DOI:10.32604/csse.2022.021980

| Computer Systems Science & Engineering DOI:10.32604/csse.2022.021980 |  |
| Article |
X-Ray Covid-19 Detection Based on Scatter Wavelet Transform and Dense Deep Neural Network
Informatics Department, Technical College of Management, Middle Technical University, Baghdad, Iraq
*Corresponding Author: Ali Sami Al-Itbi. Email: ali.sami@mtu.edu.iq
Received: 23 July 2021; Accepted: 26 August 2021
Abstract: Notwithstanding the discovery of vaccines for Covid-19, the virus's rapid spread continues due to the limited availability of vaccines, especially in poor and emerging countries. Therefore, the key issues in the present COVID-19 pandemic are the early identification of COVID-19, the cautious separation of infected cases at the lowest cost and curing the disease in the early stages. For that reason, the methodology adopted for this study is imaging tools, particularly computed tomography, which have been critical in diagnosing and treating the disease. A new method for detecting Covid-19 in X-rays and CT images has been presented based on the Scatter Wavelet Transform and Dense Deep Neural Network. The Scatter Wavelet Transform has been employed as a feature extractor, while the Dense Deep Neural Network is utilized as a binary classifier. An extensive experiment was carried out to evaluate the accuracy of the proposed method over three datasets: IEEE 80200, Kaggle, and Covid-19 X-ray image data Sets. The dataset used in the experimental part consists of 14142. The numbers of training and testing images are 8290 and 2810, respectively. The analysis of the result refers that the proposed methods achieved high accuracy of 98%. The proposed model results show an excellent outcome compared to other methods in the same domain, such as (DeTraC) CNN, which achieved only 93.1%, CNN, which achieved 94%, and stacked Multi-Resolution CovXNet, which achieved 97.4%. The accuracy of CapsNet reached 97.24%.
Keywords: Covid-19 detection; scatter wavelet transform; deep learning; dense Deep Neural Network
Ended in December 2019, a cluster of pneumonia cases with an unknown cause was linked to a seafood market in Wuhan, Hubei Province, China, and quickly becoming a pandemic [1]. The virus was isolated by Scientists from the patient’s airway epithelial cells. Furthermore, other than SARS-CoV, influenza, avian influenza, MERS-CoV, and numerous respiratory viruses, it has been formally designated as severe acute respiratory syndrome coronavirus 2 (SARSCoV2) [2,3], SARS-CoV-2 [4]. Initially, this new virus was called 2019-nCoV [5–7]. On Feb 11, 2020, the virus was renamed Covid-19 by the World Health Organization (WHO) [8]. Since the first identification in the 1960s, Coronavirus has been responsible for a large, broad proportion of human respiratory tract infections. At least seven new types of coronaviruses that can infect a human being, including the severe acute respiratory syndrome coronavirus, have been identified since 2003 and have caused significant morbidity and mortality [9,10]. Covid-19 has belonged to a coronavirus family named corona based on the outer shape, the characteristic crown-like viral particles (virions) that dot their surface [11]. Coronavirus is enveloped, single-stranded, positive-sense RNA viruses [12,13]. The RNA of the Coronavirus can be split into four different genera Alpha α, Beta β Gamma γ and Delta δ. The genera of α and β infect only mammals, while γ and δ can infect birds. α and β can spread and infect people. In addition to that, they usually cause respiratory, enteric, hepatic and neurological diseases and are widely distributed among humans, other mammals and birds [3,14–17]. It is assumed that the Covid-19 was transmitted from animals (bats) to humans for the first time while humans infected one another [18–20]. Covid-19 is an acute, life-threatening respiratory virus that mainly presents pneumonia with characteristic radiological changes of bilateral lunge [21–23]. There are many symptoms of Covid-19; people infected with Covid-19 may suffer from a dry cough, fever, trouble breathing, exhaustion, pains and aches, diarrhea and a sore throat, which are all potential symptoms in some cases [24–28]. In general, these symptoms begin mildly and gradually become more severe as the disease progresses. The true risk of Covid-19 is on older people, immunocompromised and the patients who suffer from chronic diseases along the line of cancer, high blood, pressure lung disease, diabetes, heart and chronic kidney disease. Both clinical symptoms and comprehensive laboratory tests should be evaluated to validate the diagnosis of Covid-19 [29–32].
Artificial Intelligence (AI) and related technologies are increasingly prevalent in healthcare and medical practice. It is relevant to visually-oriented specialty, such as radiography. This field has got particular attention; the reason is the wide use of x-ray examination, which is around two billion performed per year. AI techniques are also deeply employed in Magnetic Resonance Imaging (MRI) and Computed Tomography (CT) to help the medical staff diagnose and treat diseases [33–35]. Today, the world is facing a major health crisis because of the pandemic of Covid-19; the medical industry is working hard to find and utilize new technologies to help medical staff diagnose and control diseases. AI is one such technology that enables rapid monitoring of the virus's spread, classification of high-risk patients and real-time infection control. Additionally, it can forecast mortality risk by fully evaluating a patient’s prior outcomes. AI will support us in combating this illness by monitoring the community, offering medical support, issuing warnings and making infection prevention advice [36]. The Covid-19 is a serious contagious disease and rapidly spreading, and it has had disastrous consequences on both daily life and the global economy. One of the best ways to prevent and reduce it is by identifying positive cases as early as possible. Furthermore, the sooner we find those cases, the sooner we can treat them.
Real-time reverse-transcription polymerase chain reaction (RT-PCR), X-ray and CT are utilized to diagnose the Covid-19 to distinguish between infected and non-infected cases. The reports have found that RT-PCR has a lower sensitivity compering to CT of detect Covid-19 [37–39]. The DarkNet model was used as a classifier to detect and classify the chaste x-ray images for suspected COVID-19 cases. The accuracy of the system was 98.08% for binary classes [40]. An automatic framework based on deep learning has been developed to detect Covid-19 using chest CT [41]. Chest CT scan is an important and most common method that has been used to identify pneumonia. Gozes, O. et al. have been developed AI-based automated CT image analyzer tools to detecting, quantifying and tracking the Covid-19 in order to differentiate between positive and negative cases. The possible working point was 98.2% sensitivity and 92.2% specificity [42]. Both Canonical-Correlation Analysis (CCA) and Support Vector Machine (SVM) were implemented to detect Covid-19, CCA was used as a feature selection, while SVM was used as a classifier. The performance was excellent, with 98.27% accuracy, and the sensitivity was 98.93% [43].
Ahmmed K. et al. (2020) begin with preprocessing the images of x-ray by converting them into grayscales and resize them into 100 × 100. The proposed Convolutional Neural Network (CNN) begins with 32 filters, then 64 filters, and max pooling filters. The result input is in a dense neural network for the classification of the image. The chest X-ray images were obtained from the Covid-19 Radiography Database, which was considered as a primary dataset in this work. This dataset contained 1341 normal and 219 Covid-19 patient’s images. The accuracy obtained from that model was 94% [44].
The study of Asmaa A. et al. (2020) validates Decompose, Transfer and Compose (DeTraC), a deep CNN for COVID-19 chest X-ray image classification. DeTraC can cope with anomalies in the picture dataset by employing a class decomposition process to investigate its class boundaries. The experimental results demonstrated DeTraC's capacity to detect COVID-19 cases from a large image dataset, and the accuracy gained for this approach was 93% [45]. Different types of CovXNets are built and trained by Mahmud T. et al. (2020) with X-ray images of varying resolutions in the proposed method, and a stacking strategy is used to optimize their predictions. Finally, a gradient-based discriminative localization is implemented to identify the aberrant regions of X-ray pictures relating to distinct forms of pneumonia. One of the datasets included in this investigation is a collection of 5856 images obtained in Guangzhou Medical Center, China, consisting of 1583 normal X-rays, 1493 non-COVID viral pneumonia X-rays and 2780 bacterial pneumonia X-rays. Another dataset was provided by Sylhet Women's Medical College/Bangladesh. It contains 305 X-rays of distinct Covid-19, was acquired and confirmed by an expert radiologist. Accuracy gained from the proposed model is 97.4% [46]. The study of Toraman S. et al. (2020) uses chest X-ray images and capsule networks, and this study proposes a unique artificial neural network and Convolutional CapsNet, to identify COVID-19 disease. CpsNet is made up of five layers of convolutions. The first layer has 16 kernels of size 5 × 5 with a stride of 1. The first layer exit was max pooled with a size of 2 and a stride of 2. The next structure was used in the next two layers. The second and third levels’ kernel numbers are 32 and 64, respectively. The fourth layer contains 128 kernels with a stride of 1. The fifth layer is the major one, which comprises 32 capsules, each of which has applied 9 convolutional kernels with a stride of 1. The LabelCaps layer has 16-dimensional (16D) capsules for two and three classes, and all layers use the Rectified Linear Activation Function (ReLU). The dataset used in this study was collected by Cohen, and the accuracy achieved was 97.24% [47]. Several chest x-ray pictures from diverse sources are gathered by Haghanifar A. et al. (2020), and the largest publicly accessible collection was created. Finally, COVID-CXNet was developed using the transfer learning paradigm and the well-known CheXNet model. Based on relevant and meaningful variables and exact localization, this powerful model can recognize the new coronavirus pneumonia. COVID-CXNet is a step toward a COVID-19 detection system that is entirely automated and reliable. The accuracy for base model version I was 96%, and for base model version II was 98% [48]. The study of Zhang J et al. (2020) analyses the backbone network. In this article, the backbone network was an 18-layer residual CNN that was pre-trained on the ImageNet dataset. The rectangles with different colors in Fig. 1(a) depict the five stages of the backbone network. In the first stage, large convolutions with 7 × 7 kernels and a stride of 2 are utilized, followed by a 3 × 3 max-pooling layer with a stride of 2. Following that, each stage comprises two residual blocks, each with two convolutional layers and one skip connection. The input image can be encoded as a feature map with an output stride after layer-by-layer convolutional procedures. The accuracy gained was about 0.96 to detect Covid cases and 70% for detecting non-Covid cases [49]. The proposed work of Das A.K et al. (2020) uses many CNN models, including DenseNet201, Resnet50V2 and Inceptionv3. Individually, they have been trained to make predictions on their own. The models are then merged to forecast a class value using a novel weighted average ensembling technique. The study used publicly available chest X-ray pictures of COVID and normal instances to test the solution's efficacy. Training, test and validation sets were created using 538 photos of COVID patients and 468 images. The proposed method achieved an accuracy of 91.62 percent, higher than both current CNN models and the benchmark algorithm [50]. Narin A et al. (2021) have applied five different pre-trained CNN-based models, which are ResNet101, ResNet50, ResNet152, Inception-ResNetV2 and InceptionV3. For the detection of coronavirus pneumonia-infected patients, the chest X-ray radiographs were proposed in this study. The average accuracy from tests was 0.98 [51].
This paper answers two questions; first, is a scattering wavelet an effecting feature to detect Covid-19 from x-ray images? While the second question is that scattering wavelet represents a discriminant feature that can build a classifier trained with a small dataset? This research aims to build a model that can detect the infected cases of Covid-19 from X-ray images.
The rest of the paper has been classified as follows: Section 2 discusses the wavelet scattering transform, while the dense deep neural network is debated in Section 3. The proposed technique is given in Section 4. Finally, the conclusion is presented in Section 5.
2 Scattering Wavelet Transform (SWT)
Since the capability of wavelets for multi-resolution image representation, the interest in using wavelet frames for image processing has grown over recent decades. The framelet transform is close to wavelet transform Framelet has two high-frequency filters, which produce more sub bands in decomposition. Convolutional structure of the windowed scattering transform means that each layer is captured from previous by applying wavelet modulus decomposition

Figure 1: Two textures with same Fourier spectrum. (a) Textures X(u). Top: Brodatz texture. Bottom: Gaussian process. (b) Same power spectrum RX b (ω). (c) Different scattering coefficients SJ[p]X for m = 2
The SWT was early presented by Mallat [53] in 2012. The SWT constructs translation invariant, stable and informative signal representations [54]. It is stable in deformations and maintains class discrimination, especially successful in classification [55]. The SWT provides a handy classification tool [56–59]. Reliable information can be extracted at various scales by SWT, the coefficients of SWT are more descriptive than the Fourier while interacting with small deformation and rotation invariant signals [60].
The SWT network consists of two layers without training and achieves similar performance to the convolutional network. The network generates a representation Φ that is invariant to rotation, translation, color discrimination and stability to small deformations. A scattering transform is the cascading of linear wavelet transform and modulus nonlinearity.
The image is filtered using the first wavelet transform W1, Eq. (1). A complex wavelet that has been rotated and scaled, as shown in Fig. 2, then producing the modulus of each coefficient to define the first scattering layer
The initial wavelet layer is described by the base wavelet ψ1 (u), a complex function with excellent image plane localization. This wavelet is scaled by 2j1 where j is an integer and rotated by
where
This wavelet transform is a filter on image x(u), and wavelet coefficients are computed by
The spatial variable u would be subsampled by
The index

Figure 2: A 3-layer scatter network. Shows how extract any coefficients
3 Deep Dense Neural Network (DDNN)
Deep Learning (DL) has its origins in Artificial Intelligence and has emerged as a distinct and highly useful area in recent years [61]. DL architectures in Fig. 3 include models consisting of an input layer, the output layer and two or even more hidden layers of nonlinear processing units, which conduct the extraction and transformation functions that are supervised or unsupervised [62]. These layers are placed in a hierarchical pattern, with each layer’s input being the preceding layer's output, computed using a suitably chosen activation function. Each hidden layer represents a distinct image of the input space and provides a different abstraction level that is ideally more useful and closer to the eventual objective [63]. One of the most effective neural networks is Dense Deep Neural Network. It consists of several fully connected layers. It is defined as a function that ranges from

Figure 3: Example of deep dense neural network architecture
The mathematical form of a fully connected network is
Here, σ is nonlinear, and the
while the weights w(l)ij of connection lines between node j in the layer number I and another node in layer
where
Relu usually has been used in deep dense neural networks. Relu function can be defined as follows:
Both the Relu function and its derivative are consistent. If the function receives any negative input, it returns 0; however, it returns that value if the input is positive. As a result, it produces an output from a range of 0 to infinity. Fig. 4 depicts the behavior of the Relu function.

Figure 4: Relu activation function
An active and accurate Covid-19 detection has been proposed based on the Scatter Wavelet Transform and DDNN. The SWT has been employed as a feature extractor, while the DDNN is utilized as a binary classifier. Firstly, it extracts all scattering wavelet feature sets from an image pattern. The scattering wavelet list of each image consists of 7812 features saved inside a list. The system converts each x-ray image to its scattering wavelet counterpart. Fig. 5 presents the original image and its SWT.

Figure 5: (a) Represent the original image before converting it to scattering wavelet (b) represent the same x-ray image in (a) after implement scattering wavelet conversion
The converted images or the list of scattering wavelet images are divided into training and testing set of 80% and 20%, respectively. The values produced from the scattering wavelet transformation process are over 1, so these values have to be normalized and converted in decimal format between 0 and 1 to be accepted by DDNN. The DDNN classifier consists of five layers. First, the input layer that is flattening the input list of 7812 features of each x-ray image; then three hidden layers with Relu functions consist of 256,128,64 neurons, respectively; lastly, the output layer that has one neuron with sigmoid activation function, which yields wither normal lung or infected one. DDNN architecture of the proposed system is apparent in Fig. 6.

Figure 6: Deep dense neural network classifier architecture in the proposed system
This Deep Dense Neural Classifier uses Adam that is short of adaptive moment estimation, a momentum-based optimizer. The loss function used is binary cross entropy. The binary classification problems give output in the form of probability. Binary cross entropy is usually the optimizer of choice. Fig. 7 presents the block diagram of the proposed method.

Figure 7: The proposed system
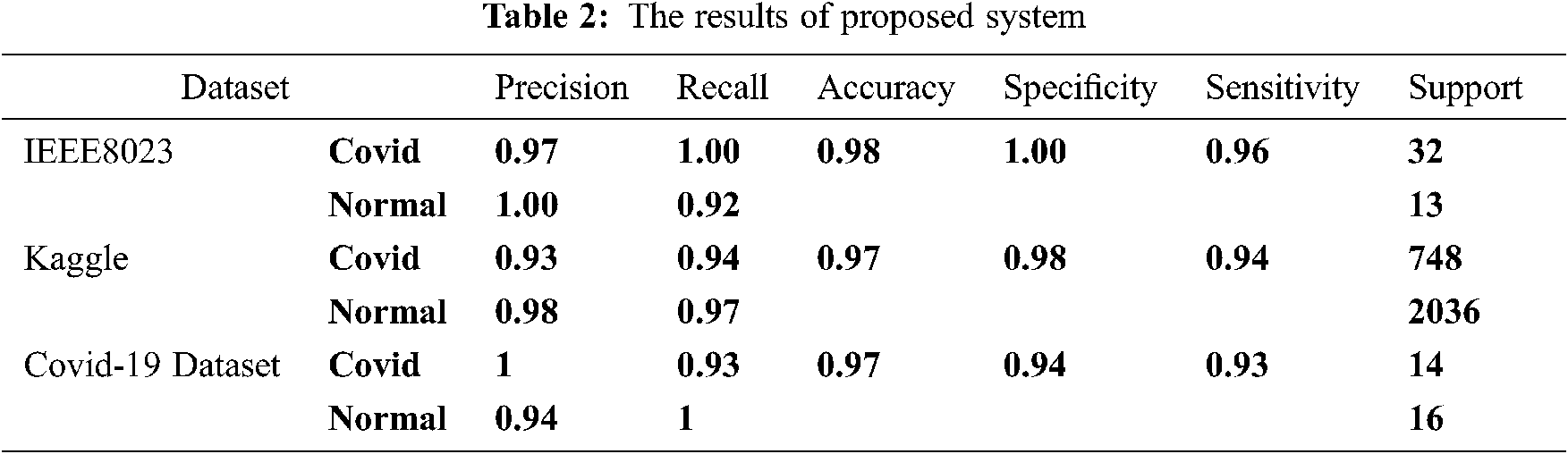
To test the proposed framework, this research has used three datasets for training, testing and evaluating the proposed method that has been developed to detect Covid-19 infections intelligently. The existing datasets have the disequilibrium issue. This issue is handled by class weighting within the training process. Tab. 1 represents the dataset with the number of images used for testing in both Covid-19 and normal classes.

The datasets vary between big datasets as Kaggle [66], smaller IEEE 80200 [67], to tiny ones the COVID-19 X-ray Image Data Sets. The reason behind choosing these three datasets was to test the proposed system in datasets with different sizes to check the efficiency of that system, which depends on scattering wavelet features as a discriminative feature in small and big datasets. Using the small datasets confirms the claims that scattering wavelets can be a good discriminative between classes for datasets with a small training/testing set.
Five metrics that measure system efficiency in this paper are: accuracy, precision, recall, sensitivity and specificity. These five metrics are derived from four parameters: true positive (TP), true negative (TN), false positive (FP) and false-negative (FN). The formulas of these metrics are in equations from 16 to 19:
The features were extracted using scattering wavelet transform, while the proposed system used DDNN as a binary classifier to classify the input x-ray images to either be detected with Covid-19 or not. The DDNN is a fully connected deep network that use a linear operation where every input is connected to every output by a wight. The system is trained using adaptive moment learning rate (ADAM) with epochs between 60 and 100 that is using the early stop in keras library that makes the training process stops when the model gets the best accuracy.
The results show that using scattering wavelet feature with deep dense neural networks classifier achieved high performance. The performance of the proposed model is clear for both small datasets like the third dataset of Covid-19 and larger ones like Kaggle and IEEE 8023. From Tab. 2, the results can be understood. For IEEE 8023 dataset, the accuracy, in general, is 0.98. The sensitivity is 0.96 in general. The sensitivity for detecting positive cases is excellent, and recall has been reached 1.00 in detecting Covid cases. The specificity registering a score of 1.00 showing an excellent in avoiding false detection or false positive. False-positive reached zero, which means that the system did not make any wrong Covid 19 alert by considering a normal case as an infected case. The specificity accommodates the value of precision of normal class detection of 1. The confusion matrix in Fig. 8b shows the number of false-negative only one from 45 x-ray pictures in the testing set. Also, false-positive is zero in Fig. 8b, which represents an indicator of the high performance of the proposed method. In the Kaggle dataset, the accuracy was 0.97 while the sensitivity was 0.94 that is a good accuracy for an imbalanced dataset with a big difference between number of normal chest images and number of infected images. This percentage is clear in the confusion matrix in Fig. 9b with 685 real detected Covid-19 cases. Specificity was also very good, with about 0.98 equal to the precision of detecting normal cases, which are the negative cases (normal cases), which is also registered in the confusion matrix in Fig. 10b with 1985 of true negative cases. Lastly, it analyzes the smallest Covid-19 online dataset. The results show a general accuracy of 97%. The sensitivity of 93% shows that the system is working efficiently to detect Covid cases, Fig. 10b confusion matrix shows that true positive is 13 while the false positive is just 1 case. The confusion matrix registers 13 cases for true positive and 19 cases for true negative, making 19 cases of 30 total testing set images. These values indicate a high performance for the proposed model trained with a small dataset of x-ray images. The sensitivity is clearly accommodating the precision of Covid detection. The specificity of 94% represents the precision of catching the normal cases. The amount of specificity of 94% may be small due to the small dataset consisting only of 64 images for training.

Figure 8: Confusion matrix for dataset IEEE 8023 (a) Normalized cm, (b) non normalized cm

Figure 9: Confusion matric of kaggle dataset (a) Normalized cm, (b) non normalized cm

Figure 10: Confusion matrix for Covid-19 online dataset (a) Normalized cm, (b) non normalized cm
In the discussion section, the proposed model has been compared with previous related works; this is shown in Tab. 3. The proposed model relying on feature extracted by scattering wavelet and deep dense neural network has shown a good accuracy of over 96.5% on average. However, when using the models that depend on only convolution neural networks individually, the accuracy was lower than the reported values, except Ozturk et al., who used Darknet CNN. Abbas et al., for example, used a small dataset of 105 Covid-19 images and 80 normal images, trained a transferred DecTrac CNN model and got an accuracy about 93.1%, which is relatively low accuracy. The CNN model defiantly needs larger datasets to be trained with acceptable accuracy, and this also tested within the papers that used pre-trained model and mixed models that used more than one pre-trained model in papers [50] that used a Merge of (Densenet201, Resnet50v2, inceptionv3) and achieved only 91% of accuracy and [51] who proposed a model of five pre-trained model that reached to the same accuracy of proposed model.
The proposed model tested with the mentioned datasets. The resulted accuracy values are answering the two mentioned research questions in the introduction section. First, is scattering wavelet is an efficient? The accuracy values were achieved in the tests indicate that scattering wavelet transform is an efficient enough to train covid-19 detection model.
The second research question was whether the scattering wavelet efficient as a discriminant feature to build the covid-19 detection system with a small training set?
The system also tested with mentioned second dataset and the accuracy was high and stable with 97%.

Because of the vast number of deaths caused by the coronavirus pandemic, healthcare services in every country have been stretched to their limits. COVID-19 can be detected early and treated more quickly, simpler and less expensively will save lives and relieve the pressure on medical providers. The image processing techniques and artificial intelligence to X-ray images will play a major role in identifying COVID-19. This research has crafted an intelligent framework for identifying COVID-19 with high accuracy and minimal complexity. The present research aims to investigate the use of scattering wavelet as a discriminative feature to detect Covid-19 from x-ray images supported by deep learning technique. The proposed system was applied to three different datasets that are varying in size. The first dataset consists of 231 pictures, the second dataset contains 13808 x-ray pictures, and the third dataset consists of 112 pictures. The proposed system shows a stable efficiency even with a small dataset. The results insisted that the scattering wavelet is efficient in providing discriminative features about the internal patterns of images even in the case of small dataset availability. Moreover, the proposed work employed the effectiveness of deep learning of classification.
These training and testing images represent all of the images from the previous experiments. The results of the present study are compared to other methods in the same domain. It is identified that the proposed method has achieved an accuracy of 98% when compared to (DeTraC) CNN, which achieved only 93.1%, CNN, which achieved 94% and stacked MultiResolution CovXNet, which achieved 97.4%. The accuracy of CapsNet reached 97.24%. For future studies, the area of detecting Covid-19 from x-ray images needs much more patient datasets, well organized, balanced and standard, because current datasets have issues like unbalancing between classes or some mistakes like normal cases images stored accidentally in Covid-19 patient classes and vice versa.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. F. He, Y. Deng and W. Li, “Coronavirus disease 2019: What we know?” Journal of Medical Virology, vol. 92, no. 7, pp. 719–725, 2020. [Google Scholar]
2. A. E. Gorbalenya, S. C. Baker, R. Baric, R. J. Groot, C. Drosten et al., “Severe acute respiratory syndrome-related coronavirus: The species and its viruses – A statement of the Coronavirus Study Group,” bioRxiv, pp. 937862, This article preprint 2020. [Google Scholar]
3. N. Zhu, D. Zhang, W. Wang, X. Li, B. Yang et al., “A novel coronavirus from patients with pneumonia in China, 2019,” New England Journal of Medicine, vol. 382, no. 8, pp. 727–733, 2020. [Google Scholar]
4. C. S. G. of the International, “The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2,” Nature Microbiology, vol. 5, no. 4, pp. 536, 2020. [Google Scholar]
5. N. Zhu, D. Zhang, W. Wang, X. Li, B. Yang et al., “A novel coronavirus from patients with pneumonia in China, 2019,” New England Journal of Medicine, vol. 382, no. 8, pp. 727–733, 2020. [Google Scholar]
6. P. Zhou, X. L. Yang, X. G. Wang, B. Hu, L. Zhang et al., “A pneumonia outbreak associated with a new coronavirus of probable bat origin,” Nature, vol. 579, no. 7798, pp. 270–273, 2020. [Google Scholar]
7. F. Wu, S. Zhao, B. Yu, Y. M. Chen, W. Wang et al., “A new coronavirus associated with human respiratory disease in China,” Nature, vol. 579, no. 7798, pp. 265–269, 2020. [Google Scholar]
8. S. Jiang, Z. Shi, Y. Shu, J. Song, G. F. Gao et al., “A distinct name is needed for the new coronavirus,” Lancet, vol. 395, no. 10228, pp. 949, 2020. [Google Scholar]
9. J. S. Kahn and K. McIntosh, “History and recent advances in coronavirus discovery,” Pediatric Infectious Disease Journal, vol. 24, no. 11, pp. S223–S227, 2005. [Google Scholar]
10. Centers for Disease Control and Prevention, “Interim guidelines for collecting, handling, and testing clinical specimens from persons for coronavirus disease 2019 (Covid-19),” Accessed 10.07.2021. Available: https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html. [Google Scholar]
11. A. Giwa and A. Desai, “Novel coronavirus COVID-19: An overview for emergency clinicians,” Emergency Medicine Practice, vol. 22, no. 2 Suppl 2, pp. 1–21, 2020. [Google Scholar]
12. B. Tang, X. Wang, Q. Li, N. L. Bragazzi, S. Tang et al., “Estimation of the Transmission Risk of the 2019-nCoV and its implication for public health interventions,” Journal of Clinical Medicine, vol. 9, no. 2, pp. 462, 2020. [Google Scholar]
13. Y. Chen, Q. Liu and D. Guo, “Emerging coronaviruses: Genome structure, replication, and pathogenesis,” Journal of Medical Virology, vol. 92, no. 4, pp. 418–423, 2020. [Google Scholar]
14. J. Cui, F. Li and Z. L. Shi, “Origin and evolution of pathogenic coronaviruses,” Nature Reviews Microbiology, vol. 17, no. 3, pp. 181–192, 2019. [Google Scholar]
15. A. M. Sahan, A. S. Al-Itbi and J. S. Hameed, “COVID-19 detection based on deep learning and artificial bee colony,” Periodicals of Engineering and Natural Sciences, vol. 9, no. 1, pp. 29–36, 2021. [Google Scholar]
16. P. I. Lee and P. R. Hsueh, “Emerging threats from zoonotic coronaviruses-from SARS and MERS to 2019-nCoV,” Journal of Microbiology Immunology and Infection, vol. 53, no. 3, pp. 365–367, 2020. [Google Scholar]
17. P. C. Y. Woo, S. K. P. Lau, C. S. F. Lam, C. C. Y. Lau, A. K. L. Tsang et al., “Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus,” Journal of Virology, vol. 86, no. 7, pp. 3995–4008, 2012. [Google Scholar]
18. Y. Guan, B. J. Zheng, Y. Q. He, X. L. Liu, Z. X. Zhuang et al., “Isolation and characterization of viruses related to the SARS coronavirus from animals in Southern China,” Science, vol. 302, no. 5643, pp. 276–278, 2003. [Google Scholar]
19. T. Singhal, “A review of Coronavirus disease-2019 (Covid-19),” Indian Journal of Pediatrics, vol. 87, no. 4, pp. 281–286, 2020. [Google Scholar]
20. N. S. AlTakarli, “Emergence of Covid-19 infection: What is known and what is to be expected-narrative review article,” Dubai Medical Journal, vol. 3, no. 1, pp. 13–18, 2020. [Google Scholar]
21. K. Chong Ng Kee Kwong, P. R. Mehta, G. Shukla and A. R. Mehta, “COVID-19, SARS and MERS: A neurological perspective,” Journal of Clinical Neuroscience, vol. 77, no. 3, pp. 13–16, 2020. [Google Scholar]
22. M. P. Wilson and A. S. Jack, “Coronavirus disease 2019 (COVID-19) in neurology and neurosurgery: A scoping review of the early literature,” Clinical Neurology and Neurosurgery, vol. 193, no. 2, pp. 105866, 2020. [Google Scholar]
23. C. Huang, Y. Wang, X. Li, L. Ren, J. Zhao et al., “Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China,” Lancet, vol. 395, no. 10223, pp. 497–506, 2020. [Google Scholar]
24. L. L. Ren, Y. M. Wang, Z. Q. Wu, Z. C. Xiang, L. Guo et al., “Identification of a novel coronavirus causing severe pneumonia in human: A descriptive study,” Chinese Medical Journal, vol. 133, no. 9, pp. 1015–1024, 2020. [Google Scholar]
25. J. F. W. Chan, S. Yuan, K. H. Kok, K. K. W. To, H. Chu et al., “A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster,” Lancet, vol. 395, no. 10223, pp. 514–523, 2020. [Google Scholar]
26. X. Yang, Y. Yu, J. Xu, H. Shu, H. Liu et al., “Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: A single-centered, retrospective, observational study,” Lancet Respiratory Medicine, vol. 8, no. 5, pp. 475–481, 2020. [Google Scholar]
27. W. Wang, J. Tang and F. Wei, “Updated understanding of the outbreak of 2019 novel coronavirus (2019-nCoV) in Wuhan, China,” Journal of Medical Virology, vol. 92, no. 4, pp. 441–447, 2020. [Google Scholar]
28. P. Zhai, Y. Ding, X. Wu, J. Long, Y. Zhong et al., “The epidemiology, diagnosis and treatment of COVID-19,” International Journal of Antimicrobial Agents, vol. 55, no. 5, pp. 105955, 2020. [Google Scholar]
29. World Health Organization, “Q&A on coronaviruses (Covid-19),” Accessed 10.05 2021. Available: https://www.who.int/news-room/q-a-detail/q-a-coronaviruses#:~:text=symptoms. [Google Scholar]
30. People with Certain Medical Conditions. Centers for Disease Control and Prevention, Accessed 10.05 2021. Available: https://www.cdc.gov/coronavirus/2019-ncov/need-extra-precautions/people-with-medical-conditions.html. [Google Scholar]
31. A. Vishnevetsky and M. Levy, “Rethinking high-risk groups in Covid-19,” Multiple Sclerosis and Related Disorders, vol. 42, no. 8, pp. 102139, 2020. [Google Scholar]
32. R. Verity, L. C. Okell, I. Dorigatti, P. Whittaker, C. Whittaker et al., “Estimates of the severity of Covid-19 disease,” medRxiv, pp. 2020.03.09.20033357,2020. [Google Scholar]
33. T. Davenport and R. Kalakota, “The potential for artificial intelligence in healthcare,” Future Healthcare Journal, vol. 6, no. 2, pp. 94–98, 2019. [Google Scholar]
34. E. J. Topol, “High-performance medicine: The convergence of human and artificial intelligence,” Nature Medicine, vol. 25, no. 1, pp. 44–56, 2019. [Google Scholar]
35. S. Kulkarni, N. Seneviratne, M. S. Baig and A. H. A. Khan, “Artificial intelligence in medicine: Where are we now?,” Academic Radiology, vol. 27, no. 1, pp. 62–70, 2020. [Google Scholar]
36. R. Vaishya, M. Javaid, I. H. Khan and A. Haleem, “Artificial Intelligence (AI) applications for COVID-19 pandemic,” Diabetes & Metabolic Syndrome: Clinical Research & Reviews, vol. 14, no. 4, pp. 337–339, 2020. [Google Scholar]
37. T. Ai, Z. Yang, H. Hou, C. Zhan, C. Chen et al., “Correlation of chest CT and RT-PCR testing for Coronavirus disease 2019 (Covid-19) in China: A report of 1014 cases,” Radiology, vol. 296, no. 2, pp. E32–E40, 2020. [Google Scholar]
38. Y. Fang, H. Zhang, J. Xie, M. Lin, L. Ying et al., “Sensitivity of chest CT for Covid-19: Comparison to RT-PCR,” Radiology, vol. 296, no. 2, pp. E115–E117, 2020. [Google Scholar]
39. H. X. Bai, B. Hsieh, Z. Xiong, K. Halsey, J. W. Choi et al., “Performance of radiologists in differentiating COVID-19 from non-COVID-19 viral pneumonia at chest CT,” Radiology, vol. 296, no. 2, pp. E46–E54, 2020. [Google Scholar]
40. T. Ozturk, M. Talo, E. A. Yildirim, U. B. Baloglu, O. Yildirim et al., “Automated detection of Covid-19 cases using deep neural networks with X-ray images,” Computers In Biology And Medicine, vol. 121, pp. 103792, 2020. [Google Scholar]
41. L. Li, L. Qin, Z. Xu, Y. Yin, X. Wang et al., “Using artificial intelligence to detect covid-19 and community-acquired pneumonia based on pulmonary CT: Evaluation of the diagnostic accuracy,” Radiology, vol. 296, no. 2, pp. E65–E71, 2020. [Google Scholar]
42. O. Gozes, M. Frid-Adar, H. Greenspan, P. D. Browning, H. Zhang et al., “Rapid ai development cycle for the coronavirus (covid-19) pandemic: Initial results for automated detection & patient monitoring using deep learning ct image analysis,” arXiv preprint arXiv:2003.05037, 2020. [Google Scholar]
43. U. Özkaya, S. Öztürk and M. Barstugan, “Coronavirus (covid-19) classification using deep features fusion and ranking technique,” in Big Data Analytics and Artificial Intelligence Against COVID-19: Innovation Vision and Approach. Springer, pp. 281–295, 2020. [Google Scholar]
44. K. Ahammed, M. S. Satu, M. Z. Abedin, M. A. Rahaman and S. M. S. Islam, “Early detection of coronavirus cases using chest x-ray images employing machine learning and deep learning approaches,” medRxiv, 2020, 10.1101/2020.06.07.20124594. [Google Scholar]
45. A. Abbas, M. M. Abdelsamea and M. M. Gaber, “Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network,” Applied Intelligence, vol. 51, no. 2, pp. 854–864, 2021. [Google Scholar]
46. T. Mahmud, M. A. Rahman and S. A. Fattah, “CovXNet: A multi-dilation convolutional neural network for automatic COVID-19 and other pneumonia detection from chest X-ray images with transferable multi-receptive feature optimization,” Computers in Biology and Medicine, vol. 122, no. 1, pp. 103869, 2020. [Google Scholar]
47. S. Toraman, T. B. Alakus and I. Turkoglu, “Convolutional capsnet: A novel artificial neural network approach to detect COVID-19 disease from X-ray images using capsule networks,” Chaos, Solitons & Fractals, vol. 140, pp. 110122, 2020. [Google Scholar]
48. A. Haghanifar, M. M. Majdabadi, Y. Choi, S. Deivalakshmi and S. Ko, “Covid-cxnet: Detecting covid-19 in frontal chest x-ray images using deep learning,” arXiv preprint arXiv:2006.13807, 2020. [Google Scholar]
49. J. Zhang, Y. Xie, Y. Li, C. Shen and Y. Xia, “Covid-19 screening on chest x-ray images using deep learning based anomaly detection, vol.27, arXiv preprint arXiv:2003.12338, 2020. [Google Scholar]
50. A. K. Das, S. Ghosh, S. Thunder, R. Dutta, S. Agarwal et al., “Automatic COVID-19 detection from X-ray images using ensemble learning with convolutional neural network,” Pattern Analysis and Applications, vol. 24, no. 3, pp. 1111–1124, 2021. [Google Scholar]
51. A. Narin, C. Kaya and Z. Pamuk, “Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks,” Pattern Analysis and Applications, vol. 24, no. 3, pp. 1207–1220, 2021. [Google Scholar]
52. J. Bruna, “Scattering representations for recognition,” Ph.D. dissertation, Ecole Polytechnique X, 2013. [Google Scholar]
53. S. Mallat, “Group invariant scattering,” Communications on Pure and Applied Mathematics, vol. 65, no. 10, pp. 1331–1398, 2012. [Google Scholar]
54. Y. Jin and Y. Duan, “Wavelet scattering network-based machine learning for ground penetrating radar imaging: Application in pipeline identification,” Remote Sensing, vol. 12, no. 21, pp. 3655, 2020. [Google Scholar]
55. Z. Liu, G. Yao, Q. Zhang, J. Zhang and X. Zeng, “Wavelet scattering transform for ECG beat classification,” Computational and Mathematical Methods in Medicine, vol. 2020, pp. 3215681, 2020. [Google Scholar]
56. J. Bruna and S. Mallat, “Classification with scattering operators,” in 2011 IEEE Conf. on Computer Vision and Pattern Recognition, Proc. of CVPR 2011, Colorado Springs, CO, USA, pp. 1561–1566, 2011. [Google Scholar]
57. J. Andén and S. Mallat, “Multiscale scattering for audio classification,” in ISMIR, 2011. Miami, FL, pp. 657–662, 2011. [Google Scholar]
58. J. Andén and S. Mallat, “Deep scattering spectrum,” IEEE Transactions on Signal Processing, vol. 62, no. 16, pp. 4114–4128, 2014. [Google Scholar]
59. R. Leonarduzzi, H. Liu and Y. Wang, “Scattering transform and sparse linear classifiers for art authentication,” Signal Processing, vol. 150, no. 6735, pp. 11–19, 2018. [Google Scholar]
60. B. Soro and C. Lee, “A wavelet scattering feature extraction approach for deep neural network based indoor fingerprinting localization,” Sensors, vol. 19, no. 8, pp. 1790, 2019. [Google Scholar]
61. L. Deng and D. Yu, “Deep learning: Methods and applications,” Foundations and Trends in Signal Processing, vol. 7, no. 3–4, pp. 197–387, 2013. [Google Scholar]
62. N. Bosch and L. Paquette, “Unsupervised deep autoencoders for feature extraction with educational data,” in Deep Learning with Educational Data Workshop at the 10th Int. Conf. on Educational Data Mining, Wuhan, Hubei, 25–28 June 2017. [Google Scholar]
63. J. Patterson and A. Gibson, Deep learning: A practitioner's approach. O'Reilly Media, Inc., 2017. [Google Scholar]
64. B. Ramsundar and R. B. Zadeh, TensorFlow for deep learning: from linear regression to reinforcement learning. O'Reilly Media, Inc., 2018. [Google Scholar]
65. Q. Mei, M. Gül and M. R. Azim, “Densely connected deep neural network considering connectivity of pixels for automatic crack detection,” Automation in Construction, vol. 110, no. 1, pp. 103018, 2020. [Google Scholar]
66. COVID-19 Radiography Database 2020. [Online]. Available: https://www.kaggle.com/tawsifurrahman/covid19-radiography-database. [Google Scholar]
67. ieee8023 / covid-chestxray-dataset, [Online]. Available: Oct 1, 2020 https://github.com/ieee8023/covid-chestxray-dataset. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |