 Open Access
Open Access
ARTICLE
CrossLinkNet: An Explainable and Trustworthy AI Framework for Whole-Slide Images Segmentation
1 Network and Data Security Key Laboratory of Sichuan Province, University of Electronic Science and Technology of China, Chengdu, 610054, China
2 Faculty of Data Science, City University of Macau, Macau, 999078, China
* Corresponding Author: Erqiang Zhou. Email:
(This article belongs to the Special Issue: Security, Privacy, and Robustness for Trustworthy AI Systems)
Computers, Materials & Continua 2024, 79(3), 4703-4724. https://doi.org/10.32604/cmc.2024.049791
Received 17 January 2024; Accepted 23 April 2024; Issue published 20 June 2024
Abstract
In the intelligent medical diagnosis area, Artificial Intelligence (AI)’s trustworthiness, reliability, and interpretability are critical, especially in cancer diagnosis. Traditional neural networks, while excellent at processing natural images, often lack interpretability and adaptability when processing high-resolution digital pathological images. This limitation is particularly evident in pathological diagnosis, which is the gold standard of cancer diagnosis and relies on a pathologist’s careful examination and analysis of digital pathological slides to identify the features and progression of the disease. Therefore, the integration of interpretable AI into smart medical diagnosis is not only an inevitable technological trend but also a key to improving diagnostic accuracy and reliability. In this paper, we introduce an innovative Multi-Scale Multi-Branch Feature Encoder (MSBE) and present the design of the CrossLinkNet Framework. The MSBE enhances the network’s capability for feature extraction by allowing the adjustment of hyperparameters to configure the number of branches and modules. The CrossLinkNet Framework, serving as a versatile image segmentation network architecture, employs cross-layer encoder-decoder connections for multi-level feature fusion, thereby enhancing feature integration and segmentation accuracy. Comprehensive quantitative and qualitative experiments on two datasets demonstrate that CrossLinkNet, equipped with the MSBE encoder, not only achieves accurate segmentation results but is also adaptable to various tumor segmentation tasks and scenarios by replacing different feature encoders. Crucially, CrossLinkNet emphasizes the interpretability of the AI model, a crucial aspect for medical professionals, providing an in-depth understanding of the model’s decisions and thereby enhancing trust and reliability in AI-assisted diagnostics.Keywords
The integration of AI into clinical practice is transforming the healthcare landscape, particularly in diagnostic pathology [1]. Accurate diagnosis is the cornerstone of effective cancer treatment, where pathologists often face the daunting task of deciphering complex patterns in whole-slide images (WSIs) to identify malignancies [2]. However, this traditional process is not without its challenges. It is labor-intensive, subject to inter-observer variability, and requires significant expertise, which may be limited in resource-constrained environments [3]. As AI technology, particularly deep learning, becomes increasingly capable of augmenting or even surpassing human performance in image recognition tasks, its potential in enhancing the speed, accuracy, and efficiency of cancer diagnosis is becoming more apparent. However, alongside these advancements, concerns have emerged about the opaque nature of AI decisions and their implications for patient safety and treatment outcomes [4].
In the sensitive realm of oncology, the trust in AI systems hinges on their ability to not only perform with high accuracy but also provide clarity on how conclusions are drawn [5]. Traditional neural networks, while proficient in various applications, often operate as “black boxes,” offering little insight into their internal decision-making processes [6]. This lack of transparency is particularly problematic in the clinical setting, where understanding the rationale behind diagnostic decisions is imperative for clinician acceptance, quality control, and ethical considerations [7]. In this context, the role of explainable AI (XAI) technologies becomes pivotal, as they bridge the gap between AI performance and human interpretability. XAI aims to make the workings of complex neural networks accessible and comprehensible, enabling clinicians to follow the AI’s logic, verify its reasoning, and, crucially, trust its outputs.
With the proliferation of digital pathology, WSIs have emerged as a standard in modern cancer diagnostics, providing an exhaustive view of tissue samples [8]. However, manual analysis of WSIs is notably laborious, time-consuming, and subject to variability due to human interpretation [9]. To mitigate these challenges, there has been a growing impetus towards incorporating explainable AI models. These models not only enhance the efficiency and accuracy of pathological analyses but also contribute to a deeper understanding of disease characteristics and progression [10]. AI’s proficiency in rapidly processing and analyzing large volumes of data has been transformative across various sectors, particularly in medical imaging [11]. In the field of pathology, AI algorithms are increasingly recognized for their potential to improve WSI interpretation, providing results that are more consistent and reproducible [12,13].
Traditional machine learning approaches for image analysis predominantly depend on manually extracted and defined features [14]. Traditional neural network approaches encounter significant challenges in medical image segmentation, particularly with whole-slide images (WSIs). Firstly, Whole Slide Images (WSIs) typically contain gigabytes of pixels, making it impossible for traditional deep neural networks to process them directly due to computational and memory constraints. This severely limits the ability to analyze WSIs comprehensively, affecting the extraction of critical diagnostic information. Furthermore, WSIs often contain tumor regions that are very small relative to the overall image size, presenting significant challenges to conventional approaches in identifying tumors and segmenting crucial minor targets. Additionally, the rigid architecture of numerous traditional neural networks, due to the lack of a modular design, restricts their adaptability. This limitation obstructs the integration of alternative feature encoders that could potentially enhance feature extraction for specific types of medical images. Lastly, the complexity of these traditional network architectures often lacks interpretability. Within clinical contexts, grasping the fundamental rationales behind diagnostic forecasts is essential for fostering trust and guiding clinical decisions.
Addressing the limitations of traditional neural network approaches, this paper introduces CrossLinkNet, which is specifically designed to address these challenges through its scalable processing of high-resolution images, enhanced sensitivity to small target regions, modular architecture for flexible feature encoding, and improved interpretability, thereby offering a significant advancement in the field of medical image segmentation.
The primary contributions of this paper are summarized as follows:
• An explainable Multi-Scale Multi-Branch Feature Encoder is proposed to enhance the network’s capability for feature extraction. The MSBE Encoder facilitates the fine-tuning of hyperparameters to customize the quantity of branches and modules. This provides flexibility in adjusting the width of the network, thereby enhancing its capability to extract features.
• A versatile medical image segmentation network architecture, CrossLinkNet, is designed to accurately identify tumor regions by leveraging cross-layer encoder-decoder connections for multi-level feature fusion, thereby enhancing feature integration and improving segmentation accuracy. Moreover, this architecture can be flexibly adapted to various tumor segmentation tasks and scenarios through the replacement of different feature encoders.
• The CrossLinkNet with an MSBE encoder demonstrates accurate segmentation results through comprehensive quantitative and qualitative experiments on two datasets, namely BOT and Kvasir. Moreover, it exhibits adaptability to various tumor segmentation tasks and scenarios by seamlessly replacing different feature encoders.
The structure of this paper is organized as follows: Section 2 provides a concise review of existing literature pertinent to pathological image analysis. Section 3 elaborates on the deep neural network proposed for pathological image segmentation. Section 4 outlines the experimental setup. Section 5 presents and analyses the experimental results obtained. Section 6 discusses the potential challenges of the proposed approach. Finnally, Section 7 offers the conclusion.
In the field of image segmentation, numerous methods have been developed over the years. Traditional segmentation techniques, such as threshold-based methods and edge detection algorithms, have laid the foundation in this domain. This section explores the evolution and intricacies of these traditional segmentation approaches and deep learning approaches.
2.1 Threshold Segmentation Method
Threshold-based image segmentation is a common technique in digital image processing and an important method for achieving image segmentation [15]. The Otsu algorithm, also known as Otsu’s method, represents a classical approach to image thresholding, initially introduced by Nobuyuki Otsu in 1979 [16]. Zhu et al. [17] developed a novel segmentation algorithm to overcome the limitations of traditional threshold-based techniques in extracting complex information. This method employs adaptive thresholds for each pixel, determined by the mean and variance of adjacent pixel values, thereby enhancing edge detection. Yang et al. [18] focused on enhancing the Otsu algorithm by examining the relationship between pixel grayscale values and cumulative pixel counts. The use of threshold-based image segmentation methods is common in digital image processing; however, they have notable limitations. One major drawback is the criticality of selecting an appropriate threshold, as different thresholds can lead to divergent segmentation outcomes.
2.2 Segmentation Method Based on an Interpretable Convolutional Neural Network
With the rapid advancement in the fields of computer vision and deep learning [19], Convolutional Neural Networks (CNNs) have emerged as a pivotal component for various visual tasks, particularly excelling in image segmentation. Kiran et al. [20] utilized ResNet for segmenting cell clusters in whole-slide cervical pathology images. Their model achieved an impressive accuracy of 99.63% on single-cell images and 96.37% on cell clusters within entire images. Lin et al. [21] introduced a deep convolutional neural network-based lesion detection system, optimized for high efficiency and sensitivity across various lesion sizes. CNN-based image segmentation algorithms show high efficiency in detecting tumors in pathological images. However, they struggle to interpret the contextual relationships among image segments, and there is a considerable gap in AI interpretability.
2.3 Segmentation Method Based on Transformer
The Vision Transformer (ViT) [22] represents a technological turning point in the field of computer vision, as it applies Transformer technology to this domain. Chen et al. [23] developed a Multimodal Co-Attention Transformer (MCAT) framework, leveraging Transformer layers for multi-instance learning to map relationships between pathological image features and genomic data. Yin et al. [24] developed the PyT2T-ViT, a streamlined Vision Transformer architecture for multi-instance learning. Transformer-based approaches demonstrate significant advantages in capturing long-range dependencies and integrating global and local information. However, their extensive model parameters require substantial hardware resources, which can result in overfitting when dealing with limited data.
The exploration of various segmentation methods in pathology imaging reveals a dynamic progression from traditional to advanced deep learning techniques. All the methods previously discussed have their limitations. With the advancement of this field, overcoming these weaknesses is essential for creating more efficient, accurate, and interpretable segmentation methods for pathological analysis.
In this paper, a universal and explainable segmentation network named CrossLinkNet is proposed, with the specific details of the network shown in Fig. 1. CrossLinkNet consists of the MSB Encoder and the CrossLinkNet Framework. The MSB Encoder is a multi-scale, multi-branch light-weight feature encoder designed for extracting multi-scale features, with its architecture illustrated in Fig. 2. Fig. 3 demonstrates the structure of the MSB Encoder in various modes. The CrossLinkNet Framework is a generic segmentation framework based on an encoder-decoder architecture with cross-layer connections, as shown in Fig. 4. By combining the CrossLinkNet Framework with the MSB Encoder, it is possible to identify more features of tumors and perform pixel-level segmentation. By substituting different feature encoders, the network can also be adapted to various medical imaging analysis tasks, ultimately achieving commendable results in segmentation accuracy. The following sections will provide detailed introductions to the Multi-Scale Multi-Branch Feature Encoder (MSBE) and the generic segmentation network architecture, CrossLinkNet.

Figure 1: CrossLinkNet architecture

Figure 2: Basic structure of MSB encoder
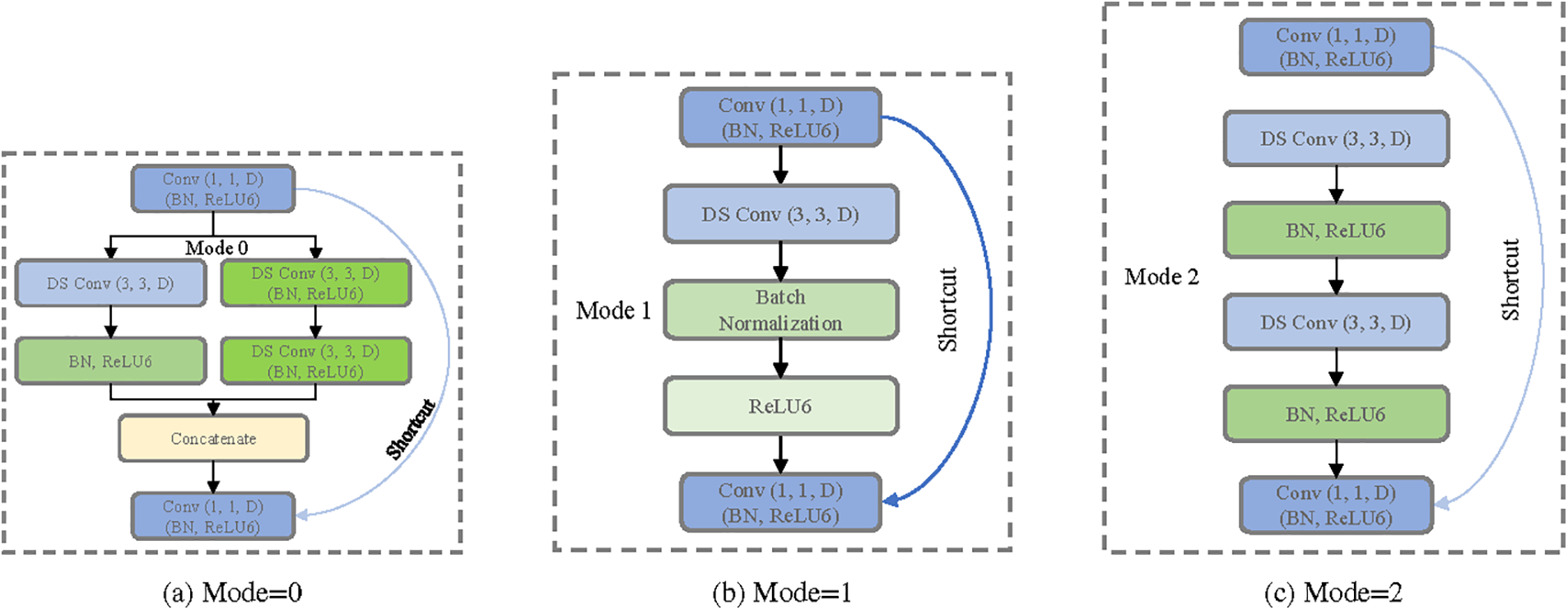
Figure 3: Different structure of MSB encoder

Figure 4: CrossLinkNet segmentation network framework
3.1 Multi-Scale Multi-Branch Feature Encoder
The extraction of multi-scale features plays a pivotal role in neural networks, particularly in tasks such as object detection and image segmentation, where it can significantly enhance the performance of the model. Traditional convolutional neural networks employ standard convolutional kernels and pooling operations of fixed sizes to extract features from images. However, this approach predominantly captures local features and is limited in its capacity to handle objects of varying sizes. To address this limitation, this section introduces a MSB encoder for feature extraction within the proposed versatile segmentation network, CrossLinkNet. The fundamental architecture of this feature encoder is depicted in Fig. 2.
The architecture of the MSB Encoder primarily encompasses dimensionality expansion, depthwise separable convolutions, multi-scale feature extraction, dimensionality reduction and skip connections. The MSB encoder initially employs a strategy of increasing dimensionality (using 1 × 1 convolutions), followed by feature extraction, and culminating in a reduction of dimensions. This approach primarily aims to bolster the network’s ability to represent complex features. Elevating the dimensionality allows the network to learn features in a higher-dimensional space, which facilitates capturing more complex feature relationships. The dimension reduction phase that follows is designed to curtail the parameter count, alleviate overfitting, and ensure the network remains efficient. The adoption of depthwise separable convolutions is based on their advantage of reducing computations and model size while still maintaining good feature extraction performance. By decomposing traditional convolutions into depthwise convolution and pointwise convolution, depthwise separable convolutions significantly reduce computational complexity and model parameters, making the model more lightweight and suitable for devices with limited computational resources. Within the strategy of multi-scale feature extraction, the application of 1 × 1, 3 × 3, and 5 × 5 convolutions caters to the necessity of discerning features across differing scales. 1 × 1 convolutions are primarily used to adjust the number of channels, reduce the number of parameters, while still retaining spatial features; 3 × 3 convolutions, a common convolution size, are effective in capturing local features; and 5 × 5 convolutions are capable of capturing features from a larger receptive field. However, the direct use of 5 × 5 convolutions involves a large number of parameters, so two stacked 3 × 3 convolutions are used as a substitute for a single 5 × 5 convolution, which not only expands the receptive field but also reduces the number of parameters, enhancing network efficiency. Finally, the integration of skip connections aims to mitigate the vanishing gradient issue encountered in training deep networks, which maintains network depth while facilitating effective information flow between layers. Skip connections allow for the direct transfer of information from shallower layers to deeper layers, aiding in the restoration of detail information and improving segmentation accuracy.
Overall, the MSBE takes into consideration multi-scale feature extraction, computational efficiency, and effective information flow. This approach is pivotal in refining the utilization of computational resources while maintaining the integrity of the model’s performance, especially critical in the segmentation of high-resolution whole slide images. Next, we will delve into Dimensionality Expansion, Depthwise Separable Convolution, Multi-Scale Feature Extraction, Dimensionality Reduction, and Skip Connections in detail.
Dimensionality Expansion: The architecture employs a standard
For an input feature map
where
Depthwise Separable Convolution: This architecture comprehensively employs depthwise separable convolutions to reduce the number of parameters and computational costs while maintaining robust feature extraction capabilities. This makes the network more lightweight and ensures efficiency without compromising on powerful representational capacity. Depthwise separable convolution involves an initial depthwise convolution, followed by a pointwise convolution. For a
where
Multi-Scale Feature Extraction: In order to capture information of various scales within the image more effectively, a multi-scale feature extraction strategy is introduced. The encoder selects different convolutional kernel sizes based on distinct patterns:
Mode = 0: Employs both
Mode = 1: Utilizes solely the
Mode = 2: Engages two consecutive
The representation of Mode 0, using a
where
Dimensionality Reduction: Following the extraction of features at different scales, a convolutional operation is applied to reduce the channel count of the output feature map, aligning it with the channel count of the input feature map in preparation for the skip connections. The dimensionality reduction can be represented using a
where
Skip Connections: The design of skip connections ensures improved information propagation through the depth of the model, guaranteeing that shallow features can be directly transmitted to deeper layers. This approach mitigates the common issue of gradient vanishing in deep networks. Additionally, skip connections facilitate easier network training and enable the construction of deeper network architectures, which, in turn, enhances the stability of model training.
The core concept of the MSB Encoder lies in the selective application of various convolutional kernel sizes based on different modes for feature extraction. Moreover, the encoder dynamically chooses the convolutional kernel size according to the mode, providing enhanced flexibility for feature extraction. The incorporation of skip connections enables the model to better maintain and convey information at deeper layers, thereby mitigating the issue of vanishing gradients and enhancing training stability. Compared to traditional convolutions, depthwise separable convolutions significantly reduce the number of parameters, thereby decreasing computational costs, but also retain impressive feature extraction capabilities. This design renders the network more lightweight while maintaining powerful representational abilities and ensuring greater efficiency. In summary, the MSB encoder not only accounts for feature extraction across varying scales but also effectively controls the number of parameters, making it a highly promising feature extractor.
3.2 Cross-Layer Connected Segmentation Network Framework
We proposed an innovative segmentation network structure, the CrossLinkNet Framework, which employs cross-layer connections to segment pathological images. The CrossLinkNet Framework’s key advantage is its ability to progressively learn and integrate information through multiple sub-modules, thereby enhancing the model’s feature extraction and contextual information recognition capability by focusing on extracting features across various dimensions. This method bolsters the model’s robustness, augmenting its tolerance to noise, variations, and perturbations present in the input data. The CrossLinkNet Framework is highly adaptable, allowing for easy customization of encoder and decoder configurations to suit different tasks by modifying sub-module structures and parameters. Each sub-module can be replaced with high-performance encoders to handle different datasets and segmentation challenges. CrossLinkNet progressively learns and integrates features across layers, enhancing segmentation results. This flexibility renders the CrossLinkNet Framework suitable for a wide array of tumor segmentation tasks and scenarios.
The CrossLinkNet architecture is a novel image segmentation network structure, where the cornerstone is the implementation of cross-layer connections that facilitate information transfer between encoders and among decoders. This design optimizes the pathways of information flow, thereby enhancing segmentation accuracy. The network consists of two principal components: The encoders and the decoders, as illustrated in Fig. 4.
The encoder, being the epicenter and novelty of the framework, aims to efficiently extract features from the input image. It employs a standard sequence of convolution, batch normalization, and activation functions to capture multi-level information from the input image, followed by max-pooling operations for downsampling. This creates feature maps at various depths, providing a rich source of information for the decoder. Diverging from conventional decoder designs, the decoder in CrossLinkNet is comprised of features from two upsampling branches and encoder features from the corresponding level. Subsequently, these upsampled feature maps are merged with the feature maps from the encoder through a concatenation operation, ensuring that cross-layer information is thoroughly harnessed during the decoding process. Such an arrangement allows the model to concurrently capture both local details and global contextual information of the image, significantly amplifying the precision of segmentation.
The encoder component of the CrossLinkNet framework is designed with remarkable flexibility, accommodating a multitude of feature extraction architectures. In this paper, the MSB encoder proposed in the preceding subsection is employed as the default encoder within the CrossLinkNet architecture, with the overall network architecture illustrated in Fig. 1. Furthermore, CrossLinkNet strategically integrates batch normalization layers post multiple convolutional layers, which can expedite the convergence of the network during the training of deep learning models and also contribute to enhancing the generalization performance of the model. This design thoroughly contemplates the balance between model training efficiency and performance.
In summary, CrossLinkNet presents an image segmentation framework utilizing cross-layer connections to optimize pathways for information flow and enhance the feature representation capabilities of the model. The flexibility of its encoder design, the innovation within its decoder structure, and the incorporation of batch normalization collectively contribute to positioning CrossLinkNet as a highly accurate solution for image segmentation tasks. It possesses vast potential for application and demonstrates exceptional ability in feature extraction.
3.3 Interpretability in MSBE and CrossLinkNet
During the design of the CrossLinkNet framework and MSBE, we specifically focused on enhancing the interpretability of the model. This is particularly vital in medical image analysis, where understanding AI’s decision-making process can significantly increase medical professionals’ trust in AI-assisted diagnostic systems. Here is how we enhanced the interpretability of CrossLinkNet and MSBE by designing specific model structural components.
Multi-Scale Feature Extraction: MSBE processes images at multiple scales using different modes (Mode = 0, Mode = 1, Mode = 2), allowing the model to capture a range of features from fine textures to macro structures. This multi-level feature extraction enhances the model’s performance and makes the decision-making process of the model more transparent. Visualizing the feature activation maps at different scales, medical practitioners can clearly see how the model makes diagnoses based on different details of the image, which offering a clear understanding about the decision-making logic of AI.
Cross-Layer Connections: The cross-layer connection design in CrossLinkNet enables high-level features to be directly associated with low-level features, which not only aids in the propagation of gradients, reducing information loss during training but also enhances the model’s interpretability. Through these connections, we can trace how high-level decisions are directly related to the original image, providing medical professionals with an intuitive way to verify the basis of the model’s decisions.
Overall, the design of CrossLinkNet and MSBE has fully taken into account the importance of interpretability in medical image analysis, especially in critical application areas like cancer diagnosis. By combining high performance with high interpretability, our model is capable of providing accurate diagnostic results, and also allows doctors to understand and trust the AI’s decision-making process, which is of significant importance for advancing the application of AI in the medical field.
This section is devoted to presenting the experimental results and analysis, conducting a thorough evaluation of the MSB encoder and the universal segmentation network CrossLinkNet using publicly available pathological image datasets. Initially, the experimental datasets and environment are introduced, followed by a detailed description of the evaluation metrics employed.
The dataset utilized in this chapter originates from the “BOT Series Competition on Pathological Slide Recognition AI Challenge”, focusing on a gastric cancer digital pathology image dataset. This dataset emphasizes the pathological characteristics of gastric cancer and is designed to support high-quality automated pathological image recognition. To ensure the dataset’s quality and accuracy, during the initial phase, each image was meticulously annotated by a medically knowledgeable pathologist and subsequently subjected to careful review by an expert panel. In later stages, images were annotated by two pathologists with extensive knowledge in the field, with their work again reviewed by experts post-completion. Such a multi-stage, multi-reviewer process guarantees the accuracy of the annotations in the dataset and minimizes errors due to subjective judgment to the greatest extent possible.
In the experimental section of the paper, the annotation files in .svg format were initially converted into binary mask images in .png format. Subsequently, the pathological and mask images, at a resolution of 2048 × 2048, were scaled down to 256 × 256 pixels for storage. To validate the model’s generalizability, the dataset was randomly split into a training set of 490 images, a validation set of 70 images, and a test set of 140 images. Each model was trained on the training set, validated on the validation set, and evaluated on the test set. The detailed distribution of the dataset is shown in Table 1.
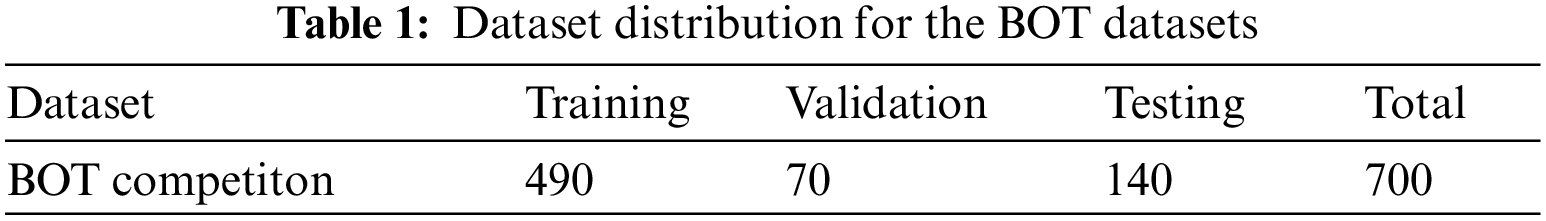
The first part of the experiment compared our proposed MSB encoder with four classic encoder architectures: VGG, ResNet34, ResNet50, and MobileNet, to explore performance differences across various backbone networks. Following this, our newly designed CrossLinkNet was compared with existing architectures like UNet and SegNet under the same encoder configurations (namely, VGG, ResNet34, ResNet50, MobileNet, and MSBE). Subsequently, to comprehensively assess the superiority of the proposed model, we conducted extensive comparative experiments of CrossLinkNet (MSBE) with a series of classical networks, such as SegNet, UNet++, as well as the renowned DeepLabV3+ and the recently introduced state-of-the-art model, PidNet. Finally, to confirm the applicability of the proposed universal segmentation network to other segmentation tasks, the CrossLinkNet (MSBE) network was also tested using the Kvasir gastrointestinal polyp segmentation dataset.
In summary, the efficacy of the MSB encoder, the performance of the universal segmentation network CrossLinkNet, the combined effect of CrossLinkNet (MSBE), and its performance on other datasets were evaluated through four different ablation studies. This multi-dimensional validation confirmed the high performance of the proposed model.
In this study, we conducted experimental validation on a high-end workstation with the following configuration: An Intel Core i7-8700k CPU, Nvidia GTX 3090 GPU, and 64 GB of system memory. The workstation was running the Ubuntu 20.04 LTS operating system. The experiments were performed using the deep learning framework Keras, with cross-entropy loss function as the foundation and stochastic gradient descent as the optimizer for the experimental model. The training was carried out for 100 epochs with a batch size set to 16. These settings enabled us to achieve favorable results within a relatively short time and allowed us to train and test more complex deep learning models.
In our experiments, the evaluation metrics include Frequency-Weighted Intersection over Union (FW_IoU), Mean Intersection over Union (mIoU), Mean Dice Coefficient (mDice), Pixel Accuracy (PA), Mean Average Precision (mAP), Mean Recall (mRecall). Through these metrics, a comprehensive understanding of the model’s performance can be gained, facilitating model assessment, improvement, and optimization.
5.1 Comparison with Different Encoders
VGG, ResNet34, ResNet50, and MobileNet are classic convolutional neural network architectures often used as feature extractors to assist other primary networks in feature extraction. In our experimental design, we developed an innovative multi-scale, multi-branch feature encoder (MSBE) module and integrated it into various baseline networks such as UNet, SegNet, and the newly proposed CrossLinkNet. We evaluated the performance of the MSB encoder in image segmentation tasks through comparative experiments. The results are shown in Table 2. For the three sets of experimental results, we used different encoders and conducted ablation studies on each encoder using different backbone network structures to accurately assess their performance in image segmentation tasks.
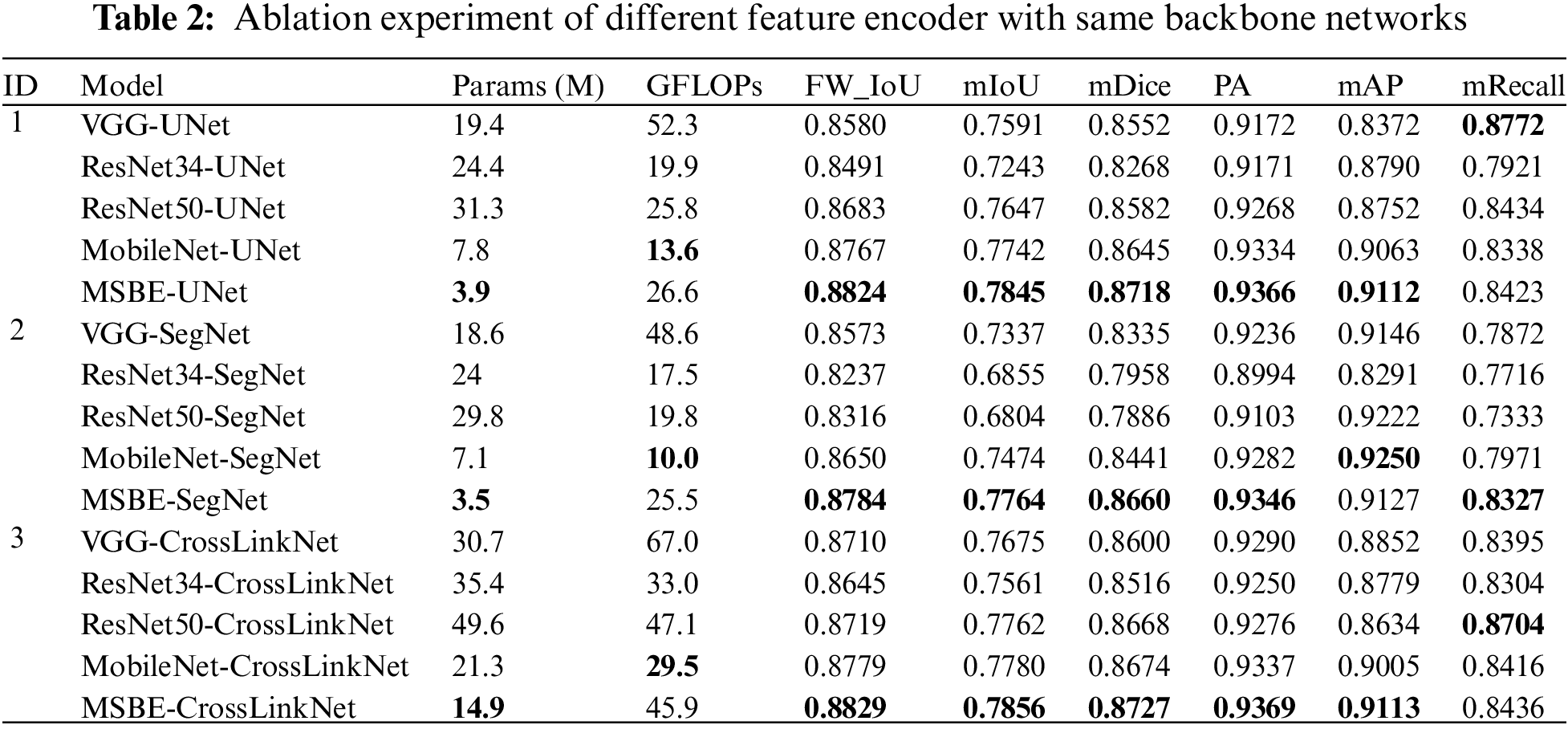
In the first set of experiments of this study, we chose UNet as the basic backbone network and combined it with five different encoder structures: VGG, ResNet34, ResNet50, MobileNet, and MSBE for ablation studies. The results showed that the UNet model, with the MSBE encoder, performed excellently on multiple performance metrics, including FW_IoU, mIoU, mDice, PA, and mAP. Specifically, MSBE-UNet achieved the best performance in terms of frequency weighted IoU, mean IoU, mean Dice coefficient, pixel accuracy, and mean precision. In terms of mIoU, MSBE-UNet was about 2.89% higher than the average of the other four encoder structures. In terms of mAP, its performance was 7.4%, 3.21%, 3.6%, and 0.48% higher than VGG, ResNet34, ResNet50, and MobileNet, respectively. However, in terms of mRecall, the performance of MSBE-UNet was slightly inferior to VGG-UNet and ResNet50-UNet. This result suggests that the MSBE encoder is more effective in feature extraction and preserving image details compared to other encoder structures.
In the second set of experiments of this study, SegNet was used as the base backbone network, and the MSBE encoder was compared with four different encoder structures: VGG, ResNet34, ResNet50, and MobileNet. The results indicated that the MSBE encoder integrated into the SegNet network exhibited superior performance compared to the other four encoders. Specifically, MSBE-SegNet surpassed the other combinations in all performance metrics except mAP. In terms of mIoU, MSBE-SegNet was 6.47% higher than the average of the other encoder structures. Also, in mRecall, MSBE-SegNet’s performance was 4.55%, 6.1%, 9.94%, and 3.55% higher than VGG, ResNet34, ResNet50, and MobileNet, respectively. Although MSBE-SegNet was slightly inferior to MobileNet-SegNet in terms of mAP, it performed better in other key performance metrics, demonstrating the effectiveness of the MSBE encoder in the SegNet framework.
In the third set of experiments of this study, we utilized the newly proposed CrossLinkNet as the base backbone network and integrated five different encoder structures: VGG, ResNet34, ResNet50, MobileNet, and MSBE for ablation studies. The results showed that the CrossLinkNet network with the MSBE encoder exhibited excellent performance, outperforming the other four encoders. Specifically, MSBE-CrossLinkNet achieved optimal performance in all five metrics: FW_IoU, mIoU, mDice, PA, and mAP, with scores of 88.29%, 78.56%, 87.27%, 93.69%, and 91.13%, respectively. In terms of mRecall, MSBE-CrossLinkNet ranked second at 84.36%, slightly behind ResNet50-CrossLinkNet’s 87.04%. These data indicate that in the CrossLinkNet-based experimental setup, MSBE-CrossLinkNet achieved the best performance in all metrics except mean recall, further confirming the superior performance of the MSBE encoder.
As demonstrated in Table 2, with different backbone networks, the model parameter quantity (Params) is generally lower when employing MSBE as the feature encoder compared to other feature encoders. When combined with UNet, SegNet, and CrossLinkNet, the parameter quantities of MSBE are merely 3.9, 3.5 and 14.9 M, respectively, significantly lower than other encoders. This highlights the parameter efficiency advantage of MSBE. These results demonstrate that MSBE efficiently extracts features with a reduced parameter count, significantly lightening the model’s storage and deployment load. Additionally, the GFLOPs of MSBE are not the lowest, but which remain at a moderate level. Specifically, in integration with CrossLinkNet, MSBE’s GFLOPs are 45.9. Although this is higher than MobileNet-UNet’s GFLOPs, it remains notably superior to the VGG and ResNet50 as encoders. MSBE achieves a reasonable computational complexity and excellent performance with fewer parameters, demonstrating its efficiency and balance between computational efficiency in feature extraction.
The comprehensive analysis of the three sets of experiments in this study shows that the MSBE encoder demonstrated the best or near-best performance in all three backbone network experiments (UNet, SegNet, and CrossLinkNet). The proposed MSBE encoder, with its theoretical multi-scale and multi-branch design advantages, has also been empirically confirmed to exhibit superior performance in segmentation tasks. Compared to traditional encoders like VGG, ResNet, and MobileNet, MSBE showed greater stability and performance across different backbone networks. This proves its vast potential for application in deep learning segmentation tasks. Subsequently, we will discuss the theoretical foundation behind MSBE and the characteristics of its network structure, further analyzing the reasons for its effectiveness.
The design philosophy of MSBE is primarily based on two foundational principles: The importance of multi-scale features and the flexibility of the network’s branching structure. In medical imaging, especially in the analysis of whole slide images, the size, shape, and texture of lesion areas can vary greatly. Conventional single-scale feature encoders are typically constrained by their fixed receptive field sizes, making it challenging to concurrently capture fine detail features and large-scale structural information. MSBE introduces multi-scale convolutional kernels, enabling the network to extract features at different scales and thus understand the image content more comprehensively. For instance, small-scale convolutional kernels can capture the fine details of an image, such as the edges and textures of cells, while large-scale kernels are able to extract broader contextual information, aiding in the understanding of the overall morphology and relative position of lesion areas. This multi-scale feature extraction strategy allows MSBE to more accurately identify and locate key areas within images. Furthermore, the multi-branch structure of MSBE enhances the network’s flexibility and adaptiveness. Deviating from the conventional linear or singular pathway structures, MSBE’s parallel branching enables the concurrent processing and integration of features across various scales. This design not only increases the network’s capacity but also enables MSBE to better utilize the multi-level information in images, while concurrently reducing the parameter overhead. In experiments, these features enable MSBE to effectively improve the accuracy and robustness of segmentation tasks without significantly increasing computational complexity.
5.2 Comparison with Different Segmentation Network Framework
UNet and SegNet are established baseline models in computer vision, with UNet being prevalently utilized in medical image segmentation and SegNet being notable for semantic segmentation tasks. In our research, a novel network termed CrossLinkNet was developed for comparative analysis with these baseline models. This comparative study, detailed in Table 3, employed ablation experiments using a consistent set of encoders (VGG, ResNet34, ResNet50, MobileNet, and MSBE) across different backbone network structures. This approach was adopted to rigorously assess the efficacy of CrossLinkNet in image segmentation tasks. The following content will provide an in-depth analysis of the results from these five distinct experimental sets.
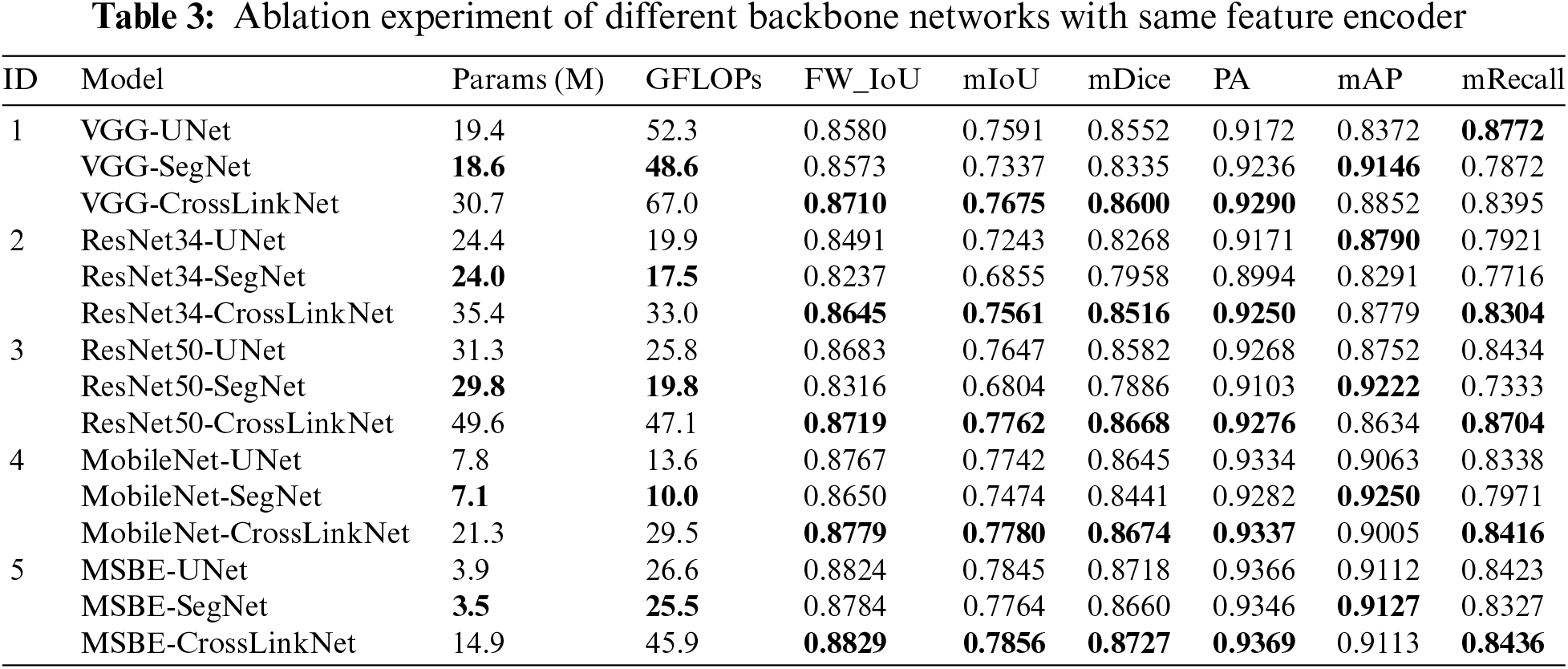
In the first set of experiments, VGG was utilized as the feature encoder, and ablation studies were conducted using three different backbone networks: UNet, SegNet, and CrossLinkNet. The results indicated that VGG-CrossLinkNet surpassed the other two comparative models across four metrics: FW_IoU, mIoU, mDice, and PA. Specifically, compared to VGG-UNet, CrossLinkNet showed an approximate 1.3% improvement in FW_IoU, about 0.85% in mIoU, around 0.5% in mDice, and a 1.2% increase in PA. Although VGG-SegNet achieved the highest score in mAP at 91.46%, its performance was poorer in other metrics. It is noteworthy that, while the mRecall of CrossLinkNet was slightly lower than that of VGG-UNet, it still represented an improvement of over 5% compared to VGG-SegNet. Overall, the performance of VGG-CrossLinkNet was superior to both VGG-UNet and VGG-SegNet.
In the second set of experiments, ResNet34 was employed as the feature encoder, and ablation studies were again conducted using the three distinct backbone networks: UNet, SegNet, and CrossLinkNet. The results demonstrated that ResNet34-CrossLinkNet achieved the best outcomes across five metrics: FW_IoU, mIoU, mDice, PA, and mRecall. Particularly in comparison with ResNet34-SegNet, ResNet34-CrossLinkNet showed superior performance in all metrics, with respective improvements of approximately 4.1%, 7.1%, 5.6%, 2.6%, 4.9%, and 5.9% in FW_IoU, mIoU, mDice, PA, mAP, and mRecall. When compared to ResNet34-UNet, ResNet34-CrossLinkNet was only marginally lower, by about 0.1%, in the mAP metric.
In the third set of experiments, ResNet50 was used as the feature encoder, with UNet, SegNet, and CrossLinkNet once again serving as the different backbone networks for ablation studies. The results indicated that ResNet50-CrossLinkNet achieved the best performance in five metrics: FW_IoU, mIoU, mDice, AP, and mAP. Notably, compared to ResNet50-SegNet, ResNet50-CrossLinkNet exhibited substantial improvements in these metrics, with increases of approximately 4%, 9.6%, 7.8%, 1.7%, and 13.7% in FW_IoU, mIoU, mDice, PA, and mRecall, respectively. Although ResNet50-UNet and ResNet50-SegNet showed slightly higher mean precision than ResNet50-CrossLinkNet, they were significantly outperformed in all other metrics by ResNet50-CrossLinkNet.
In the fourth set of experiments, MobileNet was employed as the feature encoder, with UNet, SegNet, and CrossLinkNet again used as different backbone networks for ablation studies. The results showed that MobileNet-CrossLinkNet achieved the best outcomes in five metrics: FW_IoU, mIoU, mDice, PA, and mRecall, with respective values of 87.79%, 77.8%, 86.74%, 93.37%, and 84.16%. Particularly in comparison with MobileNet-SegNet, MobileNet-CrossLinkNet demonstrated superior performance in these metrics, showing respective improvements of approximately 1.3%, 3.1%, 2.3%, 0.6%, and 4.5% in FW_IoU, mIoU, mDice, PA, and mRecall. Compared to MobileNet-UNet, MobileNet-CrossLinkNet was only marginally lower by 0.6 percentage points in mAP, but outperformed in all other evaluation metrics.
In the fifth set of experiments, MSBE was utilized as the feature encoder, with UNet, SegNet, and CrossLinkNet once again serving as the different backbone networks for ablation studies. The results indicated that MSBE-CrossLinkNet achieved the best performance across five metrics: FW_IoU, mIoU, mDice, PA, and mRecall, with respective values of 88.29%, 78.56%, 87.27%, 93.69%, and 84.36%. Notably, when compared to MSBE-UNet, MSBE-CrossLinkNet surpassed MSBE-UNet’s metrics of 88.24%, 78.45%, 87.18%, 93.66%, 91.12%, and 84.23% in all evaluation metric. Compared to MSBE-SegNet, MSBE-CrossLinkNet was only marginally lower by 0.14 percentage points in mAP, yet excelled in all other evaluation metrics.
CrossLinkNet’s foundational design is centered on boosting model performance via cross-layer connections, notably in feature fusion and information flow. This strategy, however, requires additional parameters and computations to realize such performance improvement, leading to a rise in both parameters and GFLOPs. Therefore, this can be regarded as a trade-off between model performance and computational resources.
Comparing the results from the above experiments, our proposed cross-layer linking segmentation network, CrossLinkNet, demonstrated exceptional performance under various encoders, particularly in key metrics like FW_IoU, mIoU, mDice, and PA, where it consistently showed significant advantages. Even in certain evaluation metrics (e.g., mAP) where other models may have a slight edge, CrossLinkNet still maintained a high level of overall performance, illustrating its stability and reliability in different environments. The following content will explore the network structural features of CrossLinkNet from a theoretical perspective, further analyzing the underpinnings for its effectiveness.
The primary hallmark of CrossLinkNet is its innovative cross-layer connection structure. CrossLinkNet establishes cross-layer connections among multiple levels, which effectively facilitates the fusion of low-level features with high-level features. This design enables the network to utilize deep abstract information for accurate region determination and to retain more detail information, thereby enhancing the precision of segmentation and the clarity of edges. Additionally, the cross-layer connections aid in better gradient backpropagation, minimizing the loss of information during training and thereby improving the network’s learning efficiency and stability. Furthermore, apart from cross-layer connections, CrossLinkNet adopts a modular framework. This modular architecture is crucial for a deeper analysis and comprehension of the model’s decision-making processes. Combining cross-layer connections with multi-scale feature encoding, CrossLinkNet not only excels in performance but also enhances the transparency of its network structure. In the aforementioned experiments, these advantages of CrossLinkNet enabled it to achieve superior performance.
5.3 Comparison with State-of-the-Arts Models on BOT Dataset
To validate the performance advantages of the proposed segmentation network, CrossLinkNet (MSBE), the experiment incorporated a series of classic network models for comparison, including SegNet, UNet++, DeepLabV3+, PidNet, and the latest state-of-the-art (SOTA) model, EnsNet. The experimental results, as shown in Table 4, clearly demonstrate the exceptional performance of CrossLinkNet (MSBE) across various evaluation metrics. Specifically, CrossLinkNet (MSBE) achieved an FW_IoU of 88.29%, significantly surpassing SegNet, UNet++, DeepLabV3+, PidNet, and EnsNet, and leading the second-best model, PidNet, by a margin of 0.35 percentage points. In terms of mIoU, its performance of 78.56% was also the highest, exceeding the second-placed PidNet by 0.98 percentage points. For the mDice, CrossLinkNet (MSBE) led other networks with a score of 87.27%. In PA, it reached the peak performance of 93.69%, slightly ahead of PidNet’s 93.56% by 0.13 percentage points. Although in mAP, CrossLinkNet (MSBE)’s 91.13% was slightly lower than PidNet’s 91.96%, its performance was still highly competitive compared to other networks. Finally, CrossLinkNet (MSBE) also achieved the best result in mRecall with 84.36%.
To visually demonstrate the performance of CrossLinkNet (MSBE) in image segmentation tasks, we selected three pathological images from the test set for visualization analysis, as shown in Fig. 5. The first image contains multiple small tumor regions, used to assess the model’s capability in segmenting small targets. The second image displays a large tumor area, aimed at examining the model’s effectiveness in handling large targets. The third image is a mixed case, containing both large and small tumor regions, to evaluate the model’s comprehensive segmentation performance for targets of varying sizes. In these images, the green areas represent the correctly predicted parts by the model (i.e., True Positive), the red areas indicate incorrect predictions (i.e., False Positive), and the blue areas denote the portions that the model failed to recognize (i.e., False Negative). The comparison of these three groups of images clearly demonstrates CrossLinkNet’s exceptional segmentation performance in various scenarios.

Figure 5: Comparison results of pathological image segmentation experiment
5.4 Comparison with State-of-the-Arts Models on Kvasir Dataset
To assess the versatility of the proposed segmentation network, CrossLinkNet (MSBE), across various medical datasets, we conducted a series of comparative tests using the Kvasir dataset. The Kvasir dataset is specifically designed for research in gastrointestinal (GI) imagery and videos, encompassing various GI anatomical landmarks as well as a subset dedicated to GI polyp segmentation. This experiment utilized the polyp segmentation subset of the Kvasir dataset to evaluate the performance of CrossLinkNet (MSBE) in segmenting GI tract polyps. The comparative networks used in the experiment included SegNet, UNet++, DeepLabV3+, PidNet and EnsNet, with detailed results presented in Table 5.
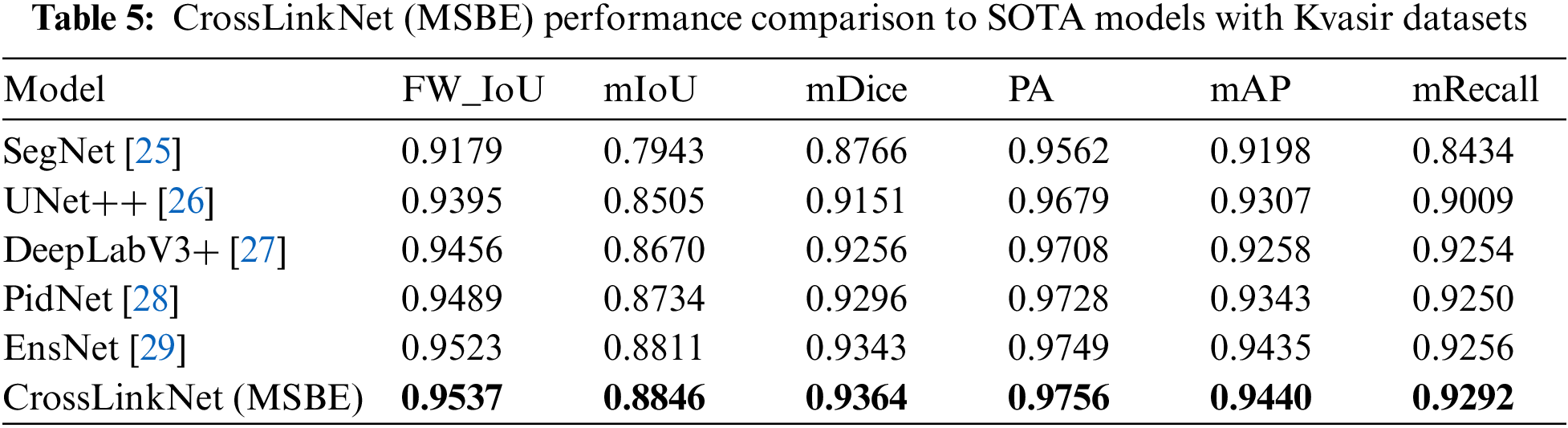
Based on the experimental results from the Kvasir dataset, it is evident that our proposed CrossLinkNet (MSBE) outperformed all other comparative network models in every evaluation metric. Specifically, CrossLinkNet (MSBE) achieved a FW_IoU of 95.37%, which is 0.14 percentage points higher than the second-ranked EnsNet and 3.58 percentage points above the lowest-ranked SegNet. In the mIoU metric, CrossLinkNet (MSBE) led with 88.46%, ahead of EnsNet’s 88.11% and surpassing SegNet’s 79.43% by 9.03 percentage points. For mDice, CrossLinkNet (MSBE) scored 93.64%, slightly higher than EnsNet’s 93.43% and more than 1 percentage point higher compared to SegNet, UNet++, and DeepLabV3+. In terms of PA, mAP, and mRecall, CrossLinkNet (MSBE) performed at 97.56%, 94.40%, and 92.92%, respectively, outperforming the second-best EnsNet by 0.07 percentage points, 0.05 percentage points, and 0.36 percentage points, while significantly surpassing the results of SegNet, UNet++, and DeepLabV3+.
To visually demonstrate the performance of the proposed CrossLinkNet model in different types of gastrointestinal (GI) tract image segmentation tasks, the experiment selected three representative images from the test set for visualization analysis, as illustrated in Fig. 6. The first image displays a GI tract image containing small polyps, used to assess the model’s segmentation effectiveness on small targets. The second image includes larger polyps, testing the model’s ability to segment larger targets. The third image is a mixed case, containing both large and small polyps, used to evaluate the model’s segmentation performance when dealing with targets of varying sizes simultaneously. In these images, green areas represent correctly predicted parts by the model (i.e., True Positive), red areas denote incorrect predictions (i.e., False Positive), and purple areas are the portions that the model failed to recognize (i.e., False Negative). The comparison of these three groups of images clearly indicates that CrossLinkNet demonstrates exceptional segmentation performance in various scenarios, validating its effectiveness and precision in medical image segmentation.

Figure 6: Comparison results of upper gastrointestinal polyp segmentation experiment
In Sections 5.3 and 5.4, we have showcased the comparative results of CrossLinkNet (MSBE) against other models on the BOT and Kvasir datasets. These experimental results demonstrate that CrossLinkNet (MSBE) surpasses other network models on several critical performance metrics. The subsequent content will analyze the reasons for the superior performance of CrossLinkNet (MSBE) from a theoretical perspective.
The superior performance of CrossLinkNet (MSBE) across diverse datasets and tasks can be attributed to a confluence of critical factors. Foremost among these is MSBE’s prowess in multi-scale feature extraction. By leveraging convolutional kernels of different scales, MSBE is capable of capturing both the detailed information and the global context of images simultaneously. This multi-scale feature extraction mechanism is particularly suitable for medical images, as they often contain lesions of varying significantly in size and morphology. Such a design enables CrossLinkNet (MSBE) to more accurately identify and segment lesion areas of various scales. Secondly, it benefits from the cross-layer connections and shortcut connections in CrossLinkNet (MSBE). The cross-layer connection structure in CrossLinkNet facilitates the fusion of features from different levels, integrating low-level details into the overarching decision-making process. This not only enhances the model’s segmentation precision but also improves the delineation of edges, which is pivotal for intricate medical image segmentation tasks. Additionally, shortcut connections tackle the gradient vanishing issue inherent in deep network training, facilitating the seamless flow of information across layers. This process is instrumental in recapturing fine details and elevating segmentation fidelity. Finally, the modular design of CrossLinkNet (MSBE) plays a pivotal role. This design approach bolsters the model’s transparency and explicability. Through cross-layer connections and multi-scale feature encoding, CrossLinkNet (MSBE) not only achieves excellent performance but also provides more comprehensible visual insights into its decision-making mechanics, which is of great significance for improving the accuracy and credibility of clinical diagnoses.
In conclusion, CrossLinkNet (MSBE)’s distinguished performance in various tasks stems from its sophisticated multi-scale feature extraction, potent feature fusion mechanisms, and a strong focus on model interpretability. These integrated features not only elevate segmentation precision and robustness but also offer an interpretable solution for medical image segmentation tasks.
The introduction of CrossLinkNet, with its innovative MSBE and cross-layer connections, marks a significant advancement in whole-slide image segmentation. However, its performance is limited by the inherent variability and complexity of medical datasets, thus it is necessary to further explore adaptive feature extraction methods and data augmentation strategies to enhance robustness. Additionally, the model’s computational intensity, necessitated by the high-resolution nature of whole-slide images, poses scalability challenges, especially in resource-constrained settings. Addressing this issue may require architectural optimizations to improve efficiency without compromising performance. Moreover, the impact of different feature encoders on the model’s effectiveness is a crucial consideration. The comparative performance of MSBE against traditional encoders like VGG, ResNet, and MobileNet within the CrossLinkNet framework warrants a detailed examination to identify the most suitable encoder for specific segmentation tasks. Future enhancements to CrossLinkNet should focus on these areas to bolster its adaptability, scalability, and overall performance in the evolving field of medical images.
In this study, we present CrossLinkNet, an innovative segmentation network for whole slide images with a focus on explainable AI, marking a significant advancement in the field of medical image analysis. The introduction of the Multi-Scale Multi-Branch Feature Encoder (MSBE) represents a pivotal innovation, enhancing the network’s ability to extract nuanced features effectively. This encoder’s adaptability in configuration through hyperparameter adjustments allows for flexibility and precision in feature extraction. Furthermore, CrossLinkNet’s unique cross-layer encoder-decoder connections markedly improve feature integration, thereby elevating segmentation accuracy. Our comprehensive experiments on datasets like BOT and Kvasir have showcased CrossLinkNet’s exceptional performance. Crucially, the incorporation of explainable AI principles within CrossLinkNet addresses the pressing need for interpretability in medical diagnostics, thereby contributing to the field of digital pathology and automated diagnostic technologies. This research not only presents innovative methodologies but also emphasizes the pivotal role of explainable AI in advancing medical imaging analysis.
Acknowledgement: The authors would like to extend their gratitude to the Network and Data Security Key Laboratory of Sichuan Province for their invaluable support. We also acknowledge the significant contributions and resources provided by the University of Electronic Science and Technology of China. Their assistance was instrumental in the completion of this research.
Funding Statement: This work was supported by the National Natural Science Foundation of China (Grant Numbers: 62372083, 62072074, 62076054, 62027827, 62002047); the Sichuan Provincial Science and Technology Innovation Platform and Talent Program (Grant Number: 2022JDJQ0039); the Sichuan Provincial Science and Technology Support Program (Grant Numbers: 2022YFQ0045, 2022YFS0220, 2021YFG0131, 2023YFS0020, 2023YFS0197, 2023YFG0148); and the CCF-Baidu Open Fund (Grant Number: 202312).
Author Contributions: The authors confirm contribution to the paper as follows: Study conception and design: Peng Xiao, Qi Zhong; dataset and experiments: Peng Xiao, Dongyuan Wu; analysis and interpretation of results: Peng Xiao, Zhen Qin; draft manuscript preparation: Peng Xiao, Jingxue Chen, Erqiang Zhou. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: There are no data and materials available to share.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. S. Shafi and A. V. Parwani, “Artificial intelligence in diagnostic pathology,” Diagn. Pathol., vol. 18, no. 1, pp. 109, 2023. doi: 10.1186/s13000-023-01375-z. [Google Scholar] [PubMed] [CrossRef]
2. B. Hunter, S. Hindocha, and R. W. Lee, “The role of artificial intelligence in early cancer diagnosis,” Cancers, vol. 14, no. 6, pp. 1524, 2022. doi: 10.3390/cancers14061524. [Google Scholar] [PubMed] [CrossRef]
3. B. Lai, J. Fu, Q. Zhang, N. Deng, Q. Jiang and J. Peng, “Artificial intelligence in cancer pathology: Challenge to meet increasing demands of precision medicine,” Int. J. Oncol., vol. 63, no. 3, pp. 1–30, 2023. doi: 10.3892/ijo.2023.5555. [Google Scholar] [PubMed] [CrossRef]
4. W. Guo, B. Tondi, and M. Barni, “Universal detection of backdoor attacks via density-based clustering and centroids analysis,” IEEE Trans. Inf. Forensics Secur., vol. 19, no. 7, pp. 970–984, 2024. doi: 10.1109/TIFS.2023.3329426. [Google Scholar] [CrossRef]
5. C. Ladbury et al., “Utilization of modelagnostic explainable artificial intelligence frameworks in oncology: A narrative review,” Transl. Cancer Res., vol. 11, no. 10, pp. 3853, 2022. [Google Scholar] [PubMed]
6. M. Farhadloo et al., “SAMCNet: Towards a spatially explainable AI approach for classifying MXIF oncology data,” in Proc. 28th ACM SIGKDD Conf. Knowl. Discov. Data Min., Washington DC, USA, Aug. 2022, pp. 2860–2870. [Google Scholar]
7. G. Fang, Y. Sun, M. Almutiq, W. Zhou, Y. Zhao and Y. Ren, “Distributed medical data storage mechanism based on proof of retrievability and vector commitment for metaverse services,” IEEE J. Biomed. Health Inform., 2023. doi: 10.1109/JBHI.2023.3272021. [Google Scholar] [PubMed] [CrossRef]
8. A. Ramírez-Mena, E. Andrés-León, M. J. Alvarez-Cubero, A. Anguita-Ruiz, L. J. Martinez-Gonzalez and J. Alcala-Fdez, “Explainable artificial intelligence to predict and identify prostate cancer tissue by gene expression,” Comput. Methods Programs Biomed., vol. 240, no. 2, pp. 107719, 2023. doi: 10.1016/j.cmpb.2023.107719. [Google Scholar] [PubMed] [CrossRef]
9. M. Khened, A. Kori, H. Rajkumar, G. Krishnamurthi, and B. Srinivasan, “A generalized deep learning framework for whole-slide image segmentation and analysis,” Sci. Rep., vol. 11, no. 1, pp. 11579, 2021. doi: 10.1038/s41598-021-90444-8. [Google Scholar] [PubMed] [CrossRef]
10. S. Alkhalaf et al., “Adaptive aquila optimizer with explainable artificial intelligence-enabled cancer diagnosis on medical imaging,” Cancers, vol. 15, no. 5, pp. 1492, 2023. doi: 10.3390/cancers15051492. [Google Scholar] [PubMed] [CrossRef]
11. G. R. Djavanshir, X. Chen, and W. Yang, “A review of artificial intelligence’s neural networks (deep learning) applications in medical diagnosis and prediction,” IT Prof., vol. 23, no. 3, pp. 58–62, 2021. doi: 10.1109/MITP.2021.3073665. [Google Scholar] [CrossRef]
12. A. L. D. Araújo et al., “Machine learning concepts applied to oral pathology and oral medicine: A convolutional neural networks’ approach,” J. Oral Pathol. Med., vol. 52, no. 2, pp. 109–118, 2023. doi: 10.1111/jop.13397. [Google Scholar] [PubMed] [CrossRef]
13. R. Chiwariro and B. Julius, “Comparative analysis of deep learning convolutional neural networks based on transfer learning for pneumonia detection,” Int. J. Res. Appl. Sci. Eng. Technol., vol. 11, no. 1, pp. 1161– 1170, 2023. doi: 10.22214/ijraset.2023.48685. [Google Scholar] [CrossRef]
14. N. S. An et al., “BlazeNeo: Blazing fast polyp segmentation and neoplasm detection,” IEEE Access, vol. 10, pp. 43669–43684, 2022. doi: 10.1109/ACCESS.2022.3168693. [Google Scholar] [CrossRef]
15. L. Roszkowiak, A. Korzyńska, D. Pijanowska, R. Bosch, M. Lejeune and C. López, “Clustered nuclei splitting based on recurrent distance transform in digital pathology images,” EURASIP J. Image Video Proc., vol. 2020, no. 1, pp. 125, 2020. doi: 10.1186/s13640-020-00514-6. [Google Scholar] [CrossRef]
16. N. Otsu, “A threshold selection method from gray-level histograms,” IEEE Trans. Syst., Man, Cybern., vol. 9, no. 1, pp. 62– 66, 1979. doi: 10.1109/TSMC.1979.4310076. [Google Scholar] [CrossRef]
17. S. Zhu, X. Xia, Q. Zhang, and K. Belloulata, “An image segmentation algorithm in image processing based on threshold segmentation,” in 2007 Third Int. IEEE Conf. Signal-Image Technol. Internet-Based Syst., Shanghai, China, Dec. 2007, pp. 673–678. [Google Scholar]
18. P. Yang, W. Song, X. Zhao, R. Zheng, and L. Qingge, “An improved OTSU threshold segmentation algorithm,” Int. J. Comput. Sci. Eng., vol. 22, no. 1, pp. 146–153, 2020. doi: 10.1504/IJCSE.2020.107266 [Google Scholar] [CrossRef]
19. W. Guo, B. Tondi, and M. Barni, “An overview of backdoor attacks against deep neural networks and possible defences,” IEEE Open J. Signal Process., vol. 3, pp. 261–287, 2022. doi: 10.1109/OJSP.2022.3190213. [Google Scholar] [CrossRef]
20. K. G. V. Kiran and G. M. Reddy, “Automatic classification of whole slide pap smear images using CNN with PCA based feature interpretation,” in Proc. IEEE/CVF Conf. Comput. Vis. Pattern Recognit. Workshops, Long Beach, CA, USA, Jun. 2019. [Google Scholar]
21. H. Lin, H. Chen, X. Wang, Q. Wang, L. Wang and P. A. Heng, “Dualpath network with synergistic grouping loss and evidence driven risk stratification for whole slide cervical image analysis,” Med. Image Anal., vol. 69, pp. 101955, 2021. doi: 10.1016/j.media.2021.101955. [Google Scholar] [PubMed] [CrossRef]
22. A. Dosovitskiy et al., “An image is worth 16 × 16 words: Transformers for image recognition at scale,” arXiv preprint arXiv:2010.11929, 2020. [Google Scholar]
23. R. J. Chen et al., “Multimodal co-attention transformer for survival prediction in gigapixel whole slide images,” in Proc. IEEE/CVF Int. Conf. Comput. Vis., 2021, pp. 4015–4025. [Google Scholar]
24. P. Yin, B. Yu, C. Jiang, and H. Chen, “Pyramid tokens-to-token vision transformer for thyroid pathology image classification,” in 2022 Eleventh Int. Conf. Image Process. Theory, Tools Appl. (IPTA), IEEE, 2022, pp. 1–6. [Google Scholar]
25. V. Badrinarayanan, A. Kendall, and R. Cipolla, “SegNet: A deep convolutional encoder-decoder architecture for image segmentation,” IEEE Trans. Pattern Anal. Mach. Intell., vol. 39, no. 12, pp. 2481–2495, 2017. doi: 10.1109/TPAMI.2016.2644615. [Google Scholar] [PubMed] [CrossRef]
26. Z. Zhou, M. M. Rahman Siddiquee, N. Tajbakhsh, and J. Liang, “UNet++: A nested U-Net architecture for medical image segmentation,” in Deep Learn. Med. Image Anal. Multimodal Learn. Clinical Decis., Granada, Spain, Sep. 20, 2018, pp. 3–11. [Google Scholar]
27. L. C. Chen, Y. Zhu, G. Papandreou, F. Schroff, and H. Adam, “Encoder decoder with atrous separable convolution for semantic image segmentation,” in Proc. Eur. Conf. Comput. Vis. (ECCV), Munich, Germany, 2018, pp. 801–818. [Google Scholar]
28. J. Xu, Z. Xiong, and S. P. Bhattacharyya, “PIDNet: A real-time semantic segmentation network inspired by pid controllers,” in Proc. IEEE/CVF Conf. Comput. Vis. Pattern Recognit., Vancouver, Canada, Jun. 2023, pp. 19529–19539. [Google Scholar]
29. M. R. Prusty, R. Dinesh, H. S. K. Sheth, A. L. Viswanath, and S. K. Satapathy, “Nuclei segmentation in histopathology images using structure-preserving color normalization based ensemble deep learning frameworks.,” Comput. Mater. Contin., vol. 77, no. 3, pp. 3077–3094, 2023. doi: 10.32604/cmc.2023.042718 [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF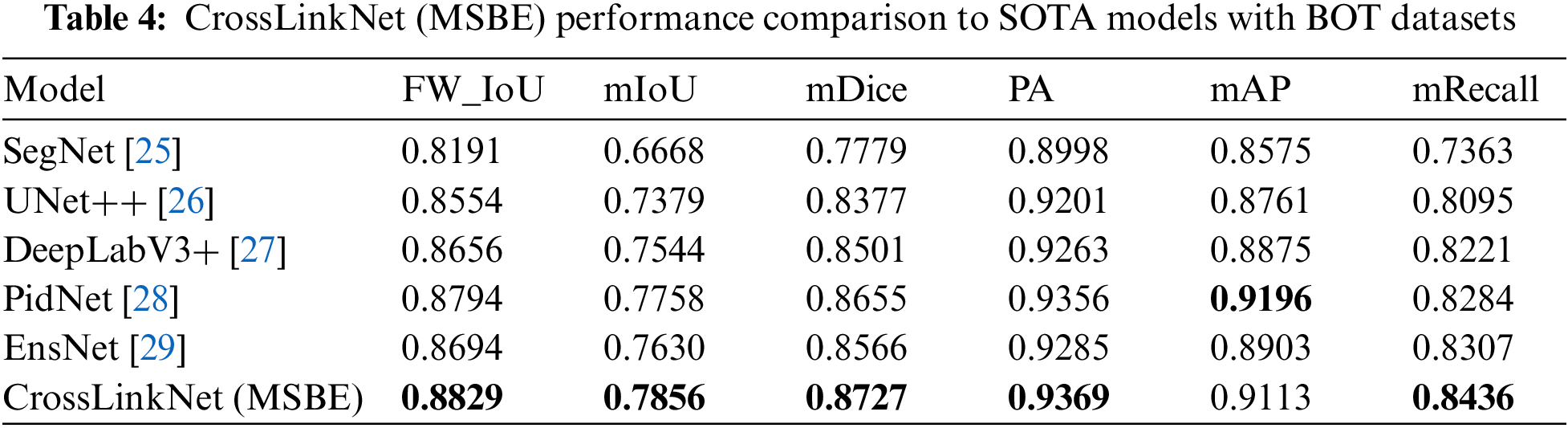
 Downloads
Downloads
 Citation Tools
Citation Tools