 Open Access
Open Access
ARTICLE
Ontology Matching Method Based on Gated Graph Attention Model
Guangxi Key Lab of Human-Machine Interaction and Intelligent Decision, Nanning Normal University, Nanning, 530100, China
* Corresponding Author: Ying Pan. Email:
Computers, Materials & Continua 2025, 82(3), 5307-5324. https://doi.org/10.32604/cmc.2024.060993
Received 14 November 2024; Accepted 11 December 2024; Issue published 06 March 2025
Abstract
With the development of the Semantic Web, the number of ontologies grows exponentially and the semantic relationships between ontologies become more and more complex, understanding the true semantics of specific terms or concepts in an ontology is crucial for the matching task. At present, the main challenges facing ontology matching tasks based on representation learning methods are how to improve the embedding quality of ontology knowledge and how to integrate multiple features of ontology efficiently. Therefore, we propose an Ontology Matching Method Based on the Gated Graph Attention Model (OM-GGAT). Firstly, the semantic knowledge related to concepts in the ontology is encoded into vectors using the OWL2Vec* method, and the relevant path information from the root node to the concept is embedded to understand better the true meaning of the concept itself and the relationship between concepts. Secondly, the ontology is transformed into the corresponding graph structure according to the semantic relation. Then, when extracting the features of the ontology graph nodes, different attention weights are assigned to each adjacent node of the central concept with the help of the attention mechanism idea. Finally, gated networks are designed to further fuse semantic and structural embedding representations efficiently. To verify the effectiveness of the proposed method, comparative experiments on matching tasks were carried out on public datasets. The results show that the OM-GGAT model can effectively improve the efficiency of ontology matching.Keywords
In the development of the Semantic Web [1], the study of ontology has a significant impact on its advancement. Currently, ontology technologies are extensively utilized across various domains, such as biomedicine, finance, engineering, law, and cultural heritage [2]. However, in the field of ontology engineering, different ontologies may use different representations and structures when describing the same concepts [3,4], which can cause a decrease in data sharing and system interoperability, and increase the complexity of knowledge integration and data fusion. This issue is referred to as ontology heterogeneity. To solve these issues, researchers have proposed the ontology matching approach [5], which aims to identify corresponding semantically identical or similar concepts across different ontologies.
In recent years, Representation Learning has become a main approach to solving problems in various domains, with models capable of automatically extracting features or representations of features [6]. Consequently, we focus on the research of ontology matching methods from the perspective of representation learning methods. Existing representation learning methods still have the following three problems in acquiring and fusing semantic and structural features of ontologies: (1) In terms of semantic embedding, existing word embedding methods such as Word2Vec [7] and GloVe [8], primarily used for natural language processing (NLP) tasks, learn the vector space representation of words by analyzing the co-occurrence of vocabulary in textual data. These methods excel in handling natural language text, but they are not directly applicable to Web Ontology Language (OWL) ontologies, as the structure and semantic characteristics of OWL ontologies significantly differ from natural language text. To address this issue, the OWL2Vec* [9] word embedding technique has been proposed, which transforms complex ontology logic and structural information into vector representations that are easily processed by machines, thereby efficiently encoding the semantic information of OWL-formatted ontologies. Its working principle involves converting OWL ontologies into graphs and then performing random walks on these graphs to generate structural documents that serve as the input corpus. Subsequently, it combines the graph structure of the ontology, logical constructors, and lexical information from textual annotations to create a comprehensive corpus. Finally, by training a Word2Vec model on the corpus, it generates embedding representations that capture the semantic information of concepts within the ontology. Although OWL2Vec takes into account the ontology’s vocabulary and logical constructors, it does not fully consider the true meaning of concepts themselves and their hierarchical relationships when encoding the semantic information of the ontology. (2) In terms of structural embedding, the Graph Convolutional Network (GCN) model can capture and utilize the graph structure information of the ontology when extracting the graph node information of the ontology [10]. The basic idea is to update the embedding representation of the current node by aggregating the characteristics of the neighboring node. However, it struggles to model and filter the importance of distant nodes associated with a central node. Furthermore, the GCN model needs to update the whole graph when updating node features, and the fusion is less efficient when there are more neighboring nodes. (3) When integrating ontological semantic and structural embedding representations, the existing direct concatenation strategies [11], while simple, fail to adequately consider the importance of semantic and structural features in the matching task.
To address the above three problems, we propose corresponding solutions from different knowledge perspectives, and the main contributions are as follows:
(1) We propose an Ontology Matching Method Based on a Gated Graph Attention Model (OM-GGAT). Firstly, in terms of semantic embedding, to enrich the contextual semantics of concepts, we employ the OWL2Vec* method to encode the semantic knowledge related to concepts in the ontology into vectors. Additionally, by embedding the path information of concepts (i.e., the complete path from the root node to the concept), we can better understand the true meaning of the concepts themselves and their hierarchical relationships with each other. Secondly, in terms of structural embedding, the ontology is transformed into the corresponding graph structure based on semantic relationships. When extracting features of the ontology graph nodes, we utilize the Graph Attention (GAT) model to assign different attention weights to each neighboring node of the central concept, adaptively aggregating the features of neighboring nodes and capturing key distant node information. Finally, a gated network is designed to effectively integrate semantic and structural embedding representations, achieving more accurate ontology matching.
(2) Unsupervised and semi-supervised ontology matching task comparison experiments on three public datasets demonstrate that OM-GGAT can effectively improve ontology matching efficiency and provide a solution for knowledge fusion. Separate experiments on semantic and structural approaches are also conducted to demonstrate that the OM-GGAT approach is effective in considering both semantic and structural approaches on the ontology matching task.
In ontology, concepts are represented by vectors constructed by feature engineering, and the ontology matching task can be transformed into the vector-based similarity calculation task between different ontologies. For example, Kolyvakis et al. [12] proposed an ontology-matching framework that uses word embedding technology to capture semantic similarity between ontologies. The real situation of semantic similarity of concepts can be distinguished by connecting the context semantics of concepts. Chen et al. [9] proposed an OWL2Vec* method that encodes semantic information in OWL ontologies by combining random walk algorithms with word embedding techniques. Xue et al. [13] proposed an Ontology Meta-Matching technique (OMM) based on deep reinforcement learning. This technology integrates multiple similarity measurement methods to discover heterogeneous entities between different ontologies. Li et al. [14] proposed a knowledge representation learning model TransO based on constraint concept types, relations, and hierarchical information, which can effectively model relations, complete the reasoning of knowledge graphs, and maintain low model complexity. However, ontology not only contains rich semantic features, but also its structural information can reveal the interrelation between concepts. Sentürk et al. [15] proposed a graph-based ontology matching framework, which converts ontology concepts and relationships into graph structures and uses subgraph mining technology to carry out effective ontology matching. These methods only rely on a single feature of the ontology, whether it is a semantic feature or structural feature, which may lead to the loss of knowledge in the matching process.
Existing research points out that it is difficult to accurately judge whether two concepts match each other using only one similarity measurement method, but combining multiple similarity strategies can significantly improve the accuracy of matching. For example, Duan et al. [11] proposed an ontology-matching method based on word embedding and structural similarity. The method mainly distinguishes semantic similarity and description association by improving word vectors. Then the ontology is transformed into a graph and the SimRank algorithm is used to calculate the structural similarity, to realize the one-to-many matching task. In recent years, researchers have begun to use Machine Learning (ML) methods to effectively integrate a variety of similar ontology matching methods. For example, Efeoglu et al. [16] proposed a Graphmatcher ontology matching system based on Graph Representation Learning. It uses Graph Representation Learning methods and Graph Attention Mechanism to compute high-level representations of classes and their related terms in ontologies to identify and align semantically similar entities in different ontologies. Xue et al. [17] proposed an ontology matching method based on an Interactive Compact Genetic Algorithm (ICGA). This method uses a compact coding mechanism and expert interaction mechanism to improve the performance and alignment quality of the algorithm. He et al. [18] proposed an ontology alignment method based on Bidirectional Encoder Representations from Transformers (BERT). In addition, to address the limitations of current suboptimal reference mappings and limited support for evaluation of machine learning-based systems, He et al. [7] proposed the DeepOnto method and an Ontology Pruning method, which could improve the relative integrity of reference mapping.
At present, many ontology matching studies focus on using information such as concept name, ontology structure, and external resources to improve matching tasks [19], however, these methods often do not take into account the deep semantic information of concepts, which leads to low accuracy of matching. In addition, although semantic-based matching methods improve accuracy by extracting semantic information, they still fail to make full use of the intrinsic logical structure of ontology to reveal the potential semantic connections between concepts. Furthermore, future research should investigate more efficient fusion strategies to integrate diverse similarity information, enhancing matching performance.
3 Ontology Matching Method Based on Gated Graph Attention Model
Since we only focus on the matching relationship between concepts in ontology. The ontology [20] is formally defined as the triplet form in Eq. (1).
where
In order to ensure the uniformity of terminology, classes, and property are collectively referred to as concepts. When the structure of an ontology is more complex, the OWL language is commonly used for its description. Therefore, in the experimental section, we will process ontologies in OWL format. Given source ontology
where
Applying the concept of attention mechanisms to ontology matching methods can further enrich the semantic knowledge of the ontology and capture the importance of neighboring nodes between different concepts. Therefore, we propose an ontology matching method based on the Gated Graph Attention Model. The model diagram is shown in Fig. 1.
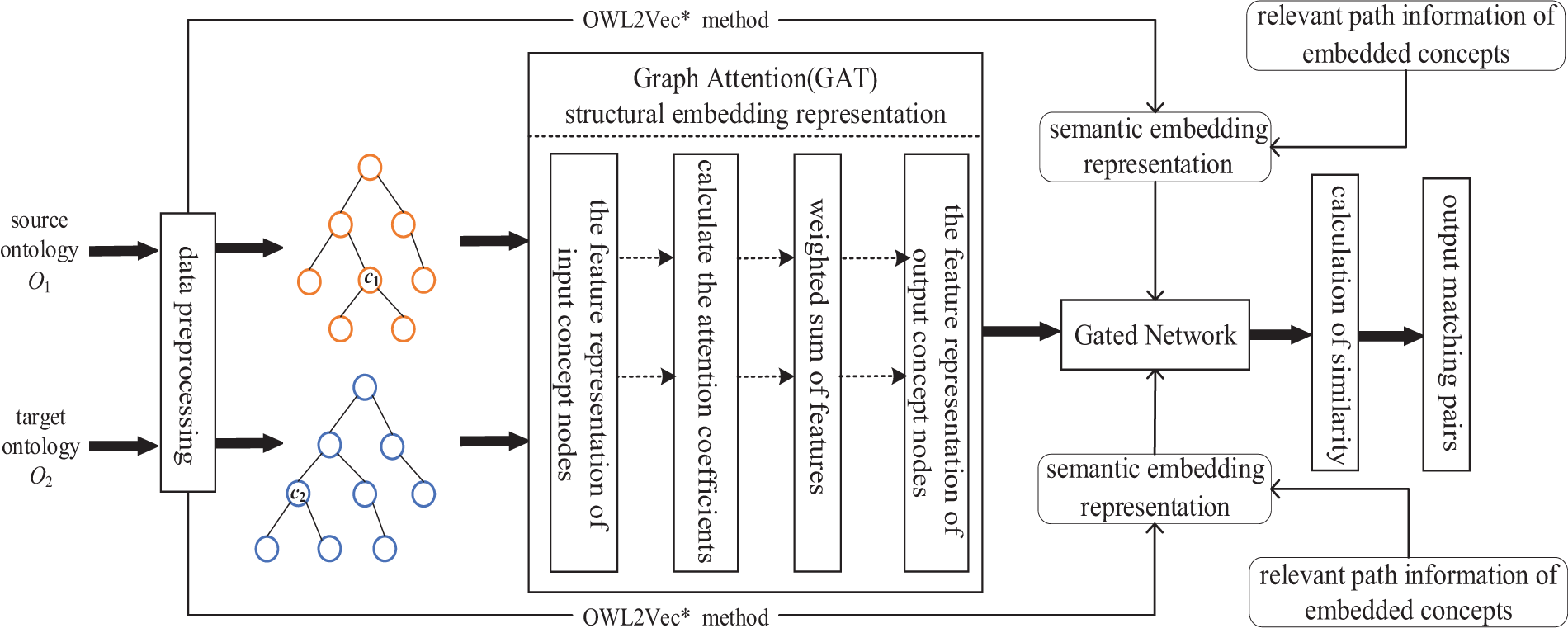
Figure 1: Overview of OM-GGAT
The model mainly includes the following four steps: (1) Semantic Embedding: Using OWL2Vec* method to encode the semantic knowledge of the ontology and embed the related path information of the concept. (2) Structural Embedding: The GAT model is used to extract node features of concepts. (3) Gated Network: Gating networks are introduced to effectively integrate the embedding of semantics and structure. (4) Ontology Matching Process: Calculate the similarity between concepts and output the final matching results in the form of matching pairs.
In order to better understand the true meaning of concepts in an ontology and the relationship between concepts, we embedded path information related to concepts. Specific steps are described as follows.
First, in the OWL ontology, the OWL2Vec* method is used to encode the semantic knowledge related to concepts into vectors. Then, the related path information of the concept is embedded, and the path refers to the sequence of concepts obtained from the “root” node of the ontology to the concept node in turn. For example, for the concept node microcephaly in the DOID ontology, the complete path information from the root node owl: Thing to the concept node microcephaly is “Thing/disease/physical disorder/microcephaly”. Therefore, the path sequence for the concept node microcephaly can be represented as (Thing, disease, physical disorder, microcephaly). The path sequence is formally described as
where
When extracting features of nodes in an ontology graph, compared to other attention mechanisms, GAT has the advantage of implicitly assigning different weights to different nodes in the neighborhood, which helps to reduce the impact of noise propagation on nodes in the graph and enhances the effectiveness of structural embedding vectors [23]. In addition, the attention score mechanism allows us to capture and filter the importance of distant nodes related to the central node, thereby further improving the performance of the model. Therefore, in order to consider the attention weight of neighbor nodes, we introduce the GAT model to learn the feature embedding of concept nodes.
First, according to the semantic relations existing in the ontology, such as subClassOf and subPropertyOf, the adjacent nodes of concepts are identified, and these concepts and their mutual relations are constructed into an undirected graph, denoted as
1. The Feature Representation of Input Concept Nodes
For graph
where
2. Calculate the Attention Coefficients
By combining node features and neighbor features, GAT can more flexibly model the semantic meaning of concepts. For example, when calculating the similarity between two concepts, GAT can focus on neighbor nodes that are semantically closer, thereby generating concept representations more accurately. Therefore, in order to represent the features of the concept node more comprehensively, we introduce the GAT model to extract the graph node features of the ontology, the working principle of which is shown in Fig. 2. Specifically, we transform the concept node vector
where
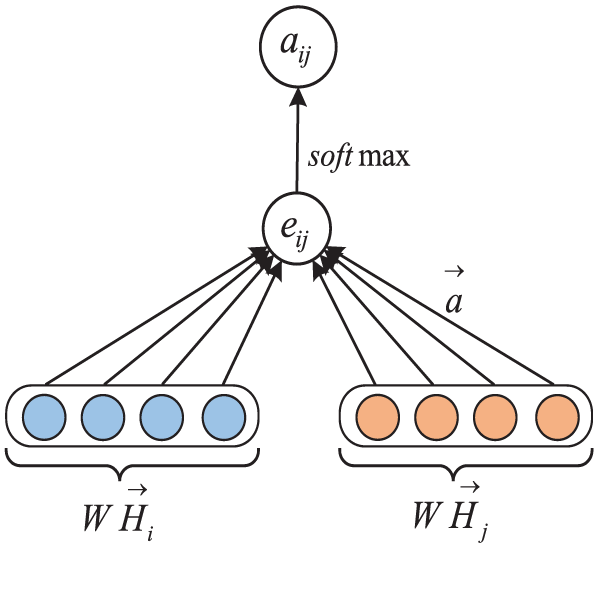
Figure 2: The working principle of GAT
For neighbor nodes, in order to better allocate the weights of different nodes, the above-calculated correlations need to be uniformly normalized. The calculation is shown in Eq. (6).
where
3. Weighted Sum of Features
After obtaining the normalized attention coefficient, it is necessary to calculate the linear combination of its corresponding features and output a new feature vector. The calculation is shown in Eq. (7).
where
4. The Feature Representation of Output Concept Nodes
In addition, to make the results of Self-Attention more stable, the concatenation method of the Multi-Head Attention mechanism is chosen. The calculation is shown in Eq. (8).
where
After the above steps, the updated embedding of the concept node will contain its own feature information and the feature information of its first-order concept neighbor node.
Compared to other fusion methods, gated networks can capture the complex interactive relationships between semantic and structural features, which increases computational complexity with additional linear transformations and non-linear activations, but offers more flexible feature selection and fusion, and the additional overhead is usually acceptable [24]. Therefore, to effectively integrate the semantic and structural embedded representations of ontologies, we introduce a gated network, whose core goal is to dynamically adjust the combination ratio of semantic information and structural information to uncover high-quality matching results. During the training process, the gating mechanism adjusts the gate values through backpropagation based on dynamically weighing the semantic and structural embeddings of the ontology, to adapt to the importance of semantic or structural features for different concept pairs, with its calculation shown as in Eqs. (9) and (10).
where
3.2.5 Ontology Matching Process
After gated network fusion, the final embedding of the concept can be obtained for the matching task. The matching process is mainly divided into the following three steps:
1. Obtain the ontology embedding. The embedding corresponding to each concept is obtained from the embedding of the source ontology
2. Calculate concept similarity. Compared to other typical distance metrics, the Manhattan distance method can accurately capture the differences between vectors, especially when there are variations in vector length or the presence of outliers [25]. This helps to avoid errors that might arise from taking approximate values. Therefore, we adopt the Manhattan distance method to further enhance the accuracy of ontology matching. The Eq. (11) is used to calculate the Manhattan Distance method between the two concepts in the L-vec and R-vec sets. The core idea is to calculate the sum of the absolute value of the coordinate difference between the two concepts on each coordinate axis.
where
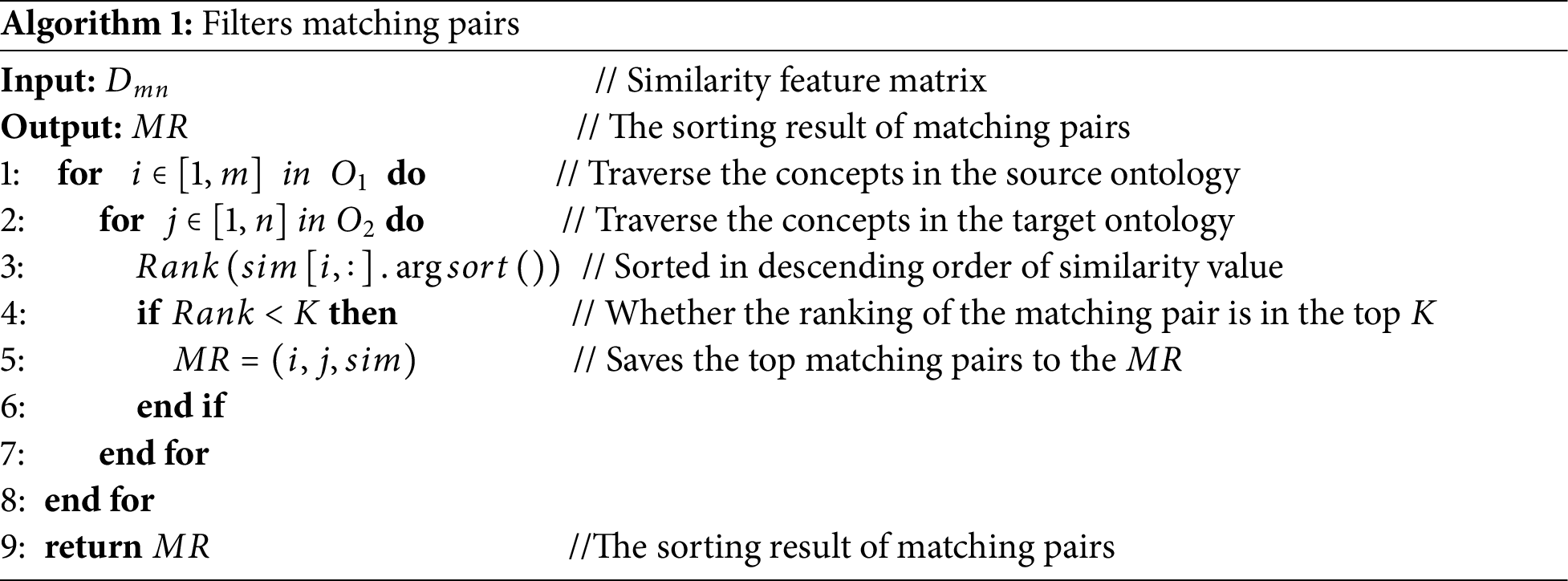
3. Filter matching pairs. Through step 2, a similarity feature matrix with
(1) Take a concept
(2) Traverse the concepts in the target ontology
4 Experimental Results and Analysis
In practical applications, the inclusion relationships between ontologies are often more numerous than equivalence relationships, and inclusion relationships play an important role in the management of ontologies and the integration of knowledge. Therefore, He et al. [7] constructed a Subsumption Matching dataset based on the Equivalence Matching relationship of the ontology. If all inferred assertions are considered at the same time, a large number of inclusion relationships are generated, which is a great challenge for matching tasks. Therefore, we mainly consider the concept of hierarchy.
We utilize the Ontology Alignment Evaluation Initiative (OAEI) 2022 Bio-ML track datasets, which included the Human Disease Ontology (DOID) [26], the Orphanet Rare Diseases Ontology (ORDO) [27], the Online Mendelian Inheritance in Man (OMIM) [28], the Systematized Nomenclature of Medicine-Clinical Terms (SNOMED CT) [29] and the National Cancer Institute Thesaurus (NCIT) [30] as the experimental datasets.
(1) DOID: The comprehensive classification set of human diseases organized by etiology provided by OAEI in 2022, mainly describing the terms, phenotypic characteristics, and related medical vocabulary of human disease concepts. The ontology has a total of more than 46,000 words for diseases and medicine, which can provide consistent terminology for the biomedical community.
(2) ORDO: This ontology aims to provide a structured vocabulary for rare diseases, primarily describing the classification of rare diseases in disease, gene-disease relationships, epidemiology, and associations between other databases (such as Reactome and UniProtKB) or classifications (such as ICD10).
(3) OMIM: A set of public terms describing the complex relationship between human genes and genetic phenotypes, derived primarily from published biomedical literature. The ontology has a structured format and is now widely used by clinicians, molecular biologists, and genome scientists.
(4) SNOMED CT (abbreviated SNOMED): A combined conceptual system based on description logic, which mainly describes the clinical terminology knowledge of diseases, diagnoses, and drugs in the form of concepts. Each concept has a unique concept code, and multiple synonyms can be used to describe the same concept.
(5) NCIT: A cancer thesaurus based on descriptive logic, which mainly describes terms related to clinical care, pharmacology, basic research, and public information. The ontology contains definitions, synonyms, and hierarchical relationships for more than 100,000 concepts, each with a unique identity.
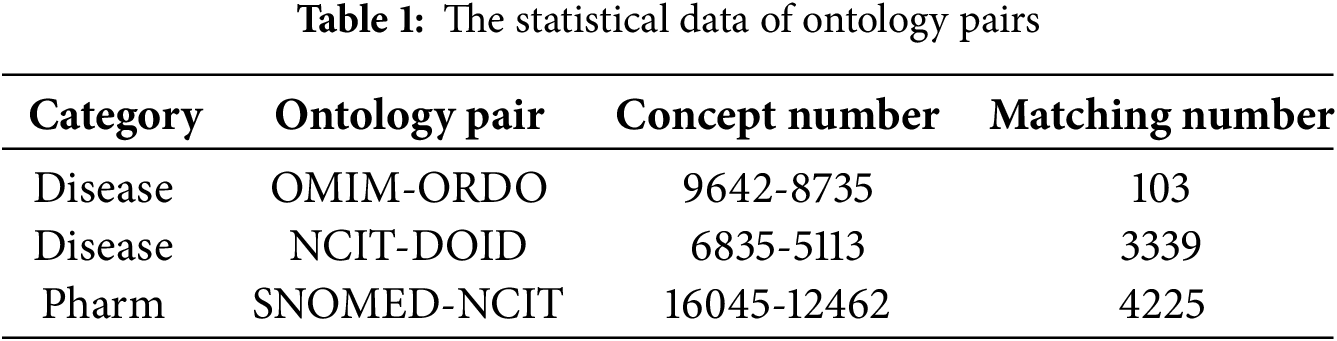
We complete the matching task on OMIM-ORDO, NCIT-DOID, and SNOMED-NCIT three datasets. The statistical data of ontology pairs is shown in Table 1. The OMIM-ORDO dataset contains only 103 matches, indicating a lower semantic similarity between the two ontologies and making the matching task more challenging. In contrast, the NCIT-DOID and SNOMED-NCIT datasets include 3339 and 4225 matches, respectively, suggesting greater conceptual overlap and relatively easier matching tasks. Furthermore, the OMIM-ORDO dataset focuses primarily on genetic terminology, while the NCIT-DOID and SNOMED-NCIT datasets cover a wider range of clinical and cancer-related terms. This discrepancy in vocabulary and semantic relationships demands more robust generalization capabilities from the model.
4.2 Experimental Evaluation Metrics
For the matching tasks in this section, the matching results are inherently incomplete. Therefore, we mainly adopt Mean Reciprocal Rank (MRR) and Hit Ratio (Hits@K) as metrics and the calculation is shown in Eqs. (12) and (13).
where
4.3 Experimental Environment and Parameter Setting
Experimental running environment: the CPU is Intel(R) Xeon(R) Gold 6133 CPU @ 2.5 GHz, the graphics GPU is NVIDIA GeForce RTX 3090, and the memory is 24 GB, written in Python language.
Parameter settings: (1) In the semantic embedding part, the random walk depth is 4, the embedding dimension is 100, the minimum word count is 1, and the training epoch is 100. (2) In the structural embedding part, the embedding dimension is 125, the heads of attention are 2, the learning rate is 0.005, the iterative learning times are 5, the training epoch of each iterative learning is 10, the batch size is 16.
4.4 Experimental Design and Comparison
This section mainly conducts experiments on three datasets: OMIM-ORDO, NCIT-DOID, and SNOMED-NCIT. Since there are relatively few comparison methods for this kind of task, we choose BERTSubs (IC) [31], Word2Vec+Random Forest (RF), and OWL2Vec*+RF [7] methods for comparison.
(1) The Word2Vec+RF method mainly encodes the contents of rdfs: label and uses the Word2Vec model trained by the 2018 Wikipedia English article. This method is simple and straightforward, relying on pre-trained word vector models and having a low computational complexity. However, since Word2Vec can only capture semantic relationships based on word context, it has limitations in expressing complex ontology semantic relationships.
(2) The OWL2Vec*+RF method uses the OWL2Vec* ontology embedding model to encode three corpora, including structure, vocabulary, and combined documents extracted from the ontology. This method fully leverages the characteristics of the ontology, capable of encoding structural and semantic information, making it more suitable for ontology matching tasks than traditional word vector models. Despite focusing on ontology features, this method does not fully model the true semantic meaning of concepts and the hierarchical relationships between them.
(3) The BERTSubs (IC) method mainly encodes the contents of rdfs: label. The architecture of BERTSubs (IC) is the same as that of BERTMap, except that the BERT model is fine-tuned using what is already declared in the ontology. Leveraging BERT’s powerful semantic representation capabilities, this method can better adapt to complex semantic tasks. However, it primarily relies on the semantic representation of class labels and may not fully consider the contextual semantic relationships between concepts.
4.4.1 Comparison and Analysis of Ontology Matching Methods
We utilize the same data partitioning method as the baseline model. For unsupervised matching tasks, the dataset is divided into a test set and a validation set according to the ratio of 9:1. The experimental results are shown in Tables 2–4. For the semi-supervised matching task, the dataset is divided into test, training, and validation sets according to the ratio of 7:2:1. The experimental results are shown in Tables 5–7.
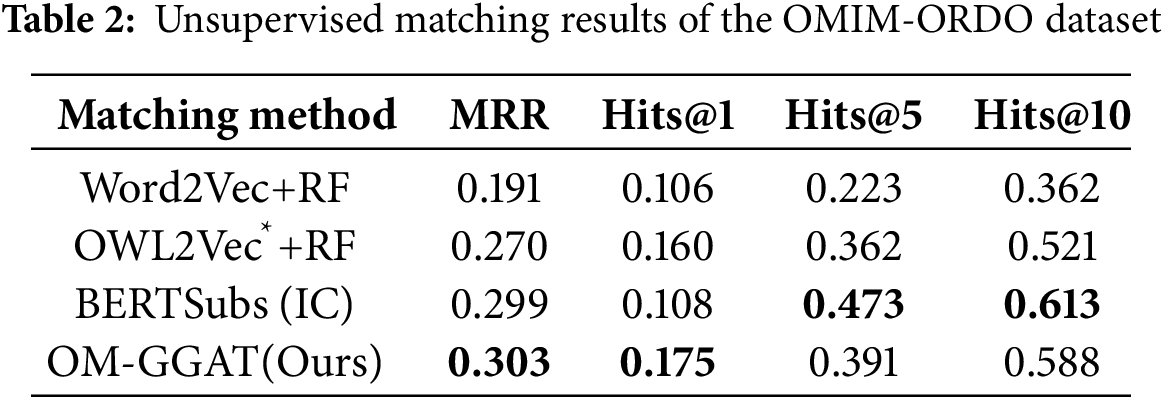
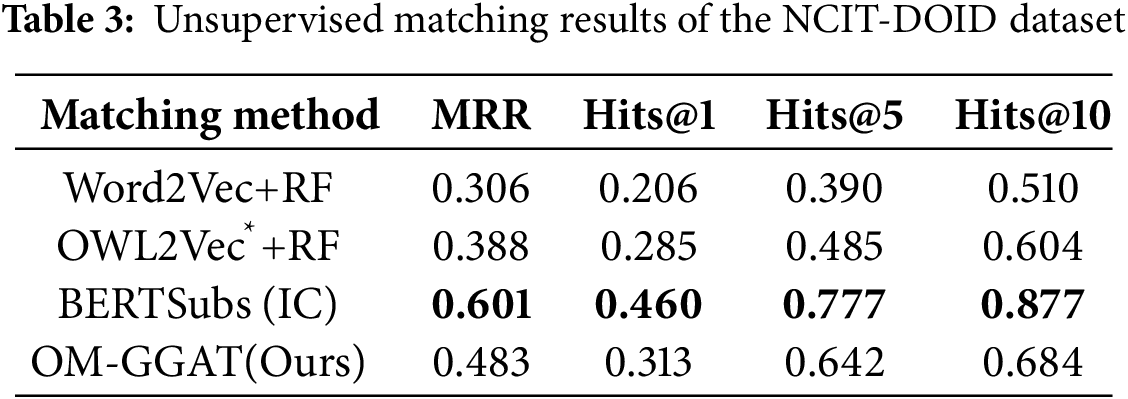
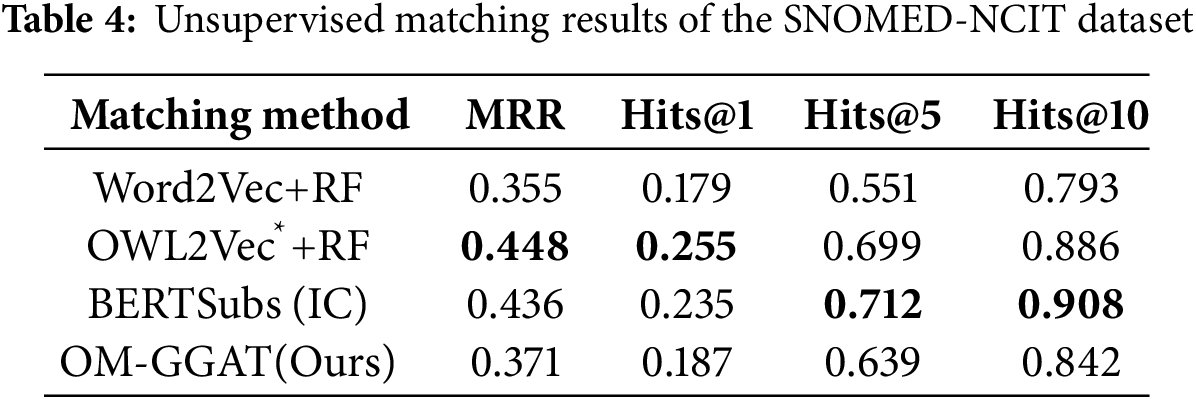
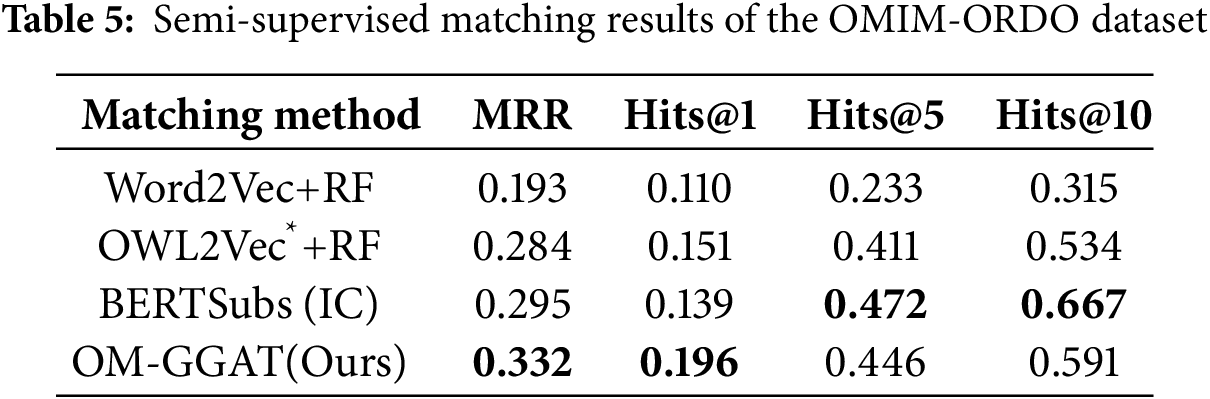
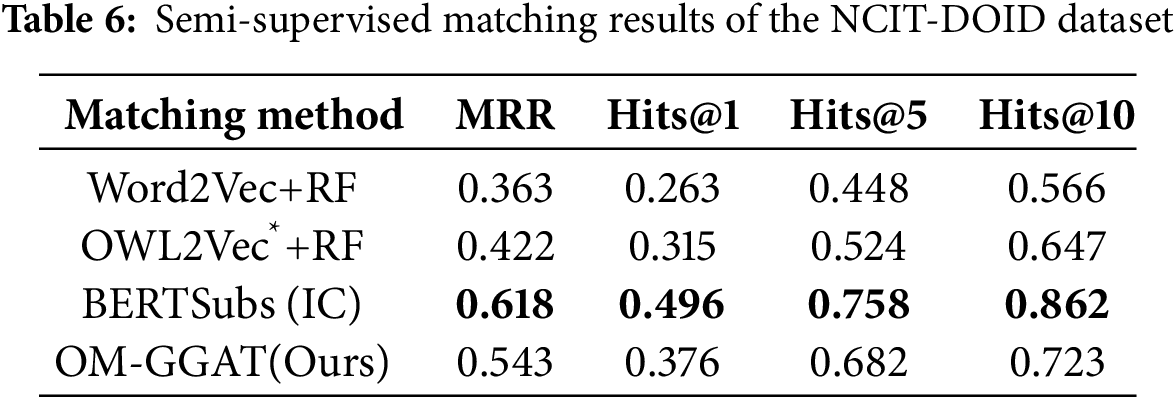
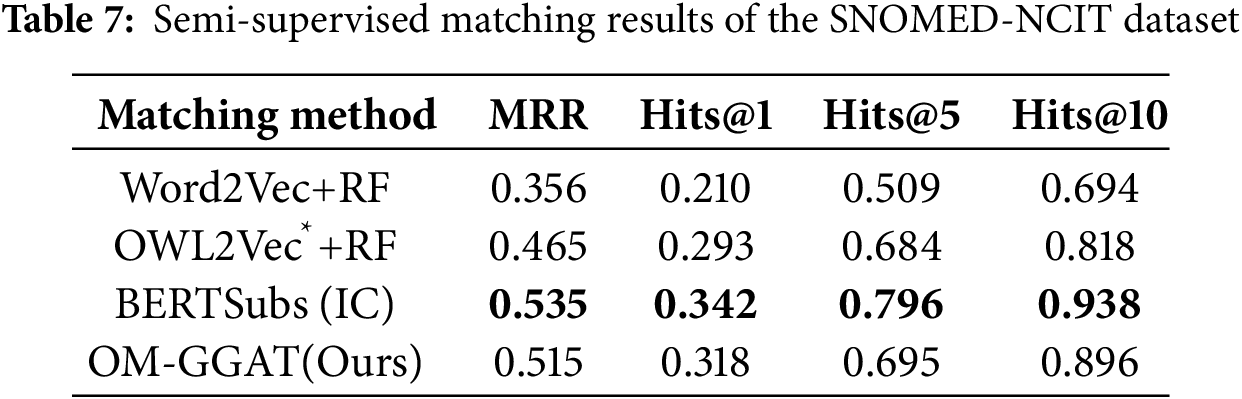
From Tables 2–7, it can be seen that OM-GGAT has certain advantages in two matching tasks on the three datasets. Specifically, the MRR and Hits@1 values of the OMIM-ORDO dataset are ranked first, and the Hits@5 and Hits@10 values are ranked second. Then, the four metrics of the NCIT-DOID dataset are better than Word2Vec+RF and OWL2Vec*+RF methods, and the four metrics of the SNOMED-NCIT dataset are better than Word2Vec+RF methods. In general, OM-GGAT can effectively improve the efficiency of ontology matching and provide solutions for knowledge fusion. The reasons for the experimental results are analyzed as follows:
(1) Analyzing the characteristics of the method. ① Word2Vec and OWL2Vec* methods mainly consider the semantic features of concepts and do not make full use of the concept hierarchy of ontology to find the corresponding relationship between concepts. Therefore, OM-GGAT has great advantages. ② BERTSubs (IC) method uses the BERT model to learn the semantic and structural features of concepts, which has a stronger semantic understanding ability and higher performance than traditional language models. Therefore, the BERTSubs (IC) method achieves better results in the matching tasks of both NNCIT-DOID and SNOMED-NCIT datasets. ③ OM-GGAT effectively exploits implicit semantic relations between concepts by embedding path information related to the concepts. Meanwhile, it leverages the word embedding model to maximize the learning of semantic embedding representations of concepts. Furthermore, OM-GGAT employs the GAT model to learn the graph node information in the ontology, which is able to assign different attention weights to each neighbor node of the concept, and thus adaptively aggregates the structural features of neighboring nodes. Compared to Word2Vec and OWL2Vec* methods, OM-GGAT has the advantage that it is able to utilize both semantic and structural features of the ontology, and its matching results are superior to them.
(2) Analyzing the characteristics of the dataset. ① The maximum depth of the concept in the OMIM ontology is 2, that is, the structure is relatively simple. Therefore, OM-GGAT’s Hits@5 value in the matching result of the OMIM-ORDO dataset in Table 2 is only 0.9% higher than that of the OWL2Vec*+RF method. ② The structure of NCIT and DOID ontology is relatively more complex and complete, and OM-GGAT has seen some performance improvement in the matching task of NCIT-DOID dataset. ③ The concept hierarchy of SNOMED and NCIT ontology can divide semantically similar concepts into the same level, which is helpful for OM-GGAT to learn the structural features of ontology better. This indicates that the OM-GGAT method has certain advantages when the concept hierarchy of the ontology is relatively complete.
4.4.2 Comparison and Analysis of Semantic and Structural Methods
The OWL2Vec* method in the comparison model only uses the corpus of ontology structure, vocabulary, and their combination, and does not use the pre-trained language model. In this paper, the OWL2Vec* method is used to extract the ontology corpus. Besides considering the true semantics of the conceptual context, the word embedding model of Word2Vec is also used to maximize the learning of the corpus. Therefore, this section will conduct ablation experiments on unsupervised and semi-supervised tasks for the semantic embedding method used, and the results are shown in Tables 8 and 9. For the convenience of expression, the original semantic embedding is denoted as
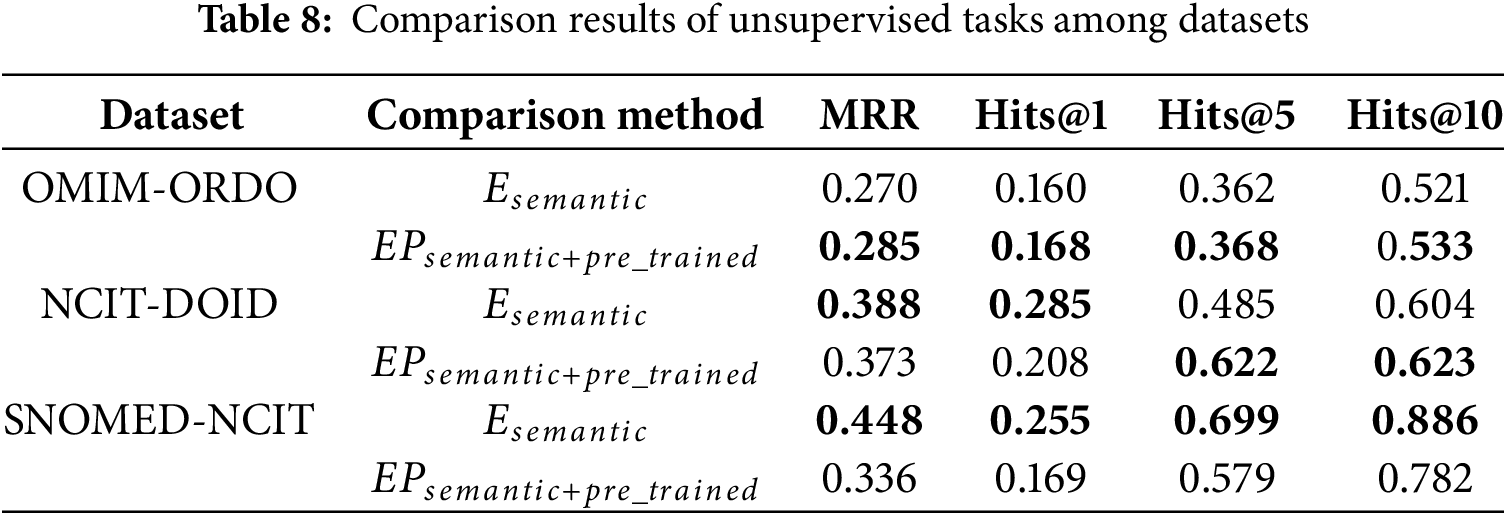
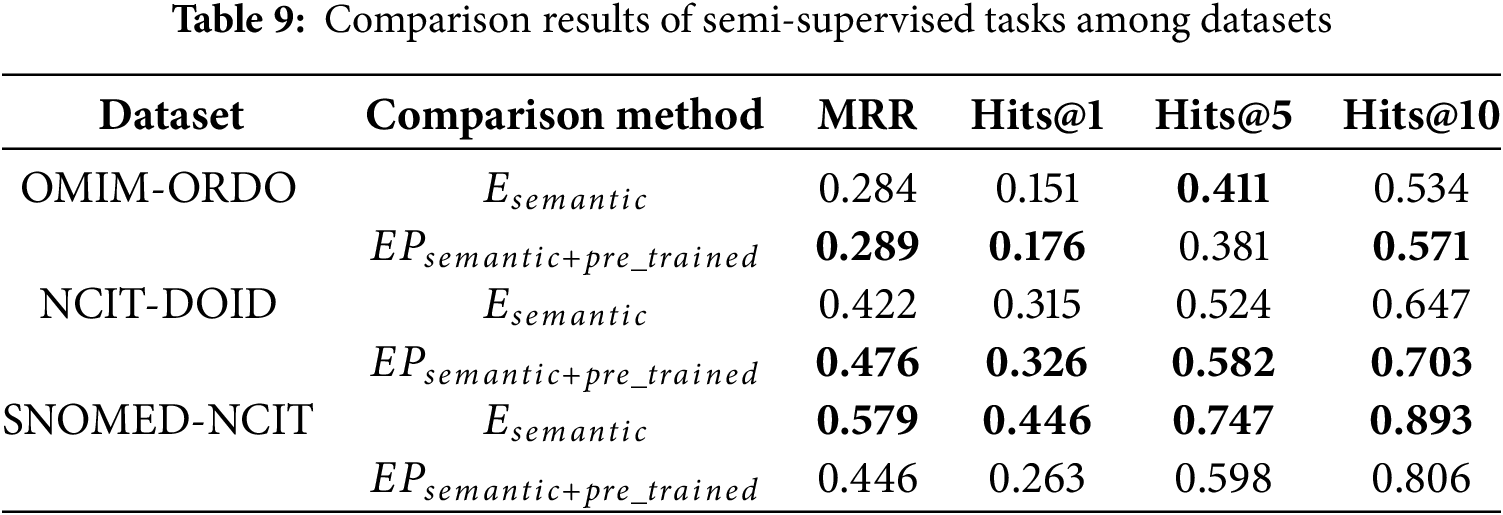
According to the experimental results in Tables 8 and 9, for OMIM-ORDO and NCIT-DOID datasets, embedding related path information of concepts and using pre-trained language models can effectively improve the matching effect. Specifically, from Table 8, the MRR, Hits@1, Hits@5, and Hits@10 values of the OM-GGAT method on the OMIM-ORDO dataset were increased by 1.5%, 0.8%, 0.6% and 1.2%, respectively. The values of Hits@5 and Hits@10 on the NCIT-DOID dataset are increased by 13.7% and 1.9%, respectively. From Table 9, the values of MRR, Hits@1, and Hits@10 on the OMIM-ORDO dataset of the OM-GGAT method are increased by 0.5%, 2.5% and 3.7%, respectively. The values of MRR, Hits@1, Hits@5 and Hits@10 on the NCIT-DOID dataset increased by 5.4%, 1.1%, 5.8% and 5.6%, respectively. The experimental results show that the embedded concept correlation path information and using pre-trained models to maximize the learning of concept embeddings allow the model to better understand the true semantics of concept contexts and the hierarchical relationships between concepts. Therefore, OM-GGAT achieves better results on the OMIM-ORDO and NCIT-DOID datasets. Moreover, according to statistics, the SNOMED dataset contains 173,408 axioms, and the NCIT dataset contains 126,381 axioms, namely, the SNOMED and NCIT ontologies are large and contain a large number of logical constructors. Since the pre-trained language model does not have good logical reasoning ability, the matching results of OM-GGAT on the SNOMED-NCIT dataset do not have a competitive advantage. On the contrary, the reasoner can be used to mine more hidden semantic knowledge of the ontology. In general, OM-GGAT is effective in mining the semantic relationships between concepts.
4.4.3 Comparison and Analysis of GAT Components
OM-GGAT not only considers the implicit semantic information of the ontology and the true semantic of the concept, but also uses the GAT model to extract the graph node information of the ontology, and assigns different attention weights to each neighbor node of the concept. Therefore, we carry out comparative experiments on unsupervised and semi-supervised tasks with Word2Vec and OWL2Vec* matching methods that do not consider ontology graph structural information. The experimental results are shown in Figs. 3–5.
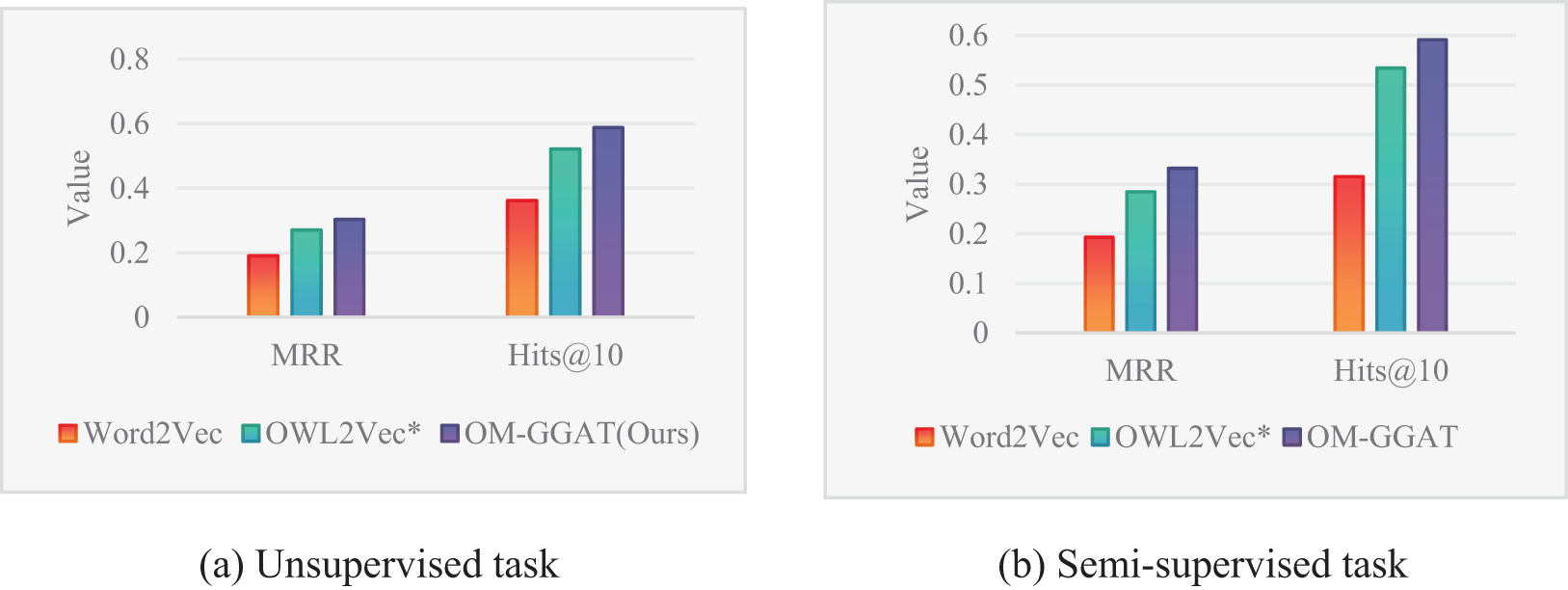
Figure 3: Results of the OMIM-ORDO dataset
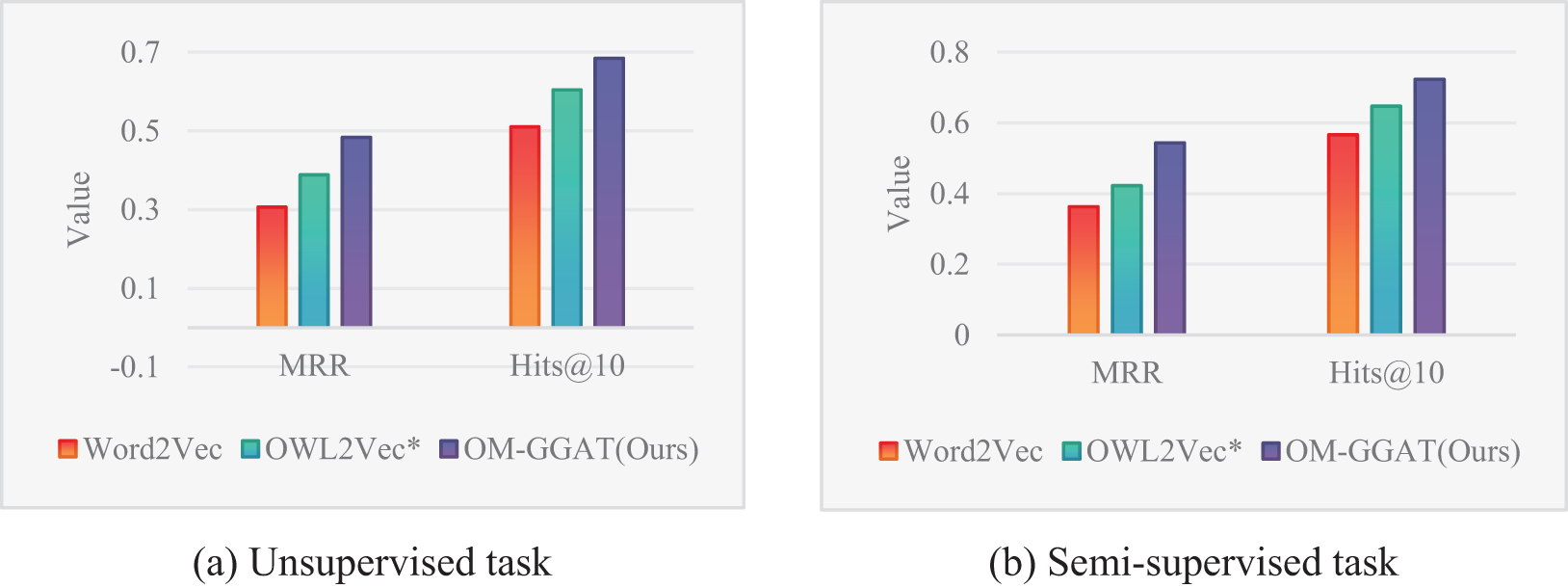
Figure 4: Results of the NCIT-DOID dataset
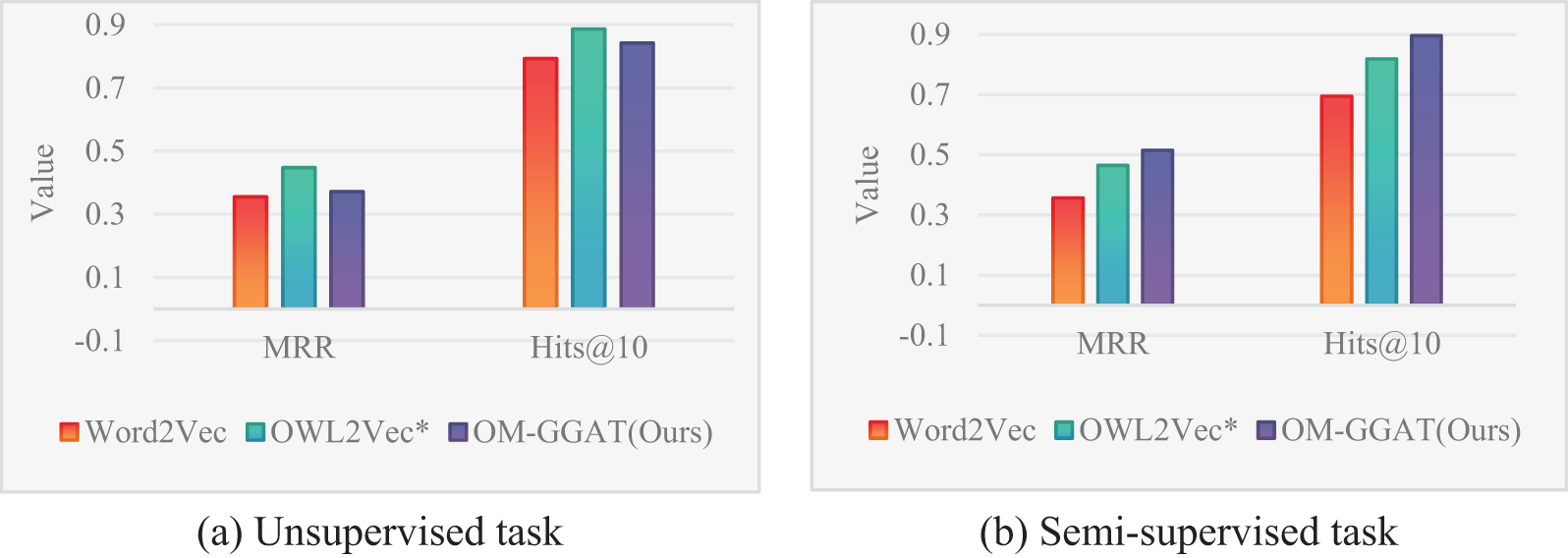
Figure 5: Results of the SNOMED-NCIT dataset
From Figs. 3–5, the MRR value and Hits@10 value of OM-GGAT are higher than those of Word2Vec and OWL2Vec* in OMIM-ORDO and NC-DOID datasets. Specifically, in the matching results of the OMIM-ORDO dataset shown in Fig. 3, the MRR and Hits@10 values of the OM-GGAT method are 3.3% and 6.7% higher than those of the OWL2Vec* method, respectively. Then, compared with Word2Vec, the MRR and Hits@10 values of the OM-GGAT method in the matching results of the NCIT-DOID dataset in Fig. 4 are 15.2% and 33% higher, respectively. These results highlight that OM-GGAT successfully combines semantic information and graph structure features of the ontology, utilizing the GAT model to assign differential weights to neighboring nodes and capture richer semantic relationships between concepts.
As the number of ontologies increases, the inter-ontology semantic relationships become increasingly complex, making it crucial to understand the true semantics of ontology-specific terms and concepts for the matching task. For ontology structural features, how to efficiently extract feature representations of concept nodes is an urgent problem that needs to be solved. Therefore, we propose the OM-GGAT method based on the idea of an attention mechanism. In terms of semantic embedding, the OWL2Vec* method is used to learn the semantic representation of concepts, and the path information of concepts is embedded to enhance the understanding of the meaning of concepts and their interrelationships. In structural embedding, the GAT model is used to adaptively aggregate the features of neighbor nodes and capture the key information of distant nodes, thereby updating and propagating effective features. Furthermore, by designing a gated network, the method effectively integrates semantic and structural embedding representations to achieve more accurate ontology matching.
OM-GGAT can effectively enhance the efficiency of ontology matching, but there is still room for further in-depth research. Currently, our research mainly focuses on the similarity issues between concepts within ontologies, while in reality, there is a substantial amount of instance knowledge within ontologies. By mining the associations between these instances and knowledge such as attributes, we can further deepen the understanding of concept relationships in ontology matching models. Therefore, in future research, we plan to incorporate instance knowledge and leverage domain-specific knowledge to parse synonyms within ontologies, in order to further enhance the quality of ontology embeddings and matching efficiency. In addition, we will continue to explore the scalability of OM-GGAT, especially its performance in handling large-scale ontologies and its application in multilingual ontology matching issues.
Acknowledgement: The authors would like to express their sincere gratitude to the editors and reviewers for their meticulous review and constructive feedback.
Funding Statement: This study is supported by the National Natural Science Foundation of China (grant numbers 62267005 and 42365008); and the Guangxi Collaborative Innovation Center of Multi-Source Information Integration and Intelligent Processing.
Author Contributions: The authors confirm their contribution to the paper as follows: study conception and design: Mei Chen, Nan Wu and Ying Pan; data collection: Nan Wu and Yunsheng Xu; analysis and interpretation of results: Mei Chen, Nan Wu and Yunsheng Xu; draft manuscript preparation: Mei Chen and Nan Wu; draft review and editing: Mei Chen and Yunsheng Xu; funding acquisition: Ying Pan. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets generated and/or analyzed during the current study are available from the OAEI 2022 Bio-ML Track repository (10.5281/zenodo.6946466) (accessed on 10 December 2024).
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Lassila O, Hendler J, Berners-Lee T. The semantic web. Sci Am. 2001;284(5):34–43. doi:10.1038/scientificamerican0501-34. [Google Scholar] [CrossRef]
2. Tudorache T. Ontology engineering: current state, challenges, and future directions. Semant Web. 2020;11(1):125–38. doi:10.3233/SW-190382. [Google Scholar] [CrossRef]
3. Wu N, Lai X, Chen M, Pan Y. Ontology matching and repair based on semantic association and probabilistic logic. IEICE Trans Inf Syst. 2024;E107-D(11):1433–43. doi:10.1587/transinf.2024EDP7028. [Google Scholar] [CrossRef]
4. Spoladore D, Pessot E, Trombetta A. A novel agile ontology engineering methodology for supporting organizations in collaborative ontology development. Comput Ind. 2023;151:103979. doi:10.1016/j.compind.2023.103979. [Google Scholar] [CrossRef]
5. Touati C, Kemmar A. Deep reinforcement learning approach for ontology matching problem. Int J Data Sci Anal. 2024;18(1):97–112. doi:10.1007/s41060-023-00425-5. [Google Scholar] [CrossRef]
6. Liu Z, Lin Y, Sun M. Representation learning for natural language processing. In: Representation learning for natural language processing. 2nd ed. Beijing, China: Springer Singapore; 2023. [Google Scholar]
7. He Y, Chen J, Dong H, Jiménez-Ruiz E, Hadian A, Horrocks I. Machine learning-friendly biomedical datasets for equivalence and subsumption ontology matching. In: Proceedings of the 21st International Semantic Web Conference; 2022. p. 575–91. [Google Scholar]
8. Pennington J, Socher R, Manning CD. GloVe: global vectors for word representation. In: Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP); 2014; Doha, Qatar. p. 1532–43. [Google Scholar]
9. Chen J, Hu P, Jimenez-Ruiz E, Holter OM, Antonyrajah D, Horrocks I. Horrocks OWL2Vec∗: embedding of OWL ontologies. Mach Learn. 2021;110(7):1813–45. doi:10.1007/s10994-021-05997-6. [Google Scholar] [CrossRef]
10. Hao Z, Mayer W, Xia J, Li G, Qin L, Feng Z. Ontology alignment with semantic and structural embeddings. J Web Semant. 2023;78:100798. doi:10.1016/j.websem.2023.100798. [Google Scholar] [CrossRef]
11. Duan H, Sun Y, Lee Y. Ontology matching method based on word embedding and structural similarity. Int J Adv Smart Convergence. 2023;12(3):75–88. doi:10.7236/IJASC.2023.12.3.75. [Google Scholar] [CrossRef]
12. Kolyvakis P, Kalousis A, Smith B, Kiritsis D. Biomedical ontology alignment: an approach based on representation learning. J Biomed Semantics. 2018;9(1):21. doi:10.1186/s13326-018-0187-8. [Google Scholar] [PubMed] [CrossRef]
13. Xue X, Huang Y, Zhang Z. Deep reinforcement learning based ontology meta-matching technique. IEICE Trans Inf Syst. 2023;106(5):635–43. doi:10.1587/transinf.2022DLP0050. [Google Scholar] [CrossRef]
14. Li Z, Liu X, Wang X, Liu P, Shen Y. TransO: a knowledge-driven representation learning method with ontology information constraints. World Wide Web. 2023;26(1):297–319. doi:10.1007/s11280-022-01016-3. [Google Scholar] [CrossRef]
15. Şentürk F, Aytac V. A graph-based ontology matching framework. New Gener Comput. 2024;42(1):33–51. doi:10.1007/s00354-022-00200-3. [Google Scholar] [CrossRef]
16. Efeoglu S. GraphMatcher: a graph representation learning approach for ontology matching. In: Proceedings of the 17th International Workshop on Ontology Matching, The 21st International Semantic Web Conference (ISWC) 2022; 2022. p. 174–80. [Google Scholar]
17. Xue X, Yang C, Mao G, Zhu H. Semi-automatic ontology matching based on interactive compact genetic algorithm. Int J Pattern Recognit Artif Intell. 2022;36(5):2257002. doi:10.1142/S0218001422570026. [Google Scholar] [CrossRef]
18. He Y, Chen J, Antonyrajah D, Horrocks I. BERTMap: a BERT-based ontology alignment system. In: Proceedings of the 36th AAAI Conference on Artificial Intelligence; 2022. p. 5684–91. [Google Scholar]
19. Portisch J, Hladik M, Paulheim H. Background knowledge in ontology matching: a survey. Semant Web. 2022;13(3):1–55. doi:10.3233/SW-223085. [Google Scholar] [CrossRef]
20. Ibrahim S, Fathalla S, Lehmann J, Jabeen H. Toward the multilingual semantic web: multilingual ontology matching and assessment. IEEE Access. 2023;11(1):8581–99. doi:10.1109/ACCESS.2023.3238871. [Google Scholar] [CrossRef]
21. Lv Z, Peng ZR. A novel periodic learning ontology matching model based on interactive grasshopper optimization algorithm. Knowl Based Syst. 2021;228(C):1–14. [Google Scholar]
22. Lv Q, Shi J, Shi H, Jiang C. A novel compact fireworks algorithm for solving ontology meta-matching. Appl Intell. 2023;53(5):5784–807. doi:10.1007/s10489-022-03618-w. [Google Scholar] [CrossRef]
23. Li J, Song D. Uncertainty-aware pseudo label refinery for entity alignment. In: Proceedings of the ACM Web Conference 2022; 2022; Lyon, France. p. 829–37. [Google Scholar]
24. Gu Y, Qu X, Wang Z, Huai B, Yuan NJ. Delving deep into regularity: a simple but effective method for Chinese named entity recognition. In: Findings of the Association for Computational Linguistics: NAACL 2022; 2022; Seattle, DC, USA. p. 1863–73. [Google Scholar]
25. Gu X, Chen X, Lu P, Lan X, Li X, Du Y. SiMaLSTM-SNP: novel semantic relatedness learning model preserving both Siamese networks and membrane computing. J Supercomput. 2024;80(3):3382–411. doi:10.1007/s11227-023-05592-7. [Google Scholar] [CrossRef]
26. Schriml LM, Mitraka E, Munro J, Tauber B, Schor M, Nickle L, et al. Human disease ontology 2018 update: classification, content and workflow expansion. Nucleic Acids Res. 2018;47(D1):D955–62. doi:10.1093/nar/gky1032. [Google Scholar] [PubMed] [CrossRef]
27. Vasant D. ORDO: an ontology connecting rare disease, epidemiology and genetic data. In: Proceeding of ISMB; 2014; Boston, MA, USA. Vol. 30. [Google Scholar]
28. Amberger JS, Bocchini CA, Schiettecatte F, Scott AF. A Hamosh OMIM.org: Online Mendelian Inheritance in Man (OMIM®an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43(D1):D789–98. doi:10.1093/nar/gku1205. [Google Scholar] [CrossRef]
29. Donnelly K. SNOMED-CT: the advanced terminology and coding system for eHealth. Stud Health Technol Inform. 2006;121:279–90. [Google Scholar] [PubMed]
30. Sioutos N, de Coronado S, Haber MW, Hartel FW, Shaiu WL, Wright LW. NCI Thesaurus: a semantic model integrating cancer related clinical and molecular information. J Biomed Inform. 2007;40(1):30–43. doi:10.1016/j.jbi.2006.02.013. [Google Scholar] [PubMed] [CrossRef]
31. Chen J, He Y, Geng Y, Jiménez-Ruiz E, Dong H, Horrocks I. Contextual semantic embeddings for ontology subsumption prediction. World Wide Web. 2023;26(5):2569–91. doi:10.1007/s11280-023-01169-9. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools