 Open Access
Open Access
ARTICLE
Leveraging EfficientNetB3 in a Deep Learning Framework for High-Accuracy MRI Tumor Classification
1 Department of Computer Science & Engineering, Faculty of Engineering and Technology, JAIN (Deemed-to-be University), Bengaluru, 562112, India
2 School of Computer Science and Engineering, VIT-AP University, Amaravati, 522241, India
3 Department of Computer Science & Technology, Madanapalle Institute of Technology & Science, Madanapalle, 517325, India
4 School of Science, Engineering and Environment, University of Salford, Manchester, M5 4WT, UK
5 School of Computer Science Engineering & Information Systems (SCORE), Vellore Institute of Technology (VIT), Vellore, 632014, India
6 Management Information System Department, College of Business Administration, King Saud University, Riyadh, 11531, Saudi Arabia
7 Adjunct Research Faculty, Centre for Research Impact & Outcome, Chitkara University, Rajpura, Punjab, 140401, India
* Corresponding Author: Surbhi Bhatia Khan. Email:
(This article belongs to the Special Issue: Medical Imaging Based Disease Diagnosis Using AI)
Computers, Materials & Continua 2024, 81(1), 867-883. https://doi.org/10.32604/cmc.2024.053563
Received 04 May 2024; Accepted 07 August 2024; Issue published 15 October 2024
Abstract
Brain tumor is a global issue due to which several people suffer, and its early diagnosis can help in the treatment in a more efficient manner. Identifying different types of brain tumors, including gliomas, meningiomas, pituitary tumors, as well as confirming the absence of tumors, poses a significant challenge using MRI images. Current approaches predominantly rely on traditional machine learning and basic deep learning methods for image classification. These methods often rely on manual feature extraction and basic convolutional neural networks (CNNs). The limitations include inadequate accuracy, poor generalization of new data, and limited ability to manage the high variability in MRI images. Utilizing the EfficientNetB3 architecture, this study presents a groundbreaking approach in the computational engineering domain, enhancing MRI-based brain tumor classification. Our approach highlights a major advancement in employing sophisticated machine learning techniques within Computer Science and Engineering, showcasing a highly accurate framework with significant potential for healthcare technologies. The model achieves an outstanding 99% accuracy, exhibiting balanced precision, recall, and F1-scores across all tumor types, as detailed in the classification report. This successful implementation demonstrates the model’s potential as an essential tool for diagnosing and classifying brain tumors, marking a notable improvement over current methods. The integration of such advanced computational techniques in medical diagnostics can significantly enhance accuracy and efficiency, paving the way for wider application. This research highlights the revolutionary impact of deep learning technologies in improving diagnostic processes and patient outcomes in neuro-oncology.Keywords
Brain tumors, emerging as abnormal cell masses within the brain’s complex structure, stand at the forefront of modern medical challenges. Their presence not only poses a significant health threat but also demands intricate diagnostic measures for effective treatment. The classification of these tumors is vital in crafting patient-specific treatment strategies and prognostic assessments. This extensive study explores the intricate field of brain tumors, focusing on their diverse classifications, diagnostic challenges, and the crucial role of Magnetic Resonance Imaging (MRI) in their detection and evaluation. Brain tumors are typically categorized into primary and secondary tumors. Primary brain tumors originate within the brain itself and encompass various types, including gliomas, meningiomas, and pituitary tumors [1,2]. Conversely, secondary brain tumors, or metastases, originate from cancer cells that have traveled from other body parts. The diversity in their biological behavior necessitates precise classification, as each tumor type requires a specific therapeutic approach.
Addressing MRI interpretation challenges involves improving imaging quality, refining radiologists’ training, and integrating AI and machine learning algorithms. AI enhances diagnostic accuracy and reduces interpretative variability. Ongoing research in imaging techniques like fMRI, DTI, and spectroscopy offers deeper insights into brain tumor pathophysiology, aiding in precise diagnosis and treatment planning [3,4].
Our method stands out due to its utilization of EfficientNetB3, known for its optimal balance of model depth, width, and resolution, which enables high performance on complex image classification tasks with reduced computational demand [5,6]. This is particularly important in medical imaging, where precision and efficiency are critical. Compared to traditional methods, our approach offers significant improvements in processing speed and diagnostic accuracy, achieving an impressive classification accuracy of 99%. This high level of performance underscores the potential of our model to provide rapid and reliable diagnostic support in clinical settings, facilitating timely and precise treatment decisions, thus highlighting its substantial benefits over existing methodologies.
The motivation behind this research is driven by the aim to enhance the accuracy and reliability of brain tumor diagnoses. Given the high stakes involved in neuro-oncology, there is a pressing need for tools that can supplement the expertise of medical professionals [7–10].
Despite significant advances in brain tumor classification using deep learning, several gaps persist: limited use of EfficientNetB3 in medical imaging, insufficient integration of diverse MRI datasets, challenges in addressing overfitting and model generalization, lack of focus on real-time diagnostic support, and a narrow focus on accuracy over comprehensive evaluation metrics. This study aims to address these gaps by leveraging EfficientNetB3 for enhanced classification, integrating diverse MRI datasets for better generalizability, implementing advanced regularization techniques to combat overfitting, emphasizing real-time diagnostic potential, and conducting a comprehensive performance evaluation using various metrics including accuracy, precision, recall, F1-score, and ROC-AUC curves.
The contributions of the proposed study are summarized as follows:
• This study introduces a cutting-edge deep learning model that incorporates the pre-trained EfficientNetB3 architecture, augmented with custom dense layers, to classify brain tumors from MRI images.
• The study demonstrates the model’s superior performance compared to existing methods and establishes a framework that can be applied in real-world clinical settings.
• Introduces a model that excels in accurately classifying gliomas, meningiomas, absence of tumors, and pituitary tumors, providing a dependable tool for clinical decision-making.
Efforts to achieve effective and precise classification of brain tumors have been a prominent focus of research in the medical field [11,12]. This literature review aims to traverse through various methodologies adopted over the years, from conventional image processing techniques to the sophisticated realm of machine learning algorithms, particularly deep learning. In Table 1, several studies are mentioned with their summary.
The literature on brain tumor classification has primarily concentrated on different deep learning models, with significant attention given to convolutional neural networks (CNNs) for their effectiveness in processing image data. Previous studies have employed architectures like VGG, ResNet, and U-Net, often yielding high accuracies but at the cost of substantial computational resources and complex tuning requirements. EfficientNetB3 tackles these challenges by utilizing a scalable architecture that optimizes the balance between depth, width, and resolution of the network, thereby improving both accuracy and computational efficiency [20]. Medina and Sánchez in their research work used the EfficientNetB0, B3, and small and used datasets that are publicly available, such as figshare, compared to this study, the proposed study proposes the use of 3 different datasets, and each feature scaled for better accuracy adaptation. Similarly, Reyes and Sánchez in their study proposed CONVNEXT and EfficientNet models, also this model was performed well but in the same, it suggested research direction leading to better feature extraction and dataset [21]. The proposed model leverages compound scaling, which allows it to systematically and uniformly scale up CNNs in a way that traditional models do not, making it uniquely suited for medical imaging tasks where both detail precision and processing speed are crucial. Through the integration of EfficientNetB3, our study advances technological capabilities in the field, presenting a method that notably enhances the speed and accuracy of brain tumor classifications compared to previous models. This addresses critical technological gaps and fulfills clinical needs more effectively.
Our methodological approach harnesses the EfficientNetB3 architecture, a testament to the convergence of engineering innovation and medical diagnostics. Through meticulous data preprocessing and augmentation strategies, we exemplify the engineering rigor applied to enhance model performance, emphasizing the synergy between computational engineering principles and healthcare applications. The workflow of the model is shown in Fig. 1.
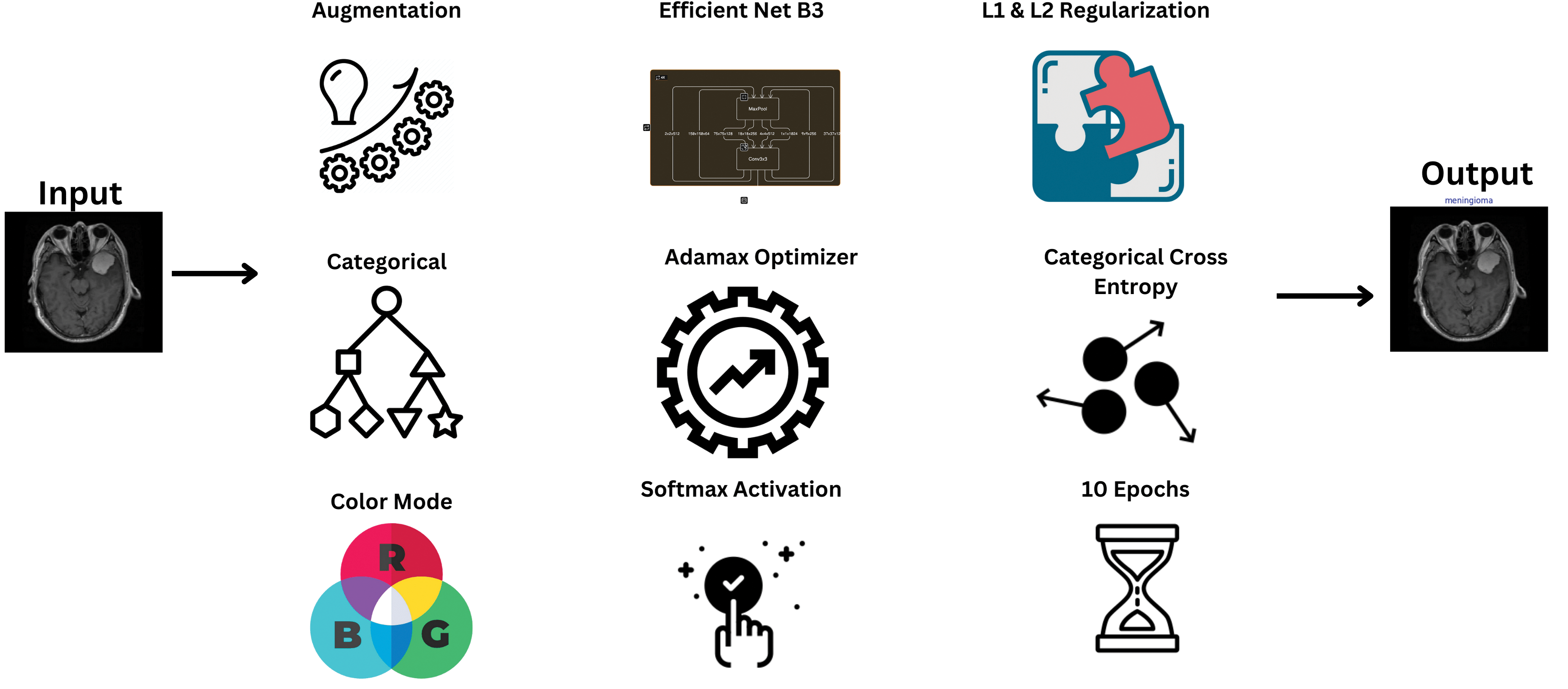
Figure 1: Model schematic diagram
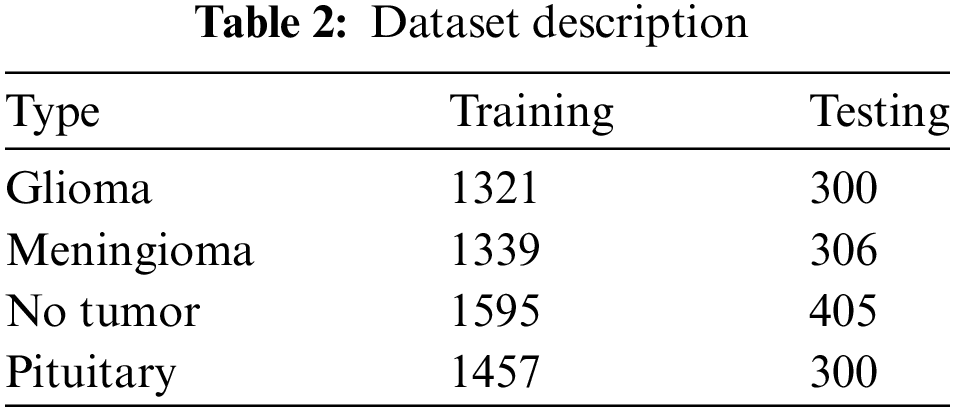
The dataset utilized in this research comprises a total of 7023 MRI pictures collected from three different sources: figshare [22], the SARTAJ dataset [23], and Br35H [24]. This compilation guarantees a comprehensive representation of tumor appearances, thereby improving the model’s resilience and applicability. Before the model was trained and evaluated, preprocessing of these pictures was indispensable. Initially, all pictures underwent resizing to a consistent dimension of 224 × 224 pixels to maintain uniformity across the dataset, which is critical for effective data handling by neural networks. This methodical distribution ensured that the model was exposed to balanced and varied data during both training and testing phases, as illustrated in Table 2.
Preprocessing is crucial in machine learning, especially for handling medical images such as MRI scans used in brain tumor classification. Fig. 2 illustrates sample images from the dataset, processed using Eq. (1).
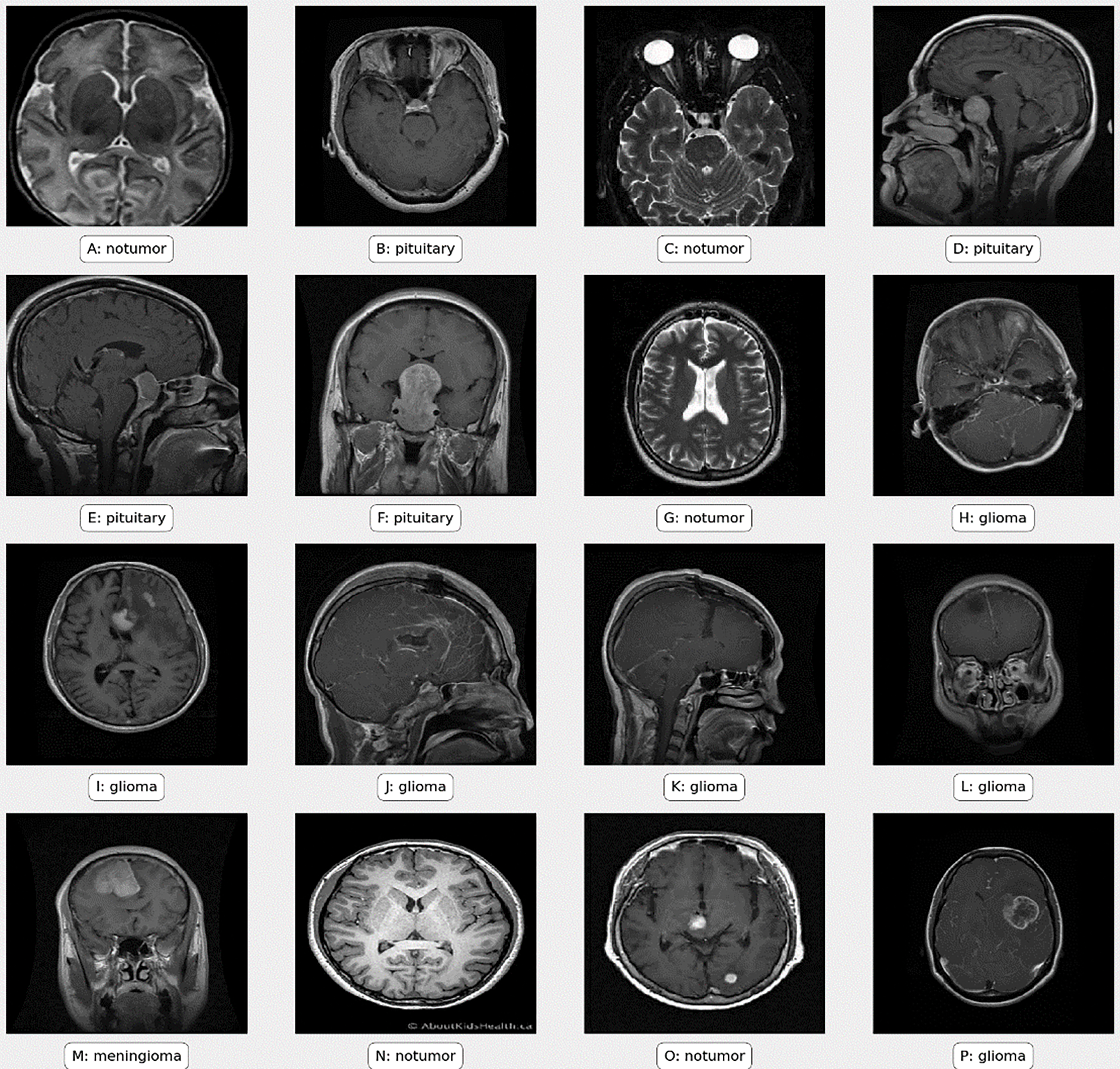
Figure 2: Sample images from the dataset
Neural networks require a consistent input format to process data efficiently. By resizing all images to the same dimensions, we ensure that each input fed into the network is of a uniform size, thereby facilitating smoother and more efficient processing and further the image is normalized using Eq. (2).
Algorithm 1 details the steps for preparing the MRI image data for model training and evaluation.

The augmentation techniques used include:
• Adjustments in Brightness and Contrast: Variations in brightness and contrast were introduced randomly. This step is particularly important as it mimics the variations that can occur in real-world imaging scenarios due to differences in MRI machine settings and conditions.
• Rotation and Flipping: The images underwent random rotation and horizontal flipping. These transformations aid the model in learning to detect tumors irrespective of their orientation within the brain.
• Scaling and Translation: Random scaling and translation (shifting) of images were also performed. This ensures that the model is not biased towards tumors of a specific size or location within the brain.
• Artificial Noise Addition: To simulate the effect of different MRI machine qualities and imaging conditions, artificial noise was occasionally added to the images. This step helps in making the model robust to noise that is often present in real MRI scans.
Further, to mimic real-world variations in MRI scans and improve the model’s adaptability, the images underwent normalization and augmentation processes. These included adjustments in brightness and contrast, as well as the application of random rotations, flips, and translations. Additionally, to simulate various qualities of MRI machines and imaging conditions, artificial noise was occasionally introduced. These preprocessing steps were vital in preparing the dataset for effective training of the deep learning model, ensuring that it not only learns to recognize patterns in tumor images but also becomes resilient to common variations and noise in clinical MRI scans.
In this research, transfer learning plays a pivotal role in enhancing the model’s accuracy in classifying brain tumors from MRI scans. We selected EfficientNetB3 for our classification model due to its exceptional efficiency and scalability, vital for medical imaging applications. EfficientNetB3, a part of the EfficientNet series, employs an innovative compound scaling approach to progressively enhance the network’s depth, width, and resolution. This strategy enables improved accuracy while simultaneously reducing the number of parameters and computational expenses. This is crucial in clinical settings with limited resources where fast processing is essential for timely diagnoses. EfficientNetB3 utilizes advanced techniques like depthwise separable convolutions to minimize computational demands without compromising performance. It has outperformed larger models such as ResNet and VGG in both accuracy and efficiency, making it ideal for medical imaging where high-quality annotated datasets are often scarce. Its ability to maintain high accuracy with limited training data and its fine-grained feature extraction capabilities are essential for accurately distinguishing subtle differences in medical images, critical for classifying brain tumors. Moreover, its robustness to variations in image quality and adaptability to different scales of input data further justify its selection, ensuring the model is powerful and versatile in handling real-world medical imaging challenges.
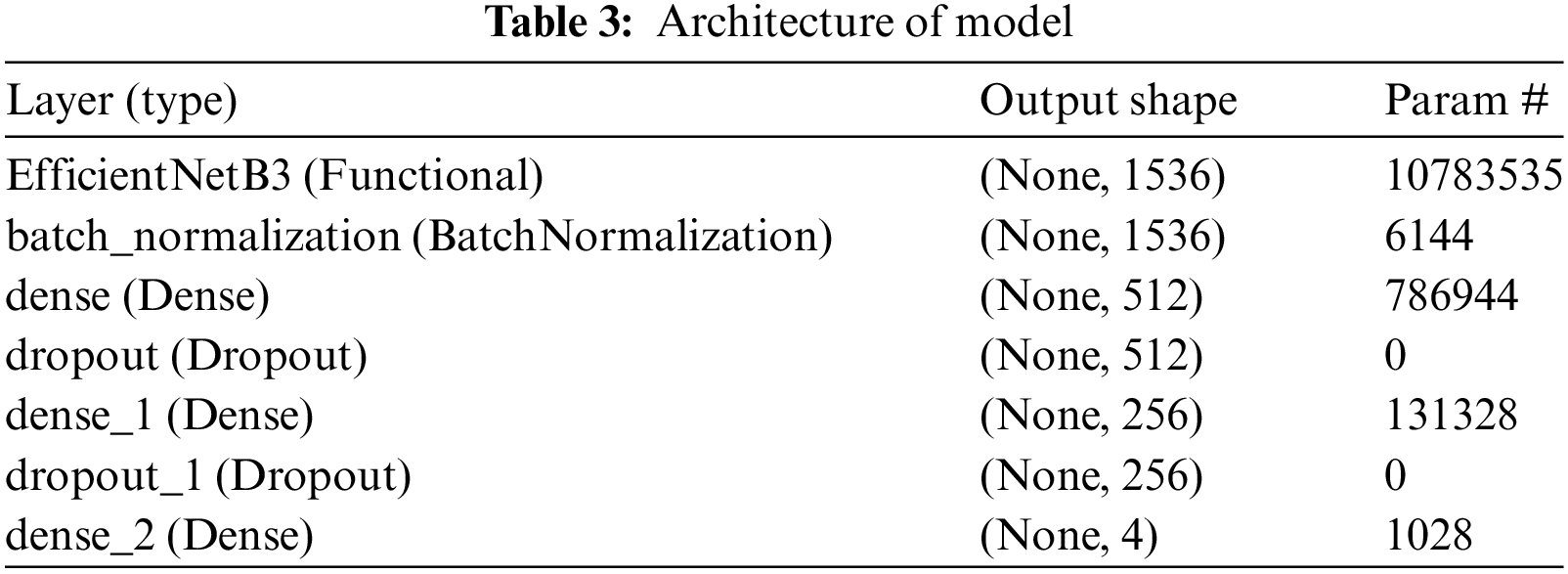
Custom layers have been added on top of EfficientNetB3 to enhance its capabilities for brain tumor classification. Following the base model, a BatchNormalization layer normalizes activations from the preceding layer, stabilizing learning and enhancing training speed. Two Dense layers with 512 and 256 units incorporate L1 and L2 regularization to counter overfitting common in medical image analysis, addressing complex patterns and inherent noise in the data. Dropout layers with rates of 0.4 and 0.2 are placed between these Dense layers to further prevent overfitting by randomly excluding features during training.
The ultimate layer is a Dense output layer that utilizes softmax activation, customized for the specific classes in the dataset (glioma, meningioma, no tumor, pituitary tumors). Softmax transforms outputs into probability scores, facilitating clinical decision-making by indicating the likelihood of each tumor type. The model is compiled with Adamax optimizer, noted for handling sparse gradients well, minimizing categorical crossentropy suitable for multi-class tasks in classification. This architecture combines advanced deep learning techniques, finely tuned for medical imaging challenges, aiming for high accuracy and reliability crucial in clinical applications.
The architecture of this model and its visual representation is given in Table 3.
Algorithm 2 outlines the construction of the neural network using the EfficientNetB3 architecture. Key steps include integrating the pre-trained EfficientNetB3 as the base, adding batch normalization and dense layers with regularization to combat overfitting, and including dropout layers for robustness, culminating in a SoftMax output layer for multi-class classification.

To customize the EfficientNetB3 model for brain tumor classification, several key changes were made. First, the original top layers were removed because they were designed for different tasks, not medical imaging. A Batch Normalization layer was added right after the base model to stabilize and speed up the learning process. Then, a series of Dense layers were included to help the model learn complex patterns specific to brain tumors. In order to mitigate overfitting, regularizers were incorporated into these Dense layers, ensuring the model does not overly specialize to the training data. Dropout layers were additionally inserted between the Dense layers to enhance prevention of overfitting, thereby decreasing dependency on specific neuron sets. These modifications enhance the model’s accuracy and robustness in diagnosing brain tumors from medical images.
3.3 Final Layer and SoftMax Activation
The final layer of the model is crucial for classifying tumors. It’s like the brain of the model, helping it decide which type of tumor it’s looking at. This layer uses something called SoftMax activation, which turns the model’s output into probabilities for each of the four tumor classes. This makes it easier to understand which type of tumor the model thinks it’s seeing. The output layer has four parts, each one representing a different tumor type, giving a score for how likely it is that the tumor belongs to each class.
3.4 Training and Validation Methodology
Training utilized the Adamax optimizer, a derivative of Adam known for effectively managing sparse gradients in noisy scenarios. A learning rate of 0.001 was chosen to strike a balance between training efficiency and convergence reliability.
Algorithm 3 outlines the training process of the model, including defining the number of epochs, utilizing real-time data from generators, and evaluating the model’s performance on new data after each epoch to encourage generalization and reduce overfitting.

During the training process, batches of images along with their corresponding labels were fed into the model. A batch size of 16 was chosen, ensuring each gradient update had ample data while remaining within the constraints of GPU memory capacity. The training and validation are shown in Fig. 3.
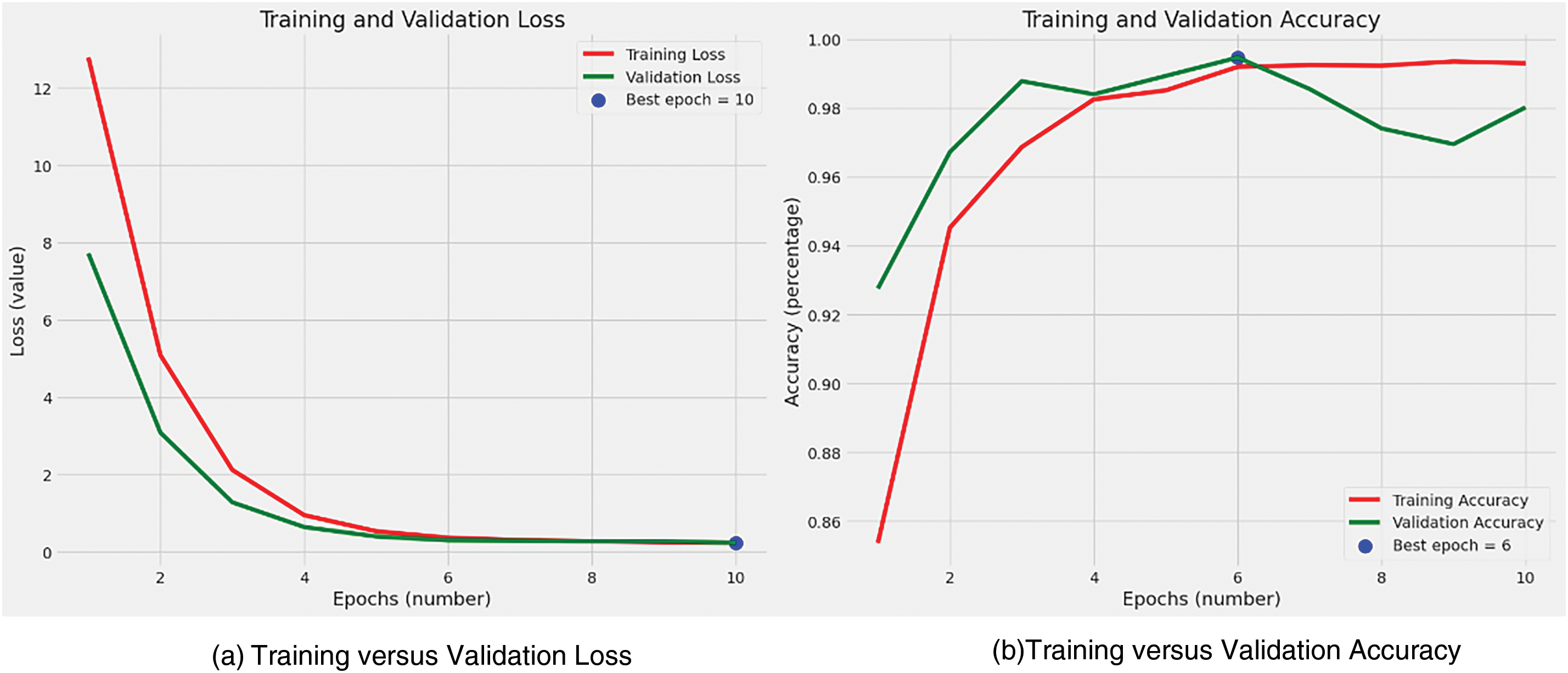
Figure 3: Training and validation
In our study of brain tumor classification using MRI images, optimizing crucial training parameters—commonly known as hyperparameters—greatly impacted the effectiveness and efficiency of the deep learning model. The choice of a learning rate, specifically configured at 0.001 using the Adamax optimizer, was meticulously made to achieve a delicate balance between rapid convergence and accuracy. This balance is especially crucial in medical imaging research. A batch size of 16 provided computational efficiency while maintaining reasonable gradient variance, helping prevent overfitting. Training for 10 epochs ensured sufficient convergence without overfitting, as indicated by stabilized loss and accuracy metrics. Dropout rates of 0.4 and 0.2 were applied in different layers to prevent overfitting and enhance robustness. L1 and L2 regularizations in the dense layers controlled overfitting and promoted model interpretability and reliability, essential in medical applications.
3.5 Model Evaluation Techniques
Assessing the model’s performance is a pivotal step in gauging its efficacy and practicality.
Accuracy: This measure is straightforward, representing the proportion of correctly predicted observations out of the total number of observations. It offers a quick evaluation of the model’s overall effectiveness and is calculated using Eq. (3).
Precision and Recall: Precision, defined as the fraction of correctly predicted positive instances out of all instances predicted as positive, can be calculated using Eq. (4). Recall, also known as sensitivity, measures the proportion of correctly predicted positive instances out of all instances that truly belong to the positive class, and is computed using Eq. (5).
F1-score: The F1-score is a metric that merges Precision and Recall into a unified value. It represents the harmonic mean of Precision and Recall, offering a balanced evaluation of a model’s performance by taking into account both false positives and false negatives. Eq. (6) delineates the computation of the F1-score.
Confusion Matrix: The confusion matrix acts as a structured table that illustrates the performance of a classification model on a defined set of test data, where the true values are already established. It provides a comprehensive breakdown of the model’s predictions compared to the actual outcomes, thus revealing the types and frequency of errors made by the model.
The results section of a research paper is pivotal in showcasing the effectiveness of the proposed model. In this study, our main goal was to evaluate a deep learning model specifically developed for classifying brain tumors from MRI images. We assessed the model’s performance using critical metrics including accuracy, precision, recall, and F1-score. These metrics are crucial for gauging the model’s effectiveness and for comparing it with existing methodologies in the field.
Accuracy: The model exhibited exceptional accuracy in distinguishing between the four types of brain tumors, achieving an impressive accuracy rate of 99% on the testing dataset. This high accuracy underscores the model’s effectiveness in precisely identifying tumor types from the MRI images provided. This achievement is particularly noteworthy due to the inherent complexity and variability present in medical imaging data, especially in MRI scans of brain tumors. Fig. 4 illustrates this finding.
• Precision, Recall, and F1-Score: The study meticulously tracked precision and recall metrics for each tumor type. The model attained a precision of 1.00 for glioma, 0.99 for meningioma, 1.00 for cases without tumors, and 0.98 for pituitary tumors. In terms of recall, the model achieved scores of 0.99 for glioma, 0.99 for meningioma, 1.00 for cases without tumors, and 1.00 for pituitary tumors. The F1-scores, which offer a balanced assessment of precision and recall, consistently demonstrated high values across all categories. These results underscore the model’s accuracy, dependability, and uniformity in classification tasks. The detailed classification report is available in Table 4.
• ROC-AUC Curve Analysis: The ROC curve visually depicts how well a binary classifier discriminates between classes as the decision threshold changes. Fig. 5 illustrates these metrics.
• Precision-Recall Curve Analysis: The precision-recall curve serves as an alternative evaluation tool for binary classifiers, especially beneficial for assessing performance in imbalanced datasets, common in medical diagnostics. Fig. 6 displays this curve.
• Confusion Matrix Analysis: The confusion matrix provided deeper insights into the model’s performance. It revealed that the model had an exceptionally low incidence of misclassification across all tumor types. For instance, the few cases where glioma was misclassified as meningioma (and vice versa) were minimal. It has been shown in Fig. 7.

Figure 4: Train loss and accuracy
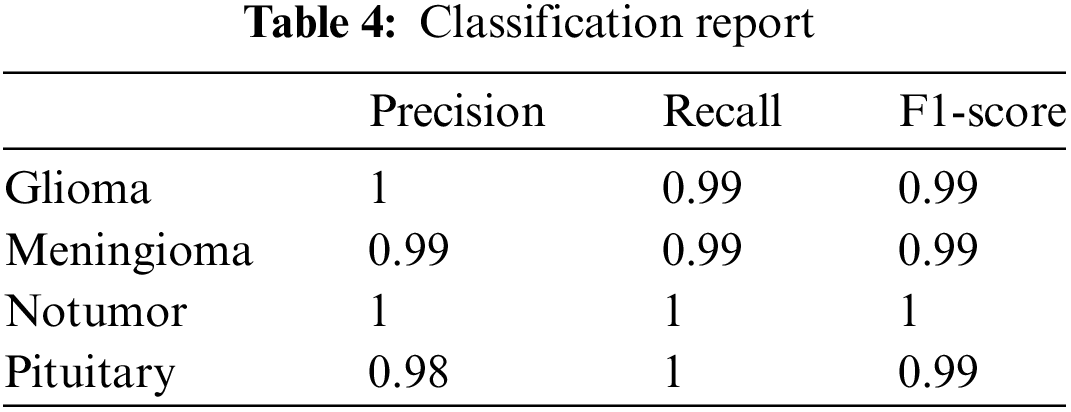

Figure 5: ROC-AUC curve

Figure 6: Precision-recall curve analysis

Figure 7: Confusion matrix
Misclassification cases primarily arose from the confusion between similar tumor types such as gliomas and meningiomas, which often share overlapping radiological features. These misclassifications could have serious clinical implications, as they might lead to inappropriate treatment planning—meningiomas typically require only surgical removal, whereas gliomas, often malignant, might need a combination of surgery, chemotherapy, and radiotherapy. Another contributing factor to misclassifications could be the image quality and preprocessing methods; artifacts like motion blur or suboptimal normalization processes might obscure crucial tumor characteristics. Additionally, model sensitivity to the variability in training data can cause overfitting, reducing the model’s ability to generalize to new, unseen images, which is a critical issue in diverse clinical environments. To minimize these risks, enhancing the dataset to include a broader spectrum of tumor appearances, implementing advanced preprocessing techniques to improve image quality, continuously evaluating and updating the model with new data, and integrating AI predictions with expert radiologist evaluations are recommended. These steps will improve the reliability of tumor classification models and ensure their practical utility in clinical settings, ultimately enhancing patient care outcomes.
In Table 5, a comparison is provided between different existing methodologies and the proposed methodology, highlighting their respective strengths and applicability.
Unlike traditional deep learning models like VGG and ResNet, which often require substantial computational resources and longer inference times due to their depth and complexity [17], EfficientNetB3 is designed to scale more efficiently. Its design employs a compound scaling technique that systematically adjusts the depth, width, and resolution of the network according to available resources, thereby substantially improving processing speed and accuracy. This makes EfficientNetB3 not only faster but also more adaptable to different computational environments, from high-end GPUs to more constrained settings, without a substantial loss in performance. The findings of this study hold significant clinical implications, particularly in enhancing the accuracy and efficiency of brain tumor diagnostics. This high level of performance can support radiologists in making more informed decisions, reducing the likelihood of misdiagnosis and ensuring that patients receive appropriate and personalized treatment plans more quickly. Additionally, the model’s efficiency and reduced computational demands make it feasible for integration into real-time clinical workflows, potentially streamlining diagnostic processes and reducing the workload on healthcare professionals.
This study introduced a deep learning model based on the EfficientNetB3 architecture to classify brain tumors from MRI images, demonstrating significant advancements over conventional approaches in medical image analysis. The study found that using the EfficientNetB3 model, brain tumors can be classified with 99% accuracy, which is a significant advancement in neuro-oncology. This precise classification can improve diagnostic accuracy and lead to more personalized and effective treatment plans for patients. The deep learning framework also enables quicker and more reliable diagnoses, easing the workload on radiologists and potentially speeding up treatment. Future research could focus on adapting and expanding the EfficientNetB3 model for other types of medical imaging, such as CT scans, PET scans, and ultrasound imaging, to enhance diagnostic accuracy across a broader range of medical conditions. Investigating the model’s performance in classifying other types of tumors and diseases could validate its versatility and robustness. Additionally, integrating multi-modal data, including clinical and genomic data alongside imaging, could provide a more comprehensive diagnostic tool. Exploring the model’s applicability in detecting early-stage diseases and its potential for predicting treatment outcomes could further advance its clinical utility.
Acknowledgement: The authors extend their appreciation to the Researchers Supporting Program at King Saud University. Researchers Supporting Project number (RSPD2024R867), King Saud University, Riyadh, Saudi Arabia.
Funding Statement: The research was supported by the Researchers Supporting Program at King Saud University. Researchers Supporting Project number (RSPD2024R867), King Saud University, Riyadh, Saudi Arabia.
Author Contributions: The authors confirm their contribution to the paper as follows: study conception and design: Mahesh Thyluru Ramakrishna and Kuppusamy Pothanaicker; data collection: Padma Selvaraj; analysis and interpretation of results; Surbhi Bhatia Khan and Vinoth Kumar Venkatesan; draft manuscript preparation: Mahesh Thyluru Ramakrishna and Kuppusamy Pothanaicker; review and editing: Saeed Alzahrani and Mohammad Alojail. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data that support the findings of this study are openly available at: https://www.kaggle.com/datasets/masoudnickparvar/brain-tumor-mri-dataset (accessed on 5 December 2023).
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. R. Asad, S. U. Rehman, A. Imran, J. Li, A. Almuhaimeed and A. Alzahrani, “Computer-aided early melanoma brain-tumor detection using deep-learning approach,” Biomedicines, vol. 11, no. 1, Jan. 2023, Art. no. 184. doi: 10.3390/biomedicines11010184. [Google Scholar] [PubMed] [CrossRef]
2. A. Aleid, K. Alhussaini, R. Alanazi, M. Altwaimi, O. Altwijri and A. S. Saad, “Artificial intelligence approach for early detection of brain tumors using MRI images,” Appl. Sci., vol. 13, no. 6, Mar. 2023, Art. no. 3808. doi: 10.3390/app13063808. [Google Scholar] [CrossRef]
3. M. Taha, D. S. B. B. Ariffin, and S. S. Abu-Naser, “A systematic literature review of deep and machine learning algorithms in brain tumor and meta-analysis,” J. Theor. Appl. Inform. Technol., vol. 101, no. 1, pp. 21–36, Jan. 2023. [Google Scholar]
4. Z. Liu et al., “Deep learning based brain tumor segmentation: A survey,” Complex Intell. Syst., vol. 9, no. 1, pp. 1001–1026, Jul. 2022. doi: 10.1007/s40747-022-00815-5. [Google Scholar] [CrossRef]
5. B. Abdusalomov, M. Mukhiddinov, and T. K. Whangbo, “Brain tumor detection based on deep learning approaches and magnetic resonance imaging,” Cancers, vol. 15, no. 16, Aug. 2023, Art. no. 4172. doi: 10.3390/cancers15164172. [Google Scholar] [PubMed] [CrossRef]
6. K. R. M. Fernando and C. P. Tsokos, “Deep and statistical learning in biomedical imaging: State of the art in 3D MRI brain tumor segmentation,” Inf. Fusion, vol. 92, no. 2, pp. 450–465, Mar. 2023. doi: 10.1016/j.inffus.2022.12.013. [Google Scholar] [CrossRef]
7. B. He et al., “A neural network framework for predicting the tissue-of-origin of 15 common cancer types based on RNA-Seq data,” Front. Bioeng. Biotechnol., vol. 8, pp. 1–11, Aug. 2020. doi: 10.3389/fbioe.2020.00737. [Google Scholar] [PubMed] [CrossRef]
8. Z. Liu et al., “Virtual formalin-fixed and paraffin-embedded staining of fresh brain tissue via stimulated Raman CycleGAN model,” Sci. Adv., vol. 10, no. 13, 2024, Art. no. 3426. doi: 10.1126/sciadv.adn3426. [Google Scholar] [PubMed] [CrossRef]
9. B. He, “A machine learning framework to trace tumor tissue-of-origin of 13 types of cancer based on DNA somatic mutation,” Biochimica et Biophysica Acta (BBA)-Mol. Basis Dis., vol. 1866, no. 11, Nov. 2020, Art. no. 165916. doi: 10.1016/j.bbadis.2020.165916. [Google Scholar] [PubMed] [CrossRef]
10. A. Anaya-Isaza, L. Mera-Jiménez, L. Verdugo-Alejo, and L. Sarasti, “Optimizing MRI-based brain tumor classification and detection using AI: A comparative analysis of neural networks, transfer learning, data augmentation, and the cross-transformer network,” Eur. J. Radiol. Open, vol. 10, Jan. 2023, Art. no. 100484. doi: 10.1016/j.ejro.2023.100484. [Google Scholar] [PubMed] [CrossRef]
11. A. Mohammed et al., “Hybrid techniques of analyzing MRI images for early diagnosis of brain tumours based on hybrid features,” Processes, vol. 11, no. 1, Jan. 2023, Art. no. 212. doi: 10.3390/pr11010212. [Google Scholar] [CrossRef]
12. S. Ullah, M. Ahmad, S. Anwar, and M. I. Khattak, “An intelligent hybrid approach for brain tumor detection,” Pak. J. Eng. Technol., vol. 6, no. 1, pp. 42–50, Jan. 2023. doi: 10.51846/vol6iss1pp34-42. [Google Scholar] [CrossRef]
13. M. Woźniak, J. Siłka, and M. Wieczorek, “Deep neural network correlation learning mechanism for CT brain tumor detection,” Neural Comput. Appl., vol. 35, no. 20, pp. 14611–14626, Oct. 2023. doi: 10.1007/s00521-021-05841-x. [Google Scholar] [CrossRef]
14. F. Demir, Y. Akbulut, B. Taşcı, and K. Demir, “Improving brain tumor classification performance with an effective approach based on new deep learning model named 3ACL from 3D MRI data,” Biomed. Signal Process. Control, vol. 81, no. 1, Jan. 2023, Art. no. 104424. doi: 10.1016/j.bspc.2022.104424. [Google Scholar] [CrossRef]
15. G. S. Tandel et al., “Role of ensemble deep learning for brain tumor classification in multiple magnetic resonance imaging sequence data,” Diagnostics, vol. 13, no. 3, Feb. 2023, Art. no. 481. doi: 10.3390/diagnostics13030481. [Google Scholar] [PubMed] [CrossRef]
16. A. Hossain et al., “A lightweight deep learning based microwave brain image network model for brain tumor classification using reconstructed microwave brain (RMB) images,” Biosensors, vol. 13, no. 2, Feb. 2023, Art. no. 238. doi: 10.3390/bios13020238. [Google Scholar] [PubMed] [CrossRef]
17. M. M. Mijwil et al., “MobileNetV1-based deep learning model for accurate brain tumor classification,” Mesop. J. Comput. Sci., vol. 2023, pp. 32–41, Jan. 2023. doi: 10.58496/MJCSC/2023/005. [Google Scholar] [CrossRef]
18. T. Ruba, R. Tamilselvi, and M. P. Beham, “Brain tumor segmentation in multimodal MRI images using novel LSIS operator and deep learning,” J. Ambient Intell. Humaniz. Comput., vol. 14, no. 10, pp. 13163–13177, Oct. 2023. doi: 10.1007/s12652-022-03773-5. [Google Scholar] [CrossRef]
19. R. Yousef et al., “Bridged-U-Net-ASPP-EVO and deep learning optimization for brain tumor segmentation,” Diagnostics, vol. 13, no. 16, Aug. 2023, Art. no. 2633. doi: 10.3390/diagnostics13162633. [Google Scholar] [PubMed] [CrossRef]
20. J. M. Medina and J. Sánchez, “High accuracy brain tumor classification with EfficientNet and magnetic resonance images,” in 5th Int. Conf. Adv. Process. Artific. Intell. International Frequency Sensor Association (IFSA) Publishing, S. L., 2023. [Google Scholar]
21. D. Reyes and J. Sánchez, “Performance of convolutional neural networks for the classification of brain tumors using magnetic resonance imaging,” Heliyon, vol. 10, no. 3, Mar. 2024, Art. no. e09812. doi: 10.1016/j.heliyon.2024.e25468. [Google Scholar] [PubMed] [CrossRef]
22. J. Cheng, “Brain tumor dataset,” figshare, 2017. Accessed: Dec. 05, 2023. [Online]. Available: https://figshare.com/articles/dataset/brain_tumor_dataset/1512427 [Google Scholar]
23. S. Bhuvaji, A. Kadam, P. Bhumkar, S. Dedge, and S. Kanchan, “Brain Tumor Classification (MRI),” Kaggle, 2020. Accessed: Dec. 10, 2023. [Online]. Available: https://www.kaggle.com/datasets/sartajbhuvaji/brain-tumor-classification-mri [Google Scholar]
24. Hamada, “Brain tumor detection,” Kaggle, 2020. Accessed: Jan. 10, 2024. [Online]. Available: https://kaggle.com/datasets/ahmedhamada0/brain-tumor-detection?select=no [Google Scholar]
25. X. Xie et al., “Evaluating cancer-related biomarkers based on pathological images: A systematic review,” Front. Oncol., vol. 11, 2020, Art. no. 763527. doi: 10.3389/fonc.2021.763527. [Google Scholar] [PubMed] [CrossRef]
26. H. Bingol and B. Alatas, “Classification of brain tumor images using deep learning methods,” Turk. J. Sci. Technol., vol. 16, no. 1, pp. 137–143, Jan. 2021. [Google Scholar]
27. R. Pillai et al., “Brain tumor classification using VGG 16, ResNet50, and Inception V3 transfer learning models,” in 2023 2nd Int. Conf. Innov. Technol. (INOCON), IEEE, 2023. [Google Scholar]
28. K. Sharma et al., “HOG transformation based feature extraction framework in modified Resnet50 model for brain tumor detection,” Biomed. Signal Process. Control, vol. 84, no. 4, Jan. 2023, Art. no. 104737. doi: 10.1016/j.bspc.2023.104737. [Google Scholar] [CrossRef]
29. L. Gaur et al., “Explanation-driven deep learning model for prediction of brain tumour status using MRI image data,” Front. Genet., vol. 13, Mar. 2022, Art. no. 822666. doi: 10.3389/fgene.2022.822666. [Google Scholar] [PubMed] [CrossRef]
30. Alshammari, “Construction of VGG16 convolution neural network (VGG16_CNN) classifier with NestNet-based segmentation paradigm for brain metastasis classification,” Sensors, vol. 22, no. 20, Oct. 2022, Art. no. 8076. doi: 10.3390/s22208076. [Google Scholar] [PubMed] [CrossRef]
31. K. K. Kumar et al., “Brain tumor identification using data augmentation and transfer learning approach,” Comput. Syst. Sci. Eng., vol. 46, no. 2, pp. 241–251, Mar. 2023. [Google Scholar]
32. Md I. Mahmud, M. Mamun, and A. Abdelgawad, “A deep analysis of brain tumor detection from mr images using deep learning networks,” Algorithms, vol. 16, no. 4, Apr. 2023, Art. no. 176. doi: 10.3390/a16040176. [Google Scholar] [CrossRef]
33. M. Islam et al., “Effectiveness of federated learning and CNN ensemble architectures for identifying brain tumors using MRI images,” Neural Process. Lett., vol. 55, no. 4, pp. 3779–3809, Apr. 2023. doi: 10.1007/s11063-022-11014-1. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF
 Downloads
Downloads
 Citation Tools
Citation Tools