 Open Access
Open Access
ARTICLE
RoBGP: A Chinese Nested Biomedical Named Entity Recognition Model Based on RoBERTa and Global Pointer
1 School of Information Science and Technology, Beijing Forestry University, Beijing, 100083, China
2 Engineering Research Center for Forestry-Oriented Intelligent Information Processing of National Forestry and Grassland Administration, Beijing, 100083, China
3 Division of Biostatistics, Department of Population Health, Grossman School of Medicine, New York University, New York, 10016, USA
* Corresponding Author: Dongmei Li. Email:
# These authors contribute equally to this work
(This article belongs to the Special Issue: Transfroming from Data to Knowledge and Applications in Intelligent Systems)
Computers, Materials & Continua 2024, 78(3), 3603-3618. https://doi.org/10.32604/cmc.2024.047321
Received 01 November 2023; Accepted 12 January 2024; Issue published 26 March 2024
Abstract
Named Entity Recognition (NER) stands as a fundamental task within the field of biomedical text mining, aiming to extract specific types of entities such as genes, proteins, and diseases from complex biomedical texts and categorize them into predefined entity types. This process can provide basic support for the automatic construction of knowledge bases. In contrast to general texts, biomedical texts frequently contain numerous nested entities and local dependencies among these entities, presenting significant challenges to prevailing NER models. To address these issues, we propose a novel Chinese nested biomedical NER model based on RoBERTa and Global Pointer (RoBGP). Our model initially utilizes the RoBERTa-wwm-ext-large pretrained language model to dynamically generate word-level initial vectors. It then incorporates a Bidirectional Long Short-Term Memory network for capturing bidirectional semantic information, effectively addressing the issue of long-distance dependencies. Furthermore, the Global Pointer model is employed to comprehensively recognize all nested entities in the text. We conduct extensive experiments on the Chinese medical dataset CMeEE and the results demonstrate the superior performance of RoBGP over several baseline models. This research confirms the effectiveness of RoBGP in Chinese biomedical NER, providing reliable technical support for biomedical information extraction and knowledge base construction.Keywords
The rapid advancement in biotechnology, medical devices, and clinical practices has led to an exponential increase in biomedical text data. These data encompass a wide range of information, including genomics, proteomics, clinical medical records, and key aspects such as disease mechanisms, drug development, and medical decision-making. Extracting valuable knowledge from this extensive biomedical text data necessitates the construction and effective management of knowledge bases. Knowledge bases serve as pivotal platforms for aggregating and integrating information from multiple sources, aiding researchers in understanding disease molecular mechanisms, drug mechanisms of action, and the implementation of personalized medicine. In knowledge base construction, tailored knowledge extraction methods are essential for processing structurally varied raw data, typically involving entity extraction and relation extraction. Named Entity Recognition (NER) is an indispensable component of knowledge extraction, used to extract and classify text-based information with special significance [1]. In the biomedical field, accurate entity recognition is essential for constructing precise knowledge bases and propelling cutting-edge research. Therefore, NER holds a significant position in the processing, analyzing, and comprehension of biomedical texts.
Currently, research in NER has covered a wide range of fields, yielding numerous markable achievements [2,3]. Compared to general fields, biomedical NER tends to be more intricate. Specifically, biomedical entities often vary greatly in length and frequently lack distinct boundaries. Furthermore, unlike typical flat entities, biomedical entities commonly feature nested structures. As shown in Fig. 1, “ ” (sympathetic nervous system involvement) in the first half of the sentence is classified as a clinical symptom entity, encompassing the nested “
” (sympathetic nervous system involvement) in the first half of the sentence is classified as a clinical symptom entity, encompassing the nested “ ” (sympathetic nervous system), recognized as a body entity. Similarly, in the second part of the sentence, “
” (sympathetic nervous system), recognized as a body entity. Similarly, in the second part of the sentence, “ ” (increased saliva secretion and sweating) is labeled as a clinical symptom entity, while the nested “
” (increased saliva secretion and sweating) is labeled as a clinical symptom entity, while the nested “ ” (saliva) and “
” (saliva) and “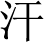 ” (sweat) are considered body entities. These intricacies present considerable challenges in biomedical NER.
” (sweat) are considered body entities. These intricacies present considerable challenges in biomedical NER.

Figure 1: An example of nested entities. The “sym” and “bod” represent recognized clinical symptom entity and body entity respectively
To cope with these challenges, deep learning techniques have become increasingly prevalent in the development and application of biomedical NER in recent years. Conventional methods treat biomedical NER as a sequence labeling task, assigning a specific label to each word or character. As a representative, Bidirectional Long Short-Term Memory with Conditional Random Fields (BiLSTM-CRF) [4] stands out for its straightforward structure and superior performance, inspiring many subsequent studies [5,6] to adopt its architecture. Advancements in research have led to the integration of advanced pretrained language models to further enhance the performance of NER. Notably, the integration with Bidirectional Encoder Representation from Transformer (BERT) [7] has become widespread. The masking mechanism of BERT enables the model to exhibit excellent semantic information extraction ability in English NER. However, when dealing with Chinese text, treating each character as a token neglects the boundary information of Chinese words, thus affecting the effectiveness of Chinese NER. Although sequence labeling methods achieve decent performance on flat NER, they falter with nested NER. An effective method is to treat nested NER as a span selection task, classifying all possible spans into predefined types in the sentence. For example, Yu et al. [8] used BERT to encode the input sequence and then adopt a biaffine attention model to assign scores to all potential spans, achieving the State-Of-The-Art (SOTA) performance on both flat and nested English NER datasets. Shen et al. [9] divided Chinese NER into a two-stage task, where the first stage aims to locate entities, and the second stage classifies entities after boundary adjustments. This method improves the model’s recognition performance for entities with longer distances through the boundary regression task. However, despite the success of these methods, they only address individual issues in Chinese biomedical NER and lack a holistic solution. To bridge these gaps, this paper focuses on addressing the three major challenges in Chinese biomedical NER during the method design phase. We fully recognize the inapplicability of conventional masking strategies in Chinese vocabulary, as well as the limitations of existing models in handling long-distance semantic information and nested entities. Consequently, we comprehensively consider these issues and aim to tackle them through the collaborative work of various modules.
Specifically, we propose a Chinese nested biomedical NER model based on RoBERTa and Global Pointer (RoBGP). RoBGP utilizes the RoBERTa-wwm-ext-large pretrained language model for obtaining word-level vectors and enhances the extraction of long-distance semantic information through the Bidirectional Long Short-Term Memory (BiLSTM) network. Simultaneously, the Global Pointer model is employed to recognize nested biomedical entities. In this paper, our main contributions are summarized as follows:
1. We propose a novel NER model called RoBGP tailored for Chinese biomedical texts. To adapt to the characteristics of the Chinese language, the model employs the RoBERTa-wwm-ext-large pretrained language model, which is based on the Chinese whole word masking (wwm) strategy to obtain more accurate word-level initial vectors. Additionally, we incorporate BiLSTM to capture contextual semantic information and enhance the model’s ability to locate long-distance entity boundaries.
2. Unlike previous sequence labeling models, we employ Global Pointer as the entity recognition module in our proposed model, effectively addressing both nested and non-nested entity recognition challenges in a unified manner.
3. We conduct extensive experiments on the publicly available Chinese medical dataset CMeEE. The experimental results validate the effectiveness of RoBGP and the importance of different modules within the model.
Section 2 of this paper provides a brief overview of related work. Section 3 elaborates on the model architecture. Section 4 describes the experimental results and provides analyses. Section 5 further discusses the results. Finally, the paper concludes with a concise summary of the entire study.
2.1 Classic Methods for Biomedical NER
Classic methods for biomedical NER can be primarily categorized into rule-based and dictionary-based methods, machine learning-based methods, and deep learning-based methods. Rule-based and dictionary-based methods were common in early NER research. For instance, Krauthammer et al. [10] proposed using dictionaries to recognize gene and protein entities. While practical, these methods faced limitations due to the continual emergence of new biomedical named entities, resulting in reduced effectiveness as dictionaries become less comprehensive. Machine learning-based methods mainly include Hidden Markov Model (HMM) [11], Maximum Entropy (ME) [12], and Conditional Random Fields (CRF) [13], which require rigorous feature selection and use features such as prefixes, suffixes, capitalization, special characters, and word stems for training. They offer improved accuracy over rule-based and dictionary-based methods, but their excessive dependency on feature selection and a singular recognition strategy limits their efficacy. With the advancement of deep learning, applying neural networks to NER has become a trend. Huang et al. [4] proposed the BiLSTM-CRF model, utilizing BiLSTM to capture contextual features and modifying the BiLSTM layer’s output through CRF by learning transition probabilities between different labels in the dataset. Ma et al. [6] employed a Convolutional Neural Networks (CNN) to extract character-level features and then used the BiLSTM-CRF model to further extract contextual features and output results. Zhang et al. [14] proposed the Lattice-LSTM model, which encoded a sequence of input characters as well as all potential words that match a lexicon. Compared with character-based methods, their model explicitly utilized word and word sequence information. Due to simplicity and effectiveness, deep learning-based methods serve as the mainstream for biomedical NER.
2.2 Pretrained Language Models in Biomedical NER
In recent years, word embedding technology has been widely applied to natural language processing tasks. Traditional word embedding models such as Word2Vec [15] and Glove [16] used static word vectors for semantic representation. However, the meaning of a word can vary in different contexts, leading to the issue of polysemy. For example, the character “ ” (clear) has completely different meanings in the two sentences of “
” (clear) has completely different meanings in the two sentences of “ ” (the patient is conscious and in good spirits) and “
” (the patient is conscious and in good spirits) and “ ” (lymph node dissection in our hospital). ELMo [17] addressed this issue to some extent but could not leverage both forward and backward context information simultaneously, which had certain limitations. BERT [7] effectively made up for the shortcomings of ELMo. For biomedical NER, we only need to set the downstream task interface and use the relevant data to fine-tune the model to obtain a more accurate embedded representation of each word in the biomedical texts. A common approach is to add a CRF layer atop the BERT output, forming the BERT-CRF model [18]. This model can use the robust semantic representations provided by BERT along with the label dependencies offered by CRF and has been proven effective on a variety of NER tasks. Zhang et al. [19] pretrained BERT on a Chinese clinical text corpus and used the resulting embeddings as input features for BiLSTM-CRF to solve the breast cancer NER problem, achieving an F1 score of 93.53%. Subsequently, Liu et al. [20] proposed an optimized model called RoBERTa, which surpassed BERT in terms of training data, batch size, and model parameters. Based on RoBERTa, Wu et al. [21] proposed a model for biomedical NER, achieving F1 scores of 93.26% and 82.87% on the CCKS2017 and CCKS2019 datasets, respectively. RoBERTa-wwm-ext-large [22] was a variant of RoBERTa that employed a Chinese whole word masking strategy when processing Chinese text. This strategy enabled the model to acquire precise word-level vectors, making it better suitable for the Chinese NER task.
” (lymph node dissection in our hospital). ELMo [17] addressed this issue to some extent but could not leverage both forward and backward context information simultaneously, which had certain limitations. BERT [7] effectively made up for the shortcomings of ELMo. For biomedical NER, we only need to set the downstream task interface and use the relevant data to fine-tune the model to obtain a more accurate embedded representation of each word in the biomedical texts. A common approach is to add a CRF layer atop the BERT output, forming the BERT-CRF model [18]. This model can use the robust semantic representations provided by BERT along with the label dependencies offered by CRF and has been proven effective on a variety of NER tasks. Zhang et al. [19] pretrained BERT on a Chinese clinical text corpus and used the resulting embeddings as input features for BiLSTM-CRF to solve the breast cancer NER problem, achieving an F1 score of 93.53%. Subsequently, Liu et al. [20] proposed an optimized model called RoBERTa, which surpassed BERT in terms of training data, batch size, and model parameters. Based on RoBERTa, Wu et al. [21] proposed a model for biomedical NER, achieving F1 scores of 93.26% and 82.87% on the CCKS2017 and CCKS2019 datasets, respectively. RoBERTa-wwm-ext-large [22] was a variant of RoBERTa that employed a Chinese whole word masking strategy when processing Chinese text. This strategy enabled the model to acquire precise word-level vectors, making it better suitable for the Chinese NER task.
2.3 Span-Based Methods in Biomedical NER
Most of the aforementioned methods are primarily based on sequence labeling and cannot directly address the issue of nested entities in biomedical NER. Earlier, a mix of rule-based and machine learning-based methods was often applied to address nesting issues, but this approach had difficulties with same-type nested entities and scaling to large datasets with long sentences. In recent years, span-based methods have risen in popularity due to their superior performance. For instance, Yu et al. [8] used ideas from graph-based dependency parsing to provide their model with a global view of the input via a biaffine model. The biaffine model scored pairs of start and end tokens in a sentence which they used to explore all spans so that the model was able to predict named entities accurately. They showed that the model worked well for both nested and flat NER through the evaluation of 8 corpora. Gu et al. [18] proposed a novel method for investigating the regularity of entity spans in Chinese NER, dubbed as Regularity-Inspired reCOgnition Network (RICON). Specifically, the proposed model consisted of two branches: a regularity-aware module and a regularity-agnostic module. The regularity-aware module captured the internal regularity of each span for better entity type prediction, while the regularity-agnostic module was employed to locate the boundary of entities and relieve the excessive attention to span regularity. An orthogonality space was further constructed to encourage two modules to extract different aspects of regularity features. Cong et al. [23] proposed a Chinese medical nested named entity recognition model based on feature fusion and a bidirectional lattice embedding graph. They introduced a medical lexicon and pinyin information to enhance the features that the model could capture for Chinese medical NER. They also considered the similarity between different entity types to improve the model’s effectiveness. Liu et al. [24] treated biomedical NER as a question-answering task, where the question is the entity type and the answer is the entity span. Our proposed method also belongs to span-based methods, but in contrast to previous research, our model predicts each character as the start or end of each span without enumerating each span, which is an efficient practice.
The architecture of RoBGP is shown in Fig. 2, which consists of three principal modules. First, each character in the biomedical text is processed to obtain its initial vector via the representation module. Subsequently, the representation module’s output feeds into the encoder module, generating enhanced feature vectors that incorporate long-distance dependencies. Finally, the recognition module is used to output the types and positions of corresponding entities.
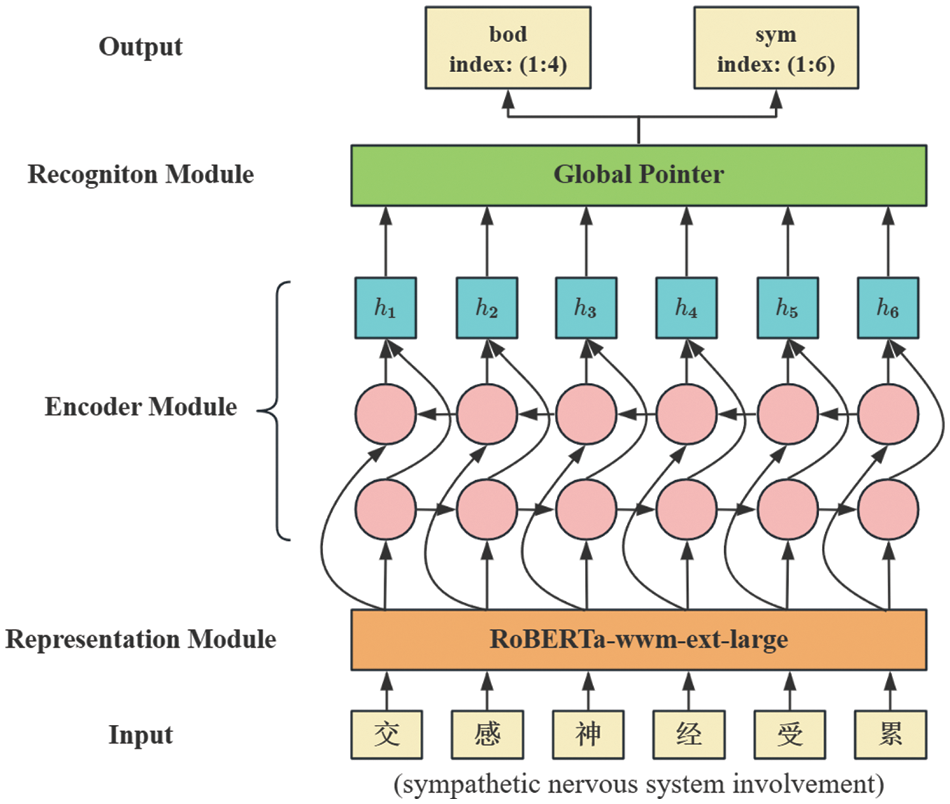
Figure 2: The architecture of RoBGP
3.1 Representation Module: RoBERTa-wwm-ext-large
Considering the unique characteristics of the Chinese language structure, we adopt RoBERTa-wwm-ext-large to obtain word-level input vectors. RoBERTa-wwm-ext-large surpasses BERT in training scope, as it is developed on a larger scale corpus and undergoes additional training steps, enhancing its ability to assimilate contextual semantics from multiple perspectives. In contrast to BERT’s character-level tokenization, RoBERTa-wwm-ext-large employs a Chinese whole word masking strategy during the pretraining phase. Specifically, the input sequence is first segmented into words, and then all Chinese characters within the same word are masked and predicted simultaneously. This strategy helps the model learn word-level semantic information. The masking strategies of BERT and RoBERTa-wwm-ext-large are shown in Table 1.

For the biomedical text shown in Table 1, the characters “ ” (saliva) and “
” (saliva) and “ ” (liquid) form a word that often appears together. However, BERT treats the character “
” (liquid) form a word that often appears together. However, BERT treats the character “ ” (liquid) as an independent unit for masking, disrupting the integral contextual structural information of the entire word. Consequently, BERT is restricted to learning semantic representation solely at the character level. In contrast, RoBERTa-wwm-ext-large considers “
” (liquid) as an independent unit for masking, disrupting the integral contextual structural information of the entire word. Consequently, BERT is restricted to learning semantic representation solely at the character level. In contrast, RoBERTa-wwm-ext-large considers “ ” (saliva) as a single unit and masks both characters simultaneously. This method allows it to capture word-level contextual semantic information, which is beneficial for the recognition of Chinese biomedical entities. Additionally, RoBERTa-wwm-ext-large employs the dynamic masking mechanism, enabling it to learn more comprehensive feature representations, thus further enhancing the model’s recognition performance.
” (saliva) as a single unit and masks both characters simultaneously. This method allows it to capture word-level contextual semantic information, which is beneficial for the recognition of Chinese biomedical entities. Additionally, RoBERTa-wwm-ext-large employs the dynamic masking mechanism, enabling it to learn more comprehensive feature representations, thus further enhancing the model’s recognition performance.
Chinese biomedical texts often feature complex syntax and lengthy named entities. To address this, we employ BiLSTM [25] for encoding the text sequences. It aids in extracting long-distance contextual semantic information and enhancing the model’s ability to locate entities at distant positions. BiLSTM comprises a forward Long Short-Term Memory (LSTM) [26] and a backward LSTM, both connected to the same output layer. LSTM is an improvement upon Recurrent Neural Networks (RNN), designing a gate mechanism to regulate information flow. It achieves selective memory through the forget gate, the input gate, and the output gate, filtering out non-essential information while preserving essential details. Additionally, it effectively solves the issue of gradient vanishing. However, unidirectional LSTM only considers past information, neglecting future information. To simultaneously capture bidirectional contextual information, we input the Chinese biomedical text sequences in both the forward and backward directions into two LSTMs for feature extraction. By utilizing the BiLSTM composed of these two LSTMs, we obtain two independent hidden states,
where
3.3 Recognition Module: Global Pointer
Pointer Networks [27] uses two independent modules to recognize the start and end positions of entities, resulting in inconsistencies between the training and prediction phases. We employ the Global Pointer (GP) model [28] as the recognition module to address this issue. Following a global normalization approach, this model treats entity boundaries as a unified whole for discrimination. For any given sentence, GP constructs one or more upper triangular matrices to traverse all valid entity spans. The number of matrices is consistent with the number of entity types, where each matrix corresponds to a specific entity type, and each cell corresponds to an entity span. As shown in Fig. 3, for the input sentence “ ” (may cause increased saliva secretion and sweating), GP constructs two upper triangular matrices. The first matrix is used to recognize body entities, while the second matrix is used to recognize clinical symptom entities.
” (may cause increased saliva secretion and sweating), GP constructs two upper triangular matrices. The first matrix is used to recognize body entities, while the second matrix is used to recognize clinical symptom entities.

Figure 3: A demo of global pointer
For positions i and j, after encoding through BiLSTM, the corresponding query vector
where
In order to further enhance the model’s ability to capture entity length and span information, we explicitly introduce relative positional encoding information in the scoring function, as shown in Eq. (5):
where
Given the fact that the biomedical field usually involves a large number of entity types, while the number of target types is relatively small, the loss function for such NER problem is defined as shown in Eq. (6):
For the NER task, it is only necessary to consider combinations where i ≤ j.
where
We conduct experiments on the publicly available Chinese medical dataset CMeEE1 [29], which is one of the largest and most comprehensive datasets for the Chinese biomedical NER task and is widely used as a benchmark for evaluating the performance of various NER models on Chinese biomedical texts. The dataset consists of 15,000 samples for training, 5,000 samples for validation, and 3,000 samples for testing. The annotated data encompasses a total of 2.2 million characters, comprising 47,194 sentences and 938 files. On average, each file contains 2,355 characters. There are 9 distinct medical entity types in the dataset, labeled as disease (dis), clinical symptom (sym), drug (dru), medical equipment (equ), medical procedure (pro), body (bod), medical laboratory test item (ite), microbiology (mic) and department (dep). Table 2 shows the labeling scheme and examples of some entities.

We use precision (P), recall (R), and F1 score to evaluate the model’s recognition performance, corresponding to Eqs. (10)–(12):
where
Before conducting the experiments, we set the maximum sentence length to 256 based on the statistics of sentence lengths in the dataset. In the pre-processing phase, we utilize a 24-layer RoBERTa-wwm-ext-large model with 16 attention heads, ultimately obtaining 1024 dimensional vectors from the dataset. To prevent overfitting in the BiLSTM layer, we apply dropout with a dropout rate of 0.5 and set the BiLSTM hidden layer size to 1024. In model training, the number of epoch is set to 10 and the batch size is set to 64. Furthermore, we train the model by the Adam optimizer [30] with an initial learning rate of 0.00003. The hyperparameters are shown in Table 3.

To validate the performance of RoBGP, we compare it with the following seven models:
1. BiLSTM-CRF [4]: A method using a BiLSTM network for capturing the contextual information of the input sequence and a CRF layer for solving sequence labeling.
2. Lattice-LSTM [14]: A variant of BiLSTM that incorporates word information into character-level inputs by constructing a word-character lattice. This model can handle the word segmentation problem and the ambiguity of Chinese characters.
3. BERT-CRF [18]: A combination of BERT and CRF. This model can utilize the rich semantic representations from BERT and the label dependencies from CRF.
4. BERT-Biaffine [8]: A model that uses BERT to encode the input sequence and a biaffine network to predict the spans and types of entities. This model can handle nested entities and exploit the interactions between entity head and tail tokens.
5. FFBLEG [23]: A feature fusion and bidirectional lattice embedding graph model that integrates character embeddings, word embeddings, and pinyin embeddings. This model can capture the polysemy of Chinese characters and the similarity between different entity types.
6. RICON [18]: A regularity-inspired recognition network consisting of two branches that use a regularity-aware module to learn the internal regularity of each span, and a regularity-independent module to localize entity spans, avoiding excessive focus on the regularity of the span.
7. FLR-MRC [24]: A machine reading comprehension framework that fuses label relations, which implicitly models the relations between different label types through graph attention networks and integrates label information with text.
Except for (1) and (2), the rest of the methods all use pretrained language models. (5), (6), and (7) are all recent SOTA models.
The performance of various models on the CMeEE dataset is displayed in Table 4. The results demonstrate that RoBGP outperforms baseline models, achieving an F1 score improvement ranging from 1.17% to 12.19%, validating the outstanding performance of RoBGP in the biomedical NER task. Upon a detailed analysis of the reasons behind this performance enhancement, we attribute it to the fact that our proposed model is more adept at recognizing entities with long-distance dependencies. Additionally, RoBGP exhibits excellent performance in handling nested entities, providing robust support for accurate localization of entity boundaries in complex contexts.

4.5.2 Comparison of Different Pretrained Language Models
To ascertain the performance of RoBERTa-wwm-ext-large [22] in RoBGP, we conduct a series of experiments using various mainstream pretrained language models, including ELMo [17], XLNet [31], BERT [7], RoBERT [20], and BERT-wwm. Notably, BERT-wwm is an enhancement of the original BERT model, incorporating a whole word masking strategy. Simultaneously, we maintain consistency in the downstream models with RoBGP, utilizing the BiLSTM-GP structure. This involves semantic encoding through BiLSTM followed by entity recognition via GP. The experimental results are shown in Table 5.

It can be observed that RoBERTa-wwm-ext-large outperforms other models in F1 score performance. In a detailed analysis, when compared to ELMo, the F1 scores of BERT, XLNet, RoBERTa, and RoBERTa-wwm-ext-large models improved by 1.03%, 2.96%, 4.35%, and 5.11%, respectively. This improvement highlights the advantage of the Transformer module in encoding, particularly in capturing contextual semantic dependencies for high-quality vector representations. In comparison to BERT-wwm, RoBERTa-wwm-ext-large’s F1 score improved by 3.24%, reflecting the beneficial effects of the dynamic masking mechanism, larger batch size, and expanded training corpora on entity recognition. In contrast to RoBERTa, RoBERTa-wwm-ext-large’s F1 score improved by 0.76%, indicating that the word-level vectors obtained through the Chinese whole word masking strategy compensate for the deficiencies of character-level vectors, making it more suitable for the Chinese NER task.
4.5.3 Comparison of Different Recognition Modules
To validate the effectiveness of the GP model, we conduct comparisons with Softmax and CRF as entity recognition modules based on RoBGP, and the experimental results are shown in Fig. 4. It can be seen that in terms of precision, recall and F1 score, CRF outperforms Softmax by 6.32%, 5.75%, and 6.1%, respectively. This indicates that during the label decoding stage, compared to Softmax, which treats each label as independent for prediction, CRF explicitly considers contextual correlations and constraints among labels, which is more conducive to improving model accuracy. In Contrast to the CRF model, the GP model achieves enhancements of 2.81%, 1.28%, and 2.13% in precision, recall, and F1 score, respectively. This demonstrates the effectiveness of GP in addressing the issue of nested entities, making it particularly suitable for the nested NER task.

Figure 4: Experimental results of different recognition modules
To further validate the effect of each module in RoBGP on model performance, we conduct ablation experiments. In each experiment, we maintained consistency in certain hyperparameters. Subsequently, we progressively eliminate one of RoBERTa-wwm-ext-large, BiLSTM, and GP. Table 6 shows the specific experimental results. Here, we add explicit names for these ablation models:
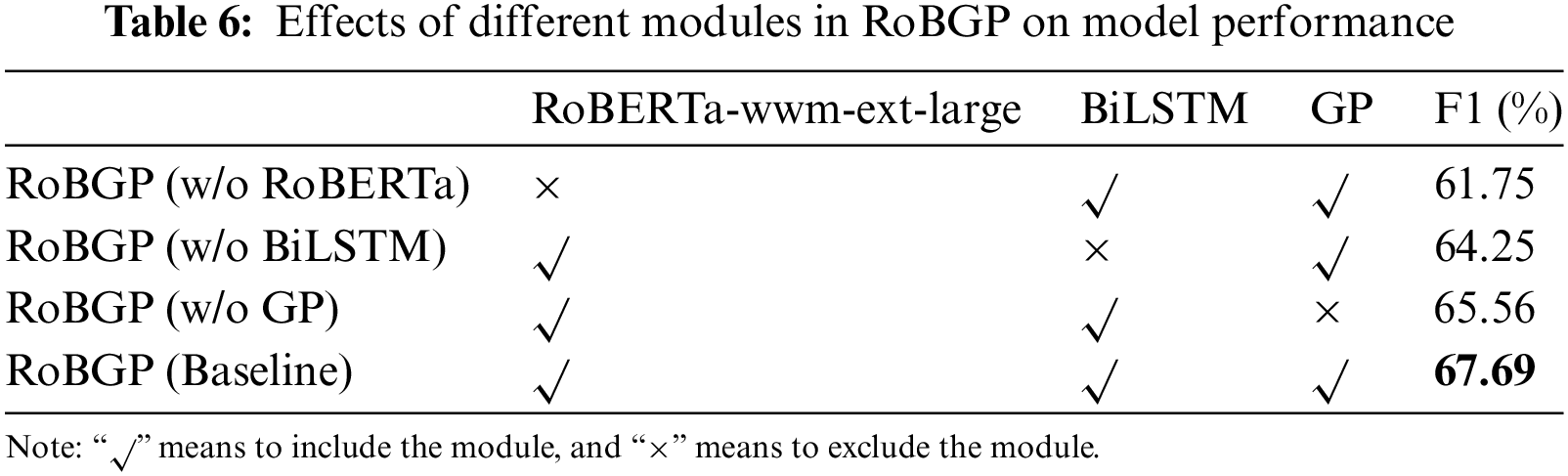
1. RoBGP (w/o RoBERTa): RoBGP model with RoBERTa-wwm-ext-large excluded.
2. RoBGP (w/o BiLSTM): RoBGP model with BiLSTM excluded.
3. RoBGP (w/o GP): RoBGP model with GP excluded.
4. RoBGP (Baseline): The complete RoBGP model, including all components.
The results indicate that all three modules play a crucial role in RoBGP, significantly enhancing its recognition performance. Specific analyses are as follows:
1. Compared to RoBGP (Baseline), the F1 score of RoBGP (w/o RoBERTa) decreases by 5.94%. This indicates that the quality of static word vectors generated by Word2Vec is inadequate, as it only establishes a one-to-one relationship between words and vectors, incapable of handling ambiguity. However, by adopting RoBERTa-wwm-ext-large, rich word-level semantic features are captured, establishing a robust foundation for the subsequent encoding stage.
2. Compared to RoBGP (Baseline), the F1 score of RoBGP (w/o BiLSTM) decreases by 3.44%. This is because BiLSTM possesses a strong ability to capture long-distance label dependency relationships, which is particularly valuable in the biomedical field. Therefore, the incorporation of BiLSTM enhances the performance of our proposed model.
3. Compared to RoBGP (Baseline), the F1 score of RoBGP (w/o GP) decreases by 2.13%. The CRF decoder exhibits good recognition performance in traditional NER tasks, but it fails to address the issue of nested named entities prevalent in the biomedical field. The GP model treats the entity’s head and tail as a whole and makes decisions within the candidate entity set, achieving remarkable performance in the nested NER task.
Our experimental results show that our proposed RoBGP can effectively utilize Chinese biomedical word information for efficient NER. We compare RoBGP with seven baselines: BiLSTM-CRF [4], Lattice-LSTM [14], BERT-CRF [18], BERT-Biaffine [8], FFBLEG [23], RICON [18] and FLR-MRC [24]. By comparing BiLSTM-CRF and Lattice-LSTM, we prove that word embedding vectors obtained through the pretrained language model are more advantageous for the downstream task. By comparing BERT-CRF and BERT-Biaffine, we show that extracting long-distance Chinese word-level information is more beneficial for biomedical NER. Additionally, GP is more suitable for recognizing nested entities compared to traditional sequence labeling models. Through the comparison with SOTA models FFBLEG, RICON, and FLR-MRC over the past 2 years, we demonstrate that RoBGP has certain advantages in the Chinese biomedical NER task.
However, we also notice that RoBGP does not achieve the highest precision, indicating some room for improvement. This shortcoming is partly attributed to the Chinese whole word masking strategy employed, which might not comprehensively process and capture character-level information, potentially leading to errors or omissions in some fine-grained entity recognition. Therefore, in our future research, we may consider exploring the use of hybrid semantic representations involving both characters and words to further refine our model.
In this paper, we propose a Chinese nested biomedical NER model called RoBGP for the fundamental task of constructing the biomedical knowledge base. The model can capture rich semantic information at the biomedical word level, extract long-distance semantic dependency relationships, and effectively recognize nested entities through the GP model. We conduct extensive experiments with our model on the publicly available Chinese medical dataset CMeEE. Alongside this, we carry out detailed comparisons with various mainstream models to evaluate our model’s performance in a comprehensive manner. The experimental results show the superiority and effectiveness of our model in the Chinese biomedical NER task. Each module plays an indispensable role in the entire framework. However, despite achieving advanced performance in experiments, our model still faces challenges and space for improvement. Particularly, we acknowledge limitations in fine-grained entity recognition. In future research, we will explore new methods and strategies to overcome this issue, aiming to enhance the quality and efficiency of knowledge base construction to fulfill the needs of the biomedical field better.
Acknowledgement: The authors extend their appreciation to all who have contributed to the field of study, and to the anonymous reviewers for their insightful comments and suggestions that have significantly enhanced the quality of this paper.
Funding Statement: This work was supported by the Outstanding Youth Team Project of Central Universities (QNTD202308) and the Ant Group through CCF-Ant Research Fund (CCF-AFSG 769498 RF20220214).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: X. H. Cui, C. Song; data collection: C. Song, D. M. Li; methodology: J. Long, Y. Yang; analysis and interpretation of results: X. H. Cui, C. Song; writing-original draft: C. Song, writing-review and editing: D. M. Li, X. L. Qu, H. C. Zhang. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Data are contained within the article. Codes are not publicly available due to copyright restrictions.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1https://tianchi.aliyun.com/dataset/95414.
References
1. D. M. Li, S. S. Luo, X. P. Zhang, and F. Xu, “Review on named entity recognition,” J. Front. Comput. Sci. Technol., vol. 16, no. 9, pp. 1954–1968, 2022. [Google Scholar]
2. J. T. Luo, S. Y. Yao, C. M. Zhao, J. Xu, and J. Feng, “A federated named entity recognition model with explicit relation for power grid,” Comput. Mater. Contin., vol. 75, no. 2, pp. 4207–4216, 2023. doi: 10.32604/cmc.2023.034439. [Google Scholar] [PubMed] [CrossRef]
3. N. Alsaaran and M. Alrabiah, “Arabic named entity recognition: A BERT-BGRU approach,” Comput. Mater. Contin., vol. 68, no. 1, pp. 471–485, 2021. doi: 10.32604/cmc.2021.016054. [Google Scholar] [PubMed] [CrossRef]
4. Z. H. Huang, W. Xu, and K. Yu, “Bidirectional LSTM-CRF models for sequence tagging,” arXiv: 1508.01991, 2015. [Google Scholar]
5. G. Lample, M. Ballesteros, S. Subramanian, K. Kawakami, and C. Dyer, “Neural architectures for named entity recognition,” in Proc. NAACL, San Diego, USA, 2016, pp. 260–270. [Google Scholar]
6. X. Z. Ma and E. Hovy, “End-to-end sequence labeling via bi-directional LSTM-CNNs-CRF,” in Proc. ACL, Berlin, Germany, 2016, pp. 1064–1074. [Google Scholar]
7. J. Devlin, M. W. Chang, K. Lee, and K. Toutanova, “BERT: Pre-training of deep bidirectional transformers for language understanding,” in Proc. NAACL, Minneapolis, USA, 2019, pp. 4171–4186. [Google Scholar]
8. J. T. Yu, B. Bohnet, and M. Poesio, “Named entity recognition as dependency parsing,” in Proc. ACL, 2020, pp. 6470–6476. [Google Scholar]
9. Y. L. Shen et al., “A two-stage identifier for nested named entity recognition,” in Proc. ACL, 2021, pp. 2782–2794. [Google Scholar]
10. M. Krauthammer, A. Rzhetsky, P. Morozov, and C. Friedman, “Using BLAST for identifying gene and protein names in journal articles,” Gene, vol. 259, no. 1–2, pp. 245–252, 2000. doi: 10.1016/S0378-1119(00)00431-5. [Google Scholar] [PubMed] [CrossRef]
11. B. T. Todorovic, S. R. Rancic, I. M. Markovic, E. H. Mulalic, and V. M. Ilic, “Named entity recognition and classification using context Hidden Markov Model,” in Proc. of the 9th Symp. on Neural Network Applications in Electrical Engineering, Belgrade, Serbia, 2008, pp. 43–46. [Google Scholar]
12. A. L. Berger, S. A. Della Pietra, and V. J. Della Pietra, “A maximum entropy approach to natural language processing,” Comput. Linguist., vol. 22, no. 1, pp. 39–71, 1996. [Google Scholar]
13. J. Lafferty, A. McCallum, and F. Pereira, “Conditional random fields: Probabilistic models for segmenting and labeling sequence data,” in Proc. ICML, Williamstown, USA, 2001, pp. 282–289. [Google Scholar]
14. Y. Zhang and J. Yang, “Chinese NER using lattice LSTM,” in Proc. ACL, Melbourne, Australia, 2018, pp. 1554–1564. [Google Scholar]
15. T. Mikolov, K. Chen, G. Corrado, and J. Dean, “Efficient estimation of word representations in vector space,” in Proc. ICLR, Scottsdale, USA, 2013, pp. 1–12. [Google Scholar]
16. J. Pennington, R. Socher, and C. D. Manning, “Glove: Global vectors for word representation,” in Proc. EMNLP, Doha, Qatar, 2014, pp. 1532–1543. [Google Scholar]
17. M. Peters et al., “Deep contextualized word representations,” in Proc. NAACL, New Orleans, Louisiana, USA, 2018, pp. 2227–2237. [Google Scholar]
18. Y. J. Gu et al., “Delving deep into regularity: A simple but effective method for Chinese named entity recognition,” in Proc. NAACL, Seattle, USA, 2022, pp. 136–145. [Google Scholar]
19. X. H. Zhang et al., “Extracting comprehensive clinical information for breast cancer using deep learning methods,” Int. J. Med. Inform., vol. 132, no. 6, pp. 103985, 2019. doi: 10.1016/j.ijmedinf.2019.103985. [Google Scholar] [PubMed] [CrossRef]
20. Y. H. Liu et al., “RoBERTa: A robustly optimized bert pretraining approach,” arXiv:1907.11692, 2019. [Google Scholar]
21. Y. Wu et al., “Research on named entity recognition of electronic medical records based on RoBERTa and radical-level feature,” Wirel. Commun. Mob. Comput., vol. 21, no. 10, pp. 1–10, 2021. doi: 10.1155/2021/2489754. [Google Scholar] [CrossRef]
22. Y. M. Cui, W. X. Che, T. Liu, B. Qin, and Z. Q. Yang, “Pre-training with whole word masking for Chinese BERT,” IEEE/ACM Trans. Audio, Speech, Language Process., vol. 29, pp. 3504–3514, 2021. doi: 10.1109/TASLP.2021.3124365. [Google Scholar] [CrossRef]
23. Q. Cong, Z. Y. Feng, G. Z. Rao, and L. Zhang, “Chinese medical nested named entity recognition model based on feature fusion and bidirectional lattice embedding graph,” in Proc. DASFAA, Tianjin, China, 2023, pp. 314–324. [Google Scholar]
24. S. Y. Liu, J. W. Duan, F. Gong, H. L. Yue, and J. X. Wang, “Fusing label relations for Chinese EMR named entity recognition with machine reading comprehension,” in Proc. ISBRA, 2022, pp. 41–51. [Google Scholar]
25. M. Schuster and K. K. Paliwal, “Bidirectional recurrent neural networks,” IEEE Trans. Signal Process., vol. 45, no. 11, pp. 2673–2681, 1997. doi: 10.1109/78.650093. [Google Scholar] [CrossRef]
26. S. Hochreiter and J. Schmidhuber, “Long short-term memory,” Neural. Comput., vol. 9, no. 8, pp. 1735–1780, 1997. doi: 10.1162/neco.1997.9.8.1735. [Google Scholar] [PubMed] [CrossRef]
27. O. Vinyals, M. Fortunato, and N. Jaitly, “Pointer networks,” in Proc. NIPS, Montreal, Canada, 2015, pp. 2692–2700. [Google Scholar]
28. J. L. Su, A. Murtadha, S. F. Pan, J. Hou, and J. Sun, “Global pointer: Novel efficient span-based approach for named entity recognition,” arXiv:2208.03054, 2022. [Google Scholar]
29. N. Y. Zhang et al., “CBLUE: A Chinese biomedical language understanding evaluation benchmark,” in Proc. ACL, Dublin, Ireland, 2022, pp. 136–145. [Google Scholar]
30. D. P. Kingma and J. Ba, “Adam: A method for stochastic optimization,” arXiv: 1412.6980, 2014. [Google Scholar]
31. Z. L. Yang, Z. H. Dai, Y. M. Yang, J. Carbonell, R. Salakhutdinov and Q. V. Le, “XLNet: Generalized autoregressive pretraining for language understanding,” in Proc. NIPS, Vancouver, Canada, 2019, pp. 5754–5764. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools