 Open Access
Open Access
ARTICLE
MCIF-Transformer Mask RCNN: Multi-Branch Cross-Scale Interactive Feature Fusion Transformer Model for PET/CT Lung Tumor Instance Segmentation
1 School of Medical Information & Engineering, Ningxia Medical University, Yinchuan, 750004, China
2 School of Computer Science and Engineering, North Minzu University, Yinchuan, 750021, China
3 Key Laboratory of Image and Graphics Intelligent Processing of State Ethnic Affairs Commission, North Minzu University, Yinchuan, 750021, China
* Corresponding Author: Huiling Lu. Email:
(This article belongs to the Special Issue: Deep Learning in Computer-Aided Diagnosis Based on Medical Image)
Computers, Materials & Continua 2024, 79(3), 4371-4393. https://doi.org/10.32604/cmc.2024.047827
Received 19 November 2023; Accepted 19 April 2024; Issue published 20 June 2024
Abstract
The precise detection and segmentation of tumor lesions are very important for lung cancer computer-aided diagnosis. However, in PET/CT (Positron Emission Tomography/Computed Tomography) lung images, the lesion shapes are complex, the edges are blurred, and the sample numbers are unbalanced. To solve these problems, this paper proposes a Multi-branch Cross-scale Interactive Feature fusion Transformer model (MCIF-Transformer Mask RCNN) for PET/CT lung tumor instance segmentation, The main innovative works of this paper are as follows: Firstly, the ResNet-Transformer backbone network is used to extract global feature and local feature in lung images. The pixel dependence relationship is established in local and non-local fields to improve the model perception ability. Secondly, the Cross-scale Interactive Feature Enhancement auxiliary network is designed to provide the shallow features to the deep features, and the cross-scale interactive feature enhancement module (CIFEM) is used to enhance the attention ability of the fine-grained features. Thirdly, the Cross-scale Interactive Feature fusion FPN network (CIF-FPN) is constructed to realize bidirectional interactive fusion between deep features and shallow features, and the low-level features are enhanced in deep semantic features. Finally, 4 ablation experiments, 3 comparison experiments of detection, 3 comparison experiments of segmentation and 6 comparison experiments with two-stage and single-stage instance segmentation networks are done on PET/CT lung medical image datasets. The results showed that APdet, APseg, ARdet and ARseg indexes are improved by 5.5%, 5.15%, 3.11% and 6.79% compared with Mask RCNN (resnet50). Based on the above research, the precise detection and segmentation of the lesion region are realized in this paper. This method has positive significance for the detection of lung tumors.Keywords
Nomenclature
| Mask RCNN | Mask Region-based Convolutional Neural Network |
| SCLC | Small Cell Lung Cancer |
| NSCLC | Non-Small Cell Lung Cancer |
| CIFEM | Cross-scale Interactive Feature Enhancement Module |
| FPN | Feature Pyramid Network |
| CIF-FPN | Cross-scale Interactive Feature fusion FPN network |
| AP | Average Precision |
| IoU | Intersection over Union |
| AR | Average Recall |
| BoT | Bottleneck Transformer |
Lung cancer is a disease in which cells proliferat randomly due to a genetic mutation. Lung cancer can be classified into central-type lung cancer and peripheral-type lung cancer according to the lesion location. Early central lung cancer is a tumor confined to the bronchial cavity or infiltrating the lobar/segmental bronchial wall, without invasion of lung parenchyma and metastasis [1]. There are early-stage peripheral lung cancer, is a tumor with its diameter ≤2 cm, and without metastasis. Compared with peripheral lung cancer, central lung cancer overlaps with hilus pulmonis and cardiovascular, so central lung cancer is more difficult to detect [2]. Lung tumors can be divided into benign tumors (such as pulmonary hemangioma) and malignant tumors (such as lung cancer) according to the lesion characters. Lung malignant tumors can be further divided into primary lung malignant tumors and metastatic lung tumors. Primary lung cancer can be further divided into small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC) [3] according to the cell difference. Therefore, lung cancer is one of the most life-threatening cancers in the world [4]. In lung tumors, lung cancer accounts for a relatively high proportion, and early detection and diagnosis are crucial to the treatment of patients.
Medical imaging technology is the routine and preferred method for the detection and staging of lung cancer. It is useful for medical research, computer-aided diagnosis, radiotherapy and evaluation of surgical results [5]. The patient pathological information can be obtained by medical images in a non-invasive way and improve the diagnosis accuracy. Medical images are divided into anatomical structure images and metabolic function images. Anatomical structure images can provide anatomical structure information of organs and location information, such as CT images, which have high spatial resolution [6]. However, it does not provide detailed molecular information, such as the metabolic function of cancer cells. Metabolic function images can show the metabolism of the human body by injecting radionuclides into the body. Because cancer cells are more active than normal cells and tissues and take up more 18F-Fluorine DeoxyGlucose (18F-FDG), these radioisotopes accumulate in cancer areas and have high metabolic properties.
Therefore, The metabolic information of the cells in the lesion area can be clearly detected, such as PET images, which have obvious contrast degree between malignant tumors and normal tissues, but the anatomical structure information cannot be provided, the lesions cannot be accurately located, and the image spatial resolution is low [6]. Hence, how to fuse PET and CT images is getting more and more important [7]. The medical imaging of lung tumors is complex. The lung cancer lesions show burrlike characteristics of different sizes, most of them have cavitation sign, air bronchiole sign, honeycomb sign, ground glass sign, and a few also have calcification sign. PET/CT images can make full use of the complementary information of CT and PET modal images, It can help doctors make more precise diagnoses and assessments. Therefore, PET/CT images have become an effective tool for tumor detection, recognition, staging and monitoring [8]. Effective fusion strategy can improve image quality [9]. However, it is a time-consuming, laborious and subjective task to extract the features of lesion lesion by hand. Instance segmentation is an effective method to solve this problem, this paper proposes a Multi-branch Cross-scale Interactive Feature fusion Transformer model (MCIF-Transformer Mask RCNN) for PET/CT lung tumor instance segmentation, which is a further development of Mask RCNN and a positive attempt for precise segmentation about lung tumors.
Image segmentation is to segment medical images into several regions of interest with specific properties and unique meanings based on some similar features in medical images. Semantic segmentation is used to predict the classification of each pixel in the input medical image. Different from the previous two, instance segmentation is based on pixel-level semantic segmentation combined with the target detection task. It is to perform instance-level segmentation and object-level recognition of organs or lesions in the detection box, and its segmentation accuracy and efficiency are better [10]. In addition, the traditional segmentation method can only deal with relatively simple scenes. Due to the complex features of medical images, blurred edges between different tissues or between tissues and lesions, irregular shape of lesion regions, and irregular beating of organs such as the heart leading to the existence of data noise, the traditional methods are prone to the problem of missed and false detections, and the segmentation effect is not ideal. In recent years, deep learning methods based on convolutional neural networks have been widely used in the field of computer vision, especially in the medical image processing fields such as CT images, X-ray images, ultrasonic images, PET images and MRI images, which can achieve accurate localization and segmentation of lesion regions. Example instance segmentation models such as Wan et al. [11], Mask RCNN [12], Cascade RCNN [13], YOLACT [14], and SOLO [15] prove that convolutional neural networks can not only achieve good results at the pixel level, but also enable instance-level learning and effectively provide object-by-object labeling of target object information. In 2023, Xinjun et al. [11] proposed a model of Multi-scale context information fusion for instance segmentation. In 2017, He et al. [12] proposed the Mask R-CNN model, which added mask prediction branch and used ROI Align instead of ROI Pooling based on Faster R-CNN to effectively solve the two-stage instance segmentation problem. In 2019, Huang et al. [16] proposed Mask Scoring R-CNN model, which based on Mask R-CNN using MaskIoU head for mask scoring, which effectively improves the accuracy of instance localization or mask segmentation. In 2019, Zhou et al. [17] proposed the Corner Mask-RCNN model, which adds corner point prediction head to fit the edge parts and effectively obtain the details of instance. In 2020, Homayounfar et al. [18] proposed the LevelSet R-CNN model, which uses Chan-Vese level set segmentation method based on the Mask R-CNN model, and obtains adaptive hyperparameters, feature tensors and symbolic distance function (TDSF) initialization by inputting ROI into a series of convolutions, which effectively solves the problem of Mask R-CNN output mask with low resolution. In 2021, Lin et al. [19] proposed S-Mask-RCNN, which adds a spatial attention mechanism on the basis of the feature extraction network ResNet. In 2021, Long et al. [20] proposed P-Mask-RCNN, which based on the probability of occurrence of pulmonary embolism, that is, to extract anchor points from candidate regions to eliminate most invalid anchor points. Transformer technology are applied to instance segmentation, Tao et al. [21] discussed the current development trend of Transformer technology in depth from the perspective of Four Secrets of Vision Transformer, which has good guidence. Although many researchers propose relate research work on instance segmentation and achieve some results, the application of instance segmentation in medical images are limited in some aspects. In addition, for non-small cell lung cancer (such as adenocarcinoma), Small lesion size leads to the lung tumor features are not obvious. The lung tumor appearances lead to changes in adjacent structures, Hence, Due to the poor ability of the model to express lesion features in medical images, the ability of the model to extract lesion features at different scales is insufficient. Aiming to these problems, this paper proposes a multi-branch cross-scale interactive feature fusion transformer model for PET/CT lung tumor instance segmentation. The main contributions are as follows:
(1) ResNet-Transformer backbone network is designed. This method combines the global and local feature information in the lung image, and the multi-head self-attention BoT module is used in the feature extraction process of lung tumors, it is good to improve the feature representation ability from multiple dimensions by establishing a long-distance dependence relationship between global pixels at the top level.
(2) A cross-scale interactive feature enhancement auxiliary network is constructed. The Cross-scale Interactive Feature Enhancement Module (CIFEM) is used to interactively associate the feature maps of different scales extracted from the backbone network. The lesion feature expression of the backbone is enhanced by aggregating adjacent context information to effectively supplement the missing details information.
(3) A cross-scale interactive feature fusion FPN network is designed. The Cross-scale Interactive Feature Fusion Module (CIFM) is used to conduct bidirectional interactive fusion between low-level detail information and high-level semantic information. The focus on tumor lesions is further enhanced by aggregating the feature information of adjacent stages.
3 MCIF-Transformer Mask RCNN Model Design
3.1 Overall Structure of MCIF-Transformer Mask RCNN Model
MCIF-Transformer Mask RCNN (Multi-branch Cross-scale Interactive feature Fusion Transformer model) is proposed in this paper. The overall structure of the model is shown in Fig. 1. The main steps of the model are as follows: 1) The ResNet-Transformer backbone network is used to extract tumor lesion features of different scales in PET/CT lung images, which is divided into five stages. In Stage 0, Stage 1, Stage 2 and Stage 3, basic residual blocks are used to extract features. In Stage 4, the multi-head self-attention BoT module is used to extract non-local feature information. 2) The cross-scale interactive feature enhancement auxiliary network is designed to supplement the shallow information into the deep features. The fine-grained attention to the lesion region is further enhanced by using the cross-scale interactive feature enhancement module. 3) The cross-scale interactive feature fusion FPN network is constructed to carry out the bidirectional interactive fusion between deep and shallow features, and more accurate and effective lesion features are obtained by enhancing low-level information such as details, location and texture in deep features. 4) The feature map is travelled by sliding window in RPN, and the foreground and background are distinguished in the corresponding anchor box and the classification probability is calculated, and the offset of coordinate points is regressed. 5) The classification, boundary box and mask information of the lesions are regressed through three prediction branches.
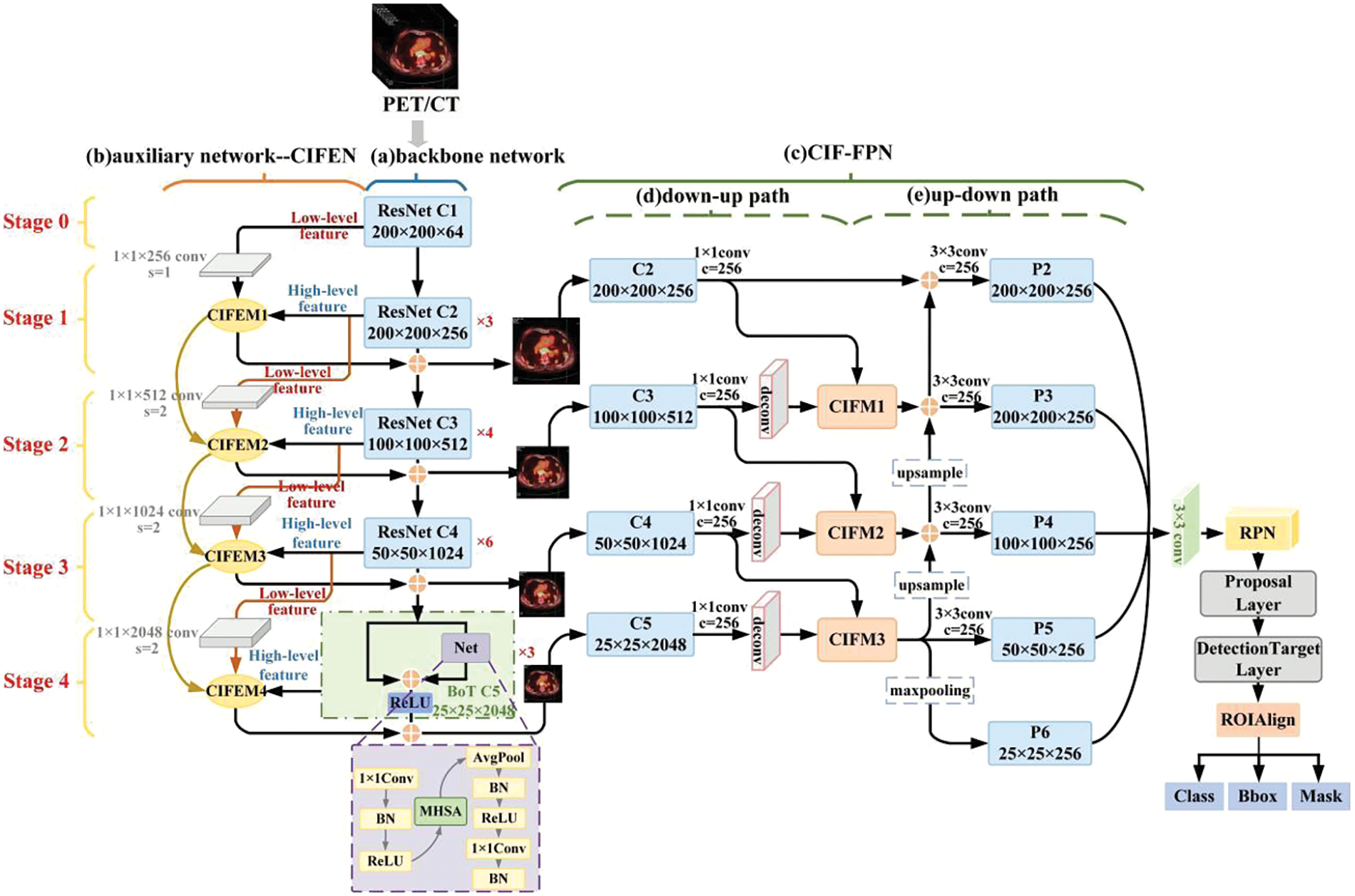
Figure 1: Structure of MCIF-transformer mask RCNN model
There are 3 parts in the MCIF-Transformer Mask RCNN model: Feature extraction, feature fusion and feature prediction. The innovation work of this paper mainly focuses on the first two steps. For the feature extraction part, the backbone and auxiliary networks are constructed to enhance the perception ability of fine-grained features of the lesion. The ResNet-Transformer backbone network is used for feature extraction of preprocessed PET/CT lung tumor images. The BoT module with global attention is used to replace the basic residual block at the top level to establish the dependency of long-distance features among pixels in non-local space. The cross-scale interactive feature enhancement auxiliary network is designed to aggregate adjacent feature maps of different resolutions and re-label the importance of spatial and channel features of lung tumors, so as to enhance the feature expression of lesions in the backbone network. In the feature fusion part, the cross-scale interactive feature fusion FPN network is designed to achieve bidirectional interactive fusion between low-level feature information and high-level semantic information of different scales features, and further enhance the feature expression ability of the model.
3.2 ResNet-Transformer Backbone Network
The resnet50 is used as the backbone of the ResNet-Transformer backbone network [22], In Stage 0 to Stage 3, the modules are composed of the basic bottleneck residual block. In Stage 4, the module is composed of the Bottleneck Transformer (BoT) [23], its structure is shown in Fig. 1. In this network, the non-local spatial information of the highest feature map is extracted by adding a multi-head self-attention module. The ability of the model to extract lesion features from PET/CT lung images is improved by focusing on the global region, so as to obtain more accurate lesion features.
The PET/CT lung tumor images are inputted into the backbone network and processed in Stage 0 to Stage 4 successively from low-level to high-level. In Stage 0, Stage 1, Stage 2 and Stage 3, lesion feature images of different sizes are extracted by basic residual blocks, and 3, 4 and 6 bottleneck blocks are stacked in series in each residual block of C2, C3, C4. This is a way to model the lesion globally and extract the global features. In Stage 4, the three bottleneck blocks in the top level are replaced with BoT modules, which are composed of 1 × 1 convolution, average pooling, multi-head self-attention (MHSA), batch normalization (BN) and ReLU activation function. MHSA is used to replace the 3 × 3 spatial convolution at the corresponding position in ResNet bottleneck block to learn the global information in high-level lung tumor feature maps, This is a way to model the lesion globally and extract the global features, and the rich lesion association features in the lung image are learned by modeling the remote semantic feature relationship. On the basis of capturing the original local lesion information, the backbone network establishes the global dependence relationship between pixels and learns the long-distance spatial correlation between pixels in the same feature map.
3.3 Cross-Scale Interactive Feature Enhancement Auxiliary Network
The feature map of superficial lung tumor has rich lesion details, such as contour, texture and location, but lacks the coarse-grained semantic information of the overall image. The feature map of deep lung tumor has semantic features, but lacks the fine-grained spatial information of the lesion region. In order to extract more lesion features and pay full attention to the fine-grained information, this paper designs an auxiliary network to achieve an interactive fusion of cross-scale shallow feature information and deep semantic information. The cross-scale interactive feature enhancement module is used to aggregate the feature maps of adjacent stages of the backbone network with different resolutions, as shown in Fig. 2. So as to enhance the attention to lesion features between channels and within each channel. Irrelevant noise except lesion is suppressed. In the feature extraction part, the backbone and auxiliary network are used to achieve feature selection of global and adjacent tumor features in lung images, so that the model could focus more on the lesion region.
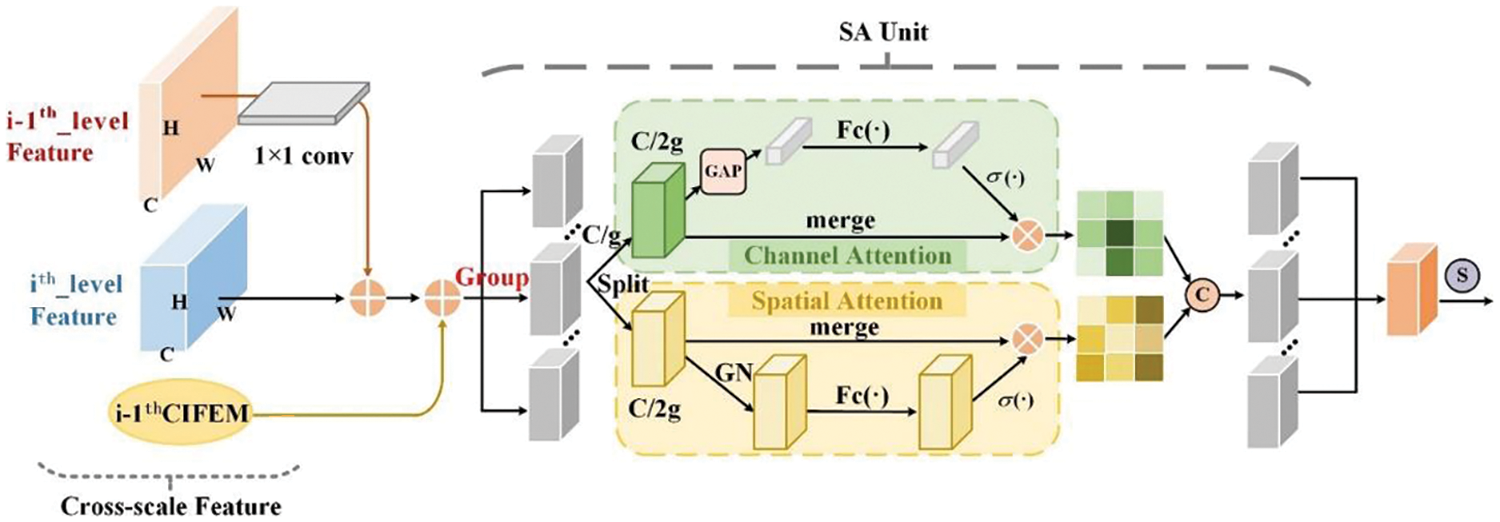
Figure 2: Cross-scale interactive feature enhancement module
The cross-scale interactive feature enhancement auxiliary network is composed of five stages (Stage 0, Stage 1, Stage 2, Stage 3, Stage 4). C1, C2, C3, C4 and C5 are feature layers that are extracted from Stage 0 to Stage 4 separately. In Stage 1, low-level feature C1 and high-level feature C2 are cross-scale fused by CIFEM1, and the results are added with high-level feature C2. The fusion results of Stage 1 are used as the input of the feature fusion network and Stage 2. In Stage 2, C2, C3 and CIFEM1 output are fused by CIFEM2. The fusion results of Stage 2 are added with high-level feature C3. The fusion results of Stage 2 are used as the input of the subsequent feature fusion network and Stage 3. Similarly, Stage 3 and Stage 4 adopted the same cross-scale feature interactive enhancement method as Stage 2.
The CIFEM is the key technology of the auxiliary network, as shown in Fig. 2. Through this cross-scale feature interaction mechanism, the features in the five stages of the network are enhanced. In the CIFEM module, this paper further optimizes the instance segmentation task of cross-scale features, and realizes the fine-grained instance segmentation and coarse-grained semantic segmentation of lung tumors. Let i = 1, there only two adjacent feature layers C1 and C2 are inputted into CIFEM1. Let i ≥ 2, there are 3 input feature maps about CIFEM2, CIFEM3 and CIFEM4, the previous feature layer, the output of the previous CIFEM and the current feature layer.
The pseudo-code of CIFEM (Algorithm 1) is expressed as:

The input of the CIFEM is composed of the adjacent feature layer of the previous feature layer (
The specific process of the CIFEM is shown in formula (1):
In formula,
3.4 Cross-Scale Interactive Feature Fusion FPN Network
In the FPN low-level network, the receptive field is small, which can capture the local features of the lesions in the lung image, and has a strong detail perception ability. In the FPN high-level network, the receptive field is large, the overall global features of lung images can be obtained, and the semantic expression ability is strong. Therefore, the fusion of local low-level information and global high-level information can effectively enhance the lesion region. For this reason, in the feature fusion part of this paper, the bidirectional interactive fusion method from low-level to high-level and from high-level to low-level is adopted, and the cross-scale complementary information between adjacent feature layers is obtained through the down-up path and up-down path, as shown in Fig. 1. Among them, the down-up path effectively strengthens the low-level feature information in the high-level feature layer, and the up-down path effectively strengthens the high-level semantic information in the low-level feature layer. The secondary fusion is used to enhance the feature expression of tumor lesions, effectively improving the accuracy of lesion recognition.
The cross-scale interactive feature fusion FPN [24] network (CIF-FPN) includes Stage 1 to Stage 4. In each stage, low-level features and adjacent high-level features are input into the cross-scale interactive feature fusion module for processing, as shown in Fig. 3. The CIFM is interactively integrated in the down-up path. The low-level information such as details, texture and location of lesions in the high-level feature layer are continuously strengthened. Then the fusion features after layer-by-layer interaction are added and fused successively by elements in the up-down path, so as to strengthen the high-level semantic information in the low-level feature layer. The ability of the model to express and identify the lesion region is enhanced by gathering the feature information of each stage.
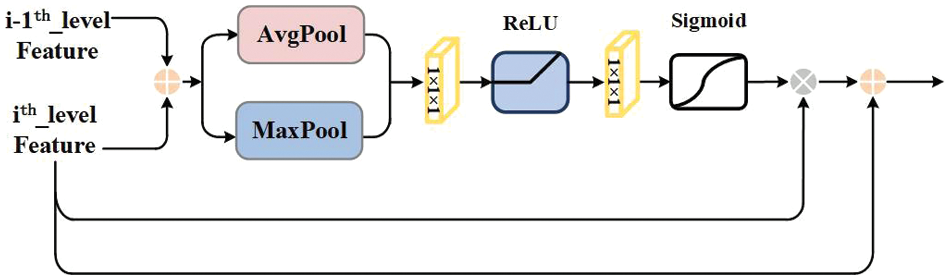
Figure 3: Cross-scale interactive feature fusion module
The feature maps from Stage 1 to Stage 4 are used as the input of CIF-FPN, which are generated by the backbone and auxiliary network. There are down-up paths and up-down paths in CIF-FPN. Convolution and deconvolution operations are shown in Fig. 1, and the basic operations will not be described here. In CIF-FPN, the first path is the down-up path. In Stage 2, C2 and C3 are inputted into CIFM1 for interactive fusion. In Stage 3, C3 and C4 are inputted into CIFM2 for interactive fusion. Similarly, in Stage 4, C4 and C5 are inputted into CIFM3 for interactive fusion. The channel number of the feature map is adjusted by 1 × 1 convolution operation to 256. The second path is the up-down path. The feature layer P5 is obtained by the first branch that the output of CIFM3 is adjusted by 3 × 3 convolution operation. The feature layer P6 is obtained by the second branch that the output of CIFM3 is adjusted by max pooling operation. The feature layer P4 are obtained by 3 × 3 convolution operation that the output of CIFM2 and the upsampled CIFM3 output are plused using pixel-by-pixel addition. The feature layer P3 is obtained by 3 × 3 convolution operation that the results of the upsampled Stage 3 and the output of CIFM1 are plused using pixel-by-pixel addition. The feature layer P2 is obtained by 3 × 3 convolution operation and the results of Stage 2 and the C2 are plused using pixel-by-pixel addition. P2-P6 feature layers are used as input for the subsequent RPN.
The pseudo-code of CIFM (Algorithm 2) is expressed as:

The input of the CIFM is the adjacent scale feature maps
The specific process of the CIFM is shown in formula (2):
In formula,
Medical imaging plays a key role in clinical applications in the fields of diagnosis, planning, surgery and radiotherapy [25]. In this paper, PET/CT medical images can be used to better locate and segment tumor lesion regions in lung images. The train datasets and test datasets of PET/CT modal medical are shown in Fig. 4, and the corresponding annotated example images are shown in Fig. 5.
Figure 4: PET/CT modal lung tumor medical images. (a) Train set and (b) test set
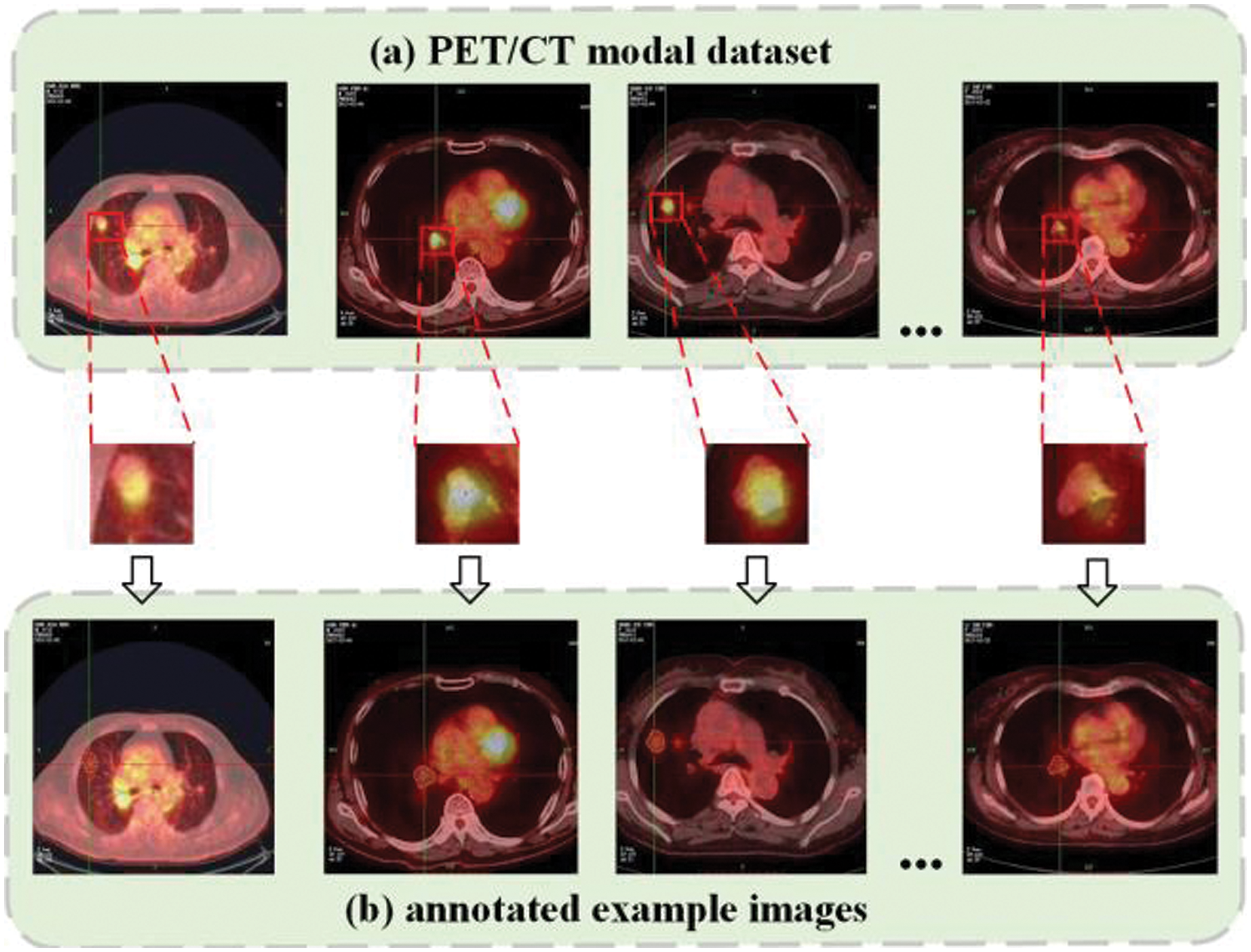
Figure 5: PET/CT lung tumor images and annotated example images
In this paper, the dataset, which is used in the experiment, is obtained from the original PET/CT images of lung cancer patients provided by the nuclear medicine department of a Grade-A tertiary hospital in Ningxia, and relevant clinicopathological diagnosis data are provided. The clinicopathological diagnosis data included textual information such as age, gender, tumor benign or malignant, medical advice examination conclusion and clinical diagnosis. The PET/CT modal dataset includes 1052 sample images of patients in the age range of 29 to 76 years from January 2018 to June 2020, of which 946 images are used as the training set and 106 images are used as the test set. Firstly, the digital imaging and communication in medicine (DICOM) files of the original PET/CT images are imported into the image processing software MicroDicom viewer and converted into JPG format data. Secondly, the lung tumor images in the form of mediastinal window and lung window are cropped using algorithms to maximize the proportion of the whole lung in the image and reduce the redundant information of the background (where the pixel value is 0). Then, the gold standard labeling of the lesion contour is completed using Labelme software under the guidance of professional radiologists. Finally, the corresponding JSON annotation file and label image are generated. The annotation file included the category label of the lesion, the coordinate value of the annotation point, the width and height of the image, and the image path.
The deep learning framework based on Pytorch is used to implement the MCIF-Transformer Mask RCNN proposed in this paper. The experimental environment is configured as the server with Intel(R) Xeon(R) Gold 6154 CPU, 256 GB memory, NVIDIA TITAN V graphics card, python 3.7, PyTorch 1.7.0 and CUDA version is 11.1.106. During network training, the epoch is set to 300, the initial learning rate is 0.0001, the batch size is 2, and the stochastic gradient descent (SGD) algorithm is used as the optimizer to optimize the model, where the parameter momentum is 0.9 and the weight decay coefficient is 1 × 10−4.
In order to comprehensively and objectively evaluate the instance segmentation performance of MCIF-Transformer Mask RCNN, the intersection over union (IoU), average precision (AP), and average recall (AR) are used as evaluation criteria to quantitatively evaluate the performance of the model [26]. In the task of lung tumor detection and segmentation, True Positive (TP) indicates that the model correctly identifies the lesion region. False Positive (FP) indicates that the normal tissue region is misdiagnosed as the lesion region. False Negative (FN) indicates that the lesion region is missed by the model as a normal tissue region. True Negative (TN) means that the model correctly identifies the normal tissue region. The definition and formula of the evaluation index are as follows:
Intersection over union is a measure of the accuracy of detecting and segmenting targets in a specific dataset. It includes two parts: The ground truth and the predicted results of the model algorithm. IoU is used to measure the correlation between the labeled box and the predicted box. The larger the value is, the higher the correlation is, and the better performance of the model. The specific calculation formula is shown in (3):
Average Precision is the percentage of the number of correctly identified targets in the total number of identified targets. It is used to measure the performance of model detectors on each category. AP50 indicates the AP value when the IoU threshold is 0.5. AP indicates that the AP value of the threshold of IoU is calculated every 0.05 in the interval of [0.50, 0.95], and then the average value of the corresponding results of 10 thresholds of IoU is calculated as the final AP value. AP_s is the number of pixels of small target objects in the image less than 32 × 32 = 1024. The specific calculation formula is shown in (4):
where, c is the detection category, which usually refers to a single category. th is the threshold value of each category, and t is the number of detection samples.
Average Recall is the percentage of the number of correctly identified targets in the test set. For the same model structure, the larger the test set size, the better the AR effect. In this paper, the IoU threshold is used to calculate the AR value every 0.05 in the interval [0.5, 0.95], and the average of all results is taken as the final result. AR1 is the average recall of 1 test; AR10 is the average recall of 10 tests. The specific calculation formula is shown in (5):
5 Experimental Results and Analysis
In order to objectively evaluate the advances and effectiveness of the MCIF-Transformer Mask RCNN, ablation experiments, comparison experiments of Mask RCNN instance segmentation models based on different backbones, and comparison experiments of different instance segmentation networks are performed on the same PET/CT lung tumor dataset. The average precision and the average recall are used for quantitative comparison and qualitative analysis, and the model performance is evaluated from detection (det) and segmentation (seg).
The first set of experiments is to explore the impact of each module on the instance segmentation performance of the model. The second set of experiments is to verify the influence of different backbone networks on the instance segmentation results of Mask RCNN. The third set of experiments is to compare the proposed model with two-stage and single-stage instance segmentation networks.
In order to verify the effectiveness and feasibility of the ResNet-Transformer backbone network, a cross-scale interactive feature enhancement auxiliary network (CIFEN) and CIF-FPN are proposed in this paper to improve the performance of the model. Mask RCNN with resnet50 backbone is used as the baseline model, and four ablation experiments are performed. Experiment 1, Mask RCNN (resnet50), Mask RCNN with resnet50 as the backbone network. The Experiment 2, CIFEN-Mask RCNN, which adds CIFEN based on resnet50 backbone network. The Experiment 3, RT+CIFEN-Mask RCNN, the backbone of the Experiment 2 is replaced by ResNet-Transformer backbone network, which replaces the basic residual block with multi-head self-attention BoT module at the top layer. Experiment 4, MCIF-Transformer Mask RCNN, which adds CIF-FPN based on the Experiment 3. Table 1 shows the quantitative index comparison of the ablation experimental results of the proposed model in this paper.
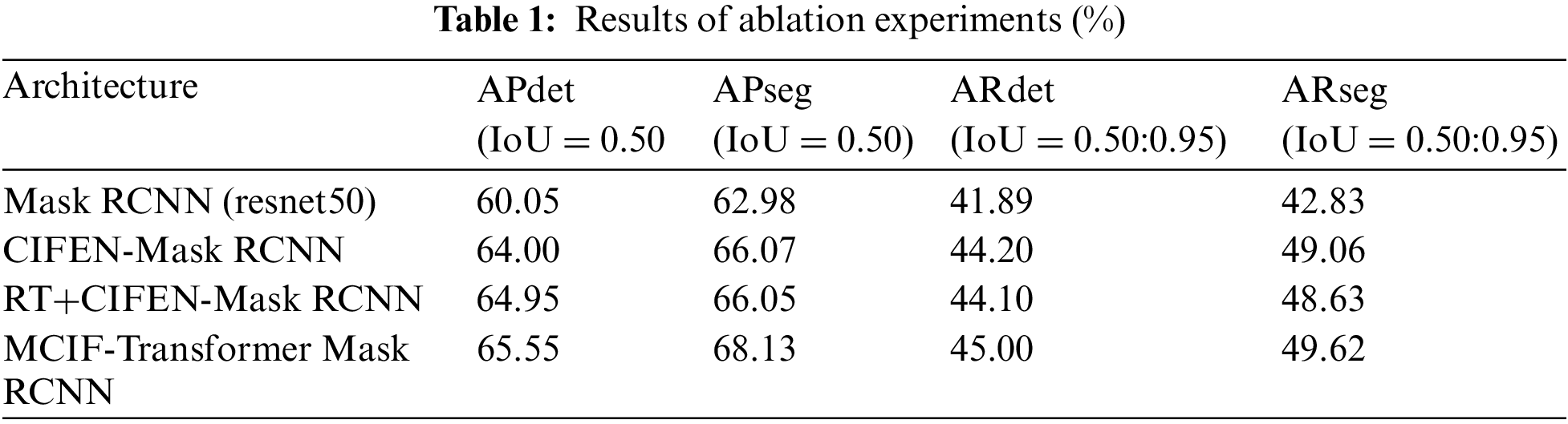
This ablation experiment quantifies the impact of submodules on the overall model detection and segmentation performance, and the experimental results are shown in Table 1. The APdet, APseg, ARdet and ARseg of the baseline model Mask RCNN (resnet50) are 60.05%, 62.98%, 41.89% and 42.83%. The baseline model is combined with CIFEM, the indicators of CIFEN-Mask RCNN are relatively improved by 3.95%, 3.09%, 2.31% and 6.23%. It shows that the auxiliary network based on CIFEM is constructed by aggregating adjacent scale features and strengthening the attention to tumor lesions in lung images. It effectively alleviates the problem of missing detection caused by the loss of information due to the small size of the lesion in the feature extraction process. The baseline model is combined with CIFEM and Bottleneck Transformer, RT+CIFEN-Mask RCNN has further improved the indicators compared with Mask RCNN (resnet50) model. Compared with CIFEN-Mask RCNN, the APdet is increased by 0.95%, and the APseg, ARdet and ARseg indicators are slightly decreased. The reason is that when training on small and medium-sized datasets, transformer lacks the inherent inductive bias of CNN, such as spatial locality and translation invariance [26]. Therefore, when the number of data sets is not sufficient, it is difficult to train the backbone network weights that introduce the BoT module, so the generalization ability is not strong and the effect is not very good. After the basic model combined with CIFEM, BoT and CIFM, the four indexes are improved, the APdet, APseg, ARdet, ARseg are 65.55%, 68.13%, 45.00%, 49.62%. It shows that the backbone network, auxiliary network and CIF-FPN proposed in this paper can enhance the feature representation and recognition ability of tumor lesions in lung images by using cross-scale adjacent high and low layer information. Shallow feature information and deep semantic information are fused to solve the problem of feature neglect. In order to more intuitively show the instance segmentation performance of the model, the radar chart is used in Fig. 6 to compare the model performance of different combination modules. It can be seen that the detection and segmentation performance of MCIF-Transformer Mask RCNN is better than that of other combination models. It shows that the introduction of cross-scale feature information interaction in the feature extraction and feature fusion stages helps to retain the detailed information of the lesion. It effectively alleviates the problem of insufficient feature extraction of the lesion region in the task of lung image instance segmentation.

Figure 6: Radar chart comparison of different module instance segmentation results of MCIF-Transformer Mask RCNN
In PET/CT lung images, there are complex conditions such as irregular beating of the heart, and similar highlighting features of other organs and lesions, which lead to the influence of noise interference data in the process of lesion identification. As the location and edge of some small lesions are not obvious, the model proposed in this paper also has false negative and false positive results, as shown in Fig. 7. The false recognition rate of this model is 39.7%. Although the model in this paper has some false detections due to the weak lesion features, the model can still focus on small lesion features and accurately locate and segment tumor regions in lung organs for most lung tumor images on the whole. Fig. 8 shows the 3D grayscale of PET/CT images and the instance segmentation results of MCIF-Transformer Mask RCNN. It can be seen that the sensitivity to weak lesions and their edges in the images is increased, and the false positive rate and false negative rate are reduced. The results show that the overall architecture and each module design of the model are reasonable, and the instance segmentation performance in the region of interest about PET/CT lung images are improved more better.
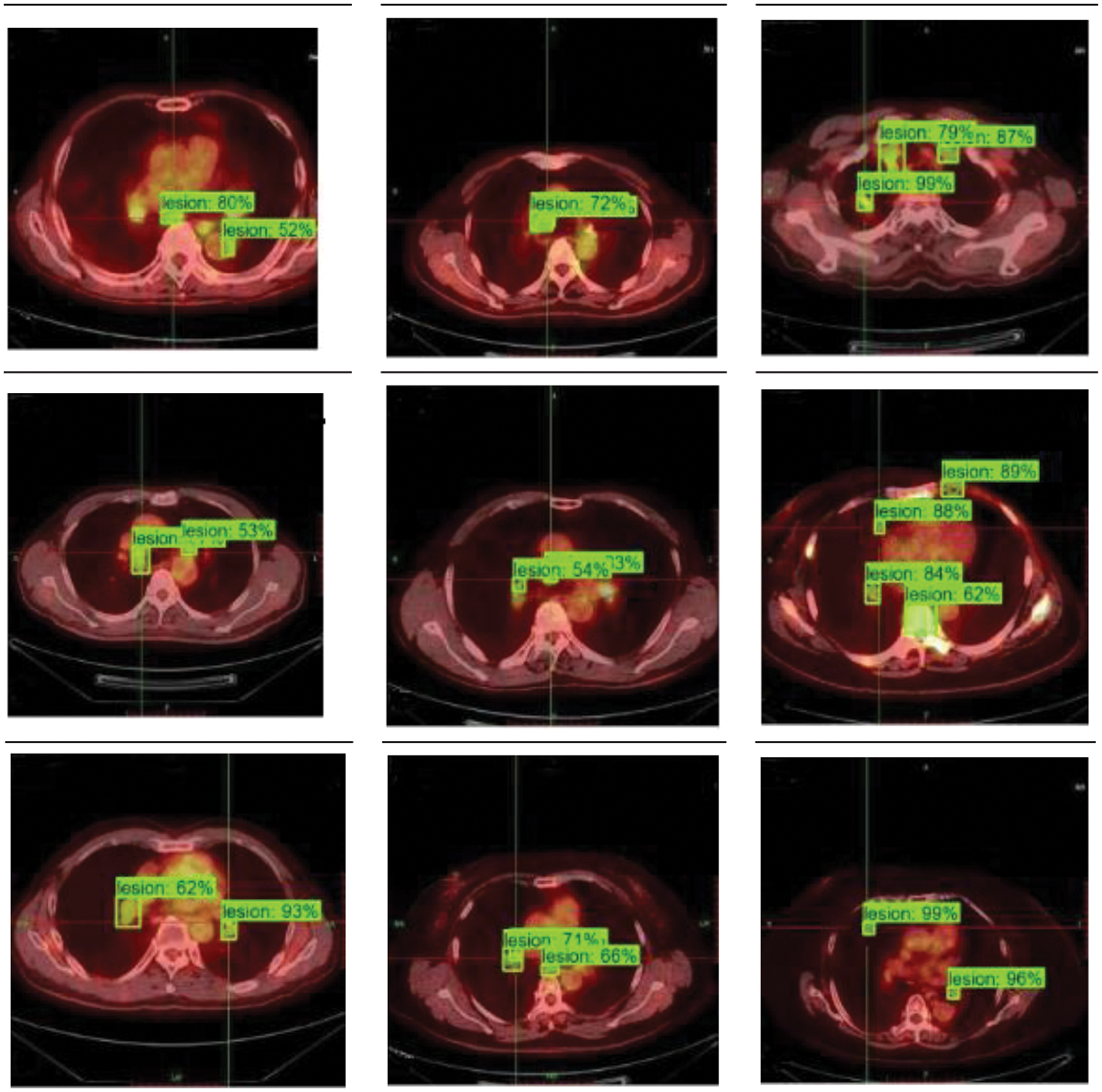
Figure 7: Instance segmentation results of partial misidentification examples of MCIF-Transformer Mask RCNN
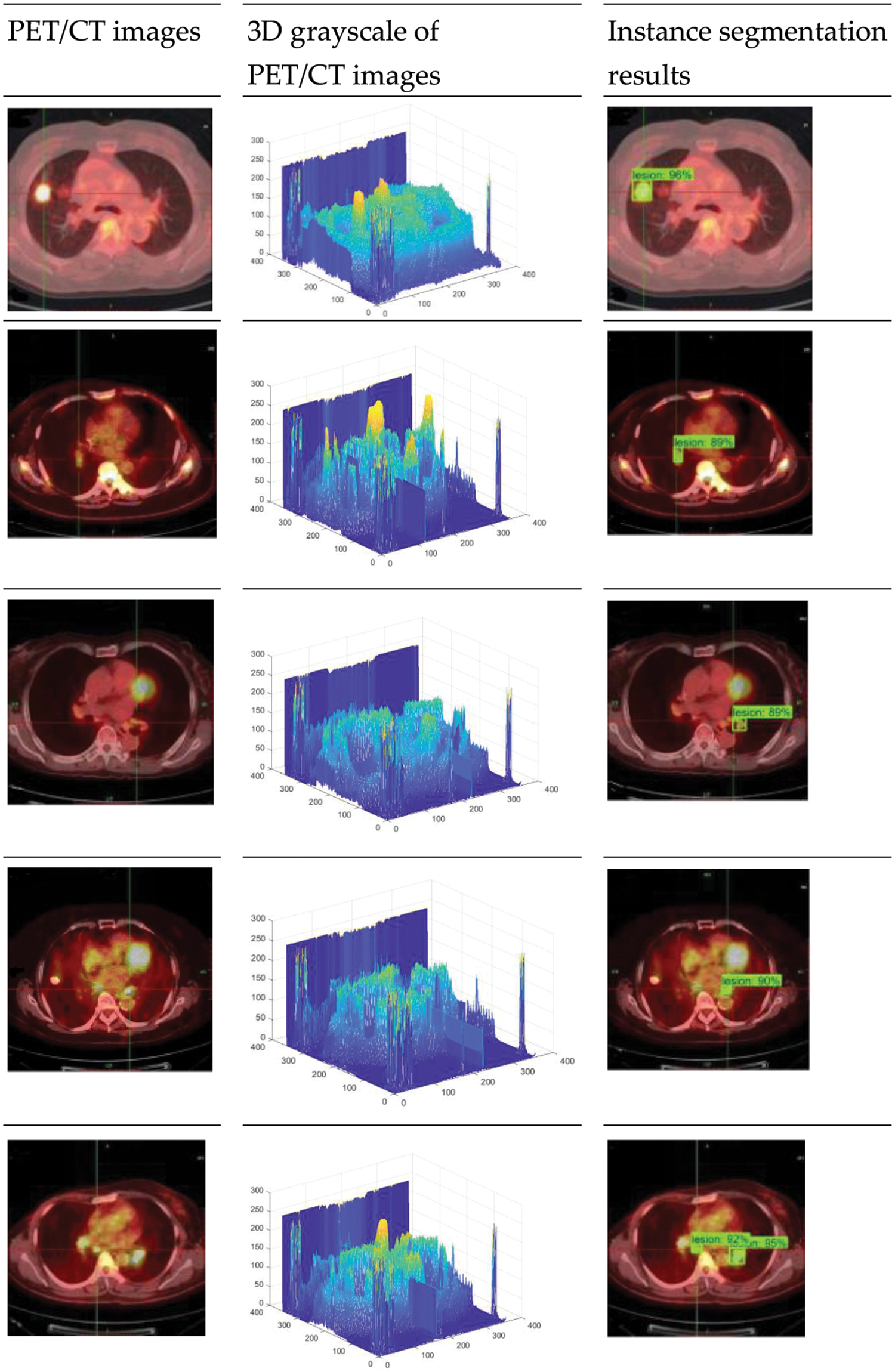
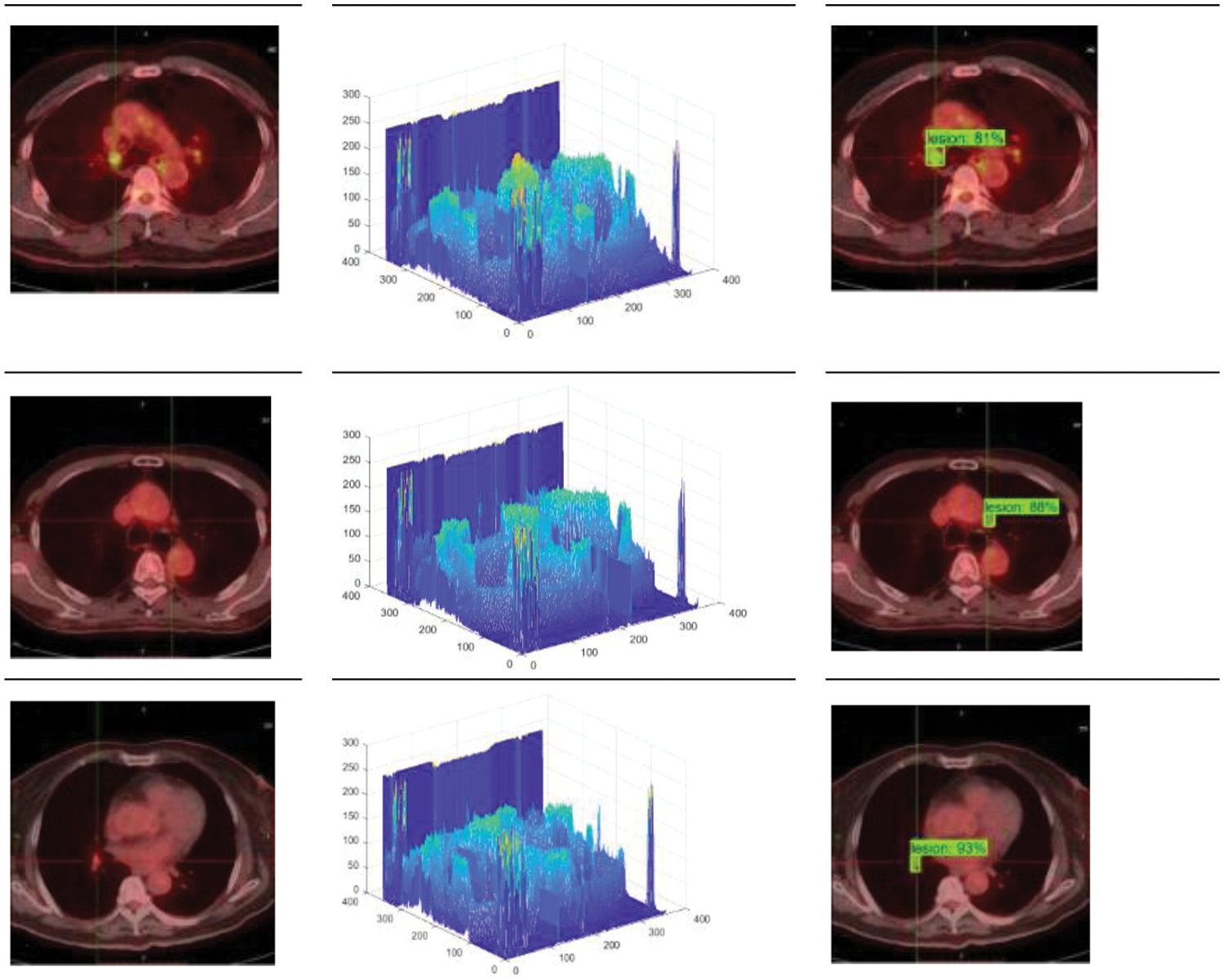
Figure 8: Instance segmentation results of MCIF-Transformer Mask RCNN
5.3 Comparison Experiments of Mask RCNN Instance Segmentation Models Based on Different Backbones
In order to verify the effectiveness of using transformer structure in the MCIF-Transformer Mask RCNN, the model in this paper is compared with the Mask RCNN model with resnet50 and resnet101 as the backbone, respectively. The detection comparison results are shown in Table 2, and the comparison of each detection index value is shown in Fig. 9.
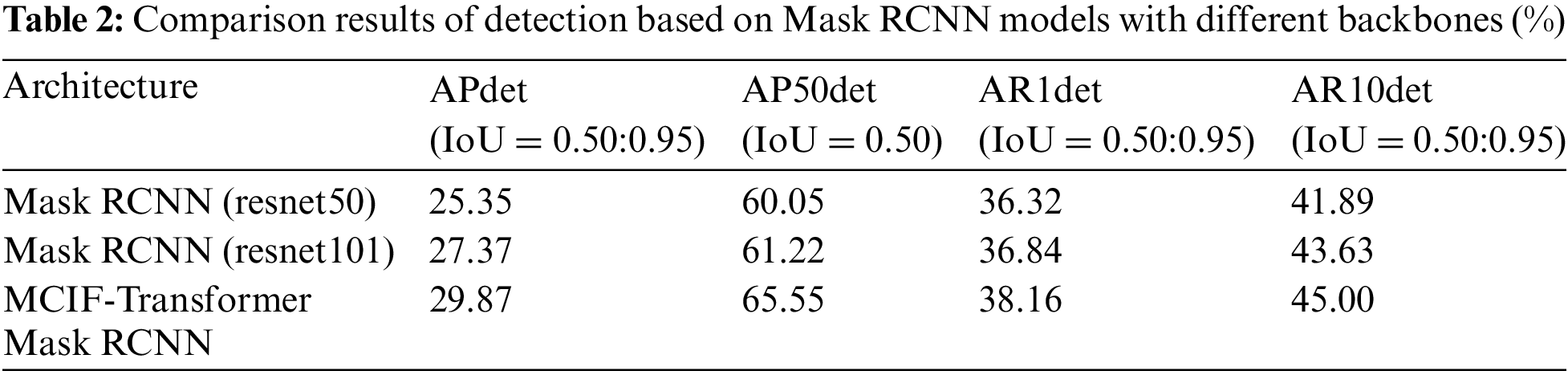

Figure 9: Comparison of detection index values of Mask RCNN instance segmentation models based on different backbones
Compared with Mask RCNN (resnet50), Mask RCNN (resnet101) increased by 2.02%, 1.17%, 0.52% and 1.74% in APdet, AP50det, AR1det and AR10det. The detection index values of MCIF-Transformer Mask RCNN proposed in this paper are 29.87%, 65.55%, 38.16% and 45.00%. Compared with Mask RCNN (resnet101), the indexes of this model are improved by 2.5%, 4.33%, 1.32% and 1.37%, respectively. It can be seen that the use of transformer structure in the backbone can effectively improve the extraction and localization of lesion features by the model.
The segmentation comparison results are shown in Table 3, and the comparison of each segmentation index value is shown in Fig. 10. It can be seen that compared with Mask RCNN (resnet50) in APseg, AP50seg, AR1seg and AR10seg, respectively, Mask RCNN (resnet101) increased by 0.70%, 0.07%, 0.71% and 2.26%. The segmentation index values of MCIF-Transformer Mask RCNN proposed in this paper are 34.07%, 68.13%, 42.08% and 49.62%. Compared with Mask RCNN (resnet101), the indexes of this model are improved by 1.94%, 5.08%, 3.12% and 4.53%. Compared with the first two networks, MCIF-Transformer Mask RCNN has a greater advantage in detecting and segmenting tumor lesions, which is due to the fact that the first two models only use convolution to obtain local lesion features, while the model in this paper learns rich correlation features in PET/CT lung images by learning them on the lowest resolution feature map based on convolution capturing local information of the image through a mixture of convolution and global self-attention in the backbone part. Further global information is acquired to enhance the localization and segmentation accuracy of lesion regions, which can better achieve feature extraction and recognition of global and local regions in lung images.
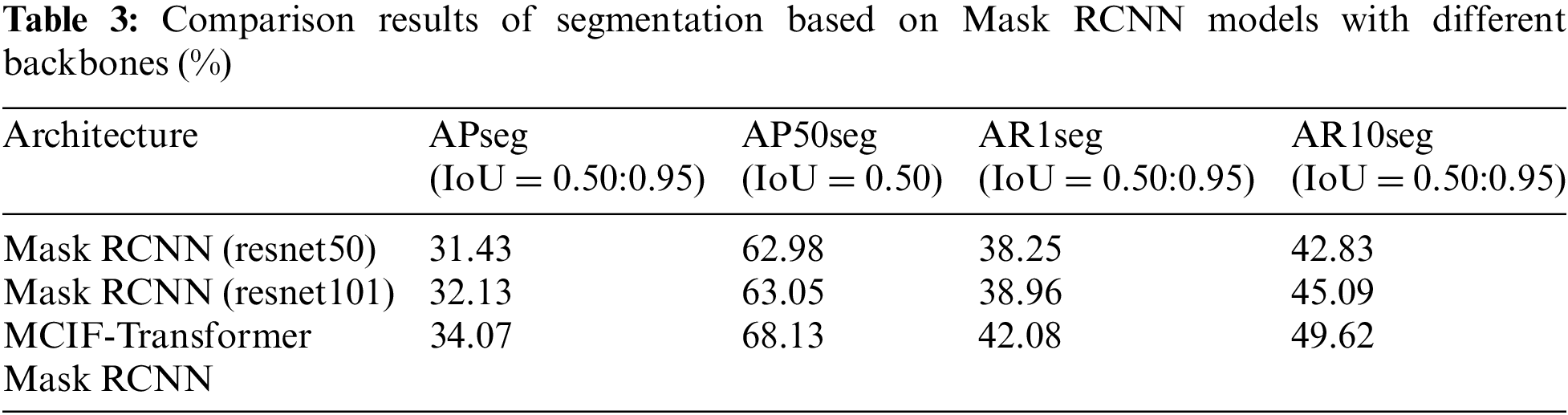
Figure 10: Comparison of segmentation index values of Mask RCNN instance segmentation models based on different backbones
5.4 Comparison Experiments of Different Instance Segmentation Networks
In order to verify that the model structure of this paper can enhance the attention to the key features of tumor lesions in lung images and improve the accuracy of instance segmentation, there are two types of experiments in this section as a whole. The first type is the comparison of two-stage instance segmentation networks, and the second type is the comparison of single-stage instance segmentation networks. Experiment 1, Mask RCNN (resnet50), resnet50 is used as the Mask RCNN of feature extraction network. Experiment 2, Mask RCNN (resnet101), resnet101 is used as the Mask RCNN of feature extraction network. Experiment 3, Cascade RCNN, resnet50 is used as the Cascade RCNN of feature extraction network. Experiment 4, Mask Scoring RCNN, resnet50 is used as the backbone of Mask Scoring RCNN. Experiment 5, YOLACT, resnet50 is used as the backbone of YOLACT. Experiment 6, MCIF-Transformer Mask RCNN, which is the model proposed in this paper. Experiments 1 to 4 are the comparison of two-stage instance segmentation networks. Experiment 5 is the comparison of single-stage instance segmentation network.
Table 4 lists the results of the comparison between the proposed model and other instance segmentation networks on the same dataset. It can be seen from the table that the APdet, APseg, APdet_s, APseg_s of the two-stage instance segmentation network Mask RCNN (resnet50) are 60.05%, 62.98%, 25.71% and 31.45%. The APdet and APseg of Mask RCNN (resnet101) are 61.22% and 63.05%. The APdet and APseg of Cascade RCNN are 62.20% and 63.70%. The APdet and APseg of Mask Scoring RCNN are 60.10% and 62.50%. The APdet and APseg of the single-stage instance segmentation network YOLACT are 57.12% and 57.48%. For the average precision of detection and segmentation tasks, MCIF-Transformer Mask RCNN is better than other networks in the four indicators, APdet reaches 65.55%, APseg reaches 68.13%, APdet_s and APseg_s respectively reaches 30.96% and 34.44%. The main reason is that compared with the mainstream networks, the MCIF-Transformer Mask RCNN can effectively express the features of tumor lesions in lung images by reasonable aggregation the spatial detail information and abstract semantic information of cross-scale features. The existing methods such as Mask RCNN ignore the interactive fusion of cross-scale feature information and focus only on the image features of the current single layer. The feature fusion part of YOLACT network lacks the feature information of transmits location, edge, and detail from low-level to high-level, which is conducive to identifying tumor lesions. In this model, the backbone and auxiliary network are used to interactively fuse adjacent shallow and deep features. The Transformer-based BoT module is used in the top layer of the backbone network to effectively capture the global information in the lung tumor image through the global multi-head self-attention mechanism. A cross-scale interactive feature enhancement auxiliary network is designed to enhance the attention to the lesion region. A cross-scale interactive feature fusion FPN network is constructed, and the down-up interactive fusion path from the low-level to the high-level is added to enhance the lesion feature information in a bidirectional interactive way. The size of the whole lung image is 356 × 356 pixels, the size of the lesion is 7 × 7 ~ 26 × 26 pixels, and the lesion accounted for 0.4% of the image. Compared with the overall PET/CT image, the data distribution of tumor lesion is uneven, and it is a target object with the feature information both weak and small. Therefore, it can be seen from the results in the sixth row of Table 4 that the detection and segmentation indexes of the model AP_s proposed in this paper are significantly improved. Other networks are tested on the same dataset, and the results show different performance, as shown in Figs. 11 and 12. Fig. 11 shows the 3D histogram of AP values of the proposed model and the comparison networks. The X-axis represents different networks. From left to right, these represent Mask RCNN (resnet50), Mask RCNN (resnet101), Cascade RCNN, Mask Scoring RCNN, YOLACT, MCIF-Transformer Mask RCNN. The Y-axis represents the average precision of detection (APdet) and the average precision of segmentation (APseg). The z-axis is the value corresponding to the two evaluation indicators. Similarly, Fig. 12 shows the 3D histogram of AP_s values for different instance segmentation networks. The results show that the proposed model achieves superior detection and segmentation performance in the PET/CT lung datasets.
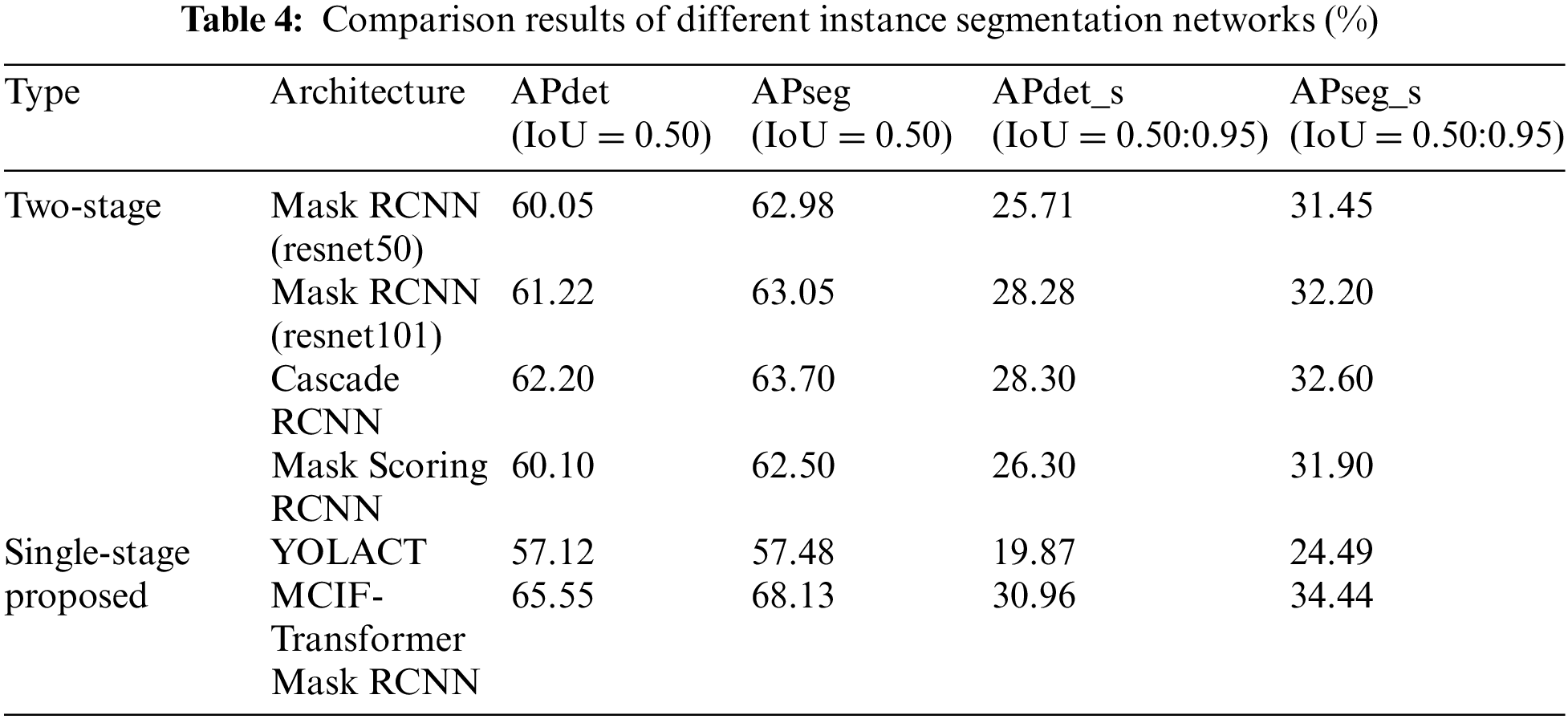
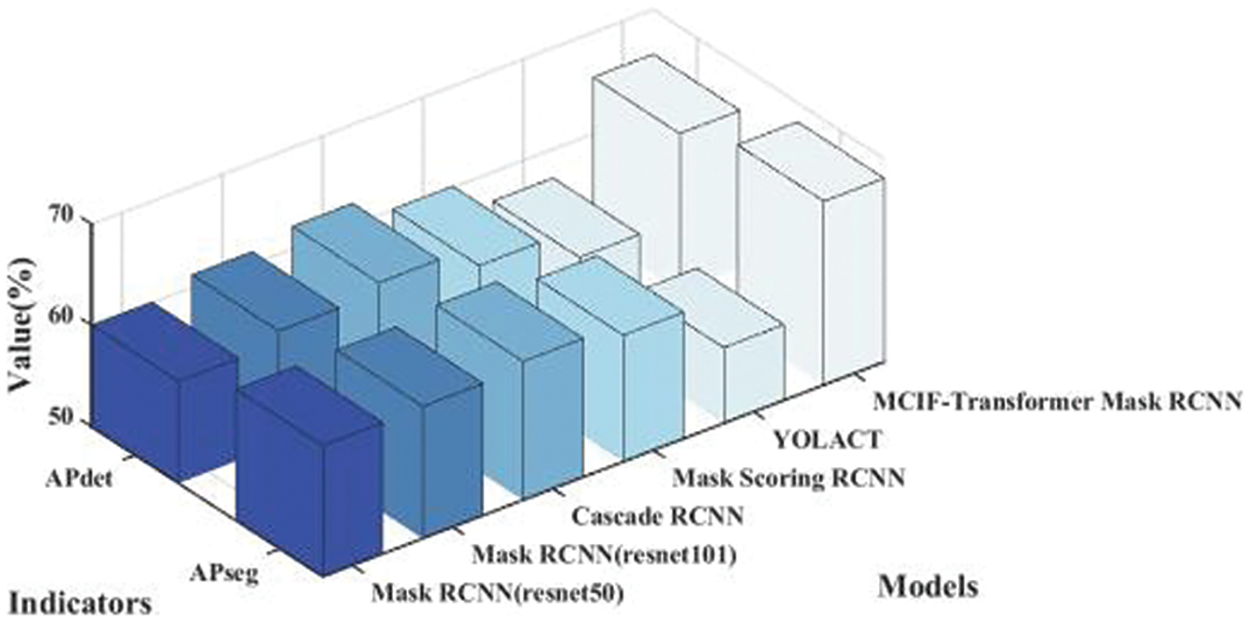
Figure 11: Comparison of 3D histogram of AP results for different instance segmentation networks
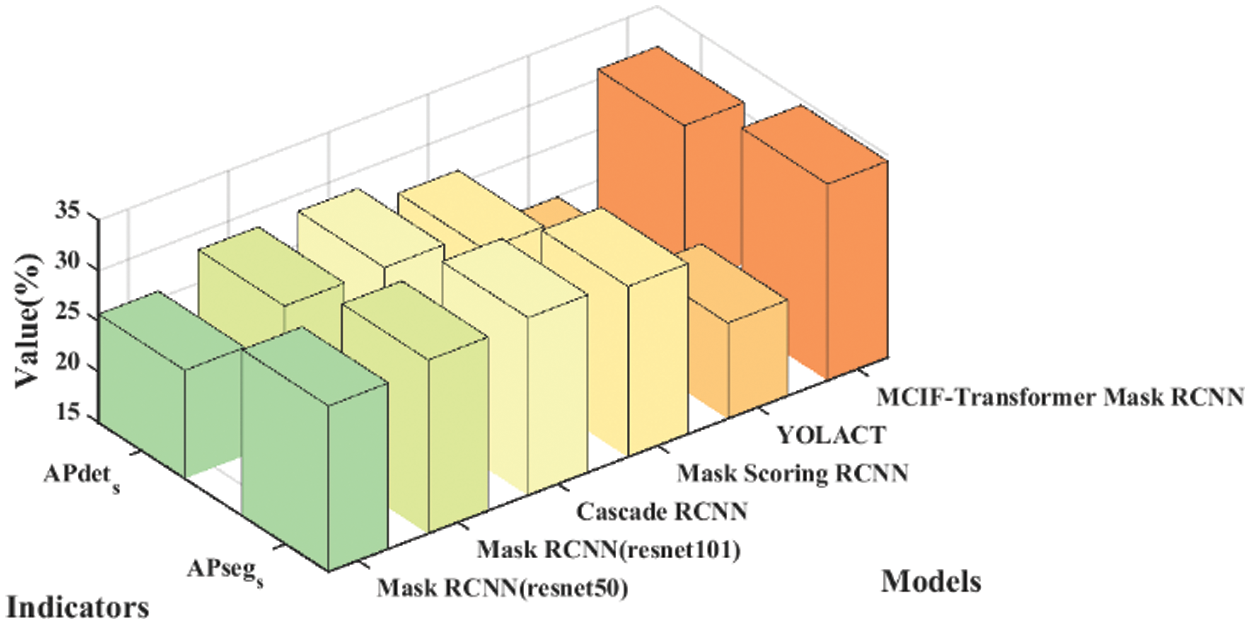
Figure 12: Comparison of 3D histogram of AP_s results for different instance segmentation networks
The detection and segmentation of tumor lesions in PET/CT lung images have important clinical significance for non-invasive diagnosis and accurate evaluation of lung cancer. In this paper, Mask RCNN is studied and MCIF-Transformer Mask RCNN model for PET/CT lung tumor instance segmentation is proposed, which is a further development of Mask RCNN and a beneficial attempt to accurately segment lung tumors. ResNet-Transformer backbone network is constructed to strengthen the non-local spatial attention to the lesion region. The recognition of lesion features is improved and richer semantic classification information is obtained. A cross-scale interactive feature enhancement auxiliary network is proposed to aggregate adjacent backbone features of different sizes, and the network is used to generate auxiliary prediction information with deep and shallow fusion features to guide the enhancement of backbone network features. A cross-scale interactive feature fusion FPN network is proposed to supplement detailed information from low-level to high-level and transfer semantic information from high-level to low-level by second fusion. The context information between adjacent feature maps with different resolutions is effectively aggregated, and the focus degree and expression ability of weak and small key features of lung tumors are enhanced. In order to verify the effectiveness and feasibility of the MCIF-Transformer Mask RCNN, the ablation experiments, the Mask RCNN instance segmentation models based on different backbones and the comparison experiments of different instance segmentation networks are conducted on the clinical dataset. Compared with Mask RCNN (resnet50), the proposed model improves APdet and APseg by 5.5% and 5.15%. The results of ablation experiments show that the detection and segmentation accuracy of the ResNet-Transformer backbone network, the cross-scale interactive feature enhancement auxiliary network and the cross-scale interactive feature fusion FPN network designed in this paper are improved compared with models of other composite modules. The results of comparison experiments show that the proposed model is significantly improved in both APdet and APseg compared with other networks. By focusing on the global information and aggregating the deep and shallow information at the global, local and adjacent aspects, the detection and segmentation of the lesion region are effectively supported. This model also has some shortcomings and limitations, such as: 1) In this paper, the multi-branch and cross-scale interactive fusion Transformer model is used to automate the tumor lesions localization and segmentation in PET/CT lung images. The results show that the model with cross-scale interactive fusion of feature information has better performance than the model that only focuses on the image features of this layer. The main reason for this result is that the shallow network has a small perception field and a small overlapping region between perception fields, which enables the network to capture local lesion information in lung images. For example, texture features, contour features, location features and other fine-grained lesion features in shallow features. The deep network has a large perception field, and the increase of overlapping regions between receptive fields enables the network to obtain the overall global features of lung images, so the deep feature map is rich in coarse-grained abstract features with strong semantics. Therefore, the integration of cross-scale local low-level features and global high-level feature are very important for the fine-grained features representation of focal areas. 2) The deep convolutional neural network achieves good results in the feature extraction task of instance segmentation. The local feature of lung image is obtained by convolutional operation, and the relationship between pixels is established in the local domain. However, modeling long distance features by stacking convolutional layers will lead to network gradient disappearance and degradation. Instance segmentation requires instance-level labeling of lesion regions, In lung images, global context information is crucial to precise localization and segment tumor lesions. Therefore, it is one of the future research directions to improve and utilize the global self-attention mechanism for medical image instance segmentation. 3) Because the model in this paper aggregates feature maps at different levels many times, the overall calculation of the model is large, which has certain limitations on the light weight of the model. In future studies, The model can be lightweight by replacing convolution with linear operations, grouping channels, and cutting unnecessary feature layers, This can effectively reduce the consumption of computing resources so that the model can be deployed on hardware devices with limited memory, which is of great significance for the popularization of computer-aided diagnosis system for lung cancer. which has certain reference significance and clinical value for lung cancer computer-aided diagnosis.
Acknowledgement: Thanks to graduate students Zhao Yannan, Shi Daozong, Wang Yaxing for their help on the paper.
Funding Statement: This research was funded by National Natural Science Foundation of China No. 62062003, Ningxia Natural Science Foundation Project No. 2023AAC03293.
Author Contributions: Huiling Lu: Data curation, Methodology, Writing–original draft, Validation, Programming, Visualization. Tao Zhou: Project administration, Writing–review & editing, Funding acquisition, Supervision.
Availability of Data and Materials: Data available on request from the authors.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. T. Tsuchida, Y. Matsumoto, T. Imabayashi, K. Uchimura, and S. Sasada, “Photodynamic therapy can be safely performed with Talaporfin sodium as a day treatment for central-type early-stage lung cancer,” Photodiagnosis Photodyn. Ther., vol. 38, pp. 102836, 2022. doi: 10.1016/j.pdpdt.2022.102836. [Google Scholar] [PubMed] [CrossRef]
2. H. Kawasaki, A. Nakamoto, N. Taira, T. Ichi, T. Yohena and T. Kawabata, “Endobronchial electrocautery wire snare prior to wedge bronchoplastic lobectomy for central-type lung cancer: A case report,” Int. J. Surg. Case Rep., vol. 10, pp. 211–215, 2015. doi: 10.1016/j.ijscr.2015.04.008. [Google Scholar] [PubMed] [CrossRef]
3. S. Najeeb and M. I. H. Bhuiyan, “Spatial feature fusion in 3D convolutional autoencoders for lung tumor segmentation from 3D CT images,” Biomed. Sig. Process. Control, vol. 78, pp. 103996, 2022. doi: 10.1016/j.bspc.2022.103996. [Google Scholar] [CrossRef]
4. A. Halder, S. Chatterjee, D. Dey, S. Kole, and S. Munshi, “An adaptive morphology based segmentation technique for lung nodule detection in thoracic CT image,” Comput. Methods Prog. Biomed., vol. 197, pp. 105720, 2020. doi: 10.1016/j.cmpb.2020.105720. [Google Scholar] [PubMed] [CrossRef]
5. J. D. Song et al., “Lung lesion extraction using a toboggan based growing automatic segmentation approach,” IEEE Trans. Med. Imag., vol. 35, pp. 337–353, 2016. doi: 10.1109/TMI.2015.2474119. [Google Scholar] [PubMed] [CrossRef]
6. S. Bai, X. Qiu, R. Hu, and Y. Wu, “A novel level set model initialized with guided filter for automated PET-CT image segmentation,” Cogn. Robot., vol. 2, pp. 193–201, 2022. doi: 10.1016/j.cogr.2022.08.003. [Google Scholar] [CrossRef]
7. T. Zhou, Q. R. Cheng, H. L. Lu, Q. Li, X. X. Zhang and S. Qiu, “Deep learning methods for medical image fusion: A review,” Comput. Biol. Med., vol. 160, pp. 106959, 2023. doi: 10.1016/j.compbiomed.2023.106959. [Google Scholar] [PubMed] [CrossRef]
8. S. Hansen, S. Kuttner, M. Kampffmeyer, T. V. Markussen, R. Sundset and Ø. S Kærnes, “Unsupervised supervoxel-based lung tumor segmentation across patient scans in hybrid PET/MRI,” Expert. Syst. Appl., vol. 16, pp. 114244, 2021. doi: 10.1016/j.eswa.2020.114244. [Google Scholar] [CrossRef]
9. T. Zhou, S. Liu, Y. L. Dong, J. Bai, and H. L. Lu, “Parallel decomposition adaptive fusion model: Cross-modal image fusion of lung tumors,” (in ChineseJ. Image Graph., vol. 28, no. 1, pp. 221–233, 2023. doi: 10.11834/jig.210988. [Google Scholar] [CrossRef]
10. A. M. Hafiz and G. M. Bhat, “A survey on instance segmentation: State of the art,” Int. J. Multimedia Inf. Retrieval, vol. 9, pp. 171–189, 2020. doi: 10.1007/s13735-020-00195-x. [Google Scholar] [CrossRef]
11. X. J. Fang, Y. Y. Zhou, M. F. Shen, T. Zhou, and F. Y. Hu, “Multi-scale context information fusion for instance segmentation,” J. Image Graph., vol. 28, no. 2, pp. 495–509, 2023. doi: 10.11834/jig.211090. [Google Scholar] [CrossRef]
12. K. He, G. Gkioxari, P. Dollar, and R. Girshick, “Mask R-CNN,” in IEEE Trans. Pattern Anal. Mach. Intell., Venice, Italy, 2017, pp. 2980–2988. doi: 10.1109/ICCV.2017.322. [Google Scholar] [CrossRef]
13. Z. Cai and N. Vasconcelos, “R-CNN: High quality object detection and instance segmentation,” IEEE Trans. Pattern Anal. Mach. Intell., vol. 43, pp. 1483–1498, 2019. doi: 10.1109/TPAMI.2019.2956516. [Google Scholar] [PubMed] [CrossRef]
14. D. Bolya, C. Zhou, F. Xiao, and Y. J. Lee, “YOLACT: Real-time instance segmentation,” in 2019 IEEE/CVF Int. Conf. Comput. Vis. (ICCV), Seoul, Korea, 2019, pp. 9156–9165. doi: 10.1109/ICCV.2019.00925. [Google Scholar] [CrossRef]
15. X. Wang, T. Kong, C. Shen, Y. Jiang, and L. Li, “SOLO: Segmenting objects by locations,” in Comput. Vis–ECCV 2020, Glasgow, UK, 2020, pp. 649–665. [Google Scholar]
16. Z. Huang, L. Huang, Y. Gong, C. Huang, and X. Wang, “Mask scoring R-CNN,” in 2019 IEEE/CVF Conf. Comput. Vis. Pattern Recog. (CVPR), California, USA, 2019, pp. 6402–6411. doi: 10.1109/CVPR.2019.00657. [Google Scholar] [CrossRef]
17. H. Zhou, L. Lei, Y. Xu, C. Peng, and Z. Shi, “Dual-supervised instance segmentation network combined with priori corner information,” in 2019 IEEE 4th Int. Conf. Image, Vis. and Comput. (ICIVC), Xiamen, China, 2019, pp. 55–60. doi: 10.1109/ICIVC47709.2019.8980993. [Google Scholar] [CrossRef]
18. N. Homayounfar, Y. Xiong, J. Liang, W. C. Ma, and R. Urtasun, “LevelSet R-CNN: A deep variational method for instance segmentation,” in Comput. Vis.–ECCV 2020, Glasgow, UK, 2020, pp. 555–571. [Google Scholar]
19. Y. Lin and Q. Zhao, “Mask-RCNN with spatial attention for pedestrian segmentation in cyber-physical systems,” Comput. Commun., vol. 180, pp. 109–114, 2021. doi: 10.1016/j.comcom.2021.09.002. [Google Scholar] [CrossRef]
20. K. Long, L. Tang, X. Pu, Y. Ren, and F. Deng, “Probability-based Mask R-CNN for pulmonary embolism detectio,” Neurocomputing, vol. 422, pp. 345–353, 2021. doi: 10.1016/j.neucom.2020.10.022. [Google Scholar] [CrossRef]
21. T. Zhou, Y. X. Niu, H. L. Lu, C. Y. Peng, Y. J. Guo and H. Y. Zhou, “Vision transformer: To discover the “Four Secrets” of image patches,” Inf. Fusion, vol. 105, pp. 102248, 2024. [Google Scholar]
22. T. Zhou, F. Z. Liu, X. Y. Ye, H. W. Wang, and H. L. Lu, “CCGL-YOLOV5: A cross-modal cross-scale global-local attention YOLOV5 lung tumor detection model,” Comput. Biol. Med., vol. 165, pp. 107387, 2023. doi: 10.1016/j.compbiomed.2023.107387. [Google Scholar] [PubMed] [CrossRef]
23. A. Srinivas, T. Y. Lin, N. Parmar, J. Shlens, and A. Vaswani, “Bottleneck transformers for visual recognition,” arXiv:2101.11605, 2021. [Google Scholar]
24. T. Y. Lin, P. Dollár, R. Girshick, K. He, B. Hariharan and S. Belongie, “Feature pyramid networks for object detection,” in 2017 IEEE Conf. on Comput. Vis. and Pattern Recogn. (CVPR), Honolulu, Hawaii, USA, 2017, pp. 936–944. doi: 10.1109/CVPR.2017.106. [Google Scholar] [CrossRef]
25. J. B. Antoine Maintz and M. A. Viergever, “A survey of medical image registration,” Med. Image Anal., vol. 2, pp. 1–36, 1998. doi: 10.1016/S1361-8415(98)80001-7. [Google Scholar] [CrossRef]
26. X. Liang, X. Lin, J. Quan, and K. Xiao, “Research on the progress of image instance segmentation based on deep learning,” Acta Electron. Sin., vol. 48, pp. 2476–2486, 2020. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools