DOI:10.32604/csse.2022.021438

| Computer Systems Science & Engineering DOI:10.32604/csse.2022.021438 |  |
| Article |
Smart COVID-3D-SCNN: A Novel Method to Classify X-ray Images of COVID-19
1College of Technological Innovation, Zayed University, AbuDhabi Campus, UAE
2School of Artificial Intelligence, Xidian University, Xian, 710071, China
3Computer Science Department, Community College, King Saud University, P.O. Box 28095, Riyadh 11437, Saudi Arabia
4Nathan campus, Griffith University, Brisbane, Australia
*Corresponding Author: Ahed Abugabah. Email: ahed.abugabah@zu.ac.ae
Received: 03 July 2021; Accepted: 07 August 2021
Abstract: The outbreak of the novel coronavirus has spread worldwide, and millions of people are being infected. Image or detection classification is one of the first application areas of deep learning, which has a significant contribution to medical image analysis. In classification detection, one or more images (detection) are usually used as input, and diagnostic variables (such as whether there is a disease) are used as output. The novel coronavirus has spread across the world, infecting millions of people. Early-stage detection of critical cases of COVID-19 is essential. X-ray scans are used in clinical studies to diagnose COVID-19 and Pneumonia early. For extracting the discriminative features through these modalities, deep convolutional neural networks (CNNs) are used. A siamese convolutional neural network model (COVID-3D-SCNN) is proposed in this study for the automated detection of COVID-19 by utilizing X-ray scans. To extract the useful features, we used three consecutive models working in parallel in the proposed approach. We acquired 575 COVID-19, 1200 non-COVID, and 1400 pneumonia images, which are publicly available. In our framework, augmentation is used to enlarge the dataset. The findings suggest that the proposed method outperforms the results of comparative studies in terms of accuracy 96.70%, specificity 95.55%, and sensitivity 96.62% over (COVID-19 vs. non-COVID19 vs. Pneumonia).
Keywords: Convolutional neural network; classification; X-ray; deep learning
COVID-19 case was reported on 19th December 2019. The disease affected the respiratory system, and within a short time, it became a pandemic globally. The significant signs of COVID-19 are headache, fever, cough, and breath problems [1]. The first case was reported in the United States of America on 20th January 2020, and quickly the infected patients were reached over 400000 in April 2020 [2]. World Health Organization (WHO) recommends maintaining a social distance to reduce the risk. The early diagnosis and automatic detection is an important tool for all countries to overcome the spread of the virus. Furthermore, systems have been created to monitor the hospitals and to identify hotspot areas [3].
These modalities, such as chest CT scan and X-ray, have played a critical role in detecting COVID-19 and monitoring the pandemic condition [4]. Typically, a radiologist uses an X-ray image as the first choice because the equipment is readily available in hospitals. During a pandemic situation, some public medical repositories collect X-ray data samples from hospitals and provide to the researchers [5]. The significant contribution of such databases is to develop a classification model that helps the doctors for screening the COVID-19 at the initial stage. The solution is to introduce a smart automatic detection system for the correct classification of images of noraml and infected persons [6]. The mathematical modeling and AI-based approaches are also helpful to diagnose COVID-19 [5].
Therefore, the deep learning approaches have provided promising results for the classification of medical images involving lung cancer, breast cancer, and Alzheimer's disease [7–10]. Accurate results have been produced by applying segmentation in the analysis. Due to remarkable achievements, deep learning is now a particular part of the medical sector [11]. Convolutional neural network (CNN) is an integral part, which extracts the efficient and useful features from input data [12]. Each layer transforms the data on a more abstract basis during analysis and learns more in-depth detail [13]. Dealing with coronavirus is one of the biggest challenges for healthcare sectors. During deep learning in the medical domain, a substantial amount of annotated data availability is challenging, which may cause overfitting [14].
Abugabah et al. [15] proposed a model with a new X-ray database. They have eight types of diseases with chest X-ray modality, including 32717 images that are used for the multi-class classification. The database includes atelectasis, nodules, cardiomegaly, infiltration, mass, Pneumonia, pneumothorax, and effusion patients. They produced high accuracy results by using deep CNN. Furthermore, they will be extending these databases for further research improvement. Zhang et al. [16] X-ray scans were used to establish a modern method for detecting COVID-19 positive and negative. It was demonstrated that the model produced 95% accuracy in terms of variety. Rajpurkar et al. [17] introduced the CheXNeXT model based on CNN using 14 different pathologies, including Chest X-ray. They introduced a deep learning approch which classified the chest infection. In [18], the researcher used four feature extraction techniques. They used these extracted features for classification by using the support vector machine (SVM). During the experiment, they applied three cross-validations, i.e., 2-fold, 5-fold, and 10- fold.
Gu et al. [6] developed 3D deep CNN with a combination of multiscale prediction to detect lung nodules accurately by using CT scans. The sensitivity of the model was 87.94% to 92.93%. Chatterjee et al. in [19] employed an AI-based detection tool for CT images to differentiate between coronavirus and non-coronavirus patients. During the analysis of the images, the AI model provided 99.6% accurate results. In [20], researcher established a weekly supervised deep learning technique with chest CT images. During preprocessing, they used image segmentation slices with the combination of grain localization strategies. For segmentation, they applied ResNet and Unit to find out the infected region of the lungs. During training the model, they used data of 110 patients and achieved 94.80% classification accuracy. In [21], the researcher proposed 3D-DCNN with a combination of multilayers for lung nodule categorization. They also provided the solution of gradient vanishing by using dense connection because the dense connection can extract the local and specific features from lung data samples. Elasnaoui et al. [22] used deep learning models that were based on CNN and produced the comparative results on COVID-19 for binary classification. The pre-trained models are VGG-16, DenseNet and Inception ResNet-V2. In training the above models, they used X-ray data samples to classify normal and infected COVID-19 persons. In conclusion, Inception-ResNet-V2 produced 92.18% classification accuracy.
In [23], the researcher proposed an automatic framework by using a neural network to categorize normal and infected person. They used 4356 CT scans from 3322 patients to train the model. The patient's average age was 45 to 60 years, and the majority of male patients were included in the training data. The dataset was collected from six different hospitals which deal with coronavirus-infected patients. Alarifi et al. [24] developed a new CAD model for lung nodule detection by using CNN. They used segmentation techniques on CT images and were divided into different patches. These patches were split into three non-nodule and three nodule types; they showed 92.8% with translation, rotation, and scaling. In [25], the researcher established an early detection model for Pneumonia, COVID-19, and ordinary persons, in which they used the 618 CT scan samples for the classification purpose and produced promising results on benchmark data samples and achieved 86.7% classification accuracy.
Chouhan et al. [26] proposed a pneumonia detection framework that was based on a deep transfer learning strategy. The main goal of their study was to overcome the issue of a large amount of training data samples and introduce a simple method to classify pneumonia patients. They adopted different architectures, i.e., GoogleNet, Alex Net, Inception V3, ResNet18, and DenseNet. These approaches are already pre-trained on the ImageNet samples. In a comparative analysis, ResNet18 outperformed and achieved 99.36% accuracy via transfer learning on pneumonia detection. Pathak et al. [4] built a state-of-the-art technique for COVID-19 by using transfer learning. In this research work ResNet-32 architecture to transfer the knowledge with CT images to classified coronavirus positive and malicious persons. In [27], the researchers proposed a model via transfer learning on COVID-19 and SARS-CoV images. They used the ResNet and Inception model for categorization and obtained 99.01% accuracy. The deep learning approach is the most successful approach of machine learning that is used for the screening of Covid-19. Jahid et al. [28] developed CNN-based approaches using augmentation Chest X-rays and attained the highest accuracy, 97.97%. They used the 6432 scans during the experimental process and divided it into 80% training and 20% testing.
Recently, several data samples are available related to pneumonia X-ray, but COVID-19 annotated data samples are not readily available. Mostly, CNN is broadly known for its capabilities for image classification, recognition, and segmentation [29]. However, the primary benefit of CNNs is that there is no need for manual feature extraction like machine learning. The present research framework is directed towards the development of COVID-3D-SCNN that is based on deep CNNs. During training, the CNN-based models produced overfitting issues. We also used the augmentation approach to overcome this issue. The primary objective of this study are as follows:
• We introduced a COVID-3D-SCNN approach to the classification of COVID-19, non-COVID-19, and pneumonia persons.
• The proposed deep learning model reduces the overfitting issue and learns from small data samples.
• Our research work is based on X-ray scans that help the early diagnosis of COVID-19 and pneumonia disease.
The rest of this paper is formed as follows: Material and methods are discussed in section 2. The experimental results of the proposed COVID-3D-SCNN and discussion are presented in Section 3, and the conclusion is presented in Section 4.
The Kaggle database was used to collect the experimental datasets. In the initial stage, we used 575 (COVID-19), 1200 (non-COVID), and 1400 (Pneumonia) images for this experiment. The pictures of the samples are shown in Fig. 2. To begin, X-ray scans are randomly divided into 85% for training and validation, and the remaining 15% for testing. Due to less number of training samples, to overcome the overfitting issue, we have used augmentation on 85% of images, as shown in the data flow chart in Fig. 1. The augmented images were used for training and validation. The final performance of the proposed model was used to check 15% testing data samples. The photos were rescaled on 224x224 before training the model.

Figure 1: Data split illustration for the experimental process

Figure 2: Illustration of three patients X-ray images (a) COVID-19, (b) Non-COVID-19, and (c) Pneumonia X-ray
In medical imaging, to maximise the number of data samples, the augmentation method is used [30]. Augmentation has some parameters that improved the images artificially. These parameters, i.e., width shift range, channel shift range, zoom, and shear range. It directly impacts a neural network to increase classification performance. In this research work, we generate five images on each available X-ray image. For training and validation, we used 13495 images.
The proposed approach is based on three significant steps, i.e., data acquisition with augmentation, feature extraction, and the classification of COVID-19 groups is the final stage. We proposed a CNN-based model inspired by the VGG family, i.e., VGG-16 and VGG-19. Our proposed approach takes 224x224 size images, and each model has five max-pooling layers, three batch normalization layers, 15 convolutional layers. In the first block, we used three Conv-layers having 64 filters with 3x3 kernel size. In the second block, 128 filters are used with three Conv layers. For the third block, 256 filters are used with the same layers and kernel size. In the fourth and fifth block, we used 512 filters with the same number of layers and kernel size. One max-pooling layer was applied in each block with 2x2 pool down-sampling, and these layers were working in a three-time parallel, as shown in Fig. 3 and details of model shown in Tab. 1. The main reason behind of proposed work that consecutive models which able to extract the more useful features together from input images [31]. It is also suitable for less number of data samples [32]. It directly impacts model performance and increases classification accuracy.

Figure 3: Proposed COVID-3D-SCNN classification model
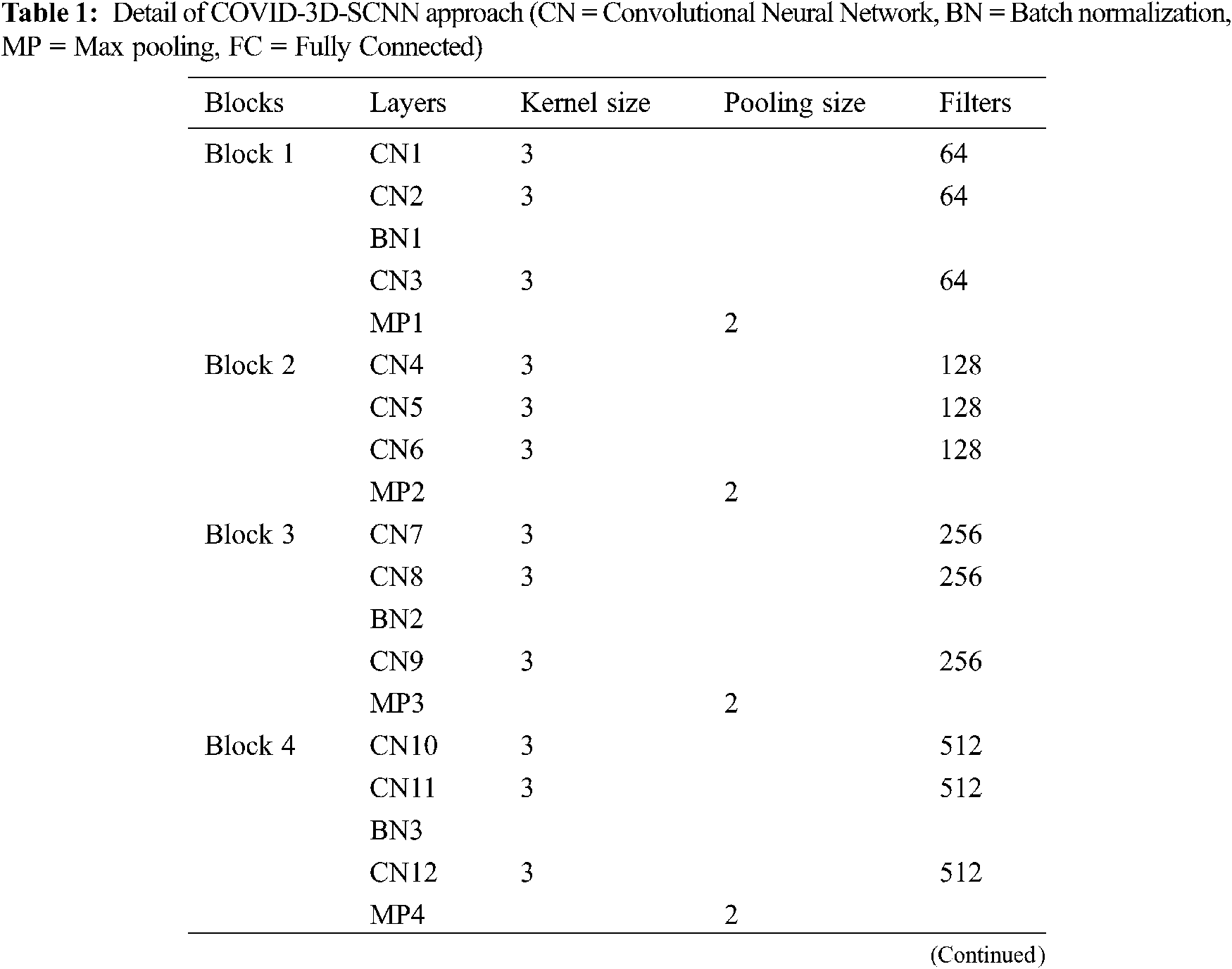
After concatenation, these three models are linked by two fully connected layers with 1024 and 512 neurons, respectively, with the ReLU activation function. Finally, the softmax layer is used for multi-class classification. We used Adam optimizer, Adamax, AdaDelta, and Nadam with categorical cross-entropy in the proposed model. Moreover, to reduce the overfitting issue, we also used the Gaussian noise.
On less annotated samples, the same models applied parallel with the same weights. During the testing of the model on input data, the siamese model extracts the more valuable features. These parallel layers directly impact on classification performance of the model.
The evaluation metrics determine how accurately the proposed approach achieved the desired target. According to the disease classification task, correctly recognized COVID-19 and pneumonia disease cases are termed as true positives (TP), and inaccurately identified instances are termed as False negatives (FN). Furthermore, if the model correctly classifies a healthy person, it is termed as real negative (TN), and an incorrectly recognized healthy person is termed as false positive (FP). In the proposed study, a significant objective is to detect COVID-19 patients, which is helpful for early diagnosis. The proposed model achieved state-of-the-art results on the multi-class classification. Overall, it measures the classification performance on metrics of accuracy, sensitivity, and specificity.
Sensitivity represents the valid positive rate. It identifies and predicts the valid positive instances which are available on the ground information. In our proposed approach, it gives the correct expected rate of the patients suffering from COVID-19 and Pneumonia. Mathematically it is given as:
where X represents the correctly recognized COVID-19 and Pneumonia, and Y represents incorrectly recognized COVID-19 and Pneumonia.
Specificity represents the true negative rate. It identifies and predicts the valid negative instances which are available on the ground information. In our proposed approach, it gives the correct predicted rate of these patients who are not suffering from COVID-19 and Pneumonia. Mathematically it is given as:
where H represents correctly recognized healthy person and UH means incorrectly recognized healthy people.
3 Experimental Results and Discussion
The proposed approach was implemented by using Keras library on Z840 workstation. To find out the effectiveness of the proposed COVID-3D-SCNN strategy that was used to classify COVID-19. We used 85% data samples for training and validation as we mention in detail in Fig. 1. For the training and validation phase, we used 100 epochs in the proposed model and attained the training accuracy 97.09%. In contrast, the validation accuracy is 96.19% by using Adam optimizer, as shown in Fig. 4. During the training process, it is observed that the proposed model is more efficient with minimum losses. The experimental results are shown in Tabs. 2 and 3 with four different optimizers. This optimizer's results demonstrated that Adam is more successful in our proposed approach.

Figure 4: Proposed model training and validation loss analysis over 100 epochs with Adam optimizer: (a) In terms of accuracy and (b) In terms of loss analysis
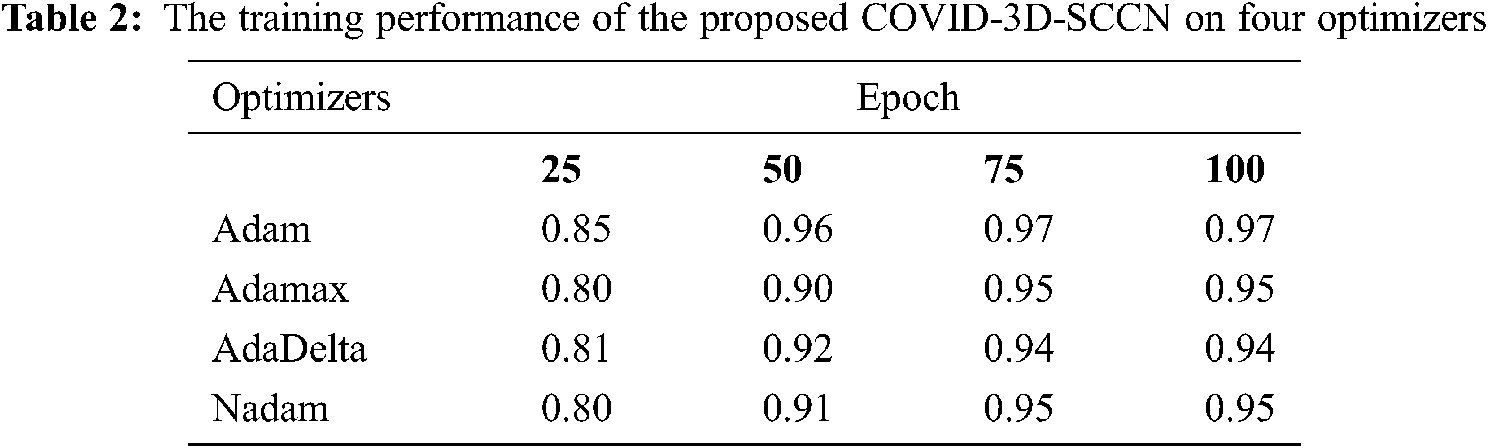
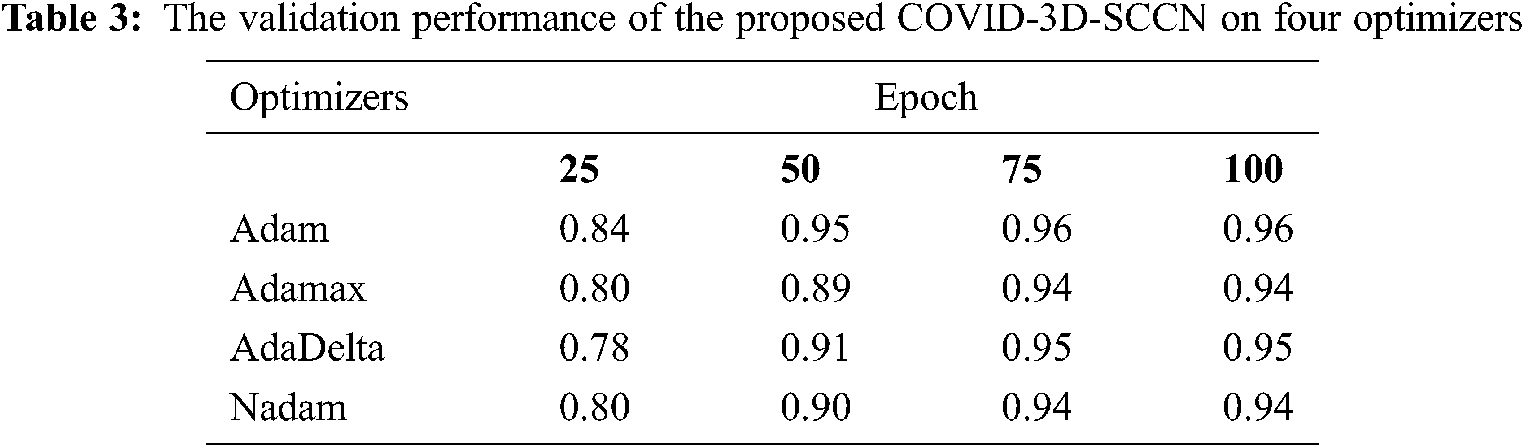
Moreover, it has a minor variation between training and validation accuracies. The augmentation approach is used to overcome the overfitting issue, and we achieved 96.70% testing accuracy, as shown in Fig. 4. The proposed COVID-3D-SCNN produces excellent results concerning the number of epochs. To diagnose serious diseases such as Pneumonia and COVID-19, it is more important to reduce false negative and false positive outcomes during model execution. It is confirmed that the proposed approach produces commendable results for three classes. Through the confusion matrix, we have shown the impact of the false negative and false positive rate in Tab. 4.
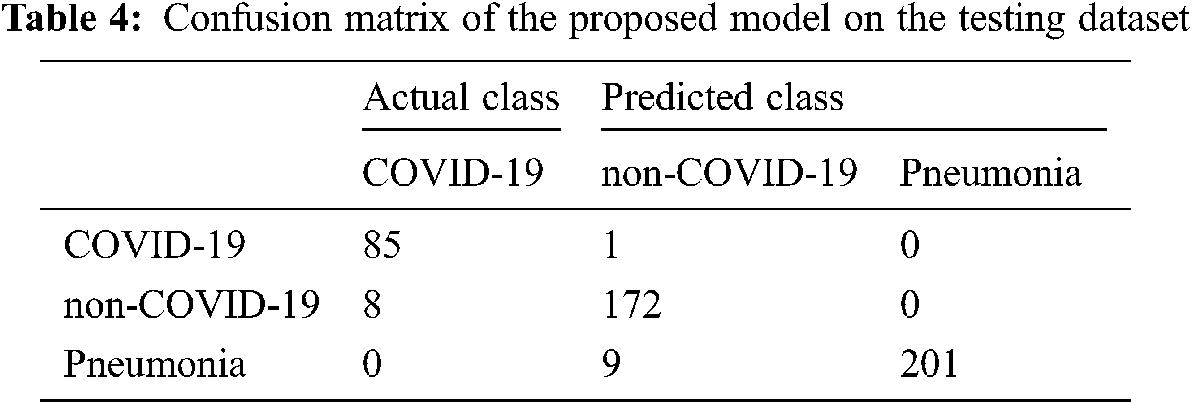
Finally, in Tab. 5, we compared our results with four state-of-the-art studies that were based on deep learning approaches with CT and X-ray images. We increased the 3.22% accuracy over state-of-the-art studies (CNN [32]) and 1.70% (over CoroNet + DCNN [33]), [34–36] respectively. Thus, COVID-3D-SCNN overcome the overfitting issue and obtained significant results on limited data samples. Effective and correct COVID-19 diagnosis is critical for early treatment. Many researchers have developed different computer-aided systems (CAS), which can be helpful in diagnosing an early stage of COVID-19. Based on the performance, it is proved that deep learning with CNNs may have the ability to extract the more useful features from X-images, which are more helpful for automatic detection of COVID-19. The results demonstrated that our proposed model produced highly accurate results in terms of classification. Our approach does not need extra feature engineering.
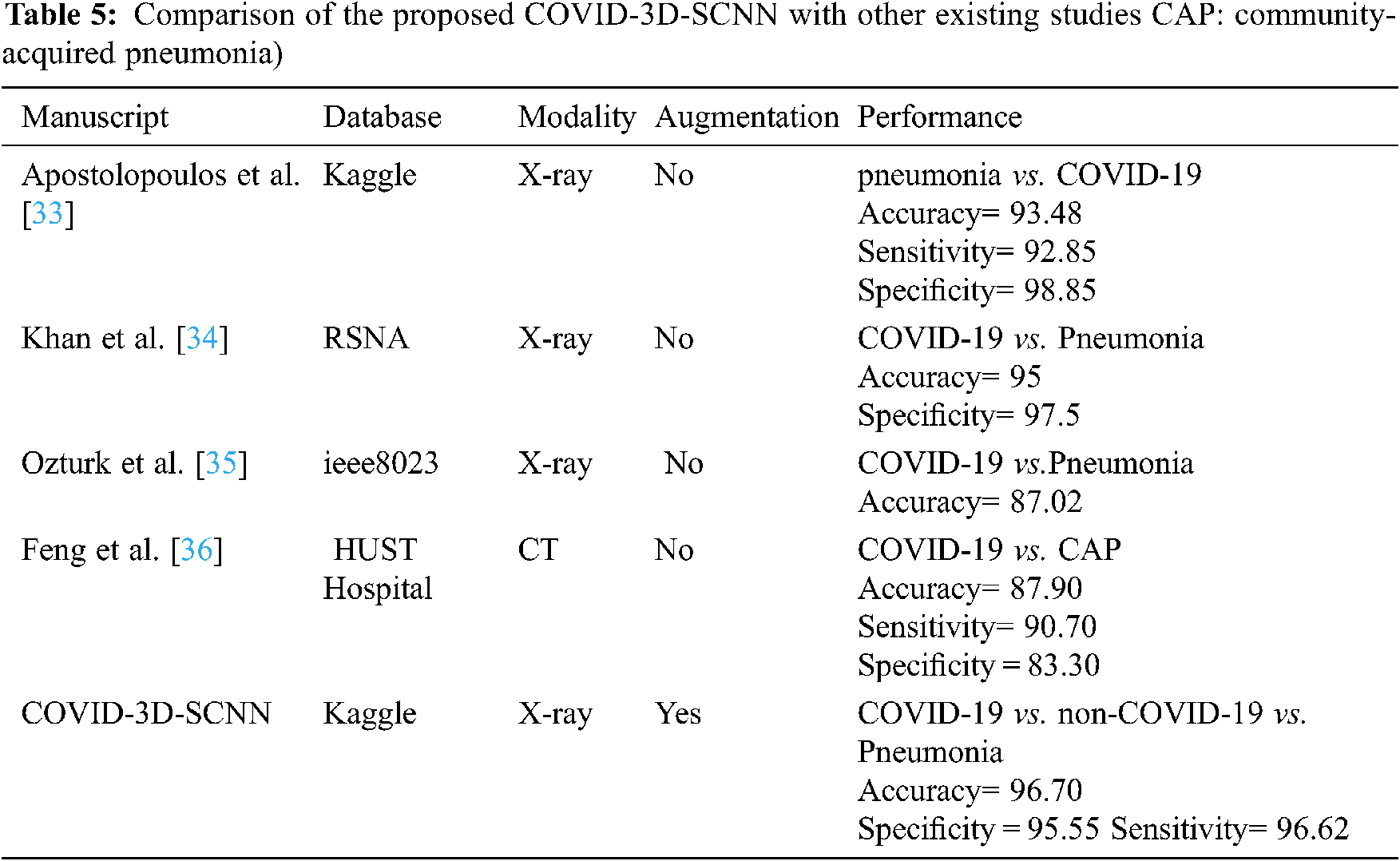
Several recent studies results are shown in Tab. 4, which are based on CT and X-ray images. In [29] research work, 1427 X-ray images were used for the experimental process that belongs to three classes: bacterial Pneumonia, COVID-19, and familiar persons. They applied CNN-based models on X-ray data and achieved 93.48% accuracy for multiclass classification. In [32], the researcher proposed CoroNet based on CNN and produced the promising result on the multiclass type. They achieved an overall 95% accuracy in three classes, i.e., Pneumonia, COVID-19, and regular. The comparative studies have some limitations due to less number of COVID-19 samples. Reference [34] introduced a CNN model based on the CNN approach. They implemented the experiment on binary classification and multi-class classification. They achieved 98.08% accuracy for binary and 87.02% for multiclassification. Our proposed approach has achieved higher accuracy as compared to other state-of-the-art studies. Deep convolutional neural network-based systems indicate that CNN plays a key role in the pandemic situation to detect COVID-19 patients.
In conclusion, COVID-19 cases are rapidly increasing on a daily basis all over the world, and different countries are facing problems due to a shortage of resources. In this situation, the detection of a single positive case is more important. In this research work, we proposed a COVID-3D-SCNN, using X-ray scans, a siamese convolutional neural network identifies COVID-19 and pneumonia persons automatically. In the developed approch, we used three consecutive models to fetch the deeper useful features from the input data. X-ray images were collected from public medical repositories. Our proposed model produced the best results in terms of classification and achieved 96.70% testing accuracy on three classes (COVID-19 vs. non-COVID-19 vs. Pneumonia), specificity 95.55%, and sensitivity 96.62%. Nevertheless, our research leads to the automated identification of COVID-19 on a smaller number of images and at a faster rate. In the future, we check this model on other medical domains such as Dementia and Alzheimer's disease. Because in AD domain also annotated data samples is significantly less. We will use this model with augmentation in AD and Dementia domain to check the performance in terms of accuracy. Furthermore, we will use new data samples of X-ray and CT scans, which can be helpful to improve the detection accuracy rate of COVID-19.
Acknowledgement: The authors would like to thank the editors and reviewers for their review and recommendations.
Data Availability: The X-ray imaging data used to support the findings of this study database (https://www.kaggle.com/prashant268/chest-xray-covid19-pneumonia).
Funding Statement: This work was supported by the Researchers Supporting Project (No. RSP-2021/395), King Saud University, Riyadh, Saudi Arabia.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. C. Huang, Y. Wang, X. Li, L. Ren, J. Zhao et al., “Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China,” Lancet, vol. 395, no. 10223, pp. 497–506, 2020. [Google Scholar]
2. M. L. Holshue, C. DeBolt, S. Lindquist, K. H. Lofy, J. Wiesman et al., “First case of 2019 novel coronavirus in the United States,” The New England Journal of Medicine, vol. 382, pp. 929–936, 2020. [Google Scholar]
3. S. Wang, Y. Zha, W. Li, Q. Wu, X. Li et al., “A fully automatic deep learning system for COVID-19 diagnostic and prognostic analysis,” European Respiratory Journal, vol. 56, no. 2, pp. 1–11, 2020. [Google Scholar]
4. Y. Pathak, P. K. Shukla, A. Tiwari, S. Stalin, S. Singh et al., “Deep transfer learning based classification model for COVID-19 disease,” IRBM, vol. 42, no. 3, pp. 1–21, 2020. [Google Scholar]
5. Y. Mohamadou, A. Halidou and P. T. Kapen, “A review of mathematical modeling, artificial intelligence and datasets used in the study, prediction and management of COVID-19,” Applied Intelligence, vol. 50, pp. 3913–3925, 2020. [Google Scholar]
6. Y. Gu, X. Lu, L. Yang, B. Zhang, D. Yu et al., “Automatic lung nodule detection using a 3D deep convolutional neural network combined with a multi-scale prediction strategy in chest CTs,” Computers in Biology and Medicine, vol. 103, pp. 220–231, 2018. [Google Scholar]
7. T. Alalifm, A. M. Tehame, S. Bajaba, A. Barnawi and S. Zia, “Machine and deep learning towards COVID-19 diagnosis and treatment: Survey, challenges, and future directions,” International Journal of Environmental Research and Public Health, vol. 18, no. 3, pp. 1–24, 2020. [Google Scholar]
8. A. A. AlZubi, A. Alarifi and M. Al-Maitah, “Deep brain simulation wearable IoT sensor device based Parkinson brain disorder detection using heuristic tubu optimized sequence modular neural network,” Measurement, vol. 161, pp. 101–124, 2020. [Google Scholar]
9. M. An and Y. Xie, “A novel CT imaging system with adjacent double X-ray sources,” Computers and Mathematical Methods in Medicine, vol. 13, pp. 1–18, 2013. [Google Scholar]
10. J. Gao, Y. Guo, Y. Sun and G. Qu, “Application of deep learning for early screening of colorectal precancerous lesions under white light endoscopy,” Computers and Mathematical Methods in Medicine, vol. 20, pp. 1–12, 2020. [Google Scholar]
11. A. Abugabah, A. Al-smadi and A. Abuqabbeh, “Data mining in health care sector: literature notes,” in Proc. of the 2019 2nd Int. Conf. on Computational Intelligence and Intelligent Systems, Bangkok, Thailand, pp. 63–68, 2019. [Google Scholar]
12. G. Litjens, T. Kooi, B. E. Bejnordi, A. A. Setio, F. Ciompi et al., “A survey on deep learning in medical image analysis,” Medical Image Analysis, vol. 42, pp. 60–88, 2017. [Google Scholar]
13. M. Yaqub, J. Feng, M. S. Zia, K. Arshid, K. Jia et al., “State-of-the-art CNN optimizer for brain tumor segmentation in magnetic resonance images,” Brain Sciences, vol. 10, no. 7, pp. 1–19, 2020. [Google Scholar]
14. N. Asiri, M. Hussain, F. Al-Adel and N. Alzaidi, “Deep learning based computer-aided diagnosis systems for diabetic retinopathy: A survey,” Artificial Intelligence in Medicine, vol. 99, pp. 101–125, 2019. [Google Scholar]
15. A. Abugabah, A. A. AlZubi, M. Al-Maitah and A. Alarifi, “Brain epilepsy seizure detection using bio-inspired krill herd and artificial alga optimized neural network approaches,” Journal of Ambient Intelligence and Humanized Computing, vol. 46, pp. 1–12, 2020. [Google Scholar]
16. J. Zhang, Y. Xie, G. Pang, Z. Liao, J. Verjans et al., “Viral pneumonia screening on chest X-ray images using confidence-aware anomaly detection,” IEEE Transactions on Medical Imaging, vol. 40, no. 3, pp. 879–890, 2020. [Google Scholar]
17. P. Rajpurkar, J. Irvin, R. L. Ball, K. Zhu, B. Yang et al., “Deep learning for chest radiograph diagnosis: A retrospective comparison of the CheXNeXt algorithm to practicing radiologists,” PLOS Medicine, vol. 15, no. 11, pp. 1–19, 2018. [Google Scholar]
18. A. M. Hussain, M. M. Al-Jawad, H. A. Jalab, H. Sahiba, R. W. Ibrahim et al., “Classification of covid-19 corona virus, pneunomina and healthy lungs in CT scans using Q-deformed entropy and deep learning features,” Entropy, vol. 22, no. 5, pp. 1–34, 2020. [Google Scholar]
19. A. Chatterjee, M. W. Gerdes and S. G. Martinez, “Statistical explorations and univariate timeseries analysis on covid-19 datasets to understand the trend of disease spreading and death,” Sensors, vol. 20, no. 11, pp. 1–17, 2020. [Google Scholar]
20. A. A. AlZubi and M. Al-Ma'aitah, “Enhanced computational model for gravitational search optimized echo state neural networks based oral cancer detection,” Journal of Medical Systems, vol. 42, no. 11, pp. 212–225, 2018. [Google Scholar]
21. H. Jung, B. Kim, I. Lee, J. Lee and J. Kang, “Classification of lung nodules in CT scans using three-dimensional deep convolutional neural networks with a checkpoint ensemble method,” BMC Medical Imaging, vol. 18, no. 1, pp. 48–59, 2018. [Google Scholar]
22. K. Elasnaoui and Y. Chawki, “Using X-ray images and deep learning for automated detection of coronavirus disease,” Journal of Biomolecular Structure and Dynamics, vol. 39, pp. 1–22, 2020. [Google Scholar]
23. L. Li, L. Qin, Z. Xu, Y. Yin, X. Wang et al., “Artificial intelligence distinguishes Covid-19 from community acquired pneumonia on chest CT,” Radiology, vol. 96, no. 2, pp. 65–71, 2020. [Google Scholar]
24. A. Alarifi and A. A. AlZubi, “Memetic search optimization along with genetic scale recurrent neural network for predictive rate of implant treatment,” Journal of Medical Systems, vol. 42, no. 11, pp. 202–213, 2018. [Google Scholar]
25. H. Ko, H. Chung, W. S. Kang, K. W. Kim, Y. Shin et al., “Covid-19 pneumonia diagnosis using a simple 2D deep learning framework with a single chest CT image: Model development and validation,” Journal of Medical Internet Research, vol. 22, no. 6, pp. 321–332, 2020. [Google Scholar]
26. V. Chouhan, S. K. Singh, A. Khamparia, D. Gupta, P. Tiwari et al., “A novel transfer learning based approach for pneumonia detection in chest X-ray images,” Applied Sciences, vol. 10, no. 2, pp. 1–16, 2020. [Google Scholar]
27. H. Benbrahim, H. Hachimi and A. Amine, “Deep transfer learning with apache spark to detect Covid-19 in chest X-ray images,” Romanian Journal of Information Science and Technology, vol. 23, pp. 117–129, 2020. [Google Scholar]
28. M. H. Jahid, M. A. Shahin and M. A. Shikhar, “Deep learning based detection and segmentation of COVID-19 pneumonia and chest x-ray image,” in IEEE Int. Conf. on Information and Communication Technology for Sustainable Development (ICICT4SDDhaka, Bangladesh, pp. 210–214, 2021. [Google Scholar]
29. K. Elasnaoui and Y. Chawki, “Using X-ray images and deep learning for automated detection of coronavirus disease,” Journal of Biomolecular Structure and Dynamics, vol. 10, no. 2, pp. 1–15, 2020. [Google Scholar]
30. A. H. Garcia and P. Konig, “Further advantages of data augmentation on convolutional neural networks,” in Int. Conf. on Artificial Neural Networks,” Slovakia, pp. 95–103, 2020. [Google Scholar]
31. A. Mehmood, M. Maqsood, M. Bashir and Y. Shuyuan, “A deep siamese convolution neural network for multi-class classification of Alzheimer disease,” Brain Sciences, vol. 10, no. 2, pp. 84–95, 2020. [Google Scholar]
32. H. Kim and Y. S. Jeong, “Sentiment classification using convolutional neural networks,” Applied Sciences, vol. 9, no. 11, pp. 1–19, 2019. [Google Scholar]
33. I. D. Apostolopoulos and T. A. Mpesiana, “Covid-19: Automatic detection from x-ray images utilizing transfer learning with convolutional neural networks,” Phyical and Engineering Science in Medicine, vol. 43, pp. 635–640, 2020. [Google Scholar]
34. A. I. Khan, J. L. Shah and M. M. Bhat, “Coronet: A deep neural network for detection and diagnosis of COVID-19 from chest x-ray images,” Computer Methods and Programs in Biomedicine, vol. 196, pp. 1–24, 2020. [Google Scholar]
35. T. Ozturk, M. Talo, E. A. Yildirim, U. B. Baloglu, O. Yildirim et al., “Automated detection of Covid-19 cases using deep neural networks with X-ray images,” Computers in Biology and Medicine, vol. 121, pp. 1–17, 2020. [Google Scholar]
36. F. Shi, L. Xia, F. Shan, B. Song, D. Wu et al., “Large-scale screening of Covid-19 from community acquired pneumonia using infection size-aware classification,” Physics in Medicine and Biology, vol. 66, no. 6, pp. 1361–1372, 2021. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |