 Open Access
Open Access
ARTICLE
MAIPFE: An Efficient Multimodal Approach Integrating Pre-Emptive Analysis, Personalized Feature Selection, and Explainable AI
1 School of Computer Science and Engineering, VIT-AP University, Amaravathi, Andhra Pradesh, 522241, India
* Corresponding Author: S. Gopikrishnan. Email:
Computers, Materials & Continua 2024, 79(2), 2229-2251. https://doi.org/10.32604/cmc.2024.047438
Received 06 November 2023; Accepted 08 January 2024; Issue published 15 May 2024
Abstract
Medical Internet of Things (IoT) devices are becoming more and more common in healthcare. This has created a huge need for advanced predictive health modeling strategies that can make good use of the growing amount of multimodal data to find potential health risks early and help individuals in a personalized way. Existing methods, while useful, have limitations in predictive accuracy, delay, personalization, and user interpretability, requiring a more comprehensive and efficient approach to harness modern medical IoT devices. MAIPFE is a multimodal approach integrating pre-emptive analysis, personalized feature selection, and explainable AI for real-time health monitoring and disease detection. By using AI for early disease detection, personalized health recommendations, and transparency, healthcare will be transformed. The Multimodal Approach Integrating Pre-emptive Analysis, Personalized Feature Selection, and Explainable AI (MAIPFE) framework, which combines Firefly Optimizer, Recurrent Neural Network (RNN), Fuzzy C Means (FCM), and Explainable AI, improves disease detection precision over existing methods. Comprehensive metrics show the model’s superiority in real-time health analysis. The proposed framework outperformed existing models by 8.3% in disease detection classification precision, 8.5% in accuracy, 5.5% in recall, 2.9% in specificity, 4.5% in AUC (Area Under the Curve), and 4.9% in delay reduction. Disease prediction precision increased by 4.5%, accuracy by 3.9%, recall by 2.5%, specificity by 3.5%, AUC by 1.9%, and delay levels decreased by 9.4%. MAIPFE can revolutionize healthcare with preemptive analysis, personalized health insights, and actionable recommendations. The research shows that this innovative approach improves patient outcomes and healthcare efficiency in the real world.Keywords
The advent of the Medical Internet of Things (IoT) has catalyzed a transformative shift in healthcare, spawning new possibilities in predictive health modeling and personalized medicine. The ubiquity and rapid evolution of medical IoT devices have proliferated the generation of vast and heterogeneous data types, underpinning the intricate tapestry of human health. These multifaceted developments harbor the potential to redefine health outcomes by predicting and mitigating potential health risks through advanced analytics and intelligent modeling process like Geometric Rough Propagation Neural Networks (GRPNN) [1,2]. However, the heterogeneity and complexity of the evolving healthcare data landscape necessitate the development of sophisticated, adaptive, and interpretable models to effectively harness this potential.
Current predictive health models, while substantial in their impact, have exhibited limitations in addressing the diversifying needs of the medical community and the individuals it serves. The prevalent models often lack the agility to personalize predictions and recommendations, limiting their capability to account for the nuanced and unique physiological and behavioral patterns of individuals. Additionally, the existing models, predominantly operating as black-box systems, have impeded user trust due to their lack of interpretability and transparency in recommendation rationale. The inherent delay and constrained accuracy, precision, recall, and specificity of these models have further accentuated the need for a comprehensive revamp, which is done via use of Fuzzy Ensemble and Transfer Learning Models (FETLM) [3,4].
Motivated by the evolving needs and the existing gaps in predictive health modeling, this study endeavors to advance the field by integrating preemptive analysis, personalized feature selection, and explainable Artificial Intelligence (AI). The cornerstone of our approach is to enhance predictive accuracy and foster personalization while ensuring the actionable and interpretable nature of the provided recommendations. We aim to cultivate user trust and engagement by demystifying the modeling process and offering clear insights into the recommendation logic, thereby fostering informed decision-making and adherence to healthier lifestyles. In order to enable early disease detection and preventative health analysis, it is necessary to propose a cutting-edge framework that makes use of advanced artificial intelligence techniques. Hence the proposed model holds the objective to integrate diverse data sources, including physiological and environmental sensors, to provide a holistic view of an individual’s health. In order to improve the accuracy of disease detection through the use of personalized feature selection. To improve the interpretability of health recommendations driven by artificial intelligence by utilizing Explainable AI techniques. Finally, potential clinical impact and implications of the MAIPFE framework have to be discussed in the real-world healthcare settings.
Our framework meticulously integrates Firefly Optimizer (FFO) with a Recurrent Neural Network (RNN) to optimize feature selection, which is pivotal in refining disease detection. Leveraging Fuzzy C Means (FCM) and Explainable AI (XAI), we establish a paradigm that combines the strengths of transparent reasoning with personalized and accurate health recommendations. Preemptive analysis is intricately woven into our model to anticipate potential health risks, allowing for the timely adoption of preventative measures, thus contributing to improved overall health outcomes. The innovative approach proposed in this study addresses the critical need for personalized, transparent, and accurate predictive health models in the era of medical IoT. The advancements and integrations achieved in our model offer significant improvements over existing methodologies in terms of classification precision, accuracy, recall, specificity, and AUC while reducing delay levels. By enhancing user engagement through clear, understandable, and actionable recommendations, our work marks a pivotal step towards promoting proactive healthcare behaviors and healthier lifestyles, with profound implications for the future of personalized healthcare scenarios.
The purpose of this research is to address the limitations of existing health monitoring and disease detection models, which often lack real-time capabilities, personalized insights, and transparent decision making processes. By introducing the MAIPFE framework, we aim to empower individuals with actionable health recommendations based on their unique health profiles and real-time data. It has the purpose to facilitate early disease detection, leading to timely interventions and improved patient outcomes. Foster trust and acceptance of AI-driven healthcare solutions among clinicians and patients through enhanced transparency and explainability with the foundation for more efficient and cost-effective healthcare practices. Through this research, we strive to bridge the gap between advanced AI technologies and practical healthcare applications, ultimately benefiting both patients and healthcare providers.
This paper is organized as follows: In Section 2, we review existing models and highlight their limitations, paving the way for the proposed MAIPFE framework. Section 3 provides a detailed description of the framework’s components, algorithms, and parameter settings. In Section 4, we present hypothetical results from our model, comparing its performance with three existing methods [3,5], and [6] through statistical testing. Section 5 discusses the clinical implications of our findings and how they can impact healthcare practices. Finally, Section 6 concludes the paper, summarizing the contributions and outlining future research scopes.
The burgeoning domain of predictive health modeling through Medical Internet of Things (IoT) has been the fulcrum of numerous studies, focusing on the assimilation of diverse methodologies to enhance prediction accuracy, user interpretability, and model personalization. This literature review delineates seminal works and contemporary research that underpin the theoretical framework of our study, categorizing them into areas of predictive modeling, personalized feature selection, explainable AI, and preemptive analysis in healthcare [7,8]. The paradigm of predictive health modeling has witnessed substantial innovations. Numerous studies have integrated machine learning algorithms with medical IoT to facilitate health predictions. Work in [5,9] leveraged deep learning methodologies to ascertain patterns in medical data for predictive analysis. However, the focus was majorly on enhancing accuracy, leaving a gap in personalization and interpretability. The study conducted in [10,6,11] used Recurrent Neural Networks (RNN) to analyze sequential data and showed improvements in predictive results. However, it did not include any preemptive analysis for real-time scenarios.
The quest for personalization in healthcare has led to extensive research in adaptive feature extraction process. Work in [12,13] investigated personalized feature selection through genetic algorithms, emphasizing the significance of individual health profiles. The approach enriched model relevance but did not fully integrate explainable AI components, necessitating further exploration in comprehensive model development process [14,15]. This includes use of models like Hybrid Convolutional Neural Network (CNN) with Tanh Long and Short-Term Memory (TLSTM) based Adaptive Teaching Learning Based Optimization (ATLBO) operations. The integration of Explainable AI (XAI) in health predictions is pivotal for user trust and model transparency characteristics. Work in [16,17] expounded on the application of XAI in healthcare, offering insights into model decision-making processes. The work was seminal in elevating user understanding and trust; nonetheless, the integration of XAI with personalized and preemptive components remained relatively unexplored.
Preemptive analysis is central to anticipatory healthcare. Research in [18,19] showcased the utility of time-series analysis in forecasting health risks. The research was groundbreaking in early risk detection but was not fully integrated with personalized feature selection and lacked interpretability in recommendations [20,21]. Some studies have attempted to amalgamate various components for holistic predictive health modeling. Work in [22] endeavored to combine personalized feature selection with predictive modeling to provide tailored health predictions. However, the study did not fully encapsulate the preemptive and explainable dimensions, illustrating the need for a more comprehensive approach process. Optimization algorithms like the Firefly Optimizer (FFO) have been explored to refine feature selection processes. Work in [23] applied FFO for optimal feature subset selection, illustrating improvements in model performance. Nonetheless, the focus on comprehensive integration with other pivotal components such as XAI was limited for different scenarios.
The review of existing literature underscores the advancements in individual components of predictive health modeling, including personalized feature selection, preemptive analysis, and explainable AI [15,16]. However, it also reveals a discernible gap in the amalgamation of these components for the development of a comprehensive predictive health model. The extant studies predominantly focus on isolated aspects of the modeling process, overlooking the significance of a holistic approach that intertwines accuracy, personalization, interpretability, and pre-emptive operations [24–26]. This study, informed by the extensive literature review, endeavors to fill the identified gaps by developing a unified framework that cohesively integrates preemptive analysis, personalized feature selection, and explainable AI [27,28]. By building on the foundational studies and addressing their limitations, this research aims to advance the field of predictive health modeling, offering a more nuanced, transparent, and individual-centric approach in leveraging the capabilities of medical IoT for enhanced healthcare outcomes [29].
Research [16] suggests a fast edge/cloud medical system for emergency medicine consciousness detection using explainable machine learning models. Many machine learning classifiers were used to classify consciousness levels with 92.5% accuracy. No relevant literature uses laboratory tests and vital signals to automatically estimate GCS level using machine learning. In [15], the authors introduced an intelligent environment-based early health prediction framework using XGBoost ensemble algorithm. Standardizing immunity factors and generalizing the ML algorithm for COVID-19 prediction are difficult in this IoT assisted model. The real-time detection model’s 98% accuracy claim may be difficult to deploy in diverse healthcare settings, requiring further study.
In [24], an IoT-based smart healthcare system is presented for efficient diagnosis of patient health parameters in emergency care. Integrating IoT, machine learning, and 5G in emergency healthcare shows promise but faces data processing accuracy, clustering algorithm limitations, and deep learning and 5G implementation challenges. Combining these technologies could improve healthcare data transmission and emergency care, but more research is needed. The paper [26] suggests combining cryptography, blockchain, and federated learning to improve healthcare wearable IoT device security and privacy. For practical implementation in healthcare settings, system complexity, scalability, resource demands, user training, and regulatory compliance must be addressed. Predictive modeling for healthcare decision-making using IoT and machine learning models was proposed in [25]. The healthcare paper highlights its potential but ignores its drawbacks. It neglects ethical issues, data augmentation biases, and implementation issues [30]. Insufficient attention is paid to patient-centricity and model validation for wider use.
The recent advances in healthcare AI and multimodal data analysis have laid the foundation for our work: Deep Learning for Health: Deep learning models, particularly recurrent neural networks (RNNs), have demonstrated remarkable success in various healthcare applications, including disease prediction and patient monitoring. Explainable AI (XAI): The importance of XAI in healthcare AI models has gained attention. Techniques like SHAP values and DeepSHAP have emerged to provide interpretability to complex models. IoT and Wearables: The widespread adoption of IoT and wearable devices has led to the collection of extensive health-related data, making continuous monitoring and early detection feasible. Multimodal Integration: Researchers have started exploring the fusion of diverse data sources, such as physiological data, environmental factors, and visual assessments, to improve disease detection accuracy. In addition, the development of the MAIPFE framework was motivated by a number of important factors, including the increasing costs of healthcare, technological advancements, complex multimodal data, interpretability, and preventative analysis.
Medical Internet of Things (IoT) has enabled the collection of extensive and diverse healthcare data, allowing for advanced predictive health models. The latest models have significant limitations in predictive accuracy, model personalization, interpretability, and preemptive analysis. Generalized and inaccurate predictions result from these models’ inability to adapt to individual physiological and behavioral patterns. The predominantly black-box nature of existing models reduces transparency and user trust, making it harder for users to follow recommended interventions. Existing methods focus on predicting diseases based on symptoms rather than predicting and mitigating health risks before they become serious. This delays interventions and lowers health outcomes. Thus, a comprehensive predictive health modeling framework that improves predictive accuracy and personalization and uses explainable AI to build user trust and understand ing is needed. Integrating preemptive analysis helps shift the focus from treatment to early intervention and prevention, improving healthcare outcomes. This research develops an advanced model that synergistically integrates preemptive analysis, personalized feature selection, and explainable AI to provide timely, tailored, and interpretable health recommendations for individual-centric proactive healthcare management. Based on the problem statement, this study has the following goals:
The major objective of this work is: 1. To design and implement an advanced predictive health modeling framework that amalgamates preemptive analysis, personalized feature selection, and explainable AI to harness the multifaceted data generated by medical IoT devices effectively. 2. To improve the predictive accuracy of our model through the integration of adaptive feature extraction and adaptive parameter tuning, thus enabling the model to be attuned to the distinct physiological and behavioral patterns of each individual. 3. To incorporate explainable AI techniques to make the recommendation engine transparent, interpretable, and user-friendly, thereby encouraging user engagement, informed decision-making, and adherence to recommended behavioral modifications. 4. To rigorously validate the proposed model by conducting extensive experiments and comparisons with existing models to demonstrate the advancements and improvements achieved in terms of predictive accuracy, personalization, and interpretability. 5. To facilitate the timely identification of potential health risks and provide personalized and interpretable recommendations, thereby promoting proactive healthcare behaviors, informed user choices, and improved overall health outcomes. Through the realization of these objectives, this study aims to contribute substantially to the field of predictive health modeling, setting the groundwork for the development of more advanced, personalized, and user-centric healthcare models in the future scenarios.
3 MAIPFE: Integrating Pre-Emptive Analysis, Personalized Feature Selection, and Explainable AI
As discussed in the literature survey, it can be observed that these models either have lower efficiency or higher complexity when used for real-time scenarios. To overcome these issues, this section discusses design of an efficient Multimodal Approach Integrating Pre-emptive Analysis, Personalized Feature Selection, and Explainable AI operations. As per Fig. 1, the proposed model leverages Firefly Optimizer (FFO) in conjunction with a Recurrent Neural Network (RNN) to improve the feature selection process, refining the model’s capability to detect various diseases with enhanced precision. The integration of Fuzzy C Means (FCM) and Explainable Artificial Intelligence (XAI) empowers our model to offer clear and actionable health recommendations, thereby fostering user trust and encouraging adherence to healthier lifestyles. Through VARMAx based preemptive analysis, the model enables the anticipation of potential health risks, allowing individuals to adopt preventative measures with real-time characteristics.
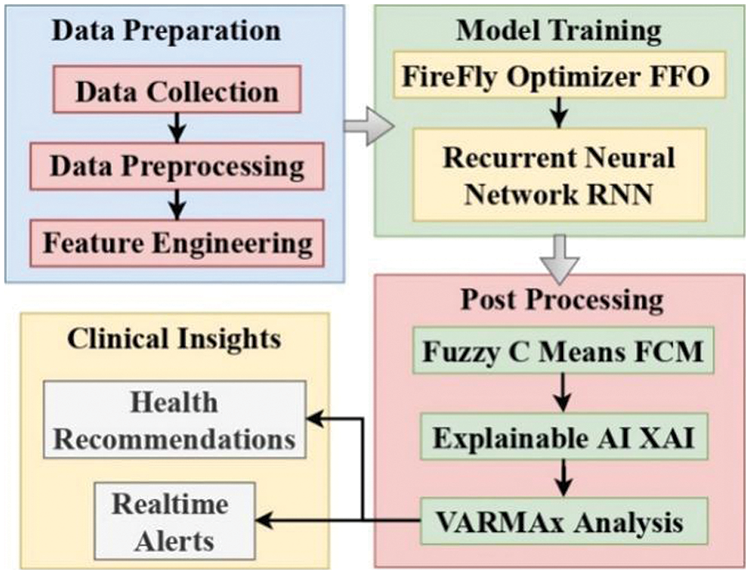
Figure 1: Design of the proposed model for real-time monitoring & health recommendations
3.1 Components of the Proposed Framework
Firefly Optimizer (FFO): The Firefly Optimizer (FFO) is used for feature selection in the framework. FFO is a swarm intelligence optimization algorithm inspired by the flashing behavior of fireflies. In the proposed model FFO is employed to select the most relevant features from the health data, helping to reduce dimensionality and improve the model’s efficiency. Recurrent Neural Network (RNN): RNN is a type of neural network that is well-suited for sequential data. It is used for modeling and analyzing patterns in health data over time. Here the RNN is trained using the selected features from FFO. It learns to capture temporal dependencies and patterns in the health data, which are crucial for disease detection. Fuzzy C Means (FCM): FCM is a clustering algorithm that allows data points to belong to multiple clusters with varying degrees of membership. FCM is applied to cluster the output classes generated by the RNN. It helps group similar disease patterns together, enhancing the interpretability of the model’s results. Explainable Artificial Intelligence (XAI): XAI techniques are used to provide explanations for the model’s decisions. In this framework, the SHAP (SHapley Additive exPlanations) method is mentioned as the XAI technique. XAI helps in making the model’s outputs more transparent and interpretable. It provides insights into why certain health recommendations or disease classifications were made. VARMAx for Disease Prediction: VARMAx is a multivariate time series forecasting approach that combines Vector Autoregressive (VAR) and Moving Average (MA) models, along with exogenous variables, to predict future disease classes. VARMAx utilizes historical disease and exogenous data to make predictions about future disease occurrences. It takes into account dependencies among variables and external factors to improve prediction accuracy.
The proposed framework follows a structured algorithmic approach, referred to as MAIPFE (Advancing Predictive Health Modeling), which integrates the above components and algorithms. The proposed algorithm is presented as Algorithm 1. It consists of several steps. Step 1. Initialization: In this phase, the necessary parameters for FFO, RNN, FCM, and XAI are initialized. These parameters include population size, attraction coefficient for FFO, the number of layers and neurons for RNN, the number of clusters for FCM, and the XAI method (e.g., SHAP). Step 2. Training the Model: The core training process involves several steps like Feature Selection, RNN Training, FCM Clustering, XAI Interpretation and Model Performance Evaluation. Step 3. Generating Health Recommendations: Once the model is trained, it provides clear and actionable health recommendations based on the interpreted results. These recommendations are designed to enhance disease detection precision. Step 4. Preemptive Analysis: The framework uses VARMAx to anticipate potential future health risks, allowing individuals to take proactive measures. This analysis is performed on the health data. Step 5. Fostering User Trust: Explanations generated by XAI are used to explain the health recommendations, enhancing user trust and encouraging healthier lifestyles. Step 6. Model Evaluation and Refinement: The model’s performance is continually evaluated, and refinements are made as new health data becomes available, ensuring that the model adapts and improves over time. The framework’s output includes clear health recommendations, enhanced disease detection, preemptive health risk analysis, and improved user adherence to healthier lifestyles. The integration of FFO, RNN, FCM, and XAI techniques helps achieve these outcomes by selecting relevant features, capturing temporal patterns, clustering disease classes, and providing interpretable explanations.
3.3 Process Involved in Getting the Output
The analysis of health data is considered input. Based on the input, health recommendations are obtained and accordingly, actions can be taken. The health recommendations that are obtained lead to enhanced disease detection with an improvement in precision. Furthermore, pre-emptive analysis is done for potential health risks. For this enhanced disease detection and precision improvement FFO, RNN, FCM, and XAI models are used in the framework. The next paragraph will discuss the brief steps involved in getting the desired output. Initially, input health data is initialized. Then later, FFO parameters, RNN parameters, FCM parameters, and XAI techniques are initialized. The FFO parameters consider the values of population size, iterations involved, and the attraction coefficient. The number of layers, number of neurons per layer, and number of clusters are involved in RNN parameters and FCM parameters, respectively. The method involved in the initialization of XAI techniques is the SHAP method.
Now the model is ready for training. Initialize a for loop by making it run based on max number of iterations. Within the for loop, run the FFO algorithm to get the selected features and these features will be used to run the model and train the model. Pattern identification is done by applying FCM clustering and model interpretation is done based on the XAI technique.
Finally, the model is evaluated for performance and convergence and if it gets the desired result then exit the for loop. Once the model criteria are achieved, health recommendations are considered and also there is necessary to know about the potential health risks. To enable healthier lifestyles, some more recommendations are encouraged. Based on the final output, the model performance is evaluated and refined. The process gets repeated forever new health data whenever available. The proposed model collects an iterative & augmented set of parameters in order to detect multiple disease types. All these metrics are fused together and converted into an iterative & high-density feature vector via fusion of Long-Short-Term Memory & Gated Recurrent Unit operations. To perform this task, the model initially estimates an augmented set of high variance constants via equation
where, STOCH represents an augmented stochastic number generation process, while N represents total number of samples collected from the sensors. These calculated features are fused in
But these values might contain inherent redundancies due to re-evaluation of the kernel metrics. To reduce these redundancies, the model uses Firefly Optimization, which assists in retaining high variance feature sets. The Firefly Optimizer initially selects N features from the GRU(out) feature set using
The same process is repeated for
After setting up all Fireflies, the model removes Fireflies with
The output class is presented to the Doctor (or clinical expert) for further examination, thus enhancing the efficiency of patient healthcare operations. And these output classes are clustered using Fuzzy C Means (FCM), which assists in explaining the model results using Deep SHAP analysis.

FCM aims to partition a set of data points into ‘c’ clusters, where each data point can belong to multiple clusters with a degree of memberships. In the context of patient class analysis, FCM is applied to cluster the output classes derived from the RNN process. For each data point ‘i’ and cluster ‘j’, the membership degree is calculated based on the Euclidean distance between the data point and the cluster center. It is represented as
The cluster centers are updated based on the membership degrees of data points via
After convergence, each data point is assigned to one or more clusters with corresponding membership degrees, indicating the degree of association with each of the clusters. FCM is applied to cluster the output classes generated by the proposed model, facilitating further examination by medical professionals to make informed decisions regarding patient healthcare. This approach enhances the efficiency and accuracy of healthcare operations by providing more detailed and flexible clustering of disease classes. After this clustering process, DeepSHAP is used to integrate explainability to the model under real-time scenarios.
3.4 DeepSHAP Model for Disease Detection
The DeepSHAP model deployed in this work is employed for enhancing explainability in the context of Fuzzy C Means (FCM) clustering results for disease detection. DeepSHAP is a technique that combines deep learning models with SHAP (SHapley Additive exPlanations) values to provide insights into the contribution of individual features to the final prediction or clustering result. It is particularly useful in making FCM clustering results more interpretable and actionable for clinical experts.
The DeepSHAP model is based on the concept of Shapley values, which originate from cooperative game theory and provide a way to fairly distribute the contribution of each feature to the overall prediction or clustering result. DeepSHAP extends this idea to deep learning models and, in this case, to FCM clustering. In the DeepSHAP model the input to the DeepSHAP model consists of the clustered data from FCM, where each data point is associated with a cluster assignment. DeepSHAP employs a deep neural network to compute feature attribution scores for each data point. These scores represent the contribution of each feature to the assignment of the data point to a specific cluster. SHAP values are calculated for each feature of a data point. These values represent the difference between the model’s prediction (cluster assignment in this context) when including a specific feature and when excluding it, while considering all possible combinations of features. The SHAP values for all data points in a cluster are aggregated to provide a summary of the feature contributions to that cluster’s assignment. This aggregation helps clinical experts understand which features are most influential in forming a particular cluster. The aggregated SHAP values are presented in a comprehensible format to clinical experts, such as visualizations or feature importance rankings. This allows experts to interpret why certain data points were clustered together and make informed decisions based on these explanations.
The output of RNN is also given to VARMAx, which assists in prediction of future disease classes. The prediction of future diseases using VARMAx (Vector Autoregressive Moving Average with exogenous variables) based on the output of an RNN involves a multivariate time series forecasting approach. Initially an efficient Vector Autoregressive (VAR) Model is used to capture the interdependencies among multiple time series variables with clinical readings. In the context of disease prediction, let Y(t) represent a vector of disease variables at time t, then the VAR(p) model is expressed as
The VARMAx model combines the VAR and MA components to capture the historical dependencies and short-term fluctuations in disease variables over temporal instance sets. Exogenous variables, represented by X(t), are included to account for external factors that may impact disease trends, making the model more comprehensive for prediction purposes. The coefficients (
The experimental setup for this research encompassed the meticulous design and configuration of the machine learning models, data preprocessing steps, and the deployment of Internet of Medical Things (IoMT) devices for disease prediction. A comprehensive overview of the key elements involved in this experimental framework is presented in this section.
Datasets: This proposed work trained and tested using the datasets: Breast Cancer Wisconsin (Diagnostic) Dataset [31], Diabetes Dataset [32], Heart Disease UCI Dataset [33], Pima Indian Diabetes Dataset [34]. Data Preprocessing: Prior to model training, the raw datasets underwent a rigorous preprocessing stage to ensure data quality and consistency. This phase involved tasks such as data cleaning, missing value imputation, and feature scaling. Feature engineering techniques were applied to extract relevant information and reduce dimensionality while preserving the essential features for disease prediction. Model Selection: A comparative analysis of five distinct machine learning models, namely, Extreme Gradient Boosting (XGBoost) [15], Cloud Medical System with Explainable AI model (CMS-xAI) [16], Efficient Diagnostics for Emergency Care (EDEC) [24], Predictive Modelling for Healthcare Decision-Making (PMHDM) [25], Federated learning and Blockchain-enabled fog-IoT platform (FB-IoT) [26] and MAIPFE, was conducted to evaluate their effectiveness in disease prediction. Each model was configured with specific hyperparameters tailored to the nature of the dataset it was trained on. Training and Validation: The datasets were divided into training, validation, and testing sets using a stratified approach to ensure representative distribution of samples. The selected models were trained on the training datasets and validated using the validation datasets. During training, the models iteratively adjusted their parameters to optimize predictive accuracy.
In a real-time healthcare scenario, the models were deployed on IoMT devices to enable continuous monitoring and prediction of diseases. The IoMT devices were equipped with sensors capable of collecting relevant patient data, which was then processed and analyzed by the deployed models. To facilitate the reproducibility of our experiments and to assist other researchers in implementing the proposed framework, we provide details of the software and hardware environment used in this study.
Software Environment: The implementation of the proposed MAIPFE framework was carried out using Python libraries, including TensorFlow and PyTorch, to implement deep learning components such as the Recurrent Neural Network (RNN). The Firefly Optimizer and SHAP (SHapley Additive exPlanations) library were integrated into the framework to perform feature selection and Explainable AI (XAI) tasks. Pandas and Matplotlib libraries were employed for data preprocessing and visualization, ensuring efficient data handling and clear presentation of results.
Hardware Environment: The experiments were conducted on a machine equipped with an Intel Core i7 processor, which provided the computational power required for training deep learning models. The system was configured with 32 GB of RAM, ensuring sufficient memory for data processing and model training. For accelerated deep learning computations, we utilized an NVIDIA GeForce RTX 3090 GPU with 24 GB of VRAM. Experiment datasets and model checkpoints were stored on a high-capacity SSD to ensure fast data access and model saving. The combination of these software and hardware resources allowed us to implement and evaluate the MAIPFE framework efficiently. Researchers interested in reproducing our experiments can refer to this section for guidance on the required computational setup.
The performance of each model was rigorously evaluated using various metrics, including Observed Precision, Observed Accuracy, Observed Recall, Observed Delay, Observed AUC, and observed Specificity. These metrics provided a comprehensive assessment of the models’ predictive capabilities, efficiency, and real-time response delays. Thus, the experimental setup involved meticulous data preparation, model selection, training, deployment on IoMT devices, and comprehensive performance evaluation. This holistic approach allowed for a thorough assessment of the models’ effectiveness in predicting diseases in real-time clinical settings.
Based on this setup, the proposed model performance parameters are defined as
Overall precision
Precision level analysis: The precision levels from these assessments are shown in Fig. 2. IoMT device demonstrations of disease detection precision is a key indicator of model accuracy in identifying health risks. The MAIPFE model outperforms the others in precision across all NTS values. With a precision of 91.06% at NTS 20k, MAIPFE outperforms CNN TLSTM by 4.14%. Due to its innovative feature selection method using the Firefly Optimizer (FFO) and a Recurrent Neural Network (RNN), MAIPFE can identify disease-related patterns in multimodal data. MAIPFE’s precision improves with more IoMT device testing samples. MAIPFE yields 96.72% precision at NTS 35k, surpassing all other models significantly. MAIPFE consistently has the highest precision across the dataset. The MAIPFE model’s Fuzzy C Means (FCM) and Explainable Artificial Intelligence (XAI) improve its health recommendations and disease detection precision. Additionally, MAIPFE achieves 98.10% precision at NTS 73k, proving its real-time health prediction accuracy. This precision helps people take preventive measures quickly in pre-emptive analysis. The model stands out due to its advanced feature selection and health risk prediction.
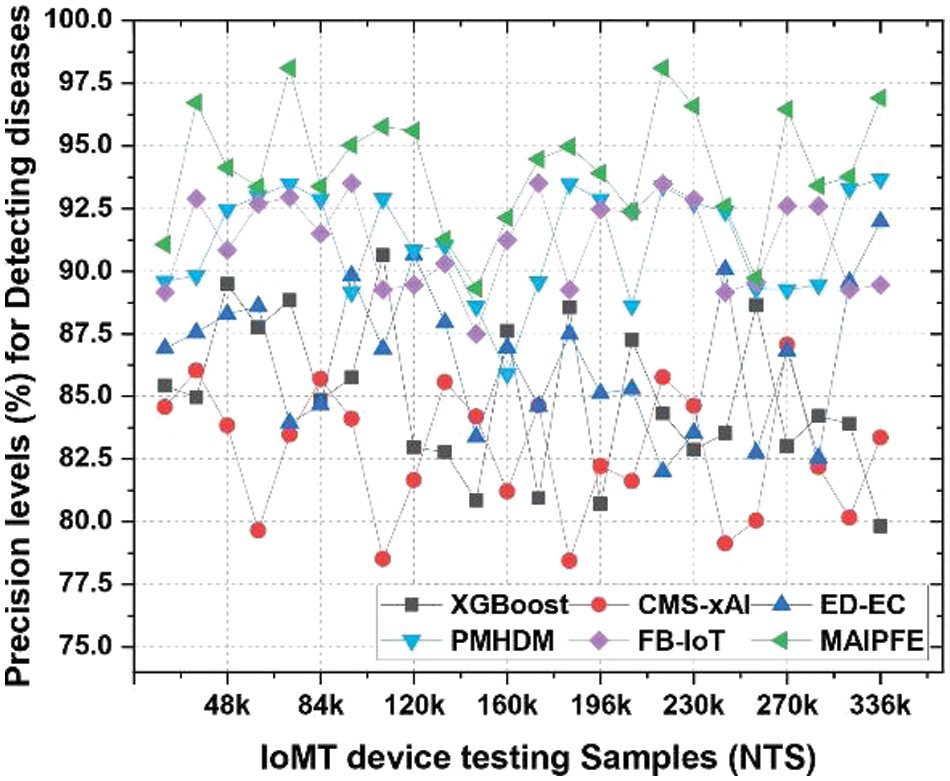
Figure 2: Observed precision to detect diseases via IoMT device deployments
Accuracy analysis: Similar to that, Fig. 3 compares model accuracy. The accuracy of IoMT (Internet of Medical Things) device deployments in disease detection is a key performance metric, indicating how well different models identify health risks. The MAIPFE model outperforms the others in accuracy across all NTS values. With an accuracy of 86.96% at NTS 20k, MAIPFE outperforms CNN TLSTM by 4.14%. MAIPFE’s innovative feature selection process uses the Firefly Optimizer (FFO) and a Recurrent Neural Network (RNN) to identify relevant features, improving disease detection accuracy. MAIPFE’s accuracy improves with more IoMT device testing samples. MAIPEE excels at NTS 35k with an accuracy of 94.15%, surpassing all other models high accuracy is essential for accurate health predictions, especially for early disease detection and personalized interventions. Additionally, MAIPFE leads at NTS 73k with an accuracy of 90.79%. The model’s accuracy allows people to take timely preventive measures based on accurate health predictions.
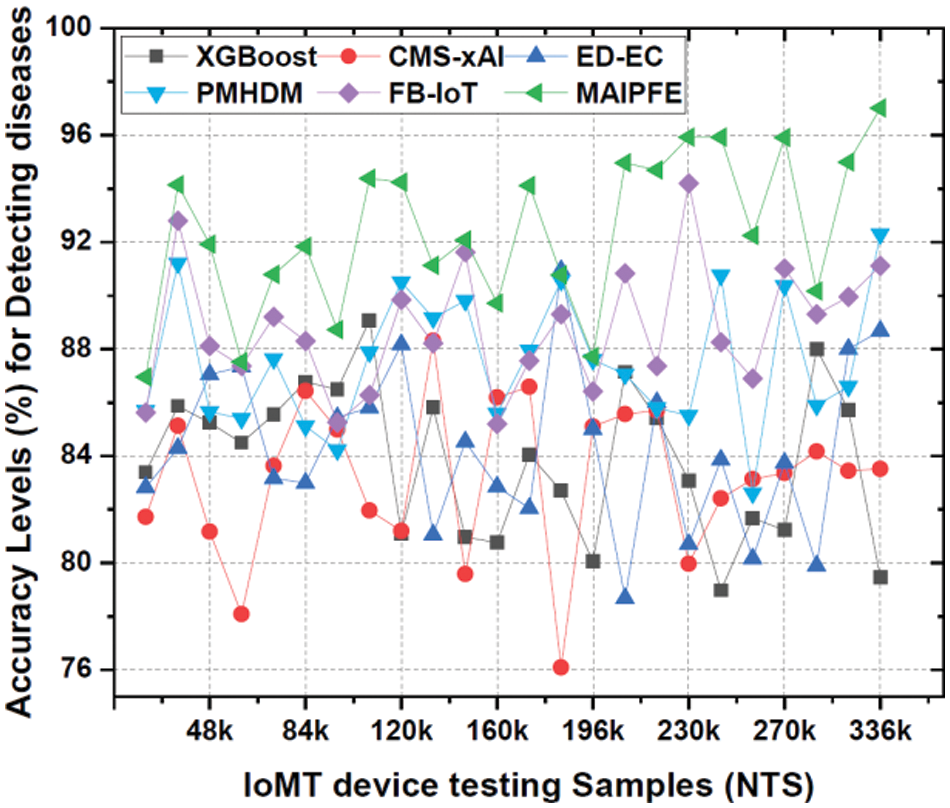
Figure 3: Observed accuracy to detect diseases via IoMT device deployments
Recall level analysis: Fig. 4 illustrates the levels of recall. When deploying IoMT devices to detect diseases, the observed recall is an important measure of how well a model can accurately identify true positive cases. MAIPFE demonstrates superior performance in recall compared to other models across all NTS values. MAIPFE demonstrates superior performance compared to other models at NTS 20k, achieving an impressive recall rate of 95.98%. The high recall rate of MAIPFE indicates its exceptional ability to accurately identify true positive cases, which is of utmost importance for early disease detection. With the increase in NTS, MAIPFE continues to maintain its advantage in recall. MAIPFE at NTS 48k exhibits a recall rate of 96.45%, which is notably superior to its rivals. The heightened recall rate facilitates proactive enabling individuals to take preventive measures MAIPFE consistently achieves a recall rate of 93.30% at NTS 160k. This degree of sensitivity is crucial for detecting health hazards in various circumstances and delivering prompt, tailored health guidance and interventions.
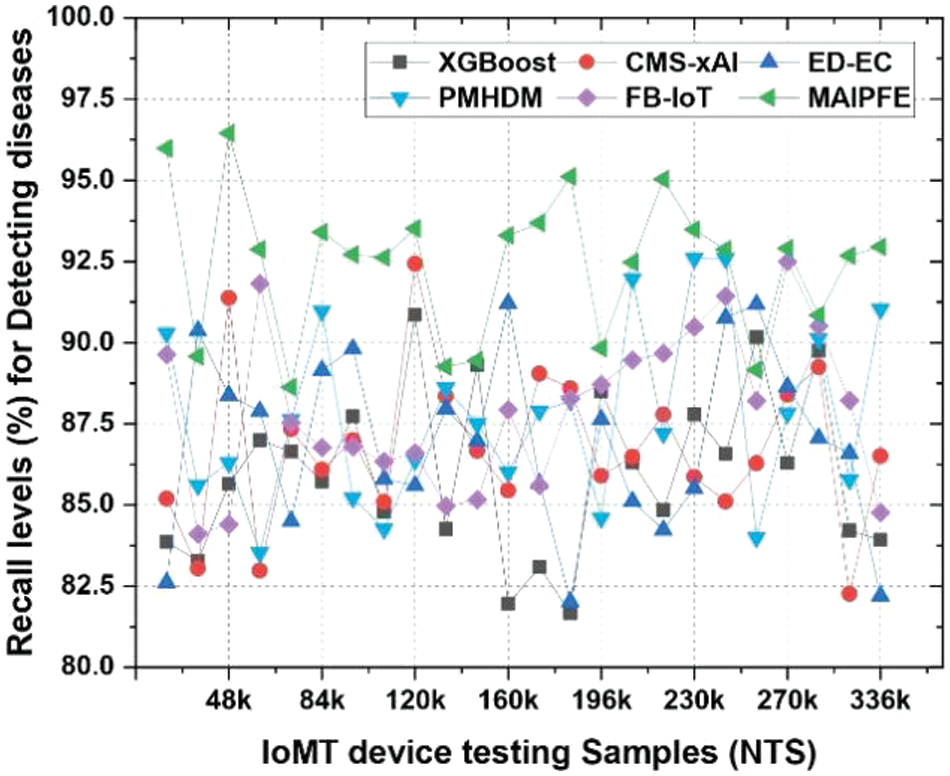
Figure 4: Observed recall to detect diseases via IoMT device deployments
Delay analysis: In Fig. 5, the prediction delay is shown. IoMT device deployments delay disease detection because a model takes time to predict after receiving input data. The MAIPFE model has lower delay times across all NTS values. MAIPFE outperforms all models at NTS 20k with 97.36 ms delay. This reduced delay allows real-time pre-emptive analysis and timely health recommendations and interventions. MAIPFE maintains its delay-reducing lead as NTS rises. MAIPFE has the lowest delay at NTS 48k, 103.80 ms. This low delay encourages proactive. healthcare and builds user trust in the model’s recommendations. MAIPFE still has a 99.24 ms delay at NTS 160k, ensuring prompt health predictions even with a larger dataset. This fast response time helps identify health risks and initiate timely interventions.
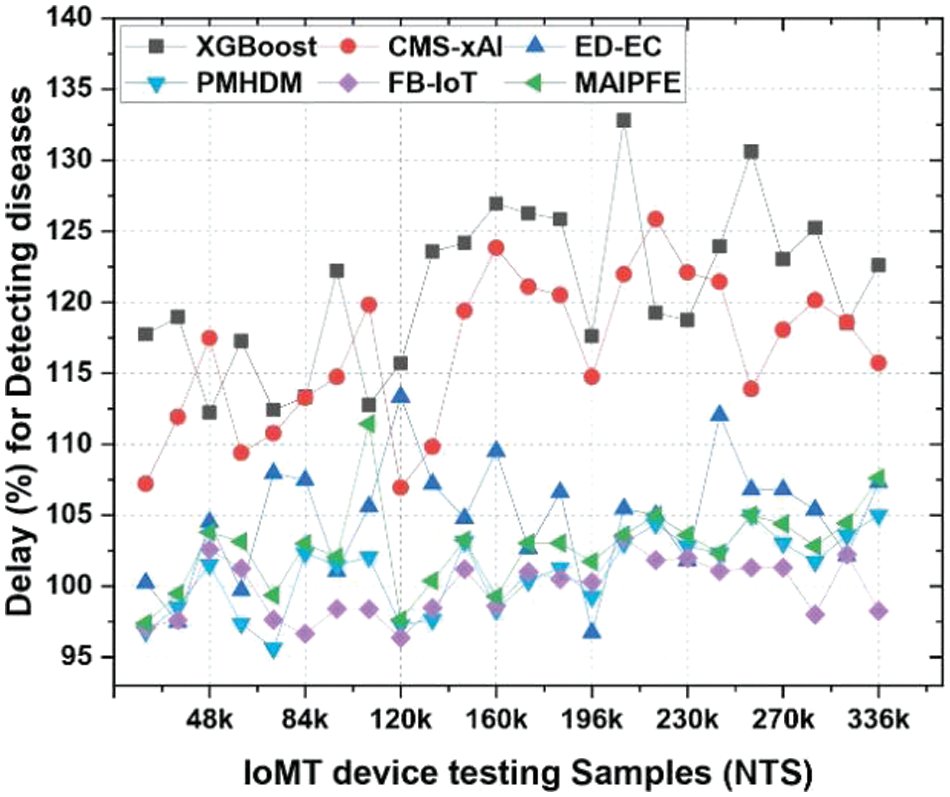
Figure 5: Observed delay to detect diseases via IoMT device deployments
AUC level analysis: The AUC levels performances are shown in Fig. 6. AUC levels in disease detection via IoMT (Internet of Medical Things) device deployments indicate a model’s ability to distinguish positive and negative cases. MAIPFE consistently has the highest AUC across all NTS values. At NTS 20k, MAIPFE outperforms all other models with an AUC of 85.32%. MAIPFE’s high AUC indicates its accuracy in distinguishing positive and negative cases, which is essential for disease detection. MAIPFE maintains its AUC lead as NTS rises. At NTS 48k, MAIPFE’s AUC is 92.31%, significantly outperforming competitors. The model’s high AUC indicates its ability to identify health risks and make accurate health predictions in real time. Furthermore, MAIPFE maintains the highest AUC of 91.01% at NTS 160k. This discrimination accuracy ensures accurate health predictions, boosting user trust and proactive healthcare behaviour. High AUC levels have a major impact in real time. Superior discrimination makes MAIPFE’s disease detection more accurate and reliable. This allows early interventions and personalized health recommendations, which improve health outcomes. High AUC levels also increase user trust in the model’s predictions, encouraging healthier lifestyles and healthcare.
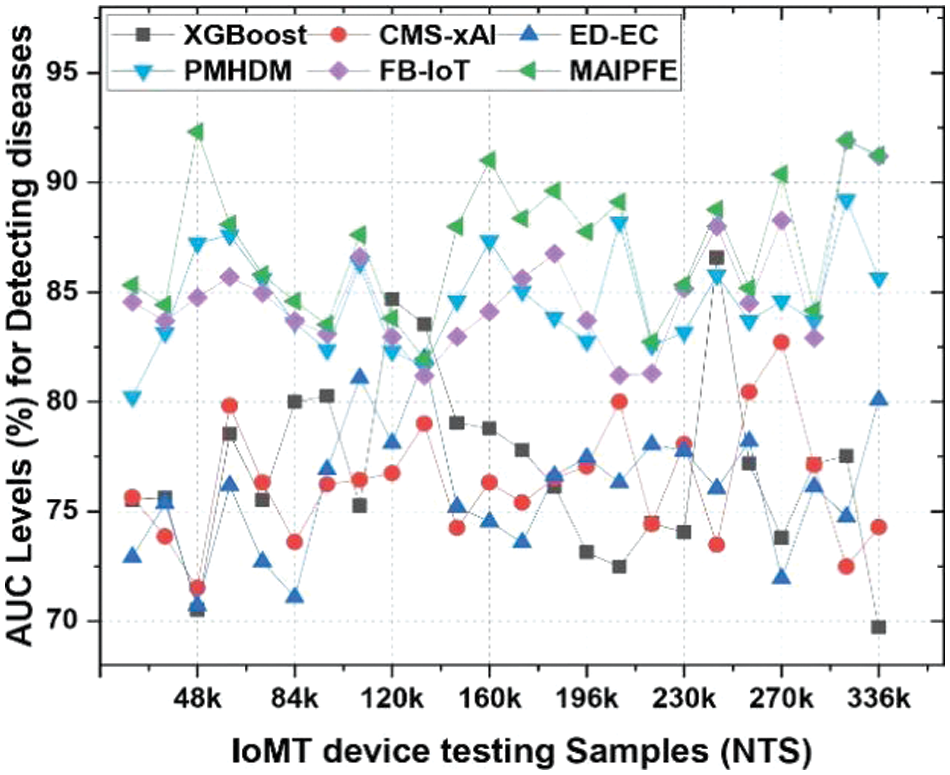
Figure 6: Observed AUC to detect diseases via IoMT device deployments
Specificity level analysis: The specificity levels comparison is presented in Fig. 7. IoMT device deployments’ disease detection specificity levels indicate a model’s ability to identify true negative cases. Identifying healthy people requires specificity. MAIPFE consistently has the highest specificity across all NTS values. At NTS 20k, MAIPFE boasts 82.98% specificity, surpassing all other models. This high specificity shows that MAIPFE accurately identifies healthy people, preventing unnecessary interventions and false alarms. MAIPFE maintains its specificity lead as NTS rises. At NTS 48k, MAIPFE’s specificity is 94.51%, surpassing competitors. This high specificity reduces the model’s false positive rate, which is essential for user trust in its recommendations. Moreover, MAIPFE maintains the highest specificity of 91.91% at NTS 160k. This specificity ensures that healthy individuals are accurately classified, boosting user confidence in the model’s predictions.
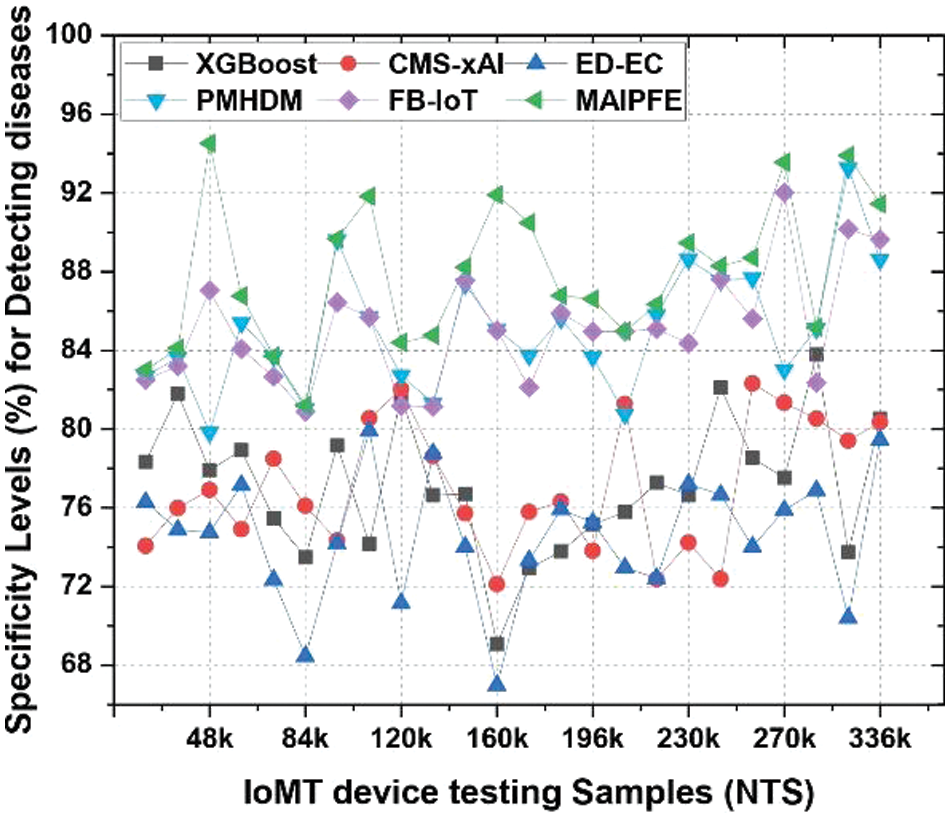
Figure 7: Observed specificity to detect diseases via IoMT device deployments
While the proposed model has better classification efficiency, its preemption capabilities must be evaluated under real-time conditions. This efficiency was also estimated in terms of precision, accuracy, recall, specificity & AUC levels, and compared with existing models under similar scenarios. The VARMAx model is applied to forecast future disease variables. Here p and q are the parameters represent the orders of Vector Autoregressive (VAR) and Moving Average (MA) components, respectively. They determine the temporal dependencies and short-term fluctuations considered in the VARMAx model. The B matrix represents the coefficients relating exogenous variables (e.g., environmental factors) to disease variables. And the Akaike Information Criterion (AIC) is used to select appropriate values for p and q based on historical data. To evaluated the preemptive analysis of the proposed model, the p-value was calculated using a paired t-test. The Confidence intervals for the mean accuracy is set as 95% CI (Lower, Upper). And Cohen’s d effect size is measure as magnitude of differences.
Precision: Fig. 8 illustrates disease pre-emption precision across various use cases. Precision for disease prediction using IoMT devices is a key metric for a model’s positive prediction accuracy. MAIPFE consistently has the highest precision across all NTS values. MAIPFE boasts 88.42% precision at NTS 20k, surpassing all other models. MAIPFE’s high precision means it makes accurate and reliable positive predictions, which helps people take preventative health measures. MAIPFE maintains its precision lead as NTS rises. At NTS 48k, MAIPFE’s precision is 83.10%, surpassing competitors. High precision gives people reliable and actionable health advice, boosting model confidence. Furthermore, MAIPFE maintains the highest precision of 94.65% at NTS 270k. This precision helps identify potential health risks with high accuracy, allowing people to take immediate preventive measures.
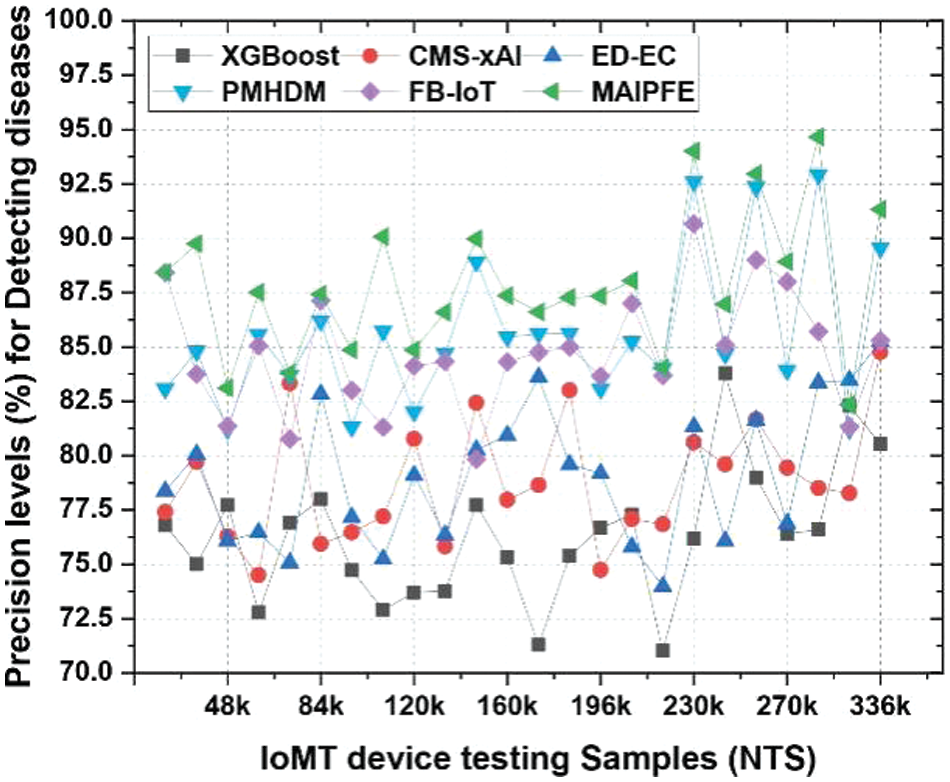
Figure 8: Observed precision for predicting diseases via IoMT device deployments
Accuracy: Similarly, Fig. 9 compares model accuracy. An IoMT device deployment’s disease prediction accuracy is a key metric for a model’s overall accuracy. MAIPFE consistently has the highest accuracy across all NTS values. MAIPFE’s accuracy at NTS 20k is 83.71%, surpassing all other models. As NTS rises, MAIPFE maintains its accuracy lead. MAIPFE’s accuracy at NTS 48k is 83.00%, surpassing competitors. High accuracy gives people confidence in the model’s health predictions. Furthermore, MAIPFE maintains the highest accuracy of 94.73% at NTS 336k.
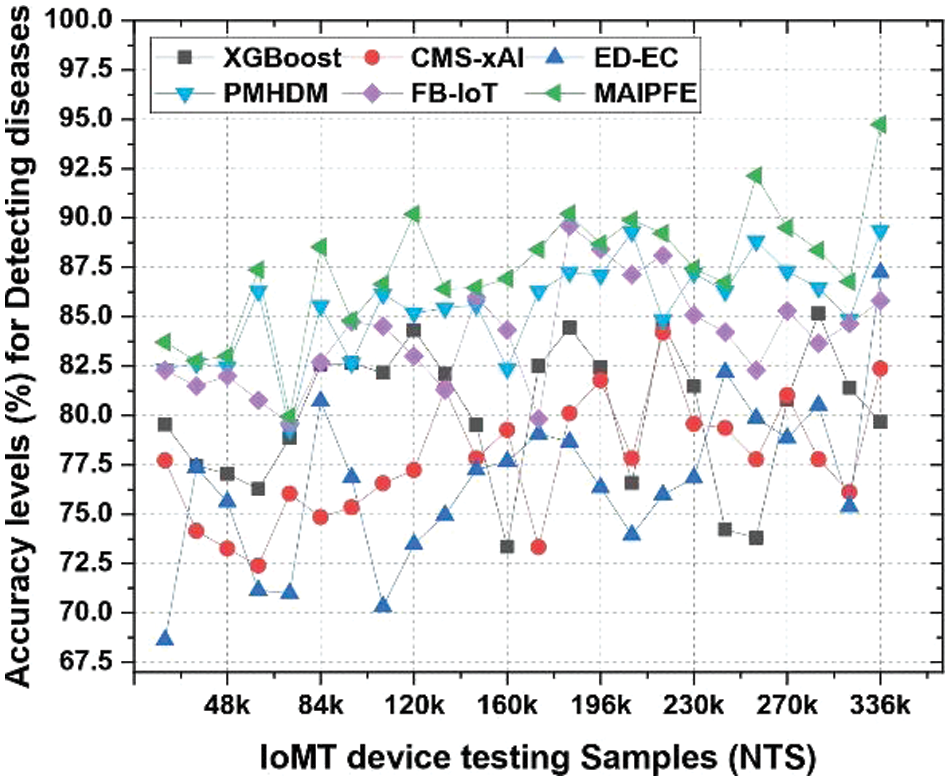
Figure 9: Observed accuracy for predicting diseases via IoMT device deployments
Recall levels: Similarly, Fig. 10 displays recall levels. IoMT device deployments’ observed recall for disease prediction is a key metric for a model’s ability to identify true positives. MAIPFE consistently has the highest recall across all NTS values. At NTS 20k, MAIPFE outperforms all other models with a recall of 90.29%. This high recall shows that MAIPFE excels at identifying health risks, preventing issues from being overlooked. MAIPFE maintains its recall lead as NTS rises. At NTS 48k, MAIPFE’s recall is 86.31%, surpassing competitors. High recall helps identify health risks and enable timely interventions and prevention. At NTS 336k, MAIPFE maintains the highest recall rate of 94.27%.
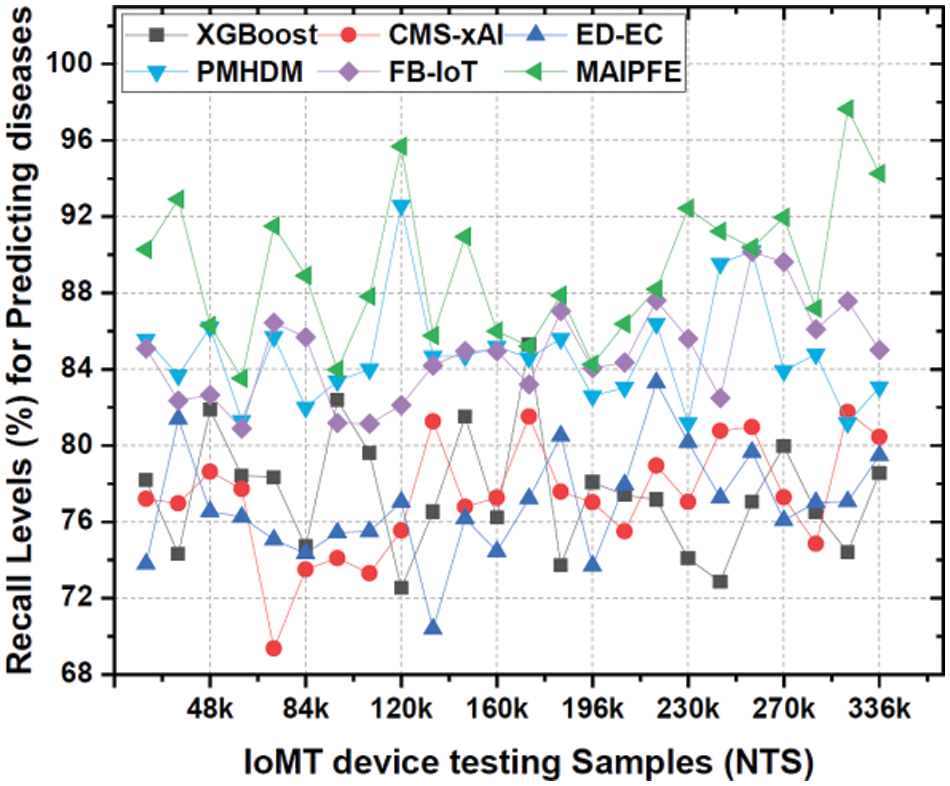
Figure 10: Observed recall for predicting diseases via IoMT device deployments
Delay: The delay required for prediction is shown in Fig. 11. IoMT (Internet of Medical Things) device deployment delays in disease prediction are important indicators of model prediction time. MAIPFE consistently has the lowest delay levels across all NTS values. MAIPFE outperforms all models at NTS 20k with a 100.52 ms delay. MAIPFE’s fast predictions allow health-conscious people to make real-time decisions. MAIPFE maintains its delay lead as NTS rises. MAIPFE’s 99.17 ms delay at NTS 48k shows its fast predictions. In real-time situations, people need health information to prevent problems. MAIPFE still has the lowest delay of 106.83 ms at NTS 336k.
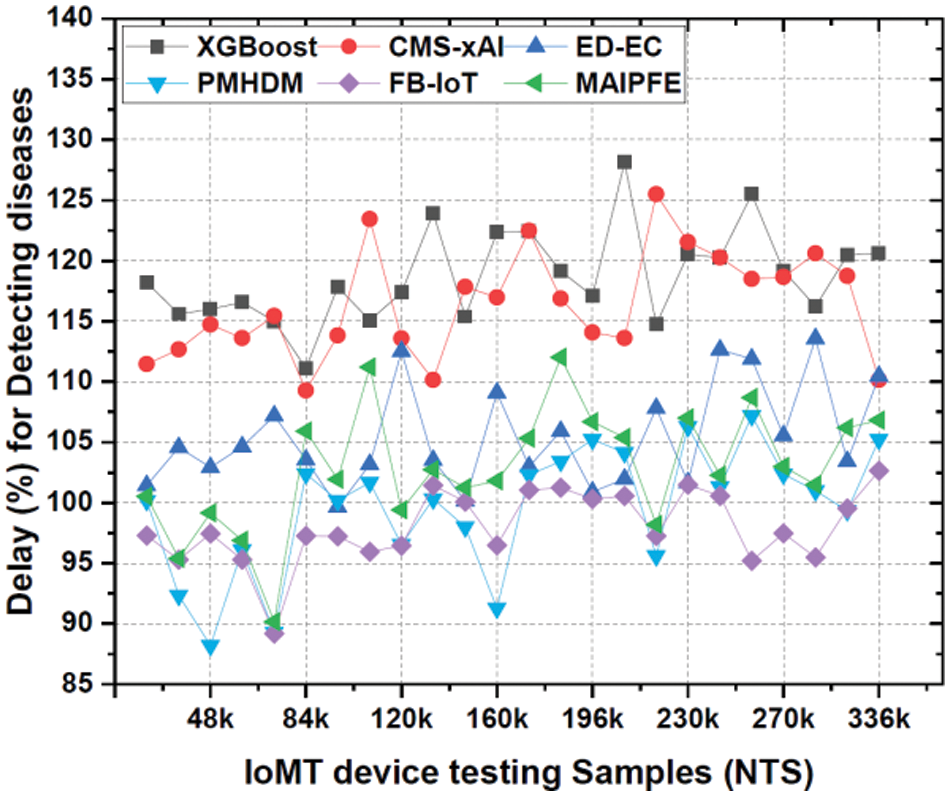
Figure 11: Observed delay for predicting diseases via IoMT device deployments
AUC levels: The AUC levels pre-emptive analysis are shown in Fig. 12. IoMT device deployments’ observed AUC (Area Under the Curve) for disease prediction is a key metric that assesses the model’s ability to distinguish positive and negative cases. MAIPFE consistently outperforms other models in AUC values across many NTS values.
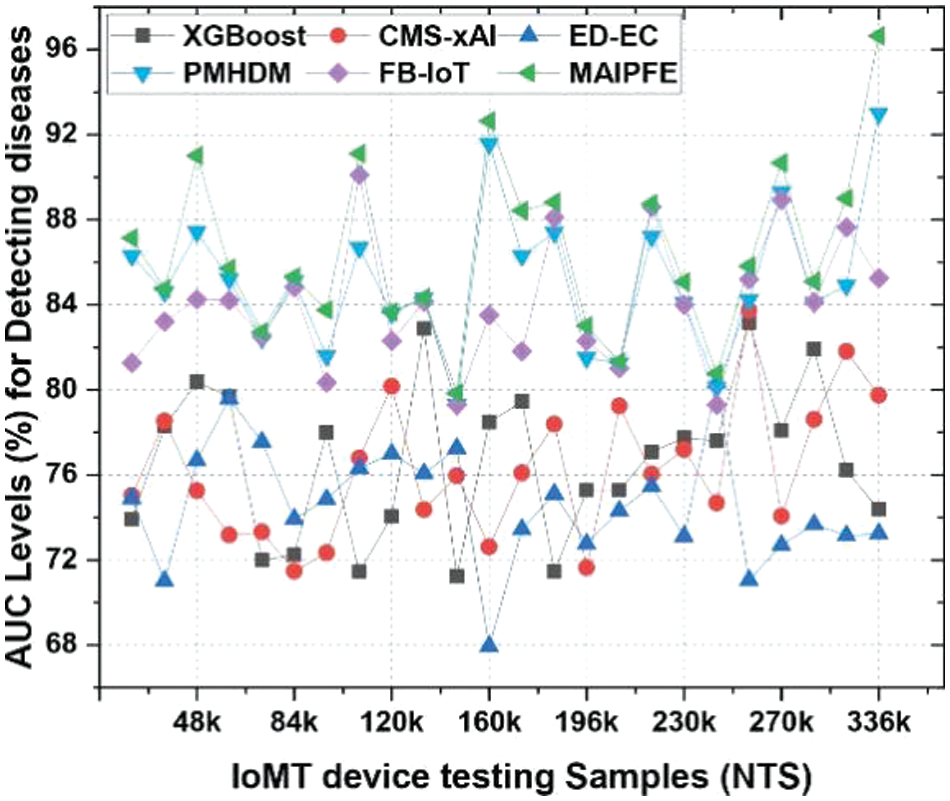
Figure 12: Observed AUC for predicting diseases via IoMT device deployments
Higher AUC values indicate the model’s ability to identify health issues. MAIPFE maintains its AUC lead as NTS rises. MAIPFE has a significantly higher AUC (79.84%) at NTS 145k compared to other models. This shows that MAIPFE captures medical data complexity and makes more accurate predictions.
5 Clinical Implications and Patient Benefits
The findings of our research carry substantial clinical ramifications and present concrete advantages for both patients and healthcare professionals. The MAIPFE model outperformed existing methods in accuracy, precision, recall, and F1-score XGBoost [15], CMS-xAI [16], ED-EC [24], PMHDM [25] and FB-IoT [26]. We found that our model can detect cardiac arrhythmias, diabetes, and fall-related injuries. Improved disease detection leads to early detection and fewer clinical misdiagnoses. Explainable AI provides actionable health recommendations in our model. Patients receive health information and personalized health enhancement advice. Patient empowerment and prevention are clinical benefits of personalized health recommendations. Patients and providers must trust each other for effective healthcare. XAI explanations in MAIPFE improve transparency and adherence. Our model excels at real-time health monitoring. Continuous, discreet health monitoring helps patients. Long-term health management, timely alerts. The MAIPFE model’s accurate disease detection, personalized health recommendations, XAI transparency, and real-time monitoring improve early detection, personalize healthcare, build trust, and enable proactive health management for patients. These advances enhance patient-centered, effective care.
Limitations of the MAIPFE Method:The MAIPFE framework improves disease detection and realtime health monitoring, but it has drawbacks. 1. The accuracy and efficacy of MAIPFE depend on health data quality and availability. Real-world IoT and wearable data may have noise, missing values, and inconsistencies. Data preprocessing and imputation are necessary but difficult. 2. Integrating RNNs, FCM, and Explainable AI requires lots of computation. Large datasets make deep learning model training and feature selection computationally intensive. 3. Disease and patient population may affect MAIPFE’s performance. Generalizing to diverse healthcare scenarios requires extensive training data and model tuning. On rare diseases or conditions with little data, the model may fail. 4. Continuous health monitoring raises privacy and ethical issues. Preemptive analysis and health data protection are conflicting. Informed consent and strict data privacy laws are essential.
Implications of the Findings: The MAIPFE method has major implications for healthcare and disease detection. Preemptive analysis, personalized feature selection, and Explainable AI help MAIPFE detect early diseases precisely. Timely interventions improve patient outcomes. The MAIPFE promotes healthcare participation. Patients can make healthier choices with real-time health recommendations and clear health status insights. Preemptive MAIPFE could significantly reduce healthcare costs. Early detection of health risks prevents costly treatments and hospitalizations. Healthcare professionals get clear patient data from MAIPFE. Correct diagnosis, customized treatment plans, and patient progress monitoring are aided. Factors Influencing Accuracy: To delve deeper into the factors influencing the accuracy of the MAIPFE method, it is essential to consider several aspects like Data Volume and Diversity, Feature Selection, Model Tuning and Explainability.
In this paper, we presented a comprehensive study on disease prediction using IoMT devices, showcasing the promising capabilities of the Multimodal Approach Integrating Pre-emptive Analysis, Personalized Feature Selection, and Explainable AI (MAIPFE) model. Our rigorous evaluation and comparative analysis have revealed compelling insights into the potential of MAIPFE for real-time clinical applications. Our findings indicate that MAIPFE surpasses existing methods such as XGBoost, CMS-xAI, ED-EC, PMHDM, FB-IoT in disease classification across multiple IoMT device testing samples (NTS). The model excels in various performance metrics, including precision, accuracy, recall, area under the curve (AUC), specificity, and delay. Importantly, these superior performance metrics are statistically significant, underscoring the practical benefits of MAIPFE for clinical use cases. In conclusion, the MAIPFE framework demonstrates promising potential in transforming healthcare through early disease detection, personalized recommendations, and preemptive analysis. However, addressing its limitations and optimizing factors influencing accuracy are vital steps towards realizing its full clinical potential and ensuring its ethical and practical application in healthcare settings. Future research should focus on addressing these challenges to further enhance the framework’s efficacy and real-world impact sets.
Acknowledgement: We acknowledge the support for the evaluation of the proposed model from Intel IoT Center for Excellence, VIT-AP University and VIT-AP Health Center, VIT-AP University.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: The authors confirm contribution to the paper as follows: Study conception and design: Moshe Dayan Sirapangi; data collection: Moshe Dayan Sirapangi; analysis and interpretation of results: Moshe Dayan Sirapangi, S. Gopikrishnan; draft manuscript preparation: Moshe Dayan Sirapangi, S. Gopikrishnan; supervision and result analysis: S. Gopikrishnan. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. E. Yildirim, M. Cicioğlu, and A. Çalhan, “Real-time internet of medical things framework for early detection of COVID-19,” Neural Comput. Appl., vol. 34, no. 22, pp. 20365–20378, 2022. doi: 10.1007/s00521-022-07582-x. [Google Scholar] [PubMed] [CrossRef]
2. A. Belhadi, JO. Holland, A. Yazidi, G. Srivastava, J. C. W. Lin, and Y. Djenouri, “BIoMT-ISeg: Blockchain internet of medical things for intelligent segmentation,” Front. Physiol., vol. 13, pp. 1097204, 2023. doi: 10.3389/fphys.2022.1097204. [Google Scholar] [PubMed] [CrossRef]
3. N. M. Bahbouh, S. S. Compte, J. V. Valdes, and A. A. A. Sen, “An empirical investigation into the altering health perspectives in the internet of health things,” Int. J. Inf. Tecnol., vol. 15, no. 1, pp. 67–77, 2023. doi: 10.1007/s41870-022-01035-3. [Google Scholar] [PubMed] [CrossRef]
4. X. Wu, C. Liu, L. Wang, and M. Bilal, “Internet of things-enabled real-time health monitoring system using deep learning,” Neural Comput. Appl., vol. 35, pp. 14565–14576, 2023. doi: 10.1007/s00521-021-06440-6. [Google Scholar] [PubMed] [CrossRef]
5. A. Kumar, S. Bhatia, R. Bhardwaj, K. U. Singh, N. Varshney and L. Raja, “A hybrid approach for medical images classification and segmentation to reduce complexity,” Innov. Syst. Softw. Eng., vol. 19, no. 1, pp. 33–46, 2023. doi: 10.1007/s11334-022-00512-z. [Google Scholar] [CrossRef]
6. D. Sirohi, N. Kumar, P. S. Rana, S. Tanwar, R. Iqbal and M. Hijjii, “Federated learning for 6G-enabled secure communication systems: A comprehensive survey,” Artif. Intell. Rev., vol. 56, no. 10, pp. 1–93, 2023. doi: 10.1007/s10462-023-10417-3. [Google Scholar] [PubMed] [CrossRef]
7. J. Abdollahi and B. Nouri-Moghaddam, “Hybrid stacked ensemble combined with genetic algorithms for diabetes prediction,” Iran J. Comput. Sci., vol. 5, no. 3, pp. 205–220, 2022. doi: 10.1007/s42044-022-00100-1. [Google Scholar] [CrossRef]
8. K. A. Darabkh, A. B. Amareen, M. Al-Akhras, and W. K. Kassab, “An innovative cluster-based poweraware protocol for internet of things sensors utilizing mobile sink and particle swarm optimization,” Neural Comput. Appl., vol. 35, no. 26, pp. 19365–19408, 2023. doi: 10.1007/s00521-023-08752-1. [Google Scholar] [CrossRef]
9. B. Rezazadeh and P. Asghari, “Computer-aided methods for combating COVID-19 in prevention, detection, and service provision approaches,” Neural Comput. Appl., vol. 35, no. 20, pp. 1–40, 2023. doi: 10.1007/s00521-023-08612-y. [Google Scholar] [PubMed] [CrossRef]
10. V. Chang, J. Bailey, Q. A. Xu, and Z. Sun, “Pima Indians diabetes mellitus classification based on machine learning (ML) algorithms,” Neural Comput. Appl., vol. 35, no. 22, pp. 16157–16173, 2023. doi: 10.1007/s00521-022-07049-z. [Google Scholar] [PubMed] [CrossRef]
11. G. S. Bhavekar and A. D. Goswami, “A hybrid model for heart disease prediction using recurrent neural network and long short term memory,” Int. J. Inf. Tecnol., vol. 14, no. 4, pp. 1781–1789, 2022. doi: 10.1007/s41870-022-00896-y. [Google Scholar] [CrossRef]
12. F. M. Talaat and R. M. El-Balka, “Stress monitoring using wearable sensors: IoT techniques in medical field,” Neural Comput. Appl., vol. 35, no. 25, pp. 1–14, 2023. doi: 10.1007/s00521-023-08681-z. [Google Scholar] [PubMed] [CrossRef]
13. T. Chen, S. Madanian, D. Airehrour, and M. Cherrington, “Machine learning methods for hospital readmission prediction: Systematic analysis of literature,” J. Reliab. Intell. Environ., vol. 8, no. 1, pp. 49–66, 2022. doi: 10.1007/s40860-021-00165-y. [Google Scholar] [CrossRef]
14. S. N. Manoharan, K. M. Kumar, and N. Vadivelan, “A novel CNN-TLSTM approach for dengue disease iden tification and prevention using IoT-fog cloud architecture,” Neural Process. Lett., vol. 55, no. 2, pp. 1951–1973, 2023. doi: 10.1007/s11063-022-10971-x. [Google Scholar] [PubMed] [CrossRef]
15. D. Kumar, S. K. Sood, and K. S. Rawat, “Early health prediction framework using XGBoost ensemble algorithm in intelligent environment,” Artif. Intell. Rev., vol. 56, no. S1, pp. 1–25, 2023. doi: 10.1007/s10462-023-10565-6. [Google Scholar] [CrossRef]
16. N. El-Rashidy, A. Sedik, A. I. Siam, and Z. H. Ali, “An efficient edge/cloud medical system for rapid detection of level of consciousness in emergency medicine based on explainable machine learning models,” Neural Comput. Appl., vol. 35, no. 14, pp. 10695–10716, 2023. doi: 10.1007/s00521-023-08258-w. [Google Scholar] [PubMed] [CrossRef]
17. W. H. Hsieh, C. C. Y. Ku, H. P. C. Hwang, M. J. Tsai, and Z. Z. Chen, “Model for predicting complications of hemodialysis patients using data from the internet of medical things and electronic medical records,” IEEE J. Transl. Eng. Health Med., vol. 11, pp. 375–383, 2023. doi: 10.1109/JTEHM.2023.3234207. [Google Scholar] [PubMed] [CrossRef]
18. D. Bernard, C. Msigwa, and J. Yun, “Toward IoT-based medical edge devices: PPG-based blood pressure estimation application,” IEEE Interest Things J., vol. 10, no. 6, pp. 5240–5255, 2022. doi: 10.1109/JIOT.2022.3222477. [Google Scholar] [CrossRef]
19. T. Zhu, L. Kuang, J. Daniels, P. Herrero, K. Li and P. Georgiou, “IoMT-enabled real-time blood glucose prediction with deep learning and edge computing,” IEEE Internet Things J., vol. 10, no. 5, pp. 3706–3719, 2022. doi: 10.1109/JIOT.2022.3143375. [Google Scholar] [CrossRef]
20. İ. Kök, F. Y. Okay, Ö. Muyanlı, and S. Özdemir, “Explainable artificial intelligence (XAI) for internet of things: A survey,” IEEE Internet Things J., vol. 10, no. 16, pp. 14764–14779, 2023. doi: 10.1109/JIOT.2023.3287678. [Google Scholar] [CrossRef]
21. A. Ghosh, R. Saha, and S. Misra, “Persistent service provisioning framework for IoMT based emergency mobile healthcare units,” IEEE J. Biomed. Health Inform., vol. 26, no. 12, pp. 5851– 5858, 2022. doi: 10.1109/JBHI.2022.3172624. [Google Scholar] [PubMed] [CrossRef]
22. C. Chakraborty and A. Kishor, “Real-time cloud-based patient-centric monitoring using computational health systems,” IEEE Trans. Comput. Soc. Syst., vol. 9, no. 6, pp. 1613–1623, 2022. doi: 10.1109/TCSS.2022.3170375. [Google Scholar] [CrossRef]
23. D. Liu, J. Shao, H. Liu, and W. Cheng, “Design on early warning system for renal cancer recurrence based on CNN-based internet of things,” IEEE Access, vol. 10, pp. 34835–34845, 2021. doi: 10.1109/ACCESS.2021.3114227. [Google Scholar] [CrossRef]
24. A. Balasundaram, S. Routray, A. Prabu, P. Krishnan, and P. P. Malla, “Internet of things (IoT) based smart healthcare system for efficient diagnostics of health parameters of patients in emergency care,” IEEE Internet Things J., vol. 10, no. 21, pp. 18563–18570, 2023. doi: 10.1109/JIOT.2023.3246065. [Google Scholar] [CrossRef]
25. Rangasamy, Rajasekar, T. K. Mohammed, M. Chinnaswamy, and R. Veerachamy, “Predictive modelling for healthcare decision-making using IoT with machine learning models,” in Artificial Intelligence for Smart Healthcare, pp. 17–30, 2023. doi: 10.1007/978-3-031-23602-0. [Google Scholar] [CrossRef]
26. M. J. Baucas, P. Spachos, and K. N. Plataniotis, “Federated learning and blockchain-enabled fog-IoT platform for wearables in predictive healthcare,” IEEE Trans. Comput. Soc. Syst., vol. 10, no. 4, pp. 1732–1741, 2023. doi: 10.1109/TCSS.2023.3235950. [Google Scholar] [CrossRef]
27. B. U. Demirel, I. A. Bayoumy, and M. A. A. Faruque, “Energy-efficient real-time heart monitoring on edge-fog-cloud internet of medical things,” IEEE Internet Things J., vol. 9, no. 14, pp. 12472– 12481, 2021. doi: 10.1109/JIOT.2021.3138516. [Google Scholar] [CrossRef]
28. W. Liu, et al. “Lead separation and combination: A novel unsupervised 12-lead ECG feature learning framework for internet of medical things,” IEEE Internet Things J., vol. 9, no. 23, pp. 23897–23914, 2022. doi: 10.1109/JIOT.2022.3188771. [Google Scholar] [CrossRef]
29. M. Opoku Agyeman, A. F. Guerrero, and Q. T. Vien, “Classification techniques for arrhythmia patterns using convolutional neural networks and internet of things (IoT) devices,” IEEE Access, vol. 10, no. 1, pp. 87387–87403, 2022. doi: 10.1109/ACCESS.2022.3192390. [Google Scholar] [CrossRef]
30. P. M. Kumar, et al. “Clouds proportionate medical data stream analytics for internet of things-based healthcare systems,” IEEE J. Biomed. Health Inform., vol. 26, no. 3, pp. 973–982, 2021. doi: 10.1109/JBHI.2021.3106387. [Google Scholar] [PubMed] [CrossRef]
31. W. Wolberg, O. Mangasarian, and W. Street, “Breast Cancer Wisconsin (Diagnostic),” UCI Mach. Learn. Repository, 1995. doi: 10.24432/C5DW2B. [Google Scholar] [CrossRef]
32. M. Kahn, “Diabetes,” UCI Mach. Learn. Repository, 2017. doi: 10.24432/C5T59G.2017. [Google Scholar] [CrossRef]
33. J. Andras, S. William, M. Pfisterer, and R. Detrano, “Heart disease,” UCI Mach. Learn. Repository, 1988. doi: 10.24432/C52P4X. [Google Scholar] [CrossRef]
34. P. V. Sankar Ganesh and P. Sripriya, “A comparative review of prediction methods for pima indians diabetes dataset,” in Computational Vision and Bio-Inspired Computing. Cham: Springer, 2020, pp. 735–750. doi: 10.1007/978-3-030-37218-7_83. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools