 Open Access
Open Access
ARTICLE
Intelligent Machine Learning Based Brain Tumor Segmentation through Multi-Layer Hybrid U-Net with CNN Feature Integration
Department of Information Technology, Faculty of Computing and Information Technology, King Abdulaziz University, P.O. Box 344, Rabigh, 21911, Saudia Arabia
* Corresponding Author: Sharaf J. Malebary. Email:
Computers, Materials & Continua 2024, 79(1), 1301-1317. https://doi.org/10.32604/cmc.2024.047917
Received 22 November 2023; Accepted 08 March 2024; Issue published 25 April 2024
Abstract
Brain tumors are a pressing public health concern, characterized by their high mortality and morbidity rates. Nevertheless, the manual segmentation of brain tumors remains a laborious and error-prone task, necessitating the development of more precise and efficient methodologies. To address this formidable challenge, we propose an advanced approach for segmenting brain tumor Magnetic Resonance Imaging (MRI) images that harnesses the formidable capabilities of deep learning and convolutional neural networks (CNNs). While CNN-based methods have displayed promise in the realm of brain tumor segmentation, the intricate nature of these tumors, marked by irregular shapes, varying sizes, uneven distribution, and limited available data, poses substantial obstacles to achieving accurate semantic segmentation. In our study, we introduce a pioneering Hybrid U-Net framework that seamlessly integrates the U-Net and CNN architectures to surmount these challenges. Our proposed approach encompasses preprocessing steps that enhance image visualization, a customized layered U-Net model tailored for precise segmentation, and the inclusion of dropout layers to mitigate overfitting during the training process. Additionally, we leverage the CNN mechanism to exploit contextual information within brain tumor MRI images, resulting in a substantial enhancement in segmentation accuracy. Our experimental results attest to the exceptional performance of our framework, with accuracy rates surpassing 97% across diverse datasets, showcasing the robustness and effectiveness of our approach. Furthermore, we conduct a comprehensive assessment of our method’s capabilities by evaluating various performance measures, including the sensitivity, Jaccard-index, and specificity. Our proposed model achieved 99% accuracy. The implications of our findings are profound. The proposed Hybrid U-Net model emerges as a highly promising diagnostic tool, poised to revolutionize brain tumor image segmentation for radiologists and clinicians.Keywords
Brain tumors are currently the most common factor in death worldwide. A brain tumor forms when abnormal cells in the brain multiply and grow out of control. Major brain tissues are harmed, and in certain cases, cancer might develop as a result [1]. It is dangerous to people, it kills, and it drastically lowers people’s standard of living. The risk of acquiring brain cancer, however, is steadily rising due to the unchecked growth of brain tumors. Due to the unchecked growth of the tumors, brain cancer has a rising incidence rate and ranks as the tenth top cause of death worldwide. Gliomas, meningiomas, and pituitary tumors are only a few of the malignancies that may affect the human brain [2–6].
Recent studies have focused mostly on the subtypes and categorization of breast cancer, as well as other malignancies, including those of the lung and colon. The processing of medical images is closely related to the timely diagnosis of patients’ conditions and subsequent rehabilitation treatment. Combining with convolutional neural networks (CNN), it can effectively enhance the accuracy of image segmentation. Due to the small amount of medical image data and unbalanced categories, it is very difficult to perform semantic segmentation of brain tumors. Moreover, the structure of brain tissue is complex, tumors may exist in various regions of the brain, the boundaries between brain tissues are difficult to distinguish, and the resolution of MRI images of brain tumors is low [7]. Many problems will greatly affect the segmentation accuracy of brain tumors and an accurate diagnosis would save the lives of many brain tumor patients.
The field of brain tumor segmentation has grown rapidly in recent years, with several segmentation methods being developed on a wide range of datasets. Three types of recently created segmentation models are clustering-based segmentation [8], supervised machine learning (ML) segmentation [9], and DL segmentation [10]. The most popular technique in medical imaging is ML based picture semantic segmentation, which is used to automatically separate and recognize different human organs and pathologies. The algorithms fully-convolutional-network (FCN) [11], SegNet [12], U-Net, DeepLab series [13], and DANet [14] are examples of common picture semantic segmentation methods. An FCN learns features from all the combinations of the features of the previous layer, however, they are incredibly computationally expensive. DeepLab cannot capture the boundaries of complex objects, although it is computationally fast. DANet is computationally expensive and requires more robustness. The U-Net network stands out among them because of its distinctive U-shaped design, which makes it more suited for the task of segmenting medical images.
Numerous researchers have made significant efforts to improve and enhance the U-Net model for medical image segmentation. It has been successfully applied to various medical characterizations, including the separation of bladder cancer cells, identification of skin lesions, gallstones, liver tumors, and brain tumors. Despite its superior segmentation performance, several challenges still need to be addressed, like the U-shaped network, which has shown promise; there is room for improvement in its structure. Many scholars have introduced auxiliary modules like attention, residual, dropout, and pyramid modules to enhance the segmentation process. However, extracting fine-grained details and coarse-grained semantics efficiently from both the encoder and decoder remains a potential area for further advancement. Moreover, the continuous improvement of the U-Net model and its variations has led to a significant increase in network parameters. While this has improved the performance of medical representation recognition, it comes at the cost of increased memory consumption and higher hardware requirements. Another problem is that the large size of the network model weight file also poses challenges in deployment, application, and upgrade on resource-limited devices like mobile terminals or embedded systems. Achieving a balance between network identification effectiveness and parameter quantity is crucial for realizing a lightweight network.
So, it is suitable to pay more attention to the actual capabilities of the U-shaped network with fewer parameters, that is, learn from each other’s strengths in the network structure, compress the parameters, and design a limited network without adding any other auxiliary modules. This study introduces the Hybrid U-Net model, which enhances MRI classification through the development of a customized U-Net and the incorporation of feature fusion based on CNN. To achieve enhanced segmentation results, a series of data preprocessing steps are employed, involving techniques such as slicing, down-sampling, and channel distribution. We used two different MRI datasets throughout the experimental phase to assess the performance of our implied model. The results of these tests demonstrated our model’s superiority to all other current state-of-the-art procedures, including cascaded and ensemble methods. This underscores the exceptional performance and effectiveness of our proposed approach within the domain of MRI data analysis. The rest of the paper is organized as follows: Section 2 briefly gives a literature review; the framework of our novel technique is shown in Section 3. Section 4 presents experimental data, comments, and comparisons with existing techniques. Finally, the article is wrapped up in Section 5.
Traditional segmentation methods use various segmentation algorithms, such as the initial segmentation algorithm, edge detection algorithm, landmark algorithm, and growing region algorithm. The details are as follows: The initial segmentation algorithm [15] is used for the segmentation process. Brain tumor MR image segmentation: Due to the signal intensity difference between brain tissue and healthy tissue, after preliminary processes such as image enhancement, tumors can be segmented according to initial measurements and morphological functions. Image segmentation methods using initial segmentation algorithms are susceptible to pre- and post-processing procedures such as homogenization and bias correction and have noise [16]. It is not easy to segment brain tumor MRI images using only information obtained from local or international measurements. Researchers often combine segmentation with other segmentation methods as a starting point. The segmentation method uses an edge detection algorithm to identify tumor edge pixels in the image by detecting changes in the image [17] and combining the pixels to obtain the area of the tumor in the final part of the image. There are many image segmentation techniques using edge detection algorithms such as Sobel, Prewitt, Roberts, and Canny [18,19], and these models have also been developed after many improvements.
Based on the improved tumor health algorithm, Moradi et al. [20] combined the image-based threshold segmentation algorithm with the Sobel operator to detect brain lesions, and then used the closed contour algorithm and object-based segmentation to detect the edges of tumor cells. Remove the tumor. Edge-based segmentation methods, although simple, sometimes produce open contours and are sensitive to thresholding. In MRI images, the boundaries of brain tumors vary widely, so boundary-based segmentation is difficult to achieve very good results. The region-growing algorithm-based segmentation approach initially selects a few seed pixels, then merges nearby comparable pixels or sub-regions into the same region in accordance with the predetermined requirements until all regions are divided, and eventually the tumor tissue is segmented. Both manually and automatically selecting the first seed are possible. A fuzzy knowledge-based seed region growth approach was put out by Lin et al. [21] for multimodal MRI imaging. This method preprocesses the seed region to identify the tumor’s original seed by exploiting fuzzy edges and similarities. The region-growing technique of segmentation is susceptible to noise and seed point initialization; low segmentation accuracy will result from choosing the wrong pixel seeds or from an image with excessive background noise [22,23].
The wavelet transform algorithm is used in the image segmentation method, which first transforms the wavelet transform concept to convert the picture histogram into wavelet coefficients [24,25]. Next, the threshold is established using the segmentation conditions and wavelet coefficients, and the tumor area is then divided according to the threshold. Chen et al. proposed a CT/MRI segmentation technology based on wavelet analysis and MRF [26] and determined the data area and data edge of the object to be partitioned. Islam et al. [27] proposed a multi-modal MRI brain segmentation technology based on wavelet analysis and machine learning, using features obtained from wavelet analysis, combining with density, distinguishing variants, and unique features, and referring to classifiers such as random forests. In general, the brain tumor MRI segmentation method based on the segmentation model generally has simple models and slow segmentation speeds and alone cannot provide good and accurate segmentation of brain tumor MRI images.
In recent years, brain tumor segmentation methods based on DL have become a popular research direction in the field of brain tumor segmentation due to their advantages of fast, efficient, and accurate extraction of tumor features. In the brain-tumor image segmentation task based on DL, commonly used segmentation algorithms are mainly CNN-based algorithms and recurrent neural networks (RNN)-based algorithms [28]. Single-path and multi-path neural network topologies are used in CNN approaches for segmenting brain tumors [29]. Convolutional layers are used to sample an input picture in a CNN, which is then followed by pooling and activation layers in a single-path neural network. Many studies have successfully segmented MRI images of brain tumors by employing single-path CNNs [30]. In contrast to single-path networks, multi-path CNNs may combine or concatenate the information derived from many processing routes at multiple scales. For brain tumor segmentation, the authors’ [31] developed a dual-channel 3D CNN with 11 layers that employs multiple, fully connected multi-path CNNs. This is the first neural network to use 3D-convolutions. After scaling the input picture, a fully linked conditional-random-field (CRF) is used to correct for any remaining imperfections. By feeding in normal-resolution picture segments, the network can learn local features of tumor images, such as texture and boundaries. Because networks are trained to learn global spatial information (such as tumor location) using low-resolution fragments as input, their segmentation performance for MRI images is poor from the standpoint of the global approach. The reliability of segmentation declines in tandem with the growth in network complexity.
Although many enhancements have been made to the original CNN, the FCN remains a masterwork of deep learning for image segmentation. Based on FCN, Badrinarayanan et al. [12] suggested an enhancement. In the SegNet model, the maximum value coordinate is recorded during the down-sampling pooling process, During the up-sampling process, the feature map values are mapped to this coordinate, and the feature values in the new feature map are set to zero. It addresses the issues of coarse segmentation and feature duplication caused by FCN’s skip connection. The Deconv-Net proposed by Noh et al. [32] added two fully connected layers behind the SegNet to make the segmentation results more accurate. The U-Net network is improved from the FCN network, and the innovative use of the U-shaped network structure can make full use of the feature information of the brain tumor image context.
U-Net has many different models, such as integration with ResNet, Dense-Net, and other modules. Increasing the number of layers in the design of neural networks can improve the performance of the network, but more layers will cause gradients to disappear and overfitting to occur. Common methods for addressing degradation issues include residual connections and dense connections. Res-U-Net [33] optimizes the network topology by substituting the residual block for the original convolution block in U-Net, albeit at the expense of segmentation accuracy. There appears to be no noticeable improvement. For the semantic segmentation challenge, Zhao et al. [34] presented the attention mechanism and developed the PSA-Net network, which learns to aggregate the contextual information of each place through the prediction of the attention map. These attention-based methods, however, need the generation of massive attention maps to calculate the association between each pixel, which is both computationally intensive and resource intensive on the GPU.
Most commonly, sequence models are processed using an RNN. Long-short-term-memory (LSTM) network and gated-recurrent-unit (GRU) structure RNN are utilized to construct temporal dependency of modulus in MRI image sequences [35] for application in brain tumor image segmentation. To better segment tumors, researchers have combined recurrent-fully-convolutional-networks (RFCN) and variational sets (VS) methods. In RFCN, the deconvolution process uses the output of the previous convolution process as the input map, and the output of the deconvolution process is used as the input of the next convolution layer. CRF based on RNN (CRF-RNN) [36] is used to process the segmentation map, and FCN feedforward is used to generate the segmentation map for image pixels. Using the original image and segmentation results as input, CRF-RNN creates an accurate segmentation image based on the intensity and location of pixels. By combining FCN with CRF-RNN, this method improves computational efficiency and shows high performance on the BraTS2015 dataset. However, deep learning-based brain segmentation still faces many challenges.
In medical literature, there is a difference between the distribution of test data and training data, which can lead to prediction or modelling problems. Deep learning models, on the other hand, focus relatively more on the distribution of pixels but ignore the differences between adjacent pixels. results in unsatisfactory segmentation results. FCN, the first application of deep learning in image semantic segmentation, incorporates end-to-end convolutional neural networks into image semantic segmentation tasks [37]. The first time to use hop connection to improve the roughness of network up sampling is proposed, images of any size can be input, based on the classical CNN network removing the fully connected layer, the deconvolutional layer is used to up sample the feature map output of its last pooling layer by 32 times, and the spatial information of the context is preserved. The output of the fusion shallow network is 8/16× up sampling, whereas the VGG16 network is 8× up sampling, but the disadvantage is that the output map obtained by the network is not fine enough and lacks the connection between pixels.
On this basis, researchers carried out many network improvement designs, such as enlarging the receptive field, extracting contextual information, introducing boundary information, adding various attention modules, and using AutoML technology. Semantic segmentation models with encoder-decoders emerged, first proposed by SegNet [38], which introduced decoders to map low-resolution encoder features to full-input resolution features for pixel-level classification. The model consists of five encoders, corresponding five decoders, and pixel-level classification layers, where each encoder contains two to three convolutional layers (containing BN and ReLU layers) and a down sampling layer [39]. Each decoder contains an up-sampling layer (with maximum pooled indexes) and two or three convolutional layers. Conventional segmentation techniques prove inadequate when confronted with the intricate nature of medical MRI images. Conversely, deep learning-based segmentation methods exhibit notable efficiency but often grapple with accuracy issues. This paper primarily concentrates on the enhancement of segmentation accuracy within the realm of brain tumor segmentation, leveraging DL methodologies.
This section is divided into several sections to explain the model step by step, where in Fig. 1 the complete methodology is shown.
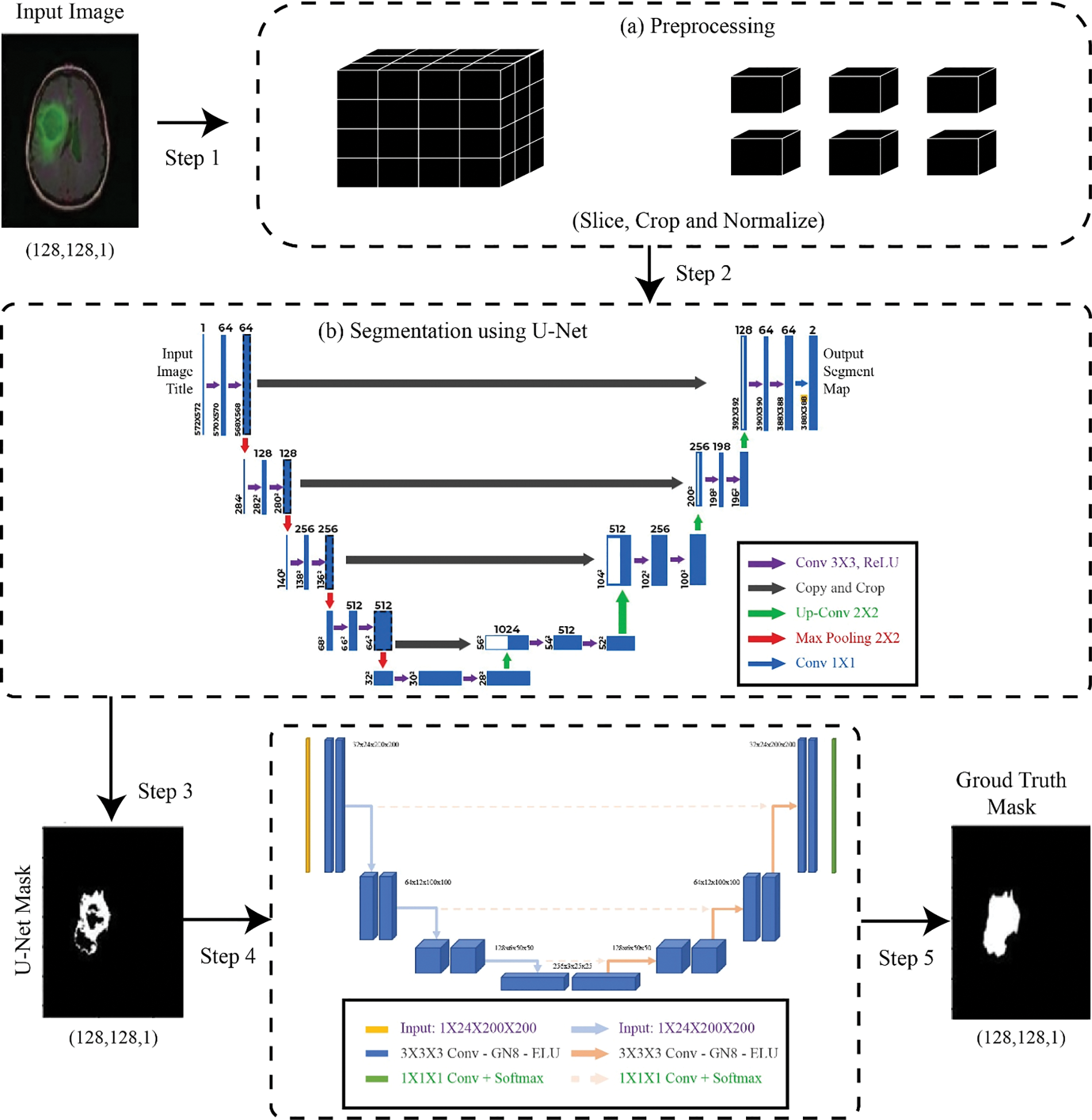
Figure 1: Proposed methodology
This study used one dataset for MRI segmentation purposes. The dataset utilized in this study is comprised of MR images with manual fluid-attenuated-inversion-recovery (FLAIR) anomaly segmentation masks from The-Cancer-Imaging-Archive. The dataset provides multi-modal images, including T1-weighted, T2-weighted, T1-contrast-enhanced, and FLAIR-MRI sequences. Each image in the dataset contains annotations of the tumor regions, providing ground truth for the training and evaluation of segmentation algorithms. There might be anything from 395 to 827 images in each category. Images from both datasets are shown in Fig. 2.

Figure 2: (a) Image (b) Image mask (c) Image + mask (d) Image-2 (e) Image-2 mask
Image preprocessing is the first step of experimental verification and the basis of subsequent experiments. The experiment’s findings are profoundly affected by the image quality. Images in the datasets range in resolution and quality since they were captured using various equipment and collection methods. Prior to conducting an experiment, picture preprocessing must be carried out to lessen the variation between data samples and the impact of equipment noise. The goal is to enhance segmentation findings by enhancing image clarity, image quality, facilitating the discovery of previously unseen elements in the image, and so on.
The basic process of preprocessing is the conversion of the image format; afterwards, images are normalized using pixel intensity modification and grayscale normalization; subsequently, contrast enhancement is performed using channel modification; and finally, data augmentation is performed using cropping, scaling, rotation, and slicing. To increase the convergence of the training network and reduce the effect of the difference between images, this experiment normalized the original images in the data set, including image size normalization and grayscale normalization. Size normalization refers to uniformly adjusting the image resolution to 128 * 128. This paper adopts the maximum and minimum normalization method for image I with normalized image I norm, and its formula is shown as:
Remove part of the black background area that does not participate in the segmentation operation to obtain a brain tumor image with a size of 128 * 128, increase the proportion of the tumor area in the image, and reduce the size of the tumor due to the small area. The problem of uneven data distribution can improve the final segmentation score of the model. The image enhancement method used here is to randomly flip the image in the left and right and front and back directions with a probability of 50%, translate the image in the X or Y direction (or both), and finally increase the amount of data in the image. Contrast-limited adaptive histogram-equalization (CLAHE) divides a given image into multiple, non-overlapping size regions and applies histogram-equalization (HE) to each region of the image to achieve an even grayscale distribution that obscures features that make images sharper pictures. The advantages of CLAHE are low noise, simple calculation, good response across image areas, preventing bright light from occurring, and making it easier to compare different areas of the image.
U-Net has immediately gained the attention of academics because of its exceptional outcomes in natural and medical picture segmentation, and it has since established itself as the industry standard. Fig. 3 displays the U-Net model’s structural layout. The network is known as an “U-Net” because it is symmetrical and resembles the letter “U” in shape. Three components make up the bulk of the model: The contracting path, expanding path, and skip connection composition. A typical CNN structure called the shrinking route, often referred to as the down sample path, consists of convolutional and pooling layers. U-Net’s decreasing route is made up of four identical modules, each of which has a 2 * 2 max pooling layer and two 3 * 3 convolutional layers. The size of the feature map is cut in half, and the number of channels is doubled each time the shrinking route executes a pooling operation.
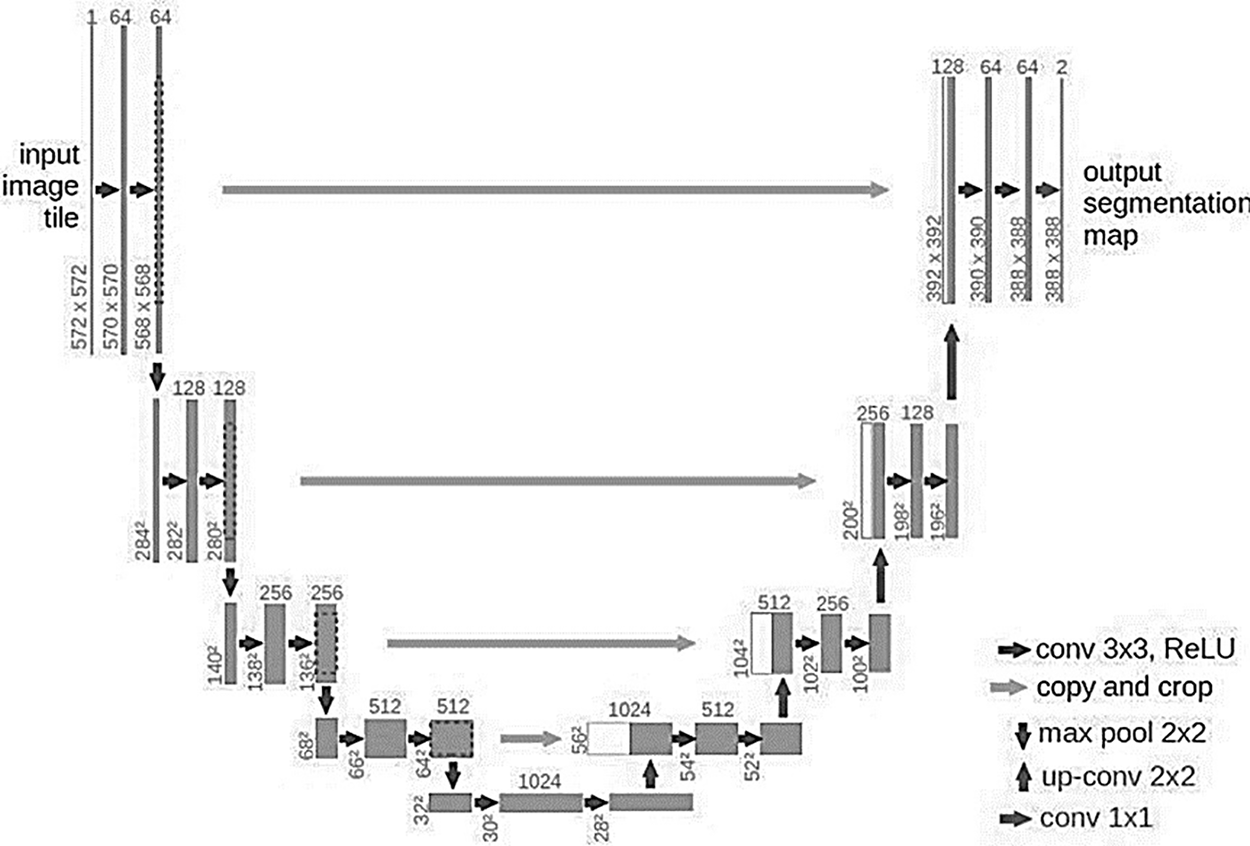
Figure 3: U-Net architecture
The resolution of a given map is to return to the highest possible level using the incremental method, also called the expansion method or incremental method. Each of the four identical parts that make up the dilation method has a 2 × 2 deconvolution-layer and two 3 × 3 convolution-layers, and the dilation method is symmetrical with the contraction method. For each deconvolution operation, the size of the feature map is doubled, and the number of channels is reduced by half. To enable depth convolution in the detail method to obtain more detailed images, cross-concatenate the shallow image in the matching method with the image in the detail method. The U-Net model always uses the binary classification function to segment cell images. A 2-channel 1 × 1 convolutional layer is connected to the last layer of the network to output the classification result. Complete symmetry is maintained in the U-Net model by the operations that map to the shrinkage route and the expansion path layer.
The U-Net model’s down sampling procedure doubles as input picture feature encoding. The most important feature information of the input image is kept, while the spatial information is compressed. The U-Net model is also known as a typical encoding-decoding network since its up-sampling method is a decoding technique that maximizes the resolution of the feature map. The U-Net model differs from conventional CNN models in that it has a deeper network structure and richer sampling layers, enabling the network to rely on a smaller training set to attain picture quality. Accurate segmentation, so it is very suitable for medical image segmentation tasks where datasets are scarce. Algorithm 1 defines the steps of the U-Net improved model. The model utilizes the Adam optimizer as well as categorical cross-entropy; moreover, 250 epochs are utilized.

3.3.1 Post-Segmentation Processing
Segmented images at the first level of model fusion were used in a CNN-based architecture for MRI classification at the second level of the model. To improve the classification accuracy of the CNN model, segmented images are used as input in the first stage. If the image is not segmented, the entire background, including borders and textures, will be visible. As a result, unwanted features are removed from low priority settings.
Convolution and max-pooling layers of the proposed CNN model, each using a different CB. For this activity, there is no padding, and a typical stride length is used. By default, the stride is set to 1, which means that for every pixel the filter is moved to the right, it will also be moved one pixel down. The first phase yields an output picture of 1822562, which is then downsized to 27272. The suggested CNN model takes the resized picture as input into the first convolution block (CB). The first CB consists of one MP layer and one convolution. The first convolution layer consists of 32 5-by-5 filters, and the MP layer is 2-by-2. The second CB is composed of two convolutional layers and one max pooling (MP) layer. The MP layer includes 16 filters in a 5-by-5 configuration, compared to 8 filters in a 5-by-5 configuration for each of the convolution layers. The third CB is composed of one MP layer and two convolution layers once more. The filters in the MP layer are 3-by-3, while the filters in the third convolution layer are 5-by-5, for a total of 16 filters. The third CB is followed by the flattening layer, which “flattens” the features by condensing the feature space into a single feature vector. Finally, feature vectors are classified into seven MRI illness classifications using three dense layers.
A typical loss function utilized in regression and machine-learning issues is mean-squared error (MSE).
Measure the mean square difference between expected and actual. The equation for MSE is shown as:
Here n are the total samples. MSE has several advantages: It is differentiable, which makes it easy to optimize using gradient descent or other optimization techniques. It is well-behaved, meaning that it is continuous, non-negative, and reaches a minimum value when the predicted values match the actual values.
Dice-similarity-coefficient (DSC) is used to evaluate the similarity between the model segmentation result image and the label image. The larger the value, the higher the similarity. The number of properly and wrongly identified test group samples may be seen in the confusion matrix. True-positive (TP) samples are located along the main diagonal, while true-negative (TN), false positive (FP), and false-negative (FN) samples are in the remaining cells. The value range is [0,1]. The equation is shown in (3).
The Jaccard-index is the ratio of the overlap between the anticipated and ground-truth segmentations to the union of those two regions. This measure can take on values between 0 and 1, with 0 denoting completely non-overlapping segmentation and 1 denoting totally overlapping segments.
One of the most crucial common measures of classification system effectiveness is the confusion matrix. The confusion matrix is a square matrix with four rows and four columns that correspond to the different categories in the data set. The following equations are used to evaluate the system’s functionality: The variables in the equations are taken from the confusion matrix. Another statistic used to judge the quality of a model is the balanced accuracy (BA), which is the ratio of correctly predicted pixels to all pixels. Based on the confusion matrix, it can be expressed by the following equation:
Precision: Indicates the proportion of pixels correctly identified as tumors to the total number of pixels identified as tumors.
Kappa, like the AUC score, assesses the agreement induced by chance; it runs from −1 (worst) to +1 (best), and a value of 0 (random classifier) represents the poorest possible performance.
Additionally, the segmentation efficiency is evaluated using the area under the receiver operation characteristic curve (AUC). If the AUC is very near to 1, then the segmentation technique is quite reliable. At a value of 0.5, it is completely unreliable and has no practical use.
The experiments are performed on a GPU with a 4 GB NVIDIA Tesla graphics card and 32 GB of RAM. The main tools and libraries that are utilized for performing experimentation are Python, Keras, PyTorch, Tensorflow, and Matplotlib.
The robustness of brain tumor segmentation was meticulously evaluated through a comprehensive comparison of the proposed hybrid U-Net model against other contemporary state-of-the-art models. The validation results, as presented in Table 1, highlight the noteworthy performance of the proposed model in various metrics compared to its counterparts. In terms of accuracy, the proposed model exhibited a remarkable achievement, approaching nearly 99%, a result on par with the widely recognized FCN. This signifies the effectiveness of the hybrid U-Net in accurately delineating brain tumors from medical images. However, what sets the proposed model apart is its balanced accuracy, which surpasses other models, including FCN, U-Net, and Atten-U-Net, by almost 4%. This indicates that the proposed model excels not only in overall accuracy but also in maintaining a balanced performance across different classes or categories of brain tumors.
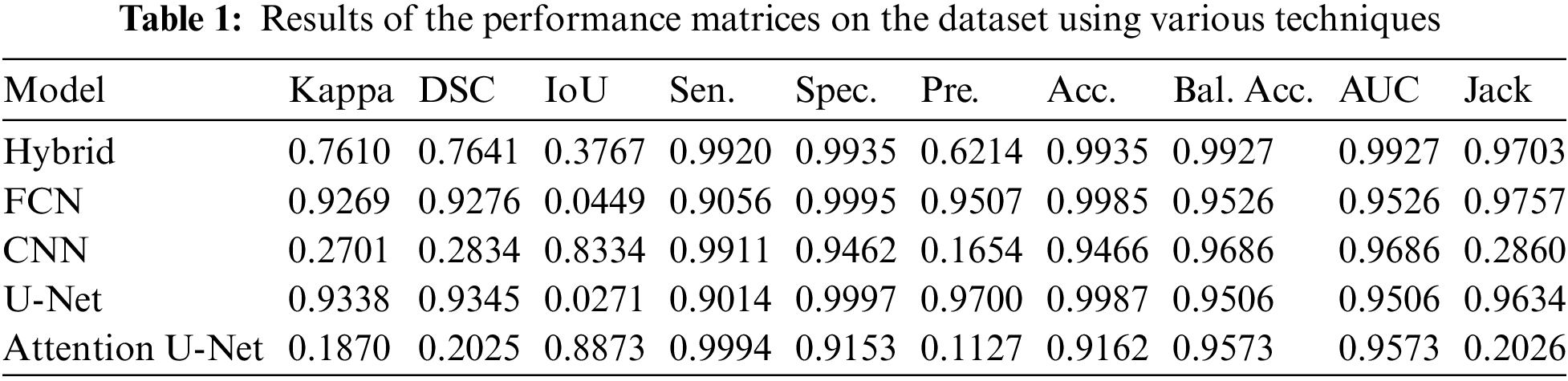
The evaluation metrics further substantiate the superiority of the hybrid U-Net. The Kappa coefficient, a measure of the positive relationship between predicted and actual segmentations, showcased a superior performance for U-Net. This implies a robust agreement between the Hybrid U-Net’s predictions and the ground truth, emphasizing its efficacy in capturing nuanced patterns and structures within the brain images. Notably, the area under the curve (AUC) metric, a key indicator of the model’s ability to discriminate between tumor and non-tumor regions, demonstrated an outstanding result of 0.99 for the proposed model. This outperforms FCN, CNN, U-Net, and Atten-U-Net, with respective AUC values of 0.95. The higher AUC underscores the hybrid U-Net’s exceptional capability to precisely identify and distinguish between normal and tumorous brain regions. Fig. 4 shows the balanced accuracy visualization for the proposed hybrid model in comparison with the other models and shows that the proposed model is far better than other models.
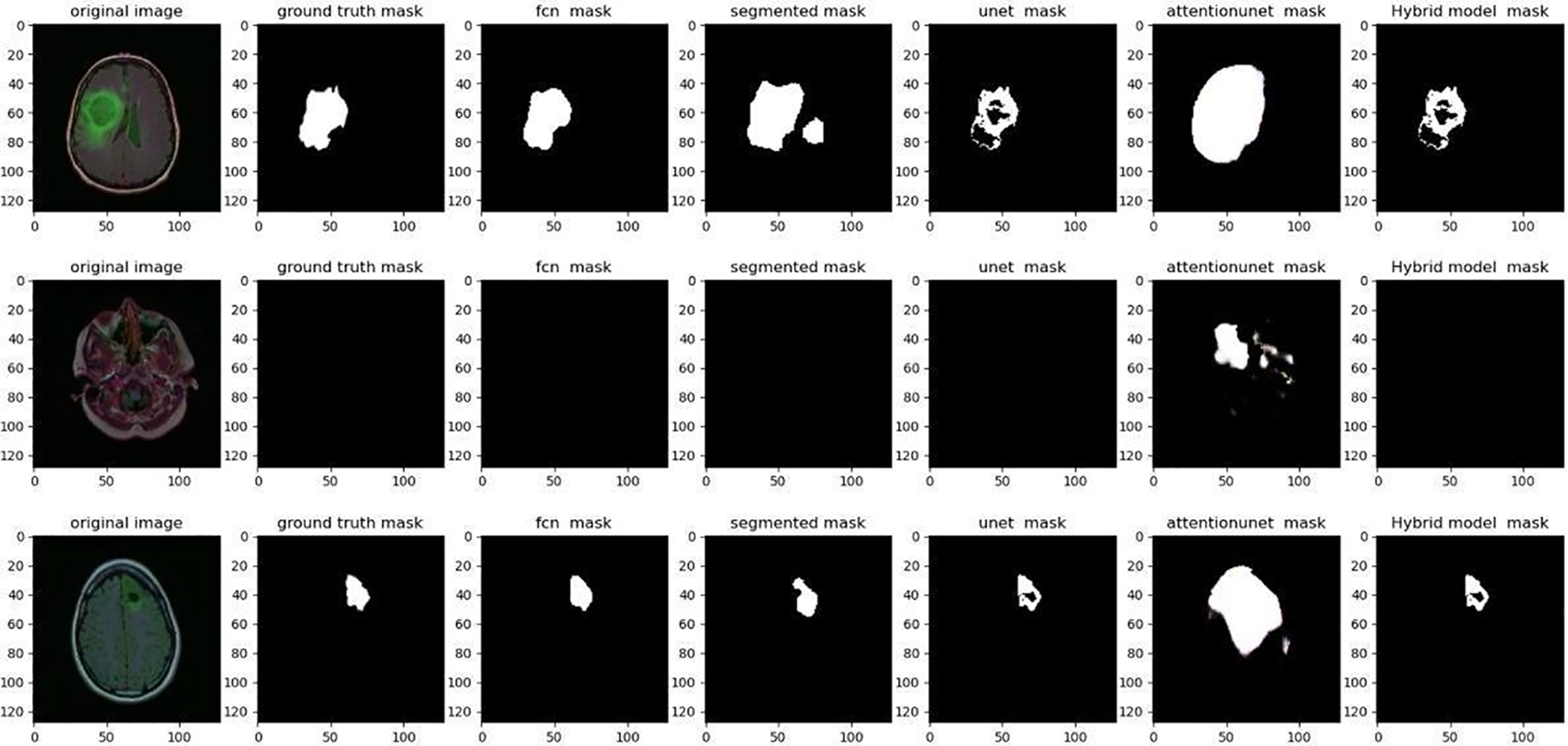
Figure 4: Comparison of several algorithms on a dataset visually
Sensitivity, a pivotal metric in evaluating image segmentation algorithms, gauges the algorithm’s ability to accurately detect true positive pixels within an image. True positive pixels represent those belonging to the target object or class, and a high sensitivity indicates that the algorithm can proficiently identify a substantial proportion of these true positive pixels. Conversely, a low sensitivity suggests that the algorithm is missing a considerable number of true positive pixels, indicating potential shortcomings in its ability to capture essential features of the target object. However, it is crucial to recognize that sensitivity alone provides an incomplete assessment of an image segmentation algorithm’s quality. High sensitivity can be achieved by erroneously classifying a surplus of pixels as belonging to the target object, leading to an elevated false-positive rate. This over-segmentation issue underscores the importance of employing multiple evaluation metrics to gain a more comprehensive understanding of the algorithm’s performance. One such complementary metric is the Jaccard coefficient, also known as the Intersection over Union (IoU).
The Jaccard coefficient assesses the accuracy of image segmentation algorithms by measuring the overlap between the predicted segmentation and the ground truth. It is calculated as the intersection of the true positive pixels divided by the union of the true positive, false positive, and false negative pixels. A higher Jaccard coefficient indicates more accurate and precise segmentation, emphasizing the algorithm’s capability to delineate the target object with fidelity. In interpreting the results, a high sensitivity coupled with a high Jaccard coefficient would signify robust performance, indicating that the algorithm not only identifies a significant portion of true positive pixels but also achieves a nuanced and accurate delineation of the target object. Conversely, a scenario where sensitivity is high, but the Jaccard coefficient is low would suggest potential issues with over-segmentation, emphasizing the need for a more refined balance between sensitivity and specificity. The proposed algorithm is almost similar in achieving the results of 0.97.
Fig. 4 displays a visual comparison of the proposed model with other algorithms; while the segmentation in our suggested hybrid model is properly apparent, the ground trust portion of the model is virtually extracted similarly to the other approaches.
The accuracy rate, a fundamental metric in assessing the efficacy of classification models, serves as the average of sensitivity and specificity across all categories in a given sample. While sensitivity and specificity focus on the true positive rate and true negative rate within individual classes, accuracy offers a holistic perspective by considering the model’s performance across all categories simultaneously. This is particularly pertinent in scenarios where the class distribution in the data is imbalanced, with certain classes having significantly fewer instances than others. Balancing test accuracy becomes crucial when faced with unequal class representation, where a minority class may be overshadowed by the majority. Relying solely on accuracy can be deceptive, as a model may achieve high accuracy by predicting the majority classes accurately but may perform poorly on the minority classes. Hence, accuracy serves as a more comprehensive metric, providing insights into the model’s overall performance by evaluating sensitivity and specificity across all categories. In the presented study, the algorithm under consideration demonstrates equal accuracy across other methods for both datasets, showcasing its robustness and effectiveness in handling diverse classes.
The improvement in classification accuracy by almost 99% is a remarkable feat, underscoring the algorithm’s proficiency in making accurate predictions across multiple categories. Fig. 5 offers a visual representation comparing the accuracy of the data, providing a clear illustration of how the proposed algorithm outperforms other methods in achieving balanced accuracy. This figure serves as a valuable visual aid for understanding the comparative performance of different algorithms. Furthermore, Fig. 6, depicting the training accuracy curve, provides insights into the algorithm’s learning dynamics. Analyzing the training accuracy curve over epochs allows for a deeper understanding of how the model refines its predictions over time. A consistent and steep increase in training accuracy suggests effective learning and adaptation to the underlying patterns in the data.
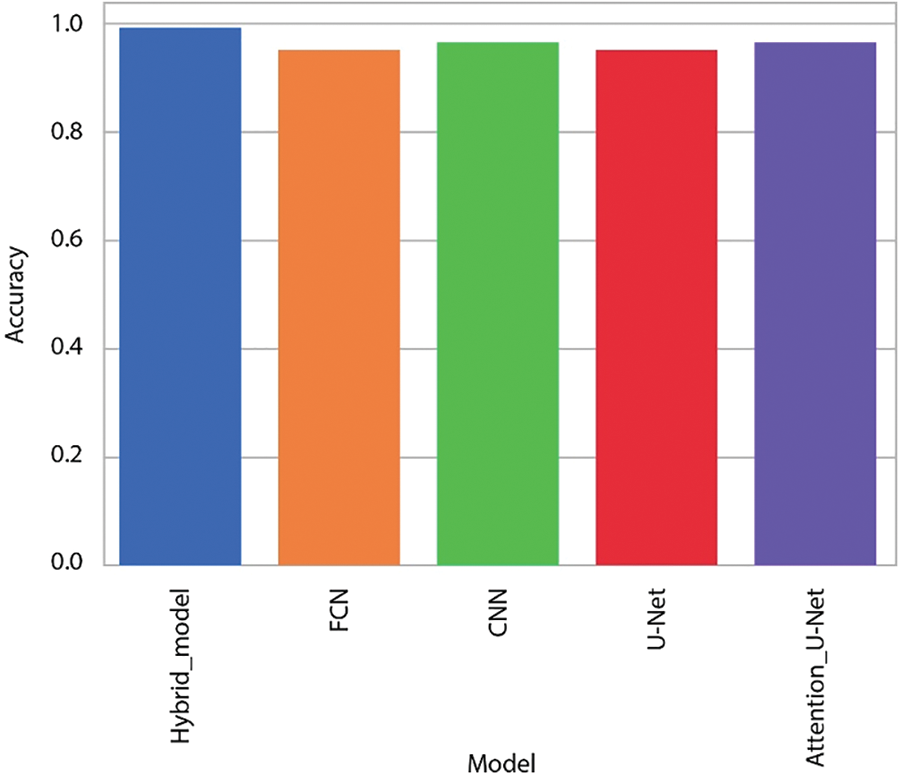
Figure 5: Intuitive comparison of various algorithms on datasets
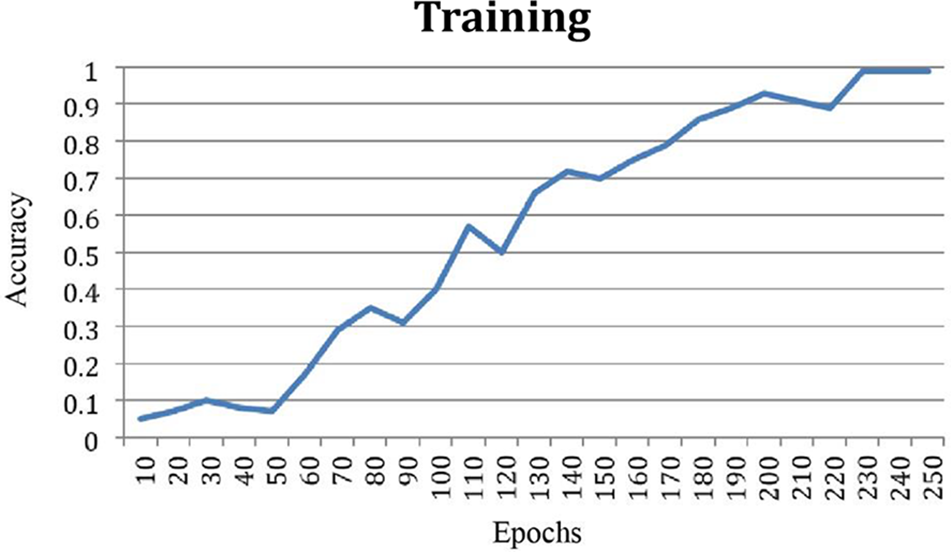
Figure 6: Training accuracy curve
The discussed method of brain tumor segmentation using a hybrid U-Net approach based on deep learning technology demonstrates promising results for improving the accuracy of tumor segmentation in MRI images. The method addresses several challenges associated with brain tumor segmentation, such as complex tumor boundaries, shape variations, unequal distribution of tumor locations and sizes, and limited availability of medical image samples. The research employs a customized layered U-Net model, a widely used architecture for semantic segmentation, to perform the segmentation task. Preprocessing techniques are applied to enhance the visibility of brain tumors in MRI images, improving the quality of the input data. The inclusion of dropout layers after each convolution block stack helps prevent over-fitting, which is essential for generalizing the model’s performance to unseen data.
The CNN process incorporates the context of brain tumor MRI images to further refine the model’s segmentation. By leveraging deep learning techniques and training on a diverse dataset, the proposed method achieves high accuracy levels, surpassing state-of-the-art models in brain tissue differentiation experiments. The reported accuracy of over 98% across all datasets, along with precision and Jaccard coefficient values exceeding 90%, indicates the effectiveness of the hybrid U-Net approach in accurately segmenting brain tumors. Although this study showed good results, some limitations are not necessary. The success of the plan depends on diversity and large enough data to train deep learning models. Additionally, the generalization of this model to different cultures and artistic processes needs to be further investigated. Future studies may focus on validating this approach on larger, more diverse data sets, as well as exploring other strategies designed to address issues such as conflict that are designed in the classroom and lead to fewer cancers. While the proposed hybrid U-Net model using U-Net and CNN for brain segmentation demonstrates results, it is important to acknowledge the limitations of this approach.
In contrast to healthy brain tissue, brain tumors may develop anywhere in the brain and take on a variety of forms. Therefore, the most accurate and sensitive segmentation method should be used. This study uses preprocessed data to improve MRI quality and then evaluates the effect of coupling the U-Net learning model-based framework (i.e., MRI boundary) on the MRI of three types of brain tumors. According to testing results, the proposed hybrid-UNet model fared better at segmenting brain tumors than earlier models. Our segmentation findings were more accurate and exact than those from rival algorithms, with better results for all performance metrics pertaining to brain tumors. The proposed method achieved 99% accuracy. The main limitations of the given research are its post-processing and pre-processing techniques, which increase the computational time.
Acknowledgement: The author would like to express sincere gratitude to the Department of Information Technology, Faculty of Computing and Information Technology, King Abdulaziz University, for their invaluable support and guidance.
Funding Statement: This research work was funded by Institutional Fund Projects under Grant No. (IFPIP: 801-830-1443). The author gratefully acknowledges technical and financial support provided by the Ministry of Education and King Abdulaziz University, DSR, Jeddah, Saudi Arabia.
Author Contributions: The author confirms contribution to the paper as follows: study conception, design, data collection, analysis, interpretation of results and draft manuscript preparation: Sharaf J. Malebary. The author reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data that support the findings of this study are available from the first and corresponding authors upon reasonable request.
Conflicts of Interest: The author declares that they have no conflicts of interest to report regarding the present study.
References
1. A. Saadon, “A smart device powered by deep learning technology to identify animals by using object detection in a live video to provide food and water to specific type of homeless animals,” Turkish J. Eng., vol. 23, no. 1, pp. 1–22, 2022. [Google Scholar]
2. P. Burman et al., “Deaths among adult patients with hypopituitarism: Hypocortisolism during acute stress, and de novo malignant brain tumors contribute to an increased mortality,” J. Clin. Endocrinol. Metabol., vol. 98, no. 4, pp. 1466–1475, 2013. doi: 10.1210/jc.2012-4059 [Google Scholar] [PubMed] [CrossRef]
3. R. Yamanaka, E. Abe, T. Sato, A. Hayano, and Y. Takashima, “Secondary intracranial tumors following radiotherapy for pituitary adenomas: A systematic review,” Cancers, vol. 9, no. 8, pp. 103–123, 2017. doi: 10.3390/cancers9080103 [Google Scholar] [PubMed] [CrossRef]
4. K. Spitler, D. Drazin, G. Hanna, A. Patel, and R. Chu, “Association of intracranial aneurysms with meningiomas, pituitary adenomas, and gliomas: Review of possible interrelationship,” Int. Sch. Res. Notices, vol. 1, no. 1, pp. 1–15, 2013. [Google Scholar]
5. M. Saeed, I. Halepoto, S. Khaskheli, and M. Bushra, “Optimization and efficiency analysis of deep learning based brain tumor detection,” Mehran Univ. Res. J. Eng. Technol., vol. 42, no. 2, pp. 188–196, 2023. doi: 10.22581/muet1982.2302.19. [Google Scholar] [CrossRef]
6. N. Grieg, L. Ries, R. Yancik, and S. Rapoport, “Increasing annual incidence of primary malignant brain tumors in the elderly,” J. Natl. Cancer Inst., vol. 82, no. 20, pp. 1621–1624, 1990. doi: 10.1093/jnci/82.20.1621 [Google Scholar] [PubMed] [CrossRef]
7. J. Liu, M. Li, J. Wang, F. Wu, T. Liu and Y. Pan, “A survey of MRI-based brain tumor segmentation methods,” Tsinghua Sci. Technol., vol. 19, no. 6, pp. 578–595, 2014. doi: 10.1109/TST.2014.6961028. [Google Scholar] [CrossRef]
8. F. Masulli and A. Schenone, “A fuzzy clustering based segmentation system as support to diagnosis in medical imaging,” Artif. Intell. Med., vol. 16, no. 2, pp. 129–147, 1999. doi: 10.1016/S0933-3657(98)00069-4 [Google Scholar] [PubMed] [CrossRef]
9. N. Burkart and M. Huber, “A survey on the explainability of supervised machine learning,” J. Artif. Intell. Res., vol. 70, no. 1, pp. 245–317, 2021. doi: 10.1613/jair.1.12228. [Google Scholar] [CrossRef]
10. S. Minaee, Y. Y. Boykov, F. Porikli, A. J. Plaza, N. Kehtarnavaz and D. Terzopoulos, “Image segmentation using deep learning: A survey,” IEEE Trans. Pattern Anal. Mach. Intell., vol. 44, no. 7, pp. 3523–3542, 2021. doi: 10.1109/TPAMI.2021.3059968 [Google Scholar] [PubMed] [CrossRef]
11. H. Lu, H. Tang, and Z. Wang, “Advances in neural networks,” in 16th Int. Symp. Neural Netw., Moscow, Russia, 2019, pp. 1–10. [Google Scholar]
12. V. Badrinarayanan, A. Kendall, and R. Cipolla, “Segnet: A deep convolutional encoder-decoder architecture for image segmentation,” IEEE Trans. Pattern Anal. Mach. Intell., vol. 39, no. 12, pp. 2481–2495, 2017. doi: 10.1109/TPAMI.2016.2644615 [Google Scholar] [PubMed] [CrossRef]
13. L. C. Chen, G. Papandreou, I. Kokkinos, K. Murphy, and A. L. Yuille, “DeepLab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs,” IEEE Trans. Pattern Anal., vol. 40, no. 4, pp. 834–848, 2017. doi: 10.1109/TPAMI.2017.2699184 [Google Scholar] [PubMed] [CrossRef]
14. J. Fu et al., “Dual attention network for scene segmentation,” in Proc. IEEE/CVF Conf. Comput. Vision Pattern Recogn., Long Beach, USA, 2019, pp. 3146–3154. [Google Scholar]
15. Z. Ma, J. Tavares, and R. Jorge, “A review on the current segmentation algorithms for medical images,” in Int. Conf. Imag. Theory Appl., Lisboa, Portugal, 2009, pp. 135–140. [Google Scholar]
16. H. Vaghela, H. Modi, M. Pandya, and M. Potdar, “A novel approach to detect chronic leukemia using shape based feature extraction and identification with digital image processing,” Int. J. Appl. Inf. Syst., vol. 11, no. 5, pp. 9–16, 2016. doi: 10.5120/ijais2016451607. [Google Scholar] [CrossRef]
17. A. Aslam, E. Khan, and M. Beg, “Improved edge detection algorithm for brain tumor segmentation,” Procedia Comput. Sci., vol. 58, no. 1, pp. 430–437, 2015. doi: 10.1016/j.procs.2015.08.057. [Google Scholar] [CrossRef]
18. Z. Qian, G. Hua, C. Cheng, T. Tian, and L. Yun, “Medical images edge detection based on mathematical morphology,” in IEEE Eng. Med. Biol. 27th Annual Conf., Las Vegas, USA, 2006, pp. 6492–6495. [Google Scholar]
19. I. Haq, S. Anwar, K. Shah, M. T. Khan, S. A. Shah and J. A. Coles, “Fuzzy logic based edge detection in smooth and noisy clinical images,” PLoS One, vol. 10, no. 9, pp. 138–162, 2015. doi: 10.1371/journal.pone.0138712 [Google Scholar] [PubMed] [CrossRef]
20. M. Moradi, P. Mousavi, and P. Abolmaesumi, “Computer-aided diagnosis of prostate cancer with emphasis on ultrasound-based approaches: A review,” Ultrasound Med. Biol., vol. 33, no. 7, pp. 1010–1028, 2007. doi: 10.1016/j.ultrasmedbio.2007.01.008 [Google Scholar] [PubMed] [CrossRef]
21. G. Lin, W. Wang, C. Kang, and C. Wang, “Multispectral MR images segmentation based on fuzzy knowledge and modified seeded region growing,” Magn. Reson. Imag., vol. 302, no. 2, pp. 230–246, 2012. doi: 10.1016/j.mri.2011.09.008 [Google Scholar] [PubMed] [CrossRef]
22. W. Chen and X. Zhang, “A new watershed algorithm for cellular image segmentation based on mathematical morphology,” in Int. Conf. Mach. Vision Human-mach. Interf., Kaifeng, China, 2010, pp. 653–656. [Google Scholar]
23. Y. Zhang, M. Brady, and S. Smith, “Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm,” IEEE Trans. Med. Imag., vol. 20, no. 1, pp. 45–57, 2001. doi: 10.1109/42.906424 [Google Scholar] [PubMed] [CrossRef]
24. G. W. Xu, Z. J. Zhang, W. H. Yuan, and L. N. Xu, “On medical image segmentation based on wavelet transform,” in Fifth Int. Conf. Intell. Syst. Des. Eng. Appl., Hunan, China, 2014, pp. 671–674. [Google Scholar]
25. J. Zhang, H. Sun, D. Gao, L. Qiao, and N. Liu, “Detection of canopy chlorophyll content of corn based on continuous wavelet transform analysis,” Remote Sens., vol. 12, no. 17, pp. 2741–2765, 2020. doi: 10.3390/rs12172741. [Google Scholar] [CrossRef]
26. B. Chen, L. Zhang, H. Chen, K. Liang, and X. Chen, “A novel extended Kalman filter with support vector machine based method for the automatic diagnosis and segmentation of brain tumors,” Comput. Methods Prog. Biomed., vol. 200, no. 2, pp. 797–815, 2021. doi: 10.1016/j.cmpb.2020.105797 [Google Scholar] [PubMed] [CrossRef]
27. M. Islam, M. Ali, A. Das, D. Duranta, and M. Alam, “Human brain tumor detection using k-means segmentation and improved support vector machine,” Int. J. Scientif. Eng. Res., vol. 11, no. 6, pp. 6–23, 2020. [Google Scholar]
28. A. Işın, C. Direkoğlu, and M. Şah, “Review of MRI-based brain tumor image segmentation using deep learning methods,” Procedia Comput. Sci., vol. 102, no. 1, pp. 317–324, 2016. doi: 10.1016/j.procs.2016.09.407. [Google Scholar] [CrossRef]
29. M. Balwant, “A review on convolutional neural networks for brain tumor segmentation: Methods, datasets, libraries, and future directions,” IRBM, vol. 43, no. 6, pp. 521–537, 2022. doi: 10.1016/j.irbm.2022.05.002. [Google Scholar] [CrossRef]
30. Y. Xue, T. Xu, H. Zhang, L. Long, and X. Huang, “SegAN: Adversarial network with multi-scale L1 loss for medical image segmentation,” Neuroinformat., vol. 16, no. 2, pp. 383–392, 2018. doi: 10.1007/s12021-018-9377-x [Google Scholar] [PubMed] [CrossRef]
31. K. Kamnitsas et al., “DeepMedic for brain tumor segmentation,” in Brainlesion: Glioma, Multiple Sclerosis, Stroke Traumatic Brain Injuries: Second Int. Workshop, BrainLes 2016, Challenges BRATS, ISLES mTOP 2016, Athens, Greece, 2016, pp. 138–149. [Google Scholar]
32. H. Noh, S. Hong, and B. Han, “Learning deconvolution network for semantic segmentation,” in Proc. IEEE Int. Conf. Comput. Vision, Santiago, Chile, 2015, pp. 1520–1528. [Google Scholar]
33. X. Xiao, S. Lian, Z. Luo, and S. Li, “Weighted Res-UNet for high-quality retina vessel segmentation,” in 9th Int. Conf. Inf. Technol. Med. Edu., Hangzhou, China, 2018, pp. 327–331. [Google Scholar]
34. H. Zhao and N. Sun, “Improved U-net model for nerve segmentation,” in Image Graphics: 9th Int. Conf., Shanghai, China, 2017, pp. 496–504. [Google Scholar]
35. J. G. Sivasai, P. N. Srinivasu, M. N. Sindhuri, K. Rohitha, and S. Deepika, “An automated segmentation of brain MR image through fuzzy recurrent neural network,” Bio-Inspired Neurocomput., vol. 22, no. 1, pp. 163–179, 2020. [Google Scholar]
36. T. Le, R. Gummadi, and M. Savvides, “Deep recurrent level set for segmenting brain tumors,” in Med. Image Comput. Comput. Assist. Intervent.–MICCAI 2018, Granada, Spain, 2018, pp. 646–653. [Google Scholar]
37. M. Yap et al., “Breast ultrasound lesions recognition: End-to-end deep learning approaches,” J. Med. Imag., vol. 6, no. 1, pp. 1007–1027, 2019. [Google Scholar]
38. P. Kumar, P. Nagar, C. Arora, and A. Gupta, “U-SegNet: Fully convolutional neural network based automated brain tissue segmentation tool,” in 5th IEEE Int. Conf. Image Process., Genova, Italy, 2018, pp. 3503–3507. [Google Scholar]
39. Z. Zhou, M. Rahman, N. Tajbakhsh, and J. Liang, “UNet++: A nested U-Net architecture for medical image segmentation,” in Deep Learn. Med. Image Anal. Multimodal Learn. Clin. Decision Support: 4th Int. Conf., Granada, Spain, 2018, pp. 3–11. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools