DOI:10.32604/csse.2021.014578

| Computer Systems Science & Engineering DOI:10.32604/csse.2021.014578 |  |
| Article |
ASRNet: Adversarial Segmentation and Registration Networks for Multispectral Fundus Images
1School of Information Science and Engineering, Shandong Normal University, Jinan, 250358, China
2Key Laboratory of Intelligent Computing & Information Security in Universities of Shandong, Shandong Provincial Key Laboratory for Novel Distributed Computer Software Technology, Institute of Biomedical Sciences, Shandong Normal University, Jinan, 250358, China
3Shandong Provincial Hospital Affiliated to Shandong University, Jinan, 250021, China
4INSA Lyon, University of Lyon, CNRS, Inserm, Villeurbanne, 69621, Cedex, France
*Corresponding Author: Yuanjie Zheng. Email: yjzheng@sdnu.edu.cn
Received: 30 September 2020; Accepted: 19 November 2020
Abstract: Multispectral imaging (MSI) technique is often used to capture images of the fundus by illuminating it with different wavelengths of light. However, these images are taken at different points in time such that eyeball movements can cause misalignment between consecutive images. The multispectral image sequence reveals important information in the form of retinal and choroidal blood vessel maps, which can help ophthalmologists to analyze the morphology of these blood vessels in detail. This in turn can lead to a high diagnostic accuracy of several diseases. In this paper, we propose a novel semi-supervised end-to-end deep learning framework called “Adversarial Segmentation and Registration Nets” (ASRNet) for the simultaneous estimation of the blood vessel segmentation and the registration of multispectral images via an adversarial learning process. ASRNet consists of two subnetworks: (i) A segmentation module S that fulfills the blood vessel segmentation task, and (ii) A registration module R that estimates the spatial correspondence of an image pair. Based on the segmention-driven registration network, we train the segmentation network using a semi-supervised adversarial learning strategy. Our experimental results show that the proposed ASRNet can achieve state-of-the-art accuracy in segmentation and registration tasks performed with real MSI datasets.
Keywords: Deep learning; deformable image registration; image segmentation; multispectral imaging (MSI)
Ophthalmologists utilize fundus photographs to monitor the progression of certain eye conditions and diseases, such as diabetic retinopathy, age-related macular degeneration (AMD), and glaucoma [1−4]. Multispectral imaging (MSI) technology, based on light emitting diode (LED) illumination across a specific wavelength range, is often used to capture a series of narrow band spectral slices of the fundus [5−8]. The wavelengths of observation are selected such that light can penetrate the entire retina and choroid, such that the resulting images are composed of light reflected from different fundus tissue components, as shown in Fig. 1a. However, eye movements may introduce spatial misalignment between the multispectral images because these images are taken at separate points in time [9,10]. Fig. 1b shows the composite color image obtained by combining the MSI-550 and MSI-660 spectral slices, which looks very similar to a conventional fundus photograph. Such fundus images are indispensable to the diagnosis of various diseases; thus, it is important to effectively estimate as well as eliminate the spatial misalignment between the MSI slices during image analysis. Moreover, multispectral fundus imaging allows for a detailed analysis of the retinal blood vessels, which can further assist ophthalmologists in diagnosis and screening of related ophthalmic and blood vessel diseases.

Figure 1: (a) Sequential MSI spectral slices of the fundus obtained using different wavelengths: 550 nm, 580 nm, 590 nm, 620 nm, 660 nm, 690 nm, 740 nm, 760 nm, 780 nm, 810 nm, and 850 nm. (b) Colored RG fundus photograph obtained by combining the green (MSI-550) and red (MSI-660) spectral slices
Image registration using MSI has two main challenges. The first is the apparent intensity difference between the multispectral images. The MSI technique uses different monochromatic wavelengths from a LED source to illuminate the fundus, which causes the incident light intensity and reflectivity at the same spatial position to vary significantly between the images. For example, as shown in Fig. 1a, the retinal blood vessels are more clearly displayed in the shorter-wavelength images, whereas the choroidal structures become more prominent at longer wavelengths. Thus, there is a significant difference between the spectral images in their overall appearance, although the retinal blood vessels remain distinguishable across the entire wavelength range. The second challenge arises due to the time difference between two consecutive images, which is enough for the eye movements to introduce a change in the effective angle of view. The non-rigid rotation of the eyeball introduces not only global displacements but also local distortions that differ across the image slices. Thus, rigid image registration is not a suitable option in this case.
To solve these challenges of multispectral fundus image registration, we propose a segmentation-driven registration network that is similar to the method discussed in Hu et al. [11]. The segmentation maps of blood vessels provide clear information regarding the underlying anatomical structure to guide the network training. The movement of the blood vessels and surrounding tissues between the images is not conducive to the training of our regression neural network; hence, we adopt the strategy of soft labels [12] to propose a weakly supervised registration network based on segmented retinal blood vessel maps for multispectral image registration. The trained model can predict the spatial correspondence between the original multispectral images directly without the blood vessel maps, and no iteration is required. However, it is difficult to obtain ground-truth blood vessel segmentation maps because it requires professional medical knowledge and thus more intensive work. Note that retinal blood vessel maps play a crucial role in helping ophthalmologists make early diagnosis of diabetes as well as several chronic cardiovascular and neurovascular diseases. Hence, the topic of retinal blood vessel segmentation has been extensively researched [13]. Deep learning-based end-to-end methods [14−16] that optimize intermediate features are used to address the problems associated with the task of blood vessel segmentation. In the context of multispectral fundus imaging, the gradual appearance of choroidal structures along with the weakening of the retinal blood vessel features at longer wavelengths further increase the difficulty of blood vessel segmentation.
Recently, several unsupervised and semi-supervised neural networks have been explored to address the challenges of obtaining ground truth in medical image processing. Both unsupervised registration [17−19] and unsupervised segmentation [20,21] learning methods define the respective tasks as a parametric function, which are then realized by optimizing a predefined objective function. Generative adversarial network (GAN) [22] based methods use adversarial learning to train the generative and discriminative modules and generate simulated datasets that can expand the training dataset [23,24]. Domain adaptation [25] is used to address the difference between the source and target data distributions; it is often used to solve the problems related to small training datasets and class imbalance in classification and segmentation tasks [26,27].
In this paper, we propose a semi-supervised deep-learning-based framework for multispectral fundus image analysis, which performs the dual tasks of registration and segmentation simultaneously. Our framework is composed of two neural networks: (1) A segmentation-driven registration network and (2) A segmentation network. Specifically, we present a scheme to better train the segmentation and registration networks by using an adversarial learning strategy. The retinal blood vessel maps generated by the segmentation network drives the training of the registration network. Based on the segmented blood vessel maps, we trained the registration network in a weakly supervised manner and obtained the spatial transform relationship between the two original retina images. Next, we compared the blood vessel map deformed by the spatial transformation layer with another map predicted by the segmentation network, and finally generated a confidence map. The confidence map provided the trustworthy regions in the segmented label map that modifies the segmentation network parameters. In this scheme, we can further adjust the segmentation and registration networks by using unlabeled data, thus countering the demand for large-scale training data. Our algorithm was applied to the tasks of multispectral fundus image segmentation and registration. Our experimental results indicate that the model proposed in this work, called “Adversarial Segmentation and Registration Nets” (ASRNet), is capable of improving the segmentation and registration accuracy significantly, compared to models that use separate algorithms for each task.
As mentioned above, GANs [22] have been proposed to train the generative network using an adversarial learning process, which has achieved great success in semi-supervised image segmentation [28]. We have also incorporated the adversarial strategy into our proposed segmentation and registration networks. As shown in Fig. 2, the proposed ASRNet consists of two subnetworks: (1) A segmentation-driven registration network (denoted as R) and (2) A segmentation network (denoted as S).
2.1.1 Segmentation-driven Registration Network
In recent years, many registration algorithms based on deep learning have been proposed [17,29−31], including multi-modality registration methods [12,32]. The study discussed in Cao et al. [32] uses pre-aligned CT and MR images to train an inter-modality registration network, while [12] describes a method to infer the dense spatial correspondence from the information contained in the manual anatomical labels. To address the intensity difference between the multi-spectral fundus images, we introduced segmentation labels in our deep-learning-based registration method. We used a regression network to predict the spatial correspondence, following which the segmented label map was deformed by the spatial transform layer to iteratively refine the image registration. In other words, we used the spatial transform relationship between the blood vessel maps to determine the corresponding relationship between the original fundus image pair. Both the manually labeled blood vessel maps and the label maps obtained from the segmentation network can be used as training data for the registration network. Our network structure was based on Fan et al. [33], which designed a regression network for image registration. The details of this step are described in Section 2.2.
A fully convolutional network (FCN) [14] as well as more improved methods such as U-Net [16], DSResUNet [34], and deep retinal image understanding (DRIU) [15] are currently the most popular models for image segmentation. To fully leverage the global information in the fundus images, our segmentation network is chosen to be a simplified U-Net [16], which effectively combines high-level and low-level features that can estimate pixel-wise blood vessel segmentation in an end-to-end manner. Our proposed network can be trained with the manually labeled blood vessel maps as well as with the possibility region map generated by the registration model. The details of this step are described in Section 2.3.

Figure 2: Flowchart of our proposed deep learning-based adversarial segmentation and registration method that consists of a segmentation-driven registration network and a segmentation network. We enable mixed supervision through two steps: (1) An image with manually labeled blood vessel maps is used for fully-supervised training of the segmentation network and weakly-supervised learning of the multispectral image registration. (2) An image without manual labeling is used for unsupervised adversarial training. Note that we use the segmentation network to output the predicted blood vessel maps for training the registration network. The deformed blood vessel maps can also be used for optimizing the segmentation network weights
2.2 Segmentation-Driven Registration Network
Deformable image registration establishes the spatial correspondence between different images. In general, the goal of deformable image registration is to optimize the energy of the form [35]

The first term  quantifies the degree of alignment between the fixed and moving images, and
quantifies the degree of alignment between the fixed and moving images, and  and
and  are the fixed and moving images. The second term
are the fixed and moving images. The second term 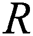 is the regularization constraint imposed on the displacement
is the regularization constraint imposed on the displacement  by inherently smooth prior knowledge.
by inherently smooth prior knowledge.
The registration network structure, as shown in Fig. 2, is composed of a regression network followed by a spatial transform layer [36]. The registration network has an encoder-decoder structure that consists of multiple layers, including convolution, pooling, and deconvolutional layers. The additional convolutional layers are placed in the gap between the encoder and the decoder to achieve balance between the low-level and high-level features [33]. By maintaining reasonable memory allocation, we reduce the number of channels in our network, similar to the algorithms presented in Balakrishnan et al. [17]. According to the energy function of deformable image registration, we propose using the following loss function that is composed of two terms such that

The training objective of the segmentation-driven registration network is to estimate a dense spatial correspondence that warps the moving image to spatially align it with the fixed image. In other words, the goal is to align the fundus image  with the corresponding blood vessel map
with the corresponding blood vessel map  , such that the dissimilarity in the blood vessel map is minimized. In particular, to utilize the edge gradient and the background of the blood vessels more effectively, we use soft label maps with a 2-D Gaussian filter (
, such that the dissimilarity in the blood vessel map is minimized. In particular, to utilize the edge gradient and the background of the blood vessels more effectively, we use soft label maps with a 2-D Gaussian filter ( ) instead of binary labels [12]. To train the segmentation-driven inter-spectral fundus image registration network, the (dis)similarity loss can be defined as the following objective function:
) instead of binary labels [12]. To train the segmentation-driven inter-spectral fundus image registration network, the (dis)similarity loss can be defined as the following objective function:

where 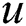 represents the pixel coordinate
represents the pixel coordinate  in the fixed label
in the fixed label 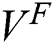 , and
, and  is the total number of pixels.
is the total number of pixels.
To ensure smoothness of the spatial transform predicted by the regression network, we propose using a composite regularization loss function to train the network, which is defined as follows:
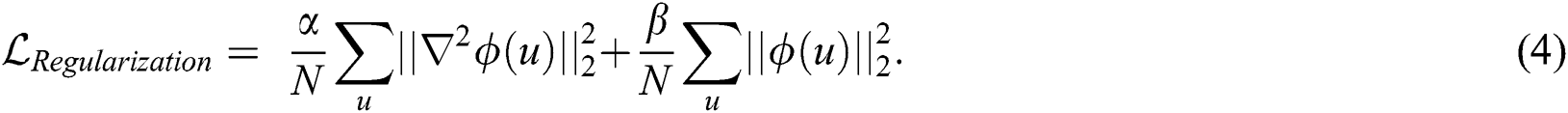
The first term represents a Laplacian ( ) operation to attain the smoothness constraint for the spatial transformation, while the second term balances the initial value of the regression model. Our experiments validate that these two constraints are indispensable for the regularization of the transformation field, where
) operation to attain the smoothness constraint for the spatial transformation, while the second term balances the initial value of the regression model. Our experiments validate that these two constraints are indispensable for the regularization of the transformation field, where  and
and  are the trade-offs characterizing the regularization parameters of the displacement. In our experiments, we set
are the trade-offs characterizing the regularization parameters of the displacement. In our experiments, we set  and
and 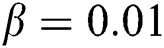 .
.
The segmentation network can be any end-to-end segmentation network. In this study, we used a convolutional neural network architecture, similar to U-Net, to segment the blood vessel maps.
Generalized dice loss: In the retinal blood vessel segmentation task of the fundus images, the blood vessel labels represent a very small fraction of the entire fundus image. To overcome this, we recommend the use of generalized dice loss [37] as the segmentation loss function to pay more attention to the pixels that are difficult to learn from. This is given by

where  is the number of semantic categories, which are only the blood vessel regions and their background in our experiments;
is the number of semantic categories, which are only the blood vessel regions and their background in our experiments;  provides the balancing weight of the prospects and the background, and we set
provides the balancing weight of the prospects and the background, and we set  ;
;  and
and  indicate the ground-truth and predicted maps from the segmentation network, respectively. The weight of the label is adjusted for volume, and the training process pays more attention to the areas that are difficult to identify, such as the edges of blood vessels and the thinner vessels.
indicate the ground-truth and predicted maps from the segmentation network, respectively. The weight of the label is adjusted for volume, and the training process pays more attention to the areas that are difficult to identify, such as the edges of blood vessels and the thinner vessels.
Adversarial loss: The segmentation network can predict the multispectral images corresponding to the blood vessel maps, and the registration network can predict the spatial correspondence between the image pairs. Thus, by warping the segmentation label using a spatial transform layer we can obtain the warped label and further generate the possibility region map. This possibility region map can be used as the ground truth to train the segmentation network using unlabeled data. The adversarial loss function has the same form as the generalized dice loss, such that:
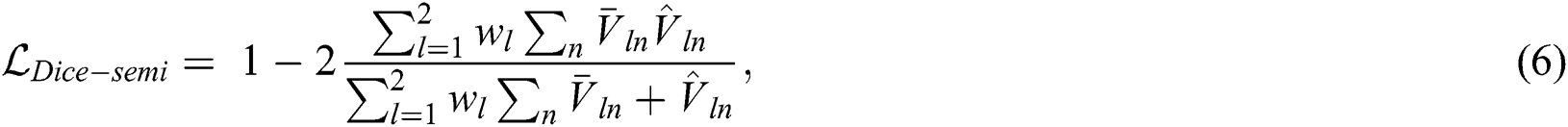
where  is the possibility region map warped by the displacement field obtained from the registration network and
is the possibility region map warped by the displacement field obtained from the registration network and 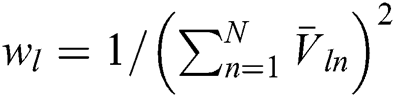 . The training of the segmentation and registration networks depends on the blood vessel labels, which optimize the respective network parameters during each iteration. This may lead to the accumulation of registration error during the adversarial training process. To avoid this, we use appropriate, unlabeled multispectral data to train the adversarial network.
. The training of the segmentation and registration networks depends on the blood vessel labels, which optimize the respective network parameters during each iteration. This may lead to the accumulation of registration error during the adversarial training process. To avoid this, we use appropriate, unlabeled multispectral data to train the adversarial network.
The MSI dataset used in this work was collected in-house. It is composed of 56 sets from 28 volunteers, with each set consisting of 11 ocular posterior pole images. The MSI images were acquired on an RHA™ instrument that performs fast imaging in the visible and near-infrared wavelengths, such that 11 images were captured with wavelengths of 550 nm, 580 nm, 590 nm, 620 nm, 660 nm, 690 nm, 740 nm, 760 nm, 780 nm, 810 nm, and 850 nm, which correspond to green, yellow, amber, red (4), and infrared (4), respectively. The image resolution is  . Next, 15 landmark features were manually selected in each image sequence by ophthalmologists using MRIcron [38]. This dataset was used to evaluate the registration performance of our method. In addition, ophthalmologists manually labeled fundus blood vessel maps for training our registration module, which in turn was used to validate our algorithm.
. Next, 15 landmark features were manually selected in each image sequence by ophthalmologists using MRIcron [38]. This dataset was used to evaluate the registration performance of our method. In addition, ophthalmologists manually labeled fundus blood vessel maps for training our registration module, which in turn was used to validate our algorithm.
The 56 image sequences containing blood vessel labels were split into four sets, such that training and validation were conducted on 24 sequences obtained from 12 volunteers; adversarial training was conducted on 16 sequences from 8 volunteers; and testing was performed on 16 sequences from 8 volunteers. During the training and validation stage, the ASRNet was optimized using both the MSI data and the corresponding retinal blood vessel maps. During the adversarial training stage, the ASRNet was optimized using only the MSI data. Note that the image data without manual labeling were used for testing. During the testing stage, no manual labeling is needed because no hints can be expected when analyzing data in reality.
The adversarial network was implemented in PyTorch [39], and it was trained on two Nvidia Tesla V100 GPUs with 32 GB of video memory. We adopted the Adam algorithm [40] to train the proposed adversarial network with an improved loss function. The two modules (S and R) were trained separately on the training dataset to obtain two initial models. Next, the two models were combined and adversarial training was performed on the entire training dataset. The training details are as follows: (1) Registration network: training epochs  , learning rate
, learning rate 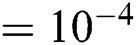 , batch size
, batch size 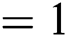 ; (2) Segmentation network: training epochs
; (2) Segmentation network: training epochs 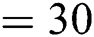 , learning rate
, learning rate  , batch size
, batch size 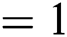 ; (3) Adversarial network: training epochs
; (3) Adversarial network: training epochs  , learning rate
, learning rate  , batch size
, batch size  . It took about
. It took about  h to train the entire network.
h to train the entire network.
To quantitatively evaluate the prediction accuracy, we utilized the mean absolute error (MAE) between the manually marked points to measure the results of registration [41]. This is defined as:
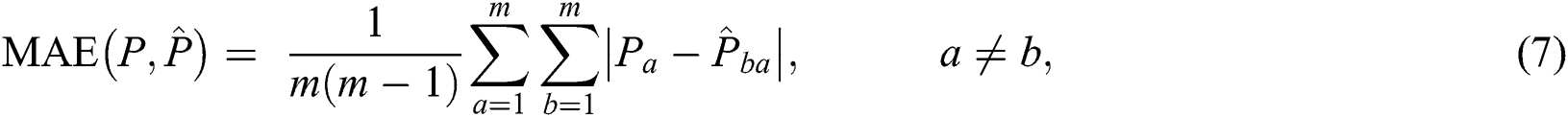
where 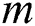 is the number of images in a group;
is the number of images in a group; 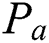 is the manually marked point;
is the manually marked point; 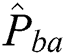 is the corresponding point in the warped moving image, and
is the corresponding point in the warped moving image, and 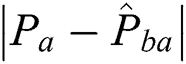 denotes the absolute error between the manually marked point and its corresponding point.
denotes the absolute error between the manually marked point and its corresponding point.
To quantitatively characterize the prediction accuracy, we utilized the Dice similarity coefficient (DSC) score [42] of the retinal blood vessel labels as a performance metric to measure the results of segmentation. This is defined as:

where  represents the ground-truth blood vessel labels and
represents the ground-truth blood vessel labels and  the predicted labels.
the predicted labels.
To demonstrate the advantage of our proposed method, in the following sections we compare our results with those of typical algorithms, as well as with the results obtained by using only the registration network ASRNet-R or only the segmentation network ASRNet-S.
Table 1: Registration results: Comparison of mean MAE values from 16 sequences of the MSI fundus image dataset obtained using methods having different strategies

In our multispectral fundus image registration experiments, all images within the set were paired with each other, such that each group contained  pairs of images. After the network was trained, we deployed it on the test dataset and evaluated the accuracy of our method by comparing with two popular registration methods: (1) VoxelMorph [43], as well as VoxelMorph with segmentation-driven strategy, named VoxelMorph (L) and (2) the label-driven weakly-supervised learning method (LDWSL) [11].
pairs of images. After the network was trained, we deployed it on the test dataset and evaluated the accuracy of our method by comparing with two popular registration methods: (1) VoxelMorph [43], as well as VoxelMorph with segmentation-driven strategy, named VoxelMorph (L) and (2) the label-driven weakly-supervised learning method (LDWSL) [11].
Tab. 1 shows the mean MAE in pixels obtained using the manually marked points in the registration results. We observe that all the algorithms that conduct training with blood vessel maps perform reasonably well for multispectral image registration. However, the performance of conventional VoxelMorph [43] without segmentation information is poor, which shows that segmentation-driven strategy is highly effective for multispectral fundus image registration. Moreover, the performance in Tab. 1 indicates that adversarial learning contributes to significantly improved results, as it provides more reference training data. Similar conclusions can also be reached by visually comparing multispectral fundus images analyzed by different methods, as shown in Fig. 3. Thus, we emphasize that our proposed ASRNet algorithm has the best performance among the registration methods discussed here, which can be attributed to its combined segmentation-driven and adversarial learning strategies. The ASRNet method is also very fast at the testing stage, automatically registering images at the rate of approximately 0.75 s second automatic registrations per image pair can be predicted on the single GPU, which is far much more efficient than traditional registration algorithms.

Figure 3: Registration results of multispectral fundus images. Top row: spectral slices corresponding to (a) MSI-550 and (c) MSI-740; and manually labeled blood vessel maps corresponding to (b) MSI-550 and (d) MSI-740. Middle row: Qualitative comparison of the fundus images using five different registration methods. Bottom row: close-ups of key areas from the corresponding fundus images in the middle row
To demonstrate the advantage of our proposed adversarial segmentation and registration network, we used the same dataset to compare our method with two classical blood vessel segmentation networks, namely, U-Net [16] and DRIU [15]. Note that U-Net [16] and ASRNet-S have the same structure and loss functions. Tab. 2 lists the mean DSC scores along with their standard deviations for the blood vessel segmentation results obtained with four different methods. We observe that the segmentation networks work well in segmenting the blood vessel maps in the shorter wavelength spectral images, but their performance deteriorates for the longer wavelength spectral images (which are often more challenging to segment due to the appearance of the choroidal structure). Our network structure is similar to that of U-Net, and we used generalized dice loss as the loss function to solve the problem of class imbalance in ocular fundus blood vessel segmentation. We see that compared to U-Net and DRIU, ASRNet-R already improves the accuracy significantly. However, ASRNet being a semi-supervised scheme using adversarial learning achieves even better segmentation results. For a visual assessment, we also present our segmentation results for the MSI fundus dataset in Fig. 4; and the results generated by different retinal blood vessel segmentation methods and their corresponding ground truths in Fig. 5. The superiority of ASRNet over U-Net and DRIU can also be estimated from the difference maps shown in Fig. 5, which were obtained between the predicted retinal blood vessel labels and the corresponding ground-truth labels.

Figure 4: Blood vessel segmentation results of multispectral fundus images. Top row: MSI spectral slices of the fundus from a patient. Middle row: Ground-truth retinal blood vessel maps. Bottom row: Segmented blood vessel maps obtained from the proposed ASRNet method

Figure 5: Comparison of the segmentation results obtained using U-Net, DRIU, ASRNet-S, and the proposed ASRNet method
Table 2: Segmentation results: comparison of DSC scores for the MSI fundus image dataset obtained by different methods. The average Dice scores are computed over retinal vessels before the parentheses, and the standard deviations are in parentheses

In this study, we presented a novel semi-supervised segmentation and registration network, ASRNet, for analyzing multispectral fundus images. This framework combines the novel segmentation-driven weakly-supervised registration method with a deep learning-based segmentation model. To address the lack of ground-truth label maps for fundus images obtained using MSI, ASRNet implements the adversarial segmentation and registration strategy to simultaneously estimate the blood vessel maps and the image-pair spatial correspondence. In this framework, the segmentation network S produces the blood vessel labels that drives the registration network; the registration network R outputs the spatial transform that is used to deform the blood vessel labels and generate the possibility region map, which can be further used to train the segmentation network. However, we should mention that, this method should not use unlabeled data excessively. Our experimental results show that the proposed ASRNet algorithm achieves simultaneous estimation of segmentation and registration and improves the accuracy of both the tasks.
Funding Statement: This work was supported by the National Natural Science Foundation of China (Grant Nos. 81871508 and 61773246); the Major Program of Shandong Province Natural Science Foundation (Grant No. ZR2019ZD04 and ZR2018ZB0419); and the Taishan Scholar Program of Shandong Province of China (Grant No. TSHW201502038).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. P. Mccluskey and R. J. Powell. (2004). “The eye in systemic inflammatory diseases,” Lancet, vol. 364, no. 9451, pp. 2125–2133. [Google Scholar]
2. M. D. Abramoff, M. K. Garvin and M. Sonka. (2010). “Retinal imaging and image analysis,” IEEE Reviews in Biomedical Engineering, vol. 3, no. 3, pp. 169–208. [Google Scholar]
3. M. D. Pinazoduran, V. Zanonmoreno, J. J. Garciamedina, J. F. Arevalo, R. Gallegopinazo et al. (2016). , “Eclectic ocular comorbidities and systemic diseases with eye involvement: A review,” BioMed Research International, vol. 2016, 6215745. [Google Scholar]
4. B. V. S. Krishna and T. Gnanasekaran. (2019). “Retinal vessel extraction framework using modified adaboost extreme learning machine,” Computers, Materials & Continua, vol. 60, no. 3, pp. 855–869. [Google Scholar]
5. N. L. Everdell, I. B. Styles, A. Calcagni, J. Gibson, J. C. Hebden et al. (2010). , “Multispectral imaging of the ocular fundus using light emitting diode illumination,” Review of Scientific Instruments, vol. 81, no. 9, 093706. [Google Scholar]
6. A. Calcagni, J. Gibson, I. B. Styles, E. Claridge and F. Orihuelaespina. (2011). “Multispectral retinal image analysis: A novel non-invasive tool for retinal imaging,” Eye, vol. 25, no. 12, pp. 1562–1569. [Google Scholar]
7. S. Li, L. Huang, Y. Bai, Y. Cheng, J. Tian et al. (2015). , “In vivo study of retinal transmission function in different sections of the choroidal structure using multispectral imaging,” Investigative Ophthalmology & Visual Science, vol. 56, no. 6, pp. 3731–3742. [Google Scholar]
8. T. Alterini, F. Diazdouton, F. J. Burgosfernandez, L. Gonzalez, C. Mateo et al. (2019). , “Fast visible and extended near-infrared multispectral fundus camera,” Journal of Biomedical Optics, vol. 24, no. 9, 096007. [Google Scholar]
9. J. Lin, Y. Zheng, W. Jiao, B. Zhao, S. Zhang et al. (2016). , “Groupwise registration of sequential images from multispectral imaging (MSI) of the retina and choroid,” Optics Express in Biomedical Engineering, vol. 24, no. 22, pp. 25277–25290. [Google Scholar]
10. Y. Zheng, Y. Wang, W. Jiao, S. Hou, Y. Ren et al. (2017). , “Joint alignment of multispectral images via semidefinite programming,” Biomedical Optics Express, vol. 8, no. 2, pp. 890–901. [Google Scholar]
11. Y. Hu, M. Modat, E. Gibson, N. Ghavami, E. Bonmati et al. (2018). , “Retinal imaging and image analysis,” in IEEE ISBI, Washington DC, USA, pp. 1070–1074. [Google Scholar]
12. Y. Hu, M. Modat, E. Gibson, W. Li, N. Ghavami et al. (2018). , “Weakly-supervised convolutional neural networks for multimodal image registration,” Medical Image Analysis, vol. 49, pp. 1–13. [Google Scholar]
13. S. Moccia, E. De Momi, S. El Hadji and L. S. Mattos. (2018). “Blood vessel segmentation algorithms—Review of methods, datasets and evaluation metrics,” Computer Methods and Programs in Biomedicine, vol. 158, pp. 71–91. [Google Scholar]
14. J. Long, E. Shelhamer and T. Darrell. (2015). “Fully convolutional networks for semantic segmentation,” in IEEE CVPR, Boston, Massachusetts, MA, USA, pp. 3431–3440. [Google Scholar]
15. K. K. Maninis, J. Pont-Tuset, P. Arbel ́aez and L. Van Gool. (2016). “Deep retinal image understanding,” in IEEE CVPR, Athens, Greece, pp. 140–148. [Google Scholar]
16. O. Ronneberger, P. Fischer and T. Brox. (2015). “U-net: Convolutional networks for biomedical image segmentation,” in Springer MICCAI, Munich, Germany, pp. 234–241. [Google Scholar]
17. G. Balakrishnan, A. Zhao, M. R. Sabuncu, J. Guttag and A. V. Dalca. (2018). “An unsupervised learning model for deformable medical image registration,” in IEEE CVPR, Salt Lake City, Utah, USA, pp. 9252–9260. [Google Scholar]
18. B. D. de Vos, F. F. Berendsen, M. A. Viergever, H. Sokooti, M. Staring et al. (2019). , “A deep learning framework for unsupervised affine and deformable image registration,” Medical Image Analysis, vol. 52, pp. 128–143. [Google Scholar]
19. Q. Fang, J. Yan, X. Gu, J. Zhao and Q. Li, “Unsupervised learning-based deformable registration of temporal chest radiographs to detect interval change,” in Medical Imaging 2020: Image Processing, vol. 11313. Houston, TX, USA: International Society for Optics and Photonics, 2020. [Google Scholar]
20. J. Chen and E. C. Frey. (2020). “An unsupervised learning model for medical image segmentation,” ArXiv Preprint ArXiv:2001.10155. [Google Scholar]
21. Z. Liu, B. Xiang, Y. Song, H. Lu and Q. Liu. (2019). “An improved unsupervised image segmentation method based on multi-objective particle swarm optimization clustering algorithm,” Computers, Materials & Continua, vol. 58, no. 2, pp. 451–461. [Google Scholar]
22. I. Goodfellow, J. Pouget-Abadie, M. Mirza, B. Xu, D. Warde-Farley et al. (2014). , “Generative adversarial nets,” in Proc. NeurIPS, Montreal, Canada, pp. 2672–2680. [Google Scholar]
23. M. Chen, T. Artières and L. Denoyer. (2019). “Unsupervised object segmentation by redrawing,” in Proc. NeurIPS, Vancouver, Canada, pp. 12726–12737. [Google Scholar]
24. X. Li, Y. Liang, M. Zhao, C. Wang and Y. Jiang. (2019). “Few-shot learning with generative adversarial networks based on woa13 data,” Computers, Materials & Continua, vol. 60, no. 3, pp. 1073–1085. [Google Scholar]
25. E. Tzeng, J. Hoffman, K. Saenko and T. Darrell. (2017). “Adversarial discriminative domain adaptation,” in IEEE CVPR, Honolulu, HI, USA, pp. 2962–2971. [Google Scholar]
26. Q. Dou, C. Ouyang, C. Chen, H. Chen and P. A. Heng. (2018). “Unsupervised cross-modality domain adaptation of convnets for biomedical image segmentations with adversarial loss,” in Proc. IJCAI, Stockholm, Sweden, pp. 691–697. [Google Scholar]
27. C. S. Perone, P. L. Ballester, R. C. Barros and J. Cohen-Adad. (2019). “Unsupervised domain adaptation for medical imaging segmentation with self-ensembling,” NeuroImage, vol. 194, pp. 1–11. [Google Scholar]
28. D. Nie, Y. Gao, L. Wang and D. Shen. (2018). “ASDNet: Attention based semi-supervised deep networks for medical image segmentation,” in Springer MICCAI, Granada, Spain, pp. 370–378. [Google Scholar]
29. H. Li and Y. Fan. (2018). “Non-rigid image registration using self-supervised fully convolutional networks without training data,” in IEEE ISBI, vol. 2018. Washington DC, USA, pp. 1075–1078. [Google Scholar]
30. B. D. D. Vos, F. F. Berendsen, M. A. Viergever, M. Staring and I. Išgum. (2017). “End-to-end unsupervised deformable image registration with a convolutional neural network,” in Springer DLMIA, Québec City, QC, Canada, pp. 204–212. [Google Scholar]
31. X. Yang, R. Kwitt and M. Styner. (2017). “Quicksilver: Fast predictive image registration–A deep learning approach,” NeuroImage, vol. 158, pp. 378–396. [Google Scholar]
32. X. Cao, J. Yang, L. Wang, Z. Xue, Q. Wang et al. (2018). , “Deep learning based inter-modality image registration supervised by intra-modality similarity,” in Springer MLMI, Granada, Spain, pp. 55–63. [Google Scholar]
33. J. Fan, X. Cao, P. T. Yap and D. Shen. (2019). “BIRNet: Brain image registration using dual-supervised fully convolutional networks,” Medical Image Analysis, vol. 54, pp. 193–206. [Google Scholar]
34. L. Yu, X. Yang, H. Chen, J. Qin and P. A. Heng. (2017). “Volumetric convnets with mixed residual connections for automated prostate segmentation from 3D MR images,” in IEEE AAAI, San Francisco, California USA, pp. 66–72. [Google Scholar]
35. A. Sotiras, C. Davatzikos and N. Paragios. (2013). “Deformable medical image registration: A survey,” IEEE Transactions on Medical Imaging, vol. 32, no. 7, pp. 1153–1190. [Google Scholar]
36. M. Jaderberg, K. Simonyan, A. Zisserman and K. Kavukcuoglu. (2015). “Spatial transformer networks,” in Proc. NeurIPS, Montreal, Canada, pp. 2017–2025. [Google Scholar]
37. C. H. Sudre, W. Li, T. Vercauteren, S. Ourselin and M. J. Cardoso. (2017). “Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations,” vol. 10553, ArXiv Preprint ArXiv:1707.03237, pp. 240–248. [Google Scholar]
38. C. Rorden and M. Brett. (2000). “Stereotaxic display of brain lesions,” Behavioural Neurology, vol. 12, no. 4, pp. 191–200. [Google Scholar]
39. A. Paszke, S. Gross, S. Chintala, G. Chanan, E. Yang et al. (2017). , “Automatic differentiation in pytorch,” in Proc. NeurIPS, Long Beach, CA, USA, pp. 1–4. [Google Scholar]
40. J. L. B. Diederik and P. Kingma. (2014). “Adam: A method for stochastic optimization,” ArXiv Preprint ArXiv: 1412. 6980. [Google Scholar]
41. J. Kim, J. Lee, J. W. Chung and Y. Shin. (2016). “Locally adaptive 2D-3D registration using vascular structure model for liver catheterization,” Computers in Biology and Medicine, vol. 70, pp. 119–130. [Google Scholar]
42. L. R. Dice. (1945). “Measures of the amount of ecologic association between species,” Ecology, vol. 26, no. 3, pp. 297–302. [Google Scholar]
43. G. Balakrishnan, A. Zhao, M. R. Sabuncu, J. Guttag and A. V. Dalca. (2019). “Voxelmorph: A learning framework for deformable medical image registration,” IEEE Transactions on Medical Imaging, vol. 38, no. 8, pp. 1788–1800. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |