 Open Access
Open Access
ARTICLE
SciCN: A Scientific Dataset for Chinese Named Entity Recognition
College of Computer, National University of Defense Technology, Changsha, 410073, China
* Corresponding Author: Shasha Li. Email:
Computers, Materials & Continua 2024, 78(3), 4303-4315. https://doi.org/10.32604/cmc.2023.035594
Received 27 August 2022; Accepted 28 September 2022; Issue published 26 March 2024
Abstract
Named entity recognition (NER) is a fundamental task of information extraction (IE), and it has attracted considerable research attention in recent years. The abundant annotated English NER datasets have significantly promoted the NER research in the English field. By contrast, much fewer efforts are made to the Chinese NER research, especially in the scientific domain, due to the scarcity of Chinese NER datasets. To alleviate this problem, we present a Chinese scientific NER dataset–SciCN, which contains entity annotations of titles and abstracts derived from 3,500 scientific papers. We manually annotate a total of 62,059 entities, and these entities are classified into six types. Compared to English scientific NER datasets, SciCN has a larger scale and is more diverse, for it not only contains more paper abstracts but these abstracts are derived from more research fields. To investigate the properties of SciCN and provide baselines for future research, we adapt a number of previous state-of-the-art Chinese NER models to evaluate SciCN. Experimental results show that SciCN is more challenging than other Chinese NER datasets. In addition, previous studies have proven the effectiveness of using lexicons to enhance Chinese NER models. Motivated by this fact, we provide a scientific domain-specific lexicon. Validation results demonstrate that our lexicon delivers better performance gains than lexicons of other domains. We hope that the SciCN dataset and the lexicon will enable us to benchmark the NER task regarding the Chinese scientific domain and make progress for future research. The dataset and lexicon are available at: .Keywords
The amount of scientific papers consistently increases as time goes on [1–4]. These papers not only condense the work of researchers but also record the trend of research attention. However, it is difficult to manually extract such critical information from so many documents, which necessitates the development of automated information extraction technologies [5,6]. As far as we know, several English scientific named entity recognition (NER) datasets have greatly promoted the development of English scientific information extraction, such as SemEval2017 [7], TMDsci [8], and SciERC [9]. Moreover, these datasets also advance the development of language models [10,11]. Several Chinese NER datasets have been published, such as MSRA [12], OntoNotes4 [13], Weibo [14], and Resume [15], but none of these datasets is specialized in the Chinese scientific domain. To the best of our knowledge, there are no Chinese scientific NER datasets until now, which hinders the development of Chinese scientific information extraction. We attribute this to the fact that annotating such datasets requires domain-specific knowledge and is time-consuming, and therefore less effort has been made to such annotations [16].
With the goal of promoting the research of Chinese scientific NER, this paper presents a large-scale Chinese scientific NER dataset–SciCN. To ensure the quality of entity annotations, we design an effective coarse-to-fine annotation procedure, including candidate generation and crowdsourcing annotation. In particular, we first collect 10,000 Chinese scientific papers of computer science from publicly available online databases. We then select 3,500 papers that are published by four well-known Chinese journals, i.e., Computer Engineering and Applications1, Journal of Computer Research and Development2, Computer Science3, and Journal of Computer Applications4. These selected papers cover diverse research topics, e.g., Machine Learning, Operating Systems, Computer Networks, Parallel Computing, Deep Learning, Computer Vision, and Pattern Recognition. We pre-define six types of entities, i.e., Method, Task, Metric, Material, Scientific Term, and Generic, and manually annotate entities contained in titles and abstracts of the 3,500 papers. Finally, we obtain a total of 62,059 entity annotations. We name the set of titles, abstracts and entity annotations as SciCN, which is the first supervised Chinese scientific NER dataset that we are aware.
In an attempt to fully investigate the characteristics of SciCN and to provide a comparable baseline for future studies, we carefully selected several previously state-of-the-art Chinese NER models and adapted them to evaluate SciCN. We roughly classify these models into two categories: character-level and lexicon-enhanced. Qualitative and quantitative experiments reveal the properties and indicate that SciCN is more challenging than other Chinese NER datasets. In addition, it has been proven that NER models can be enhanced by well-constructed lexicons. Thus, we construct a Chinese scientific lexicon, which contains 2.5 million scientific terminologies. Validation results demonstrate that our lexicon is more effective on SciCN than other lexicons. In sum, we summarize our contributions as follows:
1) We present the first Chinese scientific NER dataset–SciCN, aiming to advance the research of Chinese scientific information extraction. Moreover, SciCN contains paragraph-level abstracts, which provides more contextual information than other sentence-level NER datasets.
2) We carefully select a number of representative NER models and adapt them to evaluate SciCN, which provides comparable baselines for future research.
3) We construct a Chinese scientific domain-specific lexicon with 2.5 million scientific terminologies, which are collected from 719,938 Chinese scientific papers.
One of the primary bottlenecks in applying deep learning techniques to natural language processing (NLP) is the lack of high-quality annotated datasets [17]. It is a tedious and time-consuming process to annotate datasets manually. Thus the research community shifts its attention to annotate datasets with distant supervision and active learning [18,19]. Initially, the distant supervision relies on well-formed databases, e.g., Freebase [20], to collect examples. It then uses these examples to automatically generate distantly supervised datasets [21]. Using a set of labeled examples and a large set of unlabeled examples, active learning creates a classifier and relatively small sets of newly labeled data [22]. Although distant supervision and active learning can reduce the cost of dataset construction, they are actually complicated and often generate unbearable noise during automatic annotating [23]. Compared to these automatic methods, we manually annotate Chinese scientific papers to construct SciCN, which ensures high annotation quality.
2.2 Scientific Information Extraction
English scientific information extraction has attracted a great deal of attention [24]. And a lot of English scientific NER datasets have been proposed for the research. SemEval2017 [7] contains 500 paragraphs derived from open access papers, and it contains annotations of three types of entities, i.e., Task, Method, and Material. TDMSci [8] consists of 2,000 sentences taken from NLP papers, and it contains annotations of four types of entities, i.e., Task, Dataset, Metric, and Score. SciERC [9] is a joint entity and relation extraction dataset. It contains entity and relation annotations of 500 scientific paper abstracts. A number of Chinese NER datasets have been proposed over the years, but they are specialized in news or social media domain, such as MSRA [12], OntoNotes4 [13], Weibo [14], and Resume [15]. As far as we know, there is no dataset available for the Chinese scientific information extraction. To tackle this, we present the first Chinese scientific NER dataset, which contains manual annotations of titles and abstracts derived from 3,500 scientific papers.
Chinese NER is more challenging than English NER since Chinese sentences are not naturally segmented and have more complex morphological features. Recent studies have explored utilizing lexicon matching methods to enhance Chinese NER models. The LatticeLSTM [15] model is such a typical method, which improves the NER performance by encoding and matching words in a lexicon. SoftLexicon [25] incorporates lattice information into Chinese character embeddings through labeling and probability methods. Another classic approach is to incorporate lexicon and structural information features into the Transformer [26]. In addition, FLAT [27] exploits the flat lattice structure to capture word information via position encoding in Transformer. MECT4CNER [28] uses a novel cross-Transformer [26] method to integrate the structural information of Chinese characters with lexical information.
Accessing authoritative Chinese scientific papers is the first step in constructing a scientific NER dataset. To ensure the data quality of SciCN, we first collect 10,000 scientific papers from two online academic databases: Wanfang5 and CNKI6. We then select the papers that are relevant to computer science topics, which is achieved by filtering these papers according to their publication journals, subject classes, etc. Finally, we obtain a total of 3,500 papers published by well-known Chinese computer science journals. For each of the obtained papers, we solely annotate its title and abstract since the two parts contain the most concise information of the whole paper. Additionally, such an annotation strategy significantly reduces the burden on annotators. We report an annotation example of the title and abstract of one paper in Fig. 1. We can observe that six types of entities are annotated.
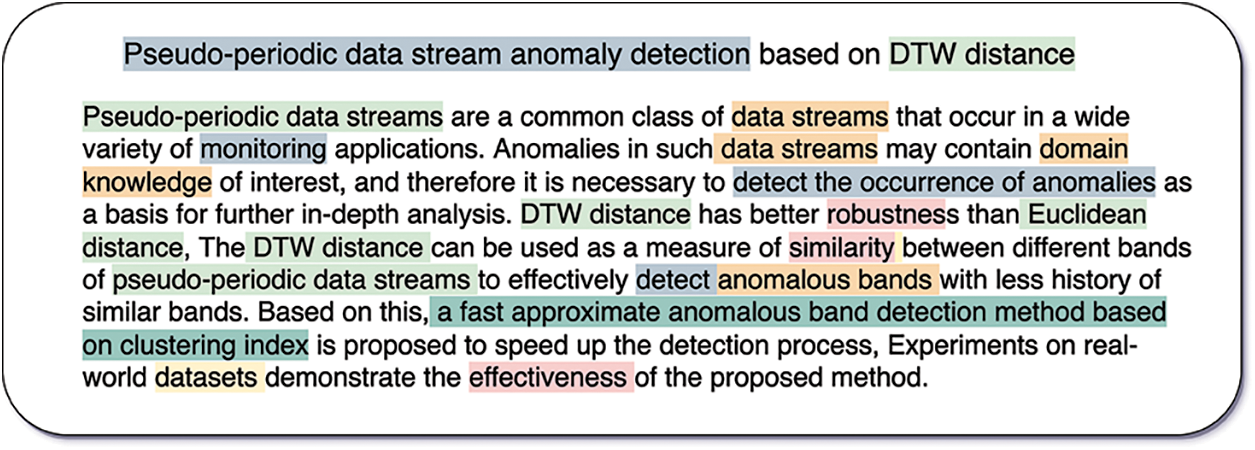
Figure 1: An annotation example of a paper’s title and abstract. Each highlight text segment denotes an entity, and its type is classified by the background color, as shown below: 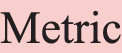 ,
,  ,
, 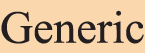 ,
,  ,
,  ,
, 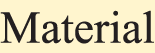
Before the start of the formal annotation, we first conduct pilot annotations and make adaptive modifications to the formal annotation schema to ensure the reliability of the annotation process. Specifically, the invited annotators perform greedy annotations for entity spans and always annotate the longer span whenever there is ambiguity. We use this strategy to remove overlapping entities. We present the complete annotation guidelines and details in Appendix A.
Annotating such a large scientific NER dataset is challenging because the annotation process is time-consuming and labor-intensive, let alone the domain-specific knowledge required [29,30]. We invite three annotators to annotate entities and two domain experts to examine the annotation quality. All the three annotators have a master’s degree in computer science, and they are trained to master the detailed and formal annotation principles. The annotators and experts conduct two annotation rounds, as shown in Fig. 2. Specifically, one of the three well-trained annotators annotates documents in the first round, and the three annotators annotate documents in parallel; in the second round, two experts check the annotations independently for possible errors or omissions and make the final decision. The experts randomly select 20% of the annotated data from each batch of submitted annotations for the quality examination. A batch with an acception ratio below 95% will be rejected, and all samples in the batch will be re-annotated. We then calculate the Cohen’s Kappa [31] to determine the agreement between annotators and experts in each batch. The average result is 80.4%, indicating a high consistency between annotators and experts. We believe that a consistent and reliable annotation process ensures the high annotation quality of SciCN.
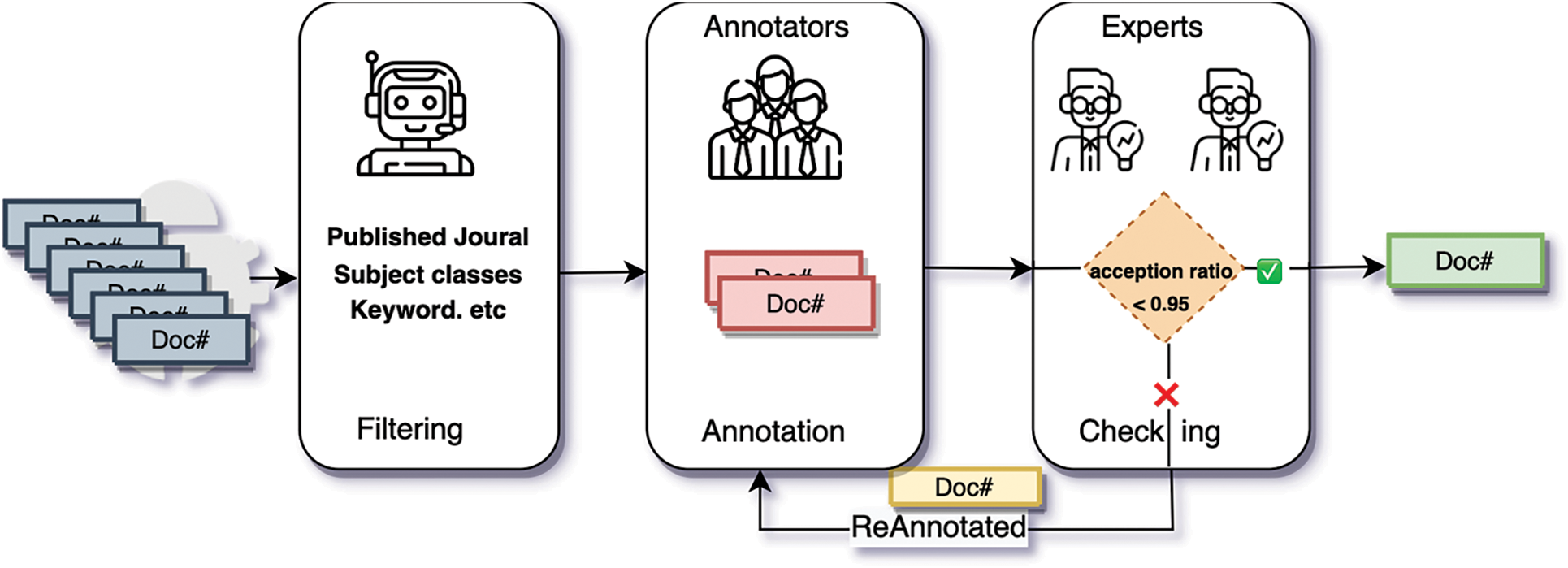
Figure 2: The overall annotation process of SciCN.  denotes the title and abstract of one paper.
denotes the title and abstract of one paper.  denotes the entity annotations of one paper, where the annotations are not examined by experts.
denotes the entity annotations of one paper, where the annotations are not examined by experts.  denotes entity annotations of one paper, where the annotations are rejected by experts,
denotes entity annotations of one paper, where the annotations are rejected by experts,  denotes the title, abstract and entity annotations of one paper, where the annotations are agreed by experts
denotes the title, abstract and entity annotations of one paper, where the annotations are agreed by experts
Using the above annotation strategy, we manually annotate entities in the titles and abstracts of the collected 3,500 papers and regard the set of titles, abstracts and entity annotations as SciCN. A total of 62,059 entities are annotated into six entity types, namely Method, Task, Metric, Material, Scientific Term, and Generic. We then make detailed data statistics for SciCN. The first is to analyze the data distributions, including the paper distribution regarding various journals and the entity distribution regarding various entity types. We visualize the paper distribution in the left part of Fig. 3, and the entity distribution in the right part. We can observe that: (1) 72.1% of the papers are from the Computer Engineering and Applications journal, and 15% are from the Computer Science journal. Papers from the other two journals only account for 12.9%. (2) The most common types of entity in SciCN are Method and Scientific Term, accounting for 35.6% and 25.7%, respectively. They are also in keeping with the special characteristics of scientific papers.
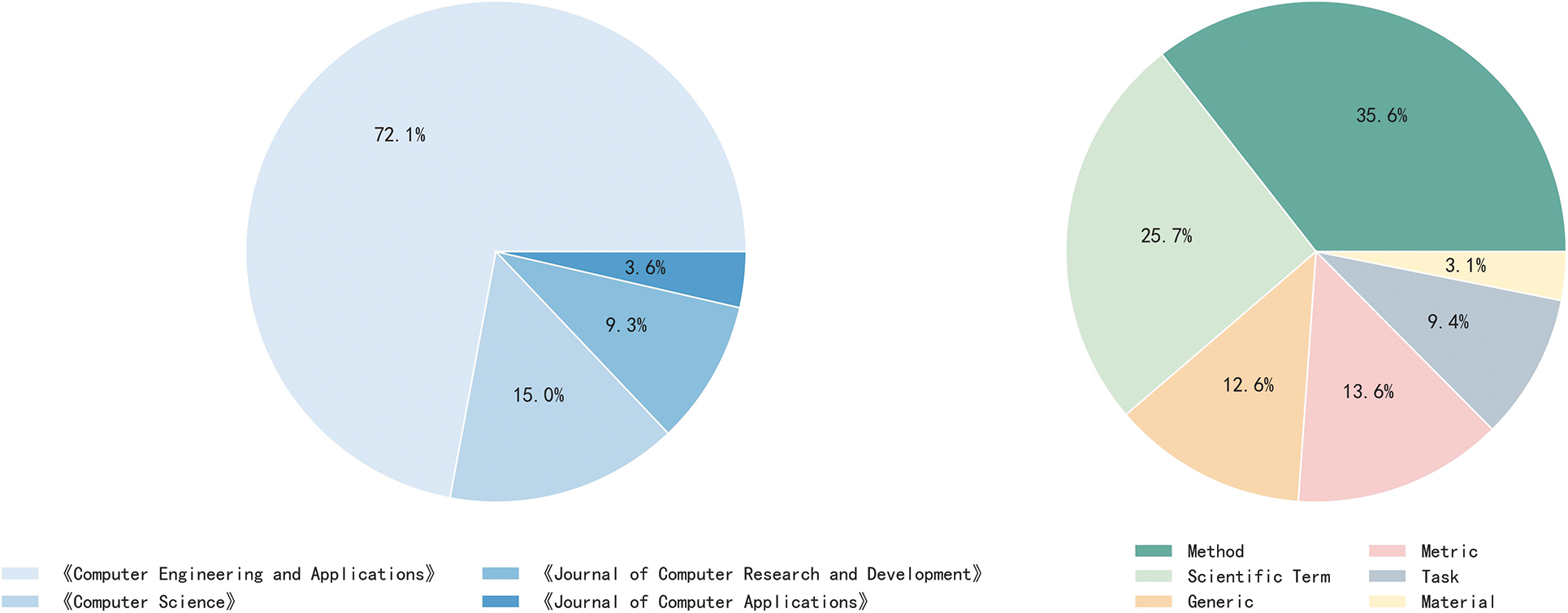
Figure 3: The left part shows paper distributions regarding various journals. The right shows entity distributions regarding various entity types
To compare our SciCN with other widely-used Chinese NER datasets, we make statistics regarding dataset domains, entity types, sentences, and characters and report the results in Table 1. We observe that MSRA [12] and OntoNotes4 [13] contain more sentences than SciCN, but they only annotate three simple entity types, i.e., Person, Location, and Organization. Although there are more entity types in Weibo [14] and Resume [15] than in SciCN, their scales are relatively small. In summary, we have designed our SciCN to provide more types of scientific entities while maintaining a sufficient dataset scale.

Most existing Chinese NER models are based on character-level semantics due to that Chinese is character-level. However, pure character-based NER models cannot fully exploit word information, so some recent studies propose to use word lexicons to enhance these character-based NER models. In order to facilitate future research on SciCN and Chinese scientific information extraction, we adapt a variety of representative Chinese NER models to evaluate SciCN, including character-level and lexicon-enhanced, as shown below (* denotes lexicon-enhanced models).
• LSTM-CRF [32] is the most classic method for the NER task. In addition to an embedding layer of input sequences, there are LSTM layers used to obtain contextual representations of input sequences, followed by a CRF layer used to learn some restrictions and rules among entity categories.
• BERT-Tagger [33] is a Transformer-based model that uses pre-training to learn from the raw datasets, and fine-tune on downstream tasks, including the NER task.
• BERT-LSTM-CRF [33] has the same model architecture as LSTM-CRF [32], with the difference that it utilizes a Transformer-based model to obtain a more comprehensive contextual representation of input sequences.
• LatticeLSTM* [15] encodes all characters and potential words recognized by a lexicon in a sentence, avoiding the error propagation of segmentation while leveraging the word information.
• SoftLexicon* [25] incorporates word lexicons into character representations for Chinese NER in a simple yet effective manner.
• FLAT* [27] converts the lattice structure into a flat structure consisting of spans. Using a well-designed position encoder to indicate the lattice structure, and each span corresponds to a character or latent word and its position in the original lattice.
• MECT4CNER* [28] uses multi-metadata embedding to fuse the information from radicals, characters, and words through a cross-Transformer network.
For each of the above baselines, we adapt it to recognize entities of SciCN by using the Chinese BERT-WWM [34] as the embedding layer, for pre-trained language models with deep Transformers (e.g., BERT [32]) have proven to be powerful encoders for NER. For the other hyper-parameters, we just keep their values the same as those reported in the original papers, with the goal of providing a fair comparison. Moreover, following the established line of work, we split the abstract paragraph into sentences and train and evaluate the baselines on them. We remain training and evaluating the baselines on paragraph-level abstracts for future work. In addition, we report model performance measured by the standard Precision, Recall, and F1 score.
As for the data splits, we randomly select 70% of the annotated papers as the training set, 10% as the development set, and 20% as the test data. We have provided detailed dataset statistic in the Table 1. Based on them, we conduct extensive comparisons with some widely-used Chinese NER datasets.
Table 2 shows the overall experimental results on the test set of SciCN. We observe that pre-trained language models and lexicon information can further improve the performance of NER models, which is in line with previous work. However, we also see that the effectiveness of the lexicon-enhanced model on SciCN is not as significant as those on the other datasets. We attribute this to the fact that the lexicon of general domain cannot fit into our SciCN well. Moreover, MECT4CNER [28] achieves the best performance on SciCN. It is worth noting that the method does not only utilize a lexicon, but is designed to integrate information about Chinese character components. To facilitate the future study, we further analyze the challenges posed by SciCN and the benefits of using lexicon and structural information in the scientific NER.

We report the performance comparisons of various NER models and Chinese NER datasets in Table 3. We observe a large performance gap between SciCN and other datasets, indicating that the existing NER models cannot generalize to the scientific domain well. We attribute this to the fact that scientific abstracts have their writing specificities. Scientific papers are intended to present specific scientific investigations or concepts within a specific research field. Thus papers are often written with domain-specific terminologies, assuming that the target audience has relevant background. Scientific papers differ from news articles in that the former contains extensive complex concepts and technical terms. A further examination of the experiment results revealed that the “unseen” terms in scientific papers can pose a significant challenge in the field of scientific information extraction.

To address the large number of scientific domain-specific terms in SciCN, we constructed a scientific dictionary containing about 2.5 million Chinese scientific terms from about 12 GB of scientific papers. We also train embeddings for these terms with Gensim [35,36], and report more details in Appendix B. To compare our scientific domain-specific lexicon with lexicons of other domains, we first define a new concept for lexicon–match ratio, which is calculated by comparing the number of entities appearing in a lexicon to the number of entities in the SciCN. We then calculate the match ratios of our scientific lexicon and other available lexicons. As shown in Table 4, the match ratio of our scientific lexicon is significantly higher than the other lexicons.
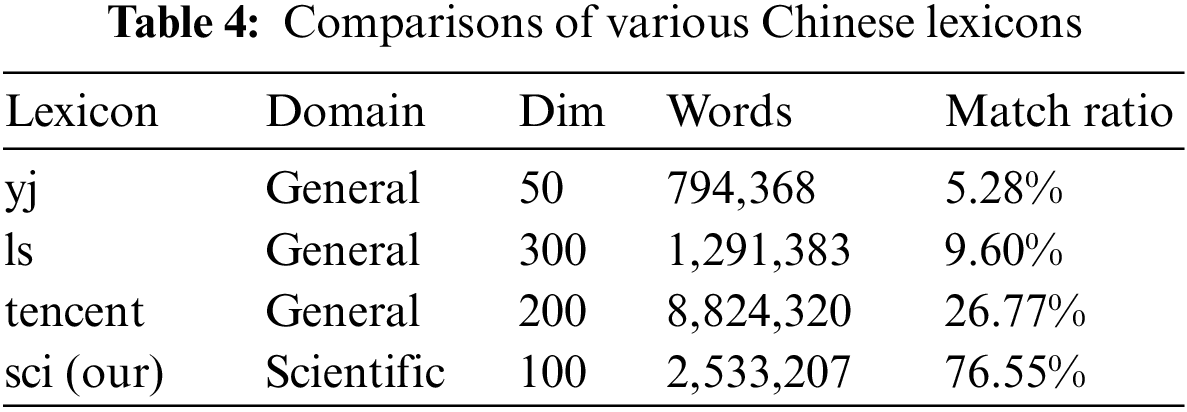
In addition, we report performance comparisons between our lexicon and the above three widely-used lexicons, i.e., yj7, ls8, and tencent9, in Table 5. We can observe that the match ratio the lexicon can affect the performance of lexicon-based NER models, for noise words in lexicons can potentially confuse character embeddings. We also find that the higher the match rate of the lexicon, the more accurate the model recognize entities. In the future, we will investigate an efficient method to enable NER models to select approximate words for model training.

In this paper, we present the first Chinese scientific NER dataset. We first illustrate the characteristics and the construction process of the high-quality and large-scale SciCN. Then we adapt a number of previous state-of-the-art Chinese NER models and use them to evaluate SciCN, with the goal of investigating the properties of SciCN, as well as providing comparable baselines for future study. In addition, we construct a Chinese scientific domain-specific lexicon with 2.5 million scientific terms, which can be used to improve the performance of NER models on SciCN. We hope the proposed Chinese scientific dataset and the scientific domain-specific lexicon can promote the research of Chinese scientific information extraction, especially the Chinese scientific NER.
Acknowledgement: Not applicable.
Funding Statement: This research was supported by the National Key Research and Development Program [2020YFB1006302].
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Jing Yang, Bin Ji; data collection: Jing Yang; analysis and interpretation of results: Shasha Li, Jun Ma, Jie Yu; draft manuscript preparation: Jing Yang, Bin Ji. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Data openly available in a public repository. The data that support the findings of this study are openly available in (https://lgithub.com/yangjingla/SciCN.git).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1http://cea.ceaj.org/CN/volumn/home.shtml
2https://crad.ict.ac.cn/CN/1000-1239/home.shtml
3http://www.jsjkx.com/CN/1002-137X/home.shtml
5https://www.wanfangdata.com.cn/index.html
7https://github.com/jiesutd/LatticeLSTM
8https://github.com/LeeSureman/Flat-Lattice-Transformer
9https://ai.tencent.com/ailab/nlp/en/data/tencent-ailab-embedding-zh-d200-v0.1.0.tar.gz
10https://github.com/pdfminer/pdfminer.six
11https://github.com/BYVoid/OpenCC
12https://github.com/fxsjy/jieba
References
1. T. Hope, A. Amini, D. Wadden, M. V. Zuylen, S. Parasa et al., “Extracting a knowledge base of mechanisms from COVID-19 papers,” in Proc. of NAACL, pp. 4489–4503, 2021. [Google Scholar]
2. K. Lo, L. L. Wang, M. Neumann, R. Kinney and D. S. Weld, “S2ORC: The semantic scholar open research corpus,” in Proc. of ACL, pp. 4969–4983, 2020. [Google Scholar]
3. D. Wright, D. Wadden, K. Lo, B. Kuehl, A. Cohan et al., “Generating scientific claims for zero-shot scientific fact checking,” in Proc. of ACL, Dublin, Ireland, pp. 2448–2460, 2022. [Google Scholar]
4. F. Dernoncourt and J. Y. Lee, “Pubmed 200k RCT: A dataset for sequential sentence classification in medical abstracts,” in Proc. of IJCNLP, Taipei, Taiwan, pp. 308–313, 2017. [Google Scholar]
5. D. Lahav, J. Saad-Falcon, B. Kuehl, S. Johnson, S. Parasa et al., “A search engine for discovery of scientific challenges and directions,” in Proc. of AAAI, Online, pp. 11982–11990, 2022. [Google Scholar]
6. J. Portenoy, M. Radensky, J. D. West, E. Horvitz, D. S. Weld et al., “Bursting scientific filter bubbles: Boosting innovation via novel author discovery,” in Proc. of CHI, New Orleans, LA, USA, pp. 1–13, 2022. [Google Scholar]
7. I. Augenstein, M. Das, S. Riedel, L. Vikraman and A. McCallum, “Semeval 2017 task 10: Scienceie-extracting keyphrases and relations from scientific publications,” in Proc. of SemEval, Vancouver, Canada, pp. 546–555, 2017. [Google Scholar]
8. Y. Hou, C. Jochim, M. Gleize, F. Bonin and D. Ganguly, “TDMSci: A specialized corpus for scientific literature entity tagging of tasks datasets and metrics,” in Proc. of EACL, pp. 707–714, 2021. [Google Scholar]
9. Y. Luan, D. Wadden, L. He, A. Shah, M. Ostendorf et al., “A general framework for information extraction using dynamic span graphs,” in Proc. of NAACL, Minneapolis, MN, USA, pp. 3036–3046, 2019. [Google Scholar]
10. I. Beltagy, K. Lo and A. Cohan, “SciBERT: A pretrained language model for scientific text,” in Proc. of EMNLP-IJCNLP, Hong Kong, China, pp. 3615–3620, 2020. [Google Scholar]
11. J. Lee, W. Yoon, S. Kim, D. Kim, S. Kim et al., “BioBERT: A pre-trained biomedical language representation model for biomedical text mining,” Bioinformatics, vol. 36, no. 4, pp. 1234–1240, 2020. [Google Scholar] [PubMed]
12. G. A. Levow, “The third international Chinese language processing bake-off: Word segmentation and named entity recognition,” in Proc. of SIGHAN, Sydney, Australia, pp. 108–117, 2006. [Google Scholar]
13. B. Roth, T. Barth, M. Wiegand and D. Klakow, “A survey of noise reduction methods for distant supervision,” in Proc. of AKBC, San Francisco, California, USA, pp. 73–78, 2013. [Google Scholar]
14. N. Peng and M. Dredze, “Named entity recognition for Chinese social media with jointly trained embeddings,” in Proc. of EMNLP, Lisbon, Portugal, pp. 548–554, 2015. [Google Scholar]
15. Y. Zhang and J. Yang, “Chinese NER using lattice LSTM,” in Proc. of ACL, Melbourne, Australia, pp. 1554–1564, 2018. [Google Scholar]
16. B. Aniek, The importance of domain-specific expertise in training customized named entity recognition models. M.S. Thesis, Utrecht University, The Netherlands, 2021. [Google Scholar]
17. R. Bernatchez, A. Durand and F. Lavoie-Cardinal, “Annotation cost-sensitive deep active learning with limited data (student abstract),” in Proc. of AAAI, Online, pp. 12913–12914, 2022. [Google Scholar]
18. A. Mandya, D. Bollegala, F. Coenen and K. Atkinson, “A dataset for inter-sentence relation extraction using distant supervision,” in Proc. of LREC, Miyazaki, Japan, pp. 1559–1565, 2019. [Google Scholar]
19. D. Shen, J. Zhang, J. Su, G. Zhou and C. L. Tan, “Multi-criteria-based active learning for named entity recognition,” in Proc. of ACL, Barcelona, Spain, pp. 589–596, 2004. [Google Scholar]
20. K. Bollacker, C. Evans, P. Paritosh, T. Sturge and J. Taylor, “Freebase: A collaboratively created graph database for structuring human knowledge,” in Proc. of SIGMOD, Vancouver, BC, Canada, pp. 1247–1250, 2008. [Google Scholar]
21. B. Y. Lin, D. H. Lee, F. F. Xu, O. Lan and X. Ren, “Alpacatag: An active learning-based crowd annotation framework for sequence tagging,” in Proc. of ACL, Florence, Italy, pp. 58–63, 2019. [Google Scholar]
22. K. Tomanek and F. Olsson, “A web survey on the use of active learning to support annotation of text data,” in Proc. of ALNLP, Boulder, Colorado, USA, pp. 45–48, 2009. [Google Scholar]
23. S. Takamatsu, I. Sato and H. Nakagawa, “Reducing wrong labels in distant supervision for relation extraction,” in Proc. of ACL, Jeju Island, Korea, pp. 721–729, 2012. [Google Scholar]
24. K. Gabor, D. Buscaldi, A. Schumann, B. QasemiZadeh, H. Zargayouna et al., “Semeval-2018 task 7: Semantic relation extraction and classification in scientific papers,” in Proc. of SE, New Orleans, Louisiana, USA, pp. 679–688, 2018. [Google Scholar]
25. R. Ma, M. Peng, Q. Zhang, Z. Wei and X. J. Huang, “Simplify the usage of lexicon in Chinese NER,” in Proc. of ACL, pp. 5951–596, 2020. [Google Scholar]
26. A. Vaswani, N. Shazeer, N. Parmar, J. Uszkoreit, L. Jones et al., “Attention is all you need,” in Proc. of NeurIPS, Long Beach, CA, USA, pp. 5998–6008, 2019. [Google Scholar]
27. X. Li, H. Yan, X. Qiu and X. J. Huang, “Flat: Chinese NER using flat lattice transformer,” in Proc. of ACL, Online, pp. 6836–6842, 2020. [Google Scholar]
28. S. Wu, X. Song and Z. Feng, “MECT: Multi-metadata embedding based cross-transformer for Chinese named entity recognition,” in Proc. of ACL, pp. 1529–1539, 2021. [Google Scholar]
29. Y. Yang, M. Zhang, W. Chen, W. Zhang, H. Wang et al., “Adversarial learning for Chinese NER from crowd annotations,” in Proc. of AAAI, New Orleans, Louisiana, USA, pp. 1627–1635, 2018. [Google Scholar]
30. J. Yang, Y. Zhang, L. Li and X. Li, “YEDDA: A lightweight collaborative text span annotation tool,” in Proc. of ACL, Melbourne, Australia, pp. 31–36, 2018. [Google Scholar]
31. J. Cohen, “A coefficient of agreement for nominal scales,” Educational and Psychological Measurement, vol. 20, pp. 37–46, 1960. [Google Scholar]
32. J. Devlin, M. W. Chang, K. Lee and K. Toutanova, “BERT: Pre-training of deep bidirectional transformers for language understanding,” in Proc. of NAACL, Minneapolis, MN, USA, pp. 4171–4186, 2019. [Google Scholar]
33. Z. Huang, W. Xu and K. Yu, “Bidirectional LSTM-CRF models for sequence tagging,” arXiv preprint arXiv:1508.01991, 2015. https://doi.org/10.48550/arXiv.1508.01991 [Google Scholar] [CrossRef]
34. Y. Cui, W. Che, T. Liu, B. Qin and Z. Yang, “Pre-training with whole word masking for Chinese BERT,” IEEE/ACM Transactions on Audio, Speech, and Language Processing, vol. 29, pp. 3504–3514, 2021. [Google Scholar]
35. R. Rehurek and P. Sojka, “Software framework for topic modelling with large corpora,” in Proc. of LREC, Valletta, Malta, pp. 45–50, 2010. [Google Scholar]
36. T. Mikolov, I. Sutskever, K. Chen, G. S. Corrado and J. Dean, “Distributed representations of words and phrases and their compositionality,” in Proc. of NeurIPS, Lake Tahoe, Nevada, USA, pp. 3111–3119, 2013. [Google Scholar]
Appendix A.
As illustrated in Table 6, we have meticulously presented the distinctions among various labels within the SciCN dataset, accompanied by plentiful examples.

1. What are the common mistakes to avoid in the annotation procedure?
Labeling errors, Labeling omission errors, Entity boundary errors, contextual entity label inconsistency.
Is nested annotation allowed?
a) Nested annotation is not allowed.
b) Please choose the granularity of annotation according to the semantic richness of the sentence.
How to annotate entities of scientific term type?
a) Entities that are commonly used in scientific papers but are not easy to categorize are usually strongly related to <method, task, material, metric>.
b) Before annotating such entities, please consider whether they can be classified as one of the above four categories (method, task, material, metric), and if not, consider annotating them as another scientific term.
What is the difference between generic entities and scientific terms?
a) There are some similarities between the two, scientific terms other scientific term should be more expressive than generic, and the priority of thought when annotating should be: scientific term, generic.
b) To distinguish between the two, scientific term should usually be related to other entities <method, task, metric, material>, or generic if no relationship exists.
c) Pronouns/infixes should be annotated as generic: e.g., our method, our system, past methods, existing research, etc.
How to annotate annotations that contain English explanations/abbreviations?
a) For example: “Schwarz-Christoffel Mapping (SCM) mapping method”.
b) It needs to be fully annotated, in this case as (Schwarz-Christoffel Mapping (SCM) Mapping Method).
Appendix B.
To train our lexicon with as many scientific papers as possible, we first extract the textual information from the PDF paper format using the API pdfminer.six10, and then we process the extracted full text. To be specific, we remove those non-utf8 chars, unify different punctuation styles and convert traditional Chinese characters into simplified characters with Open Chinese Convert (OpenCC)11. Finally, we obtain a set of scientific papers that occupy about 12 GB of disk memories. We pre-train word embeddings using Word2Vec for words that are automatically segmented by jieba12. To improve the accuracy of automatic segmentation, we additionally collect a dictionary of about 11W scientific terms from the keywords of the original paper as a jieba’s user-defined dictionary.
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools