DOI:10.32604/csse.2022.018227

| Computer Systems Science & Engineering DOI:10.32604/csse.2022.018227 |  |
| Article |
Vertex-Edge Degree Based Indices of Honey Comb Derived Network
1Centre for Advanced Studies in Pure and Applied Mathematics, Bahauddin Zakariya University, Multan, Pakistan
2College of Computer Sciences and Information Technology, Jazan University, Jazan, Saudi Arabia
*Corresponding Author: Muhammad Ibrahim. Email: mibtufail@gmail.com
Received: 01 March 2021; Accepted: 29 April 2021
Abstract: Chemical graph theory is a branch of mathematics which combines graph theory and chemistry. Chemical reaction network theory is a territory of applied mathematics that endeavors to display the conduct of genuine compound frameworks. It pulled the research community due to its applications in theoretical and organic chemistry since 1960. Additionally, it also increases the interest the mathematicians due to the interesting mathematical structures and problems are involved. The structure of an interconnection network can be represented by a graph. In the network, vertices represent the processor nodes and edges represent the links between the processor nodes. Graph invariants play a vital feature in graph theory and distinguish the structural properties of graphs and networks. In this paper, we determined the newly introduced topological indices namely, first
Keywords: Honey comb derived network; ev-degree; topological indices
A structural molecular diagram is a basic diagram in the study of structural chemical graph theory where atoms are spoken to by nodes and chemical bonds are spoken to by lines. A diagram is associated if there is an association between any pair of nodes. A network is an associated diagram that has no various lines between two nodes and loop. The number of nodes which are associated with a fixed node
The relation between the
2 The
Chellali et al. [23] gave the definition of
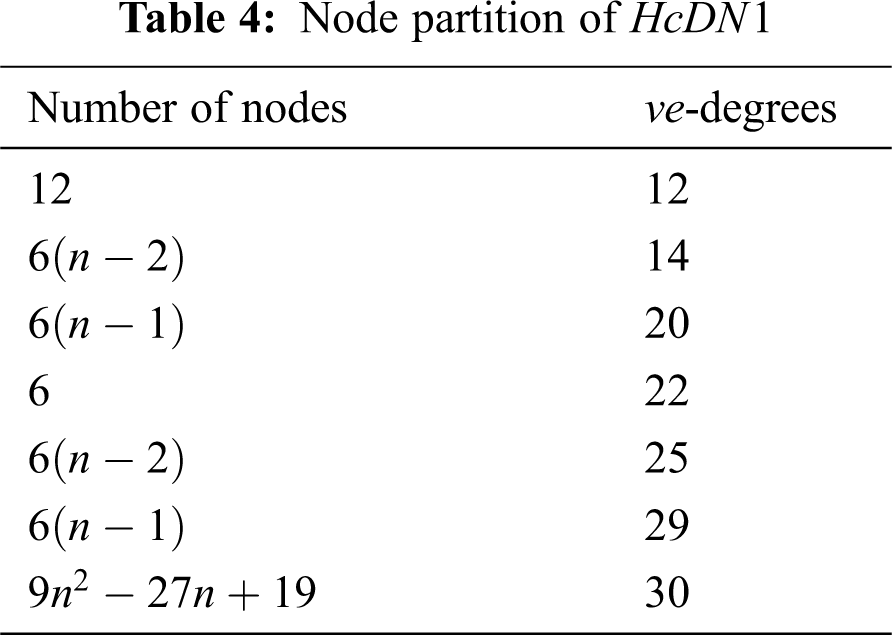
In the present section, we determined our computational results for Honey Comb derived network (see Fig. 1), which is a planar graph. The number of nodes and lines in
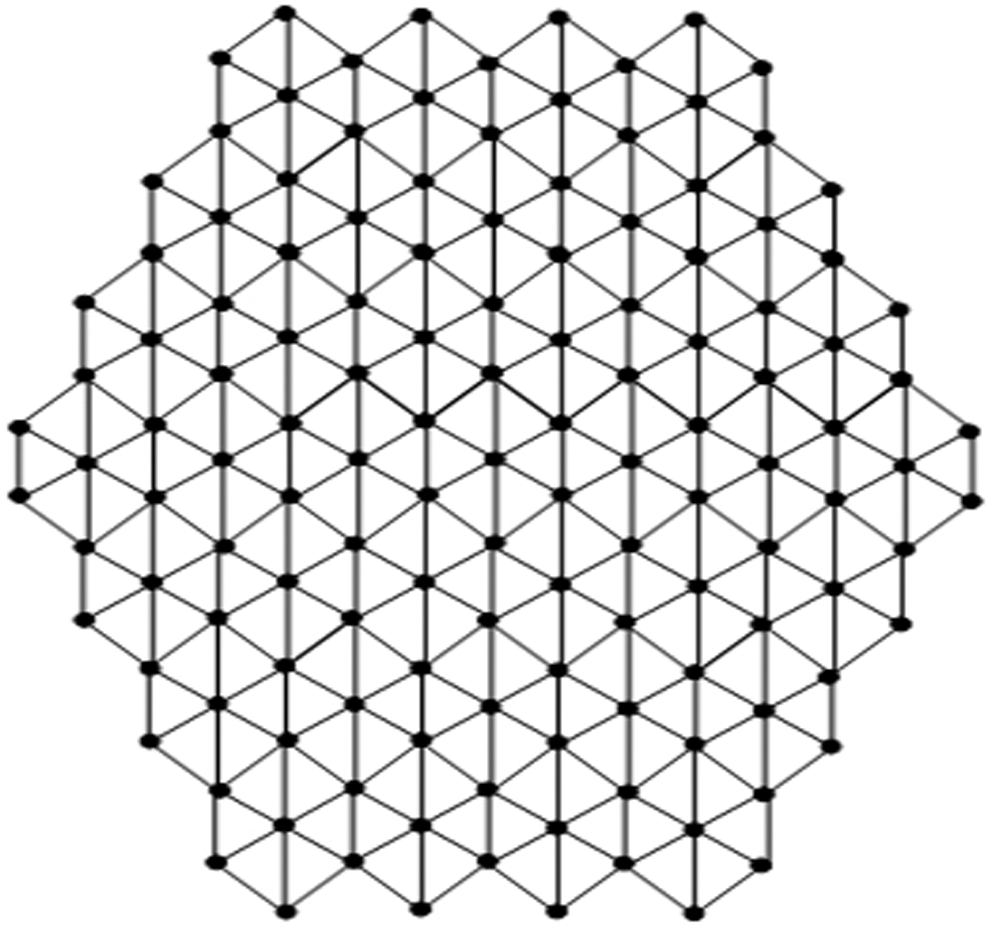
Figure 1:
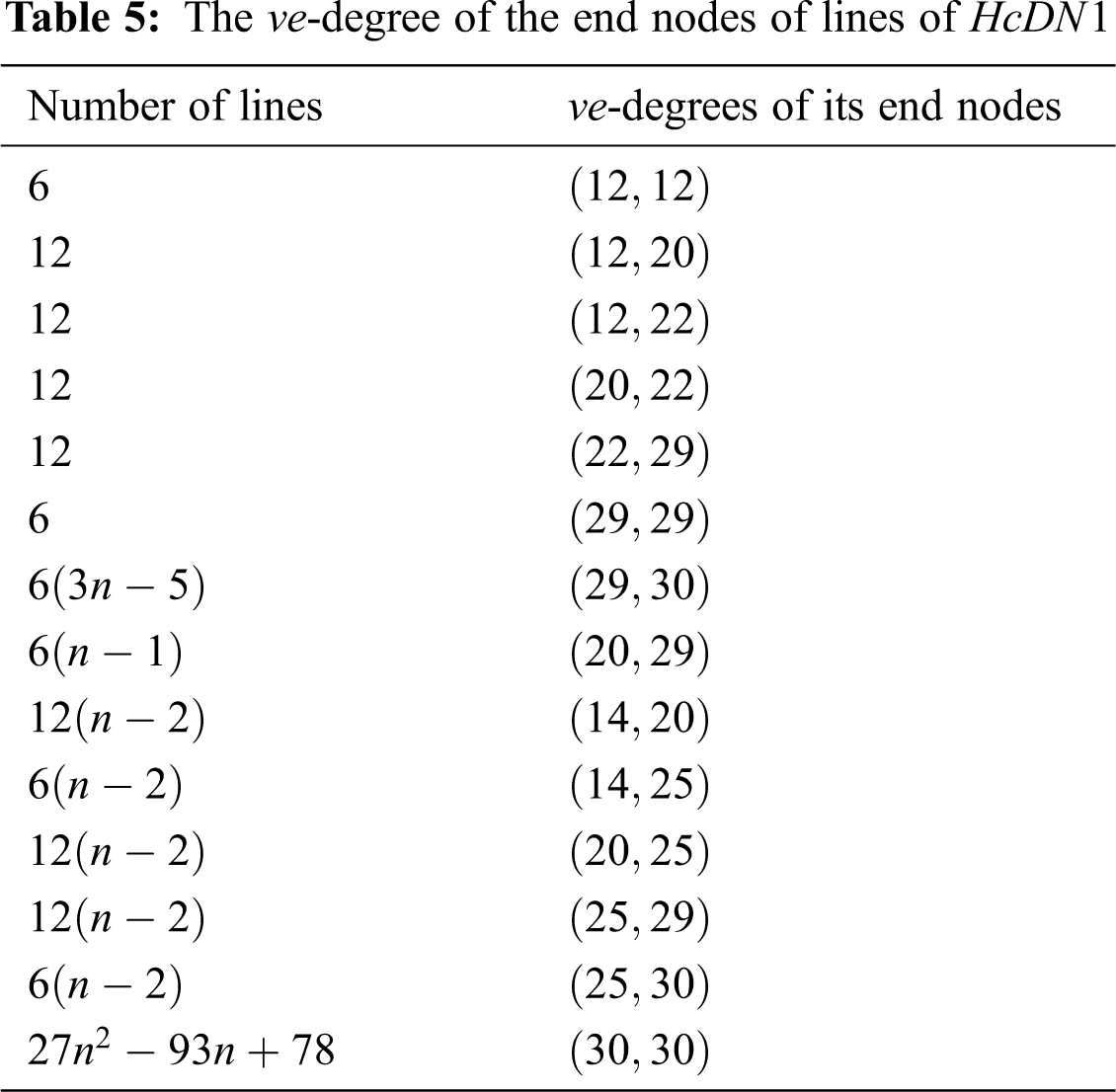
There are five types of lines in
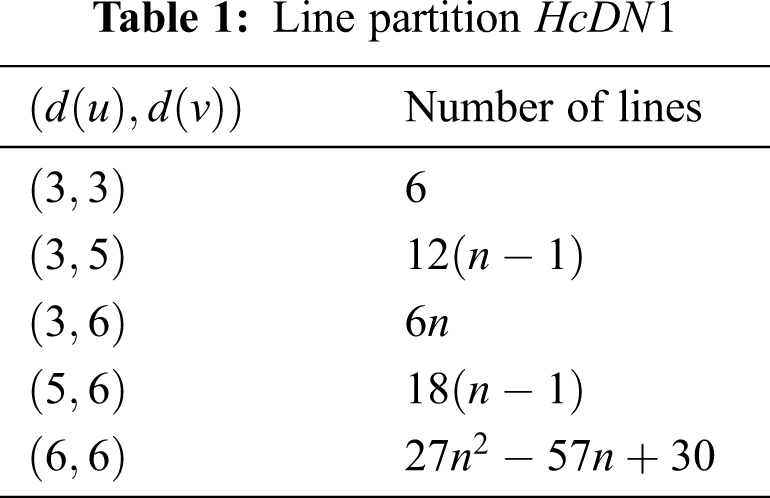
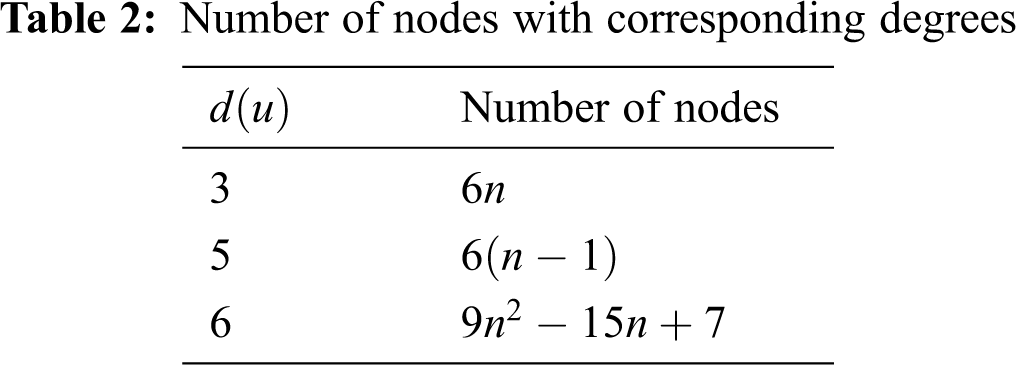
In Tab. 3, We partition the lines, based on
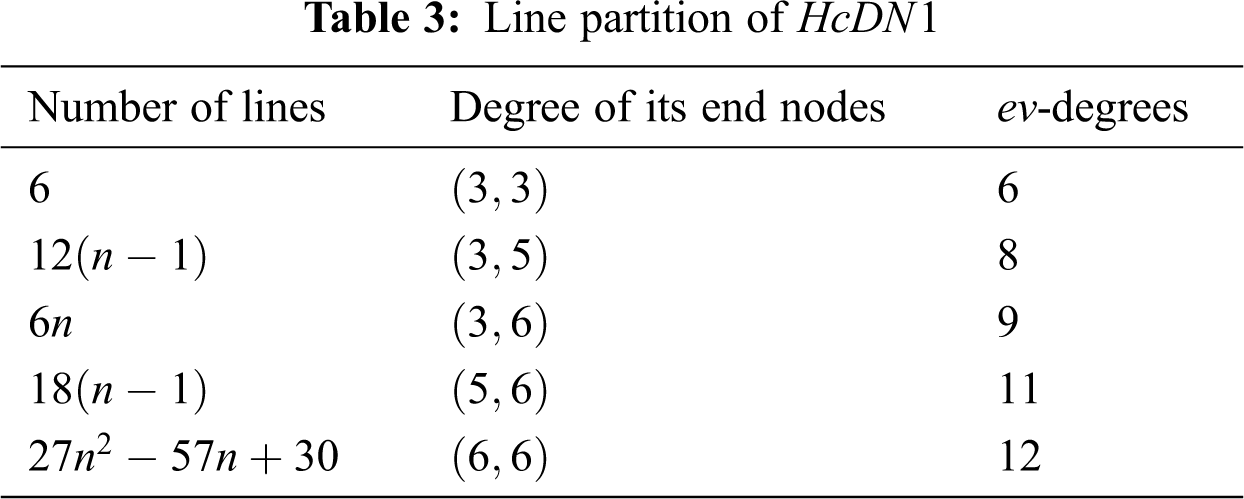
4 Computing Indices for
In this section, we will calculate
•
Now with the help of Tab. 3, we compute the
• The first
Now with the help of Tab. 4, we compute the first
• The first
Now with the help of Tab. 5, we compute the first
• The second ve-degree Zagreb index
Now with the help of Tab. 5, we compute the second
• The
Now with the help of Tab. 5, we compute the
• The
Now with the help of Tab. 3, we compute the
• The atom-bond connectivity index
Now with the help of Tab. 5, we compute the atom-bond connectivity index of
• The geometric-arithmetic index
Now with the help of Tab. 5, we compute the geometric-arithmetic index of
• The harmonic index
Now with the help of Tab. 5, we compute the Harmonic index of
• The sum-connectivity index
Now with the help of Tab. 5, we compute the Sum-connectivity index of
5 Numerical and Graphical Representation and Discussion
The
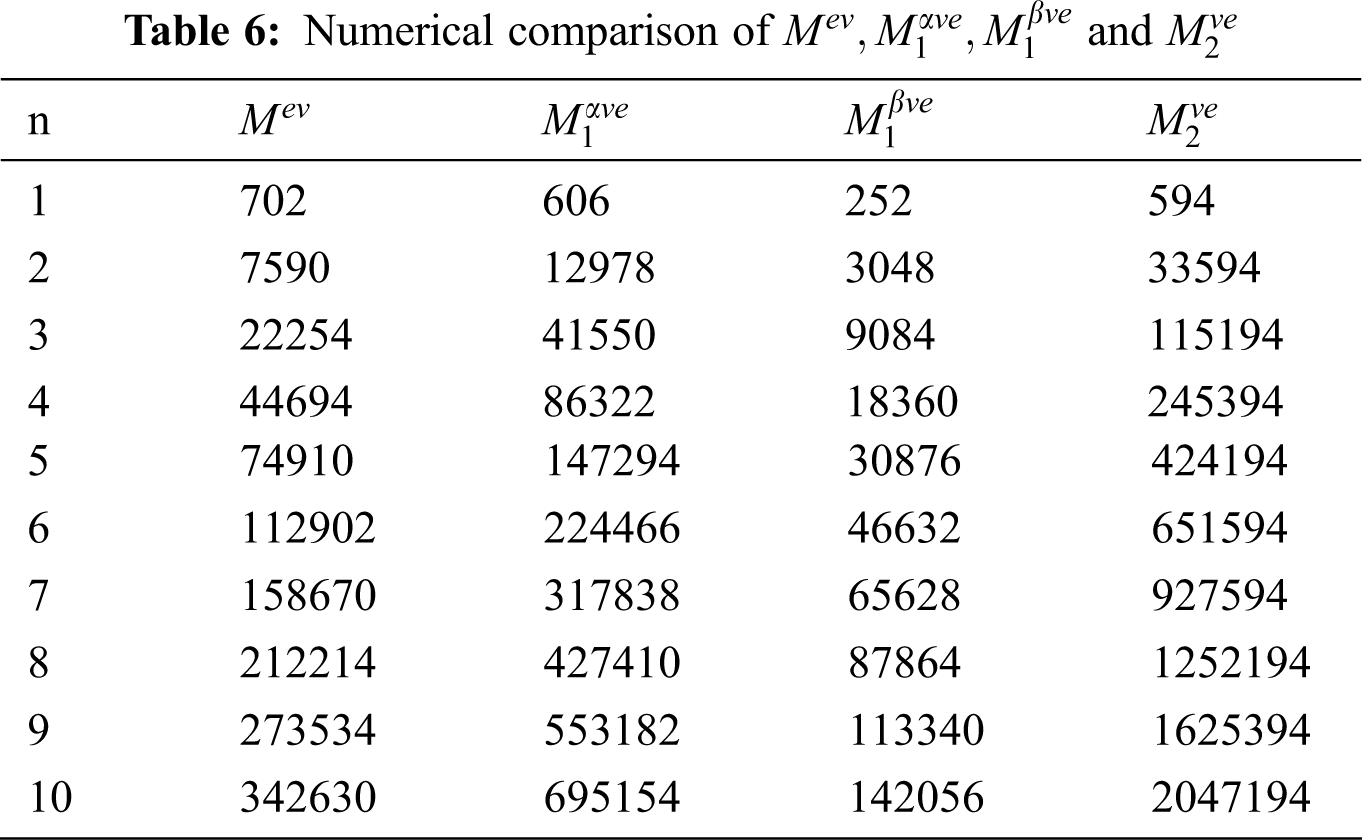
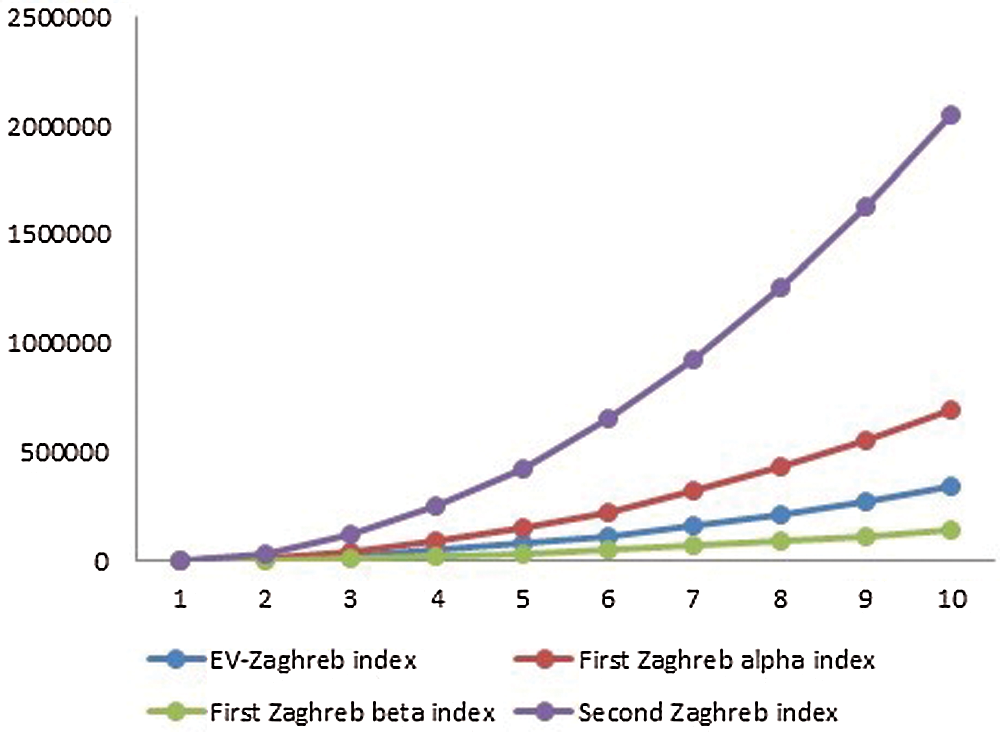
Figure 2: Graphical comparison of
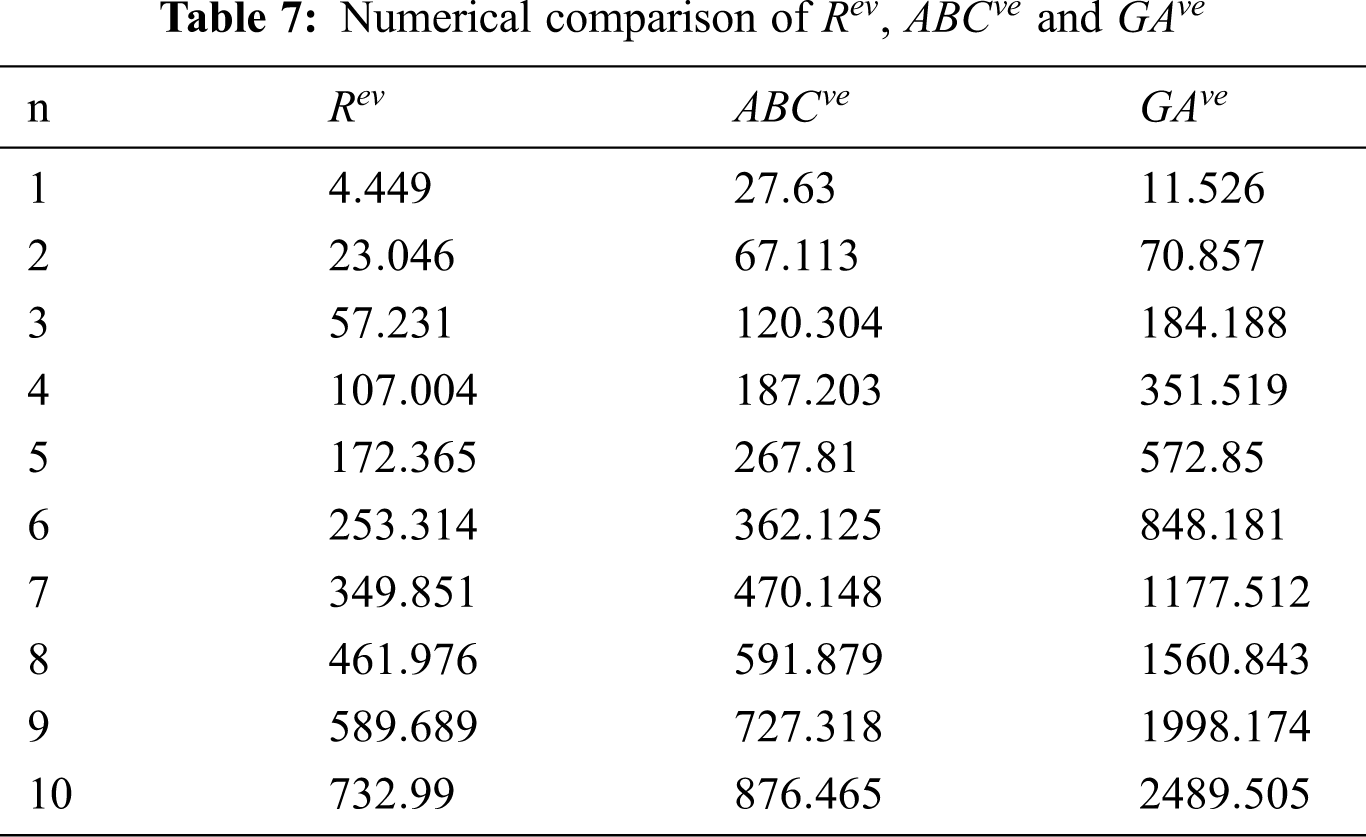
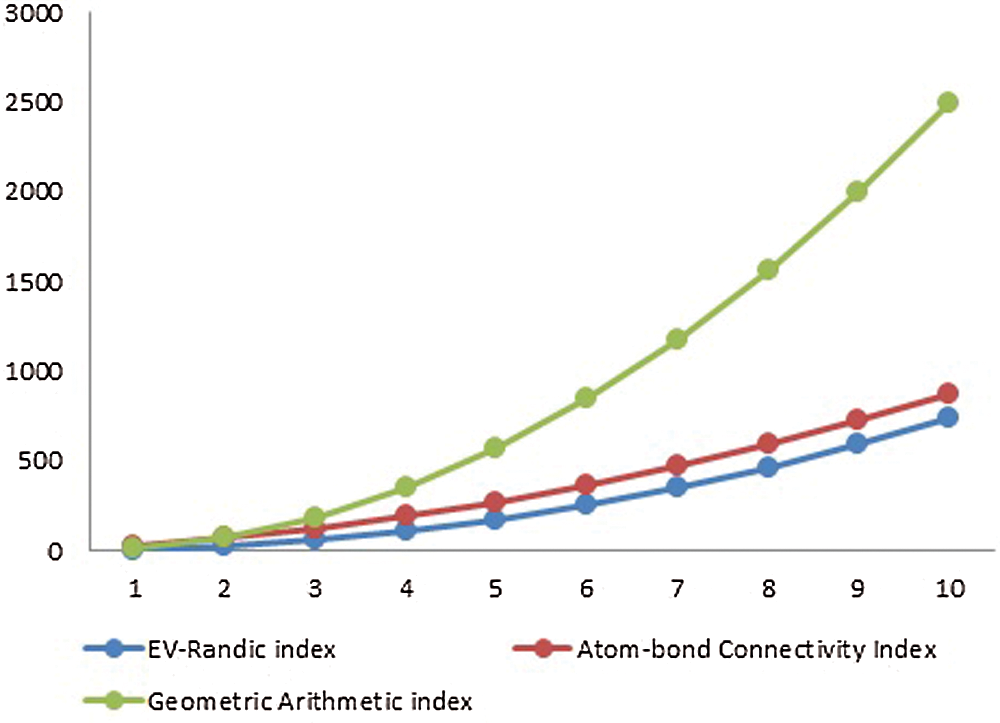
Figure 3: Graphical comparison of
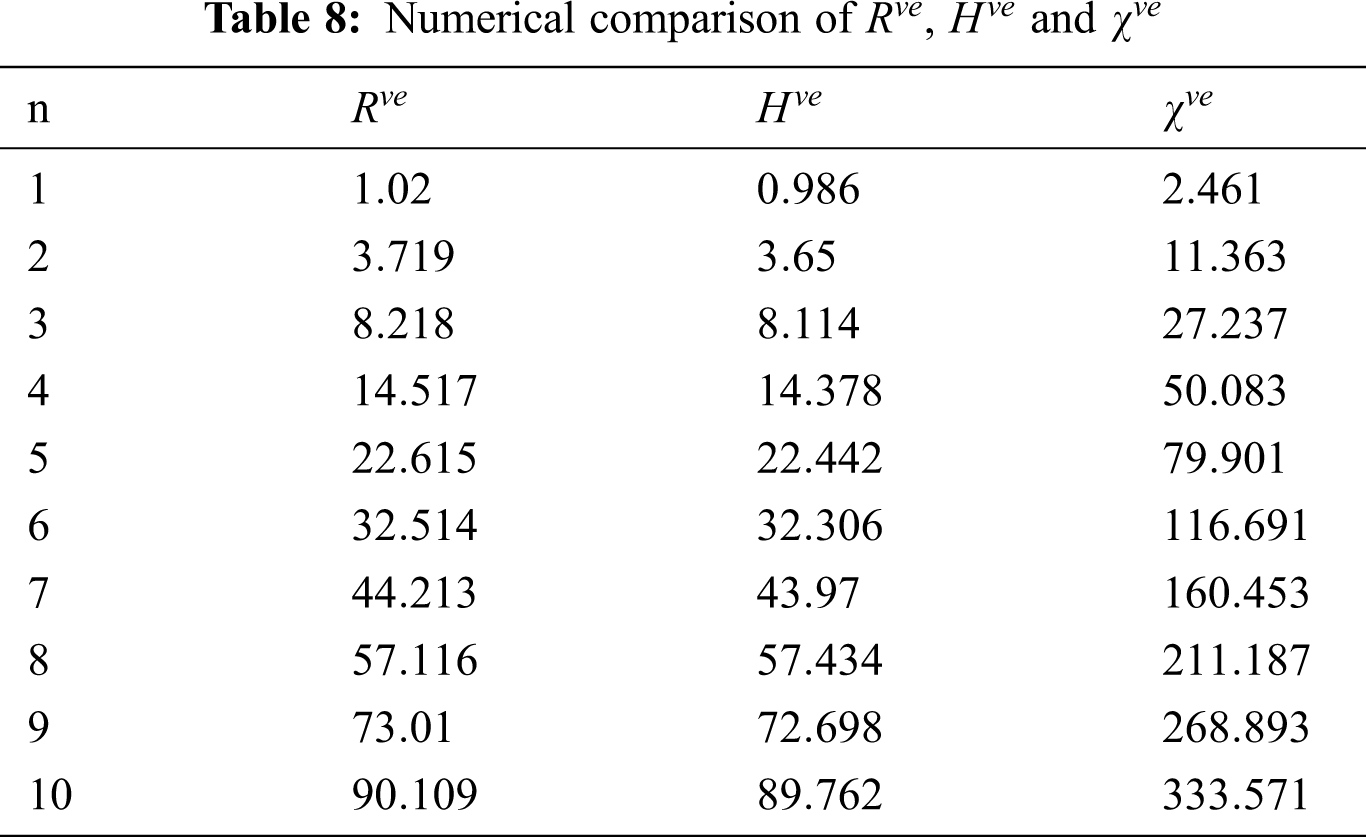
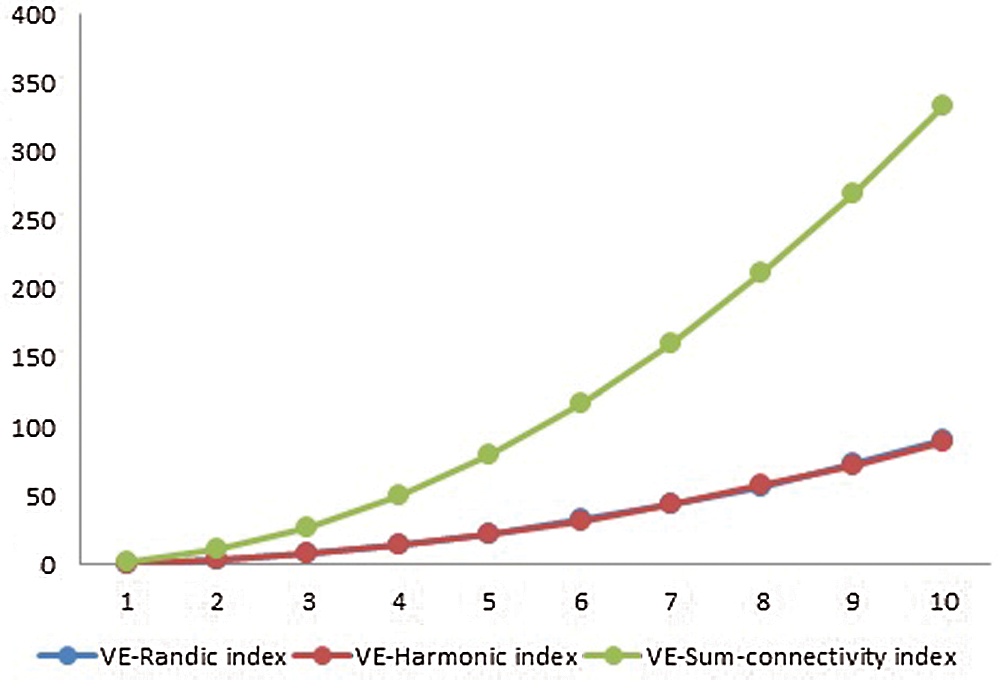
Figure 4: Graphical comparison of
There are many applications of topological descriptors in computer science, networks, agriculture and chemical graph theory etc. These descriptors help in finding the behavior of their structures. We dealt with the honey comb derived network and computed ten different types of topological descriptors which are base on
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. D. B. West, Introduction to graph theory, Second Edition. Pearson Education (Singapore) Pte. Ltd., 2002. [Google Scholar]
2. G. Rcker and C. Rcker, “On topological indices, boiling points, and cycloalkanes,” Journal of Chemical Information and Computer Sciences, vol. 39, no. 5, pp. 788–802, 1999. [Google Scholar]
3. S. Klavzar and I. Gutman, “A comparison of the Schultz molecular topological index with the Wiener index,” Journal of Chemical Information and Computer Sciences, vol. 36, no. 5, pp. 1001–1003, 1996. [Google Scholar]
4. F. M. Bruckler, T. Doslic, A. Graovac and I. Gutman, “On a class of distance-based molecular structure descriptors,” Chemical Physics Letters, vol. 503, no. 4–6, pp. 336–338, 2011. [Google Scholar]
5. H. Deng, J. Yang and F. Xia, “A general modeling of some vertex-degree based topological indices in benzenoid systems and phenylenes,” Computers and Mathematics with Applications, vol. 61, no. 10, pp. 3017–3023, 2011. [Google Scholar]
6. H. Wiener, “Structural determination of paraffin boiling points,” Journal of the American Chemical Society, vol. 69, pp. 17–20, 1947. [Google Scholar]
7. M. Randic, “Characterization of molecular branching,” Journal of the American Chemical Society, vol. 97, no. 23, pp. 6609–6615, 1975. [Google Scholar]
8. B. Bollobas and P. Erdos, “Graphs of extremal weights,” Ars Combinatoria, vol. 50, pp. 225–233, 1998. [Google Scholar]
9. D. Amic, D. Beslo, B. Lucic, S. Nikolic and N. Trinajstic, “The vertex-connectivity index revisited,” Journal of Chemical Information and Computer Sciences, vol. 38, no. 5, pp. 819–822, 1998. [Google Scholar]
10. Y. Hu, X. Li, Y. Shi, T. Xu and I. Gutman, “On molecular graphs with smallest and greatest zeroth-order general Randic index,” MATCH Communication in Mathematical and in Computer Chemistry, vol. 54, no. 2, pp. 425–434, 2005. [Google Scholar]
11. X. Li, I. Gutman and M. Randic, “Mathematical aspects of Randic-type molecular structure descriptors,”University of Kragujevac and Faculty of Science, Kragujevac, 2006. [Google Scholar]
12. M. Randic, “On history of the Randic index and emerging hostility toward chemical graph theory,” MATCH Communication in Mathematical and in Computer Chemistry, vol. 59, pp. 5–124, 2008. [Google Scholar]
13. M. Randic, “The connectivity index 25 years after,” Journal of Molecular Graphics and Modelling, vol. 20, no. 1, pp. 19–35, 2001. [Google Scholar]
14. I. Gutman and B. Furtula, “Recent results in the theory of Randic index,”University of Kragujevac and Faculty of Science, Kragujevac, 2008. [Google Scholar]
15. X. Li and Y. Shi, “A survey on the Randic index,” MATCH Communication in Mathematical and in Computer Chemistry, vol. 59, no. 1, pp. 127–156, 2008. [Google Scholar]
16. S. Nikolic, G. Kovacevic, A. Milicevic and N. Trinajstic, “The Zagreb indices 30 years after,” Croatica Chemica Acta, vol. 76, no. 2, pp. 113–124, 2003. [Google Scholar]
17. I. Gutman and K. C. Das, “The first Zagreb index 30 years after,” MATCH Communication in Mathematical and in Computer Chemistry, vol. 50, no. 1, pp. 83–92, 2004. [Google Scholar]
18. K. C. Das and I. Gutman, “Some properties of the second Zagreb index,” MATCH Communication in Mathematical and in Computer Chemistry, vol. 52, pp. 103–112, 2004. [Google Scholar]
19. I. Gutman, A. Milicevic, S. Nikolic and N. Trinajstic, “About the Zagreb Indices,” Kemija u Industriji, vol. 59, no. 12, pp. 577–589, 2010. [Google Scholar]
20. N. Idrees, R. Khalid, F. B. Farooq and S. Nasir, “Computing topological invariants of triangular chandelier lattice,” CMC-Computers, Materials & Continua, vol. 63, no. 3, pp. 1119–1132, 2020. [Google Scholar]
21. A. Milicevic, S. Nikolic and N. Trinajstic, “On reformulated Zagreb indices,” Molecular Diversity, vol. 8, no. 4, pp. 393–399, 2004. [Google Scholar]
22. A. N. A. Koam, A. Ahmad and M. F. Nadeem, “Comparative study of valency-based topological descriptor for hexagon star network,” Computer Systems Science and Engineering, vol. 36, no. 2, pp. 293–306, 2021. [Google Scholar]
23. M. Chellali, T. W. Haynes, S. T. Hedetniemi and T. M. Lewis, “On degrees and degrees in graphs,” Discrete Mathematics, vol. 340, no. 2, pp. 31–38, 2017. [Google Scholar]
24. B. Horoldagva, K. C. Das and T. A. Selenge, “On ve-degree and ev-degree of graphs,” Discrete Optimization, vol. 31, pp. 1–7, 2019. [Google Scholar]
25. S. Ediz, “A new tool for QSPR researches: ev-degree Randi index,” Celal Bayar University Journal of Science, vol. 13, no. 3, pp. 615–618, 2017. [Google Scholar]
26. B. Sahin and S. Ediz, “On ev-degree and ve-degree topological indices,” Iranian Journal of Mathematical Chemistry, vol. 9, no. 4, pp. 263–277, 2018. [Google Scholar]
27. S. Ediz, “On ve-degree molecular topological properties of silicate and oxygen networks,” International Journal of Computing Science and Mathematics, vol. 9, no. 1, pp. 1–12, 2018. [Google Scholar]
28. M. Cancan, “On ev-degree and ve-degree topological properties of Tickysim spiking neural network,” Computational Intelligence and Neuroscience, Article ID 8429120,2019. [Google Scholar]
29. A. Ahmad, “Comparative study of ve-degree and ev-degree topological descriptors for benzene ring embedded in P-type-surface in 2D network,” Polycyclic Aromatic Compounds, 2020. [Google Scholar]
30. M. Cancan, “On harmonic and ev-degree molecular topological properties of DOX, RTOX and DSL networks,” CMC-Computers, Materials & Continua, vol. 59, no. 3, pp. 777–786, 2019. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |