 Open Access
Open Access
ARTICLE
RepBoTNet-CESA: An Alzheimer’s Disease Computer Aided Diagnosis Method Using Structural Reparameterization BoTNet and Cubic Embedding Self Attention
1 College of Computer Science and Artificial Intelligence, Wenzhou University, Wenzhou, 325035, China
2 Key Laboratory of Intelligence Image Processing and Analysis, Wenzhou University, Wenzhou, 325035, China
* Corresponding Author: Zhongyi Hu. Email:
(This article belongs to the Special Issue: Deep Learning in Computer-Aided Diagnosis Based on Medical Image)
Computers, Materials & Continua 2024, 79(2), 2879-2905. https://doi.org/10.32604/cmc.2024.048725
Received 16 December 2023; Accepted 07 April 2024; Issue published 15 May 2024
Abstract
Various deep learning models have been proposed for the accurate assisted diagnosis of early-stage Alzheimer’s disease (AD). Most studies predominantly employ Convolutional Neural Networks (CNNs), which focus solely on local features, thus encountering difficulties in handling global features. In contrast to natural images, Structural Magnetic Resonance Imaging (sMRI) images exhibit a higher number of channel dimensions. However, during the Position Embedding stage of Multi Head Self Attention (MHSA), the coded information related to the channel dimension is disregarded. To tackle these issues, we propose the RepBoTNet-CESA network, an advanced AD-aided diagnostic model that is capable of learning local and global features simultaneously. It combines the advantages of CNN networks in capturing local information and Transformer networks in integrating global information, reducing computational costs while achieving excellent classification performance. Moreover, it uses the Cubic Embedding Self Attention (CESA) proposed in this paper to incorporate the channel code information, enhancing the classification performance within the Transformer structure. Finally, the RepBoTNet-CESA performs well in various AD-aided diagnosis tasks, with an accuracy of 96.58%, precision of 97.26%, and recall of 96.23% in the AD/NC task; an accuracy of 92.75%, precision of 92.84%, and recall of 93.18% in the EMCI/NC task; and an accuracy of 80.97%, precision of 83.86%, and recall of 80.91% in the AD/EMCI/LMCI/NC task. This demonstrates that RepBoTNet-CESA delivers outstanding outcomes in various AD-aided diagnostic tasks. Furthermore, our study has shown that MHSA exhibits superior performance compared to conventional attention mechanisms in enhancing ResNet performance. Besides, the Deeper RepBoTNet-CESA network fails to make further progress in AD-aided diagnostic tasks.Keywords
Alzheimer’s Disease (AD) is a neurodegenerative brain disease and a common type of dementia among middle-aged and elderly individuals [1]. It destroys the cells responsible for language and memory in the brain [2], leading to symptoms such as a decline in daily activities and cognitive abilities [3]. The initial symptoms of AD, which include implicit memory impairments, often advance to symptoms such as amnesia, spatial disorientation, impaired judgment, and abnormal language abilities over several months or years. Patients with AD often become vegetative and bedridden in the last few months or years of their lives. This disease has already deprived them of their personality and dignity [4]. Furthermore, the global incidence of AD is increasing. It is projected that the number of AD patients worldwide will surpass 100 million by 2050 [5]. More importantly, there is currently no effective treatment for AD. However, studies have shown that interventions targeting underlying diseases, psychological factors, and lifestyles can delay or prevent the development of AD symptoms [6,7]. This situation has also prompted scientists to invest more effort in exploring the pathogenesis of brain diseases such as AD, developing more accurate and effective diagnosis and treatment systems, and promoting the further development of brain projects in various countries.
Mild Cognitive Impairment (MCI) is identified as an intermediate state between Normal Control (NC) and fully developed AD [8]. Although the cognitive decline in patients with MCI is more pronounced than what is typically expected for their age, the symptoms of MCI are often mild and do not significantly affect their daily activities [9]. In a three-year multicenter clinical trial of MCI, researchers noted that MCI patients have a 16% annual risk of progressing to AD [10]. Therefore, accurate diagnosis of MCI is crucial in preventing the further development of AD symptoms. As far as we know, there are two different classification methods for the developmental stages of MCI. On the one hand, MCI can be classified into Early Mild Cognitive Impairment (EMCI) and Late Mild Cognitive Impairment (LMCI) based on the severity of memory impairment and the stage of development [11]. On the other hand, MCI can be classified into Stable Mild Cognitive Impairment (sMCI) and Progressive Mild Cognitive Impairment (pMCI) based on whether it would develop into AD. In this paper, we selected EMCI and LMCI data for the MCI-related experiments.
The neuronal loss caused by AD can be detected through Structural Magnetic Resonance Imaging (sMRI). sMRI is characterized by its non-invasive and rich image information, offering opportunities to study the pathological mechanisms of AD [12]. Since the 1990s, there have been many studies utilizing sMRI for AD analysis tasks. As a result, sMRI has become a crucial imaging biomarker for the analysis of AD and its prodromal stages [13].
Convolutional Neural Networks (CNN) are the most popular method in Deep Learning (DL), and numerous studies have utilized CNN and sMRI images for AD diagnosis. As for the network model based on Self-Attention (SA), especially the Transformer based on Multi Head Self Attention (MHSA), has become one of the state-of-the-art (SOTA) methods in Natural Language Processing (NLP) tasks [14]. In 2021, the Google research team officially introduced the Transformer network into the field of Computer Vision (CV) and proposed the Visual Transformer (ViT) model. This model transformed the 2D image problem into a 1D sequence problem and achieved state-of-the-art (SOTA) results in the ImageNet classification task, demonstrating that the Transformer structure network is highly effective in the field of Computer Vision [15]. Compared to CNN, the use of Transformer structures for AD diagnosis tasks is relatively rare due to its advanced nature.
In terms of the CNN part, it is evident that the majority of network models utilized for AD-aided diagnosis are based on the CNN structure. Conversely, there is a scarcity of research studies that have employed the Transformer structure for this purpose. Due to the inherent structural characteristics of CNN, CNN-Based network models are adept at handling local features while encountering challenges in addressing global features. This network model typically requires the stacking of multiple layers of CNN to learn certain global features, thereby enhancing the model’s receptive field to improve classification performance. However, this enhancement comes at the expense of significantly escalating computational costs. In addition, there is no absolute conflict between the CNN structure and the Transformer structure. Previous research has explored the integration of CNN and Transformer structure for CV tasks, exemplified by the BoTNet model. To tackle these issues, we adopt the BoTNet-S1 network as a baseline and optimize its structure by using structural reparametrization techniques, resulting in the RepBoTNet network. This network model integrates the capabilities of CNN structure, adept at capturing local information, and Transformer structure, skilled at incorporating global information. By doing so, it reduces prediction time and improves the classification performance of the network within the CNN part while achieving satisfactory performance metrics.
In the Transformers part, our investigation revealed that the key Multi-Head Self-Attention (MHSA) mechanism in the Transformer architecture overlooks the positional information along the channel dimension during the Position Embedding phase. While the absence of position information on the channel dimension may have minimal impact on tasks involving natural images, it can significantly affect the final classification results in medical images that contain rich channel information, such as sMRI. To address this problem, we propose CESA as a replacement for MHSA in RepBoTNet networks. CESA integrates positional information along the sMRI channel dimension, allowing the model to learn the lesions from a stereoscopic perspective in the 3D orientation. We designate this experimental model as RepBoTNet-CESA. Ultimately, the comparative experiments conducted between RepBoTNet-CESA and RepBoTNet illustrate the effectiveness of MHSA.
Based on the baseline, modifications were made to the model structure by both the CNN and Transformer parts. By fusing CNN and Transformer structures, we have developed a novel DL network for the assisted diagnosis of AD, named RepBoTNet-CESA network. The proposed network is evaluated by the ADNI public database, and the results demonstrate its effectiveness for AD diagnosis tasks. The main contributions of this study are as follows:
(1) The performance improvement of MHSA and traditional attention mechanisms on ResNet were conducted on BoTNet to supplement the original research.
(2) The performance comparison between BoTNet and BoTNet-S1 was conducted to verify the original experiments of BoTNet.
(3) Based on BoTNet-S1 as the baseline model, we proposed RepBoTNet by utilizing structural reparameterization techniques to modify its CNN structure and verified the performance improvement of the network through experiments.
(4) We proposed Cubic Embedding Self Attention (CESA) and introduced the ReoBoTNet-CESA network. CESA was compared with the baseline model’s original 2D-Position Embedding Self Attention through experiments to demonstrate the performance improvement of CESA.
(5) Following the structure of ResNet50, ResNet101, and ResNet152 networks, the initial ReoBoTNet-CE network with 50 layers was extended to 101 and 152 layers, revealing the impact of overfitting on network performance.
The remainder of this article is organized as follows: In Section 2, we provide a summary of the existing work and illustrate the feasibility of AI techniques in aiding the diagnosis of AD. In Section 3, we outline the construction methods of RepBoTNet-CESA. In Section 4, we present the experimental results that were obtained. Lastly, Section 5 ends the paper with a conclusion.
The RepBoTNet-CESA proposed in this paper involves modifications to both the CNN and Transformer structures. In the CNN part, we adhere to the design principle of structural reparameterization, as demonstrated in RepVGG, by implementing a Rep3 × 3 convolution block to improve the performance within the CNN structure. Following the construction principles of BoTNet, we have incorporated the CNN and Transformer architectures. Besides, we have enhanced the Transformer part, i.e., the MHSA, by replacing MHSA with the CESA attention mechanism as introduced in this study. This modification aims to improve the model’s performance within the Transformer structure. Subsequently, the final RepBoTNet-CESA network is developed following the design idea of the BoTNet overall network architecture. In this section, we mainly illustrate the feasibility of AI techniques in assisting the diagnosis of AD, introduce the research background concerning the structural reparameterization technology of RepVGG, and discuss the research background of combining MHSA, which is a Transformer block, with CNN structures.
2.1 AD-Aided Diagnosis Using AI Technology Is Feasible
Various AI models have been proposed for the accurate assisted diagnosis of AD. The utilization of CNN for supporting the assisted diagnosis of AD has become increasingly popular in recent years. For example, Hu et al. proposed an AD diagnosis method based on a slice voting algorithm. They utilized various coronal slices and 3D sMRI images to train the model and achieved a classification accuracy of 93.10% [16]. Gao et al. proposed the Latent Space Representation Network (LSRNet) and trained the network in two steps. In the first step, an autoencoder was utilized to extract latent high-level features and internal connections from functional connectivities (FC). In the second step, two perspective feature parses were used to extract high-level features. Long Short-Term Memory (LSTM) networks extract spatiotemporal features from a local perspective, while CNNs extract high-level features from a global perspective. Finally, the spatiotemporal features and global high-level features are fused for AD diagnosis, achieving a classification accuracy of 95.1% in the AD/NC classification task [17]. Jie et al. proposed a novel convolutional kernel, the weighted correlation kernel (wc-kernel), to measure the correlation between brain regions. By learning the weighted factors in a data-driven way, the wc-kernel can better learn the interaction between different brain regions compared to the Pearson correlation coefficient (PCC) method. Their wck-CNN extracts local (specific brain regions), global (whole-brain networks), and temporal features sequentially from the constructed Functional Connectivity Networks (FCNs) for classification. It achieves an accuracy of 85.3% for AD/NC, 81.1% for EMCI/NC, and 57.6% for AD/EMCI/LMCI/NC [18]. Inspired by the Oxford Net, Mehmood et al. proposed a Siamese Convolutional Neural Network (SCNN), which incorporates two parallel learning layers to extract more important features. They also added an extra convolutional layer to the network model to effectively learn optimal features from small datasets. By employing the Monte Carlo method to assess the significance of the classification results under optimal parameters, the researchers confirmed the effectiveness of their approach [19].
In the ongoing exploration of this field, the Transformer architecture, such as the Vision Transformer, has already showcased the efficacy of Transformer networks in tasks related to image classification [15]. However, compared to CNN, the use of Transformer for AD diagnosis tasks is relatively rare due to its advanced nature. We found that Roshanzamir et al. utilized transfer learning (TL) to train a language model based on Transformer with 500 interview data from 269 subjects in the Pitt corpus. They achieved an accuracy of 88.08% in the AD/NC classification task [20]. Sarraf et al. proposed the Optimized Vision Transformer for AD (OViTAD) model, which uses the Re-Attention mechanism to regenerate attention maps during training. This approach aims to increase the diversity of feature extraction and avoid the attention collapse phenomenon, where attention maps tend to become too similar in deeper layers. Compared to the regular ViT, OViTAD reduces the number of trainable parameters and improves the accuracy of AD diagnosis [21].
In fact, there is no absolute conflict between the CNN structure and the Transformer structure. There are some researchers who have explored the integration of CNN and Transformer structure for CV tasks, exemplified by the BoTNet model. In this paper, we adopt the BoTNet-S1 network as a baseline, modifications were made to the model structure by both the CNN and Transformer parts. Finally, we have developed a novel DL network for the assisted diagnosis of AD, named the RepBoTNet-CESA network.
The aforementioned study demonstrated the effectiveness of AI techniques such as deep learning, in aiding diagnostic tasks related to AD. Furthermore, a diverse array of AI technologies is utilized in other healthcare domains, including the field of COVID-19 diagnosis. Salman et al. used machine learning and laboratory data to predict COVID-19 patients. They compared three different classification methodologies, namely Support Vector Machines (SVM), artificial neural networks (ANN), and k-Nearest Neighbors (k-NN). The results indicated that SVM achieved the most significant classification performance [22]. Arif et al. proposed a Lasso-logistic regression model. Based on the COVID-19 clinical dataset, which enlisted the help of 78 patients from the Azizia main hospital sector, the Wasit Health Directorate, and the Ministry of Health, their method is generally accurate to 85.9% [23]. These studies illustrate the feasibility of applying AI technology in other similar medical fields beyond AD-aided diagnosis. The model we propose in this paper has the potential application in the field of another disease classification prediction, provided that the corresponding sMRI image data is accessible.
2.2 Research Background of Structural Reparameterization
Substantial advancements have been achieved in the development of CNN structures in recent years. The first typical CNN network was LeNet5, which was introduced in 1998 [24]. The initial CNN network that achieved significant success was AlexNet, leading to a direct reduction of the final error rate by almost 10% in the 2012 ILSVRC (ImageNet Large Scale Visual Recognition Challenge) [25]. Since then, CNN networks have experienced rapid development. The inception of AlexNet can be regarded as a significant milestone in the field of deep learning. VGGNet achieved higher performance by employing a simpler, deeper, and less parameterized network that utilized smaller 3 × 3 kernels [26]. ResNet introduced the residual structures and batch normalization (BN) into CNN networks, enabling the creation of very deep CNN networks and multi-branch CNN networks [27]. In contrast to the idea of increasing the depth of CNN networks in models such as ResNet, the Inception networks focused on widening CNN networks. The multi-branch and convolutions of various sizes were utilized within a single layer. Subsequently, 1 × 1 convolutions were applied to reduce dimensionality, leading to a reduction in network computational complexity and achieving higher performance [28–30]. DenseNet utilized dense connections that linked all layers to each other, ensuring that the output of each layer would serve as an additional input to all the previous layers. This approach greatly increased the network’s computational complexity but also improved its performance [31].
In the 2021 CVPR, RepVGG proposed and adopted an innovative technique for constructing CNNs known as structural reparameterization. This method aims to enhance the performance of CNNs by optimizing the network architecture. Structural reparameterization can be defined as a network construction method that first constructs a series of network structures (usually the same as the training phase, such as the multi-branch structure), and then equivalently transforms their parameters into another set of parameters (usually used in the inference phase, like converting multi-branch into single-branch). This process results in the transformation of the original structure into a different set of network structures [32]. Following this design concept, it is possible to make an original single-branch 3 × 3 convolution into a multi-branch 3 × 3 convolution in the training phase, where the multi-branch configuration includes a 3 × 3 convolution, a 1 × 1 convolution, and an Identity branch. In the inference phase, structural reparameterization is employed to equivalently transform the multi-branch into a single-branch 3 × 3 convolution. The schematic diagram illustrating this process is shown in Fig. 1. In this manner, the network model can use more complex network structures and dedicate additional training resources in the training phase to achieve improved classification outcomes, such as higher accuracy and precision. Subsequently, during the inference phase, the network model undergoes an equivalent transformation into a smaller network structure while preserving the obtained classification results (the same accuracy and precision). This approach conserves computational resources and accelerates inference speed. This design idea also imparts specific multi-scale convolutional characteristics to the network model. We call this multi-branch 3 × 3 convolution as the Rep3 × 3 convolution. Subsequently, the principle of structural reparameterization technology will be introduced.

Figure 1: Multi-branch 3 × 3 convolution in the training phase is converted to single-branch 3 × 3 convolution in the inference phase
The training phase incorporates a multi-branch structure, exemplified by ResNet, a renowned convolutional architecture. ResNet conceptualizes the information flow as y = x + f(x) and employs residual blocks to capture the mapping f. This multi-branch design allows ResNet to function as an ensemble of multiple shallow models [33]. The aggregation of multiple shallow models offers performance advantages over a single model. When each residual block contains two branches, and there are n residual blocks, the ResNet model can be conceptualized as an ensemble of 2n models.
Following the principle of structural reparameterization, similar to the operation conducted in ResNet, we have substituted all the 3 × 3 convolutions in the baseline with three-branch 3 × 3 convolutions. The three-branch convolutions consist of parallel 3 × 3 convolution, 1 × 1 convolution, and Identity branch, as shown in Fig. 1a. The information flow can be mathematically represented as y = x + g(x) + f(x). If there are n convolution blocks present, the model’s structure can be perceived as an ensemble of 3n models. If the dimensions of x and f(x) do not align, the identity branch should be eliminated. This adjustment results in the information flow becoming y = g(x) + f(x), where g(x) is implemented through a 1 × 1 convolution. In the inference phase, this multi-branch 3 × 3 convolution will be equivalently transformed into a single-branch 3 × 3 convolution, as shown in Fig. 1b. In addition, ReLU is employed in each branch, as shown in Fig. 1.
The single-branch structure of the inference phase will be elucidated in this section. It will delve into the principle of structural reparameterization, which outlines the process of transforming a multi-branch structure into an equivalent single-branch structure, as shown in Fig. 2a. Consequently, the multi-branch convolutional structure in Fig. 1a is corresponding to the initial multi-branch convolutional structure in Fig. 2a. Furthermore, Fig. 2b illustrates the changes of the convolutional parameters during the structural reparameterization process. For improved visualization outcomes, we set C2 = C1 = 2, so the 3 × 3 convolution block has four 3 × 3 parameter matrices, and the 1 × 1 convolution block has four 1 × 1 parameter matrices. For visualization purposes, only the middle parameter layer with non-zero 1 × 1 and Identity parameters within the 3 × 3 parameter layer is shown in Fig. 2b.

Figure 2: Principle of structural reparameterization
We use W(3) ∈ RC2 × C1 × 3 × 3 to represent a 3 × 3 convolution block with C1 input channels and C2 output channels, and use W(1) ∈ RC2 × C1 × 1 × 1 to represent a 1 × 1 convolution block with C1 input channels and C2 output channels. We utilize μ(3), σ(3), γ(3), and β(3) to denote the mean, standard deviation, learnable scaling factor, and bias of the BN layer following the 3 × 3 convolution block, respectively. We utilize μ(1), σ(1), γ(1), and β(1) to denote the mean, standard deviation, learnable scaling factor, and bias of the BN layer following the 1 × 1 convolution block, respectively. The terms μ(0), σ(0), γ(0), and β(0) are employed to denote the mean, standard deviation, learnable scaling factor, and bias of the BN layer on the Identity branch. Meanwhile, let M(1) ∈ RN × C1 × H1 × W1 denote the input and M(2) ∈ RN × C2 × H2 × W2 denote the output, with * symbolizing the convolution operator. When C1 = C2, H1 = H2, W1 = W2, we have:
It is important to highlight that in cases where the input and output dimensions do not match (i.e., when using the down-sampling 3D-RepVGG block), the Identity branch should be eliminated, and only the first two terms of the aforementioned equations remain valid. During the inference stage of the BN block, for any ∀1 ≤ i ≤ C2, we have:
After that, each BN block is integrated with the preceding convolution block by transforming them into convolution blocks with bias vectors. Let
Combining Eqs. (2)–(4), we obtain ∀1 ≤ i ≤ C2:
Given that the Identity branch can be considered as a 1 × 1 convolution block with a weight matrix represented by a unit matrix, the aforementioned equations are also applicable to the Identity branch. By regarding the Identity branch as a 1 × 1 convolution block, a 3D-RepVGG block will consist of one 3 × 3 weight matrix, two 1 × 1 weight matrices, and three bias vectors. The ultimate bias can be acquired by aggregating the three bias vectors. To ensure consistency in the dimensions of input and output, zero-padding is implemented on the original 1 × 1 convolution block, creating a 3 × 3 convolution block. This procedure entails the incorporation of three 3 × 3 convolution blocks that can be subsequently merged. Finally, through the integration of Eq. (5), the three branches of the 3D-RepVGG block are “fused” into a single 3 × 3 convolution block utilizing the aforementioned operation, referred to as “structural reparameterization.” The particular procedure is illustrated in Fig. 2.
2.3 Research Background of Combining MHSA with CNN
Convolutional neural networks (CNNs) provide advantages through parameter sharing and effective incorporation of local information. However, various computer vision tasks, including object detection, instance segmentation, and the field of AD-aided diagnosis addressed in this study, necessitate the acquisition of Long-Range Dependencies (LRDs) within features [34]. Acquiring knowledge in LRDs proves advantageous for a wide range of computer vision applications. For example, SENet utilizes Squeeze-and-Excitation blocks to learn dependencies within the channel dimension of features. Experimental results on the ImageNet dataset have shown the effectiveness of LRDs in the channel dimension for enhancing network performance [35]. Subsequent studies, such as CBAM [36], GENet [37], and NLNet [38], provide additional support to this perspective by learning LRDs on the entire input dataset.
CNN-based models commonly employ the strategy of stacking multiple CNN layers or increasing the CNN size to learn global features like LRDs, which leads to a notable escalation in model complexity. To tackle this issue, a more effective strategy involves employing a mechanism dedicated to learning LRDs and constructing an efficient network model that learns both local and global features. The Self-Attention (SA) mechanism within the Transformer structure is an efficient learning mechanism for LRDs. In SA, the input Query, Key, and Value of Attention all originate from the same input, and Key and Value are paired together in a pairwise manner. The attention mechanism initially computes the compatibility between the Query and Key pair, utilizing this compatibility as the weight for each corresponding Value pair. Finally, the output of the SA mechanism is determined by the weighted sum of the values. The BoTNet architecture incorporates the Multi-Head Self-Attention (MHSA) mechanism. In MHSA, Query, Key, and Value vectors undergo a transformation by a trainable linear projection H, projecting them onto a different dimension. Subsequently, multiple Attention operations are conducted across all projected dimensions to generate diverse outputs. The final output is derived by integrating all individual outputs. MHSA allows the model to learn information from distinct representation subspaces across various levels [14].
Srinivas et al. incorporated the MHSA mechanism into the ResNet architecture by introducing the Bottleneck Transformer Block (BoT block). Specifically, they substituted all 3 × 3 convolutions in stage 5 of ResNet, which comprises the final three bottleneck blocks, with MHSA. This modification led to a reduction in the total number of model parameters and an enhancement in model performance. Experiments conducted on the CoCo dataset for tasks related to instance segmentation and object detection, along with experiments on the ImageNet dataset for tasks related to image classification, have demonstrated the effectiveness of BoTNet [39]. BoTNet successfully combines the CNN structure with the Transformer structure, enabling the model to adeptly acquire both local and global features. In contrast to instance segmentation and object detection tasks, the input image size for image classification tasks is typically smaller (e.g., 256 × 256). Hence, the dimensions of the feature map size for image classification tasks (e.g., 14 × 14 or 7 × 7) are notably smaller compared to those employed in instance segmentation and object detection tasks (e.g., 64 × 64 or 32 × 32). To address this concern, downsampling was eliminated in stage 5 of BoTNet, the stride of the MHSA layer was set to 1, and the feature map size in stage 5 was increased, leading to enhancements in classification performance, and ultimately resulting in an increase in the final classification accuracy. This design is referred to as BoTNet-S1 (S1 denotes the operation in the final stage). The structural distinctions among ResNet-50, BoTNet-50, and BoTNet-S1-50 are shown in Table 1. It is noteworthy that this paper utilizes BoTNet-S1-50 as the baseline model.

3.1 Cubic Embedding Self Attention
In contrast to natural images, sMRI images exhibit a higher number of channel dimensions. For instance, the sMRI images involved in this study possess a channel size of 137. Hence, during the Position Embedding stage of Transformer-related models, we establish learnable variables Rh and Rw for the Height and Width dimensions, respectively, in addition to a learnable variable Rc for the Channel dimension. In this manner, our model incorporates not only the data pertaining to the Height and Width dimensions of the image but also integrates the information related to the Channel dimension of the image. When our model learns lesions for AD-aided diagnosis, it goes beyond merely identifying lesions solely based on the two-dimensional orientation of the sMRI image. This approach would compartmentalize lesion information along the Channel dimension, so the model learns the lesions from a three-dimensional standpoint, incorporating the three-dimensional information of the sMRI image. This method aligns more in line with real-life scenarios. We call this embedding method MHSA with Cubic Embedding Self Attention (CESA), as illustrated in Fig. 3. The operation denoted as Content-Cubic-Embedding, highlighted in red, signifies the CE (Cubic Embedding) operation executed on the data. Here, Rc, Rh, and Rw represent the learnable embedding variables set on the Channel, Height, and Width dimensions, respectively. Furthermore, it is evident that the data dimensions of both the Input and Output are consistent, with each being represented as

Figure 3: Structure of cubic embedding self attention
We used BoTNet-S1-50 as the baseline model, as previously indicated. It is important to note that the network proposed in this study aligns with BoTNet-S1-50 as it does not engage in downsampling in stage 5. Initially, all


Figure 4: Flowchart of AD-aided diagnostic tasks using ReoBoTNet-CESA
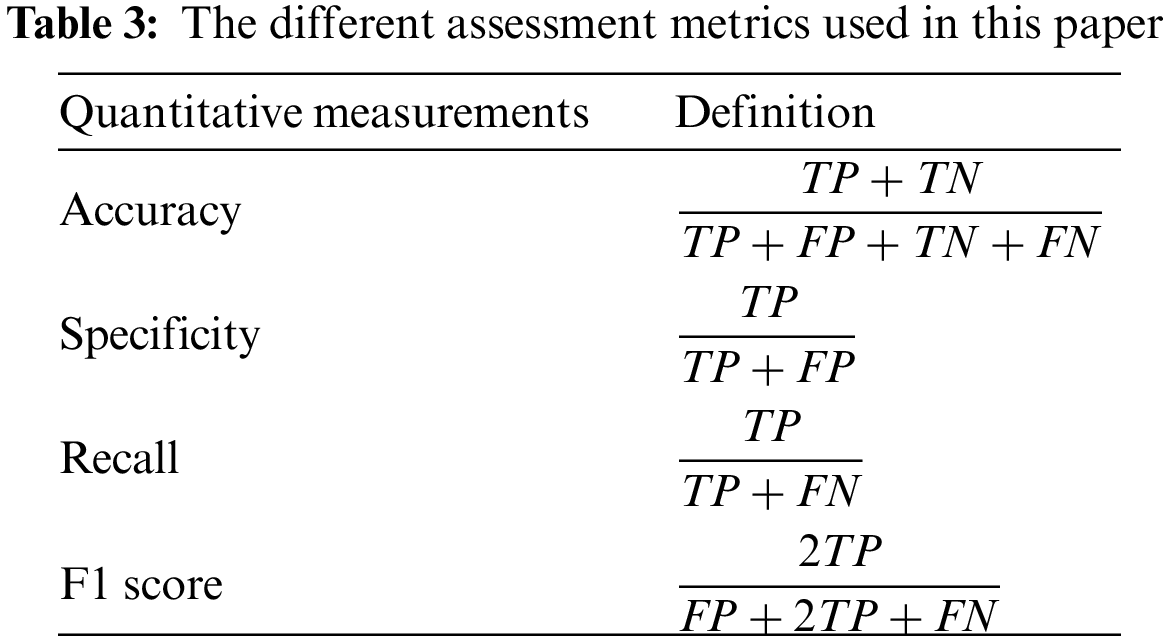
To evaluate the efficacy of various networks, a series of DL performance evaluation metrics were computed, encompassing accuracy, precision, recall, and F1 score. The definitions are shown in the table below, where TP represents the count of correctly predicted true positive subjects, FP represents the count of incorrectly predicted false positive subjects, TN represents the count of correctly predicted true negative subjects, and FN represents the count of incorrectly predicted false negative subjects. Furthermore, a confusion matrix was employed to visually represent the detailed inference outcomes of the classification task and examine the accurate partition probability across various label categories. Moreover, the evaluation metrics used in this study encompass the Receiver Operating Characteristic (ROC) curve and the Area Under Curve (AUC) of the ROC curve. These metrics offer a comprehensive evaluation of the True Positive Rate (TPR) and False Positive Rate (FPR) of the network. The calculation procedures for each experimental metric are shown in Table 3.
In this paper, we proposed the RepBoTNet-CESA network designed to support the task of AD-aided diagnosis. Firstly, preprocessing was conducted on the raw input data, primarily employing the CAT12 preprocessing operation to extract the respective gray matter images from the raw sMRI image data while removing the skull. Subsequently, the RepBoTNet-CESA network was developed, comprising 4 stages of CNN and 1 stage of CESA (Cubic Embedding Self Attention). This architecture enabled the concurrent extraction of local and global features from the input data, facilitating the derivation of output probabilities for individual labels. Finally, the performance metrics of the overall model were computed to detect the network’s effectiveness. In addition, we have illustrated that the enhancement in performance achieved through the utilization of MHSA on ResNet surpasses that of certain conventional attention mechanisms. Our study also expanded the experimental scope to include BoTNet.
To achieve AD-aided diagnosis, a series of five major steps were undertaken, encompassing data collection (S1), data preprocessing (S2), dataset partitioning (S3), training deep models (S4), and result evaluation (S5), as shown in Fig. 5.

Figure 5: Training and evaluation procedures for the AD-aided diagnostic models in this paper
Data collection S1: Obtain medical imaging data from the ADNI dataset to facilitate the execution of experiments focused on AD classification.
Data preprocessing S2: For raw AD brain neuroimages to address data variability, such as differences in brain sizes. This process aims to obtain standardized neuroimaging data and mitigate the impact of data variability on subsequent experiments. For instance, following preprocessing with SPM12, the original sMRI data yielded standard images of gray matter, white matter, and cerebrospinal fluid within the brain.
Dataset division S3: In this paper, the 5-fold cross-validation technique is used to partition the dataset into training, validation, and test datasets in a ratio of 3:1:1. The training dataset is utilized for training the classifier/model and to adjust specific weight parameters in the classifiers/models that are trainable. The test dataset is employed to assess the ultimate performance of the classifiers/models and to derive various final evaluation metrics, which are non-reusable.
Training deep models S4: After obtaining the training dataset and validation dataset. The training dataset is then utilized to train the deep learning model. After the completion of each training round, the validation dataset is employed to assess the improvement over the previous round. The optimal model parameters at that stage are then stored for subsequent evaluation of the experimental outcomes.
Result evaluation S5: The test set data derived from the dataset partition is fed into the optimal model for result assessment, leading to the calculation of the final performance evaluation metric for this AD classification model.
During the training process of the model, the epoch was set to 40, the batch size to 4, and the initial learning rate to 1e-4. The training utilized the cross-entropy loss function, employed the SGD optimizer with a momentum of 0.9 and weight decay of 1e-4, and adopted the StepLR learning rate adjustment method, where the learning rate was adjusted every 10 epochs and 0.1 times the original.
4.1 Input Data Acquisition and Data Pre-Processing
The input data for this study comprised sMRI image data obtained from the ADNI dataset. It included 98 scans from 30 AD subjects, 100 scans from 24 EMCI subjects, 105 scans from 25 LMCI subjects, and 107 scans from 26 NC subjects, totaling 433 scans from 105 subjects. The dataset was balanced, with approximately equal numbers of AD, EMCI, LMCI, and NC data. The fundamental details of the subject data are presented in Table 4. It is important to highlight that the data we downloaded from the ADNI public dataset was initially in DICOM format, necessitating conversion to NII format through the utilization of SPM12. The raw sMRI data images following these conversions are shown in Fig. 6a.


Figure 6: The sMRI image data used in this paper
In this study, the CAT12 tool was primarily utilized for the standard sMRI image preprocessing procedure. Firstly, we used SPM12 to convert the DICOM format into NII format, leading to the NII format sMRI image files in various nested folders. Subsequently, the Nibabel package was utilized to develop a Python script for the purpose of reading and storing NII format sMRI image files within a unified directory. This approach facilitated the consolidation of sMRI scan data for numerous subjects into a singular location. Next, we employed a preexisting MATLAB script for automatic AC-PC Line origin correction (https://Github.com/lrq3000/auto_acpc_reorient) to develop a MATLAB script that executed AC-PC origin correction on the sMRI images within the designated folder, encompassing all sMRI images. Then, the “Segment” operation in CAT12 was employed to segment the gray matter image of the subjects from the sMRI image. This segmented image served as the input data for our model, as shown in Fig. 6b. The “Segment” operation in CAT12 integrates image registration, segmentation, and smoothing in the preprocessing stage, streamlining the process of generating gray matter images from the sMRI images of the subjects. CAT12 also produces a quality report for the “Segment” operation, presenting various quality indices related to image and preprocessing, processing time, volumes of gray matter, white matter, and cerebrospinal fluid, along with other indices of preprocessing operation results. We ensured that the Image Quality Rating (IQR) of the average score for the quality report was equal to or higher than 70%. As shown in Fig. 7, the quality report includes details regarding the composition of blood vessels, gray matter, white matter, and cerebrospinal fluid in both the original and segmented images.

Figure 7: The quality report of the CAT12 segmentation operation provides information on the distribution of blood vessels, gray matter, white matter, and cerebrospinal fluid in the original and segmented images. The correct partitioning of the gray matter portion necessary for the experiment is demonstrated
Following preprocessing, the dimensions of the input gray matter image data were 137 × 113 × 113, which were determined to be incompatible with becoming the direct input into the RepBoTNet-CESA. Based on our experimental findings, it has been determined that the network architecture incorporating MHSA imposes particular constraints on the width and height dimensions of the input data. The original input data utilized in this paper had dimensions of 113 in width and height, which were not evenly divisible by the number of heads (set to 4) in the MHSA structure. Consequently, this mismatch prevented the MHSA from uniformly distributing the input data, thereby hindering its direct integration into the MHSA-involved structure. Hence, we maintained the channel dimension of the original input data and applied zero-padding to the width and height dimensions to extend them to 128. Consequently, the ultimate dimensions of the sMRI gray matter image input data were 137 × 128 × 128. It is important to highlight that the MHSA should ensure an equitable distribution of data by assigning distinct weights to the query, key, and value matrices. Moreover, the number of heads determines the partitioning of the data into multiple segments. In the majority of source code, the query, key, and value are typically divided directly through the use of the Reshape function. This is attributed to the equivalence of the final results obtained from these two methods, and opting for Reshape in basic division helps circumvent intricate weight calculations. In the following experiments, a 5-fold cross-validation approach was employed to divide the datasets into training and testing sets. Additionally, 25% of the data from the training set was chosen to form the validation set, resulting in a ratio of 3:1:1 for the training set, validation set, and test set, respectively. The final index results were calculated as the average of the 5-fold cross-validation.
4.2 MHSA Outperforms Traditional Attention for ResNet Performance
Prior studies have demonstrated the superior performance of utilizing MHSA in BoTNet compared to ResNet. However, there is a research gap exists in evaluating the performance improvement of MHSA in comparison to conventional attention mechanisms like SE and CBAM. The utilization of MHSA is different from traditional attention mechanisms. In the BoTNet series network, the CNN layer is substituted with the MHSA layer, with conventional attention mechanisms frequently incorporated subsequent to the CNN layer. In this study, we added the conventional attention layer following the 3 × 3 convolution layer in stage 5 of ResNet50. Subsequently, we evaluated the performance of ResNet50, ResNet50 with SE attention mechanism, ResNet50 with CBAM attention mechanism, BoTNet50, and BoTNet-S1 on the auxiliary AD classification task using data from AD patients obtained in the previous section. The results are shown in Table 5. The table illustrates that attention mechanisms have the potential to significantly enhance the model’s performance in the auxiliary diagnosis of AD using the dataset employed in this study. Compared with the ResNet50 model, the utilization of the SE attention mechanism resulted in enhancements in accuracy by 4.88%, precision by 6.46%, recall by 0.81%, and F1 score by 4.57%. Similarly, the implementation of the CBAM attention mechanism led to improvements in accuracy by 7.32%, precision by 6.98%, recall by 4.58%, and F1 score by 6.93%. The utilization of conventional attention mechanisms led to a slight increase in the model parameters; however, it significantly enhanced the classification performance of the ResNet50 network in AD/NC auxiliary diagnosis tasks. Utilizing BoTNet50, which replaces the convolutional layer in ResNet50 with the MHSA layer, led to a reduction in the model parameters and an enhancement in accuracy by 8.29%, precision by 8.45%, recall by 4.58%, and F1 score by 7.81%. This improvement surpasses the performance of conventional attention mechanisms in enhancing the ResNet50 network. BoTNet50-S1, a modified version of BoTNet50 that eliminates downsampling in stage 5, retains an equivalent number of model parameters as BoTNet50 while enhancing accuracy by 1.96%, precision by 1.05%, recall by 2.77%, and F1 score by 1.95%. Therefore, BoTNet50-S1 was selected as the baseline network for the ensuing experiments.

4.3 RepBoTNet-CESA Achieves Excellent Results on Various AD-Aided Diagnostic Tasks
To evaluate the performance of RepBoTNet-CESA on AD-aided diagnostic tasks, based on input data with dimensions of 137 × 128 × 128, the RepBoTNet-CESA model was applied to perform auxiliary diagnosis tasks for AD/NC, EMCI/NC, and AD/EMCI/LMCI/NC. Ablation experiments were also carried out. The performance comparison of each model in the AD/NC task is shown in Table 6. In comparison to the baseline, the RepBoTNet model, utilizing the structural reparameterization technique, demonstrated enhancements in accuracy by 1.95%, precision by 2.57%, recall by 0.66%, and F1 score by 1.57%. This improvement was achieved while marginally increasing the number of parameters during the training and inference phases. These results suggest that structural reparameterization contributes positively to the overall performance of the model. The introduction of structural reparameterization and CESA in RepBoTNet-CESA led to enhancements in accuracy by 0.97%, precision by 1.93%, recall by 0.25%, and F1 score by 1.11% compared to RepBoTNet. Notably, these improvements were achieved while keeping the model parameters unchanged, indicating that CESA contributes positively to the model’s performance. Table 7 shows the variance in inference speed among Baseline, RepBoTNet, and RepBoTNet-CESA models. The results suggest that structural reparameterization significantly enhances the model’s inference speed, while the incorporation of CESA structure in structural reparameterization slightly elevates the model’s inference time.


Fig. 8 shows the training loss curves derived from RepBoTNet-CESA in the AD/NC task. The figure illustrates that the training loss of RepBoTNet-CESA consistently decreases over 40 epochs, suggesting the effective learning of the training dataset by our model. Fig. 9 shows the validation loss curves derived from RepBoTNet-CESA in the AD/NC task. The figure illustrates that the validation loss of RepBoTNet-CESA rapidly converges within 15 epochs. Fig. 10 illustrates the confusion matrices derived from RepBoTNet-CESA in the AD/NC task, demonstrating the classification outcomes of the model. During the 5-fold cross-validation, the model consistently achieved a probability of correctly identifying AD and NC of over 90%. This indicates that our RepBoTNet-CESA model has effectively acquired image features that enable accurate differentiation between AD and NC. Fig. 11 shows the ROC curves and AUC values generated by RepBoTNet-CESA in the AD/NC task, providing additional evidence of the model’s exceptional classification accuracy in the AD/NC task. The images presented above are derived from fold 0 in a 5-fold cross-validation. It is important to note that the results across all five folds exhibit a similar pattern.

Figure 8: During the AD/NC task, the RepBoTNet-CESA model generated the training set loss curve for each fold of the cross-validation. Over the course of 40 epochs, the loss consistently decreased, suggesting that the neural network successfully acquired knowledge from the training dataset. The case of fold 0 is presented here, and the remaining folds exhibit similarities to fold 0

Figure 9: During the AD/NC task, the RepBoTNet-CESA model generated the validation set loss curve for each fold in the cross-validation process. The network rapidly achieved convergence on the validation set data after almost 15 epochs. The case of fold 0 is presented here, and the remaining folds exhibit similarities to fold 0

Figure 10: During the AD/NC task, the RepBoTNet-CESA model generated confusion matrix results for each fold of the cross-validation. On the test dataset, the neural network demonstrated effective prediction capabilities for samples classified as AD and NC. The case of fold 0 is presented here, and the remaining folds exhibit similarities to fold 0

Figure 11: During the AD/NC task, the RepBoTNet-CESA model generated ROC curves and calculated the AUC values for each fold of the cross-validation process. In the 5-fold cross-validation experiment, the network demonstrated outstanding classification performance consistently across all folds. The case of fold 0 is presented here, and the remaining folds exhibit similarities to fold 0
The RepBoTNet-CESA also demonstrates strong performance in tasks involving EMCI/NC and AD/EMCI/LMCI/NC. Fig. 12 illustrates the confusion matrices obtained by RepBoTNet-CESA for each fold of the AD/EMCI/LMCI/NC task, demonstrating the classification outcomes of the model. The results of the 5-fold cross-validation experiment demonstrate that the model has acquired image features capable of effectively distinguishing between AD, EMCI, LMCI, and NC. Fig. 13 shows the ROC curves and AUC values obtained by RepBoTNet-CESA in every fold of the AD/EMCI/LMCI/NC task, providing additional evidence of the model’s classification efficacy in this particular task. Table 7 shows the comparison of the inference speeds between RepBoTNet-CESA and its ablation models. The implementation of structural reparameterization has the potential to significantly decrease the inference time of the network model. The application of structural reparameterization on RepBoTNet resulted in a reduction of the inference time by 4.66 milliseconds, representing 13.75% of the original inference time. Similarly, the utilization of structural reparameterization on RepBoTNet-CESA led to a decrease in the inference time by 4.82 milliseconds, which accounted for 14.11% of the original inference time.
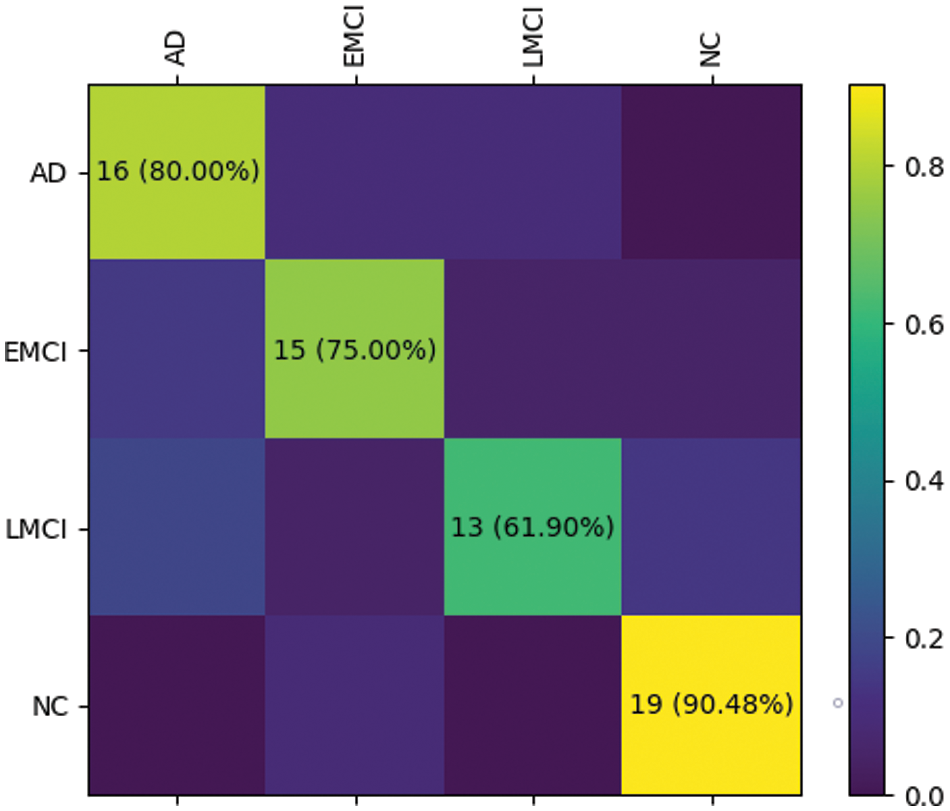
Figure 12: During the AD/EMCI/LMCI/NC task, the RepBoTNet-CESA model generated confusion matrix results for each fold of the cross-validation. On the test dataset, the neural network successfully predicted samples corresponding to AD, EMCI, LMCI, and NC groups. The case of fold 0 is presented here, and the remaining folds exhibit similarities to fold 0

Figure 13: For the task involving AD/EMCI/LMCI/NC, the RepBoTNet-CESA model generated ROC curves and AUC area results for each fold during cross-validation. In the 5-fold cross-validation experiment, the network demonstrated outstanding classification performance across all folds. The case of fold 0 is presented here, and the remaining folds exhibit similarities to fold 0
As shown in Table 8, the RepBoTNet-CESA model we propose demonstrates superior accuracy compared to alternative methods, suggesting its enhanced performance in the AD diagnostic task. This phenomenon can be ascribed to three primary factors: (1) In contrast to conventional machine learning methods, the “depth” characteristic of CNN networks enhances the network’s non-linear modeling capacity, leading to superior performance on intricate tasks. Consequently, RepBoTNet-CESA demonstrates superior performance in the AD diagnostic task when compared to approaches utilizing conventional machine learning classifiers such as RF or SVM. In comparison to Ning et al.’s approach employing the RF classifier, our model demonstrates a 3.48% enhancement in accuracy for the AD/NC task. In contrast to Zhang et al.’s method utilizing RF and LR classifiers, our model exhibits an 8.58% accuracy for the AD/NC task and a 4.65% enhancement for the EMCI/NC task. When compared to Li et al.’s technique employing the SVM classifier, our model shows a 13.44% accuracy for the EMCI/NC task. (2) In contrast to models that solely rely on Transformer networks for learning global features, our model incorporates a CNN structure to capture specific local features. Consequently, RepBoTNet-CESA demonstrates superior performance in the AD diagnostic task compared to approaches solely utilizing the Transformer network architecture. Compared with Ning et al.’s approach, which employs traditional Transformer networks like T-BERT and S-BERT for feature extraction, our model demonstrates a 12.68% enhancement in accuracy for the AD/NC task. (3) Different from models that solely rely on CNN networks for acquiring local features, our model incorporates a Transformer structure to capture specific global features. Therefore, RepBoTNet-CESA demonstrates superior performance in the AD diagnostic task compared to approaches solely utilizing the CNN network architecture. On the AD/NC task, our final accuracy is 13.3% higher than Miltiadous et al., 1.28% higher than Lyu et al.,3.48% higher than Hu et al., 4.08% higher than Shikalgar et al., 1.08% higher than Marzban et al., and 9.98% higher than Shen et al. For the EMCI/NC task, our final accuracy surpasses Jiao et al. by 1.62%, Song et al. by 7.25%, and Yu et al. by 7.33%. In the AD/EMCI/LMCI/NC task, our final accuracy exceeds Rallabandi et al. by 6.97%. Compared with Jie et al., our model demonstrates superior performance by 11.28%, 11.65%, and 23.37% on the three AD diagnostic tasks, respectively. Similarly, when compared to the research by Gao et al., our model exhibits better results by 1.48%, 8.15%, and 23.67% in the three AD diagnostic tasks, respectively. The performance metrics presented illustrate the effectiveness of our proposed RepBoTNet-CESA in addressing diverse AD diagnostic tasks, with particular emphasis on the AD/EMCI/LMCI/NC four-classification task.
4.4 Deeper RepBoTNet-CESA Network Fails to Go Further in AD-Aided Diagnostic Tasks
Apart from ResNet50, the ResNet series also includes deeper networks such as ResNet101 and ResNet152. Following the extension idea of the ResNet framework, to determine whether a deeper network could achieve better AD-aided diagnosis, we expanded the original 50-layer RepBoTNet-CESA model to 101 layers and 152 layers, identified as RepBoTNet101-CESA and RepBoTNet152-CESA, respectively. As illustrated in Table 9, the experiments conducted on the AD/NC task utilizing our input data indicated that the deeper RepBoTNet-CESA networks did not exhibit superior performance compared to the 50-layer RepBoTNet-CESA. This observation suggests the presence of overfitting, consequently resulting in diminished classification performance of the model.

The increasing age of the population will lead to a heightened burden on society due to AD. Accurate identification of AD and its prodromal stages, i.e., EMCI and LMCI, is essential for initiating early treatment and delaying disease progression. The utilization of medical image-based aided diagnosis is a crucial approach for the early identification of AD.Based on the sMRI images, this paper proposes the RepBoTNet-CESA network model for AD-aided diagnosis tasks. This network combines the benefits of CNN structures in capturing local information and Transformer structures in integrating global information. The structural reparameterization technique is employed to imbue the network with multi-scale CNN characteristics. This enhancement leads to improved classification performance on CNN, decreased computational complexity of the network, and accelerated inference speed. The CESA incorporates a three-dimensional perspective when analyzing data and integrates the positional information of the Channel dimension into the Transformer model. This enhancement has been shown to improve the network’s classification performance on the Transformer. The experiments conducted on AD-aided diagnosis using sMRI data from the ADNI dataset demonstrate the effectiveness of the proposed method.
In this study, an initial comparative experiment is carried out concerning BoTNet to ascertain the superior impact of MHSA on network performance in comparison to conventional attention mechanisms. Comparison experiments have been conducted between BoTNet and BoTNet-S1 for AD-aided diagnosis to confirm the efficacy of BoTNet-S1 in facilitating AD-aided diagnosis. This validation serves as the basis for subsequent experiments. Utilizing BoTNet-S1 as a baseline, RepBoTNet is proposed to showcase the effectiveness of Rep3 × 3 in enhancing model performance within the realm of CNN aspects. Following this, the paper introduces the CESA method, which is then utilized to develop the RepBoTNet-CESA network model. The efficacy of the model is illustrated through a comparative analysis of results in the AD-aided diagnosis task. The results of the corresponding ablation experiments conducted illustrate the effectiveness of CESA in enhancing the model performance of the Transformer part. A further experiment was conducted to compare the performance of deeper RepBoTNet-CESA models, revealing that increasing the depth of RepBoTNet-CESA did not lead to improved outcomes.
The RepBoTNet-CESA is a deep learning model with a generalized architecture. The potential application of this method extends to the classification and prediction of various diseases, provided that the relevant sMRI image data is obtainable. As shown in Table 8, RepBoTNet-CESA conducted experiments on AD/NC, EMCI/NC, and AD/EMCI/LMCI/NC tasks for AD-aided diagnosis. The range of AD-aided diagnosis tasks utilized is notably broader compared to the majority of relevant studies. Ultimately, the accuracy of the respective experiments is superior overall. This point further demonstrates the universality and validity of RepBoTNet-CESA. In the subsequent stages of developing AD-aided diagnosis platforms, RepBoTNet-CESA can serve as a highly competitive deep learning model for integration into relevant AD-aided diagnosis platforms.
While the RepBoTNet-CESA network model proposed in this study demonstrates effectiveness in the task of AD-aided diagnosis, it is important to acknowledge several limitations. (1) Beyond structural MRI (sMRI), there exist various other types of brain imaging data, such as functional MRI (fMRI) and positron emission tomography (PET). This study solely focused on AD-aided diagnosis research utilizing unimodal sMRI. With the ongoing advancements in brain imaging processing technology, the utilization of multimodal brain medical imaging data for AD-aided diagnosis has been proven to outperform the unimodal approach. The repBoTNet-CESA model does not incorporate multimodal deep learning techniques. (2) The sMRI data utilized as input in this study is comparatively less comprehensive and fewer in number than in other computer vision research domains employing deep learning techniques. (3) The issue of interpretability remains prevalent in deep learning research, presenting a complex problem that is challenging to solve. Presently, deep learning models are still regarded as black boxes, and the scientific community encounters challenges in comprehending the underlying principles governing model inference. This deficiency in comprehension results in a lack of confidence in the predictive ability of DL models, a sentiment that is similarly evident in the realm of AD-aided diagnosis.
In future research endeavors, RepBoTNet-CESA can serve as a foundational framework for subsequent related studies. Firstly, the proposed RepBoTNet-CESA can be adapted and utilized with multimodal input data to enhance the diagnostic performance. Subsequently, sMRI data from additional experimental centers can be gathered to enhance the input data scale of the model. In addition, semi-supervised methods, such as generative adversarial networks (GANs), can be utilized to supplement the available experimental data and improve the experimental outcomes.
Acknowledgement: The authors especially acknowledge Prof. Dinggang Shen of ShanghaiTech University.
Funding Statement: This work was supported by the Key Project of Zhejiang Provincial Natural Science Foundation under Grants LD21F020001, Z20F020022, and the National Natural Science Foundation of China under Grants 62072340, 62076185, and the Major Project of Wenzhou Natural Science Foundation under Grants 2021HZSY0071, ZS2022001.
Author Contributions: Study conception and design: Z. Y. Hu, X. B. Zhang; data collection: X. B. Zhang, L. Xiao; analysis and interpretation of results: X. B. Zhang, H. Huang; draft manuscript preparation: X. B. Zhang, Z. Y. Hu; software and funding: Z. Y. Hu. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All data generated or analysed during this study are available from the corresponding author upon reasonable request.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. N. Amoroso et al., “Deep learning reveals alzheimer’s disease onset in mci subjects: Results from an international challenge,” J. Neurosci. Methods., vol. 302, pp. 3–9, May 2018. doi: 10.1016/j.jneumeth.2017.12.011. [Google Scholar] [PubMed] [CrossRef]
2. P. Khan et al., “Machine learning and deep learning approaches for brain disease diagnosis: Principles and recent advances,” IEEE Access, vol. 9, pp. 37622–37655, Feb. 2021. doi: 10.1109/ACCESS.2021.3062484. [Google Scholar] [CrossRef]
3. Y. Ju and K. Y. Tam, “Pathological mechanisms and therapeutic strategies for alzheimers disease,” Neural Regener. Res., vol. 17, no. 3, pp. 543, Mar. 2022. doi: 10.4103/1673-5374.320970. [Google Scholar] [PubMed] [CrossRef]
4. R. J. Castellani, R. K. Rolston, and M. A. Smith, “Alzheimer disease,” Dis. Month, vol. 56, no. 9, pp. 484–546, Sep. 2010. doi: 10.1016/j.disamonth.2010.06.001. [Google Scholar] [PubMed] [CrossRef]
5. R. Brookmeyer, E. Johnson, K. Ziegler-Graham, and H. M. Arrighi, “Forecasting the global burden of alzheimers disease,” Alzheimer’s Dement., vol. 3, no. 3, pp. 186–191, Jul. 2007. doi: 10.1016/j.jalz.2007.04.381. [Google Scholar] [PubMed] [CrossRef]
6. C. H. V. Dyck et al., “Lecanemab in early alzheimer’s disease,” N. Engl. J. Med., vol. 388, no. 1, pp. 9–21, Jan. 2023. doi: 10.1056/NEJMoa2212948. [Google Scholar] [PubMed] [CrossRef]
7. C. Bellenguez et al., “New insights into the genetic etiology of alzheimers disease and related dementias,” Nat. Genet., vol. 54, no. 4, pp. 412–436, Apr. 2022. doi: 10.1038/s41588-022-01024-z. [Google Scholar] [PubMed] [CrossRef]
8. R. C. Petersen and S. Negash, “Mild cognitive impairment: An overview,” CNS Spectr., vol. 13, no. 1, pp. 45–53, Jan. 2008. doi: 10.1017/S1092852900016151. [Google Scholar] [PubMed] [CrossRef]
9. S. Gauthier et al., “Mild cognitive impairment,” The Lancet, vol. 367, no. 9518, pp. 1262–1270, Apr. 2006. doi: 10.1016/S0140-6736(06)68542-5. [Google Scholar] [PubMed] [CrossRef]
10. R. C. Petersen et al., “Vitamin E and donepezil for the treatment of mild cognitive impairment,” N. Engl. J. Med., vol. 352, no. 23, pp. 2379–2388, Jun. 2005. doi: 10.1056/NEJMoa050151. [Google Scholar] [PubMed] [CrossRef]
11. S. Y. Lin et al., “The clinical course of early and late mild cognitive impairment,” Front. Neurol., vol. 13, pp. 685636, May 2022. doi: 10.3389/fneur.2022.685636. [Google Scholar] [PubMed] [CrossRef]
12. M. Ewers, R. A. Sperling, W. E. Klunk, M. W. Weiner, and H. Hampel, “Neuroimaging markers for the prediction and early diagnosis of alzheimer’s disease dementia,” Trends Neurosci., vol. 34, no. 8, pp. 430–442, Jun. 2011. doi: 10.1016/j.tins.2011.05.005. [Google Scholar] [PubMed] [CrossRef]
13. S. J. Teipel, M. Grothe, S. Lista, N. Toschi, F. G. Garaci and H. Hampel, “Relevance of magnetic resonance imaging for early detection and diagnosis of alzheimer disease,” Med. Clin. North Am., vol. 97, no. 3, pp. 399–424, Feb. 2013. doi: 10.1016/j.mcna.2012.12.013. [Google Scholar] [PubMed] [CrossRef]
14. A. Vaswani et al., “Attention is all you need,” in Proc. NIPS, Long Beach, CA, USA, Dec. 2017, pp. 6000–6010. [Google Scholar]
15. A. Dosovitskiy et al., “An image is worth 16x16 words: Transformers for image recognition at scale,” in Proc. ICLR, Vienna, Austria, May 2021, pp. 11929. [Google Scholar]
16. Z. Hu, Q. Wu, C. Chen, L. Xiao, and S. Jin, “Alzheimers disease diagnosis method based on convolutional neural network using key slices voting,” in Proc. ICIST, Chengdu, Sichuan, China, May 2021, pp. 266–274. [Google Scholar]
17. L. Gao et al., “Multi-perspective feature extraction and fusion based on deep latent space for diagnosis of alzheimers diseases,” Brain Sci., vol. 12, no. 10, pp. 1348, Oct. 2022. doi: 10.3390/brainsci12101348. [Google Scholar] [PubMed] [CrossRef]
18. B. Jie, M. Liu, C. Lian, F. Shi, and D. Shen, “Designing weighted correlation kernels in convolutional neural networks for functional connectivity based brain disease diagnosis,” Med. Image Anal., vol. 63, no. 12, pp. 101709, Jul. 2020. doi: 10.1016/j.media.2020.101709. [Google Scholar] [PubMed] [CrossRef]
19. A. Mehmood, M. Maqsood, M. Bashir, and S. Y. Yang, “A deep siamese convolution neural network for multi-class classification of alzheimer disease,” Brain Sci., vol. 10, no. 2, pp. 84, Feb. 2020. doi: 10.3390/brainsci10020084. [Google Scholar] [PubMed] [CrossRef]
20. A. Roshanzamir, H. Aghajan, and M. S. Baghshah, “Transformer-based deep neural network language models for alzheimers disease risk assessment from targeted speech,” BMC Med. Inf. Decis. Making, vol. 21, no. 1, pp. 1–14, Mar. 2021. doi: 10.1186/s12911-021-01456-3. [Google Scholar] [PubMed] [CrossRef]
21. S. Sarraf, A. Sarraf, D. D. DeSouza, J. A. E. Anderson, M. Kabia and Alzheimer’s Disease Neuroimaging Initiative, “OViTAD: Optimized vision transformer to predict various stages of alzheimers disease using resting-state fMRI and structural MRI data,” Brain Sci., vol. 13, no. 2, pp. 260, Feb. 2023. doi: 10.3390/brainsci13020260. [Google Scholar] [PubMed] [CrossRef]
22. A. O. Salman and O. Geman, “Evaluating three machine learning classification methods for effective COVID-19 diagnosis,” Int. J. Math., Stat., Comput. Sci., vol. 1, pp. 1–14, Feb. 2022. doi: 10.59543/ijmscs.v1i.7693. [Google Scholar] [CrossRef]
23. Z. H. Arif and K. Cengiz, “Severity classification for COVID-19 infections based on lasso-logistic regression model,” Int. J. Math., Stat., Comput. Sci., vol. 1, pp. 25–32, Apr. 2023. doi: 10.59543/ijmscs.v1i.7715. [Google Scholar] [CrossRef]
24. Y. LeCun, L. Bottou, Y. Bengio, and P. Haffner, “Gradient-based learning applied to document recognition,” IEEE Proc., vol. 86, no. 11, pp. 2278–2324, Nov. 1998. doi: 10.1109/5.726791. [Google Scholar] [CrossRef]
25. A. Krizhevsky, I. Sutskever, and G. E. Hinton, “Imagenet classification with deep convolutional neural networks,” Commun. ACM, vol. 60, no. 6, pp. 84–90, 2017. doi: 10.1145/3065386. [Google Scholar] [CrossRef]
26. K. Simonyan and A. Zisserman, “Very deep convolutional networks for large-scale image recognition,” in Proc. ICLR, San Diego, CA, USA, May 7–9, 2015. [Google Scholar]
27. K. He, X. Zhang, S. Ren, and J. Sun, “Deep residual learning for image recognition,” in Proc. CVPR, Las Vegas, NV, USA, Jun. 2016, pp. 770–778. [Google Scholar]
28. C. Szegedy et al., “Going deeper with convolutions,” in Proc. CVPR, Boston, MA, USA, Jun. 2015, pp. 1–9. [Google Scholar]
29. C. Szegedy, S. Ioffe, V. Vanhoucke, and A. Alemi, “Inception-v4, inception-resnet and the impact of residual connections on learning,” in Proc. AAAI, San Francisco, CA, USA, Feb. 4–9, 2017. [Google Scholar]
30. C. Szegedy, V. Vanhoucke, S. Ioffe, J. Shlens, and Z. Wojna, “Rethinking the inception architecture for computer vision,” in Proc. CVPR, Las Vegas, NV, USA, Jun. 2016, pp. 2818–2826. [Google Scholar]
31. G. Huang, Z. Liu, L. V. D. Maaten, and K. Q. Weinberger, “Densely connected convolutional networks,” in Proc. CVPR, Honolulu, HI, USA, Jul. 2017, pp. 4700–4708. [Google Scholar]
32. X. Ding, X. Zhang, N. Ma, J. Han, G. Ding and J. Sun, “RepVGG: Making VGG-style ConvNets great again,” in Proc. CVPR, Nashville, TN, USA, Jun. 2021, pp. 13733–13742. [Google Scholar]
33. A. Veit, M. J. Wilber, and S. Belongie, “Residual networks behave like ensembles of relatively shallow networks,” in Proc. NIPS, Barcelona, BCN, Spain, Dec. 2016, pp. 29. [Google Scholar]
34. J. Liu, Q. Hou, M. Cheng, C. Wang, and J. Feng, “Improving convolutional networks with self-calibrated convolutions,” in Proc. CVPR, Seattle, WA, USA, Jun. 2020, pp. 10096–10105. [Google Scholar]
35. J. Hu, L. Shen, and G. Sun, “Squeeze-and-excitation networks,” in Proc. CVPR, Salt Lake City, UT, USA, Jun. 2018, pp. 7132–7141. [Google Scholar]
36. S. Woo, J. Park, J. Lee, and I. S. Kweon, “CBAM: Convolutional block attention module,” in Proc. ECCV, Munich, Bavaria, German, Sep. 2018, pp. 3–19. [Google Scholar]
37. J. Hu, L. Shen, S. Albanie, G. Sun, and A. Vedaldi, “Gather-excite: Exploiting feature context in convolutional neural networks,” in Proc. NIPS, Montreal, QC, Canada, Dec. 2018. [Google Scholar]
38. X. Wang, R. Girshick, A. Gupta, and K. He, “Nonlocal neural networks,” in Proc. CVPR, Salt Lake City, UT, USA, Jun. 2018, pp. 7794–7803. [Google Scholar]
39. A. Srinivas, T. Lin, N. Parmar, J. Shlens, P. Abbeel and A. Vaswani, “Bottleneck transformers for visual recognition,” in Proc. CVPR, Nashville, TN, USA, Jun. 2021, pp. 16519–16529. [Google Scholar]
40. A. Miltiadous et al., “DICE-net: A novel convolution-transformer architecture for Alzheimer detection in EEG signals,” IEEE Access, vol. 11, pp. 71840–71858, Jul. 2023. doi: 10.1109/ACCESS.2023.3294618. [Google Scholar] [CrossRef]
41. Y. Lyu, X. Yu, D. Zhu, and L. Zhang, “Classification of alzheimer’s disease via vision transformer: Classification of alzheimer’s disease via vision transformer,” in Proc. PETRA, New York, USA, Jun. 2022, pp. 463–468. [Google Scholar]
42. N. An, H. Ding, J. Yang, R. Au, and T. F. A. Ang, “Deep ensemble learning for alzheimer’s disease classification,” J. Biomed. Inf., vol. 105, pp. 103411, May 2020. doi: 10.1016/j.jbi.2020.103411. [Google Scholar] [PubMed] [CrossRef]
43. A. Shikalgar and S. Sonavane, “Hybrid deep learning approach for classifying alzheimer disease based on multimodal data,” in Proc. ICCET, Nagoya, Aichi-ken, Japan, Apr. 2019, pp. 511–520. [Google Scholar]
44. E. N. Marzban, A. M. Eldeib, I. A. Yassine, Y. M. Kadah, and Alzheimers Disease Neurodegenerative Initiative, “Alzheimers disease diagnosis from diffusion tensor images using convolutional neural networks,” PLoS One, vol. 15, no. 3, pp. e0230409, Mar. 2020. doi: 10.1371/journal.pone.0230409. [Google Scholar] [PubMed] [CrossRef]
45. T. Shen et al., “Predicting alzheimer disease from mild cognitive impairment with a deep belief network based on 18F-FDG-PET images,” Mol. Imaging, vol. 18, no. 1, Sep. 2019. doi: 10.1177/1536012119877285. [Google Scholar] [PubMed] [CrossRef]
46. Y. Zhang and S. Liu, “Individual identification using multi-metric of DTI in alzheimers disease and mild cognitive impairment,” Chin. Phys. B, vol. 27, no. 8, pp. 088702, 2018. doi: 10.1088/1674-1056/27/8/088702. [Google Scholar] [CrossRef]
47. Z. Jiao, Z. Xia, X. Ming, C. Cheng, and S. Wang, “Multi-scale feature combination of brain functional network for emci classification,” IEEE Access, vol. 7, pp. 74263–74273, Jun. 2019. doi: 10.1109/ACCESS.2019.2920978. [Google Scholar] [CrossRef]
48. J. Li et al., “Persistent feature analysis of multimodal brain networks using generalized fused lasso for emci identification,” in Proc. MICCAI, Lima, Peru, Oct. 2020, pp. 44–52. [Google Scholar]
49. X. Song et al., “Integrating similarity awareness and adaptive calibration in graph convolution network to predict disease,” in Proc. MICCAI, Lima, Peru, Oct. 2020, pp. 124–133. [Google Scholar]
50. S. Yu et al., “Multiscale enhanced graph convolutional network for early mild cognitive impairment detection,” in Proc. MICCAI, Lima, Peru, Oct. 2020, pp. 228–237. [Google Scholar]
51. V. P. S. Rallabandi et al., “Automatic classification of cognitively normal, mild cognitive impairment and alzheimer’s disease using structural mri analysis,” Inf. Med. Unlocked, vol. 18, no. 2, pp. 100305, Feb. 2020. doi: 10.1016/j.imu.2020.100305. [Google Scholar] [CrossRef]
52. J. Ruiz, M. Mahmud, M. Modasshir, M. S. Kaiser and Alzheimers Disease Neuroimaging Initiative, “3D densenet ensemble in 4-way classification of alzheimers disease,” in Proc. Brain Inform., Padua, Veneto, Italy, Sep. 2020, pp. 85–96. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF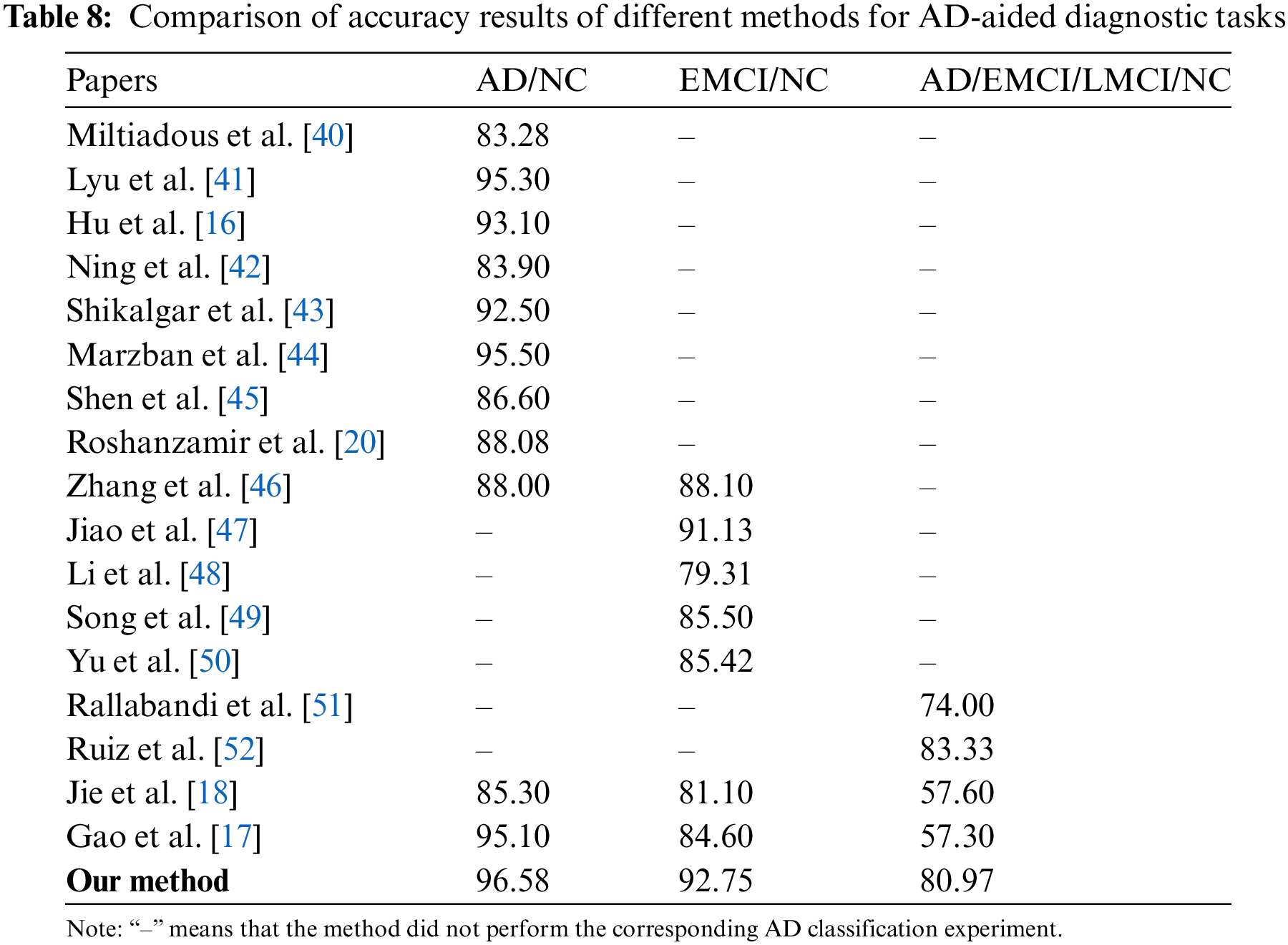
 Downloads
Downloads
 Citation Tools
Citation Tools