 Open Access
Open Access
ARTICLE
Empowering Diagnosis: Cutting-Edge Segmentation and Classification in Lung Cancer Analysis
1 Faculty of Computer Sciences & Information Technology, The Superior University, Lahore, 54000, Pakistan
2 Intelligent Data Visual Computing Research (IDVCR), Lahore, 54000, Pakistan
3 Information System Department, College of Computer and Information Sciences, Imam Mohammad Ibn Saud Islamic University (IMSIU), Riyadh, 11432, Saudi Arabia
4 School of Computer Science and Electronic Engineering (CSEE), University of Essex, Wivenhoe Park, Colchester, CO4 3SQ, UK
* Corresponding Author: Sheeraz Akram. Email:
(This article belongs to the Special Issue: Deep Learning in Medical Imaging-Disease Segmentation and Classification)
Computers, Materials & Continua 2024, 79(3), 4963-4977. https://doi.org/10.32604/cmc.2024.050204
Received 30 January 2024; Accepted 07 May 2024; Issue published 20 June 2024
Abstract
Lung cancer is a leading cause of global mortality rates. Early detection of pulmonary tumors can significantly enhance the survival rate of patients. Recently, various Computer-Aided Diagnostic (CAD) methods have been developed to enhance the detection of pulmonary nodules with high accuracy. Nevertheless, the existing methodologies cannot obtain a high level of specificity and sensitivity. The present study introduces a novel model for Lung Cancer Segmentation and Classification (LCSC), which incorporates two improved architectures, namely the improved U-Net architecture and the improved AlexNet architecture. The LCSC model comprises two distinct stages. The first stage involves the utilization of an improved U-Net architecture to segment candidate nodules extracted from the lung lobes. Subsequently, an improved AlexNet architecture is employed to classify lung cancer. During the first stage, the proposed model demonstrates a dice accuracy of 0.855, a precision of 0.933, and a recall of 0.789 for the segmentation of candidate nodules. The suggested improved AlexNet architecture attains 97.06% accuracy, a true positive rate of 96.36%, a true negative rate of 97.77%, a positive predictive value of 97.74%, and a negative predictive value of 96.41% for classifying pulmonary cancer as either benign or malignant. The proposed LCSC model is tested and evaluated employing the publically available dataset furnished by the Lung Image Database Consortium and Image Database Resource Initiative (LIDC-IDRI). This proposed technique exhibits remarkable performance compared to the existing methods by using various evaluation parameters.Keywords
The uncontrolled growth of human cells within the body is exhibited as a cancer that is extremely challenging to diagnose and identify. There are two main groups of tumor cells: Benign and malignant [1]. The non-cancerous or benign cells are not harmful and they do not spread to nearby tissues. The second type, cancer or malignant cells spreads to neighboring tissue cells and obstructs the growth of healthy cells. Even, most experienced radiologists are unable to identify these types of abnormalities in cell structure while seeking for malignancy. The most frequently occurring malignancies are breast, prostate, ovary, cervical, brain, bone, colon, and lung cancers [2]. Medical professionals need more time, resources, and expertise to segment and classify lung cancer. Traditional Computer-Aided Diagnosis (CAD) based systems rely on handcrafted features that were extracted from the lung images. These systems detect and classify pulmonary nodules from the lung images automatically [3]. Dang et al. [4] explored multiple Deep Learning (DL) architectures based on a two-layer ensemble system for the segmentation of medical images. In this work, a segmentation mask was produced by various segmentation models for each input image and then a voting strategy was applied to produce an ensemble segmentation mask in the first layer. An ensemble segmentation mask was employed as input for the final segmentation in the second layer. The results exhibited greater improvements in the field of medical segmentation by using this model.
Similarly, another study was presented by Shah et al. [5] that was based on the DL ensemble 2D Convolutional Neural Network (CNN) technique for the diagnosis of pulmonary nodules. Three deep learning models named CNN1, CNN2, and CNN3 were applied to the LUng Nodule Analysis (LUNA16) dataset. In the subsequent step, these models were combined by an ensemble 2D approach and produced 95% accuracy for the identification of lung nodules. In another study, researchers [6] presented lobe segmentation, extraction of candidate nodules, and lung cancer classification approach based on computational intelligence techniques.
Mkindu et al. [7] introduced a novel technique consisting of a 3D-CNN incorporated with channel attention mechanisms to detect pulmonary nodules in CT images. The proposed U-shaped network employed encoding and decoding for feature extraction and indicating the prediction outcomes of candidate nodules, respectively. Hybrid efficient channel attention was incorporated into the network for retaining rich information and overwhelming unusable features. Furthermore, a 3D Regional Proposal Network (RPN) consisting of three anchor boxes was used to detect multilevel candidate nodules. A 10-fold cross-validation technique was exploited on the LUNA16 dataset to validate this technique. Various advanced segmentation methods are utilized to produce enhanced segmentation tasks in the field of medical data.
Lung tumors are diagnosed by several researchers using various techniques based on computational intelligence approaches. CAD systems employ DL models that can reduce the workload of medical professionals in diagnosing different diseases, namely in segmenting, detecting, and classifying pulmonary cancer nodules. The study introduces an automated DL methodology that segments the lobes and classifies lung nodules to improve accuracy in nodule detection. Early diagnosis of lung cancer will ultimately decrease the death rate and enhance the survival rate.
The research article is structured as follows: The literature review is discussed in Section 2. The proposed methodology is shown in Section 3. Findings are analyzed in Section 4. Section 5 consists of the conclusion of this research and limitations and future work.
Lung nodule classification is an essential task for early detection of lung tumor [8]. The rapid progress of information technology has resulted in advancements in big data processing, machine learning, and, DL techniques [9]. Various computational intelligence-based approaches are widely used to segment and classify lung cancer. Delfan et al. [10] presented a model that utilized a DL framework comprising of U-Net architecture incorporated with pre-trained InceptionV3 blocks for lung tissue segmentation and obtained 0.951 of dice coefficient on LUNA16 dataset. The authors in the study [11] employed deep learning techniques to segment lung parenchyma and identify lung nodules in CT scan images. In data preprocessing phase, median filter was applied to remove noise. Recurrent Residual U-Net (R2U-Net) and watershed techniques were applied for segmentation and found that R2U-Net has the best results in segmentation. Furthermore, the segmented images obtained from R2U-Net were used to detect the nodule. Two classifiers Stacked Autoencoder (SAE) and Deep Belief Network (DBN) were implemented to classify the lung nodules and it was found that SAE classifier achieved 96.25% accuracy while DBN algorithm obtained 95.36% accuracy. Similarly, Maqsood et al. [12] demonstrated an efficient DA-Net for segmentation of lung nodules that reached 81% of dice score. Lu et al. [13] explored lung nodule segmentation consisting consisted of deep learning and reached 0.772 of dice. Fredriksen et al. [14] presented a lung tumor segmentation method based on Teacher-student technique and achieved 0.72 of dice, 0.8333 of precision, 0,8889 of recall. Similarly, another study presented by Shimazaki et al. [15] reached 0.52 of dice and 0.73 of recall. Lima et al. [16] investigated pre-trained models like Resnet50, VGG16, VGG19, Inception, and Xception and used for the automatic classification of lung tumor by using CT scan images. Pre-trained models were applied for the feature extraction from 2D slices of 3D nodules. In the subsequent step, Principal Component Analysis (PCA) approach was exploited for the reduction of dimensionality of feature vectors and produced into the same length. Next, important features were selected to combine these feature vectors and represented them into 3D nodules. For the classification of pulmonary nodules, a Random forest algorithm was used and achieved 95.34% accuracy, 90.53% sensitivity, 97.26% specificity, and 0.99 of AUC.
Soniya et al. [17] developed an automatic framework consisting of five steps including image acquisition, image enhancement, segmentation, feature extraction, and classification. In image acquisition, a total number of 250 and 1450 lung CT scans were obtained from in-house clinical and publicly available Lung Image Database Consortium and Image Database Resource Initiative (LIDC-IDRI) respectively. A weiner filter with an Unsharp masking technique was performed for noise removal from the input image in the image enhancement step. The hierarchical Random Walker with Bayes Model (HRWBM) approach was employed to enhance image sequence and the Gray Level Co-occurrence Matrix (GLCM) method was implemented to extract features. Finally, three classifiers Support Vector Machine (SVM), Feed-Forward Neural Network (FFNN), and Deep Recurrent Neural Network (DRNN) were integrated with HRWBM and implemented for the classification of lung tumor. The outcomes of the thrice classifiers demonstrated that HRWBM with DRNN has reached the best accuracy of 97.3% and 94.7% for LIDC-IDRI and in-house clinical datasets. Mothkur et al. [18] explored low-memory and lightweight deep neural networks such as vanilla 2D-CNN, 2D MobileNet, and 2D SqueezeNet for lung nodule classification. The presented Lightweight Deep Neural Network (DNN) based models achieved results as 85.21% of accuracy as well as equitable sensitivity and specificity. Mkindu et al. [19] exhibited a 3D Multi-Scale Vision Transformer (3D-MSViT) for the detection of lung nodules. The CAD system based on 3D-MSViT was used to increase multi-scale feature extraction as well as improve prediction efficiency of lung nodules and achieved a sensitivity of 97.81%.
The researchers [20] developed a framework for lung cancer diagnosis consisting of three phases: Data segregation, feature extraction, and prediction. The data segregation step was done by utilizing Butterfly optimization, second features extractions and correlation were executed by Jaya optimization and finally, auto encoder algorithm was utilized for the prediction of lung tumor. A hybrid classification method was utilized to enhance the classification accuracy. The method integrated with two deep architectures DenseNet-201 and DarkNet-53 and attained the maximum accuracy of 98.69% in terms of classification of pulmonary nodules in [21]. In another study, Elwahsh et al. [22] presented a novel deep neural learning cancer methodology for the detection of lung tumor. The method consisted of three stages: The deep network was applied to abstract features from the five cancer datasets. Next step, DNN was used to train clinical or genomic data samples and at the final stage, the model was measured in terms of detection of lung cancer at its earlier stages and achieved 93% of accuracy. The Long Short Term Memory (LSTM) was implemented for binary classification of pulmonary nodules in [23] by Gupta et al. An accuracy of 86.89% was obtained by the proposed architecture on LIDC-IDRI and LUNA16 datasets. Here is another comprehensive evaluation [24] of the experimental findings that proved that the proposed method achieved 93.2% of accuracy for the lung cancer classification based on tensor real-time transfer learning. Bhattacharjee et al. [25] applied a basic neural network for the diagnosing of lung tumor. The automatic CAD model attained 76% of accuracy, 78% of specificity, and 75% of sensitivity by using LIDC-IDRI. Similarly, another study based on the DL detection method to detect and classify lung nodules achieved an extraordinary level of accuracy. Zuo et al. [26] formed a multi-resolution CNN model integrated with knowledge transfer that was used for classifying lung nodules into cancerous or non-cancerous nodules on the publicly available LUNA16 dataset. The experimental outcomes demonstrated 0.9733 accuracy, and 0.9673 precision. Xu et al. [27] explored ensemble learning for the identification of pulmonary nodules in CT images. YOLOv3 and CNN networks were applied to private and public datasets and fused into a Logistic Regression (LR) approach for the identification of lung nodules. The experimental outcomes of the fused model were 93.82% of accuracy, 94.85% of sensitivity, and 93.82% of specificity for the public dataset while 92.31% of accuracy, 92.68% of sensitivity, and 91.89% of specificity for private dataset. The authors [28] presented an automated and intelligent approach for diagnosing of lung cancer utilizing deep ensemble learning techniques. Two standard approaches such as t-Distribution Stochastic Neighbor Embedding (t-SNE) and PCA were implemented to extract features. Furthermore, Best Fitness-based Squirrel Search Algorithm (BF-SSA) was used to select the optimal features from the extracted features. Finally, High Ranking Deep Ensemble Learning (HRDEL) based on five detection models was applied, and high-ranking classification results as final forecasted outcomes. The suggested model attained 93.15% of accuracy, 93.14% of sensitivity, and 93.16% of specificity. Mahmood et al. [29] worked on an improved CNN model for the classification of lung nodules by utilizing CT scan images from Lungx and Data Science Bowl (DSB) 2017 datasets. The proposed system consisted of AlexNet and employed three blocks for feature extractions. Softmax was used for the classification of pulmonary tumor. The suggested work by [30] consisted of handcrafted features and classification by applying five neural networks for the classification of pulmonary nodules in CT images. Various statistical parameters were measured to evaluate the performance of five classifiers. The Stochastic Gradient Descent (SGD) classifier reached the best results among all other neural classifiers. Chen et al. [31] explored a pulmonary nodules detection model consisting of multi-scale and multi-view approaches. F-Net architecture was proposed to detect candidate nodules and multi-scale was applied to share a convolutional structure model. Similarly, another study [32] focused on the usage of transfer learning and applied pre-trained VGG19 architecture for the identification of lung cancer. A tenfold cross-validation method was applied to CT images and achieved 0.82 accuracy, sensitivity 0.75, and 0.87 specificity. Lydia et al. [33] have developed an automatic lung cancer detection and classification method comprised of DL approaches. A modified regularized K-means technique was implemented to segment nodule images. Next, an improved CNN technique was applied to identify lung nodules and classify malignant nodules into S1-S4. The proposed approach attained an accuracy 96.5% when compared to other existing approaches. The researchers [34] focused on various modified AlexNet algorithms for the classification of lung nodules. Another study presented by Ye et al. [35] achieved 94.2% accuracy in classifying lung nodules by using the LIDC-IDRI dataset. Tomassini et al. [36] presented a decision support system to classify non-small cell lung cancer by using CT scans and achieved 82% of accuracy. Different methods utilized to classify lung tumor were examined in the literature review. However, these models have certain limitations such as less accuracy for classifying lung cancer as reported in Table 1. Various evaluation parameters were applied to measure the performance of the previous studies. However, these studies identified areas for improvement in classifying lung cancer.

The primary contributions of present research are as follows:
• The segmentation of CT scan images has been improved by using improved U-Net architecture.
• The candidate nodules are extracted in a well-defined way that leads to enhance the accuracy of lung nodule classification.
• Improved AlexNet architecture is implemented for the classification of the lung nodule nodules.
• Various statistical parameters are measured to evaluate the performance of the proposed Lung Cancer Segmentation and Classification (LCSC) technique and compared with existing methods.
In this work, the Lung cancer segmentation and classification model is proposed to segment the candidate nodules and detection of lung nodules, and the proposed methodology is exhibited in Fig. 1.
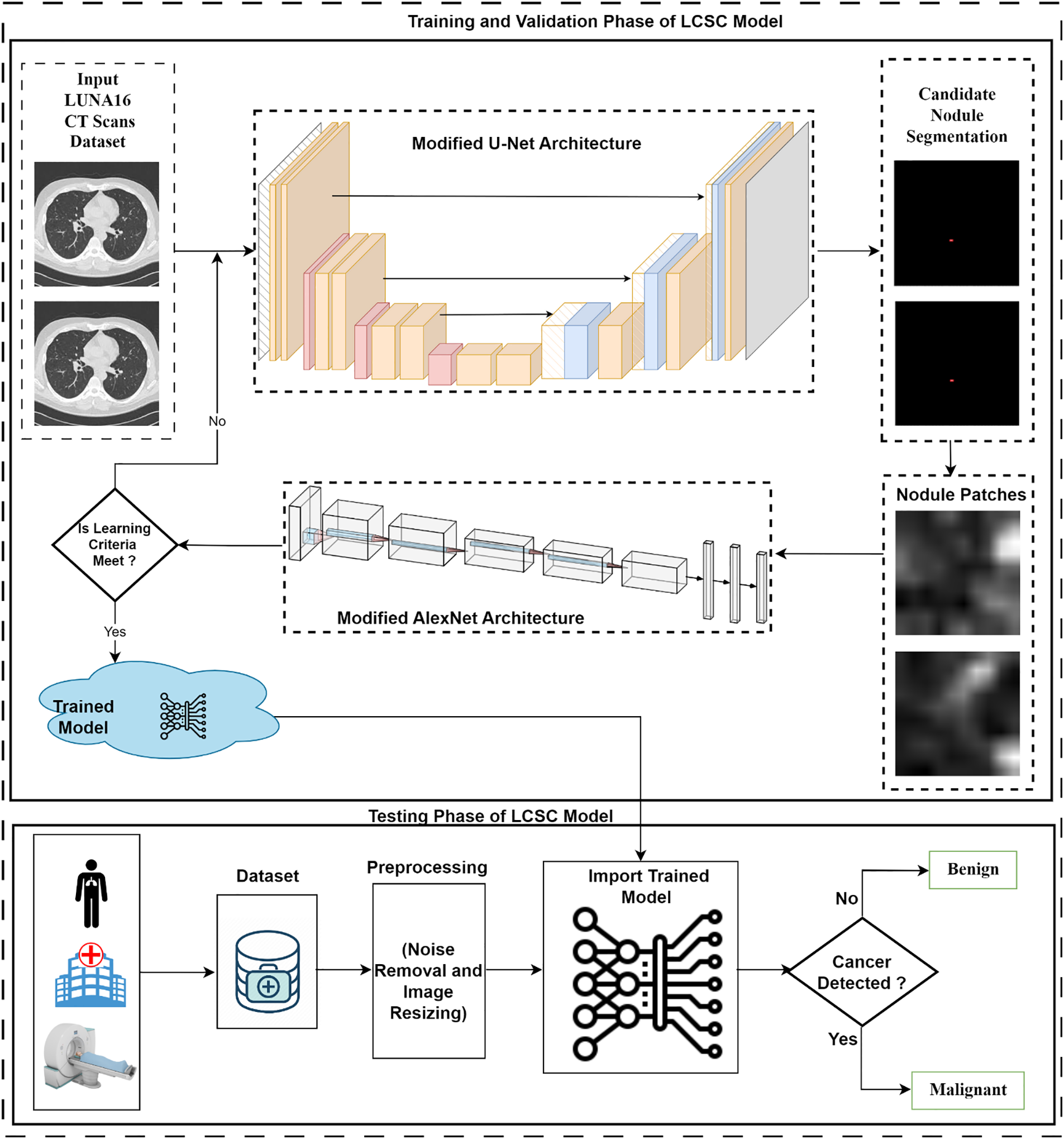
Figure 1: Workflow of the proposed Lung Cancer Segmentation and Classification (LCSC) model
The LCSC model comprises of two phases: Training and Validation phase, and the Testing phase. In the training phase, the LIDC-IDRI lung cancer dataset is utilized for training of the suggested model. Improved U-Net architecture is implemented to get segmented candidate nodules from the CT scans. In the training phase, nodule patches are used as input for the classification of lung cancer. In this phase, an improved AlexNet architecture has been implemented to extract features, and softmax function is utilized for the classification of the proposed LCSC model.
The publicly available LIDC-IDRI dataset is employed in this study. The data is stored in Digital Imaging and Communication Medicine (DICOM) images and the resolution of the acquired CT scan images is 512 × 512. The public image collection LIDC-IDRI lung cancer dataset that is offered by the National Cancer Institute of America [37] is used for the training and evaluation of the proposed strategy and technique. The LIDC-IDRI dataset comprises 1018 CT scan cases and it is annotated by four expert chest radiologists. The proposed LCSC model is trained and validated on this dataset. The CT scan images are fed to improved U-Net architecture for the segmentation of candidate nodules. A total number of 4800 CT images have been utilized for training, validation, and testing of the proposed LCSC model. CT images are divided into 80% (3840 CT images) for training, 10% (480 CT images) for validation, and 10% (480 CT images) for testing. A total number of 988 nodule slices has been used for the testing of this model.
In 2015, Ronneberger et al. [38] presented the U-Net architecture, which is based on CNN and usually employed for image segmentation tasks. The proposed U-Net architecture comprises of 4 encoder layers and 4 decoder layers which are connected by using skip connections. They designed it for the segmentation of images by predicting a pixel-wise mask representing the group of the corresponding pixel. U-Net architecture comprises of encoder and decoder network with skip connections. The encoder part or downsample is similar to ordinary CNN that contains convolutional and pooling layers, where the spatial dimensions of the input image decrease gradually as well as the number of feature maps increase. The decoder part or upsample is designed to increase the feature maps to their original spatial dimensions and produce a segmentation mask. The skip connections concatenate the encoder and decoder parts and are being used to transfer the high-resolution features of the encoder to the decoder part to enhance the segmentation accuracy. Recently, the U-Net architecture has been mostly utilized for medical image segmentation tasks by many researchers to refine segmentation output.
3.2 Improved U-Net Architecture for Segmentation
The proposed LCSC model consisting of the improved U-Net architecture is implemented to segment and extract nodule patches from the LIDC-IDRI dataset in the training phase. The proposed improved U-Net architecture is demonstrated in Fig. 2. To build the proposed LCSC model for feature extraction, improved U-Net architecture comprises of an encoder and decoder path having five layers, respectively.
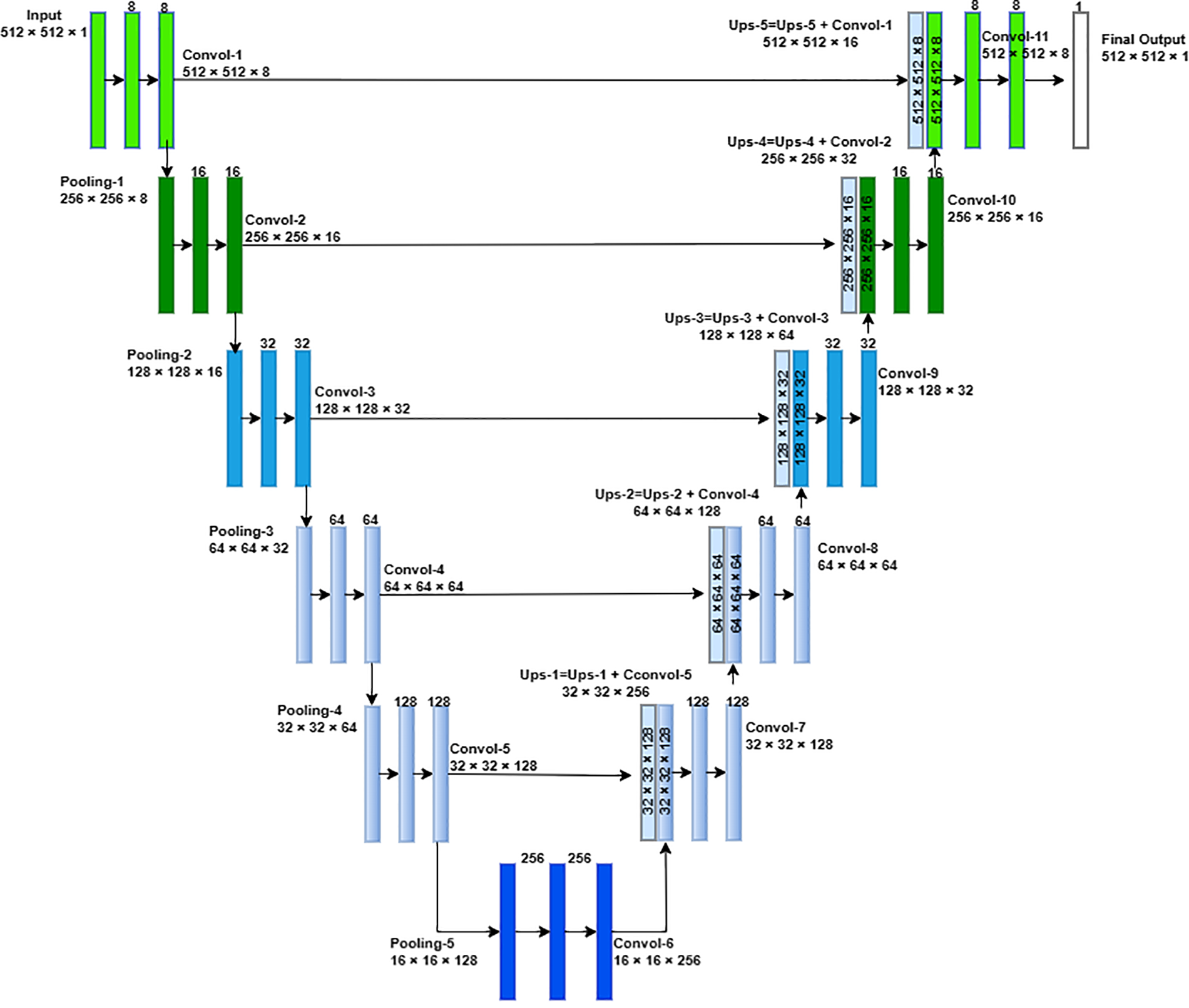
Figure 2: Improved proposed U-Net architecture for the Lung Cancer Segmentation and Classification (LCSC) model
The improved U-Net architecture produces candidate nodule segmentation with 512 × 512 × 1 as output. From these candidate nodules, we extract 48 × 48 centered at the center of candidate nodules. Now, we have selected nodule patches from 48 × 48 from the segmented candidate nodule.
The segmentation phase of the proposed LCSC model is evaluated with Dice similarity coefficient (Dice), recall, and precision, which are shown as follows:
where
3.3 Improved AlexNet Architecture for Lung Cancer Classification
The improved AlexNet architecture is demonstrated in Fig. 3. The improved AlexNet system architecture contains of six convolutional, three max pooling, and two fully connected layers. The segmented nodule patch size is 48 × 48 × 1 which is used as input in the improved AlexNet architecture. Convolutional layers are liable for extracting meaningful features from the nodule patches. The max pooling layers decrease the dimensionality of the patch size but preserve significant information.
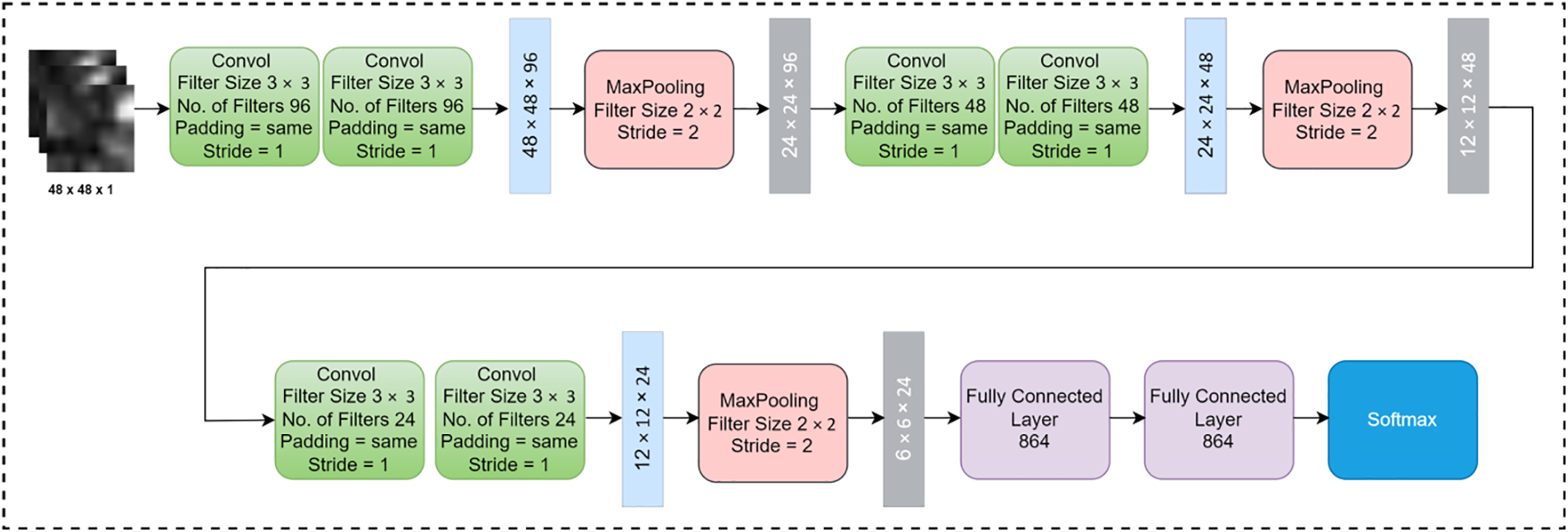
Figure 3: Improved proposed AlexNet architecture for the Lung Cancer Segmentation and Classification (LCSC) model
Fully connected layers is applied to take features from the feature extraction block and transform them into an output. Softmax was utilized to classifylung tumor into benign and malignant. In this research, Convolutional operations are performed with various numbers of filters, filter size 3 × 3, same padding, and one stride in convolutional layers. On the other hand, the max pooling operation is performed with filter size 2 × 2 and stride 2 in pooling layers. In fully connected layers, all neurons are connected to each other and finally, softmax is implemented to classify the lung tumor into malignant and benign.
The performance of the LCSC model is evaluated using a range of performance criteria. The training and testing phases were utilized in the Keras tool using Python 3.9 in this research. High sensitivity ensures early detection of lung cancer while high specificity reduces false positives, minimizing unnecessary interventions. SGD optimizer is utilized with hyperparameters such as 0.0001 of learning rate, 200 of epochs and 50 batch size.
The accuracy of positive predictions is measured by precision, whereas the completeness of positive forecast is measured by recall and the outcomes are demonstrated in Table 2. The LSCS model achieves 0.855 of Dice, 0.9333 of precision and 0.7887 of recall. LCSC model for segmentation of candidate nodule obtains better accuracy as compared to other current cutting-edge methods.

Following the segmentation of the candidate nodule, 9880 nodule slices were extracted from the 4800 CT images in the LIDC-IDRI dataset. Total number of 9880 nodule slices are further divided into 80% (7904 nodule slices) for training, 10% (988 nodule slices) for validation and 10% (988 nodule slices) for testing purpose.
The training results of proposed LCSC model for classification is demonstrated in Table 3. In the training step, a total number of 7904 are divided into 3952 benign and 3952 malignant slices for the training of classification of the proposed LCSC model, whereas, 988 nodule slices are divided into 494 benign and 494 malignant slices are used for the validation of the suggested LCSC model.

In training, LCSC model correctly predicts 3909 slices as benign and wrongly predicts 44 slices as malignant. Similarly, in malignant nodule slices, LCSC model wrongly predicts 38 sample slices and 3910 predicts correctly as malignant.
In validation phase of the proposed model, LCSC is correctly predicted 487 slices as benign and wrongly predicts 7 slices as malignant.
Similarly, in malignant nodule slices, LCSC model is wrongly predicted 15 sample slices and 479 predicted correctly as malignant. The validation outcomes of the LCSC model for classification are demonstrated in Table 4.

The testing results of proposed LCSC model for classification is reported in Table 5. In testing phase of the proposed model, LCSC correctly predicts 487 slices as benign and wrongly predicts 7 slices as malignant. Similarly, in malignant nodule slices, LCSC model wrongly predicts 15 sample slices and correctly predicts 479 slices as malignant.

The suggested LCSC model for classification of lung nodules is evaluated with various performance metrics such as accuracy, miss classification, true positive rate, positive predictive value, false positive rate, and negative predictive value. The proposed lung cancer segmentation and classification model achieves remarkable results in term of various statistical measurements. The proposed LCSC model obtains 97.06% accuracy, true positive rate 96.36%, true negative rate 97.77%, positive predictive value 97.74% and negative predictive value 96.41% in classifying lung cancer. Fig. 4 illustrates the performance metrics of proposed LCSC model to classify lung cancer.
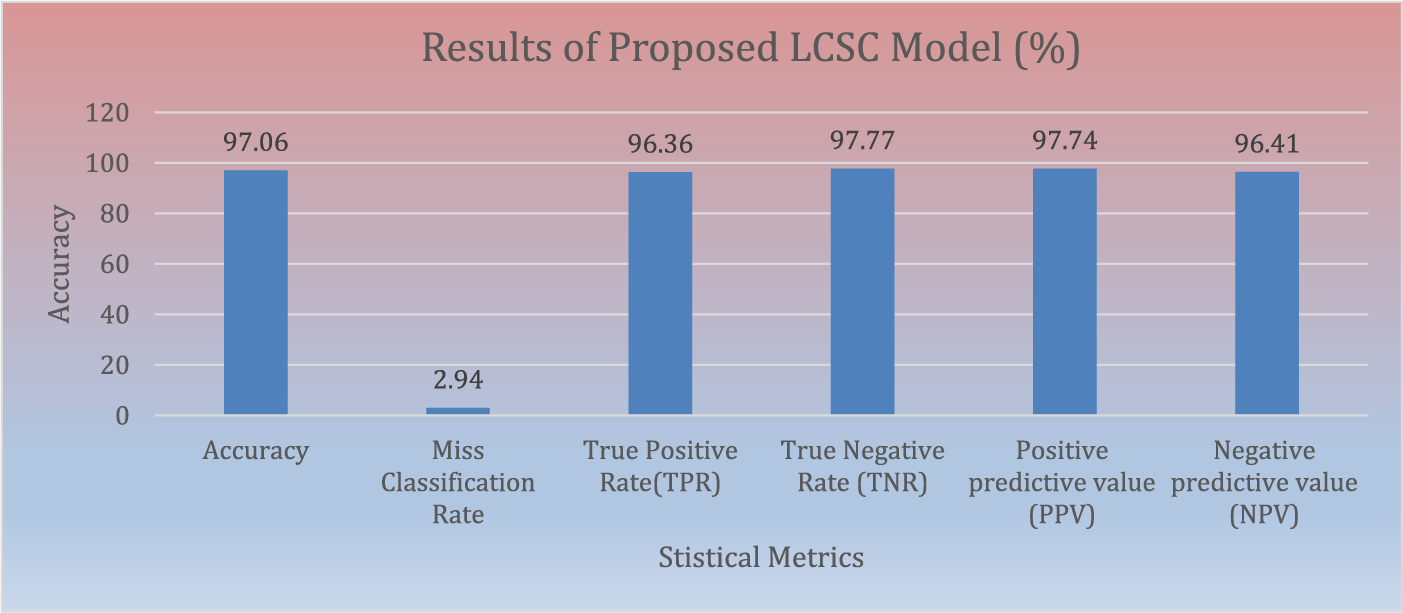
Figure 4: Results of proposed LCSC model for the classification of lung cancer
Fig. 5 demonstrates the comparison of proposed LCSC model to classify lung cancer with existing studies. The LCSC model attains 97.06% of accuracy to classify the lung cancer by utilizing LIDC-IDRI lung cancer dataset.
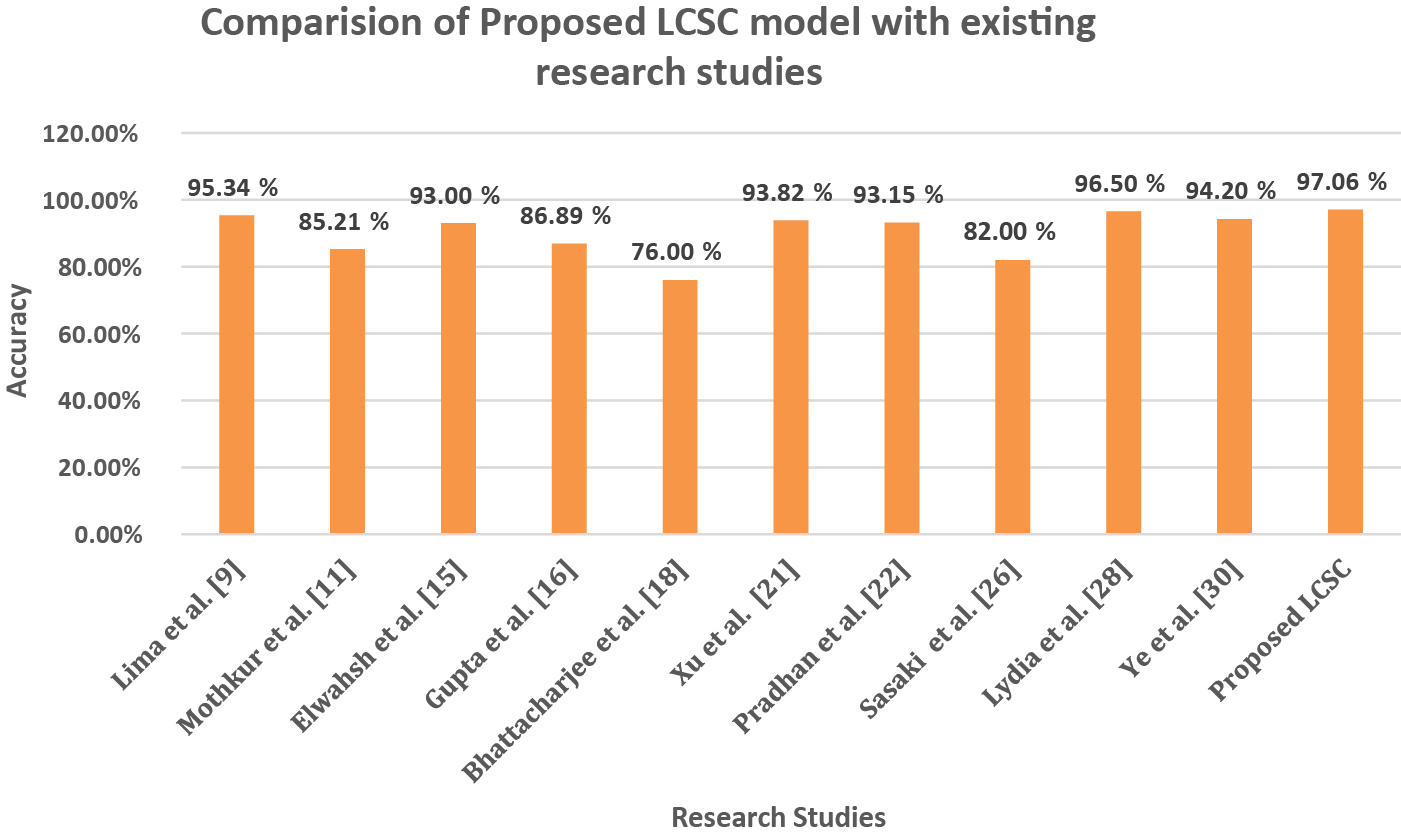
Figure 5: Comparison of accuracy between proposed LCSC model and previous studies
The present study demonstrates an effective and efficient model for lobe segmentation and lung cancer classification. The proposed LCSC model consists of two improved architectures, namely the improved U-Net architecture and the improved AlexNet architecture. An improved U-Net architecture is implemented for the segmentation of candidate nodules that are extracted from the lung lobes. Subsequently, an improved AlexNet architecture is employed to classify lung cancer. The publicly available LIDC-IDRI dataset is utilized in this research work. First, an improved U-Net architecture is implemented for the candidate nodule segmentation. The improved U-Net architecture reached 0.8550 of Dice, 0.9333 of precision, and 0.7887 of recall. Subsequently, nodule patches 48 × 48 × 1 are selected from segmented candidate nodules. Then these nodule patches are forwarded to improved AlexNet architecture to classify pulmonary nodules into benign and malignant. The proposed LCSC model reached 97.06% of accuracy, 2.94% of miss classification rate, 96.36% of the true positive rate, 97.77% of the true negative rate, 97.74% of positive predictive value, and 96.41% of negative predictive value. In comparison to other existing models, the proposed LCSC model demonstrates remarkable performance. This study aims to segment and classify lung cancer utilizing an improved U-Net and AlexNet architecture, by utilizing the LIDC-IDRI dataset. The scope of the present research is confined to a specific dataset which can be enhanced on other existing and real-time datasets. Furthermore, the LCSC model can be evaluated on other lung cancer datasets with other deep learning approaches.
Acknowledgement: We express our gratitude to Imam Mohammad Ibn Saud Islamic University (IMSIU) for their support to conduct this research.
Funding Statement: This work was supported and funded by the Deanship of Scientific Research at Imam Mohammad Ibn Saud Islamic University (IMSIU) (Grant Number IMSIU-RP23044).
Author Contributions: Study conception and design: I. Naseer, T. Masood, S. Akram, A. Jaffar, Z. Ali, A. Ahmad, S.U. Rehman; Data selected: S. Akram, Z. Ali, A. Ahmad, S.U. Rehman; Analysis and interpretation of results: I. Naseer, T. Masood, S. Akram, A. Jaffar; Draft manuscript preparation: I. Naseer, T. Masood, S. Akram, A. Jaffar. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The dataset used in this paper is publicly available can be downloaded from the corresponding author upon request.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. C. Baston, A. I. Parosanu, M. Mihai, O. Moldoveanu, I. M. Stanciu and C. Nitipir, “Tumor-to-tumor metastasis of lung cancer to kidney cancer: A review of the literature and our experience,” Diagnostics, vol. 14, no. 5, pp. 553–568, 2024. doi: 10.3390/diagnostics14050553. [Google Scholar] [PubMed] [CrossRef]
2. R. L. Siegel, A. N. Giaquinto, and A. Jemal, “Cancer statistics 2024,” CA Cancer J. Clin., vol. 74, no. 1, pp. 12–49, 2024. [Google Scholar] [PubMed]
3. M. Kawagishi et al., “A study of computer-aided diagnosis for pulmonary nodule: Comparison between classification accuracies using calculated image features and imaging findings annotated by radiologists,” Int. J. Comput. Assist. Radiol. Surg., vol. 12, no. 3, pp. 767–776, 2017. doi: 10.1007/s11548-017-1554-0. [Google Scholar] [PubMed] [CrossRef]
4. T. Dang, T. T. Nguyen, J. M. Call, E. Elyan, and C. F. M. García, “Two layer ensemble of deep learning models for medical image segmentation,” arXiv preprint arXiv:2104.04809. 2021. [Google Scholar]
5. A. A. Shah, H. A. M. Malik, A. H. Muhammad, A. Alourani, and Z. A. Butt, “Deep learning ensemble 2D CNN approach towards the detection of lung cancer,” Sci. Rep., vol. 13, no. 1, pp. 1–15, 2023. doi: 10.1038/s41598-023-29656-z. [Google Scholar] [PubMed] [CrossRef]
6. I. Naseer, S. Akram, T. Masood, M. Rashid, and A. Jaffar, “Lung cancer classification using modified u-net based lobe segmentation and nodule detection,” IEEE Access, vol. 11, no. 6, pp. 60279–60291, 2023. doi: 10.1109/ACCESS.2023.3285821. [Google Scholar] [CrossRef]
7. H. Mkindu, L. Wu, and Y. Zhao, “Lung nodule detection of CT images based on combining 3D-CNN and squeeze-and-excitation networks,” Multimed. Tools Appl., vol. 82, no. 2, pp. 25747–25760, 2023. doi: 10.1007/s11042-023-14581-0. [Google Scholar] [CrossRef]
8. I. Naseer, T. Masood, S. Akram, A. Jaffar, M. Rashid and M. Amjad Iqbal, “Lung cancer detection using modified alexnet architecture and support vector machine,” Comput. Mater. Contin., vol. 74, no. 1, pp. 2039–2054, 2023. doi: 10.32604/cmc.2023.032927 [Google Scholar] [CrossRef]
9. S. Aminizadeh et al., “The applications of machine learning techniques in medical data processing based on distributed computing and the Internet of Things,” Comput. Methods Programs Biomed., vol. 241, pp. 107745–107763, 2023. doi: 10.1016/j.cmpb.2023.107745. [Google Scholar] [PubMed] [CrossRef]
10. N. Delfan, H. A. Moghaddam, M. Modaresi, and K. Afshari, “CT-LungNet: A deep learning framework for precise lung tissue segmentation in 3D thoracic CT scans,” arXiv preprint arXiv:2212.13971, 2022. [Google Scholar]
11. G. S. Nandeesh, M. Nagabushanam, S. Pramodkumar, and S. Nandini, “Lung parenchyma segmentation and nodule detection using deep learning,” J. Opt., vol. 25, no. 4, pp. 1–8, 2023. [Google Scholar]
12. M. Maqsood, S. Yasmin, I. Mehmood, M. Bukhari, and M. Kim, “An efficient DA-Net architecture for lung nodule segmentation,” Mathematics, vol. 9, no. 13, pp. 1457–1472, 2021. doi: 10.3390/math9131457. [Google Scholar] [CrossRef]
13. D. Lu, J. Chu, R. Zhao, Y. Zhang, and G. Tian, “A novel deep learning network and its application for pulmonary nodule segmentation,” Comput. Intell. Neurosci., vol. 2022, no. 5, pp. 1–18, 2022. doi: 10.1155/2022/7124902. [Google Scholar] [PubMed] [CrossRef]
14. V. Fredriksen et al., “Teacher-student approach for lung tumor segmentation from mixed-supervised datasets,” PLoS One, vol. 17, no. 4, pp. 266147–266164, 2022. doi: 10.1371/journal.pone.0266147. [Google Scholar] [PubMed] [CrossRef]
15. A. Shimazaki et al., “Deep learning-based algorithm for lung cancer detection on chest radiographs using the segmentation method,” Sci. Rep., vol. 12, no. 1, pp. 1–10, 2022. doi: 10.1038/s41598-021-04667-w. [Google Scholar] [PubMed] [CrossRef]
16. T. Lima, D. Luz, A. Oseas, R. Veras, and F. Araújo, “Automatic classification of pulmonary nodules in computed tomography images using pre-trained networks and bag of features,” Multimed. Tools Appl., vol. 82, no. 4, pp. 42977, 2023. doi: 10.1007/s11042-023-14900-5. [Google Scholar] [PubMed] [CrossRef]
17. S. L. Soniya and T. A. B. Raj, “Lung tumor analysis using a thrice novelty block classification approach,” Sig. Image Video Process., vol. 17, no. 6, pp. 3027–3034, 2023. doi: 10.1007/s11760-023-02523-0. [Google Scholar] [CrossRef]
18. R. Mothkur and B. N. Veerappa, “Classification of lung cancer using lightweight deep neural networks,” Procedia Comput. Sci., vol. 218, no. 1, pp. 1869–1877, 2023. doi: 10.1016/j.procs.2023.01.164. [Google Scholar] [CrossRef]
19. H. Mkindu, L. Wu, and Y. Zhao, “3D multi-scale vision transformer for lung nodule detection in chest CT images,” Sig. Image Video Process., vol. 17, no. 1, pp. 2473–2480, 2023. doi: 10.1007/s11760-022-02464-0. [Google Scholar] [CrossRef]
20. M. S. Pethuraj, B. bin Mohd Aboobaider, and L. B. Salahuddin, “Developing lung cancer post-diagnosis system using pervasive data analytic framework,” Comput. Elect. Eng., vol. 105, no. 1, pp. 108528–108543, 2023. doi: 10.1016/j.compeleceng.2022.108528. [Google Scholar] [CrossRef]
21. U. Demiroğlu, B. Şenol, M. Yildirim, and Y. Eroğlu, “Classification of computerized tomography images to diagnose non-small cell lung cancer using a hybrid model,” Multimed. Tools Appl., vol. 82, no. 3, pp. 33379–33400, 2023. doi: 10.1007/s11042-023-14943-8. [Google Scholar] [CrossRef]
22. H. Elwahsh, M. A. Tawfeek, A. A. Abd El-Aziz, M. A. Mahmood, M. Alsabaan and E. El-shafeiy, “A new approach for cancer prediction based on deep neural learning,” J. King Saud Univ.–Comput. Inf,. Sci., vol. 35, no. 6, pp. 101565–101582, 2023. doi: 10.1016/j.jksuci.2023.101565. [Google Scholar] [CrossRef]
23. S. Gupta, A. Garg, V. Bishnoi, and N. Goel, “Binary classification of pulmonary nodules using long short-term memory,” in 4th IEEE Int. Conf. Art. Intell. Speech Technol., Delhi, India, 2023, pp. 1–5. [Google Scholar]
24. V. Bishnoi and N. Goel, “Tensor-RT-based transfer learning model for lung cancer classification,” J. Digit. Imaging, vol. 36, no. 4, pp. 1364–1375, 2023. doi: 10.1007/s10278-023-00822-z. [Google Scholar] [PubMed] [CrossRef]
25. A. Bhattacharjee, R. Murugan, S. Majumder, and T. Goel, “Neural network-based computer-aided lung cancer detection,” Res. Biomed. Eng., vol. 37, no. 4, pp. 657–671, 2021. doi: 10.1007/s42600-021-00173-0. [Google Scholar] [CrossRef]
26. W. Zuo, F. Zhou, Z. Li, and L. Wang, “Multi-resolution CNN and knowledge transfer for candidate classification in lung nodule detection,” IEEE Access, vol. 7, no. 3, pp. 32510–32521, 2019. doi: 10.1109/ACCESS.2019.2903587. [Google Scholar] [CrossRef]
27. Y. Xu et al., “Identification of benign and malignant lung nodules in CT images based on ensemble learning method,” Interdiscip. Sci.: Comput. Life Sci., vol. 14, no. 1, pp. 130–140, 2022. [Google Scholar]
28. K. S. Pradhan, P. Chawla, and R. Tiwari, “HRDEL: High ranking deep ensemble learning-based lung cancer diagnosis model,” Expert. Syst. Appl., vol. 213, no. 3, pp. 118956–118979, 2023. doi: 10.1016/j.eswa.2022.118956. [Google Scholar] [CrossRef]
29. S. A. Mahmood and H. A. Ahmed, “An improved CNN-based architecture for automatic lung nodule lassification,” Med. Biol. Eng. Comput., vol. 60, no. 7, pp. 1977–1986, 2022. doi: 10.1007/s11517-022-02578-0. [Google Scholar] [PubMed] [CrossRef]
30. M. A. Hussain and L. Gogoi, “Performance analyses of five neural network classifiers on nodule classification in lung CT images using weka: A comparative study,” Phys. Eng. Sci. Med., vol. 45, no. 4, pp. 1193–1204, 2022. doi: 10.1007/s13246-022-01187-3. [Google Scholar] [PubMed] [CrossRef]
31. Y. Chen, X. Hou, Y. Yang, Q. Ge, Y. Zhou and S. Nie, “A novel deep learning model based on multi-scale and multi-view for detection of pulmonary nodules,” J. Digit. Imaging, vol. 36, no. 2, pp. 688–699, 2022. doi: 10.1007/s10278-022-00749-x. [Google Scholar] [PubMed] [CrossRef]
32. Y. Sasaki, Y. Kondo, T. Aoki, N. Koizumi, T. Ozaki and H. Seki, “Use of deep learning to predict postoperative recurrence of lung adenocarcinoma from preoperative CT,” Int. J. Comput. Assist. Radiol. Surg., vol. 17, no. 9, pp. 1651–1661, 2022. doi: 10.1007/s11548-022-02694-0. [Google Scholar] [PubMed] [CrossRef]
33. M. D. Lydia and M. Parkash, “An improved convolution neural network and modified regularized k means based automatic lung nodule detection and classification,” J. Digit. Imaging, vol. 36, no. 4, pp. 1431–1446, 2023. doi: 10.1007/s10278-023-00809-w. [Google Scholar] [PubMed] [CrossRef]
34. I. Naseer, S. Akram, T. Masood, A. Jaffar, M. A. Khan and A. Mosavi, “Performance analysis of state-of-the-art CNN architectures for LUNA16,” Sensors, vol. 22, no. 12, pp. 4426–4441, 2022. doi: 10.3390/s22124426. [Google Scholar] [PubMed] [CrossRef]
35. M. Ye et al., “A classifier for improving early lung cancer diagnosis incorporating artificial intelligence and liquid biopsy,” Front Oncol., vol. 12, no. 3, pp. 853801–853810, 2022. doi: 10.3389/fonc.2022.853801. [Google Scholar] [PubMed] [CrossRef]
36. S. Tomassini et al., “On-cloud decision-support system for non-small cell lung cancer histology characterization from thorax computed tomography scans,” Comput. Med. Imaging Graph., vol. 110, pp. 102310–102322, 2023. doi: 10.1016/j.compmedimag.2023.102310. [Google Scholar] [PubMed] [CrossRef]
37. S. G. Armato et al., “The lung image database consortium and image database resource initiative: A completed reference database of lung nodules on ct scans,” Med. Phys., vol. 38, no. 2, pp. 915–931, 2011. doi: 10.1118/1.3528204. [Google Scholar] [PubMed] [CrossRef]
38. O. Ronneberger, P. Fischer, and T. Brox, “U-Net: Convolutional networks for biomedical image segmentation,” in Intern. Conf. Med. Image Comput. Comput. Assist. Interv., Munich, Germany, 2015, pp. 234–241. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools