 Open Access
Open Access
ARTICLE
Ensemble Approach Combining Deep Residual Networks and BiGRU with Attention Mechanism for Classification of Heart Arrhythmias
1 Department of Information Systems, Al-Farabi Kazakh National University, Almaty, 050040, Kazakhstan
2 Department of Mathematical and Computer Modeling, International Information Technology University, Almaty, 050040, Kazakhstan
3 Department of Software Engineering, Satbayev University, Almaty, 050013, Kazakhstan
* Corresponding Authors: Batyrkhan Omarov. Email: ,
Computers, Materials & Continua 2024, 80(1), 341-359. https://doi.org/10.32604/cmc.2024.052437
Received 07 April 2024; Accepted 03 June 2024; Issue published 18 July 2024
Abstract
This research introduces an innovative ensemble approach, combining Deep Residual Networks (ResNets) and Bidirectional Gated Recurrent Units (BiGRU), augmented with an Attention Mechanism, for the classification of heart arrhythmias. The escalating prevalence of cardiovascular diseases necessitates advanced diagnostic tools to enhance accuracy and efficiency. The model leverages the deep hierarchical feature extraction capabilities of ResNets, which are adept at identifying intricate patterns within electrocardiogram (ECG) data, while BiGRU layers capture the temporal dynamics essential for understanding the sequential nature of ECG signals. The integration of an Attention Mechanism refines the model’s focus on critical segments of ECG data, ensuring a nuanced analysis that highlights the most informative features for arrhythmia classification. Evaluated on a comprehensive dataset of 12-lead ECG recordings, our ensemble model demonstrates superior performance in distinguishing between various types of arrhythmias, with an accuracy of 98.4%, a precision of 98.1%, a recall of 98%, and an F-score of 98%. This novel combination of convolutional and recurrent neural networks, supplemented by attention-driven mechanisms, advances automated ECG analysis, contributing significantly to healthcare’s machine learning applications and presenting a step forward in developing non-invasive, efficient, and reliable tools for early diagnosis and management of heart diseases.Keywords
Nomenclature
| CNN | Convolutional Neural Network |
| BiGRU | Bidirectional Gated Recurrent Unit |
| ResNet | Deep Residual Networks |
| ECG (Electrocardiogram) | A test that measures the electrical activity of the heart and is used to identify various heart conditions |
| Arrhythmia | Irregular heartbeat patterns that can be indicative of various cardiac conditions |
In the realm of biomedical signal processing, the application of advanced machine learning techniques for the analysis of electrocardiograms (ECGs) represents a significant stride towards the enhancement of diagnostic processes, particularly in the domain of cardiovascular diseases (CVDs). Cardiovascular diseases remain the leading cause of mortality globally, underscoring the critical need for efficient, accurate, and early detection methodologies [1]. The electrocardiogram (ECG), a non-invasive test that measures the electrical activity of the heart, is pivotal in the diagnosis of various cardiac abnormalities. However, the interpretation of ECG signals demands a high level of expertise and is often time-consuming, given the complexity and variability of the signals [2].
The advent of deep learning has revolutionized the field of automated ECG analysis by providing tools capable of learning from data directly, without the need for manual feature extraction. Among these, Convolutional Neural Networks (CNNs) have shown exceptional promise in recognizing patterns and anomalies within ECG signals, attributed to their ability to capture spatial hierarchies in data [3,4]. Concurrently, the Bidirectional Gated Recurrent Unit (BiGRU), a variant of recurrent neural networks, has demonstrated its efficacy in capturing temporal dependencies and dynamics in time-series data, making it particularly suited for sequential data like ECGs [5].
However, despite these advancements, the challenge of integrating the spatial and temporal features of ECG signals in a cohesive model that maximizes diagnostic accuracy remains [6]. To address this, the proposed research introduces a novel Hybrid Deep CNN-BiGRU Model augmented with an Attention Mechanism. This model aims to harness the spatial pattern recognition capabilities of CNNs and the temporal dynamic learning of BiGRUs, while the attention mechanism focuses on the most informative parts of the ECG signal, thereby improving the overall accuracy of CVD detection.
The rationale behind combining CNNs with BiGRUs lies in their complementary strengths; while CNNs excel in analyzing visual patterns, BiGRUs are adept at understanding sequence data. The incorporation of an attention mechanism further enhances this synergy by allowing the model to dynamically weigh the importance of different features at various time steps, a critical capability given the inherent variability in ECG signals across different patients and conditions [6]. This approach aligns with the evolving landscape of machine learning in healthcare, where the integration of multiple deep learning architectures to leverage their respective strengths is becoming increasingly prevalent [7].
The significance of this study is manifold. First, it contributes to the burgeoning field of AI-enabled healthcare by presenting a robust framework for ECG analysis that could potentially streamline the diagnostic process for CVDs. Moreover, by improving the accuracy and efficiency of CVD detection, the proposed model holds the promise of facilitating early intervention, thereby reducing the morbidity and mortality associated with cardiovascular diseases [8]. Finally, this research underscores the importance of interdisciplinary collaboration in the development of AI tools for healthcare, combining insights from computer science, biomedical engineering, and clinical cardiology.
The escalating prevalence of cardiovascular diseases globally necessitates advanced diagnostic tools for precise classification and management. This work introduces an innovative ensemble approach that synergizes Deep Residual Networks (ResNets) and Bidirectional Gated Recurrent Units (BiGRU) augmented with an Attention Mechanism. The major contributions of this research include leveraging the hierarchical feature extraction capabilities of ResNets to identify intricate patterns within electrocardiogram (ECG) data, and utilizing BiGRU layers to capture the temporal dynamics essential for understanding the sequential nature of ECG signals. The integration of an Attention Mechanism further refines the model’s focus on critical segments of ECG data, ensuring a nuanced analysis that highlights the most informative features for arrhythmia classification. This combination of convolutional and recurrent neural networks, supplemented by attention-driven mechanisms, advances automated ECG analysis, contributing significantly to the development of non-invasive, efficient, and reliable tools for early diagnosis and management of heart diseases.
The exploration of automated systems for the detection and diagnosis of cardiovascular diseases (CVDs) using electrocardiogram (ECG) data has been an area of intensive research over the past decade. The utility of ECGs in diagnosing various cardiac conditions, coupled with advancements in machine learning (ML) and deep learning (DL), has paved the way for significant breakthroughs in this field [9]. This section reviews the related work, focusing on the development and application of various ML and DL models for ECG analysis, highlighting the evolution towards hybrid models and the incorporation of attention mechanisms.
Early efforts in automated ECG analysis predominantly relied on traditional machine learning algorithms, which required the manual extraction of features based on domain knowledge. For instance, Abubaker (2022) [10] employed Support Vector Machines (SVM) for classifying arrhythmias, demonstrating the potential of ML in identifying specific patterns within ECG signals. Similarly, decision trees, k-nearest neighbors (KNN), and linear discriminant analysis (LDA) have been utilized, each presenting a unique approach to the classification and interpretation of cardiac signals [11].
The advent of deep learning offered a paradigm shift in ECG analysis by eliminating the need for manual feature extraction. Convolutional Neural Networks (CNNs) emerged as a powerful tool for this purpose, given their proficiency in handling spatial data. Zhang et al. showcased the application of CNNs in detecting myocardial infarction, achieving remarkable accuracy rates [12]. Moreover, Recurrent Neural Networks (RNNs), and specifically Long Short-Term Memory (LSTM) networks, were explored for their ability to capture temporal dependencies in ECG data, further enhancing diagnostic capabilities [13].
Despite these advancements, individual models often faced limitations in capturing both spatial and temporal features comprehensively. This gap led to the exploration of hybrid models. For example, Din et al. introduced a combination of CNN and LSTM architectures, leveraging the spatial pattern recognition of CNNs with the sequence learning capability of LSTMs [14]. This hybrid approach marked a significant step towards a more holistic analysis of ECG signals, revealing the complexities of cardiac conditions more effectively.
The integration of Bidirectional Gated Recurrent Unit (BiGRU) models with CNNs further refined the analytical prowess of these hybrid systems. BiGRUs, known for their efficiency in processing time-series data, offered an improved mechanism for understanding the temporal dynamics of ECG signals. Cheng et al. demonstrated the superiority of CNN-BiGRU hybrids over their single-model counterparts, attributing this to the synergistic effect of combining spatial and temporal analysis [15].
The addition of attention mechanisms to these hybrid models introduced a new dimension to ECG signal analysis. Attention mechanisms allow models to focus on the most salient parts of the data, enhancing interpretability and accuracy. Sun presented an innovative CNN-LSTM model equipped with an attention mechanism, showcasing improved performance in detecting arrhythmic events [16]. This model underscored the potential of attention-based models in identifying subtle, yet clinically significant, features within ECG recordings.
Recent studies have further expanded on this foundation, exploring various configurations and applications of hybrid DL models. Liu et al. [17] and Oleiwi et al. [18] investigated the use of multi-channel ECG data, employing CNN-BiGRU architectures to enhance diagnostic precision across a broader spectrum of cardiovascular conditions. Furthermore, the integration of generative adversarial networks (GANs) with hybrid models has been explored for data augmentation, addressing the challenge of limited labeled ECG data in training high-performing models [19].
The significance of these developments cannot be overstated. The capacity to accurately and efficiently diagnose CVDs using ECG data holds profound implications for patient care, promising early detection and intervention. Moreover, the evolution towards models that combine the strengths of CNNs, RNNs (including LSTMs and BiGRUs), and attention mechanisms reflects a broader trend in AI research towards creating more nuanced, adaptive, and effective analytical tools [20,21].
Thus, the landscape of ECG analysis for CVD detection is rapidly advancing, driven by innovations in machine learning and deep learning. The trajectory from traditional ML algorithms to sophisticated hybrid DL models equipped with attention mechanisms illustrates a commitment to improving diagnostic accuracy and patient outcomes. As this field continues to evolve, it is anticipated that future research will further refine these models, enhancing their applicability and effectiveness in clinical settings.
Deep Learning Paradigm Shift: The emergence of Convolutional Neural Networks (CNNs) and Recurrent Neural Networks (RNNs) provided a significant leap forward in ECG analysis, eliminating the need for manual feature extraction and enabling automated recognition of spatial and temporal patterns. Hybrid models, combining CNNs with Long Short-Term Memory (LSTM) or Bidirectional Gated Recurrent Units (BiGRU), further refined this approach, capturing both spatial and temporal dynamics comprehensively.
Attention Mechanisms: The incorporation of attention mechanisms into hybrid models has added a new dimension to ECG analysis. This allows models to focus on the most salient parts of the data, enhancing interpretability and accuracy. Islam et al. showcased this with a CNN-LSTM model equipped with an attention mechanism, improving performance in detecting arrhythmic events [22].
Data Augmentation with GANs: To address the challenge of limited labeled ECG data, generative adversarial networks (GANs) have been integrated into hybrid models for data augmentation. This approach has helped expand the training datasets available to models, further improving their diagnostic precision.
Comprehensive Analysis: While hybrid models have advanced significantly, individual models still face limitations in comprehensively capturing both spatial and temporal features. Further refinements in hybrid models are needed to ensure a more holistic analysis of ECG signals.
Multi-Channel Data: The exploration of multi-channel ECG data remains an evolving area. While models such as CNN-BiGRU architectures have been employed to enhance diagnostic precision across a broader spectrum of cardiovascular conditions, more work is needed to harness the full potential of multi-channel ECG data.
Clinical Applicability: Although these advancements have shown promising results in research settings, there is a need to further develop these models to enhance their applicability and effectiveness in clinical settings. This includes ensuring their robustness and adaptability across diverse patient populations.
In conclusion, the landscape of ECG analysis for CVD detection is rapidly advancing, driven by innovations in machine learning and deep learning. The trajectory from traditional ML algorithms to sophisticated hybrid DL models equipped with attention mechanisms illustrates a commitment to improving diagnostic accuracy and patient outcomes. Future research will likely refine these models further, enhancing their applicability and effectiveness in clinical settings.
The proposed approach is structured around a sequence of fundamental phases: beginning with data setup, followed by mitigating class imbalance through data augmentation, identifying key features, and culminating in the classification of cardiovascular conditions, as illustrated in Fig. 1. The methodology initiates with an exhaustive integration of the MIT-BIH Arrhythmia database [23]. This step is succeeded by data preprocessing measures, chiefly aimed at eliminating noise from the ECG recordings. In response to the challenge presented by unequal class distributions, augmentation methods are implemented to enhance the presence of less represented classes. Following this, a process of feature extraction is undertaken, making use of a library dedicated to the extraction of features from time-series data to produce vectors that encapsulate essential attributes for the subsequent classification phase. With these preparatory activities concluded, our innovative ensemble CNN-BiLSTM model is applied to the task of classification. In this final stage, the model evaluates the feature vectors to determine the posterior probabilities associated with different classes, effectively discerning the distinct types of cardiac rhythms. The methodologies employed for feature extraction and classification will be elaborated on in the sections that follow.
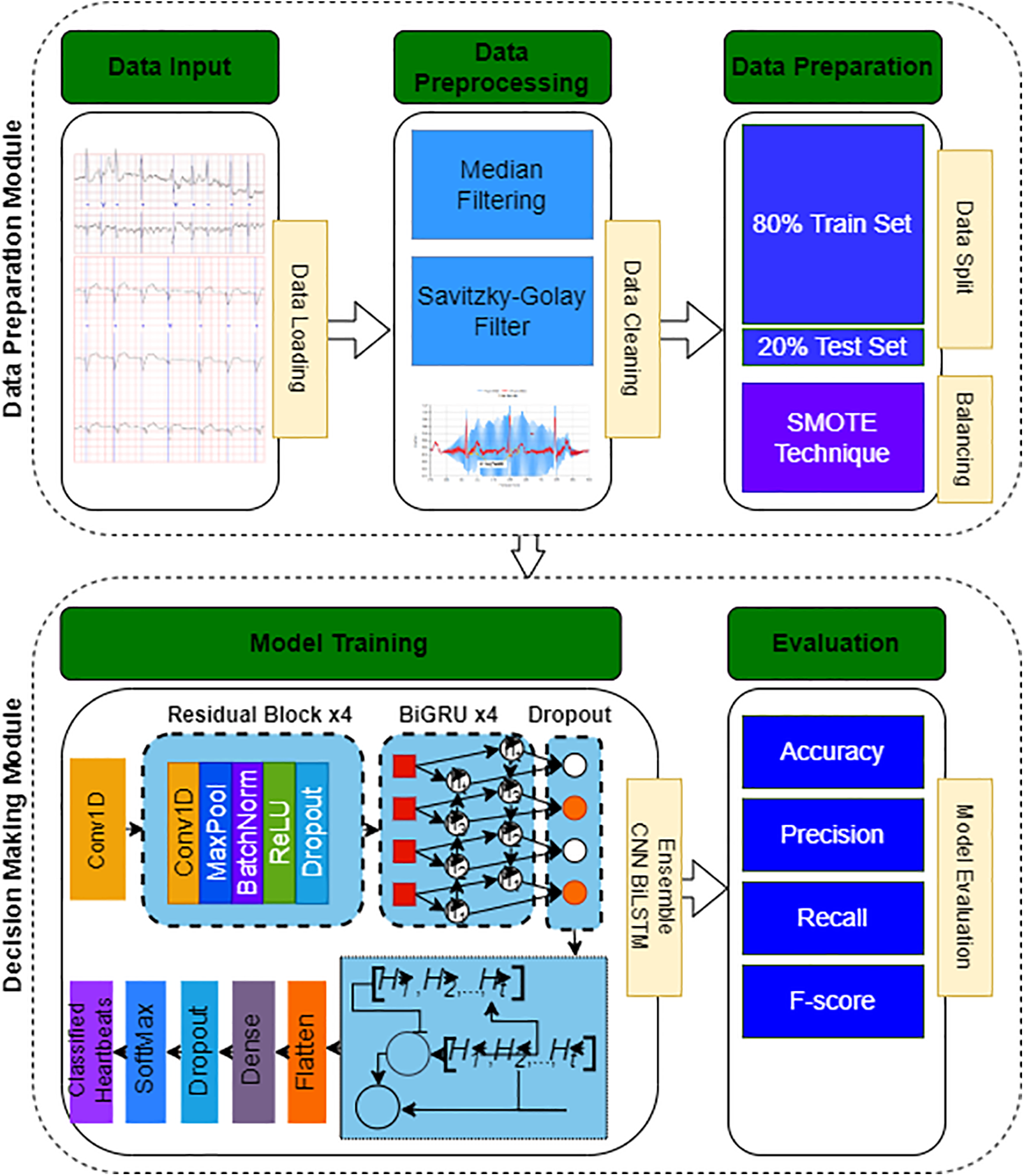
Figure 1: Flowchart of the proposed system
For our study, the MIT-BIH Arrhythmia database was utilized throughout the training and evaluation stages. Renowned in cardiac research, this database contains detailed ECG data from 47 subjects, providing 48 half-hour recordings obtained using lead II and V1 electrode positions. The data, sampled at 360 Hz and digitized with an 11-bit resolution over a 10 mV range, encompasses 109,443 heartbeats classified into five categories as per AAMI standards: Normal Beat, Supraventricular Ectopic Beat, Ventricular Ectopic Beat, Fusion Beat, and Unknown Beat. The dataset is split into training (80%) and testing (20%) segments to facilitate a comprehensive analysis across different heartbeat types.
In our study, preprocessing is essential for enhancing the quality of ECG signals by removing various noises, including muscle artifacts, motion-related disturbances, electrical noise from external sources, baseline shifts, and recording device noise [24]. We adopted two preprocessing methods to combat baseline wander and high-frequency noise. Baseline drift is reduced through median filtering with windows of 200 and 600 ms. High-frequency noise is mitigated using a Savitzky-Golay filter [25], as shown in Fig. 2. To segment the ECG signals into individual heartbeats, we utilize the Pan-Tompkins algorithm [26], known for its effectiveness in detecting R-peaks, with the MIT-BIH Arrhythmia database providing the necessary annotations. Each heartbeat is represented by a 500 ms window, allowing for consistent data analysis and interpretation.
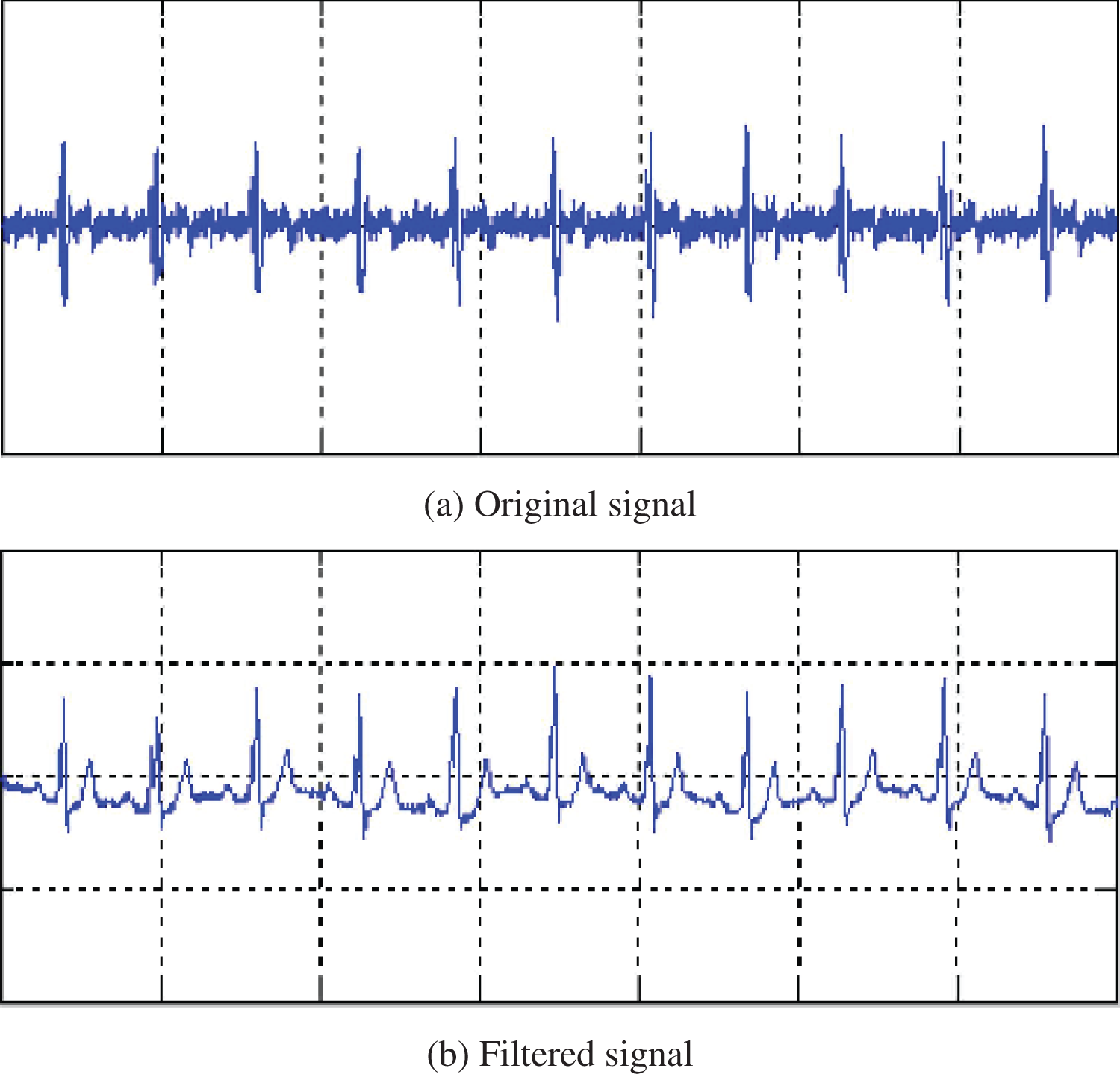
Figure 2: Original and filtered ECG
In the data preparation phase of our study, we meticulously divided the dataset into training and testing subsets, allocating 80% for training purposes and reserving 20% for evaluation. This strategic partitioning ensures comprehensive exposure to diverse data patterns during model training, while also setting aside a representative sample for rigorous testing. To address the inherent challenge of class imbalance within the dataset, we implemented the Synthetic Minority Over-sampling Technique (SMOTE). SMOTE algorithmically generates synthetic samples from the minority class, thereby equalizing the distribution across different classes. This is achieved by interpolating between neighboring examples, effectively augmenting the minority class to match the prevalence of the majority class. The formula for creating a synthetic sample
where
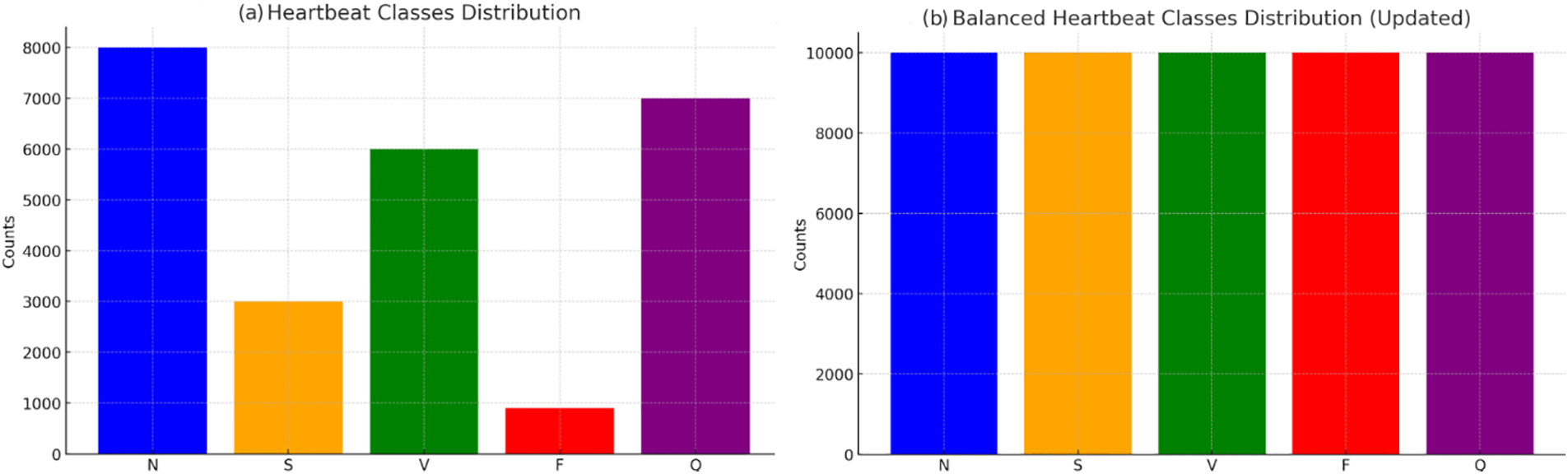
Figure 3: Applying SMOTE data balancing technique
In the feature extraction stage of our methodology, we employed the Time Series Feature Extraction Library (TSFEL) to systematically distill relevant features from the ECG signals. TSFEL is designed to automate the extraction of a comprehensive set of features, enabling the efficient analysis of time-series data. It operates by calculating a diverse array of statistical, temporal, and spectral features, thereby providing a multidimensional representation of the underlying characteristics of the ECG signals. This approach facilitates the identification of distinctive patterns and anomalies associated with various cardiovascular conditions. By leveraging TSFEL, we efficiently condensed the raw ECG data into a feature set that captures the essential dynamics and intricacies of the heart’s electrical activity. The extracted features serve as the input for the subsequent machine learning models, significantly enhancing their ability to classify and predict different types of cardiac events with high accuracy.
4 Proposed CNN-BiGRU Model with Attention Mechanism
The proposed CNN-BiGRU model with an attention mechanism is designed to leverage both convolutional and recurrent neural networks for effective feature extraction and sequence learning from ECG data, culminating in the classification of heartbeat types. This model architecture seamlessly integrates convolutional blocks for spatial feature extraction, BiGRU layers for capturing temporal dynamics, and an attention mechanism to emphasize salient features, leading to a fully connected layer that drives the classification process. Fig. 4 demonstrates the proposed model for ECG classification. In next sections, we outline the model components and the associated mathematical framework.
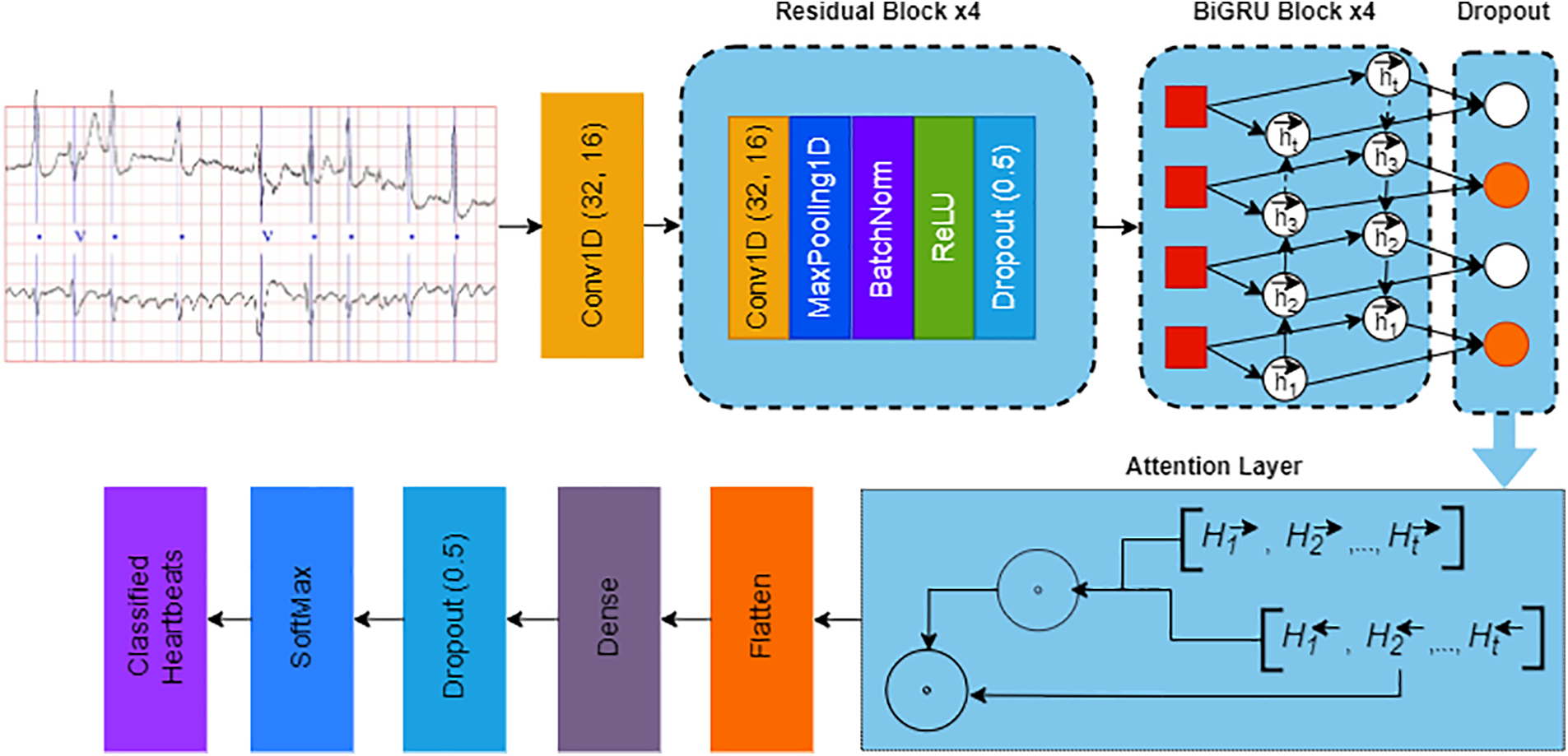
Figure 4: Architecture of the proposed CNN-BiGRU network with attention mechanism
The model’s input comprises electrocardiogram (ECG) data, encapsulated as sequences capturing the heart’s electrical activity over time. These sequences represent the foundational data from which the model discerns patterns indicative of various cardiac conditions. The input to the model consists of ECG data, represented as:
where
The Convolutional Block is designed to extract spatial features from the input data. It comprises Conv1D layers, residual blocks, and dropout for regularization, iterated four times. The Conv1D layer can be described as:
where
The BiGRU layer in the proposed network architecture plays a crucial role in capturing the temporal dynamics and dependencies within the ECG data, following the feature extraction performed by the convolutional blocks. A Bidirectional Gated Recurrent Unit (BiGRU) combines two GRU networks that process the data in opposite directions (forward and backward), allowing the model to incorporate information from both past and future contexts relative to a given time step.
A single GRU cell updates its hidden state
Update gate:
Reset gate:
where
Candidate Hidden State:
The candidate hidden state
Final Hidden State Update:
Finally, the hidden state is updated by interpolating between the old state and the candidate state, weighted by the update gate:
Bidirectional Processing
The BiGRU layer consists of two GRU networks that process the input sequence in opposite directions: one forward (
Forward hidden states:
Backward hidden states:
The final output at each time step t is the concatenation of the forward and backward hidden states:
where ⊕ denotes concatenation.
This bidirectional processing allows the network to utilize information from both the past and the future when making decisions at any given point in the sequence, enhancing its ability to capture the temporal dependencies within the ECG data effectively.
The attention mechanism weights the importance of each time step’s features:
The context vector is then flattened and passed through dense layers and a dropout layer:
Finally, the SoftMax layer outputs the probability distribution over the heartbeat classes:
where
In summary, the proposed CNN-BiGRU model with an attention mechanism represents a sophisticated approach to ECG signal analysis, optimizing the extraction and processing of both spatial and temporal features, ultimately enhancing the accuracy of heartbeat classification.
In this research, we implemented a split of training and testing data to evaluate the performance of our model using the MIT-BIH Arrhythmia Dataset. We selected 800 ECG recordings at random for the training phase, with the remaining data allocated for validation of the model’s precision. Performance was assessed through various indicators. Accuracy was determined by the ratio of accurately classified instances to the overall count. To address class imbalance, we applied the F-score, integrating precision and recall for a balanced assessment of accuracy. Precision quantifies the accuracy of positive predictions, while recall measures the model’s capacity to capture all positive instances [27–29].
Understanding the performance metrics involves recognizing true positives (TP), true negatives (TN), false positives (FP), and false negatives (FN), which depict the accurate and erroneous classifications by the model.
The graph in Fig. 5 represents the progression of performance across 80 epochs in the proposed model’s training phase. The results of this study underscore the efficacy of the proposed CNN-BiGRU model with an attention mechanism in analyzing ECG data for the detection of cardiovascular diseases. The incremental improvement in both training and validation accuracies over 80 epochs illustrates the model’s capacity to learn complex patterns inherent in ECG signals. Notably, the convergence of training and validation accuracies, with a final training accuracy of 96.5% and validation accuracy of 95.9%, signifies a robust generalization ability, minimizing the risk of overfitting. The employment of SMOTE for class balancing and TSFEL for feature extraction further contributed to the model’s performance. These results, particularly the high validation accuracy, affirm the potential of deep learning architectures in enhancing diagnostic procedures for cardiovascular conditions, promising significant implications for automated and precise ECG analysis in clinical settings.
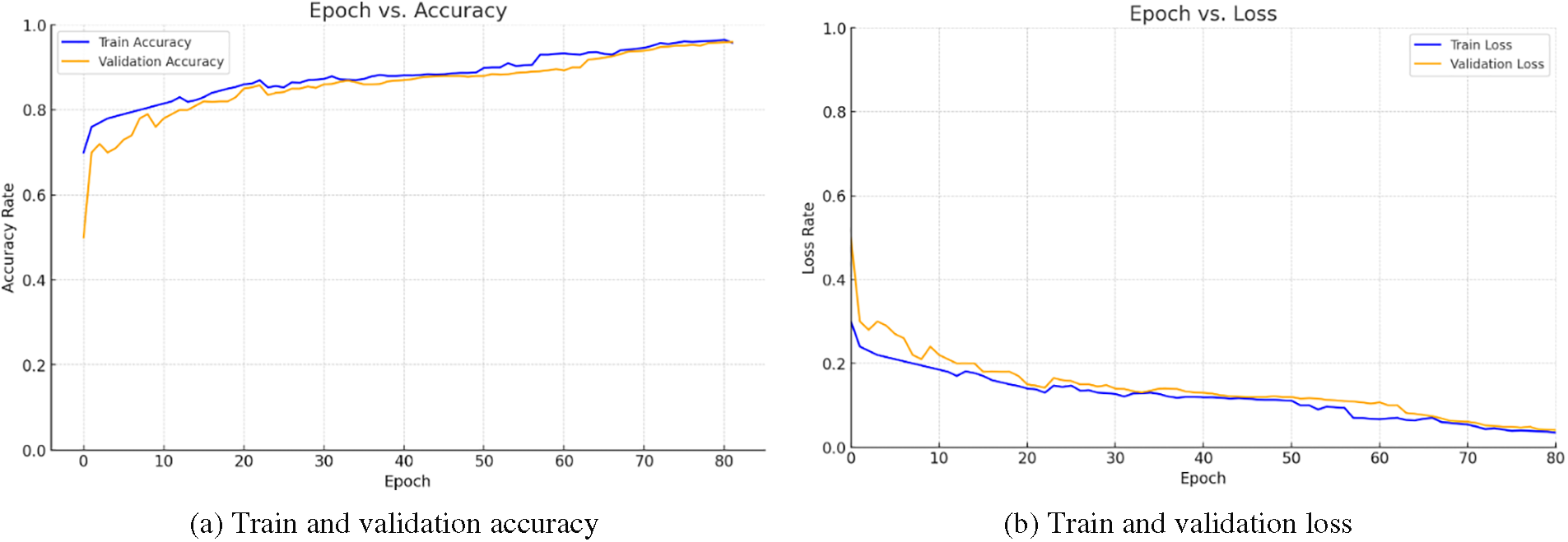
Figure 5: Train and validation accuracy of the proposed model in 80 learning epochs
Fig. 5b delineates a compelling narrative of progressive model refinement, as evidenced by the downward trajectories of both training and validation loss across 80 epochs. The consistent decrease in loss rates underscores the model’s adeptness at learning from the ECG data, refining its predictions with each epoch. The convergence of training and validation loss suggests a robust model that generalizes well to new, unseen data, thereby mitigating the risk of overfitting. This trend is indicative of the model’s effectiveness in detecting cardiac anomalies, validating the potential of the proposed CNN-BiGRU architecture with an attention mechanism in advancing cardiovascular disease diagnostics. The results underscore the model’s capacity for precise interpretation of complex ECG signals, a critical step toward enhancing clinical outcomes through early and accurate detection.
Fig. 6 presents a comprehensive evaluation of the proposed CNN-BiGRU model’s performance across multiple ECG leads, depicting confusion matrices for leads I to V6. These matrices highlight the model’s adeptness in accurately classifying heartbeats into five distinct categories: Normal (N), Supraventricular Ectopic (S), Ventricular Ectopic (V), Fusion (F), and Unknown (Q). The predominance of high values along the diagonal for each matrix reflects the model’s commendable accuracy and reliability in diagnosing various cardiac conditions. Notably, the matrices also reveal some misclassifications, particularly between closely related heartbeat types, suggesting areas for potential refinement. Overall, the consistent performance across different leads validates the model’s robustness and its applicability in clinical settings for enhancing cardiovascular disease detection and diagnosis.

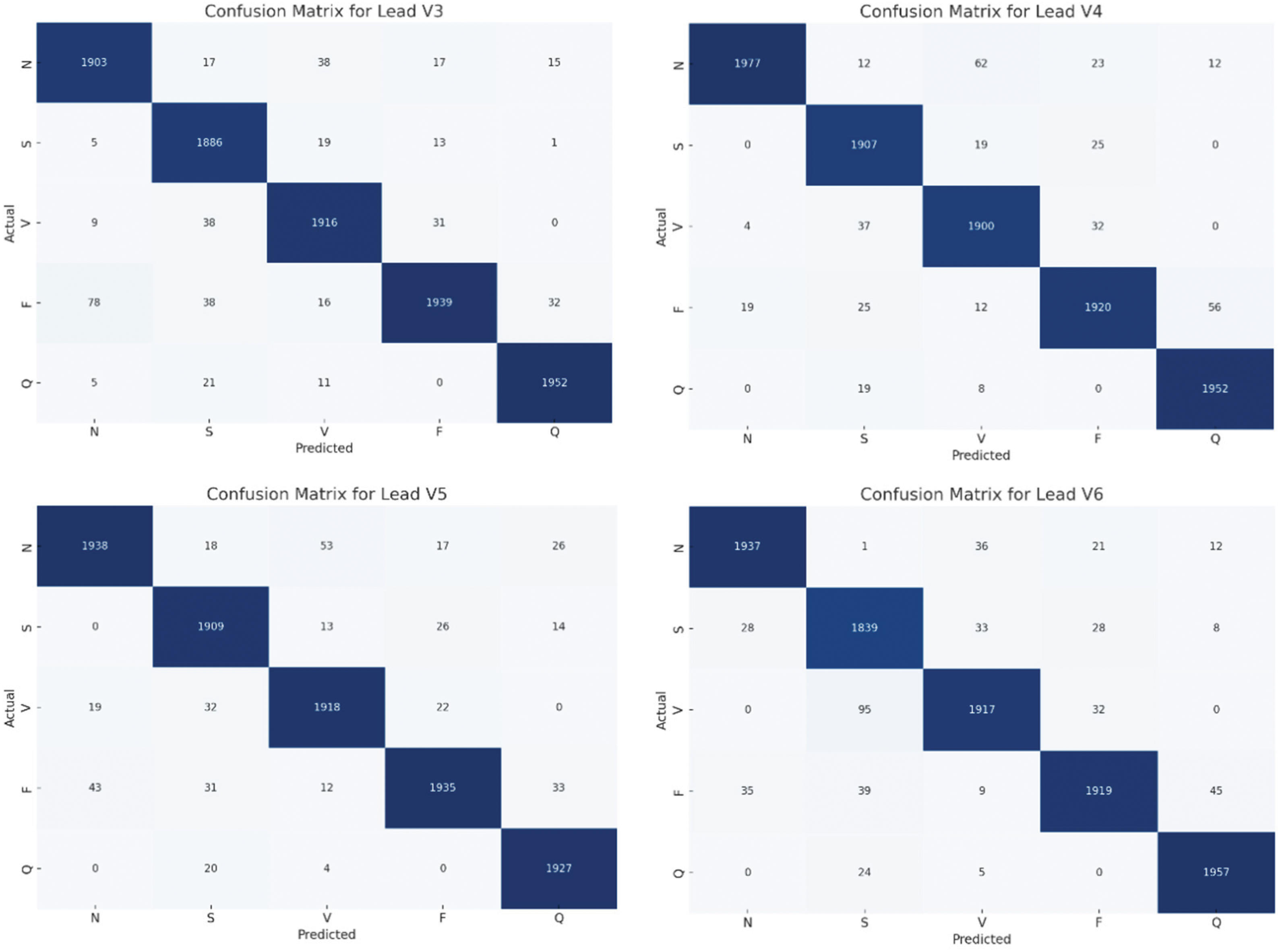
Figure 6: Confusion matrices of the proposed model for each lead
Fig. 7 exhibits the classification prowess of the proposed model through a probabilistic confusion matrix, delineating the predictive accuracies for five heartbeat categories: Normal (N), Supraventricular Ectopic (S), Ventricular Ectopic (V), Fusion (F), and Unknown (Q). Notably, the model achieves exceptional classification precision, with probabilities of correct predictions predominately above 0.9 for all categories, signifying a robust diagnostic capability. The minor probabilities of misclassification, notably for Supraventricular beats, reflect the inherent challenge in distinguishing between certain heartbeat types yet underscore the model’s substantial accuracy. This visualization encapsulates the model’s efficacy in discerning and categorizing ECG signals, emphasizing its significant potential in enhancing cardiac anomaly detection and contributing to the advancement of automated, precision-driven cardiovascular healthcare solutions.
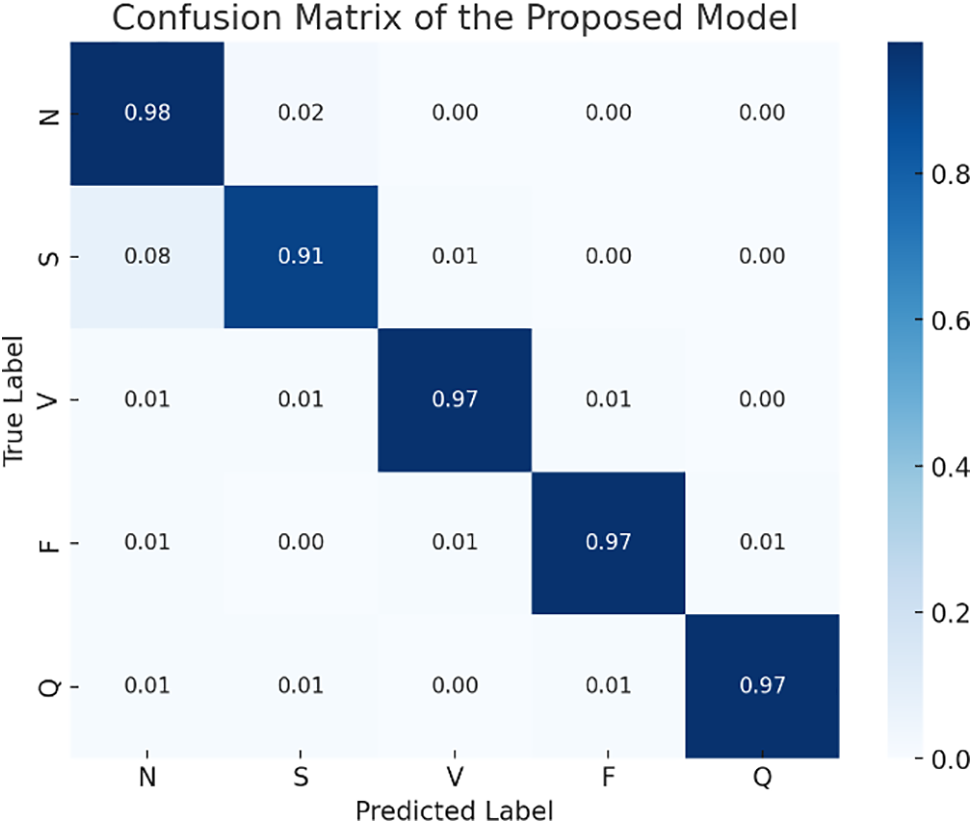
Figure 7: Confusion matrix of the proposed model in 12-lead ECG classification
Fig. 8, titled “Performance Results,” graphically delineates the accuracy metrics across three datasets: training, validation, and test, for five distinct heartbeat categories (N, S, V, F, Q). The bar chart reveals that the training accuracy (blue bars) consistently surpasses 98% across all categories, indicating the model’s profound learning capability. Validation accuracy (intense orange bars) and test accuracy (green bars) also exhibit high levels, mostly hovering around the 98% mark, albeit with a slight dip in category V, where they approach 96%. These results underscore the model’s exceptional generalization ability, affirming its potential as a reliable tool for cardiovascular disease diagnosis. The minimal discrepancy between training, validation, and test accuracies suggests that the model maintains its robustness and efficacy across unseen data, essential qualities for practical clinical application.

Figure 8: Performance results of the proposed CNN-BiGRU network with attention mechanism
Table 1 provides a comparative analysis of the proposed Ensemble CNN-BiLSTM Network against recent state-of-the-art studies in ECG analysis. The proposed model, evaluated on the MIT-BIH Arrhythmia database, achieves notable metrics with an accuracy of 98.4%, precision of 98.1%, recall of 98%, and an F-score of 98%, positioning it at the forefront of ECG diagnostic methodologies. This performance surpasses that of other recent approaches, such as the ensemble deep model by Kokubo et al. and the multiscale joint recurrence quantification analysis by Sun et al., which reported lower accuracies on different datasets. Notably, traditional models like the Naïve Bayes and Radial Basis Functions by Srinivasan et al. demonstrate competitive results but fall short of the proposed model’s benchmarks. The comparison underlines the proposed model’s superior ability to accurately diagnose cardiac conditions, affirming its potential for enhancing ECG-based diagnostics in clinical settings.
This research paper culminated in the development and validation of an Ensemble CNN-BiLSTM Network, showcasing exemplary diagnostic performance in the context of ECG analysis for cardiovascular disease detection. Employing the MIT-BIH Arrhythmia database, our proposed model achieved an impressive accuracy of 98.4%, alongside notable precision, recall, and F-score metrics, unequivocally establishing its superiority over current state-of-the-art methods. Such achievements not only highlight the robustness and efficacy of integrating convolutional and bidirectional long short-term memory networks but also underscore the critical role of advanced deep learning architectures in refining medical diagnostics. Comparative analysis with contemporaneous studies further validated our model’s advanced capabilities, outperforming other approaches with significant margins. This research advances the frontier of ECG interpretation technology, offering a promising tool for healthcare professionals to enhance the accuracy and efficiency of cardiovascular disease diagnosis. Beyond the immediate technical contributions, this work paves the way for future investigations into deep learning applications within medical imaging and diagnostics, encouraging the exploration of more complex models and diverse datasets. As we move forward, the potential integration of such AI-driven tools in clinical settings could dramatically transform patient care, enabling early detection, personalized treatment plans, and ultimately improving patient outcomes. This study serves as a foundational step towards realizing the full potential of AI in healthcare, emphasizing the importance of interdisciplinary collaboration to address the complexities of medical diagnostics.
Acknowledgement: Not applicable.
Funding Statement: This work was supported by the research project—Application of Machine Learning Methods for Early Diagnosis of Pathologies of the Cardiovascular System funded by the Ministry of Science and Higher Education of the Republic of Kazakhstan. Grant No. IRN AP13068289. The supervisor of the project is Batyrkhan Omarov.
Author Contributions: Conceptualization, Batyrkhan Omarov; methodology, Batyrkhan Omarov, Meirzhan Baikuvekov, and Daniyar Sultan; software, Daniyar Sultan; data curation, Batyrkhan Omarov; writing—original draft preparation, Batyrkhan Omarov, and Meirzhan Baikuvekov; writing—review and editing, Batyrkhan Omarov; supervision, Batyrkhan Omarov; Discussion, Maigul Zhekambayeva, Madina Suleimenova, and Nurzhan Mukazhanov. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: This research employed the MIT-BIH dataset, obtainable through the academic source cited as https://www.kaggle.com/datasets/mondejar/mitbih-database (accessed on 01/05/2024).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. M. Ferrell et al., “A terminal metabolite of niacin promotes vascular inflammation and contributes to cardiovascular disease risk,” Nat. Med., vol. 30, no. 2, pp. 424–434, 2024. doi: 10.1038/s41591-023-02793-8. [Google Scholar] [PubMed] [CrossRef]
2. M. Naz, J. Shah, M. Khan, M. Sharif, M. Raza and R. Damaševičius, “From ECG signals to images: A transformation based approach for deep learning,” PeerJ Comput. Sci., vol. 7, no. 1, pp. 1–18, 2021. doi: 10.7717/peerj-cs.386. [Google Scholar] [PubMed] [CrossRef]
3. A. Shoughi, M. Dowlatshahi, A. Amiri, M. Rafsanjani, and R. Batth, “Automatic ECG classification using discrete wavelet transform and one-dimensional convolutional neural network,” Computing, vol. 106, no. 4, pp. 1227–1248, 2023. doi: 10.1007/s00607-023-01243-0. [Google Scholar] [CrossRef]
4. H. Rai and K. Chatterjee, “Hybrid CNN-LSTM deep learning model and ensemble technique for automatic detection of myocardial infarction using big ECG data,” Appl. Intell., vol. 52, no. 5, pp. 5366–5384, 2022. doi: 10.1007/s10489-021-02696-6. [Google Scholar] [CrossRef]
5. S. Dhyani, A. Kumar, and S. Choudhury, “Arrhythmia disease classification utilizing ResRNN,” Biomed. Signal Process. Control, vol. 79, no. 13, pp. 104160, 2023. doi: 10.1016/j.bspc.2022.104160. [Google Scholar] [CrossRef]
6. Y. Jin et al., “A novel attentional deep neural network-based assessment method for ECG quality,” Biomed. Signal Process. Control, vol. 79, no. 6, pp. 104064, Jan. 2023. doi: 10.1016/j.bspc.2022.104064. [Google Scholar] [CrossRef]
7. E. Elsedimy, S. AboHashish, and F. Algarni, “New cardiovascular disease prediction approach using support vector machine and quantum-behaved particle swarm optimization,” Multimed. Tools Appl., vol. 83, no. 8, pp. 23901–23928, 2023. doi: 10.1007/s11042-023-16194-z. [Google Scholar] [CrossRef]
8. S. Dhyani, A. Kumar, and S. Choudhury, “Analysis of ECG-based arrhythmia detection system using machine learning,” MethodsX, vol. 10, no. 10, pp. 102195, 2023. doi: 10.1016/j.mex.2023.102195. [Google Scholar] [PubMed] [CrossRef]
9. B. Omarov et al., “Artificial intelligence in medicine: Real time electronic stethoscope for heart diseases detection,” Comput. Mater. Contin., vol. 70, no. 2, pp. 2815–2833, 2022. doi: 10.32604/cmc.2022.019246. [Google Scholar] [CrossRef]
10. M. Abubaker, “Detection of cardiovascular diseases in ECG images using machine learning and deep learning methods,” IEEE Trans. Artif. Intell., pp. 1, 2022. doi: 10.1109/tai.2022.3159505. [Google Scholar] [CrossRef]
11. N. Salari et al., “Detection of sleep apnea using machine learning algorithms based on ECG signals: A comprehensive systematic review,” Expert Syst. Appl., vol. 187, no. 2, pp. 115950, Jan. 2022. doi: 10.1016/j.eswa.2021.115950. [Google Scholar] [CrossRef]
12. Y. Zhang, S. Liu, Z. He, Y. Zhang, and C. Wang, “A CNN model for cardiac arrhythmias classification based on individual ECG signals,” Cardiovasc. Eng. Technol., Jan. 2022. doi: 10.1007/s13239-021-00599-8. [Google Scholar] [PubMed] [CrossRef]
13. C. Song, Z. Zhou, Y. Yu, M. Shi, and J. Zhang, “An improved Bi-LSTM method based on heterogeneous features fusion and attention mechanism for ECG recognition,” Comput. Biol. Med., vol. 169, pp. 107903, Feb. 2024. doi: 10.1016/j.compbiomed.2023.107903. [Google Scholar] [PubMed] [CrossRef]
14. S. Din, M. Qaraqe, O. Mourad, K. Qaraqe, and E. Serpedin, “ECG-based cardiac arrhythmias detection through ensemble learning and fusion of deep spatial-temporal and long-range dependency features,” Artif. Intell. Med., vol. 150, no. 25, pp. 102818, Feb. 2024. doi: 10.1016/j.artmed.2024.102818. [Google Scholar] [PubMed] [CrossRef]
15. Y. Cheng, D. Li, D. Wang, Y. Chen, and L. Wang, “Multi-label arrhythmia classification using 12-lead ECG based on lead feature guide network,” Eng. Appl. Artif. Intell., vol. 129, no. 8, pp. 107599, Mar. 2024. doi: 10.1016/j.engappai.2023.107599. [Google Scholar] [CrossRef]
16. J. Sun, “Automatic cardiac arrhythmias classification using CNN and attention-based RNN network,” Healthc. Technol. Lett., vol. 10, no. 3, pp. 53–61, Apr. 2023. doi: 10.1049/htl2.12045. [Google Scholar] [PubMed] [CrossRef]
17. K. Liu, T. Liu, D. Wen, M. Zang, S. Zhou and C. Liu, “SRTNet: Scanning, reading, and thinking network for myocardial infarction detection and localization,” Expert Syst. Appl., vol. 240, no. 10, pp. 122402, Apr. 2024. doi: 10.1016/j.eswa.2023.122402. [Google Scholar] [CrossRef]
18. Z. C. Oleiwi, E. N. AlShemmary, and S. Al-Augby, “Developing hybrid CNN-GRU arrhythmia prediction models using fast fourier transform on imbalanced ECG datasets, mathematical modelling of engineering problems,” Math. Model. Eng. Probl., vol. 11, no. 2, pp. 413–429, Feb. 2024. doi: 10.18280/mmep.110213. [Google Scholar] [CrossRef]
19. Z. Wang, S. Stavrakis, and B. Yao, “Hierarchical deep learning with generative adversarial network for automatic cardiac diagnosis from ECG signals,” Comput. Biol. Med., vol. 155, no. 8, pp. 106641, Mar. 2023. doi: 10.1016/j.compbiomed.2023.106641. [Google Scholar] [PubMed] [CrossRef]
20. Y. Wang, G. Yang, S. Li, Y. Li, L. He and D. Liu, “Arrhythmia classification algorithm based on multi-head self-attention mechanism,” Biomed. Signal Process. Control, vol. 79, no. 21, pp. 104206, Jan. 2023. doi: 10.1016/j.bspc.2022.104206. [Google Scholar] [CrossRef]
21. T. Wang, J. Sun, and Q. Zhao, “Investigating cardiotoxicity related with hERG channel blockers using molecular fingerprints and graph attention mechanism,” Comput. Biol. Med., vol. 153, pp. 106464, Feb. 2023. doi: 10.1016/j.compbiomed.2022.106464. [Google Scholar] [PubMed] [CrossRef]
22. M. Islam et al., “HARDC: A novel ECG-based heartbeat classification method to detect arrhythmia using hierarchical attention based dual structured RNN with dilated CNN,” Neural Netw., vol. 162, no. 4, pp. 271–287, 2023. doi: 10.1016/j.neunet.2023.03.004. [Google Scholar] [PubMed] [CrossRef]
23. G. B. Moody and R. G. Mark, “The impact of the MIT-BIH Arrhythmia Database,” IEEE Eng. Med. Biol. J., vol. 20, no. 3, pp. 45–50, 2001. doi: 10.1109/51.932724. [Google Scholar] [PubMed] [CrossRef]
24. J. Li, I. Tobore, Y. Liu, A. Kandwal, and L. Wang, “Non-invasive monitoring of three glucose ranges based on ECG by using DBSCAN-CNN,” IEEE J. Biomed. Health Inform., vol. 25, no. 9, pp. 3340–3350, Sep. 2021. doi: 10.1109/JBHI.2021.3072628. [Google Scholar] [PubMed] [CrossRef]
25. A. John, J. Sadasivan, and C. S. Seelamantula, “Adaptive Savitzky-Golay filtering in non-gaussian noise,” IEEE Trans. Signal Process, vol. 69, pp. 5021–5036, 2021. doi: 10.1109/TSP.2021.3106450. [Google Scholar] [CrossRef]
26. E. Y. Abd Al-Jabbar, M. M. Mohamedsheet Al-Hatab, M. A. Qasim, W. R. Fathel, and M. A. Fadhil, “Clinical fusion for real-time complex QRS pattern detection in wearable ECG using the Pan-Tompkins algorithm,” vol. 12, no. 2, pp. 172–184, Jan. 2023. doi: 10.54216/FPA.120214. [Google Scholar] [CrossRef]
27. K. C. Siontis, P. A. Noseworthy, Z. I. Attia, and P. A. Friedman, “Artificial intelligence-enhanced electrocardiography in cardiovascular disease management,” Nat. Rev. Cardiol., vol. 18, no. 7, pp. 465–478, Feb. 2021. doi: 10.1038/s41569-020-00503-2. [Google Scholar] [PubMed] [CrossRef]
28. S. K. Saini and R. Gupta, “Artificial intelligence methods for analysis of electrocardiogram signals for cardiac abnormalities: State-of-the-art and future challenges,” Artif. Intell. Rev., vol. 55, no. 2, pp. 1519–1565, Apr. 2021. doi: 10.1007/s10462-021-09999-7. [Google Scholar] [CrossRef]
29. K. Nezamabadi, N. Sardaripour, B. Haghi, and M. Forouzanfar, “Unsupervised ECG analysis: A review,” IEEE Rev. Bio. Eng., vol. 16, pp. 208–224, 2023. doi: 10.1109/RBME.2022.3154893. [Google Scholar] [PubMed] [CrossRef]
30. A. Rath, D. Mishra, G. Panda, S. C. Satapathy, and K. Xia, “Improved heart disease detection from ECG signal using deep learning based ensemble model,” Sustain. Comput.: Inform. Syst., vol. 35, no. 4, pp. 100732, Sep. 2022. doi: 10.1016/j.suscom.2022.100732. [Google Scholar] [CrossRef]
31. W. S. Admass, and G. A. Bogale, “Arrhythmia classification using ECG signal: A meta-heuristic improvement of optimal weighted feature integration and attention-based hybrid deep learning model,” Biomed. Signal Process. Control, vol. 87, no. 1, pp. 105565, Jan. 2024. doi: 10.1016/j.bspc.2023.105565. [Google Scholar] [CrossRef]
32. F. Zhou, Y. Sun, and Y. Wang, “Inter-patient ECG arrhythmia heartbeat classification network based on multiscale convolution and FCBA,” Biomed. Signal Process. Control, vol. 90, no. 1, pp. 105789, Apr. 2024. doi: 10.1016/j.bspc.2023.105789. [Google Scholar] [CrossRef]
33. C. J. Lin, H. Cheng, and C. L. Chang, “Automated detection of heart arrhythmia signals by using a convolutional Takagi-Sugeno–Kang-type fuzzy neural network,” Sens. Mater., vol. 36, no. 2, pp. 639, Feb. 2024. doi: 10.18494/SAM4682. [Google Scholar] [CrossRef]
34. T. Yoon and D. Kang, “Bimodal CNN for cardiovascular disease classification by co-training ECG grayscale images and scalograms,” Sci. Rep., vol. 13, no. 1, pp. 2308, Feb. 2023. doi: 10.1038/s41598-023-30208-8. [Google Scholar] [PubMed] [CrossRef]
35. J. Zheng, J. Zhang, S. Danioko, H. Yao, H. Guo and C. Rakovski, “A 12-lead electrocardiogram database for arrhythmia research covering more than 10,000 patients,” Sci. Data, vol. 7, no. 1, pp. 48, Feb. 2020. doi: 10.1038/s41597-020-0386-x. [Google Scholar] [PubMed] [CrossRef]
36. T. Yoon and D. Kang, “Multi-modal stacking ensemble for the diagnosis of cardiovascular diseases,” J. Pers. Med., vol. 13, no. 2, pp. 373, Feb. 2023. doi: 10.3390/jpm13020373. [Google Scholar] [PubMed] [CrossRef]
37. A. Rehman et al., “HCDP-DELM: Heterogeneous chronic disease prediction with temporal perspective enabled deep extreme learning machine,” Knowl.-Based Syst., vol. 284, pp. 111316, Jan. 2024. doi: 10.1016/j.knosys.2023.111316. [Google Scholar] [CrossRef]
38. S. Srinivasan, S. Gunasekaran, S. K. Mathivanan, M. B. Benjula Anbu Malar, P. Jayagopal and G. T. Dalu, “An active learning machine technique based prediction of cardiovascular heart disease from UCI-repository database,” Sci. Rep., vol. 13, no. 1, pp. 741, Aug. 2023. doi: 10.1038/s41598-023-40717-1. [Google Scholar] [PubMed] [CrossRef]
39. M. Bukhari et al., “A smart heart disease diagnostic system using deep vanilla LSTM,” Comput. Mater. Contin., vol. 77, no. 1, pp. 1251–1279, Jan. 2023. doi: 10.32604/cmc.2023.040329. [Google Scholar] [CrossRef]
40. A. U. Haq, J. P. Li, M. H. Memon, S. Nazir, and R. Sun, “A hybrid intelligent system framework for the prediction of heart disease using machine learning algorithms,” Mob. Inf. Syst., vol. 2018, no. 8, pp. 1–21, Dec. 2018. doi: 10.1155/2018/3860146. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF
 Downloads
Downloads
 Citation Tools
Citation Tools