 Open Access
Open Access
ARTICLE
MDCN: Modified Dense Convolution Network Based Disease Classification in Mango Leaves
1 School of Computer Science and Engineering, Vellore Institute of Technology (VIT), Chennai, India
2 Center for Cyber Physical Systems, School of Computer Science and Engineering, Vellore Institute of Technology (VIT), Chennai, India
* Corresponding Author: K. P. Vijayakumar. Email:
(This article belongs to the Special Issue: Machine Vision Detection and Intelligent Recognition)
Computers, Materials & Continua 2024, 78(2), 2511-2533. https://doi.org/10.32604/cmc.2024.047697
Received 14 November 2023; Accepted 14 December 2023; Issue published 27 February 2024
Abstract
The most widely farmed fruit in the world is mango. Both the production and quality of the mangoes are hampered by many diseases. These diseases need to be effectively controlled and mitigated. Therefore, a quick and accurate diagnosis of the disorders is essential. Deep convolutional neural networks, renowned for their independence in feature extraction, have established their value in numerous detection and classification tasks. However, it requires large training datasets and several parameters that need careful adjustment. The proposed Modified Dense Convolutional Network (MDCN) provides a successful classification scheme for plant diseases affecting mango leaves. This model employs the strength of pre-trained networks and modifies them for the particular context of mango leaf diseases by incorporating transfer learning techniques. The data loader also builds mini-batches for training the models to reduce training time. Finally, optimization approaches help increase the overall model’s efficiency and lower computing costs. MDCN employed on the MangoLeafBD Dataset consists of a total of 4,000 images. Following the experimental results, the proposed system is compared with existing techniques and it is clear that the proposed algorithm surpasses the existing algorithms by achieving high performance and overall throughput.Keywords
Revered for its rich taste, texture, and nutritional value, the mango has secured its place as one of the most widely cultivated and consumed fruits on Earth [1]. However, like many agricultural staples, it remains under the constant threat of various diseases that, if unchecked, can lead to catastrophic consequences for yield, quality, and overall economic value. In some countries such as Kenya where mango farming is a major occupation, infestation of mangoes leads to losses of more than $1 billion and more than $42 million in Africa [2]. Disease prevention, mitigation, and management in crops largely depend on the early detection and accurate classification of the disease [3,4]. Moreover, the health of a tree, especially fruit-bearing ones like the mango tree, is often mirrored in its leaves. A diseased or malnourished tree will exhibit symptoms on its leaves, making it the first line of defense and detection against potential threats. Therefore, studying these leaves will provide all the information regarding the trees’ health and whether it is affected by any disease.
For decades, the process of disease identification has predominantly been executed by human experts, utilizing their knowledge, experience, and sometimes, with the assistance of basic diagnostic tools. Nevertheless, the human-centric approach to disease detection is inherently limited by subjectivity, potential for human error, and scalability challenges, especially in vast orchards or farms. In the modern era, where technology and agriculture are blending more seamlessly than ever, the advent of machine learning and computer vision offers a promising horizon for disease detection and classification. Over the past few years, Deep Learning [5] a subset of machine learning, has showcased tremendous potential in a myriad of applications. Particularly, Deep Neural Networks (DNNs) [6] have emerged as the front-runner in tasks that require image recognition and classification. One such architecture that has garnered significant attention in the deep learning community is the Dense Network (DenseNet) [7].
What sets DenseNet apart from its contemporaries is its unique ability to establish direct connections between any two layers, ensuring maximum information flow between them. This intrinsic characteristic makes it highly efficient and reduces the vanishing gradient problem often encountered in deep networks. However, like all models, DenseNet is not devoid of challenges. While it presents a robust framework for various applications, its efficacy in specific, specifically for tasks like mango leaf disease classification remains a subject of exploration. Mango leaves can be affected by many different diseases and accurately classifying and detecting these diseases using algorithms poses many challenges. Here are some of the main problems or challenges that algorithms face in this context:
• Data imbalance: Some diseases might be more common than others, leading to an imbalance in the training dataset. Algorithms trained on imbalanced datasets might become biased toward detecting more common diseases while overlooking or misclassifying less common ones.
• Overlapping symptoms: Some diseases have symptoms that are similar or overlap with others. For example, one type of discoloration caused by one disease may look very similar to discoloration caused by another disease. This can confuse and lead to misclassification.
• Image quality and diversity: The quality of images used for training and testing algorithms is very important. Variations in lighting conditions, angles, and resolution can affect algorithm performance. Additionally, issues such as shadows, reflections, or blurriness can prevent proper detection.
• Ambient noise: Images may have a cluttered background or contain other objects, such as fruit, insects, or other plant parts. Such noise can hinder the algorithm’s ability to accurately identify diseases.
• Incomplete data: For an algorithm to be effective, it requires a significant amount of data for training. If images of certain stages or types of disease are missing, the algorithm may not be able to identify them.
• Computational complexity: Some advanced algorithms may require high computation. This becomes a challenge if real-time processing or deployment is required for devices with limited computing capabilities.
The motivation behind developing the modified algorithm is as follows: overcoming these challenges requires a combination of robust data collection, complex algorithms (like deep learning), and regular system updates. Additionally, integrating field-specific knowledge from plant pathologists can be invaluable in refining and improving the performance of algorithms. To the best of our knowledge, none of the existing systems have considered the seven diseases (Cutting Weevil, Bacterial Canker, Anthracnose, Gall Midge, Die Back, Sooty Mould, and Powdery Mildew) along with the healthy leaves class. Therefore, recognizing the above-mentioned challenges and the pressing need for advanced, accurate, and scalable solutions for mango leaf disease classification, this literature introduces a new algorithm: the MDCN (Modified Dense Convolution Network). This novel algorithm seeks to capitalize on the foundational strengths of the original DenseNet architecture while introducing custom modifications tailored explicitly for the intricate task of classifying mango leaf diseases. In doing so, it hopes to address the pitfalls of previous methods and provide a more accurate, efficient, and actionable tool for farmers, agriculturists, and the broader scientific community.
A standout feature of the Modified Dense Convolution Network is its adept utilization of transfer learning [8]. Recognizing that training deep learning models from scratch can be a resource-intensive task, transfer learning allows our algorithm to leverage pre-existing knowledge from related tasks, optimizing the model’s efficiency. This is further complemented by the integration of data loaders, which ensure efficient batch processing, thereby streamlining and accelerating the training phase. Recognizing the challenges of overfitting and unnecessary computational expenditure, our algorithm also embraces optimization strategies, most notably early stopping mechanisms. This not only conserves computational resources but also fortifies the model against potential overfitting, ensuring a balance between bias and variance. The efficiency of the proposed MDCN algorithm is expressed in terms of accuracy, F1, and precision score. Moreover, the proposed algorithm uses a new dataset called MangoLeafBD [9] dataset to train and evaluate the model since other datasets such as PlantVillage [10], PlantDoc [11], etc., are publicly available, but they do not include multiple mango leaf disease classes as present in the selected dataset, that is the different classes present in the selected dataset are as follows, Anthracnose [12], Bacterial Canker [13,14], Cutting Weevil [15], Die Back [16,17], Gall Midge [18], Powdery Mildew [19], and Sooty Mould [20]. The performance efficiency of the introduced algorithm has been assessed by comparing it with algorithms like Visual Geometry Group (VGG-16) [21], Residual Network (ResNet) [22], Traditional-DenseNet [23], and Convolution Neural Network (CNN) [24].
