 Open Access
Open Access
ARTICLE
GAN-DIRNet: A Novel Deformable Image Registration Approach for Multimodal Histological Images
1 Institute of Image Processing and Pattern Recognition, Shanghai Jiao Tong University, and Key Laboratory of System Control and Information Processing, Ministry of Education of China, Shanghai, 200240, China
2 Department of Pathology, Ruijin Hospital, Shanghai Jiao Tong University, School of Medicine, Shanghai, 200025, China
3 Department of Computer Science and Engineering, Shanghai Jiao Tong University, Shanghai, 200240, China
4 Global Institute of Future Technology, Shanghai Jiao Tong University, Shanghai, 200240, China
* Corresponding Author: Hongbin Shen. Email:
Computers, Materials & Continua 2024, 80(1), 487-506. https://doi.org/10.32604/cmc.2024.049640
Received 13 January 2024; Accepted 26 April 2024; Issue published 18 July 2024
Abstract
Multi-modal histological image registration tasks pose significant challenges due to tissue staining operations causing partial loss and folding of tissue. Convolutional neural network (CNN) and generative adversarial network (GAN) are pivotal in medical image registration. However, existing methods often struggle with severe interference and deformation, as seen in histological images of conditions like Cushing’s disease. We argue that the failure of current approaches lies in underutilizing the feature extraction capability of the discriminator in GAN. In this study, we propose a novel multi-modal registration approach GAN-DIRNet based on GAN for deformable histological image registration. To begin with, the discriminators of two GANs are embedded as a new dual parallel feature extraction module into the unsupervised registration networks, characterized by implicitly extracting feature descriptors of specific modalities. Additionally, modal feature description layers and registration layers collaborate in unsupervised optimization, facilitating faster convergence and more precise results. Lastly, experiments and evaluations were conducted on the registration of the Mixed National Institute of Standards and Technology database (MNIST), eight publicly available datasets of histological sections and the Clustering-Registration-Classification-Segmentation (CRCS) dataset on the Cushing’s disease. Experimental results demonstrate that our proposed GAN-DIRNet method surpasses existing approaches like DIRNet in terms of both registration accuracy and time efficiency, while also exhibiting robustness across different image types.Keywords
Deep learning based multi-modal histological image registration is indispensable in medical image processing [1–4]. Intraoperative navigation, 3D modeling, and fusion analysis of information from multiple sources all necessitate registration as a prerequisite step [5–8]. Existing registration methods based on deep learning are generally tailored for the registration between the computed tomography (CT) and magnetic resonance (MR) images [6,9–12]. In contrast, histological image registration presents distinct challenges compared with CT-MR registration. While CT-MR registration deals with images of the same section but different modalities, histological images require registration between different modalities of adjacent sections, significantly complicating the task of identifying corresponding anatomical relationships [13,14]. Moreover, dyeing operations inevitably introduce local fuzziness and noise interference in histological images, distorting or blurring the original features. Current CT-MR registration methods lack optimization for these challenges or integration of a priori knowledge of such deformations into their models. Furthermore, the annotation process for histological images demands more manpower and resources compared to CT/MR images [15,16]. Consequently, existing methods struggle in histological image registration. These challenges have spurred researchers to develop a novel unsupervised deep learning method for histological image registration with severe interference and deformation.
As a model of deep learning based unsupervised registration, the deformable image registration network (DIRNet) [17] can achieve results comparable to traditional registration methods in a relatively short timeframe, without requiring manual annotation of the dataset. However, DIRNet is limited to registering images of the same modality and does not account for modal differences when registering cross-modal images. Improved variants such as EDIRNet [18], MS-DIRNet [19] and Sang’s work [20] have addressed some of these limitations. Among them: EDIRNet utilizes attention mechanism to enhance the influence of edge features on registration. However, in histological images of conditions like Cushing’s disease, tissue defects and overlaps caused by operations often occur at the edges, which can limit the effectiveness of edge-based methods like EDIRNet. MS-DIRNet [19] trains a combination of global and local networks, enabling the model to capture larger abdominal movements. Sang et al. [20] implicitly model the deformation prior of the spatial changes and incorporate it into the image registration framework. However, the deformation priors caused by motion that they considered are limited by the specific situations they address. For example, the abdominal movement studied by Lei et al. [19] is mainly composed of longitudinal and transverse stretching, which may not adequately address the complex deformations present in histological images. The magnetic resonance and CT images studied by Sang et al. [20] do not exhibit significant angle deviation and partial loss as in the histological images of our study. Additionally, they do not consider the extraction of modal features from multi-modal images, further limiting their applicability to multi-modal histological image registration tasks. In Table 1, we provide detailed characteristics of the aforementioned registration methods and explain why they may not be suitable for addressing challenges encountered in such situations.
Generative Adversarial Network (GAN) [21] is a generative model for deep learning trained in the form of confrontation. The generator converts random vectors into synthetic images resembling the training images, while the discriminator distinguishes between synthetic and real images. By pitting the discriminator against the generator, GAN self-supervises the training. Building upon GAN, cyclic GAN [22] has the advantage of not requiring image pairs, but forcing the deformation field to be consistent. Mahapatra et al. [23] use GAN to generate the warped image directly and prevent unrealistic registration by leveraging the consistent forced deformation of cyclic GAN. Lei et al. [19] integrate GAN into deformable image registration (DIR) to punish unrealistically warped images with additional dense displacement vector field (DVF) regularization. However, these approaches often underutilize the feature extraction capability of the GAN discriminator.
In summary, existing methods have struggled to accomplish deformable multi-modal registration tasks on images with significant modality differences, severe noise interference, and deformation, as evidenced by datasets like Cushing’s disease [24]. To tackle these challenges, in this study, we propose a novel multimodal registration approach GAN-DIRNet, which adapts generative adversarial networks for deformable histological image registration.
Compared with previously published studies, the major contributions of this work are three-fold:
1. We designed a novel deformable multimodal image registration network based on unsupervised deep learning, which can be used for histological image registration tasks with severe noise and deformation such as the images of Cushing’s disease.
2. We proposed a GAN-DIRNet embedded with discriminators for auxiliary feature extraction.
3. We proposed a novel modal-specific feature descriptor for cross-modal histological image registration.
Traditional histological registration methods are mainly divided into three categories: Feature-point-based [25], gray-scale-based [26], metric-based [27] and transformation-based methods [12]. The registration method based on feature points [25,28,29] offers the advantages of rapid execution and low computational complexity. However, it requires manual setting of feature points, automatic search by algorithms, or a combination of both [30].
These systems often struggle to effectively manage significant disparities between modalities in pairs of histological images. They commonly face challenges in processing images of separate modalities into feature pairs. For instance, in the CRCS dataset [24], the reference images are stained to highlight adreno-cortico-tropic-hormone (ACTH) in the cytoplasm, whereas the moving images are stained to emphasize the transcription of ACTH-secreting tumor cells (t-pit) in the nucleus. Automatically identified feature points tend to focus on the nucleus of the moving image, while the corresponding positions in the fixed image are ignored due to staining defects. In essence, feature-point-based methods either necessitate meticulous manual intervention or exhibit limited reliability in handling multi-modal images.
The transformation-based approach initially involves converting one image modality to the other [31], or both modalities to a shared latent space [32]. Subsequently, registration occurs within this common feature space. Specifically, DTR-GAN [12] utilizes a bidirectional translation network to convert multi-modal images into the same modality before performing single-modal registration. Similarly, Deform-GAN [33] proposed by Zhang et al. employs the generator in GAN to transform multi-modal similarity into single-modal similarity, thereby enhancing precision. However, our images for registration employ different staining methods (one staining the nucleus and the other staining the cytoplasm), making direct conversion challenging due to substantial differences. Metric-based methods sidestep these challenges by selecting metrics less sensitive to modal variations for optimization, such as mutual information (MI) [27], regional MI, mean square error (MSE), and normalized cross correlation (NCC) [34,35].
To alleviate the need for annotation workload, unsupervised or weakly-supervised deep learning models have been deployed in multi-modal image registration. For instance, DIRNet [17], introduced by Vos et al., an unsupervised deep learning model for deformable image registration, and ADMIR [36], designed for end-to-end unsupervised affine and deformable registration of brain magnetic resonance images. A semi-supervised model proposed by Liu et al. [30] offers a method to progressively increase the number of landmarks from a limited set of annotations. However, the fundus images they analyze are relatively easy to annotate manually, unlike the histological images in our study. Among all the unsupervised models, DIRNet [17] bears the closest resemblance to our approach. Although DIRNet is adaptable to multi-modal histological image registration, it lacks distinct modules for handling moving and fixed images separately. Essentially, it overlooks the modalities’ disparities and treats them as a single-modal image registration task. While DIRNet performs adequately on the MNIST [37] dataset, our experiments reveal its subpar performance in multi-modal histological image registration. As a remedy, we propose integrating an additional feature extraction module at the outset. Subsequent experiments in later sections demonstrate that this multi-modal feature extraction module significantly enhances the accuracy of both single-modal and multi-modal image registration.
To showcase the effectiveness of our proposed method, GAN-DIRNet, we conducted evaluations on multiple datasets, including MNIST, the CRCS dataset, and the Automatic Non-rigid Histological Image Registration (ANHIR) dataset, comprising eight subsets such as breast, lung lobes, gastric, kidney, etc.
In CRCS-dataset, histological sections with adreno-cortico-tropic-hormone (ACTH) and t-pit (the transcription of ACTH-secreting tumor cells) from 35 Cushing’s disease patients was retrospectively collected. Each pair of images slated for registration had at least 5 pairs of landmarks implanted to calculate the target registration error and facilitate registration evaluation.
The ANHIR-dataset, introduced by Borovec et al. [15], encompasses eight subsets of histological images namely breast, chronic obstructive air way disease (COAD), gastric, kidney, lung lesion, lung lobes, mammary-gland, and mice-kidney respectively. It comprises a total of 481 cross-modal image pairs sourced from 18 different tissue staining operations. On average, 86 landmarks were placed on each image by 9 annotators, which can be publicly accessed together with the images.
Overall, we innovatively designed the discriminators of two GANs as embedded pre-trained parallel networks and integrated them into the input part of DIRNet as the modal feature extraction layer to enhance cross-modal registration outcomes.
The original version of DIRNet comprises three components: A convolutional neural network, a spatial transformation network, and a resampling network. Convolutional neural networks translate the input fixed and moving images into local deformation parameters. Subsequently, the spatial transformation network receives these parameters and generates a dense displacement vector field. Finally, the resampling network distorts the moving image into the warped image based on the dense displacement vector field. However, the input layers for receiving moving images and fixed images in DIRNet are identical, indicating a lack of consideration for modal differences in cross-modal image registration.
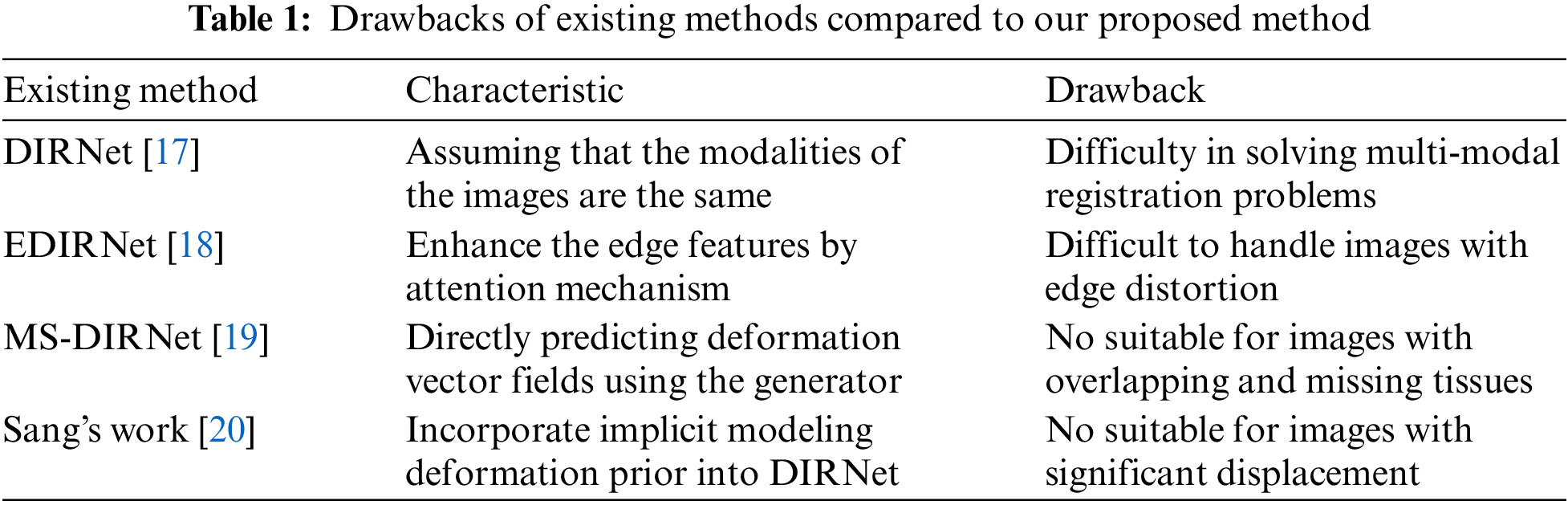
Our proposed method GAN-DIRNet comprises of three modules: GAN training, rigid registration, and deep-learning based non-rigid registration, as illustrated in Fig. 1. During the initial stage of GAN training, the training sets of moving images and fixed images are individually inputted into two identical generative adversarial networks for model training. Following adversarial training, two implicit feature extraction modules corresponding to each training set are derived from the discriminators of the two GAN networks, respectively.
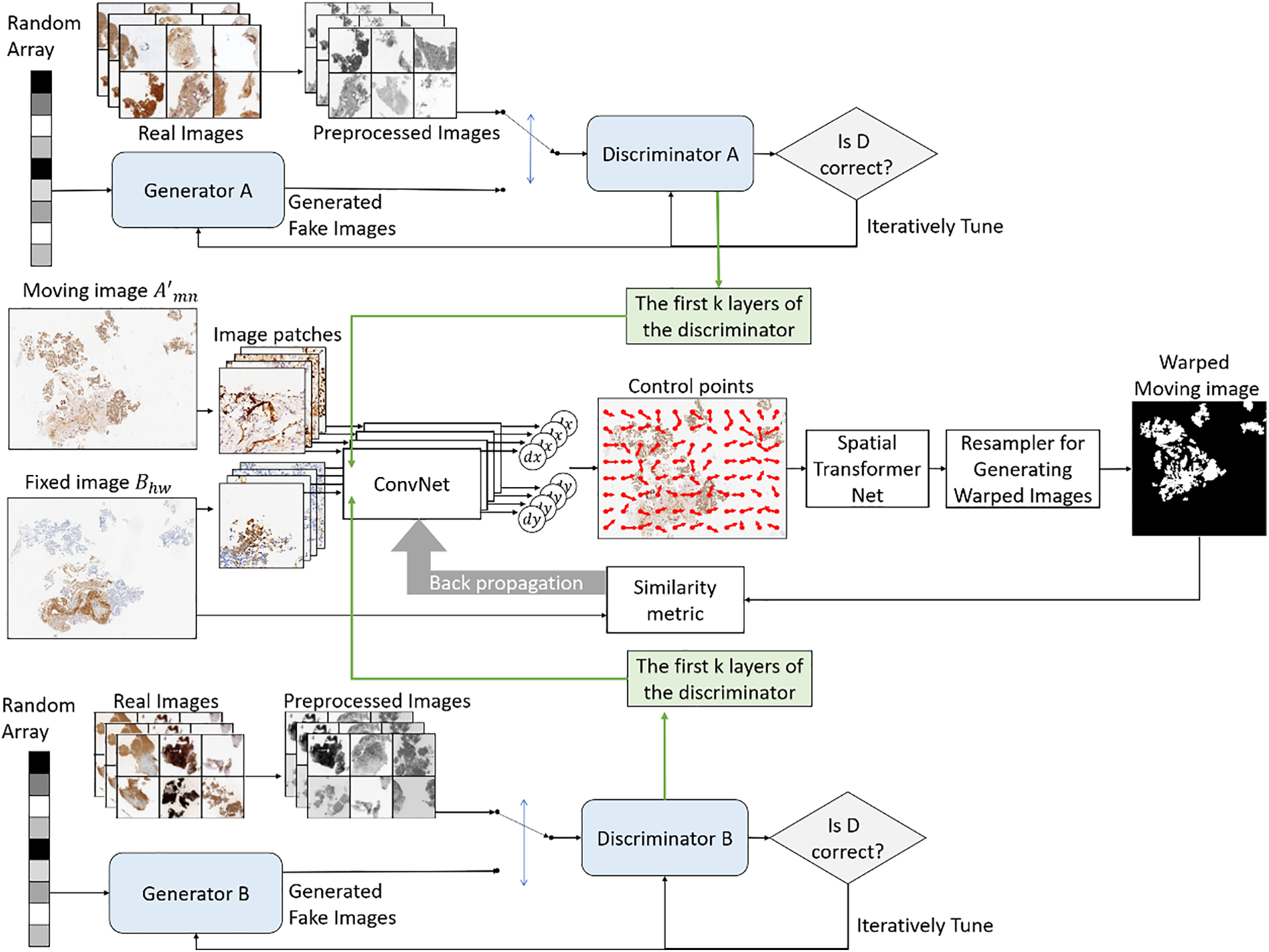
Figure 1: Schematic diagram of the principle of our proposed method
The second stage involves implementing a traditional iterative rigid registration method for the image pairs intended for registration, aiming to acquire a transformation matrix containing explicit spatial information such as translation and rotation. However, for handwritten dataset registration, this second step can be omitted since there are no rotation or translation transformations involved.
In the third stage, the first few layers of the discriminators in GAN pre-trained by moving and fixed images are embedded into the input layers of moving and fixed images in DIRNet, respectively. Subsequent to inputting the resultant image of rigid registration from the second stage, a displacement vector field (DVF) generated by GAN-DIRNet performs non-rigid deformation on the moving image via an interpolation network, with the process recorded as
1) Firstly, the moving and fixed images, preprocessed from color to grayscale, are separately inputted into GAN training. Generator A and Generator B generate fake images from random sequences that closely resemble their respective input images, while Discriminator A and Discriminator B distinguish between real images and the fake images generated by their corresponding generator. During adversarial training, the feature descriptors of the multi-modals (moving and fixed images) are extracted by the discriminators in two GANs separately. When dealing with single-modal images from the MNIST dataset, parameter k is set to 5, whereas for histological images, parameter k is set to 3. 2) The modal feature descriptors are then connected to the feature input layers of DIRNet respectively. 3) For incomplete slice images, adaptive threshold filling is conducted on the border part to mitigate the impact of feature points created by cutting and edges.
During GAN training, the training sets of moving and fixed images are individually inputted into two identical GAN networks for training, aiming to acquire the implicit feature representation of the respective image modalities. The GAN within GAN-DIRNet comprises a generator and a discriminator, both of which are constructed using convolutional neural networks (CNN), as depicted in Fig. 1.
In implementation, the generator consists of five transposed convolutional layers with a kernel size of 4 × 4, as illustrated in Fig. 2. Batch normalization [38] and rectified linear activation function (ReLU) are applied throughout the generator, except for the last layer which utilizes a tanh activation function. Conversely, the discriminator comprises five convolutional layers with the same kernel size as the generator. The final layer of the discriminator adopts a sigmoid activation function, while the other layers employ leaky ReLU. Similarly, the outputs of the middle three layers of the discriminator undergo batch normalization.
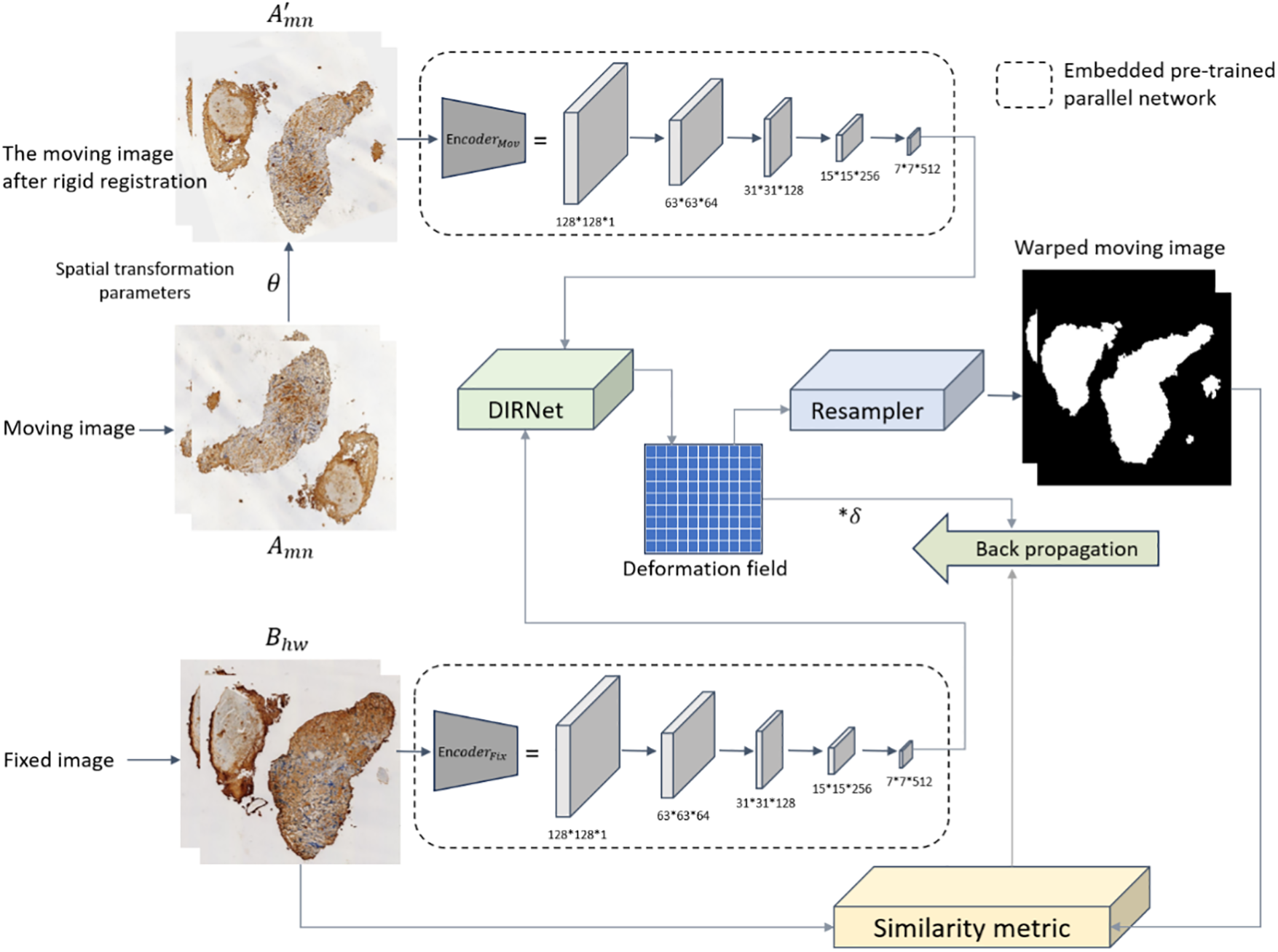
Figure 2: Schematic diagram illustrating the process of embedding the modal feature extraction module into DIRNet for model training.
The rigid registration module utilized in this article adopts the registration method outlined in the CRCS pipeline [24]. Its specific procedure is as follows. Assuming a moving image with length
The rigid registration module
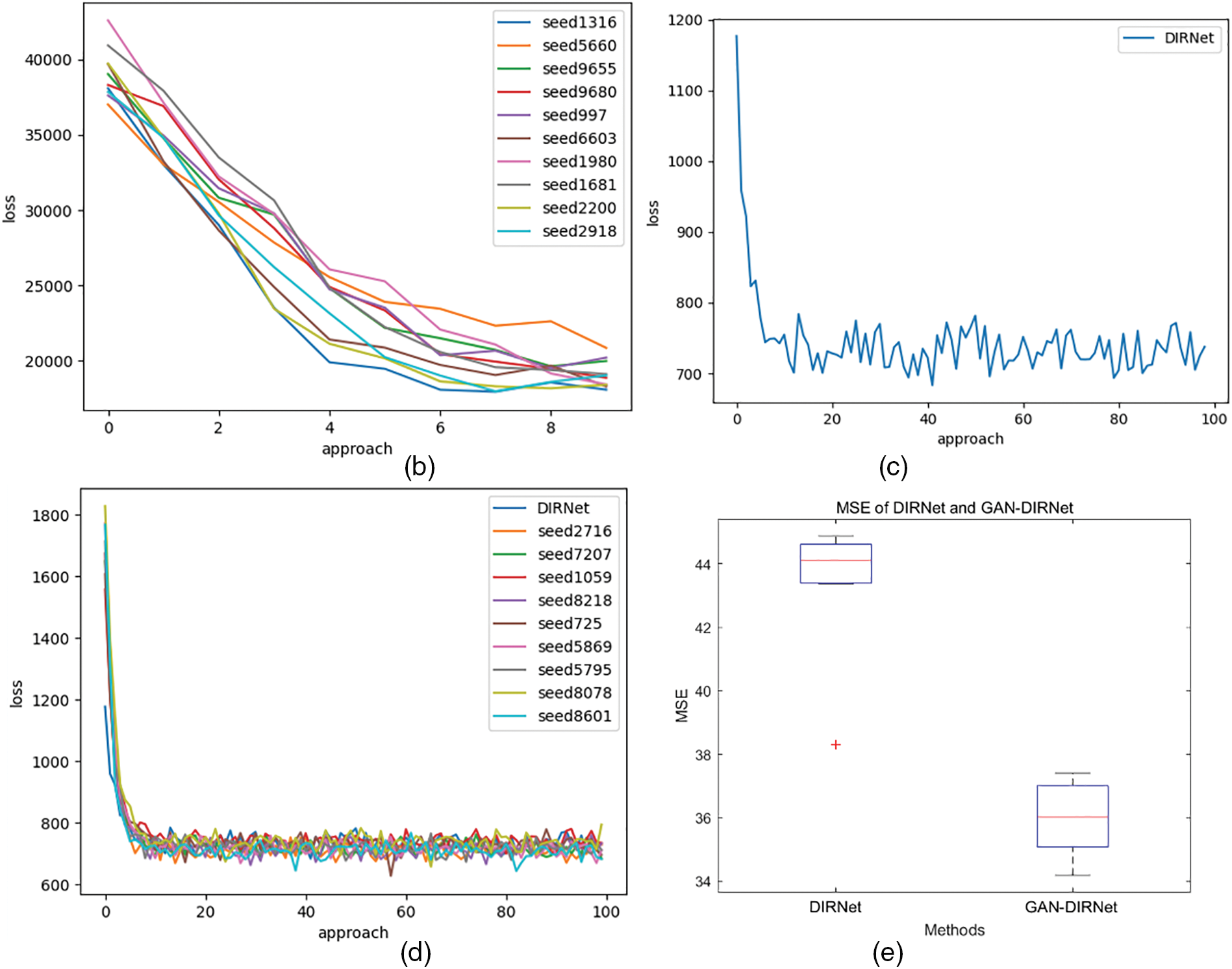
Figure 3: (a) Schematic diagram of the rigid registration module and its connection with upstream and downstream tasks. (b): The descent curves of loss during the optimization processes of GAN-DIRNet. The horizontal axis represents the number of iteration steps, while the vertical axis represents the value of the loss function, detailed in Section 3.5.2. (c): The descent curve of loss during the optimization process of DIRNet. (c) is a descent curve reproduced from the original version of the DIRNet code. In order to eliminate the impact of randomness on the results, we conducted tests on GAN-DIRNet using 10 different random seeds. Thus, there are ten descent curves in (b), whereas (c) only presents one. For comparative purposes, we retested DIRNet with 9 additional random seeds and displayed their descent curves alongside the original curve in (d). The loss function records the total loss considering the gradient accumulation and the regularization term. Its numerical value is also influenced by the image size (16 * 16 pixels for the original DIRNet, while 64 * 64 pixels for the proposed GAN-DIRNet). Therefore, the schematic diagrams of the loss function (b–d) only show their convergence rates; their values cannot be horizontally compared as in (e). (e): The boxplots represent the MSE errors on the MNIST dataset using the DIRNet and GAN-DIRNet registration models from left to right, respectively. There is a standardized measurement rule for the results of different methods before calculation, enabling horizontal comparison of their values. The sole outlier represents the sample from Vos’ work [17]. Among the ten samples from each of the two groups, even the worst performing point in the proposed model exhibits a smaller error than the best outlier in the original DIRNet
Two optimization algorithms are employed separately in the different branches of the proposed integration method: Gradient descent and one plus one evolution. Among them, mean square error (MSE) is combined with gradient descent optimization as the similarity metric, while mutual information is utilized in conjunction with the other optimization algorithm.
The specific optimization objectives for the two stages are articulated as follows:
where
Furthermore, during the registration process, various parameter settings are applied to each input pair of images to be registered. The optimal result from all the calculation branches is counted as the global optimal result
θ is a 9 × 9 matrix for rigid transformation. In the specific computation, θ transforms the pixel at coordinate
where x and y respectively denote the translation amount of the pixel in both horizontal and vertical directions, and γ represents the rigid rotation angle of the entire image.
3.5.1 Embedded Feature Encoder Module
The proposed GAN-DIRNet augments the DIRNet network architecture with a modal feature extraction module utilizing GAN’s discriminator. Unlike traditional DIRNet which treats moving and fixed images equally, our method pre-extracts modal features from both types of images and integrates them into the input layer of DIRNet. The input layers of the moving and fixed images share a similar structure derived from GAN’s discriminators.
Conceptually speaking, our network consists of encoders used to extract feature maps from a given input image. The module for extracting features of moving image is called
The simplified description of the entire process is as follows:
where
3.5.2 Mutually Reinforcing Registration and Encoder Module
During the training phase, modal feature extraction and deformable image registration are distinct processes. The modal feature modules of moving and fixed images are trained independently by two GANs with identical structures. Conversely, the image registration network is trained by simultaneously inputting images from both modalities. This setup entails optimizing the modal feature extraction model twice during the training of the image registration network.
In backpropagating through GAN-DIRNet, it is crucial to balance considerations for both similarity measures and the smoothness of the transformation field. As a result, our loss function is defined as follows:
In the defined loss function, the first term represents the similarity measure, while the second term serves as a smoothing component. The parameter
3.6 Performance Metrics for Image Registration
The quantitative indicator utilized in this article is the relative target registration error (rTRE) evaluation system [15], which is assessed following the evaluation system proposed in the Automatic Non-rigid Histological Image Registration Challenge (ANHIR). The calculation method is outlined as follows:
In principle, the image pair
The warped image is denoted as
where
Similarly, the last two indicators replace the initial step of calculating the
We initially benchmarked GAN-DIRNet on the MNIST dataset. Following the original setting in DIRNet [17], we randomly sampled the training and testing datasets from MNIST. For reproducibility and consistency, we benchmarked both DIRNet and our in-house GAN-DIRNet over ten preset random seeds, as shown in Figs. 3b–3e. Among them, Figs. 3b–3d display the comparison of the loss updates during training of DIRNet and GAN-DIRNet. Fig. 3e presents the boxplots representing the MSE errors on the MNIST dataset using the DIRNet registration model and GAN-DIRNet registration model from left to right, respectively. The solitary outlier denotes the sample code from the paper of DIRNet [17]. The red “+” outlier in Fig. 3e represents the reproduction of the original DIRNet code, which is constrained to use an initial seed of 0. We replaced 9 additional random seeds during the reproduction of DIRNet in comparison with the 10 random-seed-results of GAN-DIRNet.
The box diagram provides an intuitive observation: Out of the ten initial seeds randomly selected from each of the two groups, even the worst performance in GAN-DIRNet is on par with the best performance of the original DIRNet, amounting to a 10% improvement on average. In addition, GAN-DIRNet is 22% faster than DIRNet (as shown in Table 2), and converges much more rapidly during training. GAN-DIRNet achieves a similar performance to DIRNet after just 100 iterations compared to the 1000 iterations required by DIRNet. The experimental results are listed in Table 3.

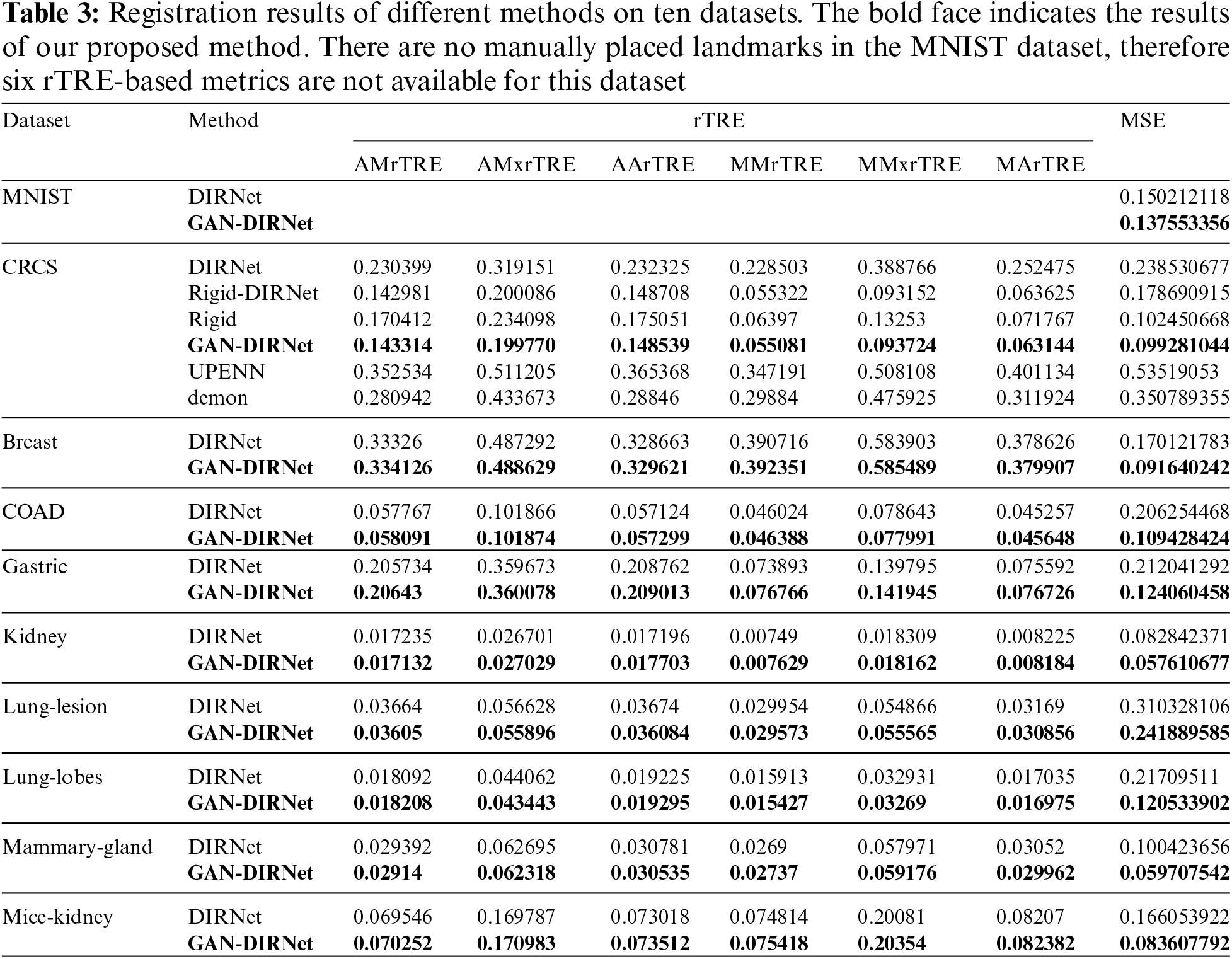
4.2 Results on Histological Image Dataset
For histological images, we firstly benchmarked both DIRNet, its variant Ridge-DIRNet, and our adaptation: GAN-DIRNet over the CRCS dataset to validate the advantages of GAN-DIRNet compared to DIRNet. Additionally, we conducted comparison with other 2 existing methods: UPENN [15] and demon [39] on CRCS, to ascertain the advantages of GAN-DIRNet compared to the most commonly used methods currently available. Specifically, UPENN is an unsupervised method that integrates multiple tools for histological image registration and achieved the second place in the ANHIR competition. While demon performs well in medical image registration such as magnetic resonance images, which also does not require manual annotation. These two methods exhibit strengths in different scenarios. As comparative experiments, they serve to confirm the unique advantages of GAN-DIRNet on histological image datasets such as CRCS from various perspectives.
Fig. 4 illustrates the registration results of the respective methods over the CRCS images. As Fig. 4 depicted, Rigid-GAN-DIRNet (simplified as GAN-DIRNet in this paper) corrected the local deformation between the registered images on the basis of rigid registration, whereas DIRNet and its variant failed to do so. Furthermore, it is evident that the latter two methods struggled to identify accurate rotation angles and translation components.
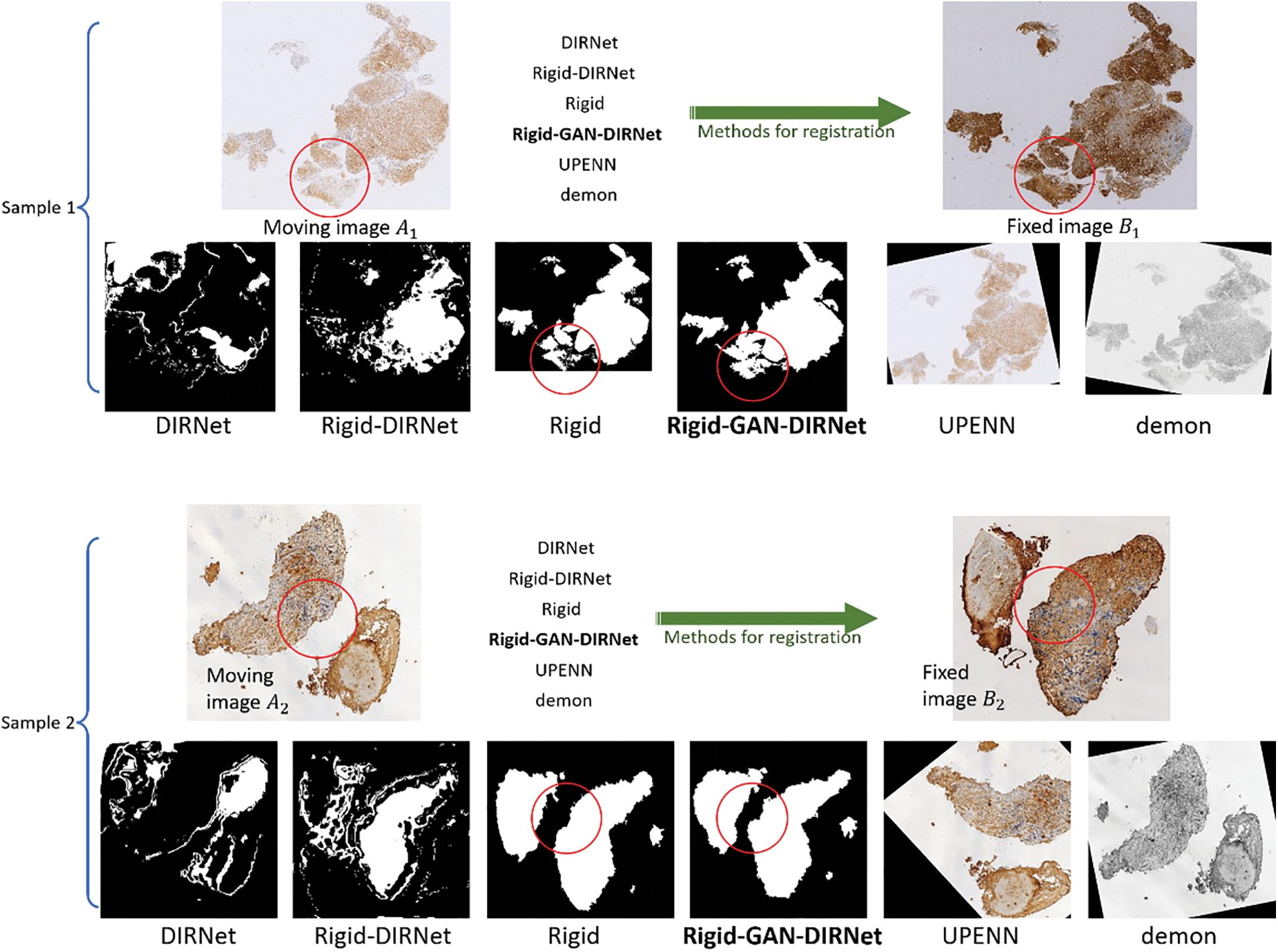
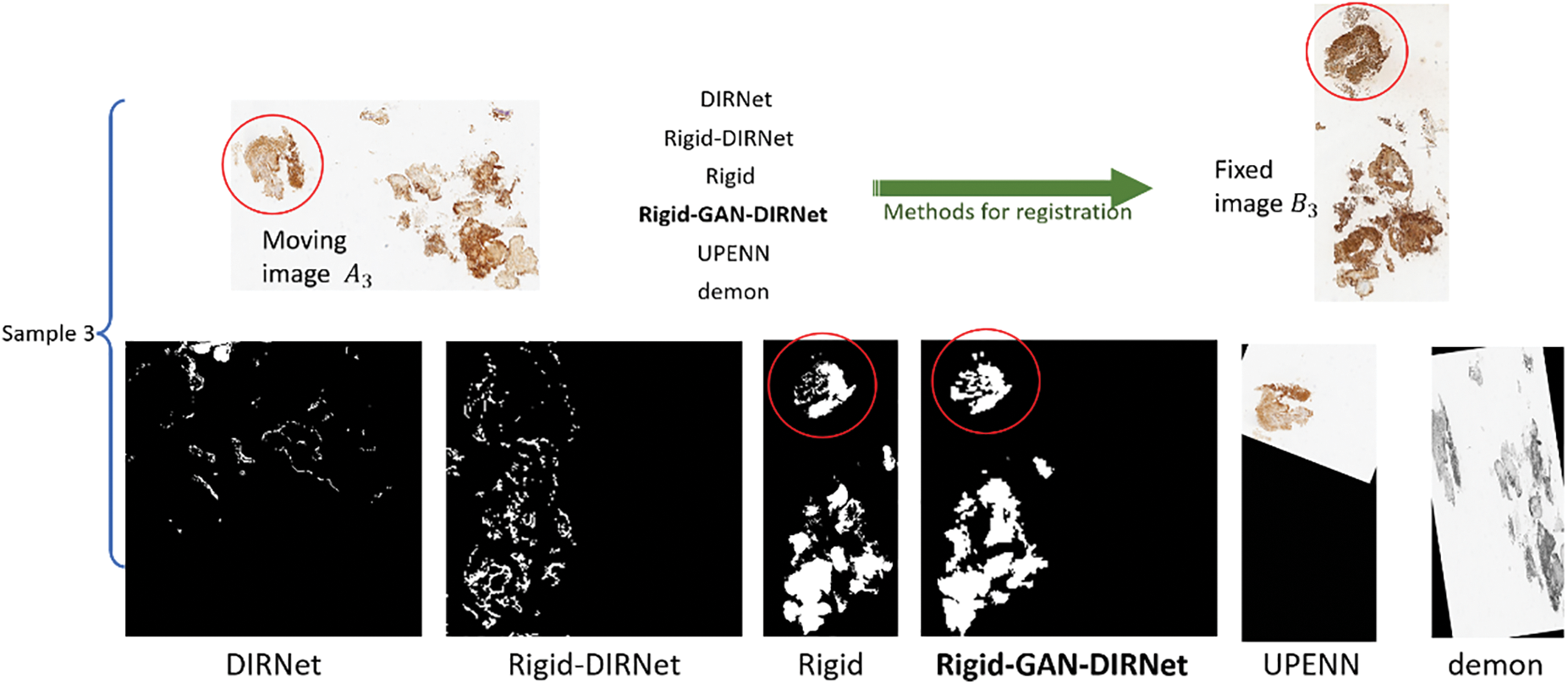
Figure 4: Comparison of the visualization results of the various registration methods. For simplicity,
In addition to visual assessment, we also conducted quantitatively analysis using two types of evaluation metrics: The landmark-based metrics (six indicators based on rTRE mentioned above) and the region-based metric, as shown in Table 3. We adopt MSE as the region-based metric by calculating the average of the sum of the squared errors obtained by subtracting a fixed image from a warped image, as illustrated in Eq. (14). The landmark-based metrics reflect the registration effect from a local perspective, while the region-based metric provides a global assessment.
where
In order to avoid inter-modal interference, the landmarks on image pairs of the CRCS dataset are manually placed at edges, as depicted in Fig. 5, which may not comprehensively reflect the registration effect. To compensate for the shortcomings of rTRE, we also introduce a universal evaluation index named mean square error (MSE) for regional area. From Table 3, it is evident that the proposed scheme demonstrates the best comprehensive performance across the ten datasets. Rigid-GAN-DIRNet achieves nearly identical performance measured by landmark-based evaluation metrics compared to Rigid-DIRNet (a method embedding only the proposed rigid registration module in DIRNet without the feature extraction module), but its advantage in the evaluation index based on regional area is obvious.
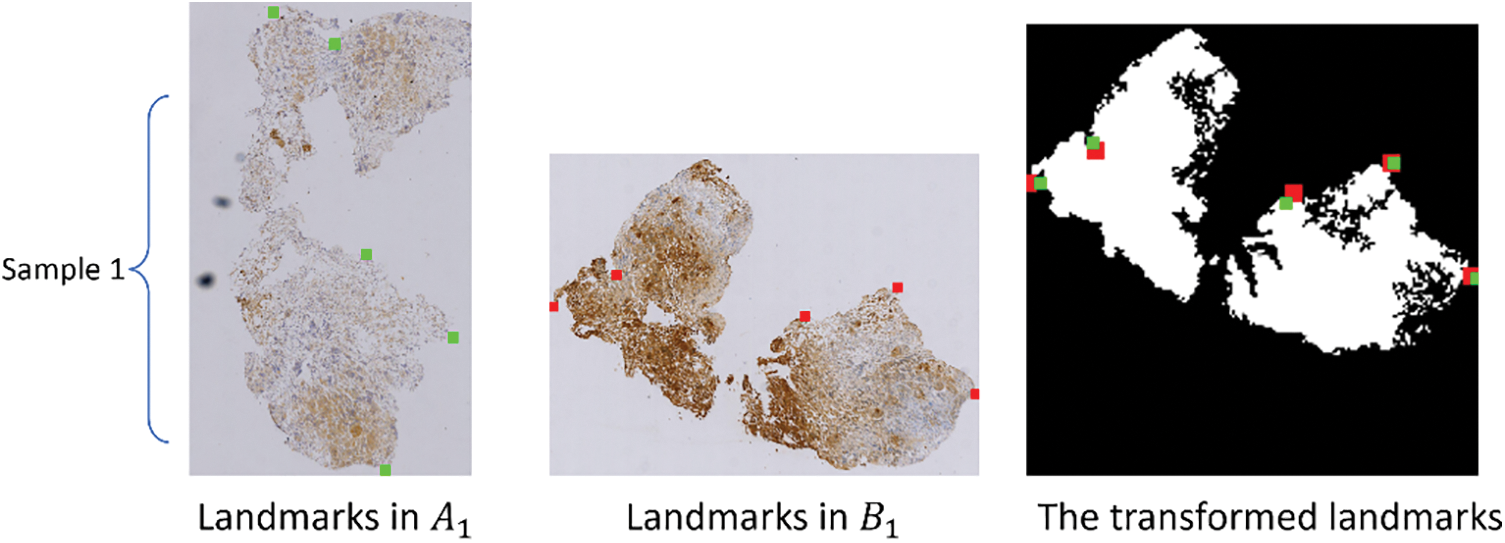
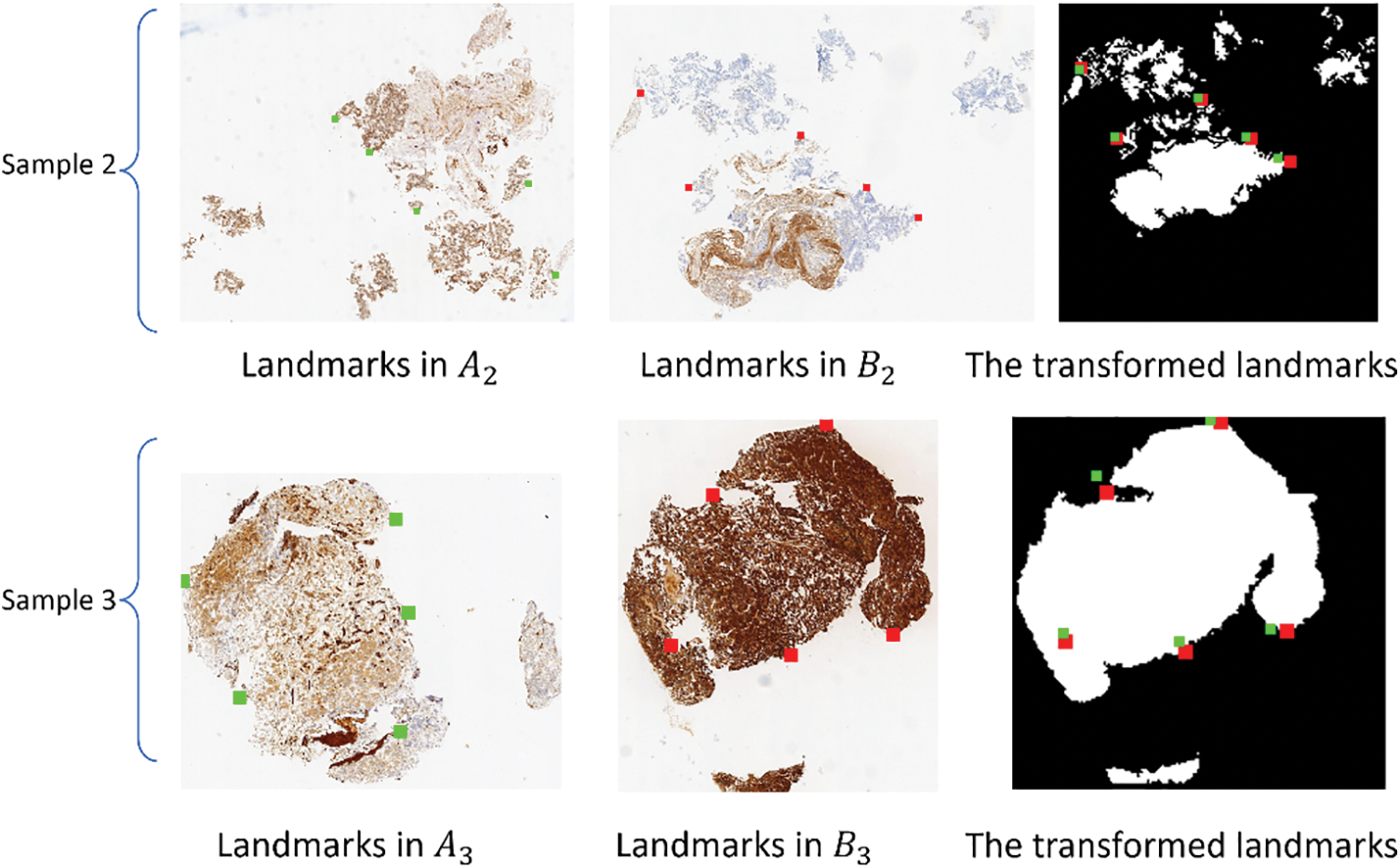
Figure 5: The first column (
In this study, we propose a novel method GAN-DIRNet to integrate the modal-specificity information and rigid registration modules into the DIRNet for deformable image registration. The results demonstrate that GAN-DIRNet achieves more accurate and faster performance than existing methods. To investigate the superiority of GAN-DIRNet, we visualize the descriptors extracted by GAN-DIRNet through t-SNE (Fig. 6). Among them, Figs. 6a and 6b represent visualizations of the test subset of the MNIST dataset and its trained descriptors through GAN-DIRNet (i.e., the output of the GAN module and the input of the DIRNet module), respectively. By comparison, it can be seen that the latter exhibits better aggregation than the former. Despite MNIST being a single-modal image dataset, the discrimination between the ten categories of the feature descriptors output by the embedded modal feature extraction layer of GAN-DIRNet is higher than that of the unprocessed original image data, which proves the powerful feature extraction capability of GAN-DIRNet.
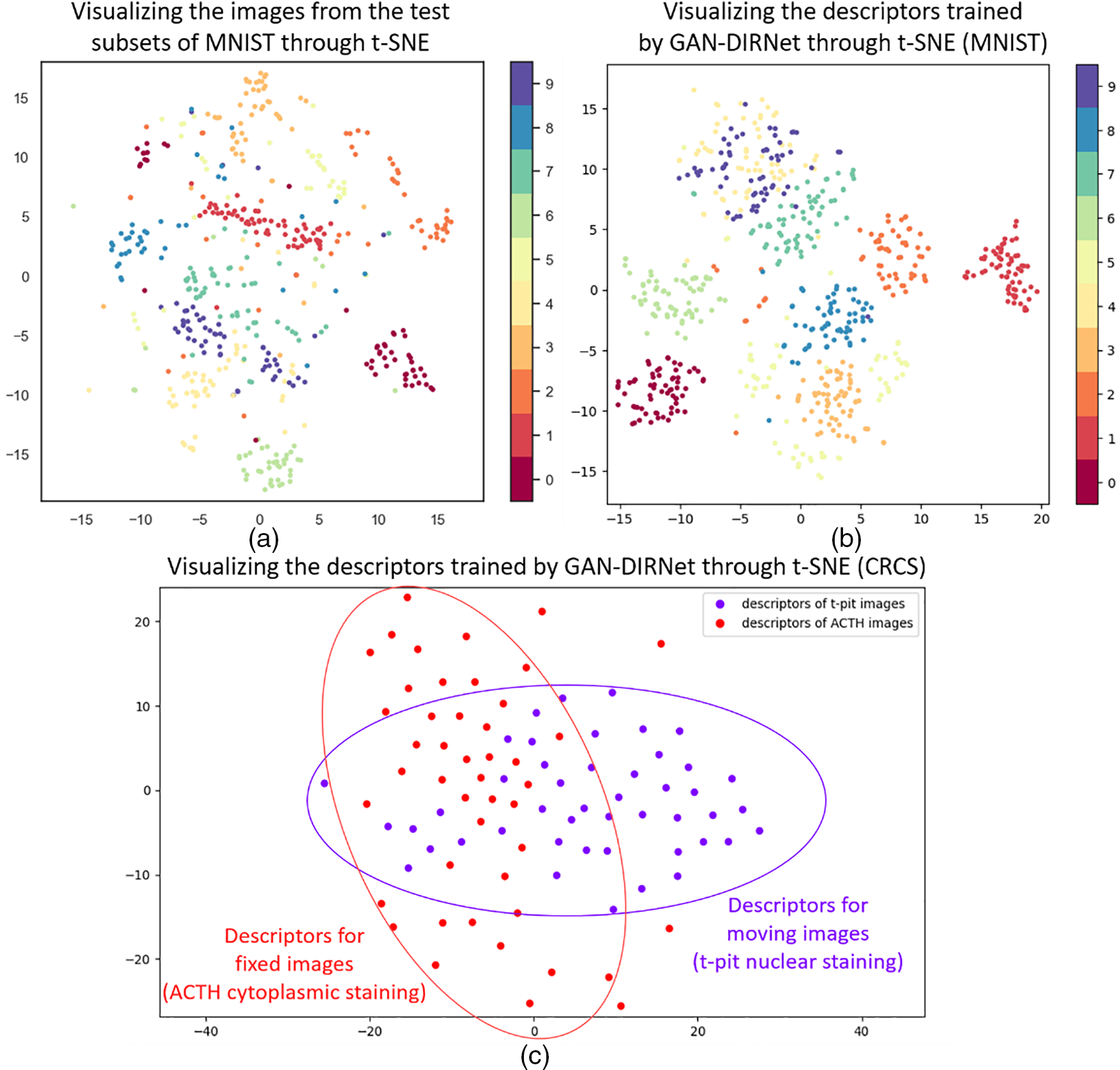
Figure 6: Visualizations through t-SNE. (a) and (b) are visualizations of the test subset of the MNIST dataset and its trained descriptors through GAN-DIRNet (i.e., the output of the GAN module and the input of the DIRNet module), respectively. By comparison, it can be seen that the latter shows better aggregation than the former. (c) is a visualization of the descriptors trained on GAN-DIRNet for the CRCS dataset
Fig. 6c illustrates a visualization of the descriptors trained on GAN-DIRNet for the CRCS dataset, where the first label represents all t-pit nuclear-staining moving images, and the second label represents all ACTH cytoplasm-staining fixed images. The difference in the distribution of the two reflects variance between the modal features of the two differently stained images. Quantitative and qualitative analysis confirms that the final registration effect of our proposed GAN-DIRNet is superior to that of the original DIRNet, which may be attributed to the function of the embedded modal feature extraction layer for differentiated processing of multi-modal images.
Multi-modal histological image registration poses one of the most challenging tasks in medical image registration. Due to the irreversibility of dyeing operations, there is inevitably non-rigid deformation and severe noise between adjacent slices, significantly affecting the registration outcome. Existing non-rigid multi-modal registration methods often struggle with histological images exhibiting substantial modality differences. In this article, we innovatively designed the discriminators of two GANs as the embedded parallel networks and integrated them into the input part of DIRNet as the modal feature extraction layer. Our proposed model, named “GAN-DIRNet”, integrates the modal-specificity information and rigid registration modules into the original DIRNet to achieve more accurate and faster performance. We conducted experimental validation on nine public datasets and a non-public clinical dataset for a rare disease to demonstrate the effectiveness of GAN-DIRNet. Finally, we performed dimensionality reduction analysis on the “intermediate results” output from the network embedding part, and analyzed the reasons why it can assist in achieving better registration results.
The advantages of GAN-DIRNet are: 1) It can adapt to non-rigid registration tasks in multimodal histological images with severe noise and artifacts; 2) It can compensate for missing tissues with non-rigid deformation; 3) It can complete convergence with fewer iterations. However, this study has limitations in the following two aspects: 1) Only 5 landmarks were placed in each image of the non-public dataset CRCS, resulting in metrics based on feature points not fully reflecting the registration effect on this dataset. 2) The rigid registration part in GAN-DIRNet adopts an integrated traditional intensity-based registration algorithm, which is computationally intensive for large-sized histological images.
The innovation of this study lies in three key points. Firstly, we establish a novel multi-modal unsupervised deep-learning based DIR for histopathology images of Cushing’s disease. Secondly, we integrate the discriminator of GAN into DIRNet to assist with image registration. Finally, we propose a novel modal-specific feature descriptor for cross-modal histological image registration.
The idea of leveraging the features of each modality to aid registration in this study may offer additional perspectives for future multi-modal registration endeavors. Importantly, both the feature extraction and rigid registration process are automated. Therefore, applying our method to the registration task between any two modal images will not incur additional workload.
Acknowledgement: Thank Mr. Yu-Xuan Chen for providing guidance on paper writing.
Funding Statement: None.
Author Contributions: Study conception and design: Haiyue Li; data collection, Haiyue Li and Jing Xie; analysis and interpretation of results: Haiyue Li, Jing Ke, Ye Yuan and Hongbin Shen; draft manuscript preparation: Haiyue Li, Xiaoyong Pan and Hongyi Xin. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: We have made our code freely available at: github.com/lhy2024/GAN-DIRNet(accessed on 20/06/2024).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Y. Ding et al., “Deep-learning based fast and accurate 3D CT deformable image registration in lung cancer,” Med. Phys., vol. 50, no. 11, pp. 6864–6880, Nov. 2023. doi: 10.1002/mp.16548. [Google Scholar] [PubMed] [CrossRef]
2. X. Deng, E. Liu, S. Li, Y. Duan, and M. Xu, “Interpretable multi-modal image registration network based on disentangled convolutional sparse coding,” IEEE Trans. Image Process., vol. 32, no. 78, pp. 1078–1091, Jan. 2023. doi: 10.1109/TIP.2023.3240024. [Google Scholar] [PubMed] [CrossRef]
3. Z. Zhu, Y. Ji, and Y. Wei, “Multi-resolution medical image registration with dynamic convolution,” presented at the 2022 IEEE Biomed. Circ. Syst. Conf. (BioCASTaipei, Taiwan, Oct. 13–15, 2022, pp. 645–649. [Google Scholar]
4. S. Rai, J. S. Bhatt, and S. K. Patra, “Deep learning in medical image analysis: Recent models and explainability,” in The Explainable AI in Healthcare, 1st ed. New York, USA: Chapman and Hall/CRC, 2024, pp. 23–49. Accessed: Jul. 17, 2023. [Online]. Available: https://www.taylorfrancis.com/books/edit/10.1201/9781003333425 [Google Scholar]
5. J. Hu, L. Li, Y. Fu, M. Zou, J. Zhou and S. Sun, “Optical flow with learning feature for deformable medical image registration,” Comput. Mater. Contin., vol. 71, no. 2, pp. 2773–2788, Dec. 2022. doi: 10.32604/cmc.2022.017916. [Google Scholar] [CrossRef]
6. M. Zou, et al., “Rigid medical image registration using learning-based interest points and features,” Comput. Mater. Contin., vol. 60, no. 2, pp. 511–525, Aug. 2019. doi: 10.32604/cmc.2019.05912. [Google Scholar] [CrossRef]
7. S. Divakaran, “An explainable method for image registration with applications in medical imaging,” in The Explainable AI in Healthcare, 1st ed. New York, USA: Chapman and Hall/CRC, 2024, pp. 68–82. Accessed: Jul. 17, 2023. [Online]. Available: https://www.taylorfrancis.com/books/edit/10.1201/9781003333425 [Google Scholar]
8. X. Wang, Z. Song, J. Zhu, and Z. Li, “Correlation attention registration based on deep learning from histopathology to MRI of prostate,” Critical Revi. TM in Biomed. Eng., vol. 52, no. 2, pp. 39–50, 2024. doi: 10.1615/CritRevBiomedEng.2023050566. [Google Scholar] [PubMed] [CrossRef]
9. M. A. Azam, K. B. Khan, M. Ahmad, and M. Mazzara, “Multimodal medical image registration and fusion for quality enhancement,” Comput. Mater. Contin., vol. 68, no. 1, pp. 821–840, Mar. 2021. doi: 10.32604/cmc.2021.016131. [Google Scholar] [PubMed] [CrossRef]
10. R. Han et al., “Deformable MR-CT image registration using an unsupervised, dual-channel network for neurosurgical guidance,” Med. Image Anal., vol. 75, no. 31, pp. 102292, Jan. 2022. doi: 10.1016/j.media.2021.102292. [Google Scholar] [PubMed] [CrossRef]
11. J. Q. Zheng, Z. Wang, B. Huang, N. H. Lim, and B. W. Papież, “Residual aligner-based network (RANMotion-separable structure for coarse-to-fine discontinuous deformable registration,” Med. Image Anal., vol. 91, no. 42, pp. 103038, Jan. 2024. doi: 10.1016/j.media.2023.103038. [Google Scholar] [PubMed] [CrossRef]
12. A. Yang, T. Yang, X. Zhao, X. Zhang, Y. Yan and C. Jiao, “DTR-GAN: An unsupervised bidirectional translation generative adversarial network for MRI-CT registration,” Appl. Sci., vol. 14, no. 1, pp. 95, Dec. 2023. doi: 10.3390/app14010095. [Google Scholar] [CrossRef]
13. B. Zitova and J. Flusser, “Image registration methods: A survey,” Image Vis. Comput., vol. 21, no. 11, pp. 977–1000, Oct. 2003. doi: 10.1016/S0262-8856(03)00137-9. [Google Scholar] [CrossRef]
14. M. Roy et al., “Deep learning based registration of serial whole-slide histopathology images in different stains,” J. Pathol. Inf., vol. 14, no. 35, pp. 100311, Oct. 2023. doi: 10.1016/j.jpi.2023.100311. [Google Scholar] [PubMed] [CrossRef]
15. J. Borovec et al., “ANHIR: Automatic non-rigid histological image registration challenge,” IEEE Trans. Med. Imaging, vol. 39, no. 10, pp. 3042–3052, Oct. 2020. doi: 10.1109/TMI.2020.2986331. [Google Scholar] [PubMed] [CrossRef]
16. Y. Zhang, J. Chen, X. Ma, G. Wang, U. A. Bhatti and M. Huang, “Interactive medical image annotation using improved Attention U-net with compound geodesic distance,” Expert. Syst. Appl., vol. 237, no. PA, pp. 121282, Mar. 2024. doi: 10.1016/j.eswa.2023.121282. [Google Scholar] [CrossRef]
17. B. D. D. Vos, F. F. Berendsen, M. A. Viergever, M. Staring, and I. Išgum, “End-to-end unsupervised deformable image registration with a convolutional neural network,” presented at the Deep Learn. Med. Image Anal. Multimod. Learn. Clinic. Deci. Supp., Québec City, QC, Canada, Sep. 14, 2017, pp. 204–212. [Google Scholar]
18. Q. Zeng, C. Yang, Q. Gan, Q. Wang, and S. Wang, “EDIRNet: An unsupervised deformable registration model for X-ray and neutron images,” Appl. Opt., vol. 62, no. 29, pp. 7611–7620, Oct. 2023. doi: 10.1364/AO.500442. [Google Scholar] [PubMed] [CrossRef]
19. Y. Lei et al., “4D-CT deformable image registration using multiscale unsupervised deep learning,” Phy. Med. & Biol., vol. 65, no. 8, pp. 085003, Apr. 2020. doi: 10.1088/1361-6560/ab79c4. [Google Scholar] [PubMed] [CrossRef]
20. Y. Sang, X. Xing, Y. Wu, and D. Ruan, “Imposing implicit feasibility constraints on deformable image registration using a statistical generative model,” J. Med. Imaging, vol. 7, no. 6, pp. 064005–064005, Nov. 2020. doi: 10.1117/1.JMI.7.6.064005. [Google Scholar] [PubMed] [CrossRef]
21. A. Creswell, T. White, V. Dumoulin, K. Arulkumaran, B. Sengupta and A. A. Bharath, “Generative adversarial networks: An overview,” IEEE Signal Process. Mag., vol. 35, no. 1, pp. 53–65, Jan. 2018. doi: 10.1109/MSP.2017.2765202. [Google Scholar] [CrossRef]
22. J. Y. Zhu, T. Park, P. Isola, and A. A. Efros, “Unpaired image-to-image translation using cycle-consistent adversarial networks,” presented at the Proc. IEEE Int. Conf. Comput. Vis., Venice, Italy, Oct. 22–29, 2017. [Google Scholar]
23. D. Mahapatra, B. Antony, S. Sedai, and R. Garnavi, “Deformable medical image registration using generative adversarial networks,” presented at the 2018 IEEE 15th Int. Symp. Biomed. Imaging (ISBI 2018), Washington, DC, USA, Apr. 4–7, 2018, pp. 1449–1453. [Google Scholar]
24. H. Li et al., “CRCS: An automatic image processing pipeline for hormone level analysis of Cushing’s disease,” Methods, vol. 222, no. 11, pp. 28–40, Feb. 2024. doi: 10.1016/j.ymeth.2023.12.003. [Google Scholar] [PubMed] [CrossRef]
25. J. M. Morel and G. Yu, “ASIFT: A new framework for fully affine invariant image comparison,” SIAM J. Imaging Sci., vol. 2, no. 2, pp. 438–469, 2009. doi: 10.1137/080732730. [Google Scholar] [CrossRef]
26. S. Bharati, M. Mondal, P. Podder, and V. Prasath, “Deep learning for medical image registration: A comprehensive review,” Int. J. Comput. Inform. Syst. & Industrial Manag. Appl., vol. 14, no. 8, pp. 173–190, Apr. 2022. doi: 10.48550/arXiv.2204.11341. [Google Scholar] [CrossRef]
27. D. Sengupta, P. Gupta, and A. Biswas, “A survey on mutual information based medical image registration algorithms,” Neurocomputing, vol. 486, no. 14, pp. 174–188, May 2022. doi: 10.1016/j.neucom.2021.11.023. [Google Scholar] [CrossRef]
28. G. Lippolis, A. Edsjö, L. Helczynski, A. Bjartell, and N. C. Overgaard, “Automatic registration of multi-modal microscopy images for integrative analysis of prostate tissue sections,” BMC Cancer, vol. 13, no. 1, pp. 1–11, Sep. 2013. doi: 10.1186/1471-2407-13-408. [Google Scholar] [PubMed] [CrossRef]
29. H. Bay, T. Tuytelaars, and L. V. Gool, “SURF: Speeded up robust features,” presented at the 9th Eur. Conf. Comput. Vis., Graz, Austria, May 7–13, 2006, pp. 404–417. [Google Scholar]
30. J. Liu, X. Li, Q. Wei, J. Xu, and D. Ding, “Semi-supervised keypoint detector and descriptor for retinal image matching,” presented at the Eur. Conf. Comput. Vis., Tel Aviv, Israel, Oct. 23–27, 2022, pp. 593–609. [Google Scholar]
31. T. Cao, C. Zach, S. Modla, D. Powell, K. Czymmek and M. Niethammer, “Multi-modal registration for correlative microscopy using image analogies,” Med. Image Anal., vol. 18, no. 6, pp. 914–926, Aug. 2014. doi: 10.1016/j.media.2013.12.005. [Google Scholar] [PubMed] [CrossRef]
32. T. C. Mok and A. Chung, “Fast symmetric diffeomorphic image registration with convolutional neural networks,” presented at the Proc. IEEE/CVF Conf. Comput. Vis. Pattern Recognit., Seattle, WA, USA, Jun. 13–19, 2020, pp. 4644–4653. [Google Scholar]
33. X. Zhang, W. Jian, Y. Chen, and S. Yang, “Deform-GAN: An unsupervised learning model for deformable registration,” arXiv preprint arXiv:2002.11430, Feb. 2020. doi: 10.48550/arXiv.2002.11430. [Google Scholar] [CrossRef]
34. S. Liu, B. Yang, Y. Wang, J. Tian, L. Yin and W. Zheng, “2D/3D multimode medical image registration based on normalized cross-correlation,” Appl. Sci., vol. 12, no. 6, pp. 2828, Mar. 2022. doi: 10.3390/app12062828. [Google Scholar] [CrossRef]
35. Y. R. Rao, N. Prathapani, and E. Nagabhooshanam, “Application of normalized cross correlation to image registration,” Int. J. Res. Eng. Technol., vol. 3, no. 5, pp. 12–16, May 2014. doi: 10.15623/ijret.2014.0317003. [Google Scholar] [CrossRef]
36. K. Tang, Z. Li, L. Tian, L. Wang, and Y. Zhu, “ADMIR-affine and deformable medical image registration for drug-addicted brain images,” IEEE Access, vol. 8, pp. 70960–70968, Apr. 2020. doi: 10.1109/ACCESS.2020.2986829. [Google Scholar] [CrossRef]
37. L. Yann, “The MNIST database of handwritten digits,” 1998. Accessed: Oct. 1, 2010. [Online]. Available: http://yann.lecun.com/exdb/mnist/ [Google Scholar]
38. S. Ioffe and C. Szegedy, “Batch normalization: Accelerating deep network training by reducing internal covariate shift,” presented at the Int. Conf. Mach. Learn., Lille, France, Jul. 6–11, 2015, pp. 448–456. [Google Scholar]
39. D. J. Kroon and C. H. Slump, “MRI modalitiy transformation in demon registration,” presented at the 2009 IEEE Int. Symp. Biomed. Imaging: From Nano to Macro, Boston, MA, USA, Jun. 28–Jul. 1, 2009, pp. 963–966. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools