 Open Access
Open Access
ARTICLE
Evaluation of Modern Generative Networks for EchoCG Image Generation
1 Department of Cybersecurity, Information Processing and Storage, Satbayev University, Almaty, 050000, Kazakhstan
2 Department of Mathematical Computer Modeling, International IT University, Almaty, 050000, Kazakhstan
3 Department of Information Systems, International IT University, Almaty, 050000, Kazakhstan
* Corresponding Author: Sabina Rakhmetulayeva. Email:
Computers, Materials & Continua 2024, 81(3), 4503-4523. https://doi.org/10.32604/cmc.2024.057974
Received 01 September 2024; Accepted 11 November 2024; Issue published 19 December 2024
Abstract
The applications of machine learning (ML) in the medical domain are often hindered by the limited availability of high-quality data. To address this challenge, we explore the synthetic generation of echocardiography images (echoCG) using state-of-the-art generative models. We conduct a comprehensive evaluation of three prominent methods: Cycle-consistent generative adversarial network (CycleGAN), Contrastive Unpaired Translation (CUT), and Stable Diffusion 1.5 with Low-Rank Adaptation (LoRA). Our research presents the data generation methodology, image samples, and evaluation strategy, followed by an extensive user study involving licensed cardiologists and surgeons who assess the perceived quality and medical soundness of the generated images. Our findings indicate that Stable Diffusion outperforms both CycleGAN and CUT in generating images that are nearly indistinguishable from real echoCG images, making it a promising tool for augmenting medical datasets. However, we also identify limitations in the synthetic images generated by CycleGAN and CUT, which are easily distinguishable as non-realistic by medical professionals. This study highlights the potential of diffusion models in medical imaging and their applicability in addressing data scarcity, while also outlining the areas for future improvement.Graphic Abstract
Keywords
Machine learning models involving deep neural networks have proven to be a key in unlocking data-driven, patient-centered diagnostic and treatment capabilities. These models can analyze vast amounts of medical data with unprecedented accuracy, identifying patterns and correlations that may be imperceptible to human clinicians, or predicting and discovering new drug interactions targeting the underlying health conditions. The ability of these models to continuously learn from new data ensures that healthcare providers can deliver increasingly personalized and effective treatments, ultimately improving patient outcomes and advancing the field of precision medicine. These advancements in ML models heavily rely on computational resources, efficient software frameworks, and availability of large amounts of high-quality data. The focus on the latter issue, as data availability remains a severe limiting factor across ML problems, particularly in the medical domain, despite the ever-growing efforts in electronic record keeping and digitization.
Upon close inspection of data scarcity in medical ML, we can broadly identify the following challenges preventing the widespread usage of in-house data collected by medical and healthcare organizations:
• Privacy concerns. Sharing medical imaging and data exposes patients to risks such as misuse, data leaks, de-anonymization, and health-related profiling which in turn can expose organizations to potential litigation. Therefore, data providers must heavily invest in privacy and security measures.
• Regulatory burden. Health-related and patient-related data in many jurisdictions require special and more rigorous handling. This necessitates a larger staff to manage the additional regulatory paperwork and compliance processes, thereby increasing operational costs and complexity.
• Data quality and quantity. The collected data may lack diversity compared to the general population, leading to significant biases in machine learning models. Additionally, there may be insufficient samples to apply proper anonymization techniques, further limiting the utility of the data.
• Data annotation and standardization. Hospitals and branches within the same organization may impose different annotation and record-keeping requirements. This lack of uniformity results in incompatible data across branches and departments, complicating the integration and utilization of the collected data.
Given the inherent limitations of readily available high-quality labeled and annotated medical data, combined with recent advancements in generative machine learning (genML/genAI), it is crucial to explore the potential of synthetic generation for medical applications. Synthetically generated images offer several advantages for medical ML, with first and foremost being that they mitigate the issues related to patient privacy and data security, as synthetic data has no real patient information. Additionally, if proper control of generation is established, the synthetic generation would allow for the creation of large-scale datasets without the need for extensive manual annotation, saving time and resources while still ensuring high-quality training material.
While synthetic data generation can be applied to various aspects of medical data—including charts, post-visit summaries, medical histories, lab work, and other imaging and screening techniques—our focus is on echocardiograms (echoCG). An echocardiogram is a heart ultrasound image used for screening and diagnosing structural and congenital heart conditions, and the leading method in detecting heart-related illnesses. Despite being a relatively inexpensive type of screening, compared to, for instance, magnetic resonance imaging (MRI), acquiring large amounts of echoCG images of high quality is not a trivial task, as such, it is an ideal candidate for controllable synthetic generation.
Our primary objectives are to address the following research questions: Is it feasible to produce high-quality heart echocardiograms that are indistinguishable from real data?, What are the current limitations of synthetically generated heart echocardiograms?, and How controllable is the synthetic generation of echoCG? Importantly, we aim to thoroughly assess the quality of generated images by conducting a multidisciplinary study involving licensed medical professionals, radiologists, surgeons, and cardiologists to provide expert evaluation. Addressing these questions and limitations is essential for advancing the practical utility of synthetic echoCG—and synthetic data in general—in medical practice, with the potential to enhance diagnostic accuracy and patient outcomes while addressing challenges related to data privacy and availability.
Paper Organization and Contributions
In Section 2, we perform an extensive survey of current art and identify three leading and, thus, most-promising image generation methods that are suitable for synthetic echoCG generation (CycleGAN, CUT, Stable Diffusion). In Section 3, we give a technical overview of these methods: CycleGAN (Section 3.2), CUT (Section 3.3), and of our procedure in adapting Stable Diffusion via LoRA (Section 3.4). In Section 4, we discuss the training details of our selected models and subsequent image generation process. In Section 5, we proceed to discuss the design of our human-subject study across medical professionals to assess perceived image quality, and also describe our survey application we developed (Section 5.2). We dedicate Section 6 to discuss our results and conclude our paper in Section 7 with our final thoughts. To summarize, we make the following contributions:
• We independently replicate the results of CycleGAN and CUT-based heart echocardiogram generation and identify the shortcomings of these methods with respect to image quality.
• We apply LoRA adaption to Stable Diffusion 1.5 to adapt the model to generate heart echocaridograms and demonstrate its utility as image generation tool.
• We conduct, first to our knowledge, a comprehensive multi-disciplinary evaluation of generated echocardiograms among cardiologists, radiologists, and heart surgeons.
The field of synthetic data generation, particularly as it pertains to medical imaging, has emerged as a dynamic and complex research area that integrates state-of-the-art methods. Here we provide an overview of the various studies, research approaches, and discoveries that have combined to influence the present understanding of the field.
2.1 Generative Adversarial Networks
The use of GANs in medical imaging has seen a significant surge in popularity within the machine learning community over the last few years. GANs have been used in the generation of a motion model from a single preoperative magnetic resonance imaging (MRI) scan [1], the upsampling of a low-resolution fundus pictures [2], the generation of synthetic computed tomography (CT) scans from brain MRIs, and the synthesis of T2-weight MRI from T1-weighted MRI (and vice versa). We give an overview of the mathematical basis behind GANs in Section 3.1.
The work of Wang et al. [3] offers a review of various image analysis methods, including generative algorithms. Their analysis contends that generative algorithms, as a subtype of analytical algorithms, have a significant distance to cover in terms of functionality, as their capabilities extend beyond replicating visual cortex functionalities. This perspective frames our understanding of the evolving landscape of generative algorithms, particularly in the context of medical imaging [4]. The work of Singh et al. [5] critically reviews methods for generating medical images. Their critique suggests that state-of-the-art image generation techniques may not currently serve as reliable sources for data saturation and preprocessing in the medical domain. Such a perspective highlights the challenges and caution required when applying generative techniques to medical image synthesis.
The work of Gilbert et al. [6] explores the generation of synthetic labeled data using slices of 3D echoCG images and actual echoCG images (EchoNET images) in unpaired fashion. The study suggested that the generated images received approval from medical professionals in the ultrasound imaging field. Additionally, GANs have been explored in paired settings as well. For example, Tiago et al. [7] used a variation of the Pix2Pix [8] network to generate two-chamber-sliced images (left ventricle and left atrium). In our study, we chose to independently replicate the work of Gilbert et al., as we believe that unpaired image translation aligns more closely with our research goals. Critically, our study differs from both of these works in two key ways: (1) we conduct a user study involving medical professionals, whereas the works of Gilbert et al. [6] and Tiago et al. [7] lack thorough medical evaluation, and (2) we compare multiple generation methods (CycleGAN, CUT, Stable Diffusion), whereas previous studies focus on a single chosen approach.
Diffusion-based models have recently established the state of the art in generating realistic and artistic images [9–11], videos [12], and other high-dimensional data. These methods work by iteratively refining a noisy image into a less noisy variant via a conditional (text, image, or other signal) or unconditional denoising network, and have deep connections to thermodynamic diffusion [13], stochastic ordinary differential equations [14], and Markov processes [15]. We provide a technical overview of diffusion models (DMs) in the context of the stable diffusion in Section 3.4.
The application of diffusion models in medical imaging is an emerging area of research; consequently, the existing body of related work remains relatively limited. Kim et al. [16] proposed augmenting diffusion models with a deformation module to better handle the dynamics of heart MRI, allowing for the generation of realistic deformations and interpolated frames. Perhaps closest to our work is the study by Özbey et al. [17], which combines pixel-level diffusion models with generative adversarial networks to enable unpaired domain translation between domains (e.g., MRI to CT), which they used to generate brain CT images. Implementation-wise, the diffusion process is guided by a source domain image (e.g., MRI) to denoise it into the target domain. Once an image is generated, cycle-consistent adversarial loss is applied, and the models are jointly trained. Similarly, Tiago et al. [18] proposed using a pixel-level diffusion process combined with adversarial loss to generate heart echoCG images. However, in contrast to Özbey et al. [17], they used a paired dataset to train and guide the generation process.
Our work differs significantly from these two approaches in the following ways: (1) we use an off-the-shelf latent diffusion model (Stable Diffusion 1.5) and fine-tune it using low-rank adaptation, which significantly reduces the training time (hours vs. days); (2) we conduct a thorough image quality assessment involving medical professionals, whereas the works of Tiago et al. [18] and Özbey et al. [17] lack such an evaluation; and (3) we use text guidance during the diffusion process, whereas previous methods used image guidance.
2.3 Quality Assessment of Synthetic Images
In image processing, various metrics such as MSE, PSNR, SSIM [19], and more recently, DNN-based metrics like LPIPS [20], have been developed for image quality assessment. However, these methods are full-reference metrics, requiring a reference image for evaluation. In the context of synthetic image generation, where a reference image often does not exist, alternative approaches are necessary.
For unpaired data generation, where no one-to-one correspondence between images can be established, two specific metrics have been developed: Inception Score (IS) [21] and Frèchet Inception Distance (FID) [22]. Both metrics assess the distance between the distribution of generated and real images in the feature space of a deep neural network, typically an Inception network. Among them, FID is considered more reliable as it accounts for both image quality and diversity. However, despite the widespread use of these metrics, human-subject evaluations remain the gold standard for assessing image generation quality, with more sophisticated methods often relying solely on human evaluation [10,11].
Based on our literature review, we selected the following models for our study, each chosen for its proven capability in medical imaging tasks:
• Cycle-consistent generative adversarial networks (CycleGANs). CycleGAN is particularly suited for medical image processing due to its ability to perform unpaired image-to-image translation, making it effective for transforming medical images between different modalities (e.g., sketch to echoCG) without the need for paired datasets. For the particular recipy we following the work of Zhu et al. [23].
• Contrastive Unpaired Translation model (CUT). CUT enhances the capabilities of models like CycleGAN by incorporating contrastive learning, which improves image quality and consistency. This is especially relevant in medical imaging, where accuracy and reliability are critical. We train CUT model following the work of Park et al. [24].
• Stable Diffusion. Stable Diffusion represents an advanced diffusion-based approach that excels in generating high-resolution, detailed images, making it highly applicable for medical scenarios that require fine-grained visual information, such as echocardiography. For our study we implement a low-rank adapted version of Stable Diffuion 1.5 model.
Generative adversarial network (GAN) is a method of generating images and consists of two models: a generative model
CycleGAN [23] is a deep learning framework that learns to translate images from a source domain
Given domains
where
In addition to the cycle consistency loss, CycleGAN uses an adversarial loss function to ensure that the generated samples are indistinguishable from real samples in both domains. This is achieved by introducing a discriminator network,
where
The overall loss function for the CycleGAN is a weighted sum of the adversarial loss and cycle consistency loss:
where
The training of CycleGAN involves minimizing the total loss function using stochastic gradient descent (SGD) or a variant thereof. The generators and discriminators are updated alternately, with the generators being updated to minimize the total loss and the discriminators being updated to maximize the adversarial loss.
Contrastive Learning for Unpaired Image-to-Image Translation [24], also known as Contrastive Unpaired Translation or CUT, is a methodology for image-to-image translation tasks without paired data. CUT is a self-supervised learning approach that leverages contrastive learning to learn a translation function between two domains without paired data. The key idea is to learn a shared representation space where the translated images from both domains are close to each other, while being far apart from the original images.
Contrastive Learning involves learning a representation by contrasting positive and negative samples. In the context of image-to-image translation, positive samples are images from the same domain, while negative samples are images from different domains. The CUT model’s goal is to find matching input-output pairs at a specific location. To achieve this, Park et al. [24] used the other parts within the input image as negative examples. They call this approach PatchNCE loss.
The final loss function is a domain-specific version of the identity loss that uses PatchNCE loss
Stable Diffusion models [10,11] are a variant of text-conditioned diffusion models operating in a semantically compressed latent space. The architecture consists of a variational autoencoder that maps images from pixel space to latent space, a text encoder to compute embeddings of text conditions, and a U-Net network used to predict a denoised version of a noisy image (latent).
The networks in the pipeline are typically trained separately to optimize different losses. In the Stable Diffution 1.5, which we use for our experiments, the text-encoder is obtained using contrastive language-image pretraining (CLIP) on a large corpus of unpaired image-text corpus, and corresponds to CLIP ViT-L/14 of Radford et al. [25]. The variational autoencoder is trained on ImageNet-1k [26] dataset to encode image
where norm
The denoiser
Despite diffusion models’ shown promise in generating realistic images, fine-tuning these models for specific tasks and datasets remains a challenging task. In this study, we use a recently proposed efficient fine-tuning method using low-rank approximation (LoRA).
While several LoRA approaches were presented in the community [28,29], they were all built upon additive low-rank finetuning. Given a weight matrix
The dataset for medical image generation was created from a selection of
For Stable Diffusion training we randomly sampled 40 images containing apical four-chamber view, with the chambers in this case being the left atrium (LA), right atrium (RA), left ventricle (LV) and right ventricle (RV) from the EchoNet dataset. Each image was labeled by a medical professional with a text description related to the shape of the heart from the medical standpoint, with the average number of words in the annotation being 22.95 (Fig. 1). In addition to the annotated data, we created a regularization dataset consisting of 400 unlabeled images from the EchoNet dataset which were used to train using a null (i.e., empty) text.

Figure 1: An example of image annotations utilized in the training process of the stable diffusion model
4.2 CycleGAN and CUT Image Generation
The images were created in accordance to the pipeline created by Gilbert et al. [6] for CycleGAN and by Park et al. [24] for CUT. The process consisted of two parts:
1. Render 2D slices from 3D anatomical model of the heart with two purposes:
• mark labels of heart chamber segments
• use it as one of the inputs for creating image in specific domain, in this case as an echocardiographic image
2. Allocate EchoNet images for creating the target domain of echocardiographic images
The training was performed for about 24 h for both CycleGAN and CUT on a single A100 GPU. We used Adam optimization method with a constant learning rate of
For CycleGAN it is also worth noting that the standard encoder-decoder generative network within model is replaced with U-Net [27] with 8 downsampling levels for increased training speed with equivalent results [6]. During the training of CycleGAN the network weights were saved every five epochs.
4.3 Stable Diffusion Generation
To generate images using diffusion models, we applied LoRA finetuning to the Stable Diffusion 1.5 model using our pre-annotated dataset. We used the LoRA weights with a rank of 8 applied to cross-attention layers on all blocks. We trained our model for 4 epochs with a batch size of 2 using the Adam optimization method with a learning rate of
For each of the methods we generated a total of 800 images. We present a randomly selected subset of those in Fig. 2. We computed the Frèchet Inception Distance (FID) score for these images against the reference EchoNet dataset. The FID scores were as follows: 75.13 for CycleGAN, 146.99 for CUT, and 168.59 for Stable Diffusion. A lower FID score typically indicates better diversity and quality, suggesting that all networks are well-trained, with CycleGAN exhibiting superior performance according to this metric. Upon visual inspection, we observed that CUT-generated images occasionally exhibit a shift in color towards orange (in less than 3% of the images), which may contribute to the slightly higher FID score. Such behavior was also noted by designers of CycleGAN and CUT [23] when generating apple-orange transitional images. In the case of Stable Diffusion, the generated images appeared less diverse to the untrained eye, which is often associated with a higher FID score. It is also worth noting that FID score could be biased during the evaluation of finite number of generated images [30]. In addition, as our user study with medical professionals demonstrates, the FID score does not necessarily correlate with the perceived quality of the model’s generated images.
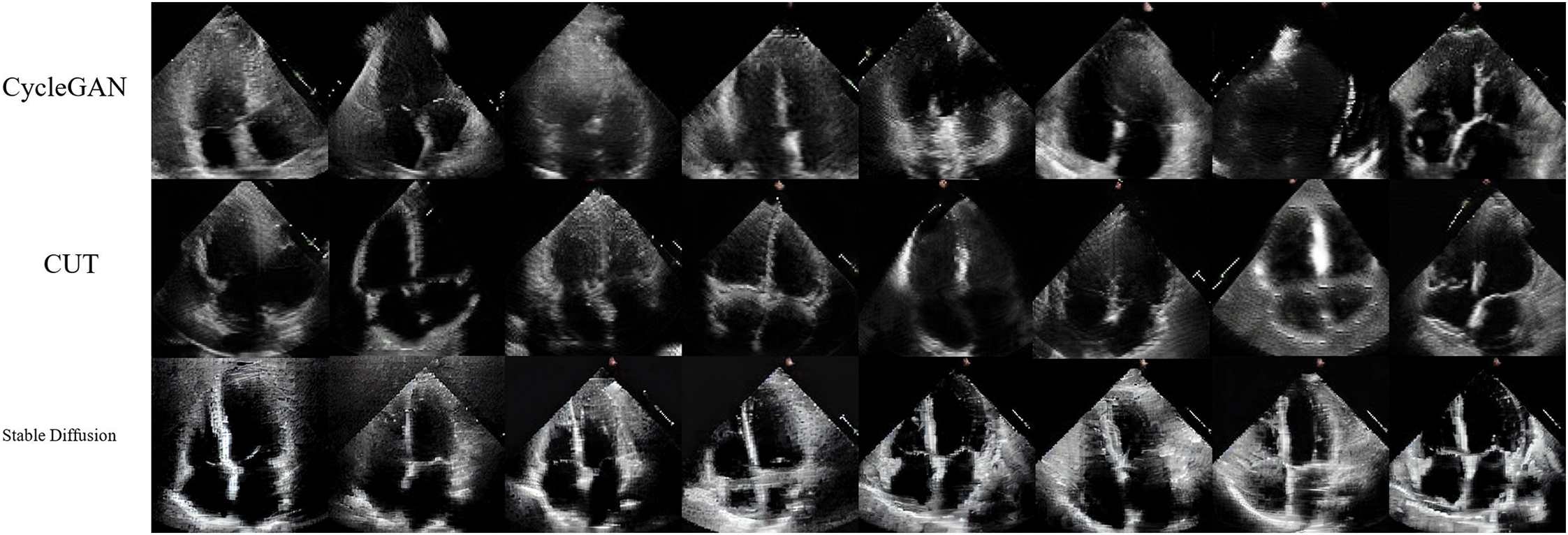
Figure 2: Examples of the generated images for each of the methods
5 User Study: Quality Evaluation
We conducted a user study involving experienced cardiologists and cardiac surgeons to assess the perceived realism and practical applicability of our selected image generation models: CycleGAN, CUT, and Stable Diffusion. The study aimed to determine how realistic the synthetically generated echocardiographic (echoCG) images appear from a medical perspective and whether any of these generation methods could convincingly replicate authentic medical images to the satisfaction of medical professionals. Prior to commencing the study, we obtained approval from the ethics committee responsible for overseeing research involving human subjects.
For the study, we recruited seventeen participants, aged between 25 and 55 years (Mean
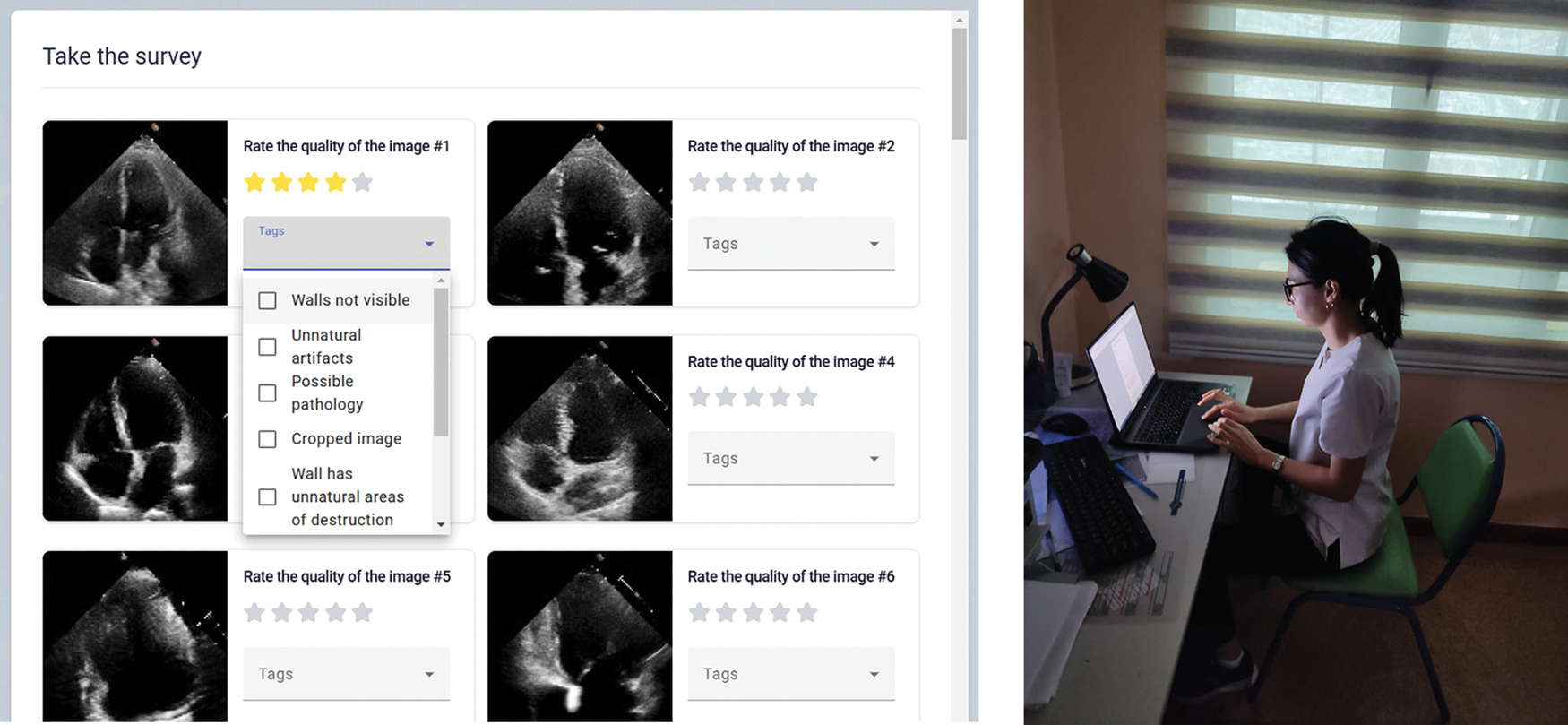
Figure 3: Left: an example of Likert scale questions in the app, right: a participant taking part in the study
We developed a custom application, named CardioQuestionnaire, running on a PC with Windows OS to collect feedback from our participants. The application comprises several key components: an application server, a data generation server, and a database server. The application server, accessed by the client application, serves as the central hub, coordinating interactions with the synthetic data and the database through its subsystems. On the right of Fig. 3, we show the screenshot of our customized application (in Russian); full English translation of the pre- and post-study questionnaires are available in Appendix A.
The software platform utilizes PostgreSQL as the database management system [31], selected for its reliability, scalability, and extensive feature set. To streamline schema modifications, we integrated Liquibase, an open-source database migration tool. Liquibase [32] facilitates systematic schema management by tracking database states and applying necessary migrations, ensuring seamless integration with PostgreSQL.
For the backend development, we selected the Java programming language. We utilized the Spring Boot framework to streamline the creation of production-ready applications, leveraging features like auto-configuration to minimize development setup time. The CardioQuestionnaire system’s frontend was designed through multiple iterations to ensure a user-friendly and ergonomic interface for our target population. Built with the Angular framework [33], it features a component-based structure that facilitates code reuse and supports the development of complex user interfaces, while a responsive layout ensures a consistent experience across devices. We present system architecture diagram in Fig. 4.
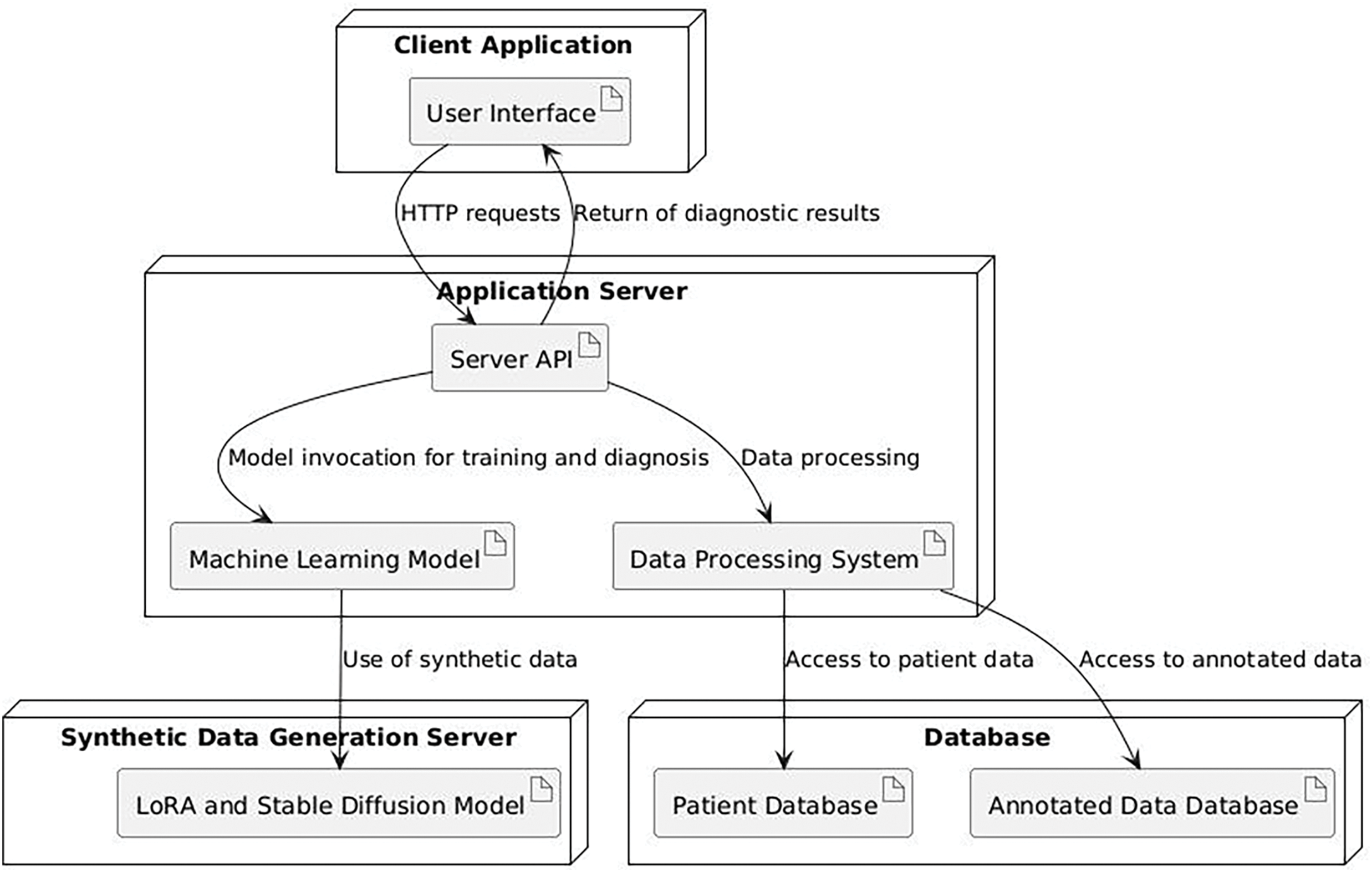
Figure 4: System architecture diagram
In this user study, we employed a within-subjects design with one independent variable: image source, which had four conditions: real images, CUT-generated images, CycleGAN-generated images, and Stable Diffusion-generated images. To control for potential order effects, conditions were systematically counterbalanced. Images were randomly selected from a pool of 400 images per condition. Each participant assessed images from all four sources and rated their perceived realism using a Likert scale ranging from 1 (very unlikely to resemble a real image) to 5 (very likely to resemble a real image).
The study was conducted in a single session, where each participant evaluated 40 images, with 10 images drawn from each of the four image source categories. In summary, the study design consisted of:
17 participants
We selected the Likert scale for this study because it provides a straightforward and reliable method for measuring subjective perceptions, which is essential for evaluating the realism of synthetic medical images. The survey was designed to capture the participants’ nuanced judgments about the realism of the images, making the Likert scale an appropriate choice for the task.
The study was conducted in controlled and standardized environments across various medical facilities. Each participant was accommodated individually. Initially, participants received an extensive briefing about the study objectives, during which the researcher addressed any questions they had about the study. Next, they were asked to complete consent forms, followed by a demographic questionnaire that inquired about age, gender, educational, and medical backgrounds. Subsequently, the main study commenced, during which the application presented participants with sets of images, comprising images generated using our selected pre-trained models: CycleGAN, CUT, and Stable Diffusion-generated images, as well as authentic medical images from the dataset. Participants were tasked with assessing the realism of each image based on their perceptions, rating each on a scale from 1 being most unrealistic to 5 being very realistic. We collected all data and evaluations, after which the researcher administered a post-study questionnaire. Additionally, any remaining questions from participants were addressed, and further comments were recorded during discussions.
A complete study session lasted approximately twenty minutes, encompassing demographic data collection, the primary study, and post-study discussions. Initially, we conducted a normality test on the data to assess its distribution, with the consolidated results presented in Table 1. We conducted four different statistical tests used to determine whether Likert scale assessments follow a normal distribution for CycleGAN, CUT, and Stable Diffusion. The findings indicated that the residuals did not follow a normal distribution. Consequently, we employed the Mann-Whitney U test for our data analysis.
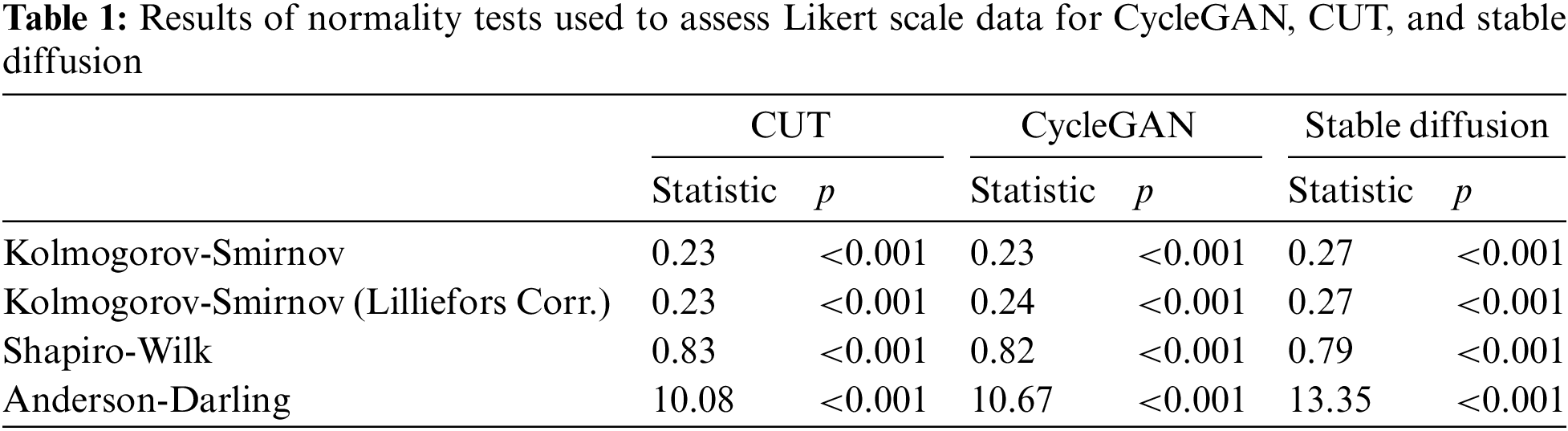
The Mann-Whitney U test, used to compare differences between two independent groups, identified a small effect of image source for the CycleGAN vs. real pair with
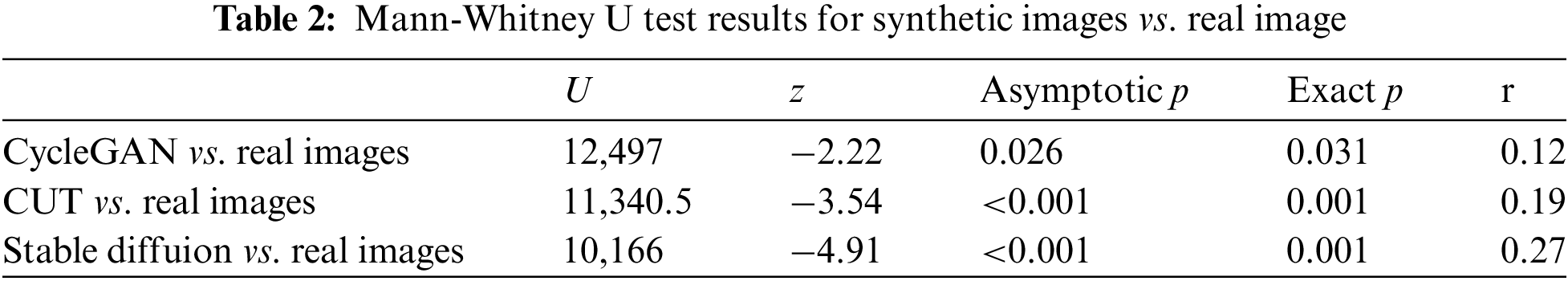
The results suggest that there is a significant statistical difference between the distributions of CycleGAN, CUT and Stable Diffusion in comparison to real images. However, for the CycleGAN vs. real image pair, the results suggest that Likert scores for CycleGAN images tend to be lower, with a mean of 2.72, a median of 3, and a standard deviation (SD) of 1.59. In contrast, the CUT vs. real and Stable Diffusion vs. real pairs reveal distinct trends: CUT images are consistently rated as less realistic, with a mean score of 2.49, a median of 2, and an SD of 4.46, while Stable Diffusion images are rated as more realistic, with a mean of 3.95, a median of 4, and an SD of 1.24. The consolidated descriptive statistics are presented in Table 3.
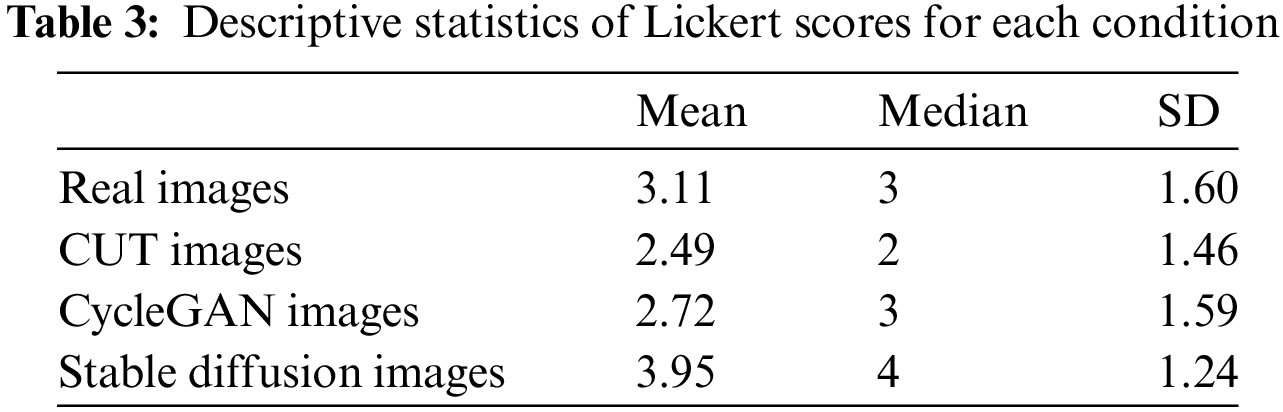
Our study suggests that a group of experienced medical professionals can readily distinguish between CycleGAN- and CUT-generated synthetic images and real ones, with CUT images receiving slightly higher scores for “realism.” This outcome is somewhat disappointing given that GAN-based methods have long dominated the computer vision field before the advent of diffusion models.
In contrast, Stable Diffusion-generated images received significantly better assessment scores (Mean = 3.95, Median = 4, SD = 1.24) compared to CUT (Mean = 2.49, Median = 2, SD = 1.46) and CycleGAN (Mean = 2.72, Median = 3, SD = 1.59), indicating that Stable Diffusion-based models are more capable of generating authentic medical images of satisfactory quality. Several factors may explain the superior performance of diffusion models. First, diffusion models refine images progressively through a denoising process, which enhances the fine details and textures crucial for accurately representing subtle structures in echocardiographic images. Second, the use of latent space for diffusion, combined with attention mechanisms in the U-Net architecture, plays a vital role in preserving the overall structural integrity and intricate details of the heart’s anatomy. In contrast, CycleGAN and CUT operate directly in pixel space and lack attention mechanisms, which, given limited training, may hinder their ability to capture medically significant features. Lastly, Stable Diffusion’s extensive pre-training on billions of realistic images, followed by fine-tuning on echocardiographic (echoCG) images using LoRA, likely conditioned the model to understand the intricacies of echoCGs at a fundamental level, further refined by fine-tuning to achieve expert-level performance.
An additional noteworthy observation is that Stable Diffusion images received a higher overall rating (Mean = 3.95, Median = 4, SD = 1.24) compared to real images (Mean = 3.11, Median = 3, SD = 1.6). One plausible explanation for this phenomenon could be the bias introduced by the necessity of prompt engineering in the Stable Diffusion image generation process. The prompts require certain patterns to be present, which doctors identify beforehand, thereby making the resulting images clearer and more analyzable. To further explore whether this phenomenon is influenced by the experience level of our participants, we plotted the average scores of each participant against their job experience and the number of weekly echoCG image readings (see Fig. 5). The analysis revealed that real images were rated as less realistic by both experienced doctors (>15 years) and novices (<8 years) alike. Similarly, the frequency of weekly echoCG readings did not significantly affect their realism ratings for real images.
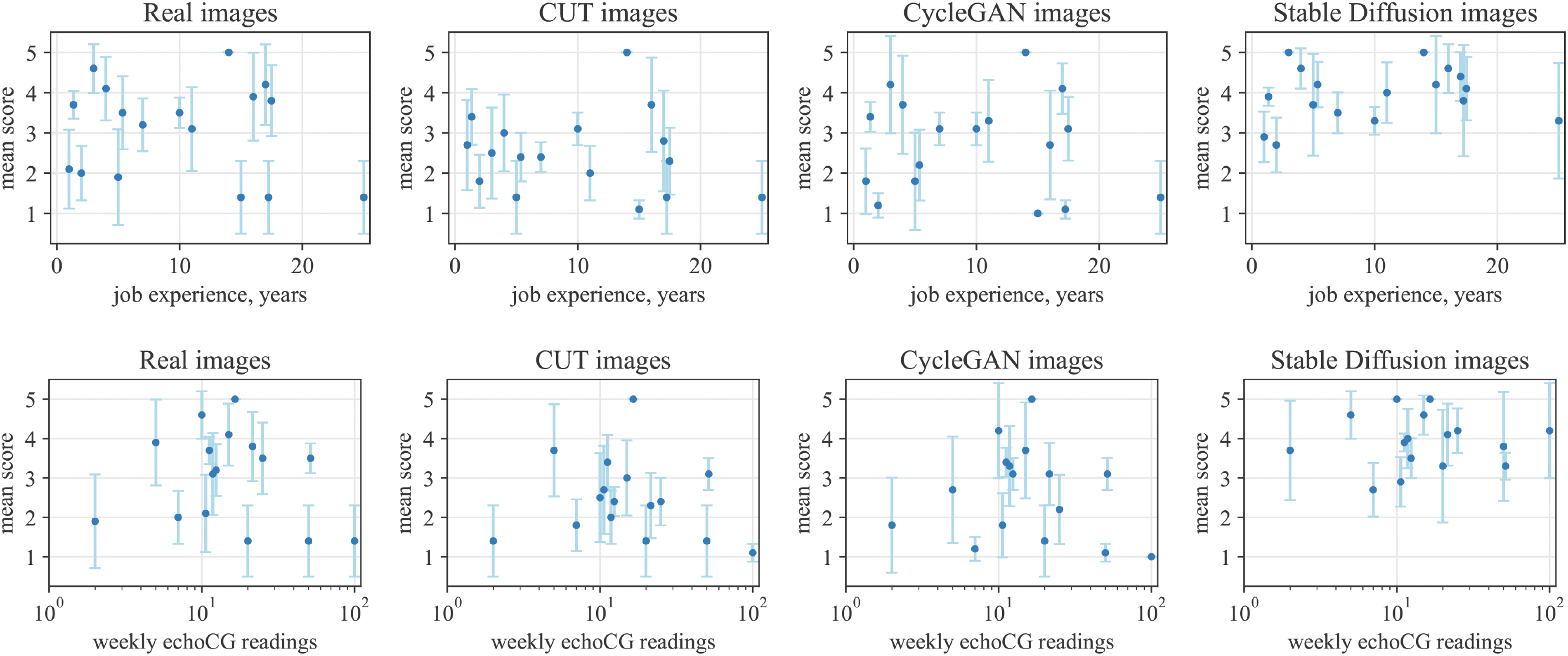
Figure 5: The mean assessment (Lickert) scores of every participants for each generation type, including real images, compared against their job experience (top) and number of weekly echoCG readings (bottom). Note that for the bottom row plots the x-axis is presented in logarithmic scale to account for the data distribution. Error bars represent the 95% confidence interval
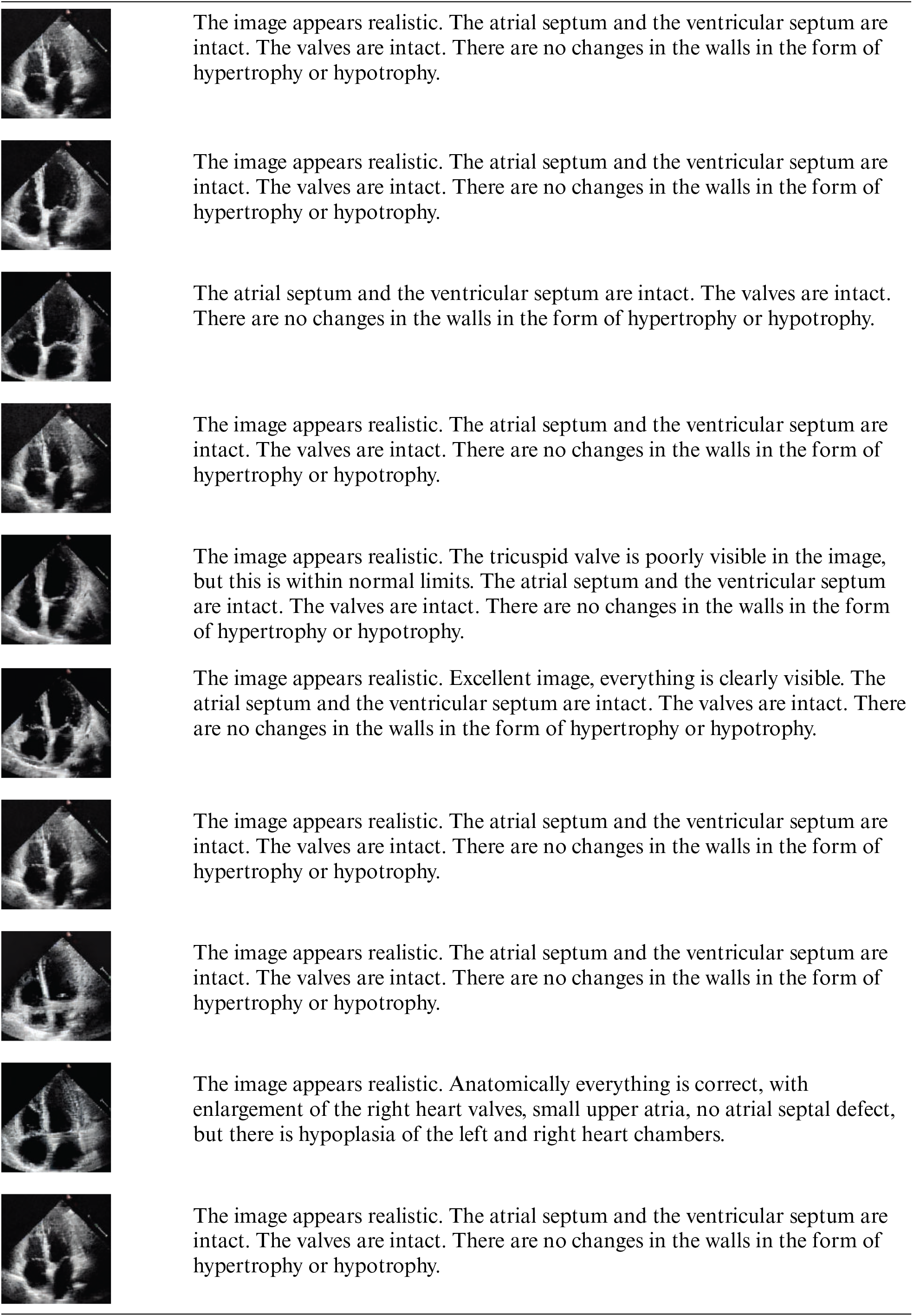
Additionally, we consulted an experienced cardiothoracic surgeon (>15 years) who interprets more than 100 images per week to annotate images from CUT, CycleGAN, and Stable Diffusion and to verbally rate their realism (see Appendix B). The surgeon rated Stable Diffusion images as very realistic, occasionally noting, “excellent image, everything is clearly visible,” whereas the overall sentiment for CUT and CycleGAN images was, “the image is not realistic; the heart is not visualized.”
In this paper, we conducted a comprehensive evaluation of CycleGAN-, CUT-, and Stable Diffusion-based methods for the synthetic generation of echocardiography (echoCG) images. The training of CycleGAN and CUT models does not require paired data, which significantly simplifies their application in medical settings, where large collections of single-modality data (such as MRIs, X-rays, and echoCGs) are commonly available. Although Stable Diffusion requires pairs of image-text descriptions, which can be cumbersome to collect, as demonstrated in Section 4, the required data for such training is minimal; only 40 annotated image-text pairs were sufficient.
Our evaluation with medical professionals (
Acknowledgement: The authors would like to express their gratitude to Dr. Yerlan Idelbayev and Dr. Gulnar Rakhmetulla for invaluable help in editing and preparing the initial draft of this paper, to Baubek Ukibassov for consultations regarding training of the models, to Dr. Nurlan Baizhigitov and Dr. Elvira Rakhmetulayeva for consultations on medical and cardiology matters.
Funding Statement: This research has been funded by the Science Committee of the Ministry of Science and Higher Education of the Republic of Kazakhstan (Grant No. AP13068032—Development of Methods and Algorithms for Machine Learning for Predicting Pathologies of the Cardiovascular System Based on Echocardiography and Electrocardiography).
Author Contributions: Conceptualization, Sabina Rakhmetulayeva; methodology, Sabina Rakhmetulayeva, Zhandos Zhanabekov, and Aigerim Bolshibayeva; software, Zhandos Zhanabekov; data curation, Zhandos Zhanabekov; writing, original draft preparation, Sabina Rakhmetulayeva, Zhandos Zhanabekov, and Aigerim Bolshibayeva; writing, review and editing, Sabina Rakhmetulayeva; supervision, Sabina Rakhmetulayeva; discussion, Sabina Rakhmetulayeva, Zhandos Zhanabekov, and Aigerim Bolshibayeva. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The study uses the EchoNet Dynamic Echocardiographic dataset, taken from 10,030 apical-4-chamber echocardiography videos from individuals who underwent imaging between 2016 and 2018 as part of routine clinical care at Stanford University Hospital. The dataset is accessible for research purposes and was downloaded in full compliance with the research user agreement [34].
Ethics Approval: This study received ethical approval from the Ethics Commission of the Republican State Enterprise on the Right of Economic Management “Institute of Genetics and Physiology” under the Science Committee of the Ministry of Science and Higher Education of the Republic of Kazakhstan. The approval was granted based on Protocol No. 3 dated 30 October 2023. All research procedures involving surveys among doctors were conducted in accordance with the ethical standards outlined and complied with relevant local and national regulations. All the doctors that participated in the study were thoroughly informed about the details of the study and gave their written consent to use the data collected in the study.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Y. Hu et al., “Intraoperative organ motion models with an ensemble of conditional generative adversarial networks,” in Int. Conf. Med. Image Comput. Comput.-Assist. Interv., 2017, pp. 368–376. doi: 10.1007/978-3-319-66185-8_42. [Google Scholar] [CrossRef]
2. D. Nie, R. Trullo, C. Petitjean, S. Ruan, and D. Shen, “Medical image synthesis with context-aware generative adversarial networks,” in Med. Image Comput. Comput. Assist. Interv.–MICCAI 2017: 20th Int. Conf., Quebec City, QC, Canada, Springer, pp. 417–425, 2017. [Google Scholar]
3. Y. Wang, X. Ge, H. Ma, S. Qi, G. Zhang and Y. Yao, “Deep learning in medical ultrasound image analysis: A review,” IEEE Access, vol. 9, pp. 54310–54324, 2021. doi: 10.1109/ACCESS.2021.3071301. [Google Scholar] [CrossRef]
4. S. Rakhmetulayeva, A. Bolshibayeva, A. Mukasheva, B. Ukibassov, Z. Zhanabekov and D. Diaz, “Machine learning methods and algorithms for predicting congenital heart pathologies,” in 2023 IEEE 17th Int. Conf. Appl. Inform. Commun. Technol. (AICT), 2023, pp. 1–6. [Google Scholar]
5. N. K. Singh and K. Raza, “Medical image generation using generative adversarial networks,” 2020. doi: 10.48550/arXiv.2005.10687. [Google Scholar] [CrossRef]
6. A. Gilbert, M. Marciniak, C. Rodero, P. Lamata, E. Samset and K. McLeod, “Generating synthetic labeled data from existing anatomical models: An example with echocardiography segmentation,” IEEE Trans. Med. Imaging, vol. 40, no. 10, pp. 2783–2794. doi: 10.1109/TMI.2021.3051806. [Google Scholar] [PubMed] [CrossRef]
7. C. Tiago et al., “A data augmentation pipeline to generate synthetic labeled datasets of 3D echocardiography images using a gan,” IEEE Access, vol. 10, pp. 98803–98815, 2022. doi: 10.1109/ACCESS.2022.3207177. [Google Scholar] [CrossRef]
8. P. Isola, J. -Y. Zhu, T. Zhou, and A. A. Efros, “Image-to-image translation with conditional adversarial networks,” 2018, arXiv:1611.07004. [Google Scholar]
9. P. Dhariwal and A. Nichol, “Diffusion models beat GANs on image synthesis,” in Proc. 35th Int. Conf. Neural Inform. Process. Syst., 2024. [Google Scholar]
10. R. Rombach, A. Blattmann, D. Lorenz, P. Esser, and B. Ommer, “High-resolution image synthesis with latent diffusion models,” in Proc. IEEE/CVF Conf. Comput. Vis. Pattern Recognit., 2022, pp. 10684–10695. [Google Scholar]
11. D. Podell et al., “SDXL: Improving latent diffusion models for high-resolution image synthesis,” 2023, arXiv:2307.01952. [Google Scholar]
12. A. Blattmann et al., “Stable video diffusion: Scaling latent video diffusion models to large datasets,” 2023, arXiv:2311.15127. [Google Scholar]
13. J. Sohl-Dickstein, E. A. Weiss, N. Maheswaranathan, and S. Ganguli, “Deep unsupervised learning using nonequilibrium thermodynamics,” in Proc. 32nd Int. Conf. Int. Conf. Mach. Learn., 2015, pp. 2256–2265. [Google Scholar]
14. Y. Song, J. N. Sohl-Dickstein, D. P. Kingma, A. Kumar, S. Ermon and B. Poole, “Score-based generative modeling through stochastic differential equations,” 2020, arXiv:2011.13456. [Google Scholar]
15. J. Ho, A. Jain, and P. Abbeel, “Denoising diffusion probabilistic models,” in Proc. 34th Int. Conf. Neural Inform. Process. Syst., Vancouver, BC, Canada, 2020. [Google Scholar]
16. B. Kim and J. C. Ye, “Diffusion deformable model for a 4D temporal medical image generation,” in Medical Image Computing and Computer Assisted Intervention–MICCAI 2022, L. Wang, Q. Dou, P. T. Fletcher, S. Speidel, S. Li, Eds. Cham: Springer Nature Switzerland, 2022, pp. 539–548. [Google Scholar]
17. M. Özbey et al., “Unsupervised medical image translation with adversarial diffusion models,” IEEE Trans. Med. Imaging, vol. 42, no. 12, pp. 3524–3539, 2023. doi: 10.1109/TMI.2023.3290149. [Google Scholar] [PubMed] [CrossRef]
18. C. Tiago, S. R. Snare, J. Šprem, and K. McLeod, “A domain translation framework with an adversarial denoising diffusion model to generate synthetic datasets of echocardiography images,” IEEE Access, vol. 11, pp. 17594–17602, 2023. doi: 10.1109/ACCESS.2023.3246762. [Google Scholar] [CrossRef]
19. Z. Wang, A. Bovik, H. Sheikh, and E. Simoncelli, “Image quality assessment: From error visibility to structural similarity,” IEEE Trans. Image Process., vol. 13, no. 4, pp. 600–612, 2004. doi: 10.1109/TIP.2003.819861. [Google Scholar] [PubMed] [CrossRef]
20. R. Zhang, P. Isola, A. A. Efros, E. Shechtman, and O. Wang, “The unreasonable effectiveness of deep features as a perceptual metric,” in 2018 IEEE/CVF Conf. Comput. Vis. Pattern Recognit., 2018, pp. 586–595. [Google Scholar]
21. T. Salimans, I. Goodfellow, W. Zaremba, V. Cheung, A. Radford and X. Chen, “Improved techniques for training gans,” in Proc. 30th Int. Conf. Neural Inform. Process. Syst., Barcelona, Spain, 2016, pp. 2234–2242. [Google Scholar]
22. M. Heusel, H. Ramsauer, T. Unterthiner, B. Nessler, and S. Hochreiter, “Gans trained by a two time-scale update rule converge to a local nash equilibrium,” in Proc. 31st Int. Conf. Neural Inform. Process. Syst., Long Beach, CA, USA, 2017, pp. 6629–6640. [Google Scholar]
23. J. -Y. Zhu, T. Park, P. Isola, and A. A. Efros, “Unpaired image-to-image translation using cycle-consistent adversarial networks,” in 2017 IEEE Int. Conf. Comput. Vis. (ICCV), 2017, pp. 2242–2251. [Google Scholar]
24. T. Park, A. A. Efros, R. Zhang, and J. Zhu, “Contrastive learning for unpaired image-to-image translation,” 2020, arXiv:2007.1565. [Google Scholar]
25. A. Radford et al., “Learning transferable visual models from natural language supervision,” in Int. Conf. Mach. Learn., PMLR, 2021, pp. 8748–8763. [Google Scholar]
26. O. Russakovsky et al., “ImageNet large scale visual recognition challenge,” Int. J. Comput. Vis., vol. 115, pp. 211–252, 2015. doi: 10.1007/s11263-015-0816-y. [Google Scholar] [CrossRef]
27. O. Ronneberger, P. Fischer, and T. Brox, “U-Net: Convolutional networks for biomedical image segmentation,” in Med. Image Comput. Comput.-Assist. Interv.–MICCAI 2015: 18th Int. Conf., Munich, Germany, Springer, 2015, pp. 234–241. [Google Scholar]
28. E. J. Hu et al., “LoRA: Low-rank adaptation of large language models,” 2021, arXiv:2106.09685. [Google Scholar]
29. S. -Y. Liu et al., “DoRA: Weight-decomposed low-rank adaptation,” 2024, arXiv:2402.09353. [Google Scholar]
30. M. J. Chong and D. Forsyth, “Effectively unbiased fid and inception score and where to find them,” in 2020 IEEE/CVF Conf. Comput. Vis. Pattern Recognit. (CVPR), 2020, pp. 6069–6078. [Google Scholar]
31. T. P. G. D. Group, “PostgreSQL: The world’s most advanced open source relational database,” 2024. Accessed: Oct. 10, 2024. [Online]. Available: https://www.postgresql.org/ [Google Scholar]
32. L. Inc., “Liquibase,” 2024. Accessed: Oct. 10, 2024. [Online]. Available: https://www.liquibase.com/ [Google Scholar]
33. G. LLC, “Angular,” 2024. Accessed: Oct. 10, 2024. [Online]. Available: https://angular.dev/api [Google Scholar]
34. D. Ouyang et al., “Video-based AI for beat-to-beat assessment of cardiac function,” Nature, vol. 580, no. 7802, pp. 252–256, Apr. 2020. doi: 10.1038/s41586-020-2145-8. [Google Scholar] [PubMed] [CrossRef]
Appendix A: Details of Questionnaire
Pre-study questionnaire Responses to the pre-study questionnaire were collected using the custom build application (see Section 5.2), with open text fields or drop-down options provided for participants to enter their answers. The questions were presented in either Russian or Kazakh, depending on the user’s choice. Below is an English translation of these questions:
1. Please enter your full name
2. Indicate your gender
3. Indicate your date of birth
4. Enter your qualification
5. Enter your years of work experience in the field of cardiology (e.g., ultrasound, cardiac surgery (pediatric, adult), cardiodiagnostics, etc.)
6. Enter how many years have passed since you graduated from medical school
7. Enter how many echocardiograms you analyze per week on average (e.g., 20)
Study questionnaire During the study, images were shown in a custom build application (see Section 5.2) and each numbered image were followed with the following question “Please evaluate the quality of the image N” in Kazakh or Russian.
Appendix B: Image Samples with Detailed Analysis
CUT images

CycleGAN images
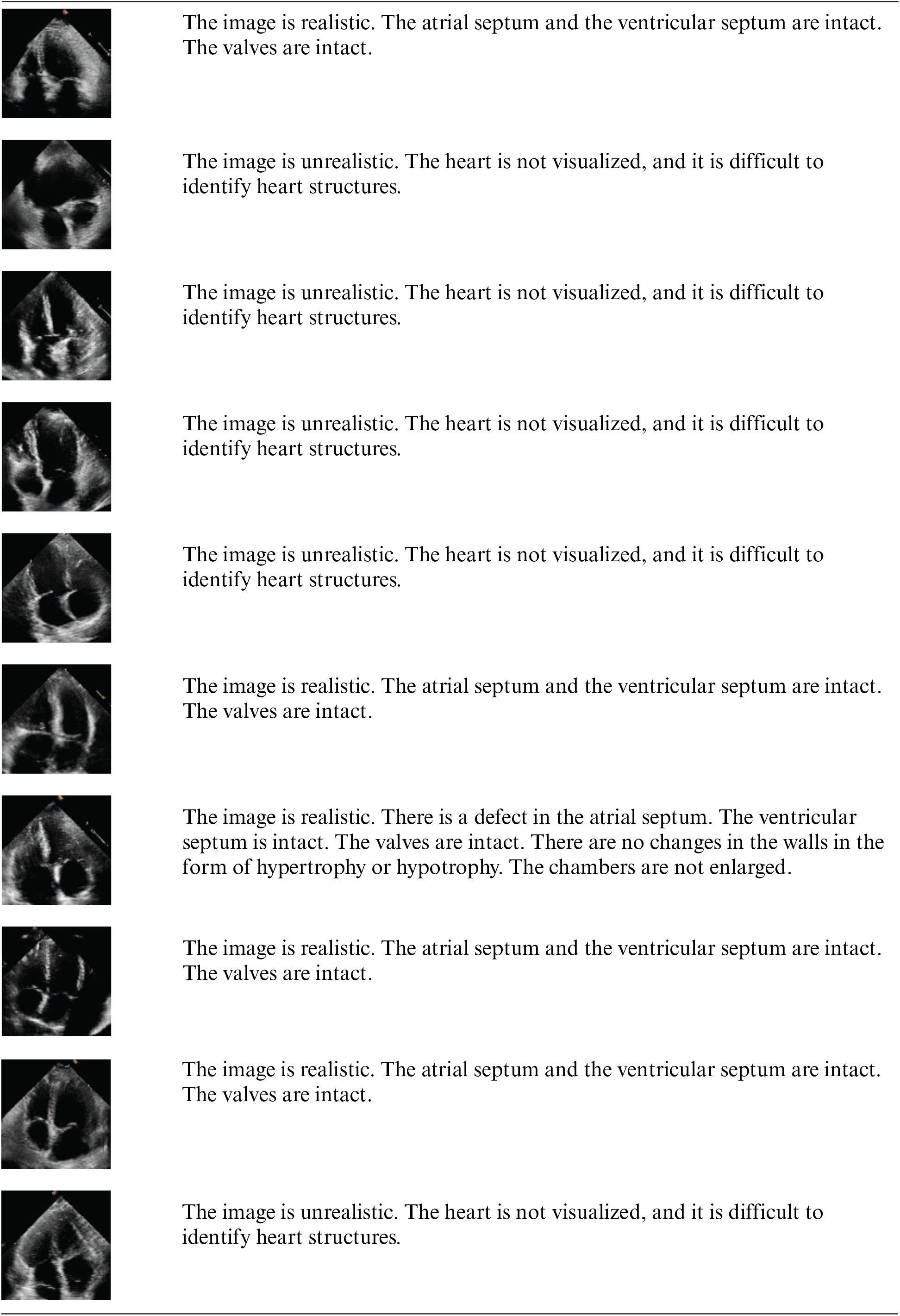
Stable Diffusion images
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools