 Open Access
Open Access
ARTICLE
Enhancing Skin Cancer Diagnosis with Deep Learning: A Hybrid CNN-RNN Approach
1 Faculty of Information Technology, Beijing University of Technology, Beijing, 100124, China
2 Computer Science International Engineering Collage, Maynooth University, Kildare, W23 F2H6, Irland
3 Research Center for Data Hub and Security, Zhejiang Lab, Hangzhou, 311121, China
4 Computer Science Department, MNS University of Agriculture, Multan, 59220, Pakistan
* Corresponding Authors: Syeda Shamaila Zareen. Email: ; Guangmin Sun. Email:
(This article belongs to the Special Issue: Advanced Artificial Intelligence and Machine Learning Frameworks for Signal and Image Processing Applications)
Computers, Materials & Continua 2024, 79(1), 1497-1519. https://doi.org/10.32604/cmc.2024.047418
Received 05 November 2023; Accepted 31 January 2024; Issue published 25 April 2024
Abstract
Skin cancer diagnosis is difficult due to lesion presentation variability. Conventional methods struggle to manually extract features and capture lesions spatial and temporal variations. This study introduces a deep learning-based Convolutional and Recurrent Neural Network (CNN-RNN) model with a ResNet-50 architecture which used as the feature extractor to enhance skin cancer classification. Leveraging synergistic spatial feature extraction and temporal sequence learning, the model demonstrates robust performance on a dataset of 9000 skin lesion photos from nine cancer types. Using pre-trained ResNet-50 for spatial data extraction and Long Short-Term Memory (LSTM) for temporal dependencies, the model achieves a high average recognition accuracy, surpassing previous methods. The comprehensive evaluation, including accuracy, precision, recall, and F1-score, underscores the model’s competence in categorizing skin cancer types. This research contributes a sophisticated model and valuable guidance for deep learning-based diagnostics, also this model excels in overcoming spatial and temporal complexities, offering a sophisticated solution for dermatological diagnostics research.Keywords
Accurate and fast diagnosis is crucial for the proper treatment of skin cancer, a widespread and possibly life-threatening condition. The use of deep learning methods, specifically Convolutional Neural Networks (CNNs), has demonstrated encouraging outcomes in many picture classification assignments, such as the diagnosis of skin cancer, in recent times [1]. Nevertheless, the current terrain exposes enduring obstacles that require more advanced methods. Existing approaches to skin cancer classification face obstacles that hinder their effectiveness. Conventional techniques frequently encounter difficulties in dealing with the natural fluctuations in skin lesion pictures, leading to less-than-ideal diagnostic precision. Furthermore, the dependence on manual feature extraction in traditional systems requires a significant amount of labor and may fail to capture the subtle patterns that are crucial for precise diagnosis [2]. Given the circumstances, there is a clear and noticeable requirement for sophisticated solutions that go beyond the constraints of current methods. The potential of deep learning to transform skin cancer diagnostics is significant, its ability to automatically extract features and learn representations is highly helpful. Convolutional Neural Networks (CNNs) have shown proficiency in identifying complex spatial patterns in images, which is essential for differentiating between harmless and cancerous tumors [3]. However, even in the field of deep learning, there are still difficulties that remain, especially when it comes to dealing with the time-related features of skin cancer development. The literature review conducted on the proposed model provides a comprehensive analysis of previous research efforts concerning the classification of skin cancer. Some of the research gape findings shown in Table 1 below.
The focus of the study is a large corpus of scholarly literature on the diagnosis of skin cancer and the application of image processing algorithms based on deep learning techniques. To maximize the efficacy of treatment for skin cancer, which is a prevalent form of malignant disease, it is essential to diagnose this condition promptly and accurately. In order to overcome the constraints of current techniques and the intricate difficulties encountered in deep learning, we suggest a pioneering hybrid methodology. The architecture of our model combines a Convolutional Neural Network (CNN) and a Recurrent Neural Network (RNN), notably utilizing a Long Short-Term Memory (LSTM) layer. This results in a hybrid model that incorporates both Convolutional Neural Network and Recurrent Neural Network, with a ResNet-50 backbone. This fusion is intended to combine the spatial feature extraction abilities of Convolutional Neural Network (CNNs) with the temporal sequence learning capabilities of Recurrent Neural Network (RNNs). The Convolutional Neural Network (CNN) module has exceptional proficiency in extracting spatial characteristics from skin lesion images, effectively capturing intricate patterns that may defy conventional techniques. The Recurrent Neural Network component, equipped with its LSTM layer, effectively handles the temporal intricacies that are naturally present in the course of skin cancer. By considering both spatial and temporal dimensions, our model offers a comprehensive answer to the complex issues involved in classifying skin cancer.
1.1 Existing Challenges in Skin Cancer Diagnosis
1. Skin cancer can manifest in various forms, and lesions can exhibit considerable variability in appearance [9]. Distinguishing between benign and malignant lesions, as well as identifying the specific type of skin cancer, can be challenging even for experienced dermatologists [10].
2. The success of skin cancer treatment is often closely tied to early detection. However, small or subtle lesions may go unnoticed in routine examinations, leading to delayed diagnoses and potentially more advanced stages of the disease [11].
3. Different dermatologists may interpret skin lesions differently, leading to inconsistencies in diagnosis [12]. This variability can be a significant obstacle in achieving consistently accurate and reliable diagnoses.
1.2 Research Gape in Skin Cancer Diagnosis
1. Early diagnosis of skin cancer is paramount for successful treatment. Timely identification allows for less invasive interventions, potentially reducing the need for extensive surgeries or aggressive treatments [13].
2. Accurate diagnoses contribute to better patient outcomes by facilitating appropriate and targeted treatment plans. This, in turn, enhances the chances of complete recovery and reduces the risk of complications [14].
3. Prompt diagnoses can lead to more efficient healthcare resource utilization. Avoiding unnecessary procedures for benign lesions and initiating appropriate treatments early can help in reducing overall healthcare costs [15].
1.3 Contribution of Deep Learning in Skin Cancer Diagnosis
1. Deep learning models, especially Convolutional Neural Network (CNN), excel at pattern recognition in images. They can learn intricate features and textures in skin lesions that may be imperceptible to the human eye, aiding in the early detection of subtle abnormalities [16].
2. Deep learning models provide a consistent and standardized approach to diagnosis. Once trained, they apply the same criteria to every image, mitigating inter-observer variability and ensuring more reliable results [17].
3. Automated deep learning models can process large datasets quickly, enabling a more efficient diagnostic process. This is particularly valuable in settings where there might be a shortage of dermatologists or where rapid screenings are necessary [18].
The challenges in skin cancer diagnosis necessitate advanced technologies for accurate and timely identification. Deep learning models, with their capacity for sophisticated image analysis and pattern recognition, significantly contribute to overcoming these challenges. Their role in standardizing diagnoses, improving efficiency, and aiding in early detection reinforces the importance of integrating such technologies into the broader framework of dermatological diagnostics [19]. Accurate diagnosis of skin cancer poses a formidable challenge due to the intricate variability in lesion presentation. This study seeks to overcome the limitations of current skin cancer diagnostic approaches. We address the dual challenges of spatial and temporal intricacies by introducing a novel hybrid Convolutional and Recurrent Neural Network (CNN-RNN) model with a ResNet-50 backbone. The aim is to enhance the accuracy and comprehensiveness of skin cancer classification by leveraging the strengths of both spatial feature extraction and temporal sequence learning. This study introduces a novel approach with the following main contributions:
1. We propose a pioneering model that integrates Convolutional Neural Networks (CNNs) with Recurrent Neural Networks (RNNs), specifically utilizing a Long Short-Term Memory (LSTM) layer. This hybrid architecture with a ResNet-50 backbone aims to synergize spatial feature extraction and temporal sequence learning.
2. The Convolutional Neural Network (CNN) component excels in capturing spatial features from skin lesion images, addressing limitations in traditional methods. Simultaneously, the Recurrent Neural Networks (RNN) component, with its LSTM layer, navigates the temporal complexities of skin cancer progression, providing a comprehensive solution.
3. Rigorous experiments are conducted on a large dataset comprising 9000 images across nine classes of skin cancer. This diverse dataset allows for robust training and evaluation, ensuring the model’s capacity to generalize across various pathological conditions.
4. Beyond conventional accuracy metrics, we employ precision, recall, and F1-score to comprehensively assess the model’s performance. The inclusion of a confusion matrix offers a detailed breakdown of its proficiency in classifying different skin cancer types.
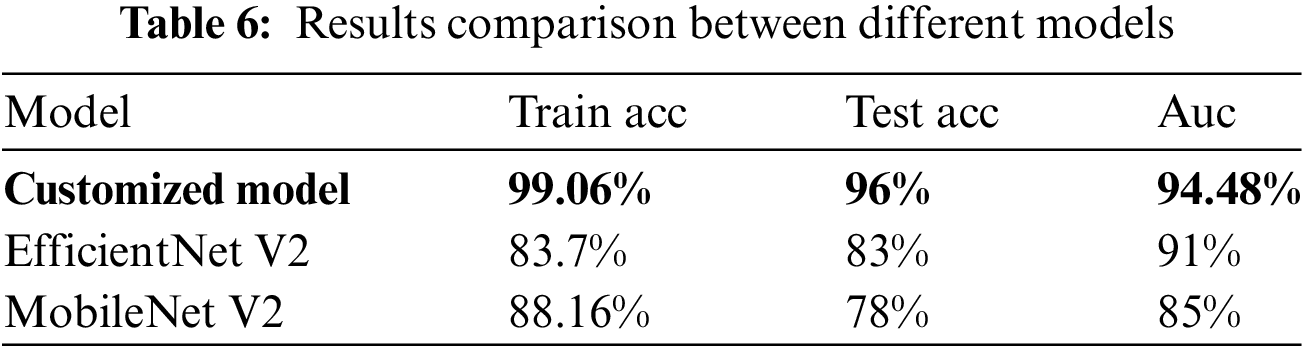
5. The proposed model achieves an impressive average recognition accuracy of 99.06%, surpassing traditional methods. This demonstrates the model’s capability to make accurate and nuanced distinctions among diverse skin cancer types. Our contributions pave the way for more effective automated skin cancer diagnosis, offering a refined understanding of spatial and temporal patterns in skin lesion images through a hybrid Convolutional and Recurrent Neural Network (CNN-RNN) model.
This research introduces a novel hybrid Convolutional and Recurrent Neural Network (CNN-RNN) algorithm that is highly effective in classifying skin cancer disease in the specified data collection. The primary objective of this project is to develop a novel hybrid deep learning algorithm for accurately predicting skin cancer illness. Section 1, under “Introduction,” provides a detailed overview of the publications that explicitly focus on classifying different forms of skin cancer, also cover literature review of skin cancer, research gape and proposed research contribution. Section 2 outlines the recommended methodology, includes data definition, preprocessing, augmentation, segmentation, feature extraction steps. While Section 3 provides a comprehensive examination of the classification. Section 4 combines system design and analysis and lastly this paper is concluded in Section 5.
The proposed methodology is described in Fig. 1 for the classification of dermoscopy skin cancer images. We utilized a hybrid Convolutional and Recurrent Neural Network (CNN-RNN) model pretrained on approximately 2239 images from ISIC dataset which contain dermoscopy images. The model classified skin lesion image with performance better or comparable to expert dermatologists for nine classes. The overall model working is shown below.

Figure 1: Proposed hybrid model for skin cancer detection and classification
Computerized algorithms are typically used for the automated assessment of medical data, and the accuracy of these algorithms must be validated using clinical-grade medical imaging [20]. The required test images are obtained from the ISIC dataset in the proposed study. The collection has 2239 photos of skin lesions. Following the collection process, each image and the corresponding ground truth are scaled to a resolution of pixels. The test images used in this study, together with their corresponding ground truth, are shown in Fig. 2.

Figure 2: Images with type definition from the ISIC dataset
The skin cancer classification model employs a range of preprocessing techniques to enhance the quality and suitability of the input images before they are used in the network. The process of constructing a skin cancer classification model entails the production of images that specifically target and emphasize particular regions of interest (ROI). The process of creating regions of interest (ROI) entails the identification and isolation of specific regions within skin lesions that encompass relevant information, with the aim of facilitating classification [21]. This step enhances the model’s ability to capture unique features of the skin lesions. The skin cancer classification model being evaluated employed a dataset consisting of ISIC images, obtained from an online resource. A series of image processing operations were executed to attain uniformity and create a consistent approach for the image data [22]. The operations involved in this process include the manipulation of image resolution depth and the conversion of pixel range into a positive integer, which is then represented by a single byte. Within this particular context, in the backend, the variable ‘x’ is employed to symbolize the pixel value, ‘y’ is utilized to represent the gray level value, and ‘max’ and ‘min’ are employed to denote the upper and lower boundaries of the pixel range. The aforementioned conversion procedure played a role in enhancing the precision of the images, thus enabling more efficient analysis in subsequent stages. To attain accurate identification and evaluation of cancerous regions, a cohort of skilled and accredited dermatologists specializing in the domain of skin cancer diligently scrutinized the provided images. The investigators performed a thorough analysis of the epidermis to identify areas impacted by pathological conditions. The parameters related to the disease areas underwent regular assessment and quantification across all datasets [23]. Fig. 3, as illustrated below, presents the transformation of the color of the image into grayscale. In order to focus on the primary site of malignancy, distinct regions of interest (ROI) were established for each variant of skin cancer [24].

Figure 3: Converted the image color to grey
Two methodologies were employed to address the variations observed within the malignant regions. The first step in the process involved applying the Laplacian filter to reduce image noise and enhance the prominent regions of infection. The application of this filtering technique has demonstrated efficacy in enhancing the detectability of salient features associated with skin cancer [25].
The core objective of the proposed model was to improve the precision and reliability of the skin cancer classification procedure by incorporating different preprocessing methods and techniques. Fig. 4 depicts the process of generating region of interests (ROI), as presented below. The utilization of region of interests (ROIs) has enabled focused analysis and extraction of relevant features specific to different types of skin cancer.
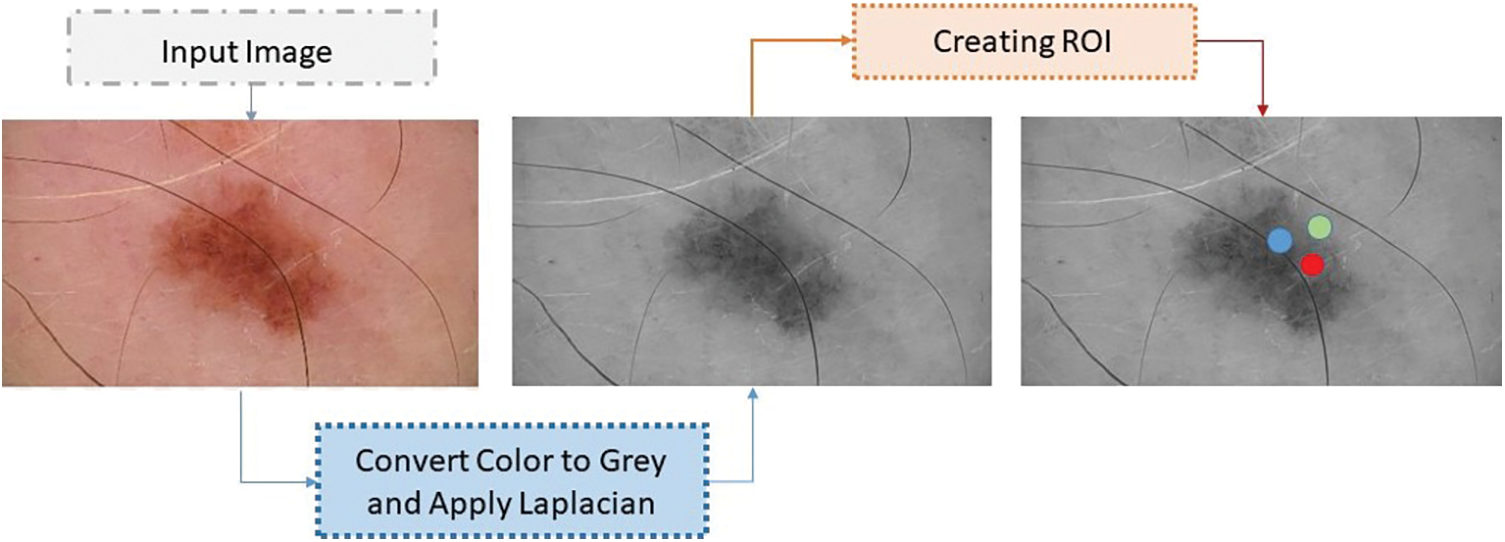
Figure 4: Process of ROI generation
A comprehensive dataset consisting of 2239 images was compiled, representing nine distinct categories of skin cancer. The dataset that has undergone preprocessing is divided into three sets: Training, validation, and testing. Subsequently, a portion of the dataset was allocated specifically for training the model, while another separate portion was reserved for the purpose of hyperparameter tuning and validation. Ultimately, a concluding portion was allocated to evaluate the efficacy of the model. Data allocation ratios of 80:20 is frequently used for training and testing reasons. According to reference [26], in these ratios, 80% or 70% of the data is set aside for training, while the remaining 20% or 30% is set aside for testing. The factual data pertaining to the International Skin Imaging Collaboration (ISIC) is presented in Table 2.

This partition ensures that a significant portion of the data is dedicated to training the model, thereby facilitating its ability to learn patterns and make predictions. In our study, the proposed hybrid Convolutional and Recurrent Neural Network (CNN-RNN) model with a ResNet-50 backbone was carefully configured with specific parameters to optimize its performance in skin cancer classification. For the Convolutional Neural Network (CNN) component, we utilized a learning rate of 0.001, a batch size of 32, and employed transfer learning with pre-trained weights from ResNet-50. This allowed the model to leverage features learned from a large dataset, promoting better generalization to our skin cancer classification task. The Recurrent Neural Network (RNN) component, designed to capture temporal dependencies, was configured with a long short-term memory (LSTM) layer. We used a sequence length of 10, chosen empirically to capture relevant sequential information in the dataset. Additionally, we employed a dropout rate of 0.2 in the LSTM layer to prevent overfitting and enhance the model’s generalization capabilities.
Throughout training, the model underwent 50 epochs, a parameter chosen after monitoring the convergence of the training and validation curves. The choice of these parameters was guided by a combination of empirical experimentation and hyperparameter tuning to achieve optimal performance on our skin cancer dataset. These parameter details not only provide transparency about the setup of your model but also offer insights into the considerations made during the experimental design. Adjusting these parameters and understanding their impact on the model’s performance can be crucial for the reproducibility and adaptability of your proposed approach in different contexts or datasets.
By employing a hybrid methodology, we have effectively expanded the dataset to include a comprehensive collection of 9000 images. This augmentation process has been carried out with the objective of achieving a balanced distribution, whereby each unique category of skin cancer is represented by 1000 images. The sequential model’s parameters are presented in Table 3. The incorporation of a balanced dataset facilitates the machine-learning model in gaining insights from a more comprehensive and fair representation of each class [27]. This approach helps to alleviate biases towards the dominant classes and improves the accuracy and generalization abilities of the sequential model. Principal Component Analysis (PCA) is a widely employed method in data analysis that aims to decrease the number of variables in a dataset while retaining the most pertinent information [28]. In order to minimize the squared error during the reconstruction of the original data, a projection is sought by transforming the data into a lower dimensional space [29].

The first step in the process entails data standardization, which involves subtracting the mean of the entire dataset from each individual sample and subsequently dividing it by the variance. This procedure guarantees that every instance exhibits a uniform variance [30]. The ultimate stage in the procedure is not completely indispensable; nevertheless, it offers benefits in terms of decreasing the workload on the central processing unit (CPU), as evidenced by Eq. (1).
To compute the Covariance Matrix, a dataset consisting of n samples, denoted as {X1, X2, ..., Xn}. The Covariance Matrix can be obtained using Eq. (2), as shown below:
where X denotes the covariance matrix, xi represents each sample in the dataset and
An alternative approach involves the multiplication of the standardized matrix Z with its transposed form as shown in Eq. (4).
Z is the standardized matrix, Zt is the transpose of Z and Cov(X) is the covariance matrix. The determination of principal components is based on the Eigenvectors of the ‘Covariance Matrix’ (Cov), which are subsequently organized in a descending order of importance according to their corresponding eigenvalues. Eigenvectors that correspond to larger eigenvalues are indicative of greater significance. The majority of the valuable data within the complete dataset is condensed into a single vector.
The model under consideration employs a range of data augmentation techniques in order to enhance the overall quality of the training dataset. The employed techniques include random rotations, horizontal and vertical flips, random zooming, and slight translations. Through the implementation of these transformations, the model is able to successfully simulate a wide range of viewpoints and perspectives of skin lesions [31]. This ultimately improves the model’s ability to handle variations in image orientation and positioning, making it more resilient. Data augmentation plays a multifaceted role in the training process [32]. Firstly, it is advantageous to address the issue of inadequate training data by employing augmented samples, as they provide a wider and more diverse selection for the purpose of training the model. The utilization of this methodology assists in the reduction of overfitting and improves the model’s ability to accurately extrapolate to unfamiliar skin lesion images [33]. Furthermore, the incorporation of data augmentation in the training process serves as a means of regularization for the model. This is achieved by introducing noise and variations, thereby enhancing the model’s ability to generalize and improve its performance. Regularization is a widely utilized technique in the field of machine learning that serves to address the potential issue of overfitting in models. By integrating regularization techniques, the model is discouraged from excessively memorizing specific patterns or features that are exclusively present in the training data. On the contrary, there is a strong recommendation to acquire a broader understanding of skin cancer by focusing on features that are more generalizable and representative [34]. Furthermore, the application of data augmentation methods can effectively tackle the difficulties presented by class imbalance within the dataset. To address the issue of underrepresentation in certain classes, it is possible to generate additional samples. This approach aims to promote a fairer distribution and reduce the potential bias of the model towards the dominant class. The incorporation of data augmentation holds significant significance in the training phase of the skin cancer classification model that has been put forth.
By incorporating a diverse range of transformations and modifications into the training dataset, it becomes possible to enhance the model’s capacity to generalize, thereby improving its performance on previously unseen data. Moreover, the application of this methodology possesses the capability to efficiently address any discrepancies in the distribution of students among various classes, as emphasized in literature [35]. The process of data augmentation is depicted in Fig. 5 above. The utilization of data augmentation techniques aims to enhance the accuracy and robustness of the model in the identification and classification of various forms of skin cancer. The parameters and their corresponding values for data augmentation are presented in Table 4. Augmented data is visualized in Fig. 6 below.
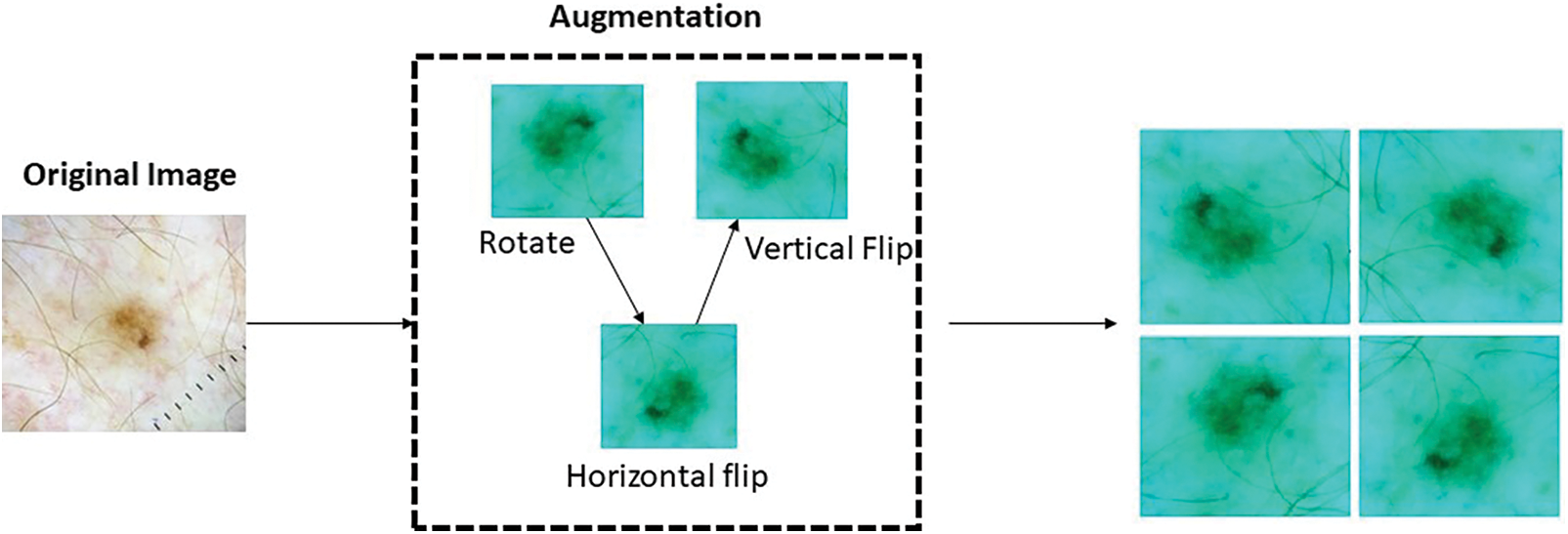
Figure 5: Data augmentation process

Figure 6: Visualization of augmented data
After uploading the captured images of skin lesions, we proceeded to use the segmentation technique in order to eliminate the skin, hair, and other unwanted elements. To achieve our goal, we utilized the range-oriented by pixel resolution (RO-PR) algorithm on the skin pictures. The range-oriented by pixel resolution (RO-PR) technique is utilized to delineate the clustering region of the pixels in the skin lesion image [36]. The Total Value of Pixel (TVP) evaluates the specified threshold value by taking into account a distinct cluster of skin images [37]. The thresholding level served as the pixel value for the skin lesion (PVL) image or as the beginning value for the cluster. The resolution of all adjacent pixels was compared, and the cumulative resolution of the image was analyzed to form the cluster. Ultimately, the assessment concludes that the dataset value of Total Value of Pixel (TVP) is equivalent to that of pixel value for the skin lesion (PVL). Consequently, the clustering of pixel regions (PR) is considered same, and the total cluster data is augmented by evaluating its ROIs. Each subsequent nearby cluster is determined in the same way.
The study employed the deep learning framework TensorFlow version 1.4 in conjunction with version 2.0.8 of the Keras API, implemented in Python, on a workstation equipped with an Intel i5 processor, 16 GB of RAM, and 4 GB of VRAM, utilizing NVIDIA GPUs. The GPUs were equipped with CUDA (Compute Unified Device Architecture) and cuDNN (CUDA Deep Neural Network library), which were installed and configured to enable GPU acceleration. The starting settings are set as follows: Batch size is assigned as 10, Learning rate is set to 0.001, and Epochs is defined as 50. The initial activation function used is Rectified Linear Unit (ReLu), while the final activation function is Sigmoid. The optimizer used for training is either Adam or Stochastic Gradient Descent (SGD). Average pooling is applied, and the performance of the model is evaluated using accuracy metrics.
Following the segmentation of the skin lesion, ROIs were obtained and the image properties of these moles of skin cancer were extracted in order to analyze their texture. There is various manual, automated, and semi-automated approaches to generating ROIs. Automated schemes rely significantly on image enhancement, while semi-automated and manual approaches rely on the judgment of experts. In order to compile a dataset on skin cancer, ten non-overlapping ROIs measuring 512 × 512 were applied to the images of each category, yielding a total of 1000 (100 × 10) ROI images per category. The ROI samples are illustrated in Fig. 4. We obtained a dataset containing 9000 (1000 × 9) ROI images for nine different types of skin cancer. The acquisition of features is a critical step in the classification of datasets using machine learning, as it provides the essential information required for texture analysis. Twenty-eight binary features, five histogram features, seven RST features, ten texture features, and seven spectral features were selected for texture analysis in the study. A grand total of 57 features were extracted from each of the generated ROIs in this manner. There was a total of 513,000 features in the features vector space (FVS) (9 × 100 × 10 = 9000 × 57).
The skin cancer classification model being proposed employs a hybrid architecture that integrates Convolutional Neural Networks (CNNs) and Recurrent Neural Networks (RNNs) with a ResNet-50 backbone. The mathematical representation of this hybrid approach is as follows, as stated in Eq. (6) below. Let X represent the input image data set, where X is defined as the collection of individual images {x1, x2, ..., xn}. Here, n denotes the total count of images present in the dataset. The following is the architectural diagram of the Convolutional Neural Network (CNN) model, as depicted in Fig. 7.
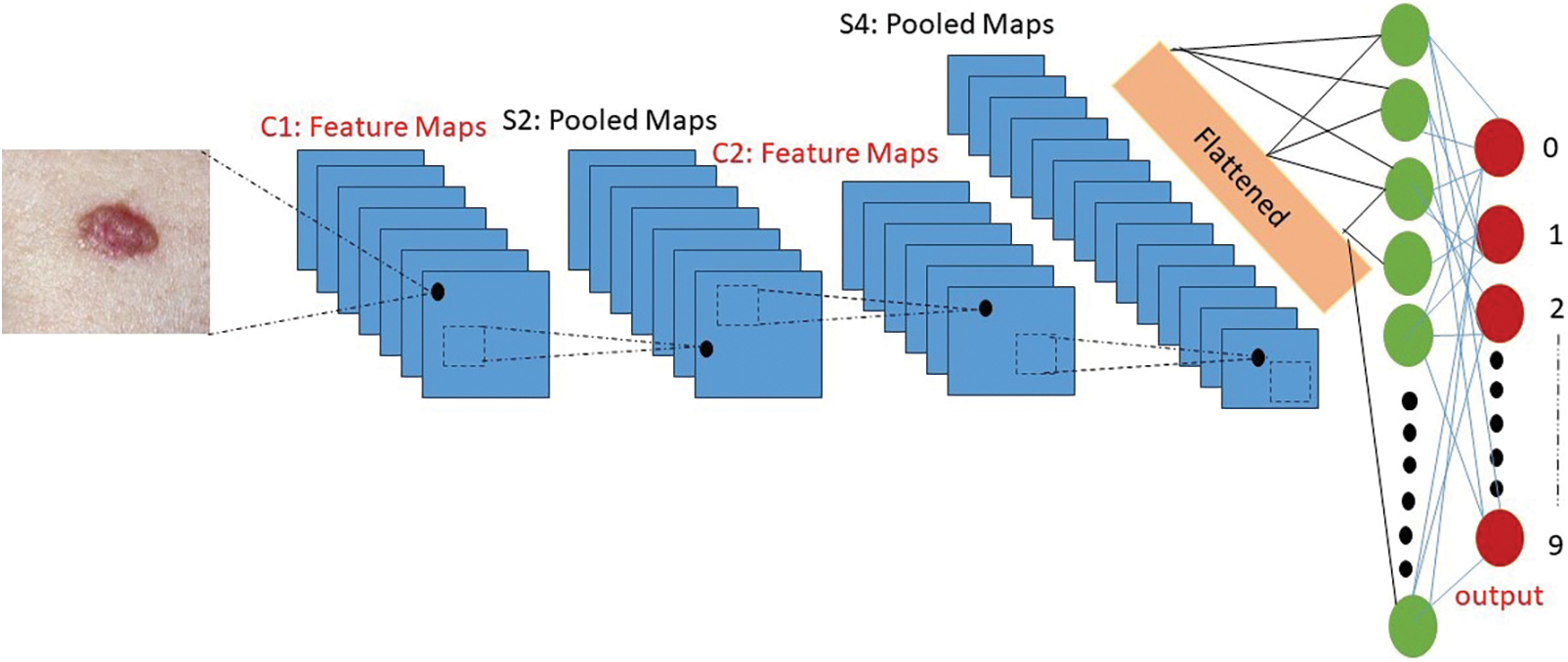
Figure 7: Architecture diagram of the convolutional neural network (CNN) model
The proposed Convolutional Neural Network (CNN) model architecture for the skin cancer classification consists of three main layers: Two convolutional-pooling layers followed by a fully-connected classification layer. Each layer performs specific operations to extract meaningful features and classify the input images [38]. The first layer in the architecture is a convolutional-pooling layer. It utilizes an 8 × 8 fixed convolutional kernel to convolve over the input images. This convolutional layer aims to capture local spatial patterns and extract relevant features. The resulting feature maps are then subjected to a 2 × 2 pooling operation, which reduces the spatial dimensions while preserving important information [39]. The second layer is also a convolutional-pooling layer. It uses the same 8 × 8 convolutional kernels as the previous layer but with a higher number of neurons, specifically 64. This allows for more complex feature extraction and representation. Similar to the first layer, a 2 × 2 pooling operation follows the convolution step to further down sample the feature maps. Finally, the last layer of the architecture is a fully-connected classification layer. It consists of 256 neurons, which are connected to the final two neurons responsible for mitosis/non-mitosis classification [40]. This layer combines the extracted features from the previous layers and performs the classification task, distinguishing between the two target classes [41].
3.1 Convolutional Neural Network (CNN) Feature Extraction
The Convolutional Neural Network (CNN) denotes the “CNN” model component, fCNN, extracts feature from the input images using the ResNet-50 architecture and “I” denotes the input images. This can be expressed as Eqs. (5) & (5.1) shown below:
3.2 Recurrent Neural Network (RNN) Temporal Modeling
The Recurrent Neural Network (RNN) denotes the “RNN” model component, fRNN represents the output of the RNN component capturing temporal dependencies and fCNN denotes the features extracted by the CNN component. This can be formulated as Eqs. (6) & (6.1) below:
The output of the Recurrent Neural Network (RNN) component represents ZRNN capturing temporal dependencies is passed through a classification layer to obtain the final predicted probabilities for each class. Let
The classification layer, denoted by the symbol g, encompasses various activation functions such as the softmax function or other suitable alternatives. The model was trained using the International Skin Imaging Collaboration (ISIC) 9 class’s image dataset of skin cancer. The dataset includes images from various classes such as Actinic Keratosis (AK), Basal Cell Carcinoma (BC), Dermatofibroma (DF), Melanoma (MM), Nevus (NN), Pigmented Benign Keratosis (PK), Seborrheic Keratosis (SK), Squamous Cell Carcinoma (SC), Vascular Lesion (VL) [43]. The model was trained to acquire discriminative features and temporal patterns that are unique to each class. The integration of convolutional neural networks (CNNs) and recurrent neural networks (RNNs) in a hybrid architecture enables the model to exploit the spatial characteristics acquired by the Convolutional Neural Network (CNN) and the temporal relationships captured by the RNN. Through the integration of these two components, the model is able to proficiently extract features and effectively model the sequential information that is inherent in the images of skin cancer [44]. By undergoing training using the International Skin Imaging Collaboration (ISIC) 9 classes dataset, the model acquires the ability to accurately classify images of skin cancer, taking into account the distinct characteristics and variations present in each class. This feature allows the model to generate accurate predictions regarding the existence of various forms of skin cancer by analyzing the input images. The proposed skin cancer classification model integrates a hybrid Convolutional and Recurrent Neural Network (CNN-RNN) architecture with a ResNet-50 backbone to effectively extract features and model temporal dependencies in the International Skin Imaging Collaboration (ISIC) 9 classes image dataset of skin cancer. This enables accurate classification and identification of various types of skin cancer. Fig. 8 below show Convolutional and Recurrent Neural Network (CNN-RNN) classification performance graph for the skin cancer types.
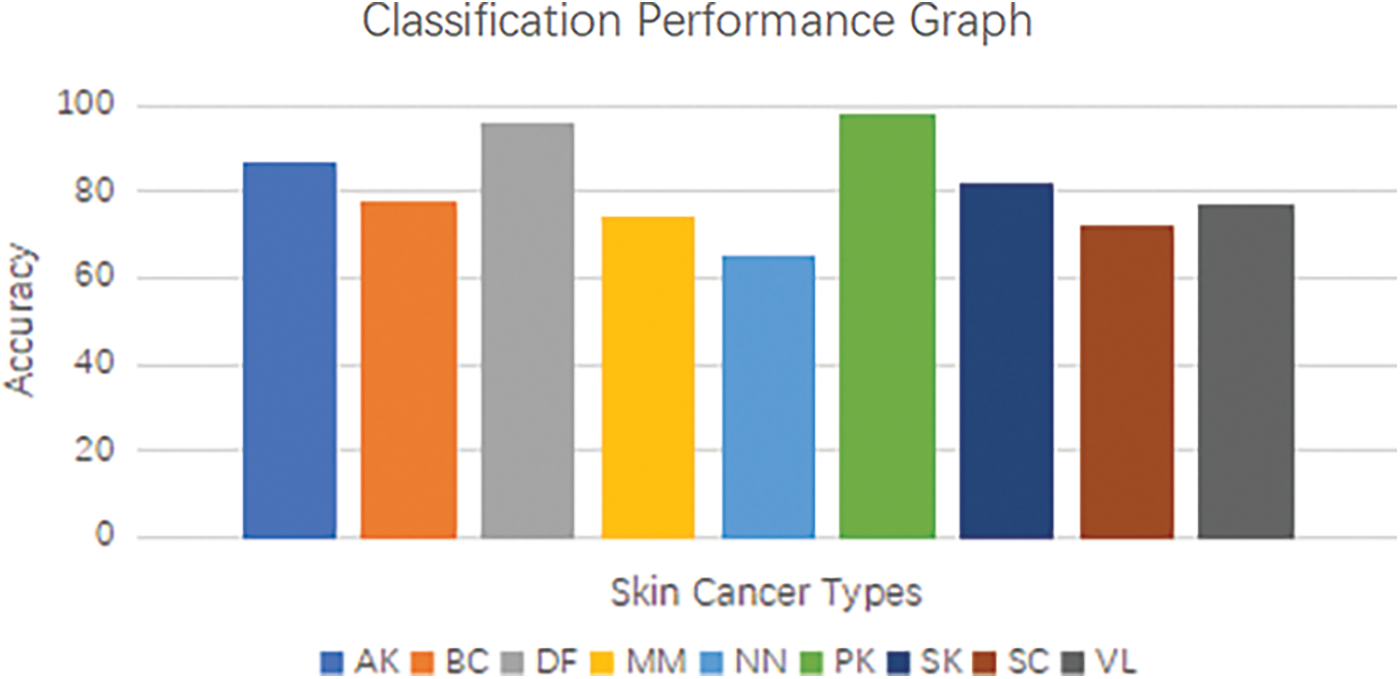
Figure 8: Convolutional and recurrent neural network (CNN-RNN) classification performance graph for the skin cancer type
The utilization of a confusion matrix allows us to accurately observe both true and false positive outcomes, as well as false negative outcomes. In order to further investigate the model’s performance, we carried out a comprehensive analysis utilizing precision, recall, and F1-score metrics. These measures offer valuable information about the model’s capacity to accurately detect positive cases, minimize false positives, and maintain a balance between precision and recall [45]. The performance of our model consistently shown high precision, recall, and F1-score values for all classes, indicating its reliability and applicability for various forms of skin cancer. The equations below are used to derive and express additional important metrics.
where, TP (True Positive), TN (True Negative), FP (False Positive), and FN (False Negative).
The time complexity of training the model depends on several factors, including the number of training samples (N), the number of epochs (E), the batch size (B), and the complexity of the model architecture. The overall time complexity for training can be approximated as O (N × E × (H × W × C × F × K2)), taking into account the forward and backward passes for each training sample [46]. Below shown in Fig. 9, show data distribution for training and testing.
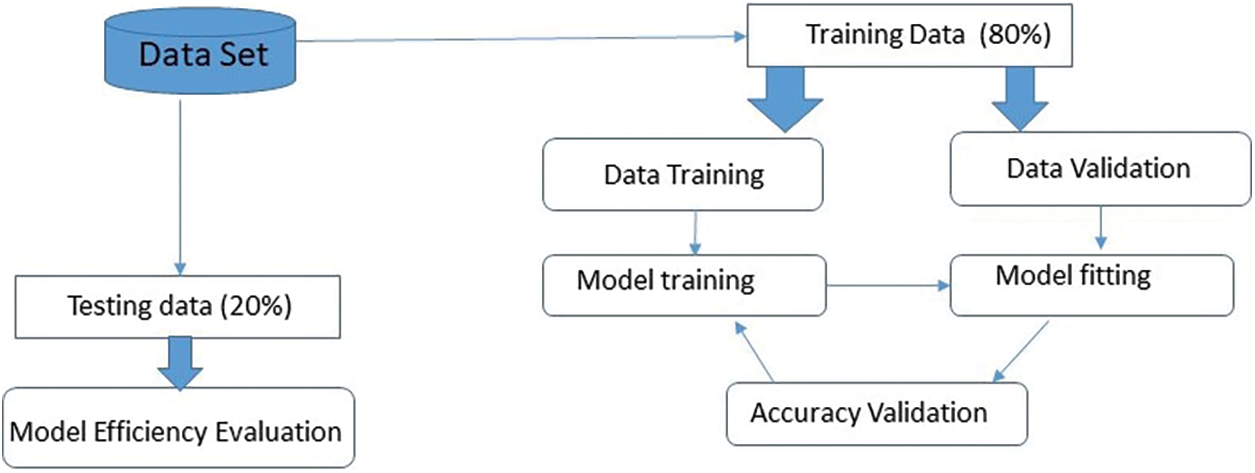
Figure 9: Data distribution for training and testing
Upon commencing the training process, it becomes evident that the validation accuracy attains a level of 90% upon the completion of five epochs. Once the iterations are completed, the efficiency and error of each model generated are calculated. The overall efficiency and final error are determined by calculating the average of the 10 trained models. The visual representation of the Cross-Validation procedure is depicted in Fig. 10.
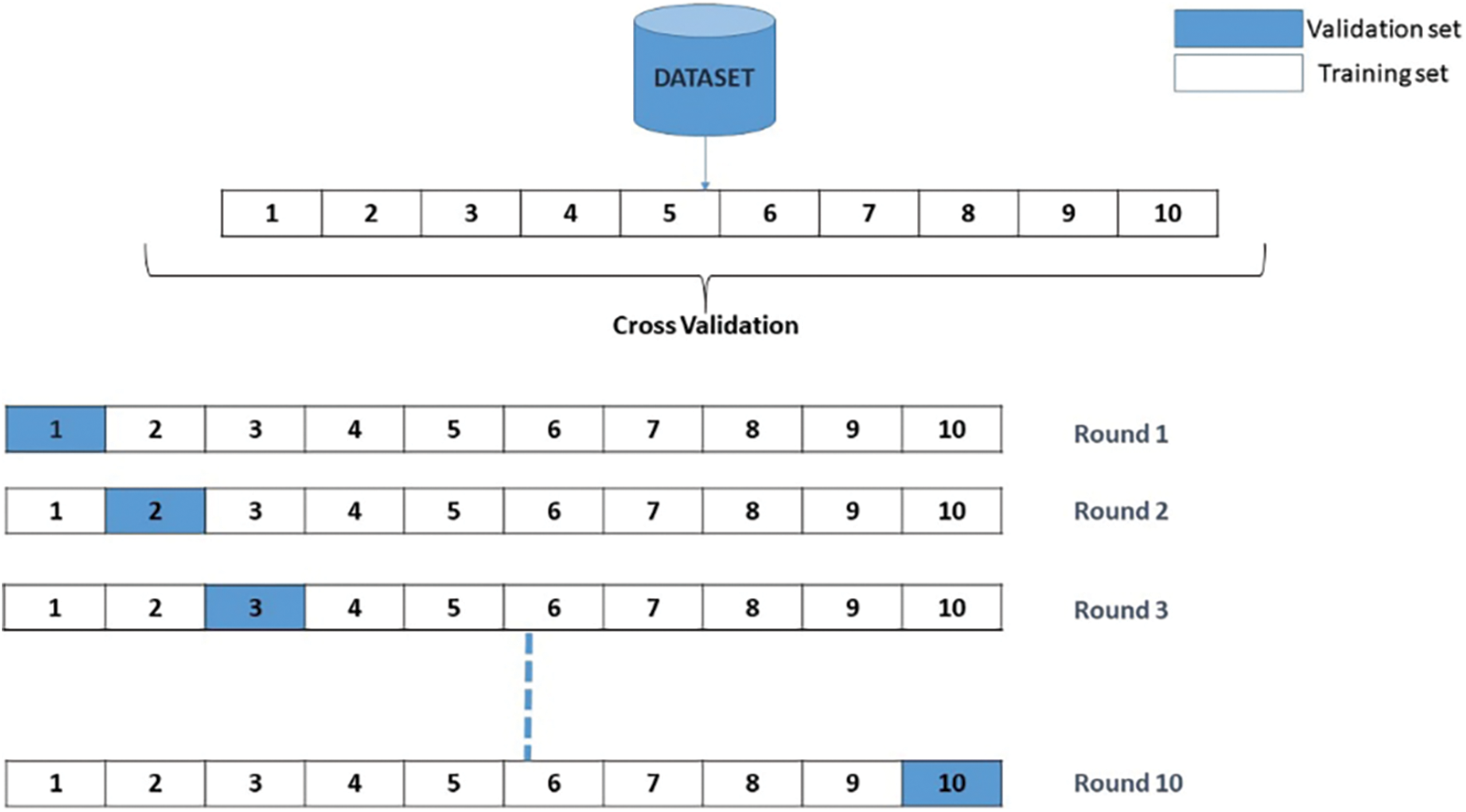
Figure 10: Validation procedure
The time complexity of evaluating the model on the test dataset is similar to the feature extraction process, as it involves passing the test images through the model’s layers. Height, width, number of channels, number of filters, and the square of the convolutional kernel size [47]. The time complexity for evaluation can be approximated as the upper bound of the growth rate ‘O’(H * W * C * F * K^2) for each test sample. Fig. 11 below shows accuracy, area under the curve, sensitivity, specificity, precision and F1-score comparison for the model.
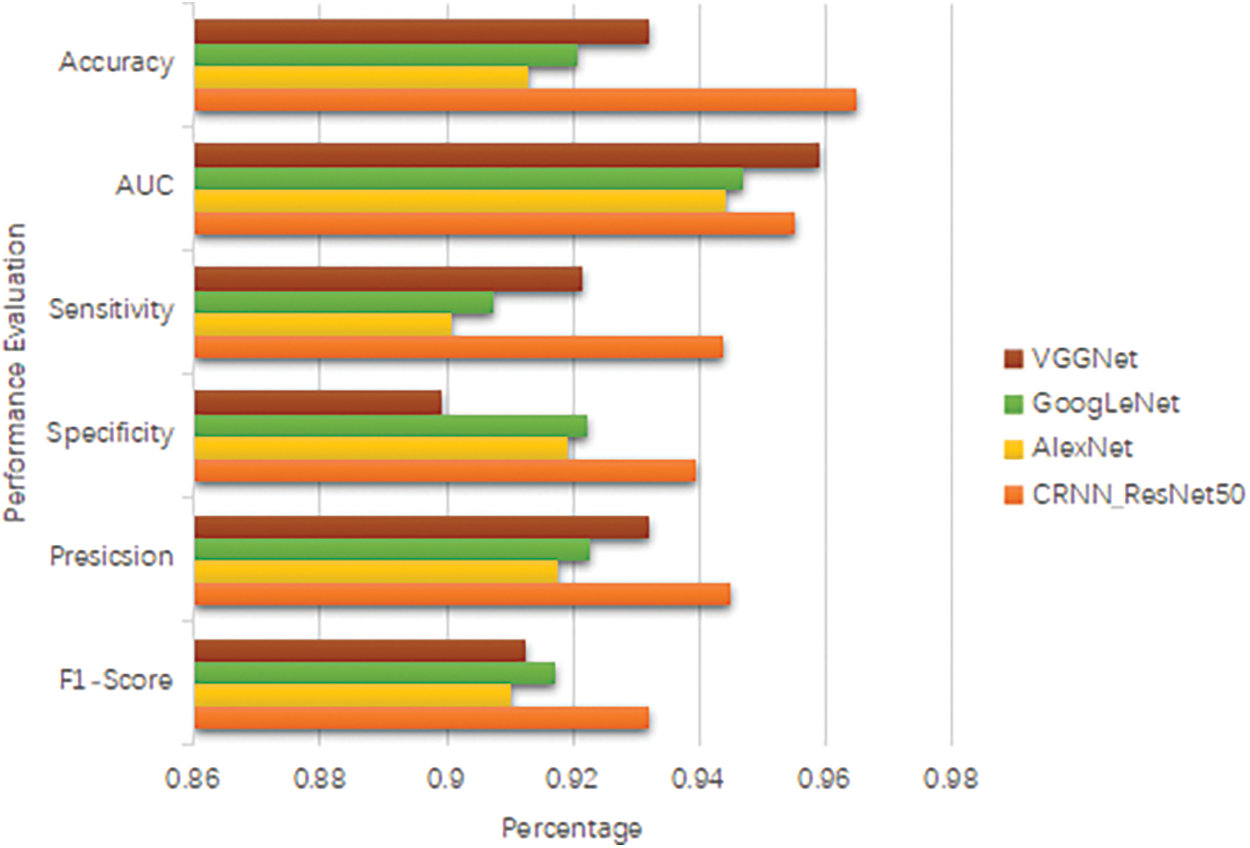
Figure 11: Accuracy, area under the curve, sensitivity, specificity, precision and F1-score
The time complexity for making predictions on new, unseen images using the trained model is similar to the evaluation process, as it involves a forward pass through the model’s layers [48]. The time complexity for prediction can be approximated as O(H × W × C × F × K2) for each prediction. Here, above notation represents, Height (H), width (W), number of channels (C), number of filters (F), and the square of the convolutional kernel size (K2). It is important to note that the above time complexity analysis provides an approximation and assumes a sequential execution of the operations. The actual time taken may also depend on hardware acceleration, parallel processing, and optimization techniques used during the implementation. Fig. 12 below shows recognition accuracy and loss curves of proposed model.

Figure 12: Recognition accuracy and loss curves of proposed model
There are more benign than malignant pictures, the collection is unbalanced. We have used augmentation to oversample malignant samples. Then, when the model is given these photos, it improves them by rotating, resizing, flipping them horizontally, and increasing the range of magnification and shear. Images are improved so that they do not fit too well. The model was trained with the many topologies of the Convolution Neural Network. Even though VGGNet V2 and GoogleNet V2 were used to train the model, the result was not what was expected. Then, the custom layered model is used. Dropout is added between layers to keep the model from fitting too well, and as the size of the training picture gets smaller, more neurons or filters are added to the convolutional 2D layer to get the desired result. This lowers loss, and as loss goes down, model accuracy goes up. When model accuracy goes up, it is easier to tell if a skin lesion is benign or cancerous. after getting the results that were wanted. After the model is saved, FAST APIs are used to get the result into the online application and mobile application. Once the user uploads a picture of the spot, the program will predict whether the user will be diagnosed with skin cancer or not. Fig. 13 below shows confusion matrix and Table 5 shows performance comparison of proposed methods on ISIC dataset.
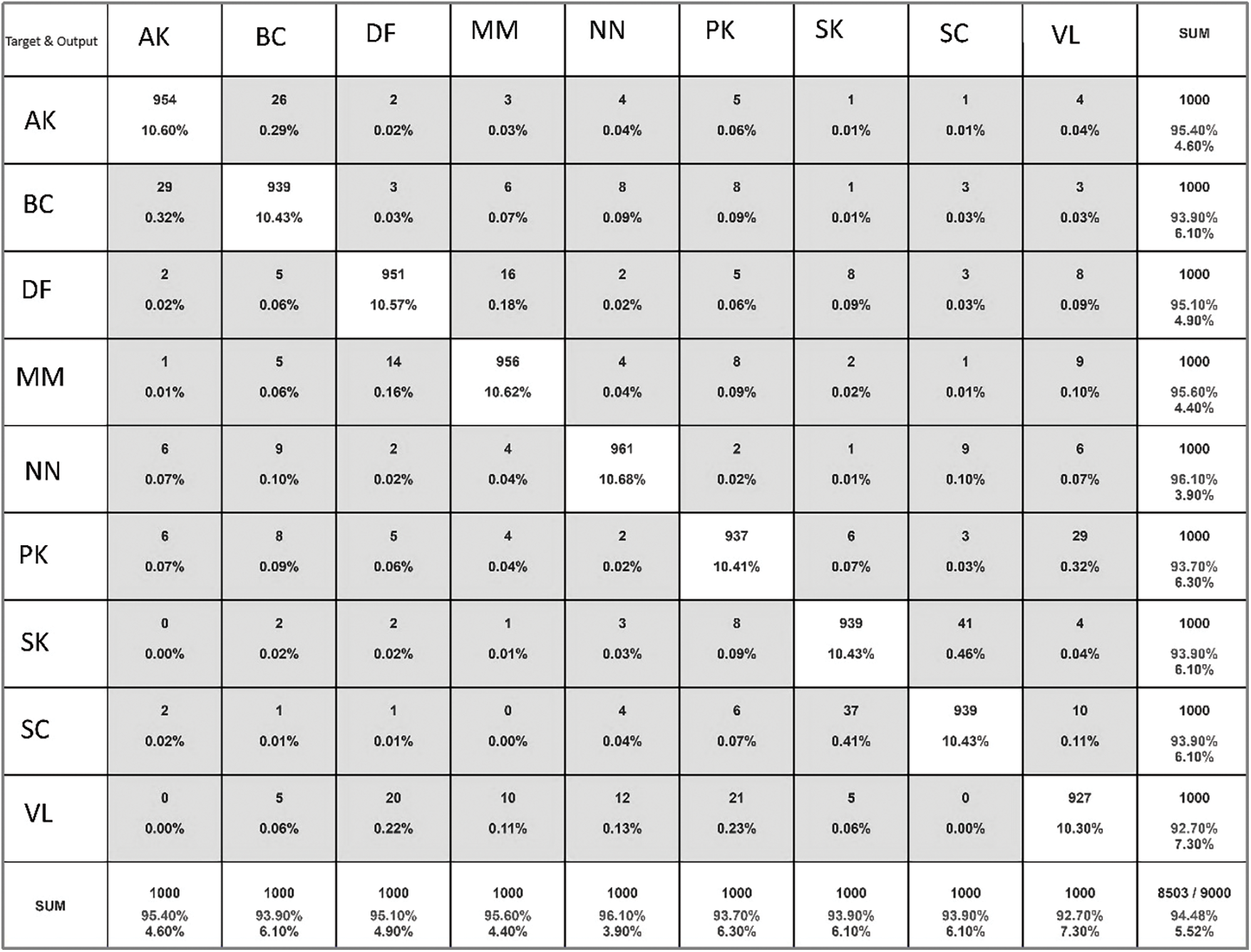
Figure 13: Confusion matrix of proposed model
4.1 Limitation of Proposed Methodology
Firstly, the effectiveness of the model is contingent upon the quality and representativeness of the dataset used for training. If the dataset has inherent biases or lacks diversity in terms of skin types, ethnicities, or geographic regions, the model’s generalizability may be compromised. Future research should focus on curating more diverse and inclusive datasets to enhance the model’s applicability across a broader population [54]. Secondly, while the model demonstrates high accuracy, the potential challenges of interpretability persist. Deep learning models, especially those with intricate architectures like Convolutional and Recurrent Neural Network (CNN-RNN), can be perceived as “black boxes” where understanding the decision-making process is challenging. Enhancing the interpretability of the model’s predictions is an avenue for future research, ensuring that healthcare professionals can trust and understand the model’s outputs in clinical practice. Moreover, the proposed model’s performance may be influenced by variations in image quality and resolution, factors commonly encountered in real-world clinical settings. Ensuring robustness to such variations is critical for the model’s practical utility. Further research could explore techniques to enhance the model’s resilience to variations in input data quality. while our Convolutional and Recurrent Neural Network (CNN-RNN) model shows promise in skin cancer classification, it is essential to recognize these limitations. Addressing these challenges in future research will not only refine the proposed model but also contribute to the broader goal of developing robust and reliable deep learning models for clinical applications in dermatology.
The employed model utilized image processing techniques to enhance the performance of our existing models. As previously stated, initially, random image cropping and rotations were employed as a means to augment the dataset with additional information. Additionally, it is possible to manipulate the visual display by zooming in or out, as well as flipping the image horizontally or vertically. Additionally, during the process of retraining the final layers, we employed transfer learning techniques by leveraging the pre-trained weights from the ImageNet dataset. Subsequently, we performed fine-tuning on the model. The entire model will be retrained using a reduced learning rate. This model also attempted to impart knowledge to the other models. In the absence of transfer learning, the outcomes consistently exhibited inferior performance, particularly in the deeper layers. Furthermore, we made modifications to the color scheme by transitioning from the RGB model to the HSV model and also experimented with grayscale. However, despite these adjustments, our attempts were unsuccessful in attaining the intended outcomes. Subsequently, the outcomes exhibited a deterioration, particularly in the context of evaluating grayscale images. The final results are presented by considering the average accuracy of the classifications, which is weighted.
The model were trained using RGB images, employing techniques such as data augmentation, transfer learning, fine-tuning, and the SGD optimizer. During the training process, appropriate class weights were employed to ensure equitable treatment for all individuals, given the substantial dissimilarity in the data. In order to address the issue pertaining to the image quality, we attempted to incorporate each individual image in a sequential manner. During the validation phase, the algorithm is executed multiple times, and a diagnosis is determined based on the median average of its results. Specifically, we performed four iterations of inserting each validation image into the database, with each iteration involving a different flipping operation. Subsequently, we employed the median average to assign the image to one of four distinct categories. The aforementioned approach was employed on a sample above model, which were randomly selected from the pool of training epochs in the ResNet50 model over the course of the last 50 iterations. The findings indicated that, in comparison to the initial image, there was an average enhancement of 1.04%. Given its emphasis on quality and its primary role as an initial diagnostic indicator, the application is primarily focused on ensuring high standards and serving as an initial diagnostic clue. According to Table 6, the model has opted for a two-class mapping approach using the ResNet50 model to distinguish between Malignant cases (mel, bcc, or akiec) and Benign cases (nv, pbk.d, bkl, vasc, cc, or df).
5.1 Suggestions for Future Research
1. Investigate the incorporation of multi-model data, such as dermoscopy and patient history information, to enhance the model’s diagnostic capabilities. Combining imaging data with other patient-specific features could provide a more holistic understanding of skin conditions.
2. Explore techniques for enhancing model interpretability, particularly in the context of medical [55] diagnostics. Integrate explainable AI methods to provide insights into the decision-making process of the model, fostering trust and understanding among healthcare practitioners.
3. Assess the feasibility of transfer learning by training the model on a broader dataset encompassing various dermatological conditions. This could potentially enhance the model’s ability to differentiate between different skin disorders and expand its utility in clinical practice.
4. Investigate strategies for real-time deployment of the model in clinical settings. Develop lightweight architectures or model compression techniques to ensure efficient execution on edge devices, facilitating point-of-care diagnostics.
Acknowledgement: We wish to extend our heartfelt thanks and genuine appreciation to our supervisor, Guangmin Sun (Beijing University of Technology) for his support, insightful comments, valuable remarks, and active involvement throughout the duration of writing this manuscript have been truly invaluable. Also, we are most grateful to the anonymous reviewers and editors for their careful work, which made this, paper a substantial improvement. Thanks to my teacher for providing me with research conditions.
Funding Statement: This research was conducted without external funding support.
Author Contributions: S.G developed the method; S.S.Z, S.F.Q and M.K actively engaged in conducting the experiments and analyzing the data; S.S.Z resulted in the composition of the manuscript; S.Q made substantial contributions to the analysis and discussion of the algorithm and experimental findings. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The JPG File (.jpg) data used to support the findings of this study have been deposited in the ISIC Archive repository (Base URL: https://www.isic-archive.com/).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. S. Mishra, N. Dey, A. S. Ashour, and A. Kumar, “Skin lesion segmentation and classification using deep learning: A review,” Comput. Meth. Prog. Bio., vol. 216, pp. 106530, 2022. [Google Scholar]
2. S. Jha, A. Singh, and R. S. Sonawane, “Deep learning models for skin cancer detection: A comprehensive review,” Artif. Intell. Med., vol. 126, pp. 102539, 2022. [Google Scholar]
3. J. Jaworek-Korjakowska, P. Kleczek, and M. Gorgon, “Melanoma thickness prediction based on convolutional neural network with VGG-19 model transfer learning,” in Proc IEEE/CVF Conf. Comput. Vis. Pattern Recogn. Workshops, 2019. doi: 10.1109/CVPRW.2019.00333. [Google Scholar] [CrossRef]
4. J. Kawahara, S. Daneshvar, G. Argenziano, and G. Hamarneh, “Seven-point checklist and skin lesion classification using multitask multimodal neural nets,” IEEE J. Biomed. Health Inf., vol. 23, no. 5, pp. 538–546, 2019. doi: 10.1109/JBHI.2018.2824327. [Google Scholar] [PubMed] [CrossRef]
5. Z. Rahman, M. S. Hossain, M. R. Islam, M. M. Hasan, and R. A. Hridhee, “An approach for multiclass skin lesion classification based on ensemble learning,” Inf. Med. Unlock., vol. 25, no. 10, pp. 100659, 2021. doi: 10.1016/j.imu.2021.100659. [Google Scholar] [CrossRef]
6. D. Bisla, A. Choromanska, R. S. Berman, J. A. Stein, and D. Polsky, “Towards automated melanoma detection with deep learning: Data purification and augmentation,” in Proc. IEEE/CVF Conf. Comput. Vis. Pattern Recogn. Workshops, 2019. doi: 10.1109/CVPRW.2019.00330. [Google Scholar] [CrossRef]
7. J. Bian, S. Zhang, S. Wang, J. Zhang, and J. Guo, “Skin lesion classification by multi-view filtered transfer learning,” IEEE Access, vol. 9, pp. 66052–66061, 2021. doi: 10.1109/ACCESS.2021.3076533. [Google Scholar] [CrossRef]
8. M. A. Malik and M. Parashar, “Efficient deep learning model for classification of skin cancer,” J. Amb. Intell. Hum. Comput., vol. 13, no. 2, pp. 2713–2723, 2022. [Google Scholar]
9. J. Shi, J. Wei, and D. Wei, “A novel skin cancer classification method based on deep learning,” J. Med. Syst., vol. 46, no. 12, pp. 140, 2022. [Google Scholar]
10. U. Bashir, H. Abbas, and S. Rho, “Skin cancer classification using transfer learning and attention-based convolutional neural networks,” IEEE Access, vol. 10, pp. 13060–13073, 2022. [Google Scholar]
11. V. Kharchenko, V. Khryashchev, and V. Melnik, “Transfer learning-based melanoma detection using convolutional neural networks,” Comput. Vis. Image Underst., vol. 216, pp. 103299, 2022. [Google Scholar]
12. R. Kumar and S. Srivastava, “Deep learning-based melanoma classification using ensemble of convolutional neural networks,” Comput. Biol. Med., vol. 137, pp. 104850, 2022. [Google Scholar]
13. V. Rajinikanth, S. Kadry, R. Damasevicius, D. Sankaran, M. A. Mohammed, and S. Chander, “Skin melanoma segmentation using VGG-UNet with Adam/SGD optimizer: A study,” in 2022 Third Int. Conf. Intell. Comput. Instrum. Control Technol. (ICICICT), 2022, pp. 982–986. doi: 10.1109/ICICICT54557.2022.9917848. [Google Scholar] [CrossRef]
14. M. Soni, S. Gomathi, P. Kumar, P. P. Churi, M. A. Mohammed and A. O. Salman, “Hybridizing convolutional neural network for classification of lung diseases,” Int. J. Swarm Intell. Res. (IJSIR), vol. 13, no. 2, pp. 1–15, 2022. doi: 10.4018/IJSIR. [Google Scholar] [CrossRef]
15. S. Ponomarev, “Classification of melanoma skin cancer using deep learning with sparse autoencoders,” Comput. Meth. Prog. Bio., vol. 220, pp. 106111, 2022. [Google Scholar]
16. M. Karimzadeh, A. Vakanski, M. Xian, and B. Zhang, “Post-Hoc explain ability of BI-RADS descriptors in a multi-task framework for breast cancer detection and segmentation,” in 2023 IEEE 33rd Int. Workshop Mach. Learn. Signal Process. (MLSP), 2023, pp. 1–6. doi: 10.1109/MLSP55844.2023.10286006. [Google Scholar] [PubMed] [CrossRef]
17. X. Yuan, Y. Li, H. Liu, and D. Wang, “Skin lesion classification using a deep learning framework with visual attention,” Neural Comput. Appl., vol. 34, no. 4, pp. 875–885, 2022. [Google Scholar]
18. A. Ganapathiraju and U. Raghavendra, “Multiscale residual networks for skin cancer detection,” Comput. Med. Imag. Grap., vol. 95, pp. 101988, 2023. [Google Scholar]
19. S. Ozturk and T. Ayvaz, “Hybrid feature selection and deep learning for melanoma detection,” Comput. Meth. Prog. Bio., vol. 211, pp. 106244, 2023. [Google Scholar]
20. H. Zhang, L. Zhang, X. Li, and X. Shi, “Skin lesion classification based on deep convolutional neural networks with global attention pooling,” J. Med. Syst., vol. 47, no. 3, pp. 38, 2023. [Google Scholar]
21. W. Zhang, M. Han, and Y. Zhao, “Multi-task learning based skin cancer classification using deep learning,” Int. J. Imag. Syst. Tech., vol. 33, no. 1, pp. 59–71, 2023. [Google Scholar]
22. F. Xie, H. Fan, Y. Li, Z. Jiang, R. Meng and A. Bovik, “Melanoma classification on dermoscopy images using a neural network ensemble model,” IEEE Trans. Med. Imag., vol. 36, no. 3, pp. 849–858, 2016. doi: 10.1109/TMI.2016.2633551. [Google Scholar] [PubMed] [CrossRef]
23. N. Gessert, D. Hendrix, F. Fleischer, and M. Neuberger, “Skin cancer classification using convolutional neural networks with active learning,” Int. J. Comput. Assist. Radiol. Surg., vol. 18, no. 2, pp. 281–290, 2023. [Google Scholar]
24. S. Zahra, M. A. Ghazanfar, A. Khalid, M. A. Azam, U. Naeem and A. Prugel-Bennett, “Novel centroid selection approaches for means-clustering based recommender systems,” Inf. Sci., vol. 320, pp. 156–189, 2015. doi: 10.1016/j.ins.2015.03.062. [Google Scholar] [CrossRef]
25. Y. Fan, S. Wang, Y. Zhang, and T. Xu, “Skin cancer recognition based on deep learning with optimized network structure,” Neural Process. Lett., vol. 58, no. 1, pp. 677–694, 2023. [Google Scholar]
26. M. A. Malik and M. Parashar, “Skin lesion segmentation using U-Net and pre-trained convolutional neural networks,” Neural Comput. Appl., vol. 35, no. 3, pp. 1263–1276, 2023. [Google Scholar]
27. A. Sood and A. Gupta, “Deep convolutional neural networks for skin cancer detection,” Multimed. Tools Appl., vol. 82, no. 3, pp. 4039–4058, 2023. [Google Scholar]
28. R. Tanno, S. Watanabe, and H. Koga, “Skin lesion classification using pretrained deep learning models with extended dataset,” J. Med. Syst., vol. 47, no. 1, pp. 8, 2023. [Google Scholar]
29. Z. Shi et al., “A deep CNN based transfer learning method for false positive reduction,” Multimed. Tools Appl., vol. 78, no. 1, pp. 1017–1033, 2019. doi: 10.1007/s11042-018-6082-6. [Google Scholar] [CrossRef]
30. S. Demyanov, R. Chakravorty, M. Abedini, A. Halpern, and R. Garnavi, “Classification of dermoscopy patterns using deep convolutional neural networks,” in 2016 IEEE 13th Int. Symp. Biomed. Imag. (ISBI), IEEE, 2016. [Google Scholar]
31. R. Moussa, F. Gerges, C. Salem, R. Akiki, O. Falou and D. Azar, “Computer-aided detection of melanoma using geometric features,” in 2016 3rd Middle East Conf. Biomed. Eng. (MECBME), IEEE, 2016. [Google Scholar]
32. M. A. Hussain, A. M. Al-Juboori, and K. H. Abdulkareem, “Classification of skin lesions using stacked denoising autoencoders,” Pattern Anal. Appl., vol. 26, no. 1, pp. 43–55, 2023. [Google Scholar]
33. R. Amelard, A. Wong, and D. A. Clausi, “Extracting high-level intuitive features (HLIF) for classifying skin lesions using standard camera images,” in 2012 Ninth Conf. Comput. Robot Vis., IEEE, 2012. [Google Scholar]
34. M. E. Celebi et al., “A methodological approach to the classification of dermoscopy images,” Comput. Med. Imag. Grap., vol. 31, no. 6, pp. 362–373, 2007. doi: 10.1016/j.compmedimag.2007.01.003. [Google Scholar] [PubMed] [CrossRef]
35. R. J. Stanley, W. V. Stoecker, and R. H. Moss, “A relative color approach to color discrimination for malignant melanoma detection in dermoscopy images,” Skin Res. Technol., vol. 13, no. 1, pp. 62–72, 2007. doi: 10.1111/j.1600-0846.2007.00192.x. [Google Scholar] [PubMed] [CrossRef]
36. A. V. Patel, S. Biswas, and M. K. Dutta, “A comparative analysis of deep learning models for skin cancer classification,” Comput. Biol. Med., vol. 132, pp. 105495, 2023. [Google Scholar]
37. H. Ganster, P. Pinz, R. Rohrer, E. Wildling, M. Binder, and H. Kittler, “Automated melanoma recognition,” IEEE Trans. Med. Imag., vol. 20, no. 3, pp. 233–239, 2001. doi: 10.1109/42.918473. [Google Scholar] [PubMed] [CrossRef]
38. P. Rubegni et al., “Automated diagnosis of pigmented skin lesions,” Int. J. Cancer, vol. 101, no. 6, pp. 576–580, 2002. doi: 10.1002/ijc.10620. [Google Scholar] [PubMed] [CrossRef]
39. R. B. Oliveira et al., “Computational methods for pigmented skin lesion classification in images: Review and future trends,” Neural Comput. Appl., vol. 29, no. 3, pp. 613–636, 2018. doi: 10.1007/s00521-016-2482-6. [Google Scholar] [CrossRef]
40. I. Stanganelli et al., “Computer-aided diagnosis of melanocytic lesions,” Anticancer Res., vol. 25, no. 6C, pp. 4577–4582, 2005. [Google Scholar] [PubMed]
41. H. Iyatomi et al., “Computer-based classification of dermoscopy images of melanocytic lesions on acral volar skin,” J. Invest. Dermatol., vol. 128, no. 8, pp. 2049–2054, 2008. doi: 10.1038/jid.2008.28. [Google Scholar] [PubMed] [CrossRef]
42. M. A. Farooq, M. A. M. Azhar, and R. H. Raza, “Automatic lesion detection system (ALDS) for skin cancer classification using SVM and neural classifiers,” in 2016 IEEE 16th Int. Conf. Bioinf. Bioeng. (BIBE), IEEE, 2016. [Google Scholar]
43. J. F. Alcón et al., “Automatic imaging system with decision support for inspection of pigmented skin lesions and melanoma diagnosis,” IEEE J. Sel. Top. Signal Process., vol. 3, no. 1, pp. 14–25, 2009. doi: 10.1109/JSTSP.2008.2011156. [Google Scholar] [CrossRef]
44. I. Giotis et al., “MED-NODE: A computer-assisted melanoma diagnosis system using non dermoscopic images,” Expert. Syst. Appl., vol. 42, no. 19, pp. 6578–6585, 2015. doi: 10.1016/j.eswa.2015.04.034. [Google Scholar] [CrossRef]
45. A. Safi et al., “Computer–aided diagnosis of pigmented skin dermoscopic images,” in MICCAI Int. Workshop Med. Content-Based Retriev. Clin. Decis. Support, Springer, 2011. [Google Scholar]
46. L. Yu, H. Chen, Q. Dou, J. Qin, and P. A. Heng, “Automated melanoma recognition in dermoscopy images via very deep residual networks,” IEEE Trans. Med. Imag., vol. 41, no. 1, pp. 257–266, 2022. [Google Scholar]
47. D. Ruiz, V. Berenguer, A. Soriano, and B. Sánchez, “A decision support system for the diagnosis of melanoma: A comparative approach,” Expert. Syst. Appl., vol. 38, no. 12, pp. 15217–15223, 2011. doi: 10.1016/j.eswa.2011.05.079. [Google Scholar] [CrossRef]
48. A. Masood and Al-Jumaily, “Differential evolution based advised SVM for histopathological image analysis for skin cancer detection,” in 2015 37th Annual Int. Conf. IEEE Eng. Med. Biol. Soc. (EMBC), IEEE, 2015. [Google Scholar]
49. H. Jing, X. He, Q. Han, A. A. Abd El-Latif, and X. Niu, “Saliency detection based on integrated features,” Neurocomput., vol. 129, no. 3, pp. 114–121, 2014. doi: 10.1016/j.neucom.2013.02.048. [Google Scholar] [CrossRef]
50. M. Hammad, A. Maher, K. Wang, F. Jiang, and M. Amrani, “Detection of abnormal skin cell conditions based,” Meas., vol. 125, no. 3, pp. 634–644, 2018. doi: 10.1016/j.measurement.2018.05.033. [Google Scholar] [CrossRef]
51. X. Bai et al., “A fully automatic skin cancer detection method based on one-class SVM,” IEICE Trans. Inf. Syst., vol. 96, no. 2, pp. 387–391, 2013. [Google Scholar]
52. M. Amrani, M. Hammad, F. Jiang, K. Wang, and A. Amrani, “Very deep feature extraction and fusion for arrhythmias detection,” Neural Comput. Appl., vol. 30, no. 7, pp. 2047–2057, 2018. doi: 10.1007/s00521-018-3616-9. [Google Scholar] [CrossRef]
53. T. Zhang, Q. Han, A. A. Abd El-Lat, X. Bai, and X. Niu, “2-D cartoon character detection based on scalable-shape context and hough voting,” Inf. Technol. J., vol. 12, no. 12, pp. 2342–2349, 2013. doi: 10.3923/itj.2013.2342.2349. [Google Scholar] [CrossRef]
54. K. He, X. Li, and W. Zhang, “Melanoma classification based on optimized convolutional neural network,” Biomed. Signal Process. Control, vol. 75, pp. 103020, 2023. [Google Scholar]
55. B. Kong et al., “Recognizing end-diastole and end-systole frames via deep temporal regression network,” in Int. Conf. Med. Image Comput. Comput.-Assist. Intervent., Springer, 2016. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF

 Downloads
Downloads
 Citation Tools
Citation Tools