 Open Access
Open Access
ARTICLE
Sepsis Prediction Using CNNBDLSTM and Temporal Derivatives Feature Extraction in the IoT Medical Environment
1 Department of Information Systems, College of Computer and Information Sciences, Princess Nourah Bint Abdulrahman University, Riyadh, 11671, Kingdom of Saudi Arabia
2 Department of Computer Science and Information Technology, Applied College, Princess Nourah Bint Abdulrahman University, Riyadh, 11671, Kingdom of Saudi Arabia
* Corresponding Author: Nurul Halimatul Asmak Ismail. Email:
Computers, Materials & Continua 2024, 79(1), 1157-1185. https://doi.org/10.32604/cmc.2024.048051
Received 26 November 2023; Accepted 20 February 2024; Issue published 25 April 2024
Abstract
Background: Sepsis, a potentially fatal inflammatory disease triggered by infection, carries significant health implications worldwide. Timely detection is crucial as sepsis can rapidly escalate if left undetected. Recent advancements in deep learning (DL) offer powerful tools to address this challenge. Aim: Thus, this study proposed a hybrid CNNBDLSTM, a combination of a convolutional neural network (CNN) with a bi-directional long short-term memory (BDLSTM) model to predict sepsis onset. Implementing the proposed model provides a robust framework that capitalizes on the complementary strengths of both architectures, resulting in more accurate and timelier predictions. Method: The sepsis prediction method proposed here utilizes temporal feature extraction to delineate six distinct time frames before the onset of sepsis. These time frames adhere to the sepsis-3 standard requirement, which incorporates 12-h observation windows preceding sepsis onset. All models were trained using the Medical Information Mart for Intensive Care III (MIMIC-III) dataset, which sourced 61,522 patients with 40 clinical variables obtained from the IoT medical environment. The confusion matrix, the area under the receiver operating characteristic curve (AUCROC) curve, the accuracy, the precision, the F1-score, and the recall were deployed to evaluate the models. Result: The CNNBDLSTM model demonstrated superior performance compared to the benchmark and other models, achieving an AUCROC of 99.74% and an accuracy of 99.15% one hour before sepsis onset. These results indicate that the CNNBDLSTM model is highly effective in predicting sepsis onset, particularly within a close proximity of one hour. Implication: The results could assist practitioners in increasing the potential survival of the patient one hour before sepsis onset.Keywords
Abbreviation
| DT | Decision tree |
| LR | Logistic regression |
| SVC | Support vector classifier |
| SVM | Support vector machine |
| MGP-AttTCN | Multitask gaussian process and attention-based deep learning model |
| GRU | Gated recurrent unit |
| RETAIN | REverse time attention model |
| DeepAISE | Deep artificial intelligence sepsis expert |
| DSPA | Deep sofa-sepsis prediction algorithm |
| SERA | Sequential element rejection and admission |
| EASP | Explainable AI sepsis predictor |
| GB | Gradient boosting |
| MARS | Multivariate adaptive regression splines |
| LASSO | Least absolute shrinkage and selection operator |
| AISE | Artificial intelligence sepsis expert |
| NB | Naïve bayes classifier |
Sepsis, a fatal medical emergency, is the most severe response of the body to an infection, which typically originates in the skin, urinary tract, lung, or gastrointestinal tract. This situation can swiftly cause organ failure, tissue damage, and death. A potential reduction in mortality and costs could be achieved through the timely and suitable management of sepsis [1]. Worldwide, sepsis remains a significant killer, with approximately 11 million deaths out of 48.9 million in the case of COVID-19 [2]. The expenses for sepsis treatment in U.S. hospitals were more than $20 billion in 2011, $23 billion in 2013, and $25 billion annually, making it the costliest disease condition to treat by a wide margin [3]. Multiple studies have shown that sepsis mortality may be lowered via prompt diagnosis and the administration of effective antibiotic treatment. Even so, it is hard to tell if someone has sepsis in its early stages because the syndrome is very different depending on changes in function, the complexity of the clinical situation, and the medicine used to treat the disease. This makes it very difficult to determine the severity of a patient’s organ failure [4].
Additionally, there is a limited, reliable method of diagnosing sepsis. Bedside assessment using various sophisticated scoring systems has been created to facilitate the early diagnosis of sepsis [5]. Currently, several clinical practice measures are in use, including “Acute Physiology and Chronic Health Evaluation (APACHE II)” [6], “Simplified Acute Physiology Score (SAPS II)” [7], “Sequential Organ Failure Assessment (SOFA)” [8], “Quick SOFA (qSOFA)” [8], “Modified Early Warning Score (MEWS)” [8], “Systemic Inflammatory Response Syndrome (SIRS)” [8], and “New Early Warning Score (NEWS)” [8], which are reliable markers of disease severity based on a combination of data from the clinic and the lab. Although these methods help predict overall deterioration or death in ICU studies, they lack the sensitivity and specificity to diagnose sepsis in an individual patient [9]. They also exhibit significant flaws in terms of methodology when applied to actual patient data [10]. The rapidly growing quantity of healthcare data will fundamentally alter the practice of medicine. Integrating laboratory data and biomarkers into clinical decision-support systems can dramatically enhance patient outcomes [11]. Most ongoing studies evaluate how well these complicated datasets may aid in making therapeutic choices.
ML-based models have demonstrated considerable utility in medicine, particularly for executing accurate and exact predictions. Several ML-based studies have built models for predicting sepsis during an ICU stay using vital signs and laboratory test data [12]. Certain studies [13] trained ML models solely on physiological characteristics as feature inputs; for example, “systolic blood pressure” (SBP), “diastolic blood pressure” (DBP), “heart rate” (HR), “respiratory rate” (RR), “temperature” (T), and “peripheral oxygen saturation” (SpO2), were employed as inputs in an XGBoost model. To anticipate the beginning of sepsis four hours in advance, reference [14] deployed a support vector machine (SVM) classifier with an AUC-ROC of 0.88. To further enhance the outcomes, their models have included patient demographics, laboratory test findings, and comorbidities [15]. The onset of sepsis was also predicted using a random forest (RF) classifier with a sensitivity score 0.8 using physiological data [16]. The authors of [17] used a fully connected LSTM-CNN model to predict early sepsis. In the time series analysis, the “insight” model [18] was expanded to include the six vital sign features, such as pH, WBC, and age. This gave an AUCROC of 0.92. Despite lacking a labeled sepsis case record, the MIMIC II and MIMIC III databases have been utilized in several investigations. Several ML and DL models have been applied for early sepsis detection using these datasets.
This study investigates the potential of employing CNNBDLSTM, a proposed hybrid DL model, for early sepsis prediction by utilizing six distinct time-frame intervals: One hour, two hours, three hours, six hours, twelve hours, and twenty-four hours before sepsis onset. A key objective was to assess the prediction method’s impact on the proposed model’s efficiency.
Based on the review of the related work in Section 2, several gaps were identified that could provide several contributions to the healthcare domain, as stated below:
• A method for anticipating the onset of sepsis is proposed by delineating six distinct time intervals aimed at forecasting the occurrence of sepsis as accurately as feasible. These time intervals are devised in accordance with the criteria outlined in the sepsis-3 standard.
• During the model classification process, both the proposed CNNBDLSTM model and various baseline models (CNN, BDLSTM, Random Forest (RF), and Extreme Gradient Boosting (XGBoost)) were trained on the MIMIC III dataset to compare their performance. Significantly, the proposed model showcased superior performance, attaining a remarkable accuracy of 99.15% and an impressive AUCROC of 99.74% when predicting sepsis onset one hour in advance.
• The classification outcomes were also contrasted with those of previous benchmark models trained on the same dataset. The findings revealed that the proposed model in the current study outperformed the benchmark models.
The following portions of this work are organized: Explain the work’s background in Section 2. Detail the available resources and processes in Section 3. Meanwhile, Section 4 will concentrate on the study’s results and discussion. Section 5 discusses the limitations. Section 6 will provide a conclusion.
This section aims to offer readers a concise overview of recent research on sepsis diagnosis and early prediction utilizing ML and DL techniques prior to the onset of infection. It is crucial to review and assess the current “state of the art” in predicting sepsis onset, given the rapid advancements in this field. By examining relevant studies published after 2020, we evaluate the features utilized for prediction, the effectiveness of various ML and DL models, the encountered challenges, and the potential future directions of research in this domain. Table 1 presents a summary of the related work concerning the three primary focus areas.
2.1 Clinical Practices-Based Sepsis Prediction
In the past, standard clinical practices employed at the bedside were utilized for predicting sepsis onset. However, these metrics were not specifically designed for sepsis prediction or used to compute sepsis risk scores consistently. In particular, the SIRS criteria, often deemed nonspecific and outdated for sepsis measurement, were commonly utilized [9–11]. Many ML-based and DL-based studies have compared their model’s achievement with the standard clinical practice. For instance, in comparison to the clinical practice score (the AUROC 0.635 for qSOFA, 0.688 for MEWS, and 0.814 for SIRS), the study conducted by [19] showed a substantially higher predictive performance with an AUCROC score of 0.931. However, it is important to note that the current study does not aim to compare against standard clinical practices. Instead, its focus is on assessing the enhancement achieved through the utilization of ML, DL, and hybrid DL with DL classifiers in predicting sepsis onset.
2.2 Machine Learning-Based Sepsis Prediction
ML methods have substantially improved the ability to diagnose and forecast sepsis. The recent related work review (Table 1) reveals that seven studies deployed ML classifiers, and two studies combined ML with DL classifiers. Reference [20] used the MIMIC III dataset to train DT, RF, and XGBoost classifiers for ML-based sepsis prediction studies. RF model appears to be the best-performing model with AUCROC of 0.92 to predict sepsis 24 h before onset. Reference [21] achieved an AUCROC of 0.86 (6 h before sepsis), while reference [22] obtained an AUCROC of 0.85 (48 h before sepsis). Both of the studies utilized the 2019 PhysioNet Challenge dataset. However, reference [23] attained better results by harnessing the Yonsei University Severance Hospital dataset to train several ML classifiers with the LR model, attaining an AUCROC of 0.86 at 6 h before sepsis. Reference [24] achieved better results (AUCROC of 0.93) than the other studies using the Tertiary academic hospital dataset, but this study does not report the hours before sepsis onset. However, reference [25] reported even more significant results using the MIMIC III dataset deploying RF, GB, and SVM. They were reported to have achieved an AUCROC of 0.96 4 h before sepsis. We will report the studies that combined ML with DL classifiers under separate sub-sections.
2.3 Deep Learning-Based Sepsis Prediction
The capability of DL to predict sepsis onset has attracted many interesting studies, particularly those that use massive datasets such as MIMIC III. In the current study, we reviewed seven DL-based studies using DL classifiers. Three studies used MIMIC III as their data source to train their proposed models, while others used different datasets, as reported in Table 1. Most of the studies harnessed CNNs, RNN, LSTM, and DNN, which showed promising results in sepsis detection. The most recent study by [26] used the Shanghai Hospital dataset to train a double fusion DL framework, which attained an AUCROC of 0.92 at 6 h before onset. They also compare their results with the clinical practice measurement. Reference [27] used MIMIC III to train their proposed model of a joint multi-task “Gaussian Process and attention-based deep learning model”. They attained an AUCROC result of 0.64 at 5 h before sepsis. Reference [28] used the 2019 DII National Data Science Challenge to train GRU, RETAIN Dipole, and LSTM. The reported result was an AUCROC score of 0.89 at 4 h before sepsis. Reference [29] utilized MIMIC III to train their proposed model, DeepAISE (Deep Artificial Intelligence Sepsis Expert). They attained a promising AUCROC result of 0.90 at 4 h before sepsis. Reference [30] also used MIMIC III to train their proposed model DSPA (“a deep learning approach for sepsis monitoring via severity score estimation”) and CNN and RF as the baseline classifiers. They obtained excellent results (AUCROC of 0.97) at 6 h before sepsis. Reference [31] achieved the lowest AUCROC score of 0.86 three hours prior to the commencement. They trained the MIMIC III dataset using DNN. Reference [32] leveraged on the 2019 PhysioNet Challenge dataset to train the fully connected LSTM-CNN (a hybrid of DL-based models). They were reported to have achieved an AUCROC result of 0.86 at 12 h before sepsis.
2.4 Hybrid Classifiers-Based Sepsis Prediction
We reviewed three studies that have been using the hybrid strategy. The 2019 PhysioNet Challenge dataset was used to train SVM, RNNLSTM, and adaptive CNN models [33]. They achieved an accuracy of 0.94 but did not report the hours before sepsis. Reference [34] leveraged the Duke University Health System dataset to train RNN, LR, and RF models. They obtained an AUCROC of 0.88 at 5 h before sepsis onset. Reference [35] combined CNN and LSTM classifiers to predict sepsis using the electronic health record CROSSTRACKS dataset. They obtained an AUCROC of 0.88 at 3 h before sepsis onset. Reference [36] reported to have used an unstructured data source from the Singapore Government Hospital dataset to train the SERA algorithm. The study attained an excellent AUCROC of 0.94 at 12 h before sepsis.
2.5 Temporal Derivative-Based Sepsis Prediction
Every hourly time frame in a patient’s medical record should have a sepsis risk assessment and a positive or negative sepsis prediction. Specifically, as stated by the World Health Organization: Sepsis, it is essential to anticipate the beginning of sepsis at least 6 h (but no more than 12 h) in advance. As such, based on this criterion in predicting early sepsis, we have to review studies that have incorporated this criterion into their studies. Among the 17 articles, 15 used “longitudinal data” for predicting early sepsis onset. The rest of the studies did not state early prediction (refer to Table 1). Four articles were reported to have predicted early sepsis at 48 to 12 h earlier with promising AUCROC results. Six articles reported sepsis onset prediction six to five hours earlier, varying results depending on the dataset used. Four studies predicted early sepsis to happen at 6 h earlier. While three articles predicted early sepsis four hours earlier. Only one article predicted sepsis onset at three hours before sepsis occurs.
According to the evaluation of relevant literature, there is a scarcity of research that, as far as we know, develops predictive models utilizing the hybrid DL approach with six observation windows, which our study proposes. A maximum of four observation windows were established prior to the onset of sepsis in the majority of investigations employing the temporal derivatives approach. In our research, we established six observation windows that adhere to the sepsis criteria: 24, 12, 6, 3, 2, and 1 h before the onset of sepsis. According to the review of seventeen papers (beginning in 2020), only three have proposed training their datasets with hybrid DL classifiers. Nevertheless, none of the six research has constructed a hybrid CNNBDLSTM model.
3.1 Proposed Research Workflow
Fig. 1 describes the workflow of the study. It consists of the data acquisition phase, the exploratory data analysis phase, the data pre-processing phase, the model classification phase, the model evaluation phase, and the final model phase.
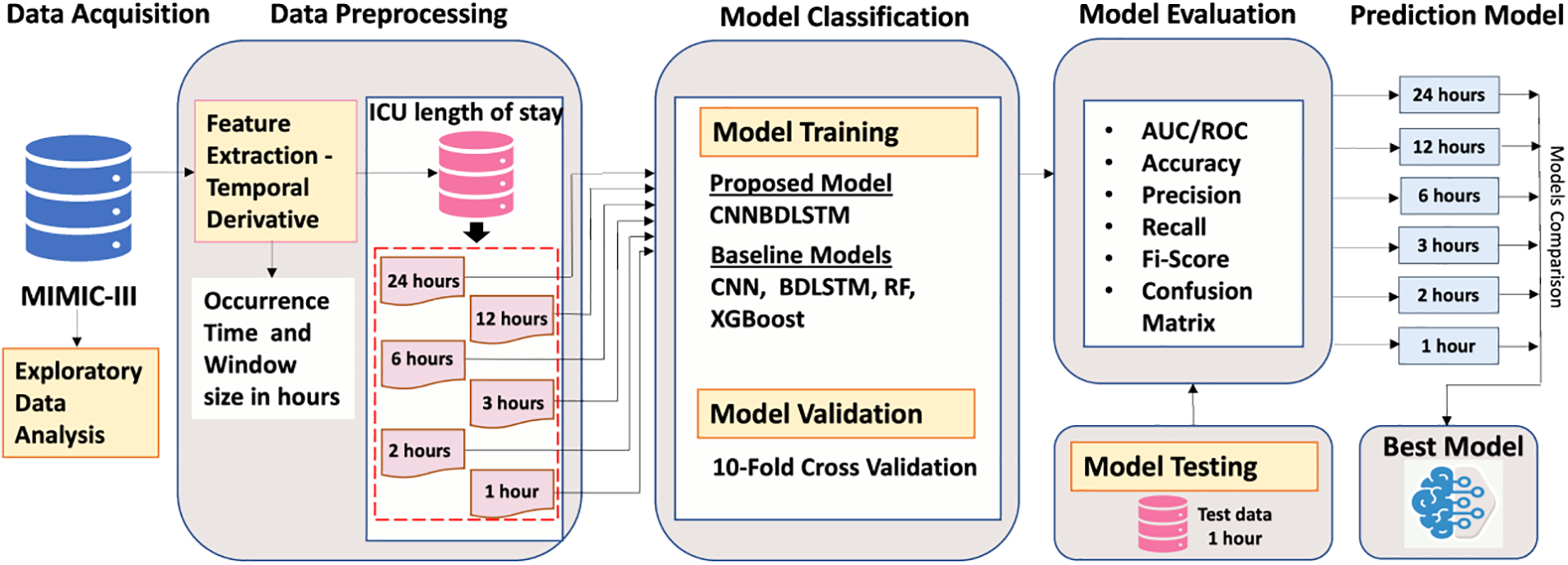
Figure 1: Proposed research workflow
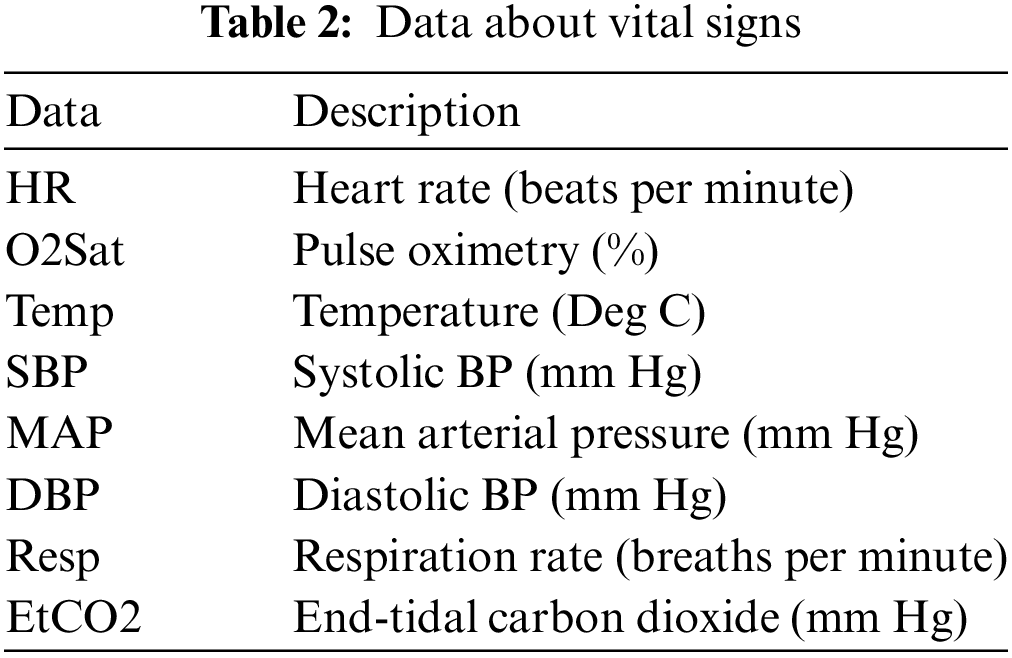
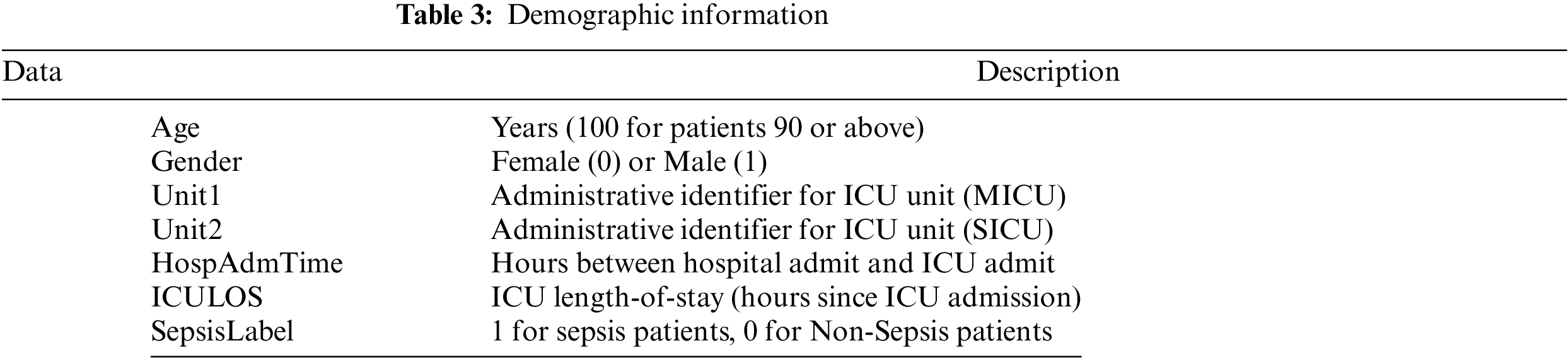
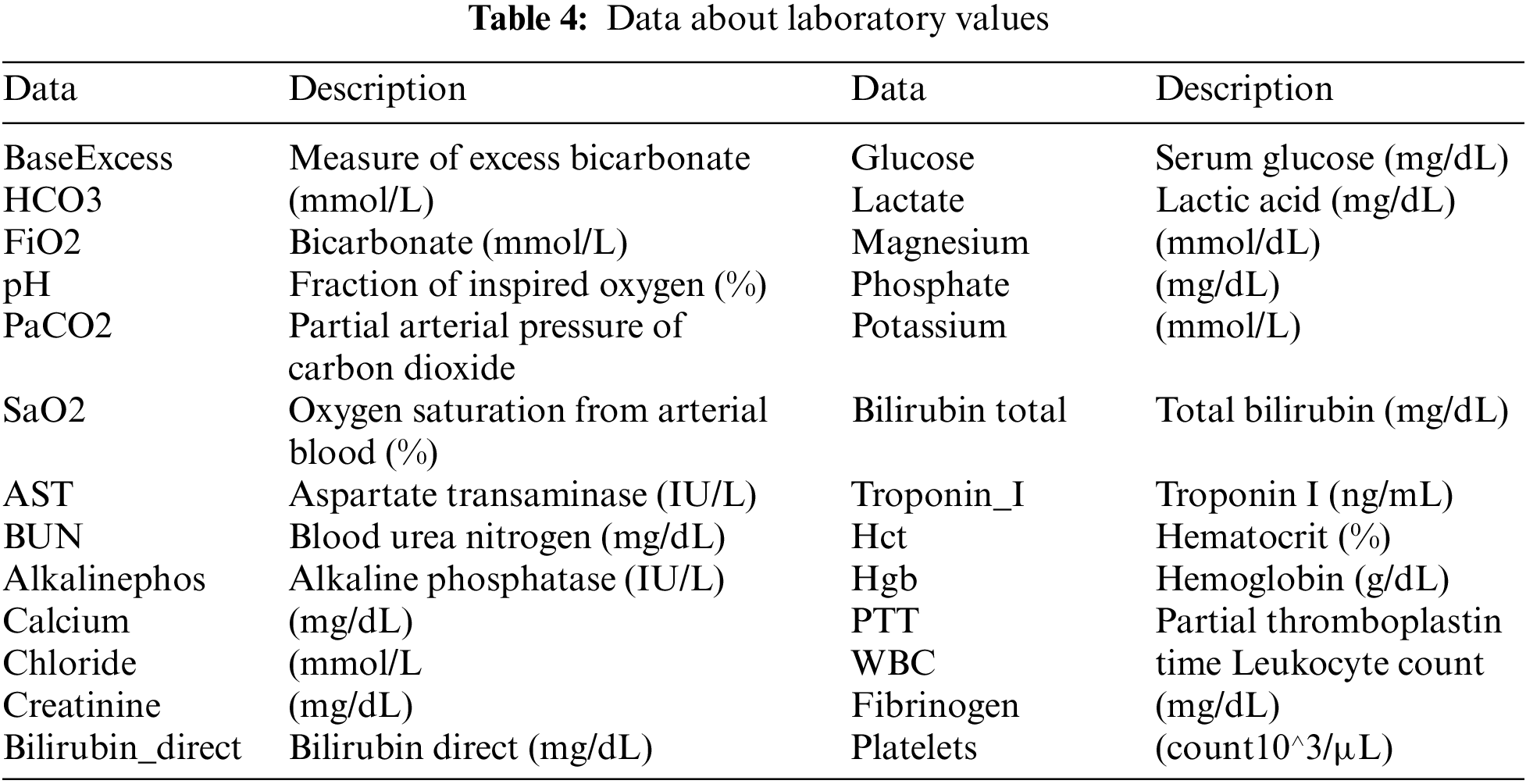
The MIMIC-III v1.4, which was a counter-public database consisting of patient clinical information about vital signs (Table 2), data about laboratory values (Table 3), and data about demographic information (Table 4), was utilized in our research [37]. The database comprises two information systems, IMDsoft MetaVision ICU and Philips CareVue Clinical, which have radically distinct data architectures. MIMIC III provides detailed data on more than 40,000 cases, or nearly 61,522 patient stays. The results are related to 53,423 distinct hospital admissions involving patients who were in the adult category.
3.3 Exploratory Data Analysis (EDA)
The EDA of the dataset is visualized in Figs. 2–4, which show the distribution of the vital signs data, the demographic data, and the laboratory data. The orange denotes the sepsis cases, while no sepsis cases are in blue.
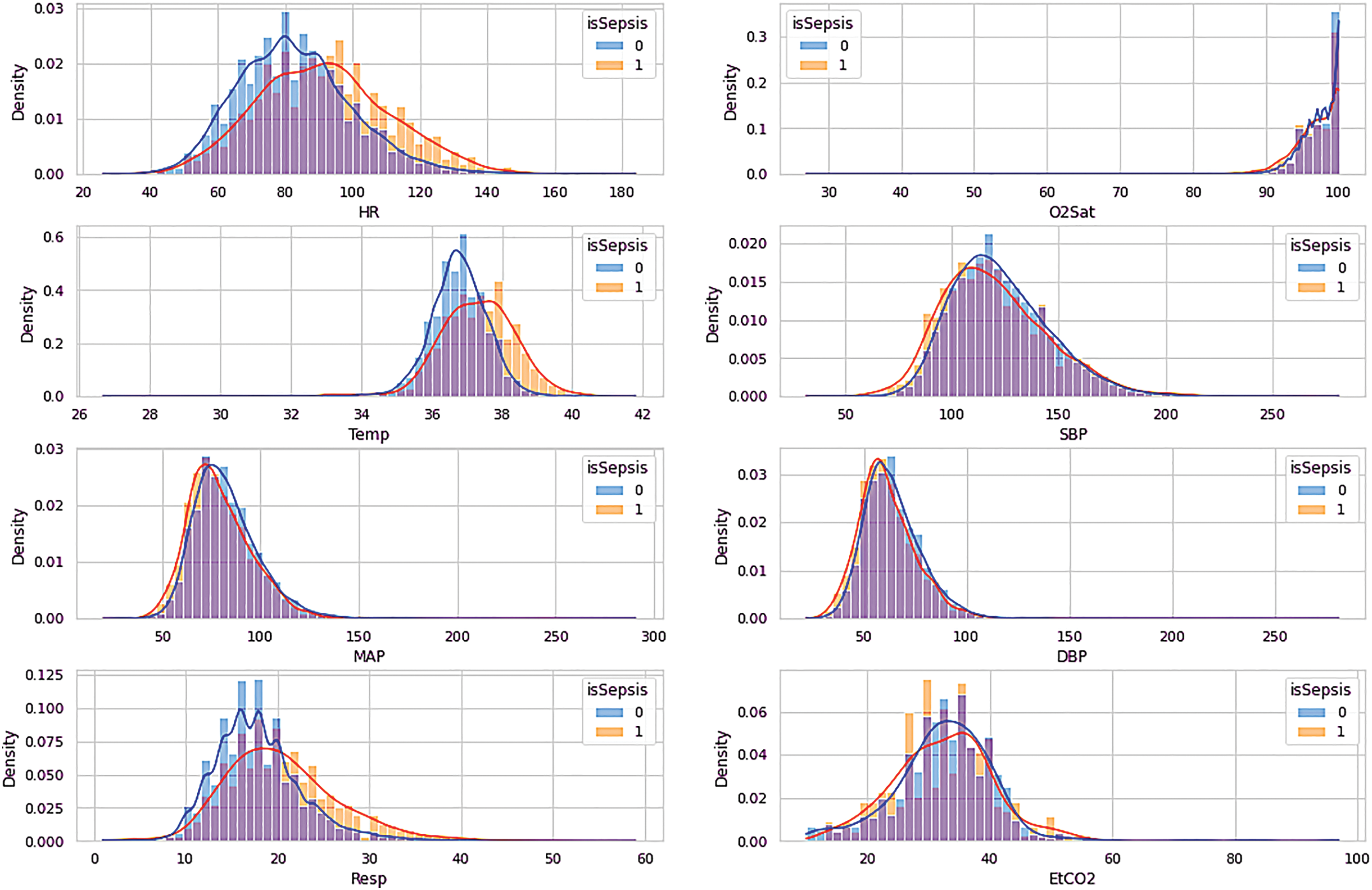
Figure 2: Visualization of vital signs data
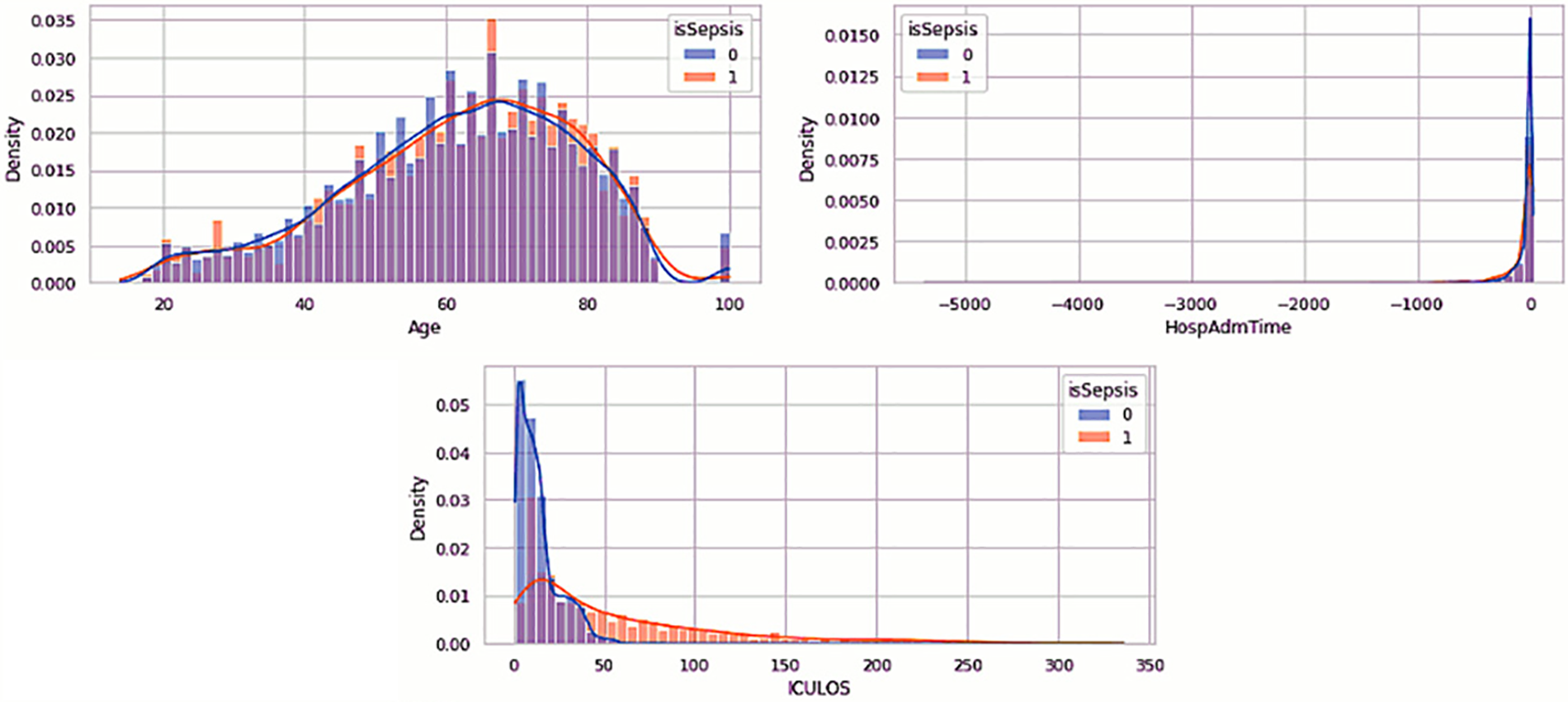
Figure 3: Visualization of demographic data
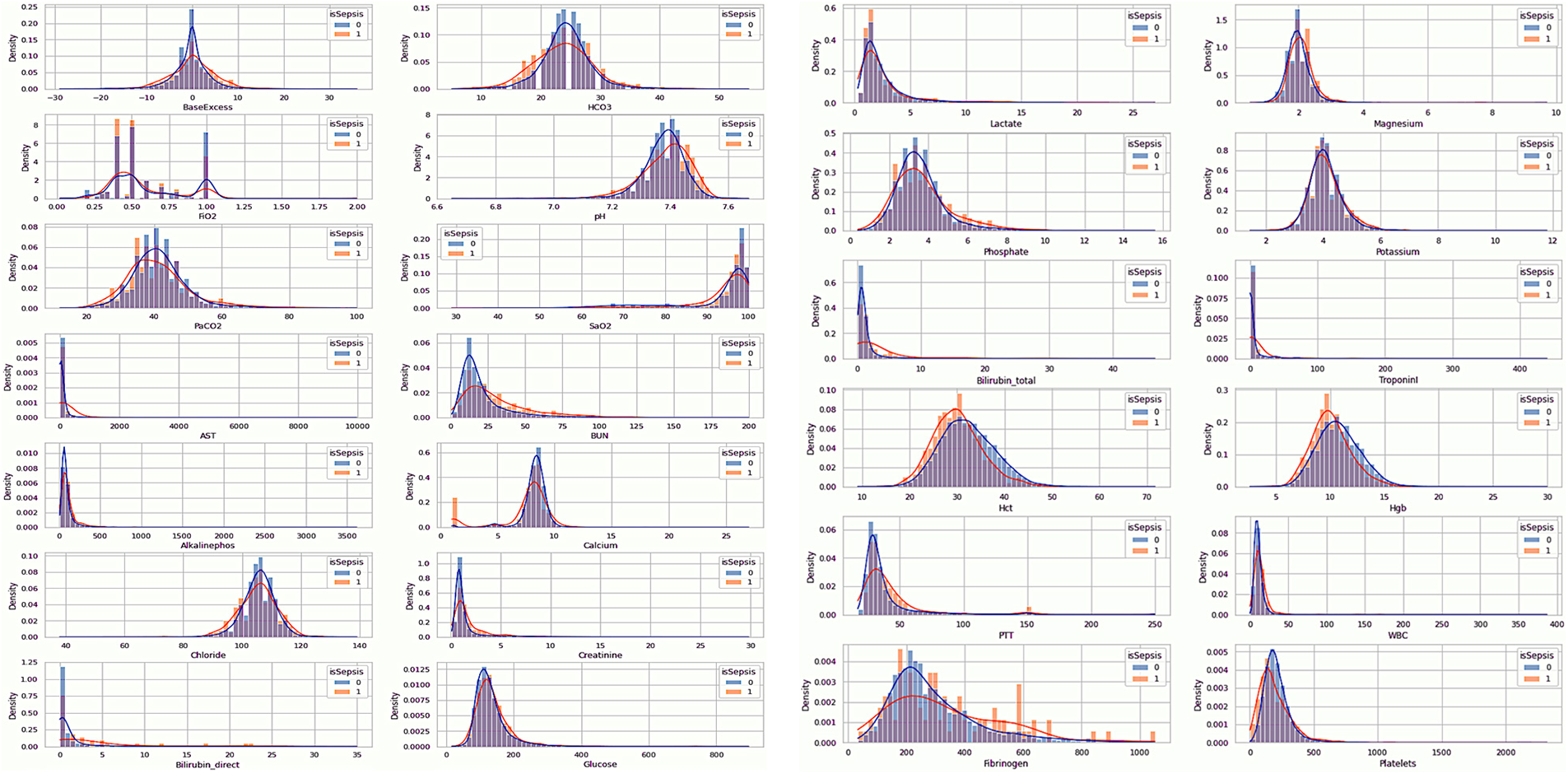
Figure 4: Visualization of laboratory data
According to the visualization in Fig. 2, HR, Temp, and Resp features differed between sepsis and non-sepsis individuals. The remaining characteristics may not affect the sepsis prediction. Table 2 provides an overview of the vital sign data.
Table 3 is a description of the demographic data. Fig. 3 suggests that age does not distinguish between patients with sepsis and those who are not. There is very little chance that a few-year age difference will result in sepsis. Age should be included in our model, potentially broken down into distinct categories, as patients aged 90 years or older have their age values set to 100. For each patient, the HospAdmTime characteristic is almost the same. Research indicates that patients who stay in the intensive care unit for an extended amount of time are more likely to get sepsis. Given that every patient receives an ICU record, it is likely that patients who did not receive an ICU record were moved from the SICU or MICU to other intensive care units (such as cardiac or trauma) (ICULOS attribute).
3.3.3 EDA of the Laboratory Data
Table 4 describes the laboratory data. The feature Base Excess, as shown in Fig. 4, exhibits a greater deviation from the mean in septic patients, although it has a similar mean. This suggests that an aberrant concentration of excess bicarbonate may be an inherent characteristic of septic patients. FiO2 is distributed bimodally and has values that are quite discrete. This feature will probably be ignored as non-representative due to the fact that only 20% of the patients have records of this value. Patients with sepsis seem to have a higher pH (more basic pH).
Additionally, it appears that sepsis-positive patients have greater BUN values. There are outliers in the calcium concentration for septic patients at shallow values despite the similar concentrations for either group of patients. Maybe this is something we should look into more. Although bilirubin deficiency appears to be higher in septic patients, it is important to remember that over 96 percent of individuals do not have this characteristic. However, bilirubin concentrations were probably only tested when medical professionals felt this characteristic was odd, and some sepsis patients had very high quantities.
Patients with sepsis also have increased bilirubin total levels. It is important to note that the sum of the direct and indirect bilirubin levels determines the total bilirubin concentration. As a result, a high correlation between this feature and Bilirubin direct is possible. Septic patients appear to have slightly lower quantities of Hct and Hgb levels. Patients with sepsis can seem to have marginally greater PTT. Patients with sepsis seem to have bimodal fibrinogen, which is marginally more concentrated than non-septic patients. We might anticipate that fibrinogen was measured for a specific reason, as around 95 percent of patients overlook this feature. Patients with sepsis may have somewhat decreased platelet counts.
Patient data preprocessing aims to enhance dataset quality, involving patient selection, dataset rebalancing, outlier removal, and handling missing data. Fig. 5 visualizes the steps of the whole process. In step one, 61,522 patients were selected for the study’s cohort. However, after excluding patients under 14 years of age, those admitted for cardio surgery, and those with missing data, the remaining number of patients is 16,984. In step two, patients admitted to the pre-ICU were removed, leaving a total of 11,811 patients. In step three, after balancing the target class, there are 1,840 patients remaining for the no-sepsis class and 537 for the sepsis class.
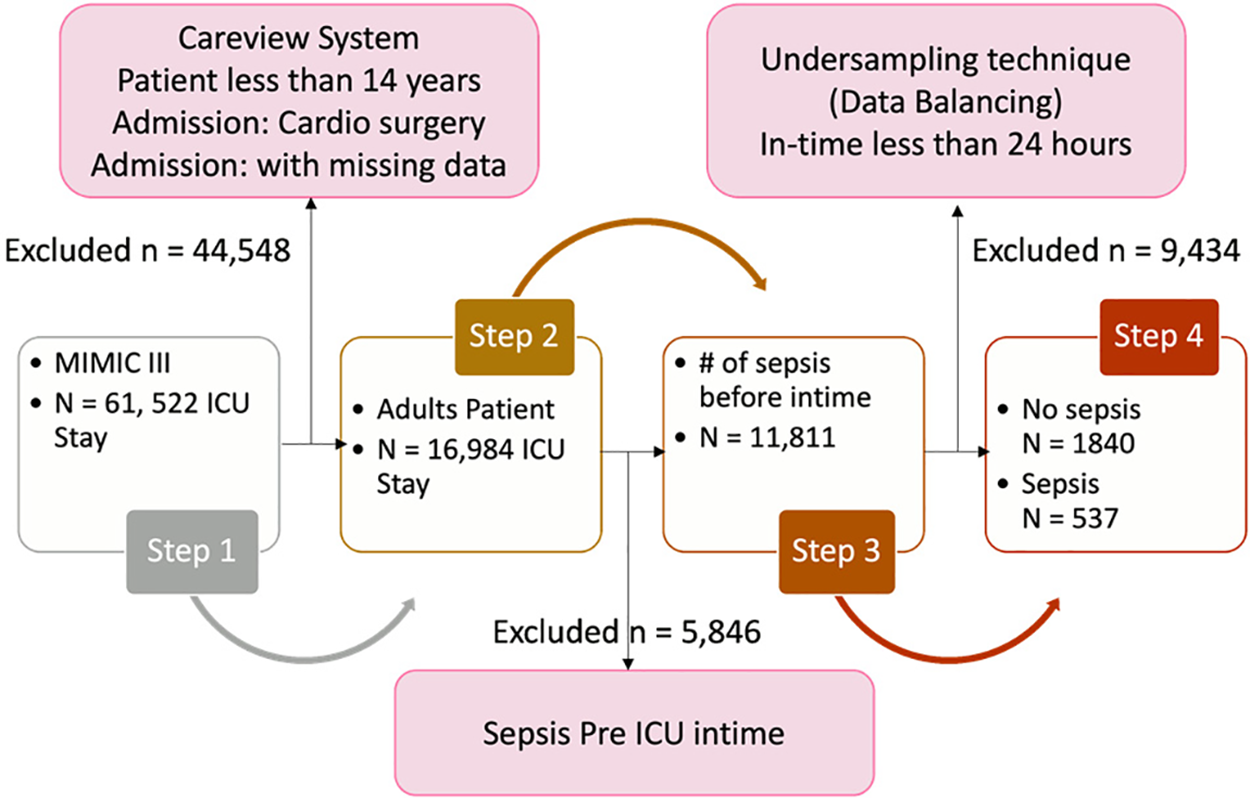
Figure 5: Visualization of the class labeling method
In this study, experiments were exclusively conducted using data from patients aged 14 years and older upon admission to the ICU. As the required patient data was not accessible in the CareVue system, the decision was made to solely utilize data compiled in MetaVision [38]. The MIMIC III dataset contains missing and highly deviated data related to ICU admission stays. This is often due to the unavailability of medical services, faulty systems, and various other factors during admission. To address this, patients with characteristics having more than 60% missing data were excluded from the analysis. Additionally, outliers in the dataset were removed as suggested by medical experts. Only patients who experienced organ malfunctions due to sepsis during their ICU stay were included for predicting sepsis onset.
3.4.2 Sepsis-3 Group Performance
Defining the sepsis cohort is crucial for building a sepsis prediction model. Therefore, a systematic approach was adopted, focusing on patients potentially impacted by infection and organ dysfunction in accordance with the sepsis-3 standard [39]. According to previous studies, this approach has resulted in a well-appropriately selected cohort. A patient potentially affected by infection is identified as someone who received antibiotics and had a body fluid sample taken within a specific timeframe. Therefore, timestamps were recorded at this specific event, and patients were classified as sepsis patients accordingly. In case one, if the body fluid sample was obtained before administering antibiotics, the medication needed to be given within 72 h. In case two, if antibiotics were administered first, the sample needed to be received within 24 h. Every timestamp for both examples in this study has to be recorded. The Postgres function detailed in Fig. 6 was used to identify organ malfunction. Finally, these patients were labeled using a SOFA measurement within the designated window span, with an increase of two or more points indicating “sepsis onset”.
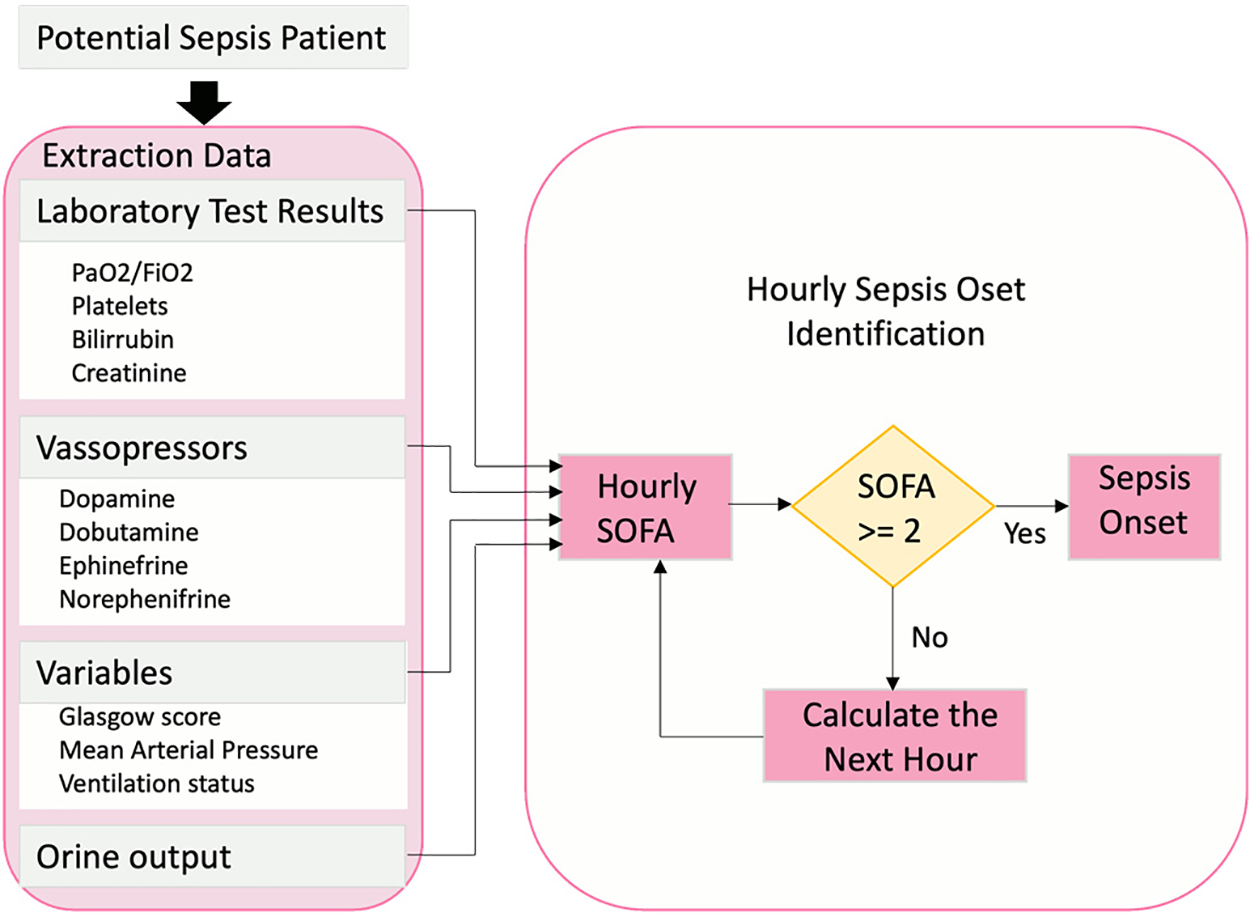
Figure 6: Sepsis onset hourly computation
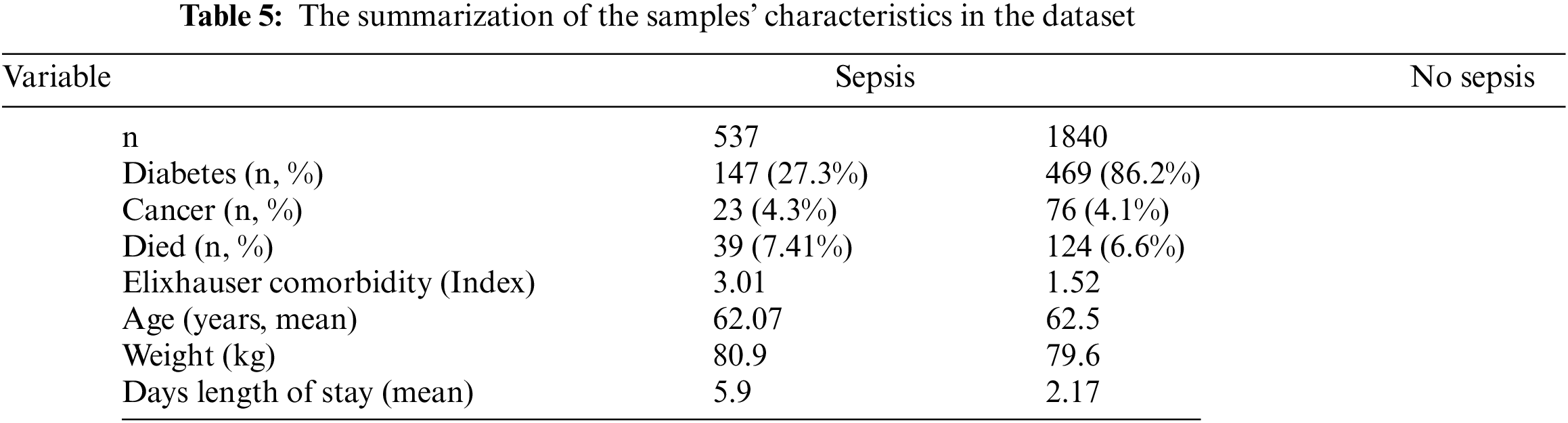
Table 5 summarizes the sample characteristics based on those with and without sepsis. 2,377 patients meeting the criteria were included. The inclusion was based on the average age of 67 years and not suffering from diabetes, cancer, or other diseases stated in the Elixhauser comorbidity index.
3.4.3 Feature Selection and Extraction
This process determines the potential features selected or excluded in the dataset, which are later used to train the prediction models. The features with the highest missing rate will be excluded. Based on expert medical advice, this study noted that 31 features (MRF) are relevant for sepsis prediction. The relevant features were clustered into three categories: The physiological data (see Table 2), the laboratory test results (see Table 3), and demographics/scores (see Table 4).
Based on the computation of the occurrence time within a window size measured in hours. (as discussed in Section 3.4.2), The temporal derivatives method proposed six distinct sampling time windows as the prediction methodology. With the knowledge that sepsis could happen at any time during the patient’s stay in the ICU, predicting the time close to sepsis onset is the aim of this study. The prediction was based on the target variable of ‘1’ for patients with sepsis and ‘0’ for patients with no sepsis cases. In order to facilitate a comprehensive comparison, it was proposed and agreed upon to utilize six extended monitoring time intervals as the “look-back” (LB) sequence of results for sepsis prediction. For each subgroup that was pulled out before, six-time frames (24, 12, 6, 3, 2, 1 h), which is the LB, become the models’ input data. In the end, six spaces—1, 2, 3, 4, 5, and 6 h—were created as the forecast times for accessing and contrasting the results of each model. The visualization of the prediction procedure is shown in Fig. 7.
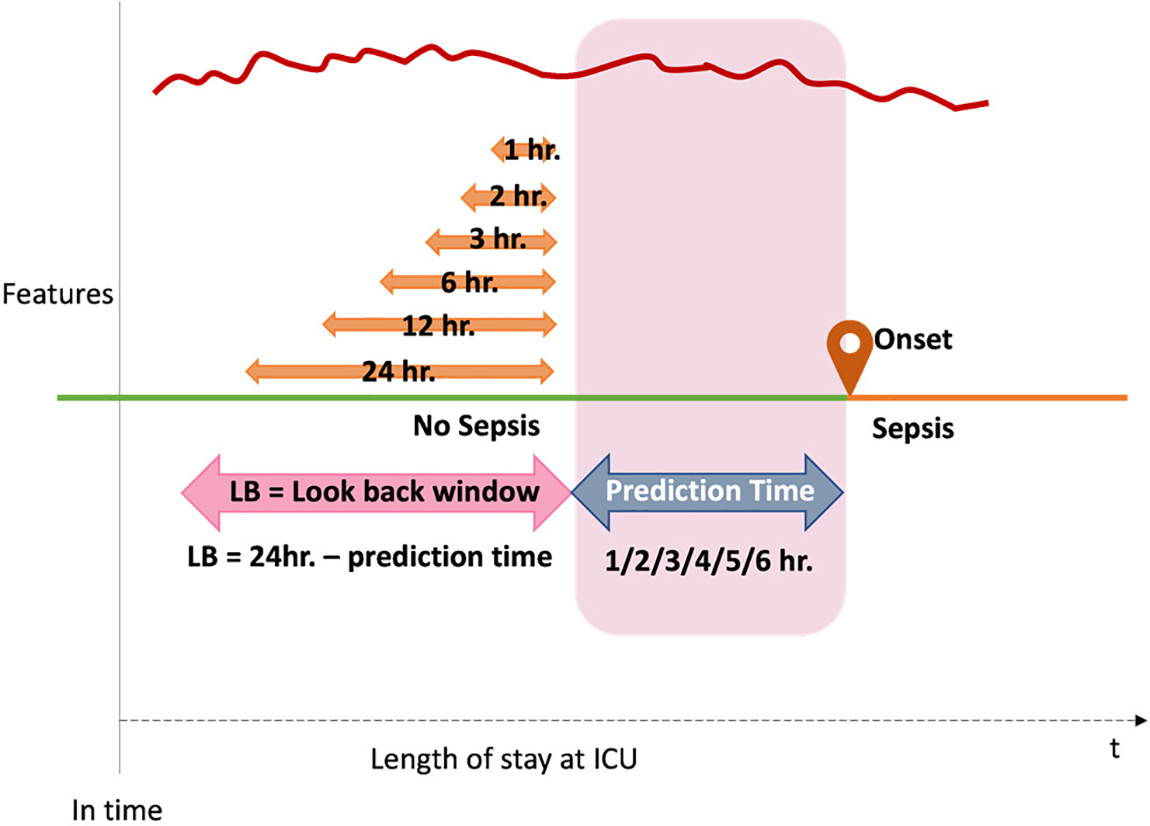
Figure 7: The proposed sepsis prediction methodology
The current work has constructed an experimental comparative study of two ML, two DL, and one hybrid DL model to predict sepsis onset. Following is a description of the classification models used.
Bi-Directional Long Short-Term Memory (BDLSTM)
This model combines two independent RNNs. Because of its architecture, the network can gather sequence information in both directions at each time step [40]. In contrast to a unidirectional LSTM, a bidirectional LSTM (BDLSTM) may store information from both the past and the future in its hidden states at the same time by running inputs in two directions: One from the past to the future and the other from the future to the past. Fig. 8 depicts the process by which the output y is determined at a given time t. Applying this algorithm to the current study, BDLSTM can store the various observation windows to predict when sepsis will begin.
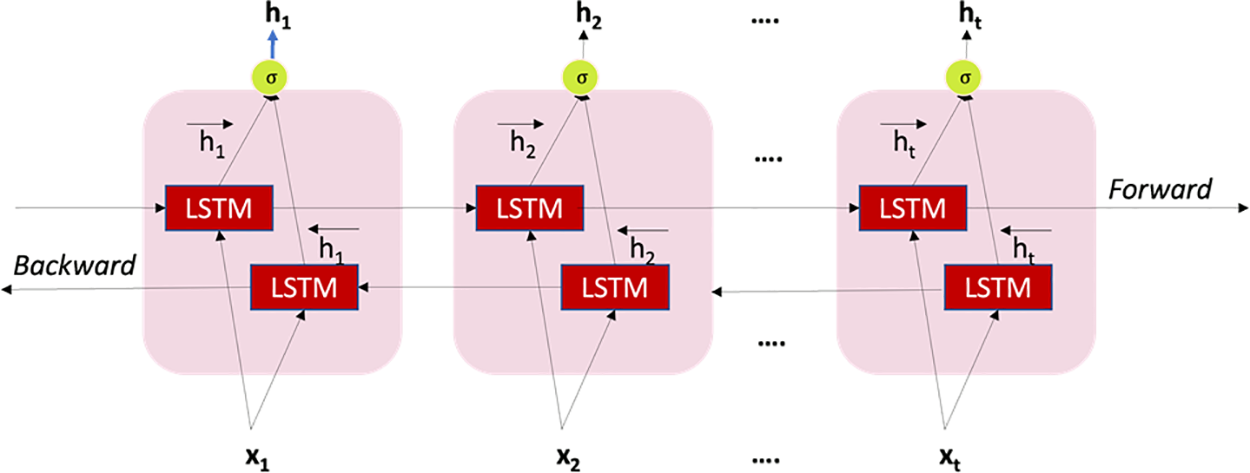
Figure 8: Structure of BDLSTM
Convolutional Neural Network (CNN)
The elements of the CNN [41] shown in Fig. 9 are the “convolutional layer,” “recalcified linear unit” (ReLU) layer, and the pooling layer, which make up the proposed CNN. The convolutional filters in the convolutional layer carry out pattern recognition. The ReLU layer maintains the same derivatives, providing nonlinearity and quick computation. The spatial and temporal ranges of features are diminished when they are combined. The convolutional layer employs a collection of convolutional filters. The filters always affect the entire feature dimension but only on a subset of the temporal region of the data. To determine whether specific patterns are present, they carry out several calculations. The filters then go across the temporal direction of the sequence, producing a series of outputs that show the locations of different patterns. When using CNN on image tasks, the filters may extend across width and height dimensions. In contrast, convolutional filters operate exclusively in the temporal domain when used on electronic health data. Using this algorithm in the current investigation would be advantageous to forecast the start of sepsis based on observations from various windows as the temporal derivatives sequence.
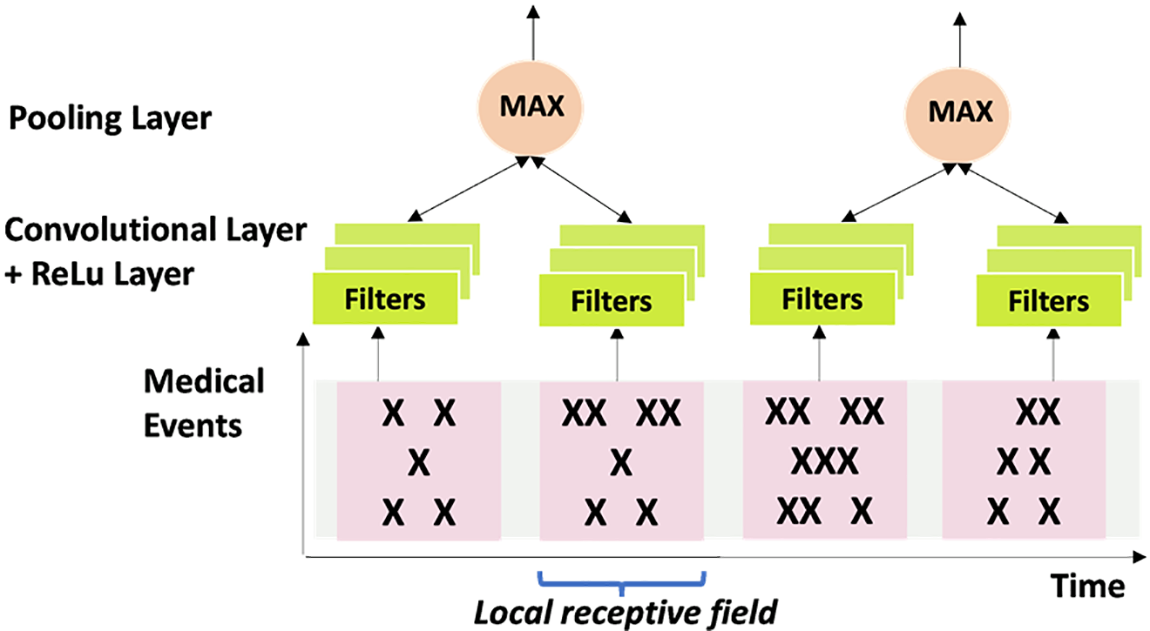
Figure 9: The structure of CNN
Simple models that predict outputs using binary splits on predictor variables are part of the tree-based structure of the RF classifier. Feature sampling is the name given to the technique [42]. Trees are the primary learners in reinforcement learning (RF). Each tree generates predictions by classifying input, subsequently used to develop a majority rule for the entire forest. We randomly select subsets of the dataset to use in each tree construction using the bagging process. The current study’s RF classifier would predict sepsis onset most accurately at a specified observation window. Fig. 10 displays the RF structure.
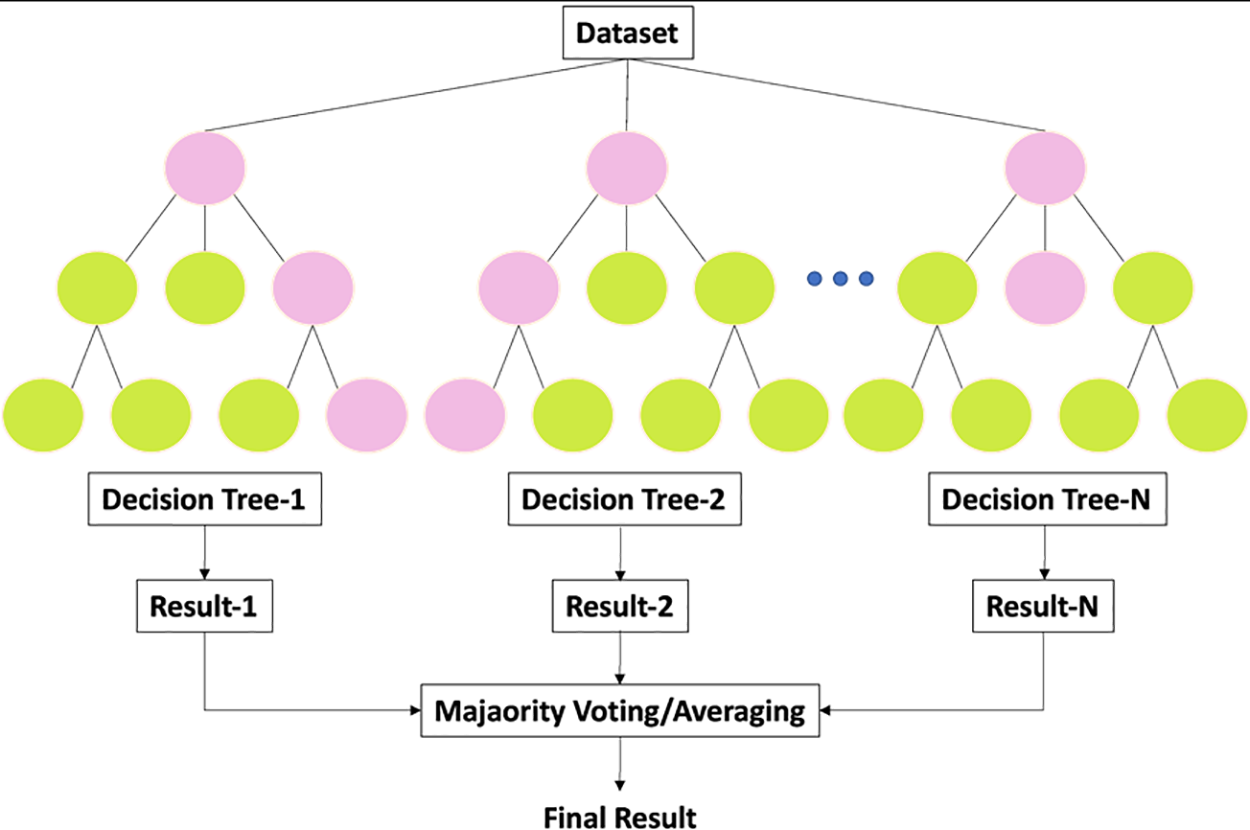
Figure 10: General architecture of RF
Extreme Gradient Boosting (XGBoost)
The XGBoost [43] model is thought to be better than the gradient tree-boosting model for learning and lowering prediction errors based on new algorithms and evaluation metrics. It is made up of more classifiers. The XGBoost model relies heavily on the tree learning method for handling scattered data. XGboost incorporates normalization to carry out the outcomes of its objective function optimization of the loss function, which considers the previous level’s outcome. Utilizing XGBoost in the current study would reduce the prediction errors by adjusting weights. The general architecture of the XGBoost algorithm is shown in Fig. 11.
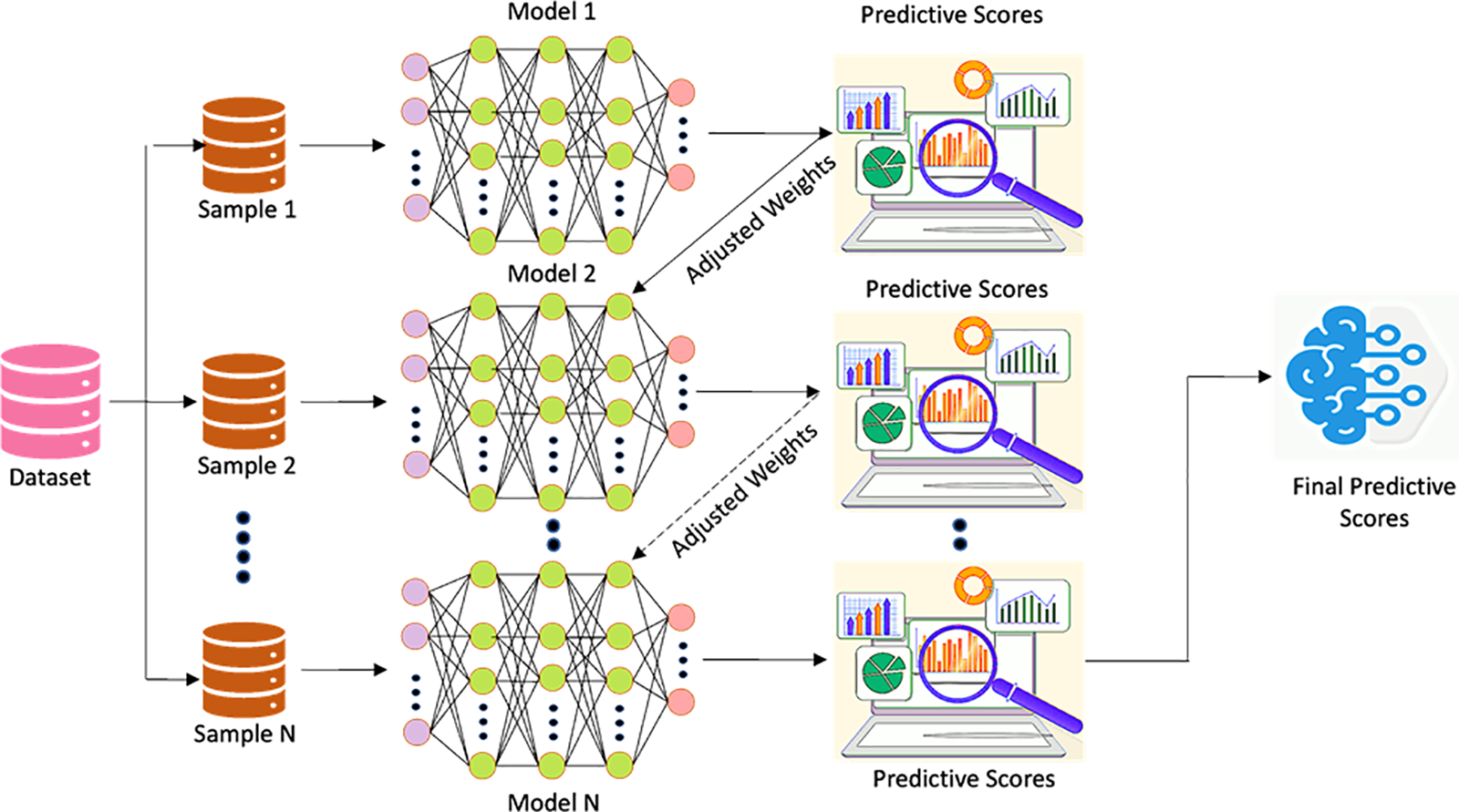
Figure 11: General architecture of XGBoost
3.6.2 The Proposed Model CNNBDLSTM Hybrid Model
The proposed model is a hybrid of BDLSTM [40] and CNN [41] constructed by harnessing the DL techniques as illustrated in Fig. 12. The following is the reason for proposing this model:
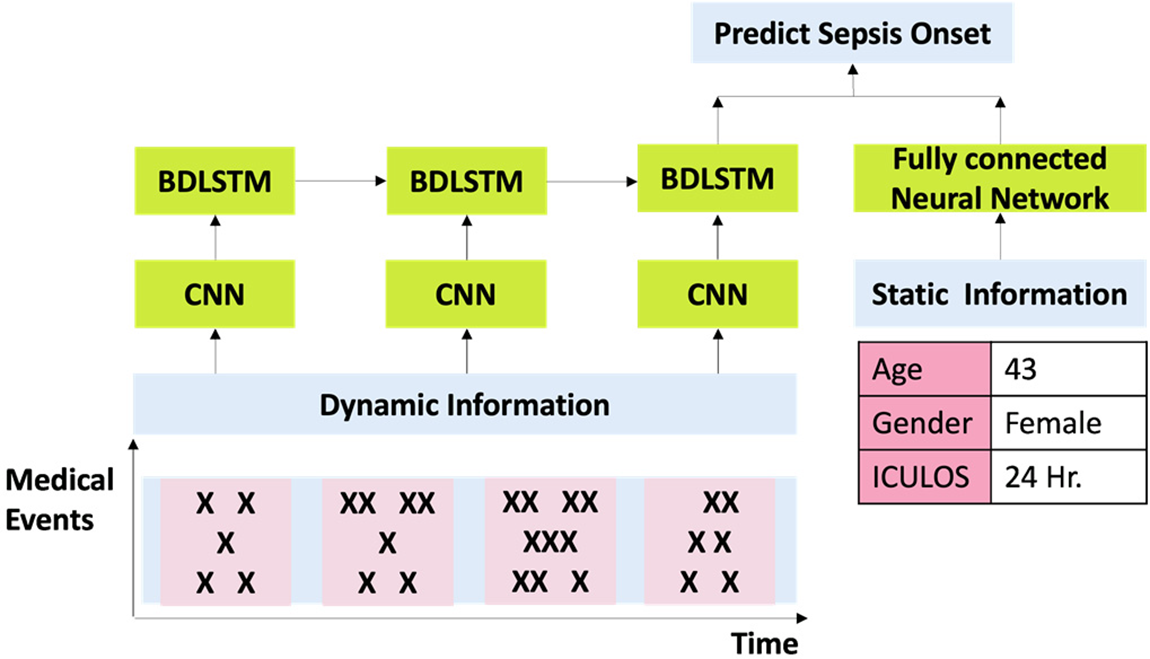
Figure 12: Structure of CNNBDLSTM
Enhanced Feature Learning: CNNs are effective at extracting spatial and temporal features from sequential data, while BDLSTMs excel in capturing long-term dependencies. By combining these two architectures, the hybrid model can leverage the strengths of both networks to learn more comprehensive and informative representations of the input data, leading to improved prediction accuracy.
Robustness to Temporal Patterns: Sepsis onset prediction often relies on recognizing subtle changes in patient data over time. The bidirectional nature of BDLSTMs allows the model to capture both forward and backward temporal dependencies in the data, making it more robust to varying temporal patterns associated with sepsis development.
Adaptability to Multimodal Data: Healthcare datasets typically contain diverse types of information, such as vital signs, laboratory results, and clinical notes. CNNs are adept at processing spatial data, such as images or spectrograms, while BDLSTMs excel at sequential data processing. This hybrid model can effectively integrate information from multiple modalities, enabling a more comprehensive analysis of patient data for sepsis prediction.
Early Prediction Capability: The combination of CNN and BDLSTM models allows for early detection of sepsis onset by leveraging both spatial and temporal information in the input data. This enables the model to identify subtle signs of sepsis development before they manifest into more severe symptoms, facilitating timely intervention and improved patient outcomes.
Overall, deploying a hybrid CNN-BDLSTM model for predicting sepsis onset offers a powerful framework that leverages the complementary strengths of both architectures, leading to more accurate and timely predictions as compared to the traditional methods.
During the implementation, the CNN layer is first trained by feeding with the dynamic data in matrix form and using ‘ReLu’ as an activation function. This adds non-linearity to the network and enables the model to learn more quickly and effectively than others. Next, CNN’s reduced matrix output is fed into the BDLSTM layer, combining static and dynamic data to create a fully connected neural network that can learn long-term dependencies. ‘ReLu’ is then used as an activation function to categorize two classes of patients into sepsis and non-sepsis based on the dynamic data (temporal derivatives of six observation windows). Later, to keep the model from overfitting, a dropout layer of 0.5 is applied; also, an attention layer is employed to give weights to the significant features while disregarding the unimportant ones. Finally, a sigmoid activation function is applied to the dense layer. Then, using binary cross-entropy as the loss function and 0.001 as the learning rate, Adam is utilized as an optimizer to minimize the loss function. The sepsis onset prediction mechanism is improved when all of these layers are combined.
This work used a 10-fold cross-validation approach to choose the optimal training model [44]. A prediction model’s efficacy may be measured with the use of cross-validation. Results from the statistical study will be extrapolated to a different dataset. To perform k-fold cross-validation, a sample is randomly split into k-equal halves. A single subsample is used to validate the model, while the remaining K-1 subsamples are utilized to train the model. The K iterations of this procedure provide validation data from a single subsample. In medical datasets, inequality between classes is a significant issue. During ICU stays, most MIMIC-III patients did not have sepsis (94.5 percent), whereas 5.5 percent of patients were diagnosed with sepsis.
Undersampling and oversampling are the two most often employed techniques for sampling data. The analysis indicates that the latter is more favorable than the former [45]. This research improved data balance by eliminating participants from the dominant group by random under-sampling (no sepsis). Therefore, the sample size was reduced to 2377 patients due to undersampling (77.4 percent no sepsis; 22.5 percent sepsis). According to [46], several metrics were acquired for each model included in this research to assess performance. Accuracy, precision, recall, F1-score, and AUC/ROC were used to evaluate the efficacy of the models. The true positive (TP) and true negative (TN) scores indicate the classifier model’s ability to predict the presence or absence of sepsis in a given patient. The false positive (FP) and false negative (FN) indicate the incorrect predictions made by the models. The accuracy is the proportion of actual positive observations to the total number of positive instances. Recall computes the total fraction of positive cases. The function’s measure specifies the average of recall and precision. The formula for the metrics is shown in Eqs. (1)–(4).
Using the AUCROC curve, Fig. 13a depicts the connection between the true positive rate (TPR) and the false positive rate (FPR) using the AUCROC function (FPR). ROC demonstrates a classifier’s ability to differentiate between its two classes (AUC). When the AUC is substantial, the model predictions are accurate.
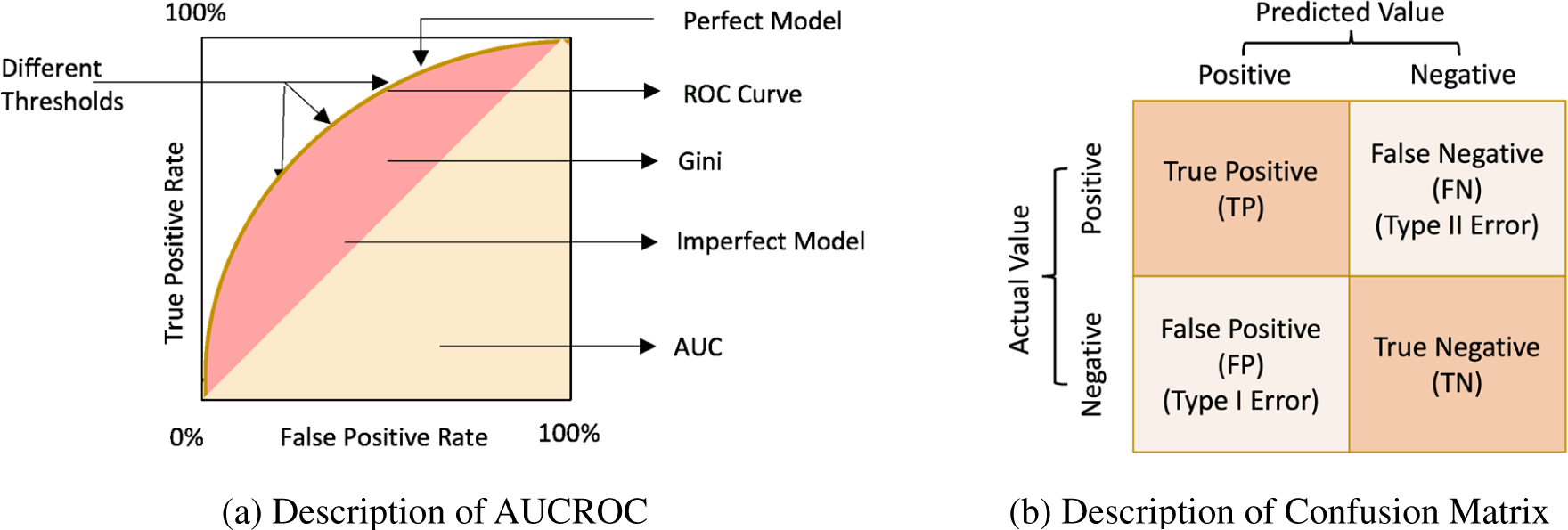
Figure 13: (a) Description of AUCROC and (b) Description of confusion matrix
Based on the following four classifications of predictions, a binary classification enables the construction of a confusion matrix, as shown in Fig. 13b.
• True positives (TP) denote that the positive samples are tagged as positive.
• False positives (FP) denote that the negative samples are tagged as positive (Type 1 error).
• True negatives (TN) denote that the negative samples are tagged as negative.
• False negatives (FN) denote that the positive samples are tagged as negative (Type 2 error).
It is a metric for assessing a classification model’s performance. Confusion matrices are superior to classification accuracy for gauging a model’s performance.
4.1 Experimental Requirements and Hyperparameter Tuning
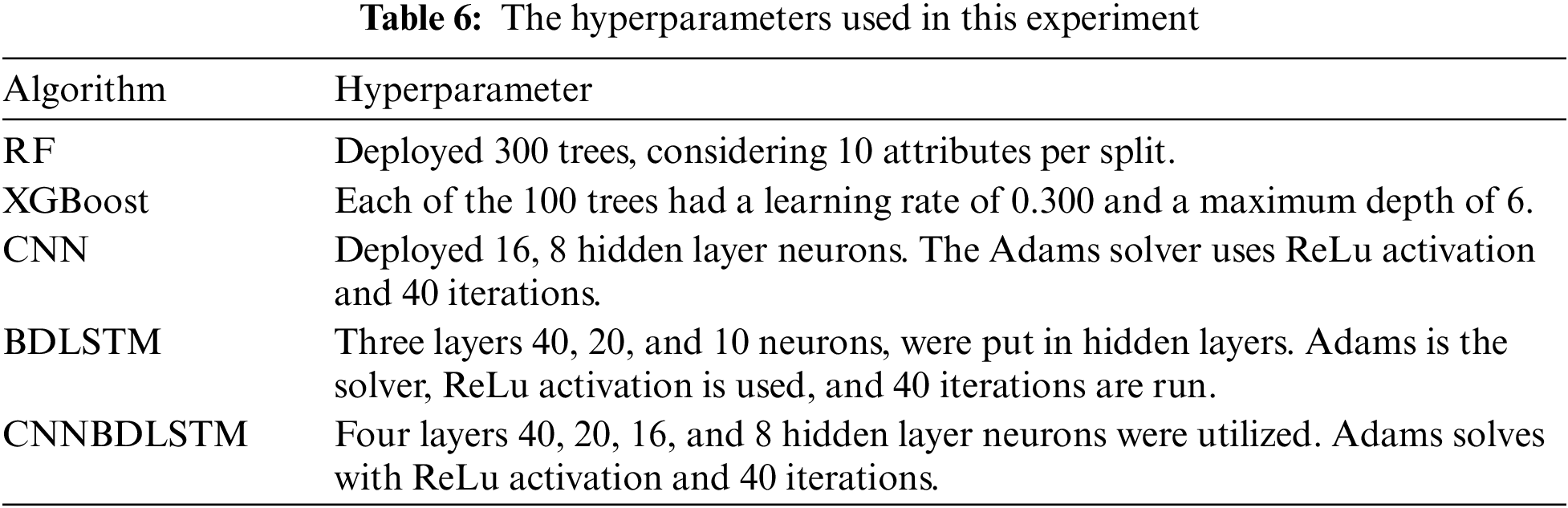
The performance of each model included in this study underwent rigorous evaluation and training through comprehensive testing, including the proposed hybrid DL model (CNNBDLSTM), the baseline models: CNN, BDLSTM, RF, and XGBoost. This study aimed to develop a reliable, efficient, and effective model for predicting early sepsis across 1, 2, 3, 6, 12, and 24-h intervals. The hyperparameters utilized to optimize and get the most favorable results for every rendered model are detailed in Table 6.
4.2 The Results of the Model Classification Performance Evaluation
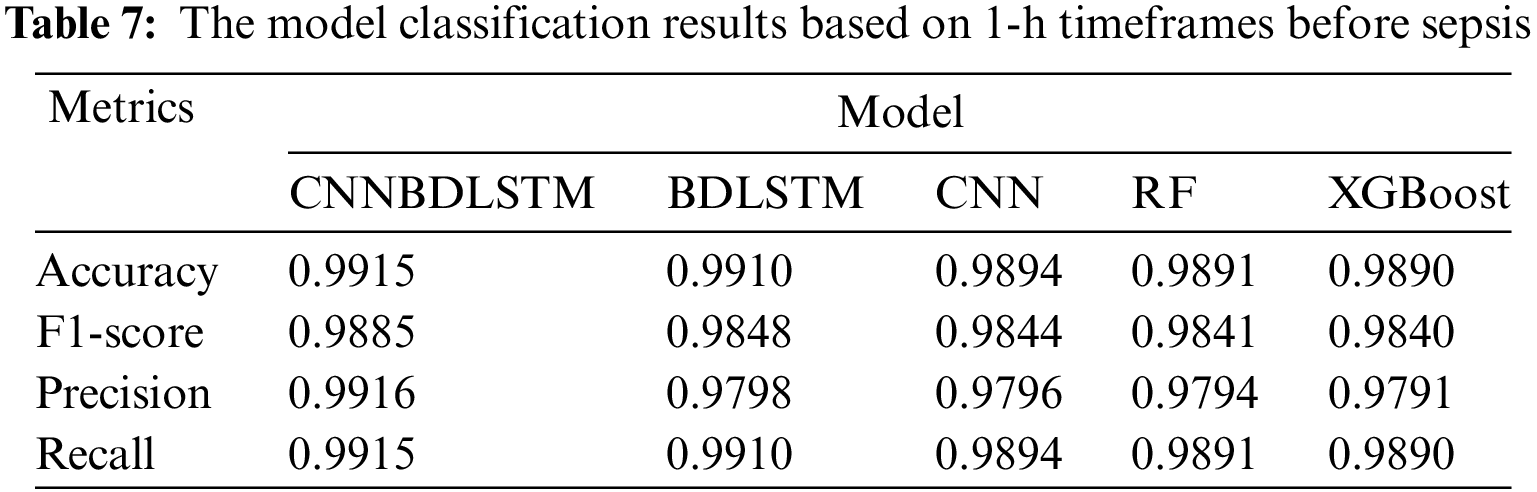
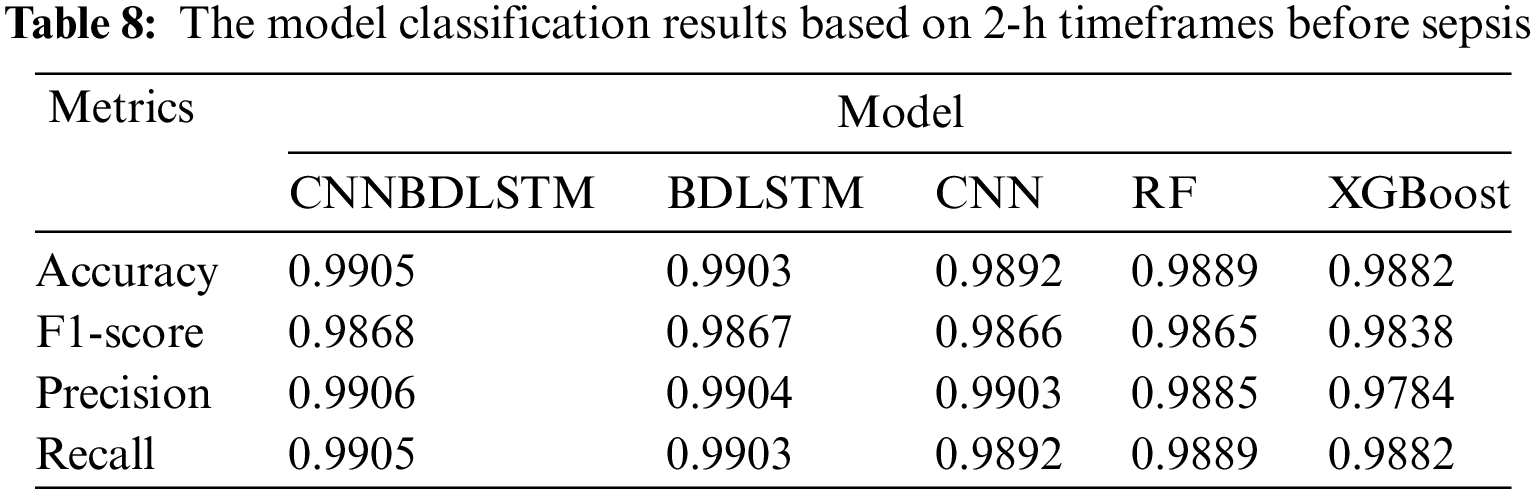
All models utilized in this study were assessed using accuracy, precision, F1-score, and recall performance metrics across various time intervals. The findings, particularly for the 1-h time frame before sepsis, reveal that CNNBDLSTM achieved the highest accuracy of 0.9915, as shown in Table 7. The results for the 2-h time frame before sepsis indicated that CNNBDLSTM achieved the highest accuracy of 0.9905, as shown in Table 8. In the 3-h time frame, CNNBDLSTM attained the highest accuracy of 0.9889, as shown in Table 9. In the 6-h time frame before sepsis, CNNBDLSTM achieved the highest accuracy of 0.9885, as shown in Table 10. For the 12-h time frame before sepsis, the results indicated that CNNBDLSTM achieved the highest accuracy of 0.9870, as shown in Table 11. In the 24-h time frame before sepsis, CNNBDLSTM achieved the highest accuracy of 0.9856, as shown in Table 12.
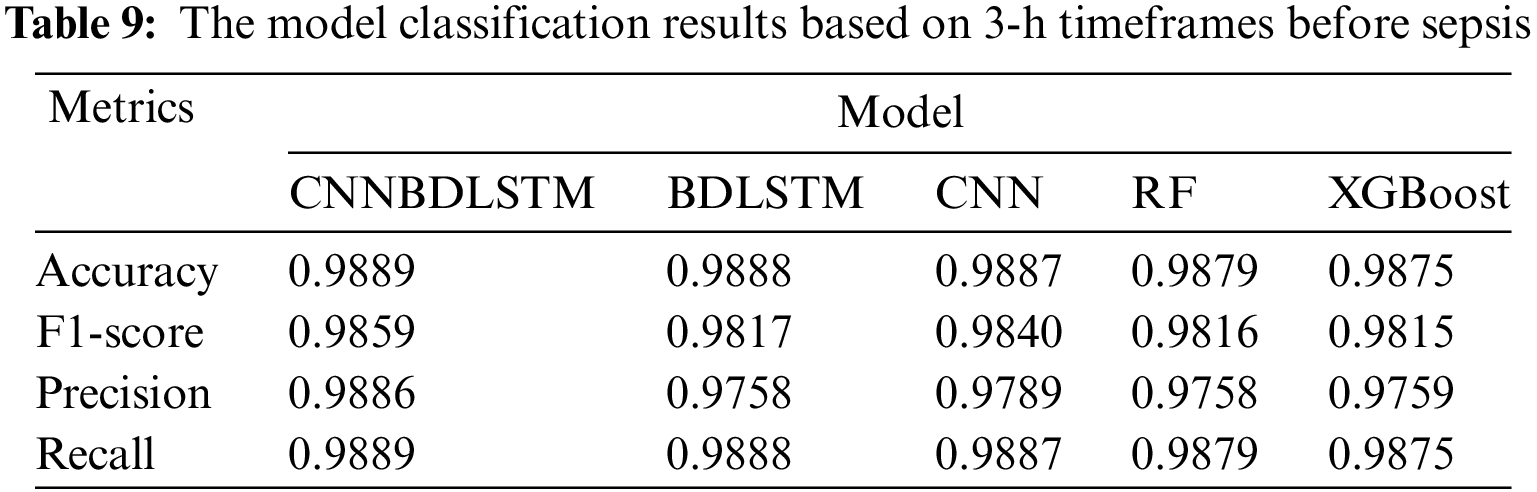
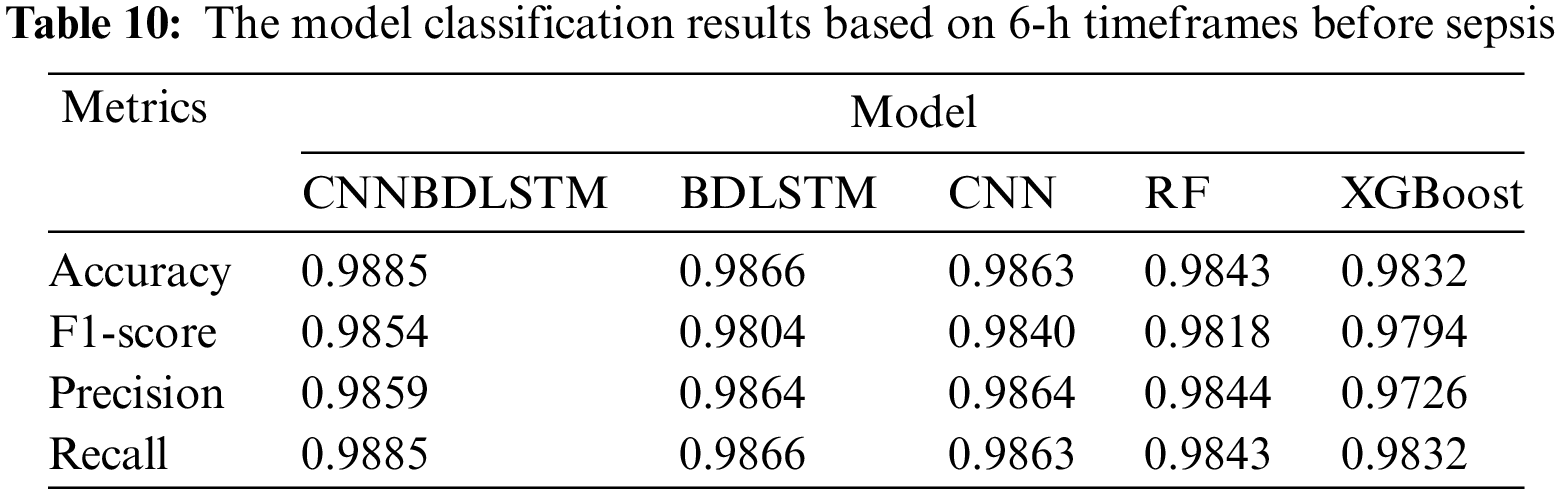
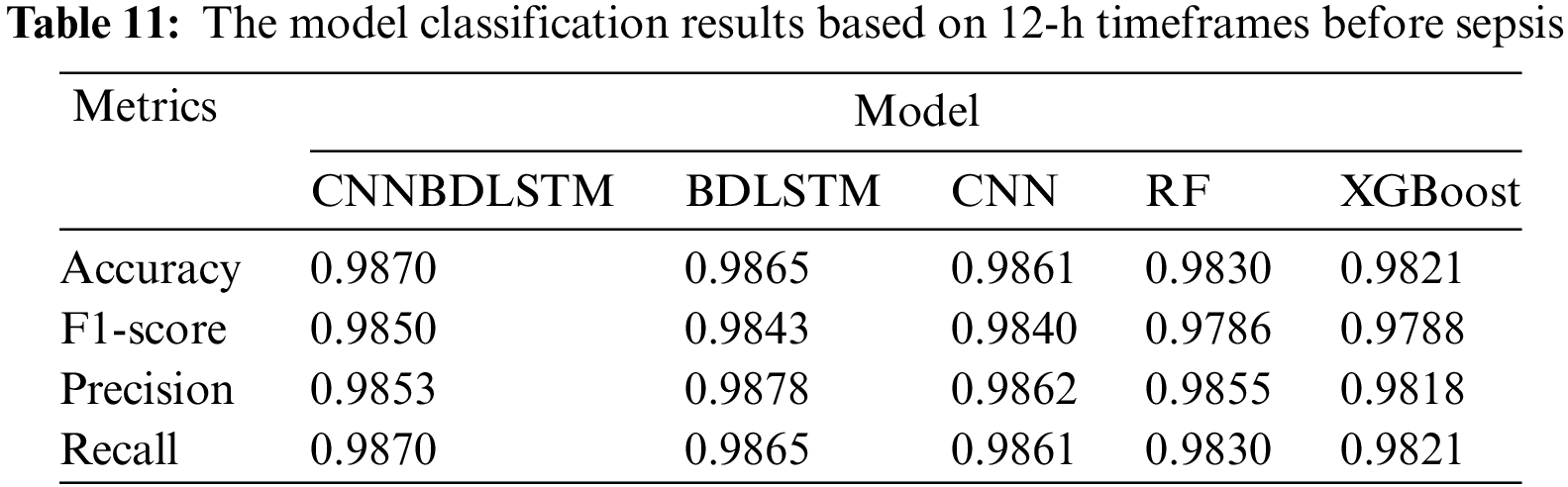
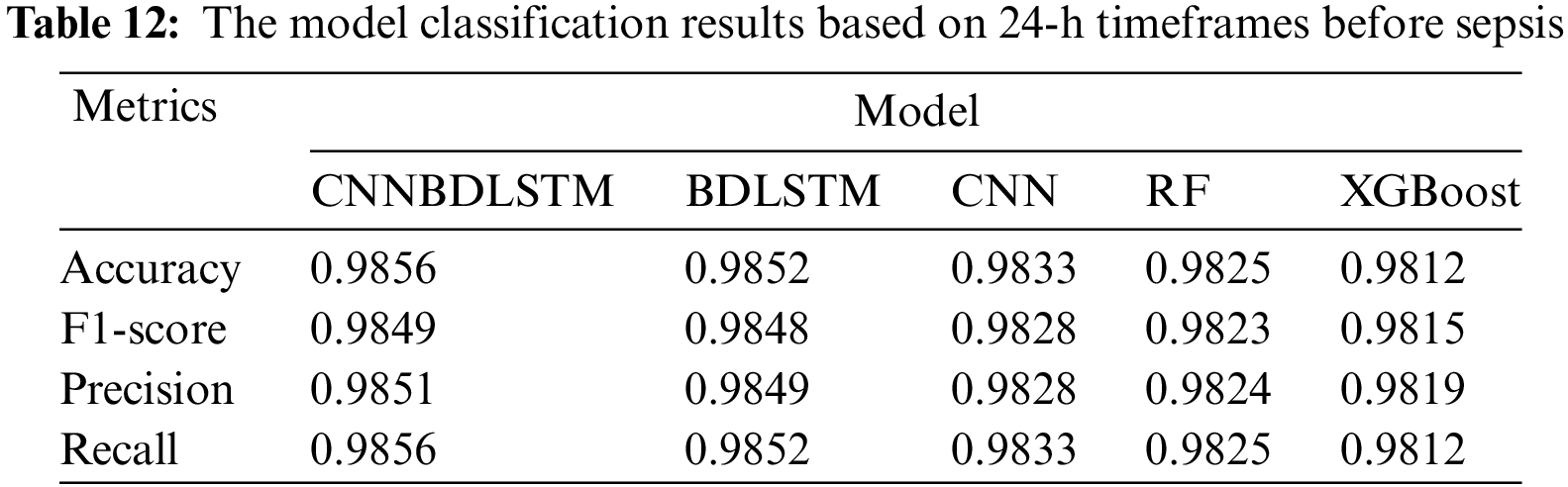
It was also noted that for the remaining performance metrics, including precision, F1-score, and recall, the models’ performance improved, with higher values observed as the timeframe approached the onset of sepsis. Once again, CNNBDLSTM achieved the highest precision values of 0.9916, 0.9906, 0.9886, 0.9859, 0.9853, and 0.9851 for the 1, 2, 3, 6, 12, and 24-h time frames before sepsis onset, respectively. In the case of the recall metric, it was observed that the proposed model once again achieved the highest recall values of 0.9915, 0.9905, 0.9889, 0.9885, 0.9870, and 0.9856 for the 1, 2, 3, 6, 12, and 24-h timeframes before sepsis onset, respectively. The comparative analysis for the F1-score metric reveals that CNNBDLSTM achieved scores of 0.9885, 0.9868, 0.9859, 0.9854, 0.9850, 0.9849, and 0.9804 for the 1, 2, 3, 6, 12, and 24-h time frames before sepsis onset, respectively.
4.3 The Results of the AUCROC Performance Evaluation
AUCROC is considered one of the most critical evaluation metrics for assessing the performance of any classification model. In this study, the performance of the proposed hybrid DL model (CNNBDLSTM) was evaluated alongside four other baseline models, namely RF, XGBoost, CNN, and BDLSTM, through hyperparameter tuning and 10-fold cross-validation. In Table 13, The AUCROC results for six-time frames before sepsis reveal that the proposed CNNBDLSTM model outperforms BDLSTM, CNN, RF, and XGBoost. The experiment focuses on predicting the time before sepsis occurs based on AUCROC evaluation metrics across the six-time frames: 24, 12, 6, 3, 2, and 1 h before sepsis onset.
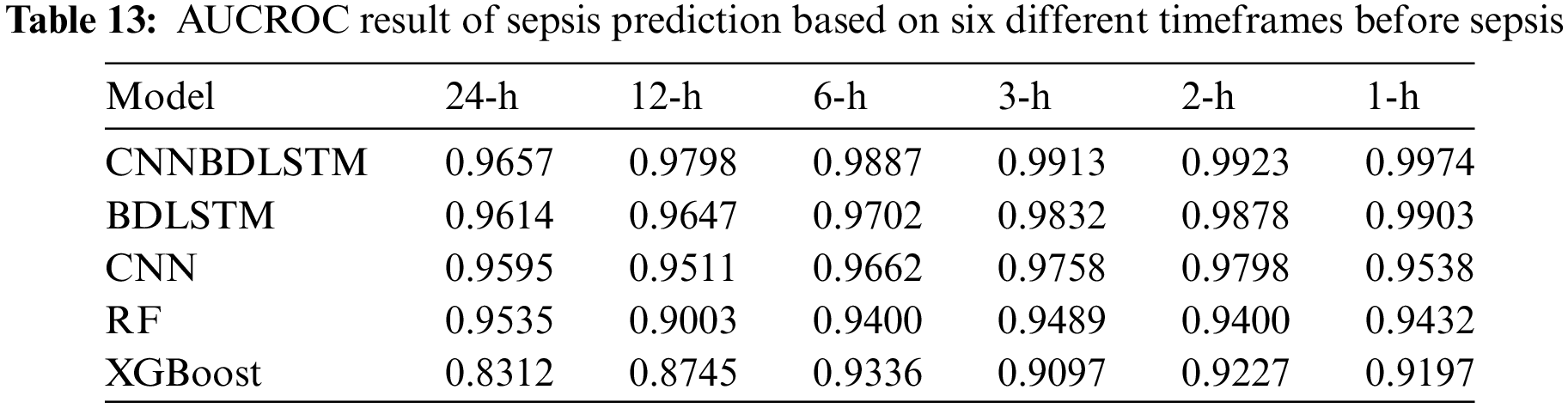
It was observed that:
• For predicting 1 h before sepsis onset, CNNBDLSTM attained the highest value of 0.9974. The second-best performing model, BDLSTM, with an AUCROC of 0.9903. CNN predicts an AUCROC of 0.9538. RF predicts 1 h before sepsis onset with a value of 0.9432. The least performed model is XGBoost, with a value of 0.9197.
• For predicting 2 h before sepsis onset, CNNBDLSTM achieved the highest prediction value of 0.9923, followed by BDLSTM with 0.9878, CNN with 0.9798, RF with 0.9400, and XGBoost with 0.9227.
• For predicting 3 h before sepsis onset, CNNBDLSTM achieved a value of 0.9913. Followed by BDLSTM 0.9832, CNN with 0.9758, RF with 0.9489, and XGBoost with 0.9097.
• For predicting 6 h before sepsis onset, CNNBDLSTM achieved the best AUCROC score of 0.9887, BDLSTM attained 0.9702, CNN obtained 0.9662, RF achieved 0.9400, and XGBoost obtained 0.9336.
• For predicting 12 h before sepsis onset, CNNBDLSTM again outperformed the rest of the models by achieving an AUCROC value of 0.9798. The next second-best model is BDLSTM, which attained a value of 0.9647. CNN achieved a value of 0.9511. RF gained a value of 0.9003. XGBoost obtained a value of 0.8745.
• For predicting 24 h before sepsis onset, CNNBDLSTM is again observed to be the best model with a value of 0.9657. BDLSTM with the value of 0.9614. CNN with the score of 0.9595. RF with a value of 0.9535 and XGBoost of 0.8312.
Fig. 14 illustrates the models’ AUCROC results predicting 1 to 3 h before sepsis onset. Fig. 15 illustrates the models’ AUCROC results of predicting 6 to 24 h before sepsis onset.
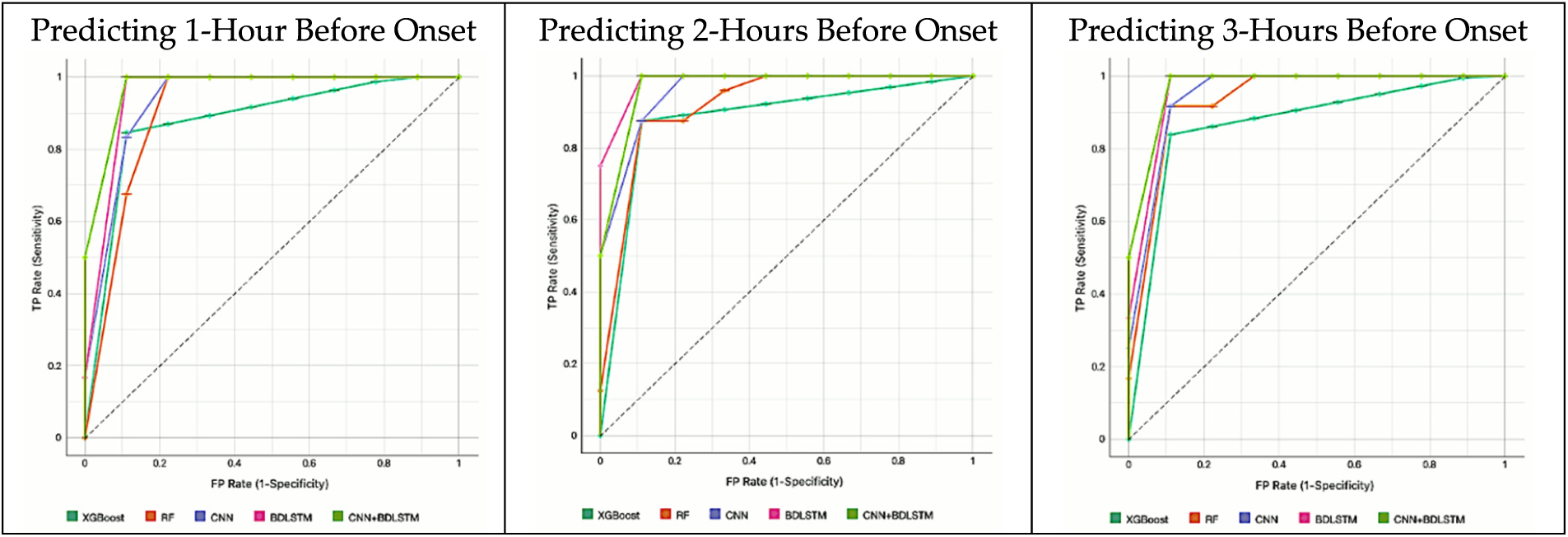
Figure 14: The results of the AUCROC of all models for predicting 1 to 3 h before sepsis
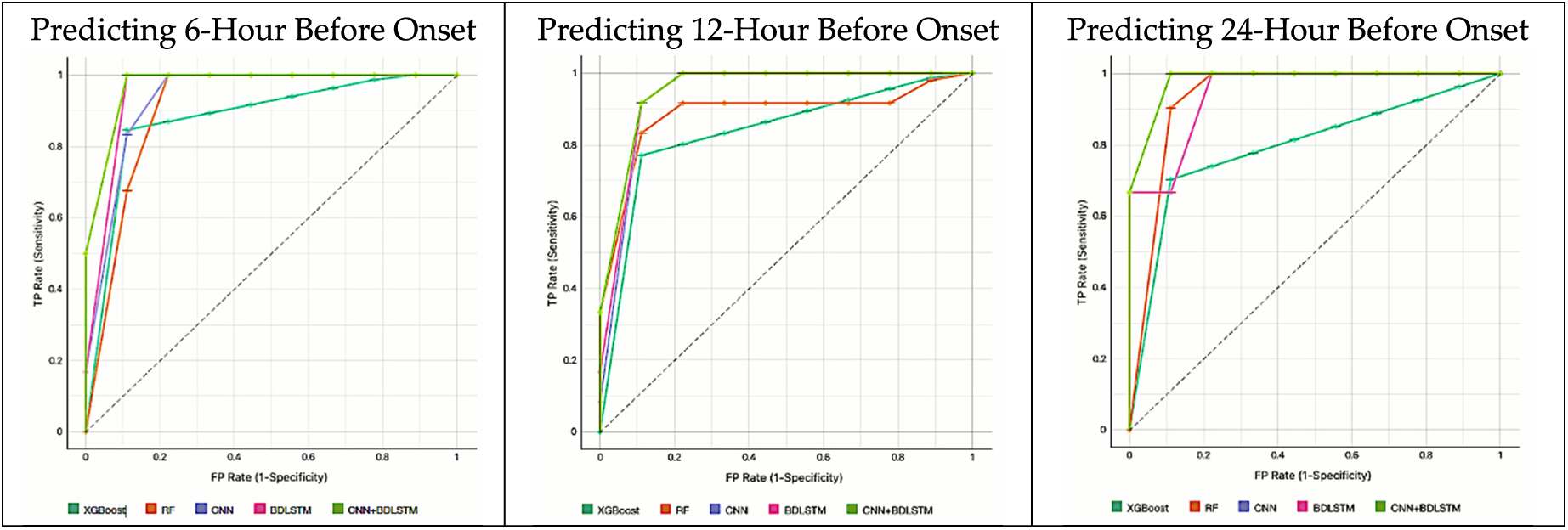
Figure 15: The results of the AUCROC of all models for predicting 6 to 24 h before sepsis
4.4 The Results of the CNNBDLSTM Confusion Matrix Performance Evaluation
This experiment aims to evaluate the performance of a classification model. The AUCROC results indicate that CNNBDLSTM is the best-performing model. In this section, we visualize and summarize the performance of the CNNBDLSTM model using the confusion matrix. The results (in Fig. 16) indicate that the CNNBDLSTM model exhibits less than 1.0% of type I errors and 0.0% of type II errors. Additionally, the model can accurately predict between 99.0% and 99.1% of true positive samples and 100% of true negative samples. These findings provide strong evidence of the model’s performance, which supports the result in Section 4.2.
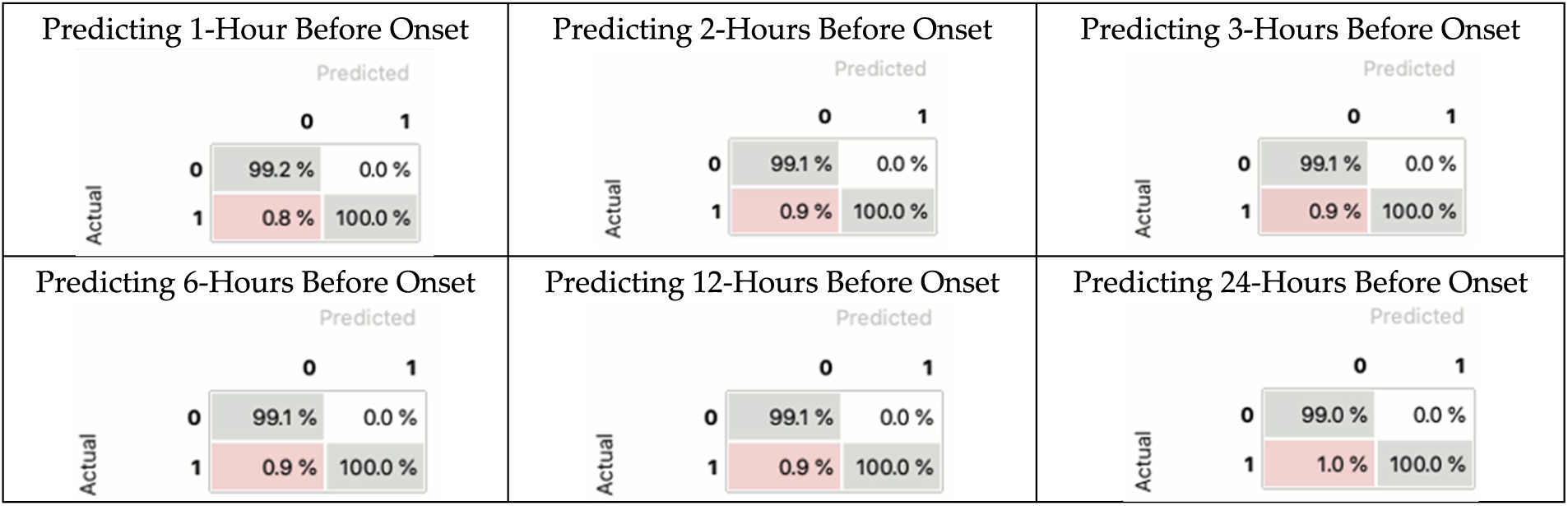
Figure 16: The result of the CNNBDLSTM confusion matrix
Based on the results presented in Tables 7 to 13, it is evident that CNNBDLSTM consistently achieved the highest accuracy and AUCROC scores across all observation window timeframes. The accuracy performance of CNNBDLSTM at each timeframe before sepsis onset is as follows: 0.9856 at 24 h, 0.9870 at 12 h, 0.9885 at 6 h, 0.9889 at 3 h, 0.9905 at 2 h, and 0.9915 at 1 h. Similarly, the AUCROC performance of CNNBDLSTM at each timeframe is as follows: 0.9657 at 24 h, 0.9798 at 12 h, 0.9887 at 6 h, 0.9913 at 3 h, 0.9923 at 2 h, and 0.9974 at 1 h before sepsis onset. These consistent results demonstrate the effectiveness of the CNNBDLSTM model in predicting sepsis across different timeframes, with particularly strong performance observed as the prediction timeframe approaches the onset of sepsis.
In the context of sepsis prediction, various factors such as data quality, feature selection, model complexity, and temporal dynamics can significantly influence the results. However, in this study, temporal dynamics emerge as the most probable factor affecting sepsis prediction accuracy.
The findings indicate a strong correlation between accuracy, AUCROC metrics, and the observation window timeframe. As the observation window timeframe approaches the onset of sepsis, there is a notable improvement in both accuracy and AUCROC scores. This suggests that models perform better when predicting closer to the time of sepsis onset, demonstrating enhanced performance with decreasing timeframes. These results underscore the importance of considering temporal dynamics in sepsis prediction models for timely intervention and improved accuracy. By closely aligning with the requirements of the sepsis-3 definition standard and emphasizing critical observation windows, this study provides valuable insights for early sepsis prediction. Specifically, the study highlights the significance of monitoring patients within the 12-h period preceding sepsis onset, further emphasizing the importance of timely intervention in improving patient outcomes.
4.6 Explanation of the Best-Performing Model
In this experiment, the ten most important features for predicting sepsis using the CNNBDLSTM model are shown in Fig. 17a. Using an explainer, Shapley additive explanation (SHAP) [47], we could decipher the features and clinical factors’ impacts and relative contributions to sepsis prediction. The results of the CNNBDLSTM models were analyzed using the tree SHAP method. The SHAP values of the tree models and ensembles may be quickly and accurately estimated using Tree SHAP. Fig. 17b shows the plot of a bee swarm, which illustrates how changing the feature value affects the model’s predictions. The color on the right indicates the feature’s value; a higher value is displayed in red, while a lower value is displayed in blue. Age, heart rate, and duration of stay in the intensive care unit were shown to be the most influential factors. The findings of laboratory tests (pH and bilirubin) and blood pressure also affect the sepsis prognosis. Results show that the CNNBDLSTM model’s effectiveness may be attributed to the factors identified.
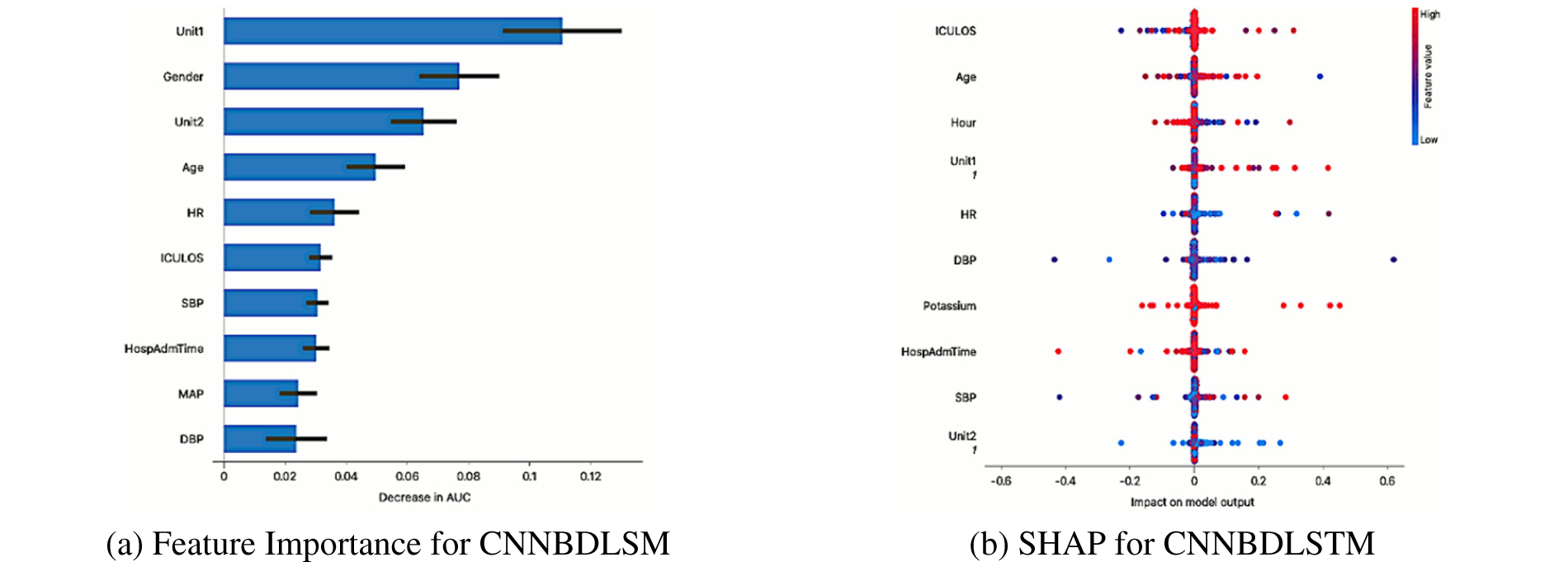
Figure 17: (a) Feature importance for CNNBDLSM and (b) SHAP for CNNBDLSTM
4.7 Comparative Analysis of the Current Study and Previous Studies
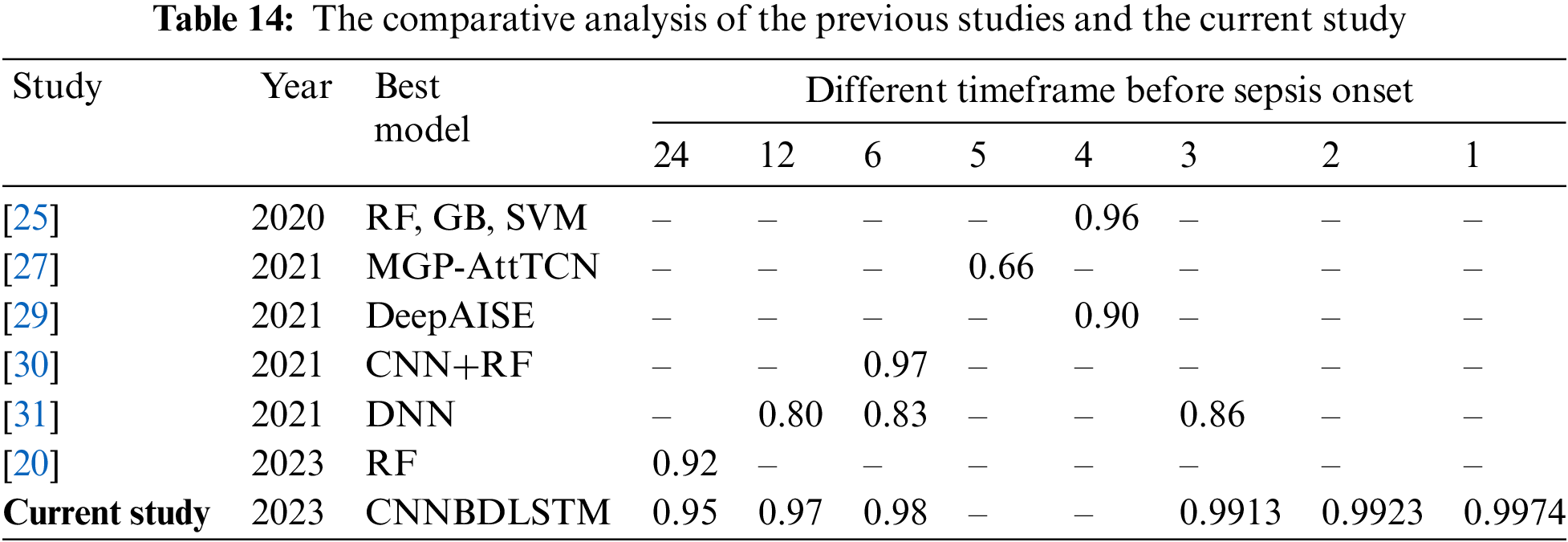
In this study, we need to benchmark our work with the previous studies to observe any improvement in the models’ performance. Thus, an extensive comparison with earlier studies that utilize the same MIMIC III dataset was performed. Given the time-dependent nature of sepsis, a reliable prediction model must allow for many validation periods. However, we discovered that only six studies conducted similar observations using temporal modalities. Before sepsis onset, the predictions were divided into nine timeframes: 24, 12, 6, 5, 4, 3, 2, and 1-h. It was observed that most studies predict the start of sepsis before the 24 to 4-h time frame. However, the current study extended the time frame to 2 and 1 h before sepsis onset, and none of the previous studies have predicted the specified time frame.
Based on the comparative analysis in Table 14, the CNNBDLSTM model in the current study achieved the highest AUCROC of 0.9974 at 1 h before the start of the sepsis. Similarly, the model in the current study predicted the highest AUCROC of 0.9923 at 2 h before early sepsis. For the case of a 6-h timeframe, the CNNBDLSTM model achieved an AUCROC of 0.98, which outperformed the previous model. The model’s efficacy improved as time approached the start of the sepsis, as seen by the trend lines of AUCROC in the six experiments. Predictions of early sepsis should be made at intervals between 1 and 2 h.
There are a few things to keep in mind about this study’s limitations. The fundamental basis of the investigation consisted exclusively of retrospective data. Based on the historical collection of the data, it is likely that numerous medical treatments, such as the implementation of warfarin in the intensive care unit, have undergone modifications over time. Furthermore, the diagnostic groups under investigation were only chosen based on electronic health data without considering imaging studies. Another limitation to be considered is that the precise time stamps were applied to time series data, substantially increasing the imported data’s complexity and security. Furthermore, it was noted that laboratory results and vital signs were rarely documented within the initial 48 h in both datasets. As a result, there was a substantial surge in the quantity of missing data across different periods. Even though the proposed model achieved high accuracy and AUCROC in predicting sepsis onset, there are some limitations that need to be considered. Both CNN and BDLSTM models can be computationally intensive, particularly when dealing with large datasets or complex architectures. Combining these two models exacerbates this issue, requiring significant computational resources and time for training and prediction. This can be a barrier in resource-constrained environments or real-time applications where efficiency is crucial.
Early diagnosis and timely administration of appropriate antibiotics for sepsis are crucial yet challenging tasks. Various prediction algorithms have been devised to evaluate the efficacy of early detection of patient deterioration, leveraging health indicators as fundamental components. While these scoring systems excel in predicting patient deterioration or mortality, they often lack precision in diagnosing sepsis for individual patients. With the availability of a vast volume and diverse range of healthcare data, it is advisable to utilize hybrid DL methods in constructing sepsis prediction models. In this study, we employed all variables available in the dataset to predict sepsis. The results demonstrate that our proposed model’s performance surpasses that of previous research models, which are often compared with traditional scoring systems for early sepsis assessment, as discussed in Section 1. Specifically, this research demonstrated superior information-gain outcomes when compared to conventional feature selection strategies. The study systematically evaluates, and contrasts models employed for early sepsis prediction utilizing extensive healthcare datasets. Given these findings, the utilization of hybrid DL models for early sepsis prediction in ICU patients is strongly recommended. The study conducted an analysis using two ML, two DL, and one hybrid DL model to predict sepsis onset across intervals ranging from 24 to 1 h. Notably, the CNNBDLSTM model emerged with exceptional performance, achieving an impressive AUCROC of 99.74% when predicting sepsis onset just one hour in advance.
In our future endeavors, it is highly promising and anticipated that incorporating additional features, such as genetic information and imaging investigations, will further enhance the predictive performance. Additionally, leveraging data obtained on the day of discharge could be instrumental in forecasting other critical outcomes, such as readmission to the intensive care unit. Our ultimate goal is to develop a secure and intelligent ML system [48] capable of delivering continuous updates on patients’ clinical status and detecting new clinical events. Such a system would serve as a valuable tool for physicians, enriching their clinical decision-making capabilities.
Acknowledgement: The authors would like to thank the Information Systems Department, College of Computer and Information Sciences, Princess Nourah bint Abdulrahman University for providing facilities to conduct the research.
Funding Statement: The authors extend their appreciation to the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia, for funding this research work through Project Number RI-44-0214.
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: S. Sakri, S. Basheer, Z.M. Zain; data collection: M. Alharaki; analysis and interpretation of results: M. Alohaly, D. Nasser. S. Basheer; draft manuscript preparation: S. Sakri. N.H.A. Ismail. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Data is available on request due to privacy and ethical restrictions.
Ethics Approval: The study was conducted with exemption approval from the Institutional Review Board of the King Abdulaziz City for Science and Technology (KACST), Kingdom of Saudi Arabia (protocol code 23-0540 on 25 July 2023).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. J. Arwyn-Jones and A. J. Brent, “Sepsis,” Surg., vol. 37, no. 1, pp. 1–8, 2019. [Google Scholar]
2. W. Alhazzani et al., “Surviving sepsis campaign: Guidelines on the management of critically ill adults with coronavirus disease 2019 (COVID-19),” Intens. Care Med., vol. 46, no. 5, pp. 854–887, 2020. doi: 10.1007/s00134-020-06022-5 [Google Scholar] [PubMed] [CrossRef]
3. C. S. Hollenbeak et al., “Costs and consequences of a novel emergency department sepsis diagnostic test: The intellisep index,” Crit. Care Explor., vol. 5, no. 7, pp. e0942, 2023. doi: 10.1097/CCE.0000000000000942 [Google Scholar] [PubMed] [CrossRef]
4. P. Arina and M. Singer, “Pathophysiology of sepsis,” Curr. Opin. Anesthesiol., vol. 34, no. 2, pp. 77–84, 2021. doi: 10.1097/ACO.0000000000000963 [Google Scholar] [PubMed] [CrossRef]
5. G. Guan, C. M. Y. Lee, S. Begg, A. Crombie, and G. Mnatzaganian, “The use of early warning system scores in pre-hospital and emergency department settings to predict clinical deterioration: A systematic review and meta-analysis,” PLoS One, vol. 17, no. 3, pp. e0265559, 2022. [Google Scholar]
6. R. Thakur, V. N. Rohith, and J. K. Arora, “Mean SOFA score in comparison with APACHE II Score in predicting mortality in surgical patients with sepsis,” Cureus., vol. 15, no. 3, pp. e36653, 2023. doi: 10.7759/cureus.36653 [Google Scholar] [PubMed] [CrossRef]
7. M. Serries, H. Zenzen, M. Heine, T. Holderried, P. Brossart and K. Schwab, “Evaluation of factors associated with survival in allogeneic stem cell-transplanted patients admitted to the intensive care unit (ICU),” Hematol., vol. 28, no. 1, pp. 2256198, 2023. doi: 10.1080/16078454.2023.2256198 [Google Scholar] [PubMed] [CrossRef]
8. R. Moreno et al., “The sequential organ failure assessment (SOFA) score: Has the time come for an update?,” Crit. Care, vol. 27, no. 1, pp. 15, 2023. doi: 10.1186/s13054-022-04290-9 [Google Scholar] [PubMed] [CrossRef]
9. A. J. Heffernan and K. J. Denny, “Host diagnostic biomarkers of infection in the ICU: Where are we and where are we going?,” Curr. Infect. Dis. Rep., vol. 23, no. 4, pp. 1–11, 2021. doi: 10.1007/s11908-021-00747-0 [Google Scholar] [PubMed] [CrossRef]
10. S. Gerry et al., “Early warning scores for detecting deterioration in adult hospital patients: Systematic review and critical appraisal of methodology,” BMJ, vol. 369, pp. m1501, 2020. doi: 10.1136/bmj.m1501 [Google Scholar] [PubMed] [CrossRef]
11. W. Walter et al., “Artificial intelligence in hematological diagnostics: Game changer or gadget?,” Blood Rev., vol. 58, no. 10, pp. 101019, 2023. doi: 10.1016/j.blre.2022.101019 [Google Scholar] [PubMed] [CrossRef]
12. D. R. Giacobbe et al., “Early detection of sepsis with machine learning techniques: A brief clinical perspective,” Front. Med., vol. 8, pp. 617486, 2021. doi: 10.3389/fmed.2021.617486 [Google Scholar] [PubMed] [CrossRef]
13. C. Y. Cheng et al., “Machine learning models for predicting in-hospital mortality in patient with sepsis: Analysis of vital sign dynamics,” Front. Med., vol. 9, pp. 964667, 2022. doi: 10.3389/fmed.2022.964667 [Google Scholar] [PubMed] [CrossRef]
14. A. Honoré, D. Forsberg, K. Adolphson, S. Chatterjee, K. Jost and E. Herlenius, “Vital sign-based detection of sepsis in neonates using machine learning,” Acta Paediatr., vol. 112, no. 4, pp. 686–696, 2023. doi: 10.1111/apa.16660 [Google Scholar] [PubMed] [CrossRef]
15. X. Li et al., “A time-phased machine learning model for real-time prediction of sepsis in critical care,” Critical Care Medicine, vol. 48, no. 10, pp. e884–e888, 2020. doi: 10.1016/j.artmed.2020.101820. [Google Scholar] [CrossRef]
16. H. F. Deng et al., “Evaluating machine learning models for sepsis prediction: A systematic review of methodologies,” iScience, vol. 25, no. 1, pp. 103651, 2022. doi: 10.1016/j.isci.2021.103651 [Google Scholar] [PubMed] [CrossRef]
17. M. Apalak and K. Kiasaleh, “Improving sepsis prediction performance using conditional recurrent adversarial networks,” IEEE Access, vol. 10, pp. 134466–134476, 2022. doi: 10.1016/j.compbiomed.2020.104110. [Google Scholar] [CrossRef]
18. M. U. Alam and R. Rahmani, “FedSepsis: A federated multi-modal deep learning-based internet of medical things application for early detection of sepsis from electronic health records using raspberry pi and jetson nano devices,” Sens., vol. 23, no. 2, pp. 970, 2023. doi: 10.3390/s23020970 [Google Scholar] [PubMed] [CrossRef]
19. N. Kijpaisalratana, D. Sanglertsinlapachai, S. Techaratsami, K. Musikatavorn, and J. Saoraya, “Machine learning algorithms for early sepsis detection in the emergency department: A retrospective study,” Int. J. Med. Inform., vol. 160, no. 10, pp. 104689, 2022. doi: 10.1016/j.ijmedinf.2022.104689 [Google Scholar] [PubMed] [CrossRef]
20. M. Gholamzadeh, H. Abtahi, and R. Safdari, “Comparison of different machine learning algorithms to classify patients suspected of having sepsis infection in the intensive care unit,” Informatics in Medicine Unlocked, vol. 38, pp. 101236, 2023. doi: 10.1016/j.imu.2023.101236. [Google Scholar] [CrossRef]
21. M. Yang et al., “An explainable artificial intelligence predictor for early detection of sepsis,” Crit. Care Med., vol. 48, no. 11, pp. e1091–6, 2020. doi: 10.1097/CCM.0000000000004550 [Google Scholar] [PubMed] [CrossRef]
22. M. A. Reyna et al., “Early prediction of sepsis from clinical data: The PhysioNet/computing in cardiology challenge 2019,” Crit. Care Med., vol. 48, no. 2, pp. 210–217, 2020. doi: 10.1097/CCM.0000000000004145 [Google Scholar] [PubMed] [CrossRef]
23. J. S. Choi et al., “Implementation of complementary model using optimal combination of hematological parameters for sepsis screening in patients with fever,” Sci. Rep., vol. 10, no. 1, pp. 273, 2020. doi: 10.1038/s41598-019-57107-1 [Google Scholar] [PubMed] [CrossRef]
24. J. Kim, H. Chang, D. Kim, D. H. Jang, I. Park and K. Kim, “Machine learning for prediction of septic shock at initial triage in emergency department,” J. Crit. Care, vol. 55, pp. 163–170, 2020. doi: 10.1016/j.jcrc.2019.09.024 [Google Scholar] [PubMed] [CrossRef]
25. Z. M. Ibrahim, H. Wu, A. Hamoud, L. Stappen, R. J. Dobson and A. Agarossi, “On classifying sepsis heterogeneity in the ICU: Insight using machine learning,” J. Am. Med. Inform. Assoc., vol. 27, no. 3, pp. 437–443, 2020. doi: 10.1093/jamia/ocz211 [Google Scholar] [PubMed] [CrossRef]
26. Y. Duan et al., “Early prediction of sepsis using double fusion of deep features and handcrafted features,” Appl. Intell., vol. 53, no. 14, pp. 17903–17919, 2023. doi: 10.1007/s10489-022-04425-z [Google Scholar] [PubMed] [CrossRef]
27. M. Rosnati and V. Fortuin, “MGP-AttTCN: An interpretable machine learning model for the prediction of sepsis,” PLoS One, vol. 16, no. 5, pp. e0251248, 2021. doi: 10.1371/journal.pone.0251248 [Google Scholar] [PubMed] [CrossRef]
28. D. Zhang, C. Yin, K. M. Hunold, X. Jiang, J. M. Caterino and P. Zhang, “An interpretable deep-learning model for early prediction of sepsis in the emergency department,” Patterns, vol. 2, no. 2, pp. 100196, 2021. doi: 10.1016/j.patter.2020.100196 [Google Scholar] [PubMed] [CrossRef]
29. S. P. Shashikumar, C. S. Josef, A. Sharma, and S. Nemati, “DeepAISE–An interpretable and recurrent neural survival model for early prediction of sepsis,” Artif. Intell. Med., vol. 113, pp. 102036, 2021. doi: 10.1016/j.artmed.2021.102036 [Google Scholar] [PubMed] [CrossRef]
30. T. Aşuroğlu and H. Oğul, “A deep learning approach for sepsis monitoring via severity score estimation,” Comput. Meth. Prog. Bio., vol. 198, no. 10219, pp. 105816, 2021. doi: 10.1016/j.cmpb.2020.105816 [Google Scholar] [PubMed] [CrossRef]
31. S. P. Oei, R. J. van Sloun, M. van der Ven, H. H. Korsten, and M. Mischi, “Towards early sepsis detection from measurements at the general ward through deep learning,” Intell.-Based Med., vol. 5, no. 8, pp. 100042, 2021. doi: 10.1016/j.ibmed.2021.100042. [Google Scholar] [CrossRef]
32. A. Rafiei, A. Rezaee, F. Hajati, S. Gheisari, and M. Golzan, “SSP: Early prediction of sepsis using fully connected LSTM-CNN model,” Comput. Biol. Med., vol. 128, pp. 104110, 2021. doi: 10.1016/j.compbiomed.2020.104110 [Google Scholar] [PubMed] [CrossRef]
33. B. Y. Al-Mualemi and L. Lu, “A deep learning-based sepsis estimation scheme,” IEEE Access, vol. 9, pp. 5442–5452, 2021. doi: 10.1109/ACCESS.2020.3043732. [Google Scholar] [CrossRef]
34. A. D. Bedoya et al., “Machine learning for early detection of sepsis: An internal and temporal validation study,” JAMIA Open, vol. 3, no. 2, pp. 252–260, 2020. doi: 10.1093/jamiaopen/ooaa006 [Google Scholar] [PubMed] [CrossRef]
35. S. M. Lauritsen et al., “Early detection of sepsis utilizing deep learning on electronic health record event sequences,” Artif. Intell. Med, vol. 104, no. 11, pp. 101820, 2020. doi: 10.1016/j.artmed.2020.101820 [Google Scholar] [PubMed] [CrossRef]
36. K. H. Goh et al., “Artificial intelligence in sepsis early prediction and diagnosis using unstructured data in healthcare,” Nat. Commun.vol. 12, no. 1, pp. 711, 2021. doi: 10.1038/s41467-021-20910-4 [Google Scholar] [PubMed] [CrossRef]
37. E. Röösli, S. Bozkurt, and T. Hernandez-Boussard, “Peeking into a black box, the fairness and generalizability of a MIMIC-III benchmarking model,” Sci. Data, vol. 9, no. 1, pp. 24, 2022. doi: 10.1038/s41597-021-01110-7 [Google Scholar] [PubMed] [CrossRef]
38. I. Persson, A. Östling, M. Arlbrandt, J. Söderberg, and D. Becedas, “A machine learning sepsis prediction algorithm for intended intensive care unit use (NAVOY SepsisProof-of-concept study,” JMIR Form. Res., vol. 5, no. 9, pp. e28000, 2021. doi: 10.2196/28000 [Google Scholar] [PubMed] [CrossRef]
39. R. Gauer, D. Forbes, and N. Boyer, “Sepsis: Diagnosis and management,” Am. Fam. Physician, vol. 101, no. 7, pp. 409–418, 2020 [Google Scholar] [PubMed]
40. S. Baral, A. Alsadoon, P. W. C. Prasad, S. Al Aloussi, and O. H. Alsadoon, “A novel solution of using deep learning for early prediction cardiac arrest in Sepsis patient: Enhanced bidirectional long short-term memory (LSTM),” Multimed. Tools Appl., vol. 80, no. 21–23, pp. 32639–32664, 2021. doi: 10.1007/s11042-021-11176-5. [Google Scholar] [CrossRef]
41. C. T. Ferraz, A. M. A. Liberatore, T. L. Yamada, and I. H. J. Koh, “A new convolutional neural network-construct for sepsis enhances pattern identification of microcirculatory dysfunction,” Intell.-Based Med., vol. 8, pp. 100106, 2023. doi: 10.1016/j.ibmed.2023.100106. [Google Scholar] [CrossRef]
42. E. A. Strickler, J. Thomas, J. P. Thomas, B. Benjamin, and R. Shamsuddin, “Exploring a global interpretation mechanism for deep learning networks when predicting sepsis,” Sci. Rep., vol. 13, no. 1, pp. 3067, 2023. doi: 10.1038/s41598-023-30091-3 [Google Scholar] [PubMed] [CrossRef]
43. A. Ullah, H. Qayyum, M. K. Khan, and F. Ahmad, “Sepsis detection using extreme gradient boost (XGBA supervised learning approach,” in Proc. 2021 Mohammad Ali Jinnah Univ. Int. Conf. Comput. (MAJICC), Karachi, Pakistan, Jul. 15–17, 2021, pp. 1–6. [Google Scholar]
44. L. Ke et al., “Identification of potential diagnostic and prognostic biomarkers for sepsis based on machine learning,” Comput. Struct. Biotechnol. J., vol. 21, no. 741, pp. 2316–2331, 2023. doi: 10.1016/j.csbj.2023.03.034 [Google Scholar] [PubMed] [CrossRef]
45. F. Alahmari, “A comparison of resampling techniques for medical data using machine learning,” J. Info. Know. Manag., vol. 19, no. 1, pp. 2040016, 2020. doi: 10.1142/S021964922040016X. [Google Scholar] [CrossRef]
46. I. Markoulidakis, I. Rallis, I. Georgoulas, G. Kopsiaftis, A. Doulamis and N. Doulamis, “Multiclass confusion matrix reduction method and its application on net promoter score classification problem,” Tech., vol. 9, no. 4, pp. 81, 2021. doi: 10.3390/technologies9040081. [Google Scholar] [CrossRef]
47. S. M. Lundberg et al., “From local explanations to global understanding with explainable AI for trees,” Nat. Mach. Intell., vol. 2, no. 1, pp. 2522–5839, 2020. doi: 10.1038/s42256-019-0138-9 [Google Scholar] [PubMed] [CrossRef]
48. A. Sundas, S. Badotra, S. Bharany, A. Almogren, E. M. Tag-ElDin and A. U. Rehman, “HealthGuard: An intelligent healthcare system security framework based on machine learning,” Sustain., vol. 14, no. 19, pp. 11934, 2022. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF
 Downloads
Downloads
 Citation Tools
Citation Tools