 Open Access
Open Access
ARTICLE
Enhancing Mild Cognitive Impairment Detection through Efficient Magnetic Resonance Image Analysis
1 Department of Computer Science and Technology, Zhejiang Normal University, Jinhua, 321002, China
2 Zhejiang Institute of Photoelectronics & Zhejiang Institute for Advanced Light Source, Zhejiang Normal University, Jinhua, 321004, China
3 Department of Data Science and Artificial Intelligence, Zarqa University, Zarqa, 13100, Jordan
4 School of Chemical and Environmental Engineering, College of Chemistry, Chemical Engineering and Materials Science, Soochow University, Suzhou, 215123, China
5 Department of Management Information Systems, College of Applied Studies and Community Service, Imam Abdulrahman Bin Faisal University, Dammam, 32244, Saudi Arabia
6 Department of Computer Science, Al Ain University, Abu Dhabi, 999041, United Arab Emirates
7 Department of Electrical Engineering, Faculty of Engineering, Minia University, Minia, 61519, Egypt
8 Healthcare Technology and Innovation Theme, Faculty Research Centre for Intelligent Healthcare, Coventry University, Coventry, CV4 8BW, UK
* Corresponding Author: Zhonglong Zheng. Email:
Computers, Materials & Continua 2024, 80(2), 2081-2098. https://doi.org/10.32604/cmc.2024.046869
Received 07 October 2023; Accepted 26 March 2024; Issue published 15 August 2024
Abstract
Neuroimaging has emerged over the last few decades as a crucial tool in diagnosing Alzheimer’s disease (AD). Mild cognitive impairment (MCI) is a condition that falls between the spectrum of normal cognitive function and AD. However, previous studies have mainly used handcrafted features to classify MCI, AD, and normal control (NC) individuals. This paper focuses on using gray matter (GM) scans obtained through magnetic resonance imaging (MRI) for the diagnosis of individuals with MCI, AD, and NC. To improve classification performance, we developed two transfer learning strategies with data augmentation (i.e., shear range, rotation, zoom range, channel shift). The first approach is a deep Siamese network (DSN), and the second approach involves using a cross-domain strategy with customized VGG-16. We performed experiments on the Alzheimer’s Disease Neuroimaging Initiative (ADNI) dataset to evaluate the performance of our proposed models. Our experimental results demonstrate superior performance in classifying the three binary classification tasks: NC vs. AD, NC vs. MCI, and MCI vs. AD. Specifically, we achieved a classification accuracy of 97.68%, 94.25%, and 92.18% for the three cases, respectively. Our study proposes two transfer learning strategies with data augmentation to accurately diagnose MCI, AD, and normal control individuals using GM scans. Our findings provide promising results for future research and clinical applications in the early detection and diagnosis of AD.Keywords
Alzheimer’s disease (AD) is a degenerative brain disorder that can lead to various symptoms, including memory loss, language problems, and cognitive disabilities. These symptoms are caused by destroying nerve cells involved in learning, thinking, and memory function. As a result of neuron damage, individuals with AD may experience difficulties performing routine body functions such as swallowing and walking [1]. Recent studies suggest that 60% to 80% of dementia cases are attributed to AD, with projections estimating a population of 88 million individuals aged 65 and above with AD by 2050. In 2019, research indicated that individuals with AD incurred an average healthcare cost of 50,201 dollars per year at 65, compared to 14,326 dollars for those without the condition. Given the demanding nature of the initial AD diagnosis, early identification is crucial for effective cost, health, and time management. Furthermore, early identification can help to reduce costs, estimated to range from 4122 dollars, as reported in a recent study [2].
Alzheimer’s disease (AD) is characterized by several stages, with mild cognitive impairment (MCI) being a common middle stage that progressively converts to dementia at a rate of 5%–10% annually. The intermediate stage is marked by the development of symptoms that pose a challenge for neurologists to classify their transformation among the stages accurately. Medical imaging techniques, such as MRI, computer tomography (CT), functional magnetic resonance imaging (fMRI), single-photon emission computed tomography (SPECT), and Positron Emission Tomography (PET), have shown promise in the domain of neuroimaging to detect structural and functional changes, as reported in recent studies [3,4]. Among these techniques, MRI imaging modality is commonly used due to the simplicity of retrieving weighted MRI sequences such as T1 and T2 scans. In recent years, the domains of machine learning and artificial intelligence have witnessed significant advancements driven by novel techniques and methodologies. Artificial Intelligence (AI) models can autonomously acquire features without requiring substantial pre-existing knowledge, and they have found extensive applications across various domains [5–8]. Among these, three key areas have gained prominence and are poised to revolutionize AI research and application landscape. First and foremost, continual learning has emerged as a crucial aspect of machine intelligence. It addresses the challenge of adapting and updating models over time to accommodate new data, allowing AI systems to continually evolve and improve their performance. Secondly, simulation enlargement and augmentation techniques have garnered attention as invaluable tools in training AI models.
On MRI scans, machine learning (ML) predicts superior results for early AD detection. Approaches depending on ML, such as support vector machine (SVM), have produced promising results on automatic detection. These techniques are further classified into three groups. When it comes to feature extraction, such as voxel-based, patch-based, and region of interest (ROI) techniques. Many computer-aided diagnosis systems (CADS) have been introduced in studies that are sometimes more complex and time-consuming. Recently, deep learning approaches in the field of computer perception overcame the issues and reduced the feature extraction cost with less time. In deep learning, Convolutional Neural Networks (CNNs) can reduce the limitations of the CAD system by extracting useful features with measurable depth and breadth. Mainly CNNs are used to solve the different image classification and feature extraction problems [9]. However, deep learning improves the results on many labeled samples. In medical imaging, the non-availability of maximum annotated datasets causes an over-fitting issue. Transfer learning is one of the most effective solutions for a condition with fewer annotated data samples. In most cases, it produces excellent results in terms of classification [10]. The brain structure of MCI and AD patients shrunk when analyzed through MRI images. Several features are used during diagnosis, but three main features are used for diagnosis after segmentation. These features are GM, white matter (WM), and cerebrospinal fluid (CSF) [11].
This article introduces two transfer learning techniques for diagnosing Alzheimer’s (AD). The first technique involves a pre-trained Deep Siamese Network (DSN) model on dementia datasets containing relative information, which is transferred to target samples of normal control (NC), mild cognitive impairment (MCI), and AD. The second method entails the creation of a cross-domain model based on the well-known VGG-16 architecture. This model undergoes a two-step training procedure, beginning with pre-training on the vast and diverse ImageNet dataset, comprising a staggering one million distinct samples. It is then fine-tuned with replacing layers to utilize a comprehensive dataset that includes samples from NC, MCI, and AD patients. This method capitalizes on the extensive knowledge and capability for feature extraction acquired through exposure to the vast ImageNet dataset. When dealing with the more specific and nuanced characteristics of NC, MCI, and AD data samples, it utilizes this broad comprehension as a foundation for further study. This knowledge transfer from a general domain to a specialized medical domain enhances the model’s ability to recognize and differentiate patterns, ultimately leading to more accurate and trustworthy diagnostic outcomes. Transfer learning techniques have shown promising results in classifying less annotated datasets, and the proposed technique utilizing transfer learning approaches achieved superior results for AD diagnosis. We handle the class imbalanced issues that directly impact on the performance of the model, in addition to resolving the other complex problems as shown below:
• We presented two transfer learning strategies: the DSN approach, the cross-dementia approach, and the second cross-domain approach for classifying AD.
• The transfer learning strategy involves leveraging knowledge from large cross-data samples and applying it to medical image classification. This approach helps overcome the limitations of training data samples in medical applications and enhances classification accuracy within the medical computing community. We believe utilizing knowledge from different configurations is critical to achieving these benefits.
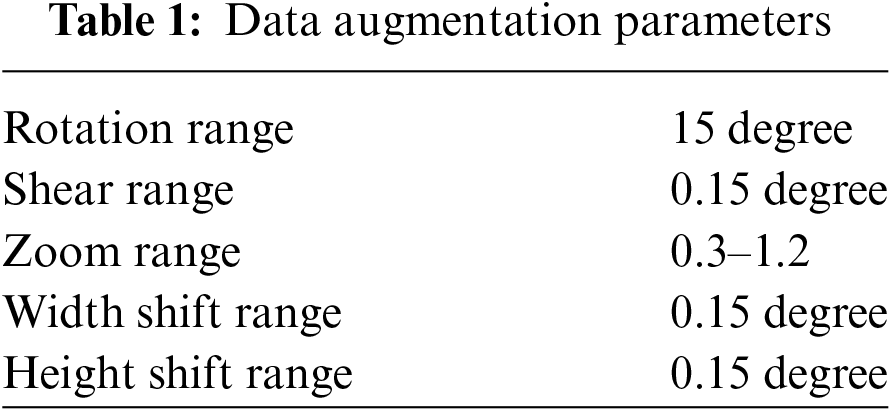
• Data augmentation was performed on training data during the training of working with the MRI data.
2.1 Machine Learning-Based Approaches
Computer-aided diagnosis methods have been developed to identify brain alterations and accelerate early diagnosis of Alzheimer’s disease (AD) using conventional machine learning algorithms. A multi-stage model was proposed, utilizing histogram feature extraction and data segmentation. During the preprocessing stage, input images were segmented into GM, WM, and CSF features, with WM images used to investigate AD as reported in a recent study [12]. Moreover, blockchain addresses contemporary difficulties associated with synchronizing patient data between various and diverse health information systems [13]. A multi-modality classification scheme was presented, utilizing a similarity measure, and a multi-modal analysis was conducted using coordinates derived from joint embedding. The study also proposed an SVM model-based approach to dementia prediction, achieving a classification accuracy of 95%. Another study presented a feature selection approach dependent on a three-dimensional (3D) displacement field, achieving an overall categorization precision of 93.05% [14]. Furthermore, a multi-layer classifier-based multi-view learning system was proposed, with the first stage focused on determining the relationship between extracted imaging features and class labels.
A recent study achieved superior results in binary classification, achieving an accuracy of 98.40%, and in multi-class classification, attaining 79.80% [15]. In another study, a discriminate sparse learning technique and a novel loss function for two methods were designed, resulting in a classification performance of 94.68% for AD vs. normal control (NC) [16]. The researchers demonstrated an approach utilizing support vector machines (SVM) and whole-brain information, comparing diagnostic accuracy between two modalities, T1-magnetic resonance imaging (T1-MRI) and Fluorodeoxyglucose Positron Emission Tomography (FDG-PET), and achieving a classification accuracy of 68%–71%, predicting variations of gray matter for AD identification. The study utilized volumes of interest (VOIs) and gray matter for categorizing NC and AD patients and employed a combination of SVM and a data fusion approach for classification, achieving an accuracy rate of 92.48% using structural MRI (sMRI) data [17]. However, many techniques still rely on handcrafted features, which necessitate the expertise of data analysts for analysis and preprocessing.
By generating synthetic hybrid features, the suggested approach is able to overcome the limitations of traditional classification methods and successfully capture additional information regarding emotional states. The methodology, based on ideas from decision science, recognizes the significant role that emotions play in human decision-making [18]. A comprehensive approach was undertaken in a separate research study involving utilizing various CNN and Recurrent Neural Network (RNN) models for both 2D and 3D MRI data analysis. A particularly noteworthy aspect of this investigation was the application of 2D CNN models to the inherently 3D MRI data. To elaborate, the study’s methodology involved the dissection of each MRI subject into multiple 2D slices. It is essential to emphasize that this segmentation was performed without considering the intricate interconnections and contextual information among the two-dimensional slices within the volumetric MRI dataset. Despite this simplification, the results achieved by the final model were auspicious, boasting an impressive classification accuracy of 96.88%. This innovative approach underscores the potential of CNN and RNN architectures in extracting valuable information from medical imaging data, even when adapting these models to handle multi-dimensional data such as 3D MRI scans. Further exploration of the interconnections within 3D MRI volumes may offer avenues for enhanced performance in future studies [19].
2.2 Deep Learning and Transfer Learning-Based Methods
Recent research has increasingly utilized neural network techniques, including deep learning (DL) models, to address machine learning (ML) problems related to the classification of Alzheimer’s disease (AD). For example, one study proposed a CNN-based strategy to improve the classification of healthy control (HC) and AD patients, using the Wilcoxon test to extract 45 radiomic features [20]. Another study introduced an approach to AD diagnosis using an SCNN based on the VGG-16 architecture, utilizing two parallel modified VGG-16 models to analyze and determine the final diagnosis. The model was tested on OASIS data, achieving an ultimate accuracy of 99.05% for multi-class classification of AD [21]. A CNN-based multi-model was also developed for multi-class classification, with the human brain network connectivity matrix extracted as input for CNN [14]. In another study, researchers proposed a DL model using 3D-CNN and LSTM, MRI and PET modalities, and achieving an overall accuracy of 94.82% [21]. Another approach recommended a combination of 2D-CNN, such as a recurrent neural network (RNN) and CNN, which learned features from the slices following the decomposition of 3D PET images [21].
Hampel et al. [22] proposed a deep CNN for the multi-class classification of AD stages based on 2D-DenseNet. They used three parallel 2D-DenseNet to analyze the MRI data and produced the final classification results. This study presented a deep learning architecture, including a stacked auto-encoder to classify the AD stages. Experimental results show that binary classification is 87.76% and multi-class classification is 47.42%. Groot et al. [23] utilized a DL methodology depending on CNN to distinguish between normal control people and two stages of MCI. They used an effective model to get more useful features that help to identify the initial stage of MCI. Deep learning models have already attained significant results in classification, tracking, and detection. However, several models have produced promising results on the maximum training dataset; many models lead to poor performance due to an inadequate number of training samples. In such a scenario, transfer learning would help create more accurate classification results. Transfer learning enhances the solution of one problem and applies it to solve the second problem, which is related to the first task. A number of transfer learning approaches have been developed for different problems of interest [24].
The created model is well-suited for pre-training in recognition and classification tasks, outperforming standard approaches in multi-class AD classification [25]. Another approach is the cross-model transfer learning technique from structural MRI (sMRI) to Diffusion Tensor Imaging (DTI). They used sMRI data for pertaining and application on DTI data for classification. They achieved 92.5% accuracy on AD vs. NC and 80.0% for MCI vs. NC. Giorgio et al. [26] developed a multi-task transfer learning model that learns from non-medical images and is applied to breast cancer diagnosis. Here, explored the efficacy of five pre-trained models for the early detection of AD and MCI. The study encompasses three distinct diagnostic NC, MCI, and AD, making it a comprehensive investigation into cognitive health. These pre-trained models were initially developed for different domains and datasets. However, the power of transfer learning by fine-tuning them on MRI datasets is pivotal for detecting AD and MCI. That approach capitalizes on the knowledge these models have accumulated from their prior training and adapts it to the intricate features and patterns found in MRI data related to cognitive health. GoogleNet emerged as the standout performer among the models evaluated, achieving a binary classification accuracy of 96.81%. This remarkable result underscores the potential of leveraging pre-trained models and fine-tuning them to enhance the accuracy and efficiency of early AD and MCI detection. Moreover, it highlights the significance of cross-domain learning in addressing complex medical diagnostic challenges [27].
Digital innovations collect patient data and enable healthcare administration beyond traditional methods, resulting in “extensive healthcare records”. Clinical repositories provide an exhaustive overview of patients’ illnesses and outcomes [28]. Our research used MRI images from the ADNI database, including MRI image data, clinical evaluations, neuropsychological testing, and mini-mental examination scores (MMSE). The ADNI is a public-private partnership established in 2003 under the leadership of Dr. Michael W. Weiner, with the primary objective of testing serial PET, MRI, other clinical assessments, and biological biomarkers for Alzheimer’s disease. Following the ADNI acquisition methodology, all datasets in the form of T1-weighted images are readily accessible. We collected MRI subjects with Multiplanar Reconstruction (MPR), B1 correction, and Grad Warp preprocessing descriptions.
Our study employed the statistical parameter mapping SPM12 for pre-processing T1-weighted images in the neuroimaging informatics technology initiative (NIFTI) format. Full-brain segmentation was applied to obtain gray matter (GM), white matter (WM), and cerebrospinal fluid (CSF) images. Specifically, we utilized GM images for our proposed technique to diagnose individuals in the standard control (NC) and mild cognitive impairment (MCI) stages. We employ bias regularization in this stage with a judiciously chosen cutoff value of 0.0001. This regularization technique helps correct image artifacts and inconsistencies that might arise during data acquisition, ensuring that our analyses are based on accurate and reliable data. Additionally, we apply a full width at half maximum (FWHM) of 60 mm, which aids in smoothing the images, reducing noise, and enhancing the discernibility of relevant features. Affine regularization is another essential component of our pre-processing pipeline. We employ the widely recognized ICBM (International Consortium for Brain Mapping) space template to achieve this. This template provides a standardized reference frame that facilitates the alignment and normalization of brain scans across all datasets. Ensuring all images are registered to a common spatial framework mitigates potential variations in brain anatomy and size, thereby enabling meaningful comparisons and analyses.
Spatial normalization is conducted using the Montreal Neurological Institute (MNI) space, a widely adopted standard in neuroimaging research. The MNI space serves as a common coordinate system, allowing us to map brain structures and regions of interest across different subjects and studies precisely. This step is crucial for achieving consistency and accuracy in our analyses, particularly when dealing with diverse datasets. We apply a Gaussian kernel for image smoothing to further refine our data. This process helps to enhance the signal-to-noise ratio in our neuroimaging data, making it easier to detect and analyze subtle changes or patterns. The choice of kernel size and smoothing parameters is carefully considered to balance noise reduction and the preservation of essential features in the images. Some samples are shown in Fig. 1.
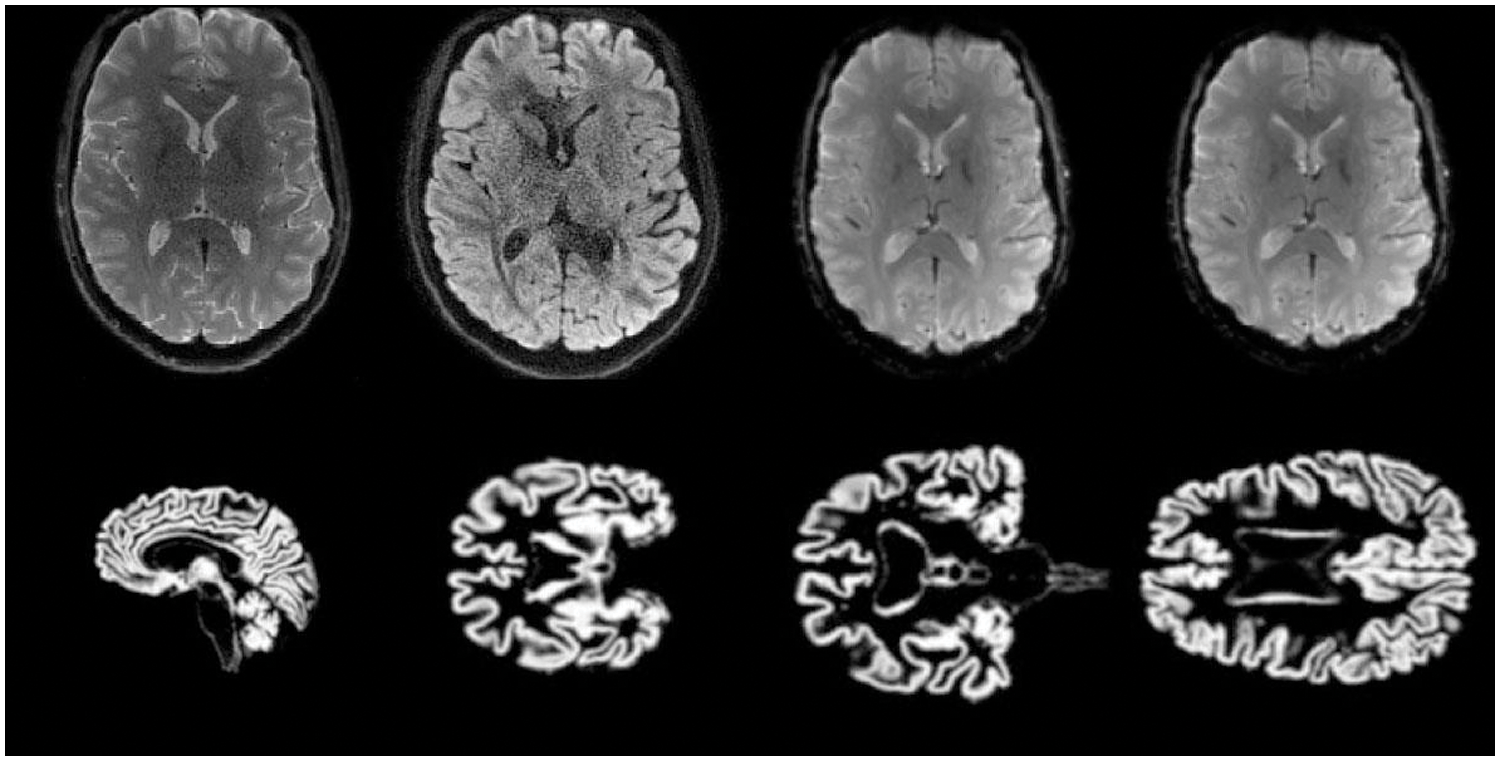
Figure 1: Training and testing MRI 2D-Images
3.2 CNNs and Transfer Learning
Convolutional Neural Networks (CNNs) are a type of deep learning model that mimics the workings of biological systems. They use convolution to extract features from processed data and pass the information to the next layer. CNNs can learn nonlinear mappings and high-dimensional features from maximum training data [29]. Pooling layers are often utilized to improve computational efficiency and preserve spatial representation. Combining convolutional and pooling layers facilitates feature extraction and reduction in input images, followed by fully connected layers that learn nonlinear relationships. Finally, a Softmax layer is used for classification. In medical applications, CNNs have been used to extract useful features from limited data samples. However, supervised deep-learning models can be susceptible to overfitting during training, leading to decreased classification performance. Moreover, deep networks typically require maximum training data [30]. Recently, transfer learning strategies have been introduced in medical image analysis to address these issues, demonstrating efficacy in reducing overfitting.
At last, the Softmax layer is used according to the classification. CNNs are used in many medical applications to learn valuable features from limited data samples. However, supervised deep learning models may result in overfitting during training, directly decreasing the classification performance. Secondly, deep networks require vast data samples for training the model [30]. Recently, many studies introduced transfer learning strategies for medical images; results demonstrate that transfer learning effectively decreases overfitting.
In [31], a model for early detection of Alzheimer’s disease was developed, which involved image representation from start to finish. Transfer learning was employed, using a pre-trained model for a new task to achieve classification results. Meanwhile, reference [32] presented a transfer learning model for Alzheimer’s disease classification, utilizing pre-trained weights from VGG-16. In Alzheimer’s disease, obtaining a large amount of labeled data is challenging due to the time and resources required for annotation, after rescaling and augmenting the input MRI scans. We proceed to incorporate the pre-trained VGG-16 model. Further elaboration on the architectural intricacies of the VGG-16 framework will be provided in the subsequent subsection for a comprehensive understanding of our methodology Nonetheless, the studies mentioned above have inspired the investigation of knowledge transfer techniques that leverage natural data to train models and convey information to medical images. This research addresses these issues using transfer learning approaches, as illustrated in Fig. 2. In our approach, we utilized the Deep Siamese Network (DSN) model, which was pre-trained on dementia scans and fine-tuned on the NC and MCI datasets. The source dataset and target dataset belong to the same domain. The target dataset was obtained from the ADNI database, and we hypothesized that features learned from different Alzheimer’s datasets with diverse configurations could be transferred.
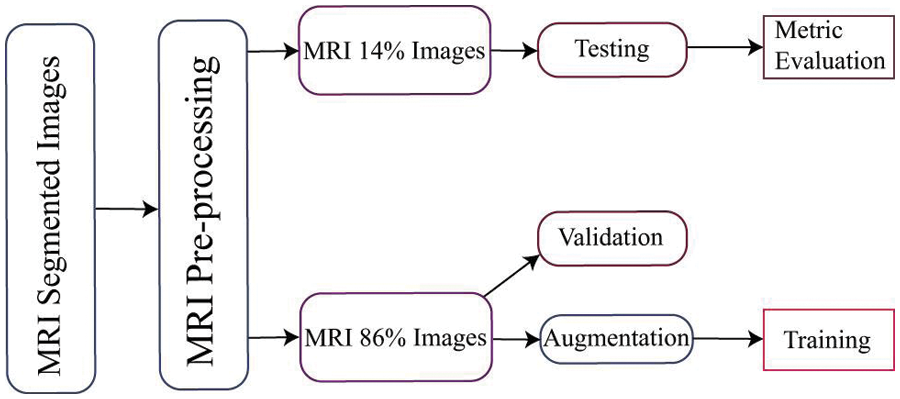
Figure 2: Proposed DSN and cross-domain approach for Alzheimer’s disease classification
The initial layers of the pre-trained model capture predominantly low-level features, while the class-specific features are located in the final layers. In light of our objective of training the network using data on AD, we decided to substitute new class-specific layers. Specifically, the initial three layers of the pre-trained model were transferred, and custom layers tailored to our particular task were introduced. We configured crucial parameters, including the weight-learning factor, bias-learning factor, and desired output size, in order to generate the entirely connected layers. In our classification problem, the output capacity of the final fully connected layer was set to correspond to the number of output classes. These parameters are crucial for optimizing the efficacy of the model. The weight-learning factor influences the model’s capacity to learn complex patterns by regulating the rate at which its weights are modified during training. The bias-learning factor regulates the rate at which the layers learn about biases, thereby enhancing the model’s capacity to detect subtleties in the data. A careful adjustment of these parameters is essential for achieving optimal convergence and precision throughout the training process. In order to evaluate the effectiveness of our training procedure, we utilize the remaining data as testing data for the trained network. This allows us to gauge the network’s performance by measuring its accuracy. We can gain insights into the network’s proficiency in classifying AD cases by conducting tests using this separate dataset.
The experimental datasets used in this research were obtained from the publicly available ADNI repository, which includes NC, MCI, and AD patients. Our study categorized the data into three binary classes: NC vs. AD, NC vs. MCI, and MCI vs. AD. To implement the cross-dementia and cross-domain strategies, we resized the images to 224 × 224. For each classification dataset, 14% of the data was allocated for testing and 86% for training, while augmentation was only applied to the training data. A data flow chart is provided in Fig. 3. We mitigated the overfitting issue by increasing the sample size, which was accomplished through data augmentation techniques, as shown in Table 1.
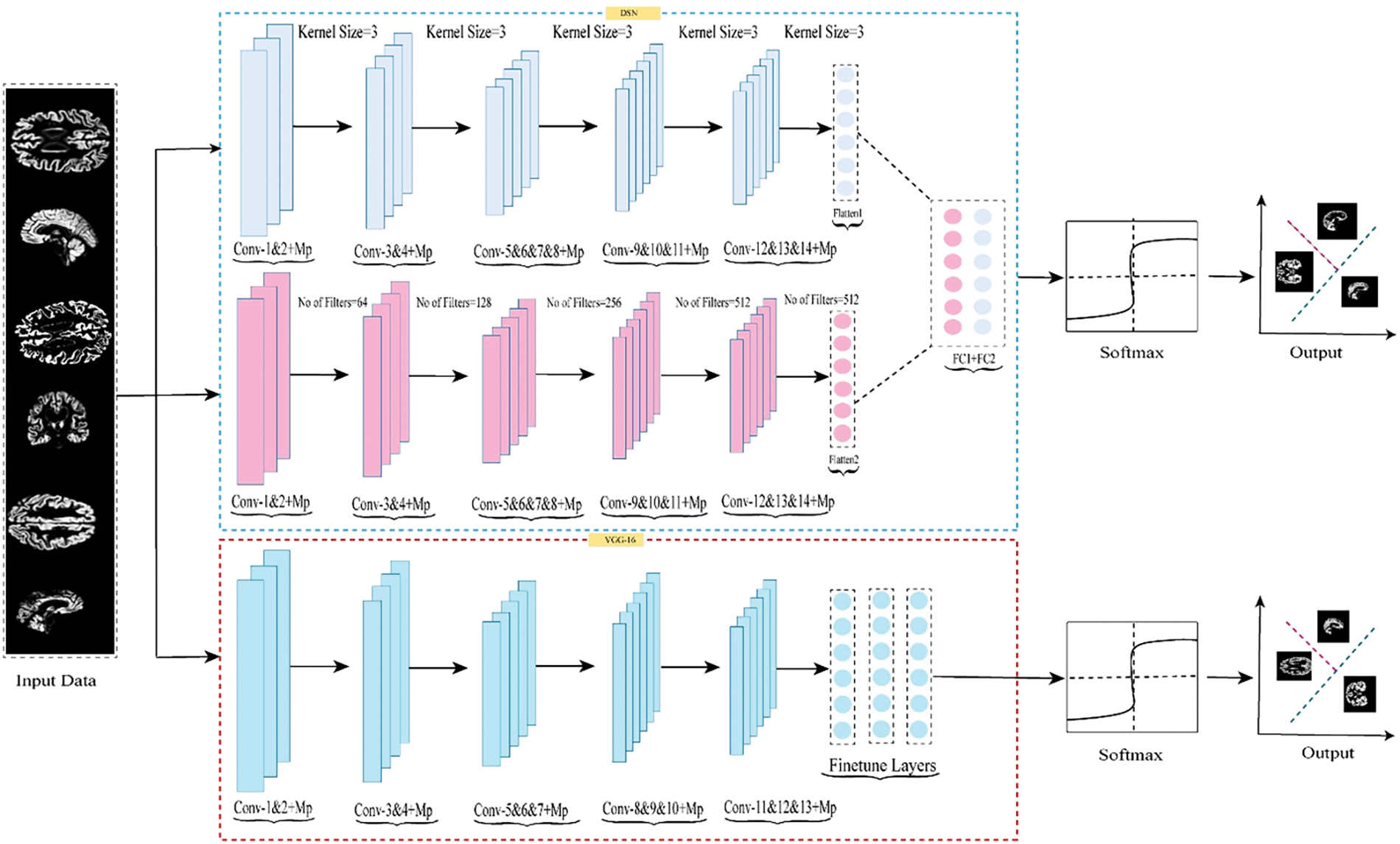
Figure 3: Schematic illustrating the suggested model’s data flow used for training, testing, and validation
The models were trained on the augmented dataset. We utilized the Z840 workstation for the entire experiment with an Intel Xeon-CPU E5-2630v3@2.40 GHz*32. We selected 270 subjects aged between 60–90 years, with varying gender distributions, as presented in Table 2.
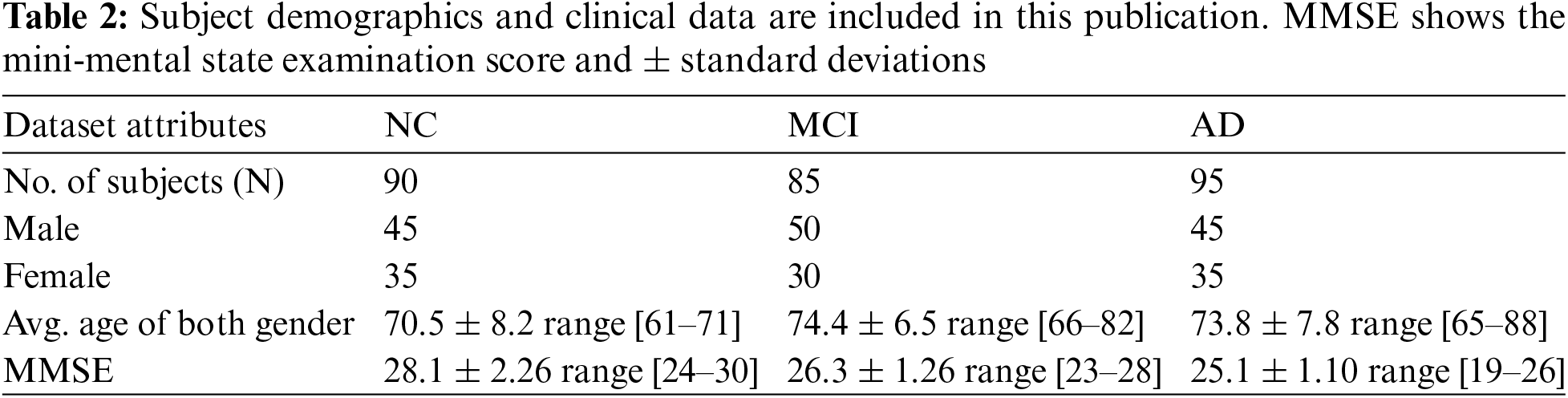
4.1 Experiment 1 with DSN Approach
In Experiment 1, we utilized the DSN model and implemented it using Keras with TensorFlow as the backend. This approach was chosen to extract more features from images, particularly when dealing with a limited number of images. The model was trained using a 0.002 learning rate and an Adamax optimizer with categorical cross-entropy. DSN was pre-trained for 20 epochs with a batch size of 64. For the cross-dementia approach, weights were transferred onto segmented images during fine-tuning. The segmented dataset comprised images from three categories: NC, MCI, and AD, with the sagittal, axial, and Coronary views utilized. The best accuracy result for NC vs. AD classification was 97.68%, with a sensitivity of 98.12% and specificity of 97.23%. For the differentiation between MCI vs. AD, the accuracy result was 94.25%, with a sensitivity of 93.63% and specificity of 94.87%. The accuracy, sensitivity, and specificity for NC vs. MCI were 92.18%, 92.41%, and 91.95%, respectively, as detailed in Table 3. The confusion matrix for the complete results is shown in Tables 4–6. Moreover, Fig. 4 shows the learning curves of the DSN model.
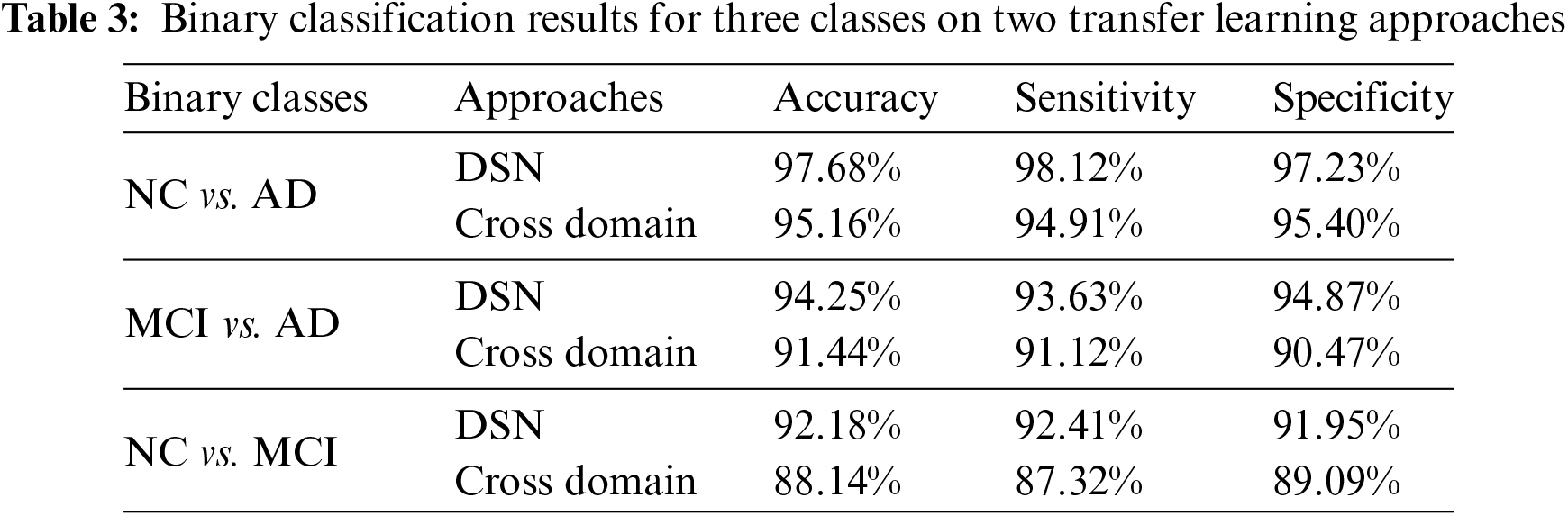

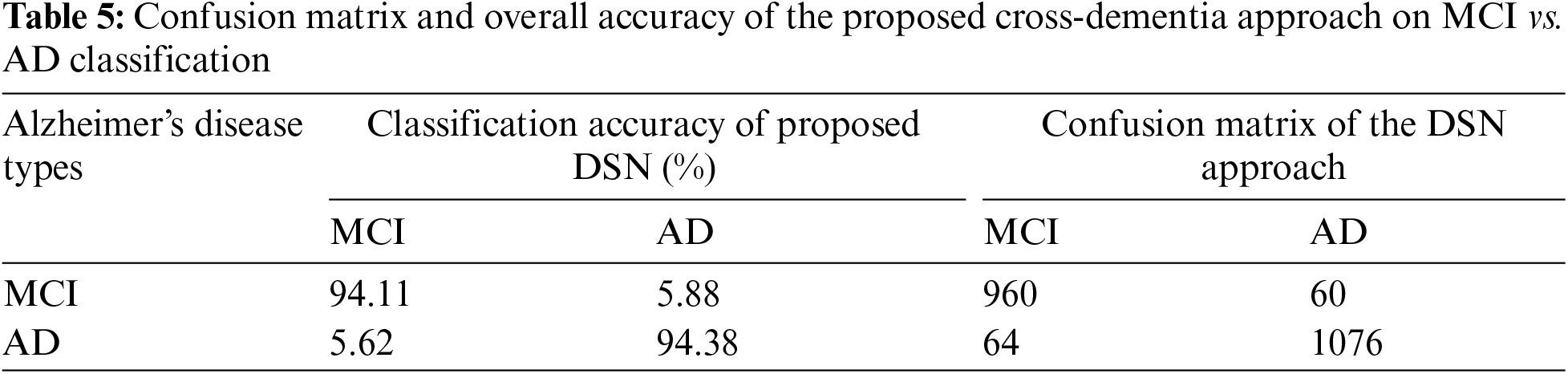
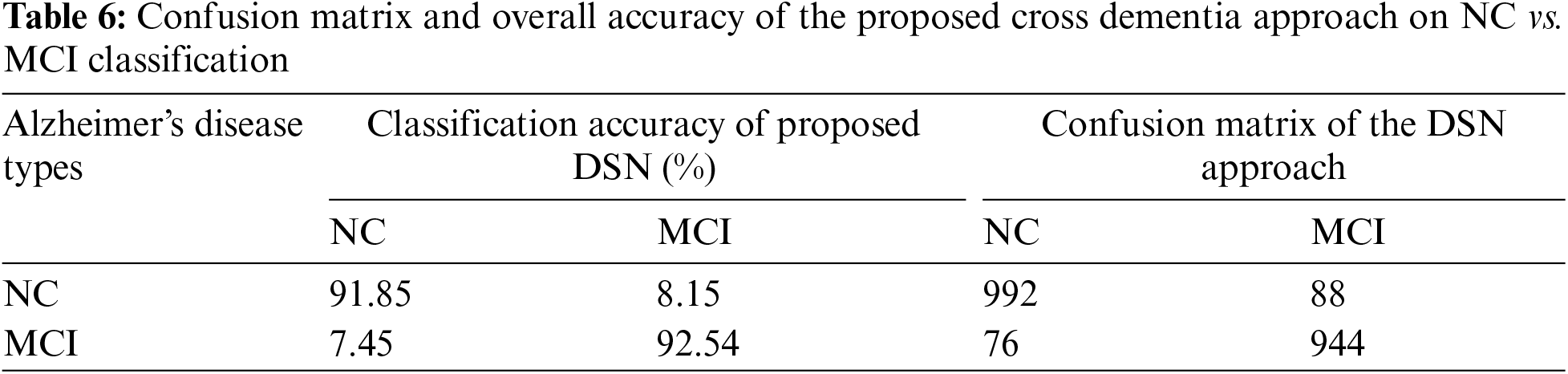
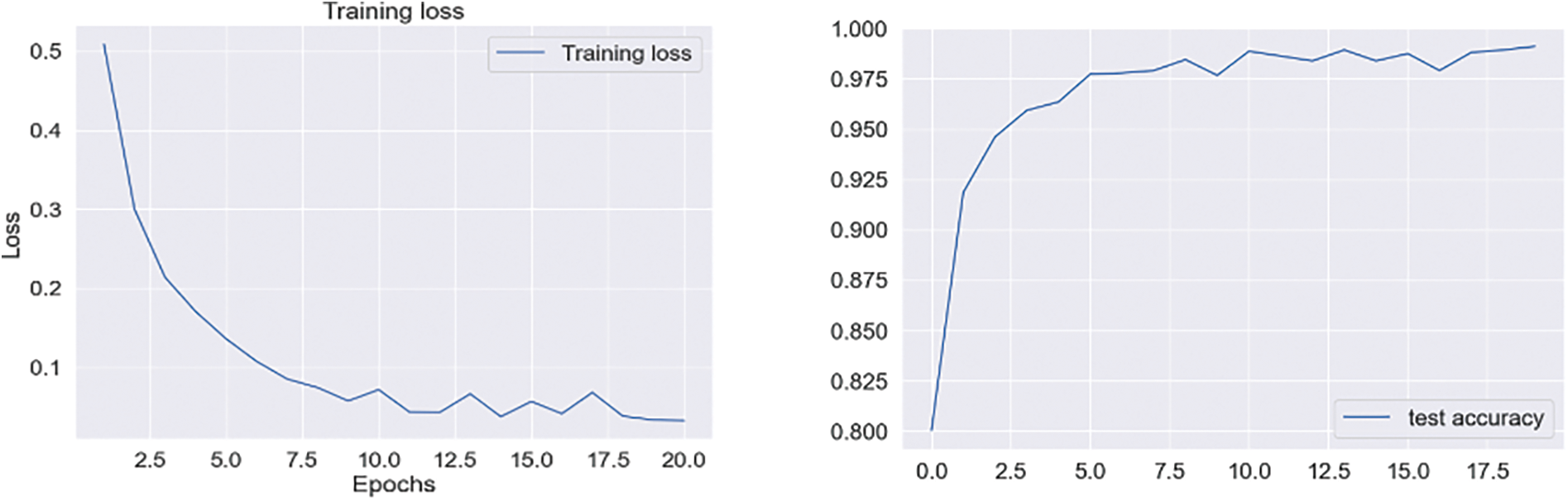
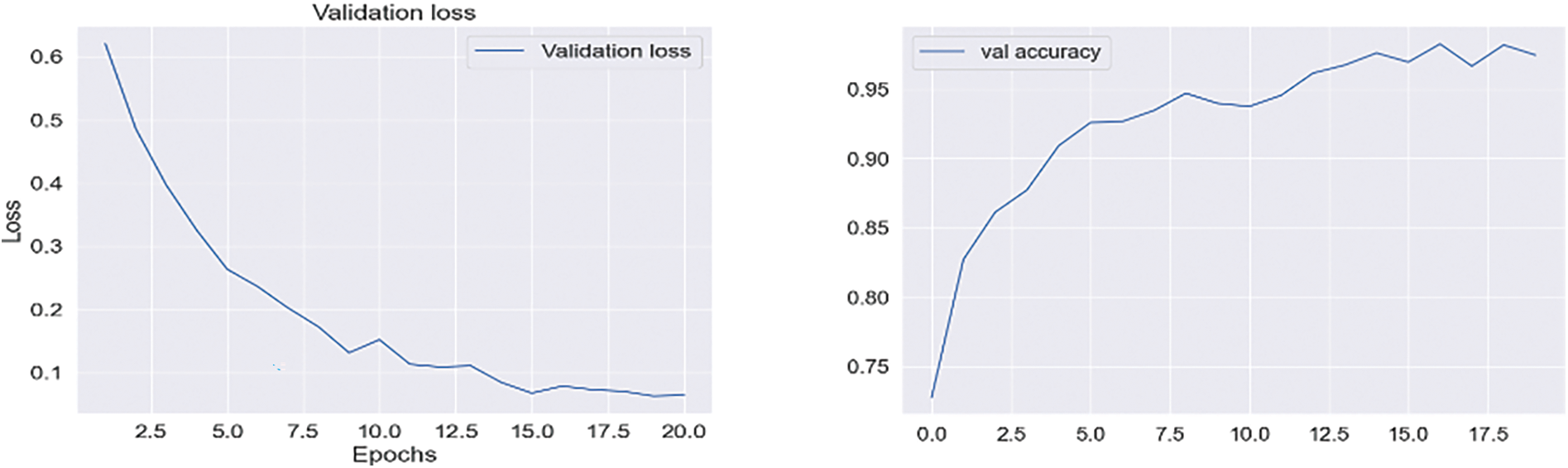
Figure 4: Proposed DSN model learning curves for training loss, validation loss, validation accuracy, and test accuracy for NC-AD
4.2 Experiment 2 with a Cross-Domain Approach
The VGG-16 pre-trained algorithm, which was trained on the ILSVRC ImageNet dataset, was utilized in our study and transferred to the segmented 3 views of the brain (i.e., coronal, sagittal, and axial datasets). The pre-training of the VGG-16 model was performed on 1.2 billion scans belonging to 1000 different domain samples. We chose this model to evaluate the classification accuracy for the cross-model and cross-dementia approaches. The initial pre-trained layers contain low-level features, while the last layers contain specific class features. Therefore, our primary objective was to fine-tune the model on a cross-domain dataset to achieve optimal categorization performance for early Alzheimer’s diagnosis. We fine-tuned the algorithm by adjusting several factors, including the learning rate and training parameters.
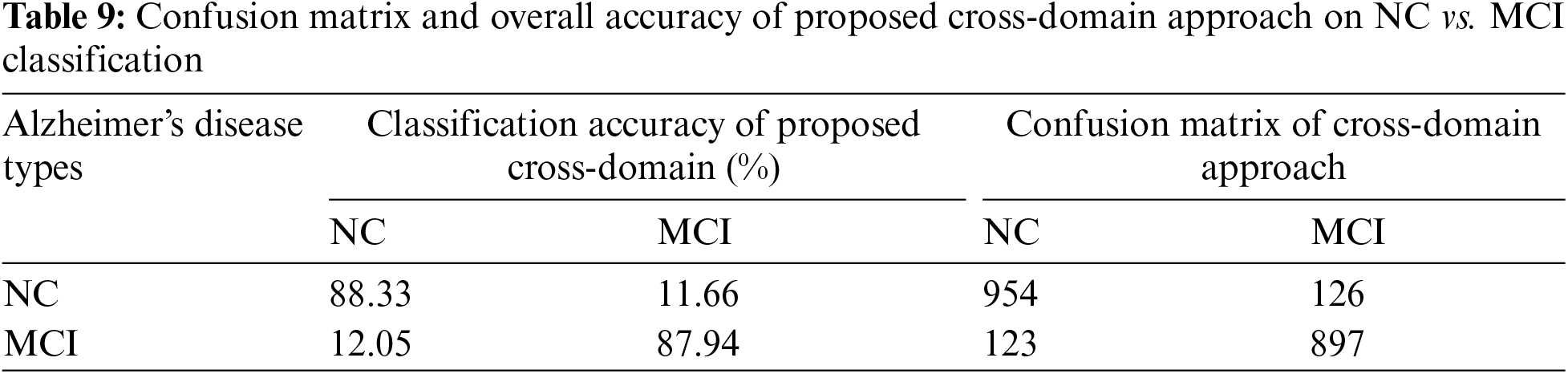
We varied the learning rate from 1e−1 to 1e−5 and adjusted the bias factor accordingly. The optimal results were achieved using a learning rate 1e−3 to distinguish between NC vs. AD, with an accuracy of 95.16% and a sensitivity of 94.91%, as presented in Table 3. On testing data, we achieved classification results of 91.44% and 88.14% for MCI vs. AD and NC vs. MCI, respectively, as illustrated in Fig. 5. The confusion matrix details for our proposed technique are provided in Tables 7–9.

Figure 5: Quantitative analysis for the binary classification results on two transfer learning approaches
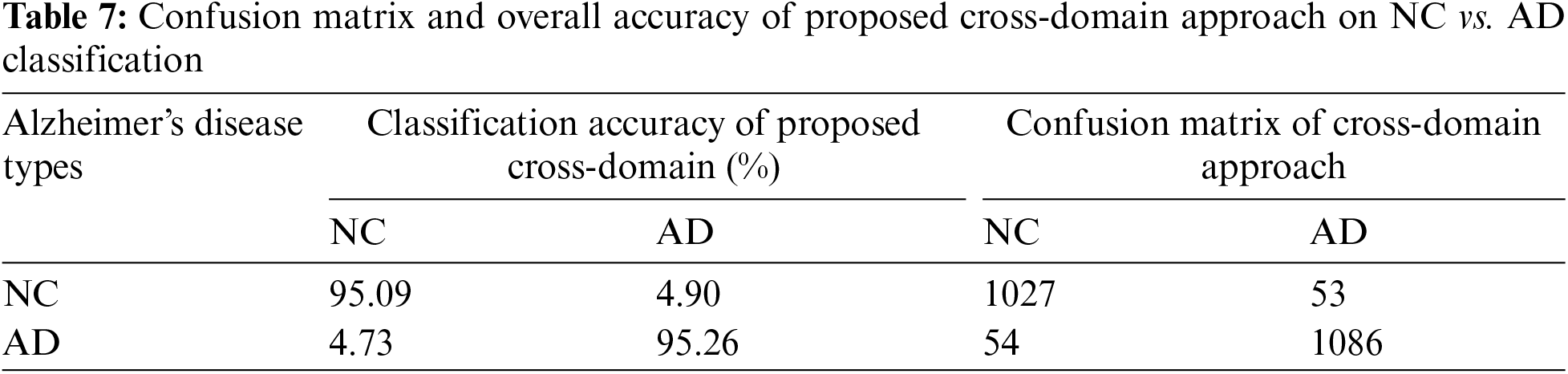
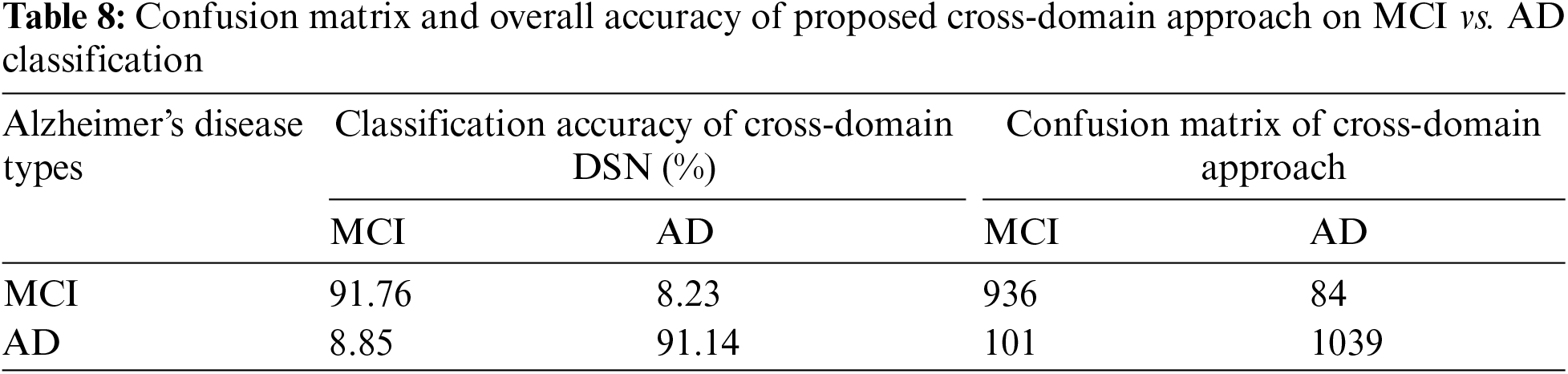
Treatment and detection of AD at an early stage are both very vital and challenging. Therefore, many researchers’ efforts introduced the models for the initial diagnosis of AD [33]. In this study, we created two approaches for the initial identification of AD, i.e., cross-dementia strategy and cross-domain strategy. The proposed models achieved high accuracy results, especially in the case of NC vs. AD classification. Furthermore, our proposed approaches produced excellent results relative to transfer learning studies in early diagnosis of AD, as shown in Table 3. A significant problem in the state of the art for all models is due to data heterogeneous issues while collecting the MRI data from different protocols that impact accuracy reduction. It can be termed a data leakage issue [34]. Some pipelines aim to reduce these issues during data acquisition and preprocessing. These programs, such as SPM12, FSL, and Clinical, can reduce the data complication during preprocessing. However, another issue is selecting brain imaging methods such as MRI, fMRI, and PET [35]. In the experiment that we planned to conduct, we used the three different data classes that were determined by the MRI modality. Also, we utilized the SPM12 preprocessing tool for data extraction. Our proposed approaches overcome the issues of less annotated data samples and outperform on a smaller number of images.
The performance of diagnostic AI models can be significantly affected by the existence of class imbalance in the datasets used. This happens when the dataset’s classes have a disproportionately small number of instances compared to other classes. To balance class representation, this difficulty must be addressed by investigating methods like oversampling, under-sampling, or creating synthetic data [7]. The authors must investigate how class imbalance affects the model’s functionality and the methods they use to lessen its consequences. Obtaining an adequate number of correctly classified fault samples can be challenging in many diagnostic applications. The lack of labeled data may make training and validating AI algorithms more difficult. This restriction might hinder the advancement and functionality of AI models [36]. On the other hand, transfer learning increased computational efficiency. Pre-trained models are especially used in clinical practice. Transfer learning improves the learning performance, decreases the overfitting problem during training the model, and attained 84.90% accuracy for the classification of NC vs. AD [37].
Our approaches are unique as we used transfer learning in two ways: cross-dementia and cross-domain strategy, which produced state-of-the-art results with minimum data samples. Continuous learning, referred to as ongoing learning or incremental learning, trains machine learning algorithms to adapt and learn from incoming data. Augmentation is improving the variety and quality of synthetic data used in machine learning, frequently by generating new data points from preexisting ones. This can involve techniques like image rotation, scaling, or noise addition. The proposed approaches attained the best results in the classification of NC vs. AD, MCI vs. AD, and NC vs. MCI. Besides, we used augmentation and GM features for classification problems. We obtained the outperformed performance through a cross-dementia approach in terms of testing accuracy of 97.68% (NC vs. AD), 94.25% (MCI vs. AD), and 92.18% (NC vs. MCI). Furthermore, the cross-domain approach produced results such as 95.16% (NC vs. AD), 91.44% (MCI vs. AD), and 88.14% (NC vs. MCI). In Table 10, we also compared the classification results of proposed approaches with state-of-the-art models that classified the NC, MCI, and AD. Noted that, in comparison to Table 10, these methods utilize DL and TL to analyze GM and WM features from many modalities, including MRI and PET. However, the proposed models have shown the highest accuracy regarding the binary classification of NC vs. AD. It is worth noting our cross-dementia model, as compared to [38], focused on NC vs. AD classification and our model improved the 2.68% accuracy.
Furthermore, if we compare it to another study [39] in which they used the ADNI data samples for classification, in that scenario, we can see our proposed model has improved the accuracy by 4.25% for (AD vs. NC). However, our proposed approach also outperforms and increases the accuracy by 7%–10%. It should be highlighted that the accuracy of our algorithms is comparable to studies conducted by earlier researchers that were based on deep learning and transfer learning employing various modalities. Two significant shortcomings that often pose challenges in the application of AI for diagnosis are class imbalance and the availability of insufficient labeled fault samples. In many real-world datasets, the distribution of data across different classes is often highly skewed. This imbalance can be particularly pronounced when dealing with medical or diagnostic datasets. For instance, consider a scenario where the majority of cases are non-pathological, while the number of cases with a specific medical condition or fault is relatively small. Class imbalance can impact the performance of AI models, as they may prioritize the majority class, leading to suboptimal detection and classification of the minority class. This is especially concerning when the minority class represents critical or rare conditions.
The efficacy of AI models is intricately tied to the quality and quantity of labeled data accessible for training purposes. In diagnostic tasks, acquiring precisely labeled fault samples poses a formidable challenge. The intricate process of collecting, annotating, and validating fault samples is both time-consuming and demands substantial resources. As a result, researchers and practitioners often find themselves in scenarios where the available labeled fault samples are insufficient to cultivate AI models that are both robust and dependable. Overcoming these limitations requires a collaborative and dedicated effort within the AI and medical communities to enhance the availability and quality of labeled fault data for more effective model training.
In this study, we investigated two approaches, namely DSN and cross-domain, using transfer learning for the early detection of AD. The first approach employed a pre-trained model based on dementia images, while the second approach involved pre-training on the ImageNet database and fine-tuning our segmented data. We evaluated the models on the publicly available ADNI datasets and achieved improved results, particularly for the cross-dementia approach in AD classification. We compared our methods with several other techniques, including transfer learning and deep learning with GM for binary classification, and found that our proposed strategies produced superior results. The classification accuracy obtained using our proposed approach was 97.68% for NC vs. AD, 94.25% for MCI vs. AD, and 92.18% for NC vs. MCI. The transfer learning-based techniques showed excellent results, which can help design models for the early identification of AD and aid practitioners in diagnosing the disease at an early stage. In the future, we plan to apply these models to categorize AD into multiple classes and fine-tune the hyper-parameters to achieve even more improved results.
Acknowledgement: Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd., and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org, accessed on 10 January 2024). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Funding Statement: Research work funded by Zhejiang Normal University Research Fund YS304023947 and YS304023948.
Author Contributions: Atif Mehmood and Zhonglong Zheng: conceptualization and formal analysis. Atif Mehmood and Farah Shahid: methodology. Atif Mehmood and Farah Shahid: writing–original draft preparation. Rizwan Khan, Mutasem K. Alsmadi and Ahmad Al Smadi: analyzed data. Ahmad Al Smadi, Shahid Iqbal, Mutasem K. Alsmadi and Zhonglong Zheng: contributed to multiple sections of the manuscript, review & editing. Mostafa M. Ibrahim, Yazeed Yasin Ghadi, and Syed Aziz Shah: review & editing, and funding. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data utilized for this article were sourced from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (https://adni.loni.usc.edu/, accessed on 10 January 2024). While the ADNI investigators contributed to the design and implementation of the ADNI and supplied the data, they did not participate in the analysis or authorship of this report. A complete listing of ADNI investigators can be found at: https://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf (accessed on 10 January 2024).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. A. Mehmood et al., “A transfer learning approach for early diagnosis of Alzheimer’s disease on MRI images,” Neuroscience, vol. 460, pp. 43–52, 2021. doi: 10.1016/j.neuroscience.2021.01.002. [Google Scholar] [PubMed] [CrossRef]
2. G. A. Pathak et al., “Two-stage bayesian gwas of 9576 individuals identifies snp regions that are targeted by mirnas inversely expressed in Alzheimer’s and cancer,” Alzheimer’s Dementia, vol. 16, no. 1, pp. 162–177, 2020. doi: 10.1002/alz.12003. [Google Scholar] [PubMed] [CrossRef]
3. X. Hao et al., “Multi-modal neuroimaging feature selection with consistent metric constraint for diagnosis of Alzheimer’s disease,” Med. Image Anal., vol. 60, pp. 101625, 2020. doi: 10.1016/j.media.2019.101625. [Google Scholar] [PubMed] [CrossRef]
4. E. Goceri, “Diagnosis of Alzheimer’s disease with sobolev gradient-based optimization and 3d convolutional neural network,” Int. J. Numer. Methods Biomed. Eng., vol. 35, no. 7, pp. e3225, 2019. [Google Scholar]
5. K. Zeng et al., “A very deep densely connected network for compressed sensing MRI,” IEEE Access, vol. 7, pp. 85430–85439, 2019. doi: 10.1109/ACCESS.2019.2924604. [Google Scholar] [CrossRef]
6. M. Sudheer et al., “DRLBTSA: Deep reinforcement learning based task-scheduling algorithm in cloud computing,” Multimed. Tools Appl., vol. 83, pp. 1–29, 2023. [Google Scholar]
7. L. Chen and H. Y. Chan, “Generative adversarial networks with data augmentation and multiple penalty areas for image synthesis,” Int Arab J Inform Technol (IAJIT), vol. 20, no. 3, pp. 428–434, 2023. doi: 10.34028/iajit. [Google Scholar] [CrossRef]
8. S. Dash, P. Parida, G. Sahu, and O. I. Khalaf, “Artificial intelligence models for blockchain-based intelligent networks systems: Concepts, methodologies, tools, and applications,” in Handbook of Research on Quantum Computing for Smart Environments. IGI Global, 2023, pp. 343–363. [Google Scholar]
9. J. Susan and P. Subashini, “Deep learning inpainting model on digital and medical images—A review,” Int. Arab. J. Inform. Technol., vol. 20, no. 6, pp. 919–936, 2023. doi: 10.34028/iajit/20/6/9. [Google Scholar] [CrossRef]
10. J. Wen et al., “Convolutional neural networks for classification of Alzheimer’s disease: Overview and reproducible evaluation,” Med. Image Anal., vol. 63, pp. 101694, 2020. doi: 10.1016/j.media.2020.101694. [Google Scholar] [PubMed] [CrossRef]
11. A. Mehmood, A. Abugabah, A. A. AlZubi, and L. Sanzogni, “Early diagnosis of Alzheimer’s disease based on convolutional neural networks,” Comput. Syst. Sci. Eng., vol. 43, no. 1, pp. 305–315, 2022. doi: 10.32604/csse.2022.018520. [Google Scholar] [CrossRef]
12. F. Ramzan et al., “A deep learning approach for automated diagnosis and multi-class classification of alzheimer’s disease stages using resting-state fmri and residual neural networks,” J. Med. Syst., vol. 44, no. 2, pp. 1–16, 2020. doi: 10.1007/s10916-019-1475-2. [Google Scholar] [PubMed] [CrossRef]
13. S. Hussain, H. Rahman, G. M. Abdulsaheb, H. Al-Khawaja, and O. I. Khalaf, “A blockchain-based approach for healthcare data interoperability,” Int. J. Adv. Soft Comput. Appl., vol. 15, no. 2, 2023. [Google Scholar]
14. M. Yaqub et al., “State-of-the-art cnn optimizer for brain tumor segmentation in magnetic resonance images,” Brain Sci., vol. 10, no. 7, pp. 427, 2020. doi: 10.3390/brainsci10070427. [Google Scholar] [PubMed] [CrossRef]
15. M. Tanveer et al., “Machine learning techniques for the diagnosis of Alzheimer’s disease: A review,” ACM Trans. Multimed. Comput., Commun., Appl., vol. 16, no. 1s, pp. 1–35, 2020. doi: 10.1145/3344998. [Google Scholar] [CrossRef]
16. A. Moscoso et al., “Time course of phosphorylated-tau181 in blood across the Alzheimer’s disease spectrum,” Brain, vol. 144, no. 1, pp. 325–339, 2021. doi: 10.1093/brain/awaa399. [Google Scholar] [PubMed] [CrossRef]
17. N. Hao et al., “Acoustofluidic multimodal diagnostic system for Alzheimer’s disease,” Biosens. Bioelectron., vol. 196, pp. 113730, 2022. doi: 10.1016/j.bios.2021.113730. [Google Scholar] [PubMed] [CrossRef]
18. O. I. Khalaf, S. R. Ashokkumar, S. Dhanasekaran, and G. M. Abdulsahib, “A decision science approach using hybrid EEG feature extraction and GAN-based emotion classification,” Adv. Decis. Sci., vol. 27, no. 1, pp. 172–191, 2023. [Google Scholar]
19. J. P. Kim et al., “Machine learning based hierarchical classification of frontotemporal dementia and Alzheimer’s disease,” NeuroImage: Clin., vol. 23, pp. 101811, 2019. doi: 10.1016/j.nicl.2019.101811. [Google Scholar] [PubMed] [CrossRef]
20. A. Mehmood, M. Maqsood, M. Bashir, and Y. Shuyuan, “A deep Siamese convolution neural network for multi-class classification of Alzheimer disease,” Brain Sci., vol. 10, no. 2, pp. 84, 2020. doi: 10.3390/brainsci10020084. [Google Scholar] [PubMed] [CrossRef]
21. M. Ghanbari et al., “Alterations of dynamic redundancy of functional brain subnetworks in Alzheimer’s disease and major depression disorders,” NeuroImage: Clin, vol. 33, pp. 102917, 2022. doi: 10.1016/j.nicl.2021.102917. [Google Scholar] [PubMed] [CrossRef]
22. H. Hampel, J. Cummings, K. Blennow, P. Gao, C. R. Jack and A. Vergallo, “Developing the ATX(N) classification for use across the Alzheimer disease continuum,” Nat. Rev. Neurol., vol. 17, no. 9, pp. 580–589, 2021. doi: 10.1038/s41582-021-00520-w. [Google Scholar] [PubMed] [CrossRef]
23. C. Groot et al., “Differential patterns of gray matter volumes and associated gene expression profiles in cognitively-defined Alzheimer’s disease subgroups,” NeuroImage: Clini, vol. 30, pp. 102660, 2021. doi: 10.1016/j.nicl.2021.102660. [Google Scholar] [PubMed] [CrossRef]
24. A. Al Smadi, A. Abugabah, A. M. Al-Smadi, and S. Almotairi, “SEL-COVIDNET: An intelligent application for the diagnosis of COVID-19 from chest X-rays and CT-scans,” Inform. Med. Unlocked, vol. 32, pp. 101059, 2022. doi: 10.1016/j.imu.2022.101059. [Google Scholar] [PubMed] [CrossRef]
25. T. Altaf et al., “Multi-class Alzheimer’s disease classification using image and clinical features,” Biomed. Signal Process. Control, vol. 43, pp. 64–74, 2018. doi: 10.1016/j.bspc.2018.02.019. [Google Scholar] [CrossRef]
26. J. Giorgio, S. M. Landau, W. J. Jagust, P. Tino, and Z. Kourtzi, “Modelling prognostic trajectories of cognitive decline due to Alzheimer’s disease,” NeuroImage: Clin, vol. 26, pp. 102199, 2020. doi: 10.1016/j.nicl.2020.102199. [Google Scholar] [PubMed] [CrossRef]
27. S. Ahmed et al., “Ensembles of patch-based classifiers for diagnosis of Alzheimer diseases,” IEEE Access, vol. 7, pp. 73373–73383, 2019. doi: 10.1109/ACCESS.2019.2920011. [Google Scholar] [CrossRef]
28. X. Xue et al., “Modelling and analysis of hybrid transformation for lossless big medical image compression,” Bioengineering, vol. 10, no. 3, pp. 333, 2023. doi: 10.3390/bioengineering10030333. [Google Scholar] [PubMed] [CrossRef]
29. K. Misquitta, M. Dadar, D. L. Collins, M. C. Tartaglia, and Alzheimer’s Disease Neuroimaging Initiative, “White matter hyperintensities and neuropsychiatric symptoms in mild cognitive impairment and Alzheimer’s disease,” NeuroImage: Clin., vol. 28, pp. 102367, 2020. doi: 10.1016/j.nicl.2020.102367. [Google Scholar] [PubMed] [CrossRef]
30. V. Nurdal, A. Wearn, M. Knight, R. Kauppinen, and E. Coulthard, “Prospective memory in prodromal Alzheimer’s disease: Real world relevance and correlations with cortical thickness and hippocampal subfield volumes,” NeuroImage: Clin., vol. 26, pp. 102226, 2020. doi: 10.1016/j.nicl.2020.102226. [Google Scholar] [PubMed] [CrossRef]
31. A. Abrol, M. Bhattarai, A. Fedorov, Y. Du, S. Plis and V. Calhoun, “Deep residual learning for neuroimaging: An application to predict progression to Alzheimer’s disease,” J. Neurosci. Methods, vol. 339, pp. 108701, 2020. doi: 10.1016/j.jneumeth.2020.108701. [Google Scholar] [PubMed] [CrossRef]
32. A. Naseer, M. Rani, S. Naz, M. I. Razzak, M. Imran and G. Xu, “Refining Parkinson’s neurological disorder identification through deep transfer learning,” Neural Comput. Appl., vol. 32, no. 3, pp. 839–854, 2020. doi: 10.1007/s00521-019-04069-0. [Google Scholar] [CrossRef]
33. Z. U. Rehman, M. S. Zia, G. R. Bojja, M. Yaqub, F. Jinchao and K. Arshid, “Texture based localization of a brain tumor from MR-images by using a machine learning approach,” Med. Hypotheses, vol. 141, pp. 109705, 2020. doi: 10.1016/j.mehy.2020.109705. [Google Scholar] [PubMed] [CrossRef]
34. M. A. Khan and Y. Kim, “Cardiac arrhythmia disease classification using LSTM deep learning approach,” Comput. Mater. Contin., vol. 67, no. 1, pp. 427–443, 2021. doi: 10.32604/cmc.2021.014682. [Google Scholar] [CrossRef]
35. J. Jiang, L. Kang, J. Huang, and T. Zhang, “Deep learning based mild cognitive impairment diagnosis using structure MR images,” Neurosci. Lett., vol. 730, pp. 134971, 2020. doi: 10.1016/j.neulet.2020.134971. [Google Scholar] [PubMed] [CrossRef]
36. Y. Gao, L. Xiaoyang, and X. Jiawei, “Fault detection in gears using fault samples enlarged by a combination of numerical simulation and a generative adversarial network,” IEEE/ASME Trans. Mechatron., vol. 27, no. 5, pp. 3798–3805, 2021. doi: 10.1109/TMECH.2021.3132459. [Google Scholar] [CrossRef]
37. S. Murugan, “DEMNET: A deep learning model for early diagnosis of Alzheimer diseases and dementia from MR images,” IEEE Access, vol. 9, pp. 90319–90329, 2021. doi: 10.1109/ACCESS.2021.3090474. [Google Scholar] [CrossRef]
38. M. Dyrba et al., “Improving 3D convolutional neural network comprehensibility via interactive visualization of relevance maps: Evaluation in Alzheimer’s disease,” Alzheimer’s Res. Ther., vol. 13, no. 1, pp. 1–18, 2021. doi: 10.1186/s13195-021-00924-2. [Google Scholar] [PubMed] [CrossRef]
39. C. V. Angkoso, H. P. A. Tjahyaningtijas, M. Purnomo, and I. Purnama, “Multiplane convolutional neural network (Mp-CNN) for Alzheimer’s disease classification,” Int. J. Intell. Eng. Syst., vol. 15, no. 1, pp. 329–340, 2022. [Google Scholar]
40. A. Ebrahimi, S. Luo, and Alzheimer’s Disease Neuroimaging Initiative, “Disease neuroimaging initiative, convolutional neural networks for Alzheimer’s disease detection on MRI images,” J. Med. Imaging, vol. 8, no. 2, pp. 4503, 2021. [Google Scholar]
41. Y. Fu’Adah, I. Wijayanto, N. Pratiwi, F. Taliningsih, S. Rizal and M. Pramudito, “Automated classification of Alzheimer’s disease based on MRI image processing using convolutional neural network (CNN) with alexnet architecture,” J. Phys.: Conf. Series, vol. 1844, no. 1, pp. 12020. [Google Scholar]
42. L. Heising and S. Angelopoulos, “Operationalising fairness in medical AI adoption: Detection of early Alzheimer’s disease with 2D CNN,” BMJ Health Care Inform., vol. 29, no. 1, pp. e100485, 2022. doi: 10.1136/bmjhci-2021-100485. [Google Scholar] [PubMed] [CrossRef]
43. M. Liu, D. Cheng, and W. Yan, “Alzheimer’s disease neuroimaging initiative. Classification of Alzheimer’s disease by combination of convolutional and recurrent neural networks using FDG-PET images,” Front. Neuroinform., vol. 12, pp. 10–3389, 2018. [Google Scholar]
44. R. Khan et al., “A transfer learning approach for multiclass classification of Alzheimer’s disease using MRI images,” Front. Neurosci., vol. 16, pp. 1050777, 2023. doi: 10.3389/fnins.2022.1050777. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF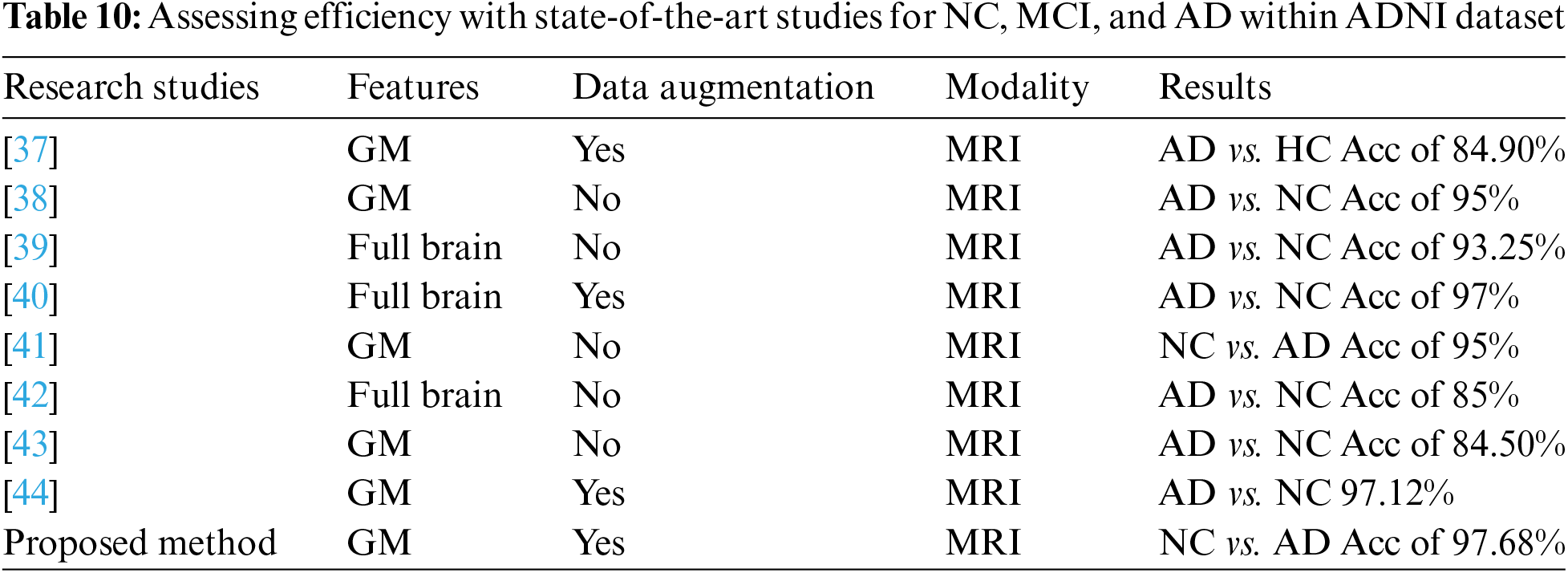
 Downloads
Downloads
 Citation Tools
Citation Tools