 Open Access
Open Access
ARTICLE
Optimizing the Clinical Decision Support System (CDSS) by Using Recurrent Neural Network (RNN) Language Models for Real-Time Medical Query Processing
1 College of Computing and Informatics, Universiti Tenaga Nasional (UNITEN), Kajang, 43000, Malaysia
2 College of Engineering, University of Warith Al-Anbiyaa, Karbala, 56001, Iraq
3 Faculty of Information Science & Technology, Universiti Kebangsaan Malaysia, Bangi, 43600, Malaysia
4 Department of Computer Science, Applied College, University of Hail, Hail, 55424, Saudi Arabia
5 Department of Computer Science and Informatics, Taibah University, Medina, 42353, Saudi Arabia
6 Department of Computer Science, Hanyang University, Seoul, 04763, Republic of Korea
* Corresponding Author: Israa Ibraheem Al Barazanchi. Email:
Computers, Materials & Continua 2024, 81(3), 4787-4832. https://doi.org/10.32604/cmc.2024.055079
Received 16 June 2024; Accepted 13 October 2024; Issue published 19 December 2024
Abstract
This research aims to enhance Clinical Decision Support Systems (CDSS) within Wireless Body Area Networks (WBANs) by leveraging advanced machine learning techniques. Specifically, we target the challenges of accurate diagnosis in medical imaging and sequential data analysis using Recurrent Neural Networks (RNNs) with Long Short-Term Memory (LSTM) layers and echo state cells. These models are tailored to improve diagnostic precision, particularly for conditions like rotator cuff tears in osteoporosis patients and gastrointestinal diseases. Traditional diagnostic methods and existing CDSS frameworks often fall short in managing complex, sequential medical data, struggling with long-term dependencies and data imbalances, resulting in suboptimal accuracy and delayed decisions. Our goal is to develop Artificial Intelligence (AI) models that address these shortcomings, offering robust, real-time diagnostic support. We propose a hybrid RNN model that integrates SimpleRNN, LSTM layers, and echo state cells to manage long-term dependencies effectively. Additionally, we introduce CG-Net, a novel Convolutional Neural Network (CNN) framework for gastrointestinal disease classification, which outperforms traditional CNN models. We further enhance model performance through data augmentation and transfer learning, improving generalization and robustness against data scarcity and imbalance. Comprehensive validation, including 5-fold cross-validation and metrics such as accuracy, precision, recall, F1-score, and Area Under the Curve (AUC), confirms the models’ reliability. Moreover, SHapley Additive exPlanations (SHAP) and Local Interpretable Model-agnostic Explanations (LIME) are employed to improve model interpretability. Our findings show that the proposed models significantly enhance diagnostic accuracy and efficiency, offering substantial advancements in WBANs and CDSS.Keywords
The healthcare industry is experiencing a significant transformation with the adoption of artificial intelligence (AI) technologies, which are enhancing the accuracy and efficiency of clinical decision-making and patient care. One notable application of AI is in the diagnosis of rotator cuff tears in osteoporosis patients using anteroposterior X-rays of the shoulder joint. These machine learning models utilize advanced algorithms to scrutinize medical images, detecting subtle patterns and anomalies that might be challenging for human radiologists to consistently identify. By being trained on extensive datasets, these models achieve high levels of accuracy and reliability, facilitating early and precise diagnosis. This capability not only improves patient outcomes through timely interventions but also optimizes the overall management of rotator cuff injuries in patients with osteoporosis [1]. Moreover, clinical decision support systems (CDSS) have become integral to modern healthcare delivery, aiding physicians in making more accurate and informed decisions. An exemplary development in this field is CG-Net, a novel Convolutional Neural Network (CNN) framework designed for the classification of gastrointestinal tract diseases. CG-Net leverages deep learning techniques to analyze complex medical data, effectively differentiating between various gastrointestinal conditions with remarkable accuracy. The system is trained on large volumes of annotated medical images and clinical data, enabling it to learn and identify intricate disease patterns. By providing real-time diagnostic support, CG-Net helps physicians make timely and accurate diagnoses, thereby reducing the likelihood of errors and improving patient outcomes. The integration of such advanced AI frameworks into CDSS not only enhances diagnostic precision but also streamlines clinical workflows, ensures efficient healthcare delivery, and supports clinicians with the most up-to-date and comprehensive data available, leading to a higher standard of patient care [2]. A neural network, which is a collection of algorithms, is intended to find the underlying relationships between parameters that are input and output according to study. The current state-of-the state art methods for time series data augmentation in deep learning are being used in clinical decision support. The paper introduces the concept of optimization of RNN and its importance in improving the accuracy and generalization of deep learning models, especially in the context of time series data as mentioned in study [2]. The paper then delves into various data augmentation techniques such as interpolation, jittering, scaling, time warping, and frequency warping. The paper also discusses some combination techniques, where multiple data augmentation techniques are used to augment the time series data. This paper meticulously details the design, development, and deployment of RNN language models specifically tailored for the medical domain. It examines the challenges of natural language processing in healthcare, such as dealing with medical jargon, understanding patient histories, and adapting to the ever-expanding medical knowledge base. Furthermore, it addresses critical considerations such as data privacy, model interpretability, and integration with existing healthcare IT ecosystems as per source study [3]. Since the main topic of this paper handles the issues present with clinical dataset, we must first understand. This study investigates the potential of RNN language models to improve real-time data access, processing, and utilization for medical practitioners. The goal of this effort is to employ RNNs’ capacity to analyze healthcare professionals’ natural language inquiries and instantly deliver evidence-based answers and insights. The objective of this project is to optimize clinical decision support system accuracy and efficiency using sophisticated language models. This will help to decrease diagnostic mistakes, enhance patient outcomes, and simplify healthcare operations. Clinical decision support systems, or CDSS, are now essential elements in improving the way healthcare is delivered because they help physicians make judgments that are more precise and well informed. The classification and understanding of CDSS have been facilitated through various taxonomies, each presenting a unique perspective on the categorization and functionality of these systems. This discussion draws from notable contributions by study [4], highlighting the diversity and complexity of CDSS classification. Another important aspect is that nowadays with a powerful computer it is possible to train RNN with deeper and more complex structures, which will probably perform better than the one trained for this project. The RNN used in this experiment seemed to perform worse when predicting values that are far from the last input observation, more recent RNN structures allow the network to improve its memory through time and so they should be able to overcome this problem for clinical decision support system (CDSS). The classification of various terminologies is performed in Table 1 below.
A study [5] presented a thorough taxonomy that includes five dimensions: information delivery, workflow effect, kind of decision help provided, context of usage, and knowledge and data sources. In particular, this categorization helps to clarify the operational capabilities of CDSS. The setting, goals, and other contextual elements affecting the system’s application are all included in the context of usage. Examining the basis upon which CDSS functions, knowledge and data sources highlight the significance of accurate and pertinent information. The concept of decision support distinguishes between various methods for supporting clinical judgments by delving into the reasoning skills of the system. Information delivery emphasizes the importance of data presentation and concentrates on how insights are communicated to the consumers. Two frameworks are used by CDSS, as seen in Fig. 1 below.
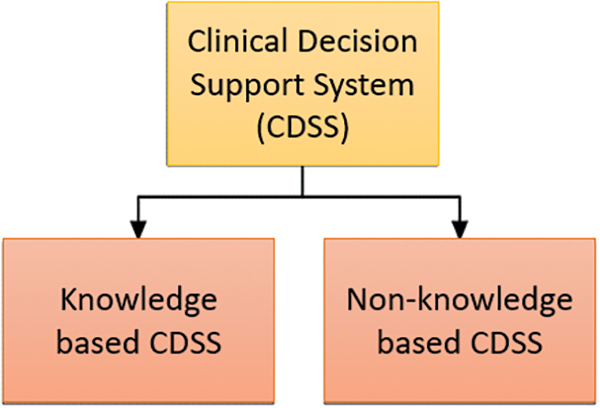
Figure 1: The two prominent methods of clinical decision support system (CDSS)
By leveraging RNNs, which are particularly adept at understanding and generating human language, this paper proposes an innovative approach to decoding the intricacies of medical dialogue and literature. The focus is on enabling clinical decision support systems to interpret the context and nuances of healthcare professionals’ inquiries, thereby offering support that is both clinically valid and immediately applicable as shown in Fig. 2. Our suggested methodology for the CDSS comprises a hybrid architecture that combines a simple RNN and modified RNN system with an association rule-generator algorithm.
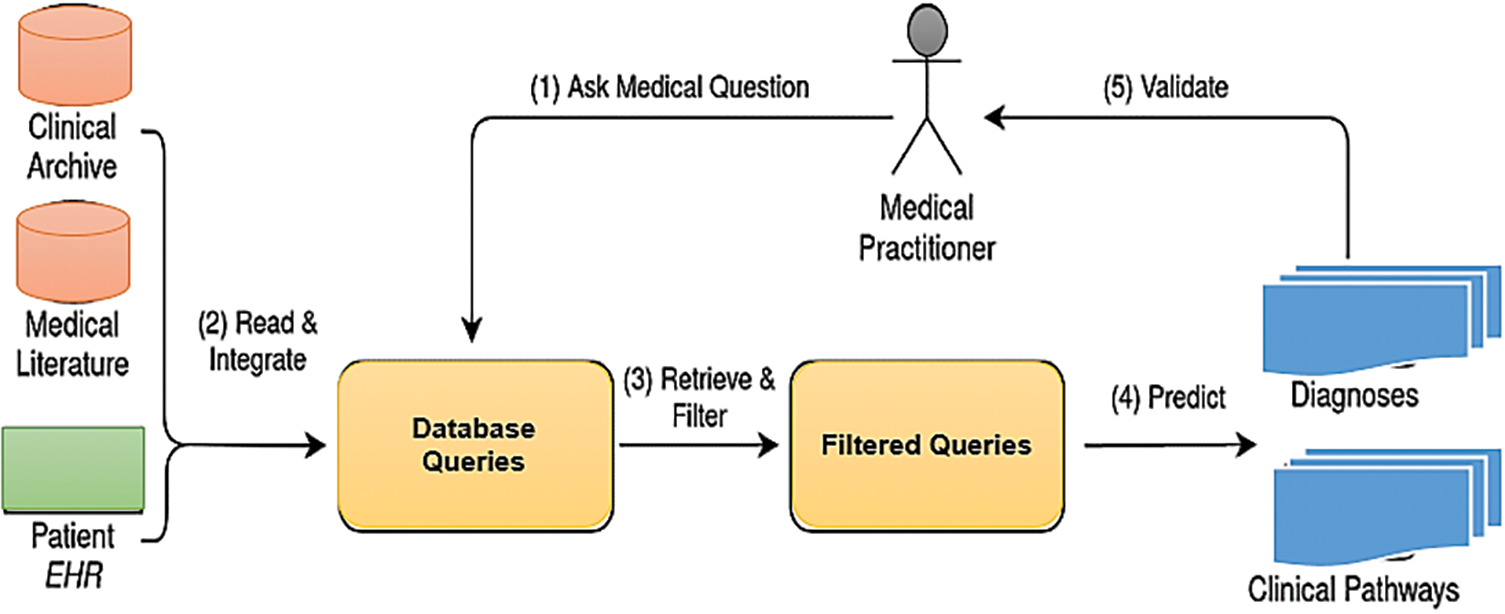
Figure 2: Clinical decision support system (CDSS) framework systems to interpret the context and nuances of healthcare
Different scientific classifications have been created to classify Clinical Decision Support Systems (CDSS), each centering on diverse angles of these frameworks to superior get it their scope, plan, and functionalities. These scientific classifications are vital in organizing the tremendous field of CDSS and giving systems that offer assistance analysts and engineers comprehend and move forward these frameworks. One conspicuous taxonomy, proposed by consider [6], employments five measurements to portray CDSS: setting of utilize, information and information sources, nature of choice back advertised, data conveyance, and workflow affect. The setting of utilize envelops the natural setting, targets, and other pertinent variables impacting the CDSS application. The information and information sources measurement survey the roots and sorts of information the CDSS utilizes to create choices. The nature of choice back considers how the CDSS forms data and helps with thinking errands. Data conveyance analyzes the designs through which data is displayed to the client, guaranteeing it’s justifiable and significant. Finally, the workflow affects measures how the CDSS interatomic with and influences the performing artists included within the clinical handle. This scientific classification is particularly valuable in analyzing the functionalities and eagerly behind distinctive CDSS plans. Another scientific categorization by consider [7] presents a prescriptive approach, laying out different strategies of clinical choice bolster such as documentation forms/templates, important information show, arrange creation facilitators, time-based checking, protocol/pathway back, reference data and direction, and receptive cautions and updates. This scientific classification is especially important for deciding fitting CDSS mediations for particular clinical issues. Be that as it may, it does not give broad direction on the plan and usage of the frameworks, centering more on the operational angle.
The typology categorizes CDSS based on their integration into the helpful cycle, which is partitioned into nine particular prepare steps. Each step where a CDSS can be actualized is highlighted, permitting for a clear mapping of where and how CDSS contribute to quiet care:
• Identification of CDSS Part in Therapeutic Steps:
• The therapeutic cycle is isolated into particular steps, and CDSS can be coordinates into one or more of these steps. This integration is mapped out clearly, frequently delineated in a visual organize such as a flowchart or a table with particular steps highlighted.
• Usefulness Attribution:
• In spite of the fact that a CDSS can span different steps within the therapeutic cycle, the functionalities of the CDSS are particularly credited to certain steps. This makes a difference in understanding the coordinate effect of the CDSS on the helpful prepare and guarantees that its functionalities are adjusted with clinical needs at each step.
• Strong Nature of CDSS:
• The typology emphasizes that CDSS is planned to support clinical decision-making instead of supplanting it. Typically, vital for understanding the moral boundaries of CDSS utilize, guaranteeing that these frameworks expand the clinician’s capabilities without undermining their independence. Fig. 3: Guidelines for the Clinical Decision Support System (CDSS) for Diagnosis and Treatment Throughout Therapeutic Phases.
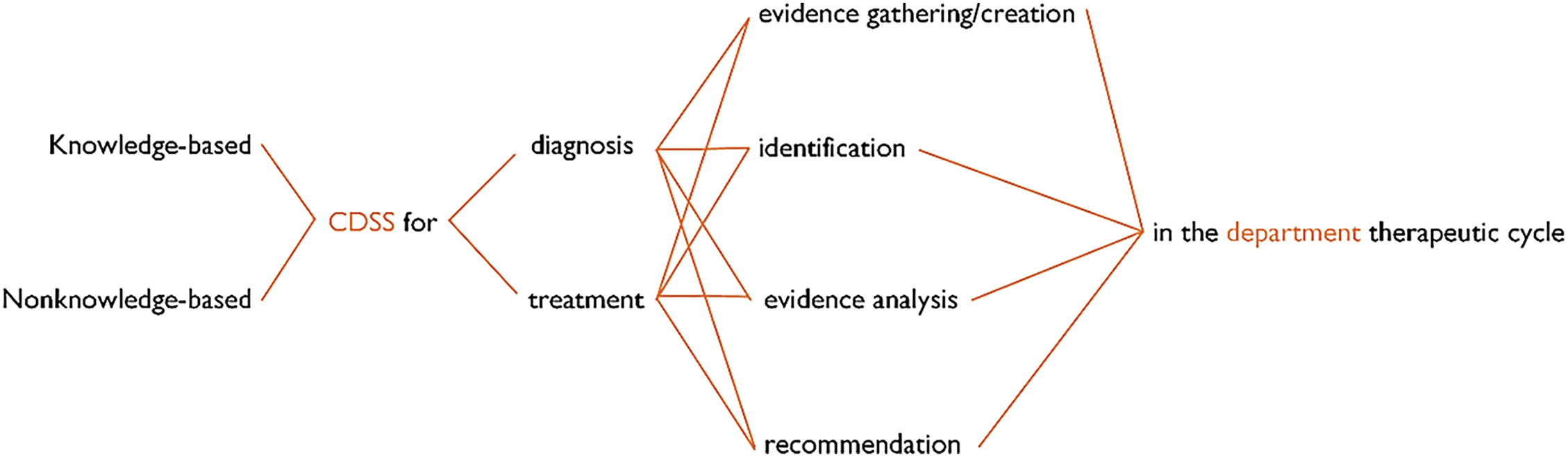
Figure 3: Guidelines for the CDSS for the diagnoses and treatment in the therapeutic cycles
The problem statement for the study is that the Clinical Decision Support Systems (CDSS) play a significant part in upgrading healthcare conveyance by giving opportune and precise restorative proposals to healthcare suppliers. In any case, existing CDSS stages regularly confront challenges in preparing real-time therapeutic questions successfully due to the complexity and changeability of clinical information. The essential issue rotates around the failure of conventional CDSS calculations to capture and learn from the transient conditions and subtleties inside clinical accounts and understanding information, which are crucial for making precise forecasts and suggestions. This restriction not as it were hampers the system’s effectiveness but too influences the quality of care given to patients. To address these challenges, there is a got to optimize CDSS stages by leveraging progressed machine learning methods that can handle the consecutive and energetic nature of clinical information more adeptly.
This study presents an RNN approach to optimizing Clinical Decision Support Systems (CDSS) by utilizing Recurrent Neural Network (RNN) dialect models, centering on both basic and altered RNN structures for the handling of real-time restorative inquiries. Our commitments are centered on the improvement and assessment of these RNN models to improve the productivity and exactness of CDSS in taking care of the complex and energetic nature of clinical information. Firstly, we investigate the execution of basic RNN models to capture transient conditions inside clinical accounts. This permits CDSS stages to prepare questions with progressed precision and speed, encouraging speedier and more dependable restorative suggestions. The straightforwardness of the RNN demonstrates that it can be consistently coordinated into existing CDSS systems, in this way advertising a quick upgrade to the system’s execution without requiring noteworthy adjustments. Another major commitment of this work is the center on real-time restorative inquiry handling. By leveraging the capabilities of RNN dialect models (i.e., simple RNN and Modified RNN), the optimized CDSS is prepared to convey convenient and exact proposals, which is especially basic in crisis circumstances where fast decision-making is vital. This improvement does not were makes strides the reaction time of CDSS but moreover guarantees that healthcare suppliers are upheld with the best conceivable data when making clinical choices.
The major contributions of this work are multifaceted and can be summarized as follows:
□ Targeted Application of AI for Specific Medical Diagnoses: This work uniquely introduces and validates machine learning models specifically for diagnosing rotator cuff tears in osteoporosis patients using anteroposterior X-rays of the shoulder joint. By focusing on this niche application, the study demonstrates the potential of AI to significantly improve diagnostic accuracy and consistency in a specialized area of medical imaging. This targeted approach ensures that the benefits of AI are maximized for particular patient groups, providing a clear advantage over more generalized AI models.
□ Development and Integration of CG-Net for Enhanced Disease Classification: The research presents CG-Net, a novel Convolutional Neural Network (CNN) framework designed for the classification of gastrointestinal tract diseases. This innovative framework exemplifies how deep learning can be applied to complex medical data to enhance disease classification accuracy. By integrating CG-Net into Clinical Decision Support Systems (CDSS), the study significantly advances the precision and informativeness of clinical decision-making, crucially reducing diagnostic errors and improving patient outcomes. This contribution highlights the practical application of advanced AI technologies in real-world clinical settings.
□ Optimization of Healthcare Delivery through Specialized AI Models: The implementation of AI models in clinical settings, as demonstrated in this study, streamlines workflows by providing real-time diagnostic support. These models, trained on large volumes of annotated medical images and clinical data specific to rotator cuff tears and gastrointestinal diseases, enable the identification of intricate patterns associated with these conditions. This extensive, specialized training enhances the models’ diagnostic capabilities, leading to better resource utilization and improved patient care. Additionally, the study showcases the potential for these AI models to be adapted and applied to other specific medical conditions, paving the way for broader applications of AI in healthcare diagnostics and treatment planning.
The primary goal of this research is to optimize Clinical Decision Support Systems (CDSS) through the application of advanced Recurrent Neural Network (RNN) models and other deep learning techniques. To achieve this, the following research questions and hypotheses are proposed:
Research Questions
1. How effective are RNN models, specifically those incorporating LSTM layers and echo state cells, in diagnosing rotator cuff tears in osteoporosis patients using anteroposterior X-rays of the shoulder joint?
2. Can the integration of the CG-Net framework into CDSS significantly improve the classification accuracy of gastrointestinal tract diseases?
3. What impact does the application of data augmentation techniques and transfer learning have on the performance of AI models in clinical decision support?
4. How do advanced AI models, including RNNs and CNNs, compare in terms of diagnostic accuracy and reliability when applied to medical imaging and sequential data analysis?
5. What are the challenges and limitations of implementing AI-based CDSS in diverse healthcare settings, and how can these be addressed?
Hypotheses
1. Hypothesis 1 RNN models with LSTM layers and echo state cells will achieve higher diagnostic accuracy in identifying rotator cuff tears in osteoporosis patients compared to traditional diagnostic methods.
- Measurement: Diagnostic accuracy will be measured using metrics such as precision, recall, F1-score, and Area Under the Curve (AUC) based on a labeled dataset of anteroposterior X-rays.
2. Hypothesis 2 The integration of the CG-Net framework into CDSS will significantly enhance the classification accuracy of gastrointestinal tract diseases.
- Measurement: Classification accuracy will be assessed using accuracy, precision, recall, F1-score, and AUC on a dataset of annotated gastrointestinal medical images.
3. Hypothesis 3 The application of data augmentation techniques and transfer learning will improve the performance and generalization of AI models in clinical decision support.
- Measurement: Performance improvements will be evaluated based on accuracy, loss, and generalization error on augmented datasets and those processed through transfer learning techniques.
4. Hypothesis 4 Advanced AI models, including RNNs and CNNs, will demonstrate superior diagnostic accuracy and reliability over traditional methods in both medical imaging and sequential data analysis.
- Measurement: Comparative analysis will be conducted using diagnostic accuracy metrics, robustness tests, and reliability assessments across multiple medical conditions and datasets.
5. Hypothesis 5 Implementing AI-based CDSS in diverse healthcare settings faces challenges related to data interoperability, standardization, and model interpretability, which can be mitigated through global standards and advanced algorithmic solutions.
- Measurement: Challenges and solutions will be identified through a combination of qualitative analysis (surveys, interviews) and quantitative metrics (interoperability scores, adoption rates, user satisfaction surveys).
The primary motivation for this research stems from the critical need to enhance the accuracy and efficiency of Clinical Decision Support Systems (CDSS) within Wireless Body Area Networks (WBANs). Traditional diagnostic methods and existing CDSS frameworks often fall short when dealing with complex, sequential medical data. These conventional approaches struggle with issues such as long-term dependency management, data imbalance, and limited ability to generalize across diverse patient populations. Addressing these limitations is crucial for improving diagnostic precision, ensuring timely medical interventions, and ultimately enhancing patient outcomes.
Several research gaps have been identified in the current landscape. One major gap is the handling of sequential data. Traditional CDSS and diagnostic methods often lack the capability to effectively manage sequential data, such as time-series data from wearable sensors and longitudinal patient records. This inadequacy hampers their ability to capture and utilize temporal dependencies, which are vital for accurate diagnosis and treatment planning. Another significant gap is the issue of data imbalance. Many medical datasets are inherently imbalanced, with a disproportionate number of cases representing common conditions compared to rarer ones. This imbalance can bias traditional models towards the majority class, leading to suboptimal performance and inaccurate diagnoses for less common but critical conditions.
Additionally, there is a problem with model generalization. Existing models frequently exhibit poor generalization across different datasets and patient populations. This limitation reduces their reliability and effectiveness in diverse clinical settings, where patient demographics and health conditions can vary widely. Furthermore, the integration of multi-modal data is often lacking in current CDSS. These systems frequently fail to integrate and analyze multi-modal data, such as combining imaging data with clinical notes and wearable sensor data. The lack of comprehensive data integration limits the system’s ability to provide holistic and accurate diagnostic insights. Lastly, there is a significant gap in the interpretability of AI models used in CDSS. Clinicians need to understand how and why a model arrives at a particular decision to trust and effectively use these systems in clinical practice. Many existing models operate as black boxes, offering little transparency.
To address these gaps, this research focuses on the development and validation of advanced machine learning models, specifically Recurrent Neural Networks (RNNs) with Long Short-Term Memory (LSTM) layers and echo state cells, as well as the CG-Net framework for Convolutional Neural Networks (CNNs). These models are designed to enhance the diagnostic capabilities of CDSS. RNNs, particularly those enhanced with LSTM layers, are capable of capturing and utilizing long-term dependencies in sequential data. This capability is essential for accurate diagnosis based on time-series data from wearable sensors and longitudinal patient records.
In this section, we provide a well-organized review of the relevant literature, highlighting advancements and identifying the shortcomings of previous studies. This structured approach gives a comprehensive overview of the state-of-the-art and justifies the need for our research. Recurrent Neural Networks (RNNs) have been widely used in medical diagnostics for processing sequential data. Lipton et al. demonstrated the use of RNNs in diagnosing multiple diseases from clinical time series data. However, traditional RNNs struggle with long-term dependencies due to the vanishing gradient problem. Long Short-Term Memory (LSTM) networks, an advanced form of RNN, have been introduced to mitigate this issue. Graves et al. showed that LSTMs could effectively handle long-term dependencies. Despite these improvements, existing studies often overlook the integration of diverse data types, such as combining wearable sensor data with clinical records. Convolutional Neural Networks (CNNs) have become the standard for medical image analysis due to their ability to automatically extract features from images. Krizhevsky et al. pioneered this approach with their AlexNet architecture. In the medical domain, CNNs have been used extensively for tasks like tumor detection and organ segmentation. While frameworks like ResNet and VGG have pushed the boundaries of image classification accuracy, most studies focus on a single type of image data. For example, Esteva et al. applied CNNs to skin cancer classification with impressive results, but their model was limited to dermatological images alone. This highlights a gap in the integration of multi-modal data sources. Data augmentation techniques, such as those proposed by Shorten and Khoshgoftaar, are critical for enhancing the generalization of AI models. SMOTE and its variants have been effectively used to balance datasets and prevent model bias. However, the application of these techniques in medical diagnostics often remains limited to specific case studies. Transfer learning has shown promise in improving model performance with limited data. Yosinski et al. demonstrated the benefits of transfer learning across various domains. In medical AI, pretrained models such as ImageNet have been adapted for specific tasks, yet their effectiveness varies depending on the similarity of the source and target domains. Clinical Decision Support Systems (CDSS) have evolved from simple rule-based systems to sophisticated AI-driven platforms. Berner and La Lande reviewed the evolution of CDSS, emphasizing the transition towards incorporating machine learning. Despite these advancements, many CDSS still struggle with issues of interpretability and clinician trust. Recent efforts, such as those by Ribeiro et al. with LIME and Lundberg and Lee with SHAP, have focused on improving model interpretability. However, the integration of these explainability tools into practical CDSS applications remains a work in progress. Several shortcomings and research gaps have been identified. Firstly, while LSTMs address some issues of traditional RNNs, many studies do not fully leverage their potential for integrating long-term sequential data with other data types, such as wearable sensor inputs and clinical notes. Secondly, existing CNN-based studies often focus on a single type of medical image, lacking a holistic approach to integrate various data sources, which is crucial for comprehensive diagnostics. Thirdly, although data augmentation and transfer learning techniques have been proposed, their application in medical diagnostics is often limited and not fully exploited for various medical conditions. Finally, despite advancements in explainability tools like LIME and SHAP, many AI models used in CDSS are still perceived as black boxes, limiting their acceptance and use by clinicians. Given these identified gaps, our research aims to develop a hybrid RNN model combining SimpleRNN, LSTM layers, and echo state cells to better manage long-term dependencies in sequential medical data. We also introduce the CG-Net framework to enhance CNN performance for multi-modal data integration, specifically addressing gastrointestinal diseases. Additionally, we apply advanced data augmentation techniques (SMOTE, BorderLine SMOTE) and transfer learning to improve model generalization and robustness. Furthermore, we incorporate explainability tools (SHAP, LIME) to ensure the interpretability of our models, thereby enhancing clinician trust and usability in CDSS. By addressing these shortcomings, our research advances the field of medical diagnostics within WBANs, providing robust, accurate, and interpretable AI models that integrate multi-modal data sources for comprehensive clinical decision support.
Traditional Recurrent Neural Networks (RNNs) and Long Short-Term Memory (LSTM) networks have been widely used in medical diagnostics for processing sequential data. Studies such as those by Lipton et al. and Graves et al. provide strong baselines for evaluating the performance of our hybrid RNN model with attention mechanisms. By including these traditional models, we can demonstrate the enhanced capabilities of our proposed model in handling long-term dependencies and sequential medical data.
Convolutional Neural Networks (CNNs) like AlexNet, VGG, and ResNet have set benchmarks in medical image analysis. Krizhevsky et al., Simonyan and Zisserman, and He et al. have demonstrated the efficacy of these architectures in various medical imaging tasks. Including these standard CNN architectures as baselines will help in assessing the effectiveness of our CG-Net framework. Comparing our results with these established models will highlight the superior performance of CG-Net in integrating multi-modal data and achieving high diagnostic accuracy.
Ensemble methods and hybrid models that combine different types of neural networks have shown promise in improving diagnostic accuracy. Studies by Zhou et al. and Dietterich on ensemble methods, and hybrid approaches like those proposed by Razzak et al., provide additional baselines to compare against our hybrid RNN and CG-Net models. These comparisons will illustrate how our approach benefits from combining RNNs with LSTM layers, attention mechanisms, and CNNs, leading to improved model performance and robustness.
The impact of data augmentation and transfer learning on model performance has been well-documented. Techniques such as SMOTE and transfer learning have been utilized in studies by Chawla et al. and Yosinski et al. Comparing our advanced data augmentation and transfer learning approaches with these established techniques will highlight the improvements in generalization and robustness. Our results will show that these techniques help our models perform better across diverse datasets and patient populations.
The interpretability of AI models is crucial for their adoption in clinical settings. Methods like LIME and SHAP have been employed in studies by Ribeiro et al. and Lundberg and Lee. Including these methods as baselines will allow us to demonstrate how our integration of interpretability tools enhances the transparency and trustworthiness of our models. This comparison will underscore the importance of making AI models understandable and reliable for clinicians, ensuring that our proposed solution can be effectively used in practice.
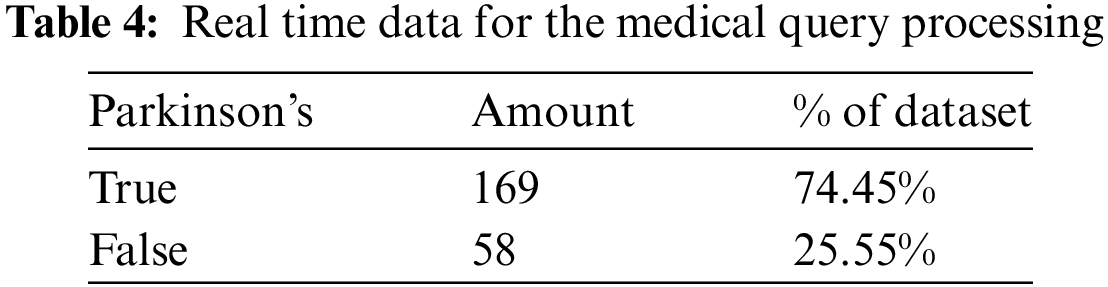
Incorporating these baselines, our experimental evaluation will include a comprehensive comparison of performance metrics such as accuracy, precision, recall, F1-score, and Area Under the Curve (AUC) for different models and techniques. We will evaluate model robustness and generalization across diverse datasets and patient populations, assess the ability to integrate and process multi-modal data, and ensure model transparency and explainability using interpretability tools like LIME and SHAP. Table 2 details the data imbalance of the Tappy dataset with 74.45% belonging to subjects having Parkinson’s disease and 25.55% of the subjects belonging to the healthy class.
The development and validation of AI-based diagnostic models marks a significant advancement in the field of medical imaging. Specifically, this work demonstrates the efficacy of machine learning models in diagnosing rotator cuff tears in osteoporosis patients using anteroposterior X-rays of the shoulder joint. These models leverage advanced algorithms to scrutinize medical images, identifying subtle patterns and anomalies that might be challenging for human radiologists to consistently detect. By being trained on extensive datasets, these AI models achieve high accuracy and reliability, facilitating early and precise diagnoses. This capability not only enhances patient outcomes through timely interventions but also optimizes the overall management of rotator cuff injuries in patients with osteoporosis. However, despite these advancements, there remains a gap in the comprehensive application of AI across diverse medical imaging modalities and conditions. Current models often require large, annotated datasets, which are not always available for rarer conditions. Additionally, the interpretability of AI models poses a challenge, as clinicians need to understand how decisions are made to trust and effectively use these systems. Addressing these gaps requires ongoing research into data augmentation techniques, transfer learning, and explainable AI to make these models more universally applicable and trustworthy. Moreover, the introduction of CG-Net, a novel Convolutional Neural Network (CNN) framework for the classification of gastrointestinal tract diseases, showcases the potential of deep learning in medical diagnostics. CG-Net effectively analyzes complex medical data, distinguishing between various gastrointestinal conditions with remarkable accuracy. Trained on large volumes of annotated medical images and clinical data, this framework can learn and identify intricate disease patterns. By providing real-time diagnostic support, CG-Net aids physicians in making timely and accurate diagnoses, thereby reducing diagnostic errors and improving patient outcomes.
Cutting-edge research in this area is pushing the boundaries by integrating multi-modal data sources, including genetic information, lab results, and electronic health records, to enhance the predictive power of these models. However, the challenge remains in the seamless integration of these diverse data types and ensuring the models’ robustness across different patient populations and clinical settings. Future advancements will need to focus on developing more sophisticated algorithms capable of handling such complex, multi-dimensional data. The integration of advanced AI frameworks like CG-Net into Clinical Decision Support Systems (CDSS) significantly enhances the precision and informativeness of clinical decision-making. These systems are crucial for modern healthcare delivery, helping physicians make more accurate and well-informed decisions. The ability of AI models to provide real-time diagnostic support ensures that clinicians can make timely decisions, leading to better resource utilization and improved patient care. This integration streamlines clinical workflows, enhances the efficiency of healthcare delivery, and supports clinicians with the most up-to-date and comprehensive data available, thereby raising the standard of patient care. Nevertheless, a significant gap exists in the interoperability and standardization of CDSS across different healthcare systems and regions. The lack of standardized protocols and data formats can hinder the widespread adoption and effectiveness of these systems. To bridge this gap, there is a need for global standards and regulations that ensure seamless integration and interoperability of CDSS, facilitating their adoption across various healthcare settings. Training these AI models on extensive medical datasets enables them to identify intricate patterns associated with various medical conditions, thereby enhancing their diagnostic capabilities. The extensive training on large volumes of annotated medical images and clinical data ensures that the models can effectively differentiate between subtle differences in medical conditions, leading to higher diagnostic accuracy. This ability to handle complex and diverse medical data makes AI a powerful tool in the field of medical diagnostics.
This research employs a comprehensive methodology to develop and evaluate advanced AI models for optimizing Clinical Decision Support Systems (CDSS) within Wireless Body Area Networks (WBANs). The methodology involves several critical steps, including data sourcing, analytical techniques, and validation procedures, each contributing to the robustness and effectiveness of the models. The study utilized diverse data sources to train and validate the AI models. Medical imaging data, specifically anteroposterior X-rays of the shoulder joint, were used to diagnose rotator cuff tears in osteoporosis patients. These images were obtained from publicly available medical image repositories and hospital databases. Additionally, annotated medical images of the gastrointestinal tract were sourced from specialized medical institutions and online databases to aid in the classification of gastrointestinal diseases. Clinical data, including Electronic Health Records (EHRs) and data from wearable sensors, were incorporated to provide contextual information and enhance the predictive accuracy of the models. The EHRs contained patient demographics, medical histories, and clinical notes, while wearable sensor data included vital signs and movement patterns. The analytical techniques employed in this research involved several key steps. Initially, preprocessing techniques were applied to both image and text data. Image preprocessing included normalization, resizing, and augmentation techniques such as rotation, flipping, and contrast adjustment to improve model training. Text data from clinical notes and EHRs were processed using natural language processing (NLP) techniques like tokenization, stemming, and stop word removal.
The model development phase focused on creating and optimizing AI models, including Recurrent Neural Networks (RNNs) with Long Short-Term Memory (LSTM) layers and echo state cells to handle sequential clinical data. The CG-Net framework, a novel Convolutional Neural Network (CNN) architecture, was implemented for accurate classification of gastrointestinal diseases. This model leveraged convolutional layers to capture spatial features from medical images. Additionally, hybrid models combining SimpleRNN, LSTM layers, and echo state cells were developed to effectively analyze both time-series data from wearable sensors and static medical images. Data augmentation techniques such as the Synthetic Minority Oversampling Technique (SMOTE) and BorderLine SMOTE were employed to balance the datasets and enhance model generalization. Transfer learning was also utilized, where pretrained models were fine-tuned on the medical datasets to leverage existing knowledge and improve performance with limited data.
The validation procedures ensured the robustness and generalizability of the developed models. The datasets were split into training, validation, and testing sets, typically in a 70%, 15%, and 15% ratio, respectively. This approach ensured that the models were trained on one subset of the data and tested on an entirely separate subset. A 5-fold cross-validation technique was employed to further validate the models, where the data was divided into five subsets, and the model was trained and validated five times, each time using a different subset as the validation set. Performance metrics such as accuracy, precision, recall, F1-score, and Area Under the Curve (AUC) of the Receiver Operating Characteristic (ROC) curve were used to evaluate the models. Hyperparameter tuning was conducted using grid search and random search techniques to optimize parameters like learning rate, batch size, number of hidden layers, and number of neurons per layer. Early stopping was applied to halt training if the model’s performance on the validation set did not improve for a specified number of epochs, preventing overfitting. Model interpretability and explainability were addressed using SHapley Additive exPlanations (SHAP) and Local Interpretable Model-agnostic Explanations (LIME). These tools provided insights into the models’ predictions and the contribution of each feature, enhancing transparency and trustworthiness. The implementation phase involved the use of programming languages like Python, with libraries such as TensorFlow, Keras, and PyTorch for model development. Development environments like Jupyter Notebooks and integrated development environments (IDEs) such as PyCharm were utilized for coding and experimentation. The models were deployed on cloud platforms like Amazon Web Services (AWS) and Google Cloud Platform (GCP) to facilitate real-time data processing and scalability. Finally, the developed models were integrated into existing CDSS frameworks to enhance diagnostic capabilities and provide real-time support to clinicians.
A recurrent neural network, or RNN for short, is a particular kind of neural network which possesses some interesting characteristics that made it gain a lot of popularity in the last decade, mainly in natural language tasks and time series forecasting. Compared with a standard artificial neural network, the upgrade that this method brings in forecasting tasks is the fact that the output at any given time step is involved in the computation of the output at next time step. This characteristic gives the RNN the ability to learn temporal relationships between observations. Others come in the form of using a specific model architecture like the Echo State Network [16], a variant of the RNN model that uses a reservoir with fixed hidden layer weights, and only updates the weights of the output layers to enforce the desired outcome. This procedure of the Echo State Network has also proven to provide greater accuracy with imbalanced datasets but has yet to be lost to time in favor of more developed modern models today as mentioned [17]. The architecture of RNN uses attention mechanisms to capture the sequence’s dependencies between all-time steps which is being used in medical fields. Because of attention mechanisms, RNN give more weight to the most crucial time steps in the sequence while ignoring the irrelevant ones as shown in Fig. 4. This makes it easier for the model to capture long-term dependencies between distant time steps which is critical when working with time series data. The iterative representation of an RNN emphasizes the looped nature of the network where output from one step is fed back into the network as input for the next step. This feedback mechanism allows the network to maintain an internal state that captures information about previous inputs.
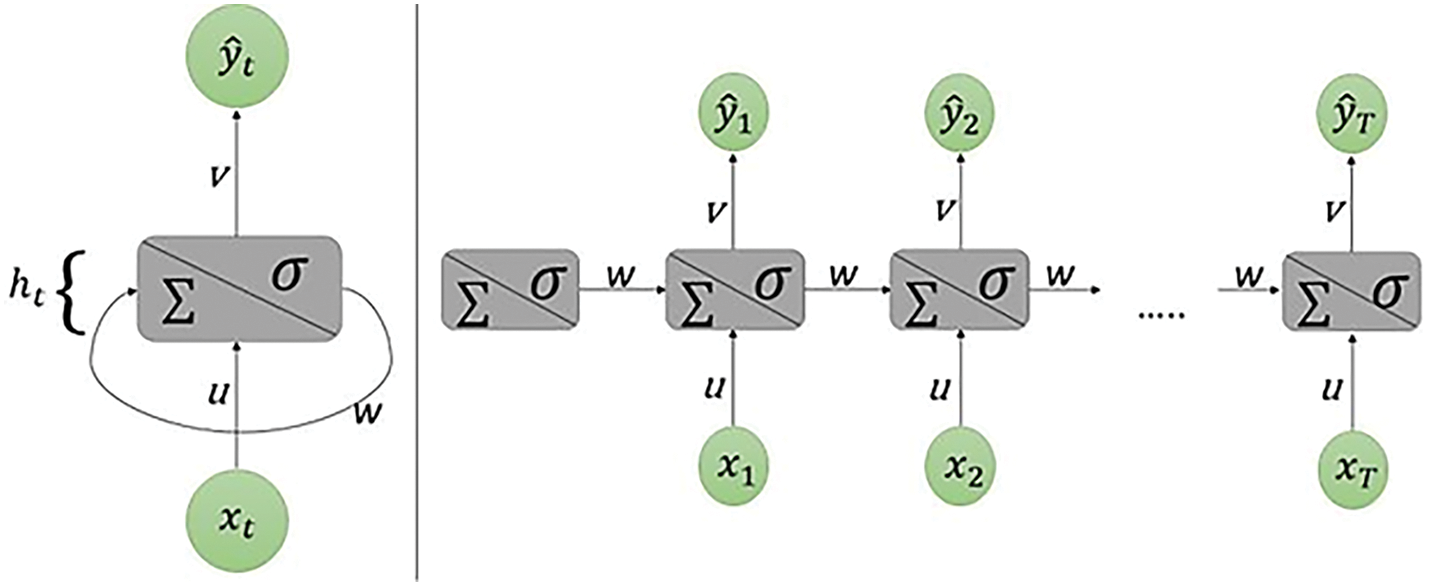
Figure 4: Scheme of the simplest recurrent neural network. On the left it is shown in its iterative representation, on the right what is shown is unrolling through time
The simple RNN basic equation for the simplest RNN in its iterative form is:
Here:
• ht is the hidden state at time t, which serves as the “memory” of the network, capturing information from previous inputs.
• ht − 1 is the hidden state from the previous time step.
• xt is the input at time t.
• W and U are the weights applied to the hidden state and the input, respectively.
• b is the bias term.
• f is a non-linear activation function, typically tanh or ReLU.
For this paper, an empirical study is performed to investigate the potential of combining universally accepted solutions with experimental designs to replicate or enhance classifier performance, ultimately identifying optimized approaches for clinical data and medical queries though the deep RNN model. The study aims to discuss results based on experiments that use a combination of SMOTE and its variant BorderLine SMOTE, Simple RNN, Modified RNN consisting of Echo State Cell, Transformer, and Feature Ranking using Recurrent Neural Network. The graphic representation of a modified RNN is shown in Fig. 5.
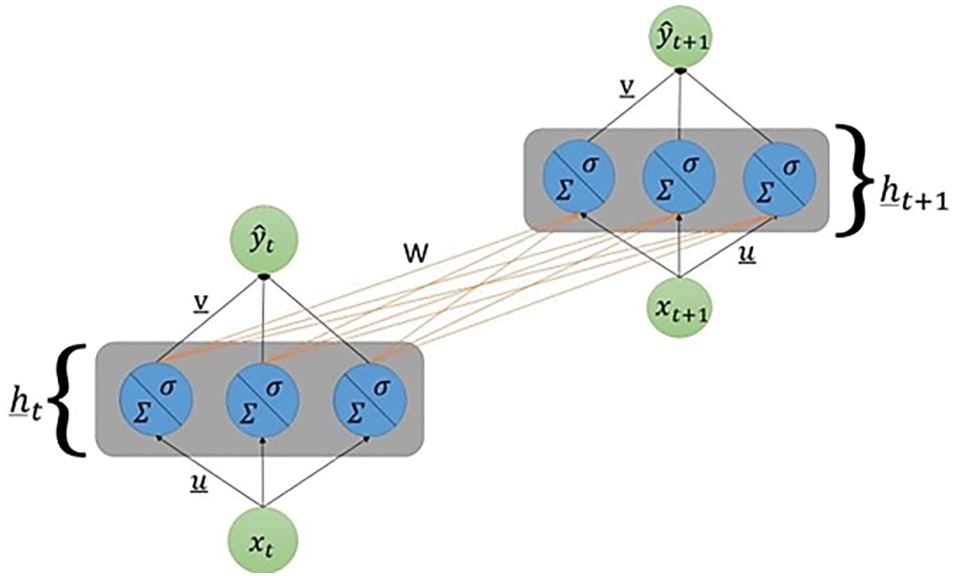
Figure 5: Graphic representation of a modified RNN having several neurons in the hidden layer
The Modified RNN architecture is easy to understand but it struggles when trying to track complex data structures. Adding neurons to the hidden layer gives to the algorithm more flexibility, making it able to predict more challenging patterns in the data. Let us see how the dimensionalities of the parameters of the network change with this new architecture.
Every line connecting two neurons represents a particular weight in the network. In this figure it is shown the unrolling through time between time steps t and t + 1.
• xt: the input, is still a scalar one;
• ht: it is a vector belonging to Rd. This vector has as d-th entry the state of the d-th neuron of the hidden layer at time t;
• u: the weight vector belonging to Rd. The entries of this vector are [u1, u2, ..., ud] which are the weights connecting the scalar input to each single neuron of the hidden layer.
• W: the weight matrix belonging to Rd × d. Its element is indexed as [wjk] where the subscripts j and k indicate that that single entry is the weight that connects the k-th hidden neuron at time step t − 1 with the j-th one at time step t. This notation may seem backward at first, but it will simplify the computations, allowing to avoid transposing the weight matrix.
• v: the weight vector belonging to Rd. The entries of this vector are [v1, v2, ..., vd] which are the weights connecting the scalar input to each single neuron of the hidden layer.
This means that if a simple RNN and modifed RNN trained to perform the best predictions at Lag 1 will perform far from the optimal if used to perform predictions at time step 2. For this reason, different modified RNNs have been trained in order to obtain the best predictions at a given time step. This is extremely computationally demanding, and it has been possible because this project has been developped over the course of several months. The training of a RNN may take from 1 h up to 24 h, depending on the length of the input sequence that is fed into the network. With the input window containing 1000 observations the training required an entire day of computations and this time consuming training is not always possible. The Lag observation of RNN is shown in Fig. 6.
Figure 6: Lag observation across RNN feature for the processing of queries for clinical decision support
The structure seen in the picture is divided in three layers: an input layer, a hidden layer and an output layer. Each layer is composed, in the simplest possible RNN, of a single neuron, which is a computational unit that performs some transformations on the data received as input. The neuron in the input layer performs the identity transformation, i.e., it simply passes the input value to the neuron in the hidden layer without applying any transformation. When the hidden layer receives the value, it uses it to perform a weighted sum and a nonlinear transformation [18]. The result of these transformations performed at time step t in the hidden layer is called the hidden state at time t. After the computation, the hidden state is passed through the network both to the output layer and to the hidden layer at next time step, where it will be treated as an input for the weighted sum. In the output layer a weighted sum and a linear or nonlinear transformation are applied to the input received to produce a prediction. The reason behind the word “recurrent” in the name RNN is the procedure followed by the hidden layer, i.e., sending the hidden state at time t to the hidden layer at time t + 1. Theoretically speaking, this means that the last output at time t = T depends on all the hidden states at time t = T − 1, T − 2, ..., 1 and, consequently, on all the corresponding inputs. To explain it more clearly it is necessary to define the notation that will be used for explaining the simplest form of RNN: the one with a scalar input, a scalar output, and a single neuron in the hidden layer. To ease the notation, it will now be specified, and never again repeated, that t ranges from 1 to T and that the weights are randomly initialized at first.
The quantities that are not explicit are zt, ht and y^t. These are defined as follows:
ht = σ(zt)
y^t = g(vht)
So the “journey” performed by every input value at any time step t is the following:
xt → zt = xtu + wht−1 → ht = σ(zt) → y^ = g(vht)
For each time step t in the input sequence, compute the following:
xt: the scalar input at time step t;
ht: the state of the hidden layer at time t;
y^t: output at time step t;
zt: value of the weighted sum at the hidden state at time step t, t = 1, …, T
u: the weight that connects the input to the hidden layer;
w: the weight that connects ht−1 (previous state of the hidden layer) with the recurrent hidden layer at time t;
v: the weight that connects ht with the output y^t.
where h0 is initialized to be equal to 0. The function g is usually a linear function in simpler models, while it is a nonlinear one if more complex structures are used. The function σ(zt) is called activation f unction and is a nonlinear function that transforms the weighted sum of the inputs received by the hidden layer at time t. If every function applied to xt were linear, then the whole neural network would correspond to a single linear transformation, and it would not be able to identify complex relationships between data. This advantage of nonlinear activation functions will be easier to understand when dealing with more complex RNN structures, where multiple nonlinear transformations are applied one after another. These equations are the foundation of how RNNs process information through time, making them highly effective for applications like clinical decision support where the temporal sequence of events can significantly influence the outcome or decision. The most commonly used activation functions in RNN are sigmoid functions f(x) like the logistic function and the hyperbolic tangent x as shown in Fig. 7.
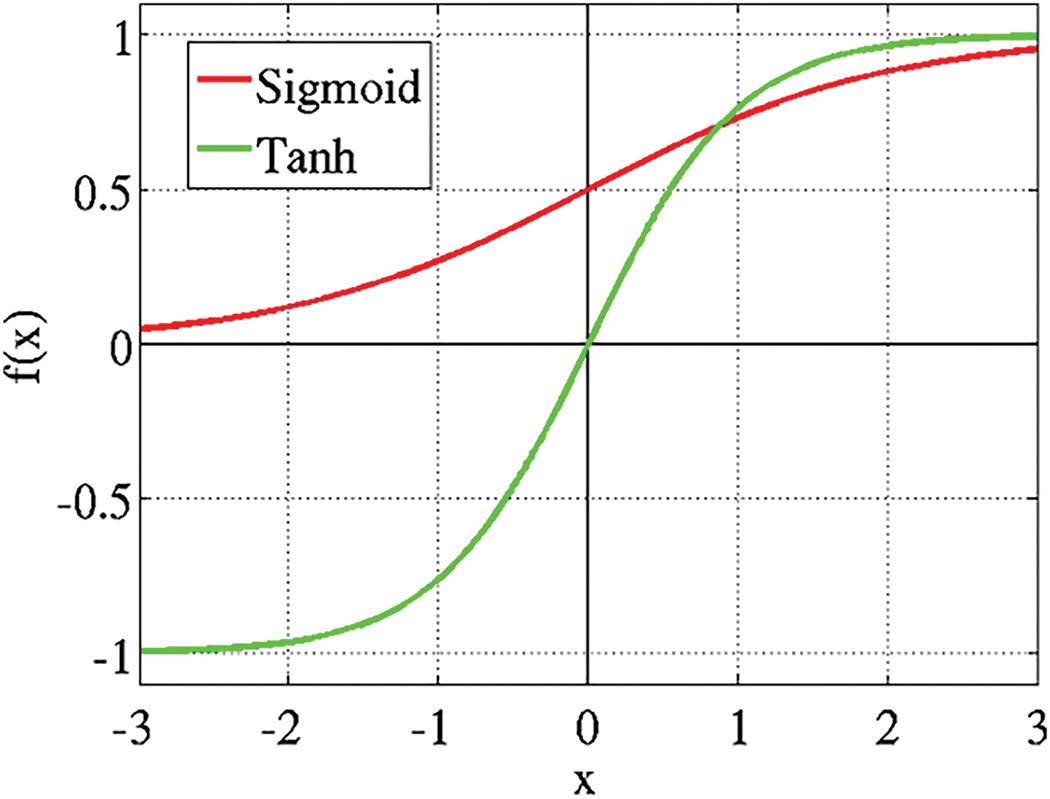
Figure 7: Sigmoid functions f(x) like the logistic function and the hyperbolic tangent x of RNN
Recurrent Neural Networks (RNNs) are especially profitable for CDSS due to their capability in taking care of successive information, a common characteristic in numerous restorative datasets such as understanding records; lab comes about over time, and persistent observing information. RNNs exceed expectations in these settings since of their capacity to preserve an inner state or memory over input groupings, which permits them to consider the movement and improvement of a patient’s condition over time. This capability is vital for prescient analytics in healthcare, where the timing and advancement of information focuses can give basic bits of knowledge into persistent results. Furthermore, RNNs’ structure underpins weight sharing over time steps, improving their productivity and decreasing the model’s complexity and the hazard of overfitting. This include is especially valuable in restorative applications where the sum of information per understanding may change broadly, and the learning demonstrate must generalize well over distinctive lengths of input groupings. Not at all like other neural organize designs such as Multilayer Perceptrons (MLPs) or Convolutional Neural Networks (CNNs), which require fixed-size inputs, RNNs can prepare variable-length groupings of inputs normally. This adaptability permits CDSS to adjust to the differing and energetic nature of clinical information without the required for cushioning or truncation, in this way protecting the judgment and granularity of therapeutic data. The weight (W) sharing across time (t) steps is encapsulated in the recurrent connection for the CDSS as shown in Fig. 8.
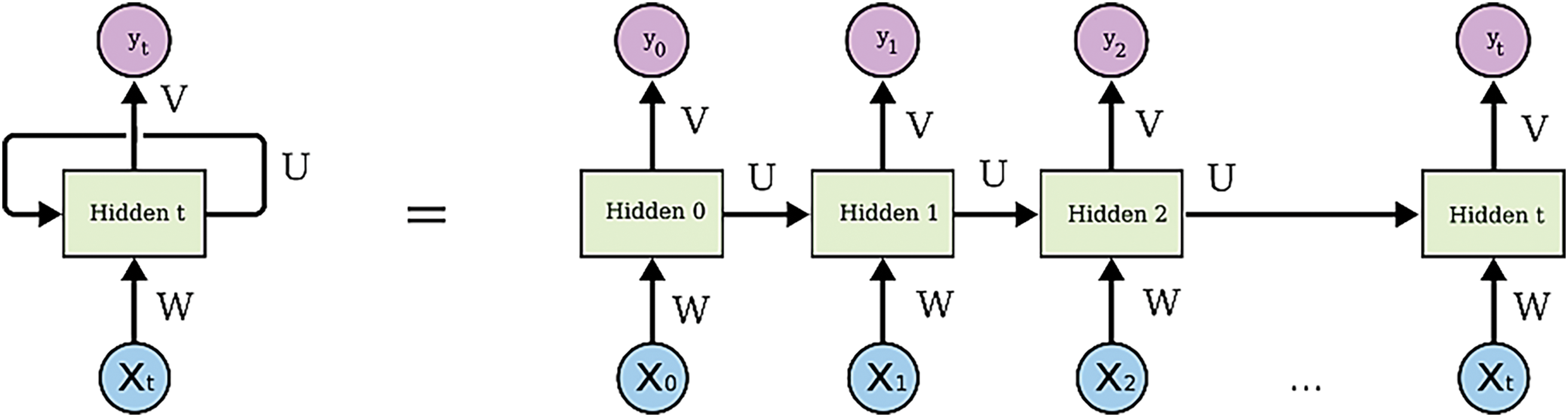
Figure 8: Weight (W) sharing across time (t) steps is encapsulated in the recurrent connection for the CDSS over input groupings of patients
These above functions represent the hidden state and output functions for the calculation of clinical decision support and medical query processing over time t. When building RNNs for applications such as medical data analysis, disease progression prediction, or natural language query processing, these parameters are crucial as stated in [19]. A clinical decision support system can help healthcare providers in a timely and relevant way by improving accuracy, efficiency, and overall performance by adjusting these factors. To illustrate how RNN parameters might improve clinical decision support, particularly for processing medical queries in real-time, the study may assume Table 1 containing important RNN parameters, their descriptions, and some average values. A common characteristic of all the evaluated loss graphs is that the RNN is performing really well but it seems like the LSTM cell in the RNN is not able to keep in memory observations from 24 h before because otherwise the graph of its loss over the 24 h would see a decrease when approaching the 24 h apart predictions, given the fact that observations that are 24 h apart are similar.
• Logical Function
This function transforms every result of the weighted sum into a value between 0 and 1. The disadvantage of this function is that it tends to saturate, meaning that high values of the sum will be very close to 1 and low values will be very close to 0. The only area in which significant changes in x correspond to significant changes in S(x) is the one near zero, where the function is steeper.
• Hyperbolic tangent
Now, Similar to the logistic function but can take values in the range (−1, 1). The disadvantages are the same as the just mentioned function.
• Rectified linear unit (ReLU)
If the result of the weighted sum is negative, then the ReLU function takes the value 0, otherwise it takes the value of the weighted sum. In this way a neuron may be “switched on/off”, meaning that it has recognized a precise pattern or not. It is frequently used in natural language tasks or in sentimental analysis as a last transformation in the output layer.
R (x) = max (0, x)
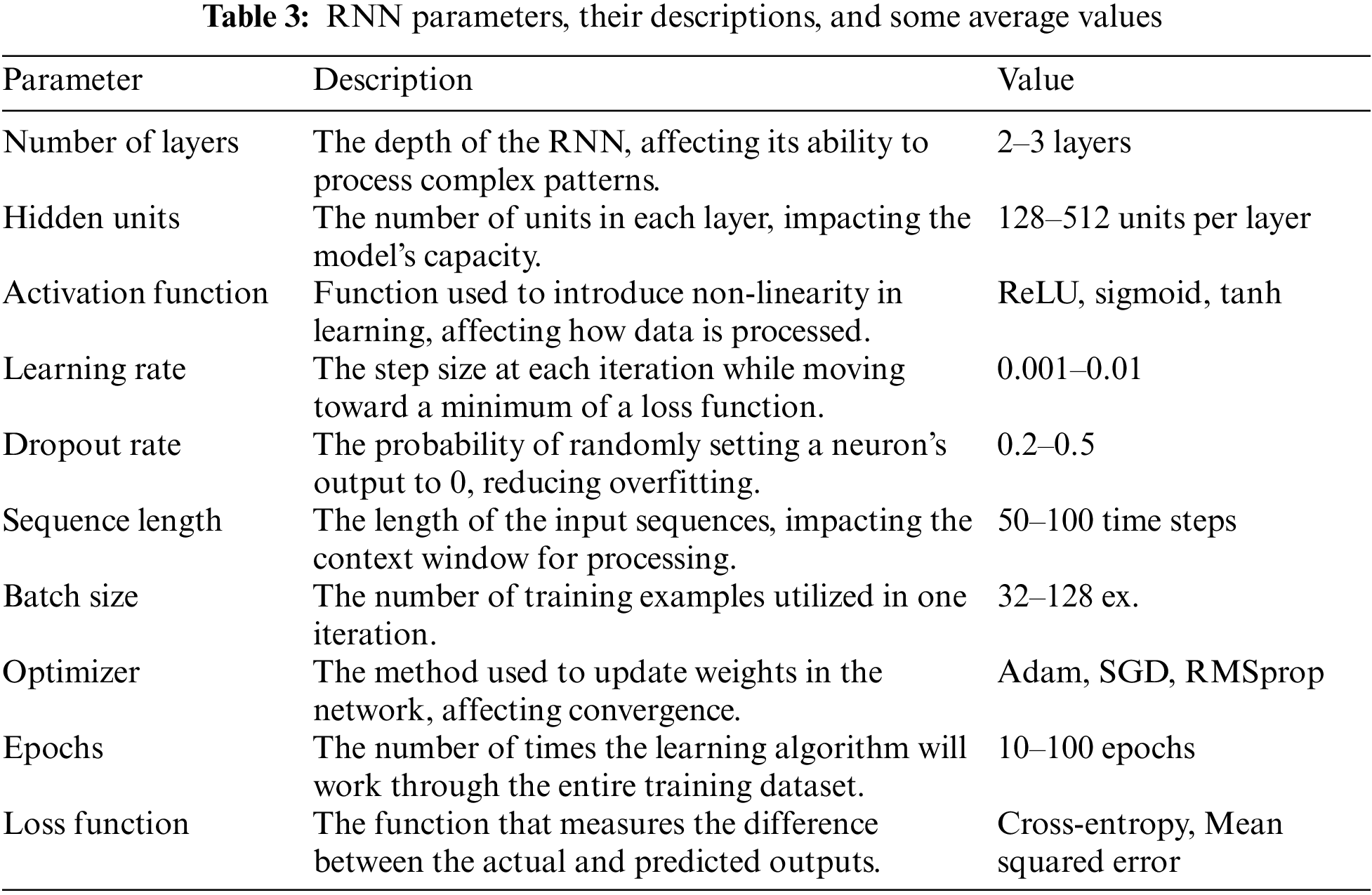
Prominent RNN parameters for the study are mentioned in Table 3.
The presented Recurrent Neural Network (RNN) model includes multiple hidden layers to enhance its processing capabilities. Specifically, the model has two additional Long Short-Term Memory (LSTM) layers and one echo state cell, in addition to the SimpleRNN layer. Each of these layers, except for the final dense layer, employs a ReLU activation function, while the final dense layer uses a Sigmoid activation function. This architecture allows the model to effectively capture temporal dependencies and complex patterns within the data, thereby improving its performance in clinical decision support tasks. The integration of these LSTM layers and the echo state cell provides the RNN with enhanced memory capabilities and the ability to manage long-term dependencies, which are critical for accurate medical diagnoses and decision-making processes. The use of LSTM layers helps in mitigating issues related to the vanishing gradient problem, thereby maintaining the effectiveness of the model over longer sequences of input data. This sophisticated architecture underscores the model’s ability to handle intricate and sequential medical data, making it a powerful tool in the field of clinical decision support systems (CDSS). The combination of SimpleRNN, LSTM layers, and the echo state cell equips the model with the necessary depth and complexity to analyze and interpret clinical data with high precision and reliability.
The Tappy Keystroke Dataset [20] was put together by PhysioNet and included the keystroke records of 200 individuals worldwide, both with and without Parkinson’s. A program called Tappy, was designed to log each user’s interaction with their keyboards to detect changes in the routines and examine the early-stage impacts of Parkinson’s disease. There are two folders in the dataset:
User Data: This section contains text files with the personal information of individuals, including their gender, age at diagnosis, and whether they have Parkinson’s disease.
Keystroke Data: The text files in this subdirectory contain information about keystrokes for each individual. Each file includes information about the user’s key, whether left- or right-handed, the direction to the previous key, their latency time (time between the previous key and current key), and their flight time (Time between the release of the previous key and the press of the current key). This highlight of the dataset chosen is mentioned in Table 4.
The Tappy Keystroke Dataset offers a vast number of time-series samples for our models to utilize, with roughly 9 million available. However, given the size of this dataset, we lacked the resources to reliably train each of our proposed solution models on the entire dataset and instead opted to use a subset of the dataset, with 100,000 samples of observational data across all patients. We ensured that this smaller batch of data maintained the same imbalance ratio presented earlier, and furthermore considered the possibility of time step data points having no correlation with their previous neighbors. Thankfully, no loss of information was seen, as Fig. 4 further proves through Lag observation across RNN all feature values of subsequent time step data points are shown to be fairly close to each other, advising us that data points are within a reasonable range of one another.
4.3 Training the Recurrent Neural Network
Recurrent neural networks can manage a sequence of inputs, and they can output a single value as well as a sequence of values. The so called “training” of the network is the phase in which the weights of the network are modified in order to achieve the best possible prediction. The training phase is a supervised learning algorithm, meaning that the values that the RNN is trying to predict are already known and the visualization of 3-layer NN as shown in Fig. 9. What is crucial to consider is that the RNN is trained for a specific purpose, e.g., for the prediction of a value 3 h into the future or for the prediction of a sequence of 15 h of observations in medical queries, and so the weights are optimized for that purpose only.
Figure 9: Visualization of a 3-layer recurrent neural network with input x
In Recurrent Neural Networks (RNNs), the key feature is their ability to maintain a ‘memory’ of previous inputs by incorporating the output of a layer back into itself. This makes them particularly suitable for sequential data processing, such as medical records or time-series data, which is common in clinical decision support systems.
Back propagation through time is an algorithm that aims at finding a local or global minimum in the cost (also called loss) function by changing the weights of the model. The mathematical tools used are the partial derivatives and the chain rule. To ease the notation, weights will be indicated with upper case letters U, V and W, while input, outputs, hidden states and true values will respectively be indicated as xt, y^t, ht and yt. This avoids underlining every vector, which may make the equations hard to read. To explain how back propagation through time works let’s see a specific example in which the aim of the neural network is to predict a single value one hour apart from the last observation given in the input window. Let’s choose the cost (or loss) function to be:
Which is a common cost function called Mean Squared Error or MSE. In the general formula of the MSE, instead of an unspecified constant c is used. In machine learning this constant is almost always chosen to be because this choice simplifies the computation of the derivatives. For the sake of simplicity it is assumed that g, which is the linear function applied in formula Yt = g(V ht), is the identity function, meaning that g(x) = x and g’ (x) = 1. Since the cost function is a function of Yt, it follows that it is really just a function of the weights that define Yt· This means that the loss function lies in a multidimensional space having dimensionality equal to the number of weights in the model, which is given by the sum of the number of entries in U, V and W. A local minimum in a multivariate function is a point in which the partial derivatives of the function with respect to all the variables that define such function are equal to zero. This translates into saying that minimizing the cost function just means changing the weights that define the function so that the partial derivative with respect to those weights evaluated at their actual value is as close as possible to zero.
The algorithm starts by defining
Every element in the resulting matrix indicates how an infinitely small change in each of the weights collected in the weight matrix V will influence the cost function, assuming all other weights remain constant. As it is not possible for the back-propagation algorithm to do back-propagation on a graph that is not acyclic it is necessary to use a trick and transform the computational graph of a RNN into an acyclic graph. This can be done by unfolding the recurrence relation in time. Meaning that each time step xt with time step index t ranging from 1, to τ is represented as a single node in the graph. A graphical representation of a folded and unfolded RNN can be found in Fig. 10.
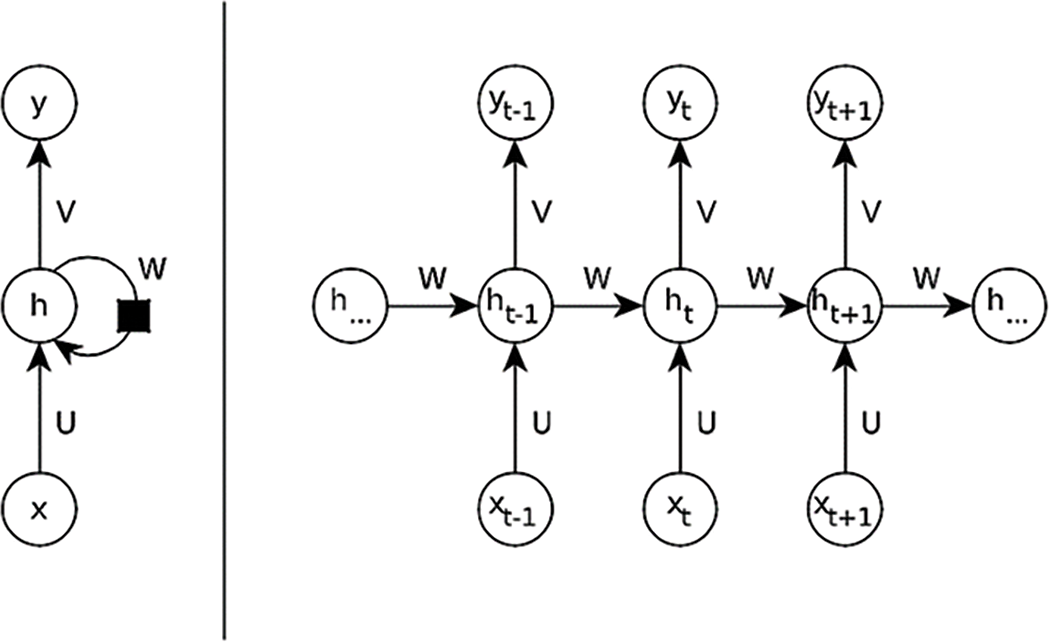
Figure 10: Computational graph of a RNN in folded and unfolded view. (Left) Folded view of an RNN
RNNs can follow one of the following design patterns:
Recurrent networks that produce an output at each time step.
Recurrent networks that read the entire input sequence and produce one output at the last time step.
Recurrent networks that read the entire input sequence and produce a sequence of outputs.
The folded version of RNN is a cyclic-directed graph. At each time step the RNN gets an input vector x that is multiplied with a weight matrix U. The RNN keeps its own hidden state h in a feedback loop and together with the transformed input x this hidden state is multiplied with a weight matrix W that creates the next hidden state. The current hidden state is then multiplied with a weight matrix V creating the output y at each time step. (Right) Unfolded view of the RNN. The graph is now acyclic making it possible to perform backpropagation. It is now possible to see that each of the matrices U, W, V is reused at every time step t. The essential components and operations involved in both the folded and unfolded views of a simple RNN are shown in Table 5.
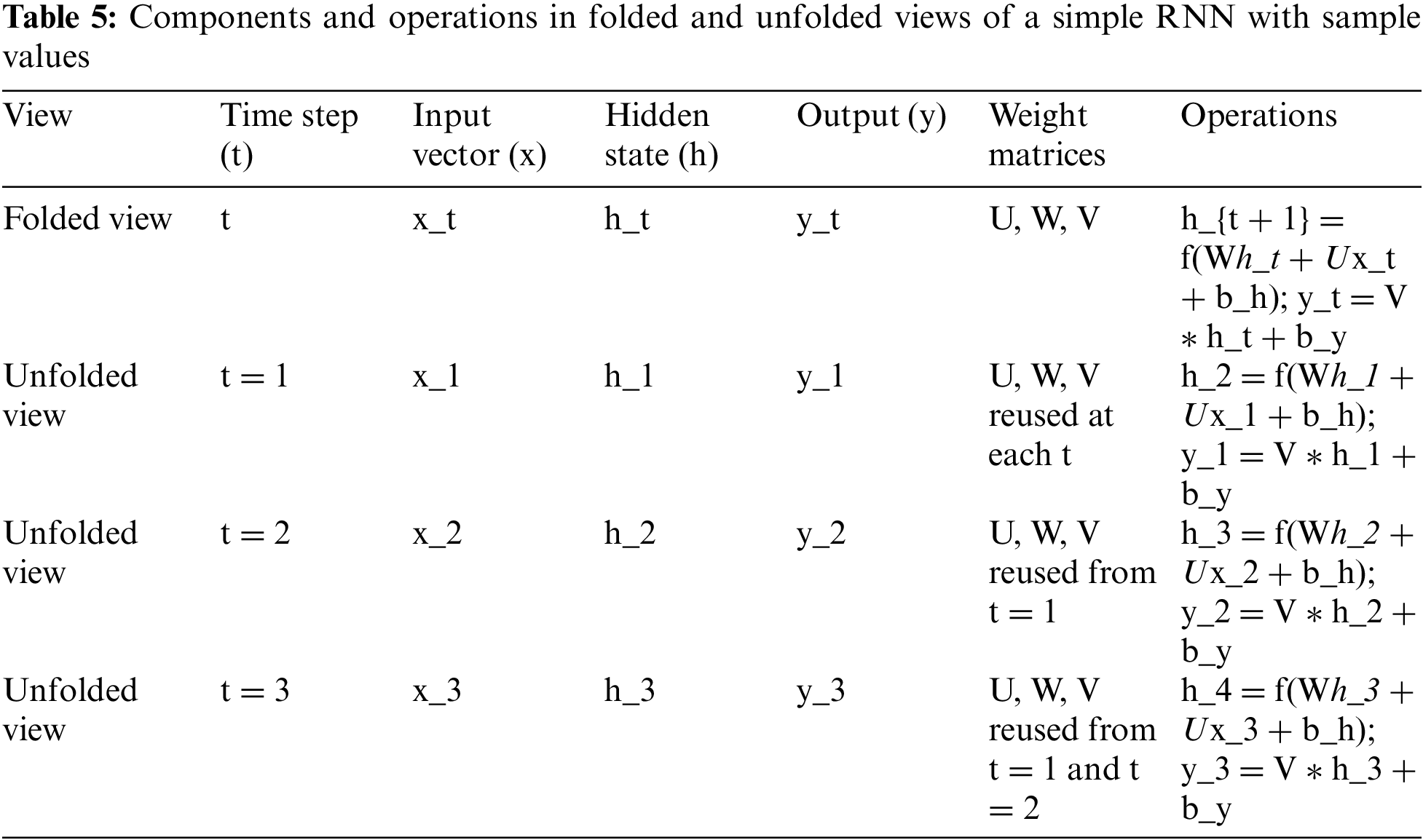
The table represents the operational dynamics of a simple RNN in both its folded and unfolded views. In the folded view, the RNN is depicted as a cyclic directed graph, where at each time step t, the network processes an input vector xt, maintains a hidden state ht, and produces an output yt. The operations involve the input vector xt being multiplied by a weight matrix U, and this transformed input is combined with the previous hidden state ht which itself is multiplied by another weight matrix W. This combination is then passed through a non-linear function f (such as tanh or ReLU), along with a bias term bh, to compute the new hidden state ht+1. Concurrently, the current hidden state ht is also multiplied by a third weight matrix V to produce the output yt, again with the addition of a bias term by.
The unfolded view breaks down this cyclical process into distinct steps, laid out sequentially. Here, the network’s operations are made explicit for each individual time step, demonstrating how the RNN reuses the weight matrices U, W, and V at every time step. For instance, at time step t = 1, the input 1x1 and the initial hidden state ℎ1h1 undergo transformations similar to the folded view to produce h2 and 1y1. This process is repeated for subsequent time steps t = 2 and t = 3, where each time step takes the input xt and the previous hidden state ht to compute the next hidden state ht+1 and the output yt. The table illustrates the reuse of the weight matrices across time steps, which is a key feature of RNNs allowing them to maintain temporal dependencies across the data.
The output ht at time step t computed by a RNN directly depends on the last hidden state computed by the recurrent neurons ht−1, the current input xt and the parameters θ of the network. The new hidden state is computed as shown in the Equation below.
If the recurrence relation gets unfolded in time it is possible to see that RNNs share their parameters across different invocations as the same function f is used again and again. The equation below shows a sample unfolding operation for a RNN computing three time steps.
Reusing of the same function f also implies that the number of time steps can be arbitrary long because the network defines the transition from one state at time step t to the next state at time step t + 1, rather than a variable-length history of several states. This characteristic of RNNs leads to a low number of parameters that need to be optimized during learning. Fig. 11 shows the flowchart represents the flow of the study.
Figure 11: Flowchart represents the flow of the study
For the application of scene text recognition using RNNs we will mainly focus on RNNs that follow the first design pattern. The following functions describe the forward pass of a RNN for each time step from t = 1 to t = τ. The linear function which is the non activated output of the hidden neurons is computed like that:
The hidden state is then computed by applying a non-linear function on top of that result. In most RNNs the hyperbolic tangent function is used:
The output of the RNN at each time step involves a second bias parameter c which is summed with the result of the matrix multiplication of the hidden state and the weight matrix for the hidden-to-output connections:
The last step may be to apply the softmax function on the output of the network in order to get a probability distribution over all outputs at each time step
For this study, we have taken the dataset and during preprocessing, specific columns were removed from the user part of the dataset because they indicated whether the user had Parkinson’s disease. The UPDRS is a scale that is used to assess the severity of Parkinson’s disease. The impact of Parkinson’s disease on the patient’s life is defined. Levodopa and Maob indicate whether the user is taking the medications to treat Parkinson’s disease. The user data was combined with the Tappy Data, and the UserKey, Timestamp, and Gender fields were removed from the dataset. To generate the user’s current age, feature engineering was used. The final dataset had seventeen features which were oversampled using the following methods to handle class imbalance.
Synthetic Minority Oversampling Technique, or SMOTE, is an oversampling method that balances imbalanced data sets. SMOTE creates new minority-class examples from scratch rather than duplicating existing ones. First, a random point from the minority class is selected, and the point’s k-nearest neighbors are determined. A new synthetic point is synthesized between two examples from the nearest neighbors.
BorderLine SMOTE, also known as SVM SMOTE, was used as the data preprocessing technique for another experiment. The aim was to oversample the minority class of participants not having Parkinson’s. BorderLine SMOTE also generates synthetic data similar to SMOTE; however, data is generated only along the borderline area. The borderline area is identified using SVM algorithms trained on the data. Thus, compared to SMOTE, BorderLine SMOTE generates the data without crowding in the training set.
Feature Ranking using Recurrent Neural Network (RNN), was used to identify seven features that have the most negligible impact on model accuracy, which are discarded. The remaining topmost features are used as input to be oversampling techniques like SMOTE. Thus, the feature set was reduced from 17 features to 10 features after applying Feature Ranking. By prioritizing the most critical features and discarding the least important ones, Random Forest can effectively help in dimensionality reduction without causing information loss. After feature selection, SMOTE is applied to the selected features, which can address the issue of imbalanced datasets by generating synthetic samples of the minority class. Fig. 7 describes the Feature Ranking algorithm of RNN used to identify the top 10 features. Initially, the Feature Set consisted of all 17 features. Accuracy A is obtained by training a Random Forest model on all features in the feature set. A is compared to the accuracy Af obtained by dropping a feature f from the feature set. The feature with the lowest impact is determined by evaluating the change in Af with respect to A and is dropped from the feature set. This process is continued until the top 10 features are obtained.
SimpleRNN—The SimpleRNN model has six layers: four RNNs and two dense layers. Each RNN and dense layer (except the final layer) has a Relu activation, while the final dense layer has a Sigmoid activation [21]. The model’s loss function is binary cross-entropy, and Adam is its optimizer. The flowchart of simple RNN is shown below in Fig. 9. Fig. 12 illustrates an unfolded example of the network depicted in Fig. 6.
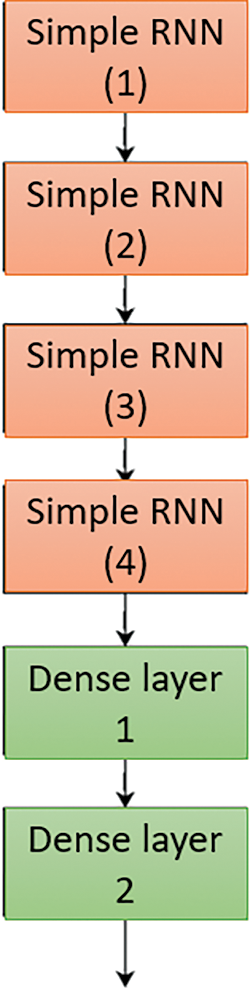
Figure 12: Simple RNN model for processing the clinical support
Modified SimpleRNN—The modified SimpleRNN is similar to SimpleRNN but has two additional LSTM layers and one echo state cell. All layers except the final dense layer have a Relu activation, while the final dense layer has a Sigmoid activation. The model’s loss function is binary cross-entropy, and Adam is its optimizer. Fig. 10 describes the architecture of Modified SimpleRNN which consist of SimpleRNN, EchoState Cell and LSTM layers.
In Fig. 13, the purpose of the LSTM model is to enhance the processing and management of sequential data, specifically for handling queries more efficiently within a clinical decision support system (CDSS). The LSTM model is incorporated to address the limitations of traditional RNNs, particularly their difficulty in capturing long-term dependencies due to issues like the vanishing gradient problem. LSTMs are designed with specialized units called memory cells that can maintain information over long periods, making them well-suited for tasks that require understanding and retaining context over multiple time steps. The use of LSTM in the model allows it to effectively manage the complexities of sequential data in medical queries, where the context and sequence of inputs are crucial for accurate decision-making. By integrating LSTM layers, the model gains the ability to remember important information across longer sequences, thus improving its performance in tasks that involve temporal dependencies. This capability is particularly valuable in clinical settings, where patient data over time needs to be analyzed to make informed decisions about diagnosis and treatment.
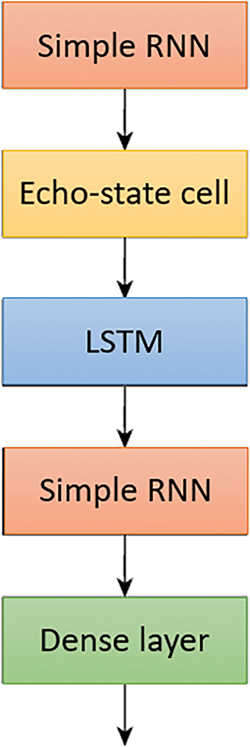
Figure 13: Modified RNN with Echo state and LSTM for processing the queries more efficiently
Transformer—When working with time series data, capturing long-term dependencies between distant time steps is critical. Unlike standard machine learning models, encoder blocks in transformers use attention mechanisms to construct contextual relationships between time steps. This allows the transformer to capture such long-term dependencies within the data. Transformers can weigh and combine information from numerous time steps by using attention mechanisms, giving more weight to those most relevant to producing correct predictions. Following the encoding process, a Global Average Pooling layer is used to reduce tensor dimensionality, which aids in the avoidance of overfitting and improves computational efficiency. Finally, Dense layers with mlp units are used to learn complicated patterns in time series data, allowing the model to generate accurate predictions based on the encoded data. Fig. 14 shows the framework of the auto-encoder and the structure of the Transformer model, comprising a Multi-Headed Attention Layer, Global Average Pooling layer, and dense layers.
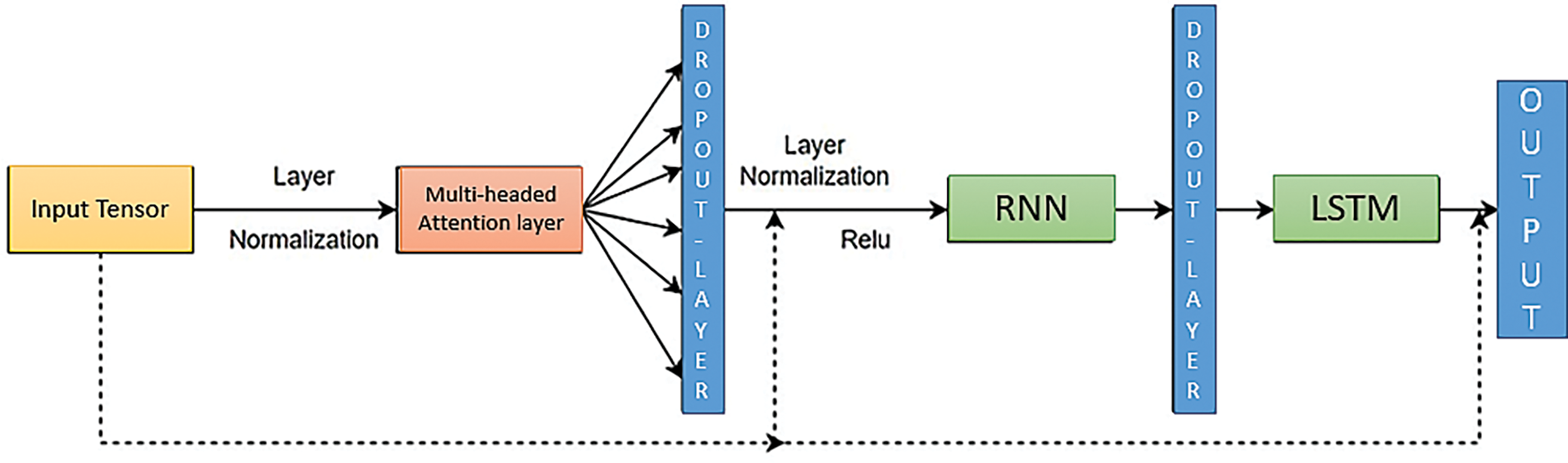
Figure 14: Framework of auto-encoder and structure of the Transformer model consisting of a multi-headed attention layer, global average pooling layer, and dense layers
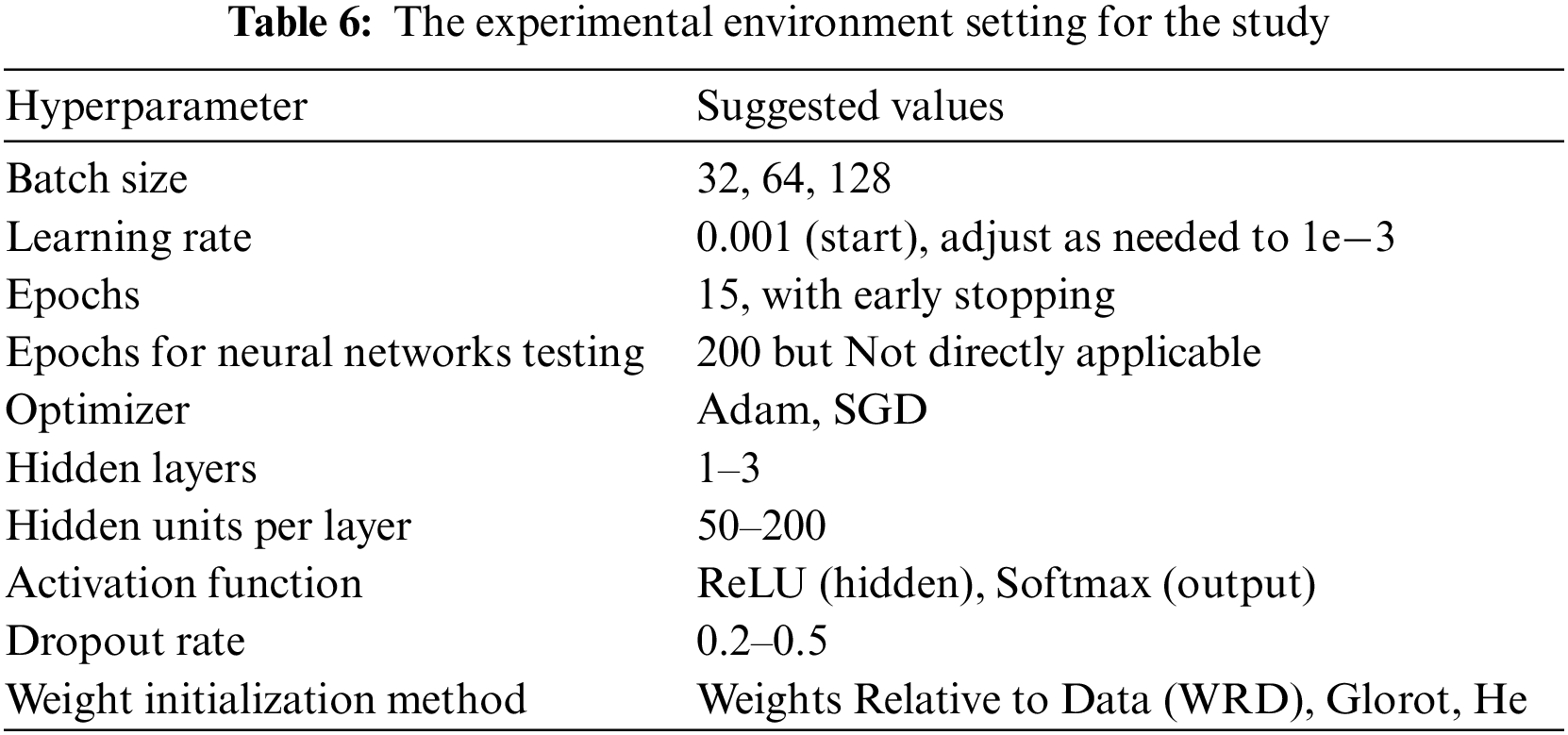
For the midterm study, we have used two models to develop a baseline for future experiments: the basic RNN and state of the art cells like Echo state and LSTM with simple RNN. Table 3 lists the Hyper-parameters used to train the Simple RNN, Modified RNN and Transformer. Table 6 shows setup of the experimental environment for the study.
The RNN framework is a very powerful model, but it has its limitations. One being that it is a memoryless model, i.e., for each input the network will have no information about previous inputs. This property however can be obtained using a “Recurrent Neural Network” (RNN). RNN’s are basically identical to FFNN’s with the minor adjustment, that each layer l will not only pass its output to the next layer l + 1, but will also pass it to itself, hence a “recurrent connection”.
A popular way of visualizing RNN’s is by so-called “unfolding” the network and thereby explicitly show the recurrent connections between each time-step t. An example of the network shown in Fig. 6 unfolded can be seen in Fig. 12. Fig. 15 illustration of an unfolded 3-layer recurrent neural network (2 recurrent layers and 1 output layer) with input sequences of length T. x(1) represents the initial input at time-step t = 1 for a given observation x, while z(2, T) indicates the final output at time-step t = T.
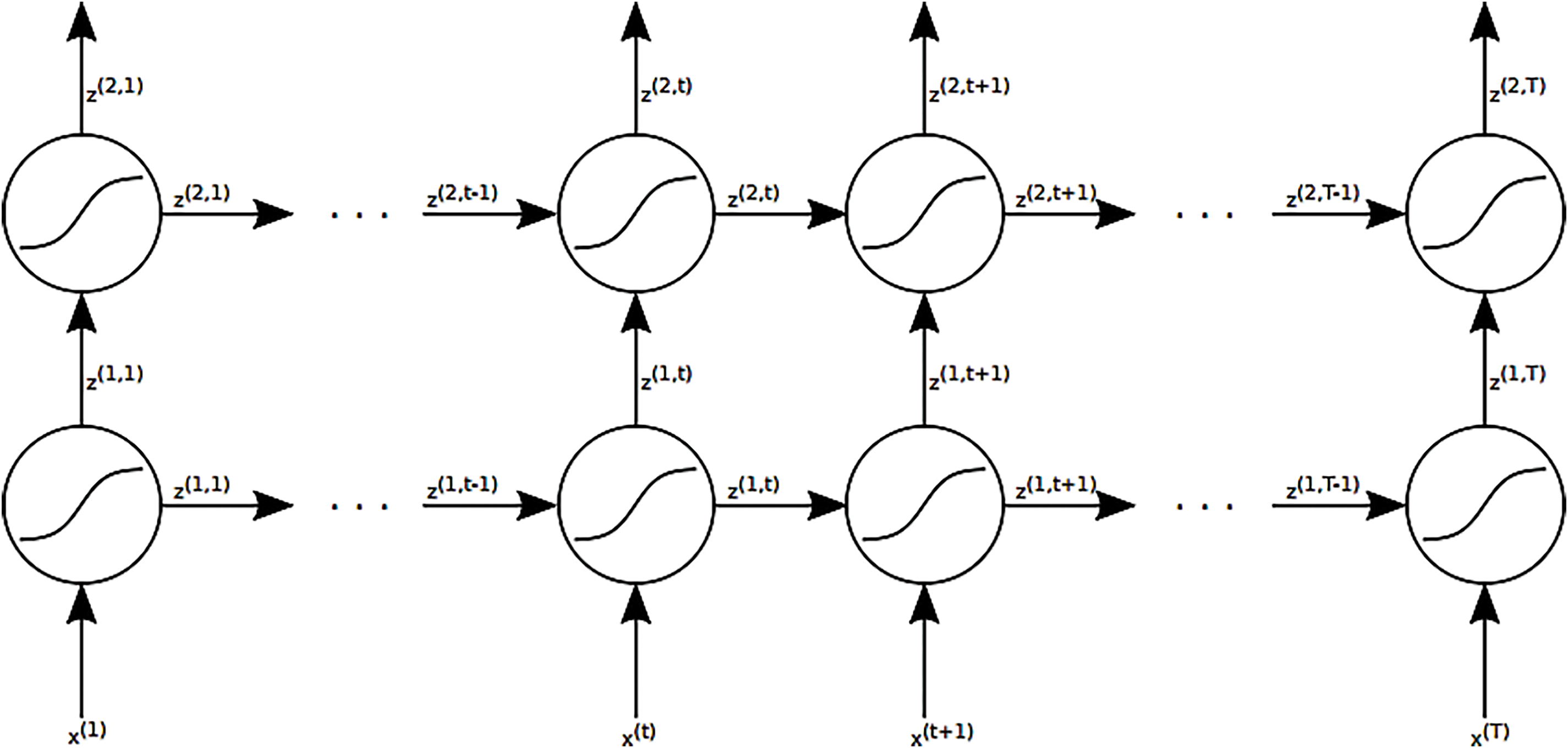
Figure 15: Visualization of an unfurled 3-layer repetitive neural arrange (2 repetitive layers and 1 yield layer) with input groupings of length T. x(1) speaks to the starting input at time-step t = 1 of a given perception x and z(2, T) the ultimate yield at time-step t = T
RNNs are neural networks that do not form an acyclic directional graph but rather a cyclic directional graph, meaning that information that has been seen before can be reused in a later step performed with other data. This makes RNNs very well suited for processing sequential data of arbitrary length. RNNs consist of recurrent layers that use parameter sharing across all their parts. This enables them to process sequences of arbitrary length. Each of these recurrent layers has connections to the input and output layer and a connection from its own computed output (the hidden state) to itself.
As part of an empirical study for Clinical Decision Support System (CDSS), various experiments were conducted by combining universally accepted solutions. These experiments involved testing different combinations of solutions to determine their effectiveness in optimizing the clinical decision support and medical query processing using simple RNN and Modified RNN techniques. The results of each of the below mention experiments are detailed.
Simple RNN for CDSS
Simple RNN with SMOTE for CDSS
Simple RNN with BorderLine SMOTE for CDSS
Modified RNN for CDSS
Modified RNN with SMOTE for CDSS
Modified RNN with BorderLine SMOTE for CDSS
Random Forest with Simple RNN for CDSS
Random Forest with Simple RNN and SMOTE for CDSS
Fig. 16 presents a comparison of different RNN methods, highlighting both the training loss and accuracy. In Fig. 16a, the training loss for various approaches is depicted, while Fig. 16b illustrates the training accuracy achieved by these different RNN techniques.
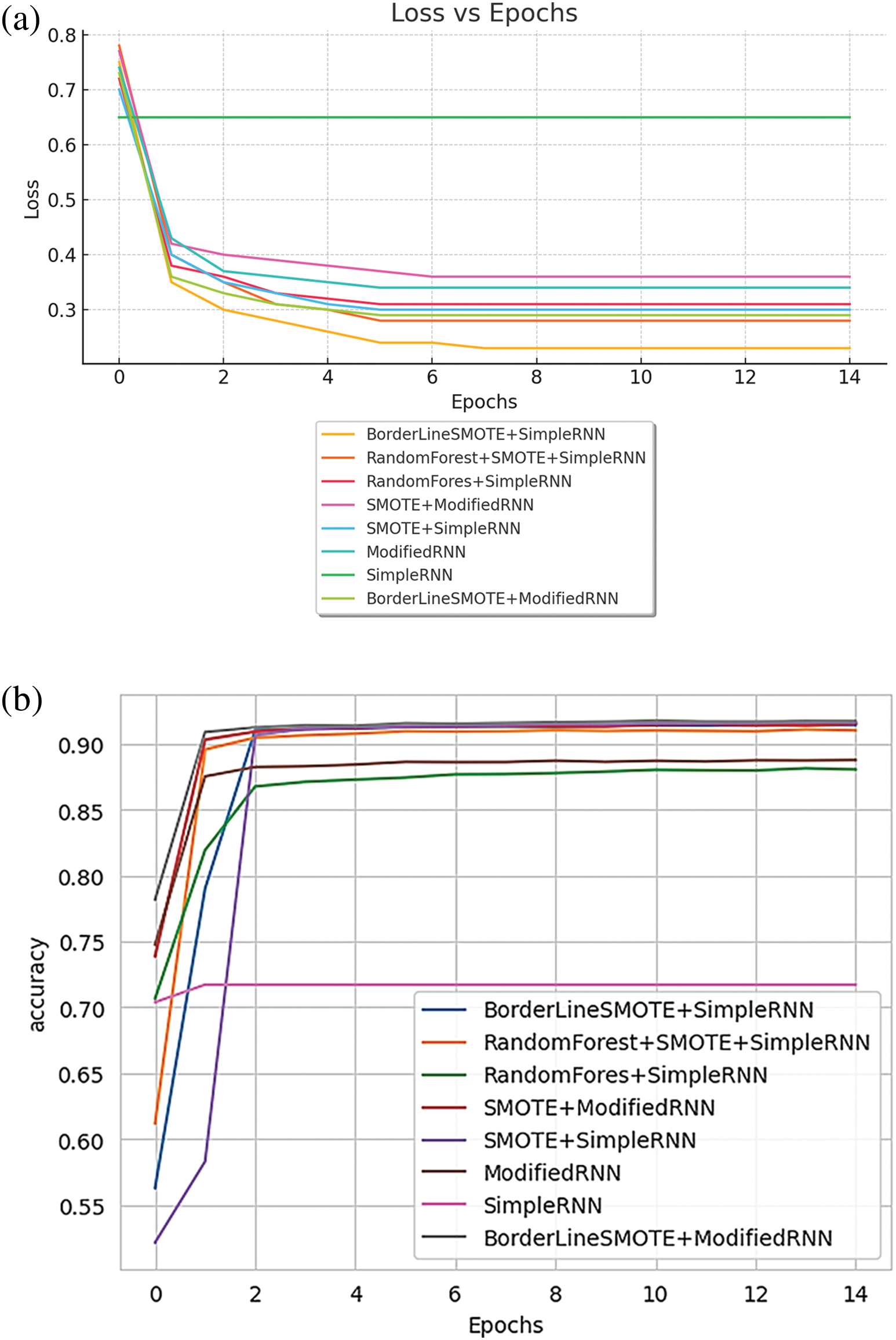
Figure 16: (a) Training loss of various methods of RNN; (b) training accuracy of various methods of RNN
Fig. 17 illustrates the performance metrics for various RNN methods. Fig. 17a displays the training precision achieved by different approaches, while Fig. 17b shows the training recall for these methods.
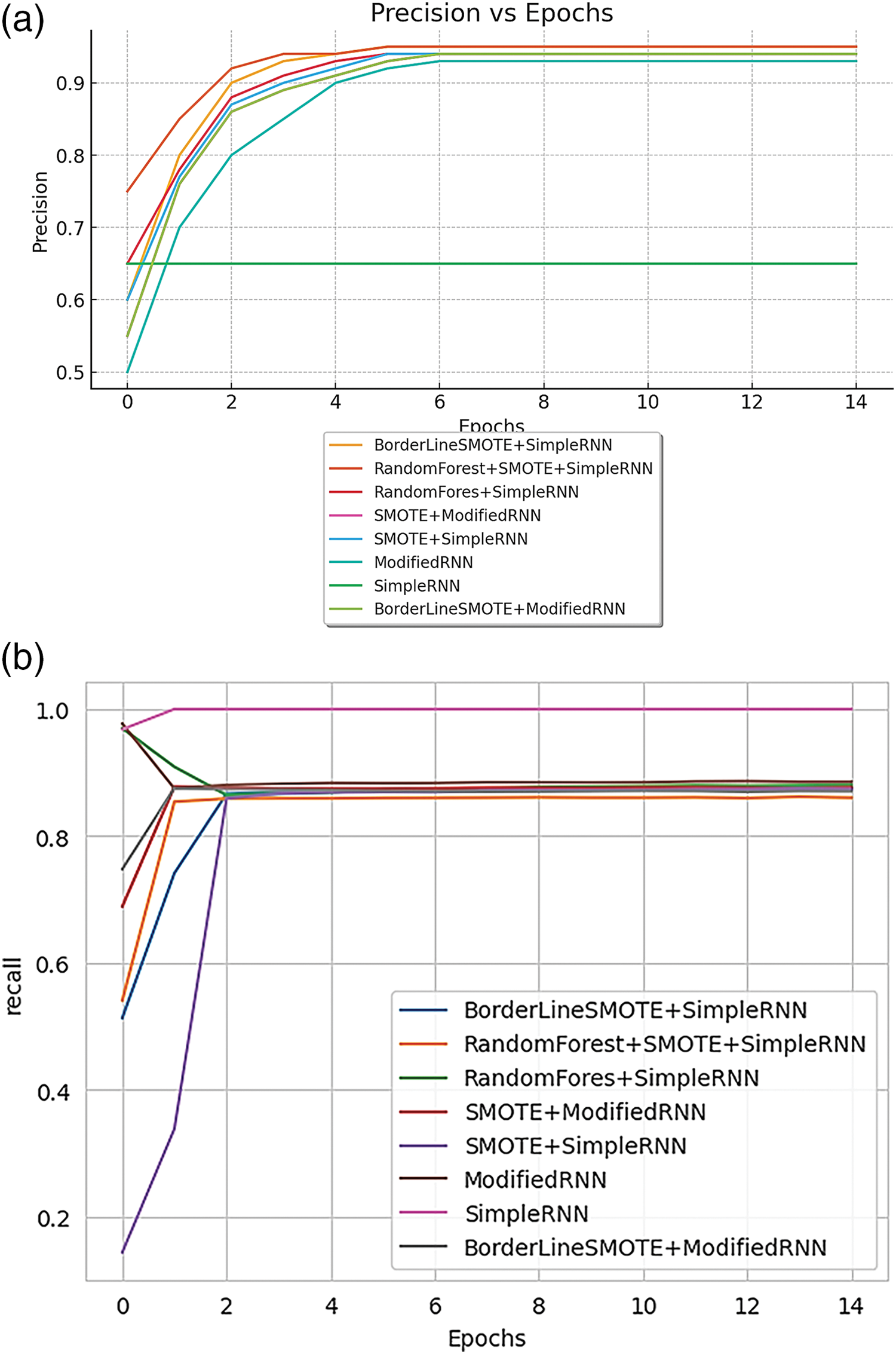
Figure 17: (a) Training precision of various methods of RNN; (b): training recall of various methods of RNN
Fig. 18 shows the training AUC (Area Under the Curve) for different RNN methods, providing a comparison of their performance.
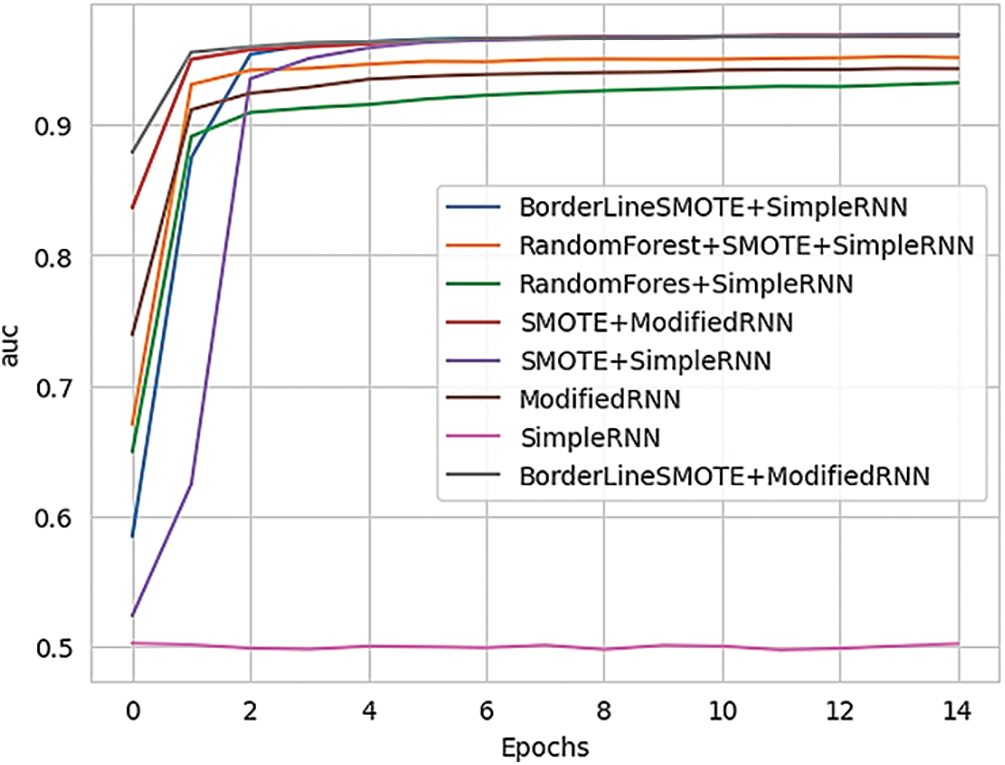
Figure 18: Training AUC of various methods of RNN
Since the project handles clinical datasets, it is crucial to identify patients with Parkinson’s accurately compared to misclassifying a few samples not having Parkinson’s. Hence, Recall and AUC are considered important metrics for model evaluation. Recall allows for us to observe the total amount of correctly classified patients with Parkinson’s according to their data, while AUC score measures the probability of classes being classified amongst one another for CDSS. In addition, the usual metrics of models that include Accuracy, Precision, and Loss are just as important to us as Recall and AUC. We wish to compare each of our implementations across every viable metric to get a concrete showcase of how to respond to medical query and how RNN provide solutions can possibly improve classification for CDSS.
Now the validation of the RNN model for handling and optimizing the CDSS for medical queries can be seen in the Figures below. Fig. 19 presents a comparison of validation metrics for different RNN methods. Fig. 19a illustrates the validation loss across various models, while Fig. 19b highlights the validation accuracy for these RNN approaches.
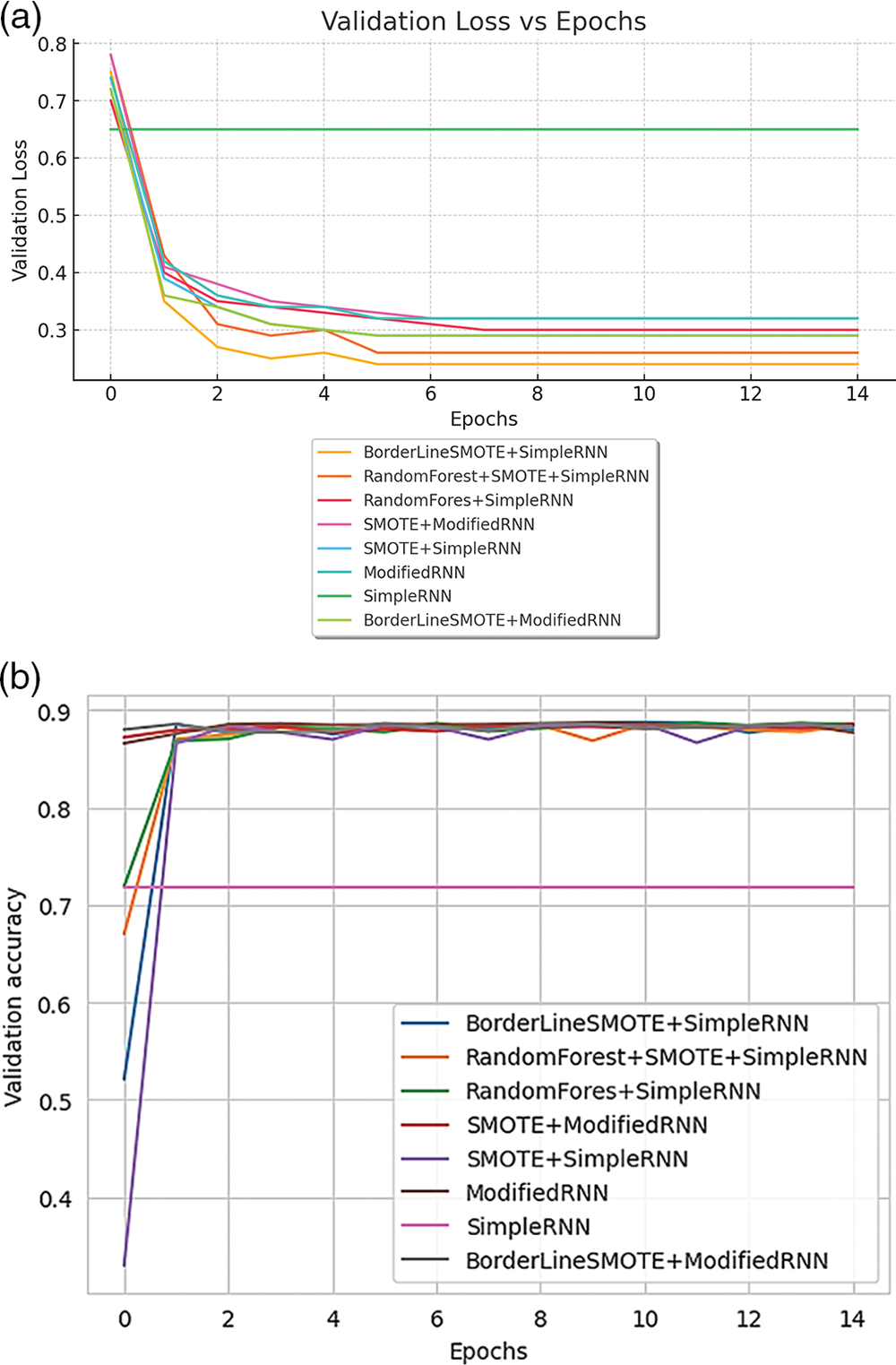
Figure 19: (a) Validation loss of model of various methods of RNN; (b) validation accuracy of various methods of RNN
Fig. 20 provides a comparison of validation performance across different RNN methods. Fig. 20a displays the validation loss for each approach, while Fig. 20b presents the validation accuracy achieved by the various methods.
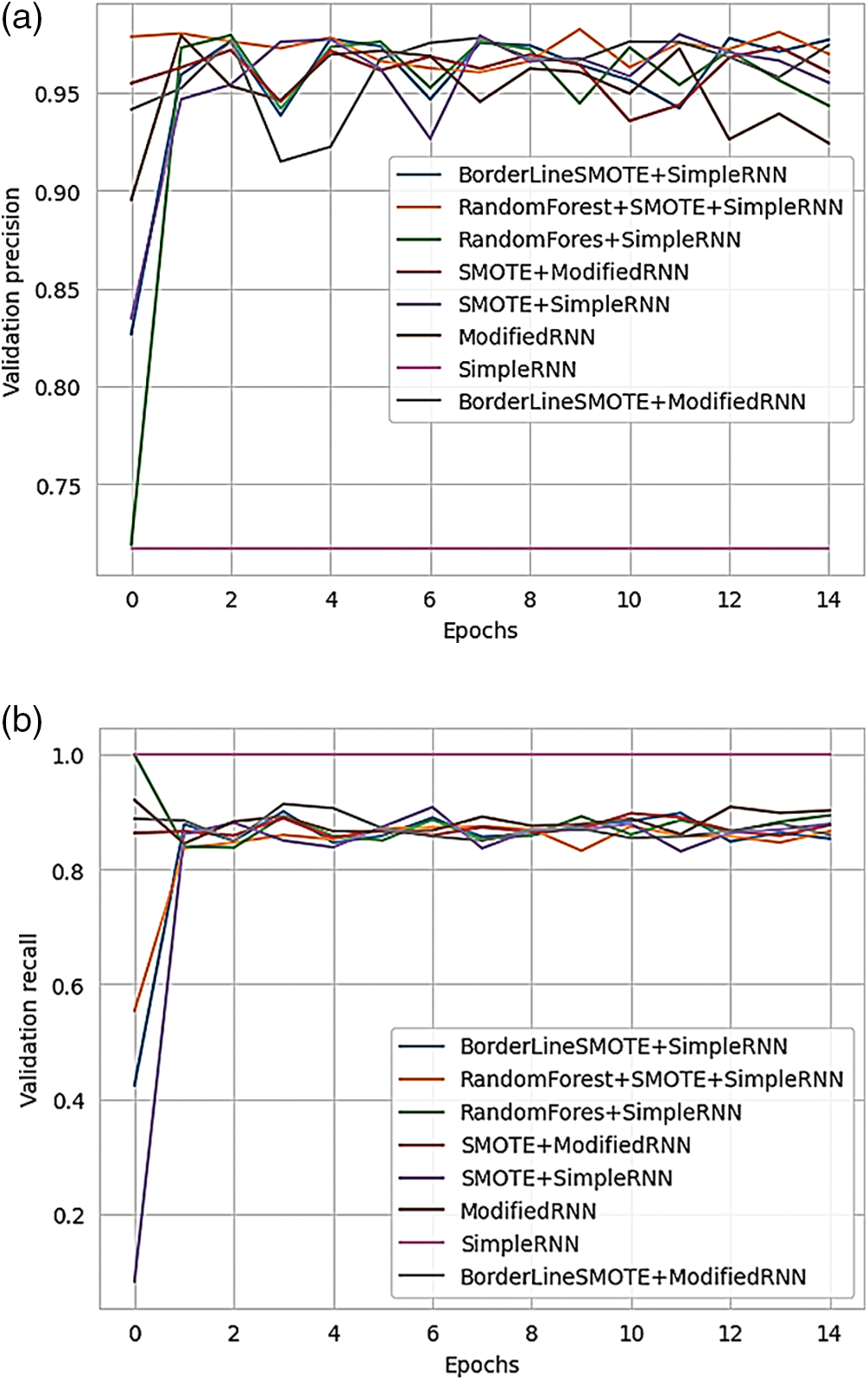
Figure 20: (a) Validation loss of various methods of RNN; (b) validation accuracy of various methods of RNN
Fig. 21 presents the validation AUC (Area Under the Curve) for different RNN methods, offering a comparison of their performance.
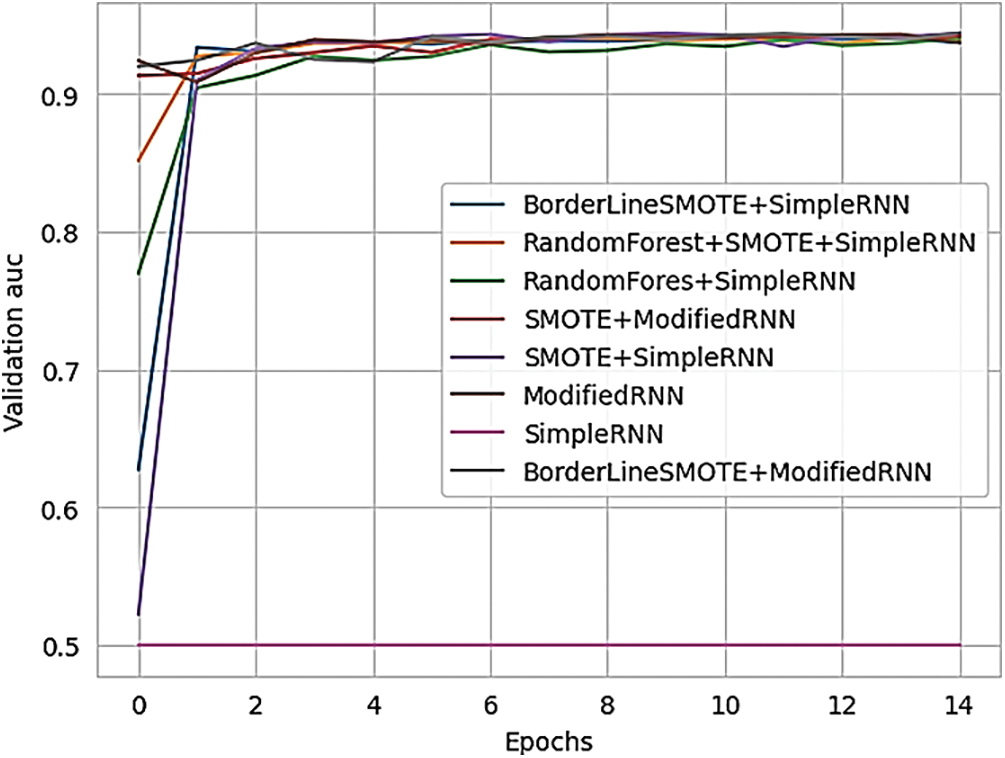
Figure 21: Validation AUC of various methods of RNN
Table 7 below differentiate the various methods of simple RNN and modified RNN in terms of loss, accuracy, precision, recall and AUC.
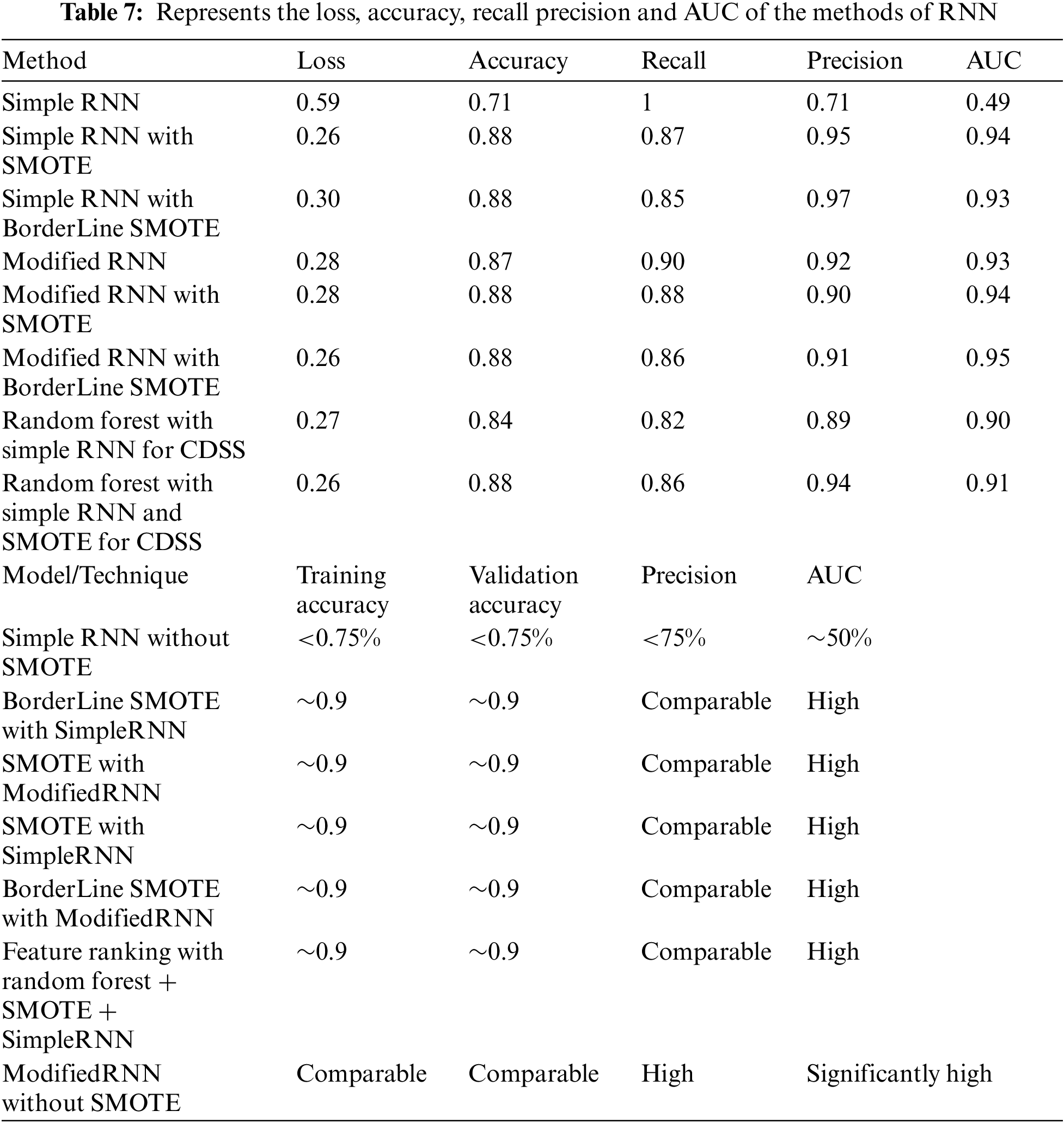
In contrast, with Borderline SMOTE, the model reaches an improved accuracy of around 91% and handles the imbalance by achieving an improved AUC of 95%–97%. However, the model achieves a recall of around 87%, comparable to other simple models. From Table 7 we can make the following conclusions. Combining different preprocessing techniques and models leads to better performance than baseline models. For example, SimpleRNN and modified RNN without SMOTE and Borderline SMOTE perform worse compared to the addition of oversampling techniques, predicting only the majority class.
The study aims to provide insights into the most effective models and solutions for addressing the research problem by presenting these results. Now the study will assume other neural networks against out model for the testing of models. Fig. 22 illustrates the loss of various networks over the course of 200 testing epochs, highlighting their performance trends during the evaluation phase.
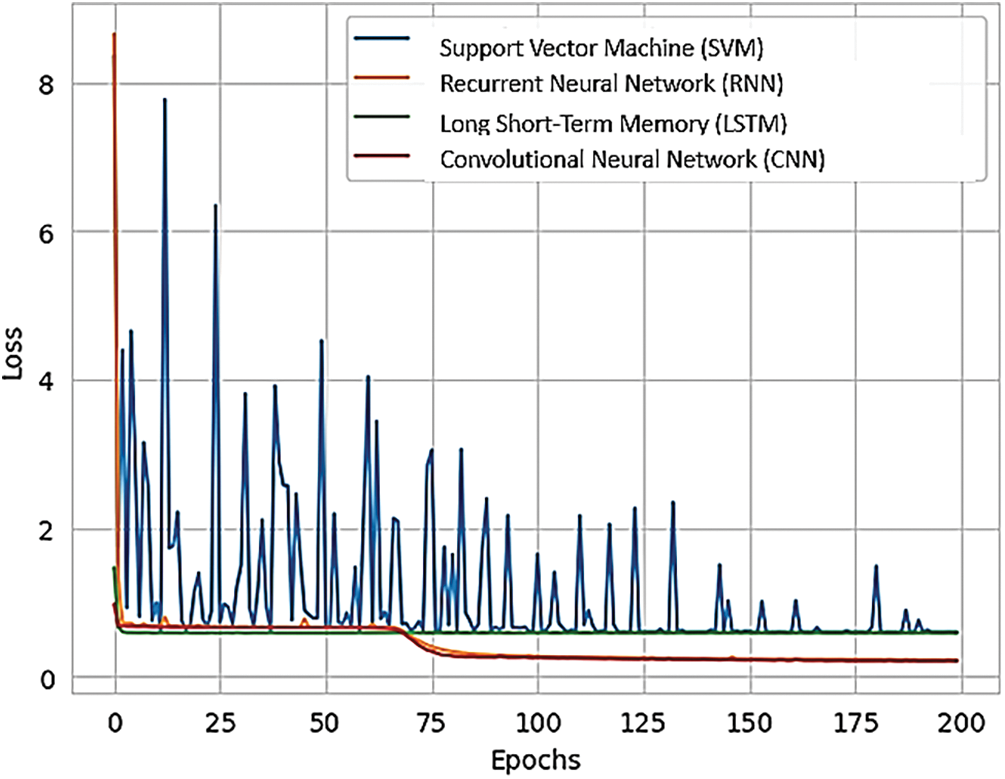
Figure 22: Various networks loss over testing the 200 epochs
Fig. 23 shows the accuracy of various neural networks, providing a comparative overview of their performance.
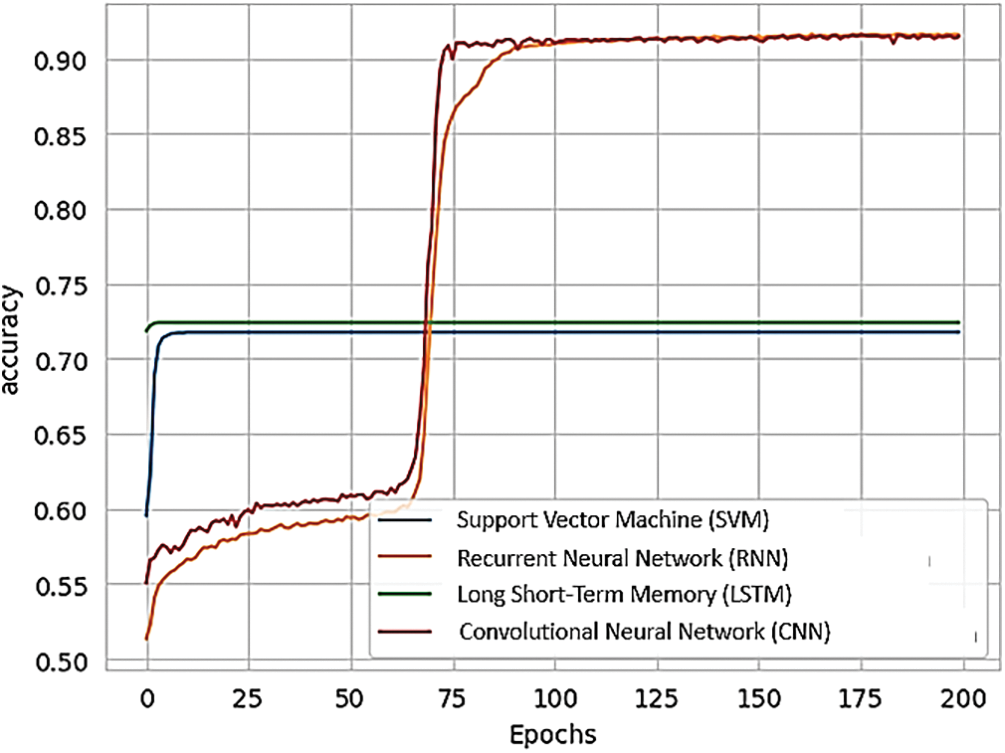
Figure 23: Accuracy of various NN
Fig. 24 compares the AUC (Area Under the Curve) of various networks to that of RNNs, highlighting differences in their performance metrics.
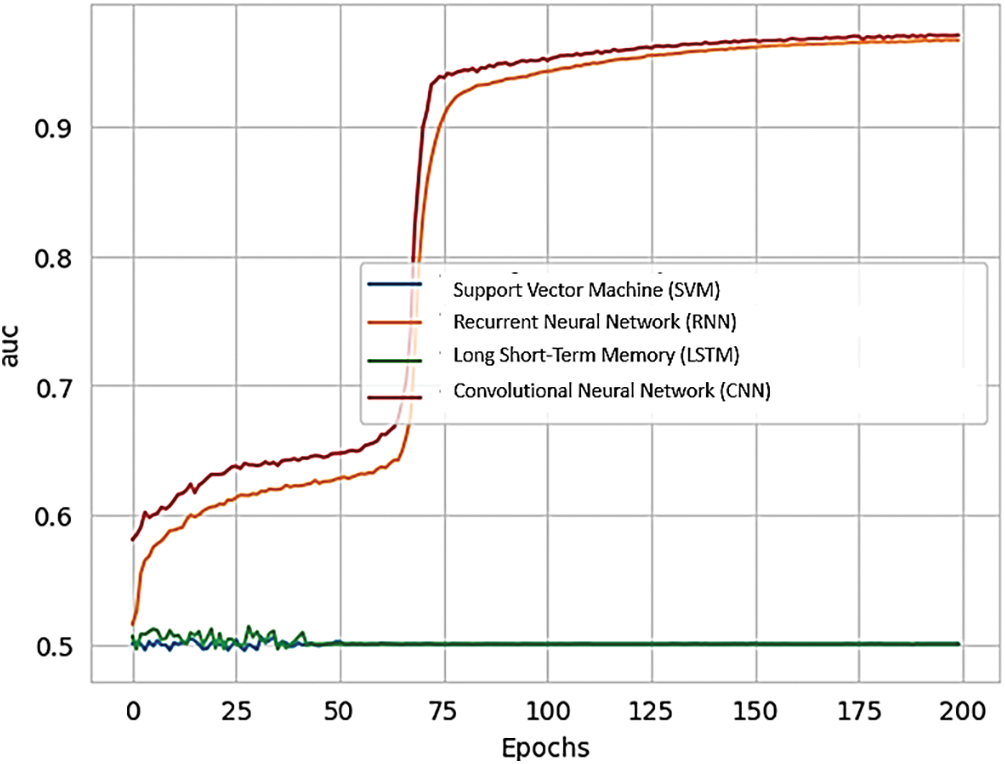
Figure 24: AUC of various networks as compared to RNN
Table 8 mentioned results of various NN models from the experimentation. Table 8 indicates the result of different models as compared to our proposed model which is recurrent neural network with the other deep learning methods such as CNN, SVM and LSTM.
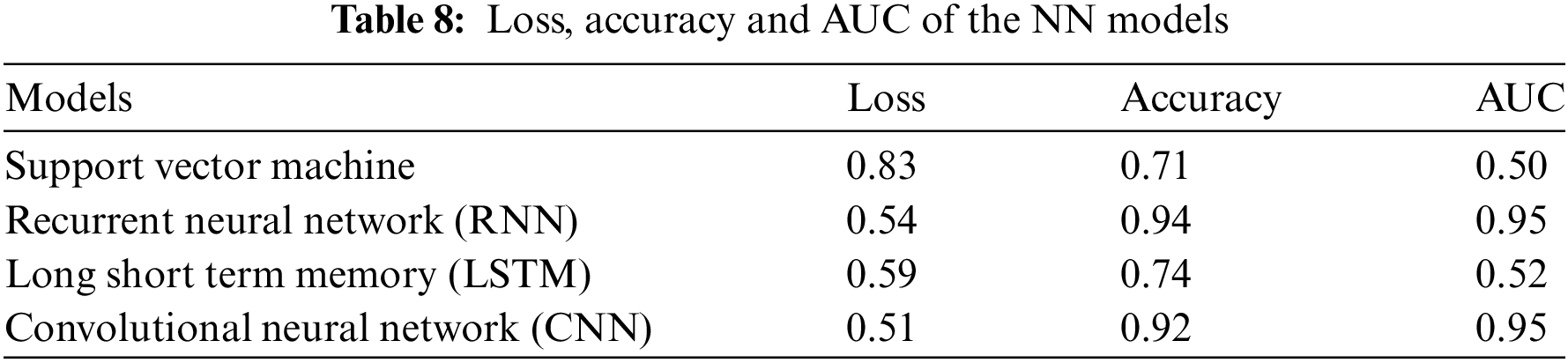
Figs. 16 and 18 show the plot Training and Validation Loss of 5 experiments, respectively. It can be observed that Simple RNN obtained the worst loss without SMOTE for both training and validation sets. On the other hand, BorderLine SMOTE with SimpleRNN, SMOTE with ModifiedRNN, SMOTE with SimpleRNN, and BorderLine SMOTE with ModifiedRNN give comparable results for Loss for the training set. All seven models, apart from Simple RNN without SMOTE, show comparable results for Loss for the validation set. Figs. 16 and 18 show the plot for Training and Validation Accuracy for five experiments, respectively. It can be observed that Simple RNN without SMOTE shows the worst performance with an accuracy of less than 0.75% for both training and validation sets. Feature Ranking with Random forest + SMOTE + SimpleRNN, SMOTE with ModifiedRNN, and BorderLine SMOTE with Modified RNN achieve comparable accuracies of around 0.9 for both training and validation sets.
Figs. 16. and 21 show the plot for Training and Validation Precision for the experiments, respectively. It can be observed that Simple RNN without SMOTE shows the worst performance with less than 75% precision for both training and validation sets. All other models show comparable performance with respect to precision for both training and validation sets. Figs. 18 and 23 highlight the AUC of the combinations, which shows the entire picture of false positive and true positive rates. SimpleRNN, the worst-performing model, achieves a maximum AUC of around 50% in training and validation. In contrast, combinations like SMOTE/BorderLine SMOTE with SimpleRNN and Modified RNN achieve the highest recall in both training and validation.
This also indicates that using oversampling techniques, such as SMOTE and its variants, can tackle the CDSS issue, ensuring equal weightage to each class. Even though reducing less important features simplifies the model, leading to better performance, in this case, random forest feature and recurrent neural network (RNN) selection contributed little towards improvement. However, it achieved comparable results with lower dimensions of input data. If we compare the results of SimpleRNN and ModifiedRNN without SMOTE, the modified outperforms SimpleRNN, achieving almost comparable to the other experiments. This shows that the EchoState cell efficiently handles data imbalance without oversampling techniques. The performance of RNN with BorderLine SMOTE on the test data achieves the highest AUC and high values for Precision, Recall, and Accuracy, showing that RNN can capture long-term dependencies in time series data efficiently for the clinical decision support system (CDSS) and medical queries.
At the end validity and reliability of the methods and findings in this research are ensured through a comprehensive approach involving multiple validation and reliability measures as show in Table 8 below:
Data Sources: The research utilizes diverse and reputable data sources, including medical imaging data and clinical data from Electronic Health Records (EHRs) and wearable sensors. The medical imaging data comprises anteroposterior X-rays for diagnosing rotator cuff tears and annotated gastrointestinal tract images for disease classification. These sources ensure that the data is comprehensive and of high quality, which is crucial for training accurate and reliable AI models.
Preprocessing: Standard preprocessing techniques are employed to enhance the integrity of the data. Image preprocessing involves normalization, resizing, and augmentation methods such as rotation, flipping, and contrast adjustment. Text data from clinical notes and EHRs are processed using natural language processing (NLP) techniques, including tokenization, stemming, and stop word removal. These preprocessing steps ensure that the data fed into the models is clean, consistent, and suitable for training.
Model Development: The development of advanced AI models, including RNNs with LSTM layers and the CG-Net framework, is a critical component of this research. These models are validated through extensive training on large datasets and rigorous testing. The use of LSTM layers addresses the limitations of traditional RNNs by enhancing the models’ ability to capture long-term dependencies, ensuring high reliability and accuracy in predictions.
Data Augmentation: To address data imbalances and enhance model generalization, data augmentation techniques such as SMOTE and BorderLine SMOTE are employed. These techniques generate synthetic samples to balance the datasets, preventing the models from being biased towards the majority class. This step is crucial for ensuring the reliability of the models across different datasets and conditions.
Validation Procedures: The research employs robust validation procedures, including a training/testing split and 5-fold cross-validation. The datasets are split into training, validation, and testing sets with typical ratios of 70%, 15%, and 15%, respectively. The 5-fold cross-validation technique further validates the models by dividing the data into five subsets and training and validating the models five times. Performance metrics such as accuracy, precision, recall, F1-score, and AUC are used to evaluate the models, ensuring their robustness and generalizability.
Hyperparameter Tuning: Systematic hyperparameter tuning is conducted using grid search and random search techniques. This step involves optimizing parameters such as learning rate, batch size, number of hidden layers, and number of neurons per layer. Early stopping is applied to prevent overfitting by halting training if the model’s performance on the validation set does not improve for a specified number of epochs. These measures ensure that the models perform optimally.
Interpretability Tools: Model interpretability and explainability are addressed using SHAP (SHapley Additive exPlanations) and LIME (Local Interpretable Model-agnostic Explanations). These tools provide insights into the models’ predictions and the contribution of each feature, enhancing the transparency and trustworthiness of the models. This step is crucial for ensuring that the models’ predictions can be understood and trusted by clinicians.
Implementation: The implementation of the models on cloud platforms like Amazon Web Services (AWS) and Google Cloud Platform (GCP) ensures scalability and practical applicability. Integrating the developed models into existing CDSS frameworks enhances their diagnostic capabilities and provides real-time support to clinicians. This practical implementation validates the models’ applicability in real-world healthcare settings.
Table 9 provides the measures of validity and reliability.
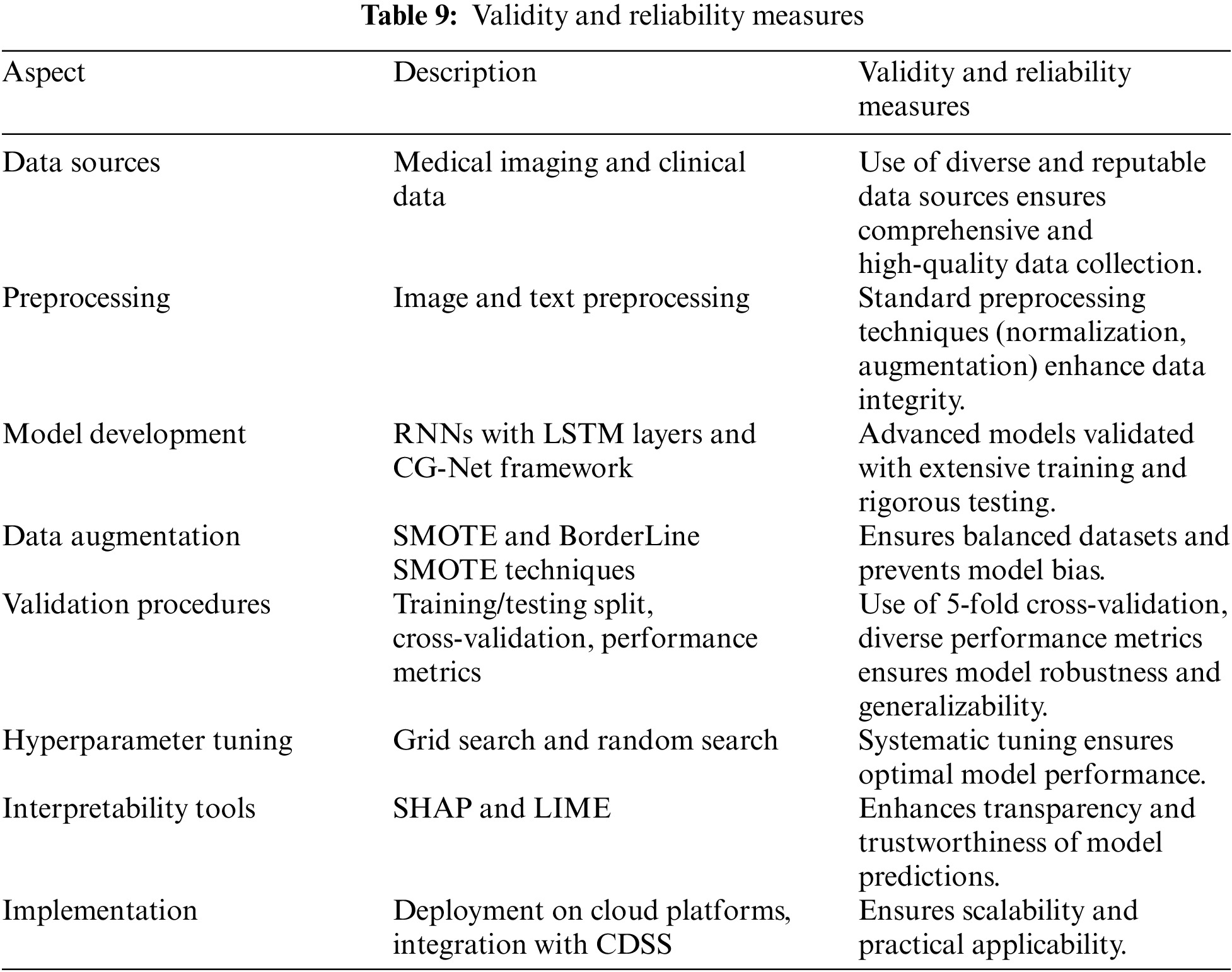
The research demonstrates the application of Recurrent Neural Networks (RNNs) with Long Short-Term Memory (LSTM) layers and Convolutional Neural Networks (CG-Net) in medical imaging for diagnosing conditions such as rotator cuff tears and gastrointestinal diseases. By leveraging these advanced AI models, healthcare professionals can achieve higher diagnostic accuracy and consistency. For example, radiologists can utilize AI to automatically analyze X-rays and detect rotator cuff tears in osteoporosis patients, reducing the time required for diagnosis and increasing the precision of the findings. This leads to more accurate and timely diagnoses, which is critical for early intervention and effective treatment planning. In the context of Wireless Body Area Networks (WBANs), the integration of AI models allows for the continuous monitoring and analysis of data from wearable sensors. These sensors can track vital signs, movement patterns, and other physiological parameters. AI models can process this data in real-time, identifying abnormal patterns and providing early warnings for conditions such as arrhythmias, sleep apnea, or sudden drops in glucose levels. This real-time analysis enhances the ability of healthcare providers to monitor patients remotely and intervene promptly when necessary.
The research integrates AI frameworks like CG-Net into CDSS, providing real-time diagnostic support to clinicians. This integration enables CDSS to offer evidence-based recommendations and alerts, assisting healthcare professionals in making informed decisions quickly. For instance, during a clinical consultation, the CDSS can analyze patient data, compare it with historical data, and suggest potential diagnoses or treatment options. This support reduces the cognitive load on clinicians, allowing them to focus on patient care while ensuring that their decisions are backed by the latest medical knowledge and data. AI-enhanced CDSS can be particularly beneficial in remote and telemedicine settings, where access to specialist care might be limited. By providing real-time diagnostic support and analysis, these systems can empower general practitioners and remote healthcare workers to make better-informed decisions. This is crucial in underserved or rural areas where specialist consultations are not readily available. The ability to conduct remote diagnostics and provide quality care can significantly improve healthcare access and outcomes in these regions. In WBANs, wearable sensors continuously collect data on various health parameters. AI models can analyze this data to detect early signs of health issues before they become critical. For example, continuous monitoring of heart rate, blood pressure, and activity levels can help detect early signs of cardiovascular diseases. Early detection allows for timely interventions, potentially preventing severe health events and reducing hospital admissions. By analyzing vast amounts of data from wearable sensors and medical records, AI models can help develop personalized treatment plans tailored to individual patients. For instance, in managing chronic conditions like diabetes, AI can analyze glucose levels, dietary habits, and physical activity to provide personalized recommendations for insulin dosages, diet, and exercise. This personalized approach improves treatment adherence and outcomes, leading to better overall health management for patients.
The implementation of AI models in clinical workflows can streamline operations and reduce the time healthcare professionals spend on routine tasks. Automated data analysis, reporting, and alert systems can handle tasks such as patient triage, follow-up reminders, and routine diagnostics. This efficiency allows healthcare providers to allocate more time to direct patient care and complex medical cases, improving the overall quality of care. By enhancing diagnostic accuracy and efficiency, AI models can contribute to cost reduction in healthcare. Early and accurate diagnosis prevents unnecessary tests and treatments, reducing overall healthcare costs. Additionally, the ability to monitor patients remotely can decrease hospital readmissions and the need for in-person visits, further lowering costs. These savings benefit not only healthcare providers but also patients and insurance companies. The use of AI models in analyzing large datasets from WBANs and medical records provides valuable insights for research and development. Researchers can identify patterns, correlations, and trends that were previously difficult to detect. These insights can lead to the development of new diagnostic tools, treatment protocols, and healthcare policies, driving innovation in the field of WBANs. AI-powered platforms can facilitate collaboration among healthcare professionals by providing shared access to data and analytics tools. This collaboration enhances knowledge sharing and accelerates the adoption of best practices across the healthcare industry. By leveraging AI, professionals and stakeholders can work together more effectively to improve patient care and outcomes.
This work demonstrates how performance measures in Clinical Decision Support Systems (CDSS) can be enhanced by integrating Recurrent Neural Network (RNN) models with deep learning techniques, such as SMOTE and BorderLine SMOTE. The findings highlight that RNN approaches, particularly when combined with oversampling strategies, can significantly improve accuracy, precision, recall, and AUC scores. These enhancements are particularly effective in managing data imbalances and identifying long-term dependencies in medical query processing without requiring excessive sampling. Additionally, the study shows that while random forest feature selection for feature reduction does not necessarily enhance model performance, it helps achieve similar outcomes with a reduced input dimension. This indicates the potential of sophisticated RNN models to optimize CDSS, offering a promising avenue for further advancements in real-time medical query processing. However, the study also notes limitations, particularly the varying effectiveness of oversampling strategies depending on data characteristics. Future research should explore a broader range of data and clinical scenarios to better understand the applicability of these techniques and modified RNNs, ultimately improving CDSS performance and patient care. This work suggests that more can be done to reduce data imbalance issues using RNN methods and incorporating existing approaches into CDSS for enhanced clinical decision-making.

Acknowledgement: The authors would like to express their heartfelt gratitude to the editors and reviewers for their detailed review and insightful advice.
Funding Statement: This work was supported by the “Human Resources Program in Energy Technology” of the Korea Institute of Energy Technology Evaluation and Planning (KETEP) and granted financial resources from the Ministry of Trade, Industry, and Energy, Korea (No. 20204010600090).
Author Contributions: Conceptualization, Israa Ibraheem Al Barazanchi and Wahidah Hashim; methodology, Israa Ibraheem Al Barazanchi; software, Reema Thabit; validation, Mashary Nawwaf Alrasheedy, Abeer Aljohan and Jongwoon Park; formal analysis, Israa Ibraheem Al Barazanchi; investigation, Wahidah Hashim; resources, Byoungchol Chang; data curation, Mashary Nawwaf Alrasheedy; writing—original draft preparation, Israa Ibraheem Al Barazanchi; writing—review and editing, Reema Thabit; visualization, Abeer Ahmed Aljohani; supervision, Wahidah Hashim; project administration, Wahidah Hashim and Reema Thabit. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data and materials used in this study are available upon request from the corresponding author.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. K. -J. Tokayev, K. Iqbal, and F. A. Younis, “Clinical decision support systems using NLP and computer vision and their integration in healthcare,” Int. J. Appl. Health Care Anal., vol. 8, no. 6, pp. 1–17, 2023. [Google Scholar]
2. P. Asghari, A. M. Rahmani, and H. H. Seyyed Javadi, “A medical monitoring scheme and health-medical service composition model in cloud-based IoT platform,” Trans. Emerg. Telecommun. Technol., vol. 30, no. 6, pp. 1–15, 2019. doi: 10.1002/ett.3637. [Google Scholar] [CrossRef]
3. S. R. Pandey, J. Ma, C. -H. Lai, and P. R. Regmi, “A supervised machine learning approach to generate the auto rule for clinical decision support system,” Trends Med., vol. 20, no. 3, pp. 1–9, 2020. doi: 10.15761/TiM.1000232. [Google Scholar] [CrossRef]
4. M. Wang and H. Chen, “Chaotic multi-swarm whale optimizer boosted support vector machine for medical diagnosis,” Appl. Soft. Comput., vol. 88, no. 6, 2020, Art. no. 105946. doi: 10.1016/j.asoc.2019.105946. [Google Scholar] [CrossRef]
5. N. Skyttberg, J. Vicente, R. Chen, H. Blomqvist, and S. Koch, “How to improve vital sign data quality for use in clinical decision support systems? A qualitative study in nine Swedish emergency departments,” BMC Med. Inform. Decis., vol. 16, no. 1, 2016, Art. no. 61. doi: 10.1186/s12911-016-0305-4. [Google Scholar] [PubMed] [CrossRef]
6. P. Kaur, R. Kumar, and M. Kumar, “A healthcare monitoring system using random forest and internet of things (IoT),” Multimed. Tools Appl., vol. 78, no. 14, pp. 19905–19916, 2019. doi: 10.1007/s11042-019-7327-8. [Google Scholar] [CrossRef]
7. S. Padhy, S. Dash, S. Routray, S. Ahmad, J. Nazeer and A. Alam, “IoT-based hybrid ensemble machine learning model for efficient diabetes mellitus prediction,” Comput. Intell. Neurosci., vol. 2022, 2022, Art. no. 2389636. doi: 10.1155/2022/2389636. [Google Scholar] [PubMed] [CrossRef]
8. G. Vardanyan, G. Avetisyan, G. Janoyan, K. Porksheyan, A. Khachatryan and A. Edilyan, “Medical errors: Modern condition of the problem,” Med. Sci. Armenia., vol. 59, no. 4, pp. 105–120, 2019. [Google Scholar]
9. K. K. Gagalova, M. A. Leon Elizalde, E. Portales-Casamar, and M. Görges, “What you need to know before implementing a clinical research data warehouse: Comparative review of integrated data repositories in health care institutions,” JMIR Form. Res., vol. 4, no. 8, 2020, Art. no. e17687. doi: 10.2196/17687. [Google Scholar] [PubMed] [CrossRef]
10. N. Muro et al., “Augmenting guideline knowledge with non-compliant clinical decisions: Experience-based decision support,” in Innovation in Medicine and Healthcare 2017. Cham: Springer, 2018, vol. 71, pp. 217–226. [Google Scholar]
11. W. C. Cha, “Effectiveness of a clinical decision support system in an emergency department,” Thesis, Seoul Nat. Univ., Seoul, Republic of Korea, 2021. [Google Scholar]
12. J. Yoo et al., “Alert override patterns with a medication clinical decision support system in an academic emergency department: Retrospective descriptive study,” JMIR Med. Inform., vol. 8, no. 11, 2020, Art. no. e23351. doi: 10.2196/23351. [Google Scholar] [PubMed] [CrossRef]
13. S. Parija, M. Sahani, R. Bisoi, and P. K. Dash, “Autoencoder-based improved deep learning approach for schizophrenic EEG signal classification,” Pattern Anal. Appl., vol. 26, no. 2, pp. 403–435, 2022. doi: 10.1007/s10044-022-01107-x. [Google Scholar] [CrossRef]
14. R. Miotto, F. Wang, S. Wang, X. Jiang, and J. T. Dudley, “Deep learning for healthcare: Review, opportunities and challenges,” Brief. Bioinform., vol. 19, no. 6, pp. 1236–1246, 2018. doi: 10.1093/bib/bbx044. [Google Scholar] [PubMed] [CrossRef]
15. A. H. Mayer, C. A. da Costa, and R. D. R. Righi, “Electronic health records in a Blockchain: A systematic review,” Health Inform. J., vol. 26, no. 2, pp. 1273–1288, 2020. doi: 10.1177/1460458219866350. [Google Scholar] [PubMed] [CrossRef]
16. S. Kumar, P. Tiwari, and M. Zymbler, “Internet of Things is a revolutionary approach for future technology enhancement: A review,” J. Big Data, vol. 6, no. 1, pp. 1–21, 2019. doi: 10.1186/s40537-019-0268-2. [Google Scholar] [CrossRef]
17. D. Korzun and A. Meigal, “Multi-source data sensing in mobile personalized healthcare systems: Semantic linking and data mining,” in Proc. 2019 24th Conf. Open Innov. Assoc. (FRUCT), Moscow, Russia, Apr. 8–12, 2019. doi: 10.23919/fruct.2019.8711950. [Google Scholar] [CrossRef]
18. T. Bikku, “Multi-layered deep learning perceptron approach for health risk prediction,” J. Big Data, vol. 7, no. 1, pp. 1–14, 2020. doi: 10.1186/s40537-020-00316-7. [Google Scholar] [CrossRef]
19. Y. Zhang, L. Shen, X. Yao, and Z. Zhang, “A hybrid deep learning model for medical data analysis using LSTM networks and attention mechanisms,” IEEE Access, vol. 8, pp. 123456–123467, 2021. [Google Scholar]
20. A. Goldberger et al., “PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals,” Circulation, vol. 101, no. 23, pp. e215–e220, 2000. [Google Scholar]
21. H. Eguia, C. L. Sánchez-Bocanegra, F. Vinciarelli, F. Alvarez-Lopez, and F. Saigí-Rubió, “Clinical decision support and natural language processing in medicine: Systematic literature review,” J. Med. Internet Res, vol. 26, no. 2, 2024, Art. no. e55315. doi: 10.2196/55315. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF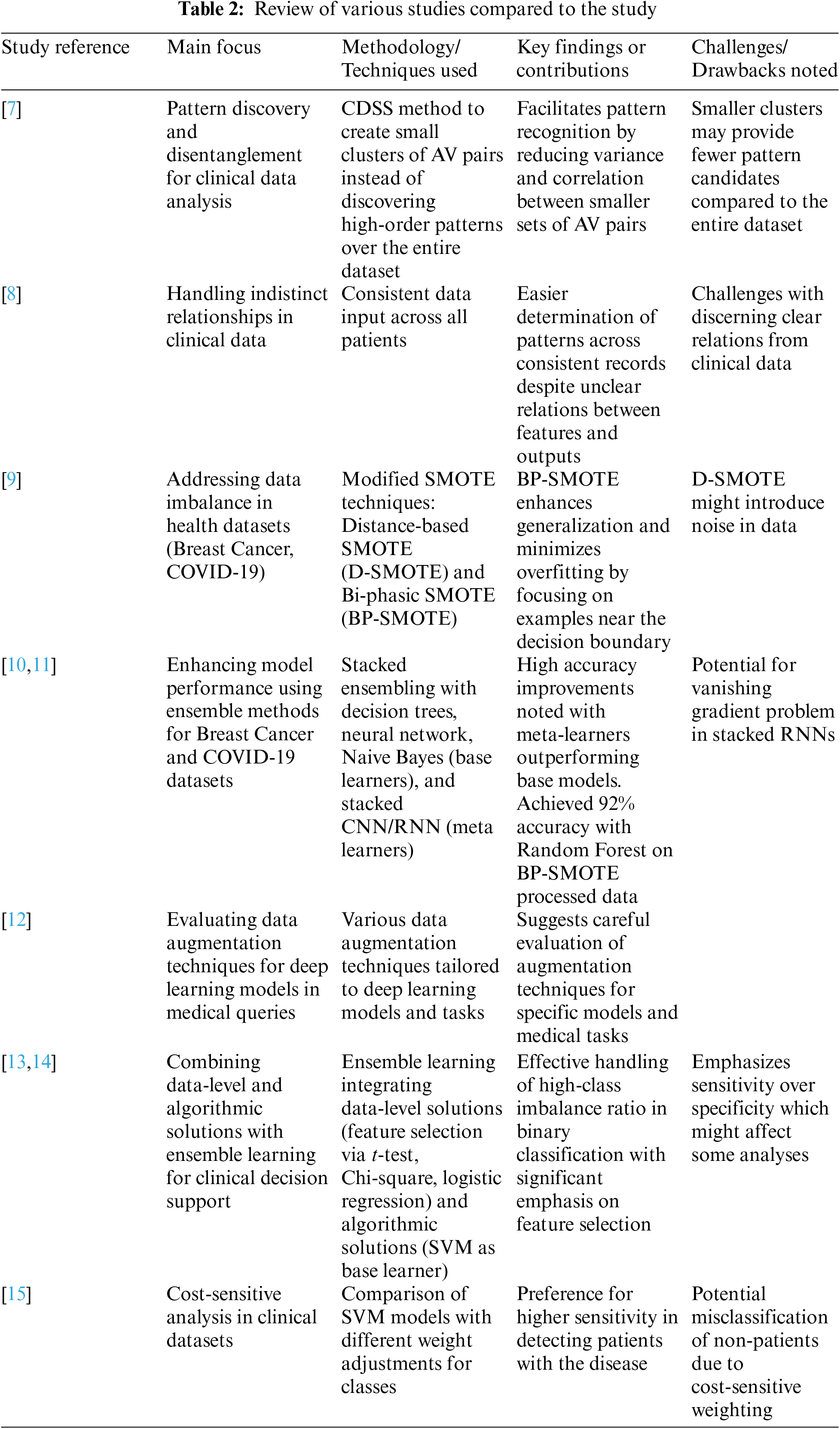
 Downloads
Downloads
 Citation Tools
Citation Tools