 Open Access
Open Access
ARTICLE
Stochastic Augmented-Based Dual-Teaching for Semi-Supervised Medical Image Segmentation
1 School of Computer Science and Engineering, Chongqing University of Technology, Chongqing, 400054, China
2 College of Artificial Intelligence, Chongqing Technology and Business University, Chongqing, 400067, China
* Corresponding Author: Yang Yuan. Email:
(This article belongs to the Special Issue: Deep Learning in Computer-Aided Diagnosis Based on Medical Image)
Computers, Materials & Continua 2025, 82(1), 543-560. https://doi.org/10.32604/cmc.2024.056478
Received 23 July 2024; Accepted 06 October 2024; Issue published 03 January 2025
Abstract
Existing semi-supervised medical image segmentation algorithms use copy-paste data augmentation to correct the labeled-unlabeled data distribution mismatch. However, current copy-paste methods have three limitations: (1) training the model solely with copy-paste mixed pictures from labeled and unlabeled input loses a lot of labeled information; (2) low-quality pseudo-labels can cause confirmation bias in pseudo-supervised learning on unlabeled data; (3) the segmentation performance in low-contrast and local regions is less than optimal. We design a Stochastic Augmentation-Based Dual-Teaching Auxiliary Training Strategy (SADT), which enhances feature diversity and learns high-quality features to overcome these problems. To be more precise, SADT trains the Student Network by using pseudo-label-based training from Teacher Network 1 and supervised learning with labeled data, which prevents the loss of rare labeled data. We introduce a bi-directional copy-paste mask with progressive high-entropy filtering to reduce data distribution disparities and mitigate confirmation bias in pseudo-supervision. For the mixed images, Deep-Shallow Spatial Contrastive Learning (DSSCL) is proposed in the feature spaces of Teacher Network 2 and the Student Network to improve the segmentation capabilities in low-contrast and local areas. In this procedure, the features retrieved by the Student Network are subjected to a random feature perturbation technique. On two openly available datasets, extensive trials show that our proposed SADT performs much better than the state-of-the-art semi-supervised medical segmentation techniques. Using only 10% of the labeled data for training, SADT was able to acquire a Dice score of 90.10% on the ACDC (Automatic Cardiac Diagnosis Challenge) dataset.Keywords
Segmenting medical images is a core task in medical image processing that plays an indispensable role in assisting diagnosis. By separating lesions or other regions of interest from the backdrop of the picture, it lays the groundwork for accurate localization and in-depth examination of the lesions. Large volumes of labeled data have enabled supervised medical image segmentation networks [1,2] to achieve impressive results. However, medical imaging variety and complexity provide difficulties. These include high annotation costs and data scarcity, which restrict the training of models for segmenting medical images. Learning with semi-supervision (SSL) strategies train models by combining scarce labeled data with abundant unlabeled data. They effectively address the issue of sparse annotations by leveraging precious labeled information to provide a wealth of a priori knowledge for unlabeled images, thereby helping to establish connections between labeled and unlabeled images. As a result, medical image segmentation techniques based on SSL have emerged as a promising area of study.
SSL has advanced significantly in the realm of medical image segmentation [3,4] in recent years. Its core strategies can be summarized as: pseudo-supervision techniques [5], prior knowledge-based techniques [4], and consistency regularization techniques [6]. Learning strategies based on the Mean Teacher structure [7–9] or cross-task consistency learning strategies [10] have demonstrated their usefulness. Recently, consistency regularization paradigms have gained more attention through the application of strong-weak augmentation [6] or the introduction of contrastive learning [11]. Strong-weak augmentation strategies involve processing the same image in different ways and use consistency loss to ensure that predictions from different views remain consistent. By calculating the distances (measured by cosine similarity) between positive and negative sample pairs taken from the data, contrastive learning seeks to reduce the gap between similar characteristics and increase the distance between dissimilar features. The significance of contrastive learning approaches is heightened in clinical settings due to the ease with which unlabeled data may be obtained.
A crucial presumption for SSL’s effectiveness is that the dataset’s labeled and unlabeled data have the same distribution. However, with a relatively little quantity of labeled data in clinical data collection, it is not viable to extrapolate the distribution of the complete dataset. An empirical distribution mismatch between labeled and unlabeled data results from this [12]. To address this issue, References [6,13] proposed a copy-paste augmentation technique, generating partially labeled images by copying small patches from labeled images and pasting them onto unlabeled images. Strictly depending on these partially labeled images means that the Teacher Network’s pseudo-labels are crucial. Confirmation bias and the widespread disregard of sparsely labeled data may result from this. Additionally, these methods perform poorly in low-contrast regions and local area segmentation.
In order to solve the aforementioned problems, we created SADT specifically for medical image segmentation tasks. In order to reduce confirmation bias, we suggest using a bidirectional copy-paste mask with progressive high-entropy filtering to weed out untrustworthy pixels in the early training phases. We introduce the use of pseudo-supervised and supervised learning to train the Student Network, addressing the issue of losing valuable labeled information. For mixed images, we design DSSCL in the feature spaces of Student Network with random feature perturbation and Teacher Network 2 to learn high-quality features, thereby improving segmentation capabilities in low-contrast and local regions.
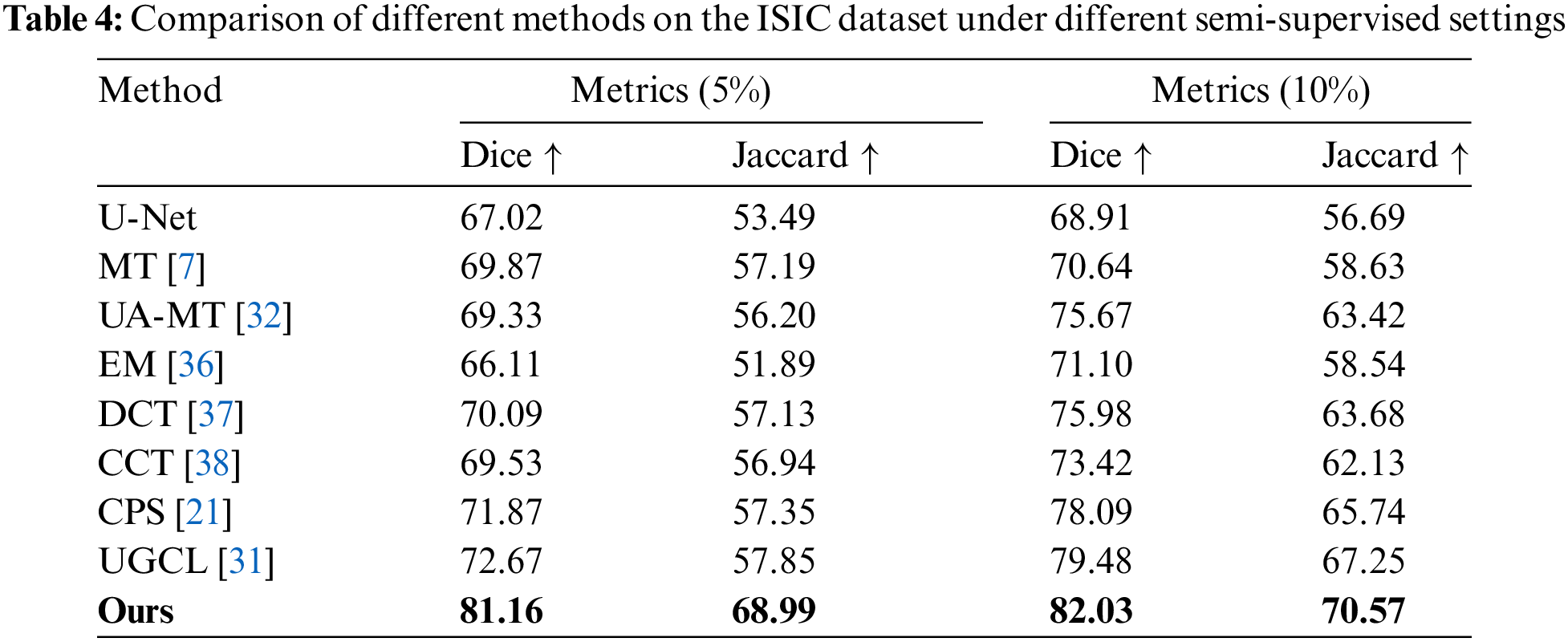
Two publicly accessible datasets, the ACDC [14] and ISIC (International Skin Imaging Collaboration) [15] datasets, were used to validate the designed SADT. Through a series of comparison trials, our offered SADT technique shows greater efficiency than existing leading methods in the semi-supervised medical image segmentation area, namely Bidirectional Copy-Paste (BCP [13]). Using the ACDC data collection, our model using just 5% labeled data achieved an 88.68% Dice score, and at 10% labeled data, it outperformed the leading BCP method by 1.26%. The distribution gap between labeled and unlabeled data is reduced by the provided SADT. It preserves scarce labeled information. It improves segmentation in low-contrast and local regions. Ablation studies were conducted to further confirm the efficiency of each component. We used Unet [1] as segmentation network baseline.
To sum up, this paper’s primary contributions are as follows:
(1) Proposing a bidirectional copy-paste mask with progressive high-entropy filtering to reduce confirmation bias caused by unreliable pixels and narrow the discrepancy in distribution between data with and without labels.
(2) Proposing a dual-teacher-assisted training strategy that combines supervision, pseudo-supervision, and contrastive learning to assist in training the Student Network.
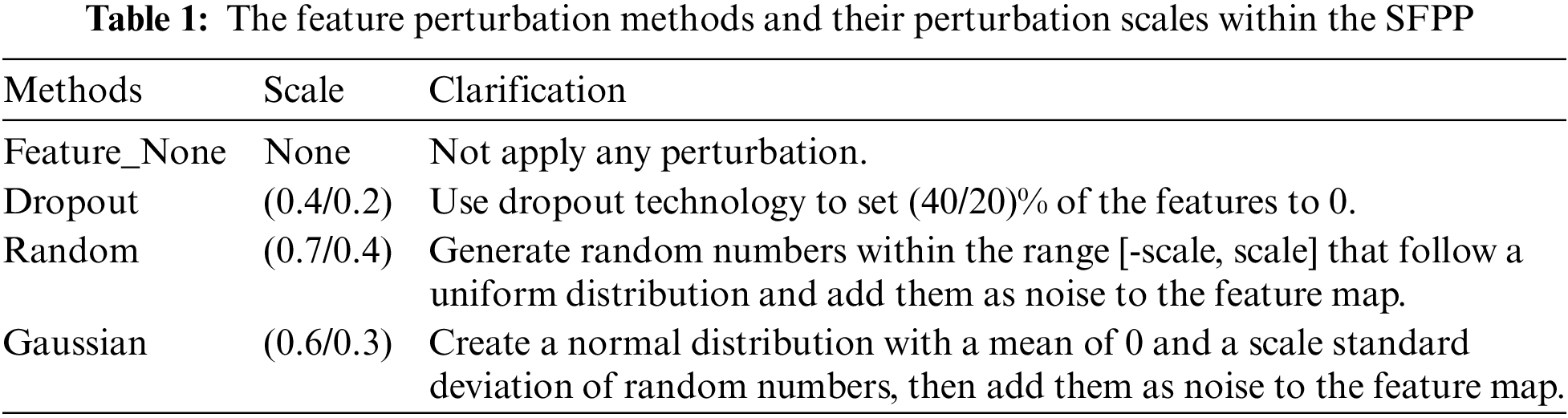
(3) Designing a Stochastic Feature Perturbation Pool (SFPP), which applies perturbations of random intensity to the features to increase feature diversity and thereby improve the capability of segmenting low-contrast areas.
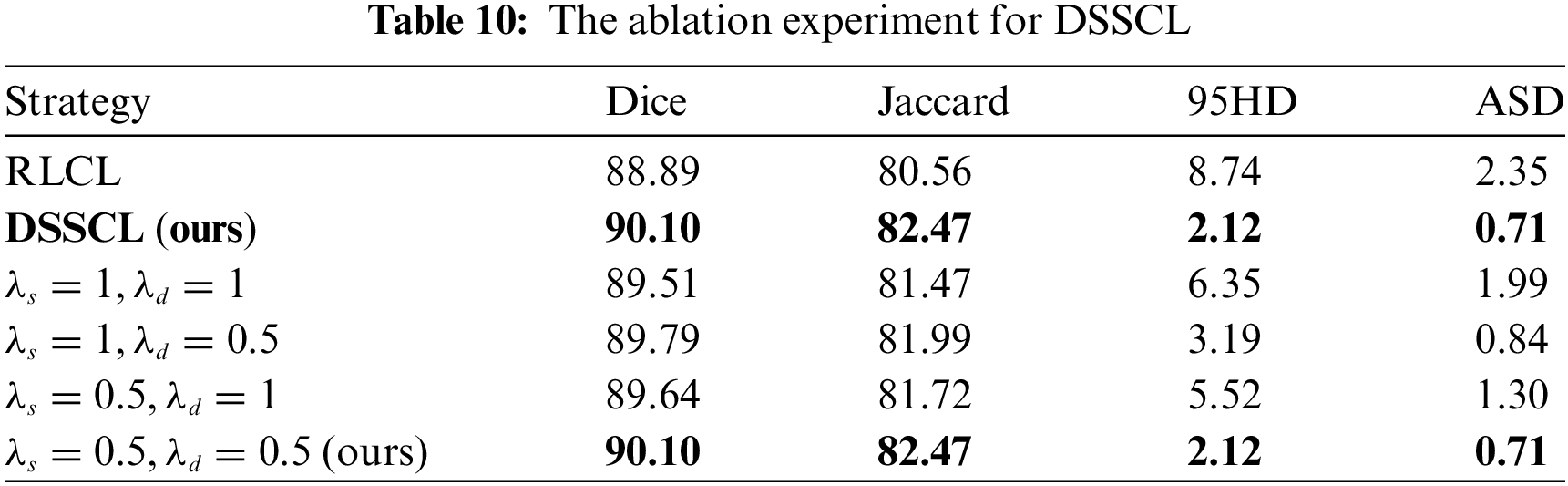
(4) Designing DSSCL to encourage the learning of high-quality features in both deep and representational layers, thereby improving the model’s low-contrast area and local segmentation performance.
(5) Proving the efficacy of the offered SADT and validating it on two public datasets.
2.1 Medical Image Segmentation
In medical image segmentation tasks, research based on the Unet model targeting various refinement directions has achieved significant improvements. Self-attention components were included by Azad et al. [16] in order to lower computing costs and improve the segmentation performance of the model. Skip connections are optimized using MultiResUnet [17] in order to include multi-scale characteristics. Rahman et al. [18,19] stated cascaded multi-scale features and attention mechanisms to address inconsistencies between local features. A universal medical image segmentation method that handled data heterogeneity and annotation variability was published by Gao et al. [20]. However, our method differs from these by leveraging an enormous volume of unlabeled data alongside scarce labeled data to train models for segmenting medical images, tackling challenges such as the difficulty of pixel-level annotations.
2.2 Semi-Supervised Learning (SSL)
SSL combines both marked and unmarked data to train a model. Unsupervised methods have higher uncertainty, while supervised training faces difficulties in data annotation. Therefore, research on SSL for semantic segmentation has rapidly developed [21–24]. Recent SSL strategies using strong and weak augmentations to enforce consistency between the results of teacher and student networks have shown good performance [25–27], where a unique method of dual-stream perturbation was presented by Yang et al. [28]. A pseudo-label correction technique was presented by Zhao et al. [29] in order to lessen the effect of noisy pseudo-labels. However, these approaches do not effectively leverage the connection between labeled and unlabeled data. Given the scarcity and diversity of medical image data, pixel-level annotation is particularly challenging. As a result, semi-supervised learning techniques for medical image segmentation have to be able to narrow the distribution gap between labeled and unlabeled images while fully using the important labeled data.
CL is fundamentally a form of metric learning that utilizes the information in a dataset to learn a powerful feature representation space. Regarding the semi-supervised segmentation of medical images, CL techniques are employed to effectively learn features [9]. Wang et al. [22] discussed use CL to move sparse area anchor features toward highly regularized key centers. ELN [23] demonstrates how the quality of features the model extracts is enhanced by patch-based per-pixel contrastive loss. The proposed DSSCL method designs a contrastive loss that considers both detailed deep features and geometric edge features across the entire batch. This approach aims to cluster similar features in the feature space, improving the quality of learned features and enhancing discrimination in low-contrast and local pixel regions.
2.4 Copy-Paste Data Enhancement Technology
Copy-paste is an effective data augmentation technique. In SSL, it helps to increase data diversity and establish connections between labeled and unlabeled data. Augmentation matters [26] adaptively copy-pastes low-confidence unlabeled samples onto labeled ones. Cut-paste consistency [6] matches unlabeled with labeled images based on color similarity and pastes more matching labeled lesion information onto unlabeled images using Gaussian blurring. Chi et al. [30] proposed an adaptive bidirectional displacement to generate new unlabeled samples, addressing the impact of perturbation methods. BCP [13] copy-pastes labeled onto unlabeled images using zero-centered masks, processing both types of images in the same way to reduce their distribution gap.
Inspired by [13,23,26,28], the proposed SADT model addresses significant loss of label information and low-quality pseudo-labels leading to confirmation bias and poor segmentation performance in low-contrast and local regions in challenges involving semi-supervised medical image segmentation using a dual-teacher assisted training strategy.
This section describes the construction of a semi-supervised learning technique for medical picture segmentation, including important terminology and symbols. Firstly, a summary of the SADT framework is given in Section 3.1. Secondly, Section 3.2 describes how pseudo-labels are copied and pasted onto true labels using a progressive high-entropy filtering mask to achieve partial supervision is described. Thirdly, in Section 3.3, the proposed SFPP strategy to enhance feature diversity in a simplified manner is detailed. Finally, in Section 3.4, the proposed DSSCL method to learn high-quality features and improve segmentation capabilities in local and low-contrast regions is described.
During the training process, the medical image dataset includes
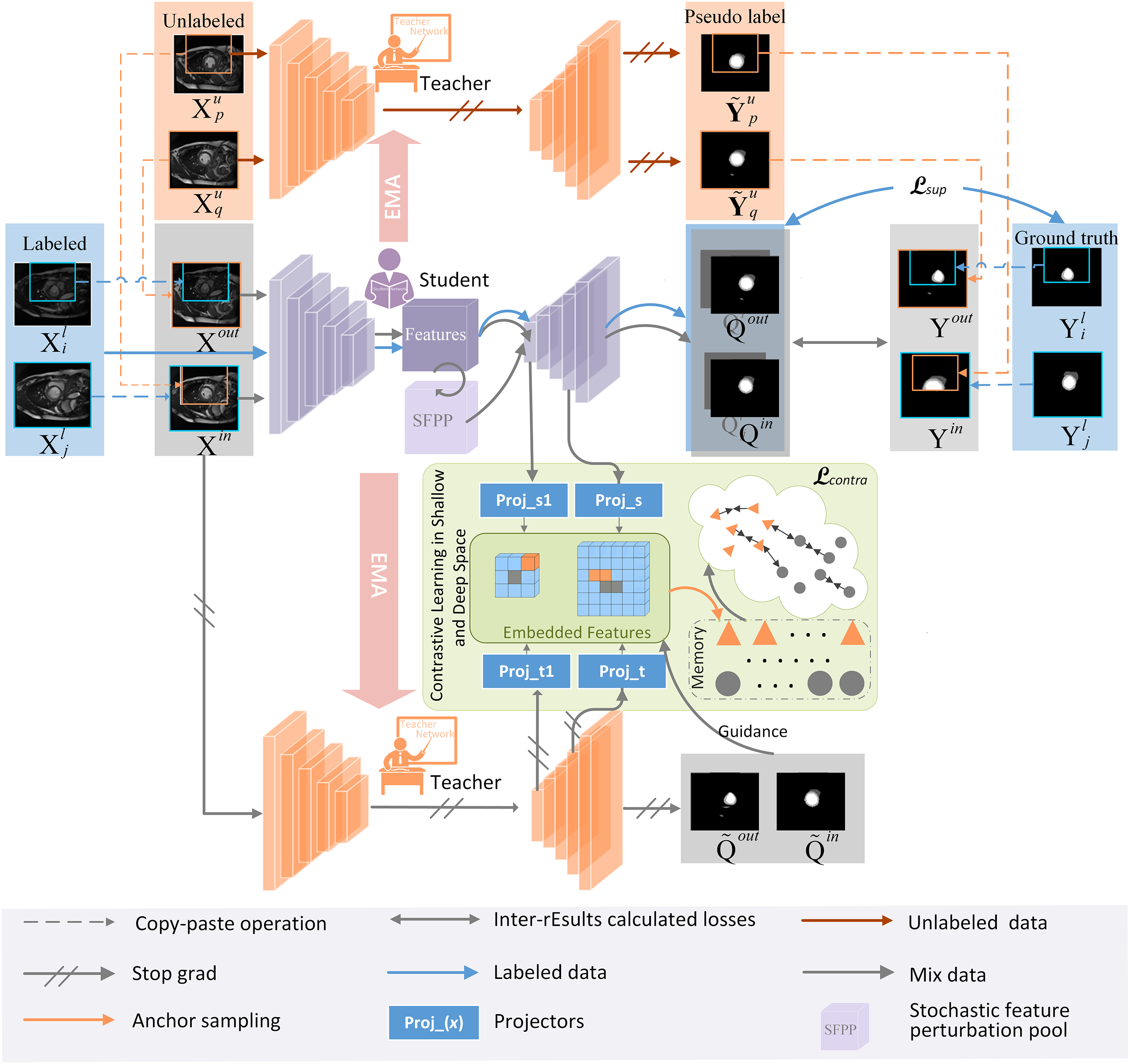
Figure 1: Overall framework
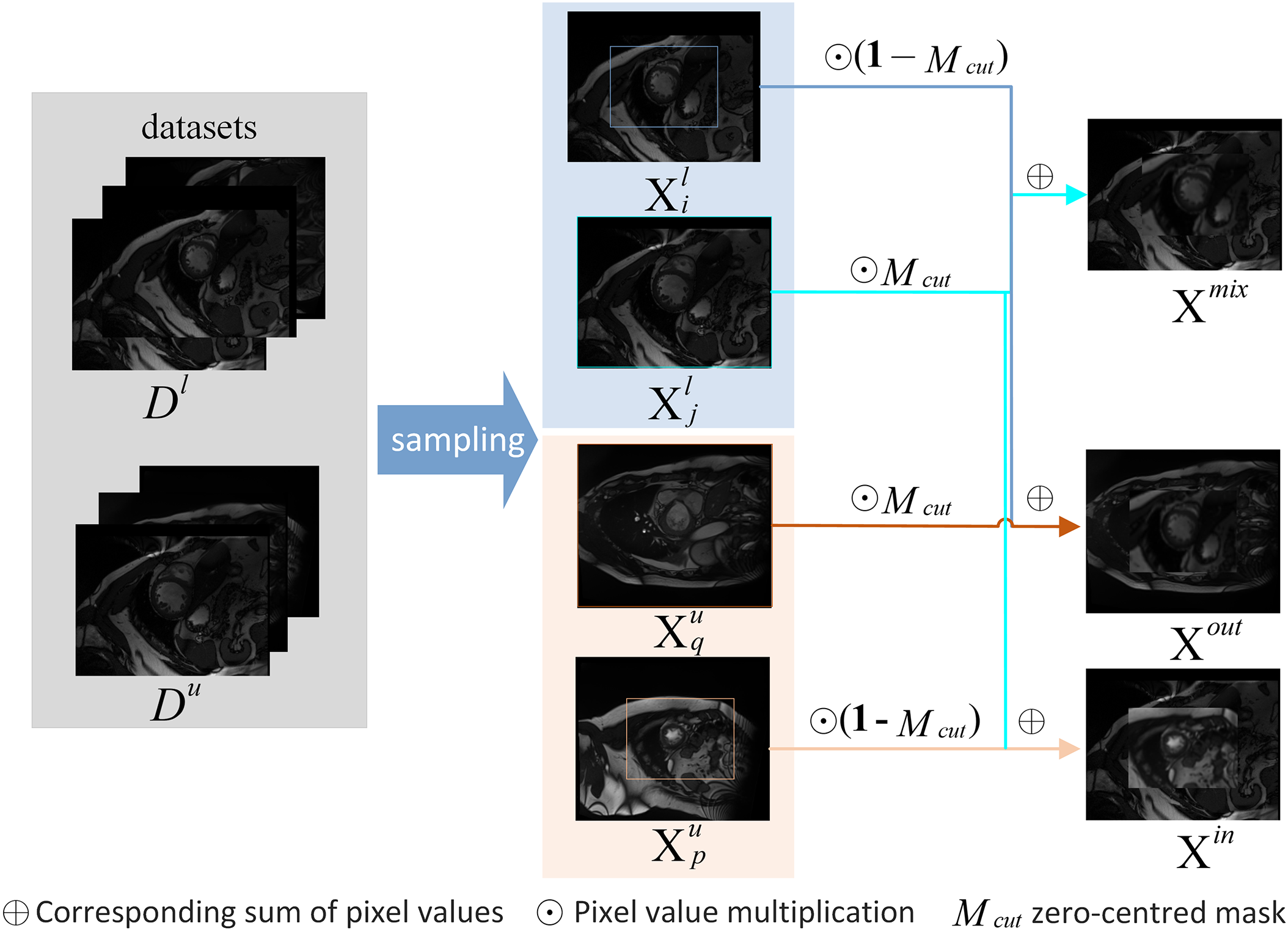
Figure 2: The detailed process for generating three types of mixed images through copy-paste
The training phase of the proposed SADT consists of two steps. The first step involves pre-training the Unet network using labeled data
A batch of tagged and unlabeled samples is obtained in each round. By reducing the supervised loss, the partial pseudo-supervised loss, and the contrastive loss, the Student Network is to be optimized. Thus, the total loss of training the Student Network is:
Pre-training In order to properly acquire previous knowledge of picture augmentation noise and labeling information, the pre-training procedure for the BCP method is enhanced.
Bidirectional copy-paste mask with progressive high-entropy filtering During the early stages of training with pseudo-supervision, some pixels are difficult to confidently classify, leading to severe confirmation bias. Therefore, we design a progressive high-entropy filtering mask for their pseudo-labeling to reduce the unreliability of pseudo-supervision. That is, in the initial stage, only pixels with low entropy in the Teacher Network’s predictions are selected. As learning progresses, the range of predicted pixels involved in loss calculation gradually expands. Specifically, firstly, obtain the prediction probabilities according to
A zero-centered mask
where
According to
The loss computation supervised by
Therefore, the partial supervised loss is computed as
3.3 Deep-Shallow Spatial Contrastive Learning (DSSCL)
Pseudo-supervision is weak in learning feature patterns of the entire dataset and in segmenting local low-contrast regions. Therefore, we introduce a new contrastive learning paradigm for mixed images—DSSCL—forming a dual-teacher-assisted training structure. As shown in Fig. 1, the Student Network and Teacher Network 2 each contain two feature projectors
In each feature embedding space, define
where V represents the set of pixels of mixed images
3.4 Stochastic Feature Perturbation Pool (SFPP)
In medical image segmentation tasks, the insufficient segmentation capability in low-contrast regions is particularly prominent. Medical images have a complicated backdrop with very little contrast between the target and background. To enhance feature diversity and improve the model’s segmentation performance in low-contrast regions, a random feature perturbation pool is designed. This method applies perturbations of random intensity to the features extracted by the Student Network. Existing feature processing uses a fixed dropout method, which limits the effectiveness of semi-supervised learning. Random feature perturbation is a simpler way to generate feature diversity, whereas fixing multiple perturbation types may overly distort the features, harming their distribution.
Specifically, it randomly selects no more than
ACDC Dataset Three kinds of annotated short-axis cardiac MR-cine (Magnetic Resonance Cine) images are included in the ACDC [14] dataset. The detailed characteristics of the dataset are shown in the first row of Table 2. ACDC is aimed at segmenting LV, RV, and Myo in cardiac dynamic magnetic resonance imaging diastolic (ED) and systolic (ES) frames. There are five cases, with 30 cases per category. The data split is conducted following the BCP method, having 100 examples established as patient scans for testing, validation, and training, respectively, of 70, 10, and 20.
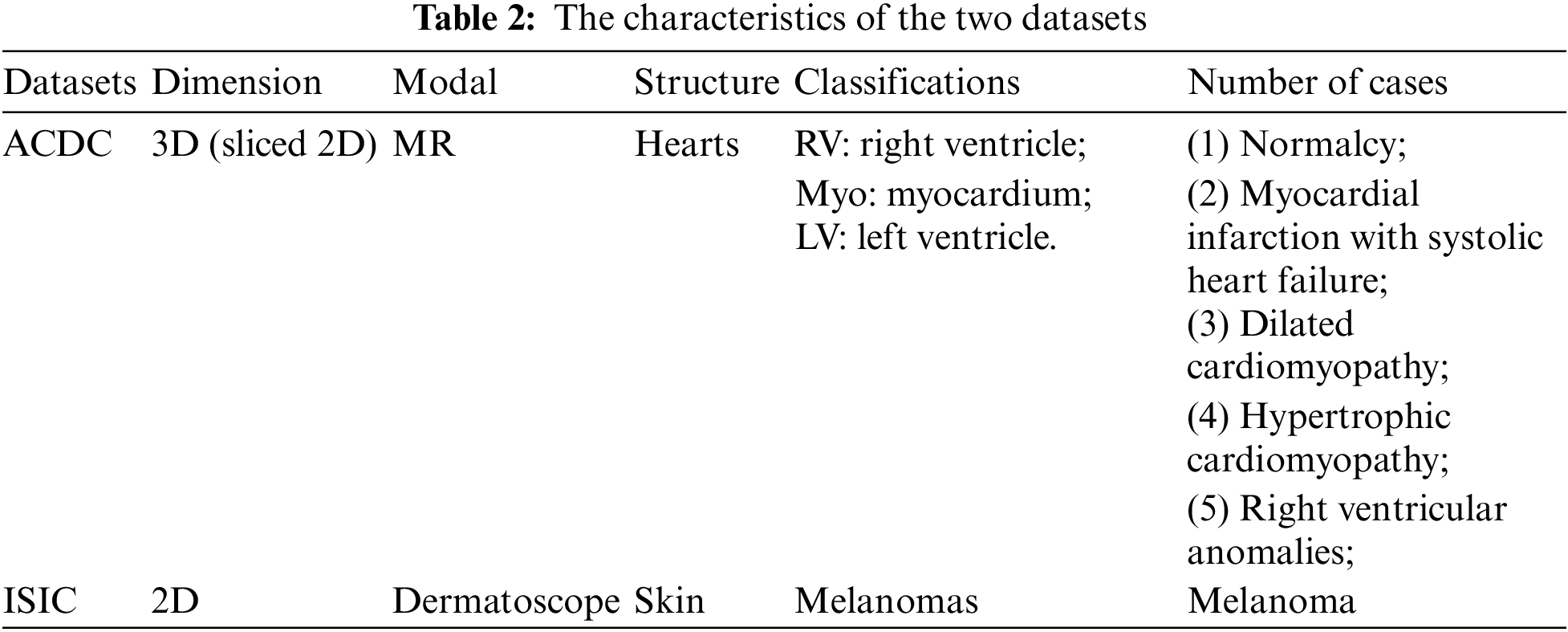
ISIC Dataset The ISIC dataset [15] is dermoscopic image dataset aimed at identifying melanoma regions through image recognition. The detailed characteristics of the dataset are shown in Table 2’s second row. The experimental design from earlier work, with 1815 images serving as the training set and 779 images serving as the validation set, to assure experiment fairness. Of the images in the training set, 5% (91) and 10% (181) include labels and are applied to various semi-supervised trial scenarios.
Four widely-used assessment measures were chosen in order to assess the effectiveness of SADT: Average Surface Distance (ASD), 95% Hausdorff Distance (95HD), Dice Score (%), and Jaccard Score (%). ASD determines the average distance between the borders, 95HD estimates the distance of the nearest point between the two regions, and Dice Score and Jaccard Score compute the proportion of overlapping areas between two target areas.
ACDC Dataset To ensure fairness, the experimental parameters were set following the BCP method, with Unet selected as the backbone network. The input picture size was set to
ISIC Dataset The experimental parameters were set following the Uncertainty-Guided Pixel Contrastive Learning (UGCL [31]) method, with Unet serving as the backbone structure. 8 labeled images were included in the batch size of 16. Pre-training and self-training iteration counts were set to 2 and 6 k, respectively, during the training phase of all trials.
We decided on the stochastic gradient descent (SGD) optimizer, which has a momentum of 0.9 and a weight decay of 0.0005. A polynomial scheduling method was used to lower the initial learning rate from 0.01 to 0.001. We trained using an NVIDIA RTX 3090 GPU and used the PyTorch package to implement the proposed approach.
4.4 Comparison with State-of-the-Art Results
ACDC Dataset The offered SADT was trained using only 5% and 10% of the labeled data, and the test was conducted on the average segmentation results performed for the four categories (background, left ventricle, right ventricle, and myocardium) using the ACDC dataset. The comparative experimental results are shown in Table 3. From the results, our method outperforms all the most recent state-of-the-art (SOTA) methods in terms of performance. The Dice score is enhanced by 1.26% as compared to the BCP approach at a labeling rate of 10%. Visualization results comparing the method to advanced methods are shown in Fig. 3. Consequently, our approach fully utilizes the labeled data while also reducing the distribution gap between labeled and unlabeled data. Furthermore, it enhances the segmentation capabilities in low-contrast and local regions, achieving more accurate segmentation of edge region pixels.
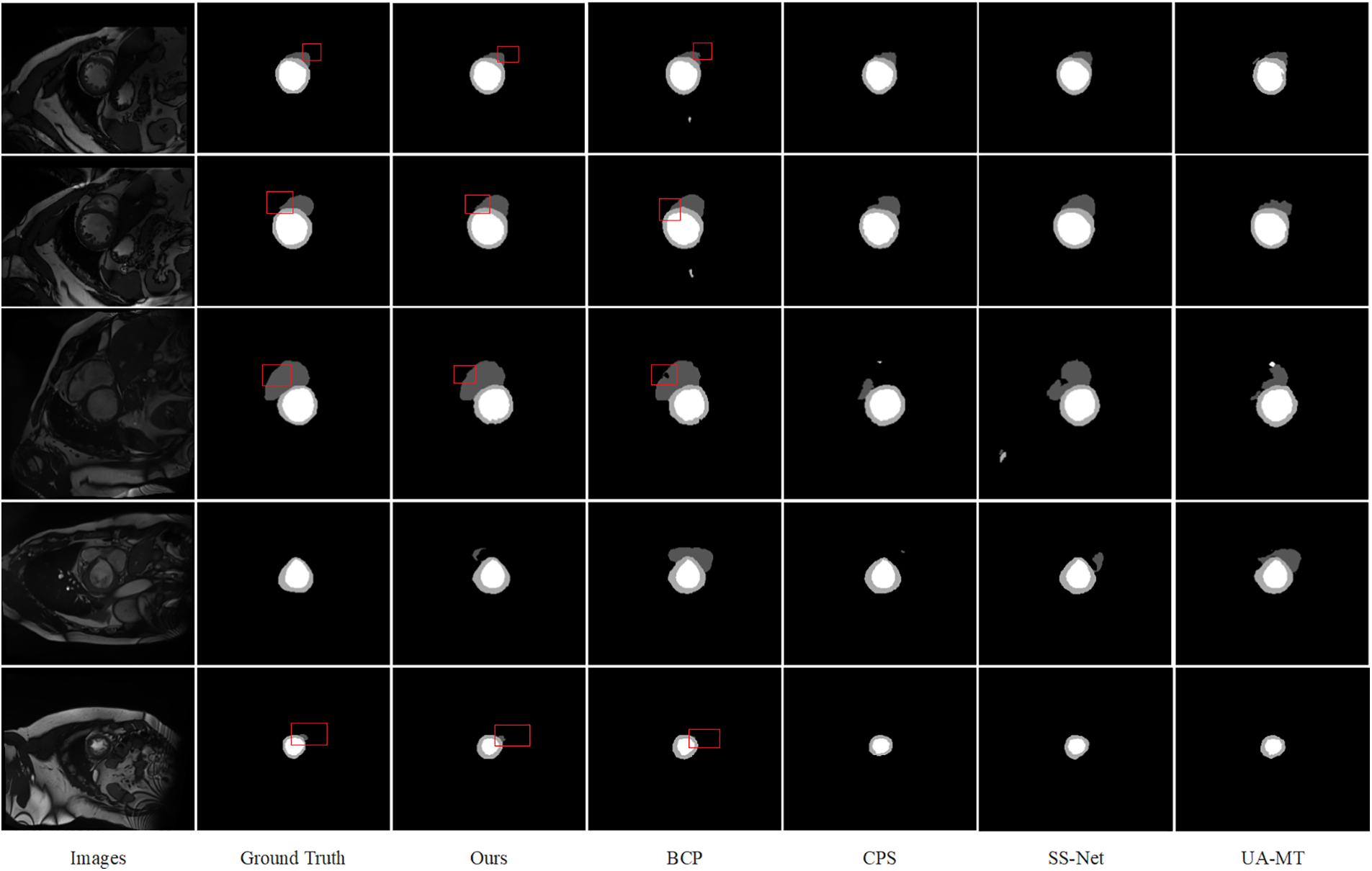
Figure 3: Comparison of several semi-supervised segmentation methods, prediction results of models trained with 10% labeled data on the ACDC dataset, and visualization of images and their labels
ISIC Dataset The proposed model was also trained on the ISIC dataset in order to confirm the model’s capacity for generalization. The average segmentation results performance for the two categories (melanoma and background) under conditions where the labeled data make up just 5% and 10% of the training data is shown in Table 4. The outcomes demonstrate that, for this dataset, our approach performs better than cutting-edge techniques. Dice and Jaccard scores were employed as assessment measures in accordance with the UGCL methodology. As demonstrated by the performance metrics when the model was trained with solely labeled data, as indicated in the first row of data in Table 4. Our proposed technique performs much better than previous methods under different semi-supervised scenarios. With a labeling ratio of 5%, the Dice score improves by 8.49% compared to the advanced UGCL method.
Through ablation experiments, the importance of each component was validated, including the improved pre-training strategy, progressively filtered bidirectional copy-paste mask, the dual-teacher structure combining supervised, pseudo-supervised, and DSSCL, and the SFPP strategy. The outcomes of these ablation tests are shown in Table 5, which also details the improvement process and considerable impact of our technique on the ACDC dataset at a 10% labeling rate.
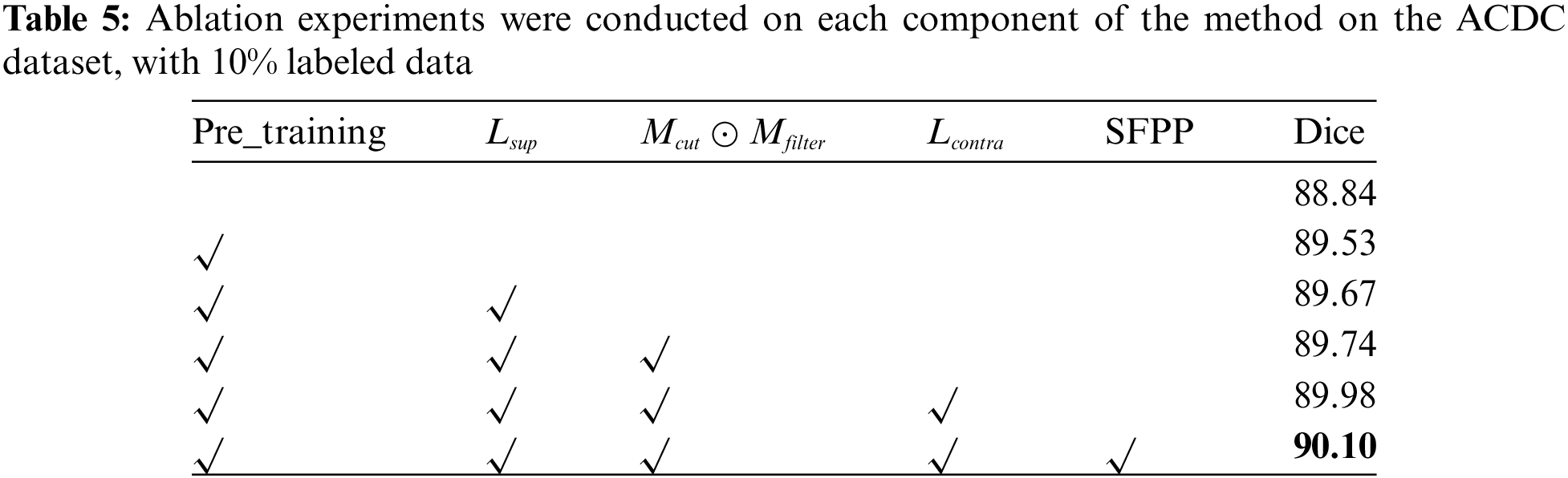
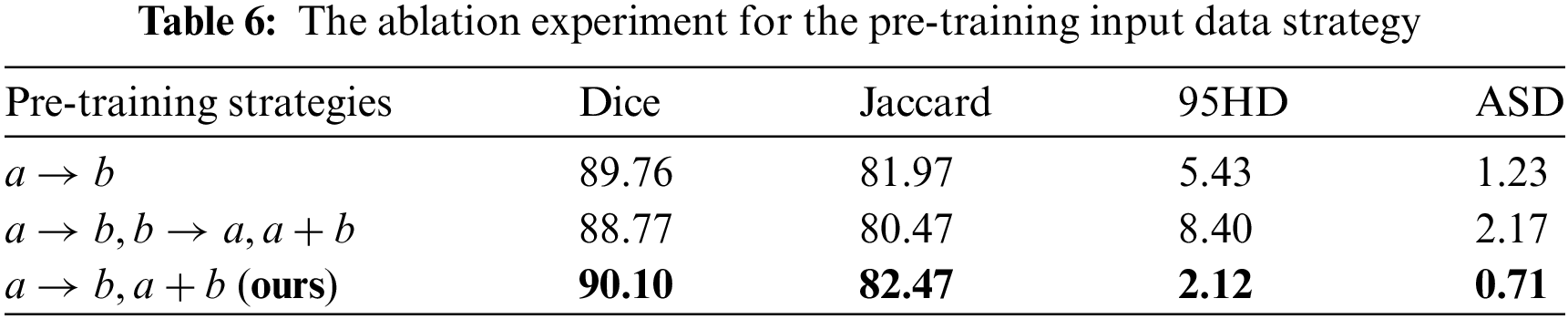
4.5.1 The Effectiveness of the Pre-Training Strategy
In the lead-up to training, the Unet was trained using the original labeled data and mixed images obtained through copy-paste operations between labeled data. Three pre-training input data schemes are designed to investigate the impact of different strategies on model training. As shown in Table 6, in each mini-batch, ‘
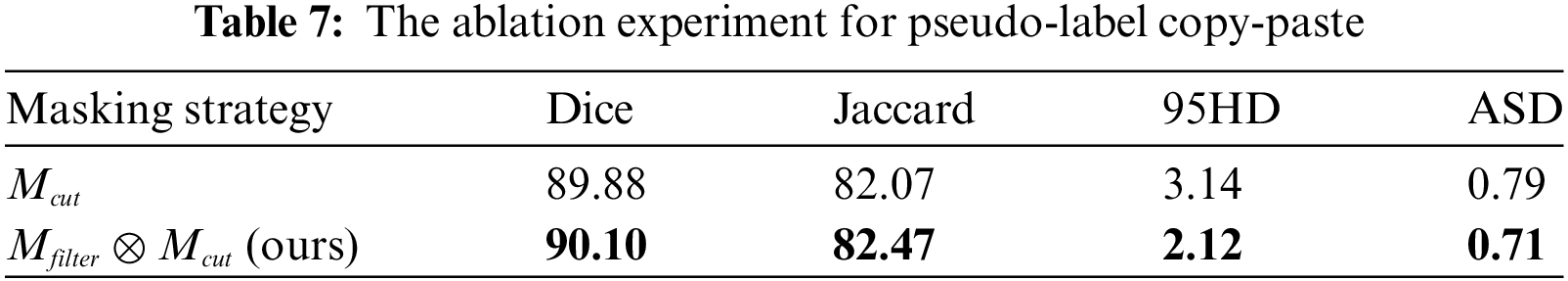
4.5.2 Validity of Filtered Pseudo-Label
The bidirectional copy-paste mask between pseudo-labels and true labels is generated through a dot product of the progressive high-entropy filter mask and the zero-centered mask. This process simultaneously reduces the gap in data distribution and the confirmation bias in pseudo-supervision. As shown in Table 7,
4.5.3 The Effectiveness of the Dual-Teacher-Assisted Training
To effectively guide the training of the Student Network, Teacher Network 1 provides pseudo-supervised learning, while Teacher Network 2 offers contrastive learning for the feature space of
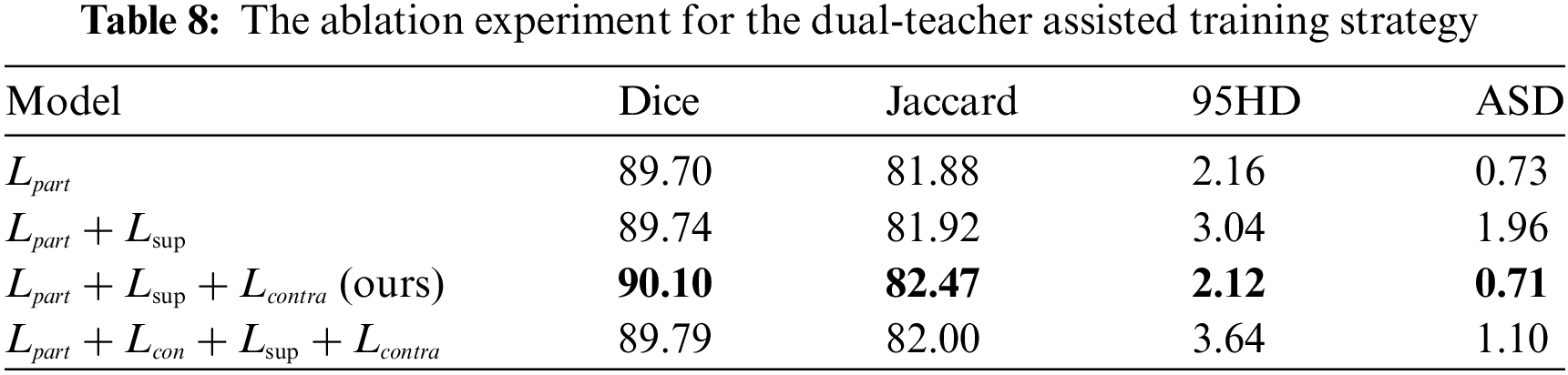
4.5.4 The Effectiveness of the SFPP
By perturbing features randomly, we aim to simplify the increase of feature diversity in contrastive learning. The effectiveness of randomly selecting up to
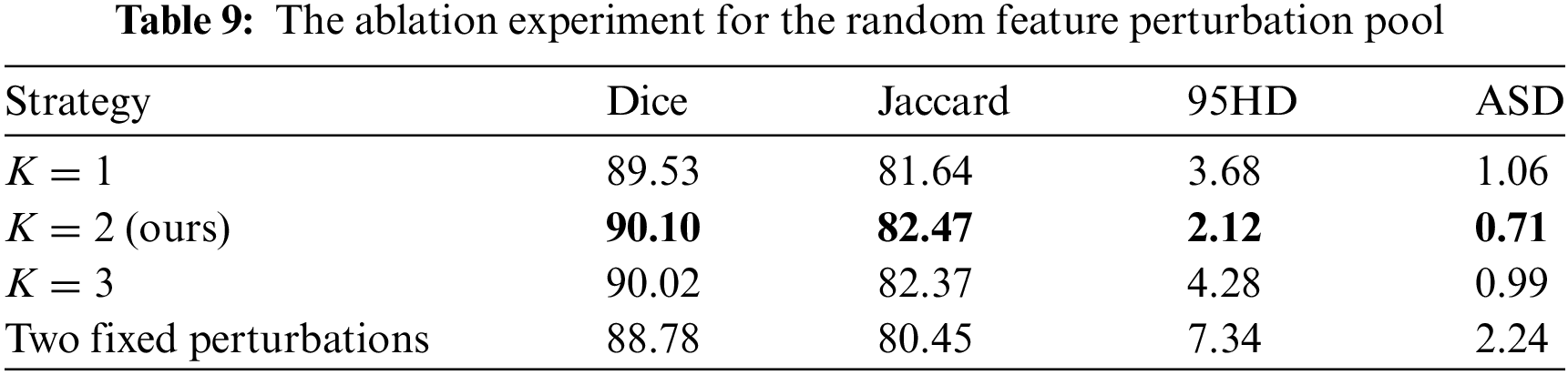
4.5.5 The Effectiveness of DSSCL
By designing contrastive learning losses at both the deep and representation layers of the decoder, the aim is to push features away from other classes and shorten the distance between comparable features. As shown in Table 10, the designed DSSCL was compared with contrastive learning at the Representation Layer alone (RLCL). The values of
We introduce SADT, a novel semi-supervised learning process, to improve medical picture segmentation accuracy. SADT combines random information enhancement techniques with DSSCL. Specifically, we adopt two teachers playing distinct roles help direct the student model’s learning process. This dual-teacher mechanism helps improve the empirical distribution matching between labeled and unlabeled data, reduces the confirmation bias of pseudo-labels, and facilitates the learning of high-quality features. Additionally, introduce the stochastic intensity-based feature perturbation pool strategy to increase feature diversity in a randomized manner. For DSSCL, we compute patch-based contrast losses at both the deep and shallow (representation) layers of the decoder, which aids in capturing local texture features and edge geometric features, thereby improving the quality of segmentation in low-contrast and local regions.
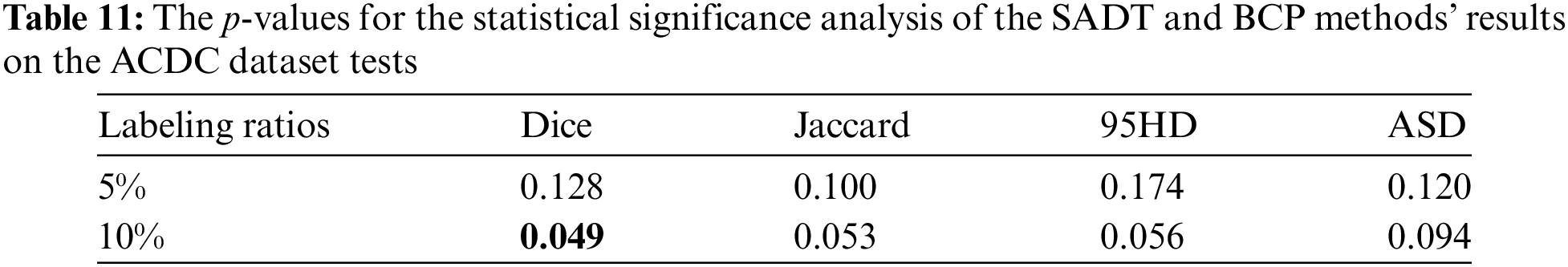
The experimental results show that with a labeling ratio of 10%, the model trained on the ACDC dataset, the SADT method improves the Dice score by 1.26% compared to the BCP method; and on the ISIC dataset, it improves by 2.55% compared to the UGCL method. These results demonstrate the superior performance of SADT. Additionally, as shown in Fig. 4, we conducted a statistical significance analysis. From the figure, it can be observed that the test result distribution of SADT is better compared to the BCP method. The difference between the highest and lowest values of the test results is smaller, the median values of the Dice and Jaccard metrics are higher than those of the BCP method, and the median values of the 95 HD and ASD metrics are lower than those of the BCP method.
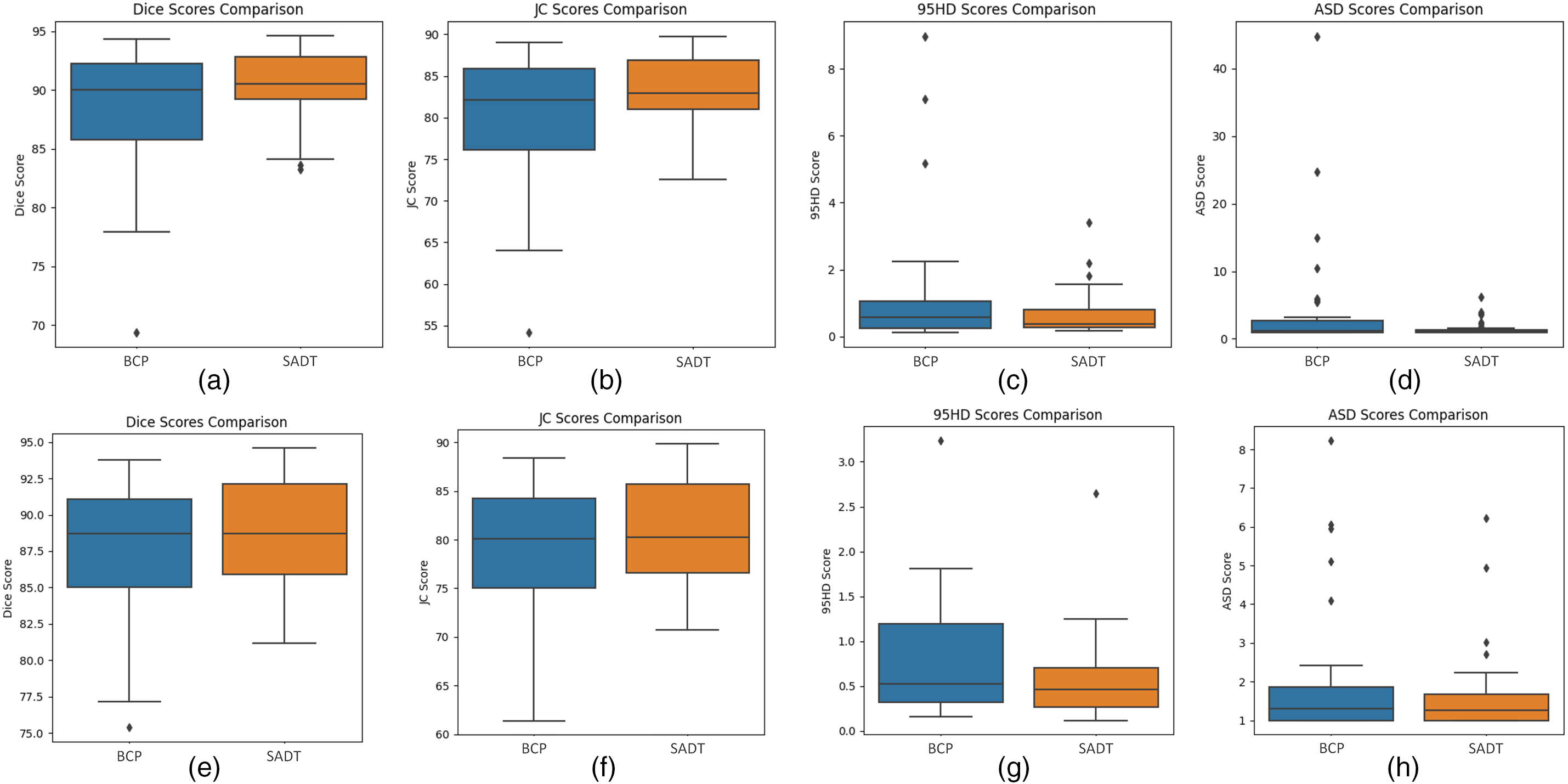
Figure 4: Boxplot comparing the distribution of metric scores for the SADT and BCP methods on the ACDC dataset test results. The labeled (a–d) represent the distribution of test scores after training the model with 10% labeled data. The box plots labeled (e–h) represent the distribution of test scores after training the model with 5% labeled data. The two approaches, BCP and SADT, are represented on the x-axis of each plot, and the evaluation metric score, with units in percentage (%), is represented on the y-axis
Although the proposed SADT method outperforms state-of-the-art semi-supervised learning strategies, there are still some limitations. Due to the use of supervised learning with real labeled data and comparative learning with mixed data in the feature space to guide the update of the student network, additional training costs are required. As shown in Table 11, the statistical t-test significance analysis results indicate that the Dice metric of the model trained with 10% labeled data shows a very significant difference compared to BCP (p-value < 0.05). However, the significance of the differences in Jaccard, 95HD, and ASD metrics weakens, with less significant advantages in outlier edge segmentation compared to BCP. The significance of differences between models trained with 5% labeled data is weaker due to the scarcity of labeled data, resulting in insignificant differences in prediction accuracy in complex and difficult-to-learn feature regions compared to the BCP method. Therefore, we will further enhance the distinctiveness of SADT compared to other advanced methods in semi-supervised medical image segmentation. In future work, we will continue to investigate the SADT framework, optimize its contrast learning strategy, and effectively apply it to different segmentation network baselines, while controlling the additional training costs.
The proposed SADT strategy enriches data augmentation with partial pseudo-supervision and contrastive learning. The distribution gap between labeled and unlabeled data is reduced by this method. It enhances segmentation capabilities in low-contrast and local regions. It also makes full use of expensive labeled information. Experimental comparisons show that the provided semi-supervised learning approach works better than the most advanced techniques, including BCP. With just 10% of the ACDC dataset’s data labeled, the Dice score reached 90.10%. On the ISIC dataset, using only 5% labeled data, the Dice score reached 81.16%, an improvement of 8.49% over UGCL.
Acknowledgement: We would like to thank the editor and the anonymous reviewers for taking the time to review and provide valuable comments.
Funding Statement: This study was supported by the Natural Science Foundation of China (No. 41804112; author: Chengyun Song).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Yang Yuan; data collection: Yang Yuan, Pengcheng Ren; analysis and interpretation of results: Yang Yuan, Hengyang Liu; draft manuscript preparation: Yang Yuan; checking and revising the manuscript preparation: Yang Yuan, Hengyang Liu, Chengyun Song, Fen Luo. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: We evaluate the semi-supervised medical image segmentation model proposed in this paper using two publicly available medical image datasets, i.e., ACDC dataset (https://humanheart-project.creatis.insa-lyon.fr/database/#collection/637218c173e9f0047faa00fb, accessed on 16 June 2023), ISIC dataset (https://challenge.isic-archive.com/data/#2018, accessed on 03 January 2024). All the data generated or analyzed in the course of this study are included in the tables of this paper.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. O. Ronneberger, P. Fischer, and T. Brox, “Unet: Convolutional networks for biomedical image segmentation,” in Med. Image Comput. Comput.-Assist. Interv.-MICCAI 2015: 18th Int. Conf., Munich, Germany, Oct. 5–9, 2015, pp. 234–241. [Google Scholar]
2. A. Tragakis, C. Kaul, R. Murray-Smith, and D. Husmeier, “The fully convolutional transformer for medical image segmentation,” in Proc. IEEE/CVF Conf. WACV, Waikoloa, HI, USA, 2023, pp. 3660–3669. [Google Scholar]
3. R. Jiao et al., “Learning with limited annotations: A survey on deep semi-supervised learning for medical image segmentation,” Comput. Biol. Med., vol. 169, 2023, Art. no. 107840. doi: 10.1016/j.compbiomed.2023.107840. [Google Scholar] [PubMed] [CrossRef]
4. F. Wu and X. Zhuang, “Minimizing estimated risks on unlabeled data: A new formulation for semi-supervised medical image segmentation,” IEEE Trans. Pattern Anal. Mach. Intell., vol. 45, no. 5, pp. 6021–6036, 2023. [Google Scholar] [PubMed]
5. Y. Wu, Z. Wu, Q. Wu, Z. Ge, and J. Cai, “Exploring smoothness and class-separation for semi-supervised medical image segmentation,” in Int. Conf. Med. Imag. Comput. Comput. Interv., 2022, pp. 34–43. doi: 10.1007/978-3-031-16443-9_4. [Google Scholar] [CrossRef]
6. B. P. Yap and B. K. Ng, “Cut-paste consistency learning for semi-supervised lesion segmentation,” in WACV, Waikoloa, HI, USA, 2023, pp. 6149–6158. [Google Scholar]
7. A. Tarvainen and H. Valpola, “Mean teachers are better role models: Weight-averaged consistency targets improve semi-supervised deep learning results,” Adv. Neural Inf. Process Syst., 2017. doi: 10.48550/arXiv.1703.01780. [Google Scholar] [CrossRef]
8. Y. Chen et al., “Dual-decoder consistency via pseudo-labels guided data augmentation for semi-supervised medical image segmentation,” 2023, arXiv:2308.16573. [Google Scholar]
9. C. You, Y. Zhou, R. Zhao, L. Staib, and J. S. Duncan, “SimCVD: Simple contrastive voxel-wise representation distillation for semi-supervised medical image segmentation,” IEEE Trans. Med. Imaging, vol. 41, no. 9, pp. 2228–2237, 2022. doi: 10.1109/TMI.2022.3161829. [Google Scholar] [PubMed] [CrossRef]
10. X. Luo, J. Chen, T. Song, and G. Wang, “Semi-supervised medical image segmentation through dual-task consistency,” in Proc. AAAI Conf. Artif. Intell., vol. 35, no. 10, pp. 8801–8809, 2021. doi: 10.1609/aaai.v35i10.17066. [Google Scholar] [CrossRef]
11. H. Basak and Z. Yin, “Pseudo-label guided contrastive learning for semi-supervised medical image segmentation,” in Proc. IEEE/CVF Conf. CVPR, Vancouver, BC, Canada, 2023, pp. 19786–19797. [Google Scholar]
12. Q. Wang, W. Li, and L. V. Gool, “Semi-supervised learning by augmented distribution alignment,” in Proc. IEEE/CVF Conf. ICCV, Seoul, Republic of Korea, 2019, pp. 1466–1475. [Google Scholar]
13. Y. Bai, D. Chen, Q. Li, W. Shen, and Y. Wang, “Bidirectional copy-paste for semi-supervised medical image segmentation,” in Proc. IEEE/CVF Conf. CVPR, Vancouver, BC, Canada, 2023, pp. 11514–11524. [Google Scholar]
14. O. Bernard et al., “Deep learning techniques for automatic MRI cardiac multi-structures segmentation and diagnosis: Is the problem solved?” IEEE Trans. Med. Imaging, vol. 37, no. 11, pp. 2514–2525, 2018. doi: 10.1109/TMI.2018.2837502. [Google Scholar] [PubMed] [CrossRef]
15. N. C. F. Codella et al., “Skin lesion analysis toward melanoma detection: A challenge at the 2017 international symposium on biomedical imaging (ISBIhosted by the international skin imaging collaboration (ISIC),” in 2018 IEEE 15th ISBI 2018, Brighton, UK, IEEE, 2018, pp. 168–172. [Google Scholar]
16. R. Azad et al., “Beyond self-attention: Deformable large kernel attention for medical image segmentation,” in Proc. IEEE/CVF Conf. WACV, Waikoloa, HI, USA, 2024, pp. 1287–1297. [Google Scholar]
17. N. Ibtehaz and M. S. Rahman, “MultiResUNet: Rethinking the U-Net architecture for multimodal biomedical image segmentation,” Neural Netw., vol. 121, no. 11, pp. 74–87, 2019. doi: 10.1016/j.neunet.2019.08.025. [Google Scholar] [PubMed] [CrossRef]
18. M. M. Rahman and R. Marculescu, “Medical image segmentation via cascaded attention decoding,” in Proc. IEEE/CVF Conf. WACV, Waikoloa, HI, USA, 2023, pp. 6222–6231. [Google Scholar]
19. M. M. Rahman, M. Munir, and R. Marculescu, “EMCAD: Efficient multi-scale convolutional attention decoding for medical image segmentation,” in Proc. IEEE/CVF Conf. CVPR, Seattle, WA, USA, 2024, pp. 11769–11779. [Google Scholar]
20. Y. Gao, “Training like a medical resident: Context-prior learning toward universal medical image segmentation,” in Proc. IEEE/CVF Conf. CVPR, Seattle, WA, USA, 2024, pp. 11194–11204. [Google Scholar]
21. X. Chen, Y. Yuan, G. Zeng, and J. Wang, “Semi-supervised semantic segmentation with cross pseudo supervision,” in Proc. IEEE/CVF Conf. CVPR, Nashville, TN, USA, 2021, pp. 2613–2622. [Google Scholar]
22. X. Wang, B. Zhang, L. Yu, and J. Xiao, “Hunting sparsity: Density-guided contrastive learning for semi-supervised semantic segmentation,” in Proc. IEEE/CVF Conf. CVPR, Vancouver, BC, Canada, 2023, pp. 3114–3123. [Google Scholar]
23. D. Kwon and S. Kwak, “Semi-supervised semantic segmentation with error localization network,” in Proc. IEEE/CVF Conf. CVPR, New Orleans, LA, USA, 2022, pp. 9957–9967. [Google Scholar]
24. A. Peláez-Vegas, P. Mesejo, and J. Luengo, “A survey on semi-supervised semantic segmentation,” 2023, arXiv:2302.09899. [Google Scholar]
25. Y. Liu, Y. Tian, Y. Chen, F. Liu, V. Belagiannis and G. Carneiro, “Perturbed and strict mean teachers for semi-supervised semantic segmentation,” in Proc. IEEE/CVF Conf. Comput. Vis. Pattern Recogn. (CVPR), New Orleans, LA, USA, 2022, pp. 4248–4257. [Google Scholar]
26. Z. Zhao, L. Yang, S. Long, J. Pi, L. Zhou and J. Wang, “Augmentation matters: A simple-yet-effective approach to semi-supervised semantic segmentation,” in Proc. IEEE/CVF Conf. CVPR, Vancouver, BC, Canada, 2023, pp. 11350–11359. [Google Scholar]
27. K. B. Nguyen, “SequenceMatch: Revisiting the design of weak-strong augmentations for Semi-supervised learning,” in Proc. IEEE/CVF WACV, Waikoloa, HI, USA, 2024, pp. 96–106. [Google Scholar]
28. L. Yang, L. Qi, L. Feng, W. Zhang, and Y. Shi, “Revisiting weak-to-strong consistency in semi-supervised semantic segmentation,” in Proc. IEEE/CVF Conf. Comput. Vis. Pattern Recogn. (CVPR), Vancouver, BC, Canada, 2023, pp. 7236–7246. [Google Scholar]
29. X. Zhao et al., “RCPS: Rectified contrastive pseudo supervision for semi-supervised medical image segmentation,” IEEE J. Biomed. Health Inform., vol. 28, no. 1, pp. 251–261, 2024. doi: 10.1109/JBHI.2023.3322590. [Google Scholar] [PubMed] [CrossRef]
30. H. Chi, J. Pang, B. Zhang, and W. Liu, “Adaptive bidirectional displacement for semi-supervised medical image segmentation,” in Proc. IEEE/CVF Conf. CVPR, Seattle, WA, USA, 2024, pp. 4070–4080. [Google Scholar]
31. T. Wang et al., “Uncertainty-guided pixel contrastive learning for semi-supervised medical image segmentation,” in Europ. Conf. Artif. Intell. (ECAI), Vienna, Austria, 2022, pp. 1444–1450. [Google Scholar]
32. L. Yu, S. Wang, X. Li, C. W. Fu, and P. A. Heng, “Uncertainty-aware self-ensembling model for semi-supervised 3D left atrium segmentation,” in Med. Image Comput. Comput. Assist. Interv.–MICCAI 2019: 22nd Int. Conf., Shenzhen, China, Oct. 13–17, 2019, pp. 605–613. doi: 10.1007/978-3-030-32245-8_67. [Google Scholar] [CrossRef]
33. S. Li, C. Zhang, and X. He, “Shape-aware semi-supervised 3D semantic segmentation for medical images,” in Med. Image Comput. Comput. Assist. Interv.–MICCAI 2020: 23rd Int. Conf., Lima, Peru, Oct. 4–8, 2020, pp. 552–561. [Google Scholar]
34. X. Luo et al., “Efficient semi-supervised gross target volume of nasopharyngeal carcinoma segmentation via uncertainty rectified pyramid consistency,” in Med. Image Comput. Comput. Assist. Interv.–MICCAI 2021: 24th Int. Conf., Strasbourg, France, Sep. 27–Oct. 1, 2021, pp. 318–329. [Google Scholar]
35. Y. Wu, M. Xu, Z. Ge, J. Cai, and L. Zhang, “Semi-supervised left atrium segmentation with mutual consistency training,” in Med. Image Comput. Comput. Assist. Interv.–MICCAI 2021: 24th Int. Conf., Strasbourg, France, Sep. 27–Oct. 1, 2021, pp. 297–306. [Google Scholar]
36. T. H. Vu, H. Jain, M. Bucher, M. Cord, and P. Pérez, “Advent: Adversarial entropy minimization for domain adaptation in semantic segmentation,” in Proc. IEEE/CVF Conf. CVPR, Long Beach, CA, USA, 2019, pp. 2517–2526. [Google Scholar]
37. S. Qiao, W. Shen, Z. Zhang, B. Wang, and A. Yuille, “Deep co-training for semi-supervised image recognition,” in Proc. ECCV, Munich, Germany, 2018, pp. 135–152. [Google Scholar]
38. Y. Ouali, C. Hudelot, and M. Tami, “Semi-supervised semantic segmentation with cross-consistency training,” in Proc. IEEE/CVF Conf. CVPR, Seattle, WA, USA, 2020, pp. 12674–12684. [Google Scholar]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF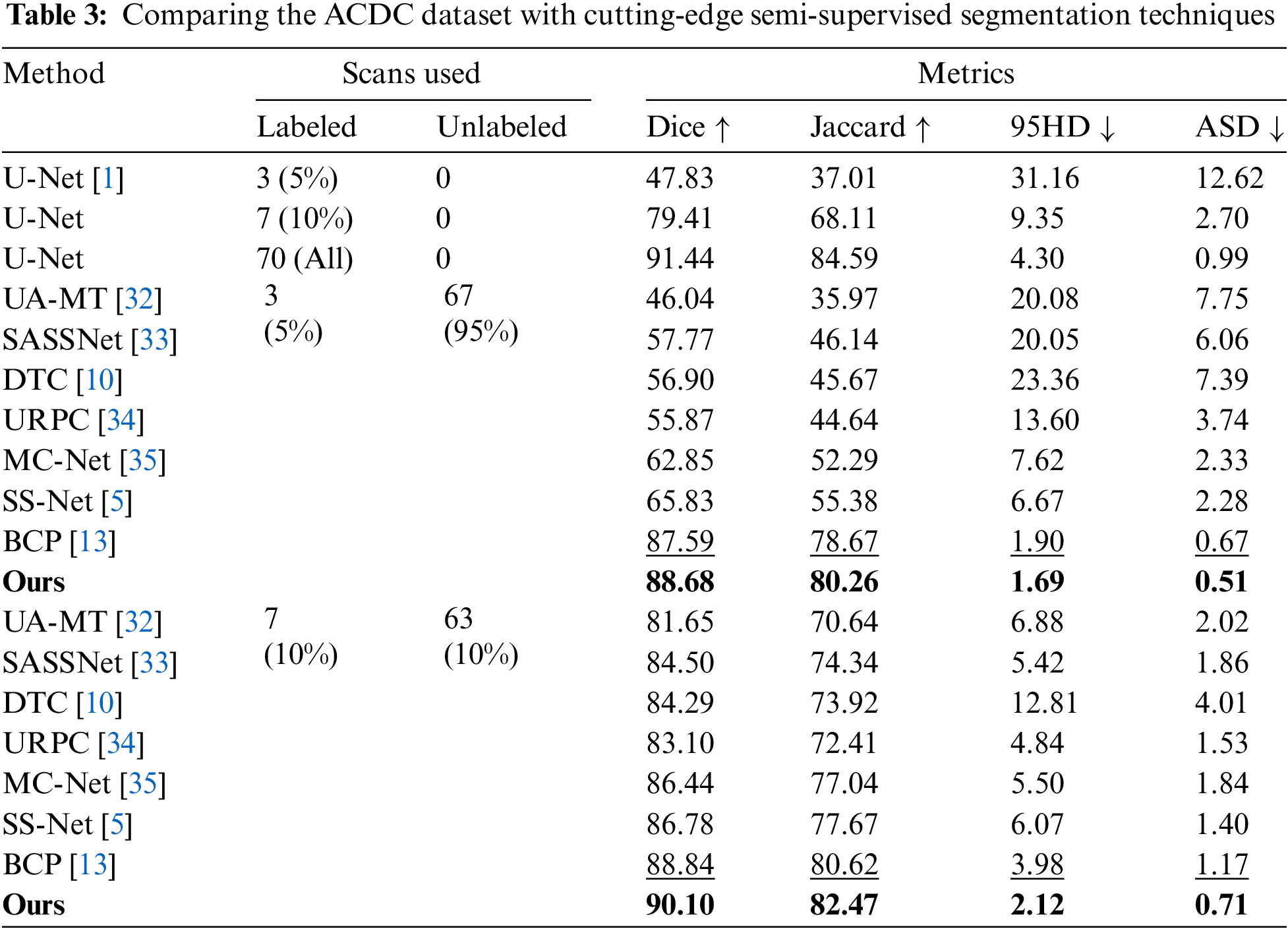
 Downloads
Downloads
 Citation Tools
Citation Tools