 Open Access
Open Access
REVIEW
Ensemble Deep Learning Approaches in Health Care: A Review
Department of Computer Science, College of Computing and Information Technology, Taif University, Taif, 21974, Saudi Arabia
* Corresponding Author: Aziz Alotaibi. Email:
(This article belongs to the Special Issue: Advancements in Machine Learning and Artificial Intelligence for Pattern Detection and Predictive Analytics in Healthcare)
Computers, Materials & Continua 2025, 82(3), 3741-3771. https://doi.org/10.32604/cmc.2025.061998
Received 19 September 2024; Accepted 16 January 2025; Issue published 06 March 2025
Abstract
Deep learning algorithms have been rapidly incorporated into many different applications due to the increase in computational power and the availability of massive amounts of data. Recently, both deep learning and ensemble learning have been used to recognize underlying structures and patterns from high-level features to make predictions/decisions. With the growth in popularity of deep learning and ensemble learning algorithms, they have received significant attention from both scientists and the industrial community due to their superior ability to learn features from big data. Ensemble deep learning has exhibited significant performance in enhancing learning generalization through the use of multiple deep learning algorithms. Although ensemble deep learning has large quantities of training parameters, which results in time and space overheads, it performs much better than traditional ensemble learning. Ensemble deep learning has been successfully used in several areas, such as bioinformatics, finance, and health care. In this paper, we review and investigate recent ensemble deep learning algorithms and techniques in health care domains, medical imaging, health care data analytics, genomics, diagnosis, disease prevention, and drug discovery. We cover several widely used deep learning algorithms along with their architectures, including deep neural networks (DNNs), convolutional neural networks (CNNs), recurrent neural networks (RNNs), and generative adversarial networks (GANs). Common healthcare tasks, such as medical imaging, electronic health records, and genomics, are also demonstrated. Furthermore, in this review, the challenges inherent in reducing the burden on the healthcare system are discussed and explored. Finally, future directions and opportunities for enhancing healthcare model performance are discussed.Keywords
Machine learning (ML) algorithms have been rapidly incorporated into many different applications due to the increase in computational power and the availability of massive amounts of data. Currently, the two leading machine learning methods are advanced deep learning and ensemble learning methods. Deep learning (DL) algorithms are a subfield of machine learning based on deep neural network structures and have gained considerable attention in recent decades due to their ability to extract complex, hidden, high-level features from inputs. These features are extracted from the raw data and allow the model to learn the data representations and patterns. The input data are processed through large layers with large quantities of neurons and parameters for automatic reasoning and decision-making. These layers receive the raw data and transform it into a nonlinear form, which is passed to the next layer. The first layer is called the input layer, the last layer is called the output layer, and the middle layers are called the hidden layers, which perform the deep learning step. Moreover, deep learning layers may include a variety of layers, such as activation layers, normalization layers, dropout layers, cropping layers, sequence layers, max pooling layers, average pooling layers, fully connected layers, combination layers, and generative adversarial network layers [1]. Deep learning algorithms have a variety of different network architectures for different regression and classification tasks, such as artificial neural networks (ANNs), convolution neural networks (CNNs), recurrent neural networks (RNNs), and generative adversarial networks (GANs), as shown in Fig. 1. However, building and training deep learning algorithms require expertise to tune the hyperparameters and massive amounts of data. Therefore, training and tuning of complex models may result in many issues, such as overfitting and gradient vanishing. Reviews of deep learning architectures have been presented in several research papers [2–4]. By utilizing recent deep learning techniques in a variety of applications, state-of-the-art performance has been achieved in several domains, including object recognition, speech recognition, genomics, and drug discovery. Ensemble learning is a reliable strategy that combines several types of baseline learning for better generalization performance. Theoretical and experimental evidence has proven that ensemble deep learning outperforms single deep learning models [5]. The effectiveness of ensemble deep learning has been widely proven in a variety of applications, such as computer vision, finance, bioinformatics, and health care [6]. Ensemble learning involves different techniques, such as bagging, boosting, and stacking. With developments in both deep learning algorithms and ensemble learning techniques, healthcare support systems are growing rapidly and widely, especially in health informatics areas, such as medical imaging, drug discovery, bioinformatics, health analytics, and public health [7]. In addition, with a large amount of available patient data [8], patient information can be collected, stored, and retrieved in real-time for several purposes, including diagnosis, monitoring, treatment, and various types of data analysis and decision-making. Utilizing an ensemble deep learning model to process patient information improves both the reliability and stability of the models by reducing the variance and avoiding overfitting [8]. Thus, ensemble deep learning has outperformed physician-level accuracy in a variety of disease diagnostic and treatment tasks, including medical imaging and cancer detection [9]. However, comparing human performance to that of artificial intelligence algorithms has led to a lack of full clinical diagnosis, as these methods use only images to perform the diagnosis. In the real world, physician readers have access to both medical records and medical images and may require additional tests for successful diagnosis. Health in general refers to a state of well-functioning behavior, and physical and mental health without any disease or health care refers to restoring health and preventing/detecting health problems in the early stage. Early disease detection can encourage people to change their bad habits, lousy eating habits, and lifestyles. Additionally, early chronic disease detection helps patients to avoid complications and expedite treatment [10]. Healthcare systems offer several high-quality services to overcome rapidly increasing and complicated issues related to elder care and chronic diseases. Common factors such as laboratory tests and experimental results, sex, age, body mass index (BMI), hypertension status, blood pressure (BP), lifestyle, diet, and exercise habits of patients are utilized to detect chronic diseases, including diabetes, heart attack, hepatitis, and kidney diseases [11]. Health specialists/physicians use medical laboratory tests and diagnoses to determine whether a patient has/suffers from a specific disease. The use of patient history and generated data from an electronic device could help specialists/physicians predict a variety of diseases in the early stages. Many intensive studies have been conducted on deep learning algorithms utilized in the healthcare field to enhance both prediction and classification performance. Esteva et al. [9] presented deep learning techniques for health care. The authors discussed deep learning in computer vision, natural language processing, reinforcement learning, and generalized methods related to health care issues but not ensemble deep learning methods. In [12], the authors reviewed Bayesian deep learning (BDL) techniques and their benefits and limitations in the healthcare field. Mahajan et al. [13] reviewed the use of ensemble learning in five highly researched diseases, i.e., diabetes, skin disease, kidney disease, liver disease, and heart conditions. Nisar et al. [1] discussed health through deep learning, including issues, challenges and opportunities. To the best of our knowledge, this paper is the first comprehensive review focusing specifically on ensemble deep learning in healthcare. This article covers only the most recent studies that integrate ensemble learning with deep learning algorithms—referred to as ensemble deep learning while excluding those that rely solely on standard ensemble learning or traditional deep learning approaches. An overview of ensemble learning is summarized. Moreover, deep learning algorithms and architectures are demonstrated. The use of ensemble deep learning algorithms in healthcare systems, including medical imaging, health data analytics, genomics, disease prediction and prevention, and drug discovery, is discussed in detail. The contributions of this review can be summarized as follows:
• This paper provides a comprehensive review of ensemble learning and deep learning algorithms and architecture, including recent models such as RNNs and GANs.
• Ensemble deep learning algorithms in health care applications, such as medical imaging and its variance, health data analytics, genomics, disease predication, and prevention and drug discovery, are covered.
• Healthcare applications based on deep ensemble learning, consisting of four significant subsections of benchmark datasets, data preprocessing, model training, and performance metrics, are summarized.
• The limitations and solutions of ensemble deep learning in health care are discussed, and future directions are provided.

Figure 1: Artificial intelligence family
The remainder of this paper is organized as follows: Section 2 and Section 3 provide an overview of ensemble learning and deep learning algorithms and architectures, respectively. In Section 4, ensemble deep learning algorithms in health care systems are comprehensively discussed. Health care applications, including datasets, data preprocessing, model training, and performance metrics, are summarized in Section 5. Section 6 provides an in-depth discussion of the limitations and future directions. Section 7 concludes this review article.
2 Overview of Ensemble Learning
This section presents an overview of ensemble learning methods and techniques for building blocks of different ensemble methods to provide efficient and reliable prediction/classification methods. Most of the reviewed research articles on ensemble learning methods attribute the discovery of the ensemble learning method to Dasarathy and Sheela (1979), who suggested partitioning the feature space using multiple classifiers [14]. Hansen et al. [15] proposed an ensemble method based on a neural network with plurality consensus that achieved better prediction performance than a single classifier. Moreover, Schapire [16] presented a method that converts a weak learner into a strong learner by combining weak classifiers, referred to as a boosting technique. Ensemble learning is a widely used machine learning technique that combines multiple base learners to form an ensemble learning model for building a robust and accurate predictive model [16]. A base learner is a single model that may easily suffer from noise, bias, and variation in the data when performing prediction. Thus, ensemble learning is applied to reduce the generalization error and enhance the classifier performance. The basic idea behind the ensemble learning framework is to aggregate the base learner classifiers C (

Figure 2: General ensemble learning method
2.1 Ensemble Learning Techniques
In this subsection, the three most popular ensemble learning techniques, i.e., bagging, boosting, and stacking, are explored and demonstrated.
Bagging, short for bootstrap aggregation, was developed by Breiman in 1996 to improve classification performance by combining and training multiple algorithms on small random subsets [18,19]. Bagging improves accuracy by creating a diverse model utilizing bootstrap sampling and aggregation steps. In bootstrap sampling, the training data sample is randomly selected from a given dataset with replacement, as shown in Fig. 3. The training data sample may be included multiple times in a subset or may not be used at all to train the model independently. In the aggregation step, the outputs from different base classifiers are aggregated to determine the final decision via majority voting. The data replacement and aggregation methods introduce diversity to create a strong base classifier. The main advantage of utilizing the bagging technique is a reduction of the variance without increasing the bias, which improves the overall prediction performance as shown in Table 1. Thus, it solves the overfitting problem introduced by the decision tree. The disadvantage of bagging is that it leads to large model complexity, loss of interpretability, and high bias. A popular bagging algorithm is random forest (RF).

Figure 3: Bagging ensemble learning process

Methods The boosting ensemble technique, which was introduced by Freund and Schapire in early 1990 [16], is based on a sequential process to correct errors made by the previous weak classifier. Boosting techniques select the misclassified data sample from the previous base classifier and assign it more weight to help the next base classifier boost the performance. However, the iterative learning technique makes boosting unsuitable for learning noisy data because the high weight assigned to noisy data is usually much greater than the weights assigned to other samples. Thus, focusing on misclassified samples causes overfitting to occur. Boosting ensemble techniques reduce the high bias and low variance. Boost ensemble algorithms include several techniques, namely, adaptive boosting (AdaBoost), gradient boosting, extreme gradient boosting (XGBoost), light gradient boosting machine (LightGBM), and CatBoost [20].
Stacking, sometimes known as stacked generalization, was introduced by Wolpert in 1990. It combines different classifier algorithms to generate a meta-model to reduce the generalization error in specific tasks. Stacking is composed of two steps. First, the base classifiers consist of different machine algorithms, such as RF, ANN, and SVM, to train the model using the original data samples. Each algorithm is trained independently. In the second step, the outputs of the other ML algorithms are used to train the metaclassifier to predict the final outputs. Utilization of more than one algorithm produces more reliable predictive models that are superior to single models [21]. Unlike bagging ensembles, which use decision tree models trained on subsets of the training data samples, stacked ensembles use different ML algorithms and are trained on the same training data samples [20]. Additionally, unlike the boosting ensemble, which sequentially trains models to correct the prediction errors of previous models, stacking trains a single model to learn how to optimally combine the predictions from the base classifiers [20].
3 Deep Learning Algorithm and Architecture
Deep learning has been used in various artificial intelligence applications. Deep learning algorithms have been applied mainly to regression, classification, and clustering tasks. Deep learning can extract high-level features through a hierarchical feature learning mechanism because it has a more complex architecture than classical machine learning algorithms such as logistic regression, support vector machines, decision trees, KNNs and naïve bays. This complex architecture requires more computational resources and more data to train and fine-tune the hyperparameters of the models. In this section, we review the most famous deep learning algorithm architectures that have been utilized in the context of ensemble learning for healthcare applications. The success of deep learning applications in health care depend on finding a suitable architecture to fit the task; thus, we explain the deep learning algorithms and architectures as follows.
Deep neural networks, which are computational models, have achieved great empirical success in several machine learning tasks, such as computer vision, speech recognition, and natural language processing. Deep neural networks consist of multiple layers, and each layer has many artificial neurons, as shown in Fig. 4. Each neuron receives features (inputs) that are multiplied by network weights. These weights are randomly initialized. Then, the sum of all multiplications is passed through an activation function (generally a nonlinear function). The output of the nonlinear function is an output of neurons. The outputs of neurons in the previous layers are used as inputs to the next layers with the formula below:
where g is the activation function,

Figure 4: Deep neural networks
The optimal weight distribution is achieved through an iterative training process using backpropagation algorithms [22].
3.2 Convolution Neural Networks
Convolutional neural networks (CNNs), introduced by Lecun et al. in 1998 and inspired by visual perception, are supervised deep learning algorithms used mainly in computer vision, NLP, drug discovery, etc. CNNs consist of three main layers, namely, convolutional layers and pooling layers, followed by fully connected layers. First, the convolutional layer utilizes many kernel (filter) weights to convolve the entire input for feature extraction, such as edges, textures, objects and lines as defined:
where
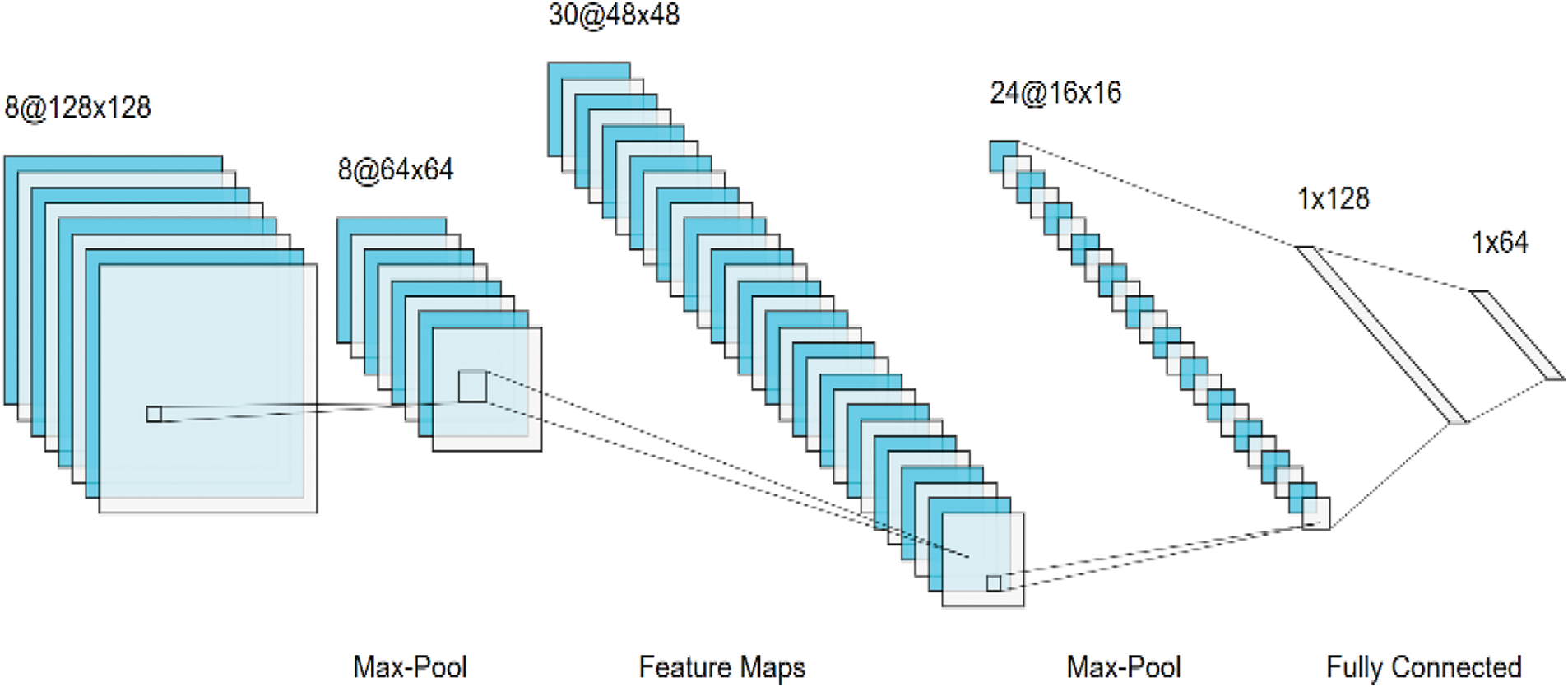
Figure 5: Convolution deep neural networks
A recurrent neural network (RNN) is a neural network with recurrent connections used to extract patterns from sequential or time-series data such as DNA, RNA, speech, video, time series, financial data, and text. A simple RNN consists of three layers: input layers, recurrent hidden layers, and output layers. The main component of RNNs is the hidden state that stores, remembers, and processes past information for long-term dependency. This hidden state is based on a memory cell, which is updated at each time step by taking two inputs: the current input and the previous hidden state. When training RNNs for long-range sequences of data, RNNs suffer from vanishing or exploding gradient problems [26]. To overcome this issue, long short-term memory (LSTM) networks and gated recurrent unit (GRU) networks have been utilized to handle long sequential learning problems. There are many RNN architectures, including one-to-one, one-to-many, many-to-one, and many-to-many.
3.4 Generative Adversarial Networks
Generative adversarial networks (GANs), introduced by Goodfellow et al. [27], are a recent type of deep learning algorithm used to capture the probability distribution of training data. GANs are designed for unsupervised learning problems, especially in state-of-the-art transfer learning, and for tackling the data imbalance challenge. GANs have been successfully applied to many generative tasks, such as image generation, natural language processing, data augmentation, and style transfer. GANs consist of two networks: a generative network and a discriminative network. The generator network learns to generate realistic data such as text or images from random noise to fool the discriminator network, where the discriminator network is a classifier that learns to distinguish between fake data and real data. There are several GAN architecture types, including vanilla GAN, conditional GAN (CGAN), deep convolutional GAN (DCGAN), StyleGAN, CycleGAN, PixelRNN, and super resolution GAN (SRGAN).
An autoencoder (AE) is an unsupervised learning algorithm used mostly for representation learning, generative modeling, denoising, dimensionality reduction (compression), feature extraction, efficient coding, anomalies, etc. An AE consists of three parts: an encoder, code, and a decoder. The encoder compresses the input data into lower dimensions through multilayer neural networks. The code, known as a latent-space representation, is a bottleneck of the network that represents compressed input data. The decoder is used to reconstruct the original input. There are several autoencoder types, namely, denoising (DAE), sparse (SAE), contractive AE (CAE), and variational (VAE). DAEs are used to remove noise from partially corrupted input where the input and output are not the same [28]. The SAE penalizes the activation function by adding a penalty term to the loss function [29]. The CAE utilizes a regularization term to make the model robust to slight changes in the input data. A VAE is a generative model that captures the underlying probability distribution of the data samples to find low-dimensional representations, perform dimensionality reduction, and extract features.
4 Ensemble Deep Learning in Healthcare System
Ensemble Deep Learning outperforms both traditional deep learning and standard ensemble learning in healthcare systems by combining predictions from multiple classifiers, resulting in improved accuracy, robustness, and generalization, particularly when handling noisy, imbalanced, and diverse healthcare datasets. It mitigates overfitting and improves sensitivity and specificity in critical diagnostic applications more effectively than standard ensemble learning. In this section, we review six of the most common healthcare tasks that utilize deep ensemble deep learning to extract useful features and enhance performance as shown in Fig. 6: medical imaging, electronic health records, genomics, disease prediction and prevention, and drug discovery.

Figure 6: (1) Low level features, (2) Middle level features, and (3) High level features
The recent success of employing ensemble deep learning has been mostly in the field of computer vision, which utilizes images and video frames for predication/classification tasks. Computer vision algorithms in health care, including radiology, ophthalmology, pathology, and dermatology, utilize medical images to improve clinical diagnosis and treatment. These medical images are acquired through different imaging modalities, including magnetic resonance imaging (MRI), computed tomography (CT), X-rays, and positron emission tomography (PET), for early diagnosis and treatment. Medical imaging modalities are classified into seven major tasks: classification, segmentation, detection, registration, reconstruction, retrieval, and enhancement [30]. Medical images, known as radiography, aid specialists/physicians in reducing workload and expediting objective opinions. In addition, specialists/physicians can easily discover hidden areas in images by performing a variety of tasks, such as object detection, classification, registration, segmentation, image reconstruction and enhancement, by utilizing advanced algorithms, such as ensemble deep learning algorithms [9,12]. Due to the wide use of both deep learning and ensemble learning technologies in medical imaging, ensemble deep learning leverages the diversity of aggregated models to significantly enhance performance accuracy, improve robustness, and effectively address challenges such as data variability, noise, and class imbalance inherent in medical imaging tasks. Fully intelligent medical imaging diagnosis and prediction health care systems are hot research topics, especially medical imaging classification, medical imaging segmentation, and medical imaging detection.
4.1.1 Medical Imaging Classification
Deep learning is applied in medical imaging classification to extract complex features from raw image data by learning hierarchical representations of input images. By incorporating ensemble deep learning with a diverse set of deep base classifiers, the approach can significantly enhance overall prediction accuracy and resilience, decreasing the possibility of incorrect classifications. This, in turn, improves the reliability of medical classification and contributes to the development of more precise and effective diagnostic outcomes. Yang et al. [31] proposed an ensemble deep learning model for medical image classification that comprises two modules: a deep tree training (DTT) scheme and a two-stage selective ensemble of CNN branches. Their proposed approach mitigates the vanishing gradient problem via DTT, which causes chain rule computation of the backpropagation. Both the CNN and DTT are split into M classifier branches, followed by two-stage selective ensemble, accuracy-based selection and diversity-based selection to select the optimal ensemble members from branch classifiers to overcome the overfitting and local optima issues. Although the proposed approach has been proven effective for medical imaging tasks, it introduces several challenges. These include the need to fine-tune numerous hyperparameters, such as the number of branches, the splitting location, and the number of selected base classifiers. This added complexity can significantly increase computational demands and the effort required for model optimization. The proposed approach was proven to be effective for medical imaging tasks. Ahn et al. [32] proposed an ensemble of different unsupervised feature learning approaches to learn feature representations to differentiate dissimilar medical images using K-means clustering and convolutional neural networks. The proposed approach jointly learns feature representations and clustering assignments in an automated end-to-end system. Their method outperformed other unsupervised methods and addressed the issue of the large volume of unlabeled datasets. Tandel et al. [25] developed an efficient computer-aided diagnostic tool for brain tumor grading using MRI sequences. The authors utilized five well-known convolutional neural models, AlexNet, VGG16, ResNet18, GoogLeNet, and ResNet50, to extract useful features. This method employed ensemble deep learning based on majority voting with relevant MRI sequence data to boost the tumor classification performance. The authors compared the proposed approach with well-known models such as AlexNet, VGG16, ResNet18, GoogLeNet, and ResNet50; the results showed a significant improvement of the proposed approach on different datasets. Gunasekaran et al. [33] presented a deep ensemble model called GIT-Net to classify gastrointestinal (GI) diseases and disorders utilizing endoscopic images. The proposed method consists of three pretrained models, ResNet50 (residual network), DenseNet201 (densely connected convolutional networks), and Inception V3, which are pretrained on the ImageNet dataset to extract complex features from the KVASIR v2 dataset with eight classes of digestive diseases. Additionally, the authors achieved accuracies of 92.96% and 95.00% through a weighted average ensemble model, which was used to reduce training time. The proposed method’s dependence on a weighted average ensemble model poses a challenge, as it significantly increases computational complexity and training time due to the integration of three large pretrained models with millions of parameters, despite achieving high accuracy. Tajammal et al. [34] proposed a deep learning-based ensembling technique to classify the six stages of Alzheimer’s disease using a pipeline of medical imaging processing. Their approach is divided into two steps. First, a custom CNN, inspired by VGG-16, is used to classify the scans of subjects into one stage. Then, VGG-16, ResNet-18, AlexNet, Inception V1, and custom CNNs are combined for multiclass classification of Alzheimer’s disease. The results show that max voting outperforms other ensemble techniques, such as stacking, blending, and averaging, with 98%, 94.2%, 92.5%, and 97.9% accuracy, respectively. However, the proposed method introduces additional complexity due to ensemble techniques like max-voting and stacking, which require meticulous tuning. Furthermore, the lack of extensive validation raises concerns about the method’s robustness and adaptability across diverse datasets. Table 2 is a summary of published articles that used ensemble deep learning for medical image classification.
4.1.2 Medical Imaging Segmentation
Automatic segmentation of medical images is crucial for accurately identifying boundaries and minimizing potential risks to a patient’s health. Ensemble deep learning techniques significantly enhance boundary detection and precision by aggregating outputs from multiple models, each specializing in different aspects of image features, such as edge detection, texture analysis, and intensity variations. Additionally, these ensemble models improve sensitivity and specificity by effectively addressing the class imbalance challenge, which is commonly encountered in detecting rare medical conditions. By leveraging the strengths of diverse models, ensemble deep learning ensures more robust and reliable segmentation outcomes, ultimately aiding in better diagnosis and treatment planning. Goyal et al. [41] proposed a skin lesion segmentation system utilizing ensemble deep learning. The authors combined Mask R-CNN and DeeplabV3+ with preprocessing and postprocessing methods to perform lesion segmentation. The proposed method outperformed other deep learning algorithms, such as FrCN, FCNs, U-Net, and SegNet. Ji et al. [42] aimed to develop automated cervical cell segmentation, including cytoplasm and nucleus segmentation, using deep ensemble learning. Six different deep learning architectures were used, namely, U-Net, U-Net++, DeepLabV3, DeepLabV3Plus, Transunet, and Segformer, which represent three different architectures, namely, encoder-decoder, dilated convolution and vision transform architectures. The ensemble learning model was initialized using a pretrained ImageNet. The final predictions were obtained by aggregating the results from multiple models using the unweighted average. The proposed method relied heavily on pre- and post-processing steps, which affected its robustness, scalability, and ease of deployment in real-world clinical applications. The authors of [43] proposed a fast and accurate autosegmentation method for organs at risk (OARs) and high-risk clinical tumor volume (HRCTV) in patients with gynecological cancer. The authors applied nnU-Net, which is an automatically adapted deep convolutional neural network based on U-Net, to segment three parts: the bladder, rectum and HRCTV on CT images. Three architectures, namely, 2D U-Net, 3D U-Net and 3D-Cascade U-Net, were utilized for fast and reproducible autosegmentation of OARs and HRCTVs in gynecological cancer. The proposed method suffers from high computational costs due to resource-intensive training and testing processes, and its task-specific nature limits generalizability across different datasets and clinical scenarios. The authors of [44] proposed two selective ensemble methods for deep learning segmentation of major vessel areas for automated quantitative coronary analysis (QCA) via invasive coronary angiography (ICA) to improve segmentation performance and reduce morphological errors in the predicted masks. The selective ensemble methods are based on the following steps: (1) obtain prediction masks from multiple segmentation models focused on different morphological features; (2) rank the prediction masks based on the morphological or estimated dice similarity coefficient (DSC); and (3) combine the prediction masks with weights that vary according to the ranks. The authors applied U-Net with DenseNet-121 to segment the three major vessels using a large database of ICA images. The proposed method outperforms the individual models, improves the segmentation performance with DSCs up to 93.07%, and provides a better delineation of coronary lesions with local DSCs of up to 93.93%. The proposed method should focus on side branch analysis for bifurcation lesion evaluation, angiographic sequence analysis for comprehensive interpretation, and reducing dependency on the best-performing individual models within selective ensembles. Rahimpour et al. [45] presented a visual ensemble selection of deep convolutional neural networks for 3D segmentation of breast tumors on dynamic contrast-enhanced MR images. The authors developed three 3D U-Net models for segmentation that were trained using different strategies: postcontrast images or a combination of postcontrast and subtraction images. The proposed method achieved segmentation results, with 77% rated as useful, demonstrating outcomes comparable to inter-radiologist agreement.
4.1.3 Medical Imaging Detection
Ensemble deep learning has transformed medical imaging detection by effectively tackling key challenges, including noise, variations in imaging modalities, class imbalance, and the demand for precise anomaly identification. By aggregating predictions from multiple models, these methods improve robustness, sensitivity, and specificity, making them highly effective for identifying intricate patterns in medical images. This approach is particularly valuable for detecting subtle anomalies, such as nodules in CT scans, lesions in MRIs, and markers of COVID-19 in chest X-rays. Zhou et al. [36] proposed an ensemble deep learning model for novel COVID-19 detection from CT images. Transfer learning was applied to the initialization parameters of the models, and three well-known convolutional neural network models were pretrained—AlexNet, GoogLeNet, and ResNet—followed by softmax as the classification layer. The proposed method achieved 99.054% detection of COVID-19 on a public dataset utilizing a relative majority vote algorithm as an ensemble classifier. Rajaraman et al. [46] proposed a modality-specific deep learning strategy to improve the generalizability of the transferred knowledge gained through modality-specific features. The authors designed and evaluated the performance of a baseline, custom, sequential CNN model for detecting tuberculosis (TB) in chest X-ray (CXR) images. Pretrained CNNs, namely, InceptionResNet-V2, Inception-V3, and DenseNet-121, were employed to learn modality-specific features. Different ensemble methods, such as majority voting, simple averaging, weighted averaging, and stacking, were used to reduce the detection variance and training data sensitivity and improve the detection performance. Tang et al. [38] proposed an ensemble deep learning model, called EDL-COVID, for detecting COVID-19 cases from chest X-ray images. Their model consists of two steps. First, multiple COVID-Net snapshot models, which are based on deep CNNs, are combined, especially for COVID-19 case detection. Second, an ensemble method called weighted averaging ensembling (WAE) is utilized. Compared with the original COVID-Net, the proposed method achieved 95% accuracy on the COVIDx dataset (93%). Alsubai et al. [39] presented a hybrid ensemble deep learning method for brain tumor detection. This ensemble model includes two deep learning algorithms—a convolutional neural network (CNN) and long short-term memory (LSTM)—for extracting features and classifying brain tumors utilizing magnetic resonance imaging (MRI) (no image detection). The proposed model achieved an accuracy of 99.1%, a precision of 98.8%, a recall of 98.9%, and an F1-measure of 99.0%. However, the variability in tumor shapes and sizes presented significant challenges. Terzi [47] proposed an ensemble of deep learning object detection models based on anatomical and pathological regions in brain MR images. The proposed detection methods used nine different state-of-the-art object detection models, namely, RetinaNet, YOLOv3, FCOS, NAS-FPN, VFNet, Faster R-CNN, Dynamic R-CNN, Cascade R-CNN, and ATSS. The proposed models were used to detect 12 anatomical and pathological regions simultaneously, including the brain ROI (brain tissue and orbital CSF), eyes, optic nerves, lateral ventricles, third ventricle peritumoral edema, contrast-enhancing region, tumor necrosis, hemorrhage, and no contrast-enhancing region. The proposed models achieved significant performance improvements, including up to 10% higher mean average precision (mAP), 18% better class-based precision for anatomical structures, and a 3.3% mAP enhancement over the best individual model. Ravi et al. [40] proposed a multichannel EfficientNet deep learning approach for detecting lung diseases such as pneumonia, tuberculosis (TB), and COVID-19 using chest X-ray images. Three multichannel pretrained models, EfficientNetB0, EfficientNetB1, and EfficientNetB2, are used for feature extraction. The stacked ensemble classification approach has two stages: first, a random forest and support vector machine are used for prediction; second, logistic regression is used for classification. Table 3 provides a summary of published articles that employ ensemble deep learning techniques for medical image detection.
Electronic health records (EHRs) are collections of patient data records that are stored electronically in both structured form (e.g., diagnosis, medication, laboratory test) and unstructured form (e.g., image scanning, free clinic notes) and are utilized through machine learning and deep learning algorithms to predict diseases from patient clinical status [59]. These records are utilized to monitor the patient’s health status, which results in a large amount of data, including personal and physical information, patient medical histories, radiological images, treatments, medication/drugs, laboratory test results, immunization dates and allergies [12]. The collected data are raw data that need to be data-mined to extract knowledge through both machine learning and deep learning to save time and lives [12]. Deep EHR learning applications can be divided into five categories: information extraction, representation learning, outcome prediction, computational phenotyping, and clinical data deidentification [60]. Among these, ensemble deep learning methods have gained significant attention for their ability to integrate multiple models, leading to enhanced predictive performance across diverse tasks. In particular, stacking is considered the most effective ensemble deep learning approach for EHR applications, as it seamlessly combines models designed for both structured and unstructured data while effectively reducing bias and variance. Murugadoss et al. [61] presented an ensemble method for automated deidentification of unstructured HERs and clinical notes. These unstructured HERs and notes often contain personally identifiable information, including names, dates of birth, phone numbers, or residential addresses of patients, which restricts their utilization in research development. The authors employed an ensemble architecture, integrating attention-based deep-learning models and rule-based methods, supported by heuristics for detecting PII in EHR data. The proposed method was tested on the i2b2 2014 and Mayo Clinic datasets and achieved a recall of 0.992% and precision of 0.979% on the former and a recall of 0.994% and precision of 0.967% on the latter. Christopoulou et al. [62] proposed an ensemble approach for relation extraction and classification between drugs and medication-related entities in EHRs. The proposed method utilized both weighted bidirectional long short-term memory (BiLSTM) networks and Walk-based models for intra- and intersentence relation extraction in EHRs and combined them using an ensemble method based on majority voting. Wang et al. [63] aimed to utilize imbalanced EHRs to predict acute kidney injury (AKI) through a fast, simple and less costly binary classification model based on an ensemble learning algorithm called the Ensemble Time Series Model (ETSM). Luo et al. [64] proposed a deep learning-based ensemble approach to automatically identify heart disease risk factors, including smoking, obesity, and diabetes, from EHRs. The proposed method utilizes bidirectional encoder representations from transformers (BERT) to extract high-level semantic information from EHRs and automates risk factor identification, which is then fed to conditional random fields (CRFs) to identify all possible risk factor indicators. The trained BERT-CRF models use majority voting. Zhang et al. [65] introduced PheME, a deep ensemble framework using multimodal data from structured EHRs and unstructured clinical notes, which is used for robust and accurate phenotype prediction. First, they employed multiple deep learning algorithms, including multilayer perceptron (MLP) and Blue-BERT algorithms, to learn reliable representations from sparse structured EHR data and clinical notes. A multimodal model then aligns multimodal features onto the same latent space to predict phenotypes. Second, the authors employed two ensemble methods, majority voting and a label model strategy, to improve phenotype prediction. Although the application of ensemble deep learning in EHRs has greatly advanced, especially in prescreening undiagnosed individuals who are more likely to have a given disease based on their available demographic, clinical and lifestyle factors, the limitations of ensemble deep learning in this field still exist [66,67].
Genomics is a branch of molecular biology concerned with mapping the structure, evolution, and function of genomes (e.g., DNA sequencing and RNA measurements) [59]. The implementation of genomics in health care can be categorized into four broad groups: infectious disease, rare disease, cancer, and common or chronic disease. These diseases can be identified in clinical use, and there is a considerable overlap in human biology. Each human has unique biological DNA, which consists of between 20,000 and 25,000 genes, where each gene is composed of between a few hundred and 2 million DNA bases. The use of genomic sequencing of pathogens for diagnosing infectious disease is rapidly increasing, especially in health care systems and medical applications. One of these applications is pharmacogenomics, which allows specialists/doctors to prescribe a specific medication and corresponding dosage based on the patient’s genetic biomarkers. Another application is clustered regularly interspaced short palindromic repeats (CRISPR), which allows for efficient gene modification in a variety of organisms and makes it possible to potentially treat chronic diseases such as cancers, HIV, β-thalassemia, and sickle cell anemia [68–70]. With the rapid development of many new ensemble deep learning approaches in several innovation and discovery domains, such as bioinformatics and genome data analysis [71], ensemble deep learning has improved performance over traditional models, increased interpretability and provided additional understanding of the structure of biological data. Boosting techniques improve feature importance estimation and predictive performance in gene expression analysis, aiding in the identification of regulatory genes. Bagging reduces variance in high-throughput sequencing data, enhancing the classification of gene functions and the prediction of regulatory elements. Stacking integrates models like CNNs, RNNs, and support vector machines through meta-models, proving effective in gene-essentiality predictions for greater accuracy. Ali Shah et al. [72] proposed an ensemble-based deep learning model, called the bidirectional encoder-decoder long short-term memory (BEDLM-CMS) model, to detect mutations in cutaneous melanoma by integrating long short-term memory (LSTM), bidirectional long short-term memory (BLSTM) and gated recurrent unit (GRU) architectures utilizing 75 types of genes. The proposed method was tested using a genomic dataset containing 2608 human samples and 6778 mutations in total along with 75 different types, outperforming other methods with an accuracy of 98%. Albaradei et al. [73] developed an ensemble of deep convolutional neural networks, called Splice2Deep models, for improving splice site (ss) prediction in genomic DNA. The authors evaluated the performance of the Splice2Deep model on five different organisms: Homo sapiens, Oryza sativa japonica, Arabidopsis thaliana, Drosophila melanogaster, and Caenorhabditis elegans. Their approach reduced the average error rates by 41.97% and 28.51% for acceptor and donor SS, respectively. Singh et al. [74] built an ensemble of two-dimensional deep neural networks and transfer learning called SPOT-RNA to predict RNA secondary structures, including noncanonical and nonnested base pairs stabilized by tertiary interactions. The proposed method was trained using an ensemble of ResNets and LSTM on the bpRNA dataset with automated annotation of the secondary structure. Le et al. [75] implemented ensemble deep learning based on a combination of five machine learning and deep learning classifiers (kNN, RF, SVM, MLP, and CNN) to identify essential genes using sequence information. The authors achieved 76.3% accuracy while identifying essential genes. The proposed method was tested on a generalized dataset by Chen et al. and achieved a sensitivity of 60.2%, a specificity of 84.6%, an accuracy of 76.3%, an MCC of 0.449, and an AUC of 0.814. Yu et al. [76] proposed SnapCCESS, an ensemble clustering framework that uses VAE and the snapshot ensemble learning technique to generate a multiview of multimodality-integrated embeddings for clustering multimodal single-cell omics data using an unsupervised ensemble deep learning framework. The proposed method outperforms other state-of-the-art multimodal embedding generation methods in integrating data modalities for clustering cells.
4.4 Disease Prediction and Prevention
Recently, global public health crises have increasingly occurred, and early prediction and prevention of such diseases can reduce the burden on the health care system [77]. Disease diagnosis refers to the process of identifying a specific disease that is associated with a person’s symptoms. Therefore, ensemble deep learning can help in anticipating a wide range of diseases and expedite early treatment based on prior training data. Due to the vast amount of data available, numerous studies have utilized ensemble deep learning approaches for disease prediction and prevention. Ensemble deep learning methods mitigate overfitting, improve generalization, and provide more reliable predictions across various medical applications like pneumonia, diabetic retinopathy, and cardiovascular diseases. These approaches incorporate advanced preprocessing techniques, transfer learning, and feature extraction to achieve state-of-the-art results in clinical decision support systems, often outperforming individual models. These studies help in early diagnosis and timely treatment to lower disease-related mortality rates. Diabetes, skin cancer, kidney disease, Alzheimer’s disease, COVID-19, and heart disease are the most common diseases that can significantly affect patient’s health [13]. Almulihi et al. [78] proposed an ensemble learning method based on a hybrid deep learning model for early heart disease prediction. Their model is based on two hybrid models with heterogeneous architectures, CNN-LSTM and CNN-GRU, which were optimized using the stacking ensemble method. The proposed model outperformed the other ML methods on two benchmark heart disease datasets. Su et al. [77] proposed an innovative deep learning model, the Whale Optimization Convolutional Neural Network (CNN), Long Short Term Memory (LSTM) and Artificial Neural Network (ANN), called the WOCLSA, which incorporates three models, ANN, CNN and LSTM. The WOCLSA model utilizes the whale optimization algorithm to optimize the neuron number, dropout and batch size parameters in the integrated ANN, CNN and LSTM models. Their model is used to predict various public health diseases and provides aid for medical disease diagnosis and prediction. Park et al. [79] aimed to build a new optimized ensemble model by blending a DNN (deep neural network) model with two ML models for disease prediction using laboratory test results. They selected 86 attributes and collected sample datasets on 5145 cases, including 326,686 laboratory samples, to investigate 39 specific diseases. Their model achieved an accuracy of 92% for the five most common diseases using different feature-selection methods. Alsekait et al. [80] developed a novel stacking ensemble deep learning method based on LSTM, CNNs, and GRUs to detect chronic kidney disease (CKD). They used multiple methods of feature selection, including mutual information, chi-squared, RFE, and tree-based (RF) methods. Their model was tested using 400 patient records from the UCI machine learning repository and achieved an accuracy of 99.69%. An et al. [81] presented stacking ensemble deep learning for Alzheimer’s disease classification. The authors utilized two sparse autoencoders for feature leaning at the voting layer and a nonlinear feature-weighted method at the stacking layers. They used a neural network as a meta classifier, which achieved great results. The authors in [82] introduced a novel deep learning model named Deep Ensemble of Adaptive Architectures, designed to address critical challenges in healthcare during the COVID-19 pandemic. The proposed method adopts a twofold approach: firstly, it employs an ensemble deep learning technique to detect COVID-19 patients; secondly, a soft robot is utilized to perform basic assessment tests on the identified patients. This method outperformed baseline approaches, including Faster-RCNN, R-FCN variants, and CNN+LSTM, in both precision and recall with an impressive accuracy of 98.32%. Zeng et al. [83] proposed efficient and accurate ear disease identification using an ensemble deep learning model. They utilized a transfer learning model based on DensNet-BC169 and DensNet-BC1615 with two ensemble classifiers and achieved an accuracy of 95.59%. Tian et al. [84] proposed ViTCNX, a deep ensemble learning model for detecting COVID-19 using lung CT images based on two advanced architectures: Vision Transformer (ViT) and ConvNeXt. The Vision Transformer was applied for robust feature extraction using self-attention mechanisms. Their method achieved outstanding results, with an accuracy of 98.21%, a recall of 99.07%, and an F1 score of 98.55%. Additionally, the authors in [85] explored ensemble deep learning techniques by combining pre-trained convolutional neural network (CNN) models with Vision Transformers (ViT) and XGBoost for carcinoma detection. They developed two ensemble models: the first model used Vision Transformers to capture long-range spatial relationships in medical images, while the second model integrated CNNs with XGBoost to enhance structured data classification. The proposed approach achieved a remarkable 98.95% accuracy on the modified CHKHC-22 dataset, demonstrating its effectiveness in carcinoma detection.
Drug discovery is the process of discovering, developing, and testing a new candidate medication through a combination of computational and experimental methods to identify therapeutically active molecules [86]. Artificial Intelligence -based approaches are increasingly being utilized in all phases of drug discovery and development as both AI technology advances and the size of drug big data expands [87]. AI has been used in various drug discovery applications, including the prediction of drug–protein interactions and the discovery of drug efficacy, ensuring the safety of biomarkers [88]. Due to the increase in the collection of pharmacological data, deep ensemble learning is expected to accelerate new drug development and innovative new drugs. It integrates diverse features, mitigates overfitting, and excels in handling high-dimensional data such as molecular fingerprints and protein structures, leading to better performance in tasks like drug-target interactions and high-throughput drug screening. Vo et al. [89]proposed a novel ensemble deep neural network model to improve the accuracy of predicting drug–drug interactions (DDI). Their proposed model can predict interactions between 86 types of drugs with an average accuracy of 93.80%. Their constructed model is based on the DDN, RF and XGBoost models, where the final output of the proposed model was the stacking result of the individual outputs. Syahid et al. [90] proposed a stacking ensemble learning framework for the accurate prediction of B-rapidly accelerated fibrosarcoma (BRAF) inhibitors. The authors utilized three machine learning algorithms, namely, extreme gradient boosting (XGB), support vector regression (SVR), and multilayer perceptron (MLP), to construct new predictive features (PFs). The first layer of the StackBRAF model receives outputs from 36 PFs and is constructed by combining 12 molecular fingerprints trained using XGB, SVR, and a deep neural network. The final layer of StackBRAF used a random forest (RF) regressor that takes the 36 PFs as input. The StackBRAF model has been proven to be a drug design algorithm for BRAF inhibitor drug discovery and drug development. Although AI technology, such as deep ensemble learning, is utilized to accelerate the development of new and innovative drugs, several types of drugs and their proteins have different structures, which makes it difficult to predict effective drug-protein combinations with high performance [91].
Deep ensemble learning techniques have surpassed other learning techniques in healthcare application tools and provide state-of-the-art solutions. In general, the history of clinical decision-making by physicians and specialists is utilized to train deep ensemble learning algorithms to develop efficient and accurate healthcare applications. Most of the published works on ensemble deep learning in healthcare applications have been intensively reviewed in this article. In this section, healthcare applications based on deep ensemble learning consist of four significant subsections for building and deploying a reliable system: benchmark datasets, data preprocessing, model training, and performance metrics.
In this subsection, we summarize the datasets most commonly used in health care applications for training deep ensemble learning models. There are several attributes that encourage researchers to utilize benchmark datasets, including sequential, image, and statistical attributes. Table 4 reviews the most utilized benchmark datasets for health care applications.
Data preprocessing is the most critical phase and plays a large role in developing robust and effective ML applications. Real data cannot be directly utilized to build and train healthcare models due to redundant, noisy, incomplete, inconsistent, and undesired data. Thus, data preprocessing is applied to enhance the data representation and generalization performance. The data preprocessing steps include missing data, noise removal, normalization, and feature selection [98] as follows.
5.2.1 Missing Data and Removing the Noise Step
Missing data is a common issue in numerical data and can affect the training process. It is extremely important to address this issue by inserting values via one of the imputation methods, such as the KNN imputation method, or deleting the entire entity [99]. Noise data are considered a negative feature that can affect model performance, especially in image processing, and they are usually removed or reduced by applying a nonlinear filter [100] or synthetic region area.
During the data preparation, the dataset may have high ranging values and independent data, which cause larger values to be weighted more and smaller values to be weighted less by the machine learning approach. Thus, scaling features to a specific range, known as normalization or standardization, is a vital step in data preprocessing and can be applied through min–max normalization, maximum absolute scaling, standardization, or robust scaling [101].
• Min–max normalization is a common linear method that transforms features between 0 and 1.
• Maximum absolute scaling: This method divides each feature by its maximum value in the dataset, which ranges between +1 and −1.
• Standardization: This is a widely used method that rescales features to a mean of 0 and a standard deviation of 1.
• Robust scaling: This method is robust to outliers by removing the median and scaling the features using the interquartile range (IQR).
Most datasets have many features for multiple purposes; not all features are relevant and helpful for a specific task, and computing many features can cause overfitting. The process of selecting appropriate features, known as feature selection, is crucial before constructing a model to eliminate irrelevant data and reduce the data dimensionality. Feature selection is helpful for avoiding noise and improving model performance [11,95,98] used different methods, such as filter methods, embedded methods, or wrapper methods.
• Data splitting: The dataset can be split into three parts: training, validation, and test data.
• Types of networks: Several networks can be utilized in health care tasks, such DNN, CNN, RNN, LSTM, GRU, GAN, and Transformers.
• Weights: The initial weight is essential for accelerating convergence and enhancing model performance via one of the following methods: random initialization, Xavier initialization, or He initialization.
• Optimizers: An optimizer is used to minimize the loss function and update weight parameters until model convergence using SGD, Adam, AdaMax, RMSprop, and Nadam.
• Loss function: This function is used to evaluate how well deep learning algorithms model a dataset. Several loss functions can be utilized for regression and classification tasks, such as the mean square error (MSE), mean absolute error (MAE), binary cross-entropy loss, categorical cross-entropy loss, and hinge loss.
• Learning rate: This is a hyperparameter used to control the size of the steps taken during the optimization.
• The number of epochs is the number of iterations needed to train the entire dataset to update the hyperparameters. Selecting an optimal number of epochs can help in generalized learning and increase performance, whereas a large number of epochs can lead to overfitting problems.
• Batch size: The entire dataset used to train the model in one epoch is called batch gradient descent, while the dataset is divided into subsets called minibatches, and a single training sample is utilized to update the parameters called stochastic batches.
• Pooling technique: This technique is used to downsample feature maps in CNNs via max pooling, average pooling, global pooling, or stochastic pooling.
• The activation function is a function that determines whether the output of the neuron should be activated, such as sigmoid, Tanh, ReLU, or softmax.
Several performance metrics are used to assess the effectiveness and robustness of the abovementioned deep ensemble learning models on unseen test data for regression and classification challenges. The most commonly used performance metrics are accuracy, precision, recall, F1 score, and MCC. All of the matrices are derived from the confusion matrix and its derived matrix, as shown in Table 5. The confusion matrices consist of four matrices: first, the true positive (TP) matrix, where the prediction and the actual outputs are both positive. Second, for a false-positive (FP), the prediction is positive, but the actual output is negative. True negative (TN) represents the number of negative cases classified correctly [102]. Finally, there is a false-negative (FN) where there is a negative prediction, while the actual result is positive.

The following is the mathematical definition of the abovementioned metrics:
Accuracy is the most common evaluation metric and is the percentage of correct classifications. It is calculated as in Eq. (8):
Precision measures the model’s ability to recognize the positive samples to a total number of classified positive samples. It is calculated as in Eq. (9):
Recall measures the model’s ability to recognize the positive samples to the actual number (correct or incorrect) of “positive” cases. It is calculated as in Eq. (10):
The F1 score is the weighted average of the precision and recall. Its value ranges between [0, 1], and the best value is 1. It is computed as in Eq. (11):
The Matthews correlation coefficient (MCC) is a correlation coefficient that is used for binary classification. It is calculated as in Eq. (12):
6 Discussion and Future Direction
This section reviews the challenges of ensemble deep learning for health care and highlights the solutions. These challenges can be categorized into three subsections as follows: ensemble deep learning model issues, health care issues, and data quality issues. Ensemble deep learning model issues can be divided into three categories: First, we address the choice of model architecture. Employing the correct deep learning model in health care is crucial for performance enhancement; however, incorporating suitable ensemble learning methods with deep learning models makes it difficult to choose a specialized model architecture that can capture hidden, complex features and perform joint predictions [28]. In addition, boosting methods are not feasible for computer vision in health care tasks and are sensitive to outliers. Bagging methods are computationally expensive [13]. Moreover, more hybrid multimodal and AutoML techniques can be utilized to overcome the choice of the best ensemble learning model architecture. Second, there is a computational expense. Ensemble deep learning models require large amounts of data to train large quantities of parameters, which consumes considerable computational resources [28]. Thus, employing advanced hardware resources such as GPTs and TPUs can help handle the computational load more efficiently. Third is interpretability. Ensemble deep learning has achieved great performance in health care classification and regression tasks due to its ability to extract complex, hidden features; however, in many health care issues, it is treated as a black box and cannot explain why such a model works perfectly on a specific task. The lack of model interpretability leads to unreliability of the results and brings risks. However, few studies have utilized explainable AI (XAI) to increase model interpretability and clinician understanding and trust [28,59,103–105]. Data quality issues: Unlike other areas where the data samples are clean and well structured, health care data samples are highly ambiguous, noisy, incomplete, and heterogeneous. Training ensemble deep learning models with massive amounts of such samples is a major challenge and requires many preprocessing steps, such as resampling, replacing missing values, and removing redundant data [59]. First, the small data sizes create challenges. Ensemble deep learning models are known for their exceptional performance with large data samples; however, most health care tasks do not have sufficient confirmed experiments for positive or negative cases, which leads to a small size of suitable data samples for training ensemble deep learning. However, transfer learning and generative processes might be used to overcome these issues [9,22,28,71]. Second, class imbalance occurs when one of the classes has more samples than the other. This causes the ensemble deep learning model to classify new samples into the majority class. Because fewer people are sick than people who are not, the number of samples from positive patients will always be less than that from negative patients; for instance, the number of samples from people without tumors is much greater than that from people with tumors [12,22,28]. Therefore, these issues could be solved through data resampling, in which a generative model is used to generate minority class samples that are close to the majority sample class. Third, because of the lack of annotations, ensemble deep learning models are trained using supervised learning and thus require large amounts of labeled samples [104]. Many health care tasks have limited availability of labeled data due to the amount of experimentally confirmed positive and negative cases, which sometimes require permissions and experts in the field [71]. In the case of large numbers of available samples, both time and experts are needed to annotate the samples to be useful, and automated annotators, which are sometimes controversial, are needed to ensure the reliability of the labeled samples. Finally, for heterogeneity, most samples used for training in health care tasks consist of diverse data types, such as statistical data, image-based data, sequential data [28,71], and synthetic samples [106]. In the future, we believe that ensemble deep learning will be further utilized to create more accurate and efficient solutions for health care. One promising direction for overcoming the limited available data is to utilize deep generative methods such as GANs to generate more samples [59]. In addition, another promising solution for large dependencies in health care is natural language processing models, including advanced models such as large language models (LLMs) and transformers. Reinforcement learning and quantum computing based on deep learning could be utilized for new discoveries and robust solutions in health care systems. In the future, researchers may consider developing both hardware requirements and software technologies to address sophisticated health care issues that may appear in enhancing ensemble deep learning models.
Ensemble deep learning has been utilized in a variety of healthcare applications to provide solutions to computer vision, natural language processing, and sequence issues. Ensemble deep learning enhances the learning generalization and prediction performance due to the utilization of multiple deep learning models. This article provides a comprehensive overview of deep learning algorithms and architectures and ensemble learning techniques in health care systems. Six common healthcare tasks have been reviewed, including medical imaging, electronic health records, genomics, disease prediction and prevention, and drug discovery. In addition, healthcare applications based on deep ensemble learning consist of four significant subsections for building and deploying a reliable system: benchmark datasets, data preprocessing, model training, and performance metrics. This review article discusses three of the most challenging issues in healthcare systems: ensemble deep learning model issues, healthcare issues, and data quality issues. It includes the high computational costs associated with training large models, the lack of interpretability of ensemble models, and the difficulties posed by heterogeneous, noisy, and imbalanced healthcare data. Recent advancements such as explainable AI (XAI), transfer learning, and generative adversarial networks (GANs) have begun to address some of these issues, enabling better model interpretability, handling class imbalance, and generating synthetic data for small datasets. In the future, researchers may explore and implement advanced technologies such as reinforcement learning, quantum computing, and large language models (LLMs) to provide more robust and effective solutions to complex healthcare issues and address computational, interpretability, and data quality issues.
Acknowledgement: The authors extend their appreciation to Taif University, Saudi Arabia, for supporting this work through project No. (TU-DSPP-2024-263).
Funding Statement: This research was funded by Taif University, Saudi Arabia, project No. (TU-DSPP-2024-263).
Availability of Data and Materials: Not applicable.
Ethics Approval: Not applicable.
Conflicts of Interest: The author declares no conflicts of interest to report regarding the present study.
References
1. Nisar DEM, Amin R, Shah NUH, Al Ghamdi MA, Almotiri SH, Alruily M. Healthcare techniques through deep learning: issues, challenges and opportunities. IEEE Access. 2021;9:98523–41. doi:10.1109/ACCESS.2021.3095312. [Google Scholar] [CrossRef]
2. Alzubaidi L, Zhang J, Humaidi AJ, Al-Dujaili A, Duan Y, Al-Shamma O, et al. Review of deep learning: concepts, CNN architectures, challenges, applications, future directions. J Big Data. 2021;8(1):53. doi:10.1186/s40537-021-00444-8. [Google Scholar] [PubMed] [CrossRef]
3. Deng L. A tutorial survey of architectures, algorithms, and applications for deep learning. APSIPA Trans Signal Inf Process. 2014;3(1). doi:10.1017/atsip.2013.9. [Google Scholar] [CrossRef]
4. Shrestha A, Mahmood A. Review of deep learning algorithms and architectures. IEEE Access. 2019;7:53040–65. doi:10.1109/ACCESS.2019.2912200. [Google Scholar] [CrossRef]
5. Pintelas P, Livieris IE. Special issue on ensemble learning and applications. Algorithms. 2020;13(6):140. doi:10.3390/a13060140. [Google Scholar] [CrossRef]
6. Khai Tran T, Thi Phan T. Deep learning application to ensemble learning—the simple, but effective, approach to sentiment classifying. Appl Sci. 2019;9(13):2760. doi:10.3390/app9132760. [Google Scholar] [CrossRef]
7. Abdel-Jaber H, Devassy D, Al Salam A, Hidaytallah L, EL-Amir M. A review of deep learning algorithms and their applications in healthcare. Algorithms. 2022;15(2):71. doi:10.3390/a15020071. [Google Scholar] [CrossRef]
8. Nguyen DK, Lan CH, Chan CL. Deep ensemble learning approaches in healthcare to enhance the prediction and diagnosing performance: the workflows, deployments, and surveys on the statistical, image-based, and sequential datasets. Int J Environ Res Public Health. 2021;18(20):10811. doi:10.3390/ijerph182010811. [Google Scholar] [PubMed] [CrossRef]
9. Esteva A, Robicquet A, Ramsundar B, Kuleshov V, DePristo M, Chou K, et al. A guide to deep learning in healthcare. Nat Med. 2019;25(1):24–9. doi:10.1038/s41591-018-0316-z. [Google Scholar] [PubMed] [CrossRef]
10. Ihnaini B, Khan MA, Khan TA, Abbas S, Daoud MS, Ahmad M, et al. A smart healthcare recommendation system for multidisciplinary diabetes patients with data fusion based on deep ensemble learning. Comput Intell Neurosci. 2021;2021:4243700. doi:10.1155/2021/4243700. [Google Scholar] [PubMed] [CrossRef]
11. Rashid J, Batool S, Kim J, Wasif Nisar M, Hussain A, Juneja S, et al. An augmented artificial intelligence approach for chronic diseases prediction. Front Public Health. 2022;10:860396. doi:10.3389/fpubh.2022.860396. [Google Scholar] [PubMed] [CrossRef]
12. Abdullah AA, Hassan MM, Mustafa YT. A review on Bayesian deep learning in healthcare: applications and challenges. IEEE Access. 2022;10:36538–62. doi:10.1109/ACCESS.2022.3163384. [Google Scholar] [CrossRef]
13. Mahajan P, Uddin S, Hajati F, Moni MA. Ensemble learning for disease prediction: a review. Healthcare. 2023;11(12):1808. doi:10.3390/healthcare11121808. [Google Scholar] [PubMed] [CrossRef]
14. Rokach L. Taxonomy for characterizing ensemble methods in classification tasks: a review and annotated bibliography. Comput Stat Data Anal. 2009;53(12):4046–72. doi:10.1016/j.csda.2009.07.017. [Google Scholar] [CrossRef]
15. Hansen LK, Salamon P. Neural network ensembles. IEEE Trans Pattern Anal Mach Intell. 1990;12(10):993–1001. doi:10.1109/34.58871. [Google Scholar] [CrossRef]
16. Schapire RE. The strength of weak learnability. Mach Learn. 1990;5(2):197–227. doi:10.1007/BF00116037. [Google Scholar] [CrossRef]
17. Mohammed A, Kora R. A comprehensive review on ensemble deep learning: opportunities and challenges. J King Saud Univ Comput Inf Sci. 2023;35(2):757–74. doi:10.1016/j.jksuci.2023.01.014. [Google Scholar] [CrossRef]
18. Breiman L. Bagging predictors. Mach Learn. 1996;24(2):123–40. doi:10.1007/BF00058655. [Google Scholar] [CrossRef]
19. Mungoli N. Adaptive ensemble learning: boosting model performance through intelligent feature fusion in deep neural networks. 2023. doi:10.48550/arXiv.2304.02653. [Google Scholar] [CrossRef]
20. Mienye ID, Sun Y. A survey of ensemble learning: concepts, algorithms, applications, and prospects. IEEE Access. 2022;10:99129–49. doi:10.1109/ACCESS.2022.3207287. [Google Scholar] [CrossRef]
21. Hwangbo L, Kang YJ, Kwon H, Lee JI, Cho HJ, Ko JK, et al. Stacking ensemble learning model to predict 6-month mortality in ischemic stroke patients. Sci Rep. 2022;12(1):17389. doi:10.1038/s41598-022-22323-9. [Google Scholar] [PubMed] [CrossRef]
22. Yu Z, Wang K, Wan Z, Xie S, Lv Z. Popular deep learning algorithms for disease prediction: a review. Clust Comput. 2023;26(2):1231–51. doi:10.1007/s10586-022-03707-y. [Google Scholar] [PubMed] [CrossRef]
23. Guo Y, Liu Y, Oerlemans A, Lao S, Wu S, Lew MS. Deep learning for visual understanding: a review. Neurocomputing. 2016;187:27–48. doi:10.1016/j.neucom.2015.09.116. [Google Scholar] [CrossRef]
24. Shamshirband S, Fathi M, Dehzangi A, Chronopoulos AT, Alinejad-Rokny H. A review on deep learning approaches in healthcare systems: taxonomies, challenges, and open issues. J Biomed Inform. 2021;113:103627. doi:10.1016/j.jbi.2020.103627. [Google Scholar] [PubMed] [CrossRef]
25. Tandel GS, Tiwari A, Kakde OG, Gupta N, Saba L, Suri JS. Role of ensemble deep learning for brain tumor classification in multiple magnetic resonance imaging sequence data. Diagnostics. 2023;13(3):481. doi:10.3390/diagnostics13030481. [Google Scholar] [PubMed] [CrossRef]
26. Wang X, Zhao Y, Pourpanah F. Recent advances in deep learning. Int J Mach Learn Cybern. 2020;11:747–50. doi:10.1007/s13042-020-01096-5. [Google Scholar] [CrossRef]
27. Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, et al. Generative adversarial nets. Adv Neural Inf Process Syst. 2014;27. [Google Scholar]
28. Cao Y, Geddes TA, Yang JYH, Yang P. Ensemble deep learning in bioinformatics. Nat Mach Intell. 2020;2(9):500–8. doi:10.1038/s42256-020-0217-y. [Google Scholar] [CrossRef]
29. Hong S, Zhou Y, Shang J, Xiao C, Sun J. Opportunities and challenges of deep learning methods for electrocardiogram data: a systematic review. Comput Biol Med. 2020;122:103801. doi:10.1016/j.compbiomed.2020.103801. [Google Scholar] [PubMed] [CrossRef]
30. Qayyum A, Qadir J, Bilal M, Al-Fuqaha A. Secure and robust machine learning for healthcare: a survey. IEEE Rev Biomed Eng. 2021;14:156–80. doi:10.1109/RBME.2020.3013489. [Google Scholar] [PubMed] [CrossRef]
31. Yang Y, Hu Y, Zhang X, Wang S. Two-stage selective ensemble of CNN via deep tree training for medical image classification. IEEE Trans Cybern. 2022;52(9):9194–207. doi:10.1109/TCYB.2021.3061147. [Google Scholar] [PubMed] [CrossRef]
32. Ahn E, Kumar A, Feng D, Fulham M, Kim J. Unsupervised feature learning with K-means and an ensemble of deep convolutional neural networks for medical image classification. 2019. doi:10.48550/arXiv.1906.03359. [Google Scholar] [CrossRef]
33. Gunasekaran H, Ramalakshmi K, Swaminathan DK, Mazzara M. GIT-Net: an ensemble deep learning-based GI tract classification of endoscopic images. Bioengineering. 2023;10(7):809. doi:10.3390/bioengineering10070809. [Google Scholar] [PubMed] [CrossRef]
34. Tajammal T, Khurshid SK, Jaleel A, Qayyum Wahla S, Ziar RA. Deep learning-based ensembling technique to classify Alzheimer’s disease stages using functional MRI. J Healthc Eng. 2023;2023:6961346. doi:10.1155/2023/6961346. [Google Scholar] [PubMed] [CrossRef]
35. Pacheco AGC, Trappenberg T, Krohling RA. Learning dynamic weights for an ensemble of deep models applied to medical imaging classification. In: 2020 International Joint Conference on Neural Networks (IJCNN); July 19–24, 2020; Glasgow, UK: IEEE; 2020. p. 1–8. doi:10.1109/ijcnn48605.2020.9206685. [Google Scholar] [CrossRef]
36. Zhou T, Lu H, Yang Z, Qiu S, Huo B, Dong Y. The ensemble deep learning model for novel COVID-19 on CT images. Appl Soft Comput. 2021;98:106885. doi:10.1016/j.asoc.2020.106885. [Google Scholar] [PubMed] [CrossRef]
37. Deshmukh PK. Improving medical image classification using ensemble learning and deep convolutional neural networks. Int J Intell Syst Appl Eng. 2024;12(4s):106–21. [Google Scholar]
38. Tang S, Wang C, Nie J, Kumar N, Zhang Y, Xiong Z, et al. EDL-COVID: ensemble deep learning for COVID-19 case detection from chest X-ray images. IEEE Trans Ind Inf. 2021;17(9):6539–49. doi:10.1109/TII.2021.3057683. [Google Scholar] [PubMed] [CrossRef]
39. Alsubai S, Khan HU, Alqahtani A, Sha M, Abbas S, Mohammad UG. Ensemble deep learning for brain tumor detection. Front Comput Neurosci. 2022;16:1005617. doi:10.3389/fncom.2022.1005617. [Google Scholar] [PubMed] [CrossRef]
40. Ravi V, Acharya V, Alazab M. A multichannel EfficientNet deep learning-based stacking ensemble approach for lung disease detection using chest X-ray images. Clust Comput. 2023;26(2):1181–203. doi:10.1007/s10586-022-03664-6. [Google Scholar] [PubMed] [CrossRef]
41. Goyal M, Oakley A, Bansal P, Dancey D, Yap MH. Skin lesion segmentation in dermoscopic images with ensemble deep learning methods. IEEE Access. 2019;8:4171–81. doi:10.1109/ACCESS.2019.2960504. [Google Scholar] [CrossRef]
42. Ji J, Zhang W, Dong Y, Lin R, Geng Y, Hong L. Automated cervical cell segmentation using deep ensemble learning. BMC Med Imag. 2023;23(1):137. doi:10.1186/s12880-023-01096-1. [Google Scholar] [PubMed] [CrossRef]
43. Li Z, Zhu Q, Zhang L, Yang X, Li Z, Fu J. A deep learning-based self-adapting ensemble method for segmentation in gynecological brachytherapy. Radiat Oncol. 2022;17(1):152. doi:10.1186/s13014-022-02121-3. [Google Scholar] [PubMed] [CrossRef]
44. Park J, Kweon J, Kim YI, Back I, Chae J, Roh JH, et al. Selective ensemble methods for deep learning segmentation of major vessels in invasive coronary angiography. Med Phys. 2023;50(12):7822–39. doi:10.1002/mp.16554. [Google Scholar] [PubMed] [CrossRef]
45. Rahimpour M, Saint Martin MJ, Frouin F, Akl P, Orlhac F, Koole M, et al. Visual ensemble selection of deep convolutional neural networks for 3D segmentation of breast tumors on dynamic contrast enhanced MRI. Eur Radiol. 2023;33(2):959–69. doi:10.1007/s00330-022-09113-7. [Google Scholar] [PubMed] [CrossRef]
46. Rajaraman S, Antani SK. Modality-specific deep learning model ensembles toward improving TB detection in chest radiographs. IEEE Access. 2020;8:27318–26. doi:10.1109/ACCESS.2020.2971257. [Google Scholar] [PubMed] [CrossRef]
47. Terzi R. An ensemble of deep learning object detection models for anatomical and pathological regions in brain MRI. Diagnostics. 2023;13(8):1494. doi:10.3390/diagnostics13081494. [Google Scholar] [PubMed] [CrossRef]
48. El Asnaoui K. Design ensemble deep learning model for pneumonia disease classification. Int J Multimed Inf Retr. 2021;10(1):55–68. doi:10.1007/s13735-021-00204-7. [Google Scholar] [PubMed] [CrossRef]
49. Alsuhibany SA, Abdel-Khalek S, Algarni A, Fayomi A, Gupta D, Kumar V, et al. Ensemble of deep learning based clinical decision support system for chronic kidney disease diagnosis in medical Internet of Things environment. Comput Intell Neurosci. 2021;2021:4931450. doi:10.1155/2021/4931450. [Google Scholar] [PubMed] [CrossRef]
50. Yoon T, Kang D. Multi-modal stacking ensemble for the diagnosis of cardiovascular diseases. J Pers Med. 2023;13(2):373. doi:10.3390/jpm13020373. [Google Scholar] [PubMed] [CrossRef]
51. Abbas S, Sampedro GA, Alsubai S, Almadhor A, Kim TH. An efficient stacked ensemble model for heart disease detection and classification. Comput Mater Contin. 2023;77(1):665–80. doi:10.32604/cmc.2023.041031. [Google Scholar] [CrossRef]
52. Arun Prakash J, Asswin CR, Ravi V, Sowmya V, Soman KP. Pediatric pneumonia diagnosis using stacked ensemble learning on multi-model deep CNN architectures. Multimed Tools Appl. 2023;82(14):21311–51. doi:10.1007/s11042-022-13844-6. [Google Scholar] [PubMed] [CrossRef]
53. Prakash JA, Ravi V, Sowmya V, Soman KP. Stacked ensemble learning based on deep convolutional neural networks for pediatric pneumonia diagnosis using chest X-ray images. Neural Comput Appl. 2023;35(11):8259–79. doi:10.1007/s00521-022-08099-z. [Google Scholar] [PubMed] [CrossRef]
54. Heisler M, Karst S, Lo J, Mammo Z, Yu T, Warner S, et al. Ensemble deep learning for diabetic retinopathy detection using optical coherence tomography angiography. Trans Vis Sci Tech. 2020;9(2):20. doi:10.1167/tvst.9.2.20. [Google Scholar] [PubMed] [CrossRef]
55. Mondal SS, Mandal N, Singh KK, Singh A, Izonin I. EDLDR: an ensemble deep learning technique for detection and classification of diabetic retinopathy. Diagnostics. 2023;13(1):124. doi:10.3390/diagnostics13010124. [Google Scholar] [PubMed] [CrossRef]
56. Olabanjo O, Wusu A, Mazzara M. Deep unsupervised machine learning for early diabetes risk prediction using ensemble feature selection and deep belief neural networks. 2023. doi:10.20944/preprints202301.0208.v1. [Google Scholar] [CrossRef]
57. Geetha G, Mohana Prasad K. Stacking ensemble learning-based convolutional gated recurrent neural network for diabetes Miletus. Intell Autom Soft Comput. 2023;36(1):703–18. doi:10.32604/iasc.2023.032530. [Google Scholar] [CrossRef]
58. Kausar N, Hameed A, Sattar M, Ashraf R, Imran AS, Abidin MZU, et al. Multiclass skin cancer classification using ensemble of fine-tuned deep learning models. Appl Sci. 2021;11(22):10593. doi:10.3390/app112210593. [Google Scholar] [CrossRef]
59. Miotto R, Wang F, Wang S, Jiang X, Dudley JT. Deep learning for healthcare: review, opportunities and challenges. Brief Bioinform. 2018;19(6):1236–46. doi:10.1093/bib/bbx044. [Google Scholar] [PubMed] [CrossRef]
60. Xie F, Yuan H, Ning Y, Ong MEH, Feng M, Hsu W, et al. Deep learning for temporal data representation in electronic health records: a systematic review of challenges and methodologies. J Biomed Inform. 2022;126:103980. doi:10.1016/j.jbi.2021.103980. [Google Scholar] [PubMed] [CrossRef]
61. Murugadoss K, Rajasekharan A, Malin B, Agarwal V, Bade S, Anderson JR, et al. Building a best-in-class automated de-identification tool for electronic health records through ensemble learning. Patterns. 2021;2(6):100255. doi:10.1016/j.patter.2021.100255. [Google Scholar] [PubMed] [CrossRef]
62. Christopoulou F, Tran TT, Sahu SK, Miwa M, Ananiadou S. Adverse drug events and medication relation extraction in electronic health records with ensemble deep learning methods. J Am Med Inform Assoc. 2020;27(1):39–46. doi:10.1093/jamia/ocz101. [Google Scholar] [PubMed] [CrossRef]
63. Wang Y, Wei Y, Yang H, Li J, Zhou Y, Wu Q. Utilizing imbalanced electronic health records to predict acute kidney injury by ensemble learning and time series model. BMC Med Inform Decis Mak. 2020;20(1):238. doi:10.1186/s12911-020-01245-4. [Google Scholar] [PubMed] [CrossRef]
64. Luo L, Wang Y, Mo DY. Identifying heart disease risk factors from electronic health records using an ensemble of deep learning method. IISE Trans Healthc Syst Eng. 2023;13(3):237–47. doi:10.1080/24725579.2023.2205665. [Google Scholar] [CrossRef]
65. Zhang S, Li H, Tang R, Ding S, Rasmy L, Zhi D, et al. PheME: a deep ensemble framework for improving phenotype prediction from multi-modal data. In: 2023 IEEE 11th International Conference on Healthcare Informatics (ICHI); June 26–29, 2023; Houston, TX, USA: IEEE; 2023. p. 268–75. doi:10.1109/ICHI57859.2023.00044. [Google Scholar] [CrossRef]
66. Stevens CA, Lyons AR, Dharmayat KI, Mahani A, Ray KK, Vallejo-Vaz AJ, et al. Ensemble machine learning methods in screening electronic health records: a scoping review. Digit Health. 2023;9:20552076231173225. doi:10.1177/20552076231173225. [Google Scholar] [PubMed] [CrossRef]
67. Xu J, Xi X, Chen J, Sheng VS, Ma J, Cui Z. A survey of deep learning for electronic health records. Appl Sci. 2022;12(22):11709. doi:10.3390/app122211709. [Google Scholar] [CrossRef]
68. Abdelhalim H, Berber A, Lodi M, Jain R, Nair A, Pappu A, et al. Artificial intelligence, healthcare, clinical genomics, and pharmacogenomics approaches in precision medicine. Front Genet. 2022;13:929736. doi:10.3389/fgene.2022.929736. [Google Scholar] [PubMed] [CrossRef]
69. Birney E, Vamathevan J, Goodhand P. Genomics in healthcare: GA4GH looks to 2022. BioRxiv. 2017:203554. [Google Scholar]
70. Zou J, Huss M, Abid A, Mohammadi P, Torkamani A, Telenti A. A primer on deep learning in genomics. Nat Genet. 2019;51(1):12–8. doi:10.1038/s41588-018-0295-5. [Google Scholar] [PubMed] [CrossRef]
71. Choi SR, Lee M. Transformer architecture and attention mechanisms in genome data analysis: a comprehensive review. Biology. 2023;12(7):1033. doi:10.3390/biology12071033. [Google Scholar] [PubMed] [CrossRef]
72. Ali Shah A, Ali Shaker AS, Jabbar S, Abbas Q, Al-Balawi TS, Celebi ME. An ensemble-based deep learning model for detection of mutation causing cutaneous melanoma. Sci Rep. 2023;13:22251. doi:10.1038/s41598-023-49075-4. [Google Scholar] [PubMed] [CrossRef]
73. Albaradei S, Magana-Mora A, Thafar M, Uludag M, Bajic VB, Gojobori T, et al. Splice2Deep: an ensemble of deep convolutional neural networks for improved splice site prediction in genomic DNA. Gene. 2020;763S:100035. doi:10.1016/j.gene.2020.100035. [Google Scholar] [CrossRef]
74. Singh J, Hanson J, Paliwal K, Zhou Y. RNA secondary structure prediction using an ensemble of two-dimensional deep neural networks and transfer learning. Nat Commun. 2019;10:5407. doi:10.1038/s41467-019-13395-9. [Google Scholar] [PubMed] [CrossRef]
75. Le NQK, Do DT, Hung TNK, Lam LHT, Huynh TT, Nguyen NTK. A computational framework based on ensemble deep neural networks for essential genes identification. Int J Mol Sci. 2020;21(23):E9070. doi:10.3390/ijms21239070. [Google Scholar] [PubMed] [CrossRef]
76. Yu L, Liu C, Yang JYH, Yang P. Ensemble deep learning of embeddings for clustering multimodal single-cell omics data. Bioinformatics. 2023;39(6):btad382. doi:10.1093/bioinformatics/btad382. [Google Scholar] [PubMed] [CrossRef]
77. Su X, Sun Y, Liu H, Lang Q, Zhang Y, Zhang J, et al. An innovative ensemble model based on deep learning for predicting COVID-19 infection. Sci Rep. 2023;13(1):12322. doi:10.1038/s41598-023-39408-8. [Google Scholar] [PubMed] [CrossRef]
78. Almulihi A, Saleh H, Hussien AM, Mostafa S, El-Sappagh S, Alnowaiser K, et al. Ensemble learning based on hybrid deep learning model for heart disease early prediction. Diagnostics. 2022;12(12):3215. doi:10.3390/diagnostics12123215. [Google Scholar] [PubMed] [CrossRef]
79. Park DJ, Park MW, Lee H, Kim YJ, Kim Y, Park YH. Development of machine learning model for diagnostic disease prediction based on laboratory tests. Sci Rep. 2021;11:7567. doi:10.1038/s41598-021-87171-5. [Google Scholar] [PubMed] [CrossRef]
80. Alsekait DM, Saleh H, Gabralla LA, Alnowaiser K, El-Sappagh S, Sahal R, et al. Toward comprehensive chronic kidney disease prediction based on ensemble deep learning models. Appl Sci. 2023;13(6):3937. doi:10.3390/app13063937. [Google Scholar] [CrossRef]
81. An N, Ding H, Yang J, Au R, Ang TFA. Deep ensemble learning for Alzheimer’s disease classification. J Biomed Inform. 2020;105:103411. doi:10.1016/j.jbi.2020.103411. [Google Scholar] [PubMed] [CrossRef]
82. Iqbal MS, Naqvi RA, Alizadehsani R, Hussain S, Moqurrab SA, Lee SW. An adaptive ensemble deep learning framework for reliable detection of pandemic patients. Comput Biol Med. 2024;168:107836. doi:10.1016/j.compbiomed.2023.107836. [Google Scholar] [PubMed] [CrossRef]
83. Zeng X, Jiang Z, Luo W, Li H, Li H, Li G, et al. Efficient and accurate identification of ear diseases using an ensemble deep learning model. Sci Rep. 2021;11:10839. doi:10.1038/s41598-021-90345-w. [Google Scholar] [PubMed] [CrossRef]
84. Tian G, Wang Z, Wang C, Chen J, Liu G, Xu H, et al. A deep ensemble learning-based automated detection of COVID-19 using lung CT images and vision transformer and ConvNeXt. Front Microbiol. 2022;13:1024104. doi:10.3389/fmicb.2022.1024104. [Google Scholar] [PubMed] [CrossRef]
85. Raju ASN, Venkatesh K, Padmaja B, Kumar CNS, Patnala PRM, Lasisi A, et al. Exploring vision transformers and XGBoost as deep learning ensembles for transforming carcinoma recognition. Sci Rep. 2024;14:30052. doi:10.1038/s41598-024-81456-1. [Google Scholar] [PubMed] [CrossRef]
86. Nag S, Baidya ATK, Mandal A, Mathew AT, Das B, Devi B, et al. Deep learning tools for advancing drug discovery and development. 3 Biotech. 2022;12(5):110. doi:10.1007/s13205-022-03165-8. [Google Scholar] [PubMed] [CrossRef]
87. Askr H, Elgeldawi E, Aboul Ella H, Elshaier YAMM, Gomaa MM, Hassanien AE. Deep learning in drug discovery: an integrative review and future challenges. Artif Intell Rev. 2023;56(7):5975–6037. doi:10.1007/s10462-022-10306-1. [Google Scholar] [PubMed] [CrossRef]
88. Dara S, Dhamercherla S, Jadav SS, Babu CM, Ahsan MJ. Machine learning in drug discovery: a review. Artif Intell Rev. 2022;55(3):1947–99. doi:10.1007/s10462-021-10058-4. [Google Scholar] [PubMed] [CrossRef]
89. Vo TH, Nguyen NTK, Le NQK. Improved prediction of drug-drug interactions using ensemble deep neural networks. Med Drug Discov. 2023;17:100149. doi:10.1016/j.medidd.2022.100149. [Google Scholar] [CrossRef]
90. Syahid NF, Weerapreeyakul N, Srisongkram T. StackBRAF: a large-scale stacking ensemble learning for BRAF affinity prediction. ACS Omega. 2023;8(23):20881–91. doi:10.1021/acsomega.3c01641. [Google Scholar] [PubMed] [CrossRef]
91. Matsuzaka Y, Uesawa Y. Ensemble learning, deep learning-based and molecular descriptor-based quantitative structure-activity relationships. Molecules. 2023;28(5):2410. doi:10.3390/molecules28052410. [Google Scholar] [PubMed] [CrossRef]
92. Müller D, Soto-Rey I, Kramer F. An analysis on ensemble learning optimized medical image classification with deep convolutional neural networks. IEEE Access. 2022;10:66467–80. doi:10.1109/ACCESS.2022.3182399. [Google Scholar] [CrossRef]
93. Suk HI, Lee SW, Shen D. Alzheimer’s disease neuroimaging initiative. Deep ensemble learning of sparse regression models for brain disease diagnosis. Med Image Anal. 2017;37:101–13. doi:10.1016/j.media.2017.01.008. [Google Scholar] [PubMed] [CrossRef]
94. Yang L, Wang SH, Zhang YD. EDNC: ensemble deep neural network for COVID-19 recognition. Tomography. 2022;8(2):869–90. doi:10.3390/tomography8020071. [Google Scholar] [PubMed] [CrossRef]
95. Si Y, Du J, Li Z, Jiang X, Miller T, Wang F, et al. Deep representation learning of patient data from Electronic Health Records (EHRa systematic review. J Biomed Inform. 2021;115:103671. doi:10.1016/j.jbi.2020.103671. [Google Scholar] [PubMed] [CrossRef]
96. Pi SW, Lee BD, Lee MS, Lee HJ. Ensemble deep-learning networks for automated osteoarthritis grading in knee X-ray images. Sci Rep. 2023;13:22887. doi:10.1038/s41598-023-50210-4. [Google Scholar] [PubMed] [CrossRef]
97. Ali Shah A, Malik HAM, Muhammad A, Alourani A, Butt ZA. Deep learning ensemble 2D CNN approach towards the detection of lung cancer. Sci Rep. 2023;13:2987. doi:10.1038/s41598-023-29656-z. [Google Scholar] [PubMed] [CrossRef]
98. Ali F, El-Sappagh S, Riazul Islam SM, Kwak D, Ali A, Imran M, et al. A smart healthcare monitoring system for heart disease prediction based on ensemble deep learning and feature fusion. Inf Fusion. 2020;63:208–22. doi:10.1016/j.inffus.2020.06.008. [Google Scholar] [CrossRef]
99. Abayomi-Alli OO, Damaševičius R, Maskeliūnas R, Misra S. An ensemble learning model for COVID-19 detection from blood test samples. Sensors. 2022;22(6):2224. doi:10.3390/s22062224. [Google Scholar] [PubMed] [CrossRef]
100. Das A. Adaptive UNet-based lung segmentation and ensemble learning with CNN-based deep features for automated COVID-19 diagnosis. Multimed Tools Appl. 2022;81(4):5407–41. doi:10.1007/s11042-021-11787-y. [Google Scholar] [PubMed] [CrossRef]
101. Md AQ, Kulkarni S, Joshua CJ, Vaichole T, Mohan S, Iwendi C. Enhanced preprocessing approach using ensemble machine learning algorithms for detecting liver disease. Biomedicines. 2023;11(2):581. doi:10.3390/biomedicines11020581. [Google Scholar] [PubMed] [CrossRef]
102. Chaturvedi SS, Tembhurne JV, Diwan T. A multi-class skin cancer classification using deep convolutional neural networks. Multimed Tools Appl. 2020;79(39):28477–98. doi:10.1007/s11042-020-09388-2. [Google Scholar] [CrossRef]
103. Zhu T, Li K, Herrero P, Georgiou P. Deep learning for diabetes: a systematic review. IEEE J Biomed Health Inform. 2021;25(7):2744–57. doi:10.1109/JBHI.2020.3040225. [Google Scholar] [PubMed] [CrossRef]
104. Li I, Pan J, Goldwasser J, Verma N, Wong WP, Nuzumlalı MY, et al. Neural natural language processing for unstructured data in electronic health records: a review. Comput Sci Rev. 2022;46:100511. doi:10.1016/j.cosrev.2022.100511. [Google Scholar] [CrossRef]
105. Syarif I, Zaluska E, Prugel-Bennett A, Wills G. Application of bagging, boosting and stacking to intrusion detection. In: Perner P, editor. Machine learning and data mining in pattern recognition. MLDM 2012. Lecture notes in computer science. Berlin/Heidelberg: Springer; 2012. Vol. 7376. [Google Scholar]
106. Ravi D, Wong C, Deligianni F, Berthelot M, Andreu-Perez J, Lo B, et al. Deep learning for health informatics. IEEE J Biomed Health Inform. 2017;21(1):4–21. doi:10.1109/JBHI.2016.2636665. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF


 Downloads
Downloads
 Citation Tools
Citation Tools