 Open Access
Open Access
ARTICLE
Model Agnostic Meta-Learning (MAML)-Based Ensemble Model for Accurate Detection of Wheat Diseases Using Vision Transformer and Graph Neural Networks
1 Department of Creative Technologies, Faculty of Computing and Artificial Intelligence, Air University, Islamabad, 44000, Pakistan
2 Department of Computer Engineering, College of Computer and Information Sciences, King Saud University, P. O. Box 51178, Riyadh, 11543, Saudi Arabia
3 Department of Computer Engineering, Bahria School of Engineering and Applied Sciences, Bahria University, Islamabad, 44000, Pakistan
4 Laboratory for Neuro & Psychophysiology, KU Leuven, Leuven, 3000, Belgium
* Corresponding Authors: Syed Muhammad Usman. Email: ; Khursheed Aurangzeb. Email:
(This article belongs to the Special Issue: Advanced Artificial Intelligence and Machine Learning Frameworks for Signal and Image Processing Applications)
Computers, Materials & Continua 2024, 79(2), 2795-2811. https://doi.org/10.32604/cmc.2024.049410
Received 06 January 2024; Accepted 29 March 2024; Issue published 15 May 2024
Abstract
Wheat is a critical crop, extensively consumed worldwide, and its production enhancement is essential to meet escalating demand. The presence of diseases like stem rust, leaf rust, yellow rust, and tan spot significantly diminishes wheat yield, making the early and precise identification of these diseases vital for effective disease management. With advancements in deep learning algorithms, researchers have proposed many methods for the automated detection of disease pathogens; however, accurately detecting multiple disease pathogens simultaneously remains a challenge. This challenge arises due to the scarcity of RGB images for multiple diseases, class imbalance in existing public datasets, and the difficulty in extracting features that discriminate between multiple classes of disease pathogens. In this research, a novel method is proposed based on Transfer Generative Adversarial Networks for augmenting existing data, thereby overcoming the problems of class imbalance and data scarcity. This study proposes a customized architecture of Vision Transformers (ViT), where the feature vector is obtained by concatenating features extracted from the custom ViT and Graph Neural Networks. This paper also proposes a Model Agnostic Meta Learning (MAML) based ensemble classifier for accurate classification. The proposed model, validated on public datasets for wheat disease pathogen classification, achieved a test accuracy of 99.20% and an F1-score of 97.95%. Compared with existing state-of-the-art methods, this proposed model outperforms in terms of accuracy, F1-score, and the number of disease pathogens detection. In future, more diseases can be included for detection along with some other modalities like pests and weed.Keywords
Wheat is an important crop which is consumed worldwide and is considered as one of the world’s major food crops. Over the past few years, the production has been increased from 697 million metric tons to 781 million metric tons due to high demand as shown in Fig. 1. The growing global demand for wheat-based foods shows the economic importance of wheat production. However, challenges such as climate change-driven weather variability, water scarcity, and crop diseases hinder consistent yields. Fungal diseases, including rusts, powdery mildew, and Fusarium head blight, significantly contribute to reduced harvests and their symptoms can help identify the occurrence of disease [1]. The timely identification and effective control of crop diseases are imperative for satisfying the growing global demand, averting potential loss in yield, and guaranteeing the enduring sustainability of agricultural practices.
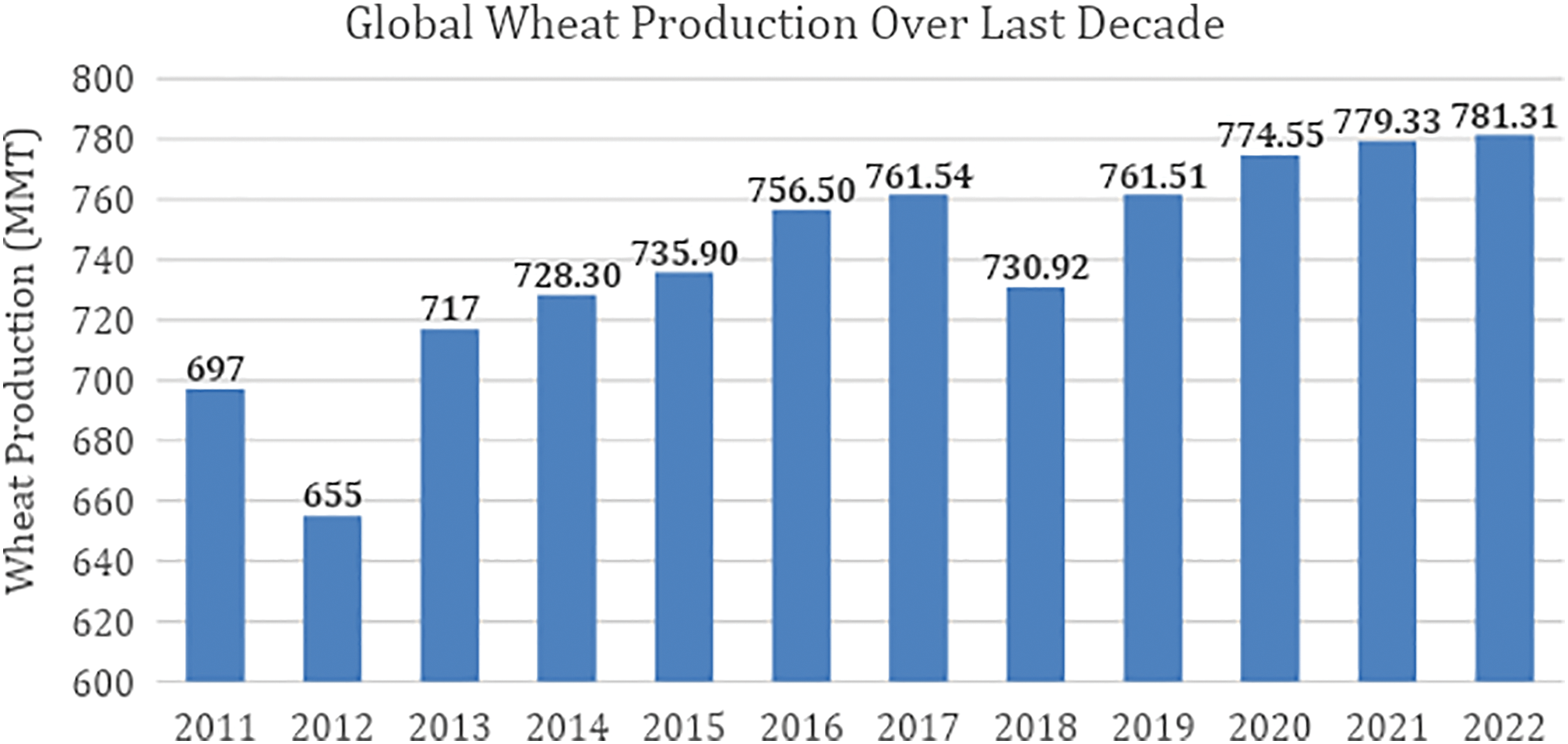
Figure 1: Global wheat production over the last decade [1]
Researchers have proposed various techniques for preprocessing [2,3], feature extraction [4], and machine learning (ML) or deep learning (DL) methods for the classification of diseased wheat plants [5–9]. Some researchers have used limited datasets to perform various classification techniques to classify wheat diseases without performing preprocessing of the dataset [10]. Various preprocessing techniques, such as Data Augmentation and Data Standardization, have been performed by researchers in the past [11,12]. Image Filtration and Histogram Equalization have also been utilized and implemented for the purpose of preprocessing by various scientists [9,13,14]. Extensive preprocessing was performed on the collected data to enhance the feature extraction and classification capability of the models being used [15]. Multiple researchers have used a Convolutional Neural Network (CNN) to classify wheat diseases [9,11]. The performance of CNN can be examined based on various factors, including accuracy and loss. The loss of CNN can be calculated by the formula as follows:
In recent years, transfer learning techniques have shown significant contribution in disease detection for wheat crops [16,17]. Deep learning algorithms have also shown good results for disease detection using thermal images [18]. Combination of traditional machine learning methods and deep learning methods have shown significant improvement in detection results, whereas, ensemble learning have further improved the accuracy of multiclass classification [19,20]. Performance of such methods have been assessed for real time applications of disease detection [21]. These methods have been applied in both open fields as well as greenhouses to recognize the wheat crop disease to mitigate the effect of disease to get increased yield of the crop [22]. Semantic segmentation is also applied by researchers to estimate the area affected by the disease on the leaf so that yield estimation can be performed and mitigate wheat stripe rust [23]. Vision Transformers (ViT) and deep learning methods have also been adopted by the researchers [16,24]. Multiple review studies have been published in recent years to highlight the potential challenges and way forward for disease detection in wheat crop [17,25–28]
To reduce wheat yield loss and close the supply-demand gap, it is necessary to diagnose the disease in the developmental phases as early as possible. The wheat disease may affect the yield of crops significantly, resulting in a shortage of supply of wheat [29], and its detection may face intense challenges. The wheat disease detection system involves the preprocessing of wheat plant images, features extraction and classification. This research makes several key contributions to the field of wheat disease detection. It addresses challenges of class imbalance in the dataset, simultaneous detection of multiple diseases and dataset images under a controlled environment. Transformer-based GANs were used to mitigate the problem of class imbalance, enhancing disease classification reliability. Feature extraction and fine-tuning employed cutting-edge techniques like ViTs and Graph Neural Networks (GNNs), enhancing the model’s capacity to capture disease-related features. Simultaneous classification of multiple disease classes was achieved through Model Agnostic Meta-Learning (MAML) with Long Short Term Memory (LSTM), Support Vector Machine (SVM), and Random Forest (RF) classifiers. Emphasizing real-world image analysis, this method is practical for application. These innovations result in significantly improved accuracy, offering a promising solution for agriculture.
The rest of the paper is organized as follows: Section 2 provides an analysis of existing research in the past few years, the proposed methodology is illustrated in Section 3, depicting the details regarding methodology and dataset, and results and observations are shown in Section 4. Section 5 includes an insight discussion regarding the existing and proposed methods, whereas the research is concluded in Section 6.
The primary step in disease detection is data acquisition, which involves gathering relevant image data. Subsequently, preprocessing is performed where the collected data is cleaned, normalized, and standardized to remove noise and inconsistencies. Feature extraction is then performed to identify relevant patterns and characteristics from the preprocessed data, which may involve techniques like dimensionality reduction or feature engineering. In the last phase, the features that are extracted are put to work for classification. This research uses ML and DL techniques to train a model adept at telling the difference between healthy samples and those with some sort of disease. Picking the right calculations and measurements for assessment is a significant piece of this cycle. Preprocessing is key, which implies cleaning up and separating the information to dispose of any commotion or superfluous subtleties. This step frequently includes techniques like normalizing the information, enlarging it, and adjusting it, as numerous scientists have performed [12,13]. When preprocessing is finished, the following stage in information examination includes taking out huge highlights from the accumulated information. This is where profound learning-based techniques become possibly the most important factor, using the strength of brain organization.
The last step includes utilizing the highlights that have been removed to sort the information into either infected or non-unhealthy classes. At this stage, an AI model was trained on a dataset that is, as of now, marked. This model is then used to foresee whether new, inconspicuous information falls into the classification of disease or not. Conventional AI procedures for this assignment incorporate calculated relapse and choice trees. This proposed methodology additionally has further developed profound learning calculations like repetitive brain organizations and convolutional brain organizations. The task for the classification model is to accurately identify the correct category for each image in the dataset, which can be either a simple yes-or-no (binary) or more complex (multi-class) challenge. In past years, much research work has been carried out by various researchers in order to develop such models which can detect and classify each diseased image of wheat crop into their respective classes with significant accuracy and efficient computational complexity.
Size normalization has been used in many studies for scaling the images into uniform sizes to perform better classification. Researchers have applied Image Filtration to preprocess the wheat images in order to filter out ineffectual dataset images [13]. Histogram Equalization has been used to preprocess the gathered dataset [14]. The technique of Data Enhancement to perform preprocessing on the used dataset has also been employed in previous work [15]. Multiple researchers have used Image Augmentation to elevate the dataset size to enhance the accuracy of classification models [12].
Feature extraction proves to be a significant step in ML or DL. Many studies have done automated feature extraction using CNN [12]. Automated feature extraction included various techniques, including Spectral Features, ResNet-50 [10], WaterShed, GrabCut, U2-Net [14], PANet, VGG, and DenseNet [24]. Classification is a very important step in DL, where the model predicts the class of the input data provided to the model. Researchers have used both machine and DL classifiers for the classification of diseased wheat crops in wheat disease detection and classification. PSO-SVM, BP and RF were employed to classify the diseased wheat images into respective diseased classes [10]. Ensemble learning, in which VGG, ResNet-101, ResNet-152, DenseNet-169, and DenseNet-210 have been used to improve the accuracy than the accuracy of all the individual classifiers [15].
Classifiers employed in the studies include PSO-SVM, BP, RF, Mask-RCNN, CNNs (such as ResNet-50, VGG-16, and EfficientNet_B0), Inception-v3, and ensemble learning [10,11,13,15]. These classifiers leverage ML and DL algorithms to classify wheat disease patterns accurately.
Furthermore, the essential focal point of the examination was on the exactness, dismissing other critical assessment boundaries like accuracy and mean normal accuracy. Additionally, the examinations were restricted to just hardly any wheat-diseased classes. Thus, the location of wheat diseases is of most extreme significance in spurring ranchers and partners to take on better yields the board rehearses. By giving opportune and precise data about the situation with their harvests, ranchers can go to suitable lengths to control the spread of infections, lessening the financial misfortunes related to these diseases.
This study proposes a four-step approach for the exact location of disease microbes for wheat crops, including information get-together and assortment, preprocessing strategies, highlight extraction, and characterization. The data is collected from various publicly available sources and combined into a single dataset of 6 classes. The information of each class displayed in Fig. 2 is then preprocessed to make it appropriate for the end goal of component extraction. The proposed approach utilized hardware that includes an Intel Core i7 with a processing speed of 2.90 GHz as the central processor unit, NVIDIA GeForce GTX 1650 for powerful graphics processing, and 16 GB of RAM to guarantee enough memory for effective data handling.
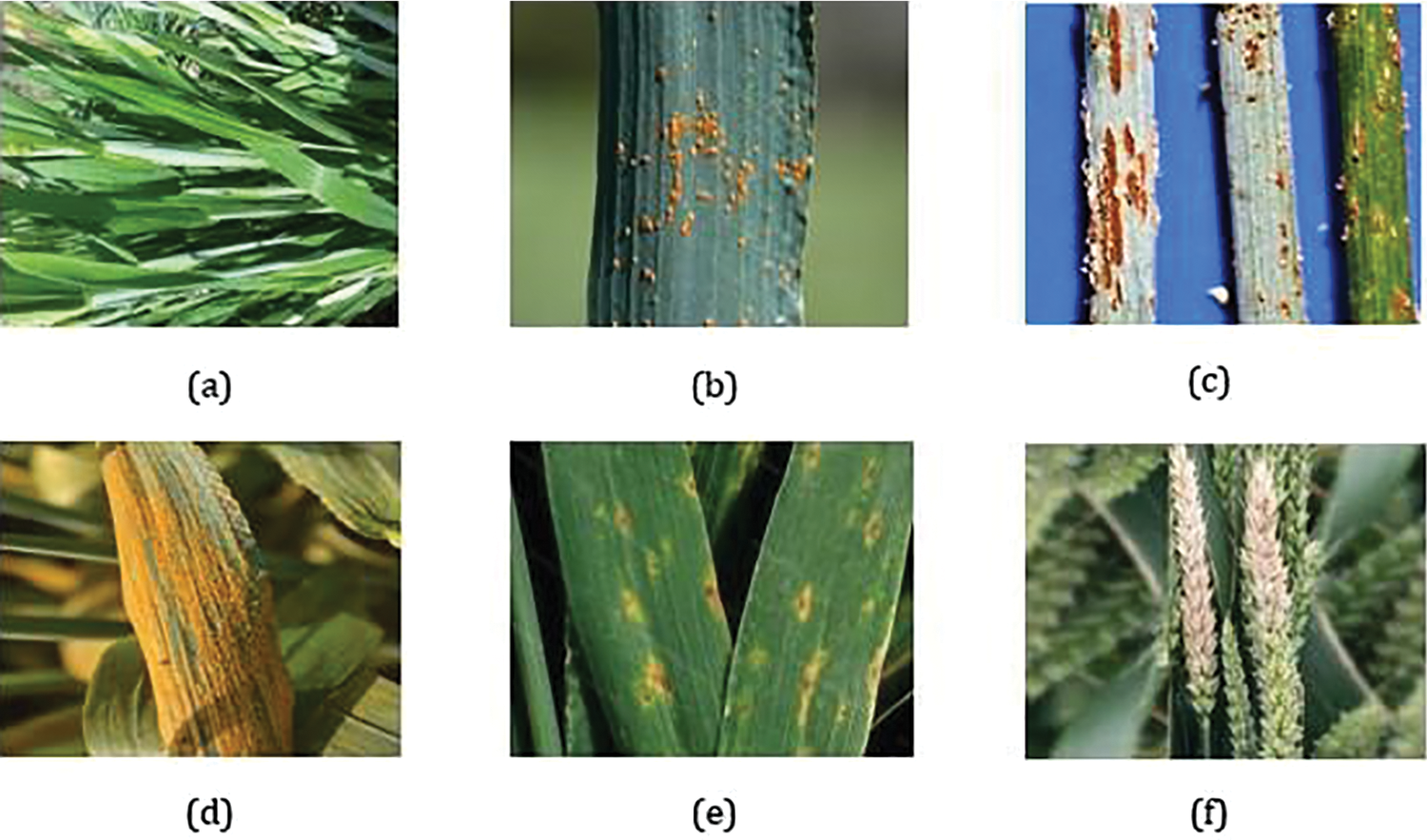
Figure 2: Wheat disease dataset samples: (a) healthy, (b) leaf rust, (c) stem rust, (d) yellow rust, (e) tan spot, (f) wheat blast
The steps and structure of proposed methodology is illustrated in Fig. 3. The features are extracted using DL model and these features are given as an input to another model for classification.
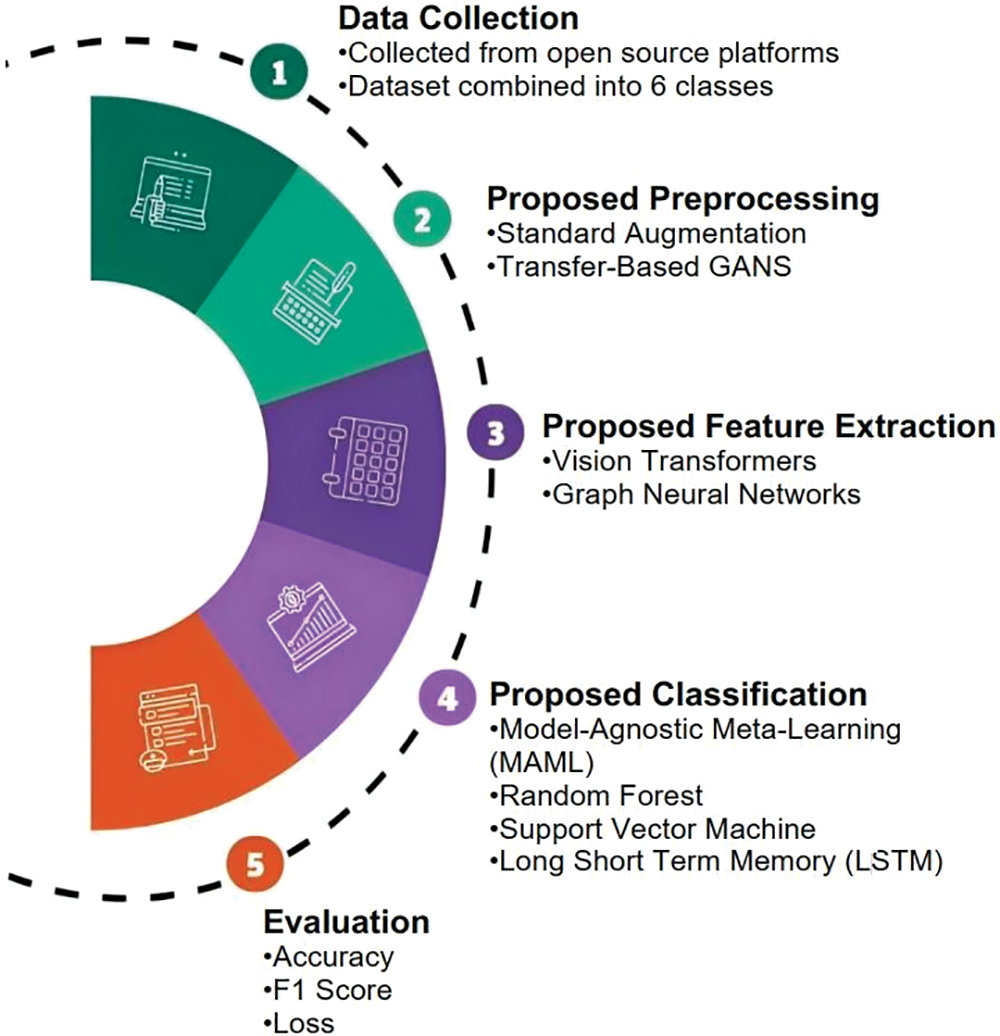
Figure 3: Proposed methodology for wheat disease detection
Picture preprocessing is fundamental in improving the quality and consistency of information for profound learning models which might incorporate picture normalization and information Expansion. Picture normalization is a critical calculate visual information examination and PC vision. Everything revolves around ensuring picture information is steady, exact, and equivalent, regardless of where it comes from, its configuration, or the circumstances under which it was obtained. Through a scope of strategies and techniques, normalizing pictures makes it more straightforward to examine information, perceive examples, and complete AI undertakings really.
In the computer vision tasks, it's not unexpected that images gathered may vary in quality, lighting, differentiation, size, and variety conveyance [30]. It can lead towards misclassification of the images at the classification phase and reduce the test accuracies. To address this, all the RGB images were resized to a uniform size of 500 pixels in width and 500 pixels. In this way, this proposed approach eliminates any size distinctions that could create problems while applying different models for classification and also make it device independent.
Image zooming, scaling and normalization also help in reducing the processing time and to increase the validation accuracies of disease detection. Preprocessing has two main targets: to avoid × CMC, 202X overfitting and to increase the validation accuracies. The basic exploratory techniques include various augmentation methods to increase and diversify the number of images in the dataset. For instance, random rotations, zooms, and contrast adjustment for each image may serve as the most frequent methods. As a result of preprocessing and data augmentation, software libraries, such as PyTorch in Python, provide a more robust dataset which is ready for application in deep learning.
Transfer Generative Adversarial Networks can be applied as a robust strategy to generate synthetic data that would resolve the dilemma of class disproportionality and data inadequacy. In one particular manner, Transfer GAN differs from previous methods by the ability to transfer features, style, or content from one image to another, which could be utilized to make ordinary images more creative. In more practical terms, there seem to be numerous opportunities due to which implementing these data augmentation techniques can noticeably increase the train dataset, ensuring a much more different and substantial amount for the deep-learning-based model to explore. Therefore, this augmentation can both diversify the data and help the classifier against overfitting to provide higher accuracies.
This examination digs into the utilization of ViT and GNN as cutting-edge techniques for highlight extraction in picture examination. The explanation that are being zeroed in on these advancements is their noteworthy abilities: They are perfect at recognizing complex examples in information, can use previous prepared models, perform outstandingly well, and upgrade the utilization of registering power. Together, these characteristics assume a significant part in supporting the exactness of identifying diseases in wheat. The objective of this proposed methodology is to take advantage of the qualities of profound advancing by utilizing ViT and GNN for taking out special and significant elements from pictures. This method is pointed toward making picture examination more exact and viable. The engineering of both ViT and GNN, with their numerous layers, is intended to get a handle on an assortment of picture portrayals. They catch everything from straightforward, low-level subtleties to complicated, significant level highlights, making it simpler to recognize complex visual examples in the pictures.
The discoveries show the viability of the models in drawing out complicated, undeniable level highlights, a capacity that essentially supports the accuracy and usefulness of picture examination frameworks. For example, the ViT’s engineering, portrayed in Fig. 4, is explicitly intended to recognize novel and significant highlights in pictures [31]. The depiction of the transformer encoder was motivated by [32]. This capacity is essential, as it permits ViT and GNN to handle difficulties like evaporating angles, which thusly empowers more compelling preparation of these models.
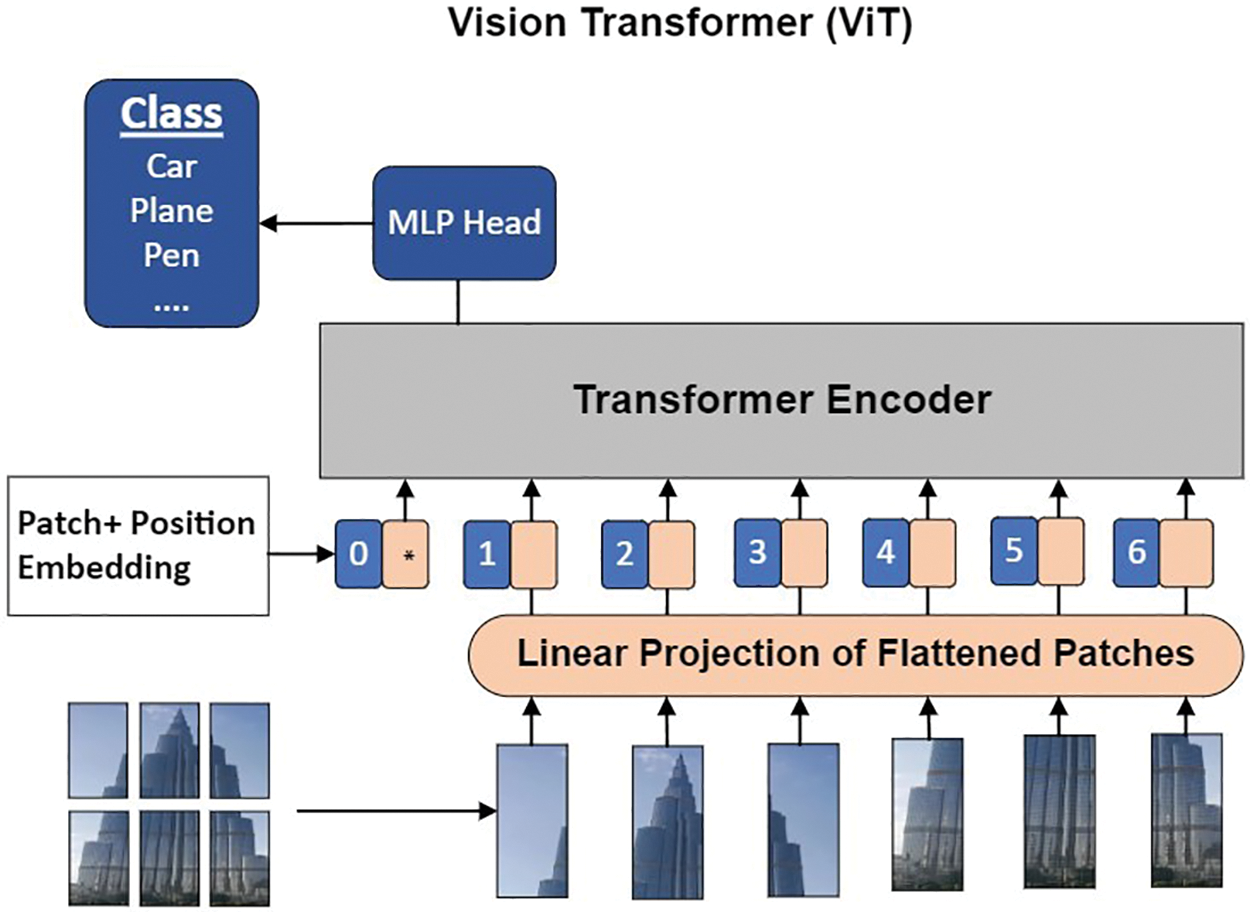
Figure 4: Architecture of vision transformer (ViT) [31]
Choosing a fitting machine or profound learning model is basic for the exact and compelling location of diseases in wheat. In this review, selection of MAML was carried out as a go-to to demonstrate for characterization. MAML hangs out in the AI people group, particularly among those zeroing in on hardly any shot and meta-learning, because of its noteworthy capacities. It succeeds in circumstances where there is a requirement for models to advance rapidly and really from limited quantities of information, for example, adjusting to new errands or acclimating to new spaces with negligible models. The flexibility and fast learning skill of MAML make it a fundamental instrument for specialists and experts who mean to foster models that can sum up well across assorted pain points. This approach is urgent in propelling the outskirts of few-shot learning and meta-learning. The design and functions of MAML are definite in Fig. 5.

Figure 5: The architecture of model agnostic meta-learning (MAML)
This research adopted a refined process specifically designed for tasks involving the classification of multiple classes. Initially, the dataset was preprocessed using Transfer-Based GANs, which significantly improved the dataset’s diversity and quality, enriching the features extracted from it. Following this, ViT and GNN are used for the intricate task of extracting features, effectively capturing detailed patterns from the images. Finally, this study utilized MAML in conjunction with three different classifiers–RF, SVM, and LSTM–to complete the classification strategy.
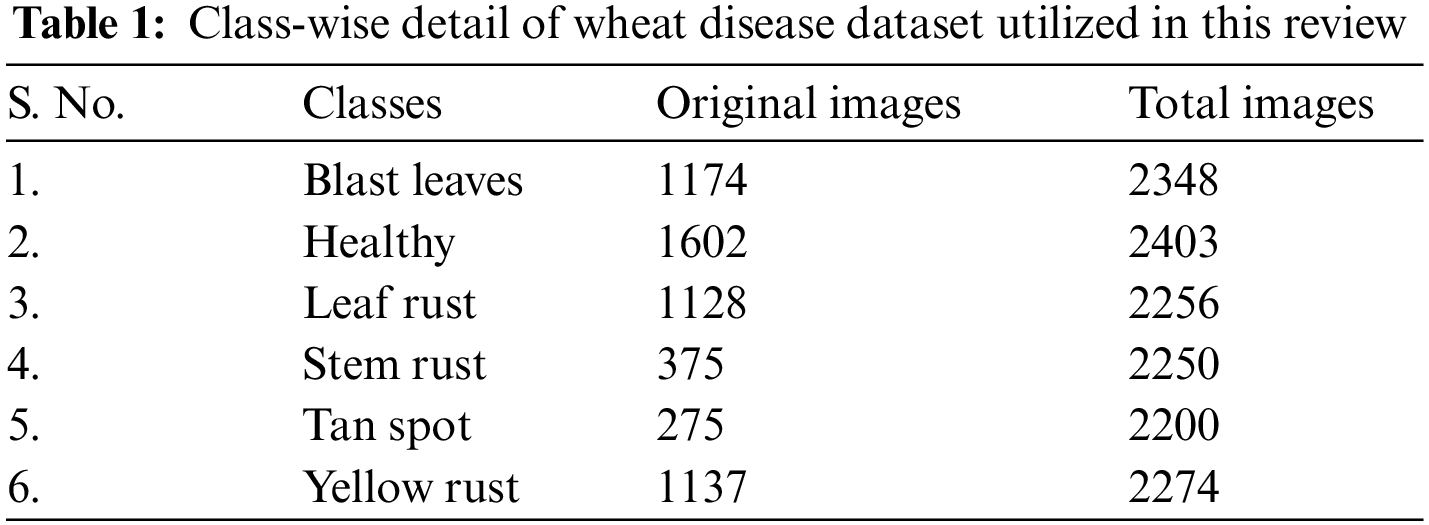
AI methods, particularly profound learning calculations, have exhibited extraordinary likely in distinguishing and sorting wheat diseases from pictures. For these techniques to be compelling, they depend on broad and shifted datasets for exact preparation and approval of the models. The quality and completeness of these datasets are critical to creating fruitful AI models for grouping plant disease. This examination ordered a nitty gritty dataset that incorporates pictures addressing six unique kinds of plant diseases: ‘sound,’ ‘leaf rust,’ ‘tan spot,’ ‘impact leaves,’ ‘stem rust,’ and ‘yellow rust,’ accumulated from various sources. The dataset at first had 5,691 pictures addressing the plant infections that is referred before: ‘sound,’ ‘leaf rust,’ ‘tan spot,’ ‘impact leaves,’ ‘stem rust,’ and ‘yellow rust.’ Then extended this assortment to 13,731 pictures. This study gives a definite outline of every diseased class, zeroing in on their particular elements, the effect they have, and the difficulties in recognizing and ordering them. Table 1 shows how these pictures are conveyed across the different disease classes. To handle the issue of class irregularity and to limit contrasts between information from different classes, this research utilized move-based GANs to expand the information.
In this research, rigorous testing was conducted in this study using a meticulously assembled image database representing a variety of wheat diseases and a training, validation, and test framework. Performance metrics evaluated the procedures applied in this study, including accuracy, precision, recall, and F1-score. The outcomes obtained demonstrated that the combination of preprocessing recommend in this study- transfer-based GAN and feature extraction through Vision Transformer and Graph Neural Network during tran- after transfer -, classification with a MAML ensured accurate and reliable wheat disease detection. This research emphasizes the need of utilizing preprocessing to improve DL model performance and crop disease detection.
The quality improvement of input data and enlarging the training dataset due to image filtration, standardization and transfer-based GANs have led to better generalization of the model and thus more accurate predictions. Additionally, augmentation was crucial for the dramatic rise in the dataset size. Vision Transformer together with Graph Neural Networks and MAML are an effective combination in what concerns the accurate identification and types classification of wheat diseases. Table 2 illustrates various possible solutions tested in order to achieve the higher and better performance of the classification model. The table depicts that the best overall configuration to achieve significant accuracy was with number of epochs being 50, with 0.2 dropout rate and a learning rate of 0.01 and in combination of batch normalization.

This study utilized numerous profound learning calculations to arrange pictures of wheat diseases and acquired critical discoveries. In the forthcoming area, the paper will introduce the outcomes from these various models. This part of the research is dedicated to explaining how each DL model contributed to the detection of wheat diseases. This research work provides an analysis of the training and testing accuracy graphs, a model’s configuration table and the evaluation metrics obtained from different models. The well-curated dataset was given as an input to ViT and GNN for the purpose of feature extraction. Subsequently various classification models were applied to attain substantial accuracy and other vital performance evaluation parameters with certain configurations given in Table 3. The softmax activation function was used which is employed as:

Results obtained using different experimental setting and confusion matrix are presented in Figs. 6 and 7, respectively.

Figure 6: Results achieved using different experimental setups: (a) VGG19 (b) Xception (c) Inception v3 (d) DenseNet121 (e) Resnet 152v2 (f) MAML ensemble

Figure 7: Confusion matrix of different experimental setups: (a) VGG19 (b) Xception (c) Inception v3 (d) DenseNet121 (e) ResNet152v2 (f) MAML ensemble
Performance metrics for several DL models employed in a classification job are shown in Table 4 below. The models are judged according to their accuracy, precision, recall, and F1-score. The MAML achieves an F1-score of 0.97 and an amazing accuracy of 99.20%, displaying exceptional precision and recall. With a respectable F1-score of 0.88 and an accuracy of 96.5%, ResNet152v2 is next, although its precision and recall are slightly lower. Inception v3, DenseNet121, and Xception exhibit competitive results across all metrics, with F1-scores ranging from 0.83 to 0.87 and accuracies between 93.96% and 94.79%. On the other hand, VGG19, while maintaining reasonable precision and recall, shows a comparatively lower F1-score of 0.81 and an accuracy of 82%.

Table 5 below presents a comprehensive summary of wheat disease detection methods and techniques, feature extraction methods, classifiers, and percentages of accuracy that each study obtained using the mentioned techniques. This critical analysis aims to weigh the merits and demerits of the reviewed techniques to underscore important findings and discoveries’ significance for wheat disease detection. The table features several feature extraction techniques, such as wavelet analysis, spectral features, image annotation, and such pre-trained models as ResNet, VGG, and AlexNet. These methods are important for extracting critical information from input data, which is critical for precise disease classification.
PSO-SVM, BP, RF, Mask-RCNN, CNNs , which include the ResNet-50, VGG-16, and EfficientNet_B0 , Inception-v3, and ensemble learning methods are some of the classifiers used in these studies. Employing machine learning and deep learning algorithms, these classifiers can detect the patterns of wheat disease precisely. There is a considerable variability in the datasets used, including data that were manually collected, publicly accessible datasets, and specialized datasets originating from specific cameras and unmanned aerial vehicles . A critical review of wheat disease detection methods uncovers significant challenges and lessons learned. A major issue is the availability of the datasets, which can impact model generalization and model robustness. Future work should focus on collecting more diverse and extensive data for improving model performance. In various other cases, images come from controlled conditions, such as greenhouse-grown plants. These data are insufficient for learning models that can work under real-world conditions. Data must capture all aspects of natural environmental variation as much as possible. The other notable issue is that most plant disease studies suffer from class imbalance. Many studies focus on one class of the disease. This negatively impacts the performance and accuracy of detecting diseases. In many other studies, only one or a few similar diseases are detected. Though high accuracy can be obtained when attempting to detect the condition of interest, the applicability of the models will be very limited.
This study denotes a huge headway in identifying wheat disease, successfully spanning basic exploration holes noted in past works. In this proposed study, various shortcomings of previous research studies have been resolved. The requirement for diverse and large-scale data is highlighted by the limited availability of datasets, which hinders generalizability and robustness which is resolved by collecting data from various sources and assembled into a single dataset of 6 classes. The images utilized in the previous research were under a controlled environment, which was in contrast to the real scenario, and this issue has been resolved as this study makes use of real-world images in uncontrolled and natural environments. Furthermore, the issue of class imbalance was faced in previous studies, disrupting the accuracy of classification models, and hence, different techniques, including GANs, have been used to resolve the issue of class imbalance. This study also helps to classify multiple diseases simultaneously, not only focusing on specific diseases as was the case in previous works.
This study tends to the basic need to recognize different diseases influencing wheat crops an element frequently neglected in the past examinations, which normally centered around individual diseases. This proposed cross-breed technique empowers the exact conclusion of five distinct wheat diseases on the double. This lines up with the mind-boggling difficulties ranchers face in the field, where harvests can be at the same time impacted by numerous diseases, affecting yield and prompting monetary misfortunes. To beat the constraints of past investigations that utilized dataset pictures from controlled conditions, this research’s exploration presents a creative picture readiness strategy. This procedure joins transfer GANs with conventional increase techniques, changing it up and adding a dash of authenticity to the dataset.
The augmentation process does not only augment the limited number of original images but also highlights the efficiency and dependability of the proposed method. The model attains an accuracy of 99.20%, which means that the proposed approach is efficient when used in a real application. The primary strength of this approach is the use of transfer GANs ensures that the dataset is expanded, thereby dealing with the deficiencies in the original images for some diseases and striving for equal representation of the diseases. The expanded dataset is critical to the improved ability of the model to differentiate the different diseases that exist in the datasets, thereby contributing to a high level of accuracy. During the classification process, the Model-Agnostic meta-Learning is used which synthesizes the classifiers outcomes, such as RF, SVM, and LSTM. The proposed method enhances generalization in detecting wheat diseases, addressing challenges such as class imbalance and data scarcity, and enabling the detection of multiple diseases concurrently.
This research examined the capacity of deep learning models in the division of wheat diseases. Model Agnostic Meta-Learning was the most viable ensemble learner in precision. This discovery emphasizes the importance of selecting the best deep learning design for certain classification tasks since the choice of model has a significant impact on results. The various and preprocessing strategies such as rescaling, zooming, shearing, flipping, and the use of Transfer GAN s improved the models’ performance by mixing training data and decreased the overfitting. The primary application of the features from powerful convolutional layers enabled the models to efficiently identify the most critical elements needed for accurate classification of various wheat diseases. This research, however, is based mainly on a data set that contains six classes of wheat diseases.
In the future, more datasets can be used in the analysis, and the number of disease types and pests covering shall be increased. This step will help understand how well these models trivialize and how strong they are. Expanding the dataset to use more wheat diseases, a wide variety of environmental conditions, and multiple stages of individual disease development will lead to full-scale assessment and the antifragility of proposed models. Moreover, it might help to use various transfer learning techniques from other crops in the grass family which are similar to wheat, such as barley or corn. This step can help enrich shareable features and disease regularities between wheat diseases detection and other crops. Lastly, researchers, various agricultural companies, and farmers should collaborate and share data. Developing extensive datasets and establishing open-access repositories through such partnerships will play a crucial role in advancing wheat disease detection efforts.
Acknowledgement: Not applicable.
Funding Statement: This Research is funded by Researchers Supporting Project Number (RSPD2024R 553), King Saud University, Riyadh, Saudi Arabia.
Author Contributions: The authors confirm contribution to the paper as follows: Study conception and design: Syed Muhammad Usman, Muhammad Zubair, Shehzad Khalid; data collection: Yasir Maqsood, Syed Muhammad Usman, Shehzad Khalid; analysis and interpretation of results: Yasir Maqsood, Syed Muhammad Usman, Musaed Alhussein, Khursheed Aurangzeb, Shehzad Khalid, Muhammad Zubair; draft manuscript preparation: Yasir Maqsood, Syed Muhammad Usman, Shehzad Khalid, Musaed Alhussein, Khursheed Aurangzeb, Muhammad Zubair. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data that support the findings of this study are available from the corresponding author, S.M.U, upon reasonable request.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. N. Chaudhary, P. Dangi, M. L. Mishra, and V. Kumar, “Wheat: Contribution to healthy diet and health,” in Handbook of Cereals, Pulses, Roots, and tubers. London, UK: CRC Press, 2021, pp. 3–34. [Google Scholar]
2. G. Veerendra, R. Swaroop, D. Dattu, C. A. Jyothi, and M. K. Singh, “Detecting plant diseases, quantifying and classifying digital image processing techniques,” Mater. Today: Proc., vol. 51, pp. 837–841, 2022. [Google Scholar]
3. Z. Iqbal, M. A. Khan, M. Sharif, J. H. Shah, M. H. ur Rehman and K. Javed, “An automated detection and classification of citrus plant diseases using image processing techniques: A review,” Comp. Electro. Agric., vol. 153, no. 2, pp. 12–32, 2018. doi: 10.1016/j.compag.2018.07.032. [Google Scholar] [CrossRef]
4. N. Ahmad, H. M. S. Asif, G. Saleem, M. U. Younus, S. Anwar, and M. R. Anjum, “Leaf image-based plant disease identification using color and texture features,” Wireless Pers. Commun., vol. 121, no. 2, pp. 1139–1168, 2021. doi: 10.1007/s11277-021-09054-2. [Google Scholar] [CrossRef]
5. H. Khan and I. U. Haq, M. Munsif, Mustaqeem, S. U. Khan and M. Y. Lee, “Automated wheat diseases classification framework using advanced machine learning technique,” Agric., vol. 12, no. 8, pp. 1226, 2022. doi: 10.3390/agriculture12081226. [Google Scholar] [CrossRef]
6. P. Fang et al., “The classification performance and mechanism of machine learning algorithms in winter wheat mapping using Sentinel-2 10 m resolution imagery,” Appl. Sci., vol. 10, no. 15, pp. 5075, 2020. doi: 10.3390/app10155075. [Google Scholar] [CrossRef]
7. K. Mochida et al., “Computer vision-based phenotyping for improvement of plant productivity: A machine learning perspective,” GigaScience, vol. 8, no. 1, pp. giy153, 2019. doi: 10.1093/gigascience/giy153. [Google Scholar] [PubMed] [CrossRef]
8. S. H. Shah, Y. Angel, R. Houborg, S. Ali, and M. F. McCabe, “A random forest machine learning approach for the retrieval of leaf chlorophyll content in wheat,” Remote Sens., vol. 11, no. 8, pp. 920, 2019. doi: 10.3390/rs11080920. [Google Scholar] [CrossRef]
9. M. A. Genaev, E. S. Skolotneva, E. I. Gultyaeva, E. A. Orlova, N. P. Bechtold and D. A. Afonnikov, “Image-based wheat fungi diseases identification by deep learning,” Plants, vol. 10, no. 8, pp. 1500, 2021. doi: 10.3390/plants10081500. [Google Scholar] [PubMed] [CrossRef]
10. L. Huang et al., “Detection of fusarium head blight in wheat ears using continuous wavelet analysis and PSO-SVM,” Agriculture, vol. 11, no. 10, pp. 998, 2021. doi: 10.3390/agriculture11100998. [Google Scholar] [CrossRef]
11. D. Kumar and V. Kukreja, “Image-based wheat mosaic virus detection with Mask-RCNN model,” in Proc. IEEE Int. Conf. Decis. Aid Sci. Appl. (DASA), Chiangrai, Thailand, 2022, pp. 178–182. [Google Scholar]
12. S. Sood and H. Singh, “An implementation and analysis of deep learning models for the detection of wheat rust disease,” in Proc. IEEE 3rd Int. Conf. Intell. Sustain. Syst. (ICISS), Thoothukudi, India, 2020, pp. 341–347. [Google Scholar]
13. T. Aboneh, A. Rorissa, R. Srinivasagan, and A. Gemechu, “Computer vision framework for wheat disease identification and classification using Jetson GPU infrastructure,” Technologies, vol. 9, no. 3, pp. 47, 2021. doi: 10.3390/technologies9030047. [Google Scholar] [CrossRef]
14. H. R. Bukhari et al., “Assessing the impact of segmentation on wheat stripe rust disease classification using computer vision and deep learning,” IEEE Access, vol. 9, pp. 164986–165004, 2021. doi: 10.1109/ACCESS.2021.3134196. [Google Scholar] [CrossRef]
15. Q. Pan, M. Gao, P. Wu, J. Yan, and M. A. AbdelRahman, “Image classification of wheat rust based on ensemble learning,” Sensors, vol. 22, no. 16, pp. 6047, 2022. doi: 10.3390/s22166047. [Google Scholar] [PubMed] [CrossRef]
16. Y. Borhani, J. Khoramdel, and E. Najafi, “A deep learning based approach for automated plant disease classification using vision transformer,” Sci. Rep., vol. 12, no. 1, pp. 11554, 2022. doi: 10.1038/s41598-022-15163-0. [Google Scholar] [PubMed] [CrossRef]
17. B. Chang, Y. Wang, X. Zhao, G. Li, and P. Yuan, “A general-purpose edge-feature guidance module to enhance vision transformers for plant disease identification,” Expert. Syst. Appl., vol. 237, no. 1, pp. 121638, 2024. doi: 10.1016/j.eswa.2023.121638. [Google Scholar] [CrossRef]
18. Ü. Atila, M. Uçar, K. Akyol, and E. Uçar, “Plant leaf disease classification using efficientNet deep learning model,” Ecol. Inform., vol. 61, pp. 101182, 2021. doi: 10.1016/j.ecoinf.2020.101182. [Google Scholar] [CrossRef]
19. G. Batchuluun, S. H. Nam, and K. R. Park, “Deep learning-based plant classification and crop disease classification by thermal camera,” J. King Saud Univ.-Comput. Inf. Sci., vol. 34, no. 10, pp. 10474–10486, 2022. doi: 10.1016/j.jksuci.2022.11.003. [Google Scholar] [CrossRef]
20. J. Chen, A. Zeb, Y. Nanehkaran, and D. Zhang, “Stacking ensemble model of deep learning for plant disease recognition,” J. Ambient Intell. Humaniz. Comput., vol. 14, no. 9, pp. 12359–12372, 2023. doi: 10.1007/s12652-022-04334-6. [Google Scholar] [CrossRef]
21. J. Eunice, D. E. Popescu, M. K. Chowdary, and J. Hemanth, “Deep learning-based leaf disease detection in crops using images for agricultural applications,” Agronomy, vol. 12, no. 10, pp. 2395, 2022. doi: 10.3390/agronomy12102395. [Google Scholar] [CrossRef]
22. J. A. Wani, S. Sharma, M. Muzamil, S. Ahmed, S. Sharma and S. Singh, “Machine learning and deep learning based computational techniques in automatic agricultural diseases detection: Methodologies, applications, and challenges,” Arch. Comput. Methods Eng., vol. 29, no. 1, pp. 641–677, 2022. doi: 10.1007/s11831-021-09588-5. [Google Scholar] [CrossRef]
23. M. Long, M. Hartley, R. J. Morris, and J. K. Brown, “Classification of wheat diseases using deep learning networks with field and glasshouse images,” Plant Pathol., vol. 72, no. 3, pp. 536–547, 2023. doi: 10.1111/ppa.13684. [Google Scholar] [PubMed] [CrossRef]
24. Y. Li et al., “Semantic segmentation of wheat stripe rust images using deep learning,” Agronomy, vol. 12, no. 12, pp. 2933, 2022. doi: 10.3390/agronomy12122933. [Google Scholar] [CrossRef]
25. D. Kumar and V. Kukreja, “Deep learning in wheat diseases classification: A systematic review,” Multimed. Tools Appl., vol. 81, no. 7, pp. 10143–10187, 2022. doi: 10.1007/s11042-022-12160-3. [Google Scholar] [CrossRef]
26. T. Sharma and G. Sethi, “Classification of image-based wheat leaf diseases using deep learning approach: A survey,” J. Sci. Res., vol. 15, no. 2, pp. 421–443, 2023. doi: 10.3329/jsr.v15i2.61680. [Google Scholar] [CrossRef]
27. R. I. Hasan, S. M. Yusuf, and L. Alzubaidi, “Review of the state of the art of deep learning for plant diseases: A broad analysis and discussion,” Plants, vol. 9, no. 10, pp. 1302, 2020. doi: 10.3390/plants9101302. [Google Scholar] [PubMed] [CrossRef]
28. M. Nagaraju and P. Chawla, “Systematic review of deep learning techniques in plant disease detection,” Int. J. Syst. Assur. Eng. Manag., vol. 11, no. 3, pp. 547–560, 2020. doi: 10.1007/s13198-020-00972-1. [Google Scholar] [CrossRef]
29. A. Ahmad, D. Saraswat, and A. El Gamal, “A survey on using deep learning techniques for plant disease diagnosis and recommendations for development of appropriate tools,” Smart Agric. Technol., vol. 3, pp. 100083, 2023. doi: 10.1016/j.atech.2022.100083. [Google Scholar] [CrossRef]
30. H. Talebi and P. Milanfar, “Learning to resize images for computer vision tasks,” in Proc. IEEE/CVF Int. Conf. Comput. Vis., 2021, pp. 497–506. [Google Scholar]
31. S. Savary, A. Ficke, J. N. Aubertot, and C. Hollier, “Crop losses due to diseases and their implications for global food production losses and food security,” Food Secur., vol. 4, no. 4, pp. 519–537, 2012. doi: 10.1007/s12571-012-0200-5. [Google Scholar] [CrossRef]
32. A. Dosovitskiy et al., “An image is worth 16x16 words: Transformers for image recognition at scale,” arXiv preprint arXiv:2010.11929, 2020. [Google Scholar]
33. Q. Hong et al., “A lightweight model for wheat ear fusarium head blight detection based on RGB images,” Remote Sens., vol. 14, no. 14, pp. 3481, 2022. doi: 10.3390/rs14143481. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF
 Downloads
Downloads
 Citation Tools
Citation Tools