 Open Access
Open Access
ARTICLE
Spatial Attention Integrated EfficientNet Architecture for Breast Cancer Classification with Explainable AI
1 Department of Electronics and Communication Engineering, Bannari Amman Institute of Technology, Sathyamangalam, 638402, India
2 School of Computer Science Engineering and Information Systems (SCORE), Vellore Institute of Technology, Vellore, 632014, India
3 School of Science, Engineering and Environment, University of Salford, Manchester, M54WT, UK
4 Department of Computer Science & Engineering, Faculty of Engineering and Technology, JAIN (Deemed-to-be University), Bengaluru, 562112, India
5 Department of Management, College of Business Administration, Princess Nourah Bint Abdulrahman University, P. O. Box 84428, Riyadh, 11671, Saudi Arabia
6 Department of Computer Engineering, College of Computer and Information Sciences, King Saud University, P. O. Box 51178, Riyadh, 11543, Saudi Arabia
7 Adjunct Research Faculty, Centre for Research Impact & Outcome, Chitkara University, Rajpura, 140401, India
* Corresponding Author: Surbhi Bhatia Khan. Email:
Computers, Materials & Continua 2024, 80(3), 5029-5045. https://doi.org/10.32604/cmc.2024.052531
Received 04 April 2024; Accepted 11 July 2024; Issue published 12 September 2024
Abstract
Breast cancer is a type of cancer responsible for higher mortality rates among women. The cruelty of breast cancer always requires a promising approach for its earlier detection. In light of this, the proposed research leverages the representation ability of pretrained EfficientNet-B0 model and the classification ability of the XGBoost model for the binary classification of breast tumors. In addition, the above transfer learning model is modified in such a way that it will focus more on tumor cells in the input mammogram. Accordingly, the work proposed an EfficientNet-B0 having a Spatial Attention Layer with XGBoost (ESA-XGBNet) for binary classification of mammograms. For this, the work is trained, tested, and validated using original and augmented mammogram images of three public datasets namely CBIS-DDSM, INbreast, and MIAS databases. Maximum classification accuracy of 97.585% (CBIS-DDSM), 98.255% (INbreast), and 98.91% (MIAS) is obtained using the proposed ESA-XGBNet architecture as compared with the existing models. Furthermore, the decision-making of the proposed ESA-XGBNet architecture is visualized and validated using the Attention Guided GradCAM-based Explainable AI technique.Keywords
Breast cancer (BC) also termed as mammary cancer, is a cruel form of cancer disease significantly affects women. BC causes breast cells to have abnormal growth and so it acquires the ability to spread progressively to other organs. Due to this, breasts are highly affected causing higher malformation among women [1]. This abnormal rupture of cells is done in an uncontrolled behavior. Based on the statistical survey of GLOBOCAN, around 19 million newer cancer cases have happened in 2020. Among these, exclusively for women, BC remains the most commonly diagnosed cancer globally. And from the above statistical survey, it is revealed that BC is not only a major source of death in developed countries but also it remains as a serious life-threatening health concern in developing nations [2]. There are several factors responsible for the incidence of breast cancer; some of them are age, gender, genetic mutations, family history, hormonal factors, and lifestyle choices [3]. Digital mammograms are obtained using mammographic procedures utilizing lower-dose X-rays as a source [4,5] used for initial cancer screening. MRI representing Magnetic Resonance Imaging offers detailed images of breast tissue and so exclusively useful for evaluating the extent of cancer in certain situations [6]. The next one is the biopsy which includes the procedure of collecting a smaller sample of tissues from the breast for examination under a microscope and this is the definitive methodology for diagnosing breast cancer [7]. In this way, the proposed research intends to design a Computer-Aided Diagnosis (CAD) based on earlier detection architecture of EfficientNet-B0 having a Spatial Attention Layer with XGBoost (ESA-XGBNet) for binary classification of mammograms.
For the application areas employing medical imaging and analysis, the initial challenge is the limited accessibility of mammogram inputs since deep learning requires a substantial amount of input images for providing effective classification performance [8]. Subsequently, the next one concerns feature engineering, that is, the deep learning model provides deep feature maps with some amount of redundancy [9]. To address these issues, the research intended to design a CAD framework using the EfficientNet-B0 model, Spatial Attention Mechanism, and XGBoost classifier. Thus, the research contributions to solving the mammogram classification are as follows:
• Mammograms from three distinct standard databases are fed as input to the proposed CAD framework. For these inputs, the work proposes an innovative framework that integrates the algorithms of deep learning and machine learning, employing pre-trained architecture for feature extrication and building a robust classifier model.
• The input mammograms are noise-removed using adaptive median filtering and contrast-enhanced using Contrast Limited Adaptive Histogram Equalization (CLAHE). The preprocessed mammograms are augmented sufficiently using appropriate data augmentation.
• The EfficientNet-B0 pre-trained model is enhanced with the spatial attention mechanism layer for computing attention weights for each spatial location.
• Flatten the above resultant feature vectors and feed as input to the XGBoost classifier.
• Finally, experimentation will be done for the classification of mammogram inputs as benign or malignant cases using the ESA-XGBNet framework.
The rest of the paper is organized as follows: Section 2 describes the background and literature study, Section 3 elaborates on the materials and methods regarding pre-processing, augmentation, and proposed transfer learning with attention mechanism (ESA-XGBNet), Section 4 describes the experimental setup, results, performance comparison with existing works and analysis, and Section 5 concludes the research with future extension of the proposed work.
The discussion on the previous sub-section states that breast tumors are serious life-threatening diseases that distress the daily lives of women all around the world. This cancer type is the most common invasive cancer among women globally, with an estimation of 2.3 million newer incidences and 685,000 mortalities annually as per the report of the World Health Organization, 2023 [10]. Earlier detection and accurate diagnosis are crucial for improving survival rates. The design of an AI-based robust CAD framework is highly helpful in earlier identification and timely interpretation of this cancer type. The CAD frameworks utilized both machine learning and deep learning techniques for providing better solutions to the classification problem [11]. The related works corresponding to the above classification problem are discussed further.
Several literature works utilized Artificial Neural Networks (ANN) for classification problems, exclusively for breast cancer. Next to this, Support Vector Machines (SVM) models were popularly and widely used for breast tumor classification. Patel et al. [12] proposed a model that integrates ANN and hybrid optimum feature selection algorithms for breast tumor classification in the year 2021. For evaluation, the work utilized the Mammographic Image Analysis Society mammogram data after suitable enhancement procedures. The paper attained a maximum of 99% accuracy in MIAS data classification. However, the work suffered from the problem of overfitting and limited data. Kumari et al. [13] proposed an innovative way of extracting features, Advanced Gray-Level Co-occurrence Matrix (AGLCM) was developed for extricating features from mammogram image sets. For preprocessing, the researchers utilized Contrast Limited Advanced Histogram Equalization (CLAHE) for enhancing the mammogram details. Texture-based features are derived for the classification phase. For this research work, the authors employed SVM, ANN, K-Nearest Neighbor (KNN), XGBoost, and Random Forest (RF) models for the classification phase. Their experimentation provided maximum classification performance for the combination of CLAHE, AGLCM, and XGBoost algorithms with around 95% accuracy. Their research has challenges in handling redundant handcrafted features and overfitting. The author of the work [14] introduced a newer and hybrid framework for the classification problem. They employed two hybridized optimizations, Harris Hawks and Crow Search Optimization with ANN and SVM algorithms. They experimented on the mammograms corresponding to the DDSM database. For this, they achieved the results of around 97% accuracy using the above hybrid optimization combination together with the ANN algorithm. They faced challenges in metaheuristic hybridization and tuning of machine learning models.
In addition to the above study, the works of [15–17] experimented with the effectiveness of radiomics feature extraction in medical images. And they revealed that a wider range of qualitative features such as information on texture, shape, and intensity characteristics of image inputs are acquired through radiomic feature extraction. This in turn helps in extracting valuable insights from the applied image inputs. However, higher dimensional feature representations with more redundant and irrelevant vectors are generated using the radiomic feature extrication [18]. This makes the above approach as inferior as compared with the deep feature extrication using CNN models. Over the past decades, the utilization of Convolution Neural Networks (CNN) for solving biomedical problems has rapidly increased for medical imaging and analysis, including breast cancer classification [19]. Out of this, the concept of transfer learning is an emerging one and is used by several researchers for their classification problems. Exclusively, in the application of breast tumor classification, recent research studies have reported an accuracy of around 98% using pretrained models namely Visual Geometry Group-19, Residual Neural Network, and Densely Connected Convolutional Networks.
In addition to the above points, transfer learning architectures foster faster training since the architectures were already trained (pre-trained) on larger databases. And can be fine-tuned easily for deployment of breast cancer classification tasks. This represents that the number of malignant cases is lower than the number of benign cases for implementation. This can lead to biased model predictions supporting only the majority classes for classification reporting to overfitting problems. For tackling this problem, researchers have proposed several strategies, namely class-weighted loss functions, data augmentation techniques, and attention mechanisms. In detail, rotation, flipping, scaling, and cropping of image augmentation will increase the amount of minority class data (e.g., malignant class images) artificially. Thus, the data augmentation leads to the generation of newer synthetic samples from the available inputs. This facilitates the classification model to have good exposure to a more diversified range of input data. And reveals that the generalization ability of the classification model is improved thereby the risk of overfitting will be reduced exclusively in the presence of class imbalance. On the other hand, the dynamic focus on informative regions on the image inputs will be increased through the use of attention mechanisms. For the employed problem of breast cancer classification, the prioritizing of crucial or malignancy regions in the mammogram inputs is done using the attention modules. This prioritization of regions is done through the assignment of adaptive weights for different regions based on their severity presence. And thus the computation and assignment of adaptive weights using attention mechanisms will support in overcoming the issue of class imbalance. In this way, the approaches of data augmentation and attention mechanism will support the research effectively for mitigating the effects of class imbalance. Thus, the proposed work makes use of the above implementation for extensive enhancement of the overall performance of the classification framework.
The paper utilized input mammograms adopted from three distinct benchmark datasets: CBIS-DDSM (Curated Breast Imaging Subset of DDSM) [16], INbreast [17], and MIAS (Mammographic Imaging Analysis Society) [18] datasets. The first dataset is a larger dataset (starting from
3.1 Pre-Processing of Digital Mammograms
To perform experimentation using digital mammograms for earlier breast cancer detection, the mammograms are preprocessed before classification. The impulse noise in the mammograms is removed using adaptive median filtering approaches where the noise-distorted pixels are processed without disturbing the noise-free pixels. This filtering approach is employed to remove any noise spikes effectively and smooth out the mammograms while preserving the crucial image features. Afterward, the Contrast Limited Adaptive Histogram Equalization (CLAHE) approach is used for enhancing the mammogram contrast adaptively. CLAHE adapts the conventional histogram equalization technique but limits the contrast amplification in local regions. Herein, the parameters of tileGridSize and clipLimit are fine-tuned as
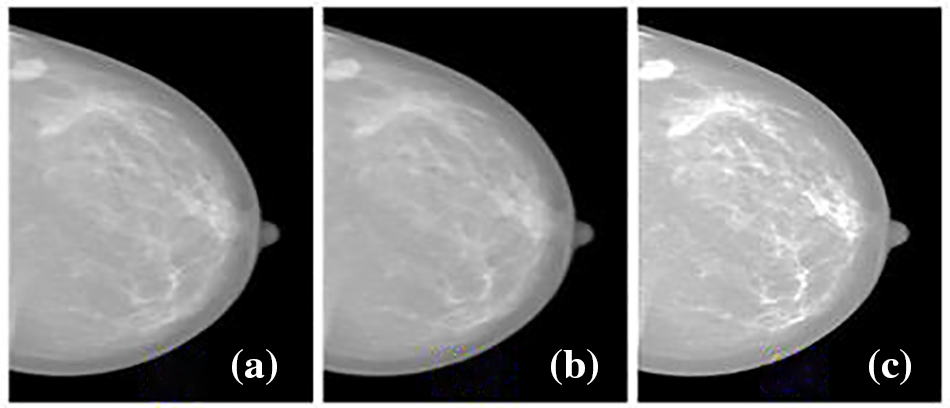
Figure 1: Mammogram preprocessing (a) original image from inbreast dataset (b) adaptive median filtered output (c) CLAHE output
3.2 Augmentation of Mammogram Inputs
The preprocessed mammograms are partitioned and stratified with a ratio of 70:30 as training and testing data together with a cross-validation partition of ten. The data partitioning before augmentation ensures that the model’s generalization ability is assessed accurately. To generate multiple amounts of mammograms with the existing images present in CBIS-DDSM, INbreast, and MIAS databases, the research makes use of augmentation. Here, the mammograms are processed with two operations involving six rotations with different degrees (45°, 90°, 135°, 180°, 234°, and 270°) and possible flipping (vertical and horizontal) of mammogram images. This facilitates the classification model to have good exposure to a more diversified range of input data. And so the generalization ability of the classification model is improved thereby the risk of overfitting will be reduced exclusively in the presence of class imbalance. The mammogram images present in the three original databases and how many mammogram images are augmented on the training data are comparatively summarized in Table 1. Here, it is revealed that the data augmentation procedure is not applied to the testing data in order to avoid data leakage and robust evaluation.
3.3 Proposed EfficientNet-B0 Integrated with Spatial Attention (ESA-XGBNet)
The research community is attempting to build several efficient CNN architectures through the balanced enhancement in width, depth, and resolution. Following this, the EfficientNet architecture provides better model performance with fewer parameters compared to conventional scaling mechanisms.
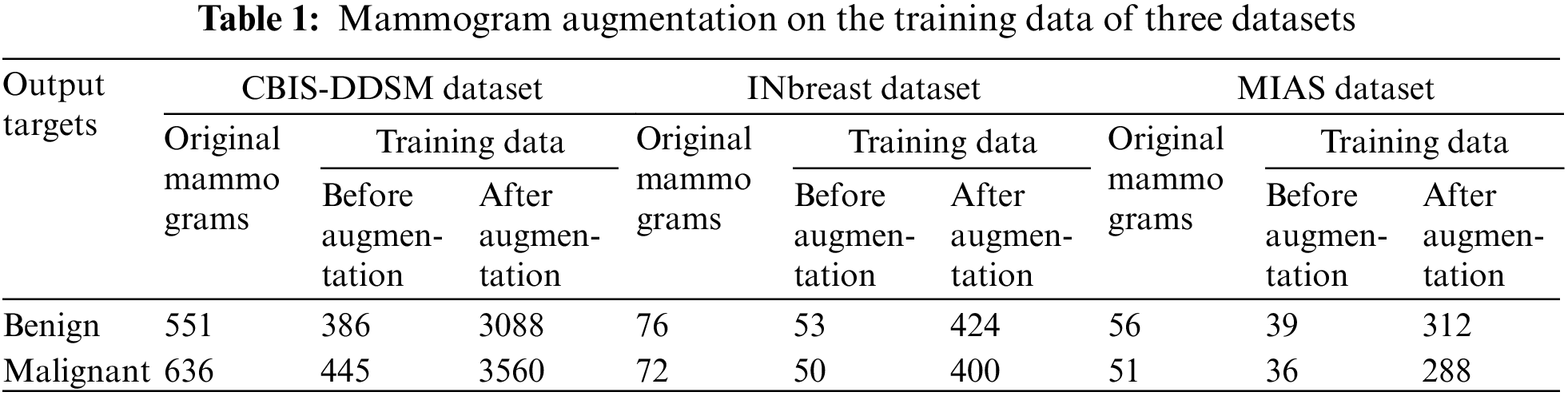
The architecture of the proposed ESA-XGBNet model is illustrated graphically in Fig. 2. The architecture of the proposed ESA-XGBNet model, depicted in Fig. 2, aims to implement EfficientNet-B0 augmented with a Spatial Attention Layer and an XGBoost model for the binary classification of mammograms, contributing to the early detection of breast tumors. The model begins with a stem convolution block responsible for initial mammogram processing, comprising convolution layers, normalization, and activation functions. Subsequent blocks consist of MBConv blocks, which are extensions of Mobile Inverted Residual Bottleneck blocks. These blocks integrate inverted residuals, linear bottlenecks, and depthwise separable convolutions, with the latter decomposing standard convolution operations into depthwise and pointwise convolutions to reduce computational costs. Additionally, the architecture employs a compound scaling approach that combines scaling factors of resolution, width, and depth in a principled manner to achieve effective and efficient model scaling.
Figure 2: Proposed ESA-XGBNet architecture for breast cancer problem
After MBConv blocks, a spatial attention layer is added to the architecture in order to take the generated feature maps from the network. After taking this, the spatial attention mechanism computes attention weights for each spatial location of the applied mammograms. For a feature map
Now, the attention weights will be obtained by passing the above global feature vector through a small dense layer (
Now, spatial attention map (
Afterward, the original feature map (
The Softmax function, as described in Eq. (2), is utilized to determine learnable weights. This process involves multiplying the original feature maps by these calculated attention weights, as outlined in Eq. (3). Consequently, highly informative regions are emphasized while less informative regions are suppressed in the preprocessed mammogram images. This operation yields attention-weighted feature maps enriched with crucial information for breast tumor classification. Following this, as depicted in Fig. 2, a Global Average Pooling (GAP) layer is introduced adjacent to the attention-weighted feature map elements. This layer is responsible for reducing spatial dimensions, generating a single value per feature map channel, thereby extracting the most relevant information for each channel. Lastly, a flatten layer is incorporated, which takes the output of GAP as inputs for the XGBoost classification model. Consequently, the attention-driven, flattened feature vectors serve as representations of the applied mammogram image inputs, enhanced through the spatial attention mechanism.
3.4 XGBoost for Tumor Classification
XGBoost (Extreme Gradient Boosting) is an ensemble learning algorithm popularly used for solving several classification tasks. The algorithm combines the predictions from multiple weak learners (decision trees) to create stronger predictive decisions. Let
In Eq. (5),
In Eq. (6), the term

4 Experimental Results and Discussion
The experimentation and its outcomes attained using the proposed ESA-XGBNet architecture evaluated with three distinct standard mammographic databases will be discussed in this section. The proposed CAD framework is evaluated using the mammograms of CBIS-DDSM, INbreast, and MIAS datasets after preprocessing. The mammographic images after CLAHE-processed is partitioned and stratified with a ratio of 70:30 as training and testing data together with a cross-validation partition of ten. The data partitioning before augmentation ensures that the model’s generalization ability is assessed accurately. Thus, the testing set should remain unchanged to accurately reflect real-world scenarios where the model encounters new, unaltered data. The adopted EfficientNet-B0 architecture takes the input mammograms of size
The comparative analysis of the proposed architecture involves experimenting with various models using the EfficientNet-B0 backbone and different classifiers. Initially, a simple EfficientNet-B0 model is used for end-to-end classification. Then, the EfficientNet-B0 model is employed for feature extraction, followed by classification using Naïve Bayes (NB), K-Nearest Neighbor (KNN), Support Vector Machines (SVM), and Decision Tree (DT) classifiers. The experimentation proceeds by integrating the EfficientNet-B0 model with the Spatial Attention (ESA) mechanism, resulting in architectures such as ESA-NB, ESA-KNN, ESA-SVM, ESA-DT, and ESA-XGBNet models. The confusion matrix demonstrating the test results obtained for ESA-XGBNet using three mammogram datasets is illustrated in Fig. 3.
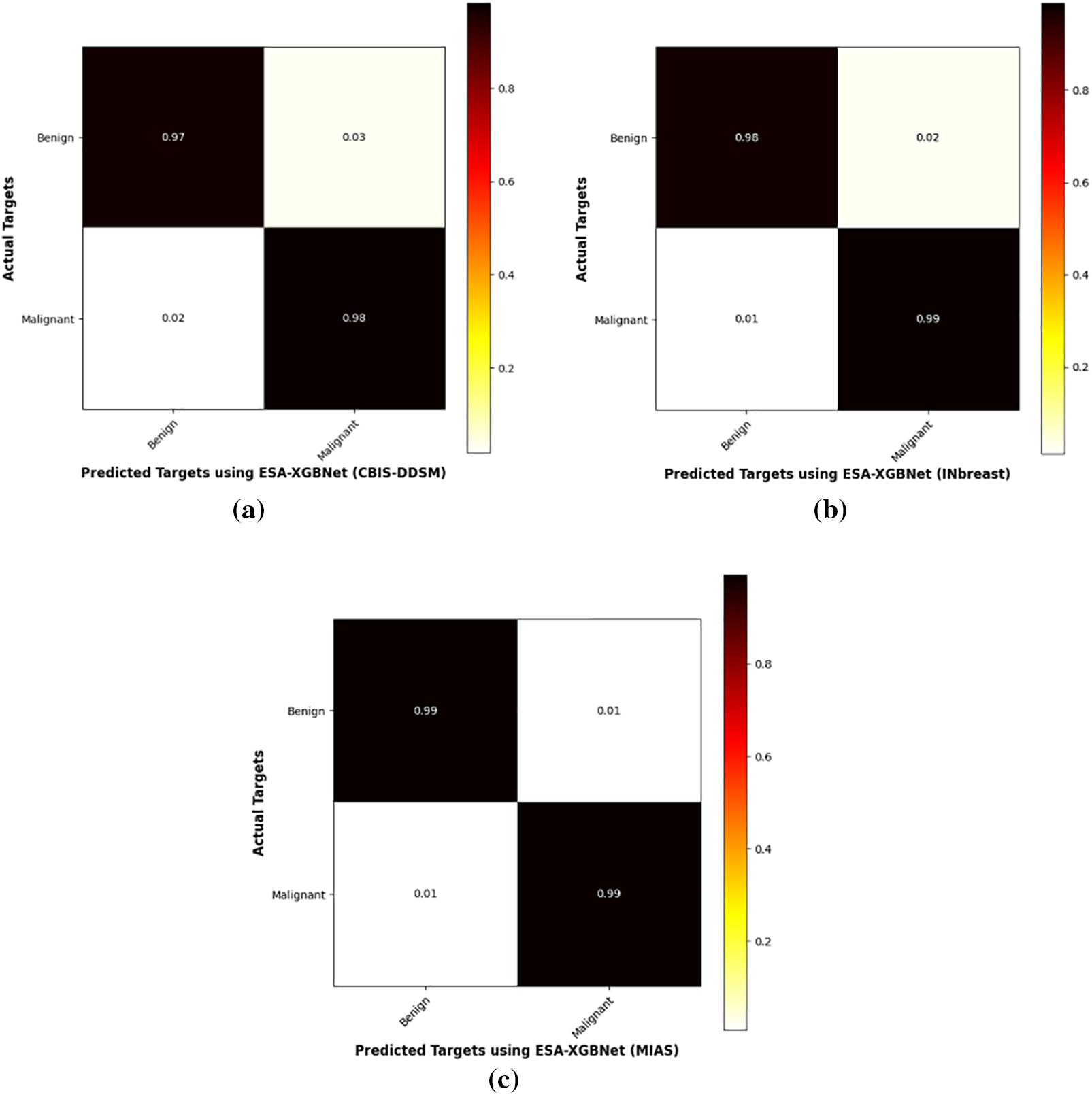
Figure 3: Confusion matrix test results obtained for ESA-XGBNet using three mammogram datasets (a) CBIS-DDSM (b) INbreast (c) MIAS datasets
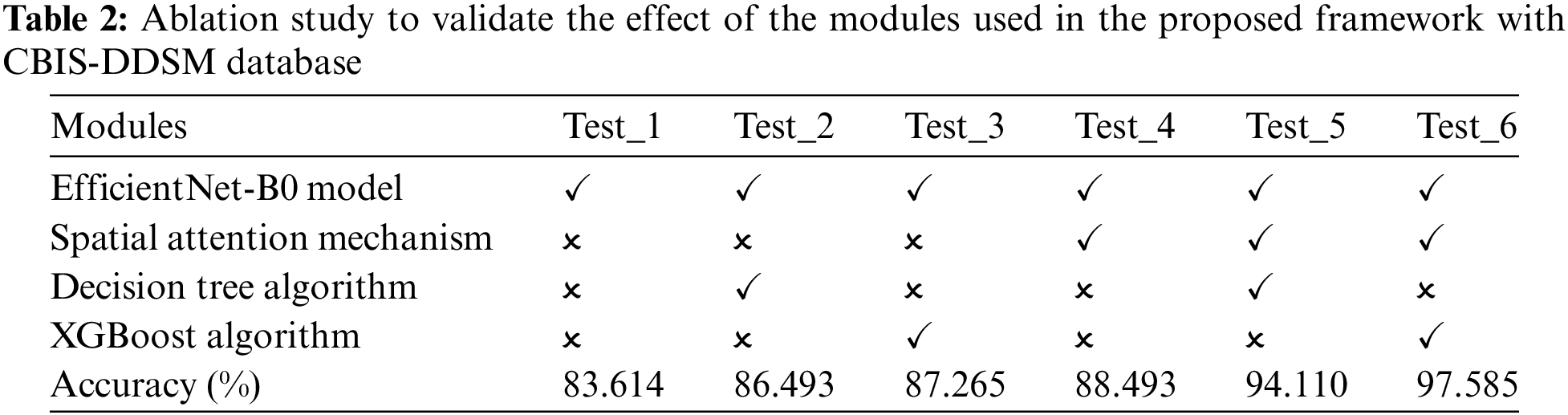
Tables 3–5 present the performance of various CNN architectures and the proposed ESA-XGBNet model applied to mammograms from CBIS-DDSM, INbreast, and MIAS databases. EfficientNet-B0 demonstrates competent classification performance, with accuracies ranging from 83.614% to 87.227% and average precision and F1 scores around 84.5% and 85.6%. Incorporating deep features from EfficientNet-B0 into different machine learning models improves classification results significantly. Support Vector Machines (SVM) yield robust accuracies ranging from 86.317% to 90.732%, with precision and F1 scores in the 87–90% range, owing to their effectiveness in capturing complex relationships among feature vectors. Decision Trees (DT) and XGBoost classifiers also demonstrate enhanced performance, with accuracies ranging from 86.493% to 93.069% for XGBoost. XGBoost particularly excels due to its ensemble learning, distributed computing, and combining multiple weak-learner predictions, resulting in improved sensitivity, specificity, precision, and F1 score compared to other models.
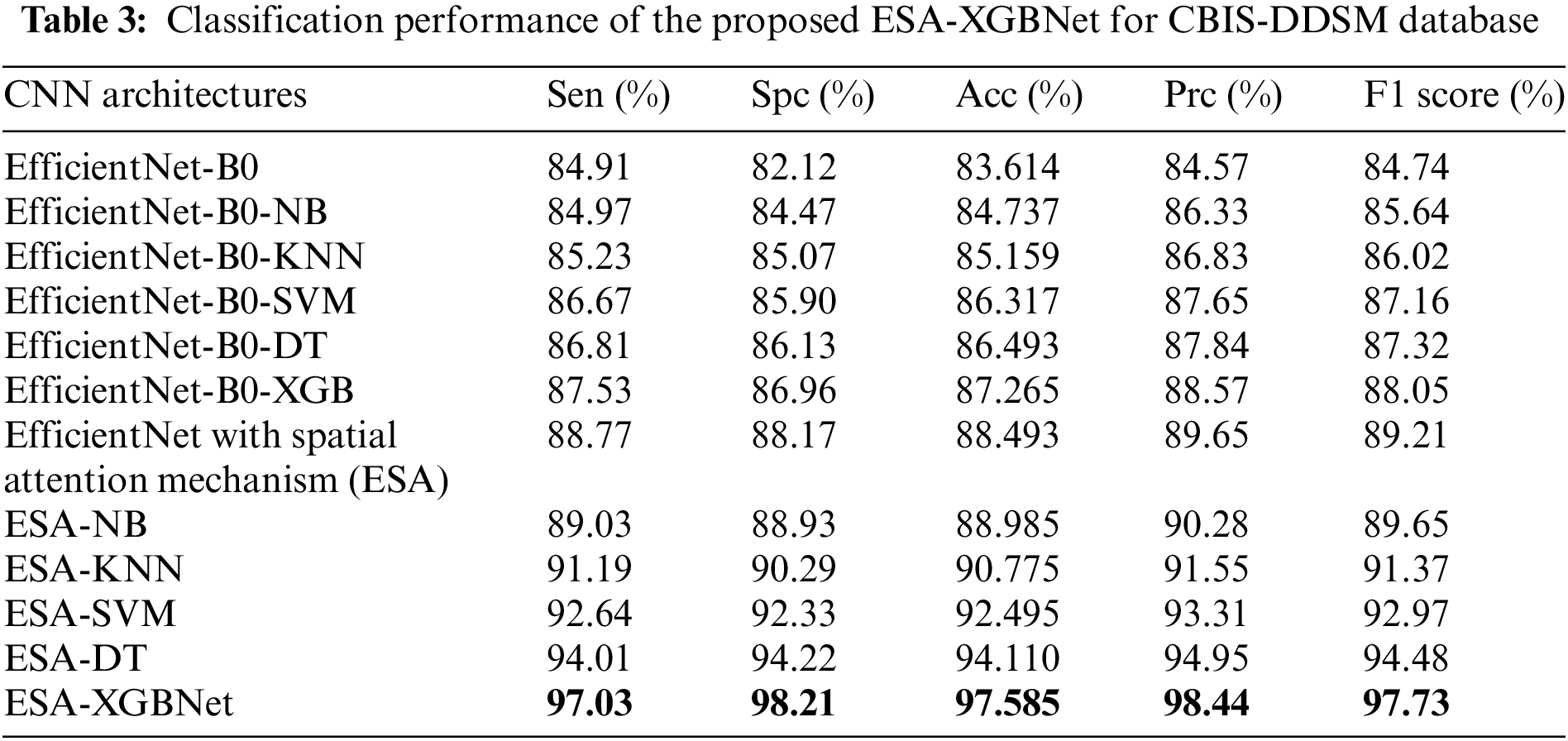
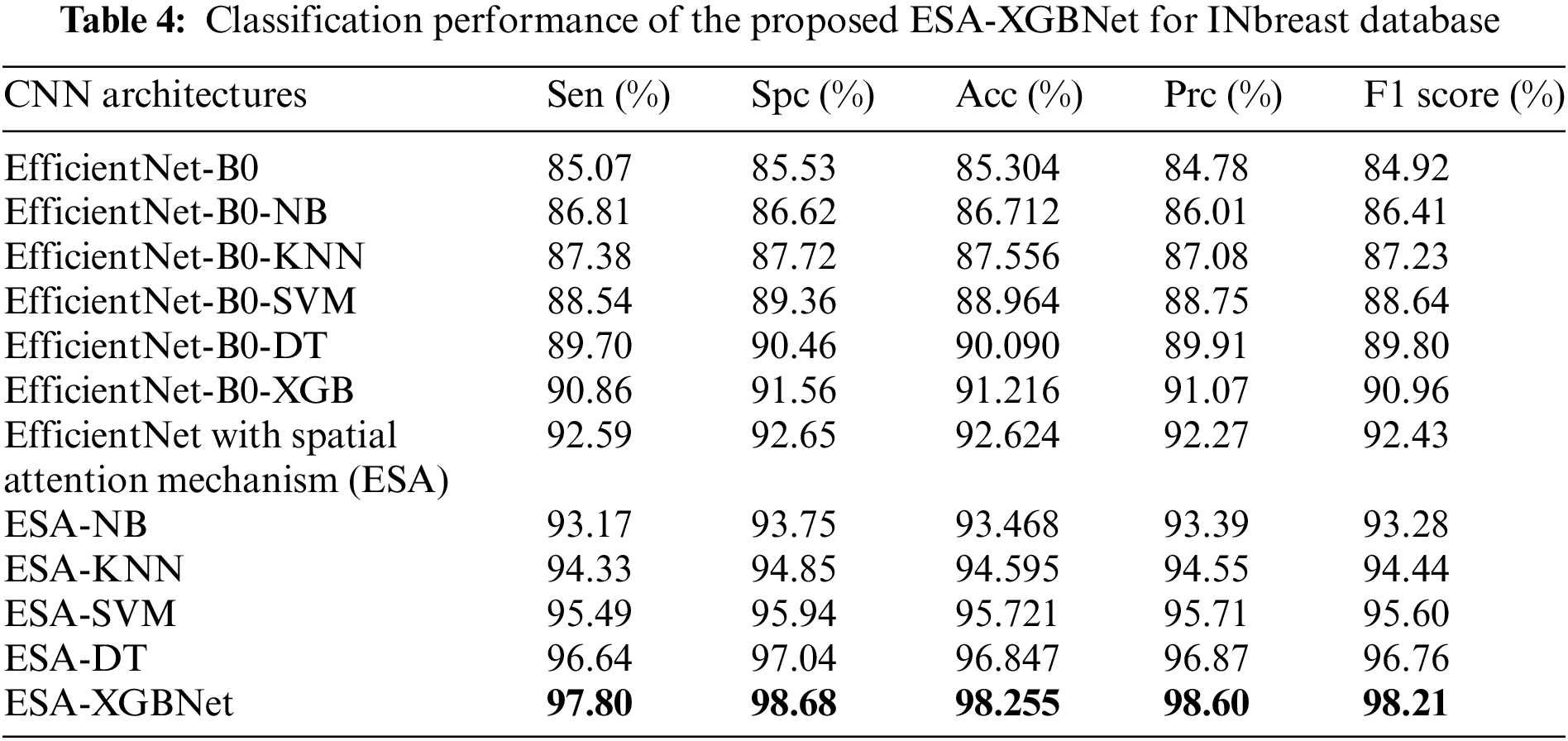
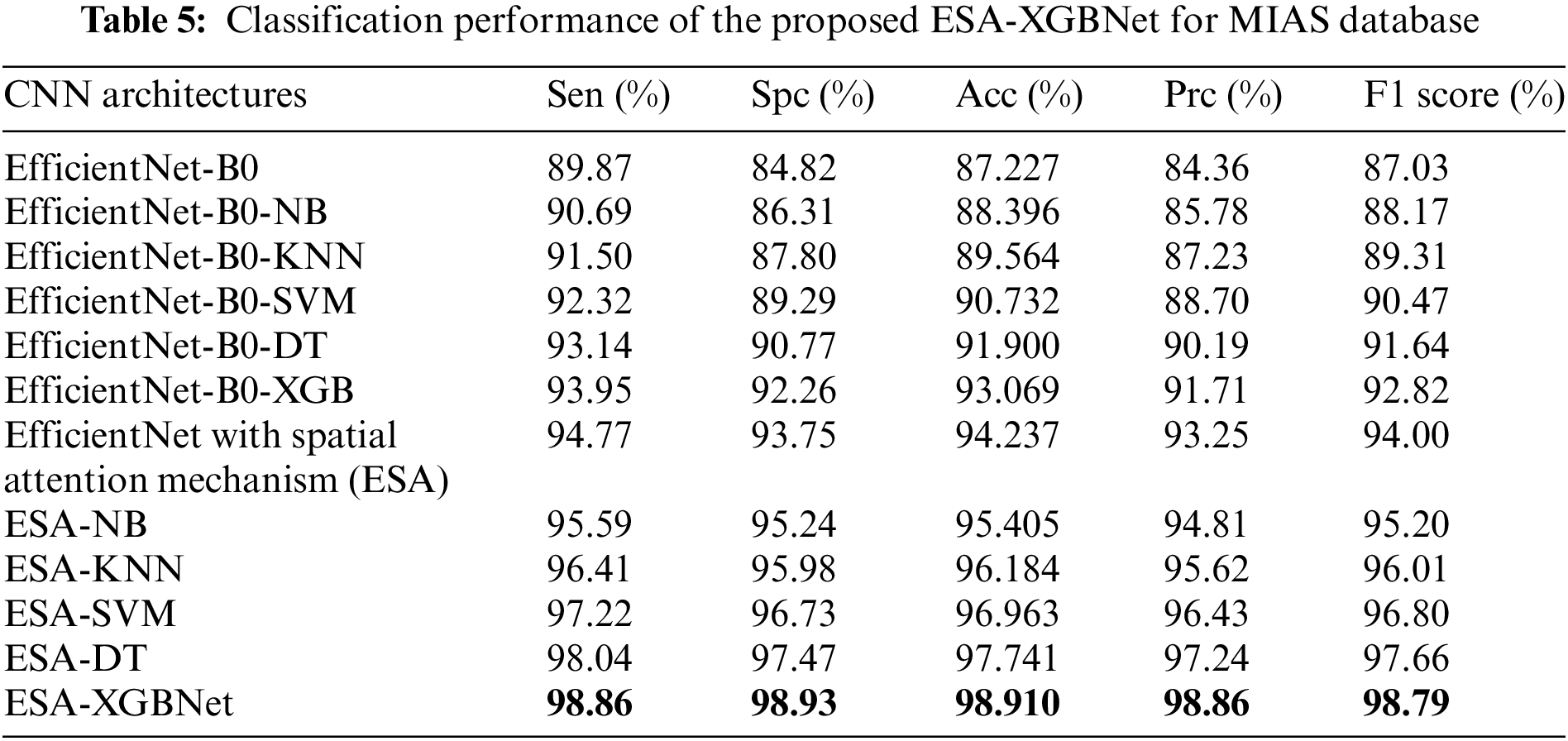
As illustrated in Fig. 2, the further direction of research is the inclusion of a spatial attention mechanism to the aforementioned experimental setup to enhance the breast tumor classification. Thus, the performance of the EfficientNet-B0 with Spatial Attention Mechanism (ESA) model is found to be more competent than the standalone architecture as given in Tables 3–5. In this way, the classification accuracies of 88.493% (CBIS-DDSM), 92.624% (INbreast), and 94.237% (MIAS) are obtained for breast tumor classification. The supreme classification performance of accuracies −97.585% (CBIS-DDSM), 98.255% (INbreast), and 98.91% (MIAS), precision scores −98.44% (CBIS-DDSM), 98.60% (INbreast), and 98.86% (MIAS), and F1 scores of 97.73% (CBIS-DDSM), 98.21% (INbreast), and 98.79% (MIAS), respectively. All the above-attained results are further validated using the Statistical Kappa (
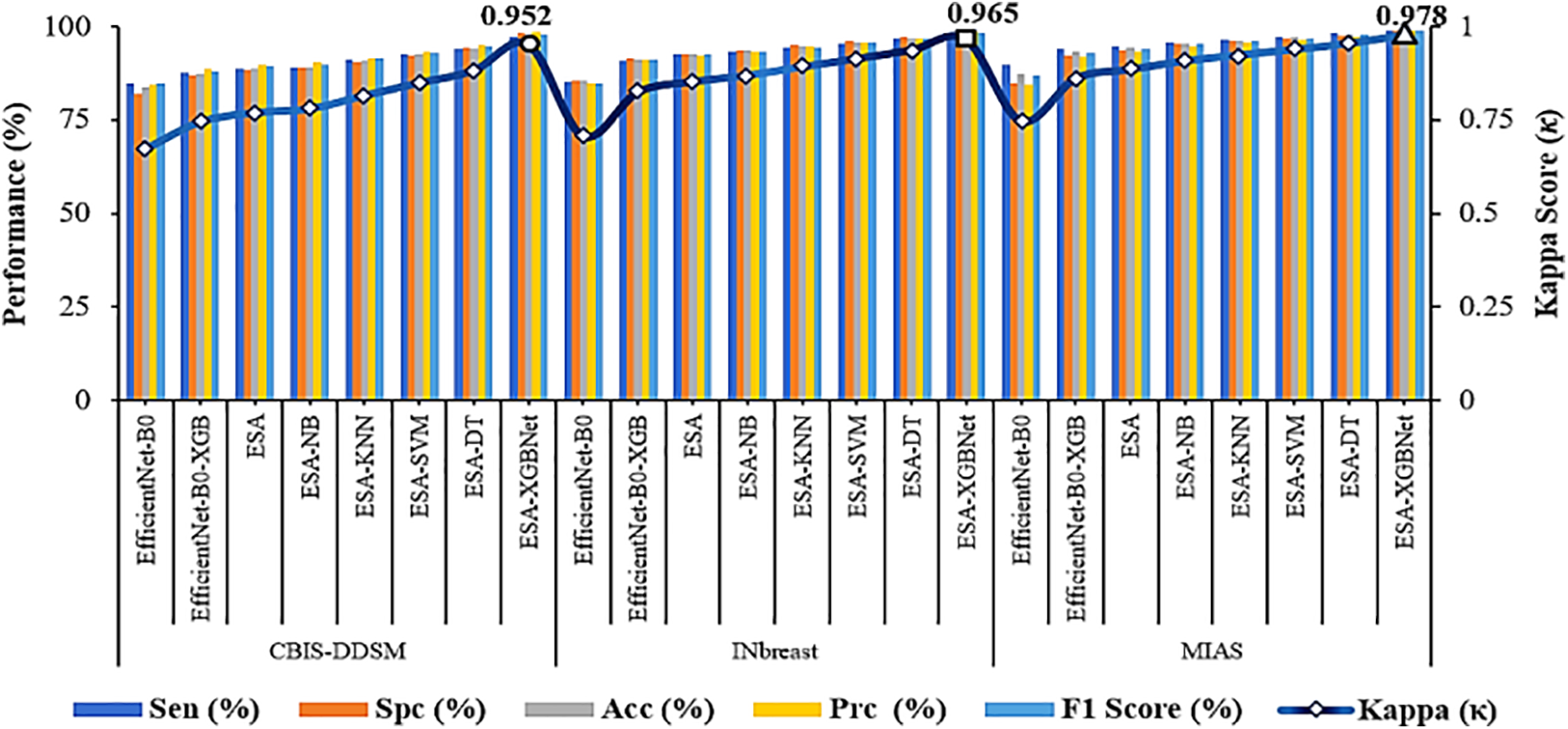
Figure 4: Performance comparison and validation of attained results–ESA-XGBNet
Even though a classification model discriminates well in identifying abnormalities, it provides a trade-off between sensitivity and specificity values Fig. 5 illustrates a plot between True Positive Rate (TPR) and False Positive Rate (FPR) for the employed classification architectures applied with three datasets. Thus, the plot of Fig. 5 illustrates how well a classification model discriminated between benign and malignant cases. Accordingly, a robust classification model will have a curve that rises steeply toward the top-right corner of the plot. In Fig. 5, the markers in each performance curve refer to the twelve employed CNN models as listed in the order of Tables 3–5 for three datasets. Accordingly, the performance curve of the proposed ESA-XGBNet steps towards the top-right for all the employed databases.
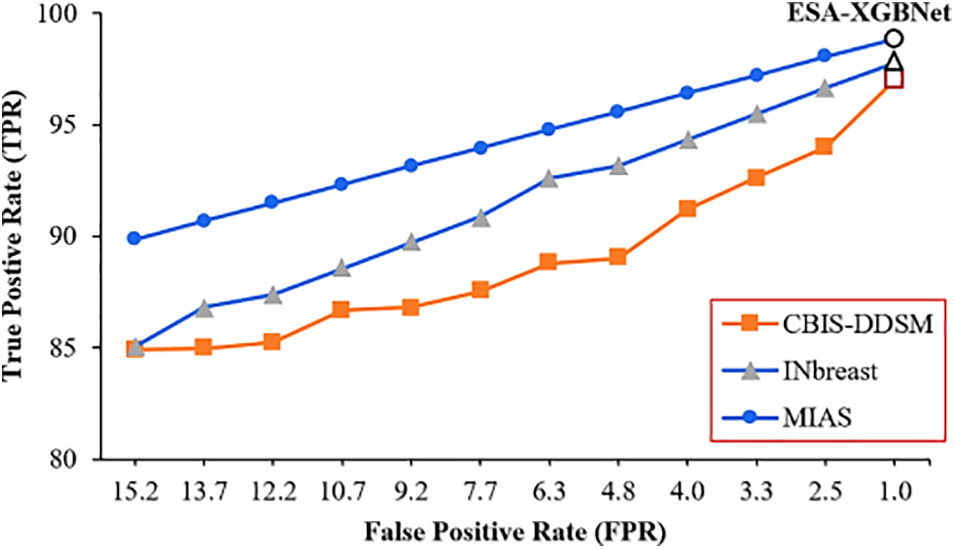
Figure 5: TPR vs. FPR plot for the proposed models applied for three datasets
Grad-CAM is a common procedure to visualize where a convolutional neural network architecture is looking for target prediction Sample mammogram (INbreast and CBIS-DDSM) visualization are given in Fig. 6 which interprets where our proposed model focused for its target prediction. Here, sample benign and malignant labeled mammograms (Fig. 6a1,a2) from the INbreast and CBIS-DDSM databases are taken. The superimposed visualizations (Fig. 6b1,b2) and their corresponding heat maps (Fig. 6c1,c2) are generated with the correct prediction labels of Benign and Malignant.
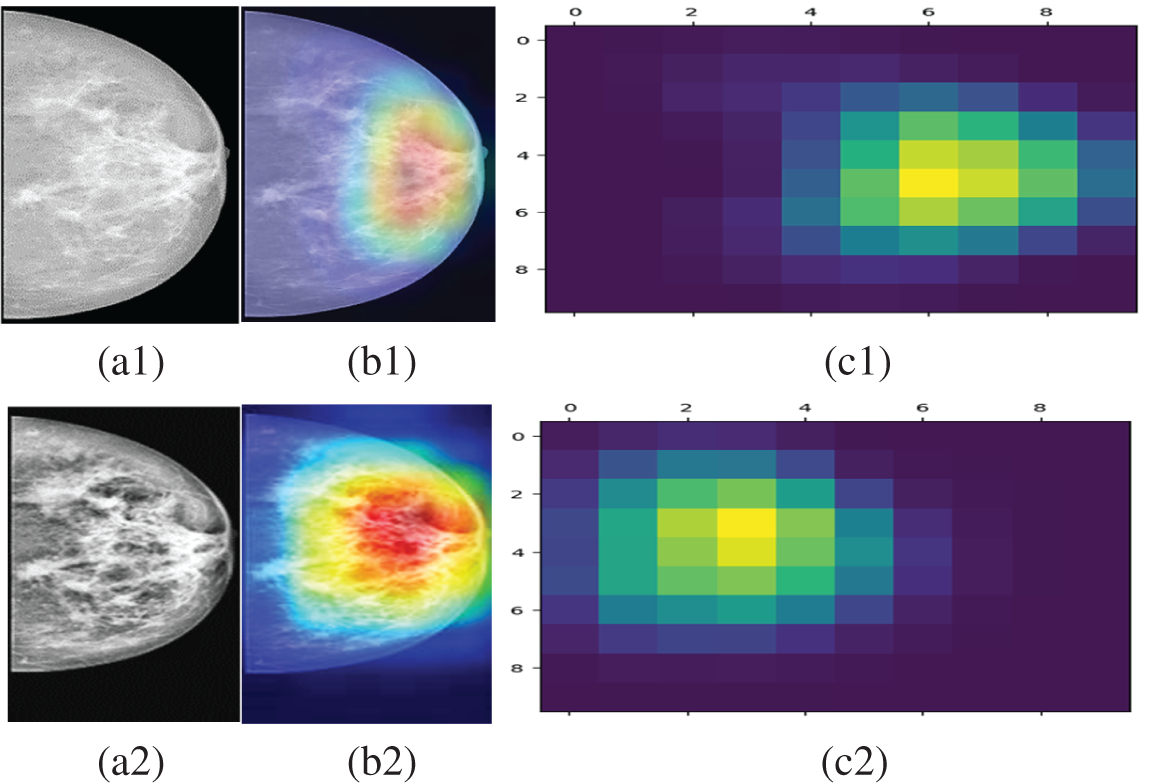
Figure 6: (a1) Preprocessed mammogram (INbreast) with a label of Benign (b1) respective superimposed visualization (c1) attention guided Grad-CAM heat maps with the correct prediction of benign label (a2) preprocessed mammogram (CBIS-DDSM) with a label of malignant (b2) respective superimposed visualization (c2) attention guided grad-CAM heat maps with the correct prediction of malignant label
As a final point of research, the comparison of existing and recent works against the proposed ESA-XGBNet architecture is summarized in Table 6. Herein, the research works were applied with diversified and different mammogram databases. And the robust classification performance of breast tumors is obtained apparently using the proposed CAD framework as depicted in Fig. 2. However, the work has the potential limitation of added complexity due to the inclusion of a spatial attention layer with EfficientNet-B0 architecture and this will be taken care in our future extension.
5 Conclusion and Future Direction
The research intended to propose a robust early-detection CAD framework for a worldwide societal disease, breast cancer. The input considers diversified mammogram images adopted from three distinct datasets: CBIS-DDSM, INbreast, and MIAS databases. The methodology involves substantial experimentation beginning with the choice of dataset and ending with a detailed performance analysis. Initially, the research preprocessed the input mammograms for their better representation in balancing contrast and exposure. After noise removal using an adaptive median filter and mammogram enhancement using the Contrast Limited Adaptive Histogram Equalization approach, the preprocessed mammogram images are then fed into the EfficientNet-B0 architecture. Here, several experiments have been conducted with the intention of obtaining robust performance for breast tumor classification. In addition to this, five machine learning models namely Naïve Bayes, K-Nearest Neighbor, Support Vector Machines, Decision Trees, and XGBoost algorithms are adopted exclusively for the classification stage. With the aim of improving the classification performance, a spatial attention layer is included in the EfficienNet-Bo model and so the weighted attention deep feature vectors are derived for ML-based classification. In this way, the robust classification performance of 97.585% (CBIS-DDSM), 98.255% (INbreast), and 98.910% (MIAS) accuracy scores are obtained using the proposed ESA-XGBNet architecture as compared with the existing models. Furthermore, the decision-making of the proposed ESA-XGBNet architecture is interpreted and validated using the kappa metric and Attention Guided GradCAM-based Explainable AI technique. The future direction of the proposed architecture will include the segmentation of tumors and utilizing breast ultrasound (BUS) clinical imageries with U-Net-based models to improve the robustness of breast cancer diagnosis. The preprocessing steps that we employed in our study have necessitated the exclusion of images that did not meet the required standards such as image quality, and annotations. In addition, demanding high computational requirements of training deep learning models, the study opted for a subset of the dataset that allowed us to conduct thorough experiments within a reasonable timeframe and resource allocation. This subset was sufficient to demonstrate the effectiveness of our proposed model. Furthermore, while the initial number of images was 1187 (CBIS-DDSM dataset), data augmentation significantly increased the effective size of our training set. This augmentation provided the model with diverse examples, enhancing its generalization capabilities without compromising the integrity of the test set. However, an extensive experimentation will be carried out using all the images of CBIS-DDSM dataset. And investigating the complexity of the proposed work with different attention mechanisms applied for distinct medical problems.
Acknowledgement: This research is supported by Princess Nourah bint Abdulrahman University Researchers Supporting Project Number (PNURSP2024R432), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Funding Statement: This research is supported by Princess Nourah bint Abdulrahman University Researchers Supporting Project, Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Sannasi Chakravarthy and Bharanidharan Nagarajan; data collection: Surbhi Bhatia Khan and Ahlam Al Musharraf; methodology: Surbhi Bhatia Khan and Ahlam Al Musharraf; analysis and interpretation of results: Vinoth Kumar Venkatesan and Khursheed Aurungzeb; draft manuscript preparation: Mahesh Thyluru Ramakrishna. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data that support the findings of this study are openly available at: https://www.cancerimagingarchive.net/collection/cbis-ddsm/, http://peipa.essex.ac.uk/info/mias.html, https://paperswithcode.com/dataset/inbreast (accessed on 18 June 2024).
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. F. Ye et al., “Advancements in clinical aspects of targeted therapy and immunotherapy in breast cancer,” Mol. Cancer, vol. 22, no. 1, 2023, Art. no. 105. doi: 10.1186/s12943-023-01805-y. [Google Scholar] [PubMed] [CrossRef]
2. M. Arnold et al., “Current and future burden of breast cancer: Global statistics for 2020 and 2040,” The Breast, vol. 66, pp. 15–23, 2022. [Google Scholar] [PubMed]
3. L. Wilkinson and T. Gathani, “Understanding breast cancer as a global health concern,” Br. J. Radiol., vol. 95, no. 1130, 2022, Art. no. 20211033. doi: 10.1259/bjr.20211033. [Google Scholar] [PubMed] [CrossRef]
4. H. Hussein et al., “Supplemental breast cancer screening in women with dense breasts and negative mammography: A systematic review and meta-analysis,” Radiology, vol. 306, no. 3, 2023, Art. no. e221785. [Google Scholar]
5. S. R. Sannasi Chakravarthy and H. Rajaguru, “SKMAT-U-Net architecture for breast mass segmentation,” Int. J. Imaging Syst. Technol., vol. 32, no. 6, pp. 1880–1888, 2022. doi: 10.1002/ima.22781. [Google Scholar] [CrossRef]
6. S. R. Sannasi Chakravarthy and H. Rajaguru, “A systematic review on screening, examining and classification of breast cancer,” in 2021 Smart Technol., Commun. Robot. (STCR), Sathyamangalam, India, 2021, pp. 1–4. doi: 10.1109/STCR51658.2021.9588828. [Google Scholar] [CrossRef]
7. A. J. A. D. Freitas et al., “Liquid biopsy as a tool for the diagnosis, treatment, and monitoring of breast cancer,” Int. J. Mol. Sci., vol. 23, no. 17, 2022, Art. no. 9952. doi: 10.3390/ijms23179952. [Google Scholar] [PubMed] [CrossRef]
8. M. Al Moteri, T. R. Mahesh, A. Thakur, V. Vinoth Kumar, S. B. Khan and M. Alojail, “Enhancing accessibility for improved diagnosis with modified EfficientNetV2-S and cyclic learning rate strategy in women with disabilities and breast cancer,” Front. Med., vol. 11, Mar. 2024. doi: 10.3389/fmed.2024.1373244. [Google Scholar] [PubMed] [CrossRef]
9. Z. Jiang et al., “Identification of novel cuproptosis-related lncRNA signatures to predict the prognosis and immune microenvironment of breast cancer patients,” Front. Oncol., vol. 12, 2022. doi: 10.3389/fonc.2022.988680. [Google Scholar] [PubMed] [CrossRef]
10. R. L. Siegel, K. D. Miller, N. S. Wagle, and A. Jemal, “Cancer statistics, 2023,” CA Cancer J. Clin., vol. 73, no. 1, pp. 17–48, 2023. doi: 10.3322/caac.21763. [Google Scholar] [PubMed] [CrossRef]
11. S. Arooj et al., “Breast cancer detection and classification empowered with transfer learning,” Front. Public Health, vol. 10, 2022, Art. no. 924432. doi: 10.3389/fpubh.2022.924432. [Google Scholar] [PubMed] [CrossRef]
12. J. J. Patel and S. K. Hadia, “An enhancement of mammogram images for breast cancer classification using artificial neural networks,” IAES Int. J. Artif. Intell., vol. 10, no. 2, 2021, Art. no. 332. doi: 10.11591/ijai.v10.i2.pp332-345. [Google Scholar] [CrossRef]
13. L. K. Kumari and B. N. Jagadesh, “A robust feature extraction technique for breast cancer detection using digital mammograms based on advanced GLCM approach,” EAI Endorsed Trans. Pervasive Health Technol., vol. 8, no. 30, 2022, Art. no. e3. doi: 10.4108/eai.11-1-2022.172813. [Google Scholar] [CrossRef]
14. S. Thawkar, “Feature selection and classification in mammography using hybrid crow search algorithm with Harris hawks optimization,” Biocybernet. Biomed. Eng., vol. 42, no. 4, pp. 1094–1111, 2022. doi: 10.1016/j.bbe.2022.09.001. [Google Scholar] [CrossRef]
15. P. Rajpurkar et al., “CheXNet: Radiologist-level pneumonia detection on chest X-rays with deep learning,” 2017, arXiv:1711.05225. [Google Scholar]
16. Lee et al., “A curated mammography data set for use in computer-aided detection and diagnosis research,” Sci. Data, vol. 4, no. 1, pp. 1–9, 2017. [Google Scholar]
17. I. C. Moreira, I. Amaral, I. Domingues, A. Cardoso, M. J. Cardoso and J. S. Cardoso, “Inbreast: Toward a full-field digital mammographic database,” Acad. Radiol., vol. 19, no. 2, pp. 236–248, 2012. doi: 10.1016/j.acra.2011.09.014. [Google Scholar] [PubMed] [CrossRef]
18. J. Suckling, J. Parker, D. Dance, S. Astley, and I. Hutt, “Mammographic image analysis society (MIAS) database v1. 21, (2015),” Accessed: Mar. 28, 2021. [Online]. Available: https://www.repository.cam.ac.uk/handle/1810/250394 [Google Scholar]
19. L. Falconí, M. Pérez, W. Aguilar, and A. Conci, “Transfer learning and fine tuning in mammogram bi-rads classification,” in 2020 IEEE 33rd Int. Symp. on Comput. Based Med. Syst. (CBMS), Rochester, MN, USA, 2020, pp. 475–480. doi: 10.1109/CBMS49503.2020.00096. [Google Scholar] [CrossRef]
20. D. A. Ragab, M. Sharkas, S. Marshall, and J. Ren, “Breast cancer detection using deep convolutional neural networks and support vector machines,” Bioinformatics Genom., vol. 7, no. 5, 2019, Art. no. e6201. doi: 10.7717/peerj.6201. [Google Scholar] [PubMed] [CrossRef]
21. A. Baccouche, B. Garcia-Zapirain, and A. S. Elmaghraby, “An integrated framework for breast mass classification and diagnosis using stacked ensemble of residual neural networks,” Sci. Rep., vol. 12, no. 1, 2022, Art. no. 12259. doi: 10.1038/s41598-022-15632-6. [Google Scholar] [PubMed] [CrossRef]
22. D. Muduli, R. Dash, and B. Majhi, “Automated diagnosis of breast cancer using multi-modal datasets: A deep convolution neural network based approach,” Biomed. Signal Process. Control, vol. 71, 2022, Art. no. 102825. doi: 10.1016/j.bspc.2021.102825. [Google Scholar] [CrossRef]
23. B. Surendiran, P. Ramanathan, and A. Vadivel, “Effect of BIRADS shape descriptors on breast cancer analysis,” Int. J. Med. Eng. Inform., vol. 7, no. 1, pp. 65–79, 2015. doi: 10.1504/IJMEI.2015.066244. [Google Scholar] [CrossRef]
24. H. Li, J. Niu, D. Li, and C. Zhang, “Classification of breast mass in two-view mammograms via deep learning,” IET Image Process., vol. 15, no. 2, pp. 454–467, 2021. doi: 10.1049/ipr2.12035. [Google Scholar] [CrossRef]
25. C. Yang, D. Sheng, B. Yang, W. Zheng, and C. Liu, “A dual-domain diffusion model for sparse-view CT reconstruction,” IEEE Signal Process. Lett., vol. 31, pp. 1279–1283, 2024. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF
 Downloads
Downloads
 Citation Tools
Citation Tools