Thus, the main contributions along with the novelty of the proposed algorithm are summarized as follows:
• MDCN uniquely integrates the synergy of transfer learning and advanced data loader. Using transfer learning, the algorithm takes advantage of pre-trained models, leveraging the wealth of knowledge available from related tasks and domains. At the same time, sophisticated data loaders streamline the data ingestion process, ensuring efficient and optimal transmission of data to the algorithm.
• The incorporation of advanced optimizer techniques ensures that MDCN converges to a solution more efficiently, reducing training times and ensuring robustness.
• While building on the DenseNet architecture, MDCN introduces innovative modifications that enhance the network’s ability to capture and differentiate subtle features of various mango leaf diseases.
• MDCN achieves an impressive accuracy of 99.56% in detecting and classifying mango leaf diseases, setting a new benchmark for such applications when compared to some of the algorithms including both traditional algorithms (VGG-16, ResNet, Traditional-DenseNet, and CNN) and state-of-the-art algorithms (CNN with three hidden layers inspired by VGG-16, VGG-19 and Unet, CCA + cubic SVM and many more).
The remaining sections of this work are as follows; Section 2 contains a review of the existing works. The introduced algorithm is formulated and discussed in Section 3. Section 4 gives a summary of the experimental findings and finally, Section 5 concludes the work.
In India, where 50% of the population relies on agriculture, microbial diseases pose a critical threat to food security. Traditional identification methods, hampered by infrastructural limits, have struggled to address this issue promptly. However, the advent of Artificial Intelligence (AI) brings a promising solution. Leveraging AI, especially deep learning and transfer learning, it is now feasible to automatically detect plant diseases from raw images. This study centers on identifying diseases in Grape and Mango leaves [25]. Utilizing a dataset of 8,438 images from the Plant Village repository and local sources, a deep convolutional neural network (CNN), specifically the Alex Net architecture, was trained for disease detection and classification. With MATLAB’s capabilities, the system achieved an impressive 99% accuracy for Grape leaves and 89% for Mango leaves. This research was materialized into a practical tool, the “JIT CROPFIX” app for Android devices, ensuring that the agricultural community can directly harness this innovative technology.
Plant diseases can profoundly hinder the growth and vitality of plant species, making the early detection of these ailments an imperative task. Historically, a plethora of Machine Learning (ML) models have been designed and utilized in the endeavor to detect and classify the myriad of plant diseases. However, with the recent strides and innovations in the realm of Deep Learning (DL)—a specialized offshoot of ML—there is been a palpable shift in the potential and promise of this domain. The enhanced accuracy that DL brings to the table has revolutionized the approach towards disease detection in plants. A multitude of refined and reconfigured DL architectures have been introduced to the research world. These architectures, combined with a variety of sophisticated visualization techniques, have the unique ability to identify and classify even the subtlest symptoms of plant diseases [26]. This comprehensive review delves deep into the intricate world of DL models, shedding light on their invaluable role in visualizing and detecting various plant diseases.
Agriculture is pivotal in India due to its rising population and increasing food needs, making it essential to boost crop yields. One major hurdle is diseases caused by bacteria, fungi, and viruses. Plant disease detection methods can address this issue. Machine learning offers effective techniques for detecting these diseases, leveraging data for precision. A review by [27] delves into various techniques used for plant disease detection using AI, both in machine and deep learning. Deep learning, especially, has shown promise in computer vision for detecting plant diseases, marking significant achievements in the field. A comparative analysis of machine vs. deep learning techniques in various studies reveals the superiority of deep learning. To mitigate crop losses, deep learning can effectively identify leaf diseases from images.
Mango crops are plagued by various diseases, impacting both yield and quality, subsequently affecting international market prices. In many countries, diagnosing these diseases is challenging due to economic constraints and insufficient infrastructure. While plant pathologists employ numerous techniques for diagnosis, these methods are often costly, time-intensive, and can provide imprecise or biased results. Over the past decade, the push towards automating mango disease diagnosis has grown, utilizing ML (Machine Learning) and DL (Deep Learning) solutions. This literature [28] categorized these results into two: those built on traditional ML algorithms and those rooted in DL. Notably, the Convolutional Neural Network (CNN), a form of DL, has garnered widespread attention for its exceptional performance in recent years. A detailed analysis of these solutions uncovers their limitations and highlights potential challenges in automated mango disease diagnosis.
Deep learning, a subset of artificial intelligence, has gained prominence due to its capabilities in automatic learning and feature extraction. Its applications span video and image analysis, natural language understanding, and voice processing. In agriculture, especially in plant protection, deep learning has emerged as a pivotal tool for tasks like plant disease identification and pest assessment. By using deep learning, the biases in manual disease feature selection can be mitigated, offering a more objective disease feature extraction. This accelerates research efficiency and expedites technological advancements. This review [29] delves into the recent advancements in utilizing deep learning for crop leaf disease identification. We highlight current trends, challenges, and the synergy between deep learning and cutting-edge imaging techniques for disease detection. This paper aims to serve as a guide for researchers navigating plant disease and pest detection while also addressing existing challenges and areas requiring further exploration.
Plant diseases significantly impact grain quality and safety. Detecting these diseases, often manifested in leaves, requires specialized knowledge that many farmers lack. Thus, there is a pressing need for automatic disease recognition in precision agriculture. While various deep learning models and computer vision have been explored, deep learning has emerged as the favored solution due to its efficacy. Authors [30] introduced a unique transformer block that employs the transformer architecture for long-range features and soft split token embedding to harness limited pixel data. The incorporation of inception architecture and cross-channel feature learning enhances fine-grained feature detection. Our model outperforms existing models, achieving 86.89% accuracy on AI2018, 99.94% accuracy on Village Plant, 77.54% accuracy on PlantDoc, and 99.22% accuracy on iBean, underscoring its superiority in plant disease identification.
Plant diseases on leaves often pose classification challenges, such as low accuracy and overfitting. For agriculture, differentiating healthy from defective products demands accurate analysis. DCNN (Deep convolutional neural networks) have proven effective for detection and classification but come with limitations like needing extensive training data and parameter tweaking. This paper [31] introduced a structure tailored for multiple plant and fruit leaf disease classifications during feature extraction. We utilize a modified deep transfer learning model and apply model engineering (ME) for feature extraction. Multiple support vector machine (SVM) models boost processing speed and feature discrimination. Kernel parameters of the RBF (radial basis function) are determined during training. The analysis involved six leaf sets from the PlantVillage and UCI datasets, including apple, corn, cotton, grape, pepper, and rice, totaling about 90,000 images. The results highlight the model’s robust classification capabilities, paving the way for advanced leaf disease diagnostic applications in agriculture.
This research in [12] introduced an NNE (neural network ensemble) for MLDR (mango leaf disease recognition) to identify diseases more effectively than traditional methods. The authors used machine learning to monitor mango leaf symptoms. Training data was derived from images of diseased leaves, which were classified based on their symptoms. The system automatically identifies mango leaf diseases by comparing new images with the trained data. The approach achieved an average accuracy of 80% in disease detection. This method ensures healthier mango plants, allowing for quicker disease identification without needing an expert, and saving time over manual checks. This advancement can lead to proper treatment, boost mango production, and cater to global market demands.
The research was conducted in the Google Colab Environment. Agriculture, essential to countries with a 70% population dependent on it like some in Asia, goes beyond mere sustenance [32]. It is pivotal for feeding vast populations. The quality and yield of crops are jeopardized by diseases, which are a major concern. Detecting these diseases can mitigate production loss. The process involves uploading leaf images, preprocessing, segmenting, extracting features, and classifying them to identify diseases. This aids farmers in discerning plant ailments. Monitoring crop health is vital for successful agriculture. This requires efficient tools and software. Image processing techniques are applied to detect leaf diseases, ensuring robust crop production in the agriculture sector.
Automatic segmentation and identification of leaf diseases are challenging due to symptom variations. Proper segmentation is crucial for computer systems to recognize mango leaf diseases like Anthracnose and apical necrosis. Authors [33] introduced a CNN-based FrCNnet (Fully-convolutional-network) model for this purpose. The FrCNnet processes each pixel of the input data after certain preprocessing techniques. It is tested using a dataset from the Mango Research Institute in Multan, Pakistan, and compared its performance with other models like VGG-16, VGG-19, and Unet. Our model achieved 99.2% accuracy with a 0.8% false negative rate, outperforming the others. The FrCNnet allows for better feature recognition, leading to enhanced disease segmentation and identification. This automation aids pathologists and mango growers in disease detection.
Mangoes are in high demand, making the rapid management of plant diseases vital for optimal returns. Manual disease detection is costly and impractical in today’s digital age due to a shortage of experts and symptom variations. A primary challenge is segmenting diseased leaf sections, crucial for accurate disease identification. This study [34] introduced a novel segmentation method based on the leaf’s vein pattern. The “leaf vein-seg” approach segments based on this pattern. Subsequently, features are taken out and merged using CCA (canonical correlation analysis)-based fusion. Finally, a cubic SVM (support vector machine) validates the results. Achieving an accuracy of 95.5%, this model aids mango growers in timely disease detection and identification. Table 1 presents the comparison performance of existing approaches.
The study reveals that all algorithms share a common goal: to improve detection rates and accurately classify images, employing various techniques. While numerous algorithms exist, each claiming superior performance, only a handful truly elevates detection and classification performance, based on an in-depth literature review. Factors like computational efficiency, scalability, and generalization ability can hamper an algorithm’s performance. To address these challenges and outshine existing algorithms, a new algorithm has been proposed. This advanced image detection and classification approach prioritizes performance and accuracy. The MDCN (Modified Dense Convolution Network) not only addresses these challenges but also optimizes training time and requires fewer epochs. This innovative algorithm builds upon the foundation of the existing Dense Net algorithm.
3.1 Mango Leaf Disease Dataset
The diagnosis of Mango Leaf Disease requires a rich and comprehensive dataset, which has been diligently curated and collated from the MangoLeafBD Dataset. With a total of 4,000 images, it provides a comprehensive overview of mango leaf health and ailments. Interestingly, out of these, 1,800 images are of distinct, individual leaves, showcasing the range and variability present in real-world scenarios. The diseases include Cutting Weevil, Bacterial Canker, Anthracnose, Gall Midge, Die Back, Sooty Mould, and Powdery Mildew. For comparative analysis and to provide a benchmark of health, the dataset also contains images of healthy mango leaves. Each of the eight categories—the seven diseases and the healthy leaves—encompasses 500 images, ensuring a balanced representation. Fig. 1 shows the leaf samples from certain classes of the disease (each of these images is resized to 240 × 240 size).

Figure 1: Sample of leaf images along with the class name after resizing
3.2 Normalization and Splitting
To improve the performance of pre-trained models, it is important to normalize the input images in the same way that the models were trained. This can be done by using a transformation sequence that converts the images to a specific format and range of values. One common transformation sequence is to read the images into PILImage format using Torchvision, and then convert them to FloatTensor format with a range of [0,1] using the ToTensor function. The normalization of the image can be done by subtracting the mean pixel value (0.5) from each image and then dividing by the standard deviation (0.5). This will convert the image values to the range of [0,1].
In Eq. (1), µ represents the mean and σ stands for the standard deviation, with both values set at 0.5. C refers to the mini-batch channel, while H signifies height and W indicates width. The minimum expected values for H and W are 224. The normalization of the MangoLeafBD dataset is performed using the mean and standard deviation of the pixel values. The mean pixel value for each channel is 0.467, 0.489, and 0.437, and the standard deviation for each channel is 0.232, 0.287, and 0.245. Images are normalized by deducting the mean pixel value from each channel and subsequently dividing it by the channel’s standard deviation. This converts the pixel values to a range of [0,1]. To mitigate the issues associated with imbalanced datasets, the data is partitioned in an 80:20 ratio for training and validation purposes, ensuring distinct datasets for each. The images are sourced from individual folders, each named after its respective class, which facilitates automatic labeling. The DataLoader then efficiently manages these labeled images, keeping track of both the Training Data (Image) and the corresponding Label (Class Name). This process organizes the dataset into eight distinct classes: one labeled as ‘normal’ representing a healthy leaf, and the other seven labeled based on the specific disease they represent. With the data classes established, the data is then directed to be processed either by CUDA (GPU) or the CPU, paving the way for model definition. Additionally, Torchvision encompasses a subset that offers model definitions for a range of tasks, covering object detection, image classification, and semantic segmentation, among others.
Transfer learning allows deep learning models previously trained on large datasets, such as the Open Images Dataset with its 600 classes, to achieve faster and more accurate results on new datasets. Rather than solely retraining the Fully Connected layers, this approach often retrains all layers, tailoring the model to specific tasks like mango leaf image classification. By refining the Convolutional Neural Network (CNN) layers, either wholly or partially, the model aligns pre-trained features with specialized features of the new dataset. Backpropagation utilizes the backward function, and the zero-grad optimizer clears past gradients, ensuring no carryover from previous loss stages. Having been pre-trained on the expansive ImageNet dataset, these models possess foundational features valuable across various computer vision tasks. This foundational knowledge, essential for feature extraction, is applied to new images even if their classes differ from the original dataset. By integrating the mango leaf images into the already established Open Images Dataset and combining both pre-trained and newly learned weights, the model evolves into an effective feature extractor for the incoming image set.
Therefore, to use the concept of transfer learning, the time-training of the proposed model will reduce the already learned foundational features required to classify a leave will be passed to the MDCN from the pre-trained CNN model in the form of parameters like weights and bias. There is another reason to use this concept, that is most of the deep-learning models experience a local minima trap, wherein a particular iteration, the model gets stuck to further optimize and produce a result due to which, the training knowledge learned during the last few iterations will be skipped, leading to low performance. By integrating this transfer-learning concept, the model will now initial the weights according to the weights and other parameters obtained from the pre-trained model, and in this process, the model’s accuracy will also increase, and thus overcome the local minima trap. The transfer learning objective is to find the optimal weight parameters
where
In Eq. (3),
Early Stopping is a vital technique in machine learning, primarily used to determine the most optimal model configuration under a given set of conditions. Essentially, during the training process, there exists a juncture at which the model’s performance on the validation dataset plateaus or even deteriorates. When the model reaches this saturation point, where there is no noticeable improvement in its output, the Early Stopping technique is used. The conditions triggering this approach often revolve around achieving the best accuracy or registering the lowest loss. For the proposed model, selecting the iteration with the lowest loss is imperative to harness the highest accuracy. While training this model, it is crucial to note that the weights of the pre-existing, pre-trained network remain untouched. The emphasis, instead, is on the freshly instituted classifier, which is meticulously trained to derive weights that best complement the new data combinations. The overarching goal of such fine-tuning is dual-pronged: first, to assimilate and adapt to novel features and, second, to seamlessly integrate and process the newly introduced mango leaf data, building upon the foundation provided by the pre-existing dataset.
In this paper, the objective is to classify mango leaf images into two categories: healthy and the associated disease type. The approach incorporates the deep learning frameworks of PyTorch and torchvision. These pre-trained models offer robust control over overfitting, ensuring optimized results from inception. As depicted in Fig. 2, DenseNet’s block diagram describes a structure with four dense blocks, each with a growth rate (k) of 32. The “121” in “DenseNet-121” indicates the total layer count within the neural network. The proposed MDCN architecture utilizes a variety of layers, comprising multiple convolution and pooling layers distributed across dense and transition blocks. The architecture includes three transition layers (with 6, 12, and 24 layers, respectively), a classification layer with 16 layers, and four DenseBlocks. DenseNet’s design facilitates feature reuse and reduces the model’s parameters, bolstering the model’s accuracy when diagnosing mango leaf diseases using the provided dataset. As mentioned earlier, the proposed model also makes use of other features like transfer learning, fine-tuning, and optimization to enhance the accuracy of the overall model.

Figure 2: Components inside a dense block
In DenseNet, each dense layer combines outputs from all preceding layers, ensuring consistency in depth. Fig. 3 demonstrates DenseNet’s workflow, showcasing how it processes mango leaf images through a mix of Dense Blocks and Transition Layers. As an image navigates through the dense blocks, feature map dimensions remain consistent, though the number of filters might vary. Following each dense block, the image progresses to a transition layer, which manages convolution and pooling. Importantly, these layers handle downsampling outside of the dense blocks. To concatenate features, feature maps within a dense block must have matching sizes. Incorporating a bottleneck convolution layer before each convolution can efficiently reduce input feature maps, optimizing computational performance. The transition layers in DenseNet are characterized by batch normalization, a convolution layer, and an average pooling layer.

Figure 3: Proposed model architecture
Fig. 4 provides an in-depth illustration of the operations within the dense block of the DenseNet architecture. This block is composed of a BN (batch normalization) layer, a ReLU (rectified linear unit) activation function, and convs (convolution operations). After the last dense block, there is a global average pooling layer, which subsequently connects to a Softmax classifier. Given that DenseNet contains L layers, there will be L (L + 1)/2 direct connections associated with these layers.

Figure 4: MDCN with four dense blocks
As depicted in Fig. 4, the layer
The combination of feature maps from layer 0 to k−1 is represented as
In Eq. (5), all the inputs for H are merged into a unified tensor during processing. Within the DenseNet structure, the sizes of feature maps can be altered via convolution and pooling operations. In the suggested model, Batch Normalization is employed, a method designed to standardize either the inputs or the activations from the preceding layer. In the development of transfer learning and convolutional neural networks, the rectified linear activation function is commonly used. DenseNet is segmented into Dense blocks, with each block having distinct filters but maintaining consistent dimensions. The Transition Layer undertakes down-sampling and employs batch normalization. Average pooling calculates the mean for specific sections of the feature map, thereby down-sampling each segment of the feature map to its average value. The model utilizes the Cross Entropy loss function and employs the Adam optimizer for weight updates. A learning rate of 1e-05 has been set, with a batch size designated as 5. This deep learning architecture consists of a total of 5,968,048 parameters.
In the outlined framework, we evaluated multiple optimizers to ascertain the maximum accuracy for the suggested model. We contrasted the performance of SGD (Stochastic Gradient Descent), RMSProp, and Adam optimizers. Adam is a fusion of Stochastic Gradient Descent and RMSProp, incorporating momentum. Notably, Adam determines the learning rate for each parameter individually. The formula used to calculate Adam is as given in Eqs. (6)–(9):
Here,
In the proposed system, experiments were carried out within a simulated environment utilizing VSCode (Visual Studio Code) as the integrated development environment, with the coding language employed being Python 3.8. on a PC with the following configuration: 12th Gen Intel(R), Core (TM) i7-12700H, 2.30 GHz, NVIDIA Corporation with the donation of the RTX3060 graphic card, and 6.00 GB RAM. Also, multiple libraries were used in the proposed model such as Tensorflow (version 2.10.0), Keras (version 2.10.0), Pandas (2.0.2), Numpy (version 1.25.2), and matplotlib (version 3.7.2).
4.2 Initialization of Variables
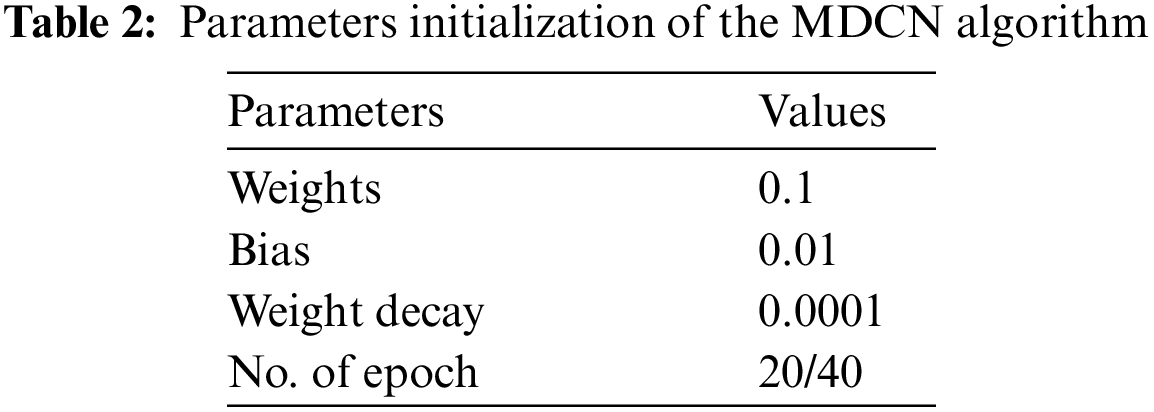
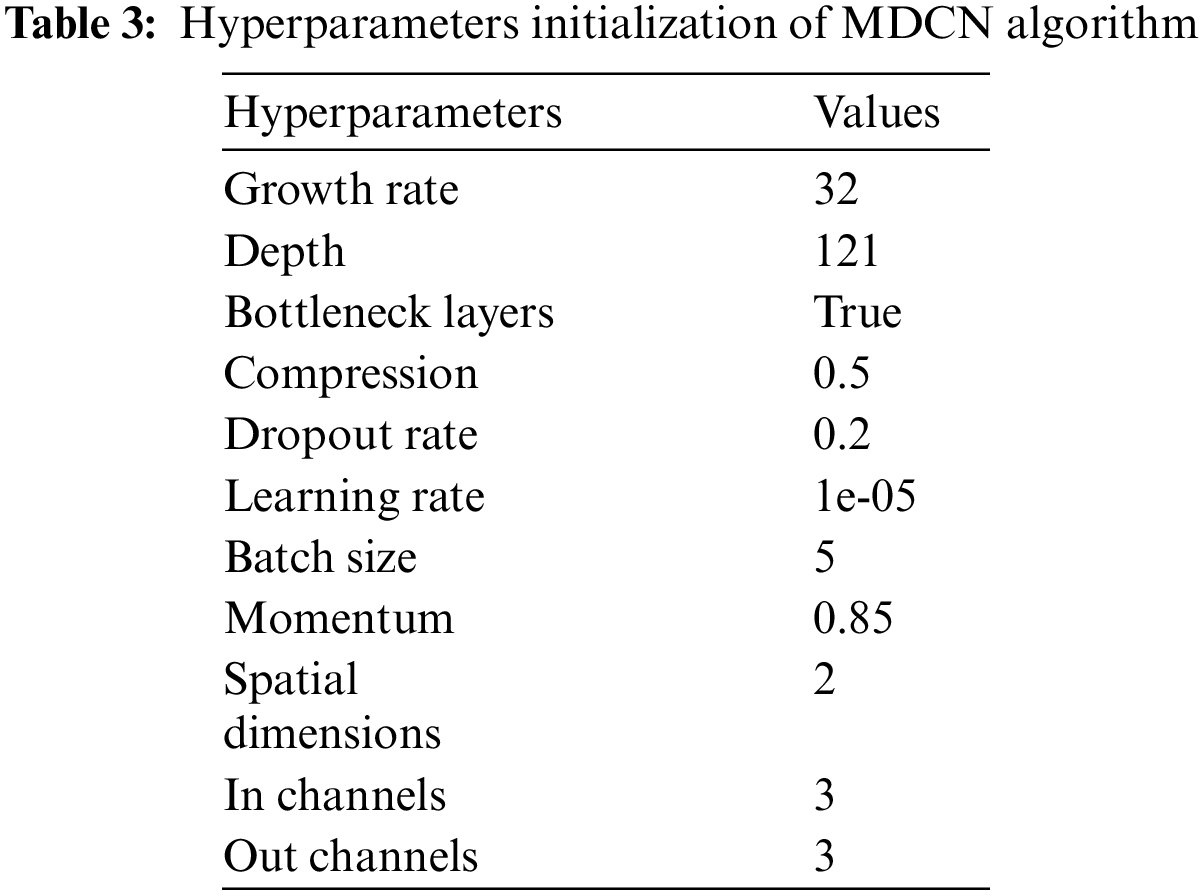
The initial settings for the parameter and hyperparameters of the proposed algorithm are detailed in Tables 2 and 3. The parameters typically represent the values derived when searching for the optimal solution set. They act as mutable variables that influence the extraction of the final set of solutions. In contrast, hyperparameters are constant and play a central role in shaping the parameter values in each iteration.
Besides identifying the image’s class label, the image visualization function will also display the image marked with its predicted class. Once this process is done, the results can be seen as illustrated in Fig. 5.

Figure 5: The real prediction result of the proposed model
The effectiveness of the suggested system is assessed using various metrics, including Precision, Accuracy, F1 score, Recall, False Positive Rate True Positive Rate. Accuracy is determined by comparing the number of correctly classified entries to the overall count, as illustrated in Eq. (10). Precision is calculated as the proportion of correctly predicted abnormal cases over the total cases that were identified as abnormal, as expressed in Eq. (11). Recall, as represented in Eq. (12), is the fraction of correctly identified abnormal cases relative to the total number of genuine abnormal cases. The F1 score provides a balanced measure between Precision and Recall, serving as their harmonic mean to evaluate the system’s accuracy, as depicted in Eq. (13):
Accuracy is further assessed using the ROC (Receiver Operating Characteristics) curve and the AUC (Area Under the ROC curve). This criterion signifies the probability that a haphazardly chosen positive instance is more likely to receive a higher positive prediction than a randomly selected negative instance.
The suggested method is tested on the MangoLeafBD dataset encompassing both normal and abnormal leaves. In the devised MDCN strategy, activation functions like ReLU and softmax are utilized. We opted for a batch size of 5, employed the Adam optimizer, and chose cross-entropy as the loss function. The training was undertaken with varying epoch values, ranging between 20 and 40. It was observed that the model achieved optimal validation accuracy and minimized loss at 40 epochs. The performance metrics of the model, namely accuracy and loss over 20 epochs, are illustrated in Figs. 6 and 7, respectively. Moreover, the time taken to train the proposed model for 20 epochs is around 1 h 29 min, and 15 s, which is less compared to other existing models.
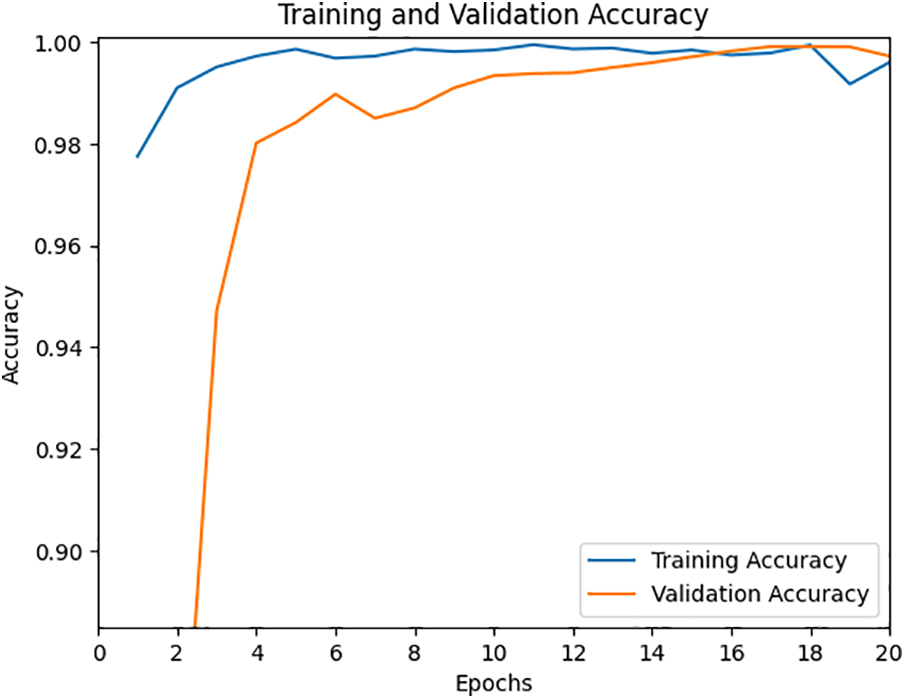
Figure 6: Accuracy during training and validation for 20 epochs

Figure 7: Loss during training and validation for 20 epochs
The proposed model’s performance in terms of accuracy and loss with 40 epochs is given in Figs. 8 and 9. Moreover, the time taken to train the proposed model for 20 epochs is around 2 h 45 min, and 31 s.

Figure 8: Accuracy during training and validation for 40 epochs

Figure 9: Loss during training and validation for 40 epochs
As depicted in Figs. 8 and 9, by employing 40 epochs, our proposed model significantly improves detection accuracy, reaching a commendable 99.56%. The empirical outcomes indicate that our system outperforms other existing algorithms in detecting various diseases. The ROC curves, showcased in Figs. 10 and 11, detail the model’s performance across different classes for both 20 and 40 epochs. Notably, with 40 epochs, the True Detection Rate (TDR) is impressively high while the False Detection Rate (FDR) remains minimal, emphasizing the model’s capability to accurately identify issues. The positioning of the ROC curve towards the upper left corner in Figs. 10 and 11 reaffirms the superior classification efficacy of our approach.

Figure 10: ROC curve for 20 epochs

Figure 11: ROC curve for 40 epochs
In the next stage of the current work, the traditional approaches such as Visual Geometry Group (VGG-16), Residual Network (ResNet), Traditional-DenseNet, and Convolution Neural Network (CNN) are considered and compared with the performance of the proposed algorithm. The performance of traditional and proposed MDCN is assessed by using evaluation metrics such as accuracy, precision, recall, and F1 score. Fig. 12 presents a performance comparison of the proposed model with the VGG-16, ResNet, DenseNet, and CNN at 20 epochs in terms of accuracy.

Figure 12: Performance comparison of MDCN with traditional approaches at 20 epochs
Fig. 13 presents a performance comparison of the proposed model with the VGG-16, ResNet, DenseNet, and CNN at 40 epochs in terms of accuracy. Tables 4 and 5 summarize the performance of the traditional approaches namely VGG-16, ResNet, DenseNet, and CNN with the proposed MDCN at 20 epochs and 40 epochs, respectively. It is observed from Tables 4 and 5, that the proposed MDCN attains better performance for detecting mango leaf diseases when compared to the traditional approaches.

Figure 13: Performance comparison of MDCN with traditional approaches at 40 epochs
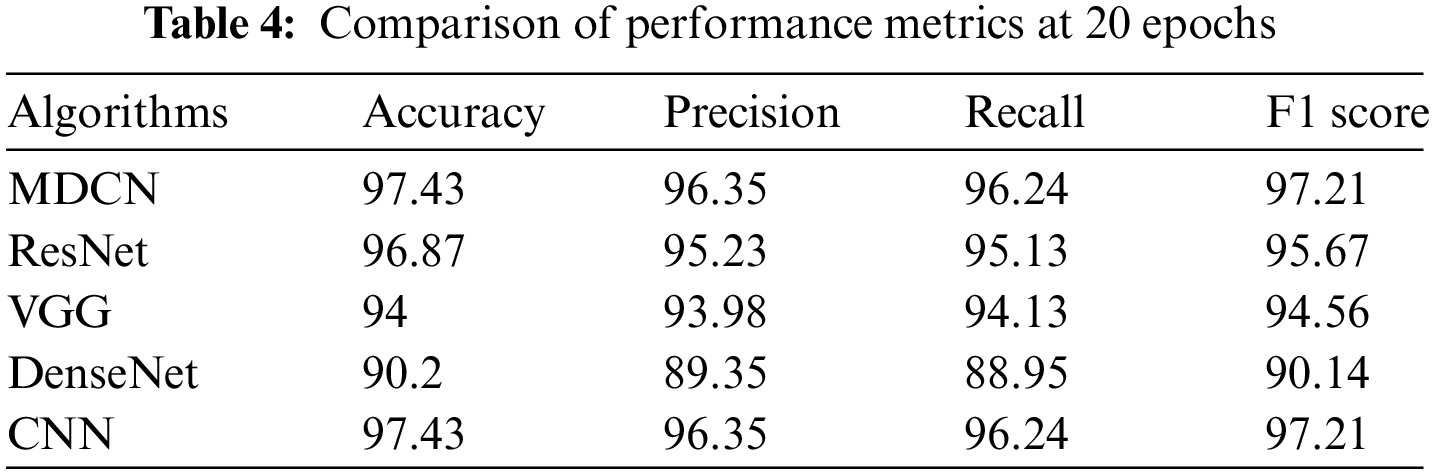

Finally, the proposed MDCN is compared with the state-of-the-art algorithms in this arena and is summarized in Table 6.
In this work, an innovative model was presented that combines the strengths of deep learning and convolutional neural networks. Specifically, this model incorporated the DenseNet architecture, a renowned deep learning structure, with the primary objective of detecting leaf diseases. The data that facilitated this study was drawn from the MangoLeafBD dataset, a comprehensive collection of images and information about mango leaf diseases. The training process for this innovative model was meticulously carried out at two different intervals, 20 epochs, and 40 epochs, to ascertain its robustness and adaptability. To further refine the performance of the system, the model integrated the concept of Early Learning. This concept, when applied, plays a pivotal role in enhancing the efficiency and overall output of deep learning systems. Transfer learning technique that repurposes pre-trained models for new tasks, was seamlessly incorporated into the DenseNet framework. This algorithm also utilized the Early Stopping mechanism, a range of optimizers, various loss functions, and a Learning Rate (LR) Scheduler, which adjusted learning rates for better convergence. The culmination of this research led to some groundbreaking findings. The newly developed MDCN algorithm displayed an impressive true detection rate while ensuring that false detections were kept to a minimum. This was particularly noteworthy when juxtaposed against pre-existing systems. When subjected to 40 epochs of training, the model achieved a staggering accuracy rate of over 99%. Furthermore, the system scored exceptionally high in terms of precision, recall, and F1 score, with average values being 97.83%, 97.73%, and 98.29%, respectively. These results unequivocally demonstrate that this model surpasses many of its contemporaries in performance.
Future efforts will involve integrating the algorithm with segmentation code to pinpoint the precise location of diseases on leaves. This integration seeks to offer a deeper understanding of the classification process. This expansion aims not only to enhance the accuracy of disease identification but also to contribute to the broader field of agricultural research, potentially aiding in the development of strategies for disease prevention in mango cultivation.
Acknowledgement: The authors wish to express their thanks to VIT management for their extensive support during this work.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: The authors confirm their contribution to the paper as follows: study conception and design: Chirag Chandrashekar, K.P. Vijayakumar, K. Pradeep and A. Balasundaram, data collection: Chirag Chandrashekar, K.P. Vijayakumar, K. Pradeep and A. Balasundaram, analysis and interpretation of results: Chirag Chandrashekar, K.P. Vijayakumar, K. Pradeep and A. Balasundaram,draft manuscript preparation: Chirag Chandrashekar, K.P. Vijayakumar, K. Pradeep and A. Balasundaram. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: MangoLeafBD Dataset, 2022, https://dx.doi.org/10.17632/hxsnvwty3r.1 (accessed on 03/08/2023).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. E. M. Yahia, J. J. Ornelas Paz, J. K. Brecht, P. García-Solís, and M. E. Maldonado Celis, “The contribution of mango fruit (Mangifera indica L.) to human nutrition and health,” Arab. J. Chem., vol. 16, no. 7, pp. 1–27, 2023. doi: 10.1016/j.arabjc.2023.104860. [Google Scholar] [CrossRef]
2. K. Mulungu, B. W. Muriithi, M. Kassie, and F. M. Khamis, “The economic performance of mango integrated pest management practices at different scales of production,” Front. Insect Sci., vol. 3, no. 1180568, pp. 1–11, 2023. doi: 10.3389/finsc.2023.1180568. [Google Scholar] [CrossRef]
3. Y. P. Wang et al., “Optimizing plant disease management in agricultural ecosystems through rational in-crop diversification,” Front. Plant Sci., vol. 12, pp. 1–10, 2021. [Google Scholar]
4. I. Buja et al., “Advances in plant disease detection and monitoring: From traditional assays to in-field diagnostics,” Sens., vol. 21, no. 2129, pp. 1–22, 2021. doi: 10.3390/s21062129 [Google Scholar] [PubMed] [CrossRef]
5. L. Alzubaidi et al., “Review of deep learning: Concepts, CNN architectures, challenges, applications, future directions,” J. Big Data, vol. 8, no. 2021, pp. 1–74, 2021. [Google Scholar]
6. V. Sze, Y. H. Chen, T. J. Yang, and J. S. Emer, “Efficient processing of deep neural networks: A tutorial and survey,” in Proc. IEEE, 2017, pp. 2295–2329. [Google Scholar]
7. T. Zhou, X. Ye, H. Lu, X. Zheng, S. Qiu and Y. Liu, “Dense convolutional network and its application in medical image analysis,” BioMed. Res. Int., vol. 2022, pp. 1–22, 2022. doi: 10.1155/2022/2384830 [Google Scholar] [PubMed] [CrossRef]
8. K. Weiss, T. M. Khoshgoftaar, and D. Wang, “A survey of transfer learning,” J. Big Data, vol. 3, no. 9, pp. 1–40, 2016. doi: 10.1186/s40537-016-0043-6. [Google Scholar] [CrossRef]
9. S. I. Ahmed et al., “MangoLeafBD: A comprehensive image dataset to classify diseased and healthy mango leaves,” Data Bri., vol. 47, no. 108941, pp. 1–11, 2023. doi: 10.1016/j.dib.2023.108941 [Google Scholar] [PubMed] [CrossRef]
10. G. Geetharamani and J. Arunpandian, “Identification of plant leaf diseases using a nine-layer deep convolutional neural network,” Comput. Electr. Eng., vol. 7, pp. 323–338, 2019. doi: 10.1016/j.compeleceng.2019.04.011. [Google Scholar] [CrossRef]
11. D. Singh, N. Jain, P. Jain, P. Kayal, S. Kumawat and N. Batra, “PlantDoc: A dataset for visual plant disease detection,” CoDS COMAD 2020: Proc. of the 7th ACM IKDD CoDS and 25th COMAD, vol. 3, pp. 249–253, 2019. doi: 10.1145/3371158.3371196. [Google Scholar] [CrossRef]
12. M. R. Mia, S. Roy, S. K. Das, and M. A. Rahman, “Mango leaf disease recognition using neural network and support vector machine,” Iran J. Comput. Sci., vol. 3, no. 3, pp. 185–193, 2020. doi: 10.1007/s42044-020-00057-z. [Google Scholar] [CrossRef]
13. J. H. Crane and R. Gazis, “Bacterial black spot (BBS) of mango in Florida,” EDIS, vol. 2020, no. 5, 2020. doi: 10.32473/edis-hs1369-2020. [Google Scholar] [CrossRef]
14. G. S. Shekhawat and P. N. Patel, “Studies on bacterial canker of mango,” J. Plant Dis. Prot., vol. 82, no. 3, pp. 129–138, 1975. [Google Scholar]
15. M. H. Rashid, H. F. El Taj, and C. Jung, “Life-table study of mango leaf cutting weevil, Deporaus marginatus Pascoe (Coleoptera: Curculionidae) feeding on four mango cultivars,” J. AsiaPac. Entomol., vol. 20, no. 2, pp. 353–357, 2017. doi: 10.1016/j.aspen.2017.02.004. [Google Scholar] [CrossRef]
16. F. H. Kamil, E. E. Saeed, K. A. El-Tarabily, and S. F. AbuQamar, “Biological control of mango dieback disease caused by Lasiodiplodia theobromae using streptomycete and non-streptomycete actinobacteria in the United Arab Emirates,” Front. Microbiol., vol. 9, no. 829, pp. 1–19, 2018. doi: 10.3389/fmicb.2018.00829 [Google Scholar] [PubMed] [CrossRef]
17. E. E. Saeed et al., “Detection and management of mango dieback disease in the United Arab Emirates,” Int. J. Mol. Sci., vol. 18, no. 2086, pp. 1–18, 2017. doi: 10.3390/ijms18102086 [Google Scholar] [PubMed] [CrossRef]
18. J. E. Peña, A. I. Mohyuddin, and M. Wysoki, “A review of the pest management situation in mango agroecosystems,” Phytoparasitica, vol. 26, no. 2, pp. 129–148, 1998. doi: 10.1007/BF02980680. [Google Scholar] [CrossRef]
19. M. Nasir, S. M. Mughal, T. Mukhtar, and M. Z. Awan, “Powdery mildew of mango: A review of ecology, biology, epidemiology and management,” Crop Prot., vol. 64, no. 2, pp. 19–26, 2014. doi: 10.1016/j.cropro.2014.06.003. [Google Scholar] [CrossRef]
20. A. Rezazadeh, “Mango tree sooty mold—UF/IFAS extension St. Lucie County. UF/IFAS extension St. Lucie County,” 2020. Accessed: Aug. 03, 2023. [Online]. Available: https://blogs.ifas.ufl.edu/stlucieco/2020/09/21/mango-tree-sooty-mold/ [Google Scholar]
21. Z. P. Jiang, Y. Y. Liu, Z. E. Shao, and K. W. Huang, “An improved VGG-16 model for pneumonia image classification,” Appl. Sci., vol. 11, no. 11185, pp. 1–19, 2021. doi: 10.3390/app112311185. [Google Scholar] [CrossRef]
22. D. Sarwinda, R. H. Paradisa, A. Bustamam, and P. Anggia, “Deep learning in image classification using residual network (ResNet) variants for detection of colorectal cancer,” Procedia Comput. Sci., vol. 179, pp. 423–431, 2021. doi: 10.1016/j.procs.2021.01.025. [Google Scholar] [CrossRef]
23. N. Hasan, Y. Bao, A. Shawon, and Y. Huang, “DenseNet convolutional neural networks application for predicting COVID-19 using CT image,” SN Comput. Sci., vol. 2, no. 5, pp. 1–11, 2021. doi: 10.1007/s42979-021-00782-7 [Google Scholar] [PubMed] [CrossRef]
24. S. Indolia, A. K. Goswami, S. P. Mishra, and P. Asopa, “Conceptual understanding of convolutional neural network—A deep learning approach,” Procedia Comput. Sci., vol. 132, no. 4, pp. 679–688, 2018. doi: 10.1016/j.procs.2018.05.069. [Google Scholar] [CrossRef]
25. U. S. Rao et al., “Deep learning precision farming: Grapes and mango leaf disease detection by transfer learning,” Glob. Transit., vol. 2, pp. 535–544, 2021. doi: 10.1016/j.gltp.2021.08.002. [Google Scholar] [CrossRef]
26. M. H. Saleem, J. Potgieter, and K. M. Arif, “Plant disease detection and classification by deep learning,” Plants, vol. 8, no. 11, pp. 1–22, 2019. doi: 10.3390/plants8110468 [Google Scholar] [PubMed] [CrossRef]
27. C. Jackulin and S. Murugavalli, “A comprehensive review on detection of plant disease using machine learning and deep learning approaches,” Meas. Sens., vol. 24, pp. 100441, 2022. doi: 10.1016/j.measen.2022.100441. [Google Scholar] [CrossRef]
28. D. Faye, I. Diop, and D. Dione, “Mango diseases classification solutions using machine learning or deep learning: A review,” J. Compu. Commun., vol. 10, no. 12, pp. 16–28, 2022. doi: 10.4236/jcc.2022.1012002. [Google Scholar] [CrossRef]
29. L. Li, S. Zhang, and B. Wang, “Plant disease detection and classification by deep learning a review,” IEEE Access, vol. 9, pp. 56683–56698, 2021. doi: 10.1109/ACCESS.2021.3069646. [Google Scholar] [CrossRef]
30. S. Yu, L. Xie, and Q. Huang, “Inception convolutional vision transformers for plant disease identification,” Internet of Things, vol. 21, no. 100650, pp. 1–18, 2023. doi: 10.1016/j.iot.2022.100650. [Google Scholar] [CrossRef]
31. M. S. Anari, “A hybrid model for leaf diseases classification based on the modified deep transfer learning and ensemble approach for agricultural AIoT-based monitoring,” Comput. Intell. Neurosci., vol. 2022, no. 6504616, pp. 1–15, 2022. doi: 10.1155/2022/6504616 [Google Scholar] [PubMed] [CrossRef]
32. N. Manoharan, V. J. Thomas, and D. A. S. Dhas, “Identification of mango leaf disease using deep learning,” in Proc. ASIANCON, Pune, India, 2021, pp. 1–8. [Google Scholar]
33. R. Saleem, J. H. Shah, M. Sharif, and G. J. Ansari, “Mango leaf disease identification using fully resolution convolutional network,” Comput. Mater. Contin., vol. 69, no. 3, pp. 3581–3601, 2021. doi: 10.32604/cmc.2021.017700 [Google Scholar] [CrossRef]
34. R. Saleem, J. H. Shah, M. Sharif, M. Yasmin, H. S. Yong and J. Cha, “Mango leaf disease recognition and classification using novel segmentation and vein pattern technique,” Appl. Sci., vol. 11, no. 11901, pp. 1–12, 2021. doi: 10.3390/app112411901. [Google Scholar] [CrossRef]
35. S. B. Ullagaddi and S. V. Raju, “Disease recognition in Mango crop using modified rotational kernel transform features,” in Proc. ICACCS, Coimbatore, India, 2017, pp. 1–8. [Google Scholar]
36. K. Srunitha and D. Bharathi, “Mango leaf unhealthy region detection and classification, computational vision and bio inspired computing,” in Lecture Notes in Computational Vision and Biomechanics. Springer, 2018, vol. 28, pp. 422–436. [Google Scholar]
37. U. P. Singh, S. S. Chouhan, S. Jain, and S. Jain, “Multilayer convolution neural network for the classification of mango leaves infected by anthracnose disease,” IEEE Access, vol. 7, pp. 43721–43729, 2019. doi: 10.1109/ACCESS.2019.2907383. [Google Scholar] [CrossRef]
38. S. Amisha, K. B. Rajneet, M. Jatinder, and S. Vinod, “Mango leaf diseases detection using deep learning,” Int. J. Knowl. Based Comput. Syst., vol. 10, pp. 40–44, 2022. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF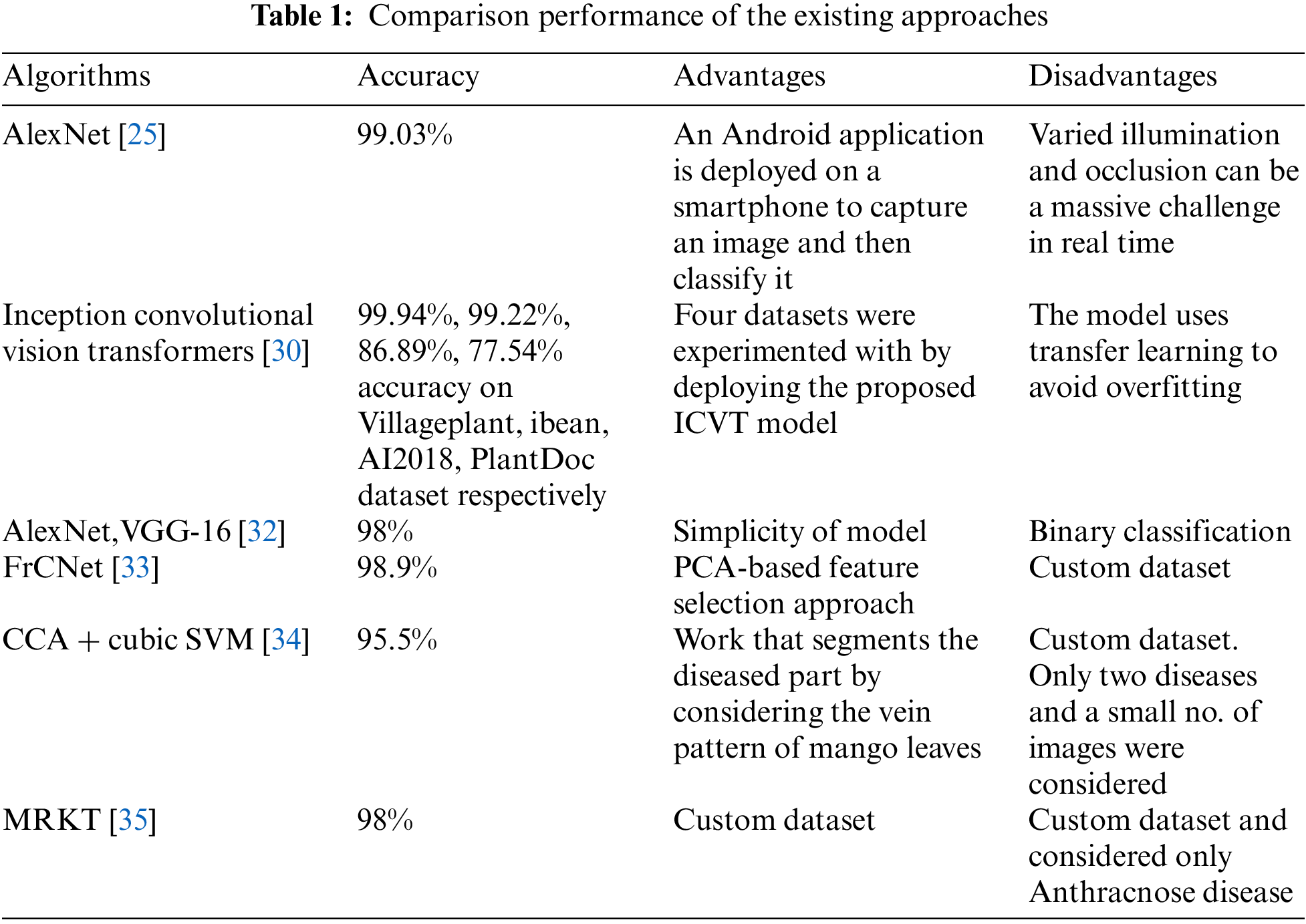

 Downloads
Downloads
 Citation Tools
Citation Tools