 Open Access
Open Access
REVIEW
A Comprehensive Review of Pill Image Recognition
1 Faculty of Information Technology, Nguyen Tat Thanh University, 300A Nguyen Tat Thanh Street, District 4, Ho Chi Minh City, 700000, Vietnam
2 Faculty of Information Technology, School of Technology, Van Lang University, 69/68 Dang Thuy Tram Street, Ward 13, Binh Thanh District, Ho Chi Minh City, 700000, Vietnam
3 Faculty of Information Technology, Ho Chi Minh City Open University, 35-37 Ho Hao Hon Street, Ward Co Giang, District 1, Ho Chi Minh City, 700000, Vietnam
* Corresponding Authors: Linh Nguyen Thi My. Email: ; Vinh Truong Hoang. Email:
(This article belongs to the Special Issue: Novel Methods for Image Classification, Object Detection, and Segmentation)
Computers, Materials & Continua 2025, 82(3), 3693-3740. https://doi.org/10.32604/cmc.2025.060793
Received 10 November 2024; Accepted 16 January 2025; Issue published 06 March 2025
Abstract
Pill image recognition is an important field in computer vision. It has become a vital technology in healthcare and pharmaceuticals due to the necessity for precise medication identification to prevent errors and ensure patient safety. This survey examines the current state of pill image recognition, focusing on advancements, methodologies, and the challenges that remain unresolved. It provides a comprehensive overview of traditional image processing-based, machine learning-based, deep learning-based, and hybrid-based methods, and aims to explore the ongoing difficulties in the field. We summarize and classify the methods used in each article, compare the strengths and weaknesses of traditional image processing-based, machine learning-based, deep learning-based, and hybrid-based methods, and review benchmark datasets for pill image recognition. Additionally, we compare the performance of proposed methods on popular benchmark datasets. This survey applies recent advancements, such as Transformer models and cutting-edge technologies like Augmented Reality (AR), to discuss potential research directions and conclude the review. By offering a holistic perspective, this paper aims to serve as a valuable resource for researchers and practitioners striving to advance the field of pill image recognition.Keywords
Pill image recognition has practical applications and benefits across various fields. In healthcare and patient care, it assists patients through mobile applications that identify pills via images, helping the elderly and those managing multiple medications by ensuring correct medication intake and reducing errors. It also supports pharmacists and doctors by verifying medications to prevent mix-ups and providing quick identification for accurate advice. In hospital medication management, it enhances inventory control by tracking and managing medication stock, and increases efficiency and accuracy through automation, minimizing the need for manual checks. For safety and legal compliance, it helps identify counterfeit or substandard medications and supports regulatory inspections and adherence to standards. Additionally, it aids the visually impaired through assistive applications that help them recognize medications and receive information via audio.
Recently, artificial intelligence (AI) has made significant strides and emerged as a powerful tool for addressing various challenges. In its early stages, pill identification was managed through various online systems that required users to manually enter multiple attributes, such as shape, color, and imprint, as shown in Fig. 1.
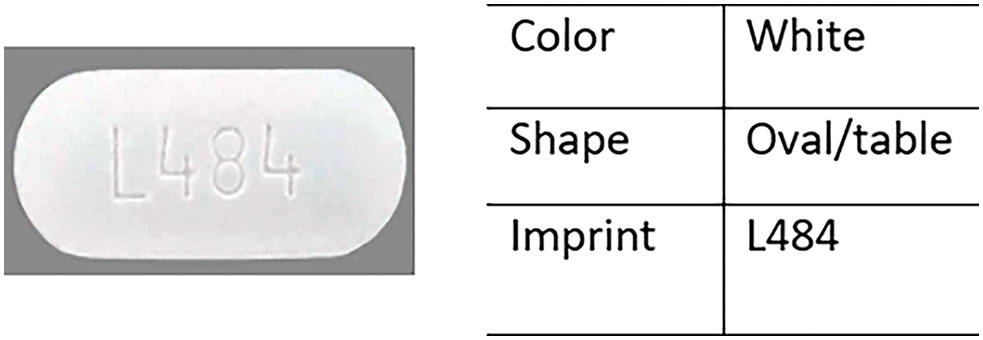
Figure 1: Shape, color, imprint of the Paracetamol pill
Websites like https://www.drugs.com/imprints.php (accessed on 15 January 2025) have made it easier for users to identify pills based on characteristics such as color, shape, and imprint. Despite this convenience, several challenges arise when relying on these platforms. For one, users must manually input various attributes, which can be difficult if the pill’s features are worn or unclear, making the process time-consuming. Additionally, the accuracy of the identification depends largely on the user’s ability to correctly describe the pill, increasing the probability of mistakes. Moreover, the website’s database may not always include the latest or less common medications, potentially leading to incomplete identification. These limitations emphasize the need for more efficient, automated solutions using pill image recognition technology.
This review highlights key ethical considerations in the development and application of pill image recognition technologies. Misidentification remains a critical risk, as inaccurate pill identification can lead to incorrect medication or dosage, potentially causing severe health consequences. To mitigate this, rigorous validation and verification processes must be implemented, alongside the use of diverse and representative datasets. Accessibility is another important factor, especially for visually impaired individuals and older adults. Developers should integrate assistive features, such as auditory feedback or simplified interfaces, to ensure inclusivity. Additionally, issues related to data privacy, security, and accountability necessitate compliance with ethical standards and regulatory frameworks. These considerations underscore the importance of ethical oversight in this field, encouraging the research community to prioritize safety, inclusivity, and trust in the development of pill image recognition systems.
In this literature review, we explore a range of methodologies employed for pill image recognition, categorizing them into four primary approaches: traditional image processing-based, traditional machine learning-based, deep learning-based, and hybrid-based methods. Traditional image processing-based methods use basic image processing techniques to extract features like shape, color, and imprint, facilitating the matching of pill images. Traditional machine learning-based methods build on these features, employing algorithms like K-Nearest Neighbors (k-NN), Support Vector Machines (SVM), and Decision Trees to classify pill images. Deep learning-based methods, powered by Convolutional Neural Networks (CNNs) and other neural network architectures, have shown remarkable performance by automatically learning intricate features from large datasets of pill images. Lastly, hybrid-based methods combine elements of the aforementioned approaches to capitalize on their respective strengths and reduce their limitations. The remainder of the article is organized as follows. Section 2 provides a comprehensive background on the foundational concepts relevant to this study. Section 3 presents a detailed literature survey. Section 4 discusses the benchmark datasets commonly used in the research community. In Section 5, a thorough comparison of various methods is conducted to highlight their respective strengths and weaknesses. Section 6 identifies open research problems. Finally, Section 7 concludes the article.
2.1 Definition of Pill Image Recognition
Pill image recognition is the process of using computer algorithms to recognize and identify pills based on visual features from photographic images of the pill. Figs. 2 and 3 illustrate the identification of one pill or multiple pills.
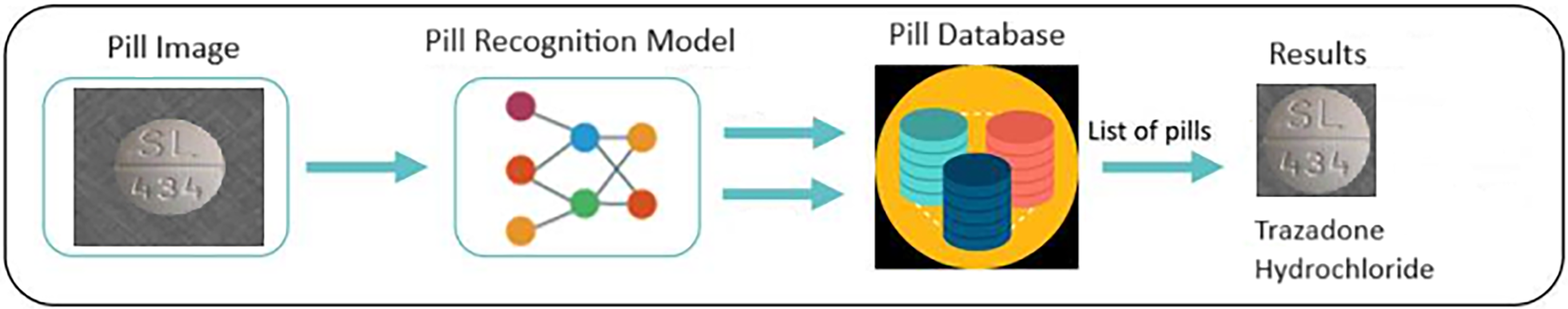
Figure 2: Defines the recognition of a pill
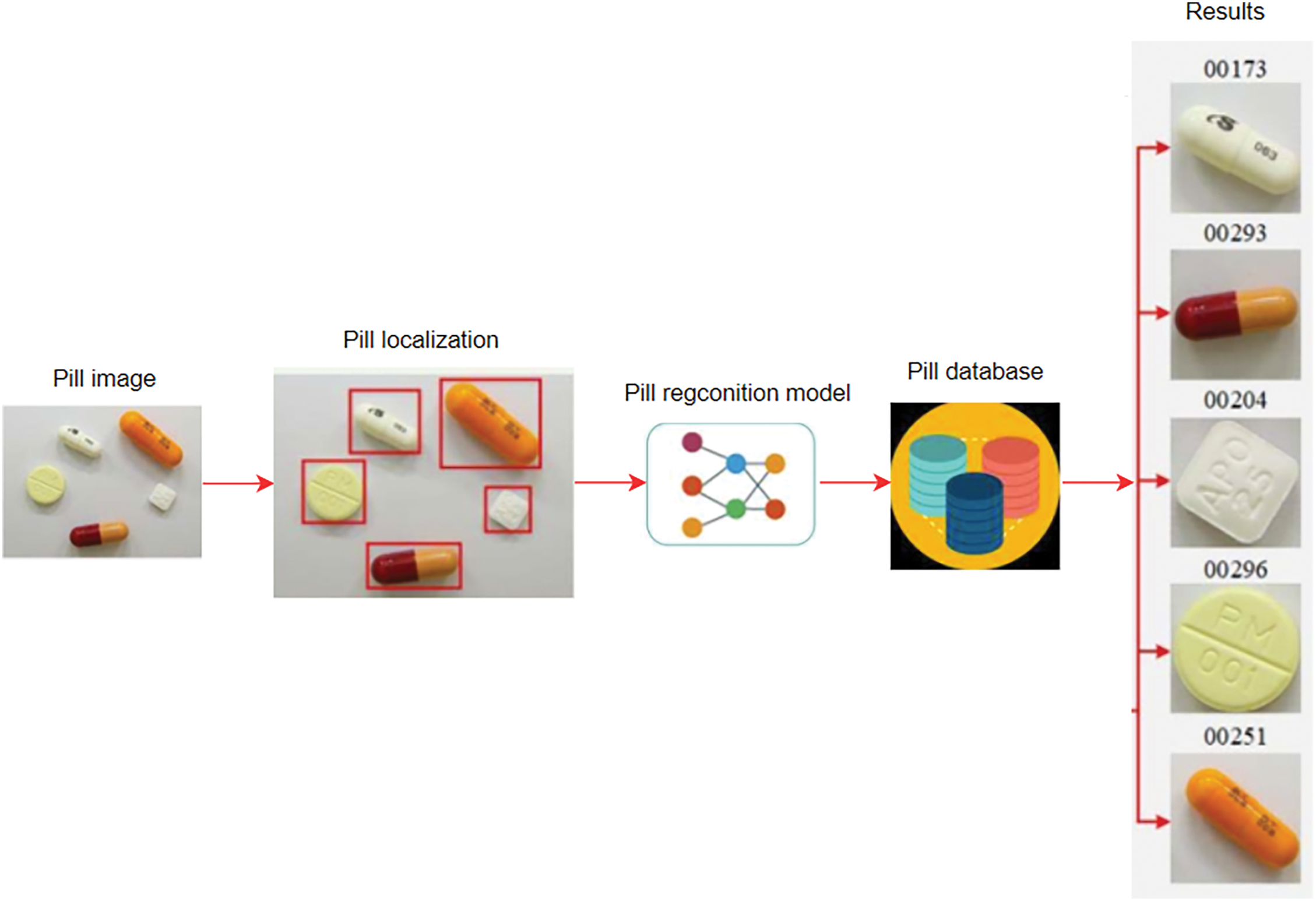
Figure 3: Defines the recognition of multiple pills
In the Fig. 2, the input is an image of a pill, including the background of the pill, the output is to identify the identity of that pill. In the Fig. 3, the input is an image of many pills, including the background of pills, the output is to identify each pill.
2.2 Definition of Pill Features
The shape of a pill refers to the geometric form or outline of the pill when viewed from above. It is one of the key physical characteristics used to identify and differentiate medications. Pill shapes can vary widely and are often designed for specific purposes related to the medication’s use, the ease of swallowing, or to avoid confusion with other pills.
The color of a pill refers to the hue or shade visible on the surface of the medication. Pill color is a significant identifying feature that helps distinguish one medication from another. It can be a single color or a combination of colors and may be used in conjunction with other physical characteristics such as shape, size, and imprint for accurate identification.
An imprint on a pill refers to any characters, symbols, logos, or patterns that are stamped, engraved, or printed on the surface of the pill. These imprints serve as unique identifiers to help distinguish one pill from another, providing essential information for identification and verification purposes. Example about shape, color, imprint of pill is shown in Fig. 1.
In this section, we review papers that utilize algorithms for pill image recognition. We categorize pill image recognition methods into four main approaches: traditional image processing-based, traditional machine learning-based, deep learning-based, and hybrid-based methods. During the data preprocessing stage, image filtering is commonly employed, with popular techniques such as averaging filters, median filters, and Gaussian filters frequently used by authors. In our summary of the methods used, we focus on the primary techniques and do not include these widely-used image filtering methods.
3.1 Traditional Image Processing-Based Methods for Pill Image Recognition
The traditional image processing-based method for pill image recognition refers to an approach that uses classic techniques in image analysis and manipulation, such as preprocessing for noise reduction and enhancement, segmentation to isolate pill features, and extraction of shape, color, and imprint attributes. This method relies on established algorithms and mathematical operations to interpret and classify pill images based on predefined visual characteristics. The main methods used by the authors in this category are summarized in Table 1.
Lee et al. [1,19] both emphasize the importance of imprint matching, using techniques like edge detection and contour analysis to extract pill imprints. These methods perform well when the pill imprints are clear and distinguishable, but their accuracy decreases when the imprint is faint or damaged. These approaches rely on predefined feature extraction algorithms, which are less flexible compared to modern learning-based approaches.
Morimoto et al. [6], Kim et al. [8] focus on matching pills based on their shape and color, utilizing features like contour descriptors and color histograms. These methods are effective for distinguishing pills with distinct shapes and colors but struggle with pills that are similar in these aspects. Shape-based recognition often suffers when pills are rotated or partially occluded, limiting the robustness of these methods in real-world conditions.
The work in Hartl et al. [9] explores the challenges of recognizing pills in unconstrained environments, such as on mobile devices, where lighting conditions and camera angles can vary significantly. The use of traditional image processing techniques here, including color and imprint analysis, shows promise, but the performance is highly dependent on controlled conditions. Variations in lighting and background can significantly affect the accuracy of recognition.
In Caban et al. [17], the shape distribution model is applied to pills by capturing the distribution of distances from a set of boundary points. This method improves the robustness to shape variations but can still be limited by the need for clear boundaries in the pill image, which might not always be available due to noise or occlusion.
Chen et al. [25] proposes a method that combines various features-shape, color, and texture-weighted dynamically depending on the context of the pill image. This method enhances the system’s flexibility to adapt to different pill appearances, though it still struggles with ambiguous or noisy images where feature distinction is difficult.
Yu et al. [36] introduces a method for recognizing pill imprints by analyzing distances between imprint features. This two-step approach improves the robustness of imprint recognition, but it is still reliant on the quality of the imprint and can struggle in cases of faint imprints.
Hema et al. [40] and Ranjitha et al. [49] emphasize feature extraction techniques, such as shape, color, and imprint, to differentiate pills. These methods are highly interpretable but lack the ability to generalize well across diverse pill types, particularly when there are subtle differences between them.
Suntronsuk et al. [43,45] tackle the challenging problem of imprint recognition, applying binarization and segmentation techniques to separate text from the pill’s surface. While effective for high-contrast imprints, these techniques face challenges when the pill’s imprint contrast is poor or the surface is reflective.
Lastly, Chokchaitam [50] addresses the issue of background interference by compensating for shadows and lighting variations. This improves the reliability of color-based features but is still sensitive to extreme lighting changes.
In summary, traditional image processing methods offer interpretability and efficiency in pill recognition but face challenges with variations in pill appearance, lighting conditions, and imprint quality. These methods are generally effective in controlled environments but struggle in real-world applications where pills may have similar colors or shapes, or where imprints are not clearly visible. As a result, while these methods laid the groundwork for pill recognition, they have been increasingly supplemented or replaced by machine learning and deep learning techniques in recent years due to the limitations in handling more complex or ambiguous pill images.
3.2 Traditional Machine Learning-Based Methods for Pill Image Recognition
The traditional machine learning-based method for pill image recognition involves using algorithms and statistical models to train on extracted features from pill images. This approach typically includes preprocessing steps to enhance image quality and feature extraction to identify key characteristics such as shape, color, and imprint. Machine learning models like Support Vector Machines (SVM), K-Nearest Neighbors (k-NN), and Random Forests are trained on these features to classify pill images based on learned patterns and similarities within a labeled dataset. The main methods used by the authors in this category are summarized in Table 2.
Cunha et al. [51] employ traditional machine learning to create a mobile-based tool for pill recognition. The system uses characteristics such as pill shape and color, employing a machine learning classifier to help elderly users identify pills. This study highlights the effectiveness of traditional methods in resource-constrained environments, particularly mobile platforms, where computational efficiency is crucial.
Yu et al. [37] focus on imprint-based features combined with traditional machine learning classifiers. Imprint information, including text or symbols on the pill surface, is extracted and processed to train models k-NN. This approach is particularly effective for pills that have similar shapes or colors but distinct imprints.
Ushizima et al. [55] explore various traditional machine learning approaches on a large-scale pill dataset. The study compares different feature extraction techniques and classifiers, including SVM and k-NN, finding that SVM performed better when combined with text and imprint features, underlining the importance of accurate feature selection.
Chupawa et al. [62] investigate the use of neural networks in pill recognition, but in the context of traditional machine learning rather than deep learning. The study employs shallow neural networks trained on features like imprints, shape, and imprint, achieving good results in recognizing pills with distinct imprints. This marks an early exploration of neural networks prior to the dominance of deep learning methods.
Vieira Neto et al. [63] introduce a novel feature extractor, CoforDes, to enhance the robustness of pill recognition systems against variations in lighting and pose. Combined with SVMs, this method provides invariance to scale and rotation, making it particularly useful in cases where pills are photographed under varying conditions.
Chughtai et al. [64] continue the exploration of shallow neural networks in pill identification. It emphasizes the effectiveness of combining hand-crafted features with a neural network classifier, demonstrating that neural networks can still produce strong results when paired with carefully designed feature sets, even without relying on deep learning.
Finally, Dhivya et al. [67] present a method that combines SVM for text recognition with an n-gram-based error correction algorithm. This method addresses challenges related to recognizing imprints on pill surfaces, particularly in low-quality images, making it a robust solution for text-based pill identification.
In summary, traditional machine learning methods for pill image recognition rely heavily on feature extraction, with classifiers such as SVM, k-NN, and shallow neural networks being commonly used. While earlier studies focused on shape and color features, more recent work has explored the use of text and imprint information as key distinguishing factors. Feature engineering remains crucial in these methods, with techniques like CoforDes and error correction algorithms enhancing robustness and accuracy. Although deep learning has overshadowed these approaches in recent years, traditional machine learning methods still provide efficient and effective solutions in scenarios with limited computational resources.
3.3 Deep Learning-Based Methods for Pill Image Recognition
The deep learning-based method for pill image recognition refers to an approach that employs deep neural networks, particularly Convolutional Neural Networks (CNNs), to automatically learn and extract features from pill images. This method eliminates the need for handcrafted features by using multiple layers of neurons to progressively learn hierarchical representations of visual data. Deep learning models are trained on large datasets of labeled pill images, enabling them to accurately classify and identify pills based on complex visual patterns and features. The main methods used by the authors in this category are summarized in Table 3.
Simonyan et al. [73] introduce the use of deep convolutional neural networks (CNNs) for image classification, laying the foundation for later work in pill image recognition. The depth of these networks allows for capturing intricate features from complex image data, which has proven beneficial for fine-grained pill identification.
Wong et al. [74] leverage a deep CNN for distinguishing between pills that have subtle visual differences. This approach demonstrates high accuracy by extracting detailed features, showing superior performance in handling pills with similar shapes or colors.
Ou et al. [79] apply a CNN to detect pills in real-world scenarios, where pills may be partially occluded or in cluttered environments. While it achieves high detection accuracy, the model’s performance degrades in scenarios with significant lighting variations.
Chang et al. [83,94] employ CNNs for real-time pill identification through wearable devices. These systems ensure rapid and accurate recognition while maintaining computational efficiency, making them suitable for resource-constrained environments like wearable smart glasses.
Larios Delgado et al. [86] emphasize computational efficiency while maintaining high accuracy. This method focuses on optimizing CNN architectures to deliver real-time pill recognition, crucial for clinical settings that require immediate results.
Cordeiro et al. [88] and Swastika et al. [89] explore multi-stream CNN architectures, where multiple networks are used to focus on different features of the pills (shape, color, imprint). The combination of these streams yields improved accuracy, especially for visually similar pills.
Usuyama et al. [91] present a benchmark for evaluating pill recognition models in low-shot settings. It highlights that models pretrained on large datasets can be adapted for pill identification with minimal additional data, making them more efficient in scenarios where labeled data is scarce.
Ou et al. [96] integrate feature pyramid networks (FPN) with CNNs to improve multiscale feature extraction. This approach achieves superior performance in detecting pills of varying sizes and in challenging backgrounds.
Marami et al. [98] address the unique challenge of recognizing discarded medications. The CNN-based system is able to identify damaged or altered pills, demonstrating the robustness of deep learning in handling real-world pill recognition scenarios.
Ling et al. [101] and Zhang et al. [130,139] focus on recognizing new pill classes with minimal data. These few-shot learning techniques enable the models to adapt to new pill types without requiring extensive retraining, making them highly adaptable in dynamic pharmaceutical environments.
Tsai et al. [107] present an innovative application where CNNs are used to automatically identify pills within a smart pillbox, ensuring that patients take the correct medications. The model’s accuracy, combined with a physical pill-dispensing system, improves patient compliance and safety.
Kwon et al. [108] apply deep learning for quality control in pharmaceutical manufacturing, where CNNs are used to inspect pills for defects. This model ensures the integrity of pills before they are distributed, highlighting the role of deep learning in ensuring quality assurance.
Lester et al. [110] evaluate CNN-based models across real-world clinical environments, showing that deep learning models generalize well when trained on diverse datasets. The study indicates that CNNs can handle varying lighting conditions and pill orientations effectively.
Ozmermer et al. [113] incorporate deep metric learning with CNNs to improve pill identification. The method outperforms traditional approaches by learning discriminative feature embeddings, enabling the model to distinguish visually similar pills more accurately.
Tan et al. [116,117] compare object detection models for pill recognition, revealing that YOLO-v3 offers the best balance between accuracy and inference speed for real-time applications. However, Faster R-CNN provides higher accuracy but at the cost of slower performance.
Tan et al. [118] propose a novel multi-stream CNN architecture that integrates different streams of information (shape, color, imprint) for pill classification. This method handles class-incremental learning more effectively, showing that it adapts to new pill classes without degrading the performance on previously learned classes.
Suksawatchon et al. [119] and Wu et al. [121] focus on shape-based recognition using deep CNNs. These models demonstrate superior performance in recognizing pills even under unconstrained conditions, such as varying orientations and partial occlusions.
Pornbunruang et al. [124] and Duy et al. [127] introduce hybrid approaches by combining CNNs with external knowledge sources, such as medical knowledge graphs. These models improve pill recognition by incorporating contextual information about the pill’s medical properties, dosage, and use cases.
Thanh et al. [128] and Duy et al. [132] integrate graph neural networks (GNNs) with CNNs to enhance pill recognition. These methods demonstrate improved accuracy by considering relationships between pills, prescriptions, and medical knowledge, providing more reliable and explainable results.
Bodakhe Sakshi et al. [129] focus on accurate and efficient pill detection using CNNs in dynamic environments, demonstrating strong performance in recognizing multiple pills in a single image.
Ashraf et al. [133] highlight the use of deep learning models that do not require extensive customization, making them more accessible for diverse clinical applications. This model performs well across different healthcare settings, demonstrating the adaptability of CNNs.
Dang et al. [135] emphasize the need for accurate real-time recognition on mobile devices. The CNN-based system ensures that visually impaired individuals can quickly and reliably identify their medications.
In conclusion, deep learning methods for pill image recognition, particularly CNN-based models, have shown significant success in both real-time and high-accuracy tasks. The creation of more efficient, flexible, and interpretable models, especially those incorporating external knowledge or few-shot learning methods, suggests a bright future for pill recognition technologies in a range of real-world applications. The diversity in architectural choices-ranging from feature pyramid networks to hybrid GNN-CNN approaches-demonstrates the growing sophistication and effectiveness of deep learning in pharmaceutical image analysis.
3.4 Hybrid-Based Methods for Pill Image Recognition
Hybrid-based methods for pill image recognition combine traditional image processing techniques with modern machine learning or deep learning approaches. This approach leverages the strengths of both methodologies to enhance accuracy and robustness in identifying pills based on their visual characteristics. The main methods used by the authors in this category are summarized in Table 4.
In Yaniv et al. [141], a hybrid method combining traditional machine learning features with convolutional neural networks (CNNs) is explored to address the challenge of pill identification. This approach aims to combine handcrafted features such as color and shape with deep learning-based features to improve accuracy, particularly for difficult cases where imprints or shapes are ambiguous.
Zeng et al. [142] present a lightweight hybrid model designed for mobile environments. This method leverages deep learning for feature extraction while incorporating post-processing techniques to handle unconstrained environments, such as varying lighting conditions and pill orientations. The hybrid approach enables efficient recognition on resource-constrained mobile devices while maintaining accuracy.
In Wang et al. [143], a hybrid approach is introduced to address the problem of limited labeled data. By combining transfer learning and semi-supervised learning, the model achieves high accuracy with minimal annotations. This method employs pre-trained deep learning models and incorporates clustering techniques like k-means to refine the classification process, demonstrating the effectiveness of hybrid systems in handling data scarcity.
Mehmood et al. [145] further explore hybrid techniques for mobile platforms, combining deep CNNs with traditional feature extraction methods to balance the computational load and accuracy. This method integrates deep learning for initial feature extraction while using clustering-based techniques to refine the identification process, optimizing the system for mobile use.
Srikamdee et al. [150] take a unique hybrid approach by combining CNNs with k-means clustering for pill identification. The CNNs extract deep features, which are then grouped using k-means clustering to handle variations in pill appearances, such as color or shape. This hybrid method allows for a more efficient search and retrieval process in real-world applications, showing its strength in large-scale pill identification systems.
Al-Hussaeni et al. [151] focus on combining CNNs with traditional image retrieval methods. The hybrid approach integrates deep learning features with image matching techniques, such as SIFT or SURF, for more accurate retrieval of similar pill images. This hybrid model aims to improve retrieval precision in scenarios where pills share similar visual characteristics but have subtle differences, like imprints or logos.
Finally, Rádli et al. [154] extend the hybrid approach by incorporating multi-stream CNNs with an attention mechanism to handle complex pill recognition tasks. The model combines multiple streams of deep learning features, such as color, shape, and imprint information, and applies attention to focus on the most discriminative features. This hybrid architecture outperforms single-stream CNNs by better capturing subtle variations in pill images and emphasizing important visual cues during classification.
In conclusion, hybrid-based methods for pill image recognition effectively combine the strengths of deep learning with traditional machine learning and clustering techniques to enhance accuracy, especially in challenging scenarios such as mobile deployment, limited data, and visually similar pills. By leveraging the complementary strengths of these methods, hybrid approaches provide a more robust solution for pill identification in diverse real-world environments.
Benchmark datasets play a crucial role in evaluating the performance of proposed methods. For pill image recognition, there is a wide range of commonly used benchmark datasets. In the methods discussed in the previous section, the authors utilized both public and private datasets. We note that some datasets are private, and we do not review these private datasets in this section. Here, we provide a brief review of the public datasets and their relevant information for pill image recognition.
4.1 National Library of Medicine (NLM)
In January 2016, the US National Library of Medicine (NLM) [141] initiated a competition aimed at developing advanced algorithms and software for accurately ranking prescription pill identifications based on images submitted by users (consumer images). The data for identification is drawn from the RxIMAGE collection (reference images).
The identification dataset was composed of three key parts: a training dataset accessible to all participants, a segregated testing dataset, and a non-segregated testing dataset reserved for evaluating the top three submissions after the competition concluded.
The training dataset contained 7000 images representing 1000 different pills. For each pill, there was one high-resolution macro photograph of each side and five consumer-quality images. For capsules, the image dataset included clear views of the pill’s imprint area with text aligned perpendicularly to the plane of the capsule. The pills included in this dataset were randomly selected from the RxIMAGE collection to ensure a representative distribution by color and shape. An example of a similar consumer-quality image and reference image in the NLM dataset for tablets and capsules is shown in Fig. 4.
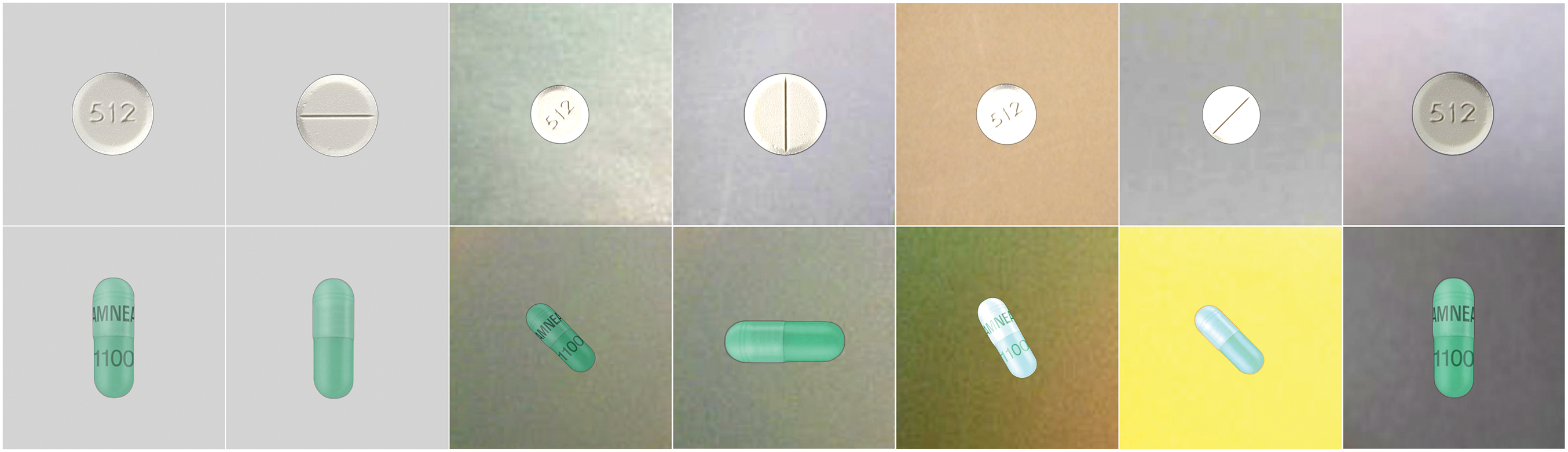
Figure 4: Similar sample images from the training dataset in the NLM dataset. The first two columns are the reference, macro photographs. The remaining pictures are consumer quality images
The distribution of pills based on their key visual properties, shape, and color in the training data is shown in Fig. 5.
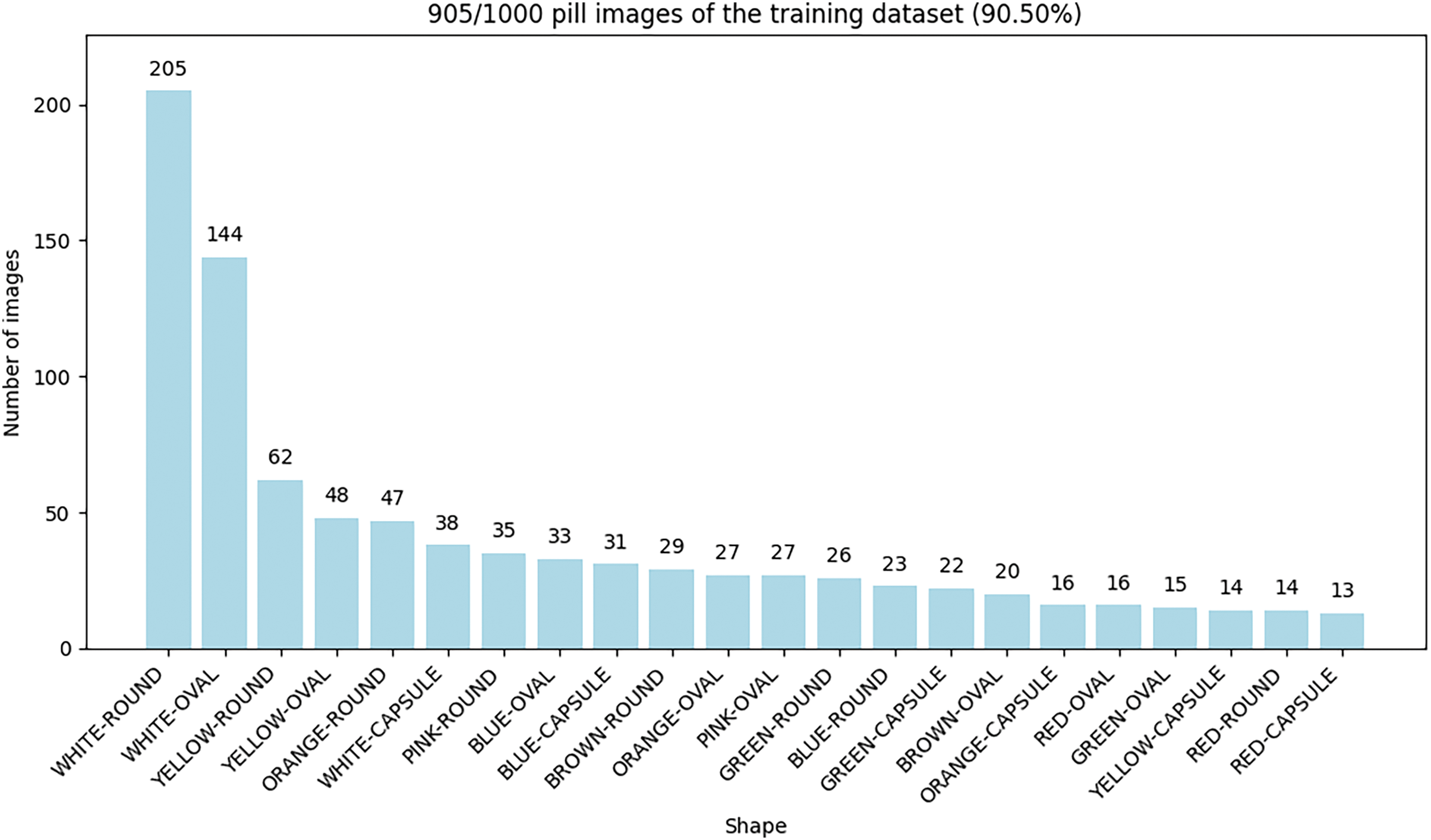
Figure 5: Distribution of pills based on their key visual properties, shape and color of training data
The segregated testing dataset was assembled using a separate set of 1000 pills that were not available to the participants during the competition. For each pill in this dataset, there were two reference images and five consumer images. The pills were randomly selected based on their shape and color to ensure their distribution was similar to that of the training set. A bar chart displaying the pill distributions by color and shape is shown in Fig. 6.

Figure 6: Distribution of pills based on their key visual properties, shape and color. The segregated testing dataset is comprised of reference and consumer quality images from products that were not part of the training set
This dataset enables the contest judges to evaluate an algorithm’s generalization ability and its alignment with their long-term goals. As the contest judges continue to acquire additional images for the RxIMAGE collection, they prefer to train an algorithm only once.
The non-segregated testing dataset included data from the same 1000 pills used in the training set. For this dataset, the contest judges used 6486 consumer-quality images that were not part of the training set. This data allows the contest judges to assess whether an algorithm is potentially overfitting to the training data, as it should generalize well to variations in consumer images similar to those it has previously encountered.
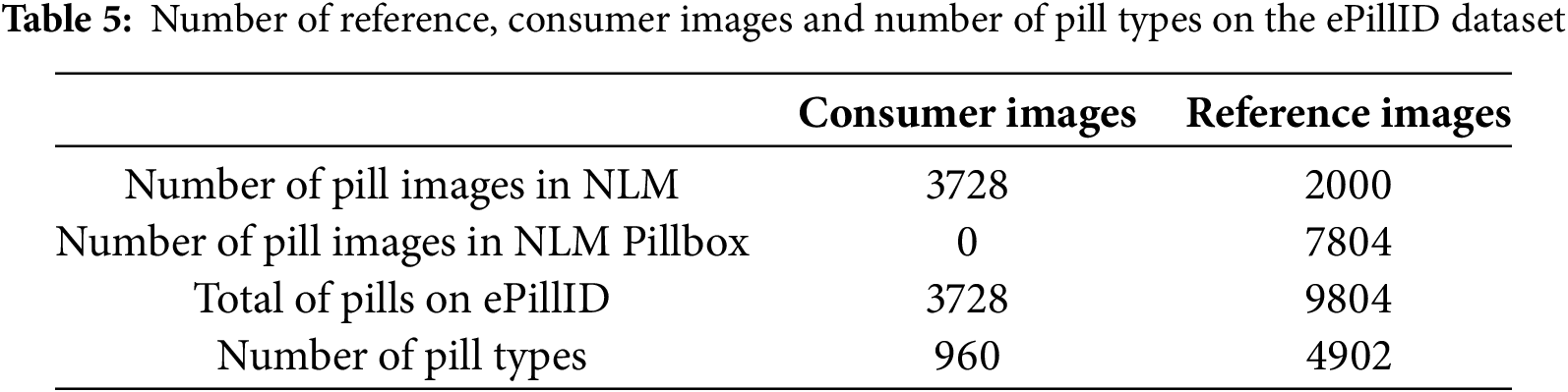
Usuyama et al. [91] used the NLM challenge dataset and the NLM Pillbox dataset to create it. The ePillID dataset includes a total of 13,532 images of 6766 pills, with each pill photographed from both sides (front and back). For each pill captured and saved as either a consumer image or a reference image, both sides are documented.
Of the 13,532 images, 3728 are consumer images and 9804 are reference images. Among these, the 3728 consumer images are sourced from the NLM dataset, while the 9804 reference images are divided into 2000 images from NLM and 7804 images from NLM Pillbox.
In the 3728 consumer images, there are 960 pill types, and in the 9804 reference images, there are 4902 pill types. Detailed descriptions of the number of images, consumer images, reference images, and pill types are shown in Table 5. These specific data are taken from the author’s article combined with the ePillID dataset that the author has published.
This dataset is particularly challenging due to its low-shot recognition setting, where most classes have only a single reference image, making it difficult for models to generalize. Various baseline models were evaluated, with a multi-head metric learning approach using bilinear features achieving the best performance. However, error analysis revealed that these models struggled to reliably distinguish between similar pill types. The paper also discusses future directions, including integrating Optical Character Recognition (OCR) to address challenges such as low-contrast imprinted text, irregular layouts, and varying pill materials like capsules and gels. Furthermore, the ePillID benchmark is set to expand with additional pill types and images, promoting further research in this critical area of healthcare.
The images in the NLM dataset have limitations related to lighting, background conditions, and equipment, among other factors. The CURE dataset [101] addresses these limitations.
This dataset contains 8973 images across 196 categories, with approximately 45 samples for each pill category, as shown in Figs. 7 and 8.
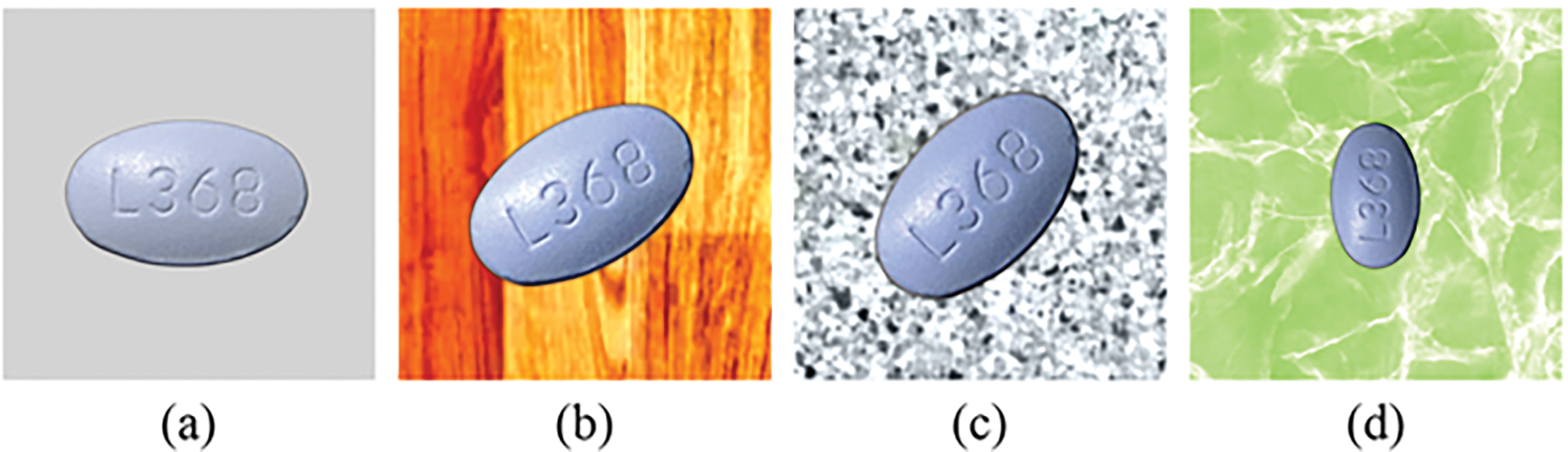
Figure 7: Similar images in the CURE dataset of one pill type: (a): reference image; (b–d): consumer images

Figure 8: Similar images in the CURE dataset of a different pill than the one in Fig. 7. (a): reference image; (b–d): consumer images
As summarized in Table 6, this dataset accounts for more challenging real-world conditions (i.e., with more diverse backgrounds, lighting, and zooming conditions), making it a better reflection of practical scenarios compared to the NLM dataset [141]. Examples of images from the dataset are shown in Figs. 7 and 8. It can be observed that: 1) images were taken under different lighting conditions, leading to significant changes in pill color (especially in Fig. 8d, where the color of the images taken under different lighting conditions with the MPI equipment varies significantly); 2) Fig. 7c,d was taken under different zooming conditions; and 3) the backgrounds in this dataset are diverse.

mCURE Dataset
To adapt the CURE dataset for the Few-shot Class-incremental Learning (FSCIL) setting, Zhang et al. [130] used a splitting strategy similar to that employed for miniImageNet in [131]. They sampled 171 classes to create the miniCURE dataset, abbreviated as mCURE. These 171 classes were divided into 91 base classes and 80 new classes. The new classes were further divided into eight incremental sessions, with the training data in each session formatted as 10-way 5-shot.
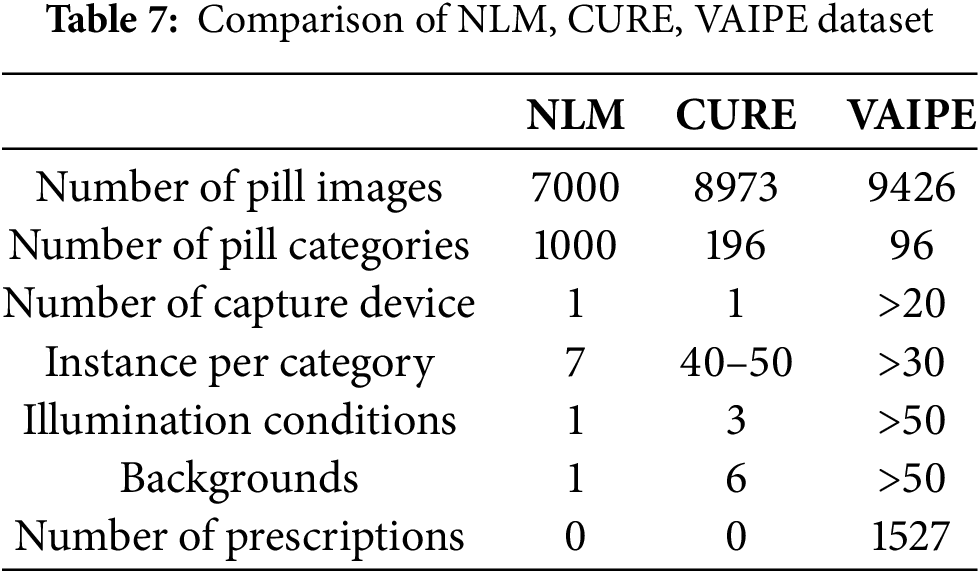
VAIPE [118] is the first real-world multi-pill image dataset, consisting of 9426 images representing 96 pill classes. The images were captured with ordinary smartphones in various settings and include prescriptions.
This dataset is designed for identifying images of multiple pills within their context, based on prescriptions. Each image containing multiple pills is annotated with a bounding box around each pill and labeled with its respective name. Prescriptions are also assigned a bounding box and labeled with the pill name and diagnosis in Vietnamese, making it suitable for the pill market in Vietnam.
The VAIPE dataset can serve as a valuable resource for training generic pill detectors. The characteristics of the VAIPE dataset, as well as comparisons with the NLM and CURE datasets, are detailed in Table 7.
VAIPE-PCIL Dataset
To facilitate research on Class-Incremental Learning (CIL) in pill image classification tasks, the authors derived a dataset version called VAIPE-PCIL (VAIPE Pill Class Incremental Learning) [118] from the original VAIPE data. VAIPE-PCIL was created by cropping pill instances from the original dataset. The authors selected only those categories that met the following criteria: 1) the number of samples should be greater than 10, and 2) the image size of samples should be at least 64

We compare the following 6 popular datasets: NLM, ePillID, CURE, mCURE VAIPE, VAIPE-PCIL in Table 9.
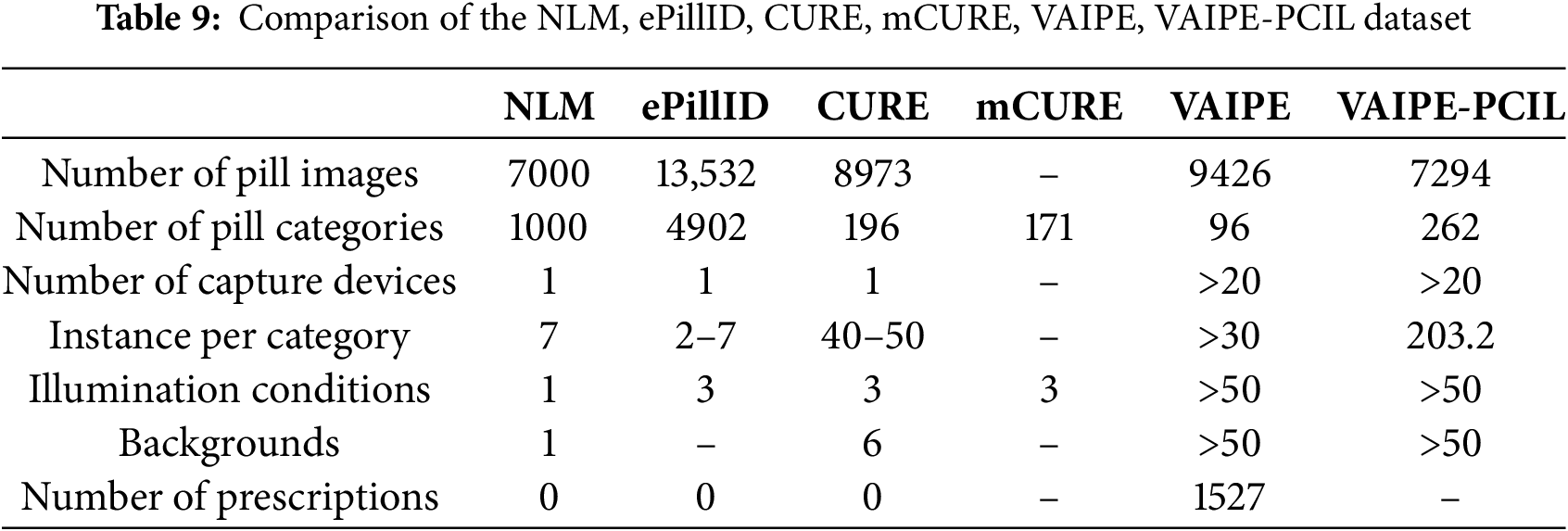
In summary, the NLM dataset is a popular source for pill identification problems, captured under ideal conditions. The ePillID dataset offers a large number of images for each pill and is more diverse, with images sourced partly from the NLM dataset and partly from the NLM Pillbox, and taken in natural conditions, is particularly challenging due to its low-shot recognition setting. The CURE dataset provides high-quality images with a variety of angles, lighting, and backgrounds, helping deep learning models handle real-life scenarios more effectively. The VAIPE dataset includes multi-pill images commonly found in the Vietnamese market, making it suitable for multi-pill image recognition with context-specific prescriptions and annotations in Vietnamese. This makes VAIPE a valuable resource for research and development in pill image recognition in Vietnam. Each dataset has its unique advantages, suited to different goals and applications in pill image recognition. Selecting the appropriate dataset depends on the user’s specific needs and the application environment.
5 Performance Comparison of Methods
In the literature survey above, there is only one article using the CURE dataset, one article using the ePillID dataset, and one article using the VAIPE-PCIL dataset. Consequently, we do not compare the evaluation metrics of the algorithms applied to these datasets. Table 10 compares the evaluation metrics of algorithms used in articles on the NLM, mCURE, and VAIPE datasets.
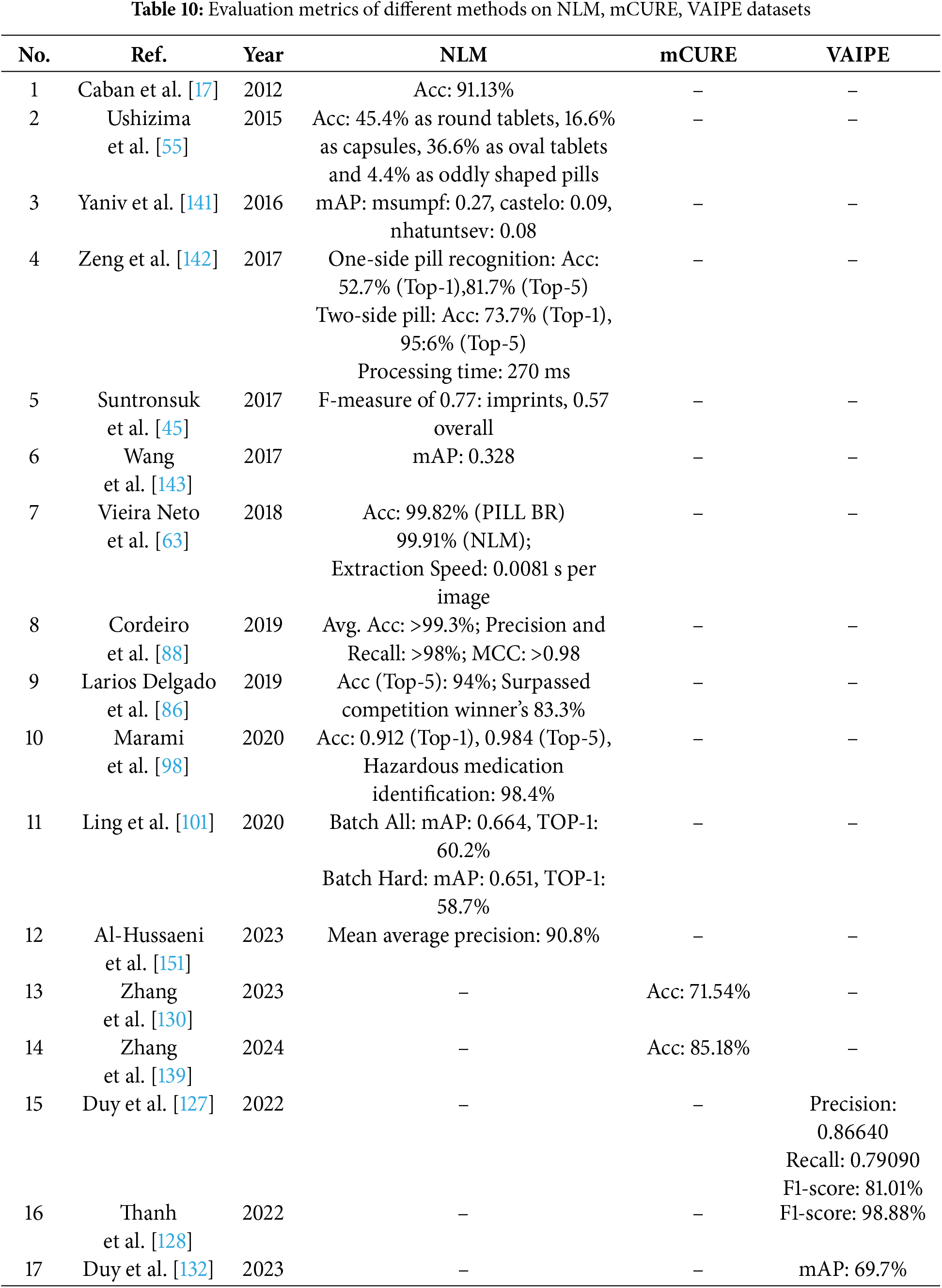
5.1 Performance Comparison of Methods on Dataset NLM
Caban et al. [17] shows strong potential for automatic medication identification. By utilizing a modified shape distribution approach that analyzes the shape, color, and imprint of pills, the system achieves 91.13% accuracy on a dataset of 568 U.S. prescription pills. This high accuracy suggests the system’s robustness against real-world variability such as lighting and camera angles. Additionally, it identifies the correct pill within the top 5 matches, even when an exact match is not found at the top. The system assumes a top-view pill image, which may limit its application in cases where different views are required. Further optimization could involve adjusting feature weights based on their relevance to recognition. Overall, the system’s performance suggests its potential for practical use in healthcare settings.
Ushizima et al. [55] discuss a pill recognition system using the NLM dataset, which demonstrates promising results but also highlights areas for improvement. The system segments pill images and extracts features based on the FDA’s recommended physical pill attributes, organizing them into four main categories: round tablets (45.4%), capsules (16.6%), oval tablets (36.6%), and oddly shaped pills (4.4%). This categorization serves as a foundational step for content-based image retrieval.
While the shape descriptors help differentiate between standard pill shapes, the system struggles to distinguish oddly shaped pills, pointing to the need for more refined feature extraction. The paper suggests incorporating additional descriptors, such as convexity points, to enhance performance in identifying irregular shapes. Future improvements include capturing more minor variations in pill contours, like contour bending energy, and incorporating lower-resolution images to ensure functionality with standard mobile cameras.
Overall, while the system provides a useful framework for classifying pills, it requires further advancements to be reliably used in real-world applications, particularly for patient safety. The ongoing research promises significant contributions to the development of practical pill recognition automation.
Yaniv et al. [141] discuss how the NLM Pill Image Recognition challenge highlights both the potential and limitations of current algorithms for pill identification using consumer-quality images. The top three submissions achieved mean average precision scores of 0.27, 0.09, and 0.08, with correct reference images retrieved in the top five results for 43%, 12%, and 11% of queries. While capsules, with distinct textual and color features, were more easily identified (74% accuracy in the top five), tablets, especially those with embossed markings, posed a challenge, with only 34% accuracy in the top five results.
The challenge revealed that improvements are needed in dataset size, feature descriptors, and image acquisition methods. The best submissions utilized deep learning, but their performance was limited by a small training dataset. To improve results, the use of data augmentation, pre-training on unrelated datasets, and more controlled image acquisition (e.g., including reference objects for pose estimation) is recommended. Overall, continued research is needed to enhance pill identification algorithms for both healthcare professionals and the public.
Zeng et al. [142] present the MobileDeepPill system, showcased in the NLM Pill Image Recognition Challenge, which demonstrates significant progress in pill recognition using mobile devices. Its architecture incorporates a triplet loss function, multi-CNNs, and a Knowledge Distillation-based model compression framework, leading to impressive recognition accuracy. In one-side pill recognition, MobileDeepPill achieved a Top-1 accuracy of 52.7% and a Top-5 accuracy of 81.7%. In two-side recognition, the system’s performance improved, reaching a Top-1 accuracy of 73.7% and a Top-5 accuracy of 95.6%. These results highlight its robustness in real-world conditions, where image quality may vary due to factors like lighting and camera angles.
MobileDeepPill is notable for its small footprint, requiring only 34 MB of runtime memory, making it suitable for mobile devices without cloud offloading. With high-end mobile GPUs, the system’s processing time has been reduced to 270 milliseconds, enabling near real-time pill identification. Although there is potential for improvement in the accuracy of one-side recognition, the strong performance in two-side recognition and overall efficiency demonstrate that MobileDeepPill is a promising tool for mobile pill identification, both in healthcare environments and for everyday use.
Suntronsuk et al. [45] present a pill imprint extraction method that offers a novel approach to pill identification by extracting text from pill imprints, which is essential for linking pill images to databases used by healthcare providers. The method involves a multi-step process, beginning with image normalization and enhancement to improve imprint contrast. Two binarization techniques, Otsu’s thresholding and K-means clustering, were evaluated, with Otsu’s method outperforming K-means in handling engraved and printed imprints.
The system achieved an F-measure of 0.77 for printed imprints, demonstrating effectiveness in this category. However, the overall F-measure of 0.57 reflects challenges in handling complex or noisy areas, particularly for engraved imprints. Despite this, the use of Tesseract for text recognition was successful with binarized images. Future work is needed to refine the binarization step, especially for engraved imprints, potentially through advanced or hybrid techniques. Overall, the method shows promise but requires further improvement for more reliable pill identification.
Wang et al. [143] propose a pill recognition system on the NLM dataset that addresses the challenges of pill identification under real-world conditions, including limited labeled data and the domain shift from controlled environments to consumer images. To overcome these issues, the authors employ data augmentation techniques to generate synthetic images, enriching the training dataset and enhancing model robustness.
The system uses a Convolutional Neural Network (CNN), specifically the GoogLeNet Inception Network, with three models trained to specialize in color, shape, and feature extraction. This ensemble approach aims to improve recognition performance. The system achieved a Mean Average Precision (MAP) score of 0.328 on the NLM dataset, which reflects its ability to recognize pills but also highlights difficulties in dealing with noise, background interference, poor lighting, and varying image quality.
Key contributions include data augmentation and model ensembling, but the relatively modest MAP score indicates the need for further improvements. The authors suggest future extensions such as dynamic model weights, expanded data augmentation (e.g., JPEG distortions), and pill segmentation to improve performance. Overall, while the system shows promise, further optimization is needed for practical use in real-world pill recognition scenarios.
Vieira Neto et al. [63] present the CoforDes feature extraction method, which demonstrates exceptional performance on the NIH NLM PIR dataset, achieving 99.82% accuracy and 99.91% specificity. This highlights its effectiveness in classifying pill images based on shape and color, outperforming traditional descriptors such as GLCM, SCM, LBP, and moments (e.g., Zernike and Hu). The method’s rapid extraction speed of 0.00810 s per image further underscores its suitability for real-time applications, making it a robust tool for pill identification in critical healthcare settings.
Additionally, the study introduces a national dataset of Brazilian pills with 100 classes, marking a valuable contribution to the field. Despite its limited size, this dataset provides a basis for future expansion, particularly given the large number of drug classes in Brazil.
Future improvements focus on two key areas: expanding the dataset to include more pill classes and incorporating texture features into CoforDes. The latter is expected to enhance its ability to handle variations not fully captured by shape and color alone, increasing robustness and accuracy in more complex scenarios.
Overall, CoforDes shows great promise as a reliable and efficient solution for pill recognition, with clear potential for real-world deployment. Its combination of high accuracy, speed, and scalability positions it as a leading approach in the field, with further advancements likely to solidify its impact.
Cordeiro et al. [88] propose a pill classification system that demonstrates outstanding performance on the NLM PIR dataset, achieving over 99.3% average accuracy, with precision and recall exceeding 98%. These metrics highlight the system’s robustness and reliability, especially in handling unbalanced classes, as evidenced by its high Matthews Correlation Coefficient (MCC) score above 0.98.
The method leverages computationally efficient machine learning algorithms, including Support Vector Machines (SVM) and Multilayer Perceptron (MLP), which classify pills based on shape and color features extracted through image processing techniques. This method demonstrates high accuracy and remains robust to translation and rotation, making it suitable for images taken under different conditions, including those captured with consumer-grade cameras.
Compared to related studies, this system offers similar or superior accuracy at a significantly lower computational cost, enhancing its suitability for real-time applications. Future directions include testing the method on images captured in less controlled settings, incorporating additional attributes such as texture and imprints, and exploring content-based image retrieval (CBIR) and legal vs. illegal pill differentiation.
Overall, the system is highly promising for practical pill classification tasks, especially in controlled environments. Further work in real-world scenarios will determine its broader applicability and potential impact.
Larios Delgado et al. [86] present a deep learning-based prescription pill identification system that demonstrated significant efficacy on the NIH NLM Pill Image Recognition Challenge dataset, achieving a top-5 accuracy of 94%, surpassing the original competition winner’s accuracy of 83.3%. This remarkable performance underscores the system’s ability to address a critical need in healthcare by reducing preventable medical errors, a leading cause of patient harm.
The study highlights the potential of artificial intelligence (AI) to transform medication reconciliation and enhance clinical workflows by leveraging deep learning for precise pill identification. The system’s capability to work with mobile images further emphasizes its practicality and accessibility for healthcare providers, promoting integration into real-world applications.
This work aligns with healthcare goals of improving patient outcomes, reducing costs, enhancing population health, and supporting providers’ work-life balance. Although the results are promising, future research should assess the system’s robustness by using diverse real-world datasets to ensure its performance across different conditions.
The 94% top-5 accuracy achieved in this study underscores the promising role of AI in reducing medication errors and improving patient safety in clinical environments.
Marami et al. [98] propose a deep learning-based method for the automatic detection and classification of prescription medications, which achieved outstanding performance, with an overall accuracy of 91.2% and a top-5 accuracy of 98.4% on the NIH NLM PIR dataset. These results underscore its robustness in identifying diverse medications, making it highly suitable for applications like drug take-back programs.
A key strength of the system is its ability to distinguish hazardous medications from non-hazardous ones with 98.4% accuracy, ensuring compliance with DEA regulations and enhancing the safety of medication disposal practices. This capability minimizes the risk of environmental and public health hazards arising from improper disposal or mixing of medications.
Beyond classification, the system offers broader utility by enabling data collection on medication wastage patterns, which can inform pharmaceutical supply chain improvements, drug monitoring, and diversion prevention. Its compatibility with mobile devices enhances accessibility, fostering greater involvement in pill take-back programs and supporting sustainable disposal practices.
In summary, the system’s high accuracy and practical applications position it as a promising tool for improving the efficiency of drug take-back programs, reducing hazardous waste, and supporting public health and environmental safety initiatives.
Ling et al. [101] propose a pill recognition system that demonstrates strong performance on the NLM dataset, effectively addressing challenges related to few-shot learning and real-world imaging conditions. A notable contribution of this study is the introduction of the CURE dataset, which provides a more extensive and diverse set of instances per class under varied conditions, enhancing the model’s generalization capabilities for complex recognition tasks.
A key innovation is the
Additionally, the model excels in processing hard samples, particularly those with imprinted text, by emphasizing high-frequency components and incorporating texture information. The use of text imprint as an auxiliary stream in the multi-stream architecture significantly improves recognition accuracy in challenging scenarios.
Overall, the proposed system outperforms state-of-the-art models on both the NLM and CURE datasets, demonstrating its robustness and effectiveness in real-world pill recognition tasks, particularly in settings with limited data and unconstrained imaging conditions. This combination of enhanced segmentation, innovative architecture, and efficient training strategies positions it as a leading solution for practical applications in pill identification.
Al-Hussaeni et al. [151] evaluate three deep learning-based pill recognition methods in this study, including two hybrid models (CNN+SVM and CNN+kNN) and the ResNet-50 architecture, to improve pill image segmentation and classification. Among these, the CNN+kNN model achieved the highest accuracy of 90.8% on the NLM dataset, outperforming existing methods by approximately 10% while maintaining a fast runtime of about 1 ms per execution.
The hybrid approach, particularly CNN+kNN, demonstrated superior accuracy over traditional methods without a significant increase in computational cost, making it suitable for practical applications. This highlights the potential of combining convolutional neural networks with powerful classifiers like k-NN for recognizing complex or incomplete pill images.
Despite its strengths, the model’s accuracy can be impacted by poor lighting conditions, which affect pill shape detection. To mitigate this, the authors proposed capturing multiple images from various angles to construct a 3D model, enhancing robustness under diverse lighting scenarios.
Overall, the study underscores the feasibility of using deep learning for pill image recognition, achieving high accuracy and rapid processing speeds. This approach shows promise for improving medication safety and accuracy, supporting error prevention in clinical and pharmaceutical settings.
In summary, deep learning-based methods have shown superior performance compared to traditional image processing and machine learning approaches in pill recognition tasks. Traditional image processing methods such as those by Caban et al. [17], Suntronsuk et al. [45] have shown significant promise in automatic pill identification, achieving high accuracy in shape, color, and imprint recognition. However, these methods often struggle with image variability such as different views, lighting conditions, and noise. In contrast, traditional machine learning approaches, such as those developed by Ushizima et al. [55] and Vieira Neto et al. [63], have achieved promising results, especially with shape and color-based classification, but still face challenges in handling irregular shapes and achieving high accuracy across diverse datasets. Hybrid approaches, such as those proposed by Yaniv et al. [141] and Zeng et al. [142], have made considerable progress by combining traditional methods with machine learning techniques, resulting in improved outcomes in real-world situations by tackling dataset limitations and image quality challenges.
However, the real advancement in performance is seen in deep learning methods. Studies by Cordeiro et al. [88], Larios Delgado et al. [86], and Ling et al. [101] have demonstrated that deep learning models, particularly those leveraging convolutional neural networks (CNNs), achieve remarkable accuracy and robustness across various pill recognition tasks. These methods not only outperform traditional and hybrid models in terms of accuracy but also show greater adaptability to real-world conditions such as varying lighting, image resolution, and complex pill features. The ability of deep learning models to process large, diverse datasets and learn distinct features like those in the CURE dataset used by Ling et al. [101] has led to improved performance in challenging scenarios, such as few-shot learning and identifying pills with imprinted text. Moreover, deep learning methods, exemplified by the work of Marami et al. [98], have achieved exceptional results, including a top-5 accuracy of 98.4%, highlighting their effectiveness in not only recognizing pills but also distinguishing hazardous medications.
In conclusion, deep learning techniques surpass traditional image processing and machine learning approaches in both accuracy and robustness, making them the most promising solution for practical pill recognition applications in healthcare and everyday use.
5.2 Performance Comparison of Methods on Dataset mCURE
The comparison of the few-shot class-incremental learning (FSCIL) frameworks introduced in Zhang et al. [130,139] reveals notable advancements in automatic pill recognition systems, particularly for dynamic and resource-constrained environments. Both studies address the challenges posed by the continuously increasing categories of pills and the limited availability of annotated data, yet their distinct architectural designs and methodological approaches lead to different levels of performance on the mCure dataset. The framework proposed in Zhang et al. [130] adopt a decoupled learning strategy that separates representation learning and classifier adaptation. The representation learning leverages a Center-Triplet (CT) loss to enhance intra-class compactness and inter-class separability, while the classifier adaptation employs a Graph Attention Network (GAT) trained on pseudo pill images to accommodate new classes incrementally. In contrast, the method presented in Zhang et al. [139] build on this foundation by integrating Discriminative and Bidirectional Compatible Few-Shot Class-Incremental Learning (DBC-FSCIL). This framework introduces forward-compatible learning, which synthesizes virtual classes as placeholders in the feature space to enrich semantic diversity and support future class updates. It also incorporates backward-compatible learning, employing uncertainty quantification to generate reliable pseudo-features of old classes, which facilitates effective Data Replay (DR) and Knowledge Distillation (KD) for balancing memory efficiency and knowledge retention. The superior performance of Zhang et al. [139] on the mCure dataset can be attributed to its more comprehensive management of the feature space and its robust approach to preserving old-class knowledge during incremental updates. By synthesizing virtual classes, the framework anticipates and incorporates future class distributions, ensuring a richer training dataset compared to the reliance on pseudo image generation in Zhang et al. [130]. Furthermore, the uncertainty-based synthesis of pseudo-features allows Zhang et al. [139] to mitigate catastrophic forgetting more effectively, addressing a critical limitation in Zhang et al. [130], which lacks explicit mechanisms for backward compatibility. Additionally, the use of DR and KD strategies in Zhang et al. [139] optimize the trade-off between performance and storage requirements, further highlighting its adaptability to real-world applications.
Overall, while both frameworks demonstrate significant advancements in FSCIL for pill recognition, the holistic design of Zhang et al. [139], balancing forward and backward compatibility, enables it to achieve superior results on mCure. This underscores the importance of simultaneously addressing feature discrimination, knowledge retention, and adaptability in designing FSCIL systems for practical scenarios.
5.3 Performance Comparison of Methods on Dataset VAIPE
The VAIPE dataset presents unique challenges for pill identification and recognition due to the high visual similarity among pills, multi-pill scenarios, and unconstrained conditions of real-world images. Several deep learning-based approaches have been proposed to address these issues, leveraging external knowledge, novel architectures, and multi-modal learning techniques. Among these, the PIKA framework, introduced by Duy et al. [127], integrates external prescription-based knowledge graphs with image features via a lightweight attention mechanism, achieving an F1-score improvement of 4.8%–34.1% over baseline methods. This approach underscores the critical role of external knowledge graphs in enhancing recognition accuracy, though its scalability is constrained by the availability of accurate prescription data. Meanwhile, the PIMA framework, proposed by Thanh et al. [128], addresses the pill-prescription matching task by leveraging Graph Neural Networks (GNNs) and contrastive learning to effectively align textual and visual representations. PIMA demonstrates notable performance gains, improving the F1-score from 19.05% to 46.95%, while maintaining efficiency with limited training costs. For multi-pill detection in real-world settings, the PGPNet framework, developed by Duy et al. [132], integrates heterogeneous a priori graphs to model co-occurrence likelihood, size relationships, and visual semantics of pills. PGPNet achieves significant improvements, with COCO mAP metrics increasing by 9.4% compared to Faster R-CNN and 12.0% over YOLO-v5, while also emphasizing robustness and explainability. However, its dependency on extensive external knowledge poses scalability challenges. Collectively, these methods address different facets of the pill recognition problem, with PIKA excelling in single-pill identification, PIMA optimizing pill-prescription matching, and PGPNet advancing multi-pill detection in complex scenarios. This comparative analysis highlights the critical importance of external knowledge, multi-modal learning, and tailored frameworks in addressing the challenges posed by the VAIPE dataset, while also identifying areas for future research, including scalability, expanded knowledge bases, and computational optimization.
5.4 Performance Comparison of Methods on Dataset VAIPE-PCIL
Nguyen et al. [118] addressed the challenge of catastrophic forgetting in pill image classification by introducing the Incremental Multi-stream Intermediate Fusion (IMIF) framework. This approach integrates an additional guidance stream, leveraging color histogram information, to enhance traditional class incremental learning (CIL) systems. The authors proposed Color Guidance with Multi-stream Intermediate Fusion (CG-IMIF), which can be seamlessly incorporated into existing exemplar-based CIL methods. Experimental evaluation on the VAIPE-PCIL dataset revealed that CG-IMIF significantly outperforms state-of-the-art methods, achieving accuracies of 76.85% (N = 5), 69.94% (N = 10), and 64.97% (N = 15) under varying task settings. These results highlight CG-IMIF’s robustness in handling incremental learning tasks in real-world pill classification scenarios, offering a promising solution for smart healthcare applications.
5.5 Performance Comparison of Methods on Dataset ePillID
Usuyama et al. [91] introduced ePillID, the largest public benchmark for pill image recognition, consisting of 13 k images representing 8184 appearance classes, corresponding to 4092 pill types with two sides. This dataset is particularly challenging due to its low-shot recognition setting, where most classes have only a single reference image. The authors evaluated various baseline models, with a multi-head metric learning approach with bilinear features yielding the best performance. Despite this, error analysis revealed that these models struggled to reliably distinguish particularly confusing pill types. The paper also highlights future directions, including the integration of Optical Character Recognition (OCR) to address challenges such as low-contrast imprinted text, irregular layouts, and pill materials like capsules and gels. This benchmark, aimed at advancing pill identification systems, also plans to expand with additional pill types and images, fostering further research in this critical healthcare task.
Based on the insights gathered from the Literature Survey, Benchmark Datasets, and the Comparison of Performance of Methods, several open research problems in the field of pill image recognition remain. In this section, we will explore these unresolved issues and discuss potential research directions that could drive future progress in the field. By identifying these open problems, we aim to provide a roadmap for researchers to focus on the most critical areas for further development and innovation in pill image recognition.
6.1 Integrating Pill Recognition Systems into Resource-Constrained Environments
In recent years, wearable devices designed to assist visually impaired individuals with pill recognition have gained significant attention. Notably, the works by Chang et al. [83,94] contribute valuable insights into this area, proposing systems that leverage deep learning models for pill identification. Despite the promising potential of these devices, several challenges hinder their real-world applicability. One major issue is the difficulty of accurately recognizing pills under adverse conditions, such as low lighting, complex backgrounds, or partial occlusion. These factors significantly reduce the effectiveness of the systems, particularly in emergency situations. Furthermore, the large diversity of medications, varying in shape, color, and imprint, presents scalability challenges, as deep learning models require extensive training datasets to ensure reliable identification across a wide range of pill types.
The computational demands of deep learning models, such as Convolutional Neural Networks (CNNs), further complicate the deployment of these systems on resource-constrained devices like smart glasses, highlighting the need for hardware optimization. Additionally, enhancing user experience is crucial, as wearable devices must provide seamless interfaces with fast response times to be practical in everyday use.
To address these challenges, future research could focus on improving recognition models by exploring advanced architectures such as Vision Transformers (ViT) or hybrid CNN-ViT models. Data augmentation methods can be used to improve the model’s robustness across different conditions, while lightweight models such as MobileNet or EfficientNet could enhance performance for wearable devices. Moreover, the integration of Augmented Reality (AR) could significantly improve user interaction, making pill recognition more intuitive. AR overlays could display crucial information, such as the pill name, dosage, and usage instructions, directly onto the user’s field of view, facilitating easy identification without the need to focus on a separate screen. The combination of AR with haptic feedback or voice prompts could further enhance usability by providing alternative modes of interaction, particularly for users with limited vision. In addition, AR could help address the challenge of recognizing pills in complex environments by emphasizing key features like shape and imprint, thereby improving identification accuracy in real-world scenarios.
Ultimately, extensive real-world testing and collaborations with healthcare organizations will be essential to ensure the seamless integration of these systems into healthcare workflows, improving safety and efficiency. The advancements in AR, along with lightweight model architectures and enhanced data processing techniques, hold the potential to transform wearable pill recognition systems into reliable tools for visually impaired individuals in their daily lives.
Several studies, including those by Hartl et al. [9], Cunha et al. [51], Dang et al. [135], Zeng et al. [142], and Mehmood et al. [145], have contributed significantly to the advancement of mobile-based pill recognition systems, providing portable, real-time solutions. However, challenges remain, particularly in adapting these systems to dynamic real-world environments with variable lighting, pill orientations, occlusions, and complex backgrounds, all of which can affect performance. Furthermore, these systems are often limited by the need for specific datasets, which pose scalability challenges due to the wide variety of pill appearances and regional differences in pill design. Mobile devices’ resource constraints also create barriers, as many models must balance computational efficiency with recognition accuracy. Additionally, the user experience, particularly for vulnerable populations like the elderly or visually impaired, requires further improvement in terms of accessibility, ease of use, and response times.
To overcome these limitations, future research should focus on enhancing algorithmic robustness through hybrid or domain adaptation techniques, as well as ensuring broader model generalization by incorporating more diverse and expansive datasets for continuous learning. Lightweight architectures like MobileNet or EfficientNet, combined with model optimization strategies, can address the computational constraints of mobile hardware. Moreover, integrating Augmented Reality (AR) into mobile pill recognition systems can significantly improve usability. AR can guide users in real-time by overlaying visual cues, such as alignment instructions or pill information, directly onto the pill in their view. This integration can be enhanced with tactile or voice feedback to further improve accessibility for users with impaired vision or limited mobility.
For these advancements to be truly effective, rigorous real-world testing and collaboration with healthcare providers will be necessary to ensure the safety, reliability, and compliance of these systems with regulatory standards. By addressing these challenges, mobile-based pill recognition systems can evolve into more robust, scalable, and user-friendly solutions, improving accessibility and patient outcomes, particularly for those in need of immediate pill identification.
6.2 Developing Datasets for Multi-Region Markets
A major challenge in pill image recognition is the creation of diverse and representative datasets that can accurately capture the wide variety of pill appearances found globally. This is particularly true for the Vietnamese market, where the pharmaceutical landscape includes both locally manufactured and imported medications. Existing international datasets often fall short in representing the full spectrum of pills available in Vietnam, leading to potential gaps in the performance and generalizability of pill recognition systems. For instance, while the VAIPE dataset [118] is useful for the Vietnamese context, it is primarily focused on prescription medications and includes a limited variety of pills, most of which are generic, with prescription information provided in Vietnamese.
To address this issue, there is an urgent need to develop a more comprehensive dataset that encompasses a wider range of pill types, reflecting the diversity of the Vietnamese pharmaceutical market.
Beyond the Vietnamese market, it is also essential to advocate for the development and use of diverse datasets that represent pills from various regions, conditions, and manufacturing standards. Such datasets should capture the global diversity of medications, enabling pill recognition systems to operate effectively across different environments and cultural contexts. This broader approach would not only improve recognition accuracy but also enhance the scalability and adaptability of systems deployed in resource-constrained settings.
The integration of pill recognition systems into resource-constrained environments, such as wearable devices for visually impaired individuals, is increasingly important. Wearable technologies, like smart glasses, are being developed to assist users with real-time pill identification. However, to ensure these systems’ effectiveness in such settings, datasets must reflect real-world variations, such as low lighting, complex backgrounds, and diverse pill types. Without a rich and diverse dataset, these systems may struggle to accurately recognize pills under challenging conditions, thereby limiting their practical utility in daily life.
Creating a robust, region-specific dataset for Vietnam, alongside the integration of international datasets, would require collaboration between local healthcare institutions, pharmaceutical companies, research organizations, and regulatory bodies. This initiative would improve the accuracy of pill recognition systems in Vietnam, facilitating their adoption in local healthcare practices and improving medication safety and patient compliance. Moreover, such a dataset would contribute to advancing the field of machine learning and computer vision within the pharmaceutical industry, providing a valuable resource for researchers and developers. Ultimately, the development of diverse and expansive pill image datasets will foster innovation and lead to better healthcare outcomes in Vietnam and beyond.
Additionally, integrating Augmented Reality (AR) into these systems could significantly enhance user interaction, making pill recognition more intuitive. AR overlays could display crucial information, such as the pill name, dosage, and usage instructions, directly onto the user’s field of view, facilitating easy identification without the need to focus on a separate screen. The combination of AR with haptic feedback or voice prompts could further improve usability, particularly for users with limited vision. Furthermore, AR could help address the challenge of recognizing pills in complex environments by emphasizing key features like shape and imprint, thereby improving identification accuracy in real-world scenarios.
Ultimately, extensive real-world testing and collaborations with healthcare organizations will be essential to ensure the seamless integration of these systems into healthcare workflows, improving safety and efficiency. The advancements in AR, along with lightweight model architectures and enhanced data processing techniques, hold the potential to transform wearable pill recognition systems into reliable tools for visually impaired individuals in their daily lives. By addressing these challenges through the development of diverse datasets and innovative technologies, we can enhance pill recognition systems’ effectiveness and expand their utility, particularly in underserved or resource-constrained settings.
6.3 Improving Performance in Pill Image Segmentation and Imprint Identification
In the methods presented in the literature review, the authors used the NLM [141], ePillID [91], VAIPE [118] and CURE [101] datasets. The NLM dataset was collected under ideal conditions, while ePillID, CURE and VAIPE were collected under natural conditions, so they are suitable for pill image recognition under real-world conditions. However, the VAIPE dataset is suitable for multi-pill image recognition in the context of prescriptions and is suitable for generic pill recognition. ePillID dataset is particularly challenging due to its low-shot recognition setting, where most classes have only a single reference image, making it difficult for models to generalize. In this section, this article focuses on the recognition of pill images of various types, in natural conditions, so this article focuses on improving the algorithms using the CURE dataset.
The accuracy and other metrics presented by Ling et al. (2020) in [101] on the CURE dataset emphasize the necessity for advancements in the pill image segmentation phase. In the above paper, the author uses the
6.3.1 Building a New Model Based on Combining
Image segmentation plays a crucial role in enhancing the performance of pill recognition systems, especially when dealing with datasets such as CURE, which contain natural conditions, complex backgrounds, and limited data. Accurate segmentation isolates the pill from distracting backgrounds, emphasizing critical features such as color, shape, and imprint. When segmentation is inaccurate, subsequent recognition stages may suffer from noise introduced by irrelevant elements, leading to reduced performance. Therefore, selecting an appropriate segmentation method is essential to improve recognition performance, particularly for datasets like CURE that are characterized by challenging visual diversity and limited training samples.
The
To address this, leveraging transfer learning using a model pre-trained on datasets with similar characteristics emerges as a viable alternative. Transfer learning not only reduces computational costs but also enhances generalization on small and complex datasets such as CURE.
The U-Net architecture is one of the most prominent and effective models for image segmentation tasks, particularly in the medical field. Its distinctive U-shaped design consists of two main parts: the encoder and the decoder. The encoder employs convolutional and pooling layers to extract complex features and reduce the spatial dimensions of the image, while the decoder restores spatial information using upsampling or transposed convolution layers to reconstruct the image to its original size. A key feature of U-Net is the use of skip connections, which directly transfer features from corresponding layers in the encoder to the decoder. This mechanism preserves detailed information and enhances accuracy, especially along object boundaries. Thanks to its powerful processing capabilities and relatively low data requirements, U-Net has become a standard for applications such as medical image segmentation.
In the context of recognizing images of pills with few data and complex backgrounds, we need U-Net-based algorithms that have been pre-trained on datasets similar to the data of pill images with complex backgrounds to optimize performance.
Popular algorithms based on U-Net that have been pre-trained on popular datasets include U-Net++, Attention U-Net, Res-U-Net,
Pill images often exhibit significant variability in shape, color, and imprint (e.g., logos, numbers, or letters), while being situated on backgrounds that range from simple to highly cluttered. Such attributes mirror the challenges present in SOD datasets. Leveraging
The architectural strengths of
6.3.2 Developing Deep TextSpotter (DTS) Pill Imprint Extraction Method Based on Improvement and Flexible Application of Existing Algorithms
Ling et al. [101] used the Deep TextSpotter (DTS) method for pill imprint extraction [104], a framework for text localization and recognition in scenes, the model is trained for both text detection and recognition in scenes in a single framework. This framework uses a region-based text detection model to identify regions in the image that are likely to contain text. This model uses methods such as Region Proposal Networks (RPN) to generate text region proposals. After identifying text regions, a recognition network is used to classify the characters in these regions. This recognition model uses Recurrent Neural Networks (RNN) and Convolutional Neural Networks (CNN) architectures to process and recognize text strings from the suggested regions.
The Deep TextSpotter framework is designed to be end-to-end trainable, meaning that both the detection and recognition models are trained together. This helps to minimize the cumulative error between the detection and recognition steps, thereby improving the overall performance of the system. However, DTS may be limited in accuracy and performance when applied to complex text cases or low-quality images. Therefore, another algorithm may be needed to effectively recognize the pill’s imprint.
Recent advancements in pill image recognition have seen the integration of Transformer-based models, which offer significant improvements over traditional convolutional neural networks (CNNs) due to their self-attention mechanism. Unlike CNNs, which function by processing local receptive fields and pooling layers, Transformers are built to capture long-range dependencies and contextual information throughout the entire image. This self-attention mechanism enables Transformers to focus on different regions of the image simultaneously, rather than sequentially, making them highly effective in recognizing fine-grained details. In the context of pill image recognition, this ability is particularly beneficial for detecting subtle variations in shape, text, and imprints, which are crucial for accurate identification. Transformers can also handle complex image conditions, such as varying backgrounds, lighting, and distortion, better than CNNs, further enhancing their suitability for pill image recognition tasks.
Two notable examples of Transformer-based models applied to pill imprint recognition are PP-OCR-v3 [160] and Robust-Scanner [161]. Both models leverage the strengths of Transformer architecture to improve the accuracy of optical character recognition (OCR) and imprint identification. In PP-OCR-v3, a combination of convolutional layers for feature extraction and Transformer-based sequence modeling for text recognition allows the model to process pill imprint images with greater precision. The Transformer encoder in this model helps capture the global context of the text imprints, enabling the system to recognize sequences of characters even in challenging conditions, such as noisy or partially obscured images. Similarly, Robust-Scanner uses a Transformer-based encoder-decoder architecture, with the encoder transforming the image into a sequence of features, while the decoder decodes these features to produce the recognized text. The self-attention mechanism within the encoder ensures that long-range relationships between different parts of the imprint, such as text or logos, are accurately captured, leading to more reliable recognition results. Both models demonstrate the effectiveness of Transformer-based architectures in pill image recognition, offering improved performance over traditional CNN-based methods, particularly in handling complex and variable image conditions.
6.3.3 Building a New Model Based on Superpixels Combined with
The integration of superpixel methods with the
Wang et al. [162] review many Superpixel segmentation algorithms, for example: SLIC [163], LSC [164], ERS [165], SEEDS [166], PB [167], CRS [168], LRW [169], TPS [170], CS [171,172], CIS [171], N-cut [173], Superpixel Lattices [174], VCells [172], Turbopixel [175], and Watershed [176].
We choose an appropriate method for pill image segmentation involves assessing the suitability of various superpixel segmentation techniques for handling the specific details and structures within pill images. Among the methods discussed, SLIC (Simple Linear Iterative Clustering) emerges as the most suitable choice for several reasons:
Firstly, SLIC is valued for its simplicity and efficiency. It uses a k-means clustering algorithm to segment pixels in both color and spatial domains. This simplicity helps minimize processing time and computational demands, which is particularly important when dealing with large pill images that contain complex details.
Secondly, SLIC produces superpixels that are uniformly sized and nearly rectangular, which helps preserve well-defined boundaries between the different regions of the pill image. This characteristic is crucial for distinguishing various components of the pill, such as the coating, core, and finer details.
Lastly, SLIC is known for its high segmentation quality, especially in applications that require detailed image analysis like pill segmentation. It consistently creates even and reliable superpixels, aiding in the accurate analysis and recognition of small structural details within the pill images.
When comparing SLIC to other methods, its advantages become clearer. LSC (Linear Spectral Clustering), while effective for spectral feature-based segmentation, might be too complex for the relatively straightforward task of pill image segmentation. ERS (Edge-Relabeling Segmentation), based on edge segmentation, may not maintain the uniformity of superpixels as well as SLIC. SEEDS (Superpixels Extracted via Energy-Driven Sampling), though it produces high-quality superpixels, can be more resource-intensive compared to SLIC. PB (Pyramid-based) segmentation is suitable for multi-resolution images but may not be ideal for pill segmentation, which generally does not require multi-scale analysis. CRS (Contour-based) methods focus on contours and might not be effective in maintaining uniform superpixels. LRW (Learning-based Recurrent Model), which uses deep learning, could achieve higher accuracy but demands significantly more computational resources. TPS (Imprint-Partitioning Segmentation), which segments based on imprint, might not align well with the specific needs of pill image details. CS (Connected Component) segmentation could fail to produce uniform superpixels. CIS (Clustered Image Segmentation) might lack the necessary uniformity for effective pill segmentation. N-cut (Normalized Cut), while useful, tends to be more complex and might not be necessary for simpler segmentation tasks. Superpixel Lattices and VCells are unlikely to provide the consistent superpixel sizes or appropriate segmentation for pill images. Turbopixel generates superpixels quickly but may not maintain the detailed boundaries as effectively as SLIC. Lastly, Watershed segmentation, using terrain concepts, may not preserve the necessary uniformity and detailed boundaries required for pill segmentation.
Therefore, SLIC stands out as the preferred method for pill image segmentation due to its balanced approach, providing high accuracy, efficiency, and maintaining crucial details within the images.
6.4 Interdisciplinary Collaboration in Pill Image Recognition
Despite the significant advancements in pill image recognition, one of the open research problems is the need for deeper interdisciplinary collaboration. As this field spans multiple domains, including computer vision, machine learning, pharmacology, and clinical medicine, research in pill image recognition would greatly benefit from the integration of knowledge and expertise from these diverse fields.
Collaboration with pharmacologists can ensure that recognition systems account for the specific characteristics of pill imprints, shapes, and colors that are critical for accurate classification. Meanwhile, clinical medicine can help refine these systems, ensuring that they are not only accurate but also useful in real-world healthcare applications such as medication verification and patient safety. Computer vision and machine learning experts can improve model performance, but understanding the complexities of medication usage and patient populations will require input from other disciplines.
Thus, fostering collaboration across these domains is not just an opportunity but a necessity for advancing the state of pill image recognition and addressing challenges like dataset limitations, algorithm accuracy, and clinical applicability. This collaboration has the potential to open new research and development opportunities, fostering the creation of more efficient, scalable, and clinically applicable systems.
In this survey, we review recent methods for pill image recognition using pill image datasets. We summarize the main techniques employed by various authors and classify them into four groups, comparing the advantages and disadvantages of these groups. We also discuss a range of benchmark datasets for pill image recognition used in recent methods. Specifically, we compare six popular datasets: NLM, ePillID, CURE, mCURE, VAIPE, and VAIPEPCIL. Additionally, we evaluate the metrics used in studies involving the NLM, mCURE, and VAIPE datasets. Finally, we suggest potential research directions for advancing pill image recognition.
Acknowledgement: We would like to thank Nguyen Tat Thanh University, Ho Chi Minh City, Vietnam, Faculty of Information Technology, School of Technology, Van Lang University, Vietnam, Faculty of Information Technology, Ho Chi Minh City Open University, Vietnam for the support of time and facilities for this study.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: The authors confirm contribution to the paper as follows: Study conception and design, collection, analysis and interpretation of results, draft manuscript preparation: Linh Nguyen Thi My. Review paper: Viet-Tuan Le, Tham Vo, Vinh Truong Hoang. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data that support the findings of this study are available from the corresponding author, upon reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Lee YB, Park U, Jain AK. PILL-ID: matching and retrieval of drug pill imprint images. In: 2010 20th International Conference on Pattern Recognition; 2010; Istanbul, Turkey: IEEE. p. 2632–5. [Google Scholar]
2. Canny J. A computational approach to edge detection. IEEE Trans Pattern Anal Mach Intell. 1986 Nov;PAMI-8(6):679–98. doi:10.1109/TPAMI.1986.4767851. [Google Scholar] [CrossRef]
3. Hu MK. Visual pattern recognition by moment invariants. IEEE Trans Inf Theory. 1962 Feb;8(2):179–87. doi:10.1109/TIT.1962.1057692. [Google Scholar] [CrossRef]
4. Jain A, Nandakumar K, Ross A. Score normalization in multimodal biometric systems. Pattern Recognit. 2005 Dec;38(12):2270–85. doi:10.1016/j.patcog.2005.01.012. [Google Scholar] [CrossRef]
5. Pill Identifier Tool. [cited 2024 Apr 17]. Available from: https://www.drugs.com/pill_identification.html. [Google Scholar]
6. Morimoto M, Fujii K. A visual inspection system for drug tablets. In: 2011 IEEE International Conference on Systems, Man, and Cybernetics; 2011; Anchorage, Alaska, USA: IEEE. p. 1106–10. [Google Scholar]
7. Wolberg G, Zokai S. Robust image registration using log-polar transform. In: Proceedings of the 2000 International Conference on Image Processing (ICIP 2000); 2000; Vancouver, BC, Canada: IEEE. Vol. 1, p. 493–6. [Google Scholar]
8. Kim D, Chun J. Drug image retrieval by shape and color similarity of the medication. In: 2011 First ACIS/JNU International Conference on Computers, Networks, Systems and Industrial Engineering; 2011; Jeju, Republic of Korea: IEEE. p. 387–90. [Google Scholar]
9. Hartl A, Arth C, Schmalstieg D. Instant medical pill recognition on mobile phones. In: IASTED International Conference on Computer Vision 2011; 2011; Vancouver, BC, Canada. p. 188–95. [Google Scholar]
10. Hartley R, Zisserman A. Multiple view geometry in computer vision. UK: Cambridge University Press; 2003. [Google Scholar]
11. Shafait F, Keysers D, Breuel TM. Efficient implementation of local adaptive thresholding techniques using integral images. In: Document recognition and retrieval XV. San Jose, CA, USA: SPIE; 2008. Vol. 6815, p. 317–22. [Google Scholar]
12. Chang F, Chen CJ, Lu CJ. A linear-time component-labeling algorithm using contour tracing technique. Comput Vis Image Understand. 2004 Feb;93(2):206–20. doi:10.1016/j.cviu.2003.09.002. [Google Scholar] [CrossRef]
13. Susstrunk SE, Holm JM, Finlayson GD. Chromatic adaptation performance of different RGB sensors. In: Color imaging: device-independent color, color hardcopy, and graphic arts VI. San Jose, CA, USA: SPIE; 2000. Vol. 4300, p. 172–83. [Google Scholar]
14. Hartl A, Arth C, Schmalstieg D. Instant segmentation and feature extraction for recognition of simple objects on mobile phones. In: 2010 IEEE Computer Society Conference on Computer Vision and Pattern Recognition-Workshops; 2010; San Francisco, CA, USA: IEEE. p. 17–24. [Google Scholar]
15. Evans AC, Thacker NA, Mayhew JE. Pairwise representations of shape. In: 1992 11th IAPR International Conference on Pattern Recognition; 1992; The Hague, Netherlands: IEEE Computer Society. Vol. 1, p. 133–6. [Google Scholar]
16. Zhu P, Chirlian PM. On critical point detection of digital shapes. IEEE Transact Pattern Analy Mach Intell. 1995;17(8):737–48. doi:10.1109/34.400564. [Google Scholar] [CrossRef]
17. Caban JJ, Rosebrock A, Yoo TS. Automatic identification of prescription drugs using shape distribution models. In: 2012 19th IEEE International Conference on Image Processing; 2012; Orlando, FL, USA: IEEE. p. 1005–8. [Google Scholar]
18. Osada R, Funkhouser T, Chazelle B, Dobkin D. Shape distributions. ACM Transact Graph. 2002 Oct;21(4):807–32. doi:10.1145/571647.571648. [Google Scholar] [CrossRef]
19. Lee YB, Park U, Jain AK, Lee SW. Pill-ID: matching and retrieval of drug pill images. Pattern Recognit Lett. 2012 May;33(7):904–10. doi:10.1016/j.patrec.2011.08.022. [Google Scholar] [CrossRef]
20. Gonzalez RC, Woods RE. Digital image processing Addison. Boston, MA, USA: Addison-Wesley Longman Publishing Co., Inc.; 1992. [Google Scholar]
21. Lowe DG. Distinctive image features from scale-invariant keypoints. Int J Comput Vis. 2004 Nov;60(2):91–110. doi:10.1023/B:VISI.0000029664.99615.94. [Google Scholar] [CrossRef]
22. Ojala T, Pietikäinen M, Harwood D. A comparative study of texture measures with classification based on featured distributions. Pattern Recognit. 1996 Jan;29(1):51–9. doi:10.1016/0031-3203(95)00067-4. [Google Scholar] [CrossRef]
23. Ojala T, Pietikainen M, Maenpaa T. Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans Pattern Anal Mach Intell. 2002 Jul;24(7):971–87. doi:10.1109/TPAMI.2002.1017623. [Google Scholar] [CrossRef]
24. Drugs.com. [cited 2024 Mar 15]. Available from: https://www.drugs.com/pill_identification.html. [Google Scholar]
25. Chen RC, Chan YK, Chen YH, Bau CT. An automatic drug image identification system based on multiple image features and dynamic weights. Int J Innov Comput Inform Cont. 2012 May;8(5):2995–3013. [Google Scholar]
26. Woods G, Gonzalez RC. Digital image processing. USA: Pearson Prentice Hall; 2002. [Google Scholar]
27. Gevers T, Smeulders AW. Color-based object recognition. Pattern Recognit. 1999;32(3):453–64. doi:10.1016/S0031-3203(98)00036-3. [Google Scholar] [CrossRef]
28. Latha D, Sheela CJJ. Enhanced hybrid CBIR based on multichannel LBP oriented color descriptor and HSV color statistical feature. Multim Tools Appl. 2022 Mar;81(17):23801–18. doi:10.1007/s11042-022-12568-x. [Google Scholar] [CrossRef]
29. Jégou H, Douze M, Schmid C. Improving bag-of-features for large scale image search. Int J Comput Vis. 2010 Aug;87(3):316–36. doi:10.1007/s11263-009-0285-2. [Google Scholar] [CrossRef]
30. MPEG-7 Overview. [cited 2024 Apr 14]. Available from: https://www.mpeg.org/standards/MPEG-7/. [Google Scholar]
31. Arif M. Evaluation of discrimination power of features in the pattern classification problem using Arif index and its application to physiological datasets. Int J Innov Comput, Inform Cont. 2011 Feb;7(2):525–36. [Google Scholar]
32. Sandler R, Lindenbaum M. Optimizing gabor filter design for texture edge detection and classification. Int J Comput Vis. 2009 Apr;84(3):308–24. doi:10.1007/s11263-009-0237-x. [Google Scholar] [CrossRef]
33. Shen LL, Zhen J. Gabor wavelet selection and SVM classification for object recognition. Acta Automatica Sinica. 2009 Apr;35(4):350–5. doi:10.1016/S1874-1029(08)60082-8. [Google Scholar] [CrossRef]
34. Zheng Y. Breast cancer detection with Gabor features from digital mammograms. Algorithms. 2010;3(1):44–62. doi:10.3390/a3010044. [Google Scholar] [CrossRef]
35. Li JB, Gao H, Pan JS. Common vector analysis of Gabor features with kernel space isomorphic mapping for face recognition. Int J Innov Comput Informa Cont. 2010 Sep;6(9):4055–64. [Google Scholar]
36. Yu J, Chen Z, Si K. Pill recognition using imprint information by two-step sampling distance sets. In: 2014 22nd International Conference on Pattern Recognition; 2014; Stockholm, Sweden: IEEE. p. 3156–61. [Google Scholar]
37. Yu J, Chen Z, Kamata S, Yang J. Accurate system for automatic pill recognition using imprint information. IET Image Process. 2015 Dec;9(12):1039–47. doi:10.1049/iet-ipr.2014.1007. [Google Scholar] [CrossRef]
38. Epshtein B, Ofek E, Wexler Y. Detecting text in natural scenes with stroke width transform. In: 2010 IEEE Computer Society Conference on Computer Vision and Pattern Recognition; 2010; San Francisco, CA, USA: IEEE. p. 2963–70. [Google Scholar]
39. Grigorescu C, Petkov N. Distance sets for shape filters and shape recognition. IEEE Trans Image Process. 2003 Oct;12(10):1274–86. doi:10.1109/TIP.2003.816010. [Google Scholar] [PubMed] [CrossRef]
40. Hema A, Anna SE. The identification of pill using feature extraction in image mining. ICTACT J Image Video Process. 2015 Feb;5(3):973–9. doi:10.21917/ijivp.2015.0143. [Google Scholar] [CrossRef]
41. Wang XY, Wu JF, Yang HY. Robust image retrieval based on color histogram of local feature regions. Multimed Tools Appl. 2009 Oct;49(2):323–45. doi:10.1007/s11042-009-0362-0. [Google Scholar] [CrossRef]
42. Bay H, Tuytelaars T, Van Gool L. SURF: speeded up robust features. In: Computer vision–ECCV 2006. Graz, Austria: Springer; 2006. p. 404–17. [Google Scholar]
43. Suntronsuk S, Ratanotayanon S. Pill image binarization for detecting text imprints. In: 2016 13th International Joint Conference on Computer Science and Software Engineering (JCSSE); 2016; Khon Kaen, Thailand: IEEE. p. 1–6. [Google Scholar]
44. Pill Identifier Tool. [cited 2024 Sep 20]. Available from: https://www.drugs.com/imprints.php. [Google Scholar]
45. Suntronsuk S, Ratanotayanon S. Automatic text imprint analysis from pill images. In: 2017 9th International Conference on Knowledge and Smart Technology (KST); 2017; Chonburi, Thailand: IEEE. p. 288–93. [Google Scholar]
46. Gonzalez RC. Digital image processing. Upper Saddle River, NJ, USA: Prentice Hall; 2009. [Google Scholar]
47. Mancas-Thillou C, Mancas M. Comparison between pen-scanner and digital camera acquisition for engraved character recognition. In: Proceeding of the 2nd. International Workshop on Camera-Based Document Analysis and Recognition; 2007; Grand Hotel Rayon, Curitiba, Brazil. Vol. 130, p. 137. [Google Scholar]
48. Smith R. An overview of the tesseract OCR engine. In: Ninth International Conference on Document Analysis and Recognition (ICDAR 2007); 2007; Curitiba, Brazil: IEEE. Vol. 2, p. 629–33. [Google Scholar]
49. Ranjitha, Kulkarni SS, Rashmi K, Athmashree A. Color and shape recognition of pills using image processing. Int J Eng Res Technol. 2019 Jun;7(8). doi:10.17577/IJERTCONV7IS08052. [Google Scholar] [CrossRef]
50. Chokchaitam S. Color compensation based on color background shadow for pill identification. In: 2021 4th International Conference on Information Communication and Signal Processing (ICICSP); 2021; Shanghai, China: IEEE. p. 399–403. [Google Scholar]
51. Cunha A, Adão T, Trigueiros P. HelpmePills: a mobile pill recognition tool for elderly persons. Procedia Technology. 2014;16(5):1523–32. doi:10.1016/j.protcy.2014.10.174. [Google Scholar] [CrossRef]
52. Structural Analysis and Shape Descriptors—OpenCV 3.0.0-dev documentation. [cited 2021 May 23]. Available from: https://docs.opencv.org/3.0.0/d3/dc0/group__imgproc__shape.html. [Google Scholar]
53. Yuen H, Princen J, Illingworth J, Kittler J. Comparative study of Hough transform methods for circle finding. Image Vis Comput. 1990 Feb;8(1):71–7. doi:10.1016/0262-8856(90)90059-E. [Google Scholar] [CrossRef]
54. Felzenszwalb PF, Huttenlocher DP. Efficient belief propagation for early vision. Int J Comput Vis. 2006 May;70(1):41–54. doi:10.1007/s11263-006-7899-4. [Google Scholar] [CrossRef]
55. Ushizima D, Carneiro A, Souza M, Medeiros F. Investigating pill recognition methods for a new national library of medicine image dataset. In: Advances in visual computing. Las Vegas, NV, USA: Springer; 2015. p. 410–9. [Google Scholar]
56. Fiji: Fiji is just imageJ. [cited 2024 Feb 24]. Available from: https://fiji.sc/Fiji. [Google Scholar]
57. Fiji-plugins: Analysis tools. [cited 2024 Feb 12]. Available from: https://imagej.net/ij/docs/menus/analyze.html. [Google Scholar]
58. Ja H. A k-means clustering algorithm. J Roy Stat Soc C Appl Stat. 1979 Mar;28(1):100–8. [Google Scholar]
59. Kohonen T. Self-organizing maps. Berlin: Springer; 2001. [Google Scholar]
60. Rousseeuw PJ. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math. 1987 Nov;20:53–65. doi:10.1016/0377-0427(87)90125-7. [Google Scholar] [CrossRef]
61. De Amorim RC, Hennig C. Recovering the number of clusters in data sets with noise features using feature rescaling factors. Inf Sci. 2015 Dec;324(1):126–45. doi:10.1016/j.ins.2015.06.039. [Google Scholar] [CrossRef]
62. Chupawa P, Kanjanawanishkul K. Pill identification with imprints using a neural network. Int J Eng Technol. 2015;1:30–5. [Google Scholar]
63. Vieira Neto MA, de Souza JWM, Reboucas Filho PP, Rodrigues AWDO. CoforDes: an invariant feature extractor for the drug pill identification. In: 2018 IEEE 31st International Symposium on Computer-Based Medical Systems (CBMS); 2018; Karlstad, Sweden: IEEE. p. 30–5. [Google Scholar]
64. Chughtai R, Raja G, Mir J, Shaukat F. An efficient scheme for automatic pill recognition using neural networks. The Nucleus. 2019;56(1):42–8. [Google Scholar]
65. Chen H, Tsai SS, Schroth G, Chen DM, Grzeszczuk R, Girod B. Robust text detection in natural images with edge-enhanced maximally stable extremal regions. In: 2011 18th IEEE International Conference on Image Processing; 2011; Brussels, Belgium: IEEE. p. 2609–12. [Google Scholar]
66. National Library of Medicine, National Institutes of Health, United States. [cited 2024 Mar 23]. Available from: https://datadiscovery.nlm.nih.gov/Drugs-and-Chemicals/Pillbox-retired-January-28-2021-/crzr-uvwg/ab out_data. [Google Scholar]
67. Dhivya AB, Sundaresan M. Tablet identification using support vector machine based text recognition and error correction by enhanced n-grams algorithm. IET Image Process. 2020 Apr;14(7):1366–72. doi:10.1049/iet-ipr.2019.0993. [Google Scholar] [CrossRef]
68. Dhivya AB, Sundaresan M. Enhancing the tablet images using noise reduction algorithms by analyzing different color models. Int J Eng Adv Technol. 2019 Dec;9(2):148–55. doi:10.35940/ijeat.B3119.129219. [Google Scholar] [CrossRef]
69. Hornik K, Mair P, Rauch J, Geiger W, Buchta C, Feinerer I. The textcat package for n-gram based text categorization in R. J Statist Softw. 2013;52(6):1–17. doi:10.18637/jss.v052.i06. [Google Scholar] [CrossRef]
70. Cavnar WB, Trenkle JM. N-gram-based text categorization. In: Proceedings of SDAIR-94, 3rd Annual Symposium on Document Analysis and Information Retrieval; 1994; Las Vegas, NV, USA: SDAIR. p. 161–75. [Google Scholar]
71. Smith R. Limits on the application of frequency-based language models to OCR. In: 2011 International Conference on Document Analysis and Recognition; 2011; Beijing, China: IEEE. p. 538–42. [Google Scholar]
72. Rychly P. A lexicographer-friendly association score. In: Proceedings of the Second Workshop on Recent Advances in Slavonic Natural Language Processing (RASLAN 2008); 2008; Czech Republic: Karlova Studánka. p. 6–9. [Google Scholar]
73. Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv:14091556. 2014. [Google Scholar]
74. Wong YF, Ng HT, Leung KY, Chan KY, Chan SY, Loy CC. Development of fine-grained pill identification algorithm using deep convolutional network. J Biomed Inform. 2017 Oct;74(1):130–6. doi:10.1016/j.jbi.2017.09.005. [Google Scholar] [PubMed] [CrossRef]
75. Yang C, Zhang L, Lu H, Ruan X, Yang MH. Saliency detection via graph-based manifold ranking. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2013; Portland, Oregon, USA: IEEE. p. 3166–73. [Google Scholar]
76. Zhou D, Weston J, Gretton A, Bousquet O, Schölkopf B. Ranking on data manifolds. Adv Neural Inform Process Syst. 2003;16:169–76. [Google Scholar]
77. Zhang Z, Luo P, Loy CC, Tang X. Learning deep representation for face alignment with auxiliary attributes. IEEE Trans Pattern Anal Mach Intell. 2015 May;38(5):918–30. doi:10.1109/TPAMI.2015.2469286. [Google Scholar] [PubMed] [CrossRef]
78. Deng J, Dong W, Socher R, Li LJ, Li K, Fei-Fei L. ImageNet: a large-scale hierarchical image database. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition; 2009; Miami, FL, USA: IEEE. p. 248–55. [Google Scholar]
79. Ou YY, Tsai AC, Wang JF, Lin J. Automatic drug pills detection based on convolution neural network. In: 2018 International Conference on Orange Technologies (ICOT); 2018; Nusa Dua, Bali, Indonesia: IEEE. p. 1–4. [Google Scholar]
80. Lin TY, Dollar P, Girshick R, He K, Hariharan B, Belongie S. Feature pyramid networks for object detection. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2017; Honolulu, HI, USA: IEEE. p. 2117–25. [Google Scholar]
81. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016; Las Vegas, NV, USA: IEEE. p. 770–8. [Google Scholar]
82. Chollet F. Xception: deep learning with depthwise separable convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2017; Honolulu, HI, USA: IEEE. p. 1251–8. [Google Scholar]
83. Chang WJ, Yu YX, Chen JH, Zhang ZY, Ko SJ, Yang TH, et al. A deep learning based wearable medicines recognition system for visually impaired people. In: 2019 IEEE International Conference on Artificial Intelligence Circuits and Systems (AICAS); 2019; Hsinchu, Taiwan: IEEE. p. 207–8. [Google Scholar]
84. Ren S, He K, Girshick R, Sun J. Towards real-time object detection with region proposal networks. IEEE Trans Pattern Anal Mach Intell. 2017 Jun;39(6):1137–49. doi:10.1109/TPAMI.2016.2577031. [Google Scholar] [PubMed] [CrossRef]
85. Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2016; Nevada, USA: Las Vegas. p. 2818–26. [Google Scholar]
86. Larios Delgado N, Usuyama N, Hall AK, Hazen RJ, Ma M, Sahu S, et al. Fast and accurate medication identification. npj Digital Med. 2019 Feb;2(1):10. [Google Scholar] [PubMed]
87. Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. In: Medical image computing and computer-assisted intervention–MICCAI 2015. Munich, Germany: Springer; 2015. p. 234–41. [Google Scholar]
88. Cordeiro LS, Lima JS, Rocha Ribeiro AI, Bezerra FN, Reboucas Filho PP, Rocha Neto AR. Pill image classification using machine learning. In: 2019 8th Brazilian Conference on Intelligent Systems (BRACIS); 2019; Salvador, Brazil: IEEE. p. 556–61. [Google Scholar]
89. Swastika W, Prilianti K, Stefanus A, Setiawan H, Arfianto AZ, Santosa AWB, et al. Preliminary study of multi convolution neural network-based model to identify pills image using classification rules. In: 2019 International Seminar on Intelligent Technology and Its Applications (ISITIA); 2019; Surabaya, Indonesia: IEEE. p. 376–80. [Google Scholar]
90. LeCun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proc IEEE. 1998;86(11):2278–324. doi:10.1109/5.726791. [Google Scholar] [CrossRef]
91. Usuyama N, Delgado NL, Hall AK, Lundin J. ePillID Dataset: a low-shot fine-grained benchmark for pill identification. In: 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW); 2020; Seattle, WA, USA: IEEE. p. 3971–7. [Google Scholar]
92. Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2017; Honolulu, HI, USA: IEEE. p. 4700–8. [Google Scholar]
93. Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, et al. Imagenet large scale visual recognition challenge. Int J Comput Vis. 2015 Apr;115(3):211–52. doi:10.1007/s11263-015-0816-y. [Google Scholar] [CrossRef]
94. Chang WJ, Chen LB, Hsu CH, Chen JH, Yang TC, Lin CP. MedGlasses: a wearable smart-glasses-based drug pill recognition system using deep learning for visually impaired chronic patients. IEEE Access. 2020;8:17013–24. doi:10.1109/ACCESS.2020.2967400. [Google Scholar] [CrossRef]
95. Liu W, Anguelov D, Erhan D, Szegedy C, Reed S, Fu CY, et al. SSD: single shot multibox detector. In: Computer vision–ECCV 2016. Amsterdam, The Netherlands: Springer; 2016. p. 21–37. [Google Scholar]
96. Ou Y, Tsai A, Zhou X, Wang J. Automatic drug pills detection based on enhanced feature pyramid network and convolution neural networks. IET Comput Vis. 2020 Jan;14(1):9–17. doi:10.1049/iet-cvi.2019.0171. [Google Scholar] [CrossRef]
97. Szegedy C, Ioffe S, Vanhoucke V, Alemi A. Inception-v4, inception-resnet and the impact of residual connections on learning. In: Proceedings of the AAAI Conference On Artificial Intelligence; 2017; San Francisco, CA, USA: AAAI Press. Vol. 31, p. 4278–84. [Google Scholar]
98. Marami B, Royaee AR. Automatic detection and classification of waste consumer medications for proper management and disposal. arXiv: 200713903. 2020. [Google Scholar]
99. Chen LC, Zhu Y, Papandreou G, Schroff F, Adam H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In: Computer vision–ECCV 2018. Munich, Germany: Springer; 2018 Sep. p. 801–18. [Google Scholar]
100. Kingma DP, Ba J. Adam: a method for stochastic optimization. arXiv:14126980. 2014. [Google Scholar]
101. Ling S, Pastor A, Li J, Che Z, Wang J, Kim J, et al. Few-shot pill recognition. In: 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2020; Seattle, WA, USA: IEEE. p. 9786–95. [Google Scholar]
102. Newell A, Yang K, Deng J. Stacked hourglass networks for human pose estimation. In: Computer vision–ECCV 2016. Amsterdam, The Netherlands: Springer; 2016. p. 483–99. [Google Scholar]
103. Hinton G, Vinyals O, Dean J. Distilling the knowledge in a neural network. arXiv:150302531. 2015. [Google Scholar]
104. Busta M, Neumann L, Matas J. Deep textspotter: an end-to-end trainable scene text localization and recognition framework. In: 2017 IEEE International Conference on Computer Vision (ICCV); 2017; Venice, Italy: IEEE. p. 2223–31. [Google Scholar]
105. Li H, Eigen D, Dodge S, Zeiler M, Wang X. Finding task-relevant features for few-shot learning by category traversal. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition; 2019; Long Beach, CA, USA: IEEE. p. 1–10. [Google Scholar]
106. Schroff F, Kalenichenko D, Philbin J. Facenet: a unified embedding for face recognition and clustering. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2015; Boston, MA, USA: IEEE. p. 815–23. [Google Scholar]
107. Tsai KL, Liau BY, Hung YM, Yu GJ, Wang YC. Development of smart pillbox using 3D printing technology and convolutional neural network image recognition. Sens Mater. 2020 May;32(5):1907–12. doi:10.18494/SAM.2020.2632. [Google Scholar] [CrossRef]
108. Kwon HJ, Kim HG, Lee SH. Pill detection model for medicine inspection based on deep learning. Chemosensors. 2021 Dec;10(1):4. doi:10.3390/chemosensors10010004. [Google Scholar] [CrossRef]
109. He K, Gkioxari G, Dollar P, Girshick R. Mask R-CNN. IEEE Trans Pattern Anal Mach Intell. 2020 Feb;42(2):386–97. doi:10.1109/TPAMI.2018.2844175. [Google Scholar] [PubMed] [CrossRef]
110. Lester CA, Li J, Ding Y, Rowell B, Yang J, Kontar RA. Performance evaluation of a prescription medication image classification model: an observational cohort. npj Digit Med. 2021 Jul;4(1):118. doi:10.1038/s41746-021-00483-8. [Google Scholar] [PubMed] [CrossRef]
111. Paszke A, Gross S, Chintala S, Chanan G, Yang E, DeVito Z, et al. Automatic differentiation in pytorch. In: NIPS 2017 Workshop on Autodiff; Long Beach, CA, USA: Curran Associates Inc.; 2017. [Google Scholar]
112. National Library of Medicine, PillBox API. [cited 2024 Apr 14]. Available from: https://lhncbc.nlm.nih.gov/RxNav/APIs/index.html. [Google Scholar]
113. Evrim Ozmermer T, Roze V, Hilcuks S, Nescerecka A. VeriMedi: pill identification using proxy-based deep metric learning and exact solution. arXiv: 210411231. 2021. [Google Scholar]
114. He K, Gkioxari G, Dollár P, Girshick R. Mask R-CNN. In: 2017 IEEE International Conference on Computer Vision (ICCV); 2017; Venice, Italy. p. 2980–8. [Google Scholar]
115. Kim S, Kim D, Cho M, Kwak S. Proxy anchor loss for deep metric learning. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition; 2020; Seattle, WA, USA: IEEE. p. 3238–47. [Google Scholar]
116. Tan L, Huangfu T, Wu L, Chen W. Comparison of RetinaNet, SSD, and YOLO v3 for real-time pill identification. BMC Med Inform Decis Mak. 2021 Nov;21(1):129. [Google Scholar]
117. Tan L, Huangfu LWT, Chen W. Comparison of YOLO v3, Faster R-CNN, and SSD for real-time pill identification. Research Square. 2021 Jul. [Google Scholar]
118. Nguyen TT, Pham HH, Nguyen PL, Nguyen TH, Do M. Multi-stream fusion for class incremental learning in pill image classification. In: Computer vision–ACCV 2022. Macao, China: Springer Nature Switzerland; 2022. p. 341–56. [Google Scholar]
119. Suksawatchon U, Srikamdee S, Suksawatchon J, Werapan W. Shape recognition using unconstrained pill images based on deep convolution network. In: 2022 6th International Conference on Information Technology (InCIT); 2022; Nonthaburi, Thailand: IEEE. p. 309–13. [Google Scholar]
120. Redmon J, Farhadi A. YOLOv3: an incremental improvement. arXiv: 180402767. 2018. [Google Scholar]
121. Wu PT, Sun TY, Lin JC, Chin CL. Round pill shape recognition system based on AY deep learning model. In: 2022 IEEE International Conference on Consumer Electronics–Taiwan; 2022; Taipei, Taiwan: IEEE. p. 553–4. [Google Scholar]
122. Chen L, Shi W, Deng D. Improved YOLOv3 based on attention mechanism for fast and accurate ship detection in optical remote sensing images. Remote Sens. 2021 Feb;13(4):660. doi:10.3390/rs13040660. [Google Scholar] [CrossRef]
123. Hurtik P, Molek V, Hula J, Vajgl M, Vlasanek P, Nejezchleba T. Poly-YOLO: higher speed, more precise detection and instance segmentation for YOLOv3. Neural Comput Appl. 2022 Feb;34(10):8275–90. doi:10.1007/s00521-021-05978-9. [Google Scholar] [CrossRef]
124. Pornbunruang N, Tanjantuk V, Titijaroonroj T. Drugtionary: drug pill image detection and recognition based on deep learning. In: Proceedings of the 18th International Conference on Computing and Information Technology (IC2IT 2022); 2022; Kanchanaburi, Thailand: Springer. p. 43–52. [Google Scholar]
125. Lin TY, Goyal P, Girshick R, He K, Dollar P. Focal loss for dense object detection. IEEE Trans Pattern Anal Mach Intell. 2020 Feb;42(2):318–27. doi:10.1109/TPAMI.2018.2858826. [Google Scholar] [PubMed] [CrossRef]
126. Duan K, Bai S, Xie L, Qi H, Huang Q, Tian Q. CenterNet: keypoint triplets for object detection. In: Proceedings of the IEEE/CVF International Conference on Computer Vision; 2019; Seoul, Republic of Korea: IEEE. p. 6569–78. [Google Scholar]
127. Nguyen AD, Nguyen TD, Pham HH, Nguyen TH, Nguyen PL. Image-based contextual pill recognition with medical knowledge graph assistance. In: Recent challenges in intelligent information and database systems. Ho Chi Minh City, Vietnam:Springer; 2022. p. 354–69. [Google Scholar]
128. Nguyen TT, Nguyen HD, Nguyen TH, Pham HH, Ide I, Nguyen PL. A novel approach for pill-prescription matching with GNN assistance and contrastive learning. In: PRICAI 2022: Trends in Artificial Intelligence: 9th Pacific Rim International Conference on Artificial Intelligence; 2022; Shanghai, China: Springer. p. 261–74. [Google Scholar]
129. Sakshi B, Amruta M, Sakshi B, Rohan B, Sayyed JI. Detection and identification pills. Int J Adv Res Innovat Ideas Edu. 2023;9(6):242–7. [Google Scholar]
130. Zhang J, Liu L, Gao K, Hu D. Few-shot class-incremental pill recognition. arXiv: 230411959. 2023. [Google Scholar]
131. Tao X, Hong X, Chang X, Dong S, Wei X, Gong Y. Few-shot class-incremental learning. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition; 2020; Seattle, WA, USA: IEEE. p. 12183–92. [Google Scholar]
132. Nguyen AD, Pham HH, Trung HT, Nguyen QVH, Truong TN, Nguyen PL. High accurate and explainable multi-pill detection framework with graph neural network-assisted multimodal data fusion. PLoS One. 2023 Sep;18(9):e0291865. doi:10.1371/journal.pone.0291865. [Google Scholar] [PubMed] [CrossRef]
133. Ashraf AR, Somogyi-Végh A, Merczel S, Gyimesi N, Fittler A. Leveraging code-free deep learning for pill recognition in clinical settings: a multicenter, real-world study of performance across multiple platforms. Artif Intell Med. 2024 Apr;150:102844. doi:10.1016/j.artmed.2024.102844. [Google Scholar] [PubMed] [CrossRef]
134. TensorFlow Lite Model Analyzer. TensorFlow. [cited 2024 May 19]. Available from: https://ai.google.dev/edge/litert. [Google Scholar]
135. Dang B, Zhao W, Li Y, Ma D, Yu Q, Zhu EY. Real-time pill identification for the visually impaired using deep learning. In: 2024 6th International Conference on Communications, Information System and Computer Engineering (CISCE); 2024; Guangzhou, China: IEEE. p. 552–5. [Google Scholar]
136. Xu K, Chen L, Wang S. A multi-view kernel clustering framework for categorical sequences. Expert Syst Appl. 2022 Jul;197(1–2):116637. doi:10.1016/j.eswa.2022.116637. [Google Scholar] [CrossRef]
137. Qin H, Zaman A, Liu X. Artificial intelligence-aided intelligent obstacle and trespasser detection based on locomotive-mounted forward-facing camera data. Proc Institut Mech Eng Part F: J Rail Rapid Transit. 2023 Feb;237(9):1230–41. doi:10.1177/09544097231156312. [Google Scholar] [CrossRef]
138. Li S, Tajbakhsh N. SciGraphQA: a large-scale synthetic multi-turn question-answering dataset for scientific graphs. arXiv:230803349. 2023. [Google Scholar]
139. Zhang J, Liu L, Gao K, Hu D. A forward and backward compatible framework for few-shot class-incremental pill recognition. arXiv:230411959. 2023. [Google Scholar]
140. Zhou DW, Wang FY, Ye HJ, Ma L, Pu S, Zhan DC. Forward compatible few-shot class-incremental learning. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition; 2022; New Orleans, LA, USA: IEEE. p. 9046–56. [Google Scholar]
141. Yaniv Z, Faruque J, Howe S, Dunn K, Sharlip D, Bond A, et al. The national library of medicine pill image recognition challenge: an initial report. In: 2016 IEEE Applied Imagery Pattern Recognition Workshop (AIPR); 2016; Washington, DC, USA: IEEE. p. 1–9. [Google Scholar]
142. Zeng X, Cao K, Zhang M. MobileDeepPill: a small-footprint mobile deep learning system for recognizing unconstrained pill images. In: Proceedings of the 15th Annual International Conference on Mobile Systems, Applications, and Services; 2017; Niagara Falls, NY, USA: ACM. p. 56–67. [Google Scholar]
143. Wang Y, Ribera J, Liu C, Yarlagadda S, Zhu F. Pill recognition using minimal labeled data. In: 2017 IEEE Third International Conference on Multimedia Big Data (BigMM); 2017; Laguna Hills, CA, USA: IEEE. p. 346–53. [Google Scholar]
144. Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, et al. Going deeper with convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2015; Boston, MA, USA: IEEE. p. 1–9. [Google Scholar]
145. Mehmood A, Yoon C, Kim S, Kim S. MobilePill: accurate pill image classification via deep learning on mobile. In: 2019 International Conference on Information and Communication Technology Convergence (ICTC); 2019; Jeju, Republic of Korea: IEEE. p. 1362–7. [Google Scholar]
146. Rother C, Kolmogorov V, Blake A. “GrabCut” interactive foreground extraction using iterated graph cuts. ACM Transact Graph. 2004 Aug;23(3):309–14. doi:10.1145/1015706.1015720. [Google Scholar] [CrossRef]
147. Jiang F, Pang Y, Lee TN, Liu C. Automatic object segmentation based on grabcut. In: Advances in computer vision. Las Vegas, NV, USA: Springer; 2019. p. 350–60. [Google Scholar]
148. Zuiderveld K, Heckbert PS. Contrast limited adaptive histogram equalization. In: Graphics gems IV. San Diego, CA, USA:Academic Press Professional, Inc.; 1994. p. 474–85. [Google Scholar]
149. Eigen D, Rolfe J, Fergus R, LeCun Y. Understanding deep architectures using a recursive convolutional network. arXiv:13121847. 2013. [Google Scholar]
150. Srikamdee S, Suksawatchon U, Suksawatchon J, Werapan W. ClinicYA: an application for pill identification using deep learning and K-means clustering. In: 2022 26th International Computer Science and Engineering Conference (ICSEC); 2022; Sakon Nakhon, Thailand: IEEE. p. 117–22. [Google Scholar]
151. Al-Hussaeni K, Karamitsos I, Adewumi E, Amawi RM. CNN-based pill image recognition for retrieval systems. Appl Sci. 2023 Apr;13(8):5050. doi:10.3390/app13085050. [Google Scholar] [CrossRef]
152. Guo G, Wang H, Bell D, Bi Y, Greer K. KNN model-based approach in classification. In: On the move to meaningful internet systems 2003: CoopIS, DOA, and ODBASE. Catania, Sicily, Italy: Springer; 2003. p. 986–96. [Google Scholar]
153. Altman NS. An introduction to kernel and nearest-neighbor nonparametric regression. Am Stat. 1992 Aug;46(3):175–85. doi:10.1080/00031305.1992.10475879. [Google Scholar] [CrossRef]
154. Rádli R, Vörösházi Z, Czúni L. Multi-stream pill recognition with attention. In: 2023 IEEE 12th International Conference on Intelligent Data Acquisition and Advanced Computing Systems: Technology and Applications (IDAACS); 2023; Dortmund, Germany: IEEE. p. 942–6. [Google Scholar]
155. Hoffer E, Ailon N. Deep metric learning using triplet network. In: Proceedings of the International Workshop on Similarity-Based Pattern Recognition; 2015; Copenhagen, Denmark: Springer. p. 84–92. [Google Scholar]
156. Tan M, Le Q. EfficientNet: rethinking model scaling for convolutional neural networks. In: Proceedings of the 36th International Conference on Machine Learning, ICML 2019; 2019; Long Beach, CA, USA: PMLR. p. 6105–14. [Google Scholar]
157. Ojala T, Pietikainen M, Harwood D. Performance evaluation of texture measures with classification based on Kullback discrimination of distributions. In: Proceedings of 12th International Conference on Pattern Recognition; 1994; Jerusalem, Israel: IEEE. Vol. 1, p. 582–5. [Google Scholar]
158. Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, et al. Attention is all you need. In: Advances in Neural Information Processing Systems 30 (NIPS 2017); 2017; Long Beach, CA, USA. Vol. 30, p. 5998–6008. [Google Scholar]
159. Qin X, Zhang Z, Huang C, Dehghan M, Zaiane OR, Jagersand M. U2-Net: going deeper with nested U-structure for salient object detection. Pattern Recognit. 2020 Oct;106(11):107404. doi:10.1016/j.patcog.2020.107404. [Google Scholar] [CrossRef]
160. Li C, Liu W, Guo R, Yin X, Jiang K, Du Y, et al. PP-OCRv3: more attempts for the improvement of ultra lightweight OCR system. arXiv:220603001. 2022. [Google Scholar]
161. Yue X, Kuang Z, Lin C, Sun H, Zhang W. RobustScanner: dynamically enhancing positional clues for robust text recognition. In: Computer vision–ECCV 2020. Glasgow, UK: Springer; 2020. p. 135–51. [Google Scholar]
162. Wang M, Liu X, Gao Y, Ma X, Soomro NQ. Superpixel segmentation: a benchmark. Signal Process: Image Commun. 2017 Aug;56:28–39. [Google Scholar]
163. Liu YJ, Yu CC, Yu MJ, He Y. Manifold SLIC: a fast method to compute content-sensitive superpixels. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2016; Las Vegas, NV, USA. p. 651–9. [Google Scholar]
164. Li Z, Chen J. Superpixel segmentation using linear spectral clustering. In: 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2015; Boston, MA, USA. p. 1356–63. [Google Scholar]
165. Liu MY, Tuzel O, Ramalingam S, Chellappa R. Entropy rate superpixel segmentation. In: CVPR 2011; 2011; Colorado Springs, CO, USA: IEEE. p. 2097–104. [Google Scholar]
166. Van den Bergh M, Boix X, Roig G, Van Gool L. SEEDS: superpixels extracted via energy-driven sampling. Int J Comput Vis. 2015 Jul;111(3):298–314. doi:10.1007/s11263-014-0744-2. [Google Scholar] [CrossRef]
167. Zhang Y, Hartley R, Mashford J, Burn S. Superpixels via pseudo-boolean optimization. In: 2011 International Conference on Computer Vision; 2011; Barcelona: IEEE. p. 1387–94. [Google Scholar]
168. Conrad C, Mertz M, Mester R. Contour-relaxed superpixels. In: Energy Minimization Methods in Computer Vision and Pattern Recognition: 9th International Conference, EMMCVPR 2013; 2013; Lund, Sweden: Springer. p. 280–93. [Google Scholar]
169. Shen J, Du Y, Wang W, Li X. Lazy random walks for superpixel segmentation. IEEE Trans Image Process. 2014 Apr;23(4):1451–62. doi:10.1109/TIP.2014.2302892. [Google Scholar] [PubMed] [CrossRef]
170. Tang D, Fu H, Cao X. Topology preserved regular superpixel. In: 2012 IEEE International Conference on Multimedia and Expo; 2012; Melbourne, Victoria, Australia: IEEE. p. 765–8. [Google Scholar]
171. Veksler O, Boykov Y, Mehrani P. Superpixels and supervoxels in an energy optimization framework. In: Computer vision–ECCV 2010; 2010; Heraklion, Crete, Greece: Springer. p. 211–24. [Google Scholar]
172. Wang J, Wang X. VCells: simple and efficient superpixels using edge-weighted centroidal Voronoi tessellations. IEEE Trans Pattern Anal Mach Intell. 2012 Jun;34(6):1241–7. doi:10.1109/TPAMI.2012.47. [Google Scholar] [PubMed] [CrossRef]
173. Shi J, Malik J. Normalized cuts and image segmentation. IEEE Transact Pattern Anal Mach Intell. 2000;22(8):888–905. doi:10.1109/34.868688. [Google Scholar] [CrossRef]
174. Moore AP, Prince SJ, Warrell J, Mohammed U, Jones G. Superpixel lattices. In: 2008 IEEE Conference on Computer Vision and Pattern Recognition; 2008; Anchorage, AK, USA: IEEE. p. 1–8. [Google Scholar]
175. Levinshtein A, Stere A, Kutulakos KN, Fleet DJ, Dickinson SJ, Siddiqi K. TurboPixels: fast superpixels using geometric flows. IEEE Trans Pattern Anal Mach Intell. 2009 Dec;31(12):2290–7. doi:10.1109/TPAMI.2009.96. [Google Scholar] [PubMed] [CrossRef]
176. Vincent L, Soille P. Watersheds in digital spaces: an efficient algorithm based on immersion simulations. IEEE Transact Pattern Anal Mach Intell. 1991 Jun;13(6):583–98. doi:10.1109/34.87344. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF



 Downloads
Downloads
 Citation Tools
Citation Tools