 Open Access
Open Access
REVIEW
AI-Driven Pattern Recognition in Medicinal Plants: A Comprehensive Review and Comparative Analysis
1 Department of Information Technology, Baba Ghulam Shah Badshah University, Rajouri, Jammu and Kashmir, 185234, India
2 Department of Computer Science, Samarkand International University of Technology, Samarkand, 141500, Uzbekistan
3 EIAS Data Science Lab, College of Computer and Information Sciences, Prince Sultan University, Riyadh, 11586, Saudi Arabia
* Corresponding Authors: Mudasir Ahmad Wani. Email: ; Muhammad Asim. Email:
(This article belongs to the Special Issue: Advances in Pattern Recognition Applications)
Computers, Materials & Continua 2024, 81(2), 2077-2131. https://doi.org/10.32604/cmc.2024.057136
Received 09 August 2024; Accepted 26 September 2024; Issue published 18 November 2024
Abstract
The pharmaceutical industry increasingly values medicinal plants due to their perceived safety and cost-effectiveness compared to modern drugs. Throughout the extensive history of medicinal plant usage, various plant parts, including flowers, leaves, and roots, have been acknowledged for their healing properties and employed in plant identification. Leaf images, however, stand out as the preferred and easily accessible source of information. Manual plant identification by plant taxonomists is intricate, time-consuming, and prone to errors, relying heavily on human perception. Artificial intelligence (AI) techniques offer a solution by automating plant recognition processes. This study thoroughly examines cutting-edge AI approaches for leaf image-based plant identification, drawing insights from literature across renowned repositories. This paper critically summarizes relevant literature based on AI algorithms, extracted features, and results achieved. Additionally, it analyzes extensively used datasets in automated plant classification research. It also offers deep insights into implemented techniques and methods employed for medicinal plant recognition. Moreover, this rigorous review study discusses opportunities and challenges in employing these AI-based approaches. Furthermore, in-depth statistical findings and lessons learned from this survey are highlighted with novel research areas with the aim of offering insights to the readers and motivating new research directions. This review is expected to serve as a foundational resource for future researchers in the field of AI-based identification of medicinal plants.Keywords
Abbreviations
| ANN | Artificial Neural Network |
| AUC | Area Under ROC (Receiver Operating Characteristic) Curve |
| BC | Bagging Classifier |
| BOF | Bag Of Features |
| BPNN | Back Propagation Neural Network |
| CART CV | Classification and Regression Tree, Computer Vision |
| CNN | Convolutional Neural Network |
| DCNN | Deep Convolutional Neural Network |
| DDLA | Dual Deep Learning Architecture |
| DT | Decision Tree |
| FAR | False Acceptance Rate |
| GLCM | Gray Level Co-Occurrence Matrix |
| GTSDM | Gray-Tone Spatial-Dependence Matrices |
| HOG | Histogram of Oriented Gradients |
| HSV | Hue saturation value |
| KNN | K-Nearest Neighbors |
| LDA | Linear Discriminant Analysis |
| LR | Logistic Regression |
| MLP | Multilayer Perceptron |
| MLPNN | Multilayer Perceptron Neural Network |
| NN | Neural Network |
| NB | Naive Bayes |
| PNN | Probabilistic Neural Network |
| PCA | Principal Component Analysis |
| RBF | Radial Base Function |
| RMSE | Root Mean Square Error |
| SVM | Support Vector Machine |
| SURF | Speeded Up Robust Features |
| RF | Random Forest |
Artificial Intelligence (AI) has emerged as a transformative technology with significant potential in various fields, including the identification and classification of medicinal plants. Traditional methods of identifying medicinal plants often rely on manual observation, which can be time-consuming, labor-intensive, and prone to errors. Moreover, the rapid loss of biodiversity and the increasing demand for herbal medicines have underscored the need for more efficient and accurate identification methods [1]. AI-based approaches offer promising solutions to these challenges by leveraging advanced algorithms automate the process of plant identification [2]. These approaches involve the development of computational models trained on large datasets of plant images, which enable them to recognize and classify plant species based on their visual characteristics such as leaf shape, color, texture, and other morphological features [3].
One of the key advantages of AI-based approaches is their ability to handle large volumes of data and process images rapidly, allowing for high-throughput screening of medicinal plants [4]. By harnessing the power of deep learning algorithms, AI systems can learn to identify patterns and extract meaningful information from complex plant images, leading to more accurate and reliable identification results [5]. Furthermore, AI-based approaches can be deployed in various settings, including botanical gardens, herbarium collections, and natural habitats, enabling researchers, botanists, and herbalists to efficiently catalog and identify medicinal plants [6]. These technologies have the potential to accelerate drug discovery processes [7], facilitate conservation efforts, and promote sustainable harvesting practices by providing valuable insights into the distribution, diversity, and medicinal properties of plant species. Overall, AI-based approaches for pattern recognition in medicinal plants represent a promising avenue for advancing botanical research, enhancing biodiversity conservation efforts, and unlocking the therapeutic potential of natural remedies for the benefit of humanity.
Plants are not only a vital resource for humans, but they also serve as the foundation of all food chains in the ecosystem. These plants encompass various botanical families, including herbs, shrubs, trees, and even certain fungi [8]. The world is abundant with a diverse array of medicinal plants, each possessing unique therapeutic properties. According to the World Health Organization, over 21,000 plant species have the potential to be used as therapeutic plants [9].
The efficacy of medicinal plants in treating various diseases is closely tied to the different parts of the plant, such as the fruits, leaves, roots, and more, and the specific manner in which these parts are utilized. Throughout history, medicinal plants have been integral to traditional healing practices across cultures and civilizations. Indigenous communities have long relied on the healing power of plants to address a wide range of health conditions, from common ailments to more serious diseases. It has been found that 80% of people treat their primary health ailments by making use of medicinal plants. Natural alternatives to allopathic medicines can be found in these medicinal plants. In comparison to modern medications, these medicinal plants have gained importance as they are less expensive, nontoxic, and have no negative effects. Medicinal plants offer a wide spectrum of health benefits, ranging from immune-boosting and anti-inflammatory properties to antimicrobial and anti-cancer effects. They are used in the prevention and treatment of various ailments, including digestive disorders, respiratory infections, cardiovascular diseases, skin conditions, and mental health disorders. Additionally, many medicinal plants possess antioxidant properties that help combat oxidative stress and promote overall well-being. Beyond their therapeutic value, medicinal plants hold cultural, economic, and ecological significance [10]. The cultivation, trade, and commercialization of medicinal plants contribute to local economies and livelihoods, particularly in rural areas where these plants are grown and harvested sustainably. Therefore, initiatives aimed at promoting sustainable cultivation, and conservation of medicinal plants is crucial for preserving biodiversity, protecting indigenous rights, and maintaining ecosystem balance. We can harness nature’s healing power to promote health, well-being, and sustainable development for present and future generations.
1.2 Manual Identification of Plants and Challenges
People from various cultures and social backgrounds have learned to identify plants and their characteristics to become a reason for the survival of some indigenous plants in various countries. This traditional way of identifying plants influenced their classification and nomenclature. However, these traditional methods are often based on oral tradition and may lack scientific rigor. This can lead to inconsistencies in classification and nomenclature, hindering efforts for systematic study and documentation [11].
A systematic categorization, identification, and naming of plants are often carried out by professional botanists (Taxonomists), who have deep knowledge of plant taxonomy. Plant taxonomists use morphological, anatomical, and chemotaxonomic approaches for the recognition and classification of plants. The manual plant identification often requires extensive fieldwork and specimen collection, which can be time-consuming, labor-intensive, and environmentally disruptive. Inaccessible or remote regions pose additional challenges, as collecting plant specimens may be logistically challenging or prohibited due to conservation concerns [3]. Every aspect of the identification depends on human perception; this results in the process of identification being complex and time-consuming. Furthermore, the subjective nature of human perception introduces another layer of complexity to plant identification. Different individuals may interpret plant characteristics differently, leading to inconsistencies in identification results. This challenge is compounded by the vast diversity of plant species and the intricate variations within each species. Many plant species exhibit morphological variations based on factors such as environmental conditions, geographical location, and genetic diversity. These variations can confound identification efforts, as plants may display different traits depending on their specific context. Additionally, certain plant species closely resemble each other, further complicating the identification process and increasing the likelihood of misclassification. Another issue is the lack of comprehensive and up-to-date reference materials for plant identification. While botanical guides and databases exist, they may not always contain the most current information or cover every plant species. This can lead to inaccuracies and gaps in knowledge, particularly for rare or newly discovered species [12]. There is also the dearth of subject experts, which gives rise to a situation called a “Taxonomic impediment” [13]. For nature enthusiasts and amateur botanists seeking to expand their knowledge of plant species, these challenges can be particularly overwhelming. Without access to specialized training or resources, accurately identifying plants becomes a significant challenge [3].
Given the numerous challenges associated with manual plant identification, such as inconsistencies in traditional practices, limited taxonomic expertise, and the subjective nature of human perception, there is a compelling need for a comprehensive review article focusing on automatic identification methodologies. Such a review will serve to consolidate the latest advancements in AI techniques applied to plant identification. By synthesizing existing research, highlighting successful methodologies, and identifying areas for improvement, the review will provide valuable insights for researchers, practitioners, and stakeholders involved in plant identification and conservation efforts. Moreover, it will help bridge the gap between traditional taxonomic practices and modern computational approaches, ultimately facilitating more accurate, efficient, and scalable methods for plant species identification.
In recent years, the scientific community has witnessed a surge in review articles/survey papers focused on plant identification using AI, reflecting the growing interest and advancements in this field. A review article by Wang et al. [11], presented various studies to explain Support Vector Machine for plant identification based on 30 leaf features (shape features = 16, color features = 4, and 11 texture features). A study [14], particularly for medicinal plant identification with machine learning, analyzed various machine learning methods including Random Forest, Multilayer Perceptron, Naive Bayes Classifier, Fuzzy Lattice Reasoning, etc., based on features namely leaf shape, venation, texture and combination of these features. Similarly, Azlah et al. [15] compared various studies based on a few machine learning algorithms namely ANN, PNN, KNN, etc. The research article [16] provided a thorough examination of various plant identification studies, particularly emphasizing the role of convolutional neural networks (CNNs) for image-based plant identification. While these reviews have significantly enriched our understanding of AI-driven plant identification methods, our study extends beyond the scope of previous research by offering in-depth insights and practical guidance that contributes to the advancement in the field of medicinal plant identification. In this work we address key challenges and provide deep understanding of AI based approaches which enhance our understanding and provide a robust framework for future studies. This study has the potential to significantly influence practical applications in the identification and utilization of medicinal plants.
The paper structure is diagrammatically presented in Fig. 1. In Section 1.1, we have briefly discussed the significance of medicinal plants, and in Section 1.2, the limitations of traditional manual identification techniques and the challenges they pose are discussed. After the introduction part, we highlight the potential of AI-based approaches to revolutionize plant identification in Section 2. This Section 2 is categorized into different subsections. The Section 2.1 presents an introduction to a few machine learning approaches and Section 2.2 introduces a few commonly used deep learning methods for plant image identification. The Section 2.3 discusses various stages of AI-based automated plant image identification. In Sections 3 and 3.1, we have discussed different dataset selection approaches followed by researchers for plant image identification. The Section 3 summarizes the studies that have utilized primary datasets and have employed different ML and DL approaches for experimentation. This section also highlights the different challenges that are faced, while using primary datasets for plant image identification. To overcome the challenges of limited primary dataset, the Section 3.1 discusses publicly available plant leaf image datasets that are typically used for automatic plant recognition. The detailed explanation of the classification Section 2.3 with the support of various plant identification studies is presented in Section 4. The Section 5 presents a comparative analysis of studies based on publicly available plant leaf image datasets. Various lessons that were learned during this literature review are presented in Section 6. In Section 6, we also present recommendations and outline promising avenues for future research in the domain of plant identification using AI, and finally the conclusion of this study is presented in Section 7. This paper comprehensively reviews widely used AI techniques for plant image identification.

Figure 1: Structure of whole work
The novel contributions of this study are as follows:
• To present consolidated knowledge on a single platform for new researchers, a comprehensive literature review was conducted to identify state-of-the-art AI techniques for medicinal plant identification.
• A detailed overview of the publicly available datasets, typically employed for automated plant identification is provided.
• A comprehensive summary of ML and DL-based studies, employing primary datasets is also presented. This includes the objective of the study, features extracted, methods/algorithms used, and the results achieved.
• For each publicly available dataset mentioned in this study, a comparative analysis of studies is conducted based on the features employed and AI techniques explored. This approach identifies successful strategies, extracts best practices, and informs more effective methodologies for medicinal plant identification.
• This study provides a road-map for future research in the evolving domain of medicinal plant identification, offering valuable insights and guidance for advancing the field.
2 AI Based Methods for Plant Identification
In this modern era, AI has greatly influenced various sectors and has proven itself the best solution for addressing complicated challenges in any area. Various studies are being carried out by researchers using AI approaches to get more insights into the identification of image data [17–21]. The most advanced AI techniques allow for the creation of frameworks and models that produce outstanding results with enhanced precision. Within AI, two key subfields, machine learning (ML) and deep learning (DL), are instrumental in automating the recognition of plant species. Employing ML and DL techniques, automatic plant species identification can be carried out optimally without facing problems caused due to manual identification approaches.
2.1 Machine Learning Algorithms
Machine learning provides computers the ability to learn without programming them explicitly. After a thorough literature study, it has been seen that researchers in the field of plant recognition are greatly interested in using machine-learning techniques for their experimentation [22]. Various commonly used ML algorithms are Support Vector Machine (SVM), Decision Tree, Random Forest Naive Bayes, Linear Regression, etc. [23]. The brief explanation of few machine-learning methods is presented as follows:
Support Vector Machine in multiclass plant image classification extend the basic binary SVM approach. One common approach to achieve this is the One-vs.-All (OvA) strategy, where K binary classifiers are trained, each distinguishing one class from the rest. Mathematically, for each class i in C = {1, 2, ..., K}, a separate binary SVM classifier is trained with samples of class i as positive examples and samples from all other classes as negative examples. During prediction, the class with the highest decision function value among all K classifiers is selected. Alternatively, the One-vs.-One (OvO) strategy trains
Decision Tree are a versatile algorithm used for multiclass plant image classification. They operate by recursively splitting the dataset based on feature values, creating a tree-like structure where each internal node represents a decision on a feature, branches represent outcomes, and leaves represent class labels. Given a dataset
Random Forest is an ensemble-learning algorithm that constructs multiple decision trees during training and outputs the class, which is the mode of the classes predicted by individual trees. For a dataset
where
where C^b is the class predicted by the b-th tree. This ensemble approach reduces overfitting and increases robustness compared to individual decision trees, making Random Forest a powerful tool for multiclass image classification tasks.
Linear regression is a machine-learning algorithm that is adapted for multiclass image identification by leveraging the one-vs.-all (OvA) or one-vs.-rest (OvR) strategy. In this approach, multiple linear regression models are trained, each focusing on distinguishing one class from the rest. Mathematically, let X represent the input features of plant images and Y denote the corresponding class labels. For K classes, Y can take values in {1, 2, ..., K}. Each linear regression model hk(x) learns a linear mapping from X to a binary outcome indicating whether the input image belongs to class k or not. The model is trained by minimizing the mean squared error (MSE) loss function represented by Eq. (4):
where N is the number of training samples, xi represents the i-th image feature vector, and yi is the binary label indicating whether the image belongs to class k (yi = 1) or not (yi = 0). Once the K linear regression models are trained, the classification of a new image involves applying each model to the input features and selecting the class with the highest predicted probability.
Naive Bayes is a probabilistic machine-learning algorithm widely used for multiclass image classification, including the identification of plant species from images. This algorithm is grounded in Bayes’ theorem and assumes that features are conditionally independent given the class label, which simplifies computations significantly. For an image represented by a feature vector x = (x1, x2,...,xn) and a class C from a set of classes {C1, C2, ..., CK}, the algorithm aims to find the class Ck that maximizes the posterior probability P (Ck| x). According to Bayes’ theorem, this can be written as Eq. (5):
Given that P(x) is constant across all classes, the focus is on maximizing the numerator P(x | Ck) P(Ck). Assuming conditional independence of the features, P(x | Ck) is decomposed into Eq. (6):
Thus, the classification decision rule simplifies to Eq. (7):
Here,
Machine learning methods for image identification often necessitate handcrafted feature engineering, a time-consuming process that may not accurately capture essential features, potentially leading to reduced accuracy. Deep learning (DL) is a discipline of machine learning that uncovers profound insights from input images. Additionally, deep learning models excel in handling vast and intricate datasets, contributing to enhanced effectiveness and efficiency. DL techniques can learn complex patterns from data without explicit feature engineering [24–27]. Deep learning algorithms have the potential to achieve outstanding outcomes in different domains like image identification, language processing, game playing, etc., as they can learn and make predictions that may be difficult for humans to discern [28]. Due to automatic feature extraction from the data, researchers prefer DL methods for plant image identification and classification. A deep learning model, Convolution Neural Network (CNN) [26], and its different architectures are widely employed by researchers for plant image classification [29]. Different Architectures of CNN that are frequently used by researchers are ResNet, DenseNet, MobileNet, VGG, AlexNet, etc. [30].
Convolutional Neural Network (CNNs) have revolutionized plant image identification by automating feature extraction and learning through convolution operations. At their core, CNNs utilize filters (or kernels) that slide over the input image to produce feature maps, highlighting essential visual characteristics like edges, textures, and patterns necessary for distinguishing different plant species. This operation is reprsented as Eq. (8):
where I(x + i, y + j) are pixel values and K (i, j) are filter values. CNNs handle large datasets efficiently, learning hierarchical feature representations through multiple layers of convolutions, pooling, and non-linear activation functions like ReLU, defined as Eq. (9):
Pooling layers, such as max pooling defined by Eq. (10):
reduce spatial dimensions, enhancing computational efficiency and reducing overfitting risks. This deep feature hierarchy allows CNNs to capture subtle differences in leaf shapes, textures, venation patterns, and colors, achieving high accuracy in plant species classification. This capability makes CNNs invaluable for biodiversity monitoring and conservation efforts.
ResNet is a type of Convolutional Neural Network (CNN), addresses the challenge of vanishing gradients in deep neural networks, enabling the training of significantly deeper architectures critical for plant image identification. The central concept of ResNet is residual learning, where the network learns residual functions with respect to layer inputs rather than directly learning unreferenced functions. This is mathematically expressed as y = F(x, {Wi}) + x, where y is the output, x is the input, and F(x, {Wi}) represents the residual mapping to be learned. ResNet incorporates shortcut connections or identity mappings to facilitate learning, allowing gradients to flow more effectively through the network during backpropagation. These connections are defined as
DenseNet (Densely Connected Convolutional Networks) is a deep learning architecture designed to address the vanishing gradient and feature reuse problems in deep neural networks. In DenseNet, each layer is connected to every other layer in a feed-forward fashion, creating dense connections between layers. This connectivity pattern enables feature reuse and encourages feature propagation throughout the network, leading to enhanced gradient flow and feature propagation compared to traditional architectures.
Mathematically, DenseNet can be represented as Eq. (11):
where
MobileNet is an innovative convolutional neural network architecture tailored to the computational constraints of mobile and embedded devices, while still achieving high accuracy in image classification tasks. Its standout feature is depthwise separable convolutions, dividing the standard convolution operation into depthwise and pointwise convolutions. Mathematically, a depthwise separable convolution is represented as Y = P (K ∗ (X ∗ D)), where X is the input feature map, K is the depthwise convolution kernel, D denotes the depthwise convolution operation, ∗ represents the convolution operation, and P is the pointwise convolution operation. By separating spatial and channel-wise convolutions, MobileNet drastically reduces computational cost and model size without sacrificing accuracy. This efficiency makes it ideal for resource-constrained environments, particularly for plant image identification in settings with limited computational resources. Moreover, MobileNet’s lightweight design facilitates rapid inference on mobile devices, enabling real-time plant species classification and biodiversity monitoring in the field.
VGG (Visual Geometry Group) network is a convolutional neural network architecture developed by the Visual Geometry Group at the University of Oxford. VGG networks are characterized by their deep structure, comprising numerous convolutional layers with small 3 × 3 filters, succeeded by max-pooling layers for downsampling. This architecture is reiterated multiple times to augment depth, resulting in VGG models with 16 or 19 layers. Mathematically, the convolutional operation in VGG can be represented as Eq. (12):
where
2.3 Automated Identification of Plant Leaf Images
Numerous initiatives have been taken to address the task of identifying plants based on different plant parts like fruits, flowers, leaves, bark, or whole plant images [20,31], utilizing various AI approaches. However, researchers often prefer leaf images, as these images are recognized as the most accessible and reliable sources of information for accurate identification of plant species. The automatic plant identification is carried out in different stages that include plant image collection, preprocessing of images, extracting features, and finally classification of plant images to their appropriate class. Fig. 2 shows a diagrammatic representation of the different stages of automated plant species identification. The detailed explanation of these stages is discussed as follows:

Figure 2: Different stages of automatic plant image identification
Image Acquisition In image acquisition, image samples of the entire plant or plant organs such as leaves are collected. Medicinal plant identification studies are often performed on primary datasets. The commonly investigated medicinal plants are Withania somnifera, Phyllanthus emblica, Azadirachta Indica (Neem) Aconitum carmichaelii, Coleus aromaticus, Andrographis paniculata, Inula recemosa, and Chromolaena odorata, which are found in India, China, Indonesia, Bangladesh and Thailand [32]. However, there is a notable dearth of research on automating the identification of these plant species. Machine learning and deep learning algorithms demand a significant number of images to achieve better classification performance [33]. Due to the scarcity of a particular medicinal plant species, it is not always possible to generate large image datasets. Therefore, the accuracy of findings in existing studies exhibits significant variations due to disparities in the number of images found within the databases used for analysis. During primary data collection, images can be captured using either the built-in cameras of mobile phones or digital cameras. Images obtained in this stage are captured in uncontrolled environments and may contain significant noise, which can hinder plant recognition accuracy. Therefore, a preprocessing stage is necessary to address these challenges.
Image Preprocessing Image processing techniques are applied to raw images to refine their quality and enhance their suitability for analysis. This involves operations such as resizing, normalization, and denoising to standardize the images and mitigate the effects of noise or variations in illumination. Image preprocessing aims to improve image data by enhancing specific features while reducing unwanted noise, which optimizes computational time and increases classification accuracy [34]. Saleem et al. [35] applied dimensionality reduction as a preprocessing step for plant species classification, using both the Flavia dataset and a primary dataset. They achieved impressive accuracy of 98.75% on Flavia and 97.25% on the primary dataset, highlighting the value of dimensionality reduction in enhancing classification accuracy. Nasir et al. [36] demonstrated a 99% detection rate with enhanced Ficus Deltoidea leaf images using PCA on 345 images from various Ficus Deltoidea variations. Barré et al. [37] introduced LeafNet, a plant identification system that uses dimensionality reduction as a preprocessing step for species identification. They conducted their research on datasets including Flavia, and Leafsnap. By applying data augmentation, they expanded the Leafsnap dataset from 26,624 to 270,161 images. The model achieved top-5 accuracies of 97.8% for Leafsnap, and 99.6% for Flavia. Kadir et al. [38] performed plant leaf classification using a Probabilistic Neural Network (PNN) on the Flavia dataset. They found that using PCA improved accuracy from 93.43% to 95% for the Flavia dataset.
Image segmentation partitions an image into distinct regions based on characteristics such as color, texture, or intensity. Gao et al. [39] applied the OTSU method [40] for thresholding, achieving an accuracy of 99% in leaf edge detection by selecting the optimal segmentation threshold based on maximum between-class variance. Complex image segmentation tasks often require advanced methods like level set segmentation, which evolves a curve within an image to precisely identify object boundaries. Nandyal et al. [41] used level set segmentation to identify 400 medicinal plant images with various ML classifiers. SVM outperformed other classifiers, achieving accuracies of 98% for trees, 97% for herbs, and 94% for shrubs. They performed preprocessing using the unsharp filter technique [42].
Husin et al. [43] emphasized resizing and edge detection in herb plant recognition, achieving a 98.9% recognition rate on a dataset of 2000 leaf images from 20 plant species. They resized images to a uniform 4800-pixel format and converted images from RGB to grayscale and then to binary to eliminate color variation. They compared edge detection techniques, finding canny edge detection [44] to have the lowest noise levels. The study used skeletonization to preserve object shape in binary images, with Singular Value Decomposition (SVD) to compress the images during feature extraction. Singh et al. [45] proposed an Artificial Neural Network (ANN) method based on geometrical features achieving an accuracy of 98.8% on leaf images from 8 regional plant species. They applied preprocessing techniques such as image resizing, RGB to grayscale conversion, grayscale to binary transformation, scaling, rotation, and denoising using wavelets. These advanced preprocessing techniques improved the robustness of the recognition system.
Feature Extraction methods are employed to identify relevant patterns and characteristics from images, aiding in creating discriminative representations. These features encapsulate crucial botanical traits such as leaf shape, texture, and color, which are instrumental in distinguishing different plant species. Feature extraction aims to minimize computational cost by reducing the dimensionality of preprocessed images without losing important information. Deep learning methods have gained substantial attention; however, these methods are computationally expensive and require an ample amount of data to automatically learn hierarchical representations. This section focuses on machine learning-based feature extraction methods, essential for extracting meaningful insights from raw images like shape, texture, color, and vein patterns. Being well versed with these techniques allows researchers to compare and evaluate different feature extraction approaches, choosing the most suitable method based on their plant images and analysis requirements. Effective feature extraction offers benefits such as reduced training time, less overfitting, and improved identification accuracy by reducing input dataset dimensionality [46]. For plant identification, researchers commonly utilize the shape, color, and texture features of leaves. Leaves are often preferred for feature extraction due to their availability and ease of collection [19]. Caglayan et al. [47] extracted shape and color features from leaf images, using area, perimeter, leaf border, aspect ratio, and rectangularity for shape features, and mean, standard deviation of intensity values, and RGB histogram for color features. Color feature extraction techniques include grey-level operators, GTSDM (Gray Tone Spatial Dependency Matrix), and LBP (Local Binary Pattern) operators. The grey-level operator assesses pixel intensities, GTSDM analyzes spatial relationships and dependencies of grey-tone values, and LBP excels in texture analysis, enhancing color feature discrimination. Arun et al. [48] used these methods on 250 high-resolution leaf images from five plant species, achieving an accuracy of 98.7%.
Combining multiple feature types enhances plant identification systems. Dahigaonkar et al. [49] combined geometric, texture, color, and shape features, achieving an accuracy of 96.66%. Kan et al. [50] used a combination of ten shape features, including three geometric features, seven Hu invariant moments [51], and five texture features, achieving a 93.33% accuracy on 240 leaf images from twelve medicinal plants. In a similar way, Jye et al. [52] developed an identification system, using SVM and ANN. The study aimed to differentiate between three visually similar Ficus species, achieving 83.3% accuracy with both ANN and SVM. Their approach included the robust Histogram Oriented Gradient (HOG) descriptor and other morphological, texture, and shape features. Vishnu et al. [34] employed KNN and ANN ML algorithms on the Flavia dataset. They focused on the HOG descriptor; achieving 98.5% accuracy using ANN. Sabu et al. [53] presented a computer vision technique for classifying 200 leaf images of 20 different plants using SURF (Speeded up Robust Features) and HOG-based techniques, achieving a 99.6% accuracy. SURF detects distinctive points within images, essential for recognizing and matching patterns, and is robust to various image transformations and lighting conditions. Elhariri et al. [54] used a multifaceted feature set, including HSV (Hue, Saturation, and Value) for color information, Gray-Level Co-occurrence Matrix (GLCM) for spatial texture relationships, and first-order texture features, achieving a 92.65% accuracy. Subsequently, Kaur et al. [55] achieved a 93.26% accuracy on the Swedish leaf dataset, using GLCM for texture feature extraction and color features derived from mean, standard deviation, kurtosis, and skewness. Local Binary Pattern (LBP) is a potent texture descriptor, and Bag of Features (BOF) aggregates local visual features efficiently, reducing dimensionality and computational complexity. Ali et al. [56] combined the features (extracted with LBP and BOF), and achieved 99.4% accuracy on the Swedish leaf dataset.
Fourier descriptors and shape-defining features (SDF) are valuable for capturing shape and contour details. Aakif et al. [57] used Fourier descriptors and SDF, achieving a 96.5% accuracy when combined with other features. In Fig. 3, we present the most important features and their extraction mechanism that are employed during the process of plant image identification.

Figure 3: Important features and various extraction methods
Classification is the final stage of AI-based plant image identification involves the classification of features extracted from the images. Typically, this classification is based on attributes such as leaf shape, texture, color, and vein patterns [58]. Researchers use either machine learning or deep learning classifiers, depending on the complexity of the problem and the size of the dataset available. Machine learning algorithms are preferred when the problem is less complex and the dataset available is small. When the dataset is large, deep neural networks like CNN and its different architectures are generally used for the classification. Notably, there are two common approaches followed for the classification process. Some researchers opt to employ a single classifier for the entire identification process, while others prefer to experiment with and compare different classifiers on their datasets. The latter approach is often used to achieve enhanced accuracy levels, as evidenced in the literature. In Fig. 4, we have mentioned the classifiers that are typically employed for the classification of plant species. The explanation of the AI-based plant classification methods is exhaustively discussed in Section 4.
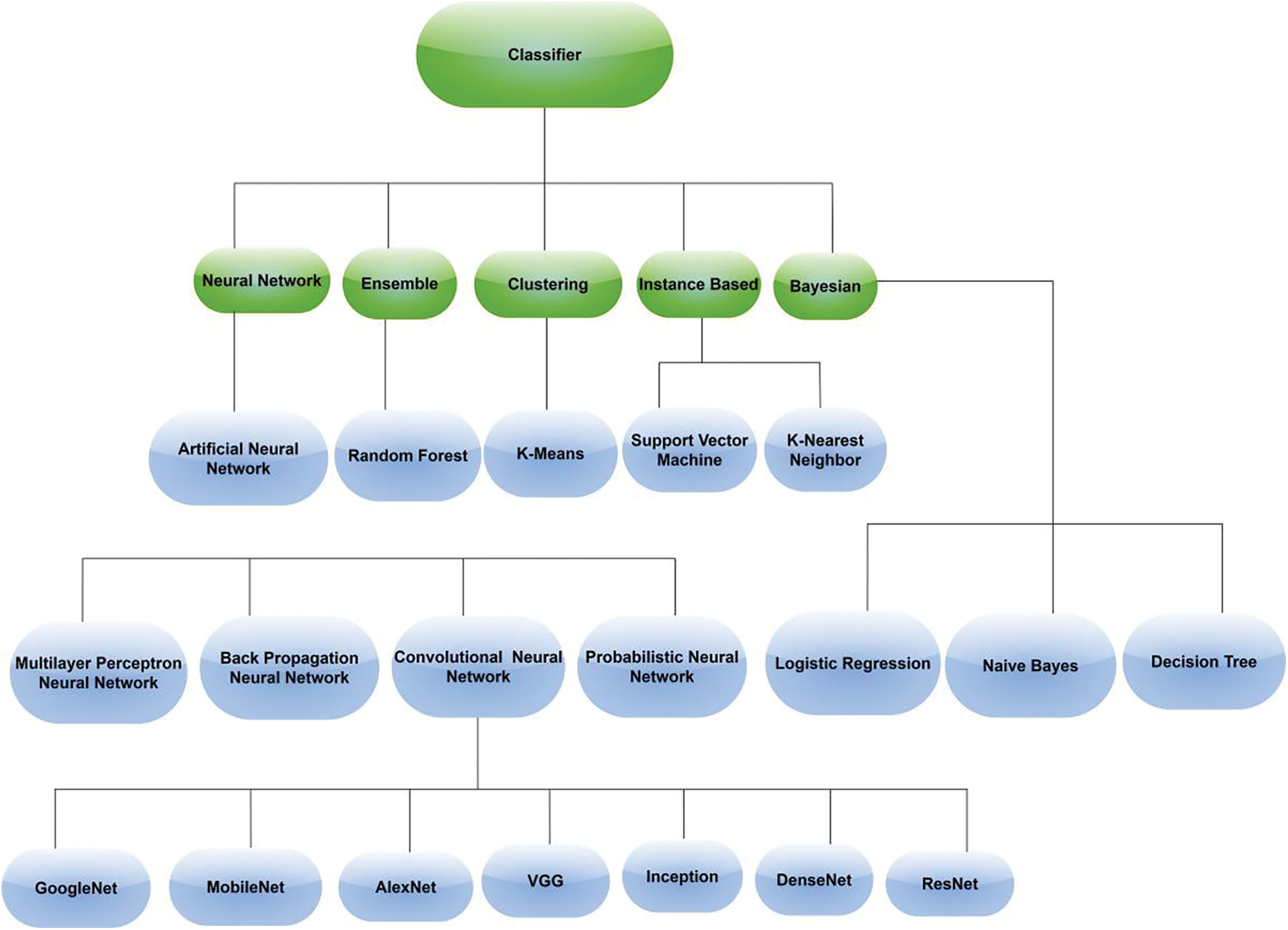
Figure 4: Important classifiers for plant identification
3 Medicinal Plant Image Datasets: Overview and Challenges
The study of medicinal plants holds immense significance as these plants, renowned for their therapeutic properties, offer a rich source of natural remedies with potential applications across diverse medical fields. In recent years, scientific research has begun to validate the efficacy of medicinal plants and unravel the mechanisms behind their therapeutic effects. For medicinal plant identification, researchers often prefer to use the primary datasets for their experimentation. Analyzing datasets serves as a critical step towards advancing the field of AI based plant image analysis. In this work we presented the range of primary dataset sizes utilized across different plant identification studies. This will provide a roadmap to future researchers into the scalability and generalizability of AI models trained on varying dataset sizes.
Table 1 presents a summary of the machine learning based plant identification studies, utilizing handcrafted features extracted from primary datasets. This comprehensive summary includes a reference (Ref.), objective of the study, methods/algorithms applied, features used, dataset details (where PD is short for Primary Dataset, T.I is short for Total images of Primary Dataset, and Sp is short for Total species in the primary dataset), and Accuracy (Presented highest accuracy achieved by a study).

In the realm of plant image identification, researchers have shown a great interest in employing deep learning techniques for conducting their research work. Table 2 presents a summary of the deep learning-based plant identification studies, utilizing primary datasets. This comprehensive summary includes a reference (Ref.), objective of the study, methods/algorithms applied, features used, and dataset details (where PD is short for Primary Dataset, T.I is short for Total images of Primary Dataset, and Sp is short for Total species in the primary dataset), and Accuracy (Presented highest accuracy achieved by a study).
The scarcity of comprehensive medicinal plant leaf image datasets poses a significant challenge in advancing the research of automated plant identification using AI. The majority of researchers train and test their models on publicly available plant datasets. Leveraging these existing datasets for automatic plant identification using AI holds great potential to address the current shortfall. Table 3 presents various publicly available datasets and their source of retrieval. These are Flavia, Swedish leaf, Leafsnap, Folio, and Mendeley Medicinal Leaf datasets. These datasets are commonly used in research studies to conduct plant identification tasks. Each of these datasets exhibits variations in the quantity of images and the specific species represented. The summary of the description of these datasets is presented as follows:
This dataset is a leaf image dataset, which contains 1907 images from 32 different plant species. There are 50 to 77 images per species. The images were collected from the Botanical Garden of Sun Yat-sen and the campus of Nanjing University, China. These leaf images were obtained using either scanners or digital cameras with a plain background. Importantly, the isolated leaf images depict leaf blades exclusively and do not include petioles [66]. Fig. 5 shows few leaf sample images from the flavia image dataset.
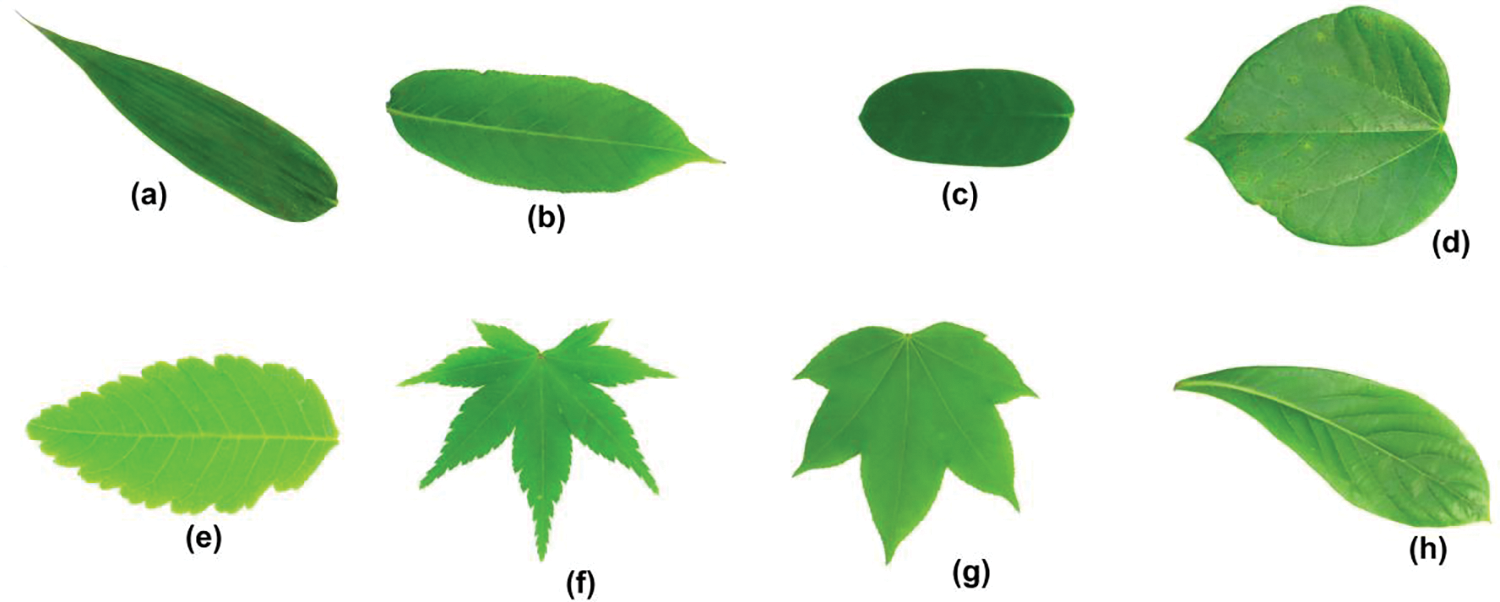
Figure 5: A few images of Flavia leaf image dataset, illustrating the diverse range of plant images incorporated for classification purposes. (a) Phyllostachys edulis (Carr.) Houz. (b) Aesculus chinensis (c) Indigofera tinctoria L. (d) Cercis chinensis (e) Koelreuteria paniculata Laxm. (f) Acer Palmatum (g) Kalopanax septemlobus (Thunb. ex A.Murr.) Koidz. (h) Phoebe nanmu (Oliv.) Ḡamble
The dataset has been created by joint efforts of the University of Linkoping and the Swedish Museum of Natural History. The dataset consists of 1125 leaf images from fifteen different Swedish tree classes and each class contains 75 images. Developing this dataset was challenging due to the significant similarity between different species within them [67]. However, researchers managed to achieve impressive accuracies by applying state-of-the art approaches for feature extraction and classification. Fig. 6 shows few leaf sample images from the Swedish leaf image dataset.
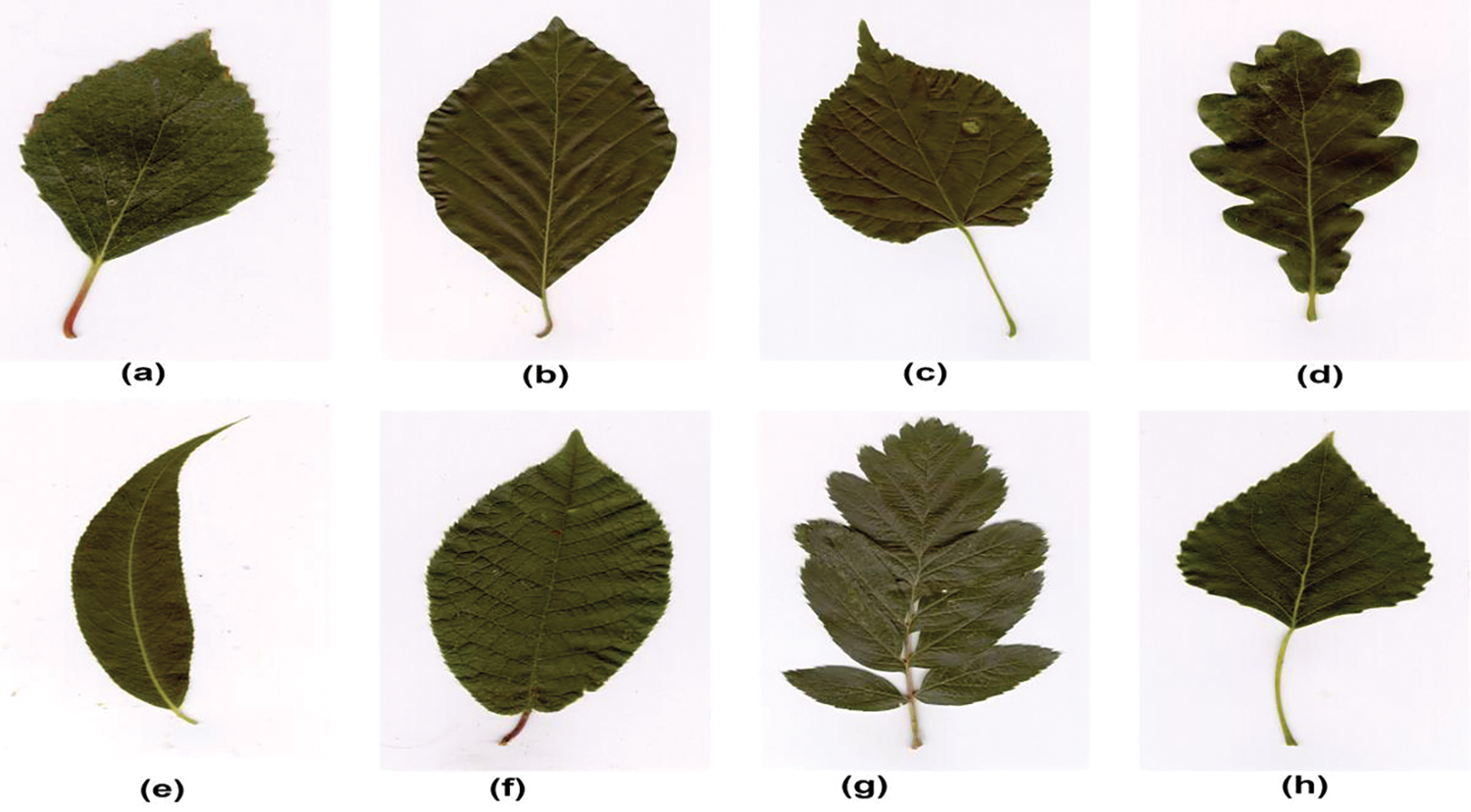
Figure 6: The sample of swedish leaf image dataset, illustrating the diverse range of plant images incorporated for classification purposes. (a) Alnus incania (b) Fagus silvatica (c) Populus (d) Quercus (e) Salix alba ‘Sericea’ (f) ASalix sinerea (g) Sorbus intermedia (h) Tilia
The dataset was created through a collaboration between computer scientists from the University of Maryland and Columbia University, along with botanists from the Smithsonian Institution in Washington. It consists of leaf images from 184 tree species captured in natural environments, referred to as field images. Fig. 7 displays several sample leaf images from the field. These images were taken under various lighting conditions and exhibit varying degrees of blur, noise, illumination differences, shadows, and other environmental factors. In addition, the dataset includes pressed leaf images from 185 plant species, captured in a controlled laboratory environment using high-quality cameras. These are referred to as lab images, with Fig. 8 providing a few examples. The comparison between Figs. 7 and 8 highlights the presence of color variations in the background, as well as the inclusion of leafstalks in the leaf images. To reduce this noise and enhance the dataset, image segmentation was performed using the Leafsnap segmentation algorithm [68]. Fig. 9 showcases a few segmented leaf images of the leafsnap dataset.
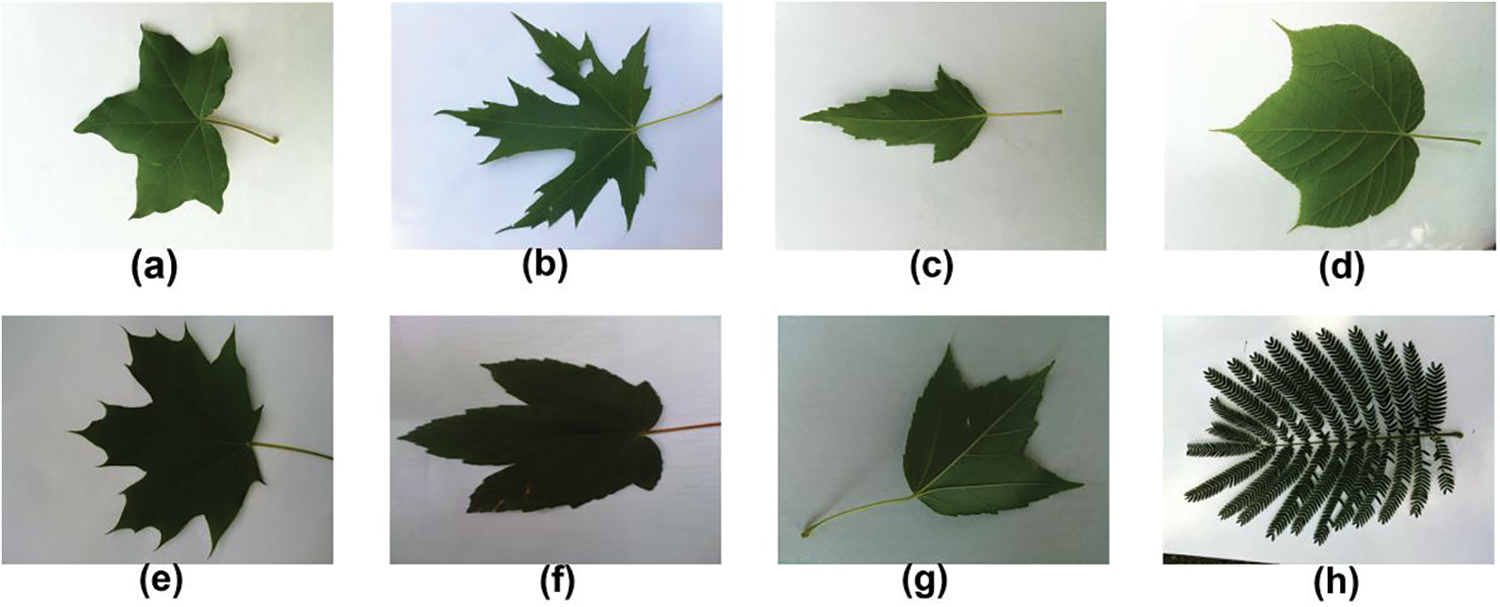
Figure 7: Some field images images from leafsnap dataset (a) Acer campestre (b) Acer saccharinum (c) Acer ginnala (d) Acer pensylvanicum (e) Acer platanoides (f) Acer pseudoplatanus (g) Acer rubrum (h) Albizia julibrissin
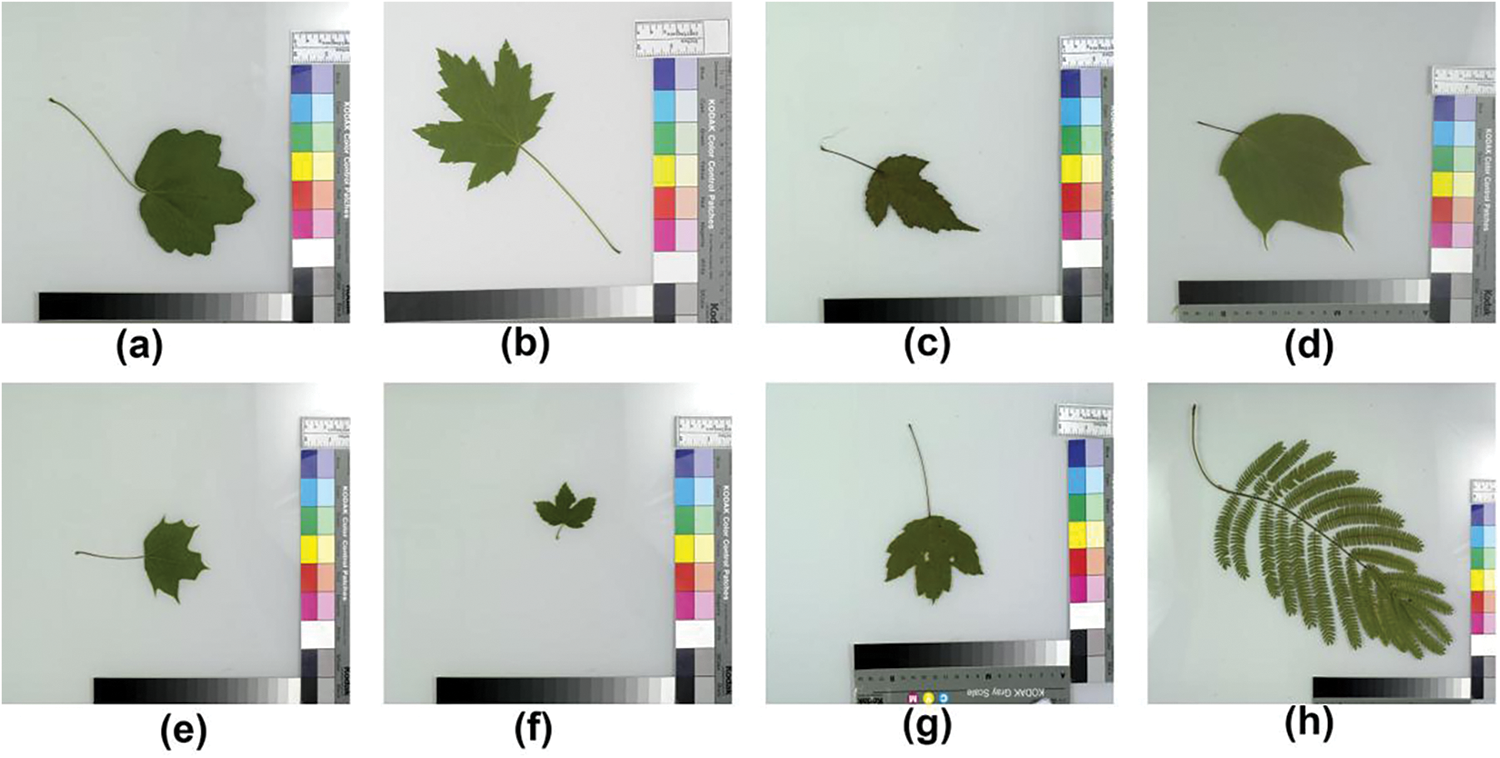
Figure 8: The lab images from leafsnap dataset, captured under a controlled lab environment (a) Acer campestre (b) Acer saccharinum (c) Acer ginnala (d) Acer pensylvanicum (e) Acer platanoides (f) Acer pseudoplatanus (g) Acer rubrum (h) Albizia julibrissin
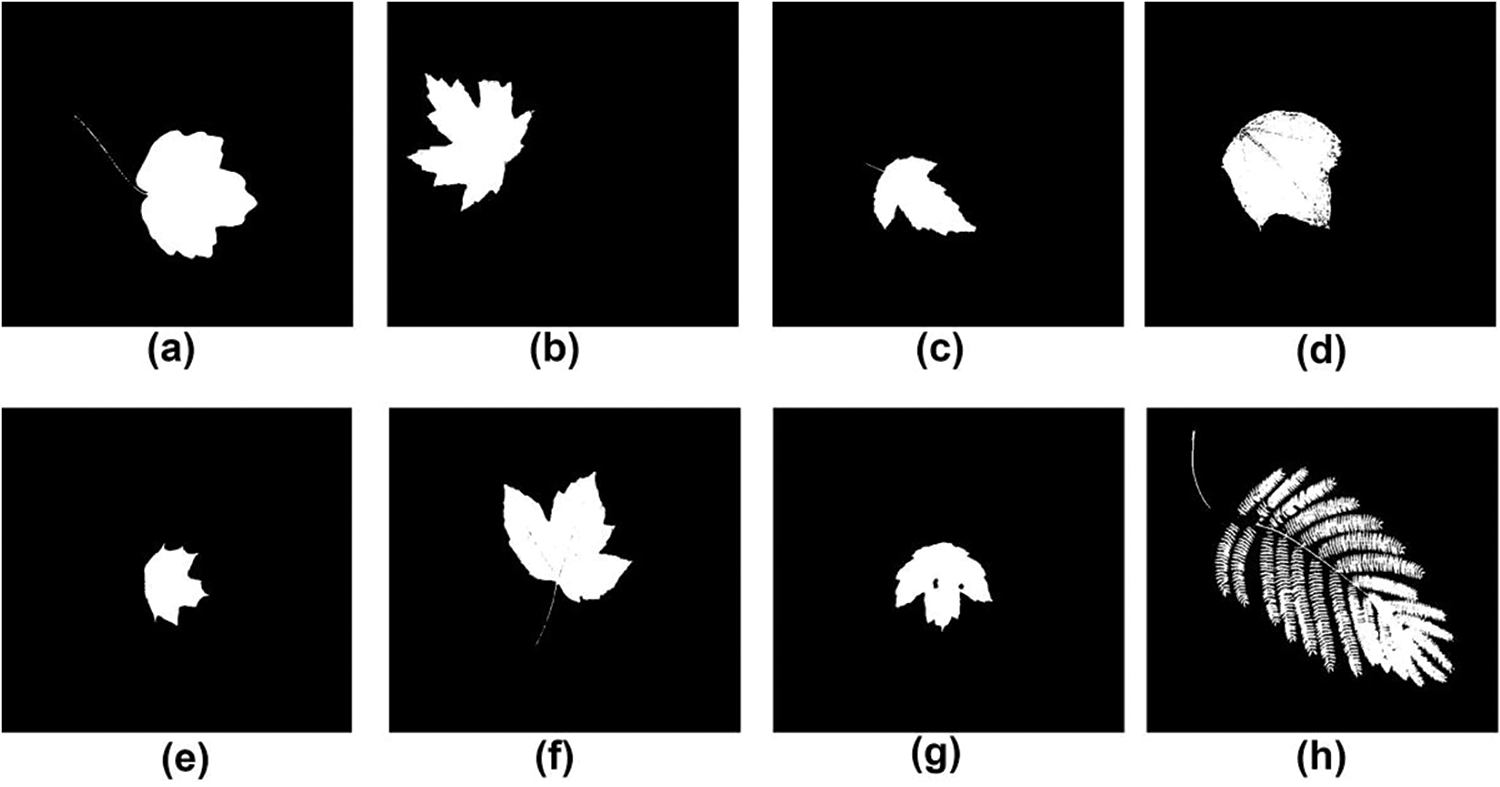
Figure 9: Few segmented leaf images of the leafsnap dataset (a) Acer campestre (b) Acer saccharinum (c) Acer ginnala (d) Acer pensylvanicum (e) Acer platanoides (f) Acer pseudoplatanus (g) Acer rubrum (h) Albizia julibrissin
This dataset contains leaf images of thirty-two different plant species photographed after placing leaves on a white background during daylight. Images were sampled from the farm of Mauritius University [69]. Fig. 10 shows a few leaf sample images from the Folio leaf image dataset.
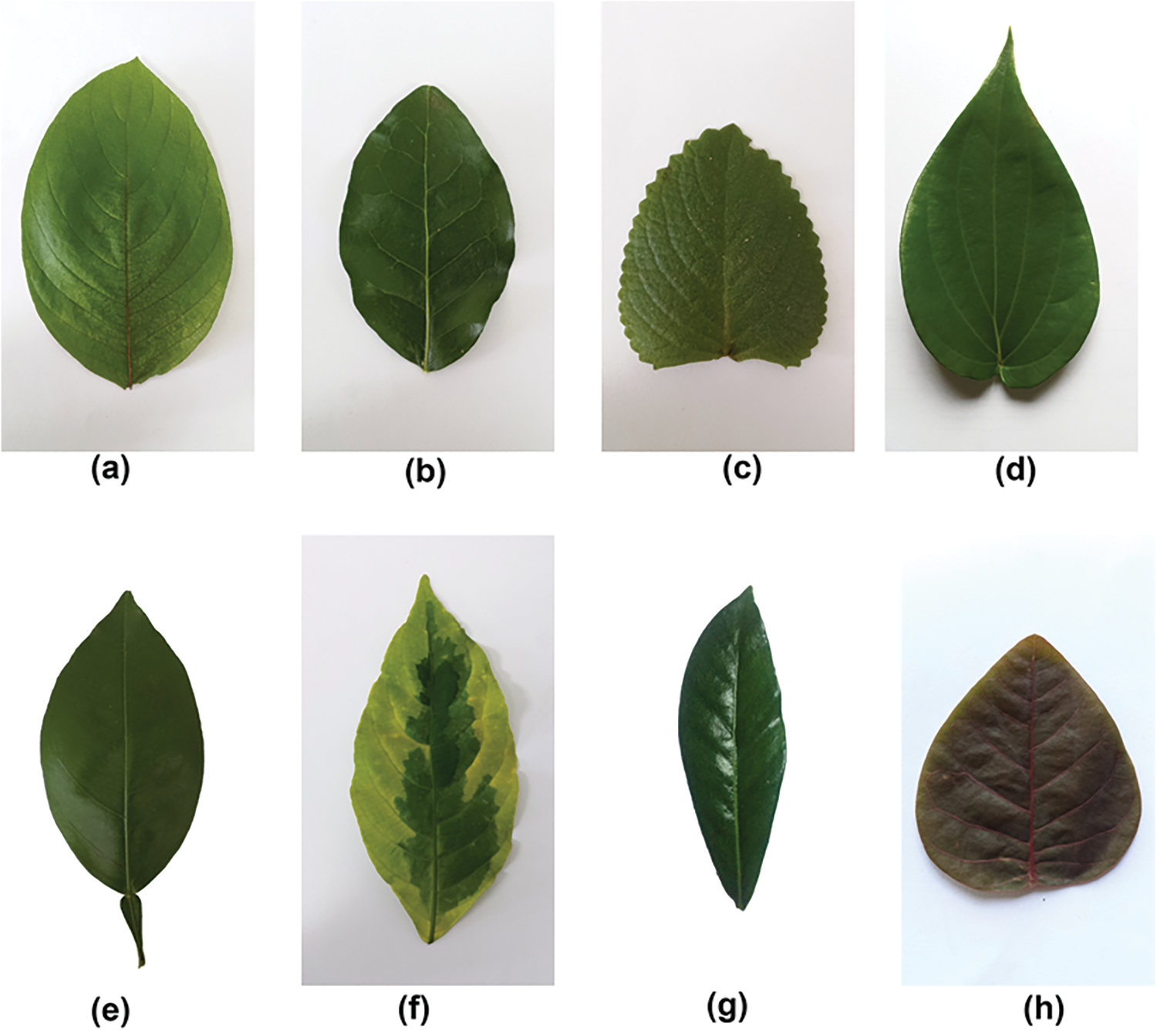
Figure 10: The sample images of folio leaf image dataset, illustrating the diverse range of plant images incorporated for classification purposes. (a) Mussaenda philippica (b) Malpighia glabra (c) Myroxylon balsamum (d) Piper betle (e) Citrus aurantium (f) Graptophyllum pictum (g) Psidium cattleianum (h) Euphorbia cotinifolia
3.5 Mendeley Medicinal Leaf Dataset
It contains a total of 1835 high-quality leaf images from thirty different medicinal plants, including Santalum album (Sandalwood), Plectranthus amboinicus (Indian Mint, Mexican mint), and Brassica juncea (Oriental mustard). Each species folder contains 60 to 100 images, labeled with the botanical/scientific names. The images were captured using a Samsung S9+ mobile camera and printed with a Canon Inkjet Printer. To enhance the dataset for training machine learning and deep learning models, the images of the leaves in the dataset were slightly rotated and tilted [32]. Fig. 11 shows a few leaf sample images from the Mendeley medicinal leaf image dataset. There are diverse health benefits associated with each plant present in this dataset. Table 4 highlights a few medicinal plant names and their associated medicinal value. This underscores the need for an intelligent automated pattern recognition system that can accurately identify medicinal plant species.
Figure 11: The sample of Mendeley medicinal leaf image dataset, illustrating the diverse range of plant images incorporated for classification purposes. (a) Alpinia galanga (Rasna) (b) Amaranthus viridis (Arive-Dantu) (c) Artocarpus heterophyllus (Jackfruit) (d) Azadirachta indica (Neem) (e) Basella alba (Basale) (f) Brassica juncea (Indian Mustard) (g) Carissa carandas (Karanda) (h) Citrus limon (Lemon)
Studies have identified bioactive compounds within plants, such as alkaloids, flavonoids, terpenoids, and phenolics, which exhibit pharmacological activities beneficial for human health. Some of the health benefits of few medicinal plants are mentioned in Table 4. Harnessing the power of AI in automatic plant identification systems, medicinal plant image datasets play a pivotal role in training and validating AI models.
4 AI in Plant Leaf Image Analysis: Diverse Approaches
Researchers in the domain of plant leaf identification have leveraged a variety of publicly available datasets, such as Flavia, Folio, Leafsnap, Swedish leaf, and Mendeley dataset, to train and evaluate their AI models. These datasets offer a diverse range of leaf images, enabling researchers to develop robust and accurate identification systems. Some researchers opt to utilize a single dataset, while others used multiple datasets to enhance the generalization and performance of their models. In their experimentations, researchers have utilized various ML and DL approaches for the plant identification tasks. Additionally, transfer learning has emerged as a valuable approach, allowing researchers to leverage pre-trained models on large-scale datasets like ImageNet and fine-tune them on plant leaf datasets to achieve improved performance. In the following subsections, we present studies that utilized these datasets and employed different approaches for various AI methodologies, including machine learning, deep learning, and transfer learning. Researchers have explored these methodologies to develop automated systems for plant leaf identification, aiming for accuracy and efficiency in classification tasks. Tables 5 and 6 present a comparison of machine learning and deep learning methods for pattern recognition in medicinal plants. Our analysis results in several recommendations and key improvements, which are discussed in Section 6.
4.1 Machine Learning Based Plant Classification
Machine learning methodologies have emerged as a cornerstone for developing accurate and efficient plant identification systems. In this subsection, we will explore studies where algorithms like Support Vector Machines, Random Forests, and K-Nearest Neighbor, etc., are used to classify plant leaves based on their features.
Pacifico et al. [73] conducted a study on medicinal plant classification using 1148 leaf images from 15 plant species, including Sambucus nigra, Illicium verum, Glycyrrhiza glabra, Bidens pilosa, Bixa Orel-Lana, Caesalpinia ferrea and others. The images were resized to 60 × 60 dimensions, and various machine-learning techniques, such as Decision Tree, K-NN, Random Forest, and Multi-Layer Perceptron with Backpropagation, were applied. They found that setting the K value in K-NN between three and five optimized results, as higher values degraded performance. The authors developed an MLP-BP algorithm with three hidden layers using a trial-and-error approach. Their experimentation included 10-fold cross-validation over 10,000 epochs, achieving an impressive 97% accuracy.
Begue et al. [59] created a recognition system for 24 medicinal plants, employing machine-learning algorithms, including Random Forest, K-Nearest Neighbor, Naive Bayes, Support Vector Machine, and Multilayer Perceptron Network, to classify images captured with a smartphone in a laboratory setting. Among these classifiers, Random Forest stood out with the highest accuracy of 90.1%, while the Multilayer Perceptron achieved 88.2% accuracy, and the k-Nearest Neighbor classifier yielded the lowest accuracy at 82.5%.
Caglayan et al. [47] developed a plant leaf recognition system by utilizing 1897 images of 32 plant species from the Flavia dataset. The study utilizes the classifiers: Support Vector Machine, K-Nearest Neighbor, Naive Bayes, and Random Forest. The research involved random sampling and 10-fold cross-validation, and through the exploration of various shape and color feature combinations, an impressive accuracy of 96.32% was achieved using the Random Forest classifier.
Gokhale et al. [74] utilized the Flavia dataset and compared machine-learning algorithms namely K-NN, Logistic Regression and Naive Bayes. The authors extracted shape, color and texture features. While testing the models Logistic Regression outperformed K-NN and Naive Bayes by having an accuracy of 83.04%. While as Munisami et al. [69] employed the Flavia dataset solely for validation. An extensive and diverse dataset named “Folio” was introduced, featuring 32 plant species. This dataset was meticulously crafted by collecting leaf images from Mauritius University farms and nearby locations. The study incorporated various preprocessing steps, including rotation, greyscaling, thresholding, and the extraction of shape and color features. K-NN classifier initially achieved 83.5% accuracy on shape features, and when shape features combined with color histogram information, the accuracy was improved by showing 87.3%. The model’s accuracy exhibited a range from 86% to 91.1% when validated on the Flavia dataset. These insights underscore the significance of dataset selection, creation of comprehensive datasets, importance of effective preprocessing techniques, and the potential variations in accuracy when validating models across different datasets. These all contribute to the ongoing refinement of plant species identification systems. Kumar [75] developed a plant classification model based on leaf morphological features by employing K-Nearest Neighbor, Decision Tree, and Multilayer Perceptron algorithms. The training and test data were split using three methods: 80:20, 3-fold, and 5-fold cross-validation. Among all the experiments, Multilayer Perceptron outperformed K-NN and Decision Tree classifiers. Results showed an accuracy of 94.89% with the 80:20 split, 94.11% with 3-fold and 95.38% with 5-fold cross-validation. A favorable accuracy of 95.42% was achieved by Multilayer Perceptron with Adaboost methodology and 5 fold cross validation. In Table 5, we provide Comparison and summary of the reviewed literature for the plant species identification based on machine learning methods/algorithms applied, Objective, dataset details which include Dataset(s) used (DU), Primary Dataset (PD), total images (T.I), Species (Sp) and Results which include accuracy (Acc.), Recall (Rec.) and precision (Pre.). Ref. means reference of the research work.
4.2 Deep Learning Based Plant Classification
The adoption of deep learning over traditional machine learning techniques signifies a shift towards more accurate identification systems. Unlike machine learning, deep learning models, including Convolutional Neural Networks (CNNs), Recurrent Neural Networks (RNNs), and renowned architectures like AlexNet, VGG, and ResNet, excel at automatically learning complex features and patterns directly from raw data. This subsection delves into this transition, highlighting how researchers leverage various datasets to train deep learning models. Several noteworthy studies based on deep learning approaches for automated plant image identification are highlighted as follows:
Jeon et al. [78] presented a CNN method for the classification of plants, using their leaf images of plants. GoogleNet was compared with its newly created variant or Model 2 by increasing the number of inceptions in the variant model. The authors resized Flavia dataset images to 229 × 229 size. Model 2 took a training time of 9 h and 18 min and showed an accuracy of 99.8% while googlenet took a training time of 8 h and 43 min and an accuracy of 99.6% was seen. Evaluation of the models was carried out for the deformed leaves and the discolored ones. Model 2 performed better in both aspects, i.e., with respect to leaf damage and discoloration, and showed an improved performance of 98.4% in the case of leaf damage and 99.65% in the case of discolored ones. It was noted that when the discoloration of leaves is increased then the recognition rate of discoloration is degraded.
Oppong et al. [79], introduced OTAMNET, a computer vision model designed for medicinal plant classification. Their approach involved the creation of a comprehensive dataset, named “MyDataset,” comprising images from 49 distinct plant species. Leveraging transfer learning with ten pre-trained networks, they identified that DenseNet201 exhibited the highest performance, achieving an accuracy of 87%, while GoogleNet displayed the least favorable performance with an accuracy of 79%. Notably, the authors enhanced DenseNet201 by integrating the Log Gabor layer, a technique known for its capacity to efficiently capture texture and structure patterns at multiple scales and orientations in images. This integration significantly boosted performance, resulting in an impressive 98% accuracy on MyDataset. Moreover, OTAMNET’s exceptional accuracy extended to other diverse datasets, achieving remarkable results such as 100% accuracy on the Swedish leaf dataset, 99% on Flavia and MD2020, and 97% on Folio. This study underscores the power of OTAMNET in achieving precise plant classification, with the Log Gabor layer playing a pivotal role in enhancing its efficacy across various datasets. Deep learning models often require pre-processing techniques and large amounts of data in order to learn effectively.
Weng et al. [60] performed data augmentation of training data using deformations like blurring, scaling, perspective transformation, up-down shift, and brightness adjustment to increase the dataset of 2200 images from 11 herb categories collected by the authors. An accuracy of 96% was achieved using 5-fold cross-validation. Bisen [80] also performed data augmentation like rotation, zoom, etc., to increase the dataset. For the proposed study, the Swedish leaf dataset was used. The authors proposed a technique with 97% accuracy to automatically recognize plants from their leaf images using Deep CNN.
The study [61] introduced a Dual Deep Learning Architecture (DDLA) for plant identification, combining feature vectors from MobileNet and DenseNet-121 models. This innovative approach incorporated lightweight yet high-performance models, and their accuracy was assessed using primary dataset named Leaf-12, as well as standard datasets like Flavia, Folio, and Swedish Leaf. Notably, DDLA paired with a Logistic Regression classifier outperformed other classification models, achieving an impressive accuracy of 99.39% on the Leaf-12 (Primary dataset). The studies highlight the advantages of leveraging lightweight yet effective deep learning models, as well as innovative feature selection techniques, to achieve remarkable accuracy in plant classification and recognition tasks.
Paulson et al. [63] developed an automatic leaf image classification system, comparing the performance of CNN, VGG-16, and VGG-19. They used CNN with a 3 × 3-kernel size and ReLU activation for feature extraction, and employed pre-trained models VGG-16 and VGG-19. The dataset included 64,000 leaf images from sixty-four medicinal plants, divided into 80% for training, 10% for validation, and 20% for testing. VGG-16 outperformed, achieving an accuracy of 97.8%. The advantage of this approach lies in the efficiency and accuracy gained through pre-trained models and advanced convolutional neural networks, making it a robust solution for medicinal plant leaf image classification.
Vo et al. [62] employed a VGG16-based approach to automatically extract features from leaf images, focusing on a diverse dataset of herbal plants from Vietnam. By modifying the VGG-16 network, they efficiently handled a dataset containing over 10,000 leaf images. The key highlight of their work lies in the adoption of a range of classification techniques, including Random Forest, KNN, SVM, AdaBoost, Logistic Regression, XGBoost, and LightGBM. Notably, the LightGBM classification method, when combined with deep learning features, emerged as a powerful tool, achieving a remarkable recognition rate of 93.6%. LightGBM, a highly efficient gradient boosting framework based on decision trees, is designed to be fast, scalable, and distributed, making it well suited for large-scale machine learning tasks. Its key features include Gradient-based One-Side Sampling (GOSS) for handling data imbalance and Exclusive Feature Bundling (EFB) for reducing memory usage. This study emphasizes the potential of utilizing deep learning features and advanced classification methods, like LightGBM, to significantly enhance plant recognition tasks.
4.3 Transfer Learning Based Plant Classification
Deep learning algorithms automatically extract insights from the input data. For image identification, these models typically demand a substantial amount of high-quality images for accurate and reliable identification. An adequate dataset may not always be accessible to researchers for experimentation, which may result in inappropriate or inefficient performance. The transfer learning approach refers to the use of efficient networking models, pre-trained on a substantial amount of data. It enables a model to learn features of a new task rather than training it from scratch on new data [81], so addresses the challenge of the limited dataset. Researchers have shown great interest in using transfer learning for plant identification, as it significantly minimizes the computational cost of the identification process [64,82,83]. The study by Kaya et al. [82] delves into the profound impact of transfer learning on plant classification, offering valuable insights into this field. Their research involved the utilization of datasets from UCI, Flavia, Swedish leaf, and PlantVillage, each with varying sample sizes, and a consistent 30% testing and 70% training data split. Initially, a CNN developed from scratch yielded the highest accuracy, achieving an impressive 97.40% on the PlantVillage dataset. Fine-tuning the CNN across datasets showcased promising cross-data accuracy, with the best result at 96.93% when PlantVillage served as the test dataset. Further enhancement came from the use of pretrained models, including Alexnet and VGG16, outperforming other methods and achieving the highest accuracy at 99.80% on the PlantVillage dataset. Notably, SVM and LDA, as classifiers, excelled when combined with VGG16, attaining an impressive accuracy of 99.1% on the Flavia dataset. Lastly, a CNN-RNN approach yielded the best result, reaching an accuracy of 99.11% on the Swedish leaf dataset. The study ultimately underscores the remarkable potential of transfer learning in significantly improving plant classification models. In Beikmohammadi et al.’s study [84], transfer learning is applied to the well known Flavia and Leafsnap datasets, demonstrating the versatility of the MobileNet architecture. With Logistic Regression as the classifier, the study reports an impressive 99.6% accuracy on the Flavia dataset, highlighting the model’s ability to precisely classify plant species.
The significance of transfer learning extraction, and advanced classification techniques are at the forefront and empower the development of robust and accurate plant identification systems [25]. Roopashree et al. [64] introduced the DeepHerb medicinal plant identification model, utilizing a dataset containing 2515 leaf images from 40 different plant species in southern India. They applied transfer learning with pre-trained networks including VGG16, VGG19, and Xception extracting features and employing ANN, SVM, and SVM with Batch Optimization (BO) as classifiers. The model demonstrated impressive real-time image accuracy of 97.5%, achieved when trained using Xception and ANN, and was deployed as a mobile application named Herbsnap, offering top-5 accuracy of 95%. In the research studies [18,85–87], all have utilized the same dataset named Mendeley Medicinal Leaf [32] for plant classification. The dataset consists of leaf images across 30 distinct classes. These studies emphasize that the choice of classification approach can significantly impact the results. In a study [86], conducted by Patil et al., various pre-trained models, including VGG-19, VGG-16, MobileNet V2, and MobileNet V1, are used to extract features. Through extensive experimentation, MobileNet-V1 stands out as the most effective feature extractor, achieving an impressive accuracy of 98% on the Mendeley medicinal leaf dataset. This study emphasizes the advantages of MobileNet-V1, which overcomes challenges like the vanishing gradient problem in deep convolutional networks, ultimately leading to improved accuracy in medicinal plant classification. The study [87] contributed to the field of medicinal plant leaf identification by exploring distinctive features, with VGG16, VGG19, and MobileNetV2. This research enhances the algorithm through fine-tuning, offering a comparative analysis of model performance with and without this strategy. During experimentation, MobileNetV2 stands out as the most successful model, achieving a testing accuracy of 81.82%.
Almazaydeh et al.’s study [85] showcased the power of Mask R-CNN, a cutting-edge framework that enhances medicinal plant classification. By utilizing the region proposal network, RoI pooling, RoI align, and various network heads, the model delivers superior results in identifying 30 medicinal plant species, surpassing classical and alternative deep learning methods. This approach not only ensures precise object detection, mask generation, and class labeling but also exhibits an average accuracy of 95.7%. The advantages of Mask R-CNN [88] become evident as it excels in extracting image features, particularly when using a ResNet with 101 layers and Feature Pyramid Network (FPN). The segmentation masks generated by the system further elevate its capabilities, and the exceptional accuracy of this technique highlights its superiority over conventional machine learning and other deep learning models. Laiali’s work with Mask R-CNN paves the way for innovative applications, including plant disease diagnosis and growth rate analysis, underscoring the remarkable potential of this method in medicinal plant identification.
The study by Hajam et al. [18] took center stage, demonstrating the efficacy of transfer learning and fine-tuning with VGG16, VGG19, and DenseNet201. To enhance predictive performance, the study employs ensembling techniques, creating four hybrid models through averaging and weighted averaging strategies. After quantitative experimentation, it was revealed that the ensemble of VGG19 + DenseNet201, with fine-tuning, excels in identification, outperforming its standalone counterparts and achieving an accuracy of 99.12% on the test set.
The researches [18,85–87] concluded that transfer learning, fine-tuning and ensemble-learning strategies significantly enhances accuracy during the validation and testing phases, providing valuable insights for the field and reinforcing the effectiveness of these techniques for medicinal plant classification.
In the study [65], Abdollahi presented a novel approach for plant identification process. The study leverages a modified MobileNetV2 architecture with transfer learning, aiming to achieve remarkable accuracy in this task. With a dataset encompassing 3000 leaf images from thirty different medicinal plants, the research embraces data augmentation techniques such as rotation and Gaussian blur to enhance the dataset’s diversity. The outcome is an impressive accuracy rate of 98.05%, marking a substantial advancement in medicinal plant species identification. Table 6 shows the comparison of the reviewed literature for the plant species identification based on deep learning methods/algorithms applied, Objective, dataset details (which include Dataset(s) used (DU), Primary Dataset (PD), total images (T.I), Species (Sp) and Results that includes accuracy (Acc.), Recall (Rec.), Precision (Pre.)). Ref. means reference of the research work.
5 Comparative Analysis of Research Studies
In the previous sections, we have discussed the wide array of approaches introduced by different studies, highlighting the diversity in experimental designs. These variations encompass differences in the plant species examined, the features analyzed, and the classification methods employed. As a result, making direct comparisons, both in terms of results and the proposed methodologies, becomes a complex task. To facilitate a more straightforward evaluation, we have selected primary studies that share a common dataset, allowing us to provide a comparative analysis of their findings.
5.1 Studies Based on Flavia Dataset
The Flavia dataset serves as a benchmark for researchers to assess and compare various methods in their studies. It comprises images of leaves from 32 distinct plant species. Primary studies made use of this dataset and a wide range of classification methods to conduct their experiments. In numerous studies, several algorithms have been recurrently employed, displaying their versatility and effectiveness in addressing classification challenges. These methods encompass Support Vector Machines (SVM) [47,89–93,83] K-Nearest Neighbors (K-NN) [35,47,75,76,93,94]. Artificial Neural Networks with backpropagation (BPNN) [34,57], Probabilistic Neural Network (PNN) [66,95], Naive Bayes [47], Random Forest [47], Decision Tree [75,94], Multilayer Perceptron (MLP) [75], Convolutional Neural Networks (CNN) [37], and various CNN architectures [61,79,82].
The lowest classification accuracy of 25.30% was achieved using SVM when leaf shape analysis was performed with Hu moments [89], indicating that the Hu descriptor is not robust when relying solely on leaf shape. Mahajan et al. [83] proposed an automatic plant species identification system using this dataset. They extracted various morphological features from the leaf images and employed multiple classifiers for classification. Their results showed that SVM, when combined with AdaBoost, outperformed other classifiers, achieving an accuracy of 94.72%.
A study by Prasad et al. [94] applied K-NN and Decision Tree algorithms on the Flavia dataset. When examining both shape and color information, K-NN outperformed Decision Tree, and the inclusion of color information significantly boosted accuracy from 84.45% (using shape features only) to 91.30% (using a combination of shape and color features). Similarly, Ali Jan Ghasab et al. [91] achieved an impressive accuracy of 96.25% using SVM by fusing shape, color, texture, and vein features. Another study [92] further improved accuracy to 97.18% by employing Zernike moments for shape feature extraction and HOG for texture feature extraction. The increase in accuracy from 96.25% to 97.18% was attributed to Zernike moments’ ability to capture robust shape information and HOG’s effectiveness in extracting local gradient texture details.
Priya et al. [93] compared SVM with the RBF kernel to K-NN based on shape and vein features and found that SVM outperformed K-NN, achieving an accuracy of 94.5% compared to K-NN’s 78%. Caglayan et al. [47] conducted experiments with four classification algorithms: K-NN, SVM, Naive Bayes, and Random Forest, utilizing both shape and color features. Their results indicated that Random Forest consistently delivered the best classification performance, with SVM achieving the lowest accuracy of 72% using only shape features. However, combining shape and color features led to significant improvements in classification results, with Random Forest achieving the highest accuracy of 96.32%.
In a study by Hossain et al. [95], shape features were used with a Probabilistic Neural Network (PNN), resulting in an accuracy of 91.40%. Aakif et al. [57] applied backpropagation neural networks (BPNN) for plant leaf classification and attained an accuracy of 96%. Another study by Saleem et al. [35] emphasized the importance of feature combination and dimensionality reduction. In their research, the K-NN classifier outperformed Decision Tree and Naive Bayes, achieving 98.75% accuracy on the Flavia dataset and 97.25% on the primary dataset.
Kumar [75] compared K-NN, Decision Tree, and Multilayer Perceptron classifiers on the Flavia dataset and found that the learning capability of the Multilayer Perceptron was significantly enhanced after incorporating AdaBoost. MLP with AdaBoost outperformed other classifiers, achieving an accuracy of 95.40%.
Several studies have explored the use of deep learning on Flavia dataset to extract discriminative features from leaf images. These studies demonstrated that convolutional neural networks (CNNs) outperform traditional handcrafted feature extraction and classification methods [37,61,79,82,84]. A state-of-the-art CNN approach proposed by [37], achieved an accuracy of 97.9% by extracting deep features from the dataset. Kaya et al. [82] utilized a transfer learning approach, employing two pretrained CNN architectures, AlexNet and VGG16, for feature extraction. In their study, classification using VGG16 combined with LDA outperformed other approaches, achieving an accuracy of 99.10%
Raj et al. [61] compared various machine learning and deep learning approaches, showing promising results. Their study revealed that concatenating features extracted with MobileNet and DenseNet121, followed by classification with logistic regression, achieved the highest accuracy of 98.71%. Oppong et al. [79] demonstrated that integrating Log-Gabor layers into the transition layers of DenseNet201 significantly enhanced feature extraction, particularly for images with complex texture patterns, resulting in an accuracy of 99% on the Flavia dataset.
Among the studies of deep learning approaches, an impressive accuracy of 99.9% was achieved when Beikmohammadi et al. [84] employed transfer learning with MobileNet for feature extraction and Logistic Regression was used as classifier. Table 7 presents a comparative analysis of the different techniques employed by primary studies using the Flavia dataset.
5.2 Studies Based on Swedish Leaf Dataset
The Swedish leaf dataset is widely used by researchers for automated plant identification tasks. Table 8 provides a comparative analysis of various techniques applied by primary studies using this dataset. The lowest accuracy, 85.7%, was reported by Xiao et al. [97], who examined the effect of removing the leafstalk (petiole) from leaf images. Initially, they achieved 93.73% accuracy, which dropped to 85.7% when the leafstalks were removed, suggesting that the leafstalk provides useful information for classification. However, the reliability of the leafstalk is debated, as its length and orientation depend on the specifics of the collection and imaging process. Despite this, the highest classification accuracies were achieved by studies [79,82,98,99].
Researchers have employed a variety of classification algorithms on this dataset. These include Support Vector Machines (SVM) [55,56,92,97,100], K-Nearest Neighbors (K-NN) [98,101], Fuzzy K-NN [99], and Convolutional Neural Networks (CNN) [79,80]. Anubha and Sathiesh [61] combined features extracted with MobileNet and DenseNet-121, achieving 99.41% accuracy with Logistic Regression as the classifier. Ling et al. [101] used the Shape Context descriptor proposed by Belongie et al. [102] to extract features and employed K-NN for classification, achieving 88.12% accuracy.
Mouine et al. [98] explored two multi-scale triangular approaches for leaf shape description: Triangle Area Representation (TAR) and Triangle Side Length Representation (TSL). They found that while TAR is affine invariant, it is computationally expensive and less precise. TSL, on the other hand, is transformation-invariant and achieved a higher classification accuracy (95.73%) than TAR (90.40%). The study also introduced Triangle-Oriented Angles (TOA), which achieved 95.20% accuracy, and a combined descriptor called Triangle Side Lengths and Angles (TSLA), which further improved accuracy to 96.53%.
Tsolakidis et al. [92] achieved 98% accuracy by combining shape and texture features, demonstrating the effectiveness of Zernike moments and Histogram of Oriented Gradients (HOG) for feature extraction. Ali et al. [56] reported an accuracy of 99.40% using SVM, with texture features extracted through Bag of Features (BOF) and Local Binary Patterns (LBP). Ren et al. [100] compared Inner Distance Shape Context (IDSC) and multi-scale overlapped block LBP. While IDSC is detailed but computationally expensive, LBP proved more efficient, achieving 96.67% accuracy with SVM using a Radial Basis Function (RBF) kernel, known for its ability to handle high-dimensional, non-linearly separable data.
Wang et al. [99] employed dual-scale decomposition and local binary descriptors (DS-LBP) for leaf classification. DS-LBP descriptors effectively integrate texture and contour information, providing invariance to translation and rotation. To address the challenge of determining an appropriate k value, typically based on error rates, they proposed a fuzzy k-Nearest Neighbors classifier (fuzzy k-NN) to enhance robustness. This demonstrates the effectiveness of DS-LBP descriptors, remaining invariant to translation and rotation and authors managed to achieve an accuracy of 99.25%. Kaur et al. [55] achieved an accuracy of 93.26% by combining texture and color features. For texture feature extraction, they used the Grey-Level Co-Occurrence Matrix (GLCM), which captures the spatial relationships of pixel intensities in an image, providing valuable texture information. Additionally, color features were extracted using statistical measures such as mean, standard deviation, kurtosis, and skewness. These statistical measures offer insights into the distribution and variation of pixel intensities, contributing to the characterization of color properties in the leaf images. The feature combination proved effective, enabling accurate multiclass classification using a Support Vector Machine (SVM).
Several studies have employed deep learning approaches for plant species classification. Raj et al. [61] achieved 94.41% accuracy by concatenating features from MobileNet and DenseNet-121 with Logistic Regression. Kaya et al. [82] used transfer learning with VGG16 and Linear Discriminant Analysis (LDA), achieving 99.11% accuracy. Bisen [80] proposed a Deep CNN with data augmentation, achieving 97% accuracy. Oppong et al. [79] demonstrated the highest accuracy of 100% by enhancing feature extraction in DenseNet201 with Log-Gabor layers. These studies collectively display effective deep learning approaches for plant species classification, with the novel technique from Stephen et al. yielding the maximum accuracy.
5.3 Studies Based on Mendeley Medicinal Leaf Dataset
The Mendeley Medicinal Leaf dataset [32] was published in a public repository on 22 October 2020. This dataset contains images with backgrounds removed, allowing machine learning and deep learning models to focus on specific identification tasks. The research studies utilizing this dataset extracted using deep convolutional neural networks. Table 9 summarizes the research studies that used this dataset for their experimentation. Fig. 12 displays the results from various studies on the Mendeley Medicinal Leaf dataset using deep learning algorithms. The graph reveals significant differences in accuracy across studies, highlighting the effectiveness of different AI methodologies in plant identification. Recent advancements, such as those by Hajam et al. [18] have achieved notable improvements in accuracy, reaching 99.12% using advanced techniques like ensembled CNNs with architectures including VGG19 and DenseNet201.

Figure 12: Performance comparision of classification methods/algorithms on Mendeley medicinal leaf dataset
5.4 Studies Based on Leafsnap and Folio Datasets
We included studies on the LeafSnap and Folio datasets for comparative analysis. However, due to the limited research conducted on these datasets, fewer insights have been derived. Table 10 and Fig. 13 summarize the research studies utilizing the LeafSnap and Folio datasets, where various machine learning and deep learning techniques were applied. The highest accuracy of 97.8% for the LeafSnap dataset was achieved by Barré et al. [37], using deep feature extraction. For the Folio dataset, the best classification results were obtained by Oppong et al. [79] who employed a Log-Gabor filter and DenseNet201. These results emphasize the importance of extracting relevant texture features from leaf images for effective leaf pattern recognition.

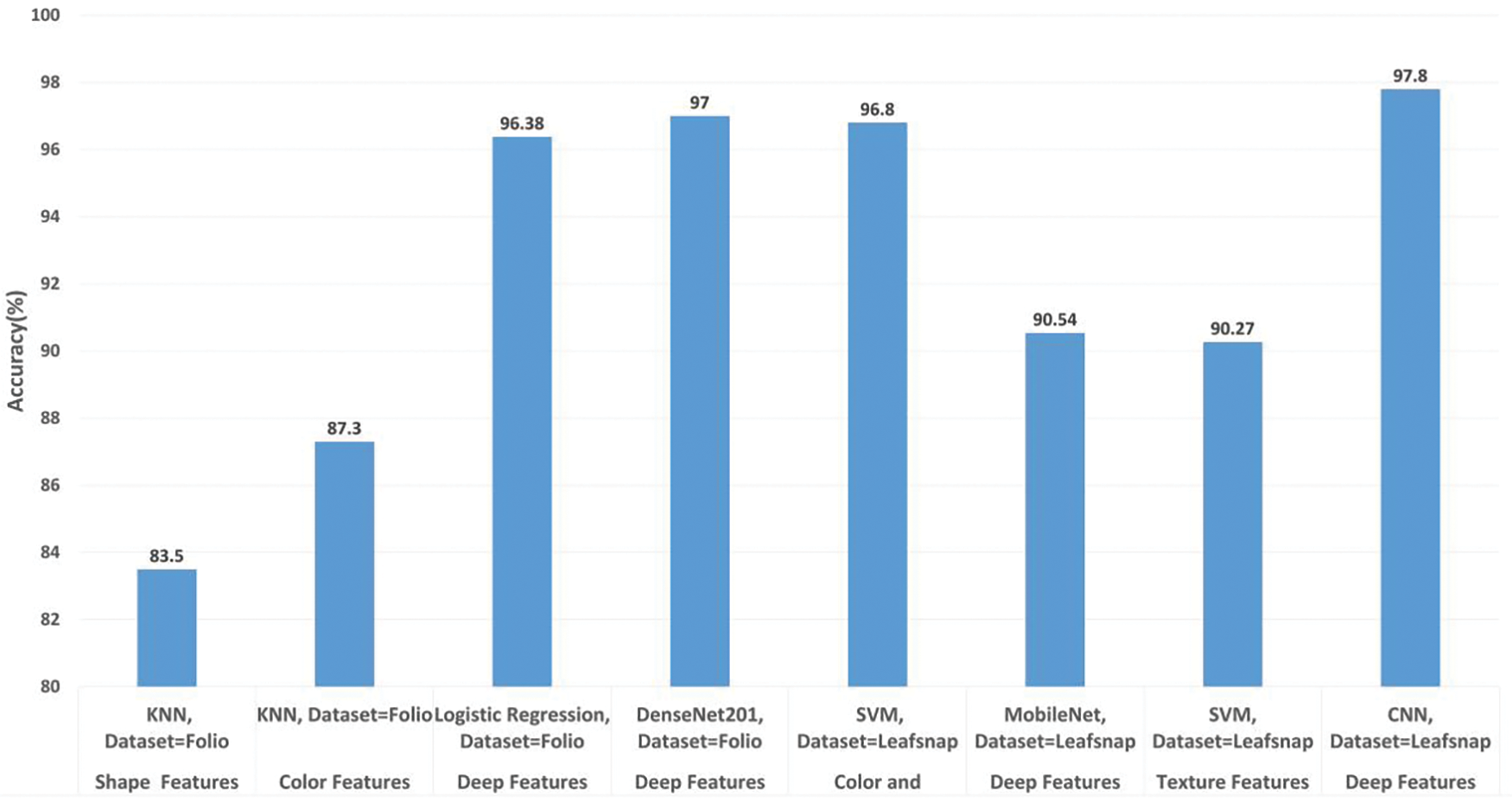
Figure 13: Studies based on leafsnap and folio datasets, employing different algorithms and features
6 Discussion and Future Research Directions
This exhaustive literature review systematically examines diverse AI approaches, emphasizing their effectiveness in the identification process and highlighting their significant role in advancing techniques for plant identification. With this foundation in place, let us now turn our attention to the valuable lessons learned from this comprehensive literature review. Our exploration will illuminate the current state of the field while outlining promising pathways for future research and development.
The principal findings of this systematic review can be summarized as follows:
• Figs. 14 and 15 highlight studies that utilize self-collected datasets for plant identification tasks. As shown in Fig. 15, only two studies have evaluated their methods on large, primary datasets with a realistic number of species [62,63]. It is crucial to develop plant identification methods capable of handling high variability of data, given that there are over 21,000 plant species with therapeutic potential. The significant increase in dataset sizes between 2017 and 2020 reflects a shift towards larger, more comprehensive datasets. This trend indicates advancements in data collection methods, improvements in computational infrastructure, and promises to enhance the performance of deep learning algorithms.
• Leaves are the most widely studied plant organ for AI based plant identification. It is because leaves are available for examination throughout most of the year, and are easy to find and to collect. Leaves can also easily be imaged compared to other plant morphological structures, such as flowers, barks, or fruits [104].
• Most of the studies utilized images with plain backgrounds, captured in controlled environments to avoid segmentation. In real-world applications, it is essential for studies to incorporate more realistic images with complex backgrounds and varying lighting conditions.
• Handcrafted feature selection mostly included shape and texture features as these features offer valuable insights into the overall morphology and structure of leaves. Shape features capture properties such as area, perimeter, circularity, and aspect ratio, which are distinctive characteristics of different plant species. Texture features encapsulate valuable information about the surface characteristics of leaves, such as roughness, smoothness, or vein patterns, which can vary significantly between different plant species. Unlike color features, these features remain relatively stable across different environmental conditions, making them reliable markers for species discrimination.
• To ensure a standardized comparison of AI methods, the highest reported accuracies by a particular AI approach is presented in Fig. 16 and in Fig. 17 for the Flavia and Swedish leaf datasets, respectively.
Figure 14: Variability in plant image primary dataset sizes among research studies employing machine learning
Figure 15: Variability in plant image primary dataset sizes among research studies employing deep learning methods
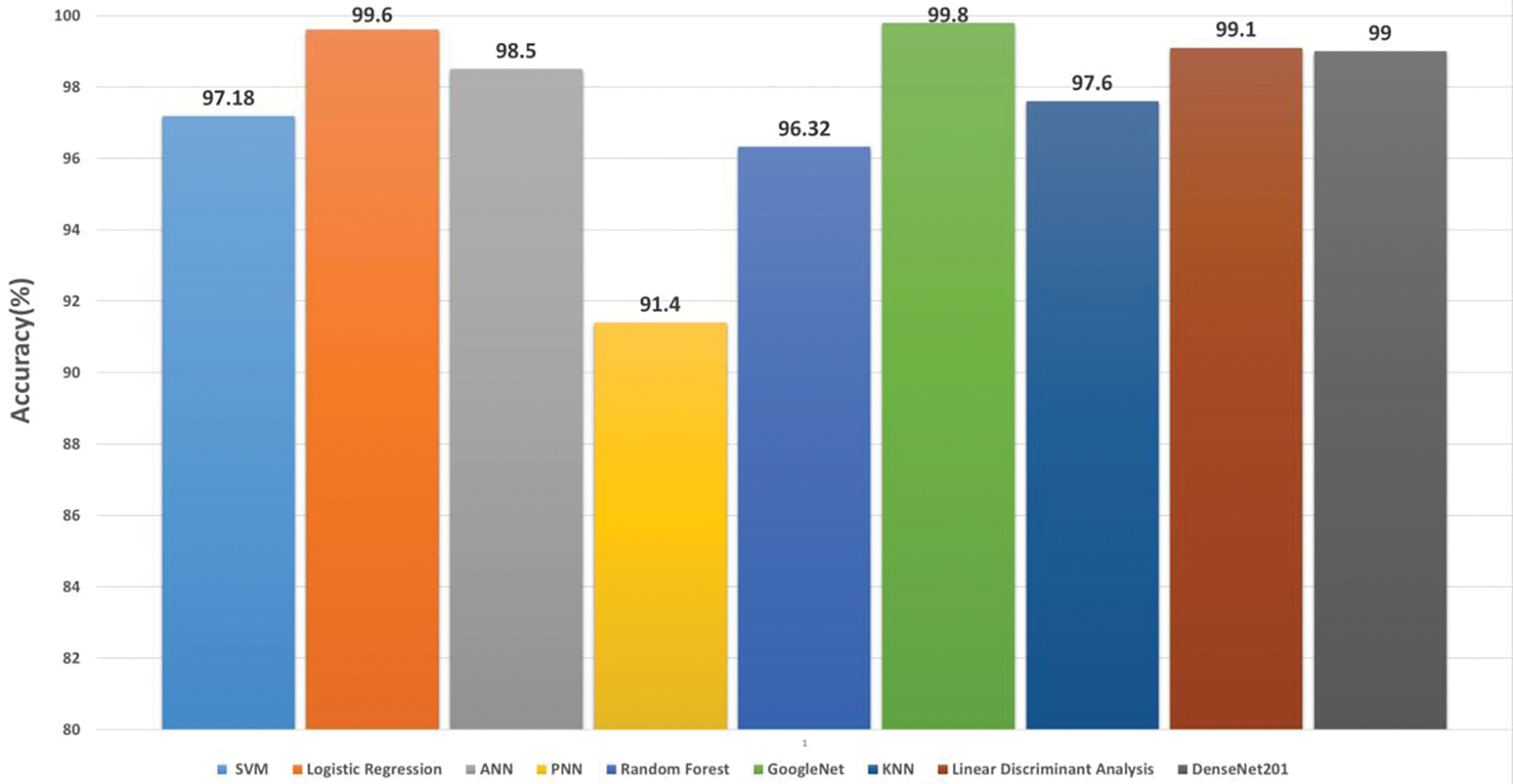
Figure 16: Top accuracies achieved by classifiers for the Flavia dataset
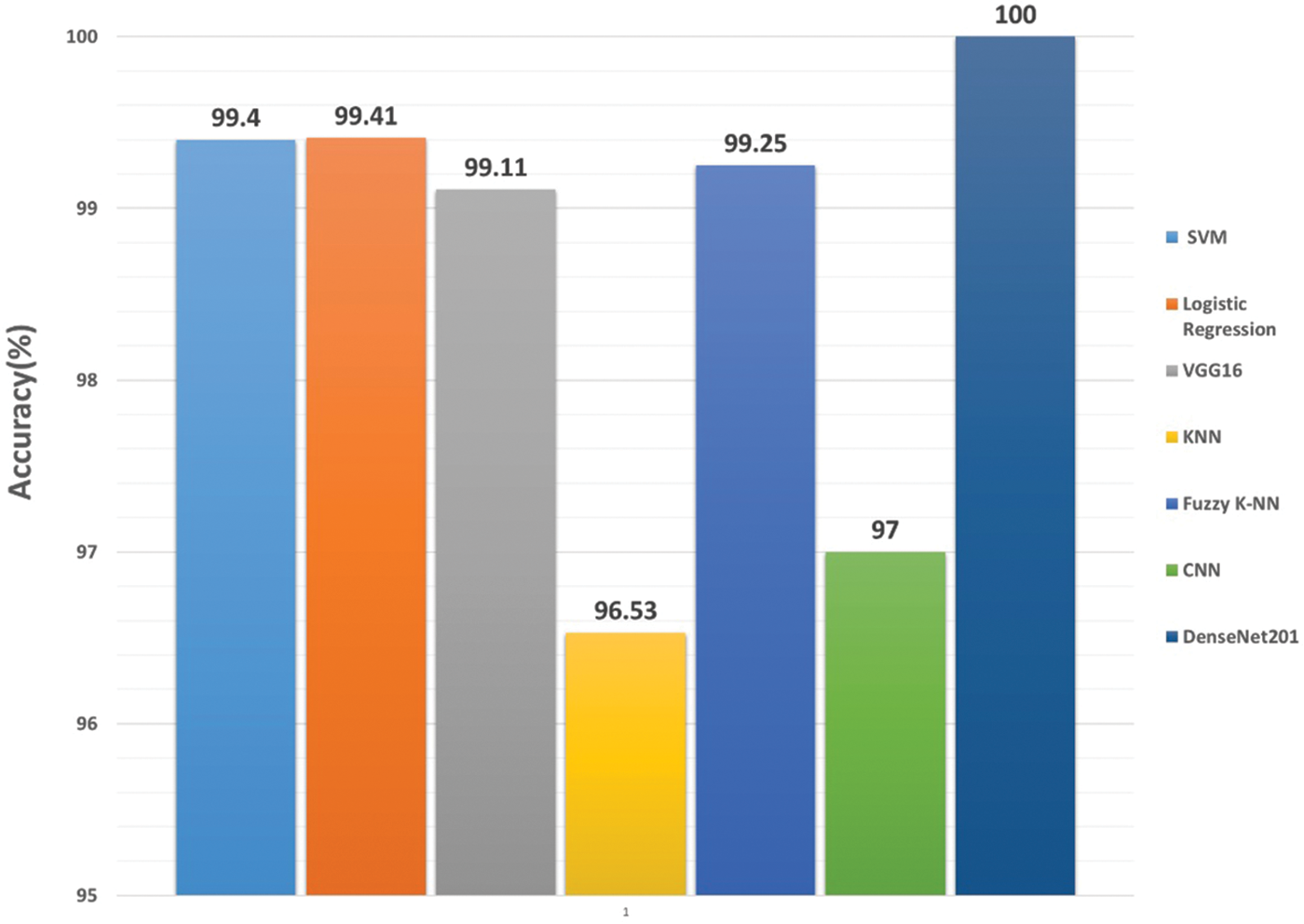
Figure 17: Top accuracies achieved by classifiers for the Swedish leaf dataset
This methodology allows for a fair assessment of their capabilities, shedding light on their potential in accurately classifying plant species. By focusing on the best-performing instance of each algorithm, researchers can gain valuable insights into their comparative effectiveness and applicability in diverse contexts. The performance of automated plant identification systems has undoubtly seen a notable improvement since the 2016 PlantCLEF challenge [105]. These systems were based on Convolutional Neural Networks (CNN). For each of publicly available, mentioned dataset, the best classification results were achieved by CNN architectures [18,37,56,79]. This is aslo clear from Fig. 18, where we provide an insightful overview of the methods used for the Flavia dataset identification. Studies employing CNN and its architectures achieved the highest accuracies in plant identification tasks [78,79,82,84]. This underscores the efficacy of deep learning methodologies, particularly convolutional neural networks, in extracting intricate features from plant images, leading to more precise and reliable classification outcomes. By leveraging CNN-based approaches, researchers can enhance the accuracy and robustness of plant identification systems, thereby advancing the field’s capabilities in species recognition and classification.
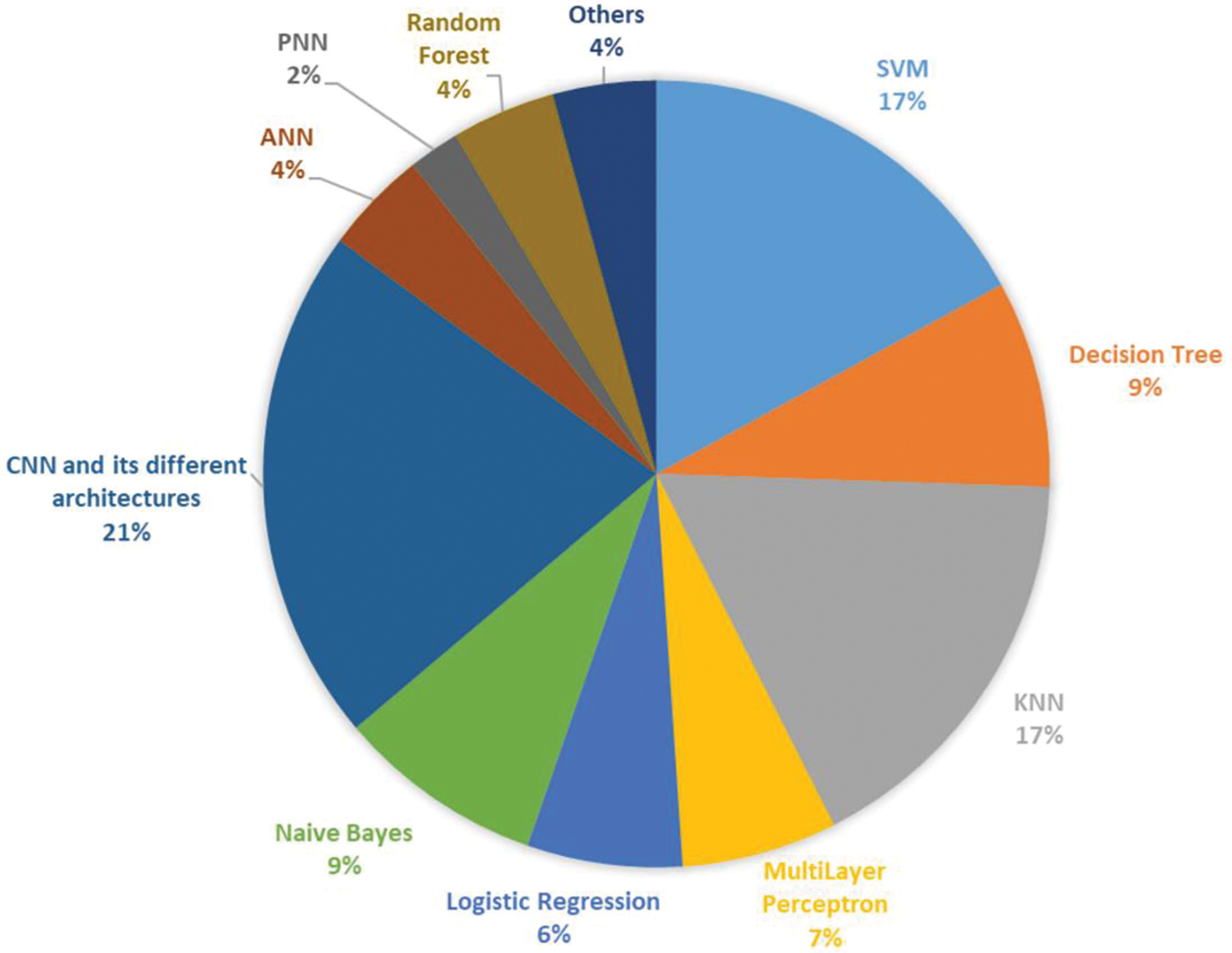
Figure 18: Algorithm/method distribution for Flavia dataset
Following a thorough comparison of machine learning and deep learning approaches, we propose several key improvements to effectively tackle the pattern recognition challenges in medicinal plant identification. To enhance the accuracy and generalization of these systems, expanding datasets is essential, as many medicinal plants remain unexplored. Video-based identification should be considered, as it captures dynamic features like 3D structure and varying lighting conditions, providing more context than static images. Furthermore, identifying plants with leaves attached to branches and including other plant parts, such as flowers, fruits or bark, can improve accuracy and reflect real-world conditions where multiple features contribute to identification. We recommend leveraging a diverse set of advanced AI methods to create a robust and accurate identification system. Convolutional Neural Networks (CNNs) are particularly well-suited for this task due to their exceptional ability to analyze and interpret visual data from plant images. CNNs excel in automatically learning and extracting features such as shapes, textures, and colors, which are crucial for distinguishing between different plant species. We also suggest incorporating transfer learning for disease detection, which not only identifies plants but also diagnoses potential issues and offers actionable treatment advice. Deep learning methods can be integrated with traditional machine learning approaches that can further enhance the system’s accuracy and efficiency. A hybrid approach can be developed which will combine CNN-extracted features with traditional classifiers—such as Support Vector Machines (SVMs) or Random Forests. This will leverage the strengths of both methods and result in improved classification performance. Also, for a more comprehensive identification system researchers can consider adopting multi-modal approaches. These systems integrate image data with other relevant sources, such as textual descriptions or environmental sensor data, to provide a richer and more nuanced understanding of the plants. This fusion of diverse data types can lead to more accurate and reliable identifications. Also, incorporating attention mechanisms into deep learning models can significantly boost the system’s capability to focus on and prioritize critical features within the plant images. Attention mechanisms can help the model to concentrate on the most relevant aspects of the data which will lead to more precise and accurate identifications. By leveraging these advanced AI techniques, we can develop a more effective, accurate, and efficient system for identifying medicinal plants.
Traditional methods of plant identification rely heavily on human expertise and cultural knowledge. These methods suffer due to complexities, time constraints, and a reliance on scarce subject experts. AI presents a promising avenue for automating plant species identification, offering more efficient and accurate results. The findings of this comprehensive review and analysis, sheds light on the current state of automated plant identification and delineating pathways for future research and development.
6.3.1 AI-Based Future Research Trends
1) Researchers are exploring various architectures like ResNet, DenseNet for plant species identification. Improvements in neural networks have led to better performance, but storage and speed issues persist, limiting their use on mobile devices. Model lightweighting, achieved through designs like MobileNet [65] and Xception [64] addresses these challenges. MobileNet, favored in plant recognition, reduces computation while maintaining accuracy. However, retraining remains time-consuming, prompting the use of transfer learning with MobileNet models.
2) A key challenge in plant species identification is the limited availability of labeled data for training AI models. Current research largely relies on limited datasets, while only a few studies utilize CNN classifiers trained on comprehensive plant image datasets [63,106], demonstrating their effectiveness in automated plant species identification systems. Due to the scarcity of training data and the computational demands of CNN training, transfer learning has become a widely adopted strategy. This involves initially pretraining a classifier on a large dataset like ImageNet and then fine-tuning it for the specific classification task using a smaller, problem-specific dataset. Many previous studies on plant species identification have employed transfer learning [18]. Once a sufficiently large plant dataset becomes available, it would be interesting to compare the classification results with those obtained from a plant identification CNN trained solely on images depicting plant taxa. Another method to address the challenge of small datasets involves employing data augmentation techniques, which often include simple alterations to images like blurring, rotation, translation, flipping, and scaling. Data augmentation has become a standard practice in computer vision to enhance the training process [60]. However, traditional augmentation metods have limitations in generating diverse variations. This has led to the exploration of synthetic data samples, which introduce greater variability and enhance the dataset to improve training. Generative Adversarial Networks (GANs) [107] offer a promising solution in this context by generating high quality, realistic, natural images.
3) AI approaches typically aim for higher classification accuracy; however, the pursuit of high classification accuracy can be effectively realized through image preprocessing and feature extraction techniques. They refine the input data and extract relevant features that contribute significantly to achieving accurate species identification. We believe that incorporating image preprocessing before feature extraction can enhance performance. This was proven by the findings of [36].
4) Edge detection simplifies images by focusing on informative edges, potentially improving accuracy of AI models for tasks where object boundaries are crucial [43]. While edge detection algorithms like Sobel [108] and Canny [44] provide initial guidance, their limitations in real-world scenarios necessitate more robust solutions. Segmentation techniques, including thresholding and level set methods [41], address these challenges by partitioning the image into distinct regions, effectively isolating plant features from the background. The study [39] highlighted optimal segmentaion technique “OTSU” [40], and achieved an accuracy of 99%.
5) Employing machine learning techniques for feature engineering, researchers utilize a diverse set of features, including shape, color, texture, and geometric features, extracted from plant images. We are of the opinion that fusing the individual features can improve the classification accuracy, compared to using them individually. For example in a study [49], an enhanced accuracy (96.66%) was seen when geometric, color, texture and shape features were used in combination. Similarly leveraging feature fusion Kan et al. [50] achieved better classification results. While traditional handcrafted feature extraction methods have been widely used, there is a growing trend towards employing deep learning techniques for automatic feature extraction. Innovative approaches, such as integrating Log-Gabor layers into deep learning architectures, have shown promise in enhancing texture-sensitive feature extraction [79]. When selecting feature extraction methods and considering deep learning models, researchers must consider the computational demands of each technique. Balancing computational intensity with accuracy is crucial for developing practical and efficient plant species recognition systems.
6) Future trends in plant identification may involve creating identification systems with intuitive user interfaces, enabling users to identify plants anytime, anywhere, under diverse conditions. Additionally, adapting the model for real-time applications holds promise for various practical applications.
6.3.2 Medicinal Plant Based Future Research Trends
A significant hurdle is the need for extensive and high-quality datasets to train AI algorithms. The availability and quality of image datasets are pivotal for the performance of automated plant identification algorithms. Yet, challenges persist, especially in identifying medicinal plant species using primary datasets, as these self collected datasets often lack standardization and comprehensiveness, particularly for rare or less-documented plant species, creating a substantial research gap. To address these data limitations, following solutions can be considered:
1. Creating Standardized Medicinal Plant Dataset Collections: Current benchmark datasets suffer from limitations in both species diversity and image quantity (refer to Section 3) due to the immense effort required for specimen collection and imaging. These datasets, primarily crafted for computer vision and machine learning applications, are typically sourced by a small group of individuals over a short timeframe and from limited geographical areas, often resulting in representation from only a few closely grown individual plants of each species. Consequently, these datasets fail to reflect real-world conditions, hindering their utility in practical applications such as image classification across different times, locations, and imaging methods. Moreover, there is a pressing need to prioritize the creation of benchmark datasets that include a comprehensive representation of medicinal plant species. While existing datasets serve various purposes, they may lack coverage of the diverse range of medicinal plant species essential for applications such as medicinal plant identification, biodiversity conservation, and herbal medicine research. Incorporating images of medicinal plants into benchmark datasets is crucial for advancing research in this domain and facilitating the development of accurate and reliable machine learning models tailored to medicinal plant identification and analysis. Therefore, efforts to enhance benchmark datasets should prioritize the inclusion of representative images from medicinal plant species, ensuring broader applicability and relevance across different fields and industries.
2. Leveraging the Extensive Array of Herbarium Samples: Herbaria worldwide have invested significant resources in collecting and preserving plant samples. Instead of conducting fieldwork for image collection, utilizing existing specimens offers a cost-effective alternative. Presently, there are over 3000 herbaria across 165 countries housing over 350 million specimens collected over centuries. Many herbaria are digitizing their collections to enhance accessibility and preservation [109]. This initiative is expected to grow substantially in the coming decade. We anticipate the creation of publically available extensive online repositories for medicinal plant images containing taxonomic information. However, analyzing herbaria specimens may pose challenges for training AI models for real-world applications due to alterations in color and structure caused by drying and imaging processes. Hence, further research is needed to develop methods for detecting and extracting features from herbaria specimens. Additionally, there is an ongoing research question regarding the adaptation of classifiers trained on herbaria specimens for use on fresh specimens.
3. Crowdsourced Medicinal Plant Image Collection: Harnessing the collective efforts of people from diverse backgrounds, including rural communities with traditional knowledge of medicinal plants, allows us to compile a rich dataset that captures the diversity and variability of medicinal flora. Crowdsourcing has notably benefited from the evolution of Web 2.0 technologies, facilitating platforms like iNaturalist and Pl@ntNET to effectively gather data through user-generated content and social media engagement [110]. Through user-friendly platforms and mobile applications, individuals can easily upload images of medicinal plants traditionally used for various ailments. This approach not only enables the rapid acquisition of a large volume of image data but also fosters community engagement and preserves indigenous knowledge about medicinal plants. However, plant image collections obtained through crowdsourcing and citizen science encounter several challenges that hamper their usefulness as training and benchmark data. For instance, while thousands of images are available for prominent taxa, less common and rare taxa are often poorly represented or entirely lacking in images. Additionally, many images in these collections do not provide sufficient detail for unambiguous identification of the displayed taxon, exhibiting issues such as blurriness or lack of clarity. Furthermore, challenges arise from inconsistent tagging of plant organs, multiple synonyms for plant species, and evolving taxonomic classifications. These challenges underscore the need forincreased efforts to maintain data quality in crowdsourced content.
4. Hybrid Deep Learning Approaches: Hybrid approaches combine the strength of traditional machine learning algorithms with the advanced capabilities of deep learning models that create a powerful synergy that enhances both accuracy and efficiency. Traditional machine learning approaches, such as decision trees, random forests, and support vector machines, have long been used for pattern recognition and classification tasks. These methods rely on feature extraction and selection that involve manual identification and selection of relevant features from the data. These approaches do not perform well on complex and high-dimensional data. On the other hand, deep learning models process and analyze large amounts of data with minimal human intervention, making them highly effective for tasks involving image recognition and classification. In medicinal plant identification deep learning models can automatically extract and learn relevant features from plant images. By integrating these two approaches, hybrid methods leverage the strengths of both traditional and deep learning techniques. Hybrid systems may use traditional machine learning algorithms for initial feature extraction and selection, followed by deep learning models for refined classification and prediction. This combination can address the limitations of each approach and enhance overall performance. In the field of medicinal plant identification, hybrid methods can significantly improve the accuracy and efficiency of identification systems. Hybrid methods can adapt to different types of data, including images, text, and sensor data, making them versatile tools for a range of applications.
5. Collaborative Efforts: The field of automated plant species identification continues to be predominantly led by academics specializing in computer vision techniques, with limited involvement from interdisciplinary teams of biologists and computer scientists over the past decade [3]. However, for the successful approaches in this domain, there is a pressing need of collaborations between biologists and computer scientists possessing significant expertise in both biology and computing science [111]. Interestingly, there is a clear call for research to transition towards more interdisciplinary endeavors. Biologists stand to enhance the application of machine learning methods with the support of computer scientists, while the latter can deepen their understanding of the problem domain by collaborating closely with biologists. This collaborative approach holds great promise for advancing automated plant species identification techniques.
Medicinal plants have a long history of serving as cost-effective and safer alternatives to allopathic medicine. This comprehensive review underscores the pivotal role of medicinal plants in allopathic medicine. Traditional manual identification of these plants, although valuable, is hindered by its time-consuming and error-prone nature, heavily dependent on human expertise and perception. In contrast, artificial intelligence (AI) techniques have emerged as a transformative solution, addressing the complexities of image recognition with notable efficacy across various domains, including agriculture, healthcare, and quality control.
This rigorous review study meticulously examined AI-driven methods for leaf image-based plant identification, encompassing a range of techniques from classical machine learning algorithms such as Support Vector Machine (SVM) and K-Nearest Neighbor (KNN) to sophisticated deep learning approaches, particularly Convolutional Neural Networks (CNNs). The analysis highlighted significant advancements and promising results achieved through these AI methodologies. Moreover, it carefully summarizes relevant research on AI algorithms; features extracted, and achieved results. Additionally, it examines widely used datasets in automated plant classification, offering deep insights into techniques for recognizing medicinal plants. Moreover, it discusses the opportunities and challenges of using AI-based approaches. Furthermore, it highlights key statistical findings and lessons learned from the survey, along with new research areas, aiming to inspire readers and guide future research.
However, despite progress, there remains a substantial scope for further research and improvement. Future work should focus on enhancing the precision and robustness of plant recognition systems by leveraging state-of-the-art deep learning techniques and exploring the potential of transfer learning. Additionally, the development of extensive and diverse digital repositories encompassing a wide array of plant species is crucial. Such repositories would provide a rich data source, significantly enhancing the training and performance of AI models. Furthermore, collaborative efforts among researchers from various disciplines, including biosciences, medicine, and informatics, are essential to drive innovation and advancement in medicinal plant identification. This review is meant to be a fundamental resource for future researchers studying AI-based identification of medicinal plants.
Acknowledgement: The authors would like to acknowledge the support of Prince Sultan University for paying the Article Processing Charges (APC) of this publication.
Funding Statement: This work was supported by the EIAS Data Science Lab, College of Computer and Information Sciences, Prince Sultan University, Riyadh, Saudi Arabia.
Author Contributions: Conceptualization, Mohd Hajam, Tasleem Arif, Akib Khanday, Mudasir Wani and Muhammad Asim; Data curation, Mohd Hajam; Formal analysis, Mohd Hajam; Funding acquisition, Muhammad Asim; Investigation, Tasleem Arif, Akib Khanday, Mudasir Wani and Muhammad Asim; Methodology, Mohd Hajam, Mudasir Wani and Muhammad Asim; Resources, Tasleem Arif; Supervision, Tasleem Arif and Akib Khanday; Validation, Akib Khanday and Mudasir Wani; Visualization, Muhammad Asim; Writing—original draft, Mohd Hajam; Writing—review editing, Tasleem Arif, Akib Khanday, Mudasir Wani and Muhammad Asim. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Not applicable.
Ethical Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. M. S. Ahmad Khan and I. Ahmad, “Herbal medicine: Current trends and future prospects,” in New Look to Phytomed.: Adv. Herb. Products Novel Drug Leads, Jan. 2019, pp. 3–13. doi: 10.1016/B978-0-12-814619-4.00001-X. [Google Scholar] [CrossRef]
2. M. Javaid, A. Haleem, I. H. Khan, and R. Suman, “Understanding the potential applications of artificial intelligence in agriculture sector,” Adv. Agrochem., vol. 2, no. 1, pp. 15–30, 2023. doi: 10.1016/j.aac.2022.10.001. [Google Scholar] [CrossRef]
3. J. Wäldchen and P. Mäder, “Plant species identification using computer vision techniques: A systematic literature review,” Arch. Comput. Methods Eng., vol. 25, no. 2, pp. 507–543, 2018. doi: 10.1007/s11831-016-9206-z. [Google Scholar] [PubMed] [CrossRef]
4. R. Gupta, D. Srivastava, M. Sahu, S. Tiwari, R. K. Ambasta and P. Kumar, “Artificial intelligence to deep learning: Machine intelligence approach for drug discovery,” Mol. Divers., vol. 25, no. 3, pp. 1315–1360, 2021. doi: 10.1007/s11030-021-10217-3. [Google Scholar] [PubMed] [CrossRef]
5. O. A. Malik, N. Ismail, B. R. Hussein, and U. Yahya, “Automated real-time identification of medicinal plants species in natural environment using deep learning models—A case study from borneo region,” Plants, vol. 11, no. 15, 2022, Art. no. 1952. doi: 10.3390/plants11151952. [Google Scholar] [PubMed] [CrossRef]
6. A. C. Tran, N. T. Nhu, Y. P. K. Thoa, N. C. Tran, and N. Duong-Trung, “Real-time recognition of medicinal plant leaves using bounding-box based models,” in Proc.-2020 Int. Conf. Adv. Comput. App., ACOMP 2020, 2020, pp. 34–41. doi: 10.1109/ACOMP50827.2020.00013. [Google Scholar] [CrossRef]
7. D. Paul, G. Sanap, S. Shenoy, D. Kalyane, K. Kalia and R. K. Tekade, “Artificial intelligence in drug discovery and development,” Drug Discov. Today, vol. 26, no. 1, pp. 80–93, 2021. doi: 10.1016/j.drudis.2020.10.010. [Google Scholar] [PubMed] [CrossRef]
8. A. Gurib-Fakim, “Medicinal plants: Traditions of yesterday and drugs of tomorrow,” Mol. Aspects Med., vol. 27, no. 1, pp. 1–93, Feb. 2006. doi: 10.1016/J.MAM.2005.07.008. [Google Scholar] [PubMed] [CrossRef]
9. Md Ariful Hassan, Md Sydul Islam, Md Mehedi Hasan, S. B. Shorif, Md Tarek Habib and M. S. Uddin, “Medicinal plant recognition from leaf images using deep learning,” Comput. Vis. Mach. Learn. Agricul., vol. 2, pp. 137–154, 2022. doi: 10.1007/978-981-16-9991-7_9. [Google Scholar] [CrossRef]
10. M. Prasathkumar, S. Anisha, C. Dhrisya, R. Becky, and S. Sadhasivam, “Therapeutic and pharmacological efficacy of selective Indian medicinal plants–A review,” Phytomed. Plus, vol. 1, no. 2, May, pp. 1–28, 2021. doi: 10.1016/j.phyplu.2021.100029. [Google Scholar] [CrossRef]
11. Z. Wang, H. Li, Y. Zhu, and T. F. Xu, “Review of plant identification based on image processing,” Arch. Comput. Methods Eng., vol. 24, no. 3, pp. 637–654, 2017. doi: 10.1007/s11831-016-9181-4. [Google Scholar] [CrossRef]
12. M. Minervini, A. Fischbach, H. Scharr, and S. A. Tsaftaris, “Finely-grained annotated datasets for image-based plant phenotyping,” Pattern Recognit. Lett., vol. 81, no. 9, pp. 80–89, 2016. doi: 10.1016/j.patrec.2015.10.013. [Google Scholar] [CrossRef]
13. M. R. de Carvalho et al., “Taxonomic impediment or impediment to taxonomy? A commentary on systematics and the cybertaxonomic-automation paradigm,” Evol. Biol., vol. 34, no. 3–4, pp. 140–143, 2007. doi: 10.1007/s11692-007-9011-6. [Google Scholar] [CrossRef]
14. K. Pushpanathan, M. Hanafi, and S. Mashohor, “Machine learning in medicinal plants recognition: A review,” Artif. Intell. Rev., vol. 54, pp. 305–327, 2020. doi: 10.1007/s10462-020-09847-0. [Google Scholar] [CrossRef]
15. M. A. F. Azlah, L. S. Chua, F. R. Rahmad, F. I. Abdullah, and S. R. W. Alwi, “Review on techniques for plant leaf classification and recognition,” Computers, vol. 8, no. 4, pp. 1–22, 2019, Art. no. 77. doi: 10.3390/Comput.s8040077. [Google Scholar] [CrossRef]
16. Y. Chen et al., “Plant image recognition with deep learning: A review,” Comput. Electron. Agricul., vol. 212, pp. 1–17, 2023, Art. no. 108072. doi: 10.1016/j.compag.2023.108072. [Google Scholar] [CrossRef]
17. A. K. Katsaggelos, “Image identification and restoration based on the expectation-maximization algorithm,” Opt. Eng., vol. 29, no. 5, pp. 436–445, 1990. doi: 10.1117/12.55612. [Google Scholar] [CrossRef]
18. M. A. Hajam, T. Arif, A. M. U. D. Khanday, and M. Neshat, “An effective ensemble convolutional learning model with fine-tuning for medicinal plant leaf identification,” Information, vol. 14, no. 11, pp. 1–20, 2023, Art. no. 618. doi: 10.3390/info14110618. [Google Scholar] [CrossRef]
19. J. Wäldchen, M. Rzanny, M. Seeland, and P. Mäder, “Automated plant species identification—Trends and future directions,” PLoS Comput. Biol., vol. 14, no. 4, 2018, Art. no. e1005993. doi: 10.1371/journal.pcbi.1005993. [Google Scholar] [PubMed] [CrossRef]
20. M. Nawaz, T. Nazir, A. Javed, S. Tawfik Amin, F. Jeribi and A. Tahir, “CoffeeNet: A deep learning approach for coffee plant leaves diseases recognition,” Expert. Syst. Appl., vol. 237, pp. 1–14, 2024, Art. no. 121481. doi: 10.1016/j.eswa.2023.121481. [Google Scholar] [CrossRef]
21. S. Ghosh, A. Singh, N. Z. Jhanjhi Kavita, M. Masud, and S. Aljahdali, “SVM and KNN based CNN architectures for plant classification,” Comput. Mater. Contin., vol. 71, no. 3, pp. 4257–4274, 2022. doi: 10.32604/cmc.2022.023414. [Google Scholar] [CrossRef]
22. S. M. Kadiwal, V. Hegde, N. V. Shrivathsa, S. Gowrishankar, A. H. Srinivasa and A. Veena, “A survey of different identification and classification methods for medicinal plants,” in Inventive Computation and Information Technologies, 2023, pp. 273–291. doi: 10.1007/978-981-19-7402-1_20. [Google Scholar] [CrossRef]
23. I. H. Sarker, “Machine learning: Algorithms, real-world applications and research directions,” SN Comput. Sci., vol. 2, no. 3, pp. 1–21, 2021, Art. no. 160. doi: 10.1007/s42979-021-00592-x. [Google Scholar] [PubMed] [CrossRef]
24. S. Panigrahi, A. Nanda, and T. Swarnkar, “Deep learning approach for image classification,” in Proc.-2nd Int. Conf. Data Sci. Business Anal., ICDSBA 2018, 2018, pp. 511–516. doi: 10.1109/ICDSBA.2018.00101. [Google Scholar] [CrossRef]
25. F. Zhuang et al., “A comprehensive survey on transfer learning,” Proc. IEEE, vol. 109, no. 1, pp. 43–76, Jan. 2021. doi: 10.1109/JPROC.2020.3004555. [Google Scholar] [CrossRef]
26. Y. LeCun, Y. Bengio, and G. Hinton, “Deep learning,” Nature, vol. 521, no. 7553, pp. 436–444, 2015. doi: 10.1038/nature14539. [Google Scholar] [PubMed] [CrossRef]
27. A. S. Almasoud et al., “Deep learning with image classification based secure CPS for healthcare sector,” Comput. Mater. Contin., vol. 72, no. 2, pp. 2633–2648, 2022. doi: 10.32604/cmc.2022.024619. [Google Scholar] [CrossRef]
28. I. H. Sarker, “Deep learning: A comprehensive overview on techniques, taxonomy, applications and research directions,” SN Comput. Sci., vol. 2, pp. 1–20, 2021, Art. no. 420. doi: 10.1007/s42979-021-00815-1. [Google Scholar] [PubMed] [CrossRef]
29. C. P. Blesslin Elizabeth and S. Baulkani, “Novel network for medicinal leaves identification,” IETE J. Res., vol. 69, no. 4, pp. 1772–1782, 2023. doi: 10.1080/03772063.2021.2016504. [Google Scholar] [CrossRef]
30. L. Alzubaidi et al., “Review of deep learning: Concepts, CNN architectures, challenges, applications, future directions,” J. Big Data, vol. 8, no. 1, pp. 1–74, 2021, Art. no. 53. doi: 10.1186/s40537-021-00444-8. [Google Scholar] [PubMed] [CrossRef]
31. D. V. Nazarenko, P. V. Kharyuk, I. V. Oseledets, I. A. Rodin, and O. A. Shpigun, “Machine learning for LC-MS medicinal plants identification,” Chemometr. Intell. Lab. Syst., vol. 156, no. 1, pp. 174–180, 2016. doi: 10.1016/j.chemolab.2016.06.003. [Google Scholar] [CrossRef]
32. S. Roopashree and J. Anitha, “Medicinal leaf dataset mendeley data,” 2020. doi: 10.17632/nnytj2v3n5.1. [Google Scholar] [CrossRef]
33. M. K. Khlifi, W. Boulila, and I. R. Farah, “Graph-based deep learning techniques for remote sensing applications: Techniques, taxonomy, and applications—A comprehensive review,” Comput. Sci. Rev., vol. 50, no. 3, pp. 1–27, Nov. 2023, Art. no. 100596. doi: 10.1016/j.cosrev.2023.100596. [Google Scholar] [CrossRef]
34. J. Vishnu Vardhan, K. Kaur, and M. Upendra Kumar, “Plant recognition using hog and artificial neural network,” Int. J. Recent Innov. Trends Comput. Commun., vol. 5, no. 5, pp. 746–750, 2017. [Google Scholar]
35. G. Saleem, M. Akhtar, N. Ahmed, and W. S. Qureshi, “Automated analysis of visual leaf shape features for plant classification,” Comput. Electron. Agric., vol. 157, no. 1, pp. 270–280, Feb. 2019. doi: 10.1016/j.compag.2018.12.038. [Google Scholar] [CrossRef]
36. A. F. A. Nasir, M. N. A. Rahman, N. Mat, and A. R. Mamat, “Automatic identification of ficus deltoidea Jack (Moraceae) varieties based on leaf,” Mod. Appl. Sci., vol. 8, no. 5, pp. 121–131, 2014. doi: 10.5539/mas.v8n5p121. [Google Scholar] [CrossRef]
37. P. Barré, B. C. Stöver, K. F. Müller, and V. Steinhage, “LeafNet: A computer vision system for automatic plant species identification,” Ecol. Inform., vol. 40, pp. 50–56, 2017. doi: 10.1016/j.ecoinf.2017.05.005. [Google Scholar] [CrossRef]
38. A. Kadir, L. E. Nugroho, A. Susanto, and P. I. Santosa, “Performance improvement of leaf identification system using principal component analysis,” Int. J. Adv. Sci. Technol., vol. 44, pp. 113–124, 2012. [Google Scholar]
39. L. Gao and X. Lin, “A method for accurately segmenting images of medicinal plant leaves with complex backgrounds,” Comput Electron. Agric., vol. 155, no. 11, pp. 426–445, 2018. doi: 10.1016/j.compag.2018.10.020. [Google Scholar] [CrossRef]
40. N. Otsu, “Threshold selection method from gray-level histograms,” IEEE Trans Syst. Man Cybern., vol. SMC-9, no. 1, pp. 62–66, 1979. doi: 10.1109/tsmc.1979.4310076. [Google Scholar] [CrossRef]
41. S. S. Nandyal, B. S. Anami, A. Govardhan, and P. S. Hiremath, “A method for identification and classification of medicinal plant images based on level set segmentation and SVM classification,” Int. J. Comput Vis Robot, vol. 3, no. 1–2, pp. 96–114, 2012. doi: 10.1504/IJCVR.2012.046417. [Google Scholar] [CrossRef]
42. E. Cernadas, L. Gomez, A. Casas, P. G. Rodríguez, and R. G. Carrión, “A method for controlling the enhancement of image feature by unsharp masking filters,” in Proc. IWISP’96, 1996, pp. 337–340. doi: 10.1016/b978-044482587-2/50075-6. [Google Scholar] [CrossRef]
43. Z. Husin et al., “Embedded portable device for herb leaves recognition using image processing techniques and neural network algorithm,” Comput. Electron. Agric., vol. 89, no. 1, pp. 18–29, Nov. 2012. doi: 10.1016/j.compag.2012.07.009. [Google Scholar] [CrossRef]
44. J. Canny, “A computational approach to edge detection,” IEEE Trans Pattern Anal. Mach. Intell., vol. 8, no. 6, pp. 679–698, 1986. doi: 10.1109/TPAMI.1986.4767851. [Google Scholar] [CrossRef]
45. S. Singh and M. S. Bhamrah, “Leaf identification using feature extraction and machine learning,” IOSR J. Electron. Commun. Eng. (IOSR-JECE), vol. 10, no. 5, pp. 134–140, 2015. doi: 10.9790/2834-1051134140. [Google Scholar] [CrossRef]
46. G. Kumar and P. K. Bhatia, “A detailed review of feature extraction in image processing systems,” in Int. Conf. Adv. Comput. Commun. Technol. (ACCT), 2014, pp. 5–12. doi: 10.1109/ACCT.2014.74. [Google Scholar] [CrossRef]
47. A. Caglayan, O. Guclu, and A. B. Can, “A plant recognition approach using shape and color features in leaf images,” in Image Analysis and Processing–ICIAP 2013, 2013, pp. 161–170. doi: 10.1007/978-3-642-41184-7_17. [Google Scholar] [CrossRef]
48. C. H. Arun and D. Christopher Durairaj, “Identifying medicinal plant leaves using textures and optimal colour spaces channel,” Jurnal Ilmu Komputer dan Informasi, vol. 10, no. 1, pp. 19–28, 2017. doi: 10.21609/jiki.v10i1.405. [Google Scholar] [CrossRef]
49. T. D. Dahigaonkar and R. T. Kalyane, “Identification of ayurvedic medicinal plants by image processing of leaf samples,” Int. Res. J. Eng. Technol., vol. 5, no. 5, pp. 351–355, May, 2018. [Google Scholar]
50. H. X. Kan, L. Jin, and F. L. Zhou, “Classification of medicinal plant leaf image based on multi-feature extraction,” Pattern Recognit. Image Anal., vol. 27, no. 3, pp. 581–587, Jul. 2017. doi: 10.1134/S105466181703018X. [Google Scholar] [CrossRef]
51. M. K. Hu, “Visual pattern recognition by moment invariants,” IRE Trans. Inf. Theor., vol. 8, no. 2, pp. 179–187, 1962. doi: 10.1109/TIT.1962.1057692. [Google Scholar] [CrossRef]
52. K. S. Jye, S. Manickam, S. Malek, M. Mosleh, and S. K. Dhillon, “Automated plant identification using artificial neural network and support vector machine,” Front. Life Sci., vol. 10, no. 1, pp. 98–107, 2017. doi: 10.1080/21553769.2017.1412361. [Google Scholar] [CrossRef]
53. A. Sabu, K. Sreekumar, and R. R. Nair, “Recognition of ayurvedic medicinal plants from leaves: A computer vision approach,” in 2017 4th Int. Conf. Image Inform. Process., ICIIP 2017, 2018, pp. 574–578. doi: 10.1109/ICIIP.2017.8313782. [Google Scholar] [CrossRef]
54. E. Elhariri, N. El-Bendary, and A. E. Hassanien, “Plant classification system based on leaf features,” in Proc. 2014 9th IEEE Int. Conf. Comput. Eng. Syst., ICCES 2014, Feb. 2014, pp. 271–276. doi: 10.1109/ICCES.2014.7030971. [Google Scholar] [CrossRef]
55. S. Kaur and P. Kaur, “Plant species identification based on plant leaf using computer vision and machine learning techniques,” J. Multimed. Inform. Syst., vol. 6, no. 2, pp. 49–60, 2019. doi: 10.33851/JMIS.2019.6.2.49. [Google Scholar] [CrossRef]
56. R. Ali, R. Hardie, and A. Essa, “A leaf recognition approach to plant classification using machine learning,” in Proc. IEEE Nat. Aerosp. Electron. Conf. (NAECON), 2018, pp. 431–434. doi: 10.1109/NAECON.2018.8556785. [Google Scholar] [CrossRef]
57. A. Aakif and M. F. Khan, “Automatic classification of plants based on their leaves,” Biosyst. Eng., vol. 139, no. 4, pp. 66–75, 2015. doi: 10.1016/j.biosystemseng.2015.08.003. [Google Scholar] [CrossRef]
58. E. Yigit, K. Sabanci, A. Toktas, and A. Kayabasi, “A study on visual features of leaves in plant identification using artificial intelligent techniques,” Comput. Electron. Agric., vol. 156, no. 2, pp. 369–377, 2019. doi: 10.1016/j.compag.2018.11.036. [Google Scholar] [CrossRef]
59. A. Begue, V. Kowlessur, U. Singh, F. Mahomoodally, and S. Pudaruth, “Automatic recognition of medicinal plants using machine learning techniques,” Int. J. Adv. Comput. Sci. Appl., vol. 8, no. 4, pp. 166–175, 2017. doi: 10.14569/issn.2156-5570. [Google Scholar] [CrossRef]
60. J. C. Weng, M. C. Hu, and K. C. Lan, “Recognition of easily-confused TCM herbs using deep learning,” in Proc. 8th ACM Multimed. Syst. Conf., MMSys 2017, Association for Computing Machinery, Inc., Jun. 2017, pp. 233–234. doi: 10.1145/3083187.3083226. [Google Scholar] [CrossRef]
61. A. P. S. S. Raj and S. K. Vajravelu, “DDLA: Dual deep learning architecture for classification of plant species,” IET Image Process, vol. 13, no. 12, pp. 2176–2182, Oct. 2019. doi: 10.1049/iet-ipr.2019.0346. [Google Scholar] [CrossRef]
62. A. H. Vo, H. T. Dang, B. T. Nguyen, and V. H. Pham, “Vietnamese herbal plant recognition using deep convolutional features,” Int. J. Mach. Learn. Comput, vol. 9, no. 3, pp. 363–367, 2019. doi: 10.18178/ijmlc.2019.9.3.811. [Google Scholar] [CrossRef]
63. A. Paulson and S. Ravishankar, “AI based indigenous medicinal plant identification,” in Proc. 2020 Adv. Comput. Commun. Technol. High Perf. Appl., ACCTHPA 2020, 2020, pp. 57–63. doi: 10.1109/ACCTHPA49271.2020.9213224. [Google Scholar] [CrossRef]
64. S. Roopashree and J. Anitha, “DeepHerb: A vision based system for medicinal plants using xception features,” IEEE Access, vol. 9, pp. 135927–135941, 2021. doi: 10.1109/ACCESS.2021.3116207. [Google Scholar] [CrossRef]
65. J. Abdollahi, “Identification of medicinal plants in ardabil using deep learning: Identification of medicinal plants using deep learning,” in 2022 27th Int. Comput. Conf., Comput. Soc. Iran (CSICC), 2022, pp. 1–6. doi: 10.1109/CSICC55295.2022.9780493. [Google Scholar] [CrossRef]
66. S. G. Wu, F. S. Bao, E. Y. Xu, Y. X. Wang, Y. F. Chang and Q. L. Xiang, “A leaf recognition algorithm for plant classification using probabilistic neural network,” in 2007 IEEE Int. Symp. Sig. Proc. Inf. Technol., 2007, pp. 11–16. doi: 10.1109/ISSPIT.2007.4458016. [Google Scholar] [CrossRef]
67. O. J. O. Söderkvist, “Computer vision classification of leaves from swedish trees,” 2001. Accessed: Nov. 30, 2022. [Online]. Available: http://urn.kb.se/resolve?urn=urn:nbn:se:liu:diva-54366 [Google Scholar]
68. N. Kumar et al., “Leafsnap: A computer vision system for automatic plant species identification,” in Lect. Not. Comput. Sci., Comput. Vis.-ECCV, 2012, pp. 502–516. doi: 10.1007/978-3-642-33709-3_36. [Google Scholar] [CrossRef]
69. T. Munisami, M. Ramsurn, S. Kishnah, and S. Pudaruth, “Plant leaf recognition using shape features and colour histogram with K-nearest neighbour classifiers,” Procedia Comput. Sci., vol. 58, no. 4, pp. 740–747, 2015. doi: 10.1016/j.procs.2015.08.095. [Google Scholar] [CrossRef]
70. K. A. Haryono and A. Saleh, “A novel herbal leaf identification and authentication using deep learning neural network,” in 2020 Int. Conf. Comput. Eng., Netw., Intell. Multimed. 2020, 2020, pp. 338–342. doi: 10.1109/CENIM51130.2020.9297952. [Google Scholar] [CrossRef]
71. R. Shailendra et al., “An IoT and machine learning based intelligent system for the classification of therapeutic plants,” Neural Process Lett., vol. 54, no. 5, pp. 4465–4493, 2022. doi: 10.1007/s11063-022-10818-5. [Google Scholar] [CrossRef]
72. A. V. Pargaien, D. Singh, M. Chauhan, H. Negi, B. Chilwal and N. Pargaien, “Identification of plant leaves having anti-diabetic property using machine learning,” in Proc. 2nd Int. Conf. Appl. Artif. Intell. Comput., ICAAIC 2023, 2023, pp. 195–200. doi: 10.1109/ICAAIC56838.2023.10140394. [Google Scholar] [CrossRef]
73. L. D. S. Pacifico, L. F. S. Britto, E. G. Oliveira, and T. B. Ludermir, “Automatic classification of medicinal plant species based on color and texture features,” in 2019 8th Brazil. Conf. Intell. Syst. (BRACIS), 2019, 741–746. doi: 10.1109/BRACIS.2019.00133. [Google Scholar] [CrossRef]
74. A. Gokhale, S. Babar, S. Gawade, and S. Jadhav, “Identification of medicinal plant using image processing and machine learning,” in Adv. Intell. Syst. Comput., Appl. Comput. Vis. Image Process., 2020, pp. 272–282. doi: 10.1007/978-981-15-4029-5_27. [Google Scholar] [CrossRef]
75. M. Kumar, “Plant species recognition using morphological features and adaptive boosting methodology,” IEEE Access, vol. 7, pp. 163912–163918, 2019. doi: 10.1109/ACCESS.2019.2952176. [Google Scholar] [CrossRef]
76. Y. G. Naresh and H. S. Nagendraswamy, “Classification of medicinal plants: An approach using modified LBP with symbolic representation,” Neurocomputing, vol. 173, no. 9, pp. 1789–1797, Jan. 2016. doi: 10.1016/j.neucom.2015.08.090. [Google Scholar] [CrossRef]
77. D. Puri, A. Kumar, J. Virmani, Kriti, “Classification of leaves of medicinal plants using laws’ texture features,” Int. J. Inf. Technol., vol. 14, no. 2, pp. 931–942, 2022. doi: 10.1007/s41870-019-00353-3. [Google Scholar] [CrossRef]
78. W. S. Jeon and S. Y. Rhee, “Plant leaf recognition using a convolution neural network,” Int. J. Fuzzy Logic Intell. Syst., vol. 17, no. 1, pp. 26–34, 2017. doi: 10.5391/IJFIS.2017.17.1.26. [Google Scholar] [CrossRef]
79. S. O. Oppong, F. Twum, J. Ben Hayfron-Acquah, and Y. M. Missah, “A novel computer vision model for medicinal plant identification using log-gabor filters and deep learning algorithms,” Comput. Intell. Neurosci., vol. 2022, no. 1, pp. 1–21, 2022. doi: 10.1155/2022/1189509. [Google Scholar] [PubMed] [CrossRef]
80. D. Bisen, “Deep convolutional neural network based plant species recognition through features of leaf,” Multimed. Tools Appl., vol. 80, no. 4, pp. 6443–6456, 2021. doi: 10.1007/s11042-020-10038-w. [Google Scholar] [CrossRef]
81. C. Tan, F. Sun, T. Kong, W. Zhang, C. Yang and C. Liu, “A survey on deep transfer learning,” in Lect. Not. Comput. Sci. Artif. Neur. Netw. Mach. Learn.–ICANN 2018, 2018, pp. 270–279. doi: 10.1007/978-3-030-01424-7_27. [Google Scholar] [CrossRef]
82. A. Kaya, A. Seydi, C. Catal, H. Yalin, and H. Temucin, “Analysis of transfer learning for deep neural network based plant classification models,” Comput Electron. Agric., vol. 158, no. 3, pp. 20–29, 2019. doi: 10.1016/j.compag.2019.01.041. [Google Scholar] [CrossRef]
83. S. Mahajan, A. Raina, X. Z. Gao, and A. K. Pandit, “Plant recognition using morphological feature extraction and transfer learning over SVM and AdaBoost,” Symmetry, vol. 13, no. 2, pp. 1–16, 2021, Art. no. 356. doi: 10.3390/sym13020356. [Google Scholar] [CrossRef]
84. A. Beikmohammadi and K. Faez, “Leaf classification for plant recognition with deep transfer learning,” in Proc. 2018 4th Iranian Conf. Sig. Process. Intell. Syst., ICSPIS 2018, 2018, pp. 21–26. doi: 10.1109/ICSPIS.2018.8700547. [Google Scholar] [CrossRef]
85. L. Almazaydeh, R. Alsalameen, and K. Elleithy, “Herbal leaf recognition using mask-region convolutional neural network (Mask R-CNN),” J. Theor. Appl. Inf Technol., vol. 100, no. 11, pp. 3664–3671, 2022. [Google Scholar]
86. S. S. Patil, S. H. Patil, A. M. Pawar, N. S. Patil, and G. R. Rao, “Automatic classification of medicinal plants using state-of-the-art pre-trained neural networks,” J Adv. Zool., vol. 43, no. 1, pp. 80–88, 2022. doi: 10.17762/jaz.v43i1.116. [Google Scholar] [CrossRef]
87. V. Ayumi et al., “Transfer learning for medicinal plant leaves recognition: A comparison with and without a fine-tuning strategy,” Int. J. Adv. Comput. Sci. Appl., vol. 13, no. 9, pp. 138–144, 2022. doi: 10.14569/issn.2156-5570. [Google Scholar] [CrossRef]
88. K. He, G. Gkioxari, P. Dollar, and R. Girshick, “Mask R-CNN,” IEEE Trans. Pattern Anal. Mach. Intell., vol. 42, no. 2, pp. 386–397, 2020. doi: 10.1109/TPAMI.2018.2844175. [Google Scholar] [PubMed] [CrossRef]
89. N. H. Pham, T. L. Le, P. Grard, and V. N. Nguyen, “Computer aided plant identification system,” in 2013 Int. Conf. Comput. Manag. Telecommun., ComManTel 2013, 2013, pp. 134–139. doi: 10.1109/ComManTel.2013.6482379. [Google Scholar] [CrossRef]
90. Q. K. Nguyen, T. L. Le, and N. H. Pham, “Leaf based plant identification system for Android using SURF features in combination with Bag of Words model and supervised learning,” in Int. Conf. Adv. Technol. Commun., 2013, pp. 404–407. doi: 10.1109/ATC.2013.6698145. [Google Scholar] [CrossRef]
91. M. Ali Jan Ghasab, S. Khamis, F. Mohammad, and H. Jahani Fariman, “Feature decision-making ant colony optimization system for an automated recognition of plant species,” Expert. Syst. Appl., vol. 42, no. 5, pp. 2361–2370, 2015. doi: 10.1016/j.eswa.2014.11.011. [Google Scholar] [CrossRef]
92. D. G. Tsolakidis, D. I. Kosmopoulos, and G. Papadourakis, “Plant leaf recognition using Zernike moments and histogram of oriented gradients,” in Lect. Not. Comput. Sci., Cham, Springer, 2014, vol. 8445, pp. 406–417. doi: 10.1007/978-3-319-07064-3_33/COVER. [Google Scholar] [CrossRef]
93. C. A. Priya, T. Balasaravanan, and A. S. Thanamani, “An efficient leaf recognition algorithm for plant classification using support vector machine,” in Int. Conf. Pattern Recognit. Inf. Med. Eng., PRIME 2012, 2012, pp. 428–432. doi: 10.1109/ICPRIME.2012.6208384. [Google Scholar] [CrossRef]
94. S. Prasad, S. K. Peddoju, and D. Ghosh, “Mobile plant species classification: A low computational aproach,” in 2013 IEEE 2nd Int. Conf. Image Inf. Process., IEEE ICIIP 2013, 2013, pp. 405–409. doi: 10.1109/ICIIP.2013.6707624. [Google Scholar] [CrossRef]
95. J. Hossain and M. A. Amin, “Leaf shape identification based plant biometrics,” in Proc. 2010 13th Int. Conf. Comput. Inf. Technol., ICCIT 2010, 2010, pp. 458–463. doi: 10.1109/ICCITECHN.2010.5723901. [Google Scholar] [CrossRef]
96. J. K. Hsiao, L. W. Kang, C. L. Chang, and C. Y. Lin, “Comparative study of leaf image recognition with a novel learning-based approach,” in Proc. 2014 Sci. Inf. Conf., SAI 2014, 2014, pp. 389–393. doi: 10.1109/SAI.2014.6918216. [Google Scholar] [CrossRef]
97. X. Y. Xiao, R. Hu, S. W. Zhang, and X. F. Wang, “HOG-based approach for leaf classification,” in Adv. Intell. Comput. Theor. App.: Aspects Artif. Intell., Berlin, Heidelberg, Springer, 2010, pp. 149–155. doi: 10.1007/978-3-642-14932-0_19. [Google Scholar] [CrossRef]
98. S. Mouine, I. Yahiaoui, and A. Verroust-Blondet, “A shape-based approach for leaf classification using multiscale triangular representation,” in Proc. 3rd ACM Int. Conf. Multimed. Retr., 2013, pp. 127–134. doi: 10.1145/2461466.2461489. [Google Scholar] [CrossRef]
99. X. Wang, J. Liang, and F. Guo, “Feature extraction algorithm based on dual-scale decomposition and local binary descriptors for plant leaf recognition,” Dig. Sig. Process.: Rev. J., vol. 34, no. 1, pp. 101–107, 2014. doi: 10.1016/j.dsp.2014.08.005. [Google Scholar] [CrossRef]
100. X. M. Ren, X. F. Wang, and Y. Zhao, “An efficient multi-scale overlapped block LBP approach for leaf image recognition,” in Intell. Comput. Theor. App. ICIC, 2012, pp. 237–243. doi: 10.1007/978-3-642-31576-3_31. [Google Scholar] [CrossRef]
101. H. Ling and D. W. Jacobs, “Shape classification using the inner-distance,” IEEE Trans. Pattern Anal. Mach. Intell., vol. 29, no. 2, pp. 286–299, 2007. doi: 10.1109/TPAMI.2007.41. [Google Scholar] [PubMed] [CrossRef]
102. S. Belongie, J. Malik, and J. Puzicha, “Shape matching and object recognition using shape contexts,” IEEE Trans. Pattern Anal. Mach. Intell., vol. 24, no. 4, pp. 509–522, 2002. doi: 10.1109/34.993558. [Google Scholar] [CrossRef]
103. S. Ghosh, A. Singh, and S. Kumar, “Identification of medicinal plant using hybrid transfer learning technique,” Indones. J. Electr. Eng. Comput. Sci., vol. 31, no. 3, pp. 1605–1615, 2023. doi: 10.11591/ijeecs.v31.i3.pp1605-1615. [Google Scholar] [CrossRef]
104. J. S. Cope, D. Corney, J. Y. Clark, P. Remagnino, and P. Wilkin, “Plant species identification using digital morphometrics: A review,” Expert. Syst. Appl., vol. 39, no. 8, pp. 7562–7573, 2012. doi: 10.1016/j.eswa.2012.01.073. [Google Scholar] [CrossRef]
105. H. Goëau, P. Bonnet, and A. Joly, “Plant identification in an open-world (LifeCLEF 2016),” in CEUR Workshop Proc., 2016. CLEF: Conf. Labs Eval. Forum, Évora, Portugal, Sep. 2016, pp. 428–439. [Google Scholar]
106. J. Carranza-Rojas, H. Goeau, P. Bonnet, E. Mata-Montero, and A. Joly, “Going deeper in the automated identification of Herbarium specimens,” BMC Evol. Biol., vol. 17, no. 1, pp. 1–14, 2017, Art. no. 181. doi: 10.1186/s12862-017-1014-z. [Google Scholar] [PubMed] [CrossRef]
107. A. Odena, C. Olah, and J. Shlens, “Conditional image synthesis with auxiliary classifier gans,” in 34th Int. Conf. Mach. Learn., ICML 2017, 2017, vol. 70, pp. 2642–2651. [Google Scholar]
108. G. Ravivarma, K. Gavaskar, D. Malathi, K. G. Asha, B. Ashok and S. Aarthi, “Implementation of Sobel operator based image edge detection on FPGA,” Mater. Today: Proc., 2021, vol. 45, no. 8, pp. 2401–2407, 2021. doi: 10.1016/j.matpr.2020.10.825. [Google Scholar] [CrossRef]
109. C. G. Willis et al., “Old plants, new tricks: Phenological research using herbarium specimens,” Trends Ecol. Evol., vol. 32, no. 7, pp. 531–546, Jul. 2017. doi: 10.1016/j.tree.2017.03.015. [Google Scholar] [PubMed] [CrossRef]
110. A. Joly et al., “Interactive plant identification based on social image data,” Ecol. Inform., vol. 23, pp. 22–34, 2014. doi: 10.1016/j.ecoinf.2013.07.006. [Google Scholar] [CrossRef]
111. K. J. Gaston and M. A. O’Neill, “Automated species identification: Why not?” Phil. Trans. R. Soc. Lond. B Biol. Sci., vol. 359, no. 1444, pp. 655–667, 2004. doi: 10.1098/rstb.2003.1442. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF

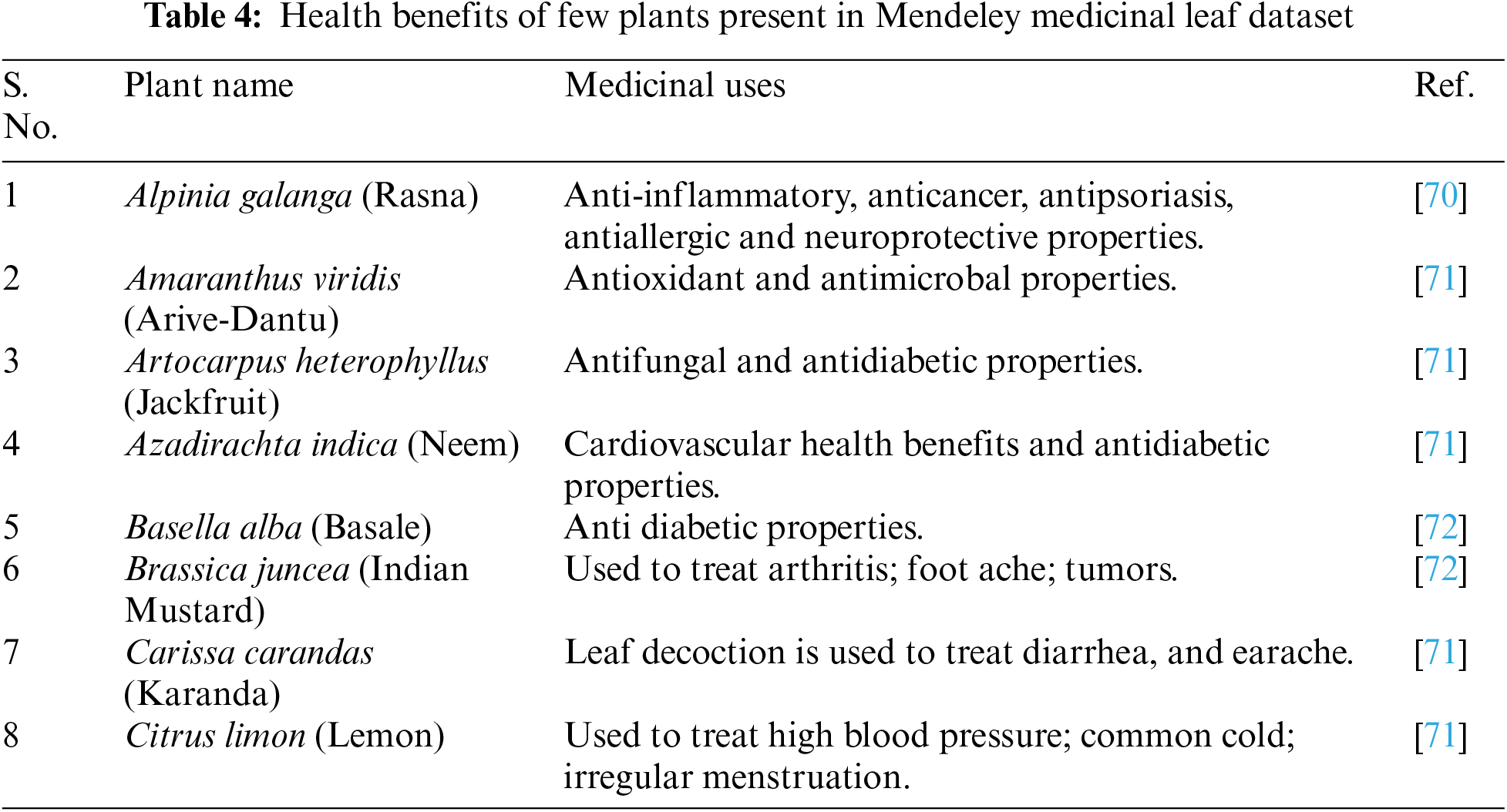





 Downloads
Downloads
 Citation Tools
Citation Tools