 Open Access
Open Access
ARTICLE
PD-YOLO: Colon Polyp Detection Model Based on Enhanced Small-Target Feature Extraction
1 New Engineering Industry College, Putian University, Putian, 351100, China
2 Engineering Research Center of Big Data Application in Private Health Medicine (Putian University), Fujian Province University, Putian, 351100, China
3 Computer College, Guangdong University of Science & Technology, Dongguan, 523668, China
* Corresponding Author: Rong-Guei Tsai. Email:
Computers, Materials & Continua 2025, 82(1), 913-928. https://doi.org/10.32604/cmc.2024.058467
Received 12 September 2024; Accepted 24 October 2024; Issue published 03 January 2025
Abstract
In recent years, the number of patients with colon disease has increased significantly. Colon polyps are the precursor lesions of colon cancer. If not diagnosed in time, they can easily develop into colon cancer, posing a serious threat to patients’ lives and health. A colonoscopy is an important means of detecting colon polyps. However, in polyp imaging, due to the large differences and diverse types of polyps in size, shape, color, etc., traditional detection methods face the problem of high false positive rates, which creates problems for doctors during the diagnosis process. In order to improve the accuracy and efficiency of colon polyp detection, this question proposes a network model suitable for colon polyp detection (PD-YOLO). This method introduces the self-attention mechanism CBAM (Convolutional Block Attention Module) in the backbone layer based on YOLOv7, allowing the model to adaptively focus on key information and ignore the unimportant parts. To help the model do a better job of polyp localization and bounding box regression, add the SPD-Conv (Symmetric Positive Definite Convolution) module to the neck layer and use deconvolution instead of upsampling. The experimental results indicate that the PD-YOLO algorithm demonstrates strong robustness in colon polyp detection. Compared to the original YOLOv7, on the Kvasir-SEG dataset, PD-YOLO has shown an increase of 5.44 percentage points in AP@0.5, showcasing significant advantages over other mainstream methods.Keywords
With the rapid advancement of medical technology, colorectal polyps, a common digestive disease, have shown an increasing incidence and detection rate year by year. According to data from the American Cancer Society, colorectal cancer (CRC) is one of the most common types of cancer in the United States and globally. Estimates suggest that 149,500 adults in the United States will receive a colorectal cancer diagnosis in 2021, with approximately 104,270 new cases of colon cancer and 45,230 new cases of rectal cancer. In 2021, the estimated number of deaths due to colorectal cancer in the United States will be over 52,980. Early detection and treatment of colorectal polyps are of enormous significance in preventing colorectal cancer. Traditional methods of colorectal polyp detection, such as colonoscopy, while enabling indirect observation of the colon’s internal condition, are limited in clinical application due to their complexity, time-consuming nature, and dependence on the experience and technical proficiency of physicians [1], often resulting in missed or false detections. Therefore, leveraging advanced technological means to enhance the accuracy and efficiency of colorectal polyp detection has become an important focus of current medical research.
The application of deep learning technology in the field of medical image processing is continuously expanding and demonstrating remarkable achievements. Traditional image processing methods often struggle to handle complex medical images. However, deep learning, by mimicking the neural network’s functioning in the human brain, particularly in the detection of colorectal polyps, is capable of automatically capturing subtle polyp features from a vast collection of colonoscopy images and precisely filtering out useful feature information [2]. This feature information not only covers intuitive features such as the shape, size, and color of polyps, but also includes deeper pathological information [3]. We can construct accurate and reliable detection models based on these features to achieve efficient and accurate detection, identification, and localization of colorectal polyp images [4,5]. In terms of advantages, deep learning not only possesses powerful feature extraction capabilities but also exhibits excellent adaptability and robustness in its models, automatically adjusting to different image qualities and lighting conditions to ensure the stability and reliability of detection results. Additionally, deep learning models have the ability to self-learn and optimize, continuously improving and optimizing models through new data to further enhance detection performance [6]. These advantages enable deep learning to have broad application prospects in the field of colorectal polyp detection, potentially providing more precise and efficient technical support for clinical diagnosis and treatment [7].
Current research in colorectal polyp detection still has some drawbacks, such as high false positive rates, missed detection rates, and poor real-time performance, undoubtedly increasing the diagnostic burden on doctors [8]. Moreover, under different lighting conditions and noise interferences, models often lack robustness, making it difficult to meet clinical requirements. Some models also struggle to achieve real-time detection speeds, impacting the feasibility of clinical applications. To address these issues, we propose a novel network model called PD-YOLO (an improved You Only Look Once model for Polyp Detection). By adding a self-attention mechanism called CBAM to help the network focus on important information and lower the number of false positives, along with a unique structure called SPD-Conv and upsampling operations, the model becomes more robust in complex situations, making it more useful. We also optimize the model structure to ensure both accuracy and inference efficiency, meeting the demand for real-time detection. The PD-YOLO model aims to enhance the automation of colonoscopy examinations, assisting doctors in rapidly and accurately identifying polyps in real-time images, reducing doctors’ workload as much as possible, and improving diagnostic accuracy and efficiency. Integrating PD-YOLO with existing imaging systems can serve as an auxiliary tool that offers a priori detection, potentially reducing misdiagnosis and missed detection rates, enabling early polyp detection, lowering the incidence of colorectal cancer, and improving patient survival rates. This tool can aid doctors in making faster and more informed decisions.
The primary contributions of this paper are listed below:
(1) The backbone section of the original YOLOv7 introduces the CBAM attention mechanism, enabling the model to concentrate more on significant regions in the image. By combining channel attention and spatial attention, the model can reduce background noise interference and improve detection accuracy.
(2) We add the SPD-Conv module to the Neck layer to improve the model’s feature extraction capability. This enhances the model’s performance when dealing with low-resolution images, particularly in detecting small polyps or weak targets.
(3) The neck layer introduces deconvolution as a replacement for the upsampling method, enabling the model to adaptively adjust the upsampling effect through learning. This enables the model to capture crucial local information on high-dimensional features, leading to better performance in detecting small polyp targets.
In recent years, colorectal polyps, a common gastrointestinal disease, have been increasing in incidence annually, with a trend towards affecting younger populations, posing a serious threat to human health. Early detection and accurate diagnosis of colorectal polyps are critical for preventing the development of colorectal cancer. With the advancement of deep learning technologies, particularly convolutional neural networks (CNNs), which have demonstrated powerful capabilities in image recognition, classification, and detection, their application in the field of medical image processing has become increasingly widespread [9]. Deep learning technologies can automatically extract polyp features by learning from a large number of colonoscopy images. This makes it possible to build accurate and reliable detection models that can be used in a wide range of situations [10].
In the field of medical image analysis, both domestic and international scholars have made significant advancements. Thomaz et al. [11] proposed a novel colon polyp detection method using GANs (Generative Adversarial Networks). This method leverages deep convolutional neural networks and data augmentation techniques to improve image detection rates and reduce false negatives in colon polyp datasets. However, challenges such as overfitting, model complexity, and slow processing speeds exist. The model’s generalization ability is limited when there are significant differences between the samples and actual data distribution. Chen et al. [12] introduced the CAF-YOLO framework based on the YOLOv8 architecture. This approach incorporates the ACFM (Attention and Convolution Fusion Module) module to enhance global and local feature modeling and optimizes multi-scale feature aggregation through the MSNN (Multi-Scale Neural Network) module. The tests show that CAF-YOLO is good at finding small, complicated lesions in biomedical images from the BCCD and LUNA16 datasets. However, the training and tuning process of this model is relatively complex, requiring additional time and computational resources. In addition to CAF-YOLO, other researchers have explored different network architectures. Liew et al. [13] suggested a way to find things that combine a better deep residual convolutional neural network with an ensemble learning method. This makes the method more sensitive and accurate. However, this method requires more data and computational resources for ensemble learning training, and it is not suitable for small-scale datasets. Nogueira-Rodriguez et al. [14] proposed a real-time deep-learning model for colon polyp detection based on the pre-trained YOLOv3 architecture. This method effectively reduces false positives through the post-processing steps of object tracking algorithms, showing promising potential in assisting diagnostic systems. However, the YOLOv3 architecture itself suffers from a lack of fine-grained loss functions, making it challenging to locate small targets. Handling dense scenes and occlusion issues limits its performance, resulting in overall network performance degradation. In their study, Tas et al. [15] suggested using convolutional neural networks to create a super-resolution method called SRCNN (Super Resolution Convolutional Neural Network) to improve the clarity of colonoscopy images. The goal was to make automatic colonoscopy diagnosis and polyp detection better. They also explored the application of CNN (Convolutional Neural Networks) models SSD and Faster R-CNN for real-time polyp detection and localization. Pre-training with the SRCNN method significantly enhanced model accuracy. However, this algorithm employs a cascaded approach, leading to substantial resource consumption and longer inference times.
In recent years, multi-task interactive networks have garnered significant attention in the field of medical image detection and analysis. Aadarsh et al. [16] looked at how well U-Net++ [17], FPN [18], DeepLabV3+ [19], PAN [20], and U-Net [21] worked at processing endoscopic images. They stressed the importance of finding a balance between accuracy, speed, and resource use. Amin et al. [22] proposed an enhanced method based on YOLOv5 for early detection of malignant colon polyps. By augmenting and properly partitioning the Kvasir-SEG dataset, this approach demonstrated high accuracy in colon polyp detection. However, YOLOv5 exhibits subpar performance in detecting small and weak targets, particularly prone to missed detections or false alarms when such targets are densely arranged. Chavarrias-Solano et al. [23] came up with a way to improve the accuracy and generalizability of lesion detection that uses knowledge distillation and a class-aware loss function, but it takes more time and computing power to train. Tamhane et al. [24] created an algorithm using Vision Transformers to find anatomical landmarks in colonoscopy images. This was done to make medical image processing more automated and accurate. An adaptive gamma correction was used to prepare the images, Vision Transformers were used to extract features, and a fully connected network classifier head was used to find important anatomical landmarks. Tummala et al. [25] proposed a combination of Vision Transformers (ViT) models for brain tumor classification in magnetic resonance imaging (MRI). This method utilized four standard ViT models that were pre-trained and fine-tuned on ImageNet for classification. Experimental results indicate that this approach can effectively enhance the overall accuracy of brain tumor type classification. However, employing the Vision Transformer structure necessitates training on larger datasets, limiting the model’s generalization capability and accuracy on smaller datasets. Malik et al. [26] developed a multi-class deep learning model for diagnosing ulcerative colitis, polyps, and dye-enhanced polyps using wireless capsule endoscopy images. This was achieved by classifying three different gastrointestinal diseases using a single model, improving classification accuracy. However, the model’s complexity increased training and inference times, making it unsuitable for real-time clinical applications. Liu et al. [27] proposed a polyp detection and localization framework for cross-device colonoscope images. This framework, which uses a hierarchical unsupervised domain adaptive detection network, improved polyp location prediction accuracy. However, the construction of the hierarchical unsupervised domain adaptive detection network requires a longer time. Wuyang et al. [28] proposed a polyp detection and segmentation method for endoscopic data using HRNet as the base network. By employing an adaptive sample selection strategy and integrating low-rank modules, they effectively boosted the model’s performance. This method achieved outstanding results in the EndoCV 2021 challenge and demonstrated excellent generalization across different datasets. However, HRNet comes with drawbacks such as high computational resource requirements and sensitivity to initial parameter settings.
In summary, existing polyp detection methods have several shortcomings. In practical applications, issues such as uneven image brightness and densely packed or partially occluded small polyp targets often lead to instances of missed detections or false alarms in current models. Additionally, models struggle to converge when working with smaller datasets, resulting in lower accuracy and poor generalization capabilities. Moreover, many existing models are complex, requiring large datasets for training, consuming significant computational resources, and exhibiting slow detection speeds, making it challenging to meet real-time detection requirements. Finding the best balance between detection accuracy, model size, accuracy, and processing speed in real-life polyp detection situations is therefore an important area for research, especially when datasets are limited.
3 Colorectal Polyp Detection Based on PD-YOLO
3.1 Overall Network Model Design
The YOLO series has always held a landmark position in the field of object detection. YOLOv7 [29] introduced a new real-time object detection architecture aimed at addressing the replacement of the reparameterization module and dynamic label assignment issues while improving accuracy through a trainable bag-of-freebies approach. Reparameterization can optimize model performance by altering the model’s structure or parameters, enhancing accuracy and efficiency. Dynamic label assignment technology resolves the problem of assigning labels to outputs from different branches in multi-output layer models. Furthermore, YOLOv7 also incorporates a trainable bag of freebies approach, a technique that enhances model performance without increasing inference costs, enabling it to meet the requirements for real-time detection while maintaining high accuracy.
This section describes a novel PD-YOLO network model tailored for colon polyps and establishes an end-to-end detection architecture. The model, based on YOLOv7, incorporates a self-attention mechanism called CBAM at the backbone layer to adaptively focus on crucial information while disregarding less important parts. We add the SPD-Conv module to the neck layer and replace the upsampling method with deconvolution to improve the model’s precision in polyp localization and bounding box regression. Fig. 1 illustrates the improved PD-YOLO network model, with the red components representing YOLO’s original elements and the darker components signifying discrepancies between the PD-YOLO network model and the original model. Fig. 2 provides a detailed exposition of the MCB and SPPCSP module structures. The following sections will offer detailed descriptions of the three added modules: CBAM, SPD-Conv, and Deconvolution.
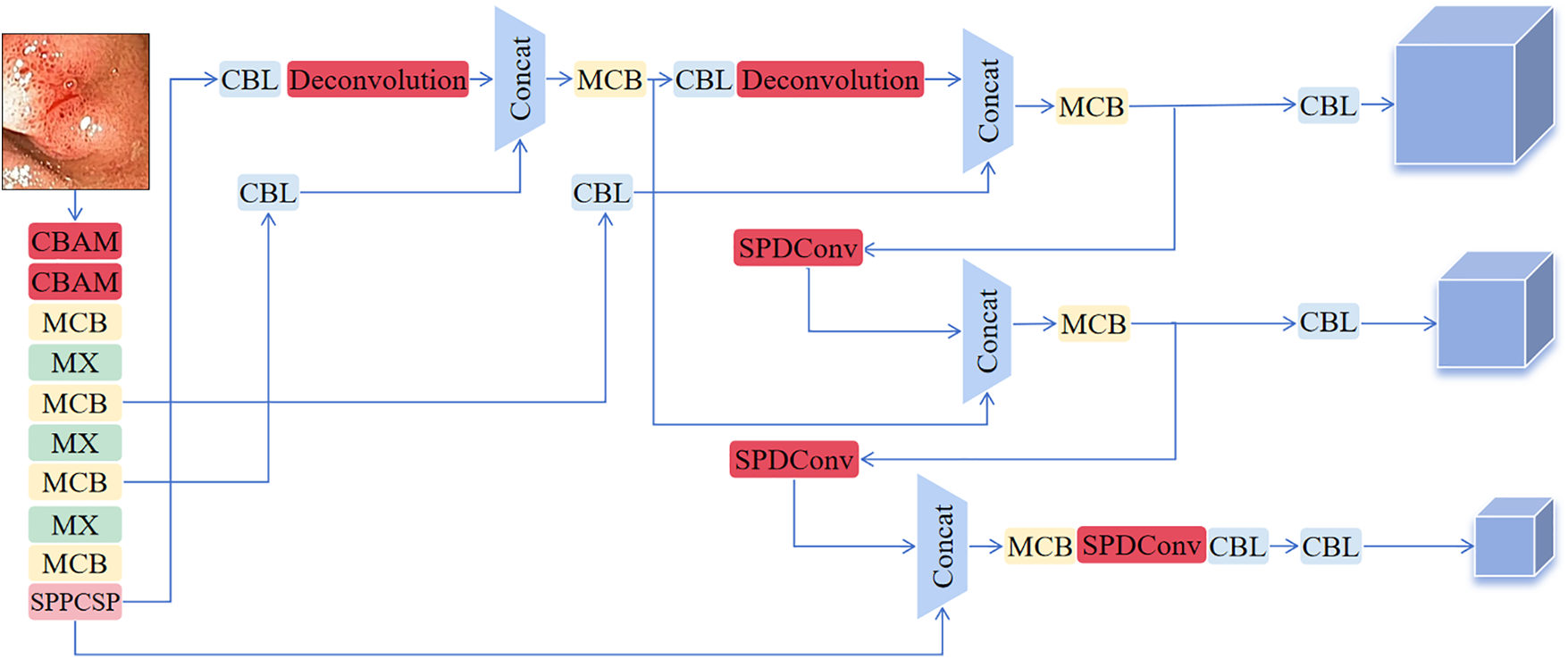
Figure 1: Design of the PD-YOLO network model
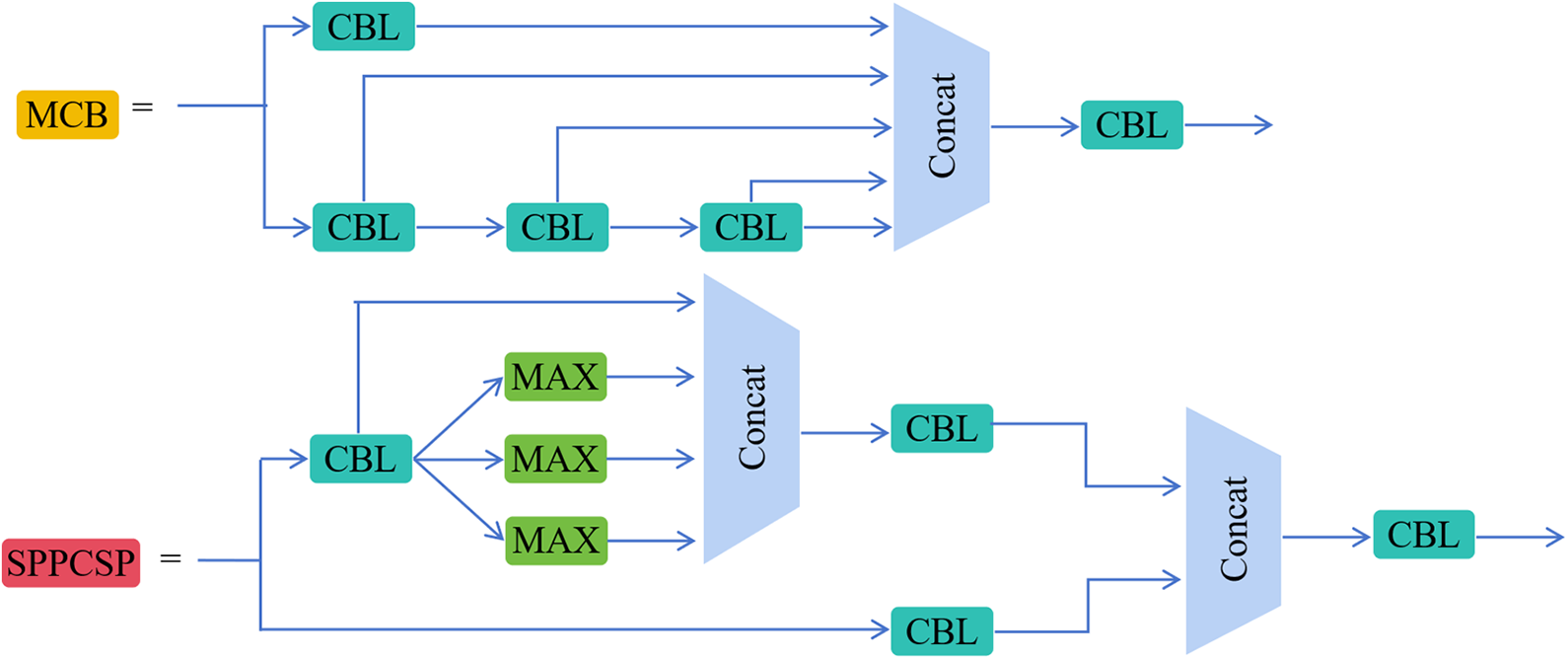
Figure 2: Schematic diagram of MCB and SPPCSP module structures
The CBAM attention mechanism significantly enhances the performance of convolutional neural networks by introducing channel and spatial attention. Its core focus lies in adaptively emphasizing key information within input features, thereby boosting the model’s perceptual capabilities. This module consists of both channel attention and spatial attention mechanisms. The channel attention module strengthens feature representation by generating channel weights through global pooling to highlight relevant channels and suppress irrelevant ones. The spatial attention module focuses on critical regions within the image, generating spatial weights through pooling operations to prioritize task-relevant areas. Fig. 3 depicts the overall process.
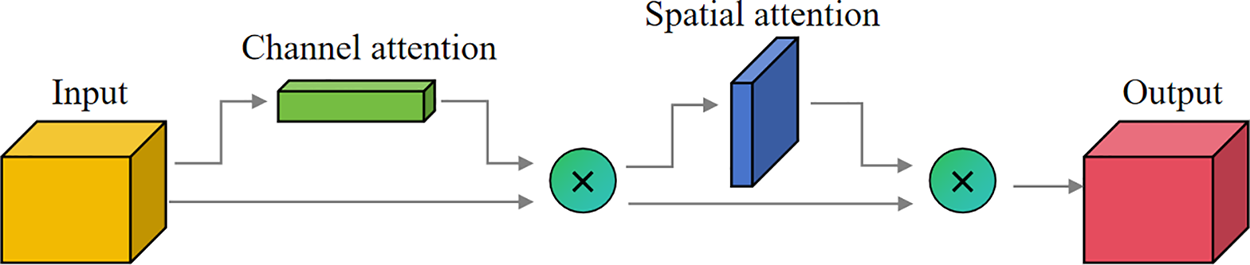
Figure 3: CBAM structure diagram
The input feature map first goes through the channel attention module of the CBAM, resulting in a feature map weighted by channel attention. Subsequently, this feature map passes through the spatial attention module to obtain the final feature map weighted by spatial attention. The mathematical reasoning process can be described as follows: given an intermediate feature mapping
In this context,
YOLOv7, as a cutting-edge one-stage object detection algorithm, excels in many scenarios; however, it still faces challenges when detecting small objects, especially when dealing with low-resolution images. To overcome this challenge, SPD-Conv significantly enhances feature representation capabilities through its unique spatial-to-depth transformation mechanism. This mechanism enables the model to more accurately capture small object features, thereby improving the overall detection performance of small objects.
Due to its capability to enhance feature representation and being a versatile building block, SPD-Conv significantly improves the model’s ability to capture minute target features. Therefore, combining SPD-Conv with YOLOv7 can notably enhance the model’s robustness. This improvement lowers the model’s sensitivity to factors such as image quality and occlusion, allowing it to maintain good detection performance in a variety of complex scenarios. In this study, the module utilizes 2x downsampling, taking into account the intermediate feature map, which has a size of
Each of the sub-feature maps is
Fig. 4 shows that this process basically involves a pooling operation and reorganizing features in the spatial dimension. Next, new representations of these reorganized features are learned through convolution.
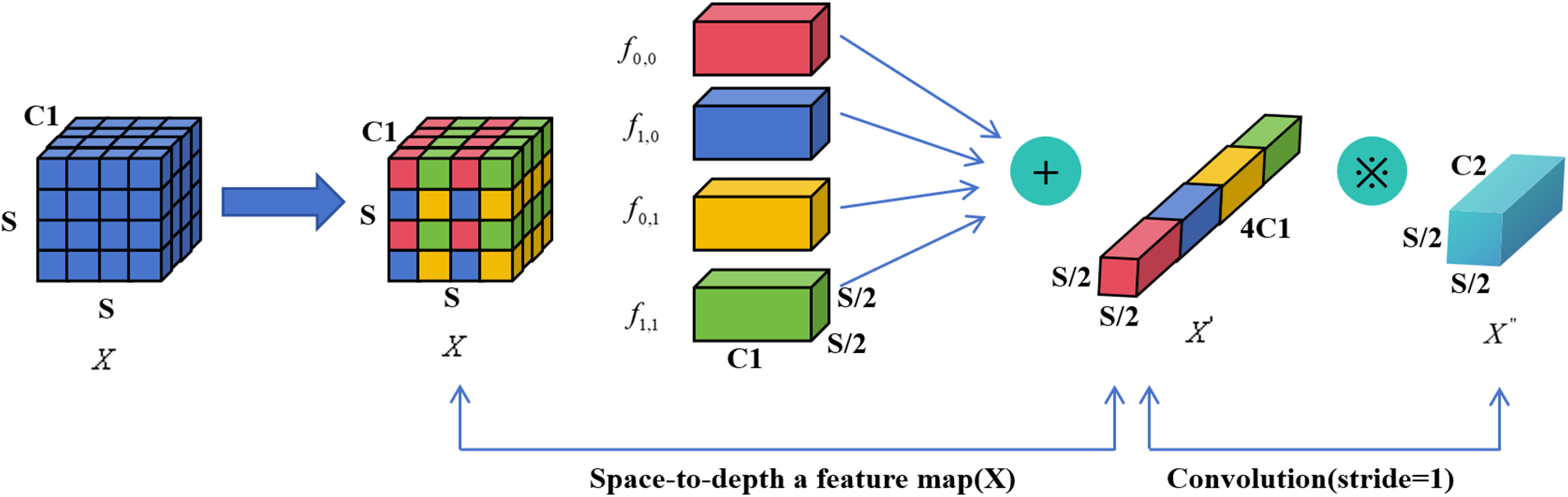
Figure 4: SPD-conv structure diagram
SPD-Conv plays a dual role in the network, enhancing feature fusion and multi-scale feature extraction. This is achieved by downsampling and reorganizing feature maps, as shown in Fig. 4, allowing the model to learn more about local and global context information at different scales. This approach helps establish richer connections between features at different levels, enhancing the model’s adaptability to object scale variations. Additionally, SPD-Conv is utilized to further refine and integrate information extracted from features at different scales. Applying SPD-Conv on feature maps of three different sizes, not only enhances feature diversity, but also promotes feature flow between scales, which is crucial for accurately detecting objects of various sizes.
Upsampling is a crucial operation that progressively restores spatial resolution during the feature extraction process, aiding the model in capturing detailed information in the image. The YOLOv7 model primarily employs nearest-neighbor interpolation, which involves filling new pixel positions by directly copying the values of the nearest neighboring pixels. While this method is advantageous in terms of speed, it can lead to issues such as discontinuities and blocking artifacts in image quality.
Deconvolution is similar to the inverse process of convolution. The forward and backward computations of a convolutional layer involve multiplication by matrices C and
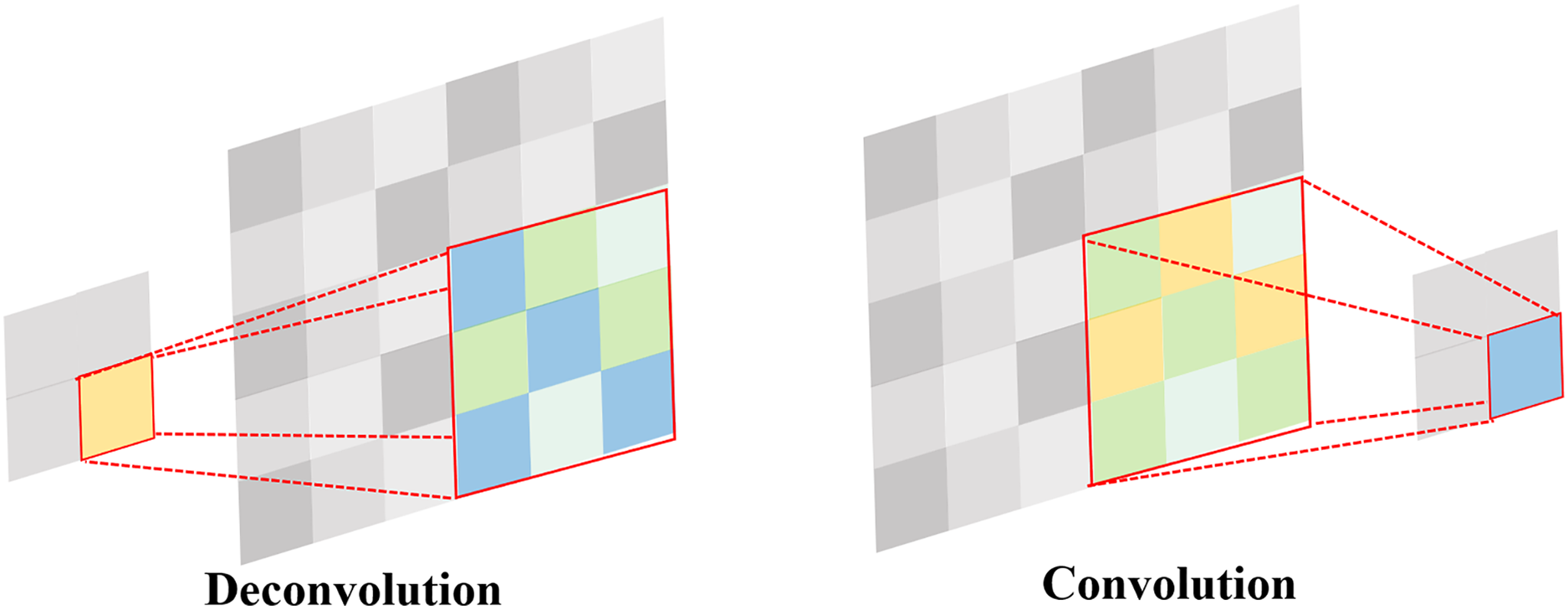
Figure 5: Comparison of convolution and deconvolution
4 Experimental Design and Results Analysis
4.1 Datasets and Evaluation Metrics
This paper utilizes the open-source Kvasir-SEG dataset and CVC-ColonDB dataset for training and validation purposes. Kvasir-SEG is a public dataset specifically designed for endoscopic image segmentation, containing images of various types of polyps and other gastrointestinal diseases. This dataset offers detailed annotations that help improve the accuracy and robustness of models in polyp segmentation tasks. CVC-ColonDB is another widely used colonoscopy image dataset, consisting of images of polyps from colonoscopy examinations. With rich annotation information, this dataset supports model training and performance evaluation, particularly excelling in polyp detection and localization. We train and debug all improvements on the Kvasir-SEG dataset, which serves as our primary dataset. We use the CVC-ColonDB dataset as supplementary data to validate the robustness of the algorithm. By combining these two datasets, the aim is to enhance the model’s generalization and accuracy on different types of endoscopic images.
The experimental software environment consisted of Python 3.9.8, PyTorch 1.12.0, and CUDA 11.6, running on hardware featuring an NVIDIA GTX3050Ti LAPTOP graphics card. We conducted all experiments under the same hyperparameters, using an input size of 640 × 640 and a batch size of 8, and the models reached stability after 150 epochs of training.
The performance evaluation metrics include precision, recall, AP@0.5, AP@0.5:0.95, GFLOPS, balanced F1-score, parameters, and FPS. Precision represents the ratio of true positive predictions to all positive predictions made by the model; recall represents the ratio of true positive predictions to all actual positive instances. AP@0.5 indicates the detection precision of the target at a threshold of 0.5; AP@0.5:0.95 represents the detection accuracy of the target within the threshold range of 0.5–0.95. GFLOPS measures algorithm complexity, while the F1-score, also known as the balanced F score, is the harmonic mean of precision and recall. We use FPS (frames per second) to measure real-time model performance. Parameters refer to the quantity of model parameters, serving as a measure of model complexity.
4.2 Ablation Experiment Design
We designed ablation experiments using YOLOv7-tiny as the base network to validate the impact of SPD-Conv, CBAM, and deconvolution on the network performance of the improved method. The Kvasir-SEG ablation experiment dataset was chosen for generalization, and the SGD (stochastic gradient descent) optimizer was used throughout the whole process. All experiments were run with the same hyperparameters.
Table 1 shows a significant improvement in the model’s performance with the addition of different network components. We can observe the impact of different combinations on the model’s performance in the ablation experiments where we added SPD-Conv, CBAM, and deconvolution modules to YOLOv7. The baseline performance of YOLOv7 was a precision of 0.9068 and a recall of 0.8837, providing a foundation for the subsequent inclusion of custom modules. After adding SPD-Conv, the model’s precision increased to 0.9367, and recall increased to 0.9147. This suggests that the SPD-Conv module significantly improved the model’s detection capabilities while adding parameters, particularly in enhancing the recognition of specific targets in complex colorectal polyp detection scenarios. The model did not converge when adding the CBAM module alone, which is why Table 1 does not display this row of data. This could be due to the model not effectively adjusting parameters or learning useful features after the introduction of CBAM, leading to no performance improvement. After adding the deconvolution module, the model achieved a precision of 0.9211 and a recall of 0.8989. This suggests that deconvolution improved detail recovery, but it did not reach the effectiveness of SPD-Conv.
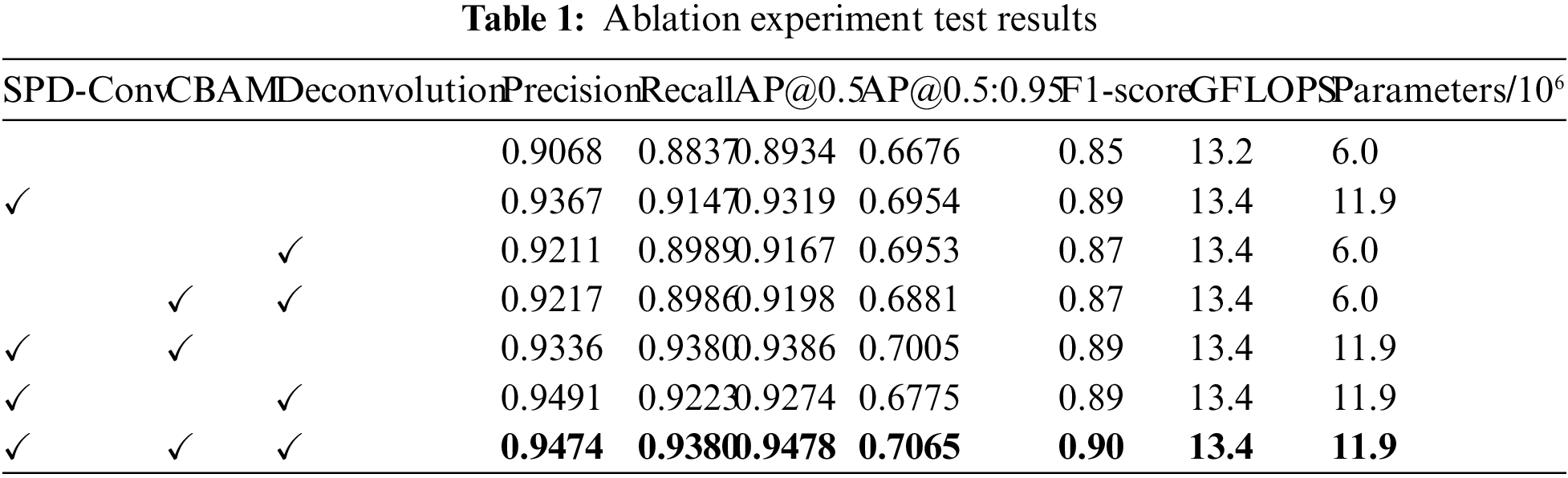
When using multiple combinations, the combination of SPD-Conv and Deconvolution exhibited outstanding model performance, with a precision of 0.9491 and a recall of 0.9223. This indicates that the combination of the deconvolution module and SPD-Conv significantly improves object detection accuracy by enhancing image detail recovery. Moreover, the model achieved a precision of 0.9474, a recall of 0.9380, an AP@0.5 of 0.9478, an AP@0.5:0.95 of 0.7065, and an F1-score of 0.90 when using all three modules (SPD-Conv, CBAM, and deconvolution) simultaneously. This combination demonstrated excellent performance. Despite CBAM not converging, the combination of SPD-Conv and deconvolution showed a significant performance boost. This enhancement did not introduce a notable increase in computational complexity, further emphasizing the cost-effectiveness of this combination.
In Fig. 6, from left to right, (a) displays the original image input, (b) YOLOv7, (c) YOLOv7+SPD-Conv, (d) YOLOv7+SPD-Conv+CBAM, (e) YOLOv7+SPD-Conv+Deconvolution, and (f) YOLOv7 +SPD-Conv+CBAM+Deconvolution forward inference diagrams. Starting from the original YOLOv7 model, we progressively introduce improvements like SPD-Conv, CBAM, and deconvolution to significantly enhance the model’s performance during the polyp detection inference process. Firstly, SPD-Conv enhances the model to learn more about local and global contextual information at different scales, aiding in establishing richer connections between features at different levels, enabling the model to perform better in detecting objects of different scales, especially when detecting small targets and processing low-resolution images. As a result, the inclusion of the CBAM attention mechanism allows the model to focus more on important regions in the image. By combining channel attention and spatial attention, the model can reduce background noise interference, further enhancing detection accuracy. Lastly, the introduction of deconvolution allows the model to generate higher-resolution feature maps. By restoring more detailed information through upsampling operations, the model achieves better results in multi-scale object detection. Observing Fig. 6, we see a significant improvement in the model’s detection performance as we gradually introduce each module. This further indicates that the SPD-Conv, CBAM, and deconvolution modules play crucial roles in enhancing model performance, particularly in handling small targets and details. Although occasional decreases in image accuracy occur, as seen in the last row, this might be due to potential information redundancy or conflicts arising from feature interactions between modules. For instance, CBAM emphasizes important features, while deconvolution may introduce artifacts or interference, compromising the model’s accuracy in target recognition. Nevertheless, the overall accuracy remains superior to the baseline model.
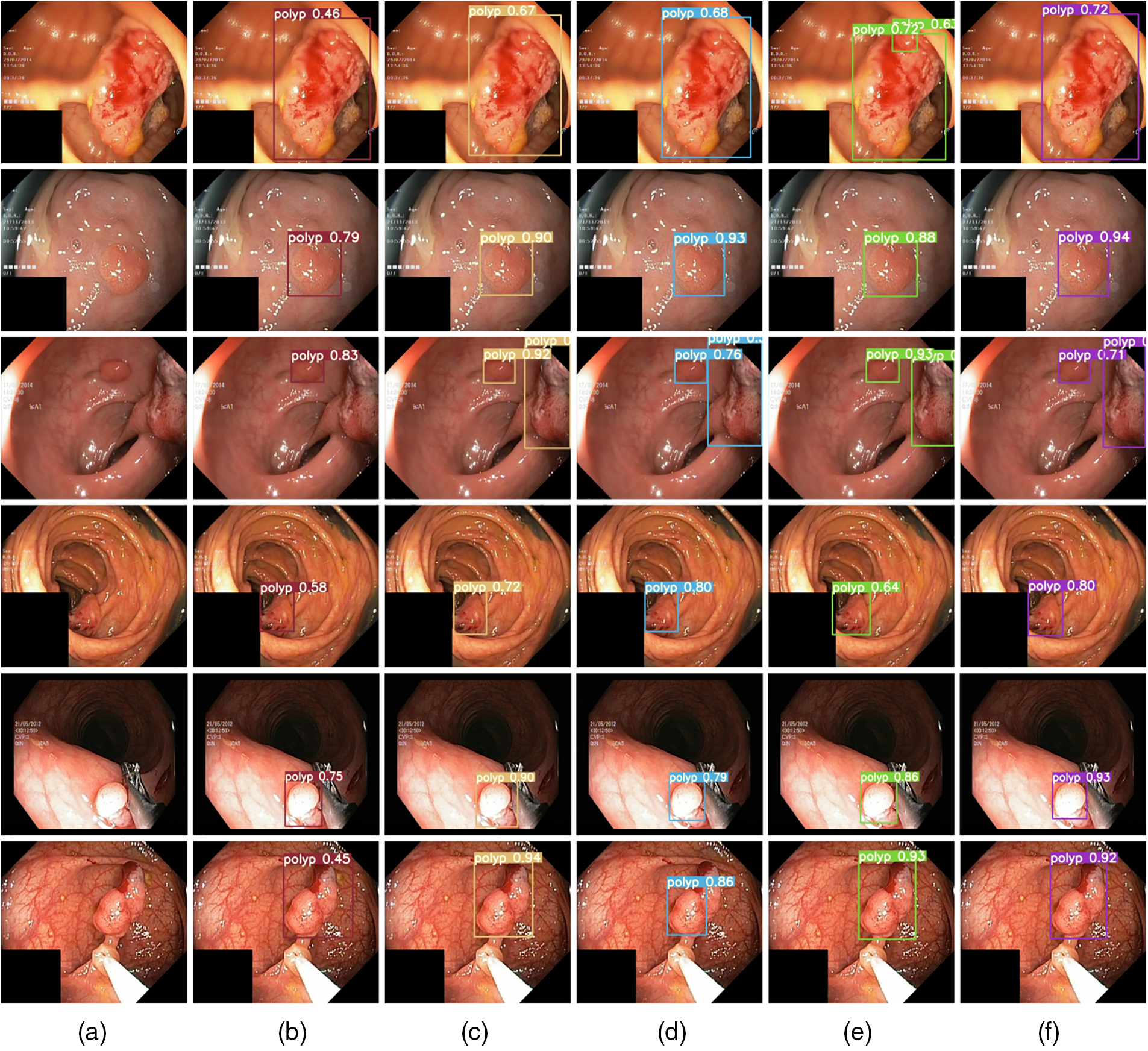
Figure 6: Forward inference results
4.3 Comparative Experiment Design
We selected YOLOv3 [30], YOLOv5 [31], the improved PP-YOLOE [32] based on the YOLO series, YOLOR [33], the non-YOLO series RT-DETR [34], Rtmdet [35], SSD [36], as well as the newly introduced YOLOv8 [37], the CAF-YOLO model, and the native YOLOv7 algorithm as comparative algorithms to evaluate the performance of the colon polyp detection method proposed in this paper. Table 2 presents the comparison results of the proposed method with other comparative algorithms on the Kvasir-SEG dataset.
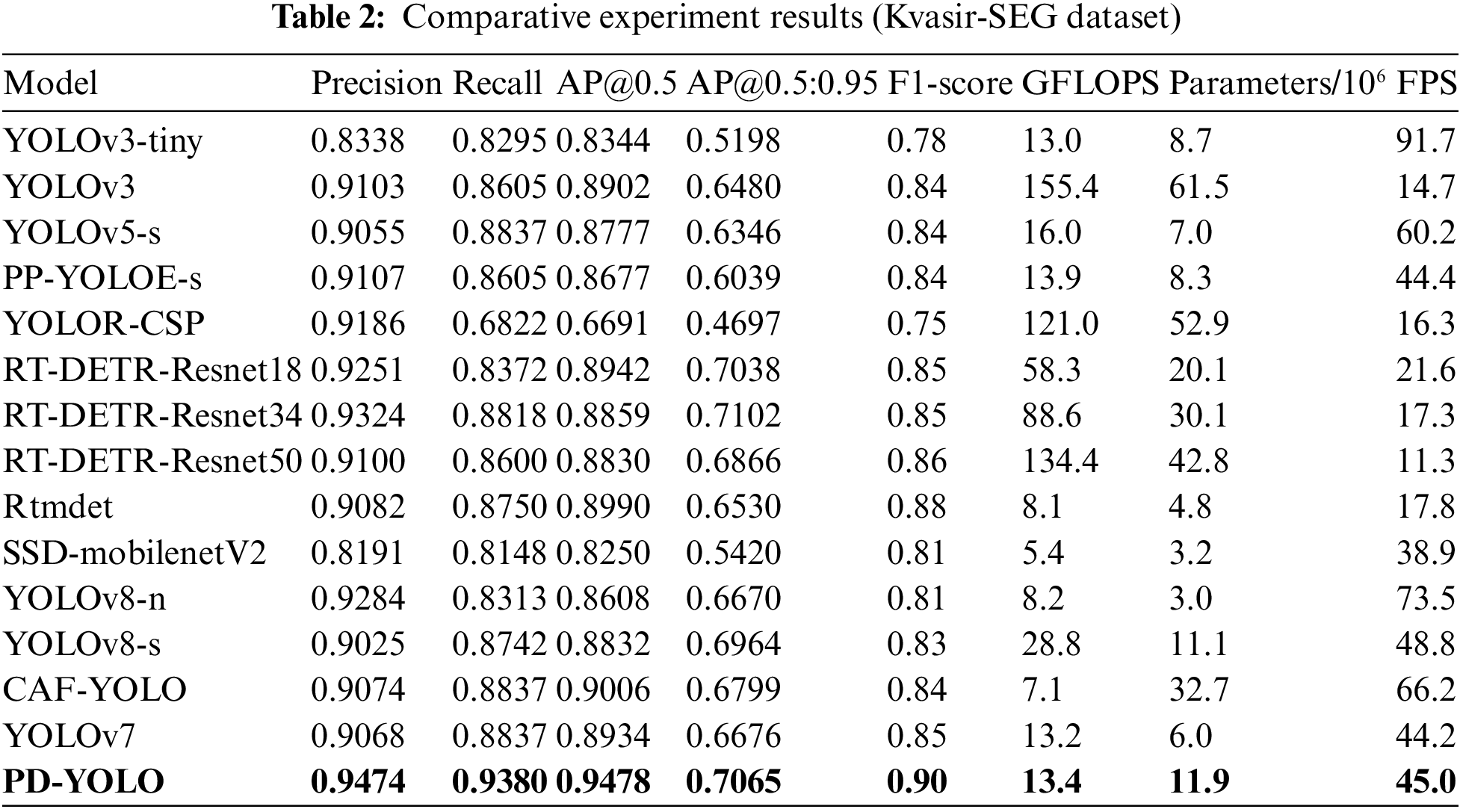
It’s clear from Table 2 that PD-YOLO does much better in the comparison tests on the Kvasir-SEG dataset, with a precision of 0.9474, a recall of 0.9380, and an AP@0.5 value as high as 0.9478 (which is 5.44 percentage points higher than the original YOLOv7’s AP@0.5 value of 0.8934). These metrics surpass those of other models, demonstrating PD-YOLO’s highly effective capability in target identification and the reduction of false positives in detection tasks. In comparison, RT-DETR-Resnet34 and RT-DETR-Resnet18 perform well, but their precision and recall are lower than PD-YOLO. Although YOLOv3-tiny excels in processing speed with an FPS of 91.7, its precision (0.8338) and recall are relatively lower, indicating shortcomings in target detection capabilities. Overall, PD-YOLO stands out with its dual advantages in precision and recall. Additionally, PD-YOLO also performs well in the F1-score, reflecting a good balance between precision and recall, making it suitable for practical scenarios in polyp detection tasks. According to the results, YOLOv8 still lags behind PD-YOLO in performance, and while CAF-YOLO has some recall capability, its lower precision limits its effectiveness in practical applications.
In Table 3, PD-YOLO does much better in the comparison tests on the CVC-ColonDB dataset, with a precision of 0.9917, recall of 0.8590, AP@0.5 as high as 0.8828, and an F1-score of 0.90. These metrics indicate that PD-YOLO can effectively identify targets and reduce false positives in object detection tasks, demonstrating its robust capabilities in practical applications. The limited sample size hindered the convergence of Rtmdet, YOLOR-CSP, and SSD, resulting in their exclusion from Table 2. In comparison, while YOLOv5-s and YOLOv7 also perform well with precisions of 0.9759 and 0.9799, respectively, and recalls close to PD-YOLO, they still slightly fall short in overall performance. RT-DETR-Resnet34 and RT-DETR-Resnet18 also perform well with precisions above 0.9862, indicating their effectiveness in handling complex objects. However, their recall rates are slightly lower and do not surpass the overall performance of PD-YOLO, especially in medical image analysis scenarios requiring high recall rates, where PD-YOLO’s high recall rate makes it more attractive in practical applications. Although PP-YOLOE approaches PD-YOLO in precision (0.9900), its recall rate is significantly lower (0.7651), impacting overall detection capabilities and suggesting potential issues with missed detections when dealing with certain objects. This difference underscores PD-YOLO’s advantages in overall performance and adaptability to application scenarios. Other models, such as YOLOv8-n and YOLOv8-s, also offer excellent performance with precisions above 0.98, but they do not fully match PD-YOLO’s recall rate and F1-score. Additionally, CAF-YOLO shows relatively weaker performance with a precision of only 0.8008, and while the recall rate is 0.8389, the overall AP and F1-score are lower than other models.
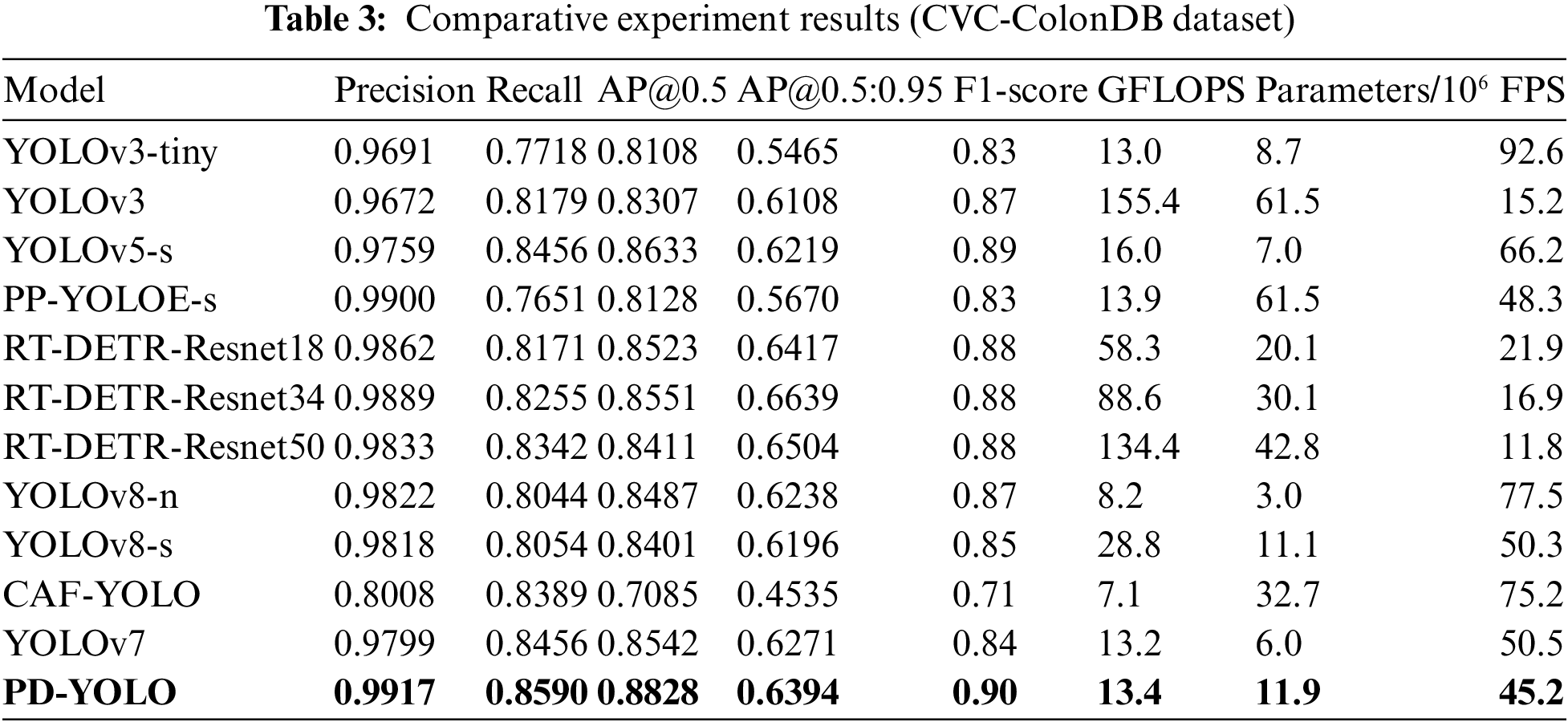
Overall, PD-YOLO demonstrates outstanding object detection capabilities on both the Kvasir-SEG and CVC-ColonDB datasets. On the Kvasir-SEG dataset, PD-YOLO achieves a precision of 0.9474, a recall of 0.9380, AP@0.5 as high as 0.9478, and an F1-score of 0.90. Compared to other models, PD-YOLO leads in several key metrics. On the CVC-ColonDB dataset, PD-YOLO’s performance is even more remarkable, with a precision reaching 0.9917, a recall of 0.8590, AP@0.5 reaching 0.8828, and an F1-score of 0.90. These metrics indicate that PD-YOLO not only effectively reduces false positives but also maintains a high level of target recognition in complex medical image environments. Compared to similar models, PD-YOLO exhibits clear advantages in precision and detection capabilities. Furthermore, PD-YOLO outperforms other models in terms of robustness, allowing it to adapt to a variety of endoscopic image features.
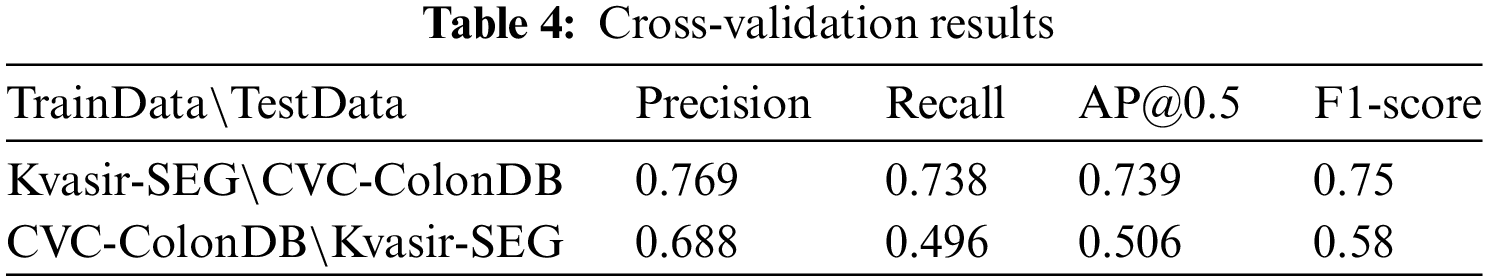
Table 4 displays the model’s performance after training on the Kvasir-SEG dataset and testing on the CVC-ColonDB dataset. It demonstrates excellent performance with a precision of 0.769, a recall of 0.738, and an F1-score of 0.75. This result indicates that the model exhibits strong generalization abilities in target recognition, effectively adapting to different types of medical image data. Particularly,when trained on the Kvasir-SEG dataset, the model can leverage its rich feature information to maintain high recognition accuracy and overall performance. However, when trained on the CVC-ColonDB dataset, the model’s performance significantly declines, with a precision of 0.688, a recall of 0.496, and an F1-score of 0.58. This decrease in performance is due to the feature differences between the datasets, leading to the model’s poor performance on new datasets, especially given the limited number of samples in the CVC-ColonDB dataset. When trained on Kvasir-SEG, the model performs better in terms of recall, whereas training on CVC-ColonDB leads to a significant increase in precision but a decrease in recall. Overall, the model exhibits balanced performance on different datasets, demonstrating excellent adaptability and generalization capabilities.
The increasing number of patients with colorectal diseases highlights the critical need for accurate detection of colorectal polyps as a key preventive measure against colorectal cancer. However, existing methods frequently face challenges, such as high false positives, due to significant imaging differences and diverse types of polyps. To get around these problems, this study introduces the PD-YOLO model, which has a self-attention mechanism called CBAM in the backbone layer to focus on important polyp features. It also has the SPD-Conv module in the neck layer to get information about multiple scales of features, and it uses deconvolution upsampling for better localization and regression. The experiment demonstrates that PD-YOLO performs excellently in colon polyp detection, with an average precision increase of 5.44 percentage points in AP@0.5 compared to YOLOv7. It shows significant advantages over other mainstream methods, offering a new and effective approach for automated colon polyp detection.
While the PD-YOLO model shows promise, there is still room for improvement. For example, when dealing with more complex or rare types of colorectal polyps, integrating other medical image analysis techniques, such as 3D reconstruction, could help capture comprehensive polyp information for more accurate diagnostic support. Future research will explore the integration of different deep learning architectures, including emerging technologies like visual transformers, to enhance detection accuracy and robustness. It is also important to use bigger and more varied training datasets to make the model more flexible in different settings, look into real-time detection applications to meet clinical or industrial needs, and think about how to combine text or audio data with visual data for multimodal fusion. Research in cross-modal fusion will further enhance the model’s contextual understanding and detection accuracy.
Acknowledgement: The authors are grateful to the anonymous reviewers and the editor for their valuable suggestions, which greatly contributed to the improved quality of this article.
Funding Statement: This research was funded by the Undergraduate Higher Education Teaching and Research Project (No. FBJY20230216), Research Projects of Putian University (No. 2023043), the Education Department of the Fujian Province Project (No. JAT220300).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Yicong Yu, Kaixin Lin; data collection: Yicong Yu, Kaixin Lin, Yuanzhi Huang; analysis and interpretation of results: Yicong Yu, Kaixin Lin, Jiajun Hong; draft manuscript preparation: Yicong Yu, Rong-Guei Tsai. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: To validate the model, we used the following public dataset: https://www.kaggle.com/datasets/debeshjha1/kvasirseg; https:www.kaggle.com/datasets/doansang/cvc-colondb (accessed on 04 March 2024).
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. X. Yang, Q. Wei, C. Zhang, K. Zhou, L. Kong and W. Jiang, “Colon polyp detection and segmentation based on improved mrcnn,” IEEE Trans. Instrum. Meas., vol. 70, pp. 1–10, 2020. doi: 10.1109/TIM.2021.3126366. [Google Scholar] [CrossRef]
2. S. Wang, Y. Yin, D. Wang, Z. Lv, Y. Wang and Y. Jin, “An interpretable deep neural network for colorectal polyp diagnosis under colonoscopy,” Knowl.-Based Syst., vol. 234, 2021, Art. no. 107568. doi: 10.1016/j.knosys.2021.107568. [Google Scholar] [CrossRef]
3. M. Misawa et al., “Artificial intelligence-assisted polyp detection for colonoscopy: Initial experience,” Gastroenterology, vol. 154, no. 8, pp. 2027–2029, 2018. doi: 10.1053/j.gastro.2018.04.003. [Google Scholar] [PubMed] [CrossRef]
4. P. Wang et al., “Real-time automatic detection system increases colonoscopic polyp and adenoma detection rates: A prospective randomised controlled study,” Gut, vol. 68, no. 10, pp. 1813–1819, 2019. doi: 10.1136/gutjnl-2018-317500. [Google Scholar] [PubMed] [CrossRef]
5. P. -J. Chen, M. -C. Lin, M. -J. Lai, J. -C. Lin, H. H. -S. Lu and V. S. Tseng, “Accurate classification of diminutive colorectal polyps using computer-aided analysis,” Gastroenterology, vol. 154, no. 3, pp. 568–575, 2018. doi: 10.1053/j.gastro.2017.10.010. [Google Scholar] [PubMed] [CrossRef]
6. A. Yamada, R. Niikura, K. Otani, T. Aoki, and K. Koike, “Automatic detection of colorectal neoplasia in wireless colon capsule endoscopic images using a deep convolutional neural network,” Endoscopy, vol. 53, no. 8, pp. 832–836, 2021. doi: 10.1055/a-1266-1066. [Google Scholar] [PubMed] [CrossRef]
7. D. Jha et al., “A comprehensive study on colorectal polyp segmentation with ResUNet++, conditional random field and test-time augmentation,” IEEE J. Biomed. Health Inform., vol. 25, no. 6, pp. 2029–2040, 2021. doi: 10.1109/JBHI.2021.3049304. [Google Scholar] [PubMed] [CrossRef]
8. R. Gong, S. He, T. Tian, J. Chen, Y. Hao and C. Qiao, “FRCNN-AA-CIF: An automatic detection model of colon polyps based on attention awareness and context information fusion,” Comput. Biol. Med., vol. 158, no. 6, 2023, Art. no. 106787. doi: 10.1016/j.compbiomed.2023.106787. [Google Scholar] [PubMed] [CrossRef]
9. K. Patel et al., “A comparative study on polyp classification using convolutional neural networks,” PLoS One, vol. 15, no. 7, 2020, Art. no. e0236452. doi: 10.1371/journal.pone.0236452. [Google Scholar] [PubMed] [CrossRef]
10. T. Rahim, S. A. Hassan, and S. Y. Shin, “A deep convolutional neural network for the detection of polyps in colonoscopy images,” Biomed. Signal Process. Control, vol. 68, no. 1, 2021, Art. no. 102654. doi: 10.1016/j.bspc.2021.102654. [Google Scholar] [CrossRef]
11. V. de Almeida Thomaz, C. A. Sierra-Franco, and A. B. Raposo, “Training data enhancements for improving colonic polyp detection using deep convolutional neural networks,” Artif. Intell. Med., vol. 111, no. 6, 2021, Art. no. 101988. doi: 10.1016/j.artmed.2020.101988. [Google Scholar] [PubMed] [CrossRef]
12. Z. Chen and S. Lu, “CAF-YOLO: A robust framework for multi-scale lesion detection in biomedical imagery,” 2024, arXiv:2408.01897. [Google Scholar]
13. W. S. Liew, T. B. Tang, C. -H. Lin, and C. -K. Lu, “Automatic colonic polyp detection using integration of modified deep residual convolutional neural network and ensemble learning approaches,” Comput. Methods Programs Biomed., vol. 206, no. 2, 2021, Art. no. 106114. doi: 10.1016/j.cmpb.2021.106114. [Google Scholar] [PubMed] [CrossRef]
14. A. Nogueira-Rodríguez et al., “Real-time polyp detection model using convolutional neural networks,” Neural Comput. Appl., vol. 34, no. 13, pp. 10375–10396, 2022. doi: 10.1007/s00521-021-06496-4. [Google Scholar] [CrossRef]
15. M. Taş and B. Yılmaz, “Super resolution convolutional neural network based pre-processing for automatic polyp detection in colonoscopy images,” Comput. Elect. Eng., vol. 90, no. 12, 2021, Art. no. 106959. doi: 10.1016/j.compeleceng.2020.106959. [Google Scholar] [CrossRef]
16. Y. G. Aadarsh and G. Singh, “Comparing UNet, UNet++, FPN, PAN and Deeplabv3+ for gastrointestinal tract disease detection,” in 2023 Int. Conf. Evol. Algor. Soft Comput. Tech. (EASCT), IEEE, 2023, pp. 1–7. [Google Scholar]
17. Z. Zhou, M. M. Rahman Siddiquee, N. Tajbakhsh, and J. Liang, “Unet++: A nested u-net architecture for medical image segmentation,” in Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, Cham: Springer, 2018, pp. 3–11. [Google Scholar]
18. T. -Y. Lin, P. Dollár, R. Girshick, K. He, B. Hariharan and S. Belongie, “Feature pyramid networks for object detection,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit., 2017, pp. 2117–2125. [Google Scholar]
19. L. -C. Chen, “Rethinking atrous convolution for semantic image segmentation,” 2017, arXiv:1706.05587. [Google Scholar]
20. H. Li, P. Xiong, J. An, and L. Wang, “Pyramid attention network for semantic segmentation,” 2018, arXiv:1805.10180. [Google Scholar]
21. O. Ronneberger, P. Fischer, and T. Brox, “U-Net: Convolutional networks for biomedical image segmentation,” in Med. Image Comput. Comput.-Assist. Interven.–MICCAI 2015, Munich, Germany, 2015, pp. 234–241. [Google Scholar]
22. M. Al Amin, B. K. Paul, and N. I. Bithi, “Real time detection and localization of colorectal polyps from colonoscopy images: A deep learning approach,” in 2022 IEEE Int. Women Eng. (WIE) Conf. Elect. Comput. Eng. (WIECON-ECE), IEEE, 2022, pp. 58–61. [Google Scholar]
23. P. E. Chavarrias-Solano, M. A. Teevno, G. Ochoa-Ruiz, and S. Ali, “Knowledge distillation with a class-aware loss for endoscopic disease detection,” in Cancer Prevention Through Early Detection, Cham: Springer, 2022, pp. 67–76. [Google Scholar]
24. A. Tamhane, T. Mida, E. Posner, and M. Bouhnik, “Colonoscopy landmark detection using vision transformers,” in MICCAI Workshop Imag. Syst. GI Endos., Springer, 2022, pp. 24–34. [Google Scholar]
25. S. Tummala, S. Kadry, S. A. C. Bukhari, and H. T. Rauf, “Classification of brain tumor from magnetic resonance imaging using vision transformers ensembling,” Curr. Oncol., vol. 29, no. 10, pp. 7498–7511, 2022. doi: 10.3390/curroncol29100590. [Google Scholar] [PubMed] [CrossRef]
26. H. Malik, A. Naeem, A. Sadeghi-Niaraki, R. A. Naqvi, and S. -W. Lee, “Multi-classification deep learning models for detection of ulcerative colitis, polyps, and dyed-lifted polyps using wireless capsule endoscopy images,” Complex Intell. Syst., vol. 10, no. 2, pp. 2477–2497, 2024. doi: 10.1007/s40747-023-01271-5. [Google Scholar] [CrossRef]
27. X. Liu, X. Guo, Y. Liu, and Y. Yuan, “Consolidated domain adaptive detection and localization framework for cross-device colonoscopic images,” Med. Image Anal., vol. 71, 2021, Art. no. 102052. doi: 10.1016/j.media.2021.102052. [Google Scholar] [PubMed] [CrossRef]
28. W. Y. Li, C. Yang, J. Liu, X. Y. Liu, X. Q. Guo and Y. X. Yuan, “Joint polyp detection and segmentation with heterogeneous endoscopic data,” in 3rd Int. Workshop Chall. Comput. Vis. Endosc. (EndoCV 2021Co-Located with the 17th IEEE Int. Symp. Biomed. Imag. (ISBI 2021), CEUR-WS Team, 2021, pp. 69–79. [Google Scholar]
29. C. -Y. Wang, A. Bochkovskiy, and H. -Y. M. Liao, “YOLOv7: Trainable bag-of-freebies sets new state-of-the-art for real-time object detectors,” in Proc. IEEE/CVF Conf. Comput. Vis. Pattern Recognit., 2023, pp. 7464–7475. [Google Scholar]
30. J. Redmon, “YOLOv3: An incremental improvement,” 2018, arXiv:1804.02767. [Google Scholar]
31. A. Karaman et al., “Robust real-time polyp detection system design based on yolo algorithms by optimizing activation functions and hyper-parameters with artificial bee colony (ABC),” Expert. Syst. Appl., vol. 221, no. 2, 2023, Art. no. 119741. doi: 10.1016/j.eswa.2023.119741. [Google Scholar] [CrossRef]
32. S. Xu et al., “PP-YOLOE: An evolved version of YOLO,” 2022, arXiv:2203.16250. [Google Scholar]
33. C. -Y. Wang, I. -H. Yeh, and H. -Y. M. Liao, “You only learn one representation: Unified network for multiple tasks,” 2021, arXiv:2105.04206. [Google Scholar]
34. Y. Zhao et al., “Detrs beat yolos on real-time object detection,” in Proc. IEEE/CVF Conf. Comput. Vis. Pattern Recognit., 2024, pp. 16965–16974. [Google Scholar]
35. C. Lyu et al., “Rtmdet: An empirical study of designing real-time object detectors,” 2022, arXiv:2212.07784. [Google Scholar]
36. L. Liu, Z. Hu, Y. Dai, X. Ma, and P. Deng, “ISA: Ingenious siamese attention for object detection algorithms towards complex scenes,” ISA Trans., vol. 143, no. 8, pp. 205–220, 2023. doi: 10.1016/j.isatra.2023.09.001. [Google Scholar] [PubMed] [CrossRef]
37. E. J. Petracchi et al., “Use of artificial intelligence in the detection of the critical view of safety during laparoscopic cholecystectomy,” J. Gastrointest. Surg., vol. 28, no. 6, pp. 877–879, 2024. doi: 10.1016/j.gassur.2024.03.018. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools