 Open Access
Open Access
ARTICLE
Olive Leaf Disease Detection via Wavelet Transform and Feature Fusion of Pre-Trained Deep Learning Models
1 Department of Information Systems, College of Computer and Information Sciences, Jouf University, Sakakah, Kingdom of Saudi Arabia
2 Department of Information Systems and Technology, Faculty of Graduate Studies and Research, Cairo University, Giza, Egypt
* Corresponding Author: Mahmood A. Mahmood. Email:
(This article belongs to the Special Issue: Deep Learning in Computer-Aided Diagnosis Based on Medical Image)
Computers, Materials & Continua 2024, 78(3), 3431-3448. https://doi.org/10.32604/cmc.2024.047604
Received 10 November 2023; Accepted 27 December 2023; Issue published 26 March 2024
Abstract
Olive trees are susceptible to a variety of diseases that can cause significant crop damage and economic losses. Early detection of these diseases is essential for effective management. We propose a novel transformed wavelet, feature-fused, pre-trained deep learning model for detecting olive leaf diseases. The proposed model combines wavelet transforms with pre-trained deep-learning models to extract discriminative features from olive leaf images. The model has four main phases: preprocessing using data augmentation, three-level wavelet transformation, learning using pre-trained deep learning models, and a fused deep learning model. In the preprocessing phase, the image dataset is augmented using techniques such as resizing, rescaling, flipping, rotation, zooming, and contrasting. In wavelet transformation, the augmented images are decomposed into three frequency levels. Three pre-trained deep learning models, EfficientNet-B7, DenseNet-201, and ResNet-152-V2, are used in the learning phase. The models were trained using the approximate images of the third-level sub-band of the wavelet transform. In the fused phase, the fused model consists of a merge layer, three dense layers, and two dropout layers. The proposed model was evaluated using a dataset of images of healthy and infected olive leaves. It achieved an accuracy of 99.72% in the diagnosis of olive leaf diseases, which exceeds the accuracy of other methods reported in the literature. This finding suggests that our proposed method is a promising tool for the early detection of olive leaf diseases.Keywords
Abbreviations
| The following abbreviations are used in this manuscript | |
| AI | Artificial Intelligence |
| CNN | Convolutional Neural Networks |
| SVM | Support Vector Machine |
| DWT | Discrete Wavelet Transform |
| LL | Low–Low |
| LH | Low–High |
| HH | High–High |
| HL | High–Low |
| Relu | Rectified Linear Unit |
| TP | True Positive |
| TN | True Negative |
| FN | False Negative |
| FP | False Positive |
Artificial intelligence (AI) is one of the fastest developing fields today, and it will affect our lives strongly. Looking at it today, AI is already being used in a number of applications, from chat assistants to driverless cars. Deep learning is becoming increasingly popular in the AI community. One type of machine learning, deep learning uses artificial neural networks to learn from data [1] and [2]. Deep learning is highly proficient in these various activities, such as disease classification and facial recognition. But there is still much we do not know about deep learning. One of the shortcomings of deep learning, however, is that it is often hard to see what these models learn [3–5]. Agriculture is one area where deep learning shows promise. Deep learning can be used to improve crop yields, detect pests and diseases, and optimize irrigation. However, some challenges remain to be addressed before deep learning can be widely adopted in agriculture [6,7]. One challenge is that deep learning models require a large amount of data to train. The data needed can be difficult and expensive to collect. Another challenge is that deep learning models can be computationally expensive to run. Despite these challenges, deep learning has the potential to revolutionize agriculture.
A nation depends on plants and agriculture for its economy. But plant diseases can sharply reduce crop yields. Because of this, diagnosing and forecasting plant diseases early is extremely important so that they can be treated as soon as possible. Improving agricultural production requires early detection and forecasting of plant diseases [8].
Olive trees are susceptible to a variety of pathogens, including bacteria. These pathogens can cause leaf diseases that can significantly reduce crop yields. Early diagnosis of leaf diseases is essential to prevent further damage. However, this can be difficult to do manually, as it requires specialized knowledge and experience. There are a few automated methods for detecting leaf diseases. These methods use image processing and machine learning to identify the symptoms of the disease. These methods are more accurate and efficient than manual methods, and they can be used to screen large numbers of trees. The use of automated methods for detecting leaf diseases is becoming increasingly important. As olive production becomes more widespread, the need for accurate and efficient methods for disease detection will grow. Recent advances in artificial intelligence and computer vision have led to the increased adoption of deep learning methods for image classification tasks [7,9].
In this paper we detect disease from olive leaf images by presenting a CNN which uses wavelet transformers to extract features. Within our CNN architecture, we use wavelet transforms along with pre-trained deep-learning models to extract features from a variety of scales in the image. This allows CNN to learn more discriminative features, which in turn can enhance the accuracy of disease detection. A wavelet transform is a technique of mathematics which allows an image to be broken down into various frequency sub-bands. Each sub-band represents a different range of frequencies in the image. After that, CNN is used to extract features from different scales on each sub-band. All sub-bands are subsequently combined with the features extracted. This is a more complete representation of the image. This method can reach 99.72% in diagnosing olive leaf diseases, and it is much more convenient than traditional techniques. This means that when the proposed model was evaluated, it turns out to have remarkably well-tuned perception results. The following is a summary of the contributions made in this research:
• A transformed-wavelet, feature-fused CNN for detecting olive leaf diseases.
• A proposed CNN architecture that combines wavelet transforms with pre-trained deep learning models to extract discriminative features from olive leaf images.
• A promising new approach to detecting olive leaf diseases. The proposed CNN architecture has the potential to be used to develop robust automated systems for early detection of olive leaf diseases, which can help olive growers minimize crop losses.
The remainder of this paper is organized as follows. Section 2 reviews the diagnosis and classification of olive diseases in recent relevant research. Section 3 presents the proposed diagnosis model for olive leaf diseases. Section 4 describes how our model was experimentally tested and analyses of the results. Section 5 presents conclusions and discusses future research.
Several studies have been conducted to develop methods for diagnosing diseases that affect plant leaves, especially olive leaves [10]. These studies have used a variety of image processing and deep learning techniques to analyze, detect, and classify various plant diseases [11,12]. Some studies have proposed deep learning models that can be used to diagnose diseases using smartphone applications [13]. Other studies have used CNN architectures and deep learning algorithms to diagnose diseases from hyperspectral images, which are images that capture a wider range of wavelengths of light than traditional RGB images [14,15].
One study [16] concerned early detection of anthracnose in olives. Anthracnose is a fungal disease that may cause serious damage to olive trees. In this study, hyperspectral images of olive leaves were classified through a CNN architecture called ResNet50. Others [17] have put forward methodologies based on deep learning for the identification and classification of plant diseases, including a hybrid model that employs both the VGG-16 machine vision architecture used by Google Images as well as MobileNet. This was then utilized to classify sunflower disease types. In yet another study [18], scientists put forward a new image analysis method to diagnose and classify olive diseases. The approach was based on the image texture properties that can be seen on olive plant leaves. The first type of analysis uses histogram thresholding and k-means segmentation to separate the infected area. The second analytical method utilizes first- to fourth-order moments which measure the relationship between infection and at least one textural feature.
Deep learning has become a popular approach to identifying and classifying plant diseases. Convolutional neural networks (CNNs) are powerful tools for extracting features from images. CNNs are effective in detection and classification tasks because they can learn features automatically and generalize well to new data. However, CNNs require a large amount of training data and a fixed set of parameters [19–21].
Several studies have addressed the challenges of using CNNs for plant disease detection. For example, researchers [22] have proposed a method for classifying olive leaf diseases using transfer learning on deep CNN architectures. The researchers tested their method on a dataset of 3,400 images of healthy leaves. Other researchers [23] have summarized the significance of deep learning as a current area of research in agricultural crop protection. They examined prior research on hyperspectral imaging, deep learning, and image processing-based methods for detecting leaf illness. Based on a review of the literature, deep learning algorithms were judged to be the best methods for identifying leaf diseases.
The amount of data gathered for use in model training has a substantial impact on the accuracy of the results obtained. Data augmentation techniques, transfer learning techniques, and CNNs are employed to alter the datasets. The problem of plant disease detection needs to be solved even when the findings of earlier research are sufficient. This is because the lack of labels in the data can impair the quality of the pixels used to depict disease symptoms.
Another study [24] used a deep learning architecture to classify a variety of plant and fruit leaf diseases. The authors used a deep transfer learning model to learn features from images. They then used radial basis function (RBF) kernels with several Support Vector Machine (SVM) models to improve feature discrimination. The results of the study suggest that deep learning is a promising approach to plant disease detection. However, more research is needed to address the challenges of using CNNs for this task. Other researchers [25] have proposed a hybrid deep-learning model for identifying olive leaf disease. The model combines three different deep learning models, including a modified vision transformer model.
Several studies recently have proposed modifications to Inception-V3. These additions have been made to increase the accuracy and speed of these CNNs. The use of data augmentation [26] is perhaps the most widespread improvement. Transfer learning is also a common kind of improvement. Transfer learning is the employment of a pre-trained CNN to seed in another new network. This technique can improve the performance of a new CNN with less training data [27]. Experiments have shown that applying data augmentation improves the performance of this model (Ksibi et al. [28], Cruz et al. [29]). The effect of data augmentation was evaluated by these authors, including how it increased performance in the model systems under study or could help to overcome closer boundaries that had been established in genetic model cells before this intervention but were in one study [30], six types of data augmentation and transfer learning methods were used to identify 39 classes of plant leaf diseases. The ResNet101 CNN model was used in another study [31] to identifiable plant diseases using hyperspectral image dataset. Another study [32] employed a genetic algorithm to determine the best epochs, those that produced accurate detection of olive leaf diseases at highest levels with various CNN models. The literature review showed the state-of-the art methods summarized in Table 1.
Our proposed model aims to achieve higher accuracy in diagnosing olive leaf diseases than methods proposed by other researchers by combining a discrete wavelet transform with EfficientNet-B7, DenseNet-201, and ResNet-152-V2, added as a feature layer in the model, and fusing all features to a CNN to determine which yields the best accuracy.
The proposed model is shown in Fig. 1. It has two main stages: data augmentation preprocessing and feature fusion from a discrete wavelet transform and pre-trained deep learning models (EffiecientNet-B7, DenseNet-201, and ResNet-152-v2).
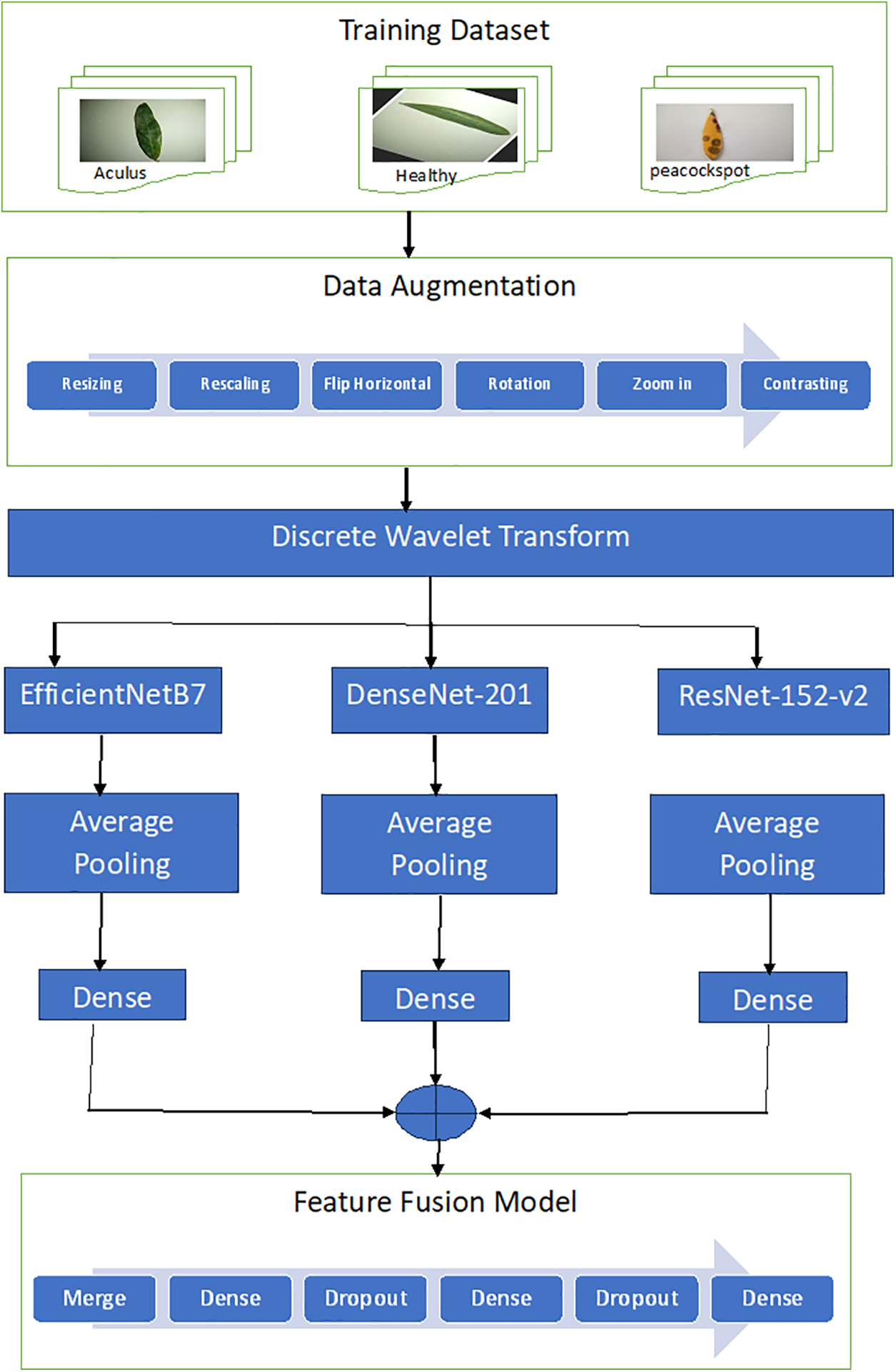
Figure 1: Proposed feature fusion model
Data augmentation is a technique whereby new data points are created from existing ones to artificially increase the size of your dataset. By performing such transformations as cropping, flipping, and rotating the data or by adding noise to them it is possible to humanize. There are two reasons why data augmentation is important. Second, it can promote the generalizability of deep learning models by solving overfitting problems. Overfitting is where a model learns the training data too well and cannot generalize to new data. Diverse data from which to learn: To prevent overfitting, a technique known as “data augmentation” can be used. Third, using data augmentation can enable us to further improve the accuracy of deep learning models, by making our dataset larger. This matters since deep learning models tend to be more accurate when trained on bigger datasets [37].
Many different data augmentation techniques can be used. Some of the most common techniques are the following:
• Cropping a portion of the image.
• Flipping the image horizontally or vertically.
• Rotating the image by a certain angle.
• Adding noise to the image.
Our model used some data augmentation techniques, specifically: (1) resizing the images to 224 × 224, (2) rescaling the images, (3) flipping the images horizontally, i.e., from left to right, (4) rotating the images clockwise 0.1 radians, (5) zooming the images out by 10%, and (6) increasing the image contrast by 10%.
3.2 Discrete Wavelet Transforms
An approximation image was extracted by means of a discrete wavelet transform (DWT). DWT is a versatile tool that enables the analysis of signals in both time and frequency. It does this by means of a windowing technique employing variable sizes, so that it is able to pick up both low-frequency and high-frequency information. A major advantage of the DWT is that it employs scale rather than frequency. This means that it can be used to analyze signals on a variety of levels. DWT, for example, can extract coarse features of an image (such as overall shape and brightness) or fine details (like individual edges and textures) [38].
DWT was used in this study to decompose a two-dimensional (2D) image into four sub-bands at each level: low–low (LL), low–high (LH), high–high (HH), and high–low (HL). The LL sub-band contains the low-frequency components of the image, while the LH, HH, and HL sub-bands contain the high-frequency components [39]. A three-level decomposition was performed, as shown in Fig. 2. The Harr wavelet was used to approximate the solid information in the image [40–43]. The original 224 × 224 image was divided into four bands: LL1 (112 × 112), LH1 (112 × 112), HH1 (112 × 112), and HL1 (112 × 112). The LL1 band was further decomposed into LL2 (56 × 56), HL2 (56 × 56), HH2 (56 × 56), and HL2 (56 × 56). The LL2 band was decomposed into LL3 (28 × 28), HL3 (28 × 28), HH3 (28 × 28), and LH3 (28 × 28). The LL3 at the lowest level contained the approximate image, which was used for further processing.
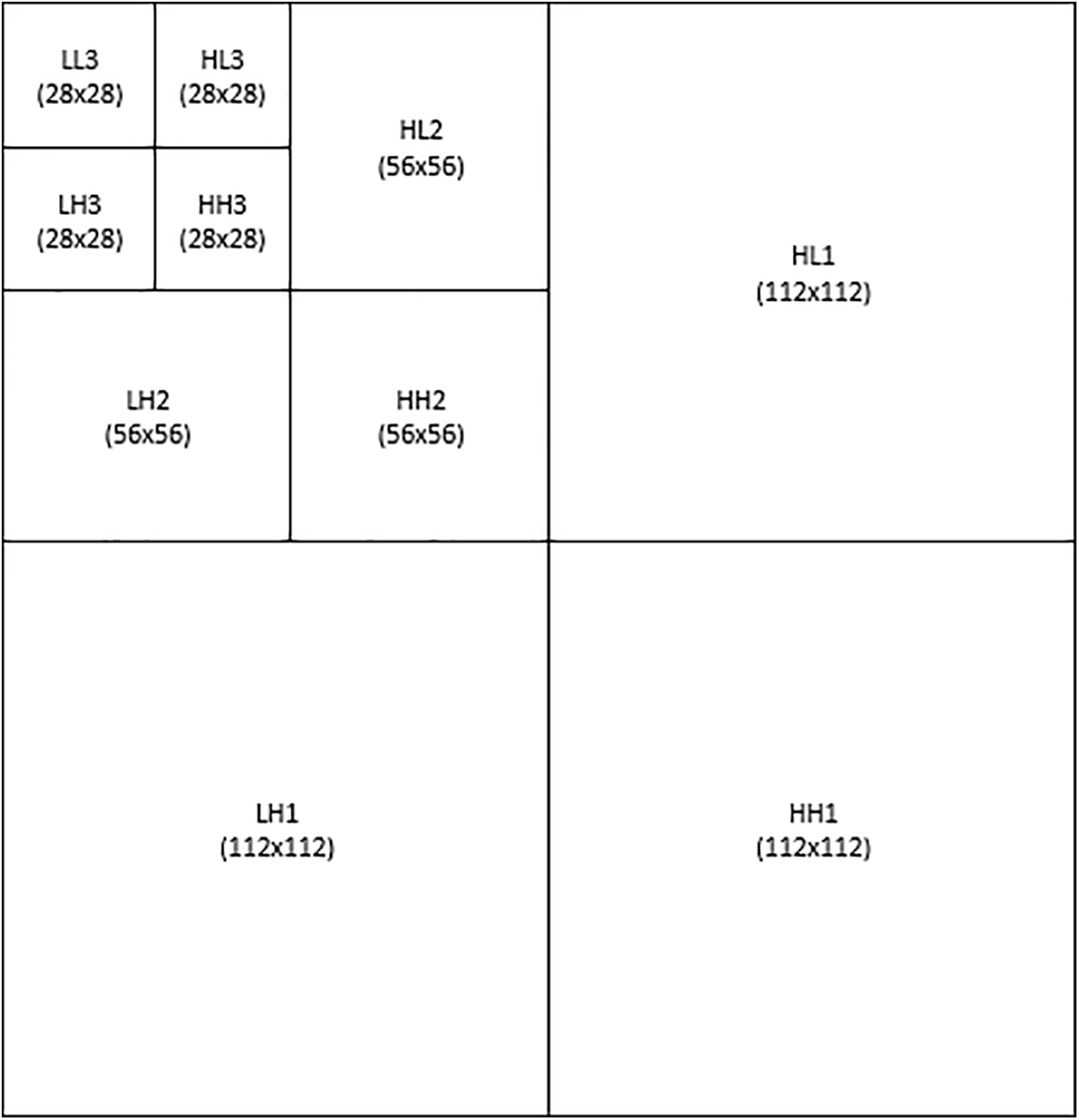
Figure 2: Three-level DWT model
DenseNet, a deep learning architecture introduced by Kumar et al. [44], has connections from every layer to every other layer. This offers several advantages, including the following:
• Elimination of the vanishing-gradient problem: The vanishing-gradient problem is a common challenge in training deep neural networks, as it can cause the network to lose its ability to learn from the data. DenseNet’s dense connectivity helps to alleviate this problem by ensuring that all layers have access to all previous feature maps.
• Improved feature propagation: DenseNet’s dense connectivity also improves feature propagation, which is the process of passing features from one layer to the next. By allowing each layer to access all previous feature maps, DenseNet can learn more complex and informative features.
• Feature reuse: DenseNet’s dense connectivity also promotes feature reuse, which means that the same features can be used by multiple layers in the network. This can help to reduce the number of parameters required in the network, which can improve performance and reduce computational cost.
DenseNet is a deep learning architecture with several advantages that make it well-suited for fruit recognition tasks. Its dense connectivity helps to alleviate the vanishing gradient problem, improve feature propagation, and promote feature reuse. This allows DenseNet to learn more complex and informative features, even for fruits with very similar features.
DenseNet-201 is a CNN with 201 layers. It belongs to the DenseNet family of models, which is known for being capable in learning rich feature representations for image classification. DenseNet-201 has been demonstrated to attain state of the art in several image classification benchmarks. As its accuracy and efficiency are both very high, it is a popular choice for image classification tasks. DenseNet-201 is a deep learning model that has been trained on the ImageNet dataset. This data set contains more than 1.2 million images from 1,00-dimension object categories and On a collection of image classification tasks (like object detection, scene classification and image retrieval) currently considered state-of-the-art, DenseNet-201 attains the best accuracy to date. DenseNet-201 compares to other CNN models in that it can attain the same rate of accuracy with fewer parameters and lower computational overhead. Just this fact makes it a suitable choice for mobile devices and other resource-constrained applications like image classification or object detection [45]. The CNN model, DenseNet-201 is very good at learning complex features from images and can function errorless even if the images are noisy or objects in them have been moved around. It is because it connects every layer to all the other layers and uses a special technique for reducing the number of parameters.
ResNet-152-V2 is the name of a 152-layer convolutional neural network (CNN) model. It belongs to the ResNet family of models, which are celebrated for their ability to learn deep residual representations suitable for image classification. It has been trained on the ImageNet dataset, which includes over 1.2 million images of objects in about a thousand different object categories. It achieves state-of-the art accuracy on several image classification tasks: object detection, scene classification and image retrieval. ResNet-152-V2 is also known for its efficiency. It is one of the most accurate CNN models (with high efficiency) [46].
EfficientNet-B7 is an accurate and efficient convolutional neural network (CNN) model. It is significantly faster and more memory-efficient than other CNN models of equal or better accuracy, achieving state-of-the art results on numerous image classification tasks. The family of EfficientNet models include Estimate-B7 and are all based on the same compound scaling method which uniformly scales depth, width and resolution. The scaling method allows EfficientNet models with high accuracy but not compromising efficiency. The ImageNet dataset on which EfficientNet-B7 is trained contains 1.2 million images in 1,000 different object categories. Trained EfficientNet-B7 is capable of handling a number of different image classification tasks, such as object detection, scene recognition and similarity searching [47].
Fig. 1 shows the proposed model, which includes a six-layer feature fusion model with merged inputs, three dense layers, and two dropout layers. The feature fusion model takes the outputs of three pre-trained models as inputs and reduces the number of output neurons to 256 and 128 in the first two dense layers using the Relu (rectified linear unit) activation function (Eq. (1)). After each dense layer, a dropout layer with a 0.2 rate is used to prevent overfitting. The last dense layer uses the softmax activation function (Eq. (2)) to reduce the number of neurons to three outputs. This feature fusion model allows the proposed model to learn more complex and informative features from the outputs of the three pre-trained models. Table 2 introduces the summary of the fusion model. The model has three input layers are DenseNet201 input layer, EfficientNetB7 input layer, and ResNet152-V2 input layer. In DenseNet152, the input layer has images with shape (28, 28, 3) augmented by some methods such as rescaling, normalization, batch normalization, activation and applying the CNN for DenseNet152. In EffiecientNetB7, the input layer consists of the images with shape (28, 28, 3) and Conv2D of DenseNet152 model. In ResNet152-V2 the input layer consists of the images with shape (28, 28, 3) and Conv2D of EfficientNetB7 model.

Traditional classification metrics are accuracy, precision, recall, and the F1 measure, which are calculated using Eqs. (3)–(6). These metrics are based on the values of true positives (TP), false positives (FP), true negatives (TN), and false negatives (FN). Fig. 3 displays the testing model.
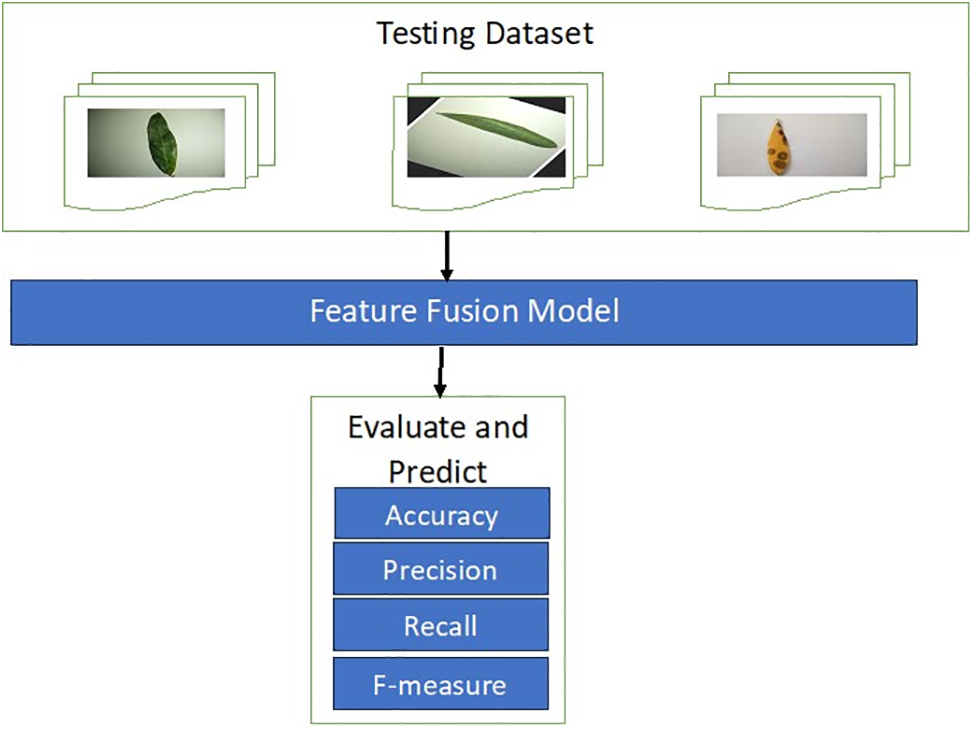
Figure 3: Testing model
4 Experimental Results and Discussion
The results of the experiments conducted using the proposed model are summarized in this section. We also evaluate the outcomes of various deep learning strategies using our own customized deep learning model. The data science server at Kaggle allowed the deployment of this model, which was created using Python. We carried out tests of EffiecientNet-B7, DenseNet-201, REsNet-152-v2, and our proposed model to gauge the applicability of the proposed model. The three subcategories that best describe our dataset are “healthy,” “peacock spot,” and “Aculus olearius”.
4.1 Dataset Description and Evaluation Metrics
The computer used for the experiments has a 64-bit operating system, an x64-based processor, an AMD Ryzen 7 7730U 2.00-GHz CPU with Radeon Graphics, and 16 GB of RAM. We evaluated the performance of our suggested ideal deep learning model using a dataset drawn from an open dataset [48]. With the help of an agricultural engineer with extensive field experience, images of 6,961 olive leaves were divided into three unique groups: healthy leaves, leaves infected with Aculus olearius, and leaves infected with olive peacock spot. Sample images of healthy and diseased olive leaves are shown in Fig. 4.

Figure 4: Dataset image samples
Table 3 shows how the dataset was split into two groups, with 70% of each group used for training and 30% used for testing. Anomalies in the distribution of datasets when they are divided into training and testing groups can have negative impacts on the CNN model’s predictions.

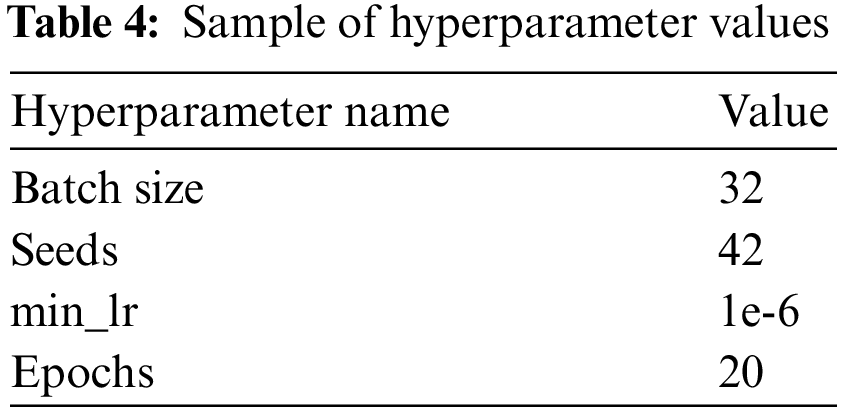
The experiment used to evaluate the proposed model had the hyperparameters shown in Table 4.
In this experiment, the training dataset images were preprocessed and augmented with several types of data augmentation. The images were resized to 224 × 224, rescaled to [0,1], flipped horizontally, rotated by 0.1 radians, zoomed in by 0.1, and contrasted by 0.1. The discrete wavelet transform (DWT) was applied to the augmented images to obtain four types of images: approximation, horizontal detail, vertical detail, and diagonal detail. The approximation images were then submitted to three pre-trained deep-learning models. The average pooling 2D layer took the outputs of the three pre-trained models as inputs and reduced the number of neurons to 512. The outputs of the pre-trained deep learning models were then merged and sent to the feature fusion model. Fig. 5 shows that the accuracy score of the validation test was 99.72% and that the loss score of the validation test was 0.0115. Fig. 6 shows that the precision score of the validation test was 0.9972. Fig. 7 shows that the recall score of the validation set was 0.9972.

Figure 5: Accuracy and loss vs. no. of epochs

Figure 6: Precision vs. no. of epochs

Figure 7: Recall vs. no. of epochs
Table 4 shows the confusion matrices for the feature-fusion pre-trained deep learning model. The test dataset had three classes: Aculus olearius, peacock spot, and healthy. Table 5 shows the feature-fusion pre-trained model with 532 samples correctly predicted out of a total of 533 samples for the Aculus olearius class. The model predicted 975 of 977 samples correctly for the peacock spot class and 576 of 578 samples correctly for the healthy class.
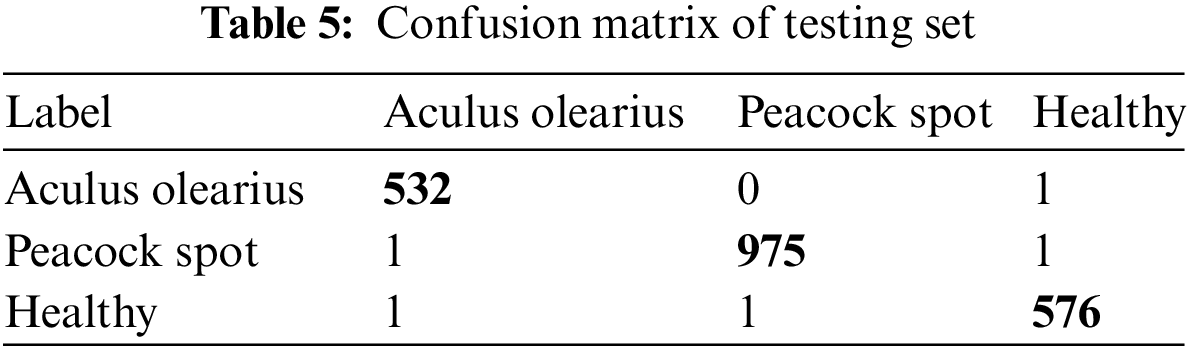
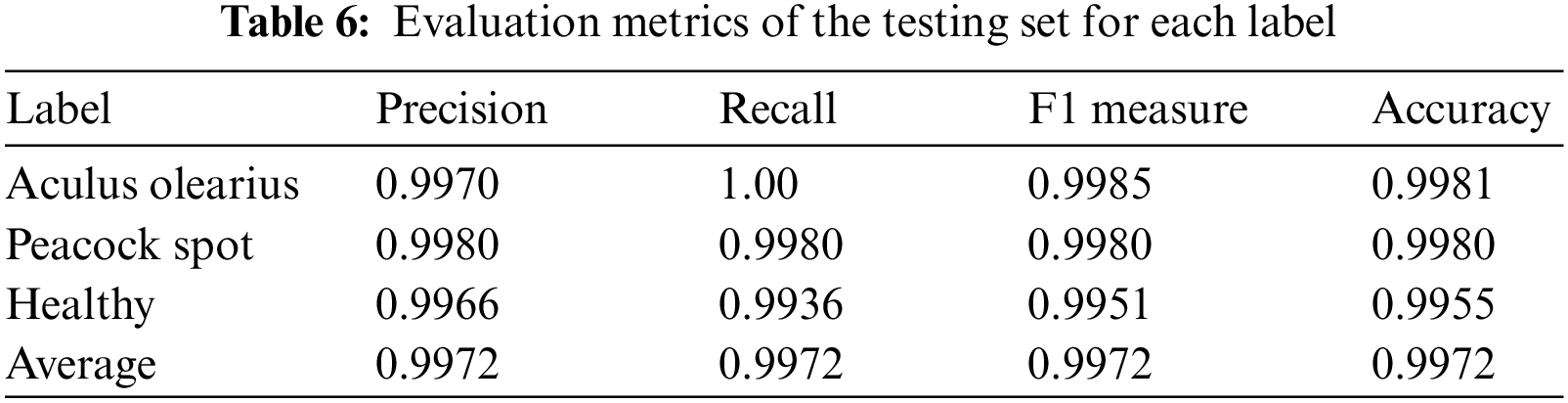
Table 5 shows the experiments’ results for the feature fusion pre-trained deep learning model. The model was applied to the collected olive leaf dataset for multiclassification. We distinguished between Aculus olearius, peacock spot, and healthy leaf images. Table 6 shows that the model had accuracy, precision, recall, and F1 measure averages of 99.72%, 99.72%, 99.72%, and 99.72%, respectively.
All of the experiments were conducted with a learning rate = 0.001, the various epoch values discussed before, a batch size = 32, and an image size = 224 × 224. The study’s dataset is available from [48]. There has been surprisingly little research on the olive plant that has been the focus of our research.
This study involved the classification of three distinct types of olive leaf diseases. The success ratio is anticipated to drop if the trials conducted on the olive leaf plant are expanded to include other illnesses that need to be detected, because diseases that affect the same type of plant may display similar symptoms. Because of the difficulties in achieving a high success rate, studies to be conducted in this field are regarded as valuable.
Our model faces three primary hurdles: complexity, overfitting, and trade-offs. Combining EfficientNet-B7, DenseNet-201, and ResNet-152-v2 leads to inherent complexity, potentially increasing computational cost, training time, and memory demands. While Kaggle’s GPU-P100 servers mitigated memory bottlenecks, addressing the remaining computational and time constraints remains an ongoing effort. Overfitting also poses a challenge, particularly with smaller datasets. To address this, we implemented data augmentation to artificially enrich the training data and combat overfitting. Finally, while ensembling models often improves accuracy, it can come at the cost of increased complexity, necessitating careful consideration of these trade-offs.
A novel deep-learning model for detecting olive leaf diseases from images using wavelet transforms and pre-trained models was developed in this study. The proposed approach leverages wavelet transforms to decompose olive leaf images into three frequency sub-bands, focusing on the third level for feature extraction. Subsequently, pre-trained deep learning models (EfficientNet-B7, DenseNet-201, and ResNet-152-v2) extract features from this sub-band, capturing information at different scales. These extracted features are then fused to create a comprehensive representation of the image, enabling the classification of olive leaf diseases into three categories: aculus olearius, peacock spot, and healthy. The experimental results demonstrate the effectiveness of the proposed model, which achieves a 99.72% testing accuracy score in disease detection. However, a limitation of this study is that the dataset involved the classification of three distinct types of olive leaf diseases. The proposed model’s performance was compared to other techniques and showed potential for enhancing other CNN architectures that warrant further investigation. Ultimately, this research aims to contribute to the development of efficient plant leaf disease detection methods, culminating in the ability to predict and select infected areas for targeted intervention.
Acknowledgement: The authors thank, recognize, and deeply appreciate the dedication of Kaggle data science website to fostering open data access, which enriched this research endeavor.
Funding Statement: No funding.
Author Contributions: Conceptualization, M.A.M., K.A.; Methodology, K.A.; Software, M.A.M.; Validation, K.A.; Resources, M.A.M., and K.A.; Data curation, M.A.M.; Formal analysis, M.A.M., and K.A.; Investigation, M.A.M.; Project administration, M.A.M.; Supervision, K.A.; Visualization, M.A.M; Writing—original draft, M.A.M., and K.A.; Writing—review & editing, M.A.M. and K.A. All authors have read and agreed to the published version of the manuscript.
Availability of Data and Materials: Not applicable.
Conflicts of Interest: The authors have no conflicts of interest to report regarding the present study.
References
1. L. Saadio et al., “Crops yield prediction based on machine learning models: Case of West African countries,” Smart Agri. Technol., vol. 2, pp. 100049, 2022. [Google Scholar]
2. R. Aworka et al., “Agricultural decision system based on advanced machine learning models for yield prediction: Case of East African countries,” Smart Agri. Technol., vol. 2, no. 11, pp. 100048, 2022. doi: 10.1016/j.atech.2022.100048. [Google Scholar] [CrossRef]
3. K. Gasmi, I. Ltaifa, G. Lejeune, H. Alshammari, L. Ammar and M. Mahmood, “Optimal deep neural network-based model for answering visual medical question,” Cybern. Syst., vol. 535, no. 5, pp. 403–424, 2022. doi: 10.1080/01969722.2021.2018543. [Google Scholar] [CrossRef]
4. O. Hrizi et al., “Tuberculosis disease diagnosis based on an optimized machine learning model,” J. Healthc. Eng., vol. 2022, no. 2, pp. 1–13, 2022. doi: 10.1155/2022/8950243. [Google Scholar] [PubMed] [CrossRef]
5. Q. Abu Al-Haija, M. Krichen, and W. Abu Elhaija, “Machine-learning-based darknet traffic detection system for IoT applications,” Electron, vol. 11, no. 556, pp. 556, 2022. doi: 10.3390/electronics11040556. [Google Scholar] [CrossRef]
6. A. Mihoub, O. Fredj, O. Cheikhrouhou, A. Derhab, and M. Krichen, “Denial of service attack detection and mitigation for Internet of Things using looking-back-enabled machine learning techniques,” Comput. Electr. Eng., vol. 98, no. 3, pp. 107716, 2022. doi: 10.1016/j.compeleceng.2022.107716. [Google Scholar] [CrossRef]
7. H. Alshammari et al., “Optimal deep learning model for olive disease diagnosis based on an adaptive genetic algorithm,” Wirel. Commun. Mob. Comput., vol. 2022, no. 4, pp. 1–13, 2022. doi: 10.1155/2022/8531213. [Google Scholar] [CrossRef]
8. M. Abdel Salam et al., “Climate recommender system for wheat cultivation in North Egyptian Sinai Peninsula,” in Proc. of the Fifth Int. Conf. on Innov. in Bio-Inspir. Comp. and Appl. IBICA 2014, 2014, pp. 121–130. [Google Scholar]
9. S. Srinivasan et al., “Deep convolutional neural network based image spam classification,” in 6th Conf. on Data Sci. Mach. Learn. Appli. (CDMA), Riyadh, Saudi Arabia, 2020, pp. 112–117. [Google Scholar]
10. M. Jasim and J. AL-Tuwaijari, “Plant leaf diseases detection and classification using image processing and deep learning techniques,” in 2020 Int. Conf. Comput. Sci. Softw. Eng. (CSASE), Duhok, Iraq, 2020, pp. 259–265. [Google Scholar]
11. K. Ferentinos, “Deep learning models for plant disease detection and diagnosis,” Comput. Electron. Agric., vol. 145, no. 6, pp. 311–318, 2018. doi: 10.1016/j.compag.2018.01.009. [Google Scholar] [CrossRef]
12. Y. Zhong and M. Zhao, “Research on deep learning in apple leaf disease recognition,” Comput. Electron. Agric., vol. 168, no. 2, pp. 105146, 2020. doi: 10.1016/j.compag.2019.105146. [Google Scholar] [CrossRef]
13. U. Mokhtar et al., “SVM-based detection of tomato leaves diseases,” Adv. Intell. Syst. Comput., vol. 323, pp. 641–652, 2014. [Google Scholar]
14. M. Alruwaili, S. Abd El-Ghany, and A. Shehab, “An enhanced plant disease classifier model based on deep learning techniques,” Int. J. Adv. Technol. Eng. Explor., vol. 19, no. 1, pp. 7159–7164, 2019. [Google Scholar]
15. S. Mohanty, D. Hughes, and M. Salath, “Using deep learning for image-based plant disease detection,” Front. Plant Sci., vol. 7, no. 1419, pp. 1–10, 2016. [Google Scholar]
16. A. Fazari, J. Oscar, V. Pellicer, and J. Gomez, “Application of deep convolutional neural networks for the detection of anthracnose in olives using VIS/NIR hyperspectral images,” Comput. Electron. Agric., vol. 187, no. 1, pp. 106252, 2021. doi: 10.1016/j.compag.2021.106252. [Google Scholar] [CrossRef]
17. A. Malik, G. Vaidya, and V. Jagota, “Design and evaluation of a hybrid technique for detecting sunflower leaf disease using deep learning approach,” J. Food Qual., vol. 2022, pp. 1–12, 2022. doi: 10.1155/2022/9211700. [Google Scholar] [CrossRef]
18. A. Sinha and R. Singh, “Olive spot disease detection and classification using analysis of leaf image textures,” Procedia Comput. Sci., vol. 167, pp. 2328–2336, 2020. doi: 10.1016/j.procs.2020.03.285. [Google Scholar] [CrossRef]
19. Y. Nanehkaran, D. Zhang, J. Chen, T. Yuan, and N. Al-Nabhan, “Recognition of plant leaf diseases based on computer vision,” J. Ambient Intell. Humaniz. Comput., vol. 10, no. 9, pp. 1–18, 2020. doi: 10.1007/s12652-020-02505-x. [Google Scholar] [CrossRef]
20. S. Kaur, S. Pandey, and S. Goel, “Plants disease identification and classification through leaf images: A survey,” Arch. Comput. Methods Eng., vol. 26, no. 2, pp. 507–530, 2019. doi: 10.1007/s11831-018-9255-6. [Google Scholar] [CrossRef]
21. M. Ji, L. Zhang, and Q. Wu, “Automatic grape leaf diseases identification via united model based on multiple convolutional neural networks,” Inf. Process. Agric., vol. 7, no. 3, pp. 418–426, 2020. doi: 10.1016/j.inpa.2019.10.003. [Google Scholar] [CrossRef]
22. S. Guz and N. Uysal, “Classification of olive leaf diseases using deep convolutional neural networks,” Neural. Comput. Appl., vol. 33, no. 9, pp. 4133–4149, 2021. doi: 10.1007/s00521-020-05235-5. [Google Scholar] [CrossRef]
23. L. Li, S. Zhang, and B. Wang, “Plant disease detection and classification by deep learning—A review,” IEEE Access, vol. 9, pp. 56683–56698, 2021. doi: 10.1109/ACCESS.2021.3069646. [Google Scholar] [CrossRef]
24. M. Saberi, “A hybrid model for leaf diseases classification based on the modified deep transfer learning and ensemble approach for agricultural AIoT-based monitoring,” Comput. Intell. Neurosci., Hindawi, vol. 2022, pp. 1–15, 2022. [Google Scholar]
25. H. Alshammari, K. Gasmi, I. Ltaifa, M. Krichen, L. Ammar and M. Mahmood, “Olive disease classification based on vision transformer and CNN models,” Comput. Intell. Neurosci., vol. 2022, no. 9, pp. 1–10, 2022. doi: 10.1155/2022/3998193. [Google Scholar] [PubMed] [CrossRef]
26. H. H. Alshammari, A. I. Taloba, and O. R. Shahin, “Identification of olive leaf disease through optimized deep learning approach,” Alex. Eng. J., vol. 72, no. 2, pp. 213–224, 2023. doi: 10.1016/j.aej.2023.03.081. [Google Scholar] [CrossRef]
27. B. Karsh, R. H. Laskar, and R. K. Karsh, “mIV3Net: Modified Inception V3 network for hand gesture recognition,” Multimed. Tools Appl., vol. 83, no. 4, pp. 1–27, 2023. doi: 10.1007/s11042-023-15865-1. [Google Scholar] [CrossRef]
28. A. Ksibi, M. Ayadi, B. O. Soufiene, M. M. Jamjoom, and Z. Ullah, “MobiRes-Net: A hybrid deep learning model for detecting and classifying olive leaf diseases,” Appl. Sci., vol. 12, no. 20, pp. 10278, 2022. doi: 10.3390/app122010278. [Google Scholar] [CrossRef]
29. A. C. Cruz, A. Luvisi, L. de Bellis, and Y. Ampatzidis, “An effective application for detecting olive quick decline syndrome with deep learning and data fusion,” Front. Plant Sci., vol. 8, pp. 1741, 2017. doi: 10.3389/fpls.2017.01741. [Google Scholar] [PubMed] [CrossRef]
30. G. Geetharamani and J. A. Pandian, “Identification of plant leaf diseases using a nine-layer deep convolutional neural network,” Comput. Electr. Eng., vol. 76, no. 3, pp. 323–338, 2019. doi: 10.1016/j.compeleceng.2019.04.011. [Google Scholar] [CrossRef]
31. A. Fazari et al., “Application of deep convolutional neural networks for the detection of anthracnose in olives using VIS/NIR hyperspectral images,” Comput. Electron. Agric., vol. 187,1, no. 1, pp. 106252, 2021. doi: 10.1016/j.compag.2021.106252. [Google Scholar] [CrossRef]
32. H. Alshammari et al., “Optimal deep learning model for olive disease diagnosis based on an adaptive genetic algorithm,” Wirel. Commun. Mob. Comput., vol. 8531213, no. 4, pp. 1–13, 2022. doi: 10.1155/2022/8531213. [Google Scholar] [CrossRef]
33. S. Uguz and N. Uysal, “Classification of olive leaf diseases using deep convolutional neural networks,” Neural. Comput. Appl., vol. 33, no. 9, pp. 4133–4149, 2021. doi: 10.1007/s00521-020-05235-5. [Google Scholar] [CrossRef]
34. A. Sinha and R. S. Shekhawat, “Olive spot disease detection and classification using analysis of leaf image textures,” Procedia Comput. Sci., vol. 167, pp. 2328–2336, 2020. [Google Scholar]
35. K. P. Ferentinos, “Deep learning models for plant disease detection and diagnosis,” Comput. Electron. Agric., vol. 145, no. 6, pp. 311–318, 2018. doi: 10.1016/j.compag.2018.01.009. [Google Scholar] [CrossRef]
36. J. M. Ponce, A. Aquino, and J. M. Andújar, “Olive-fruit variety classification by means of image processing and convolutional neural networks,” IEEE Access, vol. 7, pp. 147629–147641, 2019. doi: 10.1109/ACCESS.2019.2947160. [Google Scholar] [CrossRef]
37. M. Iman, H. R. Arabnia, and K. Rasheed, “A review of deep transfer learning and recent advancements,” Technologies, vol. 11, no. 2, pp. 40, 2023. doi: 10.3390/technologies11020040. [Google Scholar] [CrossRef]
38. X. Hao, L. Liu, R. Yang, L. Yin, L. Zhang and X. Li, “A Review of data augmentation methods of remote sensing image target recognition,” Remote Sens., vol. 15, no. 3, pp. 827, 2023. doi: 10.3390/rs15030827. [Google Scholar] [CrossRef]
39. C. Szegedy et al., “Going deeper with convolutions,” in 2015 IEEE Conf. Comput. Vis. Pattern Recognit. (CVPR), Boston, MA, USA, 2015, pp. 1–9. [Google Scholar]
40. A. Kumar, P. Chauda, and A. Devrari, “Machine learning approach for brain tumor detection and segmentation,” Int. J. Organ. Collect. Intell. (IJOCI), vol. 11, no. 3, pp. 68–84, 2021. doi: 10.4018/IJOCI. [Google Scholar] [CrossRef]
41. A. Hooda, A. Kumar, M. S. Goyat, and R. Gupta, “Estimation of surface roughness for transparent superhydrophobic coating through image processing and machine learning,” Mol. Cryst. Liq. Cryst., vol. 726, no. 1, pp. 90–104, 2021. doi: 10.1080/15421406.2021.1935162. [Google Scholar] [CrossRef]
42. A. Kumar, P. Rastogi, and P. Srivastava, “Design and FPGA implementation of DWT, image text extraction technique,” Procedia Comput. Sci., vol. 57, pp. 1015–1025, 2015. doi: 10.1016/j.procs.2015.07.512. [Google Scholar] [CrossRef]
43. A. Kumar, P. Ahuja, and R. Seth, “Text extraction and recognition from an image using image processing in Matlab,” in Conf. Adv. Commun. and Control Syst. 2013 (CAC2S 2013), 2013, pp. 429–435. [Google Scholar]
44. G. Huang, Z. Liu, L. Maaten and Q. Kilian, “Densely connected convolutional networks,” in Proc. IEEE Conf. on Comp. Vision and Pattern Recognit. (CVPR), 2017, pp. 4700–4708. [Google Scholar]
45. J. Schmeelk, “Wavelet transforms on two-dimensional images,” Math. Comput. Model., vol. 36, no. 7–8, pp. 939–948, 2002. doi: 10.1016/S0895-7177(02)00238-8. [Google Scholar] [CrossRef]
46. H. Dziri, M. A. Cherni, F. Abidi, and A. Zidi, “Fine-tuning ResNet-152V2 for COVID-19 recognition in chest computed tomography images,” in 2022 IEEE Infor. Tech. & Smart Indust. Sys. (ITSIS), Paris, France, 2022, pp. 1–4. [Google Scholar]
47. M. Tan, and Q. V. Le, “EfficientNet: Rethinking model scaling for convolutional neural networks,” in Proc. 36th Int. Conf. Mach. Learn., Long Beach, California, PMLR 97, 2019. [Google Scholar]
48. Kaggle dataset, 2021. https://www.kaggle.com/code/majedahalrwaily/olive-leaf-disease-class/input?select=Zeytin_224x224_Augmented (accessed on 01/11/2023). [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF
 Downloads
Downloads
 Citation Tools
Citation Tools