 Open Access
Open Access
ARTICLE
Attention Guided Multi Scale Feature Fusion Network for Automatic Prostate Segmentation
1 School of Information and Communication Engineering, Hainan University, Haikou, China
2 College of Computer Science and Technology, Hainan University, Haikou, China
3 Urology Department, Haikou Municipal People’s Hospital and Central South University Xiangya Medical College Affiliated Hospital, Haikou, China
4 School of Information Science and Technology, Hainan Normal University, Haikou, China
* Corresponding Author: Mengxing Huang. Email:
Computers, Materials & Continua 2024, 78(2), 1649-1668. https://doi.org/10.32604/cmc.2023.046883
Received 18 October 2023; Accepted 06 December 2023; Issue published 27 February 2024
Abstract
The precise and automatic segmentation of prostate magnetic resonance imaging (MRI) images is vital for assisting doctors in diagnosing prostate diseases. In recent years, many advanced methods have been applied to prostate segmentation, but due to the variability caused by prostate diseases, automatic segmentation of the prostate presents significant challenges. In this paper, we propose an attention-guided multi-scale feature fusion network (AGMSF-Net) to segment prostate MRI images. We propose an attention mechanism for extracting multi-scale features, and introduce a 3D transformer module to enhance global feature representation by adding it during the transition phase from encoder to decoder. In the decoder stage, a feature fusion module is proposed to obtain global context information. We evaluate our model on MRI images of the prostate acquired from a local hospital. The relative volume difference (RVD) and dice similarity coefficient (DSC) between the results of automatic prostate segmentation and ground truth were 1.21% and 93.68%, respectively. To quantitatively evaluate prostate volume on MRI, which is of significant clinical significance, we propose a unique AGMSF-Net. The essential performance evaluation and validation experiments have demonstrated the effectiveness of our method in automatic prostate segmentation.Keywords
Prostate disease is a significant disease that troubles middle-aged and older men in modern times, seriously affecting their lives and health. According to statistics, the annual incidence rate of prostate cancer is 1.41 million, accounting for 14.1% of all male cancer cases [1]. Quantitative estimation of prostate volume (PV) is very vital in the diacrisis of prostate disease. The average volume of a healthy prostate is
In the past few decades, based on traditional medical image processing technology, automatic prostate segmentation technology has shown significant progress [5]. However, owing to the complexity of prostate MRI images, accurate prostate segmentation remains challenging [2]. Many difficulties are encountered in segmentation, such as, differences in prostate size and shape between patients, imaging artifacts, unclear borders between glands and their adjacent tissues, and changes in image quality. Convolutional neural networks (CNNs) [6] are widely used in medical image analysis due to their powerful computing power and adaptive algorithms. Many CNN models have been applied to prostate segmentation for MRI images [7–9]. Furthermore, the U-Net [10] network structure further improved the accuracy of medical image segmentation. Many improved neural networks [9,11,12] based on U-Net have achieved excellent results as well. U-Net is suitable for medical image segmentation as it combines low-resolution information (providing the basis for object category identification) and high-resolution information (providing the basis for accurate segmentation and positioning). Therefore, it has become the baseline for most medical image semantic segmentation tasks, and has inspired many researchers to consider U-shaped semantic segmentation networks.
Artificial intelligence technology has been widely applied and developed in image processing [13–16]. Prostate segmentation in MRI images enables pathologists to be more time-efficient and identify more accurate treatments. Owing to the lack of clear edges and complex background textures between the prostate image and those of other anatomical structures, rendering it challenging to segment the prostate from 3D MRI images. Therefore, we propose a 3D attention-guided multi-scale feature fusion network (3D AGMSF-Net) to segment prostate MRI images. The main contributions of this study can be summarized as follows:
1. We propose a 3D AGMSF-Net, which is a fresh model applicable to segmentation challenges in 3D prostate MRI images.
2. We propose an attention mechanism for extracting multi-scale features, which is embedded in the overall jump connection of the baseline model and takes three different scale features as inputs.
3. We introduce a 3D transformer module to enhance global feature representation by adding it during the transition phase from encoder to decoder.
4. We design a feature fusion mechanism to fuse multi-scale features to express more comprehensive information.
5. On a dataset of a local hospital, we test the proposed technique. According to the results, the suggested method can segment prostate of MRI images more precisely.
The rest of this paper is organized as follows. Section 2 provides a review of previous work on prostate segmentation tasks. Section 3 introduces our dataset and provides a detailed description of the AGMSF-Net scheme and various modules designed. In Section 4, we first introduce data augmentation, implementation details, and evaluation metrics. Then we conduct a series of experiments to verify the effectiveness of our designed AGMSF-Net in prostate segmentation tasks. Finally, a comprehensive discussion and summary are shown in Sections 5 and 6.
2.1 Traditional Prostate Segmentation Algorithms
Many studies have been conducted on traditional algorithms for automatic prostate segmentation. The existing traditional prostate automatic segmentation methods are roughly divided into graph cutting [17–20], shape and atlas [21–24], deformable models [25–27], and clustering methods [28]. Qiu et al. [17] proposed a new multi-region segmentation method for simultaneously segmenting the prostate and its two central sub-regions. Mahapatra et al. [18] used random forest (RF) and graph cut methods to solve the problem of automatic prostate segmentation. Tian et al. [19,20] proposed a 3D graph cut algorithm based on superpixels for an automatic prostate segmentation method. The above methods are sensitive to noise, and are not sensitive to segmentation with insignificant differences in grayscale and overlapping grayscale values of different scales. Gao et al. [21] proposed a unified shape-based framework to extract the prostate from MRI prostate images. Ou et al. [22] proposed an automatic pipeline based on multiple atlases to the segment prostate in MRI images. Tian et al. [23] proposed a two-stage prostate segmentation method based on a fully automatic multi-atlas framework to overcome the problem of different MRI images having different fields of vision and more significant anatomical variability around the prostate. Yan et al. [24] proposed a label image constraint atlas selection method to directly calculate the distance between the test image (gray) and label image (binary) for prostate segmentation. The above methods need to obtain a segmentation map with a regional structure, which can easily cause excessive image segmentation. Toth et al. [25] proposed a segmentation algorithm based on an unmarked active appearance model, and created a deformable registration framework to generate the final segmentation. To address issues with designated landmarks, Toth et al. [26] presented a novel active appearance model solution that makes advantage of level set implementation. They found things of interest in the new image by using the registration-based technique. Rundo et al. [27] proposed a fuzzy C-mean clustering method to perform prostate MRI image segmentation. Traditional fuzzy C-means clustering algorithms do not consider spatial information and are sensitive to noise and uneven grayscale. Yanrong et al. [28] proposed a new deformable MRI prostate segmentation method by unifying deep feature learning and sparse patch matching. After the image processing using the above methods, there are still short lines and outliers that do not match the label, depending on the preprocessing work of the image.
Rundo et al. [27] proposed a new deformable MRI prostate segmentation method by unifying deep feature learning and sparse patch matching. After the image processing using the above methods, there are still short lines and outliers that do not match the label, depending on the preprocessing work of the image. Yanrong et al. [28] proposed a fuzzy C-mean clustering method to perform prostate MRI image segmentation. Traditional fuzzy C-means clustering algorithms do not consider spatial information and are sensitive to noise and uneven grayscale.
Traditional methods typically use shape priori or image priori (such as atlas) to solve the problem of weak and ambiguous boundaries of prostate cancer MRI images and the significant difference in image contrast and appearance. It is challenging to segment prostate MRI images based on traditional features or prior knowledge to achieve high accuracy, and the repeatability of the method is limited and cannot be quickly applied to medical systems.
2.2 Applications of CNNs in Prostate Segmentation
In recent years, CNNs [6] have been extensively applied to medical image analysis due to their powerful computing power and adaptive algorithms. Fully Convolutional Networks (FCNs) [29] are successful applications in automatic segmentation of medical images. Due to the complexity of medical images, more and more variants of FCNs have emerged, such as U-Net [10] and multi-scale U-Net [30]. CNNs has attracted widespread attention [31], and significant progress has been made in the automatic classification of prostate images. Cheng et al. [32] applied deep learning models to prostate MRI image segmentation and proposed a joint graph active model and CNNs. A CNN model [33] was developed to investigated the effects of encoder, decoder, and classification modules and objective functions, on the segmentation of prostate MRI. To help with prostate segmentation in MRI images, Zhu et al. [34] suggested a deep neural network model with a two-way convolution recursion layer that makes use of the context and features of the slice, views the prostate slice as a data series, and leverages the context between slices. Brosch et al. [35] proposed a novel method of boundary detection, which transforms the boundary detection formula into a regression task, in which CNN is trained to predict the distance between the surface mesh and the corresponding boundary points. Additionally, it is applied to the segmentation of an entire prostate in MRI images. The above method combines traditional methods and deep learning to segment the prostate region. The proposed model is a relatively basic convolutional neural network that relies on conventional features. Jia et al. [36] proposed a 3D adversarial pyramidal anisotropic convolution depth neural network to segment the prostate on MRI images. To fully address the challenge of insufficient data for training CNN models, a boundary-weighted adaptive model [37] was proposed. Karimi et al. [38] proposed three methods to estimate the Hausdorff distance from the segmentation probability map generated by CNN. A USE-Net [39] was proposed that combines two blocks into a U-Net. Jia et al. [40] proposed a hybrid discriminant network (HD-Net) to solve the problem of insufficient semantic recognition and spatial context modeling in prostate segmentation. Khan et al. [41] proposed evaluating the application of the four CNN model in the prostate segmentation of MRI images. Their experimental results show that CNNs with patch-wise DeepLabV3+ demonstrated the best performance. The application of pyramid, U-shaped network, attention mechanism, and other techniques for autonomous prostate segmentation has gradually increased with the advent of deep learning segmentation models. The embedding of a single module cannot extract more scale features, and the global context information is not fully expressed. Li et al. [8] proposed a pyramid mechanism fusion network (PMF-Net) to segment prostate region and pre-prostatic fat and learn global features and more comprehensive context information. They then suggested a new dual branch attention driven MRI multi-scale learning [42] and a dual attention directed 3D convolutional neural network (3D DAG-Net) [43] for the segmentation of the prostate and prostate cancer. The above methods are targeted at patients with prostate cancer, considering multi-region segmentation, such as prostate and surrounding fat, prostate and tumor segmentation results relying on correlation analysis between regions. Furthermore, we take into account data from cases other than prostate cancer, such as prostate nodules and prostate hyperplasia.
Prostate segmentation has been tackled using Deep Learning techniques, and medical image segmentation has been greatly impacted by the introduction of U-Net. Because U-Net includes high resolution and low resolution data, it is better suited for small sample segmentation. Many of the above prostate segmentation methods are according to U-Net. We believe designing a feature extraction mechanism to address the multi-scale characteristics of the prostate, and utilizing the transformer mechanism to synthesize more comprehensive contextual information. Consequently, there is a pressing need to create a transformer feature fusion, multi-scale attention, and U-Net-based precise automatic prostate region segmentation approach for MRI images.
The experimental data were obtained from the Haikou Municipal People’s Hospital and the Central South University Xiangya Medical College Affiliated Hospital, including 78 consecutive patients. All consecutive patients underwent multi-parameter MRI. They were using the GE3.0 T Signa HDX MRI scanner from the United States, with a composite 8-channel abdominal phased array coil as the receiving coil and no rectal coil used. The MRI images of the prostate were annotated by two radiologists. Before the annotation, the two doctors personally met to participate in training and practice meetings, and segmented two sample patients together. A methodology akin to that of the remaining data sets. Radiologists separately segment every component of every image. Each image is thus segmented twice. We determined that the two doctors’ segmentation findings had a correlation coefficient more than 0.95, indicating that the segmentation outcomes of the two specialists were extremely similar and that the two doctors ultimately negotiated the final manual segmentation label map.
Fig. 1 illustrated the overview of our method. We present the recommended 3D AGMSF-Net to segment prostate MRI. Our model has designed three modules on the baseline network: multi-scale attention mechanism, transformer feature extraction stage for prostate global and local information, and feature information fusion stage. To successfully preserve the continuity of space, we segmented 3D prostate MRI images using a 3D Unet as the baseline. We propose a multi-scale attention mechanism to achieve a more accurate prostate MRI image segmentation (Section 3.2). Given the difficulty in distinguishing the prostate MRI image boundary, we propose a method of 3D transformer feature extraction of prostate deeper features information to highlight the boundary features (Section 3.3). In the feature information fusion module, we fuse the extracted 3D transformer feature and the feature map generated by the 3D Unet encoder to obtain more complete detailed information (Section 3.4).
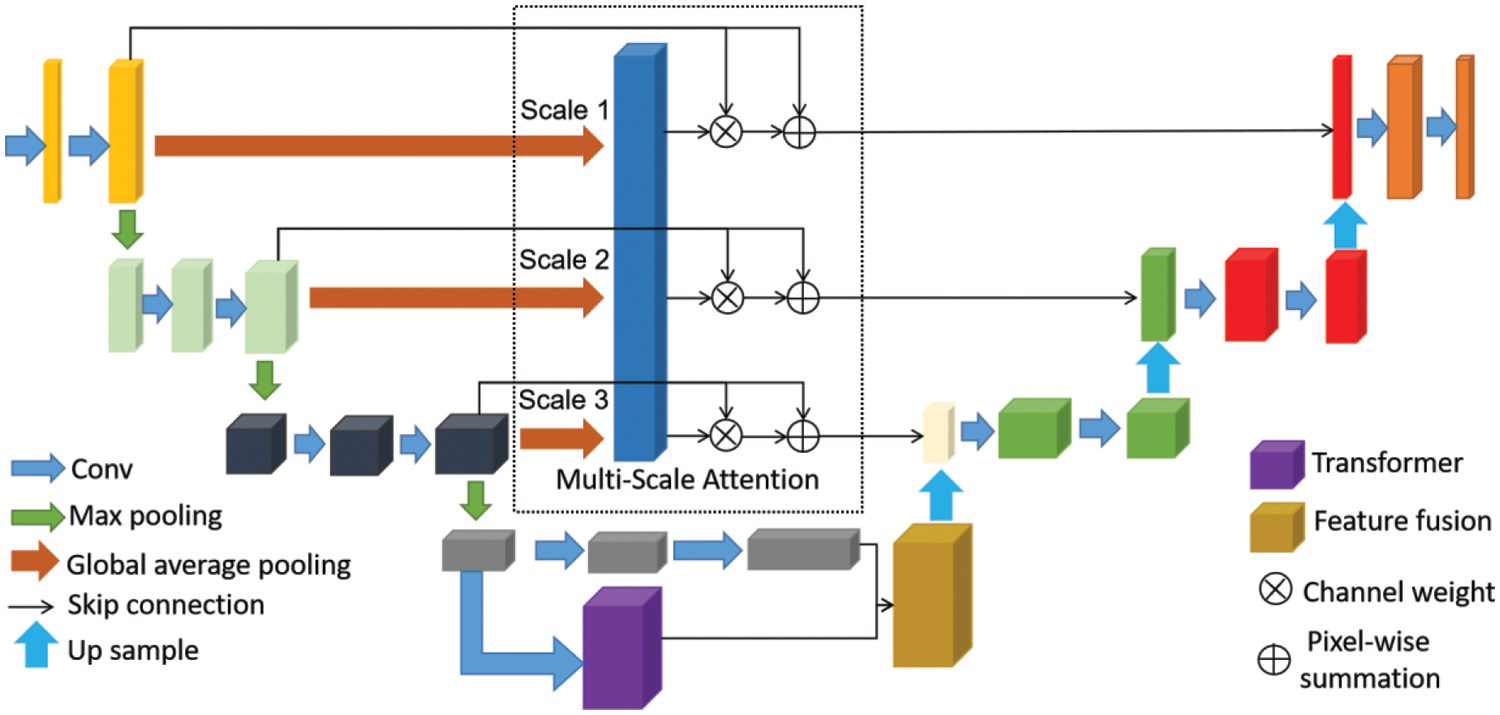
Figure 1: Our proposed model. 3D Unet as baseline, model includes multi-scale attention mechanism, transformer feature extraction and feature information fusion
3.3 Multi-Scale Attention Module
3D Unet [44] was selected as the base architecture of the 3D CNN module. The traditional 3D Unet network has demonstrated excellent achievements in image segmentation; however, many shortcomings exist in this network. Parallel skipping connections allow low-resolution features to be transmitted repeatedly; this results in fuzzy extracted image features. The high-level features extracted by the network typically contain insufficient high-resolution edge information, resulting in high uncertainties. The high-resolution edge mainly affects network decision-making (such as prostate segmentation). To solve this difficulty, inspired by Unet++ [45] and Unet3+ [46], we added an attention mechanism to extract multi-scale features for 3D Unet skip connections, as shown in Fig. 2.
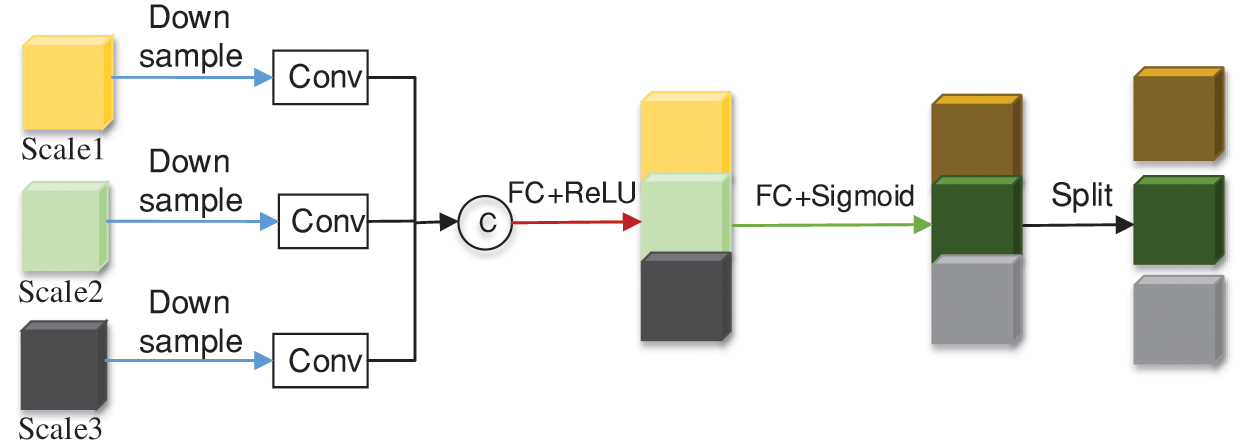
Figure 2: Overview of the multi scale attention module
Our proposed multi-scale attention module can effectively compress the feature maps of multi-layer encoders, undergoes downsampling, convolution, and concatenates them along the channel dimension. Let
Next, the spatial dimension of the feature map is reduced by using a corrected linear unit (ReLU) activation function after the convolutional layer. Two fully connected (FC) layers elongate features to enable the network to capture multi-scale information. Finally, use an S-shaped activation function
Weights are formulated as:
Weights X is partitioned according to channel numbers. Finally, we segment the weights of each layer, extend the weights to the same dimension as
There are many different implementations of attention mechanisms, including the multi-head attention mechanism. This approach has allowed transformer [47] to perform remarkably well in a variety of natural language processing. Visual transformer (ViT) [48] is a typical example of using a transformer in image processing. Although transformer lacks the inherent inductive bias of CNN, ViT using transfer learning has higher performance when pre-training uses large-scale data. Therefore, we consider using a transformer as the main feature extraction module in the network during the transition phase from encoder to decoder.
As shown in Fig. 3, we designed the 3D transformer module to introduce a self-attention mechanism to enhance global feature representation. To adjust the image input to fit the input of the transformer, ViT [48] reshaped the 2D image into a series of flat 2D blocks. To improve the situation, the transformer-based encoder was trained with low-resolution advanced features that were taken from the encoder in order to further learn global feature representation. Add positional embedding to block embedding to preserve positional information. Feed embedded features to multiple transformer layers, each model consisting of a multi-head self-attention (MSA) and a multi-layer perceptron (MLP). MSA helps networks capture richer feature information. The core idea of multi head attention is to divide the input features into multiple parts and perform independent attention calculations on each part.
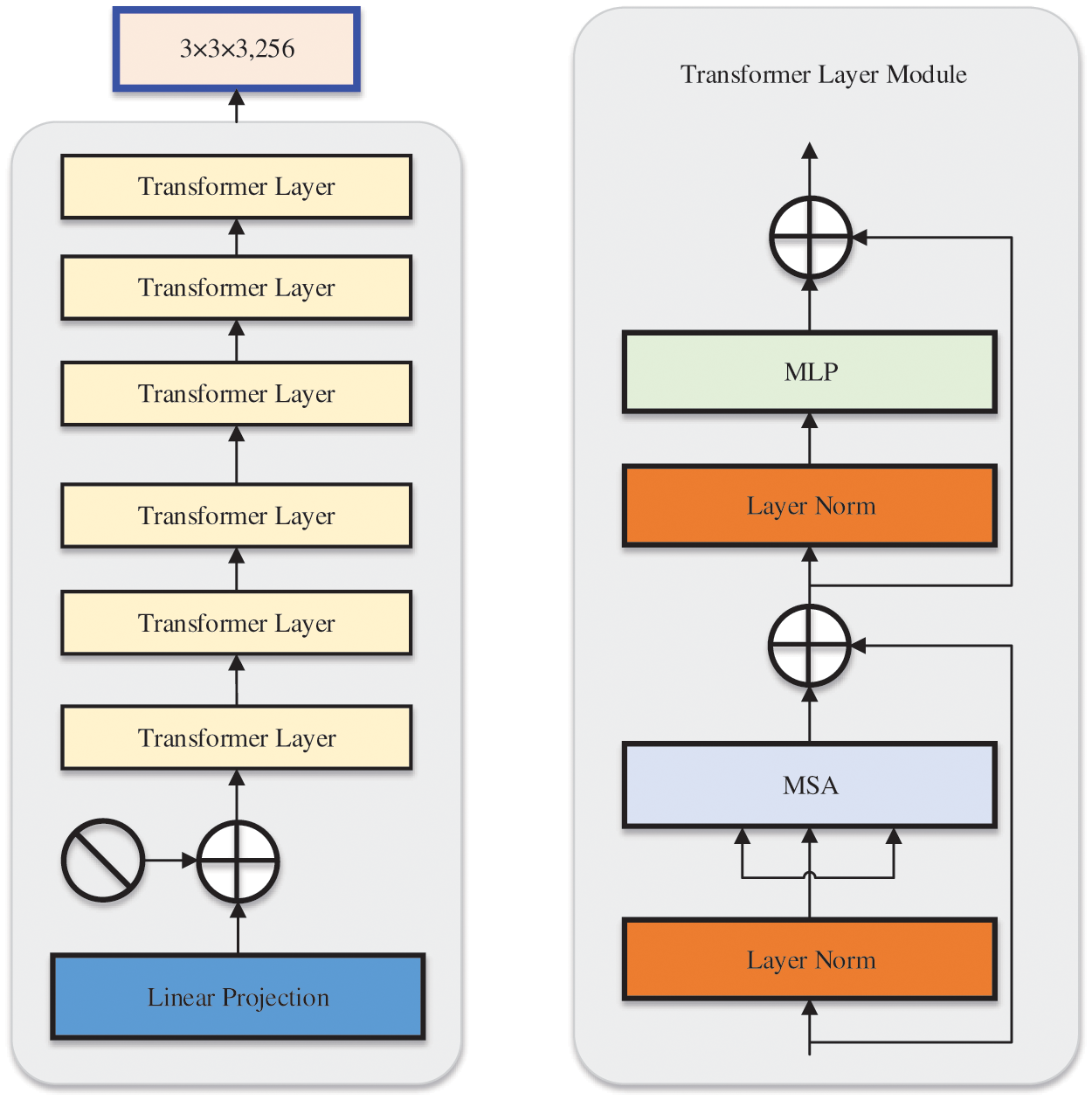
Figure 3: Overview of the transformer module. The main part consists of 6 transformer layers, each of which includes a multi-head self attention and multi-layer perceptron
After our model extracted 3D transformer features and encoder features based on the baseline encoder, we fused the two features to obtain a segmentation probability map. Fig. 4 describes the architecture of the feature fusion module. Firstly, cascade the features at the end of the baseline and 3D encoder of the 3D converter and perform average pooling and maximum pooling on the cascaded features, respectively. Then, the features obtained from the two pooling operations are added pixel by pixel and multiplied with the cascaded features. Finally, convolution with a kernel size of
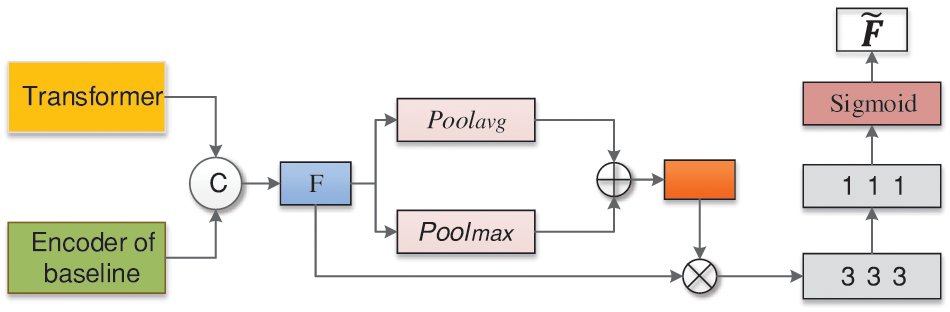
Figure 4: The framework of feature fusion module. After our model extracts transformer features and deep features based on encoder of baseline, a fusion mechanism is performed on the two features to finally obtain a segmentation probability map
Dice, which was first introduced [49] to address the category imbalance, was employed in this study as the loss function to prevent over-fitting during model training. Dice loss has since been applied extensively to medical image segmentation issues [50–52]. The following is the dice loss expressed:
where
In this section, we first introduce data augmentation used in our study. Next, we introduce the implementation details and evaluation metrics for our method. We then offered comparative experiments and ablation studies for independent analysis of our concept. We calculated the number of pixels in the corresponding region in the output of our model as the area of the prostate region for comparison with other studies.
The amount of data given by the hospital was insufficient to build a CNN model; hence, we have conducted data augmentation in our method. We performed data augmentation by successively rotating MRI picture by 90 degrees, 180 degrees, and 270 degrees, as well as flipping each image from top to bottom and left to right; The flipping performed preserved the visual structure.
One 16 GB NVIDIA Tesla V100 PCIe GPU and an Intel Xeon CPU made up the hardware setup used for the experiment. The software used was Python 3.7 and PyTorch. For our method, the 3D prostate MRI image was input into the network for training after data augmentation. Table 1 summarizes the network parameters. In the encoder part, after several
Five evaluation metrics were used to evaluate the performance of our method segmentation. Dice similarity coefficient (DSC) is one of the most commonly used evaluation metrics in medical image segmentation, defined as follows [53]:
By normalizing the intersection sizes of sets A and B to the mean of their values, the measurement assesses the degree of matching between them. The related error measure is the volumetric overlap error (VOE), which has the following definition [53]:
Relative volume difference (RVD) is defined as follows:
Furthermore, the distance error’s maximum symmetrical surface distance (MSSD) and average symmetrical surface distance (ASSD) were computed [53].
where,
The proposed network selects 3D Unet [44] as the baseline network. Multi scale attention skip connection (MSASC) was added to the baseline (B+MSASC), a transformer was added to the baseline (B+Transformer), and the transformer and feature fusion module (B+Transformer+Fusion) were aggregated in the baseline to show the accuracy of the network structure for prostate segmentation (Table 2). Adding the proposed MSASC (B+MSASC) to the baseline network dramatically improves the evaluation metrics. Compared to Baseline, the DSC for prostate segmentation increased by 1.46% with the addition of a multi-scale attention skip connection module (B+MSASC). Therefore, MSASC plays a crucial role in the proposed network structure. However, the DSC for prostate segmentation using the baseline and transformer joint module (B+Transformer) increased by 1.2%. Our method improves segmentation DSC performance by 6.79% compared to Baseline. Our developed model is able to extract both global and local features in prostate segmentation, allowing the network to learn more detailed information about the prostate.
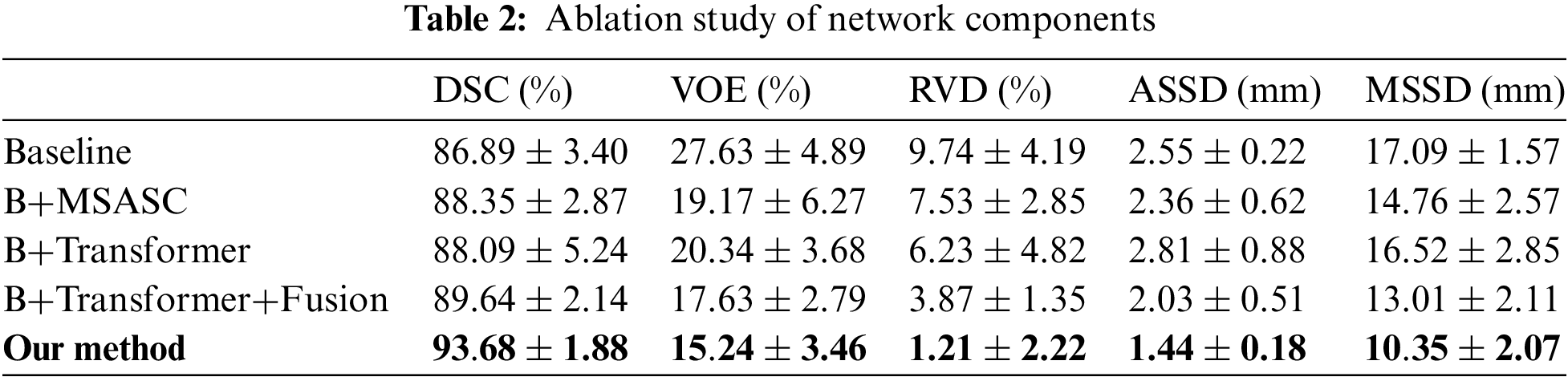
Fig. 5 shows the segmentation results of our method. The first column represents the original MRI image of the prostate. The second column represents the annotated image of the prostate by a doctor (as the ground truth). The third column represents the results of our method. The fourth column represents the comparison display of results and marks on the original drawing, the red line represents the result of our method, and the green line represents the ground truth. The fourth image’s magnified picture of the prostate’s major region is depicted in the fifth column. As illustrated in Fig. 5, our segmentation results are highly correlated with the prostate area annotated. The prostate lacks clear margins and complex background textures between the prostate and other anatomical structures, whereas the size, shape, and intensity distribution of the prostate have changed significantly. On the other hand, as column 5 of Fig. 5 shows, our approach performed well in border processing of the MRI images.
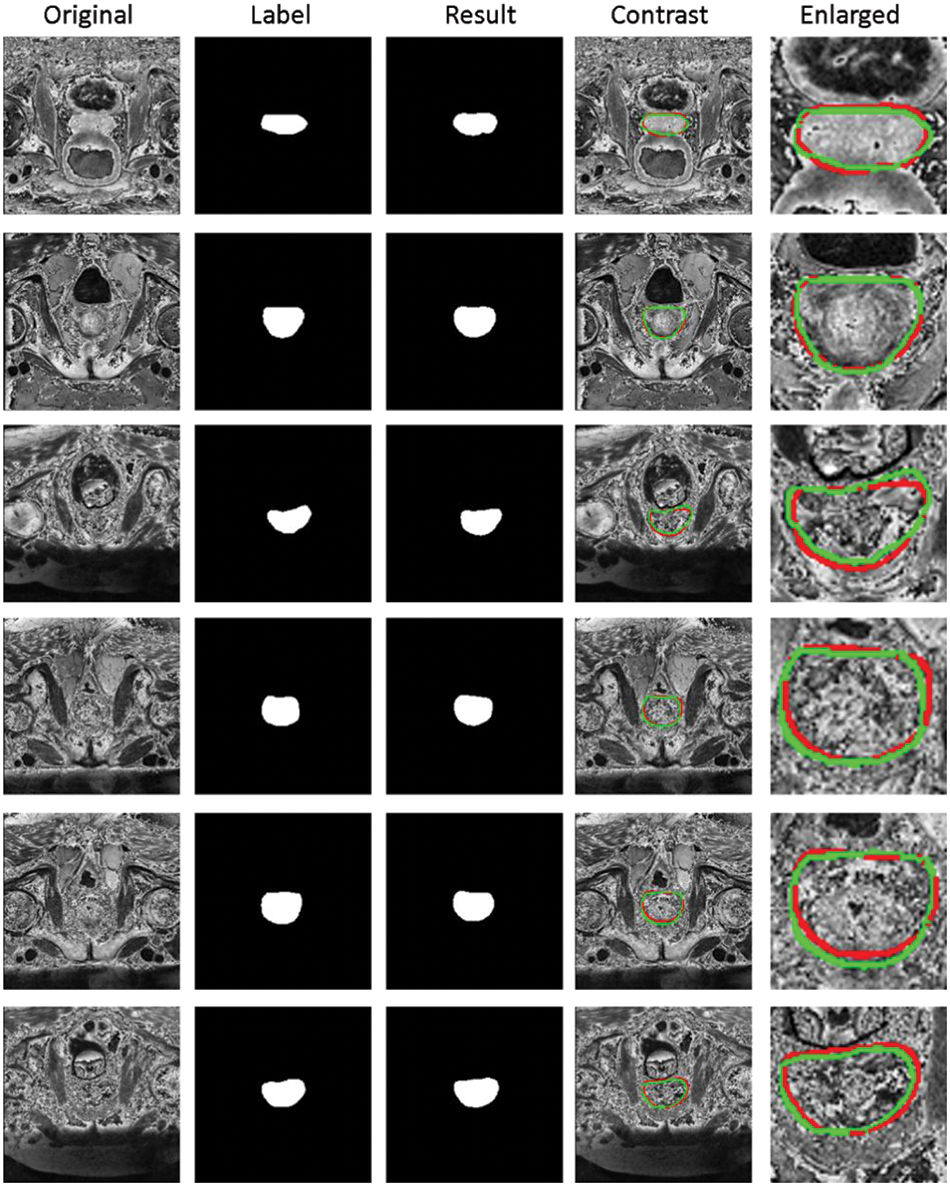
Figure 5: Segmentation results of prostate region of our method. First column: Original MRI image of prostate. Second column: The annotated image of the prostate by the doctor (as the ground truth). Third column: The results of our method. Fourth column: Comparison display of results and marks on the original drawing, the red line represents the result of our method, and the green line represents the ground truth. Fifth column: Enlarged view of the main prostate area on the fourth image
Our comparative methods include (1) Classic 3D segmentation network: 3D Unet [44], 3D Dense-Unet [54]; (2) prostate segmentation network: HD-Net [55], Patch wise DeeplabV3+ [56]; (3) transformer segmentation network: ViT [48]; and (4) recent segmentation methods: PMF-Net [8] and 3D PPU-Net [9]. 3D Unet extends the previous U-Net [10] architecture by replacing all 2D operations with 3D operations. 3D Dense-Unet [54] explores a dense attention gate based on 3D Unet to force the network to learn rich contextual information. Hybrid Discriminant Network (HD-Net) [55] is a 3D segmentation decoder that uses channel attention blocks to generate semantically consistent volumetric features and an auxiliary 2D boundary decoder that guides the segmentation network to focus on semantic discriminative on-chip features for prostate segmentation. Patch-wise DeeplabV3+ [56] studied encoding decoding CNNs for prostate segmentation in T2W MRI. ViT [48] successfully applied a transformer to image classification tasks and performed well. PMF-Net [8] and 3D PPU-Net [9] are recently proposed methods for prostate segmentation.
Table 3 shows the average quantitative score of the 5-fold cross-validation of prostate segmentation. All metric systems are described in detail in [53]. The 3D Unet [44] proposed for 3D image data and its current improved algorithm were not sensitive to the boundaries of the prostate MRI images. Compared with 3D Dense-Unet [54], HD-Net [55], Patch-wise DeeplabV3+ [56], 3D Unet [44], ViT [48], PMF-Net [8], 3D PPU-Net [9], and our method achieved better scores in terms of the DSC, VOE, RVD, ASSD, and MSSD. For the classic segmentation network, our method improves the DSC of prostate segmentation by 6.79% compared to 3D Unet and 5.93% compared to 3D Dense-Unet. For the prostate segmentation network, our method improves the DSC of prostate segmentation by 3.78% compared to HD-Net and 3.38% compared to patch-wise DeeplabV3+. For the Tranformer network, our method improves the DSC of prostate segmentation by 5.96% compared to ViT. Our method has increased the DSC value by 2.65% compared to the latest prostate segmentation method (3D PPU-Net [9]). Several other evaluation metrics have also been improved to some extent.
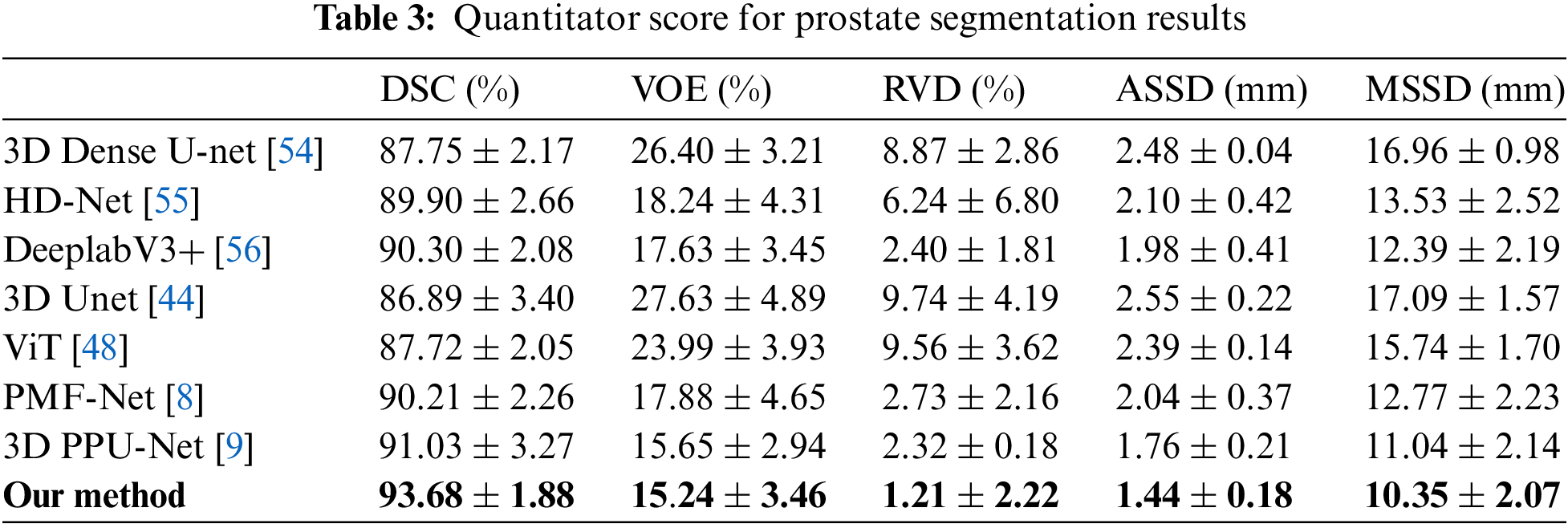
Fig. 6 presents a quantitative 5-fold cross-validation evaluation of the DSC for the 3D Dense-Unet [54], HD-Net [55], Patch-wise DeeplabV3+ [56], 3D Unet [44], ViT [48], PMF-Net [8], 3D PPU-Net [9], and our approach in the test set. HD-net solved the 3D prostate MRI image segmentation problem. Patch-wise DeeplabV3+ proposed by [56] demonstrated the best performance in prostate images. 3D Unet [44] is our network baseline and improved its skipping connection part. PMF-Net [8] and 3D PPU-Net [9] are recently proposed methods for prostate segmentation. Better DSC values for prostate segmentation are obtained by our approach thanks to the multi-scale attention module, 3D transformer, and feature fusion of encoder and transformer.
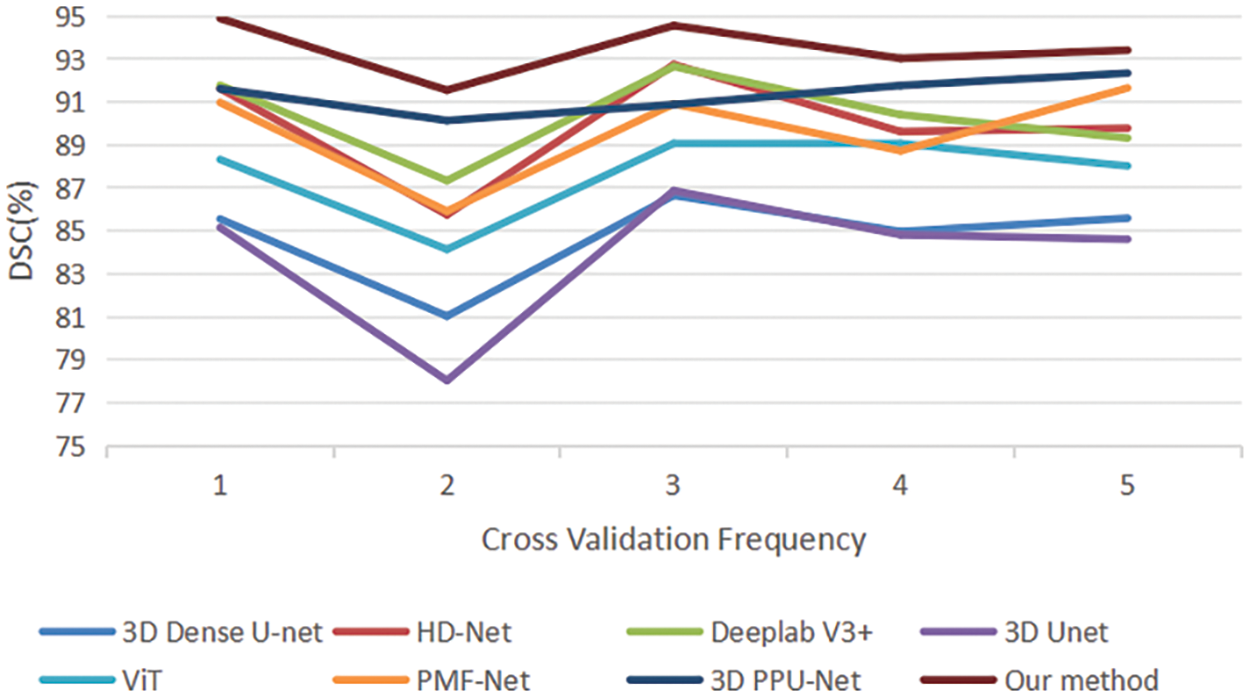
Figure 6: Quantitative comparison of 5-fold cross validation of DSC between the comparative methods and our method
In Fig. 7, prostate segmentation results are shown compared to ground truth average (red lines), 3D Dense-Unet (green lines), HD-Net (blue lines), Patch-wise DeeplabV3+ (light blue lines), 3D Unet (yellow lines), ViT (purple lines), and our method (white lines). Columns (1) and (3) in each subfigure display the segmentation findings superimposed over complete MRI images, whereas columns (2) and (4) display an expanded view of the rectangular region denoted by a black box. Some of the boundaries between prostate regions were not detectable using the other technique from the columns (2) and (4) of Fig. 7 because of the influence of inadequate contrast. On the other hand, our method yielded a better level of consistency with the usual ground facts.
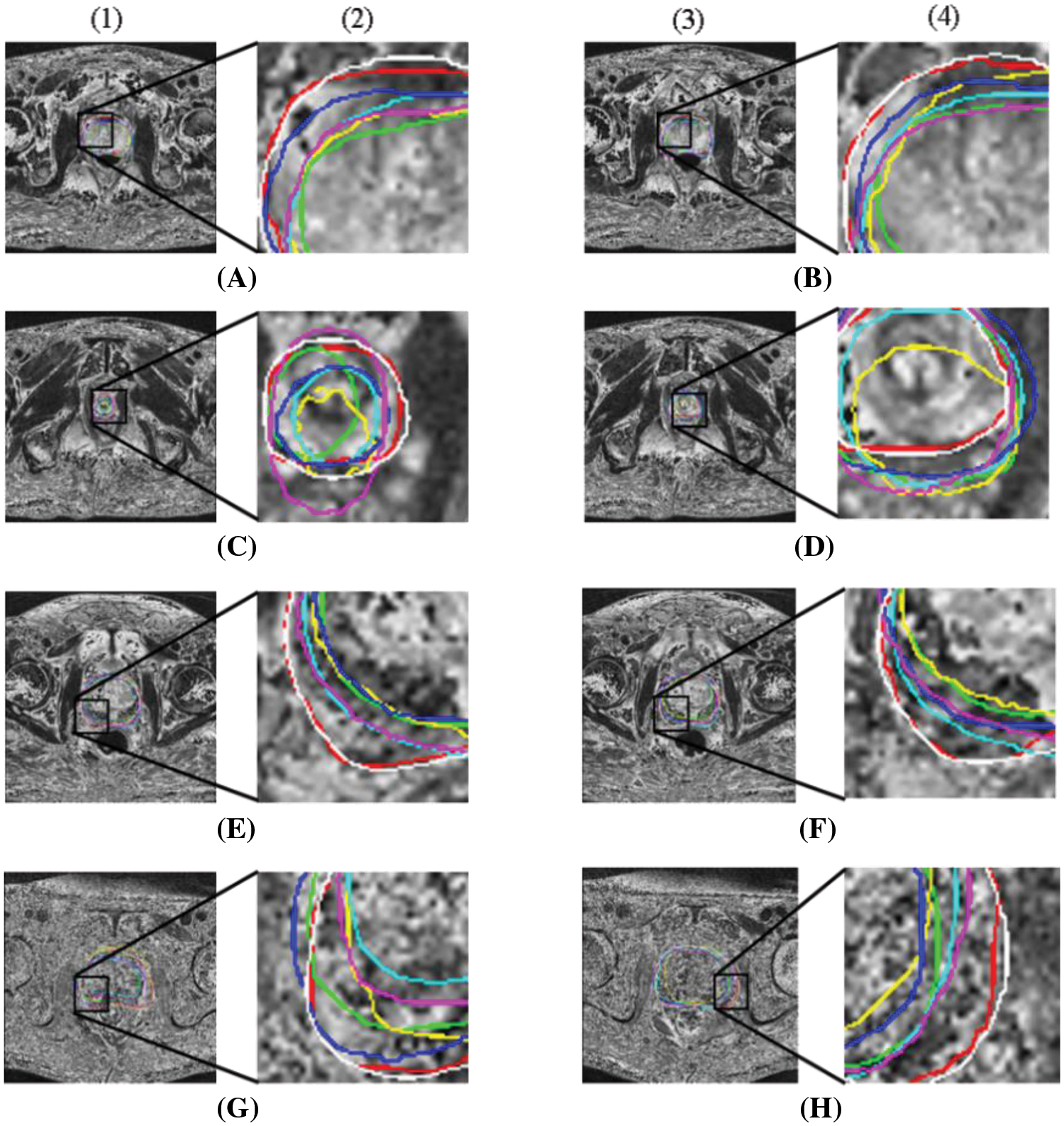
Figure 7: Comparison of segmentation results for 8 example cases. The results of different methods and the region expansion map are shown, respectively. Red line: ground truth; White line: Our method; Green line: 3D Dense-Unet [54]; Blue line: HD-Net [55]; Light blue line: Patch-wise DeeplabV3+ [56]; Yellow line: 3D Unet [44] and Purple line: ViT [48]
The 3D visualization results from the ground truth, 3D Unet [44], ViT [48], HD-Net [55], Patch-wise DeeplabV3+ [56], and 3D Dense-Unet [54], are compared with our technique in Fig. 8. The red region is the 3D prostate. As shown in Fig. 8, Our method holds a high degree of relevance between the segmentation results of prostate MRI images and the ground truth annotated by experts.
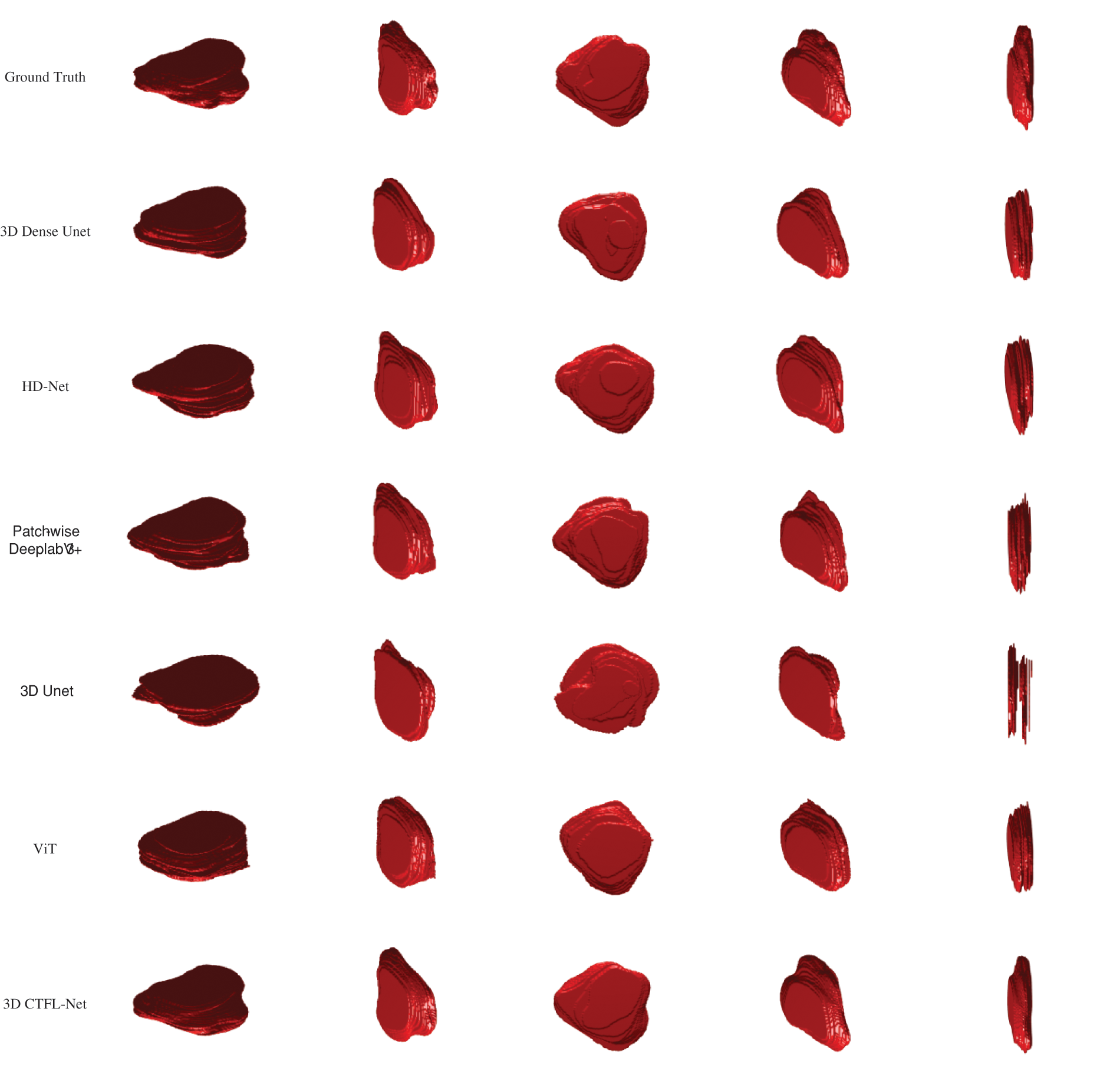
Figure 8: Comparison of 3D results between comparative methods. Based on five different perspectives of a patient, the 3D results of prostate MRI are shown
Using comparison approaches, the prostate region’s MRI image segmentation results are displayed in Fig. 9. Each column is the segmentation results of different methods: PMF-Net [8], 3D PPU-Net [9], and our method. As shown in Fig. 9, our method maintains higher consistency with the annotation than other methods. Our method has shown good segmentation results for smaller or blurry prostate regions. This depends on the multi-scale attention, transformer module, and feature fusion module we designed, which extract multi-scale information of the prostate and fuse global and local features, making the prostate segmentation results more accurate.
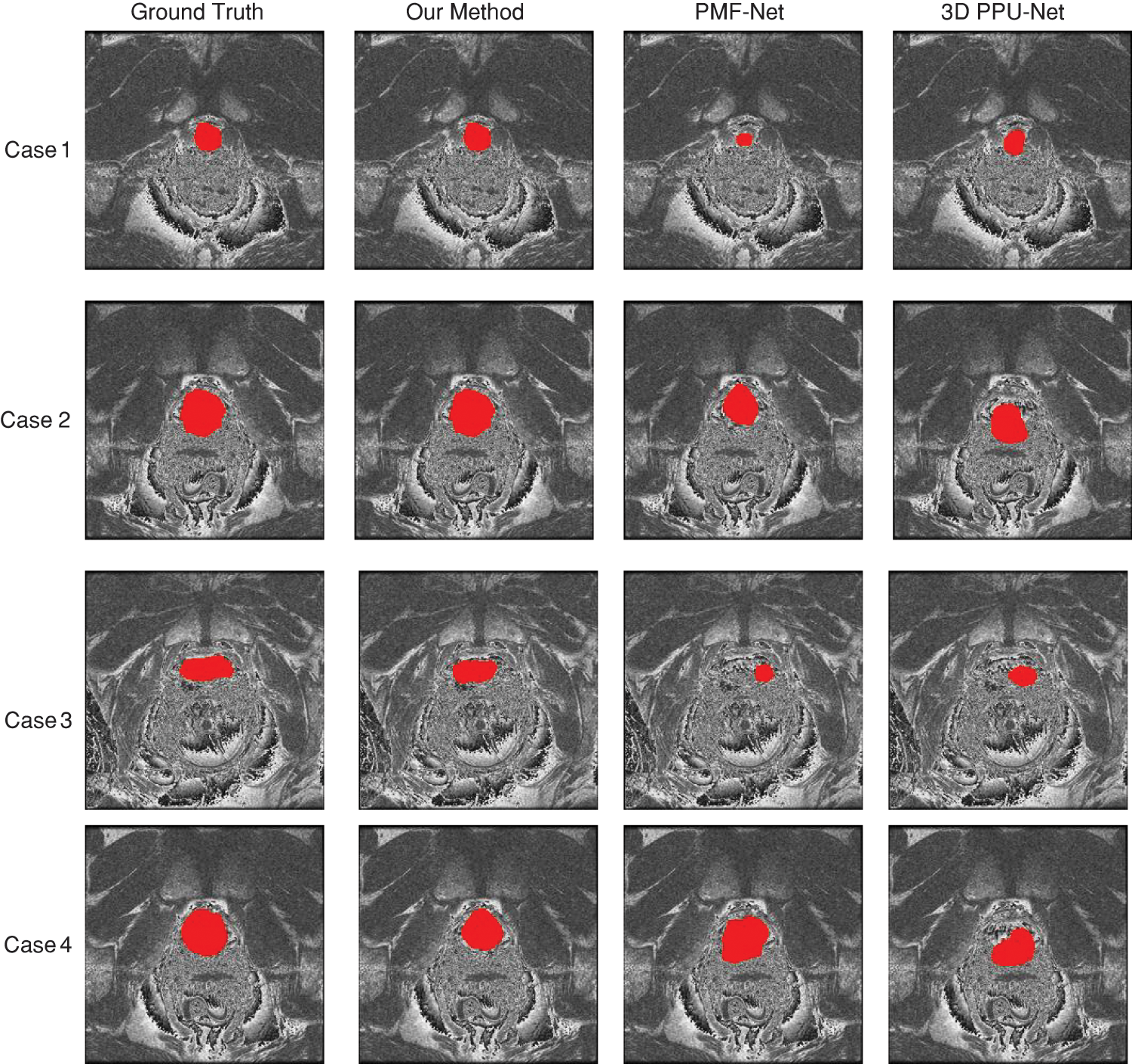
Figure 9: Comparison method for the prostate results of the MRI. Red denotes the prostate, which is a slice of the same case obtained using various techniques. Each column is the segmentation results of different methods: PMF-Net [8], 3D PPU-Net [9] and our method
In this study, we demonstrate the successful segmentation of 3D prostate MRI images using our suggested approach, 3D AGMSF-Net. Importantly, we propose that the fusion of 3D transformer features and 3D convolutional neural networks is applied to the analysis of 3D volume data, suggesting that 3D features are more suitable for 3D image segmentation by considering spatial continuity and adjacent pixel characteristics. Although some previous studies [31–43] also used CNN to solve the problem of prostate MRI image segmentation, they did not fully consider the boundary information of the prostate, making the prostate segmentation inaccurate. We propose an attention mechanism for extracting multi-scale features, which is embedded in the skip connections of the baseline model. The 3D transformer feature we proposed extracts the rich information of the prostate boundary and merges it with the deep features of the 3D CNN encoder to precisely segment prostate MRI images. The 3D transformer feature describes the regional object through global and local features, which makes up for the inaccuracy of the 3D CNN in boundary segmentation. We chose 3D Unet as the baseline of the 3D CNN module because the structure of the U-shaped network is suitable for analyzing medical images. The U-shaped network structure is a research hotspot in medical image processing. In recent years, many studies [45,46,50,54] have improved the U-net network to achieve better performance. However, none of them express enough information on multiple scales. Based on 3D Unet, each layer of decoders integrates information that is equal to or less than the scale and the more significant features of the decoder, thereby gathering comprehensive data.
We compare our method with other methods [44,48,54–56]. 3D Dense-Unet [54] achieved dense connections but does not represent semantic information at full scale. The missing information causes the extracted feature maps to be blurred. Although HD-Net [55] also considers the boundary information, that is, uses the channel attention block to generate an auxiliary 2D boundary decoder to guide the segmentation network. Still, they only consider the 2D boundary information, which is far from enough for the segmentation of 3D MRI. Patch-by-patch to segment the prostate, DeeplabV3+ [56] segmented many blocks and entered it into the previously build DeeplabV3+. The boundary as a result of the divided image, making the boundary non-continuous. We also compare our method with the transformer method ViT [48] for verification. The 5-fold cross-validation shows that our method is more consistent with the doctor’s ground truth than other algorithms. Multi-scale attention mechanism, 3D transformer, feature fusion of 3D transformer, and encoder features can effectively segment 3D prostate MRI images. The attention mechanism is embedded in the jump connection of the baseline model to extract multi-scale features of the prostate gland. The addition of a 3D transformer makes the boundary segmentation more refined. For the case of small targets (Figs. 7C and 7D), for situations where goals and backgrounds are similar (Figs. 7G and 7H), accurate segmentation can still be achieved by our method.
Although the effectiveness of the fusion of transformer features and deep features is revealed by these studies, there are also limitations. First, it takes a lot of time to extract 3D transformer features. Second, in each layer of the decoder of 3D CNN, multi-scale features are integrated, which increases the complexity of the network. We will further optimize the network in the future. In addition, we intend to investigate a better-performing method structure to solve the segmentation problem of other lesions in MRI images.
In this paper, we suggested a 3D AGMSF-Net for prostate MRI images segmentation. We propose an attention mechanism for extracting multi-scale features, which are embedded in the skip connections of the baseline model. The 3D converter module has been introduced to enhance global feature representation by adding it during the transition phase from encoder to decoder. A feature fusion mechanism has been designed to fuse multi-scale features to express more comprehensive information. We validated the proposed method on the dataset of a local hospital. The results indicated that the proposed method segmented prostate MRI images better than the improved 3D Unet method and the latest deep learning methods. Quantitative experiments have shown that our results are highly consistent with annotation. The segmentation of the prostate has a profound impact on the diagnosis of prostate cancer and lays the foundation for the automatic diagnosis and recognition of tumors. We will consider further expanding to quantitative analysis research on prostate cancer to better assist doctors in the effective treatment and prognosis of patients.
Acknowledgement: The author sincerely thanks all the participants and staff of the School of Information and Communication Engineering of Hainan University; We would also like to thank Haikou Municipal People’s Hospital and Xiangya Medical College Affiliated Hospital of Central South University for providing data support for our research.
Funding Statement: This work was supported in part by the National Natural Science Foundation of China (Grant #: 82260362), in part by the National Key R&D Program of China (Grant #: 2021ZD0111000), in part by the Key R&D Project of Hainan Province (Grant #: ZDYF2021SHFZ243), in part by the Major Science and Technology Project of Haikou (Grant #: 2020-009).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Yuchun Li, Mengxing Huang; data collection: Yu Zhang, Zhiming Bai; analysis and interpretation of results: Yuchun Li, Mengxing Huang, Yu Zhang; draft manuscript preparation: Yuchun Li, Mengxing Huang, Yu Zhang. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data that support the findings of this study are available on request from the corresponding author, Mengxing Huang and Yu Zhang. The data are not publicly available due to ethical restrictions and privacy protection.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. B. S. Chhikara and K. Parang, “Global cancer statistics 2022: The trends projection analysis,” Chemical Biology Letters, vol. 10, no. 1, pp. 451, 2023. [Google Scholar]
2. G. Litjens, R. Toth, W. van de Ven, C. Hoeks, S. Kerkskra et al., “Evaluation of prostate segmentation algorithms for MRI: The PROMISE12 challenge,” Medical Image Analysis, vol. 18, no. 2, pp. 359–373, 2014. [Google Scholar] [PubMed]
3. T. Hambrock, D. M. Somford, H. J. Huisman, I. M. V. Oort and J. O. Barentsz, “Relationship between apparent diffusion coefficients at 3.0-T MR imaging and gleason grade in peripheral zone prostate cancer,” International Journal of Medical Radiology, vol. 259, no. 2, pp. 453–461, 2011. [Google Scholar]
4. A. Tanimoto, J. Nakashima, H. Kohno, H. Shinmoto and S. Kuribayashi, “Prostate cancer screening: The clinical value of diffusion-weighted imaging and dynamic MR imaging in combination with T2-weighted imaging,” Journal of Magnetic Resonance Imaging, vol. 25, no. 1, pp. 146–152, 2007. [Google Scholar] [PubMed]
5. D. Shen, Y. Zhan and C. Davatzikos, “Segmentation of prostate bound-aries from ultrasound images using statistical shape model,” IEEE Transactions on Medical Imaging, vol. 22, no. 4, pp. 539–551, 2003. [Google Scholar] [PubMed]
6. Y. LeCun, Y. Bengio and G. Hinton, “Deep learning,” Nature, vol. 521, no. 7553, pp. 436–444, 2015. [Google Scholar] [PubMed]
7. L. Yu, Y. Xin, H. Chen, Q. Jing and P. A. Heng, “Volumetric convnets with mixed residual connections for automated prostate segmentation from 3D MR images,” in Proc. of the AAAI Conf. on Artificial Intelligence, San Francisco, California, USA, pp. 66–72, 2017. [Google Scholar]
8. Y. Li, Y. Wu, M. Huang, Y. Zhang and Z. Bai, “Automatic prostate and peri-prostatic fat segmentation based on pyramid mechanism fusion network for T2-weighted MRI,” Computer Methods and Programs in Biomedicine, vol. 223, pp. 106918, 2022. [Google Scholar] [PubMed]
9. Y. Li, C. Lin, Y. Zhang, S. Feng, M. Huang et al., “Automatic segmentation of prostate MRI based on 3D pyramid pooling Unet,” Medical Physics, vol. 50, no. 2, pp. 906–921, 2023. [Google Scholar] [PubMed]
10. O. Ronneberger, P. Fischer and T. Brox, “U-Net: Convolutional networks for biomedical image segmentation,” in Proc. of MICCAI, Munich, Germany, pp. 234–241, 2015. [Google Scholar]
11. X. M. Li, H. Chen, X. J. Qi, Q. Dou, C. W. Fu et al., “H-DenseUNet: Hybrid densely connected unet for liver and tumor segmentation from CT volumes,” IEEE Transactions on Medical Imaging, vol. 37, no. 12, pp. 2663–2674, 2018. [Google Scholar] [PubMed]
12. D. Jha, P. H. Smedsrud, M. A. Riegler, D. Johansen and H. D. Johansen, “ResUNet++: An advanced architecture for medical image segmentation,” in Proc. of ISM, San Diego, CA, USA, pp. 225–2255, 2019. [Google Scholar]
13. C. Lin, C. Qiu, H. Jiang and L. Zou, “A deep neural network based on prior-driven and structural preserving for SAR image despeckling,” IEEE Journal of Selected Topics in Applied Earth Observations and Remote Sensing, vol. 16, pp. 6372–6392, 2023. [Google Scholar]
14. Q. Chen, L. Xie, L. Zeng, S. Jiang, W. Ding et al., “Neighborhood rough residual network-based outlier detection method in IoT-enabled maritime transportation systems,” IEEE Transactions on Intelligent Transportation Systems, vol. 24, no. 11, pp. 11800–11811, 2023. https://doi.org/10.1109/TITS.2023.3285615 [Google Scholar] [CrossRef]
15. Q. Chen, W. Ding, X. Huang and H. Wang, “Generalized interval type II fuzzy rough model based feature discretization for mixed pixels,” IEEE Transactions on Fuzzy Systems, vol. 31, no. 3, pp. 845–859, 2023. [Google Scholar]
16. L. Zeng, M. Huang, Y. Li, Q. Chen and H. N. Dai, “Progressive feature fusion attention dense network for speckle noise removal in OCT images,” IEEE/ACM Transactions on Computational Biology and Bioinformatics, 2022. https://doi.org/10.1109/TCBB.2022.3205217 [Google Scholar] [PubMed] [CrossRef]
17. W. Qiu, J. Yuan, E. Ukwatta, Y. Sun, M. Rajchl et al., “Dual optimization based prostate zonal segmentation in 3D MR images,” Medical Image Analysis, vol. 18, no. 4, pp. 660–673, 2014. [Google Scholar] [PubMed]
18. D. Mahapatra and J. M. Buhmann, “Prostate MRI segmentation using learned semantic knowledge and graph cuts,” IEEE Transactions on Biomedical Engineering, vol. 61, no. 3, pp. 756–764, 2014. [Google Scholar] [PubMed]
19. Z. Tian, M. D. L.L. and B. Fei, “A supervoxel-based segmentation method for prostate MR images,” Medical Imaging: Image Processing, vol. 44, no. 2, pp. 558–569, 2017. [Google Scholar]
20. Z. Tian, L. Liu, Z. Zhang and B. Fei, “Superpixel-based segmentation for 3D prostate MR images,” IEEE Transactions on Medical Imaging, vol. 35, no. 3, pp. 791–801, 2016. [Google Scholar] [PubMed]
21. Y. Gao, R. Sandhu, G. Fichtinger and A. R. Tannenbaum, “A coupled global registration and segmentation framework with application to magnetic resonance prostate imagery,” IEEE Transactions on Medical Imaging, vol. 29, no. 10, pp. 1781–1794, 2010. [Google Scholar] [PubMed]
22. Y. Ou, J. Doshi, G. Erus and C. Davatzikos, “Multi-atlas segmentation of the prostate: A zooming process with robust registration and atlas selection,” Medical Image Computing and Computer Assisted Intervention (MICCAI) Grand Challenge: Prostate MR Image Segmentation, vol. 7, pp. 1–7, 2012. [Google Scholar]
23. Z. Tian, L. Liu and B. Fei, “A fully automatic multi-atlas based segmentation method for prostate MR images,” Medical Imaging 2015 Image Processing, vol. 9413, pp. 1067–1073, 2015. [Google Scholar]
24. P. Yan, Y. Cao, Y. Yuan, B. Turkbey and P. L. Choyke, “Label image constrained multiatlas selection,” IEEE Transactions on Cybernetics, vol. 45, no. 6, pp. 1158–1168, 2015. [Google Scholar] [PubMed]
25. R. Toth and A. Madabhushi, “Deformable landmark-free active appearance models: Application to segmentation of multi-institutional prostate MRI data,” in Proc. of MICCAI Workshop, Nice, France, 2012. [Google Scholar]
26. R. Toth and A. Madabhushi, “Multifeature landmark-free active appearance models: Application to prostate MRI segmentation,” IEEE Transactions on Medical Imaging, vol. 31, no. 8, pp. 1638–1650, 2012. [Google Scholar] [PubMed]
27. L. Rundo, C. Militello, G. Russo, A. Garufi, S. Vitabile et al., “Automated prostate gland segmentation based on an unsupervised fuzzy c-means clustering technique using multispectral T1w and T2w MR imaging,” Information, vol. 8, no. 2, pp. 49, 2017. [Google Scholar]
28. G. Yanrong, G. Yaozong and S. Dinggang, “Deformable mr prostate segmentation via deep feature learning and sparse patch matching,” IEEE Transactions on Medical Imaging, vol. 35, no. 4, pp. 1077–1089, 2016. [Google Scholar]
29. J. Long, E. Shelhamer and T. Darrell, “Fully convolutional networks for semantic segmentation,” in Proc. of CVPR, Boston, USA, pp. 3431–3440, 2015. [Google Scholar]
30. J. Li, K. V. Sarma, K. C. Ho, A. Gertych, B. S. Knudsen et al., “A multi-scale U-Net for semantic segmentation of histological images from radical prostatectomies,” in Proc. of AMIA, Washington DC, USA, pp. 1140–1148, 2017. [Google Scholar]
31. F. Altaf, S. M. S. Islam, N. Akhtar and N. K. Janjua, “Going deep in medical image analysis: Concepts, methods, challenges, and future directions,” IEEE Access, vol. 7, pp. 99540–99572, 2019. [Google Scholar]
32. R. Cheng, H. R. Roth, L. Le, S. Wang and M. J. Mcauliffe, “Active appearance model and deep learning for more accurate prostate segmentation on MRI,” in Proc. of SMI, California, USA, vol. 9784, pp. 678–686, 2016. [Google Scholar]
33. J. Mun, W. Jang, D. J. Sung and C. KimX, “Comparison of objective functions in CNN-based prostate magnetic resonance image segmentation,” in Proc. of ICIP, Beijing, China, pp. 3859–3863, 2017. [Google Scholar]
34. Q. Zhu, B. Du, T. Baris, C. Peter and P. Yan, “Exploiting interslice correlation for MRI prostate image segmentation, from recursive neural networks aspect,” Complexity, vol. 2018, pp. 1–10, 2018. [Google Scholar]
35. T. Brosch, J. Peters, A. Groth, T. Stehle and J. Weese, “Deep learning-based boundary detection for model-based segmentation with application to MR prostate segmentation,” in Proc. of MICCAI, Granada, Spain, pp. 515–522, 2018. [Google Scholar]
36. H. Jia, Y. Xia, Y. Song, D. Zhang and W. Cai, “3D APA-Net: 3D adversarial pyramid anisotropic convolutional network for prostate segmentation in MR images,” IEEE Transactions on Medical Imaging, vol. 39, no. 2, pp. 447–457, 2019. [Google Scholar] [PubMed]
37. Q. Zhu, B. Du and P. Yan, “Boundary-weighted domain adaptive neural network for prostate MR image segmentation,” IEEE Transactions on Medical Imaging, vol. 39, no. 3, pp. 753–763, 2019. [Google Scholar] [PubMed]
38. D. Karimi and S. E. Salcudean, “Reducing the hausdorff distance in medical image segmentation with convolutional neural networks,” IEEE Transactions on Medical Imaging, vol. 39, no. 2, pp. 499–513, 2019. [Google Scholar] [PubMed]
39. L. Rundo, C. Han, Y. Nagan, J. Zhang and P. Cazzaniga, “USE-Net: Incorporating squeeze-and-excitation blocks into U-Net for prostate zonal segmentation of multi-institutional MRI datasets,” Neurocomputing, vol. 365, pp. 31–43, 2019. [Google Scholar]
40. H. Jia, Y. Song, H. Huang, W. Cai and Y. Xia, “HD-Net: Hybrid discriminative network for prostate segmentation in MR images,” in Proc. of MICCAI, Shenzhen, China, pp. 110–118, 2019. [Google Scholar]
41. Z. Khan, N. Yahya, K. Alsaih, S. S. Ali and F. Meriaudeau, “Evaluation of deep neural networks for semantic segmentation of prostate in T2W MRI,” Sensors, vol. 20, no. 6, pp. 3183, 2020. [Google Scholar] [PubMed]
42. Y. Li, Y. Wu, M. Huang, Y. Zhang and Z. Bai, “Attention-guided multi-scale learning network for automatic prostate and tumor segmentation on MRI,” Computers in Biology and Medicine, vol. 165, pp. 107374, 2023. [Google Scholar] [PubMed]
43. Y. Li, M. Huang, Y. Zhang, S. Feng, J. Chen et al., “A dual attention-guided 3D convolution network for automatic segmentation of prostate and tumor,” Biomedical Signal Processing and Control, vol. 85, pp. 104755, 2023. [Google Scholar]
44. Ö. Çiçek, A. Abdulkadir, S. S. Lienkamp, B. Thomas and R. Olaf, “3D U-Net: Learning dense volumetric segmentation from sparse annotation,” in Proc. of MICCAI, Athens, Greece, pp. 424–432, 2016. [Google Scholar]
45. Z. Zhou, M. M. Rahman Siddiquee, N. Tajbakhsh and J. Lian, “UNet++: A nested u-net architecture for medical image segmentation,” in Proc. of DLMIA, Granada, Spain, pp. 3–11, 2018. [Google Scholar]
46. H. Huang, L. Lin, R. Tong, H. Hu, Q. Zhang et al., “Unet 3+: A full-scale connected unet for medical image segmentation,” in Proc. of ICASSP, Barcelona, Spain, pp. 1055–1059, 2020. [Google Scholar]
47. A. V. aswani, N. Shazeer, N. Parmar, J. Uszkoreit, L. Jones et al., “Attention is all you need,” Advances in Neural Information Processing Systems, vol. 30, pp. 5998–6008, 2017. [Google Scholar]
48. A. Dosovitskiy, L. Beyer, A. Kolesnikov, D. Weissenborn, X. Zhai et al., “An image is worth 16x16 words: Transformers for image recognition at scale,” arXiv preprint arXiv:2010.11929, 2020. [Google Scholar]
49. F. Milletari, N. Navab and S. Ahmadi, “V-Net: Fully convolutional neural networks for volumetric medical image segmentation,” in Proc. of CVPR, Las Vegas, USA, pp. 565–571, 2016. [Google Scholar]
50. H. Li, G. Jiang, R. Wang, J. Zhang, Z. Wang et al., “Fully convolutional network ensembles for white matter hyperintensities segmentation in MR images,” Neuroimage, vol. 183, pp. 650–665, 2018. [Google Scholar] [PubMed]
51. M. Drozdzal, G. Chartrand, E. Vorontsov, L. Di-Jorio, A. Tang et al., “Learning normalized inputs for iterative estimation in medical image segmentation,” in Proc. of CVPR, Salt Lake City, USA, vol. 44, pp. 1–13, 2018. [Google Scholar]
52. C. H. Sudre, W. Li, T. Vercauteren, S. Ourselin and M. J. Cardoso, “Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations,” in Proc. of DLMIA, Québec City, QC, Canada, pp. 240–248, 2017. [Google Scholar]
53. I. Voiculescu and V. Yeghiazaryan, “An overview of current evaluation methods used in medical image segmentation,” Department of Computer Science, no. RR-15-08, pp. 22, 2015. [Google Scholar]
54. T. D. Bui, L. Wang, J. Chen, W. Lin, G. Li et al., “Multi-task learning for neonatal brain segmentation using 3D Dense-Unet with dense attention guided by geodesic distance,” in Proc. of MICCAI Workshop, Shenzhen, China, pp. 243–251, 2019. [Google Scholar]
55. H. Jia, Y. Song, H. Huang, W. Cai and Y. Xia, “HD-Net: Hybrid discriminative network for prostate segmentation in MR images,” in Proc. of MICCAI, Shenzhen, China, pp. 110–118, 2019. [Google Scholar]
56. Z. Khan, N. Yahya, K. Alsaih, S. S. Ali and F. Meriaudeau, “Evaluation of deep neural networks for semantic segmentation of prostate in T2W MRI,” Sensors, vol. 20, no. 11, pp. 3183, 2020. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools