 Open Access
Open Access
ARTICLE
Joint Biomedical Entity and Relation Extraction Based on Multi-Granularity Convolutional Tokens Pairs of Labeling
1 Department of Computer Science and Technology, Shandong University of Technology, Zibo, 255000, China
2 Department of Agricultural Engineering and Food Science, Shandong University of Technology, Zibo, 255000, China
3 Department of Electrical and Information Engineering, Beijing University of Civil Engineering and Architecture, Beijing, 100044, China
* Corresponding Author: Linlin Xing. Email:
Computers, Materials & Continua 2024, 80(3), 4325-4340. https://doi.org/10.32604/cmc.2024.053588
Received 05 May 2024; Accepted 11 August 2024; Issue published 12 September 2024
Abstract
Extracting valuable information from biomedical texts is one of the current research hotspots of concern to a wide range of scholars. The biomedical corpus contains numerous complex long sentences and overlapping relational triples, making most generalized domain joint modeling methods difficult to apply effectively in this field. For a complex semantic environment in biomedical texts, in this paper, we propose a novel perspective to perform joint entity and relation extraction; existing studies divide the relation triples into several steps or modules. However, the three elements in the relation triples are interdependent and inseparable, so we regard joint extraction as a tripartite classification problem. At the same time, from the perspective of triple classification, we design a multi-granularity 2D convolution to refine the word pair table and better utilize the dependencies between biomedical word pairs. Finally, we use a biaffine predictor to assist in predicting the labels of word pairs for relation extraction. Our model (MCTPL) Multi-granularity Convolutional Tokens Pairs of Labeling better utilizes the elements of triples and improves the ability to extract overlapping triples compared to previous approaches. Finally, we evaluated our model on two publicly accessible datasets. The experimental results show that our model’s ability to extract relation triples on the CPI dataset improves the F1 score by 2.34% compared to the current optimal model. On the DDI dataset, the F1 value improves the F1 value by 1.68% compared to the current optimal model. Our model achieved state-of-the-art performance compared to other baseline models in biomedical text entity relation extraction.Keywords
The critical elements of a knowledge graph are relation facts, most of which consist of two entities connected through semantic relation. These facts take the form of (subject, relation, object) or (s, r, o) and are called relation triples. Extracting these relation triples from natural language texts is essential for constructing extensive knowledge graphs.
Early work by Zelenko et al. [1] on relational triad extraction used a pipeline approach. It first identifies all the entities in a sentence and then classifies the relation for each entity pair. This approach often encounters the error accumulation problem, as misrecognition in the early stages cannot be corrected later. To tackle this problem, subsequent research has proposed a feature-based model that jointly learns entities and relations. Examples of such work include Li et al. [2] and Yu et al. [3]. Recently, Zheng et al. [4] used a neural network-based model, which has achieved considerable success in ternary extraction tasks by replacing manually constructed features with learned representations.
In the field of biomedicine, there are relatively few studies on extracting relational triads, mainly due to the complexity and specialization of biomedical texts. These texts usually contain a large number of specialized terms and complex syntactic structures, making the extraction of relational triples particularly challenging. Although some progress has been made in this area in recent years, related research efforts are still limited compared to other fields. Li et al. [5] implemented triple extraction by separating the Named Entity Recognition (NER) and Relation Extraction (RE) subtasks through a shared parameter paradigm. Luo et al. [6] proposed an Att-BiLSTM-CRF model to jointly extract entities and relations, considering overlapping relations in the tagging scheme. As shown in Fig. 1, there are three sentence types in the biomedical corpus. The first example is the usual case where there are no overlapping triples. The second belongs to the Single Entity Overlap (SEO) case, where one entity is shared. The third case is the Entity Pair Overlap (EPO) case, where a pair of entities is shared. Biomedical texts are rich in resources and contain complex information, with a large number of domain-specific terms, such as “Phillyrin,” “Hepatocytes,” “AMPK,” etc., in the Normal-type sentences in Fig. 1. Biomedical texts differ from general domain texts in that the former contains many domain-specific terms and much overlap.
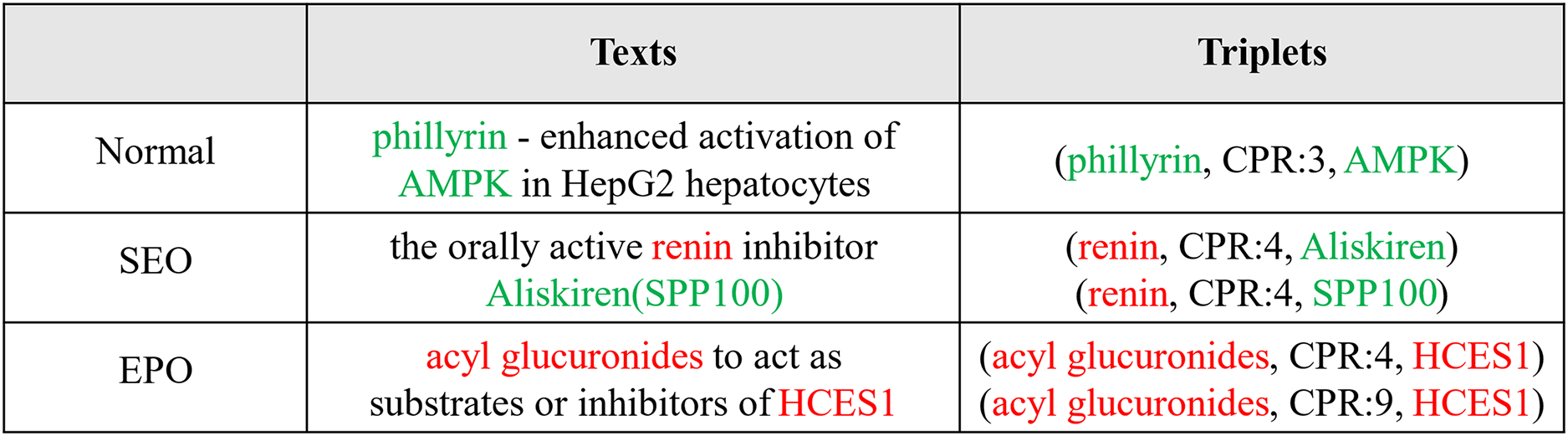
Figure 1: Examples of the normal, SEO, and EPO cases
Currently, whether it is a generic or domain-specific extraction of relational triples, the current approach ignores the nature of a ternary—its subject entity, relation, and object entity are interdependent and inseparable. In other words, extracting an element by fully sensing the information of the other two elements is reliable. In order to realize the extraction of relation triples, we use the table structure to realize the triple classification perspective to accomplish the extraction task of relation triples. For example, as shown in Fig. 1, for Normal, “phillyrin” and “AMPK” are two words in the sentence, and “CRP:3” is a predefined relation. These are all visible in training, so the triple (phillyrin, CRP:3, AMPK) can be recognized directly by judging the correctness of each triple element.
Inspired by the above ideas and utilizing the three elements of a ternary, we use all the elements of the triple, which extracts all the relation triples from it through a table structure, specifically, considering that an entity may have more than one labeled token, we transform the joint entity relation extraction task into a fine-grained triple classification problem, for a word pair
• We utilize all the elements in the relation triad to transform joint entity and relation extraction into fine-grained ternary classification and use convolution to learn the connection between relations and labels to capture the information of the subject entity, relation, and object entity simultaneously.
• We propose a multi-granularity 2D convolutional approach based on the characteristics of biomedical text for capturing semantic information between word pairs of different granularity to refine the table representation and, hence, the label classification.
• We evaluate our model on CPI and DDI datasets and show that our method outperforms state-of-the-art baselines while solving complex scenarios with overlapping relational triples.
The organization of this paper is as follows: Section 2, “Related Work,” reviews the latest techniques in joint entity and relation extraction. Section 3, “Problem Formulation,” defines the tasks involved in joint entity and relation extraction. Section 4, “Relation Tagging,” covers labeling strategies and decoding methods. Section 5, “Model,” presents the overall architecture of our proposed model. Section 6, “Experiments,” details the dataset used, the experimental setup, and the results of the experiments. Finally, Section 7, “Conclusion,” summarizes our findings and discusses future directions.
The early task of extracting entities and relations was to solve the task in a pipelined manner. Roth et al. [7] used a pipelined approach to extract sentence entities and relations. Although the pipelined approach achieved good results, it usually causes error propagation problems and ignores the correlation between the two steps. To alleviate these problems, several joint models aiming to learn entities and relations jointly have been proposed.
Ren et al. [8] noted that conventional joint models are feature-based, depend significantly on feature engineering, and demand substantial manual effort. To minimize this manual labor, recent research has explored neural network-based methods, which offer state-of-the-art performance. Zheng et al. [4] completed joint decoding by implementing a unified annotation scheme, transforming the task of extracting relational triples into an end-to-end sequence annotation problem. Zeng et al. [9] presented three models for overlapping triples and addressed the issue using a sequence-to-sequence model enhanced with a replication mechanism. Fu et al. [10] also studied the overlapping triple problem and proposed a graph convolutional network-based approach to solving the problem. Despite the initial success of the overlapping ternary problem, both approaches still need help in learning information about the overlapping parts. Wei et al. [11] proposed an innovative cascade binary labeling framework. This method first detects all potential subject entities in the sentence and then identifies all possible relational and object entities for each detected subject entity. While this approach effectively solves the problem of overlapping triples, it also presents the challenge of cascading errors. Wang et al. [12] proposed a unified labeling approach to extract entities and overlapping relations that enables single-stage joint extraction. Shang et al. [13] proposed a single-module, single-step decoding approach for joint entity and relation extraction. Zhang et al. [14] proposed a new relation-based triple labeling and a scoring model for jointly extracting entities and relations. Dai et al. [15] proposed a new method for extracting joint entities and relation, which includes a filter separator network (FSN) module. Although the model achieved good results in generalized domains, it needed to address the overlapping problem in biomedical texts by targeting the characteristics of biomedical texts.
Entity relation extraction of biomedical text is an important application of natural language processing in biomedical field, which plays a vital role in scientific research, drug discovery and many other aspects. Li et al. [16] used a bi-directional LSTM approach to design a model to extract both entities and relations simultaneously. Lai et al. [17] utilized an external knowledge augmentation model to perform joint entity and relation extraction. Zhang et al. [18] designed a tokens tagger to implement a single-stage joint biomedical entity and relation extraction to solve the biomedical text overlapping problem. Yang et al. [19] introduced a generative joint modeling approach for entity and relation extraction tasks using a medical dataset. Zhang et al. [20] proposed utilizing the medical knowledge graph to collectively extract entities and relations from Chinese medical texts.
In this paper, we propose a triple classification-based learning tag labeling method to extract relational triads from medical text and design 2D multi-granularity convolution to solve the problem of overlapping relations in the text according to the characteristics of biomedical text.
Given a sentence
For each sentence, we set all possible
Generally, we use Begin, Inside, and End to denote the token’s position in the entity. According to the existing research [11], we use the first token and the last token of the entity to complete the identification of entity boundaries, so for the extraction of a relational triple, we only need to identify the head entity and the end entity in the token labeled Begin and End. In addition, we add the [Unused1] token for each word in the sentence so that the entity for a word after the word division is still its token and has the End label.
Based on detecting subject and object boundaries to determine the labeling of word pairs, our tagging strategy uses four types of labels: (1) HB(Head-entity Begin)-TB(Tail-entity Begin). The label refers to the token pair of two entities under a specific relation, where the token pair represents the starting token of the head entity and the starting token of the tail entity. (2) HB(Head-entity Begin)-TE(Tail-entity End). The token pair corresponding to the label under a specific relation has the row label as the first token of the head entity and the column label as the last token of the tail entity. Meanwhile, the “HB-TE” label is the maximum boundary information of the two entities. (3) HE(Head-entity End)-TE(Tail-entity End). The label is similar in meaning to “HB-TB.” It refers to the token pair of two entities under a specific relation, where the token pair represents the ending token of the head entity and the ending token of the tail entity. (4) “-”. All cells are labeled as “-” except in the three cases mentioned above. For example, the sentence “phillyrin-enhanced activation of AMPK in HepG2 hepatocytes” There is a relationship “CPR:3” between the two entities “phillyrin” and “AMPK”, thus, In the table, we marked it with three related labels, “HB-TB”, “HB-TE”, and “HE-TE”, and the unrelated cells were marked with “-”.
The sparsity of the labeling matrix
Moreover, our labeling can effectively handle biomedical texts in complex semantic environments. Specifically, for SEO scenarios, two entity pairs will be labeled in different parts of
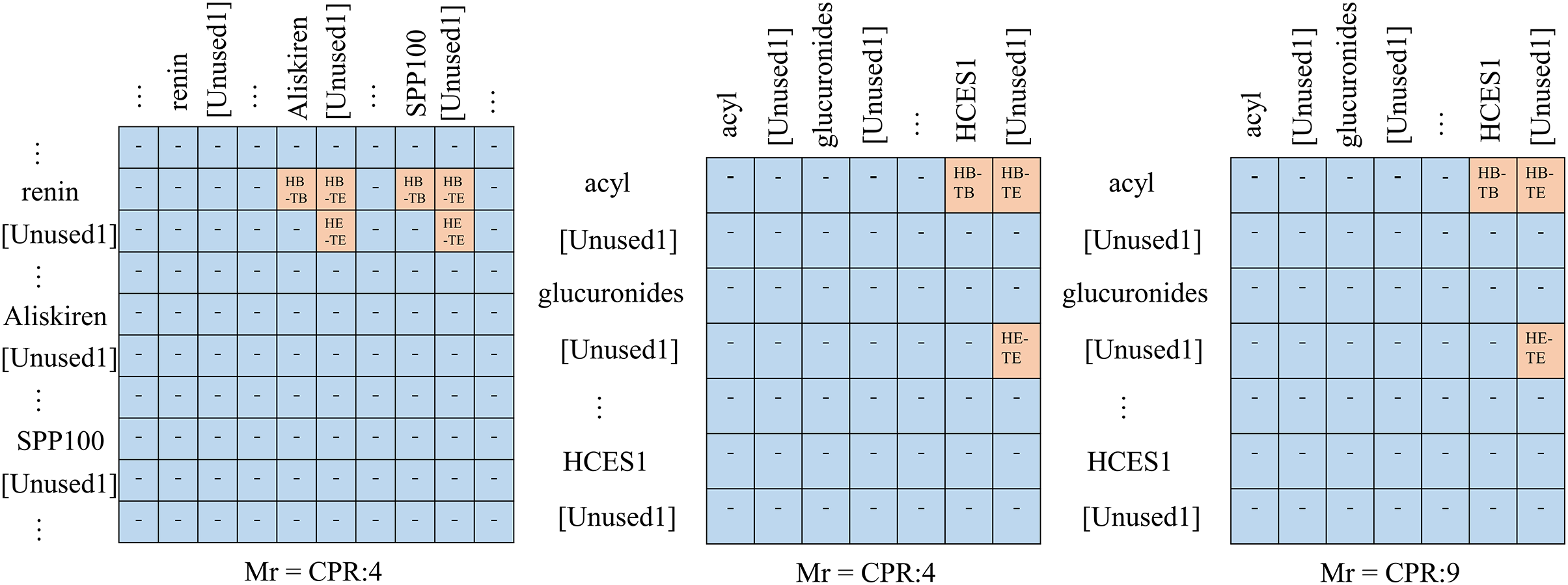
Figure 2: Example of labeling strategy. For ease of interpretation, we represent this as a two-dimensional matrix under a predefined relation, representing rows and columns as head and tail entities
The labeling matrix
We propose a Multi-granularity Convolutional Token Pairs of Labeling (MCTPL) model. Our framework, illustrated in Fig. 3, comprises three components. First, SciBERT [21] serves as the encoder to produce contextualized word embeddings from the input sentences. Multi-granularity convolution is then used to refine the representation of the form for subsequent word pair classification. Subsequently, a joint classifier is used to predict all labels.
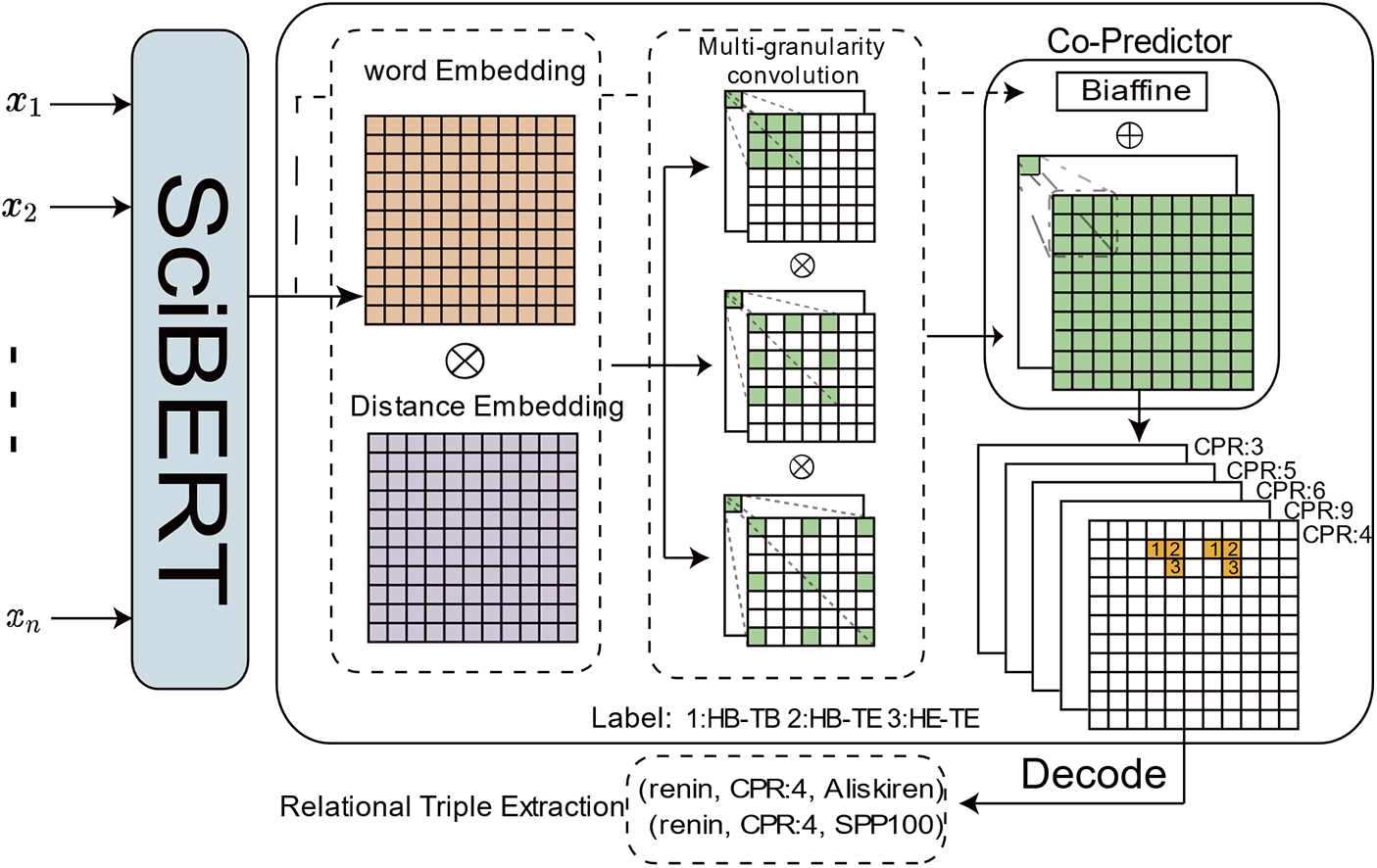
Figure 3: Overall architecture of our model.
SciBERT Encoder For sentences with input L tokens, we utilize the pre-trained SciBERT [21] as a sentence encoder to capture the word embeddings of each token:
Here,
Constructing Word Pair Tables Specifically, for each word pair
Inspired by Bert, we first construct the representation
Here,
Multi-Granularity Convolution It is well known that 2D-CNN is widely used to extract features for image classification and object detection, and it also excels in relation processing. Meanwhile, we apply a 2D-CNN by converting text into a table structure, treating the table as an image and each cell as a pixel. This corresponds to our labeling strategy, which looks for tokens for subjects and objects in a given relation. Since our model is predicting labels between word pairs, we utilize multiple 2D-CNNs with different dilation rates
where
Co-Predictor After the multi-granularity convolutional layer, we obtain the word-pair lattice representation Q and directly use the convolution to classify each pair of words. However, previous work [22] showed that the MLP predictor can be augmented by collaborating with the bipartite affine predictor for relational categorization, and here we use the convolution for categorization as well, so we use the bipartite affine predictor to collaborate with the convolution for categorization. The input to the bi-affine predictor is the output of the SciBERT encoder
where
Based on the refined table representation
where
The objective function of our model is defined as:
where
As shown in Table 1, the detailed information of the two datasets is demonstrated.
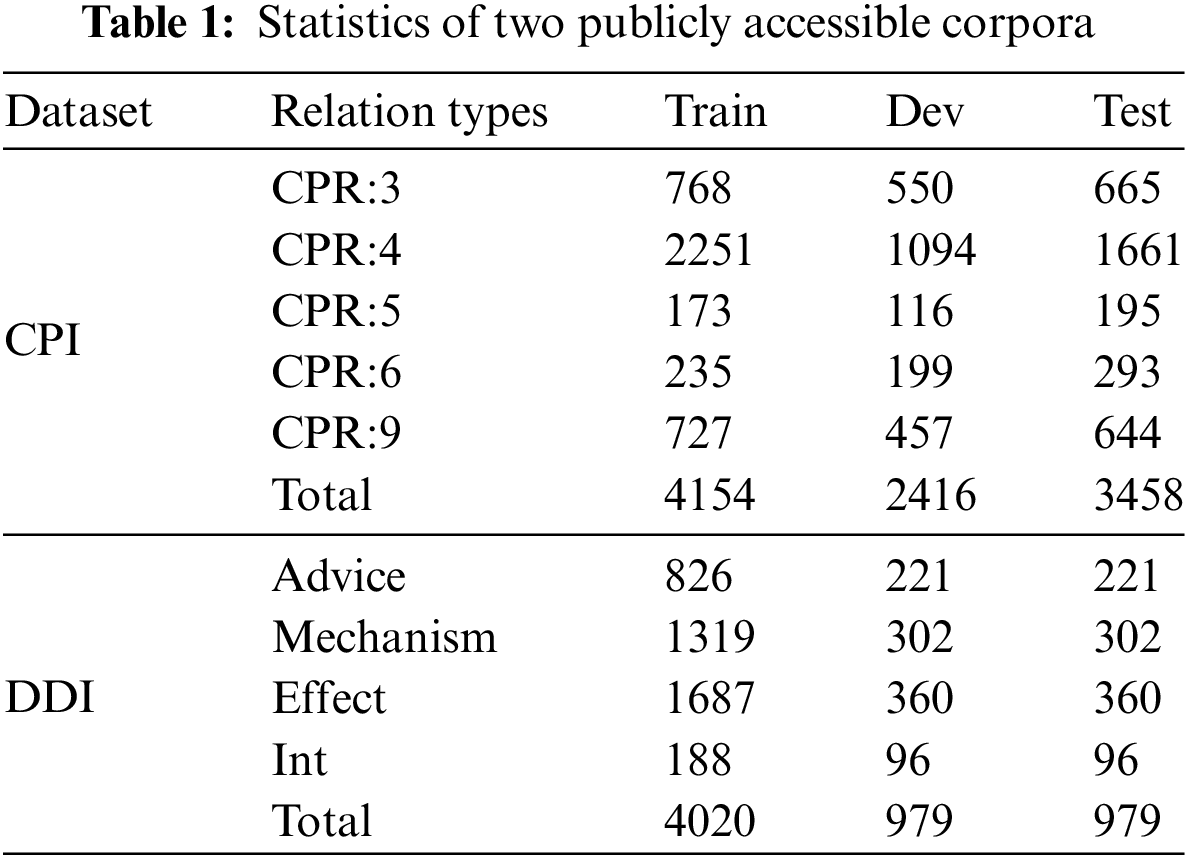
Chemistry-Protein Interactions (CPI [23]) consists of 1820 PubMed abstracts with chemistry-protein interactions for the shared task of the BioCreative VI text mining chemistry-protein interactions. We used the standard training and test sets from the ChemProt shared task and evaluated five relation.
Drug-Drug Interactions (DDI [24]) corpus is derived from 1025 documents obtained from the DrugBank database and Medline abstracts. It is considered a gold standard for evaluating information extraction methods used to identify pharmacological substances and detect drug-drug interactions in biomedical texts, as part of SemEval2013. We use 624 training and 191 test documents to evaluate performance and report on four relation types.
In addition, we employ the evaluation metrics Precision (P), Recall (R), and F1 score to analyze and assess the experimental results. The formula is expressed as:
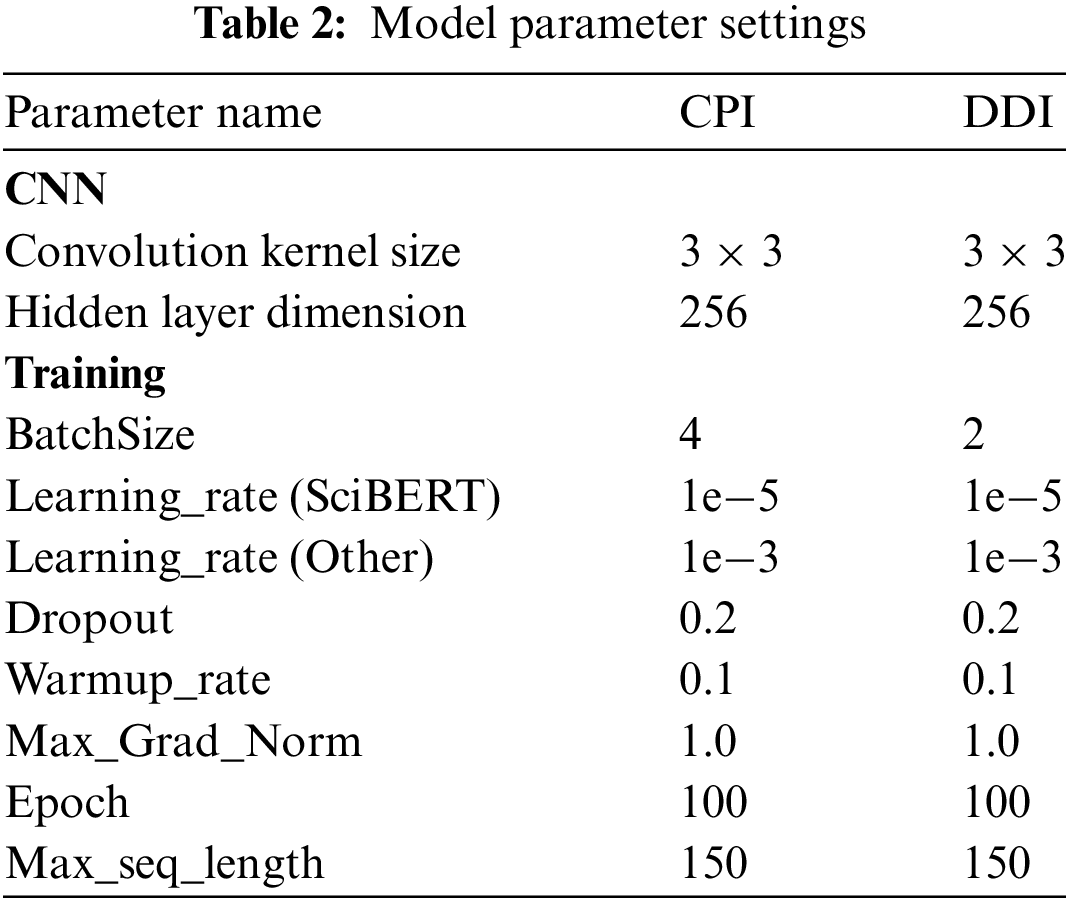
We implement the proposed model using the PyTorch framework and leverage the pre-trained SciBERT model (SciBERT-Scivocab-Uncased) available from the Hugging Face library. All parameters, except those in the pre-trained SciBERT model, are initialized randomly. The AdamW [25] algorithm was used to update the model parameters, and 100 epochs were trained in the training process. During training, the model parameters are set as shown in Table 2. All experiments used a single RTX 3090 GPU with 24 GiB of memory.
To assess our model’s performance, we compared it with various baseline models. For sequential entity and relation extraction, we used the classic model by Zhang et al. [26]. For joint entity and relation extraction, we selected several traditional models as baselines.
Zhang et al. used a hierarchical recursive neural network (RNN) to extract relation triplets by integrating shortest dependency paths (SDP) and sentence sequences.
NovelTagging (2017) transformed joint entity and relation tasks into labeling problems with a unique tagging scheme.
Graph Tagging (2018) devised a novel graph scheme to convert joint entity and relation extraction tasks into directed graphs and proposed a transfer-based directed graph incremental generation method for joint learning.
CasRel (2020) utilized a parameter-sharing stacked pointer network to identify head entities and their corresponding tail entities based on specific class relations.
PRGC (2021) approached joint entity and relation extraction by dividing it into subtasks of relation judgment, entity extraction, and subject-object alignment, and introduced a framework leveraging latent relations and global correspondences.
Luo’s method in 2020 constructed an Att-BiLSTM-CRF model to jointly extract entities and relations, considering overlapping relations in the tagging scheme.
GRTE (2021) introduced a relation triplet extraction model based on global features for table filling while handling overlapping relation extraction.
TPLinker (2020) introduced an innovative handshaking tagging scheme and a unified framework for joint entity and relation extraction to tackle exposure bias and handle complex overlapping relations.
Zhang et al. designed a token-based tagger for single-stage joint entity and relation extraction.
OneRel (2022) is a single-module, single-step decoding method for joint entity and relation extraction.
Table 3 shows how our model compares to the baseline model on CPI and DDI. On the CPI dataset, our model achieves the state-of-the-art F1 score by comparing the optimal model with a relative improvement of 1.57% in accuracy, a comparable recall to it, and a relative improvement of 2.34% in F1 value; on the DDI dataset, our model compares to the optimal model with a relative improvement of 2.69% in accuracy, a comparable recall to it, and a relative improvement in F1 value of 1.68%. We credit the outstanding performance of our model to two key advantages: (1) Our model solves the task of jointly extracting relation triples from the perspective of fine-grained triple classification, in which relations are an inseparable part of triple extraction, and the intrinsic connection between the triple information (head entity information, tail entity information, and information about relations between entities) and labels are learned during the process of learning the labeling markings, and then refine the model classification. (2) Multi-granularity 2D convolution is designed in the model. In contrast, 2D convolution is suitable for working on tables. Multi-granularity convolution can capture the interaction between word pair information at different distances in complex semantic text, refine the representation of word pairs, improve the understanding and expression ability of the model, and then dig deeper into the connection between word pairs for label classification. Based on the above two points, the model’s accuracy has improved more than the baseline model. In addition, for the recall rate is comparable to the baseline model results, our model labels the predefined relation matrix before decoding. However, the triple classification perspective allows the model to learn the intrinsic connection between the sentence and the elements in the ternary so that the model obtains more performance in terms of the precision rate. However, at the same time, it improves the precision of the model, making it learn too much semantic solid information, leading to missing some correct labeling labels.
Furthermore, due to error accumulation, joint methods generally surpass traditional pipeline models in triplet extraction. Experimental results show that single-stage extraction methods outperform two-stage combinations (such as the CasRel model). This is because two-stage joint methods struggle to effectively share information between stages, whereas single-stage approaches can manage this issue more effectively.
To further explore our method’s ability to extract overlapping triplets, we conducted experiments on various sentence types within the CPI dataset and compared the results with those of prior studies. As shown in Fig. 4, we can observe that our model achieves the best F1 scores in extracting relation triplets from different types of sentences. Furthermore, all the models we compared can handle overlapping relations, and our model outperforms them in achieving the highest F1 score for such cases. This indicates that our model excels in extracting both standard and overlapping relation triplets effectively.
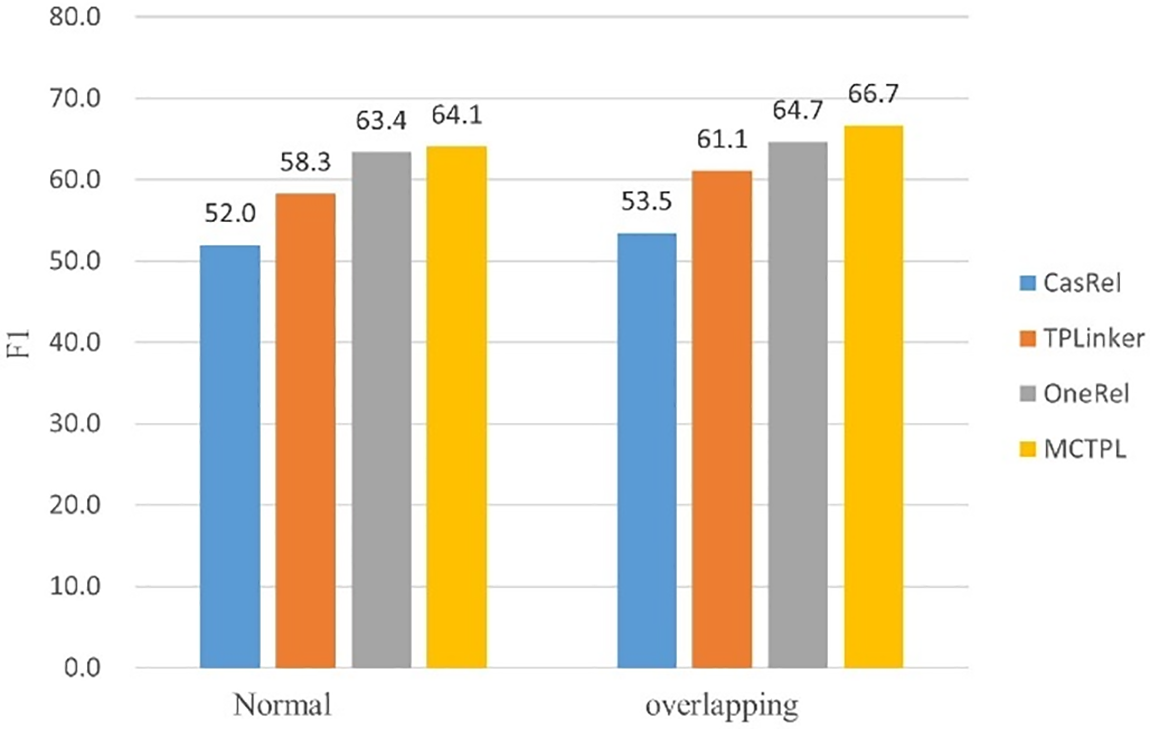
Figure 4: Extracting F1 scores for relational triples from sentences with different types
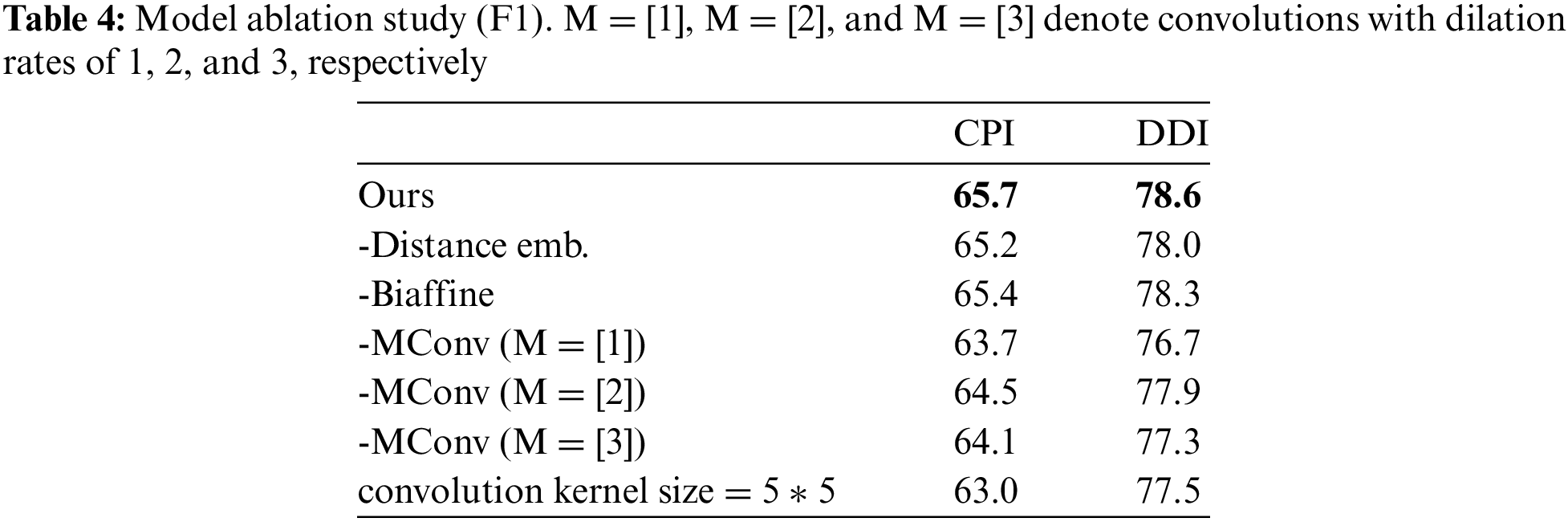
We conducted ablation studies on our model on the CPI and DDI datasets, as shown in Table 4. Firstly, in the absence of distance embeddings, we observed a slight decrease in performance on both datasets. This indicates that the distance information in our model can help improve its performance by enabling the model to learn the intrinsic relationships between entities at different distances and exploit varying degrees of semantic information from word pairs at different distances. Secondly, the Biaffine predictor can help the model learn the representation of the head and tail entities and assist in classifying the model labels. Lastly, the performance decreases upon removing fine-grained different dilation convolutions, validating the effectiveness of multi-granularity convolutions in extracting semantic information from different word pairs. It also indicates that 2D convolutions at different granularities can learn semantic information from biomedical texts, enhancing model performance and enabling more accurate and efficient extraction of relation triplets. In addition, we conducted hyperparameter experiments when the convolution kernel size was set to 5 * 5, and we found that F1 scores on CPI and DDI data sets did not increase but decreased, indicating that the convolution kernel size set to 5 * 5 made the model learn too much semantic information, resulting in poor labeling effect.
We compared the prediction results of the OneRel method and our method on the CPI dataset, as shown in Table 5.
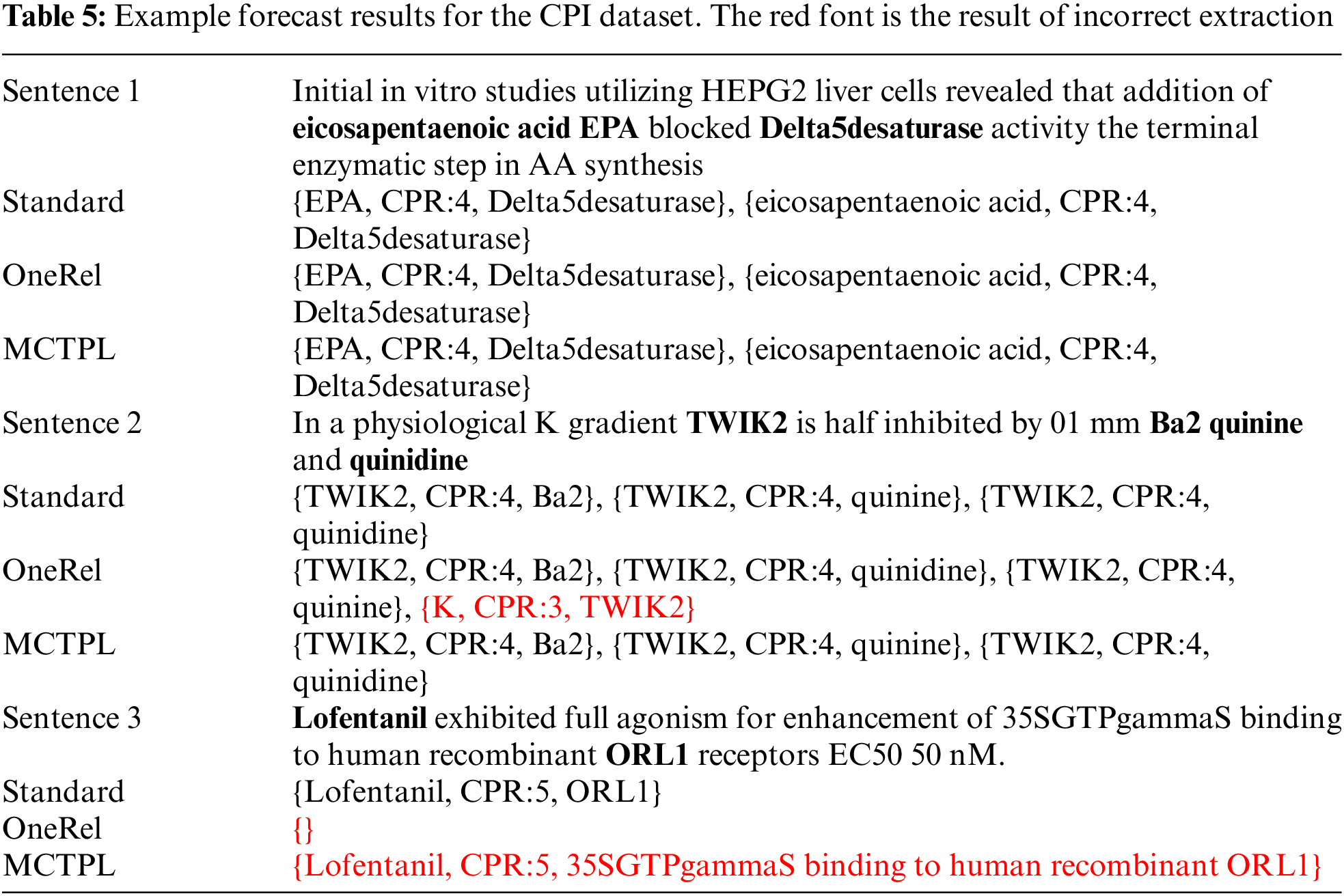
As we see from Sentence 1, Sentence 1 contains overlapping relational triples. Both OneRel’s model and our model can predict the correct relational triples, which indicates that both models are good at dealing with relational overlap; Sentence 2 has more overlapping relational triples compared to Sentence 1, and OneRel’s model, although it can extract the correct relational triples, has the additional prediction of the wrong relation triples. In contrast, our model successfully extracted the correct relational triples, which shows that our model can achieve better performance when dealing with sentences with multiple overlapping relations. This result further confirms the effectiveness of our multi-granularity convolution mechanism in digging deep semantic information of sentences; in Sentence 3, both OneRel’s model and our model do not predict correctly, indicating that there is still room for our model to improve on some relation extraction.
In this paper, we introduce a new approach for transforming joint extraction tasks into detailed triple classification problems, to fully utilize each triple element. We propose a multi-granularity 2D convolution method designed to capture interactions between neighboring and distant words, effectively addressing the challenges of extracting complex and overlapping triples in biomedical texts. Experimental results indicate that our method significantly surpasses existing techniques on the CPI and DDI datasets. Our findings and analyses reveal that our model excels in feature extraction from biomedical corpora and in handling overlapping triples.
Although our method is effective, there are still some problems that need further study. First, we use convolution to capture dependencies between word pairs, but currently we can’t intuitively analyze the information extracted by convolutional layers like in the field of computer vision. Secondly, our model needs to be improved to address the category imbalance in biomedical datasets. In recent years, many NLP researchers have improved the performance of models in information extraction tasks by introducing word pair information, which indicates that making full use of word pair information is a promising research direction. In the future, we will continue to investigate how words can be used for entity and relational joint tasks on information, and explore new ways to address category imbalances in biomedical datasets. At the same time, we also hope to find a way to explain the role of convolution in word information extraction.
Acknowledgement: The author extends gratitude to all those who have contributed to the field of this research, and appreciates the insightful comments and suggestions from anonymous reviewers, which significantly enhanced the quality of this manuscript.
Funding Statement: This work is supported by the National Natural Science Foundation of China (Nos. 62002206 and 62202373) and the open topic of the Green Development Big Data Decision-Making Key Laboratory (DM202003).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Zhaojie Sun, Linlin Xing; data collection: Zhaojie Sun, Longbo Zhang; methodology: Zhaojie Sun, Linlin Xing; analysis and interpretation of results: Zhaojie Sun; writing–original draft: Zhaojie Sun, writing—review and editing: Linlin Xing, Longbo Zhang, Hongzhen Cai, Maozu Guo. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. D. Zelenko, C. Aone, and A. Richardella, “Kernel methods for relation extraction,” in Proc. 2002 Conf. Empir. Methods Nat. Lang. Process. (EMNLP 2002), Philadelphia, PA, USA, 2002, pp. 71–78. [Google Scholar]
2. Q. Li and H. Ji, “Incremental joint extraction of entity mentions and relations,” in Proc. 52nd Annu. Meet. Assoc. Comput. Linguist., Baltimore, MD, USA, 2014, pp. 402–412. doi: 10.3115/v1/P14-1038. [Google Scholar] [CrossRef]
3. X. Yu and W. Lam, “Jointly identifying entities and extracting relations in encyclopedia text via a graphical model approach,” in Proc. 23rd Int. Conf. Comput. Linguist.: Posters, Beijing, China, 2010, pp. 1399–1407. [Google Scholar]
4. S. Zheng, F. Wang, H. Bao, Y. Hao, P. Zhou and B. Xu, “Joint extraction of entities and relations based on a novel tagging scheme,” in Proc. 55th Annu. Meet. Assoc. Comput. Linguist., Vancouver, BC, Canada, 2017, pp. 1227–1236. doi: 10.18653/v1/P17-1113. [Google Scholar] [PubMed] [CrossRef]
5. F. Li, Y. Zhang, M. Zhang, and D. Ji, “Joint models for extracting adverse drug events from biomedical text,” in Proc. Twenty-Fifth Int. Joint Conf. Artif. Intell., New York, NY, USA, 2016, pp. 2838–2844. [Google Scholar]
6. L. Luo, Z. Yang, M. Cao, L. Wang, Y. Zhang and H. Lin, “A neural network-based joint learning approach for biomedical entity and relation extraction from biomedical literature,” J. Biomed. Inform., vol. 103, 2020, Art. no. 103384. doi: 10.1016/j.jbi.2020.103384. [Google Scholar] [PubMed] [CrossRef]
7. D. Roth and Y. Chan, “Exploiting syntactico-semantic structures for relation extraction,” in Proc. 49th Annu. Meet. Assoc. Comput. Linguist.: Human Lang. Technol., Portland, OR, USA, 2011, pp. 551–560. [Google Scholar]
8. X. Ren et al., “CoType: Joint extraction of typed entities and relations with knowledge bases,” in Proc. 26th Int. Conf. World Wide Web, Perth, Australia, 2017, pp. 1015–1024. [Google Scholar]
9. X. Zeng, D. Zeng, S. He, K. Liu, and J. Zhao, “Extracting relational facts by an end-to-end neural model with copy mechanism,” in Proc. 56th Annu. Meet. Assoc. Comput. Linguist., Melbourne, Australia, 2018, pp. 506–514. doi: 10.18653/v1/P18-1047. [Google Scholar] [PubMed] [CrossRef]
10. T. Fu, P. Li, and W. Ma, “GraphRel: Modeling text as relational graphs for joint entity and relation extraction,” in Proc. 57th Annu. Meet. Assoc. Computat. Linguist., Florence, Italy, 2019, pp. 1409–1418. doi: 10.18653/v1/P19-1136. [Google Scholar] [PubMed] [CrossRef]
11. Z. Wei, J. Su, Y. Wang, Y. Tian, and Y. Chang, “A novel cascade binary tagging framework for relational triple extraction,” in Proc. 58th Annu. Meet. Assoc. Comput. Linguist., 2020, pp. 1476–1488. doi: 10.18653/v1/2020.acl-main.136. [Google Scholar] [PubMed] [CrossRef]
12. Y. Wang, B. Yu, Y. Zhang, T. Liu, H. Zhu and L. Sun, “TPLinker: Single-stage joint extraction of entities and relations through token pair linking,” in Proc. 28th Int. Conf. Comput. Linguist., 2020, pp. 1572–1582. doi: 10.18653/v1/2020.coling-main.138. [Google Scholar] [PubMed] [CrossRef]
13. Y. Shang, H. Huang, and X. Mao, “OneRel: Joint entity and relation extraction with one module in one step,” in Proc. AAAI Conf. Artif. Intell., vol. 36, no. 10, pp. 11285–11293, 2022. doi: 10.1609/aaai.v36i10.21379. [Google Scholar] [CrossRef]
14. J. Zhang, X. Jiang, Y. Sun, and H. Luo, “RS-TTS: Anovel jointentity and relation extraction model,” in Proc. 26th Int. Conf. Comput. Support. Cooperat. Work Des. (CSCWD), 2023, pp. 71–76. doi: 10.1109/CSCWD57460.2023.10152749. [Google Scholar] [CrossRef]
15. Q. Dai, W. Yang, F. Wei, L. He, and Y. Liao, “FSN: Joint entity and relation extraction based on filter separator network,” Entropy, vol. 26, no. 2, 2024, Art. no. 162. doi: 10.3390/e26020162. [Google Scholar] [PubMed] [CrossRef]
16. F. Li, M. Zhang, G. Fu, and D. Ji, “A neural joint model for entity and relation extraction from biomedical text,” BMC Bioinform., vol. 18, no. 1, pp. 1–11, 2017. doi: 10.1186/s12859-016-1414-x. [Google Scholar] [PubMed] [CrossRef]
17. T. Lai, H. Ji, C. Zhai, and Q. H. Tran, “Joint biomedical entity and relation extraction with knowledge-enhanced collective inference,” in Proc. 59th Annu. Meet. Assoc. Comput. Linguist. 11th Int. Joint Conf. Nat. Lang. Proc., 2021, pp. 6248–6260. doi: 10.18653/v1/2021.acl-long.488. [Google Scholar] [PubMed] [CrossRef]
18. L. Zhang, A. Roberts, and S. Zeki, “Joint entity and relation extraction in medical text,” in Proc. 32nd Int. Joint Conf. Artif. Intell., Macao, China, 2023, pp. 4–9. [Google Scholar]
19. Z. Yang, Y. Huang, and J. Feng, “Learning to leverage high-order medical knowledge graph for joint entity and relation extraction,” in Find. Assoc. Comput. Linguist.: ACL 2023, Toronto, ON, Canada, 2023, pp. 9023–9035. doi: 10.18653/v1/2023.findings-acl.575. [Google Scholar] [PubMed] [CrossRef]
20. Y. Zhang, J. Li, Z. Yang, H. Lin, and J. Wang, “Location-guided token pair tagger for joint biomedical entity and relation extraction,” in Proc. IEEE Int. Conf. Bioinf. Biomed. (BIBM), Las Vegas, NV, USA, 2022, pp. 666–671. doi: 10.1109/BIBM55620.2022.9995210. [Google Scholar] [CrossRef]
21. I. Beltagy, K. Lo, and A. Cohan, “SciBERT: A pretrained language model for scientific text,” in Proc. 2019 Conf. Empir. Methods Nat. Lang. Process. 9th Int. Joint Conf. Nat. Lang. Process. (EMNLP-IJCNLP), Hong Kong, China, 2019, pp. 3613–3618. doi: 10.18653/v1/D19-1371. [Google Scholar] [PubMed] [CrossRef]
22. J. Li, K. Xu, F. Li, H. Fei, Y. Ren and D. Ji, “MRN: A locally and globally mention-based reasoning network for document-level relation extraction,” in Proc. Find. Assoc. Comput. Linguist.: ACL-IJCNLP 2021, 2021, pp. 1359–1370. doi: 10.18653/v1/2021.findings-acl.117. [Google Scholar] [PubMed] [CrossRef]
23. M. Krallinger et al., “Overview of the BioCreative VI chemical-protein interaction track,” in Proc. Sixth BioCreative Chall. Eval. Workshop, Bethesda, MD, USA, 2017, pp. 141–146. [Google Scholar]
24. M. Herrero-Zazo, I. Segura-Bedmar, P. Martínez, and T. Declerck, “The DDI corpus: An annotated corpus with pharmacological substances and drug–drug interactions,” J. Biomed. Inform., vol. 46, no. 5, pp. 914–920, 2013. doi: 10.1016/j.jbi.2013.07.011. [Google Scholar] [PubMed] [CrossRef]
25. I. Loshchilov and F. Hutter, “Decoupled weight decay regularization,” in Proc. 7th Int. Conf. Learn. Represent., New Orleans, LA USA, 2019, pp. 1–19. [Google Scholar]
26. Y. Zhang, W. Zheng, H. Lin, J. Wang, Z. Yang and M. Dumontier, “Drug-drug interaction extraction via hierarchical RNNs on sequence and shortest dependency paths,” Bioinformatics, vol. 34, no. 5, pp. 828–835, 2018. doi: 10.1093/bioinformatics/btx659. [Google Scholar] [PubMed] [CrossRef]
27. S. Wang, Y. Zhang, W. Che, and T. Liu, “Joint extraction of entities and relations based on a novel graph scheme,” in Proc. Twenty-Seventh Int. Joint Conf. Artif. Intell. (IJCAI-18), Stockholm, Sweden, 2018, pp. 4461–4467. [Google Scholar]
28. H. Zheng et al., “PRGC: Potential relation and global correspondence based joint relational triple extraction,” in Proc. 59th Annu. Meet. Assoc. Comput. Linguist. 11th Int. Joint Conf. Nat. Lang. Process., 2021, pp. 6225–6235. [Google Scholar]
29. F. Ren et al., “A novel global feature-oriented relational triple extraction model based on table filling,” in Proc. 2021 Conf. Empir. Methods Nat. Lang. Process., 2021, pp. 2646–2656. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF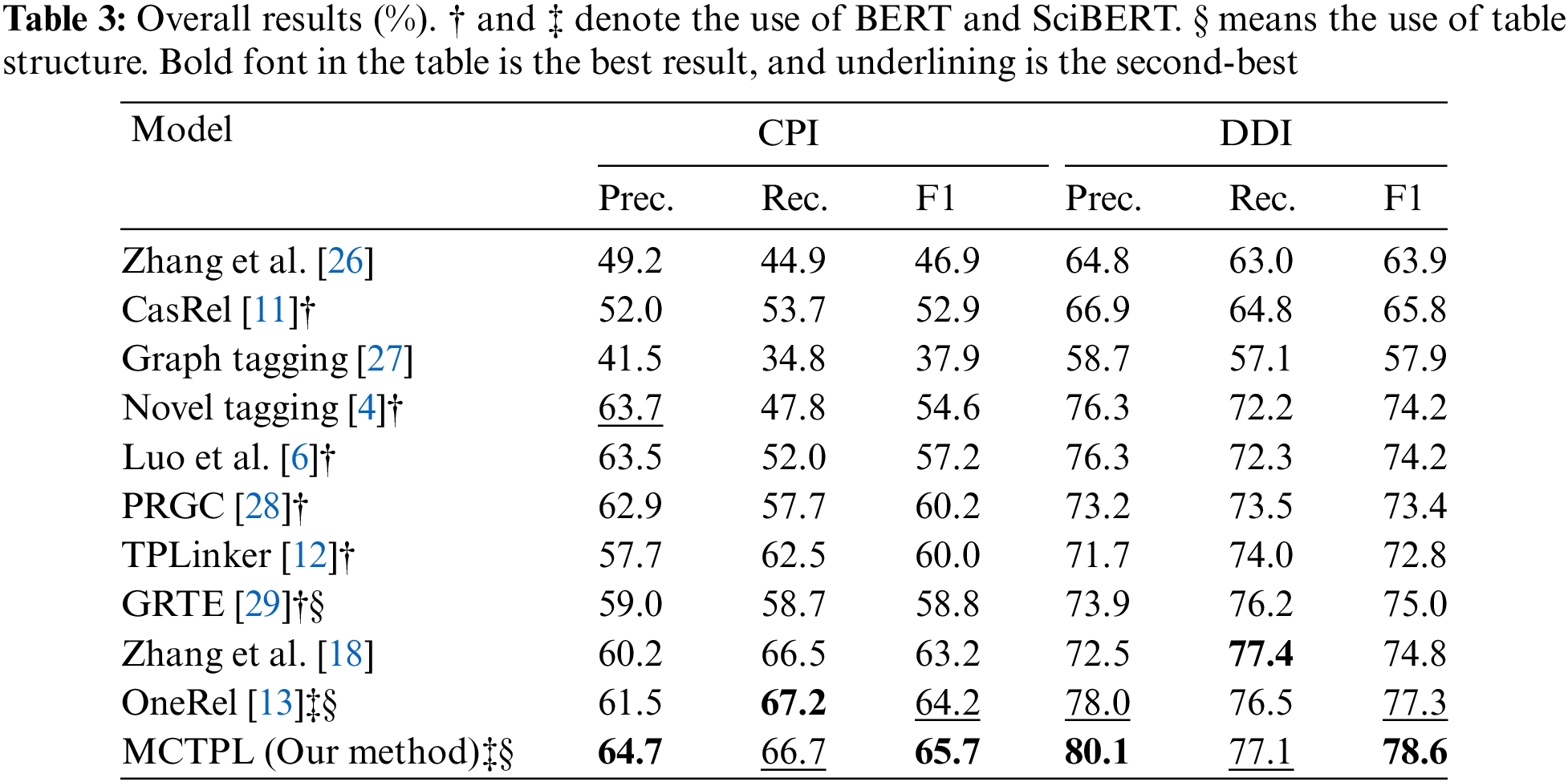
 Downloads
Downloads
 Citation Tools
Citation Tools