 Open Access
Open Access
ARTICLE
A Comprehensive Image Processing Framework for Early Diagnosis of Diabetic Retinopathy
1 College of Computer Science and Engineering, University of Ha’il, Ha’il, 81481, Saudi Arabia
2 Department of CSE, Chandigarh University, Mohali, Punjab, 140413, India
3 Department of Electronics and Communication Engineering, Amity University, Noida, 201301, India
4 Department of Information Technology, Indira Gandhi Delhi Technical University for Women, New Delhi, 110006, India
5 Chief Manager, State Bank of India, Panchkula, Haryana, 134109, India
6 Department of Computer Science and Information Engineering, Asia University, Taichung City, 413, Taiwan
7 Symbiosis Centre for Information Technology (SCIT), Symbiosis International University, Pune, 411057, India
8 Center for Interdisciplinary Research, University of Petroleum and Energy Studies (UPES), Dehradun, 248007, India
* Corresponding Author: Brij B. Gupta. Email:
(This article belongs to the Special Issue: Deep Learning in Medical Imaging-Disease Segmentation and Classification)
Computers, Materials & Continua 2024, 81(2), 2665-2683. https://doi.org/10.32604/cmc.2024.053565
Received 04 May 2024; Accepted 19 September 2024; Issue published 18 November 2024
Abstract
In today’s world, image processing techniques play a crucial role in the prognosis and diagnosis of various diseases due to the development of several precise and accurate methods for medical images. Automated analysis of medical images is essential for doctors, as manual investigation often leads to inter-observer variability. This research aims to enhance healthcare by enabling the early detection of diabetic retinopathy through an efficient image processing framework. The proposed hybridized method combines Modified Inertia Weight Particle Swarm Optimization (MIWPSO) and Fuzzy C-Means clustering (FCM) algorithms. Traditional FCM does not incorporate spatial neighborhood features, making it highly sensitive to noise, which significantly affects segmentation output. Our method incorporates a modified FCM that includes spatial functions in the fuzzy membership matrix to eliminate noise. The results demonstrate that the proposed FCM-MIWPSO method achieves highly precise and accurate medical image segmentation. Furthermore, segmented images are classified as benign or malignant using the Decision Tree-Based Temporal Association Rule (DT-TAR) Algorithm. Comparative analysis with existing state-of-the-art models indicates that the proposed FCM-MIWPSO segmentation technique achieves a remarkable accuracy of 98.42% on the dataset, highlighting its significant impact on improving diagnostic capabilities in medical imaging.Keywords
In the modern world, the most challenging task in medical image analysis is to extract critical information about the structure, shape and volume of the anatomical organs of the human body from the medical image. Medical image analysis has become a pivotal field in modern healthcare, leveraging advanced computational techniques to enhance the accuracy and efficiency of diagnosing and treating various medical conditions. Among the myriad applications within this domain, medical image segmentation stands out as a critical process. Medical image segmentation involves partitioning medical images into meaningful regions, often corresponding to different anatomical structures or pathological areas. This technique facilitates precise and automated analysis, which is crucial for accurate diagnosis, treatment planning, and monitoring of diseases.
Medical image segmentation identifies the pixels of anatomical organs or lesions from the backdrop of the medical images, such as Magnetic Resonance Imaging (MRI) and Computed Tomography (CT) images [1,2]. Medical image segmentation is an inspiring and thought-provoking emerging field of research in medical image processing. Image segmentation is fragmenting a digital image into segments based on intensity, shape, texture or color. One significant application of medical image segmentation is in the detection and management of diabetic retinopathy (DR), a leading cause of blindness worldwide. Diabetic retinopathy is a complication of diabetes that affects the retina, the light-sensitive tissue at the back of the eye. Early detection and treatment are vital to prevent vision loss, making the role of medical image analysis indispensable. Segmentation algorithms can automatically identify and delineate pathological features in retinal images, such as microaneurysms, hemorrhages, and exudates, which are indicative of diabetic retinopathy. This automation not only improves diagnostic accuracy but also enables large-scale screening programs, especially in regions with limited access to ophthalmologists. Segmentation groups pixels into many clusters based on the closeness of the intensities of the pixels in a digital image to simplify the information extraction such as shape, texture and structure [3]. It separates the images into groups of homogeneous pixels concerning some conditions. The various groups need not intersect, and neighbouring groups must be heterogeneous [4]. In general, Image segmentation assigns a label to each pixel so that the pixels with the same label correspond to certain similar properties.
Medical image segmentation plays a magnificent role in assisting doctors in the prognosis and diagnosis of severe diseases like brain tumour, breast cancer, ovarian cancer, lung cancer, liver cancer, lung nodules and Alzheimer’s disease. It accurately determines an organ’s outline or anatomical structure. The inter-observer variability in manual investigation occurs because of the variations in an individual’s skill level and medical relevance, whether curable or palliative intent, uncertainty in boundary due to volume averaging, spatial resolution and contrast levels in the medical image [5]. It leads to inaccurate and false diagnoses. Through automated medical image segmentation techniques, physicians can diagnose diseases efficiently and effectively. Deploying segmentation techniques in the medical field helps in surgical planning and tracking patient health improvement [6].
The image analysis system’s structured process flow comprises image acquisition, pre-processing, segmentation, feature extraction, and classification. Medical images are acquired from various medical imaging modalities like Magnetic Resonance Imaging, Emission Tomography, X-ray projection, Computed Tomography, thermography, Optical Coherence Tomography, Ultrasonic scanning, and tactile imaging. The acquired images from the patients are routed to the pre-processing module. While acquiring images with each imaging modality, low contrast levels and additive noise are encountered. In pre-processing, denoising through a filtering approach is done to suppress the additive noise and low contrast levels are boosted by histogram equalization. The pre-processed medical images are segmented by computer-aided segmentation methods like optimization and clustering to detect the presence and severity of the diseases and help doctors save human lives. Performance evaluation is carried out by using various performance metrics to choose the right segmentation algorithm, and this evaluation is mandatory in image analysis. For clear understanding, an efficient classification algorithm is also essential for classifying the segmented images.
Diabetic patients have a specific eye disease called diabetic retinopathy. When the diabetic sugar level in the blood increases, the swelling and leakage in blood vessels or closure of blood vessels in the retina occurs, which obstructs the blood flow in the retinal blood vessels. All these malfunctions in the retina can cause human vision loss. Hence, early detection by segmentation methods greatly helps the surgeons in early eye surgical planning to treat the patients, which may prevent blindness. Pregnancy, puberty, cataract surgery and higher body mass index (BMI) are the other risk issues of diabetic retinopathy. According to the World Health Organization (WHO), Diabetic Retinopathy (DR) accounts for 37 million cases of blindness worldwide.
Photographing a large number of diabetic patients periodically is not an easy task, and the expenses for manual retinal examination are also expensive [7]. Hence, automatic screening of retinal images can be done through the automated system, which can differentiate the retinal image as a healthy retinal image or diabetic retinopathy-affected retinal image. The workload of the ophthalmologist can be significantly reduced by examining only the disease-affected images. Non-Proliferative Diabetic Retinopathy (NPDR) and Proliferative Diabetic Retinopathy (PDR) are the two major classes of Diabetic Retinopathy. NPDR is further distinguished as mild NPDR, moderate NPDR and severe NPDR stages. Small regions of balloon-shaped swelling in the retina’s tiny blood vessels at the early stage are termed micro-aneurysms (MA’s). At the moderate stage, as the disease progresses due to poorly controlled glucose levels in the blood or due to the long-standing disease of diabetes mellitus, some MA’s in the blood vessel rupture, causing bleeding in the retina called as hemorrhages. At the advanced stage, large numbers of blood vessels are blocked and dispossess blood supply in several regions of the retina, and these spots appear as yellow or whitish cotton wool spots. At the very advanced stage called Proliferative Retinopathy, the retina triggers the growth of abnormal and fragile blood vessels. The blood leakage from these fragile walls causes severe loss of vision and even blindness.
The main intent of the proposed approach is to attain the effective identification of retinopathy from the images. The research paper presents an efficient image processing framework designed for the diagnosis of diabetic retinopathy. This framework integrates two novel algorithms: the FCM-Based Modified Inertia Weight Particle Swarm Optimization (FCM-MIWPSO) for image segmentation, and the Decision Tree-Based Temporal Association Rule (DT-TAR) for classification.
The paper is structured as follows: Section 2 discusses related works that provide a foundation for further research in this field. Section 3 details the materials and methods, including the proposed model. Section 4 presents the simulation results and provides commentary. Finally, Section 5 offers the conclusions.
As image analysis manifests its role in various applications, several studies have been conducted, including many investigations. This section presents the literature survey on the research works associated with this research work. In the related work section, we present a comparative analysis of previous studies in the field of diabetic retinopathy detection using image processing and machine learning techniques. This analysis aims to highlight the strengths and limitations of existing methods, providing a clear context for the contributions of our research. Various studies have utilized techniques such as edge detection, morphological operations, and texture analysis for the segmentation and detection of retinal abnormalities. These methods often struggle with variations in image quality and complexity of retinal structures.
A hybrid approach will provide a high level of accuracy to diagnose biomedical diseases, such as the retinal diseases, while requiring a smaller computing load Image preprocessing was used to extract retinal layers. The Firefly algorithm was used to select the best features. Machine Learning classifiers were used instead of multiclass classifications. This study utilized two public datasets, with a mean accuracy of 0.957 for the first dataset and 0.954 for the second.
Xia et al. [8] proposed a DR diagnosis scheme that combines the MPAG and the LLM. Based on the prediction score and the type of lesions in the patches, MPAG generates a weighted attention map based on patches of various sizes, fully considering the impact of lesion types on grading, solving the issue that attention maps of lesions cannot be further refined and then tailored to the final DR diagnosis task. As a result of localization, the LLM produces a global attention map. A weighted attention map and a global attention map are layered with the fundus map to enhance the attention of the classification network to the details of the lesion. This paper demonstrates the effectiveness of the proposed method through extensive experiments on the public DDR dataset, achieving an accuracy of 80.64%. Researchers can explore and achieve effective detection results for diabetic retinopathy by exploring and implementing the research findings from this study. The DRIVE (digital retinal images for vessel extraction) and STARE (structured analysis of the retina) datasets were used for this study. A performance metric including accuracy, specificity, and sensitivity was chosen to demonstrate the methodology’s effectiveness. Compared to previous studies, this study achieved higher accuracy in segmenting blood arteries using the proposed framework).
2.1 Filtering and Enhancement Techniques
In 2018, Storath et al. [9] have proposed a fast-filtering algorithm for denoising images using the unit circle values following the arc distance median. The median filter they designed is robust, value-preserving, and edge-preserving, so it is the most employed filtering tool for real-valued data smoothing. Ultrasound images are more prone to speckle noise, leading to a mixture of different regions; hence, the interpretation of lesions is incorrect. In 2017, Jain et al. [10] proposed the usage of region difference filters in ultrasound liver image segmentation. The filters assess the maximum dissimilarity of the average of two regions around the median pixel. In 2014, Strange et al. [11] implemented coherence-enhancing diffusion filtering (CED) to enhance the myofiber boundaries by extracting the eosinophilic structures and accomplishing stain decomposition from the histological muscle biopsy image while smoothing the noise within the myofiber regions.
In 2015, Rasta et al. [12] performed a comparative study on the pre-processing techniques for illumination correction and contrast enhancement in diabetic retinopathy retinal images. In their comparative study, for illumination correction, they analyzed dividing methods by median filtering and quotient method by homomorphic filtering and inspected contrast limited histogram equalization (CLAHE) and polynomial gray level transformation operator for contrast enhancement. Based on the visual and statistical evaluation, they concluded that the dividing method using median filtering is the most successful technique on the red component to estimate the background. The quotient method is more powerful on the green component for illumination correction and enhancing the retinal image’s contrast. They recommended CLAHE because of its greater accuracy and sensitivity. A comparison among the various filters, such as the box filter, Gabor filter, median filter, and Gaussian filter, for filtering the medical images. Following the estimated peak signal-to-noise ratio (PSNR) values for the Gaussian noise, the median filter has shown better outcomes in their Survey.
2.2 Segmentation and Classification Techniques Text Layout
Much research has been done on image segmentation techniques, especially for segmenting medical images to aid doctors in detecting acute diseases. The high noise-sensitive behaviour of conventional FCM is compromised by altering the membership weighting function of every cluster based on spatial neighborhood information [13]. They achieved robust noise-free segmentation for both real and artificial synthesized images. In 2016, Zou et al. [14] presented an elaborate survey on clustering-based image segmentation techniques. According to them, fuzzy membership of the matrix of FCM is consistent, as it converges to one solution at all times. In 2016, Dubey et al. [15] have taken Euclidean, Manhattan and Pearson correlation coefficients for distance calculation between cluster centres and the data point for analyzing the K-mean algorithm on the Breast Cancer Wisconsin (BCW) dataset. They proved that Euclidean distance and Manhattan performed better than Pearson correlation in distance estimation and achieved a positive% prediction accuracy of 92%.
In 2016, Zou et al. [14] developed a Particle Swarm Optimization (PSO) algorithm that is dynamic in its quantum behaviour of the swarms. Their algorithm dynamically updates the context vector every time, incorporating complete cooperation with other particles. Their experimental results have proved that their method has achieved better search ability and optimal optimization over the previous methods. The fascinating behaviour in the confinement of the best possible solution of particle swarm optimization and the fastest converging feature of FCM are combined for efficient web document clustering. Several experts have proposed modifying the inertia weight in Weighted Inertia Particle Swarm Optimization (IWPSO). In 2012, Zhang et al. [16] demonstrated an inertia weight PSO by choosing a fixed value as an inertia weight from a normally distributed random number between [0,1]. It improves the local search ability in the starting phase and global search capability at the ending phase. Their modification of inertia weight value overcomes the dependency of an increased number of iterations on inertia weight.
In 2017, Kalshetti et al. [17] presented a new algorithm that flows in two stages for segmenting medical images. A binary marker of the desired region of interest is generated using morphological functions as an image in the first stage. In the second stage, the grab-cut technique is used for segmentation, in which the binary marker image is used as a mask image. Further, the results are refined using the graphical user interface (GUI) with minimal user interaction.
In 2017, Xu et al. [18] explored a deep convolution neural network for classifying diabetic retinopathy from eye fundus color images. They demonstrated that their proposed method performed superior over traditional feature-based classification methods. In 2014, Arora et al. [3] suggested that deep learning models like Deep Neural Networks (DNN) and Convolutional Neural Networks (CNN) fix the features of retina images into various classes, such as microaneurysms, blood vessels, exudates, fluid drip and researchers suggested an approach for the detection of Alzheimer’s disease in which the reduction in extracted features like mean, eigenvector, variance, kurtosis, and standard derivation (SD) by particle swarm optimization is achieved and then to classify the brain image as Alzheimer’s disease or not, they employed decision tree classifier. The accuracy of their proposed method is limited to only 91%.
From the literature analysis, it has been identified that the image segmentation process is facing various challenges, such as (i) Utilization of a unique segmentation algorithm for all sorts of medical images, (ii) Determining the optimal objective function or correct pre-defined criterion, (iii) Particular algorithms like multi-thresholding are characterized and mathematically modelled based on complex combinatorial optimization problems. Solving the objective function of such methods can be done only by non-deterministic models, which are very difficult. (iv) Selection of proper quantitative measuring metrics to evaluate the performance of segmentation methods for different input medical images. In this research, initiation has been taken to detect diabetic retinopathy early by proposing an efficient framework comprising identified suitable pre-processing techniques and a hybridized image segmentation algorithm to increase the patient’s life expectancy and improve human life.
The contribution towards this research work is summarized below: An efficient image processing framework that includes two proposed algorithms that are FCM-Based Modified Inertia Weight Particle Swarm Optimization (FCM-MIWPSO) for segmentation and Decision Tree-Based Temporal Association Rule (DT-TAR) for classification as shown in Fig. 1. This framework consists of four important steps, as illustrated below:
1. Initially, histogram equalization, filtering and enhancement techniques were applied for pre-processing the medical images.
2. Then, pre-processed images are used for the segmentation process, which identifies the affected region of the retina. The proposed FCM-MIWPSO algorithm and existing algorithms were used in this segmentation process.
3. The segmentation process extracts various hybrid features like wavelet, run-length and histogram features.
4. After feature extraction, four existing machine learning classifiers and a proposed DT-TAR classification are employed to identify the efficient classifiers for predicting diabetic retinopathy.
The images are directly processed by histogram equalization, noise removal, and enhancement using pre-processing stages. This research work has five types of retinal fundus images, including normal patient retinal images and four stages of diabetic retinopathy such as mild, moderate, proliferative and non-proliferative. The dataset was acquired from a fundus scanning machine in Bahawal Victoria Hospital (BVH) Bahawalpur. The dataset was collected as part of a broader medical imaging project conducted at BVH, and later made accessible through Kaggle for broader use in academic and clinical research. This dataset consists of high-quality images categorized into five classes: one class representing normal retinal images and four classes representing different levels of diabetic retinopathy. The inclusion of multiple severity levels adds significant depth to the dataset, enhancing its utility for detailed analysis and classification tasks.
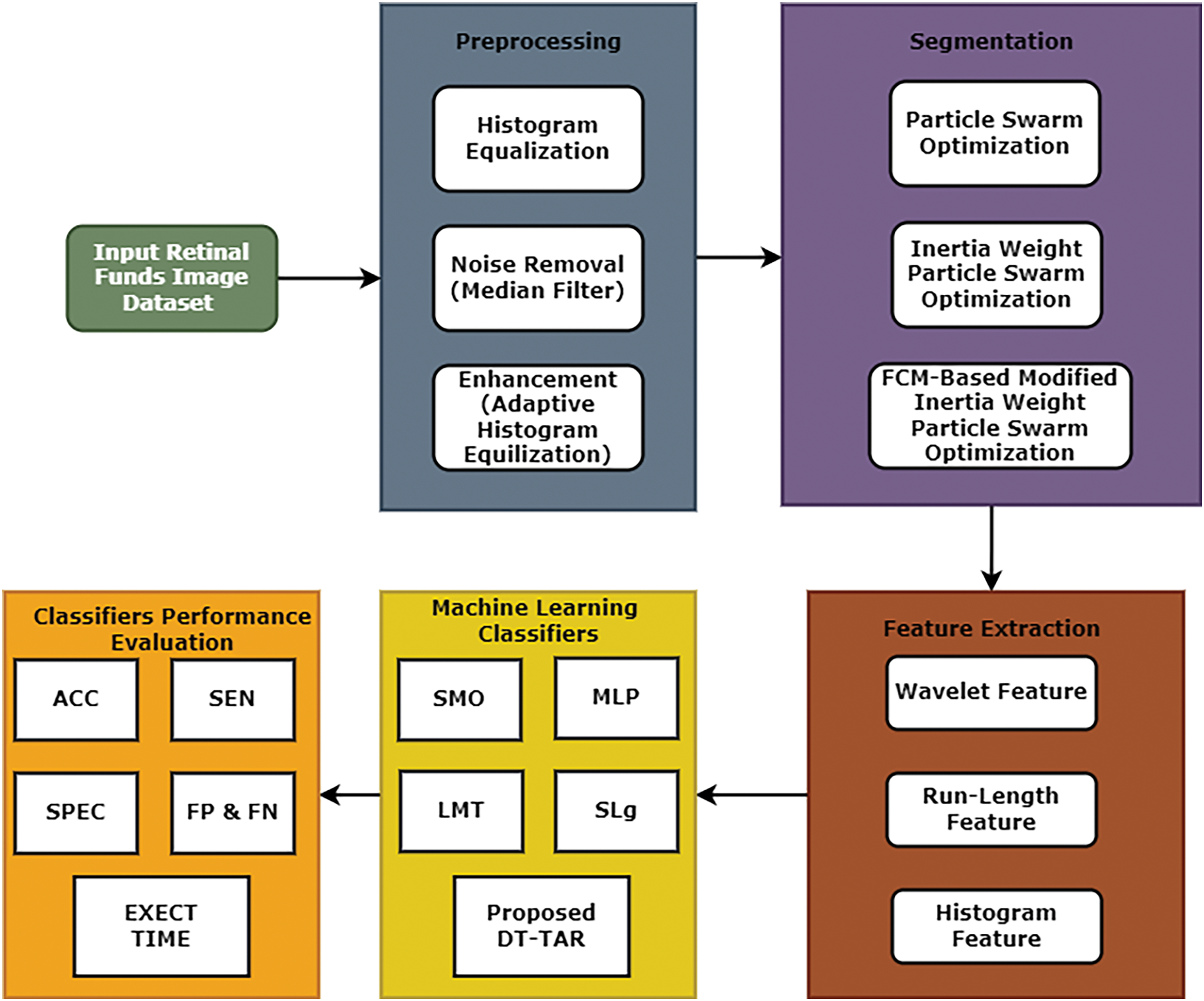
Figure 1: An efficient image processing framework
An ophthalmologist expert in the limelight of various medical examinations and biopsy reports analyzed the acquired images. The occurrence of malignancy in the image is classified by the rules generated from the testing information. Feature vectors are retrieved from the image utilized in testing and training. The classification process is highly influenced by the errors and nature of the dataset, which were rectified by the proposed technique.
Removal of noise in a medical image without disturbing the realistic features of an image is done by the filtering process. Filtering approaches are mathematically oriented and modelled based on the prior knowledge of the probability distribution of noise. The various specially designed filters considered in this research to remove the tainted noise in medical images are low pass filter, Gaussian filter, Weiner filter and median filter. The median filter conserves and maintains the edges while eliminating the corrupted noise. It is a statistical spatial domain filter that ranks high in eliminating speckle and impulse noise. The filtering process goes through pixel by pixel, replacing each pixel with the median of the neighborhood pixels. The format of neighbouring pixels is called a window, which slides pixel by pixel over the entire image. The procedure of this median filtering process is explained as follows:
1. Sort all the pixels in the image identified by an N × N mask in ascending or descending order.
2. Compute the median value of the sorted list.
3. Replace the original central pixel value with the computed median value.
4. Repeat Steps 1 to 3 until the median one replaces all the pixels.
Contrast in an image occurs due to the dissimilarity in radiance between two adjacent regions. Contrast enhancement is necessary to improve the visualization of organs, structures, edges and outlines of objects in an image. Contrast enhancement is achieved by expanding an image’s dynamic range of the gray levels. The most frequent enhancement technique used in practice is histogram equalization. Modifications have been made to the traditional histogram to enhance its performance. The image is divided into rectangular blocks; an equalizing histogram is computed for each block to find enhanced density value. Thus, the regions with different grayscale values can be simultaneously enhanced. Based on the histogram of neighbouring pixels, the histogram modification to each pixel is applied adaptively; hence, this equalization method is termed Adaptive Histogram Equalization. The contrast improvement is endorsed through the Contrast Enhancement Factor (CEF).
The study will offer a clearer understanding of the significance of preprocessing steps and substantiate the choice of noise removal and histogram equalization in the proposed framework.
3.2 Fuzzy C-Means-Based Modified Inertia Weight Particle Swarm Optimization (FCM-MIWPSO) Algorithm for Segmentation
FCM is one of the well-known algorithms used greatly in image segmentation. However, the traditional FCM, being highly sensitive to the initialization of cluster centres, falls into the local optimum solution, seriously affecting image segmentation. This study proposes a hybrid algorithm for optimizing particle swarms using enhanced Fuzzy C-Means and modified inertia weights. In the first phase, the modified inertia weight PSO is applied to find the local optimal solution. In the second phase, the final optimized solution is achieved by modified FCM with spatial features. The proposed method integrates the features of both algorithms and yields good accurate results compared to other algorithms.
In practical applications, incorporating a spatial function into the FCM algorithm results in more accurate and reliable image segmentation. This is particularly useful in medical imaging, where precise segmentation is critical for diagnosing conditions and planning treatments.
In this hybrid method, FCM could be used initially to cluster the retinal images into different groups based on pixel intensities or other features relevant to diabetic retinopathy. Then, PSO could be applied to fine-tune the parameters of the FCM algorithm, such as the number of clusters or the fuzziness coefficient, to enhance the clustering performance further. This iterative optimization process can help in finding the optimal partition of the image data, potentially leading to more accurate detection of diabetic retinopathy lesions or abnormalities.
It’s important to carefully design the hybridization of FCM and PSO, considering factors such as parameter initialization, convergence criteria, and the balance between exploration and exploitation in the optimization process. Additionally, thorough experimentation and validation using appropriate datasets with ground truth annotations are essential to assess the performance of the proposed method accurately.
The FCM-MIWPSO method for the early detection of diabetic retinopathy offers significant improvements in diagnostic accuracy, efficiency, patient outcomes, accessibility, and cost-effectiveness compared to traditional methods. These advancements can lead to better overall management of diabetic retinopathy, reducing the incidence of severe complications and improving the quality of life for patients with diabetes.
The remarkable shortfall of standard FCM is that the objective function does not incorporate the spatial information of neighbours. Hence, it is highly sensitive to noise and the segmentation results in discontinuity. In this proposed method, the spatial function is included in the membership weighting function to suppress the effect of noise. In a medical image, the nearby pixels almost hold the same feature data. So, the spatial dependency among the neighbouring pixels should be considered. In general, the spatial function can be modelled as the weighted addition of the fuzzy membership functions in the neighborhood of every pixel. It is mathematically given by the Eq. (2).
The modifications in membership function by including spatial function is given by,
where the parameter and control the significance of both functions.
In each iteration, the membership function of each pixel is computed in the spectral domain and then mapped into the spatial domain. To achieve perfect classification, new spatial function and lighting function are considered, and they are defined as,
where H(xi) represents spatial domain window centred on pixels xi.
The new spatial function comprises two parts. The coefficient αk1 in first part corrects the misclassified pixels from noisy regions and αk2 in second part quantifies the membership function. The expressions of αk1 and αk2 are,
Modified FCM segments the image by minimizing the objective function, and it is given by,
The selection of appropriate inertia weight is the primary criterion for achieving enhanced searchability for PSO. In general, the higher the value of inertia weight, the superior the algorithm performs in global search capability and inferior in local search ability; the smaller the value of inertia weight, the ability of global search and local search are just reversed. Several strategies to select a proper inertia weight have been proposed in earlier studies to balance local and global search ability trade-offs. These limitations of premature convergences and errors in accuracy and efficiency can be eliminated by proposing a modified inertia weight strategy.
The equation gives the value of inertia weight proposed in this work,
where t lies between 0 and maximum iterative time (tmax). By a suitable selection of parameters, several increasing or decreasing inertia weight strategies can be constructed with the single global minimum in [0, tmax]. Thus, this inertia weight consideration encloses a wide range of strategies. By easily changing the parameters γ, w1, w2, either better accuracy or efficiency or better accuracy and efficiency can be achieved, compromising the trade-off between efficiency and accuracy.
In this proposed method, each particle is presumed to be a candidate cluster centre and particles navigate the search space to search suitable cluster centers. Modified FCM is used to acquire cluster centres that minimize the dissimilarity function and modified inertia weight PSO attributes assigning each pixel to a cluster. Modified FCM takes these centres for better initialization. In this proposed method, instead of randomly initializing the fuzzy membership matrix, the position Pij represents the fuzzy membership matrix so that the membership matrix Uij satisfies the constraint.
where Pij represents the membership of the ith particle in the jth cluster. An n × c matrix expresses the velocity of each particle. The fitness function to be considered for modified IWPSO is given by,
where k is a constant and Jij is the cost criteria of modified FCM. For smaller the value of Jij fuzzy clustering is better and thus has a higher degree of individual fitness.
Proposed FCM-MIWPSO Algorithm
Step 1: The membership matrix with spatial features is initialized based on the position of Pij.
Step 2: Initial fitness function is evaluated as done in IWPSO.
Step 3: The velocity and position of the particles are updated, in which inertia weight is incorporated by using
Step 4: Normalize the membership matrix that includes the spatial function Uij given as,
Step 5: Compute αk1 and αk2 by using
Step 6: Calculate Sij * and Wij by using equations
Step 7: Update the membership matrices and centroids by using the equation
Step 8: Compute the objective function by equation
Step 9: Compute the new fitness function given in the equation
Step 10: Using the new fitness function, proceed with Steps 2 to 8 until the MIWPSO converges.
The dataset for this research work consists of hybrid features of diabetic retinopathy from retinal fundus images. The histogram first-order feature extraction, wavelet and run-length matrix-based second-order feature extraction techniques were used. Histogram features are utilized by selecting the object based on rows and columns. Such a binary object is utilized as a masquerade of the real image for feature extraction. Those features are identified from the intensity of every pixel for the object parts, and histogram probability P(H) is calculated as,
The wavelet transform feature can generally be considered a linear transformation implemented on a vector, where the value is a power of two, which numerically varies in quantitative values. The wavelet transform value is calculated using two sub-sampling processes: high and low pass filters.
3.4 Decision Tree-Based Temporal Association Rule (DT-TAR) Algorithm
Classifying the large set of available medical images into different classes as benign or malignant reduces physicians’ workload and time consumption as they need not examine all the test images. The Physician-to-patient ratio is very meagre in developing countries like India. So, classification methods are helpful to compromise this situation.
The DT-TAR algorithm combines the interpretability and straightforward decision-making process of decision trees with the pattern discovery capabilities of association rule mining. This hybrid approach allows the algorithm to capture both the static and dynamic relationships between features within the image data. Temporal features refer to changes in the image data over time. For medical images, these features are crucial as they can indicate the progression or regression of a condition. This work utilizes highly sophisticated classification and efficient decision-making decision tree-based temporal association rule for performing. This hybrid method integrates the characteristic features of association rule mining and decision trees. This exquisite classification algorithm uses temporal features to generate new decision trees from the training image data set. The requisite feeds for this classifier are the image features and training image data set. The most frequently occurring image features are recognized from the training image set. It formulates specially designed association rules based on temporal features between the images over a pre-defined time duration. Depending on the characteristic features like largest information gain, temporal characterization over a period and maximum information gain, the decisions for classifying the images as healthy control images and abnormal images are made.
4.1 Performance Evaluation Metrics
The premier FCM-MIWPS segmentation algorithm perfectly suited for segmenting a medical image is validated by computing various measuring metrics like accuracy, sensitivity, specificity, false positive rate, and false negative rate. When a person is subjected to medical examination, the normal person is medically termed as healthy control, and the disease-affected person is called a case. When the segmented result says positive for a disease-affected case, it is called true positive. When the segmented outcome says negative for a disease-free person, it is a true negative. When the segmentation output infers that the person is affected by the disease for healthy control, it is a false positive. When the segmentation outcome concludes that the disease-affected person has healthy control, it is a false negative.
Accuracy: This prime performance measuring metric is the ratio of true assessments to the total assessments. This image segmentation metric validates how effectively the disease-affected cases are identified as positive cases and healthy controls are identified as normal persons. The maximized performance of the segmentation algorithm is achieved when this ratio is unity or almost closer to unity, and this metric is mathematically,
Sensitivity: Sensitivity is usually termed as a true positive rate, which can be formulated as,
Specificity: Specificity is regularly called a true negative rate, which can be modelled as,
False Positive Rate: The calculative formula of false positive rate is given as,
False Negative Rate: The computational ratio of false negative rate is given as,
4.2 Experimental Result Analysis
This section demonstrates the segmented results of the employed algorithms in this research and explains the comparative analysis of performance evaluation measures for the algorithms. In this work, we have taken 20 sample images of the eye fundus image from the database. After being pre-processed by median filter and adaptive histogram equalization, these images were subjected to three different segmentation experiments using FCM, IWPSO, and our proposed method.
A confusion matrix provides a summary of prediction results on a classification problem. It shows the number of correct and incorrect predictions made by the model, broken down by each class. The matrix has dimensions equal to the number of classes, with each cell representing the count of instances. Tables 1–3 show the confusion matric in term of the false position and false negative rate. The confusion matrix, in particular, is highly informative as it displays the counts of true positive, true negative, false positive and false negative predictions. From this matrix, other metrics like precision and recall can be calculated, offering a more nuanced understanding of the model’s strengths and weaknesses, especially in handling imbalanced datasets or specific class distinctions.



The segmented output images of these methods are shown in Figs. 2–4. The comparison among the performance metric values for the algorithms used for detecting diabetic retinopathy is shown in Tables 1–3 and its graphical comparison is given in Figs. 5–7.
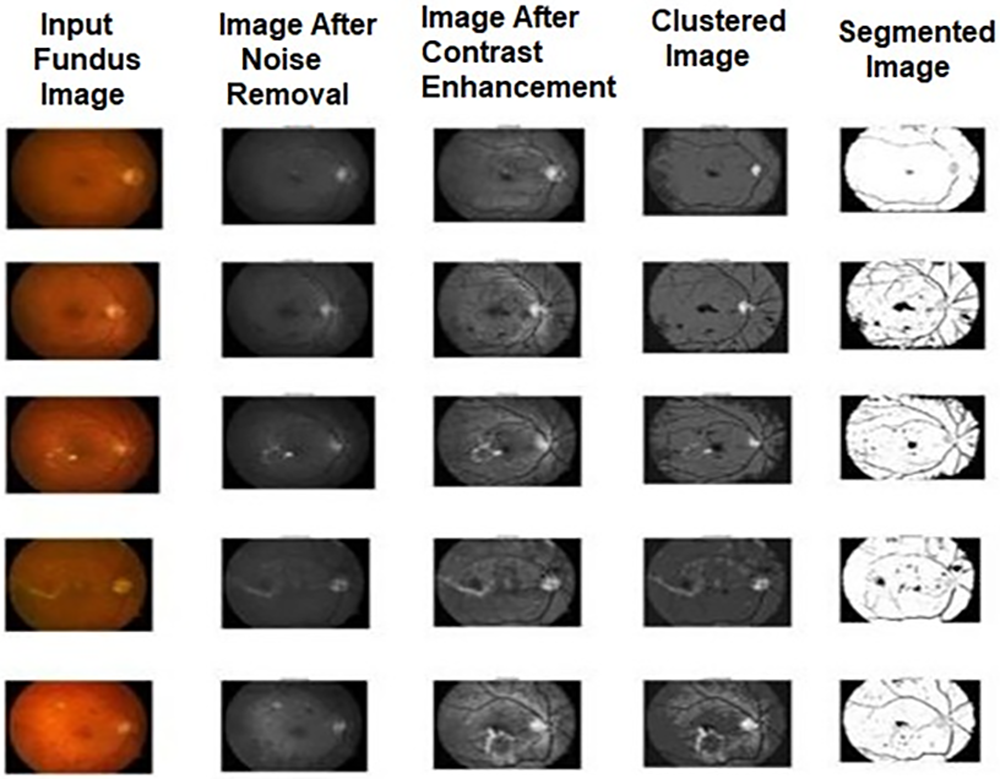
Figure 2: Sample output images of FCM for diabetic retinopathy
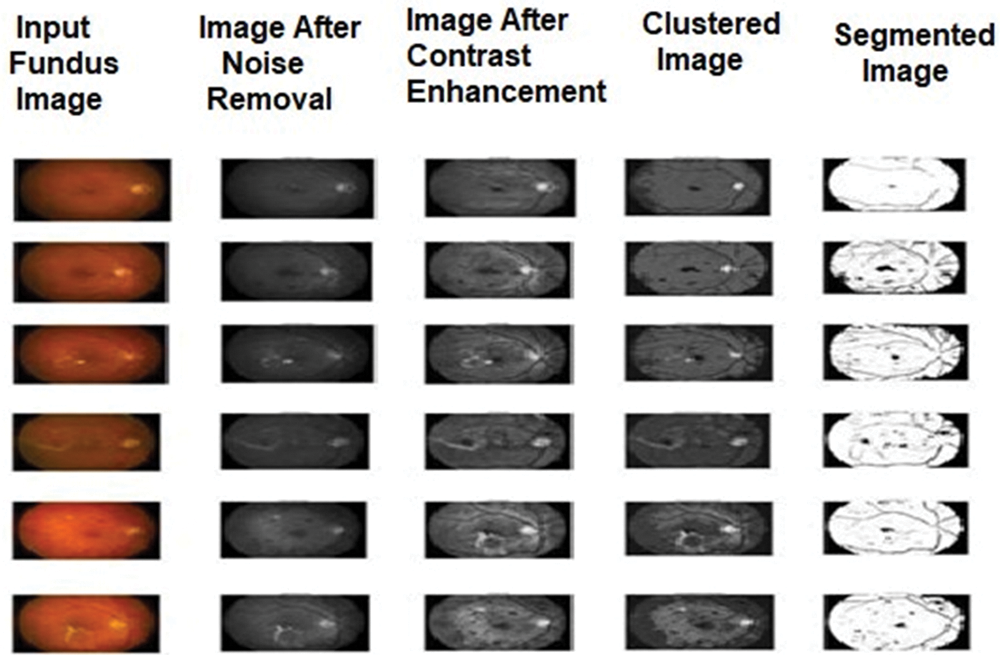
Figure 3: Sample output images of proposed FCM-MIWPSO for diabetic retinopathy
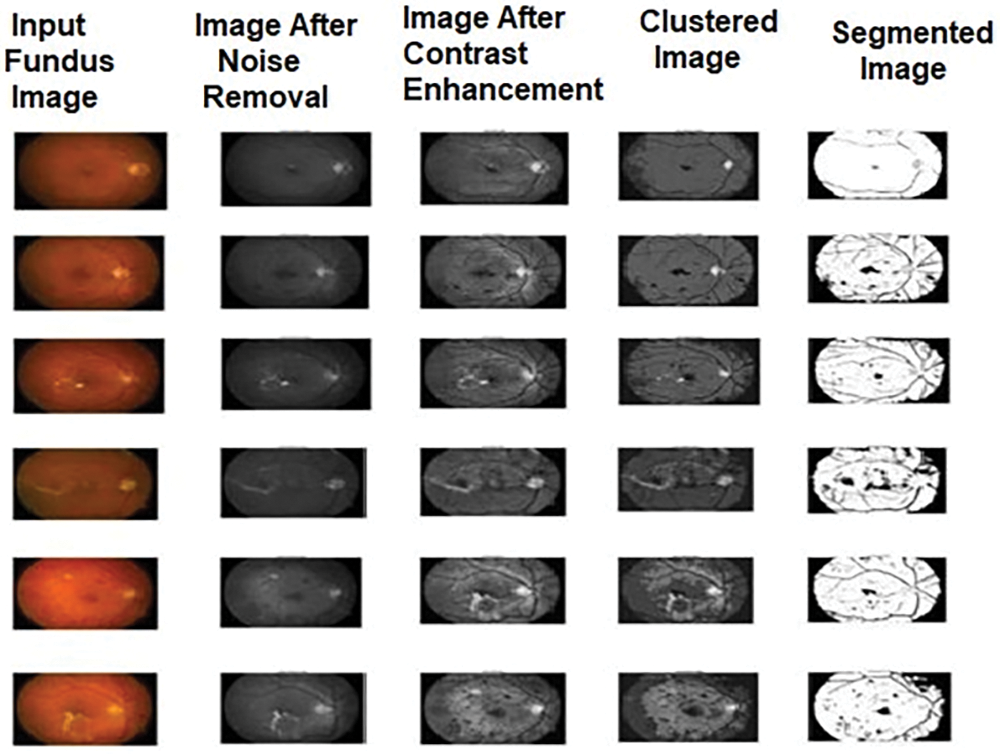
Figure 4: Sample output images of MIWPSO for diabetic retinopathy

Figure 5: Graphical comparison of performance measures of DT-TAR classification algorithm using miwpso segmented images

Figure 6: Graphical comparison of performance measures of DT-TAR classification algorithm using FCM segmented images

Figure 7: Graphical comparison of performance measures of DT-TAR classification algorithm using FCM-MIWPSO segmented images
To prove the proposed algorithm’s excellent classification, 20 samples containing normal and abnormal images were taken for our experimental consideration. These images were classified using DT-TAR and existing classifiers such as SMO, LMT, MLP, and SLg classification. These classifiers were tested using the segmented images provided by the FCM-MIWPSO segmentation algorithm, whose results are depicted in Fig. 8. From the analysis, it has been found that the proposed DT-TAR classifier performs better than the existing classifiers.
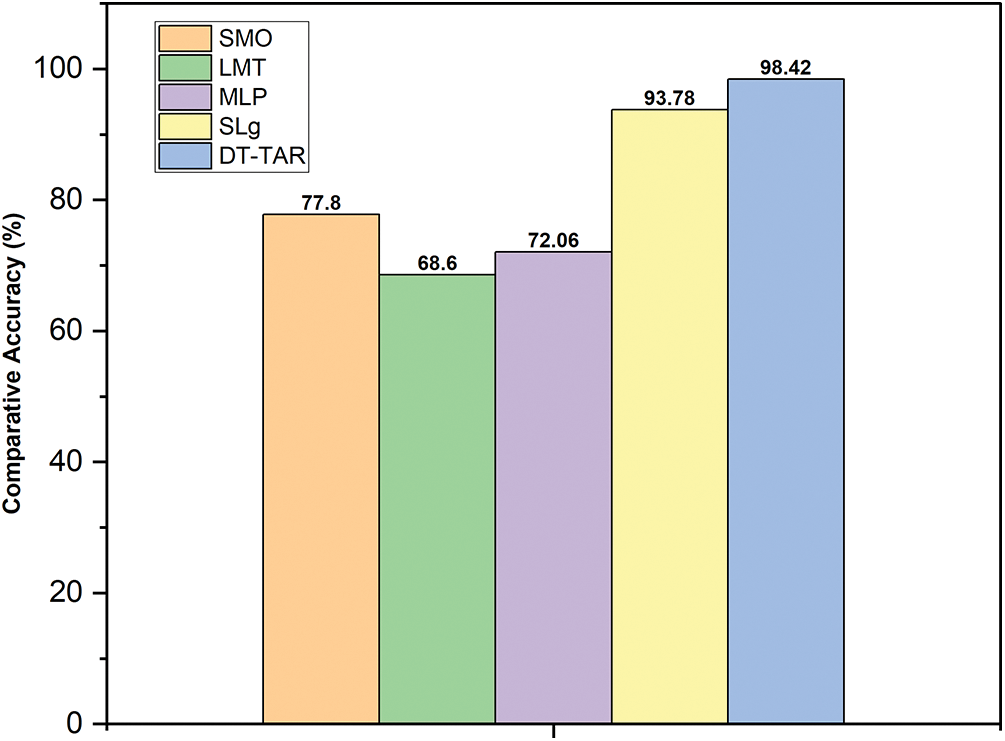
Figure 8: Graphical comparison of accuracies of existing and proposed DT-TAR classifiers using FCM-MIWPSO segmented images
The performance of the proposed model attains an accuracy of about 98.42%, which indicates the efficiency of the proposed model. The proposed model shows the higher performance as compared to the existing models as shown in Table 4.
The segmented results obtained from various segmentation algorithms, including the proposed algorithm, are thoroughly presented in this research. Performance measures for each algorithm were compared, demonstrating that the proposed method consistently achieved superior performance indicators. This makes it an optimal choice for segmenting regions of interest in medical images, particularly for detecting diabetic retinopathy. Despite the challenges of applying a single algorithm to diverse medical images, our proposed method excels in addressing these challenges, establishing it as a highly recommended segmentation algorithm for different medical imaging applications. Additionally, computational efficiency can be significantly improved by reducing the fuzzy weighting matrices and dissimilarity functions. The future scope of this research is promising, with potential applications extending beyond diabetic retinopathy detection. The proposed framework can be adapted and applied to various other medical imaging challenges, such as tumor detection, organ delineation, and vascular structure analysis. Further research could explore the integration of deep learning techniques to enhance segmentation accuracy and efficiency. Moreover, the adaptability of the proposed method to different imaging modalities, such as MRI, CT scans, and ultrasound images, could be investigated. This would broaden its applicability and impact in the medical field, ultimately contributing to more precise diagnostics and improved patient outcomes.
Acknowledgement: This project is funded by Deanship of Scientific Research at the University of Ha’il–Ha’il, Kingdom of Saudi Arabia through project number RG-21 104.
Funding Statement: Scientific Research Deanship has funded this project at the University of Ha’il–Saudi Arabia Ha’il–Saudi Arabia through project number RG-21 104.
Author Contributions: Kusum Yadav: study conception and design, analysis and interpretation of results, methodology development; Yasser Alharbi: data collection, draft manuscript preparation, figure and tables; Eissa Jaber Alreshidi: funding, supervision, resource management; Abdulrahman Alreshidi: data collection, draft manuscript preparation; Anuj Kumar Jain: methodology, study conception and design; Anurag Jain: analysis, resource management, revised menuscript; Kamal Kumar: data collection, prepossessing, data presentation; Sachin Sharma: final manuscript revision, funding, supervision; Brij B. Gupta: final manuscript revision, funding, supervision. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All data generated or analysed during this study is collected from Kaggle, which can be accessed via the following link: https://www.kaggle.com/datasets/eishkaran/diabetes-using-retinopathy-prediction (accessed on 20 March 2024).
Ethics Approval: No participation of humans takes place in this implementation process.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. R. Wang, T. Lei, R. Cui, B. Zhang, H. Meng and A. K. Nandi, “Medical image segmentation using deep learning: A survey,” IET Image Process., vol. 16, no. 5, pp. 1243–1267, 2022. doi: 10.1049/ipr2.12419. [Google Scholar] [CrossRef]
2. V. Mayya, S. Kamath, and U. Kulkarni, “Automated microaneurysms detection for early diagnosis of diabetic retinopathy: A Comprehensive review,” Comput. Methods Programs Biomed. Update, vol. 1, no. 5, 2021, Art. no. 100013. doi: 10.1016/j.cmpbup.2021.100013. [Google Scholar] [CrossRef]
3. P. Arora, P. Singh, A. Girdhar, and R. Vijayvergiya, “A state-of-the-art review on coronary artery border segmentation algorithms for intravascular ultrasound (IVUS) images,” Cardiovasc. Eng. Technol., vol. 14, no. 2, pp. 264–295, 2023. doi: 10.1007/s13239-023-00654-6. [Google Scholar] [PubMed] [CrossRef]
4. W. Burger and M. J. Burge, Digital image processing: An algorithmic introduction, Switzerland: Springer Nature, 2022. [Google Scholar]
5. P. Scheltens et al., “Alzheimer’s disease,” Lancet, vol. 397, no. 10284, pp. 1577–1590, 2021. doi: 10.1016/S0140-6736(20)32205-4. [Google Scholar] [PubMed] [CrossRef]
6. Priyanka and B. Singh, “A review on brain tumor detection using segmentation,” Int. J. Comput. Sci. Mobile Comput., vol. 2, no. 7, pp. 48–54, 2013. [Google Scholar]
7. L. Roncon, M. Zuin, G. Rigatelli, and G. Zuliani, “Diabetic patients with COVID-19 infection are at higher risk of ICU admission and poor short-term outcome,” J. Clin. Virol., vol. 127, 2020, Art. no. 104354. doi: 10.1016/j.jcv.2020.104354. [Google Scholar] [PubMed] [CrossRef]
8. Z. Xia et al., “Scheme based on multi-level patch attention and lesion localization for diabetic retinopathy grading,” Comput. Model. Eng. Sci., vol. 140, no. 1, 2024. doi: 10.32604/cmes.2024.030052. [Google Scholar] [CrossRef]
9. M. Storath and A. Weinmann, “Fast median filtering for phase or orientation data,” IEEE Trans. Pattern Anal. Mach. Intell., vol. 40, no. 3, pp. 639–652, 2018. doi: 10.1109/TPAMI.2017.2692779. [Google Scholar] [PubMed] [CrossRef]
10. N. Jain and V. Kumar, “Liver ultra sound image segmentation using region difference filters,” J. Digit. Imaging, vol. 3, no. 3, pp. 376–390, 2017. doi: 10.1007/s10278-016-9934-5. [Google Scholar] [PubMed] [CrossRef]
11. H. Strange, L. Scott, and R. Zwiggelaar, “Myofibre segmentation in H&E stained adult Sketal muscle images using coherence-enhancing diffusion filtering,” BMC Med. Imaging, vol. 14, no. 1, 2014, Art. no. 38. doi: 10.1186/1471-2342-14-38. [Google Scholar] [PubMed] [CrossRef]
12. S. H. Rasta, M. E. Partovi, H. Seyedarabi, and A. Javadzadeh, “A comparative study on preprocessing techniques in diabeticretinopathy retina image. Illumination correction and contrast enhancement,” J. Med. Signals Sens., vol. 5, no. 1, pp. 40–48, 2015. doi: 10.4103/2228-7477.150414. [Google Scholar] [CrossRef]
13. H. Shamsi and H. Seyedarabi, “Modified Fuzzy C-means clustering with spatial information for image segmentation,” Int. J. Comput. Theory Eng., vol. 4, no. 5, pp. 762–766, 2012. doi: 10.7763/IJCTE.2012.V4.573. [Google Scholar] [CrossRef]
14. Y. Zou and B. Liu, “Survey on clustering-based image segmentation techniques,” in Proc. IEEE 20th Int. Conf. Comput. Supp. Coop. Work Des., Nanchang, China, 2016, pp. 106–110. [Google Scholar]
15. A. K. Dubey, U. Gupta, and S. Jain, “Analysis of K mean clustering approach on the breast cancer Wisconsin data set,” Int. J. Comput. Assist. Radiol. Surg., vol. 11, no. 11, pp. 2033–2047, 2016. doi: 10.1007/s11548-016-1437-9. [Google Scholar] [PubMed] [CrossRef]
16. L. Zhang, L. P. Wang, and W. Lin, “Semi supervised biased maximum margin analysis for interactive image retrieval,” IEEE Trans. Image Process., vol. 21, no. 4, pp. 2294–2308, 2012. doi: 10.1109/TIP.2011.2177846. [Google Scholar] [PubMed] [CrossRef]
17. P. Kalshetti et al., “An interactive medical image segmentation framework using iterative refinement,” Comput. Biol. Med., vol. 83, no. 11, pp. 182–197, 2017. doi: 10.1016/j.compbiomed.2017.02.002. [Google Scholar] [PubMed] [CrossRef]
18. K. Xu, D. Feng, and H. Mi, “Deep convolution neural network based early automated detection of diabetic retinopathy using fundus image,” Molecules, vol. 22, no. 12, 2017, Art. no. 2054. doi: 10.3390/molecules22122054. [Google Scholar] [PubMed] [CrossRef]
19. C. Parsai, R. O’Hanlon, S. K. Prasad, and R. H. Mohiaddin, “Diagnostic and prognostic value of cardiovascular magnetic resonance in non-ischaemic cardiomyopathies,” J. Cardiovasc. Magn. Reson., vol. 14, no. 1, 2012, Art. no. 33. doi: 10.1186/1532-429X-14-54. [Google Scholar] [PubMed] [CrossRef]
20. B. Schmauch et al., “Combining a deep learning model with clinical data better predicts hepatocellular carcinoma behavior following surgery,” J. Pathol. Inf., vol. 15, no. 3, 2023, Art. no. 100360. doi: 10.1016/j.jpi.2023.100360. [Google Scholar] [PubMed] [CrossRef]
21. R. K. Jansen et al., “Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns,” Proc. Natl. Acad. Sci., vol. 104, no. 49, pp. 19369–19374, 2007. doi: 10.1073/pnas.0709121104. [Google Scholar] [PubMed] [CrossRef]
22. J. T. Oliva, H. D. Lee, N. Spolaôr, C. S. R. Coy, and F. C. Wu, “Prototype system for feature extraction, classification and study of medical images,” Expert. Syst. Appl., vol. 63, pp. 267–283, 2016. doi: 10.1016/j.eswa.2016.07.008. [Google Scholar] [CrossRef]
23. A. Romero-Corral et al., “Association of bodyweight with total mortality and with cardiovascular events in coronary artery disease: A systematic review of cohort studies,” Lancet, vol. 368, no. 9536, pp. 666–678, 2006. doi: 10.1016/S0140-6736(06)69251-9. [Google Scholar] [PubMed] [CrossRef]
24. C. S. Peri, P. Benaglia, D. P. Brookes, I. R. Stevens, and N. L. Isequilla, “E-BOSS: An Extensive stellar BOw Shock Survey: I. Methods and first catalogue,” Astron. Astrophys., vol. 538, 2012, Art. no. A108. doi: 10.1051/0004-6361/201118116. [Google Scholar] [CrossRef]
25. J. Nygård, D. H. Cobden, and P. E. Lindelof, “Kondo physics in carbon nanotubes,” Nature, vol. 408, no. 6810, pp. 342–346, 2000. doi: 10.1038/35042545. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF
 Downloads
Downloads
 Citation Tools
Citation Tools