 Open Access
Open Access
ARTICLE
GATiT: An Intelligent Diagnosis Model Based on Graph Attention Network Incorporating Text Representation in Knowledge Reasoning
School of Computer and Artificial Intelligence, Zhengzhou University, Zhengzhou, 450001, China
* Corresponding Author: Kunli Zhang. Email:
(This article belongs to the Special Issue: Graph Neural Networks: Methods and Applications in Graph-related Problems)
Computers, Materials & Continua 2024, 80(3), 4767-4790. https://doi.org/10.32604/cmc.2024.053506
Received 02 May 2024; Accepted 16 August 2024; Issue published 12 September 2024
Abstract
The growing prevalence of knowledge reasoning using knowledge graphs (KGs) has substantially improved the accuracy and efficiency of intelligent medical diagnosis. However, current models primarily integrate electronic medical records (EMRs) and KGs into the knowledge reasoning process, ignoring the differing significance of various types of knowledge in EMRs and the diverse data types present in the text. To better integrate EMR text information, we propose a novel intelligent diagnostic model named the Graph ATtention network incorporating Text representation in knowledge reasoning (GATiT), which comprises text representation, subgraph construction, knowledge reasoning, and diagnostic classification. In the text representation process, GATiT uses a pre-trained model to obtain text representations of the EMRs and additionally enhances embeddings by including chief complaint information and numerical information in the input. In the subgraph construction process, GATiT constructs text subgraphs and disease subgraphs from the KG, utilizing EMR text and the disease to be diagnosed. To differentiate the varying importance of nodes within the subgraphs features such as node categories, relevance scores, and other relevant factors are introduced into the text subgraph. The message-passing strategy and attention weight calculation of the graph attention network are adjusted to learn these features in the knowledge reasoning process. Finally, in the diagnostic classification process, the interactive attention-based fusion method integrates the results of knowledge reasoning with text representations to produce the final diagnosis results. Experimental results on multi-label and single-label EMR datasets demonstrate the model’s superiority over several state-of-the-art methods.Keywords
Recently, the medical field has been exploring the application of artificial intelligence to improve the quality and efficiency of medical services [1]. Electronic medical records (EMRs) encompass structured information detailing the patient’s diagnosis and treatment process, as well as unstructured textual data representing doctor diagnoses and treatment opinions. A physician’s clinical diagnostic process involves evaluating the likelihood of a patient having a particular disease based on their clinical manifestations and examination results. Typically, EMRs contain one or more diagnoses. For instance, a patient may receive diagnoses for both “diabetes” and “hypertension”. Viewing an EMR as a sample can pertain to multiple classifications. Thus, the problem of intelligent diagnosis can be considered a classification task, with each diagnosis in a medical record representing a distinct label [2–4].
Diagnosing and treating a patient’s disease involves using both medical knowledge and the patient’s EMR. Knowledge graphs (KGs) have emerged as a critical knowledge source in various knowledge-driven algorithms and systems [5]. KGs consist of entities and the relations between them, denoted as
Currently, the challenges of EMR-based intelligent diagnostic models are as follows:
• Current research addressing EMRs through direct encoding for text representation often ignores the heterogeneity of information across different sections of EMRs. The chief complaint information in EMRs is more important for diagnostic output compared to plain text information. Additionally, numerical information cannot be directly represented by the model. How to represent the different types of text information in EMRs is a challenge.
• Existing models improve diagnosis by incorporating external KGs introduced based on the diseases to be diagnosed, failing to notice critical information in EMRs. Furthermore, the distributed-based knowledge reasoning method is unable to effectively extract the hidden features and topologies within the KG. Therefore, it is a challenge to effectively utilize critical text information in EMRs and how to better learn implicit information about KG for diagnosis during the knowledge reasoning process.
In light of the aforementioned challenges, we consider the varying impact on diagnostic output and the diverse forms of expression. Consequently, information within EMRs can be classified into three categories: ordinary text, chief complaints, and numerical information. The chief complaint, which represents the primary pain or most apparent symptom experienced by the patient, has significance in the diagnostic process that outweighs other text information. Numerical information refers to specific test results and clinical signs recorded as numeric values in EMRs. MacBERT (MLM as correction, Mac) [10] improves the original BERT (Bidirectional Encoder Representations from Transformers) [11] model through the use of MLM (Masked Language Modeling) as a refined correction technique, optimizing its performance for more accurate natural language understanding. To address the first challenge, we incorporated the chief complaint information into the MacBERT [10] encoding and combined it with the numerical data to derive the textual representation of EMRs.
Furthermore, we construct disease subgraphs and text subgraphs to integrate EMR representation into knowledge reasoning. Disease subgraphs are identified from the KG based on the list of diseases to be diagnosed. Text subgraphs are obtained based on the entities in the EMR after retrieving them from the KG. To address the second challenge, we incorporate context nodes containing EMR information into the knowledge reasoning process and set the node category to distinguish them from the rest of the entities. The information contained within context nodes differs from that in other nodes, and the significance of entities directly extracted from EMR text varies compared to other entities. Fig. 1 illustrates a text subgraph constructed from the EMR text “In the past month, multiple measurements of elevated blood pressure, ..., worsening dizziness without apparent cause, ...”. For instance, if the final diagnosis is “hypertension”, the text subgraph should contain two entity nodes: “elevated blood pressure” and “dizziness”. Notably, the former is significantly more crucial to the final diagnosis than the latter, which is reflected in the node relevance score obtained by the model. The Graph Attention Network (GAT) [12] employed in this study utilizes an attention mechanism to assign different weights according to the relative importance among nodes, implementing various learning strategies to effectively address node disparities.
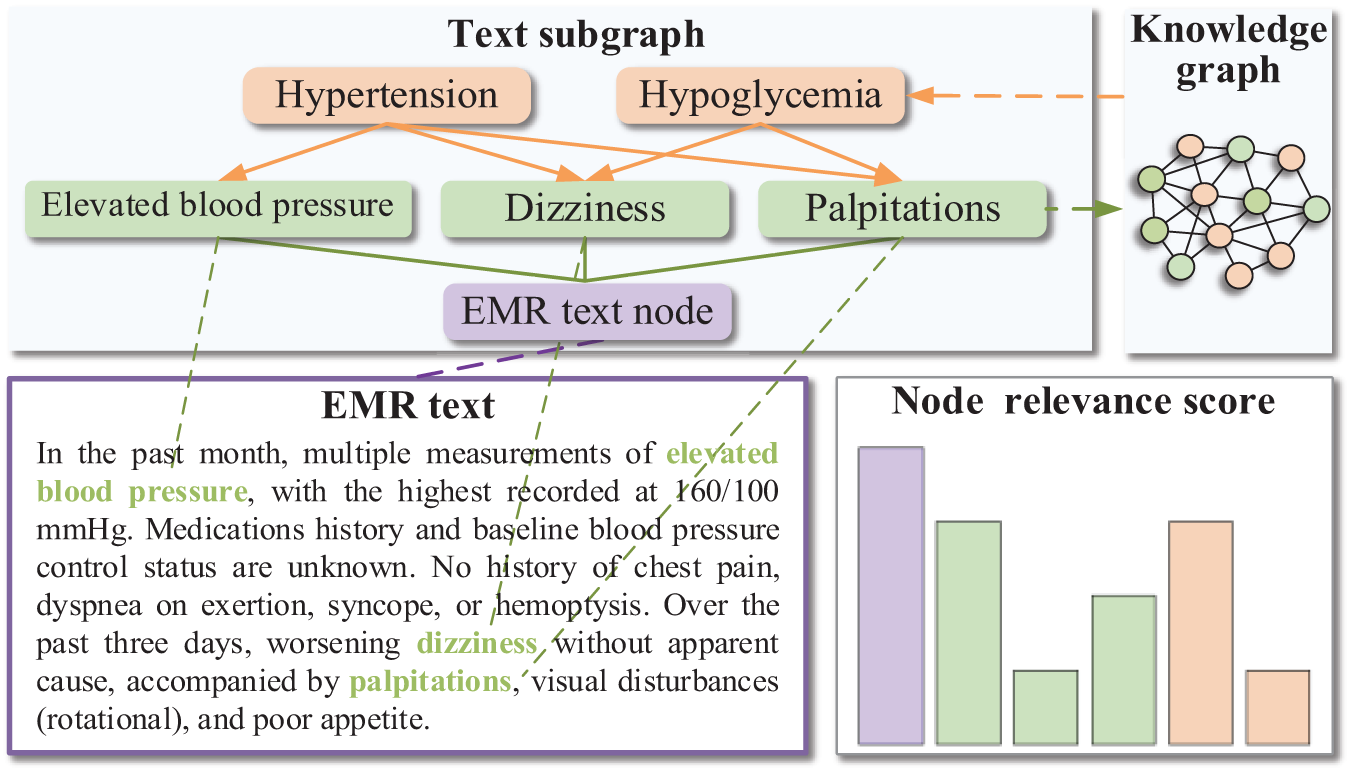
Figure 1: A sample of constructing a text subgraph
Altogether, the following are the primary contributions of our work:
• We propose GATiT, an intelligent diagnosis model based on GAT. We constructed disease subgraphs and text subgraphs, integrating disease information and EMR text information in the knowledge reasoning process. By incorporating node categories, node relevance scores, and edge relevance scores, we optimize GAT for knowledge reasoning. GAT can assign varying weights according to the relative importance of nodes and can better address the differences between nodes.
• GATiT encodes text representations of EMRs using MacBERT, which incorporates enhancements for chief complaint and numerical information. We introduce ChiefInfo Embedding, which integrates chief complaint information into the inputs of MacBERT and utilizes self-attention to fuse numerical and textual information, thereby facilitating the learning of various categories of information from EMRs.
• Comparative experiments conducted on multi-label and single-label EMR datasets using GATiT demonstrate its superiority over several state-of-the-art methods, as indicated by the analysis of experimental results.
Many researchers have studied intelligent diagnosis methods extensively. In this section, in addition to describing existing intelligent diagnosis algorithms (Section 2.1), recent work on graph neural networks is presented (Section 2.2).
Intelligent methods for disease diagnosis have received a lot of attention in recent years. Li et al. [13] proposed a deep learning framework and fine-tuned a CNN (Convolutional Neural Networks) model for intelligent-assisted diagnosis in pediatrics. The results on real-world pediatric Chinese EMR data demonstrated that this method achieved an average accuracy and F1-score of 81%. Sun et al. [14] explored a privacy-preserving medical record searching scheme (PMRSS) utilizing the ElGamal Blind Signature. This scheme allows patients to independently make medical diagnoses by securely searching and comparing previous and current medical records.
While the mentioned methods have been effective, they primarily focus on data processing and neglect the integration of medical domain knowledge crucial for physicians’ diagnostic processes. With the advancement of medical KGs, there is a growing interest in intelligent diagnosis research that incorporates external medical knowledge. Chen et al. [15] introduced a Sequence-to-Subgraph framework and devised a subgraph convolutional network and hierarchical diagnostic attentive network model (SHiDAN) based on this framework. SHiDAN organizes EMR text into densely connected subgraphs with external medical knowledge, employing hierarchical feature extraction for patient diagnosis. Yang et al. [16] proposed a time-aware KG attention approach to address the problem of knowledge decay over time and used a comprehensive representation of the local KG to select candidate global knowledge for prospective interpretation.
GNNs are extensively utilized in disease diagnosis. Yang et al. [17] proposed a feature aggregation-based intelligent diagnostic model using heterogeneous graph convolutional networks (GCNs) that emphasizes the intrinsic properties of symptoms and the multiple hidden relationships among them for effective and accurate symptom-based knowledge reasoning and disease diagnosis. Song et al. [18] incorporated disease subgraph embedding into the knowledge reasoning process using GCNs to amplify the relevance of disease-related knowledge, yet relying solely on entity subgraphs in the knowledge reasoning process presents limitations in effectively combining simulated medical knowledge with real patient cases.
According to the aforementioned studies related to deep learning-based and GNN-based intelligent diagnosis, most of the approaches have been able to obtain satisfactory results in disease diagnosis through deep learning models and disease enhancement. However, most existing intelligent diagnosis models enhance their performance by incorporating external KGs focused solely on the diseases listed in the EMR, neglecting the textual information within the EMRs. Meanwhile, most of the work introduces only EMR text and does not consider the heterogeneity of data in EMR, which leads to unsatisfactory results. Therefore, intelligent diagnosis study should focus on enhancing knowledge around EMR and external KG, and different categories of information from EMR text to obtain more accurate text representation.
The architecture of our model is shown in Fig. 2. This section describes the text representation (Section 3.1), the construction process of both the text subgraph and the disease subgraph (Section 3.2), and the knowledge reasoning (Section 3.3), and the diagnostic classification (Section 3.4).
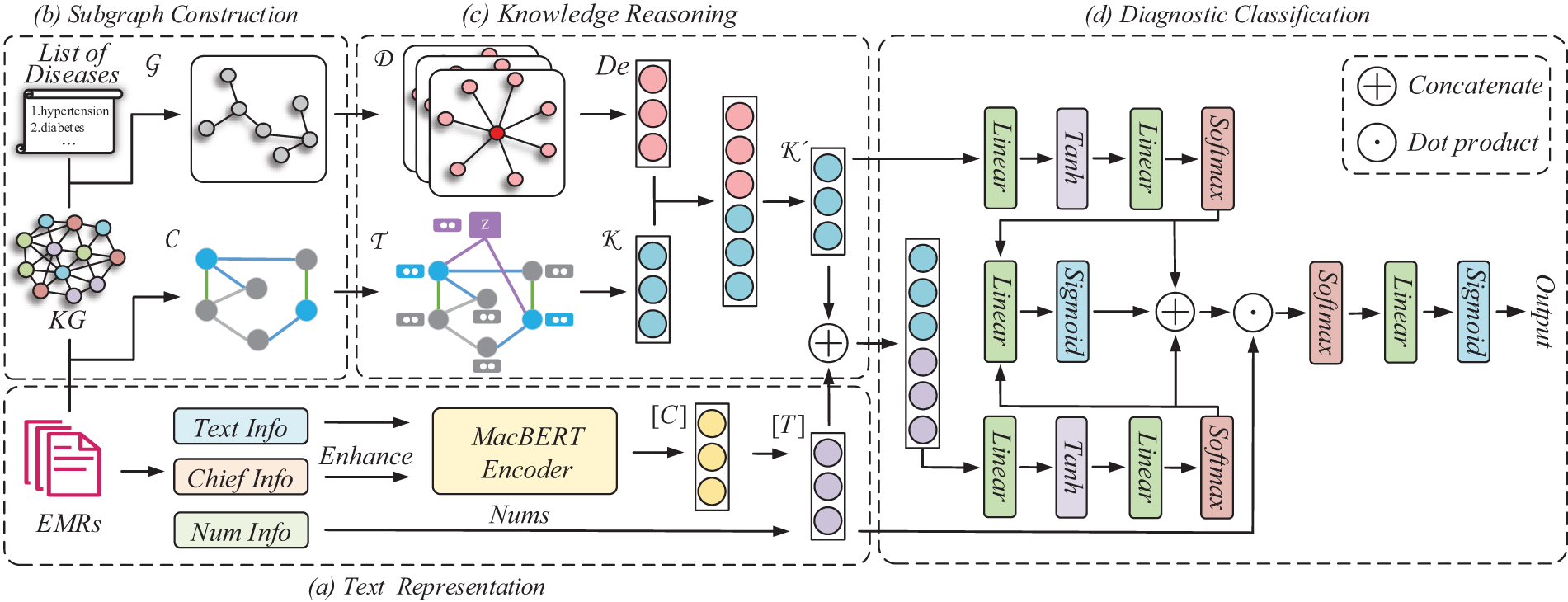
Figure 2: Illustration of GATiT. Our model is structured with text representation, subgraph construction, knowledge reasoning, and diagnostic classification. GATiT integrates knowledge from KG and text information from EMRs for intelligent diagnosis tasks. (a) In the text representation, TextInfo, ChiefInfo, and NumInfo from EMRs are encoded using MacBERT, incorporating chief complaint enhancement and numerical information injection to produce the final text representation. (b) In the subgraph construction, the disease subgraph
The text representation utilizes MacBERT and incorporates chief complaint enhancement (Section 3.1.1) and numerical information injection (Section 3.1.2) for encoding the text representation of EMRs. We define ordinary text information as TextInfo, chief complaint information as ChiefInfo, and numerical information as NumInfo.
3.1.1 Chief Complaint Enhancement
The original input of MacBERT includes Token Embedding, Segment Embedding, and Position Embedding. Since the input to GATiT is a sequence, all values in the Segment Embedding are set to zero. Position Embedding is employed to retain information about the semantic order of the input sequence or sequence pair. Building upon the achievements of Qu et al. [19] in integrating history response embedding into a dialogue system, GATiT adds Chief Embedding containing the chief complaint information to the MacBERT input. Chief Embedding labels the chief complaint information in the input sequence as
3.1.2 Numerical Information Injection
MacBERT encounters challenges in learning numerical features from EMRs. Therefore, we extract numerical features from the EMR, and the hidden layer representation
where
The process of the subgraph construction is depicted in Fig. 3. This section constructs the text subgraph (Section 3.2.1) and the disease subgraph (Section 3.2.2) within the EMR text, facilitating their application in the subsequent knowledge reasoning and establishing a close connection between the knowledge reasoning and EMR text information.
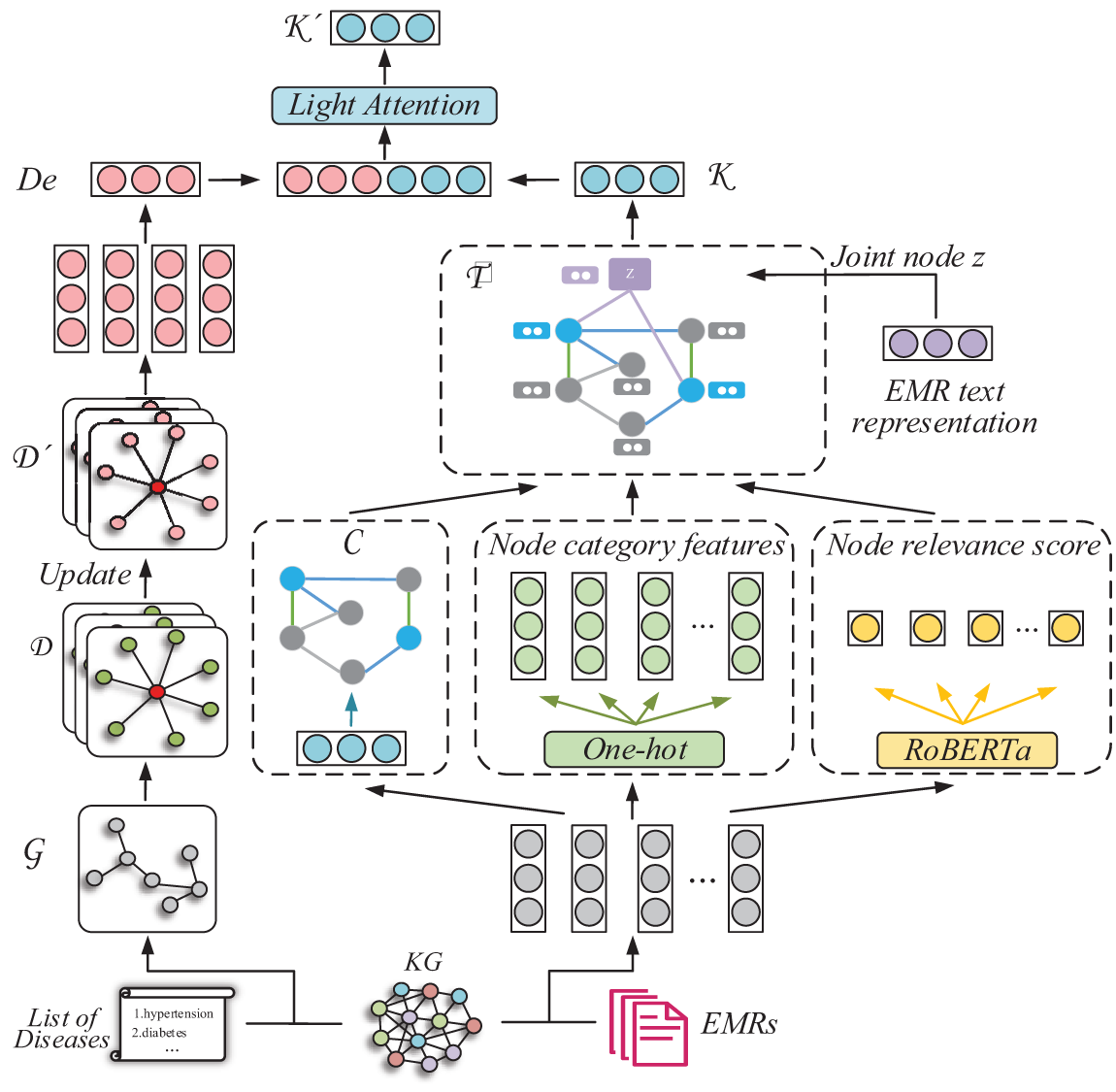
Figure 3: Illustration of the subgraph construction and the knowledge reasoning. The disease subgraphs
For a given EMR text sequence
EMR context node
where
Node category features. Following the construction process, nodes can be categorized into three types: (a) Context node, which contains the text representation of EMRs. (b) Text entity node, which corresponds to disease entities and symptom entities identified from the EMR text. (c) Other entity nodes, which represent the head entity or tail entity nodes in the triple retrieved from KGs by the text entity nodes. The category feature of a node is defined as:
where
Node relevance scores. Nodes within the same category in
where
Edge additional features. Relation type
where
where
Incorporating disease subgraph (
where
Splitting the whole graph batch gives updated
The vector representation of the final subgraph is obtained by reading
where
The architecture of knowledge reasoning is depicted in Fig. 3. Knowledge reasoning uses
Aggregation of messages between each node and neighboring nodes using an attention mechanism:
where
Unlike GAT, the message
where
The attention coefficient
where
The updating equation for the knowledge reasoning process is as follows:
where
Subgraph reasoning is the process of knowledge reasoning based on the construction of the disease subgraph and the text subgraph in Section 3.2.
Text subgraph reasoning. Construct the adjacency matrix
A self-loop is added to each node to ensure that each node’s representation influences the representation of the subsequent layer. This is accomplished by augmenting the identity matrix
The reasoning process of
where
Disease subgraph reasoning. We employ light attention to integrate disease map structure information into the reasoning process. Light attention divides the information into original global features and global features to be updated, where the original global features are defined as follows:
where
The global features to be updated are then obtained through the weight matrix
where
The final output of the knowledge reasoning is obtained by weighted addition of
where
The model can acquire
3.4.1 Reasoning Result Self-Attention Mechanism
The significance of the knowledge encapsulated within reasoning results varies in influencing the final diagnostic output. To gauge the influence of the embedded knowledge on the final diagnostic result, we introduce the reasoning result self-attention mechanism. The process unfolds as follows:
where
The knowledge noise problem emerges when a sentence veers from its original meaning due to excessive external knowledge introduction. The model gauges the correlation between the text representation and the reasoning result through reasoning result-text representation attention:
where
Employing gating mechanisms to integrate text representations and knowledge reasoning results in enhanced mitigation of knowledge noise problems:
where
The final prediction is derived by mapping the final hidden representation into the disease space:
where
Experimental evaluations utilized the multi-labeled Chinese Obstetrics EMRs (COEMRs) dataset and the publicly available single-labeled Chinese EMRs (C-EMRs) dataset. The Chinese Obstetrics Knowledge Graph (COKG) and Chinese Medical Knowledge Graph (CMeKG) serve as external knowledge sources for COEMRs and C-EMRs, respectively. Their basic statistics are provided in Tables 1 and 2. Furthermore, detailed information regarding these datasets is presented below:
• COEMRs: A multiple-label dataset comprises real EMRs from multiple hospitals, totaling 24,339 entries. Of these, 21,905 are allocated to the training set and 2434 to the testing set, following a 9:1 division ratio. The dataset retains 73 diseases for diagnosis, constituting a multi-labeled dataset. Specific samples are shown in Fig. 4.
• C-EMRs [22]: A single-label dataset comprises 18,331 EMRs. Following a 9:1 division, 16,498 records were assigned to the training set and 1833 to the test set. Due to substantial missing numerical features in C-EMRs, numerical information extraction was omitted, focusing solely on extracting chief complaint information. In contrast to COEMRs, C-EMRs present diagnostic results in a single-label format, covering ten distinct diseases, such as diabetes and hypertension.
• COKG [23]: COKG integrates medical knowledge from diverse sources, serving as a domain-specific KG focusing on common obstetric diseases. Its knowledge base comprises obstetrics textbooks, clinical guidelines, specialized medical dictionaries, and web resources, encompassing 42,616 entities and 66,261 relations.
• CMeKG [24]: A comprehensive medical domain KG that consolidates medical knowledge from various sources, including clinical documents, textbooks, encyclopedias, and medical standard nomenclatures. It focuses on diseases, drugs, diagnostic techniques, and treatment methods.



Figure 4: COEMRs sample. Chief complaint documents the patient’s most significant health problem or symptom at the time of the visit. The medical history includes the patient’s past illnesses, surgical history, and other relevant information. Admitting physical examination, obstetric examination, and auxiliary examinations document the patient’s clinical findings and serve as the primary sources of numerical information. Admitting diagnosis is the physician’s diagnosis of the patient, which serves as the categorical label for the dataset
Python is the programming language utilized for the experiment. Deep learning frameworks include PyTorch, DGL (Deep Graph Library), and Transformers. Details of the experimental environment and parameter settings are shown in Table 3. Specifically including operating system, GPU (Graphics Processing Unit), RAM (Random Access Memory), CUDA (Compute Unified Device Architecture), etc.

The selected evaluation metrics are commonly used in text classification tasks. Considering that the datasets include both single-label and multi-label instances, the evaluation metrics consist of precision (
where
4.3 Results Analysis for Comparative Experiment
To demonstrate the effectiveness of GATiT, we analyzed the experimental results. The overall performance of GATiT was first evaluated using several baseline models including Text Recurrent Neural Network (TextRNN) [25], Text Convolutional Neural Network (TextCNN) [26], Text Recurrent Convolutional Neural Network (TextRCNN) [27], TextRNN with Attention (TextRNN+Att) [28], Transformer [29], Deep Pyramid Convolutional Neural Networks (DPCNN) [30], BERT [11], MacBERT [10], KAIE (Knowledge powered Attention and Information Enhanced) [31], GSKN (Graph-based Structural Knowledge-aware Network) [32], DeGCN (Disease enhanced Graph Convolutional Network) [18]. Subsequently, we analyzed disease-specific


After preprocessing, numerical information extraction, and chief complaint information extraction, the average text length of COEMRs is 371.83. Due to the more severe lack of numerical information in C-EMRs, only chief complaint information enhancement is retained, resulting in an average text length of 352.22 for C-EMRs. The GATiT comprises 153,394,722 trainable parameters, representing a 51.71% increase compared to GSKN. The mean number of entities in the
MacBERT is trained on a large-scale corpus, significantly outperforming traditional deep learning models across all metrics, with
4.4 Results Analysis for Across Disease Groups in COEMRs
To further validate GATiT’s performance and showcase its superiority effectively, we categorized the total tags based on their occurrence frequency in COEMRS into five groups: 20–30, 31–80, 81–200, 201–1000, and >1000, as illustrated in Fig. 5.

Figure 5:
When the label occurrence frequency exceeds 200, the
4.5 Results Analysis for Each Disease in C-EMRs
Fig. 6 visualizes the
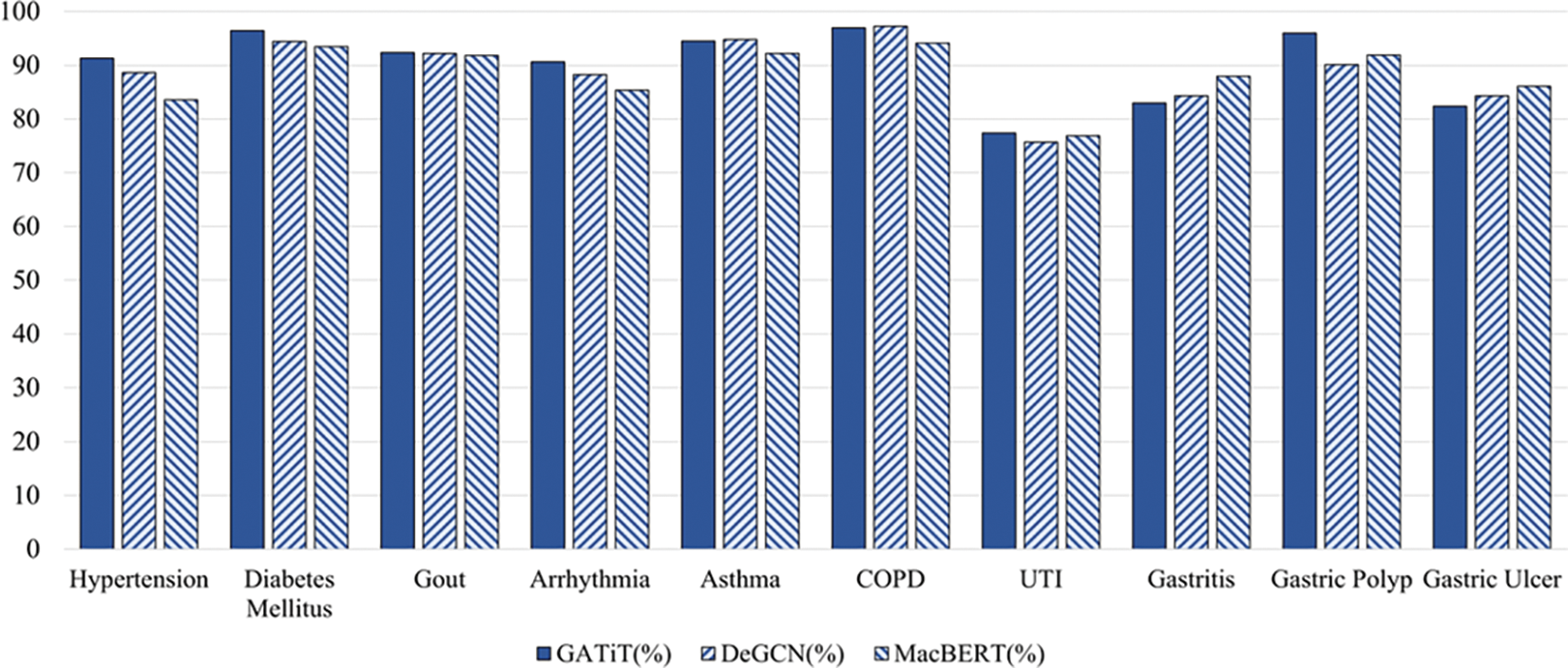
Figure 6:
4.6 Results Analysis for the Confusion Matrix
To demonstrate GATiT’s predictions across diseases, confusion matrices of GATiT and DeGCN were visualized in Figs. 7 and 8 (rounded to one decimal place, and results of 0 are not shown).

Figure 7: Visualization of DeGCN’s confusion matrix in C-EMRs

Figure 8: Visualization of GATiT’s confusion matrix in C-EMRs
4.7 Results Analysis for Each Disease in COEMRs
To validate the effectiveness of incorporating EMR text nodes and GAT into the knowledge reasoning process, we selected GATiT and DeGCN for further comparison. The specific results were analyzed by examining the

Figure 9: Disease-specific
Integrating the disease subgraph and the text subgraph helps enrich the data on low-frequency diseases, thereby improving knowledge reasoning. Additionally, incorporating EMR text nodes into the knowledge reasoning process allows the model to learn context information from EMRs, thus enhancing diagnostic accuracy.
4.8 Results Analysis for Ablation Study
To evaluate the effectiveness of GNN knowledge reasoning, light attention, and the introduced context nodes and additional features, we conducted an ablation study on COEMRs and C-EMRs. The experiments included the following conditions: (a) MHSA (Multi Head Self Attention): utilization of the multi-head self-attention mechanism in the transformer instead of light attention. (b) w/o GCN: removal of the GCN-based knowledge reasoning, with direct reading of the disease subgraph as the reasoning results. (c) w/o Att: removal of light attention. (d) w/o De: elimination of the entire disease augmentation module. (e) w/o NodeZ: removal of the context node containing the EMR information. (f) w/o Ext: elimination of node additional features and edge additional features. Complete removal of additional features leads to the degradation of GATiT to the GAT; thus, w/o Ext can be used to represent knowledge reasoning using an ordinary GAT. Specific experimental results are presented in Tables 6 and 7.


The ablation study revealed a significant decrease in all evaluation metrics of GATiT when the knowledge reasoning based on GCNs was removed. This finding underscores the necessity of incorporating knowledge reasoning based on GNNs in the intelligent diagnosis task. Substituting the light attention with the MHSA in the transformer resulted in a decrease in all evaluation metrics of the model, demonstrating the effectiveness of the light attention. The light attention utilized 4,202,512 trainable parameters and required a memory size of 16.81 MB. In contrast, during the experiment, the MHSA employed 16,789,504 trainable parameters and required a memory size of 67.58 MB. Complete removal of the attention mechanism resulted in the model’s inability to differentiate between the disease knowledge to be augmented effectively and that embedded in the textual entities, leading to respective decreases of 1.03% and 2.14% in
In multi-labeled COEMRs, where numerous diseases may be diagnosed in each medical record, removing the attention mechanism caused an 11.67% decrease in the
This paper proposes an innovative GATiT model, which incorporates the graph attention network in knowledge reasoning for medical intelligent diagnosis tasks. GATiT incorporates the disease subgraph related to the diagnosis and constructs the text subgraph of context nodes containing text information from EMRs within the knowledge reasoning. To facilitate the model’s ability to differentiate context nodes from other nodes during learning, GATiT introduces additional features. Additionally, we adapt the message-passing strategy of GAT and adjusts the calculation of attention weights to learn these features. In the text representation, we adjust MacBERT to obtain text representations of the EMR. In the diagnostic classification, we use the interactive attention-based fusion mechanism to fuse the above results to get the final output. Results from COEMRs and C-EMRs validate the effectiveness of GATiT and the importance of incorporating text information from EMRs in the knowledge reasoning process. For future work, we plan to:
• Further explore how to effectively utilize different forms of multimodal data in intelligent diagnosis research and realize the effective fusion of different modal data.
• Another focus of future work is to apply the few-shot learning technique to intelligent diagnosis research to learn the relevant features of diseases from a small amount of data.
Acknowledgement: We thank all the anonymous reviewers who generously contributed their time and efforts. Their professional recommendations have greatly enhanced the quality of the manuscript.
Funding Statement: This work was supported in part by the Science and Technology Innovation 2030–“New Generation of Artificial Intelligence” Major Project (No. 2021ZD0111000), and Henan Provincial Science and Technology Research Project (No. 232102211039).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Yu Song, Pengcheng Wu; data collection: Kunli Zhang, Yu Song; analysis and interpretation of results: Pengcheng Wu, Dongming Dai; draft manuscript preparation: Kunli Zhang, Pengcheng Wu, Mingyu Gui. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: A dataset used in this paper can be found in website. CMeKG can be accessed on http://nscc.zzu.edu.cn/know/ (accessed on 26 March 2024). The datasets C-EMRs can be downloaded from the website https://github.com/YangzlTHU/C-EMRs (accessed on 20 April 2018). Other data is available on request from the authors.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. J. H. Kamdar, J. Jeba Praba, and J. J. Georrge, “Artificial intelligence in medical diagnosis: Methods, algorithms and applications,” Mach. Learn. Health Care Perspect. Mach. Learn. Healthc., vol. 13, pp. 27–37, Mar. 2020. doi: 10.1007/978-3-030-40850-3_2. [Google Scholar] [CrossRef]
2. C. C. Aggarwal and C. Zhai, “A survey of text classification algorithms,” in Mining Text Data, 6th ed. Boston, MA, USA: Springer, 2012, pp. 163–222. doi: 10.1007/978-1-4614-3223-4_6. [Google Scholar] [CrossRef]
3. K. Zhang, H. Ma, Y. Zhao, H. Zan, and L. Zhuang, “The comparative experimental study of multilabel classification for diagnosis assistant based on Chinese obstetric EMRs,” J. Healthc. Eng., vol. 2018, no. 1, Feb. 2018, Art. no. 7273451. doi: 10.1155/2018/7273451. [Google Scholar] [PubMed] [CrossRef]
4. S. Minaee, N. Kalchbrenner, E. Cambria, N. Nikzad, M. Chenaghlu and J. Gao, “Deep learning-based text classification: A comprehensive review,” Assoc. Comput. Mach., vol. 54, no. 62, pp. 1–40, Apr. 2021. doi: 10.1145/3439726. [Google Scholar] [CrossRef]
5. A. Hogan et al., “Knowledge graphs,” ACM Comput. Surv., vol. 54, no. 71, pp. 1–37, Jul. 2021. doi: 10.1145/3447772. [Google Scholar] [CrossRef]
6. X. Hao et al., “Construction and application of a knowledge graph,” Remote Sens., vol. 13, no. 13, Jun. 2021, Art. no. 2511. doi: 10.3390/rs13132511. [Google Scholar] [CrossRef]
7. X. Chen, S. Jia, and Y. Xiang, “A review: Knowledge reasoning over knowledge graph,” Expert. Syst. Appl., vol. 141, no. 1, Mar. 2020, Art. no. 112948. doi: 10.1016/j.eswa.2019.112948. [Google Scholar] [CrossRef]
8. F. Scarselli, M. Gori, A. C. Tsoi, M. Hagenbuchner, and G. Monfardini, “The graph neural network model,” IEEE Trans. Neural Netw., vol. 20, no. 1, pp. 61–80, Dec. 2008. doi: 10.1109/TNN.2008.2005605. [Google Scholar] [PubMed] [CrossRef]
9. S. Sun et al., “Review of graph neural networks applied to knowledge graph reasoning,” J. Front. Comput. Sci. Technol., vol. 17, no. 1, pp. 27–52, Jul. 2023. [Google Scholar]
10. Y. Cui, W. Che, T. Liu, B. Qin, S. Wang and G. Hu, “Revisiting pre-trained models for Chinese natural language processing,” in Findings of the Assoc. for Comput. Linguist.: EMNLP 2020, Nov. 2020, pp. 657–668. doi: 10.18653/y1/2020.findings-emnlp.58. [Google Scholar] [CrossRef]
11. J. Devlin, M. Chang, K. Lee, and K. Toutanova, “BERT: Pre-training of deep bidirectional transformers for language understanding,” in Proc. of the 2019 Conf. of the North Am. Chapter of the Assoc. for Comput. Linguist.: Hum. Lang. Technol., Minneapolis, Minnesota, Jun. 2019, pp. 4171–4186. doi: 10.18653/v1/n19-1423. [Google Scholar] [PubMed] [CrossRef]
12. P. Veličković, G. Cucurull, A. Casanova, A. Romero, P. Liò and Y. Bengio, “Graph attention networks,” in 6th Int. Conf. on Learn. Representat., Vancouver, BC, Canada, Apr. 2018. [Google Scholar]
13. X. Li, H. Wang, H. He, J. Du, J. Chen and J. Wu, “Intelligent diagnosis with Chinese electronic medical records based on convolutional neural networks,” BMC Bioinformatics, vol. 20, no. 1, Feb. 2019, Art. no. 62. doi: 10.1186/s12859-019-2617-8. [Google Scholar] [PubMed] [CrossRef]
14. Y. Sun, J. Liu, K. Yu, M. Alazab, and K. Lin, “PMRSS: Privacy-preserving medical record searching scheme for intelligent diagnosis in IoT healthcare,” IEEE Trans. Ind. Inform., vol. 18, no. 3, pp. 1981–1990, Mar. 2022. doi: 10.1109/TII.2021.3070544. [Google Scholar] [CrossRef]
15. J. Chen, Q. Yuan, C. Lu, and H. Huang, “A novel sequence-to-subgraph framework for diagnosis classification,” in Proc. of the Thirtieth Int. Joint Conf. on Artif. Intell., Montreal, QC, Canada, Aug. 2021, pp. 3606–3612. doi: 10.24963/ijcai.2021/496. [Google Scholar] [CrossRef]
16. K. Yang et al., “KerPrint: Local-global knowledge graph enhanced diagnosis prediction for retrospective and prospective interpretations,” Proc. AAAI Conf. Artif. Intell., vol. 37, no. 4, pp. 5357–5365, Jun. 2023. doi: 10.1609/aaai.v37i4.25667. [Google Scholar] [CrossRef]
17. X. Yang, Y. Zhang, F. Hu, Z. Deng, and X. Zhang, “Feature aggregation-based multi-relational knowledge reasoning for COPD intelligent diagnosis,” Comput. Electr. Eng., vol. 114, 2024, Art. no. 109068. doi: 10.1016/j.compeleceng.2023.109068. [Google Scholar] [CrossRef]
18. Y. Song et al., “Research on double-graphs knowledge-enhanced intelligent diagnosis,” in China Health Inf. Process. Conf., Hangzhou, China, Sep. 2023, pp. 317–332. doi: 10.1007/978-981-99-9864-7_21. [Google Scholar] [CrossRef]
19. C. Qu, L. Yang, M. Qiu, W. B. Croft, Y. Zhang and M. Iyyer, “BERT with history answer embedding for conversational question answering,” in Proc. of the 42nd Int. ACM SIGIR Conf. on Res. and Dev. in Inf. Retrieval, Paris, France, Jul. 2019, pp. 1133–1136. doi: 10.1145/3331184.3331341. [Google Scholar] [CrossRef]
20. Y. Lin, Z. Liu, M. Sun, Y. Liu, and X. Zhu, “Learning entity and relation embeddings for knowledge graph completion,” in Proc. AAAI Conf. Artif. Intell., vol. 29, no. 1, Feb. 2015. doi: 10.1609/aaai.v29i1.9491. [Google Scholar] [CrossRef]
21. Y. Liu et al., “RoBERTa: A robustly optimized BERT pretraining approach,” 2019, arXiv: 1907.11692. [Google Scholar]
22. Z. Yang, Y. Huang, Y. Jiang, Y. Sun, Y. Zhang and P. Luo, “Clinical assistant diagnosis for electronic medical record based on convolutional neural network,” Sci. Rep., vol. 8, no. 1, p. 6329, Apr. 2018. doi: 10.1038/s41598-018-24389-w. [Google Scholar] [PubMed] [CrossRef]
23. K. Zhang, C. Hu, Y. Song, H. Zan, Y. Zhao and W. Chu, “Construction of Chinese obstetrics knowledge graph based on the multiple sources data,” in Workshop on Chinese Lexical Semantics, Nanjing, China, May 2021, pp. 399–410. doi: 10.1007/978-3-031-06547-7_31. [Google Scholar] [CrossRef]
24. Aodema et al., “Preliminary exploration of constructing Chinese medical knowledge graph CMeKG,” J. Chin. Inf. Process., vol. 33, no. 10, pp. 1–7, 2019. [Google Scholar]
25. S. Hochreiter and J. Schmidhuber, “Long short-term memory,” Neural Comput., vol. 9, no. 8, pp. 1735–1780, Nov. 1997. doi: 10.1162/neco.1997.9.8.1735. [Google Scholar] [PubMed] [CrossRef]
26. Y. Kim, “Convolutional neural networks for sentence classification,” in Proc. of the 2014 Conf. on Empirical Methods in Nat. Lang. Process. (EMNLP), Doha, Qata, Oct. 2014, pp. 1746–1751. doi: 10.3115/v1/D14-1181. [Google Scholar] [CrossRef]
27. S. Lai, L. Xu, K. Liu, and J. Zhao, “Recurrent convolutional neural networks for text classification,” in Proc. AAAI Conf. Artif. Intell., vol. 29, no. 1, pp. 2267–2273, 25–30 Jan., 2015. [Google Scholar]
28. P. Zhou et al., “Attention-based bidirectional long short-term memory networks for relation classification,” in Proc. of the 54th Annu. Meeting of the Assoc. for Comput. Linguist., Berlin, Germany, 2016, pp. 207–212. [Google Scholar]
29. A. Vaswani et al., “Attention is all you need,” in Adv. in Neural Inf. Process. Syst., Long Beach, CA, USA, 4–9 Dec. 2017, pp. 6000–6010. [Google Scholar]
30. R. Johnson and T. Zhang, “Deep pyramid convolutional neural networks for text categorization,” in Proc. of the 55th Annu. Meeting of the Assoc. for Comput. Linguist., Vancouver, BC, Canada, Jul. 2017, pp. 562–570. doi: 10.18653/v1/P17-1052. [Google Scholar] [PubMed] [CrossRef]
31. K. Zhang, X. Zhao, L. Zhuang, Q. Xie, and H. Zan, “Knowledge-enabled diagnosis assistant based on obstetric EMRs and knowledge graph,” in China Nat. Conf. on Chin. Comput. Linguist., Haikou, China, Springer, Oct. 2020, pp. 444–457. doi: 10.1007/978-3-030-63031-7_32. [Google Scholar] [CrossRef]
32. K. Zhang, B. Hu, F. Zhou, Y. Song, X. Zhao and X. Huang, “Graph-based structural knowledge-aware network for diagnosis assistant,” Math. Biosci. Eng., vol. 19, no. 10, pp. 10533–10549, Jul. 2022. doi: 10.3934/mbe.2022492. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools