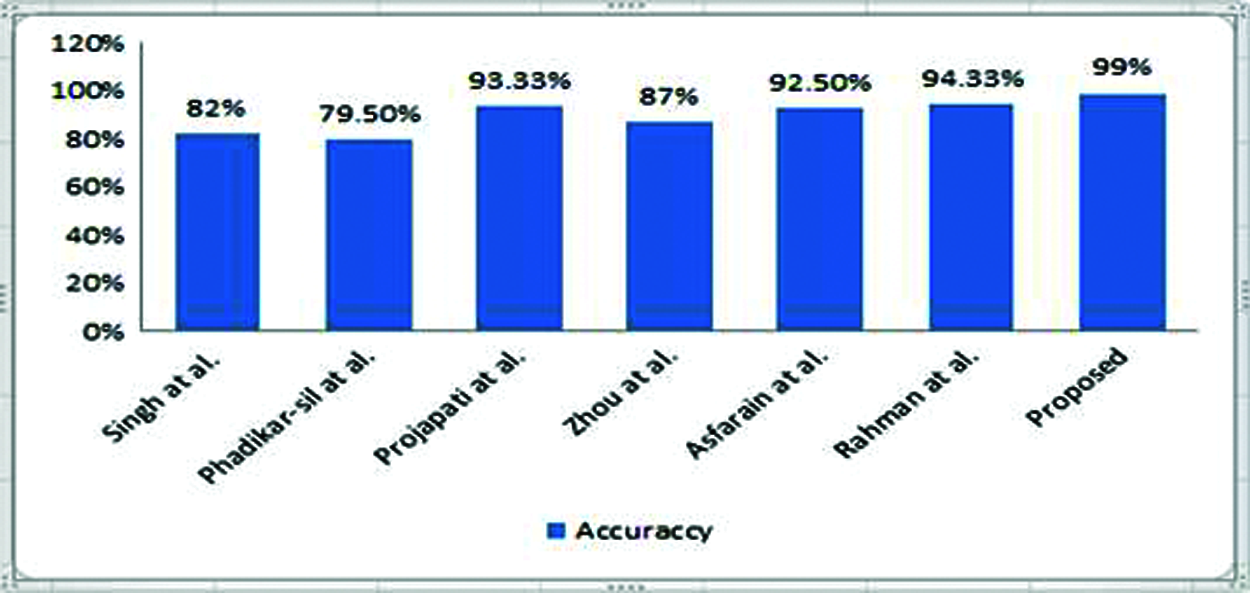
Figure 7: Comparison of proposed system accuracy with related work
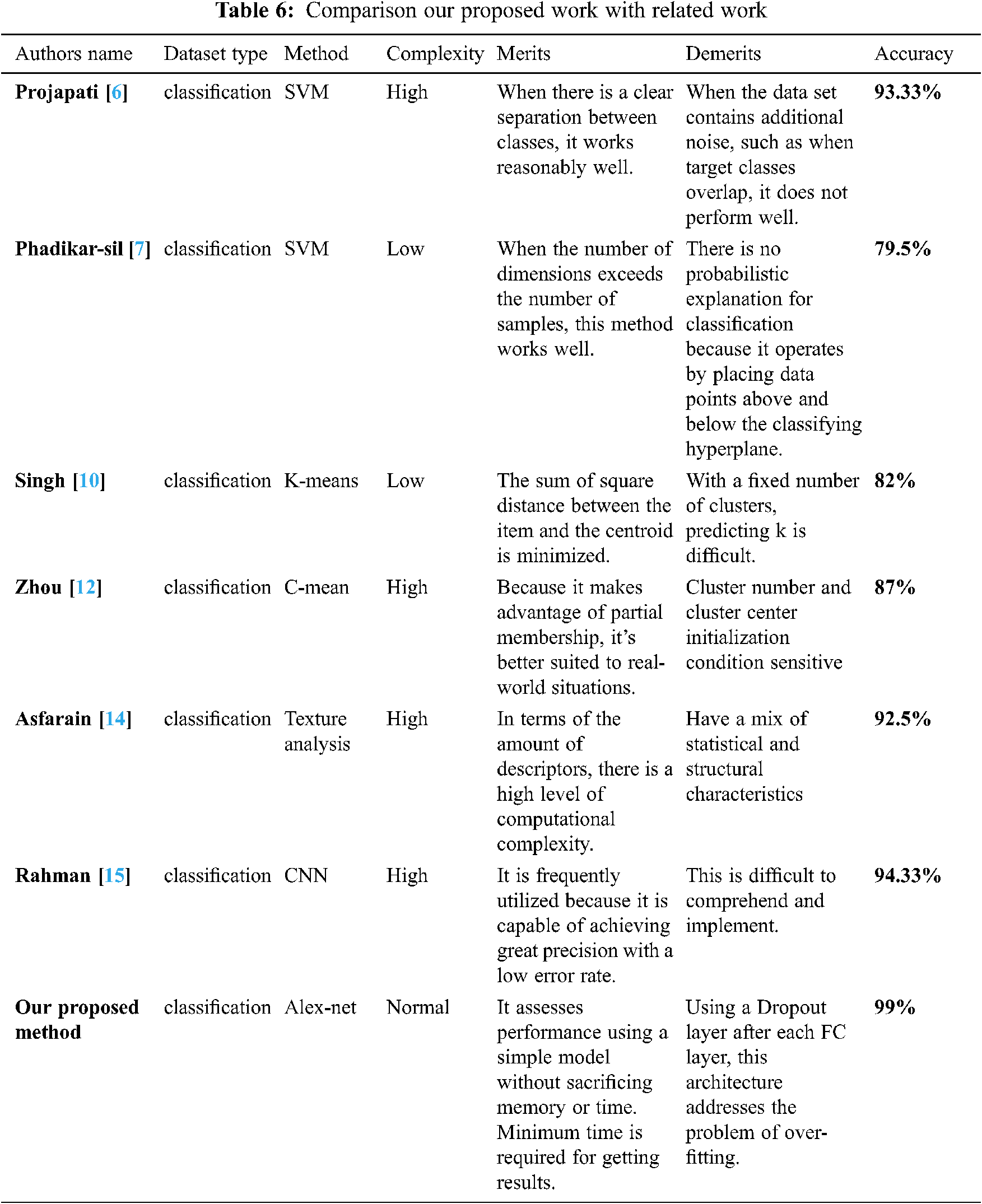
We compared our work’s accuracy to that of other studies. As can be seen in the Tab. 6, our accuracy is the best of all the research publications
In this research, we used the Alex Net neural network to detect three common rice leaf diseases, and our results are more accurate than prior works that used alternative methodologies. For picture tasks, Alex Net was preferred above CNNs in our study. This article is economically significant not only for Pakistan, but also for other rice-producing countries. We will still be able to use the K-means clustering technique, Fuzzy c-mean clustering, CART, and other fuzzy inference systems (FIS) for accuracy and process time judgment in the future.
Acknowledgement: We thank our families and colleagues who provided us with moral support.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. S. Phadikar, J. Sil and A. K. Das, “Classification of rice leaf diseases based on morphological changes,” Information and Electronics Engineering, vol. 2, no. 3, pp. 460–463, 2012. [Google Scholar]
2. R. A. D. Pugoy and V. Y. Mariano, “Automated rice leaf disease detection using color image analysis,” in Third Int. Conf. on Digital Image Processing, Chengdu, China, 8009, pp. 80090– 80097, 2011. [Google Scholar]
3. S. Weizheng, W. Yachun, C. Zhanliang and W. Hongda, “Grading method of leaf spot disease based on image processing,” in Int,. Conf. on Computer Science and Software Engineering, Hubei, 6, pp. 491–494, 2008. [Google Scholar]
4. D. A. Bashish, M. Braik and S. B. Ahmad, “A framework for detection and classification of plant leaf and stem diseases,” in Int. Conf. on Signal and Image Processing, Póvoa de Varzim, Portugal, pp. 113–118, 2010. [Google Scholar]
5. P. K. Sethy, N. K. Barpanda, A. K. Rath and S. K. Behera, “Image processing techniques for diagnosing rice plant disease: A survey,” Procedia Computer Science, vol. 167, no. 1, pp. 516–530, 2020. [Google Scholar]
6. H. B. Prajapati, J. P. Shah and V. K. Dabhi, “Detection and classification of rice plant diseases,” Intelligent Decision Technologies, vol. 11, no. 3, pp. 357–373, 2017. [Google Scholar]
7. S. Phadikar and J. Sil, “Rice disease identification using pattern recognition techniques,” in Int. Conf. on Computer and Information Technology, Sydney, Australia, 11, pp. 420–423, 2008. [Google Scholar]
8. P. Killeen and S. Behavior, “Fuzzy logic,” Journal of Experimental Psychology: Animal Behavior Processes, vol. 35, pp. 93–100, 2013. [Google Scholar]
9. B. S. Prajapati, V. K. Dabhi and H. B. Prajapati, “A survey on detection and classification of cotton leaf diseases,” International Conference on Electrical, Electronics and Optimization Techniques, vol. 10, pp. 2499–2506, 2016. [Google Scholar]
10. M. E. Pothen and M. L. Pai, “Detection of rice leaf diseases using image processing,” in Fourth Int. Conf. on Computing Methodologies and Communication, Erode, India, pp. 424–430, 2020. [Google Scholar]
11. S. D. Khirade and A. B. Patil, “Plant disease detection using image processing,” in Int. Conf. on Computing Communication Control and Automation, Pune, India, pp. 768–771, 2015. [Google Scholar]
12. Z. Zhou, Y. Zang, Y. Li, Y. Zhang, P. Wang et al., “Rice plant-hopper infestation detection and classification algorithms based on fractal dimension values and fuzzy c-means,” Mathematical and Computer Modelling, vol. 58, no. 3-4, pp. 701–709, 2013. [Google Scholar]
13. P. Sanyal and S. C. Patel, “Pattern recognition method to detect two diseases in rice plants,” The Imaging Science Journal, vol. 56, no. 6, pp. 319–325, 2008. [Google Scholar]
14. A. Asfarian, Y. Herdiyeni, A. Rauf and K. H. Mutaqin, “A computer vision for rice disease identification to support integrated pest management,” Crop Protection, vol. 61, pp. 103–104, 2014. [Google Scholar]
15. C. R. Rahman, P. S. Arko, M. E. Ali, M. A. I. Khan, S. H. Apon et al., “Identification and recognition of rice diseases and pests using convolutional neural networks,” Biosystems Engineering, vol. 194, no. 1, pp. 112–120, 2020. [Google Scholar]
16. H. Khalid, M. Hussain, M. A. A. Ghamdi, T. Khalid, K. Khalid et al., “A comparative systematic literature review on knee bone reports from MRI, x-rays and CT scans using deep learning and machine learning methodologies,” Diagnostics, vol. 10, no. 8, pp. 118–139, 2020. [Google Scholar]
17. M. A. Khan, S. Abbas, K. M. Khan, M. A. Ghamdi and A. Rehman, “Intelligent forecasting model of covid-19 novel coronavirus outbreak empowered with deep extreme learning machine,” Computers, Materials & Continua, vol. 64, no. 3, pp. 1329–1342, 2020. [Google Scholar]
18. A. H. Khan, M. A. Khan, S. Abbas, S. Y. Siddiqui, M. A. Saeed et al., “Simulation, modeling, and optimization of intelligent kidney disease predication empowered with computational intelligence approaches,” Computers Materials & Continua, vol. 67, no. 2, pp. 1399–1412, 2021. [Google Scholar]
19. G. Ahmad, S. Alanazi, M. Alruwaili, F. Ahmad, M. A. Khan et al., “Intelligent ammunition detection and classification system using convolutional neural network,” Computers Materials & Continua, vol. 67, no. 2, pp. 2585–2600, 2021. [Google Scholar]
20. B. Shoaib, Y. Javed, M. A. Khan, F. Ahmad, M. Majeed et al., “Prediction of time series empowered with a novel srekrls algorithm,” Computers Materials & Continua, vol. 67, no. 2, pp. 1413–1427, 2021. [Google Scholar]
21. S. Aftab, S. Alanazi, M. Ahmad, M. A. Khan, A. Fatima et al., “Cloud-based diabetes decision support system using machine learning fusion,” Computers Materials & Continua, vol. 68, no. 1, pp. 1341–1357, 2021. [Google Scholar]
22. Q. T. A. Khan, S. Abbas, M. A. Khan, A. Fatima, S. Alanazi et al., “Modelling intelligent driving behaviour using machine learning,” Computers Materials & Continua, vol. 68, no. 3, pp. 3061–3077, 2021. [Google Scholar]
23. S. Hussain, R. A. Naqvi, S. Abbas, M. A. Khan, T. Sohail et al., “Trait based trustworthiness assessment in human-agent collaboration using multi-layer fuzzy inference approach,” IEEE Access, vol. 9, no. 4, pp. 73561–73574, 2021. [Google Scholar]
24. N. Tabassum, A. Ditta, T. Alyas, S. Abbas, H. Alquhayz et al., “Prediction of cloud ranking in a hyperconverged cloud ecosystem using machine learning,” Computers Materials & Continua, vol. 67, no. 1, pp. 3129–3141, 2021. [Google Scholar]
25. M. W. Nadeem, H. G. Goh, M. A. Khan, M. Hussain, M. F. Mushtaq et al., “Fusion-based machine learning architecture for heart disease prediction,” Computers Materials & Continua, vol. 67, no. 2, pp. 2481–2496, 2021. [Google Scholar]
26. A. I. Khan, S. A. R. Kazmi, A. Atta, M. F. Mushtaq, M. Idrees et al., “Intelligent cloud-based load balancing system empowered with fuzzy logic,” Computers Materials and Continua, vol. 67, no. 1, pp. 519–528, 2021. [Google Scholar]
27. S. Y. Siddiqui, I. Naseer, M. A. Khan, M. F. Mushtaq, R. A. Naqvi et al., “Intelligent breast cancer prediction empowered with fusion and deep learning,” Computers Materials and Continua, vol. 67, no. 1, pp. 1033–1049, 2021. [Google Scholar]
28. R. A. Naqvi, M. F. Mushtaq, N. A. Mian, M. A. Khan, M. A. Yousaf et al., “Coronavirus: A mild virus turned deadly infection,” Computers Materials and Continua, vol. 67, no. 2, pp. 2631–2646, 2021. [Google Scholar]
29. A. Fatima, M. A. Khan, S. Abbas, M. Waqas, L. Anum et al., “Evaluation of planet factors of smart city through multi-layer fuzzy logic,” The Isc International Journal of Information Security, vol. 11, no. 3, pp. 51–58, 2019. [Google Scholar]
30. F. Alhaidari, S. H. Almotiri, M. A. A. Ghamdi, M. A. Khan, A. Rehman et al., “Intelligent software-defined network for cognitive routing optimization using deep extreme learning machine approach,” Computers, Materials and Continua, vol. 67, no. 1, pp. 1269–1285, 2021. [Google Scholar]
31. M. W. Nadeem, M. A. A. Ghamdi, M. Hussain, M. A. Khan, K. M. Khan et al., “Brain tumor analysis empowered with deep learning: A review, taxonomy, and future challenges,” Brain Sciences, vol. 10, no. 2, pp. 118–139, 2020. [Google Scholar]
32. A. Atta, S. Abbas, M. A. Khan, G. Ahmed and U. Farooq, “An adaptive approach: Smart traffic congestion control system,” Journal of King Saud University—Computer and Information Sciences, vol. 32, no. 9, pp. 1012–1019, 2020. [Google Scholar]
33. M. A. Khan, S. Abbas, A. Rehman, Y. Saeed, A. Zeb et al., “A machine learning approach for blockchain-based smart home networks security,” IEEE Network, vol. 35, no. 3, pp. 223–229, 2020. [Google Scholar]
34. M. A. Khan, M. Umair, M. A. Saleem, M. N. Ali and S. Abbas, “Cde using improved opposite-based swarm optimization for mimo systems,” Journal of Intelligent & Fuzzy Systems, vol. 37, no. 1, pp. 687–692, 2019. [Google Scholar]
35. M. A. Khan, M. Umair and M. A. Saleem, “GA based adaptive receiver for MC-CDMA system,” Turkish Journal of Electrical Engineering & Computer Sciences, vol. 23, no. 1, pp. 2267–2277, 2015. [Google Scholar]
36. S. Samir, E. Emary, K. E. Sayed and H. Onsi, “Optimization of a pre-trained alexnet model for detecting and localizing image forgeries,” Information-an International Interdisciplinary Journal, vol. 11, no. 5, pp. 275–289, 2020. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |