 Open Access
Open Access
ARTICLE
Dynamic Multi-Layer Perceptron for Fetal Health Classification Using Cardiotocography Data
1 Department of Computer Science and Engineering, Prasad V Potluri Siddhartha Institute of Technology, Vijayawada, 520007,
India
2 Amrita School of Computing, Amrita Vishwa Vidyapeetham, Amaravati, Andhra Pradesh, 522503, India
3 Department of Tele informatics Engineering, Federal University of Ceará, Fortaleza, 60455-970, Brazil
4 Department of Computer Science and Engineering, Koneru Lakshmaiah Education Foundation, Vaddeswaram, Guntur, Andhra Pradesh, 522302, India
5 Department of Computer Engineering, Chosun University, Gwangju, 61452, Republic of Korea
6 School of CSIT, Symbiosis Skills and Professional University, Pune, 412101, India
7 Department of Computer Engineering and Information, College of Engineering in Wadi Alddawasir, Prince Sattam bin Abdulaziz University, Wadi Alddawasir, 11991, Saudi Arabia
8 School of IT and Engineering, Melbourne Institute of Technology, Melbourne, 3000, Australia
* Corresponding Authors: Parvathaneni Naga Srinivasu. Email: ; Muhammad Fazal Ijaz. Email:
(This article belongs to the Special Issue: Deep Learning and IoT for Smart Healthcare)
Computers, Materials & Continua 2024, 80(2), 2301-2330. https://doi.org/10.32604/cmc.2024.053132
Received 25 April 2024; Accepted 02 July 2024; Issue published 15 August 2024
Abstract
Fetal health care is vital in ensuring the health of pregnant women and the fetus. Regular check-ups need to be taken by the mother to determine the status of the fetus’ growth and identify any potential problems. To know the status of the fetus, doctors monitor blood reports, Ultrasounds, cardiotocography (CTG) data, etc. Still, in this research, we have considered CTG data, which provides information on heart rate and uterine contractions during pregnancy. Several researchers have proposed various methods for classifying the status of fetus growth. Manual processing of CTG data is time-consuming and unreliable. So, automated tools should be used to classify fetal health. This study proposes a novel neural network-based architecture, the Dynamic Multi-Layer Perceptron model, evaluated from a single layer to several layers to classify fetal health. Various strategies were applied, including pre-processing data using techniques like Balancing, Scaling, Normalization hyperparameter tuning, batch normalization, early stopping, etc., to enhance the model’s performance. A comparative analysis of the proposed method is done against the traditional machine learning models to showcase its accuracy (97%). An ablation study without any pre-processing techniques is also illustrated. This study easily provides valuable interpretations for healthcare professionals in the decision-making process.Keywords
Nomenclature
| Acronym | Abbreviation |
| ML/DL | Machine Learning/Deep Learning |
| RF | Random Forest |
| XGB | XGBoost |
| DT | Decision Tree |
| LR | Logistic Regression |
| KNN | K-Nearest Neighborhood |
| DMLP | Dynamic Multi-Layer Perceptron Model |
| CTG | Cardiotocography Data |
Fetal health plays an important role in child’s future and healthy pregnancy. Fetal health is monitored by doctors based on the mother’s health history, physical examinations, blood tests, screening tests, and cardiotocography, which tracks heart rate and contractions to assess risks [1,2]. Regular check-ups need to be taken by the mother to know the status of the fetus’s growth and to know any potential problems. The frequency of check-ups varies from person to person based on individual health factors like high-risk pregnancies and the growth rate of the fetus. Healthy pregnancy lays the foundation for the timely development of organs and reduces the risk of chronic diseases in individuals [3,4].
AI is used in various sectors, one of which is health care. Machine learning and deep learning algorithms can be used to analyze medical records like blood test reports, ultrasound data, CTG data, etc., to detect complications in pregnant women early. AI is also useful for categorizing pregnant women (fetal health) based on their risk factors and suggesting further plans [5–7]. To process CTG data, is time-consuming and error-prone. So, AI helps analyze CTG data and provides a faster, more detailed report than human interpretation. However, AI plays a vital role in empowering healthcare professionals in the healthcare sector. Healthcare professionals with their expertise and AI-based reports, CTG data deliver optimal care for pregnant women and fetuses.
Classification of fetal health is important to ensure accurate, effective prenatal care and early detection of potential complications. Traditional methods may sometime prone to errors and inconsistencies so we need to use emerging tools such as machine and deep learning techniques. Manual processing of CTG data is time-consuming and unreliable. The available tools are not enough for fetal health classification. To enhance the diagnostic accuracy, we can use advanced neural network architectures. So, to achieve better results, we propose a Dynamic Multi-Layer Perceptron model (DMLP). We apply various pre-processing techniques, balance the dataset, scale the dataset, and extract the most important features from the CTG data of the fetus. The processed dataset is supplied as input from the traditional ML methods and the DMLP model.
Ensuring accurate and timely classification of fetal health is crucial for effective prenatal care, enabling early detection and intervention for potential complications. Traditional assessment methods, which often depend on subjective evaluation by healthcare professionals, can be prone to inconsistencies and errors. Machine Learning (ML) classifiers have emerged as promising tools to enhance diagnostic accuracy and objectivity in medical assessments. However, to push the boundaries of classification performance, it is essential to explore advanced neural network architectures. This study proposes the use of a Deep Multi-Layer Perceptron (DMLP) model for fetal health classification. By harnessing the sophisticated learning capabilities of the DMLP, we aim to improve the precision and reliability of fetal health diagnostics. Furthermore, we conduct a comprehensive comparison of the DMLP model against traditional ML classifiers to evaluate its effectiveness and identify its advantages and limitations. This research aspires to contribute significantly to the field of prenatal care by providing insights into the application of advanced neural networks, ultimately aiding in the development of more accurate and dependable diagnostic tools.
A summary of the main points of this study is as follows:
• The study presents a DMLP neural network for fetal health prediction. The process begins with a single neuron and continues through several levels, with the concealed layers and neurons being adjusted dynamically. Early stopping, batch normalization, and dropout are some of the optimization strategies that this strategy uses to improve performance deliberately.
• The study also examined classic ML methods that combined resampling, feature scaling, and feature selection; these modifications greatly enhanced the model’s performance.
• In the domain of fetal health categorization, we conducted a thorough investigation comparing the Dynamic MLP model with many classifiers, including Random Forest (RF), XGBoost (XGB), Decision Tree (DT), K-Nearest Neighborhood, and Logistic Regression (LR). To aid clinical specialists in their analysis and interpretation, we looked into the relevance of aspects related to each model.
A brief outline of the following sections, presented in chronological order, is as follows: Section 2 describes the literature review, Section 3 describes the datasets and procedures that will be utilized, and our recommended methodology. The results of the model’s execution are described in Section 4. In the last section, we will review the main points of our analysis and discuss potential avenues for further study.
Fetal health classification is important for the early detection of potential risk factors, which can be analyzed during regular check-ups. During pregnancy, the doctors observe fetal movements, heart rate, and growth using various approaches, such as blood tests and ultrasounds. When doctors identify any potential risk factors in pregnant women, timely care should be taken to avoid any miscarriage. AI-based image analysis suggests that healthcare professionals can easily make decisions. Researchers have proposed various machine and deep learning models to determine the status of the fetus. Manual processing of CTG data can sometimes be prone to errors and takes a lot of time. These models analyze the CTG data and suggest the health care professionals in decision-making. Alam et al. [8] used ML models to analyze CTG data for fetal health classification and achieved an accuracy of 97.51% using a random forest algorithm. Rahmayanti et al. [9], Noor et al. [10] have used seven algorithms ANN, LSTM, XGB, LGBM, RF, KNN, and SVM for fetal health classification using CTG data.
Akbulut et al. [11] have gathered data from 96 pregnant women through various questions and evaluations by healthcare professionals. They predicted fetal anomalies using multiple classification algorithms like DT, NN, and SVM. Abiyev et al. [12] proposed a Type2 Fuzzy Neural Network that helps in the decision-making of the fetus state in fetal health classification. Kasim [13] proposed a multi-class fetal classification using an ELM algorithm to analyze CTG data. Mandala [14] proposed LGBM for fetal health classification and achieved an accuracy of 98.31%. Shruthi et al. [15], Hoodbhoy et al. [16] focussed on regular check-ups of pregnant women; otherwise, risk factors cannot be assessed for the baby’s fetal health. Imran Molla et al. [17], Islam et al. [18] used the CTG dataset for fetal health assessment, particularly oxygen deficiency in fetuses, using a random forest algorithm, and they achieved an accuracy of 94.8%.
The current classification approaches for fetal health categorization encounter several substantial obstacles that limit their precision. A major challenge is the quality and variety of the input data in fetal health monitoring. This is because the data comes from several sources, such as ultrasound pictures, heart rate monitors, and biochemical indicators, which may differ in terms of accuracy and reliability. Furthermore, these models sometimes encounter difficulties when dealing with unbalanced datasets, in which cases of certain foetal health issues are much less common than others. This may result in predictions that are biased and lack the capacity to detect uncommon but crucial disorders accurately. Moreover, current models may not sufficiently include the ever-changing and complex nature of fetal health, in which several parameters that would associate with one other affect the final classification decision. To tackle these issues, it is necessary to create classification models that are robust to deal with imbalanced dataset, and can deal with the inconsistency during the classification.
The proposed DMLP model is proven to be efficient in dealing with complex and high dimensional data though efficient feature engineering process. DMLPs have the ability to capture complex patterns and dependencies in the data by applying multiple layers of non-linear transformations.
The manuscript will find all the required details regarding the dataset and the pre-processing methods used to improve the prediction model’s accuracy. Fig. 1 shows the workflow of the proposed approach for fetal health classification.

Figure 1: Workflow of the proposed fetal health classification model
The fetal classification dataset [19] is a publicly available dataset obtained from Kaggle with 22 columns & 2126 records extracted from cardiotocography examinations. Features such as “uterine contractions,” “light decelerations,” “prolonged decelerations,” “abnormal short-term variability,” and many more are included in the dataset. The dataset contains 3 different classes—“normal,” “suspect,” & “pathological”. The records reflect thorough assessments of various physiological parameters obtained from the cardiotocogram examinations. During prenatal therapy, it is possible to predict the fetal health state based on the assessment of these traits, which serve as the foundation for classification. The dataset’s attributes are summarized in Fig. 2, including information about the data type and possible values for each attribute.
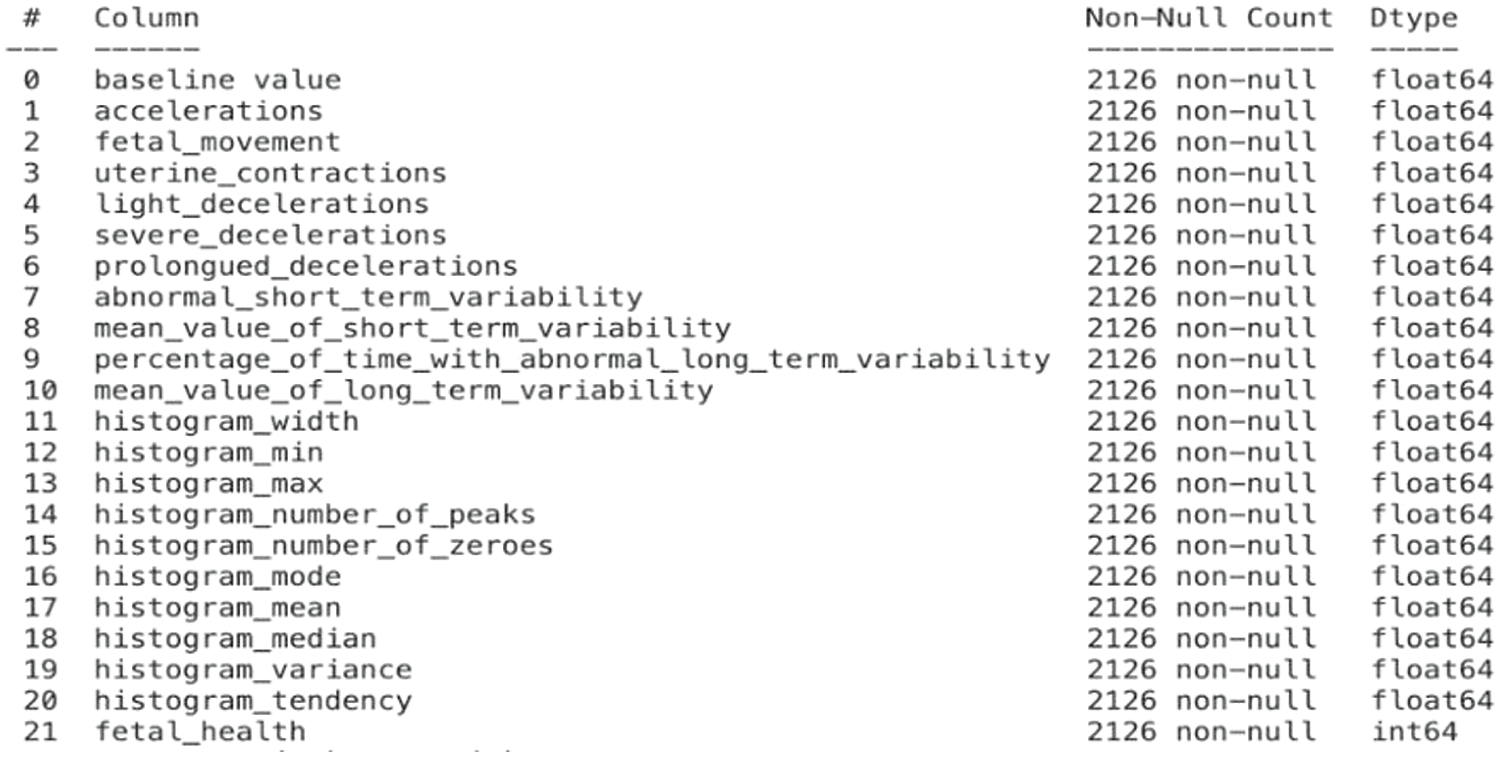
Figure 2: Features in the fetal dataset and their corresponding datatypes
A complete understanding of the distribution and uncertainty of the dataset is achieved via the study, which comprises predicting probability density and computing variances across several data points. Looking at the probability density and variances may give you a better idea of the underlying probabilistic features and how they change across different data types. In Fig. 3, a grid of Kernel Density Estimation (KDE) plots for each feature in the dataset depicts the expected probability density and variances across several ‘fetal_health’ classes (Blue-normal, orange-suspect, green-pathological). The differences in feature distributions among health classes in the color-coded charts. The exploratory data analysis (EDA) tool indispensable for this project is these KDE charts, which help with feature distribution insights, data pre-processing, feature selection, and model interpretation.

Figure 3: Graph depicting attributes in the fetal health classification dataset
Improving data quality, and reducing noise to improve the efficacy of the classification models are all responsibilities of data pre-processing. Important tasks in this phase include feature selection, cleaning, missing values management, scaling, categorical variable encoding & handling data anomalies [20]. The efficacy of data pre-processing determines the success or failure of training & evaluating models.
Using the seaborn package and matplotlib, Fig. 4 generates a heatmap to display the dataset’s correlation matrix. By looking at the size and direction of the correlations among the divergent variables in the dataset, this visualization is a great tool for understanding their relationships.

Figure 4: Heatmap of all attributes in the fetal dataset
In data analysis, dealing with outliers as shown in Fig. 5 is essential for ensuring reliable statistical models. Effective handling of outliers is achieved by using techniques including trimming, capping, transformation, and imputation, which strengthen statistical studies and improve the quality of models. We can use the following equations for capping data points (p) below the lower cap/upper cap:
p* = LC (replace p with LC), p* = UC (replace p with UC).

Figure 5: An illustration of the outliers in the dataset
3.3.2 Addressing Class Imbalance
Fig. 6 is a bar graph showing the distribution of various classes before/after SMOTE (Synthetic Minority Oversampling Technique) was applied in the fetal classification dataset. The x-axis represents the class label, which is fetal health. There are three classes: normal (represented by the number 1), suspect (represented by the number 2), and pathological (represented by the number 3). The y-axis represents the number of data points in each class. From Fig. 6 we can observe the distribution of classes is imbalanced. There are significantly more data points in the class labeled “normal” (176) than in the classes labeled “suspect” (295) and “pathological” (0). This imbalance can be a problem for machine learning algorithms, as they may favor the majority class and perform poorly on the minority classes. SMOTE [21] is an oversampling technique that can be used to address class imbalance. SMOTE works by creating synthetic data points for the minority class. These synthetic data points are created by interpolating between existing data points in the minority class. After SMOTE is applied, the distribution of classes would be more balanced. This can help to improve the performance of machine learning and deep learning algorithms on imbalanced datasets.

Figure 6: Dataset comparison pre and post SMOTE
One of the most important parts of machine learning is feature selection [22], which involves finding the right features to use for making predictions. Statistical tests such as the chi-square test, ANOVA, F-test, or mutual information score were applied. These tests determine how well each attribute correlates with the dependent variable. We choose the top
where, Oj: Observed frequency for category j; Ej: Expected frequency for category j.

Figure 7: The graphs represent the feature importance in decreasing order of their significance
To determine if ANOVA compares their means there is a statistically significant difference between more than two groups, ANOVA compared in Eq. (2).
F represents F-statistic, Mean Squares Between (MSB), and Mean Squares Within (MSW).
The mutual dependence between two variables is illustrated in Eq. (3).
G(B) is the entropy of variable B, and G(B/A) is the conditional entropy of B given A.
Because machine learning models use feature values as numerical inputs without comprehending their inherent importance, data scaling is an essential procedure in this field. Scaling is essential for making data interpretable and treating features fairly. Scaling data can be done in two main ways:
• Normalization: Features that do not have a normal (Gaussian) distribution can be handled using this method. The features are normalized when their values are adjusted to fall within a certain range and is expressed in Eq. (4).
• Standardization: Features with a normal distribution but values that vary greatly from one another are used for standardization. It normalizes the characteristics such that they all have a mean of zero and a standard deviation of one and is illustrated in Eq. (5).
Demonstration of dataset attributes post-application of SMOTE, and scaling techniques is illustrated in Fig. 8. The feature distribution determines whether the specified dataset should be normalized or standardized. For example, normalization is used when a feature’s distribution is abnormal. Standardization becomes necessary when characteristics have values that vary widely but otherwise follow a normal distribution. Notably, tree-based techniques, such as XGB and RF, may not necessitate scaling because they are less affected by feature size. Normalization is useful for some algorithms, such as LR and DMLP. The dataset is normalized using the min-max scaling strategy to increase the performance of logistic regression and DMLP algorithms.


Figure 8: Demonstration of dataset attributes post-application of SMOTE and scaling techniques
The current section of the manuscript presents the Dynamic Multi-Layer Perceptron with the other conventional classification techniques in classifying fetal health from the Cardiotocography Data. The conventional classification techniques include the RF, XGB, LR, KNN, and DT. All these methods are discussed alongside the proposed model.
By building a network of decision trees, the ensemble learning algorithm, Random Forest is useful for fetal health classification. A voting method is used during prediction to aggregate individual tree outputs and improve overall model performance after these decision trees are trained on random subsets of the data. Bootstrapping and feature selection are made more random by the algorithm at each split, which helps to improve generalization and decrease overfitting. RF evaluates feature relevance based on the decrease in impurity achieved by each feature across all trees, whereas the Gini impurity measure directs the development of decision trees. It can detect important features for reliable predictions using this approach. Complex fetal health classification tasks are well-suited to Random Forest due to its ensemble nature and feature interpretability. First, we can write the Gini impurity for an FHD in Eq. (6).
The notation
The ensemble learning algorithm XGBoost is essential for fetal health prediction because it combines regularization with gradient boosting. A loss term assessing prediction accuracy and a regularization term prohibiting overfitting make up the objective function that XGB minimizes through the sequential construction of decision trees. Computing gradients & Hessians, which stand for derivatives of the loss function, is an integral part of the optimization process. We distribute weights to trees according to their contributions, and the structure of each tree is decided by minimizing the objective function. Regularization terms control model complexity. XGB is resilient against overfitting & suitable for varied datasets; it is particularly effective for fetal health classification due to its adaptability to complex interactions, automatic management of missing values, and feature importance score. In multiclass classification, the objective function for XGB comprises a regularization term and the aggregate of the individual loss functions for each class [23]. Class probabilities are typically calculated using the softmax algorithm. Expressing the global objective function using Eq. (7) is possible.
where, n is the total instances. K is the total classes.
The predicted probability for class k is calculated using the softmax function Eq. (8).
In the above equation, the notation
A binary classification algorithm, logistic regression can be enhanced to manage jobs involving multiple classes. Regarding fetal health classification, LR models the likelihood that an instance belongs to a specific class. It is usual practice to convert the linear combination of input features into probabilities using the logistic function, often known as the sigmoid function. The softmax function allows logistic regression to be extended for multiclass classification. After calculating the probability of each class, the one with the highest probability is used to make a prediction. Solving this Eq. (9) gives the class
where,
• K is the number of classes.
• β0k, β1k,…, βnk are the coefficients associated with class k out of the K possible classes.
Training the LR model entails adjusting the coefficients while maximizing the likelihood of the observed data. The model uses the computed probabilities to forecast the class, and these coefficients define the decision boundary.
One flexible and non-parametric approach useful in fetal health classification is K-Nearest Neighbours. When applied to fetal health, KNN works on the premise that instances with comparable features should have comparable class labels. To determine a data point’s classification, the algorithm looks at the labels of its K-Nearest Neighbors, where
Here,
Using decision trees, which are robust and easily interpretable models, allows for the classification of fetal health in circumstances involving many classes. Using feature values as a basis, recursively partitioning the feature space into pieces. Each partition in a decision tree represents a node, and the last partitions, known as leaf nodes, serve as labels for the classes [25]. As part of the decision-making process, the feature space is partitioned at each node based on a given feature and a splitting criterion, such as Gini impurity or entropy. Maximizing the homogeneity of class labels inside each partition is the goal of the splitting criterion, which aims to produce as pure of divisions as feasible. With each branch in the decision tree, minimizing the impurity measure identified by the notation
where,
4.6 Dynamic Multi-Layer Perceptron Network
Predicting the fetal health status from various input features is the domain’s specialty, and the Dynamic Multi-Layer Perceptron is the customizable neural network design used for this study. Layers that reflect relevant features obtained from diagnostic tests, layers that analyze inputs using weighted connections and activation functions, and final layers that generate predictions on fetal health issues are all part of a Multi-Layer Perceptron (MLP). The DMLP design’s adaptability allows for customizing the total hidden layers and neurons according to the classification task’s complexity. Upgrading the MLP model from a one-neuron to seven neurons in a multi-feedforward layered model is the logical next step for fetal health classification. With three neurons per class to seven hidden layers, we intend to train and assess networks with diverse topologies. In the bottom hidden layer, there are 24 neurons; subsequent layers increase this amount. To avoid overfitting, use the Early Stopping call-back and choose your model and parameters based on their performance on the validation set for accuracy. The use of dropout and batch normalization significantly enhances generalizability. Also, an F1-score can be used to monitor how well the model is doing [26]. This comprehensive strategy is implemented to improve deep learning models for accurate fetal health categorization. This comprehensive strategy is being implemented to decrease concerns about overfitting. The calculation determines the weighted total for each class
where,
•
•
•
•
The softmax activation function is applied to the logits to obtain class probabilities, as illustrated in Eq. (13).
where,
It is possible to represent the objective function of a MLP network, frequently utilized for supervised learning tasks, as the reduction of a cost or loss function [27]. In the case of regression tasks, the objective function
where,
The weights and biases are updated using the gradient descent update rule. The gradient for the weights
where,
The complexity of the problem and the data determine the optimal number of hidden layers and neurons to increase the performance of DMLP. The following are a few possible benefits:
• Increasing the amount of hidden layers and neurons improves the model’s capacity to interpret complex patterns and correlations in the data. This might improve performance, especially for fetal datasets with complicated structures or non-linear relationships.
• The network’s ability to extract complicated features from input data is enhanced when more neurons are in each hidden layer.
• Hidden layers can be added to the neural network for deep learning designs.
• There is a direct correlation between the amount of model parameters and the likelihood of overfitting the training data. We delve into deeper architectures utilizing suitable regularization techniques, like dropout or weight decay.
In this section, we will examine and compare different classifiers that are used to classify fetal health. A comprehensive evaluation of these models utilized important measures like F1-score, recall, precision, and accuracy. We also looked at how scaling and other pre-processing methods affected the efficiency of these classifiers. At the same time, we explored the complexities of deep learning with the Dynamic Multi-Layer Perceptron Network, going from one hidden layer to seven.
5.1 Hyperparameters Used for Training Each Model
Machine learning model performance is greatly influenced by hyperparameters, which is why they are so important. The training procedure and the final model are affected by these external configurations, which are not learned from data. Reaching peak model performance, warding off problems like overfitting and underfitting, and improving generalization to novel, unknown data depend on correct hyperparameter tuning. Examples of common hyperparameters are parameters about the model’s architecture, regularization, and learning rate. Machine learning weakening entails experimenting with different values to discover the optimal ones. The model parameters used in the current study are shown in Table 1.
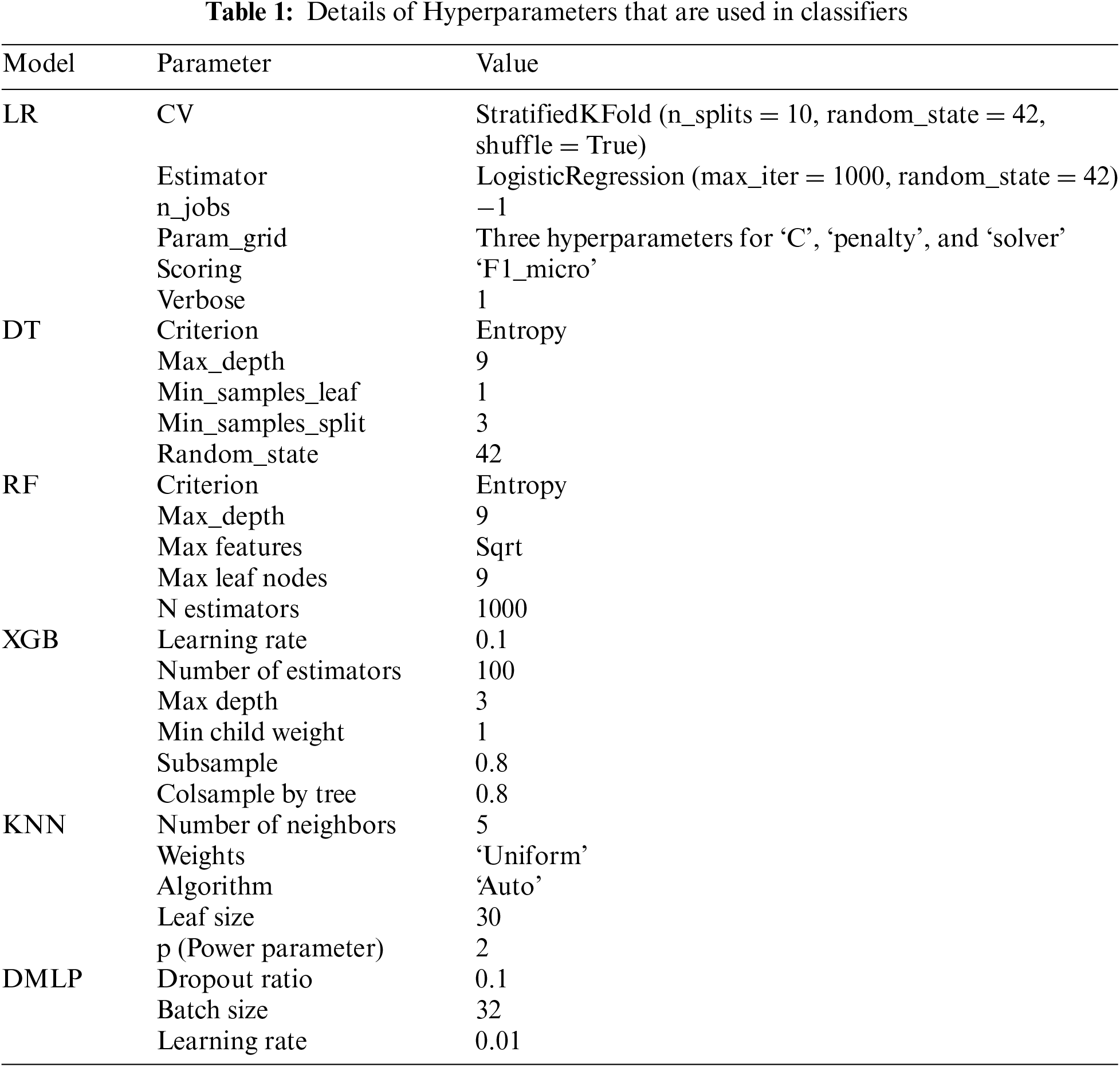
5.2 Performances of Various Classification Techniques
Various evaluation indicators were plotted across different models and setups to provide a comprehensive perspective of model performance. As a result, the study evaluated each method for fetal health classification and drew comparisons between them. In addition, we analyzed the significance of traits to learn how they contributed to classification. By providing a comprehensive assessment of the performance and interpretability of both conventional classifications, the current study seeks to shed light on their efficacy for fetal health classification. A confusion matrix is a way to summarize a machine learning model’s performance in fetal health categorization. It does this by comparing the model’s predictions with the actual outcomes. The confusion matrix appears when evaluating the results of classifications across many classes, which often happens in fetal health categorization.
Usually, there are four entries in the confusion matrix:
• First, there are cases where the model accurately predicts a positive class, such as the presence of a specific fetal health issue, these cases are called True Positives.
• In cases where the model accurately predicts a negative class—for example, the absence of a specific fetal health condition is assumed as True Negative.
• The third mistake is when the model inaccurately predicts a positive class when none exists and is assumed as a False Positive.
• When the model makes an inaccurate prediction of a negative class when one exists, it is assumed as False Negative.
Fig. 9 shows how the confusion matrix may be used to obtain several metrics for evaluating the model’s performance, including recall, accuracy, precision, and F1-score. These indicators provide insights into the model’s accuracy in identifying fetal health issues and areas for improvement.

Figure 9: Confusion matrices generated by our classifiers. (a) XGBoost (b) random forest (c) KNN (d) decision tree (e) logistic regression
In fetal health categorization, feature importance describes how each feature or variable in a dataset affects the model’s prediction ability. It helps to determine which features are crucial for the model to accurately classify cases of fetal health [28]. Understanding which traits are most crucial for model interpretation is essential, as it exposes which factors play a pivotal role in determining the outcome. Analyzing how much weight the model gave to each input feature is what feature importance analysis is all about in the context of fetal health categorization. During training, popular algorithms like DT, RF, and XGB make it easy to determine which features are most important, are illustrated in Figs. 10 and 11. Bar graphs that show feature dependency highlight which features most influence a model’s predictions by displaying their importance scores. These graphs help identify key features, simplify the model, and improve performance by focusing on the most impactful data [29].
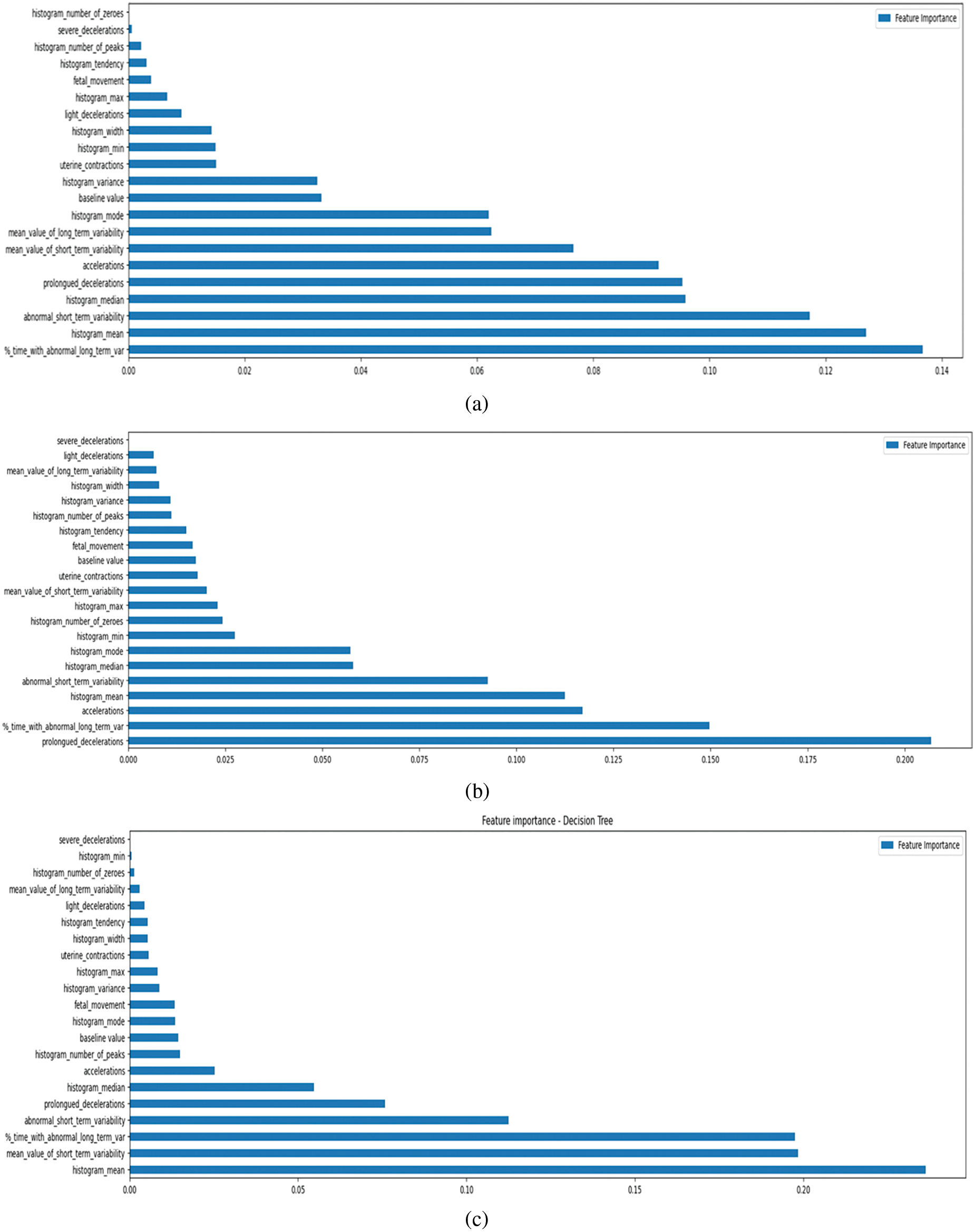

Figure 10: Importance of features as determined by our classifiers. (a) Random forest (b) XGBoost (c) decision tree (d) logistic regression (e) KNN


Figure 11: Importance of features as determined by our classifiers concerning different classes. (a) Random forest (b) XGBoost (c) decision tree (d) logistic regression (e) KNN
A feature’s impact on the accuracy of predictions is proportional to its significance score. When making decisions, the model gives more weight to features considered more important and less weight to features considered less important. Academics and practitioners can better understand the most important elements impacting fetal health outcomes through feature-importance visualizations or analyses. Improving the categorization model’s interpretability, directing more studies, and prioritizing characteristics for data gathering or enhancement can all be achieved with this information.
5.4 Assessment of the Performance of Various ML Classifiers Concerning SMOTE and Scaling
In the current study we have used the LOOCV based cross validation alongside the K-Fold validating the proposed model. A comparative analysis is also promised, encompassing three scenarios: the algorithm without any pre-processing, the algorithm with scaling, and the algorithm with the data balancing technique SMOTE. The final scenario involves applying both data balancing and scaling simultaneously. This comprehensive comparison of accuracy in Table 2 aims to provide insights into the impact of different pre-processing techniques on the algorithm’s performance, shedding light on the effectiveness of balancing and scaling individually and in combination. Analysis of various classifiers for fetal health classification is illustrated in Table 3 and Fig. 12.
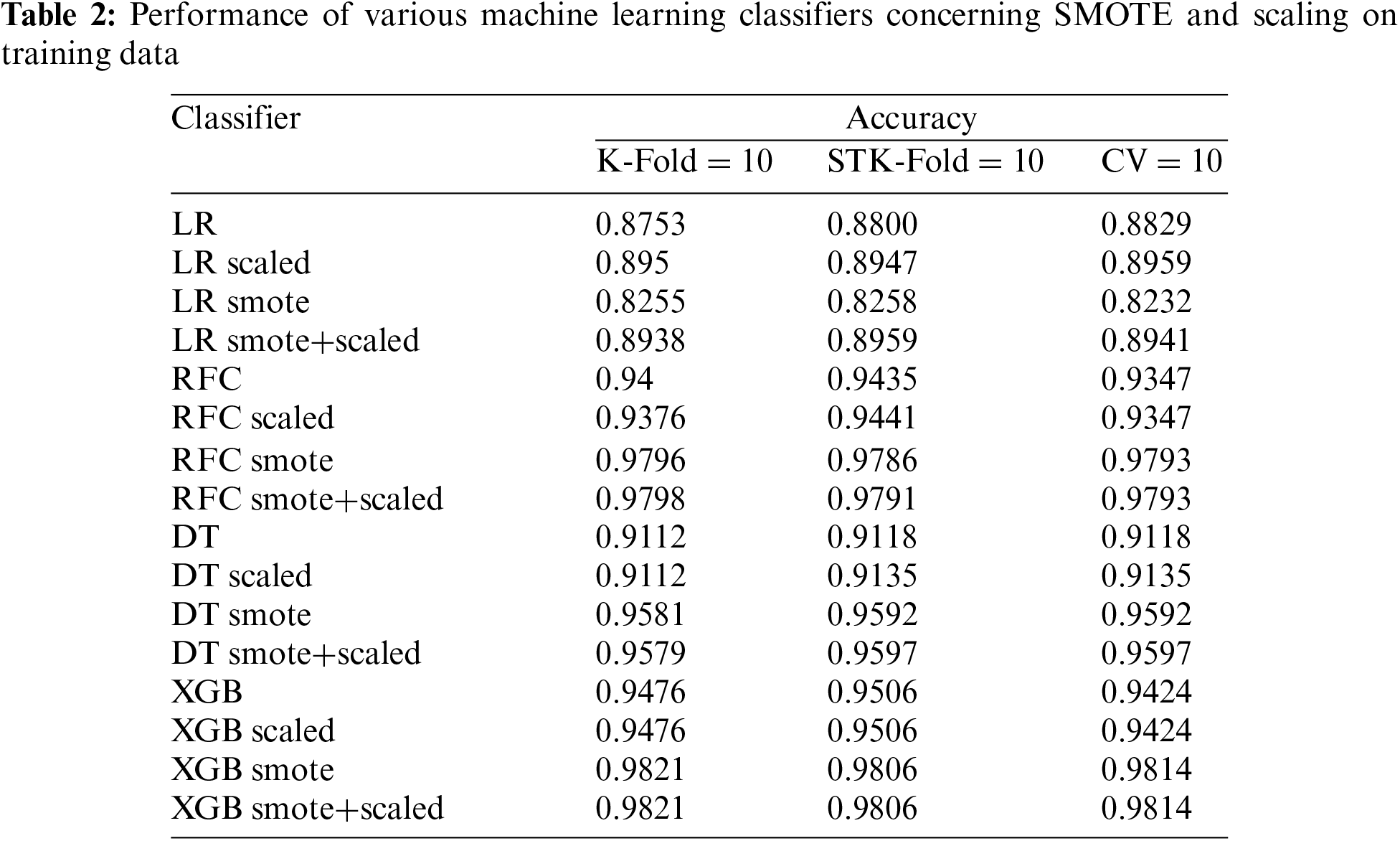
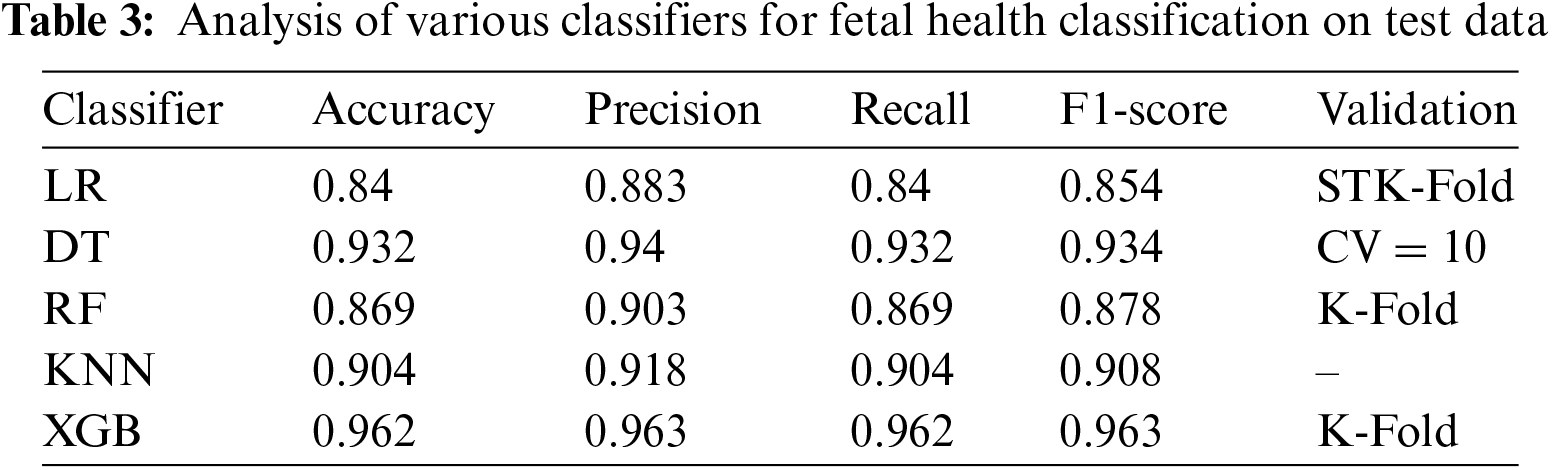


Figure 12: Comparative analysis among different classifiers
Training and validation set performance metrics are the primary emphasis of the DMLP model, which aims to analyze the connection between the model’s capacity as indicated by its layers and neurons. The dataset has been partitioned into 80-20 for training and testing. The validation set used for fine-tuning the model is the integral part of the training sample, allotted 10% of the training size. According to graphical representations, a more noticeable disparity between training and validation measures is associated with enhanced model capacity, which may indicate overperformance. With an astounding F1-score of 91.33% on the validation set and 89.95% on the testing set, the model with five hidden layers and 51,000 parameters beats others.
Nevertheless, a more robust hyperparameter search is needed because worries regarding the significance of the results arise due to the lack of statistical tests and cross-validation. Future studies should utilize comprehensive cross-validation procedures and statistical testing to reliably choose models, as larger models show lower metrics, which could indicate overfitting or randomness. Neural network analysis with emphasis on layers and neurons is illustrated in Table 4, and training accuracy, validation accuracy, training loss, and validation losses are illustrated in Fig. 13.




Figure 13: Neural network analysis with emphasis on layers and neurons
As the number of hidden layers grows, there is a general trend towards improved accuracy, recall, precision, and F1-score across training, validation, and test sets. With 6 hidden layers, the model performs at its peak, with a training accuracy of 98.90%, a validation accuracy of 94.82%, and a test accuracy of 92.96%. The performance indicators appear to reach a plateau or decline at 7 hidden layers, suggesting that overfitting is possible as the model complexity increases.
It can be observed that DMLP model had attained 97% accuracy when utilizing pre-processing techniques, whereas without pre-processing, the model attained 96% accuracy. Removing the resampling, feature scaling, and feature selection phases allowed us to better grasp the significance of data preparation in our Dynamic Multi-Layer Perceptron (DMLP) model. After removing this pre-processing, we tested the model (Table 5) on the fetal health classification task. As a result, we can gauge how much these methods improve the model’s predictive power and, maybe, learn which features were most crucial.

The practical implications associated with the current study outline the proposed model’s limitations. We used a CTG dataset from medical centre and a specific population. The demographic, genetic, and environmental differences between populations restrict the generalizability of our findings. The dataset size with limited processing records is one of the potential limitations. The feature scaling that is performed in the current study yields better accuracy, but at the same time, smoothing values would miss out on some of the significant data for analysis. The other limitation in the evaluation process is that the k-value in multi-fold validation is confined to 10, where the model yields a better accuracy compared to k=5, but needs considerable computation efforts to yield the accuracy. The tradeoff between accuracy and computational efforts may be further analyzed to improve the comprehensibility of the model. The hyperparameters are considered fixed and can be fine-tuned by further evaluating the model in different settings. The feature weights and dependencies can be further evaluated using explainable models. The ablation study can be performed by discarding some of the less significant features from the dataset for better analysis of the robustness of the model.
We present a revolutionary Dynamic Multi-Layer Perceptron neural network for fetal health categorization, which has grown from a simple one-neurons to a complex multi-layer design. The performance of the DMLP model is deliberately improved by including optimization approaches such as early stopping, batch normalization, and dropout. A comprehensive approach is successful, as shown by the synergistic study of classic machine learning approaches with the Dynamic MLP model. The predictive framework is much more effective and dependable now that resampling, feature scaling, and feature selection are part of it and an ablation study without these techniques is also presented. After comparing it to different ML classifiers, we found that the Dynamic MLP model is resilient with an accuracy of 97%. Clinical specialists can learn a lot by comparing models such as LR, XGB, DT, KNN, and RF to ensure the model can be used in real-world healthcare decision-making. These models facilitate data analysis and interpretation. We by offering a comprehensive and efficient way to improve accuracy and interpretability, our research contributes to expanding information on fetal health classification approaches.
Future research could enhance the DMLP model to allow for longitudinal analysis, which would allow for continuous monitoring of fetal health during pregnancy. Improving model transparency and fostering trust among clinic professionals can be achieved by delving more into Explainable AI techniques such as LIME and SHAP.
Acknowledgement: We thank all the authors for their research contributions.
Funding Statement: This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (NRF-2023R1A2C1005950). Jana Shafi is supported via funding from Prince Sattam bin Abdulaziz University Project Number (PSAU/2024/R/1445).
Author Contributions: The authors confirm contribution to the paper as follows: Study conception and design: Uddagiri Sirisha, Parvathaneni Naga Srinivasu, Panguluri Padmavathi, Seongki Kim; data collection: Aruna Pavate, Jana Shafi, Muhammad Fazal Ijaz; analysis and interpretation of results: Uddagiri Sirisha, Parvathaneni Naga Srinivasu, Panguluri Padmavathi, Seongki Kim, Aruna Pavate, Jana Shafi, Muhammad Fazal Ijaz; draft manuscript preparation: Uddagiri Sirisha, Parvathaneni Naga Srinivasu, Panguluri Padmavathi, Seongki Kim; supervision: Jana Shafi, Muhammad Fazal Ijaz. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Publicly available dataset is used in this study. It is available here: https://www.kaggle.com/datasets/andrewmvd/fetal-health-classification, accessed on 25 April 2024.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. A. M. Ponsiglione, C. Cosentino, G. Cesarelli, F. Amato, and M. Romano, “A comprehensive review of techniques for processing and analyzing fetal heart rate signals,” Sensors, vol. 21, no. 18, pp. 6136, 2021. doi: 10.3390/s21186136. [Google Scholar] [PubMed] [CrossRef]
2. J. Piri, P. Mohapatra, and R. Dey, “Fetal health status classification using moga-cd based feature selection approach,” in 2020 IEEE Int. Conf. Electron., Comput. Commun. Technol. (CONECCT), Bangalore, India, Jul. 2020, pp. 1–6. doi: 10.1109/CONECCT50063.2020.9198377. [Google Scholar] [CrossRef]
3. J. Li and X. Liu, “Fetal health classification based on machine learning,” in 2021 IEEE 2nd Int. Conf. Big Data, Artif. Intell. Internet Things Eng. (ICBAIE), Mar. 2021, pp. 899–902. doi: 10.1109/ICBAIE52039.2021.9389902. [Google Scholar] [CrossRef]
4. I. J. Jebadurai, G. J. L. Paulraj, J. Jebadurai, and S. Silas, “Experimental analysis of filtering-based feature selection techniques for fetal health classification,” Serbian J. Electr. Eng., vol. 19, no. 2, pp. 207–224, 2022. doi: 10.2298/SJEE2202207J. [Google Scholar] [CrossRef]
5. A. Mehbodniya et al., “Fetal health classification from cardiotocographic data using machine learning,” Expert. Syst., vol. 39, no. 6, pp. e12899, 2022. doi: 10.1111/exsy.12899. [Google Scholar] [CrossRef]
6. S. Das, H. Mukherjee, K. Roy, and C. K. Saha, “Fetal health classification from cardiotocograph for both stages of labor—A soft-computing-based approach,” Diagnostics, vol. 13, no. 5, pp. 858, 2023. doi: 10.3390/diagnostics13050858. [Google Scholar] [PubMed] [CrossRef]
7. A. Kuzu and Y. Santur, “Early diagnosis and classification of fetal health status from a fetal cardiotocography dataset using ensemble learning,” Diagnostics, vol. 13, no. 15, pp. 2471, 2023. doi: 10.3390/diagnostics13152471. [Google Scholar] [PubMed] [CrossRef]
8. M. T. Alam et al., “Comparative analysis of different efficient machine learning methods for fetal health classification,” Appl. Bionics Biomech., vol. 2022, no. 9, pp. 1–12, 2022. doi: 10.1155/2022/6321884. [Google Scholar] [PubMed] [CrossRef]
9. N. Rahmayanti, H. Pradani, M. Pahlawan, and R. Vinarti, “Comparison of machine learning algorithms to classify fetal health using cardiotocogram data,” Procedia Comput. Sci., vol. 197, no. 14, pp. 162–171, 2022. doi: 10.1016/j.procs.2021.12.130. [Google Scholar] [CrossRef]
10. N. F. M. Noor, N. Ahmad, and N. M. Noor, “Fetal health classification using supervised learning approach,” in 2021 IEEE Nat. Biomed. Eng. Conf. (NBEC), 2021, pp. 36–41. doi: 10.1109/NBEC51914.2021.9637660. [Google Scholar] [CrossRef]
11. A. Akbulut, E. Ertugrul, and V. Topcu, “Fetal health status prediction based on maternal clinical history using machine learning techniques,” Comput. Methods Programs Biomed., vol. 163, no. 6245, pp. 87–100, 2018. doi: 10.1016/j.cmpb.2018.06.010. [Google Scholar] [PubMed] [CrossRef]
12. R. Abiyev, J. B. Idoko, H. Altıparmak, and M. Tüzünkan, “Fetal health state detection using interval type-2 fuzzy neural networks,” Diagnostics, vol. 13, no. 10, pp. 1690, 2023. doi: 10.3390/diagnostics13101690. [Google Scholar] [PubMed] [CrossRef]
13. Ö. Kasim, “Multi-classification of fetal health status using extreme learning machine,” Icontech Int. J., vol. 5, no. 2, pp. 62–70, 2021. doi: 10.46291/ICONTECHvol5iss2pp62-70. [Google Scholar] [CrossRef]
14. S. K. Mandala, “Unveiling the unborn: advancing fetal health classification through machine learning,” arXiv preprint arXiv:2310.00505, vol. 1, pp. 2121, 2023. doi: 10.36922/aih.2121. [Google Scholar] [CrossRef]
15. K. Shruthi and A. S. Poornima, “A method for predicting and classifying fetus health using machine learning,” Int. J. Intell. Syst. Appl. Eng., vol. 11, no. 2, pp. 752–762, 2023. doi: 10.18201/ijisae.2023.328. [Google Scholar] [CrossRef]
16. Z. Hoodbhoy, M. Noman, A. Shafique, A. Nasim, D. Chowdhury and B. Hasan, “Use of machine learning algorithms for prediction of fetal risk using cardiotocographic data,” Int. J. Appl. Basic Med. Res., vol. 9, no. 4, pp. 226, 2019. doi: 10.4103/ijabmr.IJABMR_370_18. [Google Scholar] [PubMed] [CrossRef]
17. M. M. Imran Molla, J. J. Jui, B. S. Bari, M. Rashid, and M. J. Hasan, “Cardiotocogram data classification using random forest-based machine learning algorithm,” in Proc. 11th Nat. Tech. Seminar on Unmanned Syst. Technol. 2019: NUSYS’19, Springer Singapore, 2021, pp. 357–369. doi: 10.1007/978-981-15-5281-6_25. [Google Scholar] [CrossRef]
18. M. M. Islam, M. Rokunojjaman, A. Amin, M. N. Akhtar, and I. H. Sarker, “Diagnosis and classification of fetal health based on ctg data using machine learning techniques,” in Proc. Int. Conf. Mach. Intell. Emerg. Technol., Cham, Switzerland, Sep. 2022, pp. 3–16. doi: 10.1007/978-3-031-34622-4_11. [Google Scholar] [CrossRef]
19. “Fetal health Classification Dataset,” Kaggle. 2024. Accessed: Jun. 25, 2024.[Online]. Available: https://www.kaggle.com/datasets/andrewmvd/fetal-health-classification [Google Scholar]
20. P. Dwivedi, A. A. Khan, S. Mugde, and G. Sharma, “Diagnosing the major contributing factors in the classification of the fetal health status using cardiotocography measurements: An automl and xai approach,” in 2021,The 13th Int. Conf. Electron., Comput., Artif. Intell. (ECAI), IEEE, Jul. 2021, pp. 1–6. doi: 10.1109/ECAI52376.2021.9515070. [Google Scholar] [CrossRef]
21. J. H. Joloudari, A. Marefat, M. A. Nematollahi, S. S. Oyelere, and S. Hussain, “Effective class-imbalance learning based on SMOTE and convolutional neural networks,” Appl. Sci., vol. 13, no. 6, pp. 4006, 2023. doi: 10.3390/app13064006. [Google Scholar] [CrossRef]
22. Y. Salini, S. N. Mohanty, J. V. N. Ramesh, M. Yang, and M. M. V. Chalapathi, “Cardiotocography data analysis for fetal health classification using machine learning models,” IEEE Access, vol. 12, no. 1, pp. 26005–26022, 2024. doi: 10.1109/ACCESS.2024.3364755. [Google Scholar] [CrossRef]
23. D. Bollegala, “Dynamic feature scaling for online learning of binary classifiers,” Knowl.-Based Syst., vol. 129, no. 8, pp. 97–105, 2017. doi: 10.1016/j.knosys.2017.05.010. [Google Scholar] [CrossRef]
24. M. Mandal, P. K. Singh, M. F. Ijaz, J. Shafi, and R. Sarkar, “A tri-stage wrapper-filter feature selection framework for disease classification,” Sensors, vol. 21, no. 16, pp. 5571, 2021. doi: 10.3390/s21165571. [Google Scholar] [PubMed] [CrossRef]
25. S. Ma and J. Zhai, “Big data decision tree for continuous-valued attributes based on unbalanced cut points,” J. Big Data, vol. 10, no. 135, pp. 1328, 2023. doi: 10.1186/s40537-023-00816-2. [Google Scholar] [CrossRef]
26. T. B. Krishna and P. Kokil, “Automated classification of common maternal fetal ultrasound planes using multi-layer perceptron with deep feature integration,” Biomed. Signal Process. Control, vol. 86, no. 11, pp. 105283, 2023. doi: 10.1016/j.bspc.2023.105283. [Google Scholar] [CrossRef]
27. F. A. Bader, “An optimized single layer perceptron-based approach for cardiotocography data classification,” Int. J. Adv. Comput. Sci. Appl., vol. 13, no. 10, 2022. doi: 10.14569/IJACSA.2022.0131076. [Google Scholar] [CrossRef]
28. Y. Yin and Y. Bingi, “Using machine learning to classify human fetal health and analyze feature importance,” BioMedInformatics, vol. 3, no. 2, pp. 280–298, 2023. doi: 10.3390/biomedinformatics3020019. [Google Scholar] [CrossRef]
29. Z. Zhao, Y. Zhang, and Y. Deng, “A comprehensive feature analysis of the fetal heart rate signal for the intelligent assessment of fetal state,” J. Clin. Med., vol. 7, no. 8, pp. 223, 2018. doi: 10.3390/jcm7080223. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools